Abstract
Protease-containing ABC transporters (PCATs) couple the energy of ATP hydrolysis to the processing and export of diverse cargo proteins across cell membranes to mediate antimicrobial resistance and quorum sensing. Here, we combine biochemical analysis, single particle cryoEM, and DEER spectroscopy in lipid bilayers along with computational analysis to illuminate the structural and energetic underpinnings of coupled cargo protein export. Our integrated investigation uncovers competitive interplay between nucleotides and cargo protein binding that ensures the latter’s orderly processing and subsequent transport. The energetics of cryoEM structures in lipid bilayers are congruent with the inferred mechanism from ATP turnover analysis and reveal a snapshot of a high-energy outward-facing conformation that provides an exit pathway into the lipid bilayer and/or the extracellular side. DEER investigation of the core ABC transporter suggests evolutionary tuning of the energetic landscape to fulfill the function of substrate processing prior to export.
Similar content being viewed by others
Introduction
ABC transporters are exquisite molecular machines that traffic solutes across lipid bilayers in both directions powered by the energy of ATP hydrolysis1,2. Broadly classified into importers and exporters, functional specialization among ABC transporters led to a fascinating spectrum of molecular architectures3. A subfamily of ABC exporters specializes in the processing and translocation of entire protein domains across cell membranes. Referred to as Peptidase-Containing ABC transporters (PCATs), their architecture consists of a core homodimeric ABC exporter covalently linked to a cysteine protease domain (C39 or PEP domain)4,5,6,7. Cargos of PCATs includes bacteriocins and quorum-sensing polypeptides, fundamental bacterial defense strategies4,5,8,9.
One of the most structurally characterized PCATs is PCAT1 from the thermophilic organism C. thermocellum10,11,12. The homodimeric core ABC transporter of PCAT1 has the canonical structure of ABC Type IV exporters with two cytoplasmic nucleotide-binding domains (NBDs) and two transmembrane domains (TMDs). PCAT1 cargo protein, hereafter referred to as Sub, was identified as a 90-residue polypeptide containing an N-terminal leader sequence, which is cleaved by the PEP domain of PCAT111. Initial crystal structures of PCAT1 in two conformations11 were followed by several cryoEM snapshots under different biochemical conditions10,12. Collectively the structural record revealed the molecular architecture of PCAT1, identified elements of its ATP-dependent conformational changes, and elucidated a partial view of its interactions with Sub.
Persistently enigmatic, not only for PCAT1 but other PCATs as well13, has been the coupling mechanism of cargo protein export to the cycle of ATP turnover. CryoEM structures of Sub-bound PCAT1 identified density corresponding to Sub located in the cytoplasmic-facing chamber and tethered to the signal sequence which is bound to the PEP domain12. In contrast to these inward-facing (IF) conformations, an occluded conformation (OC) bound to ATP-γS displayed an inaccessible chamber and assembly of the NBD catalytic dimer11. Although a recent ATP-bound cryoEM structure in detergent micelles was interpreted as an outward-facing (OF) conformation10, it was not clear that the incremental opening of the extracellular side is sufficient to accommodate the export of a polypeptide the size of Sub.
Unlike solute and drug ABC exporters, PCAT1 modifies its cargo protein prior to export. Thus, alternating access of the transporter core must be coupled to the processing of the signal peptide in the PEP domain. To uncover the steps involved in this coupling, we carry out a systematic study that integrates mechanistic elements deduced from extensive ATP turnover studies, structural insight gleaned from single particle cryoEM analysis in lipid nanodiscs, and conformational dynamics from molecular dynamics (MD) simulations and double electron electron resonance (DEER)14,15 spectroscopy. CryoEM structures determined in the presence of nucleotides and/or Sub were correlated with findings from biochemical analysis of ATP hydrolysis. Furthermore, we investigate the intrinsic properties of the core ABC transporter lacking the PEP domain through a combination of cryoEM and DEER. Together, our results illuminate the structural mechanism by which the cargo protein couples to the ATP turnover cycle, an insight only accessible in the context of lipid bilayers. Along with a cryoEM structure of a bone fide OF conformation of the core transporter, our results stimulate a model of transport that departs from the canonical model of ABC transporters and contextualizes previous studies of PCAT1 in detergent micelles10.
Results
The widely accepted canonical model of ABC exporters1, which has guided much of the structural investigations in the field, posits the population of at least two intermediates. Typically, an IF state, featuring a large chamber open to the intracellular side, isomerizes to an OF state which features a splaying of the extracellular side of these transporters. Occluded conformations have been reported where the substrate binding chamber is closed on both sides of the transporter. Alternating access is powered by ATP binding and hydrolysis although most models1,2, with few exceptions16,17,18,19, assume that the former drives transition to OF whereas hydrolysis catalyzes the return to the IF. Thus, the current structural record emphasizes states stabilized by ATP, often in a background of catalytically impaired NBDs, or by trapping with vanadate following ATP hydrolysis (ADP-Vi). The latter condition has been shown to trap a high energy state populated subsequent to hydrolysis of ATP. Following this paradigm, previous investigations of PCAT1 determined Sub and ATP-bound structures, although studies into the ADP-Vi state were conspicuously absent10,11.
Modulation of the ATP turnover cycle by ADP, Mg2+ and cargo protein (Sub)
Because PCAT1 modifies its cargo prior to export, we hypothesized that this requirement imposes mechanistic constraints on its ATP hydrolysis cycle. Therefore, we examined the roles of nucleotides, Sub and Mg2+, in ATP turnover in lipid nanodiscs. Under conditions of equimolar ATP and Mg2+, we observed the expected hyperbolic isotherm with Michaelis-Menten parameters that were similar to those reported previously by us20 and others11 (Fig. 1A). However, in contrast to Lin et al. 11, we find that addition of Sub reproducibly increased Vmax with negligible effects on Km (Supplementary Table 1). Furthermore, vanadate did not inhibit ATP turnover as would be expected for a typical ABC exporter. This suggests either quick dissociation of Mg2+ thereby shortcutting formation of the vanadyl/ADP complex, or rather limited stability of the latter in the confines of the nucleotide binding sites (NBSs) that form at the interface of the Walker A motif from one NBD and the signature motif of the other NBD.
A Sub stimulates ATP hydrolysis whereas ADP competitively inhibits it (see Supplementary Table 1). Sub partly reverses the inhibition of ADP. PCAT1 turnover data is the average of 15 repeats from 8 biological repeats. PCAT1 inhibition with 2 mM ADP with 5 mM Vi is the average of 4 technical from 2 biological repeats. Turnover data with 5× Sub is derived from 5 biological repeats. B Biphasic shape of ATP turnover determined under 10 mM Mg2+ curve suggests partial inhibition by free Mg2+. The data is the average from at least 2 biological repeats. C Multiphasic dependence of Vmax on the Mg2+ to ATP ratio. The data is the average of at least two biological repeats. All data are presented as mean values ± standard deviation.
ATP turnover by PCAT1 shows an unusual dependence on free Mg2+. This was initially gleaned from ATP turnover curves obtained at constant concentration of Mg2+ (10 mM, Fig. 1B). A biphasic dependence indicated that, in the presence of free Mg2+ (below 10 mM ATP), the ATPase activity of PCAT1 is partially inhibited. To dissect the origin of this effect, we carried out Mg2+ titration at constant ATP concentrations. The resulting curves reveal distinct multiphasic behavior (Fig. 1C). As expected, low concentration of Mg2+ stimulates ATP hydrolysis reflecting the integral role of Mg2+ in the chemical reaction. However, a change in slope is observed as Mg2+ concentration increases followed by the appearance of an inhibitory effect under conditions of excess free Mg2+. As discussed below, these distinct phases may partly reflect shifts in conformational states of the transporter along with changes in nucleotide affinity.
Importantly, we find that Sub attenuates the inhibitory effects of free Mg2+. The peak Vmax observed in the presence of Sub is almost 50% higher than the one observed in the ATP turnover assay (Fig. 1C). Comparing Vmax from Fig. 1A, C indicates that partial inhibition of ATP turnover occurs even under the conditions of equimolar ATP and Mg2+ concentrations of Fig. 1A. The multiphasic characteristic of the titration curve persists in the presence of Sub including the onset of Mg2+ inhibition at high free Mg2+ concentration although the peak shifts to higher Mg2+/ATP ratios (Fig. 1C).
