Abstract
The exposome, an individual’s lifelong environmental exposure, profoundly impacts health. Somatic tissues undergo functional decline with age, exhibiting characteristic ageing phenotypes, including hair greying and cancer. However, the specific genotoxins, signals and cellular mechanisms underlying each phenotype remain largely unknown. Here we report that melanocyte stem cells (McSCs) and their niche coordinately determine individual stem cell fate through antagonistic, stress-responsive pathways, depending on the type of genotoxic damage incurred. McSC fate tracking in mice revealed that McSCs undergo cellular senescence-coupled differentiation (seno-differentiation) in response to DNA double-strand breaks, resulting in their selective depletion and hair greying, and effectively protecting against melanoma. Conversely, carcinogens can suppress McSC seno-differentiation, even in cells harbouring double-strand breaks, by activating arachidonic acid metabolism and the niche-derived KIT ligand, thereby promoting McSC self-renewal. Collectively, the fate of individual stem cell clones—expansion versus exhaustion—cumulatively and antagonistically governs ageing phenotypes through interaction with the niche.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
Microarray data were deposited in the GEO public database (accession codes GSE158611 and GSE248613). Bulk RNA-seq data were deposited in the GEO public database (accession codes GSE248614 and GSE249074). scRNA-seq data were deposited in the GEO public database (accession code GSE252031). All other data supporting the findings of the present study are available from the corresponding author upon reasonable request. Source data are provided with this paper.
Code availability
The original code is available via GitHub at https://github.com/seitalab/project_mohri_nishimura/tree/main (ref. 84).
References
Vermeulen, R., Schymanski, E. L., Barabási, A. L. & Miller, G. W. The exposome and health: where chemistry meets biology. Science 367, 392–396 (2020).
Collado, M., Blasco, M. A. & Serrano, M. Cellular senescence in cancer and aging. Cell 130, 223–233 (2007).
He, S. & Sharpless, N. E. Senescence in health and disease. Cell 169, 1000–1011 (2017).
Hernandez-Segura, A., Nehme, J. & Demaria, M. Hallmarks of cellular senescence. Trends Cell Biol. 28, 436–453 (2018).
Gorgoulis, V. et al. Cellular senescence: defining a path forward. Cell 179, 813–827 (2019).
de Magalhaes, J. P. Cellular senescence in normal physiology. Science 384, 1300–1301 (2024).
Lopez-Otin, C., Blasco, M. A., Partridge, L., Serrano, M. & Kroemer, G. Hallmarks of aging: an expanding universe. Cell 186, 243–278 (2023).
Gil, J. The challenge of identifying senescent cells. Nat. Cell Biol. 25, 1554–1556 (2023).
Cohn, R. L., Gasek, N. S., Kuchel, G. A. & Xu, M. The heterogeneity of cellular senescence: insights at the single-cell level. Trends Cell Biol. 33, 9–17 (2023).
Nishimura, E. K. Melanocyte stem cells: a melanocyte reservoir in hair follicles for hair and skin pigmentation. Pigment Cell Melanoma Res. 24, 401–410 (2011).
Maslov, A. Y., Barone, T. A., Plunkett, R. J. & Pruitt, S. C. Neural stem cell detection, characterization, and age-related changes in the subventricular zone of mice. J. Neurosci. 24, 1726–1733 (2004).
Encinas, J. M. et al. Division-coupled astrocytic differentiation and age-related depletion of neural stem cells in the adult hippocampus. Cell Stem Cell 8, 566–579 (2011).
Sousa-Victor, P. et al. Geriatric muscle stem cells switch reversible quiescence into senescence. Nature 506, 316–321 (2014).
Matsumura, H. et al. Hair follicle aging is driven by transepidermal elimination of stem cells via COL17A1 proteolysis. Science 351, aad4395 (2016).
Gross, K. M. et al. Loss of Slug compromises DNA damage repair and accelerates stem cell aging in mammary epithelium. Cell Rep. 28, 394–407 e396 (2019).
Nishimura, E. K. et al. Dominant role of the niche in melanocyte stem-cell fate determination. Nature 416, 854–860 (2002).
Tanimura, S. et al. Hair follicle stem cells provide a functional niche for melanocyte stem cells. Cell Stem Cell 8, 177–187 (2011).
Nishimura, E. K., Granter, S. R. & Fisher, D. E. Mechanism of hair graying: incomplete melanocyte stem cell maintenance in the niche. Science 307, 720–724 (2005).
