Abstract
Parkinson’s disease (PD), common neurodegenerative disorder, involves substantia nigra dopaminergic neuron loss and α-synuclein accumulation in Lewy bodies. While pathogenesis remains unclear, dysregulated urea metabolism may play a central role. This study detected elevated serum urea levels in PD patients with upregulated urea cycle enzymes. In MPTP-induced PD mice, urea accumulated in the substantia nigra and striatum, alongside increased activity of urea cycle enzymes (ODC1, ARG1, OTC) and urea transporter UT-B. Mechanistically, brain urea accumulation likely stems from imbalanced urea cycle activity and impaired UT-B-mediated clearance, with compensatory UT-B upregulation specifically in the substantia nigra. In vitro, MPTP-treated neuronal cells showed increased enzyme and UT-B expression, while high urea directly suppressed tyrosine hydroxylase (TH). Importantly, ODC1 knockdown reversed urea dysmetabolism, restored TH, and alleviated neuronal damage. These findings establish ODC1-mediated urea cycle dysregulation as a core metabolic feature of PD, proposing ODC1 or urea metabolism as novel therapeutic targets.

Similar content being viewed by others
Introduction
Parkinson’s disease (PD), the second most common neurodegenerative disease after Alzheimer’s disease (AD)1, is characterized by progressive motor dysfunction, including bradykinesia, rigidity, and resting tremor. Notably, up to 80% of PD patients develop cognitive dysfunction during disease progression2. Neuropathologically, PD is defined by the loss of substantia nigra dopaminergic neurons and the accumulation of misfolded α-synuclein into Lewy bodies that propagate across brain regions3,4. While these features are well-established, emerging evidence suggests that metabolic disturbances, particularly dysregulation of urea metabolism, may play a critical role in PD pathogenesis, a hypothesis supported by parallels in other neurodegenerative diseases.
Metabolomics studies in AD and Huntington’s disease (HD) have revealed widespread increases in brain urea levels despite their distinct clinical and genetic profiles5,6,7,8. Intriguingly, although peripheral blood urea levels in PD patients remain controversial, with reports of elevated plasma urea9, unchanged serum urea10, and reduced urea in cerebrospinal fluid11 and whole blood12, recent evidence indicates widespread urea accumulation in PD brains, mirroring observations in AD and HD13. This suggests a shared pathogenic mechanism involving urea dysregulation across neurodegenerative disorders. While the hepatic urea cycle (ornithine cycle) is the primary pathway for detoxifying ammonia into urea14, recent studies propose that the brain may harbor intrinsic urea cycle activity15,16. Key enzymes of the urea cycle, including ornithine transcarbamylase (OTC), ornithine decarboxylase 1 (ODC1), and arginase 1 (ARG1)17, are expressed in the brain, and their upregulation has been linked to neurodegenerative pathology18,19. For instance, AD brains exhibit transcriptional activation of all urea cycle enzyme genes, implicating urea cycle dysfunction in disease progression20. However, whether similar mechanisms operate in PD remains unexplored.
Elevated brain urea level may also arise from impaired urea transport. Urea transporter B (UT-B), a major facilitator of urea clearance in the brain, regulates urea homeostasis by mediating its osmotic-driven efflux. Dysfunctional UT-B has been associated with urea accumulation in the hippocampus, leading to neuronal damage21. Compensatory upregulation of UT-B in HD and AD brains further underscores its role in mitigating neurotoxic urea levels8,22, yet its involvement in PD remains unknown. Additionally, urea cycle intermediates, such as polyamines, may exert dual effects: while essential for neurotransmitter synthesis (e.g., GABA via putrescine23), their overproduction can disrupt synaptic plasticity and promote neurotoxicity24.
Therefore, this study integrates behavioral, histopathological, and molecular approaches to investigate urea metabolism in PD patients and a PD mouse model. We aim to quantify urea levels and urea cycle enzyme expression (OTC, ARG1) in PD-relevant brain regions. In addition, we explore whether dopaminergic neuron loss is mechanistically linked to urea cycle activation or UT-B dysfunction. Our findings may unveil novel pathogenic pathways and therapeutic targets for PD, bridging metabolic dysregulation with classical neuropathological hallmarks.
Results
Serum urea and urea cycle increased in PD patients, and MPTP-induced PD model mice showed significant motor dysfunction
Serum samples from PD patients and healthy controls were collected at University-Town Hospital of Chongqing Medical University. Biochemical analysis revealed significantly elevated serum urea levels in PD patients compared to controls, while serum creatinine levels showed no significant difference between groups (Fig. 1A). Western blot analysis of serum proteins further demonstrated upregulated expression of urea cycle key enzymes in PD patients. Concurrently, the expression of α-synuclein, a pathological marker of PD, was significantly increased in serum samples (Fig. 1B–C).
