Abstract
Strabismus is a common pediatric eye misalignment and has complex genetic and environmental causes. Previous genome-wide association studies (GWAS) encountered difficulties in identifying strabismus risk variants due to heterogeneity and small samples. We performed large meta-analyses of 11 European-ancestry GWAS (7 sources), analysing broad strabismus (20,464 cases, 954,921 controls) and subtypes (esotropia/exotropia). We discovered 4 loci (e.g., NPLOC4-TSPAN10-PDE6G-FAAP100, COL6A1) for strabismus and 5 additional loci (e.g., CHRNA4, MAD1L1) for strabismus subtypes and we successfully replicated the previously reported strabismus variant near NPLOC4-TSPAN10-PDE6G-FAAP100. Using mendelian randomisation, we found genetic evidence supporting maternal smoking as a causal risk factor for strabismus in offspring.
Similar content being viewed by others
Introduction
Strabismus (misalignment of the eyes) is a common ophthalmic condition with both genetic and non-genetic risk factors contributing to its aetiology. Most strabismus is comitant (or concomitant), meaning that the angle of misalignment between the two eyes remains relatively constant regardless of the direction of gaze1. Based on the direction of deviation, concomitant strabismus can be further divided into convergent/esotropia (ET, inward eye deviation) or divergent/exotropia (XT, outward eye deviation). The prevalence of ET and XT varies across populations. For example, among European regions, ET has a prevalence of 2.17%, whereas XT has a prevalence of ~1.53%. By contrast, the African region shows a prevalence of 0.13% for ET and 0.14% for XT2. The misalignment of the visual axis due to the imbalance in extraocular muscles in strabismus leads to reduced or absent binocular vision and is often associated with amblyopia3; individuals who have amblyopia face a significantly increased risk of bilateral visual impairment over their lifetime4. The pathogenesis of strabismus is poorly understood.
Previous studies suggested that various prenatal and early-life environmental factors, such as maternal smoking during pregnancy, increase the risk of strabismus5,6,7. Genetic studies have been conducted to understand the risk loci for strabismus3,8,9. The genetic contribution to strabismus has also been assessed by genome-wide association studies (GWAS). Shaaban et al.10 conducted a GWAS for strabismus and reported significant variants rs2244352 and rs912759 located within/near the WRB on chromosome 21 and ADGRL4 on chromosome 1, respectively. Plotnikov et al.11 also identified a variant rs75078292 (P = 2.24 × 10−8) within the NPLOC4-TSPAN10-PDE6G-FAAP100 gene cluster strongly associated with strabismus. Obtaining a larger sample size increases the statistical power of GWAS and is expected to lead to the discovery of further strabismus-risk variants. Here, we conducted a European ancestry meta-analysis GWAS of strabismus analyses combining 11 summary statistics from 7 sources. The meta-analysis was based on three definitions of the strabismus phenotype: broad-sense strabismus (20,464 cases and 954,921 controls), ET (5,963 cases and 588,794 controls) and XT (3998 cases and 583,468 controls). We identified 7 previously unreported risk variants that provide insights into the aetiology of strabismus.
Previous observational studies have reported an association between maternal smoking during pregnancy and strabismus in offspring5,12,13. However, conventional observational study designs cannot determine causality. In this study, we conducted Mendelian randomisation (MR) to evaluate if genetic support exists for a causal association between maternal smoking and strabismus. Further, since previous studies have reported an association between birth weight and strabismus (which may be partly mediated by maternal smoking)14, we used MR to assess support for a causal relationship between birth weight and strabismus.
Results
Meta-analysis
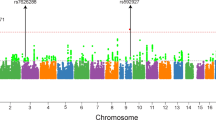
We calculated genetic correlations (rg) for three strabismus phenotypes (Supplementary Data S2). The analysis revealed that broad-sense strabismus showed a higher genetic overlap with ET (rg = 0.83, 95% CI: 0.70–0.96, P = 1.04 × 1034) compared to XT (rg = 0.60, 95% CI: 0.42–0.79, P = 1.25 × 1010). The genetic correlation of ET and XT was −0.22 (95% CI: −0.51 to 0.07, P = 0.137). We conducted GWAS meta-analyses using three strabismus definitions (Fig. 1). Using the broad non-paralytic strabismus definition, we identified 4 genome-wide significant independent variants near the NPLOC4-TSPAN10-PDE6G-FAAP100 gene cluster, COL6A1, ZNF701, and CHRNA4, respectively. Using the ET definition, we identified 4 genome-wide significant independent variants within or near UTS2, CHRNA4, DYNLRB2, and NPLOC4-TSPAN10-PDE6G-FAAP100. Using the XT definition, we identified 2 genome-wide significant independent variants near UTS2 and MAD1L1 (Table 1, Supplementary Data S4 and Fig. 2). The gene cluster NPLOC4-TSPAN10-PDE6G-FAAP100 was significantly associated with strabismus and both its sub-phenotypes. CHRNA4 was associated with broad-sense strabismus and ET (Fig. 3). UTS2 was associated with ET and XT. In total, across the different strabismus definitions, we identified 7 strabismus-associated loci where the peak SNP had P < 5 × 10−8. Only the NPLOC4-TSPAN10-PDE6G-FAAP100 locus reached genome-wide significance in previous strabismus GWAS11,15.
The red line in Manhattan plots represents the genome-wide significant threshold (P = 5 × 10−8), the green line represents the suggestive significance threshold (P = 1 × 10−5). The red line in QQ-plots represents the expected distribution of the p values, and blue/yellow/green trend represents the observed distribution. Shades represent the 95% confidence interval of the expected distribution.
Genome build for chromosome position is Homo sapiens (human) genome assembly GRCh37 (hg19) and LD (r2) is calculated from 1000 Genome European population. The blue line represents the recombination rate (cMMb). The most significant SNPs are indicated by the purple dots. The x-axis shows genes located in the genomic regions (1MB) and y-axis indicates the significance of SNP associations (−log10(P)).
Of the loci identified in the three meta-analyses, MAD1L1, UTS2, NPLOC4, CHRNA4 and ZNF701 were reported to be associated with lung function or smoking16,17,18,19, while NPLOC4-TSPAN10-PDE6G-FAAP100 and COL6A1 loci showed an association with myopia20. We looked up the pheWAS results of these lead SNPs in the GWAS Atlas21; the pheWAS results after the Bonferroni correction are listed in Tables S4–S6.
Loci associated with strabismus after adjusting for refractive error
We examined whether variants identified in our broad-sense strabismus meta-analysis were associated with published GWAS of refractive error22. The genetic correlations of the three strabismus traits and refractive error were statistically significantly different from zero, although the magnitude was modest (rg ~ 0.1–0.2, Supplementary Data S8). As would be expected because of pleiotropy between strabismus and refractive error, a conditional analysis led to a reduction in the mean test statistic across all SNPs in the genome, although three out of four (rs1996371, rs6420484 and rs8108303) from the broad-sense strabismus meta-analysis remained significantly associated with strabismus after conditioning on refractive error (P < 5 × 10−8). This suggested that these variants are specific to strabismus and are not simply associated with strabismus via their association with refractive error.
