Abstract
Evaluation of recombinant HIV-1 surface glycoproteins (Env) as vaccine candidates for Phase I human experimental trials often requires production of cGMP-grade well-ordered Env trimers. Here, we report an accelerated cGMP compatible approach for expression and purification of a stabilized HIV clade C-derived trimer ‘16055 DG4 NFL’ (for native flexibly linked). This recombinant trimer was expressed from CHO-S™ cells using a MaxCyte® VLX™ electroporation-based transient transfection process. The 16055 DG4 NFL was designed with multiple internal stabilizing mutations and, as well, deletion of four N-linked glycans (DG4) proximal to the CD4 binding site (CD4bs) engineered to improve B cell recognition of this conserved neutralizing determinant. The transient process circumvents the need to develop stable cell lines expressing the Env trimers that is often the most time-consuming step impacting vaccine development timelines. The 16055 DG4 NFL trimer was purified by immunoaffinity chromatography using the broadly neutralizing antibody (bNAb), PGT145. Following additional downstream processing steps, purified trimer was vialed, frozen and stored at –80 °C. Upon thaw and analysis, the trimer displayed homogeneity and a near-native conformation as determined by size-exclusion chromatography (SEC), negative stain and cryo-electron microscopy (EM), differential scanning calorimetry (DSC) and biolayer interferometry (BLI). The immunogenicity of the trimer was tested in rabbits with bolus, escalating dose and divided dose immunization regimens. Rabbits from all three regimens elicited tier 2 autologous neutralizing antibodies that targeted the exposed protein region at the CD4bs. The trimer is currently under investigation in a human clinical trial (NCT06332339) for safety, tolerability and as a priming candidate followed by heterologous boosting to potentially elicit cross-neutralizing antibodies.
Similar content being viewed by others
Introduction
Development of an HIV-1 vaccine targeting the surface envelope glycoprotein (Env) is facilitated by human clinical trials that historically have used recombinant Env-based proteins (the gp120 subunit) and more recently near-native Env trimers1,2,3,4,5. For the metastable HIV-1 Env trimer, structural integrity that mimics the native prefusion trimer spike is an additional hurdle compared to some less conformationally complex vaccine candidates. The clinical testing of the more traditional protein-based Env vaccines faces a more time-consuming process to produce native-like, homogenous, and stable trimers at large scale under cGMP conditions. Traditional practices for cGMP production include a time consuming (5–8 months) initial screening process for a stable Chinese Hamster Ovary (CHO) cell line that expresses the antigen of interest6,7. To accelerate the timeline from concept to clinical testing, production of smaller amounts of antigen (i.e., Env) by recombinant transfection can facilitate initial toxicity, stability, efficacy studies and Investigational New Drug filing. Transient expression of Env can also be used to assess initial efficacy in small animal models towards establishing safety, dose and tolerability in phase I clinical trials in humans. If the Phase I immunogenicity data are compelling, transient expression can be followed stable CHO cell line expression to generate sufficient protein to perform Phase II or phase III efficacy trials8. In the context of HIV-1 Env phase I candidate vaccine testing, the time-consuming step of CHO cell line development can be circumvented by using a structure-based, stabilized trimer candidate expressed at high yield and conforming to cGMP standards by 50-70 L volume transient transfection in CHO cells as we show here.
The recent success of mRNA-based vaccines against SARS-CoV2 has also sparked interest in evaluating Env trimers expressed from mRNA lipid nanoparticles9,10. Selected Env-based candidate vaccines expressed from mRNA are being evaluated in Phase I clinical trials11,12. This technology presents another alternative that may possess some advantages over the recombinant protein option discussed here. However, the initial trials have encountered unanticipated safety concerns11,12 highlighting that other time-saving approaches may be warranted to develop in parallel.
The HIV-1 Env used in this study was derived from the sequence of the Indian subtype C 16055 strain isolated following acute infection13. We first modified the native Env sequence by genetically introducing a flexible linker between the gp120 and gp41 subunits to eliminate the need for furin cleavage while maintaining native-like confirmation. This native flexibly linked (NFL) trimer design consists of a 10 amino acid residue (GGGGSGGGGS) linker between the REKR-deleted gp120 C-terminus and the unmodified gp41 N-terminus14. The trimer also contains an I559P mutation in gp41 to enhance stability by favoring the prefusion Env state15. The soluble trimer is truncated at residue 664 to eliminate hydrophobic residues in the membrane-proximal external region that contribute to aggregation16,17. The further stabilized 16055 NFL TD construct (TD for BG505 trimer-derived) possesses additional internal substitutions derived from the BG505 Env sequence E47D, K49E, V65K, E106T, I165L, E429R, R432Q, A500R that enhances trimer formation along with two helix-breaking glycine substitutions T569G and S636G that increase homogeneity and yield, and a V3 and two fusion peptide mutations N302Y, F519R and L520R that increase thermostability18,19. Additionally, a structure-guided intra-protomer disulfide I201C-A433C (CC) prevents the substantial CD4-induced conformational rearrangements usually triggered by Env engagement of human CD4. These “off-target” rearrangements expose non-neutralizing determinants not desired in a pre-fusion-based HIV Env vaccine. This trimer also has the N-glycan at residue N332 restored by a K334T mutation19,20,21 that is a major element of the N332 glycan bNAb supersite.
The HIV-1 Env trimer has naturally evolved to be extremely well-shielded by ‘self-glycans’, restricting B cell recognition of the underlying conserved neutralizing protein determinants22,23. Pre-clinically, we designed and tested several 16055 NFL trimer variants with potential N-glycan sites (PNGS) proximal to the CD4 binding site (CD4bs) selectively deleted to increase recognition by naïve B cells in vivo. Rabbit immunogenicity experiments comparing glycan-deleted to fully glycosylated NFL trimers reveal that the PNGS-deleted trimers more rapidly elicited neutralizing antibodies for CD4bs-PNGS-deleted viruses and more potent responses against fully glycosylated virus24. Part of this activity is CD4bs-directed and can be boosted with fully glycosylated trimers, indicating that targeted N-glycan deletions is a promising approach to more efficiently elicit antibodies directed toward the conserved CD4bs24,25. Accordingly, we designed the 16055 NFL DG4 variant by deleting four PNGS proximal to the CD4bs (N276Q, N301Q, N360Q, and N463Q) to genetically eliminate these N-glycans and focus B cell responses to this exposed region. Several other studies have deleted N-glycan sites and/or simultaneously enriched for shorter Man5 glycans proximal to the CD4bs to expose this region for improved immune recognition26,27,28,29. Deletion varied from a single N-glycan removal at residue N276 to removal of four glycans at positions N197, N276, N362/N363 or N460/N461/N462/N463, respectively. Glycan deletions in the 16055 DG4 NFL trimer were chosen by inspection of the fully glycosylated high-resolution Env structure to permit B-cell receptor access to the CD4bs from selected angles of approach to generate effective polyclonal antibody responses to this site. Genetic removal of N-glycan at position N301, unique to our trimer design, was engineered to allow for the potential elicitation of b12-like antibodies to the CD4bs in addition to permitting the limited angles of access revealed by VRC01-like antibodies29,30.
Here, we describe in detail the large-scale production of the 16055 DG4 NFL trimer. This process begins with transient transfection of plasmid DNA by electroporation (EP) of CHO cells and proceeds through downstream purification to cGMP standards. The production of the trimer by these transient means was guided by National Institute of Allergy and Infectious Diseases (NIAID) personnel and Advanced Bioscience Laboratories Inc. (ABL) with much of the technical guidance for downstream processes emanating from International AIDS Vaccine Initiative (IAVI). Duke Human Vaccine Initiative (DHVI) expertise also contributed to the successful trimer production reported here. The trimer has been very recently used as a prime in a fully enrolled human clinical trial (NCT06332339, HVTN 313). Using an interim process development material, we characterize the 16055 DG4 NFL trimer in vitro by multiple biophysical techniques and present the molecular structure obtained by cryo-electron microscopy (EM) to 3.8 Å resolution. We further show the in vivo antibody responses elicited by the 16055 DG4 NFL trimers in mice and rabbits using different immunization regimens. These preclinical studies confirm that bolus immunization with this recombinant material effectively primes responses to the Env trimer CD4bs for further heterologous boosting. Collectively these data guide human clinical trial design for potential elicitation of cross-neutralizing antibodies.
Results
Overall process description
The overall trimer production pipeline is divided into three parts (A) DNA production (B) upstream transfection, trimer expression and (C) downstream trimer purification, characterization and vialing. DNA synthesis of 16055 DG4 NFL was done by DNA 2.0 ATUM (Newark, CA), and large-scale plasmid DNA for transfection was prepared at Aldevron (Fargo, ND). The upstream production used a fed-batch process following an EP-based transient transfection method optimized for CHO-S™ using a MaxCyte® VLX™ large-scale transfection system. The platform was initially developed for the production of the HIV-1 CH505 gp120 immunogen for the HVTN123 clinical trial and was then adapted for the production of the 16055 DG4 NFL trimers described here31. Downstream purification used the conformationally sensitive, apex-directed broadly neutralizing antibody (bNAb), PGT145, for the immunoaffinity chromatography capture step followed by further polishing. PGT145 recognizes the quaternary structure at the apex of the trimer and was selected to specifically capture the near-native form of the 16055 DG4 NFL trimer that displays multiple broadly neutralizing determinants32. Covalently linked resin-bNAb matrix combinations were compared for the immunoaffinity capture and elution of 16055 DG4 NFL using PGT145. The downstream purification process for 16055 DG4 NFL used a purification process adapted from the GMP production of the HIV-1 BG505 SOSIP Env trimer33. A summary flowchart of the process steps for the clinical lot of 16055 DG4 NFL trimer is shown in Fig. 1.
