Abstract
The Seh1-associated complex (SEAC; GATOR in mammals) transduces amino acid signals to the Target of Rapamycin Complex 1 (TORC1), a master regulator of cell growth. The SEAC is composed of two subcomplexes, SEACIT (GATOR1), an inhibitor of TORC1 that has GAP activity against Gtr1, and SEACAT (GATOR2), which appears to regulate SEACIT. However, the molecular details of this regulation are unclear. Here we determined the cryo-electron microscopy structure of the SEAC bound to its substrate, the EGOC (Ragulator–Rag), and studied its function in TORC1 amino acid signaling. A single SEAC can interact with two EGOC molecules via SEACIT, binding exclusively to the ‘active’ version of the EGOC, without involvement of SEACAT. The GAP activity of the SEACIT is essential for the regulation of TORC1 by amino acids and its loss phenocopies the lack of Gtr1–Gtr2, establishing the SEAC–EGOC complex as an amino acid-sensing hub. Compared to other SEACAT subunits, the loss of Sea2, or its N-terminal β-propeller domain, yielded strong defects in amino acid signaling to TORC1. Our results suggest that the Sea2 β-propeller recruits a GAP inhibitor to mediate fast amino acid signaling to TORC1, with additional pathways acting with slower kinetics.
Similar content being viewed by others
Main
The Target of Rapamycin Complex 1 (TORC1) is a highly conserved master regulator of cell growth that couples nutrient availability with anabolic and catabolic processes. In yeast, amino acids and glucose regulate TORC1 activity through two small Ras-like GTPases, Gtr1 and Gtr2, in a pathway that is conserved to humans. Gtr1 and Gtr2 form a heterodimer that interacts with three other proteins, Ego1, Ego2 and Ego3, together forming the pentameric EGO complex (EGOC)1. The EGOC is anchored to the membrane of the vacuole and signaling endosomes2 via the N terminus of Ego1, which is palmitoylated and myristoylated3. Gtr1 and Gtr2 regulate TORC1 activity by changes in their nucleotide loading state: Gtr1GTP–Gtr2GDP activates TORC1 under nutrient-replete conditions, while Gtr1GDP–Gtr2GTP inactivates TORC1 during starvation4. Mechanistically, this involves, at least during changes in glucose levels, the regulated disassembly or assembly of inactive TORC1 helices (TOROIDs)5. In mammals, the Rags (RagA/B and RagC/D) are the counterparts of the Gtrs, forming a complex with Ragulator, the counterpart of Ego1–Ego2–Ego3 (Ragulator–Rag)6. As in yeast, RagA/BGTP–RagC/DGDP activates and RagA/BGDP–RagC/DGTP prevents mTORC1 activation7,8, respectively. Changes in the nucleotide loading status of the Rags/Gtrs are coupled to conformational transitions that are thought to regulate the binding to different partners. In mammals, the active Rags can recruit mTORC1 to the lysosome membrane where it can be activated by another GTPase, Rheb8.
Gtr1 and Gtr2 have low basal GTPase activity that is stimulated by a dedicated GTPase activating protein (GAP). The Lst4–Lst7 complex acts as GAP for Gtr2 (ref. 9), whereas the Seh1-associated complex (SEAC) has specific GAP activity toward Gtr1 (refs. 10,11). The SEAC can be functionally subdivided in two subcomplexes: SEACIT and SEACAT. SEACIT, which is composed of Sea1, Npr2 and Npr3, is the subcomplex that harbors the GAP activity of the complex and acts as an inhibitor of TORC1. The activity of SEACIT is believed to be counteracted in vivo by SEACAT10, which acts as an activator through an unknown molecular mechanism. In mammals, the GATOR complex, which is composed of GATOR1 (SEACIT) and GATOR2 (SEACAT)12, and FLCN–FNIP2 (Lst7–Lst4)13, act as GAPs for RagA/B and RagC/D, respectively. While specific amino acid sensors that act through GATOR2 have been described14,15, as with the SEAC, how GATOR2 regulates GATOR1 remains a major open question. The currently proposed model of regulation states that when amino acid levels are abundant, proteins that act as amino acid sensors bind their cognate amino acids, which prevents their binding to GATOR2. GATOR2 is then free to inhibit the GAP activity of GATOR1, ultimately activating mTORC1. Conversely, during amino acid starvation, ligand-free amino acid sensors bind GATOR2 and block its inhibitory action on GATOR1, inactivating mTORC1 (ref. 16). Although no amino acid sensors acting upstream of SEAC–EGOC are known in yeast, given their conservation, it is presumed that a similar mechanism of regulation occurs through species-specific sensors17. In yeast, amino acids also regulate TORC1 through Pib2, a vacuolar protein that has been proposed to act as a glutamine18 and cysteine19 sensor. How the EGOC and Pib2 pathways are interconnected remains to be resolved.
Both yeast SEACAT and mammalian GATOR2 are composed of five subunits (GATOR2 subunits in parentheses): Sea2 (Wdr24), Sea3 (Wdr59), Sea4 (Mios), Sec13 (Sec13) and Seh1 (Seh1L). Based on secondary structure predictions, initial studies identified a relationship between SEACAT subunits and coatomer complexes20 but structural information was lacking. We recently determined the cryo-electron microscopy (cryo-EM) structure of the native yeast SEAC11, revealing the interactions between SEACAT and SEACIT. SEACAT forms a central core with similar topology as COPII (SEACcore), which serves as an anchor for two flexible wings that are composed of SEACIT (SEACwing). Surprisingly, the active site of SEACIT was not occluded by SEACAT and SEACAT did not inhibit the GAP activity of SEACIT in vitro. Contrary to what has been previously suggested16, these data are not consistent with SEACAT acting through inhibition of SEACIT. How the SEAC interacts with the EGOC is not known.
The structures of GATOR1 (ref. 21) and GATOR2 (ref. 22) have been determined individually and show a remarkable conservation with the SEAC, pointing toward a conserved function and regulation. However, compared to the SEAC, where SEACAT and SEACIT form a stable complex, GATOR1 and GATOR2 require an additional complex named KICSTOR to stably interact23,24. In vivo, GATOR1 interacts with GATOR2 and KICSTOR in an amino acid-insensitive manner23. Thus, in cells, both SEACIT/GATOR1 and SEACAT/GATOR2 are constitutively bound and regulate (m)TORC1 as a single complex. Nevertheless, the only structures available of GATOR1 bound to its substrate (Ragulator–Rag) have been determined in the absence of GATOR2 or KICSTOR21, precluding a more physiological understanding of the function and regulation of the complex. Furthermore, the role of the GAP activity on TORC1 signaling and its relationship with SEAC function has not been analyzed in detail, with only one study in GATOR1 testing a single time point25. Therefore, to better understand the molecular mechanism of regulation of the SEAC/GATOR complexes, we set out to determine the structure of the native SEAC bound to the EGOC and characterize its role in amino acid sensing in detail.
Results
Cryo-EM structure of the SEAC–EGOC supercomplex
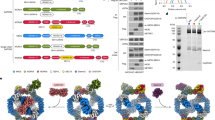
To assemble the SEAC–EGOC supercomplex, we slightly modified our native SEAC purification protocol and incubated the purified SEAC with an excess of recombinant EGOC in the presence of GDP–AlF3, which stabilizes GAP–GTPase complexes by mimicking the transition state of the GTP hydrolysis reaction26. Despite its relatively small size compared to the SEAC, extra density corresponding to the EGOC was clearly visible in the cryo-EM two-dimensional (2D) class averages (Fig. 1a). After 2D and three-dimensional (3D) classification steps, we could obtain an overall reconstruction of the complex at 3.2 Å resolution where the core, but not the wings, was well resolved (Extended Data Figs. 1 and 2a). To better resolve the wings, we expanded the particles using C2-symmetry, performed local masked 3D classification, subtracted the density of the SEACcore and performed local refinements to obtain a 3.1 Å-resolution SEACwing–EGOC reconstruction (Extended Data Figs. 1 and 2b). Similarly, to improve the resolution of regions of the SEACcore, we performed focused refinements on one monomer and on Sea2–Seh1 and Sea3–Sec13, improving the resolution to ~3.0 Å (Extended Data Figs. 1 and 2c). Finally, to aid in visualization, we combined the resulting maps to create a composite map (Fig. 1b and Extended Data Fig. 2d). We used previous structures of the SEAC11 and EGOC1 to build a model that includes all eight SEAC subunits (each present in two copies, with exception of Sea4 and Seh1 that are present in four and six copies, respectively, totaling 22 protein chains) and five EGOC subunits (two copies of each), comprising a total of 32 protein chains (Fig. 1c,d, Table 1 and Extended Data Fig. 2e,f). Of note, we observe some stretches of densities present in different parts of the wing that could not be confidently assigned, which probably correspond to part of large loops of Sea1, Npr2 and Npr3 (Extended Data Fig. 3).
