Abstract
Understanding how cells respond differently to perturbation is crucial in cell biology, but existing methods often fail to accurately quantify and interpret heterogeneous single-cell responses. Here we introduce the perturbation-response score (PS), a method to quantify diverse perturbation responses at a single-cell level. Applied to single-cell perturbation datasets such as Perturb-seq, PS outperforms existing methods in quantifying partial gene perturbations. PS further enables single-cell dosage analysis without needing to titrate perturbations, and identifies ‘buffered’ and ‘sensitive’ response patterns of essential genes, depending on whether their moderate perturbations lead to strong downstream effects. PS reveals differential cellular responses on perturbing key genes in contexts such as T cell stimulation, latent HIV-1 expression and pancreatic differentiation. Notably, we identified a previously unknown role for the coiled-coil domain containing 6 (CCDC6) in regulating liver and pancreatic cell fate decisions. PS provides a powerful method for dose-to-function analysis, offering deeper insights from single-cell perturbation data.
Similar content being viewed by others
Main
Perturbation is essential for understanding the functions of the mammalian genome that encodes protein-coding genes and non-coding elements (for example, enhancers). Single-cell profiling of cells undergoing genetic, chemical, environmental or mechanical perturbations is commonly used to examine perturbation responses at the single-cell level. Recently, high-throughput approaches of perturbation have been developed using single-cell RNA sequencing (scRNA-seq) readout, including multiplexing of perturbations and single-cell CRISPR screen (for example, Perturb-seq, CROP-seq)1,2,3,4,5,6,7. This concept has been extended to study changes in single-cell chromatin accessibility8,9, spatial transcriptomics10 on perturbations or perturbation combinations11,12,13, and other phenomena.
Decoding how perturbations lead to different cellular responses is critical for understanding fundamental biology. Technical factors, such as single-cell assays and the on-target/off-target effects of perturbations, drive differences in single-cell profiles14,15,16. In Perturb-seq experiments using CRISPR–Cas9 for knockouts, in-frame deletions16 and chromosomal losses17 can alter expression profiles and clustering patterns. More interestingly, the heterogeneity of perturbation responses is often driven by underlying biological factors (Fig. 1a). These may be cell-intrinsic (for example, the activities of coding and non-coding genomic elements, cell states or types) or cell-extrinsic (for example, environmental factors), which together define the context of a perturbation response. For example, the combined expression of transcription factors is critical for many cellular state conversions. Decoding transcription factor functions via perturbation requires accounting for the effects of the cell state and the activities of companion transcription factors. Therefore, defining the heterogeneity in perturbation responses and identifying the factors that shape these outcomes is key to understanding how cells respond to perturbations.
Current computational frameworks are inadequate for decoding the diverse outcomes of perturbations. Methods such as MUSIC18, MIMOSCA3, scMAGeCK15 and SCEPTRE19 estimate the average effects of perturbations but fall short of capturing response heterogeneity. Recently, HiDDEN20, a machine learning method, was developed to refine perturbation labels in scRNA-seq studies, although it is limited to single types of perturbation (for example, drug treatment or disease). Generative models such as SC-VAE21 separate perturbation effects from other confounding factors in Perturb-seq data. For handling technical factors, mixscape detects and mitigates confounding variations such as incomplete knockouts16, with its extension Mixscale22 modelling cellular variations in perturbation efficiency and downstream gene expression. However, these methods were primarily designed for CRISPR–Cas9 knockout and do not account for partial gene perturbations using techniques such as CRISPR interference (CRISPRi). Furthermore, they are not designed to uncover biological insights from heterogeneous perturbation outcomes, such as how partial gene perturbations affect a phenotype of interest (that is, ‘dosage’ analysis) or how biological factors influence differential perturbation responses.
Here we present a computational framework, the perturbation-response score (PS), to quantify heterogeneous perturbation outcomes in single-cell transcriptomics datasets. PS, estimated through constrained quadratic optimization, quantifies the strength of the perturbation outcome at the single-cell level. Our comprehensive benchmarks show that PS outperforms existing methods across simulated datasets, genome-scale Perturb-seq and published CRISPR-based Perturb-seq datasets. PS offers two key advances: enabling the dosage analysis of genetic perturbation and identifying biological factors that govern response heterogeneity. In essential gene Perturb-seq data, PS revealed two dosage response patterns, depending on whether moderate perturbation strongly affected downstream gene expression. PS also uncovered intrinsic and extrinsic factors governing critical gene functions in latent HIV-1 expression and pancreatic–liver development, including a previously unknown role for CCDC6 in driving duodenum cells toward liver commitment. These findings illustrate the power of PS in decoding heterogeneous perturbation outcomes from single-cell assays.
Results
PS for detecting diverse outcomes
Perturbing the same gene (or non-coding elements) may result in different phenotypic changes or transcriptional outcomes (Fig. 1a), depending on technical factors (for example, perturbation efficiency) and biological factors (for example, cell type, cell state, the activities of cofactors). Unfortunately, most existing methods are designed to detect and mitigate the effect of technical factors16, while the effects of biological factors remain unexplored. PS is built to bridge this gap by quantifying perturbation outcomes in single-cell datasets using scRNA-seq as readout, including single-cell CRISPR screens (for example, Perturb-seq), or simply multiplex scRNA-seq profiling of various perturbations (for example, sci-Plex; Fig. 1b,c). We define the PS to quantify the strength of perturbation, where PS = 0 indicates no perturbation effect and PS = 1 indicates the maximum perturbation effect observed within a dataset; for example, effects that correspond to homozygous knockouts on both gene alleles. We use the expression changes of multiple downstream targets of a perturbation to infer the (unknown) values of PS (Fig. 1b). For example, if one cell has dramatic expression changes of downstream genes, then its value of PS should be higher than cells with weak expression changes of these genes.
The PS framework consists of three steps (Fig. 1c). In the first optional step, PS identifies differentially expressed genes (DEGs) on perturbation (for example, perturbing a protein-coding gene X), by comparing the transcriptome profiles between perturbed cells and unperturbed cells. These DEGs serve as ‘signature’ genes of perturbing X. Alternatively, users may also provide their own signature genes of perturbing X (Fig. 1c). Second, PS uses a previously developed scMAGeCK15 model to estimate the average effect of perturbation on these signature genes. Third, a constraint optimization procedure is used to find the value of PS that minimizes the sum of mean squared error between predicted and measured changes of signature genes (Methods). The constraints are used such that PS is non-negative for cells with X is perturbed and is zero otherwise. Such constraints can be established on the basis of the previous information of perturbations; for example, the expression matrix of single-guide RNAs (sgRNAs).
Among published methods, mixscape16 is notable for detecting and removing technical factors, particularly incomplete gene knockouts from CRISPR–Cas9, that influence single-cell perturbation outcomes. mixscape uses a nearest-neighbour subtraction to calculate the ‘perturbation signature’ expression, then fits a Gaussian mixture model to classify cells into either ‘KO’ (fully knocked out) or ‘NP’ (non-perturbed). In contrast, PS models perturbation responses as a continuous variable ranging from 0 to 1, allowing users to specify gene lists as perturbation signature genes. This provides a more flexible framework for analysing perturbation responses, including in a cell-type-specific manner.
PS outperforms mixscape in quantifying partial perturbations
Here we compare PS with mixscape using multiple benchmark datasets. We first generated synthetic datasets, because finding a real scRNA-seq dataset that contains ground truth (that is, accurate measurements of loss-of-function on perturbation) is challenging. We used scDesign3 (ref. 23) to simulate the single-cell transcriptomic responses on perturbing different levels of Nelfb function (25, 50, 75, 100%), based on a real scRNA-seq dataset that deletes Nelfb in mouse T cells24 (Fig. 2a, Extended Data Fig. 1 and Methods). We specified different numbers of DEGs (from 10 to 500) and simulated their expression changes on perturbations of Nelfb functions. In all the cases of partial perturbation (that is, 0.25–0.75), PS outperforms mixscape in terms of the percentage of cells with correct efficiency estimation (defined as absolute error ≤0.1, Fig. 2b–e). In contrast, mixscape uniformly assigned the posterior probability of perturbation to one, leading to its better performance in quantifying 100% perturbation, but less suited to analyse partial gene perturbations. This is possibly due to the bimodal statistic model of mixscape, which only considers 100 or 0% knockout effects16.
a, Benchmark results of both PS and mixscape using simulated datasets with different settings, including perturbation effects (25–100%) and different number of DEGs from bulk RNA-seq (Nelfb knockout versus WT). For each cell, its efficiency is correctly estimated if its absolute error is no more than 0.1; that is, \(\left|{\psi }_{\mathrm{true}}-{\psi }_{\mathrm{pred}}\right| < 0.1\), where Ψtrue is the true efficiency score and Ψpred is the estimated score. b–e, The score distribution of PS and mixscape using 50 DEGs and different values of true efficiencies: DE, 50; true efficiency, 25% (b), DE, 50; true efficiency, 50% (c), DE, 50; true efficiency, 75% (d) and DE, 50; true efficiency, 100% (e). Source numerical data are available in the source data. f, Benchmark pipeline using real CRISPRi-based Perturb-seq datasets, where perturbation efficiency is directly assessed through gene expression. g–j, Benchmark results comparing PS and mixscape using a published Perturb-seq dataset. The numbers (and percentage) of genes with accurate efficiency estimation are defined by PCC < −0.1 and P value ≤0.05. The Perturb-seq experiments were conducted under low and high MOI conditions, where most cells express only one guide in low MOI and multiple guides in high MOI. The benchmark includes 23 in the low MOI dataset and 342 genes in the high MOI dataset. Correctly estimated genes in low MOI (g) and high MOI (h), and correctly estimated cells in low MOI (i) and high MOI (j). k, A representative gene (ACTB) where PS correctly estimated the efficiency of CRISPRi. A linear regression line is shown, and the shaded area indicates a 95% confidence interval of linear regression. PCC = −0.36, n = 54, and the associated Pearson’s test P value is P = 0.008. Source numerical data are available in the source data. DE, differential expression.
