Abstract
Ischemia-reperfusion injury (IRI) is one of the leading causes of acute kidney injury (AKI), predisposing patients to chronic kidney disease (CKD) due to maladaptive renal repair. Nevertheless, the molecular mechanisms and biomarkers that cause maladaptive repair remain unclear. In this study, we used single-nucleus RNA sequencing data from GEO database (GSE139107) to identify molecular markers during the transition from AKI to CKD caused by IRI. Analysis of intercellular crosstalk, trajectory and machine learning algorithms revealed hub cell clusters and genes. Proximal tubule (PT) cells, especially a new cluster (New PT2), significantly interacted with fibroblasts during the transition. The expression levels of hub genes were validated using the bulk RNA-seq data (GSE98622) and further confirmed through RT-qPCR and immunohistochemical analysis in ischemia-reperfusion injury (uIRI) mice. Ankrd1, a hub gene in New PT2, showed sustained upregulation in the proximal tubule in AKI. Compared to the sham-operated group, the expression of Ankrd1 in mice increased at 0.5 days post-reperfusion, peaked at day 1, and remained significantly elevated up to 60 days. This study indicated that the upregulation of Ankrd1 was positively associated with the progression from AKI to CKD and may potentially serve as a valuable biomarker for this transitional process.
Similar content being viewed by others
Introduction
Acute kidney injury (AKI) is a serious health risk characterized by a rapid decrease in glomerular filtration rate (GFR) and a sharp decline in renal function, making it a major contributor to chronic kidney disease (CKD) and end-stage renal failure1,2,3. Meanwhile, the emergence of CKD presents a formidable global public health challenge, affecting about 13.4% of people worldwide4,5. Prolonged pathological processes, including fibrosis, inflammation, and glomerulosclerosis in CKD, contribute to an incessant decline in renal function. Recent studies have shown that CKD can increase the risk of AKI, and AKI can also make CKD worse6. Understanding the relationship between these two conditions is important for improving patient care and preventing further kidney damage.
Ischemia-reperfusion injury (IRI) is among the most prevalent causes of AKI. Upon the initiation of IRI, there is a transient interruption in renal blood flow, followed by reperfusion. This disruption and subsequent restoration of perfusion initiate a cascade of pathophysiological changes, including cellular impairment, oxidative stress, inflammatory responses, and fibrotic processes7. In IRI, the kidney initiates a reparative process aimed at restoring normal morphology and function. However, in certain cases, maladaptive repair can lead to irreversible damage and progressive fibrosis, ultimately resulting in CKD8,9. Proximal tubule injury is a crucial characteristic of AKI and can trigger its progression into CKD10. In the early stages of AKI, proximal tubular epithelial cells undergo dysfunction, detachment, and apoptosis. Furthermore, these cells may experience G2/M cell cycle arrest while maintaining sustained production of inflammatory mediators and profibrotic cytokines11,12. The research findings have unequivocally demonstrated that tubule epithelial injury exerts a direct influence on the development of interstitial fibrosis and facilitates the accumulation of myofibroblasts through activation of the transforming growth factor-beta 1 (TGF-β1) signaling pathway13,14. These processes cause ongoing stress and a maladaptive repair response, characterized by persistent inflammation, tissue fibrosis, cell death, and aging. All of these contribute to the development of CKD6,15.
The transition from AKI to CKD is a complex process, and its pathophysiology is still poorly understood6. AKI is generally defined as a rapid increase in serum creatinine (SCr) and blood urea nitrogen (BUN) within 48 h, while CKD refers to the persistent elevation of these indicators for more than three months16. However, the molecular mechanisms and biomarkers underlying the transition from AKI to CKD remain unclear. Early studies have suggested that the late upregulation of KIM-1 and NGAL could serve as valuable markers for persistent renal injury in AKI, hypothesizing their association with CKD development17,18. Recent studies reveal an early rise in circulating fibroblast growth factor-23 levels during AKI, which remains elevated in patients with CKD and may act as a prognostic biomarker19. In CKD, associations between upregulated genes such as SOX9 and NKX6-2 and pathological features like tubulointerstitial fibrosis and tubular cell damage have been identified20. However, limited research has focused on dynamic molecular changes within proximal tubule cells during IRI-induced progression from AKI to CKD9.
In this study, we present the findings of snRNA-seq analysis conducted on the GSE139107 dataset. By employing differential expression analysis and machine learning, we have successfully identified two hub genes and investigated their dynamic expression changes during the transition from AKI to CKD with trajectory analysis. Additionally, we validated the renal tissue expression of Ankrd1 through RT-qPCR and IHC analysis (Fig. 1).
Workflow of the study. The boxes represent the study materials and methodologies. The cell clusters and genes identified by the study are indicated in red bold font. IHC, immunohistochemical; RT-qPCR, real-time quantitative polymerase chain reaction; uIRI, unilateral renal ischemia-reperfusion injury.
Results
The clustering and identification of cell types in mouse kidney subjected to IRI
After preprocessing the GSE139107 dataset, we utilized the Harmony function to eliminate the batch effect caused by sample timepoints (Fig. 2A). Following annotation, a total of 21 major cell types were identified in the kidney (Fig. 2B), and their respective proportions and cluster-enriched markers are depicted in Fig. 2C-D. The findings revealed that proximal tubule (PT) cells constituted the largest proportion (Fig. 2C). Notably, distinct cell populations characterized by dedifferentiated renal tubular cells emerged as New PT1, New PT2, New PT1/3 subtypes along with severely injured PT and proliferative PT clusters21. These five clusters exhibited associations with kidney injury and likely played pivotal roles in inducing diverse repair outcomes following ischemia-reperfusion injury21. Compared to the sham group, the New PT1 and New PT1/3 clusters showed increased proportions during the early stages of AKI and CKD, while the New PT2 cluster decreased during AKI but increased during the transition to CKD. The severe injured PT cluster was elevated only in early AKI, while the proliferative PT cluster increased sharply during the acute phase of AKI and then decreased (Fig. 2E). Therefore, severe injured PT cluster primarily participated in response to AKI whereas the other four cell types potentially contributed to AKI-CKD transition processes. Consequently, our subsequent investigations focused more on characterizing New PT1, New PT2, NewPT1/3, and proliferative PT clusters.
