Abstract
To evaluate the performance of a multi-input deep learning (DL) model in detecting two common inherited retinal diseases (IRDs), i.e. retinitis pigmentosa (RP) and Stargardt disease (STGD), and differentiating them from healthy eyes. This cross-sectional study includes 391 cases, consisting of 158 subjects with RP, 62 patients with STGD, and 171 healthy individuals. The image dataset is publicly available at http://en.riovs.sbmu.ac.ir/Access-to-Inherited-Retinal-Diseases-Image-Bank. Separate networks using the same hyperparameters were trained and tested on the dataset. Two single-input MobileNetV2 networks were employed for color fundus photography (CFP) and infrared (IR) images, and a multi-input MobileNetV2 network was applied using both imaging modalities simultaneously. The single-input MobileNetV2 achieved 94.44% diagnostic accuracy using CFP, and 94.44% accuracy employing IR images, respectively. The multi-input MobileNetV2 network outperformed both single-input networks with an accuracy of 96.3%. The impact of single-input and multi-input architectures was further evaluated on state-of-the-art neural network models and machine learning algorithms. The deep learning networks utilized in this study achieved high performance for detection of IRDs. Application of a multi-input network employing both CFP and IR image inputs improves the overall performance of the model and its diagnostic accuracy.
Similar content being viewed by others
Introduction
Inherited retinal diseases (IRDs) are rare retinal disorders that result in gradual degeneration of the retinal pigment epithelium (RPE) and photoreceptors1,2. IRD patients commonly complain of gradual loss of vision, nyctalopia or hemeralopia, color vision deficiency and restricted field of vision. Retinitis pigmentosa (RP) is the most common type of diffuse photoreceptor dystrophy, with nearly 1.5 million new cases reported annually3,4. Furthermore, Stargardt disease (STGD) is the most common type of macular dystrophy with an estimated incidence of one per 10,000 individuals3,5.
Modern procedures for the diagnosis of retinal diseases substantially rely on multimodal imaging in addition to retinal examination. Common retinal imaging including color fundus photography (CFP) and infrared (IR) imaging, could improve diagnostic accuracy by underscoring known retinal patterns. Vessel attenuation and waxy pallor of optic disc is considered the most common clinical manifestations in RP patients. In the final steps of disease progression melanin pigment containing bone spicules appear in the mid-peripheral or peripheral retina in CFP6. For STGD, pathological changes on CFP include macular atrophy and retinal flecks6.
Recently, AI-based algorithms have been widely applied in the automatic detection and grading of various diseases using medical image analysis; in particular, they have been extensively applied for retinal images in ophthalmology7,8,9,10,11. Meanwhile, to compensate for limited number of data, transfer learning techniques, VGG12,13, Inception14, and ResNet15,16 have so far been employed as state-of-the-art deep neural network architectures, especially for CFP and optical coherence tomography (OCT) images. Other lightweight architectures, such as MobileNetV217, have also been used. Fundus autofluorescence (FAF)15, OCT, and ultra-widefield images18 have been applied to detect various IRDs. In the study by Masumoto et al., high sensitivity was achieved in detecting RP on ultra-widefield pseudo color and FAF images18. The combination of convolutional neural networks (CNNs), OCT19, and FAF imaging15, has achieved high accuracy in detecting STGD. Multi-class and multi-labeled models20,21,22 have been employed to significantly improve the performance of existing methods and cover a broader range of retinal disorders. Although some studies have been predicting or detecting IRDs, none of them has simultaneously studied different types of IRDs using various retinal imaging modalities. Designing an intelligent model could improve the quality and accuracy of the health care system in order to detect patients with any type of IRDs by initial screening at the lowest expense and particularly be employed in telemedicine for remote areas. In this study, we proposed a novel method based on MobileNetV2 using a multi-input model for simultaneous processing of both CFP and IR images.
Methods
Dataset
In this study, a total of 220 patients with two types of IRDs including RP (n = 158) and STGD (n = 62) registered at the Iranian National Registry for Inherited Retinal Diseases (IRDReg®). were investigated. This combination of data was designed to replicate the real-world prevalence of these diseases (\(\:\frac{Stargardt}{RP}\cong\:\frac{1}{2.6}\))23. The gender and age distribution of patients in each of these categories are presented in Table 1. In addition, 171 healthy individuals were included as controls. There was no statistically significant difference among the study subjects.