a, The 2D class averages of the final particles used in the reconstruction. The arrows show the blurry density corresponding to the EGOC. b, A composite cryo-EM map colored according to subunits. c, A model of the SEAC–EGOC supercomplex. The predicted orientation relative to the vacuole membrane is shown on the right. d, A schematic representation of the SEAC and EGOC subunits. C-ter, C-terminal domain; NTD, N-terminal domain; CRD, C-terminal Roadblock domain; RWD, domain found in RING finger-, WD-repeat- and yeast DEAD (DEXD)-like helicase-containing proteins; DIM, domain-invasion motif.
The SEACcore is formed by Seh1, Sec13, Sea2, Sea4 and the C-terminal half of Sea3 (SEACAT portion, Sea3SAP), whereas the SEACwing is formed by Npr2, Npr3, Sea1 and the N-terminal half of Sea3 (SEACIT portion, Sea3SIP). One EGOC molecule is bound to each wing without contacting any element from the core. Accordingly, we observe virtually no change in the overall structure of the SEACcore compared to the unbound SEAC11 (Extended Data Fig. 4a,b). Binding of the EGOC ‘pushes’ the SEACwing toward the core, widening the distance between each wing by ~6 Å (Extended Data Fig. 4a,b), with the SEACwing moving as a rigid body around the Npr3–Sea4 interaction (Extended Data Fig. 4c). Superimposition of unbound and EGOC-bound wings reveals that the peripheral and flexible Sea1 is shifted slightly away from Npr2–Npr3 to accommodate binding of the EGOC (Extended Data Fig. 4d). The lack of substantial conformational change and the ability of one SEAC to bind multiple EGOC molecules simultaneously is akin to how mTORC1 binds to the Rags27 and Rag–TFEB28.
The EGOC is localized to the vacuole membrane1 and deletion of GTR1 and GTR2 reduces, but does not abolish, vacuolar localization of the SEAC11. Thus, we wished to determine whether the complex we observe is compatible with its localization in vivo. In our cryo-EM map, Ego1, Ego2 and Ego3 are not resolved as well as Gtr1 and Gtr2 (Extended Data Fig. 2b). However, the last ordered N-terminal helix of Ego1, which is next to the disordered lipidated tail that anchors the complex to the membrane, is sufficiently resolved to determine the orientation of the complex relative to the vacuole membrane, minimizing the distance of the tip of the Ego1 helix and the membrane (Fig. 1c). In this orientation, the SEACcore faces away from the membrane and is thus accessible for the binding of nutrient sensors, as previously proposed11. In turn, the SEACwing is placed adjacent to the membrane, well-positioning Sea1 for interactions with other proteins such as Vps501 (ref. 29). This orientation is also consistent with GATOR1 interacting with other lysosomal complexes such as KICSTOR30 or LYCHOS31. Given its flexibility and the lack of interactions between the EGOC and the SEACcore, each wing can interact independently with one EGOC molecule. The large degree of freedom of the SEACwing relative to the SEACcore (owing to the length of the Sea3connector, which can extend to up to 245 Å, the lack of extensive interactions between core and wing and the length of the flexible Ego1 N-terminal tail (~150 Å)) could accommodate the binding of the SEAC to one or two EGOC molecules in the context of varying membrane curvatures.
The EGOC interacts with Npr2 and Sea1
The EGOC interacts extensively with the SEACwing via Gtr1 and Gtr2 (Fig. 2a). The quality of the Coulombic potential map allowed us to unambiguously assign both nucleotides bound to Gtr1 and Gtr2: whereas Gtr1 is loaded with GDP–AlF3 (mimicking GTP), Gtr2 is loaded with GDP (Fig. 2b). Hence, the SEAC-bound EGOC represents the ‘active’ Gtr1GTP–Gtr2GDP version.
a, A model of the SEACITwing bound to the EGOC. The disordered and lipidated tail is indicated schematically. Myr, myristoylation; Palm, palmitoylation. b, The cryo-EM density of the Gtr1 and Gtr2 heterodimer. c, Binding of Gtr1 to the Npr2N-lobe. The Gtr1 Switch I and Switch II regions are indicated. d, Interactions between residues near the arginine finger and switch I of Gtr1, colored yellow. e, Binding of Gtr2 to Sea1. f, Contacts between the Sea1C-ter and the Gtr1GTPase domains. g, Interactions between the Sea1NTD and Gtr2CRD domains.
Two interfaces mediate binding of the EGOC to the SEACwing and resemble those observed in human GATOR1 (ref. 21). The first is formed by Gtr1 and Npr2–Npr3, and the second is formed by Gtr2 and Sea1. Both Npr2 and Npr3 N-lobes (which contain the Longin domain) interact with the Gtr1 GTPase domain, positioning the catalytic ‘arginine finger’ Npr2R84 next to GDP–AlF3 (Fig. 2c). Insertion of Npr2R84 into the nucleotide binding pocket of Gtr1 is stabilized by the presence of the Sea3SIP, which sits on top of the Npr2N-lobe. Several residues near the catalytic Npr2R84 make hydrogen bonds with the switch I region of Gtr1, an element that is only structured when GTP is bound (Fig. 2d). Interestingly, we observe a neighboring asparagine (Npr2N85) next to the arginine finger that interacts with the backbone of switch I, effectively stabilizing its conformation. Npr2N85 appears to be structurally analogous to the ‘auxiliary asparagine’ observed in RhoGAP32 (Extended Data Fig. 5a,b). These interactions ensure that Npr2 only interacts with Gtr1GTP. The arginine finger and neighboring asparagine are broadly conserved among species, although curiously, not in Caenorhabditis elegans (Extended Data Fig. 5c). To better understand this difference, we predicted the structure of CeGATOR1–RagA/RagC with AlphaFold 3. Interestingly, the missing asparagine in CeNprl2 is replaced by an asparagine in CeNprl3, in a loop that is positioned next to the arginine finger (Extended Data Fig. 5d). Therefore, our analysis suggests that the neighboring asparagine plays an important role in the function of GATOR/SEAC.
In contrast to the Npr2–Gtr1 interaction, Sea1 interacts with both the GTPase and C-terminal Roadblock domain (CRD) of Gtr2 on the opposite side of the nucleotide binding pocket (Fig. 2e). The binding interface between Gtr2 and Sea1 is mediated by electrostatic interactions (Extended Data Fig. 6a), which are conserved, to a lesser degree, in the RagC–DEPDC5 (ortholog of Sea1) interaction in GATOR1 (Extended Data Fig. 6b). The Sea1C-ter sits on top of the Gtr2 α5 helix (Gtr2α5), with two arginine residues (Sea1R1400 and Sea1R1404) close to negatively charged residues (Gtr2D140, Gtr2E147 and Gtr2E151) (Fig. 2f), and interactions between the Gtr2CRD and Sea1NTD include two hydrogen bonds (Fig. 2g). Interestingly, we observe an extra density near Gtr2α5 that complements the neighboring β-sheet (Gtr2β6) (Fig. 2f). Despite much effort, we were unsuccessful in our attempts to assign this density, but owing to proximity, it could correspond to part of the disordered N terminus of Sea1 (~100 amino acids are not resolved) (Extended Data Fig. 6c). This extension is not present in the DEPDC5 and, accordingly, no equivalent density is observed in the GATOR1–Rag–Ragulator cryo-EM map (Extended Data Fig. 6d).
