Abstract
Single nucleotide polymorphisms adjacent to the α-amino-β-carboxymuconate-ε-semialdehyde decarboxylase (ACMSD) gene have been associated with Parkinson’s disease (PD) in genome-wide association studies (GWAS). However, its biological validation as a PD risk gene has been hampered by the lack of available models. Using CRISPR/Cas9, we generated a zebrafish model of acmsd deficiency with marked increase in quinolinic acid. Despite this, acmsd-/- zebrafish were viable, fertile, morphologically normal and demonstrated no abnormalities in spontaneous movement. In contrast to the postulated pro-immune pathomechanism linking ACMSD to PD, microglial cells and expression of the proinflammatory cytokines cxcl8, il-1β, and mmp9 were similar between acmsd-/- and controls. The number of ascending dopaminergic neurons, and their susceptibility to MPP+, was also indistinguishable. An upregulation of kynurenine aminotransferase activity was identified in acmsd-/- zebrafish which may explain the absence of neurodegenerative phenotypes. Our study highlights the importance of biological validation for putative GWAS hits in suitable model systems.
Similar content being viewed by others
Introduction
Parkinson’s disease (PD) is a common, progressive neurodegenerative disorder with a loss of dopaminergic neurons in the substantia nigra as its pathological hallmark1. Accumulating evidence suggests an important role for neuroinflammation in PD pathogenesis2. Genome-wide association studies (GWAS) have identified over 90 genetic risk variants for sporadic PD3, with many single nucleotide polymorphisms (SNPs) occurring in or nearby genes expressed in immune cells or otherwise linked to inflammation.
α-amino-β-carboxymuconate-ε-semialdehyde decarboxylase (ACMSD) lies under a GWAS peak on chromosome 24, suggesting a possible role for ACMSD in the pathogenesis of PD. At least four SNPs in close proximity to ACMSD have demonstrated a significant association with PD, although this appears to vary by population4,5,6,7,8,9,10,11. The gene expression database GTEx Portal (gtexportal.org) suggests that ACMSD expression occurs primarily in the liver and kidneys in humans. However, expression has also been demonstrated at lower levels in the brain12. ACMSD enzymatically converts α-amino-β-carboxymuconate-ε-semialdehyde (ACMS) to α-aminomuconate semialdehyde (AMS) at a critical branching point in the catabolism of tryptophan via the kynurenine pathway (Fig. 1). In the presence of ACMSD, the kynurenine pathway terminates with the production of a neuroprotective, anti-inflammatory molecule named picolinic acid (PIC). However, in the absence of ACMSD, ACMS will dehydrate to form quinolinic acid (QUIN), a potent neurotoxin. Its neurotoxic ability is conferred via a number of mechanisms, in particular excitotoxicity via the direct activation of NMDA receptors13,14 and by increasing the release, and inhibiting the reuptake and degradation, of glutamate in synaptic regions15,16,17,18. QUIN also induces neuroinflammation. Whilst microglia and macrophages are the primary source of QUIN in the CNS, they also respond to pathological levels. QUIN directly activates astrocytes and microglia, resulting in NF-κB pathway activation and increased secretion of proinflammatory mediators, such as IL-1β, TNF-α, and COX-2, which in turn can induce neuronal cell death19,20,21,22,23. Pharmacological inhibition of the NMDA receptor is unable to prevent this activation, suggesting that it occurs via an NMDA receptor-independent mechanism24. QUIN also generates a local proinflammatory environment by enhancing oxidative stress in microglia, astrocytes and neurons19,25,26,27,28. Interestingly, the inhibition of microglial activation reduces QUIN-induced neuronal toxicity23. These data suggest that the inflammatory effect of QUIN plays an important role in QUIN-induced neurotoxicity, as opposed to direct NMDA receptor activation alone. QUIN may also have other, less well studied toxic effects including mitochondrial dysfunction19, cytoskeletal disruptions29, and disruption of the blood-brain barrier30,31.
The kynurenine pathway is involved in the catabolism of tryptophan. The production of picolinic acid is considered an anti-inflammatory branch of the pathway, whist the production of quinolinic acid is considered pro-inflammatory due to its ability to activate immune cells and induce cell death. ACMSD acts at a branching point in this pathway that determines which of these products is created. 3-HAO 3-hydroxyanthranilate oxidase, ACMSD α-amino-β-carboxymuconate-ε-semialdehyde decarboxylase, KAT kynurenine aminotransferase, KMO kynurenine-3-monooxygenase, KYUN kynureninase.
Zebrafish represent an excellent vertebrate model organism for studying gene function and neurodegenerative disease, exhibiting neurodegenerative markers such as neuronal loss and microglia activation, even during larval stages32,33,34,35,36,37,38. Zebrafish have been widely used by us and others to biologically validate genes implicated in PD, including lrrk2, gch1, gba1, pink1, dj-1, and parkin32,33,34,39,40,41,42,43,44,45.
The objective of this study was to determine the biological effect of acmsd deficiency on PD-relevant mechanisms in acmsd-/- mutant zebrafish, the first vertebrate model of ACMSD deficiency.
Results
acmsd -/- mutation was loss-of-function in zebrafish
Human ACMSD has a single orthologue in zebrafish, acmsd, which demonstrates ~80% DNA and protein homology (Figs. 2A, 3A) and conserved gene synteny between species (Fig. 2B). Using RT-PCR, we found that acmsd was expressed at increasing levels from 1 to 5dpf in whole larvae (Fig. 2C). In situ hybridisation demonstrated that this expression was largely limited to the liver, although acmsd staining was also observed in intestinal tissue (Fig. 2D–G). RT-PCR based studies demonstrated more widespread expression in adult zebrafish, with the liver and intestines demonstrating the highest expression. Lower acmsd expression levels were identified in the brain, kidney, gonads, and gall bladder (Fig. 2H).
A Exonic structure of ACMSD. Human ACMSD (ENSG00000153086) has a single orthologue in zebrafish (ENSDARG00000062549) which shares 81% sequence homology with its human counterpart (based on CLUSTAL W v1.81 data). B Synteny has been retained between species. The region comparison feature on Ensembl was used to compare a 1 Mb region around the Acmsd gene on human chromosome 2 and zebrafish chromosome 9. Not all genes in the region are included. Orthologous genes are identified by arrows. C RT-PCR demonstrated acmsd expression with an increasing concentration from 1 to 5dpf in acmsd+/+ larvae. In situ hybridisation demonstrated restricted acmsd expression to the liver (arrows) and gut at 3dpf, shown from lateral (F) and dorsal (G) views. The sense probe produced no staining (D, E). H RT-PCR demonstrated acmsd expression primarily in the liver and intestines of both adult male (shown) and adult female (not shown) zebrafish. Expression was also identified in the brain, kidney, gonad,s and gall bladder.
