Abstract
Acute myeloid leukemia (AML)-specific target antigens are difficult to identify. Here we demonstrate that HLA-DRB1 can serve as a leukemia-specific target of chimeric antigen receptor (CAR) T cells in patients with AML after allogeneic hematopoietic stem cell transplantation (allo-HCT). We identified KG2032 as a monoclonal antibody specifically bound to AML cells in about half of patients, but not to normal leukocytes other than B lymphocytes. KG2032 reacted with a subset of HLA-DRB1 molecules, specifically those in which the 86th amino acid was not aspartic acid. KG2032 reacted minimally with nonhematopoietic tissues. These results indicate that KG2032 reactivity is highly specific for AML cells in patients who carry KG2032-reactive HLA-DRB1 alleles and who received allo-HCT from a donor carrying KG2032-nonreactive HLA-DRB1 alleles. KG2032-derived CAR T or natural killer cells showed significant anti-leukemic activity in preclinical models in female mice, suggesting that they may cure patients with AML who are incurable with allo-HCT.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
All data generated during the current study are presented in this manuscript and/or are available from the corresponding author upon reasonable request. Source data are provided with this paper.
References
June, C. H. & Sadelain, M. Chimeric antigen receptor therapy. N. Engl. J. Med. 379, 64–73 (2018).
Weber, E. W., Maus, M. V. & Mackall, C. L. The emerging landscape of immune cell therapies. Cell 181, 46–62 (2020).
Liu, E. et al. Use of CAR-transduced natural killer cells in CD19-positive lymphoid tumors. N. Engl. J. Med. 382, 545–553 (2020).
Majzner, R. G. et al. GD2-CAR T cell therapy for H3K27M-mutated diffuse midline gliomas. Nature 603, 934–941 (2022).
Quintarelli, C., Del Bufalo, F. & Locatelli, F. GD2-CART01 for relapsed or refractory high-risk neuroblastoma. Reply. N. Engl. J. Med. 388, 2303–2304 (2023).
Qi, C. et al. Claudin18.2-specific CAR T cells in gastrointestinal cancers: phase 1 trial interim results. Nat. Med. 28, 1189–1198 (2022).
Adusumilli, P. S. et al. A phase I trial of regional mesothelin-targeted CAR T-cell therapy in patients with malignant pleural disease, in combination with the anti-PD-1 agent pembrolizumab. Cancer Discov. 11, 2748–2763 (2021).
Gragert, L. et al. HLA match likelihoods for hematopoietic stem-cell grafts in the US registry. N. Engl. J. Med. 371, 339–348 (2014).
Dehn, J. et al. Selection of unrelated donors and cord blood units for hematopoietic cell transplantation: guidelines from the NMDP/CIBMTR. Blood 134, 924–934 (2019).
Barker, J. N. et al. Optimal practices in unrelated donor cord blood transplantation for hematologic malignancies. Biol. Blood Marrow Transplant. 23, 882–896 (2017).
Luznik, L. et al. HLA-haploidentical bone marrow transplantation for hematologic malignancies using nonmyeloablative conditioning and high-dose, posttransplantation cyclophosphamide. Biol. Blood Marrow Transplant. 14, 641–650 (2008).
Kanakry, C. G., Fuchs, E. J. & Luznik, L. Modern approaches to HLA-haploidentical blood or marrow transplantation. Nat. Rev. Clin. Oncol. 13, 10–24 (2016).
Weisdorf, D. J. et al. Allogeneic transplantation for advanced acute myeloid leukemia: the value of complete remission. Cancer 123, 2025–2034 (2017).
Nagler, A. et al. Longitudinal outcome over two decades of unrelated allogeneic stem cell transplantation for relapsed/refractory acute myeloid leukemia: an ALWP/EBMT analysis. Clin. Cancer Res. 28, 4258–4266 (2022).
Cummins, K. D. & Gill, S. Chimeric antigen receptor T-cell therapy for acute myeloid leukemia: how close to reality? Haematologica 104, 1302–1308 (2019).
MacKay, M. et al. The therapeutic landscape for cells engineered with chimeric antigen receptors. Nat. Biotechnol. 38, 233–244 (2020).
Tambaro, F. P. et al. Autologous CD33-CAR-T cells for treatment of relapsed/refractory acute myelogenous leukemia. Leukemia 35, 3282–3286 (2021).
