Abstract
Curative strategies for human immunodeficiency virus (HIV-1) infection are hindered by incomplete characterization of the latent reservoir and limited enhancement of anti-HIV immune responses. In this study, we identify a dual role for peripheral and tissue-resident Vδ1 T cells within the gastrointestinal mucosa of virally suppressed people with HIV. Phenotypic analyses identify an increased frequency of highly differentiated, cytotoxic effector Vδ1 T cells that inhibit HIV-1 replication in vitro coinciding with increased degranulation and IFN-γ production. Conversely, we detect an enrichment of HIV-1 DNA in tissue-resident CD4 + Vδ1 T cells in situ. Despite low CD4 expression, we find circulating Vδ1 T cells also contain HIV-1 DNA which is replication-competent. We show that T cell receptor-mediated activation of peripheral Vδ1 T cells induces de novo upregulation of CD4 providing a plausible mechanism for increased permissibility to infection. These findings highlight juxtaposing roles for Vδ1 T cells in HIV-1 persistence including contribution to tissue reservoirs.
Similar content being viewed by others
Introduction
Antiretroviral therapy (ART) does not eliminate HIV-1 from the body due to a latent reservoir of cells containing provirus that maintain a quiescent transcriptional state1,2. Following the cessation of ART, viral replication is reactivated in at least a portion of this latent reservoir leading to viral rebound and disease progression3. Resting memory CD4 + T cells are considered the main contributor to the viral reservoir as they are the primary target of HIV-1. However, growing evidence suggests additional cell types such as circulating monocytes, tissue-resident macrophages, and other non-CD4 + T cells can harbor infectious provirus4,5,6,7. Improving our understanding of anti-viral immune responses and characterizing the complete latent reservoir will facilitate strategies for eradicating persistent HIV-1 infection.
Unconventional lymphocytes such as γδ T cells are among the majority of T cells that do not typically express CD4. The two main human γδ T cell subsets, Vδ1 and Vδ2, are distinguished by divergent thymic development, tissue localization, and cognate antigens8,9. Vδ1 T cells populate mucosal sites such as the gastrointestinal (GI) tract where they act as mediators of tissue homeostasis10,11. Conversely, peripheral γδ T cells are primarily comprised of innate-like Vγ9 Vδ2 T cells with minor frequencies of Vδ1 T cells that have adaptive qualities9,12,13. Peripheral Vδ1 T cells expand in response to a limited set of antigens including HIV-114. During early infection, the ratio of Vδ1:Vδ2 frequencies is inverted within the periphery and further pronounced within the GI tract7,15,16,17. Reports of their potential role in controlling viral replication in the absence of ART suggest they are a mediator of anti-viral immunity, as evidenced in recent studies which found strong correlations between Vδ1 T cell frequencies and cytotoxic effector phenotypes with the control of HIV-1 replication both in a cohort of elite controllers (EC) and a person living with HIV-1 undergoing analytical treatment interruption18,19. Vδ1 T cells can directly lyse or inhibit HIV replication through germline-encoded receptors generally associated with Natural Killer (NK) cells20,21. Whether the anti-HIV functions of Vδ1 T cell persist under viral suppression is unclear.
Few studies have investigated the susceptibility of γδ T cells to HIV-1 infection. Nevertheless, both Vδ1 and Vδ2 clones are amenable to infection in vitro and HIV-1 provirus has been quantified from isolated pan-γδ T cells by us and others22,23,24. In addition, our group identified peripheral Vδ2 T cells as a cellular reservoir capable of producing replication-competent virus upon reactivation7. We further demonstrated that activation induced upregulation of CD4 expression on Vδ2 T cells directly increasing permissibility to infection. These observations indicate that γδ T cells are not only susceptible to HIV-1 infection in vivo but may contribute to viral persistence through poorly defined mechanisms. Whether the more abundant Vδ1 subset is targeted by HIV-1 either in circulation or the GI tract remains to be elucidated.
In the present study, we found that circulating Vδ1 T cells from virally suppressed people living with HIV-1 (PWH) on stable ART displayed signatures of continuous activation marked by skewing towards a TEMRA-like (CD45RA + CD27−) phenotype with higher co-expression of cytotoxic markers. Functionally, these Vδ1 T cells retained their proliferative capacity to TCR stimulation and inhibited viral replication in vitro. Paired analysis of GI tract revealed a comparable phenotype between both compartments with a few key exceptions including a higher frequency of Vδ1 T cells expressing CD4 within the lamina propria. Using HIV DNAscope we found tissue-resident CD4 + Vδ1 T cells contain HIV DNA and constitute a previously unrecognized cellular reservoir representing around 20% of all infected CD4+ cells within the GI tract. In an independent cohort containing elite controllers (EC), HIV DNA was also quantified in Vδ1 T cells from the small and large intestine where they comprised 19% of all virally infected CD4+ cells but only 5% in EC. Despite the low ex vivo CD4 expression on circulating Vδ1 T cells, replication-competent virus was recovered from 5/11 ART-suppressed PWH. Finally, infectivity and modulation of CD4 expression on Vδ1 T cells from HIV-seronegative donors revealed de novo expression of CD4 following TCR stimulation, indicating a potential window of opportunity of increased permissibility to HIV-1 infection. These findings reveal a unique, juxtaposing role for Vδ1 T cells in HIV-1 pathogenesis with potential implications in viral persistence and cure strategies.
Results
Participant characteristics
The ART-suppressed PWH cohorts recruited at US sites were mostly male (91.3%) with a median age of 36 years old. The Spanish cohort was primarily male (100% ART PWH, 66% EC) with a median age of 40. All participants had been on ART for a minimum of 1 year (Table 1). The median age of the people without HIV-1 (HIV−) control group was 39 years and also composed of mostly male donors (59.3%).
Vδ1 T cells display a highly differentiated, cytotoxic phenotype during ART suppression
The expansion of Vδ1 T cell frequencies is maintained throughout suppressive ART16,25. In other chronic viral infections such as human cytomegalovirus (CMV), expanded Vδ1 T cells display phenotypic characteristics that resemble adaptive immunity26,27. Therefore, we hypothesized that Vδ1 T cells in ART-suppressed PWH possessed phenotypes indicative of enhanced cytotoxicity or tissue homing function. We assessed the expression of memory, cytotoxicity, and tissue homing markers in PWH and HIV controls (Supplementary Fig. 1a, b). Similar to previous reports from us and others24,25,28, peripheral Vδ1 T cells from ART-suppressed PWH were expanded compared to HIV− controls (Fig. 1a, b) and displayed an inverted Vδ1:Vδ2 ratio (Supplementary Fig. 2a). In addition, Vδ1 T cells from PWH skewed from a naive (CD45RA+ CD27+) towards a TEMRA-like (CD45RA+ CD27−) phenotype (Fig. 1c) which positively correlated with cell frequency in both groups (Supplementary Fig. 2b, c). The shift in memory phenotypes was only observed in Vδ1 T cells from PWH whereas Vδ2 T cells and conventional αβ T cells were similar between each group apart from lower naive CD4 T cells in PWH (Fig. 1d–f). In addition, Vδ1 T cells from PWH had increased expression of cytotoxic markers (CD8, CD56) as well as those involved in the recognition or lysis of HIV-infected cells (CD16, NKp30, NKG2D) (Fig. 1g). This alteration in effector phenotypes was specific to Vδ1 T cells compared to other cytotoxic cells such as Vδ2 T cells, αβ CD8 + T cells, and NK cells except for higher NKG2D expression which was present in each cell type (Fig. 1h). Excluding CCR7 that was less expressed in PWH, receptors or adhesion molecules that mediate lymphocyte migration was distributed similarly in Vδ1 T cells from PWH and HIV− controls (Supplementary Fig. 2d–f). While the PWH cohort was predominantly comprised of male donors, there were no sex-based differences between phenotypic markers within the HIV− controls (Supplementary Fig. 3a–d). To confirm persistent HIV-1 infection has a specific signature on Vδ1 T cells, we conducted a deeper phenotypic analysis in 15 PWH using a mass cytometry (CyTOF) panel previously designed by our group29. Following FlowSOM analysis, Vδ1 T cells formed a distinct cluster (cluster #4) most closely resembling highly differentiated αβ CD8 + T cells (cluster #2, CD27−CD28−CD57+) with both cell types showing higher relative expression of TNF-α and granzyme B (GzmB) compared to Vδ2 T cells and NK cells (Fig. 1i, j, Supplementary Fig. 4). Relative IFN-γ expression was the highest within Vδ2 T cells (cluster #5) which was contained to a minor frequency of cells (<1.0%), similar to previous reports of infrequent IFN-γ producing αβ CD8 T cells within ART-suppressed PWH29,30. In addition to displaying a TEMRA-like phenotype, Vδ1 T cells had elevated expression of CD160, TIGIT, and PD-1 indicative of in vivo activation occurring during the course of HIV-1 infection (Fig. 1j).
a Representative flow cytometry pseudocolor plots and b frequency of total circulating CD3 + Vδ1 and Vδ2 T cells in HIV− donors (blue, n = 28) and ART-suppressed PWH (orange, n = 21). Frequency of effector/memory subsets within c Vδ1 T cells, d Vδ2 T cells, e αβ CD8 T cells, and f αβ CD4 T cells in HIV− (n = 14) and PWH (n = 21). g Comparison of cytotoxic marker expression on Vδ1 T cells between HIV− donors (blue, n = 14) and PWH (orange, n = 21). h Radar plots displaying cytotoxic and memory marker expression between effector subsets in HIV− donors (blue, dotted line) and PWH (solid, orange line) from left to right: Vδ1 T cells, Vδ2 T cells, αβ CD8 + T cells, CD56dimCD16 NK cells, and CD56brightCD16- NK cells. i UMAP visualization of effector cell clusters identified by FlowSOM analysis of mass cytometry data from ART-suppressed PWH (n = 15) and j heatmap of relative expression of markers associated with cytotoxicity, effector/memory status, and activation/exhaustion within each cluster. k Principal component analysis of ATAC-seq data from circulating Vδ1 T cells from HIV− (blue, n = 3) and PWH (orange, n = 3). l Number of differentially accessible chromatin regions (DAR) in PWH compared to HIV− controls (absolute shrunken Log2 FC > 0.5, FDR-adjusted p value < 0.1). m Integrated genome viewer window showing reduced ATAC-seq peaks (highlighted in blue) within the ETS1 and IKZF3 loci. Two-sided Mann-Whitney U test (b, g), one-way ANOVA using the Holm-Šídák method for multiple comparisons (c–f), and Wald test using Benjamini–Hochberg correction (l). Exact p values for (b) Vδ1 p = 0.00005 and (c) TEMRA Vδ1 p = 0.00085. Mean ± S.E.M are represented. Source data are provided as a Source Data file.
