Abstract
Osteoarthritis (OA) is a common age-related disease that is correlated with a high number of senescent chondrocytes in joint tissues. Heterogeneous nuclear ribonucleoprotein D (HNRNPD) is an RNA-binding protein whose expression imbalance is associated with cell senescence, but the role of HNRNPD in the occurrence and development of OA has not been reported. In this study, HNRNPD was found to be associated with the chondrocyte senescence process. We determined the factors at the posttranscriptional level that regulated the expression of the genes that induce OA and found that HNRNPD was specifically highly expressed in OA-induced rat cartilage and in human OA cartilage. Recombinant adeno-associated virus (rAAV)-mediated HNRNPD gene overexpression alone did not significantly regulate the occurrence and development of OA in the physiological state of the joint. However, rAAV-HNRNPD significantly exacerbated experimental OA in rats subjected to destabilization of the medial meniscus. Overexpression of HNRNPD promoted mitochondrial dysfunction and the expression of FOXM1, which acts as a direct target. Furthermore, downregulation of FOXM1 in chondrocytes weakened the HNRNPD-mediated promotion of chondrocyte senescence and mitochondrial dysfunction. Our results suggest that the RNA-binding protein HNRNPD promotes chondrocyte senescence in the pathology of OA by upregulating FOXM1.
Similar content being viewed by others
Introduction
Osteoarthritis (OA) is a chronic degeneration characterized by destruction of articular cartilage stroma and mainly manifests as subchondral bone hyperplasia, synovial inflammation, and articular cartilage degeneration1,2. In the coming decades, there will be a sharp increase in the number of people suffering from OA and requiring artificial joint replacement due to OA3,4. For the elderly individual, many factors related to senescence can lead to OA. Previous studies revealed that under conditions of mitochondrial dysfunction, oxidative stress, and decreased autophagy, the function of chondrocytes changed, and abnormal phenomena such as degradation, senescence, and apoptosis occurred5,6. Therefore, in-depth study of senescence is important to elucidate the factors in OA pathogenesis and will help identify new treatment targets to delay or block the onset of OA, an achievement that would have great social significance.
Long-term survival of senescent cells leads not only to organ damage but also to OA and other age-associated diseases. Cell senescence is the basis of body aging, and in the occurrence and development of OA, chondrocytes show a variety of senescent characteristics7,8. Senescent cells exhibit nondivision, apoptosis and a frequent senescence-associated secretory phenotype (SASP); moreover, senescent cells can affect the morphology and function of the cells and tissues around them9,10. In SASP, levels of pro-inflammatory factors such as interleukin (IL)-1, IL-6 and matrix metalloproteinase (MMP)-3 have been found to increase, coinciding with the factors that promote the occurrence of OA11,12.
Studies have shown that posttranscriptional regulation of mRNA is an important molecular mechanism in the regulation of chondrocyte senescence. The regulation of mRNA stability is a crucial link influenced by both miRNAs and target genes (RNA binding proteins)13,14. The RNA-binding protein binds to the AU enrichment element in the target gene 3-untranslated-UTRs, resulting in tail translocation of PolyA, which leads to mRNA degradation15,16. In the early stage, we focused on the role of RNA binding protein in OA, and compared the expression of heterogeneous nuclear ribonuclear protein D (HNRNPD) in rat and human OA cartilage specimens, and found that the expression of HNRNPD in rat and human OA cartilage specimens was significantly higher than that in normal cartilage tissues, which was shown in Fig. 1B. HNRNPD is a kind of noncoding RNA located in cells that can bind to noncoding RNA and that plays an important role in mRNA synthesis and transport17. HNRNPD is an important molecule that regulates the stability of mRNA encoded by genes involved in the cell cycle, proliferation, senescence, and apoptosis18,19. Previous studies have shown that HNRNPD can regulate aging in skin cells20, vascular endothelial cells21, and human airway epithelium22. HNRNPD showed contradictory functions in multiple tumor model systems, promoting both pro-tumor formation events and anti-tumor formation events. The phenotypic complexity is due to the different functions of different HNRNPD subtypes in specific tissue contexts, and also because the abnormally regulated mRNA is involved in many different functions23,24. The target mRNA of HNRNPD includes both positive and negative regulators that encode the regulatory process. The complexity of these gene regulatory networks precludes a common definition of HNRNPD as a single agent that promotes or resists disease formation. There may be different regulatory modes in different cell environments and concentrations of HNRNPD, so it is of great significance to study the relevant mechanisms for specific subtypes and specific cell localization. However, no investigation has been reported on whether HNRNPD regulates mRNA and participates in the occurrence and development of OA.
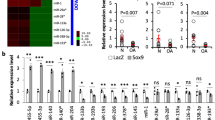
A The MRI, general and arthroscopic observation of OA cartilage tissues and corresponding non-lesion samples. Black arrowheads indicate the site of cartilage damage. B The HE, Safranin O/fast green, Alcian blue staining (scale bars; up: 600 µm, down: 240 µm), and immunohistochemical staining of HNRNPD and P21 of the cartilage tissues of OA patient and OA model rat (scale bar: 120 µm). C Immunofluorescence analysis of P16INK4AINK4a in the synovium, cartilage surface, and subchondral bone from sham and OA model rats (scale bar: 60 µm). The ratios of immunoreactive cells of P16INK4A (D), HNRNPD (E), and P21 (F) were analyzed. Data were expressed as mean ± SD (n = 5). ***P < 0.001 vs. sham or control group.
The purpose of this study was to explore the relationship between HNRNPD and the occurrence and development of OA. The results showed that the expression of HNRNPD was significantly increased in cartilage tissues of patients with OA and in OA model rats. In this research, we evaluate the effect of HNRNPD on the occurrence and development of OA and explore its regulatory mechanism. We found that recombinant adeno-associated virus (rAAV)-HNRNPD significantly accelerated the chondrocyte senescence phenotype and aggravated the occurrence of OA in a destabilization of the medial meniscus (DMM) rat model. Further studies revealed that HNRNPD regulates the expression of the FOXM1 gene and may be a key molecule in regulating the senescence phenotype of articular cartilage in OA. To the best of our knowledge, this is the first report describing a causative role for HNRNPD in chondrocyte senescence and onset of OA.
Results
Chondrocyte senescence and upregulated HNRNPD are closely associated with the progression of OA
To explore the possible association of the RNA-binding protein HNRNPD with OA pathogenesis, we first analyzed the levels of HNRNPD in osteoarthritic cartilage tissues and corresponding nonlesion samples (Fig. 1A). Prior to experimental assessment, we performed hematoxylin-eosin (H&E), Safranin O/fast green and Alcian blue staining of each of the human and rat cartilage tissue samples and detected HNRNPD protein expression using an immunohistochemical staining assay (Fig. 1B). ACLT + MMx-mediated injury reduced Safranin O and Alcian blue staining of proteoglycans and resulted in cartilage thinning, surface irregularities and joint space narrowing (specimen tissue from our previous studies25,26). The HNRNPD protein level was markedly elevated in the OA-damaged regions of human cartilage compared with that in normal cartilage. The HNRNPD protein level was also markedly increased in chondrocytes of osteoarthritic cartilage from rats with ACLT + MMx surgery (Fig. 1E), and the results demonstrated that HNRNPD was significantly upregulated in osteoarthritic cartilage.
To explore the role of cellular senescence in OA development, we examined the expression of P16INK4A and P21, typical biomarkers of senescent cells, in human and rat osteoarthritic cartilage. The levels of P16INK4A and P21 were dramatically elevated in the cartilage of the patients with OA (Fig. 1B, C). We found that compared to sham-operated rats, ACLT + MMx-mediated rats had a greater number of P16INK4A- and P21-expressing cells on the cartilage surface, as well as in the synovium and subchondral bone. Moreover, cartilage degradation was significantly aggravated during ACLT + MMx-induced OA pathogenesis, an outcome that was tightly correlated with enhanced P16INK4A and P21 expression (Fig. 1D, F). These results indicate that chondrocyte senescence and upregulated HNRNPD are tightly correlated with OA progression.
