Abstract
Bestrophin-1 (Best1) is an anion channel genetically linked to vision-threatening retinal degenerative channelopathies. Here, we identify interactions between Best1 and both isoforms of glutamic acid decarboxylases (GAD65 and GAD67), elucidate the distinctive influences of GAD65 and GAD67 on Best1’s permeability to various anions/neurotransmitters, discover the functionality of Best1 as a γ-Aminobutyric acid (GABA) type A receptor, and solve the structure of GABA-bound Best1. GAD65 and GAD67 both promote Best1-mediated Cl− currents, but only GAD65 drastically enhances the permeability of Best1 to glutamate and GABA, for which GAD67 has no effect. GABA binds to Best1 on an extracellular site and stimulates Best1-mediated Cl− currents at the nano-molar concentration level. The physiological role of GAD65 as a cell type-specific binding partner and facilitator of Best1 is demonstrated in retinal pigment epithelial cells. Together, our results reveal critical regulators of Best1 and inform a network of membrane transport metabolons formed between bestrophin channels and glutamate metabolic enzymes.
Similar content being viewed by others
Introduction
The human BEST1 gene encodes a Ca2+-activated anion channel (bestrophin-1, Best1) predominantly expressed in retinal pigment epithelium (RPE)1, and its genetic mutations, of which over 350 have been identified, cause a spectrum of at least five retinal degenerative diseases collectively known as bestrophinopathies2,3. The patients are susceptible to progressive vision loss which may eventually lead to blindness, and no treatments are currently available2,3,4. Therefore, understanding the physiological properties and cellular regulators of Best1 is critical for the development of treatment strategy for bestrophinopathies.
A clinical feature manifested by Best1 mutation carriers is an abnormal electrooculogram (EOG) light peak (LP), which represents the depolarization of the basolateral membrane of RPE due to the activation of a Cl− conductance triggered by the increase of intracellular Ca2+ concentration ([Ca2+]i)5,6,7. We previously demonstrated that Best1 is the anion channel responsible for conducting this Ca2+-dependent Cl− currents in human RPE8,9,10,11. However, several lines of evidence strongly suggest the existence of uncharacterized RPE-specific facilitator(s) of Best1 in vivo.
Firstly, although the currents in RPE cells conducted by endogenous or exogenously supplemented Best1 are similar to each other, they are significantly bigger compared to those conducted by transiently expressed Best1 in HEK293 cells8,9,12,13. Secondly, Best1 has been shown by cell-based approaches to mediate the transport of essential metabolites/neurotransmitters which are much bulkier in size compared to Cl−, such as glutamate in RPE14, as well as GABA and glutamate in astrocytes15,16,17,18,19,20. However, purified chicken Best1 is impermeable to glutamate21, while Ca2+-bound Best1 structure shows partially open channel gates along the ion conducting pathway, whose sizes are sufficient for Cl− but far insufficient for glutamate or GABA to pass through22. Therefore, we speculate that there are native Best1 facilitator(s) that promote opening of the channel gates to enhance the permeability of Best1 to Cl− and metabolites/neurotransmitters (e.g. glutamate) in RPE.
In this work, we identify both isoforms of glutamic acid decarboxylases, GAD65 and GAD67, as interacting activators of Best1. While both isoforms stimulate Best1-mediated Cl− permeation, GAD65 also increases the permeability of Best1 to neurotransmitters glutamate and γ-Aminobutyric acid (GABA). Physiologically, we demonstrate the activator role of GAD65 for Best1 in RPE cells. Moreover, we discover that a nano-molar concentration level of GABA is sufficient for stimulating Best1 function, and solve the structure of GABA-bound open-state Best1 which illustrates an extracellular allosteric modulation site. Taken together, our results reveal multiple Best1 activators and the previously unrecognized functionality of Best1 as a type A GABA receptor.
Results
GAD65 and GAD67 interact with Best1
We recently identified glutamine synthetase (GS) as a binding partner and regulator of bestrophin-2 (Best2), a paralog of Best1 within the same protein family, and found that Best2 conducts both glutamate and glutamine, the substrate and product of GS, respectively23. This raises the possibility that Best1 may interact with glutamic acid decarboxylase (GAD), the enzyme whose substrate (glutamate) and product (GABA) have both been reported to rely on the Best1 channel for transmembrane transport15,16,17,18,19,20.