Owing to the differences in the nature of the interactions in both interfaces, binding appears to be stronger to Gtr1, with Gtr2 serving to sterically restrict the binding to Gtr1 in the context of the Gtr1–Gtr2 heterodimer. This agrees with the lower local resolution of Gtr2 compared to Gtr1 (Extended Data Fig. 2b). Consequently, selectively disrupting the Npr2N-lobe–Gtr1SwitchI interface by mutating both the arginine finger and neighboring asparagine (Npr2R84AN85A) is sufficient to remove the SEAC from the vacuole, whereas a single mutation of the arginine finger is not (Extended Data Fig. 5e,f).
In the presence of GDP–AlF3, two binding modes have been observed between GATOR1 and Ragulator–Rag: one ‘GAP-productive’ binding as in our structure, and one ‘nonproductive’ binding to DEPDC5 (Sea1) (ref. 21). By contrast, we only observe a ‘GAP-productive’ binding mode—we could not obtain a stable complex when incubating the SEAC and EGOC in the presence of the nonhydrolyzable GTP analog GppNHp, which has been used to promote the ‘nonproductive’ binding21. These results are consistent with the previous proposition that the ‘nonproductive’ binding mode is not conserved in yeast11. The physiological relevance of the ‘nonproductive’ binding remains to be determined.
The SEAC is unable to bind to the ‘inactive’ EGOC
The Rag and Gtr heterodimers are known to adopt different conformations depending on their nucleotide loading status. We therefore wondered whether, in addition to the ‘active’ Gtr1GTP–Gtr2GDP state, other conformations could be accommodated by the SEAC. There is only one previous structure of the EGOC, where both Gtr1 and Gtr2 are bound to GppNHp1. Comparison of this structure with ours shows that the Gtrs adopt a very similar conformation despite the different nucleotide loading state, with only a slight shift in Gtr2 due to the binding to Sea1 (Extended Data Fig. 7a,b). As expected, the conformation of the equivalent states of the Rag heterodimer is similar (Extended Data Fig. 7c,d). Therefore, both ‘active’ and dually GTP-loaded states would, in principle, be compatible with SEAC binding.
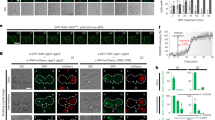
In contrast to the structures described above, there is a drastic conformational change in the RagA–RagC heterodimer in the inactive state (bound to the RagC GAP, FLCN–FNIP2 (ref. 33)) (Fig. 3a). The shift in RagCGTP compared to RagCGDP widens the distance between GTPase domains from ~37 Å in the active state to ~53 Å in the inactive state (Extended Data Fig. 7e). Superimposition of the inactive Ragulator–Rag onto our structure shows that binding of the SEAC to an inactive EGOC is not possible. When bound to GDP, the RagA switch I becomes disordered and thus the interactions with the Npr2N-lobe are lost, and retraction of the switch II produces an additional clash (Fig. 3b). Moreover, the large movement of RagC causes a severe clash with the Sea1NTD (Fig. 3c). A structure of the ‘inactive’ Gtr1GDP–Gtr2GTP is lacking, as is a structure of the ‘inactive’ RagAGDP–RagCGTP in the absence of an interactor. It is thus possible that the large conformational change observed in the inactive state is induced by FLCN–FNIP2 binding. To this end, we took advantage of the fact that deletion of GTR1 and GTR2 reduces the localization of the SEAC to the vacuole11 (Fig. 3d,e). By expressing different combinations of Gtr1 and Gtr2 from plasmids, we can test the binding of the SEAC to different versions of the Gtr1–Gtr2 heterodimer in a physiological context by determining whether the vacuolar localization of the SEAC is restored. Consistent with our structure, expression of a constitutively ‘active’ EGOC (Gtr1Q65L–Gtr2S23L) restored normal SEAC localization in Δgtr1Δgtr2 cells (Fig. 3f), whereas expression of a constitutively ‘inactive’ EGOC (Gtr1S20L–Gtr2Q66L) did not (Fig. 3g). Importantly, expression of different versions of the Gtrs did not affect the expression levels of Sea4–GFP (Extended Data Fig. 7f). Thus, the localization differences observed in the different conditions (Fig. 3h) agree with our structure-guided hypothesis and suggest that the SEAC cannot bind the inactive EGOC in vivo.
a, Comparison between the structure of Gtr1GDP–AlF3-Gtr2GDP in the SEAC–EGOC structure (‘active’) and the structure of RagAGDP–RagCGppNHp when bound to FLCN–FNIP2 (PDB: 6ulg; ‘inactive’)33. b, Structural comparison of GTP- and GDP-bound RagA/Gtr1 shows that interactions between Gtr1 and Npr2 are lost in Gtr1GDP. c, Structural comparison between RagCGppNHp and Gtr2GDP shows that movement of Gtr2 would result in a clash with Sea1. d–g, Confocal microscopy images showing the localization of the SEAC (using an endogenous GFP tag in Sea4), in otherwise WT cells (d) in the absence (e) or with expression of the ‘active’ (f) and ‘inactive’ (g) versions of Gtr1–Gtr2. Scale bars, 5 µm. h, Quantification of the ratio of Sea4–GFP intensity on the vacuole membrane versus the cytosol in strains shown in d–g. The horizontal dashed line indicates equal intensity in vacuole and cytosol. The vacuole was labeled using FM4-64. n = 60 cells per condition. Mean and standard deviation is shown. Statistical significance is shown with respect to WT cells, using Brown–Forsythe and Welch ANOVA tests. i, Cryo-EM maps of the three different classes of the Npr2latch. j, Zoom-in of the Npr2–Gtr1–Gtr2 interface in Class 2. k, Zoom-in of the Npr2–Gtr1–Gtr2 interface in Class 3. DIC, differential interference contrast.
The inability of the SEAC to bind the ‘inactive’ Gtr1–Gtr2 heterodimer is reminiscent of mTORC1, where Raptor can only bind the ‘active’ RagA–RagC heterodimer34. Both Raptor and SEAC bind to the Rag/Gtr heterodimer on the same face (Extended Data Fig. 7g,h). In addition, Raptor queries the nucleotide loading state of RagC by inserting a stretch referred to as the ‘claw’ between RagA and RagC34. The claw would clash with Switch I of RagC if structured and, hence, Raptor can only interact with RagCGDP. We noted that Npr2 contains an extended loop (~56 residues) in the vicinity of the arginine finger, which is disordered in our overall SEACwing–EGOC reconstruction. Superimposition of the AlphaFold2 prediction35 of Npr2 on our structure revealed that this loop, which we refer to as the ‘latch’, would be positioned for interactions with the Gtr1–Gtr2 heterodimer in analogous manner as the Raptor claw (Extended Data Fig. 5g). During our analysis of the cryo-EM data by 3D variability34, we observed density variation between Gtr1 and Gtr2 GTPase domains that appeared to correspond to part of the latch. Therefore, we performed 3D classification with a mask in this region and successfully resolved, to low resolution, three main classes of the Npr2latch (Fig. 3i). The first one corresponds to a flexible, disordered state and thus not visible (Class 1). The other two classes show the Npr2latch interacting exclusively with Gtr2 (Class 2; Fig. 3j) or with both Gtr1 and Gtr2 GTPase domains (Class 3; Fig. 3k). In Class 3, the Npr2latch inserts into the inter-GTPase domain space and appears to contact the tip of the α5 helix of Gtr1, while shielding the GDP bound to Gtr2. As the Npr2latch is highly flexible, our densities remain very low resolution and hence we cannot assign specific interactions. The lack of stable and defined interactions, combined with its absence in other species, argues against the Npr2latch being important for SEAC function (‘The GAP activity of the SEAC is required for TORC1 inhibition and reactivation’). Nevertheless, our analysis suggests that different components of the TORC1 signaling pathway in distinct species might reuse similar structural mechanisms to engage with the Gtrs/Rags.