16055 DG4 NFL plasmid DNA was produced from an expanded culture from an E. coli MCB. Plasmid DNA suitable for use in GMP was purified in a two-column chromatography process using Aldevron’s GMP-Source™ service (left panel). 16055 DG4 NFL was produced in CHO-S using a MaxCyte® VLX™ transfection system under GMP standards (middle panel). 16055 DG4 NFL trimers were purified from the clarified media harvest using a four-step chromatography method. Purification includes an immunoaffinity capture step using bNAb PGT145 to recover properly folded trimers, followed by three additional chromatography and further polishing steps (right panel).
The 16055 DG4 NFL trimer is expressed at large-scale following EP-mediated transient transfection
Transient transfections of CHO cells with the Env-expressing plasmid DNA were performed by EP using MaxCyte® STX™ and VLX™ transfection systems (MaxCyte, Rockville, MD). The STX™ and VLX™ are designed using a flow-based process specifically designed to facilitate a range of small- to large-scale (Fig. 2) EP processes. By design, small- to mid-scale transfections optimized on the STX™ are scalable for large-scale production using the VLX™. 16055 DG4 NFL trimer batches produced in large-scale are presented in Supplementary Table 1. Small-scale transfection verification runs were performed in shake flasks to assess the best plasmid DNA concentration for transfection in concert with cell growth characterization to determine the optimal process parameters to produce the 16055 DG4 NFL trimer. Additionally, 2, 10, and 50 L scale assessments were performed to establish the best harvest procedures, establish and verify the scalability of operational parameters in the bioreactor environment and establish acceptable parameter ranges for the cGMP production run (see Supplementary Table 2 for details). Initially, A CHO-S™ cGMP Master Cell Bank (MCB) derived from cGMP-banked suspension-adapted CHO-S™ cells was tested for identity, viability, sterility, virus-like particles and adventitious agents and subsequently released for cGMP use. Research and development grade plasmid was used for initial small-scale testing to optimize expression and established ‘best conditions’ were used for subsequent 10 and 50 L scale-up runs. GMP-Source™ grade plasmid was used for the non-GMP full-scale engineering run (70 L harvest volume) and this analysis was utilized to assess product yield, quality and process parameters prior to initiation of full-scale cGMP production (Supplementary Table 2). The cGMP run yielded 79.6 L of clarified 16055 DG4 NFL trimer harvest with a protein titer of 75.4 mg/L. The clarified harvest was partitioned into 4 L aliquots and safely stepped down to 2-8 °C and following freezing, stored at −70 to −90 °C. Comparison of the process parameters and outcomes for the 50 L Demo, 70 L engineering run, and 70 L cGMP run is included in Supplementary Table 2.
The high-density cell slurry was mixed with 16055 DG4 NFL plasmid DNA and electroporated using duplicate MaxCyte® VLX™ process assemblies. Transfected cells were rested in cell factories then expanded to 5 × 106 cells/mL in 63 L of production media. Fed batch culture was maintained for eight days, and media was harvested for downstream purification.
The PGT145 bNAb is selected over the 2G12 mAb for immunoaffinity purification
The human bNAbs PGT145 and 2G12 were previously evaluated in the context of HIV-1 Env trimers to assess their suitability for use in immunoaffinity chromatographic columns for GMP manufacturing33,34. For example, immunoaffinity purification of trimeric gp140 using 2G12 affinity capture was previously demonstrated to be suitable for GMP manufacturing33. The 2G12 antibody recognizes a complex glycan epitope on the surface of the gp120 subunit and binds with high affinity to HIV-1 Env35. However, due to the non-conformational nature of its epitope, the bNAb 2G12 does not discriminate between the desired well-ordered HIV-1 gp140 trimers from “off-target” monomeric or non-native higher-order forms. In contrast, PGT145 binds specifically to a quaternary epitope involving the apex V1/V2 regions of the HIV-1 Env trimer. This conformationally sensitive antibody binds at the center axis of the trimer, making contacts with all three gp140 monomers comprising the native trimer32, and is highly specific in recognition of near-native trimers over other disordered forms. Accordingly, PGT145- and 2G12-conjugated resins were scaled up for testing in a head-to-head comparison as a key step in the purification process for the 16055 DG4 NFL trimer. PGT145 and 2G12 were covalently conjugated onto two different support media resins using similar lysine chemistry (see Methods), Tresyl and Poros (Toyopearl AF-Tresyl-650M (Tosoh Bioscience Inc.), Poros EP-450 (ThermoFisher Scientific), respectively) to evaluate suitability for GMP manufacturing. Following conjugation, the cross-linked bNAb-resins were evaluated using purified 16055 DG4 NFL reference material (Scripps Research). Four resin-bNAb combinations, Poros-2G12, Poros-PGT145, Tresyl-2G12, and Tresyl-PGT145, were evaluated using concentrated cell culture harvest loaded at a concentration of 1 g/L in a column format (Supplementary Fig. 1a, b). The Tresyl-PGT145 affinity resin most efficiently captured soluble 16055 DG4 NFL trimers from the supernatant, with no detectable flow-through. Following elution from the resin by acidic pH buffer, the trimers from the Tresyl-PGT145 appeared to be stable and homogenous with no detectable breakdown products as determined by gel analysis (Supplementary Fig. 1a). Performance and stability of the conjugated resin were further evaluated for static and dynamic binding capacity and antibody leaching. The Tresyl-PGT145 resin was selected based on the evaluation of the product quality and yield compared to the other bNAb-resin combinations for immunoaffinity chromatography purification. Following affinity resin selection, the process was executed at a lab-scale using 24 L of transiently transfected CHO-S™ cell culture harvest to evaluate performance. The Tresyl-PGT145 affinity resin demonstrated excellent performance to purify the 16055 DG4 NFL trimer and was selected for the immunoaffinity capture step in the downstream GMP process.
The 16055 DG4 NFL trimer is purified to high homogeneity, yield and validated for viral clearance
Immunoaffinity purification was successfully performed on the 16055 DG4 NFL from a 50 L demo run harvest, followed by a 70 L engineering run harvest before cGMP production from the 70 L cGMP harvest. The demo run harvest lot was analyzed using the preliminary specifications established during the initial process development runs to characterize the overall purification processes end-to-end (see Supplementary Table 3). Purification from the 70 L engineering run was carried out to confirm scale-up parameters, specifications under GMP conditions, and provide purified trimer for the reference standard, stability, viral clearance, in vitro characterization, and immunogenicity studies in small animals. The cGMP trimer was produced to provide material for the extended stability study, proposed pre-clinical and clinical studies (HVTN 313). The process yielded 0.857 g of the trimer from a 75.40 L of clarified harvest for a 20% process recovery of the cGMP run. Comparison of trimer purification parameters and outcomes from the three runs is shown in Supplementary Table 3.
To validate the safety of the final trimer product, the cell line and raw materials were analyzed, with no contaminants detected. The purified trimer was tested at appropriate steps for contaminating viruses that were not detected above background. The capacity of the downstream processes to clear infectious viruses was also confirmed as follows. In several steps of the purification process, we ‘spiked’ murine leukemia virus to quantitatively assess the successful clearance of this virus to undetectable levels. The viral clearance study was designed to meet the requirements of the Committee for Proprietary Medicinal Products (CPMP) and regulatory authorities that recommend 3–5 logs of excess clearance for retrovirus-like particles (RVLP). The overall process clearance for the xenotropic murine leukemia virus (XMuLV), which is an enveloped virus and a model virus for RVLP, was 14.08 log10 reduction value (LRV). The process clearance for mouse minute virus (MMV), a small nonenveloped model virus, was 7.55 LRV. This analysis demonstrated that the NFL trimer downstream process resulted in an excess clearance of both endogenous RVLPs and adventitious viruses respectively exceeding the International Conference on Harmonization (ICH) guidelines (https://www.ema.europa.eu/en/documents/scientific-guideline/ich-q-5-r2-viral-safety-evaluation-biotechnology-products-derived-cell-lines-human-or-animal-origin-step-2b_en.pdf). “Trace” residual plasmid DNA was detected in the final drug product at a concentration of 7.86E + 03 copies/mL (equivalent to 5.17E-05 ng/ml). This trace DNA amount is substantially below the FDA safety limit of 10 ng/dose rendering the likelihood of undesired spontaneous transfection, DNA persistence or integration very low36,37,38,39. The final release parameters for the 16055 DG4 NFL trimer drug product are shown in Supplementary Table 4.
