Abstract
Rheumatoid arthritis (RA) is a chronic autoimmune disease characterized by joint inflammation, pain, swelling, and stiffness, with matrix metalloproteinase-9 (MMP9) playing a critical role in extracellular matrix remodelling and joint degradation. Elevated MMP9 levels are closely associated with RA severity and progression. This study aimed to identify and characterize functional and pathogenic variants of the MMP9 gene and assess their impact on RA susceptibility and severity. Various computational tools were used to identify deleterious non-synonymous single nucleotide polymorphisms (nsSNPs). These nsSNPs were further evaluated for their conservation profiles, influence on protein structure, function, and phenotypic stability. Models for all the mutant and wild type were generated and validated by ERRAT, VERIFY3D and QMEAN. Further, molecular dynamics simulations, molecular docking and gene-gene interactions were analysed. Nine damaging missense nsSNPs within the MMP9 catalytic region were identified, all of which were highly conserved, with Y262C notably resulting in the loss of a phosphorylation site. Most mutations were associated with decreased protein stability, and HOPE analysis revealed their localization in essential functional domains of MMP9. MD simulations indicated greater structural instability in these variants compared to the wild type, with Y262C, P180L, G438A, and D434N exhibiting the highest root-mean-square deviation (RMSD) values. Gene-gene interaction analysis further emphasized the significant role of MMP9 in RA-related pathways. This study offers key insights into pathogenic nsSNPs affecting MMP9 structure and function, highlighting their importance in genetic screening and potential therapeutic strategies for RA.
Similar content being viewed by others
Introduction
Rheumatoid arthritis (RA) is a chronic inflammatory joint disease characterized by synovial hyperplasia, cartilage degradation, and bone erosion1. In RA, the synovial lining undergoes dramatic changes, forming an inflammatory tissue known as the pannus that invades adjacent cartilage and bone, leading to cartilage and joint destruction2. Activated RA synovial fibroblasts (FLS) exhibit aberrant phenotypic characteristics akin to tumour cells, including increased proliferation, resistance to apoptosis, and tissue invasiveness3. Articular cartilage degradation emerges as an early hallmark of RA and is propelled by the heightened activity of proteolytic systems. Notably, RA-FLS display augmented synthesis of MMPs, comprising a zinc-dependent endopeptidase family pivotal in ECM degradation, thus contributing significantly to tissue invasion and breakdown (Fig. 1a)4. Within the MMP family, the gelatinase subgroup, encompassing MMP-2 (gelatinase A) and MMP-9 (gelatinase B), holds prominence in collagen degradation, particularly by digesting denatured collagen (gelatin) generated by collagenases. Moreover, these gelatinases target additional substrates like fibrillar collagen I and II, as well as aggrecan, predominantly found in cartilage5. Additionally, gelatinases play a pivotal role in modulating inflammation by processing cytokines and chemokines, with MMP-9 exerting stimulatory effects and MMP-2 exerting inhibitory effects6. MMP-9 remains largely undetectable in most cell types under basal conditions; however, its expression escalates in malignancies or inflammatory contexts, such as in RA, where FLS stimulate their synthesis by inflammatory cytokines7. In RA, MMP-9 levels increase in the serum and joint synovial fluid, correlating positively with disease severity and progression. Notably, MMP-9 knockout mice exhibit attenuated severity of antibody-induced arthritis, underscoring its crucial role in RA pathogenesis8.
Given the involvement of MMP9, genetic variations in MMP9 gene are intriguing candidates for analysing their impact on the susceptibility and severity of RA9. Considering the significant role of MMP9 in inflammation, studies have demonstrated that genetic variations influencing MMP9 expression can affect susceptibility and progression of various diseases including RA10. Regulation of MMP expression predominantly occurs at the transcriptional level, wherein gene promoters respond to diverse regulators such as growth factors, hormones, and cytokines. Functional polymorphisms in MMP genes have been identified and associated with susceptibility to various diseases11. In this study, we aimed to investigate the structural and functional consequences of nonsynonymous SNPs (nsSNPs) in MMP9, focusing on their potential implications for RA pathogenesis.
Pathogenesis and structure of MMP9. (a) The multifaceted role of MMP9 in the pathogenesis of RA. MMP9 contributes to key pathological processes, including ECM and cartilage degradation, tissue invasion and breakdown, immune infiltration, inflammation, and angiogenesis, which are hallmarks of RA (b) Structural representation of MMP9, highlighting its functional domains.
MMP9, encoded by the MMP9 gene on chromosome 20q13.12, encompasses 13 exons and 12 introns and contains distinct structural domains, including a catalytic domain, hemopexin-like domain, and propeptide region, critical for its enzymatic activity and regulation12. Within MMP-9, the catalytic domain comprises fibronectin type II (FN2) domains, an active site, and a region responsible for zinc binding (Fig. 1b). This protein houses two zinc ions and five calcium ions. As part of the zinc-dependent endopeptidase family, MMP-9 relies on zinc ions to execute its catalytic function13.
As MMP9 plays a crucial role in the pathology and prognosis of RA. It is vital to analyse the potential damaging effects of nsSNPs on MMP9, as these variants are most likely to have functional effects on protein structure and activity. Using a combination of computational approaches, we analyse the potential damaging effects of nsSNPs on MMP9 structure and function, stability, protein-protein interactions, and PTMs. Three-dimensional models of wild-type and mutant MMP9 proteins were generated, allowing for comparative analyses to delineate the structural alterations induced by nsSNPs, which were further corroborated by MD simulations. By predicting deleterious nsSNPs in MMP9 and elucidating their functional consequences and stability indexes by MD simulation, this study contributes to our understanding of RA pathogenesis and offers insights into potential pharmacogenomic applications, molecular diagnostics, and therapeutic interventions targeting MMP9-mediated pathways.
Methodology
Figure 2 is representing the overall methodology used in the current study.
SNP data mining
The NCBI dbSNP database was used to collect all SNPs linked to the MMP9 gene. Identification numbers (rsIDs) for nsSNPs were acquired, and the MMP9 protein sequence in FASTA format was retrieved from Uniprot14. Specifically, only missense or nonsynonymous SNPs were selected for subsequent in silico analysis15.
Identification of high-risk nsSNPs
After retrieving both the nsSNPs and protein sequence, various in silico tools were employed to predict functionally damaging nsSNPs. These tools include SIFT (Sorting Intolerant from Tolerant)16, PROVEAN (Protein Variation Effect Analyzer)17, PolyPhen2 (Polymorphism Phenotyping 2)18, PANTHER19, and SNP & GO20. Subsequently, the nsSNPs identified as damaging by these in silico tools were verified using PhD-SNP (Predictor of human Deleterious SNP)21.
Minning of nsSNP in protein domains
The InterPro server was used to classify and predict the functional domains within the MMP9 protein. The FASTA sequence was used to identify conserved domains, followed by visualization of the positions of the identified nsSNPs22.
Prediction of evolutionary conservation of MMP9
The ConSurf server was employed to predict the impact of nsSNPs on evolutionarily conserved amino acids in MMP9 by analysing phylogenetic relationships among homologous sequences using empirical Bayesian inference. The MMP9 FASTA sequence served as the input for this analysis23.
