Abstract
Non-small cell lung cancer (NSCLC) accounts for approximately 85% of all lung cancer cases and has gained considerable global attention. Epidermal growth factor receptor (EGFR) plays a key role in NSCLC treatment. NSCLC with wild-type EGFR (WT-EGFR) is not responsive to EGFR tyrosine kinase inhibitors (TKIs), and chemotherapy, such as cisplatin (cDDP), continues to be utilized as a primary clinical treatment. However, the therapeutic efficacy of cDDP remains limited. Therefore, there exists an urgent need for the development of novel therapeutic strategies. Cudratricusxanthone A (CTXA) as a natural bioactive xanthone exhibits potential antitumor activity, but the mechanism has been unknown. In this study, we aimed to evaluate the potential antitumor effect and mechanism of CTXA against NSCLC with WT-EGFR by cell experiments, molecular dynamics simulation and surface plasmon resonance (SPR) technology. Our results showed that CTXA inhibited cell proliferation in NSCLC cells by suppressing the EGFR/Erk/AKT pathway. Additionally, CTXA exhibited antitumor effects by inhibiting the migration of A549 cells, causing G1 phase arrest, and inducing cell apoptosis. Further investigation revealed that these effects were mediated by the binding of CTXA to EGFR, with a KD value of 6.302 × 10− 5 M. This interaction may lead to the suppression of EGFR phosphorylation and its downstream signaling. Moreover, the synergistic inhibition of CTXA in combination with cDDP was also found to be mediated by the suppression of the EGFR pathway and its downstream signaling. We demonstrated that CTXA exhibited antitumor activity and enhanced chemosensitivity to cDDP against NSCLC via the EGFR signaling. Our findings indicate that the utilization of CTXA alone or in combination with cDDP represents a potential novel candidate against NSCLC harboring WT-EGFR.
Similar content being viewed by others
Introduction
As the leading cause of cancer death worldwide, lung cancer is one of the global disease burdens to decreasing human life expectancy, comprising small cell lung cancer (SCLC, accounting for 15% of all lung cancers) and non-small cell lung cancer (NSCLC, accounting for 85% of all lung cancer)1,2,3,4. Epidermal growth factor receptor (EGFR) pathway is a critical signal for targeted treatment of NSCLC. Abnormal activation of EGFR signaling, which is induced by its natural ligand epidermal growth factor (EGF), affects signal transduction in its two main downstream pathways, including RAS/RAF/MEK/Erk and PI3K/AKT5,6. Among them, RAS/RAF/MEK/Erk is known to be associated with cell proliferation, migration and differentiation, and PI3K/AKT is involved in regulation of survival, cell cycle and apoptosis7,8,9. Hence, due to its multifunctional role, EGFR is regarded as an important candidate in tumorigenesis10. The advent of EGFR tyrosine kinase inhibitors (EGFR-TKIs) has successfully ameliorated the predicament faced by patients with NSCLC harboring EGFR mutations11,12,13. However, due to the absence of targetable mutations in tumors with wild-type EGFR (WT-EGFR), the efficacy of EGFR-TKIs is comparatively limited in cases with WT-EGFR, which constitutes the majority of EGFR-positive tumors overall14. An Iressa Pan-Asia Study showed that the objective response rate of gefitinib was 71.2% in patients with EGFR mutations and 1.1% in patients with WT-EGFR15. Therefore, as one of systemic therapeutic strategies, cisplatin-based chemotherapy has still been employed as the primary clinical intervention for lung cancer16. Unfortunately, NSCLC exhibits limited responsiveness to chemotherapy, and patients are subjected to a multitude of severe toxicities, including renal injury, during the course of treatment17,18. Numerous studies reported that RAS/RAF/MEK/Erk and PI3K/AKT signaling is related to both chemoresistance and chemosensitivity to cisplatin (cDDP)19,20,21. Thus, more novel therapeutic strategies need to urgently be developed for NSCLC harboring WT-EGFR.
There are several types of plants in China, including dietary and pharmaceutical sources. Natural bioactive compounds derived from plants have attracted considerable attention in recent years because of their positive efficacy in cancer treatment22,23,24,25,26. Cudratricusxanthone A (CTXA), a natural xanthone compound, is extracted from the roots of Cudrania tricuspidata Bur., which is also used in traditional Chinese medicine27,28,29. In China, the root extracts of Cudrania tricuspidata Bur. were utilized to produce an antitumor drug named Zhemu Syrup (Z31020484) for the adjuvant treatment of gastric and intestinal cancer. In addition, it has also been reported to inhibit metastasis and induce apoptosis in breast cancer cells30. However, the potential anti-lung cancer efficacy of CTXA has not been reported and warrants investigation.
In this study, we verified that CTXA inhibited cell proliferation and migration, caused cell cycle arrest, induced apoptosis, and enhanced the sensitivity to chemotherapy on NSCLC cells with WT-EGFR. More importantly, we found that the mechanism of the resulting efficacy was involving in targeting EGFR.
