Abstract
In the treatment of ovarian cancer, PARP inhibitors such as olaparib have shown promise, yet resistance in advanced cases remains a significant challenge. Our study identified SLC7A5 as a key gene associated with olaparib resistance through differential gene expression and prognostic analysis. High SLC7A5 expression was found to correlate with poor prognosis. In vivo and in vitro experiments revealed that SLC7A5 enhances olaparib resistance by stabilizing ACLY and promoting de novo lipid synthesis via the ERBB2 axis, independent of leucine. Mechanistically, SLC7A5 upregulates ERBB2 transcription through ELK1, and ERBB2 competes with CUL3 to prevent ACLY degradation. These findings suggest that targeting SLC7A5 may reverse olaparib resistance, offering new strategies for combination therapies and improving clinical outcomes in ovarian cancer treatment.

Similar content being viewed by others
Introduction
Ovarian cancer is the third most common cancer in women and accounts for ~20% of all female tumor-related deaths each year [1]. Given the high lethality, recurrence rate, and low cure rate of ovarian cancer, early diagnosis and treatment are particularly challenging. Chemotherapy remains the primary treatment modality in clinical practice. Recently, PARP inhibitors (PARPi) have emerged as a novel targeted therapy for epithelial ovarian cancer. Among them, Olaparib induces apoptosis in tumor cells via a synthetic lethal mechanism, marking a significant advancement in ovarian cancer treatment [2, 3].Unfortunately, patients with advanced ovarian cancer show strong resistance to Olaparib [2]. Overcoming the olaparib resistance is a major challenge in clinical oncology.
SLC7A5 is a highly conserved ion channel protein that is involved in cellular amino acid transport. It plays a crucial role in maintaining normal cell morphology, regulating cellular metabolism, and participating in immune regulation [4]. Studies have shown that SLC7A5 plays an important role in breast cancer treatment [5]. Leucine transported by SLC7A5 also stimulates the mTOR pathway to make cells or drug-resistant [6]. The role of SLC7A5 in Olaparib resistance in ovarian cancer remains unclear. Intriguingly, RNA-seq data revealed that A2780 Olaparib-resistance exhibited significantly higher expression levels of SLC7A5 [7]. Subsequent analysis of amino acid content in Olaparib-resistant ovarian cancer cells showed no significant changes in leucine levels, while glutamine, glycine, and aspartate levels were decreased [8]. These findings suggest that SLC7A5 may contribute to Olaparib resistance in a leucine-independent manner. Further investigation is required to elucidate the precise mechanisms underlying this resistance.
Cancer cells adapt to harsh conditions by reprogramming glycolysis and lipid metabolism, ensuring rapid energy supply. Inhibition of fatty acid oxidation (FAO) boosts olaparib’s cytotoxicity, implying that lipid metabolism remodeling could contribute to olaparib resistance [9]. However, whether lipid metabolic remodeling is directly involved in olaparib resistance and the underlying molecular mechanisms need to be further investigated. In this study, our clinical tissue analysis of ovarian cancer revealed that SLC7A5 was positively correlated with ovarian cancer stage and overexpressed in olaparib-resistant cell lines. Mechanistically, we demonstrated that SLC7A5 induced ELK1 phosphorylation and entered the nucleus to promote ERBB2 expression. Excess ERBB2 competitively binds to CUL3, thereby maintaining ACLY stability in the cell and, ultimately, metabolic remodeling. These findings provide a new drug target for the treatment of advanced ovarian cancer.
Materials and methods
Cell culture
Cells were grown in a humidified chamber at 37 °C and 5% CO2. The HEY (iCell-h364) and Caov3 (iCell-h037) cell lines were maintained in DMEM supplemented with 10% fetal bovine serum (FBS, iCell-0300) and 1% penicillin-streptomycin. SK-OV-3 (iCell-h195, RRID:CVCL_0532) and OVCAR8 (iCell-h385, RRID:CVCL_1629) cells were cultured in McCoy’s 5A and RPMI 1640 media, respectively, with identical supplements. All cell lines were purchased from Cellverse Co., Ltd.
Cell transfection and viral knockdown
These cells were seeded in 10 cm dishes at a density of 5 × 106 cells per dish 24 h prior to transfection. Opti-MEM was replaced before transfection, and the plasmids were transfected into the cells using Lipofectamine 2000 (#11668027, Invitrogen). The cells were seeded in 6-well plates at a density of 105 cells/well. The following day, cells were infected with lentivirus containing the shRNA construct at a multiplicity of infection (MOI) of 50. Replaced with a new medium 6 h later, and cells were incubated for another 48–72 h. Successful knockdown of the target gene was confirmed using qRT-PCR and western blotting.
CCK-8 assay
To measure cellular proliferation and cytotoxicity, CCK-8 experiments were conducted. The cells were plated at a density of 2000 cells/well in 96-well plates. Subsequently, 10 μL of CCK-8 solution (C0038, Beyotime) was added to each well and cultured for 2 h. The analysis was then analyzed for five consecutive days.
Drug treatments
For in vitro treatment, olaparib (HY-10162) was administered according to the cell IC50. SB204990 (HY-16450, 100 μM), TDE (HY-P2262, maximum concentration of 5 μM), and MG132 (HY-13259, 10 μM) were purchased from MCE. For the in vivo experiments, olaparib was injected intraperitoneally into mice at a concentration of 25 mg/kg. All drugs were dissolved in dimethyl sulfoxide (DMSO).
Leucine dependence assay
Cells transfected to overexpress SLC7A5 were cultured in complete medium for 48 h. Subsequently, cells were washed with phosphate-buffered saline (PBS) and resuspended in leucine-deprived medium (prepared using DMEM [Yuchun Biology, Cat# DZ2001] or RPMI-1640 [Yuchun Biology, Cat# YC-1042]). To assess leucine dependence of SLC7A5-mediated Olaparib resistance, leucine was supplemented back into the medium of designated samples.
IC50
The CCK-8 assay was also employed to determine the half-maximal inhibitory concentration (IC50) of olaparib in different cell lines. A total of 3000 cells were seeded into each well of a 96-well plate. Following cell adhesion, olaparib was added at concentrations of 0, 0.5, 1, 2, 5, 10, and 20 µM. After a 24-h treatment period, the IC50 values for the respective cell lines were evaluated using the CCK-8 assay.
Colony formation assay
Cells from various groups were inoculated and cultured in a six-well plate at a density of 103 cells per well and maintained in medium for 15 days to facilitate colony formation. Following this incubation period, the colonies were fixed using 4% PFA for 30 min and washed with phosphate-buffered saline (PBS). The colonies were then stained with 1% crystal violet for 30 min, followed by three washes with PBS to remove excess dye. After drying, photographs were taken and colonies consisting of more than 100 cells were counted. In this study, each experimental condition was replicated three times. The colony formation rate was calculated using the following formula: (number of treated colonies/number of control colonies) × 100%.