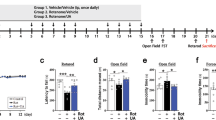
A Serum urea creatinine levels in normal and PD patients. (n = 20) (B) Western Blot of Serum protein (C) Statistical plot of Western Blot of serum protein. (n = 3) (D) MPTP model construction process. (n = 5) (E) Body weight of mice at modeling. F Paralysis tremor score of mice. G Mouse grip strength test. H Mouse rotarod test. I Mouse open field test track map and heat map. J Mouse open field test data index. Data are presented as the mean ± SEM. Significant differences were established by Student’s t-test. ***p < 0.001, **p < 0.01, and *p < 0.05.
To investigate urea metabolism in PD pathogenesis, a chronic PD model was established by intraperitoneally injection of MPTP for 8 weeks, with saline-injected mice serving as controls (Fig. 1D). Body weight monitoring throughout the modeling period (Fig. 1E) revealed no significant changes between MPTP-treated mice and controls. Behavioral analysis at the endpoint confirmed robust PD-like motor deficits in MPTP mice. Specifically, the Parkinsonism score (Fig. 1F) showed that the MPTP model mice had pronounced motor impairments. Grip strength tests (Fig. 1G), revealed a significant reduction in forelimb strength, and rotarod performance demonstrated impaired motor coordination and endurance in PD mice compared to controls (Fig. 1H). Open field test (Fig. 1I–J) provided additional insights: while total movement distance did not differ significantly between groups, MPTP mice exhibited reduced average velocity and spent markedly less time and distance traversing the center area. Collectively, these behavioral findings validated the successful induction of PD-like motor dysfunction in the MPTP model25.
Nigra striatum of MPTP-induced PD model mice showed obvious pathological manifestations
MPTP-treated and control mice revealed no gross morphological or histological abnormalities (Fig. 2A) in the kidneys, as evidenced by HE staining. However, MPTP mice exhibited a significant increase in the kidney-to-body weight ratio and elevated serum urea levels but no significant change in creatinine compared to controls (Fig. 2B). This was consistent with the previous results of serum creatinine and urea in patients with Parkinson’s disease. Immunohistochemical staining (Fig. 2C) of the substantia nigra (SN) and striatum (STR) demonstrated a marked reduction in tyrosine hydroxylase (TH) expression, a marker of dopaminergic neurons, in MPTP mice relative to controls26. Quantitative analysis confirmed decreased counts of TH-positive cells and optical density in the SN and STR of PD mice (Fig. 2D). HE and Nissl staining (Fig. 2E–H) further revealed severe neuronal loss and structural damage in the SN and STR of PD mice. Western blot analysis (Fig. 2I) corroborated these findings, showing diminished TH protein levels in the SN, though α-synuclein expression remained unchanged, consistent with the prior report27.
A Gross morphology of mouse kidney and HE staining (bar = 100 μm). B Mouse kidney-to-body weight ratio, serum urea and creatinine. C Mouse substantia nigra and striatum TH immunohistochemistry. (bar = 250 μm, bar = 1 mm). D The average optical density and the number of positive cells in the substantia nigra and striatum. E HE staining of mouse substantia nigra (short bar = 250 μm, long bar = 100 μm). F Nissl staining of mouse substantia nigra (short bar=250μm, long bar = 100 μm). G HE staining of mouse striatum (short bar = 1 mm, long bar = 100 μm). H Nissl staining of mouse striatum (short bar = 1 mm, long bar = 100 μm). I Western Blot analysis of TH, A-Synuclein in the substantia nigra and striatum of mice and protein expression and relative expression statistics. (n = 3). Data are presented as the mean ± SEM. Significant differences were established by Student’s t-test. ***p < 0.001, **p < 0.01, and *p < 0.05.
Urea cycle activation in PD brains
There is ongoing controversy regarding the activity of the urea cycle in the brain. Previous studies have found that the urea cycle in the normal brain is a metabolic pathway in the non-traditional sense, but it exhibits a normal urea cycle metabolic pathway under AD conditions20. To address controversies regarding urea cycle activity in PD, we quantified urea levels and urea cycle enzyme expression in SN and STR (Fig. 3A). MPTP-induced PD mice exhibited significantly elevated urea levels in both the SN and STR compared to controls. Western blot analysis (Fig. 3B–E) revealed upregulation of urea cycle key enzymes ODC1, ARG1 and OTC in the SN and STR of PD mice. Notably, urea transporter UT-B expression was selectively increased in the SN, but not the STR, suggesting region-specific compensatory mechanisms to mitigate urea accumulation. Additionally, we measured the intermediate by-products of the urea cycle and found that ammonia levels decreased and putrescine levels increased in the SN and STR of the MPTP group (Fig. 3F).