Solvent accessible area prediction
The NetSurfP 3.0 web server was used to predict surface-accessible amino acids within the MMP9 protein sequence, accessed on March 23, 2024. This server utilizes an artificial neural network trained on a diverse set of experimentally determined protein structures to classify amino acids as buried or exposed residues24.
Prediction of nsSNP effects on structure and function of MMP9 protein
MutPred2 was used to assess the structural and functional impact of nsSNPs. This web application predicts the pathogenicity of amino acid changes in a protein. The MMP9 protein sequence, along with details of amino acid substitutions, was submitted in the FASTA format. A significance threshold of p < 0.05 was considered “Confident,” while p < 0.01 was categorized as “Very Confident”25.
Prediction of phenotypic and structural impact of nsSNP
The structural effects of the selected deleterious nsSNPs were analysed using the HOPE online tool. HOPE integrates data from the PDB26 and UniProtKB27 and can build homology models when required. In this study, the FASTA sequence of the MMP9 protein obtained from UniProtKB was used as an input to HOPE. HOPE uses this sequence and its integrated structural databases to predict the impact of amino acid changes on the target protein. This approach enables the analysis of phenotypic and structural effects of the identified nsSNPs28.
Prediction of protein stability
To explore the impact of all deleterious nsSNPs on the stability of the MMP9 protein, i-Stable, MuPro, I-Mutant 2.0, DUET, and DynaMut were utilized. These web-based applications, based on support vector machines (SVM), predict the extent to which mutations affect protein stability. DynaMut has been specifically used to predict interatomic interactions and assess how nsSNPs influence these interactions29.
Prediction of potential PTM sites
Post-translational modifications (PTMs) play a crucial role in determining the structure, folding, and function of proteins. To identify potential PTM sites in the MMP9 protein, several in silico tools were utilized. MusiteDeep software was used to predict the methylation, phosphorylation, glycosylation, and ubiquitination sites. This online tool leverages a deep-learning framework to predict and visualize PTM sites in proteins30. Phosphorylation predictions were further validated using NetPhos 3 with a threshold of 0.5, where amino acids above this threshold were considered phosphorylated31. Glycosylation sites were predicted using NetOglyc 4.0, which operates with a threshold value of 0.5 for the predicted glycosylation score32. The input protein sequence was provided in the FASTA format, enabling the simultaneous prediction of multiple PTMs.
Protein 3D structure prediction
Phyre2, an online tool grounded on homology modelling principles, was utilized to predict 3D models for both native and mutant MMP9. The wild-type and mutant models were then compared employing the online structure alignment tool TM-Align, which provides Template Modeling scores (TM scores) and root-mean-square deviation (RMSD) values. TM scores ranged from 0 to 1, with 1 signifying perfect alignment between the two proteins. Conversely, higher RMSD values indicate significant structural variations between the mutant and wild-type structures33.
Structure validation
To validate the protein models derived from Phyre2, ERRAT, VERIFY3D, and QMEAN Expasy were utilized. The ERRAT assesses the overall quality of the model by providing a percentage score. QMEAN offers quality evaluations based on various geometric attributes and provides both overall assessments (for the entire structure) and specific evaluations (for each residue) derived from a single model. The scoring mechanism integrated six structural descriptors using a linear combination. VERIFY3D analyses the compatibility of a model with its own amino acid sequences34.
Molecular dynamic simulation
Gromacs 2020.1, was used to conduct MD simulations on both the wild-type MMP9 and its nine mutants Y262C, P180L, C329S, M422R, G183E, D434N, G306C, G438A, and C347Y. The 3D structure of wild-type MMP9, comprising 416 amino acids, was generated using Phyre2 and served as the reference structure. The simulations employed the OPLS-AA force field, a well-established force field for proteins, to ensure the accurate representation of atomic interactions. Each protein structure was placed in a cubic simulation box and solvated with SPC (simple point charge) water molecules to provide uniform solvation and maintain symmetry while ensuring sufficient space to avoid interactions with periodic images. The total number of atoms in the wild-type and mutant systems were 110,214 (WT), 124,601 (Y262C), 124,607 (P180L), 124,623 (C329S), 124,599 (M422R), 124,599 (G183E), 124,612 (D434N), 124,615 (G306C), 124,599 (G438A), and 124,633 (C347Y), respectively. Neutral pH conditions were maintained by adding appropriate numbers of Na + counter ions: 18 for WT, Y262C, P180L, C329S, D434N, G438A, and C347Y; 17 for M422R and G306C; and 19 for G183E. Energy minimization of the solvated systems was performed using 50,000 steps of the steepest descent algorithm, with minimization terminating when the maximum force dropped below 1000 kJ/mol/nm. Following energy minimization, equilibration was performed in two phases: NVT (constant volume and temperature) and NPT (constant pressure and temperature) ensembles. The temperature was maintained at 310 K using a v-rescale thermostat (modified Berendsen method), and the pressure was stabilized at 1 bar using the Parrinello-Rahman pressure coupling method. Electrostatic interactions were calculated using the particle mesh Ewald (PME) method to ensure accurate long-range interactions. Production MD simulations were run for 50 ns with a time step of 2 fs, and the trajectory data were saved at intervals of 2 ps for downstream analyses. This setup allowed us to assess the dynamic behaviour of the wild-type and mutant MMP9 proteins under near-physiological conditions. Structural deviations between the mutants and the wild-type were calculated via root-mean-square deviation (RMSD), root mean square fluctuation (RMSF), hydrogen bonds (H-bonds), and solvent accessible surface area (SASA) analysis35.
Molecular docking
For ligand-protein docking studies, the molecule Benzenesulfonamide, 2-nitro-N-phenyl (Figure S4) was selected because of its notable binding affinity within the catalytic domain of the MMP9 protein, as previously reported36. The ligand structure was sourced from PubChem. Prior to initiating the docking procedure, the protein structure was stabilized, hydrogen atoms were added, and Kollman charges were calculated for both the protein and ligand. Docking analysis targeted the active site near the zinc-binding domain, specifically the S1’ inhibitor-binding pocket37. A grid box with dimensions of 60 × 60 × 60 Å and grid spacing of 0.375 Å was defined. Molecular docking was conducted using the Lamarckian Genetic Algorithm, configured for 10 iterative steps. The docking process employed AutoDock 1.5.6 software, and the results were analysed and visualized using BIOVIA Discovery Studio 202138.
Interaction of MMP9 with other proteins
To predict the functional interactions of MMP9 with other proteins within cells, GeneMANIA and STRING (accessed on March 29, 2024) were employed to highlight the numerous interactions within the cellular environment that are crucial for their function and regulation39.
Results
Retrieval of SNP
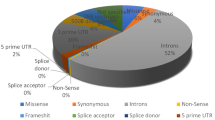
According to the dbSNP database, MMP9 contains a total of 4431 SNPs. Among these, 2530 were in intronic regions, 723 were missense mutations, and 350 were synonymous. Additionally, 11 SNPs were situated in the 5’UTR, whereas 63 were found in the 3’UTR. The remaining SNPs were located in the frameshift54, splice acceptor13, splice donor15, downstream (491), and upstream (779) regions. Our analysis was limited to missense or nsSNP SNPs. Figure 3a provides a graphical representation depicting the percentage distribution of all the SNPs.