Inomata, K. et al. Genotoxic stress abrogates renewal of melanocyte stem cells by triggering their differentiation. Cell 137, 1088–1099 (2009).
Zhang, B. et al. Hyperactivation of sympathetic nerves drives depletion of melanocyte stem cells. Nature 577, 676–681 (2020).
Johnson, R. & Jackson, I. J. Light is a dominant mouse mutation resulting in premature cell death. Nat. Genet. 1, 226–229 (1992).
Erickson, A. G., Kameneva, P. & Adameyko, I. The transcriptional portraits of the neural crest at the individual cell level. Semin. Cell Dev. Biol. 138, 68–80 (2023).
Lin, S. J. et al. Topology of feather melanocyte progenitor niche allows complex pigment patterns to emerge. Science 340, 1442–1445 (2013).
Martin, G. M. Genetic modulation of senescent phenotypes in Homo sapiens. Cell 120, 523–532 (2005).
Nasti, T. H. et al. A murine model for the development of melanocytic nevi and their progression to melanoma. Mol. Carcinog. 55, 646–658 (2016).
Lo, J. A. & Fisher, D. E. The melanoma revolution: from UV carcinogenesis to a new era in therapeutics. Science 346, 945–949 (2014).
Chou, W. C. et al. Direct migration of follicular melanocyte stem cells to the epidermis after wounding or UVB irradiation is dependent on Mc1r signaling. Nat. Med. 19, 924–929 (2013).
Modi, B. G. et al. Langerhans cells facilitate epithelial DNA damage and squamous cell carcinoma. Science 335, 104–108 (2012).
Brown, K., Buchmann, A. & Balmain, A. Carcinogen-induced mutations in the mouse c-Ha-ras gene provide evidence of multiple pathways for tumor progression. Proc. Natl Acad. Sci. USA 87, 538–542 (1990).
Chakravarti, D., Pelling, J. C., Cavalieri, E. L. & Rogan, E. G. Relating aromatic hydrocarbon-induced DNA adducts and c-H-ras mutations in mouse skin papillomas: the role of apurinic sites. Proc. Natl Acad. Sci. USA 92, 10422–10426 (1995).
Yu, L. & Liu, P. Cytosolic DNA sensing by cGAS: regulation, function, and human diseases. Signal Transduct. Target. Ther. 6, 170 (2021).
Chen, Y. G. & Hur, S. Cellular origins of dsRNA, their recognition and consequences. Nat. Rev. Mol. Cell Biol. 23, 286–301 (2022).
Motwani, M., Pesiridis, S. & Fitzgerald, K. A. DNA sensing by the cGAS–STING pathway in health and disease. Nat. Rev. Genet. 20, 657–674 (2019).
Shi, C. et al. The role of DNA damage repair (DDR) system in response to immune checkpoint inhibitor (ICI) therapy. J. Exp. Clin. Cancer Res. 41, 268 (2022).
Tudor, D. V. et al. COX-2 as a potential biomarker and therapeutic target in melanoma. Cancer Biol. Med. 17, 20–31 (2020).
Tessner, T. G., Muhale, F., Riehl, T. E., Anant, S. & Stenson, W. F. Prostaglandin E2 reduces radiation-induced epithelial apoptosis through a mechanism involving AKT activation and bax translocation. J. Clin. Invest. 114, 1676–1685 (2004).
Patterson, A. M. et al. A single radioprotective dose of prostaglandin E(2) blocks irradiation-induced apoptotic signaling and early cycling of hematopoietic stem cells. Stem Cell Rep. 15, 358–373 (2020).
Chang, C. Y. et al. NFIB is a governor of epithelial-melanocyte stem cell behaviour in a shared niche. Nature 495, 98–102 (2013).
Rabbani, P. et al. Coordinated activation of Wnt in epithelial and melanocyte stem cells initiates pigmented hair regeneration. Cell 145, 941–955 (2011).
Takeo, M. et al. EdnrB governs regenerative response of melanocyte stem cells by crosstalk with Wnt signaling. Cell Rep. 15, 1291–1302 (2016).
Nishimura, E. K. et al. Key roles for transforming growth factor beta in melanocyte stem cell maintenance. Cell Stem Cell 6, 130–140 (2010).
Yoshida, H. et al. Review: melanocyte migration and survival controlled by SCF/c-kit expression. J. Investig. Dermatol. Symp. Proc. 6, 1–5 (2001).