Pathogen-driven Vδ1 T cell activation results in chromatin remodeling which influences subsequent responses to antigen or cytokine production27. Therefore, we performed ATAC-seq analysis on purified circulating Vδ1 T cells that revealed only modest differences between PWH and HIV− controls, but a noticeable reduction in interpersonal variation (Fig. 1k). Amongst the 123 differentially accessible regions in PWH (29 open, 94 closed) (Fig. 1l), the most pronounced repressive changes occurred in regulatory regions or genes that are typically downregulated in HIV-specific effector αβ CD8 + T cells (IL7Ra, ETS-1)31,32, but also genes that regulate functional T cell exhaustion (IKZF3, ITPKB, CD5, CD6, CD96) (Fig. 1m). Among the repressed regions was NCR2 (NKp44), which potentially explains the observed downregulation at the cell surface in ART-suppressed PWH by flow cytometry analysis (Fig. 1g). Subsequent HOMER motif analysis identified transcription factor binding sites for Nuclear factor kappa B (NF-κB) in 24.3% of repressed sequences whereas 21.7% of opened regions contain sites for Krüppel-like factor 4 (Klf4) (Supplementary Fig. 5a, b) which enhances cytotoxicity in exhausted αβ CD8 + T cells33. Despite bearing some phenotypic and epigenetic resemblances to exhausted αβ CD8 + T cells, we found Vδ1 T cells from PWH have comparable proliferative responses to HIV− controls following TCR stimulation (Supplementary Fig. 5c, d). Together these data demonstrate that Vδ1 T cells in PWH display hallmarks of in vivo activation and differentiation into cytotoxic effectors despite durable viral suppression.
Vδ1 T cells from ART-suppressed PWH retain their anti-HIV functionality
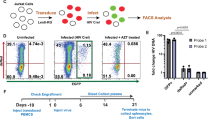
Whether Vδ1 T cell effector functions are maintained during suppressive ART is unclear. To assess anti-HIV function, we performed viral inhibition assays (VIA) previously adapted by our group (Figs. 2a and Supplementary Fig. 6)34,35. Co-culture of superinfected autologous αβ CD4 + T cells with purified Vδ1 T cells resulted in a reduction in HIVp24 production in a dose-dependent manner (mean reduction of 47.9% and 67% at effector:target (E:T) ratios of 1:1 and 1:10, respectively, Fig. 2b). Vδ1 T cells specifically degranulated in the presence of superinfected αβ CD4 + T cells compared to co-culture conditions with resting autologous αβ CD4 + T cells (Figs. 2c, d and Supplementary Fig. 7a). This coincided with a significant increase in the frequency of IFN-γ producing Vδ1 T cells (Figs. 2e, f and 7b), indicating direct recognition of cells actively producing virus.
a Schematic representation of the viral inhibition assay (VIA). αβ CD4+ cells from ART-suppressed PWH were superinfected HIV-JRCSF before co-culture with autologous Vδ1 T cells for 7 days. Created in BioRender. Mann, B. (2024) https://BioRender.com/r09k447. b HIVp24 production from day 7 supernatants in co-cultures at a 1:1 (n = 8) and 1:10 (n = 6) E:T ratio normalized to αβ CD4 T cells cultured alone. Cytotoxicity assay showing (c), representative zebra plots and (d), quantification of CD107a expression and (e), f intracellular IFN-γ production within Vδ1 T cells cultured alone, co-cultured with non-superinfected αβ CD4 + T cells, or with superinfected αβ CD4 T cells at a 1:1 ratio (n = 6). g Inhibition of viral replication from superinfected CD4 + T cells by Vδ1 T cells, Vδ2 T cells, αβ CD8 T cells, and NK cells with or without pretreatment with IL-15 (n = 5). Sample sizes reflect biological replicates corresponding to distinct ART-suppressed PWH using the two-sided Friedman test with Dunn’s correction for multiple comparisons (b, d, f, g). Individually colored points represent distinct biological replicates of HIV− donors (b, d, f, g). Mean ± S.E.M are represented. Source data are provided as a Source Data file.
To date, efforts to boost anti-HIV cellular responses against the latent reservoir have primarily focused on αβ CD8 + T cells or NK cells35,36. This includes the use of pleiotropic compounds such as the IL-15 superagonist N-803 which both reactivates viral transcription and activates cytotoxic effector cells37. To assess whether IL-15 may also enhance Vδ1 T cells, we pre-treated the cells prior to the VIA and compared the effects to matching FACS-isolated Vδ2 T cells, αβ CD8 + T cells, and NK cells. A groupwise comparison revealed IL-15 enhanced viral inhibition across all effector cell types (Fig. 2g, Supplementary Fig. 6). Specifically, pretreatment of Vδ1 T cells with IL-15 led to a greater reduction in HIVp24 production compared to untreated effector cells (mean reduction 81.4% v. 68.5% respectively). IL-15 treatment had a comparable effect on matching αβ CD8 + T cells and NK cells, but no discernible impact on Vδ2 T cells. Collectively these experiments demonstrate that not only do Vδ1 T cells retain their anti-HIV activity during ART, but also this activity may be enhanced by clinically relevant compounds being pursued for HIV-1 cure.
Circulating and Vδ1 T cells from the GI are phenotypically distinct with a higher frequency of CD4 expression in the lamina propria
Peripheral and tissue-resident Vδ1 T cells possess distinct TCR repertoires and phenotypes likely indicative of their unique roles in immunity8,38. How persistent HIV-1 infection impacts this tissue compartmentalization remains poorly understood17,18. Therefore, we conducted paired phenotypic analyses comparing circulating and Vδ1 T cells from rectosigmoid biopsies from six ART PWH. Confirming previous studies17,39, we found higher frequencies of Vδ1 T cells within the tissue compared to peripheral blood (GI mean: 7.4% v. PB mean: 4.5%, Fig. 3a, b). While peripheral Vδ1 T cells were defined by their TEMRA-like phenotype (Fig. 1c), their mucosal counterparts displayed a TEM (CD45RA-CD27−) profile (Fig. 3c). The expression of select cytotoxic markers was similar between compartments apart from lower CD16 and higher NKp44 within the tissue (Fig. 3d). Mucosal Vδ1 T cells also displayed near ubiquitous expression of CCR5 and elevated frequencies of CD161 (Supplementary Fig. 8b). Both PD-1 and TIGIT expression were comparable between compartments, however circulating Vδ1 T cells had more variable expression of each marker (Supplementary Fig. 8d).
a Representative pseudocolor plots and (b) Vδ1 and Vδ2 T cell frequency, (c) effector/memory and (d) cytotoxic marker expression on Vδ1 T cells in matched samples from peripheral blood (PB, blue) and the gastrointestinal tract (GI, orange) from ART-suppressed PWH (n = 6). e Representative pseudocolor plot and f frequency of Vδ1 (circles), Vδ2 (triangles), αβ CD4 (squares) and CD8 (diamonds) T cell subsets within the total tissue of ART-suppressed PWH (n = 6), tissue residency defined as CD103 + CD69 intraepithelial lymphocytes (IEL), CD103-CD69+ lamina propria lymphocytes (LPL), and CD103-CD69- infiltrating lymphocytes. g Representative image of IEL and LP Vδ1 T cells by phenotypic immunofluorescent analysis of CD4 (blue), CD8 (orange), and Vδ1 (purple). White arrows point to CD4 + Vδ1 + cells exclusively found in LP, findings were confirmed by two independent experiments, scale bar = 100 μm. h Comparison of CD4 expression in Vδ1 T cells between matched PB and GI from ART-suppressed PWH (n = 6). Two-sided Wilcoxon matched pairs signed-rank test (c, d, h, i). Means ± S.E.M are represented (b–d, f, h). Source data are provided as a Source Data file.
We next investigated the tissue-residency status of Vδ1 T cells compared to other lymphocyte populations based on co-expression of the αE integrin (CD103) and CD69 (Fig. 3e). Intraepithelial lymphocytes (IEL) defined as CD103+ CD69+ were primarily αβ CD8 + T cells (mean 67.9%) followed by Vδ1 T cells (mean 13.9%) with minimal frequencies of other T cell subsets (Fig. 3f). By contrast, CD103-CD69+ lymphocytes within the lamina propria (LP) and infiltrating (CD103−CD69−) cells were vastly comprised of αβ T cells with CD4+ cells representing nearly half of each population (mean 45.9% and 49.5%, respectively). Localization of Vδ1, CD4, and CD8 expressing cells within each layer of the mucosa was confirmed by conventional immunofluorescent staining of paraffin-embedded tissue samples (Fig. 3g). Our results show that mucosal Vδ1 T cells are phenotypically distinct from those in circulation with divergent patterns of immunological memory and key cytotoxic receptors. Interestingly, in situ phenotyping revealed a high proportion of Vδ1 T cells expressing CD4 that localized within the lamina propria (mean 30.4%), in contrast to Vδ1 T cells in PB which were largely CD4− (mean 1.3%, Fig. 3h). Raising the question as to whether they can be potential cellular target for HIV-1 and contribute to the latent reservoir.
Tissue-resident CD4 + Vδ1 T cells harbor HIV-1 DNA
To quantify HIV-1 DNA in tissue-resident Vδ1 T cells, we combined immunofluorescent phenotyping with the ultrasensitive next-generation HIV DNAScope assay40. We used rectosigmoid colon biopsies from ART-suppressed PWH (Fig. 3) or ileum and caecum samples from an independent cohort that included ART-suppressed PWH and EC (Table 1). Quantitative image analysis revealed variability between groups based on the frequency of total CD4+ cells which comprised a mean of 9.6% of all cells relative to the total number of nuclei in the colon compared to 51.9% in the ileum/caecum of ART-suppressed PWH and 17.6% in EC. Conversely, the frequency of total Vδ1 T cells was consistent across each group with a mean of 3.4% in the colon, 2.1% in the ileum/caecum of ART-suppressed PWH and 3.7% in EC. The frequency of CD4 + Vδ1 T cells was also similar within the colon compared to the ileum/caecum of both ART-suppressed PWH and (mean 30.3% v. 21.6% v. 20.4% respectively, Fig. 4a). Due to the high variation in total CD4+ cells, the proportion of CD4 + Vδ1 T cells to total CD4+ cells varied between each group with 10.8% in the colon and 0.9% in the ileum/caecum of ART-suppressed PWH, and 5.1% in EC (Fig. 4b).
a Quantification of CD4+ cells (orange), Vδ1 T cells (green), and CD4 + Vδ1 + T cells (blue) relative to total nuclei within the colon of ART-suppressed PWH (GW cohort, n = 5), ileum/caecum of ART-suppressed PWH (SV cohort, n = 4), and ileum/caecum of elite controllers (EC SV cohort, n = 3). b Stacked bar plots of CD4 + Vδ1 T cells (blue) and CD4 + Vδ1− cells (green) as a proportion of total CD4+ cells within each section of the GI. c Representative DNAscope image showing HIV-1 viral DNA (vDNA, red) combined with phenotypic immunofluorescent analysis of CD4 (blue) and Vδ1 (green). Arrow is pointing to one infected CD4 + Vδ1 T cell that contains vDNA, findings were confirmed by two independent experiments, scale bar = 100 μm. d Total vDNA+CD4+ cells (orange), Vδ1-CD4+ (green) and Vδ1 + CD4+ (blue) cells within the GI of two independent cohorts of ART-suppressed PWH and ECs (ART PWH Colon n = 5, ART PWH Ileum/Caecum n = 4, EC Ileum/Caecum n = 3). e Stacked bar plots representing Vδ1 + and Vδ1− cells amongst total vDNA+CD4+ cells. Frequency of total, intact and defective HIV DNA/106 cells in (f), purified, circulating Vδ1 T cells (blue) and (g) matching αβ CD4 T cells (green) from ART-suppressed PWH (n = 11) by the Intact proviral DNA Assay (IPDA). h Frequency of infected Vδ1 (blue) and αβ CD4 + T cells (green) based on the proportion of cells containing total HIV DNA. i Infectious units per million (IUPM) Vδ1 T cells represent replication-competent virus recovered in quantitative viral outgrowth assays (QVOA) from ART-suppressed PWH (n = 11). j Correlation matrix between HIV DNA species measured by IPDA, participant clinical characteristics, and phenotype of circulating Vδ1 T cells. Two-sided Spearman’s ranked correlation test (j). Mean ± S.E.M. (a, d, f–i). Source data are provided as a Source Data file.
