Abstract
Circular RNAs (circRNAs) have garnered substantial attention due to their distinctive circular structure and gene regulatory functions, establishing them as a significant class of functional non-coding RNAs in eukaryotes. Studies have demonstrated that circRNAs can interact with RNA-binding proteins (RBPs), which play crucial roles in tumorigenesis, metastasis, and drug response in cancer by influencing gene expression and altering the processes of tumor initiation and progression. This review aims to summarize the recent advances in research on circRNA-protein interactions (CPIs) and discuss the functions and mode of action of CPIs at various stages of gene expression, including transcription, splicing, translation, and post-translational modifications in the context of cancer. Additionally, we explore the role of CPIs in tumor drug resistance to gain a deeper understanding of their potential applications in the development of new anti-cancer therapeutic approaches.
Similar content being viewed by others
Introduction
Biogenesis and properties of circRNAs
In the 1970s, Sanger et al. discovered that single-stranded RNAs (ssRNAs) can exist in a circular form in plant viruses, coining the term “circRNA”1. Initially, circRNAs were thought to be byproducts of splicing errors with no significant biological function. However, advancements in high-throughput RNA sequencing (RNA-seq) over the past decade have facilitated the recognition and synthesis of circular RNAs in various organisms2,3,4,5,6,7. These circRNAs exhibit high stability, conservation, and specificity in terms of time, space, cell type, and tissue, making them promising biomarkers for cancer diagnosis and treatment8.
Precursor mRNA, consisting of introns and exons, matures into functional proteins through the action of the spliceosome. In contrast, circRNAs are produced from precursor mRNA via a selective splicing process known as “back-splicing,” where the 3’ and 5’ ends are covalently bonded to form a closed circular structure. Based on their origin, circRNAs can be categorized into four distinct types: exon circRNAs (EcircRNAs), composed solely of exons; intronic circRNAs (ciRNAs), derived exclusively from introns; exon-intron circRNAs (ElciRNAs), which incorporate both exons and introns; and intergenic circRNAs (Intergenic circRNAs), which have been recently identified in the regions between two known genes2,9. CircRNAs containing introns (EIciRNAs, ciRNAs, and intergenic circRNAs) are preferentially retained in the nucleus to regulate transcription10, while exon circRNAs (EcircRNAs) are typically enriched in the cytoplasm, where they participate in post-transcriptional regulation and act as miRNA sponges11. Lacking the conventional ‘cap’ and ‘poly(A) tail,’ circRNAs exhibit greater stability and are less susceptible to degradation by RNases. Additionally, circRNAs are not merely binary structures; they possess a specific tertiary conformation, which may confer greater affinity than their linear mRNA counterparts8.
Biological functions of circRNAs
Studies have elucidated the multifaceted roles of circRNAs in cell physiology, such as acting as miRNA “sponges,” interacting with RBPs, regulating transcription, and translating into small peptides and proteins to execute essential biological functions12. While the “miRNA sponge” mechanism has been extensively explored, the interactions between circRNAs and proteins remain less comprehensively understood.
CircRNAs interact with various RBPs to perform distinct functions and influence gene expression at multiple levels. Additionally, certain RBPs regulate circRNA biogenesis13, affect circRNA stability, and participate in circRNA transport. Recent research emphasizes the vital importance of RBPs in the promotion of tumor growth14. Furthermore, research has shown that many circRNAs can be identified in bodily fluids, and exhibit stable, high levels of expression, indicating their potential as significant cancer biomarkers15.
Given the multifunctionality and broad impact of circRNAs, comprehending their regulatory mechanisms in tumors is vital. In this review, we searched databases like PubMed, Web of Science, and Medline using keywords (“circRNA” OR “circular RNA”) AND (“cancer” OR “tumor”). We screened nearly 400 articles published in the past five years up to July 2024, focusing on the latest mechanisms and potential clinical applications of CPIs in tumor regulation. We summarized the characteristics of circRNAs and the roles of CPIs in different cancers, as shown in Fig. 1. Subsequently, we discuss how CPIs regulate tumors by affecting RBP interactions, mRNA stability, RBP subcellular localization, transcription or translation, circRNA generation, and post-transcriptional modifications, presented in Table 1, Fig. 2 and Fig. 3, respectively. Finally, we explored the role of CPIs in tumor drug resistance, summarizing six distinct resistance mechanisms (Table 2). We also examined the impact of CPIs on tumor resistance in the contexts of chemotherapy, targeted therapy, and endocrine therapy, along with their underlying mechanisms. All relevant abbreviations mentioned in the article are summarized in Table 3. Although previous studies have explored these issues, we approached them from a novel perspective of “gene expression processes.” We noticed a lack of comprehensive summaries of CPIs’ effects and mechanisms in tumors in the past three years. Our research offers a comprehensive and novel contribution to the field of CPIs, providing new insights and inspiration for further exploration. For example, how do the different effects of circRNA-RBP interactions relate to their binding sites, subcellular localization, and cellular states? What roles does the unique tertiary structure of circRNA play, distinct from other linear RNAs? These questions merit further investigation.
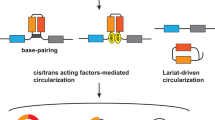
a Schematic illustration of circRNA biogenesis. Exons from pre-RNA undergo back-splicing to form four types of circular RNAs, which are then transported to the cytoplasm via the nuclear pore complex. In the cytoplasm, they perform various biological functions such as acting as miRNA sponges, scaffolding RBPs, and facilitating protein translation. Finally, they are packaged into extracellular vesicles(EVs) and released into the extracellular space. b–i Individual roles of CPIs in different cancers. The upward blue arrows indicate that CPIs promote tumor progression, while the downward arrows represent their inhibition of tumor progression.
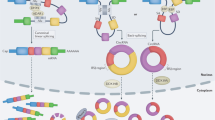
a CPIs modulating transcription. circ-DONSON recruits the NURF complex to facilitate GC progression. circRNF10 interacts with DHX15 to suppress BC’s proliferation and migration. b CPIs affecting alternative splicing. circURI1 inreracts with hnRNPM to suppress GC metastasis. circRAPGEF5 interacts with RBFOX2 to facilitate EC cells’ proliferation. c CPIs affecting translation. circPTPRA binds to IGF2BP1 to suppress tumor progression. Hsa_circ_0005358 sequesters PTBP1 to suppress cancer metastasis. d CPIs affecting RBP’s subcellular localization. circDCUN1D4 facilitates HuR’s translocation to suppress tumor metastasis. The upward blue arrows following each tumor abbreviation indicate that CPIs promote tumor progression, while the downward arrows represent their inhibition of tumor progression.
a CircNFIB binds to MEK1 to inhibit ERK phosphorylation and ICC growth. b CircNDUFB2 enhances the interaction between TRIM25 and IGF2BPs to facilitate NSCLC progression and metastasis. CircNFATC3 binds to IGF2BP3 and restricts its ubiquitination to promote the proliferation of GC. c CircAFF2 binds CAND1, enhancing its interaction with Cullin1 and inhibiting neddylation, impacting CRC radiosensitivity. d CircTLCD4-RWDD3 interacts and induces hnRNPA2B1 SUMOylation to promote lymphatic metastasis of NSCLC. e CircSPIRE1 binds both ELAVL1 and GALNT3 mRNA to facilitate E-cadherin glycosylation and suppress RCC metastasis. In each figure, circRNAs are distinguished by different colored exons. The red arrows represent the physiological processes occurring prior to the binding of circRNAs with proteins, while the blue arrows denote the subsequent processes that occur following their interaction. The upward small blue arrows following each tumor abbreviation indicate that CPIs promote tumor progression, while the downward arrows represent their inhibition of tumor progression. Please refer to the original article for detailed mechanisms.
The role of CPIs in gene regulation within cancer contexts
In 1958, Crick proposed the “central dogma,” elucidating how genetic information is transmitted from DNA to RNA and then to polypeptides, thus activating gene expression16. In tumors, the abnormal regulation of gene expression is strongly associated with the initiation and progression of cancer. Tumor cells often exhibit features of abnormal gene expression regulation, including the inactivation of tumor suppressor genes such as p53, PTEN, and the overexpression of oncogenes like MYC, RAS, HER2, BCL2, and STAT317,18,19,20,21,22. These aberrant gene expressions can lead to enhanced cell growth, proliferation, and metastatic capability, thereby promoting tumor progression. In cancer research, scientists have identified and validated the abnormal expression of circRNAs in cancer, highlighting the importance of exploring the role of CPIs in this context.
With the in-depth research on CPIs, some classic interaction forms between circRNAs and RBPs have been discovered, such as acting as protein “sponges”, “decoys”, “recruiters”, “scaffolds”. Zhou et al. categorized these interactions into three types: enhancing or disrupting the interaction between proteins A and B; forming a ternary complex with proteins A and B to bolster their interaction; and dissociating proteins A and B that normally interact with each other23. The regulatory mechanism of CPIs is intricate and multifunctional. In certain cases, circRNAs can influence the stability of RBPs, and vice versa. RBPs can also modulate the synthesis or degradation of circRNAs under specific conditions24,25. Although our understanding of CPIs is advancing, further research is necessary to fully grasp their role in tumors and to investigate the latest developments in cancer research. We will start with the first step of gene expression, transcription, to summarize the research on CPIs in tumors over the past three to five years. Furthermore, we will present examples of the extensive CPIs in human cancer and explore new targets for cancer treatment.
CPIs modulating transcription
Research indicates that circRNAs can activate the transcription of oncogenes by recruiting RBPs to promoters, thereby promoting cancer development. For instance, circ-DONSON is highly expressed in GC tissues and exhibits oncogenic effects. SOX4, a crucial transcription factor, can lead to the metastasis of various cancers when activated26. Most circRNAs are located in the cytoplasm, functioning by competitively binding with miRNAs or interacting with RBPs. However, Ding et al. demonstrated that circ-DONSON predominantly resides in the nucleus, where it recruits the NURF complex (a chromatin remodeling factor) to the SOX4 promoter, activating its transcription and promoting SOX4 expression and GC progression (Fig. 2a)10. Another example is circIPO11, which recruits topoisomerase 1 (TOP1) to the GLI1 promoter, activating its transcription, thereby promoting self-renewal of liver cancer cells and contributing to liver cancer spread27.