Predicted detrimental nsSNPs
Five computational tools were used to identify the pathogenic nature of the identified nsSNPs. SIFT used a threshold of 0.05 for the Tolerance Index (TI), classifying SNPs below this threshold as affected, resulting in the identification of 64 pathogenic nsSNPs. For PROVEAN, a threshold of -2.5 was set, resulting in 52 deleterious nsSNPs. PANTHER identified 62 nsSNPs as disease associated. PolyPhen-2 categorized 56 nsSNPs as deleterious, while SNP & Go identified 42 deleterious nsSNPs. SNPs identified as damaging by all five tools were subsequently analysed using PhD SNP. Among the total SNPs analysed, twelve were deemed deleterious by each software and were selected for additional investigation (Table 1). Figure 3b presents an overview of the outcomes obtained from all computational tools.
Exploiting nsSNPs in MMP9 domains
Interpro studies indicate that MMP9 has a total of five domains, with two main domains: a catalytic domain (Peptidase M10A) (115–444 aa) and a hemopexin-like domain (514–704 aa). Other domains, such as the fibronectin type II domain (IPR000562), Peptidase M10 (IPR001818), and peptidase (IPR006026), are encompassed within the main catalytic domain (Fig. 4). InterPro results indicated that all nonsynonymous SNPs were situated within the catalytic domain, except for A523, which was located within the hemopexin-like domain. Missense mutations were dispersed over two domains according to the InterPro server: the catalytic domain (PeptidaseM10A), containing 11 missense mutations, and the hemopexin domain harbouring only one missense mutation.
Evolutionary conservation analysis
Protein residues with high conservation scores are crucial for maintaining functional efficacy and structural integrity of proteins. The evolutionary conservation profile of MMP9 was assessed using the ConSurf analysis (Fig. 5a). This analysis revealed that 10 of the 12 nsSNPs displayed high conservation, with scores ranging from 8 to 9. Of these, six nsSNPs scored nine, indicating highly conserved residues. The presence of such conserved residues suggests their vital role in the biological function of proteins. The conservation scores for all the 12 SNPs are listed in Table 2.
Within the catalytic domain, 10 nsSNPs were identified, exhibiting high conservation scores of 8 and 9, except for Y262C, which had a score of 4. In contrast, only one nsSNP was identified in the hemopexin domain, with a score of 6. Additionally, a single nsSNP, R98L, was identified in the cysteine switch region with a conservation score of 9. InterPro predictions highlighted a loop region between the two domains that featured highly variable residues with lower conservation scores. Given the constraints of selecting both domains due to potential loop formation in structural analyses, 11 nsSNPs from the catalytic domain and the adjacent cysteine switch region were selected for further study.
Further, NetSurfP3 analysis revealed that seven of these eleven positions were deeply buried (Fig. 5b), suggesting that mutations in these residues could substantially impact the protein’s structural integrity. Additionally, two of these mutations were in alpha-helices, while the other lies in coiled regions (Table 2).
Structural and functional effects prediction by MutPred2
The 11 identified damaging nsSNPs were analysed using MutPred2 to forecast their potential impact on the structure and functionality of MMP9 protein. MutPred2 predictions encompass various alterations; detailed insights regarding these predictions are documented in Table 3.
Phenotypic and structural implications of NsSNP
The structural and functional effects of MMP9 nsSNPs owing to amino acid alterations were analysed using the HOPE tool (Figure S5) (S4 Table 9). The M422R mutation substitutes smaller, neutral methionine with larger, positively charged arginine which likely disrupt hydrophobic interactions and potentially comprised protein structure and function. The P180L variation replaces the rigid proline with a larger lysine, which may interfere with the unique backbone conformation induced by proline and affect protein interactions due to its surface location. The R98L variation alters the larger, neutral arginine to smaller, positively charged lysine, disrupting hydrogen bonds in the protein core and potentially causing misfolding. The D434N mutation replaces negatively charged aspartic acid with neutral asparagine, possibly affecting the function of a disordered region due to differences in amino acid properties. Mutations in G306C and G438A replaced the smaller, neutral glycine with a larger, hydrophobic cysteine and alanine, respectively, while G183E substituted glycine with the larger, hydrophilic glutamic acid. Glycine’s inherent flexibility is crucial for protein function; its mutation can disrupt interactions with other molecules or regions, especially since these changes occur in highly conserved, buried residues, leading to potential folding issues. The C329S mutation replaces a larger cysteine with a smaller, more flexible serine, destabilizing the rigid structure of the protein and disrupting core hydrophobic interactions, ultimately disorganizing its primary structure and function. The C347Y mutation converts cysteine to a hydrophobic tyrosine, and D390Y changes negatively charged aspartic acid to a neutral, hydrophobic tyrosine. Additionally, the Y262C mutation replaces a larger tyrosine with a smaller hydrophobic cysteine. The Y262C, C347Y, and D390Y mutations were in fibronectin type II domain 3, whereas C329S was found in domain 2, potentially compromising the function of these domains.
Predicted protein stability
Protein functionality is intrinsically linked to its stability, underscoring the significance of identifying variations in protein stability caused by nsSNPs. Five different software were used to assess the effect of harmful nsSNPs on the stability of MMP9 protein, that is, I-Mutant 2, iStable, MuPro, DynaMut, and DUET. As per the findings from iStable and MuPro, nine out of 11 nsSNPs exhibited a decrease in protein stability following mutation, with only P180L and R98L showing an increase in stability (Table 4). Conversely, results from I-Mutant indicated that all nsSNPs destabilized the protein and were classified as deleterious. D390Y was reported to stabilize the protein via both DynaMut and Duet, while all other nsSNP destabilized the protein structure according to dynaMut prediction. Duet predicted that G438A and C437Y also stabilize the protein. Since maximum of only two out of five softwares predicted the increase in stability of any variant, all 11 proteins were deemed suitable for further analysis.
The DynaMut bio-tool further predicted the effects of nsSNPs on protein stability and interatomic interactions. Analysis of the mutations, except for D390Y, which exhibited a stabilizing effect, revealed that almost all mutations destabilized interatomic hydrogen bonds and molecular interactions in the native MMP9 structure (Fig. 6). In Y262C, tyrosine formed four hydrogen bonds; however, when substituted with cysteine, only two hydrogen bonds were retained. The R98L mutation showed that arginine forms six hydrogen bonds, while leucine retained only one. In M422R, one hydrogen bond from methionine was lost due to the substitution with arginine. Similarly, in D434N, two hydrogen bonds formed by aspartic acid were replaced by other interactions after mutation to asparagine. The C437Y mutation results in the retention of only one hydrogen bond compared to the wild type. Although P180L, C329S, G183E, G306C, and G438A retained their initial hydrogen bonds, significant changes in other intermolecular interactions were observed following the mutation.
PTM predictions
PTMs add various functional groups to the side chains of proteins and play crucial roles in regulating protein folding, protein-protein interactions, and other essential protein functions. Therefore, identification of PTMs is imperative for disease research. In an investigation of potential methylated sites in MMP9, one methylation site was predicted by MusiteDeep at R98L. Both NetPhos3 and MusiteDeep predicted phosphorylation at Y262C. However, MusiteDeep and NetOgly4.0 did not predict ubiquitination or glycosylation sites.
