Abstract
Predicting disease prognosis and the efficacy of immunotherapy presents a significant challenge in the treatment of hepatocellular carcinoma (HCC). By analyzing transcriptome sequencing data from 69 patients and identifying differentially expressed immune genes, a prognostic index named the immune-related gene prognostic index (IRGPI) was established by Lasso-Cox regression. The IRGPI, which consists of six key genes, was found to be a significant predictor of poor prognosis in patients with high IRGPI scores. The model’s predictive accuracy was confirmed via receiver operating characteristic (ROC) curve analysis, with area under the curve (AUC) values of 0.85, 0.779, and 0.857 for 1-, 3-, and 5-year survival predictions, respectively. Additionally, patients with high IRGPI scores had increased levels of Treg cells and neutrophils, advanced tumor staging, microvascular invasion grading, and immune checkpoint expression. The IRGPI was also effective in predicting the efficacy of immunotherapy in the IMvigor210 dataset, demonstrating its potential as a valuable tool for assessing patient prognosis and guiding immunotherapy strategies in HCC.
Similar content being viewed by others
Introduction
Hepatocellular carcinoma (HCC) is associated with high mortality and postoperative recurrence rates, making HCC the second leading cause of cancer-related death in China1,2. Recently, immunotherapy has been shown to reduce tumor recurrence and prolong progression-free survival in various cancer types. However, owing to the unique immune microenvironment of HCC, many patients failed to respond to immunotherapy3. Thus, understanding the characteristics of the tumor microenvironment in HCC is vital for identifying predictive markers of immunotherapy.
PD-1 inhibitors such as nivolumab and pembrolizumab have emerged as second-line treatments for advanced HCC, with promising clinical outcomes4. However, predicting the efficacy of immunotherapy in patients remains an important challenge in clinical practice. Traditional predictive markers include PD-1/PD-L1 expression detected by immunohistochemistry and high-throughput assessments of high tumor mutational burden and microsatellite instability. In HCC, however, only approximately 20% of patients with high PD-L1/PD-1 expression respond well to immunosuppressive therapy5. Furthermore, a high tumor mutation burden(TMB) is rare in HCC patients6, and an Microsatellite Instability high(MSI-high) status is observed in only 2–3% of cases7. These findings suggest that conventional indicators are insufficient for predicting whether a patient with HCC is likely to benefit from immunotherapy, highlighting the need for more accurate clinical predictors of therapeutic efficacy.
Tumor-infiltrating immune cells are key effectors that mediate the cytotoxic response during immune checkpoint blockade therapy. Their composition and functional state are also critical factors that influence the efficacy of immunotherapy in cancer patients. In addition to quantifying surface markers of immune cells within the tumor microenvironment via immunohistochemistry to evaluate the immune landscape, the expression profiles of immune-related genes(IRGs) can also characterize the tumor immune microenvironment in patients. IRGs are a group of genes involved in regulating immune cell subtypes and functions8. Compared with the use of specific immunohistochemical markers, analyzing the expression profiles of IRGs in individual tumor patients provides a more comprehensive reflection of the patient’s immune microenvironment, which can subsequently guide personalized immunotherapy. Previous studies have demonstrated that a scoring system composed of five IRGs could effectively identify HCC patients who are likely to respond to immunotherapy9. Additionally, a study on lung adenocarcinoma established a scoring system based on 17 IRGs. This system was found to be associated with tumor-infiltrating follicular helper T cells, macrophages, and the tumor TMB and can also be used to predict treatment outcomes in cancer patients10. However, these studies are primarily based on publicly available clinical databases, such as the TCGA, and their applicability to the Chinese population remains to be validated.
In this study, we performed transcriptome sequencing on cancerous and adjacent noncancerous tissues from 69 patients with HCC to identify differentially expressed immune-related genes. We subsequently used Lasso-Cox regression analysis to develop a predictive model for HCC patient prognosis and immunotherapy response, which was further validated using public datasets.