A Urea levels in the substantia nigra and striatum of mice. n = 3 (B) Expressions of key urea cycle enzymes and urea channel proteins in the substantia nigra of mice by Western Blot. n = 3. C Relative expression statistics of urea cycle key enzymes and urea channel proteins in the substantia nigra of mice. n = 3. D Expressions of urea cycle key enzymes and urea channel proteins in the striatum of mice by Western Blot assay. E Western Blot analysis of relative expression statistics of key urea cycle enzymes and urea channel proteins in mouse striatum. F Measurement of ammonia and putrescine level changes in substantia nigra and striatum in the MPTP model. n = 3. G Mice dopaminergic neurons in substantia nigra co-labeled with ODC1 immunofluorescence (short bar = 100 μm, long bar = 10 μm). H Mouse dopaminergic neurons in substantia nigra co-labeled with ARG1 immunofluorescence (short bar = 100 μm, long bar = 10 μm). I Mice striatum dopaminergic neurons co-labeled with ODC1 immunofluorescence (short bar = 500 μm, long bar = 10 μm). J Mouse striate dopaminergic neurons co-labeled with ARG1 immunofluorescence (short bar = 500 μm, long bar = 10 μm). Data are presented as the mean ± SEM. Significant differences were established by Student’s t-test. ***p < 0.001, **p < 0.01, and *p < 0.05.
Immunofluorescence colocalization studies in the SN revealed reduced TH expression alongside elevated ARG1 and ODC1 expression in the PD mice. These enzymes were predominantly localized to the cytoplasm of dopaminergic neurons, implicating their direct involvement in PD pathophysiology. The spatial overlap between TH and urea cycle enzymes further supports their role in modulating dopaminergic neuron vulnerability under urea metabolic stress (Fig. 3G–J).
Urea cycle dysregulation in neuronal cell models of PD
To investigate whether urea cycle activation observed in vivo extends to cellular models, we treated three types of neurons PC12, SH-SY5Y and Neuro-2a with 2500 μM MPTP for 24 h28. Western blot analysis revealed a significant upregulation of key urea cycle enzymes and urea transporter UT-B in all three cell lines under MPTP challenge (Fig. 4A). Consistent with protein-level changes, RT-qPCR (Fig. 4B) confirmed elevated ODC1 mRNA expression in MPTP-treated neurons. Additionally, we found that MPTP-treated cells had increased urea levels, decreased ammonia levels and increased putrescine levels (Fig. 4C). To assess the direct impact of urea on neuronal physiology, PC12 neurons were exposed to different concentrations of urea. Interestingly, urea treatment reduced TH expression in PC12 neurons while upregulating urea cycle key enzyme and UT-B, mirroring the in vivo PD phenotype (Fig. 4D–E). Moreover, urea treatment also increased the level of putrescine in the cells (Fig. 4G). However, after culturing C8-D1A astrocytes with 2500 μM MPTP for 24 h26, TH expression was decreased, but the urea cycle key enzymes and urea transporter proteins showed no significant changes (Fig. 4F).
A Western Blot analysis of key urea cycle enzymes and urea channel proteins in PC12, SH-SY5-Y and Neuro-2a MPTP model. B RNA relative expression of ODC1. C Measurement of urea, ammonia and putrescine level changes in the MPTP model. n = 3. D Western Blot analysis of urea cycle key enzyme and urea channel protein expression in neurons induced by high urea. E Western Blot analysis of the relative expression of key urea cycle enzymes and urea channel proteins in neurons induced by high urea. n = 4. F Western Blot analysis of key urea cycle enzymes and urea channel proteins in C8-D1A astrocytes. G Measurement of putrescine level changes after urea treatment. Data are presented as the mean ± SEM. Significant differences were established by Student’s t-test. ***p < 0.001, **p < 0.01, and *p < 0.05.
ODC1 knockdown attenuates urea cycle activation
Given the predominant expression of ODC1 in dopaminergic neurons, we generated an ODC1- knockdown (siODC1) PC12 model to probe its mechanistic role. In MPTP-treated cells, ODC1 knockdown significantly reduced urea cycle enzyme expression and restored TH levels (Fig. 5A–B), effectively reversing PD-associated molecular changes. Immunofluorescence validated these findings, showing partial recovery of TH immunofluorescence signal in siODC1 cells (Fig. 5C). Similar rescue effects were observed in SH-SY5Y and Neuro-2a cells following ODC1 silencing (Fig. 5D–G). Moreover, the urea level decreased after ODC1 knockdown (Fig. 5H). These findings demonstrate that MPTP-induced urea cycle activation and UT-B upregulation are recapitulated in neuronal cell models, establishing a direct link between urea metabolism and dopaminergic dysfunction. The reversibility of PD-like pathology via ODC1 knockdown highlights this enzyme as a critical regulator of urea cycle activation.
A Graph of Western Blot results after knockdown of ODC1 in PC12 cells. B Statistical plot of Western Blot results after knockdown of ODC1 in PC12 cells. n = 3. C Fluorescence pattern of PC12 cells after knockdown of ODC1. (bar = 50um). D Graph of Western Blot results after knockdown of ODC1 in SH-SY5-Y cells. E Statistical plot of Western Blot results after knockdown of ODC1 in SH-SY5-Y cells. n = 3. F Graph of Western Blot results after knockdown of ODC1 in Neuro-2a cells. G Statistical plot of Western Blot results after knockdown of ODC1 in Neuro-2a cells. n = 3. H Measurement of urea levels after knockdown of ODC1. n = 3. Data are presented as the mean ± SEM. Significant differences were established by Student’s t-test. ***p < 0.001, **p < 0.01, and *p < 0.05.
