Abstract
The 2-oxoglutarate dehydrogenase complex (OGDHc) orchestrates a critical reaction regulating the TCA cycle. Although the structure of each OGDHc subunit has been solved, the architecture of the intact complex and inter-subunit interactions still remain unknown. Here we report the assembly of native, intact OGDHc from Sus scrofa heart tissue using cryo-electron microscopy (cryo-EM), cryo-electron tomography (cryo-ET), and subtomogram averaging (STA) to discern native structures of the whole complex and each subunit. Our cryo-EM analyses revealed the E2o cubic core structure comprising eight homotrimers at 3.3-Å resolution. More importantly, the numbers, positions and orientations of each OGDHc subunit were determined by cryo-ET and the STA structures of the core were resolved at 7.9-Å with the peripheral subunits reaching nanometer resolution. Although the distribution of the peripheral subunits E1o and E3 vary among complexes, they demonstrate a certain regularity within the position and orientation. Moreover, we analyzed and validated the interactions between each subunit, and determined the flexible binding mode for E1o, E2o and E3, resulting in a proposed model of Sus scrofa OGDHc. Together, our results reveal distinctive factors driving the architecture of the intact, native OGDHc.
Similar content being viewed by others
Introduction
The tricarboxylic acid (TCA) cycle is an essential pathway in aerobic metabolism. Within the TCA cycle, the 2-oxoglutarate dehydrogenase complex (OGDHc, also known as the α-ketoglutarate dehydrogenase complex (KGDHc)) catalyzes the conversion of α-ketoglutarate to succinyl-coenzyme A (CoA), which simultaneously produces NADH and releases CO21,2,3. Because this is an irreversible and rate-limiting step in the TCA cycle, OGDHc is tightly regulated according to metabolic demands4,5. Under certain pathological conditions, reactive oxygen species (ROS) generated by OGDHc have been shown to induce mitochondrial oxidative stress6,7, and decreased OGDHc activity can be plausibly linked to cancer and multiple neurodegenerative diseases such as Parkinson’s disease (PD) and Alzheimer’s disease (AD)8,9,10,11. In addition, OGDHc activity affects histone modification and regulates gene expression12,13.
OGDHc is a member of the α-ketoacid dehydrogenase family (OADHc), which also includes the pyruvate dehydrogenase complex (PDHc) and the branched-chain α-ketoacid dehydrogenase complex (BCKDHc)14. The complexes in this family usually comprise three types of metabolic enzymes, including an α-ketoacid dehydrogenase (E1), an acyltransferase (E2), and a dihydrolipoyl dehydrogenase (E3)15. Interestingly, all complexes in this family have identical E3 subunits except Pseudomonas putida16, and they all contain an E2 core with E1 and E3 as peripheral subunits bound together by non-covalent interactions17. The core of PDHc in Azotobacter vinelandii, the core of OGDHc in Escerichia coli, and the core of BCKDHc in Mycobacterium tuberculosis are all assembled from eight E2 homotrimers, forming a 24-mer cubic core18,19,20. In mammals and some other eukaryotes, E2o and E2b also assemble into a cubic core21,22,23,24, whereas E2p assembles into an icosahedral structure with 20 homotrimers25,26. Moreover, in eukaryotic PDHc, there is a special protein, E3BP (E3 binding protein), which is very similar to E2p responsible for binding E3, but the E3BP subunit is not present in OGDHc27,28. Both mammalian and bacteria E2o subunits contain a lipoyl domain (LD), an inner core domain (ICD), and other flexible linker domains required for its function15. The LD covalently binds lipoic acid to participate in the reaction, while the ICD stabilizes the core of the protein complex and catalyzes the transfer of succinyl groups to CoA29. In E2o of bacteria and yeast, there is a small peripheral subunit binding domain (PSBD) between the LD and ICD30, but no PSBD has been identified in E2o in mammals31,32,33. Instead, in mammals, some evidence suggests that E1o is participating in binding E334,35. Moreover, the dual subunit binding domain (DSBD) in E1o strongly binds to E2o and simultaneously weakly binds to E336. But so far, there are no structural data from the native complex to support this hypothesis.
Most studies to determine the OGDHc structure have used cell expression systems to express individual subunits, which are analyzed by X-ray crystallography and cryo-EM. For E. coli, the structures of E2o-LD, E2o-PSBD, a truncated ecE2o cubic core, and the trimeric form of the catalytic domain have been reported19,30,37,38, and the structures of E1o and E3 were resolved by X-ray crystallography39,40. For mammalian OGDHc, high-resolution structures of E2o and E1o were obtained by cryo-EM21,41, and the E3 structure was obtained by X-ray crystallography42, but very few studies have examined the whole structure of the OGDHc. Initially, the structure of OGDHc was thought to conform to the 2:2:1 face–edge model5,15,43, in which a single E1o dimer binds to each of the 12 edges of the E2o core while a single E3 dimer binds to each of the 6 faces; in this configuration, the E2o LDs move freely above and between the E1o, E2o-ICD and E3 subunits44. Other subunit ratios have also been proposed, including 1:1:1.5, 1:1.2:1.4, 1:2:1, and 1:1:145,46,47,48. Increasingly, the evidence points to a heterogeneous and flexible assembly of OGDHc in which the occupancy of the peripheral subunits cannot reach 100%24,49,50,51,52, indicating that E1o and E3 are flexibly distributed around the E2o core.
Owing to its heterogeneous structure, cryo-EM single-particle analysis and X-ray crystallography are inadequate to precisely determine the OGDHc overall structure. Here we combine cryo-ET, single-particle analysis (SPA), and subtomogram averaging (STA) to study the intact OGDHc, including the interactions between the E2o core and peripheral E1o and E3 subunits. On the basis of our results, we propose a model for mammalian OGDHc to report the architecture and assembly of native OGDHc.
Results
Purification and characterization of mammalian OGDHc
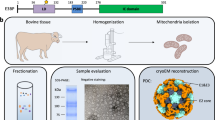
To obtain intact OGDHc, mitochondria from Sus scrofa (porcine) heart tissue were isolated and lysed. The endogenous OGDHcs within the mitochondrial matrix were extracted by polyethylene glycol precipitation and preliminarily purified using sucrose gradient fractionation (Supplementary Fig. 1a). Examination of the gradient fractions by sodium dodecyl-sulfate polyacrylamide gel electrophoresis (SDS–PAGE) suggested the presence of intact OGDHc in specific sedimentation coefficient sections, in which there was also a very small amount of co-migrating substances, corresponding to PDHc53 (Supplementary Fig. 1a). We next purified the OGDHc-containing fractions using a reformative GraFix method54 to further remove all other proteins and to stabilize the individual macromolecules of OGDHc. Results of subsequent gel exclusion chromatography combined with gel electrophoresis confirmed the purity of OGDHc-containing fractions as well as the integrity of OGDHc molecules after GraFix (Supplementary Fig. 1b). The OGDHc sample was then analyzed by mass spectrometry, and each component was identified (Supplementary Table 1). Continuous spectrophotometric measurement of the catalysis of α-ketoglutarate to generate NADH indicated that the purified OGDHcs maintained biological activity (Supplementary Fig. 1c). At the same time, the cross-corroboration of enzyme activity experiments with different substrates of OGDHc (2-oxoglutarate), PDHc (pyruvate), BCKDHc (4-methyl-2-oxovalerate) can further indicate that our purified OGDHc samples contain very small amounts of PDHcs, while BCKDHc is almost absent. The core shape of the PDHc is different from that of the OGDHc, which does not affect our subsequent manual particle selection. These results confirm that we isolated functional OGDHc in its native conformation, which was, therefore, suitable for structural analysis.
Because OGDHcs are highly flexible and heterogeneous, we combined cryo-ET and STA to reconstruct the overall structure of mammalian OGDHc (Fig. 1a–d and Supplementary Figs. 1d, 2). The OGDHc cryo-samples were prepared with 10-nm gold fiducial markers by plunge freezing. After collecting 137 tilt series using a dose-symmetry scheme, we were able to observe the intact OGDHc on reconstructed, low-passed tomograms (Fig. 1a). The tomograms were also processed by IsoNet55 (Supplementary Fig. 3a). The representative 4-fold symmetry orientation of the OGDHc cubic core and the surrounding peripheral subunits were easily identifiable, and the diameter of the whole complex was ~ 35 nm (Fig. 1a, b). In the tomogram, the number of visible peripheral subunits varied from approximately four to approximately ten. This reflects the heterogeneity of the OGDHcs24, which was consistent with the heterogeneous situations reported from E. coli51, C. thermophilum23, and bovine kidney24. Moreover, this finding suggested that the occupancy of the E2o core with peripheral subunits could not reach 100%. The peripheral subunits were mostly distributed in a spherical shell outside the cubic core. The gap between the inner core and the spherical shell was about 3 nm.
a One slide of a representative tomogram from all the 137 tomograms. One intact OGDHc was marked by the orange box. The black bar represents 100 nm. b A representative OGDHc from 2014 complexes is reconstructed by back-projecting E2o (blue), E1o (orange), and E3 (green) to the tomogram according to their refined coordinates and Euler angles. The black bar represents 3 nm. c E2o density map from subtomogram averaging. Structures are shown as transparent surfaces fitted with the atomic model of single particle E2o (this article). d E1o density map from subtomogram averaging. Structures are shown as transparent surfaces fitted with atomic model E1o (PDB:7WGR). e E3 density map from subtomogram averaging. Structures are shown as transparent surfaces fitted with atomic model E3 (PDB:6I4Q). f Statistical numbers of E1o and E3 per complex. The mean values of E1o and E3 were 4.16 and 4.10 in a complex, respectively, and the whole ratio of E1o and E3 is nearly 1:1. All of the 2014 OGDHcs were analyzed, which contain 8379 E1os and 8252 E3s. A boxplot shows the outliers, minimum, first quartile, medium, third quartile, and maximum of the data. g Occupancy of the peripheral subunits. E2o in the complex is not fully occupied, and most of the complex occupancy is lower than 50%. The largest number of reconstructed complexes are those with about 30% occupancy. h Distance distribution of the peripheral subunit. E1 and E3 both have a peak at a distance from the center of the 24-mer cubic core and their mean values are not much different. E1 distribution is more concentrated, and E3 is more diffuse.