Materials and methods
Materials
A549 and NCI-H441 cell lines were obtained from Cell Bank of Kunming (Kunming, China). Cudratricusxanthone A (CTXA) (> 96.5% purity) was obtained from BioBioPha Ltd. (Kunming, China). Cisplatin (cDDP) (> 98.5% purity) was purchased from Meilun Bio Ltd. (Dalian, China). Dimethyl sulfoxide (DMSO), which was purchased from VWR Life Science (Atlanta, United States), was used to dissolve CTXA and cDDP and then stored at − 20 °C. 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) was purchased from Sigma-Aldrich (St. Louis, United States). All primary antibodies were purchased from Cell Signaling Technology (Danvers, United States). All secondary antibodies were purchased from Thermo Fisher Scientific (Waltham, United States).
Cell culture
A549 and NCI-H441 cells were cultured in RPMI 1640 medium (MeilunBio, Dalian, China) supplemented with 10% fetal bovine serum (HyClone, CA, United States), 10 U/ml of penicillin and 0.1 mg/ml streptomycin (MeilunBio). Then, cells were maintained in a humidified incubator with 5% CO2 at 37 °C. The culture medium was replaced after approximately two days.
Cell proliferation assay
A549 or NCI-H441 cells were seeded in 96-well plates at a density of 1 × 104 cells per well and then treated with CTXA alone, cDDP alone or CTXA combined with cDDP for 24 h, respectively. MTT was then added to the cells for another 4 h. After removal of the culture medium, the remaining MTT formazan crystals were dissolved with 200 µL Dimethylsulfoxide (DMSO) and measured at 492 nm using a microplate reader. Experiments were carried out in triplicate, and the IC50 (the concentration of drug that inhibits cell growth by 50%) values were determined.
Colony formation assay
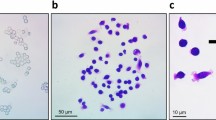
A549 cells were seeded in 60-mm plates at a density of 1 × 103 cells per plate. Cells were treated with the designated concentrations of CTXA (3, 6 or 9 µM) for 72 h. The culture medium was changed every 3 days, and the process lasted for an additional 11 days. Subsequently, the cells were fixed with paraformaldehyde and stained with a 0.01% crystal violet solution for 30 min. After capturing the images, the cells were dissolved in 10% glacial acetic acid and the absorbance was measured at 560 nm using a Multifunctional Microplate Reader.
Western blotting
A549 cells were plated in 60 mm plates at a density of 2 × 106 cells per plate and treated with CTXA, cDDP, or cDDP plus CTXA for 1 h. Total protein was extracted using RIPA buffer and PMSF (Solarbio, Beijing, China) at a ratio of 100:1. Equal amounts of protein samples were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to PVDF membranes. After the blockage of the membranes with 5% non-fat milk in TBS containing 0.1% Tween-20 for 1 h at room temperature, the membranes were incubated with primary antibodies overnight at 4 °C, and then incubated with secondary antibodies for 1 h at room temperature. Thereafter, the membranes were analyzed using a FluorChem E System (Santa Clara, CA, United States). The expression levels of proteins were quantificated by using ImageJ software version 1.52. The ratios of p-EGFR, p-Erk, and p-AKT expression relative to their respective total protein expression were quantified, while the expression levels of CyclinD1, CDK4, cleaved-caspase 9, Bcl-2, Bax, cleaved-caspase 3, and cleaved-PARP1 relative to the internal control were quantified.
Cell apoptosis assay
Cell apoptosis was analyzed by flow cytometry using an Annexin V-PE/7-AAD apoptosis detection kit (MeilunBio, Dalian, China). A549 cells were cultured in 60-mm culture plates at a density of 1 × 106 cells per plate. The culture medium was replaced with serum-free medium containing the indicated doses of CTXA (3, 6 or 9 µM). After treatment for 24 h, cells were stained with Annexin V-PE (5 µL) and 7-AAD (10 µL) for 15 min at room temperature in the dark. Finally, 1×binding buffer (400 µL) was added and the mixture was evenly mixed and detected using a flow cytometer (BD FACS Jazz, BD Biosciences, CA, United States).
Cell cycle assay
Cell cycle was assessed using a cell cycle detection kit (4 A Biotech, Beijing, China). A549 cells were seeded at a density of 2 × 106 cells/well and treated with the indicated doses of CTXA (3, 6 or 9 µM) for 24 h, and subsequently fixed with 75% chilled ethanol at − 20 °C for 24 h. Afterwards, cells were washed twice with PBS and then stained with a solution mixed with binding buffer (400 µL), 15 µL propidium iodide (PI) and 4 µL RNase A (2.5 mg/mL) for 30 min at 37 °C in the dark. The samples were analyzed using a flow cytometer (BD FACSCalibur, BD Biosciences, CA, USA).
Wound healing assay
The A549 cells were incubated at a density of 2 × 106 cells/well. A wound was created by scratching the plate with the tip. The cells were washed three times to remove any floating cell debris and then treated with various doses of CTXA (3, 6 or 9 µM). Images were acquired at 0, 6 and 24 h by using an Olympus biomicroscope (CKX41).
Molecular Docking and molecular dynamic simulation
The crystal structure of the EGFR extracellular domain (PDB code: 3NJP) was obtained from the Protein Data Bank (http://www.rcsb.org) and used as the receptor for docking. AutoDock Tools version 1.56 was used for griding and docking. The docking parameters were set to default values, except that the number of GA runs was set to 20 and the maximum number of events (medium) was set to 5,000,000. The best docking pose with the lowest binding energy was finally selected, and the receptor-ligand interaction was used for further analysis using the DiscoveryStudio visualizer and PyMol software.