Materials and methods
Patients and clinical data
A series of 69 patients with HCC who underwent surgical intervention at Mengchao Hepatobiliary Hospital of Fujian Medical University, China, between 2015 and 2020 were retrospectively enrolled in this study. All patients underwent R0 resection, and postoperative GEMOX regimens were administered to prevent recurrence. Patients who received preoperative transarterial therapies, portal embolization, or chemotherapies were excluded. To minimize the impact of perioperative factors, patients who experienced relapse or death within two months of surgery were also excluded from this study. This study was approved by the ethics committee of Mengchao Hepatobiliary Hospital of Fujian Medical University (Approval number: 2021–044-01), and informed consent was obtained from all patients to use their samples and clinical information. This study was conducted in accordance with the principles of the Declaration of Helsinki11.
Clinical data, laboratory findings, and pathological examination results, including age, sex, China Liver Cancer Staging(CNLC stage), tumor size, and serum Alpha-fetoprotein(AFP) levels, were carefully collected. Patients were followed every three months postoperatively through outpatient visits, during which they underwent serum tumor marker assessments and imaging examinations. In this study, tumor recurrence was defined as the appearance of new nodules with typical characteristics of HCC. The time interval between the date of surgery and either the date of recurrence or the last follow-up was recorded as the recurrence-free survival (RFS) time, whereas the time interval between the date of surgery and either the date of disease-related death or the last follow-up was recorded as the overall survival (OS) time.
Transcriptome sequencing
Fresh tumor and adjacent tissues (0.5 mg) were collected, and RNA was extracted using the TRIzol method. The RNA was quantified to ensure its suitability for further analysis. A total of 5 μg of RNA was enriched for poly(A)-tailed RNA using oligo(dT) magnetic beads. The RNA was then randomly fragmented and reverse-transcribed to synthesize the first strand of cDNA, followed by the synthesis of double-stranded cDNA. AMPure XP beads were used to purify and recover the double-stranded cDNA, which was subsequently end-repaired, 3' end-tailed, and ligated with sequencing adapters. The cDNA library was enriched by PCR and subjected to high-throughput sequencing on the Illumina HiSeq2500 platform. Trinity software was used for sequence assembly, and raw counts were used to quantify transcript and gene expression levels in this study.
Selection of immune-related genes and construction of the Immune-Related Gene-Based Prognostic Index (IRGPI)
Differential gene expression analysis was performed using the DEseq2(Version 1.44.0) in R software to identify genes that were differentially expressed between tumor and adjacent noncancerous tissues. A threshold of |log(FC)|> 1.5 and p < 0.05 was used to select differentially expressed genes (DEGs). Immune-related genes (IRGs) were obtained by querying the ImmPort and InnateDB databases. Genes that overlapped with the tumor-specific differentially expressed genes were identified as potential key IRGs. In this study, the potential IRGs were used as independent variables, and the 5-year survival outcome of the patients was used as the dependent variable. Lasso-Cox regression analysis was then conducted to identify key genes affecting patient prognosis, leading to the construction of an immune-related gene-based prognostic index (IRGPI). The computation of Index A entails multiplying each gene by its corresponding coefficient and subsequently aggregating these products to obtain the IRGPI. The predictive accuracy of the IRGPI for 1-, 3-, and 5-year survival was assessed by plotting ROC curves and constructing a nomogram, and calibration curves were drawn to evaluate its predictive precision.
Differences in clinicopathological features, immune infiltration, and immunotherapy prediction among different IRGPI groups
The 69 HCC patients were divided into high- and low-IRGPI groups on the basis of the median IRGPI. Differences in TNM stage, tumor size, and prognosis between the two groups were compared. The Limma package was used to analyze differential genes (p < 0.05, abs(FDR) > 1.5) between the two groups, followed by enrichment analysis with the clusterProfiler package to identify signaling pathways involved in differential gene expression. The CIBERSORT algorithm was applied to analyze immune cell infiltration in both groups, and differences in immune infiltration between the subgroups were compared. Finally, we validated the predictive value of the IRGPI for immunotherapy outcomes using the IMvigor210 dataset. We analyzed the differences in IRGPI scores between the CR + PR and SD + PD groups and plotted survival curves to explore the relationship between the IRGPI and the efficacy of immunotherapy.