IHC and HE Staining
Tumor tissues were collected from the mice and fixed in 4% paraformaldehyde for 24 h before being embedded in paraffin after the mice were sacrificed. Paraffin-embedded samples were then sectioned into 10 μm slices for staining. Prior to the staining process, the sections were deparaffinized and rehydrated, followed by heat-induced epitope retrieval using citrate buffer at 121 °C for 30 min. To inhibit endogenous peroxidase activity, sections were treated with 0.3% hydrogen peroxide in methanol for 30 min. Non-specific binding was mitigated by blocking with 10% normal bovine serum, after which the sections were incubated overnight at 4 °C with rabbit polyclonal antibodies targeting SLC7A5(28670-1-AP, Proteintech), ERBB2(84046-1-RR, Proteintech), and Ki67 (27309-1-AP, Proteintech). Following After washing with PBS, the sections were incubated with a horseradish peroxidase-conjugated secondary antibody at 37 °C for 2 h. Hematoxylin was used for counterstaining to facilitate visualization, and the operation of the tissue chip (HOvaC151Su01, SHANGHAI OUTDO BIOTECH CO.,LTD.) was the same. The chip ethics is provided by the Ethics Committee of Shanghai Superchip Biotech Co., Ltd.
Flow cytometry assay
Apoptosis was quantified using an Annexin V-APC Apoptosis Detection Kit (E-CK-A217, Elabscience). Following digestion and collection of the cells, they were resuspended in 1X binding buffer. After achieving a concentration of 105 cells/mL, annexin V- APC (5 μL) and PI (5 μL) were added, and the mixture was incubated in the dark for 20 min. The percentage of apoptotic cells was assessed using a flow cytometer (BD Biosciences) within 1 h of staining. This procedure was conducted in triplicate to ensure reliability of the results.
EDU staining assay
The cells were incubated in a medium supplemented with EdU (ST067, Beyotime, 20 mmol/L) for 2 h. Subsequently, cells were fixed with 4% PFA at for 20 min. EdU-positive cells were analyzed across various treatment groups.
RT–qPCR
RNA was extracted using the TRIzol method. RNA was reverse-transcribed into cDNA using HiScript III RT SuperMix for qPCR (R323-01, Vazyme). qPCR was performed using the PerfeCTa SYBR Green SuperMix Reaction Mixes (95054-500, QuantaBio) and the primers listed below. The Cq values were normalized to GAPDH as a reference gene. The primer sequences are listed in Table 1.
Western blots
Total protein was extracted from cells or tissues using RIPA buffer supplemented with 1 mmol/L PMSF (ST506, Beyotime) and quantified using the BCA Protein Assay Kit (P0009, Beyotime). Protein samples were separated by SDS-PAGE and transferred to a PVDF membrane (Millipore). After blocking with 5% BSA in PBST for 1 h, the PVDF membrane was incubated with primary antibodies at 4 °C overnight. The antibodies used were ACLY (15421-1-AP, Proteintech, RRID: AB_2223741), SLC7A5(28670-1-AP, Proteintech, RRID: AB_2918188), ERBB2(84046-1-RR, Proteintech), KLHL25 (67774-1-Ig, Proteintech, RRID: AB_2918539), CUL3(83619-5-RR, Proteintech), ubiquitin (10201-2-AP, Proteintech, RRID: AB_671515), GAPDH(60004-1-Ig, Proteintech, RRID: AB_2107436), and Beta Actin (866009-1-Ig, Proteintech, RRID: AB_2687938). After three washes, the membrane was treated with horseradish peroxidase-conjugated secondary antibodies for 1 h at room temperature. Finally, enhanced chemiluminescence (P0018S, Beyotime) was used to visualize the immunoblot. The intensity of the western bands was quantified using ImageJ software (RRID:SCR_003070).
co-IP
Cells were treated with the indicated drugs and collected after 8 h of treatment by washing with PBS and scraping. The pelleted cells were lysed using EBC lysis buffer (50 mM Tris, pH 8.0, 150 mM NaCl, 0.5% NP-40, 1:10,000 β-mercaptoethanol, 0.5 mM EDTA) supplemented with protease inhibitors (11836153001, Sigma-Aldrich) and phosphatase inhibitors (4906837001, Sigma-Aldrich). Cell lysates were incubated on ice for 20 min and cleared by centrifugation at 16,000 × g for 20 min. Protein extracts were quantified using BCA reagent, and then equal quantities were incubated with STING (13647S, Cell Signaling) or IgG (2729S, Cell Signaling) antibodies and magnetic protein A/G beads overnight with end-over-end agitation at 4 °C. Beads were washed five times with EBC lysis buffer before immunoprecipitated proteins were eluted using SDS sample buffer and analyzed by western blotting (described above).
Chromatin immunoprecipitation assay (ChIP)
Cells were counted 107 in native medium. According by Pierce™ Agarose ChIP Kit(26156, ThermoFisher), cells were cross-linked by 1% formaldehyde for 10 min at 37 °C before quenching with 0.125 M glycine for 5 min. Chromatin was isolated by first incubating with buffer 1, resuspending the pellet in buffer 2, and finally resuspending the cell pellet in buffer 3 and 0.5% N-laurosarcosine (L7414, Sigma-Aldrich). Isolated chromatin was then sonicated for 30 cycles of 30 s on, 30 s off using a Diagenode BioRuptor Plus sonicator on ‘high. ’ Soluble chromatin was immunoprecipitated overnight with SLC7A5(28670-1-AP, Proteintech) antibodies. Immunoprecipitated chromatin was captured using magnetic protein A/G beads (88803, Pierce). Immunoprecipitated chromatin was washed three times with low-salt buffer (0.1% SDS, (15553-035, Invitrogen), 1% Triton X-100, 2 mM EDTA pH 8, 20 mM Tris-HCl pH 8, and 150 mM NaCl), high-salt buffer three times (0.1% SDS, 1% Triton X-100, 2 mM EDTA pH 8, 20 mM Tris-HCl pH 8, 500 mM NaCl), LiCl buffer three times (0.25 M LiCl (62476, Sigma-Aldrich), 1% NP-40, 1 mM EDTA pH 8, 10 mM Tris-HCl pH 8, 1% sodium deoxycholate), and TE buffer (10 mM Tris-HCl pH 8, 10 mM EDTA pH 8) before elution in elution buffer (50 mM Tris-HCl pH 8, 10 mM EDTA, 1% SDS). The eluted chromatin was purified using the QIAquick PCR Purification kit (28106, Qiagen). Relative abundances of BMF and GATA3 promoters in immunoprecipitated chromatin were measured by qPCR using PerfeCTa SYBR Green SuperMix Reaction Mixes (95054-500, QuantaBio) and the primers listed below. The percentage of input values was calculated for each immunoprecipitation. Fold-change enrichment values were calculated by normalizing to the percentage input of the appropriate IgG control for each immunoprecipitation.
Dual luciferase reporter assay
The dual-luciferase plasmid was transfected into target cells according to the method of plasmid transfection for 48 h, and then the Dual Luciferase Reporter Gene Assay Kit II (RG029, Beyotime) was added to the lysis solution for 15 min at room temperature. Add 10 μL of the supernatant into a 96-well plate. Detection according to the instruction method and avoidance of light.
Tumour xenograft assay
Female 5–6-week-old nude mice were obtained from the Nanjing Model Animal Research Center. A tumor model was established by subcutaneously injecting 106 SK-OV-3 cells into the mice. Once the tumor volume reached ~50 mm³, the mice were randomly divided into control and experimental groups. Tumor length and width were obtained using a caliper, and tumor volumes were calculated using the formula: L × (W)2/2. At the end of the treatment period, the tumor weights were recorded. The animal ethics meets the requirements of the Harbin Medical University Institutional Research Board Approval Report.