The study protocol was approved by the Ethics Committee of the Ophthalmic Research Center affiliated to Shahid Beheshti University of Medical Sciences, Tehran, Iran (IR.SBMU.ORC.REC.1396.15) and was in accordance with the Declaration of Helsinki. The study participants signed the consent form and were assured that the gathered data would be anonymous. Dataset of the images is publicly available at http://en.riovs.sbmu.ac.ir/Access-to-Inherited-Retinal-Diseases-Image-Bank.
Initially, patients with suspected or definitive IRD diagnosis were interviewed to record their visual symptoms. Then, they underwent meticulous retinal examination by a retina specialist using a 90D lens and indirect ophthalmoscopy. In the next step, two imaging techniques including CFP (Visucam Pro NM; Carl Zeiss Meditec AG, Germany) and IR (Heidelberg Engineering, Germany), were applied for all participants.
Data preprocessing techniques, including Crop, Center, Flip (if left-eye image), and Contrast Limited Adaptive Enhancement, were applied to reduce the variance of the images and produce a more uniform image dataset. Figure 1 shows samples of the raw CFP and IR images.
Proposed method
In the present study, retinal image processing and classification were conducted using MobileNetV2. During the training process, the network learns to extract the basic features of the images and thereby, determine the type of disease according to these features. In this type of network, first, the features are extracted using convolutional layers. Then, the extracted high-level features are fed into the dense layers, the output of which is the predicted label of the input retinal images. MobileNetV2 is based on an inverted residual structure where the residual connections are between the bottleneck layers. The architecture of MobileNetV2 contains an initial fully convolutional layer with 32 filters, followed by 19 residual bottleneck layers [19].
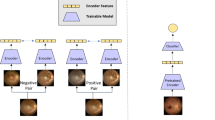
The proposed method is a novel approach that simultaneously receives CFP and IR images. As illustrated in Fig. 2, the network’s training begins with cropping the CFP and IR images, which are merged into a single image before being fed into the MobileNetV2 network. This innovative method allows the network to leverage the complementary information obtained from both types of images, enhancing its ability to predict the presence of diseases such as STDG and RP or to determine the normalcy of the individual. The network is designed to output the necessary predictions based on the integrated features derived from the combined input images.
Implementation and training
The current study utilized the TensorFlowTM24 framework as a deep learning (DL) platform. After preprocessing, images were randomly split into a training set and a test set. A CNN architecture based on the MobileNetV2 model was trained to classify the images into normal, RP, and STGD categories. Transfer learning was applied using the initial weights provided by ImageNet25. Our multi-input model was simultaneously implemented and trained on CFP and IR images. MobileNetV2 was used as a feature extractor for all of the input paths, and the same classifier layers and hyperparameters were applied. A total of 250 (79%) RP, 86 (69%) STGD, and 266 (78%) healthy eyes were utilized to train the model. Also, the remaining images were used for the test set. All models were trained for 30 epochs on the same training set and were tested on the same test set. The Adam optimizer was employed with a learning rate of \(\:{10}^{-5}\) along with a learning rate scheduler that featured a patience of 1 and a decay ratio of 0.9. The final layer utilized a SoftMax activation function, and weighted cross-entropy loss was applied during training to address the imbalance in the dataset. All models had an input shape of 224 * 224 * 3, indicating that both image modalities were loaded and processed in RGB format.
In this study, a diverse set of machine learning (ML) algorithms and DL models were employed to evaluate the performance of the proposed approach. Among the ML algorithms, the Extremely Randomized Tree26, Support Vector Machine (SVM)27, XGBoost28, Random Forest29, and LightGBM30 were utilized due to their proven effectiveness in handling structured data and their ability to capture complex patterns. Also several state-of-the-art DL models were incorporated to assess the performance on unstructured data, particularly in image-related tasks. These models include MnasNet-A131, AlexNet32, VGG1133, ShuffleNetV21 × 34, VGG1333, InceptionV335, ResNet5036, VGG1633, DenseNet12137, and MobileNetV238. Each of these architectures was chosen for its unique design and capabilities, ranging from lightweight models optimized for mobile devices to deeper networks designed for high accuracy in complex tasks. By leveraging this comprehensive set of algorithms and models, the evaluation process ensures a thorough analysis of the proposed method’s performance across various scenarios and data types.