Next, we evaluated different methods using several public CRISPRi-based Perturb-seq datasets, as CRISPRi modulates gene expression directly, allowing perturbation efficiency to be accessed from the data. Specifically, we used two published K562 CROP-seq datasets25. In the first dataset, each cell expresses only one guide RNA (gRNA) (low multiplicity of infection or MOI), while in the second dataset, multiple gRNAs are expressed per cell (high MOI). We focused on cells where the transcription start sites (TSS) of highly expressed protein-coding genes were targeted. For each gene perturbation, we excluded the expression of the perturbed gene (X) from the evaluation (Fig. 2f). PS correctly estimated CRISPRi efficiency in more than 40% of these genes (10 out of 23 for low MOI, 139 out of 342 for high MOI; Fig. 2g,h), defined as a Pearson correlation coefficient (PCC) < −0.1 and P ≤ 0.05. In contrast, mixscape correctly identified none of these genes for the low MOI dataset (Fig. 2g), or in less than 5% of all the genes for the high MOI dataset (Fig. 2h). Beyond that, PS detects a much greater number of cells that have a strong perturbation effect (PS or mixscape score >0.5, Fig. 2i,j), whose scores are strongly negatively correlated with gene expression (Fig. 2k). We also tested both methods in another CRISPRi-based Perturb-seq dataset, where sgRNAs with mismatches were introduced during the guide design, leading to partial perturbation effects26 (Extended Data Fig. 1b,c). PS has a high sensitivity and a good balance between sensitivity and specificity, evidenced by the higher areas under the receiver-operating characteristic (ROC) curve (AUC) values (Extended Data Fig. 1b) and PCC values (Extended Data Fig. 1c).
To further benchmark methods in terms of a phenotype of interest, we designed and performed a genome-scale CRISPRi Perturb-seq on both unstimulated and stimulated Jurkat, a T lymphocyte cell model (Fig. 3a), and evaluated the performances of different methods in identifying known regulators of T cell activation. We designed Perturb-seq library that contains sgRNAs targeting the TSS of 18,595 genes (4–6 guides per gene) and used a TAP-seq-based27 multiplex primer panel to detect the expressions of 374 genes with high sensitivity (Supplementary Tables 1 and 2 and Methods). We obtained high-quality scRNA-seq data on over 586,000 single cells after quality control, and the uniform manifold approximation and projection (UMAP) for dimension reduction clustering of Perturb-seq datasets clearly demonstrated the differences between stimulated and non-stimulated cells (Fig. 3b). As an independent validation, we reanalysed a published genome-scale T cell CRISPR screening dataset28, and identified 385 (and 1,297) genes that regulate (or do not regulate) T cell stimulation, respectively (Methods). To compare these genes with PS and mixscape scores, which are at a single-cell level, we calculated a ‘cumulative score’ for each gene in Perturb-seq, by summing up all PS and mixscape scores of that gene across all single cells. Because our system focuses on T cell stimulation, the cumulative score of a gene should reflect the relative importance of this gene on T cell stimulation, making it comparable with genes in pooled CRISPR screens. Indeed, both PS and mixscape identified many known positive regulators of T cell activation, such as components of the T cell receptor complex (for example, CD3D) and proximal signalling components (for example, LCK, Fig. 3c). For many positive genes, cells with higher values of PS or mixscape score are skewed towards the non-stimulating state, consistent with their negative selections in pooled CRISPR screens using T cell stimulation as readout (Fig. 3c and Extended Data Fig. 2). However, when comparing the ROC score, PS reaches a higher AUC score than mixscape (Fig. 3d), indicating its better performance in accurately separating positive from negative hits.
a, Benchmark procedure using a genome-scale Perturb-seq and a published, pooled T cell CRISPR screen. b, The distribution of unstimulated and stimulated Jurkat cells along the UMAP plot. c, The correlation of predicted cumulative scores by PS and mixscape. The cumulative score of each gene is the sum of scores of that gene across all cells, and measures the relative contribution of perturbing each gene on affecting T cell stimulation. Positive and negative genes, identified from a published, genome-scale CRISPR screen on stimulated T cells28, are marked in cyan and red, respectively. d, the ROC curve of both methods in separating positive and negative hits. e,f, Benchmark using a published ECCITE-seq where PDL1 protein expression is used as gold standard (e), and the performance of different methods in terms of predicting PDL1 protein expression (f). In f, red gene names indicate known PDL1 regulators. Source numerical data are available in the source data.
Finally, we tested different methods using a published ECCITE-seq dataset, simultaneously measuring single-cell transcriptomes, surface proteins and perturbations16. PDL1 protein expression was chosen as an independent metric for evaluation (Fig. 3e) due to its well-understood regulatory role. Of the 25 genes perturbed in the ECCITE-seq library, 17 are known regulators of PDL1 expression (Fig. 3f). We compared PS and mixscape in their ability to predict changes in PDL1 expression (Fig. 3f and Extended Data Fig. 3), alongside a naïve approach that simply used the expression of the perturbed genes. PS outperformed mixscape and the naïve method in predicting PDL1 expression for 19 out of 25 genes (76%), including 12 out of 17 (71%) known PDL1 regulators. Notably, for genes causing strong transcriptomic changes (for example, IFNGR1, IFNGR2, JAK2, STAT1), both PS and mixscape performed well, achieving AUC scores above 0.8 (Fig. 3f). However, for genes with moderate or weak perturbation effects, PS consistently outperformed mixscape, especially for confirmed PLD1 regulators (genes marked in red in Fig. 3f). Overall, these results demonstrate the outstanding performance of PS over existing methods.
Analysing dose-dependent effects of perturbation
The dosage analysis of gene or drug functions requires a careful, time-consuming adjustment of perturbation strength26,29. Since the quantifying partial gene perturbation by PS is highly accurate (for example, Fig. 2), we ask whether PS can be used to analyse dose–response perturbation responses from single-cell perturbation data. By examining ECCITE-seq data in which PDL1 expression was measured directly (Fig. 3e), we found correlations between PDL1 expression and the PS of known PDL1 regulators (Fig. 4a). The PSs of positive PDL1 regulators (for example, IFNGR1/2, STAT1; Fig. 4b) are negatively correlated with PDL1 expression, while the scores of negative regulators (for example, CUL3, BRD4) are positively correlated (Fig. 4c and Extended Data Fig. 3). One example is CUL3, which is known to destabilize and degrade PDL1 protein expression30. Consequently, higher CUL3 PSs, indicating higher CUL3 functional perturbation, correspond to higher PDL1 protein expressions (Fig. 4c). Compared with mixscape, PS more accurately predicts the quantitative changes in PDL1 expression, evidenced by stronger Pearson correlations between the two (for example, Extended Data Fig. 3).
a, The correlation between a gene’s PS and a phenotype of interest indicates positive (or negative) regulations. b,c, The correlation between PDL1 protein expression and the PS of STAT1 (b) and CUL3 (c). For STAT1 (b), PCC = −0.48, P = 7.95 × 10−27, n = 444 and for CUL3 (c), PCC = 0.44, P = 1.37 × 10−14, n = 274. The associated Pearson’s test was used to calculate P values when calculating PCC. CUL3 is a known negative regulator of PDL1, while STAT1 is a known positive regulator. d, The classification of buffered or sensitive genes using Hill equation. Here, the ED50 represents the dose (that is, perturbed gene expression) that reaches half of its maximal PS score. e, The classification of buffered or sensitive genes from published Perturb-seq datasets focusing on essential genes in K56232. f, The perturbation-expression plot of RPL5, a buffered gene. The fitted Hill curve and ED50 value are shown. g, The log fold changes of mark gene expressions (columns) on perturbing proteasome genes (rows) from the essential gene Perturb-seq dataset. Source numerical data are available in the source data.
We further investigated the relationships between perturbation efficiency and the strength of perturbation responses, which is measured by PS (Fig. 4d). We are interested in two types of gene: ‘buffered’ genes, where genes have high PSs only when higher perturbation efficiency is achieved, and ‘sensitive’ genes whose PSs are high even with moderate or weak perturbation efficiency. We use a nonlinear Hill equation to fit the values of normalized perturbed gene expression and PS (Fig. 4d and Methods), which has been previously used to determine transcription factor dosages31. The fitted Hill curve yields the empirical dosage 50 (half-maximum effective dose, ED50) value, which represents the expression value that corresponds to 50% of its maximal PS value. The Hill equation is fitted on the basis of the mean values of PS and perturbed gene expression across ten expression quantile groups, to get reliable results from noisy scRNA-seq measurements. In a published Perturb-seq32 that targets 2,285 common essential genes using CRISPRi (Fig. 4e), 488 genes underwent successful Hill equation modelling. Among them, 395 genes are buffered (ED50 ≤ 0.5), indicating high robustness to perturbation, possibly due to their essential roles in cellular functions that require buffers. Many buffered genes form protein complexes, including proteosomes (for example, PSMA3) and ribosomal subunits (for example, RPL5; Fig. 4f and Extended Data Fig. 4a,b). Ninety-three genes are sensitive genes (ED50 > 0.5), showing strong transcriptome responses even with moderate or weak efficiencies on perturbing gene expression (Extended Data Fig. 4c–f). Many sensitive genes also display buffering effects, demonstrating complex, heterogeneous responses of cells undergoing the same perturbation of essential genes. Notably, a 50% reduction of HSPA5 expression achieved near-maximal transcriptional response (and the associated growth defect), as is shown in previous studies26.
Perturbing one member of the protein complex usually leads to the expression upregulation of other members of the complex, indicating a possible mechanism for compensation (Fig. 4g). For example, perturbing proteosome subunits led to a strong expression reduction of the perturbed gene (for example, PSMA5; blue squares in Fig. 4g) and concurrent upregulation of other members of the proteosomes (for example, PSMB7, PSMD2). Similarly, perturbing genes in ribosomal subunit, mediator and RNA polymerases leads to the upregulation of members of the same functional unit (Extended Data Fig. 4g–j). To confirm our findings on a different cellular system, we examined the effects of perturbing proteasomes in our genome-scale Perturb-seq dataset (Fig. 3a). Indeed, perturbing members of the proteasome subunits lead to the upregulation of other proteosomes (Extended Data Fig. 4h), consistent with the known transcriptional feedback loop that is observed between proteasome genes33. Overall, the widespread existence of such compensatory effect may explain the perturbation-expression phenotype of buffered genes, where a strong perturbation efficiency is needed to achieve strong expression changes.
