Abstract
Adolescent major depressive disorder (AMDD) is a heterogeneous condition with rising global prevalence and limited treatment efficacy. This study integrates morphometric similarity networks (MSN) and spatial transcriptomics to identify neurobiologically distinct AMDD subtypes and their underlying molecular mechanisms. Using the HYDRA algorithm, we delineate two subtypes: AMDD1, characterized by reduced MSN strength in frontoparietal networks, heightened impulsivity, and preserved cognition; and AMDD2, marked by elevated MSN strength in limbic-visual circuits, severe emotional dysregulation, and rumination. Transcriptomic analyses reveal subtype-specific gene expression patterns, with AMDD1 associated with synaptic pruning deficits and AMDD2 linked to GABAergic inhibition deficits. Cell-type mapping highlights astrocytic dysregulation in AMDD1 and microglial activation in AMDD2, while pathway enrichment identifies distinct molecular networks, including endocannabinoid signaling in AMDD1 and MAPK-driven neuroinflammation in AMDD2. Developmental trajectory analysis uncovers critical windows for intervention, with AMDD1 showing delayed cerebellar maturation and AMDD2 exhibiting early hippocampal-striatal priming. These findings advance a precision framework for AMDD, linking spatially patterned gene expression to neurodevelopmental trajectories and offering targeted therapeutic strategies tailored to subtype-specific mechanisms. By bridging molecular, cellular, and network-level insights, this study provides a transformative approach to understanding and treating adolescent depression.
Similar content being viewed by others
Introduction
Adolescent major depressive disorder (AMDD) represents a significant global public health challenge, with rapidly increasing prevalence rates and long-term outcomes that include recurrent episodes and functional impairment into adulthood [1, 2]. Unlike adult-onset depression, AMDD is uniquely intertwined with adolescence—a period of dynamic neural reorganization. Normative developmental processes, such as synaptic pruning in the prefrontal cortex and heightened limbic reactivity, may become maladaptive in vulnerable individuals, amplifying emotional dysregulation and impulsive behaviors [3, 4]. Compounding this complexity, nearly 50% of adolescents exhibit inadequate response to first-line antidepressants [5], highlighting the need to move beyond symptom-based classifications.
Current diagnostic frameworks, such as the DSM-5, while clinically operable, reduce AMDD’s neurobiological diversity to subjective symptom clusters. For instance, irritability—a hallmark of adolescent depression—often overlaps with anxiety or behavioral disorders, leading to diagnostic delays [6]. Neuroimaging advances underscore a notable paradox: identical symptoms, such as anhedonia, map onto divergent neural mechanisms. For example, prefrontal hypometabolism may co-occur with amygdala hyperactivity in AMDD, reflecting distinct neurodevelopmental pathways [7, 8]. These inconsistencies suggested that conventional case-control designs conflate biologically distinct subgroups, which perpetuating the translational gap in AMDD research.
Transcriptomic studies in adult depression have identified corticolimbic dysregulation of synaptic plasticity (e.g., GRIN2A) and neuroinflammatory pathways (e.g., IL6R) [9]. However, adolescence, a period characterized by dynamic changes in gene expression, presents distinct vulnerabilities that are not yet fully understood. For instance, BDNF, a gene critical for synaptic pruning, peaked in expression during puberty—a developmental window coinciding with AMDD onset—but exhibited region-specific declines in prefrontal cortical hubs among affected adolescents [10, 11]. This divergence suggested that normative neurodevelopmental processes may become maladaptive when disrupted by genetic or environmental stressors. While spatial transcriptomic mapping in adults linked gene expression to structural networks [12], such approaches had not been applied to adolescents, whose rapidly evolving brains may prioritize distinct molecular pathways. Cortical regions enriched for synaptic genes in adulthood, such as the dorsolateral prefrontal cortex, could undergo localized expression shifts during adolescence, potentially amplifying vulnerability to maladaptive connectivity. Resolving this gap is imperative: if AMDD subtypes reflect spatially patterned gene expression signatures, they may unveil neurodevelopmental mechanisms that are both temporally sensitive and regionally specific, offering novel targets for timed interventions.
To bridge these gaps, we integrate morphometric similarity networks (MSN)—a structural connectivity measure reflecting synchronized neurodevelopmental processes—with spatial transcriptomics. Unlike functional connectivity, which captures transient activity states, MSN is thought to encode enduring developmental signatures, such as shared genetic influences or coordinated synaptic pruning [11]. We spatially correlate MSN-derived neuroanatomical gradients with transcriptomic profiles from the Allen Human Brain Atlas (AHBA). This approach allows us to interrogate how regionally patterned gene expression shapes structural network maturation and its dysregulation in AMDD. For example, in early-onset schizophrenia, MSN subtypes align with synaptic versus immune gene dysregulation [13], suggesting similar approaches could disentangle AMDD heterogeneity. By applying this framework to adolescents, we aim to uncover subtype-specific interactions between gene expression gradients and structural network maturation, ultimately linking molecular mechanisms to clinical phenotypes.
Guided by these advances, we address three critical questions: First, can neuroanatomical subtypes derived from MSN delineate clinically distinct AMDD subgroups with divergent symptom profiles. Second, do these subtypes reflect spatially patterned gene expression signatures. Third, how do subtype-specific trajectories interact with normative adolescent brain maturation. By addressing these questions, we aim to advance a precision framework for AMDD, linking molecular gradients to targeted interventions.
Methods
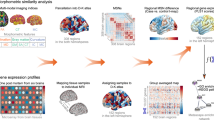
This study employed a multi-modal framework to delineate neurobiological subtypes of AMDD by integrating structural neuroimaging, transcriptomic profiling, and deep phenotyping (Fig. 1).
A: MSN construction. The MSN was computed using a 308 × 308 matrix incorporating five features (surface area, cortical thickness, gray matter volume, Gaussian curvature, and mean curvature).The MSN strength was derived by calculating the average weighted correlation coefficients between each brain region and all other regions. B: Classification and Genes analysis: The HYDRA method was applied to identify distinct AMDD subtypes based on MSN strength profiles. PLS regression was employed to determine imaging-transcriptomic associations. The relationship between whole-brain gene expression patterns and morphometric alterations across different AMDD subtypes was elucidated through PLS weight mapping, functional enrichment analysis, and cellular transcriptional signature assessment.
Participant and clinical assessment
Participants
A total of 282 adolescents aged 12–18 years were enrolled in this study, including 188 patients diagnosed with AMDD and 94 healthy controls (HC). AMDD patients were recruited from psychiatric departments at two hospitals: the First Affiliated Hospital of China Medical University (n = 163) and Shengjing Hospital of China Medical University (n = 25). HC were selected from local communities and universities through advertisements. Our study’s sample size was determined based on previous studies and enrollment capacity. This study was approved by the Ethics Committees of Shengjing Hospital of China Medical University and The First Affiliated Hospital of China Medical University. All methods were performed in accordance with the Declaration of Helsinki and relevant guidelines and regulations. Written informed consent was obtained from all participants and their legal guardians.
MDD diagnosis was confirmed by two board-certified psychiatrists who were blinded to the study hypotheses, using the Structured Clinical Interview for DSM-IV and DSM-V. Inclusion criteria for patients included:
First-time MDD diagnosis with a 17-item Hamilton Depression Rating Scale (HAMD-17) score >7 [14, 15]; (2) No comorbid Axis I/II disorders. (3) Absence of severe neurological or systemic illnesses. Exclusion criteria for all participants included (1) MRI contraindications (e.g., metallic implants) and non-right-handedness (2) Any current or prior diagnosis of a mental disorder, including but not limited to autism spectrum disorders, attention deficit hyperactivity disorder, depression, anxiety disorders, etc.; (3) Family history of mental illness; (4) severe organic brain or systemic disease. The study protocol was approved by the ethics committees of both institutions, and written informed consent was obtained from all participants and their legal guardians.
Clinical assessment
Clinical evaluations were conducted within 72 h of enrollment and included: (1) Symptom severity:HAMD-1, 14-item Hamilton Anxiety Rating Scale, Brief Psychiatric Rating Scale (BPRS), Young Mania Rating Scale (YMRS). (2) Cognitive and behavioral profiles: MATRICS Consensus Cognitive Battery, Barratt Impulsiveness Scale (BIS-11), Automatic Thoughts Questionnaire (ATQ) [16,17,18]. (3) Environmental and psychological factors: Adolescent Self-Rating Life Events Checklist (ASLEC), Egna Minnen Beträffande Uppfostran (EMBU), Cognitive Emotion Regulation Questionnaire (CERQ) [19,20,21,22,23]. (4) Suicidality assessment: Suicidal ideation (SI) was defined as affirmative responses to structured questions (e.g., “In the past two weeks, have you planned or considered suicide?”). Suicide attempts (SA) were identified via the Beck Suicide Scale, with detailed follow-up on intent and methods.
MRI data acquisition
Structural T1-weighted MRI scans were acquired using two 3.0 T scanners:
GE Sigma Scanner (n = 242): High-resolution 3D-FSPGR sequence. Siemens PRISMA Scanner (n = 22): 3D-MPRAGE sequence. (parameters in Table S1, Supporting Information 1).
