Abstract
Lipid nanoparticles (LNPs) have gained much attention after the recent launch of mRNA based Covid vaccines. For mRNA encapsulated within LNPs to be successfully translated into target proteins, the mRNA must maintain its integrity and also be free of any unintended chemical modifications. Any process or raw material related impurities—and their degradation products—pose risks of chemically modifying mRNA, thereby affecting the quality of the final product. Given its inherent chemical reactivity and close association with the LNP lipids, encapsulated mRNA is especially susceptible to modifications by reactive impurity species present in the lipids. In our recent efforts to understand mRNA-lipid interactions within LNPs, we observed that the degradants of labile lipid peroxide species in ionizable lipids react with nucleotides in mRNA, resulting in loss of mRNA’s translation efficiency in vitro. Specifically, we identified peroxide species in unsaturated dialkene groups were converted to a variety of reactive aldehyde products in mRNA LNP formulations. These findings enhance the current understanding of the adduct formation between mRNA and aldehyde species, and emphasize the critical role of deep analytical characterization of ionizable lipid stability and purity to enhance LNP product quality and shelf-life.
Similar content being viewed by others
Introduction
Remarkable advancements have been made in the development of LNPs for nucleic acid delivery. While having been successfully utilized for the transport of siRNA therapeutics, LNP technology has only recently been adopted in global usage for mRNA delivery during the COVID pandemic1. LNPs are formed by encapsulating gene cargo such as mRNA in a lipid mixture, which serves as the vehicle for cellular uptake and enables functional delivery of its cargo2. Ionizable lipids are essential components of LNPs; the design of these synthetic lipids is critical to tuning the LNP formulation, and the specific chemical structures are often proprietary. While various ionizable lipid structures and functional groups continue to be explored, generally modern designs incorporate at least one ionizable tertiary amine capable of electrostatic interactions with mRNA in a pH-dependent manner3. This crucial interaction enables the formation of LNPs and their mechanism of escape in acidified endosomes4, but colocalization of mRNA and lipids opens the possibility for reactions such as chemical modifications of mRNA to occur. In natural biological settings, various electrophilic species are recognized for forming modifications in both mRNA and DNA5,6,7,8, with lipid oxidation as a potential source7,8. Previous studies have extensively investigated this phenomenon for RNA in the context of cellular or environmental genotoxic agents6,9, though there are increasing investigations of mRNA lipid interactions in the context of mRNA therapeutics10,11,12,13.
As LNP technology continues to improve, applications are being sought requiring the delivery of longer transcripts, often longer than 10 kb, such as self-amplifying mRNA (sa-mRNA). sa-mRNA technology is a recent breakthrough for vaccine development14,15,16; unlike conventional mRNA, sa-mRNA encodes information for non-structural proteins (NSP) 1-4, which, when translated in situ, form the mRNA-dependent mRNA polymerase (RdRP) complex that serves to amplify its own transcript17. This unique feature allows sa-mRNA to achieve higher and more sustained activity with lower doses18. However, longer mRNA transcripts present increased challenges in maintaining their stability. For an LNP formulation where lipid concentrations and N/P ratios are conserved, the LNP encapsulating a longer sa-mRNA construct will have a lower number of mRNA copies per particle than one containing a shorter conventional mRNA construct. Consequently, inactivation of a single copy of a longer mRNA construct by lipid adduction or degradation can exert a greater adverse effect on compound efficacy. This increased size poses new challenges in the manufacturing of drug substances and drug products. To ensure a high-performing drug product, stringent raw material specifications informed by a thorough evaluation of reactive species and their sources are essential.
Among quality attributes, intact mRNA purity by integrity assessment is traditionally understood to be the key critical indicator for LNP stability. Recently, lipid modification of encapsulated mRNA has been shown to have a notable effect on LNP potency during storage, independent of integrity10,12. The ultra-low storage temperature required to extend LNP shelf-life poses practical limitations on their usage and come at considerable cost19. Improvement in the thermostability profile of LNPs would be an important step in reducing the cost of therapeutics while expanding their utility in different areas.
We observed unexpected potency loss independent of mRNA integrity or other quality attributes and performed an extensive investigation to characterize the phenomena compromising stability. Ion Pairing Reverse Phase chromatography (IP-RP) of extracted mRNA was a key tool to distinguish the presence and degree of the lipid adduction. Liquid Chromatography Mass Spectrometry (LC-MS) was employed to precisely elucidate chemical identities and structures of the culprit species and of adduct delta masses. These techniques underscore the importance of deep analytical characterization to deduce mechanisms of lipid adduction. The results of the investigation illustrate how oxidative stress can impact the stability of LNPs via the formation of reactive aldehydes by oxidation of unsaturated lipid groups, highlighting the need for strict lipid quality control during mRNA vaccine product development.
Results
Detection of mRNA lipid adduction in LNPs
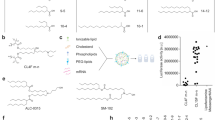
We wanted to characterize sa-mRNA encapsulated in LNPs by an LC method. We screened LNPs consisting of ~11 kb sa-mRNA, proprietary ionizable lipid, cholesterol, a phospholipid, and a polyethylene glycol (PEG) lipid, by analyzing the extracted mRNA with ion-pairing reverse phase LC-UV. Notably, for some LNPs, a distinct additional late eluting peak well resolved from the intact sa-mRNA peak was observed (Fig. 1a); the later retention time suggested a hydrophobic or adducted mRNA species. The relative area of this peak varied among different LNPs, though it was observed that certain families of ionizable lipids, with unsaturated tails, exhibited a more prominent additional peak (Supplementary Fig. 1).
a Ion Pairing-Reverse Phase (IP-RP) liquid chromatography was used to identify unadducted main peak and late eluting adducted sa-mRNA peaks using a UV detector @260 nm from a mixture of IL and sa-mRNA incubated 24 h at ambient temperature. b Analysis of LNP stability at 2–8 °C, 20 °C, and –80 °C shows mRNA lipid adduction increases over the period of 9 months. c sa-mRNA was mixed with either IL, cholesterol, a phospholipid, a PEG lipid, or with a combination of IL with one or all other lipids and lipid adduction was only observed in IL-containing mixtures. d Two lots of IL showed differential lipid adduction over a 48 h time course. e Stacked MS analysis of the two IL lots showed greater abundance of an early eluting species in lot 2, which produced more adducted mRNA.