The GAP activity of the SEAC is required for TORC1 inhibition and reactivation
Based on genetic epistasis results, it has been proposed that SEACAT/GATOR2 regulates (m)TORC1 through modulation of the GAP activity of SEACIT/GATOR1: during nutrient starvation, SEACIT/GATOR1 is active and (m)TORC1 is inhibited, whereas during amino acid repletion, SEACAT/GATOR2 inhibits SEACIT/GATOR1 and activates (m)TORC1 (refs. 10,12,16). However, there is no biochemical evidence supporting a direct inhibitory mechanism of SEACAT/GATOR2 on SEACIT/GATOR1. On the contrary, our structure shows that the SEACcore does not block EGOC binding to the SEACwing nor does it shield the active site, strongly arguing against direct inhibition of catalysis as a mechanism of regulation. Moreover, we previously showed that SEACAT does not inhibit the GAP activity of SEACIT in vitro11. Therefore, we wished to determine the consequences of abolishing the GAP activity of the SEAC on TORC1 amino acid signaling in vivo. To this end, we used cells endogenously expressing a catalytically dead SEAC mutant where the arginine finger has been mutated to an alanine (Npr2R84A; SEACCD). We have shown that this mutation abolishes the GAP activity but does not affect the integrity of the complex11, nor localization to the vacuole membrane (Extended Data Fig. 5f). We measured the phosphorylation of a TORC1-specific site on the bona fide downstream target Sch9 (ref. 36) after the removal of nitrogen and amino acids from the media and readdition of a mixture containing all amino acids (Supplementary Table 3). Given that removing both ammonium sulfate and amino acids is required to inactivate TORC1, we refer to ‘nitrogen starvation’ rather than ‘amino acid starvation’. Likewise, we chose to manipulate the levels of all amino acids because it is not known which amino acid regulates TORC1 upstream of the SEAC.
Compared to wild-type (WT) cells, where TORC1 is rapidly (~2.5 min) inactivated and reactivated after starvation and refeeding, respectively (Fig. 4a), cells with SEACCD were almost completely unresponsive to both starvation and repletion during the first 5 min (Fig. 4b). We observed a virtually identical response when each individual SEACIT subunit was deleted or when the interaction between SEACIT and EGOC was disrupted (Npr2R84A N85A) (Extended Data Fig. 8a–e). Therefore, abolishing the GAP activity recapitulates the loss of SEACIT, establishing its physiological role in amino acid signaling in yeast. Interestingly, during these time points, amino acid signaling to TORC1 appears to be mediated entirely through the GAP activity of the SEAC as Δgtr1Δgtr2 cells display the same defects as SEACCD cells (Fig. 4c). We note that all strains were still able to regulate TORC1 to some extent, with activity reaching near WT levels at around 30 min (Fig. 4d,e). This is consistent with the existence of Rag/Gtr-independent pathways that act at later time points37,38,39.
a, Immunoblots of phosphorylated and total Sch9 after starvation and repletion of amino acids (AA) in WT cells. b, Immunoblots of phosphorylated and total Sch9 after starvation and repletion of amino acids in SEACCD cells. c, Immunoblots of phosphorylated and total Sch9 after starvation and repletion of amino acids in Δgtr1Δgtr2 cells. d, Quantification of relative Sch9 phosphorylation over 30 min of starvation and subsequent amino acid repletion in different strains. Data from three independent experiments are presented as mean values ± s.d. Quantification of Δsea1 cells is shown for comparison with SEACIT-deficient strains.
We also tested whether the two extra features observed in our map, namely, the Npr2latch and the Sea1N-ter, are important for amino acid signaling. Whereas removal of the first 100 amino acids of Sea1 caused only a slight defect during inactivation, removal of the Npr2latch did not cause obvious differences to WT cells (Extended Data Fig. 9a–c,e). Mutation of two conserved arginines in the C-terminal domain of Sea1 (R4000 and R4004), which impair amino acid signaling in human GATOR1 (ref. 21), did not cause any defect (Extended Data Fig. 9d,e). Overall, these data show that, in contrast to the Npr2–Gtr1 interaction, the Sea1–Gtr2 interaction might not be critical for SEAC function.
Sea2 is essential for the function of the SEAC
Previous studies have shown that SEACAT acts as an activator of TORC1 (refs. 10,11,40). Consistent with this, we have observed increased rapamycin sensitivity (reflecting lower TORC1 activity) in Δsea2 and Δsea4 cells, with a weaker phenotype in Δsea3 cells11. This is probably because Sea3 forms part of both core and wings as deletion of the Sea3SAP caused a similar phenotype as that of Δsea2 and Δsea4 cells11. Nevertheless, despite its established role as an activator, no study so far has analyzed the individual role of Sea2, Sea3 and Sea4 in acute amino acid signaling, and their function has only been inferred from data on GATOR2.
We first asked whether, like its GATOR2 counterparts, subunits forming the SEACcore are necessary for the regulation of TORC1 by amino acids. Given our previous results11, we expected a strong reactivation defect in Δsea2 and Δsea4 cells, with a much milder phenotype in Δsea3 cells. Surprisingly, we observed a different phenotype for each strain. Upon N withdrawal and amino acid repletion, TORC1 inactivation and reactivation kinetics were nearly wild type in Δsea4 cells, slightly delayed in Δsea3 cells and strongly delayed in Δsea2 cells (Fig. 5a–e). Interestingly, the TORC1 activity kinetics of Δsea2 cells were nearly identical to those of GAP-deficient SEACIT mutants, with a delayed response in both inhibition and reactivation (Fig. 5f). These response kinetics were not due to strain-to-strain differences in protein levels (Extended Data Fig. 9f). Thus, contrary to previous suggestions40, SEACAT subunits appear not to be redundant but, as in GATOR2 (ref. 22), Sea2 plays an essential role in the SEAC. Inconsistent with our results, the loss of Mios (Sea4) in GATOR2 causes severe defects in amino acid signaling22,41. However, deletion of Mios results in degradation of Wdr24 (Sea2)22,24,41, making the interpretation of these results difficult.
a, Immunoblots of phosphorylated and total Sch9 after starvation and repletion of amino acids (AA) in WT cells. b, Immunoblots of phosphorylated and total Sch9 after starvation and repletion of amino acids in Δsea4 cells. c, Immunoblots of phosphorylated and total Sch9 after starvation and repletion of amino acids in Δsea3 cells. d, Immunoblots of phosphorylated and total Sch9 after starvation and repletion of amino acids in Δsea2 cells. These cells have a lower overall TORC1 activity and slow growth. e, Quantification of relative Sch9 phosphorylation over 30 min of starvation and subsequent amino acid repletion in WT and SEACAT deletion strains. Data from three independent experiments are presented as mean values ± s.d. f, Quantification of relative Sch9 phosphorylation over 30 min of starvation and subsequent amino acid repletion in WT, Δsea2, sea2ΔN-ter and SEACCD cells. Data from three independent experiments are presented as mean values ± s.d. g, Immunoblot of phosphorylated and total Sch9 after starvation and repletion of amino acids in the sea2ΔN-ter strain.
In both the SEACcore and GATOR2 (ref. 22), the N-terminal β-propeller of Sea2 (Sea2N-ter, Wdr24N-ter in GATOR2) protrudes out of the complex, is highly flexible (Extended Data Fig. 2a) and is the only β-propeller that does not interact with any other subunit (Fig. 1). Recent studies have found that the Wdr24N-ter is subject to nutrient-responsive post-translational modifications42,43. Moreover, in addition to serving as a binding platform for the leucine sensor Sestrin2 (ref. 22), it is also essential for GATOR2 function22. Thus, we reasoned that the Sea2N-ter plays a similar role in the SEAC. In agreement, deletion of the Sea2N-ter caused the same strong defects observed in Δsea2 cells (Fig. 5f,g), showing that, like in GATOR2, this region mediates an essential function of the complex.