Discussion
PD is a common neurodegenerative disease in the elderly29. Epidemiological data reveal an alarming tripling of worldwide PD prevalence from 2.5 million in 1990 to 6.1 million cases, which has become a serious global health challenge. Clinically, PD patients are not only accompanied by a decline in cognitive function, but also gradually aggravated movement disorders, leading to an increased risk of related pathology and death30. Massive degeneration of dopaminergic neurons and accumulation of α-synuclein are the basic pathological features of PD31; however, more and more metabolomics studies have shown that metabolism may be one of the important pathological mechanisms of PD32. Our findings revealed urea cycle activation and elevated urea levels as a central biochemical disturbance in PD pathogenesis. This biochemical anomaly aligns with prior observations in PD dementia, dementia with Lewy bodies, Alzheimer’s disease (AD), and Huntington’s disease (HD), where urea concentrations are consistently elevated across multiple brain regions, often reaching pathogenic levels comparable to uremic encephalopathy. Notably, in PD models, we observed a significant increase in urea levels within the nigrostriatal pathway, mirroring the neuropathological changes seen in human PD brains. This suggests that urea dysregulation is not merely an epiphenomenon but may directly contribute to disease pathogenesis.
Mechanistic dissection reveals paradoxical consequences of urea cycle hyperactivity. While PD models demonstrate upregulation of key enzymes (ODC1, ARG1, OTC) potentially mitigating nitrogen overload, this adaptive response generates neurotoxic byproducts. Similar to AD, where astrocytic urea cycle activation drives ammonia detoxification but paradoxically promotes cognitive decline via toxic polyamine accumulation, our data suggest that urea cycle hyperactivity in PD may exert dual roles. According to the neuron priority injury theory, dopamine neuronal injury (MPTP) in Parkinson’s disease (PD) leads to more significant disorders in the urea cycle of neurons themselves33. However, astrocytes maintain metabolic homeostasis through other pathways. Single-cell transcriptome data show that the expression of urea cycle enzymes (such as ARG1 and OTC) in astrocytes of PD patients changes by less than 2 times, while ARG1 is down-regulated by more than 5 times in neurons34. Additionally, α-syn aggregates selectively inhibit neuronal lysosomal function (affecting arginase transport), but have a weaker effect on astrocyte lysosomes, and the neuronal urea cycle is more susceptible to proteinopathies. Meanwhile, urea in astrocytes may be rapidly excreted through AQP4, leading to deviations in intracellular concentration detection. Importantly, the urea cycle of astrocytes is significantly activated only in acute ammonia poisoning and changes weakly in chronic neurodegeneration. ARG1 catalyzes the hydrolysis of arginine to produce ornithine and urea, thereby accelerating the entire urea cycle rate and driving the conversion of ammonia to urea, thereby enhancing the endogenous ammonia clearance capacity within the cells. While potentially mitigating nitrogen overload, it could also generate neurotoxic intermediates such as putrescine35, which disrupts GABAergic signaling and synaptic plasticity23,24. This dual-edged mechanism is further supported by studies in HD, where urea cycle defects and hyperammonemia precede overt neurodegeneration36, implicating urea metabolism as an early pathological trigger.
The direct consequences of excessive activation of the urea cycle also include an increase in the level of the final product urea. Elevated urea levels may extend beyond neurodegenerative pathologies to influence psychiatric disorders. Emerging evidence suggests that high urea levels may act as an independent biological factor in conditions such as depression and anxiety. Mechanistically, urea disrupts neuronal synaptic plasticity, inducing depressive-like behaviors24, and promotes abnormal myelin maintenance, which correlates with anxiety-like phenotypes37. These findings underscore the broad neurotoxic potential of urea dysregulation, potentially linking metabolic disturbances to both psychiatric and neurodegenerative manifestations.
The systemic nature of urea dysregulation in PD is underscored by clinical evidence showing elevated blood urea and creatinine levels in PD patients, correlate with disease severity and progression. In PD models, we also observed marked motor dysfunction concomitant with elevated urea levels. At the same time, the increase of urea levels can also induce PD-like pathological manifestations in neurons, so urea may be the main factor inducing motor dysfunction in PD. The parallel between urea-induced pathology in models and clinical observations in PD patients, where blood urea levels correlate with disease severity and progression7, further supports urea as a primary effector of neurodegeneration. Crucially, experimental induction of urea elevation in neurons recapitulated PD-like pathological features, including dopaminergic neuron loss and α-synuclein aggregation. In addition to the direct effect of urea on the nervous system, these peripheral biomarkers may reflect broader metabolic disruptions, including impaired renal function or altered protein catabolism, which could exacerbate central nervous system pathology.