Liao, C. P., Booker, R. C., Morrison, S. J. & Le, L. Q. Identification of hair shaft progenitors that create a niche for hair pigmentation. Genes Dev. 31, 744–756 (2017).
Botchkareva, N. V., Khlgatian, M., Longley, B. J., Botchkarev, V. A. & Gilchrest, B. A. SCF/c-kit signaling is required for cyclic regeneration of the hair pigmentation unit. FASEB J. 15, 645–658 (2001).
Kunisada, T. et al. Murine cutaneous mastocytosis and epidermal melanocytosis induced by keratinocyte expression of transgenic stem cell factor. J. Exp. Med. 187, 1565–1573 (1998).
Hyter, S. et al. Endothelin-1 is a transcriptional target of p53 in epidermal keratinocytes and regulates ultraviolet-induced melanocyte homeostasis. Pigment Cell Melanoma Res. 26, 247–258 (2013).
McGowan, K. A. et al. Ribosomal mutations cause p53-mediated dark skin and pleiotropic effects. Nat. Genet. 40, 963–970 (2008).
Murase, D. et al. The essential role of p53 in hyperpigmentation of the skin via regulation of paracrine melanogenic cytokine receptor signaling. J. Biol. Chem. 284, 4343–4353 (2009).
Chang, C. H. et al. CK1α ablation in keratinocytes induces p53-dependent, sunburn-protective skin hyperpigmentation. Proc. Natl Acad. Sci. USA 114, E8035–E8044 (2017).
Levy, C., Khaled, M. & Fisher, D. E. MITF: master regulator of melanocyte development and melanoma oncogene. Trends Mol. Med. 12, 406–414 (2006).
Widlund, H. R. & Fisher, D. E. Microphthalamia-associated transcription factor: a critical regulator of pigment cell development and survival. Oncogene 22, 3035–3041 (2003).
Shain, A. H. & Bastian, B. C. From melanocytes to melanomas. Nat. Rev. Cancer 16, 345–358 (2016).
Hayward, N. K. et al. Whole-genome landscapes of major melanoma subtypes. Nature 545, 175–180 (2017).
Krauthammer, M. et al. Exome sequencing identifies recurrent mutations in NF1 and RASopathy genes in sun-exposed melanomas. Nat. Genet. 47, 996–1002 (2015).
Dankort, D. et al. Braf(V600E) cooperates with Pten loss to induce metastatic melanoma. Nat. Genet. 41, 544–552 (2009).
Endou, M., Aoki, H., Kobayashi, T. & Kunisada, T. Prevention of hair graying by factors that promote the growth and differentiation of melanocytes. J. Dermatol 41, 716–723 (2014).
Tiger, J. B., Habeshian, K. A., Barton, D. T. & Brennick, J. B. Repigmentation of hair associated with melanoma in situ of scalp. J. Am. Acad. Dermatol. 71, e144–e145 (2014).
Lackey, A. E., Glassman, G., Grichnik, J., McDonald, J. & Correa-Selm, L. Repigmentation of gray hairs with lentigo maligna and response to topical imiquimod. JAAD Case Rep. 5, 1015–1017 (2019).
Gessler, J., Tejasvi, T. & Bresler, S. C. Repigmentation of scalp hair: a feature of early melanoma. Am. J. Med. 136, e7–e8 (2023).
Lopez-Otin, C., Pietrocola, F., Roiz-Valle, D., Galluzzi, L. & Kroemer, G. Meta-hallmarks of aging and cancer. Cell Metab. 35, 12–35 (2023).
Wang, J. et al. A differentiation checkpoint limits hematopoietic stem cell self-renewal in response to DNA damage. Cell 148, 1001–1014 (2012).
Phillips, S. et al. Cell-state transitions regulated by SLUG are critical for tissue regeneration and tumor initiation. Stem Cell Rep. 2, 633–647 (2014).
Ruzankina, Y. & Brown, E. J. Relationships between stem cell exhaustion, tumour suppression and ageing. Br. J. Cancer 97, 1189–1193 (2007).
Ungvari, A. et al. Irradiation-induced hair graying in mice: an experimental model to evaluate the effectiveness of interventions targeting oxidative stress, DNA damage prevention, and cellular senescence. Geroscience 46, 3105–3122 (2024).
Ding, L., Saunders, T. L., Enikolopov, G. & Morrison, S. J. Endothelial and perivascular cells maintain haematopoietic stem cells. Nature 481, 457–462 (2012).