HIV-1 DNA was detected in all ART-suppressed participants within both sections of the GI tract (Fig. 4c, d). The frequency of total CD4+ cells containing HIV-1 DNA was 4.6% in the colon and 0.5% in the ileum/caecum of ART-suppressed PWH. In EC, the frequency of HIV infection within total CD4 + T cells was 1.3%. HIV-1 DNA was quantified in Vδ1 T cells in 100% (5/5) of colon samples, 80% of ileum/caecum samples from ART-suppressed PWH, and 33% (1/3) of EC. HIV-1 DNA was exclusively found within Vδ1 T cells expressing the CD4 receptor which represented ~20.4% of all HIV-1 DNA + CD4+ cells within the colon and 19.2% in the ileum/caecum of ART-suppressed PWH (Fig. 4e). This was contrasted by a lower proportion of infected CD4 + Vδ1 T cells in EC (mean 5.1%, Fig. 4e). Our results indicate that not only do CD4 + Vδ1 T cells contain HIV-1 DNA, but they contribute to the latent HIV-1 reservoir across the GI tract.
Peripheral Vδ1 T cells are a latent reservoir of HIV-1
To assess whether Vδ1 T cells contribute to the peripheral latent reservoir, we quantified HIV-1 DNA within FACS-purified Vδ1 T cells by the IPDA (Supplementary Fig. 9a–d). We detected total HIV-1 DNA in Vδ1 T cells from 91% (10/11) donors including 80% (4/5 donors) of participants with positive HIV DNAscope data (Fig. 4f). Defective provirus was detected in each positive sample (Supplementary Table 2) with a lower proportion of 5’ defective provirus compared to 3’defective or hypermutated sequences. Intact provirus was not detected within Vδ1 T cells likely due to the limited number of available cells (mean of 44,350 Vδ1 cells/assay, Supplementary Fig. 9d). By contrast, total HIV-1 DNA in matched αβ CD4 + T cells run in parallel was quantified in all participants (11/11 donors) (Fig. 4g) and intact provirus within 73% (8/11) of participants along with both 5’ defective and 3’ defective/hypermutated provirus. Despite the absence of intact provirus, we found a comparably low frequency of circulating Vδ1 T cells and αβ CD4 + T cells contain total HIV-1 DNA (Fig. 4h). The IPDA requires additional processing of purified cells to isolate genomic DNA which may result in shearing intact HIV-1 genomes. This presents a unique challenge when quantifying provirus from the limited number of Vδ1 T cells available from small volume blood draws. To overcome these limitations, we conducted quantitative viral outgrowth assays (QVOA) using leukapheresis from additional 11 ART-suppressed PWH (Table 1), as we previously described7. A mean of 498,182 absolute circulating Vδ1 T cells were assayed (Supplementary Table 3) and infectious virus was recovered in 45% (5/11) of participants with a mean of 2.4 infectious units/106 cells (Fig. 4i). These results demonstrate that not only do peripheral Vδ1 T cells contribute to the latent reservoir, but also highlight additional challenges when quantifying latent HIV-1 in rare cell populations.
We next determined whether the frequency of infected Vδ1 T cells was associated with clinical characteristics of the donors (Fig. 4j). HIV-1 DNA in Vδ1 T cells was not associated with age, time since diagnosis, time on ART, or CD4 count. We also did not find any associations between HIV-1 DNA copies in matching Vδ1 T cells and αβ CD4 + T cells. Lastly, we sought to identify potential phenotypic markers on Vδ1 T cells associated with viral infection. We found positive correlations between total HIV-1 DNA in Vδ1 T cells and a TCM phenotype as well as with the expression of α4β7, both of which have previously been implicated in the identification of latently infected αβ CD4 + T cells (Fig. 4j)41,42. Interestingly, there was a strong negative association between the frequency of total HIV-1 DNA in Vδ1 T cells and the frequency of CD8 + Vδ1 T cells. Altogether, our findings demonstrate Vδ1 T cells are a previously unrecognized cellular reservoir of latent HIV-1 infection within the GI mucosa and PB. While our data indicates that CD4 expression is associated with infection of mucosal Vδ1 T cells, how peripheral Vδ1 T cells become infected is still an outstanding question due to the low or absent expression of CD4.
Infection of Vδ1 T cells is CD4−dependent
We investigated the permissibility of Vδ1 T cells to HIV-1 viral entry in vitro. Flow cytometry analysis of HIV− donors confirmed that circulating γδ T cells are predominantly the Vδ2 subset prior to infection (Fig. 5a). The majority (>90%) of both subsets did not express CD4 ex vivo although we found higher frequency and greater variability within Vδ1 T cells (Fig. 5b). CD4 expression on Vδ1 T cells was not associated with either the age or self-reported sex (Fig. 5c, d). In contrast, both Vδ1 and Vδ2 T cells had significantly higher expression of CCR5 (Fig. 5e) and CXCR4 (Fig. 5f) compared to αβ CD4 + T cells. Despite a low frequency of cells expressing CD4, successful infection of Vδ1 T cell clones lacking CD4 expression has been previously reported22. To determine the requirement of CD4 for viral entry in primary Vδ1 T cells, PHA-activated PBMCs were exposed to HIVJR-CSF in the presence or absence of a monoclonal antibody blocking CD4 prior to infection. As expected, the proportion of infected Vδ1 T cells was significantly lower than αβ CD4 + T cells (Fig. 5g, h). The majority of HIVp24 + Vδ1 T cells did not express CD4 (Fig. 5i) which is compatible with the downregulation of CD4 induced by the viral accessory proteins vpu and nef43. Nonetheless, we observed a positive correlation between CD4 expression on Vδ1 T cells prior to infection and the proportion of infected cells after seven days of infection (Fig. 5j) with CD4 blockade successfully abrogating infection (Fig. 5k). Together these data demonstrate that the primary mechanism of viral entry and infection of Vδ1 T cells is CD4−dependent.
a Frequency of total circulating CD3 + Vδ1 (blue), Vδ2 (red), and αβ CD4 + T cells (green) in HIV− donors (n = 27). b Comparison of CD4 expression on Vδ1 (blue) and Vδ2 (red) T cells which did not correlate with (c) age or (d) biological sex of HIV− donors (male, blue; female, orange, n = 27). Comparison of co-receptors (e) CCR5 and (f) CXCR4 expressed on matched Vδ1 (blue), Vδ2 (red) and αβ CD4 (green) from HIV− donors (n = 15). g Representative pseudocolor plots and (h) frequency of infected Vδ1 (blue) and αβ CD4+ (green) T cells measured as positive for intracellular HIVp24 on day 7 of PBMCs from HIV− donors infected with HIVJR-CSF ex vivo (n = 9). i Frequency of CD4+ (blue) and CD4− (orange) Vδ1 T cells positive for HIVp24 on day 7 of infection of ex vivo PBMC infection (n = 9). j Correlation between the frequency of CD4 + Vδ1 T cells prior to infection and the frequency of HIVp24 + Vδ1 on day 7 of ex vivo PBMC infection (n = 9). k Comparison of the frequency of HIVp24 + Vδ1 T cells with or without α-CD4 blockade prior to infection (n = 9). Two-sided Wilcoxon matched pairs signed-rank test (b, e, f, h, I, k). Exact p value for (e) Vδ1 v. Vδ2 p = 0.00006 and Vδ2 v. αβ CD4 p = 0.00006. Two-sided Spearman’s r and p value shown for significant correlations with simple linear regression analyses showing the line of best fit and 95% confidence intervals (c, j). Mean ± S.E.M. are represented (a, b, d–f, h, i). Source data are provided as a Source Data file.
TCR stimulation induces CD4 upregulation in a discrete subset of Vδ1 T cells
Our finding that peripheral Vδ1 T cells contain HIV-1 provirus (Fig. 4i) indicates that infection occurs in vivo despite low CD4 expression (Fig. 3h). Based on the rapid expansion and differentiation of Vδ1 T cells during HIV-1 infection, we investigated the dynamics of CD4 expression following activation in samples from HIV− donors. Within the first seven days following αCD3/αCD28 and IL-2 stimulation, the mean frequency of Vδ1 T cells expressing surface or total CD4 increased from a mean of 3.4% peaking between day three to five (mean 18.2%, Fig. 6a–c). The combination of all three stimuli consistently produced the highest frequency of CD4 + Vδ1 T cells (Fig. 6d). However, neither αCD28 co-stimulation nor IL-2 supplementation alone were sufficient to increase CD4 expression compared to unstimulated controls. Interestingly, CD4 expression increased with each successive division after αCD3/αCD28 + IL-2 stimulation (Figs. 6e, f) which was contrasted by lower proliferation of CD8 + Vδ1 T cells (Fig. 6e, g). Our data demonstrates that the induction of CD4 expression in Vδ1 T cells requires TCR-CD3 complex signaling, possibly linking modulation to antigen recognition. However, CD4 expression is excluded from CD8 + Vδ1 T cells (Fig. 6h) possibly suggesting the existence of unique mechanisms that regulate CD4 expression following activation in a discrete Vδ1 T cell subset.
a Representative zebra plots and quantification of time course, (b) surface, and c total CD4 expression on circulating Vδ1 T cells from HIV− donors (n = 5) following stimulation with αCD3/αCD28 + IL-2 (navy), IL-2 alone (yellow), or unstimulated (orange). d Comparison of CD4 expression on Vδ1 T cells from HIV− donors (n = 8) in time course analysis following stimulation with αCD3/αCD28 + IL-2 (navy), αCD3 + IL-2 (blue), αCD28 + IL-2 (green), IL-2 alone (yellow), or unstimulated (orange). e Representative histograms and frequency of (f) CD4 and (g) CD8 expressing Vδ1 T cells from HIV− donors (n = 4) across cell divisions following 5 days of stimulation with αCD3/αCD28 + IL-2. h Representative zebra plots from day 0 and day 5 following stimulation demonstrating CD4 and CD8 expression are confined to discrete Vδ1 T cell populations. i Representative zebra plots and (j) frequency of CD4 expression after depletion and subsequent expression on Vδ1 T cells from HIV− donors (n = 4) following stimulation with αCD3/αCD28 + IL-2 (navy) v. unstimulated (orange) quantified on day 0, day 3, day 5, and day 7. k UMAP of total Vδ1 T cell clusters identified by FlowSOM analysis of CD4 expression and markers associated with activation/differentiation 5 days after stimulation on Vδ1 T cells from HIV− donors (n = 6). l UMAP of relative CD4 expression overlaid onto FlowSOM clusters and (m) heatmap of relative expression of each marker. Two-way ANOVA with Holm-Šídák method for multiple comparisons (b–d, j). Exact p values for (b) surface expression p = 0.0003, (c) total expression p = 0.0008, (d) required stimuli p = 0.0001, and (j) de novo expression p = 0.004. Mean ± S.E.M. is represented. Source data are provided as a Source Data file.