To mitigate potential inter-site variability, we implemented three strategies: (1) standardized phantom calibration preceding human scans, (2) identical participant positioning protocols across sites, and (3) statistical control of site effects through covariate adjustment in all group-level analyses.
MRI image preprocessing
The T1-weighted images were preprocessed in a surface-based framework using the latest version of FreeSurfer (v7.3.2, http://surfer.nmr.mgh.harvard.edu/) [24]. The processing pipeline included robust skull stripping, bias field correction, and comprehensive tissue segmentation to delineate gray matter, white matter, and cerebrospinal fluid. In subsequent steps, each brain was partitioned into hemispheric and subcortical structures, and cortical surfaces were accurately reconstructed by generating precise gray/white matter interfaces and pial surfaces.
To ensure high data quality, images that demonstrated excessive motion artifacts or other quality issues were visually inspected and excluded from further analysis. Key quality metrics, including the Euler number and total intracranial volume (TIV) [25], were computed for each T1-weighted image to assess the integrity of the cortical reconstructions and to control for head size differences in downstream analyses. 18 cases were excluded due to poor quality.
Construction of MSN
Cortical parcellation and feature extraction
For each participant, the cortical surface was segmented into 308 spatially contiguous regions derived from the 68 cortical areas defined in the Desikan–Killiany (D-K) atlas [26, 27]. This parcellation, implemented through a backtracking algorithm, produced regions of approximately equal size (~500 mm2), thereby minimizing variability attributable to parcel size differences [28, 29]. The parcellated D-K atlas was then registered to each participant’s native cortical surface to generate individualized parcellation maps.From each of the 308 regions, five morphometric features were extracted from the T1-weighted images: surface area, cortical thickness, gray matter volume, Gaussian curvature, and mean curvature [11]. To account for differences in the distribution of these features, each morphometric feature vector was z-score normalized across regions [26, 27].
Network construction
Pairwise Pearson’s correlation analyses were subsequently performed on the normalized feature vectors, resulting in an unthresholded 308 × 308 MSN for each participant. The connectivity strength, or MSN strength, of a given region was quantified by calculating the average weighted correlation coefficient between that region and all other regions [30]. Age, sex, education, TIV and site effects were regressed out from global MSN strength.
Subtyping AMDD with HYDRA
HYDRA algorithm overview
HYDRA (Heterogeneity Through Discriminative Analysis, https://github.com/evarol/HYDRA) was employed to identify neuroanatomical subtypes of AMDD by integrating supervised classification and unsupervised clustering [31]. The algorithm operates in two stages: (1) Classification: A convex polytope is constructed using linear maximum-margin classifiers to separate AMDD patients from HCs based on MSN strength patterns. (2) Clustering: AMDD patients are grouped into subtypes according to their proximity to distinct hyperplanes (polytope faces), which represent divergent neuroanatomical trajectories.(HYDRA METHOD, Supporting Information 1).
Key parameters and stability assessment
For our analyses, HYDRA was configured with 50 iterations that alternated between hyperplane estimation and cluster assignment, complemented by 20 consensus steps to enhance clustering stability. A regularization parameter of 0.25 was employed, and tenfold cross-validation was performed to ensure the robustness of the clustering solution. To quantitatively assess the stability and consistency of the clustering, we computed the Adjusted Rand Index (ARI) [32], which corrects for chance agreement and provides a conservative measure of clustering overlap.
Case-control analysis of MSN strength for AMDD subtypes
Statistical modeling framework
To compare MSN strength between AMDD subtypes and HCs, we employed a hierarchical linear regression model (LRM): MSN strength as the dependent variable. Age, sex, education,site effect and TIV were included as covariates to control for potential confounding effects. For each participant, global MSN strength was computed as the average MSN strength across all brain regions.
To further examine regional variations, we modeled the MSN strength for each region (MSNi) using the following equation:
Two-sample t-tests comparing each AMDD subtype against HC, were performed to identify regional differences in MSN strength. A Bonferroni correction was employed to adjust for multiple comparisons, with significance defined at p < 0.05.
Functional network and developmental trajectory analysis
To examine the functional network-level abnormalities distinguishing AMDD subtypes from healthy controls, we conducted systematic analyses of MSN strength across established brain parcellations-Yeo Functional 7 Networks and von Economo atlas [33, 34]. A Bonferroni correction was employed to adjust for multiple comparisons, with significance defined at p < 0.05.
In addition, to investigate the developmental trajectories of AMDD subtypes from childhood to adolescence, we employed quadratic non-linear modeling to characterize developmental trajectories of both global MSN strength and functional network-specific MSN strength. We selected this non-linear approach based on established evidence that neurodevelopmental processes follow complex, non-linear patterns across age rather than simple linear progressions. The quadratic model allowed us to capture potential curvilinear relationships, inflection points, and rate changes in MSN development [35].
Transcriptomic correlates of MSN alterations
Regional gene expression data acquisition
Transcriptomic profiles were obtained from the AHBA (http://human.brain-map.org) [36], encompassing microarray data from 3,702 spatially annotated tissue samples across six postmortem brains (Detailed information see Supporting Information 1, Section 3). Gene expression matrices were preprocessed using the Abagen toolbox (https://github.com/rmarkello/abagen) [37] with the following steps: (1) Probe Filtering: Removed probes with expression levels below background noise in >50% of samples. (2) Probe Selection: For genes with multiple probes, retained the probe showing the highest regional specificity. (3) Spatial Registration:Allocating samples to brain regions if they were within a 2 mm Euclidean distance from the region boundary. (4) Normalization: Scaled expression values across samples using a robust sigmoid function.
Due to limited right-hemisphere coverage in AHBA, analyses focused on the left hemisphere, yielding expression data for 15,631 genes across 152 cortical regions [38].
Spatial covariation analysis
To identify transcriptional signatures associated with MSN alterations, we employed Partial Least Squares (PLS) regression: Predictors are normalized gene expression matrices (15,631 genes × 152 regions). Response Variables are case-control t-statistic maps of MSN strength differences [39]. Analysis Steps: (1) Component Extraction: The first PLS component (PLS1) was derived to maximize covariance between gene expression and MSN t-values. (2) Significance Testing: Permutation tests (10,000 iterations) assessed the statistical significance of PLS1 (p-perm < 0.0001, p-perm indicated significance assessed using standard permutation testing with 10,000 random permutations) [40]. (3) Gene Weighting: Genes with absolute Z-scores > |5| were classified as PLS1+ (positively weighted) or PLS1− (negatively weighted) [11]. Spearman’s correlations confirmed regional alignment between PLS1 scores and MSN t-maps.
Analysis of depression-related genes
We examined specific depression-associated genes from the AHBA’s 1000 Genes Characterized by ISH in the Cortex Gene Survey (help.brain-map.org/display/humanbrain/Documentation). 24 depression-associated genes were analyzed: ADRA2A, AVPR1B, CHRM2, CNR1, CREB1, CRH, CRHR1, CRHR2, CUX2, GAD2, GPR50, HTR1A, HTR1B, HTR1D, HTR3A, HTR5A, MAOA, PDE1A, SLC6A2, SLC6A4, SST, TAC1, TPH1, and TPH2 [41].
To assess their contribution to MSN alterations, we first identified overlapping genes from the 24 MDD-related genes and 15,631 background genes. We then estimated the relationship between overlapping gene expression and left hemisphere MSN case-control changes, pFDR < 0.05 was considered significant.
Functional enrichment analysis of PLS1 positive or negative genes
For functional characterization of PLS1 associated genes, we conducted comprehensive enrichment analysis: (1) Gene Selection: Applied stringent significance thresholds (Z > 5 for PLS1+ genes; Z < −5 for PLS1- genes); (2) Enrichment Analysis: Examined Gene Ontology Biological Processes (GO-BP) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways, Enriched GO terms and KEGG pathways were considered statistically significant at pFDR < 0.05; (3) Analyzed and visualized results using the online bioinformatics platform (https://www.bioinformatics.com.cn, last accessed on 10 Dec 2024) [42], which provides comprehensive data analysis and visualization capabilities.
Cell-type mapping and developmental trajectory analysis of AMDD subtype specific gene signatures
Cellular characterization of regional MSN alterations
To understand the cellular basis of MSN alterations in AMDD subtypes, we mapped PLS1-weighted genes to specific cortical cell populations. We integrated cell-type reference panels derived from five independent single-cell transcriptomic studies of human postmortem cortical tissue. This approach yielded comprehensive gene signatures for seven major cell types: endothelial cells, astrocytes, microglia, oligodendrocytes, oligodendrocyte precursor cells (OPCs), excitatory neurons, and inhibitory neurons [43].
We quantified cell-type enrichment scores for each brain region using single-sample Gene Set Enrichment Analysis (ssGSEA) implemented through the “gsva” function with Gaussian parameters in the GSVA R package [44]. This method computed enrichment scores reflecting the relative abundance of each cell type based on their characteristic gene expression profiles. We then systematically compared PLS1+ and PLS1- gene lists against these cell-type-specific reference panels to identify predominant cellular contributors to subtype-related brain changes. Statistical significance of cell-type enrichments was evaluated using permutation testing with pFDR < 0.05).