HNRNPD overexpression alone does not modulate OA pathogenesis
Based on the above findings, rAAV was used in this portion of the study to transfect HNRNPD and observe the effect on the occurrence and development of OA in vivo (Fig. 2A, B). Our previous work extensively demonstrated that rAAV9 can efficiently transport target genes to joints26. We observed that weekly rAAV-HNRNPD intra-articular (IA) injections significantly increased the expression of HNRNPD in cartilage and synovium, as confirmed by immunohistochemical staining (Fig. 2C, D). However, modified Mankin score results showed that HNRNPD overexpression did not cause cartilage destruction in sham rats, whereas injection of rAAV-HNRNPD into the articular cavity of sham rats resulted in thinning of subchondral bone, as shown by the H&E and Safranin O staining (Fig. 2E, F).
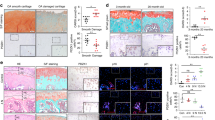
A The rAAV-HNRNPD solution was intra-articularly injected into healthy and OA rats to examine the senescense regulation. B Schematic diagram of the time course for the experiment. C SD rats received an intra-articular injection of rAAV encoding HNRNPD-specific RNA for HNRNPD or GFP into the knee. HNRNPD expression in cartilage and synovium was assessed via immunofluorescence (scale bars; up: 1200 µm, down: 30 µm). D The ratios of HNRNPD in synovium and cartilage were analyzed. Data were expressed as mean ± SD (n = 5). ***P < 0.001 versus the control group. E The general observation, H&E, and Safranin O-stained tissue histology in rat articular cartilage at 10 weeks post-surgery (scale bars; up: 600 µm, down: 120 µm). Yellow arrows indicate the subchondral bone loss. F The modified Mankin scores were assigned to tissue samples. Data were expressed as mean ± SD (n = 6). ***P < 0.001 vs. the sham-operated group; ##P < 0.01 vs. the DMM-surgical group.
For this study, the subchondral T2 signal of the knee of rats was analyzed by micro-MRI (Fig. 3A). While fat-suppressed, T2-weighted micro-MRI images from the sham-operated rats exhibited a clear, thin T2 signal consistent with the metaphyseal growth plate and cartilage surface regularity, there was no significant difference in the T2 signal between the sham-operated and rAAV-HNRNPD-injected rats.
A Pixel intensity of a sampled region of epiphyseal trabecular bone and gross morphological were assessed by micro-MRI in sham operation, rAAV-HNRNPD, DMM, and DMM + rAAV-HNRNPD groups. White arrows indicate the site of cartilage damage and subchondral bone marrow edema. B Representative micro-CT three-dimensional reconstructions of tibial and femoral subchondral bones in the rats after DMM surgery. Micro-CT images of animals in each group were used for measurements of BV/TV (C), Tb. Th (D) and Tb. Sp (E) Black arrowheads indicate sparse subchondral trabecular structure. Data were expressed as mean ± SD (n = 5). *P < 0.05 vs. sham-operated group; #P < 0.05 vs. DMM induction group.
The micro-CT technique was used to observe the changes in calcification, morphology and osteophytes in the articular cavity (Fig. 3B). Compared to the sham-operated rats, those in the rAAV-HNRNPD-injected group exhibited reductions in the quantitative morphological parameters (BV/TV) ratio (Fig. 3B) and increased the average trabecular interval (Tb. Sp) (Fig. 3D), and there was no statistically significant difference in the average trabecular thickness (Tb. Th) (Fig. 3C), which were consistent with the H&E and Safranin O staining. These results collectively indicate that overexpression of HNRNPD alone is not sufficient to cause OA pathogenesis in sham-operated rats, but it led to a decrease in subchondral bone mass with the appearance of osteoporosis.
Gene implantation of HNRNPD exacerbates OA development in a rat OA model
We further examined the effects of HNRNPD overexpression in joint tissues of rats subjected to DMM-induced OA pathogenesis. We performed gene implantation of HNRNPD in whole-joint tissues of rats via IA injection of rAAV9 in a DMM-induced rat OA model. Overexpression of HNRNPD in the joint tissues of the DMM-operated rats significantly accelerated all examined OA characteristics, as shown by the modified Mankin score, osteophyte maturity, and subchondral bone plate thickness (Fig. 2E, F). Consistent with this inhibition of osteoarthritic cartilage erosion, DMM-induced MMP-3 upregulation and Col II downregulation in damaged cartilage was strongly aggravated in the rAAV-HNRNPD-injected OA model rats (Fig. 4A, C).
A, B Immunofluorescence with antibodies to MMP-13, Col II, P53, and P16INK4A in articular cartilage from the DMM-induced OA rats with rAAV-HNRNPD treatment at 10 weeks post-surgery (scale bar: 80 µm). C, D The ratios of immunoreactive cells were quantified in articular cartilage according to immunofluorescence. Data were expressed as mean ± SD (n = 5). *P < 0.05, **P < 0.01 and ***P < 0.001 vs. the sham-operated group; #P < 0.05 and ##P < 0.01 vs. the DMM induction group.
In the area of the bone defect, the T2 signal intensity in OA model rats was significantly increased, which was consistent with the pathological changes in bone tissue, edema-like inflammation, and cartilage surface changes. The T2 signal intensity and cracked cartilage were aggravated in the DMM-operated OA model rats that had been intra-articularly injected with rAAV-HNRNPD relative to the untreated OA model rats (Fig. 3A). Compared to the sham-operated rats, those in the OA model group exhibited reductions in the BV/TV ratio (Fig. 3B) and increased Tb. Sp (Fig. 3D), and there was no statistically significant difference in Tb. Th (Fig. 3C). After the rAAV-HNRNPD gene was injected into OA rats, the above changes were further aggravated (Fig. 3B, D). These results collectively indicate that overexpression of HNRNPD in OA rats can cause obvious degeneration of articular cartilage in OA rats and promote the progression of OA.
HNRNPD accelerates chondrocyte senescence in DMM model rats
To determine the causal relationship between the overexpression of chondrocyte HNRNPD and cartilage senescence, we determined the protein expression profiles of articular cartilage from rAAV-HNRNPD-injected rats and their littermate controls. The number of P53- and P16INK4A-positive articular chondrocytes was increased in the rAAV-HNRNPD-injected rats compared with their littermate controls (Fig. 4B, D). HNRNPD overexpression in vivo promoted cartilage senescence, while in healthy cartilage, HNRNPD implantation alone could not promote the occurrence of cartilage degeneration in the short term, according to the results of cartilage morphological staining. The number of P53- and P16INK4A-positive articular chondrocytes was markedly increased in the rAAV-HNRNPD-injected DMM model rats compared with their littermate OA rats (Fig. 4B, D). HNRNPD overexpression induced chondrocyte senescence and accelerated experimental OA in DMM model rats. Collectively, these results reveal that HNRNPD overexpression in chondrocytes accelerates cell senescence and leads to markedly observable cartilage destruction in rats with traumatic OA.
