Abstract
Toxoplasma gondii is an extremely successful parasite infecting one third of the human population and numerous animals. It has a complex life cycle with multiple developmental stages that are key for its transmission and pathogenesis. But how the developmental programs are regulated is largely unknown. Here, we screen putative chromatin remodeling proteins in T. gondii and find that a novel complex containing an evolutionarily conserved ATPase SNF2L is critical for programming the parasite’s development. This complex contains four core proteins and conditional depletion of three of them leads to similar expression changes of developmentally regulated genes, including increased transcription of genes involved in sexual commitment and development. Accordingly, depletion of SNF2L causes merogony and out-budding types of division, which are otherwise only observed at the enteroepithelial stages within definitive hosts where sexual reproduction of the parasite occurs. After being recruited to target regions, SNF2L regulates gene expression by modulating local chromatin accessibility or by recruiting accessory proteins to its binding sites, thus ensuring that the gene expression and reproduction patterns are matched to the life cycle stages. Conditional depletion of SNF2L offers an opportunity to study the unique biology of the parasite during pre-sexual and sexual developments in vitro.
Similar content being viewed by others
Introduction
The Apicomplexa phylum contains a large group of unicellular pathogens that are of critical medical and veterinary importance, including Plasmodium parasites that cause malaria and Cryptosporidium species that cause diarrhea. Among them, Toxoplasma gondii is considered one of the most successful parasites because it is able to infect a wide variety of vertebrate hosts1,2,3,4. Although the definite hosts, in which the sexual reproduction of T. gondii occurs, are restricted to felines, the intermediate hosts include most warm-blooded animals and humans. Toxoplasma has a complex life cycle involving multiple developmental stages, which contribute to the transmission and pathogenesis of the parasite4,5,6. In fact, although different Apicomplexa parasites infect different hosts and use different approaches for transmission, they share similar developmental programs during their life cycles. For T. gondii, asexual proliferation in the forms of tachyzoites (acute infection stage) and bradyzoites (chronic infection stage) occurs in intermediate hosts. Depending on environmental conditions, these two forms can interconvert, which assists the transmission of parasites between intermediate hosts without completion of the life cycle and is a unique property of Toxoplasma life cycle6. The development of T. gondii in the intestinal epithelial cells of cats enables the production of oocysts, which contaminate the environments and transmit the pathogen to intermediate hosts. The development processes at enteroepithelial stages are complicated, involving a number of steps like merogony to produce merozoites, differentiation of merozoites to gametocytes, and fertilization of gametes to produce oocysts. Yet, little is known on how these steps are orchestrated7.
Throughout the life cycle of T. gondii, parasites have different life activities at different developmental stages. For instance, tachyzoites proliferate fast by endodyogeny. Bradyzoites also reproduce by endodyogeny, but they grow slowly and most parasites exit the cell cycle to stay at the G0 phase8,9. Merozoites also replicate asexually and very fast, but they do not exclusively use endodyogeny. Instead, merozoites probably use a combination of endodyogeny and endopolygeny for reproduction10,11,12. Merozoites are the pre-sexual stage of enteroepithelial development. After several rounds or asexual reproduction, some merozoites would start the sexual phase of life cycle by differentiating into male and female gametocytes, which then produce male and female gametes through gametogony. In addition to replication modes, parasites at different stages also differ significantly in gene expression. Numerous transcriptomic studies have looked at the gene expression profiles at diverse stages and revealed many stage-specific transcripts, which may serve as markers for parasite development13,14.
During the parasite’s life cycle, the development is largely unidirectional and the differentiation from one stage to the next is tightly regulated. Environmental changes seem to have important roles in developmental program decision15. Serum starvation or the use of spent media could induce sexual commitment (formation of non-replicative gametocytes) of the Plasmodium parasites in culture. More specifically, lysophosphatidylcholine, which is taken up by infected red blood cells to support the rapid growth of Plasmodium merozoites, is depleted from the serum and is the active serum component that regulates sexual commitment16. In Toxoplasma, nutrient starvation and stress conditions also promote the conversion of tachyzoites to slowly replicating bradyzoites. Alkaline medium (pH = 8.2), along with CO2 starvation, is the common way to induce bradyzoite formation in vitro17,18. But the underlying mechanisms are largely unknown. Over the years, many factors, particularly ApiAP2 family transcription factors, have been shown to regulate parasite development. AP2-G, whose expression is fine-tuned by transcription factors like AP2-G5 and chromatin modulators like heterochromatin protein 1 and gametocyte development 1, is key for sexual commitment of Plasmodium parasites19,20,21. T. gondii encodes 67 ApiAP2 factors and many of which are cell cycle or life cycle regulated and contribute to the developmental regulation. AP2IV-3 and AP2IX-9, two ApiAP2 factors expressed during early bradyzoite development, bind to the promoter regions of the same set of genes but have opposite roles in regulating their expression and bradyzoite differentiation22. AP2IX-9 suppresses bradyzoite formation, whereas AP2IV-3 activates it23. In addition to these two AP2 factors, many others, such as AP2Ib-1, AP2IV-4, AP2XI-4, AP2IX-4, and TgAP2IX-5 were also reported to regulate bradyzoite development by various mechanisms, including cooperation with the master regulator of bradyzoite development BFD1 (Bradyzoite formation deficient 1)24,25,26. Recently, it was reported that the epigenetic factors MORC and HADC3 form a complex with ApiAP2 factors to inhibit the expression of stage-specific genes, especially the ones specifically expressed in sexual developmental stages27. This complex keeps the local chromatin around its binding sites in a poised state to regulate target gene expression and presexual commitment of the parasites. Among the ApiAP2 factors interacting with MORC and HADC3, AP2XII-1 and AP2XI-2 form a heterodimer to suppress merogony and expression of enteroepithelial stage genes at the tachyzoite stage28,29,30.
Gene expression regulation by transcription factors requires the local chromatin to be in a permissive state that allows the access of specific regulators. As such, chromatin remodelers that alter the architecture of the chromatin also have important roles in regulating gene expression31,32. In model eukaryotes like yeasts, fruit flies, and mice, diverse chromatin remodeling complexes have been identified and they have key functions in cell growth, differentiation, and adaptation33,34. In parasites, the chromatin remodeling factor TbISWI in Trypanosoma brucei binds to the silent VSG (variant surface glycoprotein) expression sites and contributes to the repression of their expression35,36. Recent studies have shown that the master regulator AP2-G in malaria parasites regulates male gametocyte development through an SNF2-like chromatin remodeling ATPase37. The gametocyte SNF2 (gSNF2) is recruited to the upstream regions of male-specific genes and keeps the local chromatin in a nucleosome-free state to enable their expression37. ISWI and SNF2 are the core components of chromatin remodeling complexes with an ATPase domain that consists of DEXDc and HELICc subdomains. They use the energy of ATP hydrolysis to alter the chromatin architecture38,39.
In this work, we identify 16 putative chromatin remodeling ATPases in Toxoplasma gondii and explore the regulatory role of SNF2L. The results demonstrate that SNF2L is in a complex consisting of four core proteins. Inactivation of SNF2L, or other components of the complex changes the expression of many developmentally regulated genes, leading to altered reproduction mode and developmental programs of the parasites.
Results
Toxoplasma encodes a variety of proteins with signatures of chromatin remodelers
To investigate the possible roles of chromatin remodeling during Toxoplasma growth and development, we first sought to identify the proteins that might be involved in chromatin remodeling in Toxoplasma. For this purpose, the peptide sequences of the DEXDc and HELICc domains from known chromatin remodeling ATPases in Saccharomyces cerevisiae (Snf2, Sth1, Ino80, Swr1, Isw1, Isw2, Chd1, Fun30) were used to query the Toxoplasma gondii genome using protein BLAST searches in ToxoDB (https://toxodb.org/toxo/app, release 60), since DEXDc and HELICc domains are the key and iconic elements of chromatin remodeling ATPases38,39. The Toxoplasma hits with both DEXDc and HELICc domains (as determined by the SMART program) were individually used to BLAST against the Saccharomyces cerevisiae genome to make sure known yeast chromatin remodeling ATPases were among the top hits. Such analyses identified 16 putative chromatin remodeling ATPases in T. gondii (Fig. 1a). Eukaryotic organisms often encode multiple chromatin remodeling complexes that can be clustered to different groups, and four families (SWI/SNF, ISWI, NuRD/Mi-2/CHD, and INO80) are commonly seen. Phylogenetic analyses using representative ATPase sequences of the four canonical families from model organisms (Homo sapiens, Mus musculus, Drosophila melanogaster, and Saccharomyces cerevisiae) suggest that Toxoplasma has ATPases that belong to each of those four families (Fig. 1b) (Supplementary data 1). Like yeasts and mammals, Toxoplasma also has two ATPases (TGME49_321440 and TGME49_273870) in the ISWI family and two (TGME49_278440 and TGME49_320300) in the SWI/SNF family. For the two proteins in ISWI family, in-depth sequence comparisons suggest that TGME49_321440 has higher sequence similarities with mammalian SNF2H, whereas TGME49_273870 is more similar to mammalian SNF2L (Fig. 1b). Therefore, they were named TgSNF2H and TgSNF2L, respectively. The two proteins in the SWI/SNF family (TGME49_278440 and TGME49_320300) are also highly similar to each other (Supplementary data 1).
