Abstract
A series of new uracil derivatives/ursolic acid hybrids were designed and synthesised as potential cytotoxic agents. The uracil, thymine, 6-methyluracil and 2-thiouracil moieties were linked to ursolic acid (1) through alkyl chains of different lengths (either four or six -CH2- units). The cytotoxic activity of the synthesised conjugates was determined using the hormone-dependent breast cancer cell line MCF–7, the triple-negative breast cancer (TNBC) cell line MDA-MB-231 and normal cell lines: human skin fibroblasts (CCD-25Sk) and human bronchial epithelium (BEAS-2B). The five compounds, 4a, 5a, 6a, 7a and 9a, exhibited a significant reduction in the cell viability of human BC cell lines. Analog 6a, which demonstrated high cytotoxic activity against MCF-7 and MDA-MB-231 cell lines with IC50 values of 14.00 µM and 5.83 µM, respectively, was also antitumorigenic in all biochemical assays. It increased p53 and Bax levels in MDA-MB-231 cells as well as significantly decreased Akt kinase levels in the tested cells, and effectively inhibited collagen biosynthesis. Finally, for the selected compounds, we computationally predicted their ADME properties and performed molecular docking to Akt protein kinase. The results of computational docking indicated that the preferred binding mode for all of them was the inactive form of Akt kinase, as in the case with known Akt allosteric inhibitors.
Similar content being viewed by others
Introduction
Thymine and uracil are nucleobases found in RNA and DNA respectively, as well as in natural compounds such as willardiine, sparsomycin, and polyoxins (Fig. 1). These heterocyclic structures also play a crucial role in the normal growth and functioning of the nervous system. Changes in the regulation of purinergic transmission may lead to mental and neurodegenerative disorders1,2. Nucleobase moieties are also commonly used as building blocks in the development of nucleobase-containing drugs (NCDs) with anticancer3, antibacterial4, and antiviral properties5. Anticancer NCDs, such as 5-fluorouracil, can act as antimetabolites. This drug, approved for use in various anticancer therapies, behaves as a false building block for thymidine production, which results in decreased DNA synthesis.
One of the approaches to enhance cell specificity, overcome multidrug resistance and improve target drug delivery, are hybrid molecules of different active compounds with uracil derivatives6,7,8,9. For example, uracil and 5-FU conjugates with the natural alkaloid colchicine demonstrated a similar significant antitumor activity against adenocarcinoma (A549) cells, hepatocellular carcinoma (BEL7402) cells, and human breast carcinoma (MCF-7) cells in the MTT assay8. Kumar et al. synthesised uracil-isatin hybrids for evaluation on HeLa (cervix), MCF-7 (breast), and DU145 (prostate) human cancer lines10. In biological studies, these N, N-disubstituted uracil derivatives (uracil, 5-FU, 5-chlorouracil) revealed very similar, moderate activity against the MCF-7 breast cancer line with IC50 ranging from ca. 40 to 100 µM.
Between the structurally diverse hybrids of uracil and thymine, those containing pentacyclic terpenoids — a class of triterpenes derived from natural plants — also exhibit anticancer properties. Cheng et al. designed several oleanolic acid-uracil/thymine hybrids as potential cytotoxic agents11. Oleanolic acid and uracil/thymine were covalently fused by carbon chains of different lengths to give two types of hybrids. The first type linked the C-28 position of the phytosteroid skeleton to the N1 nitrogen atom in uracil/thymine. The second type of hybrids resulted from a double substitution of N1 and N3 nitrogen atoms in the uracil/thymine moieties. Both groups were tested in vitro against hepatocellular carcinoma Hep-G2, non-small lung cancer A549, gastric cancer BGC-823, estrogenic receptor-positive breast cancer MCF-7, and metastatic prostate cancer PC-3. The most active compounds against MCF-7 cells were identified as two uracil analogues: monosubstituted with an eight-carbon atom linker, and disubstituted with a six-carbon atom chain with IC50 below 0.1 µM. Other double-modified oleanolic acid derivatives at C-3 and C-28 were studied by Mo et al.12. The most active uracil derivative of the group, 1-[(3β)-3-(acetyloxy)-28-oxoolean-12-en-28-yl]-2,4(1 H,3 H)-pyrimidinedione, showed activity below 0.1 µM against MCF-7 breast cancer cells in vitro. Recently, the antiproliferative activity of glycyrrhetinic and ursolic acid hybrids has been studied. These acids were substituted at C-30 and C-28 positions, respectively, by carbon linkers, followed by the connection to the N1 nitrogen atom, or both the N1 and N3 atoms of uracil and thymine13. The compounds were tested in vitro on the hepatocellular carcinomas Hep-G2, A549 and HL-7702 and showed moderate or low antiproliferative activity. The glycyrrhetinic acid hybrids demonstrated a moderately better activity compared to their ursolic acid analogues. The most active compound, the glycyrrhetinic acid-thymine hybrid, exhibited an IC50 value of 10.7 µM against the A549 cell line.
Ursolic acid (UA) plays a special role among the various plant steroids present in raw materials used in natural medicine, due to its broad spectrum of biological activity, including antineoplastic properties. The C-2, C-3, or C-28 positions of the triterpenoid ring of ursolic acid have been identified as crucial elements in the targeted modification of its anticancer activity, thus finding new potential therapeutic agents (see examples in Fig. 2).
In continuation of our research on semi-synthetic derivatives of natural five-ring terpenoids with potential activity against hormone-dependent and TNBC18,19, we designed and synthesised a group of ursolic acid hybrids linked to nucleobase-type rings, namely uracil, thymine, 6-methyluracil and 2-thiouracil. Active subunits in each hybrid compound were linked at C-28 position of the steroid core to one or two nitrogen atoms from a particular nucleobase, by a 4 or 6-carbon atom bridge. This C-28 position, as well as C-3 position, had already been associated with the increased anticancer activity of ursolic acid20 (Fig. 3).
Through the utilisation of nucleobases (uracil and thymine), along with nucleobase antimetabolites (6-methyluracil and 2-thiouracil) as ursolic acid modulators, a variety of hybrid compounds were obtained for SAR, including twenty novel compounds. All structures were confirmed by NMR and HRMS, followed by an investigation of their in vitro anticancer activity on breast cancer cell lines and normal cell lines. Furthermore, additional assays were performed to determine the underlying mechanisms of action.