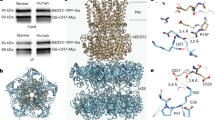
There are two isoforms of GAD enzymes in mammals, namely GAD65 and GAD67. To probe the interaction(s) between Best1 and them, Cerulean-Myc tagged human GAD65/GAD67 and Venus-His tagged human Best1 were co-transfected into HEK293 cells for co-immunoprecipitation analysis. Both GAD65 and GAD67 were co-immunoprecipitated with Best1 (Fig. 1a, Supplementary Fig. 1a), suggesting interactions between these proteins.
a Co-expressed Best1-Ven-His and GAD-Cer-Myc were detected by immunoblotting in input (top) and immunoprecipitation (IP, bottom) samples. 65, GAD65; 67, GAD67. The experiment was biologically replicated three times with similar results, and representative blots are shown. Original scans are provided in Supplementary Fig. 1a. b–d Ca2+-dependent Cl− currents conducted by Best1 (b), Best1 + GAD65 (c), and Best1 + GAD67 (d). Steady-state current density was recorded at +100 mV plotted vs. [Ca2+]i and fitted to the Hill equation; n = 5–11. I, intracellular; E, extracellular. (e, f) Representative current traces (e) and population steady-state current density-voltage (I-V) relationships (f) of Best1 alone (black) or co-expressed with GAD65 (red solid) or GAD67 (red open) at 1 μM [Ca2+]i; n = 5–11. *p < 0.05 compared to Best1 alone; Inset, voltage protocol used to elicit currents. All error bars are presented as mean values +/− SEM; p values are calculated by two-tailed unpaired Student’s t test.
GAD65 and GAD67 enhance Best1-mediated Ca2+-dependent Cl− currents
To examine the functional influences of the GAD proteins on the channel, Best1 was transfected alone or co-transfected with GAD65 or GAD67 into HEK293 cells, and the Ca2+-dependent Cl− currents were measured by whole-cell patch clamp across a range of free intracellular Ca2+ concentrations ([Ca2+]i) with Cl− as the principal anion in both external and internal patch solutions. From cells expressing Best1 alone, a plot of peak current (evoked with a + 100 mV step pulse) as a function of [Ca2+]i displayed Ca2+-dependent activation with the Ca2+ half-maximal effective concentration (EC50) at 173 µM (Fig. 1b)24. The EC50 values measured from cells co-expressing Best1 and GAD65/GAD67 are similar to that of Best1 alone (152 µM and 133 µM, respectively, Figs. 1c, d), indicating that GAD65 or GAD67 does not affect the Ca2+ sensitivity of Best1. On the other hand, the current amplitudes from cells co-expressing Best1 and GAD65 or GAD67 were significantly bigger compared to those from cells expressing Best1 alone at all tested [Ca2+]is (Figs. 1c–f), indicating a positive influence of GAD65/GAD67 on Cl− conductance of Best1. The overall and membrane-bound protein levels of Best1 were unaltered upon co-expression of GAD65 or GAD67 (Supplementary Fig. 1b), suggesting that neither GAD65 nor GAD67 promotes Best1-mediated currents by increasing the protein expression or membrane localization of the channel.
GAD65 but not GAD67 promotes Best1’s permeability to glutamate and gluconate
To test whether Best1 alone conducts glutamate, we transfected Best1 into HEK293 cells for whole-cell patch clamp. Very small currents were recorded at 1 μM [Ca2+]i (peak Ca2+) with glutamate as the principal anion in both external and internal patch solutions (Fig. 2a, black), suggesting that Best1 by itself barely conducts glutamate. To measure the relative permeability of glutamate to Cl− on Best1, currents were recorded with glutamate and Cl− as the principal anion in the external and internal solution, respectively. The reversal potential of Best1 was shifted significantly to the right (Erev = 49.7 ± 3.7 mV, Fig. 2b, black), and the relative permeability of glutamate to Cl− (PGlu/PCl) was calculated to 0.08 by the Goldman–Hodgkin–Katz equation (Fig. 2c, black).
a I-V relationships of Best1 alone (black), Best1 + GAD65 (red solid) and Best1 + GAD67 (red open), with glutamate as the principal anion in both sides of patch solutions; n = 5–7, *p < 0.05 compared to Best1 alone. b I-V relationships of Best1 alone (black), Best1 + GAD65 (red solid) and Best1 + GAD67 (red open) with external glutamate and internal Cl−; n = 6–12. (c) Relative permeability ratios of glutamate to Cl− (PGlu/PCl) in Best1 alone, Best1 + GAD65 and Best1 + GAD67; n = 6–12. *p < 0.05 compared to Best1 alone. d–f The same format as (a–c), except for gluconate replacing glutamate; n = 5-7; *p < 0.05 compared to Best1 alone. All experiments are done at 1 μM [Ca2+]i in HEK293 cells. All error bars are presented as mean values +/− SEM; p values are calculated by two-tailed unpaired Student’s t test.