Conversely, circRNAs can also inhibit tumor progression by suppressing promoter transcription. For example, p65 is a key transcription factor that facilitates the expression of the oncogene DEAH box helicase 15 (DHX15) by binding to its promoter region. However, circRNF10 can competitively bind to DHX15, inhibiting its binding to p65, thereby restraining BC progression (Fig. 2a)28.
In general, the circRNAs mentioned above primarily regulate transcription by recruiting RBPs to transcription factor promoters, enhancing or inhibiting the binding of RBPs to promoters. However, circRNAs are capable of directly interacting with transcription factors as well29. Researchers found that circRPPH1 binds to and activates transcription factor 3 (ATF3), promoting the expression of upstream frameshift 1 (UPF1) and Nestin, thus advancing glioblastoma multiforme (GBM) progression30. Additionally, circRNAs can affect the localization of histone-modifying enzymes, influencing transcription. For example, circPSD3 interacts with histone deacetylase 1 (HDAC1), retaining it in the cytoplasm, reducing its inhibition of SERPINB2, an endogenous inhibitor, thereby promoting SERPINB2 transcription and suppressing hepatocellular carcinoma metastasis31.
CPIs affecting alternative splicing
Pre-mRNA splicing
Increasing evidence indicates that circRNAs are crucial in regulating alternative splicing via different mechanisms. CircURI1 is the first circRNA found to regulate gene alternative splicing in cancer metastasis. It directly interacts with heterogeneous nuclear ribonucleoprotein M (hnRNPM), influencing the alternative splicing of metastasis-related genes like VEGFA, leading to inhibiting GC metastasis (Fig. 2b)32. Similarly, Serine and Arginine Rich Splicing Factor 1 (SRSF1) can alter the ratio of two isoforms by recognizing VEGFA pre-mRNA in GBM cells. The VEGFA gene contains eight exons, and its pre-mRNA undergoes alternative splicing to generate a pro-angiogenic isoform (VEGF-Axxxa) and an anti-angiogenic isoform (VEGF-Axxxb)33. The interaction between circSMARCA5 and SRSF1 decreases the proportion of the pro-angiogenic isoform (VEGF-AXXXa), inhibiting GBM angiogenesis34.
Another study showed that circRAPGEF5 significantly reduces the binding of RNA-binding protein fox-1 homolog 2 (RBFOX2) to transferrin receptor protein 1 (TFRC) pre-mRNA by directly interacting with RBFOX2 protein, affecting splicing activity and promoting endometrial cancer (EC) progression and iron death resistance (Fig. 2b)35.
The biogenesis of circRNA
RBPs exert a significant role in regulating circRNA generation, primarily through modulating the back-splicing process of circRNA. During back-splicing, RBPs can bind to the splice sites of pre-mRNA, promoting or inhibiting the binding of splicing factors, thereby regulating circRNA generation. RBPs such as Quaking (QKI)13, Muscleblind (MBL)24, HNRNPL36, NF90/NF11037, hnRNPF25, ESRP138, FUS39 have been found to interact with circRNA and regulate their biological functions. Additionally, RBPs can also influence circRNA generation by modulating its stability and promoting its transcription, such as USF240 and ADAR141,42. For instance, circPAPD4, which exhibits low expression in BC, is regulated by ADAR143.
CPIs affecting translation
circRNA can influence the translation of oncogenes by affecting the stability of linear mRNA. CircRNAs are produced by reverse splicing at the 5’ and 3’ splice sites after transcription by RNA polymerase II (RNA pol II) from protein-coding genes, indicating a competitive relationship with the splicing of their linear pre-mRNA24. We all know that circRNA is present in cells at a low abundance and is rarely enriched with binding sites for RBPs. So, how is it able to bind to RBPs? We identified two main reasons. First, circRNA’s unique tertiary structure enables it to competitively interact with RBPs alongside linear mRNA. Second, m6A modification may enhance the binding affinity of circRNA to RBPs.Compared to linear RNA, m6A modifications on circRNA may present more concentrated or efficient binding regions spatially44,45,46,47. Further research is necessary to elucidate these mechanisms. Numerous studies have shown that circRNAs can compete with homologous linear mRNA as well as heterologous linear mRNA. After binding with RBPs, this competitive effect is altered, leading to changes in mRNA stability. For instance, FAM120A, an oncogene associated with the AKT pathway, and Insulin-like growth factor 2 mRNA binding protein 2 (IGF2BP2), a translation suppressor, exhibit this relationship. CircFAM120A enhances FAM120A translation and promotes tumor cell proliferation by preferentially binding to IGF2BP2, preventing its binding to homologous linear mRNA48. Similarly, HuR, a well-known RBP and translation activator, binds to PABPN1 mRNA to promote translation. However, the competitive binding of circPABPN1 with HuR inhibits the translation of PABPN1, reducing cervical cancer cell proliferation49.
Research has shown two modes of interaction between circRNA and exogenous mRNA: competitive binding and enhanced binding. For example, IGF2BP1, an oncogenic factor that maintains mRNA with m6A modifications’ stability, can be disrupted by the competitive binding of circPTPRA, leading to increased mRNA decay, decreased oncogene expression, and inhibition of BCa progression (Fig. 2c)50. Similarly, hsa_circ_0005358, identified as downregulated in cervical cancer tissues, interacts with polypyrimidine tract-binding protein 1 (PTBP1), disrupting PTBP1’s binding to CUB-domain-containing protein 1 (CDCP1) mRNA. This interaction accelerates CDCP1 mRNA degradation, reducing the expression of the oncogenic gene CDCP1, thereby inhibiting cervical cancer metastasis (Fig. 2c)51.
In addition to competitive binding, some studies have focused on enhanced binding, primarily through the mechanism of acting as a protein scaffold or substrate. For instance, thioredoxin-interacting protein (TXNIP) plays a significant regulatory role in glucose and lipid metabolism. CircDCUN1D4 acts as a scaffold to facilitate the interaction between HuR and TXNIP mRNA, enhancing TXNIP mRNA stability and expression, and ultimately inhibiting LUAD metastasis and glycolysis52.
CPIs affecting RBP’s subcellular localization
CircRNAs exhibit distinct functionalities depending on their subcellular localization. In the nucleus, circRNAs serve as transcriptional regulatory factors, modulating gene expression53. In the cytoplasm, they function as “miRNA sponges” or “protein sponges,” regulating mRNA stability and translation54. The diverse localization of circRNAs significantly impacts cellular function and metabolism. Further research has revealed that the functional role of circRNAs is closely related to their regulation of the subcellular localization of specific proteins. This highlights the multidimensional importance of CPIs as regulatory elements in nucleocytoplasmic transport processes.
Several circRNAs have been reported to influence the subcellular localization of RBPs. For example, circHIF1A binds to NFIB protein, promoting its nuclear translocation and significantly increasing NFIB’s nuclear expression39. NFIB is an oncoprotein with different subcellular localization patterns in various cancers. Chen et al. discovered that circHIF1A, abundantly expressed in TNBC, predominantly locates in the cytoplasm. Their research showed that circHIF1A interacts with NFIB, leading to increased NFIB protein levels in the nucleus. Concurrently, Fused in Sarcoma (FUS) is transcriptionally regulated by NFIB, forming a positive feedback loop involving circHIF1A/NFIB/FUS, ultimately promoting TNBC progression.
These findings suggest that circRNAs can regulate RBPs’ nuclear localization by directly binding to them, as observed in other instances. For example, HuR, a nucleocytoplasmic shuttling protein, can translocate from the nucleus to the cytoplasm, where it binds to mRNA and regulates its stability and translation. The interaction between circDCUN1D4 and HuR facilitates HuR’s translocation to cytoplasm, leading to increased stability of TXNIP mRNA in the cytoplasm. Elevated TXNIP expression inhibits glycolysis and hinders the metastasis of LUAD (Fig. 2d)52. Additionally, hsa_circ_0067842 modulates CMTM6 stability by transporting nuclear HuR protein to the cytoplasm while decreasing the ubiquitination of programmed death ligand-1 (PD-L1), resulting in BC metastasis and immune evasion55. Another study revealed that circRNAs could also promote the translocation of RBPs from the cytoplasm to the nucleus. For example, the interaction between circIPO7 and YBX1 promotes YBX1 phosphorylation and nuclear translocation, allowing it to act as a transcription factor, ultimately promoting nasopharyngeal carcinoma metastasis and cisplatin resistance56.
CPIs can also impact the nuclear translocation of downstream target proteins. For instance, the interaction between circSPARC and FUS facilitates the nuclear translocation of STAT3, enhancing the expression of phosphorylated STAT3 (p-STAT3)57. STAT3, a member of the STAT family, acts as a signaling and transcription activator. Once activated, STAT3 translocates to the nucleus where it regulates gene transcription and biological processes22. CircSPARC promotes STAT3 activation by recruiting FUS, encouraging STAT3 nuclear translocation, thereby accelerating CRC progression.
CPIs affecting post-translational modifications
After transcription, splicing, and translation, circRNAs also play a critical role in post-translational modifications (PTMs) of proteins. Current research indicates that CPIs are increasingly influential in regulating protein PTMs in tumors. In Fig. 3, we illustrate the regulation of tumor PTMs mediated by CPIs.