The clustering and identification of cell types in mouse kidney subjected to ischemia-reperfusion injury. (A) The distribution of mouse IRI kidney cells in distinct sample timepoints. The data were collected at time points of 4 h, 12 h, 2 days, 14 days, and 6 weeks following ischemia-reperfusion injury (IRI) in mice compared to the Sham control group (with 4 replicates per time point). (B) UMAP plots of all datasets integrated with Harmony. (C) The proportion of cell types. (D) Expression patterns of cluster-enriched markers. (E) Alterations in the proportions of five cell clusters in PT. ATL, thin ascending limb of loop of Henle; CNT, connecting tubule; CTAL, thick ascending limb of loop of Henle in cortex; DCT, distal convoluted tubule; DTL, descending limb of loop of Henle; EC, endothelial cells; Fib, fibroblasts; ICA, type A intercalated cells of collecting duct; ICB, type B intercalated cells of collecting duct; Macro, macrophages; MTAL, thick ascending limb of loop of Henle in medulla; New PT1/2/3, new proximal tubule (PT) clusters 1/2/3; PC, principle cells of collecting duct; Pod, podocytes; PT S1/S2/S3, S1/S2/S3 segment of proximal tubule; Uro, urothelium.
The potential interaction between New PT2 and fibroblasts is revealed through intercellular crosstalk
Following AKI, some patients will experience recovery, while others may progress to renal fibrosis and chronic injury. During this process, the interaction between fibroblasts and proximal tubule cells can potentially induce maladaptive regeneration of injured tubular cells13. The number and intensity of certain signaling pathways were found to be elevated in the AKI and CKD groups compared to the sham group, indicating enhanced intercellular crosstalk (Fig. 3A). Focusing on four types of PT cells, in the sham group, they primarily exhibited autocrine behavior and potentially lacked interaction with fibroblasts. In the AKI group, fibroblasts secreted signals to New PT1 and New PT1/3 cells, but there was no corresponding feedback. Instead, New PT2 communicated with fibroblasts, while PT cells primarily interacted internally with each other. In the CKD group, the fibroblasts directly interacted with New PT2 (Fig. 3B). The signaling molecules of FGF, NCAM, NEGR, LAMININ, PTN, PDGF and COLLAGEN were significantly enriched in the CKD group (Fig. 3C). Fibroblasts were involved in sending and receiving signals, including NEGR, LAMININ and PDGF (Fig. 3D). No interactions were observed between fibroblasts and PT cells in the NEGR signaling pathway during ligand-receptor pair analyses. However, in the LAMININ signaling pathway, the interaction between laminin alpha 2 (LAMa2) and its receptors, integrin alpha V beta 8 (ITGAV & ITGB8) and dystrophin-associated glycoprotein 1 (DAG1) may potentially participate in intercellular crosstalk among New PT1/3 cells, New PT2 cells, and fibroblasts. Furthermore, the interaction between platelet-derived growth factor D (PDGFD) and platelet-derived growth factor receptor β (PDGFRβ) may play a role in the intercellular communication between all relevant PT cells and fibroblasts in the PDGF pathway (Fig. 3E). Moreover, previous research has demonstrated that epithelial cells can contribute to renal inflammation and fibrosis through PDGF signaling13. These findings suggest a potential interaction between New PT2 and fibroblasts during CKD.
Intercellular crosstalk analysis. (A-B) The interactions of various cells(A) and of 5 cell types(A) of concern (New PT1, New PT2, NewPT1/3, proliferative PT and fibroblasts) in sham, AKI (IRI_4h, IRI_12h, IRI_2d) and CKD (IRI_6w) groups. The dots represent different cell types, each depicted in a distinct color. The arrows indicate intercellular interactions, originating as ligands and terminating as receptors. Furthermore, the lines are color-matched to the cell type harboring the ligands. (C) Information flow of enriched signaling pathway in sham, AKI and CKD groups. The bold font indicates a greater proportion within the CKD group. (D) Outgoing and incoming communication patterns of 21 cell types. The cell clusters and signaling pathways of concern are indicated in bold font. (E) Significant ligand-receptor pairs within the LAMININ and PDGF signaling pathway. The terms Lama2 - Dag1 and Lama2 - (Itgav + Itgb8) denote the specific interaction between laminin alpha 2 (LAMa2) and its receptors, dystrophin-associated glycoprotein 1 (DAG1), and integrin alpha V beta 8. The term Pdgfd – Pdgfrb refers to the interaction between platelet-derived growth factor D (PDGFD) and platelet-derived growth factor receptor β (PDGFRβ).
Trajectory analysis revealed different outcomes of the New PT2 cluster in the transition from AKI to CKD
Based on pseudo-time trajectory analysis with state and sample timepoints, we inferred the cellular differentiation routines of the New PT2 cluster during the AKI-CKD transition (Fig. 4A-C). Compared to the sham group, state 5 was more likely to differentiate into PT cells that effectively undergo reparative processes following acute injury, whereas state 1 exhibited a tendency towards PT cell differentiation leading to maladaptive repair. Additionally, states 2 and 3 represented precursor PT cells prior to branching, while state 4 denoted post-injury PT cells. We found that in AKI (State 4), New PT2 cells could differentiate into cells that may then undergo regeneration (State 5) or maladaptive repair (State 1) (Fig. 4D). To search for important factors that induce different differentiation outcomes in New PT2, we performed differential expression analysis using BEAM, Findallmarkers, and Pseudobulk on State 1 and State 5 (as the control). We obtained 172 overlapping genes for subsequent analysis (Fig. 4E). Pseudo-temporal expression dynamics and the four expression patterns of 172 genes showed that cluster 1 was related to nuclear receptors and the regulation of GTPase activity. Cluster 2 was enriched in cell-substrate adhesion and kidney development. Cluster 3 may participate in ameboidal-type cell migration and positive regulation of cell motility. Cluster 4 was enriched in the regulation of cell development and cell adhesion. All four clusters were associated with kidney development and cellular status, delineating the potential role of these 172 differential genes during AKI-CKD transition (Supplementary Figure S1).
Potential differentiation outcomes in New PT2 cluster under pseudotime. (A-B) Pseudotime trajectory of New PT2 cells colored by state (A) and sample timepoints (B). (C) The distribution of cells at different time points in the samples. (D) Potential differentiation developmental direction of New PT2 cells. (E) Venn diagram showing the overlapping DEGs of two outcomes among four algorithms (BEAM, Findallmarkers, edgeR and DESeq2).