Immunohistochemistry
Formalin-fixed, paraffin-embedded tissue samples were continuously sectioned (4 μm), deparaffinized, and rehydrated. Antigen retrieval was performed in citrate buffer (pH = 6.0) at high temperature, after which endogenous peroxidase activity was blocked with 3% hydrogen peroxide solution to reduce nonspecific staining. The sections were incubated with primary antibodies at 37 ℃ for 2 h, using the following dilutions and sources: PD-1 (1:5,000, Abcam, ab137132), PD-L1[28-8] (1:5,000, Abcam, ab205921), PD-L1[SP142] (1:5,000, Abcam, ab228462), Foxp3 (1:5,000, Abcam, ab20034), CD68 (1:10,000, Abcam, ab192847), and CD4 (1:5,000, Abcam, ab133616). After washing with PBS, the appropriate secondary antibodies were added, and color development was carried out using 3,3'-diaminobenzidine (DAB). Hematoxylin was used for nuclear counterstaining. The number of positive cells in the sections was counted using Image-Pro Plus software for subsequent analysis.
Statistical analysis
Variables following a normal distribution are expressed as the means ± standard deviations, whereas categorical data are presented as percentages (%). Independent t tests were used to compare differences in continuous variables between the two groups, and the chi-square test was applied for categorical data. Pearson correlation analysis was used to examine potential correlations between two indices. The Limma package in R (version 4.3.2) was used to analyze DEGs between the two groups. Lasso Cox regression was then applied to identify key immune-related genes, which were subsequently used to construct an immune-related gene prognostic index. Kaplan‒Meier (K‒M) survival analysis, accompanied by log-rank tests, was conducted to assess prognostic differences between the two groups. Additionally, the rms package was used to plot ROC curves, and the C-index was calculated to evaluate the ability of the IRGPI to predict patient prognosis in conjunction with the construction of a nomogram model for prognosis prediction. CIBERSORT was used to quantify immune cell infiltration, and differences in immune cell composition between various IRGPI groups were compared. A p value of less than 0.05 was considered statistically significant.
Results
Transcriptome sequencing and differential gene enrichment analysis
A clear separation between cancerous and adjacent tissues was observed via principal component analysis (PCA) (Fig. 1A). Differential analysis identified 2,882 genes with a |logFC|> 1.5 (Fig. 1B). Further KEGG pathway analysis of the differentially expressed genes revealed that metabolism pathways, such as the bile acid pathway, were suppressed in tumors, whereas tumor-related pathways, such as DNA repair, were upregulated (Fig. 1C). Furthermore, ssGSEA revealed significant downregulation of immune-related signaling pathways, such as the B-cell receptor signaling pathway, in tumors, suggesting that the downregulation of immune-related pathways plays a critical role in tumorigenesis in HCC (Fig. 1D).
Transcriptomic Sequencing and Differential Gene Enrichment Analysis. (A) PCA results for normal and adjacent cancer tissues. After PCA dimensionality reduction, cancerous and adjacent tissues were clearly divided into two groups, indicating significant differences in gene expression. (B) The volcano plot for differential genes between cancerous and adjacent tissues. Differential analysis identified 2,882 genes with a |logFC|> 1.5. (C) The KEGG enrichment results of differential genes between cancerous and adjacent tissues. (D) The ssGSEA enrichment results between cancerous and adjacent tissues. ssGSEA revealed significant downregulation of immune-related signaling pathways, such as the B-cell receptor signaling pathway.