HNRNPD broadly affects the gene expression profile of chondrocytes
In this portion of the study, the pcDNA3.1-HNRNPD vector and the HNRNPD overexpression vector were constructed, and the influence on the occurrence and development of OA was explored (Fig. 5A). Compared with the control group, the pcDNA3.1-HNRNPD group exhibited increased HNRNPD protein expression (Fig. 5B). Figure5C showed that the pcDNA3.1-HNRNPD group was significantly separated from the control group and that the three biological replicates were grouped together (Fig. 5C). On the basis of the fragments per kilobase of transcript per million mapped reads (FPKM) values of each expressed gene, six association matrixes of unsupervised hierarchical clustering were established. We found that HNRNPD overexpression obviously changed the chondrocyte gene expression profile. The cluster map of differentially expressed genes revealed the close relationship between gene expression patterns and sample clustering. The heatmap plot of the FPKM values of all DEGs showed that there was a significant separation between the pcDNA3.1-HNRNPD group and the control group, and the three replicates were more consistent in the same group (Fig. 5D). Through the analysis of the differential expression profiles of the two groups, we found 70 expression profiles and 145 downregulated expression profiles (Fig. 5E). Pre-mRNA alternative splicing in the chondrocytes showed changes in the alternative 5’ splice site (A5SS) (25.06%), alternative 3’ splice site (A3SS) (23.44%), intron retention (IntronR) (21.43%), exon skipping (ES) (9.26%), and CassetteExon (6.49%) (Fig. 5F). Through Gene Ontology (GO) technology, all molecular processes, including protein binding, ATP binding, transcription factor binding, RNA binding, and cell differentiation regulation, were found to be related to HNRNPD in chondrocytes (Fig. 5G and Supplementary Fig. 1). The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses revealed that these genes were significantly related to cellular senescence, the NOD-like receptor signaling pathway, the NF-κB signaling pathway, and the PI3K-Akt signaling pathway (Fig. 5H, I and Supplementary Fig. 1). The Ras proportional thermal analysis showed that FOXM1, Smad2, Mras, Itpr1, Lin9, and Fbxw11 were important molecules in regulating cell senescence (Fig. 5J).
A Chondrocytes were co-treated by lipofectamine transfected pcDNA3.1 empty plasmid and pcDNA3.1-HNRNPD plasmid (1 μg/μL), and the RNA sequencing and AS analysis were performed. B The fluorescence expression of chondrocytes after transfection of pcDNA3.1-HNRNPD plasmid proved successful transfection (scale bar: 100 µm). C The clustering analysis of samples showed high similarity, which confirmed the correctness of the experimental design and sample sampling. D The heat map showed that the gene expression patterns and clustering relationships of the samples were similar. E Comparisons between samples showed the number of differentially expressed genes. F Histogram of the differential splicing events between chondrocytes transfected pcDNA3.1 empty and pcDNA3.1-HNRNPD plasmids. G GO analysis showed that the biological processes such as including the protein binding, ATP binding, transcription factor binding, and RNA binding were associated with HNRNPD in chondrocytes. H, I KEGG pathway analyses revealed these genes to be significantly linked to pathways including the cellular senescence, NOD-like receptor signaling pathway, NF-κB signaling pathway, and PI3K-Akt signaling pathway. J Ras ratio heatmap of cellular senescence pathway showed that many genes, including FOXM1, Smad2, Mras, Itpr1, Lin9, and Fbxw11, were closely related to cellular senescence. Data were expressed as mean ± SD (n = 5). ***P < 0.001 vs. the control group.
HNRNPD-mediated mitochondrial dysfunction leads to chondrocyte senescence
Since mitochondrial dysfunction has been identified as one of the main triggers of cellular senescence, we next tested whether mitochondrial dysfunction was involved in HNRNPD-induced cellular senescence. The unclear expression of HNRNPD in the rat chondrocytes induced by IL-1β was significantly increased, which was consistent with the expression site of the HNRNPD nuclear protein (Fig. 6A, D). The senescence-associated β-galactosidase (SA-βGal) assay confirmed that HNRNPD promoted senescence in the chondrocytes (Fig. 6B). We also found that the IL-1β or pcDNA3.1-HNRNPD plasmid induced significant mitochondrial dysfunction in the chondrocytes, and that the pcDNA3.1-HNRNPD plasmid significantly enhanced IL-1β-induced mitochondrial dysfunction. Mitochondrial membrane potential is an indicator of mitochondrial function and can be determined by using rhodamine and JC-1 staining, both of which entail fluorescent dyes sensitive to the mitochondrial membrane potential. As shown in Fig. 6, the pcDNA3.1-HNRNPD plasmid caused a gradual reduction in mitochondrial membrane potential, as observed by the decreased fluorescence intensity (Fig. 6C, F) and ratios of red/green fluorescence (Fig. 6I, J) in chondrocytes. The mitochondrial length that was determined using MitoTracker Red staining was significantly decreased after pcDNA3.1-HNRNPD plasmid treatment (Fig. 6G, H). These results suggest that HNRNPD causes mitochondrial dysfunction, which leads to cellular senescence of joints and chondrocytes in rats.
A Immunofluorescence with antibodies to HNRNPD in chondrocytes (scale bar: 100 µm). B Representative images of SA-βGal staining (scale bar: 100 µm). C Representative images of rhodamine staining (scale bar: 100 µm). The fluorescence intensities of HNRNPD (D), SA-βGal staining level (E), and fluorescence intensity of rhodamine (F) were analyzed. G Representative images of Mitotracker staining (scale bar: 100 µm). H The ratio of mitochondrial length was analyzed. I Representative images of JC-1 staining (scale bar: 100 µm). J The ratio of JC-1 fluorescence (red/green) was analyzed. Data were expressed as mean ± SD (n = 5). **P < 0.01 and ***P < 0.001 vs. control group; #P < 0.05 and ##P < 0.01 vs. IL-1β group; &P < 0.05 and &&P < 0.01 vs. HNRNPD group.
HNRNPD overexpression activates FOXM1 to promote chondrocyte senescence
Studies at the cellular level showed that overexpression of HNRNPD significantly increased FOXM1 expression in primary culture chondrocytes (Fig. 7A). To further investigate whether HNRNPD accelerated cellular senescence by activating FOXM1, we transfected chondrocytes with pcDNA3.1-HNRNPD and FOXM1 short interfering RNA (siRNA) (Fig. 7C, D). SA-βGal staining was enhanced after the HNRNPD plasmid transfection into the chondrocytes, and the enhancement effect of HNRNPD on SA-βGal staining was weakened when FOXM1 was knocked down by FOXM1 siRNA (Fig. 7E, F). We found that knockdown of FOXM1 markedly alleviated HNRNPD-induced mitochondrial dysfunction, which was further confirmed by reductions in the mitochondrial membrane potential (Fig. 7G, H), mitochondrial length (Fig. 7I, L), and ratios of red/green fluorescence using JC-1 staining (Fig. 7J, K). Taken together, these results indicate that HNRNPD overexpression by a plasmid activates FOXM1 to promote chondrocyte senescence.
A Immunofluorescence with antibodies to FOXM1 in chondrocytes (scale bar: 10 µm). B The fluorescence intensity of FOXM1 was analyzed. C Chondrocytes were treated with HNRNPD siRNA (100 nM) and non-targeting controls (100 nM) for 24 h. HNRNPD level was assessed via Western blotting (n = 3). D Tubulin was probed as a loading control and the HNRNPD to tubulin ratio was assessed. E Representative images of SA-βGal staining (scale bar: 100 µm). F The SA-βGal staining level was analyzed. G Representative images of rhodamine staining (scale bar: 100 µm). H The fluorescence intensity of Rhodamine-123 was analyzed. I Representative images of Mitotracker staining (scale bar: 100 µm). J Representative images of JC-1 staining (scale bar: 100 µm). The ratio of JC-1 fluorescence (red/green) (K) and mitochondrial length (L) were analyzed. Data were expressed as mean ± SD (n = 5). *P < 0.05 and ***P < 0.001 vs. control group; ###P < 0.001 vs. IL-1β group; &&P < 0.01 and &&&P < 0.001 vs. HNRNPD group; @@ P < 0.001 vs. IL-1β + HNRNPD group.