To further decipher the origin of the Mg2+ dependence, we investigated the effects of ADP on ATP turnover. We observed a steep competitive inhibition (Fig. 1A) that is reversed by addition of Sub (Fig. 1A), suggesting that either ADP and Sub binding is mutually exclusive, or that Sub reduces the affinity of ADP relative to ATP. Titration with ADP highlights that stimulation of PCAT1 by substrate requires ADP concentration in a physiological range (Supplementary Fig. 1) Together, the ADP and Mg2+ inhibitions (albeit at high concentration of free Mg2+) and their abrogation by Sub suggest that ADP is stabilized in the NBSs following ATP hydrolysis. This mechanism explains the lack of stimulation by Sub reported by Lin et al.11 since, in that work, they used a continuous ATP regeneration system that depleted free ADP.
Free energy calculation reveals a high affinity of PCAT1 to ADP
In support of these conclusions, we have performed calculations using the free energy perturbation (FEP) method to estimate the relative binding free energy of ATP and ADP (see methods). The simulations were performed on the ATP-bound, putatively OF structure of PCAT1 in the absence of the substrate reported by Kieuvongngam and Chen10 (PDB:7T54). Interestingly, ATP has a lower affinity as compared to ADP with a more significant difference in the presence of Mg2+ (Table 1; compare \(\Delta \Delta {G}_{{Mg}}=\Delta {G}_{{MgATP}}-\Delta {G}_{{MgADP}}\) to \(\Delta \Delta {G}_{{noMg}}=\Delta {G}_{{ATP}}-\Delta {G}_{{ADP}}\)). The results indicate that the PCAT1 favors binding ADP over ATP at least in this structure. This is in contrast with other bacterial ABC transporters such as MsbA21 (PDB:3B60) and Sav186622 (PDB: 2HYD) for which similar simulations on the OF conformation yielded negative binding free energies in all cases (Table 1), indicating that ATP is favored for binding over ADP in the absence and presence of Mg2+ with a more significant difference in the absence of Mg2+.
Structures of PCAT1 in lipid nanodiscs
The inhibitory effect of Mg2+ was not observed in detergent micelles implying that ADP competitive inhibition of ATP turnover was also substantially blunted (Supplementary Fig. 2). Therefore, we investigated the conformations of PCAT1 in lipid nanodiscs focusing primarily on previously unexplored biochemical conditions that represent intermediates in the putative transport cycle such as in the presence of ADP, ADP/Mg2+ and ADP/Sub. Regardless of biochemical conditions, cryoEM conformations of PCAT1 in lipid nanodiscs segregated into two classes that we label as IF and OC (Fig. 2, Supplementary Fig. 3, Supplementary Fig. 4, Supplementary Fig. 5, Supplementary Table 2, Supplementary Table 3). The IF conformations, which appeared very similar in the presence or absence of Sub, feature a chamber open to the cytoplasm and partially to the inner leaflet of the membrane. In contrast, the chamber was occluded to the cytoplasm in OC conformations. Moreover, the OC conformations were devoid of identifiable Sub density even when a large molar excess was added to the sample. The PEP domains in the OC conformations were disordered precluding their modeling. Finally, the conformational changes from IF to OC lead to the assembly of what is considered the pre-catalytic NBSs. Supplementary Table 4 compares our IF and OC structures to previous crystal and cryoEM structures of PCAT1 in detergent micelles10,11.
Both ADP (panel B) and ATP (panel E) stabilize an occluded (OC) conformation in the absence of Mg2+. Sub (panel C) and Mg2+ (panel A) shift the equilibrium toward the inward-facing (IF) conformation in the presence of ADP, whereas ATP/Sub bound PCAT1 remains in an OC conformation. PCAT1* denotes the cysteine-free PCAT1 (panel D).
We determined two IF structures under conditions of excess Sub: one of WT-PCAT1 and one in a protease-deficient construct where all the cysteines (PCAT1*), including at the active site of the PEP domain were replaced (Fig. 2C, D). The two structures are almost identical with a root mean square deviation (RMSD) of 1.388 Å. The substitution of the active site cysteine with serine essentially anchors Sub to the C39 domain. While we could model the leader peptide in both structures, only in PCAT1* did we observe strong Sub density in the TMD (Supplementary Fig. 6). Despite multiple attempts at local refinement we were unable to obtain structural details of the bound Sub (Supplementary Fig. 6) suggesting that Sub is disordered when bound to PCAT1 TMD as previously noted12.
Energetics of PCAT1 conformational changes
While the trapped PCAT1 conformations were either IF or OC, the ligand dependence and the implied energetics were unexpected and inconsistent with a previous10 model of PCAT1. Of central importance is the observation of an OC conformation in the presence of ADP, a finding that structurally manifests the implied high affinity of this nucleotide to PCAT1, as deduced from ATP hydrolysis studies above (Fig. 1 and Supplementary Fig. 1). Similarly, in lipid nanodiscs, an OC conformation is stabilized by the binding of ATP in the absence of Mg2+. This contrasts with the previous report where a more outward-open conformation was reported for Mg2+-free PCAT1 bound to ATP but in detergent micelles10. Supplementary Fig. 7 shows a residue by residue RMSD between the structures of ATP-bound PCAT1 in nanodiscs and detergent micelles.
The structure of ADP-bound PCAT1 in the presence of Sub provides a basis for their apparent competitive binding deduced from biochemical analysis. An IF conformation is obtained when an excess of Sub is included in the sample both in the presence or absence of Mg2+ (Fig. 2A, C). This contrasts with the persistence of the OC conformation when ATP is bound in the presence of Sub but absence of Mg2+ (Fig. 2E), suggesting that ATP binding excludes the cargo protein from the binding chamber whereas Sub stabilizes the IF even in the presence of ADP (Fig. 2A, C, D). Furthermore, previous cryoEM studies found that binding of ATP/Mg2+ and Sub resets the PCAT1 conformation to predominantly IF conformations10.
Structural basis of apparent competition between ADP and Sub
If Sub and ADP can concomitantly bind as evidence by the structures, then the apparent competition must arise from a change in relative affinity of ATP and ADP induced by Sub. We noticed that the cryoEM data set from the PCAT1/ADP/Mg2+ sample consisted of two classes of particles that differ in the extent of opening of the NBDs (Supplementary Fig. 8). In contrast, Sub-bound structure in the presence of ADP/Mg2+ consisted of one class of particles. In these structures, the leader peptide makes direct contact with residues near the A-loop (Fig. 3) suggesting a structural basis for the reversal of ADP competitive inhibition (Fig. 1A). The A-loop plays a central role in stabilizing bound nucleotides through a highly conserved Tyr, here Tyr 495, which interacts with the adenosine moiety of ATP and ADP. Comparing PCAT1 structures in the presence and absence of Sub (class 2) (Fig. 3) reveals changes in the conformation of the A-loop and the signature sequence motif (residues 623–627), which could potentially underlie the coupling between Sub and ATP/ADP binding. The disorder of the signature motif in the absence of Sub may reduce the affinity of ATP binding by destabilizing the formation of the pre-catalytic NBS.
Comparison of PCAT1_IF_ADP/Mg2+_class2 structure(light color) and PCAT1_ IF_ADP/Mg2+/Sub structure(dark color) shows that the leader peptide interacts with the A-loop altering its conformation. In addition, its binding stabilizes the signature motif as evidenced by the stronger density in that region in the IF structure determined in the presence of Sub (dark green). The cryoEM densities are shown at 4.6 σ for PCAT1_ IF_ADP/Mg2+/Sub and 7.7 σ for PCAT1_IF_ADP/Mg2+_class2.
