Abstract
Transcriptional dysregulation is a hallmark of cancer initiation and progression, driven by genetic and epigenetic alterations. Enhancer reprogramming has emerged as a pivotal driver of carcinogenesis, with cancer cells often relying on aberrant transcriptional programs. The advent of high-throughput sequencing technologies has provided critical insights into enhancer reprogramming events and their role in malignancy. While targeting enhancers presents a promising therapeutic strategy, significant challenges remain. These include the off-target effects of enhancer-targeting technologies, the complexity and redundancy of enhancer networks, and the dynamic nature of enhancer reprogramming, which may contribute to therapeutic resistance. This review comprehensively encapsulates the structural attributes of enhancers, delineates the mechanisms underlying their dysregulation in malignant transformation, and evaluates the therapeutic opportunities and limitations associated with targeting enhancers in cancer.
Similar content being viewed by others
Facts
-
Enhancers play a central role in the transcriptional mechanism of cancer, and various intracellular and extracellular factors(such as transcription factors, cytokines, and multiple signaling pathways) can act on them and drive the formation of core regulatory circuits—a highly integrated regulatory network composed of master transcription factors, their associated enhancers, and target genes.
-
Enhancer reprogramming in cancer not only unlocks the phenotypic plasticity of cancer cells, enabling traits such as proliferation, drug resistance, and metastasis but also contributes to the reshaping of the tumor microenvironment, driving cancer progression through more complex mechanisms.
-
Based on previous research, this article proposes different potential targeting therapeutic strategies, especially based on the unique mechanism of enhancers regulating the tumor microenvironment discovered recently, as well as comprehensive therapy. For instance, the combined application of chemotherapy, immunotherapy, and enhancer-targeted therapies, aiming to leverage their complementary mechanisms to overcome the limitations of single treatments and improve therapeutic outcomes (e.g. NCT06393361, NCT06563778).
Open questions
-
What key molecules or signaling pathways determine the selective activation or silencing of enhancers in cancer cells?
-
What advanced techniques and models can be utilized to capture dynamic enhancer responses during tumor progression and treatment, and how can these insights guide therapeutic strategies?
-
Based on previous research findings, we have provided some alternative targeted treatment strategies. Recent studies have explored combination strategies involving metabolic pathway inhibitors, like the use of purine synthesis inhibitors in MLL3/4-mutant cancers, and have demonstrated synergistic effects in preclinical models. Whether these comprehensive treatment strategies are feasible and can they improve patient prognosis?
Introduction
Transcription initiation is a key step in the regulation of gene expression. In eukaryotic cells, transcription begins with the binding of RNA polymerase to the promoter, which is regulated by transcription factors (TFs) [1]. Enhancers are DNA sequences located upstream or downstream of gene promoters, typically far from the promoter region, and are rich in TF binding sites (TFBS). They significantly increase gene expression in ways that are independent of direction and distance [2]. Annotation of enhancers on a genome-wide scale has been achieved through various chromatin immunoprecipitation (ChIP) techniques (e.g., ChIP-sequencing [ChIP-seq]) and chromatin accessibility analyses, including DNase-seq and the assay for transposase-accessible chromatin using sequencing (ATAC-seq). These methods were prominently utilized in the Encyclopedia of DNA Elements (ENCODE) project to define different classes of regulatory elements [3,4,5]. Additionally, high-throughput chromosome conformation capture technologies have identified the direct interaction between enhancers and promoters within the three-dimensional (3D) chromatin architecture, highlighting the functional relevance of spatial genome organization in enhancer activity [6].
Higher-order assembly of TFs—the cooperative and hierarchical interactions between TFs, co-activators, and chromatin remodelers—on enhancers is tightly regulated to ensure tissue-specific gene expression and cellular homeostasis in normal cells [7,8,9]. However, in cancerous cells, genetic mutations in TFs or chromatin regulators disrupt this balance, leading to aberrant enhancer activity [10,11,12]. Recent studies highlight the critical role of somatic mutations in enhancer dysfunction, revealing how these mutations perturb enhancer-promoter interactions [13, 14]. Mutations in epigenetic regulators like TET2 and MLL4 have been shown to disrupt enhancer-associated chromatin modifications, rebalancing key transcriptional programs in cancer [15, 16]. Moreover, structural variants, including duplications or translocations, can relocate enhancers near oncogenes, further amplifying their expression and accelerating tumor progression [17]. These insights establish a direct link between genetic mutations and enhancer dysfunction, underscoring their importance in the pathogenesis of cancer.
The deregulation of genes is a major feature of cancer, involving both genetic mutations and non-coding epigenetic reprogramming [18]. An increasing number of analyses of cancer genomes and epigenomes have shown that enhancers play a crucial role in driving the gene expression regulatory networks in cancer [19, 20]. Cancer cells hijack enhancers as platforms to integrate intracellular and extracellular signals, binding with key TFs to establish self-circulating core transcriptional axes [21]. This process dynamically confers new phenotypes to cancer cells. Cancer cells become addicted to the high transcriptional output driven by enhancers; thus, the vulnerabilities of cancer cells become potential therapeutic targets [20]. However, the specificity of targeting enhancers remains a challenge, as off-target effects or disruptions to normal enhancer activity could lead to adverse outcomes [22]. Addressing these challenges requires a deeper understanding of the molecular underpinnings of enhancer dysfunction and its links to genetic mutations, as well as the development of precision therapeutic strategies to selectively target enhancer-driven transcriptional addictions.
Structural and functional characteristics of enhancer
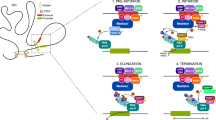
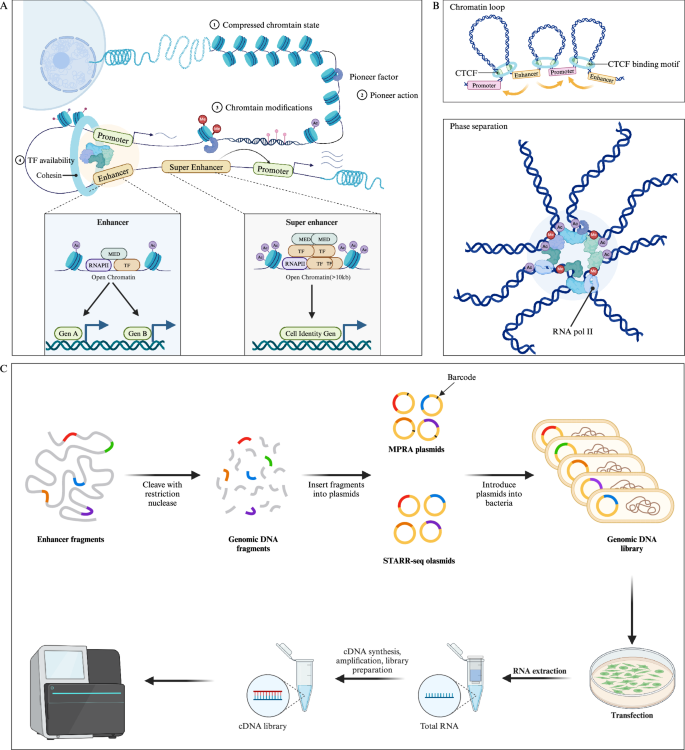
Enhancers are defined as non-coding DNA sequences capable of promoting the expression of target genes. It has been demonstrated that enhancers play crucial roles in the development and diseases [23]. In many cases, enhancers are located at significant distances from their target genes (even skipping over unrelated genes to interact with the target promoter) and are regulated by tissue-specific TFs and cofactors, such as p300/CREB-binding protein (p300/CBP), mediator (MED), and switch/sucrose non-fermentable (SWI/SNF) [24]. Through these long-range interactions, enhancers can precisely control the expression of specific genes in a temporal and spatial manner (Fig. 1) [25].
A Pioneer factors bind to and activate closed chromatin by mediating chromatin remodeling, making it accessible for other TFs. In contrast, open chromatin structures do not require this mechanism, as TFs can directly access these open enhancers. Pioneer factors recruit epigenetic modifiers to alter the local chromatin environment (such as histone methylation, acetylation, and DNA methylation), further collaborating with other TFs to activate transcription. The figure illustrates a typical enhancer and SE, with SE enriched in TFs and histone modifications (e.g., histone acetylation), driving the expression of cell identity genes. B Chromatin loop (up) and phase separation (down) model of enhancers. In the chromatin loop model, CTCF and cohesins regulate the 3D structure of chromatin through a loop extrusion mechanism, promoting enhancer-promoter interactions. In the phase separation model, the interaction between enhancers, high-density TFs and chromatin modifications forms liquid-like condensates, further modulating gene expression. C High-throughput detection of enhancer activity of DNA fragments using MRPA and STARR-seq methods. (Created with BioRender.com).
Enhancer and TFs
It is well established that the activity of enhancers is regulated by numerous transcription factors (TFs) [26]. Currently, TFs commonly found at enhancers are broadly categorized into two types: tissue-specific TFs and general TFs enhancers [27]. For example, p300/CBP, a histone acetyltransferase (HAT), is enriched at enhancers and facilitates chromatin accessibility, serving as a hallmark of enhancer activity [28]. MED1, a core component of the Mediator complex, acts as a bridge between enhancers and promoters by recruiting RNA polymerase II to initiate transcription (Fig. 1A) [29]. In various species and tissues, the transcriptional activity of enhancers differs significantly, driven by lineage-specific transcription factors (TFs) that are uniquely expressed in specific cells or tissues. For instance, FOXA1 and FOXA2 were initially identified as tissue-specific factors during foregut endoderm development [30, 31]. Interestingly, some tissue-specific TFs also serve as pioneer factors under certain conditions, and not only function within specific tissues but can also bind to closed chromatin regions and remodel them into an open state, embodying the hallmark characteristics of pioneer factors [32]. This “dual role” underscores their unique position in gene regulation.
FOXA1 and FOXA2 exhibit classic pioneer activity by binding to closed chromatin regions and opening them [33]. Their “winged-helix” DNA-binding domain enables them to penetrate nucleosomal DNA and expose chromatin regions, facilitating the binding and activation of downstream tissue-specific factors such as HNF4α in the liver and Pdx1 in the pancreas [34, 35]. Within these open chromatin regions, pioneer factors and tissue-specific TFs interact synergistically, stabilizing each other’s binding and enhancing their regulatory effects. For instance, FOXA2, by interacting with JUN, activates lineage plasticity enhancers, driving prostate cancer cells to transition from AR-dependence to a multilineage state [36]. In some cases, pioneer factors primarily open chromatin, while tissue-specific TFs determine which genes are ultimately activated. For example, SOX2 acts as a pioneer factor across various cell types, but its downstream targets are determined by co-binding with tissue-specific factors, such as OCT4 in embryonic stem cells [37, 38]. This combinatorial mechanism endows enhancers with high specificity, allowing the same pioneer factor to work with distinct tissue-specific TFs to activate unique gene expression programs in different cell types.
However, the precise mechanisms by which pioneer factors activate closed chromatin remain elusive. Some studies suggest that pioneer factors facilitate chromatin opening by inducing nucleosome repositioning [39]. For example, OCT4 binds nucleosomes through two DNA-binding domains: one anchor at the nucleosome entry site, while the other engages distal DNA, bending and exposing the DNA for nucleosome repositioning. Interestingly, OCT4 can lock DNA into specific conformations while exposing binding sites for SOX2. Moreover, H3K27ac-induced DNA sliding enhances OCT4’s multi-site binding and stabilizes the recruitment of other TFs [39].
Additionally, intrinsically disordered regions (IDRs) in pioneer factors contribute to chromatin remodeling by forming condensates or interacting with chromatin [40, 41]. For example, the IDRs of PU.1 facilitate weak interactions with chromatin components such as histone tails or non-specific DNA sequences, driving structural changes in local chromatin [40]. Similarly, the N- and C-terminal IDRs of FOXA1 enable the formation of sub-micrometer biomolecular condensates, effectively loosening compact chromatin to enhance accessibility [41]. Recent studies further indicate that the activation of closed chromatin by pioneer factors involves multi-step modifications and structural adjustments [42]. For instance, before cell division, PAX7 binds chromatin and recruits the H3K9me2 demethylase KDM1A (LSD1) and MLL, gradually converting chromatin from a repressive to a potentially active state. During cell division, enhancers dissociate from lamin B structures at the nuclear periphery, while PAX7 simultaneously recruits the SWI–SNF chromatin remodeling complex and p300, further opening enhancer regions to facilitate binding by transcription factors and co-activators [42].
Characterizing the functional syntax of enhancers
Current research highlights the critical role of histone modifications in enhancer identification and function [43]. Modifications such as H3K4me1 and H3K27ac are widely recognized as markers of active enhancers, aiding in their identification [44]. Furthermore, recent studies have revealed additional histone marks associated with active enhancers, including H3K18la [45], H4K16ac [46], H3K9ac [47, 48], H2BNTac [49], crotonylation [50], and β-hydroxybutyrylation (e.g., H3K56bhb) [51]. However, while the distribution of histone modifications provides valuable locational cues, the levels of these marks alone are insufficient to predict the functional activity of enhancers.
Enhancer function involves a complex “syntax” that integrates multiple elements beyond histone marks, including specific DNA sequences, TFBS, chromatin accessibility, and environmental cues [24, 52,53,54]. Thus, while histone modifications offer important insights into enhancer identification, a comprehensive understanding of enhancer regulation requires moving beyond modification patterns to explore the intricate molecular grammar that governs their activity.
Traditional methods for assessing enhancer activity in DNA sequences rely on reporter gene assays, such as luciferase assays, which evaluate the transcriptional activity of inserted sequences by measuring luminescence intensity [55]. However, these approaches are primarily used for validation and are not suitable for high-throughput screening. High-throughput techniques such as self-transcribing active regulatory region sequencing (STARR-seq) and massively parallel reporter assay (MPRA) have enabled genome-wide detection of potential enhancer activity (Fig. 1C) [56,57,58].
Moreover, machine learning models trained on these high-throughput sequencing datasets have proven effective in predicting active enhancers and TFBS (Table 1) [59, 60]. The combined application of ChIP-seq or ATAC-seq with STARR-seq (e.g., ChIP-STARR-seq and ATAC-STARR-seq) further advances the identification and functional validation of enhancers at a genome-wide scale, providing a powerful framework for elucidating regulatory elements and their activity [44, 61, 62].
In cancer research, by integrating eCLIP, Hi-C, and genome-wide STARR-seq, a recent study utilized ENCODE data to annotate cancer-related cell types and help construct regulatory networks that include TFs and RNA-binding proteins. This effectively describes the cell state trajectories and regulatory network reorganization during cancer transformation [63]. Regulatory single nucleotide polymorphisms associated with cancer risk were systematically identified, revealing the regulatory mechanisms of these variants on gene expression. For example, rs11055880 is associated with breast cancer by regulating the expression of ATF7IP [64]. Besides, a study that integrated DNase-seq, ChIP-seq, global run-on sequencing, STARR-seq, RNA-seq, Hi-C, and Chromatin Interaction Analysis with Paired-End-Tag (ChIA-PET) data from five cancer cell lines identified a new class of autonomous and dual SEs. These SEs regulate the transcription of highly expressed genes through long-range chromatin interactions and promote the survival of cancer cells [65]. The integration of high-throughput sequencing technologies with CRISPR can facilitate the identification of non-coding genetic variants with significant regulatory effects in cancer and link these regulatory perturbations to differences in therapeutic sensitivity. For example, in pediatric acute lymphoblastic leukemia, the combination of MPRA and CRISPR identified rs1247117 as a key variant influencing vincristine sensitivity [66].
It is important to note that both reporter assays, such as STARR-seq, and MPRA are ectopic detection methods that cannot fully recapitulate the native chromatin environment [57, 67]. Additionally, enhancer activity varies significantly across cell types, introducing inherent limitations, particularly in studies involving clinical samples [68]. A potential solution lies in leveraging single-cell ATAC-seq and CUT&Tag sequencing to gain a clearer understanding of the cell-specific nature of enhancers [69, 70]. Besides, the recent development of a single-cell MRPA method may also help address this issue [71]. These approaches preserve the original chromatin architecture and the influence of environmental factors on enhancer function. When combined with single-cell transcriptomics, they can provide deeper insights into enhancer activity within its native context.
TFs play a critical role in regulating enhancer activity. Recent studies suggest that enhancer function is not solely determined by TF binding strength but also by dynamic parameters such as TF clustering frequency, periodicity, and stability [72]. Supporting this view, enhancer function remains stable through cooperative and combinatorial TF interactions, even when evolutionary changes occur in sequences or individual TF binding sites [73]. During somatic cell reprogramming to pluripotent stem cells, the OSKM factors (Oct4, Sox2, Klf4, and c-Myc) initially bind to somatic cell enhancers, recruiting histone deacetylases (e.g., HDAC1) and redistributing somatic-specific TFs (e.g., Fra1), effectively silencing somatic enhancers. As reprogramming progresses, OSKM factors relocate to pluripotency-associated enhancers, depositing H3K27ac marks and recruiting key pluripotency TFs (e.g., Nanog and Esrrb) to support the expression of pluripotency genes [7].
In cancer, TFs such as YAP/TAZ/TEAD co-occupy enhancers with AP-1. The tumor-promoting effects of YAP/TAZ are dependent on AP-1, and AP-1 inactivation significantly impairs YAP/TAZ-driven tumor cell proliferation [74]. Additionally, FOXC1, typically absent in normal hematopoiesis, is highly expressed in certain acute myeloid leukemia (AML) subtypes. FOXC1 recruits transcriptional repressors RUNX1 and TLE3 to enhancers regulating monocyte differentiation, thereby blocking AML cell differentiation. FOXC1 deletion disrupts the binding of RUNX1 and TLE3, increasing the occupancy of differentiation-related TF CEBPA at these enhancers [75]. Similarly, SOX9, a key TF driving the transition from epidermal stem cells to hair follicle stem cells, transiently activates and redistributes chromatin regulators such as SWI/SNF and MLL3/4 to hair follicle enhancers, silencing epidermal genes. However, prolonged SOX9 activation selectively engages tumor-associated enhancers, leading to the activation of the Wnt/β-catenin signaling pathway [76]. These findings highlight the critical role of higher-order TF assemblies in the dynamic regulation of enhancers and reveal distinct TF dynamics between normal and cancer cells.
Moreover, current research suggests that enhancer RNA (eRNA) transcription is a hallmark of active enhancers [77, 78] Knockdown of eRNAs disrupts TF binding to enhancers, such as c-Jun, NF-κB, and YY1. eRNAs also interact with MED1 and RNAPII to facilitate enhancer-promoter looping. A recent study demonstrated that eRNAs directly bind the AT-hook domain of Brg1 to recruit the SWI/SNF chromatin remodeling complex to specific enhancers, simultaneously enhancing the recruitment and activation of co-activators such as MLL3/4, MED1, and p300/CBP [52]. Additionally, m6A-modified eRNAs can recruit the m6A reader protein YTHDC1, forming droplet-like condensates at enhancers. These condensates promote the clustering of co-activators like BRD4, thereby increasing enhancer activity and gene transcription [79]. These findings indicate that eRNAs are critical regulators within gene regulatory networks, playing an essential role in maintaining and modulating the dynamic balance of gene expression, although predicting enhancer activity directly from eRNAs remains challenging based on current knowledge.
The paradigm of enhancer function
The transcriptional information mediated by enhancers is transmitted through enhancer-promoter (E-P) interactions [80]. High-resolution imaging techniques have revealed complex E-P interaction networks and proposed several major E-P interaction models, such as the chromatin loop model mediated by CCCTC-binding factor (CTCF) and cohesin, phase separation, the scanning model, and the tracking model (Fig. 1B) [54, 81].
The 3D structure of the genome is organized into basic units termed topologically associating domains (TADs), maintained by boundary elements such as CTCF and cohesion. DNA sequences within these regions interact more frequently between them than with sequences in adjacent TADs [82]. In the chromatin loop model, the cohesin complex extrudes DNA loops, bringing distant enhancers and promoters into close proximity for functional communication. Of note, CTCF acts as an anchor to prevent incorrect loop formation and enhancer off-target effects [83, 84]. Despite significant support for this model, some evidence challenges its usefulness. For example, current loop structures suggest that cohesin does not affect the expression of most genes, except for those associated with SEs. Additionally, gene expression and repression coexist within some TADs. Furthermore, genome-wide deletion of CTCF does not significantly alter gene expression levels, as observed in studies on SOX2 and SOX9 [81, 85, 86].
The phase separation model is a newer theory explaining enhancer function [87]. This model is based on the concept of biomolecular condensation and phase separation, suggesting that enhancers regulate gene expression by forming droplet-like condensates [87]. For example, general TFs (bromodomain-containing protein 4 [BRD4] and MED1) can concentrate the transcription machinery at enhancers through phase separation, thereby forming condensates [88]. Dysregulation of condensates is considered a hidden driver in cancer pathology. Certain proteins in cancer cells (e.g., Yes1 associated transcriptional regulator [YAP], homeobox B8 [HOXB8], and FOS like 1 [FOSL1]) promote condensate formation, thus directly enhancing oncogene expression and cell proliferation. Additionally, condensate dysregulation can alter the chromatin structure, impacting normal gene expression and regulation [89, 90].
SEs
Through an in-depth exploration of the genome, researchers have identified enhancers exhibiting unique characteristics, termed SEs, which extend across larger genomic regions [91]. In comparison to conventional enhancers, SEs exhibit several distinctive features: (1) they span a larger genomic region, typically >10 kb; (2) they display significantly augmented histone modifications, notably H3K27ac enrichment; (3) they are intricately associated with specific cell types or states, exerting robust regulation of gene expression linked to cell characteristics such as differentiation and identity maintenance; (4) they are closely implicated in the onset and progression of diverse diseases, particularly cancer; (5) they govern lineage-specific TFs, including SOX2, KLF5, FOXA1, and FOXA2; and (6) they are more susceptible to intervention. This characterization underscores the pivotal role of SEs in gene regulation, cell identity, and disease pathology, presenting them as promising targets for intervention (Fig. 1A) [92,93,94].
Enhancer reprogramming in cancer
Enhancer reprogramming refers to the dynamic changes in chromatin state and enhancer activity during cell fate determination, differentiation processes, or disease states [95,96,97]. Numerous factors act on enhancers through complex interactions and regulatory networks, leading to widespread changes in gene expression that promote the development and progression of cancer [98,99,100]. Understanding these mechanisms is crucial for revealing the fundamental principles of gene expression regulation, as well as for the diagnosis and treatment of cancer. Based on the structural and functional characteristics of enhancers, the factors leading to enhancer reprogramming can be divided into two main categories, namely genetic mutations and epigenetic remodeling.
Somatic mutations
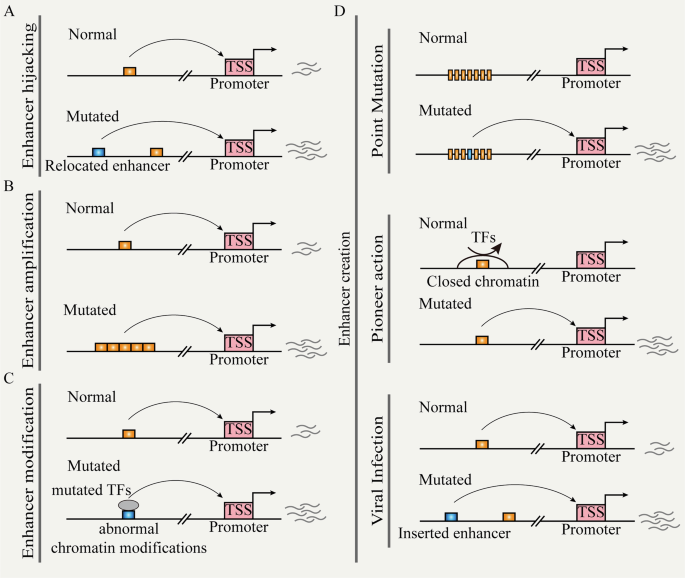
Recent research has revealed the reprogramming of enhancer functions by genetic mutations through different pathways, leading to abnormal gene expression and disease [20]. We explore several mechanisms (e.g., enhancer hijacking, enhancer creation, and enhancer modulation), illustrating their specific roles in gene regulation and discussing their impact on cancer (Fig. 2).
A Chromosomal structural variation leads to the repositioning of distal enhancers to target promoters. B Non-coding copy-number variation leads to enhancer function amplification. C Mutated TFs cause enhancer reprogramming. D Random mutation, pioneer action, and viral infection can create de novo enhancers.