Phosphorylation
CPIs regulating PTMs are mostly related to RBPs acting as kinases. CircRNAs interact with kinases to increase or prevent their binding to downstream target proteins, leading to target protein PTMs and affecting tumor initiation and progression. Researches have confirmed the interaction between circNFIB and dual-specificity mitogen-activated protein kinase kinase 1 (MEK1)58.Du and his colleagues demonstrated that circNFIB inhibits ICC progression and metastasis by interacting with MEK1 (a kinase) and preventing ERK phosphorylation, thus inhibiting ICC growth (Fig. 3a). Moreover, circNFIB exhibits a synergistic effect with the MEK inhibitor trametinib, delaying trametinib resistance and providing new hope for ICC treatment59.
Ubiquitination
Insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs) are classic RBPs involved in post-transcriptional gene regulation, acting as oncogenes in various diseases60,61,62. Recent studies have found a significant connection between IGF2BPs and ubiquitination. For instance, circNDUFB2 enhances the interaction between IGF2BPs and the E3 ubiquitin ligase TRIM25, promoting IGF2BP ubiquitination and degradation, ultimately inhibiting NSCLC progression (Fig. 3b)45. Studies have shown a connection between circRNAs and innate immune responses63, as evidenced by decreased circRNA levels in autoimmune diseases. Similarly, circNDUFB2 has been demonstrated to activate the RIG-I-MAVS pathway, recruiting immune cells into the TME, triggering immune responses, and thus suppressing tumor growth45,64. Conversely, circNFATC3 competitively binds to IGF2BP3, preventing TRIM25-mediated ubiquitination and degradation of IGF2BP3. Elevated IGF2BP3 levels subsequently promote GC cell proliferation by upregulating CCND1 mRNA expression (Fig. 3b65. Here, we found that the process of TRIM25-mediated ubiquitination and degradation of IGF2BPs is influenced by different CPI mechanisms, leading to distinct outcomes. The specific reasons for this intriguing phenomenon will be discussed in detail in the final section’ 5.1 Discussion on TRIM25-induced IGF2BPs ubiquitination regulated by different CPIs mechanisms’.
Neddylation
Neddylation, a protein modification process similar to ubiquitination, involves the covalent attachment of NEDD8 to target proteins, closely linked to tumor radiosensitivity66,67. CircAFF2, a novel m6A-regulated circRNA in CRC, enhances interaction between CAND1 and Cullin1 by binding to CAND1, inhibiting Cullin neddylation and increasing CRC radiosensitivity (Fig. 3c). This suggests circAFF2’s potential as a marker for screening radiotherapy-sensitive CRC populations or assessing CRC treatment efficacy68.
SUMOylation
SUMO modification typically occurs on the lysine residues of proteins, similar to ubiquitination, but with distinct functions and mechanisms. SUMO modification alters protein function, stability, and subcellular localization by covalently attaching the small protein SUMO (Small Ubiquitin-like Modifier)69. Research on CPIs in SUMOylation is limited. Recently, a study identified circTLCD4-RWDD3, highly upregulated in EVs of lymph node metastatic NSCLC. Mechanistically, circTLCD4-RWDD3 induces SUMOylation modification of hnRNPA2B1 by interacting with it. SUMOylated hnRNPA2B1 interacts with the SUMO-interacting motif (SIM) in ALG-2 interacting protein X (ALIX), transporting circTLCD4-RWDD3 into EVs, ultimately activating PROX1 transcription,. It is widely recognized that PROX1 serves as a fundamental regulatory factor in determining the fate of HLECs and in preserving their identity70. Therefore, these ultimately promote lymphangiogenesis and lymphatic metastasis in NSCLC cells (Fig. 3d)71.
Glycosylation
In tumor progression, glycoproteins are considered significant biomarkers. Protein glycosylations are essential in various biological processes72. Recent studies revealed that EVs secreted by renal cell carcinoma (RCC) contain a circular RNA, circSPIRE1, which can be transferred to endothelial cells, inhibiting vascular permeability and angiogenesis73. Mechanistically, circSPIRE1 forms a ternary complex with ELAV-like RNA-binding protein 1 (ELAVL1) and polypeptide N-acetylgalactosaminyltransferase 3 (GALNT3) mRNA, promoting the glycosylation and membrane localization of E-cadherin through GALNT3. Additionally, circSPIRE1 upregulates KH domain RNA-binding protein (QKI), which binds to the intronic region of circSPIRE1, enhancing its expression and forming a positive feedback loop. This loop further inhibits the mesenchymal state of RCC, suggesting circSPIRE1 as a predictive factor and potential therapeutic target for metastatic RCC (Fig. 3e)73.
CPIs mediate tumor drug resistance
Tumor drug resistance, the capacity of tumor cells to resist anticancer drugs, continues to be a major obstacle in cancer therapy74. Resistance may arise from endogenous genetic mutations or other cellular mechanisms, and it may also be due to acquired drug activation of independent survival signals in tumor cells or improved adaptability of the tumor microenvironment. It is worth noting that these mechanisms of intrinsic or acquired resistance often coexist, making tumors challenging to control and cure75,76. Currently, many circRNAs have been recognized as key regulators in tumor drug resistance77,78,79. CircRNAs fine-tune the potential mechanisms underlying the development of resistance, promoting the occurrence of intrinsic or acquired therapeutic resistance in tumor cells, either directly or indirectly. Some overexpressed circRNAs can act as tumor drivers, activating tumor cell defense pathways shortly before or after the initiation of treatment, leading to the efflux of chemotherapeutic agents80. Other circRNAs may be expressed after a certain period following treatment, creating favorable conditions for tumor cell development by activating survival signals (such as cell proliferation, apoptosis, autophagy, etc.) and modulating cellular metabolism and the immune microenvironment, which ultimately results in reduced treatment sensitivity81,82,83,84,85,86. The six resistance mechanisms involving CPIs will be outlined in Table 2. Then, this section will briefly summarize the roles of CPIs in chemotherapy resistance, targeted therapy resistance, and endocrine therapy resistance, along with the impact of drugs on tumor treatment regulated by CPIs and their potential mechanisms.
Chemotherapy resistance
Cisplatin, a widely used cytotoxic chemotherapeutic agent for various tumors, functions by binding to DNA to form cross-links, thereby inhibiting DNA replication and transcription. It can also induce programmed cell death by activating oxidative stress and pro-apoptotic signaling pathways87. Fu et al. identified a significantly upregulated circRNA, circPBX3, through transcriptome RNA sequencing in two cisplatin-resistant ovarian cancer cell lines80. Both in vitro and in vivo experiments demonstrated a positive correlation between circPBX3 and cisplatin resistance. Further investigations revealed that circPBX3 stabilizes the expression of ATP7A, a copper efflux transporter, by interacting with IGF2BP2, thereby promoting the efflux of cisplatin and leading to resistance88. Similarly, IGF2BP2, induced by circITGB6, can stabilize FGF9 (Fibroblast Growth Factor 9) mRNA and promote M2 macrophage polarization, thereby regulating the tumor immune microenvironment (TME) and contributing to cisplatin resistance in ovarian cancer89,90. This study also established that circRNAs serve as crucial regulatory factors in immune cells.
Promotion of DNA damage repair can also lead to cisplatin resistance. Researchers discovered that overexpression of circIPO7 in nasopharyngeal carcinoma cells can counteract cisplatin-induced DNA damage, thereby facilitating both nasopharyngeal carcinoma metastasis and cisplatin resistance. Mechanistically, circIPO7 interacts with YBX1 in the cytoplasm, promoting its phosphorylation and nuclear localization56. Nuclear-localized YBX1 is crucial in DNA damage repair91. Interestingly, when YBX1 binds to certain circRNAs and remains localized in the cytoplasm, it may contribute to resistance by enhancing the expression of downstream survival signaling pathway factors83.
A recent study identified a circRNA, circLIFR, that enhances the chemosensitivity of BCa cells to cisplatin92. CircLIFR modulates the sensitivity of BCa cells to cisplatin through its interaction with the human MMR gene MutS homolog 2 (MSH2). MSH2 emerged as a promising target for platinum resistance in a genome-wide CRISPR screening of BCa cell lines. Loss of MSH2 expression results in reduced apoptosis induced by cisplatin, thereby contributing to cisplatin resistance in patients with muscle-invasive bladder cancer(MIBC)93.
With the advancement of metabolomics and extensive research into cancer metabolic reprogramming, the significant roles of cellular metabolism in cisplatin resistance have become increasingly recognized94. For instance, the overexpression of circARHGAP10 can overcome cisplatin resistance in BCa cells by binding to and downregulating the stability of methionine adenosyltransferase IIa (MAT IIa, MAT2A)95. In prostate cancer, circARHGAP29 enhances the interaction between IGF2BP2 and LDHA mRNA, leading to increased LDHA expression, which promotes glycolytic metabolism and resistance to docetaxel81.
Docetaxel, a member of the taxane (PAX) drug class, exerts its mechanism by stabilizing microtubule structures, thereby inhibiting cancer cell mitosis77. AKT is a key mediator in promoting taxane resistance96. Ma et al. found that adding an AKT pathway inhibitor to PAX-resistant TCBC cells could block the enhancement of PAX resistance by circAMOTL1. This demonstrated that circAMOTL1 can bind to phosphorylated AKT protein, regulating the AKT pathway and leading to increased PAX resistance85.
Targeted therapy resistance
Targeted therapy drugs can be classified into several categories, including small molecule targeted agents, monoclonal antibodies, antibody-drug conjugates, and gene therapies. These agents achieve personalized treatment by specifically targeting certain molecules or signaling pathways in tumor cells, thereby enhancing efficacy while minimizing damage to normal cells97. Gefitinib, imatinib, sunitinib, and sorafenib are small-molecule tyrosine kinase inhibitors (TKIs). However, when tumor cells acquire mutations in target genes, effective drug binding may be impaired, leading to intrinsic resistance; for instance, NSCLC with EGFR mutations often shows poor response to TKI treatment98. The mechanisms of acquired resistance to CPIs are similar to those of chemotherapy drugs; for example, circME1 interacts with U1 snRNP to promote the transcription of its parent gene, malate dehydrogenase 1 (ME1), thereby upregulating ME1 expression, facilitating aerobic glycolysis in ccRCC, and contributing to sunitinib resistance.