Results and discussion
Chemistry
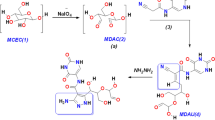
In this study, a series of uracil derivatives/ursolic acid hybrids have been synthesised, including eight ursoloil esters of N-monosubstituted uracil derivatives (4a-6a, 8a-10a, 5a', 9a') and two S-monosubstituted derivatives of thiouracil (7a, 11a). Dual products, i.e. six N, N-disubstituted uracil derivatives (4b-6b, 8b-10b) and two N, S-substituted thiouracil derivatives (7b, 11b) have also been obtained. The initial step involved the esterification of ursolic acid with an excess amount of the corresponding α,ω-dibromoalkane in the presence of potassium carbonate at room temperature to afford bromoalkyl esters 2a or 3a in high yields of 78% and 89%, respectively, while bisester by-products 2b or 3b were also isolated in minimal yields (1% and 6%, respectively, Fig. 4). Subsequently, compounds 2a and 3a were treated with uracil derivatives (uracil, thymine, 6-methyluracil, 2-thiouracil) in the presence of potassium carbonate in DMF at 50 °C providing two types of compounds: mono-products 4a-11a (8.0–92%) and bis-products 4b-11b (3.0–36.0%, Fig. 5).
For mono-derivatives 4a, 6a, 8a, 10a containing thymine and uracil moieties, only regio substitution products at the N1 nitrogen atom of the pyrimidine base were observed. In contrast, for 2-thiouracil, S-substituted products 7a and 11a were obtained with 78% and 92% yields, respectively21. In the case of substitution at the N1 nitrogen, the H-1’ protons exhibited a chemical shift within the range of 3.5-4.0 ppm, which was typical of the -CH2-N < group. Similarly, the C-1’ carbon demonstrated a characteristic chemical shift value within the range of 40–50 ppm. Conversely, the chemical shift of H-1’ protons in compounds 7a and 11a was 3.2 ppm, while that of the C-1’ approximately 30 ppm, which was more typical of the -CH2-S- group. Furthermore, the HMBC spectra for compounds 4a-6a and 8a-10a revealed a correlation between the H-1’protons and the C-2 and C-6 carbons, while for compounds 7a and 11a, the correlation between the H-1’ protons and the C-2 carbons was only observed. These findings demonstrated that, for 2-thiouracil, substitution occurred exclusively at the thione group. For the 6-methyluracil substrate, both possible nitrogen monosubstitutions were observed, i.e. on N1 (5a, 9a) or N3 (5a’, 9a’). The substitution positions were also determined by NMR for compounds derived from 6-methyluracil. While the substitution at N1 occurred, the H-1’ protons correlated with the C-2 and C-6 carbons. For the substitution at N3, the correlation between H-1’’ protons and the C-2 and C-4 carbons was evidenced.
The isolation of compounds 5a and 5a' was achieved with yields of 16% and 10%, respectively. The pair of compounds 9a and 9a' was isolated with yields of 6% and 10%, respectively. The bis-products 4b, 7b and 11b were formed in low yields (3%-7%). In the case of products 7b and 11b, we observe both S- and N3 substitution, as confirmed by NMR spectra. The use of substrates 2a or 3a, the respective uracil derivative, and an inorganic base in a molar ratio of 1:3:3, also resulted in the formation of bis-products 4b, 7b and 11b in low yields (3%-7%). To enhance the yield of the reaction, the procedure was modified by decreasing the amount of the uracil derivative, which increases the statistical probability of double substitution. Therefore, reactions were carried out in a 1:1:1 molar ratio for substrates 2a or 3a, uracil derivatives and inorganic base, yielding compounds 4b, 7b and 11b in the range from 12 to 36%. The structures of the synthesised compounds were confirmed by spectroscopic data (1H and 13C NMR, HRMS).
Cell viability and proliferation in MCF–7 and MDA–MB–231 breast cancer cells
The effect of new uracil derivatives/ursolic acid hybrids on the cell viability of MCF-7 and MDA-MB-231 human BC cell lines was evaluated using the MTT assay (Table 1). Cells were incubated with varying concentrations of the tested compounds and the reference ursolic acid (UA) and doxorubicin (DOX) for 24 hours to determine the IC50 values.
The results obtained following a 24-hour incubation of cells with the tested compounds demonstrated that five compounds 4a, 5a, 6a, 7a, and 9a caused a significant reduction in cell viability of human breast cancer cell lines. Compounds 4a, 5a, 6a and 9a exhibited slightly enhanced efficacy against TNBC MDA-MB-231 cells compared to the estrogen-dependent MCF-7 cells with IC50 values of 12.64 µM, 9.24 µM, 5.83 µM, and 12.10 µM, respectively. In the case of compound 7a, an inverse effect was observed with an IC50 of 15.96 µM against MCF-7 cells, compared to 19.43 µM against MDA-MB-231. In general, the mono-substituted products (4a, 5a, 5a’, 6a, 7a) with a shorter, four-carbon chain between both active scaffolds showed better activity against breast cancer cell lines than their counterparts with the six-carbon spacer (8a, 9a’, 10a, 11a).
Furthermore, the cytotoxicity profile of S-monosubstituted compounds 7a and 7b was found to be comparable to that of N-substituted compounds. For all bis-products (3b-11b), regardless of linker length, no anticancer activity was observed. It may be postulated that the size of these compounds (over 100 kDa) hindered their penetration through cell membranes. According to literature data, compounds with a molecular weight above 1000 Da typically exhibited low permeability through cell membranes22. Furthermore, compounds 4b-11b exceeded the commonly desired limit of 500 Da for systems (Lipiński’s rule of five23 with good oral bioavailability. However, there are many exceptions to this rule. 40% of the protein kinase inhibitors approved by the FDA have at least one Ro5 violation, including this related to molecule size (as trametinib, lapatinib, and pralsetinib, etc.)24.
Among the others compound tested, 6a exhibited the highest activity against MCF-7 and MDA-MB-231 cell lines with IC50 values of 14.00 µM and 5.83 µM, respectively. Its efficacy in reducing the viability of TNBC MDA-MB-23 cells was comparable to that of the control doxorubicin and exceeded the activity of the control ursolic acid. As a control compound, doxorubicin was more effective than ursolic acid in reducing the viability of both breast cancer lines tested.
The results indicated that the mono introduction of a preferential nucleobase substituent into the ursolic acid molecule can enhance its antitumour activity against human breast cancer lines.
To investigate the effect of new uracil derivatives/ursolic acid hybrids on the proliferation of MCF-7 and MDA-MB-231 breast cancer cell lines, the level of [3H]–thymidine incorporation into the DNA was measured (Table 2). The data indicated that the MDA-MB-231 and the MCF-7 cell lines exhibited comparable sensitivity to the compounds tested. The most potent compound 6a showed a slightly greater selectivity for reducing the proliferation of MDA-MB-231 cells. However, compounds 9a and 5a also showed similar effects on MDA-MB-231 proliferation, with IC50 values of about 10 µM.