To create a stable acmsd-/- line, we utilised CRISPR/Cas9 technology. The resulting mutant had a 1 bp insertion and 71 bp deletion in exon 6, resulting in the deletion of a 3’ splice site (Fig. 3A–C). Sequencing of homozygous mutant cDNA revealed that exon 6 underwent complete exon skipping without the retention of surrounding introns. Flanking exon sequences were indistinguishable from acmsd+/+ cDNA. In silico transcription suggested that the loss of exon 6 would result in the production of a truncated Acmsd protein (Fig. 3A). Importantly, this mutation induced a premature STOP codon in exon 8 which triggered nonsense-mediated decay of the resulting transcript in both whole larvae and adult brain tissue (Fig. 3D, E). Compared to acmsd+/+, acmsd mRNA levels were 87% lower in acmsd-/- larvae and 58% lower in acmsd-/- adult brain tissue.
A Zebrafish express a single Acmsd protein, sharing 81% protein identity to human ACMSD. A mutation was introduced into exon 6 using CRISPR/Cas9 resulting in a predicted truncated protein. Red region depicts altered amino acids in the mutated protein. B The resulting 70 bp reduction in DNA length allowed for genotyping by standard PCR. acmsd+/+ zebrafish were identified by a single 425 bp band (right), homozygous acmsd-/- mutants by a 355 bp band (left), and heterozygous acmsd+/- mutants by a double band (centre). C DNA sequence of acmsd exon 6 and intron 6. acmsd-/- zebrafish possessed a 71 bp deletion (red) and 1 bp insertion (blue) in this region, resulting in the loss of a 3’ splice site (arrows). acmsd-/- larvae demonstrated reduced acmsd expression compared to their acmsd+/+ siblings at 5dpf (D, n = 3 biological replicates (15 larvae per replicate), p = 0.0012) and in adult brain tissue (E, n = 6 biological replicates (1 brain per replicate), p = 0.0040). Statistics from two-tailed t tests using ddCt values.
acmsd -/- zebrafish do not develop overt neurodegenerative phenotypes
Despite acmsd being expressed throughout life in zebrafish, and the successful generation of a loss-of-function mutation in the acmsd gene, adult acmsd-/- zebrafish were viable, fertile, did not develop overt morphological abnormalities, and had a similar life span to their wildtype siblings (assessed up to 25mpf). Since acmsd displayed strong liver expression in the larval zebrafish, we hypothesised that acmsd-/- larvae may demonstrate abnormal liver development. However, the liver size of 5dpf acmsd-/- larvae was comparable to that of acmsd+/+ zebrafish (Fig. 4), suggesting that acmsd deficiency does not affect liver development during the larval stages. Liver size was also assessed in larvae obtained from a homozygous incross to rule out the potential contribution of maternally-derived Acmsd. This had no effect on the results (data not included).
5dpf zebrafish underwent in situ hybridisation against fatty acid-binding protein 10a (fabp10a, A, B, representative images). C There was no difference in liver area identified between genotypes (p = 0.2762, unpaired two-tailed t test). Data from three biological replicates, n = 59 per genotype. Data points represent individual fish.
We next characterised acmsd-/- for phenotypes relevant to PD which are altered in other zebrafish models of the disease, including changes in spontaneous movement, dopaminergic neuron quantification, microglial activation, and susceptibility to the classical PD neurotoxin MPP+32,33,34,39.
Spontaneous motor activity in acmsd-/- zebrafish was similar to acmsd+/+ at both larval and adult stages. acmsd-/- larvae did not demonstrate significant alterations in swimming distance compared to their acmsd+/- or acmsd+/+ siblings when exposed to alternating dark-light cycles (Fig. 5A, B, pdark = 0.5567, plight = 0.1707, pcombined= 0.1902). Similarly, adult acmsd-/- zebrafish did not move significantly less than their acmsd+/+ siblings over a 6-h period (Fig. 5C, D, p = 0.1796). These data suggest that a loss of Acmsd function does not have an effect on spontaneous movement in zebrafish.
A Following habituation, 5dpf larvae were exposed to alternating dark (black bar)-light (white bar) cycles. B Total distance travelled was indistinguishable between genotypes (Kruskal-Wallis one-way ANOVA, p = 0.1902). n = 45 acmsd-/-; 62 acmsd+/-; 35 acmsd+/+ from three biological replicates. C Following a habituation period, adult movement was recorded over 6 h at 9mpf. D. No difference in the total distance travelled was found between genotypes (unpaired two-tailed t test, p = 0.1796). n = 10 acmsd-/-; 10 acmsd+/+ (equal male/female ratio) taken at two separate time points. Graphs A + C show mean ± SEM. Graphs B + D show individual fish with mean ± SEM values.
We next investigated whether the loss-of-function mutation in acmsd resulted in a reduction in the number of ascending dopaminergic neurons in the posterior tuberculum. In situ hybridisation against th1 revealed no significant difference in the number of dopaminergic neurons between acmsd+/+ and acmsd-/- larvae at 3dpf (Fig. 6A–E, p = 0.9866). MMP+, a classical PD neurotoxin and mitochondrial complex I inhibitor, is known to reduce the number of dopaminergic neurons in larval zebrafish33. We hypothesised that acmsd-/- larvae would be more susceptible to the effects of this neurotoxin, demonstrating a greater reduction in neuronal number compared to acmsd+/+ larvae. Exposure to 3 mM MPP+ from 1 to 3dpf resulted in a 43% reduction in th1+ cells in both acmsd+/+ (p < 0.0001) and acmsd-/- larvae (p < 0.0001, Fig. 6C–E). The number of th1+ cells following MPP+ exposure was indistinguishable between genotypes (p = 0.9999), suggesting that acmsd deficiency does not alter the susceptibility of zebrafish larvae to MPP+. This experiment was also conducted on larvae obtained from a homozygous incross, similarly identifying no significant differences in acmsd-/- larvae compared to wildtype controls (data not included).