To evaluate the effect of storage temperature, LNPs made with IL, a specific proprietary ionizable lipid with unsaturated tails, were stored at various temperatures (2–8 °C, −20 °C, and −80 °C), and the relative percent area of the late peak (LP%) was calculated from the integrated peak areas. The relative area of the additional peak increased with increasing storage temperature, with a single 2–8 °C timepoint showing a substantial increase in % area over the frozen storage temperatures (Fig. 1b), indicating potential thermosensitive mRNA modifications.
The presence of a late peak occurs in IL-formulated sa-mRNA regardless of the mRNA construct sequence (Supplementary Fig. 1), suggesting a general broader issue in the mRNA-LNP system. Additionally, an increase in the relative area of this additional peak corresponded with a more pronounced decrease in protein expression than mRNA integrity as measured by capillary electrophoresis (CE) (Supplementary Fig. 2). The result of this initial exploratory study suggested potential mRNA lipid adduction affecting mRNA function independently of integrity and led to further investigations with a goal to mitigate mRNA modifications by controlling reactive species present in the LNP.
Screening of lipids in mRNA lipid adduction
To confirm that the ionizable lipid, and not other lipid components of the LNP, was responsible for mRNA lipid adduction, mixtures of sa-mRNA were made with each lipid component and combinations of IL plus lipid components at process-relevant ratios as described (Fig. 1c). Only mixtures that included sa-mRNA and IL produced a late eluting peak. Two-lipid component mixtures had similar lipid adduction levels compared to the IL control mixture. A mixture of all four lipid components with mRNA resulted in the formation of slightly less % lipid adduction, possibly due to dilution of the ionizable lipid interaction with mRNA. Additionally, we emphasize that this peak was observed when different sa-mRNA constructs were used in LNPs or a lipid and mRNA mixture (Supplementary Fig. 1), implicating IL in lipid adduction. Since IL itself is not supposed to modify mRNAs, these findings suggest that unexpected species in IL are solely responsible for forming lipid modifications on mRNA in the LNP formulation or in the simplified ionizable lipid and mRNA mixture.
Evaluation of ionizable lipid species related to lipid adduction
To better understand the profile of species in IL as they relate to lipid adduction of mRNA, we incubated sa-mRNA with two IL lots with differing manufacturing grades. Each lot was mixed with sa-mRNA using a microfluidic mixer and aliquoted for a 48 h time course at ambient temperature. mRNA was extracted at 2, 4, 24, and 48 h timepoints and stored at −80 °C. After all timepoints were collected, mRNA was analyzed by IP-RP. IL lot 2 yielded over 2 times more mRNA lipid adduction than IL lot 1 (Fig. 1d). Interestingly, mRNA lipid adduction was rapid in the first 4 h and plateaued approaching 48 h, suggesting potential reactive species were consumed at a faster rate than their formation.
LC-MS analysis was performed to identify the species that might differ in these two IL lots. Comparison of the total ion chromatograms (TIC) revealed the intact IL peak as well as the species common to both lots at various abundances (Fig. 1e). IL was eluted at 9.7 min, with an early eluting peak at 7.7 min composed mainly of +32 Da delta mass species along with lesser MS signal of +14 Da and +16 Da co-eluting species (Supplementary Fig. 3a, b). MS2 fragmentation was performed to investigate the structure and characteristic fragmentation of these lipid species (Supplementary Fig. 4). The +32 Da and +14 Da species were unique masses; for this early eluting +16 Da species, MS2 analysis showed an absence of a characteristic non-tail +16 Da fragment suggesting this species was oxidized on the tail(s) (Supplementary Fig. 5a), in contrast to other +16 Da species.
To further explore which candidate species correlated with lipid adduction, additional lots of IL (lot 3 and lot 4) were investigated, with the corresponding peak profiles exhibiting a major difference in the early eluting peak (Fig. 2a). These lots all passed the release testing for purity by Charged Aerosol Detection (CAD) with comparable purity values of 97.0%, 97.7%, 98.8%, 98.7% for the IL lots 1–4 respectively, were stored at −80 °C without solvents, and there was no noteworthy time-delay in assessment. Still, in-house MS analysis revealed differences in the abundance of additional species, indicating a potential analytical gap should be bridged to identify the species of interest. With LC-MS, we investigated the relative abundance of these potential oxide species and each of the extracted ion chromatograms (XIC) was compared to the % lipid adduction observed in extracted sa-mRNA from mixtures of sa-mRNA and each respective IL lot. The relative abundance of +32 Da species XIC (Supplementary Fig. 3c) directly corresponded to the level of late eluting species for each IL lot (Fig. 2b), indicating that this species was involved in the lipid adduction of mRNA. The levels of +16 Da and +14 Da species in the early eluting peak did not show good correspondence with % lipid adduction (Fig. 2c, d), indicating these species are not directly involved in lipid adduction. Other minor low-abundance species were present but were also not observed to correspond with % lipid adduction. Moreover, we note that it is possible there are other species that are not readily detected by LC-MS. Nevertheless, based on the observation of the salient +32 Da species observed to strongly correlate with lipid adduction for IL lots, we chose to focus the investigation on this species.
LC-MS analysis of the + 32 Da species
The lipid tails of IL resemble a polyunsaturated fatty acid (PUFA) (two double bonds at the C9 and C12 position), which are particularly susceptible to oxidation due to their electron-rich double bonds20. In MS2 analysis of peroxides of PUFA, sodium ions have been shown to enable identification of sites of peroxidation based on characteristic neutral loss products from fragmentation to identify isomers21. Based on this profiling technique, we used LC-MS to analyze individual +32 Da species of IL to search for these neutral loss products. MS2 fragmentation revealed the diagnostic sodium adduct of each of the +32 Da peroxide species with an intact mass of IL + 32 Da species and +23 Da [Na + ] (IL + 32 Da +23 Da = X Da) intact mass (Fig. 3a). Fragmentation of these peaks showed characteristic species corresponding with −140 Da and −88 Da neutral loss products of the X Da sodium adducted peroxide species, respectively, with a high mass accuracy < ±0.5 ppm (Fig. 3a, b).
a MS2 fragmentation analysis of the sodium adduct of the +32 Da species (X Da) shows a characteristic 140 Da fragment of a C9 peroxide species and b an 88 Da fragment of a C13 peroxide species. c IL has tails containing unsaturated dialkene groups that are susceptible to peroxidation at the C9 or C13 position. Dialkene tails convert to form reactive aldehyde species along with various other oxidation byproducts.