Enhancer hijacking
Chromosomal structural variations (SVs) (e.g., inversions, translocations, and fusions) directly relocate the ectopic enhancer to the vicinity of the target gene; this process is referred to as ‘enhancer hijacking’ (Fig. 2A) [17, 101, 102]. This event occurs frequently in hematologic malignancies. During acute myeloid leukemia (AML), there are recurrent and subtype-specific alterations in A/B compartments, TADs, and chromatin loops. These SVs lead to widespread enhancer-hijacking and silencer-hijacking events, thereby promoting the progression of AML [17]. In AML cells with inv (3)/t (3:3), for example, the original enhancer of GATA2 located near the ribophorin I (RPN1) gene relocated to the EVI1 locus and formed SE. This resulted in single allelic expression of GATA2 and abnormal activation of EVI1 [103]. Previous studies have established a comprehensive computational framework for predicting enhancer hijacking based on Hi-C data [104]. Recently, the integration of multi-modal analysis methods (e.g., whole-genome sequencing, Hi-C, RNA-seq, ChIP-seq, and ATAC-seq) has provided new opportunities to deeply understand the impact of SVs on gene regulation through chromatin interactions. For instance, in certain pediatric leukemia subtypes, the recurrent t(5;14) translocation results in enhancer hijacking of BCL11 transcription factor B (BCL11B), specifically activating the T cell leukemia homeobox 3 (TLX3) gene [105]. A recent study analyzing 92 cancer cell lines and clinical samples revealed that SVs lead to the formation of numerous fused TADs. This effect leads to the accumulation of a greater number of highly active enhancers and is closely associated with the activation of oncogenes, such as MYC, telomerase reverse transcriptase (TERT), and cyclin D1 (CCND1). Besides, using a deep learning-based Activity-By-Contact (ABC) model, researchers can predict which SVs are likely to activate oncogenes [106]. The machine learning tool CAPReSE has been developed for chromatin anomaly pattern recognition and size estimation. This tool accurately identifies specific SV-mediated aberrant chromatin contacts in cancer genomes. In colorectal cancer, CAPReSE identified numerous recurrent enhancer-hijacking events mediated by SVs. These events involve cell cycle and DNA processing and may play important roles in tumor progression and chemotherapy resistance [107]. Additionally, by integrating somatic copy-number variations, gene expression data, and TAD information, the cis-Expression Structural Alteration Mapping (CESAM) systematically identifies enhancer-hijacking events in pan-cancer genomes. For instance, in colorectal cancer, recurrent tandem duplications intersecting with TAD boundaries result in the formation of new 3D contact domains, encompassing insulin-like growth factor 2 (IGF2) and a lineage-specific SE, leading to high-level gene activation [108].
Enhancer creation
We define the “enhancer creation” as the process through which new regulatory elements emerge in the genome by various genetic and epigenetic mechanisms. For example, during the development of pancreatic cancer, transient precursor cells of pancreatic cancer will appear in pancreatitis, characterized by the formation of transient enhancer networks mediated by KRAS mutation. In this process, KRAS mutation stabilizes the key oncogenic activator protein 1 (AP-1) TFs (JUNB and FOSL1), locking the progenitor state to initiate tumor development [109].
Random mutations drive enhancer reprogramming by altering TFBS, rewiring promoter-enhancer connections, and modifying epigenetic marks (Fig. 2D). Enhancer activity typically relies on specific TFBS, and random mutations within enhancer regions can either create new binding sites or disrupt existing ones, leading to changes in regulatory function. For instance, in T-ALL, the insertion upstream of the TAL bHLH transcription factor 1, erythroid differentiation factor (TAL1) oncogene introduces novel MYB binding sites, resulting in the formation of SE that drives TAL1 [110]. In neuroblastoma, single nucleotide variants in the SE within the first intron of LIM domain only 1 (LMO1) eliminate the binding motif of GATA and cause LMO1 dysfunction [111]. Consistent with this, an integrated study of whole-genome and epigenome data in lung cancer revealed that enhancer mutations can create new binding sites for oncogenic TFs such as MYC or NF-κB. Conversely, other enhancer mutations disrupt binding sites for tumor-suppressive TFs, thereby weakening the transcriptional repression of oncogenes. These mutations also influence the epigenetic modifications of enhancers, leading to either increased or decreased H3K27ac levels [13]. Furthermore, random mutations can alter the 3D structure of chromatin, thereby modifying spatial contacts between enhancers and promoters. In colorectal cancer, the distant enhancer region at FADS2, marked by rs174575, recruits the transcription factor E2F1 and facilitates physical interactions between the enhancer and the promoter [112].
Copy-number amplification in non-coding regions leads to enhancer expansion or duplication, resulting in the amplification of enhancer activity (Fig. 2B). This phenomenon drives aberrant gene expression and tumorigenesis across various cancer types. For instance, in T cell acute lymphoblastic leukemia (T-ALL), the duplication of the 8q24 region leads to the amplification of a long-range acting enhancer controlled by notch receptor 1 (NOTCH1). This amplification promotes the expression of the MYC gene, thereby driving T cell development and the occurrence and progression of T-ALL [113]. Based on integrative deep whole-genome analysis, a study found that most patients with metastatic castration-resistant prostate cancer have an intergenic enhancer region amplification 624 kb upstream of AR, which correlates with increased AR expression. These hotspots also appear near MYC and are associated with long non-coding RNAs that regulate post-translational modifications of MYC [114].
Viral infections are common pathogenic factors for certain cancer types, such as liver cancer, cervical cancer, and nasopharyngeal cancer (Fig. 2D) [115]. Viral insertions activate nearby oncogenes and alter gene expression networks by introducing new regulatory sequences, thereby promoting the development and progression of cancer [116, 117]. The integration of HPV generates SEs present in HPV-human hybrid ecDNA, leading to intra-chromosomal and inter-chromosomal regulation. This process results in transcription dysregulation and oncogene expression [117]. In hepatocellular carcinoma, the insertion of hepatitis B virus enhancers into the host genome activates adjacent genes, such as TERT, KMT2B, and cyclin E1 (CCNE1). This provides a proliferative advantage to the infected cells and promotes their malignant transformation [118]. Unlike other oncogenic viruses, the EBV genome typically does not integrate into the host genome; it exists as independently replicating episomes within infected cells. Therefore, EBV promotes enhancer “creation” in a distinct manner termed “enhancer infestation”. These episomes “forcibly” convert H3K9me3-marked heterochromatin into active enhancers marked by H3K27ac and H3K4me1 in a pioneer-like manner. This conversion further activates oncogenes and promotes tumorigenesis, such as nasopharyngeal cancer and gastric cancer [119, 120].
Enhancer modulation
Enhancer activity depends on the binding of TFs and specific histone modifications. Mutations occurring in TFs or epigenetic modification enzymes can reset enhancer modifications, leading to abnormal gene expression (Fig. 2C) [121]. KMT2C and KMT2D are histone methyltransferases responsible for the monomethylation of H3K4 and are frequently mutated in cancer, such as breast cancer. They regulate estrogen receptor alpha-driven (ERα-driven) transcription by activating gene enhancers. Additionally, the loss of KMT2C and KMT2D is closely associated with genomic instability and a high mutation burden in tumors and can induce epithelial–mesenchymal transition (EMT) and metastasis, thereby promoting tumor aggressiveness [122]. The p300/CBP is a lysine acetyltransferase that regulates enhancer function through histone acetylation. Mutations in p300/CBP have been identified in various solid tumors and hematologic malignancies. These mutations often result in abnormal H3K27ac at specific enhancer regions, which are associated with cancer phenotypic markers, such as proliferation, invasiveness, and metastasis [123].
The ability of mutant TFs to alter cellular functions stems from their capacity to remodel enhancers by relocating their binding sites, modifying chromatin structure, or forming new protein complexes. For instance, the nucleoporin 98-HOXA9 (NUP98-HOXA9) fusion protein in leukemia cells drives gene expression reprogramming by forming new SE [124]. Mutant p53 directly interacts with KMT2C to regulate enhancer activity and promote oncogene expression [125]. Additionally, under the tumor necrosis factor-alpha-induced (TNF-α-induced) chronic inflammatory environment, the mutant p53 reshapes the enhancer landscape by interacting with nuclear factor-κB (NF-κB), thereby promoting cancer progression [126].
Epigenetic mechanism of enhancer reprogramming
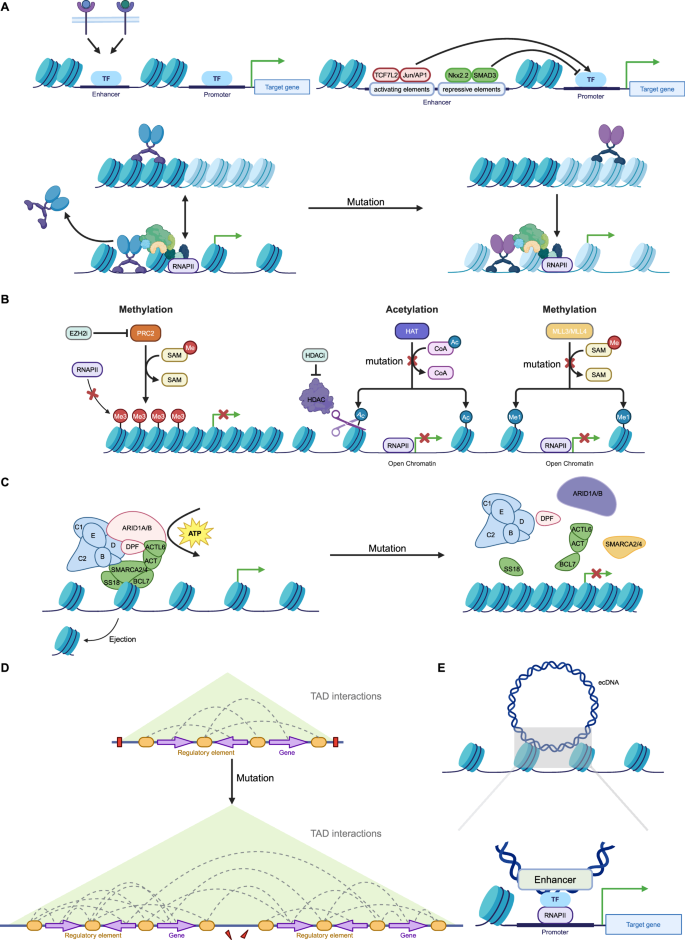
Recent evidence suggests that epigenetic regulation might control enhancer reprogramming. Indeed, the activation process of enhancers entails intricate interactions among numerous proteins, whereby any alteration in those elements can prompt enhancer reprogramming (Fig. 3) [127, 128].
A Terminal TFs from multiple signaling pathways bind to specific enhancers to respond to environmental signals. Enhancers contain activating or repressive elements, which are controlled by different TFs. However, in cancer, repressive elements are often silenced, leading to the hyperactivity of activating elements. Mutations in pioneer factors confer non-canonical functions, reshaping enhancer activity and driving cancer progression, therapeutic resistance, and phenotypic transitions. B Histone modifications, regulated by “writer” enzymes such as MLL3, MLL4, CBP, and EZH2, play a crucial role in enhancer activity and gene expression. In cancer, mutations or dysfunctions in these enzymes lead to enhancer reprogramming, promoting oncogene activation and tumor suppressor gene silencing. MLL3/MLL4 mutations impair H3K4me1 deposition, disrupting enhancer function and increasing cancer cell invasiveness. CBP/p300 mutations reduce H3K27ac, suppressing immune-related genes and enhancing oncogenic transcription. Overactive EZH2 catalyzes H3K27me3 deposition, repressing key enhancers and contributing to immune evasion and differentiation blockage. C ATP-dependent chromatin remodeling complexes, such as SWI/SNF, regulate chromatin accessibility by modifying nucleosome positioning, with mutations in their core subunits (e.g., ARID1A, SMARCA4, SMARCA2) frequently observed in cancers. These mutations compromise chromatin remodeling, altering enhancer activity and gene expression. D Disruption and rearrangement of TAD boundaries in cancer enable enhancers to form abnormal interactions with oncogenes, driving their overexpression and promoting tumor progression. E Enhancer fragments carried by ecDNA interact with target gene promoters across the genome, leading to transcriptional reprogramming. (Created with BioRender.com).
TFs and enhancer reprogramming
The high-order assemblies of TFs on enhancers allow precise regulation of target genes. This process is essential for regulating gene expression patterns during development, differentiation, and in response to environmental cues [129]. Previous research has revealed that terminal TFs associated with developmental signaling pathways, including the WNT, transforming growth factor beta (TGF-β), and leukemia inhibitory factor (LIF) pathways, demonstrate a predilection for binding to distinct enhancers, with a notable propensity for localizing to SEs (Fig. 3A) [130]. This evidence further supports the important role of TFs in manipulating enhancer function.
Recent investigations have elucidated that enhancers are multifaceted regulatory elements, offering both activating and inhibitory platforms for the interaction of diverse TFs [131]. The TF associated with the WNT signaling pathway, TCF7L2, along with binding motifs for the JUN/AP-1 and FOSL2 families, exhibit a significant enrichment in activating elements. Conversely, NK2 homeobox 2 (NKX2-2) and growth factor independent 1 (GFI1), both recognized for their cancer-suppressive roles, are preferentially associated with inhibitory elements (Fig. 3A) [132]. This dichotomy underscores the nuanced role of enhancers in dictating the intricate balance of transcriptional regulation, further implying their involvement in oncogenic processes.
MYC is overexpressed in most types of cancer and promotes oncogene transcription by binding to active promoters [133]. A recent study has shown that MYC binds to promoters, invades distal enhancers, and co-occupies them with cancer-type-specific TFs, such as ER and STAT3. MYC mediates the enhancer-specific recruitment of BRD4 through H3K9 demethylation and acetylation, thereby promoting the recruitment of RNA polymerase II and the transcription of eRNAs [134]. ER and AR are highly expressed in most types of breast and prostate cancer, driving cancer cell growth in response to hormones [135, 136]. Current results indicate that, upon hormone binding, ER and AR associate with distal enhancers and recruit numerous co-regulators and TFs to drive the expression of target genes [137]. Besides, TF mutations alter their binding affinity to enhancers and confer non-canonical functions, leading to enhancer reprogramming. For instance, mutant p53 (mutp53) reshapes the enhancer landscape in cancer cells by cooperating with NF-κB, significantly promoting the expression of tumor-associated genes and enhancing cell invasiveness [126]. Further research has revealed that mutp53 interacts with MLL4 to regulate H3K4 monomethylation at enhancers, thereby driving aberrant enhancer-mediated gene transcription [125]. Additionally, BRD4 has been shown to cooperate with mutp53 and eRNAs, facilitating chromatin accessibility and enhancer activation, which markedly upregulates inflammation-associated gene expression [138]. Another notable example is the recurrent mutation of CEBPA, which severely impairs enhancer activity in AML cells, suppressing immune-related gene expression. However, this dysfunction can be partially restored through LSD1 inhibition [139]. Further studies revealed that CEBPA mutations enhance its ability to bind and regulate the GATA2 enhancer, working in conjunction with TET2 mutations to rebalance GATA2 expression levels, thereby conferring greater competitiveness and aggressiveness to leukemia cells [15].
Moreover, the ability of pioneer factors to remodel the epigenome is also a vulnerability, as abnormal activity of pioneer factors has been detected in various types of cancer (Fig. 3A) [140, 141]. The FOXA family, known as archetypal pioneer factors, exhibits aberrant activity in various cancers [142, 143]. In prostate cancer, for instance, FOXA1 functions as a cofactor for the androgen receptor (AR), co-localizing at enhancers to coordinate hormone-regulated networks and promote cancer cell proliferation proliferation [144]. Androgen deprivation therapy (ADT) induces a redistribution of FOXA1 at enhancers, contributing to therapeutic resistance [137]. Moreover, FOXA1 mutations result in the acquisition of many non-canonical functions. In prostate cancer, FOXA1 mutations are generally categorized into three classes: Class-1 (missense mutations in the Wing2 region), Class-2 (C-terminal truncating mutations), and Class-3 (structural variants), which are predominantly associated with early-stage cancer, metastatic cancer, and advanced malignancies, respectively. In ER-positive breast cancer, mutations primarily occur in the Wing2 region and at the SY242CS site, corresponding to ER-dependent and ER-independent proliferative pathways. These mutations enable FOXA1 to bind novel, non-canonical DNA motifs, activating alternative gene expression programs and creating unique chromatin accessibility states. Additionally, mutant FOXA1 exhibits increased stability and activity at chromatin binding sites, driving stronger oncogene expression (Fig. 3A) [145,146,147]. Furthermore, in metastatic pancreatic ductal adenocarcinoma (PDA), FOXA1 becomes enriched at novel enhancer sites, driving a developmental reprogramming of PDA cells characterized by a gene expression profile resembling that of the embryonic foregut endoderm [148].
The SOX family represents another class of pioneers that bind enhancers and drive transcriptional reprogramming across various cancer types [149]. Recent studies have shown that SOX9 cooperates with TCF7L2 in gallbladder cancer to regulate each other and reprogram super-enhancers, activating multiple oncogenic signaling pathways, including ErbB and Wnt [150]. Additionally, different activation states of SOX9 during epidermal stem cell differentiation can lead to phenotypic transitions. Prolonged activation of SOX9 selectively engages tumor-associated enhancers, promoting the malignant transformation of normal cells [76]. In esophageal squamous cell carcinoma, SOX2 acquires novel genomic binding sites and collaborates with KLF5 to reprogram the epigenome. This reprogramming activates oncogenes and retroviral elements, establishing cancer cell dependency on ADAR1 [151].
While numerous studies have demonstrated that transcription factor alterations can reshape enhancers and drive phenotypic transitions in cancer cells [152], the precise mechanisms underlying these changes remain elusive. It is well established that TFs can act as both activators and repressors of transcription, but whether this dual functionality is determined by the outcome of the enhancer or the intrinsic properties of the transcription factor itself remains a topic of debate. Although some research suggests that TFs can bind different sequences within enhancers to trigger either activation or repression, the mechanisms governing this selective binding and regulation are still unclear [132]. Increasing evidence indicates that TFs often act as adaptors rather than direct regulators, exerting their effects primarily by recruiting cofactors. These cofactors influence histone modifications, chromatin remodeling, and even the 3D conformation of chromatin, thereby modulating enhancer activity and the expression patterns of target genes [153, 154]. This raises the critical question of how TFs “selectively” recruit specific cofactors to activate or repress enhancers. In fact, multiple cofactors with both activating and repressive functions coexist within cells, and their recruitment may be influenced by factors such as cell type, chromatin state, and genomic topology [155]. Some studies suggest that this selectivity may be linked to intracellular signaling pathways, chromatin accessibility, or the local environment of cis-regulatory elements [26, 156,157,158,159]. Additionally, the arrangement of TFBS, spatial conformation of enhancers, and availability of cofactors are likely to work in concert to determine the functional role of a given TF under specific conditions. Ultimately, these complex interactions construct cell-type-specific transcriptional regulatory networks, providing the molecular foundation for gene expression and phenotypic changes in cancer cells.
Chromatin modifications and enhancer reprogramming
Chromatin modifications primarily encompass DNA methylation and various histone modifications, such as acetylation, methylation, and phosphorylation, which regulate gene expression and influence cellular functions. DNA methylation typically occurs at CpG sites and represses gene expression by inhibiting transcription factor binding or altering chromatin structure. In contrast, histone modifications are dynamic, with different chemical modifications modulating chromatin compaction or relaxation, thereby affecting transcriptional activity. In cancer, the regulatory mechanisms of chromatin modifications are often disrupted, leading to widespread gene expression dysregulation and contributing to tumor development and progression (Fig. 3B) [160,161,162].
Although most enhancers are not regulated by 5mC, there is a subset of cell-type-specific enhancers that are influenced by DNA methylation. Therefore, its involvement in enhancer regions manifests in a more nuanced manner [163]. Consistent with this point, in AML, hypermethylation is associated with the silencing of enhancers, leading to the suppression of target gene expression. Although DNA hypomethylation alone is not sufficient to activate enhancers, some hypomethylated sites are associated with enhancer activation, as indicated by increased levels of H3K27ac [164]. In liver cancer, aberrant DNA methylation leads to the switching of tissue-specific enhancers during the progression of cancer, altering cell identity and affecting tumor immune surveillance [165]. The dysregulation of enhancer methylation in AML and myelodysplastic syndrome primarily manifests as hypermethylation of enhancers during myeloid lineage commitment. Mutations in tet methylcytosine dioxygenase 2 (TET2) and DNA methyltransferase 3 alpha (DNMT3A) in AML and myelodysplastic syndrome lead to either hypermethylation or hypomethylation of enhancers, thereby affecting the accessibility of specific TFBS. This epigenetic dysregulation may promote myeloid differentiation block and the development of leukemia [166].
Histone modifications constitute a highly intricate “language” of chromatin regulation [161]. In cancer, dysregulation of histone modifications is primarily driven by abnormal expression or functional alterations of epigenetic modifiers and reprogramming of metabolic pathways [98, 167]. These changes can modify the acetylation, methylation, lactylation, and other states of histones, affecting chromatin structure and gene expression. Consequently, they promote oncogene activation or tumor suppressor gene silencing, accelerating cancer progression.
MLL3 and MLL4, key “writer” enzymes responsible for depositing H3K4me1 marks at enhancers, are frequently mutated in various cancer types [168]. In breast cancer, loss of MLL3/MLL4 reduces H3K4me1 modifications at ERα target gene enhancers, impairing ERα binding and activation of these genes. This results in dysregulated expression of genes essential for cell proliferation and differentiation, increasing cancer cell invasiveness and growth [169]. Furthermore, BAP1 enhances MLL3 localization at enhancers by removing H2A ubiquitination. MLL3 mutations disrupt its interaction with BAP1, impairing enhancer localization, reducing H3K4me1 modifications, and leading to silencing of tumor suppressor genes (Fig. 3B) [170].
CBP (CREB-binding protein) and p300, histone acetyltransferases (HATs), catalyze H3K27 acetylation at enhancers to activate gene transcription [171]. Mutations in CBP/p300 represent a prominent example of enhancer reprogramming in B-cell lymphomas. These mutations lead to significant reductions in H3K27ac, resulting in sustained suppression of key enhancer functions [172,173,174]. For instance, in CBP-inactivated lymphomas, the BCL6-HDAC3 complex persistently represses immune-related genes such as MHC-II, leading to enhancer inactivation and enabling tumor cells to evade immune recognition [172]. In germinal center (GC) B cells, the inactivation of super-enhancers disrupts signaling pathways such as CD40 and BCR, impeding normal B-cell differentiation and promoting tumor transformation. Similarly, in follicular lymphoma, CBP mutations silence target genes of transcription factors such as FOXO1 and MEF2B, exacerbating enhancer inactivation and tumorigenesis [173]. Collectively, studies of cancer genomes indicate that CBP/p300 dysfunction constitutes a common enhancer reprogramming mechanism. By reducing H3K27ac levels, these mutations suppress tumor suppressor gene expression while enhancing oncogene transcription, leading to aberrant activation of genes involved in cell proliferation, survival, and immune evasion, thereby promoting tumor progression (Fig. 3B) [175].
H3K27me3, a hallmark repressive histone modification, is catalyzed by the PRC2 complex [176]. Alterations in EZH2, the catalytic subunit of PRC2, are strongly associated with various cancers [177]. In small-cell lung cancer (SCLC), overexpressed EZH2 represses CCL2 expression by depositing H3K27me3 at its enhancer, reducing macrophage infiltration into tumors and highlighting EZH2’s role in immune evasion. This epigenetic repression can be reversed by EZH2 inhibitors [178]. In breast cancer, EZH2 maintains luminal progenitor cells and restricts their differentiation by repressing enhancers of GATA3, a key TF driving luminal cell differentiation. EZH2 inhibitors remove this repressive mark, opening GATA3 enhancers, upregulating GATA3 expression, and synergizing with AKT inhibitors to induce differentiation and apoptosis in triple-negative breast cancer (TNBC) cells (Fig. 3B) [179].
ATP-dependent chromatin remodeling complexes, such as SWI/SNF, INO80, SWR1, and Mi2/chromodomain helicase DNA-binding protein (Mi2/CHD), harness the energy generated from ATP hydrolysis to induce DNA sliding over nucleosomes and histone exchange, thereby regulating chromatin accessibility [180]. Current research primarily focuses on the SWI/SNF complex and its core subunits, such as ARID1A, SMARCA4, and SMARCA2. Mutations in SWI/SNF components occur in approximately 20% of all cancer types [181], and are frequently associated with increased malignancy, poor differentiation, aggressive invasion, and therapeutic resistance [182,183,184]. These mutations compromise chromatin remodeling capabilities, particularly by altering chromatin accessibility and suppressing the expression of critical genes, thereby promoting tumor cell proliferation and survival. Notably, dual loss of SMARCA4 and SMARCA2 is prominently observed in rare, highly aggressive tumors, such as small-cell carcinoma of the ovary, hypercalcemic type (SCCOHT), and malignant rhabdoid tumors [185]. Mutations in ARID1A lead to the redistribution of enhancer-associated marks such as H3K27ac and H3K4me1, significantly increasing the expression of oncogenes (e.g., MYC, CCND1) and inflammation-related genes (e.g., IL6, TNFα), which accelerates tumor growth and facilitates immune evasion through tumor microenvironment remodeling [186, 187]. In neuroblastoma, ARID1A loss drives a transition from a neural phenotype to a mesenchymal phenotype, closely linked to enhancer reprogramming. This process activates metastasis-associated genes such as SLUG and TWIST, enhancing the invasive capabilities of tumor cells [188]. In mouse models, ARID1A mutations result in the silencing of tumor suppressor genes such as APC and TP53 while increasing the activity of oncogenic enhancers, thereby accelerating the initiation and progression of colorectal tumors [186]. In cancers with SMARCA4 mutations, SMARCA2 is often over-relied as a compensatory subunit to maintain chromatin accessibility and the expression of critical genes. It has been reported that the cooperative interaction between SMARCA2 and YAP/TEAD at enhancers is a key mechanism driving oncogene expression. Further inactivation of SMARCA2 disrupts the stability of YAP/TEAD binding at enhancer regions, thereby impairing downstream transcriptional regulatory networks (Fig. 3C) [189].