Sallman, D. A. et al. CYAD-01, an autologous NKG2D-based CAR T-cell therapy, in relapsed or refractory acute myeloid leukaemia and myelodysplastic syndromes or multiple myeloma (THINK): haematological cohorts of the dose escalation segment of a phase 1 trial. Lancet Haematol. 10, e191–e202 (2023).
Jin, X. et al. First-in-human phase I study of CLL-1 CAR-T cells in adults with relapsed/refractory acute myeloid leukemia. J. Hematol. Oncol. 15, 88 (2022).
Zhang, H. et al. Anti-CLL1 chimeric antigen receptor T-cell therapy in children with relapsed/refractory acute myeloid leukemia. Clin. Cancer Res. 27, 3549–3555 (2021).
Sauer, T. et al. CD70-specific CAR T cells have potent activity against acute myeloid leukemia without HSC toxicity. Blood 138, 318–330 (2021).
Leick, M. B. et al. Non-cleavable hinge enhances avidity and expansion of CAR-T cells for acute myeloid leukemia. Cancer Cell 40, 494–508.e495 (2022).
Jetani, H. et al. Siglec-6 is a novel target for CAR T-cell therapy in acute myeloid leukemia. Blood 138, 1830–1842 (2021).
Myburgh, R. et al. Anti-human CD117 CAR T-cells efficiently eliminate healthy and malignant CD117-expressing hematopoietic cells. Leukemia 34, 2688–2703 (2020).
Casucci, M. et al. CD44v6-targeted T cells mediate potent antitumor effects against acute myeloid leukemia and multiple myeloma. Blood 122, 3461–3472 (2013).
Perna, F. et al. Integrating proteomics and transcriptomics for systematic combinatorial chimeric antigen receptor therapy of AML. Cancer Cell 32, 506–519 e505 (2017).
Gottschlich, A. et al. Single-cell transcriptomic atlas-guided development of CAR-T cells for the treatment of acute myeloid leukemia. Nat. Biotechnol. https://doi.org/10.1038/s41587-023-01684-0 (2023).
Mandal, K. et al. Structural surfaceomics reveals an AML-specific conformation of integrin β(2) as a CAR T cellular therapy target. Nat Cancer 4, 1592–1609 (2023).
Hosen, N. et al. The activated conformation of integrin β7 is a novel multiple myeloma-specific target for CAR T cell therapy. Nat. Med. 23, 1436–1443 (2017).
Hasegawa, K. et al. Selective targeting of multiple myeloma cells with a monoclonal antibody recognizing the ubiquitous protein CD98 heavy chain. Sci. Transl. Med. 14, eaax7706 (2022).
Bonnet, D. & Dick, J. E. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat. Med. 3, 730–737 (1997).
Tzelepis, K. et al. A CRISPR dropout screen identifies genetic vulnerabilities and therapeutic targets in acute myeloid leukemia. Cell Rep. 17, 1193–1205 (2016).
Cresswell, P. Assembly, transport, and function of MHC class II molecules. Annu. Rev. Immunol. 12, 259–293 (1994).
Steimle, V., Otten, L. A., Zufferey, M. & Mach, B. Complementation cloning of an MHC class II transactivator mutated in hereditary MHC class II deficiency (or bare lymphocyte syndrome). Cell 75, 135–146 (1993).
Byrne, B. et al. Human duodenal epithelial cells constitutively express molecular components of antigen presentation but not costimulatory molecules. Hum. Immunol. 63, 977–986 (2002).
Wosen, J. E., Mukhopadhyay, D., Macaubas, C. & Mellins, E. D. Epithelial MHC class II expression and its role in antigen presentation in the gastrointestinal and respiratory tracts. Front. Immunol. 9, 2144 (2018).
Menge, A. C. & Mestecky, J. Surface expression of secretory component and HLA class II DR antigen on glandular epithelial cells from human endometrium and two endometrial adenocarcinoma cell lines. J. Clin. Immunol. 13, 259–264 (1993).
Mayer, L. et al. Expression of class II molecules on intestinal epithelial cells in humans. Differences between normal and inflammatory bowel disease. Gastroenterology 100, 3–12 (1991).
Hazenberg, M. D. & Spits, H. Human innate lymphoid cells. Blood 124, 700–709 (2014).