Detection of lipid metabolism
All indices of lipid metabolism were assessed according to the manufacturer’s guidelines. A total of 106 cells were harvested from each experimental group, and the procedures were conducted following the protocols outlined in the Free Fatty Acid (FFA) (BC0590, Solarbio), triglyceride (TG) (BC0620, Solarbio), and Total Cholesterol (TC) (BC1980, Solarbio) kits.
LD540
The cells grown on coverslips were fixed with 4% PFA for 15 min at room temperature. After washing with PBS, the dye solution was prepared according to the instructions of the LD540 Staining Kit (C2050S, Beyotime) and incubated in the dark for 20 min at room temperature. After washing twice with PBS, the film was sealed with a sealing tablet containing DAPI and images were captured using a 560 nm laser.
Statistical analysis
Statistical analyses were conducted using a two-tailed Student’s t-test or one-way ANOVA for in vitro studies, Mann-Whitney U test for in vivo studies, and log-rank test for Kaplan-Meier survival curves. less than 0.05 was considered statistically significant. Unless otherwise specified, all data were expressed as mean standard deviation, and all statistical values were considered statistically significant.
Results
SLC7A5 as a potential target for olaparib resistance in ovarian cancer
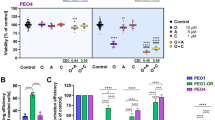
To elucidate the molecular mechanisms of olaparib resistance in ovarian cancer (OV), we performed differential gene expression analysis comparing olaparib-resistant and -sensitive A2780 cells using data from the GSE153867 database [7]. Integrating prognostic analyses of TCGA and GSE26712 datasets, we identified potential resistance-associated genes (Fig. 1A, B, Supplementary Fig. 1A, Table 2). Knockdown of these genes in A2780 cells revealed that SLC7A5 suppression significantly enhanced olaparib sensitivity, as confirmed by cell viability assays (Supplementary Fig. 1B–E). To further validate the association between SLC7A5 and Olaparib resistance, we used western blotting and PCR to compare SLC7A5 protein levels in various OV cell lines. We found higher SLC7A5 expression in HEY and SK-OV-3 cells compared to IOSE80 cells, while CAOV-3 and OVCAR-8 cells exhibited relatively lower expression (Fig. 1C). IC50 assays demonstrated that elevated SLC7A5 expression correlates with increased olaparib resistance. (Fig. 1D, E). Drug sensitivity assays revealed that SLC7A5 knockdown in HEY and SK-OV-3 cells decreased olaparib resistance, while its overexpression in CAOV-3 and OVCAR-8 cells increased resistance (Supplementary Fig. 1G). To determine if SLC7A5’s role in PARPi resistance extends beyond Olaparib, we evaluated Rucaparib sensitivity. SLC7A5-OE significantly conferred Rucaparib resistance in ovarian cancer cells, while conversely, SLC7A5-KD markedly enhanced Rucaparib sensitivity (Supplementary Fig. 1F). These findings collectively highlight SLC7A5 as a potential target of olaparib resistance in ovarian cancer, with its expression levels and genetic manipulation significantly influencing drug sensitivity.
A Venn diagram illustrating the screening logic for SLC7A5. Integrated analysis of GSE153867, GSE26712, and OS databases identified nine genes highly expressed in drug-resistant cells with intersecting prognostic values. B Gene Expression Analysis. Analysis of the GSE153867 dataset revealed elevated SLC7A5 expression in Olaparib-resistant A2780 cells (C1–C8) compared to their parental controls (O1–O8). C WB. Western blot revealed higher SLC7A5 expression in HEY and SK-OV-3 cells compared to IOSE80, and lower expression in CAOV-3 and OVCAR-8 cells. D, E Drug Resistance Assay. IC50 values showed resistance to olaparib with high SLC7A5 expression. F WB. Western blot showed significant increases in SLC7A5 expression in olaparib-resistant cells. G Colony-formation assay. Knockdown of SLC7A5 significantly reduced Olaparib resistance in resistant cells. Left. HEY-R. Right. SK-OV-3-R. Plating 1000 cells per well with drug concentrations set at their respective IC50 values. H Statistical graph of Edu Assay. Edu incorporation assay showed that SLC7A5 knockdown significantly inhibited the proliferation of resistant cells treated with Olaparib. Edu incubation time was 2 h. Blue. DAPI, Green. Edu. Statistical graph displays Edu-positive rate in all cells. I Apoptosis Assay. SLC7A5 knockdown increased sensitivity to Olaparib in resistant cells. Cell apoptosis levels were detected using Annexin V and PI. J Colony-formation assay. The addition of leucine does not affect the regulation of olaparib resistance by SLC7A5. Plating 1000 cells per well with drug concentrations set at their respective IC50 values. Leucine (1 μM) treatment for 48 h. K Apoptosis Assay. SLC7A5’s regulation of olaparib resistance is leucine-independent. Cell apoptosis levels were detected using Annexin V and PI. L Statistical graph of Edu Assay. The addition of rapamycin does not affect the regulation of olaparib resistance by SLC7A5. Rapamycin (18.74 μM) treatment for 24 h. Edu incubation time was 2 h. Blue. DAPI, Green. Edu. Statistical graph displays Edu-positive rate in all cells. M Kaplan−Meier survival curves of nude mice implanted orthotopically with SK-OV-3 R cells. In a xenograft survival study conducted in nude mice, 5 million SK-OV-3 R cells were subcutaneously injected. Mice were divided into four groups, NC + vehivel, SLC7A5 + vehivel, NC + lue and SLC7A5 + leu. Once the tumors reached a volume of 50 mm³, each group was randomly divided into control and treatment subgroups. The treatment subgroup received olaparib every other day for a total of eight doses at 25 mg/kg. In accordance with animal ethics, mice were euthanized when tumor volumes reached 1000 mm³, which was recorded as the endpoint for survival. The median survival times were as follows. 40 days for the NC + vehivel group, 42.5 days for the NC + leu group, 20 days for the SLC7A5 + vehivel group, 17.5 days for the SLC7A5 + lue group (n = 10). N In vivo experiments. In vivo experiments demonstrated that SLC7A5 knockdown enhanced the inhibitory effect of Olaparib on ovarian cancer (n = 6). Upper panel. Schematic of in vivo treatment protocol. Lower panel. Tumor images. O Tumor weight (g). All in vitro findings are derived from a minimum of three independent experiments. Error bars denote the standard deviation. Statistical significance is represented as follows. * = P < 0.05, ** = P < 0.001, *** = P < 0.0001, ns = non-significant in comparison to normal or control treatment.
SLC7A5 knockdown enhances olaparib sensitivity independently of leucine
To further investigate whether SLC7A5 influences ovarian cancer olaparib sensitivity, we established Olaparib-resistant HEY (HEY R) and SK-OV-3 (SK-OV-3 R) using a gradient of olaparib treatment. We also compared SLC7A5 expression levels between parental and resistant cells and found that the resistant cells exhibited significantly higher SLC7A5 protein levels (Fig. 1F, Supplementary Fig. 1H, I). The colony formation assay showed that SLC7A5 knockdown markedly enhanced olaparib sensitivity in HEY R and SK-OV-3 R (Fig. 1G), the result supported by the EdU assay (Fig. 1H, Supplementary Fig. 1J). As Olaparib inhibits tumor progression by inducing apoptosis through synthetic lethality, we found that SLC7A5 knockdown further increased olaparib-induced apoptosis in HEY R and SK-OV-3 R (Fig. 1I).