With the +32 Da species identified as lipid tail peroxides, it is plausible that these convert to form reactive products that can modify mRNA. Lipid peroxides, such as those occurring in PUFAs, can undergo oxidative lipid degradation that results in the formation of lipid aldehydes by fragmentation of peroxycyclization intermediates or Hock cleavage22,23,24. As IL contains unsaturated lipid tail(s) containing peroxidation sites, a representative reaction is presented with the caveat that multiple additional oxide products and therefore aldehyde species are potentially formed (Fig. 3c).
Evaluation of IL peroxide species spiking
To confirm the involvement of peroxide species in mRNA lipid adduction in a mixture of mRNA and IL, a peroxide species spiking study was performed. IL lipid was fractionated by RP-LC, and the peak containing +32 Da species was collected (Fig. 4b). Fractions were dried under a stream of nitrogen gas and resuspended in ethanol. LC-UV and LC-MS analysis of the resuspended fraction confirmed that the collected early peak contained the +32 Da species. Sa-mRNA and ionizable lipid mixtures were made and spiked with the isolated +32 Da species fraction to evaluate its effect on lipid adduction. To control for potential peroxide formation in the lipid tails of IL, a variant with fully saturated lipid tails (IL-F) was synthesized to prevent peroxide formation by removing the reactive dialkene groups (Fig. 4a). Spiking of the IL mixture with the +32 Da species resulted in a marked increase in lipid adduction, while empty fraction control spiking did not (Fig. 4c). The IL-F and sa-mRNA mixture did not produce adducted mRNA, but +32 Da species spiking into the IL-F mixture produced a corresponding increase in mRNA lipid adduction. As IL-F does not have dialkene tails that can be peroxidized and does not form adducted mRNA, this result is evidence that the unsaturated tails of IL are the source of reactive species.
a Chemical structure of IL ionizable lipid and its variants IL-F containing fully saturated tails, and IL-C containing an alternative linker, and (6Z,9Z,28Z,31Z)-heptatriaconta-6,9,28,31-tetraen-19-yl 4-(dimethylamino)butanoate (MC3). b The early eluting +32 Da peroxide species of IL was fractionated using LC-UV @214 nm and c spiked into mixtures of a high purity lot of IL and sa-mRNA, which resulted in a marked increase in adducted mRNA. Mixture of fully saturated variant IL-F and sa-mRNA did not produce a adducted mRNA but spiking with the +32 Da peroxide species markedly increased % lipid adduction.
MS analysis of lipid adduction in mRNA oligomers
To verify aldehyde products from peroxide species of IL were responsible for mRNA lipid adduction, direct MS analysis of mRNA oligomers was performed. Using a 45 mer mRNA (ODN45) with 14184.142 Da monoisotopic mass, we observed formation of lipid modifications when mixtures were made with IL and incubated for 4 days at room temperature. Additionally, we used IL-C (Fig. 4a), which is an IL variant having an additional 84 Da mass due to an alternative linker, which could be used to confirm the identity of lipid modifications. IL-C also contained +32 Da species, and IP-RP mRNA lipid adduction analysis of sa-mRNA extracted from IL-C mixtures showed equivalent lipid adduction levels as IL (Fig. 5a).
a IP-RP mRNA lipid adduction analysis of IL and variants IL-C and IL-F from sa-mRNA mixtures showing that only IL and IL-C containing dialkene groups form adducted mRNA. b Representative UV chromatogram @260 nm from LC-MS analysis of mRNA oligo incubated with ionizable lipid for 4 days at ambient temperature, showing two late eluting regions for peaks with two clusters of delta mass ranges. c Tables of the theoretical mass of tail and body modifications based on possible aldehyde products of peroxide degradation, which match observed delta masses with high accuracy and low mass error.
For mixtures with IL, late-eluting species with differing elution times and mass ranges were detected in the extracted oligomer. Two major clusters of adducts were observed: late eluting species with 6.25–7.25 min retention time were in the 100–160 Da delta mass range, while late eluting species with 8.0–8.5 min retention times ranged from 600–850 Da delta mass (Fig. 5b). Figure 5c depicts major observed adduct delta masses and a comparison to the theoretical delta masses. Among earlier late eluting species, the most abundant observed delta masses were 154.1 Da, 138.1 Da and 122.1 Da. For later eluting species, the most abundant observed delta masses were 777.6 Da, 793.6 Da, 809.6 Da, and 825.6 Da. The 4-hydroperoxy nonenal (4HNE) lipid “tail”23 is an expected product of IL peroxide degradation, and when bonded as an imine, produces a 154.1 Da species matching the observed delta mass species with 1.3 ppm mass error. Delta mass species 138.1 Da and 122.1 Da correspond with 154.1 Da −1O and −2O, respectively, which are possible oxidation variants of 4HNE. The theoretical aldehyde “body” product of IL peroxide degradation when bonded as an imine produces a 623.5 Da delta mass. The sum of the “body” and “tail” adduct masses amounts to 777.6 Da delta mass, which matches the observed 777.6 Da late eluting species with 2.2 ppm error. 793.6 Da, 809.6 Da, and 825.6 Da species correspond with 777.6 Da +1O, +2O, and +3O, respectively, which are possible “body” oxidation variants of any additional unsaturated lipid tail(s).
The prevalence of “body” + “tail” modifications from IL as dominant species on ODN45 indicates that some peroxide degradation occurred upon interaction of the ionizable lipid and mRNA, allowing both aldehyde products to be in proximity to react with the oligomer. Notably, combinations of modifications corresponding to “body” + “body” or “tail” + “tail” masses were not observed, though “body” only mass-ranged species were observed at lower abundances. Indeed, a species with mass corresponding to the “body” aldehyde peroxide degradation product was detected in IL working stocks in ethanol (Supplementary Fig. 6), indicating some aldehyde products were formed prior to mixing with mRNA.
