Abstract
Background
Since the emergence of COVID-19 at the end of 2019, the disease has led to widespread acute respiratory distress syndrome (ARDS), particularly among kidney transplant recipients (KTRs), who are at increased risk due to long-term immunosuppressive therapy. This study aims to explore the differences in immune responses between kidney transplant recipients and non-kidney transplant recipients in COVID-19-induced ARDS to identify potential therapeutic targets for improving outcomes. Single-cell RNA sequencing was performed on 108,320 cells derived from peripheral blood samples to construct a global single-cell map of COVID-19 induced ARDS in kidney transplant recipients(ARDSKT), COVID-19 induced ARDS in non transplant recipients(ARDSNKT), and healthy controls. Subsequently, using cellular clustering analysis, we obtained single-cell maps of different cell types. We employed enrichment analysis to determine the pathways involved in different subpopulations and focused on the role of key immune cells such as monocytes, megakaryocytes, B cells, and CD8+ T cells in the pathogenesis of ARDS. Significant immune differences were observed between ARDSKT and ARDSNKT. In ARDSKT, the S100A9+ MK subpopulation, which activates the NF-κB signaling pathway, was elevated, promoting inflammation. In contrast, the S100A12+ monocyte subpopulation that activates the chemokine signaling pathway was more abundant in ARDSNKT, reflecting a stronger inflammatory response, while its abundance was reduced in ARDSKT due to immunosuppression. The CXCR4+ B subpopulation, crucial for adaptive immunity, was significantly reduced in ARDSKT. Additionally, the XAF1+ Teff subpopulation, associated with apoptosis, was more abundant in ARDSKT, potentially impairing immune recovery. This study highlights the immune differences between ARDSKT and ARDSNKT, revealing the impact of immunosuppression on immune dysregulation. These findings suggest that targeting specific immune pathways can improve therapeutic strategies for ARDSKT.
Similar content being viewed by others
Background
Since its first report at the end of 2019, the coronavirus disease 2019 (COVID-19), caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has rapidly spread globally, becoming one of the most serious public health crises of this century1. Since the outbreak of COVID-19 in 2019, there have been numerous cases of acute respiratory distress syndrome (ARDS), with an incidence rate of approximately 40%2. ARDS is a complex clinical syndrome characterized by alveolar injury and an inflammatory response, which includes pathological changes such as pulmonary edema, hyaline membrane formation, and alveolar hemorrhage3. Despite the implementation of standard treatment methods, such as lung-protective ventilation strategies and conservative fluid management, the mortality rate of ARDS has decreased3. However, there remains a lack of effective pharmacological treatment options. Notably, some ARDS patients are kidney transplant recipients (KTRs) who require long-term use of immunosuppressants to reduce the risk of organ rejection4. This immunosuppression renders KTRs patients more susceptible to SARS-CoV-2 infection, thereby increasing both the incidence and mortality of ARDS, and complicating its treatment5. The purpose of this study is to investigate the immune cell response in ARDS caused by COVID-19, with a particular focus on the KTR population, in order to optimize treatment outcomes through the modulation of immunosuppressive strategies.
There are significant differences in the pathophysiological mechanisms between COVID-19-induced ARDS and traditional ARDS, which may influence treatment strategies and prognosis. Unlike ARDS caused by other factors, COVID-19-induced ARDS typically develops later (8 to 12 days post-infection), and lung compliance may be normal or even elevated, primarily due to damage to alveolar epithelial cells resulting in transparent membrane formation, interstitial edema, and fibroblast proliferation6. The pathological processes of ARDS are closely related to alterations in peripheral immune activity, characterized by a decrease in the number of T and B cells, accompanied by an increase in plasma cells7,8. Concurrently, excessive activation of inflammatory monocytes and megakaryocytes (MK) may lead to cytokine storms and subsequent tissue damage9,10. However, their specific roles in the development of COVID-19-induced ARDS remain unclear.
This study systematically investigated the differences in immune environment between ARDS non-kidney transplant recipients (ARDSNKT) and ARDS kidney transplant recipients (ARDSKT) using single-cell RNA sequencing technology. The core objective of the study is to reveal the differential role of immune cells in the pathogenesis of ARDS between these two populations and evaluate the potential application of these differences in clinical practice. This study aims to deepen our understanding of the biological characteristics of ARDSNKT and ARDSKT immune cells through in-depth analysis, and further elucidate the complexity of their immune microenvironment.
Methods
Source of material
The study adhered to the principles outlined in the Declaration of Helsinki. The participants provided written informed consent before sample collection following standard procedures. This study included 18 peripheral blood samples, including 8 COVID-19 induced ARDS patients who did not receive kidney transplantation(ARDSNKT), 5 COVID-19 induced ARDS patients who received kidney transplantation(ARDSKT), and 5 healthy individuals (Control) (Additional file 1: Table S1).
Sample pre-processing, library preparation, and sequencing
Based on our previous study11, single-cell suspensions were obtained from whole-blood samples via density centrifugation. Subsequently, the DNBelab C Series Single-Cell Library Prep Set (MGI Tech, Shenzhen, China) was used to prepare single-cell RNA sequencing (scRNA-seq) libraries, following an established method. Briefly, single-cell suspensions were subjected to droplet encapsulation, emulsion breakage, collection of mRNA-captured beads, reverse transcription, cDNA amplification, and purification, resulting in the generation of barcoded scRNA-seq libraries. Indexed sequencing libraries were constructed according to the manufacturer’s instructions. Finally, the libraries were sequenced to obtain single-cell sequencing data.