3D genome reprogramming reshapes chromatin topology by re-establishing enhancer-promoter contacts, leading to the formation of new gene regulatory circuits and the reorganization of chromatin compartments [190]. In pancreatic cancer, structural variations induce large-scale rearrangements of chromatin A/B compartments, TADs, and chromatin loops, which in turn affect the expression of cancer-driver genes such as CDKN2A and SMAD4 [191]. This 3D genome reprogramming plays a critical role not only in tumorigenesis but also in metastasis, where it is further intensified. Such reprogramming is associated with the upregulation of specific genes, including metastasis-associated genes like LIPC, whose expression is driven by metastasis-specific enhancer-promoter loops [190].
Disruption of TAD boundaries allows enhancers to rewire their connections to target genes, thereby promoting oncogene expression (Fig. 3D) [192]. In cancer, rearrangement of the TAD boundary near the IRS4 gene triggers enhancer hijacking, leading to its significantly increased expression [108]. Similarly, comprehensive epigenomic analyses have revealed widespread TAD fusion events in T-ALL, particularly involving MYC. Disruption of MYC’s TAD results in abnormal interactions between its promoter and distal enhancers, driving MYC overexpression [193]. Furthermore, chromatin interactions within TADs are significantly altered in cancer, especially between key enhancers and promoters. For instance, in T-ALL, increased chromatin interactions within TADs correlate with upregulated gene expression, and many T-ALL-specific enhancers are located within highly active TADs. Structural proteins such as CTCF and cohesin play critical roles in maintaining these interactions, enhancing chromatin interaction stability [193]. In summary, the reconfiguration of the three-dimensional chromatin structure allows enhancers to establish new contacts with promoters, leading to the formation of cancer-specific gene regulatory circuits. Disruption and rearrangement of TAD boundaries enable enhancers to bypass their original domain constraints and form aberrant interactions with oncogenes, thereby driving cancer progression.
Other factors and enhancer reprogramming
The ecDNA comprises small DNA fragments that are excised from chromosomes [194]. Recent research has identified that ecDNA carries mobile enhancers, which function as trans-acting activating elements and lead to the widespread deregulation of gene expression; thus, ecDNA correlates with cancer (Fig. 3E) [194,195,196]. A regulatory pattern similar to this mobile mechanism can also be mediated by TEs. TEs are repetitive genomic elements with binding sites for multiple TFs that exhibit enhancer characteristics [197]. Aberrant insertion of TEs into the genome can lead to dysregulation of gene expression. Furthermore, research indicates that the presence of tissue-specific TFs activates TEs across various cancer types [198].
Non-coding RNAs are integral to the orchestration of enhancer reprogramming, often acting as mediators that influence enhancer activity and gene expression. They can participate in the regulation of chromatin states by recruiting remodeling complexes, modulating the interactions between enhancers and promoters, and affecting the binding of TFs to DNA [199]. eRNAs are transcribed from active enhancer regions and might participate in various cancer signaling pathways by modulating their target genes [200]. Long non-coding RNAs can act as scaffolds for chromatin-modifying enzymes, directing them to specific genomic loci. In addition, they can sequester proteins away from chromatin, thereby influencing gene expression patterns [199].
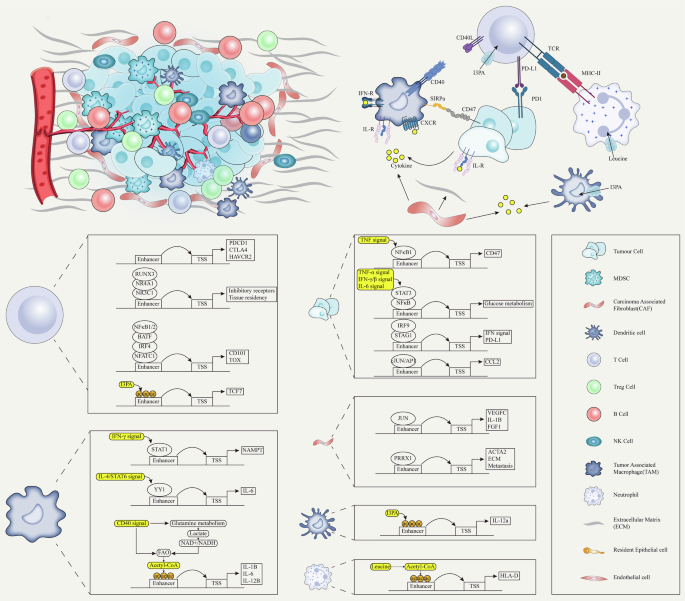
Critical roles of enhancers in cancer: unlocking phenotypic plasticity
“TFs-oncogenes-enhancer” core transcription regulatory circuitry
The enhancer-driven core regulatory circuitry (CRC) encapsulates the synergistic interplay between a constellation of core regulatory factors (CRFs) and enhancers, orchestrating the intricate gene expression patterns that underpin cellular identity and functionality [201]. This paradigm holds paramount importance for elucidating the sophisticated genetic regulatory mechanisms governing cell differentiation, ontogeny, and pathological states. At the center of the CRC lies an ensemble of CRFs—predominantly master TFs—that possess the capacity to engage with cell-type-specific enhancers, thereby catalyzing distinct gene expression profiles essential for the establishment and perpetuation of cellular identity [202]. Notably, these CRFs often engage in reciprocal positive feedback loops, reinforcing cellular states and directing cellular destiny throughout the differentiation process [201]. Nevertheless, in the context of oncogenesis, this intricate regulatory network is misused, with some components being used to facilitate the delineation of cancer subtypes, while the majority are hijacked to unlock the phenotypic plasticity inherent to malignancies. This aberrant exploitation of the CRC enables cancer cells to dynamically acquire a repertoire of capabilities essential for tumorigenesis, including unbounded proliferation, apoptosis evasion, tissue invasion, and metastatic dissemination—hallmarks that epitomize malignant cells [203, 204]. Table 2 below provides a summary of the core regulatory circuits as delineated in prevalent tumors to date in different studies.
Enhancer reprogramming drives dynamic phenotypic plasticity in cancer
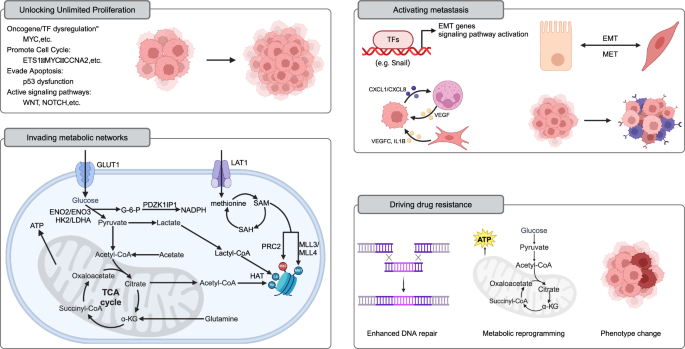
A large number of altered enhancers have been identified in cancer [124]. These oncogenic enhancers act as relay stations within the cell, integrating internal and external signals and regulating various life activities of cancer cells. They dynamically provide cancer cells with the necessary growth advantages and new phenotypes (Fig. 4) [130, 205].
Unlocking Unlimited Proliferation: MYC, as a principal regulator of cell proliferation, undergoes enhancer reprogramming widely across various tumors, driving cancer transformation and forming a shared paradigm of foundational enhancer reprogramming in cancer. Additionally, enhancer reprogramming disrupts normal growth inhibition signals by regulating the expression of cell cycle factors (e.g., MYC, CCNA2), enabling cancer cells to sustain continuous division. It also promotes tumor growth by upregulating genes like MDM2, which suppress p53 function. Enhancer reprogramming collaborates with signaling pathways such as WNT and NOTCH to sustain infinite cancer cell proliferation by forming core regulatory circuits (CRCs). For instance, in liver cancer, WNT/β-catenin signaling activates key enhancers, driving the upregulation of the DLK1/DIO3 genomic locus and promoting tumor growth and progression. Invading metabolic networks: Enhancer reprogramming disrupts metabolic pathways in tumor cells, driving significant changes in energy production, biosynthesis, and gene regulation. Enhancers upregulate glucose transporters (e.g., GLUT1) and glycolytic enzymes (e.g., HK2, LDHA), promoting the Warburg effect and supporting anabolic metabolism. They also amplify lipid synthesis by regulating factors like SREBF1 and SREBF2, meeting the high demand for fatty acids and cholesterol in cancer cells. Similarly, amino acid metabolism is enhanced through CRC-regulated genes (e.g., LAT1), promoting tumor growth and survival. Enhancer-driven NAD metabolism reprogramming supports energy production and redox balance, further fueling cancer cell proliferation. Moreover, metabolites from these metabolic pathways, such as acetyl-CoA and S-adenosylmethionine, serve as substrates for histone and DNA modifications, establishing a feedback loop between metabolism and epigenetic remodeling to sustain tumor progression. Activating metastasis: Enhancer reprogramming plays a crucial role throughout various stages of tumor metastasis by driving the epithelial–mesenchymal transition (EMT), modulating the tumor microenvironment, and enabling immune evasion and colonization at distant sites. It activates key transcription factors (e.g., Snail, Twist) and signaling pathways (e.g., TGF-β, WNT/β-catenin), enhancing cancer cell migration, invasion, and adaptation to new microenvironments. During metastasis, enhancer-driven transcriptional reprogramming promotes organ-specific gene expression programs, such as FOXA1-mediated liver metastasis in pancreatic cancer. Additionally, epigenetic memory maintained by enhancers allows tumor cells to adapt and proliferate rapidly in distant organs (such as MET). Driving drug resistance: Enhancer reprogramming plays a pivotal role in cancer chemotherapy resistance through mechanisms such as upregulation of drug resistance genes, enhanced DNA repair, metabolic reprogramming, and lineage plasticity. (Created with BioRender.com).
Unlocking unlimited proliferation
The oncogene MYC, a principal regulator of cell proliferation, is dysregulated across a broad spectrum of cancer types [133]. Recent investigations encompassing pan-cancer analyses have elucidated that the reprogramming of MYC gene enhancers is prevalent in a diverse array of human tumors. This evidence underscores the pivotal influence of “basic enhancer” reprogramming in the process of cancer transformation. Moreover, this implies that the occurrence of cancer may share a common paradigm of reprogrammed foundational enhancers [132].
Furthermore, enhancer reprogramming can precipitate alterations in the expression profiles of cell cycle regulatory factors. Such changes facilitate the dysregulation of the cell cycle, enabling cancer cells to circumvent conventional growth inhibition signals and continuously enter the cell division cycle [206]. For instance, YAP/TAZ serve as nuclear effectors orchestrating tumorigenesis via the Hippo pathway. Serving as TFs, they occupy enhancers with cofactor AP-1 and directly modulate target genes imperative for cell progression into the S phase and mitosis, such as ETS1, MYC, CCNA2, centromere protein F (CENPF) [74].
Under physiological conditions, cells respond to various growth inhibitory signals, such as p53-mediated apoptosis, to prevent uncontrolled proliferation [207]. While enhancer reprogramming does not typically induce mutations in p53, it can facilitate the expression of its antagonistic genes, such as MDM2, thereby neutralizing its function. Ubiquitin-specific peptidase 12 (USP12), a deubiquitinating enzyme, is driven by amplified SEs. The overexpression of USP12 shields MDM2 from degradation, leading to the ubiquitination and subsequent proteasomal degradation of p53 [208].
Certain signaling pathways, such as WNT, and NOTCH, play crucial roles in promoting cell proliferation [209]. Oncogenic enhancers facilitate infinite cancer cell proliferation by intercepting the terminal TFs of these pathways and forming CRCs [132]. Mutations in catenin beta 1 (CTNNB1) in cancer lead to increased stability of the β-catenin protein and sustained activation of the WNT/β-catenin signaling pathway. The sustained activation of β-catenin, in combination with the TCF4 complex, binds to the delta-like non-canonical Notch ligand 1- Wnt responsive element (DLK1-WRE) enhancer site upstream of maternally expressed 3 (MEG3), promoting chromatin opening and the deposition of H3K4me1 and H3K27ac. This drives the upregulation of the DLK1/iodothyronine deiodinase 3 (DLK1/DIO3) genomic locus, thus promoting the growth and progression of liver tumors [210].
Invading metabolic networks
Recent studies have shown that enhancer reprogramming disrupts metabolic pathways, leading to significant changes in intracellular metabolite levels. These changes alter energy production and material metabolism in tumor cells and regulate gene expression by affecting epigenetic modifications [211]. For example, the enhancer-driven MYC is closely related to glucose metabolism, lipid synthesis, and nucleotide synthesis [212].
In tumor cells, glucose is an important energy source and biosynthetic precursor [213]. Enhancers increase glucose uptake by upregulating the expression of glucose transporters, such as glucose transporter type 1 (GLUT1), and enhance the expression of glucose metabolic enzymes (such as enolase 2 [ENO2] and ENO3) [214, 215]. Additionally, enhancers activate key enzymes in the glycolysis pathway (e.g., hexokinase 2 [HK2] and lactate dehydrogenase A [LDHA]), thereby promoting the Warburg effect and increasing glycolytic flux [216, 217]. More importantly, downstream metabolites of glycolysis further support the anabolic metabolism of cancer cells and participate in the biosynthesis of ribose, amino acids, and lipids, while also regulating the intracellular redox balance. For example, SE-driven PDZK1 interacting protein 1 (PDZK1IP1) enhances the reductive capacity of colorectal cancer cells through the pentose phosphate pathway [215].
Tumor cells prefer de novo lipid synthesis, increasing the synthesis of fatty acids and cholesterol, which supports the construction of cell membranes and signal transduction [218]. SE promotes lipid synthesis pathways by regulating sterol regulatory element binding transcription factor 1 (SREBF1) and SREBF2, meeting the high demand of tumor cells for lipids [215, 219]. Furthermore, in hepatocellular carcinoma, SE-driven fatty acid synthesis-related lncRNA (FASRL) binds to acetyl-CoA carboxylase 1 (ACC1; a rate-limiting enzyme in fatty acid synthesis) and inhibits its phosphorylation, thereby promoting fatty acid synthesis [220]. The CRC formed by E74 like ETS transcription factor 3 (ELF3), KLF5, and GATA6 upregulates peroxisome proliferator-activated receptor gamma (PPARG) in esophageal cancer, leading to increased synthesis of fatty acids, phospholipids, and sphingolipids [221].
Amino acid metabolism in tumors promotes the growth and survival of cancer cells by providing energy, supporting anabolic metabolism, regulating cell signaling, and influencing epigenetic modifications [222, 223]. Of note, enhancers further amplify this effect by regulating key genes in these metabolic pathways [224]. The CRC driven by tumor protein p63 (TP63), KLF5, and SREBF1 leads to the high expression of the methionine transporter L-type amino acid transporter 1 (LAT1) in squamous cell carcinoma, significantly promoting the accumulation of methionine within the cells [225].
Nicotinamide adenine dinucleotide (NAD) is an essential coenzyme. It plays a crucial role in cellular metabolism, participating in various biochemical reactions, particularly acting as an electron carrier in redox reactions [226]. In cancer cells, NAD metabolism is significantly reprogrammed to support energy production and antioxidant defense. The increased demand for NAD+ in cancer cells drives their dependency on NAD synthesis pathways [227]. Recent studies have revealed enhancer-driven amplification of nicotinate phosphoribosyltransferase (NAPRT) and the dependency of cancer cells on NAPRT. Additionally, enhancer reprogramming has led to nicotinamide phosphoribosyltransferase (NAMPT), another rate-limiting enzyme in the NAD synthesis pathway, being counteracted by nicotinamide riboside kinase-dependent (NMRK1-dependent) NAD synthesis [228].
More importantly, metabolites produced by the enhancer-driven rapid metabolic network, such as acetyl-CoA and S-adenosylmethionine, serve as substrates for histone modifications and DNA methylation, directly affecting the epigenetic modifications of enhancers [211, 229]. Consistent with this, our team recently reported that during liver metastasis of pancreatic cancer, enhancer reprogramming promotes glycine amidinotransferase (GATM)-mediated guanidinoacetate metabolism. This process further facilitates pancreatic cancer liver metastasis through transcriptional activation and histone modification mediated by guanidinoacetate [230]. In summary, metabolism and epigenetic remodeling form a remarkably complex network of mutual regulation, thus promoting tumor growth.
Evading cell death
Apoptosis is mainly mediated through two major pathways, namely the intrinsic pathway (mitochondrial pathway) and the extrinsic pathway [231]. The permeabilization of the mitochondrial outer membrane constitutes a critical event in the intrinsic pathway, leading to the release of cytochrome c into the cytoplasm and triggering a subsequent caspase cascade, which is typically regulated by the BCL2 family [232]. In cancer, enhancer-driven overexpression of BCL2 counteracts the pro-apoptotic effects of BAX, thus protecting cells from death [233]. Notably, the use of inhibitors targeting enhancers, such as JQ1, can induce growth arrest and apoptosis in cancer cells [234]. The extrinsic pathway is initiated by extracellular signals, predominantly through the activation of death receptors, such as Fas and TNF receptors. Previous studies have reported that hypermethylation of enhancers in cancer leads to the suppression of Fas expression, thereby diminishing the sensitivity to Fas-mediated extrinsic apoptosis [235]. In addition, BRD4 exerts protective effects in various types of cancer by binding key enhancers to drive the expression of apoptosis resistance genes [236].
Furthermore, the activation of survival signaling pathways, such as NF-κB, PI3K/AKT, MAPK, and JAK/STAT, can also counteract death signals to promote the survival of cancer cells [237,238,239]. Enhancers provide a platform for the operation of various signal transduction pathways. On the other hand, in cancer, enhancer reprogramming results in transcriptional dysregulation of various oncogenes and TFs, perpetuating the activation of these pathways. For instance, in squamous cell carcinoma, TP63 and SOX2 activate the enhancer of EGFR, further stimulating the MEK/ERK1/2 and PI3K/AKT signaling [12].
Activating metastasis
The metastatic process of tumor cells commences with the invasion and migration of primary foci into adjacent tissues, in which EMT plays a significant role [240]. Research indicates that enhancer reprogramming plays a crucial role in this process by modulating the gene expression patterns of cancer cells, facilitating their transition from an epithelial to a mesenchymal phenotype. This process endows cancer cells with enhanced migratory and invasive capabilities [100]. Mechanistically, enhancer reprogramming activates key signaling pathways and TFs associated with EMT (e.g., TGF-β, WNT/β-catenin, and NOTCH) [241], as well as typical TFs (e.g., Snail, Slug, Twist, and Zeb) [242]. A recent study has demonstrated that the inhibition of FOXA2 induced by TGF-β, along with the activation of TEAD2/4, facilitates the reprogramming of a set of enhancers pre-existing within TADs, further activating the EMT process and promoting cancer cell metastasis [100]. This regulation of the gene networks intimately associated with the EMT process encompasses the suppression of epithelial markers and the expression of mesenchymal markers, the enhancement of cell adhesion, migration, and invasion, as well as the modulation of the tumor microenvironment (TME) [215, 243,244,245], thereby further promoting tumor metastasis and dissemination. For instance, in clear cell renal cell carcinoma, the formation of SEs robustly drives the expression of various CXC chemokines, such as CXCL8 and CXCL1. This process establishes an inflammatory immune microenvironment that facilitates neutrophil-dependent lung metastasis [246]. Of note, research indicates that the enhancer-driven pro-metastatic microenvironment extends beyond cancer cells. The remodeling of enhancers within other components of the TME also significantly impacts cancer progression. For instance, the activation of JUN is underscored as a pivotal factor in the activation of cancer-associated fibroblast-specific (CAF-specific) enhancers, promoting the expression of pro-metastatic genes and, thereby, augmenting breast cancer invasiveness in a non-cancer-cell-autonomous manner [247]. This suggests that enhancer reprogramming within the stromal cells of the TME plays a significant role in facilitating cancer progression and metastasis.
Notably, enhancer reprogramming amplifies the transcriptional output of metastasis-associated genes and orchestrates a complex transcriptional network that collectively enhances the metastatic potential of cancer cells, such as those associated with embryonic or stem cell-like properties, which are closely related to the metastatic capabilities of cancer cells [148, 248]. For instance, the activation of FOXA1-dependent enhancer drives an embryonic foregut endoderm transcriptional program, rendering pancreatic cancer cells more invasive and facilitating their liver metastasis [148].
Upon their departure from the primary site, cancer cells directly encounter immune cells within the circulation. Prior research has shown that cancer cells may evade immune surveillance through interactions with blood cells, such as platelets and neutrophils [249, 250]. Nonetheless, the specific role of enhancer reprogramming in this context remains to be elucidated.
Studies have demonstrated that enhancers can elevate the expression of immune checkpoint inhibitors, such as programmed cell death-ligand 1 (PD-L1), thus promoting immune escape [251]. However, the potential exploitation of this mechanism by circulating tumor cells remains under investigation. The tumor cells that survive the circulatory journey and arrive at new locations are similarly subjected to the pressures of colonization. During this phase, enhancer reprogramming facilitates the successful settlement of tumor cells and the formation of new tumors by modulating genes associated with adaptation to the microenvironment, cellular proliferation, and evasion from immune responses [243, 252]. Additionally, cancer cells that have experienced EMT may undergo a reversible process during the colonization stage, namely the mesenchymal–epithelial transition, thereby regaining epithelial characteristics to promote growth. Intriguingly, it was recently discovered that epigenetic memory explains this phenomenon. Certain enhancers, which are induced to shut down during the EMT process, do not directly enter a quiescent state; instead, they maintain a certain level of H3K4me1 modification, remaining in a primed state. When reaching a new microenvironment, these enhancers are rapidly reactivated, driving the cell transformation into an epithelial phenotype [100]. Furthermore, metastatic tumor cells may enter a state of dormancy rather than immediately forming new, a condition that might persist for years. Nevertheless, these cells are capable of rapidly resuming proliferation in response to microenvironmental signals. Recent research suggests that this phenomenon can also be explained through epigenetic memory. For instance, a set of enhancers regulated by TFs (e.g., SOX9) and located within variable chromatin structures, can activate a strong transcriptional response upon exposure to retinoic acid [252].
Cancer cells of the same type tend to metastasize to specific organs; however, the precise underlying mechanisms remain unclear [253]. Recent evidence increasingly indicates that enhancer-driven transcriptional reprogramming plays a significant role in this process. For instance, during liver metastasis of pancreatic cancer, the original pancreatic developmental program is replaced by a liver developmental program, achieved through FOXA1-driven enhancer reprogramming [148]. Similarly, in colorectal cancer cells metastasizing to the liver, enhancers acquire liver-specific TFs, FOXA2 and HNF1A, thereby activating liver-specific gene transcription. Surprisingly, further transcriptomic analyses across various cancer types revealed that similar transcriptional reprogramming occurs in distant metastases of other cancers, such as colon cancer to the lung, prostate cancer to the bone, kidney cancer to the lung, and breast cancer to the brain [254]. RUNX2 is a crucial TF associated with bone development and exhibits pioneer activity [255]. In prostate cancer, FOXO1 binds to and inhibits RUNX2-mediated bone metastasis, a finding supported by clinical data [256]. Overall, these results strongly support the notion that organotropic metastasis of cancer may be mediated by acquired enhancer-driven transcriptional reprogramming. Changes in transcriptional programs confer metastatic cancer cells with characteristics of distant organs, aiding their adaptation and colonization. These findings provide new insights into the mechanisms of cancer metastasis and lay the theoretical foundation for developing novel therapies against metastasis.
In summary, the above results underscore the regulatory role of enhancer reprogramming throughout various stages of tumor metastasis. Nonetheless, tumor metastasis remains an intricately coordinated process, involving numerous mechanisms that remain to be elucidated. For instance, extensive research has highlighted the role of exosomes in metastasis and their significance in establishing pre-metastatic niches in distant organs [257]. Concurrently, eRNAs have been identified within exosomes and are associated with the long-range regulatory functions of enhancers [258]. However, the mechanism through which enhancers regulate pre-metastatic niches has not been thoroughly investigated. Following the recent discovery of epigenetic memory, the mechanism by which tumor cells remember and respond to changes in their microenvironment at the molecular level has become clearer. This mechanism elucidates the process of tumor cell dormancy and resuscitation and reveals the process through which tumors rapidly adapt to new environments and promote proliferation upon distant colonization. Epigenetic memory, by maintaining the active state of specific enhancers, enables tumor cells to swiftly activate or suppress the expression of particular genes at opportune moments, thus playing a pivotal role during metastasis and colonization [32]. These insights provide crucial clues for the development of new therapeutic strategies targeting tumor metastasis, especially those aimed at disrupting the epigenetic memory of tumor cells to block their metastatic and colonization capabilities.
Driving drug resistance
Studies have proposed several mechanisms of chemotherapy resistance in cancer that are mediated by enhancer reprogramming, including the upregulation of drug resistance genes, enhanced cell proliferation [206], resistance to cell death [259], enhanced DNA damage repair [260], reactivation of signaling pathways [261], stemness [262], metabolic reprogramming [263], and lineage plasticity [155, 264].