To quantitatively assess the effect of Sub binding, we calculated the relative affinity of PACT1 to ATP and ADP in the IF conformation stabilized by Sub (see methods). We included MsbA as a reference since the IF structure of Sav1866 has not been determined. The results show that the binding preference of ADP over ATP calculated from the putative OF conformation is not observed in the IF state of the transporter (Supplementary Table 5). Instead, there is a preference for binding ATP both in the absence and presence of Mg2+. The preference for ATP over ADP is somewhat similar in PCAT1 and MsbA; however, the presence of substrate in PCAT1 increases this preference significantly with around 10–13 kcal/mol difference in binding affinity of ATP relative to ADP. Another interesting observation is that the presence of Mg2+ only slightly increases the affinity of ADP relative to ATP by around 3–5 kcal/mol whereas the opposite effect of around 18 kcal/mol is observed in the putative OF state. These findings indicate that the relative affinity of ATP to ADP depends on the conformation of PCAT1, particularly in the presence of substrate. Focusing on the relative Mg2+-ATP/Mg2+-ADP binding affinity, MsbA shows little conformational dependence and a persistent preference for Mg2+-ATP. In contrast, PCAT1 shows a preference for Mg2+-ATP in the inward-facing state and a strong preference for Mg2+-ADP in the outward-facing state.
A model, suggested by the data above, wherein the leader peptide modulates the relative ADP/ATP affinity and thus the ATP turnover rate would predict abrogation of ATP hydrolysis stimulation if the Sub lacks the leader peptide. In agreement with this prediction, we find that a leaderless construct of Sub does not stimulate ATP hydrolysis (Supplementary Fig. 9).
Mg2+ is required to order the NBSs
In search for insight into the complex modulation of ATP turnover by Mg2+, we compared structures obtained in the presence and absence of Mg2+. Regardless of the conformation, we found that Mg2+ binding leads to substantial conformational changes at the NBSs. Specifically, the cryoEM density corresponding to a helix consisting of residues 651–665, which packs against the Walker B motif, appears weak and uninterpretable in the absence of Mg2+, suggesting that Mg2+ contributes to the ordered arrangement of this helix (Fig. 4). Thus, the ordering of this helix may contribute to the energetics of IF stability in the presence of Mg2+, in addition to being required for triggering the hydrolysis of ATP. A structure of PCAT1 bound to Mg2+ did not reveal evidence of structural alterations beyond the unwinding of the 651–665 helix.
To further confirm the role of Mg2+ in the stability of the helix, we set out to estimate the free energy associated with the alpha-helicity of the region between T650 and L668 using well-tempered metadynamics23 simulations with a collective variable defined based on the helical content of this region. We modeled PCAT1_ADP and PCAT1_ ADP/Mg2+ on the basis of the same structure (PDB:7T54) that contains a helix in the 650–668 region. The free energy profiles indicate that the PCAT1 bound to only ADP does not have a stable helix in this region with its global minimum being around 20% helical content. In contrast, PCAT1 bound to Mg2+-ADP has a stable helix with a global minimum of around 85% helicity and another local minimum of around 65% helicity (Supplementary Fig. 10).
A non-canonical ABC signature motif modulates the relative affinity of ATP and ADP
Detailed analysis of the NBS structures reveals hints into the origin of PCAT1 affinity to ADP in the OC conformation. PCAT1’s signature motif deviates from the canonical sequence, specifically by the replacement of glycine 625 by glutamate, yielding an LSEGQ sequence. We observed that the side chain of E625 is proximal to the γ phosphate (Supplementary Fig. 11). This glutamate may also be a contributing factor to the lack of stable inhibition of ATP turnover by Vanadate.
Structure of an OF conformation of PCAT1 in lipid nanodiscs
The structures presented above as well as those obtained in detergent micelles10 unequivocally reveal an energetic bias favoring the IF conformations in the presence of Mg2+/nucleotides and when bound to Sub. Therefore, we reasoned that removing the PEP domain may alter the energetic of the conformational landscape. For this purpose, we used a transporter core construct devoid of the PEP domain, hereafter referred to as PCAT1_CC. We also substituted all the native cysteines to allow for the subsequent introduction of spin labels (see below). CryoEM structures of the core transporter bound to ADP/Mg2+ were very similar to the structures in the context of the full-length transporter (Fig. 5 and supplementary Fig. 12, Supplementary Fig. 13, supplementary Table 6). However, the OC conformation of the core transporter stands in stark contrast to full length PCAT1 where the same conditions (ADP/Mg2+) stabilizes the IF conformation.
Consistent with our prediction that the PEP domain stabilizes an IF conformation, we found that the core transporter bound to ATP in the absence of Mg2+ adopts an OF conformation. Compared to the ATP-bound structure obtained in detergent micelles of full length PCAT1 (PDBID:7T54)10, we observe large displacements of TMs 5 and 6 that further open the extracellular side of the transporter (Fig. 6). The movement of TM6 is facilitated by a hinge around residue 446 whereas TM5 undergoes a tilt without large backbone distortion. A prominent side opening to the outer leaflet of the bilayer develops, between TMs1 and 6, that may enable the exit of Sub or its more hydrophobic regions. We note that the OF conformation of the core transporter could not be trapped if the sample was incubated at 50 °C prior to the plunging of the grid.
Comparison of the putative OF conformation of PCAT1 in detergent micelles (gold) with the OF conformation of the transporter core, PCAT1_CC, in lipid nanodiscs (teal). A Side view of the two structures highlighting the opening of the latter to the outer leaflet of the membrane. B Extracellular view identifying the movement of individual helices.
Intrinsic properties of ATP turnover by the core transporter
The removal of the PEP domain increases the kcat of ATP turnover (Supplementary Table 1) as was previously reported20. In addition, the sensitivity of ATP turnover to Sub is abrogated (Supplementary Fig. 14) although we have demonstrated that Sub binds the core transporter PCAT1_CC in the TMD20. This finding reinforces our model that the leader peptide directly interacts with the A-loop to stimulate ATP hydrolysis. In nanodiscs, the multiphasic dependence of Mg2+ concentration remains intact in PCAT1_CC suggesting that it is an intrinsic property of the core transporter. The ATP turnover of the core transporter is efficiently inhibited by ADP similar to the full length PCAT1.
Conformational dynamics of the ABC transporter core
The mechanistic properties revealed above for the full-length transporter depart from the canonical model of ATP-powered alternating access of solute ABC exporters. To test if these are intrinsic properties of the core transporter, i.e. PCAT1 lacking the PEP domain, we investigated its ATP-dependent conformational changes by DEER (Supplementary Table 7). For this purpose, we measured distances between spin labels under different biochemical conditions. Spin labels were selected to report on the intracellular and extracellular sides of the TMD as well as on the assembly of the NBDs. All spin labeled mutants showed robust ATP turnover parameters (Supplementary Table 8).
Overall, we observed large distance changes (Fig. 7 and supplementary Fig. 15) spanning the membrane region, the intracellular side of the TMDs and the NBDs, most prominently in the presence of ATP-γS. Comparison of experimental and predicted distance distributions (on the basis of the ATP-γS-bound crystal structure11) reveals conformational changes that reflect the transition from an IF to an OC conformation. Figure 7 also displays predicted distance distributions on the basis of two previously10 determined IF cryoEM structures that differ by the degree of intracellular opening referred to “wide” and “narrow”. The sign of the changes in the average distances indicates a closing motion that occludes access to the Sub binding chamber. At the extracellular side, only site 202 showed distance changes, likely reporting rearrangements of the loop linking TMs 1 and 2. Deviations from predicted distributions, calculated based on the OF structure of the core transporter, are most prominent at the extracellular side. For most sites, except 372 and 703, distance distributions under apo conditions are broad and appear to span the range of distances under other conditions indicating conformational sampling between states of similar free energies.
Interplay between Sub and ADP in the conformational cycle of the core transporter
In addition to revealing the structural elements involved in the IF/OC transition, the DEER data reflect the energetic dependence of the conformational changes. In contrast to the implications from cryoEM structure of the transporter core that ADP/Mg2+ binding favors an OC conformation, analysis of distance distributions in the presence of ADP/Mg2+ suggests an IF conformation with limited structural changes relative to the Apo condition (compare Figs. 7, 8). The sites where changes are induced by ADP/Mg2+ map to the membrane segment of TM3 (residue 287) as well as to a site in the NBD selected to monitor the NBSs (residue 498). Remarkably, addition of Sub attenuates the distance changes at TM3, in agreement with the finding in the full-length PCAT1 that Sub binding attenuates ADP inhibition and stabilizes the IF conformation (Fig. 8).
