Abstract
The rise of the convergence of hypervirulence and carbapenem resistance in Klebsiella pneumoniae has been increasingly reported in recent years, however, there are few outbreak cases for these producing NDM carbapenemase. In this study, ST65-KL2 and ST11-KL64 hypervirulent and carbapenem-resistant K. pneumoniae (hvCRKP) were identified from two different outbreak cases: (1) clonal spreading of ST65-KL2 in five patients within transplantation wards spanning three months; and (2) clonal transmission of ST11-KL64 in ten patients across 10 months. The representative strains of ST65-KL2 and ST11-KL64 hvCRKP, K22877 and K56649, produced carbapenemase NDM-5 and dual carbapenemases KPC-2 and NDM-13, respectively, and both exhibited high-level carbapenem resistance. Moreover, virulent analysis showed that K22877 and K56649 were hypervirulent and the former possessed stronger virulence. Evolutionary pathways suggested ST65-KL2 and ST11-KL64 hvCRKP could be classified as CR-hvKP (hvKP acquiring carbapenem resistance) and hv-CRKP (CRKP acquiring hypervirulence), respectively. Unexpectedly, ST65-KL2 CR-hvKP showed resistance to ciprofloxacin mediated by plasmid acquisition as its spread, and ST11-KL64 hv-CRKP developed into enhanced virulence and macrophage resistance. Furthermore, compared to the ST65-KL2 CR-hvKP, the ST11-KL64 hv-CRKP tends to cause occult and persistent infection. Global genome analysis revealed ST11-KL64 hv-CRKP and ST65-KL2 CR-hvKP mainly carried blaKPC-2 and had significant differences in Ompk35/36, ybt, resistance and virulence. Effective surveillance should be implemented and novel therapeutic strategies are urgently needed to deal with refractory infections.
Similar content being viewed by others
Introduction
Klebsiella pneumoniae is a notorious opportunistic pathogen causing approximately a third of all Gram-negative bacterial infections, including bloodstream infection, pneumonia and urinary tract infection, with high morbidity and mortality1,2,3. Conventionally, it was divided into two groups: hypervirulent K. pneumoniae (hvKP) possessing high pathogenicity and capable of causing community-acquired infections, mainly in immunocompetent individuals, and classical K. pneumoniae (cKP) easily obtaining multi-drug resistance (MDR) and mainly resulting in nosocomial infections in immunocompromised patients4.
Compared to cKP, hvKP exhibits some hypervirulent features, such as capsule loci types (KL1 and KL2) and virulence plasmids (pLVPK and pK2044), usually shows hypermucoviscosity by string test and sensitivity to most antibiotics, and produces more capsular polysaccharides (CPS) and siderophore4. Furthermore, virulence plasmids usually harbor mucoid regulator genes (rmpADC or rmpA2), aerobactin genes (iutAiucABCD) and salmochelin genes (iroBCDN)5. However, the definition of hvKP remains ambiguous, as likely to cause excessive reporting in hvKP6. cKP will be easier to acquire resistance phenotypes than hvKP, owing to its lack of thick CPS, which contributes to the rise of carbapenem- and polymyxin-resistant cKP, individually or simultaneously. They commonly belong to sequence type (ST) 258, ST11, ST512, ST307 and ST157,8.
Furthermore, the increase of carbapenem-resistant K. pneumoniae (CRKP), largely due to carbapenemase production, become a global and serious threat to public health9. KPC, NDM and OXA-48 are the most popular carbapenemases for CRKP globally, and outbreak cases of CRKP harbored those carbapenemases have been documented in Europe, Asia, America, causing trouble with prevention and control10,11,12.
What is noteworthy is that the convergence of hypervirulent and carbapenem-resistant K. pneumoniae (hvCRKP) has occurred and showed great potential to cause difficult-to-treat infections13. Those strains might be generated in the following three pathways: CRKP acquiring hypervirulence (hv-CRKP), hvKP acquiring carbapenem resistance (CR-hvKP) and cKP acquiring both hypervirulence and carbapenem resistance (hvCR-KP). hv-CRKP, CR-hvKP and hvCR-KP isolated from patients have been sporadically reported within the last five years, such as ST11-KL64/KL47 hv-CRKP, ST23-KL1/ ST86-KL2 CR-hvKP and ST11-KL64 hvCR-KP14,15. The blaKPC-containing hvCRKP has been reported; however, there is limited data on blaNDM-harboring hvCRKP and its outbreaks16.
In the present study, to the best of our knowledge, we first reported two different types of fatal outbreaks, caused by ST11-KL64 hv-CRKP co-carrying blaKPC-2 and blaNDM-13 and ST65-KL2 CR-hvKP carrying blaNDM-5 in intensive care units (ICUs) and transplantation wards of a tertiary hospital from March 2023 to December 2023 and May 2019 to August 2019, respectively, in China. Whole-genome sequencing (WGS) and a series of phenotypic experiments were performed for the in-depth understanding of the above two clonal outbreaks. Dynamic evolutions of resistance and virulence were also identified in the process of dissemination. Furthermore, a thorough comparison of the genomic and clinical characteristics of the above two types of hvCRKP was also carried out.
Results
Two outbreaks of hvCRKP spreading rapidly but latently
The emergence and spread of carbapenemase-producing hvCRKP were identified by antibiotic sensitivity testing (AST), APB (3-aminophenylboronic acid) and EDTA enhancement method (APB/EDTA method), polymerase chain reaction (PCR) and WGS. In the present study, since June 2019, 15 hvCRKP strains were collected from five patients, including three kidney transplant recipients and two lung recipients, within three months (Fig. 1 and Table 1), and these belonged to the same cluster confirmed by Xbal-PFGE, cgMLST and phylogenetics (Figs. 2A, S1 and S5), revealing the outbreak of hvCRKP occurred during transplantation and hospitalization due to the monophyletic spread. Moreover, the isolation and hospitalization timelines for those hvCRKP strains revealed they derived from the two lungs and kidneys of the same donor, respectively, and then disseminated in the same ward due to close contact (Figs. 1 and 2B). Fortunately, aztreonam was an effective remedy for combating the hvCRKP and shortening the course of the disease. However, the severe complications after lung transplantation in Patients 1-1 and 1-2 brought them into critical condition, and they died in two weeks.
A Phylogenetic relatedness of the hv-CRKP (n = 21), CR-hvKP (n = 15) and reference strains (hvKP NTUH-2044 and two CRKP, HS11286 and K55397) was illustrated. Patient ID, clustering, ST, K_locus, O_locus, KPC, NDM, rmpA, rmpA2, sample type and collection date were annotated in the outbreak clone. ST sequence type, KPC K. pneumoniae carbapenemases, NDM New Delhi metallo-β-lactamases. B Timeline of outbreaks of two different hvCRKP. The left showed one chain of clonal spreading of ST65-KL2 CR-hvKP and the right showed five chains of clonal dissemination of ST11-KL64 hv-CRKP. Different chains of clonal transmission were identified by collection date of strains, initial inpatient department, bed location and transplantation surgery. The Fig. 2B was created with BioRender.com by us, and its icons derived from BioRender.