The 16055 DG4 NFL is stable and displays favorable biophysical characteristics
Stability program for the 16055 DG4 NFL trimer was established in accordance with Food and Drug Administration (FDA) and ICH guidelines. The trimer was evaluated at −80 ± 10 °C, 5 ± 3 °C and 25 ± 2 °C for strength, quality, identity, purity, safety, and potency at planned intervals (Supplementary Table 5). Available results demonstrated that the cGMP trimer is stable at -80 ± 10 °C for up to 36 months and accelerated stability studies indicated that 16055 DG4 NFL trimer stable at 5 ± 3 °C and 25 ± 2 °C for up to 1 month and 2 weeks, respectively. The stability program is currently ongoing.
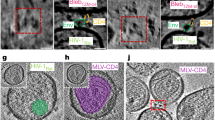
A 1 mg frozen aliquot of the 16055 DG4 NFL trimer engineering run material was thawed and characterized for trimer integrity and binding characteristics (Fig. 3). The protein consisted of a major trimer peak and a smaller peak of dimer-of trimers as resolved by SEC and Blue-native PAGE (Fig. 3A, B and Supplementary Fig. 1c). The presence of the dimer-of-trimer species was confirmed by negative stain EM (nsEM) that showed two trimer molecules associating via the C-terminal tail region (Supplementary Fig. 1d, e). The major trimer peak from SEC consists of >99% of well-ordered native-like trimers as determined by nsEM (Fig. 3C). The homogeneity of the trimer was further reflected in a sharp melting curve of the major SEC peak determined by DSC (Fig. 3D). Next, we tested the ability of the trimer to be recognized by bNAbs and non-bNAbs by BLI. The trimer was efficiently recognized by the bNAbs PGT145, PGDM1400 and PG9 (apex-directed); BG18 and PGT121 (N332 glycan patch-directed); VRC01 and VRC03 (CD4bs-directed) and 3BC315 (gp120-gp41 interface-directed) that target epitopes spanning Env major sites of vulnerability from the apex to the base (Fig. 3E). In contrast, the trimer was not recognized by the non-bNAbs F105, b6 (CD4bs-directed) and 447-52D (V3-directed, Fig. 3F). Taken together, this favorable antigenic profile indicates maintenance of the conformational integrity of the 16055 DG4 NFL trimer. In addition, the trimer maintained a favorable antigenic profile for 2 weeks when stored at 4 °C (Supplementary Fig. 2a). Due to the presence of the engineered intra-protomer disulfide I201C-A433C (CC) in the pre-receptor conformation of the 16055 DG4 NFL we confirmed that this trimer was resistant to conformational changes induced by engagement with soluble human CD4 (sCD4) as determined by the lack of binding by the bridging sheet-directed mAb, 17b (Supplementary Fig. 2b). The presence of this disulfide renders the NFL trimers resistant to CD4-induced conformational changes by human CD4 as shown previously18,20 and is an important consideration to maintain trimer integrity following inoculation into humans.
A Analytical SEC was performed using a Superdex 200 Increase column. B Blue-native PAGE (BN-PAGE). Uncropped gel image is shown in Supplemental Fig. 1c. C 2D class averages of the major trimer peak by negative stain EM. D DSC analysis showed that the trimer melts at 75 °C as a single peak. E The trimer displayed a favorable antigenic profile as determined BLI and binds to a panel of bNAbs targeting selected cross-neutralizing epitopes on the trimer from the gp120 apex toward the gp41 base. F Minimal binding was observed with a panel of non-bNAbs.
High-resolution structural and glycan analysis confirms favorable features of the 16055 DG4 NFL trimer
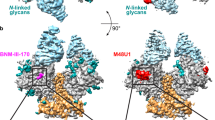
The 16055 DG4 NFL engineering run material was next assessed by cryo-electron microscopy (cryoEM). 2D and 3D data processing steps also confirmed a predominant trimer and a minor subpopulation of dimer-of-trimers but due to varying stoichiometries (single trimers or dimers-of-trimers) and apparent flexibility between the dimers, final refinements were performed using a soft mask over a single trimer, resulting in a 3.8 Å C3-symmetric reconstruction (Fig. 4 and Supplementary Table 6). The cryoEM model confirmed the presence of the engineered stabilizing mutations as well as the absence of the genetically introduced glycan deletions and the presence of the remaining natural N-linked glycans (Fig. 4D). The observed 16055 DG4 NFL stabilizing mutations included the gp120 intra-protomer disulfide bridge I201C-A433C (CC). Overall, the structure of the 16055 DG4 NFL was very similar to the near-native fully glycosylated trimer. A comparison of the cryoEM structure of 16055 DG4 NFL and crystal structure of fully glycosylated 16055 NFL trimer (PDB 5UM8, RMSD = 0.72 Å) is shown in Supplementary Fig. 3 with the positions of the four deleted glycans surrounding the CD4 binding site (CD4bs) highlighted.
A Fourier Shell Correlation resolution estimate. B local resolution estimation (in units Å) C angular distribution of observations. D Cryo-EM structure of 16055 DG4 NFL trimer with protein design elements highlighted. The three monomers comprising the trimer are shown as pink, green and gray ribbons. Protein design elements include: intra-protomer disulfide bridge I201C-A433C (CC) shown in yellow, trimer-derived (TD) substitutions shown as blue balls, proline and glycine substitutions in gp41 shown as magenta balls, asparagine to glutamine (N to Q) substitutions in the CD4 binding site shown as green balls (glycan knockouts, KO), and gp120 V3 and gp41 fusion peptide (V3-FP) mutations shown in red.
We next analyzed the glycan profile of the 16055 DG4 NFL trimer by mass spectrometry. We compared this profile to that of 16055 DG4 NFL trimers expressed in Thermo Fisher Scientific’s FreeStyle™ 293-F cells (HEK 293 F) cells and to the fully glycosylated 16055 NFL trimer expressed in HEK 293 F cells (Fig. 5). The glycan occupancy was 88, 91, and 83% respectively at N-glycosylation sites shared by all trimer-types, although the differences were not statistically significant at any of the N-glycosylation sites as analyzed by the Mann-Whitney U test with Benjamini-Hochberg correction40. We did observe statistically significant increases in the proportion of complex N-glycans and decreases in high mannose N-glycans at N88 position in the CHO cell-produced 16055 DG4 NFL compared to 16055 WT NFL. In addition, statistically significant increase in the proportion of complex N-glycans at N187 in 16055 DG4 NFL expressed from 293 F cells, compared to 16055 WT NFL was observed. Of interest, the glycan at position N625 is completely occupied in 16055 DG4 NFL expressed in CHO cells and highly occupied in trimers expressed in 293 F cells. N625 is one of the three N-glycosylation sites at the base of the HIV-1 Env trimer, and an underoccupancy of glycans in that position is known to elicit off-target binding and non-neutralizing antibodies directed towards the exposed trimer base41,42,43.
The 16055 DG4 NFL trimer is immunogenic in mice
To evaluate the in vivo immunogenicity of the 16055 DG4 NFL trimer, we inoculated mice with trimer in buffer (PBS) compared to trimers adjuvanted with either Alum (Alhydrogel, InvivoGen) or MPLA + QS21 (Avanti Polar Lipids; Desert King). As expected, no detectable binding antibodies were elicited in mice immunized with the trimer in PBS as determined by ELISA. In contrast, two weeks following the first priming immunization, mice immunized with MPLA + QS21 adjuvant elicited binding serum antibodies directed against the trimer immunogen (Supplementary Fig. 4). The magnitude of the binding antibodies increased at 4 weeks following priming immunization, further increased with a trimer boost and plateaued after the third immunization. Mice immunized with the trimer and Alum as the adjuvant elicited weak binding antibodies after two immunizations that were statistically lower than the animals in the group immunized with MPLA + QS21 adjuvant. The statistically significant difference was detected as well after the third immunization, indicating the improved responses were generated using the MPLA + QS21 adjuvant combination. The QS21 component of the adjuvant is saponin-based and such adjuvants are promising to render the heavily glycosylated Env more immunogenic44,45.
We then tested if the serum possessed neutralizing activity against the antigen-matched 16055 DG4 pseudo-virus by the standard TZM-bl assay46. Neutralization was detected in serum from two mice immunized with MPLA + QS21 after two immunizations. Following a third immunization, more potent neutralization was detected in serum from three mice from the same group (Supplementary Fig. 5). None of the animals from any of the three groups neutralized the ‘tier 1’’ MN.3 virus suggesting that the off-target V3 region was not exposed by either of the two adjuvants tested in vivo. The favorable sequestration of the V3 region can partly be attributed to the mutations designed to stabilize the trimer and expression in CHO cells that result in a homogenous, well-folded trimer as determined biophysically in vitro that remains in that state in vivo.
The 16055 DG4 NFL trimer elicits robust binding and autologous neutralizing antibodies in rabbits by multiple regimens
We evaluated the immunogenicity of the 16055 DG4 NFL trimer in rabbits as follows. When we started these experiments, the objectives were two-fold: to preclinically evaluate a potential immunization regimen for use of this trimer in an upcoming clinical trial (HVTN 313), and to switch to the use of a saponin-based adjuvant under development for the clinic, called SMNP. The ISCOM-based SMNP, containing saponin, also contains the TLR4 agonist, MPLA, incorporated into lecithin/cholesterol induced nanocages44. The rabbits were immunized with a total of 100 μg of the 16055 DG4 NFL trimer in 375 μg SMNP adjuvant over a period of 8 weeks. Rabbits were administered the trimer by three different regimens each using the same total amount of protein in each prime-boost schema for this head-to-head comparison.
