Abstract
Lung cancer is prevalent worldwide and is a major cause of cancer-related mortality. Despite being the primary model for immunotherapy research, the response rates of lung cancer patients to immunotherapy are unsatisfactory. Furthermore, research on immunogenic cell death (ICD) in lung cancer is limited, which limits the development of strategies that combine ICD-related therapies with immunotherapy. In this study, we compiled and summarized 69 genes associated with ICD and developed an IRS. Across seven independent datasets, the IRS was identified as an independent prognostic factor. IRS was positively associated with multiple tumor proliferation pathways and negatively associated with immune-related pathways. Additionally, IRS negatively correlated with the infiltration of various immune cells, supporting its association with survival outcomes. Based on the correlation between IRS and immune activity, we validated the ability of IRS to predict immunotherapy efficacy across seven immunotherapy datasets and demonstrated that patients who respond to immunotherapy tend to have a lower IRS. Moreover, utilizing single-cell RNA sequencing, we revealed the role of mast cells in the TME with the highest IRS. Through interactions with various receptors on macrophages, endothelial cells, and tumor cells, mast cells promote tumor progression, providing a comprehensive explanation for poor prognosis and lack of response to immunotherapy in patients with high IRS. Our study offers new guidance for combination therapies in lung adenocarcinoma patients and elucidated the mechanism by which mast cells contribute to cancer development within the TME.
Similar content being viewed by others
Introduction
Cancer remains the leading cause of death and a major barrier to improving life expectancy worldwide. Its prominence as the primary cause of death has become increasingly evident. Regardless of gender, lung cancer has the highest mortality rate among all cancers, significantly affecting global health1,2. Among all the pathological types, lung adenocarcinoma (LUAD) has revolutionized cancer treatment, extending the survival of patients who respond to it. However, immunotherapy remains effective only effective in a minority of patients3,4. Therefore, there is an urgent need to uncover the molecular mechanisms behind immunotherapy resistance to identify novel therapeutic strategies.
Immunogenic cell death (ICD) is a form of regulated cell death (RCD) distinct from other RCDs primarily by its ability to trigger specific antigen-mediated immune responses, which in turn leads to the formation of immunological memory. As dying cells release damage-associated molecular patterns (DAMPs), such as ATP, they recruit antigen-presenting cell (APC) precursors into the tumor microenvironment (TME) and promote the uptake of dying cancer cells by dendritic cells (DCs). This process activates a range of cytotoxic immune cells. In theory, this cascade can facilitate the strong infiltration of bone marrow-derived cells and lymphocytes into the tumor, transforming it from a “cold” to a “hot” tumor phenotype. Additionally, CD8 cytotoxic T lymphocytes (CTLs) attacking cancer cells can also initiate ICD, potentially creating a self-amplifying mechanism for antigen spread during local immune responses5,6,7,8. Given these characteristics, combining immunotherapy with ICD holds significant promise. This strategy engages T cells to target and kill tumor cells while fostering immune cell aggregation to enhance the efficacy of immunotherapy. However, a comprehensive understanding of the TME’s role in ICD, particularly in lung adenocarcinoma (LUAD) patients, remains insufficient.
To date, numerous prognostic models related to ICD have been established, but many suffer from incomplete gene sets, resulting in partial information loss9,10,11,12,13. we compiled a more comprehensive list of genes associated with ICD, constructed an ICD risk score (IRS), and validated the IRS as an independent prognostic factor across seven independent datasets. We conducted a thorough analysis of the relationships the IRS and tumor as well as immune characteristics. We compared the likelihood of ICD occurrence in LUAD patients with varying IRS levels, confirming the predictive value of IRS in immunotherapy. Additionally, using single-cell RNA sequencing technology, we found that the IRS was significantly greater IRS in the mast cells of LUAD patients. While previous research on mast cells has primarily focused on allergy-related diseases, their role in LUAD has received less attention14,15. Building on these findings, we performed a targeted analysis of mast cells within the TME, offering a more comprehensive explanation for the poor prognosis and limited efficacy of immunotherapy in patients with high IRS levels.
Materials and methods
Dataset collection and processing
Lung adenocarcinoma RNA-sequencing cohort collection
The RNA sequence expression and clinical data of LUAD patients were downloaded from The Cancer Genome Atlas (TCGA) (https://portal.gdc.cancer.gov/)16 and Gene-Expression Omnibus (GEO) (https://www.ncbi.nlm.nih.gov/geo/) (GSE1024517, GSE1481418, GSE4127119, GSE4212720, GSE6846521, GSE7209422 and GSE1321323).
Immunotherapy RNA-sequencing cohort collection
GSE93157: The data includes patients with melanoma, lung cancer, and head and neck cancer who received anti-PD-1 therapy. We selected 22 patients with lung adenocarcinoma for further analysis24.
Ravi A_2023: The dataset consists of patients with advanced non-small cell lung cancer who received anti-PD-1/PD-L1 therapy25.
GSE35640: The dataset consists of patients with advanced melanoma who received MAGE-A3 therapy26.
GSE91061: The dataset consists of patients with advanced melanoma who received anti-PD-1 therapy27.
Mariathasan S_2018: The dataset consists of patients with metastatic urothelial carcinoma who received anti-PD-L1 therapy28.
PRJEB23709: The dataset consists of patients with melanoma who received anti-PD1/CTLA4 therapy29.
phs001493: The dataset consists of patients with clear cell renal cell carcinoma who received anti-PD-1 therapy30.
Single cell RNA-sequencing cohort collection
The single-cell data for LUAD were obtained from GEO. The accession numbers are GSE17114531 (including 40,247 cells from nine LUAD patients), GSE18935732 (including 113,425 cells from nine LUAD patients) and GSE20742233 (including 92,265 cells from fifty LUAD patients who received anti-PD-1/PD-L1 therapy).
Construction of the IRS model and validation of its effect on prognosis
Among the 69 genes that were collected and curated, 54 were obtained from TCGA-LUAD. Based on these 54 genes, we performed least absolute shrinkage and selection operator (LASSO) regression analysis, and considering the model’s stability and simplicity, we constructed an ICD signature comprising 16 genes. The specific calculation method for the signature is the sum of the products of the log2-transformed expression values of each gene and the LASSO regression model coefficients. The ICD score was defined as the ICD risk score (IRS). The samples were divided into two groups, low IRS (LIRS) and High IRS (HIRS), based on the median IRS values. The R package “survival” was used to perform survival analysis. Subsequently, age, gender, and stage were included as variables, and a multivariate Cox regression model analysis was conducted to assess whether the IRS was an independent predictor in the treatment-naive cohort. These results were validated using seven LUAD datasets (TCGA-LUAD; GSE10245; GSE14814; GSE41271; GSE42127; GSE68465; and GSE72094).