A–D In situ hybridisation against th1 showed no change in staining pattern between acmsd+/+ and acmsd-/- zebrafish either before or after MPP+ exposure. E There was no difference in th1+ neuron numbers in the ventral diencephalon between acmsd+/+ and acmsd-/- zebrafish (p = 0.9866). MPP+ exposure resulted in significant reductions in th1+ cells irrespective of genotype (43.1% in acmsd+/+; 43.4% in acmsd-/-, p < 0.0001). Total cell counts between groups following exposure remained indistinguishable (p = 0.9999). n = 26–35 per group, from three biological replicates. Statistics from a two-way ANOVA with Tukey’s multiple comparisons post-hoc test. Monoaminergic neurons were visualised using the ETvmat2:GFP transgenic line at 5dpf in the full brain (F, G dorsal view) and in the DC neuronal groups (H, I) in both acmsd+/+ and acmsd-/- zebrafish. Scale bar = 50 µm. DC diencephalic neurons, Hc caudal hypothalamus, PT pretectal neural cluster, Ra raphe nucleus, Te telencephalic neurons. J No difference in the number of neuronal cell bodies in DC2 and DC4/5 was seen between acmsd+/+ and acmsd-/- zebrafish (p = 0.8126, unpaired two-tailed t test). n = 9 per genotype, from three biological replicates.No difference in the concentrations of dopamine (K, p = 0.2808) or serotonin (L, p = 0.9079) were identified in whole 11mpf brains. Data analysed using an unpaired two-tailed t test. n = 14 acmsd-/-; 12 acmsd+/+, 1 brain per replicate.
We also assessed this neuronal group at a later time point (5dpf) using a different, complementary approach, namely by crossing acmsd-/- zebrafish with the enhancer trap transgenic zebrafish line ETvmat2:GFP in which most monoaminergic neurons are labelled by green fluorescent protein (GFP) during embryonic development46. Again, there was no difference in the number of vmat2+ monoaminergic neurons between acmsd+/+ and acmsd-/- larvae (Fig. 6F–J, p = 0.8126). Additionally, in 11mpf adult brains, the levels both dopamine and serotonin remained unchanged in acmsd-/- zebrafish (Fig. 6K, L). Together, these data suggest that acmsd deficiency does not lead to dopaminergic neuronal cell loss in zebrafish.
acmsd -/- larvae do not develop an inflammatory phenotype
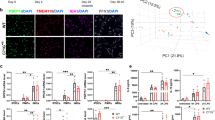
It has been postulated that ACMSD deficiency may contribute to the pathogenesis of PD through proinflammatory mechanisms47. Activated microglia are more numerous in both PD patient brains48,49,50 and zebrafish models of PD33,34. However, the number of microglia in acmsd-/- larval brains was similar to the number identified in their acmsd+/+ siblings at 5dpf (p = 0.9836, Fig. 7A–C). The percentage of activated cells was also unchanged (p = 0.8441, Fig. 7D). qPCR showed that expression of the proinflammatory cytokines cxcl8, il-1β, and mmp9 was also indistinguishable between acmsd-/- larvae and their acmsd+/+ siblings (Fig. 8). qPCR analysis was also conducted on larvae obtained from a homozygous incross, which similarly identified no difference in expression between acmsd-/- larvae and wildtype controls (data not included).
Representative images from wholemount immunohistochemistry against 4c4 in acmsd+/+ (A) and acmsd-/- (B) siblings at 5dpf (scale bar = 100 µm). C No difference was found in the number of 4c4+ cells between acmsd+/+ and acmsd-/- larvae in the forebrain, midbrain, or these combined (whole brain) (p = 0.9827 for forebrain, p = 0.9925 for midbrain, p = 0.9836 for whole brain). D No difference in microglial activation, represented as the percentage of amoeboid cells out of the total 4c4+ cell count, was identified between genotypes (p = 0.6983 for forebrain, p = 0.8307 for midbrain, p = 0.8441 for whole brain). n = 31 acmsd-/-, 27 acmsd+/+, from three biological replicates. Statistics analysed by two-way ANOVA with post hoc Sidak’s multiple comparisons test.
Metabolomics analysis revealed both increased quinolinic acid and an upregulation of kynurenine pathway enzymes
Gene duplication and compensatory upregulation of alternative pathways in CRISPR/Cas9 mutant zebrafish lines are potential confounding mechanisms in zebrafish research51. To demonstrate that the kynurenine pathway is indeed disrupted in our mutants, we undertook detailed metabolomic analysis of the kynurenine synthetic pathway in acmsd-/- zebrafish.
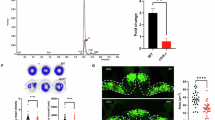
As expected, metabolomic analysis confirmed a marked increase of QUIN in acmsd-/- zebrafish. Whilst QUIN was undetectable in acmsd+/+ larvae, using the lower limit of quantification (0.42 ng/mL) we can infer that acmsd-/- larvae demonstrated at least a 34,800% increase in QUIN (Table S.1). In adult zebrafish, acmsd deficiency resulted in an average of a 71,500% increase of QUIN in the liver (Fig. 9, Table S.1) and an 11,000% increase in the brain (Table S.1). We were unable to measure PIC as it could not be separated from its isomer, nicotinic acid. Further analysis in adult liver tissue, where metabolite concentration was high enough to enable a direct comparison between genotypes, identified the upregulation of two branches of the kynurenine pathway. The kynurenic acid:kynurenine ratio demonstrated a 153% increase in homozygous mutants (Fig. 9E), suggesting increased kynurenine aminotransferase (KAT) activity, and the 3-hydroxykynurenine:kynurenine ratio demonstrated a 92% increase (Fig. 9F), suggesting an upregulation of kynurenine-3-monooxygenase (KMO).
In adult liver tissue, kynurenine was reduced in acmsd-/- zebrafish (A, p = 0.0079, Mann–Whitney test), no difference was identified between genotypes in kynurenic acid (B, p = 0.9125, unpaired, two-tailed t test) or 3-hydroxykynurenine (C, p = 0.0986, unpaired, two-tailed t test), whilst quinolinic acid was increased in acmsd-/- (D, p = 0.0079, Mann–Whitney test). The kynurenic acid: kynurenine ratio (E, p = 0.0161, unpaired, two-tailed t test) and the 3-hydroxykynurenine:kynurenine ratio (F, p = 0.0063, unpaired, two-tailed t test) were increased in acmsd-/- fish.
Discussion
We characterised a zebrafish model of acmsd deficiency for a reduction in spontaneous movement, a loss of dopaminergic neurons, an increased susceptibility to MPP+, and an increase in neuroinflammation, since these phenotypes may suggest a role of acmsd deficiency in altering the risk of developing PD. Despite a clear and very marked effect of the genetic inactivation of acmsd on enzymatic activity, as observed in our metabolomic study, we did not observe a single PD relevant abnormality in acmsd-/- larvae or adult zebrafish.