Morris, R. J. et al. Capturing and profiling adult hair follicle stem cells. Nat. Biotechnol. 22, 411–417 (2004).
Ito, M. et al. Stem cells in the hair follicle bulge contribute to wound repair but not to homeostasis of the epidermis. Nat. Med. 11, 1351–1354 (2005).
Bosenberg, M. et al. Characterization of melanocyte-specific inducible Cre recombinase transgenic mice. Genesis 44, 262–267 (2006).
Tonks, I. D. et al. Tyrosinase-Cre mice for tissue-specific gene ablation in neural crest and neuroepithelial-derived tissues. Genesis 37, 131–138 (2003).
Madisen, L. et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 13, 133–140 (2010).
Abe, T. et al. Establishment of conditional reporter mouse lines at ROSA26 locus for live cell imaging. Genesis 49, 579–590 (2011).
Mackenzie, M. A., Jordan, S. A., Budd, P. S. & Jackson, I. J. Activation of the receptor tyrosine kinase Kit is required for the proliferation of melanoblasts in the mouse embryo. Dev. Biol. 192, 99–107 (1997).
Ackermann, J. et al. Metastasizing melanoma formation caused by expression of activated N-RasQ61K on an INK4a-deficient background. Cancer Res. 65, 4005–4011 (2005).
Dankort, D. et al. A new mouse model to explore the initiation, progression, and therapy of BRAFV600E-induced lung tumors. Genes Dev. 21, 379–384 (2007).
Suzuki, A. et al. T cell-specific loss of Pten leads to defects in central and peripheral tolerance. Immunity 14, 523–534 (2001).
Kim, J., Sturgill, D., Tran, A. D., Sinclair, D. A. & Oberdoerffer, P. Controlled DNA double-strand break induction in mice reveals post-damage transcriptome stability. Nucleic Acids Res. 44, e64 (2016).
Marino, S., Vooijs, M., van Der Gulden, H., Jonkers, J. & Berns, A. Induction of medulloblastomas in p53-null mutant mice by somatic inactivation of Rb in the external granular layer cells of the cerebellum. Genes Dev. 14, 994–1004 (2000).
Lee, M. S. J. et al. B cell-intrinsic TBK1 is essential for germinal center formation during infection and vaccination in mice. J. Exp. Med. 219, e20211336 (2022).
Eshiba, S. et al. Stem cell spreading dynamics intrinsically differentiate acral melanomas from nevi. Cell Rep. 36, 109492 (2021).
Ueno, M., Aoto, T., Mohri, Y., Yokozeki, H. & Nishimura, E. K. Coupling of the radiosensitivity of melanocyte stem cells to their dormancy during the hair cycle. Pigment Cell Melanoma Res. 27, 540–551 (2014).
Joost, S. et al. Single-cell transcriptomics reveals that differentiation and spatial signatures shape epidermal and hair follicle heterogeneity. Cell Syst. 3, 221–237 (2016).
Joost, S. et al. The molecular anatomy of mouse skin during hair growth and rest. Cell Stem Cell 26, 441–457 (2020).
Yang, H., Adam, R. C., Ge, Y., Hua, Z. L. & Fuchs, E. Epithelial–mesenchymal micro-niches govern stem cell lineage choices. Cell 169, 483–496 e413 (2017).
Yamanashi, T. & Seita, J. project_mohri_nishimura. GitHub https://github.com/seitalab/project_mohri_nishimura/tree/main (2025).
Acknowledgements
We are grateful to S. Morrison (HHMI Investigator at UT Southwestern Medical Center) for the Kitl flox mice and Kitl-GFP mice. We are grateful to A. Berns (University of Amsterdam), M. McMahon (U.C. San Francisco) and P. Oberdoerffer (The National Cancer Institute) for the p53 flox mice, Braf CA mice and I-PpoI mice, respectively. The aged mice were provided by the Foundation for Biomedical Research and Innovation at Kobe through the National BioResource Project of the MEXT, Japan. We also thank R. Yajima, K. Takenouchi and T. Ando for technical support, the staff of the Laboratory Animal Research Center (IMSUT) for mouse care support and all laboratory members for helpful comments on this project and DASS Manuscript for editing. E.K.N. is supported by a JSPS Grant-in Aid for Scientific Research (S) (25H00439), an AMED CREST Project (JP22gm1710003-JP25gm1710003), an AMED Project for Elucidating and Controlling Mechanisms of Ageing and Longevity (JP17gm5010002-JP21gm5010002), an AMED SCARDA Japan Initiative for World-leading Vaccine Research and Development Centers (JP223fa627001), a JSPS Grant-in Aid for Scientific Research (A) (20H00532) and a JSPS Grant-in Aid for Scientific Research on Innovative Areas ‘Stem Cell Aging and Disease’ (26115003). Y.M is supported by a JSPS Grant-in-Aid for Young Scientists (18K15114) and a JSPS Grant-in Aid for Scientific Research (C) (25K10315). J.S. is supported by an AMED Project for Elucidating and Controlling Mechanisms of Ageing and Longevity (JP19gm5010003 JP20gm5010003) and a JSPS Grant-in Aid for Scientific Research (C) (18K08377).