Ying, Z. et al. A safe and potent anti-CD19 CAR T cell therapy. Nat. Med. 25, 947–953 (2019).
Feucht, J. et al. Calibration of CAR activation potential directs alternative T cell fates and therapeutic potency. Nat. Med. 25, 82–88 (2019).
Liu, E. et al. Cord blood NK cells engineered to express IL-15 and a CD19-targeted CAR show long-term persistence and potent antitumor activity. Leukemia 32, 520–531 (2018).
Imai, C., Iwamoto, S. & Campana, D. Genetic modification of primary natural killer cells overcomes inhibitory signals and induces specific killing of leukemic cells. Blood 106, 376–383 (2005).
Christopher, M. J. et al. Immune escape of relapsed AML cells after allogeneic transplantation. N. Engl. J. Med. 379, 2330–2341 (2018).
Lee, D. W. et al. T cells expressing CD19 chimeric antigen receptors for acute lymphoblastic leukaemia in children and young adults: a phase 1 dose-escalation trial. Lancet 385, 517–528 (2015).
Turtle, C. J. et al. CD19 CAR-T cells of defined CD4+:CD8+ composition in adult B cell ALL patients. J. Clin. Invest. 126, 2123–2138 (2016).
Park, J. H. et al. Long-term follow-up of CD19 CAR therapy in acute lymphoblastic leukemia. N. Engl. J. Med. 378, 449–459 (2018).
Maude, S. L. et al. Tisagenlecleucel in children and young adults with B-cell lymphoblastic leukemia. N. Engl. J. Med. 378, 439–448 (2018).
Jain, T. et al. Safety and feasibility of chimeric antigen receptor T cell therapy after allogeneic hematopoietic cell transplantation in relapsed/ refractory B cell non-Hodgkin lymphoma. Leukemia 33, 2540–2544 (2019).
Zhang, X. et al. Efficacy and safety of anti-CD19 CAR T-cell therapy in 110 patients with B-cell acute lymphoblastic leukemia with high-risk features. Blood Adv. 4, 2325–2338 (2020).
Dholaria, B. et al. Clinical applications of donor lymphocyte infusion from an HLA-haploidentical donor: consensus recommendations from the Acute Leukemia Working Party of the EBMT. Haematologica 105, 47–58 (2020).
Zhou, X. et al. Long-term outcome after haploidentical stem cell transplant and infusion of T cells expressing the inducible caspase 9 safety transgene. Blood 123, 3895–3905 (2014).
Ciceri, F. et al. Infusion of suicide-gene-engineered donor lymphocytes after family haploidentical haemopoietic stem-cell transplantation for leukaemia (the TK007 trial): a non-randomised phase I-II study. Lancet Oncol. 10, 489–500 (2009).
Ikeda, N. et al. Determination of HLA-A, -C, -B, -DRB1 allele and haplotype frequency in Japanese population based on family study. Tissue Antigens 85, 252–259 (2015).
Gragert, L., Madbouly, A., Freeman, J. & Maiers, M. Six-locus high resolution HLA haplotype frequencies derived from mixed-resolution DNA typing for the entire US donor registry. Hum. Immunol. 74, 1313–1320 (2013).
Han, C. et al. Desensitized chimeric antigen receptor T cells selectively recognize target cells with enhanced antigen expression. Nat. Commun. 9, 468 (2018).
Han, C. et al. Polymorphic region-specific antibody for evaluation of affinity-associated profile of chimeric antigen receptor. Mol. Ther. Oncolytics 17, 293–305 (2020).
Rose, L. M. et al. Critical Lym-1 binding residues on polymorphic HLA-DR molecules. Mol. Immunol. 36, 789–797 (1999).
Lacombe, F. et al. Flow cytometry CD45 gating for immunophenotyping of acute myeloid leukemia. Leukemia 11, 1878–1886 (1997).
Wang, X. et al. A transgene-encoded cell surface polypeptide for selection, in vivo tracking, and ablation of engineered cells. Blood 118, 1255–1263 (2011).
Weber, E. W. et al. Transient rest restores functionality in exhausted CAR-T cells through epigenetic remodeling. Science https://doi.org/10.1126/science.aba1786 (2021).
Nicholson, I. C. et al. Construction and characterisation of a functional CD19 specific single chain Fv fragment for immunotherapy of B lineage leukaemia and lymphoma. Mol. Immunol. 34, 1157–1165 (1997).