DEER distance distributions of the core transporter in lipid nanodiscs under turnover conditions (solid teal traces). Experimental and predicted distance distributions are shown in solid and dashed lines respectively. Except for 307 and 498, the distributions under turnover conditions are superimposable with those bound to ADP/Mg2+ (solid purple traces).
While most experimental and predicted distance distributions were congruent, we observed striking deviations at two sites, one in TM3 (287) and one in TM6 (442) (Fig. 8, Supplementary Fig. 15). Upon inspection, these sites appear to be in regions not well resolved in the crystal and cryoEM structures of the occluded and putative OF conformations10. It is also possible that the repacking of the spin label side chains due to the structural rearrangement contributes to the deviation from the predicted distances.
Conformations of the transporter core under turnover conditions
ATP binding or hydrolysis provides the energy input to drive conformational changes in most ABC exporters reported to date. Specifically, for many homodimeric transporters, the binding of ATP drives the population of an OF conformation. In contrast, for the transporter core of PCAT1, DEER data under turnover conditions, defined by equimolar high concentrations of ATP and Mg2+, reflect a mostly apo-like conformation (Fig. 8). These results are consistent with the conclusions from cryoEM10 of predominantly IF conformation when PCAT1 is bound to ATP/ Mg2+ and support the conclusion that the OF conformation is a high energy state that is likely to be transiently populated.
Distinct distance changes under turnover conditions observed along TM3 as well as in the NBSs mirror those observed in the presence of ADP despite a short incubation time under which ATP remains in excess. Of particular interest are the multi-population distance distributions at residue 498 where the binding of ADP/ATP elicits distinct distance changes (Fig. 8 and Supplementary Fig. 15). This residue is in proximity to the A-loop which contains the highly conserved Tyr495 that packs against the adenosine moiety (Fig. 3). Both ADP/ Mg2+ and ATP/Mg2+ shift the equilibrium away from the Apo conformation (Fig. 8). The disagreement between predicted and experimental distributions is attributed to the ambiguous cryoEM density in these regions as well as the repacking of the spin label.
Discussion
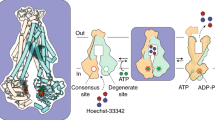
Our integrated biochemical, structural, spectroscopic, and computational investigation reveals previously unnoted elements of PCAT1 ATP turnover cycle, including the structural and energetic bases of cargo protein coupling, and complements previous structural studies10,11 by uncovering missing intermediates in the conformational cycle. Fundamental to our approach is the use of lipid nanodiscs which exposes mechanistic elements that are abrogated by solubilization in detergent micelles. Our results expound on the notion10 that PCAT1s evolved a distinct conformational energy landscape compared to canonical solute ABC transporters. They answer the central question of how the coupling between nucleotide turnover and cargo protein processing is achieved as we illustrate in the model below (Fig. 9).
The coupled cycle is initiated by binding of Sub either to the PCAT1 bound to ATP/Mg2+ in the IF conformation (1a) or to ADP-bound PCAT1 in the OC conformation (1b). The coupled cycle proceeds through processing of Sub (2) followed by ATP hydrolysis (3) to populate an OF conformation that releases leaderless Sub to the extracellular side or the outer leaflet of the membrane. Subsequently, Mg2+ dissociation (4b) could trap the transporter in an occluded conformation to reduce futile ATP hydrolysis. Alternatively, Mg2+-ADP dissociation with Mg2+-ATP binding (4a) could facilitate an IF conformation in an uncoupled cycle without Sub binding where ATP hydrolysis (5a) followed by dissociation of Mg2+ (5b) results in the OC conformation.
Interplay of ATP, ADP and Sub binding in the transport cycle
Central to our model is the discovery of tenacious competitive inhibition of ATP hydrolysis by ADP (Supplementary Fig. 1) and the abrogation of this inhibition by Sub. These findings uncover the missing link that relates the ATP hydrolysis cycle to the export of Sub. High affinity binding of ADP, following Mg2+ dissociation, may be a mechanism to reduce futile ATP hydrolysis by trapping the transporter in an occluded conformation. Weakening of ADP binding by Sub then triggers the initiation of the cycle, simultaneously driving the transporter to the IF conformation and signaling to the NBS the binding of the cargo protein thereby increasing the affinity to ATP/Mg2+. Our model posits that the modulation of the relative affinity of ATP and ADP is achieved structurally by the replacement of the highly conserved glycine by glutamate at position 625 in the canonical signature motif (Supplementary Fig. 11), which results in an electrostatic repulsion with the γ phosphate of ATP. Finally, Mg2+ plays an important role in the ATP turnover cycle. The energetic preference of IF conformation(s) upon binding of Mg2+ further ensures the kinetic coupling of ATP hydrolysis to Sub processing by reducing the probability of transition to OF conformations.
Structural model of cargo-protein coupling to ATP hydrolysis
Unlike solute ABC transporters, where the cycle entails the binding and release of unmodified substrate, PCAT1 processes its cargo protein by removing an N-terminal signal sequence prior to transport. Thus, for a productive or coupled cycle to be executed, two requirements have to be satisfied. First, ATP binding and hydrolysis must occur subsequent to the cleavage of the cargo signal sequence. Second, only the processed cargo is to be extruded. This latter requirement is likely satisfied by the strong interaction between the signal sequence and the PEP domain which anchors unprocessed Sub to the transporter likely preventing its translocation prior to processing. One of the consensus findings in this paper and by Kieuvongngam and Chen10 is the stabilization of IF conformations by Sub. In contrast, evidence have been advanced that substrates drive the formation of OC conformation or at least the formation of the pre-catalytic NBSs for some solute ABC transporters1. Sub extrusion out of the chamber requires at least the binding of ATP as evidenced by multiple structures presented here and by the Chen group10,11. Not only does ATP occlude the chamber but no evidence of Sub can be found in ATP-bound structures.
Our model allows for an uncoupled cycle in the absence of Sub. The high concentrations of ATP/Mg2+ in cells would imply that a PCAT1 population is bound to this nucleotide (Fig. 9, step 1a). However, unlike other ABC exporters, PCAT1 is predominantly in an IF conformation under these conditions, where its PEP domain is docked to the core transporter, enabling high affinity binding of the leader peptide and insertion of the cargo protein in the TMD. Binding of Sub stimulates ATP turnover by accelerating the ATP to ADP exchange of step 4a. Moreover, it funnels the ATP loaded IF of PACT1 through step 1a. Our data demonstrating that physiological levels of ADP are required for Sub stimulation rationalize the lack of stimulation by Sub in an ADP-depleting system11.
Two Sub molecules are expected to bind as previously demonstrated12 although we surmise that only one is transported through the TMD. The binding of Sub (Fig. 9, step 2) stimulates ATP hydrolysis, kinetically accelerating transition to the OF conformation (Fig. 9, step 3). Because the transition to the OF leads to disorder of the PEP domain, the binding to the now cleaved leader peptide is weakened leading to its dissociation. Although the model was formulated on the basis of binding of two Sub molecules as reported previously10, it is conceivable that an asymmetric conformation, wherein only one Sub molecule is bound, is populated. In this case, we surmise that one of the NBSs where the A-loop contact the Sub would be ATP-bound whereas the other would likely have ADP. While our model attempts to harmonize current structural and functional data, it leaves aspects of the mechanism, such as the role of Mg2+ and the ATP/ADP interplay under physiological conditions, unaccounted for.
Subsequent to ATP hydrolysis, it is likely that inorganic phosphate is quickly dissociated inducing the shift to the ADP-bound OC conformation (Fig. 9, step 4b), as demonstrated by the lack of trapping with vanadate. The ADP-bound OC conformation is then destabilized by Sub binding which primes the transporter for another cycle (Fig. 9, step 1b). Our model reinforces the role of kinetics, noted previously10, in the transport cycle of PCATs. ATP hydrolysis and the concomitant transition to the OF is the rate limiting step ensuring prior processing of the cargo protein. Multiple investigations on type IV ABC exporters have now established that while they share a similar structural mechanism of alternating access, the tuning of energetics and kinetics, as exemplified by PCAT1, represent yet another example of divergence within a conserved fold.