Screening hub genes may induce maladaptive repair in the transition from AKI to CKD
In order to further investigate the more significant genes associated with different differentiation outcomes in New PT2, our initial focus was on the expression distribution of 172 differential genes. Among these, a total of 52 genes exhibited high expression levels within the New PT2 cluster, while only 27 genes showed specific expression patterns distinct from other cell clusters (Supplementary Figure S2). The classification performances of five machine learning models (LASSO, RF-RFE, SVM, NN and XGBoost) were established and validated based on residuals, accuracy, f1_score, precision and recall indices. Among them, the RF-RFE algorithm exhibited the lowest residual and highest accuracy, f1_score, precision and recall scores in both the training and test groups (Fig. 5A-B). The RF-RFE model was constructed, and it was found that the minimum number of variables required for the Root Mean Square Error (RMSE) to reach its lowest value (0.2013 - Ankrd1, Sema5a) is two (Fig. 5C). The significance of feature genes selected from RF-RFE was demonstrated by the RMSE loss, suggesting that Ankrd1 may possess greater importance compared to Sema5a (Fig. 5D). Therefore, Ankrd1 and Sema5a were identified as pivotal candidates for subsequent analysis. However, in order to further scrutinize these two genes, integrating additional bioinformatics analyses and experimental validations is imperative to facilitate informed decisions.
Screening the hub genes by machine classifier. (A) Residual diagram of five classification. (B) Values of accuracy, f1_score, precision and recall scores to measure the prediction of classifiers. (C) The minimum number of variables was two when the RMSE at the lowest by the RF-RFE algorithm. (D) RMSE loss after feature removal from models. LASSO, least absolute shrinkage and selection operator; NN, neural network; RF-RFE, random forest-recursive feature elimination; RMSE, root mean square error; SVM, support vector machine; XGBoost, eXtreme gradient boosting.
Expression patterns and functions of subclusters for hub genes
To further explore the expression pattern and potential function of the two hub genes (Ankrd1 and Sema5a), we analyzed their expression distribution and dynamics on the pseudo-time trajectory. Interestingly, Ankrd1 and Sema5a exhibited certain differences across different branches (State 1 and State 5) (Fig. 6A-B). In single-cell data, the expression of Ankrd1 gene was significantly upregulated in different time points comparison to the sham group, while Sema5a was mainly increased during and after the transition period (Fig. 6C). And for gene expression in bulk RNA-seq data, the expression of Ankrd1 exhibited a notable biphasic pattern characterized by an initial increase followed by a subsequent decrease. Meanwhile, similar to the single-cell data, the expression of Sema5a significantly increased only during the transition period (Fig. 6D). These results suggest that Ankrd1 may play a more significant role than Sema5a from the initiation of AKI to CKD.
On the other hand, our objective was to investigate whether the two genes were expressed in specific subclusters of the New PT2 cluster and had specific potential functions. Remarkably, we observed that the expression patterns of these two genes were quite similar. Among the 9 subclusters of New PT2, Ankrd1 and Sema5a were both expressed in clusters 2, 3, 4, 5, 6, and 8 (Fig. 6E-F). Regarding the potential function of each subcluster, we first explored their distribution along a pseudo-time trajectory. The results showed that clusters 1, 2, 5, 6 and 8 were enriched in maladaptive repair branch (State 1), with clusters 5 and 6 being more significant (Fig. 6G). The cluster 1 was found to be associated with lung alveolus development, phagophore assembly site, and S-adenosylmethionine-dependent methyltransferase activity. The genes in cluster 2 primarily participate in cell junction assembly, regulation of cell morphogenesis, and response to external stimuli. Cluster 5 is involved in cellular amino acid metabolic processes, while cluster 6 exhibits epidermal growth factor receptor binding, microvillus formation, and positive regulation of response to external stimuli. Cluster 8 is associated with prespliceosome. This suggests that these five subclusters potentially participate in the regeneration of New PT2 cells by inducing an inflammatory response, regulating cell development and responses, as well as promoting prefibrosis (Fig. 6H).
Taken together, Ankrd1 is more likely to have a significant impact from the initiation of AKI to CKD, indicating its key role in abnormal regeneration of renal tubular cells after kidney injury. Conversely, Sema5a, which has higher expression in subclusters 1, 2, 5, 6 and 8, may primarily contribute to renal repair in the later stages of AKI. However, further experimental validation is required to ascertain the precise roles of these two genes in the progression of AKI-CKD.
Expression patterns and New PT2 subcluster function of hub genes. Expression distribution (A) and dynamics (B) in different branch of hub genes on the trajectories. Violin plot (the single-cell dataset GSE139107) (C) and boxplot (the bulk RNA-seq dataset GSE98622) (D) of the Ankrd1 and Sema5a genes expression at different sample timepoints. (E) UMAP displaying the clustering of New PT2 cells. (F) Expression of Ankrd1 and Sema5a genes in New PT2 subclusters. (G) Pseudotime trajectory of New PT2 cells colored by subclusters from 0 to 8. (H) Dotplot of GO enrichment analysis of New PT2 subcluster. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 vs. Sham group.
IRI-induced renal interstitial fibrosis contributes to the progression of AKI-CKD
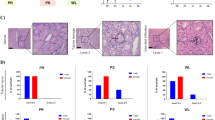
To ensure the survival rate of mice, we established a unilateral renal ischemia-reperfusion injury (uIRI) model and collected blood and left kidney tissue on days 0.5, 1, 3, 30, and 60 post-reperfusion for subsequent evaluations (Fig. 7A). Due to compensation from the contralateral normal kidney, serum creatinine (SCr) and blood urea nitrogen (BUN) concentrations did not significantly increase in the early phase after kidney reperfusion compared to the sham group; however, they began to increase at day 30 (Fig. 7B-C). The periodic acid-Schiff (PAS) staining revealed that during the early stage of IRI, the left kidney exhibited pathological alterations, including necrosis and detachment of tubular epithelial cells (TECs), loss of brush border, and cast formation. However, tubular recovery was observed at 3 days post-reperfusion (Fig. 7D). Renal interstitial fibrosis could be observed between the 30th and 60th days following reperfusion (Fig. 7D), as confirmed by Masson’s trichrome staining (Fig. 7E-F). The results demonstrated a progressive deterioration of renal interstitial fibrosis during the course of AKI-CKD, and the uIRI-induced AKI-CKD model was successfully established by clamping the left kidney.
