Abstract
Eukaryotic ribosome biogenesis is an energy-consuming process involving many ATPase-driven steps. In yeast, AAA+ protein Drg1 releases an assembly factor Rlp24, a placeholder for Rpl24, from pre-60S particles just exported to cytosol. The equivalent process in human cells involves SPATA5 (Drg1 homolog) and additional factors. However, the mechanistic details remain unclear. Here we reveal that SPATA5 forms a 4:2:2:2 complex with SPATA5L1, C1orf109, and CINP. This complex features an N-terminal ring made of C1orf109, CINP and NTDs of SPATA5/SPATA5L1, and two hexameric AAA+ ATPase rings. Intriguingly, a conserved cysteine C672 in the P-loop of SPATA5 is sulfinylated, generating an inactive conformation incompatible with ATP binding. We also obtained a cryo-EM structure of pre-60S-bound SPATA5 complex. Different from yeast, the recognition of the pre-60S particle is mediated by human-specific factor CINP, through two distinct sets of interactions: one with GTPBP4 and the other with ES27A. Taken together, these data provide structural basis for understanding the cytoplasmic maturation of the pre-60S, and reveal human-specific features that might be harnessed for therapeutic purposes.
Similar content being viewed by others
Introduction
Eukaryotic ribosome biogenesis is an intricate and energy-intensive process involving more than 200 assembly factors and around 80 snoRNAs1,2,3,4,5, including many energy-consuming enzymes, such as GTPases, RNA helicases, kinases, and ATPases6,7. Mechanical removal of certain placeholder factors is often employed as a mechanism to ensure the hierarchy of the assembly process, and to serve as quality control checkpoint. Such an example is the removal of an assembly factor Rlp24 from the pre-60S particle by AAA+ ATPase Drg1 in cytoplasm8,9,10,11,12. Rlp24 serves as a placeholder of ribosomal protein Rpl24, and this Rlp24-Rpl24 exchange is crucial for the final maturation of the pre-60S particles in the cytoplasm. Structural and biochemical studies showed that Drg1 hexamer docks onto the cytoplasmic pre-60S particles through interactions with Arx1 and ES27, captures the flexible C-terminus of Rlp24 with its conserved pore-loops, and dissociates Rlp24 from the pre-60S particle by threading the C-terminus of Rlp24, which is a prerequisite for the release of other associated assembly factors8,9,10,11,12. While this process is well studied in budding yeast, its equivalent process in human cells has remained to be elucidated. Although ribosome assembly is generally considered to be highly conserved from yeast to humans, recent studies indicated that the human process is more complex13, and an increasing number of species-specific features have been revealed14,15,16,17,18,19,20.
The human homolog of Drg1 is SPATA5 (spermatogenesis-associated protein 5), which was first suggested to be associated with spermatogenesis21. Mutations in SPATA5 have been linked to a neurodevelopmental disorder with hearing loss, seizures, and brain abnormalities, underscoring the critical importance of its physiological function22,23,24,25. In addition, mutations in the SPATA5L1, a paralogue of SPATA5, also exhibit similar clinical phenotypes26, implying that SPATA5 and SPATA5L1 may function together. These symptoms caused by SPATA5/SPATA5L1 mutations closely resemble those abnormalities in neurodevelopmental ribosomopathies, which are caused by dysregulated ribosome biogenesis5,27. A recent study revealed that SPATA5 and C1orf109 function in ribosome biogenesis through a CRISPR-based screening28. Based on previous proteomic data that CINP (CDK2-interacting protein), C1orf109, and SPATA5L1 were potential binding partners of SPATA529,30, this study further showed that these four factors likely function together to dissociate RLP24 from human pre-60S particles28. It is intriguing that four factors are necessary for this process in human while the yeast only requires one. The understanding of this difference requires a detailed mechanistic dissection of how SPATA5, SPATA5L1, C1orf109, and CINP work together to dissociate RLP24 from the human pre-60S particle.
In this study, using cryo-electron microscopy, we obtained the high-resolution structure of the SPATA5 complex and the structure of the endogenous pre-60S complex bound with the SPATA5 complex. Our data show that the mechanistic details of the RLP24 dissociation are sharply different from the equivalent process in yeast, contributing to the understanding of the increased, greater complexity in human ribosome biogenesis.
Results
SPATA5, SPATA5L1, C1orf109 and CINP assemble into a stable complex
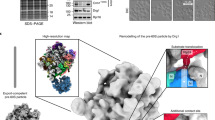
Given the potential of SPATA5, SPATA5L1, C1orf109, and CINP in promoting the dissociation of RLP24 from pre-60S ribosome in cytoplasm28, we tested the expression of these proteins individually in HEK293T cells. Unexpectedly, wild-type (WT) SPATA5 exhibited a significantly compromised expression level, compared with its Walker B mutant forms (catalytic E to Q, namely SPATA5D1EQ, SPATA5D2EQ, and double mutant SPATA52EQ) (Supplementary Fig. 1a). These mutations slow down or abolish ATP hydrolysis in most AAA+ ATPases but do not affect ATP binding10,31. Interestingly, different from SPATA5, both the WT and mutant forms of SPATA5L1 showed comparable expression levels (Supplementary Fig. 1a). Further co-overexpression tests in different combinations confirmed that the expression level of SPATA5L1 could be improved by co-expression with SPATA52EQ (Supplementary Fig. 1b), and there is also a mutual expressional promotion between C1orf109 and CINP (Supplementary Fig. 1b). To test whether these proteins could form a stable complex either alone or in combination, all the samples were purified using anti-FLAG affinity beads against the C-terminal 3 × FLAG tag of SPATA52EQ. Then, these samples were examined by size-exclusion chromatography (SEC) and negative-staining electron microscopy (nsEM). Firstly, SPATA52EQ alone showed no sharp peak, and proteins were distributed in all SEC fractions (Supplementary Fig. 1d, e). The nsEM analysis of peak fractions collected between 13.0 and 14.0 mL showed that these particles lacked the characteristic AAA+ ATPase hexameric arrangement and appeared to be aggregates (Supplementary Fig. 1f). Next, we co-expressed SPATA52EQ and SPATA5L1 and found that they also failed to form hexameric ring structures (Supplementary Fig. 1g–i). In sharp contrast, upon co-expression of SPATA52EQ, SPATA5L1, CINP and C1orf109, the SEC revealed two obviously separated peaks (Fig. 1a). The position of peak 2 indicates a molecular weight of approximately 650 kDa (Fig. 1a). SDS-PAGE and Coomassie blue staining of the fractions from the peak 2 showed that SPATA5 2EQ, SPATA5L1, C1orf109 and CINP indeed form a complex (Fig. 1b). The nsEM further demonstrated that many particles did display a hexameric ring structure (Fig. 1d), which is a typical feature of AAA+ ATPases10,31.
a Size-exclusion chromatography of the affinity-purified SPATA5 complex from cells co-expressing SPATA52EQ(E454Q/E728Q), C1orf109, SPATA5L1, and CINP, showing two separated peaks labeled as peak 1 and peak 2. b SDS-PAGE and silver staining analysis of the peak 1 sample, and coomassie blue staining analysis of the peak 2 samples. Protein bands corresponding to SPATA52EQ, C1orf109, SPATA5L1, and CINP are labeled. HSP70 is labeled with an asterisk. Source data are provided as a Source Data file. c, d Negative staining electron microscopy image of the peak 1 (c) and peak 2 (d) samples. e, f Ribosome profiling of the cell lysates co-expressing SPATA52EQ-Flag, HisSPATA5L1, StrepCINP, and HAC1orf109. Western blotting was used to confirm the presence of these proteins in the ribosomal fractions with Flag, His, Strep, and HA tag antibodies, respectively. Source data are provided as a Source Data file.
The position of the peak 1 is close to the void volume, and the nsEM showed that there are mostly ribosome-like particles in the peak 1 fractions (Fig. 1c), which is consistent with our expectation that the mutant SPATA5 complex might have been trapped on pre-60S particles. On the silver-stained SDS-PAGE gel of the fractions from peak 1, SPATA5 and SPATA5L1 bands could be readily identified (Fig. 1b). The bands of CINP and C1orf109 might have overlapped with ribosomal proteins due to the similar molecular weights (Fig. 1b). To confirm that CINP and C1orf109 are also in the ribosome-like particles, sucrose density gradient centrifugation was conducted using the cell lysates expressing these four proteins. As expected, western blotting shows that these four proteins are all present in the ribosomal fractions (Fig. 1e, f).