FOXM1 is a senescent target of HNRNPD
We used microarray analysis to examine gene expression profiles in primary-culture chondrocytes subjected to overexpression of HNRNPD via infection with the pcDNA3.1-HNRNPD plasmid. Overexpression of HNRNPD increased the levels of genes associated with senescence; among these increases, a marked increase was observed for FOXM1, which was a cellular transcription factor and a component of the cellular senescence-related transcription factor network (Fig. 5J). We assessed the expression patterns of FOXM1 in cartilage tissues from osteoarthritic cartilage tissues and corresponding nonlesion samples, and the results demonstrated that FOXM1 was significantly upregulated in osteoarthritic cartilage tissues (Fig. 8A–C), and the FOXM1 expression trend was confirmed in vitro (Fig. 8G). HNRNPD overexpression aggravated the IL-1β-induced expression of P16INK4A, P53, P21, Tom 20, and Bcl-2. FOXM1 knockdown alleviated the HNRNPD-induced expression of P16INK4A, P53, P21, Tom 20, and Bcl-2 (Fig. 8J, K). The transmission electron microscopy (TEM) results from the in vivo animal studies showed that the morphology of mitochondria in the sham group was normal, and mitochondrial ridges were clearly visible. In sham + rAAV-HNRNPD, DMM and DMM + rAAV-HNRNPD groups, the mitochondrial structure of the synovial fibroblasts was disordered, with mitochondrial swelling and blurring of the mitochondrial ridges, and the mitochondrial pathological changes were more serious in the DMM + rAAV-HNRNPD group (Fig. 8D), suggesting that the injection of rAAV-HNRNPD into the joint cavity of the sham and DMM model groups could aggravate mitochondrial damage. In vivo studies showed that the number of FOXM1-positive articular chondrocytes increased significantly in the rats injected with rAAV-HNRNPD (Fig. 8E, F). We examined whether HNRNPD regulates the FOXM1 as a direct target. We performed Coimmunoprecipitation (Co-IP) assays of chondrocytes. Endogenous HNRNPD was immunoprecipitated from chondrocyte lysates by a FOXM1 antibody. The interaction was further confirmed by immunoprecipitation of FOXM1 with HNRNPD antibody (Fig. 8I). We next verified the associations between HNRNPD and FOXM1 by conducting a RNA immunoprecipitation (RIP) assay. As shown in Fig. 8L, M, the RIP assay showed significant enrichment of HNRNPD and FOXM1 in the Ago2 antibody, and verified the correlation of HNRNPD and FOXM1. Our results collectively suggest that HNRNPD interacted with FOXM1 to induce senescence in rat articular chondrocytes.
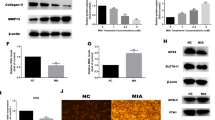
A, B Elevated FOXM1 levels closely associated with the progression of OA. Immunofluorescence analysis of FOXM1 in the cartilage surface from sham or control and OA model rat (scale bars; up: 1200 µm, down: 30 µm) or OA patient (scale bars; up: 240 µm, down: 40 µm). C The ratios of immunoreactive cells of FOXM1 were analyzed (n = 5). D The representative TEM images of rat synovium. Red arrowheads indicate mitochondrial structure (scale bars; up: 1 µm, down: 500 nm). E Immunofluorescence with antibodies to FOXM1 in articular cartilage from the DMM-induced OA rats with rAAV-HNRNPD treatment at 10 weeks post-surgery (n = 5, scale bars; up: 1200 µm, down: 30 µm). F The ratios of FOXM1 immunoreactive cells were quantified in articular cartilage according to immunofluorescence. G The protein expression of FOXM1 and HNRNPD was assessed via western blotting with tubulin as a loading control. H The ratios of FOXM1 and HNRNPD to tubulin were analyzed (n = 3). I Co-IP assay between HNRNPD and FOXM1 was performed. J The protein expressions of P16INK4A, P21, P53, Bcl-2, and Tom 20 were assessed via western blotting with tubulin as a loading control. K The ratios of P16INK4A, P21, P53, Bcl-2, and Tom 20 to tubulin were analyzed (n = 3). L, M The RIP analysis was performed to verify the correlation of HNRNPD and FOXM1 (n = 3). N The unique hydrophobic cavity of FOXM1 was large enough to bind the hydrophobic surface of HNRNPD. The hydrophobic surface of HNRNPD binds the FOXM1 with a binding energy of −53.88 kcal/mol. Data were expressed as mean ± SD. *P < 0.05 and ***P < 0.001 vs. control, sham-operated group or input group; #P < 0.05 and ##P < 0.01 vs. IL-1β or DMM induction group; &P < 0.05 and &&&P < 0.001 vs. IL-1β + HNRNPD group.
Hydrophobic interaction between HNRNPD and FOXM1
It was found that there was a hydrophobic interaction between HNRNPD and FOXM1. The binding sites of HNRNPD and FOXM1 were studied by molecular simulation. The hydrophobic cavity peculiar to FOXM1 was large enough to engage with the hydrophobic surface of HNRNPD (Fig. 8N). The spatial filling model showed that the scope of HNRNPD and the FOXM1 inhibitory pocket were closely linked. HNRNPD had a hydrophobic surface interacting with FOXM1, and the interaction energy between FOXM1 and FOXM1 was −53.88 kcal/mol.
Discussion
We demonstrate here that the RNA-binding protein HNRNPD can regulate the expression of FOXM1 and then affect the occurrence and development of OA. We found that the expression of HNRNPD in rat articular cartilage increased significantly under mechanical stimulation, such as DMM. The upregulation of HNRNPD is necessary for the development of OA since our results reveal that genetic overexpression of HNRNPD exacerbates posttraumatic OA in rats. We further report that overexpression of HNRNPD exacerbates OA pathogenesis and mitochondrial dysfunction by upregulating FOXM1. Therefore, this study proposes a new approach for inhibiting HNRNPD and provides a new avenue for the prevention and treatment of OA.
Increasing age is strongly correlated with cartilage degeneration, and the presence of senescent cells in cartilage isolated from patients undergoing total knee artificial implants has been noted. In the current study, we found that there is an obvious senescent phenomenon in the cartilage tissue of OA patients, and there are many senescent chondrocytes around the OA focus. There was no senescent phenomenon in normal healthy people, suggesting that cellular senescence is closely related to the pathogenesis of OA. However, the molecular mechanism of the correlation between senescence and the occurrence and development of OA is still unclear. A key initial finding was obtained from the observation that HNRNPD is specifically upregulated in osteoarthritic chondrocytes. Studies have shown that HNRNPD can regulate RNA variable shearing and participate in cell senescence and aging processes27,28. Here, we found that the level of HNRNPD was specifically increased in chondrocytes subjected to stimulation of OA-associated catabolic signaling. We confirmed a close relationship between HNRNPD and OA in both in vivo and in vitro studies, showing a positive correlation. The potential mechanism of this relationship is of great significance in revealing the regulation of OA pathogenesis by RNA-binding proteins, and it is worth further exploration. The subtype specificity and repetitive action of HNRNPD may be the result of its action on some mRNA target genes. In this study, we found that HNRNPD, a novel senescence regulator, was significantly elevated in osteoarthritic chondrocytes. HNRNPD is a novel type of protein that regulates senescence, and its expression is significantly increased in OA cartilage. HNRNPD regulates mitochondrial homeostasis and cellular senescence in a FOXM1-dependent manner and then affects the occurrence and development of OA.