The optimal docking model obtained from AutoDock Tools for the complex of CTXA combined with EGFR was utilized for molecular dynamics simulation on GROMACS-2020.6 package. The CHARMM36 force field and the simple point charge (SPC) water model was selected to assign CTXA and EGFR, respectively. Subsequently, the surface charges for the complex of CTXA and EGFR was configured by adding counterions (Na+ or Cl−). Subsequently, energy minimization was performed with 5000 cycles to relax all atoms, and then the minimized system was gradually heated under a normal volume and temperature (NVT) ensemble from 0 to 300 K for 1 ns. This was followed by a density equilibration ensemble at 300 K for 1 ns under normal pressure and temperature (NPT). Finally, the system was run for 30 ns with a time step of 2.0 fs.
Molecular interaction assay
Molecular interaction assays were performed using a Biacore S200 instrument (GE Healthcare, Uppsala, Sweden) based on surface plasmon resonance (SPR) technology. EGFR (30 µg/mL in 10 mM sodium acetate, pH 4.0) was coupled to a CM5 sensor chip using an amine-coupling kit (GE Healthcare, Uppsala, Sweden). Various concentrations of CTXA were injected and passed over the immobilized EGFR sensor surface. The flow rate was 30 mL/min and the binding and dissociation times were 90 s. Kinetic analyses were performed to calculate the affinity using the Biacore S200 Evaluation Software (version 1.1, GE Healthcare).
Statistical analysis
The inhibitory effects of the interaction between CTXA and cDDP were assessed by calculating the combination index (CI) values using the CompuSyn version 1.01 software (ComboSyn, Inc., Paramus, United States). The combined effect of the two agents was analyzed using the median effect analysis method according to the literature31,32. The CI values were considered to indicate whether the combined effect was synergistic (CI < 1), additive (CI = 1), or antagonistic (CI > 1).
Data from all experiments are presented as the mean ± standard error of the mean (± SEM) and were analyzed using GraphPad Prism version 5.0. All experiments were performed independently at least thrice. Statistical significance was set at p < 0.05.
Results
CTXA exhibits inhibitory activity on cell proliferation against A549 cells via EGFR signaling pathway
The chemical structure of CTXA was shown in Fig. 1A. First, we performed the MTT assay to assess the effect of CTXA on cell proliferation in NSCLC cells. As shown in Fig. 1B–E, CTXA inhibited cell viability of NSCLC cells in a dose- and time-dependent manner. When the concentration of CTXA reached 12 µM, it showed maximum antiproliferative activity on cells. The IC50 values of CTXA in A549 cells for 24 h and 48 h were 5.803µM and 3.311 µM, respectively. The IC50 values of CTXA in NCI-H441 cells for 24 h and 48 h were 6.274 µM and 5.472 µM, respectively. Due to the lower values of IC50, A549 cells were used to subsequently perform a colony formation assay, which was used to evaluate the antiproliferative potential of CTXA. As shown in Fig. 1F,G, the clonogenic potential of A549 cells was significantly inhibited by increasing doses of CTXA. These findings provide evidence for the inhibitory effect of CTXA on A549 cells.
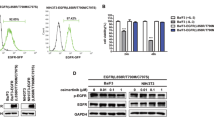
To further determine whether the inhibitory effect of CTXA was mediated by the EGFR signaling pathway, we performed a western blot assay to examine the protein expression levels of several key regulators in A549 cells. The results showed that CTXA significantly suppressed EGF-induced phosphorylation of EGFR and its downstream signaling proteins AKT and Erk after 1 h of treatment, while the total protein expression levels remained nearly constant (Fig. 2A–D). Therefore, we conclude that CTXA effectively restrained cell proliferation in A549 cells by suppressing EGFR phosphorylation and its downstream signaling in a relatively short time.
Chemical structure, and cell proliferation of CTXA treatment on A549 and NCI-H441 cells. (A) The chemical structure of CTXA. (B–E) The cell viability was detected by MTT assay. A549 cells were treated with various doses of CTXA for 24 h (B) or 48 h (C). NCI-H441 cells were treated with various doses of CTXA for 24 h (D) or 48 h (E). (F) Colony formation assay of A549 cells were treated with various doses of CTXA. (G) Quantification of clonogenic formation was analyzed. Data represent the average of three independent experiments (means ± SEM). *** p < 0.001 versus the control.
Effect of CTXA on EGFR signaling pathway in A549 cells. (A) A549 cells were treated with various doses of CTXA for 1 h and then stimulated with EGF (20 ng/ml) for 5 min. (B–D) Quantification of the p-EGFR, p-AKT and p-Erk levels versus their relative total protein levels was analyzed. Data represent the average of three independent experiments (means ± SEM). ###p < 0.001 versus the control; **p < 0.01, ***p < 0.001 versus EGF.