PS reveals factors in latent HIV and T cell activation
Reversing latent HIV-1 expression in resting CD4+ T cells is critical for curing HIV. Several genetic and epigenetic factors in CD4+ T cells have been identified as targets for latency reversal agents (LRAs), which aim to eliminate the latent HIV-1 reservoir. However, blocking these factors can also globally activate T cells, increasing LRA toxicity34,35. Therefore, understanding how key genes regulating latent HIV expression affect T cell states is crucial. We performed a Perturb-seq experiment using a previously established Jurkat HIV cell model36, which stably expresses Cas9 and is latently infected with an HIV-GFP viral vector. The Perturb-seq library targeted ten protein-coding genes (Supplementary Table 3), including known regulators of HIV-1 expression and T cell activation (for example, BIRC2), and top hits from previous CRISPR screens (for example, BRD4)36. Three Perturb-seq experiments were conducted: stimulated Jurkat cells (with phorbol 12-myristate 13-acetate (PMA) and ionomycin (I), referred to as PMA/I), followed by green fluorescent protein (GFP) sorting (GFP+ or GFP−) and unstimulated DMSO (dimethylsulfoxide)-treated cells (Fig. 5a). Single-cell transcriptomes and sgRNA expressions were obtained using the 10X Genomics Chromium platform. After quality controls, 7,063–8,811 single cells per sample were retained, with at least 69,888 reads per cell and a median of 4,744 genes expressed per cell (Extended Data Fig. 5a). sgRNAs were detected in more than 96% of cells, with 85% assigned a unique sgRNA (Supplementary Table 4). Cells clustered primarily by stimulation status (stimulated versus unstimulated, Fig. 5b).
a, The experimental design of Perturb-seq. b, The UMAP plot of single-cell transcriptome profiles. Cells are coloured by three different conditions. c, The distribution of BRD4 PS. d, The expression of HIV-GFP. e, The correlations between HIV-GFP expression and BRD4 PS that does not use HIV-GFP as the target gene. The PCC P values (calculated from Pearson’s test) are 1.40 × 10−65, 1.96 × 10−6, 6.26 × 10−29; n = 457, 179, 402 for DMSO, GFP− and GFP+ conditions, respectively. The shaded area indicates a 95% confidence interval of locally estimated scatterplot smoothing regression. f, The distribution of CCNT1 PS. g, The protein expression of HIV-GFP in response to CCNT1 knockout in different cell states (TNF versus non-stimulated). Source numerical data are available in the source data.
We investigated gene functions using our PS framework. Among the perturbed genes, BRD4 (bromodomain containing 4) showed a distinct cell state-specific pattern, where a subset of cells exhibited higher BRD4 PS values (BRD4-PS+ cells) compared with others (BRD4-PS− cells; Fig. 5c). BRD4-PS+ cells overexpress genes involved in BRD4-related functions37,38, such as NF-kB/TNF signalling, hypoxia and apoptosis (Extended Data Fig. 5b–d). These cells also have lower expression of BRD4 signature genes from an independent study39 (Extended Data Fig. 5e), indicating a stronger BRD4 perturbation. BRD4 is a known regulator of HIV transcription and activation36,40, which is consistent with the strong upregulation of HIV-GFP in BRD4-PS+ cells (Extended Data Fig. 5f). Additionally, BRD4-PS+ cells exhibit stronger GFP expression (Fig. 5d), confirming a greater BRD4 perturbation. To explore the dosage effect of BRD4 perturbation, we recalculated BRD4 PS without HIV-GFP and examined its association with HIV-GFP expression in different conditions (Fig. 5e). The correlation between BRD4 PS and HIV-GFP expression depends on cell state. In stimulated T cells (PMA/I treatment), BRD4 PS and HIV-GFP expression showed a linear, positive correlation. In unstimulated T cells (DMSO), however, a nonlinear relationship was observed, with stronger BRD4 PS (>0.5) leading to a sharp increase in HIV-GFP (Fig. 5e).
Another gene, cyclin T1 (CCNT1), also shows heterogeneity in PS distribution. Cells with CCNT1 perturbation have high PS values only in stimulated cells (Fig. 5f), and mixscape scores show a similar pattern (Extended Data Fig. 6a). In contrast, CCNT1 gene expression and guide distribution remain consistent across cell states (Extended Data Fig. 6b). Confirming our findings, the number of DEGs in cells with CCNT1 perturbation compared with cells non-targeting guides is over 100 in stimulated cells, but only one in non-stimulated cells (adjusted P < 0.001; Extended Data Fig. 6c). Notably, HIV-GFP is the strongest DEG, consistent with the known role of CCNT1 in HIV transcription activation.
CCNT1 is a key subunit of P-TEFb (positive transcription elongation factor b)/CDK9 complex that drives RNA transcription, including HIV. Transcription elongation control of P-TEFb/CDK9 is regulated by multiple mechanisms, such as T cell signalling pathways (for example, NF-kB), translation control and epigenetic modification41. These factors vary between T cell states (for example, NF-kB; Extended Data Fig. 6d), probably explaining the cell state-specific differences in CCNT1 PS. Despite this dependency, CCNT1 PS weakly correlates with HIV-GFP within a single-cell state, unlike BRD4 PS (Fig. 5e and Extended Data Fig. 6e).
To further validate that CCNT1 perturbation response is cell state-dependent, we stimulated Jurkat cells using TNF. We measured HIV-GFP expression as an indicator of CCNT1 perturbation, because CCNT1 knockout strongly reduces HIV-GFP expression42,43. With TNF stimulation, CCNT1 knockout reduced HIV-GFP expression by more than 50%, compared with less than 5% in unstimulated cells (Fig. 5g), confirming that the T cell state is critical for CCNT1 function.
Cell states, including T cells, often exist in a continuous space, which can be captured through scRNA-seq. We investigated whether PS can identify factors regulating continuous T cell states, beyond the discrete states of stimulated versus non-stimulated cells (Fig. 5b). CD69 messenger RNA (mRNA) expression was used as a marker for continuous T cell stimulation (Extended Data Fig. 6f), as CD69 is an early T cell activation marker44. The PS of CD247, a top hit from pooled CRISPR and TAP-seq screens (Fig. 3), showed a strong negative correlation with CD69 expression (Wilcox test P = 1.26 × 10−51; Extended Data Fig. 6g,h), consistent with the essential role of CD247 in T cell activation (Fig. 3). In contrast, the PS of PDCD1 (PD-1), a checkpoint protein that inhibits T cell activation, was positively correlated with CD69 expression (Wilcox test P = 1.3 × 10−3; Extended Data Fig. 6i,j). These findings highlight the power of PS as a computational framework for identifying cofactors that drive transcriptomic responses across both discrete and continuous cell states.
PS uncovers gene functions in pancreatic differentiation
To study the functions of lineage regulators during human pancreatic differentiation, we used an established in vitro human embryonic stem (hES) cell pancreatic differentiation system to generate cells corresponding to early-stage (definitive endoderm, DE) and middle-stage (pancreatic progenitor, PP) pancreas development. We generated ten clonal hES cell lines with the homozygous knockout of four genes (Supplementary Table 5), including two known pancreatic lineage regulators (HHEX, FOXA1) and two uncharacterized candidate regulators from previous genetic screens (OTUD5, CCDC6)45,46. These clones were labelled with distinct LARRY (lineage and RNA recovery) DNA barcodes47, pooled and differentiated into DE and PP stages using established protocols45. Single-cell gene expression was then profiled using the 10X genomics Chromium platform (Fig. 6a), with clone identity determined from LARRY barcodes.
a, Experimental design of multiplexing scRNA-seq on the knockout clones of different genes. b, The UMAP plot of single-cell transcriptome profiles, coloured by different clusters (left) or clones (right). c, The PS distribution of HHEX. d, The percentage of cells in PP/LV/DUO cell types from different clones. e, The correlations of CCDC6 PSs calculated from different cell types. The PCC is calculated from all cells with CCDC6 knockouts and is shown as numbers on the heatmap. f, Two different distribution patterns of CCDC6 PSs. g, The top enriched gene ontology terms of DEGs from PP/PP in transition. Enrichr was used to perform enrichment analysis and calculate the associated adjusted P value. h, The percentage of cells in PP/LV/DUO cell types from CCDC6 clones. i, The percentage of cells with PDX1+ (a PP marker) or HNF4A+ (a LV marker) by flow cytometry sorting. The data are based on two CCDC6 knockouts (KO1, KO2) and one WT control. For PDX1+, the adjusted P value for WT versus CCDC6 KO1 is 0.002 and for WT versus CCDC6 KO2 is 0.011. For HNF4A+, the adjusted P value for WT versus CCDC6 KO1 is 0.031 and for WT versus CCDC6 KO2 is 0.015. Three independent replicates are performed for each condition. The multiple comparison-adjusted P value is calculated by one-way analysis of variance test. *P < 0.05, **P < 0.01. Source numerical data are available in the source data.
Among 26,286 single cells that passed the quality control, over 97% (25,694 of 26,286) had at least one barcode detected, and over 80% (20,678 of 25,694) were identified as singlets, which were retained for downstream analysis. UMAP clustering revealed different known cell types during pancreatic differentiation on the basis of the expression of cell-type-specific markers (Fig. 6b and Extended Data Fig. 7). These included DE, PP, liver/duodenum progenitor (LV/DUO), endocrine precursor and cells in transition stages (for example, DE in transition, PP in transition).
Among the knockout genes, HHEX showed high PS in cells transitioning between PP and LV/DUO stages (Fig. 6c and Extended Data Fig. 7), consistent with the known function of HHEX as a key determinant of cell fate decision. HHEX deletion drives DE cells towards LV/DUO lineage rather than PP45. This was reflected by the reduced percentage of PP-annotated cells in the HHEX knockout condition (Fig. 6d). Similarly, the PS of FOXA1, another key transcription factor during PP differentiation, was high in DE and PP cells, consistent with the specific expression pattern of FOXA1 in these cell types (Extended Data Fig. 8a–c).