MS analysis of lipid adduction in DLin-MC3-DMA/oligomer mixtures
Having identified peroxide degradation products as the primary source of reactive aldehydes involved in mRNA lipid adduction by IL, we wanted to further verify this mechanism in a well-known ionizable lipid. DLin-MC3-DMA (MC3) was evaluated due to having unsaturated tails. As in IL, LC-MS analysis of MC3 revealed an early eluting species containing dominant +32 Da species among others and a late eluting peak corresponding to adducted mRNA was observed in mixtures of MC3 and sa-mRNA using the IP-RP mRNA lipid adduction assay (Supplementary Fig. 7). ODN45 extracted from a MC3 mixture showed the presence of the 154.1 Da “tail” 4HNE species, and the later eluting species corresponded to the mass of a potential oxidation variant of MC3 “body” (Fig. 5c). This is different from ODN45 extracted from mixtures with IL or IL-C which had modifications corresponding to “body” + “tail” masses. Despite this difference, the presence of discrete “body” and “tail” sized modifications aligns with the mass of presumed reactive aldehyde products of MC3.
Discussion
In this study, we identified peroxide-derived ionizable lipid species involved in sa-mRNA lipid adduction, finding characteristic MS2 fragments in diagnostic sodium adducts. We additionally evaluated sa-mRNA lipid adduction in LNPs and mixtures with ionizable lipids using IP-RP chromatography and characterized lipid modifications formed on various mRNA constructs using LC-MS. We show that ionizable lipids with peroxidized unsaturated lipid tails undergo cleavage to form reactive aldehyde products, which directly modify mRNA and can compromise mRNA activity. Theoretical mRNA lipid adduction calculations in this study were based upon a hypothetical imine bond with primary amine-containing nucleotides, though conjugations such as an enamine bond, Michael addition, or other mechanisms, are not ruled out25. It should also be noted that other electrophiles not readily detected by current LC-MS methods may also react with nucleophiles in LNPs. The studies in this manuscript primarily involved IL with dialkene in the tail(s), and lipid variants with the same type of tail(s), but the results serve as a broader warning regarding lipid peroxidation and reactive aldehyde mechanisms and their impact on lipid delivery systems for genetic medicines.
Understanding the factors influencing mRNA stability in LNPs remains vital for the safe and effective development of vaccines and therapeutics. In this context, examining lipid species associated with mRNA lipid adduction is critical to establishing appropriate manufacturing controls for raw materials, gaining a comprehensive understanding of the phenomena, and devising effective mitigation strategies. To date, mechanisms of nucleic acid lipid adduction6,7,8,10,12,26 and peroxidation of PUFAs27,28 have been previously characterized, particularly in the context of cellular and environmental factors. To our knowledge, this is the first work demonstrating mRNA lipid adduction by ionizable lipids via aldehyde cleavage products of peroxide species in the context of LNP stability, ionizable lipid quality control, and vaccine product development.
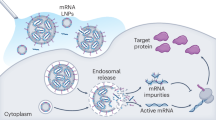
Based on the results in this study, we propose the following model for mRNA lipid adduction to occur by ionizable lipid peroxide species. mRNA phosphates are brought near ionizable lipids through interactions with ionizable tertiary amines1. Hydrophobic tails of IL are normally segregated from the surrounding aqueous environment due to immiscibility. Peroxides on lipid tails, with potentially greater exposure to the aqueous environment, can undergo degradation by various mechanisms including Hock cleavage, peroxyradical cleavage, or hydroxyradical β-scission23 (Fig. 6b). The resulting aldehyde products can interact with nucleophilic amines found in mRNA bases facilitated by the proximity of mRNA at the hydrophobic/hydrophilic interface (Fig. 6c). This is exemplified in our observation that mixtures containing mRNA and highly hydrophobic dodecyl aldehyde (DA) lacking an ionizable tertiary amine do not produce detectable mRNA lipid adduction, however when DA is spiked into mixtures of sa-mRNA with IL, lipid adduction is observed (Supplementary Fig. 8). Our results imply that the amphipathic nature of the ionizable lipid with its charged tertiary amine and lipid tails play a role in facilitating the adduction of the encapsulated mRNA25. In summary, reactive species, including but not limited to lipid aldehydes, which may not readily react with mRNA, are effectively brought into proximity with mRNA by the ionizable lipid acting as a bridge between lipid components and hydrophilic mRNA.
Lipid oxidation by reactive oxygen species (ROS) is a known source for producing bioactive compounds, of which aldehydes are critical due to their ability to amplify and propagate damage29. With the aldehyde-generating capacity of lipid peroxides combined with the bridging effect provided by the ionizable lipid, mRNAs in LNPs are vulnerable to these damaging oxide products. Alkene bonds—due to their weak π bonds—are generally regarded as “hot spots” for oxidative reactions30, highlighting the importance of lipid designs with functional groups that are resistant to oxidation by ROS.
As we have demonstrated, peroxidized lipids can be a crucial contributor to lipid adduction in LNP formulations. It has been shown that peroxidation in unsaturated tails can occur by auto-oxidation and photo-oxidation20,21. However, it is crucial to address potential specific contaminants that promote ROS generation and subsequent peroxidation8,31. For example, species such as Copper are transition metals that can catalyze the formation of peroxides32. We observe that IL lots with greater levels of peroxide species produce more adducted mRNA and have correspondingly higher levels of transition metals (Supplementary Table 1). Due to the damaging effects that transition metals can have on lipids as well as mRNA, their levels must be stringently controlled in their feed stocks as well as excipients, which are combined with the drug product.
To improve the purity of critical LNP components, such as the ionizable lipids, it is essential to develop analytical methods for monitoring specific species of interest. These methods will help ensure lipid quality and reduce the risk of mRNA lipid adduction during LNP production and storage. In this study, we conducted a thorough investigation into the underlying mechanisms of mRNA lipid adduction using an array of analytical techniques such as IP-RP-LC and LC-MS. These tools enable the identification of oxidation species and related reactive aldehyde species, which play a role in mRNA lipid adduction. The insights inform the design of appropriate controls and specifications for raw material manufacturing, which can mitigate the risk of reactive species to enable the robust development of mRNA-based LNP vaccines and therapies with greater stability and efficacy.