The first regimen was done as ‘bolus’’ injections (prime and boost immunization, 50 μg each at 0 and 8 weeks), the second by ‘low-dose escalation’’ (seven low-dose to high-dose inoculations; as described elsewhere as escalating dose) and the third by what we termed a ‘divided dose’’ regimen (five inoculations of 20 μg each, two weeks apart)47,48. The immunization intervals and dose details are shown in Supplementary Table 7. The amount of adjuvant was adjusted accordingly to the quantity of the immunogen protein dose when applicable. Test bleeds were analyzed for binding antibodies and serum neutralization every two weeks following the initial priming inoculation.
Two weeks after the first administration, 23/24 rabbits elicited binding antibodies against 16055 DG4 NFL trimer indicating that the antigen is immunogenic in rabbits when formulated with SMNP adjuvant (Fig. 6). The binding titers revealed a trend that showed escalating dose > divided dose > bolus dose but the differences between the three regimens were not statistically significant. However, at the next three sampling points up to week 8, the binding titers revealed a trend divided dose > escalating dose > bolus dose. The binding titers of the divided dose group were statistically significantly higher than the bolus group (week 6, 8) and escalating dose (week 6) respectively. To note, the divided dose group of rabbits were immunized with 60 μg and 80 μg of the trimer by week 6 and 8 respectively in contrast to 50 μg of trimer for the bolus and escalating dose groups, consistent with the relative binding titers. Two weeks following the final protein/adjuvant boost in this comparison (week 10, all three groups), binding titers were similar across all the regimens. The escalating dose mean ED50 was slightly higher than bolus or divided dose, but differences between the immunization regimens were not statistically significant (Fig. 6).
In parallel, we assessed the ability of the rabbit sera to neutralize both the four CD4bs-glycan-deleted 16055 and 16055 wild-type pseudo-viruses. No neutralization of either virus was detected in the pre-bleeds and bleeds at 2 weeks following the initial inoculation. Weak to moderate neutralization activity was detected in rabbits from all the groups by week 4 (ID50 values; Fig. 7). Like the binding titers, the neutralization titers followed the trend divided dose > escalating dose > bolus group at weeks 4, 6, and 8, respectively. These patterns are likely explained by the differences in the inoculated total dose and timing of antigen administered to animals in each group. Two weeks after the final immunization (week 10), the neutralization titers of the bolus and escalating dose increased to similar levels to those of the divided dose group. We investigated if any of the antibodies elicited by the 16055 DG4 NFL trimers also neutralized the glycan restored wild-type pseudovirus. Two rabbits, one in each of bolus and divided dose groups weakly neutralized the wild-type pseudo virus at weeks 6 and 8 (Supplementary Fig. 6). Neutralization improved following the boosting immunization with sera from five or six rabbits from all three groups registering weak to moderate ID50 titers at 2 weeks following immunization.
Immunizations are shown as spheres. The neutralization titers for the escalating dose and divided dose groups were significantly higher than the bolus group at weeks 6 and 8 (p < 0.01); the divided dose group is significantly higher than the bolus group at week 8 (p < 0.05). At week 10, two weeks after the boosting dose at week 8, all the groups registered improved but statistically non-significantly different ID50 neutralization titers. We further analyzed neutralization titers at weeks 18 and 20, detecting decreases in the ID50 values (10 and 12 weeks following the last boost).
Discussion
The soluble HIV-1 Env is a metastable trimer that has been the focus of pre-clinical and clinical vaccine design and development49. The successful expression and purification of the well-ordered trimers to high homogeneity over the last two decades was made possible by numerous structured-based stabilizing mutations in both the gp120 and gp41 subdomains15,16,18,50. Recent efforts to produce a stabilized clade A-based BG505 SOSIP gp140 trimer for clinical studies involved the development of stable 293 T and CHO cell lines33,51. The SOSIP trimer design requires co-transfection (and optimization) of furin to obtain full cleavage of the gp120 and gp41 domains and subsequent well-ordered conformation. The “single chain” NFL platform used here has the advantage of being furin/cleavage-independent and more suited for large-scale production by transient transfection of a single gene. Here, we show that the stabilized 16055 DG4 NFL trimer design facilitated the successful cGMP large-scale expression and purification following transient transfection using the electroporation method. The homogeneity and stability of the trimer expressed in a large scale is remarkable, as it is likely expressed from multiple independent cells in parallel following the transient transfection. The frozen and then thawed trimer maintains conformational integrity and can be stored for at least another 2 weeks at room temperature (RT) with no loss of structural integrity. The improved shelf life has favorable practical implications as a clinical candidate in developing countries with limited access to cold chain storage.
Once a candidate vaccine antigen has been identified and developed, sponsors and vaccine developers face several critical decisions, including selection of vaccine modality, time to production, and manufacturing costs for early development of Phase I GMP material. One of the unique opportunities the COVID-19 pandemic presented was the ability to compare multiple vaccine platforms, including viral vectors, mRNA and subunit vaccines as well as developing strategies to accelerate the production of investigational vaccines. While multiple vaccine types proved to be efficacious for severe diseases, no single vaccine modality met all the ideal target product profile characteristics for manufacturing and immunogenicity; each platform presented strengths and weaknesses, suggesting a breadth of options will be necessary to develop future countermeasures. For example, mRNA and viral vectors were better suited for rapid deployment; however longitudinal studies have shown that inactivated virus and recombinant subunit vaccines may have improved safety and immunologic responses over the aforementioned strategies. Protein-based vaccines can result in the generation of long-lived plasma blasts and elicit increased breadth of the response providing years of protection52,53,54,55. Additionally, subunit-based vaccines have a historical track record with government regulatory agencies due to it safety profile, thus it is likely to remain an integral part in the toolkit of vaccine strategies. Manufacturing logistics and cost problems should not impede the evaluation of novel recombinant vaccine candidates, as these have engineering solutions. The transient transfection by electroporation and immunoaffinity purification process developed for the 16055 DG4 HIV-1 Env glycoprotein program presented here builds on the work of Dey et al. validating the viability of this approach for the rapid production of HIV-1 Env recombinant subunit vaccines for Phase I clinical trial evaluation and can serve as a paradigm to produce future HIV-1 Env trimer-based vaccines33. While transient transfection is unsuitable for Phase 3 and commercial scale manufacturing, this niche approach can be quickly deployed for the evaluation of novel HIV-1 Env trimer vaccine constructs or iterations of the same protein with unique design features and can be used to quickly assess immunogenicity and safety of the target antigen(s) in preclinical and Phase I studies. The process developed here takes advantage of a single CHO-S MCB that can be transfected with different plasmid DNA HIV-1 Env constructs in a ‘plug-and-play’ manner to produce multiple HIV-1 Env glycoproteins. This process eliminates the need to characterize and release multiple MCBs. We expect the overall vaccine production timeline could be reduced from a minimum of twelve months for stable cell line or six months for stable cell pool development to about four months for the transient electroporation method described here and associated cost savings of > $500 K. The bulk of timeline savings is due to elimination of pool selection, Research Cell Bank production, stability, testing and release steps (see the accompanying Commentary by M. Pensiero). Electroporation is a relatively automated process; transfected protein production occurs over the course of days and purification can be performed within two weeks. Selection of PGT145 for the downstream process was critical to the success of this program, as this single step provided greater than 90% purity and achieved five logs of viral clearance for XMuLV, which can be challenging for products derived from cell lines of animal origin. The stabilized 16055 DG4 NFL trimer was amenable to elution at the relatively low pH of 4.0 when followed by immediate neutralization with buffer at pH. A cGMP-suitable, non-affinity purification process using anion exchange chromatography was recently developed to purify well-ordered HIV-1 Env trimers from clades A and C, respectively, providing an alternate means of purifying the trimeric spike56. However, development of bNAb-based affinity resins for purification of Env trimers in parallel during transient transfection would be beneficial to the field. It would be advantageous to develop a more cost-effective ligand, i.e. nanobody, that recognizes the native-like trimer epitope but could be manufactured or, better yet, sourced at a fraction of the cost. While successful in meeting program goals, multiple areas in the production process could be improved to increase yield and reduce costs. Improvements in plasmid design, feed strategies and the use of high cell density perfusion cultures could be investigated. The traditional path for scaling up goes from process development scale, engineering full scale to cGMP. Because the scalability of the process has been shown to be consistent from small scale to large batches, it is possible to go from a process development scale of 50 L to ~200 L of cGMP harvest without the need to produce an engineering run wherever warranted with proper controls in place.