CCDC6 is one of the top hits of CRISPR screens, and its perturbation hinders PP differentiation45,48. However, its precise role during pancreatic differentiation remains largely unknown. CCDC6 appears to have distinct functions across different cell types, as the DEGs between these cell types show minimal overlap (Extended Data Fig. 8d–f). To account for cell type-specific effects, we calculated PSs on the basis of the DEGs in four main cell types: DE in transition, DE, PP/PP in transition and LV/DUO. Unbiased clustering of these CCDC6 PSs revealed two distinct patterns across cell types (Fig. 6e). PS from late-stage cell types (PP/PP in transition/LV/DUO; ‘pattern 1’) were distinct from those of early-stage cell types (DE in transition/DE; ‘pattern 2’, Fig. 6f and Extended Data Fig. 9a,b).
In early-stage cell types, PS correlates with factors such as POU5F1 and SOX17, which are associated with continuous cell state transitions in DE (Extended Data Fig. 9c,d). DEGs in these stages were enriched in targets of stem cell transcription factors (for example, SOX2, POU5F1, NANOG) and cell cycle regulation genes (Extended Data Fig. 9e–g), consistent with the known function of CCDC6 as a cell cycle regulator49,50. In contrast, DEGs in late-stage cell types were primarily targets of HNF4A, a key transcription factor driving LV/DUO differentiation (Fig. 6g and Extended Data Fig. 9h). HNF4A was upregulated in late-stage cells on CCDC6 (Extended Data Fig. 8e). Compared with wild-type (WT) cells, CCDC6 knockout cells showed a notable decrease in the percentage of PP cells and an increase in LV/DUO cells (Fig. 6h). These results indicate that CCDC6 plays distinct roles in different cell types, including regulating the differentiation between LV/DUO and PP cell types.
To further validate these predictions, we performed flow cytometry to evaluate the effects of CCDC6 knockout on late-stage cell types (PP/LV/DUO). We measured the percentage of HNF4A+ cells (LV marker) and PDX1+ cells (PP marker). As predicted, CCDC6 knockout significantly reduced the PDX1+ population and increased the HNF4A+ population in three biological replicates (Fig. 6i and Extended Data Fig. 9i), confirming the enrichment of CCDC6 PS in LV/DUO populations (Fig. 6g–h).
Discussion
Understanding cellular responses to perturbations is a central task in modern biology, from studying tumour heterogeneity to developing personalized medicine. These perturbations may be genetic (for example, gene knockouts), chemical (for example, small molecules), mechanical (for example, pressure) or environmental (for example, temperature changes). Single-cell genomics profiles of perturbations are commonly used to investigate the mechanisms of perturbations. Many technologies, including Perturb-seq and sci-Plex, allow multiplexing of multiple perturbations in a high-throughput manner within a single experiment. However, a major bottleneck is the lack of a computational model to fully unlock the potential of high-content perturbation, especially for discovering new biological insights. Here we introduce the PS framework to model the heterogeneous transcriptomic responses, enabling key biological discoveries from this complexity.
Partial gene perturbation is common in experiments due to factors such as dose-controlled drug treatment, incomplete gene knockout (for example, RNA or CRISPR interference, epigenome editing) or random DNA editing from CRISPR–Cas9. These partial perturbations add complexity to biological processes, such as distinguishing between haploinsufficient genes that cause disease when partially disrupted, and haplosufficient genes that require complete knockout for functional disruption. The PS framework excels in quantifying these perturbations and enables detailed dosage analysis across multiple datasets. In addition to measuring perturbations, PS reveals cell-intrinsic and extrinsic factors that influence perturbation responses, making it a powerful functional genomics tool in various areas, such as T cell activation, essential gene function, latent HIV-1 expression, pancreatic differentiation and so on. Current methods for studying partial gene functions often rely on complex CRISPR designs. In contrast, PS provides a versatile and systematic approach for analysing partial perturbations across various methods (for example, CRISPRi or CRISPR–Cas9) and assays (for example, Perturb-seq or multiplex scRNA-seq), enabling the exploration of dosage effects across diverse biological contexts.
PS provides a general framework to analyse several main determinants of perturbation heterogeneity: the strength of perturbation per se (for example, Figs. 2 and 5d; BRD4 in Fig. 5c); compensatory mechanisms in response to perturbations, especially on essential genes (for example, proteosomes; Fig. 4g) and cell type or state specificity (for example, T cell states in Fig. 5; differentiation cell types in Fig. 6). Cell type or state is linked to perturbation responses in three distinct ways: it may change because of perturbation (for example, CCDC6 and HHEX in Fig. 6), it may serve as a critical context that defines perturbation outcomes (for example, T cell states in Fig. 5f,g) or it could act as a confounding factor (for example, BRD4 perturbation heterogeneity in Fig. 5c). PS offers a flexible framework for analysing the heterogeneity of perturbation responses from all these aspects.
PS is also valuable for identifying drug targets in genetic perturbations (for example, CRISPR–Cas9). While titrating pharmaceutical interventions (for example, varying drug doses) is relatively straightforward, precisely controlling the dose of genetic perturbations is more challenging. PS provides a convenient alternative for dose-dependent perturbation analysis, which is critical for drug design. For example, BRD4 is the primary target of bromodomain inhibitor (BETi), a promising class of LRAs for reactivating latent HIV-1 expression. Our analysis indicates stronger BRD4 perturbations are needed to induce HIV-GFP expression (Fig. 5c,d). This finding aligns with observations that many cells escape BRD4 perturbation by CRISPR–Cas9 (ref. 16), limiting BETi efficacy due to efficiency and associated toxicity (as BRD4 is an essential gene). Our previous study36 also demonstrated that significantly higher doses of BETi (for example, JQ1) are needed to induce latent HIV-1 expression at levels similar to other potent LRAs. These results emphasize the need for synergistic drug combinations to mitigate the narrow therapeutic window of BETi.
Confounding factors are a major source of variation in single-cell perturbation studies. These factors can be explicitly modelled using generalized linear models if the confounding source is known or corrected using statistical approaches such as matrix factorization (for example, GSFA51), or independent component analysis (for example, CINEMA-OT52). PS does not directly model confounding factors but can combine with these methods to remove them or detect their influence on perturbation effects (for example, Fig. 5c). In some cases, what are considered confounding factors may reveal biological insights, such as perturbation efficiency or cell state. The orthogonal design of PS allows it to be integrated with existing methods to both correct for confounders and measure perturbation strength.
There are two limitations of PS. First, PS uses two separate stages: initially estimating the effect sizes of perturbations (step 2 in Fig. 1c), and then estimating the PS per cell (step 3). The first stage assumes that all cells have the same effect size (that is, PS = 1), which may not hold true in all scenarios. A more robust approach, such as the expectation-maximization algorithm or Bayesian inference method, could jointly estimate both effect sizes and PS more accurately. Second, perturbations on essential gene functions can affect cellular viability53,54, but single-cell profiling only captures cells that survive perturbations. This ‘survival bias’ means PS may only reflect the perturbation responses in a subset of cells rather than capturing the full range of effects. To address this, PS could be combined with recent prediction methods that account for uneven distribution between perturbed and non-perturbed cells55. Notably, PS complements a recently developed tool, Mixscale22, which also addresses cellular variations in perturbation efficiency, and provides methodology for optimized differential expression analysis and molecular pathway signature reconstruction. Both PS and Mixscale provide valuable computational methods to understand cellular responses to various types of perturbation.
Methods
The PS framework
PS estimation follows three steps (Fig. 1c): target gene identification (step 1), average perturbation effect estimation using scMAGeCK (step 2) and PS estimation using constrained optimization (step 3).
Step 1: target gene identification
We performed differential expression analysis between cells with certain perturbation (for example, knocking out gene X) and negative control cells. In Perturb-seq, control cells are typically those with non-targeting gRNAs, while in pooled scRNA-seq, they are WT cells. In high MOI conditions, control cells may also include those without the specific perturbation. The Wilcoxon rank sum test (via Seurat) identified and ranked DEGs, with top genes selected as potential targets. By default, an absolute log fold change threshold of 0.1 is used, and if a minimum number of target genes (default of ten, Extended Data Fig. 10a) is not met, the threshold is iteratively decreased (up to three times) to include more genes. Users can also provide their own target gene list, skipping this step if previous knowledge is available.
Step 2: average perturbation effect estimation
We used the linear regression module in scMAGeCK (scMAGeCK-LR) to estimate the average perturbation effect. scMAGeCK-LR takes the expressions of target genes (identified in step 1) across all cells as input and outputs a β score, conceptually similar to log fold change. The β score offers two advantages: it supports high MOI Perturb-seq datasets, where cells may express multiple guides, and allows simultaneous estimation of multiple perturbations (for example, genome-scale perturbations) in one step, unlike standard DEG analysis.
The mathematical model of scMAGeCK-LR is as follows. Let Y represent the log-transformed expression matrix of M cells and N target genes, which are the union of all target genes identified in step 1 for K perturbations. Let D be the binary cell identity matrix, where djX = 1 if cell j contains sgRNAs targeting gene X and djX = 0 otherwise. The matrix Β contains the β scores, where βXA > 0 indicates gene X is positively affects gene A’s expression and βXA < 0 indicates a negative effect.
The expression matrix Y is modelled as:
where Y0 is the basal expression in an unperturbed state and ϵ is Gaussian noise. Y0 can be estimated from negative control cells or neighbouring negative control cells, as in mixscape16. The value of Β can be estimated using ridge regression:
where I is the identity matrix and λ is a small positive value (default 0.01).
Step 3: PS estimation using constrained optimization
We revise equation (1) to incorporate PS. Here, the log-transformed expression matrix Y is modelled as:
where Ψ is the non-negative, raw PS matrix with the same size as D in step 2 (M × K). Each element ΨjX in Ψ indicates the raw PS of cell j of perturbing gene X. Here, B is the matrix of β scores estimated in step 2. To find Ψ, we minimize the squared error between predicted and observed gene expressions across all cells:
subject to the following constraints:
Here, U is the upper bound of raw Ψ values, dik is from the binary cell identity matrix in step 2, 1 ≤ j ≤ M is the index of single cells, 1 ≤ i ≤ N is the index of target genes and 1 ≤ k ≤ K is the index of perturbations.
Since Ψ has non-negative constraints, we simplify the objective function as follows:
This results in a constrained quadratic optimization problem, solvable using methods such as Newton’s method. The final normalized PS is scaled to [0,1] as follows:
The choice of λ
By default, λ is set to 0.01, but we also provide a method to choose λ to control the false-positive rate. This involves randomly selecting control cells (for example, cells expressing non-targeting gRNAs) and calculating the percentage of these cells with PS ≥ 0.5, which represents the false-positive rate. λ can then be chosen on the basis of the desired false-positive rate (for example, 0.1; Extended Data Fig. 10b). This is implemented in the scmageck_best_lambda function in our source code.