Processing of scRNA-seq data
The Seurat package based on the R language12 was used for quality control (QC) of scRNA-seq data, filtering out the 1% of cells with the highest and lowest levels of gene expression and those with mitochondrial gene expression of > 10%. Subsequently, the results obtained from QC were integrated and clustered, the clustering results were identified, and a comprehensive single-cell atlas was generated. The IntegrateData function was used to integrate all single-cell data. Next, a Uniform Manifold Approximation and Projection (UMAP) algorithm was used for graph-based clustering and nonlinear dimension reduction to visually display similar cell populations13.
Cell and cell subpopulation clustering analyses
Based on cell marker-related databases, laboratory identifications, and classical cell markers identified in previous studies, the captured cell clusters were annotated as several major cell types8,14, including B cells, CD8+ T cells, erythroids, monocytes, MK, naive T cells, and innate lymphoid cells (ILC). Subsequently, the Seurat package based on the R language12 was used to perform dimension reduction clustering of different cell types to analyze the unique variations of each subpopulation. We manually annotated the subpopulations by combining disease-related gene markers to obtain detailed subpopulation information. The clustering results were uniformly visualized using UMAP.
Functional and gene set enrichment analysis
This analysis was performed using the clusterProfiler package15 in R. The Kyoto Encyclopedia of Genes and Genomes (KEGG) was used for enrichment analysis of each cell subpopulation to determine their potential biological functions16. Enriched pathways were considered statistically significant at P < 0.05.
Pseudotime analysis
The Monocle3 software package based on R17 was used to reveal cell trajectories based on gene expression changes to study and understand the processes of cell development, differentiation, and transcriptional dynamics. The results were visualized using the UMAP algorithm based on cell clusters for dimension reduction and visualization.
Gene regulatory network (GRN) analysis
The GRN determines and maintains the transcriptional specificity of cell types, aiding in understanding how cell characteristics are established, maintained, and disrupted in diseases18. To explore the vital regulatory factors driving and maintaining cell state behaviors, we analyzed and reconstructed a GRN centered on transcription factors (TFs) using the Python module tool pySCENIC19.
Intercellular communication analysis
The iTALK package20, based on the R language, was used to identify highly credible ligand-receptor interactions between cell subpopulations. This tool prioritizes the identification of highly or differentially expressed genes in cell clusters, which are subsequently matched using a ligand-receptor database (https://doi.org/10.1101/507871) to identify crucial intercellular communication events. For further analysis, we divided the gene pairs into four groups, namely, cytokines, immune checkpoint genes, growth factors, and others.
Data analysis and statistics
All bioinformatic analyses in this study were conducted using the BioinforCloud platform (http://www.bioinforcloud.org.cn). Statistical analyses were performed using the R software (version 4.0.5) for all studies. Student’s t-test was used to analyze the statistical differences between the three groups, and correlation coefficients were calculated using Spearman analysis21. Differences were considered statistically significant at P value < 0.05.
Results
scRNA-seq overview and identification of major cell types in ARDS
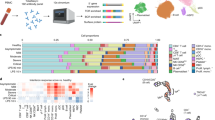
The workflow of the study is illustrated in Fig. 1A. We identified 108,320 high-quality single-cell transcriptomes using scRNA-seq and clustered them into 35 distinct cell clusters to construct a comprehensive single-cell atlas for COVID-19. This dataset comprises 35,505 cells from 8 samples of ARDS non-kidney transplant recipients (ARDSNKT), 23,236 cells from 5 samples of ARDS kidney transplant recipients (ARDSKT), and 49,575 cells from 5 control samples. Based on their unique gene markers, we identified seven distinct cell types: B cells, CD8+ T cells, erythroid cells, monocytes, MK, naive T cells, and ILC) (Fig. 1B-C). We further examined the expression levels and localization of known marker genes to support the annotation of each cell type (Fig. 1D-E). Compared to the control group, we observed increased abundances of B cells, MK, and monocytes, while the abundance of T cells decreased in both patient groups. Notably, ARDSNKT exhibited higher abundances of CD8+ T cells, B cells, MK, and naive T cells than ARDSKT (Fig. 1F), suggesting that ARDSKT may exhibit a diminished immune response. In summary, we observed significant differences in the abundance of cell types among different groups and conducted in-depth exploration of the expression patterns of these cell types.
Single-cell RNA sequencing overview and identification of major cell types in Acute Respiratory Distress Syndrome (ARDS). (A) Flowchart. (B) Mapping of single-cell atlas groups. (C) Mapping of various cell subpopulations in the single-cell atlas. (D) Bubble chart showing cell type annotating genes. (E) Single-cell density map showing cell type specific marker genes. (F) Stacked bar chart showing differences in cell abundance of different groups.