Trastuzumab is an effective monoclonal antibody for treating HER2-positive BC, but resistance remains a concern99. While the use of PI3K and MEK pathway inhibitors can enhance trastuzumab sensitivity100, circCDYL2 poses challenges to this treatment. Yun et al. demonstrated that circCDYL2 acts as a scaffold, facilitating the interaction between growth factor receptor-bound protein 7 (GRB7) and focal adhesion kinase 1 (FAK), thereby sustaining the activity of downstream AKT and ERK1/2, which enhances trastuzumab resistance in HER2-positive BC cells. Moreover, in vivo experiments indicate that inhibitors of FAK or GRB7 can reverse trastuzumab resistance induced by circCDYL2, suggesting the potential biomarker role of the circCDYL2/GRB7/FAK pathway101. Additionally,another study found that the small molecule ferroptosis inducer erastin effectively restores the antitumor activity of trastuzumab. Wang et al. found that circBGN binds and enhances the interaction between OTUB1 and SLC7A11, stabilizing SLC7A11 protein and inhibiting ferroptosis, leading to trastuzumab resistance in HER2-positive BC84.
Endocrine therapy resistance
Endocrine therapy is a critical treatment strategy for estrogen receptor (ER)-positive BC patients. Dysregulation of ERα signaling is the fundamental cause of tamoxifen resistance in these patients102. In this context, circRNA may exert a role through the competitive endogenous RNA (ceRNA) mechanism by promoting ERα signaling transduction, leading to the activation of ERα and enhancing tamoxifen resistance in BC. For instance, a study discovered that circPVT1, which is significantly upregulated in ER-positive BC, enhances the expression of estrogen receptor 1 (ESR1) and ERα target genes through the ceRNA mechanism. Furthermore, it inhibits type I interferon (IFN) signaling by interacting with mitochondrial antiviral signaling protein (MAVS), thereby suppressing antitumor immunity. Targeting circPVT1 holds promise as a diagnostic biomarker and therapeutic target for ER-positive BC79.
Discussion and future perspectives
CPIs in TRIM25-induced IGF2BPs Ubiquitination
As previously discussed in the ubiquitination section above, the process of IGF2BPs being ubiquitinated and degraded by TRIM25 is regulated by different CPI mechanisms, leading to varying outcomes. CircNDUFB2 acts as a scaffold here, forming the circNDUFB2/IGF2BPs/TRIM25 ternary complex, which promotes TRIM25-dependent ubiquitination and degradation of IGF2BPs. In contrast, circNFATC3 functions as a ceRNA, competing with TRIM25 for binding sites on IGF2BP3, thereby reducing the ubiquitination and degradation of IGF2BP3 and maintaining its protein stability.
We analyze this for two main reasons. First, in the process of IGF2BPs being ubiquitinated and degraded by TRIM25, circNDUFB2 serves as a “necessary factor,” while circNFATC3 acts as a “regulatory factor.” Researchers have found that the effective ubiquitination of IGF2BPs by TRIM25 is achieved with circNDUFB2 as a scaffold. The m6A modification of circNDUFB2 enhances its binding to the C-terminal KH domains of IGF2BPs and facilitates the formation of the TRIM25/circNDUFB2/IGF2BPs ternary complex through its interaction with the PRY/SPRY RBD of TRIM25, mediating the ubiquitination and degradation of IGF2BPs. Knockdown or overexpression of circNDUFB2 can respectively decrease or increase the ubiquitination of IGF2BPs. When the RBD of TRIM25 is mutated, TRIM25 is unable to bind to circNDUFB2, resulting in no changes in the ubiquitination of IGF2BPs.
For circNFATC3, RIP and co-IP experiments have shown that it competes with TRIM25 for binding to the N- and C-terminal domains of IGF2BP3, reducing the binding between TRIM25 and IGF2BP3 and thereby increasing the stability of IGF2BP3. Moreover, knockdown of TRIM25 can rescue the increase in IGF2BP3 ubiquitination caused by the knockdown of circNFATC3. This indicates that circNFATC3 regulates TRIM25-induced degradation of IGF2BP3 through its interaction with IGF2BP3 protein.
Second, m6A-modified circRNAs are more likely to bind with IGF2BPs and TRIM25 to form ternary complexes. As we know, the ceRNA mechanism is widely recognized as the primary way circRNA regulates cellular functions, where circRNAs specifically bind to miRNAs or RBPs to competitively regulate downstream gene expression, participating in vital biological processes such as cell proliferation, differentiation, and tumorigenesis. This mechanism is characterized by its broad applicability, stability, and specificity in biology. In contrast, research on circRNAs as part of ternary complexes is relatively limited, likely due to the complexity of their interactions and research constraints. IGF2BPs, as an oncogenic m6A “reader” protein, can specifically recognize and bind to m6A-modified RNAs, regulating their stability and translation60. The m6A modification of circNDUFB2 may enhance its affinity for IGF2BPs and influence TRIM25’s ubiquitin ligase activity towards IGF2BPs. Another CLIP-seq experiment in HeLa cells showed that TRIM25 prefers to bind to RNAs rich in “G” and “C” sequences103, while the sequence of m6A-modified circNDUFB2 contains the “GGAC” N6-methyladenosine (m6A) core motif, which may explain why circNFATC3 cannot bind to TRIM25.
TRIM25 and IGF2BPs are both classic RBPs. IGF2BPs contain six classic RNA-binding domains (RBDs), consisting of two N-terminal RRM domains and four C-terminal KH domains. The KH domain is critical for IGF2BPs to bind RNA, as it can enhance IGF2BP1’s affinity for target mRNA by specifically binding to RNA sequences, thereby regulating the stability of the RNA-protein(RNP) complex. Most RBPs can target multiple RNAs through sequence-dependent or structure-specific mechanisms14,104. We speculate that IGF2BPs can also simultaneously bind to multiple RNA motifs to form stable protein-RNA complexes. Notably, IGF2BP1 can recognize RNAs with specific nucleotide sequences (such as 3’ UTR) or specific three-dimensional structures105, which may explain why circRNAs, despite lacking a 3’ UTR, can still bind to IGF2BP1.
TRIM25’s RBD is primarily reflected in the PRY/SPRY domain, which endows it with the ability to specifically recognize and bind to RNA. Through its interactions with RNA, TRIM25 regulates various cellular processes, serving an essential function particularly in antiviral immunity and signal transduction. The integrity of this domain is essential for TRIM25 to effectively mediate ubiquitination.
By exploring the interactions between TRIM25 and IGF2BPs with different circRNAs, we have gained insights into the diverse roles that CPI mechanisms play under various conditions in different cancers. Future research should not only further elucidate the regulatory differences of these interactions across various cancer types but also focus on how different circRNAs selectively modulate specific RBP families. Additionally, the ways in which circRNAs influence processes such as immune response and cellular metabolism through different pathways merit further investigation.
Future studies will also need to develop more precise tools to capture and analyze the interactions between circRNAs and RBPs. New technologies, such as RNA immunoprecipitation sequencing (RIP-seq) and cross-linking immunoprecipitation sequencing (CLIP-seq), may provide strong support for revealing insights in this field.
Implications of CPIs in cancer therapeutics and future directions for drug development
CircRNAs, as closed-loop non-coding RNAs, possess a significantly longer half-life compared to linear RNAs. After interacting with cancer-promoting RBPs (such as HuR, FUS, and IGF2BPs), circRNAs exhibit the potential of CPIs as therapeutic targets and diagnostic tools by regulating tumorigenesis, progression, and therapeutic resistance. The high or low expression of specific circRNAs can be associated with early cancer detection or disease progression. For example, circEIF3I is significantly upregulated in PDAC tissues, where it promotes PDAC metastasis and poor prognosis by interacting with SMAD3 and APA21106. Consequently, circEIF3I may serve as a promising biomarker for predicting PDAC metastasis and adverse outcomes. Further investigation of the functions of CPIs in different cancers not only promotes the development of tumor biomarkers but also provides new perspectives for targeted cancer therapies, which is crucial for clinical applications.
Targeting CPIs is expected to become a novel cancer treatment strategy. Designing antibodies or small molecule drugs that can interfere with CPIs may effectively inhibit tumor progression. For instance, in the previously mentioned TRIM25-mediated ubiquitination degradation of IGF2BPs, mutations in the PRY/SPRY RBD of TRIM25 can significantly impair the binding of circNDUFB2. We speculate that it may be possible to design a molecule that induces mutations in the RBD of RBPs bound by highly expressed circRNAs in tumors, potentially reversing the pro-cancer effects caused by circRNA-RBP binding.
Additionally, RBP targeting can be employed to inhibit the functions of CPIs. In HER2(+) BC cells, inhibitors of FAK or GRB7 have been found to reverse trastuzumab resistance caused by the overexpression of the circCDYL2-GRB7-FAK complex101. Both GRB7 and FAK are signaling proteins that, together with circCDYL2, regulate downstream classical pathways (PI3K/AKT, RAS/ERK) to enhance trastuzumab sensitivity. However, targeting a single downstream pathway protein with molecular inhibitors may be ineffective against tumor resistance, as tumor resistance often arises from the interplay of multiple pathways (PI3K/AKT, RAS/ERK, Wnt/β-catenin, NF-κB, etc.). Selecting RBPs that can regulate multiple pathways may significantly improve drug sensitivity.