To probe the influence of GAD65/GAD67 on Best1’s glutamate permeability, Best1 was co-transfected with GAD65 or GAD67 into HEK293 cells and subjected to patch clamp analysis under the same conditions. Strikingly, robust glutamate currents were recorded in cells co-expressing Best1 and GAD65 (Fig. 2a, red solid), but not in those co-expressing Best1 and GAD67 (Fig. 2a, red open). Moreover, the relative permeability of glutamate to Cl− on Best1 was drastically increased to 0.94 upon GAD65 co-expression, but remained low at 0.13 upon GAD67 co-expression (Fig. 2c, red).
To further validate these results, the same set of patch clamp experiments were performed with gluconate replacing glutamate in the patch solutions. Consistently, tiny gluconate currents were recorded in cells expressing Best1 alone or co-expressing Best1 and GAD67, in sharp contrast to the robust gluconate currents in cells co-expressing Best1 and GAD65 (Figs. 2d, e). Meanwhile, PGluc/PCl on Best1 was drastically increased from 0.07 to 0.98 by GAD65, but not altered by GAD67 (Fig. 2f).
Together, our results suggest that although both isoforms of GAD can interact with Best1 and promote its Cl− currents, only GAD65 stimulates Best1 to conduct large anions such as glutamate and gluconate.
Involvement of the neck and aperture
The gating of Best1 channel is primarily mediated by two Ca2+-dependent gates, the neck and the aperture, in the ion conducting pathway22,25,26,27. We previously found that Ca2+-binding only partially opens the neck and aperture in wild-type (WT) Best1 to a radius of ~2.0 Å, which is insufficient to accommodate large anions such as glutamate and gluconate (both with a radius of ~3.5 Å). Consistently, Best1 exhibits a very low permeability to glutamate or gluconate (Fig. 2). To investigate the contributions of the neck and aperture in restricting glutamate, we utilized two previously reported Best1 mutants, one with triple alanine substitutions at the neck (I76A/F80A/F84A, or 3A for abbreviation), and the other with a single alanine substitution at the aperture (I205A)26. The 3A and I205A mutants are specifically deficient for gating at the neck and aperture, respectively, due to replacement of constriction-forming residues with the shorter side-chained alanine, which mimics a constantly open state22,25,26. Therefore, the involvement of the aperture and neck in glutamate permeation can be separately examined.
Patch clamp was performed using HEK293 cells transiently expressing Best1-3A or Best1-I205A alone or co-expressing Best1-3A/Best1-I205A plus GAD65/GAD67, with Cl− or glutamate as the principal anion in both internal and external solutions. While both isoforms of GAD caused a similar increase of Cl− currents (Figs. 3a, e), only GAD65 significantly enhanced glutamate currents mediated by either Best1-3A or Best1-I205A (Figs. 3b, f). Then, currents were recorded with glutamate and Cl− as the principal anion in the external and internal solution, respectively. Under this bi-ionic condition, the reversal potential was measured as 50.4 ± 7.2 mV and 16.2 ± 1.8 mV for Best1-3A and Best1-I205A, respectively (Figs. 3c, g, black), corresponding to a relative permeability of glutamate to Cl− as 0.1 and 0.5, respectively (Figs. 3d, h, black). Moreover, the relative permeability of glutamate to Cl− of Best1 was not significantly affected by GAD67 but drastically increased by GAD65: from 0.1 to 1.0 for Best1-3A, and from 0.5 to 0.9 for Best1-I205A (Figs. 3d, h).
a, b I-V relationships of Best1-3A alone (black), Best1-3A + GAD65 (red solid) and Best1-3A + GAD67 (red open), with Cl− (a) or glutamate (b) as the principal anion in both sides of patch solutions; n = 5-6. *p < 0.05 compared to Best1-3A alone. c, d I-V relationships (c) and PGlu/PCl relative permeability ratios (d) with external glutamate and internal Cl− in Best1-3A alone (black), Best1-3A + GAD65 (red solid) and Best1-3A + GAD67 (red open); n = 5. *p < 0.05 compared to Best1-3A alone. e–h The same format as (a–d) for Best1-I205A; n = 5–7; *p < 0.05 compared to Best1-I205A alone. All experiments are done at 1 μM [Ca2+]i in HEK293 cells. All error bars are presented as mean values +/- SEM; p values are calculated by two-tailed unpaired Student’s t test.