IRI-induced renal interstitial fibrosis in AKI-CKD progression. (A) Experimental design. The red arrow indicates that the left kidney underwent ischemia for 45 min and began reperfusion. The numbers on the tinmeline indicate the time of kidney reperfusion and animal sacrifice. (B and C) Changes in serum creatinine (B) and blood urea nitrogen (C) over the reperfusion time. (D) Representative morphological changes of kidney stained with periodic acid-Schiff. Representative images of kidney stained with Masson (E) and the percentage of the fibrotic area (F). Scale bar: 50 μm; magnification: 400×; n = 4 mice in each group; *P < 0.05, ***P < 0.001, ****P < 0.0001 vs. Sham group. AKI, acute kidney injury; CKD, chronic kidney disease; IRI, ischemia-reperfusion injury; PAS, periodic acid–Schiff.
Ankrd1 is a potential biomarker for the progression of AKI-CKD induced by IRI
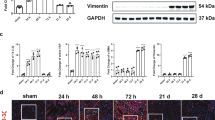
The expression of hub genes Ankrd1 and Sema5a in the kidney during AKI-CKD was evaluated using real-time quantitative polymerase chain reaction (RT-qPCR) to investigate the findings mentioned above (Fig. 8A-B). The results from RT-qPCR demonstrated that, compared to the sham group, Ankrd1 expression increased at 0.5th day post-reperfusion, peaked at 1th day, and remained significantly elevated up to 60 days. However, Sema5a expression only showed a significant increase at 3th day post-reperfusion, with no notable differences observed at other time points. The immunohistochemical (IHC) analysis data demonstrated a significant expression of Ankrd1 in renal tissue following IRI, thereby providing further validation for the RT-qPCR results (Fig. 8C-D). The collective findings indicate a positive correlation between the upregulation of Ankrd1 and the progression of AKI-CKD induced by IRI. Additionally, double immunofluorescence staining confirmed robust expression of Ankrd1 in proximal tubule cells (PTCs), which coexpressed with the PTC marker lotus tetragonolobus lectin (LTL) (Fig. 8E). Furthermore, we observed minimal colocalization between Ankrd1 and WT1-positive glomeruli (Fig. 8F).
Validated of hub genes in vivo experments. The RT-qPCR analysis of Ankrd1 (A) and Sema5a (B) in the progression of AKI-CKD. Images of Ankrd1 immunohistochemistry staining from mouse kidney sections (D) and the percentage of the positive area (C). Scale bar: 50 μm; magnification: 200×; (C-D) Histological and immunohistochemical analysis for proximal tubule cells marker LTL (E) and glomeruli cells marker WT1 (F) in uIRI kidney tissue. Scale bar: 20 μm; magnification: 400×. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 vs. Sham group; n = 4. IRI, ischemia-reperfusion injury; LTL, Lotus Tetragonolobus Lectin.
Discussion
The findings of various studies have suggested that the AKI-CKD progression may be attributed to the repetitive and severe damage inflicted on the proximal renal tubules, accompanied by maladaptive repair, thus highlighting their crucial role in the underlying cellular mechanisms10,22,23. Therefore, the aim of this study is to identify potential biomarkers within proximal tubule cells that can serve as indicators of AKI-CKD progression, employing various bioinformatics analyses. Firstly, we identified the interaction between the new proximal tubule cell cluster 2 (New PT2) and fibroblasts. Then, we determined two potential hub genes in New PT2 through trajectory analysis, differential expression analysis, and five machine learning algorithms. Finally, we further validated that Ankrd1 may serve as a superior biomarker compared to Sema5a in the progression of acute kidney injury to chronic kidney disease using RT-qPCR and IHC analysis.
The progression of renal fibrosis plays a crucial role in the transition from AKI to CKD10,14,17. A critical step in this fibrotic process is the conversion of quiescent fibroblasts into activated myofibroblasts24. In CKD, the activation of myofibroblasts and pericytes plays a crucial role in the deposition of the extracellular matrix (ECM) in the kidney and the promotion of renal fibrosis25,26,27. Both fibroblast-to-myofibroblast and pericyte-to-myofibroblast transitions are mainly initiated by injured tubular epithelial cells, which secrete platelet-derived growth factor (PDGF) and TGF-β1, concurrently triggering the process of epithelial-mesenchymal transition (EMT)28,29,30,31. Our study also confirmed fibroblast-PT cell interactions in the PDGF signaling pathway (Fig. 3E). Overall, injured tubular epithelial cells undergo partial EMT, leading to a compromised regenerative capacity. This process results in increased levels of proinflammatory and profibrotic factors (TGF-β1, PDGF), which promote the transformation of fibroblasts and pericytes into myofibroblasts. Moreover, given the interactions between different PT segments and fibroblasts, as well as their involvement in maladaptive repair, exploring biomarkers in specific PT segments can aid in the development of therapeutic strategies for the transition from AKI to CKD32.
Ankyrin repeat domain 1 (Ankrd1), also known as cardiac ankyrin repeat protein, belongs to the family of ankyrin repeat proteins33. The Ankrd1 protein functions as a profibrotic transcriptional co-regulator, playing a crucial role in the regulation of fibrosis, cardiac hypertrophy, damage repair, and wound healing34,35. Research shows that increased Ankrd1 levels in PAI-1 knockout cardiac tissues may significantly initiate cardiac fibrogenesis36. Meanwhile, Ankrd1 is also a target gene for Yap/Taz, which promotes fibroblast activation and kidney fibrosis37,38, while TGF-β1 stimulation can upregulate Yap/Taz and its target gene Ankrd1 in renal cells39,40. Furthermore, studies have demonstrated that TGF-β1 can induce the expression of Ankrd1 in vascular smooth muscle cells41. In turn, Ankrd1 is also associated with the TGF-β/Wnt signaling pathway, which plays a crucial role in promoting EMT42. Consequently, Ankrd1 may be implicated in fibroblast activation and EMT induction during the progression from AKI to CKD. However, the direct link between Ankrd1 expression and progressive renal fibrosis remains unclear and requires further experimental verification. Future research may focus on elucidating the functional role of inhibiting or silencing Ankrd1 expression, exploring the potential of gene therapy targeting Ankrd1 in the treatment of AKI, and bringing new directions and breakthroughs to clinical practice.