Our studies revealed that high expression of HNRNPD can promote the pathological process of OA in DMM-induced OA model rats, and the high expression of HNRNPD in OA chondrocytes is closely related to the pathological process of OA. OA is a whole-joint disease that involves multiple cell types and multiple pathological changes in all joint tissues29,30. In the process of the occurrence and development of OA, targeted gene transfection aiming at the whole-joint will become a new avenue for the treatment of OA31,32. After delivery through IA administration, the rAAV9 vectors expressing transgenes did not induce cell-mediated immunity or inflammation, indicating a viable means of treating OA26,33. We injected rAAV9 into the knee cavity in vivo and efficiently transfected it into the articular cartilage and synovial tissue of rats, resulting in overexpression of the target gene, thus successfully simulating the local genetic modification of the articular cavity. In this study, the injection of rAAV9 into the knee joint of rats significantly increased the expression level of HNRNPD, which was confirmed by immunofluorescence detection. However, the high expression of HNRNPD in normal rat joints does not lead to cartilage structural damage or synovitis. This phenomenon occurs because under physiological conditions, the excessive expression of HNRNPD does not cause normal cartilage degeneration, especially in terms of morphology. In normal cartilage tissue, the genetic modification of HNRNPD in rat articular cartilage can promote the senescence phenotype of chondrocytes, and the chondrocytes are in a state of mitosis obstruction, which is an abnormal function of chondrocytes at the microscopic level, but does not cause pathological changes, such as articular cartilage degeneration at the macroscopic level. Compared with OA model group, HNRNPD overexpression induced by rAAV9 injection did not cause morphological damage to normal cartilage, indicating that HNRNPD overexpression in normal cartilage tissue may lead to sub-health of articular cartilage tissue, and further development may lead to abnormal cartilage morphology and OA, which requires longer observation. However, we found subchondral bone loss, which indicates that the early effects of rAAV on bone and joint were concentrated in subchondral bone. The cartilage was protected by a dense cartilage matrix and did not show significant morphological damage. The subchondral bone showed lacunar trabecular structure and was prone to morphological changes. This also fully reflects that the pathological changes in the process of rAAV acting on normal knee joints first occur in subchondral bone, and then affects the characteristics of articular cartilage. Meanwhile, the proteins related to catabolism and senescence phenotype in the sham + rAAV group were higher than those in the sham group, suggesting that the injection of rAAV into normal joints could induce acceleration of cartilage catabolism and senescence phenotype. The morphological changes of cartilage were not only reflected in the cellular and molecular level but more importantly affected the integrity of cartilage matrix, so the cellular level changes in cartilage tissue were earlier than the morphological changes.
Further studies showed that HNRNPD overexpression not only caused senescence of cartilage tissue but also accelerated degeneration of cartilage tissue in rats with DMM-induced OA model. What is the progression of cartilage degeneration after HNRNPD overexpression under simulated pathological conditions? We found that the direct delivery of rAAV9-HNRNPD into the articular cartilage in the DMM-induced rat OA model enhanced HNRNPD expression and significantly increased the modified Mankin scores compared to those of OA model animals. With the implantation of HNRNPD genes, the pathological degree of chondrocyte senescence was further accelerated, presenting a gradually worsening cascade of damaging effects and presenting more severe morphological abnormalities in the articular cartilage structure. The differences in the functional effects of HNRNPD in the physiological state and pathological manifestations of articular cartilage are based on the scope of the timeline. If beyond the timeline, HNRNPD will also have a positive promoting effect on the morphological impact of cartilage in the physiological state. These results indicate that HNRNPD overexpression has pathological effects on cartilage loss that occurs in preexisting OA disease conditions. However, the morphological effects of HNRNPD on the physiological state of articular cartilage require further observation.
Because HNRNPD can regulate the degradation of target mRNA and then regulate the biological behavior of cells, this study aims to explore the mechanism of OA cartilage injury. After HNRNPD overexpression, a series of genes were found to undergo significant changes, indicating that HNRNPD is involved in the transcriptional regulation of many genes. Using GO/KEGG and other bioinformatics analyses, it was found that HNRNPD is related to the NOD-like receptor signaling pathway, the NF-κB signaling pathway, and the PI3K-Akt signaling pathway in the process of cell senescence. Notably, the two regulated genes, FOXM1 and Smad2, have been closely associated with cellular senescence in OA. We found that FOXM1 is a direct target of HNRNPD in chondrocytes. Of the mRNAs altered by the pcDNA3.1-HNRNPD plasmid-mediated overexpression of HNRNPD in chondrocytes, the mRNAs encoding FOXM1 showed marked increases. FOXM1 is upregulated in osteoarthritic cartilage compared with normal cartilage. Although accumulating evidence indicates that FOXM1 exerts regulatory effects on cellular senescence34, previously little was known about the upstream signaling molecules responsible for regulating FOXM1 expression. This study will lay a foundation for elucidating the regulatory effect of FOXM1 on the function of chondrocytes.
Many studies have confirmed that mitochondria play an important role in the occurrence and development of OA35. Analysis of mitochondrial function in osteoarthritic chondrocytes showed decreased activity of mitochondrial respiratory complexes II and III and increased mitochondrial mass compared to healthy chondrocytes36. In OA, mitochondrial damage is an important cause of cartilage degeneration37,38. Studies have shown that abnormal mitochondrial aggregation is closely related to aging and senile diseases39. Mitochondrial dynamics play an important role in the natural aging process, senescence and aging, and potentially age-related diseases40,41. HNRNPD plays a key role in the maintenance of mitochondrial homeostasis42,43. To further elucidate the mitochondrial changes corresponding to HNRNPD overexpression in OA, we used plasmid-transfected chondrocytes to analyze mitochondrial function after HNRNPD overexpression. Our study revealed a loss of mitochondrial membrane potential, mitochondrial fragmentation, and disruption of mitochondrial dynamics. Consistent with the functional decrease in the mitochondrial membrane potential, the amount of ATP production also decreased with HNRNPD overexpression. Our study showed that HNRNPD promotes chondrocyte senescence, characterized by mitochondrial dysfunction in chondrocytes, morphological swelling of mitochondria, matrix deformation, and loss of mitochondrial cristae in synovial fibroblasts. FOXM1 can be increased by HNRNPD as a downstream target gene of HNRNPD, synergistically promoting cellular senescence. HNRNPD targets the regulation of FOXM1-mediated chondrocyte senescence, which is caused by promoting mitochondrial dysfunction, causing mitochondrial membrane potential and morphological abnormalities in both in vivo and in vitro, ultimately leading to sustained degeneration of articular cartilage. HNRNPD, as a key and important RNA binding protein in the pathogenesis and progression of OA, transcriptionally regulates FOXM1-mediated mitochondrial dysfunction, metabolic abnormalities, and the senescent phenotype, which is an important mechanism by which HNRNPD promotes the occurrence and development of OA. We found that HNRNPD affects the mitochondrial metabolism of OA through the FOXM1 signaling pathway, which in turn affects the senescence process of OA.
This study lays a theoretical foundation for elucidating the molecular mechanism by which HNRNPD participates in the occurrence and development of OA. HNRNPD carries out these functions by regulating mitochondrial homeostasis and promoting senescent phenotypes in a FOXM1-dependent manner (Fig. 9). In terms of mechanism, activation of HNRNPD is driven by FOXM1 in mitochondrial dysfunction, such as mitochondrial membrane potential and mitochondrial morphology abnormality, exacerbating IL-1β-induced chondrocyte senescence. Therefore, HNRNPD promotes IL-1β-induced cell senescence by activating FOXM1 and inhibiting HNRNPD may be an attractive strategy for preventing or treating OA. This study reveals a new mechanism by which HNRNPD regulates the occurrence and development of OA and lays a theoretical foundation for exploring new targets for the prevention and treatment of OA.
HNRNPD promotes chondrocyte senescence by upregulating FOXM1 and inhibiting HNRNPD may be an attractive strategy for preventing of treating OA (scale bar: 120 µm). Reprinted with the permission from ref. 47. Copyright Elsevier.