3D modelling of the mutations of MMP9
The complete 3D structure of MMP9 is currently unavailable in the PDB. Hence, the sequence from UniProt served as the basis for conducting structural homology modelling of MMP9. According to the findings of Consurf and Interpro, the majority of conserved mutations lie within the catalytic domain. Consequently, structural investigations have focused only on nsSNPs in the catalytic domain. The homologous model was generated using the template c1L6JA. Among the 707 residues in MMP9, 405 were included in the projected model. Most remarkably, this projected structure included every highly conserved non-synonymous mutation. To provide a better conformational overview, the 3D structure of WT MMP9 protein is shown in Fig. 7a. The model was visualized using BIOVIA Discovery Studio and color-coded for clarity. Green represents the propeptide region (residues 20–109), red denotes the catalytic domain (residues 110–215 and 390–440), and pink highlights the three fibronectin type II domains (residues 216–389). The reliability of the generated protein model was validated using three distinct structural assessment tools: ERRAT, VERIFY3D, and QMEAN. The wild-type MMP9 model demonstrated high-quality structural parameters with a QMEAN score of 0.85, ERRAT score of 73.284, and VERIFY3D score of 96.15%, confirming the structural accuracy and robustness of the model. The 3D structures of all the mutants were predicted using Phyre2. For all mutant models, the ERRAT server’s prediction of the overall quality factor was > 70. Moreover, it was observed that in all models, more than 95% of the residues exhibited an average 3D-1D score ≥ 0.1, thus confirming the quality of all models (Table 5). Subsequently, TM-Align was used to compare the predicted protein models and determine RMSD and TM-scores. While RMSD measurements show the average distance between the backbone atoms of the mutant and wild proteins, TM-score offers information about the topological similarities between proteins. Notably, the mutant models of D390Y and R98L exhibited RMSD values of 0, leading to their elimination from further consideration. As a result, Pymol was used to choose and superimpose only those mutant structures (i.e., 9) that had RMSD values larger than 0.7 over the wild type (Fig. 7b). The RMSD and TM-scores for each model are listed in Table 4.
Structural visualization. (a) 3D structure of MMP9 protein generated by Phyre2. Green represents the propeptide region, red denotes the catalytic domain, and pink highlights the three fibronectin type II domains. (b) Superimposed structures of wild and mutant models. Yellow colour indicates the wild type while red magenta and blue colours depict the mutations.
Molecular dynamic simulations
Protein stability was evaluated using RMSD analysis (Fig. 8A). The results indicated a significant variation in RMSD patterns between the wild type and mutant proteins. The nine mutant variants exhibited higher fluctuations than the wild type, although their RMSD values eventually stabilized with a relatively lower fluctuation rate of approximately 0.49 nm. For the wild-type protein, a sharp increase in RMSD was observed between 15 and 25 ns, which stabilized after 30 ns, followed by two minor fluctuations at 40 and 45 ns. The average RMSD values for all variants (Table 6) revealed the following order of instability: Y262C > P180L > D434N > G438A > G183E > C329S > G306C > M422R > Wild > C347Y. Notably, the mutants exhibited higher average RMSD values compared to the native protein, indicating reduced stability. Among the mutants, Y262C, P180L, G438A, and D434N were the most unstable, with average RMSD values of 0.754, 0.685, 0.507, and 0.513 nm, respectively. These findings suggested that mutations can significantly alter the structural stability of proteins. Since proper protein function depends on a stable structure, it can be inferred that these mutations may compromise both the stability and activity of the protein.
RMSF analysis was conducted to evaluate the differences in structural flexibility among specific amino acid residues in the proteins (Fig. 8b). The RMSF values for both the wild type and mutant proteins were calculated to assess whether the mutations impacted the dynamic behaviour of the residues. Distinct fluctuation patterns were observed between the wild type and mutants, with four mutant simulations showing higher average RMSF values than the wild type. The mutants were ranked in the following order: Y262C > G438A > D434N > C347Y > Wild > G306C > M422R > P180L > G183E > C329S (Table 6). Significant fluctuations were particularly evident in the Y262C, G438A, and D434N variants, with average RMSF values of 0.295 nm, 0.226 nm, and 0.224 nm, respectively. These variants display increased residue fluctuations in multiple regions. The Y262C mutant exhibited notable fluctuations at residues 60–63, 130–131, 160–161, 282, 293–298, 306–308, and 317–325, with some fluctuations exceeding 0.5 nm. Similarly, the G438A mutant showed higher fluctuations at residues 29, 224, 296, 307–309, 354–357, and 367, whereas the D434N mutant showed fluctuations at residues 60, 61, 296, and 306. These findings suggest that the mutations affected the flexibility of the entire protein and not just individual residues. Overall, the results indicated that the mutations altered both the flexibility and activity of the proteins in this study. The RMSF findings were consistent with the RMSD results, further supporting this conclusion.
Intramolecular hydrogen bonds are critical for maintaining protein structure stability. To understand the relationship between structural flexibility and NH bond formation, the H-bond frequencies between the native and mutant proteins were analysed (Fig. 8c). The wild structure exhibited H-bonds from ~ 270 to ~ 240 throughout the simulation period. However, Y262C, P180L, D434N, G183E, and G306C were stable and showed slight variations around ~ 250 to ~ 240. Meanwhile, G438 and C329S fluctuated from ~ 260 to ~ 245, whereas C347Y and M422R fluctuated from ~ 260 to ~ 230. These variations highlight the destabilizing effects of these mutations on the MMP9 3D structure.
The solvent-accessible surface area (SASA) reflects the portion of the protein surface exposed to the surrounding solvent and plays a vital role in protein stability and residue rearrangement during folding. SASA values for both the wild-type and mutant proteins are illustrated in Fig. 8d. A lower SASA value indicates a compact protein structure, while a higher value suggests a more diffuse structure. Changes in SASA values indicate alterations in the protein’s structural conformation. To assess how mutations affected the structure of native proteins, the SASA values for the wild-type and nine mutant proteins were analysed. The average SASA values, shown in Table 6, reveal that the wild-type protein has a smaller average SASA value than the mutants. The ranking of the average SASA values was as follows: G183E > D434N > P180L > C347Y > G306C > G438A > Y262C > Wild > C329S > M422R. The wild-type protein exhibited SASA values ranging from ~ 222.5 to 212.5 nm². In comparison, slight variations were observed in the Y262C, P180L, and C329S mutants, with SASA values ranging from ~ 225–220 nm², ~ 227.5–220 nm², and ~ 223–213 nm², respectively. However, more pronounced changes were noted in the D434N, G183E, C347Y, G306C, G438A, and M422R mutants, with SASA values ranging from ~ 230–220 nm², ~ 230–215 nm², ~ 215–230 nm², ~ 225–210 nm², ~ 212–226 nm², and ~ 220–210 nm², respectively. These findings suggest that mutations can significantly alter the tertiary structures of protein. The increased SASA values in the mutants indicated that their structures were more expanded than those of the native proteins. The SASA results are consistent with the RMSD analysis, further supporting these observations.