Unlike Sea3 and Sea4, Sea2 does not interact with the SEACwing and is located far from the active site, with the Sea2N-ter being at least ~180 Å away from Npr2R84. Therefore, it is unlikely that Sea2 acts directly on Npr2 in neither the same nor an adjacent SEA complex (‘Discussion’). To clarify their epistatic relationship, we generated Δsea2 npr2R84A (SEACCD Δsea2) double-mutant cells in which we assessed TORC1 activity following nitrogen starvation and subsequent amino acid repletion. Intriguingly, despite retaining the inhibition defects of the single mutants at early time points (<15 min), at later time points, TORC1 activity levels in SEACCDΔsea2 cells more closely resembled those of WT cells (Fig. 6a–c). Notably, the loss of GAP activity in npr2R84A cells suppressed the rapamycin sensitivity of Δsea2 cells (Extended Data Fig. 9g). These results indicate that fixing GAP activity on, as is presumably the case in Δsea2 cells, or off, as in npr2R84A cells, blocks acute amino acid signaling to TORC1 while other factors still function, semi-independently of the SEAC, to relay amino acid-derived signals to TORC1 at later time points.
a, Immunoblots of phosphorylated and total Sch9 after starvation and repletion of amino acids (AA) in WT, SEACCDΔsea2, SEACCD and Δsea2 cells. b, Quantification of relative Sch9 phosphorylation over 30 min of starvation and amino acid repletion in WT, SEACCDΔsea2 and SEACCD cells. Data from three independent experiments are presented as mean values ± s.d. c, Quantification of relative Sch9 phosphorylation over 30 min of starvation and amino acid repletion in WT, SEACCDΔsea2 and Δsea2 cells. Data from three independent experiments are presented as mean values ± s.d. d, A model of regulation of the SEAC on the vacuolar membrane. We propose that the GAP activity regulates the GTP loading of the EGOC, which self-limits localization of the SEAC to the vacuole membrane. The interplay between the N-terminal propeller of Sea2 and the catalytic arginine finger of Npr2 remains unclear and, if it exists, would probably require the function of an as of yet unknown factor.
Discussion
The regulation of SEAC/GATOR function remains an important unresolved question in nutrient sensing by (m)TORC1. Here, we show that (1) the SEACAT module does not affect binding of EGOC to the SEACIT module, supporting the lack of inhibitory effect on the GAP activity previously observed in vitro11; (2) the SEAC selectively interacts with the Gtr1GTP–Gtr2GDP heterodimer; (3) the GAP activity is essential for inhibition and reactivation of TORC1 at early time points after changes in amino acid levels; and (4) Sea2 plays a predominant role in the function of the SEAC.
A key missing piece in the regulation of mTORC1 by amino acids is how GATOR2 functions upstream of GATOR1. A recent study suggested that GATOR2 can ubiquitinate and promote the degradation of GATOR1 (ref. 41), although there is contradictory evidence against this22. Our structure, combined with our previous observations, suggests that the SEAC acts as a single entity in the regulation of TORC1, with SEACAT acting as a scaffolding platform rather than directly inhibiting SEACIT. It is highly likely that this is conserved in GATOR, as several studies have shown that amino acids do not modulate the interaction between GATOR1 and GATOR2 (refs. 23,24,44), and structures of each module are highly conserved. Thus, as we11 (and more recently others44) have proposed, SEAC/GATOR should be viewed as a single regulatory complex composed of two functional modules, rather than two separate complexes.
The prevailing model in the literature is that binding of ligand-free amino acid sensors to GATOR2 prevents it from inhibiting the GAP activity of GATOR1 (ref. 16). In this scenario, GATOR2 possesses an intrinsic ability to inhibit the GAP activity of GATOR1. However, purified SEAC, containing both the GATOR2 (SEACAT) and GATOR1 (SEACIT) counterparts, has robust GAP activity in vitro11. These results demonstrate that SEACAT does not intrinsically inhibit the GAP activity of SEACIT. It is likely that GATOR2 functions similarly as known amino acid sensors bind far away from the predicted interface with GATOR1 (that is the N terminus of Wdr59) and their binding causes only minor changes in the overall structure of GATOR2 (refs. 45,46) and does not affect the binding of GATOR2 to GATOR1 (ref. 46). Nevertheless, mutations affecting SEACAT, and Sea2 in particular, present phenotypes consistent with high GAP activity; however, given the distance between the active site and Sea2, direct regulation either in cis or in trans (on the vacuole membrane) is unlikely. A parsimonious explanation for these observations is that SEACAT, and probably the Sea2 β-propeller, functions by recruiting a factor that antagonizes the GAP activity of SEACIT (Fig. 6d).
Curiously, even in mutants where amino acid-mediated regulation of TORC1 was perturbed at early time points, TORC1 was still regulated at later time points, suggesting that another pathway communicates amino acid-derived signals to TORC1 (Fig. 6d). This delayed regulation is reminiscent of the previously observed ‘sustained TORC1 activity’—TORC1 activity that was similarly regulated independently of Gtr1/2 (ref. 37). Whereas Pib2 appears to regulate TORC1 in an amino acid-specific manner in parallel to the Gtrs, it still does so at early time points18,19. More recently, the Teleman group has reported strikingly similar, fast and slow amino acid signaling upstream of mTORC139. In this work, they suggest that while some amino acids (for example, leucine and arginine) have dedicated sensors (that is, Sestrin/Castor) that act on mTORC1 with fast kinetics via the GATOR complex, most amino acids are sensed and signaled with slower kinetics to mTORC1 via the stress-activated kinase Gcn2. Intriguingly, this Gcn2 signaling to mTORC1 also appeared to be partially dependent on the GATOR complex. Future work will be aimed at assessing the roles of Gcn2 and other factors in signaling amino acid levels to TORC1 along with the SEAC.
To conclude, while in mammalian systems, where specific amino acids regulate binding of factors to GATOR2 and GATOR1 to regulate mTORC1 activity, in yeast, we propose that amino acid cues regulate binding of factors to Sea2 of SEACAT to ultimately inhibit the GAP activity of SEACIT. These mechanisms enable relatively fast regulation of (m)TORC1 activity but, in both systems, other slower mechanisms additionally act at later time points.
Methods
Yeast strains
The yeast strains used in this study are of TB50 background. Classical recombination-based or CRISPR-based techniques were used to construct different strains. All strains were verified by PCR, sequencing or western blot (where applicable). The yeast strains used in this study are listed in Supplementary Table 1 and plasmids in Supplementary Table 2.
Native SEAC purification
Native SEAC was purified using a TAP tag in Sea4, essentially as previously described11, with the only modification being that the complex was eluted in EGOC binding buffer mixed with TEV protease (20 mM HEPES pH 7.4, 150 mM NaCl, 5 mM dithiothreitol (DTT), 0.8 g l−1 EGOC, 1 mM GDP, 2 mM MgCl2, 2 mM AlCl3 and 20 mM NaF). The complex was prepared freshly for cryo-EM grid preparation.
EGOC purification
Recombinant full-length EGOC was purified as previously described11. Plasmids encoding WT Gtr1 and Gtr2 were cotransformed with a plasmid containing full-length Ego1, Ego2 and Ego3, where Ego1 was N-terminally tagged with a histidine tag. Protein expression was induced by 0.1 mM isopropyl-β-d-thiogalactopyranoside at 18 °C overnight. Cells were collected by centrifugation and resuspended in lysis buffer (50 mM Tris HCl pH 7.4, 300 mM NaCl, 5% glycerol, 20 mM imidazole, 0.15% CHAPS and 1 mM MgCl2), supplemented with 1 mM PMSF, cOmplete EDTA-free Protease Inhibitor Cocktail (Roche), DNase and lysozyme. Cells were lysed using an Emulsiflex (Avestin) and centrifuged at 15,000 rpm for 45 min at 4 °C. The supernatant was loaded into a HiTrap FF Crude column (Cytiva) and eluted with 250 mM imidazole. The buffer was exchanged to 25 mM HEPES–NaOH pH 7.4, 150 mM NaCl, 10% glycerol and 2 mM DTT using a HiPrep 26/10 column. Then, the sample was injected into a MonoQ FF column (Cytiva) and eluted with a gradient from 150 mM to 1 M NaCl. To remove bound nucleotides, the fractions containing the complex were incubated overnight with 20 mM EDTA at 4 °C. Afterward, the sample was concentrated and loaded on a Superdex S200 Increase (Cytiva) equilibrated with 25 mM HEPES pH 7.4, 150 mM NaCl, 10% glycerol, 2 mM DTT and 2 mM EDTA. Peak fractions were collected and concentrated, flash frozen in liquid nitrogen and stored at −80 °C until use.
Cryo-EM grid preparation
For cryo-EM grid preparation, 5 μl of SEAC–EGOC mix was deposited on 1.2/1.3 Quantifoil Au grids and plunge frozen in a Leica GP2 instrument.