Materials and methods
Reagents and antibodies
FOXM1 antibody (#48295) was obtained from Signalway Antibody, Maryland, USA. Antibodies HNRNPD (#ab259895) and type II collagen (Col II, #ab307674) were obtained from Abcam (Cambridge, UK). Anti-P53 (#21891-1-ap), TOM20 (#11802-1-ap), and P16INK4A (#10883-1-ap) were purchased from Proteintech Group, Inc (Wuhan, China). β-tubulin (#T0023) was obtained from Affinity Biosciences (Cincinnati, OH, USA). Mitochondrial membrane potential assay kit with Rhodamine-123 and JC-1, MitoTracker Red CMXRos, and SA-β-gal assay kit were purchased from Beyotime Institute of Biotechnology, China. Fetal bovine serum (FBS) and penicillin/streptomycin were obtained from Gibco BRL (Maryland, USA). All other reagents were of analytical grade.
Human samples
Human OA cartilages were obtained from patients undergoing total knee arthroplasty (n = 5). Normal control cartilages were obtained from traffic accident patients with no history of arthritis (n = 5). Cartilages were excised from the tibial plateau and femoral condyles during total knee replacement surgery. Cartilage samples were fixed in 4% paraformaldehyde, decalcified in 15% EDTA, and paraffin-embedded for further histological analysis.
All human samples were obtained from Renmin Hospital of Wuhan University, Wuhan, China. According to Kellgren & Lawrence grading criteria, patients with OA belong to grade III-IV. This study was approved by the Ethics Committee of Renmin Hospital of Wuhan University (2022K-K228).
RNA-sequencing and bioinformatics analysis
RNA samples were used for RNA sequencing to ensure a sufficient amount of RNA and to reduce the heterogeneity. IlluminaHiSeq4000 was used in this portion of the study to conduct RNA sequencing. Six transcriptome sequencing samples (HNRNPD_1st, HNRNPD_2nd, HNRNPD_3rd, Ctrl_1st, Ctrl_2nd, and Ctrl_3rd) were obtained. The FPKM was used to refer to the expression of a given gene. The RNA marker was confirmed by an Agilent2100 bioanalyzer (LABX, Midland, Canada). For the determination of the RNA sequence, the template used was “RNA compatibility number” 6.5. Through the determination of the gene expression profile, low-quality environmental contaminants and high-pollution substances containing a large amount of base (N) noise were screened. Afterward, the profile was compared with the reading value of the reference gene (NCBI Rnor6.0) by using Bowtie244. Then, the differentially expressed genes were screened by DEseq2. MeV (https://sourceforge.net/projects/mev-tm4/) was used to establish a heatmap to evaluate the DEG levels. For determination of the gene functions, DEGs were used as inputs to the GO and KEGG databases.
Animal experiment
Male Sprague-Dawley (SD) rats (weight: 160–180 g) were obtained from the Hunan SJA Laboratory Animal Co., Ltd., Changsha, China. All experiments were approved by the Animal Care and Use Committee of the Medical School at Wuhan University. The experimental rats were placed in a routine environment for one week and randomly divided into four groups with 6 rats in each group. These groups were as follows: a sham operation group, a rAAV-HNRNPD + sham group, an OA induction group, and an OA + rAAV-HNRNPD group. The rats were anesthetized by inhalation of isoflurane oxygen mixture (2–5% isoflurane in 100% oxygen). OA was induced in the right knee joint of rats by transecting the DMM45. For the rAAV-HNRNPD group, the animals received a weekly IA injection into the knee with rAAV encoding a sequence-specific for HNRNPD (Shanghai Genechem Co., Ltd, 1 × 1010 drp/30 μl/joint) at 4 weeks postoperation. The sham-operated and OA-induced control groups received an injection of 30 μl of phosphate-buffered saline (PBS) into the right knee joint. At 10 weeks after surgery, the animals were euthanized by cardiac bloodletting, and samples were taken for further testing.
In vivo micro-MRI
The right knee joint of each rat was evaluated with 9.4 T high magnetic field micro-MRI (BioSpec 70/30 USR, Germany). Fat suppression, spin-echo, and T2-weighted scan modes were used to observe the possibility of bone marrow lesion-like/edema-like phenomena. The sagittal position of the medial condyle of the femur was selected from each group, and ImageJ software (National Institutes of Health, Bethesda, MD) was used for quantitative analysis of water signals in these bones.
Micro-CT examination
Images of the knee joint were collected by micro-CT (Bruker MicroCT N.V., Skyscan1176) provided by Kontich, Belgium, with 4000 × 2672 pixels and 9 μm voxel size in the same direction. Through the comprehensive evaluation of the image, the BV/TV, the Tb. Th and the Tb. Sp were measured by a three-dimensional morphological technique.
TEM
The 2 mm3 synovial sample came from the iliac ligament and was fixed with 2.5% glutaraldehyde 0.1 M PBS solution of pH 7.3. The sample was washed with PBS, postfixed at 4 °C with 1% osmium tetroxide, washed with dH2O after 3 h, dried with different concentrations of alcohol, and then encapsulated with epoxy resin. It was stained twice by H-600 IV TEM (Hitachi, Japan), and the mitochondria and lysosomes of the synovial fibroblasts in the surface layer of the synovial tissue were observed.
Histological analysis
The knee joint of each rat was removed and fixed with 4% paraformaldehyde for 24 h, followed by decalcification for 6 weeks, and then was embedded in paraffin. Sagittal sections (5 µm) were prepared, and H&E and Alcian blue staining were used. The two observers conducted their observations double-blinded, and the modified Mankin scoring system46 was used for the pathophysiological study. Moreover, the articular cartilage of the rats was observed by the Safranin O green fixation method.
Immunohistochemistry and immunofluorescence assay
The different concentrations of streptavidin-peroxidase were determined by immunohistochemical techniques. In short, these tissues were treated with a moderate resistance and then observed under a light microscope. The proportion of positive cells was detected by immunohistochemistry method. Immunofluorescence-positive cell ratios of HNRNPD, P16INK4A, P21, MMP3, Col II, P53, and FOXM1 were assessed using Image-Pro Plus 6.0 (Media Cybernetics Co., USA). The fluorescent-conjugated secondary antibody was cultured in the dark for 1 h and then detected by fluorescence microscopy (AX10, Carl Zeiss).
Isolation and culture of cells
The tissue culture technique of primary knee joint cartilage from the SD rats in a 5-day growth period was used. This procedure was carried out by collecting slices that were washed three times with PBS and cut into thin slices of 0.5–1 mm3. The cells were then digested with 0.25% trypsin and 0.02% EDTA for 1 h, transferred to a culture plate containing 0.2% type II collagenase, incubated at 37 °C for 4–5 h, centrifuged for 5 min at 1000 rpm, and finally added to complete DMEM/F12 (containing 10% fetal bovine serum and penicillin/streptomycin). The third passage chondrocytes were cultured in a 37 °C wet 5% CO2 incubator and used in the following cell experiment study.
Synthesis of siRNA and cell stimulation
Dharacon Accell siRNA and chaos control for FOXM1 (OBio Technology, Shanghai) were adopted in this study. The siRNAs had the following-sequences: 5′- CCUCAGAGGAAGAAGCAAAGATT-3′ (sense) and 3′- UCUUUGCUUCUUCCUCUGAGGTT-5′ (antisense). For preparation, the concentration of siRNAs was 100 nm. In complete medium containing 10 μg/mL HNRNPD plasmids (Shanghai Genechem Co., Ltd) and 100 nM FOXM1 siRNA, third-generation chondrocytes were cultured for 24 h, and then 10 ng/mL IL-1β was added for 24 h.