We then picked 2014 OGDHc core subunits (E2o) and 17,513 peripheral subunits (E1o and E3) from the tomograms and performed classification and STA, which yielded a 7.9-Å E2o core map, a 9.7-Å E1o map, and a 12.2-Å E3 map (Fig. 1c–e and Supplementary Figs. 1d, 2). Among the refined peripheral subtomograms, approximately 50% were E1o subunits and 50% were E3 subunits (see “Methods”). After back-projecting the picked subunits to the tomograms, we reconstructed the whole complex structure (Fig. 1b and Supplementary Fig. 3b). In each reconstructed complex map, we could clearly observe the position and orientation of the peripheral subunits around the E2o core, and the results indicated that the assembly of the OGDHc is not consistent with the face–edge model5,15. We further speculated that E1o and E3 subunits were connected to the core domain of E2o via a linker whose density could not be seen in the displayed map owing to its flexible swinging. An average of 4.16 ± 1.82 E1o subunits and 4.10 ± 2.13 E3 subunits were found to be randomly distributed on each E2o core (Fig. 1f). The maximum number of E1o and E3 subunits reached 12 and 13, respectively, while the minimum number for both subunits was 0. This large variation in subunit stoichiometry is probably one of the main factors driving the structural heterogeneity of OGDHcs. We considered OGDHcs with 0 E1o or E3 subunits to be not fully assembled, and this result may also indicate that E1o and E3 subunits can independently bind the E2o core without the other peripheral subunit. By stipulating two conditions where either all 24 of the E2o monomers were bound to peripheral subunits (100% occupied) and where none of the E2o monomers was bound to peripheral subunits (0% occupied), we determined that the core occupancy of S. scrofa OGDHc was roughly concentrated between 25%–50% (Fig. 1g), which is a little higher than the situation in E. coli OGDHcs with about 2–8 peripheral subunits51, indicating that OGDHc core occupancy varies between species. More interestingly, although the overall E1o:E3 ratio is nearly 1:1, the E1o:E3 ratio in each complex was highly variable among OGDHcs, suggesting a complicated binding mode for E1o and E3 in the complex (Supplementary Fig. 4a). This ratio diverges from the 2:2:1 ratio of E1o:E2o:E3 proposed in the original face-edge model as well as from other proposed stoichiometries of 1:1:1.5 and 1:2:121.
In situ characterization of mammalian OGDHc
In order to verify that the peripheral subunits do not dissociate during purification and sample preparation, which could introduce some artifacts, we plunged freezing the porcine mitochondria and then directly imaged the samples and collected cryo-ET tilt series. Endogenous proteins within the mitochondria are protected within the bilayer membrane and are, therefore, not affected by purification and sample preparation procedures. We collected 28 tilt series and reconstructed the tomograms by compensating the missing wedge with IsoNet. The mitochondria outer membrane (OMM) and cristae, as well as the cubic core in the mitochondrial matrix, are clearly visible in the tomogram (Fig. 2a). We also estimated the size of a cubic core complex, which is very similar to the purified OGDHc, with a scale of about 35 nm (Fig. 1a). However, both OGDHc and BCKDHc core are cubic morphologies in the mitochondria of the porcine heart tissue. Therefore, we can’t distinguish whether the cube core is OGDHc or BCKDHc only based on the tomograms.
a One slide of a representative mitochondria tomogram from all 28 tomograms. Complex with the cubic core in the mitochondria matrix was marked by the orange box. The black bar represents 100 nm. b Statistical numbers of E1o and E3 per cubic-core complex. The mean values of E1o and E3 were 4.19 and 4.36 in a complex, respectively, and the whole ratio of E1o and E3 is nearly 1:1. All of the 86 complexes were analyzed, which contain 361 E1os and 375 E3s. A boxplot shows the outliers, minimum, first quartile, medium, third quartile, and maximum of the data. c Occupancy of the peripheral subunits. The cubic-core complex is not fully occupied, and most of the complex occupancy is lower than 50%. The largest number of particles in our reconstructed complexes are those with about 30% occupancy, which is the same as the purified OGDHc.
We manually picked 86 OGDHc/BCKDHc cubic cores, as well as 1054 initial peripheral subtomograms (need to remove duplicates) around these 86 cores (Supplementary Fig. 5). The number of the peripheral subtomograms proves that the occupancy of the complex is still not high (1054/(86 ∗ 24) = 51%) in the mitochondria without subtomogram averaging. We subsequently refined the cubic core to 14.4 Å. At this resolution it is impossible to distinguish between the OGDHc and BCKDHc due to the subtle structural differences between them (Supplementary Fig. 5). Next, we use E1o (PDB:7WGR), E1b (PDB:2J9F) and E3 (PDB:6I4Q) as initial references for multi-reference 3D classification (Supplementary Fig. 5c). The results showed that 507, 547 and 0 particles were classified into the E1o, E3, and E1b reference class, respectively. Our overall classification results seem to suggest that the number of BCKDHc in our selected mitochondria is much lower than that of OGDHc. However, there is currently no direct biochemical approach to identify and cross-validate BCKDHc and OGDHc in the in situ mitochondrial environment. Thus we consider the cubic-core complex to be OGDHc and E1 to be E1o based on the existing technology. This does not lead to appreciable statistical errors in the number and occupancy of the peripheral subunits.
In mitochondria, each cubic-core complex has an average of 4.19 ± 2.03 E1, and an average of 4.36 ± 1.63 E3 (Fig. 2b). Among these complexes, a minimum of three peripheral subunits were present, and in addition, there exist complexes with 0 E1os, the same as in the purified samples, suggesting that there were still not fully assembled complexes in vivo. We calculated the occupancy and found that, similar to the in vitro purified OGDHc, the occupancy was still mostly less than 50%, and most of the occupancy is concentrated in 30%–40% (Fig. 2c). Meanwhile, the same situation as in the purified OGDHc, the overall E1:E3 ratio of the cubic-core complex in mitochondria is nearly 1:1, but E1:E3 ratio in each complex was also highly variable among complexes (Supplementary Fig. 4b). The statistics of the number and occupancy of in situ mitochondria samples proved that the peripheral subunits did not dissociate and lead to artifacts during the purification and sample preparation process, which is essential to support the analysis of the purified samples. Therefore, subsequent statistical analyses were performed based on our purified OGDC samples.
Distribution of peripheral subunits around the core
Screening of all the reconstructed OGDHcs showed that the distribution of peripheral E1o and E3 subunits, as well as the position and orientation, were variable between each complex (Figs. 1h, 3). Statistical analyses revealed that, among the 2014 screened OGDHcs, E1o and E3 both approximately followed the normal distribution. On average, E1o and E3 distributed 153.97 ± 34.4 Å and 148.90 ± 51.00 Å, respectively, from the center of the cubic core (Fig. 1h). The mean value of the distance between the mass of E1o/E3 and the core is about the same but the peak shape of the distance distribution for E1o was sharper compared with that of E3, which indicates that E1o subunits are more clustered. Besides, compared with the native OGDHcs from C. thermophilum23, our results indicate that although the mean value of the distance of E1o and E3 retain approximately equal from the core, the standard deviation of the E1o’s and E3’s distance is not about the same. By subtracting E1o and E3’s own size (Fig. 1b), it seems that E1o’s edges are positioned a little closer to the outer circumference of the cubic core, which is somewhat inconsistent with the previous results obtained with in vitro recombinant E. coli OGDHc, showing the E3 gap was farther than the E1o gap51. Moreover, we use the swinging distance, which is defined as the distance between E1o’s or E3’s contour boundary (to subtract the geometric dimension size) and the N-terminal ICD residue L221 to characterize the putative binding position of the E1o/E3 on E2o linker (Methods details). This linker region contains poly-A and poly-P peptides that don’t appear to form a stable and fixed conformation and exist as the loop form, which is one of the most important factors in the high heterogeneity of OADHc family proteins. We found that the swing distances of E1o and E3 were 95.20 ± 35.59 Å and 98.39 ± 49.99 Å, respectively. (Supplementary Fig. 6a, b). To further depict the position distribution of the peripheral subunit intuitively, we defined a new coordinate system, with the 3-fold axis of the cube’s diagonal as the z-axis and the center of the vertex as the origin, and we calculated the relative position of each E1o or E3 with respect to its nearest adjacent vertex and projected them onto the x-y plane (Fig. 3a, d, “Methods” details). Statistically, the equilateral triangular distribution shows a triple symmetry feature corresponding to the E2o trimer form of each vertex. The result showed that, although the distribution of E1o and E3 varied among each complex, the positions of the peripheral subunits were not completely random; rather, there appeared to be some constraints that determined the binding pattern and forced the subunits to swing according to statistical laws.
a Relative position of a E1o group to one vertex. The points represent individual projections of the mass of E1o along the Z-axis. The positive direction of the z-axis is defined as the direction from the origin of the core to the center of the vertex. The characteristics of 3-fold symmetry in the x-y plane are demonstrated. b Orientation of E1o around the core. E1o prefers a tilt of 30° relative to the E2 center-E1o center line. c Comprehensive distance and angle distribution of E1o. The horizontal axis and the vertical axis represent the angles and distance of each E1o relative to E2o. The shade of color represents the frequency of the corresponding distance and angle. d Relative position of E3 to one vertex. e Orientation of E3 around the core. E3 distributes diffusely in the tilt of 60–120°. f Comprehensive distance and angle distribution of E3. g Illustration of angle tilt of E1o and E3’s orientation in the complex. The orientation angle is defined as the angle between the C2 axis of the E1o/E3 and the E2o-E1o/E3 line.