Besides, semaphorin 5a (Sema5a), a member of the semaphorin family, has been reported to augment angiogenesis by promoting endothelial cell proliferation and inhibiting apoptosis. Additionally, it may potentially mitigate myocardial ischemia-reperfusion injury following cardiac surgery43,44. Our data indicated that the expression level of Sema5a exhibited a significant elevation exclusively during the later stages of AKI, without any substantial differences thereafter. Conversely, Ankrd1 demonstrated a continuous increase from the initiation of AKI to CKD, suggesting that Ankrd1 may serve as a superior biomarker compared to Sema5a for monitoring the progression from AKI to CKD.
In summary, our inference suggests that the upregulation of Ankrd1 is associated with the progression of AKI-CKD induced by IRI. Following an IRI attack, proximal tubule cells undergo injury and secrete profibrotic factors such as PDGF and TGF-β1, which may contribute to the upregulation of Ankrd1. The upregulation of Ankrd1 is linked to fibroblast activation and occurrence of EMT, thereby promoting renal fibrosis and significantly increasing the likelihood of AKI progressing into CKD (Fig. 9). The AKI-CKD progression was found to be accompanied by a significant upregulation of Ankrd1 in renal cells, as demonstrated by RT-qPCR and IHC analysis. However, further investigation is still required to fully understand the underlying mechanisms of action for Ankrd1. Furthermore, considering the potential influence of AKI severity, future studies should explore the effects of varying stages of AKI on CKD progression45. Additionally, it is crucial to investigate whether repeated AKI hits have cumulative effects or involve distinct pathophysiological mechanisms46,47. Such studies will contribute to a deeper understanding of the complex processes underlying the transition from AKI to CKD.
Methods
Data collection and snRNA-seq dataset clustering
The single-nucleus RNA sequencing (snRNA-seq) dataset was obtained from the GEO database (http://www.ncbi.nlm.nih.gov/geo). The dataset GSE139107 used in this study includes gene expression profiles at various time points after IRI in mice compared to the sham control (4 biological replicates per group). Based on the histological changes and BUN levels reported in the original literature of the dataset, we defined the time points for AKI and CKD as follows: IRI_4h and IRI_12h represented the early stages of AKI, IRI_2d corresponded to the acute phase of AKI, IRI_14d marked the transition from AKI to CKD, and IRI_6w represented the CKD stage8. Quality control analysis was performed on the data using the Seurat R package, with a filter set at min.genes = 200 and max.genes = 1200 per cell. Additionally, filtering based on the number of unique molecular identifiers (UMIs) (> 6,000) and mitochondrial percentage (< 10%) was applied. The top 5000 variable genes were identified using the FindVariableFeatures function followed by principal component analysis (PCA) for dimension reduction with pc.num = 29. Data integration was then conducted using the RunHarmony function. Furthermore, distinct clusters were identified utilizing the FindClusters function with a resolution parameter of 0.9. Subsequently, these clusters were visualized through uniform manifold approximation and projection (UMAP), implemented via the RunUMAP function.
To validate the expression levels of hub genes during the progression from AKI to CKD, we selected the external bulk RNA-seq dataset GSE98622 from the GEO database (http://www.ncbi.nlm.nih.gov/geo). This dataset compiled renal tissue samples from mice at 2 h, 4 h, 24 h, 48 h, 72 h, 7 days, 14 days, 28 days, and 6 months post-AKI, as well as the sham group.
Intercellular crosstalk analysis
The CellChat object was generated using the createCellChat function, followed by preprocessing, computation of communication probability, and visualization with netVisual_circle. Subsequently, the CellChat objects were combined and compared between each group to identify unique pathways among them. The interaction pairs belonging to these unique pathways were selected for evaluating intercellular crosstalk.
Trajectory analysis and differential expression analysis
Differential analysis of multiple time points in New PT2 cells enabled the identification of genes with distinct developmental trajectories. Subsequently, Monocle2, an algorithm available in the monocle R package, was employed to map cells onto a pseudotime trajectory. Branch-dependent genes were then obtained using branched expression analysis modeling (BEAM) provided by Monocle2, and differential comparisons were performed between two selected branches. Simultaneously, we performed a joint analysis using the edgeR and DESeq2 algorithms within the FindAllMarkers and Pseudobulk functions to identify differentially expressed genes associated with two distinct developmental outcomes. The resulting differentially expressed genes were visualized through heatmaps, categorized into clusters based on their expression patterns, and subjected to Gene Ontology (GO) enrichment analysis. To visualize the expression patterns of hub genes over pseudotime, we used the plot_cell_trajectory function in conjunction with the plot_genes_branched_pseudotime function.
Machine algorithm evaluation and screening for hub genes
The training and test groups were subjected to five machine learning algorithms, namely Random Forest-Recursive Feature Elimination (RF-RFE), Least Absolute Shrinkage and Selection Operator (LASSO), Neural Network (NN), Support Vector Machine (SVM), and eXtreme Gradient Boosting (XGBoost). These algorithms utilized the DALEX R package to calculate residuals and feature importance. To evaluate classifier predictions, accuracy, f1_score, precision, and recall scores were computed. Subsequently, hub gene selection was based on the lowest residual value along with the four highest scores. Additionally, a 10-fold cross-validation approach using the randomForest R package was employed to construct the Random Forest-Recursive Feature Elimination (RF-RFE) algorithm in order to identify hub genes.
Clustering and functional analysis of cellular subclusters
The new proximal tubule cell cluster 2 (New PT2) was isolated for subsequent analysis and underwent clustering without Harmony integration. A total of 3000 highly variable genes were selected for dimension reduction using PCA (pc.num = 3), followed by clustering with the RunUMAP function, resulting in the identification of nine distinct subclusters. Subsequently, the FindAllMarkers function was employed to identify genes specifically expressed within each subcluster. Finally, GO enrichment analysis was performed for each cellular subcluster using the clusterProfiler R package.