We also applied the above procedures to the results of the GWAS for ET and for XT. All four XT lead SNPs and two of four ET lead SNPs (rs228636 and rs8070929) retained genome-wide significance in the conditional analysis (Supplementary Data S9).
eQTL look-up
To assess the functional relevance of the lead strabismus loci, we evaluated the eQTLs associated with 4 lead SNPs (rs2150458, rs1996371, rs6420484, and rs8070929) from the broad-sense strabismus GWAS meta-analysis. By filtering associations based on an eQTL FDR < 0.05, the 4 lead GWAS SNPs were mapped to 231 significant eQTLs. No specific tissue type dominated the eQTL associations, but blood cells (BIOSQTL) had the largest proportion of eQTLs. SNPs rs2150458, rs1996371, rs6420484, and rs8070929 were linked to 15, 75, 128 and 13 eQTLs, respectively (Supplementary Data S10).
TWAS
We conducted a cross-tissue TWAS to detect strabismus-risk genes. The tissue weights from GTEx were applied in the UTMOST framework23. The cross-tissue analysis examined 17290 genes across 44 GTEx tissues. After accounting for multiple testing (P < 0.05/17290 = 2.89 × 10−6), nine significant loci were identified in the broad-sense strabismus GWAS meta-analysis: ADAMTS7, ALYREF, C17orf70, COL6A2, CREB3L3, DCXR, OSER1, PDE6G, SLC16A3. A further eight loci were identified in the ET TWAS analysis: CALU, CREB3L3, FBXL18, KXD1, NLRP9, NPAS4, TNRC18, TSACC, and three in the XT TWAS analysis (FTSJ2, MAD1L1, RAB3A).
Replication of previously published loci
We examined three previously published strabismus variants, rs2244352, rs912759 and rs75078292, in our meta-analysis (Table 2). The lead non-accommodative ET variant, rs2244352, identified in the ET GWAS reported by Shaaban et al.10, reached nominal significance, but was not genome-wide significantly associated with strabismus (P > 5 × 10−8) in any of our meta-analyses. The accommodative ET variant rs912759 from the same paper demonstrated no association in our meta-analyses. The locus at rs75078292 reported by Plotnikov et al.11 was identified as genome-wide significant in the broad-sense strabismus and ET meta-analysis and reached nominal significance in the XT meta-analysis.
Association of maternal cigarette smoking with strabismus
Previous observational studies have suggested maternal smoking may be associated with strabismus risk5,6. Using a SNP, which has been shown to index maternal smoking (rs16969968)24, we performed an MR analysis to evaluate the genetically inferred causal link between smoking and three strabismus phenotypes (strabismus, ET and XT; Table 3). The MR results provided evidence to support a causal effect where maternal smoking increased the risk of strabismus (P < 0.05 for broadly defined strabismus as well as for ET and XT) (Table 3). A sensitivity analysis in which additional SNPs were used as instrumental variables for maternal smoking produced a similar result (Supplementary Results and Data S11).
Association of birth weight with strabismus
Low birth weight has been reported as a risk factor for strabismus5,14,24. Also, a lower birth weight could potentially be a mediator in the causal pathway from maternal smoking to strabismus. Hence, we conducted a second MR analysis to investigate the genetic association between birth weight and strabismus. This MR analysis indicated that, for each 500-g increase (~1 standard deviation) in offspring birth weight, ORs for strabismus, ET, and XT risk were 1.05, 1.10, and 1.02, respectively (all P > 0.05, as presented in Table 4). To validate our MR findings, we compared them with results from a published observational study25 (Fig. 4). While our MR results did not yield strong evidence (P > 0.05) to support a causal link between birth weight and strabismus, the confidence intervals of ORs were relatively wide and overlapped with the observational results. For instance, when considering a 500 g increase in birth weight from 3500–3999 g to 4000–4499 g, the MR OR overlapped with observational results.
We compared odds ratios (ORs) change from our Mendelian randomisation (MR) analysis of birth weight on esotropia (a) and exotropia (b) with observational results published by Torp-Pedersen et al. The x-axis contains different groups based on per 500 g birth weight increase. The birth weight changes were labelled as 3000–3499 g, 3500–3999 g and 4000–4499 g in the original paper, we re-labelled them to ‘500 g increase from 2500 g’, ‘500 g increase from 3000 g’ and ‘500 g increase from 3500 g’ and present the ORs change per 500 g increase on birth weight (green points in a, yellow points in b). Error bars are the 95% confidence interval of ORs change. The red point represents the MR OR per 500 g increase in birth weight, and the red error bars represent the 95% confidence interval of MR OR.
We then assessed whether there was evidence for causality in both directions. We performed MR analyses to assess the effect of strabismus/ET/XT on birth weight using GWAS-significant SNPs identified from the current study as the instruments. We found no evidence for an effect of strabismus risk on birth weight using the IVW method (OR = 0.98 per doubling odds of strabismus; 95% CI = 0.94–1.02; P = 0.57; OR = 1.02 per doubling odds of ET; 95% CI = 1.00–1.04; P = 0.43; OR = 0.96 per doubling odds of XT; 95% CI = 0.93–1.00; P = 0.27).
Discussion
In this study, we have conducted the largest genome-wide meta-analysis for strabismus. We identified seven genetic variants significantly associated with strabismus using different definitions of strabismus. We performed MR using the well-established maternal smoking variable rs16969968 and showed that genetically-proxied maternal smoking increases the risk of offspring strabismus/ET; this adds genetic evidence to the existing conventional observational studies, bolstering the case for there being a causal relationship between maternal smoking and strabismus risk.
Strabismus is a heterogeneous condition; although some genetic factors are common across various subtypes, others are unique to specific forms26. Our results indicate that the statistical power of meta-analysis varies depending on the definition of strabismus used in the clinical data, suggesting that subtype-specific genetic factors may influence the susceptibility to different forms of strabismus. The low genetic correlation (rg = −0.22, Supplementary Data S2) between ET and XT suggests that these sub-phenotypes have different biological mechanisms. The broad-sense strabismus meta-analysis identified the same number of independent SNPs as the ET meta-analysis, despite the difference in sample size (Ncase = 20,464 for the broad-sense strabismus analysis; Ncase = 5963 for the ET meta-analysis). Future studies should endeavour to collect more detailed phenotype information to better dissect this heterogeneity, although large sample sizes will be required. Restricting the age range of cases may also help increase the accuracy of some future strabismus GWAS. In this study, we opted not to use ICD data in the UKB sample as these data were collected from older people whose ICD records did not reflect their childhood disease status.
The NPLOC4-TSPAN10-PDE6G-FAAP100 locus on chromosome 17 was associated with both broad-sense strabismus and ET. This locus was first reported in a UKB strabismus GWAS11 and replicated in the FinnGen cohort27. Our findings verified that this locus has a strong association with strabismus. However, this locus did not reach the genome-wide significance level in XT analysis, again supporting potential divergent biological mechanisms underlying ET and XT.
Genetic loci identified in previous strabismus GWAS have been associated with a diverse range of ocular phenotypes, including myopia or refractive errors10,11,28. Some of our findings may have been influenced by pleiotropy between strabismus and refractive errors in the current GWAS analyses. We were unable to include refractive error as a covariate due to the lack of access to individual-level data for the bulk of the input data. Instead, we compared the genetic correlation between our strabismus GWAS and published GWAS of refractive error22 (Supplementary Data S8), and we applied the mtCOJO approach to screen for SNPs associated with strabismus after adjusting for their effects on refractive error (Supplementary Data S9). Consistent with modest pleiotropy between strabismus and refractive error (rg ~ 0.1–0.2), we found that the number of genome-wide significant lead SNPs associated with strabismus reduced after conditioning on refractive error.