Another outbreak case was caused by ST11-KL64 hvCRKP, the most prominent ST-KL in China, which was first discovered in the blood and bronchoalveolar lavage fluid (BALF) of a patient suffering from acute exacerbation of chronic obstructive pulmonary disease and resulted in septic shock and death. The hvCRKP, co-carrying blaKPC-2 and blaNDM-13, attracted our great attention owing to its hypervirulence and MDR. Over the following ten months, the hvCRKP was sequentially isolated from nine patients with kidney transplantation, tumor, cholangitis and intracerebral hemorrhage (Table 1). All 21 ST11-KL64 hvCRKP, five chains of clonal transmission, were categorized into a single cluster by the same methods as the ST65-KL2 hvCRKP (Fig. 2A). Treatment effectiveness against the ST11-KL64 hvCRKP was limited, which caused recurrent infections and death, although the strains showed sensitivity to the combination of aztreonam and ceftazidime-avibactam (ATM-CZA). Patients 2-2, 2-3, 2-4, 2-6, 2-8, and 2-10 suffering chronic renal failure have undergone kidney transplantation in the same ward, and had close contact in time and space within seven months, implying nosocomial spread (Figs. 1 and 2B). Of note, occult infections due to the retention of hvCRKP in Patients 2-2 and 2-6 have existed during the initial hospitalization and both appeared clinical infective symptoms in post-discharge. Interestingly, after admission, hvCRKP was immediately isolated from four patients, Patients 2-1, 2-5, 2-7 and 2-9, hinting that hvCRKP have disseminated regionally.
Resistance phenotype and molecular characterization of hvCRKP
ST11-KL64 hvCRKP strain K56649 and ST65-KL2 hvCRKP strain K22877 were taken as the representative strains for further multidrug resistance research.
AST showed ST11-KL64 K56649 expressed resistance to most beta-lactam antibiotics, including carbapenems, aztreonam and CZA, aminoglycosides, fluoroquinolones and sulfamethoxazole and trimethoprim (SXT), but was sensitive to colistin and tigecycline (Table 2). A series of resistance genes were detected in K56649 by WGS and were consistent with resistance phenotypes to beta-lactams (blaKPC-2, blaNDM-13, blaCTX-M-65, blaTEM-1B and blaLAP-2), aminoglycosides (rmtB), fluoroquinolones (qnrS1) and SXT (dfrA14, sul1 and sul2). Nanopore sequencing and S1-PFGE revealed K56649 harbored one chromosome and five plasmids, designated as pK56649-1, pK56649-2-KPC, pK56649-3-VIR, pK56649-4-NDM and pK56649-5 (Table S1). Replicons of those plasmids were ColRNAI, IncFII(pHN7A8)/IncR, IncHI1B/IncFIB(K), IncI1 and undetected replicon, respectively. pK56649-2-KPC harbored blaKPC-2 and its replicon was popular in blaKPC-2-carrying plasmids. The surrounding environment of blaKPC-2, located on an IncFII(pHN7A8)/IncR plasmid pK56649-2-KPC, was IS26-hp-hp-klcA-korC-ΔISKpn6-blaKPC-2-ISKpn27-Tn3tnpR-IS26, which shared the core structure with two of the most popular blaKPC-2-harboring genetic environments in China, United States and Europe, Tn1721-based structure and Tn4401 (Fig. 3A). As previously reported, the blaKPC-2-harboring environment has a potential spread to other plasmids in the same strains by two sides of IS26 via transposable recombination, although transposable elements released inside the cytoplasm were not found17. Another carbapenemase gene blaNDM-13 was carried by IncI1 plasmid pK56649-4-NDM and first reported in plasmid pIOMTU558-NDM of Escherichia coli. To the best of our knowledge, blaNDM-13 harbored by K. pneumoniae, was first found in this study. Importantly, plasmid pNDM13-SR33 of Salmonella enterica SR33 was identical to pK56649-4-NDM (Fig. 3B), which suggested pK56649-4-NDM might be transferred to K. pneumoniae from S. enterica. Compared to NDM-1, NDM-13 with two substitutions (D95N and M154L, Fig. S2) displayed significant hydrolytic activity against cefotaxime and no changes for carbapenems. Genetic environment of blaNDM-13 was IS1294-ΔISAba125-blaNDM-13-ΔbleMBL-trpF. Two specific opposite sequences (CTTG) flanking IS1294 revealed IS1294 inserted ISAba125 by rolling-circle transposition. Moreover, the downstream sequence of blaNDM-13 also existed many characteristic target sites (GTTC), probably indicating blaNDM-13 was mobilized by IS1294, as previously reported18. Furthermore, conjugative prediction and conjugation experiments showed blaNDM-13-harboring pK56649-4-NDM have complete conjugative regions and capability of spread to K. pneumoniae KP54 and E. coli J53 (Fig. S3). However, blaKPC-2 only could be transferred to K. pneumoniae KP54 with the help of pK56649-4-NDM by plasmid transfer since pK56649-2-KPC lacked the genes encoding relaxase and type IV coupling protein (T4CP).
A Linear alignment of blaKPC-2-bearing structure on pK56649-2-KPC (plasmid of ST11-KL64 hv-CRKP) and two classical surroundings, Tn1724-based structure and Tn4401. ORFs are indicated by arrows. Sequences of shared homology are marked by grey shading. B Linear alignment of blaNDM-13-bearing pK56649-4-NDM of K56649 (the ST11-KL64 hv-CRKP) and pNDM13-SR33 of Salmonella enterica SR33. C Circular alignment analysis of plasmid pK56649-3-VIR (plasmid of ST11-KL64 hv-CRKP), pK22877-1-VIR (plasmid of ST65-KL2 CR-hvKP) and two classical hypervirulent plasmids, pLVPK and pK2044. Virulence genes were marked by red shading. D Maximum-likelihood phylogenetic tree of pK56649-2-KPC and its homological plasmids. E Maximum-likelihood phylogenetic tree of pK56649-4-NDM and its homological plasmids. Circles from the inside out showed species, carbapenemase, country and year, and the number in the circle was tree scale.
Compared to ST11-KL64 hvCRKP strain K56649, ST65-KL2 strain K22877 remained susceptible to aztreonam, aminoglycosides and quinolones, which offered more choices to combat infections. blaNDM-5 was the only β-lactamase resistance gene identified in K22877 using WGS (Table 2), as might explain why it was sensitive to aztreonam. Moreover, genome analysis revealed that K22877 carried one chromosome and two plasmids, named pK22877-1-VIR and pK22877-2-NDM, belonging to IncFIB(K)/IncHI1B and IncX3, respectively. pK22877-2-NDM harbored blaNDM-5 and its replicon was a common replicon of blaNDM-harboring plasmid in Enterobacteriales. Moreover, the genetic environment of blaNDM-5 was ΔIS3000-IS5-ΔISAba125-blaNDM-5-bleMBL-trpF-dsbD-ΔcutA-IS26-ISKox3. Conjugation assays indicated pK22877-2-NDM carrying blaNDM-5 could be transferred to E. coli J53 and K. pneumoniae KP54, identified by PCR, to cause the spread of carbapenem resistance (Fig. S3).