Based on the concept of synthetic lethality, poly (ADP-ribose) polymerase (PARP) inhibitors (PARPi) have been extensively utilized in the treatment of tumors carrying BRCA mutations (e.g., breast and pancreatic cancers) to target cancer cells with homologous recombination repair (HRR) deficiencies [265]. However, resistance to PARPi partially limits their clinical application, with the principal mechanism of resistance being the restoration of HRR [266]. A recent study in ovarian cancer has demonstrated the presence of SEs at the KLF5 locus and formed CRC. Elevated expression of KLF5 promotes the expression of key HRR pathway genes, including RAD51, checkpoint kinase 1 (CHEK1), RAD54 like (RAD54L), essential meiotic structure-specific endonuclease 1 (EME1), and Bloom syndrome (BLM), thereby reversing the HRR suppression mediated by PARPi. Moreover, epigenetic targeting of KLF5 can restore sensitivity to PARPi [260].
Dysfunction in signaling pathways emerges as a critical mechanism underlying acquired drug resistance. Targeted therapeutic approaches designed to inhibit specific signaling pathways are significantly challenged by the reactivation of these pathways, which leads to drug resistance [267]. Serving as hubs for multiple signaling pathways, enhancers contribute to tumor growth recovery by reactivating targeted signaling pathways or activating alternative bypass routes. For instance, the downregulation of MAPK pathway negative regulators, mediated by enhancer reprogramming, leads to the reactivation of the MAPK pathway, resulting in resistance to MEK inhibitors. However, the combination of histone deacetylase (HDAC) inhibitors (i.e., HDACi), can effectively overcome this resistance [261].
Prolonged use of fibroblast growth factor receptor (FGFR) inhibitors may lead to the suppression of SWI/SNF, thereby activating YAP-dependent enhancers. This, in turn, alters the mechanistic target of the rapamycin kinase-mediated (mTOR-mediated) amino acid metabolic pathway, leading to tumor growth [263]. Additionally, under BRAF/MEK inhibitor treatment, enhancers regulated the formation of new transcriptional states and activated oxidative phosphorylation as the primary energy source for drug-resistant myeloma cells [268]. The above evidence supports the connection between metabolic reprogramming and cancer drug resistance, highlighting the role of enhancers in this process.
Lineage plasticity refers to the evolution of cells from one differentiated state to another. Recent studies have discovered that such transformations can determine therapeutic responses [269]. Therapy-induced shifts in enhancer landscapes facilitate phenotypic transitions in cancer cells, leading toward drug resistance [270]. During the treatment of lung adenocarcinoma with KRAS inhibitors, a subset of patients exhibits adeno-to-squamous transition, which is associated with the activation of the enhancer for the squamous lineage TF ΔNp63. The upregulation of ΔNp63 activates signals for the adeno-to-squamous transition plasticity signature, thereby promoting KRAS inhibitor resistance [264]. In breast cancer, the ERα, along with other oncogenic TFs (e.g., GATA3 and AP-1), drives global enhancer reprogramming and the reconfiguration of transcriptional networks. This leads to the transdifferentiation of breast cancer cells from a luminal/epithelial state to a basal/mesenchymal phenotype, thereby acquiring endocrine resistance [155]. Similarly, in prostate cancer, the inhibition of the AR pathway activates changes in the FOXA2-dependent transcriptional pattern, driving the adeno-to-neuroendocrine transition, thereby causing castration resistance [271].
Taken together, the multifaceted role of enhancer reprogramming in cancer resistance not only enhances our understanding of complex resistance mechanisms but also provides a foundation for developing new treatment strategies. These strategies may include direct interventions targeting enhancer activity, or reversing drug resistance by altering epigenetic status, providing new directions for cancer treatment.
Maintaining stemness
CSCs are typically characterized by functions such as sustaining self-renewal, initiating tumor development, and mediating drug resistance. Recent reviews have delved into the epigenetic regulatory mechanisms involved in the formation of CSCs [272].
Typically, enhancers maintain stemness by driving the expression of stem cell TFs, such as SOX2, OCT4, c-MYC, and KLF4 [203, 273]. These TFs are pivotal in sustaining the core regulatory network essential for stem cell identity, self-renewal capabilities, and pluripotency. Enhancer reprogramming further modulates the chromatin landscape, rendering it more permissive for the transcriptional machinery to access key stemness genes, such as TP63, MET, FOSL1, and X-box binding protein 1 (XBP1) [248, 274, 275]. This ensures the continuous expression of genes that are critical for maintaining a stem-like state and allows for the rapid adaptation of cancer cells to environmental stresses or therapeutic interventions. Additionally, this reprogramming supports the phenotypic plasticity of cancer cells, enabling transitions between different cellular states, for instance, in breast cancer, overexpression of MYC triggers phenotypic transitions and induces a stem cell-like state [276]. Therefore, the role of enhancers extends beyond the mere activation of individual genes; it encompasses the orchestration of complex gene networks that collectively uphold the stem cell-like characteristics within tumors, presenting a significant challenge and opportunity for cancer therapy.
Enhancer-based subtypes of cancer
The specific tissue distribution is among the most remarkable features of enhancers. Most enhancers are inactive in a given tissue or organ, and only a few that determine cell identity are activated and involved in cell fate determination [54]. This characteristic is maintained in tumors. Genome-wide studies have shown that different cancer subtypes show significantly varied enhancer maps. In other words, different enhancer landscapes drive the emergence of different cancer subtypes [277].
Many studies have revealed the potential of enhancers in profiling cancer subtypes. Wu et al. previously summarized the relevant reports, and we made some additions on this basis [277] (Table 3).
Enhancer reprogramming and TME
The TME is a highly complex ecosystem, consisting of a variety of cellular and non-cellular components, which profoundly influences the evolution of cancer [278]. Extensive research has unveiled the critical impact of enhancer-driven TME remodeling on tumor progression, treatment response, and resistance to therapy. Within this biological framework, enhancer reprogramming serves as a mechanism driving the intrinsic transformation of cancer cells and directly shapes the dynamic changes in the TME. This regulatory mechanism encompasses aspects ranging from immune cell modulation and extracellular matrix remodeling to angiogenesis (Fig. 5) [279,280,281].
Different cells in the tumor microenvironment respond to various signals (such as cytokines, intercellular receptor-ligand interactions, metabolites, etc.) to regulate enhancer-driven transcriptional programs. These signals modulate enhancer activity, driving the reprogramming of cancer cell transcriptional programs and remodeling of the tumor microenvironment.
Despite the substantial infiltration of T cells within the TME, a significant portion inevitably progresses toward an exhausted state (exhausted T cells [TEX]) [282]. A recent study reported on the epigenetic mechanisms underlying the heterogeneous function and dysfunction status of tumor-infiltrating lymphocytes. Specifically, six groups of TME-infiltrated CD8+ T cells were defined based on their chromatin status and connected to specific gene and enhancer/promoter pairs. Clusters 1 and 2 were associated with exhaustion, characterized by high programmed cell death 1 (PD-1) expression and activity of exhaustion markers, such as hepatitis A virus cellular receptor 2 (HAVCR2), thymocyte selection associated high mobility group box (TOX), TOX2, and layilin (LAYN). Cluster 3 was associated with progenitor TEX (TPEX), while clusters 4 and beyond represented memory, cytotoxicity-associated, and heat-shock response genes, indicating diverse functional states within the T cell population [283]. Using single-cell ATAC-seq technology to analyze tumor-infiltrating T cells, 19 subpopulations were identified. In TEX cells, enhancers associated with inhibitory receptor genes were discovered, such as the +5 kb enhancer of PDCD1 (encoding PD-1), which showed specific accessibility in TEX. Similarly, distal enhancers at the cytotoxic T-lymphocyte associated protein 4 (CTLA4) and HAVCR2 gene loci also exhibited specific activity in TEX, further supporting the notion that the exhausted state may be regulated by state-specific enhancers. Additionally, TEX undergoes two stages during differentiation. The first stage (intermediate exhaustion stage) involves increased accessibility of cis-regulatory elements near inhibitory receptor genes, while the second stage (terminal exhaustion stage) involves increased accessibility of cis-regulatory elements near genes associated with terminal T cell dysfunction, such as CD101 and TOX [280]. Notably, the exhausted state of T cells appears to be reversible. The indole-3-propionic acid (I3PA) produced by the cooperation between Lactobacillus johnsonii and Clostridium sporogenes in the gut increases the H3K27ac at the SE of TCF7 in TPEX, reactivates TPEX, and maintains stemness in CD8+ T cells [284]. Additionally, I3PA promotes the production of IL12a by increasing the deposition of H3K27ac at the IL12a enhancer in dendritic cells, thereby enhancing the function of tumor-infiltrating CD8+ T cells [285]. Furthermore, enhancer reprogramming within cancer cells can also promote the expression of PD-L1 on the cell surface, thereby mediating immune escape [251, 286, 287].
The interactions between tumor-associated macrophages (TAMs) and cancer cells, as well as stromal cells within the TME, through mechanisms of enhancer reprogramming, can further sustain and exacerbate various characteristics of cancer [288]. Local IFN-γ signaling activates STAT1 and manipulates the intronic enhancer of nicotinamide phosphoribosyl transferase (NAMPT). Subsequently, NAMPT influences the glucose metabolism and tricarboxylic acid cycle of TAMs, driving the anti-tumor reprogramming of TAMs [289]. Similarly, the activation of the CD40 signal also leads to the activation of enhancers at pro-inflammatory gene loci in TAMs through metabolic reprogramming, thereby coordinating the polarization of the anti-tumor phenotype [290]. Conversely, enhancer-driven C-C motif chemokine ligand 2 (CCL2) production in pancreatic ductal adenocarcinoma leads to the recruitment of TNF-α-positive macrophages and triggers the transition from a “classical” subtype to a more aggressive “basal-like” subtype [288]. Intriguingly, drug-induced enhancer activation can reprogram the M2-type into the M1-type, thereby restoring the anti-tumor effects of macrophages [291]. In summary, these results highlight the crucial role of enhancer reprogramming in modulating the interactions between TAMs and TME, thereby significantly impacting cancer progression.
Within the complex ecosystem of the TME, neutrophils (tumor-associated neutrophils [TANs]) emerge as pivotal players whose functions extend beyond traditional roles in inflammation and immune defense [292]. A recent report identified 10 states of TANs, demonstrating their tissue and phenotypic plasticity. Diversity in metabolic pathways may be associated with this phenomenon, further suggesting a complex interplay between metabolic reprogramming and the multifaceted roles of TANs within the TME. Intriguingly, among them, leucine metabolism facilitates the activation of the SE of major histocompatibility complex-II (MHC-II) through the production of acetyl-CoA, thereby enhancing the antigen presentation mechanism and effective activation of T cells [293]. Besides, cancer cell-derived cytokines could shape an inflammatory TME, thereby triggering neutrophil-dependent metastasis [246]. These studies suggest that the activity and behavior of TANs are intricately regulated by epigenetic mechanisms, among which enhancer reprogramming stands out as a key modulator.
Additionally, CAFs stand out as key architects in the stromal component and contribute to the creation of a protective niche for cancer cells through epigenetic modulation [294]. For example, cytokines secreted by CAFs, such as IL6 and IL8, can promote the phosphorylation of the BRD4 protein. In turn, BRD4 binds to enhancers, inducing chromatin remodeling and supporting the transcriptional mechanisms within cancer cells [295]. Within CAFs, enhancer reprogramming is one of the key mechanisms driving the transition from normal fibroblasts to promoters of cancer progression. For example, in breast cancer, studies have found that the activation of the JUN TF binding to specific enhancers in CAFs drives the reprogramming of these enhancers, thus activating the expression of a series of genes that promote tumor invasiveness and metastasis [247]. Paired related homeobox 1 (PRRX1) has been identified as a master TF controlling the myofibroblastic lineage progression in CAFs. Through integrated gene expression and chromatin accessibility analyses, studies have found that PRRX1 drives CAF activation by reprogramming enhancer activity, such as upregulating actin alpha 2, smooth muscle (ACTA2), and other genes involved in extracellular matrix remodeling and cell migration [296].
The presence of inflammatory clues is another important feature of the TME [297]. The TNF-NFκB1 signaling pathway directly regulates the expression of CD47 by binding to a constituent enhancer within a breast cancer-specific SE. In turn, CD47 inhibits the phagocytic activity of immune cells (e.g., macrophages) by interacting with the SIRPa receptor on these cells [298]. Mutations in stromal antigen 2 (STAG2) activate interferon regulatory factor 9 (IRF9) by regulating 3D genome organization, thereby enhancing type I interferon signaling and increasing PD-L1 expression. This process is associated with the replacement by STAG1, as well as increased H3K27ac signaling and the formation of new E-P loops [299]. Additionally, TNF-α, IFN-γ, and IL6 signals induce SE formation in colorectal cancer, with inflammatory signaling further directly regulating their activity through NF-κB and STAT3 [215].
Overall, these findings provide a comprehensive overview of the crucial role of enhancer reprogramming in the TME, impacting the dynamics between cellular and non-cellular components. It highlights the complexity of the TME, where cellular and epigenetic mechanisms (including enhancer reprogramming) drive significant changes in immune cell function, thereby contributing to cancer progression, immune escape, and therapy resistance.
Targeting carcinogenic enhancers: promising strategies for transcription network normalization
Targeting carcinogenic enhancers presents a groundbreaking approach in the quest to normalize the transcription networks that are pivotal for cancer progression. Dysregulation of these regulatory elements is instrumental in activating oncogenic pathways and sustaining the malignant phenotype of cancer cells. By specifically addressing these enhancers, researchers aim to disrupt the aberrant transcriptional programs that drive tumor growth and resistance mechanisms. This strategy holds the potential to precisely curtail the expression of oncogenes and restore the normal regulatory circuits of the cell, offering a promising direction for cancer therapy that targets the genetic underpinnings of the disease. As we delve deeper into the molecular intricacies of enhancer function within cancerous cells, the opportunity to redefine therapeutic interventions becomes increasingly tangible, marking a significant shift toward more targeted and effective cancer treatments.
The therapeutic strategies targeting enhancers that have been proposed to date are based on various stages of enhancer activation, as well as the ablation of the transcription machinery [300]. Given that terminal TFs of multiple signaling pathways frequently occupy enhancers, the use of pathway inhibitors to block the transmission of external signals to enhancers is an effective strategy [132]. Using the JAK pathway inhibitor ruxolitinib to inhibit IL6/STAT3 signaling disrupts the ER-FOXA1-pSTAT3 enhancer-driven transcriptional program of target genes in breast cancer and suppresses the invasive capacity of cancer cells [301]. Similarly, NOTCH inhibitors antagonize the function of ETS-related gene-dependent (ERG-dependent) enhancers and inhibit the growth and invasiveness of prostate cancer cells [302].
Another important approach is targeting the epigenetic modifications of enhancers. Some drugs disrupt enhancer histone modifications by inhibiting DNMTs, HDACs, and HATs [161]. Several small molecule inhibitors of CBP/p300 have been developed [303]. A-485 binds to the catalytic active sites of p300 and CBP, competitively inhibiting the binding of acetyl-CoA, thereby significantly impairing acetylation deposition. A-485 demonstrates lineage-specific antiproliferative activity in 124 cancer cell lines, particularly showing significant effects in hematologic malignancies and AR+ prostate cancer [304]. Similarly, inobrodib (CCS1477) reduces the gene expression driven by AR and C-MYC in prostate cancer [305]. Additionally, a novel chemical degrader of CBP/p300, namely dCBP-1, eliminates key enhancers driving MYC expression and significantly inhibits the growth of multiple myeloma cells [306]. Compared with CBP/p300, several HDACi have been approved for the treatment of cancer [307]. Recent studies have also examined novel HDACi and combined therapy, demonstrating improved efficacy in cancer therapy [308,309,310,311]. Targeting DNMT can remove abnormal DNA hypermethylation and, consequently, rescue the genes that have been silenced. In cancer cells that enter a dormant state in response to TGF-β, the stimulator of interferon genes (STING) promoter and enhancer exhibit high methylation and chromatin suppression, leading to reduced STING activity. Using DNMT inhibitors can rescue STING expression, triggering the expression of interferons and pro-inflammatory chemokines, thereby enhancing immune recognition and clearance [312]. Enhancer of zeste 2 polycomb repressive complex 2 subunit (EZH2) is the catalytic subunit of the polycomb repressive complex 2 (PRC2) and is responsible for H3K27me3. EZH2 is overexpressed or dysfunctional in numerous types of cancer, and EZH2 inhibitors exert their anti-tumor effects by blocking its methyltransferase activity, thereby reactivating silenced genes [176]. IHMT-337 covalently binds to EZH2 and degrades it through a ubiquitin-dependent pathway, thereby inhibiting the proliferation of breast cancer and diffuse large B-cell lymphoma [313]. Currently, several EZH2 inhibitors, such as valemetostat tosilate (Ezharmia) and tazemetostat (Tazverik), have been approved for the treatment of cancer [314, 315].
In cancer therapy, targeting the SWI/SNF complex shows great potential, as disrupting its key subunits can significantly impact chromatin structure and gene expression [316]. AU-15330 is a highly selective degrader targeting SMARCA2 and SMARCA4, which induces a specific loss of chromatin accessibility at enhancers in prostate cancer cells driven by AR and FOXA1. This disruption affects the core enhancer circuits that are dependent on these TFs, thereby eliminating the downstream oncogenic gene programs [317]. During SWI/SNF ablation, enhancers remain persistently repressed; however, the accessibility and transcriptional activity of many promoters are restored, possibly due to the compensatory action of the EP400/TIP60 coactivator complex. Simultaneous inhibition of EP400/TIP60 can enhance the sensitivity to SWI/SNF inhibition [318].
Although the effects of TFs on enhancers are well established, directly targeting TFs remains a challenge [319]. However, TFs participate in the assembly of the transcription machinery by recruiting cofactors, such as CDKs and BRD4. These approaches have been thoroughly summarized in previous articles, hence will not be elaborated further here [320,321,322]. Instead, building on this foundation, we aim to explore innovative avenues that extend beyond traditional interventions.
Combination therapy can achieve higher efficiency for tumor cell clearance in a shorter period of time [323]. For example, cotreatment with bromodomain antagonist, HDACi, and CDK4/6 inhibitors can synergistically induce apoptosis in ibrutinib-resistant mantle cell lymphoma cells [324]. The combined use of HDACi overcomes the resistance to MEK inhibitors caused by the downregulation of MAPK pathway negative regulators [261]. More importantly, increasing evidence suggests that enhancer perturbation can activate immune responses, potentially involving interferon signaling pathways, tumor immunogenicity, and pyroptosis [281].
Therefore, combined immunotherapy is a promising treatment strategy(Table 4). A recent study combining PD-1 monoclonal antibodies, HDACi, and vascular endothelial growth factor (VEGF) monoclonal antibodies showed improved prognosis in colorectal cancer [308]. A phase II clinical study combining anti-PD-1 and HDACi in peripheral T cell lymphoma is also currently underway [325].
Enhancers are not unique to cancer cells but are co-opted from normal cells [326]. Hence, any therapy that directly targets enhancers could potentially impact the transcriptional mechanisms of normal cells (although cancer cells seem to be more sensitive to this intervention). This may somewhat limit its clinical utility. A potential approach to overcoming this limitation is to precisely deliver drugs to tumor tissues through drug delivery systems, such as a magnetic drug delivery system. This allows for the precise targeting of diseased tissues under the influence of an external magnetic field, thereby reducing peripheral uptake [327].
Metabolic reprogramming supports the growth of tumor cells and has been recognized as a vulnerability to cancer [328]. Combining the targeting of metabolic pathways and enhancers may provide a powerful strategy for improving cancer treatment. Under sufficient oxygen conditions, cancer cells prefer to utilize glycolysis; this preference provides a potential way to distinguish normal cells from tumor cells [329]. Certain key glycolytic enzymes, such as HK, are driven by enhancers and highly expressed in cancer cells [217]. 2-Deoxy-D-glucose (2-DG) is a HK2 inhibitor with a similar structure to that of glucose and has been used as an adjuvant therapy for chemotherapy in various tumors [330]. Recent research has shown that enhancer reprogramming induced by KMT2D deficiency leads to the activation of the glycolytic pathway, which can be inhibited by targeted drugs (e.g., 2-DG). This evidence provides strong support for this therapeutic strategy [16]. Additionally, under MLL3/4 mutation, MLL1 (another member of the COMPASS family) compensates by regulating the expression of purine metabolism-related genes, leading to a significant increase in cellular dependence on purine and pyrimidine metabolism and heightened sensitivity to purine synthesis inhibitors, such as Lometrexol [331]. In conclusion, these findings provide new insights into the connection between epigenetic regulation and metabolic dependencies in cancer, while proposing potential novel strategies for cancer therapy.
Synthetic lethality offers a new possibility for precisely targeting enhancers in cancer cells [332]. Epigenetic modifying enzymes in cancer cells often undergo recurrent mutations that are absent in normal cells. By identifying and targeting these mutated cancer cells, it is possible to achieve specific interference with enhancers [333]. For instance, targeting p300 in cancer cells carrying CBP mutations results in synthetic lethality [334]. The double mutations of SMARCA4 and SMARCA2 have been linked to strong synthetic lethality. When SMARCA2 is mutated, targeting the degradation of SMARCA4 using pharmacological inhibitors (e.g., BRM014) leads to a widespread loss of chromatin accessibility and H3K27ac at enhancer regions [335, 336]. The EP400/TIP60 activity induced by SWI/SNF inhibition is associated with transcriptional recovery; thus, simultaneous ablation of EP400/TIP60 results in a synthetic lethality effect [318]. By utilizing genomic and epigenomic data from patients with cancer, active enhancers, and related synthetic lethal gene pairs can be identified in cancer cells obtained from a specific patient. Based on this information, personalized treatment plans can be designed, selecting the most suitable enhancer inhibitors and other synthetic lethal target inhibitors for combination therapy. In summary, targeting cancer cell enhancers based on the concept of synthetic lethality holds promise for providing an efficient and highly selective strategy for the treatment of cancer.
Antibody-drug conjugate (ADC) drugs provide another powerful way for precisely targeting enhancers. Cancer cells typically express high-specificity and high-abundance antigens on their surface, which are expressed at markedly lower levels in normal cells. This allows ADC drugs to use specific antibodies to recognize and bind to these antigens on the surface of cancer cells. This ensures the delivery of toxic drugs directly into the cancer cells, thereby minimizing damage to normal cells [337]. Additionally, it is possible that ADC drugs can carry traditional chemotherapeutic agents, as well as be combined with epigenetic regulators and metabolic inhibitors, to enhance their anticancer effects. For example, ADC drugs that target enhancer reprogramming can use specific antibodies to deliver epigenetic regulators or metabolic inhibitors precisely to cancer cells, effectively inhibiting cancer cell growth and metastasis. Although the use of ADC drugs has achieved significant clinical success, research on targeting enhancers remains scarce [338]. Future research may focus on optimizing drug carriers, combination therapies, and personalizing ADC treatment plans based on the genomic and epigenomic characteristics of patients and cancer cell surface antigen expression.
Simultaneously, the discovery of high-resolution maps of tumor-infiltrating immune cells has unveiled potential strategies for manipulating the TME. The immunosuppressive nature of TME is, to some extent, driven by enhancer reprogramming, with the potential for reversal [280]. This has been illustrated by recent findings on the interplay between gut microbiota and enhancers. These insights suggest that altering the gut microbiome could modify enhancer activity within the TME, thereby transforming an immunosuppressive environment into one that supports immune-mediated tumor suppression [284]. This emerging understanding opens new avenues for therapeutic intervention, in which the modulation of enhancer landscapes through dietary changes, probiotic supplementation, or targeted microbial therapies(e.g., fecal microbiota transplantation) could become integral components of comprehensive strategies for the treatment of cancer [339, 340].
Additionally, recent advancements have led to the development of an epigenetic modification tool based on CRISPR gene editing technology, termed CRISPRon and CRISPRoff. This tool enables heritable editing of enhancers, paving the way for targeted manipulation of tissue-specific gene expression components and the potential eradication of cancer transcription networks [341]. A recent study utilized CRISPR-Cas9 technology to target the erythroid-specific enhancer of BCL11A, aiming to restore fetal hemoglobin (HbF) expression as a therapeutic strategy for transfusion-dependent β-thalassemia (TDT) and sickle cell disease (SCD). By generating and infusing edited CD34+ hematopoietic stem and progenitor cells (CTX001), both patients demonstrated significantly increased HbF levels and alleviation of disease-related symptoms during follow-up. The TDT patient achieved transfusion independence, while the SCD patient experienced no recurrence of vaso-occlusive crises. These findings suggest that enhancer-targeted gene editing offers an effective and durable treatment approach for these hemoglobinopathies.
Despite the promising potential of enhancer-targeting therapies, several challenges remain unresolved. One significant challenge is the emergence of acquired resistance. For instance, the dual EZH1/EZH2 inhibitor valemetostat initially shows strong therapeutic efficacy; however, resistant clones often emerge with prolonged treatment. This resistance may be driven by mutations in key subunits of the PRC2 complex, such as EZH2 or EED, which reduce the inhibitory efficiency of valemetostat and restore H3K27me3 levels. Additionally, resistance may develop through compensatory mechanisms involving DNA methylation. Tumor cells can upregulate DNMT3A or lose TET2 function, substituting DNA methylation for H3K27me3 marks to re-silence critical tumor suppressor genes and restore a malignant phenotype [314]. These mechanisms suggest that monotherapy targeting enhancers may be insufficient to overcome the epigenetic adaptability of enhancer-dependent cancers.