To investigate how the structure and function of the protein were affected by these mutations, we analysed the superimposed structures of the wild-type and mutant proteins at various time intervals based on the RMSD values (Fig. 9). These findings revealed that mutations caused significant disturbances in several loop regions of the protein, particularly within the fibronectin type II domain. For instance, mutations at positions Y262C, P180L, G183E, M422R, D434N, and G438A occur within the catalytic site of the protein and induce notable alterations in the specific loop region (residues Arg424–Pro430) that form the catalytic site. Additionally, mutations at positions G306C, C329S, and C347Y are in the fibronectin type II domain (residues 216–389), which is critical for regulating protein-protein interactions and ligand binding to the MMP9 protein. These results suggest that point mutations can significantly affect protein stability, leading to structural changes that ultimately affect protein functionality.
Molecular docking
Molecular docking was employed to evaluate the impact of potential mutations in MMP9 on its interaction with a selected drug molecule. Based on a comprehensive literature review, the chosen molecule was identified as a novel inhibitor for modulating MMP activity in RA and was therefore used as the reference drug in this study. Docking analysis was performed for both wild-type MMP9 and its mutant forms (Y262C, P180L, C329S, M422R, G183E, D434N, G306C, G438A, and C347Y), as illustrated in Fig. 10. Corresponding 2D interaction maps were generated to visualize the binding interactions. The binding affinity of wild-type MMP9 with the drug molecule was calculated to be − 11.47 kcal/mol, with interacting amino acids aligned well with those reported in the literature. In contrast, the binding affinities for the mutant forms showed slight variations, calculated as − 11.99 kcal/mol for Y262C, − 11.98 kcal/mol for P180L, − 11.96 kcal/mol for M422R, − 11.98 kcal/mol for G438A, − 11.98 kcal/mol for D434N, − 11.94 kcal/mol for G183E, − 12.01 kcal/mol for G306C, − 11.97 kcal/mol for C329S, and − 11.98 kcal/mol for C347Y. Table 7 provides the specific amino acid residues in the active site that interacted with the drug molecule for both wild-type and mutant structures. For wild-type MMP9, the interacting residues included LEU418, ARG424, HIS401, VAL398, TYR423, and LEU397. Mutations Y262C, P180L, C329S, and C347Y showed identical interaction profiles to the wild type, with no new interactions observed. However, mutations M422R, G438A, D434N, G183E, and G306C introduced an additional hydrogen bond interaction with GLU416, alongside the residues observed in the wild type. This new interaction with GLU416 observed in five of the mutants underscores the structural alterations induced by these mutations, which may influence the binding dynamics and potentially modulate the activity of MMP9. These findings provide critical insights into how specific mutations in MMP9 can alter drug-binding properties, thereby offering a deeper understanding of the molecular mechanisms relevant to RA.
Gene-gene interaction
GeneMANIA and STRING were employed to anticipate how MMP9 interacts with other genes in the cell. Figure 11a presents an illustration of the STRING findings, revealing interactions with ten proteins: TIMP1, TIMP2, TIMP3, LCN2, MMP1, THBS1, CD44, CTSG, ELN, and DMP1. GeneMANIA predicted physical interactions of MMP9 with RECK, CXCL6, CD44, TGFB1, MEPIA, TIMP1, MMP15, COL12A1, MMP1, MMP14, CXCL6, MMP11, LCN2, MMP10, CXCL5, and PRSS2. Additionally, GeneMANIA identified genes that were co-expressed with MMP9, including CXCL5, PRSS2, LCN2, MMP10, MMP14, MMP1, MMP15, MMP8, COL12A1, and TIMP1. Pathway analysis revealed associations between TXLNG and HBFGF, whereas co-localization was observed with MMP8 and TGFB1. Furthermore, GeneMANIA predicted the shared domains between MMP9 and MMP1, MMP8, MMP10, MMP11, MMP14, and MMP15. The predictions generated by GeneMANIA are shown in Fig. 11b. Common genes between both software were TIMP1, MMP1, LCN2, and CD44.
Discussion
RA is a chronic autoimmune disease characterized by excessive inflammation, synovial hyperplasia, immune cell infiltration, pannus formation, and progressive joint damage40. MMP9, expressed in inflammatory cells and osteoclasts, is implicated in various pathological conditions, including cancer, placental malaria, and immune disorders41. In RA, MMP9 contributes to ECM degradation42, with elevated serum levels associated with decreased collagen synthesis43. This study examined MMP9 polymorphisms, identifying 4431 allelic variants from NCBI, including 723 missense variants. To validate the prediction of deleterious nsSNPs, a comprehensive bioinformatics approach was employed using SIFT, PROVEAN, PolyPhen2, PANTHER, and SNP & GO. These tools assess mutations based on evolutionary conservation, structural attributes, and sequence homology. To enhance reliability, PhD-SNP was used for further validation, leveraging machine-learning models trained on experimentally verified data. This multi-tool strategy minimizes biases and ensures robust identification of high-risk nsSNPs44. Using six predictive tools, 12 highly deleterious nsSNPs were identified with strong confidence scores, aligning with methodologies previously applied by Irfan et al.,45.
Domain analysis of MMP9 identified two key domains: a catalytic domain (residues 115–444) and a hemopexin domain (residues 514–704). The intervening loop region (445–513) lacked homologous structural templates in the PDB, precluding its computational modelling46. To ensure structural fidelity and predictive reliability, only the catalytic domain was selected for further analysis. This strategy is consistent with previous methodologies by Iraj et al. and Mehmood et al., where single-domain selection was necessitated by the absence of structural data, minimizing the risk of generating an unstable model47,48.
Evolutionary conservation analysis using ConSurf revealed that highly conserved amino acids were primarily located within the catalytic domain. Functionally significant residues tend to be highly conserved, making mutations at these sites potentially disruptive to protein function45,49. Mutations in buried residues may affect structural stability, while those in exposed regions could impact protein activity50. Among the 11 analysed mutations, seven were buried, and four were exposed. Notably, nsSNPs P180L, G183E, and G306C were highly conserved yet exposed, suggesting functional relevance. In contrast, D390Y, C329S, M422R, R98L, D434N, G438A, and C347Y were conserved and buried, indicating potential structural disturbances.
MutPred2 and HOPE analyses revealed that all nsSNPs induced structural, functional, and phenotypic changes in MMP9, highlighting their potentially deleterious nature51. Stability assessments using iStable, MuPro, I-Mutant 2.0, DynaMut, and DUET consistently showed that these mutations decrease protein stability. At least three of these tools confirmed reduced stability for each mutation, suggesting that these point mutations negatively impacts the stability of the MMP952.
PTMs are critical for protein folding, interactions, and disease progression53. Phosphorylation, a crucial PTM, modifies protein structure by activating or deactivating its function. Analysis using NetPhos and MusiteDeep showed that the Y262C mutation deleted a phosphorylation site at position 262. Despite having a conservation score of 4 and being exposed, this mutation may still influence protein structure and function. Mutations that disrupt phosphorylation sites can cause deleterious effects. Additionally, MusiteDeep identified the loss of a methylation site at residue 98 due to the R98L mutation, though it did not significantly alter protein structure, as indicated by stable RMSD values.