Recent studies with HIV-1 Env trimers have shown an advantage of a escalating dose regimen performed over ~2 weeks compared to the more traditional single-dose bolus inoculation47,48. In these studies, the escalating dose regimen resulted in increased germinal centers, somatic hypermutation and neutralizing antibody titers compared to bolus. Accordingly, in anticipation of a human clinical trial, we tested the efficacy of 16055 DG4 NFL trimer as a prime and boost (100 μg total) in rabbits with SMNP adjuvant via bolus, escalating dose and divided dose immunization regimens (Supplementary Table 7). The trimer-specific binding titers (EC50) and pseudo virus neutralization titers (ID50) of the antigen matched DG4 virus reveal the divided dose of rabbits displayed significant improved antibody responses over the other two regimens at weeks 6 and 8. This improvement can be attributed to the higher dosage of the trimer and equally spaced-out immunizations that continuously primes the rabbits in the divided dose group. However, the boosting immunization at week 8 for the bolus and escalating dose groups resulted in a significant improvement of antibody titers at similar levels with the divided dose group. These results indicate that the traditional bolus prime:boost regimen with the 16055 DG4 NFL trimer in SMNP is similar to the other two regimens when tested in rabbits. Accordingly, the 16055 DG4 NFL trimer was administered as a priming candidate in clinical trial HVTN 313 (45/45 participants enrolled) with two bolus immunizations separated by 2 months followed by heterologous boosting with trimers from two different clades. In addition, one group of participants will be administered the 16055 DG4 NFL trimer only in five immunizations over 12 months to potentially elicit antibodies against the CD4bs, the gp120-gp41 or gp120-120 interfaces or other sites of Env trimer vulnerability25.
Methods
Manufacturing process development overview
The goal of the manufacturing process development effort was to produce 16055 DG4 NFL trimer glycoprotein for a human clinical trial using a transient transfection manufacturing process developed by ABL Inc. Overall developed manufacturing process, suitable for cGMP manufacturing, was a result of collaborative effort. DNA 2.0 ATUM (Newark, CA) performed DNA synthesis of designed 16055 DG4 NFL trimer plasmid. Aldevron (Fargo, ND) produced the 16055 DG4 NFL trimer plasmid and the corresponding MCB. Catalent performed upstream and downstream manufacturing process development and production for PGT145 antibody and Polymun Scientific performed conjugation of PGT145 antibody with AF-Tresyl-650M resin for immunoaffinity capture resin production. ABL performed 16055 DG4 NFL trimer upstream process development and production, IAVI performed downstream purification process development, and DHVI GMP program performed downstream process scale-up and production of 16055 DG4 NFL trimer. Final fill finish of the trimer was performed at Vetter Pharma.
Manufacture of PGT145 antibody
Antibody-based immunoaffinity chromatography resins used in cGMP manufacturing constitute a critical reagent, and their production is guided by the principles outlined in the Food and Drug Administrations (FDA) “Guidance for Industry, Monoclonal Antibodies Used as Reagents in Drug Manufacturing (March 2001).” ABL partnered with Catalent Pharma Solutions Inc. (Catalent, Somerset, NY) to produce PGT145 mAb reagent. A stable cell line producing PGT145 was engineered by retroviral transduction (GPEx system) using DNA provided by Scripps Research in CHO cells (clone 43-4 from the GCHO LC2/HC3; Project 3682). The PGT145 expressing MCB (Lot# 16045) was subsequently manufactured at Catalent from the stable cell line. ABL was granted permission by Scripps Research to utilize the MCB and the production process developed for Project 3682 to produce PGT145 reagent (Scripps Consortium for HIV/AIDS Vaccine Development [CHAVD]). PGT145 was purified using a three-step purification process: clarification, capture on MabSelect™ (Cytiva, Marlborough, MA) chromatography in one cycle with a low pH hold for viral inactivation, and anion exchange chromatography (Mustang Q XT) in two cycles. The purification resulted in a 70% overall yield and 138 g PGT145.
Manufacture of resin-bNAb combinations
Conjugation of bNAbs (2G12 and PGT145) to two different resins was performed as per the manufacturer recommendations. The Tresyl and Poros resins bind covalently to the free amino or thiol groups on the surface of the antibody. Trial conjugations were performed with 100 µl of resin and ~10 mg of bNAb/ml of resin at 1:1 volume ratio at 2 °C–8 °C and RT, respectively, in PBS buffer supplemented with 0.5 M NaCl, pH 7.14. The conjugation efficiency of the bNAb to the resin was calculated based on measuring the amount of free bNAb in the supernatant post conjugation. Effective conjugation (>90%) was obtained at RT with both the resins and 2G12 and PGT145 respectively. Tresyl-PGT145 immunoaffinity resin suitable for the purification of HIV-1 Env under cGMP was manufactured at Polymun Scientific, GmbH (Polymun, Klosterneuburg, Austria). PGT145 reagent was coupled to Tresyl resin at a 5.7 L scale. The antibody was coupled at a bNAb density of 6.7 mg/mL, and the binding capacity was determined to be 0.95 mg/mL. The resin met the specifications for particle size, antibody leaching, endotoxin, and bioburden (Supplementary Table 8). The batch was manufactured using certified animal-free components and materials and in compliance with the European Pharmacopeia “Guidance on minimizing the risk of transmitting animal spongiform encephalopathy agents via human and veterinary medicinal products (EMA/410/01 rev.3)” and the (FDA) “Guidance for Industry, Monoclonal Antibodies Used as Reagents in Drug Manufacturing (March 2001).” The resin was stored in 0.05% sodium azide storage buffer. 2.5 L of this resin was utilized in the successful cGMP purification of 16055 DG4 NFL.
Trimer upstream production
CHO-S™ cells were expanded from a single MCB vial in growth media (CD OptiCHO™ [Thermo Fisher Scientific, Waltham, MA] + 6 mM Glutamax™ [Thermo Fisher Scientific]) at 37 °C in shake flasks for 12 days (Fig. 2). Subsequently, the cell culture was further expanded in a 200 L stirred tank bioreactor (STR) to 55.8 L for three days, then expanded to 150 L for one day until the target cell density of 5.0–6.0 x 106 cells/mL was reached. After expansion, 2.2 L cell suspension was prepared for electroporation. 100 L CHO-S™ cell culture was concentrated 50-fold and buffer exchanged into MaxCyte’s proprietary electroporation (EP) buffer by Ksep® (Sartorius, Goettingen, Germany) continuous centrifugation. CHO-S™ cells were electroporated using a MaxCyte® VLX™ with plasmid DNA at 275 µg/mL of concentrated cell suspension. The prepared cell suspension was equally divided into two VLX® processing assemblies and electroporated using a predesigned MaxCyte program optimized for electroporating CHO-S™ cells. Electroporated cells were rested for 45 min at 37 °C in 10-layer Cell Factories (approximately 550 mL of cell suspension/cell factory). Following recovery, cells were transferred to 200 L STR at a target seeding density of ~5 x 106 cells/mL in 63 L production media (CD OptiCHO™+ 2 mM Glutamax™) and supplemented with 3% culture volume of Feed 1 (63.8% CHO CD Efficient Feed™ A + 42.5 g/L Difco™ TC Yeastolate). Transfected cells were cultured at 37 °C for 18 h. At 18 h post-transfection, Valproic Acid (VPA) was added to 1 mM final concentration, and the temperature was shifted to 32 °C. VPA is an inhibitor of histone deacetylase activity and is added since it has been shown to increase recombinant mRNA and protein concentration in transiently transfected CHO cultures57. Culture was supplemented with 3% culture volume of Feed 2 (63.8% CHO CD EfficientFeed™ A + 42.5 g/L Difco™ TC Yeastolate + 8.3 mM GlutaMax™) on days 2, 4, and 6. Throughout the process, glucose was supplemented to a concentration of 2.5 g/L whenever culture glucose concentration fell below 2 g/L. Samples were taken daily for viable cell density, viability, metabolites, and titer (starting on day 3 through day 8) assessment. Bulk harvest was collected on day 8. A two-step clarification process was performed using D0HC (primary) and A1HC (secondary) Millistak+® pod depth filters (Millipore Sigma, Burlington, MA) to clarify the harvest material, followed by sterile filtration using a Millipore Express® SHC 0.5/0.2 µm cartridge filter. Following clarification, 79.6 L of 16055 DG4 NFL trimer harvest was recovered. Clarified harvest was partitioned into 4 L aliquots in 5 L biotainers. The harvest material was then safely stepped down to 2 °C–8 °C, frozen and stored at –70 to –90 °C.
