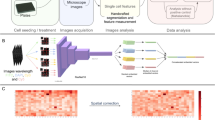
Abstract
Self-organised three-dimensional (3D) cell cultures, collectively called 3D-oids, include spheroids, organoids and other co-culture models. Systematic evaluation of these models forms a critical new generation of high-content screening (HCS) systems for patient-specific drug analysis and cancer research. However, the standardisation of working with 3D-oids remains challenging and lacks convincing implementation. This study develops and tests HCS-3DX, a next-generation system for HCS analysis in 3D imaging and image evaluation. HCS-3DX is based on three main components: an automated Artificial Intelligence (AI)-driven micromanipulator for 3D-oid selection, an HCS foil multiwell plate for optimised imaging, and image-based AI software for single-cell data analysis. We validated HCS-3DX directly on 3D tumour models, including tumour-stroma co-cultures. Our data demonstrate that HCS-3DX achieves a resolution that overcomes the limitations of current systems and reliably and effectively performs 3D HCS at the single-cell level. Its application will enhance the accuracy and efficiency of drug screening processes, support personalised medicine approaches, and facilitate more detailed investigations into cellular behaviour within 3D structures.
Similar content being viewed by others
Introduction
For decades, evaluation of drug effects has relied on two-dimensional (2D) cell cultures as model systems. However, 2D cell cultures cannot accurately capture the complex physiological characteristics of tissues and tumour microenvironments1. Three-dimensional (3D) cell cultures are gaining more attention as a relevant model system for drug testing2,3,4. In particular, the so-called “3D-oids” (3D models including spheroids, organoids, tumouroids, and assembloids) have been broadly tested and proven to mimic in vivo conditions. Indeed, 3D-oids can maintain tissue structure which makes them highly relevant for numerous biological research areas, including drug discovery, regenerative medicine, tumour biology, and immunotherapy5,6,7.
Since 2010, the continuously increasing attention on 3D cell cultures has resulted in almost 40,000 articles (based on the search terms “spheroid, organoid, tumoroid, and assembloid”) and more than 168,000 publications discussing high-content systems. However, only 1% of these publications present high-content screening (HCS) analyses based on imaging systems, and only 76 contributions refer to HCS imaging of 3D-oids (Supplementary Table 1). These small numbers show that there are challenges in developing 3D HCS platforms, including 3D-oid generation, handling, imaging, and analysis.
At the level of 3D-oid generation, there are concerns about: (a) morphological variability8,9, (b) penetration properties of compounds, including specific stains10, (c) inner distribution and biological characteristics of the cells11,12. While seminal work has been conducted on standardising generation protocols13, the nature of 3D models means that high variability is present when screening large numbers of 3D-oids14. To handle 3D-oids while ensuring experimental reproducibility and standardisation, the use of Artificial Intelligence (AI)-driven systems was proposed in order to manipulate and select similar 3D-oid aggregates15,16,17. For instance, AI-driven micromanipulators combining morphological pre-selection with automated pipetting systems reduce time and ensure reliability when transferring spheroids to the imaging plates18.
Complex drug screening analyses require single-cell phenotyping, necessitating imaging at the highest resolution19. Light-sheet fluorescence microscopy (LSFM) is able to visualise large samples at the cellular level with high imaging penetration, minimal phototoxicity and photobleaching20,21. The available LSFM HCS systems for 3D-oids have different drawbacks (e.g. different sample preparation, light penetration, imaging time) (Supplementary Table 2). In addition, the amount of generated data is typically vast and heterogeneous, making data analysis a time-consuming and computationally demanding procedure requiring automation22. Several pipelines and tools have been introduced for the analysis of 3D data, but no standard has been defined for quantitative tasks, including segmentation, classification, and feature extraction23,24,25.
In this work, we present an HCS system combining engineering innovations, advanced imaging, and AI technologies. Precisely, we described HCS-3DX, a customisable HCS system for 3D imaging and analysis of 3D-oids at a single-cell level (Fig. 1a). HCS-3DX includes (I) selection and transfer of morphologically homogeneous 3D spheroids using a custom-developed tool, called SpheroidPicker (Fig. 1b); (II) single-cell LSFM imaging using a custom Fluorinated Ethylene Propylene (FEP) foil multiwell plate (Fig. 1c); (III) an AI-based custom 3D data analysis workflow developed in Biology Image Analysis Software (BIAS, Single-Cell Technologies Ltd., Szeged, Hungary)26 (Fig. 1d). We present a multitude of experiments to show that HCS-3DX can be reliably used for single-cell 3D HCS. First, morphological analysis of tumour spheroids under different conditions was executed to define the best setup for 3D-oid selection using the SpheroidPicker, an AI-guided 3D cell culture delivery system18. Then, the accuracy of 2D features at multiple levels was validated (Fig. 2). Before extending the feature analysis to 3D cultures, we specifically quantified screening performance and image quality while using the designed HCS foil multiwell plate. Finally, the effectiveness of the HCS-3DX system was proven on monoculture and co-culture tumour models via quantitative evaluation of tissue composition at single-cell resolution (Fig. 2).
a HCS-3DX workflow includes 8 essential steps: (I) 3D cell culture generation, (II) AI-driven 2D object detection, feature analysis and selection, (III) automated sample transfer from the source culture vessel, (IV) transfer to a custom-developed HCS imaging plate, (V) LSFM screening, (VI) generation of massive 3D image datasets, (VII) image analysis with BIAS, (VIII) feature extraction and single-cell classification. b Step II-IV - working principle of the SpheroidPicker, an AI-guided, 3D cell culture delivery system using a light microscope for 2D morphology-based spheroid screening and analysis, enabling semi- or fully-automated transfer of selected spheroids with predefined morphology features. c Step IV - selected spheroids are transferred to the HCS foil multiwell plate compatible with LSFM to obtain single-cell resolution in 3D. d Step VII - a representative image of a nuclei-labelled spheroid analysed and visualised by BIAS, with single cells indicated by green bounding boxes.
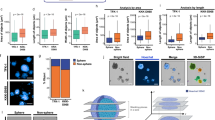
a Representative images of the 2D annotated dataset, including monoculture (purple) and co-culture (green) spheroids with the smallest and largest volumes, and least and greatest sphericity values. The scale bar represents 50 µm. b Representative image of a monoculture spheroid composed of HeLa Kyoto cells (green - EGFP-alpha-tubulin; red - H2B-mCherry; grey - actin). Monoculture spheroids were generated in 3 independent experiments by 3 experts, resulting in n = 223 for 2D and n = 110 for 3D. The scale bar represents 50 µm. c Representative image of a co-culture spheroid comprising HeLa Kyoto and MRC-5 cell lines (blue - DAPI, green – EGFP-alpha-tubulin; red - H2B-mCherry; grey - actin). Co-culture spheroids were generated in 3 independent experiments by 3 experts, resulting in n = 203 for 2D and n = 114 for 3D. The scale bar represents 50 µm. d Assembled HCS plate in use. e 3D printed heat-resistant mould element that was used to form the FEP foil. f Vacuum-formed transparent FEP foil where each cuvette is suitable for one sample. For visualisation, each cuvette was filled with cell culture medium. g The insert element that fits into the cuvettes of the foil. The insert element only secures the position of the samples at the bottom of the cuvettes. h The grid element that fits into the base prevents the movement of the FEP foil after filling up with the detection solution. Two versions are available. i The base of the plate is an additional component that provides volume for the detection solution and space inside the plate for the water immersion objectives. Two versions are available. j Representative co-culture spheroids showing a spherical and an irregular example from the dataset. Images from left to right show the brightfield images with the corresponding annotation, the maximum intensity projection (MIP) images acquired with the light-sheet microscope, whole spheroid segmentation, and cytoplasm and nucleus segmentation combined with machine learning-based classification. The MRC-5 cells are illustrated in orange, while the HeLa Kyoto cells are illustrated in purple. The scale bar represents 50 µm.
Results
Concept of HCS-3DX
The developed HCS-3DX platform overcomes many limitations of the current analysis of 3D-oids. HCS-3DX comprises all the steps required to evaluate 3D cell cultures at a single-cell level by enabling reliable, fast and automatic high-content imaging of multiple 3D-oids with a high penetration depth (Supplementary Table 3 and Supplementary Movie 1). Consequently, the proposed concept is advantageous for a wide range of research purposes, including industrial drug screening, personalised medicine, or basic research.