Given the function of SPATA5 in dissociating RLP24 from pre-60S particles28, we overexpressed the mutant SPATA5 complex in HEK293T cells to assess the impact of the ATP-hydrolysis-deficient mutation of SPATA5. As expected, SPATA52EQ leads to a strong defect in RLP24 dissociation (Supplementary Fig. 2a), indicative of a possibility that the mutant SPATA5 complex could be stuck with its substrate on the pre-60S particle. Furthermore, there was an apparent nucleolar retention of RPL28 (Supplementary Fig. 2b–f), implying that the sequestration of RLP24 in the cytoplasm should have further caused defects in the nucleolar steps of ribosome assembly. These observations have phenocopied the mutant HEK293T cells with genes for individual components of the SPATA5 complex knocked out28.
Structural organization of the SPATA5 complex
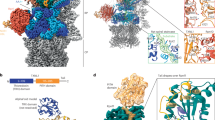
Cryo-EM analysis of the SPATA5 complex initially generated a density map for the overall complex at a global resolution of 3.8 Å (Supplementary Figs. 3a, b, 4a and Supplementary Table 1). Due to the flexible connection between the NTD-ring and the D1-ring of the SPATA5 complex, local refinement was performed, focusing on the NTD-ring and the D1/D2 rings separately, resulting in improved maps at resolutions of 3.4 Å and 3.5 Å, respectively (Supplementary Fig. 4b, c). An atomic model was built using the AlphaFold-predicted structures of these four proteins as initial templates (Fig. 2c, d and Supplementary Fig. 5a–c).
a, b Cryo-EM map of the SPATA5 complex, viewed from the side (a), the top, and the bottom (b) directions. Two SPATA5 conformers are labeled as SPATA51 (hot pink) and SPATA52 (steel blue). SPATA5L1, C1orf109, and CINP are colored orange, cyan, and spring green, respectively. c, d Atomic model of the SPATA5 complex, viewed from the side (c), and from the top and bottom (d) directions. e Domain organization of SPATA5, SPATA5L1, CINP and C1orf109. The NTD insertion of SPATA5 and the D2 domain insertion of SPATA5L1 are highlighted. The red arrows indicate the mutation sites for D1 and D2 of SPATA5.
The dimensions of the SPATA5 complex measure about 141 Å in height and 133 Å in width (Fig. 2a). This complex consists of SPATA5, SPATA5L1, C1orf109, and CINP in a stoichiometric ratio of 4:2:2:2 (Fig. 2a–d). Both SPATA5 and SPATA5L1 contain an N-terminal domain (NTD, composing of NTD1 and NTD2 subdomains) (Supplementary Fig. 6a), and a long insertion sequence (residues 215–314) is found in the NTD2 of SPATA5 (Fig. 2e). Vice versa, an insertion sequence of residues 604–621 lies in the D2 domain of SPATA5L1, following the Inter-Subunit Signaling motif (ISS) (Fig. 2e and Supplementary Fig. 7). These two insertions are flexible and not resolved in the cryo-EM density map. With regard to CINP and C1orf109, they exhibit similar secondary structures, both consisting of five α-helices (Fig. 2e).
From the top view, two sets of the “T”-shaped C1orf109-CINP heterodimer are arranged in an apparent C2-symmetric manner (Fig. 3a). Within the “T”-shaped heterodimer, the loop between α1-α2 helices of C1orf109 interacts with CINP through extensive hydrophobic interactions (Fig. 3b, c). The NTD of one SPATA5 (named SPATA51) binds at the “T”-junction of C1orf109-CINP heterodimer in a vertical orientation, relative to the NTD-ring (Supplementary Fig. 6b), and interacts with both C1orf109 and CINP within this heterodimer (Fig. 3a, b). The NTDs of the other SPATA5 (named SPATA52) and SPATA5L1 adopt vertical and horizontal orientations respectively (Supplementary Fig. 6b), and they form a subcomplex to bridge together the two C1orf109-CINP heterodimers (Fig. 3a). Specifically, the NTD of SPATA52 binds between C1orf109 of one C1orf109-CINP heterodimer and CINP of the other C1orf109-CINP heterodimer (Fig. 3a, f, h), whereas the NTD of SPATA5L1 only binds to C1orf109 (Fig. 3a, j). Through this network of inter-domain interactions, the NTD-ring of the SPATA5 complex forms a funnel-like structure, and the helical bundles of CINP and C1orf109 constitute the inner wall of the funnel (Fig. 3a and Supplementary Fig. 6b). This strategic location of CINP and C1orf109 immediately suggests that they likely function as adapters to mediate the interaction between the SPATA5/5L1 hexamer and the substrate.
a Top view of the atomic model of the SPATA5 complex NTD-ring. The components are color-coded, as in Fig. 2. b Zoomed-in view of the interfaces among CINP, C1orf109, and SPATA51. c–e Close-up view of the interactions between C1orf109 with CINP (c), between SPATA51 and C1orf109 (d), and between SPATA51 and CINP (e). Interacting residues are shown and labeled, with disease-relevant residues labeled in bold (e.g., R84 and G185 of SPATA5). f Zoomed-in view of the interface between SPATA52 and CINP. g Close-up view of the interactions between SPATA52 and CINP. h Zoomed-in view of the interface between C1orf109 and SPATA52. i Close-up view of the interactions between SPATA52 and C1orf109. j Zoomed-in view of the interfaces among SPATA52, C1orf109, and SPATA5L1. k–m Close-up views of the interactions between SPATA5L1 and SPATA52 (k), and between SPATA5L1 and C1orf109 (l, m).
Many naturally occurring mutations in SPATA5 and SPATA5L1 have been linked to a disease condition, named NEDHSB (neurodevelopmental disorder with hearing loss, seizures, and brain abnormalities) (Supplementary Data 1)22,23,24,25,26. A previous study revealed that some of these mutations predominantly affect structural stability or mediates protein-protein interaction32. Given the potential of the NTD-ring in the complex assembly, we further analyzed those mutations located in the NTDs of SPATA5 and SPATA5L in the structural context and found that many of them are located at the subunit interfaces. For instance, R84 within the NTD1 of SPATA5 interacts with both E147 of CINP (Fig. 3e) and D199 of C1orf109 (Fig. 3i), contributing to the interactions with these two factors. The R84Q mutation of SPATA5 would disrupt the polar interactions at both interfaces (Fig. 3e, i). For another example, the G185E mutation within the NTD2 of SPATA5 is located at both the SPATA51-C1orf109 (Fig. 3b) and the SPATA52-CINP interfaces (Fig. 3f), and this mutation may perturb the two interfaces due to the introduction of a large negatively charged side chain (Fig. 3d, g). Similarly, disease-relevant mutations of SPATA5L1 are also located at the subunit interfaces. The NTD of SPATA5L1 and C1orf109 have an extensive interface (Fig. 3j, l, m), and a pathogenic mutation T26A of SPATA5L1 is located in a polar micro-environment between the NTD1 of SPATA5L1 and the α3 of C1orf109 (Fig. 3l). Moreover, three mutations, R64W, D66Y and F71L of SPATA5L1, are concentrated at the interface between the NTDs of SPATA5L1 and SPATA52 (Fig. 3j, k), underscoring the importance of this NTD subcomplex in bridging the two CINP-C1orf109 heterodimers. Altogether, the locations of these above-mentioned NTD mutations firmly demonstrate that the interaction network that forges the NTD-ring in the SPATA5 complex are critically important for its physiological function.
Unique structural features of the SPATA5 complex and functional differentiation between SPATA5 and SPATA5L1
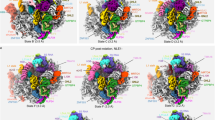
SPATA5 and SPATA5L1 constitute the hexamer in a stoichiometric ratio of 4:2 (Fig. 2c, d), which has not been reported in other AAA+ ATPase hexamers. More interestingly, the complex displays different deviations from typical AAA+ hexamers at different axial positions. At the NTD-ring, the structure is in a square-like shape (Supplementary Fig. 6b). In sharp contrast, the complex is hexagonal at the D1-ring (Fig. 4a) and deformed into an elongated hexagon at the D2-ring (Fig. 4b). As a result, the overall structure displays a two-fold symmetry, with the four SPATA5 subunits exhibiting two different conformations (Supplementary Fig. 8a, b). Notably, the differences between SPATA51 and SPATA52 lie in both the orientations between the D1 and D2 domains, and between the large and small subdomains of the D2 domain (Supplementary Fig. 8a, b).
a, b Atomic models of D1-ring (a) and D2-ring (b) of the SPATA5 complex from the top view. Proteins are shown in licorice style, and ATP molecules are displayed in sphere style. c–e Close-up views of ATPase sites in the D1-ring. The ATP molecules of SPATA51, SPATA52, and SPATA5L1 are shown with their density maps. f–h Close-up views of functionally important residues within the three ATPase sites. Residues of Walker A (khaki), Walker B (cyan), sensor I (green), and arginine fingers (magenta) are displayed in ball-and-stick style. i ATPase assay of the SPATA5WT, SPATA5D1EQ, SPATA5D2EQ, SPATA5L1D1EQ, and SPATA5L1D2EQ complexes in the presence (+) or absence (−) of substrate (residues 85–163 of RLP24). Buffer served as a control. (N = 3 biological replicates; individual data points shown as dots, data are presented as mean ± SD, ** p < 0.01, ***p < 0.001, see Supplementary Data 4). j (Upper panel) Western blot analysis of crude ribosomes from cells expressing wild-type (WT) and mutant SPATA5 complexes for 60 h. NC denotes cells treated with transfection reagents, whereas WT, 5D1EQ, 5D2EQ, 5L1D1EQ, and 5L1D2EQ indicate the SPATA5WT, SPATA5E454Q, SPATA5E728Q, SPATA5L1E304Q, and SPATA5L1E565Q complexes, respectively. (Lower panel) Quantification of RLP24 retention relative to RPL28 in the crude ribosomes (N = 3 biological replicates; data are presented as mean ± SD, *p < 0.05, see Supplementary Data 5). Source data are provided as a Source Data file.