These results raised the possibility that pre-existing CD4 + Vδ1 T cells may have more robust proliferative responses than CD4− cells. Therefore, we depleted CD4+ cells from PBMCs prior to stimulation and analyzed CD4 expression kinetics. Upon activation of Vδ1 + CD4− cells, we observed de novo CD4 expression in a mean frequency of 15.7% with similar kinetics to the activation of whole PBMCs (Fig. 6i, j). We next performed ATAC-seq analysis from Vδ1 T cells isolated ex vivo revealing that the Cd4 locus was largely repressed within both HIV− individuals and virally suppressed PWH (Supplementary Fig. 10). CD4 expression is regulated through multiple epigenetic or transcriptional mechanisms throughout the development of αβ T cells including DNA methylation44. Therefore, using a methylation-sensitive restriction enzyme-based assay, we were able to directly detect methylation of CpG dinucleotides within the Cd4 promoter region of a portion of Vδ1 T cells in both unstimulated and TCR-stimulated conditions (Supplementary Fig. 11a–c). To determine the role of methylation in CD4 expression, we utilized the DNA methyltransferase I (DNMT1) inhibitor 5-Azacytidine (5-Aza) which prevents the carryover of homeostatic methylation in daughter cells following cell division. Inclusion of 5-Aza in PBMC cultures stimulated with αCD3/αCD28 and IL-2 had a consistent additive effect on the frequency of CD4 + Vδ1 T cells five days after activation compared to mock treated controls (Supplementary Fig. 11c, d). Together, these data show that a subpopulation of Vδ1 T cells that express CD4 follows discrete epigenetic programs that differentiate it from other subpopulations such as CD8 + Vδ1 T cells.
Next, we assessed whether CD4 + Vδ1 T cells constitute a unique subpopulation potentially associated with a discrete activation and differentiation profile. Within 5 days of stimulation, high frequencies of Vδ1 T cells expressed CD69, CD25, and HLA-DR showing a spectrum of cells within early, mid, and late-stage activation respectively (Supplementary Fig. 12a). While there were minimal changes in co-stimulatory molecules CD27 and CD28, significant proportions of Vδ1 T cells had downregulated naive markers CD45RA and IL-7 receptor α (IL-7Rα) and upregulated lymphoid homing receptors CD62L and CCR7 (Supplementary Fig. 12b–d). To better understand the distribution of CD4 expression within different Vδ1 T cell subsets, we performed FlowSOM analysis. CD4 expression was concentrated in a distinct cluster (cluster #5) featuring the highest relative expression of CD25 and intermediate expression of CD27, CD28, and CD62L with low expression of CD45RA and IL-7Rα (Fig. 6k–m, Supplementary Fig. 12e). On the contrary, CD4−Vδ1 T cells (cluster #2) had the lowest expression of CD25, CD27, CD28, and CD62L along with the highest expression of CD45RA (Fig. 6m). Our findings show that a specific subset of Vδ1 T cells upregulates CD4 following activation and may represent a distinct population that become susceptible to HIV-1 infection in vivo.
Discussion
γδ T cells are not only a central component to immunosurveillance of mucosal tissue, but also play a crucial role in response to microbial infections45. HIV-1 infection permanently reshapes the γδ T cell compartment with the most pronounced effects occurring in the Vδ1 subset both within the periphery and the GI tract17,39. In the present study, we reveal a dichotomy for circulating and mucosal Vδ1 T cell subpopulations with discrete functional contributions in ART-suppressed PWH. A subset of highly differentiated, cytotoxic effectors that inhibits HIV-1 replication and a second, possibly distinct subset expressing the CD4 receptor that contributes contribution to the latent HIV-1 reservoir.
In agreement with previous studies, we found ART-suppressed PWH had elevated frequencies of peripheral Vδ1 T cells that skewed towards a TEMRA-like phenotype coinciding with higher expression of cytotoxic receptors18,25. Although γδ T cells have historically been considered innate-like, recent evidence suggests that certain subsets can undergo clonal expansion and assume properties more closely aligned with adaptive αβ CD8 + T cells responses12. Our data revealed a positive correlation between the proportion of CD8 + Vδ1 T cells and both total circulating frequencies and TEMRA-like Vδ1 T cells suggesting a link between CD8 expression and in vivo effector differentiation. The specific expansion of CD8 + Vδ1 T cells observed in other diseases such as cardiovascular disease or chronic antigen exposure that occurs in bacterial (mycobacterium tuberculosis) or viral (CMV) infections as well as cancer may be a byproduct of prolonged exposure to inflammatory conditions46. The proposed etiology of Vδ1 T cell expansion in PWH has ranged from recognition of gut-derived microbial antigens to HIV-mediated impairment of tissue homing, but a definitive mechanism has yet to be described47,48. IL-7 signaling was recently implicated as the main driver behind the expansion of thymic derived CD8 + Vδ1 T cells49. Plasma IL-7 levels are elevated in untreated HIV-1 infection and subsequently restored along with IL-7Rα expression on αβ T cells during ART50. Instead, we observed persistent downregulation of IL-7Rα on Vδ1 T cells suggesting continuous exposure to antigen may occur despite suppressive ART.
Few studies have investigated the functional consequences to the phenotypic changes in Vδ1 T cells during ART suppression25. Our study provides a comparative analysis of both phenotype and anti-HIV function between matched circulating γδ T cell subsets, αβ CD8 + T cells, and NK cells. We found Vδ1 T cells from ART-suppressed individuals displayed a higher frequency of cells expressing NKp30 and CD16 compared to HIV− controls similar to NK cells. Similarly, both cell types co-expressed CD160 and TIGIT indicating potential shared regulatory mechanisms of their effector functions28,51. Despite these similarities to NK cells, unsupervised clustering analysis indicated that Vδ1 T cells are most similar to antigen-experienced αβ CD8 + T cells with the highest relative expression of TNF-α, GzmB, and PD-1. In contrast to exhausted αβ CD8 T cells, we found circulating Vδ1 T cells in ART-suppressed PWH retained their proliferative responses to in vitro TCR stimulation despite elevated PD-1 expression similar to reports of tissue-resident Vδ1 T cells within the context of solid tumors52. While we did not observe any differences in either PD-1 or TIGIT expression between matched circulating and mucosal Vδ1 T cells, interpersonal variation was lower in tissue-resident cells possibly highlighting divergent regulation of inhibitory receptor expression between anatomical compartments. Collectively these data suggest expression of such inhibitory receptors on Vδ1 T cells is likely indicative of an activated phenotype rather than one strictly associated with functional exhaustion. Perhaps more importantly, we demonstrate that circulating Vδ1 T cells inhibited HIV-1 replication of superinfected αβ CD4 + T cells. Inhibition coincided with increased degranulation and IFN-γ production which suggests direct recognition of infected target cells. While these are surrogate markers for direct cytotoxicity, the secretion of β chemokines such as MIP-1α, MIP-β, and RANTES by Vδ1 T cells likely contribute to viral inhibition as previously reported21. Several innate receptors have been implicated in the recognition of HIV-1 infected cells by Vδ1 T cells including NKp30 and NKG2C20,21. Additionally, NKG2D could potentially mediate recognition as we and others have previously reported for the Vδ2 subset7,28. Whether the TCR plays a role and identification of potential cognate ligands is an intriguing question. This may require the use of cutting-edge CRISPR-Cas9 knockout screenings which have previously identified the genetic determinants of γδ TCR recognition53,54. Furthermore, we found Vδ1 T cell anti-HIV function to be enhanced by pretreatment with the γc cytokine IL-15 similar to αβ CD8 + T cells and NK cells. The positive effects of IL-15 on γδ T cell cytotoxicity, and in particular effector Vδ1 T cells, is well established26,55. Our data show that Vδ1 T cells in ART-suppressed PWH retain anti-HIV responses which warrants further attention in clinical studies aimed at eradicating the latent HIV-1 reservoir.
A major finding from our study is the high prevalence of CD4 + Vδ1 T cells across the GI tract that contained HIV-1 DNA. Immune cell frequencies naturally vary across different sections of the GI tract; however, HIV specifically depletes intestinal αβ CD4 + T cells during untreated infection and it is unclear if this population is reconstituted during long-term ART56,57. While total CD4+ cell counts showed variation between donor groups and sections of the GI tract, both total Vδ1 and CD4 + Vδ1 T cell frequencies were consistent relative to total nuclei. CD4 + Vδ1 T cells constituted up to 30% of all infected CD4+ cells within the colon and the small intestine despite their low phenotypic frequencies which averaged <10% of total CD4+ cells and <1% of total cells within each tissue. Interestingly, HIV-1 DNA was also quantified in only one out of three samples from the GI tract of EC, which is compatible with a reported strong negative correlation between the percentage of pro-inflammatory Vδ1 T cells and viral load within the GI tract of EC18. Tissue-resident Vδ1 T cells primarily localize within the epithelium where they are mediators of tissue surveillance8 or in smaller fractions within the lamina propria where they play a role in crosstalk with commensal microbiota58. Both the presence of CD4 expression and HIV-1 DNA were limited to LP Vδ1 T cells whereas IEL Vδ1 T cells displayed similar expression of cytotoxic markers to their circulating counterparts. Our data suggests an opposing role for Vδ1 T cells based on tissue localization during persistent infection.
Lastly, we found that circulating Vδ1 T cells harbor both HIV-1 DNA and replication-competent provirus similar to the Vδ2 subset previously characterized by our group7. We quantified HIV-1 DNA within 91% of participants and infectious virus was recovered from 45% of participants indicating a high prevalence of in vivo infection despite low frequencies of circulating CD4 + Vδ1 T cells. Modulation of CD4 on Vδ1 T cells has previously been described in the context of other viral infections, including in vitro infection with human herpesvirus 659. Our group has also demonstrated that Vδ2 T cells transiently upregulate CD4 following activation via ex vivo antigen stimulation. Moreover, despite the extremely low/absent expression of CD4 in ART-suppressed individuals, Vδ2 T cells from PWH displayed increased CD4 expression of up to 20% during the acute phase of infection, which results in systemic immune activation7. Given the robust activation and expansion of Vδ1 T cells during early infection, we hypothesized that they may transiently upregulate CD4 similar to Vδ2 T cells thereby increasing their permissibility to HIV-1 entry. We show that the Cd4 locus is repressed ex vivo and that stimulation through the TCR-CD3 complex resulted in de novo expression of CD4 on a discrete Vδ1 T cell subset. This subset was distinct from CD8 + Vδ1 T cells, highly proliferative, and associated with high CD25 expression which is an indicator of TCR signaling strength60. Furthermore, our detection of CpG methylation within the Cd4 promoter region of only a portion of Vδ1 T cells suggest divergent epigenetic programs exist for cells that upregulate the receptor versus those that do not. These findings define two specific Vδ1 T cell subsets, one that becomes permissible to HIV-1 infection and directly contributes to the latent reservoir and one cytotoxic subset potentially resistant to HIV-1 infection.