Trimer downstream purification
The 16055 DG4 NFL downstream purification process is outlined in Fig. 1. Clarified cell culture harvest stored at −70 °C to −90 °C was thawed at 2 °C–8 °C, filtered with 0.22 µm PES Sartopore 2 filter (Sartorius) and concentrated 5-fold by ultrafiltration/diafiltration (UFDF) using a 300 kDa Pellicon® 3 filter (Millipore Sigma). The concentrated harvest was subsequently virally inactivated by non-ionic detergent treatment. For the detergent viral inactivation, 10% Triton X-100 solution was spiked into concentrated harvest to achieve final concentration of 0.5% (w/v). After 60 min incubation at RT, harvest was loaded onto a PGT145-Tr immunoaffinity column equilibrated with 20 mM Sodium Phosphate, 150 mM NaCl, pH 7.4 and purified in bind and elute mode. The column was loaded at an equivalent of 1 g trimer/L resin and performed in multiple cycles. The target molecule was eluted by pH shift in 100 mM Citrate, pH 4.0, and the eluate was adjusted rapidly upon collection to pH 8.0 using 1 M Tris, pH 9.0. Eluates from PGT145-Tr cycles were pooled and subsequently buffer-exchanged by UFDF into 20 mM Tris, 75 mM NaCl pH 8.0 using a 100 kDa Pellicon® 3 filter (Millipore Sigma). To remove residual PGT145, material was loaded onto a MabSelect SuRe™ Protein-A column (Cytiva) operated in a flow-through mode. The flow-through was next adjusted to pH 6.0 with 1 M Tris-HCl, pH 4.0, diluted to 17.0 mS/cm with water and loaded onto a Nuvia™ cPrime™ mixed-mode cation exchange column (Bio-Rad Laboratories, Inc., Hercules, CA) operated in a flow-through mode. For virus removal, the Nuvia™ cPrime™ flow-through was nanofiltered using Viresolve® Shield H and Viresolve Pro Modus 1.3 Device virus filter train (Millipore Sigma). The nanofiltrate was concentrated to ~3 mg/mL by UFDF using a 30 kDa Pellicon® 3 filter (Millipore Sigma). A final polishing step was performed using tandem 17.8 x 25 cm preparative Superdex 200 PG size-exclusion chromatography columns (Cytiva) equilibrated to 20 mM Tris, 75 mM NaCl, pH 8.0. The tandem columns were loaded at an equivalent of 2.0% column volume and performed in eight cycles. The 16055 DG4 NFL trimer peak fractions were pooled together, concentrated to ~1 mg/mL, and diafiltered into Tris-buffered saline as the final formulation buffer (20 mM Tris, 100 mM NaCl, pH 7.5) using a 30 kDa Pellicon® 3 filter (Millipore Sigma). A final sterilization was performed using a 0.22 µm PES Sartopore 2 filter before bulk fill for subsequent storage at −70 °C to −90 °C for pre-clinical and clinical use. The overall process yielded 0.857 g of the trimer from a 75.40 L of clarified harvest for a 20% process recovery. Viral clearance studies for the final product were guided by CPMP, Note for Guidance on Virus Validation Studies: The Design, Contribution and Interpretation of Studies Validating the Inactivation and Removal of Viruses, CBER/FDA: Points to Consider in the Characterization of Cell Lines Used to Produce Biologicals, ICH Topic Q5A: Quality of Biotechnological Products: Viral Safety Evaluation of Biotechnology Products Derived From Cell Lines of Human or Animal Origin, and CBER/FDA: Points to Consider in the Manufacture and Testing of Monoclonal Antibody Products for Human Use. Four downstream unit operations were assessed for viral clearance: Triton X-100 viral inactivation, virus removal by two unique chromatography steps (Tresyl-PGT145 Immuno-affinity and MMC Nuvia cPrime Polishing) and virus removal via Viresolve Pro. Scale down models were shown to be comparable to the process run at manufacturing scale in the Duke cGMP Manufacturing Facility (DGF). Virus spiking studies were executed for each step at BioReliance. Known amounts of model virus were spiked into the load for each process step, the remaining virus in the product pool after completion of the process step was measured, and the LRV was calculated. The overall process LRV was then estimated by adding the individual LRV’s from steps that demonstrated virus reduction (>1 log). The final fill finish of the trimer product was performed at Vetter Pharma (Skokie, IL); 2 mL glass vials were filled at 0.75 mL/vial and stored at –70 to –90 °C.
Electron microscopy (EM)
Negative stain EM was performed by diluting 16055 DG4 NFL engineering run material to ~0.03 mg/mL in Tris-buffered saline and adsorbing onto a carbon-coated, glow discharged copper mesh grid. The sample was then stained for 45 s with 2% (w/v) uranyl formate before imaging on an FEI Tecnai T12 Spirit operating at 120 keV and equipped with an FEI 4 K Eagle CCD camera. Data collection was automated using Leginon and 2D class averages generated using Appion suite58,59.
CryoEM was performed using the same 16055 DG4 NFL engineering run material (at a concentration of 3.4 mg/mL). The sample was briefly mixed with lauryl maltose neopentyl glycol (LMNG; final concentration 0.005 mM; Anatrace) to aid with particle dispersion and a 3.5 µl drop was applied to carbon foil Quantifoil 1.2/1.3 holey grids. Sample vitrification was performed using the Vitrobot Mark IV (Thermo Scientific). Data collection was performed on an FEI Titan Krios equipped with a Gatan K2 Summit direct electron detector (300 keV, 1.045 Å/pixel counting mode). Leginon58 was used for automated data collection, frame alignment was performed using MotionCor260 and subsequent data processing was performed using cryoSPARC v361 following standard workflows. 2D class averages and 3D ab initio models suggested a mixture of free trimers and dimer-of-trimers. To aid with alignment, a soft mask was generated over the trimer with strongest signal and used for downstream refinement steps. The final reconstruction was generated using the Non-uniform 3D Refinement job with C3 symmetry imposed. Estimated resolution of obtained structure based on Fourier shell correlation cutoff of 0.143 is 3.8 Å. Model building was performed by docking a crystal structure of 16055 NFL (PDB 5UM8) into the cryo-EM map in UCSF Chimera62, manually introducing mutated residues, building and refinement in Coot 0.9.863, and real space refinement using Rosetta64 and Phenix65. Final models were validated using MolProbity and EMRinger in the Phenix suite. A summary of data collection, processing, and model building statistics is summarized in Fig. 4 and Supplementary Table 6. The cryoEM map and model have been deposited in the EM Data Bank and Protein Data Bank, respectively, under the accession codes listed in Supplementary Table 6.
Glycan analysis
DeGlyPHER was used to ascertain site-specific glycan occupancy and processivity on the examined glycoproteins40. HIV-1 Env glycoproteins were exchanged to water using Microcon Ultracel PL-10 centrifugal filter. Glycoproteins were reduced with 5 mM tris(2-carboxyethyl)phosphine hydrochloride (TCEP-HCl) and alkylated with 10 mM 2-Chloroacetamide in 100 mM ammonium acetate for 20 min at RT. Initial protein-level deglycosylation was performed using 250 U of Endo H for 5 µg trimer, for 1 h at 37 °C. Glycoproteins were digested with 1:25 Proteinase K (PK) for 30 min at 37 °C. PK was denatured by incubating at 90 °C for 15 min, then cooled to RT. Peptides were deglycosylated again with 250 U Endo H for 1 h at 37 °C, then frozen at −80 °C and lyophilized. 100 U PNGase F was lyophilized, resuspended in 20 µl 100 mM ammonium bicarbonate prepared in H218O, and added to the lyophilized peptides. Reactions were then incubated for 1 h at 37 °C, subsequently analyzed by LC-MS/MS.
Samples were analyzed on an Q Exactive HF-X mass spectrometer. Samples were injected directly onto a 25 cm, 100 μm ID column packed with BEH 1.7 μm C18 resin. Samples were separated at a flow rate of 300 nL/min on an EASY-nLC 1200 UHPLC. Buffers A and B were 0.1% formic acid in 5% and 80% acetonitrile, respectively. The following gradient was used: 1–25% B over 160 min, an increase to 40% B over 40 min, an increase to 90% B over another 10 min and 30 min at 90% B for a total run time of 240 min. Column was re-equilibrated with solution A prior to the injection of the sample. Peptides were eluted from the tip of the column and nanosprayed directly into the mass spectrometer by application of 2.8 kV at the back of the column. The mass spectrometer was operated in a data-dependent mode. Full MS1 scans were collected in the Orbitrap at 120,000 resolution. The ten most abundant ions per scan were selected for HCD MS/MS at 25 NCE. Dynamic exclusion was enabled with an exclusion duration of 10 s and singly charged ions were excluded.
Protein and peptide identification were done with Integrated Proteomics Pipeline. Tandem mass spectra were extracted from raw files using RawConverter and searched with ProLuCID against a database comprising UniProt reviewed (Swiss-Prot) proteome for Homo sapiens (UP000005640), UniProt amino acid sequences for Endo H (P04067), PNGase F (Q9XBM8), and Proteinase K (P06873), amino acid sequences for the examined proteins, and a list of general protein contaminants66,67. The search space included no cleavage-specificity. Carbamidomethylation (+57.02146 C) was considered a static modification. Deamidation in presence of H218O (+2.988261 N), GlcNAc (+203.079373 N), oxidation (+15.994915 M), and N-terminal pyroglutamate formation (–17.026549 Q) were considered differential modifications. Data was searched with 50 ppm precursor ion tolerance and 50 ppm fragment ion tolerance. Identified proteins were filtered using DTASelect2 and utilizing a target-decoy database search strategy to limit the false discovery rate to 1%, at the spectrum level68,69. A minimum of 1 peptide per protein and no tryptic end per peptide were required and precursor delta mass cut-off was fixed at 15 ppm. Statistical models for peptide mass modification (modstat) were applied. Census2 label-free analysis was performed based on the precursor peak area, with a 15 ppm precursor mass tolerance and 0.1 min retention time tolerance70. “Match between runs” was used to find missing peptides between runs. Data analysis using GlycoMSQuant was implemented to automate the analysis40. GlycoMSQuant summed precursor peak areas across replicates, discarded peptides without NGS, discarded misidentified peptides when N-glycan remnant-mass modifications were localized to non-NGS asparagines and corrected/fixed N-glycan mislocalization where appropriate.