Author information
Authors and Affiliations
Contributions
Y.M. performed the majority of the experiments with support from J.N., H. Morinaga, T.A., H. Matsumura and S.K. Y.M., T.K., T.Y. and J.S. performed and analysed the single-cell RNA-seq experiments. D.N. and H. Matsumura established Dct-CreERT2 mice. K.K. and K.J.I. provided TBK1 flox mice. T.N., M.H. and T.S. collected and provided the human melanoma samples. Y.M. and E.K.N. designed the study and wrote the paper. E.K.N supervised the project.
Corresponding author
Ethics declarations
Competing interests
E.K.N. is a co-founder and shareholder of EADERM Co., Ltd., which is not related to the content of this study. The other authors declare no competing interests.
Peer review
Peer review information
Nature Cell Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
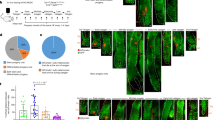
Extended Data Fig. 1 IR-induced p21 expression is coupled with hair cycle progression.
a, Appearance of young (2 months old) and of old (23 months old) C57BL/6N mice with some gray hairs. White arrowheads indicate gray hairs. b, Graph showing the percentage of gray hairs per 100 hairs (Young; n=3, Old; n=10, biological independent experiments). Data are presented as means ± SEM. Mann-Whitney’s U test. c, Immunohistological analysis of the bulge of Dct-H2B-GFP KI mice at the indicated time points with or without IR. Arrowheads indicate p21+ McSCs (GFP+ cells). Scale Bar = 50 μm. Dp; Hair depilation.
Extended Data Fig. 2 Carcinogens promote McSC self-renewal.
a, Experimental scheme for DMBA treatment (upper). Macroscopic time-course images of the dorsal skin of C57BL/6 mice treated with Vehicle or DMBA at the indicated time points. Immunohistological analysis of the dorsal skin of Dct-H2B-GFP KI mice showing the distribution of GFP+ melanocytes (arrows) at 12 weeks post-onset DMBA treatment. The two panels of acetone and DMBA, respectively, show different hair follicles. Scale Bar = 60 μm. Dp; Hair depilation. b, Coat colors in the 2nd hair cycle after treatment with or without IR and UVB. Immunohistological analysis of hair follicles of Dct-LacZ Tg mice at 5 days in the 2nd hair cycle (2nd-D5). Arrowheads indicate McSCs (LacZ+ cells). The dotted lines show the Bg/sBg (Bulge/sub-Bulge). Scale Bar = 50 μm.
Extended Data Fig. 3 Induction of DSBs in McSCs induces their depletion and hair graying.
a, Diagram illustrating the induction of melanocyte-specific DSBs with I-PpoI endonuclease by Cre-mediated recombination and TAM-dependent transfer into the nucleus. b, Experimental scheme for Tyr-Cre/I-PpoI mice (upper). Coat colors of WT and Tyr-Cre/I-PpoI mice in the 2nd hair cycle and immunohistological analysis of KIT+ cells in the bulge at 5 days in the 3rd hair cycle (3rd-D5). Scale Bar = 30 μm. The dotted lines show the Bg/sBg (Bulge/sub-Bulge). Dp; Hair depilation. c, Immunohistological analysis of γH2AX in McSCs of Dct-H2B-GFP KI mice at 24 h after treatment with different types of genotoxins. Scale Bar = 20 μm. Bg=Bulge, Hg=Hair germ, DP=Dermal papilla. Graph showing the percentage of γH2AX+ McSCs (n=3, biological independent experiments). Data are presented as means ± SEM. One-way ANOVA with Dunnett’s post-hoc tests. d, FACS sorting analysis of McSCs in Dct-H2B-GFP murine skin at telogen. The 7AAD(-)/Singlets/GFPhigh/SSClow population was used as McSCs. e, Graph showing the relative expression of melanocyte-related genes in McSCs at telogen. Data are from one of two separate experiments. f, Identification of DMBA-induced CAA → CTA mutations in codon 61 of the H-Ras gene (H-RasQ61L) in McSCs by digital droplet PCR (left). McSCs were sorted from the skin of Dct-H2B-GFP KI mice at 28 days after vehicle or DMBA treatment. One-dimensional (1D) amplitude plots are shown with the associated manual threshold gating (pink line) for HEX (WT, green) and FAM (mutant, blue) amplitudes. Genomic DNAs collected from melanocytic lesion (bulk) developed by the DMBA/TPA method and mutated plasmid (H-RasQ61L) are used as positive controls. DDW is also used as a negative control. Graph showing the percentages of H-RasQ61L within McSCs (right). Data are from one of two separate experiments.