Materials and methods
LNP synthesis
Briefly, sa-mRNA (1.34 g/L; CSL Seqirus) in sodium citrate (50 mM; pH 6.0; Teknova) and lipids (25.6 mM) in ethanol were mixed in a 3:1 aqueous to ethanol volume ratio (N/P = 7) using a microfluidic mixer (Precision Nanosystems). Lipids used in the formulation include a phospholipid 1,2-distearoyl-sn-glycero-3-phosphocholine (DSPC; Avanti), cholesterol (Sigma-Aldrich), a PEG lipid 1,2-dimyristoyl-rac-glycero-3-methoxypolyethylene glycol-2000 (DMG-PEG; NOF American Corporation), and IL (WuXi STA, the full structure beyond the tail(s) is proprietary). LNPs were immediately diluted in-line with sodium citrate buffer and concentrated using tangential-flow filtration (TFF) and subsequently diafiltered with a cryo buffer (Cytiva). LNPs were filtered with a 0.8/0.2 µm guard filter (Sartorius) followed by a 0.45/0.2 µm sterile filter (Sartorius) and stored at −80 °C.
RNA/ionizable lipid mixtures
For sa-mRNA mixtures with IL, sa-mRNA (0.053 mg/mL) in water and IL lipid (1.919 mg/mL; WuXi STA) in ethanol were mixed in a 2:1 aqueous to ethanol ratio in 50 µL in sodium acetate (37.5 mM; pH 5.5; Thermo Fisher Scientific; AM9740). Mixtures were stored at ambient temperature for 2 h. For the 48 h stability study, sa-mRNA (0.053 mg/mL) in citrate buffer (50 mM; pH 6.0) and IL (1.919 mg/mL) in ethanol were mixed using a Benchtop NanoAssemblr (Precision Nanosystems) at a 2:1 flow rate ratio and 12 mL/min total flow rate. For mRNA oligomer mixtures with IL, mRNA oligomer ODN45 (0.053 mg/mL) with sequence 5’-rUrUrUrUrUrArArCrArUrUrUrCrArArArArArArArArArArArArArArArArArArArArArArArArArArArArArArA-3’ (IDTDNA) and IL, IL-C, IL-F (Wuxi AppTec) in ethanol were mixed in a 3:1 aqueous to ethanol ratio to 0.047 mg/mL oligomer and 2.559 mg/mL ionizable lipid in sodium acetate (37.5 mM; pH 5.5). For mRNA oligomer mixtures with MC3 (Fisher Scientific), 0.047 mg/mL oligomer and 3.544 mg/mL ionizable lipid in sodium acetate (37.5 mM; pH 5.5) were mixed in a 3:1 aqueous to ethanol ratio. Mixtures were stored at ambient temperature for 4 days. For sa-mRNA mixtures made with combinations of a phospholipid (Avanti), cholesterol (Sigma-Aldrich), a PEG lipid (NOF America), and IL (WuXi STA), sa-mRNA in water and lipids in ethanol were mixed at a 3:1 aqueous to ethanol ratio in the following combinations: sa-mRNA, IL; sa-mRNA, phospholipid; sa-mRNA, cholesterol; sa-mRNA, PEG lipid; sa-mRNA, IL, cholesterol; sa-mRNA, IL, phospholipid; sa-mRNA, IL, PEG lipid; sa-mRNA, IL, phospholipid, cholesterol, PEG lipid. Mixtures were made in sodium acetate (37.5 mM; pH 5.5) and stored at ambient temperature for 48 h.
RNA Extraction from LNPs and mRNA/ionizable lipid mixtures
LNPs and mixtures of mRNA and lipids were extracted in a 1:10 dilution in 60 mM ammonium acetate in isopropyl alcohol (IPA) and centrifuged at 4 °C for 20 min at 14,000 rpm. Pellets were washed with 1 mL of IPA and centrifuged at 4 °C for 5 min at 14,000 rpm. Pellets were air dried and resuspended in RNAse-free water (Promega), and mRNA concentrations were measured with a Nanodrop LITE (Thermo Scientific).
IP-RP-LC-UV mRNA lipid adduction assay
Chromatographic separation of samples was performed on a Vanquish UHPLC (Thermo Scientific) using a reverse-phase column at 0.2 mL/min flow rate at 55 °C. Mobile phase A (MPA) was triethylammonium acetate (TEAA)(100 mM; Millipore-Sigma) in water, and mobile phase B (MPB) was TEAA (100 mM) in acetonitrile (Millipore-Sigma). Separation method MPB%: 10% hold for 3 min, 10–30% in 2 min, 30–60% in 0.5 min, 60–100% in 2.5 min, 100% hold for 1 min, 100–10% in 0.1 min, and 10% hold for 2.9 min. Extracted mRNA samples were injected at 200 ng, and eluted peaks were detected at 260 nm wavelength using a UV detector.
Fractionation of IL with LC
Lipid fractionation was performed on a Vanquish UPLC (Thermo Scientific) using an XBridge BEH C18 OBD Prep Column, 130 Å, 5 µm, 10 × 100 mm (Waters) column at 50 °C with 0.95 mL/min flow rate. Mobile phases used were as follows: MPA was trifluoracetic acid (TFA) (0.1%; Millipore-Sigma) in water, and MPB was TFA (0.1%) in isopropanol (Thermo Fisher Scientific). Samples were separated using the following MPB% %: 30% hold for 5 min, 30–70% in 0.1 min, 70–77% in 6.9 min, 77–100% in 0.1 min, 100% hold for 2.9 min, 100–30% in 0.1 min, and 30% hold for 3.9 min. Approximately 100 µg (5 µg/µL) of lipid was injected per run for 12 runs, and eluted peaks were detected at 214 nm wavelength, and peaks of interest were collected using a fraction collector. Fractions were dried using a stream of dry nitrogen gas and resuspended in ethanol (Koptec). Fractionated samples were resuspended in the combined volume of ethanol used for 6 sample injections, and recovery was estimated at 80%. Positive mode IP-RP LC-MS of fractions was performed to confirm the masses of isolated species.