There are three unique interfaces that mediate the formation of the SPATA5 hexamer. Typically, AAA+ ATPases function primarily by utilizing their conserved pore loop motifs to bind to their substrates33,34,35, and they rely on the sequential hydrolysis of ATP on different protomers to provide the energy for the substrate translocation through a hand-over-hand mechanism36. As exhibited in the multiple sequence alignment (Supplementary Fig. 7), the PL-Is (pore loop I) of SPATA5 D1 and D2 domains possess an aromatic residue, F427 and Y701, respectively, similar to its yeast homolog Drg1 (Supplementary Fig. 7). In contrast, the D1 PL-I of SPATA5L1 lacks an aromatic residue, and a positively charged residue R274 is in this equivalent position (Supplementary Figs. 7, 8c). Although the D1 PL-Is of human p97 and yeast Cdc48 also contain a non-aromatic residue, they are still hydrophobic methionine or leucine (Supplementary Fig. 7). This difference between SPATA5 and SPATA5L1 implies that they might have been functionally differentiated in the substrate processing.
Next, we examined the ATP binding sites within the SPATA5 complex. In the D1-ring, all six ATPase sites are occupied by ATP molecules (Fig. 4a). The two-fold symmetry leads to the formation of three distinct ATPase centers for the D1-ring. In the SPATA52: SPATA51 site, the arginine fingers from SPATA51 are distant from ATP (Fig. 4c, f), suggesting that this site is not optimally configured for ATP hydrolysis37. The sites of SPATA5L1: SPATA52 and SPATA51: SPATA5L1 are similarly arranged in an inactive state (Fig. 4d, e, g, h). In addition, ATP in the SPATA51: SPATA5L1 center appears to be in a sub-stoichiometric ratio (Fig. 4d).
We further investigate why the entire SPATA5 complex is in an inactive configuration for ATP-hydrolysis. The ATPase activity of the Drg1 complex is triggered by its substrate8. We speculate that the substrate is also important for the ATPase activity of the SPATA5 complex. The expression level of WT SPATA5 was very low even when co-expressed with SPATA5L1, CINP, and C1orf109 (Supplementary Fig. 1c). We found that the fusion of a GFP to the C-terminus of SPATA5 (SPATA5WT-GFP) could improve its expression. Thus, to measure the ATPase activities of different ATPase centers on SPATA5 and SPATA5L1, we purified the WT and various forms of mutant SPATA5 complexes (four components were co-expressed), carrying a mutated Walker B motif (E to Q) in either the D1 or D2 domain of SPATA5 or SPATA5L1 (5D1EQ, 5D2EQ, 5L1D1EQ, 5L1D2EQ). The in vitro ATP hydrolysis assay using the different SPATA5 complexes showed that the ATPase activity critically depends on the substrate, the C-terminal fragment (residues 85–163) of RLP24. In the absence of the substrate, neither the SPATA5WT-GFP nor these mutant complexes exhibited measurable ATPase activity (Fig. 4i). Upon addition of the substrate, ATPase activity increased significantly for all WT and mutant complexes. As expected, the four mutant complexes all showed reduced ATPase activity compared to the SPATA5WT-GFP complex (Fig. 4i), indicating that the ATPase centers on the SPATA5 complex all contribute to the overall ATP-hydrolysis activity. Next, we assessed the retention of RLP24 on the pre-60S particles in HEK293T cells upon expressing various forms of mutant SPATA5 complexes. The crude ribosomes (including immature ribosomal subunits) were first obtained by sucrose cushion centrifugation, and the RLP24 retention was examined by western blot analysis (RPL28 was used as a control). The results show that the SPATA5D2EQ complex exhibited the most pronounced retention of RLP24 (Fig. 4j). In contrast, the SPATA5D1EQ, the SPATA5L1D1EQ, and the SPATA5L1D2EQ complexes only showed minor inhibitory effects on RLP24 dissociation. These findings suggest that the D2 ATPase center of SPATA5 might be more important for the function of the SPATA5 complex in RLP24 dissociation.
The oxidative modification of the P-loop cysteine residue of SPATA5 inactivates its ATPase activity
With regard to the D2 domains, very surprisingly, only two SPATA5L1 subunits exhibit ATP-bound states (Fig. 4b and Supplementary Fig. 8d, e). The ATP-binding P-loop/Walker A motif of SPATA5 is folded completely differently, incompatible with ATP binding (Fig. 5a–c). Structural comparisons of the SPATA5 with SPATA5L1 P-loops indicate that a cysteine residue C672 in the P-loop of SPATA5 is in a distinct orientation and sterically clashes with the ATP molecule (Fig. 5d–f, h). Consequently, the D2-ring only possesses two ATPase centers in two SPATA5L1 subunits. Further examination of the density map revealed that C672 of SPATA5 displays a much larger side-chain density (Fig. 5b, c), indicative of a post-translational modification. Notably, the equivalent positions of C672 on Drg1 and p97 are also cysteines (Fig. 5g), and the P-loop residue C522 of Drosophila Cdc48 was previously shown to be subjected to oxidative modification, leading to the inactivation of the ATP hydrolysis38. Therefore, we performed post-translational modification profiling of the purified SPATA5 complex using mass spectrometry and found that almost all SPATA5 proteins are sulfinylated (Cys-SO2H) at the C672 position (Supplementary Fig. 9a, c, Supplementary Data 2 and Supplementary Table 2).
a–c The P-loops of SPATA5/SPATA5L1 D2 domains, with their densities shown. Only the SPATA5L1 D2 domain contains an ATP molecule. d–f The atomic models of the P-loops of the SPATA5/SPATA5L1 D2 domains showing the different conformations of the Cysteine residues in SPATA5 and SPATA5L1. g Sequence alignment of the P-loops of classical type II AAA+ ATPases, including human SPATA5, SPATA5L1, NSF, and p97, as well as yeast Drg1 and Cdc48, and Drosophila p97. h Structural alignments of the P-loops of SPATA5L1 D2 domain and SPATA51/SPATA52 D2 domains, demonstrating the steric clashes between the P-loops of SPATA51/SPATA52 D2 domain and ATP molecules.
To further validate whether this C672 modification occurs on endogenous WT SPATA5, we established a CRISPR knock-in cell line (GFP-strep-SPATA5) and purified the endogenous SPATA5 complex. Consistent with the data on recombinant proteins, we also detected the sulfinylation at the C672 site (Supplementary Fig. 9b, c, Supplementary Data 2 and Supplementary Table 2), with an overall modification level of approximately 70% (Supplementary Table 2). Since we have included 1 mM TCEP in all purification buffers, this modification should not be introduced during purification. Together, these data unveil an interesting mechanism, which might be universal for many AAA+ ATPases, that the cysteine oxidation in the P-loop serves as a regulatory switch to turn off the ATPase activity.