Extended Data Fig. 4 Activation of p53 and cytosolic DNA/RNA sensing pathways by IR.
a, Enrichment plot for KEGG pathways. b, Heatmap of expressed genes ranked by GSEA. Colors range from dark red to dark blue representing the highest and lowest expression of each gene, respectively. Circles indicate representative core enrichment genes. c, Coat colors of irradiated C57BL/6 mice with or without dmPGE2 in the 1st hair cycle after irradiation with 5 Gy IR. d, Immunohistological analysis of the bulge of irradiated Dct-H2B-GFP KI mice with or without dmPGE2 at 5 days in the 1st hair cycle. Scale Bar = 50 μm. e, Quantification of the relative mRNA expression level of p21 in NHEMs (in vitro) at 3 h after IR treatment with Vehicle or PGE2 (n=3, biological independent experiments). One-way ANOVA with Tukey’s post-hoc tests. Rpl11 is used as an internal control for the qPCR analysis.
Source data
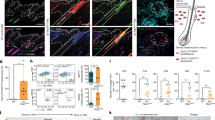
Extended Data Fig. 5 Kitl deficiency in the niche for McSCs causes McSC depletion and resultant hair graying.
a, FACS sorting analysis of HFSCs/Hg in K15-EGFP Tg murine skin. The singlets/7AAD−/GFPhigh/ITGA6high/ScaI− population was used as niche cells for McSCs. b, Immunohistological analysis of KITL in the bulge (KRT15+) and hair germ (P-CAD+) of Kitl-GFP KI mice at telogen. Scale Bar = 30 μm. Bg=Bulge, Hg=Hair germ. c, Immunohistological analysis of KITL in the dorsal skin of Kitl-GFP KI mice at 3 days after treatment with Vehicle or DMBA. Arrows indicate GFP+ cells in Bg/sBg (Bulge/sub-Bulge) and arrowheads indicate GFP+ cells in the IFE. Scale Bar = 200 μm. d, Experimental scheme for Kitl cKO mice. Dp; Hair depilation. e,f,g, Coat colors of WT and K15CrePR; Kitlfx/fx (Kitl cKO) mice at 3 weeks (e), at 7 weeks (f) and at 28 days in the 1st hair cycle (1st-D28) (g). h, Immunohistological analysis of McSCs in the bulge of WT and Kitl cKO mice at 5 days in the 2nd hair cycle (2nd-D5). McSCs are visualized with Dct-LacZ Tg mice. Graph showing quantification of McSCs in the bulge (WT; n=3, Kitl cKO; n=4, biological independent experiments). Scale Bar = 30 μm. Two-tailed Student’s t-test. i, Coat colors of WT and K14-Kitl Tg mice after irradiation with 0 Gy or 5 Gy IR. Immunohistological analysis of McSCs in the bulge of WT and K14-Kitl Tg mice. McSCs are visualized using Dct-LacZ Tg mice. Scale Bar = 50 μm. Graph showing the frequency of hair follicles still containing McSCs in the bulge (n=3, biological independent experiments). Two-tailed Student’s t-test. j, Coat colors of Tyr-Cre; I-PpoI (I-PpoI) and K14-Kitl; Tyr-Cre; I-PpoI (K14-Kitl/I-PpoI) mice and immunohistological analysis of KIT+ cells in the bulge. The dotted lines show the Bg/sBg. Scale Bar = 30 μm. k, Experimental scheme for microarray analysis with FACS-purified McSCs. McSCs are labeled using the Dct-H2B-GFP KI allele. l, GSEA Enrichment plot for KEGG Arachidonic acid metabolism, RIG-I like receptor signaling pathway and Cytosolic DNA sensing pathway. m, Quantification of the relative mRNA expression level of COX2 in vitro using NHEMs (n=3, biological independent experiments). One-way ANOVA with Dunnett’s post-hoc tests. Rpl11 is used as an internal control for the qPCR analysis. Data are presented as means ± SEM (h,i,m).