Establishment of the immune-related gene-based prognostic index (IRGPI)
To further analyze which immune-related genes (IRGs) might play a key role in tumor development and progression, we downloaded 2,501 IRGs from the ImmPort database. These genes intersected with the 2,882 differentially expressed genes (DEGs) identified from the differential expression analysis, resulting in 181 IRGs with significant differential expression. These 181 IRGs were then subjected to LASSO analysis (Fig. 2A). LASSO regression was used to select key parameters, and after the variation characteristics of these variable coefficients were calculated, tenfold cross-validation was applied to iterative analyses, resulting in an optimal model with minimal error and the best performance (Fig. 2B). The IRGs selected by LASSO regression included NDRG1, MMP12, CTSE, COLEC12, RELB, and KLRD1. A Cox regression model was further developed on the basis of the parameters selected through LASSO regression. These six genes constitute a formula for calculating IRGPIs. The IRGPIs = [0.0486 × mRNA expression level of COLEC12] + [0.1655 × mRNA expression level of CTSE] + [−0.0271 × mRNA expression level of KLRD1] + [0.0908 × mRNA expression level of MMP12] + [0.0115 × mRNA expression level of NDRG1] + [0.0588 × mRNA expression level of RELB]. Base on the median expression of IRGPIs, the patients was divided into IRGPI-high and IRGPI-down group in order to investigate the expression pattern of the six genes in the the two groups. It was demostrated that patients in the IRGPI-high had worse prognoses (cutoff = −0.19). A trend toward higher expression of RELB, NDRG1, MMP12, CTSE, and COLEC12 was observed in patients with a poor prognosis, whereas KLRD1 expression was higher in patients with a better prognosis (Fig. 2C). Multivariate Cox regression analysis revealed that the IRGPI, which is composed of these six genes, is an independent risk factor affecting patient prognosis(Fig. 2D). Survival curve analysis revealed that collectin subfamily member 12(COLEC12), cathepsin E (CTSE), killer cell lectin-like receptor D1(KLRD1), matrix metalloproteinase 12(MMP12), N-myc downstream-regulated gene 1(NDRG1), and RELB proto-oncogene(RELB) were significantly associated with poor prognosis, with high expression of KLRD1 linked to better outcomes (Fig. 2E). COLEC12 is a a member of the C-type lectin family that functions as a scavenger receptor, binding to carbohydrate antigens on microbes to facilitate their recognition and removal, as well as mediating the recognition, internalization, and degradation of oxidized low-density lipoproteins by endothelial cells. CTSE encodes an aspartic protease that plays a significant role in antigen processing and presentation . KLRD1 can be expressed on NK cells and participates in the regulation of NK cell functions . MMP12 is an well-known extracellular matrix-degrading enzyme involved in tissue remodeling, wound healing, atherosclerosis progression, and tumor invasion. NDRG1 is associated with cell differentiation, organ formation, embryonic development, and maintaining cellular differentiation levels. Its expression is influenced by various factors, including hypoxic conditions, and it may serve as a marker and diagnostic tool for tumor progression. NF-kB subunit, is involved in the negative regulation of lymphocyte differentiation and interferon-β production, and is related to breast cancer and immune deficiencies. All these six genes have been identified to be associated with the tumor microenvironment (TME) and prognosis of various cancers, which demostrated that IRGPI can be used to forecast the prognosis and immunotherapy outcomes in HCC.
Establishment of the IRGPI. (A) Venn diagram of differentially expressed genes and immune-related genes. The intersection of 2,882 differentially expressed genes (DEGs) and 2,501 immune-related genes (IRGs) resulted in 181 overlapping genes, which were used for further analysis. (B) LASSO Cox results demonstrating genes closely associated with prognosis: LASSO Cox regression identified six key immune-related genes—COLEC12, CTSE, KLRD1, MMP12, NDRG1, and RELB—that were significantly associated with patient prognosis. (C) Risk factor association plot for the IRGPI. The risk factor association plot demonstrates the relationship between the IRGPI score and the expression levels of the six genes, indicating that higher IRGPI scores are linked to worse prognosis. RELB, NDRG1, MMP12, CTSE, and COLEC12 were more highly expressed in high-risk patients, whereas KLRD1 was expressed at lower levels in those with better prognoses. (D) Multivariate Cox regression analysis demonstrating that the IRGPI was an independent prognostic factor. Multivariate Cox regression analysis confirmed that the IRGPI is an independent risk factor influencing patient prognosis, with a significant impact on survival outcomes. (E) Survival curve analysis of the six genes. Kaplan–Meier survival curves for COLEC12, CTSE, KLRD1, MMP12, NDRG1, and RELB demonstrated their significant associations with poor prognosis, where high expression of KLRD1 was correlated with better outcomes.
IRGPI as a predictor of prognosis in patients with HCC
Patients were divided into high and low groups on the basis of the median IRGPI value. Kaplan–Meier (K-M) survival curves revealed that patients with a high IRGPI had poorer outcomes (Fig. 3A). ROC curve analysis further demonstrated the prognostic value of the IRGPI, with AUC values of 0.85, 0.779, and 0.857 for 1-, 3-, and 5-year predictions, respectively. The C-index values were 0.912, 0.829, and 0.813, indicating high diagnostic value (Fig. 3B, C). The AUC values and C-index values exceeded 0.7 and 0.8 respectively, which meant that diagnostic model was considered to perform well. A nomogram model was constructed to facilitate the clinical application of the IRGPI (Fig. 3D).