CTXA induces cell cycle arrest in A549 cells
Based on the above results, we further evaluated whether CTXA affected cell cycle distribution in A549 cells. The cells were incubated with the assigned doses of CTXA, stained with propidium iodide (PI), and analyzed using flow cytometry. As shown in Fig. 3A,B, increasing doses of CTXA resulted in an obvious increase in the number of cells in the G1 phase. A subsequent western blot assay was conducted to examine the expression levels of relative proteins, which are key regulators of the G1 phase. We observed that the expression levels of CylinD1 and CDK4 were downregulated in a concentration-dependent manner compared to those in the control (Fig. 3C–E).
Regulation of CTXA on cell cycle distribution in A549 cells. (A) Cells were treated with various doses of CTXA (3, 6 or 9 µM) for 24 h. The cell cycle distribution (G0/G1, S and G2/M) was detected by flow cytomery and analyzed by FlowJo software. (B) Quantification of cell proportions in each phase of the cell cycle were analyzed. (C) Expression levels of Cyclin D1 and CDK4 were examined by Western blot assay. (D,E) Quantification of CyclinD1 and CDK4 expression levels was analyzed. **p < 0.01, ***p < 0.001 versus the control.
CTXA inhibits migration and induce apoptosis against A549 cells
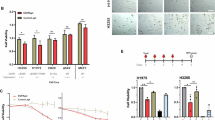
We treated A549 cells with 3, 6 or 9 µM CTXA for 6–24 h using a wound healing assay. As shown in Fig. 4A,B, the migration capacity of A549 cells was inhibited by CTXA in a concentration-dependent manner at 6 h and 24 h. In addition, we treated A549 cells with various concentrations of CTXA (3, 6, 9 µM) for 24 h, and then subjected them to Annexin V-PE and 7-AAD double staining to test the apoptotic cells by using flow cytometry. As shown in Fig. 4C,D, compared to the control group, the rate of apoptosis was significantly increased in a concentration-dependent manner with respect to CTXA. We also assessed the expression levels of apoptotic markers using western blotting. The results shown in Fig. 4E–J indicate that the expression levels of cleaved-caspase9, Bcl-2, Bax, cleaved-caspase3 and cleaved-PARP1 showed a significant increase with increasing doses of CTXA. Together, the above results indicates that CTXA exerts its anti-lung cancer effects on A549 cells by inhibiting migration and promoting apoptosis.
CTXA inhibited cell migration and induced apoptosis in A549 cells. (A) Scratches were created and then A549 cells were treated with assigned concentrations of CTXA (3, 6 or 9 µM) for 6–24 h. (B) Quantification of the ratio of the recovered wound area was analyzed. (C) Cells were treated with assigned doses of CTXA (3, 6 or 9 µM) for 24 h, and then analyzed by flow cytometry. (D) The proportions of apoptotic cells were expressed as percentages. (E) The expression levels of relative apoptotic proteins were determined by using Western blot. (F–J) Quantification of cleaved-caspase9, Bcl-2, Bax, cleaved-caspase3 and cleaved-PARP1 expression levels was analyzed. *p < 0.05, **p < 0.01, ***p < 0.001 versus the control.
CTXA directly binds to EGFR extracellular domain
To investigate the binding mode of CTXA interacting with the EGFR, molecular docking simulation was implemented. As shown in Fig. 5A–C, CTXA was docked in domain II of EGFR extracellular domain, which is an important region for EGFR dimerization, forming two hydrogen bonds with the amino acid residues of GLY-259 and TYR-261. The value of docking energy is − 4.59 kcal/mol.
The binding model between CTXA and EGFR. (A) Binding mode of CTXA to EGFR extracellular domain (PDB ID: 3NJP) was constructed by molecular docking. Domain I, II, III and IV of EGFR extracellular domain and EGF were respectively depicted by yellow, green, blue, pink and red cartoon. (B) The details of binding between CTXA and EGFR extracellular domain was analyzed. Key residues were displayed as sticks and colored by element type (C, green; O, red; N, blue; polar H, white.), whereas hydrogen bonds were shown as yellow dash lines. (C) 2D model of CTXA interaction of EGFR extracellular domain was shown.
Subsequently, to access the stability of the predicted binding structure for CTXA interacting with EGFR extracellular domain, the root means square deviation (RMSD) analysis was performed by using molecular dynamic simulation. The results of Fig. 6A showed that the overall RMSD values for the complex of CTXA and EGFR extracellular domain stabilized no fluctuations, compared with free EGFR, which were observed with less than 0.2 nm deviations. The overall radius of gyration (Rg) values of the complex of CTXA-EGFR decreased compared to those of free EGFR, suggesting the complex of CTXA-EGFR formed a more compact structure (Fig. 6B). In addition, the SASA values of free EGFR and the complex of CTXA-EGFR were further analyzed, indicating that the interaction between CTXA and EGFR became more stability (Fig. 6C). To better analyze the values of root means square fluctuation (RMSF), the trajectory during 20 ns to 30 ns was extracted, revealing a similar overall distribution in both free EGFR and the complex of CTXA-EGFR (Fig. 6D). As observed in Fig. 6E, hydrogen bonds, playing a crucial role in the interaction between ligand and receptor, gradually formed, with the number of bonds ranging from 1 to 3 during the 30 ns simulation. These results indicated that our predicted model of CTXA interacted with EGFR extracellular domain is credible. Hence, to further verify the interaction between CTXA and EGFR extracellular domain, we performed a molecular interaction assay using surface plasmon resonance (SPR) technology. It was observed that CTXA directly bound to the EGFR extracellular domain with a KD value of 6.302 × 10− 5 M (Fig. 6F).