Taken together, these results are consistent with our previous finding that both the neck and aperture play critical roles in channel gating and ion selectivity, and suggest that GAD65 helps open up both the neck and aperture, presumably to a fully open state, for conducting glutamate, whereas GAD67 increases the open probability of both the neck and aperture but they are still in a partially open state to restrict glutamate.
The C-terminus of Best1 is critical for GAD-mediated activation
Bestrophin paralogs are highly conserved in the transmembrane region, which occupies the first ~2/3 length of the protein, but very diverse in the C-terminal ~1/3 portion, which constitutes a long intracellular tail. We previously reported that Best11-405, which contains the first 405 residues of Best1, conducts Ca2+-dependent Cl− currents similar to those from the WT Best1 in transiently transfected HEK293 cells22. To examine the involvement of the C-terminal tail in the interactions between Best1 and GAD65/GAD67, Venus-tagged Best11-405 was co-transfected with Cerulean-tagged GAD65 or GAD67 into HEK293 cells, and subjected to patch clamp and co-immunoprecipitation. Remarkably, Cl− currents from Best11-405 were not affected by GAD65 or GAD67 (Fig. 4a), and Best11-405 was not co-immunoprecipitated with GAD65 or GAD67 (Fig. 4b, Supplementary Fig. 1a). These results strongly suggest that the C-terminal tail of Best1 is necessary for the interaction with GAD65 and GAD67, as well as the impact of GAD proteins on Best1.
a I-V relationships of Best11-405 alone (black), Best11-405 + GAD65 (red solid) and Best11-405 + GAD67 (red open), with Cl− as the principal anion in both sides of patch solutions; n = 6–15. b Co-expressed Best11-405-Ven-His and GAD-Cer-Myc were examined by immunoblotting in input (top) and immunoprecipitation (IP, bottom) samples. The experiment was biologically replicated three times with similar results, and representative blots are shown. Original scans are provided in Supplementary Fig. 1a. c, d Bar chart showing the steady-state current densities at +100 mV from cells transiently expressing the indicated Best1 C-terminal truncation constructs alone (black) and in the presence of GAD65 (c, red) or GAD67 (d, red) with Cl− as the principal anion in both sides of patch solutions; n = 5–15. e The same format as (c), but with glutamate as the principal anion in both sides of patch solutions; n = 5–6. All experiments are done at 1 μM [Ca2+]i in HEK293 cells. All error bars are presented as mean values +/− SEM.
To further delineate the GAD65/GAD67 effector region(s) on Best1, we generated C-terminal serial truncations (with the Venus-His tag) containing the first 450 (Best11-450), 490 (Best11-490), or 545 (Best11-545) residues of Best1, respectively. These truncation mutants were transfected alone or individually co-transfected with Cerulean-Myc tagged GAD65/GAD67 into HEK293 cells for whole-cell patch clamp. As expected, each of them alone conducted Ca2+-dependent Cl− currents similar to those from full-length (FL) Best1 (Best11-585) and Best11-405 (Figs. 4c, d, Supplementary Fig. 2). Upon co-expression of GAD65 or GAD67, currents from Best11-490 and Best11-545 were gradually increased compared to those from Best11-405, but still smaller than those from FL Best1, while currents from Best11-450 were unaffected just like Best11-405 (Figs. 4c, d, Supplementary Fig. 2). Moreover, none of the Best1 truncations conducted elevated glutamate currents in the presence of GAD65 (Fig. 4e). These results suggest that residues 451–585 in the C-terminus of Best1 are critical for the GAD65/GAD67-mediated stimulation.