Previous observational studies have reported that maternal smoking during pregnancy has significant effects on offspring’s vision health5,6,7,29,30. MR uses genetic data to infer causality in a framework that is typically subject to different sources of confounding bias compared to observational studies and thus provides an additional source of evidence. However, the limited sample size of existing strabismus GWAS has hindered the use of MR to investigate the causal relationship between maternal smoking and strabismus. Here we report the first MR study to examine the relationship between maternal smoking and strabismus. We used a well-established maternal smoking proxy instrumental variable, rs16969968, to show that maternal smoking during pregnancy is linked to the risk of broadly defined strabismus as well as ET and XT (Table 3).
To confirm the single SNP MR result, we used all top variants from Saunders et al.31 as instrument variables for maternal smoking and found concordant results (Supplementary Data S11, Supplementary Results). However, the specific ORs from the multiple SNP analysis are more difficult to reliably interpret because they are based on the simplifying assumption that offspring genotype reflects maternal genotype31. As the age of onset for strabismus (in early childhood) occurs earlier than the average age of smoking initiation (around 15 years old or later)32,33, it is very likely that the SNPs instrumenting smoking behaviour index maternal smoking rather than indexing risk relating to the offspring’s smoking behaviour.
There are several limitations to consider in the MR analysis. First, although the SNP (rs16969968) used in our primary analysis as a genetic proxy for maternal smoking plays a well-established role in smoking behaviour, it remains possible the SNP had pleiotropic effects on maternal traits other than smoking initiation (or pleiotropic effects on a maternal trait that is a confounder of the maternal smoking-offspring strabismus relationship); such a pleiotropic effect could have biased the direction or magnitude of the MR result. However, examining rs16969968 in Open Targets Genetics34, there is no evidence that this SNP affects traits other than smoking and directly related traits (such as lung cancer). Second, although we estimated the magnitude of the risk of strabismus conferred by maternal smoking, accurately estimating a specific OR using MR is difficult35,36, especially in the case of maternal exposure’s effect on an offspring outcome. Ideally, an MR analysis would condition maternal genetic effects on the offspring genotype37, but this approach was not possible with the data available. Third, for our MR sensitivity analysis based on multiple SNP IVs, our estimates were derived assuming that the offspring genotype for each SNP was indicative of maternal smoking risk. Irrespective of the precise magnitude of the risk, our MR analyses provide an independent line of evidence for a link between maternal smoking and strabismus. In comparison to previous observational studies5,7, MR studies are subject to different sources of confounding bias. Therefore, combining past evidence from observational studies5 and the additional evidence from our MR study, there is consistent evidence of a causal relationship between maternal smoking and strabismus. Fourth, our study focused on individuals of European descent. Due to the lack of samples, we were unable to seek to replicate our GWAS lead loci in non-European cohorts. Although maternal smoking rates differ across countries2,38, maternal smoking is associated with strabismus in conventional observational studies across various ancestry groups5,7,39. Our MR findings in Europeans provide support for a causal link between maternal smoking and strabismus. Future genetic studies should be conducted in a wider range of ancestries to expand the scope of gene mapping and MR studies.
Notably, although we inferred the relationship between genetically-proxied maternal smoking and strabismus was causal, the actual mechanism underlying this association remains unclear. Given the well-known link between maternal smoking and birth weight, we conducted a secondary MR analysis to investigate the genetic association between birth weight and strabismus (Supplementary Results, Fig. S4); the confidence intervals on our MR estimates overlapped with those from a previous observational study of babies in the middle of the weight range (3000–4000 g)25. An advantage of observational studies is that they allow simple dissection of the effect of a particular increase in birth weight across a range of birth weights (e.g. low, medium, high), with a previous study showing that, for example, a 500 g change in birth weight was associated with strabismus among small babies (~2000 g), but that this effect was not be seen in larger babies (~4000 g)14,25. However, various confounders can influence conventional observational studies, which may introduce biases and distort the observed associations. A key advantage of our MR estimates is that they are less likely than observational to be affected by confounding. A disadvantage of our MR analysis is that, like most MR studies, we assume a linear relationship, which may only partially capture how birth weight affects strabismus across the full range of birth weights in the population. In the future, this could be revisited by applying MR to subsets of babies of low birth weight (e.g. <2000 g), although much larger sample sizes than we currently have would be required for adequate power.
Strengths of our study include conducting GWAS meta-analyses with multiple large cohorts for different types of strabismus. We also compared the GWAS of ET and XT and indicated the different biological mechanisms underlying these two subtypes. Furthermore, we conducted MR and showed evidence for a causal effect of maternal smoking during pregnancy on strabismus risk in offspring, consistent with conventional observational studies.
In summary, GWAS meta-analyses of strabismus and two of its sub-phenotypes identified a total of seven genome-wide significant genetic variants, six of which were unreported findings. The identification of these genetic loci associated with strabismus susceptibility enhances our understanding of its biological mechanisms. In addition, we obtained strong genetic evidence supporting a causal link between maternal smoking and strabismus. Thus, this work augments ongoing public health efforts aimed at reducing the rate of maternal smoking.
Methods
This study complies with all relevant ethical regulations. All participants provided informed consent, and individual-level data were anonymized and analysed in accordance with the approved protocols.
Datasets
We included 7 sources with 11 sets of GWAS summary statistics for adult strabismus (Table 1). These included a GWAS based on clinical strabismus data from the Kaiser Permanente Genetic Epidemiology Research on Adult Health and Aging (GERA) cohort, Finngen, and the Estonian Biobank (EstBB), along with a GWAS based on self-reported strabismus from UK Biobank (UKB), Lifelines, the Australian Genetics of Depression Study (AGDS) and a published ET GWAS using USA/Australia samples10.
UK Biobank
The UKB is a large-scale United Kingdom biomedical database containing in-depth genetic and phenotypic data from ~500,000 participants who were between the ages of 40 and 69 years at recruitment. Approximately 488,000 participants were genotyped on high-density SNP arrays. The genotype data underwent quality control and imputation procedures as previously described (Bycroft et al.40). Approximately 96 million variations were imputed utilising resources of the Haplotype Reference Consortium (HRC) and UK10K haplotype and 487,409 individuals were retained after genotyping quality control. To validate the ancestral background from UKB self-report ethnicity (Data-Field 21000), we used the k-means clustering method and clustered the top 20 principal components (PCs) into 20 clusters. The PCA clusters were compared with the self-report ethnicity. UKB individuals who had consistent European self-report ethnicity and genetic clusters were used in GWAS (mainly white British, N = 438,637). We included 2744 self-reported strabismus cases who gave this as their ‘reason for glasses/contact lenses’ (Data-Field 6147). Controls were 306,683 participants without self-reported strabismus, diagnosed strabismus (ICD-10 code H49 and H50), and no history of eye surgeries in loss of vision (Data-Field 5181, 5324, 5325, 5326, 5327 and 5328) (UKB phenotype Sep 2021 update).
We conducted a GWAS for strabismus in UKB using the software Regenie (version 2.2.4) (Mbatchou et al.41), adjusting for sex, age and the top 10 PCs. SNPs with MAF > 0.01 and imputation quality score (INFO score) > 0.8 were retained in the following analysis.