We explored whether leucine participates in SLC7A5-mediated olaparib resistance by overexpressing SLC7A5 and culturing cells for 24 h in either normal or leucine-rich medium before treating them with olaparib. Interestingly, leucine addition did not significantly affect SLC7A5-regulated resistance (Fig. 1J, K, Supplementary Fig. 1K). Further experiments with rapamycin, an mTOR inhibitor, revealed that mTOR inhibition did not alter SLC7A5-mediated olaparib resistance (Fig. 1L). In subsequent experiments, we employed a nude mouse model to assess the influence of leucine on SLC7A5-mediated olaparib resistance. Nude mice were subcutaneously inoculated with SK-OV-3, and olaparib treatment (25 mg/kg) was commenced upon reaching a tumor volume of 50 mm³. Tumors were allowed to grow to 1000 mm³ before the mice were humanely sacrificed. The experiment demonstrated that overexpression of SLC7A5 substantially enhanced olaparib resistance. Notably, the intake of leucine did not further amplify this effect (Fig. 1M). In vivo experiments involving subcutaneous implantation of SK-OV-3 R cells in nude mice, followed by olaparib treatment (25 mg/kg) once tumors reached 50 mm2, demonstrated that SLC7A5 knockdown significantly enhanced the inhibitory effects of olaparib on ovarian cancer (Fig. 1N, O). Based on these findings, we hypothesize that in ovarian cancer, SLC7A5 may regulate olaparib resistance in a leucine-mTOR-independent manner.
SLC7A5 promotes ovarian tumorigenesis in vitro and in vivo
Our earlier experiments indicated that SLC7A5 induces olaparib resistance in ovarian cancer independently of its leucine transport function. Clinical observations indicate that Olaparib resistance is predominantly associated with advanced-stage patients [10, 11]. Accordingly, we performed tissue microarray analysis to examine the expression of SLC7A5. Interestingly, SLC7A5 expression was positively correlated with the pathological grade, with higher expression levels observed in more advanced stages (Fig. 2A, B, Table 3). Additionally, survival analysis revealed that increased SLC7A5 expression was associated with a poorer prognosis and shorter survival (Fig. 2C, Supplementary Fig. 2A, B). This discovery has implications for drug development and clinical applications, warranting further investigation into SLC7A5’s role in ovarian cancer progression.
A Immunohistochemistry (IHC). IHC revealed elevated SLC7A5 expression in late-stage patients, which was positively correlated with pathological grade. B Statistical graph of IHC. The correlation between SLC7A5 expression and tumor staging was analyzed in tissue microarray samples. Results showed that SLC7A5 expression increased with tumor progression. Stage I (3/8), Stage II (9/34), Stage III (36/64), and Stage IV (25/26). This suggests a potential role of SLC7A5 in advanced tumor development. C Survival Analysis. Kaplan-Meier analysis indicated that patients with high SLC7A5 expression had poorer prognoses and shorter survival times. D, E WB. The high expression efficiency of SLC7A5 and the knockdown efficiency using lentiviral shSLC7A5 #1 and #2 were detected in HEY-R and SK-OV-3-R cells. Overexpression was achieved using SLC7A5 plasmid in CAOV-3 and OVCAR-8 cells. F, G Colony-formation assay. SLC7A5 knockdown significantly inhibited cell proliferation, whereas overexpression promoted it. F HEY-R and SK-OV-3-R, G Caov-3 and Ovcar-8. Plating 2000 cells per well. H, I Apoptosis Assays. SLC7A5 knockdown increased apoptosis in cells, whereas overexpression SLC7A5 inhibited it. H HEY-R and SK-OV-3-R, I Caov-3 and Ovcar-8. J–L In vivo experiments. Subcutaneous tumor growth in nude mice with SK-OV-3-R cells showed that SLC7A5 knockdown inhibited tumor growth. J Bioluminescence Imaging. K Tumor volume (mm3). M IHC Analysis. IHC showed a significant decrease in Ki67 expression following SLC7A5 knockdown (n = 6). All the in vitro data were obtained from at least three independent experiments. Error bars represent the standard deviation. Statistical significance is denoted as follows. * P < 0.05, ** P < 0.001, *** P < 0.0001, ns non-significant compared to the normal or control treatments.
Assessing the impact of SLC7A5 on tumor progression in vitro involved knocking down SLC7A5 in HEYR and SK-OV-3R cells, and overexpressing it in CAOV-3 and OVCAR-8 cells to evaluate its effects on cellular functions. (Fig. 2D, E, Supplementary Fig. 2C). The colony formation assay revealed that SLC7A5 knockdown significantly inhibited the proliferation of HEY-R and SK-OV-3-R cells (Fig. 2F, Supplementary Fig. 2D), while SLC7A5 overexpression in CAOV-3 and OVCAR-8 cells enhanced proliferation (Fig. 2G) Apoptosis assays showed that SLC7A5 knockdown increased apoptosis (Fig. 2H), whereas overexpression decreased apoptosis(Fig. 2I). In vivo experiments with subcutaneous SK-OV-3R cell implantation in mice demonstrated that SLC7A5 knockdown significantly suppressed tumor growth (Fig. 2J–M). These results demonstrate that SLC7A5 facilitates the progression of ovarian cancer.
In summary, Identifying SLC7A5 as a pivotal regulator of olaparib resistance and ovarian cancer progression, the data presented demonstrate its operation independent of the leucine-mTOR pathway, underscoring its significance as a potential therapeutic target and prognostic indicator.
SLC7A5 stimulates the de novo synthesis of lipid metabolism
Previous studies have implicated SLC7A5 in the regulation of chemoresistance through the leucine-mTOR signaling pathway in breast cancers [6]. In contrast, our investigation into olaparib resistance in ovarian cancer has revealed that neither leucine supplementation nor mTOR pathway inhibition affects SLC7A5-mediated resistance (Fig. 1J–M). This finding suggests that in ovarian cancer, SLC7A5 may engage alternative pathways to modulate resistance to olaparib. To investigate the mechanism by which SLC7A5 regulates olaparib resistance, a Gene Set Enrichment Analysis (GSEA) was conducted using The Cancer Genome Atlas (TCGA) database for ovarian cancer (Supplementary Fig. 3A). The analysis revealed that SLC7A5 upregulates multiple lipid metabolism-related pathways (Supplementary Fig. 3B). Based on these findings, we hypothesize that SLC7A5 may influence de novo lipid synthesis. Subsequently, we performed a correlation analysis between SLC7A5 and the expression of genes involved in the de novo lipid synthesis pathway. SLC7A5 significantly enhances the activation of the de novo lipid synthesis pathway (Fig. 3A, B). ELISA measurements demonstrated that knocking down SLC7A5 significantly suppressed levels of Free Fatty Acids, Total Cholesterol, and Triglycerides (Fig. 3C). Conversely, overexpression of SLC7A5 led to increased levels of these lipid components (Supplementary Fig. 3C). Lipid droplet assays conducted SLC7A5 knockdown showed a pronounced suppression of lipid droplet accumulation in cells (Fig. 3D). Concurrently, immunohistochemistry (IHC) analysis indicated a significant reduction in the protein levels of ACLY upon SLC7A5 knockdown (Fig. 3E). These experimental findings collectively demonstrate that SLC7A5 knockdown markedly inhibits the activation of the lipid metabolic synthesis pathway.