Molecular and phenotypic characterizations of hypervirulence in ST11-KL64 and ST65-KL2 hvCRKP
Genomic analysis indicated all ST11-KL64 (n = 21) and ST65-KL2 (n = 15) hvCRKP harbored hypervirulent-like plasmid, pK56649-3-VIR and pK22877-1-VIR, respectively. The two hypervirulent-like plasmids have >90% coverage and >99% identity with the typical hypervirulent plasmid pLVPK and pK2044 (Fig. 3C). Replicons of the four plasmids were identical and belonged to IncHI1B/IncFIB. pK22877-1-VIR carried a series of hypervirulent markers, including rmpA, iutAiucABCD and iroBCDN, the same as pLVPK and pK2044 (Table S1).
Compared to pK22877-1-VIR, pK56649-3-VIR did carry rmpA, rmpA2 and iutAiucABCD, but did not carry iroBCDN. Interestingly, the promotor of rmpA in plasmid pK56649-3-VIR, was truncated by ISKpn26 in the position -6 region of rmpA, causing the block of transcription (Fig. S4).
Likely, the same truncations in ST11-KL64 strains of spread chains I, II and V were found. Of note is that the promotor of rmpA in ST11-KL64 strains of spread chain III and IV was cut off by ISKpn74 in –2 region of rmpA except for K60365 of chain IV. The promotor of rmpA of K60365 showed deletion of 2802 bp in –2 region of rmpA. However, the rmpA2 and its promoter in the ST11-KL64 hvCRKP were intact, compared to the incomplete promotor of rmpA. The rmpA2 was identical to that of pK2044, a hypervirulent plasmid of KL1 hvKP, which significantly enhanced hypervirulence in K. pneumoniae. Opposite to pK56649-3-VIR, pK22877-1-VIR harbored a complete and wild promotor P11T of rmpA, and did not contain rmpA2. Additionally, K22877 and K56649 contained other virulence factors located in chromosome, including mrkABCDFHIJ, fimABCDEFGHIK, complete iron uptake system (entABCDEFS, fepABCDG) and a series of T6SS genes.
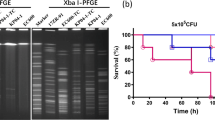
To verify the hypervirulence of two hvCRKP, a battery of phenotype experiments was carried out. K22877 was found to be hypermucoviscous by string test in a blood plate medium, whereas K56649 was not. G. mellonella infection model and murine model were performed to identify the virulence of K56649 and K22877(Fig. 4A–C). Analysis suggested mortality of the larvae (injection with 1 × 105 CFU) and the mice (injection with 2 × 107 CFU) infected with K56649 and K22877, respectively, was significantly higher compared to hypovirulent strain ATCC 700603 (P < 0.01), and comparable to hypervirulent strain NTUH-K2044 (P > 0.05), using Kaplan–Meier survival curve, which implied K56649 and K22877 were hypervirulent CRKP.
A The survival rates of K56649 (ST11-KL64 hv-CRKP), K22877 (ST65-KL2 CR-hvKP) and K60365 (ST11-KL64 hv-CRKP) in G. mellonella infection model. B, C Murine model infected with K56649, K22877 and K60365 using different dose (2 × 106 and 2 × 107 CFU, respectively). D Mucoviscosity difference between K56649 and K60365. E, F Comparison of relative thickness and uronic acids content of bacteria capsular between K56649 and K60365. Relative thick was calculated by total diameter/bacterial diameter. G Survival rates of K22877, K56649 and K60365 resistance to macrophage phagocytosis. H Survival rates of K56649 and K60365 inside of macrophage. I Comparison of biofilm formation among K22877, K56649, K60365 and control strains.
Evolutionary pathways of ST11-KL64 and ST65-KL2 hypervirulent CRKP
The KPC-2- and NDM-13-producing hvCRKP belonged to ST11-KL64, a classical ST-KL of cKP, and kept a closer phylogenetic distance with cKP but not hvKP (Fig. 2A). Therefore, the ST11-KL64 hvCRKP might have originated from cKP, not hvKP. Moreover, homological plasmids with pK56649-2-KPC hinted almost of all plasmids, dating back to 2010, were carried by K. pneumoniae, and most of them carried blaKPC-2 and were isolated from China (Fig. 3D). On the contrary, the homology plasmids of pK56649-4-NDM were carried by other Gram-negative bacteria, mainly including E. coli, S. enterica, but not K. pneumoniae, and from Europe, America, Asia, and Africa, tracing back to 1976 (Fig. 3E). Two of those homology plasmids also harbored blaNDM-13, including an identical plasmid pNDM13-SR33 (CP092912.1) in S. enterica with pK56649-4-NDM. In addition, conjugation experiments verified pK56649-4-NDM was a self-conjugative plasmid, and pK56649-2-KPC, a non-self-conjugative plasmid, could be mobilized with the help of pK56649-4-NDM. Therefore, we inferred pK56649-4-NDM probably originated from S. enterica, and pK56649-2-KPC was acquired earlier than pK56649-4-NDM for the hvCRKP co-carrying blaKPC-2 and blaNDM-13. Owing to the considerable existence of ST11-KL64 blaKPC-2-carrying hv-CRKP, hypervirulent plasmid pK56649-3-VIR was obtained after acquiring pK56649-2-KPC or pK56649-4-NDM. Moreover, strain 16ZR-60 (GCF_016750625.1), isolated from pus in Hong Kong, carried blaKPC-2 and the same virulent genes with K56649, and is the closest relative to K56649, only 317 SNPs difference (Fig. S6). In summary, the above analysis showed ST11-KL64 hvCRKP belonged to hv-CRKP and pK56649-2-KPC was obtained prior to pK56649-4-NDM (Fig. S3).
Compared to hv-CRKP, the ST65-KL2 hvCRKP carrying pLVPK-like plasmid, belonged to mainstream ST of hvKP and was classified as hvKP by clustering with representative hvKP, which indicated pK22877-2-NDM was obtained by the hvKP and the hvCRKP was CR-hvKP. More than five hundred homology plasmids of the pK22877-2-NDM were identified on NCBI and mainly harbored in E. coli (46.0%) and K. pneumoniae (21.0%). Of note, the IncX3 blaNDM-5-carrying plasmid pK22877-2-NDM showed no difference in size and structure with seven plasmids in Klebsiella spp., E. coli, E. hormaechei and Raoultella ornithinolytica from human and poultry in different countries, including America, Australia, China and Denmark, which showed that blaNDM-5-harboring IncX3 plasmid could transfer between different species and have spread worldwide.