The key role of S100A9+ MK subpopulation in ARDS inflammatory response
MK and their progeny, platelets, play critical roles in various biological processes, including blood clotting, hemostasis, inflammation, angiogenesis, and innate immunity22. MK are essential for both innate and adaptive immunity in patients with ARDS. Through cluster analysis, we identified eight MK subpopulations (Fig. 2A). Compared to the control group, S100A12+ MK subpopulation exhibited increased abundance in the disease groups, particularly in ARDSNKT, while S100A9+ MK subpopulation showed increased abundance in ARDSKT (Fig. 2B). Single-cell density maps demonstrated the enrichment of relevant markers in these MK subpopulations (Fig. 2C). Through enrichment analysis, we found that the S100A9+ MK subpopulations were significantly enriched in the regulation of immune effector processes, positive regulation of chemokine production, and other biological processes, and this subpopulation was also significantly enriched in KEGG signaling pathways such as the NF-κB signaling pathway and NOD-like receptor signaling pathways (Fig. 2D). Most importantly, this subpopulation significantly activated the NF-κB signaling pathway (Fig. 2E), which is widely recognized as a key pro-inflammatory signaling pathway23. Additionally, we studied the expression levels of inflammatory cytokines and found that in the S100A9+ MK subpopulation, the expression levels of cytokines such as CXCL8, IL-10, and TNF were significantly upregulated (Fig. 2F), indicating that the S100A9+ MK subpopulation may play a key role in promoting the formation of cytokine storms. Pseudotime analysis of MK subpopulations revealed a differentiation and developmental trajectory from the initial LTBP1+ MK subpopulation to the terminal S100A9+ MK subpopulation(Fig. 2G). Through GRN analysis, we found that these MK subpopulations were divided into two modules regulated by TFs, such as KLF1, TAL1, E2F6, AR, SRF, SPIB, RUNX3, and ZNF503, and further explored their activities (Fig. 2H-I).
The potential role of S100A9+ megakaryocytes (MK) subpopulation in modulating the inflammatory response in ARDS. (A) Single-cell atlas displaying the single-cell landscape of MK . (B) Dot-line graph illustrating the abundance changes of different subpopulations within the MK group. (C) Density plot showing the markers of MK cell subpopulations. (D) Bar chart displaying the significantly enriched BP and KEGG signaling pathways of the MK cell subpopulation. (E) The gene set enrichment analysis revealed the activated signaling pathways in the MK subpopulation. (F) Stacked violin plots showing the expression levels of inflammatory cytokines by S100A9+ MK subpopulation. (G) Single-cell atlas mapping of MK progression trajectories and Monocle3 pseudotime values. (H) Gene regulatory network (GRN) graph showing the module of marker genes in MK subtypes. (I) The activity of transcription factors (TFs) in MK subtypes.
In summary, we conclude that compared to the ARDSNKT group, the increased abundance of the S100A9+ MK subpopulation in the ARDSKT group may promote the formation of cytokine storms by activating the NF-κB signaling pathway and promoting the production of inflammatory cytokines, ultimately exacerbating the progression of ARDS in renal transplant patients.
Immunosuppression in ARDSKT reduces the inflammatory response caused by S100A12+ mono subpopulation
Monocytes are a subpopulation of white blood cells that play a crucial role in maintaining homeostasis, pathogen recognition and clearance, and the inflammatory response24. To explore monocyte heterogeneity, we conducted cluster analysis and identified nine distinct subpopulations (Fig. 3A). In the ARDSNKT group, the abundance of the S100A12+ Mono subpopulation was significantly higher than in the control group. Conversely, in the ARDSKT group, the abundance of the S100A12+ Mono subpopulation was significantly lower (Fig. 3B). This difference suggests that the S100A12+ Mono subpopulation may play a key role in the pathogenesis of ARDS, and its function may be influenced by the status of kidney transplantation. Single-cell density plots displays the expression patterns of specific markers within these subpopulations (Fig. 3C). Further analysis indicated that the S100A12+ Mono subpopulation was significantly enriched in biological processes such as cytokine secretion and regulation of inflammatory responses, and also significantly enriched in KEGG signaling pathways such as the chemokine signaling pathway and the Toll-like receptor signaling pathway (Fig. 3D). Additionally, this subpopulation significantly activated the chemokine signaling pathway (Fig. 3E). Chemokines, as key chemotactic cytokines, play a central role in regulating cell migration and positioning; they not only guide immune effector cells to sites of infection or inflammation but also coordinate interactions between immune cells25. In the S100A12+ Mono subpopulation, inflammatory cytokines CXCL8, IL-6 showed high expression (Fig. 3F), suggesting that this subpopulation may play a critical role in driving inflammatory responses during the worsening of ARDS. Pseudotime analysis indicated that MS4A7 + monocytes were the starting point of differentiation and gradually differentiated into the terminal states of S100A12+ Mono subpopulation (Fig. 3G). GRN analysis revealed that the monocytes subtype markers clustered into two modules, with S100A12+ Mono subpopulation being regulated by ETS transcription factor 7 (ETV7) (Fig. 3H). Subsequently, we assessed the activity of TFs that regulate monocytes subtypes (Fig. 3I).
