Abstract
The claudin protein family plays key roles in maintaining normal structure and function of epithelial and endothelial tight junctions. While several prior studies have addressed the expression of claudin in adipocytes that do not form tight junctions, here we demonstrate that CLDN5 is selectively expressed in non-thermogenic adipocytes within adipose tissue. Ablation of CLDN5 in adipocyte impairs thermogenesis and energy expenditure. CLDN5 deficiency also significantly increases diet-induced fat mass in mice, accompanied with glucose intolerance and insulin resistance. Mechanistically, CLDN5 affects the subcellular localization of Y-box protein 3, which directly regulates IL10 expression via binding to its promoter and specific sites in 3′-untranslated region, thereby acts in a paracrine manner to signal through IL10R in neighbouring thermogenic adipocytes. These findings expand our understanding about location and function of the extra-tight junction claudin proteins and provide molecular insights into signaling mechanisms underlying adipose thermogenesis that could inform future therapy.
Similar content being viewed by others
Introduction
Claudins (CLDNs) are transmembrane proteins that assemble into tight junction strands and play an essential role in regulating paracellular ion transport and membrane polarity in epithelia and endothelia1,2. Some CLDNs can have a signaling role in cells without tight junctions, where they often appear to represent a signaling hotspot in different cell types that recruits a range of different pathways involved in the regulation of cell behavior and gene expression through the interaction of their cytoplasmic adaptor proteins with transcription factors, regulators of Rho family GTPases, and other signaling proteins3,4,5,6,7. CLDN5 is unique among CLDN family members as its predominant expression in the endothelium and non-redundant function in the control of vascular permeability8. Recent studies documented the presence of CLDN5 in adipocytes of fat tissue2, although adipocytes themselves don’t contain tight junction structure and are interconnected via gap junctions9. However, the functional roles of CLDN5 in adipose biology and energy homeostasis remain to be charted.
We and others have previously shown that CLDN5 binds to the scaffolder ZO1, which directly interacts with the tight-junction-specific transcription factor ZO1-associated nucleic acid binding protein (ZONAB), leading to the sequestration of ZONAB at the tight junction10,11,12. ZONAB is a member of the Y-box protein family, which are DNA/RNA-binding proteins belonging to a large family of proteins with the cold shock domain and contain three members, including Y-box protein 1 (YBX1), Y-box protein 2 (YBX2), and Y-box protein 3 (YBX3, also known as ZONAB). Cold shock domain was originally identified in cold shock proteins (CSPs) in bacteria where some CSPs were induced in response to a sudden decrease in temperature to initiate an adaptive response13. Studies have shown that cold shock domain is conserved from bacteria to human in binding RNA or DNA, which confers a wide variety of functions to CSPs, including their involvement in adaptive responses to cold stress14. YBX1 increases in response to cold in thermogenic adipose tissues and contributes to the induction of thermogenic gene expression15,16,17,18. The expression of YBX2 increases during white fat browning and brown adipogenesis, playing a regulatory role in adipocyte thermogenesis by stabilizing Pgc1α mRNA19. In addition to thermogenic regulation, YBX2 promotes the mRNA translation of glycolytic genes by directly binding to their 5′-UTRs, which facilitates brown adipocytes glycolysis in response to thermogenic stimuli20. These results strongly suggest that Y-box protein family retain an evolutionarily conserved role during cellular stress to facilitate cold adaptation. Thus, we speculated that the CLDN5-ZO1-YBX3 axis may contribute to regulation of energy balance in adipose tissue.
In this work, we identify a previously unrecognized function of CLDN5 in regulating energy expenditure in both brown adipose tissue (BAT) and inguinal white adipose tissue (iWAT). CLDN5 is selectively and abundantly expressed in non-thermogenic adipocytes. Ablation of CLDN5 in adipose tissue impairs thermogenesis and energy expenditure. CLDN5 deficiency also significantly increases diet-induced fat mass in mice, accompanied with glucose intolerance and insulin resistance. Mechanistically, CLDN5 deploys paracrine effector IL10 to actively regulate IL10R signaling in neighboring thermogenic adipocytes. We also provide mechanistic insights into the regulation of IL10 by CLDN5 by showing that CLDN5 affects the subcellular localization of YBX3, which directly regulates IL10 expression via binding to its promoter and to its specific sites in the 3′-untranslated region (3’-UTR).
Results
CLDN5 is selectively enriched in non-thermogenic adipocytes
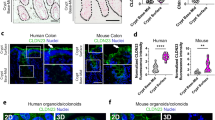
Both the human protein atlas (HPA) transcriptomic datasets and the single cell type transcriptomics map of human tissues clearly revealed that CLDN5 displayed high expression level in fat tissue and in adipocytes (Fig. 1a and Supplementary Fig. 1a)21,22. To further investigate the distribution of CLDN5 in adipose tissue, we took advantage of a transgenic C57BL/6 mouse strain expressing E (enhanced) GFP under the control of the Cldn5 promoter, denoted CLDN5-GFP. GFP fluorescence was seen in capillaries, and counterstaining for the endothelial cell marker CD31 confirmed that the bulk of CLDN5 was in blood capillaries of both BAT and iWAT (Fig. 1b). Only 14% in the BAT and 4% in the iWAT of total adipocytes were labeled as GFP+ cells (Supplementary Fig. 1b), and these cells exhibit little to no colocalization with UCP1 (Fig. 1c). We next isolated GFP+ and GFP- adipocytes from BAT and iWAT of CLDN5-GFP mice via flow sorting (Supplementary Fig. 1c). qRT-PCR analysis demonstrated enrichment of Cldn5, Fabp4, and Cd36 in GFP+ adipocytes, whereas Ucp1 was enriched in GFP− adipocytes (Fig. 1d, e). Previous work demonstrated the existence of two brown adipocyte subpopulations, brown adipocytes with high thermogenic activity (BA-H) and brown adipocytes with low thermogenic activity (BA-L). The brown adipocytes within the BA-L subpopulation had substantially lower UCP1 and ADIPOQ expression and were enriched in CD36 and FABP4 as well as tight junction protein CLDN5, and our study confirmed these omics data (Fig. 1f and Supplementary Fig. 1d). In iWAT, both GFP+ cells and GFP- cells exhibited high expression of Cd36 and Fabp4, with significantly higher expression in GFP+ cells than in GFP- cells. Furthermore, Ucp1 was expressed at very low levels in GFP+ cells. Immunofluorescence staining revealed colocalization of CLDN5 with FABP4 and CD36 (Fig. 1g and Supplementary Fig. 1e). These findings collectively demonstrate that CLDN5 is distributed in a CD36-high, FABP4-high, and UCP1-low cell subpopulation in iWAT.
a CLDN5 mRNA expression across various normal tissues obtained from the NCBI HPA RNA-seq dataset (project PRJEB4337). b, c Immunofluorescence staining of CD31 (b) and UCP1 (c) in BAT and iWAT from CLDN5-GFP mice. d, e qRT-PCR analysis of indicated genes in both GFP+ and GFP- adipocytes isolated from BAT and iWAT of CLDN5-GFP mice (n = 3). f, g Immunofluorescence staining of FABP4 and CD36 in BAT (f) and iWAT (g) from CLDN5-GFP mice. h Immunofluorescence staining of CD68 in BAT and iWAT from CLDN5-GFP mice. For b, c, f–h, each experiment was repeated three times with consistent results. Data represent the mean ± SEM. Statistical analyses were performed using two-sided unpaired t test. Scale bar, 50 μm. Source data are provided as a Source Data file.
To further verify these findings, we generated tissue-specific Cldn5 knockout mice by crossing Cldn5-floxed mice with three commonly used Cre lines that are driven by the mouse Ucp1, Adipoq, or Fabp4 gene promoter. In agreement with the finding that CLDN5 was enriched in the UCP1 and ADIPOQ low-expressing brown adipocyte subpopulation described above, expression of CLDN5 in BAT did not show any significant difference between the Cldn5flox/flox and Cldn5flox/flox; Ucp1-Cre mice (Supplementary Fig. 1f, g), or Cldn5flox/flox and Cldn5flox/flox; Adipoq-Cre mice (Supplementary Fig. 1i, j). In WAT, expression of CLDN5 in iWAT, eWAT, isolated inguinal mature adipocytes, and isolated stromal vascular fraction (SVF) cells was similar between Cldn5flox/flox and Cldn5flox/flox; Ucp1-Cre mice despite robust Ucp1 promoter activity in beige adipocytes (Supplementary Fig. 1f–h). These results suggest that CLDN5 is not typically expressed in UCP1-positive beige adipocytes under normal conditions. Although there was no significant difference in the expression of CLDN5 in iWAT and eWAT from Cldn5flox/flox and Cldn5flox/flox; Adipoq-Cre mice (Supplementary Fig. 1i, j), the Cldn5 mRNA level was markedly reduced by 54% in isolated inguinal mature adipocytes, while no differences were observed in isolated SVF cells (Supplementary Fig. 1k). These findings indicate that besides a portion of mature adipocytes, CLDN5 is also expressed in specific types of SVF cells within the inguinal fat pad. Next, we isolated the GFP+ and GFP- SVF cells from iWAT of CLDN5-GFP mice by flow sorting. qRT-PCR analysis revealed high expression levels of Pref-1 and Icam1 (marker of preadipocytes) and Cd31 (marker of endothelial cells) in GFP+ SVF cells (Supplementary Fig. 1l), indicating CDLN5 is also expressed on preadipocytes in addition to mature adipocytes and endothelial cells in iWAT.
Western blot and quantitative PCR analysis confirmed the efficient deletion of CLDN5 in the BAT, iWAT, and eWAT of Cldn5flox/flox; Fabp4-Cre mice (Supplementary Fig. 1m, n). Fabp4-Cre activity has been also reported in other cell types and tissues, such as macrophages and endothelial cells23. Here, GFP exhibited little to no colocalization with the macrophage-specific marker CD68, indicating adipose tissue macrophages do not express CLDN5 (Fig. 1h). Furthermore, immunofluorescence staining, as indicated by the lack of CLDN5 colocalization with CD31 but appropriate signal in the endothelial cells of Cldn5flox/flox; Fabp4-Cre adipose tissues, confirmed that the Fabp4-Cre–mediated Cldn5 deletion was largely confined to adipocytes (Supplementary Fig. 1o, p). Since Fabp4-Cre mice result in efficient Cldn5 knockout in both BAT and WAT, we used Cldn5flox/flox and Cldn5flox/flox; Fabp4-Cre mice as the control and adipocyte Cldn5 KO mice in the following experiments.
Adipocyte CLDN5 expression is regulated by thermogenic stimulus
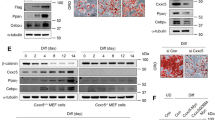
We next test whether CLDN5 expression is regulated by thermogenic stimulus. We first examined BAT, iWAT, and eWAT from animals housed under room temperature (RT, 22 °C) or cold conditions (4 °C). Seven days of cold exposure caused an increase in CLDN5 mRNA and protein expression in BAT and iWAT of wild-type mice (Supplementary Fig. 2a–c). These results demonstrated a selective induction of CLDN5 in thermogenic adipose depots (BAT and iWAT) in response to cold environment. When CLDN5-GFP mice were housed in a cold environment (4 °C), increase of GFP fluorescence intensity on individual adipocytes and the percentages of GFP+ adipocytes were observed with increased expression of UCP1 in BAT and iWAT (Fig. 2a, b). Thus, upregulation of CLDN5 may be a consequence of increase in the level of CLDN5 on GFP+ cells and the increased number of GFP+ cells. Furthermore, we observed the colocalization of GFP and UCP1 within adipocytes at 4 °C (Fig. 2a, b), which should be due to the conversion of non-thermogenic cells to thermogenic cells in response to cold stimulation. Additionally, challenging fasted mice with acute cold stimulation at 4 °C for 6 h significantly increased the expression of CLDN5 (Fig. 2c–e). Treatment with β3-selective adrenergic agonist CL-316,243 for 10 days also resulted in a significant increase in the expression of CLDN5 as well as UCP1 in both BAT and iWAT (Fig. 2f–h). qRT-PCR, Western blot, and immunofluorescence analyses showed that PGC1α, UCP1, PPARγ, and FABP4 mRNA and protein levels were significantly upregulated in response to thermogenic stimuli, corroborating the increase in CLDN5 expression is associated with thermogenic activation (Fig. 2c, d, f, g and Supplementary Fig. 2a, b, d–h). These observations suggest that CLDN5 has a role in the regulation of energy metabolism.
a, b Immunofluorescence staining of UCP1 in BAT (a) and iWAT (b) from CLDN5-GFP mice housed at room temperature (RT) or 4 °C for 7 days. Each experiment was repeated three times with consistent results. c-e Expression of CLDN5 and markers associated with thermogenic function and adipocyte differentiation assessed by qRT-PCR analysis (c, n = 5 for RT, n = 6 for 4 °C) and Western blot analysis with densitometric quantification (d, n = 3), and immunofluorescent staining of CLDN5 with mean fluorescent intensity (MFI) quantification analysis (e, n = 5) in BAT and iWAT from wild-type mice challenged at RT or 4 °C for 6 h. f–h Expression of CLDN5 and markers associated with thermogenic function and adipocyte differentiation assessed by qRT-PCR analysis (f, n = 6 for saline, n = 5 for CL-316,243) and Western blot analysis with densitometric quantification (g, n = 3), and immunofluorescent staining of CLDN5 with MFI quantification analysis (h, n = 5) in BAT and iWAT from wild-type mice treated with saline or 1 mg/kg/day CL-316,243 for 10 days. Data represent the mean ± SEM. Statistical analyses were performed using two-sided Mann-Whitney tests (for Cldn5 and Ucp1 in iWAT, c) or two-sided unpaired t test (for other genes in c and all data in d–h). Scale bar, 50 μm. Source data are provided as a Source Data file.