Analysis of immunological characteristics and pathway enrichment
Based on the msigdb 50 hallmark gene set34, GSVA was used to compute the corresponding scores. The R package “MCP-counter” was employed to assess immune infiltration reflected by the abundance of microenvironment cell populations (MCPs)35. The cytolytic activity (CYT) scores were calculated using the gene expression of two cytolytic markers, GZMA and PRF136. The T-cell-inflamed gene expression profile (GEP) score of each sample was computed based on the GEP gene signature37, using GSVA. The Vall d’Hebron Institute of Oncology (VHIO) gene expression signature (VIGex) score for each sample was computed based on the VIGex gene signature38, through GSVA. The tumor mutational burden (TMB), and neoantigen information were obtained from Thorsson et al.39. Genes with log2FC > 0.5 and padj < 0.05 were selected as differentially expressed genes for Gene Ontology (GO) enrichment analysis.
Calculation of the mast cell score in the lung adenocarcinoma RNA-sequencing cohort
Based on the following two gene sets, we calculated the score for each sample using GSVA.
TNF+Mast: TPSB2, TPSAB1, KIT, CPA3, TNF.
VEGF+Mast: TPSB2, TPSAB1, KIT, CPA3, VEGFA.
The D-score was defined as the GSVA score (VEGFA + MC) minus the GSVA score (TNF + MC).
Lung adenocarcinoma single-cell RNA-sequencing procession
The “Seurat” R package (version 4.3.0) was used for conduct quality control according to the parameters specified in the literature31,32, resulting in 40,247 and 113,425 cells for further analysis. After selecting 2000 highly variable genes through the FindVariableFeatures function in Seurat, principal component analysis (PCA) was performed using these genes. Select the first 30 PCs to run the FindNeighbors and RunUMAP functions. The “Harmony” R package (version 0.1.1) was used to remove batch effects. The IRSs of all cells were calculated using Addmodulscore based on the ICD-signature genes. Monocle 2 were used for trajectory40.
Distinguishing between benign and malignant epithelial cells
To identify malignant cells, the copy number variation (CNV) of each subcluster of the epithelial cell cluster was analyzed using the “infercnv” R package (version 1.14.2)41. Fibroblasts, endotheliocytes (GSE189357), and B cells (GSE171145) were used as reference cells. For infercnv analysis, we used a cutoff value of 0.1 as the minimum average read count for each gene in the reference cells.
Cell-cell communication in single-cell data
To explore the potential interactions of mast cells with other cell types in the TME, we utilized the R package “CellChat” (version 1.6.1)42 and R package “nichenetr” (version 2.1.0)43.
Statistical analysis
For partial datasets, the “surv_cutpoint” function from the R package “survminer” was used to determine the optimal cutoff point and mitigate batch effects in calculations. Pearson’s correlation was employed to compute the correlation coefficient between the IRS and immune or hallmark features. The Wilcoxon rank-sum test was utilized to determine significant differences in the IRS between different groups. For multiple comparisons, the p-values were adjusted using the BH method for multiple testing. Statistical nonsignificance was set at p-values > 0.05.
Results
Construction of the IRS and its impact on LUAD patient survival
To investigate the role of ICD in LUAD patients, we identified 69 relevant genes from the literature. Among these, 54 were derived from the TCGA-LUAD dataset. Subsequent analyses focused on these 54 genes (Table 1)5,6,7,8,44. Initially, NMF clustering was applied to categorize TCGA-LUAD patients into two clusters based on the expression profiles of these genes, with an optimal k of 2 (Fig. 1A). The clustering results revealed that patients in cluster B exhibited a more favorable prognosis (Fig. 1B; HR = 0.6821 (95% CI: 0.5018–0.9227), p = 0.014). Next, we employed LASSO regression to identify 16 key genes for predicting survival. Among these, seven were identified as risk factors (HR > 0) and nine as protective factors (HR < 0) (Fig. 1C, Supplemental Fig. 1A, B). These genes were integrated into the construction of an ICD-related prognostic signature. The ICD score of Group A was significantly higher than that of Group B, leading us to hypothesize that patients with lower scores may have better survival outcomes (Fig. 1D).
By investigating the prognostic performance of the 16-gene ICD signature and stratifying all samples into high and low groups based on the median, we found that patients with higher ICD scores had worse overall survival (OS) and progression-free survival (PFS) (Fig. 1E, F; OS: HR = 2.49 (95% CI: 1.8185–3.4095), p < 0.0001; PFS: HR = 1.6223 (95% CI: 1.2274–2.1441), p = 6e−04). The ICD score was defined as the ICD risk score (IRS), and the TCGA-LUAD samples were divided into two groups: low IRS (LIRS) and high IRS (HIRS). Additionally, multivariate Cox regression analysis revealed that the IRS remained a significant predictor of survival, even after adjusting for well-established clinical factors such as age, gender, tumor stage, race, gene mutations, treatment history, and smoking status (Fig. 1H, Supplemental Fig. 2A). Furthermore, the prognostic value of the IRS was validated in six independent cohorts. Compared with HIRS, LIRS was associated with better OS (Fig. 1G, GSE10245: HR = 4.5691 (95% CI: 1.4656–14.2445), p = 0.0042; GSE14814: HR = 2.4201 (95% CI: 1.2116–4.8343), p = 0.01; GSE41271: HR = 1.8732 (95% CI: 1.1522–3.0454), p = 0.01; GSE42127: HR = 2.294 (95% CI: 1.2222–4.3057), p = 0.0079; GSE68465: HR = 1.4858 (95% CI: 1.1482–1.9227), p = 0.0024; GSE72094: HR = 1.9133 (95% CI: 1.3067–2.8016), p = 0.00071), and multivariate Cox regression analysis incorporating clinical factors confirmed these findings (Fig. 1H, Supplemental Fig. 2B–G). By comparing the ROC curve and calibration curve, the effectiveness of IRS is significantly superior to that of the classic biomarkers for lung adenocarcinoma (Supplemental Fig. 3A–F).
In summary, the IRS can predict the prognosis of LUAD patients. Based on the median IRS, we divided all the samples into HIRS and LIRS groups for further analyses.