Kaiser Permanente GERA cohort
The GERA cohort contains genome-wide genotype, clinical, and demographic data of over 110,000 adult members of the Kaiser Permanente Northern California (KPNC) Medical Care Plan42. The Institutional Review Board of the Kaiser Foundation Research Institute has approved all study procedures. Patients with strabismus were diagnosed by a Kaiser Permanente ophthalmologist and were identified from clinical diagnoses captured in the KPNC electronic health records (EHR) system. These clinical diagnoses were recorded in the EHR system as International Classification of Diseases, Ninth or Tenth Revision (ICD-9 or ICD-10) codes. In GERA, strabismus cases were defined based on diagnosis codes (ICD-9: 378.0x, 378.1x, 378.31x, 378.32x, and 378.9x; or ICD-10 codes equivalent: H50.0x, H50.1x, H50.2x, and H50.9x). After excluding subjects who had any evidence of strabismus based on ICD-10 codes (H49 and H50), our control group included all the non-cases. All controls had at least one vision exam recorded in the KPNC EHR system. In total, 5763 ‘broad’ strabismus cases (or 1582 ET cases; or 1018 XT cases) and 59,797 controls from the GERA non-Hispanic white sample were included in this study. Protocols for participant genotyping, data collection and quality control have been described in detail42. Briefly, GERA participants’ DNA samples were extracted from Oragene kits (DNA Genotek Inc., Ottawa, ON, Canada) at KPNC and genotyped at the Genomics Core Facility of UCSF. DNA samples were genotyped at over 665,000 genetic markers on four ethnic-specific Affymetrix Axiom arrays (Affymetrix, Santa Clara, CA, USA) optimised for European, Latino, East Asian, and African American individuals43. Genotype quality control (QC) procedures and imputation were conducted on an array-wise basis44. For imputation, we additionally removed variants with call rates <90% by array. Genotypes were then pre-phased with Eagle (v2.3.2)45, and then imputed with Minimac3 (v2.0.1)46, using two reference panels. Variants were preferred if present in the EGA release of the HRC (N = 27,165; no indels) reference panel45, and from the 1000 Genomes Project Phase III release if not (N = 2504; including indels)47.
In GERA, GWA analyses were conducted for three strabismus phenotypes (‘broad’ strabismus, ET, and XT) using logistic regression models adjusting for age, sex, and ancestry PCs. GWASs were conducted using PLINK v1.9 (www.cog-genomics.org/plink/1.9/).
FinnGen
The FinnGen project (https://www.finngen.fi/en) is a nationwide biobank project launched in 2017. FinnGen plans to collect ~500,000 biobank samples in Finland over 6 years (~10% of the population). The variants were genotyped using the ThermoFisher Axiom custom array v2 that contains 723,376 probesets for 664,510 markers. In addition to the core GWAS markers (about 500,000), it contains about 116,000 coding variants enriched in Finland (https://www.finngen.fi/en/researchers/genotyping). Genotype imputation was conducted by using the population-specific SISu v4.2 imputation reference panel (which contains 8554 whole-genome sequencing (WGS) data of Finnish individuals). The detailed QC and imputation procedures have been described at https://finngen.gitbook.io/documentation/.
We downloaded the GWAS summary statistics from Data Freeze 8 for three strabismus definitions: broad-sense strabismus (coded as ‘Other strabismus’ by FinnGen), convergent concomitant strabismus (ET) and divergent concomitant strabismus (XT). The FinnGen GWAS of ‘Other strabismus’ (primarily comprising ICD-10 H50) included 5604 cases and 297,342 controls; the GWAS of convergent concomitant strabismus (primarily comprising ICD-10 H50.0) included 1368 cases and 297,342 controls; the GWAS of divergent concomitant strabismus (primarily comprising ICD-10 H50.1) included 1863 cases and 297,342 controls.
Estonian Biobank
The EstBB is a population-based biobank with 212,955 participants in the current data freeze (2022v1). All biobank participants have signed a broad informed consent form and information on ICD codes is obtained via regular linking with the National Health Insurance Fund and other relevant databases, with the majority of the EHR collected since 200448.
The EstBB GWAS for the ICD-10 H50* strabismus phenotype included 2,818 cases and 195,861 controls; the GWAS of convergent concomitant strabismus (comprising ICD-10 H50.0) contains 1057 cases and 197,622 controls; the GWAS of divergent concomitant strabismus (comprising ICD-10 H50.1) included 926 cases and 197,753 controls.
All EstBB participants have been genotyped at the Core Genotyping Lab of the Institute of Genomics, University of Tartu, using Illumina Global Screening Array v3.0_EST. Samples were genotyped and PLINK format files were created using Illumina GenomeStudio v2.0.4. Individuals were excluded from the analysis if their call rate was < 95% or if sex based on heterozygosity of the X chromosome did not match sex in phenotype data. Before imputation, variants were filtered by call rate < 95%, Hardy–Weinberg equilibrium p value < 1 × 10−4 (autosomal variants only), and minor allele frequency < 1%. Variant positions were in build 37 and all variants were changed to be from the TOP strand using GSAMD-24v1-0_20011747_A1-b37.strand.RefAlt.zip files from https://www.well.ox.ac.uk/~wrayner/strand/ webpage. Pre-phasing was performed using the Eagle v2.3 software45. The number of conditioning haplotypes Eagle2 used when phasing each sample was set to: --Kpbwt = 20,000, and imputation was performed using Beagle 5.4 (v.18May20.d20) with effective population size Neff = 20,00049. A population-specific reference panel consisting of 2297 WGS samples was used for imputation50. Based on PC analysis, samples of non-European ancestry and samples that were twins or duplicates of included samples were removed.
Association analysis in the EstBB was carried out for all variants with an INFO score >0.7 using the additive model as implemented in Scalable and Accurate Implementation of Generalized mixed model (SAIGE v1.0.7)51, with a saddle point approximation to calibrate unbalanced case-control ratios. Logistic regression was carried out with LOCO = TRUE setting and was adjusted for current age, age-squared, sex and 10 PCs as covariates, analysing only variants with a minimum minor allele count of 2.
Lifelines
Lifelines is a large, multigenerational cohort study that includes over 167,000 participants (10%) from the northern population of the Netherlands. The study included participants from three generations, who will be followed for at least 30 years, to obtain insight into healthy ageing. Detailed population characterisation was described by Scholtens et al.52 and Sijtsma et al.53.
Lifelines samples were genotyped in two separate stages. The first stage used the Illumina Cyto SNP12 v2 chip (~15,000 samples) and the second stage the Illumina Global Screening Array (GSA) chip (~35,000 samples). For the purpose of this analysis, the CytoSNP and GSA datasets were treated as separate analyses (CytoSNP samples that were duplicated or had close relatives in the GSA dataset were excluded beforehand). SNP data obtained from the array were imputed using human reference genomes, including the Genome of The Netherlands (GoNL) release 554 and the 1000 Genomes phase 1 v3 reference panels55, using Minimac (version 2012.10.3.9)56. Prior to imputation, SHAPEIT2 was employed for genotype pre-phasing57, and the Genotype Harmonizer was used to align the genotypes with the reference panels to address strand issues58. Cleaned pedigree files and in- and output files for imputation algorithms were created in PLINK59. The imputation analysis was conducted using Beagle (version 3.1.0.8)60.
Strabismus cases were identified based on self-report of strabismus surgery or self-report of strabismus as the ‘reason to start using glasses/contact lenses’ and were under the age of 8 when they started wearing glasses or contact lenses.