Adipocyte Cldn5 ablation represses thermogenesis and energy expenditure
To dissect the role of CLDN5 in metabolic homeostasis, we analyzed 6-week-old Cldn5 knockout mice with a C57BL/6 background. Despite the robust Cldn5 reduction in adipose tissue of Cldn5flox/flox; Fabp4-Cre mice, there was no distinguishable difference in body weight, adipose tissue weight, and morphology as revealed by H&E staining (Supplementary Fig. 3a, b), indicating CLDN5 is dispensable for adipose tissue development in mice. Glucose tolerance, insulin tolerance, and serum levels of T-CHO, TG, HDL-C, LDL-C, and free fatty acid (FFA) did not differ between Cldn5flox/flox and Cldn5flox/flox; Fabp4-Cre mice (Supplementary Fig. 3c, d). To study the effect of Cldn5 knockout at the molecular level, we performed RNA sequencing (RNA-seq) on BAT from chow-fed, 5-week-old Cldn5flox/flox and Cldn5flox/flox; Fabp4-Cre mice. As shown in Fig. 3a, 123 genes were downregulated, and 103 genes were upregulated. Gene Ontology analysis showed that genes linked to thermogenic pathways, including NADH dehydrogenase complex assembly, oxidative phosphorylation (OXPHOS), ATP metabolic process, and respiratory electron transport chain were significantly enriched (Fig. 3b). qRT-PCR analysis further confirmed the expression of genes related to thermogenesis and OXPHOS was dramatically decreased in Cldn5flox/flox; Fabp4-Cre mice, such as Adrb1, Pgc1α, Ucp1, Prdm16, Cox5b, Ndufa2, Atp5e, Uqcr11, Uqcrq, Cox7a1, Cox11, Cox6a1, Ndufb6, Ndufs6 (Fig. 3c). At the protein level, the knockout of CLDN5 in BAT markedly diminished the induction of key thermogenic proteins UCP1 and PGC1α (Fig. 3d), as well as components of complexes I-V (Fig. 3e). In line with the reduced expression of PGC1α, a decrease in mitochondrial counts was observed (Fig. 3f). Transmission electron microscopy (TEM) revealed that the deletion of CLDN5 in BAT led to abnormal mitochondrial morphology (Fig. 3g). The deletion of CLDN5 in iWAT resulted in substantial alterations in pathways associated with thermogenesis, as unveiled by RNA-seq analysis (Supplementary Fig. 3e, f). This perturbation was further confirmed by the downregulation of genes related to both thermogenesis and OXPHOS (Supplementary Fig. 3g, h), along with noticeable anomalies in mitochondrial counts (Supplementary Fig. 3i). In contrast, the deletion of CLDN5 in eWAT did not induce alterations in genes associated with mitochondrial respiration (Supplementary Fig. 3j–m). Therefore, these data suggest that the absence of CLDN5 may affect the thermogenic function in BAT and iWAT.
a Volcano plot of RNA-seq data in BAT showing gene expression differences between Cldn5flox/flox and Cldn5flox/flox; Fabp4-Cre mice, analyzed by the DESeq2 R package (1.16.1). b Top enriched biological processes of RNA-seq data in BAT determined by Gene Ontology analysis using clusterProfiler 3.8.1. c–g qRT-PCR analysis of indicated genes related to thermogenesis and oxidative phosphorylation (OXPHOS) (c, n = 5), Western blot analysis of PGC1α, UCP1 (d, n = 3) and OXPHOS proteins (e, n = 3), mitochondrial DNA (mtDNA) copy number (f, n = 8), and transmission electron microscopy (g) of BAT from Cldn5flox/flox and Cldn5flox/flox; Fabp4-Cre mice. h–k Energy expenditure (h), food intake (i), physical activity (j), and respiratory exchange rate (RER, k) of Cldn5flox/flox (n = 4) and Cldn5flox/flox; Fabp4-Cre mice (n = 5). l, m Rectal temperature (l), infrared thermal images and quantification of BAT interscapular temperature (m) of Cldn5flox/flox (n = 14) and Cldn5flox/flox; Fabp4-Cre mice (n = 13) challenged at 4 °C for 4 h. n–p Rectal temperature (n, n = 7), infrared thermal images and quantification of BAT interscapular temperature (o, n = 7), and free fatty acid (FFA) levels (p, Cldn5flox/flox, n = 5; Cldn5flox/flox; Fabp4-Cre, n = 6) in Cldn5flox/flox and Cldn5flox/flox; Fabp4-Cre mice kept at 4 °C for 7 days. q, r qRT-PCR analysis (q) and Western blot analysis (r, n = 3) of PGC1α and UCP1 in Cldn5flox/flox and Cldn5flox/flox; Fabp4-Cre mice housed at RT or 4 °C for 7 days. q, BAT: Pgc1α (n = 7), Ucp1 (n = 6); iWAT: Pgc1α (n = 5), Ucp1 (n = 7). s, t qRT-PCR analysis (s, n = 5) and Western blot analysis (t, n = 3) of PGC1α and UCP1 in Cldn5flox/flox and Cldn5flox/flox; Fabp4-Cre mice treated with saline/CL-316,243. u, v HE staining with droplet size quantification in the mice shown in panels n–r (u) and s, t (v), n = 5. Scale bar, 5 μm (g) or 50 μm (u, v). Data represent the mean ± SEM. Statistical analyses were performed using Mann-Whitney test (Atp5e in c, o), unpaired t test (other genes in c; d–f, l–n), two-way ANOVA with Tukey’s post-hoc test (q–v), ANCOVA (h, i), or ANOVA (j, k). All tests were two-sided. N, nucleus; LD, lipid droplet. Source data are provided as a Source Data file.
Next, we probed the influence of CLDN5 deficiency on energy homeostasis. Underwent a 2-day acclimation period in the metabolic cages, the energy expenditure, food intake, physical activity, and respiratory exchange ratio (RER) of chow-fed mice at 6-8 weeks of age were continuously monitored for 24 hours. We found Cldn5flox/flox; Fabp4-Cre mice displayed reduced energy expenditure (Fig. 3h and Supplementary Fig. 3n), without observable changes in food intake as well as physical activity compared with Cldn5flox/flox controls (Fig. 3i, j), indicating a promoting action of CLDN5 in maintaining energy homeostasis. Additionally, there were no significant changes in RER levels between the two groups during either day or night, indicating that the fat-specific loss of CLDN5 did not affect metabolic fuel preference (Fig. 3k). Given that cold exposure increases CLDN5 expression in BAT and iWAT, we hypothesized that CLDN5 might be required for cold-induced thermogenesis in adipocytes. Core body temperature was measured at RT as well as during acute exposure to 4 °C. Cldn5flox/flox and Cldn5flox/flox; Fabp4-Cre male mice exhibited a similar body temperature at RT (Fig. 3l). However, Cldn5flox/flox; Fabp4-Cre mice were less capable of defending body temperature during acute cold stress (Fig. 3l, m), indicating impaired acute cold-induced thermogenesis. To dissect if CLDN5 may regulate BAT activity and browning of WAT in response to chronic cold exposure or the β-adrenergic agonist, Cldn5flox/flox; Fabp4-Cre mice and Cldn5flox/flox control mice were exposed to 4 °C for seven days or injected with CL-316,243 for ten days. Cldn5flox/flox; Fabp4-Cre mice kept at 4 °C for 7 days exhibited lower body temperatures and BAT temperatures (Fig. 3n, o) and higher serum FFA levels, indicating impaired thermogenesis in these mice (Fig. 3p). Expression of UCP1 and PGC1α, the key mitochondrial protein in thermogenesis, was markedly reduced in BAT and iWAT of Cldn5flox/flox; Fabp4-Cre mice, after either chronic cold exposure (Fig. 3q, r and Supplementary Fig. 3o) or multiple injection of CL-316,243 (Fig. 3s, t and Supplementary Fig. 3p). Histological analyses also showed larger and more lipid droplets in adipocytes of BAT and iWAT (Fig. 3u, v) of Cldn5flox/flox; Fabp4-Cre mice. These data indicate that adipocyte-specific deletion of Cldn5 prevents the cold-induced increase in UCP1 and PGC1α expression and impairs the BAT activity and browning of WAT in response to chronic cold exposure or the β-adrenergic agonist. Note that UCP1 was already decreased in Cldn5flox/flox; Fabp4-Cre mice without cold stimulation, which was in accordance with the decreased systemic energy expenditure (Fig. 3q–t and Supplementary Fig. 3o, p). Similar to male mice, female knockout mice also exhibited impaired thermogenesis in response to acute and chronic cold exposure (Supplementary Fig. 4a–d), larger and more numerous lipid droplets in adipocytes (Supplementary Fig. 4e), and reduced expression of UCP1 and other thermogenesis markers in BAT and iWAT (Supplementary Fig. 4f, g).
Adipocyte Cldn5 abrogation mice exhibit an obesity-prone phenotype
To examine the requirement of CLDN5 for maintaining energy balance during energy overload, we next investigated whether CLDN5 KO mice are sensitive to high-fat diet (HFD)-induced obesity. 8 weeks old Cldn5flox/flox; Fabp4-Cre and Cldn5flox/flox control male mice were fed a HFD for 16 weeks, and body weight was measured weekly. We found that Cldn5flox/flox; Fabp4-Cre mice gained significantly more body weight than Cldn5flox/flox controls starting at the 8th week of HFD feeding and persisting thereafter, indicating their increased susceptibility to diet-induced obesity as compared with their control littermates (Fig. 4a). Moreover, the size and weight of white and brown fat pads were elevated in HFD-fed Cldn5flox/flox; Fabp4-Cre mice than their Cldn5flox/flox control littermates (Fig. 4b–f). Histological analyses showed adipocyte hypertrophy in the BAT, iWAT, and eWAT of Cldn5flox/flox; Fabp4-Cre mice (Fig. 4g). Furthermore, Cldn5flox/flox; Fabp4-Cre mice displayed significantly lower energy expenditure compared with their relative Cldn5flox/flox controls during both dark and light cycles (Fig. 4h). Although CLDN5 KO mice showed a tendency toward reduced locomotor activity and increased food intake during the night— warranting further investigation to fully explore the significance and underlying mechanisms of this trend—neither food intake nor locomotor activity reached significant differences between the genotypes (Fig. 4i, j). This suggests that the greater body weight gain in Cldn5flox/flox; Fabp4-Cre mice was due to decreased energy expenditure. Similar to normal chow, HFD-induced Cldn5flox/flox; Fabp4-Cre mice had similar RER levels to Cldn5flox/flox mice (Fig. 4k). The expression levels of thermogenic and OXPHOS genes, along with the mitochondrial copy number, exhibited a significant decrease in BAT of Cldn5flox/flox; Fabp4-Cre mice (Fig. 4l–o). The results of iWAT were consistent with BAT except for complex III in OXPHOS proteins (Fig. 4p–r). The majority of OXPHOS gene expression and the mitochondrial copy numbers were also downregulated in eWAT of Cldn5flox/flox; Fabp4-Cre mice (Supplementary Fig. 5a–c). Elevated serum glucose levels (Supplementary Fig. 5d) and exacerbated glucose intolerance and insulin resistance (Fig. 4s, t) were observed in Cldn5flox/flox; Fabp4-Cre mice after 16 weeks of HFD consumption. There was no difference in serum lipid levels except for HDL between the two groups of mice (Supplementary Fig. 5e). Similar results were obtained in female mice subjected to HFD, including body weight (Supplementary Fig. 5f, g), fat tissue weight (Supplementary Fig. 5h, i), adipocyte hypertrophy (Supplementary Fig. 5j), serum lipid levels (Supplementary Fig. 5k), glucose intolerance and insulin resistance (Supplementary Fig. 5l), serum glucose level (Supplementary Fig. 5m), energy expenditure, food intake, locomotor activity and RER (Supplementary Fig. 5n–q). There was no significant difference in body weight between normal diet treated mice (Supplementary Fig. 5r, s). These data collectively demonstrate that Cldn5flox/flox; Fabp4-Cre mice were more prone to HFD-induced obesity.
a Body weights of Cldn5flox/flox mice (n = 12) and Cldn5flox/flox; Fabp4-Cre mice (n = 10). b Gross appearance of representative mice. c Representative image of adipose tissues. d, e Tissue weights (d) and tissue weights normalized by tibial length (e) of BAT, iWAT, and eWAT of Cldn5flox/flox mice (n = 12) and Cldn5flox/flox; Fabp4-Cre mice (n = 10). f Representative MRI images in axial and coronal planes (n = 4). A, anterior; R, right; L, left; P, posterior; S, superior; I, inferior. Scale bar, 5 mm. g HE staining with lipid droplet size quantification of BAT, iWAT, and eWAT (n = 5). h–k Energy expenditure (h), food intake (i), locomotor activity (j), and RER (k) assessed by using CalR software, n = 5. l-o qRT-PCR analysis of indicated genes related to thermogenesis and OXPHOS (l, n = 5), Western blot analysis with densitometric quantification of PGC1α and UCP1 (m, n = 3), Western blot analysis with densitometric quantification of OXPHOS proteins (n, n = 3), and mtDNA copy number (o, n = 8) in BAT. p–r qRT-PCR analysis of indicated genes related to thermogenesis and OXPHOS (p, n = 5), Western blot analysis and densitometric quantification of OXPHOS proteins (q, n = 3), and mtDNA copy number (r, n = 5) in iWAT. s Glucose tolerance test (GTT) and quantitation of AUC (n = 10). t Insulin resistance test (ITT) and quantitation of AUC (n = 8). Data are expressed as the mean ± SEM. Statistical analyses were performed using two-sided unpaired t test (a, d–g, l-t), two-sided ANCOVA (h, i), or two-sided ANOVA (j, k). AUC, area under the curve. Source data are provided as a Source Data file.