Another major challenge is the highly dynamic nature of enhancer function, which can vary significantly across cell types, developmental stages, and disease states. This heterogeneity arises not only from genetic background differences but also from multiple layers of regulation, including chromatin state, epigenetic marks, and transcription factor binding dynamics. Single-cell ATAC and CUT&Tag studies have revealed substantial variation in enhancer accessibility and epigenetic modifications across different clones, directly impacting the expression of key genes and functional differentiation of tumors [68, 280, 342, 343]. For example, single-cell ATAC sequencing has shown differences in enhancer networks within tumor-infiltrating immune cells that influence therapeutic responses [342], while single-cell CUT&Tag has clarified how H3K27me3 marks are redistributed in different clones to repress or activate specific genes [68]. This heterogeneity may result in inconsistent therapeutic outcomes among patient groups and contribute to the emergence of treatment resistance due to dynamic tumor adaptation [68].
During tumor progression, enhancers can undergo reprogramming, activating or repressing specific gene networks through transcription factor reassembly or chromatin state alterations, thereby driving phenotypic plasticity to adapt to new microenvironments or therapeutic pressures [155]. For example, in pancreatic cancer, PRMT1 modulates the methylation state of enhancers to restrict gemcitabine-induced transcription factor binding (e.g., MAFF and BACH1), reprogramming enhancer activity and promoting chemotherapy resistance [344].
Although epigenomics and single-cell technologies have provided critical insights into the dynamic changes in enhancer activity, their clinical application is hindered by high costs, technical complexity, and time consumption. Real-time monitoring of enhancer activity changes in patients and correlating these with therapeutic outcomes remains a significant technical and logistical challenge.
These challenges underscore the need for innovative strategies and tools to enhance the efficacy and specificity of enhancer-targeting therapies while addressing resistance and heterogeneity in cancer treatment.
Conclusion and prospect
Enhancer reprogramming constitutes a fundamental mechanism underpinning the transcriptional dysregulation in cancer, influencing cell fate decisions, and contributing to the phenotypic plasticity of cancer cells [98]. The intricate interaction between enhancers and the transcriptional machinery dictates the cellular identity and functionality, rendering enhancers critical determinants of cancer progression and viable targets for therapeutic intervention [94]. The reprogramming of enhancers in cancer is a double-edged sword, driving oncogenesis while offering a strategic target for disrupting the aberrant transcriptional networks that sustain cancer cells. The therapeutic potential of targeting enhancer dysregulation holds promise for the development of innovative cancer treatments. This approach offers a path to disrupt the addiction of cancer cells to aberrant enhancer-driven transcriptional programs. Enhancer-driven transcription networks are frequently remodeled during cancer resistance and metastasis, providing an opportunity to predict patient prognosis, while showing potential advantages in assessing the likelihood of metastasis [148, 301].
Despite this promise, targeting enhancers for therapy presents significant challenges that need to be addressed. Enhancers are often tissue- and cell-type-specific, which increases the difficulty of achieving specificity without off-target effects [54]. Off-target activity could disrupt normal transcription programs in healthy cells, leading to unintended consequences [22, 345, 346]. Furthermore, cancer cells may develop resistance to enhancer-targeting therapies, either through the rewiring of transcriptional networks or the activation of alternative enhancers, which necessitates a deeper understanding of resistance mechanisms and the development of strategies to overcome them [347].
Due to the lack of drugs specifically targeting cancer cell enhancers, future research may focus more on precision therapy strategies, such as those based on the concept of synthetic lethality or the use of ADC drugs [348, 349]. Additionally, the abnormal metabolic pathways exhibited by cancer cells also present a potential vulnerability. Synthetic biology techniques have also been utilized to design and construct artificial enhancers for the precise regulation of gene expression and the development of novel gene therapy strategies. However, these approaches face technical and biological limitations, including achieving spatial and temporal control of enhancer activity and avoiding unintended activation of oncogenic programs.
Although it is established that enhancers exhibit different activities in various tissues and cell types, the specific underlying molecular mechanisms remain unclear. For example, the factors that determine the activity of an enhancer in a particular tissue remain unknown. This is significant in deciphering the abnormal activity of enhancers in cancer. For instance, the circumstances under which FOXA1 is activated, leading to liver metastasis of pancreatic cancer, are also unknown [148]. Current research mainly focuses on aberrantly activated enhancers; there is limited knowledge regarding enhancers that are silenced in cancer. Understanding this mechanism could help researchers manipulate those silenced enhancers to reactivate normal transcription programs. Despite the known importance of TFs in controlling enhancer activity, the mechanisms by which they are repositioned on enhancers remain unexplained. Further investigation into enhancer specificity, resistance mechanisms, and their interplay with the broader transcriptional network will enrich our understanding of enhancers and provide new perspectives and methods for gene regulation, biological research, and clinical applications.
References
Haberle V, Stark A. Eukaryotic core promoters and the functional basis of transcription initiation. Nat Rev Mol Cell Biol. 2018;19:621–37.
Andersson R, Sandelin A. Determinants of enhancer and promoter activities of regulatory elements. Nat Rev Genet. 2020;21:71–87.
Van Nostrand EL, Freese P, Pratt GA, Wang X, Wei X, Xiao R, et al. A large-scale binding and functional map of human RNA-binding proteins. Nature. 2020;583:711–9.
Moore ConsortiumEP, Purcaro JE, Pratt HE MJ, Epstein CB, Shoresh N, et al. Expanded encyclopaedias of DNA elements in the human and mouse genomes. Nature. 2020;583:699–710.
Meuleman W, Muratov A, Rynes E, Halow J, Lee K, Bates D, et al. Index and biological spectrum of human DNase I hypersensitive sites. Nature. 2020;584:244–51.
Robson MI, Ringel AR, Mundlos S. Regulatory landscaping: how enhancer-promoter communication is sculpted in 3D. Mol Cell. 2019;74:1110–22.
Chronis C, Fiziev P, Papp B, Butz S, Bonora G, Sabri S, et al. Cooperative binding of transcription factors orchestrates reprogramming. Cell. 2017;168:442–59.e20.
Zhou, Zhang P, Sethi Y, Ye I, Trembley MA L, Cao Y, et al. GATA4 regulates developing endocardium through interaction With ETS1. Circ Res. 2022;131:e152–68.
Brennan KJ, Weilert M, Krueger S, Pampari A, Liu HY, Yang AWH, et al. Chromatin accessibility in the Drosophila embryo is determined by transcription factor pioneering and enhancer activation. Dev Cell. 2023;58:1898–916.e9.
Long HK, Prescott SL, Wysocka J. Ever-changing landscapes: transcriptional enhancers in development and evolution. Cell. 2016;167:1170–87.
Bhagwat AS, Lu B, Vakoc CR. Enhancer dysfunction in leukemia. Blood. 2018;131:1795–804.
Jiang Y, Jiang YY, Xie JJ, Mayakonda A, Hazawa M, Chen L, et al. Co-activation of super-enhancer-driven CCAT1 by TP63 and SOX2 promotes squamous cancer progression. Nat Commun. 2018;9:3619.
Hariprakash JM, Salviato E, La Mastra F, Sebestyen E, Tagliaferri I, Silva RS, et al. Leveraging tissue-specific enhancer-target gene regulatory networks identifies enhancer somatic mutations that functionally impact lung cancer. Cancer Res. 2024;84:133–53.
Zhao Z, Aoi Y, Philips CN, Meghani KA, Gold SR, Yu Y, et al. Somatic mutations of MLL4/COMPASS induce cytoplasmic localization providing molecular insight into cancer prognosis and treatment. Proc Natl Acad Sci USA. 2023;120:e2310063120.
Heyes E, Wilhelmson AS, Wenzel A, Manhart G, Eder T, Schuster MB, et al. TET2 lesions enhance the aggressiveness of CEBPA-mutant acute myeloid leukemia by rebalancing GATA2 expression. Nat Commun. 2023;14:6185.
Alam H, Tang M, Maitituoheti M, Dhar S, Kumar M, Han C. et al. KMT2D deficiency impairs super-enhancers to confer a glycolytic vulnerability in lung cancer. Cancer Cell. 2020;37:599–617.e7.
Xu J, Song F, Lyu H, Kobayashi M, Zhang B, Zhao Z, et al. Subtype-specific 3D genome alteration in acute myeloid leukaemia. Nature. 2022;611:387–98.
Hanahan D. Hallmarks of cancer: new dimensions. Cancer Discov. 2022;12:31–46.
Wu S, Ou T, Xing N, Lu J, Wan S, Wang C, et al. Whole-genome sequencing identifies ADGRG6 enhancer mutations and FRS2 duplications as angiogenesis-related drivers in bladder cancer. Nat Commun. 2019;10:720.
Sur I, Taipale J. The role of enhancers in cancer. Nat Rev Cancer. 2016;16:483–93.
Pan X, Li X, Sun J, Xiong Z, Hu H, Ning S, et al. Enhancer methylation dynamics drive core transcriptional regulatory circuitry in pan-cancer. Oncogene. 2022;41:3474–84.
Liang T, Wang F, Elhassan RM, Cheng Y, Tang X, Chen W, et al. Targeting histone deacetylases for cancer therapy: trends and challenges. Acta Pharm Sin B. 2023;13:2425–63.
Kvon EZ, Waymack R, Gad M, Wunderlich Z. Enhancer redundancy in development and disease. Nat Rev Genet. 2021;22:324–36.
Chen Z, Snetkova V, Bower G, Jacinto S, Clock B, Dizehchi A, et al. Increased enhancer-promoter interactions during developmental enhancer activation in mammals. Nat Genet. 2024;56:675–85.
Schoenfelder S, Fraser P. Long-range enhancer-promoter contacts in gene expression control. Nat Rev Genet. 2019;20:437–55.
Lambert SA, Jolma A, Campitelli LF, Das PK, Yin Y, Albu M, et al. The human transcription factors. Cell. 2018;172:650–65.
Tang S, Xue Y, Qin Z, Fang Z, Sun Y, Yuan C, et al. Counteracting lineage-specific transcription factor network finely tunes lung adeno-to-squamous transdifferentiation through remodeling tumor immune microenvironment. Natl Sci Rev. 2023;10:nwad028.
Narita T, Ito S, Higashijima Y, Chu WK, Neumann K, Walter J, et al. Enhancers are activated by p300/CBP activity-dependent PIC assembly, RNAPII recruitment, and pause release. Mol Cell. 2021;81:2166–82.e6.
Ramasamy S, Aljahani A, Karpinska MA, Cao TBN, Velychko T, Cruz JN, et al. The mediator complex regulates enhancer-promoter interactions. Nat Struct Mol Biol. 2023;30:991–1000.
Strazzabosco M. Foxa1 and Foxa2 regulate bile duct development in mice. J Hepatol. 2010;52:765–7.
Gao N, LeLay J, Vatamaniuk MZ, Rieck S, Friedman JR, Kaestner KH. Dynamic regulation of Pdx1 enhancers by Foxa1 and Foxa2 is essential for pancreas development. Genes Dev. 2008;22:3435–48.
Balsalobre A, Drouin J. Pioneer factors as master regulators of the epigenome and cell fate. Nat Rev Mol Cell Biol. 2022;23:449–64.
Liu N, Wang A, Xue M, Zhu X, Liu Y, Chen M. FOXA1 and FOXA2: the regulatory mechanisms and therapeutic implications in cancer. Cell Death Discov. 2024;10:172.
Sekiya S, Suzuki A. Direct conversion of mouse fibroblasts to hepatocyte-like cells by defined factors. Nature. 2011;475:390–3.
Lantz KA, Kaestner KH. Winged-helix transcription factors and pancreatic development. Clin Sci. 2005;108:195–204.
Wang Z, Townley SL, Zhang S, Liu M, Li M, Labaf M, et al. FOXA2 rewires AP-1 for transcriptional reprogramming and lineage plasticity in prostate cancer. Nat Commun. 2024;15:4914.
Mamun MMA, Khan MR, Zhu Y, Zhang Y, Zhou S, Xu R, et al. Stub1 maintains proteostasis of master transcription factors in embryonic stem cells. Cell Rep. 2022;39:110919.
Wang Z, Oron E, Nelson B, Razis S, Ivanova N. Distinct lineage specification roles for NANOG, OCT4, and SOX2 in human embryonic stem cells. Cell Stem Cell. 2012;10:440–54.
Sinha KK, Bilokapic S, Du Y, Malik D, Halic M. Histone modifications regulate pioneer transcription factor cooperativity. Nature. 2023;619:378–84.
Frederick MA, Williamson KE, Fernandez Garcia M, Ferretti MB, McCarthy RL, Donahue G, et al. A pioneer factor locally opens compacted chromatin to enable targeted ATP-dependent nucleosome remodeling. Nat Struct Mol Biol. 2023;30:31–7.
Ji D, Shao C, Yu J, Hou Y, Gao X, Wu Y, et al. FOXA1 forms biomolecular condensates that unpack condensed chromatin to function as a pioneer factor. Mol Cell. 2024;84:244–60.e7.
Gouhier A, Dumoulin-Gagnon J, Lapointe-Roberge V, Harris J, Balsalobre A, Drouin J. Pioneer factor Pax7 initiates two-step cell-cycle-dependent chromatin opening. Nat Struct Mol Biol. 2024;31:92–101.
Field A, Adelman K. Evaluating enhancer function and transcription. Annu Rev Biochem. 2020;89:213–34.
Barakat TS, Halbritter F, Zhang M, Rendeiro AF, Perenthaler E, Bock C, et al. Functional dissection of the enhancer repertoire in human embryonic stem cells. Cell Stem Cell. 2018;23:276–88.e8.
Galle E, Wong CW, Ghosh A, Desgeorges T, Melrose K, Hinte LC, et al. H3K18 lactylation marks tissue-specific active enhancers. Genome Biol. 2022;23:207.
Pal D, Patel M, Boulet F, Sundarraj J, Grant OA, Branco MR, et al. H4K16ac activates the transcription of transposable elements and contributes to their cis-regulatory function. Nat Struct Mol Biol. 2023;30:935–47.
Chang CH, Liu F, Militi S, Hester S, Nibhani R, Deng S, et al. The pRb/RBL2-E2F1/4-GCN5 axis regulates cancer stem cell formation and G0 phase entry/exit by paracrine mechanisms. Nat Commun. 2024;15:3580.
Aseem SO, Jalan-Sakrikar N, Chi C, Navarro-Corcuera A, De Assuncao TM, Hamdan FH, et al. Epigenomic evaluation of cholangiocyte transforming growth factor-beta signaling identifies a selective role for histone 3 lysine 9 acetylation in biliary fibrosis. Gastroenterology. 2021;160:889–905.e10.
Narita T, Higashijima Y, Kilic S, Liebner T, Walter J, Choudhary C. Acetylation of histone H2B marks active enhancers and predicts CBP/p300 target genes. Nat Genet. 2023;55:679–92.
Fang Y, Xu X, Ding J, Yang L, Doan MT, Karmaus PWF, et al. Histone crotonylation promotes mesoendodermal commitment of human embryonic stem cells. Cell Stem Cell. 2021;28:748–63.e7.
Qin F, Li B, Wang H, Ma S, Li J, Liu S, et al. Linking chromatin acylation mark-defined proteome and genome in living cells. Cell. 2023;186:1066–85.e36.
Saha D, Animireddy S, Lee J, Thommen A, Murvin MM, Lu Y, et al. Enhancer switching in cell lineage priming is linked to eRNA, Brg1’s AT-hook, and SWI/SNF recruitment. Mol Cell. 2024;84:1855–69.e5.
Liao J, Ho J, Burns M, Dykhuizen EC, Hargreaves DC. Collaboration between distinct SWI/SNF chromatin remodeling complexes directs enhancer selection and activation of macrophage inflammatory genes. Immunity. 2024;57:1780–95.e6.
Yang JH, Hansen AS. Enhancer selectivity in space and time: from enhancer-promoter interactions to promoter activation. Nat Rev Mol Cell Biol. 2024;25:574–91.
Liu B, He Y, Wu X, Lin Z, Ma J, Qiu Y, et al. Mapping putative enhancers in mouse oocytes and early embryos reveals TCF3/12 as key folliculogenesis regulators. Nat Cell Biol. 2024;26:962–74.
Bergman DT, Jones TR, Liu V, Ray J, Jagoda E, Siraj L, et al. Compatibility rules of human enhancer and promoter sequences. Nature. 2022;607:176–84.
Arnold CD, Gerlach D, Stelzer C, Boryn LM, Rath M, Stark A. Genome-wide quantitative enhancer activity maps identified by STARR-seq. Science. 2013;339:1074–7.
Kwon SB, Ernst J. Investigating enhancer evolution with massively parallel reporter assays. Genome Biol. 2018;19:114.
Smith GD, Ching WH, Cornejo-Paramo P, Wong ES. Decoding enhancer complexity with machine learning and high-throughput discovery. Genome Biol. 2023;24:116.
Avsec Z, Agarwal V, Visentin D, Ledsam JR, Grabska-Barwinska A, Taylor KR, et al. Effective gene expression prediction from sequence by integrating long-range interactions. Nat Methods. 2021;18:1196–203.
Wang X, He L, Goggin SM, Saadat A, Wang L, Sinnott-Armstrong N, et al. High-resolution genome-wide functional dissection of transcriptional regulatory regions and nucleotides in human. Nat Commun. 2018;9:5380.
Hansen TJ, Hodges E. ATAC-STARR-seq reveals transcription factor-bound activators and silencers across the chromatin accessible human genome. Genome Res. 2022;32:1529–41.
Zhang J, Lee D, Dhiman V, Jiang P, Xu J, McGillivray P, et al. An integrative ENCODE resource for cancer genomics. Nat Commun. 2020;11:3696.
Liu S, Liu Y, Zhang Q, Wu J, Liang J, Yu S, et al. Systematic identification of regulatory variants associated with cancer risk. Genome Biol. 2017;18:194.
White SM, Snyder MP, Yi C. Master lineage transcription factors anchor trans mega transcriptional complexes at highly accessible enhancer sites to promote long-range chromatin clustering and transcription of distal target genes. Nucleic Acids Res. 2021;49:12196–210.
Bhattarai KR, Mobley RJ, Barnett KR, Ferguson DC, Hansen BS, Diedrich JD, et al. Investigation of inherited noncoding genetic variation impacting the pharmacogenomics of childhood acute lymphoblastic leukemia treatment. Nat Commun. 2024;15:3681.
Deng C, Whalen S, Steyert M, Ziffra R, Przytycki PF, Inoue F, et al. Massively parallel characterization of regulatory elements in the developing human cortex. Science. 2024;384:eadh0559.
Wu SJ, Furlan SN, Mihalas AB, Kaya-Okur HS, Feroze AH, Emerson SN, et al. Single-cell CUT&Tag analysis of chromatin modifications in differentiation and tumor progression. Nat Biotechnol. 2021;39:819–24.
Bartosovic M, Kabbe M, Castelo-Branco G. Single-cell CUT&Tag profiles histone modifications and transcription factors in complex tissues. Nat Biotechnol. 2021;39:825–35.
Regner MJ, Wisniewska K, Garcia-Recio S, Thennavan A, Mendez-Giraldez R, Malladi VS, et al. A multi-omic single-cell landscape of human gynecologic malignancies. Mol Cell. 2021;81:4924–41.e10.
Zhao S, Hong CKY, Myers CA, Granas DM, White MA, Corbo JC, et al. A single-cell massively parallel reporter assay detects cell-type-specific gene regulation. Nat Genet. 2023;55:346–54.
Kawasaki K, Fukaya T. Functional coordination between transcription factor clustering and gene activity. Mol Cell. 2023;83:1605–22.e9.
Khoueiry P, Girardot C, Ciglar L, Peng PC, Gustafson EH, Sinha S, et al. Uncoupling evolutionary changes in DNA sequence, transcription factor occupancy and enhancer activity. eLife. 2017;6:e28440.
Zanconato F, Forcato M, Battilana G, Azzolin L, Quaranta E, Bodega B, et al. Genome-wide association between YAP/TAZ/TEAD and AP-1 at enhancers drives oncogenic growth. Nat Cell Biol. 2015;17:1218–27.
Simeoni F, Romero-Camarero I, Camera F, Amaral FMR, Sinclair OJ, Papachristou EK, et al. Enhancer recruitment of transcription repressors RUNX1 and TLE3 by mis-expressed FOXC1 blocks differentiation in acute myeloid leukemia. Cell Rep. 2021;36:109725.
Yang Y, Gomez N, Infarinato N, Adam RC, Sribour M, Baek I, et al. The pioneer factor SOX9 competes for epigenetic factors to switch stem cell fates. Nat Cell Biol. 2023;25:1185–95.
Kim J, Diaz LF, Miller MJ, Leadem B, Krivega I, Dean A. An enhancer RNA recruits KMT2A to regulate transcription of Myb. Cell Rep. 2024;43:114378.
Rothschild G, Basu U. Lingering questions about enhancer RNA and enhancer transcription-coupled genomic instability. Trends Genet. 2017;33:143–54.
Lee JH, Wang R, Xiong F, Krakowiak J, Liao Z, Nguyen PT, et al. Enhancer RNA m6A methylation facilitates transcriptional condensate formation and gene activation. Mol Cell. 2021;81:3368–85.e9.
Hsieh TS, Cattoglio C, Slobodyanyuk E, Hansen AS, Darzacq X, Tjian R. Enhancer-promoter interactions and transcription are largely maintained upon acute loss of CTCF, cohesin, WAPL or YY1. Nat Genet. 2022;54:1919–32.
Uyehara CM, Apostolou E. 3D enhancer-promoter interactions and multi-connected hubs: organizational principles and functional roles. Cell Rep. 2023;42:112068.
Davidson IF, Bauer B, Goetz D, Tang W, Wutz G, Peters JM. DNA loop extrusion by human cohesin. Science. 2019;366:1338–45.
Dixon JR, Gorkin DU, Ren B. Chromatin domains: the unit of chromosome organization. Mol Cell. 2016;62:668–80.
Chang LH, Ghosh S, Papale A, Luppino JM, Miranda M, Piras V, et al. Multi-feature clustering of CTCF binding creates robustness for loop extrusion blocking and topologically associating domain boundaries. Nat Commun. 2023;14:5615.
Chakraborty S, Kopitchinski N, Zuo Z, Eraso A, Awasthi P, Chari R, et al. Enhancer-promoter interactions can bypass CTCF-mediated boundaries and contribute to phenotypic robustness. Nat Genet. 2023;55:280–90.
Despang A, Schopflin R, Franke M, Ali S, Jerkovic I, Paliou C, et al. Functional dissection of the Sox9-Kcnj2 locus identifies nonessential and instructive roles of TAD architecture. Nat Genet. 2019;51:1263–71.
Hnisz D, Shrinivas K, Young RA, Chakraborty AK, Sharp PA. A phase separation model for transcriptional control. Cell. 2017;169:13–23.
Sabari BR, Dall'Agnese A, Boija A, Klein IA, Coffey EL, Shrinivas K, et al. Coactivator condensation at super-enhancers links phase separation and gene control. Science. 2018;361:eaar3958.
Mehta S, Zhang J. Liquid-liquid phase separation drives cellular function and dysfunction in cancer. Nat Rev Cancer. 2022;22:239–52.
Lu B, Zou C, Yang M, He Y, He J, Zhang C, et al. Pharmacological inhibition of core regulatory circuitry liquid-liquid phase separation suppresses metastasis and chemoresistance in osteosarcoma. Adv Sci (Weinh). 2021;8:e2101895.
Du M, Stitzinger SH, Spille JH, Cho WK, Lee C, Hijaz M, et al. Direct observation of a condensate effect on super-enhancer controlled gene bursting. Cell. 2024;187:331–44.e17.
Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–19.
Sengupta S, George RE. Super-enhancer-driven transcriptional dependencies in cancer. Trends Cancer. 2017;3:269–81.
Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-Andre V, Sigova AA, et al. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–47.
Kai Y, Li BE, Zhu M, Li GY, Chen F, Han Y, et al. Mapping the evolving landscape of super-enhancers during cell differentiation. Genome Biol. 2021;22:269.
Kochat V, Raman AT, Landers SM, Tang M, Schulz J, Terranova C, et al. Enhancer reprogramming in PRC2-deficient malignant peripheral nerve sheath tumors induces a targetable de-differentiated state. Acta Neuropathol. 2021;142:565–90.
Yu M, Hu X, Pan Z, Du C, Jiang J, Zheng W, et al. Endogenous retrovirus-derived enhancers confer the transcriptional regulation of human trophoblast syncytialization. Nucleic Acids Res. 2023;51:4745–59.
Herz HM, Hu D, Shilatifard A. Enhancer malfunction in cancer. Mol Cell. 2014;53:859–66.
Fagnocchi L, Poli V, Zippo A. Enhancer reprogramming in tumor progression: a new route towards cancer cell plasticity. Cell Mol Life Sci. 2018;75:2537–55.