The results of the first set of experiments (cytotoxicity, cell proliferation) led to the selection of compounds 4a, 5a’, 5a, 6a, 7a, 9a’and 9a for further biological studies. Cytotoxicity analysis of selected compounds against normal cell lines was performed. Further biological assays (collagen biosynthesis, proapoptotic activity, Akt concentration) were performed in TNBC MDA-MB-231 breast cancer cell line, which seems to be more sensitive to the tested agents, where we found lower IC50 for the most examined compounds.
Cytotoxicity analysis of selected compounds against normal lines
Cytotoxicity testing of the new compounds was conducted on two cell lines: human skin fibroblasts (CCD-25Sk) and human bronchial epithelium (BEAS-2B), using the MTT assay. The cells were exposed to compounds for 24 h. IC50 values were determined to assess toxicity to normal cells. Values exceeding 100 µM suggest low or no toxicity under experimental conditions (Table 3).
Ursolic acid (UA) showed moderate toxicity against both cell lines, reaching an IC50 of 32.28 ± 2.46 µM for fibroblasts and 16.02 ± 1.76 µM for BEAS-2B cells. Some derivatives, such as 4a, 5a’, 8a and 9a, had comparable or slightly higher toxicity to normal cells, with IC50 values in the range of about 18–35 µM (fibroblasts) and 15–18 µM (BEAS-2B). The compound 6a possessed weaker cytotoxic activity against normal cells (almost five times lower against human skin fibroblasts with IC50 of 26.88 ± 1.69 and ten times lower against BEAS-2B with IC50 of 60.22 ± 3.77) in comparison with TNBC breast cancer cell line. In contrast, compounds such as 5a, 7a and 9a’ did not show significant toxicity against any of the cell lines tested - IC50 values exceeded 100 µM, which may indicate a favorable safety profile for these substances in the context of normal cells.
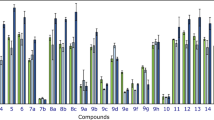
To evaluate the selectivity of the cytotoxic effects of the tested compounds, the selectivity index (SI) was determined by comparing the IC50 values against breast cancer cells (MCF-7 and MDA-MB-231) and normal cells (skin fibroblasts and BEAS-2B), according to the formula: SI = IC50 (normal cell line) / IC50 (cancer cell line). Regarding the compounds analyzed, 5a and 6a showed the highest selectivity especially against MDA-MB-231 cancer cell line, reaching SI values > 10, which is considered a high level of selectivity. The remaining compounds had lower SI values (mostly < 3), indicating limited selectivity. It is worth noting that the selectivity index of ursolic acid (UA), the reference compound, was comparable to most of the tested derivatives (Fig. 6).
Collagen biosynthesis
Proline metabolism has been associated with cancer, affecting the regulation of tumour cell growth and survival. Cancer-associated fibroblasts (CAFs) show a high rate of proline biosynthesis for collagen production to form the pro-cancer extracellular matrix (ECM). Reduced proline metabolism in CAFs causes inhibition of cancer growth and metastasis25. Studies on breast cancer xenografts showed that tumour collagen was produced from glutamine in CAFs (Fig. 7). The study also proved the high expression of the key enzyme for proline synthesis PYCR1 in the stroma of patients with breast cancer and CAFs26. PYCR1 also modulates the process of apoptosis in various cancers. In the Bcl-2 family, silencing PYCR1 significantly decreases the expression levels of the anti-apoptotic proteins Bcl-2 and Bcl-xl27 and increases the level of the pro-apoptotic Bax marker protein28.
PYCR1 expression and proline metabolism are also up-regulated in poorly differentiated breast cancer, and high levels of PYCR1 in TNBC tumours are correlated with poor survival and a higher risk of metastasis and recurrence29. In MDA-MB-231 cells with PYCR1-knockdown, sphere-formation assays showed a significant reduction in the number and diameter of spheroids29.
Since inhibition of collagen synthesis may be the small-molecule mechanism of action30 against breast cancer cells, we investigated the incorporation of radioactive precursor (5-[3H]-proline) into cell proteins, after labelling cells in a growth medium with different concentrations of the tested compounds. Most of the compounds showed inhibitory effects on collagen biosynthesis in the MDA-MB-231 line in a dose-dependent manner (Fig. 8). The highest degree of inhibition was observed for compound 5a. At concentrations of 10 µM, 25 µM and 100 µM, the incorporation of the radioactive precursor to 5a was 52%, 16% and 2%, respectively. Additionally, compound 6a also demonstrated high inhibitory potential and effectively inhibited collagen biosynthesis in the tested cell line. IC50 values reached about 10 µM for compounds 5a, 6a and 9a. In comparison, ursolic acid demonstrated a weaker effect on collagen biosynthesis than the most active compounds.
Proapoptotic activity
Apoptosis can be triggered by extracellular death signals, lack of survival signals, and genetic or toxicological damage31. The process of apoptosis is associated with the activation of caspases, cysteine proteases responsible for initiating and executing programmed cell death (PCD). This process occurs through two main pathways: the intrinsic and extrinsic pathways. The intrinsic pathway, the mitochondrial-dependent activation of apoptosis, is mainly mediated by proteins of the Bcl-2 family32,33. This family includes about twenty members, both proapoptotic, such as Bax, Bak, and BNIP3, which facilitate mitochondrial permeability, and anti-apoptotic, such as Bcl-2 and Bcl-x, which inhibit cellular effects or cytochrome c (cyt c) release in mitochondria. In the intrinsic pathway, the tumour suppressor protein p53 represents a crucial element, as it mediates Bax transactivation and influences Bcl-2 inhibition. Bax and other members of the Bcl-2 family, such as Bak and Bid, are involved in protein-protein interactions that release cytochrome c from mitochondria to the cytosol34. This promotes the formation of the apoptotic complex by binding to apoptotic protease-activating factor 1 (Apaf-1) and forming an apoptosome that activates caspase-9, followed by caspase-3 activation35,36. Finally, caspase-3 executes apoptosis by cleaving the inhibitor of caspase-activated DNase (ICAD), resulting in DNA degradation or fragmentation37 (see Fig. 9).
We investigated the levels of different critical proteins in the p53 signalling pathway to evaluate this signal transduction as a potential target for the compounds tested. Initially, the impact on the levels of the suppressor protein p53 was examined. As shown in Fig. 10, compounds 6a, 9a and 9a’ significantly increased p53 levels in the tested concentration, i.e. 20 µM. We detected 3.46, 2.97 and 4.05 ng/mL. Compounds 4a and the reference ursolic acid increased the p53 concentration, but the values were statistically insignificant.