2D features variability facilitates definition of 3D-oid pre-selection parameters
In the first experiment, a comparative study was executed for the pre-selection of spheroids generated under the same conditions. To verify the ideal screening parameters in 2D, brightfield images obtained by various objectives were compared by measuring radiomic features of co-culture spheroids. The same spheroids were imaged at different magnifications (Supplementary Table 4). Specifically, 50 spheroids were imaged with 2.5x, 5x, 10x, and 20x objectives resulting in 200 images (Supplementary Fig. 1a). The images were manually annotated and 2D morphological features were extracted using BIAS and ReViSP (a specific tool for estimating the volume of spheroids using a single 2D brightfield image27). On average, for all the extracted features (i.e. Diameter, Perimeter, Area, Volume 2D, Circularity, Sphericity 2D, and Convexity), the relative differences between different magnifications (considering 20x as a reference) were less than 5%, except for Volume 2D that reached almost 6.5% (Fig. 3a and Supplementary Table 5). Perimeter, Sphericity 2D, Circularity, and Convexity showed significant differences between the 2.5x and the 5x/10x objectives since the lower image resolution resulted in a less accurate representation (Supplementary Fig. 1b, c). The 2.5x objective resulted in the fastest imaging, including finding and focusing; however, it showed the least accurate feature extraction. Both 5x and 10x objectives were ideal for imaging spheroids since they increased the imaging speed by ~45% or ~20% while providing relatively accurate values. The 20x objective provided the highest image resolution (Supplementary Fig. 1a) but required more time to find and focus on the spheroids. Nevertheless, 20x objective was used as a reference for relative feature comparisons for all further experiments.
a Spider plot representation of the relative differences of features displayed as a percentage, acquired by annotating the same spheroids, imaged by 4 different objectives with different magnifications. Each objective was compared to the annotations that were executed on the images acquired with the 20x objective. After manual annotation, features such as Diameter, Perimeter, Area, Volume 2D, Circularity, Sphericity 2D, and Convexity were displayed (n = 50). b Beeswarm plot of the monoculture (grey) and co-culture (light blue) spheroids divided by experts (vertical distribution) and experiments (orange - Experiment 1, green - Experiment 2, purple - Experiment 3), indicating the volume and sphericity of each sample. Overall, 426 spheroids were screened. Each circle represents a spheroid on a volume axis showing the sphericity values by the size and intensity (bigger and more intensive circles represent the most spherical spheroids). The number of spheroids evaluated in each batch is displayed, with different colours representing individual experiments and black indicating the total count. c Boxplot visualisation of the monoculture (purple, n = 223) and co-culture (orange, n = 203) spheroids based on Diameter (µm), Area (µm2), and Circularity (−). Among the three measurements, statistically significant differences were observed in two cases (p < 0.001, ***), and one case showed significance with p < 0.0255 (*). d Size comparison of spheroids for each experiment (n ≥ 16, see Fig. 3b). Significant differences are not visualised. Dashed lines show the overall average of monoculture and co-culture experiments. e Shape comparison of spheroids for each experiment (n ≥ 16, see Fig. 3b). For the statistical analysis, a two-sided non-parametric Kruskal-Wallis test was conducted, followed by Dunn’s multiple comparison test. When comparing only two groups, the two-sided Kolmogorov-Smirnov test was utilised. *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001. Box plots were visualised using Tukey’s method, with medians shown as central lines, boxes representing interquartile ranges, and whiskers extending to 1.5× interquartile range. Abbreviations: M - monoculture, C - co-culture, Exp - experiment. Source data are provided as a Source Data file.
Analysis of spheroid model variability
In order to measure tumour model heterogeneity, mono- and co-culture spheroids generated by 3 experts with extensive daily experience in 3D cell cultures were compared. The experts repeated the same experiments 3 times (Fig. 3b–e and Fig. 4a). A total of 426 spheroids, 223 mono- and 203 co-cultures were generated. For the monoculture spheroids, each expert generated samples of 100 HeLa Kyoto human cervical cancer cells per well in a 384-well U-bottom cell-repellent plate and incubated for 48 h before fixation. In the case of co-cultures, 40 HeLa Kyoto cells were seeded first, and after 24 h 160 MRC-5 human fibroblast cells were added and incubated for another 24 h before fixation. Each spheroid was manually annotated to extract the 2D features.
a Distribution of monoculture (n = 223) and co-culture (n = 203) spheroids based on the Diameter, Area, Solidity, and Circularity features. Each circle represents a spheroid, and each colour corresponds to different experts (green - Expert 1, purple - Expert 2, and orange - Expert 3). The central lines display the average value for all the samples, whilst the dashed lines display the plus and minus one standard deviation. Based on the standard deviation, 9 different regions with a corresponding percentage of the number of spheroids are shown. Spearman correlation was used to evaluate the relationship between the variables. For cases showing either positive or negative correlation, scatter plots were generated using a fitted regression line with shaded bands indicating the standard error of the mean (SEM). Source data are provided as a Source Data file.
Although all the experts used the same equipment within the same environment and followed exactly the same protocol, the inter-operator comparison showed great variability regarding the size and shape of the generated spheroids (Fig. 3b–e and Fig. 4a). The direct comparison of mono- and co-culture spheroids showed significant differences whilst evaluating Diameter, Circularity, and Area (Fig. 3c). By evaluating the 2D features of the monoculture spheroids, Expert 1 generated significantly bigger spheroids (Fig. 3b, d). Although the range of Area and Volume 2D is wide, Circularity and Sphericity 2D values were independent of spheroid size and there were no significant differences between the experts and batches (Fig. 3b–e). We observed increased variability between experts and between batches when generating the co-culture spheroids with two different cell lines (Fig. 3d). Compared to monocultures, the co-culture spheroids had twice as many seeding cells and, on average, smaller Volume 2D, indicating more compact spheroids. Visualising the individual spheroids based on 2D parameters and their respective standard deviation revealed the distribution, and the number representing the most similar samples differed (Fig. 4a). The data showed stronger correlation and a greater number of spheroids when arranged according to Circularity and Diameter: −0.69 and 60.5% for monoculture and −0.54 and 55.1% for co-cultures. Using Area instead of Diameter resulted in no correlation. Furthermore, spheroids were less similar, resulting in a smaller quantity of spheroids for Circularity and Area and Solidity and Area (49.7% and 50.2% for monoculture and 51.2% and 54.6% for co-culture). The results indicate that the number of ideal spheroids changes even up to 10% when different pairs of features are used. Overall, feature selection for spheroid characterization is crucial in terms of outcomes. In particular, evaluations should include one morphological and one size-related feature.
Design of an optimised HCS imaging plate for LSFM
An HCS multi-well plate with predefined positions for the samples was designed to allow fast and automatic multiple object screenings using an LSFM (Fig. 2d–i, and Supplementary Data 1). The plate was developed and tested on the Leica TCS SP8 DLS upright LSFM (Fig. 5a–f). To validate the resulting image quality and measure the screening time, T-47D spheroids were generated with a diameter under 200 µm. This cell line was previously used in different experiments due to its favourable optical properties28,29,30. Two different groups of 5 spheroids were defined: as recommended by Leica, the first 5 were embedded into agarose in U-shape glass capillaries inserted in Petri dishes; the other 5 spheroids were placed into the HCS plate. Regarding the image quality, the difference between the groups was minimal and not noticeable to the naked eye (Fig. 5a). Therefore, we measured the quality of the images using the intensity variance metric, which can characterise the general blurriness of an image qualitatively29. Considering the average values, no significant differences were observed, suggesting similar image quality (Fig. 5b). Although the spheroids screened with the HCS plate showed slightly better scores, this phenomenon can be explained by the size and morphology differences of randomly selected spheroids.
a Fluorescent images of T-47D spheroids stained with DRAQ5 were used as a reference to compare image quality. Randomly selected spheroids screened in the HCS plate (left image) and in Petri dish (right image) were visualised where images were selected from the top, middle, and bottom regions. Scale bars represent 100 µm. b To characterise the general blurriness of the whole z-stack, the intensity variance metric was used both for the inside circle and for the whole spheroid. Results are shown in a boxplot, showing that no significant differences were measured between the HCS plate and the Petri. n = 5 for each group. The Kolmogorov-Smirnov test was used. *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001. c Preparation and screening times were measured for both techniques and displayed on a stacked bar plot. Starting from the darkest colour the 4 categories are Sample preparation, Change sample (this category does not apply to the HCS plate), Calibration, and Imaging. The total time was calculated for 10 samples for each method. d Example image of a cleared T-47D spheroid labelled with DRAQ5 for nuclei (purple) and Phalloidin 488 for actin (green) and optically cleared with Sucrose protocol. A total of 5184 nuclei were segmented using BIAS. Scale bar represents 100 µm. e By co-culturing T-47D breast cancer cells with MRC-5 fibroblasts and EA.hy926 endothelial cells, a hydrogel-based tumour model was created to track the tumour microenvironment in real time. CellTracker dyes (orange, deep red, and green) are used to detect cells. The tumour, endothelial, and fibroblast cells are marked by red, green, and white labels, respectively. Scale bar represents 50 µm. f A ground truth dataset of HeLa Kyoto - MRC-5 co-cultures was created using the HCS plate. Scale bar represents 50 µm. Box plots were visualised using Tukey’s method, with medians shown as central lines, boxes representing interquartile ranges, and whiskers extending to 1.5× interquartile range.