Immunosuppression in ARDSKT reduces the inflammatory response caused by S100A12+ Mono subpopulation. (A) Single-cell atlas displaying the single-cell landscape of Mono. (B) Dot-line graph illustrating the abundance changes of different subpopulations within the Mono group. (C) Density plot showing the markers of Mono cell subpopulations. (D) Bar chart displaying the significantly enriched BP and KEGG signaling pathways of the Mono cell subpopulation. (E) The gene set enrichment analysis revealed the activated signaling pathways by S100A12+ Mono subpopulation. (F) Density chart plots showing the expression levels of inflammatory cytokines in Mono subpopulation. (G) Single-cell maps showing the progression trajectory and Monocle 3 pseudotime values of monocytes. (H) GRN plot shows the marker group modules of monocytes. (I) Activity of TFs in monocyte subtypes.
Based on these findings, we believe that an increase in S100A12+ Mono subpopulation in the ARDSNKT group may activate inflammatory responses and accelerate the development of ARDS. But in kidney transplant patients, due to immunosuppressive therapy, this inflammatory response may be suppressed, thereby reducing inflammation and protecting patients.
The decrease in the abundance of the CXCR4+ B subpopulation mediates the exacerbation of ARDSKT
B cell dysfunction can lead to prolonged systemic inflammation characterized by a cytokine storm, contributing to the development and progression of ARDS26. To investigate B cell heterogeneity, we performed subclustering analysis and identified nine distinct B cell subpopulations with unique specific markers (Fig. 4A-B). Among these subpopulations, we paid special attention to the differential expression of specific markers associated with B cells and plasma cells. A subpopulation characterized by high expression of CD79A and CD79B was clearly identified as B cells, while those with higher expression levels of PRDM1, XBP1, MZB1, and SSR4 were classified as plasma cells (Fig. 4C). Among these subpopulations, we focused particularly on the CXCR4+ B subpopulation, as it was abundant in the control group but significantly reduced in ARDS patients, especially in those with ARDSKT (Fig. 4D). Enrichment analysis results showed that the CXCR4+ B subpopulation was significantly enriched in key biological processes such as B cell-mediated immunity and viral defense responses, and also significantly enriched in KEGG signaling pathways such as the B cell receptor signaling pathway and antigen processing and presentation (Fig. 4E). Additionally, the CXCR4+ B subpopulation significantly activated the B cell receptor signaling pathway (Fig. 4F), which is crucial for normal B cell development and adaptive immune responses27. Through pseudotime analysis, we identified the developmental trajectory of the cell subtypes and discovered that B cells originated from a CXCR4+ B subpopulation starting state and differentiated into an intermediate state of HSPA5+ Plasma subpopulation (Fig. 4G). Subsequently, we explored the TFs regulating B and plasma cell subtypes and found that CXCR4+ B subpopulation were primarily regulated by SPI1 and that CEBPA primarily regulated HSPA5+ Plasma subpopulation (Fig. 4H-I).
The decrease in the abundance of the CXCR4+ B subpopulation mediates the exacerbation of ARDS after renal transplantation. (A) Single-cell atlas displaying the single-cell landscape of B. (B) Density plot showing the markers of B cell subpopulations. (C). Stacked violin plots showing the expression of markers distinguishing B cell subpopulations. (D) Dot-line graph illustrating the abundance changes of different subpopulations within the B group. (E) Bar chart displaying the significantly enriched BP and KEGG signaling pathways of the B cell subpopulation. (F) The gene set enrichment analysis revealed the activated signaling pathways by CXCL4+ B subpopulation. (G) Single-cell atlas mapping of B cell progression trajectories and Monocle3 pseudotime values. (H) GRN graph showing the module of marker genes in B cell subtypes. (I) Activity of TFs in B cell subtypes.
Our findings reveal a significant reduction in the abundance of the CXCR4+ B subpopulation in ARDS patients, particularly in those who have undergone renal transplantation. The reduced abundance of the CXCR4+ B cell subpopulation in transplant patients may affect the BCR signaling pathway, thereby influencing B cell-mediated immune functions and indirectly promoting the development of ARDS in these patients.
XAF1+ Teff subpopulation promotes cell apoptosis, mediating the formation of ARDSKT inflammation
CD8+ T cell differentiation, activation, and functionality are heightened in patients with ARDS, playing critical roles in immune activation and cytotoxic responses to foreign antigens28,29. To investigate the heterogeneity of CD8+ T cells across different populations, we performed clustering analysis and identified nine distinct subpopulations (Fig. 5A). Further exploration focused on the enrichment of relevant markers within these CD8+ T cell subpopulations (Fig. 5B). Among these subpopulations, the cell clusters expressing PRF1, CCL5, and IL-2RB were identified as effector T cells (Teff), while those expressing IL-7R and SELL were classified as central memory T cells (Tcm) (Fig. 5C). Notably, compared with the control group and ARDSNKT group, the abundance of the XAF1+ Teff subpopulation was significantly increased in the ARDSKT group and became the main component of this group (Fig. 5D). Through enrichment analysis, we found that this subpopulation was significantly enriched in key biological processes such as signal transduction after DNA damage and inflammatory cell apoptosis processes, as well as KEGG signaling pathways such as cell apoptosis and NOD-like receptor signaling pathways (Fig. 5E). Additionally, the XAF1+ Teff subpopulation significantly activated the cell apoptosis process (Fig. 5F), and pro-apoptotic genes BCL2L11, BID, CASP9, FAS, FASLG, TNFSF10 were highly expressed in this subpopulation (Fig. 5G). Pseudotime analysis was used to determine the differentiation trajectory of CD8+ T cells, revealing that XAF1+ Teff subpopulation was in an intermediate state derived from the early Teff differentiation (Fig. 5H). GRN analysis revealed a module regulated by TFs, such as LEF1, KLF3, and STAT2, with XAF1+ Teff subpopulation being regulated by ETV7 (Fig. 5I). The activities of TFs that regulate CD8+ T cell subpopulations were further explored (Fig. 5J).