Finally, tumor progression can be inhibited by targeting circRNAs associated with cancer. Antisense oligonucleotides (ASOs) can modulate the biological functions of circRNAs by binding to specific sequences79. However, designing ASOs that target circRNAs and effectively delivering them to target cells poses significant challenges. The CRISPR-Cas13 technology has also been shown to knock out circRNAs that are highly expressed in tumor cell lines, offering efficiency and specificity, although it has drawbacks related to off-target effects48. Future research should focus on how to stably knock out highly expressed circRNAs in tumors without affecting the related homologous linear mRNAs. For circRNAs that are lowly expressed in tumors, specific circRNAs can be synthesized and packaged into exosomes for delivery into cancer cells to achieve tumor suppression.
In the future, research targeting CPIs remains of great significance. As far as we know, circRNA-based biomarkers have not yet found application in clinical practice, and future research should consider the following aspects: First, m6A modifications affect RNA stability and translation efficiency; targeted knockout of specific circRNA m6A modifications could inhibit the progression of tumors related to m6A-associated CPIs. Second, directional mutations in the binding regions of circRNAs, particularly key binding sites on RBPs, may lead to their dissociation from proteins, thereby affecting the biological functions of RBPs. Lastly, targeting the ubiquitination and degradation of pro-cancer proteins is an effective strategy to suppress cancer progression. By reducing the stability of pro-cancer proteins, key signaling pathways in tumor cells can be disrupted, inhibiting their growth and spread. Future studies need to further explore these mechanisms to develop more precise targeted therapies.
Conclusion
In this review, we summarized the biological characteristics, functions, interaction mechanisms with proteins, and the impact on tumor drug resistance of CPIs. We highlighted the significant contributions of circular RNAs and RBPs in tumor development over the past three to five years, focusing on the “central dogma” perspective and presenting a comprehensive and novel mechanism of “CPIs” across various steps of gene expression.
There are still many aspects of circRNA and tumor research that warrant further exploration. For instance, some circRNAs are capable of encoding proteins, which can activate cancer by producing truncated proteins36. Additionally, scientists have discovered that circRNAs within exosomes are integral to various mechanisms of chemotherapy resistance in cancer, owing to their enrichment and stability facilitated by exosome protection83,107,108.These specific mechanisms require further investigation.
In conclusion, circRNA-protein interactions exerts a crucial role in tumor biology by influencing various cellular processes related to tumor development, metastasis, and drug response. Understanding CPIs in tumors opens up exciting possibilities for their potential applications in tumor therapy, including the inhibition of tumor drug resistance. Due to their distinctive circular structure that confers high stability, circRNAs hold promise as diagnostic markers for early cancer detection and prognostic indicators for disease progression109. These aspects necessitate further validation through more animal models and clinical trials.
Note added to proof
All figures in the manuscript were independently created by the authors without the use of any third-party images or tools such as Biorender. Therefore, no permission statements are required.
References
Sanger, H. L. et al. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc. Natl Acad. Sci. 73, 3852–3856 (1976).
Gao, Y., Wang, J. & Zhao, F. CIRI: an efficient and unbiased algorithm for de novo circular RNA identification. Genome Biol. 16, 4 (2015).
Jeck, W. R. et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 19, 141–157 (2013).
Rybak-Wolf, A. et al. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol. Cell 58, 870–885 (2015).
Talhouarne, G. J. S. & Gall, J. G. Lariat intronic RNAs in the cytoplasm of Xenopus tropicalis oocytes. RNA 20, 1476–1487 (2014).
Lu, S. et al. RNA-Seq Revealed a Circular RNA-microRNA-mRNA Regulatory Network in Hantaan Virus Infection. Frontiers in Cellular and Infection. Microbiology 10, 97 (2020).
Zhu, Z. et al. Gut microbiota regulate tumor metastasis via circRNA/miRNA networks. Gut Microbes 12, 1788891 (2020).
Liu, C. X. & Chen, L. L. Circular RNAs: Characterization, cellular roles, and applications. Cell 185, 2016–2034 (2022).
Li, J. et al. Circular RNAs in cancer: biogenesis, function, and clinical significance. Trends Cancer 6, 319–336 (2020).
Ding, L. et al. Circular RNA circ-DONSON facilitates gastric cancer growth and invasion via NURF complex dependent activation of transcription factor SOX4. Mol. Cancer 18, 45 (2019).
Ma, N. et al. Circular RNA circNFATC3 acts as a miR-9-5p sponge to promote cervical cancer development by upregulating SDC2. Cell. Oncol. 44, 93–107 (2021).
Kristensen, L. S. et al. The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 20, 675–691 (2019).
Conn, S. J. et al. The RNA binding protein quaking regulates formation of circRNAs. Cell 160, 1125–1134 (2015).
Wang, S. et al. RNA-binding proteins and cancer metastasis. Semin. Cancer Biol. 862, 748–768 (2022).
Su, M. et al. Circular RNAs in Cancer: emerging functions in hallmarks, stemness, resistance and roles as potential biomarkers. Mol. Cancer 18, 90 (2019).
Crick, F. Central dogma of molecular biology. Nature 227, 561–563 (1970).
Hassin, O. & Oren, M. Drugging p53 in cancer: one protein, many targets. Nat. Rev. Drug Discov. 22, 127–144 (2023).
Valabrega, G., Montemurro, F. & Aglietta, M. Trastuzumab: mechanism of action, resistance and future perspectives in HER2-overexpressing breast cancer. J. Eur. Soc. Med. Oncol. 18, 977–984 (2007).
Worby, C. A. & Dixon, J. E. PTEN. Annu. Rev. Biochem. 83, 641–669 (2014).
Dang, C. V. MYC on the path to cancer. Cell 149, 22–35 (2012).
Roberts, A. W., Wei, A. H. & Huang, D. C. S. B. C. L. 2 and MCL1 inhibitors for hematologic malignancies. Blood 138, 1120–1136 (2021).
Zou, S. et al. Targeting STAT3 in cancer immunotherapy. Mol. Cancer 19, 145 (2020).
Zhou, W. Y. et al. Circular RNA: metabolism, functions and interactions with proteins. Mol. Cancer 19, 172 (2020).
Ashwal-Fluss, R. et al. circRNA biogenesis competes with Pre-mRNA splicing. Mol. Cell 56, 55–66 (2014).
Wang, S. et al. Circular RNA circPFKP promotes cell proliferation by activating IMPDH2 in prostate cancer. Cancer Lett. 524, 109–120 (2022).
Wang, N. et al. CXCL1 derived from tumor-associated macrophages promotes breast cancer metastasis via activating NF-κB/SOX4 signaling. Cell Death Dis. 9, 880 (2018).
Gu, Y. et al. Circular RNA circIPO11 drives self-renewal of liver cancer initiating cells via Hedgehog signaling. Mol. Cancer 20, 132 (2021).
Zheng, W. et al. CircRNF10-DHX15 interaction suppressed breast cancer progression by antagonizing DHX15-NF-κB p65 positive feedback loop. Cell. Mol. Biol. Lett. 28, 34 (2023).
Zhang, R. et al. circ_SIRT1 upregulates ATG12 to facilitate Imatinib resistance in CML through interacting with EIF4A3. Gene 893, 147917 (2024).
Xu, J. et al. UPF1/circRPPH1/ATF3 feedback loop promotes the malignant phenotype and stemness of GSCs. Cell Death Dis. 13, 645 (2022).
Xu, L. et al. circPSD3 is a promising inhibitor of uPA system to inhibit vascular invasion and metastasis in hepatocellular carcinoma. Mol. Cancer 22, 174 (2023).
Wang, X. et al. CircURI1 interacts with hnRNPM to inhibit metastasis by modulating alternative splicing in gastric cancer. Proc. Natl Acad. Sci. USA 118, e2012881118 (2021).
Ye, X. et al. Altered ratios of pro‐ and anti‐angiogenic VEGF‐A variants and pericyte expression of DLL4 disrupt vascular maturation in infantile haemangioma. J. Pathol. 239, 139–151 (2016).
Barbagallo, D. et al. CircSMARCA5 regulates VEGFA mRNA splicing and angiogenesis in glioblastoma multiforme through the binding of SRSF1. Cancers 11, 194 (2019).
Zhang, J. et al. CircRAPGEF5 interacts with RBFOX2 to confer ferroptosis resistance by modulating alternative splicing of TFRC in endometrial cancer. Redox. Biology 57, 102493 (2022).
Li, Y. et al. HNRNPL circularizes ARHGAP35 to produce an oncogenic protein. Adv. Sci. (Weinh.) 8, 2001701 (2021).
Li, X. et al. Coordinated circRNA Biogenesis and Function with NF90/NF110 in Viral Infection. Mol. Cell 67, 214–227.e7 (2017).
Pan, X. et al. Circular RNA circ-TNPO3 inhibits clear cell renal cell carcinoma metastasis by binding to IGF2BP2 and destabilizing SERPINH1 mRNA. Clin. Transl. Med. 12, e994 (2022).
Chen, T. et al. CircHIF1A regulated by FUS accelerates triple-negative breast cancer progression by modulating NFIB expression and translocation. Oncogene 40, 2756–2771 (2021).
Wang, X. et al. The circACTN4 interacts with FUBP1 to promote tumorigenesis and progression of breast cancer by regulating the expression of proto-oncogene MYC. Mol. Cancer 20, 91 (2021).
Zhang, J. et al. ADAR1 regulates vascular remodeling in hypoxic pulmonary hypertension through N1-methyladenosine modification of circCDK17. Acta Pharm. Sin. B 13, 4840–4855 (2023).
Ivanov, A. et al. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep. 10, 170–177 (2015).
Zhou, B. et al. CREBZF mRNA nanoparticles suppress breast cancer progression through a positive feedback loop boosted by circPAPD4. J. Exp. Clin. Cancer Res. 42, 138 (2023).
Chen, R. X. et al. N6-methyladenosine modification of circNSUN2 facilitates cytoplasmic export and stabilizes HMGA2 to promote colorectal liver metastasis. Nat. Commun. 10, 4695 (2019).