Co-IP assay
The regulatory effect of HNRNPD on FOXM1 protein was analyzed by Co-IP. Chondrocytes were dissolved in a precooled buffer containing protease inhibitors, and incubated with anti-HNRNPD and anti-FOXM1 antibodies at 4 °C overnight. They were then combined with protein A Sepharose, cultured for 1 h at 4 °C, and rinsed with water three times. Then, western blotting was performed.
RIP
RIP assay was performed by adopting the Magna RIP kit (Sigma-Aldrich) following the manufacturer’s protocols. RIP was used to verify the binding relationship between HNRNPD and FOXM1. Whole-cell lysates were incubated overnight at 4 °C with RIP buffer containing magnetic beads (GenScrip, NJ, USA) coupled with anti-Ago2 antibody (Proteintech, Wuhan, China) or negative control immunoglobulin G. The beads were washed, and RNA was extracted. Co-precipitated RNA was evaluated by qRT-PCR.
SA-β-gal assay staining
After 24 h of treatment, the chondrocytes were fixed with 4% formaldehyde (Rich Joint, Shanghai) to assess cell vitality. SA-β-gal staining assays using an SA-β-gal staining kit were performed according to the protocol issued by the manufacturer.
Immunofluorescence method
An in vitro culture system with 6 pores was used to achieve a fusion rate of 70%, and serum protein starvation was carried out. Then, the cells were fixed with 4% paraformaldehyde for 20 min, impregnated for 5 min, and blocked with 1% BSA for 10 min. The chondrocytes were incubated with monoclonal antibodies against HNRNPD and FOXM1 for 2 h. The chondrocytes were redeveloped and incubated with secondary antibody (Bost Bioengineering Company, Wuhan, China) for 1 h. 4’,6-Diamidino-2-phenylindole (DAPI) was used for nuclear staining, and a fluorescence microscope (AX10, Carl Zeiss) was used for imaging.
Mitochondrial membrane potential assay
A mitochondrial membrane potential analysis kit was used according to the manufacturer’s instructions. The chondrocytes were stained with 10 µM rhodamine-123 or 800 μL JC-1 working solution at 37 °C for 25 min. Then, after removing the staining solution, 2 mL DMEM containing 10% FBS was added to the chondrocytes. A fluorescence microscope (Olympus, Japan) was used to observe the cells, and the fluorescence intensity and the ratio of JC-1 fluorescence were measured.
MitoTracker Red staining
The chondrocytes were cultured with 50 nM MitoTracker Red at 37 °C for 20 min. DAPI was used as a fluorescence quenching agent, and the cells were fixed on 4% paraformaldehyde for 20 min. The morphology of the podocyte mitochondria was observed by laser scanning electron microscopy (Olympus, Japan). The length ratio of mitochondria was measured by using the Image-Pro Plus 6.0 quantitative analysis system (Bio-Rad Laboratories, USA).
Western blotting
A whole protein extraction kit was used to separate proteins from chondrocytes according to the manufacturer’s instructions. The protein content was determined by a biological colorimetric protein test (Bio-Rad Laboratories, Inc., Hercules, CA). Approximately 30 μg of protein was analyzed by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS‒PAGE, Sigma‒Aldrich). The separated protein was transferred onto a film, which was blocked with 5% skim milk and treated at room temperature for 1 h. Antibodies against HNRNPD (1:1000), P21 (1:1000), P16INK4A (1:1000), P53 (1:5000), Bcl-2 (1:1000), FOXM1 (1:1000), and TOM20 (1:5000) were used for 12 h. The cell membrane was probed with anti-tubulin, which served as a loading control.
Molecular docking
FOXM1 (PDB: 7FJ2, resolution: 3.098 Å) and HNRNPD (PDB: 5IM0, resolution 1.7 Å) were downloaded from the RCSB protein database (www.rcsb.org/). The X-ray crystallization protein-protein simulation was carried out by using the ZDOCK3.0.2 program. For the combination of HNRNPD and FOXM1, two docking positions were selected. HNRNPD was calculated by using the Tripos force field and the Gestager-Hackl charge spectrum. The docking simulation of HNRNPD and FOXM1 was carried out by using Sybyl software. Finally, the 3D view was graphically processed by UCSFPyMoL.
Statistical analysis
All trials were conducted with specimens provided by 3–5 donors. Significant differences between groups were determined by the t-test. A one-way ANOVA and postoperative Bonferroni adjustment were used to analyze three or more groups. The results are expressed as the mean ± standard deviation (SD). There were significant differences among all indexes (P < 0.05).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
DEGs annotated to GO and KEGG databases are provided in the Supplementary Fig. 1. Uncropped blot images are provided as Supplementary Fig. 2 in the Supplementary Information. The numerical source data to all graphs are provided in the Supplementary Data 1 file. The RNA sequencing and bioinformatics analysis datasets provided in this study are available at NCBI-Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra/). The accession code for deposited data is PRJNA1193849.
References
Sanchez-Lopez, E., Coras, R., Torres, A., Lane, N. E. & Guma, M. Synovial inflammation in osteoarthritis progression. Nat. Rev. Rheumatol. 18, 258–275 (2022).
Yue, L. & Berman, J. What is osteoarthritis? Jama 327, 1300 (2022).
Katz, J. N., Arant, K. R. & Loeser, R. F. Diagnosis and treatment of hip and knee osteoarthritis: a review. Jama 325, 568–578 (2021).
Wakale, S. et al. How are aging and osteoarthritis related? Aging Dis. 14, 592–604 (2023).
Sun, K., Jing, X., Guo, J., Yao, X. & Guo, F. Mitophagy in degenerative joint diseases. Autophagy 17, 2082–2092 (2021).
Coryell, P. R., Diekman, B. O. & Loeser, R. F. Mechanisms and therapeutic implications of cellular senescence in osteoarthritis. Nat. Rev. Rheumatol. 17, 47–57 (2021).
Liu, Y., Zhang, Z., Li, T., Xu, H. & Zhang, H. Senescence in osteoarthritis: from mechanism to potential treatment. Arthritis Res. Ther. 24, 174 (2022).
Chen, X. et al. METTL3-mediated m(6)A modification of ATG7 regulates autophagy-GATA4 axis to promote cellular senescence and osteoarthritis progression. Ann. Rheum. Dis. 81, 87–99 (2022).
Birch, J. & Gil, J. Senescence and the SASP: many therapeutic avenues. Genes Dev. 34, 1565–1576 (2020).
Roh, K. et al. Lysosomal control of senescence and inflammation through cholesterol partitioning. Nat. Metab. 5, 398–413 (2023).
Arra, M., Swarnkar, G., Alippe, Y., Mbalaviele, G. & Abu-Amer, Y. IκB-ζ signaling promotes chondrocyte inflammatory phenotype, senescence, and erosive joint pathology. Bone Res. 10, 12 (2022).
Ko, P. Y. et al. Cross-linked hyaluronate and corticosteroid combination ameliorate the rat experimental tendinopathy through anti-senescent and -apoptotic effects. Int. J. Mol. Sci. 23, 9760 (2022).
Gebauer, F., Schwarzl, T., Valcárcel, J. & Hentze, M. W. RNA-binding proteins in human genetic disease. Nat. Rev. Genet. 22, 185–198 (2021).
Corley, M., Burns, M. C. & Yeo, G. W. How RNA-binding proteins interact with RNA: molecules and mechanisms. Mol. Cell 78, 9–29 (2020).
Webster, M. W., Stowell, J. A. & Passmore, L. A. RNA-binding proteins distinguish between similar sequence motifs to promote targeted deadenylation by Ccr4-Not. eLife 8, e40670 (2019).
Assabban, A. et al. Tristetraprolin expression by keratinocytes protects against skin carcinogenesis. JCI insight 6, e140669 (2021).