Our study has several limitations. Our cohorts were primarily male with a narrow range of ages, which may influence γδ T cell frequencies and phenotype as previously reported by us and others9,61. Second, our inability to detect intact provirus with the IPDA due to a limited number of cells precludes a more accurate quantification of latently infected Vδ1 T cells. However, the proportion of intact and defective provirus within αβ CD4 + T cells was comparable to previous studies and we did not observe amplification failure, which can occur due to interpersonal sequence variation within the viral reservoir62,63. Furthermore, while we successfully recovered replication-competent virus from 5/11 donors, the anti-HIV capabilities of Vδ1 T cells may present an additional confounding factor that limits viral outgrowth in a traditional QVOA. Given the rarity of αβ CD4 + T cells with intact provirus (~100 copies per 106 cells)64 and even less frequent inducible provirus (~1 per 106 cells)65, these findings underscore important considerations for assay selection when attempting to quantify rare cellular reservoirs.
In summary, our study has revealed a previously unknown role for circulating and mucosal Vδ1 T cells in HIV-1 persistence. Unlike other chronic viral infections26,27,66, our work highlights the unique immunological circumstance of persistent HIV-1 infection, characterized by Vδ1 T cells that have opposing roles in controlling and contributing to latent infection. Determining the specificity of these cytotoxic Vδ1 T cells and characterizing the complete contribution of CD4 + Vδ1 T cells as a latent reservoir will improve our understanding of the role of γδ T cells in the greater context of anti-viral immunity and could prove fruitful for HIV-1 cure strategies.
Methods
Study subjects
Participants were enrolled and written informed consent was obtained through approved protocols from the Office of Human Research for the George Washington University (GW), the Office of Human Research Ethics for the University of North Carolina (UNC) Institutional Review Boards (IRBs), and the Ethical Committee for Virgen del Rocio University Hospital, Seville, Spain. Peripheral blood mononuclear cells (PBMCs) were isolated from 53 virally suppressed PWH enrolled at either GW or UNC (Table 1). Additionally, 3–5 mm3 biopsy samples were harvested from the rectosigmoid colon within six participants for paired analyses of tissue-resident Vδ1 T cells from PWH enrolled at GW. Embedded biopsies from both the small and large intestine of an independent cohort including ART-suppressed PWH (3 ileum, 1 caecum) and EC (2 ileum, 1 caecum) enrolled at the Institute of Biomedicine of Seville were included in experiments quantifying HIV DNA within the GI mucosa. Participants enrolled at GW and UNC were compensated for their time and commitment to the project.
Buffy coats were purchased from the Gulf Coast Regional Blood Center without personal identifiers. PBMCs isolated from 27 people living without HIV-1 (HIV-) were used as controls in appropriate experiments.
Gastrointestinal biopsy tissue preparation
Sixteen 3–5 mm3 biopsies were obtained via sigmoidoscopy and colonoscopy procedure with single-use, EndoJaw Jumbo forceps with a 3.7 mm channel (Olympus medical) from 10 ART-suppressed PWH and 3 EC. Biopsy specimens were placed in RPMI (GI-RPMI) supplemented with 20% pooled Human AB serum (Innovative Research, Inc.), 2% Penicillin/Streptomycin (Sigma-Aldrich), 250 ng/mL Gentamicin (Gibco), and 5 ng/mL Amphotericin B (Gibco) then placed on ice until further processing. Four whole biopsies from each participant were fixed in 10% neutral buffered formalin at room temperature for 24 h then washed in increasing concentrations of ethanol and cleared with xylene prior to paraffin embedding. The remaining 12 biopsies were rinsed in RPMI, and then the tissue was dissociated using a three-step procedure to isolate cells for phenotyping: (1) Disruption of the epithelial layer: biopsies were placed in 20 mL of media (ER Solution) containing Hanks Buffered Salt Solution (Gibco), 10% fetal bovine serum (FBS), 10 mM EDTA (Invitrogen), and 5 mM DTT (Sigma-Aldrich) then incubated at 37 °C for 20 min within an orbital shaker set to 130 rpm. The media was then decanted through a 70 μm cell strainer and biopsies were transferred to a fresh tube containing 20 mL of ER solution for a second round of incubation. The biopsies were then rinsed in 20 mL of RPMI containing 10 mM HEPES (FisherScientific), and 5% FBS and incubated at 37 °C for 20 min within an orbital shaker set to 130 rpm. Media from each incubation was pooled and centrifuged at 600 × g for 10 min to collect intraepithelial lymphocytes. (2) Digestion of the lamina propria: Following epithelial disruption, biopsies were placed in 1 mL of RPMI with 0.5 mg/mL of collagenase D (Sigma-Aldrich), 0.1 mg/mL DNase I (StemCell Technologies), and 10 mM HEPES (FisherScientific) then incubated at 37 °C for 30 min in a water bath. Samples were vortexed every 3–4 min throughout the incubation. (3) Mechanical disruption: Following enzymatic digestion, biopsies were mechanically disrupted through a 70 μm cell strainer using a 16-gauge blunt-ended syringe (StemCell Technologies. The strainer was rinsed with 50 mL of GI-RPMI and centrifuged at 600 × g for 10 min. The supernatant was aspirated, and cells isolated from both epithelium disruption (IEL fraction) and enzymatic digestion (LP fraction) were combined and incubated in 1 mL RPMI with 0.1 mg/mL DNase I and 10 mM HEPES at room temperature for 30 min. Following the final wash, cells were resuspended in GI-RPMI, counted three times with a LUNA-FL cell counter (Logos Biosystems), and incubated at 37 °C overnight prior to phenotyping.
Flow cytometry immunophenotyping and cell sorting
PBMCs were isolated by Ficoll-gradient, and viably stored in 20% DMSO and FBS in Liquid Nitrogen until further use. Cells were thawed and counted three times with a LUNA-FL cell counter (Logo Biosystems). Phenotyping of PBMCs and dissociated tissue was conducted using a core panel of titrated fluorochrome-conjugated monoclonal antibodies (all from Biolegend unless otherwise stated) targeting lineage defining markers: CD3 (clone SK7), Vδ1 (clone REA173, Miltenyi), Vδ2 (clone B6), CD4 (clone SK3), and CD8 (clone SK1) followed by additional markers split into three separate panels describing immunological memory: CD45RA (clone HI100), CD27 (clone M-T271), CCR7 (clone G043H7), cytotoxicity: CD56 (clone 5.1H11), CD16 (clone 3G8), NKp30 (clone P30-15), NKp44 (clone P44-8), NKG2D (clone 1D11), and tissue homing: CD103 (clone Ber-ACT8), α4β7 (clone Hu117, R&D Systems), CCR5 (clone J418F1), CD161 (clone DX12, BD Biosciences), and CXCR5 (clone J252D4). Cells were harvested, washed, and stained with a viability dye (Zombie Aqua fixable viability kit, BioLegend) followed by surface staining with each respective panel for 20 min in the dark at 4 °C. Cells were then washed and fixed in 2% PFA. Samples were acquired in an LSRFortessa X-20 (BD Biosciences) and analyzed using FlowJo software v10.10.0 (BD Biosciences). Either Vδ1 T cells or other effector cells (Vδ2 T cells, αβ CD8 + T cells, NK cells) were directly sorted from PBMCs (gating strategies shown in Supplementary Figs. 6 and 9) using a FACSAria II (BD Biosciences) or SH800 (Sony Biotechnology). PBMCs were depleted of αβ T cells (StemCell Technologies) prior to sorting Vδ1 T cells to enrich the pre-sort sample.
Mass cytometry (CyTOF) and clustering and dimensionality reduction analysis
Additional phenotyping was conducted on thirteen ART PWH using mass cytometry29. Three million PBMCs were stained with a monoclonal antibody targeting CD45 conjugated to non-radioactive yttrium (89Y) and spiked with a single control donor labeled with Indium 115 (115In). Cells were then stained with monoisotopic cisplatin 198 to discriminate viability per manufacturer suggestions (Fluidigm) followed by incubation with heavy metal conjugated antibodies (Fluidigm) targeting markers for CD14 (clone M5E2, Lutetium-175), CD33 (clone WM53, Gadolinium-158), CD11c (clone 3.9, Neodymium-150), CD123 (clone 6H6, Neodymium-143), CD19 (clone HIB19, Neodymium-142), CD3 (clone UCHT1, Praseodynmium-141), Vδ1 (clone REA173, Europium-151), Vδ2 (clone 123R3, Ytterbium-171), CD8 (clone RPA-T8, Samarium-152), CD56 (clone NCAM16.2, Samarium-149), CD16 (clone 3G8, Bismuth-209), TNF-α (clone Mab11, Neodymium-144), IFN-γ (clone B27, Ytterbium-172), GrzmB (clone GB11, Erbium-167), CD27 (clone O323, Dysprosium-161), CD28 (clone CD28.2, Neodymium-148), CD45RA (clone HI100, Neodymium-146), IL-7Rα (clone A019D5, Erbium-168), CCR7 (clone G043H7, Erbium-170), CCR5 (clone NP-6G4, Gadolinium-156), CD69 (clone FN50, Dysprosium-164), CD25 (clone 2A3, Thulium-169), HLA-DR (clone L243, Ytterbium-174), CD160 (clone 688327, Samarium-154), TIGIT (clone MBSA44, Europium-153), PD-1 (clone EH12.2H7, Gadolinium-155), and CD57 (clone HCD57, Ytterbium-176) in cell staining buffer (CSB) for 30 min at room temperature. Cells were then washed twice in CSB, fixed in 2% paraformaldehyde for 60 min, then washed once with Perm/Wash Buffer (Biolegend). Cells were incubated for 30 min at 37 °C with the intracellular antibody cocktail in Perm/Wash Buffer. Stained cell suspensions were barcoded (20-Plex Pd Barcoding Kit, Fluidigm), pooled, resuspended in 62.5 nM Cell-ID Intercalator-Ir (Fluidigm) in Maxpar Fix and Perm Buffer (Fluidigm) and incubated overnight at 4 °C. Immediately prior to data acquisition, samples were centrifuged, washed in CSB, followed by a wash in Cell Acquisition Solution (CAS, Fluidigm), filtered (40 μm Cell Strainer, FlowMi), and cell concentration adjusted in CAS to 0.5 ×106 cells/mL with addition of 10% (v/v) EQ Four Element Calibration Beads (Fluidigm). Samples were acquired on a Helios mass cytometer (Fluidigm) equipped with a WB injector.
CyTOF data was initially prepared in FlowJo v10.10.0 (BD Biosciences) by gating on events based on singlets, viability, DNA content, and 87Y staining to differentiate samples from spiked in control. Myeloid cell populations (Monocytes, Macrophages, Dendritic cells) and B cells were excluded by gating on events negative for CD14, CD33, CD11c, CD123, and CD19. Effector cell populations were determined based on gating strategies defined in Supplementary Fig. 1a. The populations of interest from each donor were concatenated and exported as raw value CSV files. Computational analysis of data was performed using the Spectre R package67. Values were arcsinh transformed using a co-factor of 10 to redistribute the data on a linear scale and compress low end values near zero. The data was then merged and unbiased hierarchical clustering performed using the FlowSOM algorithm (metacluster k = 5)68. Cell types were annotated based on analysis of marker expression within each metacluster. Subsequently, the data was downsampled to 50,000 events and visualized by the Uniform Manifold Approximation and Projection (UMAP).