XAF1+ Teff subpopulation promotes cell apoptosis, mediating the formation of ARDS inflammation after renal transplantation. (A) Single-cell atlas displaying the single-cell landscape of CD8+ T. (B) Stacked violin plots showing the expression of markers distinguishing CD8+ T subpopulations. (C) Density plot showing the markers of CD8+ T subpopulations. (D) Dot-line graph illustrating the abundance changes of different subpopulations within the CD8+ T group. (E) Bar chart displaying the significantly enriched BP and KEGG signaling pathways of the CD8+ T subpopulation. (F) The gene set enrichment analysis revealed the activated signaling pathways by XAF1+ Teff subpopulation. (G) Density plots showing the expression of proapoptotic genes in the CD8+ T subpopulation. (H) Single-cell atlas mapping of CD8+ T cell progression trajectories and Monocle3 pseudotime values. (I) GRN graph showing the module of marker genes in CD8+ T subpopulations. (J) Activity of TFs in CD8+ T cell subpopulations.
Based on these findings, we hypothesize that in ARDS patients after renal transplantation, the increased abundance of the XAF1+ Teff subpopulation may promote Teff cell apoptosis. This apoptotic process may weaken the ability of Teff cells to clear viruses, hinder the normal process of immune reconstitution, thereby promoting the development of chronic inflammation and exacerbating the progression of ARDS after transplantation.
Global analysis of intercellular communication in COVID-19 and construction of ligand-receptor interaction network
In order to understand the interactions between different cell populations, we explored the mechanisms of interaction between overall cell subpopulations in immune checkpoints, cytokines, growth factors, and other factors, and inferred a hypothesized cell interaction network. In the field of immune checkpoint research, we observed that multiple cell subpopulations in two different groups interact through the BTL4-TNFSF14 signaling axis(Fig. 6A-B). The interaction between TNFSF14 and BTLA activates the inflammatory signaling pathway, promoting the production of pro-inflammatory cytokines, which is crucial for regulating ADRS immune responses30. In the field of cytokine research, we noticed that the MKs in both groups mainly act on CXCR2 receptors expressed on lymphocytes and monocyte subpopulations by secreting PPBP ligands(Fig. 6C-D). Activated platelets release PPBP, which induces the aggregation and activation of neutrophils and other immune cells, playing an important role in inflammatory diseases. In recent years, a large number of studies have shown that the PPBP-CXCR2 axis is abnormally expressed in various inflammatory diseases and has a strong correlation with the pathogenesis31. In the field of growth factor research, we observed that the S100A9+ MK subpopulations in both groups mainly interact with multiple cell subpopulations through the TGFB1-CXCR4 signaling axis (Fig. 6E-F). Finally, in other aspects, we found that S100A12+ MK and S100A12+ monocytes significantly enriched the ligand S100A8, which acts on themselves and S100A9+ MK and S100A12+ monocytes through the S100A8-TLR4 ligand receptor axis (Fig. 6G-H). In summary, these research findings reveal a complex network of interactions between cell subpopulations and provide a new perspective for understanding the regulatory mechanisms of immune responses.
Global analysis of intercellular communication in COVID-19 and construction of ligand-receptor interaction network. (A,B) The circular and bubble plots illustrate the communication events between cell subpopulations in different groups regarding immune checkpoints. (C,D) The circular and bubble plots illustrate cytokine communication events between subpopulations of cells in different groups. (E,F) The circular and bubble plots illustrate growth factor communication events between subpopulations of cells in different groups. (G,H) The circular and bubble plots illustrate communication events between subpopulations of cells in different groups regarding other aspects.
Discussion
This study, through scRNA-seq analysis, constructed a single-cell immune landscape of COVID-19-induced ARDS patients, covering both kidney transplant recipients, non-kidney transplant patients, and healthy controls. We focused on four immune subpopulations, including MK, monocytes, B cells, and CD8+ T cells, systematically exploring the differences in these cells within the immune environments of COVID-19-induced ARDS in kidney transplant and non-kidney transplant patients, as well as their roles in the pathogenesis of ARDS.
MK is a large cell type that produces platelets. Systemic inflammatory responses promote the formation of MK in platelets, and MK plays a role in hemostasis, immunity, and inflammation28,29. This study found that the abundance of the S100A9+ MK subpopulation was significantly increased in the ARDSKT group compared to the ARDSNKT group. S100A9 is a member of the S100 protein family and can form heterodimers known as calprotectin32. Studies have shown that calprotectin primarily regulates inflammation by inducing cytokines, chemokines, reactive oxygen species (ROS), and nitric oxide (NO), leading to cytokine storms33, which are considered key factors in the progression of ARDS induced by COVID-19. We further discovered that the S100A9+ MK subpopulation significantly activates the NF-κB signaling pathway and highly expresses inflammatory factors. NF-κB is a family of dimeric transcription factors that play a central role in coordinating inflammatory responses34. This finding suggests that the S100A9+ MK subpopulation may act as a key factor driving cytokine storms, and its increased abundance in the ARDSKT group may exacerbate the overreaction of the immune system, thereby promoting disease progression. Additionally, cell communication analysis showed that the S100A9+ MK subpopulation regulates the immune functions of T cells and B cells through the TGFB1-CXCR4 axis. Studies have reported that excessive release of TGFB1 can impair lymphocyte regulation and reduce lymphocyte counts, leading to persistent focal inflammation35, which further supports the pro-inflammatory role of this subpopulation in the immune microenvironment.