Prognostic value of the IRGPI for patients with HCC. (A) OS was shorter in the IRGPI-high group than in the IRGPI-low group. Patients in the IRGPI-high subgroup had significantly shorter overall survival than those in the IRGPI-low subgroup did, indicating that a higher IRGPI score is associated with a worse prognosis. (B) ROC curves for 1-, 3-, and 5-year prognostic prediction by the IRGPI. The ROC curves demonstrate the predictive accuracy of the IRGPI for 1-, 3-, and 5-year survival outcomes. The area under the curve (AUC) values were 0.850 for 1-year survival, 0.779 for 3-year survival, and 0.857 for 5-year survival, indicating strong predictive performance of the IRGPI over these timeframes. (C) Calibration curves for the ability of the IRGPI to predict 1-, 3-, and 5-year prognoses. The calibration curves demonstrated the agreement between the predicted and observed survival outcomes at 1, 3, and 5 years. The C-index values for IRGPI prediction were 0.912, 0.829, and 0.813 for 1-, 3-, and 5-year survival, respectively, indicating a high degree of accuracy in prognostic prediction. (D) Nomogram for predicting 1-, 3-, and 5-year prognoses on the basis of the IRGPI and MVI. A nomogram incorporating the IRGPI and other prognostic factors, such as microvascular invasion (MVI), was constructed to predict patient survival at 1, 3, and 5 years. The nomogram provides a visual tool for clinicians to estimate individual patient survival probability on the basis of these combined factors.
Differences in clinical characteristics and immune infiltration between different IRGPI groups
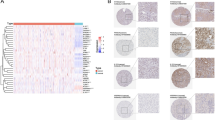
To further investigate the differences in the tumor immune microenvironment between the IRGPI groups, CIBERSORT was used to quantify immune infiltration. The analysis revealed that patients in the IRGPI-high group exhibited significantly lower levels of naive CD4+ T cells, which is typically associated with proinflammatory and antitumor responses, than those in the IRGPI-low group. Conversely, the IRGPI-high subgroup had significantly greater proportions of Treg cells, neutrophils, and other immune cells linked to an immunosuppressive microenvironment. These results suggest that the IRGPI-high group is characterized by a typical immunosuppressive microenvironment, potentially contributing to their worse prognosis (Fig. 4A). Additionally, a comprehensive heatmap analysis demonstrated that patients with high IRGPIs had significantly greater tumor thrombus and microvascular invasion (MVI) levels than did those with low IRGPIs (The exact clinical data of each patients was upload as supplementary table 1). These findings further underscore the critical role of the IRGPI in assisting in the evaluation of prognosis (Fig. 4B).
Analysis of clinical characteristics and immune infiltration in different IRGPI groups. (A) Immune cell infiltration analysis in patients with different IRGPIs. Significant differences in the level of naive CD4 + T cells, were observed in the IRGPI-low group. Conversely, higher levels of Treg cells and neutrophils were present in the IRGPI-high group than in the IRGPI-low group, indicating an immunosuppressive TME. The lower panel of Fig. 4A showed the representative immunohistochemical (IHC) results for CD4, CD68, and Foxp3. (B) Complex heatmap of clinical characteristics between different IRGPI groups. Patients in the IRGPI-high group had greater tumor thrombus and MVI levels, reflecting the prognostic value of the IRGPI. ns, not significant; *, p < 0.05; ***, p < 0.001; na, not existed according to the ssGSEA results.
Prediction of the efficacy of immunotherapy in patients with advanced bladder cancer or HCC via the IRGPI
We used data from the IMvigor210CoreBiologies package to further assess the predictive value of the IRGPI for immunotherapy outcomes. Tumor tissues from 10 patients were randomly selected, and immunohistochemical staining with PD-L1 (SP142, 28–8) and PD-1 antibodies was performed to evaluate the differences in immune checkpoint expression between the two groups. The results revealed a trend toward a decreased IRGPI in patients with complete or partial response (CR/PR) compared with those with progressive disease or stable disease (PD/SD) (Fig. 5A). In the IMvigor210 cohort, patients with a high IRGPI had poorer prognosis, which was consistent with our study cohort(Fig. 5B). According to the relevant regulations of the US FDA, when patients are planned to be treated with pembrolizumab, the PD-L1 (22C3) clone antibody should be used to detect PD-L1 expression. The PD-L1 (28–8) clone antibody is used for nivolumab, and the PD-L1 (SP263) or PD-L1 (SP142) clone antibody is used for atezolizumab. Therefore, we chose PD-L1 (SP142, 28–8) for immunohistochemical analysis to assess whether the IRGPI can aid in the evaluation of PD-L1 expression in HCC. The immunohistochemistry results demonstrated that patients in the IRGPI-high group had higher PD-L1 (SP142, 28–8) and PD-1 expression than did those in the IRGPI-low group (Fig. 5C).