Construction of animal model of kidney injury
Healthy male C57BL/6J mice (wild type, 6 weeks old) were obtained from Guangdong Sijiajingda Biotechnology Co., Ltd (China) and housed under controlled conditions with a constant room temperature on a 12-hour light/dark cycle. The mice had ad libitum access to food and water. All animal experimental procedures were approved by the Medical Ethics Committee of South China University of Technology (No.2022019), and all experiments were performed in accordance with relevant guidelines and regulations. For the AKI-CKD model, uIRI was induced in the left kidney of mice as previously described48. Briefly, mice were anesthetized with a 1% sodium pentobarbital solution (50 mg/kg) administered via intraperitoneal injection, before being immediately transcardiacally perfused with phosphate-buffered saline (PBS). The left kidney was exposed and non-invasive arterial clips were used to clamp the renal pedicles for 45 min. The sham group underwent only exposure of the left kidney without clamping. After clip removal, mice were returned to their cages and allowed to recover for a period ranging from 0.5th to 60th days. At the end of the experiment, mice (n = 4 per uIRI group) were perfused with PBS through the left cardiac ventricle until their kidneys were cleared of blood after obtaining a 500 µL blood sample. Kidney tissues and blood samples were collected for subsequent experiments, respectively. SCr and BUN levels were measured using an automatic biochemistry analyzer (Hitachi, Japan).
Histological and immunohistochemical analysis
The mouse kidney tissue was fixed in 4% paraformaldehyde, dehydrated using an ethanol gradient, and subsequently embedded in paraffin. Sections that were three microns thick were then prepared and stained with PAS following the manufacturer’s instructions. Furthermore, Masson’s trichrome staining was conducted to evaluate the degree of renal fibrosis49. Quantitative scoring of interstitial fibrosis was conducted using Image J software. For IHC staining, sections were incubated with Ankrd1 antibodies (1:200; Bioss; bs-8074R) overnight at 4 °C and subsequently processed according to the instructions of the UltraSensitiveTM SP IHC Kit (Maixin, China). Double immunofluorescence staining was performed for Ankrd1/WT1 (1:100; Servicebio; GB11382) or Ankrd1/Lotus Tetragonolobus Lectin (LTL; 1:400; Vector; B-1325-2), with primary antibody incubation carried out separately for each antibody. Specifically, each section was first incubated overnight at 4 °C with the Ankrd1 primary antibody, followed by a second primary antibody LTL or anti-WT1 and a subsequent 2-hour incubation. The sections were subsequently washed three times with PBS prior to incubation at 37 °C for one hour with Cy3-conjugated secondary antibody (1:200; Bioss; bs-8074R) or Alexa Fluor 488-conjugated secondary antibody (1:100, Servicebio; GB11382). Subsequently, DAPI was utilized for nuclear DNA staining, and the images were acquired utilizing a Nikon Eclipse Ti2-E fluorescence microscope. The obtained images from each section underwent blind analysis and quantification through the utilization of Image J software.
RNA isolation and real-time quantitative polymerase chain reaction
The Tissue RNA Purification Kit PLUS (EZBioscience, USA) was used to extract total RNA from kidneys, followed by reverse transcription to obtain cDNA. RT-qPCR experiments were performed on a LightCycler 96 instrument (Roche, Switzerland), using Color SYBR Green qPCR Mix (EZBioscience). Each sample underwent 45 cycles during the experiment. All procedures strictly adhered to the manufacturer’s instructions. The data were normalized and analyzed using the 2 − ΔΔCt method, with gene expression being normalized to β-actin. The primer sequences are provided below: mouse-Ankrd1: forward, GCTTAGAAGGACACTTGGCGATC and reverse, GACATCTGCGTTTCCTCCACGA; mouse-β-actin: forward, CATGTACGTTGCTATCCAGGC and reverse, CTCCTTAATGTCACGCACGAT; mouse-Sema5a: forward, GACTTGCTAGGCCCGAGAC and reverse, TCTTTGTAAGAGACGACAGGGT.
Statistical analyses
The mean ± SD was utilized for expressing all of the data. Data processing, statistical analysis, and plotting were performed using R 4.2.2 software, Figdraw (https://www.figdraw.com), Generic Diagramming Platform (https://gdp.rjmart.cn), and ProcessOn (https://www.processon.com) respectively. The statistical tests consisted of the two-sided unpaired Student’s t-test and two-way ANOVA with a significance level set at p < 0.05.
Data availability
The datasets analyzed in this study are available in the GEO repository, [http://www.ncbi.nlm.nih.gov/geo]. The names of the repository/repositories and accession number(s) can be found in the Methods.
Abbreviations
- AKI:
-
Acute kidney injury
- BUN:
-
Blood urea nitrogen
- CKD:
-
Chronic kidney disease
- IRI:
-
Ischemia–reperfusion injury
- LASSO:
-
Least absolute shrinkage and selection operator
- LTL:
-
Lotus tetragonolobus lectin
- NN:
-
Neural network
- PAS:
-
Periodic acid–Schiff
- PBS:
-
Phosphate buffer saline
- PT:
-
Proximal tubule
- RF-RFE:
-
Random forest-recursive feature elimination
- RMSE:
-
Root mean square error
- SCr:
-
Serum creatinine
- SVM:
-
Support vector machine
- XGBoost:
-
EXtreme gradient boosting
References
Zhang, M. et al. Single cell dissection of epithelial-immune cellular interplay in acute kidney injury microenvironment. Front. Immunol. 13, 857025. https://doi.org/10.3389/fimmu.2022.857025 (2022).
Gao, L., Zhong, X., Jin, J., Li, J. & Meng, X. M. Potential targeted therapy and diagnosis based on novel insight into growth factors, receptors, and downstream effectors in acute kidney injury and acute kidney injury-chronic kidney disease progression. Signal. Transduct. Tar. 5, 9. https://doi.org/10.1038/s41392-020-0106-1 (2020).
Guo, R. Z. et al. The Road from AKI to CKD: Molecular mechanisms and therapeutic targets of Ferroptosis. Cell Death Dis. 14, 426. https://doi.org/10.1038/s41419-023-05969-9 (2023).
Lv, J. C. & Zhang, L. X. Prevalence and disease burden of chronic kidney disease. Adv. Exp. Med. Biol. 1165, 3–15. https://doi.org/10.1007/978-981-13-8871-2_1 (2019).
Ni, Y. F. et al. RfxCas13d-mediated inhibition of alleviates renal fibrosis via PI3K/AKT signaling pathway. Ren. Fail. 46, 2331612. https://doi.org/10.1080/0886022x.2024.2331612 (2024).