Next, we calculated the angle between the z-axis of E1o/E3 and the E2o-E1o/E2o-E3 line to reflect the angle change of the peripheral subunits (Fig. 3g). The tilt angle distribution suggested that E3 has a large angular rotation range, which was almost entirely distributed between 60° and 120° (Fig. 3e), while the E1o angle was relatively fixed, and most E1o subunits concentrated around 30° (Fig. 3b). Similar findings were observed when we analyzed the angle between the z-axis of E1o/E3 and its nearest vertex–E1o/E3 line (Supplementary Fig. 6c, d). Our comprehensive analyses of distance and angle distribution (Fig. 3c, f) suggested that E1o and E3 subunits were both distributed flexibly around the E2o core, but E1o is distributed more concentratedly compared with E3, and E3 may have more degrees of freedom and a more complicated binding mode in OGDHcs. The aggregated distribution of E1o indicates that most E1os were less likely affected by and insensitive to the swinging of the linker. By projecting the refined E1o/E3’s subtomograms in two dimensions with a larger box size (144 pixels), we categorized the projections into 20 classes (Supplementary Fig. 7) and can see that, since the reconstructing parameters for E1o can bring out the approximate shape of E2o, the majority of E1os seemed to be anchored to the surface of E2o doing slight oscillations. This suggested that for most E1o dimers, both sides are fixed, whereas, on E2o, there may be more than one binding site for E1o dimer, which seems to allow reasonable constraints on the framework. In the case of E3, most of E3’s subtomograms parameters cannot bring out E2o’s geometry (class 1), and even in the classes where E2o’s signal was displayed, the common characteristics of the core signals shown are not as distinctive as those of E1o, which seemed that most of E3 were not slight vibrations outside the core, but may be connected with the linker, thus randomly oscillating around the core.
Native structure of each OGDHc component
To obtain a high-resolution map of the OGDHc core, we acquired a dataset consisting of 6,587 micrographs of the OGDHc sample without gold fiducial markers (Supplementary Fig. 8a), and we solely extracted the interior components to resolve the E2o core structure via single-particle analysis. Using 2D class average evaluation, we validated the regular square-shaped features of the core frame (Supplementary Fig. 8b), and we subsequently imposed octahedral symmetry during 3D reconstruction to resolve the S. scrofa OGDHc core structure to a resolution of 3.3 Å based on 183,521 selected particles (Supplementary Figs. 8, 9). The cubic shape of the core comprised 24 E2o units in total and was demonstrated by distinct symmetric views with two-, three-, and four-fold axes (Fig. 4a). Thus, our data indicates that the core scaffold of S. scrofa OGDHc shares significant structural similarities with that of both bovine BCKDHc and A. vinelandii PDHc18,22.
a Cryo-EM density map of the cubic core. The blue highlights the E2o symmetric units. b The monomer model for the E2o core domain. The secondary structure is labeled in the figure. H: helix, β: β-strand, T: turn. c Interactions in one homotrimer. The head interaction site (HI), side interaction site (SI), waist interaction site (WI), and tail interaction site (TI) are circled in the atomic model. d Interactions between two homotrimers. The hydrophobic pocket is circled in the interaction region. e Extra interactions in one homotrimer. f Extra interactions between two homotrimers. g E1o subtomogram averaging maps fitted with PDB: 7WGR. Densities of the linker region in E1o (S41-L128, D590 to S616, V362 to R379) are highlighted in the insets.
For each monomer, the atomic model of the E2o inner core domain (residues 221–454) could be properly built according to the density map (Fig. 4b), while other parts of the E2o subunits such as LD (residues 71–145) and the linker region (residues 146–220) remained invisible. Within the structure of the E2o inner core, we identified conserved secondary structures, which included six α helixes (H1–H6), two short turns (T1, T2), and a central β sheet (β2, β5-β7, β9-β11) (Fig. 4b). All these structural components showed good agreement between the amino acid sequence and the respective side chain densities (Supplementary Fig. 9c). E2o ICDs are arranged as trimers to constitute 8 vertexes of the core frame with substantial hydrogen bonds (Fig. 4c, d). These interaction sites are highly consistent with those recently reported for the E2o trimer of bovine OGDHc24 and are mainly located in four regions: (a) N-terminal interaction (NI) sites at the N-terminal β1 of ICD; (b) head interaction (HI) sites around three H1 helix at the top of the vertex; (c) waist interaction (WI) sites at the β2 and β8 in the vertex flank; and d) tail interaction (TI) sites of R439 and F379’ at the vertex end (Fig. 4c). Notably, despite very high similarity among species, stronger binding was observed in the N-terminal and the head of the vertex in S. scrofa E2o trimers compared to the recombinant human-E2o21. For instance, extra side-chain interactions (D318–R222’, Q234–K309’) and side-chain to main-chain interactions (K228–E310’) were detected at the NI sites in addition to the typical main-chain to main-chain hydrogen bonds (Fig. 4e). Likewise, additional side-chain to main-chain bindings (K241–Q244’, K241–T246’) observed at HI sites to maintain the interaction of three H1 helixes (Fig. 4e). Collectively, these findings indicate E2o trimer in S. scrofa OGDHc seems to interact tightly in the N terminal compared with that of other mammals.
Between neighboring E2o trimers, a palindromic pattern is formed in which hydrophobic interactions play a vital role in vertex interactions (Fig. 4d). L453 on the C-terminal end of E2o, with its contiguous counterpart, forms a prominent arm inserted into the opposite hydrophobic pocket (Fig. 4d), as described in several 2-oxoglutarate dehydrogenase complexes21,24. In addition, hydrogen bond interactions were also participating in the stabilization between E2o trimers. The extra side-chain interactions (D263–R449’), and side-chain to main-chain binding (L452–N341’) in the atomic model were identified compared to human E2o (Fig. 4f). In contrast, the catalytic and substrate binding residues as well as the active center remain conserved in the mitochondrial dihydrolipoamide acyltransferases of OGDHc, which was previously reported21.
The native structure of peripheral subunits is determined by STA. E1o homodimers adopt a transketolase fold. Due to relatively high homology and sequence identity (Supplementary Fig. 10), we embedded the cryo-EM structure of human E1o (PDB: 7WGR) into the 9.7-Å STA density map (Fig. 4g). Most secondary structures in each domain could be well fitted to the map, which indicated that the overall structure of the native protein is similar to that of the overexpressed purified protein41. H17, H6, and H16 in the α/β1 domain is the best-resolved density (Supplementary Fig. 2a). We also found that the excess density of the linker region (residues 590–616) in the STA map partially connected the α/β1 and α/β2 domains (Fig. 4g), which could not be modeled due to its flexibility in the atomic model. Similarly, the small linker in the α/β1 domain (residues 362–379) was not modeled for the same reason. In addition, we resolved some excess densities at the edges of the N-terminal domain (Fig. 4g), which could not correspond to the atomic model because of their flexibility.
The STA structure of E3 is bone-shaped, and we docked the structure of the human E3 homodimer (PDB: 6I4Q) into the 12.2-Å STA density map (Fig. 1e). Because of limited resolution, we could not visualize each individual secondary structure, but we could discern the outline of the density of the corresponding domain in E3 (Fig. 1e). In sum, the reconstruction of each native OGDHc subunit was highly similar to each corresponding high-resolution structure, which demonstrates that the subunits in OGDHc are conserved among species. This is consistent with the critical role of OGDHc in regulating the TCA cycle.
Interaction among OGDHc components
Analysis of each OGDHc we reconstructed showed that there is a certain amount of E1os and E3s bound around the E2o core, and the distribution of E1o and E3 in the outer ring suggests that the assembly and binding of the subunits is diverse and complicated. Our structural analysis and statistical results indicate that E1o is distributed more concentrated around the inner core while E3 is distributed more diffusely. In addition, there is no PSBD in mammalian E2o to bind E1o and E3. Therefore, it remains unclear how the peripheral subunits bind around the core to form functional OGDHc. Interestingly, there is some evidence that the DSBD in E1o can bind E2o and E3 simultaneously36. After screening all of our tomograms, we found some E1o in the complex is likely to bind to E3 (Fig. 5a); however, this close proximity between E1o and E3 in OGDHc is atypical.
a A representative complex containing tightly adjacent E1o and E3. The orange dot marked particle is identified as E1o, and the green triangle marked particle is identified as E3. b Organization of S. scrofa E2o and E1o domains. c Western blot analysis for subunit interaction detection between each OGDHc component. The first row is for anti-E1o detection and anti-E2o-ICD detection, and the second row is for anti-E3 detection. All of the experiments were performed in triplicate. Source data are provided in the Source Data file. d Surface plasmon resonance was used to detect the interaction between E1o and E3. The real-time binding curves were obtained by injecting different concentrations (49nm-25μM) of the analytes over the ligands-immobilized SPR chip. The curves were analyzed to calculate the equilibrium dissociation constant (KD = 1.84 μM). e Assembly of a structural model of intact eukaryotic OGDHC. The architecture model contains 6 binding modes (i) E1o-E2o_linker binding state, (ii) E1o-E2o_ICD binding state, (iii) E1o-E2o_LD binding state, iv)E3-E2o_liker binding state, v) E3-E2o_LD binding state, (vi) E1o-E3 binding state. Structure models were built on the basis of our single-particle E2o core (blue) in the center with E1o (orange, PDB:7WGR) and E3 (green, PDB:6I4Q) as peripheral subunit, LD (brown, PDB:1LAB) as E2o component. The flexible linker between the LD and the ICD in E2o and the N terminal linker in E1o is presented as the blue and yellow curves, respectively.