Predictive value of the IRGPI for immunotherapy. (A) Distribution of IRGPI scores in the IMvigor210 cohort. A trend toward a decreased IRGPI in patients with complete or partial response (CR/PR) compared with those with progressive disease or stable disease (PD/SD) was observed. (B) OS was longer in patients with high IRGPI scores in the IMvigor210 cohort. (C) Immunohistochemistry results for PD-L1 (SP142, 28–8) and PD-1 expression. ***, p < 0.001.
Discussion
The expression of immune-related genes (IRGs) can characterize the tumor immune microenvironment, and tumor-infiltrating lymphocytes are key factors influencing the effectiveness of immunotherapy. In this study, key IRGs were identified through transcriptomic sequencing of HCC and adjacent tissues, leading to the establishment of the immune-related gene prognostic index (IRGPI). These findings indicated that this index could assist in the prognostic assessment of patients. Additionally, analysis of the IMvigor210 dataset and differences in PD-L1 and PD-1 expression among the different IRGPI groups further suggested that the IRGPI can be utilized to guide immunotherapy decisions for patients.
Transcriptome sequencing and bioinformatics analyses revealed significant downregulation of immune-related pathways in tumors, indicating that HCC may promote immune escape by downregulating immune pathways. In this study, the IRGPI is composed of six genes: COLEC12, CTSE, KLRD1, MMP12, NDRG1, and RELB. These six genes are different from those in former research, which might be explained by the differences of the patients chosen. COLEC12 encodes a member of the C-type lectin family involved in various immune functions, including pathogen recognition, phagocytosis, and immune response regulation. Studies suggest that COLEC12 expression is positively correlated with immune infiltration, indicating a potential role for COLEC12 in inhibiting the tumor immune response in gastric cancer12. CTSE encodes a pancreatic enzyme involved in antigen processing and presentation and has been identified as a key gene in a bladder cancer prognostic model13. KLRD1 encodes a surface receptor on NK cells and some T cells and has been found to be a protective factor in bladder cancer prognosis14. MMP12, which encodes matrix metalloproteinase-12, is involved in extracellular matrix degradation and remodeling; elevated MMP12 plasma levels have been associated with shorter overall survival in patients with lung squamous cell carcinoma15,16. NDRG1 regulates cell growth, differentiation, the stress response, and cancer development. In pancreatic ductal adenocarcinoma, NDRG1 overexpression was found to promote MHC-I upregulation on tumor cells, enhancing T-cell infiltration and activity and thereby boosting antitumor immunity17. RELB, encoding a transcription factor in the NF-κB family, plays crucial roles in the immune response, inflammation, cell survival, and development.
The composition of immune cells in the tumor microenvironment is both a prognostic factor and an indicator of immunotherapy decisions. High expression of inhibitory molecules such as PD-1/PD-L118, TIM-319, and LAG-320or infiltration by immunosuppressive cells such as Tregs, M2 macrophages, and neutrophils often indicates a poor prognosis due to effective immune escape by tumor cells. However, these inhibitory proteins and cells may also suggest better responses to immunotherapy21. CIBERSORT analysis revealed that patients in the IRGPI-high group had significantly lower levels of inflammatory cells, such as naive CD4+ T cells, and significantly greater proportions of immunosuppressive cells, such as Treg cells and neutrophils, than those in the IRGPI-low group. These results suggest that the IRGPI-high subgroup has a typical immunosuppressive microenvironment, which may partly explain the poorer prognosis of patients in this subgroup.