Australian genetics of depression cohort study
The AGDS release 11 contains 20,689 participants who have been recruited through the Australian Department of Human Services and a media campaign. Participants completed an online questionnaire that consisted of a compulsory module that assessed self-reported psychiatric disease history and other traits related to psychopathology. By September 2018, DNA samples from 15,792 participants had been collected using saliva kits. The detailed sample recruitment information has been previously described61. The genotyping of AGDS data was performed using the Illumina Global Screening Array (GSA). The genotype data were imputed via TOPMed Imputation Server62, and the SNPs were dropped by high missingness (>1%), deviation from Hardy–Weinberg equilibrium (<1 × 10−6), and low minor allele frequency (<1%). We also rejected individuals with missing rates >0.01 and kinship coefficients greater than 0.2.
There were 233 participants of European ancestry with self-reported strabismus and 15,117 controls without strabismus. We conducted a GWAS via Regenie (version 2.2.4) (Mbatchou et al. 2021), adjusting for sex, age and the top 20 PCs. SNPs with MAF > 0.01 and INFO score >0.8 were retained.
Previously published USA/Australia/UK GWAS
GWAS for ET was conducted by Shaaban et al.10 examined a white European American cohort as the discovery cohort containing non-accommodative (826 cases and 2991 controls) or accommodative (224 cases and 749 controls) ET samples. The replication cohorts involved non-accommodative (689 cases and 1448 controls) or accommodative (66 cases and 264 controls) ET samples from white European, Australian and United Kingdom populations. These four groups were included as separate cohorts in our meta-analysis.
The sample collection and GWAS analysis procedures have been described in detail in the paper10. Briefly, 337,204 SNPs were genotyped using OmniExpress arrays and passed QC. These SNPs were imputed against 1000 Genomes phase 1 v3 European reference panels55 using IMPUTE2 programme (version 2)63. Phenotyping of data was based on participant examinations by an ophthalmologist, optometrist, or orthoptist; participant questionnaires and reviews of additional medical records.
The authors applied a mixed linear additive model for the 2018 Shaaban study, rather than logistic regression. Therefore, we applied the equation to convert the SNP effect sizes from linear scale beta values to the odds ratios:
Where k = Ncases/Nsamples.
Phenotype definition
We considered three phenotype definitions: broad-sense strabismus (based primarily on ICD-10 code H50, non-paralytic strabismus), ET (based primarily on ICD-10 code H50.0) and XT (based primarily on ICD-10 code H50.1).
For datasets with high-quality ICD-10 data (Finngen, GERA and Estonian Biobank), ICD-10 data were used. For UKB, self-report phenotypes were used in preference to the ICD-10 codes, because the medical records did not cover the relevant early-life period for these participants. For Finngen, we used ‘Other strabismus’ (primarily comprising ICD-10 H50, https://risteys.finngen.fi/endpoints/H7_STRABOTH), convergent concomitant strabismus (primarily comprising ICD-10 H50.0, https://risteys.finngen.fi/endpoints/H7_CONVERSTRAB) and divergent concomitant strabismus (primarily comprising ICD-10 H50.1, https://risteys.finngen.fi/endpoints/H7_DIVERGSTRAB). To maximise the sample size of meta-analyses for broad-sense strabismus, the GERA, EstBB and FinnGen ICD-10-based data were combined with the other GWAS summary statistics based on self-report strabismus data (which did not distinguish between ET and XT) from UKB (field 6147), AGDS and Lifelines. For the broad-sense strabismus and the ET analysis, we also included a published ET GWAS10, which contains four summary statistics involving: accommodative ET (based on combined discovery and replication data) and non-accommodative ET (based on combined discovery and replication data). For the XT meta-analysis, we combined XT GWAS from Finngen, GERA and the Estonian Biobank. The sample sizes of the input datasets are listed in Supplementary Data S1.
Statistical analyses
GWAS meta-analyses
We combined the 11 summary statistics using the weighted-sum scheme (METAL software: 5th May 2020) (Willer et al.64). Following the METAL document, we computed the effective sample size for each input data, where Neff = 4/(1/Ncases + 1/Nctrls). Any variants in the input GWAS with INFO score < 0.3 and MAF < 0.01 were removed prior to the meta-analysis. For the requirements of post-GWAS analysis, we calculated the effect size (logOR) and standard error using the equation described by65:
where
Freq is the allele frequency. Weight is proportional to the square root of the effective sample size, as per the METAL document http://genome.sph.umich.edu/wiki/Metal_Documentation (Willer et al. 201064). Given the variation in QC and imputation strategies across the input GWAS summary statistics, variants that were not present in all datasets were included without establishing a specific threshold for the number of studies. Besides, due to X chromosome was excluded in some GWAS sources, we limited our analysis to autosomes to keep the input GWAS consistent.
Conditional analysis
We performed a conditional test using GCTA-COJO (v. 1.94.4; Yang et al.66) to identify statistically independent variants associated with strabismus (window of 1 megabase (MB)). Some of the previously identified strabismus genes may influence strabismus via their effect on refractive error/myopia67, therefore, we applied the mtCOJO approach in GCTA (v. 1.94.4)68 to assess the effect of each SNP on strabismus, accounting for the effect of refractive error. This method performs a conditional analysis where the effect of SNPs on a disease is conditioned upon the disease status. The LD reference for GCTA-COJO and mtCOJO included 4,990 randomly selected individuals of White British ancestry in UKB66.
Genetic correlation analyses
We conducted LD score regression on the GWAS summary statistics for all three strabismus meta-analyses to first estimate the SNP-based heritability and to calculate genetic correlation across strabismus phenotypes69,70. We used a similar procedure to estimate the genetic correlation between three strabismus traits and refractive error22.
Post-GWAS analyses
The Open Targets Genetics platform (https://genetics.opentargets.org/) was used to annotate independent variants by their nearby genes for each independently-associated strabismus variant34. Since the ET GWAS identified the largest number of genome-wide significant variants, it was used as the primary analysis in the post-GWAS analysis. The effect of the lead ET meta-analysis variants on genes expression was investigated using eQTL data from GTEx (V6, V7 and V8, muti-tissue)71,72,73, BIOSQTL (blood cells)74,75, The Brain eQTL Almanac (Braineac, Brain)76, CommonMind Consortium (CMC, Brain)77, Database of Immune Cell Expression, Expression quantitative trait loci (eQTLs) and Epigenomics (DICE, immune cells)78, eQTLcatalogue (muti-tissue)79, eQTLGen (muti-tissue)80, EyeGEx (eye)81, PsychENCODE (brain)82 and xQTLServer (dorsolateral prefrontal cortex)83 through FUMA platform (https://fuma.ctglab.nl/). We conducted a cross-tissue TWAS using UTMOST (v2.0)84. The UTMOST analysis performed single-tissue association tests for 44 GTeX V6 tissues. This was followed by a cross-tissue association test combining 44 gene-trait associations through the joint generalised Berk-Jones (GBJ) test.
The lead SNPs from the PheWAS analysis were cross-referenced against the GWAS Atlas21 (https://atlas.ctglab.nl/PheWAS). The results were filtered by Bonferroni correction (Number of GWASs considered).