Simulated datasets
Twenty simulated datasets were generated by the simulator scDesign3 (ref. 23) (v.1.1.1) with modifications for Perturb-seq. The simulation uses scDesign3’s parametric model to capture the characteristics of the reference scRNA-seq data, which includes Nelfb-perturbed (knockout, KO) and unperturbed (WT) mouse T cells24. The datasets were generated under 20 different settings, combining two parameters: the number of downstream genes affected by Nelfb’s knockout (10, 50, 100, 200 and 500) and the perturbation efficiency (25, 50, 75 and 100%). The simulation steps (detailed in steps 1–4 below) use downstream genes identified from the bulk RNA-seq data (Supplementary Data 1 from the original publication24), resulting in a total of 20 simulated datasets.
Before running the simulation, we preprocess the scRNA-seq dataset and the bulk DE gene rank list. First, we apply the same quality control as the original publication24, retaining cells with 1,000–5,000 detected genes and less than 12% mitochondrial unique molecular identifier counts. Second, we impute and amplify the WT mouse cell gene-by-cell count matrix to enhance perturbation effects. Using the R package scImpute56 (v.0.0.9), we impute the WT count matrix to reduce sparsity and then multiply it by an amplification factor of ten to extend the range of gene expression levels. Third, we combine the imputed WT count matrix and the knockout count matrix to create a gene-by-cell matrix. This matrix has dimensions (p + 1) × N, representing p + 1 genes (Nelfb and p other genes) and N cells, split into NWT WT cells and NKO knockout cells. Fourth, we extract Nelfb’s counts as a vector (C) and denote the remaining gene counts as a p × N matrix Y. Fifth, we refine the bulk DE gene list by excluding genes with zero rows in Y. Last, we reduce the computation by using the scran57 package (v.1.28.2) to select 3,000 highly variable genes in Y. The final Y matrix, after including the refined bulk DE genes, has dimensions 3,390 × N.
We know which cells have Nelfb perturbed, represented by an N-dimensional binary vector K, where Kj = 0 indicates a WT cell and Kj = 1 indicates a knockout cell. K and C are used as two covariate vectors, and Y is used as the reference count matrix for scDesign3. We modify scDesign3 to use Y, C, K, the refined DE genes, the number of Nelfb’s downstream genes and the perturbation efficiency to simulate data in the following four steps:
Step 1: modelling each gene’s marginal distribution independently
For each gene i, if it is a downstream gene of Nelfb, Yij, conditional on Cj, follows a zero-inflated negative binomial (ZINB) distribution with mean μij, dispersion ɸi and zero-inflation probability νij. For a non-downstream gene, Yij, follows a ZINB distribution with mean μi, dispersion ɸi and zero-inflation probability νi. These distributions are specified using a generalized additive model for location, scale and shape. The first D genes in Y (where D \(\in \{0,\,10,\,50,\,100,\,200,\,500\}\)) represent the top DE genes, defined as Nelfb’s downstream genes. scDesign3 is modified so that downstream gene’s marginal distributions depend on Cj, while non-downstream gene’s distributions are independent of Cj.
For Nelfb’s downstream gene \(i=1,\,\ldots ,D\):
For Nelfb’s non-downstream gene \(i=D+1,\,\ldots ,p\):
After parameter estimation by the R package gamlss (v.5.4-12), the fitted distribution of \({Y}_{ij}{|}{\bf{C}}_{j}\), for \(i=1,\,\ldots ,D\), is denoted as \(\text{ZINB}(\,{\hat{\mu }}_{{ij}},\,{\hat{\phi }}_{i},\,{\hat{\upsilon }}_{{ij}})\) with the cumulative distribution function (CDF) \({\hat{F}}_{{ij}}\); the fitted distribution of Yij, for \(i=D+1,\,\ldots ,p\), is denoted as \(\text{ZINB}(\,{\hat{\mu }}_{i},\,{\hat{\phi }}_{i},\,{\hat{\upsilon }}_{i})\) with the CDF \({\hat{F}}_{i}\). The other parameters including ɑi, βi, γi and ηi are estimated as \({\hat{\alpha }}_{i}\), \({\hat{\beta }}_{i}\), \({\hat{\gamma }}_{i}\) and \({\hat{\eta }}_{i}\) for each i respectively.
Step 2: modelling genes’ joint distribution using the Gaussian copula
To approximate pairwise gene–gene correlations in the reference dataset, scDesign3 uses the Gaussian copula. Based on the marginal distributions from step 1, scDesign3 approximates the joint distribution of the p genes in cell j as
where \({{\Phi}^{-{1}}}({\cdot})\) is th e inverse CDF of the standard Gaussian distribution, 0 is the p-dimensional zero vector and \({\hat{\bf{R}}}\left(\bf{K}_{j}\right)\) is the estimated p × p gene–gene correlation matrix of the Gaussian copula, conditional on Kj. Since Kj is binary, we estimate two gene–gene correlation matrices: one for the WT cells (Kj = 0) and one for the knockout cells (Kj = 1). A technique called distributional transform is used to make the CDFs continuous (see ref. 58 for details).
Step 3: modifying the fitted parameters
To generate synthetic datasets with different perturbation efficiencies, we adjust the mean parameters for Nelfb’s downstream genes in the knockout cells on the basis of the user-specified efficiency. We assume the first \({\bf{N}}^{\text{KO}}={\sum }_{j=1}^{N}I({\bf{K}}_{j}=1)\) cells are knockout cells and update the mean parameters \({\hat{\mu }}_{{ij}}\) for downstream genes (\({i}\in \{1,\,\ldots ,D\}\), \(j\in \{1,\,\ldots ,\,{\bf N}^{\text{KO}}\}\)) as follows. For the 50% perturbation efficiency: We randomly sample NKO values from Cj in WT cells \((j\in \{{\bf N}^{\text{KO}}+1,\,\ldots ,{\bf N}\,\}\}\), scale them by 0.5 and store them as \(\bf{C}^*={\left(\bf{C}_{1}^{* },\ldots ,\bf{C}_{{\bf N}^{\text{KO}}}^{* }\right)}^{T}\). We then modify the mean parameters for the downstream genes in synthetic knockout cells as \({\hat{\mu }}_{{ij}}={\hat{\alpha }}_{i}+{\hat{\beta }}_{i}\times{\bf{C}}_{j}^{* }\). For the 100% perturbation efficiency, we set C* to a zero vector of length NKO and modify \({\hat{\mu }}_{{ij}}\) similarly. We do not modify mean parameters for WT cells, non-downstream genes, dispersion parameters or zero-inflation probability. The N-dimensional vector S represents Nelfb’s counts in the synthetic cells, with \({\bf{S}}_j={\bf{C}}_j^*\) for \(j\in \{1,\,\ldots ,\,{\bf N}^{\text{KO}}\}\) (knockout cells) and Sj = Cj for \(j\in \{{\bf N}^{\text{KO}}+1,\,\ldots ,{\bf N}\}\) (WT cells).
Step 4: generating synthetic data with the fitted model and modified parameters
First, we independently sample NWT Gaussian vectors of length p from the p-dimensional multivariate Gaussian distribution \({\bf N}\left({\bf 0},{{\bf {\hat R}}}\left({{\bf{K}}}_{j}=0\right)\right)\) and NKO Gaussian vectors from \({\bf N}\left({\bf 0},\bf{\hat{{{R}}}}\left({\bf K}_{j}=1\right)\right)\). These vectors are stacked by row into a p × N Gaussian matrix \(\widetilde{{{Z}}}\). Next, using the parameter estimates (modified or not) from step 3, we convert the Gaussian matrix \(\widetilde{{{Z}}}\) into a p × N ZINB count matrix \(\widetilde{{{Y}}}\) as
Last, we combine \(\widetilde{{{Y}}}\) with S by row to form a (p + 1) × N synthetic count matrix (\(\left[\begin{array}{c}\widetilde{{{Y}}}\\ S\end{array}\right]\)).
Dosage analysis of essential gene perturbations
Data preprocessing
The essential gene Perturb-seq dataset on K562 cells is downloaded from the original study. The normalized gene expression, whose processing method is described in the original study, is used directly. We further normalize the gene expression of each perturbed gene to [0,1], where 0 is the minimum expression of that gene and 1 represents the median gene expression of the perturbed gene in control cells expressing non-targeting guides. To address the noisy and sparse nature of scRNA-seq datasets, we only focus on genes whose corresponding gRNAs are expressed in at least 100 cells. For each perturbed gene, we further define ten expression bins on the basis of the expression quantiles of each gene (0, 10 to 100%), and calculate the average PS value and gene expression for these ten bins. These values will be used to fit the Hill equation as described below.
Hill equation
We use Hill equation to fit the expression-PS curve (Fig. 3d) and classify genes into either buffered or sensitive categories31. A similar approach has been applied to classify sensitive or buffered enhancers bound by SOX9. We use the four-parameter log–logistic function (LL.4) in the R package drc (v.3.0-1) for Hill curve fitting. The four-parameter log–logistic function is defined as:
where x is the normalized expression of the gene being perturbed, and c, d represents the lower and upper bound, respectively. b and e are two other parameters of the Hill curve. In particular, x = e indicates the value where half of the maximum PS score is reached (that is, (c + d)/2), and is defined as the ED50 value of the Hill curve. Buffered (or sensitive) genes are defined as those whose ED50 ≤ 0.5 (or >0.5), respectively, representing whether a greater (or smaller) than 50% expression reduction of the perturbed gene is needed to reach 50% perturbation effect.
Filtering
Because Hill curve fitting does not always generate desired curves, we only keep genes that satisfy the following condition: Hill (x = 0) − Hill(x = 1.0) ≥ 0.2. This heuristic filtering assumes a gene with strong expression perturbation (x = 0.0) should have a larger PS value than a gene with little or no expression perturbation (x = 1.0).