Therefore, ST11-KL64 hvCRKP and ST65-KL2 hvCRKP belonged to hv-CRKP and CR-hvKP, respectively, from the opposite evolutionary pathways (Fig. S3).
Virulence evolution of ST11-KL64 hv-CRKP and resistance evolution of ST65-KL2 CR-hvKP
Interestingly, K60365, another ST11-KL64 hv-CRKP, was isolated from the blood of the patient 2-7, and showed a positive string test in China blue agar, compared with K56649 (Fig. S7). Mucoviscosity, relative thickness of bacterial capsular and uronic acids also revealed that K60365 possessed more CPS than K56649 (P < 0.0001, P < 0.01, and P < 0.001, respectively, Fig. 4D–F). Moreover, K60365 exhibited stronger resistance to macrophage phagocytosis and intracellular survival than K56649 (P < 0.05, Fig. 4G, H). Therefore, the above-suggested K60365 has a higher level of virulence. Moreover, competitive growth showed K60365 had better growth than K56649 (w = 5.59 ± 0.59). However, biofilm formation analysis demonstrated that the mucoid isolate K60365 had less biofilm than the non-mucoid strain K56649 (Fig. 4I), and growth curve also showed K60365 had fitness cost (Fig. S8A).
Moreover, WGS and S1-PFGE revealed K23016 and K23033 acquired an IncQ resistance plasmid, harboring aac(6’)-Ib-cr, aph(3’)-Ia, aadA1, aadA5, blaOXA-1, blaTEM-1B, blaCTX-M-3, fosA3 and catB3, in the process of spread (Fig. S1). Therefore, resistance evolution showed ciprofloxacin resistance was mediated by aac(6’)-Ib-cr in K23016 and K23033, compared to K22866 (Table 2). There is no significant difference in biological fitness cost between K22877 and K23016 (Fig. S8B).
Similarity and diversity between the two types of hvCRKP
Extensive analysis was conducted to gain a better understanding of the threat that the two hvCRKPs pose to human health (Table 3). ST65-KL2 CR-hvKP originated monophyletically and disseminated only within one hospital across three months. Compared to the CR-hvKP, ST11-KL64 hv-CRKP showed more chains of transmission across different hospitals and continually spread in the hospital within ten months. For resistance, the hv-CRKP contained more resistance genes than the CR-hvKP, which resulted in the dilemma of treating the hv-CRKP. More importantly, infection with hv-CRKP was difficult to eliminate and sometimes was occult. With regard to virulence, the mortality of mice infected with hv-CRKP (1 × 108 CFU/mL) unit was significantly increased compared with ATCC 700603, while the mortality of mice infected with the hv-CRKP (1 × 107 CFU/mL) showed no statistical significance compared with ATCC 700603 (Fig. 4B, C). However, the CR-hvKP continually maintained higher mortality of mice with different doses strains (106, 107 and 108 CFU/mL)19. Correspondently, the virulence of CR-hvKP was stronger than that of hv-CRKP. Moreover, the LD50 of mice injected with hv-CRKP also showed lesser virulence than that of CR-hvKP, according to the summary of the previous studies (Fig. S9A and Table S2). Therefore, we concluded that hv-CRKP generally possessed medium virulence. However, according to clinical outcomes, the hv-CRKP has higher mortality than CR-hvKP (60.0%, 6/10 VS 40.0%, 2/5, respectively).
To better understand the differences of resistance and virulence between ST65-KL2 CR-hvKP and ST11-KL64 hv-CRKP, ST11-KL64 (n = 2360) hv-CRKP and ST65-KL2 CR-hvKP (n = 56) were retrieved from NCBI, up to December 31, 2023 (Supplementary Data 1 and 2). Carbapenemase genes detection showed ST11-KL64 hv-CRKP was dominated by blaKPC-2 (96.9%, 2286/2360), followed by blaKPC-2 and blaNDM-1 (1.1%, 27/2360) and blaKPC-2 and blaNDM-5 (0.8%, 18/2360), while ST65-KL2 CR-hvKP was dominated by blaKPC-2 (67.9%, 38/56), followed by blaNDM-5(10.7%, 6/56), blaIMP-6 (3/56;5.4%) and blaIMP-4 (5.4%,3/56). The majority of ST11-KL64 hv-CRKP (96.1%, 2270/2360) had ybt 9 and ICEKp3, whereas the majority of ST65-KL2 CR-hvKP (69.6%, 39/56) had ybt 17 and ICEKp10. Most of ST11-KL64 hv-CRKP (97.9%, 2311/2360) is likely to have OmpK35 truncation/OmpK36GD, according to the OmpK35/36 analysis, while most of ST65-KL2 CR-hvKP (94.6%, 53/56) have normal Ompk35/36, which made the former show higher MICs for carbapenems. The number of resistance genes and their classes hinted that ST11-KL64 hv-CRKP presented an advantage over ST65-KL2 CR-hvKP to acquire exogenous genes easily, but the virulence score is the opposite (Fig. S9B–D). Furthermore, the ratio of wild rpmA promotor P11T of ST11-KL64 hvCRKP was 31.9% and significantly lower than that of ST65-KL2 hvCRKP (31.9% VS 71.4%, Fig. S9E).
Discussion
In the past few years, hvCRKP has been increasingly reported globally, particularly in Asia, Europe and America, since the first report of ST23-KL1 blaKPC-2-harboring CR-hvKP isolated from an 85-year-old American with acute myeloid leukemia in 201420. We also reported the emergence of the ST65-KL2 NDM-5-producing hvCRKP, isolated from the BALF of a lung transplant patient in June 201919. Between 2019 and 2023, more than 200 publications concerning hvCRKP in China, accounting for two-thirds of the total worldwide, could be retrieved from PubMed, suggesting hvCRKP was prevalent throughout China and required immediate monitoring and management.
To date, several disseminations of hvCRKP in different hospitals and environments were reported in China, and these strains largely produced carbapenemases KPC and carried hypervirulent plasmids, including ST11-KL47/KL64 hv-CRKP, ST859-K19 hv-CRKP, etc.14,21. The proliferation and development of NDM-producing hv-CRKP and CR-hvKP, however, have been rarely documented. Herein, our findings have revealed two outbreaks of divergent evolutionary hvCRKP, ST11-KL64 hv-CRKP and ST65-KL2 CR-hvKP, isolated from patients in critical or transplantation situations in China. All 21 KPC- and NDM-co-producing ST11-KL64 hv-CRKP from the same hospital, belonging to the same clonal cluster, were polyphyletic in origin, and presumably have disseminated regionally due to their specimen collection in the initial hospitalization (≤24 h) from different wards. It was noteworthy that the infections caused by the hv-CRKP either progressed rapidly or recurrently happened in those ten patients. Unlike ST11-KL64 hv-CRKP, whose spreading pathway remained elusive, NDM-producing ST65-KL2 CR-hvKP, a typical hvKP, originated from donor organ and transmitted among five recipients, causing the adverse outcomes of lung transplant recipients and alerting the urgency of anti-infection for patients with the infection of CR-hvKP after solid organ transplantation.