Next, we analysed the effectiveness of the HCS plate. To measure the average screening time, 10 spheroids were separated into Petri dishes and embedded into agarose, while the other 10 spheroids were placed into the HCS plate. Comparing the total screening time, the HCS plate performed twice as fast (48 ± 7 min) whilst the screening time in the Petri dishes took approximately 102 ± 9 min (Fig. 5c). The largest difference was observed during sample preparation: manually plating the spheroids into the HCS plate took 14 ± 2 min, while Petri embedding the spheroids into agarose took approximately 37 ± 5 min. Another significant difference appeared during the calibration process, where replacing and calibrating each spheroid took approximately 26 ± 4 min. This does not apply to the HCS plate since all 10 spheroids were separated and placed into individual cuvettes on the same plate. Other calibration and imaging processes took 30 ± 3 min for both approaches. However, finding samples within the plate is easier, as all positions are predefined and one calibration at the beginning is sufficient. Thus, our HCS plates achieved image quality that matched previous studies28,29 while showing greater effectiveness that will optimise screening. Overall, the custom-developed HCS plate is easy to use, reduces imaging times, and ensures the same image quality as the method recommended by the manufacturer. The plate is compatible with fixed and optically cleared spheroids with a diameter of 350 µm (Supplementary Movie 2), live-cell imaging of hydrogel-based multi-cellular models, and creating high quality 3D datasets (Fig. 5d–f and Supplementary Movie 3).
2D and 3D feature comparison
From the spheroids used for the 2D imaging, 110 monoculture and 114 co-culture spheroids were randomly selected, and fluorescence images were acquired by LSFM using the HCS plate. Images were imported to the BIAS software to measure 3D features at the nucleus, cell, and spheroid levels (Supplementary Note 1). First, the correlations were analysed between the 2D and 3D features independently for each dataset (Fig. 6a). Among the tested features, Solidity showed the highest correlation when compared to other shape features both 2D and 3D. There was no correlation between the size and shape descriptors. In general, the 2D estimation resulted in smaller spheroids, whilst each shape descriptor reached higher values and showed a significant difference compared to 3D (Fig. 6b). Monoculture spheroids showed a significant difference between Volume 2D and Volume 3D, but there was no significant difference for the co-culture spheroids. Percentage differences from the average values of the 2D and 3D features were measured where Volume and Solidity reached the lowest values (18.6% and 18.8% for the monoculture; 4% and 21.4% for the co-culture) (Fig. 6c). Although the co-culture spheroids showed a particular 4% difference for the direct comparison, the whole dataset showed a 19.6% difference (Supplementary Fig. 2b).
a Heatmap representation of correlations between features. Correlation between the selected features was tested using Spearman’s correlation method. Purple colour represents the monoculture dataset (n = 223 for 2D and n = 110 for 3D) and the orange colour represents the features collected from the co-culture dataset (n = 203 for 2D and n = 114 for 3D). b Boxplot representation of the 2D and 3D features. Thirty-seven spheroids were selected and verified that the 2D measurements correspond to the same objects in 3D. Features such as Volume 3D (µm3), Solidity (−), Sphericity (−), and Extent (−) were extracted from both the 2D and 3D images and used for comparison. Results are displayed as Monoculture 2D (light purple, M-2D), Monoculture 3D (dark purple, M-3D), Co-culture 2D (light orange, C-2D), and Co-culture 3D (dark orange, C-3D). The Kolmogorov-Smirnov test was used. *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001. Each group showed a statistically significant difference (p < 0.001). c Percentage difference from the average values between the 2D and 3D features are shown. Purple colour represents the monoculture dataset, orange colour represents the co-culture dataset (n = 37). d Boxplot representation of the number of nuclei in the monoculture dataset for each experiment (n ≥ 7). Significant differences were not included. Correlation was measured between the number of nuclei, Volume 3D, and Area. e Boxplot representation of the number of nuclei in the co-culture dataset for each experiment (n ≥ 7). Significant differences were not included. Correlation is measured between the number of nuclei, Volume 3D, and Area. Box plots were visualised using Tukey’s method, with medians shown as central lines, boxes representing interquartile ranges, and whiskers extending to 1.5× interquartile range. All scatter plots were generated using a fitted regression line, with shaded bands indicating the standard error of the mean (SEM). Source data are provided as a Source Data file.
To provide additional information relevant to phenotypic screening, we examined nucleus segmentation using the HCS system (Fig. 6d, e). For the monocultures, the average number of nuclei was 358, while the biggest spheroid (585) had 2.76 times more nuclei than the smallest (212 nuclei) (Supplementary Fig. 3). For the co-culture spheroids, the average number of nuclei was 280, and the biggest spheroid (423) had 2.32 times more nuclei than the smallest (185) (Supplementary Fig. 4). While both datasets showed a high positive correlation between the number of cells and the corresponding Volume 3D, only a moderate positive correlation was observed by changing volume to Area (Fig. 6d, e). No correlation was measured for Solidity and the cell number (Supplementary Fig. 2d). Further divergence between the cell number and Area was noted when the percentage difference was calculated for each experiment (Supplementary Fig. 2c). In both cases, co-culture spheroids reached a higher correlation (0.90 for Volume 3D and 0.62 for Area) than monocultures (0.83 for Volume 3D and 0.53 for Area) (Fig. 6d, e).
Next, we selected a spherical and an irregular spheroid from co-cultures as representative cases (Fig. 2j and Fig. 7a). The spherical sample revealed that MRC-5 cells were grouped in the middle of the spheroid and surrounded by the HeLa Kyoto cells (Fig. 8a–c). This structure, a fibroblast core covered by the tumour cells, was more common and spheroids with a more spherical shape showed similar distributions. Meanwhile, the structure of the irregular spheroid had multiple MRC-5 cores and HeLa Kyoto cells were more evenly distributed. The position of the cells within a spheroid proved to be a good indication of the structure, however, since the point of intersection is a characteristic feature due to size and the total number of cells.
a Vertical projection of a spherical and irregular spheroid that displays the number of HeLa Kyoto (purple) and MRC-5 (orange) cells. The centre of each spheroid was defined using whole spheroid segmentation. Starting from the centre position, spheres with increasing radius were used to create shells and count the number of objects in each section. Line plots show the number of cells per class based on the shell analysis. b Spider plot representation of the Spearman’s correlation between 2D and 3D features. Paired spheroids ranked by Solidity 3D (grey, n = 34) are divided into a Spherical (dark blue, n = 18) and Irregular (red, n = 16) groups. c Correlation between 2D and 3D features (n = 34). Previously displayed spherical (dark blue) and irregular (red) spheroid, along with the paired samples are displayed. Outliers (black and red) were not used for the evaluation. All scatter plots were generated using a fitted regression line, with shaded bands indicating the standard error of the mean (SEM). Source data are provided as a Source Data file.
a 3D visualisation of the Spherical and Irregular spheroids where each circle represents a cell displayed based on their position within the spheroid and class (HeLa Kyoto - purple and MRC-5 - orange). Each class is also visualised separately. b Collected 2D, 3D, and Neighbourhood (NH) features for both spheroids are displayed. c 3D representation of the Spherical and Irregular co-culture spheroids. Both top and side views show the nuclei of the HeLa Kyoto (purple) and MRC-5 (orange) cells. For visualisation, a square was placed around the spheroids indicating the side view. The scale bar represents 50 µm for the top and 100 µm for the side view. Leica’s LAS X microscope software was used to display the images.
Among the total spheroid population, 37 spheroids (both for mono- and co-cultures) were identified both in 2D and 3D. This was important to compare the 2D and 3D features measured exactly on the same spheroids. Accordingly, this subpopulation was the one considered to create a ranking using Solidity 3D and to divide the dataset into Spherical and Irregular groups to understand which morphology is more predictable based on the 2D features. Volume, Circularity/Volume 3D, and Solidity showed the highest positive correlations for the spherical group, 0.78, 0.78, and 0.72. Sphericity and Circularity/Sphericity 3D showed a moderate positive correlation (Fig. 7b). In general, the Spherical group always reached a higher correlation than the Irregular group (i.e. amorph spheroids), indicating that the more regular shape resulted in better predictability. Visualising spheroids by their shape (Solidity, Sphericity, Extent), two outliers (one of them is the Irregular spheroid) were distinguished and removed from the analysis (Fig. 7c). However, relying only on the size information, the removed outliers cannot be separated from the other samples. Due to the nature of 3D-oids, comparing hundreds of samples may always show high heterogeneity. Utilising 2D parameters while excluding 3D considerations allows faster and less demanding analysis resulting in overall greater similarity between samples. Perfectly spherical models are ideal for estimating 3D properties (e.g. Volume 2D and Solidity) since other 3D-oids may show irregular shapes that reduce predictability and comparability. For any 2D analysis, using at least 2 non-correlating features, such as size and shape descriptors, potentially helps to remove strong outliers. However, a 2D approach is insufficient to select the most similar spheroids in 3D.