The evidence of ACMSD being involved in the pathogenesis of PD is less strong than for other putative or confirmed PD risk genes. A regulatory effect of the SNP rs6430538, located upstream of the ACMSD gene, and eight other SNPs found to be in linkage disequilibrium with rs6430538, on ACMSD expression has been postulated but remains unproven. Furthermore, although the existing data is not clear, there is evidence to suggest that there may also be multiple genes at this locus. Of note, the lead ACMSD SNP rs57891859 is actually intronic to TMEM1633, an understudied transporter protein which may be involved in lysosomal zinc handling3,52.
A disease-segregating mutation in the ACMSD gene (pTrp26Stop) has been reported in a Spanish family with cortical myoclonus, epilepsy, and parkinsonism. Only one of the six affected family members had parkinsonism, a then 53-year-old woman who first developed postural tremor of both hands at the age of 17 and generalised seizures since the age of 20. She only developed symptoms and signs of parkinsonism aged 49, including additional motor features, such as marked postural tremor and a significant memory deficit (MoCA score 19/30). A brain MRI showed abnormalities not in keeping with the diagnosis of PD, including cerebellar atrophy, high intensity brain stem signals in T2, and other changes suggestive of Wallerian degeneration53. Unfortunately, response to dopaminergic medication was not reported and focussed imaging of the nigrostriatal dopaminergic system, such as a DATScan—which would have confirmed the postulated loss of dopaminergic neurons—was not undertaken. A missense mutation (p.Glu298Lys) of unknown functional significance was also detected in a single late-onset patient with sporadic PD54. Biochemical studies of the kynurenine pathway in serum or CSF from people with sporadic PD have also given conflicting results (see Table 1).
We unexpectedly identified the upregulation of two branches of the kynurenine pathway (Fig. 10). The neuroprotective pathway (kynurenic acid production) was increased to a greater extent than the canonical pathway, suggesting a compensatory upregulation of kynurenic acid, an NMDAR antagonist. This may protect against QUIN-induced toxicity and explain the absence of a phenotype in acmsd-/- zebrafish.
QUIN was historically thought to only activate NMDARs with NR2A/NR2B subunits13, which are less widespread in the zebrafish CNS than in humans55. However, this selectivity has since been questioned56 and QUIN-induced toxicity has been demonstrated to act via non-NMDAR pathways15,16,17,18,19,20,21,24,25,26,27,28,29,30,31. Kynurenic acid, an NMDAR antagonist, also offers protection from QUIN-induced toxicity via NMDAR-independent mechanisms24,57. Conditioned media from QUIN-activated microglia can induce neuronal death whilst the inhibition of microglial activation reduces QUIN-induced toxicity23, suggesting the proinflammatory effect of QUIN may play a larger role in neuronal death than excitotoxicity.
Zebrafish are sensitive to QUIN; exogenous exposure in larvae results in stunted growth, cardiac effects, and seizures58,59, and in adults, direct injection into the telencephalon causes cell death and microglial recruitment60. Therefore, high QUIN concentrations, such as those seen throughout life in our mutant, would likely cause measurable effects in the absence of additional protective mechanisms. This further supports the hypothesis that the increased activity of the neuroprotective branch of the kynurenine pathway may be protecting acmsd-/- zebrafish against toxic levels of QUIN.
It is important to note the limitations of the genetic zebrafish model, in particular that genetic compensation in CRISPR-Cas9 mutants can mask the expected functional effect of particular gene defects. However, the absence of any acmsd paralogues reduces this likelihood significantly in our acmsd mutant zebrafish line. Direct genetic compensation also appears implausible due to the observed marked increase in QUIN in both acmsd-/- larvae and adults. We did, however, see a clear functional compensation, namely the increased activity of KAT. It is not clear whether this occurs at the pre- or post- translational level, and it would be interesting to see whether this gene is expressed at an increased level in acmsd-/- zebrafish. Future experiments could include KAT morpholino injection into acmsd-/- mutants to reveal a more significant role for Acmsd deficiency in zebrafish.
In summary, acmsd deficiency does not result in a PD-relevant phenotype in zebrafish. The results of our study, the limited genetic evidence, and the conflicting data from biochemical studies in sporadic PD patient tissue emphasise the need for further clarification of the possible role of ACMSD as a possible PD risk gene and of the importance of the kynurenine pathway in the pathogenesis of PD.
Methods
Zebrafish husbandry
Adult zebrafish were maintained at 28 °C in the Biological Services Aquarium at the University of Sheffield and subject to a light-dark cycle of 14 and 10 h, respectively. Embryos were obtained by pair mating and maintained in E3 media with methylene blue. 0.003% phenylthiourea [fluorochem] was used to prevent pigmentation and 4.2% tricaine used for anaesthesia. Regulated procedures followed the UK Home Office Animals (Scientific Procedures) Act 1986 under project licence PP6422743 (Professor Oliver Bandmann). All authors complied with the ARRIVE guidelines. acmsd loss-of-function mutants (acmsd-/-, allele code sh619) were created using the CRISPR/Cas9 system. A single guide RNA [Sigma-Aldrich] targeting 5′-ACTGCTCGCTATTTGTGCAT(CCA)-3′ in exon 6 of the acmsd gene was injected with tracrRNA and Cas9 [NEB] to single-cell stage zebrafish embryos. The line was genotyped using the following primers 5′-CCCCAGAGCTGTTTCCTGTC-3′ and 5′-CCGTGAGCAAAGCAGACCTT-3′ [Integrated DNA Technologies] as described previously61,62 and maintained as heterozygous colonies in the AB background. All experiments were conducted on zebrafish obtained from a heterozygous incross, unless stated otherwise.
Quantitative polymerase chain reaction (qPCR)
RNA was extracted from homogenised adult organs or whole larvae using TRIzol Reagent [Invitrogen]. RNA concentrations were determined using the NanoDrop 1000 Spectrophotometer [Thermo Scientific] and cDNA created using the Verso cDNA Synthesis Kit [Thermo Scientific]. Transcript levels were quantified using a CFX96 Touch Real-Time PCR Detection System [BioRad] and Brilliant III Ultra-Fast SYBR Green qPCR Master Mix [Agilent Technologies]. rps29 served as a reference gene to which expression was normalised and data were analysed using the delta-delta Ct method. Primers are outlined in Table 2. These primers were also used for reverse transcription-PCR (RT-PCR).