In the present study, the resistance phenotype and genotype analysis revealed ST11-KL64 hv-CRKP, co-carrying blaKPC-2 and blaNDM-13, only showed sensitivity to colistin, tigecycline and the combined administration of CZA and aztreonam. However, colistin and tigecycline were not recommended as first-line regimens in CRKP treatment22, Therefore, in China, considering the accessibility of novel antimicrobial agents, the combination of CZA and aztreonam became the essential medication to address the KPC- and NDM-producing hvCRKP. Compared to ST11-KL64 hv-CRKP, ST65-KL2 CR-hvKP only carried blaNDM-5, and presented simpler resistance genotype. It’s worth noting that, in contrast to some blaKPC-2-carrying CR-hvKP, which displayed low-level resistance to carbapenems23, blaNDM-5-carrying CR-hvKP often demonstrated high-level resistance. Furthermore, blaNDM of the two hvCRKP types, namely, CR-hvKP and hv-CRKP, could be transferred into E. coli and K. pneumoniae via plasmid, and its genetic surroundings have a potential transferability to cause horizontal spread by forming mobile genetic elements. Although the majority of hvCRKP carried blaKPC and showed good responses to CZA24,25, the transformation of KPC subtypes, e.g., from blaKPC-2 to blaKPC-33, and the acquisition of NDM could cause resistance to CZA during treatment with CZA26,27.
Both phenotype and genotype are required to identify the hypervirulence in CRKP strains. A series of virulence experiments, including G. mellonella infection, murine and macrophage phagocytosis assay, showed both CR-hvKP and hv-CRKP belonged to hypervirulent strains. Generally, the majority of research used different virulence genes and their combinations to define the hypervirulence of K. pneumoniae, but lacking experimental validation thus might overstate the incidence of hvCRKP. Accurate and practicable definition of the hypervirulence of hvCRKP remains challenging. Truncation and mutations of hypervirulent genes and the absence and difference of their promotor, such as rmpA and rmpA2, rendered them non-functional, and the hypervirulent phenotypes will be lost16. Comprehensive analysis of the LD50 from previous studies (Table S2) and existing murine model results, revealed CR-hvKP has a more significant virulence phenotype than hv-CRKP. Therefore, compared to CR-hvKP, hv-CRKP might potentially pose a lesser risk of infection and disease severity, which might be coined as “sub-hypervirulent” CRKP. Of course, its reasonability requires further study. Virulent plasmid rarely spreads independently, either collectively transmitted with the help of other self-transferable plasmids that had the maintenance of origin of transfer site (oriT) or evolved into self-transferable plasmids by plasmid infusion, including some blaKPC-2-carrying plasmids. It might promote the formation of self-transferrable both carbapenem-resistant and hypervirulent plasmids to accelerate dissemination of hvCRKP13,23,28.
Evolutionary pathways showed the acquisition of blaNDM-carrying plasmid and hypervirulent plasmid occurred in an opposing manner for the carbapenem-resistant and hypervirulent convergence of the two different hvCRKP. Most CRKP carried blaKPC-2-harboring plasmids in China, and the vertical spread of KPC-producing CRKP and the horizontal spread of conjugative blaKPC-harboring plasmids led to the rise and predominance of CRKP strains in hospitals. Owing to the thick CPS and other constraints, CRKP is more likely to acquire virulence genes than hvKP to obtain resistance genes29. Moreover, compared to ST11-KL47, previous studies reported ST11-KL64 hv-CRKP is gradually replacing the former and exhibited greater virulence30,31. Compared to CR-hvKP, therefore, hv-CRKP strains, particularly ST11-KL64 hv-CRKP, were better adapted for survival in hospital environments and became prominent hvCRKP32. Accurate identification of hvCRKP is a prerequisite of effective treatment strategies. AST and resistance genes help us to use valid antimicrobial agents against hvCRKP. Moreover, hypervirulence alerts the timely and sufficient treatment. In this study, enhanced resistance and virulence were found in the ST65-KL2 CR-hvKP and ST11-KL64 hv-CRKP, respectively, in the process of dissemination. Therefore, timely detection for the change of resistance and virulence is essential, especially for those in critical conditions.
Potential therapeutic approaches and infection control measures should be urgently explored and implemented. Though the ST11-KL64 hv-CRKP in the present study was sensitive to ATM-CZA, it continually existed in bacterial killing assay and proliferated inside macrophages, leading to its intractable removal. Therefore, we should not only use antibiotics against these refractory infections with hvCRKP. In the future, the combined use of immunological regulation, phage therapy, and antibiotics, where much work remains to be done to assist in designing workable solutions, will be more efficient in addressing hvCRKP33,34,35. Besides, a series of prevention and control for infections should be strictly implemented, including physical isolation, handwashing, wearing masks and effective clearance for hvCRKP in the environment and equipment. Effective surveillance strategies for hvCRKP population in healthcare sectors should be rigorously carried out, including active screening for the patients in ICU and transplantation wards.
The present study is limited by several factors. First, the size of ST65-KL2 CR-hvKP and ST11-KL64 hv-CRKP outbreak cases is relatively small within the study period, even though the small sample size is not uncommon in this type of study, therefore reducing the robustness of our study. The conclusions should be interpreted with caution. Second, persistent infection of the ST11-KL64 hv-CRKP was a significant and interesting issue, requiring further study.
In conclusion, the ST65-KL2 CR-hvKP and ST11-KL64 hv-CRKP, having simultaneously hypervirulent and multidrug-resistant, pose an intense ability to disseminate and cause high mortality. Resistance and virulence evolution accelerate a substantial threat to human health. Control measures should be put in place to prevent further dissemination throughout the community and hospital setting. More researches should be focused on the virulence and resistance evolution caused by persistent infection in host and its mechanism in the future.
Methods
Outbreak investigation and its global surveillance for hv-CRKP and CR-hvKP
During our surveillance of CRE isolates, two suspected outbreaks were noted: CR-hvKP strains (n = 15), showing positive string test and encoding blaNDM-5, were collected from five patients from June 2019 to August 2019, and hv-CRKP strains (n = 21), co-encoding blaKPC-2 and blaNDM-13, were collected from ten patients from March 2023 to December 2023. Furthermore, when CRKP strains from the same sample type (e.g., blood, urine, BLAF) of a patient, had identical morphological characteristics and antibiotic susceptibility profiles, only the first isolate was chosen for further study. The demographic characteristics of the patients and detailed information of the above strains were collected.
As described previously36, species identification was determined by matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) (Bruker Daltonik, Bremen, Germany). Furthermore, to investigate the global prevalence and differences of the above two different types of hvCRKP, ST11-KL64 hv-CRKP (n = 2360) and ST65-KL2 CR-hvKP (n = 56) were retrieved up to December 31, 2023 from GenBank of the national center for biotechnology information (NCBI). The hvCRKP is genotypically defined as containing the following genes simultaneously: carbapenemase genes, rmpA or rmpA2 and iutAiucABCD or iroBCDN.