Evaluation metrics
The performance of the models was evaluated based on various metrics, including accuracy, balanced accuracy, precision, F1 score, sensitivity, specificity, and negative predictive value (NPV). The Receiver Operating Characteristics (ROC) and Precision-Recall (PR) curves of all classification models were reported for each approach. Furthermore, Grad-CAM visualization of the class activation functions was implemented to enhance the interpretability of the final multi-input model. This representation shows the regions and areas of the image that the model is concentrating on which helps identify the feature the model considers relevant to its prediction. This step can ensure that the model detects meaningful elements in an image and makes predictions based on meaningful patterns.
Results
The classification performance of various ML algorithms and DL networks is summarized in Table 2, highlighting the superiority of DL models over traditional ML approaches. Among the ML algorithms, LightGBM achieved the highest accuracy (77.16%) and balanced accuracy (67.92%), outperforming other ML methods such as Random Forest and XGBoost. However, the DL models demonstrated significantly better performance, with MobileNetV2 achieving the highest accuracy (96.3%) and balanced accuracy (95.48%), along with an AUC of 99.31%, surpassing all other models. Inception V3 and VGG16 also performed exceptionally well, with equal accuracy accuracies of 95.68% and AUC values of 99.37% and 98.52%, respectively. The results indicate that DL models, particularly MobileNetV2, provide more robust and reliable classification performance compared to traditional ML methods, making them better suited for the automated detection of inherited retinal diseases.
As shown in Table 3, the Multi-input models, which combine CFP and IR images, outperform the Single-input models (using either CFP or IR alone). This superiority stems from the complementary information provided by the two modalities: CFP captures detailed color-based retinal features, while IR highlights deeper structures and vascular patterns. For instance, in the MobileNetV2 model, the Multi-input configuration achieved an accuracy of 96.3% and an AUC of 99.31%, whereas the Single-input models (CFP and IR) both achieved an accuracy of 94.44%. Similarly, in the Inception V3 model, the Multi-input setup achieved an accuracy of 95.68% and an AUC of 99.37%, outperforming CFP (92.59%) and IR (93.21%). These results demonstrate that combining CFP and IR in Multi-input models enhances sensitivity, accuracy, and the ability to detect more complex retinal disease features, offering a significant advantage over Single-input approaches.
The Precision-Recall (PR) curves and Receiver Operating Characteristic (ROC) curves for all three models are presented in Fig. 3. These curves illustrate the superior performance of the Multi-input model, which consistently achieved the highest precision and recall across all disease categories. The ROC curves further confirm the Multi-input model’s ability to effectively balance sensitivity and specificity, achieving near-perfect AUC values for all three classes (RP, STGD, and healthy eyes).
To enhance the interpretability of the proposed model, Grad-CAM visualizations were generated to identify the regions of the retinal images that contributed most to the model’s predictions. As shown in Fig. 4, the Multi-input model accurately focused on disease-relevant features, such as bone spicules and vascular attenuation in RP and macular atrophy and retinal flecks in STGD. These visualizations validate that the model makes predictions based on meaningful retinal features, increasing its reliability for clinical applications.
The performance details of the proposed method are summarized in Table 4, which presents the metrics for detecting RP, STGD, and normal eyes. The Multi-input model, which combines both CFP) and IR images, significantly outperformed the Single-input models in overall performance. Notably, the precision for detecting normal eyes improved to 100%. Furthermore, the specificity values for RP and STGD detection increased to 95.83% and 98.39%, respectively, as indicated in Table 4. These improvements reflect the benefits of integrating complementary information from CFP and IR imaging modalities.