The Phyre2-generated model encompassed residues Val29 to Gly444, representing the 3D structure of the first domain. Standard structural validation tools were utilized to assess model reliability. The model achieved an ERRAT score of 73.28, indicating reasonable reliability in non-bonded atomic interactions compared to the crystal structure score of 88.6054. The QMEAN score of 0.85 closely aligned with the crystal structure score of 0.89, demonstrating high structural consistency48. Additionally, the VERIFY3D score of 96.15% closely matched the crystal structure score of 97.04%, confirming the accuracy of the residue environment55. These validation metrics, previously applied by Udosen et al., have been established as reliable for homology model assessment56. Similarly, mutant models were evaluated using these tools, all of which exhibited robust structural validation scores, supporting their accuracy. RMSD analysis was conducted to quantify structural deviations between mutant and wild-type models. Greater divergence between the mutant alpha carbon backbone and its wild-type counterpart was indicated by higher RMSD values. Conversely, minimal RMSD values suggest that the mutated structure closely resembles that of the wild type, with minimal impact on the structural chemistry of the protein57. Among the results, Y262C, P180L, C329S, M422R, G183E, D434N, G306C, G438A, and C347Y exhibited the highest deviation, while D390Y and R98L displayed the least deviation from the wild type, suggesting minimal impact of MMP9 polymorphism at these positions on the protein structure. Consequently, only nine mutant structures with high RMSD were superimposed onto the wild type for enhanced visualization of these nsSNPs. MD simulation results revealed the profound impact of these nsSNPs on the structural stability, flexibility, and solvent accessibility of MMP9. RMSD analysis demonstrated significant structural perturbations in the mutants compared to the wild type, with mutations in critical regions, such as the catalytic site and fibronectin type II domain, causing persistent fluctuations and reduced stability46. Several loop regions in MMP9 contribute to its flexibility and stability, which are integral to its function58. For instance, the S-shaped double loop stabilizes Zn²⁺ and Ca²⁺ ions near the active-site cleft, and any perturbation in this region can significantly affect the catalytic domain’s stability59. Similarly, the contact loop in the fibronectin II domain (residues 306–311) exhibits high flexibility, influencing domain interactions, while the specificity loop (residues Arg424–Pro430) impacts substrate binding and specificity, making it highly sensitive to structural changes60. Mutations, such as Y262C, disrupted native interactions in these loops, increasing flexibility and RMSD fluctuations. RMSF analysis corroborated these findings, highlighting heightened flexibility in loop regions crucial for structural integrity, particularly in mutants like Y262C, G438A, and D434N. SASA analysis indicated a more expanded and solvent-accessible conformation in the mutants, suggesting disrupted folding and stability. The superimposition of wild-type and mutant structures highlights disturbances in these key loop regions, indicating potential impairments in enzymatic activity and ligand binding61,62. The docking results complemented these findings by shedding light on the functional implications of these structural changes. While the wild-type protein exhibited a strong binding affinity (− 11.47 kcal/mol) with the drug molecule, mutants maintained comparable binding affinities, preserving the overall binding capability. However, structural rearrangements in the binding pocket, particularly in mutants like G438A and D434N, led to the emergence of a new hydrogen bond with GLU416. These changes aligned with the higher RMSD values and increased flexibility observed during simulations, indicating altered spatial positioning of residues in the binding pocket. Conversely, mutants with lower fluctuations, such as C329S and C347Y, did not exhibit this additional interaction, which is consistent with their relatively stable RMSD profiles. The increased SASA observed in mutants supports the docking findings, suggesting an expanded conformation that may enhance drug adaptability to altered binding pockets, albeit at the cost of structural destabilization. The heightened flexibility and disruption in critical regions observed in simulations underscore the potential impairment of enzymatic activity in these mutants despite preserved drug binding. Combined MD simulation and docking analyses revealed the dual impact of nsSNPs on protein stability and drug interactions. These findings emphasize the significance of integrating computational approaches to understand the structural and functional effects of genetic variations. Consistent with Mehmood et al.63, our results provide valuable insights for developing targeted therapeutic strategies for both wild-type and mutant MMP9.
To find the MMP9 protein’s interacting partners, a gene-gene interaction analysis was performed64. Mutations within ligand-binding domains and motifs may disrupt these interactions, potentially contributing to disease development. Using STRING and GeneMANIA databases, MMP9 was found to interact with 10 and 20 proteins, respectively. TIMP1, MMP1, LCN2, and CD44 were common interaction partners in both databases, with TIMP1 showing the strongest interaction. A similar approach was previously used by Irfan et al.,45.Tissue inhibitors of metalloproteinases (TIMPs) play a crucial role in regulating MMP activity and maintaining ECM balance65. Their involvement extends to various conditions, including vascular diseases, coronary syndromes, immune disorders, and heart failure66.
The four nsSNPs, Y262C, P180L, G438A, and D434N, identified in MMP9 open new avenues, as their association with human diseases remains unexplored. Our in-silico analysis revealed that these mutations reduce protein stability and cause significant structural deviations compared to the wild-type MMP9. Mutations in MMP9 are known to play a critical role in arthritis progression by affecting key processes such as ECM remodelling, inflammatory signalling, angiogenesis, cell migration, and tissue damage. Notably, imbalances in the MMP9/TIMP1 ratio have been linked to inflammatory arthritis67. Since MMP9 relies on ligand binding to regulate immune responses. Mutations, especially those affecting binding domains may disrupt this interaction, potentially contributing to cancer and autoimmune diseases.
While this study provides a comprehensive analysis of pathogenic nsSNPs in the MMP9 gene through an array of bioinformatics tools, it is important to acknowledge certain limitations. The absence of experimental validation is a significant limitation, as computational predictions, although robust and multidimensional, cannot fully capture the complexity of biological systems. In vitro assays and in vivo models are essential to confirm the functional consequences of the predicted mutations and validate their relevance to disease susceptibility and progression. Furthermore, this study does not currently address the population-specific prevalence of these nsSNPs, which is crucial for understanding their broader clinical significance. Certain variants may have distinct allele frequencies across populations, thereby influencing their contribution to RA pathology. Future work will focus on integrating experimental approaches to substantiate these findings while analysing population-specific allele frequencies using genetic databases. This will help to bridge the gap between in silico predictions and real-world biological implications.
Conclusion
Our comprehensive bioinformatics analysis identified nine nsSNPs in MMP9 gene, predominantly within the catalytic domain, with potential detrimental effects on protein function and stability. Notably, the Y262C mutation’s loss of a phosphorylation site and the structural destabilization observed in key variants underscore their potential as biomarkers for RA susceptibility. Further functional studies are imperative to elucidate the biological mechanisms of these polymorphisms in RA pathogenesis. Additionally, population-based investigations are crucial for assessing their prevalence and informing targeted therapeutic strategies tailored to the affected individuals.
Data availability
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
References
Yan, M., Su, J. & Li, Y. Rheumatoid arthritis-associated bone erosions: evolving insights and promising therapeutic strategies. Biosci. Trends. 14 (5), 342–348 (2020).
Weyand, C. M. & Goronzy, J. J. The immunology of rheumatoid arthritis. Nat. Immunol. 22 (1), 10–18 (2021).