Importantly, our findings raise critical questions about causality. While elevated urea levels are associated with PD-like motor deficits and neuronal pathology in models, whether urea accumulation directly drives neurodegeneration or arises secondary to other metabolic insults remains unclear. The shared urea cycle abnormalities across diverse neurodegenerative diseases, despite their distinct clinical and neuropathological features, suggest a convergent pathway of urea metabolism dysregulation. This could stem from mitochondrial dysfunction, oxidative stress, or protein aggregation, all of which are known to disrupt cellular homeostasis in PD7,38. Additionally, the role of transcriptional dysregulation, as seen in HD where mutant huntingtin disrupts C/EBPα-mediated urea cycle enzyme expression7, may offer parallels for PD pathogenesis.
The increase in urea levels is not only related to the abnormally active urea metabolism, but also to the abnormal transport and clearance in the brain. Urea transporters (UTs) are a group of membrane channel proteins responsible for selective permeability to urea across cellular membranes39,40. Among them, UT-B is widely expressed in kidney, heart, brain, testis, urinary tract and other tissues41. Our previous study on UT-B knockout mice found that UT-B deficiency leads to urea accumulation in the brain, which causes neuromorphological changes, such as neuronal loss24,37,42. At the same time, through single-cell RNA sequencing and morphological observation, we found that UT-B was highly expressed in substantia nigra and other neurons, suggesting that UT-B may play an important role in the excretion of urea in substantia nigra neurons21. Therefore, this study found that the expression of UT-B in the nigrostriatal region, which regulates motor function, was enhanced in the PD model, suggesting that UT-B expression plays an important role in the regulation of motor function and may be an important mechanistic target in the pathogenesis of PD. Furthermore, the interaction between urea and neuroinflammation warrants attention. For instance, inhibition of urea transporter UT-B reduces pro-inflammatory mediators (e.g., TNF-α, NO) in microglia43, suggesting that urea transport may modulate neuroinflammatory cascades, a hallmark of PD and other neurodegenerative diseases.
However, there are some limitations of this study and some have not been fully determined. While our study establishes a strong association between urea cycle activation and PD, longitudinal studies are needed to determine whether urea dysregulation precedes or follows neurodegeneration. The translational relevance of animal models to human PD also requires validation, particularly regarding regional specificity of urea accumulation and its interaction with α-synuclein pathology. Furthermore, therapeutic strategies targeting urea cycle enzymes (e.g., ODC1 silencing) or polyamine metabolism, as demonstrated in AD17, should be explored in PD animal models to assess their potential to mitigate motor and cognitive deficits. Meanwhile, we should use A53T mice for the same experimental verification.
In conclusion, urea cycle activation and high urea emerge as a unifying metabolic signature in neurodegenerative diseases, with profound implications for PD pathogenesis. The dual role of urea, acting both as a compensatory mechanism for nitrogen detoxification and a source of neurotoxic intermediates, highlights its central yet paradoxical contribution to disease progression. Elucidating the interplay between urea metabolism and protein aggregation may unlock novel therapeutic avenues, emphasizing the need for biomarker development and targeted interventions in PD and related disorders.
Methods
Animals and drugs
Male C57BL/6 J mice (8 weeks old; 22–25 g) were used for experiments. The animals were housed in the Laboratory Animal Center of Chongqing Medical University, with 12-h light and dark cycles and the same sleeping conditions. The animal experiments conducted in this study were approved by the Chongqing Medical University Animal Care and Use Committee (approval number: IACUC-CQMU-2025-0611) and performed following institutional and national guidelines. All mice were euthanized under isoflurane anesthesia.
After 1 week of animal adaptation, mice were intraperitoneally injected with 1-methyl-4-phenyl-1,2,3, 6-tetrahydropyridine (MPTP; ST1020-100mg, Beyotime, Chongqing, China) at a dose of 20 mg/kg, once a day for 7 days; mice in the control group were given equal volume normal saline once a day for 7 days. During the modeling period, the changes in body weight and the tremor state were monitored and recorded. The modeling method was consistent with the conventional Parkinson’s modeling method26. After the end of behavioral study, mouse eye venous blood was collected in a centrifugal tube and serum was isolated. At the same time, the brain tissue was partitioned and sampled.
For human samples, all biochemical tests were performed at the Department of Neurology in the University-Town Hospital of Chongqing Medical University. Blood samples were collected by venipuncture into the heparin treated centrifuge tubes and placed statically for 30 min. Plasma was separated by centrifugation at 3000 rpm for 15 min. Serum chemistry values, which including serum creatinine level, urea level, were measured using serum biochemistry analyzer. All experimental operations were approved by the Ethics Sub-committee of the University Town Hospital affiliated to Chongqing Medical University (LL-202440).
Behavior analysis
Behavioral tests were performed on all mice after modeling. The grip strength test assessed forelimb strength by suspending mice by the tail, allowing them to grip a force transducer with their forepaws, and recording the peak force (in grams, g). For the open-field test, mice were acclimatized in the testing room for 30 min. The mice were then placed in the center of a 40 × 40 cm opaque plexiglass arena for 5 min under 500 lux illumination. Locomotor activity was recorded using SMART video tracking software. In the rotarod test, mice were placed on a rotating rod accelerating from 4 to 40 rpm over 5 min. After 3 days of training (4 rpm, 5 min/day), latency to fall was averaged over three trials with 1-h intervals.