Discussion
The current study demonstrated the excellent performance of a DL model in detecting RP and STGD based on CFP and IR images. These two entities were selected due to being the most prevalent types of IRDs in different populations and the fact that the clinical diagnosis of RP may not be easily possible very early or advanced stages of the disease due to similarities to other retinal pathologies. Furthermore, this automated model might improve reaching a clinical diagnosis given the widespectrum of causative genes reported in RP.
Based on recommendations of the American Academy of Ophthalmology (AAO)39, non-invasive retinal imaging is promising in the diagnosis of IRDs. Herein, we investigated the utility of IR images alongside CFP for detection of STGD and RP; this approach has not been previously reported. As shown in Table 5, although numerous studies have investigated automated detection of different types of IRDs using different retinal images17,18,19,40,41, none of them has investigated the use of IR images.
In the current study, the MobileNetV2 architecture was trained simultaneously using CFP and IR images to diagnose RP and STGD. The single-input analysis using CFP had an acceptable accuracy of 94.44%, AUC of 98.83%, and average precision of 94.38%. Corresponding values utilizing a single input model with IR images achieved an overall accuracy of 94.44%, AUC of 99.15%, and average precision of 94.43%. The single-input IR model had higher sensitivity for STGD and higher specificity for RP detection. Numerous experiments showed that the proposed multi-input approach resulted in higher diagnosis accuracy than classical models that use either CFP or IR retinal images as their single input.
In the current study, RP detection using a single-input CFP model showed higher sensitivity compared to the model used by Chen et al.40 conducted on CFP images from a large sample of 1153 RP and 517 healthy eyes. These authors utilized a fine-tuned Xception architecture model to distinguish RP patients from normal eyes with the sensitivity of 95.71%, AUC-ROC of 96.74% and accuracy of 96%. Chen et al. [37] trained and validated their model based on registered images in the Taiwan inherited retinal degeneration project employing various instruments and technicians. Uur data were collected from the IRDReg® program in which all patients were examined by one retina specialist (N.D) and one optometrist (H.S), and a single experienced technician conducted all CFP and IR imaging.
Another study on RP patients conducted by Masumoto et al.18 investigated the processing of ultrawide- field pseudo color and ultra-wide field autofluorescence images using a VGG16 model for RP detection. They found that both imaging modalities achieved high (> 99%) values for AUC-ROC, sensitivity, and specificity metrics. This slight difference with our figures can be attributed to the extensive field of data capture by ultrawide- field pseudo color and autofluorescence imaging.
Shah et al.19 specifically performed a study on patients with STGD regarding disease detection and severity utilizing two methods: VGG19 architecture and a customized LeNet model42. Their model was capable of distinguishing STGD patients from healthy controls using optical coherence tomography (OCT) images and yielding accuracy of 99%, sensitivity of 99.8%, and specificity of 98% for the first model. This level of performance may be attributed to this fact that OCT is focused on retinal structures in the macula, the area most severely involved in STGD.
An automated model was developed by Fujinami-Yokokawa et al.41 for prediction of the most common causative genes in RP, STGD andOccult macular dystrophy by analyzing CFP and FAF images. The overall accuracy, sensitivity, and specificity of CFP was 88.2%, 88.3%, and 97.4%, respectively with Inception V3 architecture, trained and, tested, with 4-fold cross validation method, while our model could automatically detect the RP and STGD more accurately. It could be concluded that the sensitivity of > 90% was achieved by analyzing of the CFP images in RP patients with mutation in RP1L1 gene and high specificity (> 90%) was identified in detection of RP and STGD patients having mutation in ABCA4, EYS and RP1L1. In a different study published by the same author analyzing the OCT images on a small population, the mean accuracy and sensitivity of 100% for ABCA4, > 78% for RP1L1, and > 89% for EYS were identified. The analysis of the causative genes by automated models is going to be investigated in the future studies of IRDReg® program.
In a study conducted by Miere et al.15 on patients with RP, STGD and Best disease by automated analyzing of \FAF images, ehe overall accuracy was 95% for all studied IRDs. The sensitivity, specificity, and AUC-ROC were obtained greater than 96% for detection of RP and STGD. In our study, the sensitivity and specificity of IR and CFP images for detection of RP and STGD were more than 90%. This difference in results might be attributed to the different imaging techniques, variation in parameters and architectures used for classification.