Definition
In the study, hypervirulent and carbapenem-resistant K. pneumoniae was defined as hvCRKP. Afterwards we classified hvCRKP as CR-hvKP (hvKP acquiring carbapenem resistance), hv-CRKP (CRKP acquiring hypervirulence) and hvCR-KP (cKP acquiring both hypervirulence and carbapenem resistance), respectively.
Antibiotic sensitivity testing (AST)
Microdilution broth method (bio-KONT, Ltd China) was used to determine the minimal inhibitory concentrations (MICs) of imipenem, meropenem, ceftazidime/avibactam, aztreonam/avibactam, tigecycline and colistin, with E. coli ATCC 25922 as the quality control strain37. Meanwhile, N335 susceptibility cards and Vitek-2 system (bioMérieux, France) were applied for the MICs of other drugs, including amikacin, aztreonam and ciprofloxacin. The breakpoint of tigecycline was interpreted by the US FDA on AST38. The results of other antimicrobial agents were defined following the standards of the Clinical Laboratory Standards Institute (CLSI, 2021)39. The production of carbapenemases was evaluated using the carbapenemase inhibitor APB (3-aminophenyl boronic acid) and EDTA enhancement method (APB/EDTA method), as recommended by CLSI 202139. Moreover, High-level carbapenem resistance was defined as the MIC value of meropenem was ≥16 µg/mL, according to the previous study40.
Whole-genome sequencing and bioinformatic analysis
WGS was performed using Illumina HiSeq 2500 platform for all strains and nanopore sequencing method on MinION flow cells only for K22877, K56649 and K60365. The filtration of raw reads and De novo assembly were conducted using skewer41, SPAdes Genome Assembler v3.13.142 and Unicycler43. The antimicrobial resistance, virulence genes, K type and multilocus sequence types (MLST) were identified by Kleborate v2.4.144. The result of Kleborate can be found in Supp Data 3. Plasmid replicon was determined by PlasmidFinder45 on the CGE server (https://cge.cbs.dtu.dk/services/). The single nucleotide polymorphism (SNP) was performed using Snippy (https://github.com/tseemann/snippy). Linear alignments of blaKPC-2- and blaNDM-13-bearing structures were generated using Easyfig_win_2.146. The comparison of virulence plasmids was determined by the BLAST Ring Image Generator (BRIG) v0.95.2247. The transferability of blaKPC-2- and blaNDM-13-carrying plasmids was performed by oriTfinder48. Core genome MLST (cgMLST, 2358 core genes, pairwise ignoring missing values, Ridom SeqSphere+9.0.9 software) and Xbal-Pulsed-field gel electrophoresis (PFGE)49 were performed to confirm the homology of those strains in the two outbreaks. To trace the origin of pK56649-2-KPC and pK56649-4-NDM, the maximum-likelihood phylogenetic tree of their homological plasmids in this study was built by RaxML50 from the alignment generated by mafft v751. The homological plasmids from global strains were defined that plasmids from nucleotide BLAST database (nr/nt) of NCBI have ≥95% identity and coverage with pK56649-2-KPC and pK56649-4-NDM via online sequence blasting. The iTOL (https://itol.embl.de) was used for visualization. To understand the diversities of two different types of hvCRKP, rmpA and its promotor were abstracted from genomes of ST11-KL64 hv-CRKP (n = 2360) and ST65-KL2 CR-hvKP (n = 56), and were compared with that of hypervirulent pLVKP (AY378100.1) by using BLAST + 2.14.0.
Plasmid conjugation assays and fitness cost
Plasmid conjugation experiments were performed for K. pneumoniae strains K22877 and K56649. Azide-resistant E. coli J53 and amikacin-resistant K. pneumoniae KP54 were applied as the recipient strains. Briefly, both donor and recipient were adjusted to a McFarland standard of 0.5 and mixed at a ratio of 1:3, and after 18-h incubation at 37 °C, the mixture was selected on China blue agar (CBA) plates containing both meropenem (1 mg/L) and amikacin (16 mg/L) or azide (120 mg/L). The above transconjugants were confirmed by PCR and AST.
To evaluate the fitness cost of the evolutionary strains, the growth curve assay was performed. Growth curves of K22877 with K23033 and K56649 with K60365 were measured during the exponential growth phase as the maximum increase in optical density over time, as previously described52. Competitive growth test was used to estimate relative fitness of K56649 and K60365 with different mucoid phenotypes on CBA53. Briefly, equal volumes (10 μL) of overnight cultures of hypermucoid K60365 and regular-mucoid K56649 were mixed and added into 5 mL fresh LB broth to grow competitively for 24 h. Then 20 µl of overnight culture was added into 5 mL fresh LB broth to grow again for 24 h. The cultures were serially diluted using PBS and grown on antibiotic-free CBA agar. Bacteria with different mucoid phenotypes were distinguished by morphologies and string tests. Relative fitness (w) was calculated using this formula, w = log10(N148/N10)/log10(N248/N20), where N1 and N2 are defined as the number of K60365 and K56649, respectively. w < 1 suggests a fitness cost, while w > 1 suggests a fitness advantage. Each experiment was repeated in triplicate.
Virulence evaluation
Galleria mellonella infection model and murine model were performed as described previously54. Briefly, for G. mellonella, exponential growth phase bacteria cultures (37 °C, 200 rpm, 4 h) were washed with sterile PBS and then adjusted to a McFarland standard of 0.5. Suspension was thereafter diluted to 1 × 107 CFU/mL. Ten larvae were injected with 10 μL above bacterial suspension and PBS as control, and then incubated at 37 °C in darkness for 72 h. The dead larvae were counted per 12 h. For murine model, single colonies of K. pneumoniae isolates, including hypervirulent strain NTUH-K2044 and hypovirulent strains ATCC 700603, were grown in LB broth, and diluted to 1 × 107 and 1 × 108 CFU/mL with sterile PBS. Female BALB/c mice (6–8 weeks,18–20 g and each group of four mice) were injected with the above stains at 200 μL by intraperitoneal injection. The dead mice were counted for 7 days after infection. The experiment was performed in triplicate. The log-rank test was performed using GraphPad Prism 6. Ethics oversight: All animal experiments were conducted following institutional ethics requirements under China-Japan Friendship Hospital (CJFH) approved this study (2022-KY-054). We have complied with all relevant ethical regulations for animal use.
To estimate the differences of virulence between K56649 and K60365, mucoviscosity assay, capsular thickness, biofilm formation and uronic acids, were conducted as performed previously54. Briefly, mucoviscosity assay was performed by measuring the OD600 of bacteria supernatant, which was obtained by centrifuging bacteria cultures overnight (MCF = 14.0) for 5 min at 2000 rpm. Capsular was distinguished by ink staining, and relative capsular thickness was measured by total diameter/bacterial diameter. Uronic acids was extracted as performed previously54 and was measured by OD520. Moreover, biofilm of bacteria culture in 96-well polystyrene plates was stained with 0.1% crystal violet at room temperature for 20 min, as described previously54.