Construction of the ICD risk model and its impact on LUAD patients survival. (A) NMF clustering heatmap divided the TCGA-LUAD cohort into clusters A and B. (B) Kaplan-Meier curves showed the survival outcomes of patients classified as A and B in TCGA-LUAD. (C) LASSO regression analysis was used to construct gene coefficients for the ICD risk score (IRS), where coefficients > 0 represent risk genes and coefficients < 0 represent protective genes. (D) The distribution of IRS across cluster A and cluster B. (E,F) Kaplan-Meier curves demonstrated the survival outcomes of HIRS and LIRS patients in the TCGA-LUAD. (E) Overall-survival (OS), (F) Progression-free survival (PFS). (G) Kaplan-Meier curves demonstrated the survival outcomes of HIRS and LIRS patients in the GEO-LUAD cohort (GSE10245, GSE14814, GSE41271, GSE42127, GSE68465, and GSE72094). (H) Forest plot showed that the IRS is an independent prognostic factor for lung adenocarcinoma patients across seven datasets, as determined by multivariate Cox regression analysis (*p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001).
IRS exhibited significant correlations with tumor and immune characteristics
To investigate the factors contributing to the significant differences in prognosis between LIRS and HIRS patients, we analyzed the correlations between IRS and tumor as well as immune characteristics. The IRS of late-stage (stage III/IV) patients was significantly higher than that of early-stage (stage I/II) patients (Fig. 2A). In terms of tumor differentiation, highly differentiated (G1/G2) patients had a lower IRS compared to poorly differentiated (G3) patients (Fig. 2B). The molecular subtypes of LUAD can be divided into terminal respiratory unit (TRU), proximal-proliferative (PP), and proximal-inflammatory (PI) subtypes. The TRU subtype is associated with a more favorable prognosis45,46, showing a lower IRS than both the PP and PI subtypes (Fig. 2C). Age does not appear to significantly correlated with IRS, as inconsistent results were observed across multiple datasets (Supplemental Fig. 4A–F). In terms of gender, although some datasets suggest that males have higher IRS, these differences were not statistically significant (Supplemental Fig. 4G–M). Analysis based on the GSE72094 dataset reveals that IRS is significantly higher in smokers compared to non-smokers (Supplemental Fig. 3N–P). This difference may be related to increased infiltration of CD8+ T cells and NK cells induced by smoking47,48. Additionally, we examined key LUAD mutations, including EGFR, KRAS, and TP53 (Supplemental Fig. 5A–C). No significant difference in IRS was observed between patients with or without EGFR/KRAS mutations, but patients with TP53 mutations exhibited a significantly higher IRS compared to those with wild-type TP53.
We conducted a comparative analysis of the correlation between the IRS and hallmark pathways across six datasets. Our findings revealed a positive correlation between IRS and cell proliferation-related pathways, such as “G2M_CHECKPOINT”, “MYC_TARGETS_V2”, and “DNA_REPAIR”. In contrast, immune-related pathways, including “INTERFERON_GAMMA_RESPONSE”, “IL6_JAK_STAT3_SIGNALING”, and “IL2_STAT5_SIGNALING”, showed a significantly negative correlation with the IRS (Fig. 2D). Furthermore, we observed that the infiltration of antitumor immune cells, such as T cells and dendritic cells, was negatively correlated with IRS across seven independent datasets (Fig. 2E). Subsequently, we assessed the correlation between IRS and the three immune cytotoxicity scores. The results demonstrated a negative correlation between IRS and CYT, GEP, and VIGex across multiple datasets (Fig. 2F–H). These three immune-related signatures were positively correlated with immune activity. Patients exhibiting higher scores were more likely to respond to immunotherapy and had a better prognosis.
The IRS is correlated with multiple clinical and immune features. (A) The distribution of the IRS across early (I/II) and late (III/IV) TNM stages. (B) The distribution of the IRS across high (G1/G2) and low (G3) differentiation group. (C) The distribution of the IRS across the TRU, PI, and PP. (D) Correlation analysis between the GSVA scores of the 50 Hallmark pathways and the IRS, the lung adenocarcinoma cohort comprised the TCGA-LUAD, GSE10245, GSE41271, GSE42127, GSE68465, and GSE72094). (E) Correlation analysis between immune cell infiltration levels and the IRS. Immune cell infiltration was computed using MCP-counter, and the lung adenocarcinoma cohort comprised the TCGA-LUAD, GSE10245, GSE14814, GSE41271, GSE42127, GSE68465, and GSE72094 cohorts. (F, G) Bar plot showing the correlation of CYT (F), GEP (G), VIGex (H) and the IRS, the LUAD cohort comprised the TCGA-LUAD, GSE10245, GSE14814, GSE41271, GSE42127, GSE68465, and GSE72094 cohorts (*p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001).
LIRS can be combined with ICD inducers and immunotherapy
ICD requires specific conditions, including immunogenicity (adjuvanticity, antigenicity) and the tumor immune microenvironment (TIME)5. In the absence of suitable antigens, cell death triggers inflammatory responses that do not engage the adaptive immunity. Antigenicity alone, without adjuvanticity, may lead to immune tolerance. Ultimately, the TIME plays a critical role in determining whether T cells can effectively eliminate tumor cells5. We compared the expression of DAMPs ligand receptors between the two groups and observed that LIRS increased the expression of multiple genes, particularly the ATP receptors P2RX7 and P2RY2 (Supplemental Fig. 5D). Furthermore, gene ontology (GO) enrichment analysis revealed that genes highly expressed in LIRS were enriched in purine nucleotide receptors and their associated downstream pathways (Supplemental Fig. 5E). In LIRS, ATP and its receptors are likely to activate immune cells, and the binding of ATP to its receptors is a crucial step in initiating ICD5. These findings suggest that, when considering only adjuvants, LIRS has a greater likelihood of inducing ICD. Regarding antigenicity, no significant difference were observed in TMB or neoantigen levels between the two groups (Supplemental Fig. 5F–H)39. However, LUAD displays a higher mutation frequency than other tumor types49. These generally correlates with the production of tumor-specific antigens, suggesting that patients with both HIRS and LIRS exhibit elevated immunogenicity. Immune cells directly involved in ICD (T cells, CD8+ T cells, cytotoxic lymphocytes, NK cells, and dendritic cells) were more abundant in LIRS patients (Fig. 2E). In addition, LIRS exhibited increased levels of CYT36, GEP37,and VIGex38 (Fig. 2F–H), indicating a stronger immune activation state. A comprehensive analysis suggests that, within the TIME, LIRS has a greater likelihood of facilitating ICD occurrence. Given the three critical conditions for ICD, LIRS appears to have a higher potential to promote ICD.