Adipocyte CLDN5 deficiency affects thermogenic function through paracrine mechanisms
Although CLDN5 is relatively enriched in the non-thermogenic adipocyte subpopulation, the absence of CLDN5 affects the thermogenic function of BAT and iWAT. We speculated that these CLDN5-deficient cells interact with thermogenic cells through a paracrine factor to modulate whole-organ thermogenesis. To test this hypothesis, we co-cultured differentiated brown and beige adipocytes with primary CLDN5-expressing adipocytes of Cldn5flox/flox; Fabp4-Cre and Cldn5flox/flox mice using a transwell system. Considering the potential expression of CLDN5 in SVF cells and the targeting capability of Fabp4-Cre for such cells, we isolated mature adipocytes for sorting the CLDN5-expressing cell population from both BAT and iWAT utilizing the anti-CD36 magnetic beads. We used SVF cells from BAT and iWAT of wild-type mice to differentiate into brown adipocytes and beige adipocytes, respectively (Fig. 5a). We found that co-culturing with primary CLDN5-expressing adipocytes from Cldn5flox/flox; Fabp4-Cre mice led to decreased expression of UCP1 and PGC1α (Fig. 5b, c) in differentiated brown and beige adipocytes, along with downregulated basal and maximal respiration (Fig. 5d, e), compared to co-culturing with Cldn5flox/flox control adipocytes. To mimic cold exposure in vitro, we added membrane-permeable cAMP analogue dibutyryl cAMP (dbcAMP) during the co-culture. The dbcAMP treatment data demonstrated that CLDN5 deficiency affected the expression of thermogenic markers and mitochondrial respiration under stimulated conditions (Fig. 5f–i). To address the question of whether an increase in CLDN5 expression would cause the opposite phenotype, we infected primary CLDN5-expressing adipocytes from BAT and iWAT with adeno-associated virus (AAV) that overexpresses Cldn5 (Fig. 5j), which led to a fourfold overexpression of Cldn5 (Fig. 5k). We observed higher levels of UCP1 and PGC1α expression in differentiated brown and beige adipocytes co-cultured with AAV-CLDN5-transfected cells, while lower levels were seen in those co-cultured with AAV-CTL-transfected cells (Fig. 5l, m), both in the absence and presence of dbcAMP.
a–e Schematic illustration of the co-culture of differentiated brown and beige adipocytes with CD36+ primary mature adipocytes isolated from BAT and iWAT of Cldn5flox/flox and Cldn5flox/flox; Fabp4-Cre mice (a). qRT-PCR analysis of Pgc1α and Ucp1 (b, n = 4), Western blot analysis with densitometric quantification of of PGC1α and UCP1 (c, n = 3), and oxygen consumption rate (OCR) (d and e) of brown and beige adipocytes. Data in d represent n = 4 for Cldn5flox/flox mice and n = 5 for Cldn5flox/flox; Fabp4-Cre mice. Data in e represent n = 4. f-i qRT-PCR analysis (f, n = 4) and Western blot analysis with densitometric quantification (g, n = 3) of PGC1α and UCP1, along with measurements of oxygen consumption rate (OCR) (h and i, n = 4) performed in differentiated brown and beige adipocytes treated with dbcAMP while co-cultured with CD36+ primary adipocytes isolated from BAT and iWAT of Cldn5flox/flox and Cldn5flox/flox; Fabp4-Cre mice. j–m Schematic illustration of the co-culture of differentiated brown and beige adipocytes with AAV-transfected CD36+ primary mature adipocytes isolated from BAT and iWAT of wild-type mice (j). qRT-PCR analysis of Cldn5 (k, n = 4), qRT-PCR analysis of Pgc1α and Ucp1 (l, n = 4), and Western blot with densitometric quantification of PGC1α and UCP1 (m, n = 3) of brown and beige adipocytes with or without dbcAMP treatment. Panel a was created in BioRender89. Panel b was created in BioRender90. Data are expressed as the mean ± SEM. Statistical analyses were performed using two-sided Mann-Whitney test (Pgc1α in brown adipocytes, b) or two-sided unpaired t-tests (other genes in b and all data in c-i, k–m). Ads, Adipocytes. dbcAMP, dibutyryl cAMP. CTL, control. Source data are provided as a Source Data file.
Since cold exposure could convert low-thermogenic brown adipocytes into high-thermogenic cells and white adipocytes into beige adipocytes, another possibility is loss of CLDN5 may disturb the conversions between adipocyte subpopulations. However, our in vitro adipocyte culture data showing that UCP1+ cell number and UCP1 expression level induced by CL-316,243 were not affected by CLDN5 deletion (Supplementary Fig. 6a–c), indicate that CLDN5 is unlikely to be involved in the adipocyte conversion. Together, these data demonstrate that the suppressed thermogenic capacity of BAT and iWAT is likely attributed to a paracrine interaction between cells deficient in CLDN5 and thermogenic cells.
CLDN5 modulates adipocyte YBX3 nuclear translocation and subsequent IL10 production
CLDN5 is known to interact with ZO1 and YBX3. Notably, ZO1 were dominantly expressed in GFP+ adipocytes from the BAT and iWAT at a significantly higher level than GFP- adipocytes (Fig. 6a). Recent studies identified YBX family was involved in adipose tissue homeostasis16,19,24. Accordingly, we hypothesized that CLDN5 forms a complex with ZO1 and YBX3 in adipocytes, which modulates the subcellular targeting and distribution of YBX3. We confirmed the endogenous interaction between CLDN5, ZO1, and YBX3 in adipocyte tissue (Fig. 6b). Confocal imaging showed membrane, cytoplasmic, and weak nuclear YBX3 labeling in Cldn5flox/flox mouse adipocytes and distinct colocalization of YBX3 with DAPI in Cldn5flox/flox; Fabp4-Cre mice (Fig. 6c), which indicated that CLDN5 deletion in adipocytes increased YBX3 nuclear localization. Subcellular fractionation of the BAT and iWAT, followed by Western blot analysis, confirmed the increased nuclear expression of YBX3 in Cldn5flox/flox; Fabp4-Cre mice (Fig. 6d). These observations support that CLDN5 forms a complex with ZO1 and YBX3 in normal adipocytes under physiological conditions, whereas absence of CLDN5 causes unbound YBX3 to translocate to the nucleus. Beyond its role as a transcription factor, YBX3 acts as an RNA-binding protein that modulates mRNA abundance via affecting multiple RNA processing steps. We next examined the mRNA abundance of factors which can be secreted from both brown and white adipocytes25,26,27,28 in Cldn5flox/flox and Cldn5flox/flox; Fabp4-Cre mice. Systematic profiling of 139 secreted factors in Cldn5flox/flox versus Cldn5flox/flox; Fabp4-Cre mice revealed significant dysregulation of 22 adipokines (5 upregulated, 17 downregulated), with Il10 displaying the most dramatic change among the deregulated genes having been reported to be involved in thermogenesis and energy expenditure (Fig. 6e and Supplementary Data 1). Notably, cold exposure under both acute and chronic conditions induced significant downregulation of IL10 accompanied by concurrent upregulation of CLDN5 (Fig. 6f, g). Complementary in vitro studies demonstrated that dbcAMP stimulation triggered an inverse temporal relationship, with progressive CLDN5 induction occurring alongside persistent IL10 suppression (Fig. 6h–k), indicating these pathways may be mechanistically connected in thermogenic regulation.
a qRT-PCR analysis of Zo1 in GFP+ and GFP- adipocytes from BAT/iWAT of CLDN5-GFP mice (n = 3). b Co-IP showing that endogenous CLDN5 interacts with endogenous ZO1/YBX3 in BAT/iWAT. IP/Blot antibodies as indicated. *Lanes show 10% of the input amount of other lanes. c Immunofluorescence staining of YBX3 in BAT/iWAT from Cldn5flox/flox and Cldn5flox/flox; Fabp4-Cre mice. Scale bar, BAT (20 μm) and iWAT (50 μm). d Western blot analysis of the subcellular localization of YBX3 in adipocytes isolated from BAT/iWAT of Cldn5flox/flox and Cldn5flox/flox; Fabp4-Cre mice. ATP1A, GAPDH, and Histone H3 serving as membrane fraction (M), cytosolic fraction (C), and nuclear fraction (N) controls. e qRT-PCR analysis of Il10 in BAT from Cldn5flox/flox and Cldn5flox/flox; Fabp4-Cre mice, n = 6. f, g qRT-PCR analysis (f, in BAT, n = 7 for RT or 7 d, n = 8 for 6 h; in iWAT, n = 8.) and Western blot analysis with densitometric quantification (g, n = 3) of IL10 in BAT and iWAT of wild-type mice housed at RT, 4 °C for 6 h or 7 days (7 d). h–k qRT-PCR (h and j, n = 3) and Western blot (i and k, n = 3) analysis of IL10 in dbcAMP-stimulated brown/beige adipocytes from wild-type mice harvested at the indicated time points. l-m Association of YBX3 with Il10 mRNA as measured by RIP and qRT-PCR analysis from wild-type mice using an YBX3/IgG antibody (l) or from Cldn5flox/flox and Cldn5flox/flox; Fabp4-Cre mice using an anti-YBX3 antibody (m), n = 3 biological replicates. n–p Luciferase assays in HEK293 cells. Il10 3’UTR/promoter activity with YBX3 overexpression (n and p); 3’UTR deletion mutants with YBX3 overexpression (o); n = 4 independent experiments. Statistical analyses were performed using two-sided unpaired t test (a, e, l and m), two-sided one-way ANOVA with Dunnett’s post-hoc test (f–h, j) or Tukey’s post-hoc test (n–p). For b–d, each experiment was repeated three times with consistent results. Data are expressed as the mean ± SEM. Ads, Adipocytes. Source data are provided as a Source Data file.
YBX3 has been previously demonstrated to bind an RNA Y-box protein recognition sequence (YRS) ([UAC][CA]CA[UC]C[ACU])29, and sequence analysis revealed that mouse Il10 transcript has two potential binding sites in its 3′-UTR (ACACCT and CCACCT). We next examined whether YBX3 directly interacted with Il10 mRNA in adipocyte tissue. RNA immunoprecipitation (RIP) analysis using an anti-YBX3 antibody followed by measurement of Il10 mRNA levels by qRT-PCR analysis in the immunoprecipitated material revealed that Il10 mRNA associated with YBX3, as Il10 mRNA was highly enriched in the YBX3 immunoprecipitate relative to the IgG immunoprecipitate (Fig. 6l). In line with the elevated nuclear YBX3 levels in Cldn5flox/flox; Fabp4-Cre mice, there was a seven-fold increase in immunoprecipitated Il10 compared with the control group (Fig. 6m). Next, we generated the reporter constructs containing the entire mouse Il10 3′-UTR sequence cloned downstream of Renilla luciferase gene. The reporter was transfected with YBX3 into HEK293 cells. Luciferase activities were increased following co-transfection with YBX3 expression vectors, and this increase in activity was attenuated by removing the predicted site ACACCT, but not site CCACCT, in Il10 3′-UTR (Fig. 6n, o), indicating that YBX3 acts as an activator of IL10 expression via targeting sequence ACACCT of Il10 3′-UTR. YBX3 has also been reported to control gene expression as a transcription factor. To evaluate whether YBX3 plays a transcriptional role on Il10 promoter, HEK293 cells were transiently transfected with a Firefly luciferase reporter plasmid driven by Il10 promoter fragments together with YBX3 expression vectors or its vector control. Overexpression of YBX3 led to a significant increase in luciferase activity (Fig. 6p), indicating that YBX3 played a crucial role in the regulation of Il10 promoter activity. Taken together, these findings suggest that YBX3 serves as an RNA-binding protein as well as a transcription factor to regulate IL10 expression.
CLDN5 deficiency activates IL10 signaling to limit thermogenesis in vitro
Based on the finding that CLDN5 deficiency regulates IL10 expression, we hypothesize that CLDN5 modulates adipose tissue thermogenesis through IL10-mediated mechanisms. While previous studies have reported variable thermogenic effects of IL1030,31,32,33,34,35,36,37,38, our systematic investigation clarifies its inhibitory role. In wild-type mice, BAT or iWAT injection of recombinant IL10 significantly reduced tissue temperature, downregulated thermogenic genes, and increased lipid droplet size (Supplementary Fig. 7a–h). Conversely, AAV-mediated Il10 knockdown in BAT or iWAT upregulated thermogenic genes and reduced lipid accumulation, though cold tolerance remained unaffected (Supplementary Fig. 7i–p). Complementary in vitro experiments demonstrated that IL10 treatment suppressed both UCP1 and PGC1α levels in differentiated brown and beige adipocytes (Supplementary Fig. 8a–d). These consistent findings establish IL10 as a potent thermogenic inhibitor in adipose tissues.
We observed enhanced IL10 mRNA and protein abundance in CLDN5-expressing adipocytes of Cldn5flox/flox; Fabp4-Cre mice (Fig. 7a–c), accompanied by increased mRNA expression of the well-established IL10 target genes, such as Ucp1 (Fig. 5b, c, f, g), Cidea, Cox8b, Elovl3, and Prdm16 (Fig. 7d, e and Supplementary Fig. 9a), in co-cultured differentiated brown and beige adipocytes with or without dbcAMP. Conversely, CLDN5 overexpression via AAV-mediated gene transfer decreased IL10 secretion in CLDN5-expressing cells from BAT and iWAT (Fig. 7f). To test directly the causal link between CLDN5-dependent IL10 regulation and the altered IL10 signaling in Cldn5-deficient adipocytes, we investigated whether the effects of Cldn5 deletion on IL10 signaling activation could be blunted by IL10 neutralizing antibody. The addition of neutralizing antibody of IL10 to co-culture medium significantly attenuated the regulatory effect of CLDN5 deficiency on Ucp1, Cidea, Cox8b, Elovl3, and Prdm16 gene expression (Fig. 7g, h and Supplementary Fig. 9b) and upregulated the basal and maximal respiration in differentiated brown and beige adipocytes with or without dbcAMP (Fig. 7i, j and Supplementary Fig. 9c). AAV-mediated IL10 knockdown in CD36+ adipocytes from Cldn5flox/flox; Fabp4-Cre mice before co-culture enhanced UCP1 and PGC1α expression in differentiated brown and beige adipocytes (Supplementary Fig. 9d–k). Furthermore, the downregulation of these target genes was abolished when IL10Rα was blocked using an IL10Rα blocking antibody in differentiated brown and beige adipocytes co-cultured with CLDN5-expressing adipocytes from Cldn5flox/flox; Fabp4-Cre mice, both in the absence and presence of dbcAMP (Fig. 7k, l and Supplementary Fig. 9l). These results indicate that CLDN5 may promote the activation of thermogenesis mainly through inhibiting the IL10-IL10R signaling pathway.
a qRT-PCR analysis of Il10 in iWAT from Cldn5flox/flox and Cldn5flox/flox; Fabp4-Cre mice, n = 8. b Western blot analysis with densitometric quantification of IL10 in BAT and iWAT from Cldn5flox/flox and Cldn5flox/flox; Fabp4-Cre mice (n = 3). c IL10 ELISA assays in culture medium collected from adipocytes isolated from BAT and iWAT of Cldn5flox/flox and Cldn5flox/flox; Fabp4-Cre mice (n = 5). d, e qRT-PCR analysis of IL10 target genes in differentiated brown (d) and beige (e) adipocytes treated with dbcAMP while co-cultured with CD36+ primary adipocytes isolated from BAT and iWAT of Cldn5flox/flox and Cldn5flox/flox; Fabp4-Cre mice (n = 4). f IL10 ELISA assays in culture medium collected from AAV-transfected adipocytes isolated from wild-type mice (n = 4). g–j qRT-PCR analysis of IL10 target genes (g and h, n = 3) and OCR (i and j) in differentiated brown and beige adipocytes treated with dbcAMP along with either an IL10 neutralizing antibody or control IgG while co-cultured with CD36+ primary adipocytes isolated from BAT and iWAT of Cldn5flox/flox; Fabp4-Cre mice. i, IgG (n = 3), anti-IL10 (n = 4). j, IgG (n = 4), anti-IL10 (n = 5). k, l qRT-PCR analysis of IL10 target genes in differentiated brown (k) and beige (l) adipocytes treated with dbcAMP along with either an IL10Rα blocking antibody or control IgG while co-cultured with CD36+ primary adipocytes isolated from BAT and iWAT of Cldn5flox/flox; Fabp4-Cre mice (n = 3). Data are expressed as the mean ± SEM. Statistical analyses were performed using two-sided Mann–Whitney test (BAT in c) or two-sided unpaired t test (a, b, iWAT in c, d–l). Ads, Adipocytes. Source data are provided as a Source Data file.