Molecular dynamics simulation and molecular interaction analysis of CTXA interacted with EGFR. (A) The RMSD analysis for free EGFR and the complex of CTXA and EGFR was shown. (B) The Rg analysis for free EGFR and the complex of CTXA and EGFR was observed. (C) The SASA analysis for free EGFR and the complex of CTXA and EGFR was observed. (D) The RMSF analysis for free EGFR and the complex of CTXA and EGFR was observed. (E) The number of hydrogen bonds between CTXA and EGFR extracellular domain was depicted during 30 ns. (F) Interaction between CTXA and EGFR extracellular domain was assessed by SPR technology.
CTXA enhances chemosensitivity to cisplatin in A549 cells
Platinum-based chemotherapy, including cisplatin (cDDP), is still employed as a primary clinical intervention for NSCLC harboring WT-EGFR, whereas the therapeutic effect of chemotherapy remains unsatisfactory. Based on this, we evaluated whether CTXA could cause an increase on effect of cDDP aginst A549 cells. As shown in Fig. 7AA, 3–15 µM of cDDP did not affect the viability of A549 cells. After treated with CTXA (3 µM) and cDDP (12 or 15 µM), the combinations of CTXA and cDDP were found to significantly inhibit the viability of A549 cells compared to CTXA or cDDP alone (Fig. 7B,C). We subsequently utilized the CompuSyn software to analyze the combination index (CI) values of the two agents, and it was found that the combination of CTXA and cDDP exerted synergistic inhibitory effects with CI < 1 (Table 1).
To confirm whether the synergistic mechanism might be derived from the EGFR signaling pathway, we conducted a western blot assay to determine the expression levels of key proteins after treatment with 3 µM of CTXA and 15 µM of cDDP, or a combination of the two agents for 1 h. As shown in Fig. 7D–G, CTXA combined with cDDP significantly suppressed the phosphorylation of EGFR, Erk and AKT, compared to CTXA or cDDP alone. These results indicated that CTXA could increase the sensitivity of A549 cells to cDDP, and the synergistic effect might be mediated by the EGFR signaling pathway.
The synergistic inhibition of CTXA combined with cDDP on cell growth of A549 cells. (A) A549 cells were treated with various doses of cDDP. (B) A549 cells were incubated with CTXA (3 µM) and cDDP (12 µM) in combination or individually for 24 h. (C) A549 cells were incubated with CTXA (3 µM) and cDDP (15 µM) in combination or individually for 24 h. (D) A549 cells were incubated with CTXA (3 µM) and cDDP (15 µM) in combination or individually for 1 h, and then expression levels of EGFR pathway were detected by Western blot. (E–G) Quantification of the p-EGFR, p-Erk and p-AKT levels versus their relative total protein levels was analyzed. Data represent the average of three independent experiments (means ± SEM). ##p < 0.01, ###p < 0.001 versus the control; ***p < 0.001 versus EGF.
Discussion
NSCLC has become a major obstacle to increasing human health because of the high mortality. Compared to NSCLC harboring EGFR-activating mutations, tumors harboring WT-EGFR are comparatively insensitive to EGFR-TKIs14. As a conventional therapy strategy, platinum-based chemotherapy, including cDDP, remains the first-line treatment for most patients with advanced NSCLC, but its efficacy is limited by inherent or acquired resistance18,33,34. Natural bioactive compounds extracted from plant resources have attracted considerable attention because of their structures and potential anticancer activities. However, these agents are difficult to utilize and develop until the molecular mechanism is understood8,35. Among these agents, CTXA is a natural xanthone derived from the roots of Cudrania tricuspidata Bur. with potential antitumor effects30. However, the activity and mechanism of CTXA against NSCLC was limited by sufficient evidences to support. Therefore, in this study, we investigated the antitumor effects of CTXA in NSCLC with WT-EGFR and elucidated the underlying mechanism of the resulting efficacy. Our findings provide a theoretical basis for the design and development of novel drugs.
EGFR, as a key signaling molecule for targeted treatment of NSCLC, is a transmembrane protein that is mediated by binding with its natural ligand EGF. Usually, phosphorylation of EGFR mediated by EGF induced RAS/RAF/MEK/ERK and PI3K/AKT5,6. The two main downstream signaling pathways of EGFR play critical roles in the regulation of tumor occurrence, proliferation, migration, cell cycle and apoptosis for NSCLC8,36. In this study, CTXA was found to inhibit cell proliferation by suppressing EGF-induced phosphorylation of EGFR, Erk and AKT. Therefore, to further confirm the anti-tumor efficacy and mechanism of CTXA, we investigated the effect of CTXA on cell cycle, migration and apoptosis in A549 cells. To some extent, CTXA also exhibited a pro-apoptotic effect. The apoptotic ratio, which was associated with the expression levels of apoptotic proteins, showed a significant increase with increasing doses of CTXA. However, the pro-apoptotic effect of CTXA is relatively weaker than its anti-proliferatie effect. This suggests that CTXA may primarily inhibit cell proliferation to exert its antitumor efficacy. Furthermore, CTXA could inhibit cell migration, induce G1-phase arrest by modulating CyclinD1 and CDK4. These finding proved that CTXA exerts an anti-tumor activity against NSCLC harboring WT-EGFR.