Western blot
Appropriate amounts of brain tissue and cells that were rinsed with PBS were weighed and homogenized in lysis buffer containing protease inhibitors. After sonication, the tissue was placed on ice and centrifuged at 12,000 × g at 4 °C for 15 min. The supernatant was collected and the concentration was measured using the BCA kit (Beyotime, P0010). 25 μg aliquots of protein were separated by SDS-PAGE (Epizyme, PG122) and transferred to PVDF (Epizyme, WJ002) membranes. After incubation with Quick Block Western solution (Biosharp, BL506A), membranes were incubated with the following primary antibodies: anti-ARG1 (1:1000; Proteintech, #16001-1-AP), anti-ODC1 (1:1000; Abcam, ab193338), anti-OTC (1:1000; Abcam, ab203859), anti-UT-B (1:1000; CST, #44897S), anti-TH (1:1000; Proteintech, #25859-1-AP), anti-α-synuclein (1:1000; Proteintech, #66412-1-Ig), and anti-β-actin (1:1000; Abmart, P60709). Membranes were then incubated with secondary antibody (1:10000; anti-goat IgG, Proteintech, SA00001-2) at room temperature for 1.5 h. Membranes were extensively washed with TBST and protein bands were detected using a chemiluminescence imaging system. The band intensities of the target proteins were quantified using image analysis software ImageJ. The images were background subtracted to eliminate the influence of nonspecific signals. Regions of interest were delineated consistently. Normalization was performed by dividing the integrated density of target proteins by that of β-actin. If technical replicates were made for the same bio-sample, the average of their normalized values was taken as the final representative value for that bio-sample. These mean values were included in subsequent statistical analyses based on biological replicates. Normalized values were analyzed using statistical software.
HE and Nissl staining
Brain tissue was fixed with 4% paraformaldehyde, embedded in paraffin, and sectioned at 5 μm thickness for histological examination. Sections were dewaxed, rehydrated, stained with hematoxylin and eosin (HE) using a staining kit (Bioss, C02-04004) and Nissl stain using a separate kit (Bioss, S0198), then examined by light microscopy.
Immunohistochemical and immunofluorescence analysis
The following primary antibodies were used for immunohistochemical (IHC) and immunofluorescence (IF) analyses: anti-ODC1 (1:200; Abcam, ab193338), anti-OTC (1:200; Abcam, ab203859), anti-ARG1 (1:200; Proteintech, #16001-1-AP), anti-TH (1:200; Proteintech, #25859-1-AP), and anti-UT-B (1:200; CST, #44897S). Sections were incubated with primary antibodies at 4 °C overnight, washed three times with PBS, then incubated with secondary antibody (1:500) at 37 °C for 50 min. For IHC: Detection was performed using the biotin-streptavidin HRP system, followed by coverslipping. For IF: After secondary antibody incubation, sections were coverslipped with an antifade mounting medium containing DAPI (Beyotime, P0131). Fluorescence was imaged using a Nikon confocal laser scanning microscope with FITC and TRITC channels.
Urea, Ammonia and Putrescine assay
According to the kit (BioAssay, DIUR-500) instructions, urea levels in mouse brain tissue and cells were measured using BioAssay Systems. Following the kit (UpingBio, YPD1091) instructions, ammonia levels in mouse brain tissues and cells were measured. According to the kit (Gelatins, JLC_K6002-96T) instructions, Putrescine levels in mouse brain tissues and cells were measured.
Cell culture
PC12, SH-SY5Y, and Neuro-2a cells were cultured in high-glucose DMEM containing 10%FBS and 1% penicillin/streptomycin in a 37 °C, 5% CO2 incubator. C8-D1A cells were cultured in high-glucose DMEM medium containing 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin in a 37 °C, 5%CO2 incubator. The medium was refreshed every 24 h. When the cell confluence reached more than 90%, detached with 0.25% trypsin, the subsequent experiments were performed. The experiment was divided into two groups, a control group and an experimental group treated with 2500 μM MPTP, and further assays were conducted after 24 h of culture, respectively. At the same time, neurons were treated with different concentrations of urea for 24 h. PC12 cells were transfected with siODC1 for 72 h, and then the corresponding experiments were performed. Knockdown sequences were provided by Sangon Biotech Co., Ltd. (Shanghai, China), accession number: NM_013614.
RNA extraction and RT-qPCR
Total RNA was extracted from the cells for concentration and purity determination, cDNA was synthesized using a reverse transcription kit (Vazyme, R412-01) following the manufacturer’s protocol, and RT-qPCR was performed on a Bio-Rad system. The primers, provided by Sangon Biotech Co., Ltd. (Shanghai, China), are listed in Table 1.
Statistical analysis
All the data in the figure came from at least three independent repeated experiments. These data were expressed as mean ± SEM, and P < 0.05 was considered significant. *p < 0.05, **p < 0.01, ***p < 0.001. All statistical analyses were performed using GraphPad Prism 8.0 (GraphPad Software, Inc., San Diego, CA, USA).