Li, B. et al. circNDUFB2 inhibits non-small cell lung cancer progression via destabilizing IGF2BPs and activating anti-tumor immunity. Nat. Commun. 12, 295 (2021).
Du, W. W. et al. Identifying and characterizing circRNA-protein interaction. Theranostics 7, 4183 (2017).
An, Y. & Duan, H. The role of m6A RNA methylation in cancer metabolism. Mol. Cancer 21, 14 (2022).
Li, S. et al. Screening for functional circular RNAs using the CRISPR–Cas13 system. Nat. Methods 18, 51–59 (2021).
Abdelmohsen, K. et al. Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol. 14, 361–369 (2017).
Xie, F. et al. CircPTPRA blocks the recognition of RNA N6-methyladenosine through interacting with IGF2BP1 to suppress bladder cancer progression. Mol. Cancer 20, 68 (2021).
Cen, Y. et al. hsa_circ_0005358 suppresses cervical cancer metastasis by interacting with PTBP1 protein to destabilize CDCP1 mRNA. Molecular Therapy. Nucleic Acids 27, 227–240 (2022).
Liang, Y. et al. circDCUN1D4 suppresses tumor metastasis and glycolysis in lung adenocarcinoma by stabilizing TXNIP expression. Mol. Ther. Nucleic Acids 23, 355–368 (2021).
An, M. et al. Aberrant nuclear export of circNCOR1 underlies SMAD7-mediated lymph node metastasis of bladder cancer. Cancer Res. 82, 2239–2253 (2022).
Tsitsipatis, D. et al. AUF1 ligand circPCNX reduces cell proliferation by competing with p21 mRNA to increase p21 production. Nucleic Acids Res. 49, 1631–1646 (2021).
Li, J. et al. Circular RNA hsa_circ_0067842 facilitates tumor metastasis and immune escape in breast cancer through HuR/CMTM6/PD-L1 axis. Biol. Direct 18, 48 (2023).
Hong, X. et al. CircIPO7 promotes nasopharyngeal carcinoma metastasis and cisplatin chemoresistance by facilitating YBX1 nuclear localization. Clin. Cancer Res. 28, 4521–4535 (2022).
Wang, J. et al. The circular RNA circSPARC enhances the migration and proliferation of colorectal cancer by regulating the JAK/STAT pathway. Mol. Cancer 20, 81 (2021).
Fang, J. Y. & Richardson, B. C. The MAPK signalling pathways and colorectal cancer. Lancet Oncol. 6, 322–327 (2005).
Du, J. et al. CircNFIB inhibits tumor growth and metastasis through suppressingMEK1/ERK signaling in intrahepatic cholangiocarcinoma. Mol. Cancer. 21, 18 (2022).
Bell, J. L. et al. Insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs): post-transcriptional drivers of cancer progression?. Cell. Mol. Life Sci. 70, 2657–2675 (2013).
Lederer, M. et al. The role of the oncofetal IGF2 mRNA-binding protein 3 (IGF2BP3) in cancer. Semin. Cancer Biol. 29, 3–12 (2014).
Gutschner, T. et al. Insulin-like growth factor 2 mRNA-binding protein 1 (IGF2BP1) is an important protumorigenic factor in hepatocellular carcinoma. Hepatology 59, 1900–1911 (2014).
Liu, C. X. et al. Structure and degradation of circular RNAs regulate PKR activation in innate immunity. Cell 177, 865–880.e21 (2019).
Chen, Y. G. et al. Sensing self and foreign circular RNAs by intron identity. Mol. Cell 67, 228–238.e5 (2017).
Yang, F. et al. CircNFATC3 promotes the proliferation of gastric cancer through binding to IGF2BP3 and restricting its ubiquitination to enhance CCND1 mRNA stability. J. Transl. Med. 21, 402 (2023).
Zheng, S., Wu, Y. & Li, Z. Integrating cullin2-RING E3 ligase as a potential biomarker for glioblastoma multiforme prognosis and radiosensitivity profiling. Radiother. Oncol.: J. Eur. Soc. Ther.Radiol. Oncol. 154, 36–44 (2021).
Kerscher, O., Felberbaum, R. & Hochstrasser, M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu. Rev. Cell Dev. Biol. 22, 159–180 (2006).
Shao, Y. et al. ALKBH5/YTHDF2-mediated m6A modification of circAFF2 enhances radiosensitivity of colorectal cancer by inhibiting Cullin neddylation. Clin. Transl. Med. 13, e1318 (2023).
Chang, H. M. & Yeh, E. T. H. SUMO: From bench to bedside. Physiol. Rev. 100, 1599–1619 (2020).
Johnson, N. C. et al. Lymphatic endothelial cell identity is reversible and its maintenance requires Prox1 activity. Genes Dev. 22, 3282–3291 (2008).
Diao, X. et al. SUMOylation-triggered ALIX activation modulates extracellular vesicles circTLCD4-RWDD3 to promote lymphatic metastasis of non-small cell lung cancer. Signal Transduct. Target. Ther. 8, 426 (2023).
Pinho, S. S. & Reis, C. A. Glycosylation in cancer: mechanisms and clinical implications. Nat. Rev. Cancer 15, 540–555 (2015).
Shu, G. et al. Exosomal circSPIRE1 mediates glycosylation of E-cadherin to suppress metastasis of renal cell carcinoma. Oncogene 42, 1802–1820 (2023).
Nussinov, R., Tsai, C. J. & Jang, H. Anticancer drug resistance: An update and perspective. Drug Resistance Updates: Reviews and commentaries in antimicrobial and anticancer. Chemotherapy 59, 100796 (2021).
Gottesman, M. M. Mechanisms of cancer drug resistance. Annu. Rev. Med. 53, 615–627 (2002).
Gottesman, M. M. et al. Toward a better understanding of the complexity of cancer drug resistance. Annu. Rev. Pharmacol. Toxicol. 56, 85–102 (2016).
Chen, Z. et al. A novel Trojan horse nanotherapy strategy targeting the cPKM‐STMN1/TGFB1 axis for effective treatment of intrahepatic cholangiocarcinoma. Adv. Sci. 10, 2303814 (2023).
Zhang, Z., Gao, Z., Fang, H., Zhao, Y. & Xing, R. Therapeutic importance and diagnostic function of circRNAs in urological cancers: from metastasis to drug resistance. Cancer Metastas. Rev. 43, 867–888 (2024).
Yi, J. et al. CircPVT1 promotes ER-positive breast tumorigenesis and drug resistance by targeting ESR1 and MAVS. The. EMBO J. 42, e112408 (2023).
Fu, L. et al. Circular RNA circPBX3 promotes cisplatin resistance of ovarian cancer cells via interacting with IGF2BP2 to stabilize ATP7A mRNA expression. Hum. Cell 35, 1560–1576 (2022).
Jiang, X. et al. EIF4A3-Induced circARHGAP29 promotes aerobic glycolysis in docetaxel-resistant prostate cancer through IGF2BP2/c-Myc/LDHA signaling. Cancer Res. 82, 831–845 (2022).
Wen, C. et al. CircSETD3 mediates acquired resistance to gefitinib in non-small lung cancer cells by FXR1/ECT2 pathway. The. Int. J. Biochem. Cell Biol. 154, 106344 (2023).
Xu, J. et al. CircRNA-SORE mediates sorafenib resistance in hepatocellular carcinoma by stabilizing YBX1. Signal Transduct. Target. Ther. 5, 298 (2020).
Wang, S. et al. A novel circular RNA confers trastuzumab resistance in human epidermal growth factor receptor 2-positive breast cancer through regulating ferroptosis. Environ. Toxicol. 37, 1597–1607 (2022).
Ma, J. et al. Posttranscriptional regulation of AKT by circular RNA angiomotin- like 1 mediates chemoresistance against paclitaxel in breast cancer cells. Aging 11, 11369–11381 (2019).
Lv, Y. et al. Noncoding RNAs as sensors of tumor microenvironmental stress. J. Exp. Clin. Cancer Res. 41, 224 (2022).
Romani, A. M. P. Cisplatin in cancer treatment. Biochem. Pharmacol. 206, 115323 (2022).
Zhu, S. et al. A role for the ATP7A copper transporter in Tumorigenesis and Cisplatin resistance. J. Cancer 8, 1952–1958 (2017).
Li, H. et al. CircITGB6 promotes ovarian cancer cisplatin resistance by resetting tumor-associated macrophage polarization toward the M2 phenotype. J. Immunother. Cancer 10, e004029 (2022).
Singla, D. K. et al. Fibroblast growth Factor-9 enhances M2 macrophage differentiation and attenuates adverse cardiac remodeling in the infarcted diabetic heart. PLoS ONE 10, e0120739 (2015).
Marchesini, M. et al. ILF2 is a regulator of RNA splicing and DNA damage response in 1q21-amplified multiple myeloma. Cancer Cell 32, 88–100.e6 (2017).
Zhang, H. et al. CircLIFR synergizes with MSH2 to attenuate chemoresistance via MutSα/ATM-p73 axis in bladder cancer. Mol. Cancer 20, 70 (2021).
Goodspeed, A., Jean, A. & Costello, J. C. A Whole-genome CRISPR Screen Identifies a Role of MSH2 in Cisplatin-mediated cell death in muscle-invasive bladder cancer. Eur. Urol. 75, 242–250 (2019).
Cocetta, V., Ragazzi, E. & Montopoli, M. Links between cancer metabolism and cisplatin resistance. Int. Rev. Cell Mol. Biol. 354, 107–164 (2020).
Yang, C. et al. Methionine orchestrates the metabolism vulnerability in cisplatin resistant bladder cancer microenvironment. Cell Death Dis. 14, 525 (2023).
Mabuchi, S. et al. Inhibition of phosphorylation of BAD and Raf-1 by Akt sensitizes human ovarian cancer cells to paclitaxel. J. Biol. Chem. 277, 33490–33500 (2002).