Functional pathway analysis of cell-type-specific gene signatures
To elucidate the biological mechanisms in cell-type-specific gene signatures, we conducted functional enrichment analysis. For the overlapping genes, functional enrichment analysis was performed using bioinformatics platform (https://www.bioinformatics.com.cn, last accessed on 10 Dec 2024) [42]. Enriched GO terms and KEGG pathways were considered statistically significant at pFDR < 0.05.
Developmental expression mapping of AMDD-associated genes
For developmental trajectory analysis, subtype-specific gene lists (PLS1± genes) were cross-referenced with the Brainspan developmental transcriptome atlas using the cell-type specific expression analysis (CSEA) tool (http://doughertytools.wustl.edu/CSEAtool.html) [45]. Enrichment scores were computed for each gene set across 15 developmental stages (prenatal to adulthood) and 12 brain regions. The tool integrates human data from the Brainspan collection to provide parallel analysis of transcripts enriched in specific human brain regions and/or developmental windows.We investigated the correlation between PLS weighted genes and developmental time spans across different brain regions in distinct AMDD subtypes through developmental gene expression enrichment analysis.
Results
Demographic and clinical characteristics
The AMDD group (n = 179) and HC (n = 85) were matched for age and education but differed significantly in sex distribution (70.95% female in AMDD vs. 51.76% in HC, χ2 = 9.30, p = 0.002), aligning with epidemiological reports of higher female vulnerability to adolescent depression [46]. Clinically, AMDD patients exhibited elevated depressive (HAMD-17, p < 0.001), anxiety (HAMA-14, p < 0.001), and cognitive impairment scores (MCCB, p < 0.001), alongside heightened impulsivity (BIS-11, p < 0.001) and negative automatic thoughts (ATQ, p < 0.001). Environmental stressors further distinguished the groups, with AMDD participants reporting greater academic pressure (ASLEC, p < 0.001) and paternal rejection (EMBU, p = 0.011).Notably, 55.87% of AMDD patients endorsed suicidal ideation, with 36.31% reporting prior suicide attempts—a subgroup demonstrating significantly higher impulsivity scores than non-attempters (p < 0.001). (Table S4, Supporting Information 1). Homogeneity of variance was assessed using Levene’s test prior to conducting t-tests.
Identification of AMDD subtypes using HYDRA
The HYDRA algorithm identified two distinct neurobiological subtypes within the AMDD cohort, demonstrating robust clustering stability with an ARI of 0.83 across 50 iterations. (Fig. S1, Supporting Information 1). Comparative model evaluation confirmed the two-cluster solution as optimal, balancing biological interpretability and statistical rigor, with AMDD1 (n = 86) and AMDD2 (n = 93) representing phenotypically divergent subgroups. This classification emerged from tenfold cross-validation, where regularization parameters (λ = 0.25) and consensus clustering minimized overfitting while preserving inter-subtype heterogeneity.
Clinical features across AMDD subtypes
The neurobiological differentiation of AMDD subtypes translated into distinct clinical phenotypes. AMDD2 exhibited heightened emotional dysregulation, marked by elevated depressive (HAMD-17: p = 0.020) and anxiety (HAMA-14: p = 0.001) severity compared to AMDD1. In contrast, AMDD1 demonstrated pronounced behavioral disinhibition, with higher impulsivity (BIS-11: p = 0.013) and negative cognitive distortions (ATQ: p = 0.025), despite preserved global cognitive performance (MCCB: p = 0.030). This dissociation suggests divergent neural substrates for emotional and cognitive regulation.
Coping strategies further distinguished the subtypes: AMDD2 relied heavily on maladaptive internalizing behaviors, including rumination (p = 0.011) and self-blame (p = 0.038), whereas AMDD1 reported greater externalizing stressors, such as academic pressure (p = 0.008) and paternal punishment (p < 0.001). Suicidal ideation prevalence was comparable between subtypes (AMDD1: 53.5% vs. AMDD2: 58.1%, χ2 = 0.38, p = 0.538), yet suicide attempt rates trended higher in AMDD2 (39.8% vs. 32.6%, χ2 = 1.01, p = 0.315), paralleling their elevated emotional distress. There were no statistically significant differences in sex, age, and education between AMDD1 and AMDD2. (Tables 1, 2).
MSN in AMDD subtypes
Neuroanatomical divergence between subtypes
AMDD1 exhibited reduced MSN strength in the dorsolateral prefrontal cortex (dlPFC; left lateral occipital cortex) and anterior cingulate cortex (ACC; left precentral gyrus), networks central to executive function and motor planning. In contrast, AMDD2 demonstrated elevated MSN strength in emotion-processing hubs, notably the amygdala (right pericalcarine cortex) and parahippocampal gyrus (right parahippocampal part2). This hyperconnectivity extended to visual association areas such as the cuneus (right cuneus part3) and lingual gyrus (right lingual part4). (Tables S1, S2; Supporting Information 2).
Compared to HC, AMDD1 showed widespread MSN weakening in frontoparietal networks, including the superior frontal gyrus (right superior frontal part1) and inferior parietal lobule (left inferior temporal part4), regions essential for attentional control.Conversely, AMDD2 displayed selective hyperconnectivity in limbic-striatal circuits, such as the nucleus accumbens (left lateral occipital part5) and hippocampus (right lingual part6) (Fig. 2a–c).
In addition, we divided the individuals with AMDD into two subgroups: drug-naive (n = 81) and drug-used groups (n = 98), to explore the medication effects on MSN. We found that irrespective of medication status, similar patterns to the case-control t-map were observed. (Fig. S4, Supporting Information 1), detailed medication records (n = 45) of specific medications and dosages were shown in Table S3.
Spatial correlation with healthy controls
The spatial correlation between MSN strength in HC and case-control differences in AMDD subtypes revealed distinct neurodevelopmental patterns. For AMDD1, the MSN strength of HC exhibited a positive spatial correlation with case-control t-values across cortical regions (r(308) = 0.88, p-spin < 0.0001; p-spin indicates significance assessed using spin permutation testing that accounts for spatial auto correlation in brain maps), indicating that regions with higher baseline connectivity in HC showed amplified reductions in AMDD1. Specifically, 24% of regions with positive MSN strength in HC demonstrated hyperdifferentiation in AMDD1, while 12% of regions with negative MSN strength in HC displayed decoupling. AMDD2 exhibited a negative spatial correlation between HC MSN strength and case-control t-values (r(308) = −0.676, p-spin < 0.0001), where regions with lower baseline connectivity in HC showed marked hyperconnectivity in AMDD2. 15% of regions with negative MSN strength in HC demonstrated hyperscoping, while 48% of regions with positive MSN strength in HC exhibited decoupling (Fig. 2d).
Functional network abnormalities
In Yeo 7 functional networks, AMDD1 exhibited reduced MSN strength in the somatomotor network and visual network compared to HC, whereas AMDD2 showed increased MSN strength in the visual network.
For the von Economo atlas, AMDD1 showed reduced MSN strength in both the primary motor network and primary sensory network. Conversely, AMDD2 displayed significantly increased MSN strength in the primary motor. (Fig. S5, Supporting Information 1).
Developmental trajectories
For the developmental trajectories of AMDD subtypes from childhood to adolescence. The somatomotor network of AMDD1 showed a rapid increase followed by a sustained decline (r = −0.24, p = 0.027). AMDD2’s ventral attention networks declined consistently during childhood and stabilized during adolescence. (Fig. S6, Supporting Information 1).
Transcriptomic correlates of MSN alterations
PLS regression analysis revealed significant spatial covariation between regional gene expression profiles and MSN alterations across AMDD subtypes. In AMDD1, PLS1 explained 46.93% of the variance in case-control MSN t-values (p-perm < 0.0001), while in AMDD2, PLS1 accounted for 30.22% of the variance (p-perm < 0.0001), underscoring distinct transcriptomic contributions to neuroanatomical changes. (Fig. S8, Supporting Information 1). The spatial distribution of PLS1-weighted gene expression maps demonstrated robust correlations with MSN strength patterns: AMDD1 exhibited a strong positive association (r = 0.66, p-spin < 0.0001), whereas AMDD2 showed a moderate yet significant correlation (r = 0.48, p-spin < 0.0001), reflecting subtype specific gene-brain structure interaction.