The above analysis revealed a significant negative correlation between the IRS and immune activity. The correlation between IRS and the expression levels of immuno-oncology target genes was analyzed. Some data showed a positive correlation between IRS and CD274, while a negative correlation was observed with CTLA4 (Fig. 3A). To further investigate the relationship between IRS, immunotherapy efficacy, and prognosis, we computed the IRS across multiple immunotherapy datasets. We found that patients who responded to immunotherapy had a lower IRS compared to non-responders (Fig. 3B–D). Additionally, a greater proportion of treatment responders than non-responders were observed in the LIRS group (Fig. 3E–G, (E) GSE93157: p = 0.036; (F) GSE35640: p = 0.022; (G) GSE91061: p = 0.008). Survival analysis further demonstrated that patients in the LIRS group had significantly better survival outcomes than those in the HIRS group (Fig. 3H–L, (H) GSE93157: HR = 5.3611 (95% CI: 1.394–20.6182), p = 0.0069; (I) Ravi A_2023: HR = 1.7474 (95% CI: 1.0691–2.856), p = 0.024; (J) Mariathasan S_2018: HR = 1.3391 (95% CI: 1.0154–1.766), p = 0.038; (K) PRJEB23709: HR = 4.0618 (95% CI: 1.2348–13.3618), p = 0.014; (L) phs001493: HR = 3.7218 (95% CI: 1.3791–10.0444 ), p = 0.0056). Collectively, these results indicate that the IRS possesses independent prognostic ability and predictive power for immunotherapy efficacy.
The role of the IRS in predicting the efficacy of immunotherapy. (A) The correlation between IRS and the expression of immuno-oncology target genes in seven independent lung adenocarcinoma datasets. (B–D) The distribution of IRS across different immune therapy responses (NR, nonresponder; R, responder). (B) GSE93157. (C) GSE35640. (D) GSE91061. (E–G) Stacked bar plots showed the proportions of responsive and nonresponsive patients in HIRS and LIRS. (E) GSE93157. (F) GSE35640. (G) GSE91061. (H–L) Kaplan-Meier curves showed the survival outcomes of patients classified as HIRS or LIRS in the immunotherapy cohort. (H) GSE93157. (I) Ravi A_2023. (J) Mariathasan S_2018. (K) PRJEB23709. (L) phs001493 (*p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001).
Mast cells mediate immune suppression
To investigate the factors contributing to immune suppression in HIRS patients, we conducted a detailed cellular-level analysis using two single-cell datasets from LUAD. By examining the expression patterns of specific markers for each cell subtype, we categorized all cells into seven major subgroups: myeloid cells (LYZ, CD68), fibroblasts (DCN, LUM), B cells (CD79A, MZB1), T cells (CD3D, CD3E), epithelial cells (KRT8, EPCAM), endothelial cells (CLDN5, RAMP2), and mast cells (CPA3, KIT) (Fig. 4A, C, Supplemental Fig. 6A–D). Next, we calculated the IRS for each cell type and observed that mast cells exhibited a significantly higher IRS compared to other cell types (Fig. 4B, D). Mast cells can be further categorized into two subtypes: TNF + MCs and VEGFA + MCs. TNF + MCs, which are linked to antitumor immunity and a favorable prognosis, and VEGFA + MCs, which are associated with a poorer prognosis50. Both independent single-cell datasets revealed that mast cells expressed high levels of VEGFA, while only a few cells expressed TNF (Fig. 4E–H).
According to TCGA-LUAD data, the HIRS group exhibited a greater D-score (Fig. 5A). A larger difference between VEGFA + MCs and TNF + MCs correlated with worse patient prognosis (Fig. 5B, HR = 1.4927 (95% CI: 1.1122–2.0034), p = 0.0072). After further annotation (Supplemental Figs. 7, 8), we conducted an in-depth analysis using CellChat to investigate the interactions between mast cells and other cell types, aiming to elucidate their critical roles in shaping the TME. The analysis revealed that mast cells primarily function as signal senders, engaging in pathways that promote tumorigenesis, such as the VEGF, CSF, and EGF pathways, (Fig. 5C–F, Supplemental Fig. 9A–E). Mast cell-derived VEGFA promotes angiogenesis by acting on endothelial cells, thereby facilitating tumor progression and reducing immune infiltration51,52 (Fig. 5D, F, Supplemental Fig. 9C, F). In the CSF pathway, mast cells almost entirely initiate signaling functions that have a strong macrophage-recruiting effect53. Compared to M1 macrophages, mast cells demonstrated a more significant recruitment effect on M2 macrophages (Fig. 5E, F, Supplemental Fig. 9D, F). Besides its role in recruiting macrophages, CSF1-CSF1R enables macrophages to acquire the immunosuppressive and tumor-promoting M2 subtype54,55. Additionally, mast cells can release migration inhibitory factor (MIF), which acts on macrophages (Fig. 5G). Mast cells recruit macrophages through multiple pathways and promote their polarization toward the M2 phenotype, thereby creating an immunosuppressive microenvironment. Furthermore, mast cells play a critical role in the EGF pathway by acting as signal senders through the release of AREG, which activates the EGFR pathway and directly promotes tumor proliferation56,57,58. (Fig. 5E, F, Supplemental Fig. 9E, F).
Single-cell transcriptomic analysis revealed that VEGFA is a characteristic biomarker of VEGFA + mast cells, with its expression level significantly higher in tumor-associated mast cell subsets compared to other cell types (Fig. 5G, H, Supplemental Fig. 9G, H). Notably, ligand-receptor interaction analysis using nichenetR demonstrated that among all potential ligands in the tumor microenvironment, VEGFA exhibited the highest binding potential to its receptor (Fig. 5I), suggesting its key role in mediating signaling pathways. Pseudotime analysis further confirmed a dynamic upregulation of VEGFA expression along the mast cell differentiation trajectory (Supplemental Fig. 9I, J). Based on this multi-dimensional evidence, we propose that VEGFA is a critical effector molecule in VEGFA + mast cells that promotes tumor progression. Based on the analysis of single-cell sequencing data from lung cancer patients treated with immune checkpoint inhibitors, the expression level of VEGFA in mast cells was significantly upregulated in the Non-Major Pathological Response (Non-MPR) group compared to the MPR group (Fig. 5J), while there was no statistical difference in TNF expression between the two groups (Supplemental Fig. 9K). These findings suggest that the key mechanism underlying the poor response to immunotherapy in patients with elevated IRS may involve the secretion of mast cell-derived VEGFA.
Through the analysis of the single-cell dataset, we identified that the key cell associated with poor prognosis in HIRS is the VEGFA + mast cell. Furthermore, this study uncovered the critical mechanisms by which these cells promote tumor progression and induce resistance to immunotherapy.
Mast cells exhibited the highest IRS in the LUAD single-cell cohort. (A, C) UMAP plots showed the major cell types. (A) GSE189357. (C) GSE171145. (B, D) The distribution of IRSs across different cell types. (B) GSE189357. (D) GSE171145. (E, G) Feature plots showed the expression of subtype markers for mast cells (TNF and VEGFA). (E) GSE189357. (G) GSE171145. (F, H) Violin plot showed the expression of subtype markers for mast cells (TNF and VEGFA). (F) GSE189357. (H) GSE171145 (*p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001).