ATAC-seq and data analysis
Isolated cryopreserved Vδ1 T cells were sent to Active Motif (Carlsbad, CA) to perform the Fixed Cell ATAC-seq assay. The cells were processed according to the Fixed Cell ATAC-Seq Kit instruction (Active Motif cat# 53151) except for 30 min of cell fixation incubation time with cell fixation buffer. Resulting material was quantified using iSeq (Illumina) and sequenced with PE42 on the NovaSeq6000 (Illumina). Reads were aligned using the BWA algorithm (mem mode; default settings). Duplicate reads were removed, only reads mapping as matched pairs and only uniquely mapped reads (mapping quality ≥1) were used for further analysis. Alignments were extended in silico at their 3’-ends to a length of 200 bp and assigned to 32-nt bins along the genome. The resulting histograms (genomic “signal maps”) were stored in bigWig files. Peaks were identified using the MACS v2.1.0 algorithm at a cutoff of p value 1e-7, without control file, and with the –nomodel option. Peaks that were on the ENCODE blacklist of known false ChIP-Seq peaks were removed. Signal maps and peak locations were used as input data to Active Motifs proprietary analysis program. HOMER motif analysis was performed using BED files of differentially accessible peaks as input for the findMotifsGenome.pl command with a 200 bp search area centered around the midpoint of the differential region (+100 bp, −100 bp). The resulting identified motifs were enriched across all sequences whereas individual peak regions are not annotated with specific motifs.
Generation of HIV-1 viral stock
HEK293T cells (ATCC) were seeded and cultured to 50–80% confluency prior to transfection with Fugene6 (Promega) and HIVJR-CSF plasmid DNA (NIH AIDS Reagent Program) at a ratio of 3:1. Twenty-four hours later, supernatant was removed and replenished with fresh RPMI + 10% fetal bovine serum. After an additional 24 hours, supernatant was harvested and filtered through a 0.45 μm polyethylsulfone membrane to remove cellular debris and frozen at −80 °C. Viral titers were quantified by HIVp24 ELISA (Perkin Elmer).
In vitro HIV-1 infection of PBMCs and isolated CD4 + T Cells
PBMCs from HIV-seronegative donors were activated with 3 μg/mL PHA and 100 U/mL IL-2 for 48 hours. Cells were then washed twice and infected with the viral strain HIVJR-CSF by spinoculation at 2000 × g for 2 hours. Cells were washed twice to remove unbound virus, resuspended in cRPMI supplemented with 100 U/mL IL-2 (Peprotech), and cultured in 96-well plates for 7 days. Infection was confirmed by flow cytometry, cells were harvested, washed, and stained with a viability dye (Zombie Aqua fixable viability kit, BioLegend) followed by surface staining with monoclonal antibodies (Biolegend) against CD3 (clone SK7), Vδ1 (clone REA117, Miltenyi), and CD4 (clone SK3). Cells were then fixed and permeabilized with Cytofix/CytoPerm (BD Biosciences) for 20 min at 4 °C in the dark. Cells were then washed in 1× Perm/Wash Buffer (BD Biosciences) and stained for intracellular HIVp24 (clone KC-57, Beckman Coulter) for 20 min at 4 °C in the dark. Cells were then washed twice and analyzed on a Fortessa LSR X-20 (BD Biosciences) and analyzed using the FlowJo software v10.10.0 (BD Biosciences). For viral inhibition assays, isolated resting αβ CD4 + T cells from ART-suppressed PWH were directly spinnoculated with HIVJR-CSF at 2000 × g for 4 hours in the presence of 8 μg/mL polybrene prior to co-culture with effector cells.
Viral inhibition assay
We performed an adaptation of the conventional viral inhibition assay34,35. Fifty thousand FACS-sorted Vδ1 T cells from ART-suppressed PWH were used as effectors and co-cultured with HIV-JR-CSF superinfected CD4 + T cells as targets at a 1:1 or 1:10 (Effector: Target) ratio. Superinfected CD4+ cells cultured alone were used as a control, and each experimental condition was cultured in triplicate. Culture supernatants were harvested on day 7 following co-culture and stored at −80 °C until HIVp24 ELISA quantification (ABL, Inc.) was performed. Results are expressed as the percentage of viral inhibition normalized to superinfected CD4 + T cells cultured alone. Additional head-to-head assays were performed to compare viral inhibition from matching Vδ1, Vδ2, αβ CD8 T cells, and NK cells. To test the effect of IL-15 treatment on boosting viral inhibition for each effector cell type, only effectors were pre-treated with 25 ng/mL IL-15 (Peprotech) for 24 hours, then washed prior to co-culture with superinfected CD4 + T cells. Viral inhibition of IL-15 treated effectors was compared to that of untreated effectors for each cell type.
Degranulation and IFN-γ assay
FACS-isolated Vδ1 T cells were co-cultured with either HIVJR-CSF–infected autologous CD4+ cells as targets. CD4 + T cells were infected following the approach described above. At least 100,000 effector cells were co-cultured at a 1:1 ratio with CD4+ target cells in 96-well plates for 5 hours in the presence of GolgiStop (BD Biosciences) and a CD107a monoclonal antibody (clone H4A3, BD Biosciences). Cells were then harvested, washed with staining buffer, stained with CD3 (clone SK7, Biolegend) and Vδ1 (clone REA117, Miltenyi) for 20 min on ice in the dark, washed twice, resuspended in staining buffer. In parallel cultures, cells were fixed and permeabilized with Cytofix/CytoPerm (BD Biosciences) for 20 min at 4 °C in the dark. Cells were then washed in 1× Perm/Wash Buffer (BD Biosciences) and stained for intracellular IFN-γ (clone B27, Biolegend) for 20 min at 4 °C in the dark. All samples were acquired in the Attune Acoustic Focusing Cytometer (Applied Biosystems) and analyzed using the FlowJo software v10.10.0 (BD Biosciences).
Quantitative viral outgrowth assay (QVOA)
QVOA was performed using isolated cell subpopulations from leukapheresis products from ART-suppressed PWH (Table 1)7,41. Sorted Vδ1 T cells were cultured in limiting dilution from 100,000 to 5000 cells in replicates of 2 to 9, depending on cell availability and activated with 2.5 µg/mL PHA and 100 U/mL IL-2 for 24 hours. After washing, isolated and activated CD4 + T cells from uninfected individuals were added to the culture for viral outgrowth and cells were kept in culture for 23 days harvesting the supernatant for HIVp24 detection by ELISA (ABL Inc.) at days 15, 19 and 23. Results are shown as Infectious Units per million cells (IUPMs) that represent a maximum-likelihood estimate of the inducible reservoir with 95% confidence intervals69.
Intact proviral DNA assay (IPDA)
Genomic DNA was isolated from either FACS-purified Vδ1 T cells or negative magnetic selection (StemCell Technologies) of αβ CD4 + T cells using the QIAmp DNA mini kit (Qiagen). Additional precautions were taken to minimize DNA shearing during isolation including the use of wide-bore pipette tips (Thermo Fisher Scientific) and minimizing the use of vortex mixing. DNA concentrations and purity were measured using a Nanodrop spectrophotometer (Thermo Fisher Scientific). Intact and defective HIV-1 copies/million cells were determined by droplet digital PCR using the IPDA64. Genomic DNA was combined with ddPCR Supermix for Probes (no dUTPs, BioRad), primers (final concentration 900 nM, Integrated DNA Technologies), probe(s) (final concentration 250 nM, Thermo Fisher Scientific) and nuclease free water. Primer sequence targeting HIV DNA were as follows:
HIV-1 Ψ Forward Primer CAGGACTCGGCTTGCTGAAG
HIV-1 Ψ Reverse Primer- GCACCCATCTCTCTCCTTCTAGC
HIV-1 Env Forward Primer- AGTGGTGCAGAGAGAAAAAAGAGC
HIV-1 env Reverse Primer- GTCTGGCCTGTACCGTCAGC
Probe sequences targeting each amplicon were as follows:
HIV-1 Ψ Probe- FAM- TTTTGGCGTACTCACCAGT- MGBNFQ
HIV-1 env Probe- VIC-CCTTGGGTTCTTGGGA- MGBNFQ
HIV-1 Hypermutant Probe- CCTTAGGTTCTTAGGAGC- MGBNFQ.
HIV-1 copies were adjusted for shearing and normalized to cell counts based on separate, parallel reactions targeting the RPP30 gene63. Primer sequences targeting RPP30 were as follows:
RPP30 Forward Primer- GATTTGGACCTGCGAGCG
RPP30 Reverse Primer- GCGGCTGTCTCCACAAGT
RPP30 Shear Forward Primer- CCATTTGCTGCTCCTTGGG
RPP30-Shear Reverse Primer- CATGCAAAGGAGGAAGCCG.
Probes targeting each RPP30 amplicon were as follows:
RPP30 Probe- VIC-CTGACCTGAAGGCTCT- MGBNFQ
RPP30 Shear Probe- FAM- AAGGAGCAAGGTTCTATTGTAG- MGBNFQ
Droplets were formed using an Automated Droplet Generator and and cycled at 95 °C for 10 min, then 45 cycles of (94 °C for 30 seconds and 59 °C for 1 min), then 98 °C for 10 min. Following amplification, droplets were analyzed on a QX200 Droplet Reader (BioRad) using QuantaSoft software v1.7.4. Each reaction was run in quadruplicate and thresholds for positive signal were determined using both no template and HIV-seronegative controls. Genomic DNA isolated from JLat J89 cells which contain a single integrated copy of HIV-1 was used as a positive control in each assay.