Movement analysis
5dpf larvae obtained via a heterozygous incross were transferred to 48-well plates in 500 µL E3 and movement analysed using the ZebraBox [Viewpoint Life Sciences]. Following a 30-min habituation period, movement during alternating 5-min dark-light cycles was recorded over 30 min. Siblings were genotyped using sacrificed whole larvae. For adult movement analysis, 9mpf fish of known sex and genotype were placed in individual tanks and movement recorded for 7 h using the ViewPoint software. The first hour was omitted as a habituation period.
MPP+ exposure
Dechorionated 3dpf larvae were exposed to 1-methyl-4-phenylpyridinium (MPP+) [Sigma] for 48 h via constant immersion in 3 mL E3 with daily media changes. Following exposure, larvae were fixed in 4% paraformaldehyde, transferred into methanol by serial dilution (30%; 50%; 70%; 100% methanol in PBSTw (PBS, 0.1% Tween-20)), and stored at −20 °C for at least 24 h.
Wholemount in situ hybridisation (WISH)
Digoxigenin-labelled riboprobes were generated from cDNA using standard PCR and the following primers; th1 forward 5′-AGTGCACCTGTCGGATGTTA-3′ and th1 reverse 5′-GCGTCCACAAAGCTTTCTGA-3′; acmsd forward 5′- GATCCAGAGGCTCGGATTCG-3′ and acmsd reverse 5′-CCAGAGCATTTCCAGCAAGC-3′; fabp10a forward 5′-AGCTTCTCCAGAAAGCATGG-3′ and fabp10a reverse 5′-TCCTGATCATGGTGGTTCCT-3′. A T7 polymerase site was added to the reverse primers to generate anti-sense probes, and to the forward primers to generate sense probes. PCR products were cleaned up using the Minelute Reaction Clean-Up kit [QIAGEN] and RNA transcribed using T7 polymerase and digoxigenin-labelled NTPs at 37 °C overnight. Probes were diluted to 1 ng/µL in hybridisation buffer A and WISH performed as previously described63.
Quantification of dopaminergic neurons from DC2, DC4, and DC5 subpopulations
Following WISH against th1, 3dpf zebrafish heads were mounted in glycerol and visualised on an AxioPlan microscope with a Plan-NEOFLUAR 20× Ph2 objective [Zeiss]. Tails were retained for genotyping. th1+ neurons from DC2 and DC4/5 populations, as described by Rink and Wullimann64, were counted blinded to genotype and treatment group. For assessment at 5dpf, monoaminergic neurons were labelled using the fluorescent vesicular monoamine transporter 2 (vmat2) transgenic line [allele code sh237Tg, 46]. acmsd+/-;ETvmat2:GFP zebrafish were crossed to either acmsd+/+ or acmsd-/- zebrafish, genotyped, and imaged using an AiryScan confocal microscope [Zeiss] with a 10× objective lens.
Microglial analysis
Immunohistochemistry against 4c4 was conducted as previously described65, using mouse anti-4c4 (gifted from Dr Alexander McGown, The University of Sheffield, 1:50; antibody:block) for 3 days and AlexaFluor 488 [Invitrogen] secondary antibody (1:200; antibody:PBSTw) for 3 days. Imaging and analyses were performed as previously described33. Cells were assigned to one of two distinct groups; active (amoeboid) or inactive (ramified with at least one visible process).
High-performance liquid chromatography (HPLC)
Whole brains from 11mpf zebrafish of known genotype were homogenised in 300 µL homogenisation solution. Dopamine and serotonin concentrations were measured by HPLC as described earlier66. Measured concentrations were adjusted by tissue weight and neurotransmitters reported as tissue content (ng/g tissue).
Kynurenine pathway metabolite analysis
Kynurenine pathway metabolite analysis was carried out by Charles River Laboratories using mass spectrometry. Five biological replicates were conducted for each experiment. Five 5dpf larvae obtained from a homozygous incross were combined and homogenised in 100 µL extraction solvent using sonication. Homogenates were centrifuged, the supernatants stored as larval extract, and metabolites reported as extract concentrations. For adult tissue analysis, brains and livers were extracted from 9mpf zebrafish obtained from a heterozygous incross and homogenised using a Precellys homogeniser in 5 µL extraction solvent per mg of tissue. Homogenates were centrifuged, the supernatants stored as extracts, and metabolites reported as tissue content (ng/g tissue).
Statistics
All experiments were completed in at least triplicate, with each biological replicate utilising embryos from separate parental batches. Graphs and statistical analyses were conducted in GraphPad Prism 7. Graphs are shown as means ± standard error of the mean (SEM). Where relevant, normality was assessed using a Shapiro-Wilk test. Tests used are described per graph. Significance was determined as p < 0.05.
Data availability
The datasets generated during the current study are available from the corresponding author on reasonable request.
Abbreviations
- ACMS:
-
α-amino-β-carboxymuconate-ε-semialdehyde
- ACMSD:
-
α-Amino-β-Carboxymuconate-ε-Semialdehyde Decarboxylase
- AMS:
-
α-aminomuconate semialdehyde
- ANOVA:
-
Analysis of variance
- bp:
-
Basepairs
- Cas9:
-
CRISPR-associated protein 9
- cDNA:
-
Complementary DNA
- CNS:
-
Central nervous system
- COX-2:
-
Cyclooxygenase-2
- CRISPR:
-
Clustered, Regularly-Interspaced, Short, Palindromic Repeats
- CSF:
-
Cerebral Spinal Fluid
- CXCL8:
-
C-X-C Motif Chemokine Ligand 8
- DNA:
-
Deoxyribonucleic acid
- dpf:
-
Days post fertilisation
- FABP10a:
-
Fatty acid binding protein 10a
- GBA1:
-
Glucocerebrosidase 1
- GCH1:
-
Guanosine triphosphate Cyclohydrolase 1
- GFP:
-
Green Fluorescent Protein
- GWAS:
-
Genome-wide association studies
- IL-1β:
-
Interleukin-1β
- KAT:
-
Kynurenine aminotransferase
- LRRK2:
-
Leucine-rich repeat kinase 2
- Mmp9:
-
Matrix metallopeptidase 9
- MoCA:
-
Montreal Cognitive Assessment
- mpf:
-
Months post fertilisation
- MPP+:
-
1-methyl-4-phenylpyridinium
- mRNA:
-
Messenger RNA
- NF-κB:
-
Nuclear factor kappa B
- NMDA:
-
N-Methyl-D-aspartic acid or N-Methyl-D-aspartate
- PBS:
-
Phosphate buffered saline
- PBStw:
-
Phosphate buffered saline with 0.01% tween
- PCR:
-
Polymerase chain reaction
- PD:
-
Parkinson’s disease
- PIC:
-
Picolinic acid
- PINK1:
-
PTEN induced putative kinase 1
- qPCR:
-
Quantitative polymerase chain reaction
- QUIN:
-
Quinolinic Acid
- RNA:
-
Ribonucleic acid
- RPS29:
-
Ribosomal Protein S29
- RT-PCR:
-
Reverse Transcriptase PCR
- SEM:
-
Standard error of the mean
- SNPs:
-
Single nucleotide polymorphisms
- TH1:
-
Tyrosine Hydroxylase 1
- TMEM163:
-
Transmembrane Protein 163
- TNF-α:
-
Tumour Necrosis Factor - alpha
- VMAT2:
-
Vesicular monoamine transporter 2
- WISH:
-
Wholemount in situ hybridisation
- WT:
-
Wildtype
References
Postuma, R. B. et al. MDS clinical diagnostic criteria for Parkinson’s disease. Mov. Disord. 30, 1591–1601 (2015).