VEGFAhi mast cells promote tumor progression through various cellular interactions. (A) The distribution of D-score between HIRS and LIRS. The D-score was defined as the GSVA score (VEGFA + MC) minus the GSVA score (TNF + MC). (B) Kaplan–Meier curve showed the survival outcomes of patients with high and low D-score in the TCGA-LUAD cohort. (C–E) Heatmaps showed the relative importance of each cell population based on the computed network centrality measures of different signaling pathway networks. (C) VEGF. (D) CSF. (E) EGF. (F) Bubble plot showed representative cell-cell signaling pathways from mast cells to other cells in GSE189357. (G,H) The expression levels of VEGFA in seven cell types. (G) GSE171145; (H) GSE189357. (I) Heatmap showing the predicted ligand activity by NicheNet on genes highly expressed in mast cell. (J) Box plot showing the expression levels of VEGFA in mast cells between Non-MPR (Major Pathological Response) and MPR samples.
Discussion
In this study, we established an IRS model to explore the association between the IRS and various tumor and immune characteristics. Patients with HIRS exhibited stronger tumor proliferation capabilities, whereas patients with LIRS demonstrated a more active immune status, which explains the association between IRS and poorer prognosis. Considering the three conditions of ICD: adjuvanticity, antigenicity, and the tumor immune microenvironment, we believe that LIRS is more likely to induce ICD. Although the IRS was not initially designed to predict the immune therapy response, our analysis of CYT, GEP, VIGex, and immune infiltration suggested that patients in the LIRS group were more likely to benefit from immune therapy. We validated this finding using multiple immune therapy datasets and found that the IRS can predict immune therapy efficacy, with a lower IRS indicating a greater likelihood of responding to immunotherapy. Integrated with single-cell data analysis, we found that mast cells had the highest IRS. Mast cells recruit macrophages and direct them toward the M2 phenotype, promoting angiogenesis and tumor cell proliferation. The above analysis was based on retrospective data, and further validation through prospective studies would add significant value.
We compared the distribution of IRS multiple clinical datasets. Our findings showed that later stages of the disease were associated with higher IRS, while poorly differentiated samples had lower IRS. These results align with the poor prognostic outcome typically associated with high IRS, indicating that a higher IRS corresponds to a greater clinical risk. Additionally, we examined the clear etiological factors of lung cancer, including smoking status. In a lung cancer dataset, we found that the IRS was significantly higher in smokers compared to non-smokers. This difference may be related to increased infiltration of CD8+ T cells and NK cells induced by smoking47,48, In the smoking group, both PD-L1 expression and tumor mutational burden (TMB) were significantly higher than in the non-smoking group59. Smokers were more likely to respond to immunotherapy, and our analysis also found that higher IRS (LIRS) was associated with an increased likelihood of response to immunotherapy. Regarding gender, IRS levels were generally higher in males than in females across multiple datasets, although this difference was not statistically significant. There were no significant differences in IRS based on age distribution. Currently, there is no available data on patients’ complications and environmental living conditions in relation to lung cancer, so we are unable to analyze any potential correlation between these factors and IRS.
ICD holds significant potential in cancer therapy. Following ICD activation, various immune cells infiltrate into the tumor, converting its phenotype from “cold” to “hot”. This aligns well with the principles of immunotherapy, suggesting that combining immunotherapy with ICD inducers could produce synergistic effects. In this study, we analyzed for the first time the potential for different patients to develop ICD. The occurrence of ICD depends on three conditions: adjuvanticity, antigenicity, and an suitable tumor immune microenvironment5. Data analysis reveals that LUAD patients have a high TMB, which is typically associated with the production of tumor-specific antigens49, suggesting that patients with both HIRS and LIRS exhibit elevated immunogenicity. During ICD, dying cells release ATP into the extracellular space. When ATP binds to its receptors, it promotes the infiltration and activation of various immune cells. Mature DCs present tumor-specific antigens to cytotoxic T cells, activating them to kill tumor cells, thereby triggering ICD5,6,7,8,60,61. However, ATP that does not bind to receptors is converted into adenosine by extracellular nucleotidases, such as NT5E and ENTPD1, which exert potent immunosuppressive functions62,63. Our analysis revealed that both ATP receptors (P2RX7 and P2RY2) were highly expressed in the LIRS group. Furthermore, enrichment analysis revealed that genes highly expressed in LIRS were enriched in purine nucleotide receptors and their associated downstream pathways. These results indicate that ATP in the LIRS group is more likely to bind to its receptors, P2RX7 and P2RY2, thereby initiating ICD. In terms of the tumor immune microenvironment, T cells, DCs, and cytotoxic lymphocytes were negatively correlated with the IRS in multiple datasets, and several immune-related pathways also showed a negatively correlated with the IRS. Based on the analysis of the three conditions required for ICD to occur, we hypothesize that the LIRS group has a greater potential for ICD. Further analysis of multiple immunotherapy datasets revealed that patients who responded to immunotherapy had lower IRS scores, and the proportion of responders in the LIRS group was significantly higher than that in the HIRS group. Survival curves indicated that the LIRS group exhibited better OS and PFS. Therefore, the LIRS group demonstrates a greater potential for ICD occurrence and is more likely to respond to immunotherapy. In the LIRS group, the combination of ICD inducers with immunotherapy could enhance the efficacy of both treatments. In this study, we included two cohorts of lung cancer patients receiving anti-PD-1/PD-L1 therapy. Owing to the limited number of lung cancer immunotherapy cohorts, we further expanded our analysis to incorporate multiple cohorts of patients with other tumor types who also received anti-PD-1/PD-L1 therapy. IRS showed a strong negative correlation with CTLA4; however, due to data limitations, we were unable to draw any conclusions regarding the potential of IRS as a predictor for anti-CTLA4 therapy response.