Methods
Cloning and site-directed mutagenesis
Codon-optimized wild-type PCAT1 derived from C. thermocellum (GenScript) was cloned into the pET19b vector encoding an N-terminal decahistidine tag under control of an inducible T7 promoter. A cysteine-deficient variant of PCAT1 (designated as PCAT1-CL or PCAT1*) was engineered, entailing the substitution of native cysteines with the following amino acids: C12S, C21S, C25S, C129F, C171A, C581A, C687A, and C713L. Additionally, we constructed a variant lacking the C39 peptidase domain (termed PCAT1_CC), featuring the following cysteine mutations: C171A, C581A, C687A, and C713L. All primers used for mutagenesis has been listed in the excel supplemental file.
PCAT_CC was used as the template to introduce cysteine and background mutations via site-directed mutagenesis using complementary oligonucleotide primers. Substitution mutations were generated using a single-step PCR wherein the entire template plasmid was replicated from a single mutagenic primer. PCAT_CC mutants were sequenced using both T7 forward and reverse primers to confirm mutagenesis and the absence of aberrant changes. Mutants are designated by the native residue and primary sequence position followed by the mutant residue.
The substrate of PCAT1, Cthe_0535, herein referred to as Sub, was previously identified as a 90-residue protein containing an N-terminal signal sequence and encoded in the same operon as PCAT111, was codon-optimized and synthesized for expression in Escherichia coli by cloning into PET19b vector similarly to PCAT1 with an N-terminal decahistidine tag.
Expression, purification, and labeling of PCAT1
E. coli C43 (DE3) cells were freshly transformed with the pET19b vector harboring the PCAT1 constructs. A single transformant colony was utilized to inoculate Luria–Bertani (LB) media (LabExpress) supplemented with 0.1 mg/mL ampicillin (Gold Biotechnology). This culture was grown overnight (∼15 h) at 34 °C, which was then used to inoculate 4 L of minimal medium A at a 1:50 dilution. The cultures were incubated with shaking at 37 °C until they achieved an absorbance at 600 nm (Abs600 nm) of approximately 0.8. PCAT_CC expression was induced by the addition of 1 mM IPTG (Gold Biotechnology). The cultures were incubated overnight (∼15 h) at 18 °C and then harvested by centrifugation.
The resulting cell pellets were resuspended in 80 mL of resuspension buffer (50 mM Tris⋅HCl, pH 7.4, 150 mM NaCl, 10% [vol/vol] glycerol) with 10 mM DTT. The cells were lysed by five passes through an Avestin C3 homogenizer. Cell debris was removed by centrifugation at 9000 × g for 10 min. Membranes were subsequently isolated from the supernatant by centrifugation at 200,000 × g for 1.5 h.
The isolated membrane pellets were solubilized in the resuspension buffer containing 20 mM β-DDM (Anatrace) and 0.5 mM DTT and incubated on ice with stirring for 1 h. Insoluble material was cleared by centrifugation at 200,000 × g for 30 min. The clarified extract was bound to 1.0 mL (bed volume) of Ni-NTA Superflow resin (Qiagen) at 4 °C for 2 h. After washing with 5 bed volumes of buffer containing 50 mM imidazole, PCAT1 was eluted with buffer containing 300 mM imidazole.
Cysteine mutants of PCAT1 were labeled with two rounds of 20-fold molar excess 1-oxyl-2,2,5,5-tetramethylpyrroline-3-methyl methanethiosulfonate (Enzo Life Sciences) per cysteine at room temperature in the dark over a 4 h period, after which the sample was placed on ice at 4 °C overnight (∼15 h) to yield the spin label side chain R1. Excess spin label was removed by size exclusion chromatography over a Superdex200 Increase 10/300 GL column (GE Healthcare) into 50 mM Tris, pH 7.4, 50 mM NaCl, 1 mM β-DDM, and 10% (vol/vol) glycerol buffer. Peak fractions of purified PCAT1 were combined and concentrated using an Amicon Ultra 100,000 MWCO filter concentrator (Millipore), and the final concentration was determined by A280 measurement (ε = 107,500 M − 1⋅cm−1 for PCAT-CL and WT, ε = 73,160 M − 1⋅cm−1 for PCAT-CC,) in preparation for subsequent experimental applications.
Reconstitution of PCAT into nanodiscs
E. coli polar lipids and l-α phosphatidylcholine (PC) (Avanti Polar Lipids) were combined in a 3:2 (wt/wt) ratio, solubilized in chloroform, evaporated to dryness on a rotary evaporator, and desiccated overnight under vacuum in the dark. The lipid mixture was rehydrated in 50 mM MOPS pH 7.4, 50 mM NaCl buffer to a final concentration of 20 mM, homogenized by freezing and thawing for 10 cycles, and stored in small aliquots at −80 °C.
MSP1D1 and MSP1D1E3 proteins were expressed and purified as described in previous studies [3,4] and dialyzed into 50 mM Tris/MES pH 7.5 buffer. Purified MSP1D1/MSP1D1E3 was concentrated using a 10,000 MWCO filter concentrator, with final protein concentration determined by A280 measurement (MSP1D1, ε = 21,430 M−1 ⋅ cm−1; MSP1D1E3 ε = 29,910 M−1 ⋅ cm−1).
For reconstitution into nanodiscs purified PCAT1 or mutants thereof in β-DDM micelles were combined with hydrated E. coli polar lipids/PC lipid mixture, MSP, and sodium cholate at the following molar ratios: lipid:MSP, 50:1; MSP:PCAT1 at 10:1; and detergent:lipid, 3:1. The mixtures were incubated at 4 °C for one hour. Detergent removal was facilitated by two additions of 0.1 g/mL Biobeads (Bio-Rad), each followed by incubation at 4 °C. Subsequently, 0.2 mg/mL Biobeads were added and mixed overnight, with an additional hour of mixing the next day after adding another 0.2 mg/mL Biobeads. Biobeads were then removed using a 0.45 µm filter. Size exclusion chromatography was employed to segregate nanodiscs with PCAT1 from empty nanodisc into 50 mM MOPS, pH 7.4, 50 mM NaCl buffer with 10% (vol/vol) glycerol. The PCAT1-containing nanodiscs were concentrated using an Amicon ultra 100,000 MWCO filter concentrator and assessed via SDS/PAGE to verify reconstitution and estimate reconstitution efficiency. Quantification of spin-labeled mutants in nanodiscs was performed as previously described, comparing the intensity of their integrated continuous-wave electron paramagnetic resonance (CW-EPR) spectrum to that in detergent micelles24.
CW-EPR and DEER spectroscopy
Continuous-wave electron paramagnetic resonance (CW-EPR) spectra of spin-labeled PCAT1 samples were collected at room temperature on a Bruker EMX spectrometer (X-band, 9.5 GHz) at an incident power of 10 mW and 1.6 Gauss modulation amplitude. Scan width was 120–150 Gauss.
Double Electron-Electron Resonance (DEER) spectroscopy was conducted using an Elexsys E580 EPR spectrometer at a Q-band frequency of 33.9 GHz. This was performed employing a dead-time free four-pulse sequence at 83 K, with pulse lengths of 10 to 14 ns (π/2) and 20 to 28 ns (π) for the probe pulses, and 40 ns for the pump pulse. The frequency separation between the observe and pump pulses was 63 MHz. To ascertain the role of nucleotide and Sub in the conformational cycling of the transporter, DEER samples were prepared with 10 mM MgATPγS, 10 mM MgATP, or 10 mM MgADP with and without 5-fold molar excess of Sub. Samples for DEER were cryoprotected with 24% (vol/vol) glycerol, incubated at 50 °C for 20 mins (2 min for MgATP) and flash-frozen in liquid nitrogen.
Analysis of primary DEER decays was performed using proprietary software developed in-house, operating within the MATLAB (MathWorks) environment as previously described25. Briefly, the software conducts a global analysis of DEER decays under varying conditions for the same spin-labeled pair. It assumes that the distance distribution comprises a sum of Gaussian functions, where the number and proportion of these functions are established based on a statistical criterion. For few sites (207, 303, 226 and 324), the DEER decay was shorter than optimal due to signal to noise consideration. However, confidence band analysis supports our interpretation.
For the prediction of distance distributions on the PCAT1 structures, in silico 1 ns molecular dynamics simulations were conducted. These simulations utilized dummy spin labels and followed default parameters as outlined in the DEER Spin-Pair Distributor at the CHARMM-GUI website26,27.