Structure of the pre-60S particles co-purified with the mutant SPATA5 complex
The ribosome-like particles of the peak 1 fractions were also subjected to cryo-EM analysis (Supplementary Fig. 10a and Supplementary Table 1). As expected from the function of SPATA5, these particles turned out to be large subunit assembly intermediates and two major states were revealed through 3D classification. The first state was resolved at the final resolution of 2.7Å (Supplementary Fig. 10b), closely resembling the previously described human pre-60S structures, in which nuclear assembly factor GNL2 has dissociated15. In the structure of the state 1, the ITS2 has been removed and most of the rRNAs have assumed their mature-like conformations. For instance, the 5S RNP has rotated, and the peptidyl transferase center helices H68-H69 have been stabilized (Fig. 6a). Regarding the assembly factors, TMA16, GTPBP4, eIF6, ZNF593, and RLP24 are present (Fig. 6a). The state 2 is, in general, similar to the state 1, except that ZNF593 is in sub-stoichiometric occupancy and H68-71 is flexible and not yet in its mature-like conformation (Supplementary Fig. 10c, d). These structural observations indicate that they are cytoplasmic pre-60S particles. Although all four components of the SPATA5 complex are present in the sample (Fig. 1b, e, f), the SPATA5 complex was not stably resolved in the final density map. Nevertheless, the examination of nsEM images indeed found many SPATA5 complex-bound pre-60S particles (Fig. 6b). These data collectively suggest that the SPATA5 complex is indeed involved in the maturation of the cytoplasmic pre-60S particles.
a Cryo-EM map of the pre-60S particle co-purified with the mutant SPATA5 complex. Assembly factors TMA16, GTPBP4, eIF6, ZNF593, RLP24 and 5S RNA and 28S RNA H68-H69 are color-coded based on pdb:6LSS. b Representative particles of the pre-60S bound with the mutant SPATA5 complex from the negative staining EM. The position of the SPATA5 complex is annotated using the dashed circle. c Cryo-EM map of the pre-60S particle bound with the SPATA5 complex. Assembly factors GTPBP4, RLP24, PA2G4, the SPATA5 complex and 28S rRNA ES27A are color-coded. The two interfaces between the pre-60S particle and the SPATA5 complex are indicated with red arrows. The cartoon model of the SPATA5 complex illustrates its orientation relative to the pre-60S particle. d Cryo-EM map of the yeast pre-60S bound with Drg1 hexamer (EMD-14471). Assembly factors Nog1, Rlp24, Arx1, Drg1 complex, and the ES27 of 25S rRNA are color-coded. The interface between the pre-60S particle and the Drg1 complex is indicated with a red arrow. The cartoon model of the Drg1 complex illustrates its orientation relative to the pre-60S particle. e The CINP-GTPBP4 interface on the pre-60S particle. f The CINP-ES27A interface on the pre-60S particle. g The cross-linking analysis of the pre-60S-SPATA5 complex. The cross-linking between RLP24 with SPATA5, C1orf109, and CINP are indicated and colored blue, cyan, and lime, respectively. h Western blot analysis of input and GST IP samples probed with GST, Strep, or HA antibodies. Non-specific bands were labeled with an asterisk. Source data are provided as a Source Data file.
PA2G4 stabilizes ES27A and subsequently facilitates the stabilization of the SPATA5 complex on the pre-60S particle
In yeast, Drg1 binds to pre-60S particles through direct interactions with Arx1 and ribosomal RNA expansion segment ES2712. Given the role of Arx1 in interacting with Drg1, it is reasonable to speculate that its human homolog PA2G4 may also contribute to the stabilization of the SPATA5 complex. However, in the density maps of the pre-60S particles co-purified with the SPATA5 complex, we did not detect the binding of PA2G4. It is known that the binding of PA2G4 is critical for maintaining a stable conformation of ES27A15. Similarly, the lack of PA2G4 in our maps has resulted in smeared densities for ES27A. Therefore, we sought to increase the stability of the SPATA5 complex on the pre-60S particle by supplying excessive recombinant PA2G4 proteins before cryo-sample preparation. At first, we expressed and purified recombinant PA2G4 using E. coli cells (Supplementary Fig. 11a), and tested its binding to the SPATA5 complex in vitro. However, PA2G4 did not co-migrate with the SPATA5 complex on the SEC profile (Supplementary Fig. 11b). Same as Arx1, PA2G4 contains a similar surface used for interacting with ES27A of human 28S rRNA (Supplementary Fig. 11c, d)15,39,40. Nevertheless, structural alignment shows that PA2G4 is considerably different from Arx1 in its Drg1-interacting region (Supplementary Fig. 12). Three loops of Arx1 are responsible for interacting with Drg1 in the yeast pre-60S structure12. Interestingly, two of them are absent in PA2G4, and the only existing one shows a totally different secondary structure (Supplementary Figs. 11e, f and Supplementary Fig. 12). These analyses imply that PA2G4 and the SPATA5 complex may not have direct interactions. Given the role of ES27 in the stabilization of Drg139,40, we reasoned that the introduction of PA2G4 to the peak 1 sample might still be able to indirectly stabilize the SPATA5 complex on human pre-60S particle through conformational stabilization of ES27A.
This strategy was proven to be effective, and the subsequent cryo-EM analysis resolved a pre-60S class with relatively solid density for both PA2G4 and the SPATA5 complex (Fig. 6c and Supplementary Fig. 13a). Further 3D classification and refinement could improve the pre-60S map and the local map of the bound SPATA5 complex to resolutions of 3.0 Å and 6.9 Å, respectively (Supplementary Fig. 13b, c and Supplementary Table 1), in which the secondary structure features of the NTD-ring began to appear and could accommodate the atomic model of the NTD-ring very well (Supplementary Fig. 14g). In contrast, the densities of the ATPase rings are highly fragmented and the low-pass filtered version is still feature-less (Fig. 6c). This is in agreement with the fact that connection between the NTD-ring and D1-ring is highly flexible.
CINP and C1orf109 are adapter proteins for the SPATA5 complex
Very interestingly, a comparison with the structure of the yeast pre-60S-Drg1 complex reveals a substantially different orientation of the SPATA5 complex on the pre-60S particle, compared with Drg1 (Fig. 6d and Supplementary Fig. 14d, e). Drg1 relies on one major site to interact with the pre-60S particle, directly interacting with Arx1 and ES2712. In contrast, there are two interfaces between the SPATA5 complex and the pre-60S particle (Fig. 6c). The first interface is protein only, involving CINP and GTPBP4 (Fig. 6c, e). This binding interface is consistent with the model of the C1orf109-CINP-GTPBP4 complex predicted by AlphaFold Multimer41, and K22, K36, L122, and Y125 of CINP show direct interactions with the residues 452–474 of GTPBP4 (Supplementary Fig. 14f). To further validate this interface, we carried out immunoprecipitation experiments using GST-tagged fragment of GTPBP4 (residues 426–500), HA-tagged C1orf109 and Strep-tagged CINP. In agreement with the structural observations, GTPBP4 was able to immunoprecipitate CINP, but not C1orf109 (Fig. 6h), confirming a direction interaction between GTPBP4 and CINP.
The other interface is between ES27A and the NTD-ring of the SPATA5 complex. Specifically, CINP and the NTD of SPATA51 directly contact ES27A (Fig. 6c, f). In this interface, ES27A is sandwiched by PA2G4 and the SPATA5 complex, but no direct interaction was observed between PA2G4 and the SPATA5 complex (Fig. 6f), which is consistent with our in vitro binding experiment (Supplementary Fig. 11b). It has to be noted that this pattern of interactions is different from that of Drg1, which binds to both Arx1 and ES27. These data indicate that in human cells, the interface provided by GTPBP4 plays an essential role in docking the SPATA5 complex onto the pre-60S particle. Importantly, CINP contributes to both the interfaces between the SPATA5 complex and the pre-60S particle, while C1orf109 does not interact with the pre-60S particle directly.
In this arrangement of the SPATA5 complex on the pre-60S particle, the C-terminal end of RLP24 would be placed above the funnel entrance of the CINP-C1orf109 tetramer (Supplementary Fig. 14a). The distance between the last resolved C-terminal residue (R135) of RLP24 and the PL-I of the D1-ring is about 65 Å (Supplementary Fig. 14b), and the unresolved C-terminal region (residues 136–163) of RLP24 contains another helix (~ 60 Å in length) (Supplementary Fig. 14b), indicating that the very C-terminal end of RLP24 is capable of reaching the PL-I of the D1-ring. To verify whether the C-terminal region of RLP24 could directly interact with CINP or C1orf109, we used AlphaFold Multimer to predict the potential interfaces. The prediction shows that the C-terminal end of RLP24 is inserted into the funnel, and the last two helices (residues 80–163) of RLP24 are able to bind to both CINP and C1orf109 (Supplementary Fig. 14c). Consistently, our immunoprecipitation experiments show that a fragment (residues 85–163) of RLP24 was indeed able to immunoprecipitate both CINP and C1orf109 (Fig. 6h).
To further validate our structural model, we performed crosslinking mass spectrometry. We identified several highly informative crosslinked peptides involving RLP24, which show that the C-terminal region of RLP24 is spatially close to CINP, C1orf109, and the NTD of SPATA5 (Fig. 6g). More importantly, K141 of RLP24 was detected to be crosslinked with K434, which is located in the α2 helix of the D1 domain of SPATA5 and close to the PL-I, indicating that the C terminal tail of RLP24 could indeed be threaded into the central pore of the SPATA5 complex. Altogether, these data collectively support that CINP and C1orf109 are adapter proteins to mediate the interaction between the SPATA5 complex and its substrate RLP24.