Immunization studies
C57/BL6 mice (female, six per group) were immunized each with 5 μg of 16055 DG4 NFL trimer adjuvanted with 100 μg Alum or 10 μg of MPLA + 10 μg of QS21. Control mice were immunized with trimer lacking adjuvant. Immunizations were performed at weeks 0, 4 and 10 by the intramuscular route; the dose was split equally to each hind leg to access more draining lymph nodes. Bleeds were collected on the day and 2 weeks after each immunization. All immunization and bleeds were performed under anesthesia with administration of 1–3% isoflurane. At the conclusion of the experiment, mice were euthanized by administration of 5–10% isoflurane in an inhalation chamber. All protocols for animal handling were approved by IACUC of Scripps Research. For the immunogenicity studies in rabbits, New Zealand female white rabbits were immunized with 16055 DG4 NFL trimer formulated in SMNP adjuvant. SMNP was prepared as previously described44. Three groups of rabbits (8 animals per group) were primed and boosted with the trimer as a bolus dose, escalating dose, or divided dose regimen. Details on the immunization regimens are described in Supplementary Table 7. Immunizations were performed by intramuscular route injections split equally on each hind leg without anesthesia. The dose of adjuvant was adjusted to maintain a protein-to-adjuvant ratio of 50 μg trimer to 375 μg adjuvant during all immunizations. The rabbits were bled to obtain serum for antibody analysis by the marginal ear veins without anesthesia. The rabbits were returned to the colony at the end of the study. Immunization protocols were approved by IACUC of ProSci Inc (Poway, CA).
ELISA analysis
For the IgG binding ELISA we used a His-tagged version of the 16055 DG4 NFL trimer as the target. The 96-well plates (half-well from Corning Inc. and 50 μl reagent volume for all reagent incubations) were coated with mouse anti-His mAb at 2 μg/mL for 1 h. The plates were blocked for 1 h with 150 μl PBS buffer comprising 2% nonfat milk and 5% fetal bovine serum. The plates were washed four times with 150 μl of PBS buffer supplemented with 0.02% Tween 20 between each of the subsequent incubation steps. 16055 DG4 NFL trimer was captured on the plates by the C-terminal polyhistidine-tag for 1 h. The plates were incubated for 1 h with 5-fold serial dilutions of serum with a starting dilution of 1:10. The plates were incubated with horseradish peroxidase-coupled anti-mouse or anti-rabbit IgG at 1:5000 dilution for 1 h and developed with TMB substrate solution. The plates were visually examined for development of the colorimetric substrate and the reaction was stopped with 0.3 N sulfuric acid and the absorbance was measured at 450 nm. For the sCD4-induced conformational change binding assay, the 16055 DG4 NFL trimer (with the 201–433 CC disulfide) and BG505 NFL (no CC disulfide) were captured on the plates by a C-terminal His-tag. The trimers were incubated with 4-fold molar excess of sCD4 prior to the addition of 5-fold serial dilutions of mAbs at a starting concentration of 50 μg/ml. Anti-human IgG-peroxidase at 1:5000 dilution was used as the secondary antibody, followed by addition of the TMB substate solution. Half-maximum effective concentration serum binding titers (ED50) were obtained by fitting the data in GraphPad Prism software v9.4.1.
Serum neutralization assay
Inhibition of entry of HIV-1 Env pseudotyped viruses into standard TZM-bl cells was used to determine the neutralization capacity of serum46. Sera dilution that resulted in 50% reduction (ID50) in relative light units was determined by fitting the neutralization dose-response curves by non-linear regression using a 5-parameter hill slope equation.
Data availability
Cryo-EM map has been deposited into the EM Data Bank under accession code EMD-44474 and Cryo-EM model of 16055 DG4 NFL trimer has been deposited to the Protein Data Bank under accession code 9BE9. All other relevant data generated, analyzed, and presented in this manuscript are available on request from the corresponding author.
References
Flynn, N. M. et al. Placebo-controlled phase 3 trial of a recombinant glycoprotein 120 vaccine to prevent HIV-1 infection. J. Infect. Dis. 191, 654–665 (2005).
Houser, K. V. et al. Safety and immunogenicity of an HIV-1 prefusion-stabilized envelope trimer (Trimer 4571) vaccine in healthy adults: a first-in-human open-label, randomized, dose-escalation, phase 1 clinical trial. EClinicalMedicine 48, 101477 (2022).
Leggat, D. J. et al. Vaccination induces HIV broadly neutralizing antibody precursors in humans. Science 378, eadd6502 (2022).
Pitisuttithum, P. et al. Randomized, double-blind, placebo-controlled efficacy trial of a bivalent recombinant glycoprotein 120 HIV-1 vaccine among injection drug users in Bangkok, Thailand. J. Infect. Dis. 194, 1661–1671 (2006).
Rerks-Ngarm, S. et al. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N. Engl. J. Med. 361, 2209–2220 (2009).
Hunter, M., Yuan, P., Vavilala, D. & Fox, M. Optimization of protein expression in mammalian cells. Curr. Protoc. Protein Sci. 95, e77 (2019).
Schmieder, V. et al. Towards maximum acceleration of monoclonal antibody development: leveraging transposase-mediated cell line generation to enable GMP manufacturing within 3 months using a stable pool. J. Biotechnol. 349, 53–64 (2022).
Bolisetty, P., Tremml, G., Xu, S. & Khetan, A. Enabling speed to clinic for monoclonal antibody programs using a pool of clones for IND-enabling toxicity studies. MAbs 12, 1763727 (2020).
Baden, L. R. et al. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N. Engl. J. Med. 384, 403–416 (2021).
Polack, F. P. et al. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N. Engl. J. Med. 383, 2603–2615 (2020).
Riddler, S. A. et al. High frequency of chronic urticaria following an investigational HIV-1 BG505 MD39.3 trimer mRNA vaccine in a phase 1, randomized, open-label clinical trial (HVTN 302). Ann. Intern. Med. 10, 02701 (2025).
Willis, J. R. et al. Vaccination with mRNA-encoded nanoparticles drives early maturation of HIV bnAb precursors in humans. Science 21, eadr8382 (2025).
Kulkarni, S. S. et al. Highly complex neutralization determinants on a monophyletic lineage of newly transmitted subtype C HIV-1 Env clones from India. Virology 385, 505–520 (2009).
Sharma, S. K. et al. Cleavage-independent HIV-1 Env trimers engineered as soluble native spike mimetics for vaccine design. Cell Rep. 11, 539–550 (2015).
Sanders, R. W. et al. Stabilization of the soluble, cleaved, trimeric form of the envelope glycoprotein complex of human immunodeficiency virus type 1. J. Virol. 76, 8875–8889 (2002).
Sanders, R. W. et al. A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog. 9, e1003618 (2013).
Klasse, P. J. et al. Influences on trimerization and aggregation of soluble, cleaved HIV-1 SOSIP envelope glycoprotein. J. Virol. 87, 9873–9885 (2013).
Guenaga, J. et al. Structure-guided redesign increases the propensity of HIV Env to generate highly stable soluble trimers. J. Virol. 90, 2806–2817 (2015).
Guenaga, J. et al. Glycine substitution at helix-to-coil transitions facilitates the structural determination of a stabilized subtype C HIV envelope glycoprotein. Immunity 46, 792–803.e793 (2017).
Kwon, Y. D. et al. Crystal structure, conformational fixation and entry-related interactions of mature ligand-free HIV-1 Env. Nat. Struct. Mol. Biol. 22, 522–531 (2015).
Nguyen, H. T., Alsahafi, N., Finzi, A. & Sodroski, J. G. Effects of the SOS (A501C/T605C) and DS (I201C/A433C) disulfide bonds on HIV-1 membrane envelope glycoprotein conformation and function. J. Virol. 93, 00303 (2019).
Berndsen, Z. T. et al. Visualization of the HIV-1 Env glycan shield across scales. Proc. Natl Acad. Sci. USA 117, 28014–28025 (2020).
Stewart-Jones, G. B. et al. Trimeric HIV-1-Env structures define glycan shields from clades A, B, and G. Cell 165, 813–826 (2016).
Dubrovskaya, V. et al. Targeted N-glycan deletion at the receptor-binding site retains HIV Env NFL trimer integrity and accelerates the elicited antibody response. PLoS Pathog. 13, e1006614 (2017).
Dubrovskaya, V. et al. Vaccination with glycan-modified HIV NFL envelope trimer-liposomes elicits broadly neutralizing antibodies to multiple sites of vulnerability. Immunity 51, 915–929.e917 (2019).
Caniels, T. G. et al. Germline-targeting HIV vaccination induces neutralizing antibodies to the CD4 binding site. Sci. Immunol. 9, eadk9550 (2024).
Crooks, E. T. et al. Effects of partially dismantling the CD4 binding site glycan fence of HIV-1 Envelope glycoprotein trimers on neutralizing antibody induction. Virology 505, 193–209 (2017).
LaBranche, C. C. et al. HIV-1 envelope glycan modifications that permit neutralization by germline-reverted VRC01-class broadly neutralizing antibodies. PLoS Pathog. 14, e1007431 (2018).