Chawla, L. S., Eggers, P. W., Star, R. A. & Kimmel, P. L. Acute kidney Injury and chronic kidney disease as interconnected syndromes. New. Engl. J. Med. 371, 58–66. https://doi.org/10.1056/NEJMra1214243 (2014).
Han, S. J. & Lee, H. T. Mechanisms and therapeutic targets of ischemic acute kidney injury. Kidney Res. Clin. Prac. 38, 427–440. https://doi.org/10.23876/j.krcp.19.062 (2019).
Kirita, Y., Wu, H. J., Uchimura, K., Wilson, P. C. & Humphreys, B. D. Cell profiling of mouse acute kidney injury reveals conserved cellular responses to injury. P Natl. Acad. Sci. USA. 117, 15874–15883. https://doi.org/10.1073/pnas.2005477117 (2020).
Dong, Y. et al. Ischemic duration and frequency determines AKI-to-CKD progression monitored by dynamic changes of tubular biomarkers in IRI mice. Front. Physiol. 10, 153. https://doi.org/10.3389/fphys.2019.00153 (2019).
Takaori, K. et al. Severity and frequency of Proximal Tubule Injury determines renal prognosis. J. Am. Soc. Nephrol. 27, 2393–2406. https://doi.org/10.1681/Asn.2015060647 (2016).
Chen, J. et al. Cellular senescence of renal tubular epithelial cells in acute kidney injury. Cell. Death Discov. 10, 62. https://doi.org/10.1038/s41420-024-01831-9 (2024).
Lee, K., Gusella, G. L. & He, J. C. Epithelial proliferation and cell cycle dysregulation in kidney injury and disease. Kidney Int. 100, 67–78. https://doi.org/10.1016/j.kint.2021.03.024 (2021).
Wu, C. F. et al. Transforming growth factor β-1 stimulates profibrotic epithelial signaling to activate pericyte-myofibroblast transition in obstructive kidney fibrosis. Am. J. Pathol. 182, 118–131. https://doi.org/10.1016/j.ajpath.2012.09.009 (2013).
Fan, Z. Y. et al. Immune-related SERPINA3 as a biomarker involved in diabetic nephropathy renal tubular injury. Front. Immunol. 13, 979995. https://doi.org/10.3389/fimmu.2022.979995 (2022).
Guerrero-Mauvecin, J. et al. Regulated necrosis role in inflammation and repair in acute kidney injury. Front. Immunol. 14, 1324996. https://doi.org/10.3389/fimmu.2023.1324996 (2023).
Kellum, J. A. et al. Acute kidney injury. Nat. Rev. Dis. Primers. 7, 52. https://doi.org/10.1038/s41572-021-00284-z (2021).
Ko, G. J. et al. Transcriptional analysis of kidneys during repair from AKI reveals possible roles for NGAL and KIM-1 as biomarkers of AKI-to-CKD transition. Am. J. Physiol-Renal. 298, F1472–F1483. https://doi.org/10.1152/ajprenal.00619.2009 (2010).
Bansal, A., Nigoskar, S. & Thalquotra, M. Comparison of BTP, NGAL, KIM-1, & ADMA biomarkers in CKD and non-CKD subjects. Int. J. Biochem. Mol. Biol. 14, 32–39 (2023).
Lu, Y. H., Xu, S. T., Tang, R., Han, C. & Zheng, C. X. A potential link between fibroblast growth factor-23 and the progression of AKI to CKD. Bmc Nephrol. 24, 87. https://doi.org/10.1186/s12882-023-03125-1 (2023).
Nakagawa, S. et al. Molecular markers of Tubulointerstitial Fibrosis and tubular cell damage in patients with chronic kidney disease. Plos One. 10, e136994. https://doi.org/10.1371/journal.pone.0136994 (2015).
Lu, Y. A. et al. Single-nucleus RNA sequencing identifies new classes of proximal tubular epithelial cells in kidney fibrosis. J. Am. Soc. Nephrol. 32, 2501–2516. https://doi.org/10.1681/Asn.2020081143 (2021).
Endo, T. et al. Exploring the origin and limitations of kidney regeneration. J. Pathol. 236, 251–263. https://doi.org/10.1002/path.4514 (2015).
Rudman-Melnick, V. et al. Single-cell sequencing dissects the transcriptional identity of activated fibroblasts and identifies novel persistent distal tubular injury patterns in kidney fibrosis. Sci. Rep-Uk. 14, 439. https://doi.org/10.1038/s41598-023-50195-0 (2024).
LeBleu, V. S. et al. Origin and function of myofibroblasts in kidney fibrosis. Nat. Med. 19, 1047–1054. https://doi.org/10.1038/nm.3218 (2013).
Niculae, A. et al. Pathway from Acute kidney Injury to chronic kidney disease: molecules involved in Renal Fibrosis. Int. J. Mol. Sci. 24, 14019. https://doi.org/10.3390/ijms241814019 (2023).
Xu, C. et al. Regulation of pericyte metabolic reprogramming restricts the AKI to CKD transition. Metabolism 145, 155592. https://doi.org/10.1016/j.metabol.2023.155592 (2023).
Hu, L. M. et al. Cathepsin G promotes arteriovenous fistula maturation by positively regulating the MMP2/MMP9 pathway. Ren. Fail. 46, 2316269. https://doi.org/10.1080/0886022x.2024.2316269 (2024).
Vega, G., Alarcón, S. & San Martín, R. The cellular and signalling alterations conducted by TGF-β contributing to renal fibrosis. Cytokine 88, 115–125. https://doi.org/10.1016/j.cyto.2016.08.019 (2016).
Zhang, Y. et al. IRF1/ZNF350/GPX4-mediated ferroptosis of renal tubular epithelial cells promote chronic renal allograft interstitial fibrosis. Free Radical Bio Med. 193, 579–594. https://doi.org/10.1016/j.freeradbiomed.2022.11.002 (2022).
Guo, J. Q. et al. PDGF-BB/PDGFRβ promotes epithelial-mesenchymal transition by affecting PI3K/AKT/mTOR-driven aerobic glycolysis in Wilms’ tumor G401 cells. Cell. Biol. Int. 46, 907–921. https://doi.org/10.1002/cbin.11780 (2022).