For the MDA-MB-231 line, the effects induced by the selected compounds at 20 µM concentration were shown in Fig. 11. The most effective 6a increased Bax levels from 653 ng/mL in the control sample to 751 pg/mL. Ursolic acid remained the most active compound at the evaluated concentration and increased Bax level to 760 pg/mL. In conclusion, the results demonstrated a consistent trend of increased Bax and p53 levels for compounds 4a, 6a, 9a and 9a’ in the MDA-MB-231 cell line at a 20 µM concentration. Therefore, it can be postulated that the induction of apoptosis in these compounds depends on the p53 protein. This contrasts with the action of compounds 5a, 5a’ and 7a, where decreased proapoptotic Bax and p53 concentrations were observed.
Akt activity
The serine/threonine kinase Akt, also known as protein kinase B (PKB), plays an essential role in regulating diverse cellular activities such as cell proliferation, growth, survival, metabolism, and migration. Akt is involved in the control of signaling protein synthesis and immune cell function38. It influences many aspects of cancer biology, as hyperactivation of Akt increases cell survival, proliferation, and invasion. It is therefore clinically relevant to the outcome of cancer therapy, e.g. by increasing resistance to chemotherapy and radiotherapy39,40. Kale et al. demonstrated that activation of the Akt pathway is associated with a reduced sensitivity to chemotherapy. An increase in Akt elevates the phosphorylation of Bax, which converts this protein from a proapoptotic to an anti-apoptotic signaling transducer (Fig. 12)41. Therefore, for over two decades, Akt has been investigated as a potential therapeutic target in cancer treatment. Various compounds are currently being obtained as Akt inhibitors and some of them are being tested in the clinic as anticancer drugs in monotherapy and combination therapy42.
Phosphorylated Bax protects cells from apoptosis. Akt phosphorylates (P) Bax at position S184 in cells with increased Akt signaling. Phosphorylation prevents Bax from entering the outer mitochondrial membrane, allowing binding of the activator BH3, and making cells resistant41.
Therefore, the Akt kinase concentration was also measured in MDA-MB-231 cells, after a 24-hour incubation. The results shown in Fig. 13 indicated that treatment with compounds 4a, 5a, 5a’, 6a, 7a and 9a resulted in a statistically significant decrease in Akt levels. The most effective compounds were 4a, 5a’ and 6a, which reduced the Akt level from 8.22 U/mL in the control to 1.6 U/mL at a concentration of 20 µM. Ursolic acid, as a reference compound, also decreased Akt concentration to 4.49 U/mL.
ADME properties
The selected key ADME properties of the compounds studied are presented in Table 4. All ursolic acid derivatives were outside the commonly desired limit of 500 Da for systems with good oral bioavailability43, but within the modern limit of < 700 Da44. All also showed additional violations of Lipinski’s rule of five, due to relatively high logP values. The predicted logP values of > 6 may be problematic in the context of their bioavailability, as well as a high probability to accumulate in the organism. However, computational toxicity studies predicted that all compounds had LD50 values in the range of 1100–2000 mg/kg, similar to other known drugs, and can exhibit respiratory toxicity and immunotoxicity.
Molecular docking
The results of computational docking of all the ursolic acid derivatives studied indicated that the preferred binding mode for all of them was to the inactive form of Akt, like allosteric Akt inhibitors. The predicted binding site of all derivatives was virtually identical, as were the ligand orientations and interactions with residues that stabilized the derivatives in the binding pocket. The binding site was also virtually identical to the binding site of miransertib from the 5KCV crystal structure, although predicted binding poses were stabilized by different residues. In the case of the 5KCV crystal structure miransertib is stabilized mainly by hydrogen bonds to peptide bond of THR211, carboxylic group of ASP274 and van der Waals interaction with Trp80 (Fig. 14a). For the ursolic acid, the predicted binding pose involved a hydrogen bond between the hydroxyl group of the ligand and SER205, a hydrogen bond and a salt bridge between the carboxyl group of the ligand and both ASN54 and ARG273 (Fig. 14b). In the case of the ursolic acid derivatives, the former interaction remained intact, similarly to the hydrogen bond between the carbonyl group and ASN54. However, the interaction with ARG273 was disrupted due to structural modifications of the carboxyl group (Fig. 14c). For some derivatives, ARG273 was predicted to rotate away from the ligand and not form any relevant interactions, while for others it was predicted to form hydrogen bonds with atoms of the pyrimidinedione moiety. This was possible due to the high flexibility of the alkyl linker connecting this group to the ursolic acid core. In the case of derivative 4a, estimated to have the Gibbs free energy of binding as high as ursolic acid, the long alkyl linker positions the terminal pyrimidine moiety close to LYS14 and forming with it an additional hydrogen bond (Fig. 14c). In addition, the values obtained for the Gibbs free energies of binding were quite similar, given the expected accuracy of the computational approach of less than 1 kcal/mol. Based on the results presented in Table 5, it can be suggested that the predicted Gibbs free energy of binding was very similar for ursolic acid and its derivatives 4a, 5a, 5a’, 6a, and 7a, and significantly lower for 9a and 9a’. Additionally, the lowest (most favourable) binding was found for ursolic acid and its derivative 4a with the values of -13.5 kcal/mol, which were higher (less favourable) than the corresponding Gibbs free energy value of miransertib in the crystal structure pose. These results suggested that the binding of ursolic acid and the 4a derivative was weaker, and therefore their inhibitory potential was likely to be lower approximately by a factor of 10.
(a) The predicted (cyan) and experimental (green) binding pose of miransertib in the 5KCV crystal structure; (b) The predicted binding pose of the ursolic acid (1) in the 5KCV crystal structure; (c) The predicted binding pose of 4a in the 5KCV crystal structure; (d) The predicted binding site of derivative 5a’ in the inactive form of Akt (4GV1 crystal structure). Figure 14 has been created using The PyMOL Molecular Graphics System ver. 1.3 (https://pymol.org).
As mentioned above, the Gibbs free energies of all compounds studied to the active conformation (based on the 4GV1 crystal structure) or to the PH domain only (based on the 1UNQ crystal structure) were estimated to be lower. However, for selected ligands, the differences between these values for the active and inactive conformations were of the order of ~ 1 kcal/mol, therefore, we could not exclude binding to the active conformation. The lowest binding energy for the active conformation was found for 5a’, with a Gibbs free energy binding of -13.1 kcal/mol. The binding pose of this derivative aligned well with the binding pose of capivasertib (stabilized in the binding pocket via interactions with GLU228, GLU234 and MT281), and the ligand was predicted to be mainly stabilised by van der Waals interactions and a single hydrogen bond between LYS179 and an oxygen atom of the pyrimidinedione moiety (Fig. 14d).