A GSEA. The enrichment analysis results of gene SLC7A5 in the fatty acyl-CoA biosynthesis pathway highlight its potential significance. With a normalized enrichment score (NES) of 1.5658, a P-value of 0.031, and a false discovery rate (FDR) of 0.0927, it suggests that SLC7A5 may play a crucial role in this biosynthetic process, warranting further investigation and study. B Correlation Analysis. Analysis of TCGA database revealed a positive correlation between SLC7A5 and de novo lipid metabolism pathway-associated proteins ACLY, ACACA, and FASN, suggesting that SLC7A5 is involved in lipid metabolism remodeling. C ELISA. Measurements of Tri-Glycerides, Free Fatty Acids, and Total Cholesterol showed that SLC7A5 knockdown significantly inhibited lipid metabolism in HEY and SK-OV-3 cells. Cells (5 × 106, lysed by sonication. D Lipid Droplet Staining. LD540 staining revealed reduced lipid droplets upon SLC7A5 knockdown. Red fluorescence was observed using a fluorescence microscope at an excitation wavelength of 537 nm. E Immunohistochemistry (IHC). IHC analysis of tumor samples from mice showing reduced ACLY expression following SLC7A5 knockdown. F ELISA. ELISA analysis revealed that SB 204990, an ACLY inhibitor, significantly inhibited lipid metabolism remodeling caused by SLC7A5 overexpression in CAOV-3 and OVCAR-8 cells. Cells were treated with 100 µM SB 204990 for 24 h. G Lipid Droplet Staining. LD540 staining shows a significant reduction in lipid droplet accumulation induced by SLC7A5 overexpression following treatment with SB 204990. H Western Blot Analysis. SB 204990 inhibited the upregulation of ACLY, ACACA, and FASN proteins induced by SLC7A5 overexpression. I–K SB 204990 treatment markedly suppressed the increased Olaparib resistance in CAOV-3 and OVCAR-8 cells caused by SLC7A5 overexpression. I Colony formation assay. J Cell viability assay. K Apoptosis Assay. All in vitro data were derived from at least three independent experiments. Error bars represent standard deviation. Statistical significance is indicated by the following symbols. * P < 0.05, ** P < 0.001, *** P < 0.0001, ns non-significant compared to the normal or control treatments.
SLC7A5-activated de novo lipid metabolism pathway is critical for olaparib resistance
To determine whether SLC7A5 regulates olaparib resistance through de novo lipid metabolism in ovarian cancer, we treated cells with SB204990, the ACLY inhibitor, and assessed its impact on cell viability and olaparib resistance. The results showed that SB204990 not only significantly inhibited the cellular levels of Free Fatty Acids, Total Cholesterol, and Triglycerides (Fig. 3F) but also reduced intracellular lipid droplet levels (Fig. 3G) and suppressed the expression of lipid metabolism-associated enzymes (Fig. 3H). Interestingly, under the influence of SB204990, the effect of SLC7A5 on the regulation of olaparib resistance diminished (Fig. 3I). The IC50 assay (Fig. 3J) and apoptosis (Fig. 3K) results supported this conclusion. The findings suggest that SLC7A5 may modulate olaparib resistance via the de novo lipid synthesis pathway.
SLC7A5 regulates de novo lipid synthesis in an ERBB2-dependent manner
Previous experiments have demonstrated that SLC7A5 can regulate de novo lipid synthesis, which is not entirely dependent on the mTOR signaling pathway. To elucidate the mechanism by which SLC7A5 regulates de novo lipid synthesis, we identified pathways potentially involved in this process from the TCGA database (Supplementary Fig. 4A). Our findings indicate that ERBB2 may be implicated in the regulation of de novo lipid synthesis by SLC7A5 (Fig. 4A, B). Although ERBB2 has been reported to promote Platinum resistance in TP53-positive ovarian cancer, its role in olaparib resistance remains unclear [12, 13]. Additionally, we utilized the STRING database to screen for potential interacting proteins of ACLY, ACACA, and FASN, and performed an intersection analysis with pathway molecules. We observed that ERBB2 interacts with CUL3 (Table 4), and the CUL3-KLHL25 complex ubiquitinates and degrades ACLY [14]. Thus, we hypothesize that ERBB2 may competitively bind to the CUL3-KLHL25 complex, preventing ACLY from being subjected to proteolytic degradation. To validate the hypothesis, we initially treated ERBB2-knockdown cells with MG132 to determine if ACLY protein levels would change. The experimental results demonstrated that treatment with MG132 could restore ACLY protein levels (Fig. 4C), suggesting that ERBB2 regulates ACLY via the proteasome pathway. Knockdown of ERBB2 in HEY and SK-OV-3 cells led to shortened ACLY half-life (Fig. 4D) and increased ubiquitination (Fig. 4E). Co-IP experiments with CUL3 antibody under conditions of ERBB2 overexpression showed that CUL3 preferentially binds to ERBB2, inhibiting the formation of the CUL3-KLHL25 complex (Fig. 4F). In further experiments, we knocked down CUL3 and KLHL25 alongside ERBB2 and observed that the knockdown of CUL3 and KLHL25 significantly reversed the degradation of ACLY by ERBB2 (Fig. 4G, H). All results to date substantiate that ERBB2 modulates ACLY protein levels through the CUL3-KLHL25 complex.
A GSEA. The analysis shows a significant enrichment with a high NES of 2.0577, a P-value less than 0.001, and an FDR of 0.0119, indicating a strong association between SLC7A5 and this signaling pathway. B Correlation Analysis. TCGA database analysis revealed a positive correlation between ERBB2 and de novo lipid metabolism pathway-associated proteins ACLY, ACACA, and FASN, suggesting ERBB2 is involvement in lipid metabolism remodeling. C Proteasome Inhibition Assay. Knockdown of ERBB2 significantly reduces ACLY levels (line 2), which are restored upon treatment with 50 µM MG-132 for 6 h (line 4). D CHX Half-life Assay. ERBB2 knockdown shortens the half-life of ACLY (Left). Nuclear-cytoplasmic fractionation revealed that ERBB2 signaling selectively regulates ACLY activity/modification in the cytoplasmic compartment (Right). CHX. 10 µg/mL. E Denaturing IP Assay. Overexpression of ERBB2 decreases ACLY ubiquitination. F Co-Immunoprecipitation (Co-IP) Assay. Overexpression of ERBB2 enhances binding between CUL3 and ERBB2, while reducing binding between CUL3 and KLHL25. All in vitro data are derived from at least three independent experiments. G Proteasome Inhibition Assay. SLC7A5 knockdown increases ACLY (line 2), but ERBB2 knockdown further decreases ACLY levels (line 3). This decrease is inhibited by 50 µM MG-132 for 6 h (line 6). H Denaturing IP Assay. ERBB2 knockdown increases ACLY ubiquitination (line 2), which is reduced upon additional knockdown of CUL3(line 3) or KLHL25 (line 4). I Proteasome Inhibition Assay. ERBB2 knockdown decreases ACLY (line 2), but this decrease is reversed by knockdown of CUL3(line 3) or KLHL25 (line 4). MG-132 treatment (50 µM for 6 h) restores ACLY expression (line 7, 8). J Denaturing IP Assay. SLC7A5 overexpression reduces ACLY ubiquitination (line 2), which is increased upon ERBB2 knockdown (line 3). K ELISA. ELISA measurements of Tri-Glycerides, Free Fatty Acids, and Total Cholesterol showed that ERBB2 knockdown reverses the lipid metabolism remodeling caused by SLC7A5 overexpression. L Lipid Droplet Staining. LD540 staining demonstrated a reduction in lipid droplet accumulation induced by SLC7A5 following ERBB2 knockdown. Error bars represent standard deviation. Statistical significance is denoted as follows. * = P < 0.05, ** = P < 0.001, *** = P < 0.0001, ns = non-significant compared to normal or control treatment.