Zola, H. et al. Preparation and characterization of a chimeric CD19 monoclonal antibody. Immunol. Cell Biol. 69, 411–422 (1991).
Yokoi, T. et al. Identification of a unique subset of tissue-resident memory CD4(+) T cells in Crohn’s disease. Proc. Natl Acad. Sci. USA 120, e2204269120 (2023).
Acknowledgements
The authors thank the Kinki Cord Blood Bank and Hyogo Cord Blood Bank for CB samples and T. Maeda, N. Nishiura (Suita Municipal Hospital), M. Kanou, F. Mima (Osaka International Cancer Institute), H. Shibayama (Osaka National Hospital), Y. Yamashita (Ashiya Municipal Hospital), T. Kida (Toyonaka Municipal Hospital), Y. Morikawa (Sumitomo Hospital), M. Kawakami (Nippon Life Hospital) and Y. Azenishi (Minoo Municipal Hospital) for AML samples. We also thank K. Terasaki and M. Yamaguchi for technical assistance, and E. Morii for fruitful discussions. This work was supported in part by the Japan Agency for Medical Research and Development (AMED) (20cm0106361h0002, 21bm0404076h0001, 22cm0106381h0002, 23ck0106819h0001 and 24bk0104170h0001 to N.H.); the Japan Society for the Promotion of Science (JSPS) KAKENHI (JP19K16799 and JP21K15487 to K.H. and JP26461404 and JP20H03710 to N.H.); grants from the Chemo-Sero- Therapeutic Research Institute (to N.H.) and Takeda Science Foundation (to N.H.). This research was supported in part by the Research Support Project for Life Science and Drug Discovery (Basis for Supporting Innovative Drug Discovery and Life Science Research) from AMED (JP23ama121011 to J.T.). The funders had no role in the study design, data collection and analysis, decision to publish or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
S.I., K.H., Y.K. and N.H. designed the research study, performed the experiments, analyzed the data, and wrote the paper. T.A., R.K., T.W., M.Y., Y.I., H.U., M. Matsubara, M.T., M.S., S. Kida, K.S., K. Tsutsumi, K.F., J.F., T.U., S. Kusakabe, A. Hino, M. Ichii, A. Hirose, H.N., M.H., T.N., M. Inoue, K. Yoshihara, S.Y., S.U., T. Tachi, H. Kuroda, K.M., N.K., H. Kishima, E.I., M. Murakami, T. Takiuchi, Tadashi Kimura, T.H., Toru Kimura, Y.S., C.I., K. Yusa, R.M., T.O., H.E., K. Takeda, Y.O., A.K. and J.T. performed the experiments.
Corresponding author
Ethics declarations
Competing interests
N.H. has applied for a patent entitled ‘Antibody against acute myeloid leukemia’ (PCT/JP2023/042029) through the Osaka University Office for University–Industry Collaboration. The other authors declare no competing interests.
Peer review
Peer review information
Nature Cancer thanks Leo Luznik, Katayoun Rezvani and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
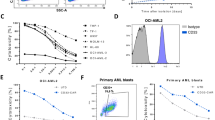
Extended Data Fig. 1 Distinct reactivity of KG2032 toward AML cells was detected in approximately half of patients with AML.
a, b. Flow cytometric analysis of KG2032 reactivity to AML cells in the BM of patients with AML. A total of 32 patients were analyzed. Results from patients with high reactivity to KG2032 and those of patients with low or no reactivity to KG2032 are shown separately in panels a and b. Analyses of the expression of CD34/CD38 or CD45/CD34 on CD3− BM cells are shown. AML cells were identified according to the expression of these markers. In a and b, a single analysis was performed for each sample. c. Frequencies of KG2032-positive cells in each subpopulation of normal PBMCs (n = 6) or AML cells (n = 32). d. Flow cytometric analyses of the reactivity of KG2032 or L243 (a known anti-HLA-DR mAb) to CD34+CD38− leukemic stem cells (LSCs) or CD34+CD38+ AML cells. Representative results are shown. e. Frequencies of KG2032-positive cells in CD34+CD38− LSCs or CD34+CD38+ AML cells (n = 4). f. Mean fluorescence intensity (MFI) of KG2032 in AML samples before treatment (n = 14) or those in relapse (n = 4). Data are expressed as means ± s.e.m. Statistical difference was determined by two-tailed Welch’s t-test.