To determine the interactions among the OGDHc subunits, we separately overexpressed the S. scrofa E2o LD (residues 71–145), linker region (residues 146–220), the di-domain containing both the LD and the linker region (residues 71–220) and the ICD (residues 221–455) of the E2o (Fig. 5b). As for peripheral subunits, we overexpressed the full-length E1o, the E1o linker region (residues 41–128), the DSBD (residues 58–80), the E1o main region (residues 129–1038) (Fig. 5b) and full-length E3. For the non-core region, in vitro pull-down assays were performed by using the GST-LD, GST-E2o-linker, GST-di, GST-E1o, GST-E3 as bait proteins to separately pull-down the E1o and E3 as prey proteins (Fig. 5c). For the core region’s interaction, GST-E1o, GST-E1o-linker, GST-E1o-DSBD and GST-E1o-main region were used as bait proteins to pull down ICD. Western blot analysis showed that the di-domain is indeed combined with E1o and E3, respectively (Fig. 5c). It is consistent with our cryo-ET and STA results in which the peripheral subunits can bind to E2o core via the invisible region in cryo-EM map. Further analysis showed that both LD and the linker region of E2o can bind E1o and E3, but the binding strength is weak compared with the input, especially for E3. This indicates that, in mammalian OGDHc, there is indeed no PSBD to strongly bind E1o and E3, such as is present in PDHc. However, the linker region has similar functions to PSBD in combining the peripheral E1o and E3 subunits, but these interactions are much weaker than those of PSBD, which is the main difference between E2o and E2p. Furthermore, the binding of E3 to E2o is weaker than the binding of E1o and E2o, which is similar to previously reported data36. For the core region, we found that E2o-ICD can bind to E1o but not to E3, which can reveal that why a significant portion of E1os concentrates near the core to a certain extent. More specifically, the E2o-ICD binds to the N-terminal linker region of E1o, which provides an alternative mode of E1o and E2o’s binding in addition to the interaction between E1o and E2o-linker. The E1o-main region seems to have no contribution to the E2o core’s binding. In addition, quantitative method surface plasmon resonance (SPR) was also used to detect interactions between the critical subunits (Fig. 5d and Supplementary Fig. 11). The GST-tag was excised from the E2o-LD, E2o-linker, E1o, E3, ICD, E1o-linker, and E1o-main region, and the results show that ICD does bind to E1o and E1o-linker, but not bind to E3 and E1o-main-region (Supplementary Fig. 11a–d), which corroborate with our western-blot results. At the same time, the binding of LD to E1 and E3 is indeed very weak, but the binding of E2o-linker to E1o is stronger than E2o-LD to E1o and is also stronger than that of E2o-linker and E3 (Supplementary Fig. 11e–h) and their affinity is weaker than E2o-ICD to E1o.
Importantly, we also detected the interactions between E1o and E3 subunits when using GST-E1o, GST-E1o-linker, GST-E1o-DSBD, and GST-E1o-main-region as bait proteins to pull down E3. This result shows that E1o and E3 can combine with each other, which can also be concluded from SPR results at a dissociation constant (KD) of 1.84 μM (Fig. 5c, d). Meanwhile, the SPR results indicated that the affinity of E1o and E3 was not much different from that of E1o and E2o-linker (Fig. 5c and Extended Fig. 11f). What’s more importantly, E3 and E1o’s affinity is much stronger (~ 100 times) than E3 and E2o-linkers (Fig. 5c and Extended Fig. 11g, h), which indicates that most E3s may participate in the complex via E1o rather than E2o. Moreover, E3 was bound to E1o-linker, but not to E1-main-region (Extended Fig. 11i, j), suggesting that the position of E3 relative to E1o is also oscillating, consistent with our western-blot results and the same conclusion can be drawn from cryo-ET. We calculated the distance between each E1o and its nearest adjacent E3 in our tomograms and found that most of the distances between the corresponding E1o and E3 were still relatively far, farther than the maximum distance that the rigidity density can stick to each other (100-Å) (Supplementary Fig. 12a). And when E1o and E3 that were close, the angles between them did not seem to be fixed (Supplementary Fig. 12b). We screened each of our back-projected complexes in previous cryo-ET reconstruction and found a variety of different patterns between E1o and E3 subunits that were close enough to potentially interact (Supplementary Fig. 12c). Then we extracted the subtomograms of potentially interacted E1o and E3 (distance less than 100 Å) and their 2D projections by regarding E1o as the center. 2D classification of the projections showed only a diffuse region around E1o in the class average (Supplementary Fig. 12d). Meanwhile, we directly reconstructed and classified the E1o-E3 subtomograms with the Euler angle obtained from previous E1o-STA refinement. The results showed that E1o was reconstructed well in the center; however, there was no fixed density of E3, only some faint density around the reconstructed E1o subunit (Supplementary Fig. 12e). A similar finding was obtained for E3 as the center when the Euler angle of E3 was used to reconstruct (Supplementary Fig. 12f). This indicates that in OGDHc, E1o and E3 do not combine in a fixed pattern determined by the putative binding E1o and E3. Further experimental results showed that the DSBD did interact with E3 (Fig. 5c), corresponding to the previous result36. Collectively, the binding of E1o and E3 to the complex is attributed to the integrated interactions detected between the subunits (Fig. 5c, d, and Supplementary Fig. 11), and because of this, the entire OGDHc is an intact and stable complex, even in the absence of PSBD.
Discussion
Elucidating the overall structure and the assembly mechanism of the OGDHc family represents an important goal for structural biologists, and the resolution of the intact, native complex is of utmost importance. Here, we established an assay to obtain endogenous mammalian OGDHc from S. scrofa heart tissue. Compared with protein complexes expressed in vitro, protein complexes isolated from tissues can more accurately reflect the structure and the tested enzyme activity can reflect biological significance. By combining single-particle analysis, cryo-ET, and STA, we obtained an overall structure of native mammalian OGDHc and validated our findings by in vitro pull-down assay and SPR.
Although the overall structure of OGDHc is highly flexible, the distribution of each of the subunits occurs within certain parameters. Interestingly, the spatial distribution of E1o in OGDHc seems to have fewer degrees of freedom and more constraints than E3. The unobserved di-domain was identified to be critical for E1o/E3–E2o interactions. However, the relatively weak binding between E2o-E1o and E2o-E3 may be due to the fact that there is no PSBD in mammalian E2o, which may also explain, to some extent, why the occupancy of OGDHc is not as high as that of PDHc. The 2D classification results, the GST-pull down assay, and SPR all indicate that both the E2o-linker and E2o-ICD can bind E1o, although E2o-ICD is the stronger one. Therefore, we deduced that in the portion of E1o near the core surface, one monomer of E1o’s dimer binds to the E2o-linker and the other monomer binds to the ICD and that the putative binding site for E1o on the E2o-linker may not far from the ICD. This kind of both-end-combination mode on two E1o’s monomers results in a very restricted range of E1o’s motion around the core. At the same time, the results of the back-projecting complex and the two-dimensional classification also showed that there seemed to be different interaction manners for E1o on the surface of the core. However, due to the constraints of cryo-ET, we cannot exclude the possibility that a part of the E1o dimers may occupy the linker or ICD of two E2o monomers. If such a situation exists, the binding site of E2o may be underutilized, which may also be another contribution to the low occupancy of OGDHc.
In the complexes we reconstructed, some E1os were far from the core, and the swinging of the binding region in the linker domain could not reach these distant subunits; therefore, the LD may be required to combine some E1o relatively far from the core. For these E1os, each homodimer contains two LD binding sites, suggesting that one plays an active role in the reaction, while the other can maintain a relatively homeostatic combination to hold the E1o in place. This situation may be similar for some E3 far away from the core, in which this part of E3 binds LD to participate in the complex. However, in our result, E3 did not bind to ICD. Thus, for the portion of E3 close to the core, they are not directly bound to the ICD but are bound to the E2o-linker or LD, indicating that they have a relatively large swinging range to reach the edge of the core. This may be the main factor contributing to the diffuse distribution of E3. Moreover, E1o–E3 binding in OGDHcs is distinct compared with other OADHc members, and our cryo-ET results suggest that E1o–E3 binding in S. scrofa OGDHc is variable, indicating that E3 binds the flexible region in the N terminus of E1o. This agrees with our western blot, SPR results, and the previous work showing that E1o binds E3 via the DSBD in the N terminus36. Interestingly, the stronger bind of E3-E1o compared with E3-E2o-linker and E3-E2o-LD seems to indicate that similarly to E2o, E1o can be regarded as another “core” in the complex, insofar as it binds other subunits (E2o/E3) via the flexible region in the N terminus and randomly oscillates. Meanwhile, it is possible for E2o–E1o–E3–E2o to form a closed-loop structure in which all three subunits interact, substantially increasing the stability of the OGDHc in different cellular milieus.
Based on our cryo-ET and biochemical experiments, we proposed a complete architectural model of native, intact mammalian OGDHc (Fig. 5e). This model contains 6 binding modes among each component: E1o–E2o_linker binding state, E1o–E2o_ICD binding state, E1o–E2_LD binding state, E3–E2o_linker binding state, E3–E2o_LD binding state, and E1o–E3 binding state. Although the number and proportion of peripheral subunits in each complex are different, we chose a 1:1 ratio and a mean value of 4 E1o homodimers and 4 E3 homodimers to build the model. The E2o ICD is centered as the core of the complex, and E1o binds the E2o linker region and ICD close to the core as well as binds the E2o LD relatively far from the core. E3 does not interact with the ICD, and it only binds LD and E2o-linker. Meanwhile, E1o binds E3 by its N-terminal linker. E1o-LD binding is based on the putative LD binding site resolved in the E1o cryo-EM single-particle structure41, whereas E1o–E2o linker binding and E1o–E3 binding are based on the N-terminal of E1o. And E3-LD and E3-linker are based on the binding method predicted by Alphafold2Complex56.
A recently identified protein KGD4 (Ymr31/MRPS36), which was previously described as a subunit of the mitochondrial ribosome, is thought to be a molecular adapter to bind E3 in OGDHc57. A model was proposed in which KGD4 binds to the E1o–E2o to recruit the E3 subunit into the complex, functionally replacing the PSBD58. In OGDHc from C. thermophilum, it has been suggested that there exists E3BPo, tethering E3 to E2o core23. However, in our research, we did not observe an obvious KGD4 or E3BPo band in our SDS-PAGE analysis (Supplementary Fig. 1c), and it is difficult to identify it by cryo-EM due to its potentially flexible structure. Meanwhile, our results from STA do not indicate any portion of the density where KGD4 may exist, although this could be due to the resolution limitations of STA and the low SNR of the tomograms and particles. Also, it seems that, even if KGD4 is a component in the complex, the amount of KGD4 is very low, and the binding ratio of KGD4 to E2o and E3 is well below 1:1 according to our results. Therefore, our data suggest that E3 is likely to be involved in the complex through E2o and E1o, even if the bond between E3 and E2o is not strong on account of the absence of the PSBD. Another possible explanation is that the expression of KGD4 and assembly of OGDHc is specific in different tissues and species, and it is not evident in our S. scrofa heart-derived OGDHc.