IL10Rα is known to trigger the activation of signal transducer and activator of transcription 3 (STAT3)39,40,41, we next assessed whether CLDN5 deletion affects thermogenesis through the STAT3 signaling pathway. We co-cultured CLDN5-expressing primary adipocytes with differentiated brown or beige adipocytes and observed that the phosphorylation of STAT3 in these adipocytes was significantly elevated when co-cultured with CLDN5-expressing adipocytes isolated from Cldn5flox/flox; Fabp4-Cre mice compared to those co-cultured with CLDN5-expressing adipocytes isolated from Cldn5flox/flox mice (Supplementary Fig. 9m). When we added the specific STAT3 inhibitor C188-9 to the co-culture medium of adipocytes isolated from Cldn5flox/flox; Fabp4-Cre mice, the phosphorylation level of STAT3 was inhibited, leading to the restored expression of UCP1 and other thermogenesis-related genes, both in the absence and presence of dbcAMP (Supplementary Fig. 9n–q). These results indicate that CLDN5 may regulate thermogenesis by modulating the STAT3 signaling pathway.
CLDN5 deficiency activates IL10 signaling to limit thermogenesis in vivo
In order to determine whether the effect of Cldn5 deficiency on energy expenditure is attributable to increased IL10-IL10R signaling in vivo, we performed a loss-of-function study using multiple sites in situ injections of siRNA targeting Il10ra (si-Il10ra) in BAT and iWAT (Fig. 8a, b). Direct injection of si-Il10ra into the BAT of Cldn5flox/flox; Fabp4-Cre mice led to a reduction in IL10Rα mRNA and protein levels by 48.8% and 43.8%, respectively, compared to mice receiving a scrambled control siRNA (Fig. 8c, d). Protein levels of PGC1α and UCP1 (Fig. 8d) and mRNA levels of Pgc1α, Ucp1, Cidea, Cox8b, Elovl3, and Prdm16 were also markedly enhanced in the BAT of Cldn5flox/flox; Fabp4-Cre mice that were treated with si-Il10ra (Fig. 8e). We also noted that lipid content was reduced in BAT (Fig. 8f). Mice receiving si-Il10ra were better able to withstand a cold challenge relative to mice that received the control siRNA (Fig. 8g). Consistently, mice administered multiple orthotopic injections of si-Il10ra in iWAT exhibited diminished levels of IL10Rα mRNA and protein (Fig. 8h, i), elevated expression of Ucp1, Pgc1α, Cidea, Cox8b, Elovl3 and Prdm16 (Fig. 8i, j), reduced lipid droplet accumulation (Fig. 8k), and enhanced cold challenge responsiveness (Fig. 8l), in comparison to mice receiving the control siRNA.
a, b Schematic of experimental design in Cldn5flox/flox; Fabp4-Cre mice, with in situ BAT (a) or iWAT (b) injections of Il10ra siRNA (si-Il10ra) or control siRNA (si-Ctrl), maintained at 4 °C for 7 days. c–g Analysis in BAT after siRNA injections: qRT-PCR analysis of Il10ra (c, n = 5), Western blot analysis with densitometric quantification of PGC1α, IL10Rα, and UCP1 (d, n = 3), qRT-PCR analysis of IL10 target genes (e, n = 5), HE staining with lipid droplet size quantification (f, n = 5), and rectal temperature (g, n = 7 for si-Ctrl, n = 9 for si-Il10ra). h-l Analysis in iWAT after siRNA injections: qRT-PCR analysis of Il10ra (h, n = 5), Western blot with densitometric quantification analyses of PGC1α, IL10Rα, and UCP1 (i, n = 3), qRT-PCR analyses of IL10 target genes (j, n = 5), HE staining with lipid droplet size quantification (k, n = 5), and rectal temperature (l, n = 5 for si-Ctrl, n = 7 for si-Il10ra). m, n Schematic of experimental design in Cldn5flox/flox; Fabp4-Cre mice, with in situ BAT (m) or iWAT (n) injections of AAV-sh-Ctrl or AAV-sh-Il10, housed for 3 weeks before 7 days at 4 °C. o-s Analysis in BAT AAV-injected mice: qRT-PCR analysis of Il10 in BAT, liver, and muscle (o, n = 6), Western blot analysis with densitometric quantification of IL10, PGC1α, and UCP1 (p, n = 3), qRT-PCR analysis of IL10 target genes (q, n = 6), HE staining with lipid droplet size quantification (r, n = 5), rectal temperature (s, n = 10). t–x Analysis in iWAT AAV-injected mice: qRT-PCR analysis of Il10 in iWAT, liver, and muscle (t, n = 6), Western blot analysis with densitometric quantification of IL10, PGC1α, and UCP1 (u, n = 3), qRT-PCR analysis of IL10 target genes in iWAT (v, n = 6 for AAV-sh-Ctrl, n = 7 for AAV-sh-Il10), HE staining with lipid droplet size quantification (w, n = 5), and rectal temperature (x, n = 10). Data are expressed as the mean ± SEM. Statistical analyses were performed using two-sided unpaired t test. Source data are provided as a Source Data file. Panels a and m were created in BioRender91. Panels b and n were created in BioRender92.
To further test the causal relationship between loss of CLDN5, IL10 upregulation, and mouse phenotype, we treated Cldn5flox/flox; Fabp4-Cre mice with recombinant adeno-associated virus serotype 2/9 (rAAV2/9) containing the FABP4 promoter to drive Il10 shRNA expression. Cldn5flox/flox; Fabp4-Cre mice received multiple orthotopic injections of 6.4 × 1010 vg particles of either AAV-sh-Ctrl or AAV-sh-Il10 in the BAT or iWAT pads on both sides and were then kept for an additional 3 weeks prior to cold exposure. (Fig. 8m, n). qPCR studies indicated that the AAV-sh-Il10 significantly inhibited IL10 expression in BAT, but did not affect Il10 transcript levels in other thermogenic organs, including liver and muscle (Fig. 8o, p). Under cold conditions, we found that Il10 knockdown significantly upregulated the expression of thermogenic genes (Fig. 8p, q), reduced lipid droplet accumulation (Fig. 8r), and improved the cold intolerance (Fig. 8s) in Cldn5flox/flox; Fabp4-Cre animals following in situ injection in the BAT pads. Consistently, mice that received AAV-sh-Il10 in iWAT consistently demonstrated reduced levels of IL10 mRNA and protein, while expression levels in muscle and liver remained unchanged (Fig. 8t, u). Additionally, these mice exhibited increased expression of Ucp1, Pgc1α, Cidea, Cox8b, Elovl3, and Prdm16 (Fig. 8u, v), along with decreased lipid droplet accumulation (Fig. 8w) and improved responsiveness to cold challenges (Fig. 8x) compared to those receiving AAV-sh-Ctrl. These findings suggest that inhibiting the IL10-IL10R signaling pathway in mice can mitigate the impaired thermogenesis caused by CLDN5 deficiency in BAT and iWAT.
Cldn5 deletion using Ucp1-Cre and Adipoq-Cre mice recapitulates findings in Cldn5 flox/flox; Fabp4-Cre mice
Fabp4-Cre mice have been reported to exhibit quite broad expression of Cre recombinase in both adipose and non-adipose tissues such as brain, muscle, macrophages, and intestine42. In adipose tissues, Fabp4-Cre is expressed in mature adipocytes as well as adipo-progenitors43. To further examine whether the defective in cold induced thermogenesis and obesity-prone phenotype in the Cldn5flox/flox; Fabp4-Cre mice was due to loss of CLDN5 in adipocytes or not, we deleted the Cldn5 gene in adipocytes using Ucp1-Cre and Adipoq-Cre mouse lines. It has been proved that the BA-L adipocytes with low expression of Ucp1 could convert into UCP1 high-expressing cells upon cold temperature exposure2. Meanwhile, UCP1 is highly induced in iWAT after cold exposure, due to the widespread increase in beige adipocytes. In agreement with the induction of UCP1 by cold stimuli, the Cldn5flox/flox; Ucp1-Cre mice displayed significantly reduced expression of CLDN5 in BAT and iWAT after one week of housing at 4 °C (Fig. 9a, b). Furthermore, the thermogenesis of Cldn5flox/flox; Ucp1-Cre mice was also dampened as indicated by intolerance to cold challenge (Fig. 9c), decreased UCP1 expression (Fig. 9d, e), and less multilocular lipid droplets in BAT and iWAT by HE staining (Fig. 9f). Additionally, Cldn5flox/flox; Ucp1-Cre female mice gained more weight and consumed less energy than the Cldn5flox/flox control mice on HFD as indicated by body weight (Fig. 9g, h), size and weight of fat tissue (Fig. 9i–l), adipocyte hypertrophy (Fig. 9m), energy expenditure (Fig. 9n). Furthermore, no difference in food intake, locomotor activity and RER was observed between the two groups of mice (Fig. 9o–q). Similarly, CLDN5 expression was significantly reduced in BAT and iWAT of Cldn5flox/flox; Adipoq-Cre mice at 4 °C for 1 week (Fig. 9r, s). Accordingly, Cldn5flox/flox; Adipoq-Cre female mice were also more sensitive to cold-induced hypothermia (Fig. 9t) and HFD-induced obesity (Fig. 9u) compared with Cldn5flox/flox control mice. Thus, both the Cldn5flox/flox; Adipoq-Cre and Cldn5flox/flox; Ucp1-Cre mice recapitulated our findings in Cldn5flox/flox; Fabp4-Cre mice. Collectively, these data clearly demonstrate that CLDN5 directly targets adipocytes to promote thermogenesis and protect against obesity.
a–e qRT-PCR analysis (a, n = 6) and Western blot analysis (b, n = 3) of CLDN5 in BAT/iWAT, rectal temperature and interscapular BAT temperature (c, n = 7), qRT-PCR analysis (d, n = 5 for Cldn5flox/flox mice, n = 6 for Cldn5flox/flox; Ucp1-Cre mice) and Western blot analysis of UCP1 (e, n = 3) in BAT/iWAT of Cldn5flox/flox and Cldn5flox/flox; Ucp1-Cre mice kept at 4 °C. f HE staining with lipid droplet size quantification in BAT/iWAT of Cldn5flox/flox and Cldn5flox/flox; Ucp1-Cre mice kept at RT or 4 °C (n = 5). g–q Body weights (g), gross appearance of representative mice (h), gross appearance, weights and weights normalized by tibial length of BAT, iWAT, and eWAT (i–k), representative MRI images shown in axial and coronal planes (l), HE staining with average lipid droplet size quantification (m, n = 5), energy expenditure (n), food intake (o), locomotor activity (p) and RER (q) in Cldn5flox/flox and Cldn5flox/flox; Ucp1-Cre mice fed HFD for 16 weeks. g, j and k, n = 13 for Cldn5flox/flox mice, n = 14 for Cldn5flox/flox; Ucp1-Cre mice. Quantification of MRI images is expressed as a percentage of total fat area over total area (n = 4). n–q, n = 5. r-t qRT-PCR analysis (r, BAT: n = 7 for Cldn5flox/flox mice, n = 6 for Cldn5flox/flox; Adipoq-Cre mice; iWAT: n = 7 per genotype) and Western blot analysis (s, n = 3) of CLDN5 in BAT/iWAT, rectal temperature, and interscapular BAT temperature (t, n = 12 for Cldn5flox/flox mice, n = 10 for Cldn5flox/flox; Adipoq-Cre mice) of Cldn5flox/flox and Cldn5flox/flox; Adipoq-Cre mice kept at 4 °C. u Body weights of the Cldn5flox/flox (n = 7) and Cldn5flox/flox; Adipoq-Cre (n = 9) mice fed a HFD for 16 weeks. Data are expressed as the mean ± SEM. Statistical analyses were performed using two-sided Mann-Whitney test (g, BAT in j and k, axial view in l, u), two-sided unpaired t test (a–f, iWAT in j and k, coronal view in l, m, r–t), two-sided analysis of ANCOVA (n, o) or two-sided ANOVA (p, q). A, anterior; R, right; L, left; P, posterior; S, superior; I, inferior. Scale bar: 5 mm (l); 100 μm (f, m). Source data are provided as a Source Data file.