Monocytes play a crucial role not only in host defense but also in tissue repair and fibrosis36. In this study, we found that the abundance of the S100A12+ Mono subpopulation was significantly increased in the ARDSNKT group and significantly decreased in the ARDSKT group. S100A12, as a molecule secreted by monocytes, has multiple extracellular activities, including chemotaxis, induction of oxidative stress, and promotion of inflammatory responses37. Inflammation further activates monocytes, inducing the expression and release of S100A12 and promoting the expression of other inflammatory genes38. This activation may lead to excessive inflammatory responses and tissue damage, especially in COVID-19-induced ARDS. We further discovered that the S100A12+ Mono subpopulation significantly activated the chemokine signaling pathway and highly expressed various inflammatory factors, indicating the pro-inflammatory role of this subpopulation in immune responses. However, due to the long-term use of immunosuppressants in kidney transplant patients, this immunosuppressive environment may inhibit the pro-inflammatory response of S100A12+ Mono subpopulation, leading to a significant decrease in the abundance of this subpopulation in the ARDSKT group. This may, to some extent, alleviate the inflammatory response, thereby providing immunological protection for kidney transplant patients.
B cells play a crucial role in adaptive immune responses, particularly in the production of antibodies and the establishment of immune memory. Our study found that in patients with ARDS caused by COVID-19, the abundance of the CXCR4+ B subpopulation was significantly reduced compared to the healthy control group, with an even more pronounced decrease in the ARDSKT group. C-X-C chemokine receptor type 4 (CXCR4) plays an essential role in the migration of B cells to germinal centers, differentiation into plasma cells, and entry into the bloodstream and secondary lymphoid organs39. We found that the CXCR4+ B subpopulation significantly activated the B cell receptor signaling pathway, which is crucial for the normal development of B cells and adaptive immune responses27. The reduction in the abundance of CXCR4+ B subpopulation may weaken the immune response and antibody production against SARS-CoV-2, especially in kidney transplant patients, where the use of immunosuppressants further inhibits the function of the CXCR4+ B subpopulation, thereby affecting B cell-mediated immune responses, which may contribute to the exacerbation of ARDS.
In adaptive immunity, CD8+ T cells are crucial in controlling COVID-19 virus infections through their cytotoxic activity and the release of effector cytokines40. We found that in patients with COVID-19-induced ARDS, the abundance of the XAF1+ Teff subpopulation was significantly increased, particularly in the ARDSKT group. XIAP-associated factor 1 (XAF1) is an interferon-stimulated gene that has been shown to enhance apoptosis41. It has been reported that the overexpression of XAF1 induced by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection promotes cellular apoptosis42,43. Furthermore, our study found that the XAF1+ Teff subpopulation significantly activated apoptosis and highly expressed pro-apoptotic genes, which is consistent with previous reports. Based on these findings, we believe that the increased abundance of the XAF1+ Teff subpopulation may exacerbate the apoptosis of Teff cells, thereby reducing the immune system’s ability to clear the virus, promoting the formation of chronic inflammation, and further worsening the condition of post-transplant ARDS.
Unlike previous single-cell studies on ARDS, this study is the first to compare the functional status and role of various immune cells, including B cells and CD8+ T cells, in COVID-19-induced ARDS patients with and without kidney transplantation. Previous studies have not systematically explored the role of these specific immune cells in ARDS patients after kidney transplantation. This study fills this research gap and reveals the key role of these cells in disease severity by comprehensively analyzing the immune regulatory mechanisms of immune cells in ARDS.
However, this study still has some limitations. First, the small and single-source sample size may affect the generalizability of the results. Second, the use of immunosuppressants by kidney transplant patients may interfere with the assessment of immune responses. Lastly, this study only analyzed peripheral blood PBMCs and did not cover the local immune microenvironment. Therefore, future studies should expand the sample size, combine animal models with long-term follow-up, in order to fully understand the immune mechanisms.
Conclusions
This study, through single-cell RNA sequencing, reveals functional differences in immune cell subpopulations in patients with COVID-19-induced ARDS, particularly among kidney transplant and non-kidney transplant patients. We identified several key immune cell subpopulations, including MK, monocytes, B cells, and CD8+ T cells, and discussed their different roles in immune responses and disease progression. This study found that the S100A9+ MK subpopulation was significantly elevated in ARDSKT patients, potentially exacerbating the condition by promoting cytokine storms and enhancing inflammatory responses. The significant increase in the abundance of S100A12+ Mono subpopulation in the ARDSNKT group may be closely related to excessive inflammatory responses. Additionally, the reduction in the CXCR4+ B subpopulation and the increase in the XAF1+ Teff subpopulation suggest that adaptive immune function is suppressed, which may affect antiviral immunity and immune recovery. Overall, this study reveals the key role of immune cell subpopulations in the progression of ARDS, especially immune dysregulation in kidney transplant patients, providing new ideas for immune-modulating treatments that may improve the prognosis of these patients.