Next-generation DNAscope in situ hybridization combined to IFA and quantitative image analysis
We utilized next-generation, ultrasensitive in situ hybridization technology for the detection of both HIV DNA (DNAscope) with quantitative image analysis40. We scanned and analyzed full tissue sections (2 sections per sample) to maximize the size of tissue to be assessed. Viral DNA detection (DNAscope) was performed by utilizing the sense probe targeting the reverse strand (coding strand- catalog# 425531, Advanced Cell Diagnostics, Inc. (ACD)). ACD brown kit was used to amplify and detect viral DNA. Pretreatments 1 (endogenous peroxidase block; ACD), 2 (1× RNAscope Target Retrieval Reagents, ACD), and 3 (protease digestion solution; 2.5 ug/mL, ACD) were performed as recommended by the manufacturer ACD. Slides are washed 3 times in TBS Tween-20 (0.05% v/v) and then dispensed pre-warmed probe diluent to prevent carrying water into hybridization step. Then pre-warmed probe was dispensed and hybridized overnight (between 10 to 18 hours) at 40 °C. Amplification and detection were performed according to the manufacturer’s instruction, with the only major modification in the protocol was using 0.5× wash buffer for the first wash step right after overnight probe incubation instead of 1× wash buffer to wash out any nonspecific probe binding. Amplification steps were performed following ACD instructions. After amplification 6 the detection of the DNA was done using TSA reagent (Thermo Fisher Scientific) at 1:2000 for 5 min at room temperature. To phenotype the cells harboring vDNA we combined DNAscope with an immunofluorescence approach using antibodies directed to CD4 (polyclonal Goat IgG, 1:500, Abcam), CD8a (polyclonal Rabbit IgG, 1:100, Thremo Fisher Scientific) and/or γδTCR (clone H-41 Mouse IgG, 1:500, Santa Cruz). Slides were washed and then incubated with secondary donkey anti-goat IgG-Alexa Fluor 488, donkey anti-mouse IgG-Alexa Fluor 594 and anti-rabbit IgG-Alexa Fluor 647 (all from Molecular Probes/Thermo Fisher Scientific) for 1 hour at room temperature and washed three times for 5 min in TBS + Tween (0.05% v/v). All slides were counterstained with DAPI (ready-to-use (RTU), Advanced Cell Diagnostics) for 10 min, washed and cover slipped with #1.5 GOLD SEAL cover glass (Electron Microscopy Sciences) using Prolong Gold reagent (Invitrogen). Slides were then scanned using the Akoya Fusion microscope at ×40, acquisition for all slides were made using identical parameters with exposure for DAPI: 1 ms, 488: 75 ms, 594: 100 ms and 647: 110 ms. All image analysis was performed using QuPath software, v0.4.370. All biopsies were analyzed for up to whole three tissue sections except for folded or damaged tissue areas. Parameters used for positive cell detection were as follows: background radius 8 µm, median filter radius 0 µm, sigma 1.5 µm, minimum area 10 µm2, maximum area 400 µm2 and cell expansion 3 µm, and each threshold was adjusted from 15 to 1.5 based on intensity of each marker. Representative pictures were selected to show lamina propria showing cells of interest and were extracted using ImageJ. No manipulation was performed on representative data aside of slight adjustment of intensity for easier visualization.
Vδ1 T cell activation and cell culture
Cells were either stimulated with 3 μg/mL of plate-bound αCD3 (clone OKT3), 5 μg/mL αCD28 (clone 28.2), and 100 U/mL of IL-2 or left unstimulated for a total of 7 days with half of the media replenished on day 3. Depending on the experiment, cells were harvested at select time points, washed, and stained with a viability dye (Zombie Aqua fixable viability kit, BioLegend) followed by surface staining with monoclonal antibodies (Biolegend) against CD3 (clone SK7), Vδ1 (clone REA117, Miltenyi), CD4 (clone SK3), and CD8 (clone SK1). In some experiments, cells were additionally stained for CD69 (clone FN50), CD25 (clone BC96), HLA-DR (clone L243), CD27 (clone M-T271) CD28 (clone CD28.2), CD45RA (clone HI100), IL-7Rα (clone A019D5), CD62L (clone DREG-56), and CCR7 (clone G043H7). Samples were acquired in a Fortessa LSR X-20 (BD Biosciences) and analyzed using the FlowJo software v10.10.0 (BD Biosciences).
Proliferation assay
Ten million PBMCs from PWH and HIV-seronegative controls were incubated with 2 μM carboxyfluorescein succinimidyl ester (CFSE) in 1× PBS for 20 min at 37 °C in the dark with continuous shaking. Five times the volume of cRPMI was then added to quench the staining. Cells were then centrifuged at 600 × g for 10 min, aspirated, and resuspended at a concentration of 106 cells/mL in cRPMI. Cells were then activated by αCD3/αCD28 + IL-2 stimulation or left unstimulated for a total of 7 days with half of the media replenished on day 3. On day 7, cells were harvested, washed, and stained with a viability dye (Zombie Aqua fixable viability kit, BioLegend) followed by surface staining with monoclonal antibodies against CD3 (clone SK7), Vδ1 (clone REA117), CD4 (clone SK3), and CD8 (clone SK1). Samples were acquired in a Fortessa LSR X-20 (BD Biosciences) and analyzed using the FlowJo software v10.10.0 (BD Biosciences).
COBRA assay
Methylation at three potential CpG dinucleotides within the Cd4 promoter of Vδ1 T cells was determined using Combined Bisulfite Restriction Analysis (COBRA)71. Fifty thousand Vδ1 T cells from HIV-seronegative donors were FACS-isolated from stimulated PBMCs (αCD3, αCD28, and IL-2) or unstimulated controls following 5 days in culture. Genomic DNA was directly bisulfite converted using the EZ DNA Methylation-Direct Kit (Zymo Research). Converted DNA was then mixed with 1× PCR Buffer, 1.5 mM MgCl2, 0.2 mM dNTP mix, 0.2 μM Primers, and Platinum Taq (Thermo Fisher Scientific). Primer sequences:
CD4 Bis Forward Primer- GGGGGTGTTAAAGATTATATTTAATTTA
CD4 Bis Reverse Primer- AAACTAAACAAAAAATATCTAAAACACC
Reactions were cycled at 94 °C for 2 min, then 35 cycles of (94 °C for 30 seconds, 54.5 °C for 30 seconds, 72 °C for 1 min). Amplicons were then digested with either methylation-sensitive restriction enzyme AccI, HinfI, or Esp3I (New England Biolabs) corresponding to a specific potential CpG site. Digested products were heat-inactivated and directly visualized on a 4% agarose gel with SybrSafe (Thermo Fisher Scientific). Universally non-methylated and methylated human genomic DNA (Zymo Research) were used as negative and positive controls respectively.
Inhibition of DNA methylation
Total γδ T cells were isolated from one-hundred million PBMCs from HIV-seronegative donors by negative magnetic selection (StemCell Technologies). Cells were then activated by αCD3/αCD28 + IL-2 stimulation in the presence of 500 nM/mL 5-Azacytidine (5-Aza) or 1× PBS as mock treatment for 5 days. 5-Aza or mock treatment was replenished each day and IL-2 was replenished on day 3 of culture. On day 5, cells were harvested, washed, and stained with a viability dye (Zombie Aqua fixable viability kit, BioLegend) followed by surface staining with monoclonal antibodies against CD3 (clone SK7), Vδ1 (clone REA117), CD4 (clone SK3), and CD8 (clone SK1). Samples were acquired in a Fortessa LSR X-20 (BD Biosciences) and analyzed using the FlowJo software v10.10.0 (BD Biosciences).
Statistical analyses
Statistical analyses were performed in GraphPad Prism or with the R programming language. Two-tailed Mann-Whitney U test was used to test for differences between groups whereas two-tailed Wilcoxon Matched Pairs Signed-Rank test was used for paired analyses. Significant correlations between Vδ1 T cell phenotypes, HIV-1 DNA, and participant characteristics were calculated with two-tailed Spearman’s ranked correlation test. Two-way ANOVA was used to determine differences between multiple groups over time. Technical replicates were averaged, and outliers were excluded from analysis when greater than ± 2 S.D from the mean.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The raw and processed ATAC-seq data in this study have been deposited in the Gene Expression Omnibus (GEO) public database under accession code GSE273603. The source data that supports the findings of this study are available as a Source Data file. Source data are provided with this paper.
References
Finzi, D. et al. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science 278, 1295–1300 (1997).
Palmer, S. et al. Low-level viremia persists for at least 7 years in patients on suppressive antiretroviral therapy. Proc. Natl. Acad. Sci. USA 105, 3879–3884 (2008).
Davey, R. T. Jr. et al. HIV-1 and T cell dynamics after interruption of highly active antiretroviral therapy (HAART) in patients with a history of sustained viral suppression. Proc. Natl Acad. Sci. USA 96, 15109–15114 (1999).
Veenhuis, R. T. et al. Monocyte-derived macrophages contain persistent latent HIV reservoirs. Nat. Microbiol. 8, 833–844 (2023).
Ganor, Y. et al. HIV-1 reservoirs in urethral macrophages of patients under suppressive antiretroviral therapy. Nat. Microbiol. 4, 633–644 (2019).
Yukl, S. A. et al. The distribution of HIV DNA and RNA in cell subsets differs in gut and blood of HIV-positive patients on ART: implications for viral persistence. J. Infect. Dis. 208, 1212–1220 (2013).
Soriano-Sarabia, N. et al. Peripheral Vgamma9Vdelta2 T cells are a novel reservoir of latent HIV infection. PLoS Pathog. 11, e1005201 (2015).
Ribot, J. C., Lopes, N. & Silva-Santos, B. gammadelta T cells in tissue physiology and surveillance. Nat. Rev. Immunol. 21, 221–232 (2021).
Sanz, M. et al. Deep characterization of human gammadelta T cell subsets defines shared and lineage-specific traits. Front Immunol. 14, 1148988 (2023).
Di Marco Barros, R. et al. Epithelia use butyrophilin-like molecules to shape organ-specific gammadelta T cell compartments. Cell 167, 203–218.e217 (2016).
Mikulak, J. et al. NKp46-expressing human gut-resident intraepithelial Vdelta1 T cell subpopulation exhibits high antitumor activity against colorectal cancer. JCI Insight 4, https://doi.org/10.1172/jci.insight.125884 (2019).
Davey, M. S., Willcox, C. R., Baker, A. T., Hunter, S. & Willcox, B. E. Recasting human Vdelta1 lymphocytes in an adaptive role. Trends Immunol. 39, 446–459 (2018).
Davey, M. S. et al. The human Vdelta2(+) T-cell compartment comprises distinct innate-like Vgamma9(+) and adaptive Vgamma9(-) subsets. Nat. Commun. 9, 1760 (2018).
Autran, B. et al. T cell receptor gamma/delta+ lymphocyte subsets during HIV infection. Clin. Exp. Immunol. 75, 206–210 (1989).
Enders, P. J. et al. HIV-mediated gammadelta T cell depletion is specific for Vgamma2+ cells expressing the Jgamma1.2 segment. AIDS Res Hum. Retroviruses 19, 21–29 (2003).
De Maria, A. et al. Selective increase of a subset of T cell receptor gamma delta T lymphocytes in the peripheral blood of patients with human immunodeficiency virus type 1 infection. J. Infect. Dis. 165, 917–919 (1992).
Poles, M. A. et al. Human immunodeficiency virus type 1 induces persistent changes in mucosal and blood gammadelta T cells despite suppressive therapy. J. Virol. 77, 10456–10467 (2003).
Olson, G. S. et al. Increased frequency of systemic pro-inflammatory Vdelta1(+) gammadelta T cells in HIV elite controllers correlates with gut viral load. Sci. Rep. 8, 16471 (2018).
Climent, N. et al. Immunological and virological findings in a patient with exceptional post-treatment control: a case report. Lancet HIV 10, e42–e51 (2023).
Fausther-Bovendo, H. et al. NKG2C is a major triggering receptor involved in the V[delta]1 T cell-mediated cytotoxicity against HIV-infected CD4 T cells. Aids 22, 217–226 (2008).
Hudspeth, K. et al. Engagement of NKp30 on Vdelta1 T cells induces the production of CCL3, CCL4, and CCL5 and suppresses HIV-1 replication. Blood 119, 4013–4016 (2012).
Wallace, M. et al. Functional gamma delta T-lymphocyte defect associated with human immunodeficiency virus infections. Mol. Med. 3, 60–71 (1997).
Imlach, S., Leen, C., Bell, J. E. & Simmonds, P. Phenotypic analysis of peripheral blood gammadelta T lymphocytes and their targeting by human immunodeficiency virus type 1 in vivo. Virology 305, 415–427 (2003).