Materials and methods
Chemical synthesis
Ursolic acid was obtained from ATOMOLE, China. Other materials, solvents, and reagents were of commercial origin and were used without additional operations. Reactions were monitored on silica gel TLC plates 60 F254 (Merck, Darmstadt, Germany). Visualizations were performed with UV light (254 and/or 365 nm) and then with CeMo stain and subsequent charring45. The melting points were determined using the Melting Point System (Mettler Toledo MP70). Solvents were evaporated under reduced pressure at 40 °C on the Büchi Rotavapor. Flash column chromatography was performed on silica gels (200–300 mesh). The synthesis of compounds and their NMR, HRMS characterization were described in Supporting Information.
Cell culture
Cell culture MCF-7 and MDA-MB-231 human breast cancer cells as well as normal cell lines: human skin fibroblasts (CCD-25Sk CRL-1474 ™) and human bronchial epithelium (BEAS-2B, CRL-3588™) were purchased from the ATCC—American Type Culture Collection. Human breast cancer MCF-7 and MDA-MB-231 cells were maintained in complete growth medium DMEM supplemented with 10% FBS and 1% antibiotics (penicillin/streptomycin). Cells were cultured in Costar flasks at 37 °C with 5% CO2 until they reached sub-confluence (90–95%). Sub-confluent cells were treated with 0.05% trypsin and 0.02% EDTA in calcium free phosphate buffered saline, counted in a hemocytometer and seeded in 6-well plates (Nunc) in 2 mL of growth medium (DMEM without phenol red with 10% CPSR1). Cells which reached about 80% of confluency, were used for the assays46.
Cell viability assay
Cell growth was evaluated in MCF-7, MDA-MB-231, human skin fibroblasts (CCD-25Sk, CRL-1474 ™) and human bronchial epithelium (BEAS-2B, CRL-3588 ™) following treatment with the tested compounds using MTT (3-(4,5-dimethylthiazole-2-yl)-2,5-diphenyltetrazolium bromide) assay46,47. Absorbance of converted dye in living cells was measured at a wavelength of 570 nm. Cell viability of breast cancer cells cultured in the presence of ligands was calculated as a per cent of control cells.
[3H]thymidine incorporation assay
The incorporation of [3H]thymidine into DNA was used as a measure of cell proliferation. MCF-7 and MDA-MB-231 cells were seeded in 6-well tissue culture plates at a density of 5 × 105 well− 1 in complete growth medium and grown as described above. Cells were treated with different concentrations of compounds. Cells were incubated with compounds for 24 h at 37 °C before 0.5 µC, [3H]thymidine was added to each well for a 4 h period to measure the incorporation of radioactive component into the DNA. Radioactivity was quantitated in a scintillation counter. [3H]thymidine incorporation was expressed as dpm well− 1. Each experiment was repeated at least three times48.
Collagen production
Incorporation of radioactive precursor into proteins was measured after labeling of the cells in growth medium with varying concentrations of the tested compounds for 24 h with 5-[3H]proline (5 µCi ml− 1, 28 Ci mmol− 1). Incorporation of tracer into collagen was determined by digesting proteins with purified Clostridium histolyticum collagenase49,50. Results are shown as combined values for cell plus medium fractions.
Determination of Akt, Bax, p53 concentration
High sensitivity assay kits (Abcam, Invitrogen) were used to determine the concentrations of selected proteins in cell lysates after 24 h of incubation with novel compounds and reference drug in 10 µM and 20 µM concentrations. After trypsinization, cells were washed thrice with cold PBS and centrifuged at 1000× g for 5 min at 4 °C. Then, cells (1.5 × 106) were suspended in lysis buffer for whole-cell lysates. After the second centrifugation, cellular supernatants were immediately frozen at − 70 °C. Untreated cancer cells were taken as a control51.
The microtiter plates included in the kits were pre-coated with a specific antibody for the antigen being analysed. The tests were conducted following the manufacturer’s protocols.
Statistical analysis
The obtained results are presented as mean ± SEM from three independent experiments performed in duplicate. Statistical analysis was performed using GraphPad Prism Version 9.3 (San Diego, CA, USA). The one–way ANOVA with Tuckey multiple comparison post–hoc test was used to show differences between control and cancer cells exposed to varying concentrations of UA derivatives and the reference drug. A statistically significant differences were defined at * p ≤ 0.0518.
Computational details
Models of all studied ligands, ursolic acid and its derivatives, were prepared based on the structure of ursolic acid (PubChem CID: 64945), similarly to our previous work18,52. As there are two known major modes of Akt inhibitors binding to Akt and two different binding sites, depending on the conformation of Akt and the orientation of its kinase domain versus the PH domain, we selected three different crystal structures for molecular docking42. First, we considered the active conformation and separate models of the kinase domain (PDB code: 4GV1) and the PH domain (PDB code: 1UNQ) and we also considered the inactive conformation of the entire protein (PDB code: 5KCV)53,54,55. In the molecular docking part, we used a standard flexible ligand protocol as implemented in the AutoDock Vina ver. 1.1.2. software56. In the first stage, we performed flexible docking of all of the studied derivatives to rigid models of the three protein models to verify if there are any additional potential binding sites. However, in all cases, all studied ligands docked preferentially to the binding site of capivasertib (in the model based on the 4GV1 structure) and miransertib (in the model based on the 5KCV structure). In the case of docking to the PH domain (PDB code: 1UNQ) all investigated ligands also docked preferentially to one particular site, defined in detail later. In the second stage for each protein system studied, we selected flexible amino acid residues that were located within 4.0 Å of ursolic acid in the first stage of docking. The flexible amino acid residues consisted of the following sets: LEU156, LYS179, MET227, ALA230, GLU234, LYS276, GLU278 and PHE438 for 4GV1 model, LYS14, GLU17, TYR18, ARG23, ARG25, LYS39, LEU52, ASN53, ASN54, GLN79, and ARG86 for the 1UNQ model, and ASN53, ASN54, GLN79, TRP80, THR82, ILE84, SER205, LEU210, LEU264, LYS268, VAL270, VAL271, ARG273, ASP274 and ASP292 for the 5KCV model; all other residues were kept rigid. For molecular docking, we used standard AutoDock Vina parameters with 20 × 20 × 20 Å docking boxes centred either at the entire protein (first stage) or at the potential binding site (second stage) and the exhaustiveness parameter set to 18. We also performed molecular docking of miransertib to the model based on 5KCV structure to verify whether our algorithm can reproduce experimental findings and found that the predicted pose is very similar to the experimental one (rmsd of 1.6 Å). In the analysis of final poses, for hydrogen bond determination we used the standard criteria: the donor-acceptor distance below 3.0 Å and the donor-hydrogen-acceptor angle above 150°, while for hydrophobic interactions we uses a distance criterion of 3.5–5 Å. Additionally, we verified the ADME properties of the studied compounds using QikProp 4.6 software (Schrödinger Inc., New York, NY, USA) with default options, while toxicity was predicted using the ProTox-3.0 server57.