Ellis, L. M. & Hicklin, D. J. Resistance to targeted therapies: refining anticancer therapy in the era of molecular oncology. Clin. Cancer Res. 15, 7471–7478 (2009).
Santoni-Rugiu, E. et al. Intrinsic resistance to EGFR-Tyrosine kinase inhibitors in EGFR-mutant non-small cell lung cancer: differences and similarities with acquired resistance. Cancers 11, 923 (2019).
Romond, E. H. et al. Trastuzumab plus adjuvant chemotherapy for operable HER2-positive breast cancer. N. Engl. J. Med. 353, 1673–1684 (2005).
Serra, V. et al. PI3K inhibition results in enhanced HER signaling and acquired ERK dependency in HER2-overexpressing breast cancer. Oncogene 30, 2547–2557 (2011).
Ling, Y. et al. circCDYL2 promotes trastuzumab resistance via sustaining HER2 downstream signaling in breast cancer. Mol. Cancer 21, 8 (2022).
Osborne, C. K. Tamoxifen in the treatment of breast cancer. N. Engl. J. Med. 339, 1609–1618 (1998).
Choudhury, N. R. et al. RNA-binding activity of TRIM25 is mediated by its PRY/SPRY domain and is required for ubiquitination. BMC Biol. 15, 105 (2017).
Tenenbaum, S. A. et al. Identifying mRNA subsets in messenger ribonucleoprotein complexes by using cDNA arrays. Proc. Natl Acad. Sci. USA 97, 14085–14090 (2000).
Wächter, K. et al. Subcellular localization and RNP formation of IGF2BPs (IGF2 mRNA-binding proteins) is modulated by distinct RNA-binding domains. Biol. Chem. 394, 1077–1090 (2013).
Zhao, Z. et al. circEIF3I facilitates the recruitment of SMAD3 to early endosomes to promote TGF-β signalling pathway-mediated activation of MMPs in pancreatic cancer. Mol. Cancer 22, 152 (2023).
Guo, X. et al. Exosomal circular RNAs: A chief culprit in cancer chemotherapy resistance. Drug Resist. Updates: Rev. Comment. Antimicrob. Anticancer Chemother. 67, 100937 (2023).
Pan, Y., Lin, Y. & Mi, C. Cisplatin-resistant osteosarcoma cell-derived exosomes confer cisplatin resistance to recipient cells in an exosomal circ_103801-dependent manner. Cell Biol. Int. 45, 858–868 (2021).
Chen, L. & Shan, G. CircRNA in cancer: Fundamental mechanism and clinical potential. Cancer Lett. 505, 49–57 (2021).
Chen, Q. et al. Circular RNA ACTN4 promotes intrahepatic cholangiocarcinoma progression by recruiting YBX1 to initiate FZD7 transcription. J. Hepatol. 76, 135–147 (2022).
Jie, M. et al. CircMRPS35 suppresses gastric cancer progression via recruiting KAT7 to govern histone modification. Mol. Cancer 19, 56 (2020).
Yang, F. et al. Circ-HuR suppresses HuR expression and gastric cancer progression by inhibiting CNBP transactivation. Mol. Cancer 18, 158 (2019).
Wang, L. et al. Circular RNA circRHOT1 promotes hepatocellular carcinoma progression by initiation of NR2F6 expression. Mol. Cancer 18, 119 (2019).
Li, H. et al. Therapeutic targeting of circ-CUX1/EWSR1/MAZ axis inhibits glycolysis and neuroblastoma progression. EMBO Mol. Med. 11, e10835 (2019).
Yang, Y. et al. circCAPRIN1 interacts with STAT2 to promote tumor progression and lipid synthesis via upregulating ACC1 expression in colorectal cancer. Cancer Commun. 43, 100–122 (2023).
Huang, S. et al. Mechanism of tumor-derived extracellular vesicles in prostatic cancer progression through the circFMN2/KLF2/RNF128 axis. Apoptosis: Int. J. Program. Cell Death 28, 1372–1389 (2023).
Razpotnik, R. et al. Circular RNA hsa_circ_0062682 binds to YBX1 and promotes oncogenesis in hepatocellular carcinoma. Cancers 14, 4524 (2022).
Zhang, J. Y. et al. ebv-circRPMS1 promotes the progression of EBV-associated gastric carcinoma via Sam68-dependent activation of METTL3. Cancer Lett. 535, 215646 (2022).
Sui, X., Wang, Y. & Liu, H. hsa_circ_0101119 facilitates the progression of cervical cancer via an interaction with EIF4A3 to inhibit TCEAL6 expression. Mol. Med. Rep. 24, 654 (2021).
Feng, Y. et al. Circular RNA circ0005276 promotes the proliferation and migration of prostate cancer cells by interacting with FUS to transcriptionally activate XIAP. Cell Death Dis. 10, 792 (2019).
He, T. et al. Extracellular vesicle-circEHD2 promotes the progression of renal cell carcinoma by activating cancer-associated fibroblasts. Mol. Cancer 22, 117 (2023).
Guo, S. et al. Deregulated expression and subcellular localization of CPSF6, a circRNA-binding protein, promote malignant development of esophageal squamous cell carcinoma. Chin. J. Cancer Res.u 34, 11–27 (2022).
Wang, G. et al. CircRNA_100290 promotes GC cell proliferation and invasion via the miR-29b-3p/ITGA11 axis and is regulated by EIF4A3. Cancer Cell Int. 21, 324 (2021).
Chen, Z. et al. CircPLCE1 facilitates the malignant progression of colorectal cancer by repressing the SRSF2-dependent PLCE1 pre-RNA splicing. J. Cell. Mol. Med. 25, 7244–7256 (2021).
Shen, Y. et al. CircPDIA4 Induces gastric cancer progression by promoting ERK1/2 activation and enhancing biogenesis of oncogenic circRNAs. Cancer Res. 83, 538–552 (2023).
Sun, Y. M. et al. circMYBL2, a circRNA from MYBL2, regulates FLT3 translation by recruiting PTBP1 to promote FLT3-ITD AML progression. Blood 134, 1533–1546 (2019).
Wang, X., Chen, M. & Fang, L. hsa_circ_0068631 promotes breast cancer progression through c-Myc by binding to EIF4A3. Molecular Therapy. Nucleic Acids 26, 122–134 (2021).
Liu, H. et al. Circular RNA circDLC1 inhibits MMP1-mediated liver cancer progression via interaction with HuR. Theranostics 11, 1396–1411 (2021).
Wei, W. et al. Circ0008399 interaction with WTAP promotes assembly and activity of the m6A Methyltransferase complex and promotes cisplatin resistance in bladder cancer. Cancer Res. 81, 6142–6156 (2021).
Zheng, R. et al. Exosomal circLPAR1 functions in colorectal cancer diagnosis and tumorigenesis through suppressing BRD4 via METTL3-eIF3h interaction. Mol. Cancer 21, 49 (2022).
Liu, X. et al. CircMYH9 drives colorectal cancer growth by regulating serine metabolism and redox homeostasis in a p53-dependent manner. Mol. Cancer 20, 114 (2021).
Zhu, T. et al. Oncogenic circTICRR suppresses autophagy via binding to HuR protein and stabilizing GLUD1 mRNA in cervical cancer. Cell Death Dis. 13, 479 (2022).
Liu, J. et al. circIPO7 dissociates caprin-1 from ribosomes and inhibits gastric cancer cell proliferation by suppressing EGFR and mTOR. Oncogene 42, 980–993 (2023).
Chen, M. et al. METTL3-Modulated circUHRF2 Promotes Colorectal Cancer Stemness and Metastasis through Increasing DDX27 mRNA Stability by Recruiting IGF2BP1. Cancers 15, 3148 (2023).
Tan, L. M. et al. Circular RNA XRCC5 aggravates glioma progression by activating CLC3/SGK1 axis via recruiting IGF2BP2. Neurochem. Int. 166, 105534 (2023).
Li, K. et al. A circular RNA activated by TGFβ promotes tumor metastasis through enhancing IGF2BP3-mediated PDPN mRNA stability. Nature. Communications 14, 6876 (2023).
Zhang, Y. et al. CircASPH enhances exosomal STING to facilitate M2 macrophage polarization in colorectal cancer. Inflamm. Bowel Dis. 29, 1941–1956 (2023).
Tong, W. et al. CircREOS suppresses lipid synthesis and osteosarcoma progression through inhibiting HuR-mediated MYC activation. J. Cancer 14, 916–926 (2023).
Huang, C., Xu, R. & Zhu, X. Jiang H. m6A-modified circABCC4 promotes stemness and metastasis of prostate cancer by recruiting IGF2BP2 to increase stability of CCAR1. Cancer Gene Ther. 30, 1426–1440 (2023).
Wu, T. et al. Hsa_circ_0006232 promotes laryngeal squamous cell cancer progression through FUS-mediated EZH2 stabilization. Cell Cycle 20, 1799–1811 (2021)..
Lu, L. et al. EIF4a3-regulated circRABL2B regulates cell stemness and drug sensitivity of lung cancer via YBX1-dependent downregulation of MUC5AC expression. Int. J. Biol. Sci. 19, 2725–2739 (2023).
Huang, Q. et al. A novel circular RNA, circXPO1, promotes lung adenocarcinoma progression by interacting with IGF2BP1. Cell Death Dis. 11, 1031 (2020).
Luo, P. et al. CircKIF4A combines EIF4A3 to stabilize SDC1 expression to activate c-src/FAK and promotes TNBC progression. Cell. Signal. 108, 110690 (2023).
Deng, S. et al. Circular RNA ARHGAP5 inhibits cisplatin resistance in cervical squamous cell carcinoma by interacting with AUF1. Cancer Sci. 114, 1582–1595 (2023).
Wang, Y. et al. Downregulation of hsa_circ_0074854 suppresses the migration and invasion in hepatocellular carcinoma via interacting with HuR and via suppressing exosomes-mediated Macrophage M2 polarization. Int. J. Nanomed. 16, 2803–2818 (2021).