The activation of EGFR begins with the binding of EGF to the extracellular domain of EGFR37,38. Binding of EGF to the extracellular domain of EGFR facilitates the formation of EGFR dimers. Dimerization triggers activation of the EGFR kinase domain, leading to EGFR phosphorylation and signal transduction in downstream pathway39,40. In this study, it was observed that CTXA rapidly suppressed EGFR phosphorylation. This finding suggests that CTXA may directly interact with EGFR, rather than exert its effects through transcriptional regulation. Based on it, we performed a molecular docking simulation and found that CTXA might bound to the EGFR extracellular domain. Then, the RMSD analysis calculated by molecular dynamic simulation was utilized to access the stability for the binding structure of CTXA interacted with the EGFR extracellular domain. The results indicated that the predicted complex model is credible. Subsequently, the results of the molecular interaction by SPR technology further suggested that CTXA directly interacts with the EGFR extracellular domain, exhibiting rapid binding and dissociation kinetics. Despite the relatively high KD value compared to the IC50 value, the interaction exhibited rapid binding kinetics, suggesting that the binding of CTXA to the extracellular domain of EGFR is rapid and direct. Collectively, as shown in Fig. 8, this interaction between CTXA and EGFR might suppress the phosphorylation of EGFR and its downstream pathways, ultimately exerting antitumor effects.
Due to the limitations of EGFR-TKIs, platinum-based chemotherapy, including cDDP, has been used as one of the primary treatments for most advanced NSCLC patients. However, the efficacy of platinum-based chemotherapy is not always satisfactory. It has been reported that Erk may be positively correlated with multidrug resistance in many cancers41,42. Numerous studies have suggested that downregulation of Erk signaling may reverse drug resistance and enhance sensitivity to cDDP in NSCLC cells19,20. Furthermore, we found that (−)-Epigallocatechin-3-gallate derivatives in combination with cDDP exhibited synergistic inhibition in NSCLC cells by targeting EGFR in our previous studies17,43. Therefore, in this study, we evaluated whether CTXA may enhance chemosensitivity to cDDP in A549 cells through the EGFR/Erk pathway. These results suggest that CTXA combined with cDDP exerts synergistic inhibitory effects on A549 cells via suppression of p-EGFR and p-Erk. However, the above results were based on an in vitro study and there is no evidence that CTXA could also exert similar effects in vivo. A more detailed in vivo study elucidating the mechanism of action is ongoing, and will be reported in due course.
Conclusions
In summary, our study, for the first time, demonstrated that CTXA exerts its antitumor effect and enhances chemosensitivity to cDDP in vitro by binding to the EGFR extracellular domain and suppressing EGFR and its downstream signaling. These findings will provide a scientific basis for further study and development of novel treatments for NSCLC harboring WT-EGFR.
Data availability
All data generated or analyzed during this study are included in this manuscript.
References
Siegel, R. L., Miller, K. D., Wagle, N. S. & Jemal, A. Cancer statistics, 2023. Cancer J. Clin. 73 (1), 17–48. https://doi.org/10.3322/caac.21763 (2023).
Siegel, R. L., Miller, K. D., Fuchs, H. E. & Jemal, A. Cancer statistics, 2021. Cancer J. Clin. 71 (1), 7–33. https://doi.org/10.3322/caac.21654 (2021).
Chen, Z., Fillmore, C. M., Hammerman, P. S., Kim, C. F. & Wong, K. K. Non-small-cell lung cancers: a heterogeneous set of diseases. Nat. Rev. Cancer. 14 (8), 535–546. https://doi.org/10.1038/nrc3775 (2014).
Siegel, R. L., Miller, K. D. & Jemal, A. Cancer statistics, 2019. Cancer J. Clin. 69 (1), 7–34. https://doi.org/10.3322/caac.21551 (2019).
Marmor, M. D., Skaria, K. B. & Yarden, Y. Signal transduction and oncogenesis by erbb/her receptors. Int. J. Radiat. Oncol. Biol. Phys. 58 (3), 903–913. https://doi.org/10.1016/j.ijrobp.2003.06.002 (2004).
Reungwetwattana,T., Weroha, S. J. & Molina, J. R. Oncogenic pathways, molecularly targeted therapies, and highlighted clinical trials in non-small-cell lung cancer (NSCLC). Clin. Lung Cancer. 13 (4), 252–266. https://doi.org/10.1016/j.cllc.2011.09.004 (2012).
Schlessinger, J. Receptor tyrosine kinases: legacy of the first two decades. Cold Spring Harb. Perspect. Biol. 6 (3), a008912. https://doi.org/10.1101/cshperspect.a008912 (2014).
Kang, D. Y. et al. The inhibitory mechanisms of tumor PD-L1 expression by natural bioactive Gallic acid in Non-Small-Cell lung cancer (NSCLC) cells. Cancers. 12 (3), 727. https://doi.org/10.3390/cancers12030727 (2020).