Altered CLDN5 expression in human adipose tissue associates with metabolic disorders
To provide a translational perspective on our findings, we explored the GEO database (https://www.ncbi.nlm.nih.gov/geo/)44,45 to obtain and analyze CLDN5 expression data in human adipose tissue across various conditions. Analysis of the GSE962446 revealed that CLDN5 expression was elevated in the omental adipose tissue of children with obesity compared to normal-weight children (Supplementary Fig. 10a). Further analysis of the GSE250847 showed a significant increase in CLDN5 expression in subcutaneous adipose tissue from non-diabetic female Pima Indians with obesity compared to non-diabetic lean female subjects (Supplementary Fig. 10b). Additionally, analyses of the GSE15690648 and GSE2095049 demonstrated a significant decrease in CLDN5 levels in subcutaneous adipose tissue from metabolically unhealthy individuals with obesity compared to metabolically healthy individuals with obesity, as well as a significant reduction in insulin-resistant patients with obesity compared to insulin-sensitive patients with obesity (Supplementary Fig. 10c, d). Furthermore, the GSE2795150 indicated that patients with obesity with type 2 diabetes exhibited lower CLDN5 expression levels in subcutaneous adipose tissue compared to lean patients (Supplementary Fig. 10e). Overall, these findings indicate that CLDN5 may play a critical role in obesity and metabolic diseases, warranting further investigation into its mechanisms and potential therapeutic implications.
Discussion
While CLDN5 is most commonly and prominently expressed by epithelial or endothelial cells, whereby it performs barrier-type functions, CLDN5 has also been implicated in fat tissue and adipocytes2. Here, we have outlined an unexpected role for CLDN5 in the modulation of adipocyte thermogenesis. Genetic ablation of Cldn5 in mice impaired thermogenic function, at least in part due to elevated expression of IL10, a direct target of activation by YBX3. Human adipose tissue also expresses CLDN5, suggesting CLDN5 may play a similar pro-thermogenic function in humans as well.
First, CLDN5 expression was found in mouse WAT and BAT, implying a functional significance of CLDN5 in adipose tissue. Immunofluorescence analyses indicated that CLDN5 is mainly associated with the plasma membranes of adipocytes. Adipocytes in BAT and WAT can be classified into three distinct cell types: white adipocytes, responsible for energy storage; brown adipocytes, which efficiently burn energy for heat production through non-shivering thermogenesis; and a related, less thermogenically efficient population known as beige adipocytes51. Recent studies employing single cell RNA-seq or single-nuclei RNA-seq have provided insights into diverse subpopulations of adipocytes with distinct molecular and metabolic features within a single adipose depot52. By using transgenic mice expressing a GFP under the control of the Cldn5 promoter, we observed CLDN5 was expressed in only a small subset of cells within both white and brown adipose tissues. In BAT, CLDN5 is relatively enriched in non-thermogenic adipocytes with low expression levels of UCP1 and ADIPOQ, and its expression is influenced by environmental temperature. In WAT, CLDN5 is expressed in a small proportion of non-thermogenic mature adipocytes as well as in preadipocytes.
To directly investigate the role of CLDN5 in adipose tissues, we have generated three mouse models with the CLDN5 gene deletion mediated by Fabp4-Cre, Adipoq-Cre, or Ucp1-Cre, respectively. Fabp4-Cre is expressed in mature adipocytes as well as adipocyte progenitor cells, while recombination by Adipoq-Cre primarily occurs in mature adipocytes23,53. By contrast, Ucp1-Cre mice exhibit abundant Cre expression in the thermogenic brown adipocytes of the BAT and beige adipocytes of the iWAT54. However, it’s worth noting that all three Cre lines are not specifically restricted to adipose tissue. For example, Adipoq-Cre causes recombination in some osteoblasts55 and reproductive accessory glands56, whereas Fabp4-Cre causes widespread recombination due to a broader tissue expression23. The activity of Ucp1-Cre was observed in brown and inguinal adipose tissues, as well as in the kidney, adrenal glands, thymus, and hypothalamus54. We thus investigated the phenotype of Cldn5 conditional deletion mice using all three Cre lines. Here, we obtained different Cldn5 knockout efficiency in adipose tissue among three mouse models at room temperature with a normal chow diet. In BAT, Fabp4-Cre efficiently deleted Cldn5, whereas Adipoq-Cre and Ucp1-Cre failed to induce Cldn5 deletion. This finding further supports the enrichment of CLDN5 within the brown adipocyte subpopulation characterized by low levels of UCP1 and ADIPOQ. In WAT, both Fabp4-Cre and Adipoq-Cre induced efficient deletion of Cldn5 in adipocytes, whereas Ucp1-cre failed to catalyze the recombination, in agreement with the distribution of CLDN5 in non-thermogenic adipocytes. We also found that Ucp1-Cre-induced Cldn5 knockout in BAT and iWAT occurred when mice were housed at 4 °C for 1 week, consistent with the induction of Ucp1 by cold stimuli.
After validating efficient Cldn5 knockout in both BAT and WAT using Fabp4-Cre, we performed phenotypic characterization of these knockout mice. To circumvent potential effects of gene deletion by Fabp4-Cre in tissues other than adipocytes, we supplemented our analysis with Cldn5 knockout mice mediated by Adipoq-Cre and Ucp1-Cre, as well as in vitro analysis. Cldn5 KO mice that maintained on a normal diet showed significantly reduced energy expenditure without altered food intake. Consistent with their decreased energy expenditure, mice deficient in CLDN5 were more susceptible to diet-induced obesity and glucose intolerance than WT mice. To determine whether the effect of CLDN5 deficiency on energy expenditure is attributable to decreased heat production, we next analyzed adaptive thermogenesis. We performed a cold tolerance test and pharmacological β3-adrenergic stimulation and found that Cldn5 KO mice were hypersensitive to cold-induced hypothermia, providing strong evidence that Cldn5 KO mice are defective in energy expenditure and adaptive thermogenesis compared to WT mice. Abolishing Cldn5 in adipocytes with Ucp1-Cre or Adipoq-Cre mirrored the phenotype observed in mice with Fabp4-Cre, including the blunting of cold-induced metabolic improvements as indicated by intolerance to cold challenge, decreased UCP1 expression and less multilocular lipid droplets, as well as the predisposed hypersensitivity to HFD-induced obesity. However, these differences were less pronounced than in Cldn5 ablation mice using the Fabp4-Cre, suggesting that CLDN5 on other cell types beyond mature adipocytes also exerts a functional role. Collectively, these data suggest that CLDN5 is relatively enriched in the non-thermogenic adipocyte subpopulation; however, absence of CLDN5 affects the thermogenic function of BAT and iWAT.
Adipose tissue is composed of mature adipocytes, adipocyte progenitors, immune cells, endothelial cells, smooth muscle cells, pericytes, neurons, and Schwann cells57. In addition to the intercellular crosstalk between adipocytes and other cell types within adipose tissue, adipocytes secrete numerous factors to modulate the thermogenic activity of adjacent adipocytes or themselves via paracrine pathways51. By using CLDN5-expressing primary mature adipocytes co-cultured with brown/beige adipocytes differentiated from SVF, we found that CLDN5 regulates thermogenesis through paracrine signaling and its actions on thermogenic adipocytes. The impaired thermogenesis and energy expenditure observed in Cldn5 KO mice are, at least in part, attributed to the impact on secretion of IL10, an anti-inflammatory cytokine which inhibits adrenergic signaling and adipose thermogenesis through its receptor IL10Rα expressed in mature adipocytes30. Forced expression of Il10 siRNA in adipocyte tissue or AAV-mediated Il10 knockdown in adipocytes is sufficient to reverse the thermogenic defects caused by Cldn5 deficiency. The capacity for adipocytes to produce IL10 is intriguing, given that myeloid cells such as macrophages and dendritic cells are important IL10 producers. However, the expression of IL10 in adipocytes has also been reported previously33,58. Thus, these findings expand our understanding of the complexity of regulatory links between thermogenic adipocytes and non-thermogenic adipocytes. Further investigation is required to determine whether the heightened production of adipocyte-derived secreted IL10 could exert a systemic effect on other tissues, thus contributing to the consequences of Cldn5 ablation.
We provide both in vitro and in vivo evidence that IL10 is a downstream target of CLDN5 in adipocytes and CLDN5 deficiency increased mRNA and protein levels of IL10, which has been demonstrated to directly suppress thermogenesis in adipocytes via a STAT3-dependent signaling pathway30,31,32,33. Interestingly, several studies have observed the opposite of these results; namely, IL10 shows a pro-thermogenic action34,35,36,37,38. While the mechanisms underlying these divergent observations remain unclear, potential influencing factors may include genetic background variations, metabolic states, and the inflammatory status of experimental models59,60,61,62,63. While our study focused on IL10, however, it is important to consider other potential factors and pathways that might be involved in the observed changes in thermogenic capacity due to CLDN5 deficiency. The deletion of CLDN5 also perturbs the expression of both well-characterized thermogenic adipokines, including PHOSPHO1 and GNAS64,65,66, along with 18 additional factors whose metabolic functions remain uncharacterized. Although these identified candidates may contribute to the observed phenotypic effects, their biological roles require further experimental validation. Moreover, considering the fundamental developmental and functional differences between brown and beige adipocytes, subsequent investigations should systematically examine potential depot-specific functions of CLDN5 to comprehensively elucidate its tissue-dependent regulatory mechanisms.
Apart from two canonical functions of known as fence and gate, several proteins of the tight junction have been found to participate in the regulation of gene expression67. The tight junction adaptor protein ZO1 interacts with the transcription factor YBX3 and controls accumulation of YBX3 in the nucleus by cytoplasmic sequestration11,68. YBX3 has been reported to be involved in transcriptional activation and repression, as well as in post-transcriptionally translation and mRNA stability69. We show here that the existence of a complex containing CLDN5, ZO1, and YBX3 in adipocytes. The loss of CLDN5 is likely to disrupt this membrane protein complex, resulting in the translocation of YBX3 into the nucleus. This translocation leads to the upregulation of IL10 expression, facilitated by YBX3 binding to specific sites in the 3’-UTR of IL10 mRNA as well as the IL10 promoter, thereby regulating its expression. In fact, this is not the first report of the regulatory role of YBX3 in cytokine production. YBX3 was recently documented to promote the production of various proinflammatory cytokines by stabilizing NFKBIZ, thereby contributing to YBX3-mediated proviral effects for SARS-CoV-270.
Our observations indicate that CLDN5 plays a role in the response to cold shock through the regulation of ZO1–YBX3 signaling pathway in adipocytes. In line with this, we observed increased expression of CLDN5 upon cold exposure or treatment with the adrenergic receptor β−3 agonist CL-316,243, and the stress-induced CLDN5 might increase the interaction with ZO1 and results in cytoplasmic sequestration of YBX3. Interestingly, the mRNA level of YBX3 was also quickly elevated following cold exposure in a separate study71,72. Cold exposure thus leads to two opposed mechanisms that regulate YBX3 activation: a pathway that inhibits YBX3 nuclear translocation via increasing the expression of CLDN5 and a pathway that increase the mRNA expression of YBX3 by external stimuli. It will be important to identify additional YBX3 upstream regulators to fully comprehend its role in response to environmental stress. Furthermore, our finding that YBX3 activation contributes to impaired thermogenic capacity by upregulating the expression of IL10 would suggest that YBX3 functions differently from its family members YBX1 and YBX2. However, at this time, we cannot completely rule out its role in other processes involving cold adaption, and future comprehensive studies will be necessary to directly investigate its functional relationship to this stress.
In summary, these findings expand our understanding about the location and function of the extra-tight junction CLDN proteins and provide molecular insights into signaling mechanisms underlying adipose thermogenesis which will undoubtedly lead to identification of pathways controlling thermogenic adipocyte activity, depicted in in Fig. 10. The altered expression of CLDN5 in human adipose tissue, as demonstrated by our analysis of multiple datasets, suggests that CLDN5 may play a significant role in the development of obesity and other metabolic disorders. Targeting CLDN5 could therefore have potential therapeutic implications for metabolic diseases. In addition, considering the central role that tight junction transmembrane proteins play in mechano-sensing and mechano-transduction, coupled with the mechanosensitive and mechanoresponsive properties that adipocytes possess73,74, it is conceivable that alterations in mechanical stress and cell volume, influenced by physical stimuli or nutritional conditions, could impact adipocyte physiology by affecting tight junction transmembrane proteins. Furthermore, CLDN5 was originally identified as one of the deleted genes in 22q11 deletion syndrome (22q11DS), also known as DiGeorge syndrome or Velocardiofacial syndrome characterized genetically by microdeletions within chromosome 22 which can comprise up to 40 genes75. The prevalence of obesity and type 2 diabetes was significantly greater in 22q11.2DS than in the general population76,77, however, contributors to obesity have not been determined. Our findings also provide valuable information for better understanding the phenotypes and support the involvement of CLDN5 in predisposing an increased risk of obesity in individuals with 22q11DS.
CLDN5 is highly enriched in the UCP1 low-expressing adipocytes subpopulation. CLDN5 forms a complex with ZO1 and YBX3 in normal adipocytes under physiological conditions. Mice lacking CLDN5 in adipocytes have reduced heat production, reduced energy consumption, and an obesity-prone phenotype. CLDN5 deletion causes YBX3 to enter the nucleus, promotes the expression and secretion of IL10, and then affects the expression of thermogenic genes in neighboring thermogenic cells through IL10-IL10Ra signaling. The graphic was created in BioRender93.
Limitations of the Study
First, although we observed an increase in CLDN5 in BAT and iWAT tissue of mice during cold adaptation, a detailed understanding of the mechanisms by which cold is perceived by these cells, as well as the identification of signals that control CLDN5 transcription to drive thermogenesis, remains elusive. Second, because CLDN5 was found to be enriched in the CD36+ cell subpopulation, this study utilized CD36-expressing adipocytes as a proxy for CLDN5-expressing adipocytes in the in vitro analysis of iWAT. However, it is crucial to acknowledge that CD36 is not exclusively specific to CLDN5-expressing cells; it is observed in both CLDN5+ and CLDN5- cells. Third, macrophages express high levels of IL10R and have been suggested to be the important target of IL10 effects78,79. Whether macrophages are also the target for the metabolic effects of CLDN5/IL10 pathway to facilitate or inhibit adipose thermogenesis remains to be established.