Discussion
In this work, we found that SPATA5, SPATA5L1, C1orf109, and CINP can form a stable AAA+ ATPase complex, the SPATA5 complex. Within this complex, the ATPase domains of SPATA5 and SPATA5L1 constitute the hexameric D1-D2 rings in a stoichiometric ratio of 4:2, and the NTDs of SPATA5 and SPATA5L1 along with two C1orf109-CINP heterodimers form the NTD-ring. The fact that pre-60S particles could be co-purified with the mutant SPATA5 complex suggests that the SPATA5 complex may function in ribosome assembly, which is consistent with previously reported data28. The structure of the pre-60S particles bound with the mutant SPATA5 complex further shows that SPATA5, SPATA5L1, C1orf109, and CINP act in concert as bona fide ribosome assembly factors to facilitate the release of RLP24 in the cytoplasm. Both GTPBP4 and ES27A contribute to the recruitment of the SPATA5 complex onto the pre-60S particle, and the adapter CINP also mediates the interactions with the pre-60S particle. Of note, there is no interaction observed between PA2G4 and the SPATA5 complex. All these data demonstrate that the recruitment of the SPATA5 complex is distinct from the equivalent process in yeast, in which ES27 and Arx1 form a docking platform for Drg1 hexamer, and no adapter proteins are required12. A comparison of Arx1 and PA2G4 shows that PA2G4 is considerably smaller (about 200 amino acids shorter) and lacks the important Drg1-interacting loops on Arx1 (Supplementary Figs. 11, 12). Sequence alignment of RLP24/Rlp24 across multiple model organisms revealed that only the yeast version contains a unique highly positively charged C-terminus (Supplementary Fig. 15a). Furthermore, SPATA5L1 homologs only exist in species where RLP24 lacks this C-terminal tail (Supplementary Fig. 15b), indicating a possible co-evolution of these factors. The shortening of this C-terminal tail of RLP24 likely necessitates that the dissociation machinery has to be positioned closer to the substrate or operates through alternative mechanisms to approach and bind to the substrate. According to our IP and cross-linking data (Fig. 6h and Supplementary Data 3), CINP and C1orf109 should have a direct role as adapter proteins to initiate substrate recognition. These observations highlight a few interesting species-specific features of the SPATA5 complex in the human pre-60S assembly and explain the molecular role of these extra adapters in the SPATA5 complex.
Based on our high-resolution structure of the SPATA5 complex (Fig. 2a–d), we analyzed the reported pathogenic mutations of SPATA5/5L1 in the context of the inter-subunit interactions (Fig. 3). A majority of mutations are not at the interaction interface, and they likely impact the protein folding and stability. Several mutations are found in functional motifs, such as arginine finger and ATP binding pocket of the ATPase domain (Supplementary Data 1), suggesting an apparent effect on the ATPase activity of the SPATA5 complex. Importantly, many mutations of SPATA5/SPATA5L1 are localized in the subunit interfaces at the NTD-ring (Supplementary Data 1), manifesting an important role of the NTD-ring in the assembly and function of the SPATA5 complex. In addition, some mutations might affect the conformational signal transduction within the SPATA5 complex. For example, M669R of SPATA5L1 is at the hydrophobic interface between the small subdomain of SPATA5L1 and the large subdomain of SPATA52 (Supplementary Fig. 8f), implying that it may impair the stabilization of the hexamer or conformational signal transduction through this interface.
Very intriguingly, we found that there is a high-level modification at the C672 residue within the P-loop of SPATA5. This oxidative modification (sulfinylation) not only occurs on overexpressed mutant SPATA52EQ but is also present in the majority of WT endogenous SPATA5 proteins. This modification results in a completely different conformation for the P-loop, incompatible with ATP binding. Additionally, it also induces dramatic structural changes on the D2 domain of SPATA51, leading to an inward rotation of the small subdomain toward the large subdomain (Supplementary Fig.8h, i). Consequently, this rotation causes the D2-ring of the SPATA5 complex to deviate from the typical hexameric arrangement and become an elongated hexagon (C2 symmetry). Very recently, a cryo-EM structure of the WT SPATA5 complex was reported32. This structure, named 55LCC complex by the authors, is nearly identical to ours in the NTD region (Supplementary Fig. 8j). But there are significant differences in the D1 and D2 rings (Fig. 4a, b and Supplementary Fig. 8g–i). In our structure, the D1-ring adopts a more relaxed conformation compared to the 55LCC complex (Fig. 4a and Supplementary Fig. 8g). The most prominent difference lies in the arrangement of the D2-ring. Sharply different from ours, the 55LCC complex exhibits a six-fold configuration (Supplementary Fig. 8h). Because the 55LCC complex was obtained by heterologous expression in insect system and may lack native post-translational modification, which we believe is the reason for the observed structural differences.
Although sulfinylation (Cys-SO2H) on proteins was historically considered to be irreversible under physiological conditions42, sulfiredoxin enzymes that reduce sulfinic acid to sulfenic acid have been identified in both yeast and human cells43,44,45, and an increasing number of protein substrates for sulfiredoxin enzymes have been reported. In addition, there was also experimental evidence showing the presence of yet-to-be-discovered cysteine sulfinic acid reductases in the cells46. Therefore, the high-level sulfinylation at the C672 site on the endogenous SPATA5 proteins raises the possibility of a reversible modification, suggesting an under-recognized regulatory mode for the SPATA5 complex through cellular redox signaling pathways.
During eukaryotic ribosome assembly, shuttling assembly factors, such as Nmd347, eIF648, Bud2049, Rlp2411, and Nog150 incorporate into the pre-60S particles in the nucleus, which are necessary for early steps of subunit construction. After nuclear export of the pre-60S particles, they are released in the cytoplasm and recycled back to the nucleus4. The failure in their release and recycling causes immediate disruption to the following cytoplasmic maturation steps of the pre-60S particles, and because of their cytoplasmic sequestration, the next rounds of subunit assembly in the nucleus is also hampered. In the present work, the mutations of the SPATA5 impaired the dissociation of RLP24 (Supplementary Fig. 2a). The compromised recycling of RLP24 leads to abnormal ribosome assembly and nucleolar stress51,52, indicated by the nucleolar retention of RPL28 upon the introduction of Walker B mutations on the SPATA5 complex (Supplementary Fig. 2b–f). These observations have phenocopied many yeast shuttling assembly factors. Given the role of the SPATA5 complex in RLP24 dissociation, the NEDHSB may be considered an under-characterized neurodevelopmental ribosomopathy5,27.
In addition to its role in ribosome biogenesis, CINP was also reported to participate in the DNA replication process53 and might play a role in response to DNA damage for genome stability maintenance54. During the manuscript preparation, a recent work proposed that SPATA5, together with SPATA5L1, CINP, and C1orf109, functions in genome integrity maintenance during DNA replication through responding to replication fork damage32. Although the DNA/RNA binding properties and the DNA/RNA-enhanced ATPase activity of the SPATA5 complex have been assayed in vitro, no directed biochemical link has been established between the SPATA5 complex and the components of the replisome and related factors32. It has been well documented that the defects in ribosome assembly, such as the failed incorporation of RPL11, RPL5, 5S RNA into pre-60S particles, could cause nucleolar stress and activate the p53 pathway (RP-MDM2-p53 axis), which would elicit subsequent events such as cell cycle arrest and apoptosis55,56,57. It is plausible that the replication stress and severe chromosome instability reported upon dysfunction of the SPATA5 complex may be secondary effects of ribosome assembly defects. In the present work, we determined the structure of the SPATA5 complex bound with the endogenous pre-60S particle, providing a solid and direct functional connection of this complex to ribosome assembly.
Methods
Purification of the SPATA5 complex
For the purification of WT or mutant SPATA5 complexes, HEK293F cells were co-transfected with plasmids encoding the WT or mutant SPATA5 complexes using polyethylenimine (PEI) as the transfection agent. After transfection, the cells were cultured for 48 h to allow for protein expression, followed by cell harvesting. The harvested cell pellets were resuspended in buffer A, consisting of 40 mM HEPES, pH 7.5, 150 mM NaCl, 1 mM DTT, 10% glycerol, 10 mM MgCl2, and a protease inhibitor cocktail (Mei5 Biotech, MF182-plus-01). To promote cell lysis, 0.1% IGEPAL was added to the cell suspension, and the mixture was incubated at 4 °C for 30 min. The lysates were then subjected to centrifugation to separate the supernatant. The supernatant was incubated with Flag affinity beads (Smart-Lifesciences SA042025) for 2.5 h at 4 °C to selectively capture the SPATA5 complex through the Flag tag on various forms of SPATA5. After incubation, the beads were washed multiple times with buffer A to remove non-specifically bound proteins. The proteins bound to the beads were eluted using buffer B, which is buffer A supplemented with 0.5 mg/mL 3 × Flag peptides. Further purification was conducted using size-exclusion chromatography with a Superose 6 Increase 10/300 GL Column (Cytiva), which was equilibrated with buffer C (40 mM HEPES, pH 7.5, 150 mM NaCl, 1 mM DTT, 5% glycerol and 10 mM MgCl2).