Scherer, H. U., Häupl, T. & Burmester, G. R. The etiology of rheumatoid arthritis. J Autoimmun [Internet]. ;110:102400. (2020). Available from: https://www.sciencedirect.com/science/article/pii/S0896841119308431
Grillet, B. et al. Proteoform analysis of matrix metalloproteinase-9/gelatinase B and discovery of its citrullination in rheumatoid arthritis synovial fluids. Front. Immunol. 12, 763832 (2021).
Salomão, R. et al. Involvement of matrix metalloproteinases in COVID-19: molecular targets, mechanisms, and insights for therapeutic interventions. Biology (Basel). 12 (6), 843 (2023).
Yin, N. et al. A novel indomethacin/methotrexate/MMP-9 SiRNA in situ hydrogel with dual effects of anti-inflammatory activity and reversal of cartilage disruption for the synergistic treatment of rheumatoid arthritis. Nanoscale 12 (15), 8546–8562 (2020).
Koss, C. K. Macrophage Phenotypes in Lung Fibrosis. ; (2021).
Kar, S. et al. Interleukin-9 facilitates osteoclastogenesis in rheumatoid arthritis. Int. J. Mol. Sci. 22 (19), 10397 (2021).
Varghese, A., Chaturvedi, S. S., Fields, G. B. & Karabencheva-Christova, T. G. A synergy between the catalytic and structural Zn (II) ions and the enzyme and substrate dynamics underlies the structure–function relationships of matrix metalloproteinase collagenolysis. JBIC J. Biol. Inorg. Chem. 26, 583–597 (2021).
Li, S. et al. COL3A1 and MMP9 serve as potential diagnostic biomarkers of osteoarthritis and are associated with immune cell infiltration. Front. Genet. 12, 721258 (2021).
Minyaylo, O., Ponomarenko, I., Reshetnikov, E., Dvornyk, V. & Churnosov, M. Functionally significant polymorphisms of the MMP-9 gene are associated with peptic ulcer disease in the Caucasian population of central Russia. Sci. Rep. 11 (1), 13515 (2021).
Sohn, E. H. et al. Genetic association between MMP9 and choroidal neovascularization in age-related macular degeneration. Ophthalmol. Sci. 1 (1), 100002 (2021).
Škereňová, M. et al. Common gene haplotypes of gelatinases and their tissue inhibitors in abdominal aortic aneurysm. Gen. Physiol. Biophys. 39(1), 37–41 (2020).
Consortium, U. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 47 (D1), D506–D515 (2019).
Sherry, S. T. et al. DbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 29 (1), 308–311 (2001).
Sim, N-L. et al. SIFT web server: predicting effects of amino acid substitutions on proteins. Nucleic Acids Res [Internet]. ;40(W1):W452–7. (2012). Available from: https://doi.org/10.1093/nar/gks539
Choi, Y. & Chan, A. P. PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics 31 (16), 2745–2747 (2015).
Adzhubei, I., Jordan, D. M. & Sunyaev, S. R. Predicting functional effect of human missense mutations using PolyPhen-2. Curr. Protoc. Hum. Genet. 76 (1), 7–20 (2013).
Thomas, P. D. et al. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 13 (9), 2129–2141 (2003).
Capriotti, E. et al. WS-SNPs&GO: a web server for predicting the deleterious effect of human protein variants using functional annotation. BMC Genom. 14, 1–7 (2013).
Capriotti, E. & Fariselli, P. PhD-SNPg: a webserver and lightweight tool for scoring single nucleotide variants. Nucleic Acids Res. 45 (W1), W247–W252 (2017).
Paysan-Lafosse, T. et al. InterPro in 2022. Nucleic Acids Res. 51 (D1), D418–D427 (2023).
Rubin, M. & Ben-Tal, N. Using consurf to detect functionally important regions in RNA. Curr. Protoc. 1 (10), e270 (2021).
Høie, M. H. et al. NetSurfP-3.0: accurate and fast prediction of protein structural features by protein Language models and deep learning. Nucleic Acids Res. 50 (W1), W510–W515 (2022).
Pejaver, V. et al. Inferring the molecular and phenotypic impact of amino acid variants with MutPred2. Nat. Commun. 11 (1), 5918 (2020).
Burley, S. K. et al. Protein data bank (PDB): the single global macromolecular structure archive. Protein Crystallogr. Methods Protoc. 1607, 627–641 (2017).
Boutet, E. et al. UniProtKB/Swiss-Prot, the manually annotated section of the UniProt knowledgebase: how to use the entry view. Plant. Bioinforma Methods Protoc. 1347, 23–54 (2016).
Nagin, D. S. Project HOPE: does it work? Criminol. Pub Pol’y. 15, 1005 (2016).
Rodrigues, C. H. M., Pires, D. E. V. & Ascher, D. B. DynaMut: predicting the impact of mutations on protein conformation, flexibility and stability. Nucleic Acids Res. 46 (W1), W350–W355 (2018).
Wang, D. et al. MusiteDeep: a deep-learning based webserver for protein post-translational modification site prediction and visualization. Nucleic Acids Res. 48 (W1), W140–W146 (2020).
Poon, A. & Predicting Phosphorylation A critique of the netphos program and potential alternatives. Stanf. Biochem. 218, 582–585 (2004).
Shinwari, K. et al. In-silico assessment of high-risk non-synonymous SNPs in ADAMTS3 gene associated with Hennekam syndrome and their impact on protein stability and function. BMC Bioinformatics [Internet]. ;24(1):251. (2023). Available from: https://doi.org/10.1186/s12859-023-05361-6
Zhang, Y. & Skolnick, J. TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 33 (7), 2302–2309 (2005).
Dym, O. VERIFY3D Dym 2008. Int. Union Crystallogr. F, 521 (2006).
Van Der Spoel, D. et al. GROMACS: fast, flexible, and free. J. Comput. Chem. 26 (16), 1701–1718 (2005).
Sharif, M., John, P., Bhatti, A., Paracha, R. Z. & Majeed, A. Evaluation of the inhibitory mechanism of Pennisetum glaucum (pearl millet) bioactive compounds for rheumatoid arthritis: an in vitro and computational approach. Front Pharmacol., 7(November) (2024).
Liu, N. et al. Computational study of effective matrix metalloproteinase 9 (MMP9) targeting natural inhibitors. Aging (Albany NY). 13 (19), 22867–22882 (2021).
Sharma, S., Sharma, A. & Gupta, U. Molecular Docking studies on the Anti-fungal activity of Allium sativum (Garlic) against mucormycosis (black fungus) by BIOVIA discovery studio visualizer 21.1. 0.0. Ann. Antivirals Antiretrovir. 5 (1), 28–32 (2021).
Franz, M. et al. GeneMANIA update 2018. Nucleic Acids Res. 46 (W1), W60–W64 (2018).
Figus, F. A., Piga, M., Azzolin, I., McConnell, R. & Iagnocco, A. Rheumatoid arthritis: extra-articular manifestations and comorbidities. Autoimmun. Rev. 20 (4), 102776 (2021).
Zhang, H. et al. MMP9 protects against LPS-induced inflammation in osteoblasts. Innate Immun. 26 (4), 259–269 (2020).