Negative mode LC-MS for mRNA oligomer/ionizable lipid mixtures
ESI-MS was performed using an Orbitrap Exploris 240 (Thermo Fisher Scientific) in negative ion mode using nitrogen sheath and auxiliary gas. Analysis was performed with ion transfer tube temperature 325 °C, vaporizer temperature at 325 °C, negative voltage of 2500 V, sheath gas 45 Arb, aux gas 15 Arb, sweep gas 2 Arb. Orbitrap Resolution was 120,000, with scan range 570–2000 m/z and 70% RF lens. AGC Target was Standard, with a custom injection time of 100 ms. 200 ng of extracted mRNA oligomer was separated using an ACQUITY Premier Oligonucleotide C18 2.1 ×50 mm column with 130 Å pore size and 1.7 µm particle size (Waters) at a 0.2 mL/min flow rate at 55 °C using a column pre-heater on a Vanquish UHPLC (Thermo Scientific). MPA was triethylamine (10 mM; TEA) (Millipore-Sigma) and hexafluoro-2-propanol (HFIP) (50 mM; Sigma-Aldrich) in water, and MPB was TEA (10 mM) and HFIP (50 mM) in methanol (50%; Millipore-Sigma). Separation method MPB %: 2-10% in 3 min, 10-30% in 2 min, 30–60% in 0.5 min, 60–100% in 2.5 min, 100% hold for 1 min, 100-2% in 0.1 min, and 2% hold for 2.9 min. UV was also collected at 260 nm. Peak deconvolution was performed in BioPharma Finder Intact Mass mode.
LC-MS structural elucidation of lipids and unknown species
ESI-MS was performed using an Orbitrap Exploris 240 (Thermo Scientific) in positive ion mode using nitrogen sheath and auxiliary gas. Analysis was performed with ion transfer tube temperature 275 °C, vaporizer temperature at 300 °C, positive voltage of 3250 V, sheath gas 40 Arb, aux gas 10 Arb, sweep gas 1 Arb. The MS1 resolution was 120,000 with scan range 105–1200 m/z and 70% RF lens. AGC Target was Standard, with a custom injection time of 50 ms. Standard intensity and dynamic exclusion filters were applied. Data-dependent MS2 was performed with an isolation window of 1.5 m/z, Orbitrap resolution of 30,000, and stepped normalized HCD collision energy of 25/30%. The scan range was defined by the first mass of 40 m/z. AGC Target was Standard, with a custom injection time of 54 ms. Approximately 1.125 µg (1125 ng/µL) of lipid was injected, and samples were separated using an ACQUITY UPLC BEH C18 2.1 ×50 mm column with 130 Å pore size and 1.7 µm particle size (Waters) at a 0.25 mL/min flow rate. MPA is TFA (0.1%) in water. MPB is TFA (0.1%) in isopropanol. The Vanquish UHPLC system had a column temperature of 50 °C using a column pre-heater and oven (Thermo Scientific). Lipid was separated using the following MPB %: 30% hold for 5 min, 30-70% in 0.1 min, 70-77% in 6.9 min, 77–100% in 0.1 min, 100% hold for 2.9 min, 100-30% in 0.1 min, and 30% hold for 3.9 min. UV was also collected at 214 nm. Data analysis was performed in FreeStyle and Skyline33,34 software.
sa-mRNA activity assay
BHK-21 cells (ATCC; catalog #BHK-21 [C-13]) were cultured in growth media (DMEM + 1% Pen/Strep + 5% FBS + 1% Glutamax (L-glutamine)). A population of 1×106 cells in 250 µL in Opti-MEM (Gibco; catalog #31985062) was electroporated with a Genepulser (BioRad, catalog #11668019) with 100 ng extracted mRNA. Cells were transferred to 6-well plates with outgrowth media (DMEM + 1% Pen/Strep + 5% FBS + 1% Glutamax) and incubated for 18 h at 37 °C before staining with anti-H1/H5 and anti-N1 antibodies, followed by analysis with an Accuri flow-cytometer (BD). An sa-mRNA reference standard was included as an assay control (Supplementary Fig. 2). The gating strategy is exemplified in Supplementary Fig. 9.
Capillary electrophoresis
The samples were analyzed on a Sciex PA800 Plus system with a laser-induced fluorescence detector. The extracted mRNA was diluted to approximately 25 µg/mL and combined with Formamide (Thermo Scientific) at a ratio of 1:1 and SYBR Green II (Thermo Scientific) in dimethyl sulfoxide (Fisher Chemical) at a ratio of 1:500. The sample was heated at 70 °C for 2 min and transferred to ice. After cooling for 5 min, the samples were briefly centrifuged and degassed. The gel and dye mixture consisted of 1% polyvinylpyrrolidone, 1x Tris (89 mM)-buffered borate (89 mM) with 2 mM EDTA (Invitrogen), 4 M urea (Sigma-Aldrich), and 1:5000 SYBR Green II. Pressure injection of 0.5 psi for 5 s was used. Separation proceeded at 5.0 kV for 17 min. Data was analyzed using 32 Karat software.
ICP-MS analysis of lipids
ICP-MS analysis of IL was performed on an Agilent 7700x in He mode. To 100 mg of each sample in a tared microwave digestion vessel, 200 μL nitric acid (HNO3) and 1000 μL of water were added. A digestion blank was prepared without the addition of a sample. Vessels were digested at 190 °C for 10 min in a microwave digestion system and brought to 10 mL final volume in water after cooling to ambient conditions. A single-point calibration reference standard was at 100 ppb by pipetting 100 μL of a multi-element custom standard into 10 mL final volume with HNO3 (2%; v/v) (Supplementary Table 1).