Tansey, M. G. et al. Inflammation and immune dysfunction in Parkinson disease. Nat. Rev. Immunol. https://doi.org/10.1038/s41577-022-00684-6 (2022).
Nalls, M. A. et al. Identification of novel risk loci, causal insights, and heritable risk for Parkinson's disease: a meta-analysis of genome-wide association studies. The Lancet Neurology. 18, 1091–1102 (2019).
Nalls, M. A. et al. Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson’s disease. Nat. Genet. 46, 989–993 (2014).
Bandrés-Ciga, S. et al. Genome-wide assessment of Parkinson’s disease in a Southern Spanish population. Neurobiol. Aging 45, 213.e213–213.e219 (2016).
Chang, K. -H., Chen, C. -M., Chen, Y. -C., Fung, H. -C. & Wu, Y. -R. Polymorphisms of ACMSD-TMEM163, MCCC1, and BCKDK-STX1B are not associated with Parkinson’s disease in Taiwan. Parkinson’s. Dis. 2019, 3489638 (2019).
Lill, C. M. et al. Comprehensive research synopsis and systematic meta-analyses in Parkinson’s disease genetics: the PDGene database. PLoS Genet. 8, e1002548 (2012).
Nalls, M. A. et al. Imputation of sequence variants for identification of genetic risks for Parkinson’s disease: a meta-analysis of genome-wide association studies. Lancet 377, 641–649 (2011).
Pihlstrøm, L. et al. Supportive evidence for 11 loci from genome-wide association studies in Parkinson’s disease. Neurobiol. Aging 34, 1708.e1707–1713 (2013).
Tejera-Parrado, C. et al. A replication study of GWAS-genetic risk variants associated with Parkinson’s disease in a Spanish population. Neurosci. Lett. 712, 134425 (2019).
Wang, L. et al. Association of three candidate genetic variants in ACMSD/TMEM163, GPNMB and BCKDK/STX1B with sporadic Parkinson’s disease in Han Chinese. Neurosci. Lett. 703, 45–48 (2019).
Fukuoka, S. et al. Identification and expression of a cDNA encoding human alpha-amino-beta-carboxymuconate-epsilon-semialdehyde decarboxylase (ACMSD). A key enzyme for the tryptophan-niacine pathway and “quinolinate hypothesis”. J. Biol. Chem. 277, 35162–35167 (2002).
de Carvalho, L. P. Bochet, P. & Rossier, J. The endogenous agonist quinolinic acid and the non endogenous homoquinolinic acid discriminate between NMDAR2 receptor subunits. Neurochem. Int. 28, 445–452 (1996).
Stone, T. W. & Perkins, M. N. Quinolinic acid: a potent endogenous excitant at amino acid receptors in CNS. Eur. J. Pharm. 72, 411–412 (1981).
Tavares, R. G., Schmidt, A. P., Abud, J., Tasca, C. I. & Souza, D. O. In vivo quinolinic acid increases synaptosomal glutamate release in rats: reversal by guanosine. Neurochem. Res. 30, 439–444 (2005).
Tavares, R. G. et al. Quinolinic acid stimulates synaptosomal glutamate release and inhibits glutamate uptake into astrocytes. Neurochem. Int. 40, 621–627 (2002).
Tavares, R. G. et al. Quinolinic acid inhibits glutamate uptake into synaptic vesicles from rat brain. Neuroreport 11, 249–253 (2000).
Ting, K. K., Brew, B. J. & Guillemin, G. J. Effect of quinolinic acid on human astrocytes morphology and functions: implications in Alzheimer’s disease. J. Neuroinflammation 6, 36 (2009).
Hosoi, R. et al. Evaluation of intracellular processes in quinolinic acid-induced brain damage by imaging reactive oxygen species generation and mitochondrial complex I activity. EJNMMI Res. 11, 99 (2021).
Minghetti, L. et al. Effects of the adenosine A2A receptor antagonist SCH 58621 on cyclooxygenase-2 expression, glial activation, and brain-derived neurotrophic factor availability in a rat model of striatal neurodegeneration. J. Neuropathol. Exp. Neurol. 66, 363–371 (2007).
Moresco, R. M. et al. Quinolinic acid induced neurodegeneration in the striatum: a combined in vivo and in vitro analysis of receptor changes and microglia activation. Eur. J. Nucl. Med. Mol. Imaging 35, 704–715 (2008).
Guillemin, G. J., Croitoru-Lamoury, J., Dormont, D., Armati, P. J. & Brew, B. J. Quinolinic acid upregulates chemokine production and chemokine receptor expression in astrocytes. Glia 41, 371–381 (2003).
Feng, W. et al. Microglia activation contributes to quinolinic acid-induced neuronal excitotoxicity through TNF-α. Apoptosis 22, 696–709 (2017).
Pierozan, P. et al. Kynurenic acid prevents cytoskeletal disorganization induced by quinolinic acid in mixed cultures of rat striatum. Mol. Neurobiol. 55, 5111–5124 (2018).
Braidy, N., Grant, R., Adams, S., Brew, B. J. & Guillemin, G. J. Mechanism for quinolinic acid cytotoxicity in human astrocytes and neurons. Neurotox. Res. 16, 77–86 (2009).