Expression and purification of Sub
The expression and purification of Sub was performed as previously outlined with slight modifications20. Briefly, sub was expressed in E. coli BL21 (DE3) Gold competent cells. Cells were grown in 1 L Terrific Broth at 37 °C with shaking to an OD600 of 1.8–2.0, induced with 1 mm IPTG and allowed to grow for an additional 4 h. The cultures were harvested by centrifugation at 6000 × g for 15 min and the cell pellet was subsequently resuspended in lysis buffer composed of 50 mM Tris, pH 7.5, 150 mM NaCl, 5 mM EDTA, with a protease inhibitor cocktail (Roche). Cells were lysed by sonication (30 cycles of 10 s on/25 s off, 40% amplitude). The lysate was centrifuged at 17,000 × g for 15 min to sediment unbroken cells and inclusion bodies containing Sub.
The cell pellets were washed in 25 ml of wash buffer containing 50 mM Tris, pH 7.5, 150 mM NaCl, 0.5 mM EDTA, 0.5 mM DTT, 1% (w/v) Triton X-100. The suspension was centrifuged at 17,000 × g for 15 min, and the cell pellet wash was repeated with the same buffer without Triton. Pellet containing the inclusion bodies was solubilized by resuspension in the same wash buffer (2.5 mL) supplemented with 10 mM DTT and 6 M urea under constant stirring at room temperature for 30 mins or until dissolved. After centrifugation at 17,000 × g for 15 min, the solution was rapidly diluted using buffer containing 50 mM Tris, pH 7.5, 150 mM NaCl, 0.01% β-DDM to initiate refolding. The sample was further centrifuged to remove insoluble material and nickel-affinity chromatography was performed to isolate Sub. Sub was further purified using a 5 mL Hi-Trap desalting column in GF buffer to remove imidazole. Sub was then concentrated in an Amicon Ultra 3000 MWCO filter concentrator (Millipore) and stored in small aliquots at −80 °C for future experiments. The purity of Sub was confirmed via SDS-PAGE analysis.
ATPase assay
ATPase activity for PCAT1 and PCAT1 mutants was determined by a calorimetric inorganic phosphate assay as previously described20. Briefly, nanodisc reconstituted PCAT1 in 50 mM MOPS, pH 7.4, 50 mM NaCl buffer was incubated with increasing concentrations of ATP at 50 °C for 8 min. The reaction was halted upon returning to ice and the addition of an equal volume of 12% SDS. The color was subsequently developed using a 1:1 solution of ammonium molybdate (2% in 1 M HCl) and ascorbic acid (12% in 1 M HCl). The absorbance of samples was measured at a wavelength of 850 nm on a BioTek Synergy H4 microplate reader. The amount of phosphate released was determined by comparison to a standard curve generated from inorganic phosphate. The maximal rate (Vmax) of PCAT1 and its variants was derived using the Levenberg-Marquart nonlinear least squares fitting approach in Origin (OriginLab). To determine the effect Mg2+ on ATP turnover, Mg2+ was added at equimolar concentrations of ATP or increasing Mg2+ concentration with constant ATP concentration.
Electron microscopy sample preparation and data acquisition
Nanodisc reconstituted PCAT1 wild type, PCAT1-CL, and PCAT1-CC with MSP1D1 scaffold protein under different biochemical conditions were prepared at a final concentration of 0.5 mg/ml. Samples were applied to glow discharged Quantifoil UltraAU Foil holey grids (1.2/1.3, 300 mesh). For both wild type and cysteine-less PCAT1 (PCAT1*) samples with ADP in a Mg2+-free environment, a final concentration of 10 mM ADP, 1 mM EDTA, 0.5 mM EGTA, and/or Sub (at five times the protein concentration) was mixed with the protein. These mixtures were incubated at 50 °C for 10 min prior to application to the grids. In the case of PCAT1 with ADP/Mg2+, the samples contained a final concentration of 10 mM ADP and 10 mM MgCl2, along with optional Sub, and followed the same incubation procedure. For wild type PCAT1 in an Mg2+ environment, only 10 mM MgCl2 was included in the sample, which was then incubated at 50 °C for 10 min. Similarly, for wild type PCAT1 with ATP in a Mg2+-free environment, the final concentration comprised 5 mM ATP, 1 mM EDTA, 0.5 mM EGTA, and 5x substrate, followed by incubation at 50 °C for 1 min before grid application.
PCAT1_CC samples in MSP1D1 nanodiscs, along with nucleotides, substrate, and additives at 1.4 mg/ml, was applied to glow-discharged Quantifoil holey carbon grids (1.2/1.3, Cu, 300 mesh). For PCAT1_CC with ADP/Mg2+, the final concentration included 10 mM ADP, 10 mM MgCl2, and optional Sub, incubated at 50 °C for 10 min. For vanadate trapping PCAT1-CC sample, the final concentrations were 6 mM ATP, 6 mM MgCl2, and 5 mM Vi, also incubated at 50 °C for 10 min. The OF PCAT1-CC sample in a Mg2+-free environment consisted of 10 mM ATP, 1 mM EDTA, and 0.5 mM EGTA, incubated on ice for 30 min.
Grids were blotted for 3.0–4.5 s with a blotting force between 10–12 and 100% relative humidity at 8 °C. They were then plunge-frozen in liquid ethane cooled by liquid nitrogen using a Vitrobot System Mark IV (Gatan). CryoEM data collection occurred at liquid nitrogen temperature using a 300 kV TEM with Gatan K3 BioQuantum and ThermoScientific Falcon3 direct electron detectors. All cryoEM movies were recorded with a 20 eV slit width from the energy filter, at a nominal magnification of 130kx, corresponding to a calibrated pixel size of 0.647 Å/pix. Additional details are available in the supplementary material.
Image processing, model building and refinement
Image processing for all data sets was conducted using CryoSparc v4.2.128. We initiated the process by applying Patch Motion Correction and Patch CTF estimation to the imported movies. Depending on the image quality, a manual curation of exposures was occasionally necessary to eliminate poor-quality images, thereby enhancing the resolution of the resulting maps.
Particle selection was conducted using the Blob Picker, with diameters ranging from 100 Å to 200 Å. This was followed by a 2D classification process, employing a box size of 360 pixels, to acquire templates for subsequent template picking using the Template Picker. The particles selected by this method were extracted with a box size of 360 pixels. We then conducted a 2D classification, with class numbers varying between 50 and 200.
The selected particles were subsequently utilized for Ab-Initio Reconstruction, with the number of classes set between 2 and 4. The quality of the maps and the number of particles were manually assessed, guiding the decision to apply 2 to 4 maps for Heterogeneous Refinement. Optimization of the final map was achieved through homogeneous refinement, non-uniform refinement, and local refinement, applying C2 symmetry where applicable.
Structure generation commenced with Model Angelo29, followed by manual inspection and refinement using Phenix 1.20.130. PDB validation confirmed that all structures were free from major issues. For visualization, figures were generated utilizing UCSF Chimera31 and Pymol (The PyMOL Molecular Graphics System, Version 1.2r3pre, Schrödinger, LLC.).
Molecular dynamics simulations
All-atom MD simulations were performed on PCAT1, MsbA, and Sav1866 systems. For the PCAT1 simulations in the putative OF state, we used the cryoEM structure of ATP-bound PCAT1 reported by Kieuvongngam and Chen10 (PDB:7T54). Magnesium ions were introduced into the system by aligning the nucleotides of the 7T54-based structure with the equivalent nucleotides in PDB:7T57 (which contains Mg2+), and then substituting the magnesium-complexed nucleotides into our structure. By removing the Mg2+ from these structures, we then generated the magnesium-free nucleotide structures. We therefore made 4 models of outward-facing PCAT1 including PCAT1_ATP, PCAT1_ADP, PCAT1_ ATP/Mg2+ and PCAT1_ ADP/Mg2+. Based on PDB: 2HYD22 and PDB:3B6021, we generated similar models for Sav1866 and MsbA, respectively. In addition, we used the cryoEM structure reported here (PDB: 8VP3) for the inward-facing structure of PCAT1 in the presence of ADP/Mg2+and Sub. By replacing the ADP by ATP and removing the Mg2+ in either model, we generated 4 models for PCAT1/Sub in the inward-facing state. The same 4 models were then used, after removing the Sub to generate another 4 models for substrate-free PCAT1 in the inward-facing state. In addition, an inward-facing MsbA structure based on PDB:3B5W21 was created and used within the same strategy to dock ATP, ADP, ATP/Mg2+, or ADP/Mg2+.