Purification of PA2G4 and substrate RLP24
The expression of His-tagged PA2G4 and His-MBP-tagged RLP24 (residue 85–163) in E. coli Transetta (DE3) cells (TransGen Biotech) was induced by the addition of 1 mM IPTG when the OD600 reached 0.6 to 0.8. The induction was carried out at 18 °C overnight. After induction, bacterial cells were harvested via centrifugation and resuspended in lysis buffer containing 40 mM HEPES, pH 7.5, 500 mM NaCl, 10% glycerol, 1 mM DTT, 1 mM PMSF, and 20 mM imidazole. The cells were then lysed using sonication, and the lysate was clarified by centrifugation at 34,960 g for 40 min at 4 °C. The clear supernatant was collected, and the Ni-NTA beads were added and incubated for 1.5 h at 4 °C. After incubation, the beads were harvested by centrifugation and washed four times with the lysis buffer to remove non-specifically bound proteins. The protein was eluted from the beads using a lysis buffer containing 500 mM imidazole. The eluted proteins were then concentrated and further purified using a Superdex 200 Increase gel filtration column, and peak fractions were collected and concentrated.
Polysome profiling and fractionation
Cultured cells were harvested and washed twice with ice-cold PBS. The cell pellets were then resuspended and lysed in buffer containing 40 mM HEPES, pH 7.5, 150 mM NaCl, 1 mM DTT, 10 mM MgCl2, protease inhibitor cocktail (Mei5 Biotech, MF182-plus-01), RNase inhibitor and 0.1% IGEPAL for 30 min. The resulting lysates were clarified by centrifugation. The clarified lysates were carefully layered onto a linear sucrose gradient ranging from 5% to 50%. Centrifugation was performed at 260,800 × g for 2.5 h at 4 °C using a SW41 rotor in a Beckman ultracentrifuge. After centrifugation, fractions were collected, and their absorbance at 260 nm was measured. Subsequently, these fractions were analyzed by SDS-PAGE and Western blot to identify and quantify the target proteins.
Cryo-EM sample preparation
The affinity-purified mutant SPATA52EQ complexes were first incubated with 5 mM ATP on ice for 30 min, followed by SEC (Superose 6 Increase 10/300 GL) with 5% glycerol. The SEC fractions corresponding to the mutant SPATA5 complex were then supplemented with 1 mM BS3 for cross-linking, which was stopped by adding 1 M Tris-HCl (pH 7.5) to achieve a final Tris-HCl concentration of 20 mM.
In the chamber of an FEI Vitrobot, which was maintained at 4 °C and 100% humidity, 4 μl (~ 1.2 mg/mL) aliquot of the SPATA52EQ complexes was applied to Quantifoil R1.2/1.3 Au grids, which had been subjected to glow discharge for 60 s in a hydrogen and oxygen gas mixture in advance. After waiting for 5 s, the grids were blotted for 1 second using a blot force of – 1.
For cryo-EM sample preparation of the ribosomal particles, the peak 1 fractions in Fig. 1a were firstly assessed using negative staining electron microscopy. With or without the addition of recombinant PA2G4 proteins, these samples were subjected to cryo-EM sample preparation using the identical procedures as for the SPATA5 complex, except that the Quantifoil R1.2/1.3 Cu grids (+ 2 nm carbon film) were used to increase the binding of the ribosomal particles to the girds.
Initial screening of all cryo-EM grids was conducted using an FEI Talos Arctica microscope equipped with an FEI Ceta camera, operated at 200 kV.
Data collection and image processing
For the SPATA5 complex, movies were captured using a Titan Krios G4 microscope operated at 300 kV, with a Gatan K3 Summit camera and a GIF Quantum energy filter. Data collection was performed using SerialEM software58 at a magnification of 81,000 ×, achieving a pixel size of 1.092 Å. The defocus ranges from − 1.2 to − 2.0 μm. Each recorded movie consists of 32 frames in a total exposure time of 2.98 seconds. Three batches of movies were collected and subjected to similar processing procedures in RELION 4.059. Initial processing steps included motion correction with MotionCorr260 and Contrast Transfer Function (CTF) estimation with CTFind461. Particle picking was performed using a model trained with Topaz software62. Subsequent 2D classification was carried out to eliminate low-quality and noisy particles. Through multiple rounds of 3D classification, 542,141 high-quality particles were selected and subjected to further processing in CryoSPARC63. Ab initio reconstruction and heterogeneous refinement were used with C1 symmetry applied, and the final dataset contains 194,789 particles. Owing to the inherent flexibility of the SPATA5 complex, non-uniform refinement with C1 symmetry was employed to improve the density map, achieving an overall resolution of 3.80 Å. Due to the flexible connections between the NTD-ring and ATPase rings, local refinements with C1 symmetry were carried out separately on these two regions. As a result, the final resolution of the NTD ring reached 3.39 Å, and the resolution of the ATPase rings reached 3.51 Å.
Data collection for the peak 1 sample was performed using the same set of equipment and parameters. In total, 2211 movie stacks were collected and subjected to motion correction and CTF estimation using MotionCorr2 and CTFind4, respectively. Subsequent particle picking generated a dataset of 289,998 particles. Through several rounds of 3D classification, 110,778 pre-60S particles were selected. Further refinement and additional rounds of classification revealed two main conformations for the pre-60S particles.
Data collection for the peak 1 sample supplemented with PA2G4 was carried out on a 300 kV Titan Krios G2 microscope, equipped with a Gatan K3 camera and a Quantum energy filter. The microscope was set to a magnification of 81,000 ×, achieving a pixel size of 1.1 Å. The defocus range was maintained consistently from − 1.2 to − 2.0 μm, and each movie contains 32 frames, with an exposure time of 2.57 seconds. The initial 382,686 particles were subjected to rounds of 2D and 3D classification, generating a final set of 56,844 pre-60S particles for further processing. These particles were recentered to extra densities between ES27A and RLP24 and reextracted. Subsequent 3D classification utilized a mask diameter of 150 Å without angular alignment, focusing on these extra densities, revealing a class with solid densities for the SPATA5 complex.
Atomic model building and refinement
For the modeling of the SPATA5 complex, the resolution of the obtained map was sufficient to clearly distinguish the individual protein components of the complex. The initial templates for the NTD-ring were derived from the AlphaFold protein structure database64. The predicted 3D models of CINP, C1orf109, SPATA5-NTD, and SPATA5L1-NTD (obtained by truncating the N-terminal domains (NTDs) from the full-length predicted structures of SPATA5 and SPATA5L1, respectively) were fitted into the maps using rigid-body fitting in UCSF Chimera65, and adjusted manually in COOT66. For the model building of the ATPase rings of the SPATA5 complex, the same procedures were applied. In addition, ATP molecules were modeled into the D1 domain of SPATA5/5L1 and the D2 domain of SPATA5L1.
For each part of the SPATA5 complex, multiple rounds of model refinement were conducted using real-space refinement in PHENIX67, with secondary structure and geometry constraints applied. The final refined models were evaluated using MolProbity68. The maps and models were visualized using UCSF Chimera and ChimeraX69 for analysis.
Co-IP (Immunoprecipitation) assay
GST co-IP was performed by co-expressing GST-RLP24 (85–163) or GST-GTPBP4 (426–500) with Strep-CINP or HA-C1orf109. GST alone was co-expressed with Strep-CINP or HA-C1orf109 as a negative control. After 48 h of transfection, the cells were harvested and washed using PBS. The cell pellets were lysed in lysis buffer (40 mM HEPES, pH 7.5, 150 mM NaCl, 1 mM DTT, 10% glycerol, 10 mM MgCl2, 0.1% IGEPAL, and 1 × protease inhibitor cocktail) for 30 min at 4 °C. Cell lysates were then centrifuged at 21,130 × g for 30 min at 4 °C. The supernatant was collected and incubated with GST beads for 2.5 h at 4 °C. The GST beads were washed four times with lysis buffer (without IGEPAL). Then, the collected GST beads were boiled in 2×SDS-PAGE loading buffer at 100 °C for 10 min. The samples were subjected to SDS-PAGE and western blot using GST, Strep, or HA antibodies, respectively.
Generation of RPL28-Flag-GFP and GFP-Strep-SPATA5 CRISPR knock-in cell lines
HEK293T cells were maintained in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and cultured at 37 °C in a humidified atmosphere containing 5.5% CO2. To achieve genomic insertion of C-terminal RPL28-Flag-GFP and N-terminal GFP-Strep-SPATA5 fusion, we employed standard CRISPR/Cas9 techniques70. Guide RNAs were designed using the crispor website. Specific guide RNA sequences (5’-CTCCCCAGAGCGTTTCAAAA-3’ and 5’-TTGAAGCGCGCACATTGAGT-3’) for RPL28 and SPATA5 were cloned into the pX330 vector, respectively. Two donor DNA fragments were designed to contain the RPL28-Flag-GFP/GFP-Strep-SPATA5 sequences, flanked by approximately 800 bp homology arms.