Xu, Z. H. et al. Continuous genetic monitoring of transient mesenchymal gene activities in distal tubule and collecting duct epithelial cells during renal fibrosis. J. Cell. Biochem. 125, e30541. https://doi.org/10.1002/jcb.30541 (2024).
Wen, Y. M. et al. Analysis of the human kidney transcriptome and plasma proteome identifies markers of proximal tubule maladaptation to injury. Sci. Transl Med. 15, eade7287. https://doi.org/10.1126/scitranslmed.ade7287 (2023).
Ling, S. S. M., Chen, Y. T., Wang, J., Richards, A. M. & Liew, O. W. Ankyrin Repeat Domain 1 protein: a functionally pleiotropic protein with Cardiac Biomarker potential. Int. J. Mol. Sci. 18, 1362. https://doi.org/10.3390/ijms18071362 (2017).
Bujor, A. M., Adili, E., Parvez, F., Marden, A., Trojanowska, M. & G. & Fli1 downregulation in scleroderma myeloid cells has profibrotic and proinflammatory effects. Front. Immunol. 11, 800. https://doi.org/10.3389/fimmu.2020.00800 (2020).
Samaras, S. E., Almodóvar-García, K., Wu, N. J., Yu, F. & Davidson, J. M. Global deletion of results in a wound-healing phenotype associated with dermal fibroblast dysfunction. Am. J. Pathol. 185, 96–109. https://doi.org/10.1016/j.ajpath.2014.09.018 (2015).
Ghosh, A. K., Murphy, S. B., Kishore, R. & Vaughan, D. E. Global gene expression profiling in PAI-1 knockout murine heart and kidney: molecular basis of Cardiac-Selective Fibrosis. Plos One. 8, e63825. https://doi.org/10.1371/journal.pone.0063825 (2013).
Chen, J. C. & Harris, R. C. Interaction of the EGF receptor and the hippo pathway in the diabetic kidney. J. Am. Soc. Nephrol. 27, 1689–1700. https://doi.org/10.1681/Asn.2015040415 (2016).
Liang, M. et al. Yap/Taz deletion in gli cell-derived myofibroblasts attenuates fibrosis. J. Am. Soc. Nephrol. 28, 3277–3289. https://doi.org/10.1681/Asn.2015121354 (2017).
Szeto, S. G. et al. YAP/TAZ are mechanoregulators of TGF-β-Smad signaling and renal fibrogenesis. J. Am. Soc. Nephrol. 27, 3117–3128. https://doi.org/10.1681/Asn.2015050499 (2016).
Gui, Y. et al. Yap/Taz mediates mTORC2-stimulated fibroblast activation and kidney fibrosis. J. Biol. Chem. 293, 16364–16375. https://doi.org/10.1074/jbc.RA118.004073 (2018).
Umeda-Yano, S. et al. The regulation of gene expression involved in TGF-β signaling by ZNF804A, a risk gene for schizophrenia. Schizophr Res. 146, 273–278. https://doi.org/10.1016/j.schres.2013.01.026 (2013).
Zhou, M. et al. Role of epithelial-mesenchymal transition in Retinal Pigment Epithelium Dysfunction. Front. Cell. Dev. Biol. 8, 501. https://doi.org/10.3389/fcell.2020.00501 (2020).
Du, Y. et al. Elevated semaphorin5A in systemic lupus erythematosus is in association with disease activity and lupus nephritis. Clin. Exp. Immunol. 188, 234–242. https://doi.org/10.1111/cei.12924 (2017).
Wu, T. et al. Circulating small extracellular vesicle-encapsulated SEMA5A-IT1 attenuates myocardial ischemia-reperfusion injury after cardiac surgery with cardiopulmonary bypass. Cell. Mol. Biol. Lett. 27, 95. https://doi.org/10.1186/s11658-022-00395-9 (2022).
Ostermann, M. et al. Controversies in acute kidney injury: Conclusions from a kidney disease: improving global outcomes (KDIGO) Conference. Kidney Int. 98, 294–309. https://doi.org/10.1016/j.kint.2020.04.020 (2020).
Acosta-Ochoa, I. et al. Second, Third, and more hits: Impact of successive aki episodes on Ckd progression and Aki To Ckd transition. Nephrol. Dial Transpl. 38, I492–I493 (2023).
Vanmassenhove, J., Kielstein, J., Jörres, A. & Van Biesen, W. Management of patients at risk of acute kidney injury. Lancet 389, 2139–2151. https://doi.org/10.1016/S0140-6736(17)31329-6 (2017).
Chen, Y. L. et al. Pyruvate kinase M2 regulates kidney fibrosis through pericyte glycolysis during the progression from acute kidney injury to chronic kidney disease. Cell. Proliferat. 57, e13548. https://doi.org/10.1111/cpr.13548 (2024).
Zager, R. A., Johnson, A. C. M., Andress, D. & Becker, K. Progressive endothelin-1 gene activation initiates chronic/end-stage renal disease following experimental ischemic/reperfusion injury. Kidney Int. 84, 703–712. https://doi.org/10.1038/ki.2013.157 (2013).
Acknowledgements
We thank all the participants of this study and the researchers who upload the original data to GEO database. We also thank Figdraw (www.figdraw.com) for the assistance in creating mechanistic map.
Funding
This work was funded by the National Natural Science Foundation of China (grant numbers 31870928(QHL), 32271360(QHL), 82170761(ML) and 81770677(ML)), the Natural Science Foundation of Guangdong Province, China (grant numbers 2021A1515010040(QHL) and 2023A1515010829(QHL)).
Author information
Authors and Affiliations
Contributions
HLL, CQZ, LMH, ML and QHL designed the study. CQZ collected and analyzed the data. LMH designed and performed experiments. YK provided guidance on data analysis. HLL searched relevant literature, generated partial figures and drafted the initial manuscript. QHL and ML reviewed and edited the manuscript. All authors reviewed the manuscript and approved the submitted version.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
The study protocol was reviewed and approved by the Medical Ethics Committee of South China University of Technology (No.2022019). All methods in the study were reported in accordance with ARRIVE guidelines (https://arriveguidelines.org). Written informed consent was obtained from all the participants.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Li, H., Hu, L., Zheng, C. et al. Ankrd1 as a potential biomarker for the transition from acute kidney injury to chronic kidney disease. Sci Rep 15, 4659 (2025). https://doi.org/10.1038/s41598-025-88752-4
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-88752-4