Conclusions
In conclusion, we designed and synthesised 20 hybrids of ursolic acid with uracil, thymine, 6-methyluracil and 2-thiouracil, in which ursolic acid was linked to the respective nucleobases via alkyl linkers of different carbon chain lengths (n = 4 or n = 6). Five compounds 4a, 5a, 6a, 7a and 9a, significantly reduced cell viability and inhibited the proliferation of the MCF-7 and MDA-MB-231 breast cancer lines. Bis-products (3b-11b), regardless of the length of the carbon linker, showed no anticancer or antiproliferative effects. In comparison to both normal cell lines, compounds 5a and 6a demonstrated the highest degree of cytotoxic selectivity against the TNBC MDA-MB-231 cell line. The measured SI for compound 5a was above 10 for both tested normal cell lines. For compound 6a, this result was observed when comparing the cytotoxic effect against the TNBC and BEAS-2B cell lines.
Furthermore, the effects of the compounds on collagen biosynthesis, measured by 5-[3H]-proline incorporation, were studied to identify that the 5a and 6a analogues as the most effective in this assay. Analysis of changes in the levels of p53 proteins (p53, Bax) indicated that compounds 4a, 6a, 9a and 9a’ may act through the mechanism of mitochondria-dependent activation of apoptosis in the MDA-MB-231 cancer cell line. All compounds were also found to reduce Akt levels in MDA-MB-231 cells, but the most potent were 4a, 5a’ and 6a. Theoretical approaches were also used to predict the potential biological action of the new hybrids based on their physical properties. ADME data showed that all ursolic acid derivatives were within the modern limit of molecular weight < 700 Da and showed no other violations of the Lipinski rule of five. Computational docking results indicated that the preferred binding mode for all hybrids was the inactive form of Akt, like known allosteric Akt inhibitors. The Gibbs free energies of binding to the active conformation or the PH domain predicted the lowest binding energy for the active conformation of 5a’, with its binding position matching that of capivasertib. Analogue 6a showed high cytotoxic and antiproliferative activity in both breast cancer cell lines tested. It also revealed selectivity against TNBC MDA-MB-231 cells and demonstrated antitumorigenic properties in the biochemical assays. It increased the levels of p53 and Bax in MDA-MB-231 cells, significantly decreased Akt in MCF-7 cells, and effectively inhibited collagen biosynthesis (Fig. 15).
Although compound 6a exhibited activity in vitro, additional research is necessary to fully evaluate its therapeutic potential. The decision to proceed with in vivo studies depends on the completion of in vitro studies, which are an ethical and scientific criterion. Therefore, we plan to investigate the mechanism of action of compound 6a in additional carcinogenesis inhibition models and examine its pharmacokinetic properties. We anticipate that the results will justify conducting in vivo studies, including those using PDX models of breast cancer.
Data availability
All data generated or analysed during this study are included in this published article and its supplementary information files.
References
Fumagalli, M., Lecca, D., Abbracchio, M. P. & Ceruti, S. Pathophysiological role of purines and pyrimidines in neurodevelopment: unveiling new Pharmacological approaches to congenital brain diseases. Front. Pharmacol. 8, 941. https://doi.org/10.3389/fphar.2017.00941 (2017).
Micheli, V. et al. Neurological disorders of purine and pyrimidine metabolism. Curr. Top. Med. Chem. 11 (8), 923–947. https://doi.org/10.2174/156802611795347645 (2011).
Tsesmetzis, N., Paulin, C. B. J., Rudd, S. G. & Herold, N. Nucleobase and nucleoside analogues: resistance and Re-Sensitisation at the level of pharmacokinetics, pharmacodynamics and metabolism. Cancers 10 (7), 240. https://doi.org/10.3390/cancers10070240 (2018).
Thomson, J. M. & Lamont, I. L. Nucleoside analogues as antibacterial agents. Front. Microbiol. 10, 952. https://doi.org/10.3389/fmicb.2019.00952 (2019).
Wong, X. K., Ng, C. S. & Yeong, K. Y. Shaping the future of antiviral treatment: spotlight on Nucleobase-Containing drugs and their revolutionary impact. Bioorg. Chem. 144, 107150. https://doi.org/10.1016/j.bioorg.2024.107150 (2024).
Pałasz, A. & Cież, D. In search of uracil derivatives as bioactive agents. Uracils and fused uracils: synthesis, biological activity and applications. Eur. J. Med. Chem. 97, 582–611. https://doi.org/10.1016/j.ejmech.2014.10.008 (2015).
Sanduja, M., Gupta, J. & Virman, T. Recent advancements in uracil and 5-Fluorouracil hybrids as potential anticancer agents: A review. J. Appl. Pharm. Sci. 10 (02), 129–146. https://doi.org/10.7324/JAPS.2020.102019 (2020).
Lihong, S. et al. Synthesis and biological evaluation of novel uracil and 5-fluorouracil-1-yl acetic acid-colchicine conjugate. Chem. Res. Chin. Univ. 31, 367–371. https://doi.org/10.1007/s40242-015-4445-3 (2015).
Li, X. Y. et al. Design, synthesis and biological evaluation of N-phenyl-(2,4-dihydroxypyrimidine-5-sulfonamido)benzoyl Hydrazide derivatives as thymidylate synthase (TS) inhibitors and as potential antitumor drugs. Eur. J. Med. Chem. 154, 267–279. https://doi.org/10.1016/j.ejmech.2018.05.020 (2018).
Kumar, K., Sagar, S., Esau, L., Kaur, M. & Kumar, V. Synthesis of novel 1H-1,2,3-triazole tethered C-5 substituted uracil–isatin conjugates and their cytotoxic evaluation. Eur. J. Med. Chem. 58, 153–159. https://doi.org/10.1016/j.ejmech.2012.10.008 (2012).
Cheng, K. G. et al. Synthesis and cytotoxic evaluation of several oleanolic acid–uracil/thymine conjugates. Med. Chem. Commun. 7, 972–981. https://doi.org/10.1039/C6MD00061D (2016).
Mo Wb, S., Ch, Huang, Jy, Liu, J., Chen, Z. & Kg, C. Synthesis of acyl oleanolic acid-uracil conjugates and their anti-tumor activity. Chem. Cent. J. 10, 69. https://doi.org/10.1186/s13065-016-0217-5 (2016).
Sun, L. et al. Antiproliferative activity of ursolic acid/glycyrrhetinic acid-uracil/thymine hybrids. Med. Chem. Res. 28, 892–899. https://doi.org/10.1007/s00044-019-02344-2 (2019).