Qiao Y, Wang Z, Tan F, Chen J, Lin J, Yang J, et al. Enhancer reprogramming within pre-existing topologically associated domains promotes TGF-beta-induced EMT and cancer metastasis. Mol Ther. 2020;28:2083–95.
Ottema S, Mulet-Lazaro R, Erpelinck-Verschueren C, van Herk S, Havermans M, Arricibita Varea A, et al. The leukemic oncogene EVI1 hijacks a MYC super-enhancer by CTCF-facilitated loops. Nat Commun. 2021;12:5679.
Montefiori LE, Bendig S, Gu Z, Chen X, Polonen P, Ma X, et al. Enhancer hijacking drives oncogenic BCL11B expression in lineage-ambiguous stem cell leukemia. Cancer Discov. 2021;11:2846–67.
Groschel S, Sanders MA, Hoogenboezem R, de Wit E, Bouwman BAM, Erpelinck C, et al. A single oncogenic enhancer rearrangement causes concomitant EVI1 and GATA2 deregulation in leukemia. Cell. 2014;157:369–81.
Wang X, Xu J, Zhang B, Hou Y, Song F, Lyu H, et al. Genome-wide detection of enhancer-hijacking events from chromatin interaction data in rearranged genomes. Nat Methods. 2021;18:661–8.
Botten GA, Zhang Y, Dudnyk K, Kim YJ, Liu X, Sanders JT, et al. Structural variation cooperates with permissive chromatin to control enhancer hijacking-mediated oncogenic transcription. Blood. 2023;142:336–51.
Xu Z, Lee DS, Chandran S, Le VT, Bump R, Yasis J, et al. Structural variants drive context-dependent oncogene activation in cancer. Nature. 2022;612:564–72.
Kim K, Kim M, Lee AJ, Song SH, Kang JK, Eom J, et al. Spatial and clonality-resolved 3D cancer genome alterations reveal enhancer-hijacking as a potential prognostic marker for colorectal cancer. Cell Rep. 2023;42:112778.
Weischenfeldt J, Dubash T, Drainas AP, Mardin BR, Chen Y, Stutz AM, et al. Pan-cancer analysis of somatic copy-number alterations implicates IRS4 and IGF2 in enhancer hijacking. Nat Genet. 2017;49:65–74.
Li Y, He Y, Peng J, Su Z, Li Z, Zhang B, et al. Mutant Kras co-opts a proto-oncogenic enhancer network in inflammation-induced metaplastic progenitor cells to initiate pancreatic cancer. Nat Cancer. 2021;2:49–65.
Mansour MR, Abraham BJ, Anders L, Berezovskaya A, Gutierrez A, Durbin AD, et al. Oncogene regulation. An oncogenic super-enhancer formed through somatic mutation of a noncoding intergenic element. Science. 2014;346:1373–7.
Oldridge DA, Wood AC, Weichert-Leahey N, Crimmins I, Sussman R, Winter C, et al. Genetic predisposition to neuroblastoma mediated by a LMO1 super-enhancer polymorphism. Nature. 2015;528:418–21.
Tian J, Lou J, Cai Y, Rao M, Lu Z, Zhu Y, et al. Risk SNP-mediated enhancer-promoter interaction drives colorectal cancer through both FADS2 and AP002754.2. Cancer Res. 2020;80:1804–18.
Herranz D, Ambesi-Impiombato A, Palomero T, Schnell SA, Belver L, Wendorff AA, et al. A NOTCH1-driven MYC enhancer promotes T cell development, transformation and acute lymphoblastic leukemia. Nat Med. 2014;20:1130–7.
Quigley DA, Dang HX, Zhao SG, Lloyd P, Aggarwal R, Alumkal JJ, et al. Genomic hallmarks and structural variation in metastatic prostate cancer. Cell. 2018;174:758–69.e9.
Krump NA, You J. Molecular mechanisms of viral oncogenesis in humans. Nat Rev Microbiol. 2018;16:684–98.
Luebeck J, Coruh C, Dehkordi SR, Lange JT, Turner KM, Deshpande V, et al. AmpliconReconstructor integrates NGS and optical mapping to resolve the complex structures of focal amplifications. Nat Commun. 2020;11:4374.
Tian R, Huang Z, Li L, Yuan J, Zhang Q, Meng L, et al. HPV integration generates a cellular super-enhancer which functions as ecDNA to regulate genome-wide transcription. Nucleic Acids Res. 2023;51:4237–51.
Li CL, Li CY, Lin YY, Ho MC, Chen DS, Chen PJ, et al. Androgen receptor enhances hepatic telomerase reverse transcriptase gene transcription after hepatitis B virus integration or point mutation in promoter region. Hepatology. 2019;69:498–512.
Mizokami H, Okabe A, Choudhary R, Mima M, Saeda K, Fukuyo M, et al. Enhancer infestation drives tumorigenic activation of inactive B compartment in Epstein-Barr virus-positive nasopharyngeal carcinoma. EBioMedicine. 2024;102:105057.
Okabe A, Huang KK, Matsusaka K, Fukuyo M, Xing M, Ong X, et al. Cross-species chromatin interactions drive transcriptional rewiring in Epstein-Barr virus-positive gastric adenocarcinoma. Nat Genet. 2020;52:919–30.
Li J, Chin CR, Ying HY, Meydan C, Teater MR, Xia M, et al. Loss of CREBBP and KMT2D cooperate to accelerate lymphomagenesis and shape the lymphoma immune microenvironment. Nat Commun. 2024;15:2879.
Tinsley E, Bredin P, Toomey S, Hennessy BT, Furney SJ. KMT2C and KMT2D aberrations in breast cancer. Trends Cancer. 2024;10:519–30.
Attar N, Kurdistani SK. Exploitation of EP300 and CREBBP lysine acetyltransferases by cancer. Cold Spring Harb Perspect Med. 2017;7:a026534.
Ahn JH, Davis ES, Daugird TA, Zhao S, Quiroga IY, Uryu H, et al. Phase separation drives aberrant chromatin looping and cancer development. Nature. 2021;595:591–5.
Rahnamoun H, Hong J, Sun Z, Lee J, Lu H, Lauberth SM. Mutant p53 regulates enhancer-associated H3K4 monomethylation through interactions with the methyltransferase MLL4. J Biol Chem. 2018;293:13234–46.
Rahnamoun H, Lu H, Duttke SH, Benner C, Glass CK, Lauberth SM. Mutant p53 shapes the enhancer landscape of cancer cells in response to chronic immune signaling. Nat Commun. 2017;8:754.
Abatti LE, Lado-Fernandez P, Huynh L, Collado M, Hoffman MM, Mitchell JA. Epigenetic reprogramming of a distal developmental enhancer cluster drives SOX2 overexpression in breast and lung adenocarcinoma. Nucleic Acids Res. 2023;51:10109–31.
Alcala-Vida R, Awada A, Boutillier AL, Merienne K. Epigenetic mechanisms underlying enhancer modulation of neuronal identity, neuronal activity and neurodegeneration. Neurobiol Dis. 2021;147:105155.
Zeller P, Yeung J, Vinas Gaza H, de Barbanson BA, Bhardwaj V, Florescu M, et al. Single-cell sortChIC identifies hierarchical chromatin dynamics during hematopoiesis. Nat Genet. 2023;55:333–45.
Hnisz D, Schuijers J, Lin CY, Weintraub AS, Abraham BJ, Lee TI, et al. Convergence of developmental and oncogenic signaling pathways at transcriptional super-enhancers. Mol Cell. 2015;58:362–70.
Shlyueva D, Stampfel G, Stark A. Transcriptional enhancers: from properties to genome-wide predictions. Nat Rev Genet. 2014;15:272–86.
Chen PB, Fiaux PC, Zhang K, Li B, Kubo N, Jiang S, et al. Systematic discovery and functional dissection of enhancers needed for cancer cell fitness and proliferation. Cell Rep. 2022;41:111630.
Solvie D, Baluapuri A, Uhl L, Fleischhauer D, Endres T, Papadopoulos D, et al. MYC multimers shield stalled replication forks from RNA polymerase. Nature. 2022;612:148–55.
Jakobsen ST, Jensen RAM, Madsen MS, Ravnsborg T, Vaagenso CS, Siersbaek MS, et al. MYC activity at enhancers drives prognostic transcriptional programs through an epigenetic switch. Nat Genet. 2024;56:663–74.
Tan MH, Li J, Xu HE, Melcher K, Yong EL. Androgen receptor: structure, role in prostate cancer and drug discovery. Acta Pharmacol Sin. 2015;36:3–23.
Hanker AB, Sudhan DR, Arteaga CL. Overcoming endocrine resistance in breast cancer. Cancer Cell. 2020;37:496–513.
Linder S, Hoogstraat M, Stelloo S, Eickhoff N, Schuurman K, de Barros H, et al. Drug-induced epigenomic plasticity reprograms circadian rhythm regulation to drive prostate cancer toward androgen independence. Cancer Discov. 2022;12:2074–97.
Rahnamoun H, Lee J, Sun Z, Lu H, Ramsey KM, Komives EA, et al. RNAs interact with BRD4 to promote enhanced chromatin engagement and transcription activation. Nat Struct Mol Biol. 2018;25:687–97.
Liu M, Du M, Yu J, Qian Z, Gao Y, Pan W, et al. CEBPA mutants down-regulate AML cell susceptibility to NK-mediated lysis by disruption of the expression of NKG2D ligands, which can be restored by LSD1 inhibition. Oncoimmunology. 2022;11:2016158.
Sun Y, Zhou B, Mao F, Xu J, Miao H, Zou Z, et al. HOXA9 reprograms the enhancer landscape to promote leukemogenesis. Cancer Cell. 2018;34:643–58.e5.
Deng Q, Natesan R, Cidre-Aranaz F, Arif S, Liu Y, Rasool RU, et al. Oncofusion-driven de novo enhancer assembly promotes malignancy in Ewing sarcoma via aberrant expression of the stereociliary protein LOXHD1. Cell Rep. 2022;39:110971.
Baca SC, Takeda DY, Seo JH, Hwang J, Ku SY, Arafeh R, et al. Reprogramming of the FOXA1 cistrome in treatment-emergent neuroendocrine prostate cancer. Nat Commun. 2021;12:1979.
Goglia AG, Alshalalfa M, Khan A, Isakov DR, Hougen HY, Swami N, et al. Pan-cancer genomic analysis reveals FOXA1 amplification is associated with adverse outcomes in non-small cell lung, prostate, and breast cancers. J Natl Cancer Inst. 2025;117:188–97.
Li M, Liu M, Han W, Wang Z, Han D, Patalano S, et al. LSD1 inhibition disrupts super-enhancer-driven oncogenic transcriptional programs in castration-resistant prostate cancer. Cancer Res. 2023;83:1684–98.
Parolia A, Cieslik M, Chu SC, Xiao L, Ouchi T, Zhang Y, et al. Distinct structural classes of activating FOXA1 alterations in advanced prostate cancer. Nature. 2019;571:413–8.
Arruabarrena-Aristorena A, Maag JLV, Kittane S, Cai Y, Karthaus WR, Ladewig E, et al. FOXA1 mutations reveal distinct chromatin profiles and influence therapeutic response in breast cancer. Cancer Cell. 2020;38:534–50.e9.
Adams EJ, Karthaus WR, Hoover E, Liu D, Gruet A, Zhang Z, et al. FOXA1 mutations alter pioneering activity, differentiation and prostate cancer phenotypes. Nature. 2019;571:408–12.
Roe J, Hwang C, Somerville T, Milazzo J, Lee E, Da Silva B, et al. Enhancer reprogramming promotes pancreatic cancer metastasis. Cell. 2017;170:875–88.e20.
Dodonova SO, Zhu F, Dienemann C, Taipale J, Cramer P. Nucleosome-bound SOX2 and SOX11 structures elucidate pioneer factor function. Nature. 2020;580:669–72.
Yan S, Liu Z, Wang T, Sui Y, Wu X, Shen J, et al. Super-enhancer reprograming driven by SOX9 and TCF7L2 represents transcription-targeted therapeutic vulnerability for treating gallbladder cancer. Adv Sci. 2024;11:e2406448.
Wu Z, Zhou J, Zhang X, Zhang Z, Xie Y, Liu JB, et al. Reprogramming of the esophageal squamous carcinoma epigenome by SOX2 promotes ADAR1 dependence. Nat Genet. 2021;53:881–94.
Bi M, Zhang Z, Jiang Y, Xue P, Wang H, Lai Z, et al. Enhancer reprogramming driven by high-order assemblies of transcription factors promotes phenotypic plasticity and breast cancer endocrine resistance. Nat Cell Biol. 2020;22:701–15.
Kim S, Wysocka J. Deciphering the multi-scale, quantitative cis-regulatory code. Mol Cell. 2023;83:373–92.
Spitz F, Furlong EE. Transcription factors: from enhancer binding to developmental control. Nat Rev Genet. 2012;13:613–26.
Bi M, Zhang Z, Jiang YZ, Xue P, Wang H, Lai Z, et al. Enhancer reprogramming driven by high-order assemblies of transcription factors promotes phenotypic plasticity and breast cancer endocrine resistance. Nat Cell Biol. 2020;22:701–15.
Stadhouders R, Filion GJ, Graf T. Transcription factors and 3D genome conformation in cell-fate decisions. Nature. 2019;569:345–54.
Neumayr C, Haberle V, Serebreni L, Karner K, Hendy O, Boija A, et al. Differential cofactor dependencies define distinct types of human enhancers. Nature. 2022;606:406–13.
Bell CC, Balic JJ, Talarmain L, Gillespie A, Scolamiero L, Lam EYN, et al. Comparative cofactor screens show the influence of transactivation domains and core promoters on the mechanisms of transcription. Nat Genet. 2024;56:1181–92.
Chen L, Zhang Z, Han Q, Maity BK, Rodrigues L, Zboril E, et al. Hormone-induced enhancer assembly requires an optimal level of hormone receptor multivalent interactions. Mol Cell. 2023;83:3438–56.e12.
Calo E, Wysocka J. Modification of enhancer chromatin: what, how, and why? Mol Cell. 2013;49:825–37.
Zhao S, Allis CD, Wang GG. The language of chromatin modification in human cancers. Nat Rev Cancer. 2021;21:413–30.
Kubo N, Chen PB, Hu R, Ye Z, Sasaki H, Ren B. H3K4me1 facilitates promoter-enhancer interactions and gene activation during embryonic stem cell differentiation. Mol Cell. 2024;84:1742–52.e5.
Kreibich E, Kleinendorst R, Barzaghi G, Kaspar S, Krebs AR. Single-molecule footprinting identifies context-dependent regulation of enhancers by DNA methylation. Mol Cell. 2023;83:787–802.e9.
Qu Y, Siggens L, Cordeddu L, Gaidzik VI, Karlsson K, Bullinger L, et al. Cancer-specific changes in DNA methylation reveal aberrant silencing and activation of enhancers in leukemia. Blood. 2017;129:e13–25.
Li J, Li Y, Li W, Luo H, Xi Y, Dong S, et al. Guide positioning sequencing identifies aberrant DNA methylation patterns that alter cell identity and tumor-immune surveillance networks. Genome Res. 2019;29:270–80.
Benetatos L, Vartholomatos G. Enhancer DNA methylation in acute myeloid leukemia and myelodysplastic syndromes. Cell Mol Life Sci. 2018;75:1999–2009.
Na F, Pan X, Chen J, Chen X, Wang M, Chi P, et al. KMT2C deficiency promotes small cell lung cancer metastasis through DNMT3A-mediated epigenetic reprogramming. Nat Cancer. 2022;3:753–67.
Fagan RJ, Dingwall AK. COMPASS ascending: emerging clues regarding the roles of MLL3/KMT2C and MLL2/KMT2D proteins in cancer. Cancer Lett. 2019;458:56–65.
Gala K, Li Q, Sinha A, Razavi P, Dorso M, Sanchez-Vega F, et al. KMT2C mediates the estrogen dependence of breast cancer through regulation of ERalpha enhancer function. Oncogene. 2018;37:4692–710.
Wang L, Zhao Z, Ozark PA, Fantini D, Marshall SA, Rendleman EJ, et al. Resetting the epigenetic balance of Polycomb and COMPASS function at enhancers for cancer therapy. Nat Med. 2018;24:758–69.
Hogg SJ, Motorna O, Cluse LA, Johanson TM, Coughlan HD, Raviram R, et al. Targeting histone acetylation dynamics and oncogenic transcription by catalytic P300/CBP inhibition. Mol Cell. 2021;81:2183–200.e13.
Jiang Y, Ortega-Molina A, Geng H, Ying H, Hatzi K, Parsa S, et al. CREBBP inactivation promotes the development of HDAC3-dependent lymphomas. Cancer Discov. 2017;7:38–53.
Zhang J, Vlasevska S, Wells VA, Nataraj S, Holmes AB, Duval R, et al. The CREBBP acetyltransferase is a haploinsufficient tumor suppressor in B-cell lymphoma. Cancer Discov. 2017;7:322–37.
Garcia-Ramirez I, Tadros S, Gonzalez-Herrero I, Martin-Lorenzo A, Rodriguez-Hernandez G, Moore D, et al. Crebbp loss cooperates with Bcl2 overexpression to promote lymphoma in mice. Blood. 2017;129:2645–56.
Chen Q, Yang B, Liu X, Zhang XD, Zhang L, Liu T. Histone acetyltransferases CBP/p300 in tumorigenesis and CBP/p300 inhibitors as promising novel anticancer agents. Theranostics. 2022;12:4935–48.
Duan R, Du W, Guo W. EZH2: a novel target for cancer treatment. J Hematol Oncol. 2020;13:104.
Verma A, Singh A, Singh MP, Nengroo MA, Saini KK, Satrusal SR, et al. EZH2-H3K27me3 mediated KRT14 upregulation promotes TNBC peritoneal metastasis. Nat Commun. 2022;13:7344.
Zheng Y, Wang Z, Wei S, Liu Z, Chen G. Epigenetic silencing of chemokine CCL2 represses macrophage infiltration to potentiate tumor development in small cell lung cancer. Cancer Lett. 2021;499:148–63.
Schade AE, Perurena N, Yang Y, Rodriguez CL, Krishnan A, Gardner A, et al. AKT and EZH2 inhibitor skill TNBCs by hijacking mechanisms of involution. Nature. 2024;635:755–63.
Clapier CR, Iwasa J, Cairns BR, Peterson CL. Mechanisms of action and regulation of ATP-dependent chromatin-remodelling complexes. Nat Rev Mol Cell Biol. 2017;18:407–22.
Kadoch C, Crabtree GR. Mammalian SWI/SNF chromatin remodeling complexes and cancer: mechanistic insights gained from human genomics. Sci Adv. 2015;1:e1500447.
Yoshida A, Kobayashi E, Kubo T, Kodaira M, Motoi T, Motoi N, et al. Clinicopathological and molecular characterization of SMARCA4-deficient thoracic sarcomas with comparison to potentially related entities. Mod Pathol. 2017;30:797–809.
Yavas A, Ozcan K, Adsay NV, Balci S, Tarcan ZC, Hechtman JF, et al. SWI/SNF complex-deficient undifferentiated carcinoma of the pancreas: clinicopathologic and genomic analysis. Mod Pathol. 2024;37:100585.
Xue Y, Morris JL, Yang K, Fu Z, Zhu X, Johnson F, et al. SMARCA4/2 loss inhibits chemotherapy-induced apoptosis by restricting IP3R3-mediated Ca(2+) flux to mitochondria. Nat Commun. 2021;12:5404.
Field NR, Dickson KA, Nassif NT, Marsh DJ. SMARCA4 and SMARCA2 co-deficiency: An uncommon molecular signature defining a subset of rare, aggressive and undifferentiated malignancies associated with defective chromatin remodeling. Cancer Lett. 2024;605:217282.
Mathur R, Alver BH, San Roman AK, Wilson BG, Wang X, Agoston AT, et al. ARID1A loss impairs enhancer-mediated gene regulation and drives colon cancer in mice. Nat Genet. 2017;49:296–302.
Xu C, Huang KK, Law JH, Chua JS, Sheng T, Flores NM, et al. Comprehensive molecular phenotyping of ARID1A-deficient gastric cancer reveals pervasive epigenomic reprogramming and therapeutic opportunities. Gut. 2023;72:1651–63.
Shi H, Tao T, Abraham BJ, Durbin AD, Zimmerman MW, Kadoch C, et al. ARID1A loss in neuroblastoma promotes the adrenergic-to-mesenchymal transition by regulating enhancer-mediated gene expression. Sci Adv. 2020;6:eaaz3440.
Kotagiri S, Blazanin N, Xi Y, Han Y, Qudratullah M, Liang X, et al. Enhancer reprogramming underlies therapeutic utility of a SMARCA2 degrader in SMARCA4 mutant cancer. Cell Chem Biol. 2024;31:2069–2084.e9.
Ren B, Yang J, Wang C, Yang G, Wang H, Chen Y, et al. High-resolution Hi-C maps highlight multiscale 3D epigenome reprogramming during pancreatic cancer metastasis. J Hematol Oncol. 2021;14:120.
Du Y, Gu Z, Li Z, Yuan Z, Zhao Y, Zheng X, et al. Dynamic interplay between structural variations and 3D genome organization in pancreatic cancer. Adv Sci. 2022;9:e2200818.
Luo, Zhu H, Eshelman MA G, Fung TK, Lai Q, Wang F, et al. HOTTIP-dependent R-loop formation regulates CTCF boundary activity and TAD integrity in leukemia. Mol Cell. 2022;82:833–51.e11.
Kloetgen A, Thandapani P, Ntziachristos P, Ghebrechristos Y, Nomikou S, Lazaris C, et al. Three-dimensional chromatin landscapes in T cell acute lymphoblastic leukemia. Nat Genet. 2020;52:388–400.
Hung KL, Yost KE, Xie L, Shi Q, Helmsauer K, Luebeck J, et al. ecDNA hubs drive cooperative intermolecular oncogene expression. Nature. 2021;600:731–6.
Zhu Y, Gujar AD, Wong CH, Tjong H, Ngan CY, Gong L, et al. Oncogenic extrachromosomal DNA functions as mobile enhancers to globally amplify chromosomal transcription. Cancer Cell. 2021;39:694–707.e7.
Helmsauer K, Valieva ME, Ali S, Chamorro Gonzalez R, Schopflin R, Roefzaad C, et al. Enhancer hijacking determines extrachromosomal circular MYCN amplicon architecture in neuroblastoma. Nat Commun. 2020;11:5823.
Meng S, Liu X, Zhu S, Xie P, Fang H, Pan Q, et al. Young LINE-1 transposon 5’ UTRs marked by elongation factor ELL3 function as enhancers to regulate naive pluripotency in embryonic stem cells. Nat Cell Biol. 2023;25:1319–31.
Karttunen K, Patel D, Xia J, Fei L, Palin K, Aaltonen L, et al. Transposable elements as tissue-specific enhancers in cancers of endodermal lineage. Nat Commun. 2023;14:5313.
Zhang T, Xia W, Song X, Mao Q, Huang X, Chen B, et al. Super-enhancer hijacking LINC01977 promotes malignancy of early-stage lung adenocarcinoma addicted to the canonical TGF-beta/SMAD3 pathway. J Hematol Oncol. 2022;15:114.
Palazzo AF, Koonin EV. Functional long non-coding RNAs evolve from junk transcripts. Cell. 2020;183:1151–61.
Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–56.
Li M, Huang H, Li L, He C, Zhu L, Guo H, et al. Core transcription regulatory circuitry orchestrates corneal epithelial homeostasis. Nat Commun. 2021;12:420.
Jiang YY, Jiang Y, Li CQ, Zhang Y, Dakle P, Kaur H, et al. TP63, SOX2, and KLF5 establish a core regulatory circuitry that controls epigenetic and transcription patterns in esophageal squamous cell carcinoma cell lines. Gastroenterology. 2020;159:1311–27.e19.
Decaesteker B, Louwagie A, Loontiens S, De Vloed F, Bekaert SL, Roels J, et al. SOX11 regulates SWI/SNF complex components as member of the adrenergic neuroblastoma core regulatory circuitry. Nat Commun. 2023;14:1267.
Zhang J, Wang Q, Qi S, Duan Y, Liu Z, Liu J, et al. An oncogenic enhancer promotes melanoma progression via regulating ETV4 expression. J Transl Med. 2024;22:547.
Ye J, Cai S, Feng Y, Li J, Cai Z, Deng Y, et al. Metformin escape in prostate cancer by activating the PTGR1 transcriptional program through a novel super-enhancer. Signal Transduct Target Ther. 2023;8:303.
Liu Y, Su Z, Tavana O, Gu W. Understanding the complexity of p53 in a new era of tumor suppression. Cancer Cell. 2024;42:946–67.
McClurg UL, Chit N, Azizyan M, Edwards J, Nabbi A, Riabowol KT, et al. Molecular mechanism of the TP53-MDM2-AR-AKT signalling network regulation by USP12. Oncogene. 2018;37:4679–91.
Fendler A, Bauer D, Busch J, Jung K, Wulf-Goldenberg A, Kunz S, et al. Inhibiting WNT and NOTCH in renal cancer stem cells and the implications for human patients. Nat Commun. 2020;11:929.