3D single-cell analysis of co-culture spheroid model
A total of 114 HeLa Kyoto/MRC-5 spheroids were analysed at a single-cell level through segmentation and classification using a custom-developed analysis pipeline in BIAS (Fig. 9a–d and Supplementary Movie 4). Results on cell ratio showed that only 25% of the 114 spheroids predominantly consisted of HeLa Kyoto cells (hereafter called HeLa Kyoto Majority: H-M group), while 63% primarily contained MRC-5 cells (hereafter called MRC-5 Majority: M-M group) (Fig. 9a). Only 14 spheroids showed equal numbers of the 2 cell lines (hereafter called E group). The dataset included 32012 segmented objects where 18659 were classified as MRC-5 and 13353 HeLa Kyoto cells. By dividing spheroids based on their class (H-M, E, and M-M groups), higher total cell number was identified for the M-M group (Fig. 9a). Cytoplasm segmentation showed that the tumour cell line has significantly bigger volume, almost double the size of the fibroblast cells (Fig. 9b). Features extracted from the cellular microenvironment (i.e. neighbourhood features) further support the size differences since the distances between the nuclei and the cells are increased in favour of HeLa Kyoto cells. Considering the size difference between the cell lines, we wanted to test whether the different cell ratio changes the morphology of the spheroids. The H-M group showed significantly smaller spheroids and there was no significant difference for Solidity (Fig. 9c and Supplementary Fig. 2e). Although each spheroid is supposed to have a similar cell number, significant differences were measured concerning the number of cells for the different compositions (Supplementary Fig. 2f). Correlation between the total cell number and Volume 3D showed the highest correlation for the E group (Rs: 0.91), followed by the M-M (Rs: 0.86) and H-M (Rs: 0.79) (Fig. 9d). As a final experiment, we were interested in whether examining 2D features alone is enough to select ideal spheroids. The 36 paired spheroid images ranked by Solidity 3D and separated into 2 groups were used for this analysis. We plotted and highlighted Spherical and Irregular spheroids using the highest correlating 2D features (Area, Solidity 2D, and Circularity) for comparison. The scatter plot showed that the 2 groups are indistinguishable and data points cannot be visually separated. Relying on Circularity, spheroids from the Spherical group are outside of the standard deviation (Fig. 10a). Although Solidity 2D showed slightly better separation of the 2 groups compared to Circularity, the data points are intermixed and cannot be visually distinguished.
a Cell ratio for the 114 co-culture spheroids showed 72 spheroids with MRC-5 cells in the majority (orange), 28 spheroids containing a majority of HeLa Kyoto (purple), and 14 spheroids with an equal number (grey). Ratio of classified objects with the exact number are displayed. The 114 spheroids were divided into 3 groups based on the cell type majority (i.e. HeLa Kyoto Majority, Equal, MRC-5 Majority). The size of the doughnut charts and the numbers in their centre represent the average cell numbers for each group. The average number of cells for each class is displayed with orange for HeLa Kyoto and purple for MRC-5. b Comparison of the two cell lines based on Volume 3D (µm3), the average inter-nuclear, and cell distance of the same class in µm. In this experiment, 114 co-culture spheroids were utilised, and average values were derived by aggregating the results across all constituent cells. Each group showed a statistically significant difference (p < 0.001). c Features (Solidity, Volume 3D, number of cells) collected from the spheroids with different cell ratio were displayed based on the 3 groups (HeLa Kyoto majority n = 28, Equal n = 14, MRC-5 majority n = 72). Dark grey colour represents the average value of the whole dataset (n = 114). Statistically significant differences were identified in two cases, with p-values of <0.0023 (**) and <0.001 (***), respectively. d Spearman’s correlation of the total number of cells and Volume 3D were visualised based on HeLa Kyoto majority (purple), MRC-5 majority (orange), and Equal (grey) groups separately. The variation in sample size (n) across groups was a direct outcome of the cell ratio experiment. n = 114. For the statistical analysis, a two-sided non-parametric Kruskal-Wallis test was conducted, followed by Dunn’s multiple comparison test. When comparing only two groups, the two-sided Kolmogorov-Smirnov test was utilised. *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001. Box plots were visualised using Tukey’s method, with medians shown as central lines, boxes representing interquartile ranges, and whiskers extending to 1.5× interquartile range. Source data are provided as a Source Data file.
a 36 spheroids ranked by Solidity 3D, are separated into Spherical (dark blue) and Irregular (red) groups. These spheroids along with the total dataset (grey) were plotted in a scatter plot using only 2D features (Circularity, Solidity, and Area). Each circle represents a spheroid and dashed lines represent the standard deviation. n = 223 and n = 203 for the monoculture and co-culture dataset. Source data are provided as a Source Data file.
Discussion
This study has developed, validated and presented HCS-3DX, a customisable 3D imaging and HCS system for analysing 3D-oids at a single-cell level. This innovative system addresses several challenges associated with 3D HCS, including the pre-selection, handling, imaging, and analysis of 3D-oids.
A critical aspect of this work was developing an HCS plate that can separate and allow fast, automated screening of multiple objects on the same plate, where the position of the samples is predefined. The plate is optimised to overcome time-consuming sample preparation processes. Nevertheless, it is highly adaptable and customisable, allowing modifications to be made within hours to suit individual needs. The plate was developed and tested on an upright light-sheet microscope to achieve single-cell resolution in spheroids. However, any type of 3D cell culture can be screened.
We proved that the designed multiwell plate doubles screening speed and provides high penetration depth, 250 or even 350 µm for optically cleared tumour spheroids28,29 using a Leica TCS SP8 DLS microscope. Since FEP foil allows gas exchange and LSFM offers reduced phototoxicity and increased imaging speed, we found this combination the most ideal for long-term live imaging of the specimen. Furthermore, compatibility with a multi-pipette or a pipetting robot, SpheroidPicker, allows AI-based pre-selection and an automated transfer process (Supplementary Note 2). Several previous publications report that pre-selecting spheroids is crucial in many aspects but for many reasons, reproducibility is the most essential15,18,31,32,33,34.
Our understanding of a 3D-oid depends on the extracted features. Morphological features (e.g. Volume and Sphericity) influence functional endpoints (e.g. viability and necrosis)9. However, we demonstrated that relying only on 2D features limits the analysis, leading to an incomplete representation of the data. We compared features extracted from brightfield (2D) and fluorescence (3D) images of spheroid models to quantify and understand the importance of losing one dimension. Considering image-based features, certain 2D parameters are more important than others when pre-selection is an option. Most publications describe 3D-oids using Diameter, Perimeter, Area, Volume, Circularity, Sphericity, and Convexity, estimated using images acquired at various magnifications32,33,35. While the choice of different objectives depends on many aspects (size, speed, working distance, etc.), we wanted to see whether the variability causes a problem for comparing different studies. Comparing 4 different objectives ranging from 2.5x-20x, the average differences were less than 5%, and significant differences were measured between the 2.5x, the 5x and 10x objectives. Among the tested features, we observed that Perimeter, Convexity, Sphericity, and Circularity showed significant differences. The higher magnification images resulted in more precise contours that follow the edge of the objects accurately, while annotations on images taken with lower magnification objectives were less accurate. Thus, annotating images taken at different magnifications caused discrepancies for Perimeter and for all the features that rely on it (e.g. Circularity, Sphericity, and Convexity). In addition to the objective’s physical parameters (such as magnification, numerical aperture, and spherical aberration), the lower image resolution and the accuracy of the annotator caused the general inconsistency between the objectives.
We suggest that a 10x objective provides a fair compromise for general feature reliability if 20x is unavailable or has an excessively narrow field of view, even though the 2.5x or 5x objectives would not distort the entire analysis. In addition, available AI-based segmentation models can improve the consistency of object detection and reduce time-consuming annotation tasks.
As 3D-oids pre-selection is often recommended, it is important to decide which features are ideal for selection. By discarding one dimension, using Diameter instead of Area, the spheroids are numerically more similar to each other. In addition, Diameter and Circularity showed moderate negative correlations (Rs: −0.69 and −0.54), indicating that smaller spheroids were more circular and more spheroids were found to be ideal. Such correlation is not general since different 3D models may show opposite results36. Plotting spheroids based on Area and Circularity showed no correlation and less spheroids as ideal. Volume estimated using 2D features resulted in slightly smaller values but proved to be more accurate when spheroids were more rounded. Other 2D shape descriptors like Solidity, Sphericity, and Extent showed significantly higher values compared to the measured 3D values. By measuring exactly the same spheroids both in 2D and 3D, Volume, Area/Volume 3D, and Solidity showed the highest correlations (i. e. 0.78, 0.78, and 0.72, respectively). However, the highest correlation is only possible for the most spherical samples, and irregular objects decrease the predictability. Correlation between the number of segmented objects to Volume 3D showed a strong positive correlation (0.83 and 0.90), whilst Area showed weaker correlation (0.53 and 0.62). In conclusion, AI-based nucleus segmentation accurately represents spheroids, whilst estimating the number of cells based on the Area of the sample is not accurate.