Ryu, J. K., Kim, S. U. & McLarnon, J. G. Blockade of quinolinic acid-induced neurotoxicity by pyruvate is associated with inhibition of glial activation in a model of Huntington’s disease. Exp. Neurol. 187, 150–159 (2004).
Vandresen-Filho, S. et al. Cerebral cortex, hippocampus, striatum and cerebellum show differential susceptibility to quinolinic acid-induced oxidative stress. Neurol. Sci. 36, 1449–1456 (2015).
Pláteník, J., Stopka, P., Vejražka, M. & Štípek, S. Quinolinic acid—Iron(II) complexes: slow autoxidation, but enhanced hydroxyl radical production in the Fenton reaction. Free Radic. Res. 34, 445–459 (2001).
Pierozan, P. et al. Signaling mechanisms downstream of quinolinic acid targeting the cytoskeleton of rat striatal neurons and astrocytes. Exp. Neurol. 233, 391–399 (2012).
Reynolds, D. S. & Morton, A. J. Changes in blood-brain barrier permeability following neurotoxic lesions of rat brain can be visualised with trypan blue. J. Neurosci. Methods 79, 115–121 (1998).
St’astný, F., Skultétyová, I., Pliss, L. & Jezová, D. Quinolinic acid enhances permeability of rat brain microvessels to plasma albumin. Brain Res. Bull. 53, 415–420 (2000).
Flinn, L. J. et al. TigarB causes mitochondrial dysfunction and neuronal loss in PINK1 deficiency. Ann. Neurol. 74, 837–847 (2013).
Larbalestier, H. et al. GCH1 deficiency activates brain innate immune response and impairs tyrosine hydroxylase homeostasis. J. Neurosci. JN-RM-0653-0621, https://doi.org/10.1523/jneurosci.0653-21.2021 (2021).
Keatinge, M. et al. Glucocerebrosidase 1 deficient Danio rerio mirror key pathological aspects of human Gaucher disease and provide evidence of early microglial activation preceding alpha-synuclein-independent neuronal cell death. Hum. Mol. Genet. 24, 6640–6652 (2015).
Keatinge, M. et al. Unexpected phenotypic and molecular changes of combined glucocerebrosidase and acid sphingomyelinase deficiency. Dis. Model. Mech. 16, https://doi.org/10.1242/dmm.049954 (2023).
Hu, Y. et al. Mis-localization of endogenous TDP-43 leads to ALS-like early-stage metabolic dysfunction and progressive motor deficits. Mol. Neurodegener. 19, 50 (2024).
Fleming, A. et al. Unexpected phenotype reversion and survival in a zebrafish model of multiple sulfatase deficiency. Front. Cell Dev. Biol. 10, 843079 (2022).
Burgess, H. A. & Burton, E. A. A critical review of zebrafish neurological disease models-1. the premise: neuroanatomical, cellular and genetic homology and experimental tractability. Oxf. Open Neurosci. 2, kvac018 (2023).
Flinn, L. et al. Complex I deficiency and dopaminergic neuronal cell loss in parkin-deficient zebrafish (Danio rerio). Brain 132, 1613–1623 (2009).
Bretaud, S., Allen, C., Ingham, P. W. & Bandmann, O. p53-dependent neuronal cell death in a DJ-1-deficient zebrafish model of Parkinson’s disease. J. Neurochem. 100, 1626–1635 (2007).
Prabhudesai, S. et al. LRRK2 knockdown in zebrafish causes developmental defects, neuronal loss, and synuclein aggregation. J. Neurosci. Res. 94, 717–735 (2016).
Suzzi, S. et al. Deletion of lrrk2 causes early developmental abnormalities and age-dependent increase of monoamine catabolism in the zebrafish brain. PLoS Genet. 17, e1009794 (2021).
Anichtchik, O. et al. Loss of PINK1 function affects development and results in neurodegeneration in zebrafish. J. Neurosci. 28, 8199–8207 (2008).
Xi, Y. et al. Impaired dopaminergic neuron development and locomotor function in zebrafish with loss of pink1 function. Eur. J. Neurosci. 31, 623–633 (2010).
Sallinen, V. et al. Dopaminergic cell damage and vulnerability to MPTP in Pink1 knockdown zebrafish. Neurobiol. Dis. 40, 93–101 (2010).
Wen, L. et al. Visualization of monoaminergic neurons and neurotoxicity of MPTP in live transgenic zebrafish. Dev. Biol. 314, 84–92 (2008).
Thirtamara-Rajamani, K. et al. Is the enzyme ACMSD a novel therapeutic target in Parkinson’s disease? J. Parkinsons Dis. 7, 577–587 (2017).
Imamura, K. et al. Distribution of major histocompatibility complex class II-positive microglia and cytokine profile of Parkinson’s disease brains. Acta Neuropathol. 106, 518–526 (2003).
Gerhard, A. et al. In vivo imaging of microglial activation with [11C](R)-PK11195 PET in idiopathic Parkinson’s disease. Neurobiol. Dis. 21, 404–412 (2006).
Ouchi, Y. et al. Microglial activation and dopamine terminal loss in early Parkinson’s disease. Ann. Neurol. 57, 168–175 (2005).
El-Brolosy, M. A. et al. Genetic compensation triggered by mutant mRNA degradation. Nature 568, 193–197 (2019).
Cuajungco, M. P. & Kiselyov, K. The mucolipin-1 (TRPML1) ion channel, transmembrane-163 (TMEM163) protein, and lysosomal zinc handling. Front Biosci. 22, 1330–1343 (2017).
Martí-Massó, J. F. et al. The ACMSD gene, involved in tryptophan metabolism, is mutated in a family with cortical myoclonus, epilepsy, and parkinsonism. J. Mol. Med. 91, 1399–1406 (2013).
Vilas, D. et al. A novel p.Glu298Lys mutation in the ACMSD gene in sporadic Parkinson’s disease. J. Parkinsons Dis. 7, 459–463 (2017).
Cox, J. A., Kucenas, S. & Voigt, M. M. Molecular characterization and embryonic expression of the family of N-methyl-D-aspartate receptor subunit genes in the zebrafish. Dev. Dyn. 234, 756–766 (2005).
Pittaluga, A., Pattarini, R., Feligioni, M. & Raiteri, M. N-methyl-D-aspartate receptors mediating hippocampal noradrenaline and striatal dopamine release display differential sensitivity to quinolinic acid, the HIV-1 envelope protein gp120, external pH and protein kinase C inhibition. J. Neurochem 76, 139–148 (2001).