In another study conducted by Miere et al.43, the single input ResNet101 model findings of FAF images with highest accuracy of 92% and AUC-ROC of 98.1% were reported for differential diagnosis of advanced age- related macular degeneration (AMD) with geographic atrophy from late onset STGD and pseudo- Stargardt pattern dystrophy. This study mainly focused on the differential diagnosis of AMD comparison with the above mentioned IRD types and no results were reported in detection of STGD disease for further comparison.
Deep neural networks have been extensively used for medical image analysis and diagnosis. The complex architecture of such networks allows them to achieve a level of abstraction where high-level information is extracted from raw images. This high-level information is then used for classification of the input image. MobileNetV2 was chosen over other network architectures in this study due to the highest accuracy and precision compared with the other popular architectures, such as DenseNet121, ResNet50, InceptionV3, and VGG16.
The increasing amount of data for diagnosis can be time-consuming and overwhelming for medical professionals. This issue is under extensive investigation by ongoing studies and AI applications can be particularly be beneficial in medicine, particularly in the field of ophthalmology. AI-based automated models enable more accurate diagnosis for referring IRD patients by analyzing their condition. This could also be influential in the early diagnosis of other family members suspicious of IRD diseases with less dependency on a retina specialist40,41.
Automated AI models can be effective in decision support systems to telescreen and detect patients with different IRDs in remote locations with no access to retina specialists. These AI modalities may also lead to more accurate IRD diagnosis, foster personalized medicine, assist possible gene therapy, and eventually help provide better therapeutic outcomes17.
The current study demonstrated that the suggested multi-input MobileNetV2 outperformed its single-input alternatives where only IR or CFP images were used separately. Its efficiency was comparable to other state-of-the-art neural networks and machine learning algorithms. One of the strengths of the current study was that all images were collected from the patients in the Iranian National Registry for Inherited Retinal Diseases (IRDReg®) in which the retinal imaging was conducted by a single experienced technician using a same imaging device. The proposed multi-input approach seems to enjoy the advantage of high diagnostic accuracy and acceptably low amount of data required to train the network. By merging features of IR and CFP images in the model, RP and SGDT were diagnosed more accurately than using single input IR and CFP image models.
One of the limitations of the present study was that the DL model could not grade disease severity in RP and STGD. This model may have the potential to get improved and diagnose of other types of IRDs, detect disease progression over time and also classify disease severity. In the future studies, other imaging modalities can be investigated alongside CFP and IR images.
Conclusion
We proposed a multi-input deep learning neural network model based on MobileNetV2 to classify IRDs. In this approach, each individual’s CFP and IR images were fed into the model to get a prediction. Our results indicate that by accumulating visual features extracted from both of these imaging modalities, the multi-input model outperforms the single-input models separately trained on each set of CFP or IR images. This configuration makes use of the complimentary data of the two imaging modalities to help diagnose the condition.
Data availability
Data could be available by sending a request to the corresponding author.
References
Sabbaghi, H. et al., The first inherited retinal disease registry in Iran: research protocol and results of a pilot study. (2020).
Ben-Yosef, T. Inherited retinal diseases. MDPI 23, 13467 (2022).
Sabbaghi, H. et al. Choroidal thickness in different types of inherited retinal dystrophies. J. Ophthalmic Vis. Res. 15, 351 (2020).
Ayton, L. N., Guymer, R. H. & Luu, C. D. Choroidal thickness profiles in retinitis pigmentosa. Clin. Exp. Ophthalmol. 41, 396–403 (2013).
Sodi, A. et al. EDI OCT evaluation of choroidal thickness in Stargardt disease. Plos One 13, e0190780 (2018).
Pichi, F., Abboud, E. B., Ghazi, N. G. & Khan, A. O. Fundus autofluorescence imaging in hereditary retinal diseases. Acta Ophthalmol. 96, e549–e561 (2018).
Jagan Mohan, N., Murugan, R., Goel, T., Mirjalili, S. & Roy, P. A novel four-step feature selection technique for diabetic retinopathy grading. Phys. Eng. Sci. Med. 44, 1351–1366 (2021).