Upon further investigation, we overexpressed SLC7A5 and simultaneously knocked down ERBB2, and observed that ACLY ubiquitination decreased with reduced ERBB2 levels (Fig. 4I, J). This finding indicated that SLC7A5 regulation of ACLY is dependent on ERBB2 protein levels. Collectively, these results demonstrate that ERBB2 maintains ACLY protein stability by competitively binding to CUL3, thereby regulating lipid metabolic remodeling. ELISA (Fig. 4K) and lipid droplet assays (Fig. 4L) showed that ERBB2 knockdown significantly inhibited de novo lipid synthesis induced by SLC7A5 overexpression. Collective experimental evidence indicates that SLC7A5 regulates de novo lipid metabolism by modulating ACLY stability via ERBB2, a leucine-mTOR-independent mechanism newly reported. This insight offers a novel perspective for investigating olaparib resistance attributed to lipid metabolism.
SLC7A5-regulated olaparib resistance depends on ERBB2
Previously, we established that SLC7A5 influences de novo lipid synthesis via ERBB2, yet its involvement in olaparib resistance remains unclear. To test this hypothesis, we investigated the role of ERBB2 in SLC7A5-mediated Olaparib resistance by co-regulating SLC7A5 and ERBB2 (Supplementary Fig. 4B). Colony-formation and apoptosis assays showed that SLC7A5 overexpression increased ovarian cancer tolerance to olaparib, which was significantly inhibited by ERBB2 knockdown (Fig. 5A, B). Additionally, the cell viability assays following olaparib treatment corroborated these findings (Fig. 5C). Subsequently, we intend to investigate the impact of ERBB2 on the promotion of olaparib resistance by SLC7A5 using animal survival models [15]. We subcutaneously inoculated 5 million CAOV-3 cells into nude mice. Once the tumor volume reached 50 mm³, we initiated olaparib treatment (administered every other day for a total of eight doses at 25 mg/kg each). These results indicated that overexpression of SLC7A5 rendered the tumor tissue relatively insensitive to olaparib treatment. Specifically, the survival extension in the NC drug-treated group was 63% (P = 0.0076), whereas that in the SLC7A5 overexpression group was 33.3% (P = 0.2654). Notably, ERBB2 knockdown significantly suppressed the drug resistance conferred by SLC7A5 overexpression, with the survival extension in the SLC7A5 + shERBB2 group reaching 58.3% (P = 0.0003) (Fig. 5D, E, Supplementary Fig. 4F). Collectively, these results suggest that SLC7A5’s regulation of Olaparib resistance may be dependent on the ERBB2 protein.
A–C Cell Function Experiments. ERBB2 knockdown reverses olaparib resistance induced by SLC7A5 overexpression. A Clonogenic Assay. B Apoptosis Assay. C Cell Viability Assay. D, E Kaplan−Meier survival curves of nude mice implanted orthotopically with CAOV-3 cells. In a xenograft survival study conducted in nude mice, 5 million CAOV-3 cells were subcutaneously injected. D Mice were divided into three groups, NC + shCtrl, SLC7A5 + shCtrl, and shSLC7A5 + shERBB2, with 20 mice per group. Once the tumors reached a volume of 50 mm³, each group was randomly divided into control and treatment subgroups. The treatment subgroup received olaparib every other day for a total of eight doses at 25 mg/kg, while the control group received an equivalent volume of DMSO. In accordance with animal ethics, mice were euthanized when tumor volumes reached 1000 mm³, which was recorded as the endpoint for survival. The median survival times were as follows. 27.5 days for the (NC + shCtrl + DMSO) group, 15 days for the (SLC7A5 + shCtrl + DMSO) group, 30 days for the (SLC7A5 + shERBB2 + DMSO) group, 45 days for the (NC + shCtrl + Olaparib) group, 20 days for the (SLC7A5 + shCtrl + Olaparib) group, and 47.5 days for the (NC + shCtrl +Olaparib) group (n = 10). E The relative extension of survival was statistically analyzed following olaparib treatment. All the in vitro data were obtained from at least three independent experiments. Error bars represent standard deviation. * = P < 0.05, ** = P < 0.001, *** = P < 0.0001, ns = non-significant compared to normal or control treatment. F Clonogenic Assay. The knockdown of ERBB2 inhibits the proliferative effects induced by the overexpression of SLC7A5. G–K In vivo experiments. Subcutaneous implantation of CAOV-3 cells showed that ERBB2 inhibits SLC7A5-mediated tumor promotion. G Tumor. H Tumor Volume(mm3). I Tumor Weight (g). J Immunohistochemistry (IHC). K Western Blot Analysis. All in vitro data points are derived from at least three independent experiments. Error bars represent the standard deviation. Statistical significance is indicated by the following symbols. * = P < 0.05, ** = P < 0.001, *** = P < 0.0001, ns = non-significant compared to normal or control treatment.
SLC7A5-ERBB2 axis promotes ovarian tumorigenesis
Here, we aim to demonstrate whether SLC7A5’s role in ovarian cancer onset is ERBB2-dependent, executing a series of ERBB2 blockade experiments. We hypothesized that SLC7A5 promotes ovarian tumorigenesis in an ERBB2-dependent manner. Based on this hypothesis, we overexpressed SLC7A5 and knocked down ERBB2 in CAOV-3 and OVCAR-8 cells. The results indicated that ERBB2 knockdown inhibited the proliferation-promoting effects of SLC7A5 overexpression (Fig. 5F). In vivo experiments implantation of CAOV-3 cells co-regulated by SLC7A5 and ERBB2 in nude mice revealed that SLC7A5 overexpression alone promoted ovarian cancer progression, which was inhibited by ERBB2 knockdown (Fig. 5G–I). IHC (Fig. 5J) and western blot (Fig. 5K) analyses also confirmed that SLC7A5 affects ERBB2 protein expression. Collective experimental evidence has demonstrated that the SLC7A5-ERBB2 axis influences both the ovarian tumorigenesis. and the development of olaparib resistance, independently of the mTOR signaling pathway. Consequently, future clinical trials may consider combining olaparib with ERBB2 inhibitors for the treatment of patients exhibiting olaparib resistance.