Abbadi, D., Yang, M., Chenette, D. M., Andrews, J. J. & Schneider, R. J. Muscle development and regeneration controlled by AUF1-mediated stage-specific degradation of fate-determining checkpoint mRNAs. Proc. Natl Acad. Sci. USA 116, 11285–11290 (2019).
Zhang, Q. et al. Nuclear speckle specific hnRNP D-like prevents age- and AD-related cognitive decline by modulating RNA splicing. Mol. Neurodegener. 16, 66 (2021).
Tsitsipatis, D. et al. AUF1 ligand circPCNX reduces cell proliferation by competing with p21 mRNA to increase p21 production. Nucleic Acids Res. 49, 1631–1646 (2021).
Yu, D. et al. Role of AUF1 in modulating the proliferation, migration and senescence of skin cells. Exp. Ther. Med. 23, 45 (2022).
He, J. et al. Targeting of AUF1 to vascular endothelial cells as a novel anti-aging therapy. J. Geriatr. Cardiol. JGC 14, 515–523 (2017).
Salvato, I. et al. Expression of targets of the RNA-binding protein AUF-1 in human airway epithelium indicates its role in cellular senescence and inflammation. Front. Immunol. 14, 1192028 (2023).
Wei, Z. et al. CircUBE3A(2,3,4,5) promotes adenylate-uridylate-rich binding factor 1 nuclear translocation to suppress prostate cancer metastasis. Cancer Lett. 588, 216743 (2024).
Lu, J. et al. N(6) -methyladenosine-modified circSTX6 promotes hepatocellular carcinoma progression by regulating the HNRNPD/ATF3 axis and encoding a 144 amino acid polypeptide. Clin. Transl. Med. 13, e1451 (2023).
Jiang, H. et al. Innate/inflammatory bioregulation of surfactant protein D alleviates rat osteoarthritis by inhibiting toll-like receptor 4 signaling. Front. Immunol. 13, 913901 (2022).
Zhou, Y. et al. Berberine-mediated up-regulation of surfactant protein D facilitates cartilage repair by modulating immune responses via the inhibition of TLR4/NF-ĸB signaling. Pharmacol. Res. 155, 104690 (2020).
Han, Y. M. et al. β-hydroxybutyrate prevents vascular senescence through hnRNP A1-mediated upregulation of Oct4. Mol. cell 71, 1064–1078 e1065 (2018).
Bampton, A. et al. HnRNP K mislocalisation is a novel protein pathology of frontotemporal lobar degeneration and ageing and leads to cryptic splicing. Acta Neuropathol. 142, 609–627 (2021).
Ching, K., Houard, X., Berenbaum, F. & Wen, C. Hypertension meets osteoarthritis—revisiting the vascular aetiology hypothesis. Nat. Rev. Rheumatol. 17, 533–549 (2021).
Yao, Q. et al. Osteoarthritis: pathogenic signaling pathways and therapeutic targets. Signal Transduct. Target. Ther. 8, 56 (2023).
Son, Y. O., Kim, H. E., Choi, W. S., Chun, C. H. & Chun, J. S. RNA-binding protein ZFP36L1 regulates osteoarthritis by modulating members of the heat shock protein 70 family. Nat. Commun. 10, 77 (2019).
Choi, W. S. et al. The CH25H-CYP7B1-RORα axis of cholesterol metabolism regulates osteoarthritis. Nature 566, 254–258 (2019).
Schrenker, S. et al. In vivo rAAV-mediated human TGF-β overexpression reduces perifocal osteoarthritis and improves osteochondral repair in a large animal model at one year. Osteoarthr. Cartil. 31, 467–481 (2023).
Xiang, X. et al. Cellular senescence in hepatocellular carcinoma induced by a long non-coding RNA-encoded peptide PINT87aa by blocking FOXM1-mediated PHB2. Theranostics 11, 4929–4944 (2021).
Liu, D. et al. Mitochondrial quality control in cartilage damage and osteoarthritis: new insights and potential therapeutic targets. Osteoarthr. Cartil. 30, 395–405 (2022).
Zhang, Y. et al. Reprogramming of mitochondrial respiratory chain complex by targeting SIRT3-COX4I2 axis attenuates osteoarthritis progression. Adv. Sci. 10, e2206144 (2023).
Jiang, W. et al. Mechanisms linking mitochondrial mechanotransduction and chondrocyte biology in the pathogenesis of osteoarthritis. Ageing Res. Rev. 67, 101315 (2021).
Zhu, S. et al. Sirt3 promotes chondrogenesis, chondrocyte mitochondrial respiration and the development of high-fat diet-induced osteoarthritis in mice. J. Bone Miner. Res. Off. J. Am. Soc. Bone Miner. Res. 37, 2531–2547 (2022).
D’Amico, D. et al. Urolithin A improves mitochondrial health, reduces cartilage degeneration, and alleviates pain in osteoarthritis. Aging cell 21, e13662 (2022).
Yu, B. et al. Mitochondrial phosphatase PGAM5 modulates cellular senescence by regulating mitochondrial dynamics. Nat. Commun. 11, 2549 (2020).
Campos, J. C. et al. Exercise preserves physical fitness during aging through AMPK and mitochondrial dynamics. Proc. Natl Acad. Sci. USA 120, e2204750120 (2023).
Latorre, E., Torregrossa, R., Wood, M. E., Whiteman, M. & Harries, L. W. Mitochondria-targeted hydrogen sulfide attenuates endothelial senescence by selective induction of splicing factors HNRNPD and SRSF2. Aging 10, 1666–1681 (2018).
Abbadi, D., Andrews, J. J., Katsara, O. & Schneider, R. J. AUF1 gene transfer increases exercise performance and improves skeletal muscle deficit in adult mice. Mol. Ther. Methods Clin. Dev. 22, 222–236 (2021).
Gómez, R., Villalvilla, A., Largo, R., Gualillo, O. & Herrero-Beaumont, G. TLR4 signalling in osteoarthritis-finding targets for candidate DMOADs. Nat. Rev. Rheumatol. 11, 159–170 (2015).
Zhang, L. et al. Characteristics of sensory innervation in synovium of rats within different knee osteoarthritis models and the correlation between synovial fibrosis and hyperalgesia. J. Adv. Res. 35, 141–151 (2022).
Han, B. et al. Differentiated activities of decorin and biglycan in the progression of post-traumatic osteoarthritis. Osteoarthr. Cartil. 29, 1181–1192 (2021).
Hodgkinson, T., Kelly, D. C., Curtin, C. M. & O’Brien, F. J. Mechanosignalling in cartilage: an emerging target for the treatment of osteoarthritis. Nat. Rev. Rheumatol. 18, 67–84 (2022).
Acknowledgements
This project was funded by National Natural Science Foundation of China (grant number: 82272528) and Natural Science Foundation of Hubei Province (grant number: 2022CFB117).
Author information
Authors and Affiliations
Contributions
Conceptualization and writing-original draft: H.J., YB.Z., G.H., and P.J.; Data curation, formal analysis, and visualization: J.M. and Y.L.; Methodology, project administration, and writing-review & editing: Y.Z. All authors reviewed the results and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Ethics statement
This study was approved by the Animal Care and Use Committee of Medical School, Wuhan University (WDRM20220603C) and the Ethics Committee of Renmin Hospital of Wuhan University (2022K-K228).
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editors: Martina Rauner and Joao Valente.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Jiang, H., Zhang, Y., Hu, G. et al. RNA-binding protein HNRNPD promotes chondrocyte senescence and osteoarthritis progression through upregulating FOXM1. Commun Biol 7, 1695 (2024). https://doi.org/10.1038/s42003-024-07407-8
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s42003-024-07407-8