Mohan, N. J., Murugan, R., Goel, T. & Roy, P. Fast and robust exudate detection in retinal fundus images using extreme learning machine autoencoders and modified Kaze features. J. Digit. Imaging. 35, 496–513 (2022).
Mohan, N. J., Murugan, R., Goel, T. & Roy, P. DRFL: Federated learning in diabetic retinopathy grading using fundus images. IEEE Trans. Parallel Distrib. Syst. 34, 1789–1801 (2023).
Mohan, N. J. et al. Optimal hybrid feature selection technique for diabetic retinopathy grading using fundus images. Sādhanā 48, 102 (2023).
Mohan, N. J., Murugan, R., Goel, T., Tanveer, M. & Roy, P. An efficient microaneurysms detection approach in retinal fundus images. Int. J. Mach. Learn. Cybernet. 14, 1235–1252 (2023).
Gómez-Valverde, J. J. et al. Automatic glaucoma classification using color fundus images based on convolutional neural networks and transfer learning. Biomedical Opt. Express. 10, 892–913 (2019).
Nagasato, D. et al. Deep-learning classifier with ultrawide-field fundus ophthalmoscopy for detecting branch retinal vein occlusion. Int. J. Ophthalmol. 12, 94 (2019).
Li, F. et al. Automatic detection of diabetic retinopathy in retinal fundus photographs based on deep learning algorithm. Transl. Vis. Sci. Technol. 8, 4–4 (2019).
Miere, A. et al. Deep learning-based classification of inherited retinal diseases using fundus autofluorescence. J. Clin. Med. 9, 3303 (2020).
Hemelings, R. et al. Accurate prediction of glaucoma from colour fundus images with a convolutional neural network that relies on active and transfer learning. Acta Ophthalmol. 98, e94–e100 (2020).
Guo, C., Yu, M. & Li, J. Prediction of different eye diseases based on fundus photography via deep transfer learning. J. Clin. Med. 10, 5481 (2021).
Masumoto, H. et al. Accuracy of a deep convolutional neural network in detection of retinitis pigmentosa on ultrawide-field images. PeerJ 7, e6900 (2019).
Shah, M., Roomans Ledo, A. & Rittscher, J. Automated classification of normal and Stargardt disease optical coherence tomography images using deep learning. Acta Ophthalmol. 98, e715–e721 (2020).
Nagasawa, T. et al. Accuracy of ultrawide-field fundus ophthalmoscopy-assisted deep learning for detecting treatment-naïve proliferative diabetic retinopathy. Int. Ophthalmol. 39, 2153–2159 (2019).
Sahlsten, J. et al. Deep learning fundus image analysis for diabetic retinopathy and macular edema grading. Sci. Rep. 9, 10750 (2019).
Redd, T. K. et al. Evaluation of a deep learning image assessment system for detecting severe retinopathy of prematurity. Br. J. Ophthalmol. 103, 580–584 (2019).
Arai, Y. et al., Retinitis pigmentosa with EYS mutations is the most prevalent inherited retinal dystrophy in Japanese populations. J. Ophthalmol. 2015 (2015).
Abadi, M. et al. Tensorflow: Large-scale machine learning on heterogeneous distributed systems. arXiv preprint arXiv:1603.04467 (2016).
Deng, J. et al. Imagenet: A large-scale hierarchical image database. in IEEE conference on computer vision and pattern recognition 248–255 (IEEE, 2009).
Geurts, P., Ernst, D. & Wehenkel, L. Extremely randomized trees. Mach. Learn. 63, 3–42 (2006).
Hearst, M. A., Dumais, S. T., Osuna, E., Platt, J. & Scholkopf, B. Support vector machines. IEEE Intell. Syst. Their Appl. 13, 18–28 (1998).
Chen, T., Guestrin, C. & Xgboost A scalable tree boosting system. in Proceedings of the 22nd acm sigkdd international conference on knowledge discovery and data mining 785–794 (2016).
Breiman, L. Random forests. Mach. Learn. 45, 5–32 (2001).
Ke, G. et al. Lightgbm: A highly efficient gradient boosting decision tree. Adv. Neural Inf. Process. Syst. 30 (2017).