Among 24 MDD associated genes, 13 overlapped with our PLS1 gene sets. In AMDD1, five genes showed significant spatial correlations with MSN t-values (|r|>0.35, pFDR < 0.05): TAC1 (r = 0.58, p-spin < 0.0001), CNR1 (r = 0.51, p-spin < 0.0001), SST (r = 0.39, p-spin < 0.0001), CUX2 (r = −0.54, p-spin < 0.0001). For AMDD2, only GAD2 (r = 0.38, pFDR = 0.002) exhibited significant associations, highlighting divergent molecular mechanisms. (Fig. 3c–d, Table S4 Supporting Information 2).
a. The distribution of differences in MSN strength and PLS1 scores in the left hemisphere of the AMDD1and AMDD subtype. b. Scatterplots showing the significant spatial correlation between PLS1 scores and the case-control t-value maps of MSN strength in both AMDD subtypes; AMDD1, Spearman’s r = 0.66, p-spin < 0.0001; AMDD2, Spearman’s r = 0.48, p-spin < 0.0001. c, d The expression of MDD-related genes from ISH datasets was positively or negatively associated with regional changes in MSN, including 6 positive genes (i.e., TAC1, CNR1, SST1 HTR1A and GAD2) and one negative genes (CUX2). All r values were determined by Spearman’s correlation analysis, and p values were obtained from spatial correlation tests and adjusted with FDR correction.
Subtype-specific molecular pathways
Using univariate Z-tests, we identified 2072 PLS1+ genes and 1071 PLS1− genes for AMDD1, alongside 1807 PLS1+ genes and 6 PLS1− genes for AMDD2 (all pFDR < 0.0001, Table S2, 3 Supporting Information 2). These gene sets encapsulated transcriptional signatures strongly linked to MSN alterations.
To delineate the biological significance of PLS1± gene sets, functional enrichment analysis was performed. In AMDD1, PLS1+ genes were enriched in neurotransmitter regulation (e.g., alcohol metabolic process) and synaptic protein homeostasis (ubiquitin-mediated proteolysis) (Fig. 4a, b). These pathways implicate astrocytic detoxification deficits and disrupted synaptic pruning—mechanisms converging on impulsive phenotypes (Fig. 4a, b). For AMDD2, PLS1+ genes dominated chromatin remodeling (histone deacetylation) and stress-responsive signaling (MAPK pathway) (Fig. 4c, d). PLS1− genes uniquely associated with efferocytosis, suggesting microglial phagocytic dysfunction may sustain neuroinflammation, amplifying rumination in this subtype (Fig. 4c, d).
a, c: GO and KEGG pathway enrichment for AMDD1 a and AMDD2 subtypes c. The bar chart displays significantly enriched BP and pathways, with red bars representing PLS+ genes and blue bars representing PLS- genes. Numbers inside bars indicate gene counts in each pathway. Statistical significance is shown on the x-axis as -log10(p-value), with longer bars indicating stronger significance (pFDR < 0.05). b, d: Chord diagram illustrating the relationships between enriched pathways and genes in the AMDD1b and AMDD2 d. Node size and color represent gene count and pathway category, respectively.
Functional and cellular specificity of MSN-associated genes
Cell-type mapping of AMDD subtypes
Cell-type mapping revealed subtype-specific molecular mechanisms underlying MSN alterations (Fig. 5a, Table S6, 7 Supporting Information 2). In AMDD1, PLS1+ genes showed pronounced enrichment in astrocytes (230 genes, p-perm = 0.0069) and excitatory neurons (177 genes, p-perm = 0.0157), with pathways implicating synaptic plasticity (e.g., glutamatergic transmission, pFDR < 0.001) and metabolic detoxification. These findings align with AMDD1’s frontoparietal hypoconnectivity, suggesting that astrocytic dysregulation disrupts synaptic pruning—a process critical for adolescent cognitive maturation (Fig. 5b). Conversely, PLS1− genes in AMDD1 were enriched in excitatory (148 genes, pperm = 0.0034) and inhibitory neurons (114 genes, pperm = 0.0035) (Fig. 5d).
a. The distribution of regional gene expression maps of seven brain cells using ssGSEA scores. b. The number of overlapping genes with AMDD1 PLS1 + weighted genes for each cell type. c. Enrichment results for AMDD1 overlapped PLS1+ genes. d. The number of overlapping genes with AMDD1 PLS1 - weighted genes for each cell type. e. Enrichment results for AMDD1 overlapped PLS1- genes. f. The number of overlapping genes with AMDD2 PLS1 + weighted genes for each cell type. g. Enrichment results for AMDD2 overlapped PLS1+ genes. Asterisks (*) indicate statistical significance (p-perm < 0.05) for cell-type enrichment.
In contrast, AMDD2’s PLS1+ genes were uniquely enriched in inhibitory neurons (131 genes, p-perm = 0.0069), with functional terms highlighting GABAergic synapse dysfunction (GO:0098982, pFDR = 0.002) and neuroinflammatory responses (KEGG:04668, pFDR = 0.004) This inhibitory neuron signature correlates with the limbic hyperconnectivity observed in AMDD2, potentially amplifying emotional salience through disrupted interneuron-mediated feedforward inhibition (Fig. 5f).
Functional enrichment analysis of cell-type-specific genes
Functional analysis of cell-type-specific genes revealed distinct molecular pathways underlying MSN alterations in each subtype (Table S8, Supporting Information 2). For AMDD1, PLS1+ genes overlapped with astrocytes and excitatory neurons were enriched in synaptic plasticity pathways such as glutamatergic synaptic transmission and PPAR signaling, suggesting a coordinated role of glia-neuron interactions in maintaining network resilience. In contrast, PLS1− genes in AMDD1, predominantly localized to inhibitory neurons, were linked to calcium ion transport and ubiquitin-mediated proteolysis, pointing to dysregulated calcium signaling and impaired protein homeostasis as drivers of cognitive-impulsive dissonance. For AMDD2, PLS1+ genes overlapped with inhibitory neurons showed enrichment in glutamatergic synapse and long-term potentiation pathways, reflecting maladaptive synaptic strengthening in limbic circuits that may amplify emotional hypersynchrony. The absence of significant functional enrichment in AMDD2 PLS1− genes (Fig. 5c, e, g).
Developmental trajectory analysis for subtype-specific gene expression
Developmental trajectory analysis uncovered critical spatiotemporal windows for subtype-specific gene expression. In AMDD1, PLS1+ genes exhibited pronounced enrichment in the amygdala during adolescence and the cortex during late fetal stages. These genes also peaked in the cerebellum during middle-late childhood, suggesting delayed motor coordination development. Conversely, PLS1− genes in AMDD1 were dynamically active in the cerebellum across childhood and young adulthood. For AMDD2, PLS1+ genes showed robust expression in the hippocampus during late mid fetal stages and the striatum during adolescence. Notably, both subtypes shared transient enrichment in the late fetal amygdala, indicating early-life origins of emotional dysregulation that diverge in later developmental trajectories (Fig. 6).
The dot plots represent the results of the enrichment analysis for different developmental stages and brain regions, comparing the AMDD1 and AMDD2 groups. The plots are organized into four panels. Red dots indicate significant enrichment (pFDR < 0.05), while blue dots indicate non-significant results. Circle size corresponds to the statistical significance level, with larger circles representing stronger significance as indicated by -log10(p-value). Empty space indicates lack of significant enrichment.The scale ranges from 0.20–4.00 as shown in the legend.
Discussion
Our study delineated two neurobiologically distinct subtypes of adolescent major depressive disorder characterized by divergent structural network alterations, molecular signatures, and clinical trajectories. AMDD1 exhibited reduced MSN strength in frontoparietal networks crucial for cognitive control, accompanied by heightened impulsivity and negative automatic thoughts despite preserved general cognitive function. In contrast, AMDD2 demonstrated elevated MSN strength in limbic-visual circuits involved in emotion processing, alongside more severe depressive and anxiety symptoms with maladaptive coping strategies centered on rumination and self-blame. These findings challenge traditional symptom-based classifications of depression, instead proposing a neurodevelopmental framework where spatially patterned gene expression interacts with maturational processes to shape disease heterogeneity.
AMDD1 exhibited a pattern of reduced MSN strength in frontoparietal regions critical for cognitive control and executive function. This pattern likely reflected disrupted neurodevelopmental processes rather than conventional connectivity deficits. For instance, diminished MSN strength in the supra-frontal region may reflect aberrant synaptic pruning during a critical developmental window. When adolescents with this frontoparietal pattern face emotional challenges, their diminished top-down regulatory capacity may manifest as difficulty suppressing maladaptive behavioral responses, despite intact cognitive understanding of consequences. The somatomotor developmental trajectory in AMDD1 showed an atypical pattern—rapid early increase followed by sustained decline—contrasting with normative adolescent development where motor networks typically stabilize post-puberty [47].
Conversely, AMDD2 presented a different neurobiological profile. These patients exhibited increased morphometric similarity between limbic and visual processing regions, alongside clinical symptoms dominated by rumination and emotional reactivity. This pattern suggested aberrant developmental synchronization between regions processing emotional and sensory information—a finding consistent with the subjective experience of being unable to disengage from negative emotional content that characterizes rumination [48]. Visual imagery became tightly coupled with emotional content, creating a self-reinforcing circuit of negative thought patterns. Toenders et al. directly observed this phenomenon, finding that rumination in adolescent depression correlates with heightened amygdala-visual cortex connectivity during emotional face processing [49]. Meanwhile, the differences in HAMD-17 scores between subtypes appeared modest, their clinical significance became apparent when considering the distinct symptom clusters. These divergent symptom patterns reflected different pathophysiological mechanisms, as evidenced by their correlation with distinct MSN alterations and gene expression profiles.