Ellert-Miklaszewska, A., Kaminska, B. & Konarska, L. Cannabinoids down-regulate PI3K/Akt and Erk signalling pathways and activate proapoptotic function of bad protein. Cell. Signal. 17 (1), 25–37. https://doi.org/10.1016/j.cellsig.2004.05.011 (2005).
Sigismund, S., Avanzato, D. & Lanzetti, L. Emerging functions of the EGFR in cancer. Mol. Oncol. 12 (1), 3–20. https://doi.org/10.1002/1878-0261.12155 (2018).
Cohen, M. H., Williams, G. A., Sridhara, R., Chen, G. & Pazdur, R. FDA drug approval summary: gefitinib (ZD1839) (Iressa) tablets. Oncologist. 8 (4), 303–306. https://doi.org/10.1634/theOncologist.8-4-303 (2003).
Herbst, R. S., Morgensztern, D. & Boshoff, C. The biology and management of non-small cell lung cancer. Nature. 553 (7689), 446–454. https://doi.org/10.1038/nature25183 (2018).
Zhou, C. et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): a multicentre, open-label, randomised, phase 3 study. Lancet Oncol. 12 (8), 735–742. https://doi.org/10.1016/S1470-2045(11)70184-X (2011).
Cuan, X. D. et al. Antitumor effects of erlotinib in combination with Berberine in A431 cells. BMC Pharmacol. Toxicol. 24 (1), 29 (2023).
Mok, T. S. et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N. Engl. J. Med. 361 (10), 947–957. https://doi.org/10.1056/NEJMoa0810699 (2009).
Ghosh, S. Cisplatin: The first metal based anticancer drug. Bioorg. Chem. 88, 102925. https://doi.org/10.1016/j.bioorg.2019.102925 (2019).
Wang, J. et al. (-)-Epigallocatechin-3-gallate derivatives combined with cisplatin exhibit synergistic inhibitory effects on non-small-cell lung cancer cells. Cancer Cell Int. 19, 266. https://doi.org/10.1186/s12935-019-0981-0 (2019).
Kryczka, J., Kryczka, J., Czarnecka-Chrebelska, K. H. & Brzeziańska-Lasota, E. Molecular mechanisms of chemoresistance induced by cisplatin in NSCLC cancer therapy. Int. J. Mol. Sci. 22 (16), 8885. https://doi.org/10.3390/ijms22168885 (2021).
Liu, Z. L. et al. Apatinib resensitizes cisplatin-resistant non-small cell lung carcinoma A549 cell through reversing multidrug resistance and suppressing ERK signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 21 (23), 5370–5377. https://doi.org/10.26355/eurrev_201712_13922 (2017).
Chai, D. et al. Total saponins from Paris Forrestii reverse multidrug resistance of MCF-7/ADM cells by suppression of P-gp via ERK signaling pathway. Biol. Pharm. Bull. 43 (12), 1823–1830. https://doi.org/10.1248/bpb.b20-00014 (2020).
Yu, M. L., Qi, B. Q., Wu, X. X., Xu, J. & Liu, X. L. Baicalein increases cisplatin sensitivity of A549 lung adenocarcinoma cells via PI3K/Akt/NF-κB pathway. Biomed. Pharmacother. 90, 677–685. https://doi.org/10.1016/j.biopha.2017.04.001 (2017).
Majolo, F., Knabben, D. O. B. D. L., Marmitt, D. J., Bustamante-Filho, I. C. & Goettert, M. I. Medicinal plants and bioactive natural compounds for cancer treatment: important advances for drug discovery. Phytochem. Lett. 31, 196–207. https://doi.org/10.1016/j.phytol.2019.04.003 (2019).
Jung, S. K. et al. Butein, a novel dual inhibitor of MET and EGFR, overcomes gefitinib-resistant lung cancer growth. Mol. Carcinog. 54 (4), 322–331. https://doi.org/10.1002/mc.22191 (2015).
Li, J. et al. Pristimerin induces apoptosis and inhibits proliferation, migration in H1299 lung cancer cells. J. Cancer. 11 (21), 6348–6355. https://doi.org/10.7150/jca.44431 (2020).
Kumar, S. et al. The anticancer potential of flavonoids isolated from the stem bark of erythrina suberosa through induction of apoptosis and Inhibition of STAT signaling pathway in human leukemia HL-60 cells. Chemico-Biol. Interact. 205 (2), 128–137. https://doi.org/10.1016/j.cbi.2013.06.020 (2013).
Xu, S. et al. A novel potent anticancer compound optimized from a natural Oridonin scaffold induces apoptosis and cell cycle arrest through the mitochondrial pathway. J. Med. Chem. 60 (4), 1449–1468. https://doi.org/10.1021/acs.jmedchem.6b01652 (2017).
Lee, B. W., Gal, S. W., Park, K. M. & Park, K. H. Cytotoxic Xanthones from cudrania tricuspidata. J. Nat. Products. 68 (3), 456–458. https://doi.org/10.1021/np030481a (2005).
Yoon, C. S. et al. A prenylated xanthone, cudratricusxanthone A, isolated from cudrania tricuspidata inhibits Lipopolysaccharide-Induced neuroinflammation through Inhibition of NF-κB and p38 MAPK pathways in BV2 microglia. Molecules (Basel Switzerland). 21 (9), 1240. https://doi.org/10.3390/molecules21091240 (2016).