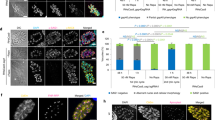
a Domain structures of the putative chromatin remodeling ATPases in Toxoplasma identified by BLAST searches. DEXDc: DEAD-like helicases superfamily (SMART #: SM00487); HELICc: helicase superfamily c-terminal domain (SMART #: SM00490); SANT: SANT SWI3, ADA2, N-CoR and TFIIIB” DNA-binding domains (SMART #: SM00717); AT_hook domain: DNA binding domain with preference for A/T rich regions (SMART # SM000384); SnAC: Snf2-ATP coupling, chromatin remodeling complex (SMART #: SM01314); zf-PARP: Poly(ADP-ribose) polymerase and DNA-Ligase Zn-finger region (SMART #: SM01336); RING: Ring finger (SMART #: SM00184); UBA_4: UBA-like domain (Pfam #: PF14555); COM: Chromatin organization modifier domain (SMART #: SM000298); DUF: DUF4208 domain (SMART #: SM001176); PHD: PHD zinc finger (SMART #: SM000249); BROMO: bromo domain (SMART #: SM000297); SNF2_N: SNF2_N domain (Pfam #: PF00176). b Phylogenetic analyses of the putative chromatin remodeling ATPases in Toxoplasma, which were done in MEGA 11 using the maximum likelihood algorithm. c IFA checking the localization of putative chromatin remodeling ATPases in Toxoplasma, as well as the depletion of their expression by IAA treatments, using strains with each of the ATPases tagged with an mAID degron (which contains an HA tag) at their C-termini in the endogenous gene loci. d Impact of chromatin remodeling ATPases depletion on parasite growth, as determined by plaque assays using mAID tagged strains with or without IAA treatment. The parental strain ME49-Tir1 was included as a control. e Impact of the depletion of chromatin remodeling ATPases on parasite division. All patterns that are different from the regular endodyogeny are classified as “Abnormal”. Means ± SD of n = 3 independent experiments, ****p < 0.0001, two-tailed unpaired Student’s t-test. Source data are provided as a Source Data file.
Next, we focused on the six proteins (TGME49_320300, TGME49_278440, TGME49_321440, TGME49_273870, TGME49_280800, and TGME49_258240) in Toxoplasma that belong to the four canonical chromatin remodeling subfamilies. First, a mini-AID (auxin inducible degron) tag (mAID) was inserted to the C-termini of each of these proteins at their endogenous gene loci (Supplementary Fig. 1a) in the type II strain ME49 Tir1, to allow auxin dependent depletion of these proteins for functional tests. Clonal strains of the tagged lines were examined by diagnostic PCRs to confirm the correct integration of the AID tag (Supplementary Fig. 1b). Through probing the expression of HA (which was fused to mAID) by immunofluorescence assays (IFA), it was evident that all six proteins were localized in parasite nuclei. In addition, treating parasites with indole-3-acetic acid (IAA), a natural auxin, efficiently depleted the expression of these proteins to undetectable levels (Fig. 1c). Using a plaque assay that estimates the overall fitness of parasites, it was found that except TGME49_258240 (TgCHD1), depletion of the other five proteins resulted in complete lack of plaque formation, indicating essential roles during parasite propagation (Fig. 1d and Supplementary Fig. 1c, d). TgCHD1 depletion also reduced parasite growth, as indicated by smaller plaques formed by the IAA-treated 258240-mAID mutants (Fig. 1d and Supplementary Fig. 1c, d). Interestingly, except TgCHD1, depletion of any of the other five putative ATPases led to altered division patterns of the parasites. Mutants lacking TGME49_273870 (SNF2L) had the most severe changes (Fig. 1e) after 24 h of IAA treatment, since 75% of the parasites displayed abnormal (any pattern that was not classic endodyogeny) division patterns that resembled certain stages of enteroepithelial development in cats. Therefore, we decided to investigate this protein further to examine its roles during parasite growth and development.
SNF2L is a conserved protein widely present in eukaryotes, from yeast to plants and humans (Supplementary Fig. 2a). These proteins contain DEXDc and HELICc motifs at the N-terminal part to make the ATPase domain, whereas the C-terminal part typically contains DNA binding and nucleosome recognition modules (Supplementary Fig. 2b). To further examine its expression patterns in Toxoplasma, we constructed a SNF2L-3*HA strain, whose endogenous SNF2L was tagged with three tandem HA tags (Supplementary Fig. 2c, d). IFA suggested that SNF2L was expressed at all stages of the cell cycle of tachyzoites, and its protein level did not seem to fluctuate significantly during the cell cycle (Supplementary Fig. 2e).
SNF2L depletion altered the reproduction modes of tachyzoites
To study the physiological roles of SNF2L, the SNF2L-mAID (273870-mAID, also called iSNF2L) strain was used. Consistent with the IFA results (Fig. 1c), IAA efficiently depleted SNF2L to undetectable levels after 6 h of treatment, as determined by Western blotting (Fig. 2a). The lack of plaque formation after SNF2L depletion (Fig. 1d) prompted us to check whether parasite replication was affected. Thus, the proliferation rates of the SNF2L-mAID strain cultured in the presence or absence of IAA for 24 h were compared. Although IAA treatment did not affect the replication of the parental strain (Fig. 2b), it significantly reduced the replication of iSNF2L, as indicated by the reduced number of parasites in each parasitic vacuole (PV) (Fig. 2b). More importantly, SNF2L seemed to alter the reproduction modes of the parasites. Wildtype tachyzoites replicate through endodyogeny and the PVs typically contain 2n parasites (1, 2, 4, 8, 16 etc.) (Fig. 2b). However, this was not the case in SNF2L depleted mutant, whose PVs could contain any number (1, 2, 3, 4, 5 etc.) of parasites (Fig. 2b), a phenomenon that was also observed in Toxoplasma mutants lacking AP2XII-1 or AP2XI-2, which were thought to propagate by merogony28,29. These results suggest that SNF2L depleted mutant might also proliferate by merogony. To further check the reproduction mode, the parasites were subjected to IFA analyses using GAP45 or IMC7 and IMC1 to probe the mother parasites and newborn daughters, respectively28,40. Without IAA treatment, nearly all dividing parasites of the iSNF2L strain underwent endodyogeny (Fig. 2c, d). In contrast, the division of IAA-treated parasites was significantly altered (Fig. 2c–e). Six hours after treatment, the earliest time point at SNF2L was depleted to undetectable levels, no change in division patterns could be observed. With 12 h of IAA treatment, early signs of endopolygeny started to emerge. When depletion was induced for 24 h, endopolygeny became common and early signs of out-budding could be observed (Fig. 2c). After 36 h IAA treatment, out-budding was frequently seen (Fig. 2c, d). These altered division patterns often led to daughter parasites that differed significantly in size and sometimes even anucleated parasites (Fig. 2d, e). With IAA treatment for 36 h, roughly 75% of the dividing parasites displayed abnormal (any form other than endodyogeny) division patterns (Fig. 2d). Among them, about 11% of the dividing parasites underwent merogony (Fig. 2c–e), characterized by three or more nuclei in a dividing mother cell. Moreover, over 38% of them were in the process of out-budding, in which a daughter parasite budded off from the surface of a mother parasite (Fig. 2c–e). The out-budding type of division is somewhat similar to microgametocytogenesis in wildtype parasites that produces male gametes during sexual development10,11,12. Nonetheless, we did not see mature male gametes formation in the SNF2L-depleted parasites, since no parasites with obvious flagella were detected. Together, these results suggest that SNF2L depletion leads to reproduction modes that resemble the enteroepithelial stages in definitive hosts.
a Western blotting demonstrating the depletion of SNF2L expression by IAA treatment in the SNF2L-mAID (iSNF2L) strain. ALD was included as a loading control. b Intracellular replication assay illustrating the reduced growth rates and chaotic division patterns of the SNF2L depletion mutants. The number of parasites in each PV of the iSNF2L and ME49-Tir1 strains treated with or without IAA for 24 h (with 12 h of ± IAA pretreatment before the replication assay) was counted. Means ± SEM of n = 3 independent experiments, ***p < 0.0001, Kolmogorov-Smirnov test. SNF2L depletion leads to altered modes of parasite reproduction, as revealed by IFA staining of IMC1, IMC7, or GAP45 in the iSNF2L/IMC7-Myc (c) or iSNF2L (d) strains. The +IAA parasites in (d) were treated with IAA for 36 h in total (12 h of pretreatment in T25 flasks followed by 24 h of treatment on coverslips before imaging). Arrowheads in (c) indicate parasites with replicating/segregating nuclei but no daughter cell formation, whereas arrows indicating endopolygeny-like division. Yellow arrows in (d) indicate splitting of mother parasite or its nucleus, whereas arrowheads indicate parasites without nuclear DNA. e Percentage of SNF2L-depleted parasites undergoing indicated types of reproduction described in (d). More than 100 vacuoles with identifiable mode of reproduction were analyzed in n = 3 independent experiments. Data are presented as means ± SD. Source data are provided as a Source Data file.
To further check the unusual division patterns of the SNF2L-depleted parasites, transmission electron microscopy was used. Without IAA treatment, essentially all dividing parasites underwent endodyogeny, with two daughter cells emerging internally from a mother parasite and each with an intact nucleus (Fig. 3a). In contrast, endodyogeny was barely observed in SNF2L depleted parasites. Instead, the out-budding type of division at different stages was frequently detected (Fig. 3b–g). During this process, a portion of the nuclear materials of the mother cell was “split” into the daughter cell, and then the daughter cell was pinched off the surface of the mother cell to generating a newborn parasite (Fig. 3b–g). It is worth noting that the segregation of mother nuclear materials to daughter parasites in this out-budding division did not guarantee an intact nucleus for the daughter cell. In fact, the amount of nuclear DNA inherited by daughter parasites seemed to be random, and some daughters had little to no nuclear DNA. Out-budding could occur in mother parasites without fully segregated nuclei, with nuclear splitting and daughter cell budding occurred simultaneously (Fig. 3c). It also occurred in mother parasites with fully segregated nuclei and one daughter cell took one separated nucleus during budding (Fig. 3e). Interestingly, some of the SNF2L depleted parasites were found to contain multiple mitochondria (Fig. 3g), instead of just one in wildtype tachyzoites. Multiple mitochondria were also described in male gametes and mature gametocytes6,12, which further supports that the out-budding type of division in SNF2L depleted mutants is similar to the microgametocytogenesis process6,11,12. Together, these data indicate that SNF2L-depleted mutants exhibited morphological and division characteristics of enteroepithelial stage parasites.
a endodyogeny of the iSNF2L strain without IAA treatment. Each mother cell contained two daughter cells (D), and each daughter cell had a single nucleus (Nu). b–g reproduction patterns and morphologies of the iSNF2L strain treated with IAA, which displayed out-budding (bud) type of division. In addition, some parasites contained multiple mitochondria (Mi). Source data are provided as a Source Data file.