Data from the IMvigor210 cohort are commonly used to assess the predictive efficacy of models for immunotherapy outcomes22. The analysis revealed a trend toward increasing IRGPI scores in patients with effective treatment compared with those with disease progression. Furthermore, the different IRGPIs in the IMvigor210 dataset revealed significant prognostic differences, further underscoring the value of the IRGPI in guiding immunotherapy decisions for patients. However, the lack of statistical significance between the groups may be due to the limited generalizability of the results from HCC to other solid tumors. Further analysis of the role of the IRGPI in HCC patients receiving immunotherapy is needed. In clinical practice, the immunohistochemical detection of PD-1/PD-L1 expression is a common predictor for guiding immunotherapy in various cancers. Therefore, immune checkpoint expression was compared between different IRGPI groups. Statistical analysis revealed that the PD-L1 CPS was greater in patients with a high IRGPI, with a more significant difference in PD-1 expression. The manual interpretation of indicators such as PD-1 and PD-L1 can be challenging, and immunohistochemical detection also carries the risk of false-positives and false-negatives. Compared with conventional immunohistochemical markers, the IRGPI can be used to quantify the immune microenvironment composition, offering a more comprehensive reflection of a patient’s immune status, thereby guiding immunotherapy more effectively.
This study also has some limitations. Owing to the small numbers of HCC patients, we couldn’t divide them into the train and validation cohort to verify the reliability of the IRGPI, which can be verified in our further study. Meanwhile, these patients in our study didn’t receive immunotherapy after operation, therefore we couldn’t have the chance to compare their genes expression differences, but instead turned to the public dataset IMvigor210 cohort to validate the potential of IRGPIs in guiding immunotherapy. Subsequent study should be carried to evaluate the efficacy IRGPIs in predicting the efficacy of immunotherapy in IRGPI-low and IRGPI-high patients. Furthermore, future researches should focus on conducting multi-center validations, and exploring the application of IRGPI in other types of tumors.
In conclusion, this study established a 6-gene prognostic model based on transcriptomic data and the ImmPort database, which effectively evaluated patient prognosis. Additionally, this model may guide decisions regarding whether HCC patients should receive immunotherapy.
Data availability
The datasets generated and analyzed during the current study are available in the Genome Sequence Achieve of National Genomics Data Center (NGDC) [https://ngdc.cncb.ac.cn/gsa-human/browse/HRA000464].
References
Yang, C. et al. Evolving therapeutic landscape of advanced hepatocellular carcinoma. Nat. Rev. Gastroenterol. Hepatol. 20, 203–222. https://doi.org/10.1038/s41575-022-00704-9 (2023).
Yang, X. et al. Precision treatment in advanced hepatocellular carcinoma. Cancer Cell 42, 180–197. https://doi.org/10.1016/j.ccell.2024.01.007 (2024).
Gao, X. et al. Immunotherapy and drug sensitivity predictive roles of a novel prognostic model in hepatocellular carcinoma. Sci. Rep. 14, 9509. https://doi.org/10.1038/s41598-024-59877-9 (2024).
Stark, M. C., Joubert, A. M. & Visagie, M. H. Molecular farming of pembrolizumab and nivolumab. Int. J. Mol. Sci. https://doi.org/10.3390/ijms241210045 (2023).
Sia, D. et al. Identification of an immune-specific class of hepatocellular carcinoma. Based Mol. Featur. Gastroenterol. 153, 812–826. https://doi.org/10.1053/j.gastro.2017.06.007 (2017).
Li, L. et al. Implications of driver genes associated with a high tumor mutation burden identified using next-generation sequencing on immunotherapy in hepatocellular carcinoma. Oncol. Lett. 19, 2739–2748. https://doi.org/10.3892/ol.2020.11372 (2020).
Le, D. T. et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 357, 409–413. https://doi.org/10.1126/science.aan6733 (2017).
Hu, B., Yang, X. B. & Sang, X. T. Molecular subtypes based on immune-related genes predict the prognosis for hepatocellular carcinoma patients. Int. Immunopharmacol. 90, 107164. https://doi.org/10.1016/j.intimp.2020.107164 (2021).
Gu, X. et al. Model based on five tumour immune microenvironment-related genes for predicting hepatocellular carcinoma immunotherapy outcomes. J. Transl. Med. 19, 26. https://doi.org/10.1186/s12967-020-02691-4 (2021).