Stojanovic, S. K., Stamenkovic, B. N., Cvetkovic, J. M., Zivkovic, V. G. & Apostolovic, M. R. A. Matrix Metalloproteinase-9 level in synovial Fluid—Association with joint destruction in early rheumatoid arthritis. Med. (B Aires). 59 (1), 167 (2023).
Mousavi, M. J. et al. Transformation of fibroblast-like synoviocytes in rheumatoid arthritis; from a friend to foe. Autoimmun. Highlights. 12, 1–13 (2021).
Joshi, I. et al. Artificial intelligence, big data and machine learning approaches in genome-wide SNP-based prediction for precision medicine and drug discovery. In Big Data Analytics in Chemoinformatics and Bioinformatics. (eds. Basak, S. C. & Vračko, M.) 333–357 (Elsevier, 2023).
Irfan, M., Iqbal, T., Hashmi, S., Ghani, U. & Bhatti, A. Insilico prediction and functional analysis of nonsynonymous SNPs in human CTLA4 gene. Sci Rep [Internet]. ;12(1):1–11. (2022). Available from: https://doi.org/10.1038/s41598-022-24699-0
Mondal, S., Adhikari, N., Banerjee, S., Amin, S. A. & Jha, T. Matrix metalloproteinase-9 (MMP-9) and its inhibitors in cancer: A minireview. Eur J Med Chem [Internet]. 194:112260. (2020). Available from: https://www.sciencedirect.com/science/article/pii/S0223523420302270
Mehmood, A., Nawab, S., Jin, Y., Kaushik, A. C. & Wei, D. Q. Mutational impacts on the N and C terminal domains of the MUC5B protein: A transcriptomics and structural biology study. ACS Omega 8 (2022).
Ahmed, I., John, P. & Bhatti, A. Association analysis of Vascular Endothelial Growth Factor-A (VEGF-A) polymorphism in rheumatoid arthritis using computational approaches. Sci Rep [Internet]. ;13(1):1–15. (2023). Available from: https://doi.org/10.1038/s41598-023-47780-8
Kaushik, A. C. et al. Mining Cancer Cell Line-Based Drugs to Benefit KRAS(G12D)Pancreatic Adenocarcinoma Patients. Proc – 2021 IEEE Int Conf Bioinforma Biomed BIBM. 2021;2423–8. (2021).
Savojardo, C., Manfredi, M., Martelli, P. L. & Casadio, R. Solvent accessibility of residues undergoing pathogenic variations in humans: from protein structures to protein sequences. Front. Mol. Biosci. 7, 626363 (2021).
Nawar, N. et al. Structure analysis of deleterious NsSNPs in human PALB2 protein for functional inference. Bioinformation 17 (3), 424 (2021).
Marabotti, A., Scafuri, B. & Facchiano, A. Predicting the stability of mutant proteins by computational approaches: an overview. Brief. Bioinform. 22 (3), bbaa074 (2021).
Zhong, Q. et al. Protein posttranslational modifications in health and diseases: functions, regulatory mechanisms, and therapeutic implications. MedComm 4 (3), e261 (2023).
Mahmud, S. et al. Designing a multi-epitope vaccine candidate to combat MERS-CoV by employing an immunoinformatics approach. Sci Rep [Internet]. ;11(1):15431. (2021). Available from: https://doi.org/10.1038/s41598-021-92176-1
Reddy, P. P. Homology modeling of microbial nattokinase enzyme, an anti-blood clotting (Fibrinolytic) agent using computational tools. Res. J. Pharm. Technol. 13 (9), 4135–4138 (2020).
Udosen, B. et al. In-silico analysis reveals druggable single nucleotide polymorphisms in angiotensin 1 converting enzyme involved in the onset of blood pressure. BMC Res Notes [Internet]. ;14(1):457. (2021). Available from: https://doi.org/10.1186/s13104-021-05879-z
Zolfaghari, M. et al. Resistance mechanism of plutella Xylostella (L.) associated with amino acid substitutions in Acetylcholinesterase-1: insights from homology modeling, Docking and molecular dynamic simulation. Insects 15 (3), 144 (2024).
Vandooren, J., Van den Steen, P. E. & Opdenakker, G. Biochemistry and molecular biology of gelatinase B or matrix metalloproteinase-9 (MMP-9): The next decade. Crit Rev Biochem Mol Biol [Internet]. ;48(3):222–72. (2013). Available from: https://doi.org/10.3109/10409238.2013.770819
Tochowicz, A. et al. Crystal Structures of MMP-9 Complexes with Five Inhibitors: Contribution of the Flexible Arg424 Side-chain to Selectivity. J Mol Biol [Internet]. ;371(4):989–1006. (2007). Available from: https://www.sciencedirect.com/science/article/pii/S0022283607007085
Elkins, P. A. et al. Structure of the C-terminally truncated human ProMMP9, a gelatin-binding matrix metalloproteinase. Acta. Crystallogr. Sect. D [Internet]. 58(7), 1182–1192. (2002). https://doi.org/10.1107/S0907444902007849
Wang, Q. et al. Computational screening and analysis of lung cancer related non-synonymous single nucleotide polymorphisms on the human Kirsten rat sarcoma gene. Molecules 24(10), 1–20 (2019).
Mehmood, A. et al. Structural dynamics behind clinical mutants of PncA-Asp12Ala, Pro54Leu, and His57Pro of Mycobacterium tuberculosis associated with Pyrazinamide resistance. Front. Bioeng. Biotechnol. 7 (December), 1–16 (2019).
Mehmood, A., Kaushik, A. C., Wang, Q., Li, C. D. & Wei, D. Q. Bringing structural implications and deep Learning-Based drug identification for KRAS mutants. J. Chem. Inf. Model. 61 (2), 571–586 (2021).
Dhivyadharshini, J. & Rajasekar A. Gene-gene interaction interactome of periodontal disease & and pancreatic cancer lead by angiogenesis pathway and epidermal growth factor receptor signaling pathway. Int. J. Med. Dent. 27(2), 189 (2023).
Gomez, D. E., Alonso, D. F., Yoshiji, H. & Thorgeirsson, U. P. Tissue inhibitors of metalloproteinases: structure, regulation and biological functions. Eur. J. Cell. Biol. 74 (2), 111–122 (1997).
Niño, M. E. et al. TIMP1 and MMP9 are predictors of mortality in septic patients in the emergency department and intensive care unit unlike MMP9/TIMP1 ratio: Multivariate model. PLoS One [Internet]. ;12(2):e0171191. (2017). Available from: https://doi.org/10.1371/journal.pone.0171191
Giannelli, G. et al. MMP-2, MMP-9, T I M P-1 and TIMP-2 levels in patients with rheumatoid arthritis and psoriatic arthritis. BRIEFPAPER Clin. Exp. Rheumatol. 22, 335–338 (2004).
Author information
Authors and Affiliations
Contributions
M.S. is the main authors of the paper. K.R helped in data analysis. P.J. helped in designing the methodology and he is the research supervisor and corresponding author of this article. A.B. co-supervised and reviewed this article.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Sharif, M., Rasool, K., John, P. et al. Decoding pathogenic MMP9 variants in rheumatoid arthritis using computational and molecular dynamics approaches. Sci Rep 15, 33743 (2025). https://doi.org/10.1038/s41598-025-92553-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-92553-0