SNF2L depletion results in altered expression of developmentally regulated genes
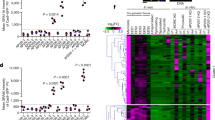
To understand how and why the deletion of SNF2L led to changes in the reproduction modes of the parasites, we first performed transcriptomic analyses to examine the global gene expression changes after SNF2L depletion. The SNF2L-mAID strain treated with IAA for 0 h, 12 h and 24 h was subjected to RNA-Seq analyses (Supplementary Fig. 3a). Using DESeq2 to call for differentially expressed genes (DEGs) with the criteria of adjusted p (Q) value < 0.05 and fold change > 2, a substantial number of DEGs were identified (Supplementary Fig. 3b–e). Moreover, the gene expression changes became more profound following longer depletion of SNF2L (Supplementary Fig. 3f). After 12 h of IAA treatment to deplete SNF2L, 301 parasite genes were found to be upregulated and 193 were downregulated (Supplementary data 2). With 24 h of IAA treatment, 976 genes were upregulated and 646 were downregulated (Supplementary data 2). Importantly, 79.4% (239 / 301) and 59.1% (114/193) of genes that were up- or down-regulated after 12 hours of IAA treatment were also up- or down-regulated after 24 hours of treatment (Supplementary Fig. 3f). We then focused our analyses on samples treated for 12 h, to minimize secondary effects of SNF2L depletion on gene transcription. Of these 494 DEGs (12 h/0 h), 82.8% of them are developmentally regulated, meaning that they have different expression levels during different stages of the life cycle. Notably, 81% (244 out of 301) of the genes with increased mRNA abundance upon SNF2L depletion matched those upregulated during enteroepithelial or chronic infection stages compared to tachyzoites41,42,43. Among these 244, the majority (135) were genes upregulated in merozoites (Fig. 4a). To further demonstrate the similarities in gene expression change upon SNF2L depletion and during parasite development, we examined the transcription patterns of the 301 genes that were upregulated in SNF2L deficient parasites across different developmental stages in wildtype parasites44. The results showed that, nearly 46.5% of the genes were upregulated in bradyzoites compared to tachyzoites, whereas 67.8% were increased in enteroepithelial stages (EES1-EES5) and 35.9% were upregulated in both bradyzoites and enteroepithelial stages (Fig. 4b). When the top 30 upregulated genes (excluding hypothetical proteins) in the SNF2L depleted mutants were subjected to similar analyses, it was evident that the majority of them were merozoite specific genes, including merozoite markers like GRA11B and AAH245,46 (Fig. 4c). In addition, SNF2L depletion also resulted in the increased transcription of gamete-specific genes, such as PF16, HAP2, and TGME49_30633847,48,49 (Fig. 4d). Quantitative RT-PCR on selected genes further confirmed their increased transcript levels in SNF2L depletion mutants (Supplementary Fig. 3g–m). To check whether the changes in transcript levels affect the protein abundance of the corresponding genes, TMT (Tandem Mass Tags)-based quantitative proteomics was used to compare the protein level changes upon SNF2L depletion (12 h of IAA treatment). The results from three repeated experiments indicated that a number of genes with increased mRNA levels also showed increased protein abundances, including merozoite markers like PAN (TGME49_209920), GRA11B, and MIC17B in SNF2L depleted parasites (Fig. 4e). Furthermore, a Ty tag was inserted to the C terminus of endogenous PF16, a microgamete specific gene upregulated in the SNF2L depletion mutants27, to construct the iSNF2L PF16-Ty strain (Supplementary Fig. 4). IFA results suggest that PF16-Ty could be detected in the absence of SNF2L, but not in its presence (Fig. 4f), suggesting that SNF2L depletion did increase the protein abundance of proteins like PF16.
a Venn diagram showing the overlap of genes upregulated (≥2-fold) in the SNF2L-depleted mutants with the genes that are upregulated in wildtype merozoites (Mz), bradyzoites (Bz), or sporozoites (Sp) compared with tachyzoites (Tz). Data for the latter are from published datasets available in ToxoDB. ***p < 0.0001, Pearson’s Chi-squared tests indicating genes upregulated in the SNF2L depletion mutant were significantly enriched in gene sets associated with bradyzoites, merozoites, or sporozoites. b For the 301 genes that were upregulated in the SNF2L-depleted mutants, their expression patters in wildtype strain during the life cycle were plotted as a heatmap using data from ToxoDB. c Life cycle expression patterns of the top 30 upregulated genes (excluding hypothetical proteins) in the SNF2L-depleted mutants were plotted as a heatmap. d expression patterns HAP2, PF16, and TGME49_30633839 during the parasites’ life cycle, as well as in iSNF2L mutants with or without IAA treatment. e Volcano plot showing the abundance change of proteins in the iSNF2L strain before and after IAA treatment for 12 h, as determined by TMT-based quantitative proteomics. Data from three biological replicates were plotted. Raw data are shown in Supplementary data 3. f Expression of PF16 in the SNF2L-depleted mutant, as determined by IFA on the iSNF2L/PF16-Ty strain with or without IAA treatment for 60 h. Source data are provided as a Source Data file.
Among the 193 genes downregulated in the SNF2L depleted mutants after 12 h of IAA treatment, 85.5% (165/193) were also developmentally regulated (Supplementary Fig. 5a). However, in contrast to the 301 upregulated genes, which were mostly upregulated during enteroepithelial or chronic infection stages, the 193 downregulated genes did not show a uniform pattern of expression change across development. Roughly half of them were upregulated during enteroepithelial or chronic infection stages, whereas the other half were downregulated during those stages (Supplementary Fig. 5b–e). Nonetheless, taken together, these results clearly show that SNF2 has a critical role in controlling the expression of developmentally regulated genes.
SNF2L binds to promoter regions of target genes and alters local chromatin accessibility
To elucidate the underlying mechanisms responsible for the gene expression changes in SNF2L depletion mutants, CUT&Tag followed by DNA sequencing was used to analyze the binding sites of SNF2L in the Toxoplasma genome. Chromatin bound by SNF2L was precipitated by an HA antibody (a naïve mouse IgG was used side by side as a negative control) from the lysate of the SNF2L-3*HA strain. Subsequent DNA sequencing showed that SNF2L was enriched near the transcription start sites (TSS) of genes, with 91.7% of the binding sites located in the promoter regions (Fig. 5a–c). Although the SNF2L binding sites were scattered across the chromosomes (one example is given in Fig. 5b for chromosome XII), a closer look suggested that many of the genes upregulated in the SNF2L depletion mutants, such as GRA80, GRA82 and MIC17A, were targeted by SNF2L near their promoter regions (Fig. 5d). To further examine the specific binding of SNF2L to chromatin sites, similar CUT&Tag analyses on the iSNF2L strain were performed, using parasites that were treated with or without IAA for 12 h (the +IAA group served as negative controls). The results showed that the SNF2L binding sites revealed by these two CUT&Tag experiments (one done with the SNF2L−3*HA strain and the other with the iSNF2L strain) were quite similar (Fig. 5b). About 80% of the genes identified by CUT & Tag in iSNF2L were also identified by CUT & Tag in SNF2L-3*HA. Together, these results suggest that CUT & Tag did identify the specific binding sites of SNF2L on the chromosomes.
a Enrichment of SNF2L near the transcription start site (TSS) (from −3 Kb to 3 Kb region centered around TSS), as determined by CUT&Tag in the SNF2L-3HA strain using an HA antibody. Pull-down with a naive mouse IgG was included as a control. b Genome browser view of the position and intensity of DNA sequences bound by SNF2L. Data were derived from (a) and similar CUT&Tag experiments done in the iSNF2L strain with or without 12 h IAA treatment. Chromosome XII was shown as an example. c Distribution of SNF2L on different types of genetic elements, derived from the analysis of data in (a). d Enrichment of SNF2L at the upstream regions of selected genes, using data from (a). Meanwhile, the accessibility of chromatin revealed by ATAC-Seq at these regions in the iSNF2L strain treated with or without IAA was also included for comparison. e, f Overall chromatin accessibility in the iSNF2L strain treated with or without IAA, as determined by ATAC-Seq (e). Chromosome XII was shown as an example (f). g accessibility of different types of genetic elements in the iSNF2L strain with and without IAA treatment, derived from the analysis of data in (e). h accessibility of the upstream regions of genes that are upregulated (upper panel) or downregulated (lower panel) in the SNF2L-depleted parasites. Means ± SEM were plotted. Source data are provided as a Source Data file.
Since SNF2L is predicted to be a chromatin remodeler, we sought to test whether it had a role in modifying the chromatin architecture. For this purpose, ATAC-seq (Assay for Transposase-Accessible Chromatin with high-throughput sequencing) was used to determine the chromatin accessibility in the SNF2L-mAID mutant before and after IAA treatment (12 h). The results showed that the overall chromatin accessibility was significantly reduced in the SNF2L-depleted parasites, and the reduction mostly occurred at the TSS regions (Fig. 5e–g). Moreover, >54% of the genes with reduced chromatin accessibility were identified as targets of SNF2L by CUT&Tag in both SNF2L−3*HA and iSNF2L strains. The reduced chromatin accessibility after SNF2L depletion is consistent with the predicted role of SNF2L as a chromatin remodeler. To more precisely assess how gene transcription might be affected by SNF2L, the chromatin accessibility of genes that were up- or down-regulated in the SNF2L-depleted mutants was analyzed separately. The ATAC-seq data suggest that although the chromatin accessibility around both sets of genes was decreased after SNF2L depletion, the decrease seemed to be more profound in downregulated genes (Fig. 5h), suggesting that SNF2L might affect the expression of these genes by different mechanisms. For upregulated genes like GRA80 and MIC17A, SNF2L depletion reduced chromatin accessibility by 40% around their promoter regions, but they were still reasonably accessible (Fig. 5h, d). In contrast, the chromatin accessibility was reduced by nearly 70% around the promoters of genes downregulated in the SNF2L depletion mutants (Fig. 5h).