Lugo-Huitrón, R. et al. On the antioxidant properties of kynurenic acid: free radical scavenging activity and inhibition of oxidative stress. Neurotoxicol. Teratol. 33, 538–547 (2011).
Cassar, S. et al. Measuring drug absorption improves interpretation of behavioral responses in a larval zebrafish locomotor assay for predicting seizure liability. J. Pharm. Toxicol. Methods 88, 56–63 (2017).
Majewski, M., Kasica, N., Jakimiuk, A. & Podlasz, P. Toxicity and cardiac effects of acute exposure to tryptophan metabolites on the kynurenine pathway in early developing zebrafish (Danio rerio) embryos. Toxicol. Appl. Pharm. 341, 16–29 (2018).
Skaggs, K., Goldman, D. & Parent, J. M. Excitotoxic brain injury in adult zebrafish stimulates neurogenesis and long-distance neuronal integration. Glia 62, 2061–2079 (2014).
Wilkinson, R. N., Elworthy, S., Ingham, P. W. & van Eeden, F. J. A method for high-throughput PCR-based genotyping of larval zebrafish tail biopsies. Biotechniques 55, 314–316 (2013).
Keatinge, M. et al. CRISPR gRNA phenotypic screening in zebrafish reveals pro-regenerative genes in spinal cord injury. PLoS Genet. 17, e1009515 (2021).
Thisse, C. & Thisse, B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat. Protoc. 3, 59–69 (2008).
Rink, E. & Wullimann, M. F. Development of the catecholaminergic system in the early zebrafish brain: an immunohistochemical study. Brain Res. Dev. Brain Res. 137, 89–100 (2002).
Inoue, D. & Wittbrodt, J. One for all–a highly efficient and versatile method for fluorescent immunostaining in fish embryos. PLoS ONE 6, e19713 (2011).
Sallinen, V. et al. MPTP and MPP+ target specific aminergic cell populations in larval zebrafish. J. Neurochem 108, 719–731 (2009).
Widner, B., Leblhuber, F. & Fuchs, D. Increased neopterin production and tryptophan degradation in advanced Parkinson’s disease. J. Neural Transm. 109, 181–189 (2002).
Schulte, E. C. et al. Alterations in lipid and inositol metabolisms in two dopaminergic disorders. PLoS One 11, e0147129 (2016).
Han, W., Sapkota, S., Camicioli, R., Dixon, R. A. & Li, L. Profiling novel metabolic biomarkers for Parkinson’s disease using in-depth metabolomic analysis. Mov. Disord. 32, 1720–1728 (2017).
Oxenkrug, G., van der Hart, M., Roeser, J. & Summergrad, P. Peripheral tryptophan - kynurenine metabolism associated with metabolic syndrome is different in Parkinson’s and Alzheimer’s diseases. Endocrinol. Diabetes Metab. J. 1. http://researchopenworld.com/wp-content/uploads/2017/11/EDMJ-2017-113-Gregory-F-Oxenkrug-USA.pdf (2017).
Havelund, J. F. et al. Changes in kynurenine pathway metabolism in Parkinson patients with L-DOPA-induced dyskinesia. J. Neurochem. 142, 756–766 (2017).
Sorgdrager, F. J. H. et al. Age- and disease-specific changes of the kynurenine pathway in Parkinson’s and Alzheimer’s disease. J. Neurochem. 151, 656–668 (2019).
Heilman, P. L. et al. Tryptophan metabolites are associated with symptoms and nigral pathology in Parkinson’s disease. Mov. Disord. 35, 2028–2037 (2020).
Klatt, S. et al. A six-metabolite panel as potential blood-based biomarkers for Parkinson’s disease. NPJ Parkinsons Dis. 7, 94 (2021).
Luan, H. et al. LC-MS-based urinary metabolite signatures in idiopathic Parkinson’s disease. J. Proteome Res. 14, 467–478 (2015).
Lewitt, P. A. et al. 3-hydroxykynurenine and other Parkinson’s disease biomarkers discovered by metabolomic analysis. Mov. Disord. 28, 1653–1660 (2013).
Iwaoka, K. et al. Impaired metabolism of kynurenine and its metabolites in CSF of Parkinson’s disease. Neurosci. Lett. 714, 134576 (2020).
Ogawa, T. et al. Kynurenine pathway abnormalities in Parkinson’s disease. Neurology 42, 1702–1706 (1992).
López Nadal, A., Peggs, D., Wiegertjes, G. F. & Brugman, S. Exposure to antibiotics affects saponin immersion-induced immune stimulation and shift in microbial composition in zebrafish larvae. Front. Microbiol. 9, 2588 (2018).
Bower, N. I. et al. Vegfd modulates both angiogenesis and lymphangiogenesis during zebrafish embryonic development. Development 144, 507–518 (2017).
Acknowledgements
We would like to thank Dr Alexander McGown for providing the 4c4 antibody. We would also like to thank all Biological Services Aquarium staff who care for our zebrafish, in particular Lisa van Hateren for providing the genotyping service. Metabolomics (mass spectrometry) was conducted by Charles River Laboratories. In situ hybridisation and ETvmat2:GFP live imaging was carried out in the Wolfson Light Microscopy Facility, University of Sheffield. This work was supported by funding from NZP/Dextra, the Medical Research Council (MRC, MR/R011354/1 and MR/M006646/1) and a University of Sheffield Batelle PhD studentship. The funders played no role in study design, data collection, analysis and interpretation of data, or the writing of this manuscript. Freek van Eeden was funded by a Biotechnology and Biological Sciences Research Council (BBSRC) grant - grant number BB/R015457/1.
Author information
Authors and Affiliations
Contributions
E.F.: Conceptualisation, Methodology, Formal Analysis, Investigation, Writing—Original Draft, Writing—Review & Editing; M.K.: Conceptualisation, Methodology (production of mutant), Writing—Review & Editing; O.P.: Writing—Review & Editing; P.P.i.: Investigation (HPLC), Writing—Review & Editing; P.P.a.: Investigation (HPLC), Writing—Review & Editing; F.v.E.: Supervision, Writing—Review & Editing; R.B.M.: Supervision, Writing—Review & Editing; O.B.: Conceptualisation, Writing—Review & Editing, Supervision, Funding acquisition.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Fargher, E., Keatinge, M., Pearce, O. et al. A zebrafish model of acmsd deficiency does not support a prominent role for ACMSD in Parkinson’s disease. npj Parkinsons Dis. 11, 118 (2025). https://doi.org/10.1038/s41531-025-00940-1
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41531-025-00940-1