Methods
Animals
Mice were housed in a standard environment which was characterized by 12 h light/dark cycle, 22-25 °C, and 40-60% humidity with free access to water and chow (Jiangsu Xietong Pharmaceutical Bio-engineering Co., Ltd, XTI01FZ-003). All animals were maintained in specific pathogen-free facilities.
Cldn5flox/flox mice were generated by constructing a targeting vector with an SDA-flanked neomycin cassette and flox sites around the first exon of Cldn511. After crossing with C57BL/6 J mice to remove the Neo transgene, heterozygous Cldn5flox/+ mice were intercrossed to produce homozygous Cldn5flox/flox mice. Cldn5flox/flox mice were crossed with Fabp4-cre (Jackson Laboratory, 018965), Adipoq-cre (Jackson Laboratory, 028020), or Ucp1-cre (Jackson Laboratory, 024670) to generate conditional knockout mice. Homozygous with Cre negative littermates served as controls.
C57BL/6 J mice were obtained from Vital River Laboratory (China). The transgenic CLDN5-GFP mouse was kindly provided by Dr. Betsholtz and Dr. Gaengel from Uppsala University. Our study examined male and female animals, and similar findings are reported for both sexes. Female and male mice were used unless otherwise specified.
Cell lines
HEK293 cells (CRL-1573) were obtained from ATCC (USA) and cultured according to the distributor’s recommendations.
Plasmids
Mouse Ybx3 transcript variant 1 (pCMV6-Ybx3-V1, NM_139117) and Ybx3 transcript variant 2 (pCMV6-Ybx3-V2, NM_011733) expression plasmids were purchased from Origene Technologies (USA).
Antibodies
The antibodies used in this study are summarized in Supplementary Table 1.
Cold exposure studies
For the 4 °C cold exposure experiment, 8-10-week-old mice were individually or doubly housed in a non-bedded cage at 4 °C with access to food and water for the time points indicated in the figure legend. Rectal temperatures were measured using a digital thermometer (KEW BASIS, China). Interscapular BAT temperatures were recorded using a FLIR T540 infrared camera (FLIR Systems, USA) and analyzed with a specific software package (FLIR Tools Software). For each animal per group, three pictures were taken and analyzed. At the end of the experiment, mice were sacrificed to collect samples.
CL-316,243 treatment
8-10-week-old Cldn5flox/flox and Cldn5flox/flox; Fabp4-Cre mice were intraperitoneally injected with CL-316,243 (MedChemExpress, HY-116771A) at a dose of 1 mg/kg body weight or an equal volume of saline daily. After 10 days, mice were sacrificed to collect samples.
HFD feeding
8-week-old Cldn5flox/flox; Fabp4-Cre mice, Cldn5flox/flox; Ucp1-Cre mice, Cldn5flox/flox; Adipoq-Cre mice and their corresponding littermate Cldn5flox/flox mice were fed a HFD (Jiangsu Xietong Pharmaceutical Bio-engineering Co., Ltd, XTHF60-1). The composition of the diet includes a protein mass ratio of 23.25%, a fat mass ratio of 34.55%, and a carbohydrate mass ratio of 27.2%. The corresponding energy contributions are as follows: 18.14% from protein, 60.65% from fat, and 21.22% from carbohydrates. Body weight was measured weekly and belly fat distribution was determined using Bruker Biospec MRI system (Bruker BioSpin). At the end of the experiment, serum and tissue samples were collected for further analysis.
Il10ra siRNA treatment
The injection of siRNA into mice for gene knockdown has been widely reported80,81,82, involving local administration, systemic administration, and target administration. In this study, each interscapular BAT from Cldn5flox/flox; Fabp4-Cre mice, aged 8-10 weeks, received multiple sites in situ injections with si-Il10ra (3 nmol, RiboBio) or its negative siRNA control (si-Ctrl, RiboBio) in 50 μL saline twice a week. For iWAT administrations, another set of Cldn5flox/flox; Fabp4-Cre mice, aged 8-10 weeks, were injected with 3 nmol si-Il10ra or si-Ctrl on both sides of the iWAT pads twice a week. After the second injection, mice were transferred to a 4 °C environment. The third injection was given four days later, and body temperature was measured after seven days in the cold chamber. At the end of the treatment, mice were euthanized, and various tissues were collected for subsequent experiments. The Il10ra siRNA targeted sequence: GCATCTTAGTCATATCTATGC.
rAAV2/9 injection to BAT and iWAT
The recombinant AAV2/9 vector expressing Il10-targeting shRNA under the control of the adipocyte-specific FABP4 promoter (rAAV2/9-Fabp4-ZsGreen-miR30-Il10 shRNA, AAV-sh-Il10) and its corresponding control vector (rAAV2/9-Fabp4-ZsGreen, AAV-sh-Ctrl) were designed and synthesized by Hanbio Co. Ltd. Eight-week-old wild-type and Cldn5flox/flox; Fabp4-Cre mice received multi-point injections of either AAV-sh-Il10 or AAV-sh-Ctrl (6.4 × 10^10 viral genomes in 40 μL PBS per injection) into the interscapular BAT or bilateral iWAT depots. Three weeks post-injection, the mice were transferred to a 4 °C environment for seven days, after which their body temperature were measured. At the conclusion of the treatment, the mice were euthanized, and various tissues were collected for subsequent experiments. The targeted sequence for the Il10 shRNA was GAATGAATTTGACATCTTCAT.
Recombinant IL10 Administration in Adipose Tissue
10-week-old wild-type mice were randomly allocated into four experimental groups and received bilateral, multi-point injections of either PBS (vehicle control) or recombinant mouse IL10 (SinoBiological, 50245-MNAE) at doses of 50, 250, or 500 ng in 40 μL PBS into the interscapular BAT or bilateral iWAT depots. Injections were administered twice weekly for one week to ensure sustained IL10 exposure. During the treatment period, all mice underwent a 7-day cold challenge at 4 °C to activate thermogenic pathways prior to terminal analysis. Interscapular BAT/iWAT temperatures were monitored throughout the cold exposure period using a FLIR T540 infrared camera, and tissues were harvested for molecular and histological assessments.
Glucose tolerance tests (GTT) and Insulin tolerance tests (ITT)
GTT was conducted after a 12-hour fasting period. Blood glucose levels were measured at 0, 15, 30, 45, 60, 90, and 120 minutes following the intraperitoneal injection of D-glucose (Sigma, G7021; 1.25 g/kg for HFD-induced mice and 1.5 g/kg for normal chow-induced mice). ITT was performed after 4 hours of fasting, with blood glucose levels measured at 0, 15, 30, 45, 60, 90, and 120 minutes following the intraperitoneal injection of insulin (Biological Industries, 41-975-100; 1.2 U/kg for HFD-treated mice and 0.8 U/kg for normal diet-treated mice). Blood glucose concentrations were measured by tail vein bleeding using a blood glucose meter (Yuwell, China) at the designated time points after injection.
Lipid and IL10 Measurement
After a 12-hour fasting period, mice were subjected to blood sample collection into a 1.5 mL tube, followed by incubation at room temperature for 2 hours. The blood was then centrifuged at 2000 x g for 20 minutes, and the serum was collected and stored at −80 °C. Serum levels of total cholesterol (T-CHO), HDL cholesterol (HDL-C), LDL cholesterol (LDL-C), triglycerides (TG), and free fatty acids (FFA) were measured using commercial kits (Nanjing Jiancheng, China; T-CHO, A111-1-1; HDL-C, A112-1-1; LDL-C, A113-1-1; TG, A110-1-1; FFA, A042-2-1) in accordance with the manufacturer’s instructions. IL10 levels in tissues or cell culture medium were determined using a mouse IL10 ELISA kit (MLBIO, China, mlC50274-1) following the manufacturer’s instructions.
Metabolic cages
The energy consumption and expenditure of mice were assessed using the Comprehensive Laboratory Animal Monitoring System (CLAMS, Columbus Instruments). Mice were individually housed in chambers under a 12-hour light-dark cycle, with free access to food and water. Prior to data recording, the mice underwent a 2-day acclimation period in the metabolic cages. Energy expenditure, food intake, and physical activity were continuously monitored for 24 hours.
Stromal vascular fractions (SVF) culture and adipocyte differentiation
White and brown SVF cells were isolated and differentiated into beige and brown adipocytes, respectively, according to standard procedures as previously described83. Briefly, BAT or iWAT was excised from mice, minced, and placed in the digestion buffer containing 1.5 mg/mL collagenase type II (Worthington-Biochem, LS004176) and dispase II (Gibco, 17105-041) at 37 °C for 30 minutes. The digested mixture was passed through 100 μm cell strainer and centrifuged at 500 x g for 10 mins at room temperature to pellet SVF cells. SVF cell pellets were resuspended in DMEM medium containing high glucose (VivaCell, C3113-500), 10% fetal bovine serum (FBS; VivaCell, C04001-500), and 1% penicillin/streptomycin (Meilun Biotech Co., Ltd., MA0110) and seeded on collagen-coated dishes. For differentiation assays, 95-97% confluent SVF cells were changed with induction medium containing 62.5 μM indomethacin (Sigma, I7378), 0.5 mM 3-isobutyl-1-methylxanthine (IBMX; Sigma, I5879), 2 μg/mL dexamethasone, 1 μM insulin (Biological Industries, 41-975-100), 1 nM 3,3ʹ,5-triiodo-L-thyronine (T3; Sigma, T2877), and 1 μM rosiglitazone (Sigma, R2408) for 3 days. The medium was then changed to maintenance medium (high-glucose DMEM with 10% FBS, 1 μM insulin, 1 nm T3, and 1 μM rosiglitazone), and then cultured for 7 days. The medium was replaced with new maintenance medium every other day. For fluorescence-activated cell sorting (FACS), SVF cells isolated from BAT and iWAT of CLDN5-GFP mice were sorted using a BD FACS Aria™ III Cell Sorter and BD FACSDiva software 7.0 (BD Biosciences).
In vitro adipocyte stimulation assays
To model cold-induced thermogenic activation in vitro, differentiated adipocytes were treated with 0.5 mM dibutyryl-cAMP (dbcAMP; Selleck Chemicals, S7858), a membrane-permeable cAMP analog. Time-course experiments (0, 1, 3, 6, 12, and 24 h exposure) were conducted to profile the dynamic regulation of CLDN5 and IL10 during thermogenic stimulation.
For IL10 functional studies, wild-type-derived brown and beige adipocytes were exposed to recombinant mouse IL10 (PeproTech, 210-10-10UG) at graded concentrations (0–500 ng/mL) for 16 h. Quantitative PCR and immunoblotting were subsequently performed to assess thermogenic gene expression.
Primary adipocyte isolation, culture, and sorting
BAT or iWAT was dissected, minced, and placed in a digestion buffer containing 1.5 mg/mL of collagenase type II and dispase II. The mixture was incubated in a shaking water bath at 37 °C and 190 rpm for 30 minutes. After digestion, 5 mL DMEM medium (containing high glucose, 10% FBS, and 1% penicillin/streptomycin) was added to halt the process. The digest was then strained through a 200 μm filter and centrifuged at 500 x g for 10 minutes at room temperature. The floating adipocytes were collected, resuspended in DMEM containing high glucose, 10% FBS, and 1% penicillin/streptomycin, and subsequently seeded in dishes.
For fluorescence-activated cell sorting (FACS), adipocytes isolated from the BAT and iWAT of CLDN5-GFP mice were labeled with PE-conjugated anti-CD31 (BioLegend, 102407) and APC-conjugated anti-CD45 antibodies (Cell Signaling Technology, 41104) for 30 minutes at room temperature, then washed and analyzed by flow cytometry. Cell aggregates and debris were excluded using side scatter-area (SSC-A) versus forward scatter-area (FSC-A) plots. GFP+CD31-CD45- and GFP-CD31-CD45- adipocytes were then sorted using BD FACS Aria™ III Cell Sorter and BD FACSDiva software 7.0 (BD Biosciences). Gating strategy for flow cytometry and cell sorting was shown in Supplementary Fig. 1c.
For magnetic bead cell sorting, adipocytes isolated from BAT and iWAT of Cldn5flox/flox and Cldn5flox/flox; Fabp4-Cre mice were incubated with biotin anti-CD36 antibody (BioLegend, 102604), and subsequently with magnetic beads conjugated to anti-biotin antibodies (Pierce™ Streptavidin Magnetic Beads, Invitrogen, 88816) to separate collect the CD36+ adipocytes.
Co-culture experiments
SVF cells from BAT and iWAT were differentiated into brown and beige adipocytes, respectively. The isolated CD36+ primary adipocytes were cultured in complete DMEM medium. Co-culture was performed using a transwell chamber system (0.4-μm pores, Corning, 3460) in a 12-well dish, with CD36+ primary adipocytes plated in the inserts and differentiated brown and beige adipocytes seeded in the wells. After each treatment, the differentiated brown and beige adipocytes were collected for further analysis.
For CLDN5 overexpression experiments, freshly isolated CD36+ adipocytes isolated from 8-10-week-old wild-type mice were seeded in a 12-well plate. AAV expressing the mouse Cldn5 gene (AAV-CLDN5) or ZsGreen, serving as a control (prepared by Hanbio Biotechnology Inc, China), at a dose of 7 × 10^9 vg, was added to each well. After 12 hours of AAV infection, the transduction mixture was removed, and the cells were maintained in 1 mL of fresh culture medium.
In order to determine whether CLDN5-deficient adipocytes communicate with thermogenic cells through paracrine substances, CD36+ primary adipocytes isolated from Cldn5flox/flox and Cldn5flox/flox; Fabp4-Cre mice were co-cultured with the differentiated brown and beige adipocytes for 48 h. To assess whether CLDN5-depleted cells influence thermogenic cells through IL10, IL10 neutralizing antibody (Abcam, AB9969; 1 μg/mL) or IL10Rα blocking antibody (Novus, NBP3-07330; 1 μg/mL), along with their corresponding negative control antibodies IgG (1 μg/mL) were introduced into the CD36+ adipocyte culture medium.
In the co-culture of brown adipocytes with CD36+ primary adipocytes, 1 mM dbcAMP was added for 4 hours prior to sample collection. In the co-culture of beige adipocytes with CD36+ primary adipocytes, 0.5 mM dbcAMP was added for 24 hours prior to sample collection84,85,86,87.