Methods
Ethical statement
All animal maintenance and experimental procedures used in the current study were carried out according to guidelines set forth by the Institutional Animal Care and Use Committee (IACUC) of Tsinghua University, Beijing, China. Fresh heart tissues were collected from eight-month-old male domestic pigs purchased from the market at Tsinghua University.
Isolation of mitochondria from S. scrofa heart
All steps to isolate mitochondria were performed at 4 οC. To isolate mitochondria, 300 g S. scrofa heart tissue was chopped into small pieces, washed three times with ultrapure water to remove blood, and suspended in 200 ml buffer A (100 mM Tris pH 7.4, 225 mM sorbitol, 75 mM sucrose, 1 mM EGTA, and 0.1% BSA). Then, the minced tissue was homogenized in a high-speed blender for 300 s, and the homogenate was centrifuged twice at 3000 × g for 20 min to remove cell debris. Supernatants were pooled and centrifuged again at 20,000 × g for 30 min. The pellets containing crude mitochondria were collected. Crude mitochondria were suspended in 100 mL buffer B (10 mM Tris pH 7.4, 250 mM sucrose, 40% Percoll, and 1 mM EGTA) and centrifuged at 60,000 × g for 50 min. The clear mitochondrial band was aspirated carefully and diluted with buffer C (100 mM Tris pH7.4, 100 mM KCl, 1 mM EGTA). The pure mitochondria were obtained after the diluted sample was centrifuged for 30 min at 20,000 × g.
Purification of OGDHc
The pure mitochondria were collected and resuspended in buffer K (25 mM potassium phosphate, pH 6.5; 10 mM sodium chloride, 1% PMSF) and disrupted using a high-pressure crusher three times. The mitochondrial lysate was centrifuged at 60,000 × g for 30 min and de-precipitated to retain the mitochondrial matrix. Then, 15% (w/v) PEG-6000 was added dropwise to the supernatant, and the solution was incubated for 10 min at room temperature to extract OGDHcs. The crude extract was collected by ultracentrifugation at 150,000 × g for 30 min at 4 οC and suspended in 50 ml buffer D (50 mM MOPs, pH 7.2; 1 mM magnesium chloride; 20 mM DTT). After dissolving at 4 °C for 12 h, the resulting supernatant was concentrated to a volume of 6 ml using a 100-kDa-cutoff centrifuge filter (Millipore). Next, the sample was divided into 6 1-ml aliquots, which were centrifuged on 0.3–1.0-M sucrose cushions in 11.5 ml buffer S (25 mM Tris, pH 7.4; 20 mM potassium chloride) at 150,000 × g for 2.5 h at 4 °C using a SW41 rotor (Beckman Colter). Gradients were fractionated (1 ml per gradient) and analyzed seriatim on 10% SDS–PAGE gels. Fractions containing OGDHc components were identified and pooled. Next, for further purification by GraFix, the OGDHc fractions were re-concentrated to 6 ml and centrifuged in 12 ml pre-prepared 0.5–1.5 M sucrose gradient (0.1% EDC) at 120,000 × g for 17 h. Then, GraFix gradients were split into 1 ml fractions and evaluated seriatim by SDS–PAGE. Finally, OGDHc fractions were subjected to gel exclusion chromatography using a micro S6 column (GE Healthcare) to obtain endogenous OGDHcs.
In vitro enzyme activity assay
OGDHc (0.6 mg/mL final concentration) were added to a 96-well black, clear-bottom plate (Costar 3603) within 190 μL buffer D. NAD+, MgCl2, ThDP, CoA were added at 3 mM, 2 mM, 2 mM, 0.5 mM final concentration while the different substrates in each group (2-oxoglutarate, pyruvate and 4-methyl-2-oxovalerate) were added at 2 mM final concentration. The final volume was controlled at 200 μL each group. Reactions were incubated for 1 h at 37 °C while being read on an Enspire device (PerkinElmer) once every 60 s. By measuring the absorption of 340 nm light, the concentration of NADH can be indirectly reflected. Each group experiment was repeated three times and the figures were created by GraphPad.
Mass spectrometry
The protein solution was subjected to an in-depth analytical examination after the digestion process, by using the Thermo Scientific Vanquish Neo UHPLC system. A 60-minute gradient elution method was performed at a flow rate of 0.300 ml/min. Following the chromatographic separation, the system interfaced seamlessly with the Thermo Orbitrap Ascend Tribrid mass spectrometer. For this analysis, an analytical column, which consisted of a fused quartz capillary (100 mm inner diameter, 40 cm in length) packed with C-18 stationary phase material (pore size 120 Å, particle size 1.9 µm, sourced from Dr. Maisch, Ammerbuch, Germany) was utilized. The mobile phases, identified as solvents A and B, were characterized by the following compositions: solvent A was a solution of 0.1% formic acid, whereas solvent B was a mixture containing 100% acetonitrile and 0.1% formic acid. The Orbitrap Ascend Tribrid mass spectrometer was set to operate in a data-dependent acquisition mode, facilitated by the Xcalibur 4.6 software suite. Mass spectrometric analysis was conducted in a full-scan mode, scanning a mass range from 350 to 1600 m/z at a resolving power of 240,000. The Orbitrap was also employed for the efficient execution of high-speed MS/MS scans. For protein identification, the S. scrofa database was retrieved from UniProt and utilized in conjunction with Proteome Discoverer software (Version 3.0, Thermo Fisher Scientific, USA). The MS/MS spectra generated from each LC-MS/MS analysis were collected and analyzed. The search criteria were stringent, requiring strict trypsin cleavage specificity and allowing for a maximum of two missed cleavage sites. Mass tolerances were precisely defined; a 20 ppm tolerance was set for precursor ions detected by the Orbitrap mass spectrometer, and a 0.02 Da tolerance was applied for the mass measurements of fragment ions in the MS2 spectra. The assessment of the false discovery rate (FDR) for peptide identifications was performed using the percolator algorithm, which is an integral module of the Proteome Discoverer software suite. The FDR calculation was based on peptide-spectrum matches (PSMs) resulting from database searches against a decoy database. Peptides that mapped to a specific protein group which were identified as unique hits. To ensure the robustness of protein identifications, a stringent FDR threshold of 0.01 was applied.
Cryo-EM sample preparation and data collection
OGDHcs were prepared for cryo-EM analysis by applying 4 μl aliquots of purified OGDHcs (0.15 mg/ml) to glow-discharged 400-mesh Quantifoil R1.2/1.3 grids (Quantifoil, Micro Tools, Germany). Grids were blotted for 1.5 s and plunged into liquid ethane using an FEI Mark IV Vitrobot operated at 4 °C and 100% humidity. High-resolution images were collected on an FEI Titan Krios microscope operated at a voltage of 300 kV with a Gatan K2 direct electron detector (FEI). Automated single-particle data acquisition was performed using the EPU package (FEI), with a pixel size of 1.097 Å at the object scale and with defocus ranging from −1.5 μm to −2.5 μm. The dose rate on the detector was ~ 50 electrons per pixel per second, with a total exposure time of 1.0 s. Each micrograph stack contained 24 frames. A total of 6587 micrographs were acquired, among which 1935 and 1581 micrographs were obtained from the sample stage tilted at 20° and 30°, respectively, to prevent dominant orientation.
Cryo-EM image processing
The workflow for single-particle analysis is summarized in Supplementary Fig. 8d. For each micrograph in the dataset, the collected stacks were processed by subregion motion correction and dose weighting using MotionCor259. Summed micrographs were further subjected to CTFFIND460 for estimating contrast transfer function (CTF) parameters. Approximately 1500 particles were manually picked and processed by two-dimensional (2D) classification to produce auto-picking templates from the resulting 2D averages. Then, particles were auto-picked using RELION 3.061. A total of 244,587 particles were subjected to a two-round 2D classification to exclude noise and other bad particles. The qualified particles (183,521) from 2D averages were sub-selected for further 3D analysis. To achieve higher resolution, we imposed two rounds of 3D classification and obtained good particles with good signals. The selected particles were then re-centered and processed by auto-refine with a soft mask using RELION 3.0. Gctf62 was used to refine the local defocus parameters. After post-processing, we generated a 3.3-Å resolution map with octahedral symmetry. The local resolution map was calculated using ResMap63 and displayed in Chimera64.
3D model building
Initial atomic modeling was performed using CryoNet65. The 3.3-Å cryo-EM map and C-terminal 234 amino acid residues (221–454) of E2o were used as input owing to the lack of N-terminal density. The output model composed of 24 E2o core-forming domains was manually adjusted in Coot-0.9.766. After steric clashes and sidechain rotamer conformations were improved, we used real-space refinement and obtained the final model using Phenix67. Visualization of the cryo-EM density map was accomplished in UCSF Chimera64, and the structural model was analyzed and illustrated using PyMol.
Cryo-ET sample preparation and data collection
For the purified OGDHc samples, OGDHc was mixed with 10 nm colloidal gold in a 1:1 ratio at a concentration of 0.15 mg/ml, and 4 μL of the solution was added to the glow-discharged grid at the same as the cryo-EM sample preparation. Grids were blotted for 1.5 s and plunged into liquid ethane using an FEI Mark IV Vitrobot operated at 4 °C and 100% humidity, then stored in liquid nitrogen until imaging. The grids were loaded into a 300-KV FEI Titan Krios transmission electron microscope with an energy filter (slit width, 20 eV). The samples were imaged using a Gatan K3 Summit direct-detection camera in super-resolution mode at the nominal magnification (64,000 ×), with a calibrated pixel size of 0.6795 Å. A tilt series was collected from 0° using a dose-symmetry scheme68 with a 3° angular increment between −60° and +60° by SerialEM69. At each angle, a 10-frame movie was recorded with an exposure time of 0.27 s, giving a dose of 123e/Å2 per tilt series. The defocus range was from −2 to −4.5 μm.