Genome-scale Perturb-seq on Jurkat cells
Perturb-seq
We performed genome-scale Perturb-seq on Jurkat E6 cells expressing dCas9-KRAB. Cells were transduced with a genome-wide CRISPRi CROP-seq library at a high MOI and split into untreated and activated populations (stimulated with anti-TCR and anti-CD28 antibodies for 24 h). Cells were labelled with cell hashing antibodies (Supplementary Table 6) and loaded into 16 channels of a 10X Chromium X instrument, with 115,000 cells per channel and expected recovery rate of 60,000 cells per channel (including 24% multiples). Samples were pooled unequally (10% untreated, 90% treated) and sequenced using NovaSeq S4 PE100 with asymmetric read mode (R1, 28 cycles; R2, 172 cycles at 9,000–10,000 input reads per cell.
sgRNA library design
The genome-wide CRISPRi library targeted the TSS coordinates, calculated from publicly available FANTOM CAGE peaks data. In total, 18,595 genes were targeted, with four sgRNAs per gene. An additional library targeting 3,220 was designed using Jurkat-specific TSS, which were calculated from public Jurkat CAGE-seq datasets. The final library contained 3,220 genes with eight sgRNAs per gene and 15,375 genes with four sgRNAs per gene.
Data preprocessing
We obtained high-quality scRNA-seq data from more than 586,000 single cells, with a median of 13 guides detected per cell and an average of 400 cells per gene perturbation. Transcriptomic reads were mapped using STAR and STAR solo (v.2.7.10a) against a custom gtf annotation (gencode.v34, hg38). Mapping was also performed against custom references for sgRNAs and hash labels. Filtering was done using EmptyDrops, and an initial Seurat object was created with min.cells = 5 and min.features = 10. Outliers were removed on the basis of mitochondrial and mRNA content. Cell labels were assigned using the deMULTIplex method, keeping only cells with exactly one known label. sgRNA calling was conducted using a binomial test using a 0.05 threshold on Benjamini–Hochberg corrected P values. Data from 16 channels were merged, normalized and scaled using Seurat functions, followed by cell cycle scoring, principal components analysis (PCA) and UMAP.
Comparison with pooled CRISPR screens
We analysed published genome-scale T cell CRISPR screens28 using MAGeCK59 (v.0.5.9.5). Positive genes (affecting T cell stimulation) were defined by a false discovery rate of <0.01 (negative selection), and negative genes (not affecting T cell stimulation) were defined by robust ranking aggregation (RRA) scores >0.5 and median log fold changes <0.1. Genes affecting Jurkat growth and/or viability, extracted from DepMap60 Jurkat CRISPR screen, were excluded. A total of 385 positive and 1,297 negative genes were identified for further analysis.
HIV latency Perturb-seq
We used a previously established Jurkat cell line model of HIV latency36, where an HIV vector links GFP to the LTR promoter to measure viral transcription reactivation and HIV latency reversal. Cas9-expressing cells were transduced with a lenti-sgRNA library (MilliporeSigma; LV14, U6-gRNA-10x:EF1a-Puro-2a-BFP) targeting ten genes (three gRNAs per gene). The library contained five positive regulators (NFKB1, CCNT1, PRKCA, TLR1, MAP3K14) and five negative regulators (NFKBIA, NELFE, HDAC2, BRD4, BIRC2) of HIV transcription, along with non-targeting controls. Then 850,000 cells were transduced at an MOI of 0.3 using 8 μg ml−1 polybrene in 2 ml of Roswell Park Memorial Institute medium containing 10% FBS and 1% penicillin–streptomycin. Media was replaced after 24 h, and 2 days posttransduction, cells were selected with 1.5 μg ml−1 puromycin for 5 days. Postselection, cells were split into three groups: unstimulated, and two-thirds stimulated with PMA/I (50 ng ml−1 PMA and 1 μM Ionomycin) for 16 h. Stimulated cells were sorted into GFP+ and GFP− populations, and all three groups were analysed using 10X Genomics single-cell sequencing protocol. Sequencing data (gene expression and CRISPR guide capture) were processed with Cell Ranger (v.6.1.2), and feature-barcode matrices from the three groups were merged for analysis using Seurat (v.4.3.1). Cells expressing more than 7,500 or fewer than 200 genes, or those with >15% mitochondrial reads were excluded. Cells with multiple sgRNAs (due to multiplet droplets or transductions) were also removed. After quality control, data were normalized and scaled. PCA was performed on the top 2,000 highly variable genes, followed by clustering and UMAP embeddings. Biological significance of clusters was explored using Enrichr61.
Pancreatic KO clones and pooled scRNA-seq
Culture of hES
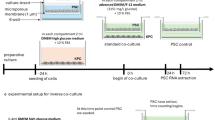
Experiments were performed using H1 (NIHhESC-10-0043) and HUES8 (NIHhESC-09–0021) hES cell lines, following National Institutes of Health (NIH) guidelines and Tri-SCI ESCRO Committee approval. Generation of KO hES cells was described in published studies, including HHEX KO H1 and HUES8 cell lines45, FOXA1 KO HUES8 cell lines62, OTUD5 KO HUES8 cell lines and CCDC6 KO H1 cell lines46. Cells were regularly confirmed to be mycoplasma-free by the MSKCC Antibody & Bioresource Core Facility. KO and WT hES cells were maintained in Essential 8 (E8) medium (Thermo Fisher, A1517001) on vitronectin (Thermo Fisher, A14700) precoated plates at 37 °C with 5% CO2. The ROCK inhibitor Y-27632 (5 µM; Selleck Chemicals, S1049) was added to E8 medium for 1 day after passaging or thawing.
hES cell-directed pancreatic differentiation
hES cells were seeded at 2.3 × 105 cells per cm2 on vitronectin-coated plates in E8 medium with 10 µM Y-27632. After 24 h, cells were washed with PBS and differentiated through a four-stage protocol45. In stage 1 (definitive endoderm, 3 days), cells were cultured in S1/2 medium supplemented with 100 ng ml−1 Activin A (Bon Opus Biosciences) and 5 µM CHIR99021 (04-0004-10, Stemgent) for 1 day, followed by 100 ng ml−1 Activin A for the next 2 days. In stage 2 (primitive gut tube, 2 days), cells were cultured in S1/2 medium supplemented with 50 ng ml−1 human fibroblast growth factor (KGF/FGF-7, AF-100-19, PeproTech) and 0.25 mM Vitamin C (VitC, Sigma-Aldrich, A4544). For stage 3 (PP1, 2 days), cells were cultured in S3/4 medium supplemented with 50 ng ml−1 KGF, 0.25 mM VitC and 1 µM retinoic acid (R2625, MilliporeSigma). In stage 4 (PP2, 4 days), cells were cultured in S3/4 medium supplemented with 50 ng ml−1 KGF, 0.1 µM retinoic acid, 200 nM LDN193189 (Stemgent, 04-0019), 0.25 µM SANT-1 (Sigma, S4572), 0.25 mM VitC and 200 nM TPB (Cell-permeable PKC activator, EMD Millipore, 565740). The base medium formulations were as follows: S1/2 medium consisted of 500 ml of MCDB 131 (15-100-CV, Cellgro), supplemented with 2 ml of 45% glucose (G7528, MilliporeSigma), 0.75 g of sodium bicarbonate (S5761, MilliporeSigma), 2.5 g of bovine serum albumin (BSA) (68700, Proliant) and 5 ml of GlutaMAX (35050079, Invitrogen). S3/4 medium consisted of 500 ml MCDB 131 supplemented with 0.52 ml of 45% glucose, 0.875 g of sodium bicarbonate, 10 g of BSA, 2.5 ml ITS-X and 5 ml of GlutaMAX.
Cell infection with LARRY barcode virus
LARRY barcode constructs (Addgene, 140024) were transfected into 293T cells to generate lentivirus. KO and WT hES cell clones were infected with unique barcodes at low MOI. GFP+ cells were sorted and cultured in E8 medium.
Pooled scRNA-seq
One day before differentiation, ten barcoded hES cell clones were mixed in equal numbers and seeded at 2.3 × 105 cells per cm2. Cells were collected at DE and PP2 stages, frozen and later thawed for scRNA-seq. GFP+ cells were collected and processed using the 10X Genomics platform. Complementary DNA and LARRY barcode libraries were generated separately using specific primers (forward CTACACGACGCTCTTCCGATCT; reverse GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTtaaccgttgctaggagagaccataT).
Data analysis
Transcriptome and barcode libraries were processed with Cell Ranger (v.6.1.2) and analysed using Seurat (v.4.3.1). Cells with >7,000 or <200 genes, or >20% mitochondrial reads were excluded. Singlets were identified on the basis of feature-barcode counts, and cells with multiple barcodes were removed, resulting in 20,678 cells for analysis. Normalization, PCA, clustering and UMAP were performed on the top 2,000 highly variable genes.
Flow cytometry
Cells were dissociated using TrypLE Select and resuspended in fluorescence activated cell sorting buffer (5% FBS, 5 mM EDTA in PBS). Live/Dead Fixable Violet cell stain (Invitrogen, L34955) was used to discriminate dead cells from live cells. Permeabilization and fixation was performed at room temperature for 1 h, followed by antibody in permeabilization buffer. Antibodies for this study include HNF4A, Novus Biologicals, NBP2-67679, 1:200; PDX1, R&D Systems, AF2419, 1:500, Donkey anti-Rabbit IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Thermo Fisher, 1:500; Donkey anti-goat IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Thermo Fisher, 1:500. Cells were then analysed using BD LSRFortessa. Data analysis and figures were generated using FlowJo (v.10), with the gating strategy shown in Extended Data Fig. 10c.
Statistics and reproducibility
Sample sizes (cell numbers) were not predetermined using statistical methods. Experiments were not randomized, and investigators were not blinded during allocation or outcome assessment. Cells with multiple barcodes were excluded, and rigorous quality control measures were applied to remove low-quality cells or empty droplets. Statistical methods for calculating P values are detailed in the figure legends or methods section.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All data generated or analysed in this study are included in this article and its Supplementary Tables. Sequencing data, including genome-scale Perturb-seq, HIV Perturb-seq and pooled scRNA-seq, are available at Gene Expression Omnibus under accession number GSE247601. Reanalysed previously published data are available under the accession numbers: GSE120861, GSE132080, GSE153056, GSE182862 and SRP158611. Source data are provided with this paper.
Code availability
We implemented this framework as part of the scMAGeCK pipeline15. The PS source code, documentation and tutorials can be found on GitHub (https://github.com/davidliwei/PS).