Shape and size features showed moderate or no correlation, thus the number of outliers cannot be distinguished using only one feature. Comparing spheroids in 2D and 3D showed that describing the population of spheroids requires at least two parameters relevant to the size and the shape (Supplementary Fig. 5). Whilst pre-selection certainly helps to remove strong outliers, it does not select the most similar spheroids in 3D.
The heterogeneity of spheroids is influenced by the type of cells, the environment, and the formation method, indicating the complexity of generating 3D cell cultures13. To measure the variability of the spheroid models, 3 experts repeated the same experiments 3 times generating 426 monoculture (223) and co-culture (203) spheroids that were analysed in 2D and 3D. Despite using identical equipment and working in the same environment, the comparison of the manual plating revealed great heterogeneity in the size and morphology of spheroids. Furthermore, significant differences were measured by comparing the performance of the various experts. By generating spheroids from a single cell line, we demonstrated that a higher number of replicates and a proper 2D feature quantification combined with pre-selection is recommended.
A particularly insightful evaluation showed a significant difference in size but no difference in Circularity, indicating that spheroid shape was unaffected by size changes. As expected, cell type and ratio had a major impact on spheroid formation. Although co-cultures were seeded with twice as many cells, those spheroids were significantly smaller. First, HeLa Kyoto cells exhibit a significantly higher proliferation rate compared to MRC-5 cells37. Accordingly, when starting with 100 HeLa Kyoto cells and incubating for two days, the resulting spheroids contained an average of 358 cells per spheroid. In contrast, the co-culture spheroids reached an average of only 280 cells per spheroid. Second, MRC-5 cells show a denser arrangement at the central region of the spheroid, resulting in an average intercellular distance of 11.2 ± 0.5 µm, whereas HeLa Kyoto cells show an average distance of 14.2 ± 1.1 µm.
Thus, the cell type and ratio have a significant effect on spheroid formation. Interestingly, co-culture spheroids showed a higher positive correlation for the size and number of cells than monocultures, even though the 2 different cell types showed unequal sizes in spheroids.
Increasing the complexity of the biological model introduces more bias into the experiment, potentially leading to incorrect conclusions. Apart from size, shape, and number of cells, the cell ratio and structural information are also needed to describe a multi-cell line spheroid. For our co-culture model, we selected the 1:4 ratio for the HeLa Kyoto and MRC-5 spheroids based on literature data reporting increased stroma content are often associated with poorer patient outcomes38. Achieving single-cell resolution and applying 3D analysis (including segmentation, classification, and feature extraction), 114 co-culture spheroids were analysed and compared at a cellular level. After evaluating the individual cells composing the spheroids, we observed that spheroids can be categorised into 3 groups according to the various cell ratios and total cell numbers. Although the groups showed significant differences in size, there was no inequality in shape. The total number of cells revealed that spheroids with more cells contained more MRC-5 cells than HeLa Kyoto.
In this large-scale, single-cell analysis experiment, we demonstrated that complex models relying on interactions between various cell types result in high variety. However, the consistency of morphologically similar spheroids also varies with the expert conducting the experiment. Interestingly, co-culture models generated with identical protocols showed varying cell type dominance; 63%, 25% and 12% of spheroids showed the majority of MRC-5 cells, the majority of HeLa cells, or equal cell numbers, respectively. This phenomenon can be explained by the cumulative effect of a series of technical factors and also by the influence of the tumour-stroma interactions. Delinasios et al., for example, described that interactions between proliferating fibroblasts and HeLa cells in vitro drastically influences each other’s growth pattern, and survival depending on the fibroblast/HeLa ratio, cell topology (cell-cell contact and the architectural pattern developed during co-culture) and frequent medium change37.
Pre-selection methods for 2D spheroids did not bring the promised impact in this field, however it is still essential to reduce the model heterogeneity and the high variability resulting from the generation process35. It should be pointed out that 2D pre-selection applied to more complex models may eliminate one specific type of spheroid with different cell ratio rather than selecting the most similar ones. In addition, selecting the spheroids using a single 2D image leads to risk of misclassification or data misinterpretation in case of irregular spheroids. Therefore, 3D data and structural differences between 3D-oid models emphasises the need for a single-cell analysis pipeline to avoid incorrect data interpretations and false conclusions.
Over the years, only a few methods were published for 3D HCS. Widefield and confocal fluorescence systems are more frequently applied since such systems are compatible with standard plate formats, thus more commonly used for screening. Widefield fluorescence imaging offers high screening speed and compatibility with microfluidic systems, but it has low penetration depth and only allows limited feature extraction39,40. On the other hand, confocal fluorescence microscopy systems have subcellular resolution but with a much slower image acquisition and higher phototoxicity12,41,42,43. In addition, due to the limited light penetration, HCS methods are usually demonstrated on smaller 3D-oids using standard but more expensive imaging plates44.
HCS systems designed for LSFM overcome most of the disadvantages offering single-cell resolution images with fast image acquisition, however, complex sample preparation and the usage of special plates is required24,45,46,47. Furthermore, such systems are usually specific to the imaging setup resulting in limitations (i. e. size or type specific imaging plate, manufacturing price and protocol of the plate, modifications of the microscope, availability and complexity of code).
Working with 3D-oids has resulted in significant variability, thus the developments aimed at evaluating the samples must also be flexible. Imaging and image analysis systems are needed that ensure the interchangeability of individual components, transparency in data reporting, and accessibility of the code. A comprehensive method review is presented in Supplementary Table 2 and 6.
Biological limitations of this study include the reduced relevance of the spheroid models. To more accurately mimic the tumour microenvironment, future models should incorporate multicellular spheroids48 and include extracellular matrix components to better reflect in vivo conditions. Also, the spheroids were imaged consistently at 48 h but only at a single time point. As a result, dynamic morphological changes over time were not monitored or analysed.
In this work, we developed HCS-3DX, a versatile HCS system which comprises all the steps needed to evaluate 3D cell cultures from the beginning to the end at a single-cell level. By integrating AI-driven solutions and advanced imaging techniques such as LSFM, HCS-3DX enables precise, automated, and high-throughput analyses at a single-cell level. Accordingly, it represents a substantial leap forward in 3D HCS, offering a comprehensive solution that bridges the gap between traditional 2D models and complex in vivo environments. Its application will enhance the accuracy and efficiency of drug screening processes, support personalised medicine approaches, and facilitate more detailed investigations into cellular behaviour within 3D structures. In conclusion, its robustness and versatility make HCS-3DX a valuable tool for researchers and clinicians, paving the way for future innovations in biomedical research.
Methods
Spheroid models
Spheroid monocultures were generated using HeLa Kyoto EGFP-alpha-tubulin/H2B-mCherry cervical cancer cells (Cell Lines Service; CLS-300670, Eppelheim, Germany). Cells were maintained in a HeLa Kyoto medium consisting of DMEM (Lonza, Basel, Switzerland), 10% Fetal Bovine Serum (FBS, Euroclone, Milan, Italy), 2 mM L-glutamine (Lonza), 0.5 mg/ml G418 (Gibco, Montana, United States), and 0.5 µg/ml puromycin (Sigma-Aldrich Chemie GmbH, Steinheim, Germany). To generate uniform spheroids, 100 cells were seeded into each well in U-bottom cell-repellent 384-well plates (Greiner Bio-One, Kremsmünster, Austria) for 48 h at 37 °C and 5% CO2. After 48 h, spheroids were collected and then washed 3 times with Dulbecco’s Phosphate Buffered Saline (DPBS), and fixed with 4% Paraformaldehyde (PFA) for 60 min. Spheroids were washed with DPBS 3 times and stored at 4 °C in DPBS until imaging. Before imaging, spheroids were incubated in 0.1% Triton X-100 overnight at room temperature and washed 3 times with DPBS. For actin labelling, spheroids were stained with 1:200 Flash Phalloidin NIR 647 (Biolegend, San Diego, California) for 60 min. Before the imaging, spheroids were washed with DPBS 3 times. Monoculture experiments were independently conducted 3 times by 3 separate experts (n = 223). T-47D spheroids used for the image quality test were generated according to previous publication28. The T47-D human breast cancer cell line (ATCC, HTB-133, Lot: 63542749) was cultured following the recommended ATCC protocols. Multicellular spheroids were generated using the SphericalPlate 5D system (Kugelmeiers Ltd., Erlenbach, Switzerland) in accordance with the manufacturer’s instructions, with 750 cells seeded per spheroid. The incubation time was optimised for 7 days, with the culture medium refreshed every other day. Upon maturation, spheroids were washed twice with DPBS, fixed in 4% PFA for one hour at room temperature, and subsequently washed twice with DPBS before storage in DPBS at 4 °C. Uniform spheroids based on size and morphology were manually selected and stained with DRAQ5 (ThermoFisher, USA) to label cell nuclei.