SNF2L forms a complex with novel nuclear factors
Chromatin remodeling complexes typically contain multiple subunits, with an ATPase as the core. To check whether Toxoplasma SNF2L is also in a complex with other proteins, co-immunoprecipitation (Co-IP) was performed in the SNF2L-3*HA strain to precipitate SNF2L and its interacting proteins, which were subsequently identified by mass spectrometry (MS). A number of proteins, including AP2X-4 and two uncharacterized proteins TGME49_203980 (herein named SLIF1 for SNF2L Interacting Factor 1) and TGME49_217480 (herein called SLIF2 for SNF2L Interacting Factor 2) were identified as top hits, because peptides derived from these proteins were enriched in the experimental group with HA antibody for Co-IP, compared to the control group using naïve mouse IgG (Fig. 6a). To further check whether these identified proteins do form a complex, we first tagged the C termini of SLIF1 and SLIF2 with an mAID (which contained an HA tag) epitope at their endogenous gene loci (Supplementary Fig. 6a–c). Subsequently, a similar Co-IP approach using the mAID-tagged lines (including AP2 X-4-mAID that was constructed previously50) was used to identify binding partners for each of these proteins. The reciprocal Co-IP and MS results show that SNF2L did form a complex with SLIF1, SLIF2, and AP2X-4 (Fig. 6b–d). Except that there was probably no direct interaction between AP2X-4 and SLIF2, the other components of the complex were always identified as top hits in any given Co-IP/MS experiments (Fig. 6a, d and Supplementary data 4). Analyzing the Co-IP/MS results with the SFINX (Straightforward Filtering IndeX) program51,52 suggest that SNF2L, SLIF1, SLIF2, and AP2X-4 form the core of the complex (Fig. 6e and Supplementary data 5).
a–d Protein hits from co-IP experiments that were used to identify binding partners for the prey proteins (marked in red). The number of unique peptides derived from untagged (Ctrl) and HA tagged (HA) strains was used to generate these graphs. e co-IP results analyzed by the Straightforward Filtering IndeX (SFINX) to show the possible protein interactions in the SNF2L-containing complex. Raw data are shown in Supplementary data 5. f–h co-IP and Western blotting examining the interactions between SNF2L, SLIF1, and SLIF2. i Cartoons showing the SNF2L/ISW2 chromatin remodeling complexes in indicated organisms. j–l Heat maps showing the life cycle expression patterns of genes upregulated (≥2-fold) in SLIF1, SLIF2, or AP2X-4 depleted mutants. Data were derived from ToxoDB. m Venn diagram showing the overlap of genes upregulated (≥2-fold) in SNF2L, AP2X-4, and SLIF1 depleted mutants. Source data are provided as a Source Data file.
To further validate the protein interactions within the SNF2L-containing complex, each of the subunits was precipitated from the corresponding HA or mAID-tagged line with an HA antibody, then Western blotting was used to detect the presence of other subunits, using antibodies raised against them. As examples, the interactions between SNF2L and SLIF1, as well as SLIF2, were confirmed (Fig. 6f–h). These data suggest that SNF2L, SLIF1, SLIF2, and AP2X-4 do form a complex. Interestingly, except humans (which have three), the ISWI family chromatin remodeling complexes in other model organisms like yeast and fruit fly all have four proteins as core components (Fig. 6g), suggesting a conserved working mechanism. On the other hand, except SNF2L, which is highly conserved, the other three components (SLIF1, SLIF2, and AP2X-4) of the Toxoplasma complex have very low sequence similarities with proteins outside apicomplexa. This suggests that the targets regulated by the Toxoplasma complex may have different sequence properties or chromatin modifications than the SNF2L targets in other eukaryotes.
Similar to SNF2L, AP2X-4 has been shown before to be critical for tachyzoite proliferation and proper expression of life cycle-regulated genes50. To check the function of the two uncharacterized components (SLIF1 and SLIF2) in the complex, the mAID-tagged strains were used for phenotypic analyses (Supplementary Fig. 6a–c). IAA treatment for one hour could efficiently induce the degradation of both SLIF1 and SLIF2 in corresponding tagged lines (Supplementary Fig. 6d, e). Plaque assays showed that SLIF1 was essential for tachyzoite growth, since IAA-treated SLIF1-mAID failed to produce any visible plaques. On the other hand, SLIF2 seemed to be dispensable, as IAA treatment did not affect plaque formation of the SLIF2-AID strain (Supplementary Fig. 6f–k). Intracellular replication assays showed that depletion of SLIF1 caused slower proliferation and altered mode of reproduction of the parasites (Supplementary Fig. 7a–c), similar to the depletion of SNF2L. Whereas the replication rates and modes of reproduction of the SLIF2-depleted mutants were indistinguishable from those of the parental strain (Supplementary Fig. 7b–d). Depletion of AP2X-4 also did not alter the division pattern, as the parasites still proliferated by endodyogeny (Supplementary Fig. 7e).
RNA-Seq was used to assess the gene transcription changes upon depletion of SLIF1, SLIF2, or AP2X-4. Similar to the SNF2L-deficient mutants, parasites lacking any of the three genes displayed altered expression of developmentally regulated genes. In particular, depletion of SLIF1 led to increased mRNA levels of 131 genes, 67% of which are the ones upregulated in bradyzoites and enteroepithelial stages during normal development of wildtype parasites (Fig. 6j). Depletion of AP2X-4 increased the transcript levels of 1129 genes and 82% of them are upregulated in bradyzoites and enteroepithelial stages (Fig. 6k). Relatively speaking, disruption of SLIF2 resulted in smaller gene expression changes. Of the 19 genes upregulated in the SLIF2-depleted mutants, 94% are upregulated in bradyzoites and enteroepithelial stages during normal development (Fig. 6I). Interestingly, 70.8% of the genes upregulated in the SNF2L depletion mutants were also upregulated in the AP2X-4 deficient mutants. Similarly, 62.6% of the genes upregulated in the SLIF1 depleted mutants were also upregulated in the AP2X-4 deficient mutants (Fig. 6m). Moreover, majority of the genes upregulated in all three mutants (lacking SNF2L, AP2X-4 or SLIF1) were merozoite specific markers, such as GRA11A, GRA11B, and GRA82 (Fig. 6m). These results are consistent with that these factors are in a complex to control the expression of developmentally regulated genes.
Discussion
Toxoplasma and other apicomplexan parasites have similar developmental programs that are tightly regulated during their life cycles. Previous studies have shown that transcriptional and epigenetic regulations play key roles in controlling the parasites’ developmental programs1,7,53. In this work, we find that the chromatin architecture is also a crucial factor that regulates parasites’ life cycle progression. Like other eukaryotes, T. gondii encodes multiple chromatin remodeling complexes with distinct ATPases. Among these complexes, we find that the SNF2L-containing one has critical roles in regulating parasites’ developmental programs, including reproduction modes and gene expression patterns. Loss of the core component SNF2L promoted tachyzoites to proliferate by merogony and out-budding, suggesting commitment to enteroepithelial stage development. This is the first time out-budding was observed in parasites cultured in vitro since this type of proliferation normally only occurs during microgametocytogenesis that produces male gametes from microgametocytes11,12. In our SNF2L-depleted mutants, over 35% of the dividing parasites were undergoing out-budding (Fig. 2e). In this regard, SNF2L depletion offers a unique opportunity to study this process in vitro, which is otherwise not possible.
Like the homologous ISW2 complex in Saccharomyces cerevisiae, the SNF2L-containing chromatin remodeling complex of Toxoplasma gondii also consists of four subunits, suggesting conserved modes of action54. During chromatin remodeling, the Itc1 and Dpb4 subunits of the yeast ISW2 complex bind extranucleosomal DNA, and Dpb4 serves as the anchor point for the complex on DNA. Dls1, on the other hand, forms a dimer with Dpb4 to promote the nucleosome spacing activity of the ISW2 complex55,56,57. The SLIF1, SLIF2, and AP2X-4 proteins in the T. gondii SNF2L complex may have similar modes of function as the subunits in the ISW2 complex, but require further investigations. In addition to structural similarities, the T. gondii SNF2L complex and the yeast ISW2 complex may also have similar functions. In response to environmental changes like nutrient starvation, S. cerevisiae cells quit mitotic proliferation and initiate a differentiation process that leads the formation of haploid spores through meiosis58. During mitotic proliferation, ISW2 is recruited to specific regions by the Ume6 repressor to repress the expression of early meiotic genes, thus inhibiting spore formation59,60. Similarly, the SNF2L complex in T. gondii was critical for repressing the expression of developmentally regulated genes, especially enteroepithelial stage-specific genes, at the tachyzoite stage to ensure the parasites proliferate rapidly by endodyogeny. Such structural and functional similarities between the ISWI family chromatin remodeling complexes in different organisms suggest their conservation during evolution. On the other hand, except the core ATPases SNF2L, other subunits of the Toxoplasma SNF2L complex do not have homologs in model eukaryotes like yeasts or humans. In fact, homologs of SLIF1, SLIF2, and AP2X-4 can only be found in Coccidia parasites (Supplementary data 1), suggesting that they may have specific functions in regulating these parasites’ lifestyle. Interestingly, the SNF2L remodeler in Plasmodium falciparum was reported very recently, and it was also found to interact with AP2 transcription factors and epigenetic factors like MORC, similar to SNF2L in Toxoplasma. In addition, SNF2L in P. falciparum was also found to regulate the sexual and asexual developments of the parasites61. Similar functions of SNF2L in Toxoplasma and Plasmodium suggest that the roles of SNF2L may be conserved in Apicomplexa parasites. Nonetheless, the composition of the SNF2L complexes may be different in these organisms, since the SLIF1 and SLIF2 proteins that are core of the Toxoplasma complex do not seem to be present in P. falciparum (Supplementary data 1). Interestingly, Pachano et al. also examined the role of SNF2L in Toxoplasma recently, and they found similar binding partners of SNF2L like we did. They also showed that SNF2L was critical for parasite growth. However, depletion of SNF2L in their study did not affect chromatin accessibility and had little effect on gene expression, which is different than what we saw. Such discrepancy is likely due to the incomplete depletion of SNF2L in the TgSNF2L-mAID strain in the Pachano et al. study. After 48 h IAA treatment, SNF2L could still be detected by Western blotting62. In contrast, treating our iSNF2L strain with IAA for 6 h depleted SNF2L to undetectable levels (Fig. 2a).