References
Bock, C. et al. High-content CRISPR screening. Nat. Rev. Methods Primers 2, 9 (2022).
Srivatsan, S. R. et al. Massively multiplex chemical transcriptomics at single-cell resolution. Science 367, 45–51 (2020).
Dixit, A. et al. Perturb-seq: dissecting molecular circuits with scalable single-cell RNA profiling of pooled genetic screens. Cell 167, 1853–1866.e17 (2016).
Adamson, B. et al. A multiplexed single-cell CRISPR screening platform enables systematic dissection of the unfolded protein response. Cell 167, 1867–1882.e21 (2016).
Jaitin, D. A. et al. Dissecting immune circuits by linking CRISPR-pooled screens with single-cell RNA-seq. Cell 167, 1883–1896.e15 (2016).
Datlinger, P. et al. Pooled CRISPR screening with single-cell transcriptome readout. Nat. Methods 14, 297–301 (2017).
Xie, S., Duan, J., Li, B., Zhou, P. & Hon, G. C. Multiplexed engineering and analysis of combinatorial enhancer activity in single cells. Mol. Cell 66, 285–299.e5 (2017).
Rubin, A. J. et al. Coupled single-cell CRISPR screening and epigenomic profiling reveals causal gene regulatory networks. Cell 176, 361–376.e17 (2019).
Liscovitch-Brauer, N. et al. Profiling the genetic determinants of chromatin accessibility with scalable single-cell CRISPR screens. Nat. Biotechnol. 39, 1270–1277 (2021).
Dhainaut, M. et al. Spatial CRISPR genomics identifies regulators of the tumor microenvironment. Cell 185, 1223–1239.e20 (2022).
Norman, T. M. et al. Exploring genetic interaction manifolds constructed from rich single-cell phenotypes. Science 365, 786–793 (2019).
Replogle, J. M. et al. Combinatorial single-cell CRISPR screens by direct guide RNA capture and targeted sequencing. Nat. Biotechnol. 38, 954–961 (2020).
Wessels, H.-H. et al. Efficient combinatorial targeting of RNA transcripts in single cells with Cas13 RNA Perturb-seq. Nat. Methods 20, 86–94 (2023).
Hill, A. J. et al. On the design of CRISPR-based single-cell molecular screens. Nat. Methods 15, 271–274 (2018).
Yang, L. et al. scMAGeCK links genotypes with multiple phenotypes in single-cell CRISPR screens. Genome Biol. 21, 19 (2020).
Papalexi, E. et al. Characterizing the molecular regulation of inhibitory immune checkpoints with multimodal single-cell screens. Nat. Genet. 53, 322–331 (2021).
Tsuchida, C. A. et al. Mitigation of chromosome loss in clinical CRISPR-Cas9-engineered T cells. Cell 186, 4567–4582.e20 (2023).
Duan, B. et al. Model-based understanding of single-cell CRISPR screening. Nat. Commun. 10, 2233 (2019).
Barry, T., Wang, X., Morris, J. A., Roeder, K. & Katsevich, E. SCEPTRE improves calibration and sensitivity in single-cell CRISPR screen analysis. Genome Biol. 22, 344 (2021).
Goeva, A. et al. HiDDEN: a machine learning method for detection of disease-relevant populations in case-control single-cell transcriptomics data. Nat. Commun. 15, 9468 (2024).
Tu, X. et al. A supervised contrastive framework for learning disentangled representations of cell perturbation data. In Proc. 18th Machine Learning in Computational Biology Meeting Vol. 240 (eds Knowles, D. A. & Mostafavi, S.) 90–100 (PMLR, 2024).
Jiang, L. et al. Systematic reconstruction of molecular pathway signatures using scalable single-cell perturbation screens. Nat. Cell Biol. https://doi.org/10.1038/s41556-025-01622-z (2025).
Song, D. et al. scDesign3 generates realistic in silico data for multimodal single-cell and spatial omics. Nat. Biotechnol. 42, 247–252 (2024).
Wu, B. et al. RNA polymerase II pausing factor NELF in CD8+ T cells promotes antitumor immunity. Nat. Commun. 13, 2155 (2022).
Gasperini, M. et al. A genome-wide framework for mapping gene regulation via cellular genetic screens. Cell 176, 1516 (2019).
Jost, M. et al. Titrating gene expression using libraries of systematically attenuated CRISPR guide RNAs. Nat. Biotechnol. 38, 355–364 (2020).
Schraivogel, D. et al. Targeted Perturb-seq enables genome-scale genetic screens in single cells. Nat. Methods 17, 629–635 (2020).
Shifrut, E. et al. Genome-wide CRISPR screens in primary human T cells reveal key regulators of immune function. Cell 175, 1958–1971.e15 (2018).
Wessels, H.-H. et al. Prediction of on-target and off-target activity of CRISPR-Cas13d guide RNAs using deep learning. Nat. Biotechnol. 42, 628–637 (2024).
Zhang, J. et al. Cyclin D–CDK4 kinase destabilizes PD-L1 via cullin 3–SPOP to control cancer immune surveillance. Nature 553, 91–95 (2018).
Naqvi, S. et al. Precise modulation of transcription factor levels identifies features underlying dosage sensitivity. Nat. Genet. 55, 841–851 (2023).
Replogle, J. M. et al. Mapping information-rich genotype-phenotype landscapes with genome-scale Perturb-seq. Cell 185, 2559–2575.e28 (2022).
Radhakrishnan, S. K. et al. Transcription factor Nrf1 mediates the proteasome recovery pathway after proteasome inhibition in mammalian cells. Mol. Cell 38, 17–28 (2010).
Bullen, C. K., Laird, G. M., Durand, C. M., Siliciano, J. D. & Siliciano, R. F. New ex vivo approaches distinguish effective and ineffective single agents for reversing HIV-1 latency in vivo. Nat. Med. 20, 425–429 (2014).
Mbonye, U., Leskov, K., Shukla, M., Valadkhan, S. & Karn, J. Biogenesis of P-TEFb in CD4+ T cells to reverse HIV latency is mediated by protein kinase C (PKC)-independent signaling pathways. PLoS Pathog. 17, e1009581 (2021).
Dai, W. et al. Genome-wide CRISPR screens identify combinations of candidate latency reversing agents for targeting the latent HIV-1 reservoir. Sci. Transl. Med. 14, eabh3351 (2022).
Yin, M. et al. Potent BRD4 inhibitor suppresses cancer cell-macrophage interaction. Nat. Commun. 11, 1833 (2020).
Tan, Y.-F., Wang, M., Chen, Z.-Y., Wang, L. & Liu, X.-H. Inhibition of BRD4 prevents proliferation and epithelial-mesenchymal transition in renal cell carcinoma via NLRP3 inflammasome-induced pyroptosis. Cell Death Dis. 11, 239 (2020).
Shu, S. et al. Synthetic lethal and resistance interactions with BET bromodomain inhibitors in triple-negative breast cancer. Mol. Cell 78, 1096–1113.e8 (2020).
Li, Z., Guo, J., Wu, Y. & Zhou, Q. The BET bromodomain inhibitor JQ1 activates HIV latency through antagonizing Brd4 inhibition of Tat-transactivation. Nucleic Acids Res. 41, 277–287 (2013).
Mbonye, U., Kizito, F. & Karn, J. New insights into transcription elongation control of HIV-1 latency and rebound. Trends Immunol. 44, 60–71 (2023).
Wei, P., Garber, M. E., Fang, S.-M., Fischer, W. H. & Jones, K. A. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell 92, 451–462 (1998).
Peng, J., Zhu, Y., Milton, J. T. & Price, D. H. Identification of multiple cyclin subunits of human P-TEFb. Genes Dev. 12, 755–762 (1998).
Cibrián, D. & Sánchez-Madrid, F. CD69: from activation marker to metabolic gatekeeper. Eur. J. Immunol. 47, 946–953 (2017).
Yang, D. et al. CRISPR screening uncovers a central requirement for HHEX in pancreatic lineage commitment and plasticity restriction. Nat. Cell Biol. 24, 1064–1076 (2022).
Rosen, B. P. et al. Parallel genome-scale CRISPR screens distinguish pluripotency and self-renewal. Nat. Commun. 15, 8966 (2024).
Weinreb, C., Rodriguez-Fraticelli, A., Camargo, F. D. & Klein, A. M. Lineage tracing on transcriptional landscapes links state to fate during differentiation. Science 367, eaaw3381 (2020).
Li, Q. V. et al. Genome-scale screens identify JNK-JUN signaling as a barrier for pluripotency exit and endoderm differentiation. Nat. Genet. 51, 999–1010 (2019).
Thanasopoulou, A., Stravopodis, D. J., Dimas, K. S., Schwaller, J. & Anastasiadou, E. Loss of CCDC6 affects cell cycle through impaired intra-S-phase checkpoint control. PLoS ONE 7, e31007 (2012).
Morra, F. et al. FBXW7 and USP7 regulate CCDC6 turnover during the cell cycle and affect cancer drugs susceptibility in NSCLC. Oncotarget 6, 12697–12709 (2015).
Zhou, Y., Luo, K., Liang, L., Chen, M. & He, X. A new Bayesian factor analysis method improves detection of genes and biological processes affected by perturbations in single-cell CRISPR screening. Nat. Methods 20, 1693–1703 (2023).
Dong, M. et al. Causal identification of single-cell experimental perturbation effects with CINEMA-OT. Nat. Methods 20, 1769–1779 (2023).
Hart, T., Brown, K. R., Sircoulomb, F., Rottapel, R. & Moffat, J. Measuring error rates in genomic perturbation screens: gold standards for human functional genomics. Mol. Syst. Biol. 10, 733 (2014).
Morgens, D. W., Deans, R. M., Li, A. & Bassik, M. C. Systematic comparison of CRISPR/Cas9 and RNAi screens for essential genes. Nat. Biotechnol. 34, 634 (2016).
Bunne, C. et al. Learning single-cell perturbation responses using neural optimal transport. Nat. Methods 20, 1759–1768 (2023).
Li, W. V. & Li, J. J. An accurate and robust imputation method scImpute for single-cell RNA-seq data. Nat. Commun. 9, 997 (2018).