For IL10 knockdown experiments, freshly isolated CD36+ adipocytes from Cldn5flox/flox and Cldn5flox/flox; Fabp4-Cre mice were seeded in six-well plates and transduced with either AAV-sh-Il10 or AAV-sh-Ctrl at a dose of 1.6 × 1010 power viral genomes per well. After 12 hours of viral exposure, the virus-containing medium were replaced with fresh cell medium. These transfected adipocytes were then co-cultured with fully differentiated brown and beige adipocytes for 48 hours prior to analysis.
To investigate whether the STAT3 signaling pathway is involved in the impaired thermogenesis resulting from CLDN5 deletion, differentiated brown and beige adipocytes were treated with either vehicle or 10 μM of the STAT3 inhibitor C188-9 (Selleck Chemicals, S8605) for 30 minutes, followed by co-culture with CD36+ primary adipocytes isolated from the BAT and iWAT of Cldn5flox/flox; Fabp4-Cre mice.
Flow cytometry analysis
UCP1+ cell proportion were detected by flow cytometry analysis. The isolated CD36+ cells were stimulated with 1 μM CL-316,243 or vehicle (water) in DMEM complete medium for 12 hours. Subsequently, the cells were fixed with 0.01% formaldehyde, permeabilized with 0.1% Tween-20, and incubated with anti-UCP1 antibody in the dark at 4 °C for 30 minutes. After removing unbound primary antibodies, cells were stained with FITC-conjugated donkey anti-rabbit IgG (Invitrogen, A32795). Following the wash-off of unbound secondary antibodies, cells underwent flow cytometry analysis using a flow cytometer (BD Bioscience). Gating strategy for flow cytometry was shown in Supplementary Fig. 6a.
Mitochondrial respiration assay
The oxygen consumption rate (OCR) was measured using a Seahorse XF24 extracellular flux analyzer (Seahorse Bioscience) following the manufacturer’s protocol for mitochondrial stress testing. To detect uncoupled respiration, maximal respiration, and non-mitochondrial respiration, 2.5 μM oligomycin, 2 μM carbonyl cyanide-p-trifluoromethoxyphenylhydrazone (FCCP), and 1 μM rotenone/antimycin were delivered, respectively. All reagents were obtained from the Seahorse XF Cell Mito Stress Test Kit (Agilent, 103015-100). The OCR values were normalized by total protein.
Whole mount confocal microscopy
Whole-mount confocal microscopy was conducted as previously described88. For whole-mount staining of adipose tissue, BAT or iWAT was sectioned into tissue blocks measuring 4 mm × 4 mm × 2 mm on ice, followed by fixation in 1% paraformaldehyde. After 20 minutes, tissues were blocked with 10% FBS in PBS (containing 0.2% Tween 20), incubated with the appropriate primary antibody overnight, stained with a fluorophore-conjugated secondary antibody, and counterstained with DAPI. Subsequently, the entire adipose tissue was scanned using a laser-scanning confocal microscope (Zeiss LSM 880). Immunofluorescence images were captured and processed with Zen Software v2.3 (blue edition). Tissue samples incubated with secondary antibodies alone (without the primary antibody) served as negative controls. Negative controls for immunofluorescence staining were shown in Supplementary Fig. 11.
RNA sequencing
Total RNA was extracted from BAT, iWAT and eWAT of 5 weeks old Cldn5flox/flox and Cldn5flox/flox; Fabp4-Cre mice for RNA-seq analysis (Novogene). RNA-Seq was performed on the Annoroad Genome Illumina Novaseq platform. Five independent samples were analyzed per group. Paired-end clean reads were aligned to the mouse reference genome (Ensemble_GRCm38.90) using TopHat (version 2.0.12), and mRNA expression was quantified by aligned reads using HTSeq-Count (version 0.6.1). Differential expression analysis between the two groups was performed using the DESeq2 R package (1.16.1), and the resulting P-values obtained from the Wald test were corrected using the Benjamini-Hochberg method to control the false discovery rate. Genes with an adjusted P-value < 0.05 and a fold change > 2.0 were considered differentially expressed.
Quantitative real-time PCR (qRT-PCR)
The samples were homogenized in Trizol reagent (Ambion, USA). Total RNA was isolated and reverse transcribed using a reverse transcription system kit following the manufacturer’s instructions (Takara, Japan). cDNA was quantified by qRT-PCR using SYBR Green PCR Master Mix (ABI, USA) on a Thermo Fisher QuantStudio3 system instrument. The expression of each gene was calculated after normalization to the expression of β-actin. The oligonucleotide primer sequences are provided in Supplementary Table 2.
For the quantification of mtDNA copy number, total DNA was isolated from BAT, iWAT, and eWAT. The relative mtDNA copy number was determined using primers specific for the mitochondrial COX2 gene and normalized to the nuclear DNA-encoded β-actin gene. The primer sequences can be found in Supplementary Table 2.
Histologic analysis
Tissues were harvested and fixed with a 4% paraformaldehyde (PFA) solution for 24 hours and then embedded in paraffin or OCT. Sections of 5 μm thickness were collected and subjected to HE staining for histological evaluation. The average size of lipid droplets was analyzed by ImageJ software, with at least 5 fields examined for each mouse.
Transmission electron microscopy (TEM) was performed on glutaraldehyde-fixed, epoxy-embedded adipose tissue samples and stained with uranyl acetate and lead citrate. TEM images were collected by RADIUS Software v2.1.
Western blot analysis
For protein extraction, tissues were homogenized in RIPA lysis buffer supplemented with a complete protease inhibitor cocktail (Sangon Biotech, C600387-0005). Cellular fractions (membrane, cytosolic, and nuclear) obtained from BAT and iWAT were extracted using Qproteome Cell Compartment Kit (Qiagen, 37502) following the manufacturer’s instructions. Protein expression was assessed by SDS/PAGE under reducing conditions, and immunoblotting was conducted with specific antibodies. HRP-conjugated secondary antibodies (ABclonal), followed by ECL (Thermo Fisher, 34580) incubation, enabled protein band detection. Integration of all blot images was performed using Adobe Illustrator 2022. Image J v1.8.0 was employed for quantifying Western blot results.
Co-immunoprecipitation (Co-IP)
Adipose tissue was lysed in 50 mM Tris (pH 8.0) by 25–30 repeated passages through a 25-gauge needle, followed by centrifugation at 5000 × g. The lysate membranes were extracted using CSK buffer (150 mM NaCl; 1% Triton X-100; 50 mM Tris, pH 8.0; and protease inhibitors). The membrane extract was precleared by incubation with protein A/G-sepharose (Proteintech, PR40025) before Co-IP. The precleared membrane extract was incubated with anti-CLDN5, anti-ZO1, anti-YBX3, and anti-rabbit IgG (as a negative control) antibodies at 4 °C for 16 hours. The antibody-bound material was precipitated with protein A/G-Sepharose, washed three times with CSK buffer, and detected by immunoblotting.
RNA immunoprecipitation (RIP)
The RNA-immunoprecipitation (RIP) was performed with the Magna RIP RNA-binding protein immunoprecipitation kit (Millipore, 17-701) in accordance with the manufacturer’s instructions. Isolated lysates of BAT were incubated overnight at 4 °C with gentle rotation in the presence of excess (10 μg) IP antibody (IgG, anti-YBX3). RNA in IP materials was used in RT reactions followed by qPCR analysis to detect Il10 mRNA levels.
Luciferase reporter assay
The mouse Il10 gene promoter (2 kb before start codon of the Il10 open reading frame, synthesized by Shanghai Sango Biotechnology) was cloned into pGL3-Basic luciferase reporter vector (Promega) with NheI and HindIII sites. The pGL3 reporter, the pGL4.74 Renilla luciferase control vector (Promega), and pCMV6-Ybx3 vector were co-transfected into HEK293 cells in 96-well culture dishes using lipofectamine-2000. The wild-type 3′-UTR and mutant 3′-UTR (the deletion of the predicted YBX3 binding sites CCACCT and ACACCT was designated as Del1 and Del2, respectively) of mouse Il10 gene (synthesized by Shanghai Sango Biotechnology) were inserted into the psiCHECK-2 (Clontech) downstream of the luciferase gene using XhoI/NotI, respectively. The wild-type or mutant psiCHECK-2-Il10: 3′-UTR and pCMV6-Ybx3 vector were co-transfected into HEK293 cells in 96-well culture dishes using lipofectamine 2000. Twenty-four hours after transfection, firefly and renilla luciferase activities were measured using a chemiluminescence reporter assay system—Dual Glo (Promega, E2980) in Fluostar Omega (BMG). Luciferase activities were recorded using Omega Software v5.50 R4.
Study approval
All animal experimental protocols in this study were approved by the Institutional Animal Care and Use Committee of Binzhou Medical University (Document No. 2020-20) and conducted in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals. Appropriate measures were taken to reduce the pain or discomfort of experimental animals.
Statistical analysis
All the replicate experiments were repeated at least three times. All analyses were performed by Prism 8 (GraphPad Software Inc.). All data are represented as mean ± SEM. Outliers were excluded based on Grubbs’ outlier test (GraphPad, p < 0.05). Shapiro-Wilk test was conducted to assess normality, and two-sided Mann-Whitney test (non-parametric) were employed when normality was violated. For normally distributed data, comparisons between two groups were analyzed using two-sided unpaired t-test (two-tailed) and multiple groups by two-sided one-way ANOVA or two-way ANOVA with Tukey’s post-hoc test or Dunnett’s post-hoc test. Metabolic cage data of mice were analyzed by two-sided ANCOVA (data of energy expenditure and food intake) or ANOVA (data of locomotor activity and RER) using CaIR software (version 1.3; https://calrapp.org/). P < 0.05 was considered statistically significant.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The data supporting the findings from this study are available within the manuscript and its supplementary information. The raw RNA-seq data have been deposited at NCBI sequence read archive (SRA) database with accession numbers PRJNA1070960. The CLDN5 expression data in human adipose tissue analyzed in this study were obtained from previously published datasets deposited in the NIH Gene Expression Omnibus·(GEO) database under the accession codes.GSE9624 [https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE9624], GSE2508, GSE156906, GSE20950 and GSE27951. Source data are provided with this paper.
References
Meoli, L. & Günzel, D. The role of claudins in homeostasis. Nat. Rev. Nephrol. 19, 587–603 (2023).
Song, A. et al. Low- and high-thermogenic brown adipocyte subpopulations coexist in murine adipose tissue. J. Clin. Invest 130, 247–257 (2020).
Lu, Z. et al. A non-tight junction function of claudin-7-Interaction with integrin signaling in suppressing lung cancer cell proliferation and detachment. Mol. Cancer 14, 120 (2015).
Tiwari-Woodruff, S. K. et al. OSP/claudin-11 forms a complex with a novel member of the tetraspanin super family and beta1 integrin and regulates proliferation and migration of oligodendrocytes. J. Cell Biol. 153, 295–305 (2001).
Zhang, Y. et al. A novel function of claudin-5 in maintaining the structural integrity of the heart and its implications in cardiac pathology. Biochim Biophys. Acta Mol. Basis Dis. 1870, 167274 (2024).
Jiang, S. et al. Cardiac-specific overexpression of claudin-5 exerts protection against myocardial ischemia and reperfusion injury. Biochim Biophys. Acta Mol. Basis Dis. 1868, 166535 (2022).
Dhawan, P. et al. Claudin-1 regulates cellular transformation and metastatic behavior in colon cancer. J. Clin. Invest 115, 1765–1776 (2005).
Richards, M. et al. Claudin5 protects the peripheral endothelial barrier in an organ and vessel-type-specific manner. eLife 11, https://doi.org/10.7554/eLife.78517 (2022).
Zhu, Y. et al. Connexin 43 mediates white adipose tissue beiging by facilitating the propagation of sympathetic neuronal signals. Cell Metab. 24, 420–433 (2016).
Beutel, O., Maraspini, R., Pombo-García, K., Martin-Lemaitre, C. & Honigmann, A. Phase separation of zonula occludens proteins drives formation of tight junctions. Cell 179, 923–936.e911 (2019).
Sun, H. et al. Loss of CLDN5 in podocytes deregulates WIF1 to activate WNT signaling and contributes to kidney disease. Nat. Commun. 13, 1600 (2022).
Balda, M. S. & Matter, K. The tight junction protein ZO-1 and an interacting transcription factor regulate ErbB-2 expression. EMBO J. 19, 2024–2033 (2000).
Chaudhary, A. et al. Correlating multi-functional role of cold shock domain proteins with intrinsically disordered regions. Int. J. Biol. macromolecules 220, 743–753 (2022).
Lindquist, J. A. & Mertens, P. R. Cold shock proteins: from cellular mechanisms to pathophysiology and disease. Cell Commun. Signal.: CCS 16, 63 (2018).
Park, J. H. et al. A multifunctional protein, EWS, is essential for early brown fat lineage determination. Developmental cell 26, 393–404 (2013).
Rabiee, A. et al. White adipose remodeling during browning in mice involves YBX1 to drive thermogenic commitment. Mol. Metab. 44, 101137 (2021).
Wu, R. et al. RNA-binding protein YBX1 promotes brown adipogenesis and thermogenesis via PINK1/PRKN-mediated mitophagy. FASEB J. 36, e22219 (2022).
Wu, R. et al. Transcriptional and post-transcriptional control of autophagy and adipogenesis by YBX1. Cell Death Dis. 14, 29 (2023).
Xu, D. et al. RNA binding protein Ybx2 regulates RNA stability during cold-induced brown fat activation. Diabetes 66, 2987–3000 (2017).
Zhao, Q. et al. Phosphorylated YBX2 is stabilized to promote glycolysis in brown adipocytes. iScience 26, 108091 (2023).
Fagerberg, L. et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol. Cell Proteom. 13, 397–406 (2014).
Karlsson, M. et al. A single-cell type transcriptomics map of human tissues. Science advances 7, https://doi.org/10.1126/sciadv.abh2169 (2021).
Lee, K. Y. et al. Lessons on conditional gene targeting in mouse adipose tissue. Diabetes 62, 864–874 (2013).