Extended Data Fig. 6 KITL promotes the repair of DSBs and ROS scavenging.
a, Representative images showing DSBs caused by IR in NHEMs pre-treated with or without KITL in vitro. Scale Bar = 200 μm. Graph showing the quantification of the percentage of cells with a comet tail. Three biological independent experiments were performed (n=9 fields, total: > 70 cells). Two-way ANOVA with Tukey’s post-hoc tests. b, Quantification of the relative mRNA expression levels of the indicated genes in NHEMs following the addition of KITL (n=3, biological independent experiments). One-way ANOVA with Dunnett’s post-hoc tests compared to 0 h. c, FACS data of NHEMs showing changes in fluorescence intensity of H2DCFDA, an indicator dye of oxidative stress, caused by IR pretreated with or without KITL in vitro. Normalized Mean Fluorescence Intensities (MFI) are shown in the right graph (n=7, biological independent experiments). Two-way ANOVA with Tukey’s post-hoc tests. d, Quantification of the relative mRNA expression levels of the indicated genes in NHEMs following the addition of KITL (n=3, biological independent experiments). One-way ANOVA with Dunnett’s post-hoc tests compared to 0 h. Rpl11 is used as an internal control for the qPCR analysis (b,d). Data are presented as means ± SEM (a-d).
Extended Data Fig. 7 Niche-derived KITL promotes melanomagenesis.
a, Immunohistological analysis of the distribution of GFP+ melanocytes in the epidermis of WT and of Kitl cHetero mice at 4 weeks post-onset DMBA treatment. GFP+ melanocytic lineages are visualized with Dct-H2B-GFP KI mice. Arrows indicate melanocytes in the IFE. Graph showing the number of GFP+ cells in the epidermis (WT; n=5, Kitl cHetero; n=4, biological independent experiments). The experimental scheme for DMBA treatment is shown in Extended Data Fig. 2a. Scale Bar = 100 μm. Two-tailed Student’s t-test. b, Macroscopic views of the dorsal skin of WT and of Kitl cHetero mice at 16 weeks post-onset DMBA treatment. Graph showing the number of melanocytic pre-cancerous lesions (indicated by arrowheads) on the dorsal skin (WT; n=8, Kitl cHetero; n=5, biological independent experiments). The experimental scheme for DMBA treatment is shown in Extended Data Fig. 2a. Two tailed Student’s t-test. c, Line graph showing the number of melanocytic precancerous lesions on the dorsal skin at the indicated time points after DMBA treatment (Ink4a/Arf−/−; K14-Kitl; n=3, Ink4a/Arf−/−; n=3, Ink4a/Arf+/−; K14-Kitl; n=12, Ink4a/Arf+/−; n=6, Ink4a/Arf+/+; K14-Kitl; n=5, Ink4a/Arf+/+; n=9, biological independent experiments). Data are presented as means ± SEM (a-c). d, Macroscopic view of dorsal skin showing melanocytic tumors (arrow) in Mc1re/e; Ink4a/Arf+/−; K14-Kitl mice at the indicated time points.
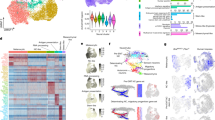
Extended Data Fig. 8 Defining the main epidermal clusters by lineage-specific marker genes.
a, UMAP plot of all 61,787 single-cell transcriptomes visualized for individual mice. b, Expression of group-specific marker genes projected onto the UMAP plot.
Extended Data Fig. 9 Decreased expression of niche-derived cytokines and down-regulation of the p53 pathway in an aged niche for McSCs.
a, Graphs showing the expression of the indicated genes in FACS-purified HFSCs/Hg at early anagen (n=3, biological independent experiments). Two-tailed Student’s t-tests. b,c, Pathways down-regulated (b) or up-regulated (c) in FACS-purified HFSCs/Hg at early anagen analyzed by GSEA using the c2.cp.biocarta.v7.5 gene set collection (permutation test). d, Enrichment plot for Biocarta P53_pathway (Young, early anagen vs Old, early anagen). e, Heatmap showing the Biocarta P53_pathway gene set core signature. Colors range from dark red to dark blue representing the highest and lowest expression of each gene, respectively. f, Graphs showing the expression of p53, Cdkn1a (p21) and Cdkn2a (p16) in HFSCs/Hg at telogen and early anagen (n=3, biological independent experiments). Two-tailed Student’s t-tests. g, Enrichment plot for KEGG P53_signaling_pathway (Vehicle, early anagen vs DMBA, early anagen). Data are presented as means ± SEM (a,f).