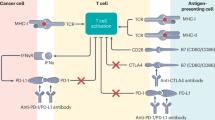
By analyzing two independent single-cell datasets, we identified mast cells that exhibited the highest IRS. Mast cells are tissue-resident immune cells that play a critical role in allergic inflammation through IgE-mediated mechanisms. They regulate the immune function by releasing stored granules and cytokines14,15,64. To date, research on mast cells has focused primarily on allergic diseases, with their role in tumors remaining incompletely understood. Previous studies classified mast cells into two groups: TNF + MCs (mast cells producing TNF) and VEGFA + MCs (mast cells overexpressing VEGFA)50. However, the specific mechanisms underlying these mast cell groups in the TME remain elusive. In our study, we found that mast cells with the highest IRS generally overexpressed VEGFA, which was associated with poor prognosis. Through cellular interaction analysis, we discovered that VEGFA + MCs recruit macrophages and promote their polarization toward the M2 phenotype via CSF1 and MIF. M2 macrophages play a strong immunosuppressive role in the TME. CSF1R inhibitors are widely used in clinical cancer therapy; inhibiting CSF1-CSF1R pathway reduces macrophage infiltration into tumors, diminishes M2 polarization, and improves patient survival54,55,65. Additionally, VEGFA + MCs interact with endothelial cells via VEGFA-VEGFR signaling, VEGFA promotes abnormal blood vessel growth, which obstructs effective blood flow and hampers immune cells, thereby reducing immune cell infiltration. Additionally, VEGFA negatively affects the maturation of dendritic cells and their antigen-presenting capacity, which limits the activation of tumor-specific T cells, which limits the activation of tumor-specific T cells57,66,67. This effect of VEGFA partly explains the significant negative correlation between IRS and immune cell infiltration57. Furthermore, VEGFA + MCs enhance tumor cell proliferation by binding AREG to EGFR in tumor cells. (Fig. 6). The above analysis is based on retrospective data, and the lack of experimental validation of the results is a clear limitation of our study. In future research, we will integrate multi-omics data with various experimental techniques to validate our analytical results.
In this study, we identified that VEGFA + mast cells play multiple roles in promoting tumor progression. However, research on mast cell-targeted therapies in oncology remains limited to date. We believe that targeting mast cells holds significant potential in anticancer treatment. Currently, numerous clinical trials have shown promising results when combining VEGF-targeting drugs with anti-PD1/PDL1 therapies68,69. Based on our findings, blocking VEGFA + mast cells partially inhibited abnormal angiogenesis and reduced M2 macrophage infiltration, thereby alleviating the strong immune suppression within the tumor microenvironment. Additionally, tumor cell proliferation was suppressed. We believe that combining the blockade of VEGFA + mast cells with existing immune therapies could substantially enhance treatment outcomes and benefit more patients. Nevertheless, mast cells in the tumor microenvironment have a dual function: they can both promote tumor growth and suppress tumor progression. Further research is needed to explore VEGFA + mast cells in greater detail, identify specific blocking targets, and minimize potential adverse effects. In the next phase of our research, we plan to conduct more in-depth studies on mast cells.
Conclusions
Immunogenic cell death can serve as a prognostic factor for survival and immunotherapy response. VEGFA + mast cells promote tumor progression through various pathways. Patients with different IRS may benefit from tailored drug treatments to improve survival outcomes.
Data availability
All data generated or analysed during this study are included in this published article.
References
Siegel, R. L., Giaquinto, A. N. & Jemal, A. Cancer statistics, 2024. CA Cancer J. Clin. 74, 12–49. https://doi.org/10.3322/caac.21820 (2024).
Sung, H. et al. Global Cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71, 209–249. https://doi.org/10.3322/caac.21660 (2021).
Morad, G., Helmink, B. A., Sharma, P. & Wargo, J. A. Hallmarks of response, resistance, and toxicity to immune checkpoint Blockade. Cell 184, 5309–5337. https://doi.org/10.1016/j.cell.2021.09.020 (2021).
Guo, H., Hou, Y., Wang, C. & Ding, J. How to optimize the immune checkpoint blockade therapy for cancers? Oncologie 26, 343–348. https://doi.org/10.1515/oncologie-2024-0037 (2024).
Kroemer, G., Galassi, C., Zitvogel, L. & Galluzzi, L. Immunogenic cell stress and death. Nat. Immunol. 23, 487–500. https://doi.org/10.1038/s41590-022-01132-2 (2022).
Galluzzi, L., Buque, A., Kepp, O., Zitvogel, L. & Kroemer, G. Immunogenic cell death in cancer and infectious disease. Nat. Rev. Immunol. 17, 97–111. https://doi.org/10.1038/nri.2016.107 (2017).
Galluzzi, L. et al. Consensus guidelines for the definition, detection and interpretation of Immunogenic cell death. J. Immunother Cancer. 8. https://doi.org/10.1136/jitc-2019-000337 (2020).
Fucikova, J. et al. Detection of Immunogenic cell death and its relevance for cancer therapy. Cell. Death Dis. 11, 1013. https://doi.org/10.1038/s41419-020-03221-2 (2020).
Wang, X. et al. An Immunogenic cell death-related classification predicts prognosis and response to immunotherapy in head and neck squamous cell carcinoma. Front. Immunol. 12, 781466. https://doi.org/10.3389/fimmu.2021.781466 (2021).
Liu, Z. et al. An Immunogenic cell death-related signature predicts prognosis and immunotherapy response in stomach adenocarcinoma. Apoptosis 28, 1564–1583. https://doi.org/10.1007/s10495-023-01879-5 (2023).
Yang, J. et al. Integration of single-cell RNA sequencing and bulk RNA sequencing to reveal an Immunogenic cell death-related 5-gene panel as a prognostic model for osteosarcoma. Front. Immunol. 13, 994034. https://doi.org/10.3389/fimmu.2022.994034 (2022).
Tian, Z., Li, X. & Jiang, D. Analysis of immunogenic cell death in atherosclerosis based on scRNA-seq and bulk RNA-seq data. Int. Immunopharmacol. 119, 110130. https://doi.org/10.1016/j.intimp.2023.110130 (2023).
Gan, X., Tang, X. & Li, Z. Identification of Immunogenic cell-death-related subtypes and development of a prognostic signature in gastric cancer. Biomolecules 13. https://doi.org/10.3390/biom13030528 (2023).
Wilcock, A., Bahri, R., Bulfone-Paus, S. & Arkwright, P. D. Mast cell disorders: from infancy to maturity. Allergy 74, 53–63. https://doi.org/10.1111/all.13657 (2019).
Falduto, G. H., Pfeiffer, A., Luker, A., Metcalfe, D. D. & Olivera, A. Emerging mechanisms contributing to mast cell-mediated pathophysiology with therapeutic implications. Pharmacol. Ther. 220, 107718. https://doi.org/10.1016/j.pharmthera.2020.107718 (2021).
Cancer Genome Atlas Research. The Cancer genome atlas Pan-Cancer analysis project. Nat. Genet. 45, 1113–1120. https://doi.org/10.1038/ng.2764 (2013).
Kuner, R. et al. Global gene expression analysis reveals specific patterns of cell junctions in non-small cell lung cancer subtypes. Lung Cancer 63, 32–38. https://doi.org/10.1016/j.lungcan.2008.03.033 (2009).