Macrophage phagocytosis assay was performed to evaluate the phagocytosis resistance of hvCRKP with slight modification54. Briefly, RAW264.7 macrophages in DEME were infected with 2 × 106 (MOI = 5) hvCRKP and the control strains NTUH-K2044 and ATCC 700603. After incubation with the above bacteria for 10 min, 30 min, 90 min and 180 min, respectively, the cells were then gently washed three times with PBS and lysed with 0.2% Triton X-100, and the survival bacteria were diluted and cultured onto CBA agar overnight to count, as described previously54.
Bacterial survival rate was calculated as follows: = Number of bacteria in the experiment group/Number of bacteria in the control group.
Intracellular bacteria killing was also performed. The incubation with RAW264.7 and K56649 or K60365 for 30 min was exposed to tigecycline (128 mg/L) to kill extracellular bacteria for one hour. The above incubation was washed three times with PBS and then cultured one hour to compare the relative survival rate. The experiments were conducted in triplicates.
Statistics and reproducibility
GraphPad Prism 8.2.1 was used for data analyses. Data are expressed as mean ± standard deviation. Significant differences were assessed using a one-way analysis of variance, with P < 0.05 being considered statistically significant.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Complete sequences of K22877, K56649 and K60365 were deposited to National Center for Biotechnology Information (NCBI) in BioProject: PRJNA1102845. Supplementary data to this article can be found in the appendix as separate Excel files.
References
Navon-Venezia, S., Kondratyeva, K. & Carattoli, A. Klebsiella pneumoniae: a major worldwide source and shuttle for antibiotic resistance. FEMS Microbiol. Rev. 41, 252–275 (2017).
Cassini, A. et al. Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: a population-level modelling analysis. Lancet Infect. Dis. 19, 56–66 (2019).
Paniagua-Garcia, M. et al. Attributable mortality of infections caused by carbapenem-resistant Enterobacterales: results from a prospective, multinational case-control-control matched cohorts study (EURECA). Clin. Microbiol. Infect. 30, 223–230 (2024).
Russo, T. A. & Marr, C. M. Hypervirulent Klebsiella pneumoniae. Clin. Microbiol. Rev. 32, e00001–19 (2019).
Russo, T. A. et al. Identification of biomarkers for differentiation of hypervirulent Klebsiella pneumoniae from classical K. pneumoniae. J. Clin. Microbiol. 56, e00776–18 (2018).
Kochan, T. J. et al. Klebsiella pneumoniae clinical isolates with features of both multidrug-resistance and hypervirulence have unexpectedly low virulence. Nat. Commun. 14, 7962 (2023).
Wyres, K. L., Lam, M. M. C. & Holt, K. E. Population genomics of Klebsiella pneumoniae. Nat. Rev. Microbiol. 18, 344–359 (2020).
Walker, K. A. & Miller, V. L. The intersection of capsule gene expression, hypermucoviscosity and hypervirulence in Klebsiella pneumoniae. Curr. Opin. Microbiol. 54, 95–102 (2020).
Heng, H. et al. Global genomic profiling of Klebsiella pneumoniae: a spatio-temporal population structure analysis. Int. J. Antimicrob. Agents 63, 107055 (2024).
Emeraud, C. et al. Emergence and rapid dissemination of highly resistant NDM-14-producing Klebsiella pneumoniae ST147, France, 2022. Eur. Surveill. 28, 2300095 (2023).
Lapp, Z. et al. Regional spread of blaNDM-1-containing Klebsiella pneumoniae ST147 in post-acute care facilities. Clin. Infect. Dis. 73, 1431–1439 (2021).
Di Pilato, V. et al. Resistome and virulome accretion in an NDM-1-producing ST147 sublineage of Klebsiella pneumoniae associated with an outbreak in Tuscany, Italy: a genotypic and phenotypic characterisation. Lancet Microbe. 3, e224–e234 (2022).
Yang, X., Dong, N., Chan, E. W., Zhang, R. & Chen, S. Carbapenem resistance-encoding and virulence-encoding conjugative plasmids in Klebsiella pneumoniae. Trends Microbiol. 29, 65–83 (2021).
Gu, D. et al. A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. Lancet Infect. Dis. 18, 37–46 (2018).
Liu, L. et al. Chasing the landscape for intrahospital transmission and evolution of hypervirulent carbapenem-resistant Klebsiella pneumoniae. Sci. Bull. 68, 3027–3047 (2023).
Yang, X. et al. Molecular epidemiology of carbapenem-resistant hypervirulent Klebsiella pneumoniae in China. Emerg. Microbes Infect. 11, 841–849 (2022).
Tang, Y., Li, G., Shen, P., Zhang, Y. & Jiang, X. Replicative transposition contributes to the evolution and dissemination of KPC-2-producing plasmid in Enterobacterales. Emerg. Microbes Infect. 11, 113–122 (2022).
Ma, K., Feng, Y., McNally, A. & Zong, Z. Hijacking a small plasmid to confer high-level resistance to aztreonam-avibactam and ceftazidime-avibactam. Int. J. Antimicrob. Agents 62, 106985 (2023).
Zhao, J. et al. Characterization of an NDM-5-producing hypervirulent Klebsiella pneumoniae sequence type 65 clone from a lung transplant recipient. Emerg. Microbes Infect. 10, 396–399 (2021).
Cejas, D. et al. First isolate of KPC-2-producing Klebsiella pneumonaie sequence type 23 from the Americas. J. Clin. Microbiol. 52, 3483–3485 (2014).
Zhu, J., Jiang, X., Zhao, L. & Li, M. An Outbreak of ST859-K19 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese Teaching Hospital. mSystems 7, e0129721 (2022).
Tamma, P. D. et al. Infectious Diseases Society of America 2024 guidance on the treatment of antimicrobial-resistant gram-negative infections. Clin. Infect. Dis. 7, ciae403 (2024).
Zhou, Y. et al. Characterization difference of typical KL1, KL2 and ST11-KL64 hypervirulent and carbapenem-resistant Klebsiella pneumoniae. Drug Resist. Updat. 67, 100918 (2023).
Guo, M. Q. et al. Genomic epidemiology of hypervirulent carbapenem-resistant Klebsiella pneumoniae at Jinshan local hospital, Shanghai, during 2014-2018. J. Microbiol. Immunol. Infect. 57, 128–137 (2024).
Wu, Y. et al. Global evolution and geographic diversity of hypervirulent carbapenem-resistant Klebsiella pneumoniae. Lancet Infect. Dis. 22, 761–762 (2022).