For the co-culture models, the 1:4 HeLa Kyoto and MRC-5 ratio with a total number of 200 cells per well was selected based on previous optimisations. Co-culture spheroids were generated using the same HeLa Kyoto cells and MRC-5 fibroblasts (ATCC, CCL-171, Lot: 63405646). The manufacturer’s instructions were followed for the maintenance of the cell cultures. To generate spheroids, a co-culture medium consisting of DMEM, 10% FBS, 1% L-glutamine (2 mM), and 1% Penicillin-Streptomycin-Amphotericin B mixture (all from Lonza) was used. 40 HeLa Kyoto cells were seeded into each well in U-bottom cell-repellent 384-well plates at 37 °C and 5% CO2. After 24 h of incubation, 160 MRC-5 cells per well were added onto the HeLa Kyoto cells and the co-cultures were incubated for one more day. After 24 h, the co-culture spheroids were collected from each well and washed 3 times with DPBS. 4% PFA was used for 60 min to fix the samples, then washed again with DPBS 3 times and stored at 4 °C in DPBS until imaging. Before imaging, spheroids were incubated in 0.1% Triton X-100 and 1 µg/ml DAPI overnight at room temperature and washed 3 times with DPBS. Spheroids were stained with 1:200 Flash Phalloidin NIR 647 for 60 min. Finally, spheroids were washed with DPBS 3 times. Co-culture experiments were independently conducted 3 times by 3 separate experts (n = 203).
Hydrogel-based multicellular human tumour models were generated by co-culturing CellTracker Orange CMTMR (Invitrogen)-stained T-47D ductal carcinoma and stroma cells, i.e. CellTracker Deep Red-stained MRC-5 fibroblasts, and CellTracker Green CMFDA-stained EA.hy926 endothelial cells in hydrogel matrix (TrueGel3D, Sigma-Aldrich) directly in the HCS plate.
HCS foil multiwell plate
A customisable 3D imaging multiwell plate was designed for the screening of 3D-oids at a single-cell level. The plate includes: (I) a 3D printed base element that retains the detection fluid; (II) a FEP foil for separating the position of the samples; (III) an insert element to secure the samples’ position within the foil; and (IV) a 3D printed grid element to fix the FEP foil position (Fig. 2d–i). This designed plate is suitable for examining a large number of samples with an LSFM, with a single sample in each cuvette.
FEP foil
FEP foils are fully transparent, with a refractive index of 1.341–1.347. FEP is chemically inert and resistant to organic solvents, acids, and bases, similar to the majority of fluorocarbon plastics. Additionally, the material satisfies FDA (21CFR.177.1550) and EU (2002/72/EC) requirements20. FEP-films (Holscot Europe, Netherlands) with a 50 µm thickness were cut into 15 × 15 cm pieces and cleaned with 70% alcohol. Each foil was placed and clamped into the frame of a JT-18 vacuum-forming machine (Yuyao Jintai Machine Company, China). To get small and uniform shapes, the heater needs to raise the temperature to a level near the glass transition temperature of the FEP-foil (260–280 °C). Once the heater reaches the desired temperature, the positive mould is quickly placed onto the vacuum-forming machine, and the foil is quickly pressed onto the mould whilst the vacuum suction is switched on. The FEP foil that has been extruded is then carefully taken out of the mould and cleaned with an isopropyl alcohol bath for 5 min to remove the excess resin components.
Insert element
To fix the samples’ position inside the cuvettes, hydrogels or a 3D-printed insert element are suitable (Supplementary Fig. 6a). While hydrogels are optimal for smaller samples, the insert element offers an alternative solution to secure the position within the foil. The insert element contains the same pyramid-like structure as the foil, however, there is no sphere at the top. Inserting the element inside the cuvettes secures all samples in the spheres without deforming or harming the cells. To elevate the samples from the bottom to the same z-positions, the FEP foil with hydrogel or insert element should be turned over and placed inside of the base element. To provide precise positioning and a locking mechanism of the cuvettes inside of the base, the insert contains 4 magnets that attach to the 4 other magnets on the base.
Base element
The base components of the plate were printed with the Prusa i3 MK3S + 3D printer with PETG (3DP-PETG1.75-01-BK; Gembird, Shenzhen, China) in two different size versions (Supplementary Fig. 6b) with the corresponding grid elements (Supplementary Fig. 6c, d). The bigger version is 85 / 127 / 15.4 mm (average size of a 96-well plate) which is suitable for screening 56 samples, while the smaller version 85 / 73.8 / 15.4 mm was designed only for 28 samples. To create a waterproof base for the samples, the 24 / 64 mm coverslips (Menzel Gläser, Germany) were secured with silicone grease (ThermoFisher, USA) to the bottom of the bases. The cover glass provides a transparent bottom for any inverted microscopy setup while allowing the freedom of using dipping objectives. Models were designed in Blender and STL files (stereolithography files) were exported to PrusaSlicer v 2.7.1. All models were 3D printed with the default Prusa PETG filament profile and with “0.4 Quality print settings” using a 0.4 nozzle.
Grid element
To secure the foil’s position in the plate, we printed a secure element that allows to fix the position of the FEP foil within the base (Supplementary Fig. 6c, d). This compartment was printed with the Prusa i3 MK3S+ printer using PETG. The grid element can be slid into predefined spaces, securing the foil to the bottom of the plate.
Positive mould
A positive mould for the spikes was created using Blender 3.0. Each mould was designed to fit perfectly into the base of the plate (Supplementary Fig. 7a). For printing, the 3D models were exported to a stereolithographic file format (.stl) and then imported into PrusaSlicer v 2.7.1 (Prusa Research, Czech Republic). Each heat-resistant mould was printed with the Prusa SL1 printer, which is a stereolithography (SLA) resin-based 3D printer that forms objects layer by layer using a liquid resin that is ultraviolet (UV)-cured. For printing, the DruckWege Type D High Temp resin (TDH-VIO-500, Groningen, The Netherlands) was used. This printer provides high-resolution printing capabilities enabling the production of intricate and detailed objects with smooth surface finishes. The mechanical and thermal characteristics of heat-resistant resin enable it to endure the vacuum-forming process. The positive moulds are UV-cured and washed with isopropanol to remove excess resin from the surface. Then the moulds were examined under a stereomicroscope and cleaned by submerging them in an ultrasonic bath before being used.
Assembly of the HCS Plate
First, biological samples should be placed precisely into the cuvettes of the FEP foil. An additional 3D printed element which is not part of the HCS plate can be used to prevent the movement of the foil while transferring the samples (Supplementary Fig. 7c). Prefilling the cuvettes of the foil with liquid before sample transfer can help remove air bubbles. The sample transfer process can be done manually, using a single or multi-channel pipette (8 channels) or a pipetting robot. After pipetting, a visual check is necessary because all the samples should sink to the bottom of the cuvette and positioned in the middle. If a sample is not in the middle of the cuvette, a gentle shake of the foil or pipetting the samples to the bottom may resolve this issue. If all the samples reach the bottom of the cuvettes of the FEP foil and there are no air bubbles next to the samples, then the position of the samples should be secured. Removing excess mounting media from the cuvettes allows the user to pipette a low melting point agarose and thus fix the position of the 3D sample inside the foil. The alternative solution is to gently push the insert element into the FEP foil until each cuvette is secured. By utilising either method, samples should be ready to be placed into the base element by turning over the whole FEP foil containing all the samples. At this point, all the samples should be at the top of the cuvettes covered by the transparent foil. Next, the FEP foil with the samples should be placed into the base element, where each cuvette is positioned within the coverslip. After checking the alignment of the cuvettes, the user must carefully fill the plate with the detection solution by removing all the air bubbles under the FEP foil. Before filling up completely, the grid element should be placed on top of the FEP foil to stabilise and prevent additional movements of the samples. Finally, before inserting the base into the microscope, the plate with the detection solution must be carefully filled to proceed with the calibration process (Supplementary Fig. 7b).
Microscopes
For brightfield imaging, the fixed monoculture and co-culture spheroids were placed into a 35/10 mm cell culture dish with a glass bottom (627965, Cellview, Austria) in PBS. Brightfield images were taken with the Leica TCS SP8 using 4 different objectives: 2.5x/0.07, 5x/0.15, 10x/0.32, and 20x/0.4 (Supplementary Table 4).
The HCS foil multiwell plate was validated on the Leica TCS SP8 Digital LightSheet (DLS) microscope, exploiting a standard 96-well plate insert. A 25x/0.95 detection objective with a mounted 2.5 mm mirror device was used to capture fluorescence DLS images, illuminated by the 5x/0.15 objective. Images with a resolution of 2048 × 2048 pixels and a pixel size of 0.144 µm were captured with the sCMOS DFC9000 Leica camera. A 2 µm gap distance between the images was used in each z-stack. dH2O mounting medium was used for every spheroid. A laser with a wavelength of 638 nm and an exposure time of 200 ms with 20% laser intensity (maximum laser intensity 350 mW) was used. Laser intensity was manually adjusted for each channel (i. e. 405, 488, 552, and 638 nm wavelength).