Dodecyl aldehyde (DA) spiking of mRNA/ionizable lipid mixtures
For sa-mRNA mixtures with IL, sa-mRNA (0.075 mg/mL) in water and IL lipid (1.5 mg/mL) in ethanol were mixed in a 3:1 aqueous to ethanol ratio in 40 µL in sodium acetate (37.5 mM; pH 5.5). For DA (Sigma-Aldrich) spiked mixtures, DA was diluted in ethanol and mixed with IL at 5%, 25%, or 50% by weight of IL before being mixed with sa-mRNA. For DA-spiked control samples, DA was mixed with sa-mRNA at 5%, 25%, or 50% by weight of IL. Mixtures were stored at ambient temperature in the dark for 48 h and extracted as described (Supplementary Fig. 8).
Statistics and reproducibility
Assay measurements were taken from distinct samples. Formal statistical analyses were not performed as this is an observational descriptive study. Evaluation of late eluting species were performed using RP-IP with UV data for initial identification of the lipid modifications, and stability in a time course are representative results which were replicated > 60 times with similar results (Fig. 1). Screening of lipid components of LNPs and their combinations in mixtures with IL were replicated 2 times with similar results (Fig. 1). Experiments analyzing the levels of species in different lots of IL using XIC data versus the capacity of the IL lot to produce adducted mRNA using the lipid adduction assay were replicated 3 times with similar results (Fig. 2). Fragmentation analysis of IL species using LC-MS2 was replicated 3 times with similar results (Fig. 3). Experiments where peroxide species were fractionated by RP-LC and spiked into mixtures of IL and sa-mRNA were replicated 3 times with similar results (Fig. 4). Experiments using LC-MS to analyze lipid modification mass on mRNA oligomer from mixtures with IL were replicated 4 times with similar results and with IL-C and MC3 experiments were replicated 2 times with similar results (Fig. 5).
For data shown in the supplementary information, the lipid adduction assay with late peak observation has been performed > 60 times, and representative testing results were chosen to summarize screening studies (Supplementary Fig. 1). Experiments evaluating relative mRNA integrity by lipid adduction assay and CE versus activity were replicated 3 times with similar results (Supplementary Fig. 2). MS analysis of early eluting peak in IL lots was performed 3 times with similar results (Supplementary Fig. 3 and 4). Analysis of the presence or absence of characteristic MS2 mass fragments of IL in two different +16 Da species was performed 3 times with similar results (Supplementary Fig. 5). Analysis of characteristic MS2 fragments to identify IL and peroxide degradation products of peroxidized IL was performed 3 times with similar results (Supplementary Fig. 6). Experiments assessing adducted mRNA in mixtures of MC3 and sa-mRNA using IP-RP mRNA lipid adduction assay and species in MC3 lipid using LC-MS were performed 2 times with similar results (Supplementary Fig. 7). Experiments where DA was spiked into mixtures of sa-mRNA and IL and extracted sa-mRNA was analyzed using the IP-RP mRNA lipid adduction assay were repeated 3 times with similar results (Supplementary Fig. 8). Evaluation of elemental species in IL batches using ICP-MS was performed once (Supplementary Table 1).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The source data supporting the graphs and charts are available in the Supplementary Data file. The data within this paper and other findings of this study are available from the corresponding author upon reasonable request.
References
Semple, S. C., Leone, R., Barbosa, C. J., Tam, Y. K. & Lin, P. J. C. Lipid nanoparticle delivery systems to enable mRNA-based therapeutics. Pharmaceutics 14, 398 (2022).
McKenzie, R. E., Minnell, J. J., Ganley, M., Painter, G. F. & Draper, S. L. mRNA synthesis and encapsulation in ionizable lipid nanoparticles. Curr. Protoc. 3, e898 (2023).
Han, X. et al. An ionizable lipid toolbox for RNA delivery. Nat. Commun. 12, 7233 (2021).
Hajj, K. A. & Whitehead, K. A. Tools for translation: non-viral materials for therapeutic mRNA delivery. Nat. Rev. Mater. 2, 17056 (2017).
Garaycoechea, J. I. et al. Alcohol and endogenous aldehydes damage chromosomes and mutate stem cells. Nature 553, 171–177 (2018).
Zhou, M., Wang, D. O., Li, W. & Zheng, J. RNA adduction derived from electrophilic species in vitro and in vivo. Chem. Biol. Interact. 351, 109748 (2022).
Sevilla, C. L. et al. Development of monoclonal antibodies to the malondialdehyde−deoxyguanosine adduct, pyrimidopurinone1. Chem. Res. Toxicol. 10, 172–180 (1997).
Marnett, L. J. Lipid peroxidation—DNA damage by malondialdehyde. Mutat. Res. Fundam. Mol. Mech. Mutagen. 424, 83–95 (1999).
Shukla, R. K., Badiye, A., Vajpayee, K. & Kapoor, N. Genotoxic potential of nanoparticles: structural and functional modifications in DNA. Front. Genet. 12, 728250 (2021).
Packer, M., Gyawali, D., Yerabolu, R., Schariter, J. & White, P. A novel mechanism for the loss of mRNA activity in lipid nanoparticle delivery systems. Nat. Commun. 12, 6777 (2021).
Birdsall, R. E. et al. Monitoring stability indicating impurities and aldehyde content in lipid nanoparticle raw material and formulated drugs. J. Chromatogr. B 1234, 124005 (2024).
Hashiba, K. et al. Overcoming thermostability challenges in mRNA–lipid nanoparticle systems with piperidine-based ionizable lipids. Commun. Biol. 7, 556 (2024).
Aliakbarinodehi, N. et al. Interaction kinetics of individual mRNA-containing lipid nanoparticles with an endosomal membrane mimic: dependence on pH, protein corona formation, and lipoprotein depletion. ACS Nano 16, 20163–20173 (2022).
Sheridan, C. First self-amplifying mRNA vaccine approved. Nat. Biotechnol. 42, 4–4 (2024).
Chang, C. et al. sa-mRNA influenza vaccine raises a higher and more durable immune response than mRNA vaccine in preclinical models. Vaccine 51, 126883 (2025).
Chang, C. et al. Self-amplifying mRNA bicistronic influenza vaccines raise cross-reactive immune responses in mice and prevent infection in ferrets. Mol. Ther. Methods Clin. Dev. 27, 195–205 (2022).
Malone, B., Urakova, N., Snijder, E. J. & Campbell, E. A. Structures and functions of coronavirus replication-transcription complexes and their relevance for SARS-CoV-2 drug design. Nat. Rev. Mol. Cell Biol. 23, 21–39 (2022).