Sanceau J, Poupel L, Joubel C, Lagoutte I, Caruso S, Pinto S, et al. DLK1/DIO3 locus upregulation by a beta-catenin-dependent enhancer drives cell proliferation and liver tumorigenesis. Mol Ther. 2024;32:1125–43.
Zhou Z, Li J, Ousmane D, Peng L, Yuan X, Wang J. Metabolic reprogramming directed by super-enhancers in tumors: an emerging landscape. Mol Ther. 2024;32:572–9.
Nguyen TTT, Zhang Y, Shang E, Shu C, Torrini C, Zhao J, et al. HDAC inhibitors elicit metabolic reprogramming by targeting super-enhancers in glioblastoma models. J Clin Invest. 2020;130:3699–716.
Hay N. Reprogramming glucose metabolism in cancer: can it be exploited for cancer therapy? Nat Rev Cancer. 2016;16:635–49.
Hsieh MH, Choe JH, Gadhvi J, Kim YJ, Arguez MA, Palmer M, et al. p63 and SOX2 dictate glucose reliance and metabolic vulnerabilities in squamous cell carcinomas. Cell Rep. 2019;28:1860–78.e9.
Zhou RW, Xu J, Martin TC, Zachem AL, He J, Ozturk S, et al. A local tumor microenvironment acquired super-enhancer induces an oncogenic driver in colorectal carcinoma. Nat Commun. 2022;13:6041.
Yan T, Shen C, Jiang P, Yu C, Guo F, Tian X, et al. Risk SNP-induced lncRNA-SLCC1 drives colorectal cancer through activating glycolysis signaling. Signal Transduct Target Ther. 2021;6:70.
Maitituoheti M, Keung EZ, Tang M, Yan L, Alam H, Han G, et al. Enhancer reprogramming confers dependence on glycolysis and IGF signaling in KMT2D mutant melanoma. Cell Rep. 2020;33:108293.
Cheng C, Geng F, Cheng X, Guo D. Lipid metabolism reprogramming and its potential targets in cancer. Cancer Commun. 2018;38:27.
Li LY, Yang Q, Jiang YY, Yang W, Jiang Y, Li X, et al. Interplay and cooperation between SREBF1 and master transcription factors regulate lipid metabolism and tumor-promoting pathways in squamous cancer. Nat Commun. 2021;12:4362.
Peng JY, Cai DK, Zeng RL, Zhang CY, Li GC, Chen SF, et al. Upregulation of superenhancer-driven LncRNA FASRL by USF1 promotes de novo fatty acid biosynthesis to exacerbate hepatocellular carcinoma. Adv Sci. 2022;10:e2204711.
Ma S, Zhou B, Yang Q, Pan Y, Yang W, Freedland SJ, et al. A transcriptional regulatory loop of master regulator transcription factors, PPARG, and fatty acid synthesis promotes esophageal adenocarcinoma. Cancer Res. 2021;81:1216–29.
Mossmann D, Muller C, Park S, Ryback B, Colombi M, Ritter N, et al. Arginine reprograms metabolism in liver cancer via RBM39. Cell. 2023;186:5068–83.e23.
Bian Y, Li W, Kremer DM, Sajjakulnukit P, Li S, Crespo J, et al. Cancer SLC43A2 alters T cell methionine metabolism and histone methylation. Nature. 2020;585:277–82.
Shen Q, Wang R, Liu X, Song P, Zheng M, Ren X, et al. HSF1 stimulates glutamine transport by super-enhancer-driven lncRNA LINC00857 in colorectal cancer. Cancers. 2022;14:3855.
Nam C, Li LY, Yang Q, Ziman B, Zhao H, Hu B, et al. A druggable cascade links methionine metabolism to epigenomic reprogramming in squamous cell carcinoma. Proc Natl Acad Sci USA. 2024;121:e2320835121.
Munk SHN, Merchut-Maya JM, Adelantado Rubio A, Hall A, Pappas G, Milletti G, et al. NAD(+) regulates nucleotide metabolism and genomic DNA replication. Nat Cell Biol. 2023;25:1774–86.
Navas LE, Carnero A. NAD(+) metabolism, stemness, the immune response, and cancer. Signal Transduct Target Ther. 2021;6:2.
Chowdhry S, Zanca C, Rajkumar U, Koga T, Diao Y, Raviram R, et al. NAD metabolic dependency in cancer is shaped by gene amplification and enhancer remodelling. Nature. 2019;569:570–5.
Ren J, Ren B, Liu X, Cui M, Fang Y, Wang X, et al. Crosstalk between metabolic remodeling and epigenetic reprogramming: A new perspective on pancreatic cancer. Cancer Lett. 2024;587:216649.
Yang J, Ren B, Ren J, Yang G, Fang Y, Wang X, et al. Epigenetic reprogramming-induced guanidinoacetic acid synthesis promotes pancreatic cancer metastasis and transcription-activating histone modifications. J Exp Clin Cancer Res. 2023;42:155.
Bertheloot D, Latz E, Franklin BS. Necroptosis, pyroptosis and apoptosis: an intricate game of cell death. Cell Mol Immunol. 2021;18:1106–21.
Vandenabeele P, Bultynck G, Savvides SN. Pore-forming proteins as drivers of membrane permeabilization in cell death pathways. Nat Rev Mol Cell Biol. 2023;24:312–33.
Bal E, Kumar R, Hadigol M, Holmes AB, Hilton LK, Loh JW, et al. Super-enhancer hypermutation alters oncogene expression in B cell lymphoma. Nature. 2022;607:808–15.
Dawson MA, Prinjha RK, Dittmann A, Giotopoulos G, Bantscheff M, Chan WI, et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature. 2011;478:529–33.
Petak I, Danam RP, Tillman DM, Vernes R, Howell SR, Berczi L, et al. Hypermethylation of the gene promoter and enhancer region can regulate Fas expression and sensitivity in colon carcinoma. Cell Death Differ. 2003;10:211–7.
Hu J, Pan D, Li G, Chen K, Hu X. Regulation of programmed cell death by Brd4. Cell Death Dis. 2022;13:1059.
Fan Y, Mao R, Yang J. NF-kappaB and STAT3 signaling pathways collaboratively link inflammation to cancer. Protein Cell. 2013;4:176–85.
Ullah R, Yin Q, Snell AH, Wan L. RAF-MEK-ERK pathway in cancer evolution and treatment. Semin Cancer Biol. 2022;85:123–54.
Hoxhaj G, Manning BD. The PI3K-AKT network at the interface of oncogenic signalling and cancer metabolism. Nat Rev Cancer. 2020;20:74–88.
Huang Y, Hong W, Wei X. The molecular mechanisms and therapeutic strategies of EMT in tumor progression and metastasis. J Hematol Oncol. 2022;15:129.
Cui S, Wu Q, Liu M, Su M, Liu S, Shao L, et al. EphA2 super-enhancer promotes tumor progression by recruiting FOSL2 and TCF7L2 to activate the target gene EphA2. Cell Death Dis. 2021;12:264.
Peng L, Jiang B, Yuan X, Qiu Y, Peng J, Huang Y, et al. Super-enhancer-associated long noncoding RNA HCCL5 is activated by ZEB1 and promotes the malignancy of hepatocellular carcinoma. Cancer Res. 2019;79:572–84.
Wang L, Wang E, Prado Balcazar J, Wu Z, Xiang K, Wang Y, et al. Chromatin remodeling of colorectal cancer liver metastasis is mediated by an HGF-PU.1-DPP4 axis. Adv Sci. 2021;8:e2004673.
Liu M, Zhou J, Liu X, Feng Y, Yang W, Wu F, et al. Targeting monocyte-intrinsic enhancer reprogramming improves immunotherapy efficacy in hepatocellular carcinoma. Gut. 2020;69:365–79.
Zhang C, Wei S, Sun WP, Teng K, Dai MM, Wang FW, et al. Super-enhancer-driven AJUBA is activated by TCF4 and involved in epithelial-mesenchymal transition in the progression of hepatocellular carcinoma. Theranostics. 2020;10:9066–82.
Nishida J, Momoi Y, Miyakuni K, Tamura Y, Takahashi K, Koinuma D, et al. Epigenetic remodelling shapes inflammatory renal cancer and neutrophil-dependent metastasis. Nat Cell Biol. 2020;22:465–75.
Li Q, Lv X, Han C, Kong Y, Dai Z, Huo D, et al. Enhancer reprogramming promotes the activation of cancer-associated fibroblasts and breast cancer metastasis. Theranostics. 2022;12:7491–508.
Dong J, Li J, Li Y, Ma Z, Yu Y, Wang CY. Transcriptional super-enhancers control cancer stemness and metastasis genes in squamous cell carcinoma. Nat Commun. 2021;12:3974.
Liu X, Song J, Zhang H, Liu X, Zuo F, Zhao Y, et al. Immune checkpoint HLA-E:CD94-NKG2A mediates evasion of circulating tumor cells from NK cell surveillance. Cancer Cell. 2023;41:272–87.e9.
Szczerba BM, Castro-Giner F, Vetter M, Krol I, Gkountela S, Landin J, et al. Neutrophils escort circulating tumour cells to enable cell cycle progression. Nature. 2019;566:553–7.
Fan Z, Wu C, Chen M, Jiang Y, Wu Y, Mao R, et al. The generation of PD-L1 and PD-L2 in cancer cells: From nuclear chromatin reorganization to extracellular presentation. Acta Pharm Sin B. 2022;12:1041–53.
Michelatti D, Beyes S, Bernardis C, Negri ML, Morelli L, Bediaga NG, et al. Oncogenic enhancers prime quiescent metastatic cells to escape NK immune surveillance by eliciting transcriptional memory. Nat Commun. 2024;15:2198.
Azubuike UF, Tanner K. Biophysical determinants of cancer organotropism. Trends Cancer. 2023;9:188–97.
Teng S, Li YE, Yang M, Qi R, Huang Y, Wang Q, et al. Tissue-specific transcription reprogramming promotes liver metastasis of colorectal cancer. Cell Res. 2020;30:34–49.
Hojo H, Saito T, He X, Guo Q, Onodera S, Azuma T, et al. Runx2 regulates chromatin accessibility to direct the osteoblast program at neonatal stages. Cell Rep. 2022;40:111315.
Zhang H, Pan Y, Zheng L, Choe C, Lindgren B, Jensen ED, et al. FOXO1 inhibits Runx2 transcriptional activity and prostate cancer cell migration and invasion. Cancer Res. 2011;71:3257–67.
Morrissey SM, Zhang F, Ding C, Montoya-Durango DE, Hu X, Yang C, et al. Tumor-derived exosomes drive immunosuppressive macrophages in a pre-metastatic niche through glycolytic dominant metabolic reprogramming. Cell Metab. 2021;33:2040–58.e10.
Pefanis E, Wang J, Rothschild G, Lim J, Kazadi D, Sun J, et al. RNA exosome-regulated long non-coding RNA transcription controls super-enhancer activity. Cell. 2015;161:774–89.
Shang S, Yang J, Jazaeri AA, Duval AJ, Tufan T, Lopes Fischer N, et al. Chemotherapy-induced distal enhancers drive transcriptional programs to maintain the chemoresistant state in ovarian cancer. Cancer Res. 2019;79:4599–611.
Wu Y, Chen S, Shao Y, Su Y, Li Q, Wu J, et al. KLF5 promotes tumor progression and parp inhibitor resistance in ovarian cancer. Adv Sci. 2023;10:e2304638.
Liu S, Zou Q, Chen JP, Yao X, Guan P, Liang W, et al. Targeting enhancer reprogramming to mitigate MEK inhibitor resistance in preclinical models of advanced ovarian cancer. J Clin Invest. 2021;131:e145035.
Wang SM, Lin WC, Lin HY, Chen YL, Ko CY, Wang JM. CCAAT/Enhancer-binding protein delta mediates glioma stem-like cell enrichment and ATP-binding cassette transporter ABCA1 activation for temozolomide resistance in glioblastoma. Cell Death Discov. 2021;7:8.
Li Y, Qiu X, Wang X, Liu H, Geck RC, Tewari AK, et al. FGFR-inhibitor-mediated dismissal of SWI/SNF complexes from YAP-dependent enhancers induces adaptive therapeutic resistance. Nat Cell Biol. 2021;23:1187–98.
Tong X, Patel AS, Kim E, Li H, Chen Y, Li S, et al. Adeno-to-squamous transition drives resistance to KRAS inhibition in LKB1 mutant lung cancer. Cancer Cell. 2024;42:413–28.e7.
Petropoulos M, Karamichali A, Rossetti GG, Freudenmann A, Iacovino LG, Dionellis VS, et al. Transcription-replication conflicts underlie sensitivity to PARP inhibitors. Nature. 2024;628:433–41.
Li H, Liu ZY, Wu N, Chen YC, Cheng Q, Wang J. PARP inhibitor resistance: the underlying mechanisms and clinical implications. Mol Cancer. 2020;19:107.
Morris EJ, Jha S, Restaino CR, Dayananth P, Zhu H, Cooper A, et al. Discovery of a novel ERK inhibitor with activity in models of acquired resistance to BRAF and MEK inhibitors. Cancer Discov. 2013;3:742–50.
Waldschmidt JM, Kloeber JA, Anand P, Frede J, Kokkalis A, Dimitrova V, et al. Single-cell profiling reveals metabolic reprogramming as a resistance mechanism in BRAF-mutated multiple myeloma. Clin Cancer Res. 2021;27:6432–44.
Quintanal-Villalonga A, Chan JM, Yu HA, Pe’er D, Sawyers CL, Sen T, et al. Lineage plasticity in cancer: a shared pathway of therapeutic resistance. Nat Rev Clin Oncol. 2020;17:360–71.
Song B, Park SH, Zhao JC, Fong KW, Li S, Lee Y, et al. Targeting FOXA1-mediated repression of TGF-beta signaling suppresses castration-resistant prostate cancer progression. J Clin Invest. 2019;129:569–82.
Han M, Li F, Zhang Y, Dai P, He J, Li Y, et al. FOXA2 drives lineage plasticity and KIT pathway activation in neuroendocrine prostate cancer. Cancer Cell. 2022;40:1306–23.e8.
Feng Y, Liu X, Pauklin S. 3D chromatin architecture and epigenetic regulation in cancer stem cells. Protein Cell. 2021;12:440–54.
Liu SX, Wang C, Lin RB, Ding WY, Roy G, Wang HB, et al. Super-enhancer driven SOX2 promotes tumor formation by chromatin re-organization in nasopharyngeal carcinoma. EBioMedicine. 2023;98:104870.
Zhang M, Hoyle RG, Ma Z, Sun B, Cai W, Cai H, et al. FOSL1 promotes metastasis of head and neck squamous cell carcinoma through super-enhancer-driven transcription program. Mol Ther. 2021;29:2583–600.
Zhou J, Wang S, Nie D, Lai P, Li Y, Li Y, et al. Super-enhancer landscape reveals leukemia stem cell reliance on X-box binding protein 1 as a therapeutic vulnerability. Sci Transl Med. 2021;13:eabh3462.
Poli V, Fagnocchi L, Fasciani A, Cherubini A, Mazzoleni S, Ferrillo S, et al. MYC-driven epigenetic reprogramming favors the onset of tumorigenesis by inducing a stem cell-like state. Nat Commun. 2018;9:1024.
Wu T, Huang H, Wang X. Dissecting super-enhancer heterogeneity: time to re-examine cancer subtypes? Trends Genet. 2022;38:1199–203.
Elhanani O, Ben-Uri R, Keren L. Spatial profiling technologies illuminate the tumor microenvironment. Cancer Cell. 2023;41:404–20.
Evan GI, Hah N, Littlewood TD, Sodir NM, Campos T, Downes M, et al. Re-engineering the pancreas tumor microenvironment: a “regenerative program” hacked. Clin Cancer Res. 2017;23:1647–55.
Satpathy AT, Granja JM, Yost KE, Qi Y, Meschi F, McDermott GP, et al. Massively parallel single-cell chromatin landscapes of human immune cell development and intratumoral T cell exhaustion. Nat Biotechnol. 2019;37:925–36.
Ning H, Huang S, Lei Y, Zhi R, Yan H, Jin J, et al. Enhancer decommissioning by MLL4 ablation elicits dsRNA-interferon signaling and GSDMD-mediated pyroptosis to potentiate anti-tumor immunity. Nat Commun. 2022;13:6578.
Cao T, Zhang W, Wang Q, Wang C, Ma W, Zhang C, et al. Cancer SLC6A6-mediated taurine uptake transactivates immune checkpoint genes and induces exhaustion in CD8(+) T cells. Cell. 2024;187:2288–304.e27.
Riegel D, Romero-Fernandez E, Simon M, Adenugba AR, Singer K, Mayr R, et al. Integrated single-cell profiling dissects cell-state-specific enhancer landscapes of human tumor-infiltrating CD8(+) T cells. Mol Cell. 2023;83:622–36.e10.
Jia D, Wang Q, Qi Y, Jiang Y, He J, Lin Y, et al. Microbial metabolite enhances immunotherapy efficacy by modulating T cell stemness in pan-cancer. Cell. 2024;187:1651–65.e21.
Zhang Q, Zhao Q, Li T, Lu L, Wang F, Zhang H, et al. Lactobacillus plantarum-derived indole-3-lactic acid ameliorates colorectal tumorigenesis via epigenetic regulation of CD8(+) T cell immunity. Cell Metab. 2023;35:943–60.e9.
Wang Q, Zhang J, Wen Y, Qi S, Duan Y, Liu Q, et al. The pleiotropic enhancer enh9 promotes cell proliferation and migration in non-small cell lung cancer via ERMP1 and PD-L1. Biochim Biophys Acta Mol Basis Dis. 2024;1870:167015.
Wang J, Ge J, Wang Y, Xiong F, Guo J, Jiang X, et al. EBV miRNAs BART11 and BART17-3p promote immune escape through the enhancer-mediated transcription of PD-L1. Nat Commun. 2022;13:866.
Tu M, Klein L, Espinet E, Georgomanolis T, Wegwitz F, Li X, et al. TNF-α-producing macrophages determine subtype identity and prognosis via AP1 enhancer reprogramming in pancreatic cancer. Nat Cancer. 2021;2:1185–203.
Huffaker TB, Ekiz HA, Barba C, Lee SH, Runtsch MC, Nelson MC, et al. A Stat1 bound enhancer promotes Nampt expression and function within tumor associated macrophages. Nat Commun. 2021;12:2620.
Liu PS, Chen YT, Li X, Hsueh PC, Tzeng SF, Chen H, et al. CD40 signal rewires fatty acid and glutamine metabolism for stimulating macrophage anti-tumorigenic functions. Nat Immunol. 2023;24:452–62.
Sun X, Zhou L, Wang Y, Deng G, Cao X, Ke B, et al. Single-cell analyses reveal cannabidiol rewires tumor microenvironment via inhibiting alternative activation of macrophage and synergizes with anti-PD-1 in colon cancer. J Pharm Anal. 2023;13:726–44.
Que H, Fu Q, Lan T, Tian X, Wei X. Tumor-associated neutrophils and neutrophil-targeted cancer therapies. Biochim Biophys Acta Rev Cancer. 2022;1877:188762.
Wu Y, Ma J, Yang X, Nan F, Zhang T, Ji S, et al. Neutrophil profiling illuminates anti-tumor antigen-presenting potency. Cell. 2024;187:1422–39.e24.
Niu N, Shen X, Wang Z, Chen Y, Weng Y, Yu F, et al. Tumor cell-intrinsic epigenetic dysregulation shapes cancer-associated fibroblasts heterogeneity to metabolically support pancreatic cancer. Cancer Cell. 2024;42:869–84.e9.
Wang W, Tang YA, Xiao Q, Lee WC, Cheng B, Niu Z, et al. Stromal induction of BRD4 phosphorylation results in chromatin remodeling and BET inhibitor resistance in colorectal cancer. Nat Commun. 2021;12:4441.
Lee KW, Yeo SY, Gong JR, Koo OJ, Sohn I, Lee WY, et al. PRRX1 is a master transcription factor of stromal fibroblasts for myofibroblastic lineage progression. Nat Commun. 2022;13:2793.
Greten FR, Grivennikov SI. Inflammation and cancer: triggers, mechanisms, and consequences. Immunity. 2019;51:27–41.
Betancur PA, Abraham BJ, Yiu YY, Willingham SB, Khameneh F, Zarnegar M, et al. A CD47-associated super-enhancer links pro-inflammatory signalling to CD47 upregulation in breast cancer. Nat Commun. 2017;8:14802.
Chu Z, Gu L, Hu Y, Zhang X, Li M, Chen J, et al. STAG2 regulates interferon signaling in melanoma via enhancer loop reprogramming. Nat Commun. 2022;13:1859.
Thandapani P. Super-enhancers in cancer. Pharmacol Ther. 2019;199:129–38.
Siersbaek R, Scabia V, Nagarajan S, Chernukhin I, Papachristou EK, Broome R, et al. IL6/STAT3 signaling hijacks estrogen receptor alpha enhancers to drive breast cancer metastasis. Cancer Cell. 2020;38:412–23.e9.
Kron KJ, Murison A, Zhou S, Huang V, Yamaguchi TN, Shiah YJ, et al. TMPRSS2-ERG fusion co-opts master transcription factors and activates NOTCH signaling in primary prostate cancer. Nat Genet. 2017;49:1336–45.
He ZX, Wei BF, Zhang X, Gong YP, Ma LY, Zhao W. Current development of CBP/p300 inhibitors in the last decade. Eur J Med Chem. 2021;209:112861.
Lasko LM, Jakob CG, Edalji RP, Qiu W, Montgomery D, Digiammarino EL, et al. Discovery of a selective catalytic p300/CBP inhibitor that targets lineage-specific tumours. Nature. 2017;550:128–32.
Welti J, Sharp A, Brooks N, Yuan W, McNair C, Chand SN, et al. Targeting the p300/CBP axis in lethal prostate cancer. Cancer Discov. 2021;11:1118–37.
Vannam R, Sayilgan J, Ojeda S, Karakyriakou B, Hu E, Kreuzer J, et al. Targeted degradation of the enhancer lysine acetyltransferases CBP and p300. Cell Chem Biol. 2021;28:503–14.e12.
Simon RP, Robaa D, Alhalabi Z, Sippl W, Jung M. KATching-up on small molecule modulators of lysine acetyltransferases. J Med Chem. 2016;59:1249–70.
Wang F, Jin Y, Wang M, Luo HY, Fang WJ, Wang YN, et al. Combined anti-PD-1, HDAC inhibitor and anti-VEGF for MSS/pMMR colorectal cancer: a randomized phase 2 trial. Nat Med. 2024;30:1035–43.
Bachy E, Camus V, Thieblemont C, Sibon D, Casasnovas RO, Ysebaert L, et al. Romidepsin plus CHOP versus CHOP in patients with previously untreated peripheral T-cell lymphoma: results of the Ro-CHOP Phase III Study (Conducted by LYSA). J Clin Oncol. 2022;40:242–51.
O’Connor OA, Horwitz S, Masszi T, Van Hoof A, Brown P, Doorduijn J, et al. Belinostat in patients with relapsed or refractory peripheral T-cell lymphoma: results of the pivotal Phase II BELIEF (CLN-19) Study. J Clin Oncol. 2015;33:2492–9.
Xu B, Zhang Q, Hu X, Li Q, Sun T, Li W, et al. Entinostat, a class I selective histone deacetylase inhibitor, plus exemestane for Chinese patients with hormone receptor-positive advanced breast cancer: a multicenter, randomized, double-blind, placebo-controlled, phase 3 trial. Acta Pharm Sin B. 2023;13:2250–8.
Hu J, Sanchez-Rivera FJ, Wang Z, Johnson GN, Ho YJ, Ganesh K, et al. STING inhibits the reactivation of dormant metastasis in lung adenocarcinoma. Nature. 2023;616:806–13.
Mei H, Wu H, Yang J, Zhou B, Wang A, Hu C, et al. Discovery of IHMT-337 as a potent irreversible EZH2 inhibitor targeting CDK4 transcription for malignancies. Signal Transduct Target Ther. 2023;8:18.
Yamagishi M, Kuze Y, Kobayashi S, Nakashima M, Morishima S, Kawamata T, et al. Mechanisms of action and resistance in histone methylation-targeted therapy. Nature. 2024;627:221–8.
Kim KH, Roberts CW. Targeting EZH2 in cancer. Nat Med. 2016;22:128–34.
Mittal P, Roberts CWM. The SWI/SNF complex in cancer - biology, biomarkers and therapy. Nat Rev Clin Oncol. 2020;17:435–48.
Xiao L, Parolia A, Qiao Y, Bawa P, Eyunni S, Mannan R, et al. Targeting SWI/SNF ATPases in enhancer-addicted prostate cancer. Nature. 2022;601:434–9.
Martin BJE, Ablondi EF, Goglia C, Mimoso CA, Espinel-Cabrera PR, Adelman K. Global identification of SWI/SNF targets reveals compensation by EP400. Cell. 2023;186:5290–307.e26.
Henley MJ, Koehler AN. Advances in targeting ‘undruggable’ transcription factors with small molecules. Nat Rev Drug Discov. 2021;20:669–88.
Donati B, Lorenzini E, Ciarrocchi A. BRD4 and cancer: going beyond transcriptional regulation. Mol Cancer. 2018;17:164.