Cryo-EM data collection
Grids were screened for quality and preliminary data collection was performed on a 200 kV Talos Arctica equipped with a Falcon 3 direct electron detector (at the Dubouchet Center for Imaging (DCI) Geneva). Final data were collected on a 300 kV Titan Krios equipped with a Selectris X energy filter and a Falcon 4i direct electron detector in the DCI (Lausanne). A total of 11,177 untilted and 7,253 35° tilted movie frames were collected with a total exposure of 40e− Å−2 and a nominal pixel size of 0.726 Å.
Cryo-EM data processing
The cryo-EM processing pipeline is detailed in Extended Data Fig. 1b. Movies in EER format were preprocessed (Patch Motion correction and Patch CTF estimation) on the fly with CryoSPARC Live. All subsequent steps were performed in cryoSPARC47. Ultimately, from 1,649,067 template picks, we selected 208,039 well-aligning SEAC–EGOC particles that were refined with C2-symmetry to 3.2 Å resolution (consensus map). To improve different regions of the complex, particles were symmetry expanded, particle subtracted (to eliminate signal from the other monomer and wings) and subjected to local (with nonuniform regularization) refinements on different regions of the core. After local refinement on the wing (which was not resolved in the consensus map), focused 3D classification on this region yielded two classes with good EGOC density. We performed an additional 3D classification with a high-resolution threshold to select better-aligning particles, signal subtracted the rest of the SEAC and obtained a reconstruction at 3.1 Å resolution from 133,077 particles. We noted weak density between the Gtr1 and Gtr2 GTPase domains that could correspond to a disordered loop of Npr2 (Npr2latch). We therefore used a mask to perform focused 3D classification on this region and obtained three different reconstructions with variations in this density, which we called Classes 1–3. We individually locally refined each class.
All maps (except the three Npr2latch classes) were modified using DeepEMhancer48. To create a composite map, all focused maps were aligned in UCSF Chimera49 to the consensus map, regions around 4 Å were selected and merged using the ‘vop maximum’ command. Local resolution was estimated with Blocres50 within cryoSPARC. Figures were made using UCSF ChimeraX51.
Model building
The structure of the SEAC (Protein Data Bank (PDB):8ADL) was used as a starting model. For the EGOC, AlphaFold252 predictions and a published crystal structure (PDB:6JWP)1 were used as starting models. Initial rigid-body fitting was performed in Chimera and subsequent manual building was performed in Coot53. Models were real space refined iteratively in Phenix54. Some visible stretches of density were not built owing to our inability to unambiguously assign them (Extended Data Fig. 3). Attempts to use automated model building tools were also unsuccessful. Thus, we preferred to not interpret them.
Immunoblots
Amino acid starvation and repletion experiments were performed as follows. Yeast was grown to an OD600 of 0.8–1 in complete synthetic media (CSM; 2% glucose, yeast nitrogen base without amino acids with ammonium sulfate (USBiological, Y2025) and drop-out mix (USBiological, D9515)). After taking the first time point (0), cultures were filtered on 0.2 μm filter membranes and resuspended in an equal volume of CSM–amino acids–ammonium sulfate (2% glucose, yeast nitrogen base without amino acids or ammonium sulfate). After 30 min, the drop-out mix was added to the culture at 1× final concentration. The drop-out mix contains all 20 amino acids, plus adenine, uracil, p-aminobenzoic acid and inositol (Supplementary Table 3).
For each time point, 10 ml of culture was mixed with cold TCA (final concentration 6%) and kept on ice. Cells were pelleted, washed with acetone and dried on a SpeedVac. Dried samples were processed or kept at −80 °C. Cells were lysed using a bead beater in lysis buffer (25 mM Tris pH 6.8, 6 M urea and 1% SDS) and mixed with 2× SDS–sample buffer (25 mM Tris pH 6.8, 20% glycerol, 2% SDS, 0.02% bromophenol blue and 200 mM DTT). Samples were resolved on a 4–20% gradient TGX gel (Bio-Rad) and transferred to nitrocellulose membranes on an iBlot 2 (Thermo Fisher). Membranes were blocked with 1× TBS buffer with 5% BSA for 30–60 min and incubated with primary antibodies overnight at 4 °C (1:5,000, anti-Sch9; 1:25,000 anti-Sch9S758; custom-made). After washing with 1× TBS, secondary antibodies (1:10,000, IRDye antibodies) were incubated for 1 h at room temperature and the blot was imaged on a LiCOR system. Quantification was performed on ImageStudio 5.5 (LiCOR) and plotted and analyzed using GraphPad Prism. The same blotting protocol was used for GFP (sc-9996, Santa Cruz; 1:2,000), actin (Sigma A5441; 1:2,500) and Hog1 (sc-165978, Santa Cruz; 1:5,000). Note that data from WT and other strains are replotted in multiple figure panels for comparative purposes.
Confocal microscopy
Strains were grown on CSM to an OD600 of about 0.7–1. Z stacks were collected on a Zeiss LSM800 confocal laser scanning microscope and processed using ImageJ 1.52p. The ratio of vacuole-to-cytosol GFP intensity was essentially done as previously11, using FM4-64 to delineate the vacuole membrane. Statistical analysis was performed in GraphPad.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Cryo-EM maps and models have been deposited in the Electron Microscopy Database (EMDB) and PDB, respectively, with accession codes EMD-51887 and PDB 9H5K for the full SEAC–EGOC, and EMD-51867 and PDB 9H4Q for the SEACwing–EGOC. The consensus SEAC–EGOC map has been deposited with accession code EMD-51904, with the following codes for focused maps on the core: EMD-51905 (monomer), EMD-51906 (Sea3–Sec13–Sea2–Seh1), EMD-51907 (Sea3–Sec13) and EMD-51908 (Sea2–Seh1). Maps of the three different Npr2latch classes have been deposited as additional maps for the SEACwing–EGOC. Data and materials can be obtained from the corresponding authors upon request. Source data are provided with this paper.
References
Zhang, T. et al. Structural insights into the EGO-TC-mediated membrane tethering of the TORC1-regulatory Rag GTPases. Sci. Adv. 5, eaax8164 (2019).
Hatakeyama, R. et al. Spatially distinct pools of TORC1 balance protein homeostasis. Mol. Cell 73, 325–338 e328 (2019).
Kogan, K., Spear, E. D., Kaiser, C. A. & Fass, D. Structural conservation of components in the amino acid sensing branch of the TOR pathway in yeast and mammals. J. Mol. Biol. 402, 388–398 (2010).
Binda, M. et al. The Vam6 GEF controls TORC1 by activating the EGO complex. Mol. Cell 35, 563–573 (2009).
Prouteau, M. et al. EGOC inhibits TOROID polymerization by structurally activating TORC1. Nat. Struct. Mol. Biol. 30, 273–285 (2023).
Sancak, Y. et al. Ragulator–Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell 141, 290–303 (2010).
Kim, E., Goraksha-Hicks, P., Li, L., Neufeld, T. P. & Guan, K. L. Regulation of TORC1 by Rag GTPases in nutrient response. Nat. Cell Biol. 10, 935–945 (2008).
Sancak, Y. et al. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 320, 1496–1501 (2008).
Peli-Gulli, M. P., Sardu, A., Panchaud, N., Raucci, S. & De Virgilio, C. Amino acids stimulate TORC1 through Lst4–Lst7, a GTPase-activating protein complex for the Rag family GTPase Gtr2. Cell Rep. 13, 1–7 (2015).
Panchaud, N., Peli-Gulli, M. P. & De Virgilio, C. SEACing the GAP that nEGOCiates TORC1 activation: evolutionary conservation of Rag GTPase regulation. Cell Cycle 12, 2948–2952 (2013).
Tafur, L. et al. Cryo-EM structure of the SEA complex. Nature 611, 399–404 (2022).
Bar-Peled, L. et al. A tumor suppressor complex with GAP activity for the Rag GTPases that signal amino acid sufficiency to mTORC1. Science 340, 1100–1106 (2013).
Tsun, Z. Y. et al. The folliculin tumor suppressor is a GAP for the RagC/D GTPases that signal amino acid levels to mTORC1. Mol. Cell 52, 495–505 (2013).
Wolfson, R. L. et al. Sestrin2 is a leucine sensor for the mTORC1 pathway. Science 351, 43–48 (2016).