For the mitochondria samples, the pure mitochondria were washed twice using 5 ml buffer C, and 4 μL of the sample was added to the glow-discharged grid (Quantifoil R2/2, Au, 200 mesh). Grids were blotted for 8 s, waited for 60 s, and then plunged into liquid ethane using an FEI Mark IV Vitrobot operated at 22 °C and 100% humidity, then stored in liquid nitrogen until imaging. The grids were loaded into a 300-KV FEI Titan Krios transmission electron microscope with an energy filter (slit width, 20 eV). The samples were imaged using a Gatan K3 Summit direct-detection camera in super-resolution mode at the nominal magnification (81,000 ×), with a calibrated pixel size of 0.75 Å. Tilt series were collected from 0° using a dose-symmetry scheme with a 3° angular increment between − 60° and + 60° by SerialEM. At each angle, an 8-frame movie was recorded with a dose of 123e/Å2 per tilt series. The defocus range was from − 2 to − 4.5 μm.
Cryo-ET data processing
For the purified OGDHc samples, the beam-induced motion and the defocus of the frames were corrected and estimated in Warp70. The tilt series with gold fiducial markers were combined and aligned by fiducial alignment in IMOD71. Tilt series with less than three fiducial markers and more than 2 pixels residues were discarded. Then, the processed stacks were CTF-corrected using NovaCTF72. Finally, 137 tomograms were generated by weighted-back-projection (WBP) in IMOD. Tomograms were 4 × binned and low-pass filtered to 80 Å in EMAN 2.373 and were generated by IsoNet55 for visualization.
For the mitochondria samples, all the micrographs were motion-corrected and CTF-estimated in Warp. The tilt series were aligned by AreTomo74 and CTF-corrected by NovaCTF. 28 tomograms were generated by IMOD and were submitted to IsoNet to compensate for the missing wedge.
Particle picking and subtomogram averaging
For the OGDHc tomograms, particle picking was performed in Dynamo75. In total, 2014 OGDHcs containing 2014 E2o subunits and corresponding 17,513 peripheral subunits (E1o + E3) were manually picked. Each particle was clicked on its center. All of the picked particles were saved in a Dynamo catalog75.
STA was performed in RELION 3.061. For E2o, 2014 subtomograms were firstly extracted by 4 × downsampling into a 48 × 48 × 48 box, then our single-particle E2o structure was used as a template, low-pass filtered to 30 Å for 3D alignment with C1 symmetry. The resolution was restricted to 20 Å, and we re-extracted the bin2 subtomograms into a 96 × 96 × 96 box for auto-refining, which used gold standard alignment with O symmetry imposed to reach the final resolution of 8.4 Å. A criterion of 0.143 for the Fourier shell correlation was used to estimate the resolution76. The final post-processed map achieved 7.9-Å resolution, and universal empirical B-factors of −390 were applied to sharpen the map. The local resolution map was calculated using ResMap63 and displayed in Chimera64.
For E1o and E3 peripheral subunits, 17,513 subtomograms were extracted from bin4 tomograms into a 36 × 36 × 36 box. Considering the heterogeneity and complexity of the particles, two rounds of multireference classification were performed with E1o (PDB:7WGR) single-particle structure and E3 (PDB:6I4Q) crystal structures, low-passed to 30 Å, as templates, which finally yielded 8,995 E1o and 8,518 E3 particles. Coordinates of the two classes were used to extract the bin2 particles into a 72 × 72 × 72 box. The E1o and E3s bin2 particles were further refined by imposing C2 symmetry using golden standard alignment. After removing duplicates, 8379 E1o particles were refined and reached a resolution of 9.7 Å, and 8,252 E3 particles were refined and reached a resolution of 12.2 Å. Universal empirical B-factors of –750 and –200 were applied to sharpen the E1o and E3 maps, respectively. The local resolution map was calculated using ResMap and displayed in Chimera.
For mitochondria tomograms, particle picking was performed in Dynamo and 86 complexes were selected, containing 86 E2 cubic cores and 1054 peripheral subunits. The E2 was processed the same as purified OGDHc, which was extracted firstly into a 48 × 48 × 48 box. The subtomograms were all aligned using the STA map (low-passed to 30 Å) generated from the purified OGDHcs’ E2o with C1 symmetry. After that, bin2 particles were extracted into a 96 × 96 × 96 box and refined at the resolution of 14.4 Å. For peripheral subunits, 1054 subtomograms were extracted from bin4 tomograms into a 36 × 36 × 36 box. The multireference classification was conducted with E1o (PDB:7WGR), E3 (PDB:6I4Q), and E1b (PDB:2J9F), low-passed to 30 Å as the initial template, which results in 507 particles in E1o class, 547 particles in E3 class, and no particles in E1b class. The selected E1o class and E3 class were re-extracted from bin2 tomograms, and after removing duplicates, they were both refined to 18 Å. A criterion of 0.143 for the Fourier shell correlation was used to estimate the resolution.
Subunit back-projection and complex reconstruction
In total, 2014 OGDHcs from OGDHc tomograms were reconstructed by projecting all E2o, E1o, and E3 subunits according to their refined coordinates and Euler angles. Plugin subtomo2Chimera (code repository https://doi.org/10.5281/zenodo.6820118) was used to apply the inverse transformations on the E1o, E2o, E3 to position it in the original tomogram. UCSF Chimera64 and ChimeraX77 were used to display the results.
Complex statistical analysis
Statistical analyses were performed to analyze the regularities of E1o, E3, and E2o. The number of E1o and E3 subunits in each complex was counted (Fig. 1f), from which the proportion (Fig. 1g) and the E1/E3 ratio (Supplementary Fig. 4) were derived. All 2,014 OGDHcs were included in the analysis except the E1/E3 ratio, from which we excluded the complexes with 0 E1o or 0 E3 subunits. For statistical analyses of the distances between subunits, the mass centers of E1o, E3, and E2o were used to determine the distance and relative position (Figs. 1h, 3a, c, d, f and Supplementary Fig 12a). The tilt angles of E1o and E3 with respect to E2o were defined as the angle between the z-axis (C2 axis with N-terminal as positive direction) on the E1o/E3 map and the E2o–E1o/E3 vector (Fig. 3b, c). To analyze the vertex, we selected the coordinates of the most N-terminal residue of the E2–ICD (L221) in our atomic model as the vertex center to calculate the corresponding distance and relative position of the peripheral subunits. For all E1o and E3 subunits, the vertex closest to them was selected, and they were classified into 8 groups. Each vertex and its corresponding peripheral subunits were rotated to the positive z-axis through a rotation matrix to calculate the position relative to the most adjacent vertex (Fig. 3a, d). The swinging distance was defined as the distance of the length of the tangent from the vertex center to the sphere with E1o/E3 as the center, and half of the sum of the longest and shortest widths was set as the diameter (Supplementary Fig. 6a, b). In addition, to test the interactions between E1o and E3, we analyzed the distances between each E1o and the most adjacent E3 in each complex and calculated the angles between the z-axis of the corresponding E1o and E3 (Supplementary Fig. 6c, d).
Plasmid construction and gene cloning
The gene encoding the full-length E1o (OGDH, UniProt ID: H0V0P6), E2o (DLST, Uniprot ID: Q9N0F1), and E3 (DLD, UniProt ID: P09623) were synthesized and cloned into the prokaryotic expression vector pGEX-6p-1. For E2o-LD (residues 71–145), E2o-linker (residues 146–220), and E2o-di (residues 71–220), the indicated fragment was amplified by PCR using the synthesized E2o gene as template, while E1o-linker (residues 41–128), E1o-main region (residues 129–1038) and E1o-DSBD (residues 58–80) using E1o gene as template. The amplified fragments were cloned into pGEX-6p-1 to produce the GST fusion protein. All constructs were confirmed by DNA sequencing. The recombinant plasmids were transformed into E. coli strain BL21 for protein expression.
Subunit expression and purification
E. coli cells were grown in LB medium supplemented with 50 μg/ml ampicillin at 37 °C until the optical density (OD) reached 0.6. Protein expression was induced by adding IPTG to a final concentration of 1 mM, and the induced culture was then incubated for 18 h at 16 °C with shaking. The cells were then harvested and resuspended in lysis buffer (300 mM KCl, 50 mM KH2PO4, 1 mM MgCl2, 1 mM PMSF pH 8.0 for E1o and E1o-main region; 50 mM MOPS, 147 mM NaCl, 1 mM MgCl2, 1 mM PMSF pH 8.0 for E2o-LD, E2o-di, E2o-linker, E3, E1o-linker and DSBD; 100 mM Tris, 150 mM NaCl, 1 mM EDTA pH 8.0 for E2o-ICD) and crushed by a high-pressure homogenizer. Cell debris was removed by centrifuging at 105,000 × g for 90 min at 4 °C. Expressed proteins were isolated using a GST-affinity column to adsorb the protein, and GST-E1o, GST-E3, GST-E2o-ICD were digested with PreScission Protease overnight at 4 °C. The amount of beads in the column was strictly controlled to ensure the protein was overloaded on the beads. The eluant containing E1o, E3, or E2o-ICD was concentrated and loaded onto a HiLoad 16/60 Superdex 200 Prep Grade column (GE Healthcare) equilibrated with the lysis buffer. The purified proteins were all analyzed by SDS − PAGE.