The subtype-specific patterns of environmental stress responsivity further illuminate these neurobiological differences. AMDD1’s heightened sensitivity to academic pressure and paternal punishment suggests that frontoparietal network disruptions impair stress buffering without directly intensifying emotional experience. This aligns with Mulders et al.‘s meta-analysis showing that cognitive control network dysfunction in depression primarily affects emotional regulation rather than generation [50]. AMDD2’s paradoxical pattern—higher symptom severity despite lower reported stressors—revealed how limbic connectivity amplifies emotional responses to even moderate environmental challenges, driving maladaptive rumination and self-blame.
Our molecular analyses identified potential genetic correlates of the observed morphometric alterations. PLS analysis revealed TAC1 as a gene with strong positive weighting in AMDD1. TAC1 encodes substance P and related neuropeptides, and correlates strongly with frontoparietal hypoconnectivity [51]. Mechanistically, substance P modulates prefrontal glutamatergic signaling and synaptic plasticity, with elevated levels disrupting the excitatory-inhibitory balance needed for cognitive control [52]. TAC1 overexpression in animal models produces impulsivity and reward dysregulation by impairing prefrontal inhibition of striatal pathways—precisely the clinical profile observed in AMDD1 [53, 54]. Similarly, CNR1 encoding cannabinoid receptor 1, regulates presynaptic neurotransmitter release during synaptic pruning. Disrupted endocannabinoid signaling during adolescence impairs the selective elimination of redundant synapses in frontoparietal circuits [55], leading to inefficient network connectivity despite intact cognitive processing—explaining AMDD1’s preserved cognition alongside elevated impulsivity [56]. In addition, CUX2 is a transcription factor critical for neuronal maturation and synaptic pruning, showed a strong negative correlation with MSN strength in AMDD1. Reduced CUX2 expression may impair synaptic refinement in frontoparietal region [57, 58]. This finding suggests that CUX2 dysregulation, potentially exacerbated by environmental stressors, may further disrupt the developmental trajectory of cognitive control networks in AMDD1. Although our current analyses controlled for sex to minimize confounding, preliminary evidence suggested that sex hormones differences may associate to divergent neurodevelopmental trajectories and stress responses between males and females.
AMDD2’s association with GAD2—the gene encoding glutamic acid decarboxylase—pointed to fundamentally different pathophysiology centered on GABAergic inhibitory dysfunction [59]. GAD2 catalyzes GABA synthesis primarily in limbic interneurons that normally dampen emotional reactivity [60]. Fee et al. directly observed reduced GABA levels in the anterior cingulate cortex of depressed adolescents correlating with rumination severity [61]. This mechanism differs markedly from adult depression models dominated by microglial inflammation and monoamine deficiency [62], highlighting adolescence-specific vulnerabilities in inhibitory circuit maturation [63]. Moreover, sex differences may regulate emotional processing and GABAergic inhibitory functions, consequently affecting the operation of negative affect systems in AMDD2. Longitudinal studies showed accelerated cortical thinning precedes adolescent depression onset [64], while animal models demonstrate stress-induced transcriptional changes correlate with connectivity alterations [65], providing plausible mechanisms for the morphometric alterations we observed.
The spatial covariation of PLS1-weighted gene expression with MSN alterations revealed distinct molecular pathways driving subtype-specific pathology. In AMDD1, PLS1+ genes showed significant enrichment in cellular metabolism, neurotransmission, and alcohol metabolic processes, suggesting alterations in detoxification functions crucial for synaptic environment maintenance during neurodevelopment [66]. Disruption of these processes could impair the precisely timed mechanism of synaptic pruning in frontoparietal regions, potentially contributing to the reduced MSN strength we observed in AMDD1.Most compelling was the enrichment of retrograde endocannabinoid signaling among AMDD1’s PLS1+ genes. This system played a pivotal role in regulating synaptic pruning during adolescence, with CB1 receptors abundantly expressed in frontoparietal regions undergoing active remodeling [67]. Disruption of this pathway could impair the adaptive neural circuits while compromising redundant connection elimination, potentially explaining the reduced morphometric similarity in cognitive control networks and linking AMDD1’s gene expression profile to its clinical impulsivity manifestation.
The pathway enrichment findings in AMDD2 reveal a molecular landscape dominated by stress-responsive signaling and epigenetic regulation. The significant enrichment of MAPK signaling among PLS1+ genes is particularly noteworthy. This pathway serves as a critical transducer of stress signals, activating downstream transcriptional machinery that can fundamentally reshape neural circuit function [68]. In the context of adolescent brain development, MAPK hyperactivation could alter the trajectory of limbic circuit maturation through persistent modifications to the epigenetic landscape, as suggested by the concurrent enrichment in histone deacetylation pathways. The unique enrichment of efferocytosis pathways in AMDD2’s PLS1− genes adds another dimension to its biological signature. Efferocytosis—the process by which microglia clear apoptotic cells—plays a crucial role in maintaining neural homeostasis. Impairments in this cleaning function could lead to accumulated cellular debris and sustained inflammatory signaling in emotion-processing circuits [69]. This finding aligns with evidence implicating neuroinflammation in depressive disorders, particularly those characterized by persistent rumination.
The cellular specificity analysis further clarified these mechanisms by identifying the cell populations driving subtype pathology. AMDD1’s PLS1+ genes showed significant enrichment in astrocyte markers, positioning glia as central players in frontoparietal network disruption. During adolescence, astrocytes orchestrate synaptic pruning through phagocytosis of weak synapses and regulation of extracellular glutamate [70]. Dysfunction in these astrocyte-mediated processes could impair the precise elimination of redundant connections needed for mature cognitive control networks. In contrast, AMDD2’s selective enrichment in inhibitory neuron signatures reinforced GABAergic dysfunction as the primary driver of limbic hyperreactivity. During adolescence, GABAergic interneurons undergo substantial maturation, with disruption of this process leading to improper emotion regulation [71]. McKlveen et al. demonstrated that stress-induced alterations in prefrontal inhibitory interneurons lead to emotional dysregulation in rodent models—a process potentially accelerated in AMDD2 by developmental vulnerability [72].
The developmental trajectory analysis reveals critical temporal windows that illuminate both etiology and intervention timing. AMDD1-associated genes peak in the amygdala during adolescence, aligning with the critical role of this region in pubertal stress reactivity [73, 74]. The amygdala’s reciprocal connections with prefrontal regions—undergoing active pruning during this period. This mechanism contrasted with adult models where chronic inflammation, not developmental pruning errors, dominates pathology [40]. AMDD2-associated genes, conversely, showed strongest expression in the hippocampus during late mid-fetal development and striatum during adolescence, indicating early developmental vulnerabilities that emerge clinically during adolescent stress exposure. This temporal pattern suggested fundamentally different developmental origins: AMDD1 represented contemporaneous disruption of adolescent brain maturation, while AMDD2 involved early-life priming that manifests during adolescent stress exposure. The subtypes’ divergent trajectories—AMDD1’s delayed cerebellar maturation versus AMDD2’s fetal hippocampal-striatal priming—aligned with preclinical models where prenatal stress accelerates limbic development at the expense of regulatory circuits [75], creating vulnerability that emerges during adolescent stress exposure [76].
Given these subtype-specific mechanisms, a pressing challenge emerged: How can we leverage developmental timing to optimize interventions? For AMDD1, targeting pubertal synaptic plasticity through cognitive or pharmacological means may recalibrate frontoparietal networks. Cognitive remediation therapies designed to strengthen prefrontal-striatal connectivity have shown promise in improving impulse control in similar neuropsychological profiles. Motter et al. demonstrated that targeted cognitive exercises improved executive function and reduced depressive symptoms in patients with prefrontal hypoconnectivity [77]. Pharmacologically, the TAC1 pathway offers a novel target: neurokinin-1 receptor (NK1R) antagonists, which block substance P signaling, could potentially normalize prefrontal inhibitory control networks. Animal studies show that NK1R antagonists reduce impulsivity and improve prefrontal function during stress—directly addressing AMDD1’s core pathophysiology [78]. While NK1R antagonists such as aprepitant had established safety profiles in adolescents for chemotherapy-induced nausea [79], limited evidence exists for psychiatric indications. Developmental considerations, including potential impacts on neural plasticity and brain maturation, required careful evaluation before these agents could be considered for adolescent mood disorders.
For AMDD2, our findings pointed toward dual therapeutic targeting of GABAergic and inflammatory mechanisms. GABAergic modulators might address the core inhibitory deficit, while anti-inflammatory agents—particularly JNK inhibitors targeting the MAPK pathway—could attenuate the stress-responsive signaling cascade that appears to drive this subtype’s pathology [80]. However, JNK inhibitors remain largely investigational with minimal adolescent safety data, necessitating rigorous preclinical studies in developmental models before clinical translation. Novel GABA-enhancing compounds that specifically upregulate GAD2 activity, rather than acting directly on GABA receptors, could offer mechanistic precision with fewer side effects in adolescents [81, 82]. Furthermore, with its emotion regulation difficulties, AMDD2 might benefit more from interventions addressing rumination and emotional reactivity, such as mindfulness-based approaches or interpersonal therapy [83, 84].