GABA’s poor permeability and trans promotive effect on Best1
Since GABA is the product of GAD65/GAD67, we tested if Best1 is permeable to GABA by patch clamp using transiently transfected HEK293 cells with GABA as the principal passing ion in the external solution and Cl− as the only anion in the internal solution. The reversal potential of Best1 was drastically shifted to the right (Erev = 51.7 ± 6.2 mV, Fig. 5a, black), corresponding to a low relative permeability of GABA to Cl− (PGABA/PCl) of 0.09 (Fig. 5b, black). Under the same patch conditions, the PGABA/PCl remained low at 0.05 for Best1-3A but increased to 1.01 for Best1-I205A (Figs. 5c–f, black), indicating that the aperture but not the neck plays a critical role in GABA selectivity.
a I-V relationships of Best1 alone (black), Best1 + GAD65 (red solid) and Best1 + GAD67 (red open) with Cl- and GABA as the principal anion in the internal and external solution, respectively; n = 6–9. b Relative permeability ratios of GABA to Cl- (PGABA/PCl) in Best1 alone (black), Best1 + GAD65 (red solid) and Best1 + GAD67 (red open); n = 5-9. *p < 0.05 compared to Best1 alone. c, d I-V relationships (c) and PGABA/PCl (d) measured for Best1-3A in the same format as a, b, respectively; n = 5; *p < 0.05 compared to 3A alone. e, f I-V relationships (e) and PGABA/PCl (f) measured for Best1-I205A in the same format as a, b, respectively; n = 5–11. g, h Relative ion conductance ratios of external GGABA/GCl (g) and internal GCl/GCl (h) in Best1 when GABA replacing Cl- in the external solution; n = 5–11. All electrophysiology experiments are done at 1 μM [Ca2+]i in HEK293 cells. All error bars are presented as mean values +/− SEM; p values are calculated by two-tailed unpaired Student’s t test.
Notably, although Best1 is barely permeable to GABA (Figs. 5a, g), the inward currents (Cl− efflux) under the bi-ionic condition of external GABA with internal Cl− were elevated compared to those when Cl− is the only anion on both sides of the patch solutions (Figs. 1f, 5a, h). These results indicate that GABA on the external side of the Best1 channel promotes the outward movement of intracellular Cl− in trans, suggesting an involvement of GABA in channel gating despite its poor permeability. Moreover, this trans effect was impaired in the 3A mutants but retained in the I205A mutant (Fig. 5h), suggesting that GABA promotes Best1 gating through the neck.
GABA-bound Best1 structure
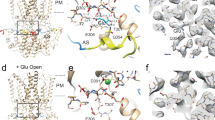
To elucidate the structural basis of GABA-mediated trans promotive effect on Best1, we incubated purified Best1 with 20 mM GABA prior to cryo-EM grid preparation and solved the GABA-bound Best1 structure at 2.4–2.5 Å resolution (Fig. 6a, Supplementary Fig. 3, Supplementary Table 1). The GABA binding site on Best1 is extracellular (Fig. 6a), which was previously identified as a Cl− binding site by X-ray anomalous diffraction studies with chicken Best128.
a Side view of Best1 in the GABA-bound fully open state with the protein depicted as transparent ribbons and GABA molecules as blue space-filling spheres. The inset indicates the location for the zoomed in region in (b). b Left, close-up of GABA binding site with hydrogen bonds depicted as dotted lines and interacting residue side chains labeled. Middle and right, the model and map (shown at sigma 5) of the GABA binding site on Best1 in two views (45-degrees rotation) with local residues labeled. c Top views of Best1 in different conformations (from the extracellular side of the membrane) with GABA molecules shown as blue spheres, I76 as brown spheres, and F80/F84 as pink spheres for the fully open state (FFFFF, left), intermediate state 1 (PCCCC, second from left), intermediate state 2 (PCPCC, third from left), and fully closed state (CCCCC, right). d, e I-V relationships of Best1 with 20 mM GABA substituting 20 mM Cl− in the standard external (d) or internal (e) solution under the 1 μM [Ca2+]i condition; n = 5–6. f Normalized steady-state current density at +100 mV (Y axis) plotted vs. different concentrations of external GABA (X axis) and fitted to the Hill equation; n = 5–6. All error bars are presented as mean values +/− SEM.