Zhou, T. et al. Quantification of the impact of the HIV-1-glycan shield on antibody elicitation. Cell Rep. 19, 719–732 (2017).
Pritchard, L. K. et al. Glycan clustering stabilizes the mannose patch of HIV-1 and preserves vulnerability to broadly neutralizing antibodies. Nat. Commun. 6, 7479 (2015).
Wolfe, L. S. et al. Development of a platform-based approach for the clinical production of HIV gp120 envelope glycoprotein vaccine candidates. Vaccine 39, 3852–3861 (2021).
Lee, J. H. et al. A broadly neutralizing antibody targets the dynamic HIV envelope trimer apex via a long, rigidified, and anionic beta-hairpin structure. Immunity 46, 690–702 (2017).
Dey, A. K. et al. cGMP production and analysis of BG505 SOSIP.664, an extensively glycosylated, trimeric HIV-1 envelope glycoprotein vaccine candidate. Biotechnol. Bioeng. 115, 885–899 (2018).
Cupo, A. et al. Optimizing the production and affinity purification of HIV-1 envelope glycoprotein SOSIP trimers from transiently transfected CHO cells. PLoS One 14, e0215106 (2019).
Sanders, R. W. et al. The mannose-dependent epitope for neutralizing antibody 2G12 on human immunodeficiency virus type 1 glycoprotein gp120. J. Virol. 76, 7293–7305 (2002).
Alderton, M. R., Gray, P. J. & Prolf, D. F. Genetic vaccination: can plasmid DNA deliver its expectations? J. Milit. Veterans’ Health 10, 95681947 (2001).
Davis, H. L. & McCluskie, M. J. DNA vaccines for viral diseases. Microbes Infect. 1, 7–21 (1999).
Khan, A. S. Characterization and qualification of cell substrates for manufacturing viral vaccines in the United States. BioProcess J. 8, 8–12 (2009).
Shroff, K. E. et al. Induction of HSV-gD2 specific CD4(+) cells in Peyer’s patches and mucosal antibody responses in mice following DNA immunization by both parenteral and mucosal administration. Vaccine 18, 222–230 (1999).
Baboo, S. et al. DeGlyPHER: an ultrasensitive method for the analysis of viral spike N-glycoforms. Anal. Chem. 93, 13651–13657 (2021).
Brouwer, P. J. M. et al. Immunofocusing and enhancing autologous Tier-2 HIV-1 neutralization by displaying Env trimers on two-component protein nanoparticles. NPJ Vaccines 6, 24 (2021).
Cottrell, C. A. et al. Mapping the immunogenic landscape of near-native HIV-1 envelope trimers in non-human primates. PLoS Pathog. 16, e1008753 (2020).
Derking, R. et al. Enhancing glycan occupancy of soluble HIV-1 envelope trimers to mimic the native viral spike. Cell Rep. 35, 108933 (2021).
Silva, M. et al. A particulate saponin/TLR agonist vaccine adjuvant alters lymph flow and modulates adaptive immunity. Sci. Immunol. 6, eabf1152 (2021).
Sliepen, K. et al. Interplay of diverse adjuvants and nanoparticle presentation of native-like HIV-1 envelope trimers. NPJ Vaccines 6, 103 (2021).
Li, M. et al. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J. Virol. 79, 10108–10125 (2005).
Cirelli, K. M. et al. Slow delivery immunization enhances HIV neutralizing antibody and germinal center responses via modulation of immunodominance. Cell 177, 1153–1171 (2019).
Lee, J. H. et al. Long-primed germinal centres with enduring affinity maturation and clonal migration. Nature 609, 998–1004 (2022).
Kim, J., Vasan, S., Kim, J. H. & Ake, J. A. Current approaches to HIV vaccine development: a narrative review. J. Int AIDS Soc. 24, e25793 (2021).
Binley, J. M. et al. A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion-associated structure. J. Virol. 74, 627–643 (2000).
Chung, N. P. et al. Stable 293 T and CHO cell lines expressing cleaved, stable HIV-1 envelope glycoprotein trimers for structural and vaccine studies. Retrovirology 11, 33 (2014).
Bayani, F. et al. An overview of the vaccine platforms to combat COVID-19 with a focus on the subunit vaccines. Prog. Biophys. Mol. Biol. 178, 32–49 (2023).
Palin, A. C. et al. The persistence of memory: defining, engineering, and measuring vaccine durability. Nat. Immunol. 23, 1665–1668 (2022).
Heinz, F. X. & Stiasny, K. Distinguishing features of current COVID-19 vaccines: knowns and unknowns of antigen presentation and modes of action. NPJ Vaccines 6, 104 (2021).
Vashishtha, V. M. & Kumar, P. The durability of vaccine-induced protection: an overview. Expert Rev. Vaccines 23, 389–408 (2024).
Gulla, K. et al. A non-affinity purification process for GMP production of prefusion-closed HIV-1 envelope trimers from clades A and C for clinical evaluation. Vaccine 39, 3379–3387 (2021).
Wulhfard, S., Baldi, L., Hacker, D. L. & Wurm, F. Valproic acid enhances recombinant mRNA and protein levels in transiently transfected Chinese hamster ovary cells. J. Biotechnol. 148, 128–132 (2010).
Cheng, A. et al. Leginon: new features and applications. Protein Sci. 30, 136–150 (2021).
Lander, G. C. et al. Appion: an integrated, database-driven pipeline to facilitate EM image processing. J. Struct. Biol. 166, 95–102 (2009).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Punjani, A., Zhang, H. & Fleet, D. J. Non-uniform refinement: adaptive regularization improves single-particle cryo-EM reconstruction. Nat. Methods 17, 1214–1221 (2020).
Pettersen, E. F. et al. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Casanal, A., Lohkamp, B. & Emsley, P. Current developments in coot for macromolecular model building of electron cryo-microscopy and crystallographic data. Protein Sci. 29, 1069–1078 (2020).
Conway, P., Tyka, M. D., DiMaio, F., Konerding, D. E. & Baker, D. Relaxation of backbone bond geometry improves protein energy landscape modeling. Protein Sci. 23, 47–55 (2014).
Afonine, P. V. et al. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. D. Struct. Biol. 74, 531–544 (2018).
He, L., Diedrich, J., Chu, Y. Y. & Yates, J. R. III Extracting Accurate Precursor Information for Tandem Mass Spectra by RawConverter. Anal. Chem. 87, 11361–11367 (2015).
Xu, T. et al. ProLuCID: An improved SEQUEST-like algorithm with enhanced sensitivity and specificity. J. Proteom. 129, 16–24 (2015).
Tabb, D. L., McDonald, W. H. & Yates, J. R. III DTASelect and contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J. Proteome Res. 1, 21–26 (2002).
Peng, J., Elias, J. E., Thoreen, C. C., Licklider, L. J. & Gygi, S. P. Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: the yeast proteome. J. Proteome Res. 2, 43–50 (2003).
Park, S. K., Venable, J. D., Xu, T. & Yates, J. R. III A quantitative analysis software tool for mass spectrometry-based proteomics. Nat. Methods 5, 319–322 (2008).
Acknowledgements
Manufacturing of the 16055 DG4 NFL trimer was supported by the NIH NIAID DAIDS VTRB “NIAID Preclinical Development Support” IDIQ contract HHSN272201100021I awarded to ABL Inc. The work was also supported by HIVRAD grants P01 AI104722, P01 AI157299 (SB, JG, ED, RW, and RTW), grant P01 AI124337 (RTW), James B. Pendleton Charitable Trust, grants R01 AI0988602, R01 AI145055, Scripps CHAVD UMI AI144462 (RTW, JRY, JCP and ABW) and funding from the IAVI (JG and RTW). The full list of donors for IAVI can be found on the website http://www.iavi.org. We thank Michael Pensiero (DAIDS) for technical inputs and overseeing the project. We thank Sal Butera (Scripps, CHAVD) for permission to use PGT145 for affinity purification. We thank Dr. Christina Mayer for her contributions to the IND submission. The funders played no role in study design, data collection, analysis and interpretation of data, or the writing of this manuscript.
Author information
Authors and Affiliations
Contributions
S.B., J.G., V.A. and R.T.W. designed the research studies. J.G. and R.T.W. designed the trimer construct. E.G., V.A., S.W., L.H., and T.F. oversaw the protein production of the clinical lot. S.B., E.D.D., and R.W. performed immunization studies and serum analysis. W-H.L. and G.O. performed the EM analysis and determined the cryo-EM structure. A.B.W. oversaw the structural analysis. S.B. and J.K.D. performed glycan analysis. J.R.Y. and J.C.P. supervised the glycan analysis. E.B-A., K.A.R., and D.J.I. provided the adjuvant. S.B., E.G., V.A., J.G., S.B., S.W., L.H., G.O., and R.T.W. wrote the paper. T.F. and R.T.W. supervised the study. All authors reviewed the results. All authors read and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
D.J.I. is an inventor on a patent related to SMNP adjuvant. All other authors declare no competing interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Bale, S., Gustchina, E., Guenaga, J. et al. Accelerated cGMP production of near-native HIV-1 Env trimers following electroporation transfection and immunogenicity analysis. npj Vaccines 10, 198 (2025). https://doi.org/10.1038/s41541-025-01218-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41541-025-01218-6