The temporal origins of these subtypes demand developmentally-timed prevention strategies. For AMDD2, fetal hippocampal gene enrichment suggests that interventions during childhood—before adolescent symptom emergence—could buffer limbic circuit development in high-risk populations, such as offspring of mothers with prenatal depression [85]. Early life stress-reduction programs have demonstrated lasting effects on amygdala reactivity and emotion regulation [86, 87], potentially preventing the emergence of AMDD2-like phenotypes. For AMDD1, preventive interventions during early adolescence focusing on executive function development could potentially normalize frontoparietal maturation before impulsivity becomes entrenched [88]. Recent clinical trials confirm that cognitive training during early adolescence significantly reduces subsequent depression risk in vulnerable youth [89], supporting this developmental approach.
Our findings extended existing theoretical frameworks in key ways. The Cognitive Control Deficit Model identified frontoparietal dysfunction in adolescent depression [90], but our AMDD1 subtype implicated TAC1/CNR1 pathways may as specific molecular drivers. Similarly, while the Affective Processing Bias Model emphasized emotional reactivity [91], our AMDD2 findings challenged its glutamatergic focus by demonstrating primary GABAergic dysfunction in adolescents. Within the RDoC framework [92], these subtypes demonstrated how similar clinical presentations arose from different domain disruptions—AMDD1 primarily affecting Cognitive Systems and AMDD2 predominantly disrupting Negative Valence Systems—explaining why symptom-based approaches often failed to yield consistent neurobiological correlates. Furthermore, translating these neurobiological patterns to individual prediction represents an important bridge toward clinical application. This translation requires developing validated classification algorithms for reliable subtype assignment, simplifying assessment through targeted biomarkers like peripheral blood biomarkers, and conducting prospective studies confirming that subtype classification meaningfully predicts treatment outcomes.
Limitations and future directions
There are still some limitations in our study. (1) The cross-sectional design limited causal inferences regarding subtype trajectories and treatment responses. Validating potential subtype-specific interventions would require longitudinal studies tracking subtype stability, target engagement studies confirming pathway modulation, and stratified clinical trials evaluating differential treatment responses. (2) AHBA represented only six adult neurotypical donors and could not distinguish finer neuron subtypes. As age-appropriate transcriptomic and single-cell technologies evolve, future studies may better elucidate precise neuronal circuits affected in adolescent depression. (3) While we controlled for sex as a covariate, sex-stratified analyses were not performed. Given known sex differences in depression pathophysiology and treatment response, future research should explicitly examine neurodevelopmental trajectories and molecular pathways by sex to inform targeted interventions. (4) Our study lacked peripheral biomarker data that could enhance clinical translation. Future research should integrate blood markers with neuroimaging to identify accessible signatures. (5) Finally, although depression mechanisms show conservation across populations, environmental and sociocultural factors may influence subtype expression, necessitating international replications to validate these subtypes across diverse populations.
Conclusion
In conclusion, our study advanced adolescent depression heterogeneity by identifying two neurobiologically distinct subtypes with differential morphometric network patterns, clinical presentations, and molecular signatures. By integrating structural neuroimaging with spatial transcriptomics, we provided novel insights into the molecular architecture underlying these network disruptions, highlighting subtype-specific cellular processes and developmental windows that may inform targeted therapeutic approaches. As we continue to unravel the complex interplay between genes, brain, and behavior during adolescent development, such integrated approaches may ultimately transform our ability to diagnose, treat, and prevent the devastating impact of depression during this critical developmental period.
Data availability
(1) Human Gene Expression Data: All human gene expression data supporting this study are available in the Allen Brain Atlas under “Complete normalized microarray datasets” (https://human.brain-map.org/static/download).
(2) MDD-related Gene List from ISH: The MDD-related genes obtained from in situ hybridization (ISH) can be accessed via http://help.brain-map.org/download/attachments/2818165/HBA_ISH_GeneList.pdf?version=1&modificationDate=1348783035873&api=v2.
(3) Cell-Specific Gene Set List: The compiled cell-specific gene set list, derived from large-scale single-cell studies of the adult human cortex, is available from the raw dataset provided by Seidlitz et al. (https://static-content.springer.com/esm/art%3A10.1038%2Fs41467-020-17051-5/MediaObjects/41467_2020_17051_MOESM8_ESM.xlsx).
(4) Neuroimaging Preprocessing: We utilized the FreeSurfer software suite (version 7.3.2), which is freely available at http://surfer.nmr.mgh.harvard.edu/.
(5) MSN Analysis and PLS Code: The codes for constructing the MSN and performing the PLS analysis are openly shared on GitHub at https://github.com/SarahMorgan/Morphometric_Similarity_SZ.
(6) HYDRA for Clustering Analysis: The HYDRA software, which we used for clustering analyses, is also publicly available at https://github.com/evarol/HYDRA.
Code availability
(1) Human Gene Expression Data: All human gene expression data supporting this study are available in the Allen Brain Atlas under “Complete normalized microarray datasets” (https://human.brain-map.org/static/download).
(2) MDD-related Gene List from ISH: The MDD-related genes obtained from in situ hybridization (ISH) can be accessed via http://help.brain-map.org/download/attachments/2818165/HBA_ISH_GeneList.pdf?version=1&modificationDate=1348783035873&api=v2.
(3) Cell-Specific Gene Set List: The compiled cell-specific gene set list, derived from large-scale single-cell studies of the adult human cortex, is available from the raw dataset provided by Seidlitz et al. (https://static-content.springer.com/esm/art%3A10.1038%2Fs41467-020-17051-5/MediaObjects/41467_2020_17051_MOESM8_ESM.xlsx).
(4) Neuroimaging Preprocessing: We utilized the FreeSurfer software suite (version 7.3.2), which is freely available at http://surfer.nmr.mgh.harvard.edu/.
(5) MSN Analysis and PLS Code: The codes for constructing the MSN and performing the PLS analysis are openly shared on GitHub at https://github.com/SarahMorgan/Morphometric_Similarity_SZ.
(6) HYDRA for Clustering Analysis: The HYDRA software, which we used for clustering analyses, is also publicly available at https://github.com/evarol/HYDRA.
References
Avenevoli S, Swendsen J, He JP, Burstein M, Merikangas KR. Major depression in the national comorbidity survey-adolescent supplement: prevalence, correlates, and treatment. J Am Acad Child Adolesc Psychiatry. 2015;54:37–44.e2.
Organization WH. Adolescent mental health. World Health Organization, Geneva, Switzerland, 2023.
Insel TR, Wang PS. Rethinking mental illness. JAMA. 2010;303:1970–1.
March JS, Silva S, Petrycki S, Curry J, Wells K, Fairbank J, et al. The treatment for adolescents with depression study (TADS): long-term effectiveness and safety outcomes. Arch Gen Psychiatry. 2007;64:1132–43.
Zhou X, Teng T, Zhang Y, Del Giovane C, Furukawa TA, Weisz JR, et al. Comparative efficacy and acceptability of antidepressants, psychotherapies, and their combination for acute treatment of children and adolescents with depressive disorder: a systematic review and network meta-analysis. Lancet Psychiatry. 2020;7:581–601.
Zisook S, Lesser I, Stewart JW, Wisniewski SR, Balasubramani GK, Fava M, et al. Effect of age at onset on the course of major depressive disorder. Am J Psychiatry. 2007;164:1539–46.
Psychogiou L, Navarro MC, Orri M, Côté SM, Ahun MN. Childhood and adolescent depression symptoms and young adult mental health and psychosocial outcomes. JAMA Netw Open. 2024;7:e2425987.
Leaf PJ, Alegria M, Cohen P, Goodman SH, Horwitz SM, Hoven CW, et al. Mental health service use in the community and schools: results from the four-community MECA study. Methods for the epidemiology of child and adolescent mental disorders study. J Am Acad Child Adolesc Psychiatry. 1996;35:889–97.
Gandal MJ, Haney JR, Parikshak NN, Leppa V, Ramaswami G, Hartl C, et al. Shared molecular neuropathology across major psychiatric disorders parallels polygenic overlap. Science. 2018;359:693–7.
Dow-Edwards D, MacMaster FP, Peterson BS, Niesink R, Andersen S, Braams BR. Experience during adolescence shapes brain development: From synapses and networks to normal and pathological behavior. Neurotoxicol Teratol. 2019;76:106834.
Morgan SE, Seidlitz J, Whitaker KJ, Romero-Garcia R, Clifton NE, Scarpazza C, et al. Cortical patterning of abnormal morphometric similarity in psychosis is associated with brain expression of schizophrenia-related genes. Proc Natl Acad Sci USA. 2019;116:9604–9.
Romero-Garcia R, Whitaker KJ, Váša F, Seidlitz J, Shinn M, Fonagy P, et al. Structural covariance networks are coupled to expression of genes enriched in supragranular layers of the human cortex. Neuroimage. 2018;171:256–67.