Recently, inactivation of a number of factors in T. gondii, such as MORC, HDAC3, AP2XII-1, and AP2XI-2, has been shown to increase the transcription of enteroepithelial stage genes, like the depletion of SNF2L did27,28,29. Indeed, when the gene expression patterns of different mutants were compared, it was clear that most (77.4%) of the genes upregulated in the SNF2L depletion mutant were also upregulated in the MORC depletion mutant. Many of these genes also showed increased transcription in AP2XII-1 or AP2XI-2 depleted mutants (Supplementary Fig. 8). These results suggest that there are probably functional connections between these factors. Interestingly, MORC and HDAC3 were identified as potential interacting proteins of SNF2L and AP2X-4 (Supplementary data 4), respectively, suggesting interactions between the MORC complex and the SNF2L complex. Following these observations, we propose a model which the MORC and the SNF2L complexes work together to ensure the correct expression patterns of developmentally regulated genes at the tachyzoite stage. The SNF2L complex is placed to the upstream regions of the developmentally regulated genes, likely through AP2X-4 or the two novel SNF2L-interacting factors SLIF1 or SLIF2. Then, for the genes that will be upregulated along bradyzoite and/or enteroepithelial development, the chromatin remodeling activity of the SNF2L complex makes their upstream regions accessible for binding by factors like AP2XII-1 and AP2XI-2, which further recruit MORC/HDAC3 to inhibit the transcription of these genes at the tachyzoite stage. At these sites, there is likely a direct interaction between the MORC complex and the SNF2L complex, as the coIP/MS results indicated. As such, depletion of SNF2L resulted in release of AP2XII-1, AP2XI-2, and MORC/HDAC3 suppressors from the target sites, leading to increased expression of target genes. For the genes that will be downregulated along bradyzoite and/or enteroepithelial development, loading of the SNF2L complex to their promoter regions makes them accessible to the transcriptional machinery for active transcription. Accordingly, removal of SNF2L complex from those sites would decrease the chromatin accessibility and inhibit the transcription of corresponding genes. In consistent with this hypothesis, CUT&Tag results from our work and others showed that, although SNF2L was found at the upstream regions of genes that were both up- and down-regulated in the SNF2L depletion mutants, AP2XII-1, AP2XI-2, MORC and HDAC3 were only found at the upstream regions of genes upregulated in SNF2L depletion mutants (Supplementary Fig. 9). These results suggest that the up- and down-regulated genes are controlled by SNF2L through different mechanisms.
Our results show that SNF2L prevents out-budding or other types of reproduction and controls the expression of developmentally regulated genes in tachyzoites to ensure rapid parasite proliferation by endodyogeny. During life cycle progression, the activity of the SNF2L complex must be modulated so that the parasites can transit from one developmental program to another. To assess how SNF2L may be regulated, we estimated the binding partner differences of SNF2L under different conditions. The SNF2L−3*HA strain was cultured under normal tachyzoite conditions, or induced with alkaline medium for bradyzoite formation. Then, proteins bound to SNF2L were co-immunoprecipitated and identified by mass spectrometry. The results showed that under all tested conditions, the four core subunits of the SNF2L complex could always be co-immunoprecipitated. On the other hand, the accessory proteins binding to the SNF2L complex did vary under different growth conditions. For example, during induction of bradyzoite transition, SNF2L seemed to bind to different ApiAP2 and epigenetic factors (Supplementary Fig. 10 and Supplementary data 6). The change of binding partners may be a way to regulate the activity of the SNF2L complex during parasite development, which deserves further investigation.
Methods
Parasite cultures
All strains used in this study are listed in Supplementary Tables 1. They were propagated in human foreskin fibroblasts (ATCC, USA) that were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Sigma-Aldrich, USA) supplemented with 10% fetal bovine serum (Gibco, USA), 2 mM glutamine, and 1% penicillin-streptomycin. The cultures were tested (by qualitative PCR) monthly to make sure they were free of mycoplasma contamination. IAA (Sigma-Aldrich, USA) at a final concentration of 500 μM was used to induce degradation of mAID-tagged proteins, when needed63,64.
Plaque and intracellular replication assays
Freshly egressed tachyzoites were harvested, purified, and resuspended in DMEM medium. For the plaque assays, purified parasites were used to infect HFF monolayers seeded in 6-well plates (about 200 parasites per well) and cultured for 10 days under standard tachyzoite growth conditions. The samples were then fixed with 4% paraformaldehyde and stained with 0.1% crystal violet to visualize the plaques65. For intracellular replication assays, purified parasites were used to invade HFF monolayers seeded on coverslips in 24-well plates for 10 min. Then, noninvaded parasites were washed away with DMEM, and the invaded ones were cultured for indicated amount of time. Subsequently, the samples were then fixed with 4% paraformaldehyde, and the number of parasites in each PV was determined by IFA staining using rabbit anti ALD65. Each experiment was repeated three times independently.
Reagents
The following antibodies were used in the immunofluorescence, immunoblotting, co-IP, and CUT&Tag assays: Rabbit anti ALD polyclonal and mouse anti Ty monoclonal antibodies (provided by Prof. David Sibley at Washington University in St. Louis), mouse anti HA tag (the TANA2 clone (for CUT&Tag), MBL International Corporation, Japan; 12CA5 (for all rest applications), Abcam, China), rabbit anti IMC1 (provided by Dr. Qun Liu from China Agricultural University), mouse anti GAP45 (homemade), mouse anti Centrin1 (the 20H5 clone, Sigma-Aldrich, USA), mouse anti SNF2L (homemade), mouse anti SLIF1 (homemade), HRP-labeled goat anti-rabbit or goat anti-mouse IgG (Beyotime Biotechnology, China), Alexa Fluor 488 or 594 conjugated goat anti-mouse or goat anti-rabbit IgG (Fisher Scientific. USA).
Identification and phylogenetic analyses of putative chromatin remodeling ATPases in Toxoplasma gondii
The sequences of the DEXDc (SMART ACC: SM000487) and HELICc (SMART ACC: SM000490) domains of Saccharomyces cerevisiae chromatin remodeling ATPases (Swi2/snf2, Sth1, Isw1, Isw2, Ino80, Swr1, Chd1 and Fun30) were identified by SMART, and were then used as queries to do protein BLAST searches in the ToxoDB database (https://toxodb.org/toxo/app/). Hits were individually analyzed by SMART, and the ones with both DEXDc and HELICc domains were kept for further analyses. Each of the Toxoplasma proteins with both DEXDc and HELICc domains was used as queries to BLAST against Saccharomyces cerevisiae in NCBI. Only the ones that could identify known Saccharomyces cerevisiae chromatin remodeling ATPases as top hits were considered chromatin remodeling ATPases in T. gondii. For phylogenetic analyses, selected sequences were first aligned in Clustal W. Then, phylogenetic analyses were performed in MEGA 11, using the Maximum Likelihood algorithm based on the Jones-Taylor-Thornton (JTT) model. Protein sequences were extracted from Uniport and their access numbers are as follow. Saccharomyces cerevisiae proteins: Snf2, P22082; Sth1, P32597; Isw1, P38144; Isw2, Q08773; Ino80, P53115; Swr1, Q05471; Chd1, P32657. Drosophila melanogaster proteins: Brm, P25439; ISWI, Q24368; INO80, Q9VDY1; Domino, Q9NDJ2; CHD1, Q7KU24. Mouse proteins: SMCA2, Q6DIC0; SMCA4, Q3TKT4; SNF2H, Q91ZW3; SNF2L, Q6PGB8; INO80, Q6ZPV2; EP400, Q8CHI8; CHD1, P40201. Human proteins: SMCA2, P51531; SMCA4, P51532; SNF2H, O60264; SNF2L, P28370; INO80, Q9ULG1; EP400, Q96L91; SRCAP, Q6ZRS2; CHD1, O14646. Toxoplasma gondii proteins: TGME49_320300, A0A125YUI6; TGME49_278440, S8GAJ1; TGME49_273870, S8F5C2; TGME49_321440, S8GK42; TGME49_280800, S8GMT5; TGME49_258240, S8F110.
Construction of plasmids and transgenic parasite strains
All plasmids used in this study, as well as the methods for their construction, are listed in Supplementary Table 2. All primers used were synthesized by a commercial provider (Tsingke Biotechnology Co., Ltd, China) and are listed in Supplementary Table 3. The locus-specific CRISPR plasmids were generated according to previously described protocols63,66. All other plasmids were constructed using the ClonExpress MultiS Cloning Kit (Vazyme Biotech, China) for multi-fragment cloning. The pUC19-ME49SNF2L-mAID-HXGPRT plasmid was constructed by recombining the fragments of mAID-3HA-HXGPRT, 5′ and 3′ homologous arms of SNF2L into the plasmid pUC19. The mAID construct was amplified from pTUB1::YFP-mAID-3HA-HXGPRT (Addgene plasmid no. 87259), whereas the 5′ and 3′ homologous arms were amplified from genomic DNA of the ME49 Δhxgprt TIR1 strain67. The pUC19-ME49SNF2L−3HA-DHFR plasmid was constructed by cloning the fragments of 3HA-DHFR, the 5′ and 3′ homologous arms of SNF2L, into the plasmid pUC19. The 5′ and 3′ homologous arms were amplified from genomic DNA of ME49. The pUC19-PAN-Ty-DHFR*, pUC19-TGME49_306338-Ty-DHFR*, pUC19-HAP2-Ty-DHFR*, pUC19-PF16-Ty-DHFR*, and pUC19-IMC7-MYC-DHFR* fragments (which were amplified from pUC19-Ty-DHFR*) and the homologous arms of corresponding genes (which were amplified from the genomic DNA of the ME49 strain) into pUC19 by multi-fragment cloning.
All transgenic strains were constructed by CRISPR/Cas9-mediated site-specific gene editing66,68. Locus-specific CRISPR plasmids and homology templates were co-transfected into the purified tachyzoites of the corresponding strains and selected with 25 µg/ml mycophenolic acid and 50 µg/ml xanthine (Sigma-Aldrich, USA) (for strains with the HXGPRT marker) or 1 µM pyrimethamine (Sigma-Aldrich, USA) (for strains with the DHFR* marker) or 10 µM 5-fluoro-2’-deoxyruridine (Sigma-Aldrich, USA) (for strains with inserts that disrupted the UPRT locus). Individually, clones were obtained by limiting dilution and screened by diagnostic PCRs (primers are listed in Supplementary Table 3) and immunofluorescent assays before use.
Immunofluorescent assays (IFA)
IFA was performed as described previously63,66,69. In brief, Toxoplasma gondii-infected HFF cells grown on coverslips were fixed in 4% formaldehyde (Biosharp, China) at room temperature for 20 min, followed by permeabilization with 0.1% (v/v) Triton X-100 for 15 min. The samples were then blocked in phosphate-buffered saline (PBS) containing 3% (w/v) bovine serum albumin (BSA). After incubating with the primary antibodies for 30 min, the samples were washed with PBS and then incubated with secondary antibodies in dark for 30 min. Cell nuclei were stained with Hoechst for 10 min at room temperature. After washing three times with PBS, the coverslips were mounted onto microscope slides (Citotest, China) containing anti-fade mounting medium (Beyotime, China) and imaged under an Olympus FluoView FV1000 Confocal Microscope (Olympus Life Science, Japan).