Data availability
All authors agreed to be accountable for all aspects of work ensuring integrity and accuracy. Data is provided within the manuscript or supplementary information files.
References
Erkkinen, M. G., Kim, M. O. & Geschwind, M. D. Clinical neurology and epidemiology of the major neurodegenerative diseases. Cold Spring Harb. Perspect Biol. 10, a033118 (2018).
Hely, M. A. et al. The Sydney multicenter study of Parkinson’s disease: the inevitability of dementia at 20 years. Mov. Disord. 23, 837–844 (2008).
Braak, H. et al. Staging of brain pathology related to sporadic Parkinson’s disease. Neurobiol. Aging 24, 197–211 (2003).
Jellinger, K. A. Is Braak staging valid for all types of Parkinson’s disease?. J. Neural Transm. (Vienna) 126, 423–431 (2019).
Xu, J. et al. Graded perturbations of metabolism in multiple regions of human brain in Alzheimer’s disease: snapshot of a pervasive metabolic disorder. Biochim. Biophys. Acta 1862, 1084–1092 (2016).
Patassini, S. et al. Identification of elevated urea as a severe, ubiquitous metabolic defect in the brain of patients with Huntington’s disease. Biochem Biophys. Res. Commun. 468, 161–166 (2015).
Patassini, S. et al. Metabolite mapping reveals severe widespread perturbation of multiple metabolic processes in Huntington’s disease human brain. Biochim Biophys. Acta 1862, 1650–1662 (2016).
Handley, R. R. et al. Brain urea increase is an early Huntington’s disease pathogenic event observed in a prodromal transgenic sheep model and HD cases. Proc. Natl. Acad. Sci. USA 114, E11293–e302 (2017).
Glaab, E. et al. Integrative analysis of blood metabolomics and PET brain neuroimaging data for Parkinson’s disease. Neurobiol. Dis. 124, 555–562 (2019).
Hatano, T. et al. Identification of novel biomarkers for Parkinson’s disease by metabolomic technologies. J. Neurol. Neurosurg. Psychiatry 87, 295–301 (2016).
Trezzi, J. P. et al. Distinct metabolomic signature in cerebrospinal fluid in early parkinson’s disease. Mov. Disord. 32, 1401–1408 (2017).
Troisi, J. et al. A metabolomic signature of treated and drug-naïve patients with Parkinson’s disease: a pilot study. Metabolomics 15, 90 (2019).
Scholefield, M. et al. Severe and regionally widespread increases in tissue urea in the human brain represent a novel finding of pathogenic potential in Parkinson’s disease dementia. Front Mol. Neurosci. 14, 711396 (2021).
Morris, S. M. Jr. Regulation of enzymes of the urea cycle and arginine metabolism. Annu Rev. Nutr. 22, 87–105 (2002).
Bensemain, F. et al. Evidence for induction of the ornithine transcarbamylase expression in Alzheimer’s disease. Mol. Psychiatry 14, 106–116 (2009).
Prokop, J. W. et al. Emerging role of ODC1 in neurodevelopmental disorders and brain development. Genes (Basel) 12, 470 (2021).
Wang, H., Ran, J. & Jiang, T. Urea. Subcell. Biochem 73, 7–29 (2014).
Hansmannel, F. et al. Is the ornithine transcarbamylase gene a genetic determinant of Alzheimer’s disease?. Neurosci. Lett. 449, 76–80 (2009).
Neill, M. A. et al. Quantitative RT-PCR comparison of the urea and nitric oxide cycle gene transcripts in adult human tissues. Mol. Genet Metab. 97, 121–127 (2009).
Ju, Y. H. et al. Astrocytic urea cycle detoxifies Aβ-derived ammonia while impairing memory in Alzheimer’s disease. Cell Metab. 34, 1104–1120.e8 (2022).
Huang, B. et al. Expression of urea transporter B in normal and injured brain. Front Neuroanat. 15, 591726 (2021).
Castillo, E. et al. Comparative profiling of cortical gene expression in Alzheimer’s disease patients and mouse models demonstrates a link between amyloidosis and neuroinflammation. Sci. Rep. 7, 17762 (2017).
Sequerra, E. B. et al. Putrescine as an important source of GABA in the postnatal rat subventricular zone. Neuroscience 146, 489–493 (2007).
Wang, H. et al. High urea induces depression and LTP impairment through mTOR signalling suppression caused by carbamylation. EBioMedicine 48, 478–490 (2019).
Si, X. et al. Matrix metalloproteinase-9 inhibition prevents aquaporin-4 depolarization-mediated glymphatic dysfunction in Parkinson’s disease. J. Adv. Res 56, 125–136 (2024).
Jackson-Lewis, V. & Przedborski, S. Protocol for the MPTP mouse model of Parkinson’s disease. Nat. Protoc. 2, 141–151 (2007).
Vila, M. et al. Alpha-synuclein up-regulation in substantia nigra dopaminergic neurons following administration of the Parkinsonian toxin MPTP. J. Neurochem 74, 721–729 (2000).
Yao, Y. et al. Effect of eleutheroside E on an MPTP-induced Parkinson’s disease cell model and its mechanism. Molecules 28, 3820 (2023).