Extended Data Fig. 2 KG2032 reacts with a subset of polymorphic HLA-DRB1 molecules, specifically those in which the 86th amino acid was not aspartic acid.
a. Flow cytometric analysis of the reactivity of KG2032 and L243 (a known anti-HLA-DR mAb) toward K562 cells expressing different HLA-DRB1 molecules together with HLA-DRA. KG2032-reactive HLA-DRB1 alleles are shown in red and KG2032-nonreactive alleles are shown in blue. Data are representative of two independent experiments. b. Comparison of the amino acid sequences of HLA-DRB1*04:05 and DRB1*15:02. c. Amino acid sequences of region 2 in the KG2032-reactive or nonreactive HLA-DRB1 alleles.
Extended Data Fig. 3 KG2032 reactivity in various subsets of normal hematopoietic cells.
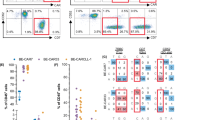
a. Flow cytometric analysis of the reactivity of KG2032 or L243 (a known anti-HLA-DR mAb) to subsets of PBMCs from healthy donors. KG2032-reactive HLA-DRB1 alleles are shown in red and KG2032-nonreactive alleles are shown in blue. A total of 10 donors were analyzed. Six donors carried KG2032-reactive HLA-DRB1 alleles and 4 donors did not. The results of two representative donors in each group are shown (donors 3 and 4: KG2032-reactive, donors 5 and 6: KG2032-nonreactive). The analysis of another donor in each group is shown in Fig. 3a. b. Flow cytometric analyses of the reactivity of KG2032 or L243 to each subpopulation of normal BM cells. HSC: hematopoietic stem cells, HPC: hematopoietic progenitor cells. A total of 3 donors were analyzed and the result of a representative donor is shown. The analysis of another donor is shown in Fig. 3d. In a and b, a single analysis was performed for each sample. c. Frequencies of KG2032- or L243-positive cells in each subpopulation of normal BM cells (n = 3). d. Flow cytometric analyses of the reactivity of KG2032 or L243 to each subpopulation of normal PBMCs. cDC: conventional dendritic cells, pDC: plasmacytic dendritic cells. e. Frequencies of KG2032- or L243-positive cells in each subpopulation of normal PBMCs (n = 3). f. Flow cytometric analyses of the reactivity of KG2032 or L243 to CD34+ CB cells stimulated with the indicated concentrations of IFN-γ for 24 h. g. Frequencies of KG2032- or L243-positive cells in CD34+ CB cells stimulated with the indicated concentrations of IFN-γ (n = 3). h. Flow cytometric analyses of the reactivity of KG2032 or L243 to subpopulations of PBMCs stimulated with the indicated concentrations of IFN-γ for 24 h. i. Frequencies of KG2032- or L243-positive cells in each subpopulation after stimulation with the indicated concentrations of IFN-γ (n = 3). In b–i, three donors carrying various KG2032-reactive HLA-DRB1 alleles were analyzed, and representative flow cytometric analyses are shown. In c, e, g and i, data are presented as means ± s.e.m.
Extended Data Fig. 4 KG2032 binds minimally to nonhematopoietic tissues carrying KG2032-reactive HLA-DRB1 alleles, even under severe inflammation.
a. Flow cytometric analysis of KG2032 or L243 reactivity to intestinal epithelial cells or lamina propria B cells obtained from the nontumorous regions of surgically resected small bowels of patients with colon cancer. SB, small bowel, pt., patient. b. Flow cytometric analysis of KG2032 or L243 reactivity to microglial cells obtained from surgically resected brain tissues of patients with nonmalignant brain diseases. (Pt. 2: focal cortical dysplasia, Pt. 3: hippocampal sclerosis) c. Immunohistochemistry of KG2032 or L243 on frozen sections of normal human lung or endometrial tissues. Representative data from one of three patient samples for each tissue are shown. Scale bars, 20 mm. d. KG2032 or L243 reactivity to lung epithelial cell line H1975 treated with IFN-γ for 48 h. Data are representative of two independent experiments. e. Flow cytometric analysis of KG2032 or L243 reactivity to intestinal epithelial cells obtained from surgically resected small bowels of patients with inflammatory bowel disease (IBD: Crohn’s disease). In a, b and e, data are representative of three donors or patients. Analysis of another donor and patient is shown in Fig. 3e, f, h. f, g. Flow cytometric analysis of KG2032 or L243 reactivity to Lin−CD127+CD161+ cells, which are enriched with innate lymphoid cells, in an unaffected region of surgically resected colon (f) before and (g) after IFN-γ stimulation for 24 h. Data are representative of two samples. KG2032-reactive HLA-DRB1 alleles are shown in red and KG2032-nonreactive alleles are shown in blue.