HEK293T cells were seeded in 6-well plates at 80% confluency and co-transfected with the guide RNA vector and the donor vector at a 1:1 mass ratio using polyethylenimine (PEI) as the transfection agent. After transfection, the cells were cultured for one week. GFP-positive cells were identified using flow cytometry (Astrios EQ) and seeded into 96-well plates to develop clonal populations, which were routinely inspected under a fluorescence microscope to remove any multiclonal or false-positive clones. Genomic integration of the RPL28-Flag-GFP/GFP-Strep-SPATA5 insert was verified by PCR analysis of the extracted genomic DNA. The positive clones were further validated through DNA sequencing and assessed by western blotting to confirm protein expression and ensure accurate tagging.
Cross-linking mass spectrometry (XL-MS)
Approximately 35 µL of ribosomal samples co-purified with the SPATA5 complex (0.18 µM), 10 µL of the SPATA5 complex (2.4 µM), and 4 µL of PA2G4 proteins (42 µM) were mixed and divided into three equal portions. These portions were cross-linked with BS3 at final concentrations of 0.5 mM, 1 mM, and 3 mM, respectively, in the presence of 1 mM ATP. The cross-linking reaction was conducted at room temperature for 1 h. To terminate the reaction, 100 mM NH₄HCO₃ was added, and the mixture was incubated at room temperature for 15 min. Subsequently, 10 µL of 2 × SDS-PAGE loading buffer was added, and the samples were boiled at 100 °C for 8 min. Finally, 8 µL of each sample was subjected to SDS-PAGE.
After SDS-PAGE and Coomassie blue staining, the desired protein band was excised, and the gel was dehydrated with acetonitrile. The proteins were then reduced and alkylated before undergoing tryptic digestion at 37 °C overnight. The resulting peptides were analyzed using a Thermo Fusion Lumos mass spectrometer coupled with the EASY-nLC 1200 system. Peptide mixtures were loaded onto a C18 trap column with Buffer A (0.1% formic acid) and separated using a gradient of Buffer B (0.1% formic acid, 80% acetonitrile) at a flow rate of 280 nL/min (2 min: 4% to 8% B; 68 min: 8% to 32% B; 5 min: 32% to 90% B; 15 min: 90% B). Data were acquired in data-dependent acquisition (DDA) mode. For MS1 scanning, the Orbitrap resolution was set to 60,000, with a mass range of 350–1,500 m/z. The AGC (Automatic Gain Control) target was set to 400,000, with a maximum injection time of 50 ms. For MS2 scanning, fragmentation was performed using higher-energy collision-induced dissociation (HCD) at stepped energy levels of 25%, 30%, and 35%. The Orbitrap resolution for MS2 was set to 30,000, with an AGC target of 100,000 and a maximum injection time of 150 ms. The precursor ion isolation window was set to 1.6 m/z.
The combined LC-MS/MS data (0.5 mM, 1 mM, and 3 mM BS3 cross-linked samples) were processed using pLink software with the following parameters: analysis was performed against the UniProt Homo sapiens custom protein sequence database. BS3 was used as the cross-linker, and trypsin was selected as the protease for enzymatic digestion. A maximum of 3 missed cleavages was allowed. The fixed modification included carbamidomethylation of cysteine residues. Variable modifications considered were the oxidation of methionine and acetylation of the protein N-terminus. The precursor ion mass tolerance and fragment ion mass tolerance were both set to 20 ppm. The minimum peptide length for analysis was set to 6 amino acids, and the PSM-level false discovery rate (FDR) was set to 5%.
In vitro ATPase activity assay
Approximately 0.2 μM of wild-type (WT) or mutant complex samples were incubated with 1 mM ATP at 37 °C for 30 min in enzyme activity buffer (40 mM HEPES, pH 7.5, 150 mM NaCl, 10 mM MgCl₂, 5% glycerol, 1 mM TCEP), in the presence or absence of RLP24 (residues 85–163). Following incubation, 120 μL of enzyme activity buffer was added to dilute the reaction mixture, reducing the final ATP concentration to below 0.25 mM. For activity measurement, 80 μL of the diluted sample was mixed with 20 μL of malachite green dye buffer (Sigma, MAK307). The mixture was incubated at room temperature for 30 min, and absorbance was measured at 620 nm. Each sample was analyzed with three biological replicates, each containing three independent technical replicates, ensuring the accuracy and reproducibility of the results.
Statistics and Reproducibility
All experiments were performed with appropriate replicates to ensure statistical validity. Data presented in Fig. 1b–d, f are based on three independent experiments. Data in Fig. 6h are based on two independent experiments.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The cryo-EM map and atomic coordinate of the SPATA5 complex generated in this study have been deposited in the EMDB and PDB databases under accession codes EMD-63069 and PDB-9LGO [https://doi.org/10.2210/pdb9LGO/pdb], respectively. The cryo-EM maps of the pre-60S ribosomal subunit (state1/state2) have been deposited in the EMDB under accession codes EMD-63085 and EMD-63087. The cryo-EM map of the pre-60S-SPATA5 complex has been deposited in the EMDB under accession code EMD-63206. The AP-MS/MS proteomics data generated in this study have been deposited in the ProteomeXchange database under accession code [PXD054917]. Source data generated in this study are provided in the Source Data file associated with this paper. Source data are provided in this paper.
References
Warner, J. R. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 24, 437–440 (1999).
Klinge, S. & Woolford, J. L. Jr. Ribosome assembly coming into focus. Nat. Rev. Mol Cell Biol. 20, 116–131 (2019).
Bassler, J. & Hurt, E. Eukaryotic ribosome assembly. Annu. Rev. Biochem. 88, 281–306 (2019).
Woolford, J. L. Jr. & Baserga, S. J. Ribosome biogenesis in the yeast Saccharomyces cerevisiae. Genetics 195, 643–681 (2013).
Bohnsack, K. E. & Bohnsack, M. T. Uncovering the assembly pathway of human ribosomes and its emerging links to disease. EMBO J. 38, e100278 (2019).
Kressler, D., Hurt, E. & Bassler, J. Driving ribosome assembly. Biochim. Biophys. Acta 1803, 673–683 (2010).
Watkins, N. J. & Bohnsack, M. T. The box C/D and H/ACA snoRNPs: key players in the modification, processing and the dynamic folding of ribosomal RNA. Wiley Interdiscip. Rev. RNA 3, 397–414 (2012).
Kappel, L. et al. Rlp24 activates the AAA-ATPase Drg1 to initiate cytoplasmic pre-60S maturation. J. Cell Biol. 199, 771–782 (2012).
Loibl, M. et al. The drug diazaborine blocks ribosome biogenesis by inhibiting the AAA-ATPase Drg1. J. Biol. Chem. 289, 3913–3922 (2014).
Ma, C., Wu, D., Chen, Q. & Gao, N. Structural dynamics of AAA + ATPase Drg1 and mechanism of benzo-diazaborine inhibition. Nat. Commun. 13, 6765 (2022).
Pertschy, B. et al. Cytoplasmic recycling of 60S preribosomal factors depends on the AAA protein Drg1. Mol. Cell. Biol. 27, 6581–6592 (2007).
Prattes, M. et al. Visualizing maturation factor extraction from the nascent ribosome by the AAA-ATPase Drg1. Nat. Struct. Mol. Biol. 29, 942–953 (2022).
Tafforeau, L. et al. The complexity of human ribosome biogenesis revealed by systematic nucleolar screening of Pre-rRNA processing factors. Mol. Cell 51, 539–551 (2013).
Ameismeier, M., Cheng, J., Berninghausen, O. & Beckmann, R. Visualizing late states of human 40S ribosomal subunit maturation. Nature 558, 249–253 (2018).
Liang, X. et al. Structural snapshots of human pre-60S ribosomal particles before and after nuclear export. Nat. Commun. 11, 3542 (2020).
Zhang, Y. et al. Visualizing the nucleoplasmic maturation of human pre-60S ribosomal particles. Cell Res. 33, 867–878 (2023).
Singh, S., Vanden Broeck, A., Miller, L., Chaker-Margot, M. & Klinge, S. Nucleolar maturation of the human small subunit processome. Science 373, eabj5338 (2021).
Cheng, J. et al. 90S pre-ribosome transformation into the primordial 40S subunit. Science 369, 1470–1476 (2020).
Ameismeier, M. et al. Structural basis for the final steps of human 40S ribosome maturation. Nature 587, 683–687 (2020).
Vanden Broeck, A. & Klinge, S. Principles of human pre-60S biogenesis. Science 381, eadh3892 (2023).
Liu, Y., Black, J., Kisiel, N. & Kulesz-Martin, M. F. SPAF, a new AAA-protein specific to early spermatogenesis and malignant conversion. Oncogene 19, 1579–1588 (2000).