Yao G, Zou T, Luo J, Hu S, Yang L, Li J, et al. Cortical structural changes of morphometric similarity network in early-onset schizophrenia correlate with specific transcriptional expression patterns. BMC Med. 2023;21:479.
Drysdale AT, Grosenick L, Downar J, Dunlop K, Mansouri F, Meng Y, et al. Resting-state connectivity biomarkers define neurophysiological subtypes of depression. Nat Med. 2017;23:28–38.
Zimmerman M, Martinez JH, Young D, Chelminski I, Dalrymple K. Severity classification on the hamilton depression rating scale. J Affect Disord. 2013;150:384–8.
Liang S, Yu W, Ma X, Luo S, Zhang J, Sun X, et al. Psychometric properties of the MATRICS consensus cognitive battery (MCCB) in Chinese patients with major depressive disorder. J Affect Disord. 2020;265:132–8.
Li X, Phillips M, Xu D, Zhang Y, Yang S, Tong Y, et al. [Reliability and validity of an adapted Chinese version of the Barratt Impulsiveness Scale] Chinese {use Share Publication ‘download’ function to view Chinese article}. Chin Ment Health J. 2011;25:610–5.
Hollon SD, Kendall PC. Cognitive self-statements in depression: development of an automatic thoughts questionnaire. Cognit Ther Res. 1980;4:383–95.
Garnefski N, van den Kommer TN, Kraaij V, Teerds J, Legerstee JS, Onstein EJ. The relationship between cognitive emotion regulation strategies and emotional problems: comparison between a clinical and a non‐clinical sample. Eur J Pers. 2002;16:403–20.
Li Z, Wang L, Zhang L. Exploratory and confirmatory factor analysis of a short-form of the EMBU among Chinese adolescents. Psychol Rep. 2012;110:263–75.
Perris C, Jacobsson L, Lindström H, von Knorring L, Perris H. Development of a new inventory assessing memories of parental rearing behaviour. Acta Psychiatr Scand. 1980;61:265–74.
Liu XC, Oda S, Peng X, Asai K. Life events and anxiety in Chinese medical students. Soc Psychiatry Psychiatr Epidemiol. 1997;32:63–67.
Liu X, Kurita H, Uchiyama M, Okawa M, Liu L, Ma D. Life events, locus of control, and behavioral problems among Chinese adolescents. J Clin Psychol. 2000;56:1565–77.
Fischl B. FreeSurfer. Neuroimage. 2012;62:774–81.
Rosen A, Roalf DR, Ruparel K, Blake J, Seelaus K, Villa LP, et al. Quantitative assessment of structural image quality. Neuroimage. 2018;169:407–18.
Desikan RS, Ségonne F, Fischl B, Quinn BT, Dickerson BC, Blacker D, et al. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage. 2006;31:968–80.
Seidlitz J, Váša F, Shinn M, Romero-Garcia R, Whitaker KJ, Vértes PE, et al. Morphometric similarity networks detect microscale cortical organization and predict inter-individual cognitive variation. Neuron. 2018;97:231–47.e7.
Romero-Garcia R, Atienza M, Clemmensen LH, Cantero JL. Effects of network resolution on topological properties of human neocortex. Neuroimage. 2012;59:3522–32.
Whitaker KJ, Vértes PE, Romero-Garcia R, Váša F, Moutoussis M, Prabhu G, et al. Adolescence is associated with genomically patterned consolidation of the hubs of the human brain connectome. Proc Natl Acad Sci USA. 2016;113:9105–10.
King DJ, Wood AG. Clinically feasible brain morphometric similarity network construction approaches with restricted magnetic resonance imaging acquisitions. Netw Neurosci. 2020;4:274–91.
Varol E, Sotiras A, Davatzikos C, Alzheimer’s Disease Neuroimaging Initiative. HYDRA: revealing heterogeneity of imaging and genetic patterns through a multiple max-margin discriminative analysis framework. Neuroimage 2017; 145:346–64.
Varol E, Sotiras A, Davatzikos C. MIDAS: Regionally linear multivariate discriminative statistical mapping. Neuroimage. 2018;174:111–26.
Schaefer A, Kong R, Gordon EM, Laumann TO, Zuo XN, Holmes AJ, et al. Local-global parcellation of the human cerebral cortex from intrinsic functional connectivity MRI. Cereb Cortex. 2018;28:3095–114.
Banovac I, Sedmak D, Judaš M, Petanjek Z. Von economo neurons - primate-specific or commonplace in the mammalian brain. Front Neural Circuits. 2021;15:714611.
Baum GL, Cui Z, Roalf DR, Ciric R, Betzel RF, Larsen B, et al. Development of structure-function coupling in human brain networks during youth. Proc Natl Acad Sci USA. 2020;117:771–8.
Braun E, Danan-Gotthold M, Borm LE, Lee KW, Vinsland E, Lönnerberg P, et al. Comprehensive cell atlas of the first-trimester developing human brain. Science. 2023;382:eadf1226.
Markello RD, Arnatkeviciute A, Poline JB, Fulcher BD, Fornito A, Misic B. Standardizing workflows in imaging transcriptomics with the abagen toolbox. eLife. 2021;10:e72129.
Arnatkeviciute A, Fulcher BD, Fornito A. A practical guide to linking brain-wide gene expression and neuroimaging data. Neuroimage. 2019;189:353–67.
Abdi H, Williams LJ. Partial least squares methods: partial least squares correlation and partial least square regression. Methods Mol Biol. 2013;930:549–79.
Li J, Seidlitz J, Suckling J, Fan F, Ji GJ, Meng Y, et al. Cortical structural differences in major depressive disorder correlate with cell type-specific transcriptional signatures. Nat Commun. 2021;12:1647.
Zeng H, Shen EH, Hohmann JG, Oh SW, Bernard A, Royall JJ, et al. Large-scale cellular-resolution gene profiling in human neocortex reveals species-specific molecular signatures. Cell. 2012;149:483–96.
Tang D, Chen M, Huang X, Zhang G, Zeng L, Zhang G, et al. SRplot: a free online platform for data visualization and graphing. PLoS ONE. 2023;18:e0294236.
Seidlitz J, Nadig A, Liu S, Bethlehem R, Vértes PE, Morgan SE, et al. Transcriptomic and cellular decoding of regional brain vulnerability to neurogenetic disorders. Nat Commun. 2020;11:3358.
Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics. 2013;14:7.
Dougherty JD, Schmidt EF, Nakajima M, Heintz N. Analytical approaches to RNA profiling data for the identification of genes enriched in specific cells. Nucleic Acids Res. 2010;38:4218–30.
Salk RH, Hyde JS, Abramson LY. Gender differences in depression in representative national samples: Meta-analyses of diagnoses and symptoms. Psychol Bull. 2017;143:783–822.
Petanjek Z, Judaš M, Šimic G, Rasin MR, Uylings HB, Rakic P, et al. Extraordinary neoteny of synaptic spines in the human prefrontal cortex. Proc Natl Acad Sci USA. 2011;108:13281–6.
Hamilton JP, Etkin A, Furman DJ, Lemus MG, Johnson RF, Gotlib IH. Functional neuroimaging of major depressive disorder: a meta-analysis and new integration of base line activation and neural response data. Am J Psychiatry. 2012;169:693–703.
Toenders YJ, Schmaal L, Nawijn L, Han L, Binnewies J, van der Wee N, et al. The association between clinical and biological characteristics of depression and structural brain alterations. J Affect Disord. 2022;312:268–74.
Mulders PC, van Eijndhoven PF, Schene AH, Beckmann CF, Tendolkar I. Resting-state functional connectivity in major depressive disorder: a review. Neurosci Biobehav Rev. 2015;56:330–44.
Schwarz MJ, Ackenheil M. The role of substance P in depression: therapeutic implications. Dialogues Clin Neurosci. 2002;4:21–29.
Ebner K, Muigg P, Singewald G, Singewald N. Substance P in stress and anxiety: NK-1 receptor antagonism interacts with key brain areas of the stress circuitry. Ann N Y Acad Sci. 2008;1144:61–73.
Yan TC, McQuillin A, Thapar A, Asherson P, Hunt SP, Stanford SC, et al. NK1 (TACR1) receptor gene ‘knockout’ mouse phenotype predicts genetic association with ADHD. J Psychopharmacol. 2010;24:27–38.
Deslauriers J, Larouche A, Sarret P, Grignon S. Combination of prenatal immune challenge and restraint stress affects prepulse inhibition and dopaminergic/GABAergic markers. Prog Neuropsychopharmacol Biol Psychiatry. 2013;45:156–64.
Rubino T, Prini P, Piscitelli F, Zamberletti E, Trusel M, Melis M, et al. Adolescent exposure to THC in female rats disrupts developmental changes in the prefrontal cortex. Neurobiol Dis. 2015;73:60–69.
Schneider M. Puberty as a highly vulnerable developmental period for the consequences of cannabis exposure. Addict Biol. 2008;13:253–63.