Lun, A. T. L., Bach, K. & Marioni, J. C. Pooling across cells to normalize single-cell RNA sequencing data with many zero counts. Genome Biol. 17, 75 (2016).
Sun, T., Song, D., Li, W. V. & Li, J. J. scDesign2: a transparent simulator that generates high-fidelity single-cell gene expression count data with gene correlations captured. Genome Biol. 22, 163 (2021).
Li, W. et al. MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol. 15, 554 (2014).
Corsello, S. M. et al. Discovering the anti-cancer potential of non-oncology drugs by systematic viability profiling. Nat. Cancer 1, 235–248 (2020).
Kuleshov, M. V. et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44, W90–W97 (2016).
Lee, K. et al. FOXA2 is required for enhancer priming during pancreatic differentiation. Cell Rep. 28, 382–393.e7 (2019).
Acknowledgements
We thank all members from the Li and Huangfu laboratory for comments and discussions. We thank J. P. Taylor-King for discussions. This study is supported by grant numbers NIH R01 HG010753 and HL168174 (to W.L.), District of Columbia Center for AIDS (DC-CFAR) Research Transitioning Investigator Award (grant number AI117970, to W.L.), pilot award for Children’s National Hospital and Virginia Tech (CNH-VT) Research Collaborations on AI (to W.L.) and startup support from the Center for Genetic Medicine Research at Children’s National Hospital (to W.L.). D.H is supported by grant numbers NIH UM1 HG012654 and U01 HG012051. J.J.L. is supported by National Science Foundation grant numbers DBI-1846216 and DMS-2113754, NIH R35 GM140888, a Johnson and Johnson WiSTEM2D Award, a Sloan Research Fellowship, a UCLA David Geffen School of Medicine W.M. Keck Foundation Junior Faculty Award and Chan-Zuckerberg Initiative Single-Cell Biology Data Insights (Silicon Valley Community Foundation grant number 2022-249355). W.D. and R.F.S. are supported by the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Contributions
W.L. conceived the project. W.L. and B.S. developed the method. W.L., B.S. and D.L. designed and performed the experiments and analysed the data. W.D., N.F.M., H.Z. and B.S. performed and analysed HIV Perturb-seq under the supervision of J.D.S., R.F.S. and W.L. B.S., Q.W. and D.S. performed synthetic experiments under the supervision of W.L. and J.J.L. D.L., D.Y., B.W., B.R. and H.T.K generated pancreatic differentiation dataset and performed validations under the supervision of D.H. A.K., A.V., N.U. and A.L. generated and analysed genome-scale Perturb-seq under the supervision of T.B. X.C., L.C. and Y.D. performed the analysis and interpretation of the results. W.L. and B.S. wrote the paper with input from all the authors. W.L., T.B., J.J.L., R.F.S. and D.H. supervised the study.
Corresponding author
Ethics declarations
Competing interests
T.B. is a cofounder and Managing Director of Myllia Biotechnology. A.K., A.V., N.U. and A.L. are employees of Myllia Biotechnology. The other authors declare no competing interests.
Peer review
Peer review information
Nature Cell Biology thanks Xin He, Fabian Theis and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Benchmark different methods using simulated and real datasets.
a, Steps to generate simulated datasets using scDesign3 from a real scRNA-seq dataset that knocks out Nelfb gene. b-c, Benchmark results of different methods on another published CRISPRi-based Perturb-seq, where mismatches are introduced into guides to attenuate perturbation effects. The Area Under the Curve (AUC) values between the predicted scores and predicted sgRNA activities (b) and the Pearson Correlation Coefficient (PCC) values between the predicted scores of each method and the expressions of perturbed genes are reported for every perturbed (c), using the prediction methods provided in the original study26. Source numerical data are available in source data.
Extended Data Fig. 2 A genome-scale Perturb-seq.
a-b, The distribution of PS and mixscape predicted scores of top hits including CD247 (a) and LCK (b) in the pooled screen. c-d, The correlation between PSs and perturbed gene expression. For CD247 (c), PCC = -0.56, p = 0, n = 4,772, and for LCK (d), PCC = -0.46, p = 2.41e-140, n = 2,728.
Extended Data Fig. 3 Predictions of PDL1 protein expression from a published ECCITE-seq dataset.
The figure displays the ROC curve (first panel), correlations between PS results and PDL1 protein expression (middle panel), and correlations between mixscape results and PDL1 protein expression (third panel) for each gene. The correlations are separated by cell classifications: NP (non-perturbed), defined as mixscape score <= 0.5, and KO (knockout), defined as mixscape score > 0.5. For a fair comparison, mixscape classification results were used to plot PSs (middle panel). For CUL3, the PCC p-values for PS are p = 1.63e-10 for NP cells and p = 8.28e-11 for KO cells, while for mixscape, the p-values are p = 0.57 for NP cells and p = 0.003 for KO cells. For IFNGR1, the PCC p-values for both PS and mixscape are p = 0 for both NP and KO cells. For MYC, the PCC p-values for PS are p = 0.011 for NP cells and p = 0.014 for KO cells, and for mixscape, the p-values are p = 0.849 for NP cells and p = 0.872 for KO cells. For BRD4, the PCC p-values for PS are p = 0.011 for NP cells and p = 0 for KO cells, while for mixscape, the p-values are p = 0.767 for NP cells and p = 0.054 for KO cells. Source numerical data are available in source data.
Extended Data Fig. 4 Buffered genes and sensitive genes.
a, the mean expression and PS value, calculated for each of the 10 expression quantiles, are used for fitting the Hill equation in RPL5, a buffered gene. b, The distribution of PS scores within each of the 10 expression quantiles. c-e, The Hill equation curve and the distribution of PS score for HSPA5, a sensitive gene. f, A gene (BRD4) whose expression has no correlation with PS (p = 0.41). g-j, the log fold changes of gene expressions upon perturbing genes within the same protein complex, including ribosomal subunits (g), proteosome (from genome-scale Perturb-seq in Fig. 4a; h), RNA polymerase (i) and mediator complex (j). Data in g-j come from essential gene Perturb-seq.
Extended Data Fig. 5 Characterization of BRD4-mediated transcriptional responses in HIV Perturb-seq.
a, The number of genes (nFeature_RNA), UMI counts (nCount_RNA) and the fraction of mitochondrial RNAs in three different conditions. b, Clustering results. c, Enriched Gene Ontology (GO) terms of cluster 8. d, The distribution of BRD4-targeting gRNAs. e, The expression distribution of BRD4 signature genes in cluster 8 vs other clusters (p = 4.63e-20). Only cells express BRD4-targeting gRNAs are included. f, Differential expression results between BRD4 PS+ cells vs BRD4 PS- cells. Source numerical data are available in source data.
Extended Data Fig. 6 CCNT1’s Effect on HIV Transcription in Perturb-seq.
a, The distribution of CCNT1 mixscape score, calculated by mixscape. b, The expressions of CCNT1 (left) and CCNT1-targeting gRNAs (right). c, Differential expression results between CCNT1-targeting cells and non-targeting control cells in two different cell states. Dotted horizonal line indicates the position corresponding to adjusted p-val=0.001. d, The expressions of NFKB1. e, The quantitative perturbation-expression relationship between GFP and CCNT1 PS, similar with Fig. 6e. The PCC p-values (calculated from Pearson’s test) are 0.37, 5.9e-6, and 0.04 for DMSO, GFP- and GFP+ conditions, respectively. f, CD69 mRNA expression correlates with the continuous, stimulated state of T cells. g-j, distribution of PS values in CD247 (g) and PDCD1 (i), and the cumulative distribution PS, grouped by CD69 expression quantiles (h,j). Wilcox test p values for CD247 is 1.26e-51 and for PDCD1 is 1.3e-3.
Extended Data Fig. 7 Cell type assignment based on known expression markers of different cell types in pancreatic differentiation scRNA-seq.
Cell types are assigned based on known expression markers in single-cell RNA-seq data from pancreatic differentiation. The figure shows the identification of various cell types using specific marker genes, helping to categorize different stages of pancreatic cell development.
Extended Data Fig. 8 DEG analysis.
a-b, The distribution of FOXA1 PSs across two different clones. c, The expression pattern of FOXA1. d-e, The DEG analysis results of CCDC6 knockout clones vs. wild-type clones in different cell types. f, Overlap of statistically significant DEGs (adj p < 0.05, |log2FC | >0.25) in DE and LV/DUO cell types.
Extended Data Fig. 9 Different CCDC6 functions.
a-b, The two patterns of CCDC6 PSs in LV/DUO (a) and DE in transition (b) cell types. c-d, The distribution of PS across different expression quantiles of POU5F1 (PCC = 0.22 p = 7.19e-66) and SOX17 (PCC = -0.19, p = 1.75e-46), two known factors that capture continuous cell state during DE in transition. For each gene, 5 expression quantiles are used, and 1 indicates the lowest expression quantile of that gene. e-h, Additional enriched terms using Enrichr on DEGs of CCDC6 knockout. i, Flow cytometry analysis of PDX1 and HNF4A expression upon CCDC6 knockout. One representative plot of three biological replicates is shown.
Extended Data Fig. 10 Choosing the best parameters in PS and gating strategy.
a, The effect of the number of input genes in PS on the % correct cells in simulation datasets using different perturbation efficiencies and different number of DEGs. b, The effect of different lambda values on false positive rate. The false positive rates of different lambda values in two datasets are shown. c, Gating strategy for FACS sorting.
Supplementary information
Supplementary Tables 1–6
Table 1. Genome-scale Perturb-seq sgRNA library genes Table 2. Genome-scale Perturb-seq targeted primer panel genes Table 3. HIV Perturb-seq library design Table 4. Sequencing quality control summary of HIV Perturb-seq Table 5. Genotype summary of 10-clone scRNA-seq pancreatic differentiation dataset Table 6. Hash oligos for genome-scale Perturb-seq
Source data
Source Data Figs. 2–6 and Extended Data Figs. 1–6 and 9
Statistical source data.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Song, B., Liu, D., Dai, W. et al. Decoding heterogeneous single-cell perturbation responses. Nat Cell Biol 27, 493–504 (2025). https://doi.org/10.1038/s41556-025-01626-9
Received:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s41556-025-01626-9
This article is cited by
-
Systematic reconstruction of molecular pathway signatures using scalable single-cell perturbation screens
Nature Cell Biology (2025)