Extended Data Fig. 5 The modified KG2032 CAR T cells react with AML cells, but not with normal epithelial cells even when stimulated with IFN-γ.
a. Constructs of VHVL-KG2032 CAR. b. Flow cytometric analysis of CAR transduction efficiencies in CAR T cells. c. Secretion of IFN-γ by the indicated CAR T cells after co-culture with KG1a AML cells. Mock-transduced T cells were used as controls (n = 2 technical replicates). d. Constructs of the VHVL86-KG2032 CAR e,f. The same analysis as b and c, comparing the VHVL-KG2032 CAR and VHVL86-KG2032 CAR. g. Constructs of the VHVL86-1XXKG2032 CAR h,i. The same analysis as b and c, comparing the VHVL86-KG2032 CAR and VHVL86-1XX-KG2032 CAR. j. Flow cytometric analysis of KG2032 reactivity to the indicated cells. k. 51Cr release assay using HLA-DRB1*04:05-expressing U937 cells as target (n = 3 technical replicates from a single experiment, repeated two times with similar results). E/T, effector/target. l. 51Cr release assay using primary AML patients’ BM cells as target (n = 5 technical replicates from a single experiment). m. Cytotoxicity assay performed via flow cytometry using either the CD4+ or CD8+ fraction of the modified KG2032 CAR or control T cells. Results of an experiment are shown. n. KG2032 or L243 reactivity to KO52 cells treated with IFN-γ for 48 h. Data are representative of two independent experiments. o-q. Secretion of IFN-γ and IL-2 by modified KG2032 CAR T cells after co-culture with (o) normal intestinal epithelial cells purified from the unaffected region of small bowel in patients with colon cancer (n = 3 technical replicates from a single experiment), (p) HT29 colon epithelial cell line or H1975 lung epithelial cell line stimulated with IFN-γ for 48 h (n = 3 technical replicates from a single experiment, repeated two times with similar results), or (q) intestinal organoid–derived epithelial cells stimulated with IFN-γ for 48 h (n = 1 for donor 1; n = 3 technical replicates for donor 2 from a single experiment for each). In each case, co-culture with KG1a cells was performed as a positive control.
Extended Data Fig. 6 Modified KG2032 CAR T cells exerted a significant anti-AML effect in vivo.
The experimental design is shown in Fig. 5i. A total of five experiments were performed. The results of another experiment are shown in Fig. 5j, k. a–d. Bioluminescence imaging of mice infused with the indicated cells. In c, quantitation of whole-body luminescence is shown below, along with the body weight of mice. In d, flow cytometric analysis of CD19 expression in KG1a cells is shown. (n = 7 per group in a; n = 4 per group in b; n = 5 per group in c and d). Data are expressed as mean ± s.e.m. P values were determined by two-tailed Welch’s t-test (a–d).
Extended Data Fig. 7 Modified KG2032 CAR T cells eradicated primary AML cells engrafted in immunodeficient mice.
The experimental design for a,b is shown in Fig. 5i. a. Bioluminescence imaging of mice infused with the indicated cells. Quantitation of whole-body luminescence is shown below, along with the body weight of mice (n = 5 per group). b. Human IFN-γ or mouse IL-6 levels in the serum of mice 1 day after infusion of unmodified or modified KG2032 CAR T cells (n = 5 per group). The experimental design for c and d is shown in Fig. 6d. c. Flow cytometric analysis of BM cells from mice transplanted with primary AML cells (UPN11) and then infused with either modified KG2032 CAR T cells or mock-transduced (control) T cells. Analysis was performed 31 d after transplantation of AML cells (n = 6 per group; a representative result for each group is shown in Fig. 6e). BM cells were aspirated from the tibia. Plots are pre-gated as PI−mTer119−mCD45− cells. AML cells were identified as mCD45−hCD3−hCD45+CD34+ cells. d. Flow cytometric analysis of BM cells in the 4 mice that survived 6 months after the infusion of modified KG2032 CAR T cells. The plots are pre-gated as PI−mTer119−mCD45− cells. In c and d, the experiment was performed with AML cells from a single patient (UPN11). In a and b, data are presented as means ± s.e.m. P values were determined by two-way ANOVA with the Bonferroni post hoc test (a) or two-tailed Welch’s t-test (b).