Geall, A. J. et al. Nonviral delivery of self-amplifying RNA vaccines. Proc. Natl Acad. Sci. USA 109, 14604–9 (2012).
Kis, Z., Kontoravdi, C., Shattock, R. & Shah, N. Resources, production scales and time required for producing RNA vaccines for the global pandemic demand. Vaccines 9, 3 (2020).
Ito, J. et al. Evaluation of lipid oxidation mechanisms in beverages and cosmetics via analysis of lipid hydroperoxide isomers. Sci. Rep. 9, 7387 (2019).
Ito, J., Mizuochi, S., Nakagawa, K., Kato, S. & Miyazawa, T. Tandem mass spectrometry analysis of linoleic and arachidonic acid hydroperoxides via promotion of alkali metal adduct formation. Anal. Chem. 87, 4980–4987 (2015).
Kaur, K., Salomon, R. G., O’Neil, J. & Hoff, H. F. Carboxyalkyl pyrroles in human plasma and oxidized low-density lipoproteins. Chem. Res. Toxicol. 10, 1387–1396 (1997).
Spickett, C. The lipid peroxidation product 4-hydroxy-2-nonenal: advances in chemistry and analysis. Redox Biol. 1, 145–152 (2013).
Esterbauer, H., Schaur, R. J. & Zollner, H. Chemistry and biochemistry of 4-hydroxynonenal, malonaldehyde and related aldehydes. Free Radic. Biol. Med. 11, 81–128 (1991).
Minko, I. G. et al. Chemistry and biology of DNA containing 1,N 2-deoxyguanosine adducts of the α,β-unsaturated aldehydes acrolein, crotonaldehyde, and 4-hydroxynonenal. Chem. Res. Toxicol. 22, 759–778 (2009).
Zhu, P., Lee, S. H., Wehrli, S. & Blair, I. A. Characterization of a lipid hydroperoxide-derived rna adduct in rat intestinal epithelial cells. Chem. Res. Toxicol. 19, 809–817 (2006).
Ayala, A., Muñoz, M. F. & Argüelles, S. Lipid peroxidation: production, metabolism, and signaling mechanisms of malondialdehyde and 4-hydroxy-2-nonenal. Oxid. Med. Cell. Longev. 2014, 360438 (2014).
Lee, S. H. & Blair, I. A. Characterization of 4-Oxo-2-nonenal as a Novel Product of Lipid Peroxidation. Chem. Res. Toxicol. 13, 698–702 (2000).
Hill, B. G. & Bhatnagar, A. Beyond reactive oxygen species: aldehydes as arbitrators of alarm and adaptation. Circ. Res. 105, 1044–1046 (2009).
Shen, Q. et al. Triacylglycerols are preferentially oxidized over free fatty acids in heated soybean oil. npj Sci. Food 5, 7 (2021).
Dix, T. A. & Aikens, J. Mechanisms and biological relevance of lipid peroxidation initiation. Chem. Res. Toxicol. 6, 2–18 (1993).
Knight, J. A. & Voorhees, R. P. Peroxidation of linolenic acid-catalysis by transition metal ions. Ann. Clin. Lab. Sci. 20, 347–52 (1990).
MacLean, B. et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26, 966–968 (2010).
Adams, K. J. et al. Skyline for small molecules: a unifying software package for quantitative metabolomics. J. Proteome Res. 19, 1447–1458 (2020).
Acknowledgements
Payel Sil, John Pryor, and Greg Bacola supported in vitro protein expression assays. LNPs, binary mixtures of sa-mRNA and IL, and sa-mRNA were made by Chrissy Hofmann, Nick Murphy, Sonic (Xiaojun) Li, Jackie Long, Cal Stubbs, Gray Wyatt, Julia Bryant, Valerie Costeniuc, Carmen Murray, Chase Hullender, and Andrew Beam. PPD performed ICP-MS analysis of lipids. We thank Con Panousis, Yingxia Wen, Ania Senczuk, Nick Manzo, and Younghoon Kim for insightful discussions about the lipid adduction project.
Author information
Authors and Affiliations
Contributions
R.K. identified the peroxidation pathway for covalent lipid addition in synthetic oligos, performed lipid/mRNA mixture and spiking studies, preparative lipid species fractionation, designed and carried out LC-MS and CE analysis, and wrote the manuscript with input from all authors. Z.W. identified the differences in ionizable lipid impacting LNP quality, designed and carried out LC-MS studies and data analysis, and revised the manuscript with input from all authors. Z.W. contributed to identifying the peroxidation pathway, to LC-MS and IP-RP mRNA lipid adduction method development, and to shaping and writing the manuscript. R.K., Z.W., H.S. and J.Q. contributed to the design of experiments. H.S. contributed to IP-RP mRNA lipid adduction assay development, performed lipid/mRNA mixture and peroxide spiking studies and IP-RP-LC, supported MS analysis, and contributed to the manuscript. Z.C. performed early work characterizing ionizable lipid species by LC-MS. Y.Z. contributed to the project management and provided critical feedback to the manuscript. J.Q. hypothesized the formation of lipid peroxides and their subsequent degradation in IL. Their presence was later confirmed by R.K. and Z.W. J.Q. provided strategic directions for the project, supervised the work in this study, and contributed to shaping, writing, and revising the manuscript.
Corresponding author
Ethics declarations
Competing interests
Z.W., H.S., Z.C., Y.Z. and J.Q. are employees of CSL. R.K. was an employee of Aditi Consulting at the time of work and is currently an employee of Autonomous Therapeutics in Rockville Maryland at the time of manuscript submission. Some authors may own stocks of CSL. The authors declare no other competing interests.
Peer review
Peer review information
Communications Chemistry thanks Yusuke Sato and the other, anonymous, reviewer for their contribution to the peer review of this work. Peer review reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Kang, R.S., Wang, Z., Sapa, H. et al. Spotlighting the criticality of lipid quality control through a mechanistic investigation of mRNA activity loss in lipid nanoparticles. Commun Chem 8, 313 (2025). https://doi.org/10.1038/s42004-025-01699-5
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s42004-025-01699-5