Li, W. et al. A novel synthetic ursolic acid derivative inhibits growth and induces apoptosis in breast cancer cell lines. Oncol. Lett. 15 (2), 2323–2329. https://doi.org/10.3892/ol.2017.7578 (2018).
Tian, T. et al. Synthesis of novel oleanolic acid and ursolic acid in C-28 position derivatives as potential anticancer agents. Arch. Pharm. Res. 40 (4), 458–468. https://doi.org/10.1007/s12272-016-0868-8 (2017).
Dar, B. A., Lone, A. M., Shah, W. A. & Qurishi, M. A. Synthesis and screening of ursolic acid-benzylidine derivatives as potential anti-cancer agents. Eur. J. Med. Chem. 23, 111:26–32. https://doi.org/10.1016/j.ejmech.2016.01.026 (2016).
Friedrich., Serbian, I. et al. Synthesis and cytotoxic evaluation of malachite green derived oleanolic and ursolic acid piperazineamides. Med. Chem. Res. 29, 926–933. https://doi.org/10.1007/s00044-020-02536-1 (2020).
Michalak, O. et al. Biological activity, ADME and molecular Docking studies of novel ursolic acid derivatives as potent anticancer agents. Int. J. Mol. Sci. 24 (10), 8875. https://doi.org/10.3390/ijms24108875 (2023).
Michalak, O. et al. Synthesis and anti-tumour, Immunomodulating activity of Diosgenin and Tigogenin conjugates. J. Steroid Biochem. Mol. Biol. 198, 105573. https://doi.org/10.1016/j.jsbmb.2019.105573 (2020).
Similie, D. et al. An update on pentacyclic triterpenoids ursolic and oleanolic acids and related derivatives as anticancer candidates. Antioxidants 13 (8), 952. https://doi.org/10.3390/antiox13080952 (2024).
Prachayasittikul, S. et al. Synthesis and structure-activity relationship of 2-thiopyrimidine-4-one analogs as antimicrobial and anticancer agents. Eur. J. Med. Chem. 46 (2), 738–742. https://doi.org/10.1016/j.ejmech.2010.12.009 (2011).
Matsson, P. & Kihlberg, J. How big is too big for cell permeability?? J. Med. Chem. 9 (5), 1662–1664. https://doi.org/10.1021/acs.jmedchem.7b00237 (2017).
Lipinski, C. A., Lombardo, F., Dominy, B. W. & Feeney, P. J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv Rev. 1 (1–3), 3–26. https://doi.org/10.1016/s0169-409x(00)00129-0 (2001).
Roskoski, R. Jr Rule of five violations among the FDA-approved small molecule protein kinase inhibitors. Pharmacol. Res. 191, 106774. https://doi.org/10.1016/j.phrs.2023.106774 (2023).
Kay, E. J., Zanivan, S. & Rufini, A. Proline metabolism shapes the tumor microenvironment: from collagen deposition to immune evasion. Curr. Opin. Biotechnol. 84, 103011. https://doi.org/10.1016/j.copbio.2023.103011 (2023).
Kay, E. J. et al. Cancer-associated fibroblasts require proline synthesis by PYCR1 for the deposition of pro-tumorigenic extracellular matrix. Nat. Metab. 4, 693–710. https://doi.org/10.1038/s42255-022-00582-0 (2022).
Cai, F. et al. Pyrroline 5 carboxylate reductase 1 promotes proliferation and inhibits apoptosis in non-small cell lung cancer. Oncol. Lett. 15, 731–740. https://doi.org/10.3892/ol.2017.7400 (2018).
Ye, Y., Wu, Y. & Wang, J. Pyrroline-5-carboxylate reductase 1 promotes cell proliferation via inhibiting apoptosis in human malignant melanoma. Cancer Manag Res. 10, 6399–6407. https://doi.org/10.2147/CMAR.S166711 (2018).
Cui, B. et al. Pyrroline-5-carboxylate reductase 1 reprograms proline metabolism to drive breast cancer stemness under psychological stress. Cell. Death Dis. 14, 682. https://doi.org/10.1038/s41419-023-06200-5 (2023).
Baldari, S., Di Modugno, F., Nisticò, P. & Toietta, G. Strategies for efficient targeting of tumor collagen for cancer therapy. Cancers 14 (19), 4706. https://doi.org/10.3390/cancers14194706 (2022).
Nair, P., Lu, M., Petersen, S. & Ashkenazi, A. Apoptosis initiation through the cell-extrinsic pathway. Methods Enzymol. 544, 99–128. https://doi.org/10.1016/B978-0-12-417158-9.00005-4 (2014).
Shoshan-Barmatz, V., Arif, T. & Shteinfer-Kuzmine, A. Apoptotic proteins with non-apoptotic activity: expression and function in cancer. Apoptosis 28, 730–753. https://doi.org/10.1007/s10495-023-01835-3 (2023).
Lossi, L. The concept of intrinsic versus extrinsic apoptosis. Biochem. J. 479 (3), 357–384. https://doi.org/10.1042/BCJ20210854 (2022).
Sekar, G., Ojoawo, A. & Moldoveanu, T. Protein-protein and protein-lipid interactions of pore-forming BCL-2 family proteins in apoptosis initiation. Biochem. Soc. Trans. 50 (3), 1091–1103. https://doi.org/10.1042/BST20220323 (2022).
Wang, H. et al. Mechanism of Heshouwuyin inhibiting the cyt c/Apaf-1/Caspase-9/Caspase-3 pathway in spermatogenic cell apoptosis. BMC Complement. Med. Ther. 20 (1), 180. https://doi.org/10.1186/s12906-020-02904-9 (2020).
Vodovotz, Y. et al. Inflammatory modulation of hepatocyte apoptosis by nitric oxide: in vivo, in vitro, and in Silico studies. Curr. Mol. Med. 4 (7), 753–762. https://doi.org/10.2174/1566524043359944 (2004).
Bagci, E. Z., Vodovotz, Y., Billiar, T. R., Ermentrout, G. B. & Bahar, I. Bistability in apoptosis: roles of bax, bcl-2, and mitochondrial permeability transition pores. Biophys. J. 90 (5), 1546–1559. https://doi.org/10.1529/biophysj.105.068122 (2006).
Szymonowicz, K., Oeck, S., Malewicz, N. M. & Jendrossek, V. New insights into protein kinase b/akt signaling: role of localized Akt activation and Compartment-Specific target proteins for the cellular radiation response. Cancers 10 (3), 78. https://doi.org/10.3390/cancers10030078 (2018).