Western blot analysis
Immunoblot analysis of protein was performed as described previously63. Briefly, collected parasites were lysed in a protein lysis buffer (10 mM Tris-HCl, pH 6.8, 0.5% SDS (v/v), 10% glycerol (v/v), 1 mM EDTA, and protease inhibitor cocktail) for 10 min, and then boiled for 10 min. Proteins were separated using SDS-PAGE and transferred to nitrocellulose membranes (Cytiva, USA). The blots were then incubated with primary and secondary antibodies, respectively, followed by detection with BeyoECL Moon Kit (Beyotime, China). The blots were eventually scanned by an Amersham Typhoon 5 imager (GE Healthcare, UK).
Transmission electron microscopy
HFF monolayers infected with ME49 iSNF2L tachyzoites were cultured in T75 flasks with or without IAA for 24 h. Then, cells were scraped off and collected by centrifugation at 500 × g for 6 min, followed by fixation with 0.25% (wt/vol) glutaraldehyde in 0.1 mol/L PBS at 20 °C for 4 h. Subsequently, cells were spun down at 500 g for 10 min and collected for sample preparation as previously described28,70. Samples sectioning (60–70 nm thick) was done using the EM UC6 Ultramicrotome (Leica, Germany) system. Then, the samples were stained with 2% (vol/vol) uranyl acetate and examined under a Hitachi 80 kV transmission electron microscope (H-7650 TEM; Hitachi High-Tech Group, Japan).
Quantitative Proteomics using TMT
Parasites of the SNF2L-mAID strain treated with or without IAA for 24 h (three biological replicates for each condition) were purified by filtration (through membranes with 3 μm pore size), proteins were reduced by incubation with tris (2-carboxyethyl) phosphine (TCEP, final concentration was 10 mM) (Thermo Scientific, USA) at 55 °C for 1 h and then alkylated with iodoacetamide (final concentration is 18.75 mM) (Sigma-Aldrich, USA) for 30 min at room temperature in dark. After that, six volumes of pre-chilled acetone were added to precipitate the proteins overnight. The protein pellet was air-dried for 2−3 min. The pellet was subsequently resuspended with 100 mM HEPES pH 8.5 and digested with trypsin at 1:40 (w/w) (Promega, USA) overnight at 37 °C. The concentration of digest proteins was measured with Quantitative Colorimetric Peptide Assay (Thermo Scientific, USA). Then, 80 μg peptides of each sample (three samples for the +IAA group and three for the −IAA group) were subjected to TMT-6plex labeling, according to the manufacturer’s instructions. The TMT-labeled samples were then vacuum centrifuged to near dryness and desalted on a MonoSpinTM C18 (GL Science, Japan).
A quarter of the pooled TMT-labeled peptides was resuspended in 300 μL 10% buffer C (50 mM ammonium hydroxide, pH 10). Separation was performed using Agilent 1260 pump and a 4.6 mm × 250 mm BEH C18, 3.5 μm column (Waters, USA) with a 58 min gradient from 5% to 80% buffer B (100% acetonitrile) at a flow rate of 0.5 ml/min while buffer C was running constantly at 10% of total flow rate during the process. Buffer A was 100% water. Fractions were collected over 75 min at 2 min intervals after the start of the gradient in 1.5 ml Eppendorf tubes. A total of 50 fractions were collected, which were then consolidated into 25. Samples were subsequently vacuum centrifuged to near dryness and reconstituted in 5% formic acid for LC-MS/MS analysis.
All 25 fractions were analyzed by a home-made 30 cm-long pulled-tip analytical column (75 μm ID packed with ReproSil-Pur C18-AQ 1.9 μm resin, Dr. Maisch GmbH). The column was placed in line with an Easy-nLC 1200 nano HPLC (Thermo Scientific, San Jose, CA) for mass spectrometry analysis. The analytical column temperature was set at 55 °C during the experiments. The mobile phase and elution gradient used for peptide separation were as follows: 0.1% formic acid in water as buffer A and 0.1% formic acid in 80% acetonitrile as buffer B, 0–1 min, 1%-8% B; 1–104 min, 8–35% B; 104–114 min, 35%–50% B, 114-115 min, 50%–100% B, 115–120 min, 100% B. Data-dependent tandem mass spectrometry (MS/MS) analysis (2 s cycle time) was performed with an Orbitrap Eclipse Tribrid mass spectrometer (Thermo Scientific, San Jose, CA). Peptides eluted from the column were directly electro-sprayed into the mass spectrometer with the application of a distal 2.3-kV spray voltage. MS1 spectra were acquired in the Orbitrap (R = 120k; AGC target: standard; MaxIT: auto; RF Lens = 40%; mass range = 400–1600). Dynamic exclusion was employed for 60 s, excluding all charge states for a given precursor. MS2 spectra were collected in the Orbitrap (R = 30k; first mass = 110 m/z; AGC target = 250,000; MaxIT = 54 ms). MS scan functions and LC solvent gradients were controlled by the Xcalibur data system (Thermo Scientific, USA).
RNA-Seq analysis
Tachyzoites of the iSNF2L, iSLIF1, and iSLIF2 strains treated with IAA for 0, 12, or 24 h (three replicates were independently prepared and analyzed for each condition) were purified and collected. Total RNA was extracted using the TRIzol reagent (Invitrogen, USA) according to the manufacturer’s protocol. RNA purity and concentration were evaluated using the NanoDrop 2000 spectrophotometer (Thermo Scientific, USA). RNA integrity was assessed using the Agilent 2100 Bioanalyzer (Agilent Technologies, USA). Then, the libraries for sequencing were constructed using the VAHTS Universal V6 RNA-seq Library Prep Kit according to the manufacturer’s instructions. The mRNA sequencing and analyses were conducted by OE Biotech Co., Ltd. (Shanghai, China). The libraries were sequenced on the illumina Novaseq 6000 platform, and 150 bp paired-end reads were generated. Raw reads were first processed using fastp, and low-quality reads were removed to obtain the clean reads71. The clean reads were mapped to the reference genome of ME49 using HISAT272. FPKM of each gene was calculated, and the read counts of each gene were obtained by HTSeq-count73. PCA analysis was performed using R (v 3.2.0) to evaluate the biological duplication of samples. Differential expression analysis was performed using the DESeq274. Adjusted P (Q) value ≤ 0.05 and fold-change ≥ 2 were set as the threshold for differential expression genes (DEGs). Raw data have been deposited to the GEO database with the accession numbers GSE268892, GSE268652, and GSE268750.
CUT&Tag
Intracellular parasites of the iSNF2L (treated with or without IAA for 12 h) or SNF2L-3HA strains were mechanically released from host cells by needle passage and purified by filtration. Each strain or condition contained three biological replicates. Approximately 5 × 105 tachyzoites were collected and processed using Illumina’s Hyperactive Universal CUT&Tag Assay Kit (Vazyme Biotech, China), according to the manufacturer’s instructions. Mouse anti-HA was used as primary antibody for immunoprecipitation. For samples from the SNF2L−3HA strain, parallel experiments using naive mouse IgG were included as controls. For the iSNF2L samples, the IAA-treated samples were used as controls for those without IAA treatment. The resulting DNA was amplified using the TD 202 TruePrep Index Kit V2 for Illumina (Vazyme Biotech, China) for library construction. The libraries were enriched, quantified, and sequenced using the PE150 model on a NovaSeq 6000 sequencer (Illumina). High-throughput sequencing and data analysis were performed by Yingzi Gene Technology Co., Ltd (Wuhan, China). Briefly, raw sequencing data were initially filtered using Cutadapt (version 2.5) to remove low-quality reads and trim adapter-contaminated sequences. Clean readings were then mapped to T. gondii reference genome (ME49) using Bowtie 2 (version 2.3.4.1) with default parameters. Library quality was assessed by calculating insert size using Samtools (version 1.12) and analyzing read distribution around transcription start sites (TSS) using DeepTools (version 3.5.1). Peak identification was performed using MACS 2 version 2.1.1. Bigwig is generated using bigWigMerge and bedGraphToBigWig (version 4.11.1). Peaks were annotated with ChIPseeker (version 1.32.1). Visualization of peaks in genomic regions was performed using IGV Genome Browser. The original data have been deposited in the GEO database under accession numbers GSE293095 and GSE293409.
ATAC-Seq
Fresh tachyzoites of the ME49 SNF2L-mAID strain treated with or without IAA for 12 h were harvested, filtered through 3 µm polycarbonate membranes, and centrifuged at 1000 × g for 8 min. The tachyzoites that were collected were then ground in liquid nitrogen for a period of ~30 min. Then, 980 μL of cell lysis buffer (including 50 mM HEPES (Life technologies, USA), 140 mM NaCl (Invitrogen, USA), 1 mM EDTA (Invitrogen, USA), 10% glycerol (Sigma-Aldrich, USA), 0.5% NP-40 (Thermo Scientific, USA), and 0.25% Triton X-100 (Thermo Scientific, USA)) and 20 μL protease inhibitor (50×) (Roche, Switzerland) were added to each sample and vortexed for 1 min. Nuclei from the lysed parasites were enriched following the previously described ATAC-seq method75. Then, 50 μL of transposition reaction mixture (ATAC-seq library prep kit, Yingzi Gene, China) was added and incubated at 37 °C for 1 h. The products were then recovered using a DNA purification and concentration kit (Jian Shi Biotechnology, China). The recovered products were subjected to PCR amplification using NEB Next High-Fidelity 2X PCR Master Mix (NEB, USA) and adapter primers. The PCR products were purified and size-selected using KAPA Pure Beads (KAPA BIOSYSTEMS, USA). The prepared libraries were sequenced on the Illumina Nova6000 PE150 platform. The bigwig files containing enriched peaks were generated and visualized as described above in the CUT&Tag section. Sequencing and data analyses were provided by Wuhan Yingzi Gene Technology Co., Ltd The original ATAC-Seq data have been deposited to the GEO database and can be accessed through the accession number GSE268890.