Data availability
The datasets presented in this study can be found online at the National Genomics Data Center under accession numbers HRA004752 and HRA005498.
Abbreviations
- ARDS:
-
Acute respiratory distress syndrome
- KTR:
-
Kidney transplant recipient
- MK:
-
Megakaryocytes
- COVID-19:
-
Coronavirus Disease 2019
- QC:
-
Quality control
- UMAP:
-
Uniform Manifold Approximation and Projection
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- GRN:
-
Gene regulatory network
- TFs:
-
Transcription factors
- Teffs:
-
Effector T cells
- ROS:
-
Reactive oxygen species
- TLR4:
-
Toll-like receptor 4
- TGFB1:
-
Transforming growth factor beta
- NO:
-
Nitric oxide
- CXCR4:
-
C-X-C chemokine receptor type 4
- XAF1:
-
XIAP-associated factor 1
- IFN:
-
Interferon
References
Muralidar, S., Ambi, S. V., Sekaran, S. & Krishnan, U. M. The emergence of COVID-19 as a global pandemic: Understanding the epidemiology, immune response and potential therapeutic targets of SARS-CoV-2. Biochimie 179, 85–100 (2020).
Rumende, C. M., Susanto, E. C. & Sitorus, T. P. The management of pulmonary fibrosis in COVID-19. Acta Med. Indones 53 (2), 233–241 (2021).
Xu, H., Sheng, S., Luo, W., Xu, X. & Zhang, Z. Acute respiratory distress syndrome heterogeneity and the septic ARDS subpopulation. Front. Immunol. 14, 1277161 (2023).
Bakr, M. A., Nagib, A. M. & Donia, A. F. Induction immunosuppressive therapy in kidney transplantation. Exp. Clin. Transpl. 12 (Suppl 1), 60–69 (2014).
Azzi, Y. et al. COVID-19 infection in kidney transplant recipients at the epicenter of pandemics. Kidney Int. 98 (6), 1559–1567 (2020).
Gosangi, B. et al. COVID-19 ARDS: a review of imaging features and overview of mechanical ventilation and its complications. Emerg. Radiol. 29 (1), 23–34 (2022).
Roltgen, K. & Boyd, S. D. Antibody and B cell responses to SARS-CoV-2 infection and vaccination. Cell. Host Microbe 29 (7), 1063–1075 (2021).
Wilk, A. J. et al. A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat. Med. 26 (7), 1070–1076 (2020).
Knoll, R., Schultze, J. L. & Schulte-Schrepping, J. Monocytes and macrophages in COVID-19. Front. Immunol. 12, 720109 (2021).
Montazersaheb, S. et al. COVID-19 infection: an overview on cytokine storm and related interventions. Virol. J. 19 (1), 92 (2022).
Lu, J. et al. Immune characteristics of kidney transplant recipients with acute respiratory distress syndrome induced by COVID-19 at single-cell resolution. Respir Res. 25 (1), 34 (2024).
Stuart, T., Srivastava, A., Madad, S., Lareau, C. A. & Satija, R. Single-cell chromatin state analysis with Signac. Nat. Methods 18 (11), 1333–1341 (2021).
Hozumi, Y., Wang, R., Yin, C. & Wei, G. W. UMAP-assisted K-means clustering of large-scale SARS-CoV-2 mutation datasets. Comput. Biol. Med. 131, 104264 (2021).
Zhu, L. et al. Single-Cell sequencing of peripheral mononuclear cells reveals distinct immune response landscapes of COVID-19 and influenza patients. Immunity 53 (3), 685–696e683 (2020).
Yu, G., Wang, L. G., Han, Y. & He, Q. Y. ClusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16 (5), 284–287 (2012).
Zeng, X. et al. Screening of key genes of Sepsis and septic shock using bioinformatics analysis. J. Inflamm. Res. 14, 829–841 (2021).
Trapnell, C. et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 32 (4), 381–386 (2014).
Badia, I. M. P. et al. Gene regulatory network inference in the era of single-cell multi-omics. Nat. Rev. Genet. 24 (11), 739–754 (2023).
Pratapa, A., Jalihal, A. P., Law, J. N., Bharadwaj, A. & Murali, T. M. Benchmarking algorithms for gene regulatory network inference from single-cell transcriptomic data. Nat. Methods 17 (2), 147–154 (2020).
Jin, S. et al. Inference and analysis of cell-cell communication using cellchat. Nat. Commun. 12 (1), 1088 (2021).
Lu, J. et al. Documenting the immune response in patients with COVID-19-induced acute respiratory distress syndrome. Front. Cell. Dev. Biol. 11, 1207960 (2023).
Gelon, L., Fromont, L. & Lefrancais, E. Occurrence and role of lung megakaryocytes in infection and inflammation. Front. Immunol. 13, 1029223 (2022).
Lawrence, T. The nuclear factor NF-kappaB pathway in inflammation. Cold Spring Harb Perspect. Biol. 1 (6), a001651 (2009).