Chantranupong, L. et al. The CASTOR proteins are arginine sensors for the mTORC1 pathway. Cell 165, 153–164 (2016).
Goul, C., Peruzzo, R. & Zoncu, R. The molecular basis of nutrient sensing and signalling by mTORC1 in metabolism regulation and disease. Nat. Rev. Mol. Cell Biol. 24, 857–875 (2023).
Liu, G. Y., Jouandin, P., Bahng, R. E., Perrimon, N. & Sabatini, D. M. An evolutionary mechanism to assimilate new nutrient sensors into the mTORC1 pathway. Nat. Commun. 15, 2517 (2024).
Ukai, H. et al. Gtr/Ego-independent TORC1 activation is achieved through a glutamine-sensitive interaction with Pib2 on the vacuolar membrane. PLoS Genet. 14, e1007334 (2018).
Zeng, Q., Araki, Y. & Noda, T. Pib2 is a cysteine sensor involved in TORC1 activation in Saccharomyces cerevisiae. Cell Rep 43, 113599 (2024).
Dokudovskaya, S. et al. A conserved coatomer-related complex containing Sec13 and Seh1 dynamically associates with the vacuole in Saccharomyces cerevisiae. Mol. Cell. Proteomics 10, M110 006478 (2011).
Egri, S. B. et al. Cryo-EM structures of the human GATOR1–Rag–Ragulator complex reveal a spatial-constraint regulated GAP mechanism. Mol. Cell 82, 1836–1849 e1835 (2022).
Valenstein, M. L. et al. Structure of the nutrient-sensing hub GATOR2. Nature 607, 610–616 (2022).
Wolfson, R. L. et al. KICSTOR recruits GATOR1 to the lysosome and is necessary for nutrients to regulate mTORC1. Nature 543, 438–442 (2017).
Peng, M., Yin, N. & Li, M. O. SZT2 dictates GATOR control of mTORC1 signalling. Nature 543, 433–437 (2017).
Shen, K., Valenstein, M. L., Gu, X. & Sabatini, D. M. Arg-78 of Nprl2 catalyzes GATOR1-stimulated GTP hydrolysis by the Rag GTPases. J. Biol. Chem. 294, 2970–2975 (2019).
Wittinghofer, A. Signaling mechanistics: aluminum fluoride for molecule of the year. Curr. Biol. 7, R682–685 (1997).
Anandapadamanaban, M. et al. Architecture of human Rag GTPase heterodimers and their complex with mTORC1. Science 366, 203–210 (2019).
Cui, Z. et al. Structure of the lysosomal mTORC1–TFEB–Rag–Ragulator megacomplex. Nature 614, 572–579 (2023).
Goyal, S. et al. Vps501, a novel vacuolar SNX–BAR protein cooperates with the SEA complex to regulate TORC1 signaling. Traffic 23, 192–207 (2022).
Padi, S. K. R. et al. Phosphorylation of DEPDC5, a component of the GATOR1 complex, releases inhibition of mTORC1 and promotes tumor growth. Proc. Natl Acad. Sci. USA 116, 20505–20510 (2019).
Shin, H. R. et al. Lysosomal GPCR-like protein LYCHOS signals cholesterol sufficiency to mTORC1. Science 377, 1290–1298 (2022).
Hakoshima, T., Shimizu, T. & Maesaki, R. Structural basis of the Rho GTPase signaling. J. Biochem. 134, 327–331 (2003).
Shen, K. et al. Cryo-EM structure of the human FLCN–FNIP2–Rag–Ragulator complex. Cell 179, 1319–1329 e1318 (2019).
Rogala, K. B. et al. Structural basis for the docking of mTORC1 on the lysosomal surface. Science 366, 468–475 (2019).
Punjani, A. & Fleet, D. J. 3D variability analysis: resolving continuous flexibility and discrete heterogeneity from single particle cryo-EM. J. Struct. Biol. 213, 107702 (2021).
Urban, J. et al. Sch9 is a major target of TORC1 in Saccharomyces cerevisiae. Mol. Cell 26, 663–674 (2007).
Stracka, D., Jozefczuk, S., Rudroff, F., Sauer, U. & Hall, M. N. Nitrogen source activates TOR (target of rapamycin) complex 1 via glutamine and independently of Gtr/Rag proteins. J. Biol. Chem. 289, 25010–25020 (2014).
Meng, D. et al. Glutamine and asparagine activate mTORC1 independently of Rag GTPases. J. Biol. Chem. 295, 2890–2899 (2020).
Figlia, G. et al. mTORC1 senses glutamine and other amino acids through GCN2. EMBO J. 44, 4825–4866 (2025).
Panchaud, N., Peli-Gulli, M. P. & De Virgilio, C. Amino acid deprivation inhibits TORC1 through a GTPase-activating protein complex for the Rag family GTPase Gtr1. Sci. Signal. 6, ra42 (2013).
Jiang, C. et al. Ring domains are essential for GATOR2-dependent mTORC1 activation. Mol. Cell 83, 74–89 e79 (2023).
Dai, X. et al. AMPK-dependent phosphorylation of the GATOR2 component WDR24 suppresses glucose-mediated mTORC1 activation. Nat. Metab. 5, 265–276 (2023).
Yin, S. et al. CDK5–PRMT1–WDR24 signaling cascade promotes mTORC1 signaling and tumor growth. Cell Rep. 42, 112316 (2023).
Valenstein, M. L. et al. Rag–Ragulator is the central organizer of the physical architecture of the mTORC1 nutrient-sensing pathway. Proc. Natl Acad. Sci. USA 121, e2322755121 (2024).
Jansen, R. M. et al. Structural basis for mTORC1 regulation by the CASTOR1–GATOR2 complex. Nat. Struct. Mol. Biol. 32, 1980–1988 (2025).
Su, M. Y. et al. Cryo-EM structures of amino acid sensors bound to the human GATOR2 complex. Cell Rep. 44, 116088 (2025).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Sanchez-Garcia, R. et al. DeepEMhancer: a deep learning solution for cryo-EM volume post-processing. Commun. Biol. 4, 874 (2021).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Cardone, G., Heymann, J. B. & Steven, A. C. One number does not fit all: mapping local variations in resolution in cryo-EM reconstructions. J. Struct. Biol. 184, 226–236 (2013).
Pettersen, E. F. et al. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010).
Rittinger, K., Walker, P. A., Eccleston, J. F., Smerdon, S. J. & Gamblin, S. J. Structure at 1.65 A of RhoA and its GTPase-activating protein in complex with a transition-state analogue. Nature 389, 758–762 (1997).
Acknowledgements
We thank the DCI Geneva (https://cryoem.unige.ch) and Lausanne (https://dci-lausanne.ch) for their support. This work was funded by the European Research Council (AdG TENDO), the Swiss National Science Foundation and the Canton of Geneva (to R.L.) and by grant PID2023-147101NA-I00 funded by MCIU/AEI/10.13039/501100011033 and by the European Union, ERDF ‘A way of making Europe’ (to L.T.). The funders had no role in the study design, data collection and analysis, decision to publish or preparation of the paper.
Author information
Authors and Affiliations
Contributions
L.T. initiated the project. L.T. performed protein purification and complex reconstitution, cryo-EM sample preparation and optimization, data collection and processing, model building and interpretation, strain construction, confocal microscopy and immunoblots. L.B. and Y.Z. performed strain construction and immunoblots. C.G. performed protein purification. R.L. and L.T. supervised the work and prepared the paper with input from the other authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Structural & Molecular Biology thanks Constantinos Demetriades, Riko Hatakeyama and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available. Primary Handling Editor: Katarzyna Ciazynska and Melina Casadio, in collaboration with the Nature Structural & Molecular Biology team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Cryo-EM data processing pipeline.
a, Representative denaturing gel of the SEAC mixed with the EGOC, as used for cryo-EM analysis, and EGOC alone with TEV used for the elution of the SEAC. This experiment was performed at least 3 times. b, Representative micrographs from untilted and tilted datasets. Data collection was only performed once from this sample. Scale bar = 50 nm c, Processing pipeline for the analysis of cryo-EM data.