Zhu, C. Q. et al. Prognostic and predictive gene signature for adjuvant chemotherapy in resected non-small-cell lung cancer. J. Clin. Oncol. 28, 4417–4424. https://doi.org/10.1200/JCO.2009.26.4325 (2010).
Sato, M. et al. Human lung epithelial cells progressed to malignancy through specific oncogenic manipulations. Mol. Cancer Res. 11, 638–650. https://doi.org/10.1158/1541-7786.MCR-12-0634-T (2013).
Tang, H. et al. A 12-gene set predicts survival benefits from adjuvant chemotherapy in non-small cell lung cancer patients. Clin. Cancer Res. 19, 1577–1586. https://doi.org/10.1158/1078-0432.CCR-12-2321 (2013).
Director’s Challenge Consortium for the Molecular Classification of Lung Adenocarcinoma. Gene expression-based survival prediction in lung adenocarcinoma: a multi-site, blinded validation study. Nat. Med. 14, 822–827. https://doi.org/10.1038/nm.1790 (2008).
Schabath, M. B. et al. Differential association of STK11 and TP53 with KRAS mutation-associated gene expression, proliferation and immune surveillance in lung adenocarcinoma. Oncogene 35, 3209–3216. https://doi.org/10.1038/onc.2015.375 (2016).
Tomida, S. et al. Relapse-related molecular signature in lung adenocarcinomas identifies patients with dismal prognosis. J. Clin. Oncol. 27, 2793–2799. https://doi.org/10.1200/JCO.2008.19.7053 (2009).
Prat, A. et al. Immune-related gene expression profiling after PD-1 Blockade in non-small cell lung carcinoma, head and neck squamous cell carcinoma, and melanoma. Cancer Res. 77, 3540–3550. https://doi.org/10.1158/0008-5472.CAN-16-3556 (2017).
Ravi, A. et al. Genomic and transcriptomic analysis of checkpoint Blockade response in advanced non-small cell lung cancer. Nat. Genet. 55, 807–819. https://doi.org/10.1038/s41588-023-01355-5 (2023).
Ulloa-Montoya, F. et al. Predictive gene signature in MAGE-A3 antigen-specific cancer immunotherapy. J. Clin. Oncol. 31, 2388–2395. https://doi.org/10.1200/JCO.2012.44.3762 (2013).
Riaz, N. et al. Tumor and microenvironment evolution during immunotherapy with nivolumab. Cell 171, 934–949. https://doi.org/10.1016/j.cell.2017.09.028 (2017). e916.
Mariathasan, S. et al. TGFbeta attenuates tumour response to PD-L1 Blockade by contributing to exclusion of T cells. Nature 554, 544–548. https://doi.org/10.1038/nature25501 (2018).
Gide, T. N. et al. Distinct immune cell populations define response to anti-PD-1 monotherapy and anti-PD-1/anti-CTLA-4 combined therapy. Cancer Cell 35, 238–255 e236. https://doi.org/10.1016/j.ccell.2019.01.003 (2019).
Miao, D. et al. Genomic correlates of response to immune checkpoint therapies in clear cell renal cell carcinoma. Science 359, 801–806. https://doi.org/10.1126/science.aan5951 (2018).
Yang, L. et al. Single-cell transcriptome analysis revealed a suppressive tumor immune microenvironment in EGFR mutant lung adenocarcinoma. J. Immunother. Cancer 10. https://doi.org/10.1136/jitc-2021-003534 (2022).
Zhu, J. et al. Delineating the dynamic evolution from preneoplasia to invasive lung adenocarcinoma by integrating single-cell RNA sequencing and Spatial transcriptomics. Exp. Mol. Med. 54, 2060–2076. https://doi.org/10.1038/s12276-022-00896-9 (2022).
Hu, J. et al. Tumor microenvironment remodeling after neoadjuvant immunotherapy in non-small cell lung cancer revealed by single-cell RNA sequencing. Genome Med. 15, 14. https://doi.org/10.1186/s13073-023-01164-9 (2023).
Liberzon, A. et al. The molecular signatures database (MSigDB) hallmark gene set collection. Cell. Syst. 1, 417–425. https://doi.org/10.1016/j.cels.2015.12.004 (2015).
Becht, E. et al. Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol. 17, 218. https://doi.org/10.1186/s13059-016-1070-5 (2016).
Rooney, M. S., Shukla, S. A., Wu, C. J., Getz, G. & Hacohen, N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell 160, 48–61. https://doi.org/10.1016/j.cell.2014.12.033 (2015).
Ayers, M. et al. IFN-gamma-related mRNA profile predicts clinical response to PD-1 Blockade. J. Clin. Investig. 127, 2930–2940. https://doi.org/10.1172/JCI91190 (2017).
Hernando-Calvo, A. et al. A pan-cancer clinical platform to predict immunotherapy outcomes and prioritize immuno-oncology combinations in early-phase trials. Medicine 4, 710–727 e715. https://doi.org/10.1016/j.medj.2023.07.006 (2023).
Thorsson, V. et al. The immune landscape of cancer. Immunity 48, 812–830 e814. https://doi.org/10.1016/j.immuni.2018.03.023 (2018).
Qiu, X. et al. Single-cell mRNA quantification and differential analysis with census. Nat. Methods 14, 309–315. https://doi.org/10.1038/nmeth.4150 (2017).
Venteicher, A. S. et al. Decoupling genetics, lineages, and microenvironment in IDH-mutant gliomas by single-cell RNA-seq. Science 355. https://doi.org/10.1126/science.aai8478 (2017).
Jin, S. et al. Inference and analysis of cell-cell communication using cellchat. Nat. Commun. 12, 1088. https://doi.org/10.1038/s41467-021-21246-9 (2021).
Browaeys, R., Saelens, W. & Saeys, Y. NicheNet: modeling intercellular communication by linking ligands to target genes. Nat. Methods 17, 159–162. https://doi.org/10.1038/s41592-019-0667-5 (2020).
Garg, A. D., De Ruysscher, D. & Agostinis, P. Immunological metagene signatures derived from Immunogenic cancer cell death associate with improved survival of patients with lung, breast or ovarian malignancies: A large-scale meta-analysis. Oncoimmunology 5, e1069938. https://doi.org/10.1080/2162402X.2015.1069938 (2016).
Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature 511, 543–550. https://doi.org/10.1038/nature13385 (2014).
Hanley, C. J. et al. Single-cell analysis reveals prognostic fibroblast subpopulations linked to molecular and immunological subtypes of lung cancer. Nat. Commun. 14, 387. https://doi.org/10.1038/s41467-023-35832-6 (2023).
Saetta, M. et al. CD8+ T-lymphocytes in peripheral airways of smokers with chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 157, 822–826. https://doi.org/10.1164/ajrccm.157.3.9709027 (1998).