Poewe, W. et al. Parkinson disease. Nat. Rev. Dis. Prim. 3, 17013 (2017).
Jankovic, J. & Tan, E. K. Parkinson’s disease: etiopathogenesis and treatment. J. Neurol. Neurosurg. Psychiatry 91, 795–808 (2020).
Reich, S. G. & Savitt, J. M. Parkinson’s disease. Med Clin. North Am. 103, 337–350 (2019).
Li, S. M. et al. Neurotransmitter accumulation and Parkinson’s disease-like phenotype caused by anion channelrhodopsin opto-controlled astrocytic mitochondrial depolarization in substantia nigra pars compacta. MedComm (2020) 5, e568 (2024).
Surmeier, D. J., Obeso, J. A. & Halliday, G. M. Selective neuronal vulnerability in Parkinson disease. Nat. Rev. Neurosci. 18, 101–113 (2017).
Morrison, J. K. et al. Single-cell transcriptomics reveals conserved cell identities and fibrogenic phenotypes in zebrafish and human liver. Hepatol. Commun. 6, 1711–1724 (2022).
Liu, P., Jing, Y. & Zhang, H. Age-related changes in arginine and its metabolites in memory-associated brain structures. Neuroscience 164, 611–628 (2009).
Natesan, V., Mani, R. & Arumugam, R. Clinical aspects of urea cycle dysfunction and altered brain energy metabolism on modulation of glutamate receptors and transporters in acute and chronic hyperammonemia. Biomed. Pharmacother. 81, 192–202 (2016).
Huang, B. et al. High urea induces anxiety disorders associated with chronic kidney disease by promoting abnormal proliferation of OPC in amygdala. Eur. J. Pharm. 957, 175905 (2023).
Mattson, M. P. & Arumugam, T. V. Hallmarks of brain aging: adaptive and pathological modification by metabolic states. Cell Metab. 27, 1176–1199 (2018).
Stewart, G. The emerging physiological roles of the SLC14A family of urea transporters. Br. J. Pharm. 164, 1780–1792 (2011).
Lucien, N. et al. UT-B1 urea transporter is expressed along the urinary and gastrointestinal tracts of the mouse. Am. J. Physiol. Regul. Integr. Comp. Physiol. 288, R1046–R1056 (2005).
Li, X., Chen, G. & Yang, B. Urea transporter physiology studied in knockout mice. Front Physiol. 3, 217 (2012).
Zhao, X. et al. High urea promotes mitochondrial fission and functional impairments in astrocytes inducing anxiety-like behavior in chronic kidney disease mice. Metab. Brain Dis. 40, 186 (2025).
Jones, A. C. et al. Inhibition of urea transporter (UT)-B modulates LPS-induced inflammatory responses in BV2 microglia and N2a neuroblastoma cells. Neurochem Res. 46, 1322–1329 (2021).
Acknowledgements
The authors acknowledge the support received from the foundation. This study was supported by the Key projects of Chongqing Key Laboratory of Development and Utilization of Genuine Medicinal Materials in Three Gorges Reservoir Area (KFKT2022001,to Jianhua Ran; KFKT2022010, to Boyue Huang); the National Natural Science Foundation of China grants (81770738, 82370739, to Jianhua Ran); Chongqing Science and Technology Commission(CSTB2023NSCQ-MSX0510,to Jianhua Ran); Science Foundation of Chongqing Education Commission (KJQN202400417, to Boyue Huang); Chongqing Natural Science Foundation (Chongqing Science and Technology Development Foundation) (CSTB2024NSCQ-KJFZMSX0075, to Boyue Huang); the Key Project of Chongqing Key Laboratory of Development and Utilization of Genuine Medicinal Materials in Three Gorges Reservoir Area (Sys20210008 to Dilong Chen); Innovative Research Group Project of Natural Drug Antitumor of Chongqing Municipal Education Commission (CXQT20030 to Dilong Chen); Chongqing Talent Plan Project (cstc2022ycjh-bgzxm0226 to Dilong Chen); The Science and Technology Research Program of Chongqing Municipal Education Commission (KJZD-M202202701, to Dilong Chen).
Author information
Authors and Affiliations
Contributions
Shengyao Zhang: writing—original draft, formal analysis, visualization. Guoran Wan: investigation, formal analysis. Yu Qiu: investigation. Meng Zhang: investigation, formal analysis. Hongmei Deng: formal analysis. Qiongfang Wang: investigation. Junyi Hu: formal analysis. Jie Gui: formal analysis. Dilong Chen: visualization, funding acquisition. Boyue Huang: visualization, funding acquisition, writing—review and editing, supervision. Jianhua Ran: visualization, funding acquisition, supervision, conceptualization. All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhang, S., Wan, G., Qiu, Y. et al. Urea cycle dysregulation drives metabolic stress and neurodegeneration in Parkinson’s disease. npj Parkinsons Dis. 11, 237 (2025). https://doi.org/10.1038/s41531-025-01099-5
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41531-025-01099-5