In all cases, once the preliminary structure of protein and nucleotide with or without Mg2+ had been obtained, CHARMM-GUI26,32 was then used to build the simulation systems. Using Membrane Builder33, structures were embedded in a membrane bilayer consisting of 45% POPE, 45% POPG, and 10% cardiolipin (TOCL2), oriented using PPM2.034, and then solvated using TIP3P35 water along with 0.15 M NaCl and neutralizing counterions. The approximate final size of each transporter system was ~450,000 atoms. In addition, we simulated solvated the ATP, ADP, Mg2+-ATP, or Mg2+-ADP in water to perform simulations in the absence of transporters.
All simulations were run with NAMD2.1436 using periodic boundary conditions and the CHARMM36m forcefield37,38. The default CHARMM-GUI equilibration for membrane systems begins with a 10,000-step energy minimization using the conjugate gradient method, followed by a 6-step restraining regimen which was performed over 22.5 ns in an NVT ensemble. A further 20 ns of production was then performed for each system in an NPT ensemble under equilibrium conditions. All simulations up to this point were carried out at 310 K using a Langevin integrator with a damping coefficient of γ = 1.0 ps-1 and a 2 fs timestep. A pressure of 1 atm was maintained using the Nosé–Hoover Langevin piston method39,40. The cutoff distance for non-bonded interactions was set to 12 Å, and long-range electrostatics were computed with the Particle Mesh Ewald (PME)41 method.
Free energy perturbation simulations
A free energy perturbation (FEP)42 approach was used to calculate the relative binding free energy of ADP/ATP. The FEP process consisted of gradually annihilating (forward process) or returning (backward process) the λ-phosphates for the systems in question to simulate the hydrolysis of bound ATP into ADP in the presence and absence of the Mg2+ ion. Each simulation comprised 20 windows (Δλ = 0.05 per window), with each window consisting of 10 ps of alchemical equilibration followed by 240 ps of data collection (5 ns total). Every window for a given simulation was initialized with the same initial conformation coming from the previously described equilibrated trajectories, but with the phosphates already partially annihilated, allowing all windows to be run simultaneously in parallel. To circumvent the endpoint problem for van der Waals interactions43,44, a soft-core potential was used such that the electrostatic interactions were fully decoupled from the system by λ = 0.5. Free energy differences were calculated using the Bennett Acceptance Ratio (BAR)45 method. The resulting data from FEP calculations were statistically analyzed using the VMD46 ParseFEP plugin47 to obtain the relative binding free energies.
Well-tempered metadynamics simulations
Following the equilibrium simulations, well-tempered metadynamics23 simulations were conducted for PCAT1_ADP and PCAT1_ADP/Mg2+ systems in the outward-facing state for 100 ns with the same parameters used for the equilibrium simulations. The collective variable “alpha” (α)48 was utilized to investigate the α-helical propensity of residues between T650 and L668.
Collective variable “alpha” (\(\alpha\)) is defined as a one-dimensional scalar to quantify the \(\alpha\)-helical propensity of a peptide of length N:
where \({\theta }_{n}\) is the angle formed by \({C}_{\alpha }^{\left(n\right)}-{C}_{\alpha }^{\left(n+1\right)}-{C}_{\alpha }^{\left(n+2\right)}\) and \({d}_{n}\) is distance between \({O}^{(n)}\) and \({N}^{(n+4)}.\) \(f(\theta )\) and \(g(d)\) are score functions (both ranging from 0 to 1) quantifying the likelihood of \(\theta\) and \({d}\) being associated with an \(\alpha\)-helix. More specifically:
where \({\theta }_{0}\) and \(\delta \theta\) are 88 and 15 degrees, respectively and \({d}_{0}\) is 3.3 \(\mathring{\rm A}\). For the well-tempered metadynamics, the height and width of the gaussians were 1 kcal/mol and 0.05, respectively, and the pseudo temperature was 1200 K.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The data that support this study are available from the corresponding authors upon request. All cryoEM 3D density maps of the PCAT1 in nanodiscs in Fig. 2, Fig. 5, supplementary table 2 and supplementary table 6 have been deposited in the Electron Microscopy Data Bank under accession codes EMD-43405 (PCAT1_OC_ADP); EMD-43404 (PCAT1_IF_ADP/Sub); EMD-43406 (PCAT1_IF_ADP/Mg2+_Class2); EMD-43400 (PCAT1_IF_ADP/Sub/Mg2+); EMD-43401 (PCAT1*_IF_ADP/Sub); EMD-43402 (PCAT1_IF_Mg2+_Class2); EMD-43403 (PCAT1_OC_ATP/Sub); EMD-43394 (PCAT1_CC_ADP/Mg2+); EMD-43396 (PCAT1_CC_ADP/Mg2+/Sub); EMD-43393 (PCAT1_CC_ATP/Mg2+/Vi); EMD-43398 (PCAT1_CC_ATP); EMD-44015 (PCAT1_IF_ADP/ Mg2+_Class1); EMD-44402 (PCAT1_IF_ Mg2+_Class1)(supplementary table 2 and supplementary table 6). Atomic coordinates for the atomic models have been deposited in the Protein Data Bank under accession codes 8VPA (PCAT1_OC_ADP); 8VP9 (PCAT1_IF_ADP/Sub); 8VPB (PCAT1_IF_ADP/Mg2+_Class2); 8VP3 (PCAT1_IF_ADP/Sub/Mg2+); 8VP5 (PCAT1*_IF_ADP/Sub); 8VP6 (PCAT1_IF_Mg2+_Class2); 8VP8 (PCAT1_OC_ATP/Sub); 8VOX (PCAT1_CC_ADP/Mg2+); 8VOZ (PCAT1_CC_ADP/Mg2+/Sub); 8VOW (PCAT1_CC_ATP/Mg2+/Vi); 8VP1 (PCAT1_CC_ATP); 9AZL (PCAT1_IF_ADP/Mg2+_Class1); 9BAA (PCAT1_IF_ Mg2+_Class1). The MD simulation data necessary to reproduce the data including equilibration and FEP simulation input files are deposited to the Zenodo repository [https://doi.org/10.5281/zenodo.13864175]. Previous published structures referred in the manuscript is 7T54. A source data file is available. Source data are provided with this paper.
References
Thomas, C. & Tampe, R. Structural and mechanistic principles of ABC transporters. Annu Rev. Biochem. 89, 605–636 (2020).
Locher, K. P. Mechanistic diversity in ATP-binding cassette (ABC) transporters. Nat. Struct. Mol. Biol. 23, 487–493 (2016).
Thomas, C. et al. Structural and functional diversity calls for a new classification of ABC transporters. FEBS Lett. 594, 3767–3775 (2020).
Gebhard, S. A. B. C. transporters of antimicrobial peptides in Firmicutes bacteria - phylogeny, function and regulation. Mol. Microbiol. 86, 1295–1317 (2012).
Gebhard, S. & Mascher, T. Antimicrobial peptide sensing and detoxification modules: unravelling the regulatory circuitry of Staphylococcus aureus. Mol. Microbiol. 81, 581–587 (2011).
Kanonenberg, K., Schwarz, C. K. & Schmitt, L. Type I secretion systems - a story of appendices. Res. Microbiol. 164, 596–604 (2013).
Lenders, M. H., Reimann, S., Smits, S. H. & Schmitt, L. Molecular insights into Type I secretion systems. Biol. Chem. 394, 1371–1384 (2013).
Havarstein, L. S., Diep, D. B. & Nes, I. F. A family of bacteriocin ABC transporters carry out proteolytic processing of their substrates concomitant with export. Mol. Microbiol. 16, 229–240 (1995).
Hiron, A., Falord, M., Valle, J., Debarbouille, M. & Msadek, T. Bacitracin and nisin resistance in Staphylococcus aureus: a novel pathway involving the BraS/BraR two-component system (SA2417/SA2418) and both the BraD/BraE and VraD/VraE ABC transporters. Mol. Microbiol. 81, 602–622 (2011).