Extended Data Fig. 10 Dct-CreERT2 mice enable the specific targeting of McSCs in hair follicles, allowing the controlled induction of invasive melanoma.
a, Experimental scheme for tracing of McSC-lineage cells. b, Distribution of GFP+ cells in the dorsal skin of Dct-CreERT2; Rosa-H2B-GFPKI/+ or Tyr-CreERT2; Rosa-H2B-GFPKI/+ mice at 4 weeks post-TAM injection. GFP+ cells in the dermis of Tyr-CreERT2; Rosa-H2B-GFPKI/+ mice are indicated by a yellow arrowhead. Scale Bar = 60 μm. c, Distribution of GFP+ cells in hair follicles of Dct-H2B-GFP KI mice at telogen and anagen. GFP+ cells in the bulge represent McSCs. Scale Bars = 60 μm. d, The percentage of GFP+ cells located in the dorsal skin of Tyr-CreERT2; Rosa-H2B-GFPKI/+ mice (Tyr promoter) and of Dct-H2B-GFP KI mice (Dct promoter). A total of 388 GFP+ cells and 269 GFP+ cells were counted for the Tyr promoter (n=2) and Dct promoter (n=3), respectively (biological independent experiments). e, Distribution of tdTomato+ cells in hair follicles of Dct-CreERT2; Rosa-tdTomato mice. Arrowheads indicate tdTomato+/KIT+ cells in the hair follicle. Scale Bars = 20 μm. f, Tyr-CreERT2; BrafV600E; Ptenfx/fx mice were depilated and injected with TAM for 3 consecutive days. Macroscopic view of Tyr-CreERT2; BrafV600E; Ptenfx/fx mice at 1 month after the 1st TAM injection. Expansion of GFP+ melanocytic lineages in the dermis of the mice was visualized using the Dct-H2B-GFP transgene. g, Macroscopic views of the dorsal skin of WT and of Dct-CreERT2; BrafV600E; Ptenfx/fx (DBP) mice at the indicated time points post-onset TAM injection. One-half of the dorsal skin was depilated. Yellow arrows indicate dome-like unpigmented nodules that mimic human Nodular-type melanomas. Graphs showing the quantification of melanomas (shown by yellow arrows) on the dorsal skin at 4 weeks (WT; n=20, DBP; n=11, biological independent experiments) and at 8 weeks (WT; n=8, DBP; n=6, biological independent experiments). Data are presented as means ± SEM. Mann-Whitney’s U-tests. h, Dct-H2B-GFP mice or Rosa-tdTomato mice were used to trace the fate of McSC-lineage cells. Immunohistological analysis of the dorsal skin of Dct-H2B-GFP and of DBP mice at the indicated time points post-onset TAM injection. The two panels of DBP mice at 4 weeks post-onset TAM injection show different hair follicles of the same skin. Scale Bars = 100 μm. i, Macroscopic images of lymph nodes (LN).
Supplementary information
Supplementary Information
Supplementary Fig. 1.
Supplementary Tables
Supplementary Table 1. Selected genes associated with melanogenesis in the HFSC/Hg (Telogen). Genes that show a significant change in expression (P < 0.05) are highlighted in yellow. Supplementary Table 2. Selected genes associated with melanogenesis in the HFSC/Hg (early anagen). Genes that show a significant change in expression (P < 0.05) are highlighted in yellow. Supplementary Table 3. Primer sequences.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 9
Statistical source data.
Source Data Extended Data Fig. 10
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Mohri, Y., Nie, J., Morinaga, H. et al. Antagonistic stem cell fates under stress govern decisions between hair greying and melanoma. Nat Cell Biol 27, 1647–1659 (2025). https://doi.org/10.1038/s41556-025-01769-9
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41556-025-01769-9
This article is cited by
-
Hair Graying as an Evolutionary Checkpoint against Malignancy: a Stem Cell Perspective
Stem Cell Reviews and Reports (2026)
-
Melanomagenesis antagonizes stem cell ageing
Nature Reviews Cancer (2025)