Mendelian randomisation of maternal smoking and strabismus
We leveraged our large-scale genetic data on strabismus to test the hypothesis that maternal smoking is causally associated with offspring strabismus risk using MR. We used the well-validated smoking SNP rs1051730/rs16969968 located in the nicotine receptor gene cluster CHRNA5–CHRNA4–CHRNB3 as an instrumental variable85. Each additional allele of rs16969968 in an offspring has been shown to be associated with a 1.02-higher odds of maternal smoking24. The detailed description of this instrument has been described by Yang et al.24. Here we use offspring genotype as a predictor of (i) maternal smoking in pregnancy24 and (ii) strabismus risk in the offspring (the effect size is estimated from the strabismus GWAS in this study). The Wald-ratio test from the TwoSampleMR package in R86 was used to evaluate the association of maternal smoking on offspring strabismus risk. Given that strabismus is a binary outcome, ORs were converted (by multiplying logORs by 0.693 (log2) and then exponentiating) to ORs per doubling in odds, to reflect the average change in the strabismus per doubling increase in the risk of maternal smoking. We also tested whether there was evidence for a causal link between smoking and strabismus using the offspring’s genome (see Supplementary Results and Supplementary Data S11).
Mendelian randomisation of birth weight and strabismus
To investigate if there is genetic evidence to support a causal relationship between birth weight and strabismus, we utilised MR analyses, using birth weight as an exposure14,24. We performed MR using SNPs associated with birth weight87, selecting SNP instruments with only foetal effects to avoid potential horizontal pleiotropic effects on strabismus through the maternal genotype (in cases where >1 SNP was chosen from a single locus, we selected only 1 SNP with the smallest P value so that SNPs chosen were uncorrelated). Given that these SNPs in the maternal genome do not affect offspring birth weight, they are less likely to influence other offspring outcomes. We excluded SNPs where the structural equation model had indicated a potential issue with model convergence (Supplementary Data S6 in Warrington et al). We conducted a two-sample MR analysis between birth weight and three strabismus phenotypes in our analysis using the TwoSampleMR package86, and applied MR-Egger intercept and MR-PRESSO to check the pleiotropy. We compared the MR OR with the ORs change per 500 g increase from a published observational study25. Changes in ORs and their confidence intervals were derived from the ratio of ORhigh-weight to ORlow-weight.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Individual-level data from the UK Biobank were accessed under application number 25331. UK Biobank data are available by request through the UK Biobank Access Management System at https://www.ukbiobank.ac.uk/. FinnGen Data are available by request via https://www.finngen.fi/en. The GWAS summary statistics for strabismus, esotropia and exotropia are available at https://doi.org/10.5281/zenodo.16918155. The Kaiser Permanente GERA, Estonian Biobank, AGDS and Lifelines data are not publicly available as this could compromise research participant privacy/consent.
Code availability
The code used for generating results and visualisation is available at 10.5281/zenodo.15462034. Other scripts used in this study are available upon request to the corresponding authors. The following software packages were used for data analyses: LOCUSZOOM (v. 0.14.0): http://locuszoom.sph.umich.edu/; LD score regression software: https://github.com/bulik/ldsc; METAL software (5th May 2020 release): http://csg.sph.umich.edu/abecasis/Metal/; PLINK software (v.1.9): www.cog-genomics.org/plink/1.9/; R: https://cran.r-project.org/; REGENIE software (v 2.2.4): https://rgcgithub.github.io/regenie/overview/; GCTA-COJO (v. 1.94.4): https://yanglab.westlake.edu.cn/software/gcta/#COJO; GCTA-mtCOJO (v. 1.94.4): https://yanglab.westlake.edu.cn/software/gcta/#mtCOJO; TwoSampleMR: https://github.com/MRCIEU/TwoSampleMR; CMplot: https://github.com/YinLiLin/CMplot.
References
Donahue, S. P. Pediatric Strabismus. N. Engl. J. Med. 356, 1040–1047 (2007).
Hashemi, H. et al. Global and regional prevalence of strabismus: a comprehensive systematic review and meta-analysis. Strabismus 27, 54–65 (2019).
Michaelides, M. & Moore, A. T. The genetics of strabismus. J. Med. Genet. 41, 641–646 (2004).
Harrad, R. & Williams, C. Risk, causes and outcomes of visual impairment after loss of vision in the non-amblyopic eye, a population-based study. Surv. Ophthalmol. 48, 235–236 (2003).
Yang, Y. et al. Maternal smoking during pregnancy and the risk of strabismus in offspring: a meta-analysis. Acta Ophthalmol. 97, 353–363 (2019).
Ponsonby, A. L. et al. The association between maternal smoking in pregnancy, other early life characteristics and childhood vision: the Twins Eye Study in Tasmania. Ophthalmic Epidemiol. 14, 351–359 (2007).
Cotter, S. A. et al. Risk factors associated with childhood strabismus: the multi-ethnic pediatric eye disease and Baltimore pediatric eye disease studies. Ophthalmology 118, 2251–2261 (2011).
Graeber, C. P., Hunter, D. G. & Engle, E. C. The genetic basis of incomitant strabismus: consolidation of the current knowledge of the genetic foundations of disease. Semin. Ophthalmol. 28, 427–437 (2013).
Khan, A. O. et al. Potential linkage of different phenotypic forms of childhood strabismus to a recessive susceptibility locus (16p13.12-p12.3). Mol. Vis. 17, 971–976 (2011).
Shaaban, S. et al. Genome-wide association study identifies a susceptibility locus for comitant esotropia and suggests a parent-of-origin effect. Investig. Ophthalmol. Vis. Sci. 59, 4054–4064 (2018).
Plotnikov, D. et al. A commonly occurring genetic variant within the NPLOC4-TSPAN10-PDE6G gene cluster is associated with the risk of strabismus. Hum. Genet. 138, 723–737 (2019).
Hakim, R. B. & Tielsch, J. M. Maternal cigarette smoking during pregnancy. A risk factor for childhood strabismus. Arch. Ophthalmol. 110, 1459–1462 (1992).
Torp-Pedersen, T. et al. In-utero exposure to smoking, alcohol, coffee, and tea and risk of strabismus. Am. J. Epidemiol. 171, 868–875 (2010).
Gulati, S. et al. Effect of gestational age and birth weight on the risk of strabismus among premature infants. JAMA Pediatr. 168, 850–856 (2014).
Choquet, H. et al. A large multiethnic GWAS meta-analysis of cataract identifies new risk loci and sex-specific effects. Nat. Commun. 12, 3595 (2021).
Liu, M. et al. Association studies of up to 1.2 million individuals yield new insights into the genetic etiology of tobacco and alcohol use. Nat. Genet. 51, 237–244 (2019).
Karlsson Linnér, R. et al. Genome-wide association analyses of risk tolerance and risky behaviors in over 1 million individuals identify hundreds of loci and shared genetic influences. Nat. Genet. 51, 245–257 (2019).
Thorgeirsson, T. E. et al. Sequence variants at CHRNB3-CHRNA6 and CYP2A6 affect smoking behavior. Nat. Genet. 42, 448–453 (2010).
Barton, A. R., Sherman, M. A., Mukamel, R. E. & Loh, P.-R. Whole-exome imputation within UK Biobank powers rare coding variant association and fine-mapping analyses. Nat. Genet. 53, 1260–1269 (2021).
Pickrell, J. K. et al. Detection and interpretation of shared genetic influences on 42 human traits. Nat. Genet. 48, 709–717 (2016).
Watanabe, K. et al. A global overview of pleiotropy and genetic architecture in complex traits. Nat. Genet. 51, 1339–1348 (2019).
Hysi, P. G. et al. Meta-analysis of 542,934 subjects of European ancestry identifies new genes and mechanisms predisposing to refractive error and myopia. Nat. Genet. 52, 401–407 (2020).