Cubelos B, Sebastián-Serrano A, Kim S, Moreno-Ortiz C, Redondo JM, Walsh CA, et al. Cux-2 controls the proliferation of neuronal intermediate precursors of the cortical subventricular zone. Cereb Cortex. 2008;18:1758–70.
Nieto M, Monuki ES, Tang H, Imitola J, Haubst N, Khoury SJ, et al. Expression of Cux-1 and Cux-2 in the subventricular zone and upper layers II-IV of the cerebral cortex. J Comp Neurol. 2004;479:168–80.
Pehrson AL, Sanchez C. Serotonergic modulation of glutamate neurotransmission as a strategy for treating depression and cognitive dysfunction. CNS Spectr. 2014;19:121–33.
Soumier A, Sibille E. Opposing effects of acute versus chronic blockade of frontal cortex somatostatin-positive inhibitory neurons on behavioral emotionality in mice. Neuropsychopharmacology. 2014;39:2252–62.
Fee C, Banasr M, Sibille E. Somatostatin-positive gamma-aminobutyric acid interneuron deficits in depression: cortical microcircuit and therapeutic perspectives. Biol Psychiatry. 2017;82:549–59.
Wohleb ES, Franklin T, Iwata M, Duman RS. Integrating neuroimmune systems in the neurobiology of depression. Nat Rev Neurosci. 2016;17:497–511.
Datta D, Arnsten A. Loss of prefrontal cortical higher cognition with uncontrollable stress: molecular mechanisms, changes with age, and relevance to treatment. Brain Sci. 2019;9:113.
Whittle S, Lichter R, Dennison M, Vijayakumar N, Schwartz O, Byrne ML, et al. Structural brain development and depression onset during adolescence: a prospective longitudinal study. Am J Psychiatry. 2014;171:564–71.
Bagot RC, Cates HM, Purushothaman I, Lorsch ZS, Walker DM, Wang J, et al. Circuit-wide transcriptional profiling reveals brain region-specific gene networks regulating depression susceptibility. Neuron. 2016;90:969–83.
Chung WS, Welsh CA, Barres BA, Stevens B. Do glia drive synaptic and cognitive impairment in disease. Nat Neurosci. 2015;18:1539–45.
Heng L, Beverley JA, Steiner H, Tseng KY. Differential developmental trajectories for CB1 cannabinoid receptor expression in limbic/associative and sensorimotor cortical areas. Synapse. 2011;65:278–86.
Reul JM, Chandramohan Y. Epigenetic mechanisms in stress-related memory formation. Psychoneuroendocrinology. 2007;32:S21–25.
Keren-Shaul H, Spinrad A, Weiner A, Matcovitch-Natan O, Dvir-Szternfeld R, Ulland TK, et al. A unique microglia type associated with restricting development of Alzheimer’s disease. Cell. 2017;169:1276–90.e17.
Chung WS, Allen NJ, Eroglu C. Astrocytes control synapse formation, function, and elimination. Cold Spring Harb Perspect Biol. 2015;7:a020370.
Caballero A, Tseng KY. GABAergic function as a limiting factor for prefrontal maturation during adolescence. Trends Neurosci. 2016;39:441–8.
McKlveen JM, Morano RL, Fitzgerald M, Zoubovsky S, Cassella SN, Scheimann JR, et al. Chronic stress increases prefrontal inhibition: a mechanism for stress-induced prefrontal dysfunction. Biol Psychiatry. 2016;80:754–64.
Tyborowska A, Volman I, Niermann H, Pouwels JL, Smeekens S, Cillessen A, et al. Early-life and pubertal stress differentially modulate grey matter development in human adolescents. Sci Rep. 2018;8:9201.
Kang HJ, Kawasawa YI, Cheng F, Zhu Y, Xu X, Li M, et al. Spatio-temporal transcriptome of the human brain. Nature. 2011;478:483–9.
Entringer S, Buss C, Rasmussen JM, Lindsay K, Gillen DL, Cooper DM, et al. Maternal cortisol during pregnancy and infant adiposity: a prospective investigation. J Clin Endocrinol Metab. 2017;102:1366–74.
Teicher MH, Samson JA. Annual research review: enduring neurobiological effects of childhood abuse and neglect. J Child Psychol Psychiatry. 2016;57:241–66.
Motter JN, Pimontel MA, Rindskopf D, Devanand DP, Doraiswamy PM, Sneed JR. Computerized cognitive training and functional recovery in major depressive disorder: a meta-analysis. J Affect Disord. 2016;189:184–91.
Yan TC, Hunt SP, Stanford SC. Behavioural and neurochemical abnormalities in mice lacking functional tachykinin-1 (NK1) receptors: a model of attention deficit hyperactivity disorder. Neuropharmacology. 2009;57:627–35.
Meyer TA, Habib AS, Wagner D, Gan TJ. Neurokinin-1 receptor antagonists for the prevention of postoperative nausea and vomiting. Pharmacotherapy. 2023;43:922–34.
Geng M, Shao Q, Fu J, Gu J, Feng L, Zhao L, et al. Down-regulation of MKP-1 in hippocampus protects against stress-induced depression-like behaviors and neuroinflammation. Transl Psychiatry. 2024;14:130.
Lopez B, Schwartz SJ, Prado G, Campo AE, Pantin H. Adolescent neurological development and its implications for adolescent substance use prevention. J Prim Prev. 2008;29:5–35.
Fuchs C, Abitbol K, Burden JJ, Mercer A, Brown L, Iball J, et al. GABA(A) receptors can initiate the formation of functional inhibitory GABAergic synapses. Eur J Neurosci. 2013;38:3146–58.
Dunning DL, Griffiths K, Kuyken W, Crane C, Foulkes L, Parker J, et al. Research Review: The effects of mindfulness-based interventions on cognition and mental health in children and adolescents - a meta-analysis of randomized controlled trials. J Child Psychol Psychiatry. 2019;60:244–58.
Eckshtain D, Kuppens S, Ugueto A, Ng MY, Vaughn-Coaxum R, Corteselli K, et al. Meta-Analysis: 13-Year Follow-up of Psychotherapy Effects on Youth Depression. J Am Acad Child Adolesc Psychiatry. 2020;59:45–63.
Van den Bergh B, van den Heuvel MI, Lahti M, Braeken M, de Rooij SR, Entringer S, et al. Prenatal developmental origins of behavior and mental health: the influence of maternal stress in pregnancy. Neurosci Biobehav Rev. 2020;117:26–64.
Luby JL, Barch DM, Belden A, Gaffrey MS, Tillman R, Babb C, et al. Maternal support in early childhood predicts larger hippocampal volumes at school age. Proc Natl Acad Sci USA. 2012;109:2854–9.
Tottenham N, Hare TA, Quinn BT, McCarry TW, Nurse M, Gilhooly T, et al. Prolonged institutional rearing is associated with atypically large amygdala volume and difficulties in emotion regulation. Dev Sci. 2010;13:46–61.
Diamond A, Lee K. Interventions shown to aid executive function development in children 4 to 12 years old. Science. 2011;333:959–64.
Brent DA, Brunwasser SM, Hollon SD, Weersing VR, Clarke GN, Dickerson JF, et al. Effect of a cognitive-behavioral prevention program on depression 6 years after implementation among at-risk adolescents: a randomized clinical trial. JAMA Psychiatry. 2015;72:1110–8.
Kerestes R, Davey CG, Stephanou K, Whittle S, Harrison BJ. Functional brain imaging studies of youth depression: a systematic review. Neuroimage Clin. 2014;4:209–31.
Disner SG, Beevers CG, Haigh EA, Beck AT. Neural mechanisms of the cognitive model of depression. Nat Rev Neurosci. 2011;12:467–77.
Cuthbert BN, Insel TR. Toward the future of psychiatric diagnosis: the seven pillars of RDoC. BMC Med. 2013;11:126.
Acknowledgements
This work was supported by the Joint Fund for Regional Innovative Development (U24A20700, to YT) and the Science and Technology Innovation 2030-Major Project on Brain Science and Brain-like Research (2021ZD0200700, to YT). We extend our sincere gratitude to all participants for their invaluable contributions to this study. We also thank the research teams at Shengjing Hospital of China Medical University and the First Affiliated Hospital of China Medical University for their support in participant recruitment and data collection. Finally, we acknowledge the technical support provided by the Allen Institute for Brain Science and the Brainspan Consortium for their publicly available transcriptomic data, which were instrumental in this study.
Author information
Authors and Affiliations
Contributions
YT and PW conceptualized and designed the study. PW performed data analysis, wrote and revised the manuscript. LK and YZ contributed to patients recruitment and clinical assessments. YT obtained funding and supervised the project. PW, CD, ZW, YS, LW, ZT, YW, YL, QS and YNZ contributed to clinical and MRI data collection.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Wu, P., Kong, L., Zhou, Y. et al. Neurobiological subtypes of adolescent depression: a multimodal integration of morphometric similarity network and spatial transcriptomics. Mol Psychiatry 30, 5448–5462 (2025). https://doi.org/10.1038/s41380-025-03133-7
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s41380-025-03133-7