Image analysis
All 2D and 3D raw images were saved as an *.xlef file format using the LAS X software (Leica, Germany), then images were directly imported into the BIAS software (Single-Cell Technologies Ltd., Szeged, Hungary). Image quality of the HCS multiwell plate was evaluated using intensity variance, a metric implemented in Spheroid Quality Measurement (SQM), an open-source ImageJ/Fiji plugin29. Due to the nature of the data, different workflows were created for the 2D and 3D datasets (Supplementary Fig. 9). Therefore, these were handled separately during the analysis.
2D workflow
The AnaSP49 software was used to manually annotate brightfield images in order to obtain the ReViSP27 based volume features (e.g. Volume 2D) and binary masks. The original images with their corresponding masks were imported into BIAS software26. The import process is the only manual task that is required in the workflow, the rest of the pipeline can be executed automatically (Supplementary Fig. 9a). To import and visualise the segmentation results, the Segmentation module was used. Next step is the List creator, an intermediate module that links results of different segmentations into a list of meta-objects that serve as an input for the future processing steps in the pipeline, such as the Feature Extraction module that extracts various features, e.g. intensity, shape, size of these meta-objects (spheroid in this case). Using the Exporters module, all features were exported as a *.csv file.
3D workflow
For the 3D dataset, 2D U-Net50 and a StarDist51 models were used to segment the whole spheroid (using only the grey channel - actin) and nuclei (using only the blue channel - DAPI) in 3D using the Execute module (Supplementary Note 1 and Supplementary Fig. 9b). This module allows the execution of third-party applications or scripts as a part of a BIAS pipeline. Generated masks were visualised in the 3D segmentation module. Mask operator/exclude function was used to discard all objects outside of the spheroid’s volume and keep objects on the border and within the spheroid. In case of the nuclei mask, the Mask operator/dilate function with a 4 µm dilatation was utilised to increase the volume and to collect cell information. Object linker module has a similar functionality as the List Creator and it was used to create the meta-objects with the corresponding spheroid, cell, and nuclei information and the generated output was used to collect features with the Feature extraction 3D. By using a supervised machine learning algorithm from the Machine learning classification module, cells of the co-culture spheroids were classified either as a HeLa Kyoto or an MRC-5. To distinguish the cells’ type, the intensity channels 2 and 3 (green - EGFP-alpha-tubulin; red - H2B-mCherry) were used to create a training dataset with 750 classified objects in each category. A Multi-Layer Perceptron (MLP) classifier was used to classify all the cells with a combined average accuracy of 98.59% according to a K-Fold cross-validation (K=10) (where classes reached 98.40% for the HeLa Kyoto and 98.78% for the MRC-5 classes). Using the Statistics module, segmented objects with a particularly small size and missing features were removed from further analysis (NUCLEUS CONTOUR STAT MEAN > ‘20’ and CELL CONTOUR STAT MEAN > ‘30’). To measure the closest distance between cells, Neighbourhood features/K-neighbours ‘1’ was used for both classes separately. Finally, all features were exported as a *.csv file using the Exporters module.
Statistical analysis
Statistical analyses were conducted using GraphPad Prism 8 software. Box plots were visualised using Tukey’s method, with medians shown as central lines, boxes representing interquartile ranges, and whiskers extending to 1.5× interquartile range. The Kolmogorov-Smirnov test was applied to assess normal distribution. For analysing the results of the 2D features, the two-sided non-parametric Kruskal-Wallis test followed by Dunn’s multiple comparisons was used. The significance level was set at α = 0.05 with a 95% confidence interval, and p-values were adjusted for multiple comparisons.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. For further details and data referenced in this article, please visit the links provided below: Monoculture and co-culture spheroid image dataset: https://doi.org/10.6084/m9.figshare.c.7658858.v1. Optically cleared spheroids: https://doi.org/10.6084/m9.figshare.c.5051999.v1. HCS of tumour-stroma spheroid multicultures: https://doi.org/10.6084/m9.figshare.c.7357135. Annotated 3D image dataset: https://doi.org/10.6084/m9.figshare.c.7020531.v1. Source data are provided with this paper.
Code availability
SpheroidPicker: Source code52: https://doi.org/10.5281/zenodo.16536353. The model files (3D printed elements)53: https://doi.org/10.5281/zenodo.14679243. Original and the improved segmentation models54: https://doi.org/10.5281/zenodo.14675683. Annotated dataset55: https://doi.org/10.5281/zenodo.14679303BIAS software: https://www.sct.bio/custom-bias-links. Additional data related to this paper may be requested from the corresponding author.
References
Duval, K. et al. Modeling physiological events in 2D vs. 3D cell culture. Physiol. (Bethesda) 32, 266–277 (2017).
Carragher, N. et al. Concerns, challenges and promises of high-content analysis of 3D cellular models. Nat. Rev. Drug Discov. 17, 606 (2018).
Ascheid, D. et al. A vascularized breast cancer spheroid platform for the ranked evaluation of tumor microenvironment-targeted drugs by light sheet fluorescence microscopy. Nat. Commun. 15, 3599 (2024).
Li, L., Zhou, Q., Voss, T. C., Quick, K. L. & LaBarbera, D. V. High-throughput imaging: Focusing in on drug discovery in 3D. Methods 96, 97–102 (2016).
Sawant-Basak, A. & Obach, R. S. Emerging models of drug metabolism, transporters, and toxicity. Drug Metab. Dispos. 46, 1556–1561 (2018).
Sant, S. & Johnston, P. A. The production of 3D tumor spheroids for cancer drug discovery. Drug Discov. Today Technol. 23, 27–36 (2017).
Vogt, N. Assembloids. Nat. Methods 18, 27 (2021).
In Vitro three-dimensional (3D) cell culture tools for spheroid and organoid models. SLAS Discovery 28, 119–137 (2023).
Zanoni, M. et al. 3D tumor spheroid models for in vitro therapeutic screening: a systematic approach to enhance the biological relevance of data obtained. Sci. Rep. 6, 19103 (2016).
Huang, B.-W. & Gao, J.-Q. Application of 3D cultured multicellular spheroid tumor models in tumor-targeted drug delivery system research. J. Control. Release 270, 246–259 (2018).
LeSavage, B. L., Suhar, R. A., Broguiere, N., Lutolf, M. P. & Heilshorn, S. C. Next-generation cancer organoids. Nat. Mater. 21, 143–159 (2022).
Mysior, M. M. & Simpson, J. C. An automated high-content screening and assay platform for the analysis of spheroids at subcellular resolution. PLoS One 19, e0311963 (2024).
Peirsman, A. et al. MISpheroID: a knowledgebase and transparency tool for minimum information in spheroid identity. Nat. Methods 18, 1294–1303 (2021).
Chalkley, A. S., Mysior, M. M. & Simpson, J. C. A robust method for the large-scale production of spheroids for high-content screening and analysis applications. J. Vis. Exp. https://doi.org/10.3791/63436 (2021).
Wang, Y. & Jeon, H. 3D cell cultures toward quantitative high-throughput drug screening. Trends Pharmacol. Sci. 43, 569–581 (2022).
Chen, Z. et al. Automated evaluation of tumor spheroid behavior in 3D culture using deep learning-based recognition. Biomaterials 272, 120770 (2021).
Akshay, A. et al. SpheroScan: a user-friendly deep learning tool for spheroid image analysis. Gigascience 12, (2022).
Grexa, I. et al. SpheroidPicker for automated 3D cell culture manipulation using deep learning. Sci. Rep. 11, 14813 (2021).
Swinney, D. C. & Anthony, J. How were new medicines discovered?. Nat. Rev. Drug Discov. 10, 507–519 (2011).
Hötte, K. et al. Ultra-thin fluorocarbon foils optimise multiscale imaging of three-dimensional native and optically cleared specimens. Sci. Rep. 9, 17292 (2019).
Stelzer, E. H. K. et al. Light sheet fluorescence microscopy. Nat. Rev. Methods Primers 1, (2021).
Ruan, X. et al. Image processing tools for petabyte-scale light sheet microscopy data. Nat. Methods https://doi.org/10.1038/s41592-024-02475-4 (2024).
Piccinini, F. et al. Software tools for 3D nuclei segmentation and quantitative analysis in multicellular aggregates. Comput. Struct. Biotechnol. J. 18, 1287–1300 (2020).
de Medeiros, G. et al. Multiscale light-sheet organoid imaging framework. Nat. Commun. 13, 4864 (2022).
Piccinini, F., Tesei, A., Zanoni, M. & Bevilacqua, A. ReViMS: Software tool for estimating the volumes of 3-D multicellular spheroids imaged using a light sheet fluorescence microscope. Biotechniques 63, 227–229 (2017).
Mund, A. et al. Deep Visual Proteomics defines single-cell identity and heterogeneity. Nat. Biotechnol. 40, 1231–1240 (2022).