Bennett, J. P. et al. Standardization of dual-energy x-ray visceral adipose tissue measures for comparison across clinical imaging systems. Obesity, https://doi.org/10.1002/oby.23885 (2023).
Wang, G. X., Zhao, X. Y. & Lin, J. D. The brown fat secretome: metabolic functions beyond thermogenesis. Trends Endocrin Met 26, 231–237 (2015).
Villarroya, J., Cereijo, R., Giralt, M. & Villarroya, F. Secretory proteome of brown adipocytes in response to cAMP-mediated thermogenic activation. Front Physiol. 10, 67 (2019).
Pogodzinski, D., Ostrowska, L., Smarkusz-Zarzecka, J. & Zysk, B. Secretome of adipose tissue as the key to understanding the endocrine function of adipose tissue. Int J. Mol. Sci. 23, 2309 (2022).
Deshmukh, A. S. et al. Proteomics-based comparative mapping of the secretomes of human brown and white adipocytes reveals EPDR1 as a novel batokine. Cell Metab. 30, 963–975.e967 (2019).
Kleene, K. C. Y-box proteins combine versatile cold shock domains and arginine-rich motifs (ARMs) for pleiotropic functions in RNA biology. Biochemical J. 475, 2769–2784 (2018).
Rajbhandari, P. et al. IL-10 Signaling Remodels adipose chromatin architecture to limit thermogenesis and energy expenditure. Cell 172, 218–233.e217 (2018).
Beppu, L. Y. et al. Tregs facilitate obesity and insulin resistance via a Blimp-1/IL-10 axis. JCI Insight 6, https://doi.org/10.1172/jci.insight.140644 (2021).
Yuan, Y. et al. Linker histone variant H1.2 is a brake on white adipose tissue browning. Nat. Commun. 14, 3982 (2023).
Rajbhandari, P. et al. Single cell analysis reveals immune cell-adipocyte crosstalk regulating the transcription of thermogenic adipocytes. Elife 8, https://doi.org/10.7554/eLife.49501 (2019).
de-Lima-Junior, J. C. et al. Abnormal brown adipose tissue mitochondrial structure and function in IL10 deficiency. EBioMedicine 39, 436–447 (2019).
de Freitas, R. A. et al. Interleukin-10 deficiency induces thoracic perivascular adipose tissue whitening and vascular remodeling. J. Mol. Histol. 55, 527–537 (2024).
Rahman, S. M. et al. CCAAT/enhancer-binding protein beta (C/EBPbeta) expression regulates dietary-induced inflammation in macrophages and adipose tissue in mice. J. Biol. Chem. 287, 34349–34360 (2012).
Nga, H. T. et al. Interleukin-10 attenuates liver fibrosis exacerbated by thermoneutrality. Front Med (Lausanne) 8, 672658 (2021).
Choi, E. W. et al. Fas mutation reduces obesity by increasing IL-4 and IL-10 expression and promoting white adipose tissue browning. Sci. Rep. 10, 12001 (2020).
Beguelin, W. et al. IL10 receptor is a novel therapeutic target in DLBCLs. Leukemia 29, 1684–1694 (2015).
Coomes, S. M. et al. CD4(+) Th2 cells are directly regulated by IL-10 during allergic airway inflammation. Mucosal Immunol. 10, 150–161 (2017).
Schmetterer, K. G. & Pickl, W. F. The IL-10/STAT3 axis: Contributions to immune tolerance by thymus and peripherally derived regulatory T-cells. Eur. J. Immunol. 47, 1256–1265 (2017).
Heffner, C. S. et al. Supporting conditional mouse mutagenesis with a comprehensive cre characterization resource. Nat. Commun. 3, 1218 (2012).
Liu, J., Xu, Z., Wu, W., Wang, Y. & Shan, T. Cre recombinase strains used for the study of adipose tissues and adipocyte progenitors. J. Cell Physiol. 232, 2698–2703 (2017).
Edgar, R., Domrachev, M. & Lash, A. E. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30, 207–210 (2002).
Barrett, T. et al. NCBI GEO: archive for functional genomics data sets-update. Nucleic Acids Res 41, D991–D995 (2013).
Aguilera, C. M. et al. Genome-wide expression in visceral adipose tissue from obese prepubertal children. Int J. Mol. Sci. 16, 7723–7737 (2015).
Lee, Y. H. et al. Microarray profiling of isolated abdominal subcutaneous adipocytes from obese vs non-obese Pima Indians: increased expression of inflammation-related genes. Diabetologia 48, 1776–1783 (2005).
Fuchs, A. et al. Associations among adipose tissue immunology, inflammation, exosomes and insulin sensitivity in people with obesity and nonalcoholic fatty liver disease. Gastroenterology 161, 968–981.e912 (2021).
Hardy, O. T. et al. Body mass index-independent inflammation in omental adipose tissue associated with insulin resistance in morbid obesity. Surg. Obes. Relat. Dis. 7, 60–67 (2011).
Keller, P. et al. Gene-chip studies of adipogenesis-regulated microRNAs in mouse primary adipocytes and human obesity. BMC Endocr. Disord. 11, 7 (2011).
Sun, W. et al. snRNA-seq reveals a subpopulation of adipocytes that regulates thermogenesis. Nature 587, 98–102 (2020).
Maniyadath, B., Zhang, Q., Gupta, R. K. & Mandrup, S. Adipose tissue at single-cell resolution. Cell Metab. 35, 386–413 (2023).
Inouye, K. E. et al. Endothelial-derived FABP4 constitutes the majority of basal circulating hormone and regulates lipolysis-driven insulin secretion. JCI insight 8, https://doi.org/10.1172/jci.insight.164642 (2023).
Claflin, K. E. et al. Conditional gene targeting using UCP1-Cre mice directly targets the central nervous system beyond thermogenic adipose tissues. Mol. Metab. 55, 101405 (2022).
Mukohira, H. et al. Mesenchymal stromal cells in bone marrow express adiponectin and are efficiently targeted by an adiponectin promoter-driven Cre transgene. Int Immunol. 31, 729–742 (2019).
Rinaldi, V. et al. Evidence for RNA or protein transport from somatic tissues to the male reproductive tract in mouse. Elife 12, https://doi.org/10.7554/eLife.77733 (2023).
Shamsi, F., Wang, C. H. & Tseng, Y. H. The evolving view of thermogenic adipocytes - ontogeny, niche and function. Nat. Rev. Endocrinol. 17, 726–744 (2021).
Acosta, J. R. et al. Human-specific function of IL-10 in adipose tissue linked to insulin resistance. J. Clin. Endocrinol. Metab. 104, 4552–4562 (2019).
Vitali, A. et al. The adipose organ of obesity-prone C57BL/6J mice is composed of mixed white and brown adipocytes. J. Lipid Res 53, 619–629 (2012).
Guerra, C., Koza, R. A., Yamashita, H., Walsh, K. & Kozak, L. P. Emergence of brown adipocytes in white fat in mice is under genetic control. Effects on body weight and adiposity. J. Clin. Invest 102, 412–420 (1998).
Collins, S., Daniel, K. W., Petro, A. E. & Surwit, R. S. Strain-specific response to beta 3-adrenergic receptor agonist treatment of diet-induced obesity in mice. Endocrinology 138, 405–413 (1997).
Simon, M. M. et al. A comparative phenotypic and genomic analysis of C57BL/6J and C57BL/6N mouse strains. Genome Biol. 14, R82 (2013).
Soccio, R. E. et al. Targeting PPARgamma in the epigenome rescues genetic metabolic defects in mice. J. Clin. Invest 127, 1451–1462 (2017).
Jiang, M., Chavarria, T. E., Yuan, B., Lodish, H. F. & Huang, N. J. Phosphocholine accumulation and PHOSPHO1 depletion promote adipose tissue thermogenesis. Proc. Natl Acad. Sci. USA 117, 15055–15065 (2020).
Allu, P. K. R. et al. Role of mTORC2 in biphasic regulation of brown fat metabolism in response to mild and severe cold. J. Biol. Chem. 296, 100632 (2021).
Zeng, L. et al. Loss of cAMP signaling in CD11c immune cells protects against diet-induced obesity. Diabetes 72, 1235–1250 (2023).
Zihni, C., Mills, C., Matter, K. & Balda, M. S. Tight junctions: from simple barriers to multifunctional molecular gates. Nat. Rev. Mol. cell Biol. 17, 564–580 (2016).
Ruan, Y. C. et al. CFTR interacts with ZO-1 to regulate tight junction assembly and epithelial differentiation through the ZONAB pathway. J. cell Sci. 127, 4396–4408 (2014).
Cooke, A. et al. The RNA-binding protein YBX3 controls amino acid levels by regulating SLC mRNA abundance. Cell Rep. 27, 3097–3106.e3095 (2019).
Zhao, H. et al. SARS-CoV-2 RNA stabilizes host mRNAs to elicit immunopathogenesis. Mol. Cell 84, 490–505.e499 (2024).
Chen, L. Y. et al. RNA-binding protein YBX3 promotes PPARgamma-SLC3A2 mediated BCAA metabolism fueling brown adipogenesis and thermogenesis. Mol. Metab. 90, 102053 (2024).
Wang, W. Q., Gao, X. L., Gong, Y. F. & Feng, K. YBX3 regulates thermogenesis and energy expenditure by regulating UCP1 in brown adipose tissue. J. Shandong Univ. (Health Sci.) 62, 38–47 (2024).
Schwayer, C. et al. Mechanosensation of tight junctions depends on ZO-1 phase separation and flow. Cell 179, 937–952.e918 (2019).
Zwick, R. K., Guerrero-Juarez, C. F., Horsley, V. & Plikus, M. V. Anatomical, physiological, and functional diversity of adipose tissue. Cell Metab. 27, 68–83 (2018).
Greene, C. et al. Dose-dependent expression of claudin-5 is a modifying factor in schizophrenia. Mol. psychiatry 23, 2156–2166 (2018).
Voll, S. L. et al. Obesity in adults with 22q11.2 deletion syndrome. Genet. Med.: Off. J. Am. Coll. Med. Genet. 19, 204–208 (2017).
Van, L. et al. 22q11.2 microdeletion and increased risk for type 2 diabetes. EClinicalMedicine 26, 100528 (2020).
Zigmond, E. et al. Macrophage-restricted interleukin-10 receptor deficiency, but not IL-10 deficiency, causes severe spontaneous colitis. Immunity 40, 720–733 (2014).
Sun, J. et al. IL-10R inhibition reprograms tumor-associated macrophages and reverses drug resistance in multiple myeloma. Leukemia, https://doi.org/10.1038/s41375-024-02391-8 (2024).
Zhang, H. et al. Aldehyde Dehydrogenase 2 Mediates alcohol-induced colorectal cancer immune escape through stabilizing PD-L1 expression. Adv. Sci. 8, 2003404 (2021).
He, J. et al. Sulfiredoxin-1 attenuates injury and inflammation in acute pancreatitis through the ROS/ER stress/Cathepsin B axis. Cell Death Dis. 12, 626 (2021).
Zhang, H. G. et al. LncRNA NEAT1 controls the lineage fates of BMSCs during skeletal aging by impairing mitochondrial function and pluripotency maintenance. Cell Death Differ. 29, 351–365 (2022).
Wang, Y. et al. METTL3 is essential for postnatal development of brown adipose tissue and energy expenditure in mice. Nat. Commun. 11, 1648 (2020).
Ohno, H., Shinoda, K., Spiegelman, B. M. & Kajimura, S. PPARgamma agonists induce a white-to-brown fat conversion through stabilization of PRDM16 protein. Cell Metab. 15, 395–404 (2012).
Wang, L. et al. A MicroRNA linking human positive selection and metabolic disorders. Cell 183, 684–701.e614 (2020).
Ma, X., Xu, L. & Mueller, E. Forkhead box A3 mediates glucocorticoid receptor function in adipose tissue. Proc. Natl Acad. Sci. USA 113, 3377–3382 (2016).
Uldry, M. et al. Complementary action of the PGC-1 coactivators in mitochondrial biogenesis and brown fat differentiation. Cell Metab. 3, 333–341 (2006).
Berry, R. et al. Imaging of adipose tissue. In Methods in enzymology 47–73 (Academic Press, 2014).
Gong, Y. https://BioRender.com/d60u193 (2025).
Gong, Y. https://BioRender.com/a27s791 (2025).
Gong, Y. https://BioRender.com/6tfm2cw (2025).
Gong, Y. https://BioRender.com/o1vsyk0 (2025).
Gong, Y. https://BioRender.com/kc0s4dx (2025).
Acknowledgements
We express our gratitude to Dr. Gaengel and Dr. Betsholtz from Uppsala University for generously providing the CLDN5-GFP mice. We thank the Medicine and Pharmacy Research Center of Binzhou Medical University for assistance on electron microscopy imaging and FACS sorting. This work was funded by the National Natural Science Foundation of China (81670620, 81870485 and 32371172 to Y.G., 82370706 and 81970578 to S.Z., 81971281 to H.S., 82200514 to K.F., and 32200758 to C.W.), Taishan Scholars Program of Shandong Province (ts20190953 to Y.G.), Natural Science Foundation of Shandong Province (ZR2024QH437 to K.F., ZR2021QH115 to Y.A. and ZR2021QC105 to C.W.), Shandong First Medical University Academic Promotion Program (2020LI001 to S.Z.), and the Major Basic Research Program of the Shandong Province Natural Science Foundation (ZR202206080014 to S.Z).
Author information
Authors and Affiliations
Contributions
K.F. and W.W. designed and conducted in vivo and in vitro experiments, performed data analysis, and wrote the manuscript. X.G., H.Y., M.X., B.F., Q.J., J.Y., Y.L,. and Q.X. assisted in performing in vivo studies. X.G., C.W., Y.A., P.J., M.W., and H.S. performed mouse genotyping and histologic analysis. Y.G., S.Z., and F.K. jointly designed and supervised the study, analyzed the data, and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Feng, K., Wang, W., Gao, X. et al. Adipocyte CLDN5 promotes thermogenesis and energy expenditure through regulation of IL10 expression. Nat Commun 16, 6151 (2025). https://doi.org/10.1038/s41467-025-61371-3
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-61371-3