SLC7A5-regulated ERBB2 transcription via ELK1
To clarify how SLC7A5 modulates ERBB2 protein expression, we knocked down SLC7A5 in SK-OV-3 and HEY cells, and RT-PCR and Western blot analyses revealed a significant reduction in ERBB2 mRNA (Fig. 6A). Based on these results, we explored SLC7A5 transcriptional regulatory effect on ERBB2 expression. Initially, we identified SLC7A5-interacting proteins using databases and intersected them with ERBB2 transcription factors. This analysis identified five potential factors: MYC, CEBPB, EP300, ELK1, and ESR1 (Table 5). Subsequent shRNA-mediated knockdown of these proteins followed by RT-PCR analysis revealed that ELK1 knockdown significantly reduced ERBB2 expression (Fig. 6B, Supplementary Fig. 4C). Western blot analysis revealed that ELK1 knockdown abrogated SLC7A5-mediated regulation of ERBB2 protein levels (Fig. 6C). Endogenous co-IP and chIP experiments confirmed the molecular interaction between SLC7A5 and ELK1 (Fig. 6D, Supplementary Fig. 4D, E). Collectively, these results demonstrate that SLC7A5 promotes ELK1-mediated transcription of ERBB2.
A RT-PCR and WB Analysis. The knockout of SLC7A5 significantly inhibited the expression of ERBB2. Line left. HEY, Line right. SKOV3. B RT-PCR Analysis. RT-PCR showed that ELK1 knockdown significantly inhibits SLC7A5-mediated ERBB2 transcription. C Western Blot Analysis. Western blot demonstrated that ELK1 knockdown reverses the increase in ERBB2 protein levels caused by SLC7A5. D Nuclear and Cytoplasmic Fractionation. Nuclear and cytoplasmic fractionation combined with Western blot showed that SLC7A5 overexpression significantly promotes the nuclear translocation of pELK1. E Co-Immunoprecipitation (Co-IP). Co-IP confirmed SLC7A5- ELK1 protein-protein interactions. F Dual Luciferase Reporter Assay. This assay showed that SLC7A5 enhances ELK1 binding to the ERBB2 transcript in a dose-dependent manner. H RT-PCR Analysis. RT-PCR demonstrated that TDE significantly inhibits SLC7A5-mediated ERBB2 protein expression. G Dual Luciferase Reporter Assay. This assay showed that TDE significantly inhibits ELK1 binding to the ERBB2 transcript in a dose-dependent manner. I Chromatin Immunoprecipitation (ChIP) Assay. ChIP analysis showed that TDE significantly inhibits ELK1 binding to the ERBB2 transcript. J Dual Luciferase Reporter Assay. This assay demonstrated that ELK1 binds to the ERBB2 promoter sequence “CCTTCCATC”. Left. Schematic of dual luciferase reporter site mutations. Right. Dual luciferase assay results. K–M Proliferation Assay. TDE significantly mitigates the Olaparib resistance induced by SLC7A5 overexpression. K Edu Assay. L Colony-formation assay. M Cell Viability Test. All in vitro data are derived from at least three independent experiments. Error bars represent standard deviation. Statistical significance is denoted as follows. * = P < 0.05, ** = P < 0.001, *** = P < 0.0001, ns = non-significant compared to normal or control treatment.
Further investigation is needed to determine how SLC7A5 affects ELK1-mediated ERBB2 transcription. Overexpression of SLC7A5 significantly enhance ELK1 binding to the ERBB2 promoter region (Fig. 6E). Nuclear-cytoplasmic fractionation experiments demonstrated that SLC7A5 overexpression facilitated ELK1 nuclear translocation (Fig. 6F). Furthermore, treatment of cells overexpressing SLC7A5 with the ELK1 inhibitor TDE resulted in dose-dependent inhibition of SLC7A5 regulation of ERBB2 (Fig. 6G). Dual-luciferase reporter gene and chIP assays further confirmed that TDE could block ELK1 binding to the promoter region (Fig. 6H). These results suggest that ELK1 transcriptional activation of ERBB2 depends on SLC7A5-mediated phosphorylation and nuclear translocation. Additionally, by constructing dual-luciferase plasmids with mutated promoter sequences, it was confirmed that ELK1 could bind to the ERBB2 promoter sequence CCTTCCATC (Fig. 6I, J). Consequently, we have elucidated the transcriptional mechanism of ERBB2 regulation by SLC7A5.
ELK1 inhibition reverses SLC7A5-induced olaparib resistance
To further investigate the role of ELK1 in SLC7A5-mediated Olaparib resistance, SLC7A5-overexpressing cells were treated with TED. Cell proliferation assays (Fig. 6K–M) revealed that TED significantly inhibited Olaparib resistance induced by SLC7A5 overexpression. Collectively, these findings suggested that SLC7A5 enhances olaparib tolerance by promoting ELK1 function, thereby increasing ERBB2 transcription.
SLC7A5 induces olaparib resistance in ovarian cancer via a non-canonical pathway
Our study indicates that resistance to olaparib in patients with OV is closely associated with the overexpression of SLC7A5, regardless of leucine levels. To clarify SLC7A5’s role in olaparib resistance, we analyzed the correlation between SLC7A5 expression in ovarian cancer cells and their resistance to olaparib, and found that cells with elevated SLC7A5 expression exhibited increased resistance. Furthermore, analysis of TCGA database suggested that SLC7A5 may be involved in the de novo lipid synthesis. The experimental results indicated that in cells with high SLC7A5 expression, lipid metabolism was significantly activated and ERBB2 expression was markedly upregulated. Mechanistic studies revealed that SLC7A5 promotes ERBB2 expression through transcriptional regulation and that excessive ERBB2 maintains the stability of the ACLY protein by binding to CUL3. Overactivation of the SLC7A5-ERBB2 axis provides potential guidance for chemotherapy in patients with advanced ovarian cancer (Fig. 7). Our findings confirmed that SLC7A5 can enhance de novo lipid synthesis and confer resistance to olaparib.
Discussion
Our study demonstrates that SLC7A5 modulates ovarian cancer resistance to olaparib via a leucine-independent pathway. SLC7A5 predominantly facilitates transmembrane transport of glutamine and leucine. Consequently, overexpression of SLC7A5 may lead to remodeling of glutamine metabolism [4, 16], thereby affecting the oxidative stress state of cells. Olaparib exerts its cytotoxic effects by inhibiting PARP enzyme activity, blocking DNA damage repair pathways, and inducing cancer cell apoptosis while also triggering ROS production to enhance its cytotoxicity [17]. Studies have shown that antioxidants such as N-acetylcysteine (NAC) can mitigate the enhanced DNA damage induced by olaparib [18], suggesting the need for further investigation into whether SLC7A5 modulates cellular oxidative stress to reduce sensitivity to olaparib. From the perspective of amino acid transport, inhibiting glutamine uptake, suppressing mitochondrial glutaminase GLS2 from converting glutamine to glutamate, and preventing the conversion of glutamate to α-ketoglutarate via GOT1 can counteract ferroptosis [19, 20]. ACLY influences cellular redox state by regulating NADPH production NADPH, which acts as a regenerator of antioxidants such as glutathione, can reduce ROS levels, thereby protecting cells from oxidative damage [21]. In addition, lipid metabolism is crucial for sensitivity to ferroptosis. Metabolites of polyunsaturated fatty acids (PUFA) can increase ferroptosis sensitivity, whereas the metabolism of monounsaturated fatty acids (MUFA) can limit lipid peroxidation and ferroptosis [22]. Since there is currently no evidence that SLC7A5 participates in the regulation of ferroptosis through these mechanisms to affect cellular sensitivity to olaparib, further research is required.