Yu, T. et al. Circular RNA circ-TNPO3 suppresses metastasis of GC by acting as a protein decoy for IGF2BP3 to regulate the expression of MYC and SNAIL. Molecular Therapy. Nucleic Acids 26, 649–664 (2021).
Xia, B. et al. A circular RNA derived from MMP9 facilitates oral squamous cell carcinoma metastasis through regulation of MMP9 mRNA stability. Cell Transplant. 28, 1614–1623 (2019).
Zhao, W. et al. Circular RNA (circ-0075804) promotes the proliferation of retinoblastoma via combining heterogeneous nuclear ribonucleoprotein K (HNRNPK) to improve the stability of E2F transcription factor 3 E2F3. J. Cell. Biochem. 121, 3516–3525 (2020).
Zhong, C. et al. M6A-modified circRBM33 promotes prostate cancer progression via PDHA1-mediated mitochondrial respiration regulation and presents a potential target for ARSI therapy. Int. J. Biol. Sci. 19, 1543–1563 (2023).
Tan, J. et al. Hsa_circ_0005050 interacts with ILF3 and affects cell apoptosis and proliferation by disrupting the balance between p53 and p65. Chem.-Biol. Interact. 368, 110208 (2022).
Liu, Y. et al. Hypoxia-induced FUS-circTBC1D14 stress granules promote autophagy in TNBC. Adv. Sci. 10, e2204988 (2023).
Zhao, H., Chen, S. & Fu, Q. Exosomes from CD133+ cells carrying circ-ABCC1 mediate cell stemness and metastasis in colorectal cancer. J. Cell. Biochem. 121, 3286–3297 (2020).
Hou, Y. et al. The circular RNA circ_GRHPR promotes NSCLC cell proliferation and invasion via interactions with the RNA-binding protein PCBP2. Clin. Exp. Pharmacol. Physiol. 48, 1171–1181 (2021).
Liang, Y. et al. CircIMMP2L promotes esophageal squamous cell carcinoma malignant progression via CtBP1 nuclear retention dependent epigenetic modification. Clin. Transl. Med. 11, e519 (2021).
Cheng, C. et al. Smoking-induced M2-TAMs, via circEML4 in EVs, Promote the progression of NSCLC through ALKBH5-regulated m6A modification of SOCS2 in NSCLC cells. Adv. Sci. (Weinh., Baden.-Wurtt., Ger.) 10, e2300953 (2023).
Wu, Z. et al. Jin H. m6A-Modified circTET2 Interacting with HNRNPC regulates fatty acid oxidation to promote the proliferation of chronic lymphocytic leukemia. Adv. Sci. (Weinh.) 10, e2304895 (2023).
Chen, Y. et al. Circular RNA circAGO2 drives cancer progression through facilitating HuR-repressed functions of AGO2-miRNA complexes. Cell Death Differ. 26, 1346–1364 (2019).
Wang, X. et al. CircRNA-CREIT inhibits stress granule assembly and overcomes doxorubicin resistance in TNBC by destabilizing PKR. J. Hematol. Oncol. 15, 122 (2022).
Gu, L. et al. circCYP24A1 facilitates esophageal squamous cell carcinoma progression through binding PKM2 to regulate NF-κB-induced CCL5 secretion. Mol. Cancer 21, 217 (2022).
Ju, H. et al. A novel intronic circular RNA, circGNG7, inhibits head and neck squamous cell carcinoma progression by blocking the phosphorylation of heat shock protein 27 at Ser78 and Ser82. Cancer Commun. 41, 1152–1172 (2021).
Yang, H. et al. CircPTK2 (hsa_circ_0005273) as a novel therapeutic target for metastatic colorectal cancer. Mol. Cancer 19, 13 (2020).
Hu, X. et al. circGSK3β promotes metastasis in esophageal squamous cell carcinoma by augmenting β-catenin signaling. Mol. Cancer 18, 160 (2019).
Chen, J. et al. Circ-GLI1 promotes metastasis in melanoma through interacting with p70S6K2 to activate Hedgehog/GLI1 and Wnt/β-catenin pathways and upregulate Cyr61. Cell Death Dis. 11, 596 (2020).
Lin, J. et al. Hypoxia-induced exosomal circPDK1 promotes pancreatic cancer glycolysis via c-myc activation by modulating miR-628-3p/BPTF axis and degrading BIN1. J. Hematol. Oncol. 15, 128 (2022).
Xu, Z. et al. Circular RNA circPOLR2A promotes clear cell renal cell carcinoma progression by facilitating the UBE3C-induced ubiquitination of PEBP1 and, thereby, activating the ERK signaling pathway. Mol. Cancer 21, 146 (2022).
Chen, H. et al. Hypoxia-inducible CircPFKFB4 promotes breast cancer progression by facilitating the CRL4DDB2 E3 ubiquitin Ligase-mediated p27 degradation. Int. J. Biol. Sci. 18, 3888–3907 (2022).
Xu, J. et al. CircCDR1as mediates PM2.5-induced lung cancer progression by binding to SRSF1. Ecotoxicol. Environ. Saf. 249, 114367 (2023).
Li, P. et al. Characterization of circSCL38A1 as a novel oncogene in bladder cancer via targeting ILF3/TGF-β2 signaling axis. Cell Death Dis. 14, 59 (2023).
Wang, Z. et al. Circular RNA MTCL1 promotes advanced laryngeal squamous cell carcinoma progression by inhibiting C1QBP ubiquitin degradation and mediating beta-catenin activation. Mol. Cancer 21, 92 (2022).
Chen, S. et al. circNEIL3 inhibits tumor metastasis through recruiting the E3 ubiquitin ligase Nedd4L to degrade YBX1. Proc. Natl Acad. Sci. USA 120, e2215132120 (2023).
Yang, R. et al. Hypoxia-induced circWSB1 promotes breast cancer progression through destabilizing p53 by interacting with USP10. Mol. Cancer 21, 88 (2022).
Deng, J. et al. Specific intracellular retention of circSKA3 promotes colorectal cancer metastasis by attenuating ubiquitination and degradation of SLUG. Cell Death Dis. 14, 750 (2023).
Li, Z. et al. circRNA-SFMBT2 orchestrates ERα activation to drive tamoxifen resistance in breast cancer cells. Cell Death Dis. 14, 482 (2023).
Cui, Z. et al. O-GlcNAcylated LARP1 positively regulated by circCLNS1A facilitates hepatoblastoma progression through DKK4/β-catenin signalling. Clin. Transl. Med. 13, e1239 (2023).
Song, W. et al. Sevoflurane suppresses colorectal cancer malignancy by modulating β-catenin ubiquitination degradation via circSKA3. Cell Signal 114, 110987 (2024).
He, Y. et al. circPTPN22 attenuates immune microenvironment of pancreatic cancer via STAT3 acetylation. Cancer Gene Ther. 30, 559–566 (2023).
Xie, B. et al. CircXRN2 suppresses tumor progression driven by histone lactylation through activating the Hippo pathway in human bladder cancer. Mol. Cancer 22, 151 (2023).
Zhang, X. et al. Essential roles of exosome and circRNA_101093 on ferroptosis desensitization in lung adenocarcinoma. Cancer Commun. 42, 287–313 (2022).
Yuan, G. et al. Upregulated circRNA_102231 promotes gastric cancer progression and its clinical significance. Bioengineered 12, 4936–4945 (2021).
Li, X. et al. circEXOC6B interacting with RRAGB, an mTORC1 activator, inhibits the progression of colorectal cancer by antagonizing the HIF1A-RRAGB-mTORC1 positive feedback loop. Mol. Cancer 21, 135 (2022).
Li, X. et al. circCDYL2, overexpressed in highly migratory colorectal cancer cells, promotes migration by binding to Ezrin. Front. Oncol. 11, 716073 (2021).
Zhang, X. et al. Circ SMARCA5 Inhibited Tumor Metastasis by Interacting with SND1 and Downregulating the YWHAB Gene in Cervical Cancer. Cell Transplant. 30, 963689720983786 (2021).
Wang, C. et al. CircHIPK3 negatively regulates autophagy by blocking VCP binding to the Beclin 1 complex in bladder cancer. Discover. Oncology 14, 86 (2023).
Chen, C., Zhang, M. & Zhang, Y. Circ_0000079 decoys the RNA-binding protein FXR1 to interrupt formation of the FXR1/PRCKI complex and decline their mediated cell invasion and drug resistance in NSCLC. Cell Transplant. 29, 963689720961070 (2020).
Zhang, M. X. et al. CircME1 promotes aerobic glycolysis and sunitinib resistance of clear cell renal cell carcinoma through cis-regulation of ME1. Oncogene 41, 3979–3990 (2022).
Acknowledgements
This study was supported by the National Natural Science Foundation of China (No. 82072887) and Provincial Doctoral Research Start-up Fund Project:The mechanism of circNFATC3’s multiple regulation of SDC2 in promoting cervical cancer progression and metastasis(Project number:2022-BS-133).
Author information
Authors and Affiliations
Contributions
N.Z. contributed to the manuscript and form. XJW depicted the figure. Y.W.L. and C.C.S. modified the grammar. YL and YMS summarized the molecular mechanisms of CPIs. N.Y.M. and Y.S.J. contributed to the conception and design. All authors have read and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editors: Kaliya Georgieva. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhang, N., Wang, X., Li, Y. et al. Mechanisms and therapeutic implications of gene expression regulation by circRNA-protein interactions in cancer. Commun Biol 8, 77 (2025). https://doi.org/10.1038/s42003-024-07383-z
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s42003-024-07383-z
This article is cited by
-
CircRNAs: functions and emerging roles in cancer and immunotherapy
BMC Medicine (2025)
-
Emerging role and clinical applications of circular RNAs in human diseases
Functional & Integrative Genomics (2025)