GST pull-down and western blot
Equal amounts of overloaded E2o-LD, E2o-di, E2o-linker, and E1o beads were incubated with the same volume and same concentration of E3, equal amounts of overloaded E2o-LD, E2o-di, E2o-linker, and E3 beads were incubated with the same volume and concentration of E1o, equal amounts of overloaded E1o, E3, E1o-linker, E1o-main region and DSBD beads were incubated with the same volume and concentration of E2o-ICD and equal amounts of overloaded E1o-linker, E1o-DSBD, and E1o main region were incubated with the same volume and concentration of E3 at 4 °C for 12 h. The incubation protein and the beads overloaded with GST were used as positive and negative controls, respectively. After thoroughly washing off the incubation protein with the E1o/E3/E2o-ICD lysis buffer, equal amounts of the beads with GST-tagged protein interacting with E1o/E3/E2o-ICD were analyzed on to a 4–20% SDS–PAGE gel (GenScript) and transferred onto a polyvinylidene difluoride (PVDF) membrane. Membranes were washed in PBST and blocked with 10% milk for 1 h, then incubated overnight at 4 °C with primary antibodies specific for E1o (Bioswamp, Cat # PAB31066; diluted 1:5000 in 10% milk), E3 (Cloud-Clone Corp, Cat # PAA919Hu01; diluted 1:5000 in 10% milk) or E2o-ICD (Invitrogen, Cat # PA5-22239; diluted 1:5000 in 10% milk). Next, the membranes were washed 6 times in PBST, incubated with horseradish peroxidase-conjugated anti-rabbit antibodies at 1:5000 dilution at room temperature for 1 h, and washed again 6 times in PBST. Densitometric analysis was conducted, and the blot shown is representative of three independent experiments. Uncropped, unprocessed blots are presented in the Source Data.
Protein binding affinity measurement
GST-E1o, GST-E3, GST-E2o-ICD, GST-E2o-LD, GST-E2o-linker, GST-E1o-linker and GST-E1o-main-region were digested with PreScission Protease overnight at 4 °C. The binding kinetics of related proteins were analyzed by Surface plasmon resonance (SPR) assay using Biacore 8 K plus (Cytiva) at room temperature (25 °C). E1o, E3, or E2o-ICD proteins were immobilized on the chip CM5 surface (pH = 4.0 for E1o and E2o-ICD, whereas pH = 5.5 for E3), and other proteins were applied as the analyte via serial dilution. Proteins used were exchanged into PBS buffer and serially diluted with PBST (PBS containing 0.05% Tween-20) as indicated in figures, sequentially injected into the chip at a flow rate of 30 μL/min for 2 min, and then allowed to dissociate. Data were analyzed using Biacore insight evaluation software (Cytiva).
Protein-protein interaction model
Protein-protein interactions were modeled using AlphaFold2Complex56 prediction (version 1.4.0, template database: 2 August 2022). First, the network models from AF2 developed for E3, E2o-LD, E2o-di, and E2o-linker single protein sequences were predicted. Then, the stoichiometry of the complex, including the presence of E1o and E3 as dimers, was provided to generate the models.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The 3D cryo-electron microscopy density map of E2o has been deposited in the Electron Microscopy Data Bank under accession code EMD-37965. The coordinates of the atomic model of E2o have been deposited in the Protein Data Bank 8X02. Cryo-ET subtomogram averaging maps were deposited in Electron Microscopy Data Bank under accession codes: EMD-37966 (E1o), EMD-37967 (E2o), and EMD-37968 (E3). In situ cryo-ET maps were deposited in Electron Microscopy Data Bank under accession codes: EMD-61087 (E1o), EMD-61089 (E2o), and EMD-61090 (E3). PDB codes of previously published structures used in this study are 7WGR, 2J9F, 6I4Q, and 1LAB. Source data are provided as a Source Data file. Correspondence and requests for materials should be addressed to M.Y. (maojunyang@mail.tsinghua.edu.cn). Source data are provided in this paper.
References
Sheu, K. F. R. & Blass, J. P. The alpha-ketoglutarate dehydrogenase complex. Ann. N. Y. Acad. Sci. 893, 61–78 (1999).
Massey, V. The composition of the ketoglutarate dehydrogenase complex. Biochim. Biophys. Acta 38, 447–460 (1960).
Reed, L. J. Multienzyme complexes. Accounts Chem. Res. 7, 40–46 (1974).
Waskiewicz, D. E. & Hammes, G. G. Elementary steps in the reaction-mechanism of the alpha-ketoglutarate dehydrogenase multienzyme complex from escherichia-coli - kinetics of succinylation and desuccinylation. Biochemistry 23, 3136–3143 (1984).
Gibson, G. E., Park, L. C. H., Sheu, K. F. R., Blass, J. P. & Calingasan, N. Y. The alpha-ketoglutarate dehydrogenase complex in neurodegeneration. Neurochem. Int. 36, 97–112 (2000).
Starkov, A. A. et al. Mitochondrial α-ketoglutarate dehydrogenase complex generates reactive oxygen species. J. Neurosci. 24, 7779–7788 (2004).
Tretter, L. & Adam-Vizi, V. Generation of reactive oxygen species in the reaction catalyzed by α-ketoglutarate dehydrogenase. J. Neurosci. 24, 7771–7778 (2004).
Chang, L. C., Chiang, S. K., Chen, S. E. & Hung, M. C. Targeting 2-oxoglutarate dehydrogenase for cancer treatment. Am. J. Cancer Res. 12, 1436–1455 (2022).
Gibson, G. E., Blass, J. P., Beal, M. F. & Bunik, V. The alpha-ketoglutarate-dehydrogenase complex - A mediator between mitochondria and oxidative stress in neurodegeneration. Mol. Neurobiol. 31, 43–63 (2005).
Gibson, G. E. et al. Alpha-ketoglutarate dehydrogenase complex-dependent succinylation of proteins in neurons and neuronal cell lines. J. Neurochem. 134, 86–96 (2015).
Chen, H. et al. Reductions in the mitochondrial enzyme α-ketoglutarate dehydrogenase complex in neurodegenerative disease - beneficial or detrimental? J. Neurochem. 139, 823–838 (2016).
Wang, Y. et al. KAT2A coupled with the α-KGDH complex acts as a histone H3 succinyltransferase. Nature 552, 273–277 (2017).
Huang, F. et al. Control of histone demethylation by nuclear-localized α-ketoglutarate dehydrogenase. Science 381, https://doi.org/10.1126/science.adf8822 (2023).
Patel, M. S., Roche, T. E. & Harris, R. A. in MCBU Molecular and Cell Biology Updates (2017).
Perham, R. N. Domains, motifs, and linkers in 2-oxo acid dehydrogenase multienzyme complexes - a paradigm in the design of a multifunctional protein. Biochemistry 30, 8501–8512 (1991).
Burns, G., Sykes, P. J., Hatter, K. L. & Sokatch, J. R. Isolation of a third lipoamide dehydrogenase from Pseudomonas putida. J. Bacteriol. 171, 665–668 (1989).
Patel, M. S. & Roche, T. E. Molecular-biology and biochemistry of pyruvate-dehydrogenase complexes. Faseb J. 4, 3224–3233 (1990).
Mattevi, A. et al. Atomic-structure of the cubic core of the pyruvate-dehydrogenase multienzyme complex. Science 255, 1544–1550 (1992).
Knapp, J. E. et al. Crystal structure of the truncated cubic core component of the Escherichia coli 2-oxoglutarate dehydrogenase multienzyme complex. J. Mol. Biol. 280, 655–668 (1998).
Bruch, E. M. et al. Actinobacteria challenge the paradigm: A unique protein architecture for a well-known, central metabolic complex. Proc. Natl. Acad. Sci. USA 118, e2112107118 (2021).
Nagy, B. et al. Structure of the dihydrolipoamide succinyltransferase (E2) component of the human alpha-ketoglutarate dehydrogenase complex (hKGDHc) revealed by cryo-EM and cross-linking mass spectrometry: Implications for the overall hKGDHc structure. Biochim. Biophys. Acta Gen. Subj. 1865, https://doi.org/10.1016/j.bbagen.2021.129889 (2021).
Kato, M. et al. A synchronized substrate-gating mechanism revealed by cubic-core structure of the bovine branched-chain α-ketoacid dehydrogenase complex. EMBO J. 25, 5983–5994 (2006).
Skalidis, I. et al. Structural analysis of an endogenous 4-megadalton succinyl-CoA-generating metabolon. Commun. Biol. 6, 552 (2023).
Liu, S., Xia, X., Zhen, J., Li, Z. & Zhou, Z. H. Structures and comparison of endogenous 2-oxoglutarate and pyruvate dehydrogenase complexes from bovine kidney. Cell Discov. 8, 126 (2022).
Yu, X. et al. Structures of the human pyruvate dehydrogenase complex cores: A highly conserved catalytic center with flexible N-terminal domains. Structure 16, 104–114 (2008).
Kyrilis, F. L. et al. Integrative structure of a 10-megadalton eukaryotic pyruvate dehydrogenase complex from native cell extracts. Cell Rep. 34, https://doi.org/10.1016/j.celrep.2021.108727 (2021).
De Marcucci, O. & Lindsay, J. G. Co.mponent X. An immunologically distinct polypeptide associated with mammalian pyruvate dehydrogenase multi-enzyme complex. Eur. J. Biochem. 149, 641–648 (1985).
Brautigam, C. A. et al. Structural insight into interactions between dihydrolipoamide dehydrogenase (E3) and E3 binding protein of human pyruvate dehydrogenase complex. Structure 14, 611–621 (2006).
Bunik, V. I., Wagner, T. & Bellinzoni, M. in Encyclopedia of Biological Chemistry III 259–271 (2021).
Robien, M. A. et al. Three-dimensional solution structure of the E3-binding domain of the dihydrolipoamide succinyltransferase core from the 2-oxoglutarate dehydrogenase multienzyme complex of Escherichia coli. Biochemistry 31 13, 3463–3471 (1994).
Nakano, K. et al. Purification and molecular cloning of succinyltransferase of the rat alpha-ketoglutarate dehydrogenase complex. Absence of a sequence motif of the putative E3 and/or E1 binding site. J. Biol. Chem. 266, 19013–19017 (1991).
Nakano, K. et al. Isolation, characterization and structural organization of the gene and pseudogene for the dihydrolipoamide succinyltransferase component of the human 2-oxoglutarate dehydrogenase complex. Eur. J. Biochem. 224, 179–189 (1994).
Nakano, K. et al. Isolation and sequence analysis of the rat dihydrolipoamide succinyltransferase gene. DNA Seq. 13, 363–367 (2009).