Hu, Y. et al. A statistical framework for cross-tissue transcriptome-wide association analysis. Nat. Genet. 51, 568–576 (2019).
Yang, Q., Millard, L. A. C. & Davey Smith, G. Proxy gene-by-environment Mendelian randomization study confirms a causal effect of maternal smoking on offspring birthweight, but little evidence of long-term influences on offspring health. Int. J. Epidemiol. 49, 1207–1218 (2020).
Torp-Pedersen, T. et al. Perinatal risk factors for strabismus. Int. J. Epidemiol. 39, 1229–1239 (2010).
Martinez Sanchez, M. et al. Presence of copy number variants associated with esotropia in patients with exotropia. JAMA Ophthalmol. 142, 243–247 (2024).
Plotnikov, D., Pärssinen, O., Williams, C., Atan, D. & Guggenheim, J. A. Commonly occurring genetic polymorphisms with a major impact on the risk of nonsyndromic strabismus: replication in a sample from Finland. J. AAPOS 26, e1–12.e6 (2022).
UK Biobank—Neale lab. http://www.nealelab.is/uk-biobank/.
Schellhas, L. et al. Maternal and child genetic liability for smoking and caffeine consumption and child mental health: an intergenerational genetic risk score analysis in the ALSPAC cohort. Addiction 116, 3153–3166 (2021).
Nita, M. & Grzybowski, A. Smoking and eye pathologies. A systemic review. Part I. Anterior eye segment pathologies. Curr. Pharm. Des. 23, 629–638 (2017).
Saunders, G. R. B. et al. Genetic diversity fuels gene discovery for tobacco and alcohol use. Nature 612, 720–724 (2022).
Reitsma, M. B. et al. Spatial, temporal, and demographic patterns in prevalence of smoking tobacco use and initiation among young people in 204 countries and territories, 1990-2019. Lancet Public Health 6, e472–e481 (2021).
Kanukollu, V. M. & Sood, G. Strabismus (StatPearls Publishing, 2022).
Ghoussaini, M. et al. Open Targets Genetics: systematic identification of trait-associated genes using large-scale genetics and functional genomics. Nucleic Acids Res. 49, D1311–D1320 (2021).
Gage, S. H. et al. Assessing causality in associations between cannabis use and schizophrenia risk: a two-sample Mendelian randomization study. Psychol. Med. 47, 971–980 (2017).
Burgess, S. & Labrecque, J. A. Mendelian randomization with a binary exposure variable: interpretation and presentation of causal estimates. Eur. J. Epidemiol. 33, 947–952 (2018).
Evans, D. M., Moen, G.-H., Hwang, L.-D., Lawlor, D. A. & Warrington, N. M. Elucidating the role of maternal environmental exposures on offspring health and disease using two-sample Mendelian randomization. Int. J. Epidemiol. 48, 861–875 (2019).
Lange, S., Probst, C., Rehm, J. & Popova, S. National, regional, and global prevalence of smoking during pregnancy in the general population: a systematic review and meta-analysis. Lancet Glob. Health 6, e769–e776 (2018).
Zhang, X. J. et al. Prevalence of strabismus and its risk factors among school aged children: The Hong Kong Children Eye Study. Sci. Rep. 11, 13820 (2021).
Bycroft, C. et al. The UK Biobank resource with deep phenotyping and genomic data. Nature 562, 203–209 (2018).
Mbatchou, J. et al. Computationally efficient whole-genome regression for quantitative and binary traits. Nat. Genet. 53, 1097–1103 (2021).
Banda, Y. et al. Characterizing race/ethnicity and genetic ancestry for 100,000 subjects in the Genetic Epidemiology Research on Adult Health and Aging (GERA) Cohort. Genetics 200, 1285–1295 (2015).
Hoffmann, T. J. et al. Design and coverage of high throughput genotyping arrays optimized for individuals of East Asian, African American, and Latino race/ethnicity using imputation and a novel hybrid SNP selection algorithm. Genomics 98, 422–430 (2011).
Kvale, M. N. et al. Genotyping informatics and quality control for 100,000 subjects in the Genetic Epidemiology Research on Adult Health and Aging (GERA) Cohort. Genetics 200, 1051–1060 (2015).
Loh, P.-R. et al. Reference-based phasing using the Haplotype Reference Consortium panel. Nat. Genet. 48, 1443–1448 (2016).
Das, S. et al. Next-generation genotype imputation service and methods. Nat. Genet. 48, 1284–1287 (2016).
Birney, E. & Soranzo, N. Human genomics: the end of the start for population sequencing. Nature 526, 52–53 (2015).
Leitsalu, L. et al. Cohort Profile: Estonian Biobank of the Estonian Genome Center, University of Tartu. Int. J. Epidemiol. 44, 1137–1147 (2015).
Browning, S. R. & Browning, B. L. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am. J. Hum. Genet. 81, 1084–1097 (2007).
Mitt, M. et al. Improved imputation accuracy of rare and low-frequency variants using population-specific high-coverage WGS-based imputation reference panel. Eur. J. Hum. Genet. 25, 869–876 (2017).
Zhou, W. et al. Efficiently controlling for case-control imbalance and sample relatedness in large-scale genetic association studies. Nat. Genet. 50, 1335–1341 (2018).
Scholtens, S. et al. Cohort Profile: LifeLines, a three-generation cohort study and biobank. Int. J. Epidemiol. 44, 1172–1180 (2015).
Sijtsma, A. et al. Cohort Profile Update: Lifelines, a three-generation cohort study and biobank. Int. J. Epidemiol. 51, e295–e302 (2022).
Genome of the Netherlands Consortium. Whole-genome sequence variation, population structure and demographic history of the Dutch population. Nat. Genet. 46, 818–825 (2014).
1000 Genomes Project Consortium et al. A global reference for human genetic variation. Nature 526, 68–74 (2015).
Howie, B., Fuchsberger, C., Stephens, M., Marchini, J. & Abecasis, G. R. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat. Genet. 44, 955–959 (2012).
Delaneau, O., Zagury, J.-F. & Marchini, J. Improved whole-chromosome phasing for disease and population genetic studies. Nat. Methods 10, 5–6 (2013).
Deelen, P. et al. Genotype harmonizer: automatic strand alignment and format conversion for genotype data integration. BMC Res. Notes 7, 901 (2014).
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
Browning, B. L. & Browning, S. R. A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am. J. Hum. Genet. 84, 210–223 (2009).
Byrne, E. M. et al. Cohort profile: the Australian genetics of depression study. BMJ Open 10, e032580 (2020).
Taliun, D. et al. Sequencing of 53,831 diverse genomes from the NHLBI TOPMed Program. Nature 590, 290–299 (2021).
Howie, B. N., Donnelly, P. & Marchini, J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 5, e1000529 (2009).
Willer, C. J., Li, Y. & Abecasis, G. R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26, 2190–2191 (2010).
Zhu, Z. et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat. Genet. 48, 481–487 (2016).
Yang, J. et al. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat. Genet. 44, S1–S3 (2012).
Tang, S. M. et al. Refractive errors and concomitant strabismus: a systematic review and meta-analysis. Sci. Rep. 6, 35177 (2016).
Zhu, Z. et al. Causal associations between risk factors and common diseases inferred from GWAS summary data. Nat. Commun. 9, 224 (2018).
Brown, B. C. Asian Genetic Epidemiology Network Type 2 Diabetes Consortium, Ye, C. J., Price, A. L. & Zaitlen, N. Transethnic genetic-correlation estimates from summary statistics. Am. J. Hum. Genet. 99, 76–88 (2016).