James, K. S. et al. Measuring the contribution of gammadelta T cells to the persistent HIV reservoir. AIDS 34, 363–371 (2020).
Pihl, R. M. F. et al. Distinct subsets of Vdelta1 effector and Vdelta2 gammadelta T cells shift in frequency and are linked to plasma inflammatory markers during ART-suppressed HIV infection. J. Infect. Dis. https://doi.org/10.1093/infdis/jiae091 (2024).
Davey, M. S. et al. Clonal selection in the human Vdelta1 T cell repertoire indicates gammadelta TCR-dependent adaptive immune surveillance. Nat. Commun. 8, 14760 (2017).
McMurray, J. L. et al. Transcriptional profiling of human Vdelta1 T cells reveals a pathogen-driven adaptive differentiation program. Cell Rep. 39, 110858 (2022).
Field, K. R., Wragg, K. M., Kent, S. J., Lee, W. S. & Juno, J. A. gammadelta T cells mediate robust anti-HIV functions during antiretroviral therapy regardless of immune checkpoint expression. Clin. Transl. Immunol. 13, e1486 (2024).
Sanz, M. et al. Aminobisphosphonates reactivate the latent reservoir in people living with HIV-1. Front Immunol. 14, 1219250 (2023).
Xu, Y. et al. Reliable estimation of CD8 T cell inhibition of in vitro HIV-1 replication. Front. Immunol. 12, 666991 (2021).
Wherry, E. J. et al. HIV-specific CD8 T cells express low levels of IL-7Ralpha: implications for HIV-specific T cell memory. Virology 353, 366–373 (2006).
Grenningloh, R. et al. Ets-1 maintains IL-7 receptor expression in peripheral T cells. J. Immunol. 186, 969–976 (2011).
Nah, J. & Seong, R. H. Kruppel-like factor 4 regulates the cytolytic effector function of exhausted CD8 T cells. Sci. Adv. 8, eadc9346 (2022).
Garrido, C. et al. gammadelta T cells: an immunotherapeutic approach for HIV cure strategies. JCI Insight 3, https://doi.org/10.1172/jci.insight.120121 (2018).
Garrido, C. et al. Interleukin-15-stimulated natural killer cells clear HIV-1-infected cells following latency reversal ex vivo. J. Virol. 92, https://doi.org/10.1128/JVI.00235-18 (2018).
Jones, R. B. & Walker, B. D. HIV-specific CD8(+) T cells and HIV eradication. J. Clin. Invest. 126, 455–463 (2016).
Miller, J. S. et al. Safety and virologic impact of the IL-15 superagonist N-803 in people living with HIV: a phase 1 trial. Nat. Med. 28, 392–400 (2022).
Chowers, Y., Holtmeier, W., Harwood, J., Morzycka-Wroblewska, E. & Kagnoff, M. F. The V delta 1 T cell receptor repertoire in human small intestine and colon. J. Exp. Med. 180, 183–190 (1994).
Cimini, E. et al. Primary and chronic HIV infection differently modulates mucosal Vdelta1 and Vdelta2 T-cells differentiation profile and effector functions. PLoS One 10, e0129771 (2015).
Deleage, C. et al. Defining HIV and SIV reservoirs in lymphoid tissues. Pathog. Immun. 1, 68–106 (2016).
Soriano-Sarabia, N. et al. Quantitation of replication-competent HIV-1 in populations of resting CD4+ T cells. J. Virol. 88, 14070–14077 (2014).
Jimenez-Leon, M. R. et al. Vedolizumab and ART in recent HIV-1 infection unveil the role of alpha4beta7 in reservoir size. JCI Insight, https://doi.org/10.1172/jci.insight.182312 (2024).
Chen, B. K., Gandhi, R. T. & Baltimore, D. CD4 down-modulation during infection of human T cells with human immunodeficiency virus type 1 involves independent activities of vpu, env, and nef. J. Virol. 70, 6044–6053 (1996).
Sellars, M. et al. Regulation of DNA methylation dictates Cd4 expression during the development of helper and cytotoxic T cell lineages. Nat. Immunol. 16, 746–754 (2015).
Vantourout, P. & Hayday, A. Six-of-the-best: unique contributions of gammadelta T cells to immunology. Nat. Rev. Immunol. 13, 88–100 (2013).
Roy Chowdhury, R. et al. NK-like CD8(+) gammadelta T cells are expanded in persistent Mycobacterium tuberculosis infection. Sci. Immunol. 8, eade3525 (2023).
Harris, L. D. et al. Mechanisms underlying gammadelta T-cell subset perturbations in SIV-infected Asian rhesus macaques. Blood 116, 4148–4157 (2010).
Poggi, A. et al. Migration of V delta 1 and V delta 2 T cells in response to CXCR3 and CXCR4 ligands in healthy donors and HIV-1-infected patients: competition by HIV-1 Tat. Blood 103, 2205–2213 (2004).
Sumaria, N. et al. Perinatal thymic-derived CD8alphabeta-expressing gammadelta T cells are innate IFN-gamma producers that expand in IL-7R-STAT5B-driven neoplasms. Nat. Immunol. https://doi.org/10.1038/s41590-024-01855-4 (2024).
Colle, J. H. et al. CD127 expression and regulation are altered in the memory CD8 T cells of HIV-infected patients-reversal by highly active anti-retroviral therapy (HAART). Clin. Exp. Immunol. 143, 398–403 (2006).
Belkina, A. C. et al. Multivariate computational analysis of gamma delta T Cell inhibitory receptor signatures reveals the divergence of healthy and ART-suppressed HIV+ aging. Front. Immunol. 9, 2783 (2018).
Davies, D. et al. PD-1 defines a distinct, functional, tissue-adapted state in Vdelta1(+) T cells with implications for cancer immunotherapy. Nat. Cancer 5, 420–432 (2024).
Deseke, M. et al. A CMV-induced adaptive human Vdelta1+ gammadelta T cell clone recognizes HLA-DR. J. Exp. Med. 219, https://doi.org/10.1084/jem.20212525 (2022).
Mamedov, M. R. et al. CRISPR screens decode cancer cell pathways that trigger gammadelta T cell detection. Nature 621, 188–195 (2023).
García, V. E. et al. IL-15 enhances the response of human gamma delta T cells to nonpeptide [correction of nonpetide] microbial antigens. J. Immunol. 160, 4322–4329 (1998).
Estes, J. et al. Collagen deposition limits immune reconstitution in the gut. J. Infect. Dis. 198, 456–464 (2008).
Asowata, O. E. et al. Irreversible depletion of intestinal CD4+ T cells is associated with T cell activation during chronic HIV infection. JCI Insight 6, https://doi.org/10.1172/jci.insight.146162 (2021).
Papotto, P. H., Yilmaz, B. & Silva-Santos, B. Crosstalk between gammadelta T cells and the microbiota. Nat. Microbiol. 6, 1110–1117 (2021).
Lusso, P., Garzino-Demo, A., Crowley, R. W. & Malnati, M. S. Infection of gamma/delta T lymphocytes by human herpesvirus 6: transcriptional induction of CD4 and susceptibility to HIV infection. J. Exp. Med. 181, 1303–1310 (1995).
Shatrova, A. N. et al. Time-dependent regulation of IL-2R alpha-chain (CD25) expression by TCR signal strength and IL-2-induced STAT5 signaling in activated human blood T lymphocytes. PLoS One 11, e0167215 (2016).
Cairo, C. et al. Impact of age, gender, and race on circulating gammadelta T cells. Hum. Immunol. 71, 968–975 (2010).
Lee, G. Q. et al. HIV-1 subtype A1, D, and recombinant proviral genome landscapes during long-term suppressive therapy. Nat. Commun. 15, 5480 (2024).
Kinloch, N. N. et al. HIV-1 diversity considerations in the application of the intact proviral DNA assay (IPDA). Nat. Commun. 12, 165 (2021).
Bruner, K. M. et al. A quantitative approach for measuring the reservoir of latent HIV-1 proviruses. Nature 566, 120–125 (2019).
Ho, Y. C. et al. Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell 155, 540–551 (2013).
Fujishima, N. et al. Skewed T cell receptor repertoire of Vdelta1(+) gammadelta T lymphocytes after human allogeneic haematopoietic stem cell transplantation and the potential role for Epstein-Barr virus-infected B cells in clonal restriction. Clin. Exp. Immunol. 149, 70–79 (2007).
Ashhurst, T. M. et al. Integration, exploration, and analysis of high-dimensional single-cell cytometry data using Spectre. Cytom. A 101, 237–253 (2022).
Van Gassen, S. et al. FlowSOM: using self-organizing maps for visualization and interpretation of cytometry data. Cytom. A 87, 636–645 (2015).
Rosenbloom, D. I. et al. Designing and interpreting limiting dilution assays: general principles and applications to the latent reservoir for human immunodeficiency virus-1. Open Forum Infect. Dis. 2, ofv123 (2015).
Bankhead, P. et al. QuPath: open source software for digital pathology image analysis. Sci. Rep. 7, 16878 (2017).
Xiong, Z. & Laird, P. W. COBRA: a sensitive and quantitative DNA methylation assay. Nucleic Acids Res. 25, 2532–2534 (1997).
Acknowledgements
This research was funded by the National Institute of Allergy and Infectious Diseases (NIAID) of the National Institutes of Health (NIH) under award numbers R01-AI125097 and R21-AI157864 to NSS, and facilitated by the services provided by the District of Columbia Center for AIDS Research under an NIH funded program (P30AI117970), the Martin Delaney Collaboratory CARE (UM1 AI126619), the University of North Carolina Mass Cytometry Core and Cancer Research Center Core Support (P30CA016086), and from the NCI, NIH, under Contract No. 75N910D00024/HHSN261201500003I, and by the Instituto de Salud Carlos III, (Fondo Europeo de Desarrollo Regional, FEDER, “a way to make Europe”, research contracts CD23/00187 to JV, and research projects PI19/01127, and PI22/01796) to ERM. ERM has a contract from the Spanish National Research Council (CSIC). We thank Dr. Chiappinelli for her guidance in assessing DNA Methylation and providing 5-Azacytidine, Dr. Cresswell and the GWU flow cytometry core, and Dr. Chandler for acquiring tissue biopsies.
Author information
Authors and Affiliations
Contributions
N.S.S. designed and supervised the study and wrote and edited the manuscript. B.T.M. designed, conducted, and analyzed experiments and wrote and edited the manuscript. M.S. (Marta Sanz), J.V., E.R.M., M.S. (Marc Siegel), and C.D. reviewed the manuscript. M.S. (Marta Sanz), C.M.S., A.C., M.A.I., and M.L.C. conducted experiments. K.L., M.S. (Marc Siegel), J.V., and E.R.M. provided clinical samples for the study. C.D. designed, conducted, and analyzed HIV DNAscope experiments. All authors have approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Andrew McMichael, Bruno Silva-Santos and the other, anonymous, reviewer for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Mann, B.T., Sanz, M., Clohosey, M.L. et al. Dual role of circulating and mucosal Vδ1 T cells in the control of and contribution to persistent HIV-1 infection. Nat Commun 16, 5817 (2025). https://doi.org/10.1038/s41467-025-57260-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-57260-4