Extended Data Fig. 8 KG2032 CAR NK cells recognized and killed AML cells in vitro and in vivo.
a. CD107a degranulation measured by flow cytometry in CAR NK cells or control (non-transduced) NK cells upon co-culture with the indicated cells (n = 5 technical replicates from a single experiment). Experiments were performed with three different AML samples and those with two samples are shown. The result of the experiments with another AML sample is shown in Fig.7d. b. Cytotoxicity assay performed via flow cytometry. AML cells were co-cultured with either KG2032 CAR NK cells or control NK cells at an effector:target ratio of 3:1 for 48 h, and the percentages of AML cells were measured by staining with anti-CD34 or anti–CLL1 mAb. A single experiment was performed. Each combination of target and effector cells was tested in technical triplicate and one representative result is shown. c. 51Cr release assay to measure specific lysis of primary AML cells by KG2032 CAR NK cells or control NK cells (n = 5 technical replicates from a single experiment). E/T, effector/target. Experiments were performed with three different AML samples and those with two samples are shown. The result of the experiments with another AML sample is shown in Fig.7e. d, e. KG2032 CAR NK cells established from CB cells carrying KG2032-reactive HLA-DRB1 alleles. Data are representative of two independent experiments using different donors. d. Representative flow cytometric analysis of the expression of CD56, CD3, and CAR on KG2032 CAR NK cells. e. 51Cr release assay to measure specific lysis of the indicated target cells by KG2032 CAR NK cells shown in d or control NK cells (n = 3 technical replicates from a single experiment, repeated with similar results using different donors).
Extended Data Fig. 9 KG2032 CAR NK cells eradicated primary AML cells engrafted in immunodeficient mice.
The experimental design for a–e is shown in Fig. 7f. a–e. Bioluminescence imaging of mice infused with the indicated cells. Quantitation of whole-body luminescence is shown below. A total of six independent experiments were performed with different CB donors, and the results of all mice in each experiment are shown separately in a–e (a. n = 6 in the control NK group and n = 5 in the KG2032 CAR NK group, b. n = 5 per group, c. n = 4 per group, d. n = 6 per group, e. n = 6 per group). The results of another experiment are shown in Fig. 7g, h. f. Experimental design for g–i. g. Bioluminescence imaging of mice infused with the indicated cells. h. Quantification of whole-body luminescence. i. Mouse survival curves. In f-i, results of an experiment are shown. j. The experimental design is shown in Fig. 8d. Flow cytometric analysis of BM cells from mice infused with the indicated cells 29 d after transplantation of AML cells. (n = 5 per group). Analyses of other mice are shown in Fig. 8e. AML cells: mCD45−hCD34+, CAR NK cells: mCD45−hCD34−hCD56+CAR+. Data are expressed as means ± s.e.m. P values were determined by two-tailed Welch’s t-test (a–e, h) and log-rank test (i).
Supplementary information
Supplementary Information
Supplementary Figs. 1 and 2 and uncropped scans of the gel in Supplementary Fig. 2.
Supplementary Tables 1–3
Supplementary Table 1: Patient characteristics. Supplementary Table 2: The frequency of KG2023-reactive HLA-DRB1 alleles in Japanese, European white and African American populations. Supplementary Table 3: Summary of antibodies used in this study.
Source data
Source Data Figs. 1–8 and Extended Data Figs. 1–9
Statistical source data
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ikeda, S., Hasegawa, K., Kogue, Y. et al. CAR T or NK cells targeting mismatched HLA-DR molecules in acute myeloid leukemia after allogeneic hematopoietic stem cell transplant. Nat Cancer 6, 595–611 (2025). https://doi.org/10.1038/s43018-025-00934-1
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s43018-025-00934-1
This article is cited by
-
Exploiting HLA-DR mismatches for CAR therapy in acute myeloid leukemia
Nature Cancer (2025)