Vivanco, I. & Sawyers, C. L. The phosphatidylinositol 3-Kinase AKT pathway in human cancer. Nat. Rev. Cancer. 2 (7), 489–501. https://doi.org/10.1038/nrc839 (2002).
Altomare, D. A. & Testa, J. R. Perturbations of the AKT signaling pathway in human cancer. Oncogene 24 (50), 7455–7464. https://doi.org/10.1038/sj.onc.1209085 (2005).
Kale, J. et al. Phosphorylation switches Bax from promoting to inhibiting apoptosis thereby increasing drug resistance. EMBO Rep. 19 (9), e45235. https://doi.org/10.15252/embr.201745235 (2018).
Pervanidis, K. A., D’Angelo, G. D., Weisner, J., Brandherm, S. & Rauh, D. Akt inhibitor advancements: from Capivasertib approval to Covalent-Allosteric promises. J. Med. Chem. 67 (8), 6052–6063. https://doi.org/10.1021/acs.jmedchem.4c00075 (2024).
LipinskiCA Lead- and drug-like compounds: the rule-of-five revolution. Drug Discov Today Technol. 1 (4), 337–341. https://doi.org/10.1016/j.ddtec.2004.11.007 (2004).
Matsson, P., Doak, B. C., Over, B. & Kihlberg, J. Cell permeability beyond the rule of 5. Adv. Drug Deliv Rev. 101, 42–61. https://doi.org/10.1016/j.addr.2016.03.013 (2016).
Pirrung, M. C. The Synthetic Organic Chemist’s Companionpp. 171–172 (John Wiley & Sons, Inc., 2006).
Gornowicz, A. et al. Cytotoxic efficacy of a novel dinuclear platinum(II) complex used with anti-MUC1 in human breast cancer cells. Mol. Cell. Biochem. 392, 161–174. https://doi.org/10.1007/s11010-014-2018-2 (2014).
Carmichael, J., DeGraff, W. G., Gazdar, A. F., Minna, J. D. & Mitchell, J. B. Evaluation of a tetrazolium-based semiautomated colorimetric assay: assessment of chemosensitivity testing. Cancer Res. 47 (4), 936–942 (1987).
Lepiarczyk, M. et al. Cytotoxic activity of octahydropyrazin[2,1-a:5,4-a′]diisoquinoline derivatives in human breast cancer cells. Arch. Pharm. Res. 38, 628–641. https://doi.org/10.1007/s12272-014-0444-z (2015).
Peterkofsky, B., Palka, J., Wilson, S., Takeda, K. & Shah, V. Elevated activity of low molecular weight insulin-like growth factor-binding proteins in Sera of vitamin C-deficient and fasted Guinea pigs. Endocrinology 128 (4), 1769–1779. https://doi.org/10.1210/endo-128-4-1769 (1991).
Gornowicz, A., Szymanowski, W., Czarnomysy, R., Bielawski, K. & Bielawska, A. Anti-HER2 monoclonal antibodies intensify the susceptibility of human gastric cancer cells to Etoposide by promoting apoptosis, but not autophagy. PLoS One. 16 (8), e0255585. https://doi.org/10.1371/journal.pone.0255585 (2021).
Buzun, K. et al. 2-{5-[(Z,2Z)-2-Chloro-3-(4-nitrophenyl)-2-propenylidene]-4-oxo-2-thioxothiazolidin-3-yl}-3-methylbutanoic acid as a potential Anti-Breast cancer molecule. Int. J. Mol. Sci. 23 (8), 4091. https://doi.org/10.3390/ijms23084091 (2022).
Kim, S. et al. PubChem in 2021: new data content and improved web interfaces. Nucleic Acids Res. 49 (D1), D1388–D1395. https://doi.org/10.1093/nar/gkaa971 (2021).
Addie, M. et al. Discovery of 4-amino-N-[(1S)-1-(4-chlorophenyl)-3-hydroxypropyl]-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidine-4-carboxamide (AZD5363), an orally bioavailable, potent inhibitor of Akt kinases. J. Med. Chem. 56 (5), 2059–2073. https://doi.org/10.1021/jm301762v (2013).
Milburn, C. C. et al. Binding of phosphatidylinositol 3,4,5-trisphosphate to the pleckstrin homology domain of protein kinase B induces a conformational change. Biochem. J. 375 (Pt 3), 531–538. https://doi.org/10.1042/BJ20031229 (2003).
Lapierre, J. M. et al. Discovery of 3-(3-(4-(1-Aminocyclobutyl)phenyl)-5-phenyl-3H-imidazo[4,5-b]pyridin-2-yl)pyridin-2-amine (ARQ 092): an orally bioavailable, selective, and potent allosteric AKT inhibitor. J. Med. Chem. 59 (13), 6455–6469. https://doi.org/10.1021/acs.jmedchem.6b00619 (2016).
Trott, O. & Olson, A. J. AutoDock vina: improving the speed and accuracy of Docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31 (2), 455–461. https://doi.org/10.1002/jcc.21334 (2010).
Banerjee, P., Kemmler, E., Dunkel, M. & Preissner, R. ProTox 3.0: a webserver for the prediction of toxicity of chemicals. Nucleic Acids Res. 52(W1), W513-W520 (2024). https://doi.org/10.1093/nar/gkae303
Acknowledgements
The authors wish to acknowledge our colleague, Ewa Dominiczak, for her contribution to the synthesis and purification of the compounds.
Funding
This research was funded by the Polish Ministry of Science and Higher Education under the framework of a Łukasiewicz–Industrial Chemistry Institute statutory project (84133301) and Medical University of Bialystok, grant No. B.SUB.25.364.
Author information
Authors and Affiliations
Contributions
Conceptualization, O.M., M.C., A.G.; methodology, O.M., M.C., A.G.; software, D.M. and B.T.; validation, A.G., N.F. and Y.K.; formal analysis, M.C, O.M., S.Ż., A.G. and M.K.; investigation, O.M., A.G., M.C., N.F., Y.K., S.Ż., P.R. and M.K.; resources, O.M., B.T. and A.G.; data curation, D.M., B.T., A.G. and M.K.; writing-original draft preparation, O.M., M.C., A.G., P.R. and B.T.; writing-review and editing O.M, M.C., A.G. and B.T.; visualization, D.M., B.T., P.R. and A.G.; supervision, O.M., A.B. and B.T.; project administration, O.M, M.C. and A.G.; funding acquisition O.M., M.C. and A.G. All authors have read and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Michalak, O., Cybulski, M., Kubiszewski, M. et al. Uracil derivatives/ursolic acid hybrids - naturally derived compounds as anticancer agents. Sci Rep 15, 28803 (2025). https://doi.org/10.1038/s41598-025-14351-y
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-14351-y