Co-immunoprecipitation (Co-IP) and mass spectrometry
Freshly egressed tachyzoites of indicated strains were collected and purified through 3 μm polycarbonate membranes. Then, 5 × 108 parasites of each strain were harvested, washed three times with ice-cold PBS, and lysed with 400 μl ice-cold IP lysis buffer (500 μL NP-40 Lysis Buffer with 10 μl PMSF) (Beyotime, China). During lysis, the samples were placed in a water bath sonicator and sonicated for 15 min in iced water. The lysates were then centrifuged at 1800 × g for 5 min, and the supernatants were collected for subsequent co-IP experiments. For co-IP, the supernatants of parasite lysates were incubated with mouse IgG pretreated Protein G magnetic beads (ABclonal, China) at 4 °C for 1 h to remove proteins that bound the beads or mouse IgG. The cleared supernatants were then added to Protein G magnetic beads loaded with mouse anti-HA antibody (MBL International Corporation, Japan) (or mouse IgG antibody as control) and incubated at 4 °C for 10 h. Subsequently, the beads were washed three times with cold TBS. Bound proteins were then sent for mass spectrometry analyses.
Beads were first washed with 200 µl 100 mM Tris-Cl pH 8.5 three times, then dissolved 50 µl 8 M urea with 100 mM Tris-Cl (pH 8.5). 5 mM Tris(2-Carboxyethyl)-Phosphine HCl (TCEP, Thermo Scientific) for reduction and 10 mM iodoacetamide (IAA, Sigma) for alkylation were added, sonicated, and incubated at room temperature in dark for 30 min, respectively. The protein mixture was diluted four times with 100 mM Tris-Cl and digested with Trypsin at 1:50 (w/w) (Promega). The digestion was stopped by 5% Formic Acid (FA, Thermo Scientific), and the peptide mixture was desalted by MonoSpinTM C18 column (GL Science). Desalted mixture was dried with a SpeedVac and resuspended in 0.1% FA for MS analysis.
The peptide mixture was analyzed by a home-made 30 cm-long pulled-tip analytical column (75 μm ID packed with ReproSil-Pur C18-AQ 1.9 μm resin, Dr. Maisch GmbH), and the column was then placed in line with an Easy-nLC 1200 nano HPLC (Thermo Scientific) for mass spectrometry analysis. The analytical column temperature was set at 55 °C during the experiments. The mobile phase and elution gradient used for peptide separation were as follows: 0.1% formic acid in water as buffer A and 0.1% formic acid in 80% acetonitrile as buffer B, 0–1 min, 2%–10% B; 1–81 min, 10–35% B; 81–96 min, 35%–60% B, 96–111 min, 60%–100% B, 111–120 min, 100% B. The flow rate was set as 300 nl/min.
Data-dependent MS/MS analysis was performed with a Q Exactive Orbitrap mass spectrometer (Thermo Scientific). Peptides eluted from the LC column were directly electrosprayed into the mass spectrometer with the application of a distal 2.0-kV spray voltage. A cycle of one full-scan MS spectrum (m/z 300-1800) was acquired, followed by top 20 MS/MS events, sequentially generated on the first to the twentieth most intense ions selected from the full MS spectrum at a 28% normalized collision energy. Full scan resolution was set to 70,000 with automated gain control (AGC) target of 3e6. MS/MS scan resolution was set to 17,500 with isolation window of 1.8 m/z and AGC target of 1e5. The number of microscans was one for both MS and MS/MS scans, and the maximum ion injection time was 50 and 100 ms, respectively. The dynamic exclusion settings used were as follows: charge exclusion, 1 and >7; exclude isotopes, on; and exclusion duration, 15 s. MS scan functions and LC solvent gradients were controlled by the Xcalibur data system (Thermo Scientific, USA).
The acquired MS/MS data were analyzed against ToxoDB using Proteome Discoverer 2.4 (Thermo Scientific). Mass tolerances for precursor ions were set at 20 ppm, and for fragments were set at 0.08 Da. Trypsin was defined as cleavage enzyme; Cysteine alkylation by iodoacetamide was specified as fixed modification with mass shift 57.02146; Methionine oxidation was set as dynamic modification with mass shift 15.9949. A decoy database containing the reversed sequences of all the proteins was appended to the target database to accurately estimate peptide probabilities and false discovery rate (FDR), and FDR was set at 0.01.
Statistical analyses
Except the RNA-Seq, CUT&Tag, and co-IP/MS results, all other data were analyzed using Prism 9 (GraphPad Software Inc., USA). Sample size was determined based on similar experiments published in previous literature. Statistical analyses were performed using unpaired two-tailed Student’s t-test or Kolmogorov-Smirnov test, as indicated in the figure legends. Chi-squared tests were performed in SPSS (version 27, IBM Inc., USA) to see the degree of gene enrichment of DEGs in SNF2L-depleted parasites to different categories, as indicated in figure legends. All immunofluorescence assays (IFA) and Western blots were independently repeated at least twice, and similar results were obtained. Only representative images were shown.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The raw sequencing data generated in this study have been deposited in the GEO database of NCBI under the following accession codes: GSE268892, GSE268750, GSE268652 (RNA-Seq), GSE293409, GSE293095 (CUT&Tag), and GSE268890 (ATAC-Seq). The TMT quantitative proteomics data have been deposited in the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository with the identifier number PXD055403. Other RNA-Seq data analyzed in this study are publicly available from ToxoDB or NCBI, as indicated in figure legends. All data are included in the article or the Supplementary Information. Source data are provided with this paper.
References
Kim, K. The epigenome, cell cycle, and development in toxoplasma. Annu. Rev. Microbiol. 72, 479–499 (2018).
White, M. W. & Suvorova, E. S. Apicomplexa cell cycles: something old, borrowed, lost, and new. Trends Parasitol. 34, 759–771 (2018).
Montoya, J. G. & Liesenfeld, O. Toxoplasmosis. Lancet 363, 1965–1976 (2004).
Robert-Gangneux, F. & Dardé, M. L. Epidemiology of and diagnostic strategies for toxoplasmosis. Clin. Microbiol. Rev. 25, 264–296 (2012).
Attias, M. et al. The life-cycle of Toxoplasma gondii reviewed using animations. Parasit. Vectors 13, 588 (2020).
Dubey, J. P., Lindsay, D. S. & Speer, C. A. Structures of Toxoplasma gondii tachyzoites, bradyzoites, and sporozoites and biology and development of tissue cysts. Clin. Microbiol. Rev. 11, 267–299 (1998).
Farhat, D. C. & Hakimi, M. A. The developmental trajectories of Toxoplasma stem from an elaborate epigenetic rewiring. Trends Parasitol. 38, 37–53 (2022).
Francia, M. E. & Striepen, B. Cell division in apicomplexan parasites. Nat. Rev. Microbiol. 12, 125–136 (2014).
Ferguson, D. J., Hutchison, W. M., Dunachie, J. F. & Siim, J. C. Ultrastructural study of early stages of asexual multiplication and microgametogony of Toxoplasma gondii in the small intestine of the cat. Acta Pathol. Microbiol Scand. B Microbiol. Immunol. 82, 167–181 (1974).
Tomasina, R. & Francia, M. E. The structural and molecular underpinnings of gametogenesis in toxoplasma gondii. Front. Cell Infect. Microbiol. 10, 608291 (2020).
Dubey, J. P. & Frenkel, J. K. Cyst-induced toxoplasmosis in cats. J. Protozool. 19, 155–177 (1972).
Speer, C. A. & Dubey, J. P. Ultrastructural differentiation of Toxoplasma gondii schizonts (types B to E) and gamonts in the intestines of cats fed bradyzoites. Int J. Parasitol. 35, 193–206 (2005).
Nardelli, S. C. et al. Genome-wide localization of histone variants in Toxoplasma gondii implicates variant exchange in stage-specific gene expression. BMC Genomics 23, 128 (2022).
Garfoot, A. L., Wilson, G. M., Coon, J. J. & Knoll, L. J. Proteomic and transcriptomic analyses of early and late-chronic Toxoplasma gondii infection shows novel and stage specific transcripts. BMC Genomics 20, 859 (2019).
Abdi, A. I. et al. Plasmodium falciparum adapts its investment into replication versus transmission according to the host environment. Elife 12, https://doi.org/10.7554/eLife.85140 (2023).
Brancucci, N. M. B. et al. Lysophosphatidylcholine regulates sexual stage differentiation in the human malaria parasite plasmodium falciparum. Cell 171, 1532–1544.e1515 (2017).
Hammouda, N. A., Ibrahim, I. R., Elkerdany, E. D., Negm, A. Y. & Allam, S. R. Experimental conversion of virulent RH Toxoplasma gondii tachyzoites in vitro. East Mediterr. Health J. 7, 181–188 (2001).
Salimi, M., Shojaee, S., Keshavarz, H. & Mohebali, M. Cyst formation from virulent RH strain of toxoplasma gondii tachyzoite: in vitro cultivation. Iran. J. Parasitol. 11, 81–85 (2016).
Russell, A. J. C. et al. Regulators of male and female sexual development are critical for the transmission of a malaria parasite. Cell Host Microbe 31, 305–319.e310 (2023).
Poran, A. et al. Single-cell RNA sequencing reveals a signature of sexual commitment in malaria parasites. Nature 551, 95–99 (2017).
Brancucci, N. M. B. et al. Heterochromatin protein 1 secures survival and transmission of malaria parasites. Cell Host Microbe 16, 165–176 (2014).
Hong, D. P., Radke, J. B. & White, M. W. Opposing Transcriptional Mechanisms Regulate Toxoplasma Development. mSphere 2, https://doi.org/10.1128/mSphere.00347-16 (2017).
Radke, J. B. et al. ApiAP2 transcription factor restricts development of the Toxoplasma tissue cyst. Proc. Natl. Acad. Sci. USA 110, 6871–6876 (2013).
Radke, J. B. et al. Transcriptional repression by ApiAP2 factors is central to chronic toxoplasmosis. PLoS Pathog. 14, e1007035 (2018).
Waldman, B. S. et al. Identification of a Master Regulator of Differentiation in Toxoplasma. Cell 180, 359–372.e316 (2020).
Huang, S. et al. Toxoplasma gondii AP2IX-4 regulates gene expression during bradyzoite development. mSphere 2, https://doi.org/10.1128/mSphere.00054-17 (2017).