Piccinini, F., Tesei, A., Arienti, C. & Bevilacqua, A. Cancer multicellular spheroids: volume assessment from a single 2D projection. Comput. Methods Prog. Biomed. 118, 95–106 (2015).
Diosdi, A. et al. Cell lines and clearing approaches: a single-cell level 3D light-sheet fluorescence microscopy dataset of multicellular spheroids. Data Brief. 36, 107090 (2021).
Diosdi, A. et al. A quantitative metric for the comparative evaluation of optical clearing protocols for 3D multicellular spheroids. Comput. Struct. Biotechnol. J. 19, 1233–1243 (2021).
Blondeel, E. et al. Sequential orthogonal assays for longitudinal and endpoint characterization of three-dimensional spheroids. Nat. Protoc. https://doi.org/10.1038/s41596-025-01150-y (2025).
Zieger, V. et al. Towards automation in 3D cell culture: Selective and gentle high-throughput handling of spheroids and organoids via novel pick-flow-drop principle. Adv. Healthc. Mater. 13, 2303350 (2024).
Shahbazi, A. S. et al. A multi-stage weakly supervised design for spheroid segmentation to explore mesenchymal stem cell differentiation dynamics. BMC Bioinforma. 26, 20 (2025).
Sampaio da Silva, C. et al. High-throughput platform for label-free sorting of 3D spheroids using deep learning. Front. Bioeng. Biotechnol. 12, 1432737 (2024).
SpAi: A Machine-learning Supported Experimental Workflow for high-throughput spheroid production and analysis. Biosensors and bioelectronics: X 100588 (2025).
Pong, K. C. C. et al. Automated uniform spheroid generation platform for high throughput drug screening process. Biosensors 14, 392 (2024).
Hwang, H. J., Oh, M.-S., Lee, D. W. & Kuh, H.-J. Multiplex quantitative analysis of stroma-mediated cancer cell invasion, matrix remodeling, and drug response in a 3D co-culture model of pancreatic tumor spheroids and stellate cells. J. Exp. Clin. Cancer Res. 38, 258 (2019).
Delinasios, J. G. et al. Proliferating fibroblasts and HeLa cells co-cultured in vitro reciprocally influence growth patterns, protein expression, chromatin features and cell survival. Anticancer Res 35, 1881–1916 (2015).
Li, X. et al. Intravoxel incoherent motion MR imaging of early cervical carcinoma: correlation between imaging parameters and tumor-stroma ratio. Eur. Radiol. 28, 1875–1883 (2018).
Mittler, F. et al. High-Content Monitoring of Drug Effects in a 3D Spheroid Model. Front Oncol. 7, 293 (2017).
Lee, S.-Y., Doh, I., Nam, D.-H. & Lee, D. W. 3D Cell-Based High-Content Screening (HCS) Using a Micropillar and Microwell Chip Platform. Anal. Chem. 90, 8354–8361 (2018).
Miniaturized and multiplexed high-content screening of drug and immune sensitivity in a multichambered microwell chip. Cell Rep. Methods 2, 100256 (2022).
Cutrona, M. B. & Simpson, J. C. A high-throughput automated confocal microscopy platform for quantitative phenotyping of nanoparticle uptake and transport in spheroids. Small 15, e1902033 (2019).
Muckom, R. et al. High-throughput 3D screening for differentiation of hPSC-derived cell therapy candidates. Sci. Adv. 6, eaaz1457 (2020).
Development of an automated 3D high content cell screening platform for organoid phenotyping. SLAS Discovery 29, 100182 (2024).
Eismann, B. et al. Automated 3D light-sheet screening with high spatiotemporal resolution reveals mitotic phenotypes. J. Cell Sci. 133, (2020).
Beghin, A. et al. Automated high-speed 3D imaging of organoid cultures with multi-scale phenotypic quantification. Nat. Methods 19, 881–892 (2022).
De Beuckeleer, S. et al. An agarose fluidic chip for high-throughput organoid imaging. Lab Chip 25, 235–252 (2025).
Diosdi, A. et al. Single-cell light-sheet fluorescence 3D images of tumour-stroma spheroid multicultures. Sci. Data 12, 492 (2025).
Piccinini, F. AnaSP: a software suite for automatic image analysis of multicellular spheroids. Comput. Methods Prog. Biomed. 119, 43–52 (2015).
Ronneberger, O., Fischer, P. & Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015 234–241 (2015).
Weigert, M., Schmidt, U., Haase, R., Sugawara, K., & Myers, G. Star-convex polyhedra for 3D object detection and segmentation in microscopy. In Proceedings of the IEEE/CVF winter conference on applications of computer vision, 3666–3673 (2020).
Grexa, I. HCS-3DX: a next-generation AI-driven automated 3D-oid high-content screening system. SpheroidPicker: v1. 0. 0-beta Zenodo. https://doi.org/10.5281/zenodo.16536353 (2025).
Grexa, I., Diosdi, A. & Horvath, P. HCS-3DX: a next-generation AI-driven automated 3D-oid high-content screening system. SpheroidPicker parts Zenodo. https://doi.org/10.5281/zenodo.14679243 (2025).
Grexa, I., Horvath, P. & Diosdi, A. HCS-3DX: a next-generation AI-driven automated 3D-oid high-content screening system. SpheroidPicker segmentation models. Zenodo https://doi.org/10.5281/zenodo.14675683 (2025).
Grexa, I., Horvath, P. & Diosdi, A. HCS-3DX: a next-generation AI-driven automated 3D-oid high-content screening system. 2D spheroid dataset [Data set]. Zenodo https://doi.org/10.5281/zenodo.14679303 (2025).
Acknowledgements
The authors would like to thank Lilla Pintér (HUN-REN BRC, Szeged, Hungary) for her technical support. A.D., T.T., I.G., B.S., N.H., and P.H. acknowledge generous support from the Lendület BIOMAG grant (no. 2018–342), TKP2021-EGA09, HUNRENTECH (TECH-2024-34), Horizon-BIALYMPH, Horizon-SYMMETRY, Horizon-SWEEPICS, H2020-Fair-CHARM, HAS-NAP3, TKCS-2024/73; PerMel-AI (EPPERMED2024−590), the HUNTER-Excellence 2024, and grant from OTKA-SNN no. 139455/ARRS and NKKP (ADVANCED_24_151202). P.H. gratefully acknowledge support from the FIMM High Content Imaging and Analysis Unit (FIMM-HCA; HiLIFE-HELMI), and Finnish Cancer Society. F.Pi. acknowledges support from the MAECI Science and Technology Cooperation Italy-South Korea Grant Years 2023–2025 by the Italian Ministry of Foreign Affairs and International Cooperation (CUP project: J53C23000300003); M.H. from the János Bolyai Research Scholarship of the Hungarian Academy of Sciences.
Author information
Authors and Affiliations
Contributions
A.D., M.H., F.Pi., and P.H. contributed to the conceptualization of the study. A.D., T.T., I.G., B.S., and M.H. were responsible for developing the methodology. F.K. and A.K. provided software support. A.D., T.T., I.G., and N.H. conducted validation. A.D., M.H., F.Pi., and P.H. performed the formal analysis. A.D., T.T., I.G., B.S., N.H., and F.Pa. carried out the investigation. A.D., I.G., F.K., A.K., M.H., K.B., F.Pi., and P.H. contributed essential resources. A.D., T.T., I.G., B.S., and M.H. handled data curation. A.D., M.H., and F.Pi. were responsible for drafting the original manuscript. T.T., I.G., B.S., N.H., F.K., A.K., K.B., F.Pa., and P.H. reviewed and edited the manuscript. A.D. and B.S. created the visualisations. K.B., F.Pa., and P.H. supervised the research. A.D., K.B., F.Pi., and P.H. managed project administration. K.B., F.Pi., and P.H. secured funding. All authors reviewed and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
Akos Diosdi and Peter Horvath declare a competing interest. The present technical solution is disclosed by AD and PH in detail in International Publication Pamphlet No. WO2023/242604 A1 (published on 21 December 2023); national phase patent application nos. US 18/875,683 and EP 23758709.2 are pending before the United States Patent and Trademark Office (USPTO) and the European Patent Office (EPO), respectively. Any use of the publication document or the technical solution disclosed therein requires prior written authorization or approval from the applicants. Akos Diosdi, Istvan Grexa, Ferenc Kovacs, Andras Kriston, and Peter Horvath are employed by Single-Cell Technologies Ltd., Szeged, Hungary, which is developing the BIAS (Biology Image Analysis Software) software. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interests.
Peer review
Peer review information
Nature Communications thanks Ali Hashemi Gheinani and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Diosdi, A., Toth, T., Harmati, M. et al. HCS-3DX, a next-generation AI-driven automated 3D-oid high-content screening system. Nat Commun 16, 8897 (2025). https://doi.org/10.1038/s41467-025-63955-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-63955-5