Zhang, Y., Tian, X., Fan, F., Wang, X. & Dong, S. The dynamic evolution and IS26-mediated interspecies transfer of a bla(NDM-1)-bearing fusion plasmid leading to a hypervirulent carbapenem-resistant Klebsiella pneumoniae strain harbouring bla(KPC-2) in a single patient. J. Glob. Antimicrob. Resist. 35, 181–189 (2023).
Zhang, P. et al. Emergence of bla(KPC-33)-harboring hypervirulent ST463 Pseudomonas aeruginosa causing fatal infections in China. J. Infect. 85, e86–e88 (2022).
Xu, Y. et al. Mobilization of the nonconjugative virulence plasmid from hypervirulent Klebsiella pneumoniae. Genome Med. 13, 119 (2021).
Wyres, K. L. et al. Distinct evolutionary dynamics of horizontal gene transfer in drug resistant and virulent clones of Klebsiella pneumoniae. PLoS Genet. 15, e1008114 (2019).
Xie, M. et al. Clinical use of tigecycline may contribute to the widespread dissemination of carbapenem-resistant hypervirulent Klebsiella pneumoniae strains. Emerg. Microbes Infect. 13, 2306957 (2024).
Zhou, K. et al. A point mutation in recC associated with subclonal replacement of carbapenem-resistant Klebsiella pneumoniae ST11 in China. Nat. Commun. 14, 2464 (2023).
Tian, D. et al. Prevalence of hypervirulent and carbapenem-resistant Klebsiella pneumoniae under divergent evolutionary patterns. Emerg. Microbes Infect. 11, 1936–1949 (2022).
Fajardo-Lubian, A. & Venturini, C. Use of bacteriophages to target intracellular pathogens. Clin. Infect. Dis. 77, S423–S432 (2023).
Li, J. et al. Development of phage resistance in multidrug-resistant Klebsiella pneumoniae is associated with reduced virulence: a case report of a personalised phage therapy. Clin. Microbiol. Infect. 29, e1601–e1607 (2023).
Eskenazi, A. et al. Combination of pre-adapted bacteriophage therapy and antibiotics for treatment of fracture-related infection due to pandrug-resistant Klebsiella pneumoniae. Nat. Commun. 13, 302 (2022).
Lu, B. et al. Listeriosis cases and genetic diversity of their L. monocytogenes Isolates in China, 2008-2019. Front. Cell Infect. Microbiol. 11, 608352 (2021).
Zhao, J. et al. Convergence of MCR-8.2 and chromosome-mediated resistance to colistin and tigecycline in an NDM-5-Producing ST656 Klebsiella pneumoniae isolate from a lung transplant patient in China. Front. Cell Infect. Microbiol. 12, 922031 (2022).
Marchaim, D. et al. Major variation in MICs of tigecycline in Gram-negative bacilli as a function of testing method. J. Clin. Microbiol. 52, 1617–1621 (2014).
CLSI. Performance standards for antimicrobial susceptibility testing. 34sted. CLSI supplement M100. Wayne, PA: Clinical and Laboratory Standards Institute. (2024).
Satlin, M. J. et al. Multicenter clinical and molecular epidemiological analysis of bacteremia due to carbapenem-resistant enterobacteriaceae (CRE) in the CRE epicenter of the United States. Antimicrob. Agents Chemother. 61, e02349–16 (2017).
Jiang, H., Lei, R., Ding, S. W. & Zhu, S. Skewer: a fast and accurate adapter trimmer for next-generation sequencing paired-end reads. BMC Bioinform. 15, 182 (2014).
Prjibelski, A., Antipov, D., Meleshko, D., Lapidus, A. & Korobeynikov, A. Using SPAdes De Novo assembler. Curr. Protoc. Bioinform. 70, e102 (2020).
Wick, R. R., Judd, L. M., Gorrie, C. L. & Holt, K. E. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 13, e1005595 (2017).
Lam, M. M. C. et al. A genomic surveillance framework and genotyping tool for Klebsiella pneumoniae and its related species complex. Nat. Commun. 12, 4188 (2021).
Carattoli, A. et al. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 58, 3895–3903 (2014).
Sullivan, M. J., Petty, N. K. & Beatson, S. A. Easyfig: a genome comparison visualizer. Bioinformatics 27, 1009–1010 (2011).
Alikhan, N. F., Petty, N. K., Ben Zakour, N. L. & Beatson, S. A. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12, 402 (2011).
Li, X. et al. oriTfinder: a web-based tool for the identification of origin of transfers in DNA sequences of bacterial mobile genetic elements. Nucleic Acids Res. 46, W229–W234 (2018).
Wang, X. et al. Emergence of a novel mobile colistin resistance gene, mcr-8, in NDM-producing Klebsiella pneumoniae. Emerg. Microbes Infect. 7, 122 (2018).
Stamatakis, A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014).
Katoh, K., Rozewicki, J. & Yamada, K. D. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 20, 1160–1166 (2019).
Gao, H. et al. The transferability and evolution of NDM-1 and KPC-2 co-producing Klebsiella pneumoniae from clinical settings. eBioMedicine 51, 102599 (2020).
Zhang, B. et al. Comparison of two distinct subpopulations of Klebsiella pneumoniae ST16 co-occurring in a single patient. Microbiol. Spectr. 10, e0262421 (2022).
He, J. et al. Opposite evolution of pathogenicity driven by in vivo wzc and wcaJ mutations in ST11-KL64 carbapenem-resistant Klebsiella pneumoniae. Drug Resist. Updat. 66, 100891 (2023).
Acknowledgements
The authors would like to acknowledge the support of National Center for Respiratory Medicine. This work was supported by Beijing Natural Science Foundation (7242124) and the CAMS Innovation Fund for Medical Sciences (CIFMS; No.2021-I2M-1-030).
Author information
Authors and Affiliations
Contributions
Z.F.L.: Writing - original draft & review & editing, Software, Methodology, Formal analysis and Data curation. L.Z.H. & L.Z.Y.: Software and Methodology. L.X.M. & L.Z.C.: Data curation and Methodology. Z.X.X. and Y.X.R.: Methodology. Z.J.K. & Z.Y.L.: Writing - review & editing. L.H.B.: Writing - review & editing, Visualization, Supervision, Project administration, Conceptualization and Funding acquisition.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval
The Ethics Committee of the China-Japan Friendship Hospital (CJFH) approved this study (2022-KY-054) and waived the necessity of informed consent due to the observational nature of the study with the guarantee of anonymous data collection. All in vivo animal studies were in line with ethically and regulatory requirements and approvals.
Peer review
Peer review information
Communications Biology thanks Varun Shamanna, Polly Yap and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: Tobias Goris.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhang, F., Li, Z., Li, Z. et al. Two outbreak cases involving ST65-KL2 and ST11-KL64 hypervirulent carbapenem-resistant Klebsiella pneumoniae: similarity and diversity analysis. Commun Biol 7, 1602 (2024). https://doi.org/10.1038/s42003-024-07310-2
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s42003-024-07310-2