Yi, M. et al. Immune signature-based risk stratification and prediction of immune checkpoint inhibitor’s efficacy for lung adenocarcinoma. Cancer Immunol. Immunother. : CII 70, 1705–1719. https://doi.org/10.1007/s00262-020-02817-z (2021).
World Medical Association Declaration of Helsinki. ethical principles for medical research involving human subjects. Jama 310, 2191–2194. https://doi.org/10.1001/jama.2013.281053 (2013).
Li, G. Z., Deng, J. F., Qi, Y. Z., Liu, R. & Liu, Z. X. COLEC12 regulates apoptosis of osteosarcoma through Toll-like receptor 4-activated inflammation. J. Clin. Lab. Anal. 34, e23469. https://doi.org/10.1002/jcla.23469 (2020).
Chen, H., Yang, W., Li, Y., Ma, L. & Ji, Z. Leveraging a disulfidptosis-based signature to improve the survival and drug sensitivity of bladder cancer patients. Front. Immunol. 14, 1198878. https://doi.org/10.3389/fimmu.2023.1198878 (2023).
Shi, W., Dong, J., Zhong, B., Hu, X. & Zhao, C. Predicting the prognosis of bladder cancer patients through integrated multi-omics exploration of chemotherapy-related hypoxia genes. Mol. Biotechnol. https://doi.org/10.1007/s12033-024-01203-9 (2024).
Kerzeli, I. K. et al. Elevated levels of MMP12 sourced from macrophages are associated with poor prognosis in urothelial bladder cancer. BMC Cancer 23, 605. https://doi.org/10.1186/s12885-023-11100-0 (2023).
Zhang, W. et al. MMP12 serves as an immune cell-related marker of disease status and prognosis in lung squamous cell carcinoma. PeerJ 11, e15598. https://doi.org/10.7717/peerj.15598 (2023).
Zhang, Z. et al. NDRG1 overcomes resistance to immunotherapy of pancreatic ductal adenocarcinoma through inhibiting ATG9A-dependent degradation of MHC-1. Drug Res. Updat. : Rev. & Comment. Antimicrob. & Anticancer Chemother. 73, 101040. https://doi.org/10.1016/j.drup.2023.101040 (2024).
Tang, B. et al. Targeted xCT-mediated Ferroptosis and Protumoral Polarization of Macrophages Is Effective against HCC and Enhances the Efficacy of the Anti-PD-1/L1 Response. Adv. Sci. 10, e2203973. https://doi.org/10.1002/advs.202203973 (2023).
Sauer, N. et al. TIM-3 as a promising target for cancer immunotherapy in a wide range of tumors. Cancer Immunol. Immunothe. : CII 72, 3405–3425. https://doi.org/10.1007/s00262-023-03516-1 (2023).
Sauer, N. et al. LAG-3 as a potent target for novel anticancer therapies of a wide range of tumors. Int. J. Mol. Sci. https://doi.org/10.3390/ijms23179958 (2022).
Yi, M. et al. Exploiting innate immunity for cancer immunotherapy. Mol. Cancer 22, 187. https://doi.org/10.1186/s12943-023-01885-w (2023).
Xie, T. et al. Multi-cohort validation of Ascore: an anoikis-based prognostic signature for predicting disease progression and immunotherapy response in bladder cancer. Mol. Cancer 23, 30. https://doi.org/10.1186/s12943-024-01945-9 (2024).
Acknowledgements
We would like to express our sincere gratitude to Springer Nature Author Services (SNAS) for their expert language editing of our manuscript.
Funding
This study was sponsored by Fuzhou Science and Technology Bureau (2022-S-011), Fujian Provincial Health Commission Medical Innovation Research Project(2023CXA051).
Author information
Authors and Affiliations
Contributions
Chen Y and Chen LH conceived and designed the experiments. Liu XL, Wang M and Zhang YB performed the experiments. You PY analyzed the data. Chen Y and Chen LH wrote the paper. All authors contributed to the article and approved the submitted version.
Corresponding authors
Ethics declarations
Competing interests
The authors have no conflicts of interest to disclose.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
You, P., Liu, X., Wang, M. et al. Development and validation of an Immune-related Gene-based model for predicting prognosis and immunotherapy outcomes in hepatocellular carcinoma patients. Sci Rep 15, 6618 (2025). https://doi.org/10.1038/s41598-025-90183-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-90183-0