Stolberg, V. R. et al. Role of CC chemokine receptor 4 in natural killer cell activation during acute cigarette smoke exposure. Am. J. Pathol. 184, 454–463. https://doi.org/10.1016/j.ajpath.2013.10.017 (2014).
Lawrence, M. S. et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 499, 214–218. https://doi.org/10.1038/nature12213 (2013).
Cheng, S. et al. A pan-cancer single-cell transcriptional atlas of tumor infiltrating myeloid cells. Cell 184, 792–809 e723. https://doi.org/10.1016/j.cell.2021.01.010 (2021).
Goel, H. L. & Mercurio, A. M. VEGF targets the tumour cell. Nat. Rev. Cancer 13, 871–882. https://doi.org/10.1038/nrc3627 (2013).
Sun, X. et al. Direct extracellular NAMPT involvement in pulmonary hypertension and vascular remodeling. Transcriptional regulation by SOX and HIF-2alpha. Am. J. Respir Cell. Mol. Biol. 63, 92–103. https://doi.org/10.1165/rcmb.2019-0164OC (2020).
Noy, R. & Pollard, J. W. Tumor-associated macrophages: from mechanisms to therapy. Immunity 41, 49–61. https://doi.org/10.1016/j.immuni.2014.06.010 (2014).
Pyonteck, S. M. et al. CSF-1R Inhibition alters macrophage polarization and blocks glioma progression. Nat. Med. 19, 1264–1272. https://doi.org/10.1038/nm.3337 (2013).
Cannarile, M. A. et al. Colony-stimulating factor 1 receptor (CSF1R) inhibitors in cancer therapy. J. Immunother. Cancer 5, 53. https://doi.org/10.1186/s40425-017-0257-y (2017).
Stahler, A. et al. Amphiregulin expression is a predictive biomarker for EGFR Inhibition in metastatic colorectal cancer: combined analysis of three randomized trials. Clin. Cancer Res. 26, 6559–6567. https://doi.org/10.1158/1078-0432.CCR-20-2748 (2020).
Bu, M. T., Chandrasekhar, P., Ding, L. & Hugo, W. The roles of TGF-beta and VEGF pathways in the suppression of antitumor immunity in melanoma and other solid tumors. Pharmacol. Ther. 240, 108211. https://doi.org/10.1016/j.pharmthera.2022.108211 (2022).
Levantini, E., Maroni, G., Del Re, M. & Tenen, D. G. EGFR signaling pathway as therapeutic target in human cancers. Semin. Cancer Biol. 85, 253–275. https://doi.org/10.1016/j.semcancer.2022.04.002 (2022).
LoPiccolo, J., Gusev, A., Christiani, D. C. & Janne, P. A. Lung cancer in patients who have never smoked - an emerging disease. Nat. Rev. Clin. Oncol. 21, 121–146. https://doi.org/10.1038/s41571-023-00844-0 (2024).
Kepp, O. et al. ATP and cancer immunosurveillance. EMBO J. 40, e108130. https://doi.org/10.15252/embj.2021108130 (2021).
Di Virgilio, F., Sarti, A. C., Falzoni, S., De Marchi, E. & Adinolfi, E. Extracellular ATP and P2 purinergic signalling in the tumour microenvironment. Nat. Rev. Cancer. 18, 601–618. https://doi.org/10.1038/s41568-018-0037-0 (2018).
Boison, D. & Yegutkin, G. G. Adenosine metabolism: emerging concepts for cancer therapy. Cancer Cell 36, 582–596. https://doi.org/10.1016/j.ccell.2019.10.007 (2019).
Moesta, A. K., Li, X. Y. & Smyth, M. J. Targeting CD39 in cancer. Nat. Rev. Immunol. 20, 739–755. https://doi.org/10.1038/s41577-020-0376-4 (2020).
Marshall, J. S. Mast-cell responses to pathogens. Nat. Rev. Immunol. 4, 787–799. https://doi.org/10.1038/nri1460 (2004).
Joyce, J. A. & Pollard, J. W. Microenvironmental regulation of metastasis. Nat. Rev. Cancer. 9, 239–252. https://doi.org/10.1038/nrc2618 (2009).
Ferrara, N. & Adamis, A. P. Ten years of anti-vascular endothelial growth factor therapy. Nat. Rev. Drug Discov. 15, 385–403. https://doi.org/10.1038/nrd.2015.17 (2016).
Lee, W. S., Yang, H., Chon, H. J. & Kim, C. Combination of anti-angiogenic therapy and immune checkpoint Blockade normalizes vascular-immune crosstalk to potentiate cancer immunity. Exp. Mol. Med. 52, 1475–1485. https://doi.org/10.1038/s12276-020-00500-y (2020).
Park, S. et al. Phase III, randomized study of Atezolizumab plus bevacizumab and chemotherapy in patients with EGFR- or ALK-mutated non-small-cell lung cancer (ATTLAS, KCSG-LU19-04). J. Clin. Oncol. 42, 1241–1251. https://doi.org/10.1200/JCO.23.01891 (2024).
Socinski, M. A. et al. IMpower150 final overall survival analyses for Atezolizumab plus bevacizumab and chemotherapy in first-line metastatic nonsquamous NSCLC. J. Thorac. Oncol. 16, 1909–1924. https://doi.org/10.1016/j.jtho.2021.07.009 (2021).
Acknowledgements
Not applicable.
Funding
This work was supported by National Natural Science Foundation of China (82273215, 82472986), China Postdoctoral Science Foundation Funded Project (2021M693558, 2022T150741), Innovation-Driven Project of Central South University (2020CX043), Outstanding Youth Fund of Natural Science Foundation of Hunan Province (2021JJ20092), Project of scientific research plan of Hunan Provincial Health Commission (202204022468).
Author information
Authors and Affiliations
Contributions
Meng Zhang: Conceptualization, Methodology, Software, Formal analysis, Investigation, Writing-original draft, Writing-review & editing, Visualization; Guowei Zhou: Methodology, Software, Validation, Investigation; Yantao Xu: Software, Validation, Investigation; Benliang Wei: Methodology, Software, Validation, Investigation; Qian Liu : Methodology, Software, Validation; Guanxiong Zhang: Conceptualization, Methodology, Software, Resources, Data Curation, Writing-review & editing, Supervision; Ruimin Chang: Conceptualization, Resources,Data Curation, Writing-review & editing, Supervision, Project administration, Funding acquisition.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhang, M., Zhou, G., Xu, Y. et al. Immunogenic cell death signature predicts survival and reveals the role of VEGFA + Mast cells in lung adenocarcinoma. Sci Rep 15, 7213 (2025). https://doi.org/10.1038/s41598-025-91401-5
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-91401-5