Farhat, D. C. et al. A MORC-driven transcriptional switch controls Toxoplasma developmental trajectories and sexual commitment. Nat. Microbiol 5, 570–583 (2020).
Fan, F., Xue, L., Yin, X., Gupta, N. & Shen, B. AP2XII-1 is a negative regulator of merogony and presexual commitment in Toxoplasma gondii. mBio 14, e0178523 (2023).
Antunes, A. V. et al. In vitro production of cat-restricted Toxoplasma pre-sexual stages. Nature 625, 366–376 (2024).
Wang, J. L. et al. The transcription factor AP2XI-2 is a key negative regulator of Toxoplasma gondii merogony. Nat. Commun. 15, 793 (2024).
Kadoch, C. & Crabtree, G. R. Mammalian SWI/SNF chromatin remodeling complexes and cancer: mechanistic insights gained from human genomics. Sci. Adv. 1, e1500447 (2015).
Wu, J. I., Lessard, J. & Crabtree, G. R. Understanding the words of chromatin regulation. Cell 136, 200–206 (2009).
Clapier, C. R. & Cairns, B. R. The biology of chromatin remodeling complexes. Annu Rev. Biochem 78, 273–304 (2009).
Shang, J. Y. & He, X. J. Chromatin-remodeling complexes: Conserved and plant-specific subunits in Arabidopsis. J. Integr. Plant Biol. 64, 499–515 (2022).
Stanne, T. et al. Identification of the ISWI Chromatin Remodeling Complex of the Early Branching Eukaryote Trypanosoma brucei. J. Biol. Chem. 290, 26954–26967 (2015).
Hughes, K. et al. A novel ISWI is involved in VSG expression site downregulation in African trypanosomes. Embo J. 26, 2400–2410 (2007).
Kaneko, I., Nishi, T., Iwanaga, S. & Yuda, M. Differentiation of Plasmodium male gametocytes is initiated by the recruitment of a chromatin remodeler to a male-specific cis-element. Proc. Natl. Acad. Sci. USA 120, e2303432120 (2023).
Clapier, C. R., Iwasa, J., Cairns, B. R. & Peterson, C. L. Mechanisms of action and regulation of ATP-dependent chromatin-remodelling complexes. Nat. Rev. Mol. Cell Biol. 18, 407–422 (2017).
Lin, A., Du, Y. & Xiao, W. Yeast chromatin remodeling complexes and their roles in transcription. Curr. Genet. 66, 657–670 (2020).
Gaskins, E. et al. Identification of the membrane receptor of a class XIV myosin in Toxoplasma gondii. J. Cell Biol. 165, 383–393 (2004).
Hehl, A. B. et al. Asexual expansion of Toxoplasma gondii merozoites is distinct from tachyzoites and entails expression of non-overlapping gene families to attach, invade, and replicate within feline enterocytes. BMC Genomics 16, 66 (2015).
Pittman, K. J., Aliota, M. T. & Knoll, L. J. Dual transcriptional profiling of mice and Toxoplasma gondii during acute and chronic infection. BMC Genomics 15, 806 (2014).
Fritz, H. M. et al. Transcriptomic analysis of toxoplasma development reveals many novel functions and structures specific to sporozoites and oocysts. PLoS ONE 7, e29998 (2012).
Ramakrishnan, C. et al. An experimental genetically attenuated live vaccine to prevent transmission of Toxoplasma gondii by cats. Sci. Rep. 9, 1474 (2019).
Ramakrishnan, C., Walker, R. A., Eichenberger, R. M., Hehl, A. B. & Smith, N. C. The merozoite-specific protein, TgGRA11B, identified as a component of the Toxoplasma gondii parasitophorous vacuole in a tachyzoite expression model. Int J. Parasitol. 47, 597–600 (2017).
Wang, Z. T., Verma, S. K., Dubey, J. P. & Sibley, L. D. The aromatic amino acid hydroxylase genes AAH1 and AAH2 in Toxoplasma gondii contribute to transmission in the cat. PLoS Pathog. 13, e1006272 (2017).
Feix, A. S. et al. Progression of asexual to sexual stages of Cystoisospora suis in a host cell-free environment as a model for Coccidia. Parasitology 148, 1475–1481 (2021).
Walker, R. A. et al. RNA Seq analysis of the Eimeria tenella gametocyte transcriptome reveals clues about the molecular basis for sexual reproduction and oocyst biogenesis. BMC Genomics 16, 94 (2015).
Straschil, U. et al. The Armadillo repeat protein PF16 is essential for flagellar structure and function in Plasmodium male gametes. PLoS ONE 5, e12901 (2010).
Zhang, J., Fan, F., Zhang, L. & Shen, B. Nuclear factor AP2X-4 governs the expression of cell cycle- and life stage-regulated genes and is critical for toxoplasma growth. Microbiol Spectr. 10, e0012022 (2022).
Long, S., Anthony, B., Drewry, L. L. & Sibley, L. D. A conserved ankyrin repeat-containing protein regulates conoid stability, motility and cell invasion in Toxoplasma gondii. Nat. Commun. 8, 2236 (2017).
Titeca, K. et al. SFINX: straightforward filtering index for affinity purification-mass spectrometry data analysis. J. Proteome Res. 15, 332–338 (2016).
Cui, L. & Miao, J. Chromatin-mediated epigenetic regulation in the malaria parasite Plasmodium falciparum. Eukaryot. Cell 9, 1138–1149 (2010).
Hargreaves, D. C. & Crabtree, G. R. ATP-dependent chromatin remodeling: genetics, genomics and mechanisms. Cell Res. 21, 396–420 (2011).
Donovan, D. A. et al. Basis of specificity for a conserved and promiscuous chromatin remodeling protein. Elife 10, https://doi.org/10.7554/eLife.64061 (2021).
Ohya, T., Maki, S., Kawasaki, Y. & Sugino, A. Structure and function of the fourth subunit (Dpb4p) of DNA polymerase epsilon in Saccharomyces cerevisiae. Nucleic Acids Res. 28, 3846–3852 (2000).
Casari, E. et al. Dpb4 promotes resection of DNA double-strand breaks and checkpoint activation by acting in two different protein complexes. Nat. Commun. 12, 4750 (2021).
Morrison, A. J. Chromatin-remodeling links metabolic signaling to gene expression. Mol. Metab. 38, 100973 (2020).
Goldmark, J. P., Fazzio, T. G., Estep, P. W., Church, G. M. & Tsukiyama, T. The Isw2 chromatin remodeling complex represses early meiotic genes upon recruitment by Ume6p. Cell 103, 423–433 (2000).
Yadon, A. N., Singh, B. N., Hampsey, M. & Tsukiyama, T. DNA looping facilitates targeting of a chromatin remodeling enzyme. Mol. Cell 50, 93–103 (2013).
Watzlowik, M. T. et al. Plasmodium blood stage development requires the chromatin remodeller Snf2L. Nature 639, 1069–1075 (2025).
Pachano, B. et al. An ISWI-related chromatin remodeller regulates stage-specific gene expression in Toxoplasma gondii. Nat. Microbiol. 10, 1156–1170 (2025).
Li, Y. et al. Rapid metabolic reprogramming mediated by the AMP-activated protein kinase during the lytic cycle of Toxoplasma gondii. Nat. Commun. 14, 422 (2023).
Chen, P. et al. A pyruvate transporter in the apicoplast of apicomplexan parasites. Proc. Natl. Acad. Sci. USA 121, e2314314121 (2024).
Yang, X. et al. Essential role of pyrophosphate homeostasis mediated by the pyrophosphate-dependent phosphofructokinase in Toxoplasma gondii. PLoS Pathog. 18, e1010293 (2022).
Shen, B., Brown, K. M., Lee, T. D. & Sibley, L. D. Efficient gene disruption in diverse strains of Toxoplasma gondii using CRISPR/CAS9. mBio 5, e01114 (2014).
Brown, K. M., Long, S. & Sibley, L. D. Plasma Membrane Association by N-Acylation Governs PKG Function in Toxoplasma gondii. mBio 8, https://doi.org/10.1128/mBio.00375-17 (2017).
Shen, B., Brown, K., Long, S. & Sibley, L. D. Development of CRISPR/Cas9 for Efficient Genome Editing in Toxoplasma gondii. Methods Mol. Biol. 1498, 79–103 (2017).
Xia, N. et al. Pyruvate homeostasis as a determinant of parasite growth and metabolic plasticity in toxoplasma gondii. mBio. 10, https://doi.org/10.1128/mBio.00898-19 (2019).
Nwafor, C. C. et al. Genetic and biochemical investigation of seed fatty acid accumulation in arabidopsis. Front. Plant Sci. 13, 942054 (2022).
Chen, S., Zhou, Y., Chen, Y. & Gu, J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890 (2018).
Kim, D., Langmead, B. & Salzberg, S. L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360 (2015).
Roberts, A., Trapnell, C., Donaghey, J., Rinn, J. L. & Pachter, L. Improving RNA-Seq expression estimates by correcting for fragment bias. Genome Biol. 12, R22 (2011).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Zhang, Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008).
Acknowledgements
We thank Dr. Yue Yin from the Mass Spectrometry System at the National Facility for Protein Science in Shanghai (NFPS) for protein mass spectrometry analyses. We also thank Dr. Jian Zhao from the electron microscopy facility of the State Key Laboratory of Agricultural Microbiology in Huazhong Agricultural University for his help in EM analyses. This work was supported by the Guangdong Major Project of Basic and Applied Basic Research (2020B0301030007, to BS), the National Natural Science Foundation of China (32325048 to BS), the National Key Research and Development Program of China (2022YFD1800200 to BS), and the fundamental research funds for the central universities (2662025DKPY004, to BS).
Author information
Authors and Affiliations
Contributions
Y.Z. and B.S. designed the research; Y.Z., B.F., H.X., Y.L., X.L., L.X., L.W., F.F., X.Z., and Y.C. performed the experiments; Y.Z., S.Q., and B.S. analyzed the data; Y.Z. and B.S. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhu, Y., Fan, B., Xu, H. et al. Regulation of the developmental programs in Toxoplasma by a novel SNF2L-containing chromatin remodeling complex. Nat Commun 16, 5757 (2025). https://doi.org/10.1038/s41467-025-60795-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-60795-1