Kieuvongngam, V. & Chen, J. Structures of the peptidase-containing ABC transporter PCAT1 under equilibrium and nonequilibrium conditions. Proc Natl Acad Sci USA 119, https://doi.org/10.1073/pnas.2120534119 (2022).
Lin, D. Y., Huang, S. & Chen, J. Crystal structures of a polypeptide processing and secretion transporter. Nature 523, 425–430 (2015).
Kieuvongngam, V. et al. Structural basis of substrate recognition by a polypeptide processing and secretion transporter. Elife 9, https://doi.org/10.7554/eLife.51492 (2020).
Yu, L. et al. Structural basis of peptide secretion for Quorum sensing by ComA. Nat. Commun. 14, 7178 (2023).
Claxton, D. P., Kazmier, K., Mishra, S. & McHaourab, H. S. Navigating membrane protein structure, dynamics, and energy landscapes using spin labeling and EPR spectroscopy. Methods Enzymol. 564, 349–387 (2015).
McHaourab, H. S., Steed, P. R. & Kazmier, K. Toward the fourth dimension of membrane protein structure: insight into dynamics from spin-labeling EPR spectroscopy. Struct. (Lond., Engl.: 1993) 19, 1549–1561 (2011).
Dastvan, R., Mishra, S., Peskova, Y. B., Nakamoto, R. K. & McHaourab, H. S. Mechanism of allosteric modulation of P-glycoprotein by transport substrates and inhibitors. Science 364, 689–692 (2019).
Mishra, S. et al. Conformational dynamics of the nucleotide binding domains and the power stroke of a heterodimeric ABC transporter. Elife 3, e02740 (2014).
Tang, Q. et al. Asymmetric conformations and lipid interactions shape the ATP-coupled cycle of a heterodimeric ABC transporter. Nat. Commun. 14, 7184 (2023).
Verhalen, B. et al. Energy transduction and alternating access of the mammalian ABC transporter P-glycoprotein. Nature 543, 738–741 (2017).
Rahman, S. & McHaourab, H. S. ATP-dependent interactions of a cargo protein with the transmembrane domain of a polypeptide processing and secretion ABC transporter. J. Biol. Chem. 295, 14678–14685 (2020).
Ward, A., Reyes, C. L., Yu, J., Roth, C. B. & Chang, G. Flexibility in the ABC transporter MsbA: alternating access with a twist. Proc. Natl Acad. Sci. USA 104, 19005–19010 (2007).
Dawson, R. J. P. & Locher, K. P. Structure of a bacterial multidrug ABC transporter. Nature 443, 180–185 (2006).
Barducci, A., Bussi, G. & Parrinello, M. Well-tempered metadynamics: a smoothly converging and tunable free-energy method. Phys. Rev. Lett. 100, 020603 (2008).
Jagessar, K. L., Claxton, D. P., Stein, R. A. & McHaourab, H. S. Sequence and structural determinants of ligand-dependent alternating access of a MATE transporter. Proc. Natl Acad. Sci. 117, 4732–4740 (2020).
Stein, R. A., Beth, A. H. & Hustedt, E. J. A straightforward approach to the analysis of double electron-electron resonance data. Methods Enzymol. 563, 531–567 (2015).
Jo, S., Kim, T., Iyer, V. G. & Im, W. CHARMM-GUI: a web-based graphical user interface for CHARMM. J. Comput. Chem. 29, 1859–1865 (2008).
Qi, Y. et al. CHARMM-GUI DEER facilitator for spin-pair distance distribution calculations and preparation of restrained-ensemble molecular dynamics simulations. J. Comput. Chem. 41, 415–420 (2020).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Jamali, K. et al. Automated model building and protein identification in cryo-EM maps. bioRxiv, https://doi.org/10.1101/2023.05.16.541002 (2023).
Liebschner, D. et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr D. Struct. Biol. 75, 861–877 (2019).
Pettersen, E. F. et al. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–12 (2004).
Lee, J. et al. CHARMM-GUI input generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM simulations using the CHARMM36 additive force field. J. Chem. Theory Comput. 12, 405–413 (2016).
Jo, S., Kim, T. & Im, W. Automated builder and database of protein/membrane complexes for molecular dynamics simulations. PLoS One 2, e880 (2007).
Lomize, A. L., Pogozheva, I. D. & Mosberg, H. I. Anisotropic solvent model of the lipid bilayer. 2. Energetics of insertion of small molecules, peptides, and proteins in membranes. J. Chem. Inf. Model 51, 930–946 (2011).
Jorgensen, W. L., Chandrasekhar, J., Madura, J. D., Impey, R. W. & Klein, M. L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926–935 (1983).
Phillips, J. C. et al. Scalable molecular dynamics with NAMD. J. Comput. Chem. 26, 1781–1802 (2005).
Best, R. B. et al. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone φ, ψ and side-chain χ(1) and χ(2) dihedral angles. J. Chem. Theory Comput. 8, 3257–3273 (2012).
Huang, J. et al. CHARMM36m: an improved force field for folded and intrinsically disordered proteins. Nat. Methods 14, 71–73 (2017).
Feller, S. E., Zhang, Y., Pastor, R. W. & Brooks, B. R. Constant pressure molecular dynamics simulation: the Langevin piston method. J. Chem. Phys. 103, 4613–4621 (1995).
Martyna, G. J., Tobias, D. J. & Klein, M. L. Constant pressure molecular dynamics algorithms. J. Chem. Phys. 101, 4177–4189 (1994).
Darden, T., York, D. & Pedersen, L. Particle mesh Ewald: An N⋅log(N) method for Ewald sums in large systems. J. Chem. Phys. 98, 10089–10092 (1993).
Zwanzig, R. W. High‐temperature equation of state by a perturbation method. I. nonpolar gases. J. Chem. Phys. 22, 1420–1426 (1954).
Beutler, T. C., Mark, A. E., van Schaik, R. C., Gerber, P. R. & van Gunsteren, W. F. Avoiding singularities and numerical instabilities in free energy calculations based on molecular simulations. Chem. Phys. Lett. 222, 529–539 (1994).
Zacharias, M., Straatsma, T. P. & McCammon, J. A. Separation‐shifted scaling, a new scaling method for Lennard‐Jones interactions in thermodynamic integration. J. Chem. Phys. 100, 9025–9031 (1994).
Bennett, C. H. Efficient estimation of free energy differences from Monte Carlo data. J. Comput. Phys. 22, 245–268 (1976).
Humphrey, W., Dalke, A. & Schulten, K. VMD - Visual Molecular Dynamics, J. Mol. Graph. 14, 33–38 (1996).
Liu, P., Dehez, F., Cai, W. & Chipot, C. A Toolkit for the Analysis of Free-Energy Perturbation Calculations. J. Chem. Theory Comput. 8, 2606–2616 (2012).
Fiorin, G., Klein, M. L. & Hénin, J. Using collective variables to drive molecular dynamics simulations. Mol. Phys. 111, 3345–3362 (2013).
Acknowledgements
This work was supported by NIH grant R35 GM152382 to Hassane S. Mchaourab and NIH grant R35GM147423 to Mahmoud Moradi. EM data collections were conducted at the Center for Structural Biology Cryo-EM Facility at Vanderbilt University. We acknowledge the use of the Glacios cryo-TEM, which was acquired by NIH grant S10 OD030292-01. The simulations were performed on Frontera at the Texas Advanced Computing Center (TACC) through LRAC award CHE21003, made possible by the National Science Foundation award OAC-1818253.
Author information
Authors and Affiliations
Contributions
R.Z. performed the cryoEM experiments and the ATPase assays. K.J. performed DEER measurement in collaborations with R.S. A.P. and M.B. performed the MD simulation under the supervision of M.M. E.K. provided advice for structure determination and analysis. H.M. designed the research. R.Z., R.S. and H.M. analyzed, and interpreted the structures. H.M. wrote the manuscript with input from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhang, R., Jagessar, K.L., Brownd, M. et al. Conformational cycle of a protease-containing ABC transporter in lipid nanodiscs reveals the mechanism of cargo-protein coupling. Nat Commun 15, 9055 (2024). https://doi.org/10.1038/s41467-024-53420-0
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-024-53420-0