Extended Data Fig. 2 Map and model validation statistics.
a, Local resolution estimate, half-map FSC plot and orientation distribution from the final refinement of the full consensus reconstruction. b, Local resolution estimate, half-map FSC plot and orientation distribution from the final refinement of the SEACwing-EGOC reconstruction. c, Local resolution estimates and half-map FSC plots for the focused refinements on the SEACcore. d, Composite map fitted with full SEAC-EGOC model. e, Model-map FSC plots for the Full SEAC-EGOC and the SEACwing-EGOC. f, Representative densities for the cryo-EM maps.
Extended Data Fig. 3 Unassigned densities in the SEAC wing.
a, Unsharpened cryo-EM map of the SEAC wing, colored according to its subunits. Unassigned densities are shown in grey. b, Extra density around the Npr2-Npr3 N-lobes. c, Extra density between the Sea1NTD and Gtr2GTPase. d, Extra density between Npr2, Npr3 and Sea1. e, Extra density between the Sea4N-ter and Npr2C-lobe. f, Missing regions in Sea1. g, Missing regions in Npr3. h, Missing regions in Npr2.
Extended Data Fig. 4 Comparison between unbound and EGOC-bound SEAC structures.
a, Structure of the unbound SEAC (PDB: 8adl)11. b, Structure of the EGOC-bound SEAC. Arrows indicate a small opening of the wings relative to the core compared to the unbound structure. c, Comparison of the relative position of the SEACwing between unbound (grey) and EGOC-bound SEAC. The wing rotates around a pivot on Npr3-Sea4, which causes a displacement of the active site of ~11 Å (distance between Npr2 arginine finger in both models). Models were superimposed on the Sea4 copies that form the back interface, which is adjacent to the wings and interacts with Npr3. d, Comparison of the superimposed SEACwing between unbound and EGOC-bound SEAC. There is a small shift in Sea1 that serves to accommodate Gtr2.
Extended Data Fig. 5 Comparison between the SEACwing, RhoGAP and GATOR1.
a, Interaction between Npr2 and Gtr1, showing the positions of the arginine finger (Arg84) and neighbouring asparagine (Asn85). b, Structure of RhoGAP and RhoA (PDB: 1tx4)55, indicating the arginine finger (Arg85) and auxiliary asparagine (Asn194). c, Sequence alignment of Npr2 in different species, showing that the Npr2 neighbouring asparagine is specifically absent in C.elegans. d, AlphaFold 3 prediction of CeGATOR1-RagA-RagC interaction shows that an asparagine from CeNpr3 (N53) structurally replace the lacking asparagine in CeNpr2. e, Confocal images of Npr2R84AN85A cells expressing SEA4-GFP. f, Confocal images of Npr2R84A cells expressing SEA4-GFP. Scale bar = 5 μm. Representative image of 3 replicates.
Extended Data Fig. 6 Binding between Sea1 and Gtr2.
a, Electrostatic potential of the Gtr2-Sea1 binding interface. b, Electrostatic potential of the RagC-DEPDC5 binding interface (PDB: 7t3b)21. c, Unassigned cryo-EM densities between Gtr2 and Sea1 (left panel) and the last visible N-terminal residue of Sea1 (right panel, Ala101). Due to the position, the density next to Gtr2 likely corresponds to the disordered first 100 amino acids of Sea1. d, Cryo-EM map of the GATOR1-Ragulator-Rag structure (EMD:25653)21 showing the absence of density next to Gtr2, even at a lower threshold. This is consistent with the lack of the N-terminal extension in DEDPC5.
Extended Data Fig. 7 Comparison between Gtr1-Gtr2 and RagA-RagC in different states, and binding of the Rag/Gtr heterodimer to Raptor and SEAC.
a, Structure of the Gtr1GDP-AlF3-Gtr2GDP heterodimer bound to the SEAC. b, Structure of Gtr1GppNHp-Gtr2GppNHp heterodimer in the EGOC crystal structure (PDB: 6jwp)1. c, Structure of the RagAGTP-RagGDP heterodimer bound to Raptor (PDB:6u62)34. d, Structure of the RagAGDP-AlF3-RagCGDP-AlF3 heterodimer bound to GATOR1 (PDB:7t3b)21. e, Structure of the RagAGDP-RagGppNHp heterodimer bound to FLCN-FNIP2 (PDB: 6ulg)33. The distance between equivalent residues on RagA-RagC and Gtr1-Gtr2 is indicated for each structure, showing that the Gtr heterodimer bound to the SEAC has a very similar conformation as the Raptor-bound Rag heterodimer. Likewise, the inactive RagA-RagC shows a drastically different conformation compared to the other structures. f, Western blot against GFP, with actin as a loading control, in Δgtr1Δgtr2 cells expressing different versions of the Gtr1-Gtr2 heterodimer. g, Structure of Raptor-Ragulator-Rags, with the stretch called the “claw” indicated (PDB: 6u62)34. h, AlphaFold2 predicted model for Npr2 superimposed onto the SEAC-EGOC structure. Npr2 contains a large, disordered stretch (“Latch”) that is positioned in the vicinity of the arginine finger.
Extended Data Fig. 8 TORC1 amino acid signalling in SEACIT deletion strains.
a, Immunoblot of phosphorylated and total Sch9 after starvation and repletion of amino acids in wild type cells. b, Immunoblot of phosphorylated and total Sch9 after starvation and repletion of amino acids in Δsea1, Δnpr2 and Δnpr3 cells. c, Immunoblot of phosphorylated and total Sch9 after starvation and repletion of amino acids in Npr2R84AN85A cells. d, Quantification of relative Sch9 phosphorylation over 30 minutes of starvation and amino acid repletion in wild type and SEACIT deletion strains. Data from three independent experiments, except for Δnpr3 (two independent experiments), are presented as mean values +/- SD. e, Quantification of relative Sch9 phosphorylation over 30 minutes of starvation and amino acid repletion in wild type, Δsea1 and Npr2R84AN85A cells. Data from three independent experiments are presented as mean values +/- SD.
Extended Data Fig. 9 Effects of SEACIT mutants and growth assays.
a, Immunoblot of phosphorylated and total Sch9 after starvation and repletion of amino acids in wild type cells. b, Immunoblot of phosphorylated and total Sch9 after starvation and repletion of amino acids in sea1ΔN-ter cells. c, Immunoblot of phosphorylated and total Sch9 after starvation and repletion of amino acids in npr2Δlatch cells. d, Immunoblot of phosphorylated and total Sch9 after starvation and repletion of amino acids in wild type and Sea1R4000/4004A cells. The wild type panel is repeated from Fig. 4a. e, Quantification of relative Sch9 phosphorylation over 30 minutes of starvation and amino acid repletion in wild type, sea1ΔN-ter, npr2Δlatch and Sea1R4000/4004A cells. Data from two independent experiments, as no difference between mutant and wild type cells were observed. f, Side-by-side comparisons of Sch9 and phospho-Sch9 levels at basal (0 min) and starved (30 min) time points amongst mutant strains. g, Growth assays on complete synthetic media, minus and plus rapamycin.
Supplementary information
Source data
Source Data Fig. 3
Confocal microscopy full stack images.
Source Data Fig. 3
Quantification.
Source Data Fig. 4
Uncropped western blots.
Source Data Fig. 4
Quantification.
Source Data Fig. 5
Uncropped western blots/quantification.
Source Data Fig. 5
Quantification.
Source Data Fig. 6
Uncropped western blots/quantification.
Source Data Fig. 6
Quantification.
Source Data Extended Data Fig. 1/Table 1
Uncropped SDS–PAGE.
Source Data Extended Data Fig. 5/Table 5
Confocal microscopy full stack images.
Source Data Extended Data Fig. 7/Table 7
Uncropped western blots.
Source Data Extended Data Fig. 8/Table 8
Uncropped western blots/quantification.
Source Data Extended Data Fig. 8/Table 8
Quantification.
Source Data Extended Data Fig. 9/Table 9
Uncropped western blots.
Source Data Extended Data Fig. 9/Table 9
Quantification.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Tafur, L., Bonadei, L., Zheng, Y. et al. Structure and function of the yeast amino acid-sensing SEAC–EGOC supercomplex. Nat Struct Mol Biol (2026). https://doi.org/10.1038/s41594-026-01746-2
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41594-026-01746-2