Cormican, S. & Griffin, M. D. Human monocyte subset distinctions and function: insights from gene expression analysis. Front. Immunol. 11, 1070 (2020).
Sokol, C. L. & Luster, A. D. The chemokine system in innate immunity. Cold Spring Harb Perspect. Biol. 7(5). (2015).
Yao, C. et al. Cell-Type-Specific immune dysregulation in severely ill COVID-19 patients. Cell. Rep. 34 (1), 108590 (2021).
Burger, J. A. & Wiestner, A. Targeting B cell receptor signalling in cancer: preclinical and clinical advances. Nat. Rev. Cancer 18 (3), 148–167 (2018).
Anft, M. et al. COVID-19-Induced ARDS is associated with decreased frequency of activated memory/effector T cells expressing CD11a(+). Mol. Ther. 28 (12), 2691–2702 (2020).
Mo, J. et al. Single-cell analysis reveals dysregulated inflammatory response in peripheral blood immunity in patients with acute respiratory distress syndrome. Front. Cell. Dev. Biol. 11, 1199122 (2023).
Carreras, J. et al. High TNFRSF14 and low BTLA are associated with poor prognosis in follicular lymphoma and in diffuse large B-cell lymphoma transformation. J. Clin. Exp. Hematop 59 (1), 1–16 (2019).
Wu, Q., Tu, H. & Li, J. Multifaceted roles of chemokine C-X-C motif ligand 7 in inflammatory diseases and Cancer. Front. Pharmacol. 13, 914730 (2022).
Vazquez-Alejo, E. et al. Persistent exhausted T-Cell immunity after severe COVID-19: 6-Month evaluation in a prospective observational study. J. Clin. Med. 12(10). (2023).
He, D. et al. Single-Cell RNA sequencing and transcriptome analysis revealed the immune microenvironment and gene markers of acute respiratory distress syndrome. J. Inflamm. Res. 16, 3205–3217 (2023).
Mitchell, S., Vargas, J. & Hoffmann, A. Signaling via the NFkappaB system. Wiley Interdiscip Rev. Syst. Biol. Med. 8 (3), 227–241 (2016).
Jacquemont, L. et al. Terminally differentiated effector memory CD8(+) T cells identify kidney transplant recipients at high risk of graft failure. J. Am. Soc. Nephrol. 31 (4), 876–891 (2020).
Huang, D. Y. et al. Megakaryocytes in pulmonary diseases. Life Sci. 301, 120602 (2022).
Jukic, A., Bakiri, L., Wagner, E. F., Tilg, H. & Adolph, T. E. Calprotectin: from biomarker to biological function. Gut 70 (10), 1978–1988 (2021).
Ren, X. et al. COVID-19 immune features revealed by a large-scale single-cell transcriptome atlas. Cell 184 (7), 1895–1913 (2021). e1819.
Franitza, S. et al. TGF-beta1 enhances SDF-1alpha-induced chemotaxis and homing of Naive T cells by up-regulating CXCR4 expression and downstream cytoskeletal effector molecules. Eur. J. Immunol. 32 (1), 193–202 (2002).
Allardet-Servent, J. et al. The association of low CD4 expression on monocytes and low CD8 + T-Cell count at hospital admission predicts the need for mechanical ventilation in patients with COVID-19 pneumonia: A prospective monocentric cohort study. Crit. Care Explor. 4 (12), e0810 (2022).
Liu, B. Q. et al. XAF1 prevents hyperproduction of type I interferon upon viral infection by targeting IRF7. EMBO Rep. 24 (1), e55387 (2023).
Han, Y. et al. XAF1 protects host against emerging RNA viruses by stabilizing IRF1-Dependent antiviral immunity. J. Virol. 96 (17), e0077422 (2022).
Kuang, M. et al. XAF1 promotes anti-RNA virus immune responses by regulating chromatin accessibility. Sci. Adv. 9 (33), eadg5211 (2023).
Funding
This study was supported by the Joint Project on Regional High-Incidence Diseases Research of the Guangxi Natural Science Foundation (2023GXNSFDA026023), the Key Research and Development Program of Guangxi (Guike AB23026012 and GuiKe AB22080088), the National Natural Science Foundation of China (82360374 and 82360372), and the Guangxi Medical and Health Key Discipline Construction Project.
Author information
Authors and Affiliations
Contributions
J.L, X.Z and J.Z conceived the study and designed the study. J.P, J.H, J.Y, B.L, S.W, W.C, X.X, J.Y, Y.Y and J.M performed data collection and data analysis. J.P, J.H, J.Y, J.L and X.Z drafted the manuscript. J.Z supervised manuscript preparation. All authors read and approved the final manuscript for publication.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
This study was approved by the Institutional Review Board and Ethics Committee of the Second Affiliated Hospital of Guangxi Medical University. The studies involving human participants complied with the Declaration of Helsinki and were reviewed and approved by the Ethics Committee of the Ethics Committee of the Second Affiliated Hospital of Guangxi Medical University, and samples were obtained from all patients which informed consent were obtained from all participants and /or their legal guardians.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Pang, J., Huang, J., Yu, J. et al. Immune dysregulation in COVID-19 induced ARDS in kidney transplant recipients revealed by single-cell RNA sequencing. Sci Rep 15, 6895 (2025). https://doi.org/10.1038/s41598-025-91439-5
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-91439-5