Tan, M. et al. Mnasnet: Platform-aware neural architecture search for mobile. in Proceedings of the IEEE/CVF conference on computer vision and pattern recognition 2820–2828 (2019).
Krizhevsky, A., Sutskever, I. & Hinton, G. E. Imagenet classification with deep convolutional neural networks. Adv. Neural Inf. Process. Syst. 25 (2012).
Simonyan, K. & Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409.1556 (2014).
Ma, N., Zhang, X., Zheng, H. T. & Sun, J. Shufflenet v2: Practical guidelines for efficient cnn architecture design. in Proceedings of the European conference on computer vision (ECCV) 116–131 (2018).
Szegedy, C., Vanhoucke, V., Ioffe, S., Shlens, J. & Wojna, Z. Rethinking the inception architecture for computer vision. in Proceedings of the IEEE conference on computer vision and pattern recognition 2818–2826 (2016).
He, K., Zhang, X., Ren, S. & Sun, J. Deep residual learning for image recognition. in Proceedings of the IEEE conference on computer vision and pattern recognition 770–778 (2016).
Huang, G., Liu, Z., Van Der Maaten, L. & Weinberger, K. Q. Densely connected convolutional networks. in Proceedings of the IEEE conference on computer vision and pattern recognition 4700–4708 (2017).
Sandler, M., Howard, A., Zhu, M., Zhmoginov, A. & Chen, L. C. Mobilenetv2: Inverted residuals and linear bottlenecks. in Proceedings of the IEEE conference on computer vision and pattern recognition 4510–4520 (2018).
Stuart, A. Inherited Retinal Diseases. American Academy of Ophthalmology. https://www.aao.org/eyenet/article/inherited-retinal-diseases (2018).
Chen, T. C. et al. Artificial intelligence–assisted early detection of retinitis pigmentosa—the most common inherited retinal degeneration. J. Digit. Imaging. 34, 948–958 (2021).
Fujinami-Yokokawa, Y. et al. Prediction of causative genes in inherited retinal disorder from fundus photography and autofluorescence imaging using deep learning techniques. Br. J. Ophthalmol. 105, 1272–1279 (2021).
LeCun, Y., Bottou, L., Bengio, Y. & Haffner, P. Gradient-based learning applied to document recognition. Proc. IEEE86, 2278–2324 (1998).
Miere, A. et al. Deep learning-based classification of retinal atrophy using fundus autofluorescence imaging. Comput. Biol. Med. 130, 104198 (2021).
Acknowledgements
This article has been taken from the disease registry, titled “The Iranian National Registry for Inherited Retinal Dystrophy (IRDReg®)” and the code number of IR.SBMU.ORC.REC.1396.15, supported by the Deputy of Research and Technology at Shahid Beheshti University of Medical Sciences (http://dregistry.sbmu.ac.ir).
Author information
Authors and Affiliations
Contributions
F.J.: Data analysis; Software development; Manuscript writing. H.A: Patient examination; Data interpretation; Manuscript writing. F.S: Data analysis; Software development; Manuscript writing. A.K.: Data analysis; Manuscript writing. N.D.: Patient examination; Data interpretation; Manuscript writing. S.F.: Patient examination. T.M.: Patient examination. M.N.: Patient examination. B.K.D.A.: Software development. A.S.T.: Software development. F.K.: Software development. H.B.J.: Manuscript writing. K.B.: Manuscript writing. S.S.: Patient examination. M.Y.: Software development. T.S.: Software development. K.S.: English editing. H.G.K.: Data analysis; Software development; Manuscript writing. M.S.P.: Data analysis; Software development; Manuscript writing. H.S.: Data analysis; Software development; Manuscript writing.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Jafarbeglou, F., Ahmadieh, H., Soleimani, F. et al. A deep learning model for diagnosis of inherited retinal diseases. Sci Rep 15, 22523 (2025). https://doi.org/10.1038/s41598-025-04648-3
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-04648-3
Keywords
This article is cited by
-
A self-supervised learning method for detection of retinitis pigmentosa and Stargardt disease
Scientific Reports (2025)
-
The role of artificial intelligence and machine learning in human disease diagnosis: a comprehensive review
Iran Journal of Computer Science (2025)