Tanaka, A. J. et al. Mutations in SPATA5 are associated with microcephaly, intellectual disability, seizures, and hearing loss. Am. J. Hum. Genet. 97, 457–464 (2015).
Buchert, R. et al. SPATA5 mutations cause a distinct autosomal recessive phenotype of intellectual disability, hypotonia and hearing loss. Orphanet. J. Rare Dis. 11, 130 (2016).
Puusepp, S. et al. Compound heterozygous SPATA5 variants in four families and functional studies of SPATA5 deficiency. Eur. J. Hum. Genet. 26, 407–419 (2018).
Zanus, C. et al. Description of a peculiar alternating ictal electroclinical pattern in a young boy with a novel SPATA5 mutation. Epileptic Disord. 22, 659–663 (2020).
Richard, E. M. et al. Bi-allelic variants in SPATA5L1 lead to intellectual disability, spastic-dystonic cerebral palsy, epilepsy, and hearing loss. Am. J. Hum. Genet. 108, 2006–2016 (2021).
Hetman, M. & Slomnicki, L. P. Ribosomal biogenesis as an emerging target of neurodevelopmental pathologies. J. Neurochem. 148, 325–347 (2019).
Ni, C. et al. Labeling of heterochronic ribosomes reveals C1ORF109 and SPATA5 control a late step in human ribosome assembly. Cell Rep. 38, 110597 (2022).
Huttlin, E. L. et al. Architecture of the human interactome defines protein communities and disease networks. Nature 545, 505–509 (2017).
Szklarczyk, D. et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47, D607–D613 (2019).
Lo, Y. H. et al. Cryo-EM structure of the essential ribosome assembly AAA-ATPase Rix7. Nat. Commun. 10, 513 (2019).
Krishnamoorthy, V. et al. The SPATA5-SPATA5L1 ATPase complex directs replisome proteostasis to ensure genome integrity. Cell 187, 2250–2268 (2024).
Chang, C. W., Lee, S. & Tsai, F. T. F. Structural elements regulating AAA+ protein quality control machines. Front. Mol. Biosci. 4, 27 (2017).
Gates, S. N. & Martin, A. Stairway to translocation: AAA+ motor structures reveal the mechanisms of ATP-dependent substrate translocation. Protein Sci. 29, 407–419 (2020).
White, S. R. & Lauring, B. AAA+ ATPases: achieving diversity of function with conserved machinery. Traffic 8, 1657–1667 (2007).
Puchades, C., Sandate, C. R. & Lander, G. C. The molecular principles governing the activity and functional diversity of AAA+ proteins. Nat. Rev. Mol Cell Biol. 21, 43–58 (2020).
Miller, J. M. & Enemark, E. J. Fundamental Characteristics of AAA+ Protein Family Structure and Function. Archaea 2016, 9294307 (2016).
Noguchi, M. et al. ATPase activity of p97/valosin-containing protein is regulated by oxidative modification of the evolutionally conserved cysteine 522 residue in walker a motif. J. Biol. Chem. 280, 41332–41341 (2005).
Bhaskar, V. et al. Dynamic association of human Ebp1 with the ribosome. RNA 27, 411–419 (2021).
Wild, K. et al. MetAP-like Ebp1 occupies the human ribosomal tunnel exit and recruits flexible rRNA expansion segments. Nat. Commun. 11, 776 (2020).
Evans, R. et al. Protein complex prediction with AlphaFold-Multimer. Preprint at https://doi.org/10.1101/2021.10.04.463034 (2022).
Jacob, C., Holme, A. L. & Fry, F. H. The sulfinic acid switch in proteins. Org. Biomol. Chem. 2, 1953–1956 (2004).
Biteau, B., Labarre, J. & Toledano, M. B. ATP-dependent reduction of cysteine-sulphinic acid by S. cerevisiae sulphiredoxin. Nature 425, 980–984 (2003).
Woo, H. A. et al. Reversible oxidation of the active site cysteine of peroxiredoxins to cysteine sulfinic acid. Immunoblot detection with antibodies specific for the hyperoxidized cysteine-containing sequence. J. Biol. Chem. 278, 47361–47364 (2003).
Chang, T. S. et al. Characterization of mammalian sulfiredoxin and its reactivation of hyperoxidized peroxiredoxin through reduction of cysteine sulfinic acid in the active site to cysteine. J. Biol. Chem. 279, 50994–51001 (2004).
Akter, S. et al. Chemical proteomics reveals new targets of cysteine sulfinic acid reductase. Nat. Chem. Biol. 14, 995–1004 (2018).
Ho, J. H., Kallstrom, G. & Johnson, A. W. Nmd3p is a Crm1p-dependent adapter protein for nuclear export of the large ribosomal subunit. J. Cell Biol. 151, 1057–1066 (2000).
Biswas, A. et al. Opposing action of casein kinase 1 and calcineurin in nucleo-cytoplasmic shuttling of mammalian translation initiation factor eIF6. J. Biol. Chem. 286, 3129–3138 (2011).
Bassler, J. et al. The conserved Bud20 zinc finger protein is a new component of the ribosomal 60S subunit export machinery. Mol. Cell. Biol. 32, 4898–4912 (2012).
Kallstrom, G., Hedges, J. & Johnson, A. The putative GTPases Nog1p and Lsg1p are required for 60S ribosomal subunit biogenesis and are localized to the nucleus and cytoplasm, respectively. Mol. Cell. Biol. 23, 4344–4355 (2003).
Sloan, K. E., Bohnsack, M. T. & Watkins, N. J. The 5S RNP couples p53 homeostasis to ribosome biogenesis and nucleolar stress. Cell Rep. 5, 237–247 (2013).
Nicolas, E. et al. Involvement of human ribosomal proteins in nucleolar structure and p53-dependent nucleolar stress. Nat. Commun. 7, 11390 (2016).
Grishina, I. & Lattes, B. A novel Cdk2 interactor is phosphorylated by Cdc7 and associates with components of the replication complexes. Cell Cycle 4, 1120–1126 (2005).
Lovejoy, C. A. et al. Functional genomic screens identify CINP as a genome maintenance protein. Proc. Natl. Acad. Sci. USA 106, 19304–19309 (2009).
Aspesi, A. & Ellis, S. R. Rare ribosomopathies: insights into mechanisms of cancer. Nat. Rev. Cancer 19, 228–238 (2019).
Farley-Barnes, K. I., Ogawa, L. M. & Baserga, S. J. Ribosomopathies: Old concepts, new controversies. Trends Genet. 35, 754–767 (2019).
Pecoraro, A., Pagano, M., Russo, G. & Russo, A. Ribosome biogenesis and cancer: Overview on ribosomal proteins. Int. J. Mol. Sci. 22, https://doi.org/10.3390/ijms22115496 (2021).
Mastronarde, D. N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005).
Kimanius, D., Dong, L., Sharov, G., Nakane, T. & Scheres, S. H. W. New tools for automated cryo-EM single-particle analysis in RELION-4.0. Biochem. J. 478, 4169–4185 (2021).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Rohou, A. & Grigorieff, N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015).
Bepler, T. et al. Positive-unlabeled convolutional neural networks for particle picking in cryo-electron micrographs. Nat. Methods 16, 1153–1160 (2019).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Pettersen, E. F. et al. UCSF Chimera–a visualization system for exploratory research and analysis. J Comput. Chem. 25, 1605–1612 (2004).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Afonine, P. V. et al. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. D Struct. Biol. 74, 531–544 (2018).
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010).
Pettersen, E. F. et al. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021).
Ran, F. A. et al. Genome engineering using the CRISPR-Cas9 system. Nat. Protocols 8, 2281–2308 (2013).
Acknowledgements
We thank the Core Facilities at the School of Life Sciences, Peking University (PKU) for help with negative staining EM; the PKU Cryo-EM Platform and J. Wang., J. Zhao. and B. Xu. in PKU Institute of Advanced Agricultural Sciences for cryo-EM data collection; the PKU High-performance Computing Platform for help with computation; the National Center for Protein Sciences (PKU Branch) for assistance with flow cytometry and mass spectrometry. The work was supported by the National Science Foundation of China (32230051 to N.G.).
Author information
Authors and Affiliations
Contributions
Y.D. prepared the samples of the SPATA5 complex and pre-ribosomal particles collected the cryo-EM data. Y.Z. and Y.D. performed EM analysis (with the help of D.W., N.L., and C.M.). Y. D. established the CRISPR KI cell lines and performed the biochemical and cellular experiments. N.G., Y.Z., and Y.D. performed cryo-EM model building and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Dai, Y., Wu, D., Li, N. et al. Cryo-EM structure of the AAA+ SPATA5 complex and its role in human cytoplasmic pre-60S maturation. Nat Commun 16, 3806 (2025). https://doi.org/10.1038/s41467-025-58894-0
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-58894-0