Bulik-Sullivan, B. et al. An atlas of genetic correlations across human diseases and traits. Nat. Genet. 47, 1236–1241 (2015).
GTEx Consortium. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 348, 648–660 (2015).
Gibbs, J. R. et al. Abundant quantitative trait loci exist for DNA methylation and gene expression in human brain. PLoS Genet. 6, e1000952 (2010).
GTEx Consortium. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science 369, 1318–1330 (2020).
Bonder, M. J. et al. Disease variants alter transcription factor levels and methylation of their binding sites. Nat. Genet. 49, 131–138 (2017).
Zhernakova, D. V. et al. Identification of context-dependent expression quantitative trait loci in whole blood. Nat. Genet. 49, 139–145 (2017).
Ramasamy, A. et al. Genetic variability in the regulation of gene expression in ten regions of the human brain. Nat. Neurosci. 17, 1418–1428 (2014).
Hoffman, G. E. et al. CommonMind Consortium provides transcriptomic and epigenomic data for Schizophrenia and Bipolar Disorder. Sci. Data 6, 180 (2019).
Schmiedel, B. J., et al. Impact of genetic polymorphisms on human immune cell gene expression. Cell 175, 1701–1715.e16 (2018).
Kerimov, N. et al. A compendium of uniformly processed human gene expression and splicing quantitative trait loci. Nat. Genet. 53, 1290–1299 (2021).
Võsa, U. et al. Large-scale cis- and trans-eQTL analyses identify thousands of genetic loci and polygenic scores that regulate blood gene expression. Nat. Genet. 53, 1300–1310 (2021).
Ratnapriya, R. et al. Retinal transcriptome and eQTL analyses identify genes associated with age-related macular degeneration. Nat. Genet. 51, 606–610 (2019).
Wang, D. et al. Comprehensive functional genomic resource and integrative model for the human brain. Science 362, eaat8464 (2018).
Ng, B. et al. An xQTL map integrates the genetic architecture of the human brain’s transcriptome and epigenome. Nat. Neurosci. 20, 1418–1426 (2017).
Rodriguez-Fontenla, C. & Carracedo, A. UTMOST, a single and cross-tissue TWAS (Transcriptome Wide Association Study), reveals new ASD (Autism Spectrum Disorder) associated genes. Transl. Psychiatry 11, 256 (2021).
Munafò, M. R. et al. Association between genetic variants on chromosome 15q25 locus and objective measures of tobacco exposure. J. Natl. Cancer Inst. 104, 740–748 (2012).
Hemani, G. et al. The MR-Base platform supports systematic causal inference across the human phenome. eLife 7, e34408 (2018).
Warrington, N. M. et al. Maternal and fetal genetic effects on birth weight and their relevance to cardio-metabolic risk factors. Nat. Genet. 51, 804–814 (2019).
Acknowledgements
We would like to acknowledge the data resource used in this meta-analysis, including Kaiser Permanente GERA Cohort, the UK Biobank Resource (application number 25331), the FinnGen study, Lifelines, Australian Genetics of Depression Cohort Study and USA/Australia/UK cohorts used in ET GWAS. This research has been conducted using the UK Biobank Resource (www.ukbiobank.ac.uk) under the Application number 25331. We are grateful to the KPNC members who have generously agreed to participate in the Kaiser Permanente Research Program on Genes, Environment, and Health. Genotyping of the Kaiser Permanente GERA cohort was funded by a grant from the National Institute on Aging, National Institute of Mental Health, and National Institute of Health Common Fund (RC2AG036607). Support for GERA participant enrolment, survey completion, and biospecimen collection for the Research Program on Genes, Environment and Health was provided by the Robert Wood Johnson Foundation, the Wayne and Gladys Valley Foundation, the Ellison Medical Foundation, and Kaiser Permanente Community Benefit Programs. The GERA genotype data are available upon application to the KP Research Bank (https://researchbank.kaiserpermanente.org/). HC is supported by the National Eye Institute (NEI) (R01 EY027004 and R01 EY033010). We want to acknowledge the participants of the Estonian Biobank (EstBB) for their contributions. The Estonian Biobank analyses were partially carried out in the High Performance Computing Center, University of Tartu. The work of the Estonian Genome Center, University of Tartu, was funded by the European Union through Horizon 2020 research and innovation programme under grants #810645 and #894987, through the European Regional Development Fund projects GENTRANSMED (2014-2020.4.01.15-0012), MOBEC008, MOBERA21 and Estonian Research Council Grant PRG1291. K.Õ. and L.M. acknowledge Estonian Research Council Grant PRG471 and PRG2040. The activities of the EstBB are regulated by the Human Gene Research Act, which was adopted in 2000 specifically for the operations of EstBB. Individual-level data analysis in EstBB was carried out under ethical approval 1.1-12/624 from the Estonian Committee on Bioethics and Human Research (Estonian Ministry of Social Affairs), using data according to release application 6-7/GI/1457 from the Estonian Biobank. The Estonian Biobank Research Team (estbbresearch@ut.ee) was responsible for data collection, genotyping, quality control and imputation and consisted of Andres Metspalu (andres.metspalu@ut.ee), Mait Metspalu (mait.metspalu@ut.ee), Lili Milani (lili.milani@ut.ee), Reedik Mägi (reedik.magi@ut.ee), Mari Nelis (mari.nelis@ut.ee) and Georgi Hudjashov (georgi.hudjashov@ut.ee). The Lifelines Biobank initiative has been made possible by funding from the Dutch Ministry of Health, Welfare and Sport, the Dutch Ministry of Economic Affairs, the University Medical Center Groningen (UMCG, the Netherlands), the University of Groningen and the Northern Provinces of the Netherlands. The generation and management of GWAS genotype data for the Lifelines Cohort Study is supported by the UMCG Genetics Lifelines Initiative (UGLI). UGLI is partly supported by a Spinoza Grant from NWO, awarded to Cisca Wijmenga. The authors wish to acknowledge the services of the Lifelines Cohort Study, the contributing research centres delivering data to Lifelines, and all the study participants. S.M. and D.A.M. acknowledge Program Grant (#1150144) and Centre of Research Excellence (#1116360) funding from the Australian National Health and Medical Research Council (NHMRC). P.G. (#1173390) and J.S.O. (#2018420) are supported by NHMRC Investigator Grants. L.D.H. is supported by an Australian Research Council Discovery Early Career Researcher Award (DE240100014). S.M. and D.A.M. are supported by research fellowships from NHMRC. E.C.E. is a Howard Hughes Medical Institute investigator.
Author information
Authors and Affiliations
Consortia
Contributions
Conception and design: W.H., J.S.O., L.D.H., P.G., S.M., Y.W. Analysis and interpretation: W.H., S.M. Data collection: W.H., S.M., P.V.D.M., M.S.M., J.V., K.K., L.M., K.O., E.A., N.M., D.P., C.J., R.M., H.S., E.P., Kuldar K., J.G., D.M., E.E., H.C., T.P. Obtained funding: S.M., D.M., P.G. Overall responsibility: W.H., S.M.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
He, W., van der Most, P.J., Ong, JS. et al. Large-scale GWAS of strabismus identifies risk loci and provides support for a link with maternal smoking. Nat Commun 16, 7890 (2025). https://doi.org/10.1038/s41467-025-62456-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-62456-9