This study revealed that the SLC7A5/ERBB2 pathway enhances cellular resistance to olaparib through the reconfiguration of lipid metabolism. However, the detailed mechanisms by which SLC7A5 affects the pharmacological effects of olaparib have not been fully elucidated, and may involve interference with cholesterol, cellular oxidative stress status, and cell death pathways. Therefore, further investigations are required. To clarify resistance targets and reduce the toxic side effects of clinical drug use, future research should focus on how SLC7A5 modulates the efficacy of olaparib, analyzing the impact of SLC7A5 on the sensitivity of ovarian cancer to olaparib from aspects such as oxidative stress and ferroptosis.
In summary, SLC7A5 is typically overexpressed in olaparib-resistant patients and is significantly associated with poor prognosis. SLC7A5 inhibition can significantly enhance the sensitivity of ovarian cancer cells to olaparib. Our research suggests that SLC7A5 may serve as a novel target for predicting prognosis and addressing olaparib resistance.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
Lheureux S, Braunstein M, Oza AM. Epithelial ovarian cancer: evolution of management in the era of precision medicine. CA Cancer J Clin. 2019;69:280–304.
LaFargue CJ, Dal Molin GZ, Sood AK, Coleman RL. Exploring and comparing adverse events between PARP inhibitors. Lancet Oncol. 2019;20:e15–28.
Don SD. PARP inhibitors for targeted treatment in ovarian cancer. Lancet. 2017;390:1929–30.
Zhang L, Sui C, Yang W, Luo Q. Amino acid transporters: emerging roles in drug delivery for tumor-targeting therapy. Asian J Pharm Sci. 2020;15:192–206.
Saito Y, Li L, Coyaud E, Luna A, Sander C, Raught B, et al. LLGL2 rescues nutrient stress by promoting leucine uptake in ER+ breast cancer. Nature. 2019;569:275–9.
Xu M, Sakamoto S, Matsushima J, Kimura T, Ueda T, Mizokami A, et al. Up-regulation of LAT1 during antiandrogen therapy contributes to progression in prostate cancer cells. J Urol. 2016;195:1588–97.
Tan J, Zheng X, Li M, Ye F, Song C, Xu C, et al. C/EBPβ promotes poly(ADP-ribose) polymerase inhibitor resistance by enhancing homologous recombination repair in high-grade serous ovarian cancer. Oncogene. 2021;40:3845–58.
Murali R, Balasubramaniam V, Srinivas S, Sundaram S, Venkatraman G, Warrier S, et al. Deregulated metabolic pathways in ovarian cancer: cause and consequence. Metabolites. 2023;13:560.
Artibani M, Masuda K, Hu Z, Rauher PC, Mallett G, Wietek N, et al. Adipocyte-like signature in ovarian cancer minimal residual disease identifies metabolic vulnerabilities of tumor-initiating cells. JCI Insight. 2021;6:e147929.
Abdel-Razeq H, Sharaf B, Khater S, Baidoun HJ, Bani Hani H, Taqash A, et al. Clinical outcomes of patients treated with ribociclib in combination with aromatase inhibitors or fulvestrant for HR-positive, HER2-negative metastatic breast cancer, real-world data from a low-resourced country. Immunotargets Ther. 2024;13:501–12.
Coulson-Gilmer C, Morgan RD, Nelson L, Barnes BM, Tighe A, Wardenaar R, et al. Replication catastrophe is responsible for intrinsic PAR glycohydrolase inhibitor-sensitivity in patient-derived ovarian cancer models. J Exp Clin Cancer Res. 2021;40:323.
Ilson DH, van Hillegersberg R. Management of patients with adenocarcinoma or squamous cancer of the esophagus - ClinicalKey. Gastroenterology. 2018;154:437–51.
Waddell N, Pajic M, Patch AM, Chang DK, Kassahn KS, Bailey P, et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature. 2015;518:495–501.
Zhang C, Liu J, Huang G, Zhao Y, Yue X, Wu H, et al. Cullin3–KLHL25 ubiquitin ligase targets ACLY for degradation to inhibit lipid synthesis and tumor progression. Genes Dev. 2016;30:1956–70.
Sepporta MV, Praz V, Bourloud KB, Joseph JM, Jauquier N, Riggi N, et al. TWIST1 expression is associated with high-risk neuroblastoma and promotes primary and metastatic tumor growth. Communications Biol. 2022;5:42.
Liu T, Han C, Fang P, Ma Z, Wang X, Chen H, et al. Cancer-associated fibroblast-specific lncRNA LINC01614 enhances glutamine uptake in lung adenocarcinoma. J Hematol Oncol. 2022;15:141.
Okazaki A, Gameiro PA, Christodoulou D, Laviollette L, Schneider M, Chaves F, et al. Glutaminase and poly(ADP-ribose) polymerase inhibitors suppress pyrimidine synthesis and VHL-deficient renal cancers. J Clin Investig. 2017;127:1631–45.
Osborne HC, Larrosa I, Schmidt CK. Sesquiterpene lactones potentiate olaparib-induced DNA damage in p53 wildtype cancer cells. Int J Mol Sci. 2022;23:1116.
Song X, Liu J, Kuang F, Chen X, Zeh HJ, Kang R, et al. PDK4 dictates metabolic resistance to ferroptosis by suppressing pyruvate oxidation and fatty acid synthesis. Cell Rep. 2021;34:108767.
Hassannia B, Vandenabeele P, Vanden Berghe T. Targeting ferroptosis to iron out cancer - ScienceDirect. Cancer Cell. 2019;35:830–49.
Icard P, Wu Z, Fournel L, Coquerel A, Lincet H, Alifano M. ATP citrate lyase: a central metabolic enzyme in cancer. Cancer Lett. 2020;471:125–34.
Pope LE, Dixon SJ. Regulation of ferroptosis by lipid metabolism. Trends Cell Biol. 2023;33:1077–87.
Acknowledgements
We would like to thank all the members who participated in this study. Thank you for your efforts, especially for your assistance with experimental design, pathological analysis, and data processing.
Funding
This study was supported by the Heilongjiang Provincial Natural Science Foundation of China (No. LH2020H124).
Author information
Authors and Affiliations
Contributions
Shuang Liu: conceptual design, data generation, analysis and interpretation; manuscript preparation, review, and editing. Yunlong He: Data curation, formal analysis, Methodology, Software, Validation, Writing—review, and editing. Xin Ning: Conceptualization, Data curation, formal analysis, Methodology, Software, Writing the original draft. Wei Xu: Formal analysis, Writing—original draft. Xiaowei Hu: Conceptualization, Data curation, Methodology, Software. Ling Qin: Data curation, Methodology, Resources, Writing—original draft. Yupeng Yang: manuscript preparation, review and editing. Xihai Chen: Formal analysis, Methodology, Supervision, Writing—original draft, writing—review, and editing. Rong Ma: Data curation, formal analysis, funding acquisition, Methodology, Project administration, Supervision, Writing—review, writing—review, and editing.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical compliance
This study was conducted in compliance with all relevant guidelines and regulations. The experimental protocol was approved by the Ethics Committee of Shanghai Superchip Biotech Co., Ltd. (Shanghai, China).
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Liu, S., He, Y., Ning, X. et al. SLC7A5-ERBB2 axis drives olaparib resistance via de novo lipid synthesis in ovarian cancer. Oncogene 45, 140–263 (2026). https://doi.org/10.1038/s41388-025-03584-w
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41388-025-03584-w