McCartney, R. G. et al. Subunit interactions in the mammalian α-ketoglutarate dehydrogenase complex. J. Biol. Chem. 273, 24158–24164 (1998).
Rice, J. E., Dunbar, B. & Lindsay, J. G. Sequences directing dihydrolipoamide dehydrogenase (E3) binding are located on the 2-oxoglutarate dehydrogenase (E1) component of the mammalian 2-oxoglutarate dehydrogenase multienzyme complex. EMBO J. 11, 3229–3235 (1992).
Zhou, J. et al. A multipronged approach unravels unprecedented protein–protein interactions in the human 2-oxoglutarate dehydrogenase multienzyme complex. J. Biol. Chem. 293, 19213–19227 (2018).
Ricaud, P. M., Howard, M. J., Roberts, E. L., Broadhurst, R. W. & Perham, R. N. Three-dimensional structure of the lipoyl domain from the dihydrolipoyl succinyltransferase component of the 2-oxoglutarate dehydrogenase multienzyme complex of Escherichia coli. J. Mol. Biol. 264, 179–190 (1996).
Knapp, J. E. et al. Expression, purification, and structural analysis of the trimeric form of the catalytic domain of the Escherichia coli dihydrolipoamide succinyltransferase. Protein. Sci. 9, 37–48 (2000).
Frank, R. A. W., Price, A. J., Northrop, F. D., Perham, R. N. & Luisi, B. F. Crystal structure of the E1 component of the escherichia coli 2-oxoglutarate dehydrogenase multienzyme complex. J. Mol. Biol. 368, 639–651 (2007).
Chandrasekhar, K. et al. Insight to the interaction of the dihydrolipoamide acetyltransferase (E2) core with the peripheral components in the Escherichia coli pyruvate dehydrogenase complex via multifaceted structural approaches. J. Biol. Chem. 288, 15402–15417 (2013).
Zhong, Y. et al. Structural basis for the activity and regulation of human α-ketoglutarate dehydrogenase revealed by Cryo-EM. Biochem. Biophys. Res. Commun. 602, 120–126 (2022).
Szabo, E. et al. Crystal structures of the disease-causing D444V mutant and the relevant wild type human dihydrolipoamide dehydrogenase. Free Radic. Bio. Med. 124, 214–220 (2018).
Reed, L. J. et al. Reconstitution of the Escherichia coli pyruvate dehydrogenase complex. Proc. Natl. Acad. Sci. USA 72 8, 3068–3072 (1975).
Berg, A. & deKok, A. 2-oxo acid dehydrogenase multienzyme complexes. The central role of the lipoyl domain. Biol. Chem. 378, 617–634 (1997).
Hirashima, M., Hayakawa, T. & Koike, M. Mammalian alpha-keto acid dehydrogenase complexes. II. An improved procedure for the preparation of 2-oxoglutarate dehydrogenase complex from pig heart muscle. J. Biol. Chem. 242 5, 902–907 (1967).
Angelides, K. J. & Hammes, G. G. Structural and mechanistic studies of the alpha-ketoglutarate dehydrogenase multi-enzyme complex from Escherichia-coli. Biochemistry 18, 5531–5537 (1979).
Bunik, V., Kaehne, T., Degtyarev, D., Shcherbakova, T. & Reiser, G. Novel isoenzyme of 2-oxoglutarate dehydrogenase is identified in brain, but not in heart. FEBS J. 275, 4990–5006 (2008).
Dekok, A., Kornfeld, S., Benziman, M. & Milner, Y. Subunit Composition and Partial Reactions of the 2-Oxoglutarate Dehydrogenase Complex of Acetobacter-Xylinum. Eur. J. Biochem. 106, 49–58 (1980).
Wagenknecht, T., Francis, N. & Derosier, D. J. Alpha-ketoglutarate dehydrogenase multi-enzyme complex may be heterogeneous in quaternary structure. Biophys. J. 37, A7–A7 (1982).
Wagenknecht, T., Francis, N. & Derosier, D. J. Alpha-ketoglutarate dehydrogenase complex may be heterogeneous in quaternary structure. J. Mol. Biol. 165, 523–539 (1983).
Wagenknecht, T., Grassucci, R. & Schaak, D. Cryoelectron microscopy of frozen-hydrated alpha-ketoacid dehydrogenase complexes from Escherichia-coli. J. Biol. Chem. 265, 22402–22408 (1990).
Murphy, G. E. & Jensen, G. J. Electron cryotomography of the E. coli pyruvate and 2-oxoglutarate dehydrogenase complexes. Structure 13, 1765–1773 (2005).
Chen, M. et al. Molecular architecture of mammalian pyruvate dehydrogenase complex. Protein Cell https://doi.org/10.1093/procel/pwae044 (2024).
Kastner, B. et al. GraFix: sample preparation for single-particle electron cryomicroscopy. Nat. Methods 5, 53–55 (2007).
Liu, Y.-T. et al. Isotropic reconstruction for electron tomography with deep learning. Nature Communications 13, https://doi.org/10.1038/s41467-022-33957-8 (2022).
Gao, M., Nakajima An, D., Parks, J. M. & Skolnick, J. AF2Complex predicts direct physical interactions in multimeric proteins with deep learning. Nat. Commun. 13, https://doi.org/10.1038/s41467-022-29394-2 (2022).
Heublein, M. et al. The novel component Kgd4 recruits the E3 subunit to the mitochondrial α-ketoglutarate dehydrogenase. Mol. Biol. Cell 25, 3342–3349 (2014).
Hevler, J. F. et al. MRPS36 provides a structural link in the eukaryotic 2-oxoglutarate dehydrogenase complex. Open Biol. 13, https://doi.org/10.1098/rsob.220363 (2023).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Rohou, A. & Grigorieff, N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015).
Zivanov, J. et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. ELife 7, https://doi.org/10.7554/elife.42166 (2018).
Zhang, K. Gctf: Real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016).
Kucukelbir, A., Sigworth, F. J. & Tagare, H. D. Quantifying the local resolution of cryo-EMEM density maps. Nat. Methods 11, 63−+ (2014).
Pettersen, E. F. et al. UCSF Chimera?A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Xu, K., Wang, Z., Shi, J., Li, H. & Zhang, Q. C. A2-Net: Molecular Structure Estimation from Cryo-EM Density Volumes. Proceedings of the AAAI Conference on Artificial Intelligence, vol. 33, 1230–1237 (2019).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. Sect. D Biol. Crystallogr. 60, 2126–2132 (2004).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. Sect. D Biol. Crystallogr. 66, 213–221 (2010).
Hagen, W. J. H., Wan, W. & Briggs, J. A. G. Implementation of a cryo-electron tomography tilt-scheme optimized for high resolution subtomogram averaging. J. Struct. Biol. 197, 191–198 (2017).
Mastronarde, D. N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005).
Tegunov, D. & Cramer, P. Real-time cryo-electron microscopy data preprocessing with Warp. Nat. Methods 16, 1146–1152 (2019).
Kremer, J. R., Mastronarde, D. N. & McIntosh, J. R. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116, 71–76 (1996).
Turoňová, B., Schur, F. K. M., Wan, W. & Briggs, J. A. G. Efficient 3D-CTF correction for cryo-electron tomography using NovaCTF improves subtomogram averaging resolution to 3.4 Å. J. Struct. Biol. 199, 187–195 (2017).
Galaz-Montoya, J. G., Flanagan, J., Schmid, M. F. & Ludtke, S. J. Single particle tomography in EMAN2. J. Struct. Biol. 190, 279–290 (2015).
Zheng, S. et al. AreTomo: An integrated software package for automated marker-free, motion-corrected cryo-electron tomographic alignment and reconstruction. J. Struct. Biol. X 6, https://doi.org/10.1016/j.yjsbx.2022.100068 (2022).
Castaño-Díez, D., Kudryashev, M., Arheit, M. & Stahlberg, H. Dynamo: A flexible, user-friendly development tool for subtomogram averaging of cryo-EM data in high-performance computing environments. J. Struct. Biol. 178, 139–151 (2012).
Scheres, S. H. W. & Chen, S. Prevention of overfitting in cryo-EM structure determination. Nat. Methods 9, 853–854 (2012).
Goddard, T. D. et al. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018).
Acknowledgements
We thank the Tsinghua University Branch of China National Center for Protein Sciences (Beijing) for providing the cryo-EM facility support. We thank Dr. XiaoMin Li, Dr. Fan Yang, and Yutong Song for the data collection; The computation was completed on the Yanglab GPU workstation. This work was supported by the National Key R&D Program of China (2022YFA1302701), the National Natural Science Foundation of China (32030056), the scientific project of Beijing Life Science Academy (2023300CA0090), the King Abdullah University of Science and Technology (KAUST) Office of Sponsored Research (OSR) under Award (OSR-2020-CRG9-4352), and Office of Research Administration (ORA) under Award No URF/1/4352-01-01, FCC/1/1976-44-01, FCC/1/1976-45-01, REI/1/5234-01-01, REI/1/5414-01-01.
Author information
Authors and Affiliations
Contributions
M.Y. directed the study. Y.Z., M.C. and M.Z. did the protein purification. Y.Z. and M.C. performed cryo-EM and cryo-ET sample preparation and data collection. Y.Z. did the structural determination with the help of M.C. and S.Z.; X.C. and Z.Y. performed the SPR experiments with the help of Y.Z. and J.Y.; M.Y., Y.Z., M.C., and S.Z. built the model and drew the figures. M.Y., Y.Z., M.C., S.Z., J.Y., and X.G. wrote the manuscript. All authors contributed to the discussion of the data and editing of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhang, Y., Chen, M., Chen, X. et al. Molecular architecture of the mammalian 2-oxoglutarate dehydrogenase complex. Nat Commun 15, 8407 (2024). https://doi.org/10.1038/s41467-024-52792-7
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-024-52792-7
This article is cited by
-
Protein lipoylation in cancer: metabolic reprogramming and therapeutic potential
Cell Death Discovery (2025)
-
Advantages and Limitations of AlphaFold in Structural Biology: Insights from Recent Studies
The Protein Journal (2025)