Kim, T. J. et al. Cudratricusxanthone A isolated from the root bark of cudrania tricuspidata inhibits the proliferation of vascular smooth muscle cells through the suppression of PDGF-receptor beta tyrosine kinase. Biol. Pharm. Bull. 30 (4), 805–809. https://doi.org/10.1248/bpb.30.805 (2007).
Jeon, S. M., Lee, D. S. & Jeong, G. S. Cudraticusxanthone A isolated from the roots of cudrania tricuspidata inhibits metastasis and induces apoptosis in breast cancer cells. J. Ethnopharmacol. 194, 57–62. https://doi.org/10.1016/j.jep.2016.08.042 (2016).
Chou, T. C. & Talalay, P. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv. Enzyme Regul. 22, 27–55. https://doi.org/10.1016/0065-2571(84)90007-4 (1984).
Shen, Y., Du, Y., Zhang, Y. & Pan, Y. Synergistic effects of combined treatment with Simvastatin and exemestane on MCF-7 human breast cancer cells. Mol. Med. Rep. 12 (1), 456–462. https://doi.org/10.3892/mmr.2015.3406 (2015).
Galluzzi, L. et al. Molecular mechanisms of cisplatin resistance. Oncogene. 31 (15), 1869–1883. https://doi.org/10.1038/onc.2011.384 (2012).
Yang, L. & Wang, Z. Natural products, alone or in combination with FDA-Approved drugs, to treat COVID-19 and lung cancer. Biomedicines 9 (6), 689. https://doi.org/10.3390/biomedicines9060689 (2021).
Sun, P. et al. Wighteone exhibits an antitumor effect against EGFR L858R/T790M mutation non-small cell lung cancer. J. Cancer. 12 (13), 3900–3908. https://doi.org/10.7150/jca.54574 (2021).
Johnson, G. L. & Lapadat, R. Mitogen-activated protein kinase pathways mediated by ERK, JNK, and p38 protein kinases. Science. 298 (5600), 1911–1912. https://doi.org/10.1126/science.1072682 (2002).
Berger, C., Krengel, U., Stang, E., Moreno, E. & Madshus, I. H. Nimotuzumab and cetuximab block ligand-independent EGF receptor signaling efficiently at different concentrations. J. Immunother. 34 (7), 550–555. https://doi.org/10.1097/CJI.0b013e31822a5ca6 (2011).
Yarden, Y. & Schlessinger, J. Epidermal growth factor induces rapid, reversible aggregation of the purified epidermal growth factor receptor. Biochemistry. 26 (5), 1443–1451. https://doi.org/10.1021/bi00379a035 (1987).
Gadella, T. W. Jr & Jovin, T. M. Oligomerization of epidermal growth factor receptors on A431 cells studied by time-resolved fluorescence imaging microscopy. A stereochemical model for tyrosine kinase receptor activation. J. Cell Biol. 129 (6), 1543–1558. https://doi.org/10.1083/jcb.129.6.1543 (1995).
Sun, P. et al. Coccinic acid exhibits anti-tumor efficacy against NSCLC harboring EGFR L858R/T790M mutation via the EGFR/STAT3 pathway. Bioorg. Chem. 154, 108038. https://doi.org/10.1016/j.bioorg.2024.108038 (2024).
Brozovic, A. & Osmak, M. Activation of mitogen-activated protein kinases by cisplatin and their role in cisplatin-resistance. Cancer Lett. 251 (1), 1–16. https://doi.org/10.1016/j.canlet.2006.10.007 (2007).
He, Y. et al. PGK1 contributes to tumorigenesis and Sorafenib resistance of renal clear cell carcinoma via activating CXCR4/ERK signaling pathway and accelerating Glycolysis. Cell Death Dis. 13 (2), 118. https://doi.org/10.1038/s41419-022-04576-4 (2022).
Zi, C. T. et al. Synthesis of (-)-epigallocatechin-3-gallate derivative containing a Triazole ring and combined with cisplatin/paclitaxel inhibits NSCLC cancer cells by decreasing phosphorylation of the EGFR. J. Chem. Res. 44 (9–10), 586–591. https://doi.org/10.1177/1747519820910390 (2020).
Funding
This research was funded by the Development and Technical Services for Homology of Food and Medicine Project (202500043110184), the Project of the Yunnan Province Agricultural Basic Research Joint Foundation (202401BD070001-056) and the Project of Yunnan International Science and Technology Specialists (202403AK140041).
Author information
Authors and Affiliations
Contributions
J.S., J.W. and P.S. designed the study. P.S., Z.W., R.Z., B.X. and R.S. performed experiments. Z.W., P.S., R.Z. and X.L. analyzed the data. P.S., Z.W. and J.W. wrote the manuscript. J.S., J.W. and P.S. reviewed the manuscript. All authors reviewed and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Sun, P., Wang, Z., Zhang, R. et al. Cudratricusxanthone A exhibits antitumor activity and enhances chemosensitivity to cisplatin against NSCLC via targeting EGFR. Sci Rep 15, 29425 (2025). https://doi.org/10.1038/s41598-025-14889-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-14889-x