Zhang J, Liu W, Zou C, Zhao Z, Lai Y, Shi Z, et al. Targeting super-enhancer-associated oncogenes in osteosarcoma with THZ2, a covalent CDK7 inhibitor. Clin Cancer Res. 2020;26:2681–92.
Liu Q, Guo L, Lou Z, Xiang X, Shao J. Super-enhancers and novel therapeutic targets in colorectal cancer. Cell Death Dis. 2022;13:228.
Plana D, Palmer AC, Sorger PK. Independent drug action in combination therapy: implications for precision oncology. Cancer Discov. 2022;12:606–24.
Sun B, Shah B, Fiskus W, Qi J, Rajapakshe K, Coarfa C, et al. Synergistic activity of BET protein antagonist-based combinations in mantle cell lymphoma cells sensitive or resistant to ibrutinib. Blood. 2015;126:1565–74.
A phase II study of anti-programmed death-1(PD-1) antibody sintilimab plus histone deacetylase(HDAC) inhibitor chidamide in patients with relapsed/ refractory peripheral T-cell lymphoma [Internet]. 2020. Available from: https://clinicaltrials.gov/study/NCT04512534.
Jindal GA, Farley EK. Enhancer grammar in development, evolution, and disease: dependencies and interplay. Dev Cell. 2021;56:575–87.
Farzin A, Etesami SA, Quint J, Memic A, Tamayol A. Magnetic nanoparticles in cancer therapy and diagnosis. Adv Healthc Mater. 2020;9:e1901058.
Faubert B, Solmonson A, DeBerardinis RJ. Metabolic reprogramming and cancer progression. Science. 2020;368:eaaw5473.
Liao M, Yao D, Wu L, Luo C, Wang Z, Zhang J, et al. Targeting the Warburg effect: a revisited perspective from molecular mechanisms to traditional and innovative therapeutic strategies in cancer. Acta Pharm Sin B. 2024;14:953–1008.
Zhang D, Li J, Wang F, Hu J, Wang S, Sun Y. 2-Deoxy-D-glucose targeting of glucose metabolism in cancer cells as a potential therapy. Cancer Lett. 2014;355:176–83.
Zhao Z, Cao K, Watanabe J, Philips CN, Zeidner JM, Ishi Y, et al. Therapeutic targeting of metabolic vulnerabilities in cancers with MLL3/4-COMPASS epigenetic regulator mutations. J Clin Invest. 2023;133:e169993.
Lord CJ, Ashworth A. PARP inhibitors: synthetic lethality in the clinic. Science. 2017;355:1152–8.
Huang A, Garraway LA, Ashworth A, Weber B. Synthetic lethality as an engine for cancer drug target discovery. Nat Rev Drug Discov. 2020;19:23–38.
Ogiwara H, Sasaki M, Mitachi T, Oike T, Higuchi S, Tominaga Y, et al. Targeting p300 addiction in CBP-deficient cancers causes synthetic lethality by apoptotic cell death due to abrogation of MYC expression. Cancer Discov. 2016;6:430–45.
Schick S, Grosche S, Kohl KE, Drpic D, Jaeger MG, Marella NC, et al. Acute BAF perturbation causes immediate changes in chromatin accessibility. Nat Genet. 2021;53:269–78.
Hoffman GR, Rahal R, Buxton F, Xiang K, McAllister G, Frias E, et al. Functional epigenetics approach identifies BRM/SMARCA2 as a critical synthetic lethal target in BRG1-deficient cancers. Proc Natl Acad Sci USA. 2014;111:3128–33.
Fu Z, Li S, Han S, Shi C, Zhang Y. Antibody drug conjugate: the “biological missile” for targeted cancer therapy. Signal Transduct Target Ther. 2022;7:93.
Godwin CD, Gale RP, Walter RB. Gemtuzumab ozogamicin in acute myeloid leukemia. Leukemia. 2017;31:1855–68.
Zhou CB, Zhou YL, Fang JY. Gut microbiota in cancer immune response and immunotherapy. Trends Cancer. 2021;7:647–60.
Alexander JL, Wilson ID, Teare J, Marchesi JR, Nicholson JK, Kinross JM. Gut microbiota modulation of chemotherapy efficacy and toxicity. Nat Rev Gastroenterol Hepatol. 2017;14:356–65.
Nunez JK, Chen J, Pommier GC, Cogan JZ, Replogle JM, Adriaens C, et al. Genome-wide programmable transcriptional memory by CRISPR-based epigenome editing. Cell. 2021;184:2503–19.e17.
Long Z, Sun C, Tang M, Wang Y, Ma J, Yu J, et al. Single-cell multiomics analysis reveals regulatory programs in clear cell renal cell carcinoma. Cell Discov. 2022;8:68.
Janssens DH, Meers MP, Wu SJ, Babaeva E, Meshinchi S, Sarthy JF, et al. Automated CUT&Tag profiling of chromatin heterogeneity in mixed-lineage leukemia. Nat Genet. 2021;53:1586–96.
Nguyen CDK, Colon-Emeric BA, Murakami S, Shujath MNY, Yi C. PRMT1 promotes epigenetic reprogramming associated with acquired chemoresistance in pancreatic cancer. Cell Rep. 2024;43:114176.
Longley J, Johnson PWM. Epigenetics of indolent lymphoma and how it drives novel therapeutic approaches-focus on EZH2-targeted drugs. Curr Oncol Rep. 2021;23:76.
Elfiky AMI, Ghiboub M, Li Yim AYF, Hageman IL, Verhoeff J, de Krijger M, et al. Carboxylesterase-1 assisted targeting of HDAC inhibitors to mononuclear myeloid cells in inflammatory bowel disease. J Crohns Colitis. 2022;16:668–81.
Kazansky Y, Cameron D, Mueller HS, Demarest P, Zaffaroni N, Arrighetti N, et al. Overcoming clinical resistance to EZH2 inhibition using rational epigenetic combination therapy. Cancer Discov. 2024;14:965–81.
Morel D, Almouzni G, Soria JC, Postel-Vinay S. Targeting chromatin defects in selected solid tumors based on oncogene addiction, synthetic lethality and epigenetic antagonism. Ann Oncol. 2017;28:254–69.
Lee ECY, Reichl KD, Gopalsamy A. Synthetic lethality: targeting the SMARCA2 bromodomain for degradation in SMARCA4-deficient tumors - a review of patent literature from 2019-June 2023. Expert Opin Ther Pat. 2024;34:211–29.
Fernandez M, Miranda-Saavedra D. Genome-wide enhancer prediction from epigenetic signatures using genetic algorithm-optimized support vector machines. Nucleic Acids Res. 2012;40:e77.
Beer MA. Predicting enhancer activity and variant impact using gkm-SVM. Hum Mutat. 2017;38:1251–8.
Erwin GD, Oksenberg N, Truty RM, Kostka D, Murphy KK, Ahituv N, et al. Integrating diverse datasets improves developmental enhancer prediction. PLoS Comput Biol. 2014;10:e1003677.
Rajagopal N, Xie W, Li Y, Wagner U, Wang W, Stamatoyannopoulos J, et al. RFECS: a random-forest based algorithm for enhancer identification from chromatin state. PLoS Comput Biol. 2013;9:e1002968.
Khamis AM, Motwalli O, Oliva R, Jankovic BR, Medvedeva YA, Ashoor H, et al. A novel method for improved accuracy of transcription factor binding site prediction. Nucleic Acids Res. 2018;46:e72.
Min X, Zeng W, Chen S, Chen N, Chen T, Jiang R. Predicting enhancers with deep convolutional neural networks. BMC Bioinform. 2017;18:478.
Alipanahi B, Delong A, Weirauch MT, Frey BJ. Predicting the sequence specificities of DNA- and RNA-binding proteins by deep learning. Nat Biotechnol. 2015;33:831–8.
Yang J, Ma A, Hoppe AD, Wang C, Li Y, Zhang C, et al. Prediction of regulatory motifs from human Chip-sequencing data using a deep learning framework. Nucleic Acids Res. 2019;47:7809–24.
Kelley DR, Snoek J, Rinn JL. Basset: learning the regulatory code of the accessible genome with deep convolutional neural networks. Genome Res. 2016;26:990–9.
de Almeida BP, Reiter F, Pagani M, Stark A. DeepSTARR predicts enhancer activity from DNA sequence and enables the de novo design of synthetic enhancers. Nat Genet. 2022;54:613–24.
Avsec Z, Weilert M, Shrikumar A, Krueger S, Alexandari A, Dalal K, et al. Base-resolution models of transcription-factor binding reveal soft motif syntax. Nat Genet. 2021;53:354–66.
Nguyen QH, Nguyen-Vo TH, Le NQK, Do TTT, Rahardja S, Nguyen BP. iEnhancer-ECNN: identifying enhancers and their strength using ensembles of convolutional neural networks. BMC Genomics. 2019;20:951.
Gonzalez-Avalos E, Onodera A, Samaniego-Castruita D, Rao A, Ay F. Predicting gene expression state and prioritizing putative enhancers using 5hmC signal. Genome Biol. 2024;25:142.
Chen S, Gan M, Lv H, Jiang R. DeepCAPE: a deep convolutional neural network for the accurate prediction of enhancers. Genom Proteom Bioinform. 2021;19:565–77.
Dibaeinia P, Sinha S. Deciphering enhancer sequence using thermodynamics-based models and convolutional neural networks. Nucleic Acids Res. 2021;49:10309–27.
Bu H, Hao J, Gan Y, Zhou S, Guan J. DEEPSEN: a convolutional neural network based method for super-enhancer prediction. BMC Bioinformatics. 2019;20:598.
Zhang TH, Flores M, Huang Y. ES-ARCNN: predicting enhancer strength by using data augmentation and residual convolutional neural network. Anal Biochem. 2021;618:114120.
Quang D, Xie X. DanQ: a hybrid convolutional and recurrent deep neural network for quantifying the function of DNA sequences. Nucleic Acids Res. 2016;44:e107.
Yang B, Liu F, Ren C, Ouyang Z, Xie Z, Bo X, et al. BiRen: predicting enhancers with a deep-learning-based model using the DNA sequence alone. Bioinformatics. 2017;33:1930–6.
Li J, Pu Y, Tang J, Zou Q, Guo F. DeepATT: a hybrid category attention neural network for identifying functional effects of DNA sequences. Brief Bioinform. 2021;22:bbaa159.
Basith S, Hasan MM, Lee G, Wei L, Manavalan B. Integrative machine learning framework for the identification of cell-specific enhancers from the human genome. Brief Bioinform. 2021;22:bbab252.
Chen D, Cao Y, Tang H, Zang L, Yao N, Zhu Y, et al. Comprehensive machine learning-generated classifier identifies pro-metastatic characteristics and predicts individual treatment in pancreatic cancer: a multicenter cohort study based on super-enhancer profiling. Theranostics. 2023;13:3290–309.
Sanda T, Lawton LN, Barrasa MI, Fan ZP, Kohlhammer H, Gutierrez A, et al. Core transcriptional regulatory circuit controlled by the TAL1 complex in human T cell acute lymphoblastic leukemia. Cancer Cell. 2012;22:209–21.
Ott CJ, Federation AJ, Schwartz LS, Kasar S, Klitgaard JL, Lenci R, et al. Enhancer architecture and essential core regulatory circuitry of chronic lymphocytic leukemia. Cancer Cell. 2018;34:982–95.e7.
Harada T, Heshmati Y, Kalfon J, Perez MW, Xavier Ferrucio J, Ewers J, et al. A distinct core regulatory module enforces oncogene expression in KMT2A-rearranged leukemia. Genes Dev. 2022;36:368–89.
Tsuzuki S, Yasuda T, Kojima S, Kawazu M, Akahane K, Inukai T, et al. Targeting MEF2D-fusion oncogenic transcriptional circuitries in B-cell precursor acute lymphoblastic leukemia. Blood Cancer Discov. 2020;1:82–95.
Jin Y, Chen K, De Paepe A, Hellqvist E, Krstic AD, Metang L, et al. Active enhancer and chromatin accessibility landscapes chart the regulatory network of primary multiple myeloma. Blood. 2018;131:2138–50.
Chen L, Huang M, Plummer J, Pan J, Jiang YY, Yang Q, et al. Master transcription factors form interconnected circuitry and orchestrate transcriptional networks in oesophageal adenocarcinoma. Gut. 2020;69:630–40.
Pan J, Silva TC, Gull N, Yang Q, Plummer JT, Chen S, et al. Lineage-specific epigenomic and genomic activation of oncogene HNF4A promotes gastrointestinal adenocarcinomas. Cancer Res. 2020;80:2722–36.
Cancer Genome Atlas Research N, Analysis Working Group, Asan U, Agency BCC, Brigham, Women’s H, et al. Integrated genomic characterization of oesophageal carcinoma. Nature 2017;541:169–75.
Hao JJ, Lin DC, Dinh HQ, Mayakonda A, Jiang YY, Chang C, et al. Spatial intratumoral heterogeneity and temporal clonal evolution in esophageal squamous cell carcinoma. Nat Genet. 2016;48:1500–7.
Lin DC, Wang MR, Koeffler HP. Genomic and epigenomic aberrations in esophageal squamous cell carcinoma and implications for patients. Gastroenterology. 2018;154:374–89.
Cao W, Wu W, Yan M, Tian F, Ma C, Zhang Q, et al. Multiple region whole-exome sequencing reveals dramatically evolving intratumor genomic heterogeneity in esophageal squamous cell carcinoma. Oncogenesis. 2015;4:e175.
Zhang T, Song X, Zhang Z, Mao Q, Xia W, Xu L, et al. Aberrant super-enhancer landscape reveals core transcriptional regulatory circuitry in lung adenocarcinoma. Oncogenesis. 2020;9:92.
Du K, Sun S, Jiang T, Liu T, Zuo X, Xia X, et al. E2F2 promotes lung adenocarcinoma progression through B-Myb- and FOXM1-facilitated core transcription regulatory circuitry. Int J Biol Sci. 2022;18:4151–70.
Fournier M, Bourriquen G, Lamaze FC, Cote MC, Fournier E, Joly-Beauparlant C, et al. FOXA and master transcription factors recruit mediator and cohesin to the core transcriptional regulatory circuitry of cancer cells. Sci Rep. 2016;6:34962.
Kong R, Patel AS, Sato T, Jiang F, Yoo S, Bao L, et al. Transcriptional circuitry of NKX2-1 and SOX1 defines an unrecognized lineage subtype of small-cell lung cancer. Am J Respir Crit Care Med. 2022;206:1480–94.
Riddick G, Kotliarova S, Rodriguez V, Kim HS, Linkous A, Storaska AJ, et al. A core regulatory circuit in glioblastoma stem cells links MAPK activation to a transcriptional program of neural stem cell identity. Sci Rep. 2017;7:43605.
Garancher A, Lin CY, Morabito M, Richer W, Rocques N, Larcher M, et al. NRL and CRX define photoreceptor identity and reveal subgroup-specific dependencies in medulloblastoma. Cancer Cell. 2018;33:435–49.e6.
Boulay G, Awad ME, Riggi N, Archer TC, Iyer S, Boonseng WE, et al. OTX2 activity at distal regulatory elements shapes the chromatin landscape of group 3 medulloblastoma. Cancer Discov. 2017;7:288–301.
Lin CY, Erkek S, Tong Y, Yin L, Federation AJ, Zapatka M, et al. Active medulloblastoma enhancers reveal subgroup-specific cellular origins. Nature. 2016;530:57–62.
Singh S, Abu-Zaid A, Jin H, Fang J, Wu Q, Wang T, et al. Targeting KDM4 for treating PAX3-FOXO1-driven alveolar rhabdomyosarcoma. Sci Transl Med. 2022;14:eabq2096.
Gryder BE, Pomella S, Sayers C, Wu XS, Song Y, Chiarella AM, et al. Histone hyperacetylation disrupts core gene regulatory architecture in rhabdomyosarcoma. Nat Genet. 2019;51:1714–22.
Boeva V, Louis-Brennetot C, Peltier A, Durand S, Pierre-Eugene C, Raynal V, et al. Heterogeneity of neuroblastoma cell identity defined by transcriptional circuitries. Nat Genet. 2017;49:1408–13.
Decaesteker B, Denecker G, Van Neste C, Dolman EM, Van Loocke W, Gartlgruber M, et al. TBX2 is a neuroblastoma core regulatory circuitry component enhancing MYCN/FOXM1 reactivation of DREAM targets. Nat Commun. 2018;9:4866.
Durbin AD, Zimmerman MW, Dharia NV, Abraham BJ, Iniguez AB, Weichert-Leahey N, et al. Selective gene dependencies in MYCN-amplified neuroblastoma include the core transcriptional regulatory circuitry. Nat Genet. 2018;50:1240–6.
Wang L, Tan TK, Durbin AD, Zimmerman MW, Abraham BJ, Tan SH, et al. ASCL1 is a MYCN- and LMO1-dependent member of the adrenergic neuroblastoma core regulatory circuitry. Nat Commun. 2019;10:5622.
Ran L, Chen Y, Sher J, Wong EWP, Murphy D, Zhang JQ, et al. FOXF1 defines the core-regulatory circuitry in gastrointestinal stromal tumor. Cancer Discov. 2018;8:234–51.
Hemming ML, Lawlor MA, Zeid R, Lesluyes T, Fletcher JA, Raut CP, et al. Gastrointestinal stromal tumor enhancers support a transcription factor network predictive of clinical outcome. Proc Natl Acad Sci USA. 2018;115:E5746–55.
Peng L, Jiang Y, Chen H, Wang Y, Lan Q, Chen S, et al. Transcription factor EHF interacting with coactivator AJUBA aggravates malignancy and acts as a therapeutic target for gastroesophageal adenocarcinoma. Acta Pharm Sin B. 2024;14:2119–36.
Shi X, Zheng Y, Jiang L, Zhou B, Yang W, Li L, et al. EWS-FLI1 regulates and cooperates with core regulatory circuitry in Ewing sarcoma. Nucleic Acids Res. 2020;48:11434–51.
Murakami S, White SM, McIntosh AT, Nguyen CDK, Yi C. Spontaneously evolved progenitor niches escape Yap oncogene addiction in advanced pancreatic ductal adenocarcinomas. Nat Commun. 2023;14:1443.
Zheng ZZ, Xia L, Hu GS, Liu JY, Hu YH, Chen YJ, et al. Super-enhancer-controlled positive feedback loop BRD4/ERalpha-RET-ERalpha promotes ERalpha-positive breast cancer. Nucleic Acids Res. 2022;50:10230–48.
Shi W, Zhong B, Dong J, Hu X, Li L. Super enhancer-driven core transcriptional regulatory circuitry crosstalk with cancer plasticity and patient mortality in triple-negative breast cancer. Front Genet. 2023;14:1258862.
Prutsch N, He S, Berezovskaya A, Durbin AD, Dharia NV, Maher KA, et al. STAT3 couples activated tyrosine kinase signaling to the oncogenic core transcriptional regulatory circuitry of anaplastic large cell lymphoma. Cell Rep Med. 2024;5:101472.
Chen Y, Xu L, Mayakonda A, Huang ML, Kanojia D, Tan TZ, et al. Bromodomain and extraterminal proteins foster the core transcriptional regulatory programs and confer vulnerability in liposarcoma. Nat Commun. 2019;10:1353.
Takeda T, Yokoyama Y, Takahashi H, Okuzaki D, Asai K, Itakura H, et al. A stem cell marker KLF5 regulates CCAT1 via three-dimensional genome structure in colorectal cancer cells. Br J Cancer. 2022;126:109–19.
Li QL, Lin X, Yu YL, Chen L, Hu QX, Chen M, et al. Genome-wide profiling in colorectal cancer identifies PHF19 and TBC1D16 as oncogenic super enhancers. Nat Commun. 2021;12:6407.
Bleu M, Gaulis S, Lopes R, Sprouffske K, Apfel V, Holwerda S, et al. PAX8 activates metabolic genes via enhancer elements in renal cell carcinoma. Nat Commun. 2019;10:3739.
Sin-Chan P, Mumal I, Suwal T, Ho B, Fan X, Singh I, et al. A C19MC-LIN28A-MYCN oncogenic circuit driven by hijacked super-enhancers is a distinct therapeutic vulnerability in ETMRs: a lethal brain tumor. Cancer Cell. 2019;36:51–67 e7.
Neyret-Kahn H, Fontugne J, Meng XY, Groeneveld CS, Cabel L, Ye T, et al. Epigenomic mapping identifies an enhancer repertoire that regulates cell identity in bladder cancer through distinct transcription factor networks. Oncogene. 2023;42:1524–42.
McKeown MR, Corces MR, Eaton ML, Fiore C, Lee E, Lopez JT, et al. Superenhancer analysis defines novel epigenomic subtypes of non-APL AML, including an RARalpha dependency targetable by SY-1425, a potent and selective RARalpha agonist. Cancer Discov. 2017;7:1136–53.
Ho SWT, Sheng T, Xing M, Ooi WF, Xu C, Sundar R, et al. Regulatory enhancer profiling of mesenchymal-type gastric cancer reveals subtype-specific epigenomic landscapes and targetable vulnerabilities. Gut. 2023;72:226–41.
Tanaka Y, Chiwaki F, Kojima S, Kawazu M, Komatsu M, Ueno T, et al. Multi-omic profiling of peritoneal metastases in gastric cancer identifies molecular subtypes and therapeutic vulnerabilities. Nat Cancer. 2021;2:962–77.
Lomberk G, Blum Y, Nicolle R, Nair A, Gaonkar KS, Marisa L, et al. Distinct epigenetic landscapes underlie the pathobiology of pancreatic cancer subtypes. Nat Commun. 2018;9:1978.
Yuan C, Chen H, Tu S, Huang HY, Pan Y, Gui X, et al. A systematic dissection of the epigenomic heterogeneity of lung adenocarcinoma reveals two different subclasses with distinct prognosis and core regulatory networks. Genome Biol. 2021;22:156.
Sato T, Yoo S, Kong R, Sinha A, Chandramani-Shivalingappa P, Patel A, et al. Epigenomic profiling discovers trans-lineage SOX2 partnerships driving tumor heterogeneity in lung squamous cell carcinoma. Cancer Res. 2019;79:6084–100.
Gartlgruber M, Sharma AK, Quintero A, Dreidax D, Jansky S, Park YG, et al. Super enhancers define regulatory subtypes and cell identity in neuroblastoma. Nat Cancer. 2021;2:114–28.
Stelloo S, Nevedomskaya E, Kim Y, Schuurman K, Valle-Encinas E, Lobo J, et al. Integrative epigenetic taxonomy of primary prostate cancer. Nat Commun. 2018;9:4900.
Xu L, Chen Y, Huang Y, Sandanaraj E, Yu JS, Lin RY, et al. Topography of transcriptionally active chromatin in glioblastoma. Sci Adv. 2021;7:eabd4676.
Huang H, Hu J, Maryam A, Huang Q, Zhang Y, Ramakrishnan S, et al. Defining super-enhancer landscape in triple-negative breast cancer by multiomic profiling. Nat Commun. 2021;12:2242.
Orouji E, Raman AT, Singh AK, Sorokin A, Arslan E, Ghosh AK, et al. Chromatin state dynamics confers specific therapeutic strategies in enhancer subtypes of colorectal cancer. Gut. 2022;71:938–49.
Nassar AH, Abou Alaiwi S, Baca SC, Adib E, Corona RI, Seo JH, et al. Epigenomic charting and functional annotation of risk loci in renal cell carcinoma. Nat Commun. 2023;14:346.
Iyyanki T, Zhang B, Wang Q, Hou Y, Jin Q, Xu J, et al. Subtype-associated epigenomic landscape and 3D genome structure in bladder cancer. Genome Biol. 2021;22:105.
Alvarez-Benayas J, Trasanidis N, Katsarou A, Ponnusamy K, Chaidos A, May PC, et al. Chromatin-based, in cis and in trans regulatory rewiring underpins distinct oncogenic transcriptomes in multiple myeloma. Nat Commun. 2021;12:5450.
Funding
National Natural Science Foundation of China (81972321 and 82273455to LY, 82103016 and 62133006 to GY), the CAMS Innovation Fund for Medical Sciences (CIFMS) (2021-I2M-1-002 to YZ), the National High Level Hospital Clinical Research Funding (No. 2022-PUMCH-D-001), the Non-proft Central Research Institute Fund of Chinese Academy of Medical Sciences (No. 2018PT32014), and the National Multidisciplinary Cooperative Diagnosis and Treatment Capacity Building Project for Major Diseases NSFC(81970763).
Author information
Authors and Affiliations
Contributions
JY, FZ, and XL designed the article, wrote the original manuscript, and created the figures. YF, XW, XL, RX, DJ, and YT helped revise the manuscript and provide interpretations of the relevant articles. GY, LY, and YZ contributed to the manuscript review, supervised the project, and funding acquisition. All authors have read and approved the final submission.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Yang, J., Zhou, F., Luo, X. et al. Enhancer reprogramming: critical roles in cancer and promising therapeutic strategies. Cell Death Discov. 11, 84 (2025). https://doi.org/10.1038/s41420-025-02366-3
Received:
Revised:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41420-025-02366-3
This article is cited by
-
Decoding osteosarcoma from heterogeneity to precision therapy
Discover Oncology (2025)
-
Enhancer regulation in cancer: from epigenetics to m6A RNA modification
Archives of Pharmacal Research (2025)