There are several points of contact between GABA and Best1 (Fig. 6b): one oxygen atom of the GABA carboxyl group forms hydrogen bonds with the side chain hydroxyl of Y68 (2.8 Å) and the backbone nitrogen atoms of V275 (3.1 Å) and F276 (3.1 Å), while the other carboxyl oxygen atom forms a hydrogen bond with the side chain hydroxyl of T277 (3.2 Å) and is in close proximity to the backbone nitrogen of T277 (3.5 Å). The alpha carbon (C2) forms a van der Waals contact with the beta carbon of P274 (3.5 Å), while the GABA beta carbon forms a van der Waals contact with the C3 carbon of the ring of the Y72 sidechain (3.4 Å); the nitrogen atom of the GABA molecule is in close proximity to the hydroxyl group of the Y72 sidechain (3.6 Å, Fig. 6b). Strikingly, 18% of particles in the Best1 + GABA data set display a fully open neck (FFFFF, Fig. 6c, Supplementary Figs. 3d, 4b and 5a), which was previously obtained only with truncated or mutant Best1. It illustrates a distinctly larger opening of the neck compared to that in the Ca2+-bound GABA-free Best1 (PPPPP, Supplementary Fig. 5b), and is very similar to the conformation seen with a truncated Best11-345 lacking the C-terminal autoinhibitory segment (AS) in our previous studies (Root Mean Square Deviation, RMSD 0.4 from 8D1O)22. In this GABA-bound open state, one GABA molecule sits within the pocket formed at the N-terminal helix dipole of helix S4a of each Best1 protomer (Figs. 6a–c). By sharp contrast, 10% of Best1 particles from the same sample are in a fully closed state (CCCCC, Fig. 6c, Supplementary Figs. 3f, 4a and 5c). Besides the fully open (FFFFF) and closed (CCCCC) states, we also identified two intermediate states: one with four closed protomers plus one partially open protomer (PCCCC, 32%), and one with three closed plus two partially open protomers (PCPCC, 31%) (Fig. 6c, Supplementary Figs. 3g, h). Notably, only four of the five Best1 protomers within each pentameric assembly are associated with GABA-like densities (thus four GABA molecules per channel) in these intermediate states (Supplementary Fig. 6), in contrast to all five protomers bound with GABA in the fully open state. Taken together, our results indicate that extracellular GABA-binding strongly promotes Best1 neck opening, and full GABA occupancy of all five protomers is required for full opening of the Best1 neck.
The aperture of GABA-bound Best1 is indistinguishable from that of GABA-free Best1 (Supplementary Fig. 4), suggesting that the binding of GABA to Best1 does not affect the structural conformation of the aperture.
Best1 behaves like a GABA receptor
The identification of an extracellular GABA-binding site on Best1 prompted us to further examine the influence of GABA on Best1 channel function by adding GABA to the standard Cl− patch solutions during patch clamp. Consistent with the structural results, both inward and outward currents from HEK293 cells expressing Best1 were significantly increased when 20 mM GABA was used to substitute Cl− in the external solution (Fig. 6d), but substituting the same amount of Cl− with GABA in the internal solution had no effect (Fig. 6e). To evaluate the dose dependency of GABA, we performed patch clamp analysis with different concentrations of GABA in the external solution. The plot of outward current density (Cl− influx) vs. extracellular GABA concentration was fitted to the Hill equation, and the EC50 concentration of extracellular GABA required for activating Best1 was measured as 371 nM (Fig. 6f).
Taken together, our results suggest that GABA binds to Best1 on the extracellular side and promotes Best1-mediated Cl− current at the level of nanomolar concentrations, but is much less permeable on Best1 compared to Cl−. As these features resemble the characteristics of GABA type A (GABAA) receptors, we conclude that Best1 is a GABA receptor.
GAD65 but not GAD67 promotes Best1’s permeability to GABA
To probe the influence of GAD65/GAD67 on Best1’s GABA permeability, Best1 was co-transfected with GAD65 or GAD67 into HEK293 cells and subjected to patch clamp analysis with GABA as the principal passing ion in the external solution and Cl− as the only anion in the internal solution. The relative permeability of GABA to Cl− remained at 0.09 in the presence of GAD67, but drastically increased to 0.97 in the presence of GAD65 (Erev = 48.7 ± 4.0 mV and Erev = 0.8 ± 0.9 mV, respectively, Figs. 5a, b). These results strongly suggest that GAD65 but not GAD67 promotes the permeability of Best1 to GABA, which is consistent with the enhancement of glutamate and gluconate permeation on Best1 by GAD65 but not by GAD67 (Fig. 2). As expected, both GAD65 and GAD67 constantly increased inward Cl− currents under this condition (Fig. 5a), recapturing the promotive effect of both GAD proteins on Best1-mediated Cl− conductance.
To investigate how GADs affect Best1 channel gates for GABA permeation, we performed the same set of experiments with the Best1-3A and Best1-I205A mutants. With Best1-3A, the relative permeability of GABA to Cl− was not significantly affected by GAD67 but dramatically increased from 0.05 to 0.98 by GAD65 (Figs. 5c, d), suggesting that GAD65 but not GAD67 fully opens the aperture to accommodate GABA. By contrast, with Best1-I205A, neither GAD65 nor GAD67 had any significant influence on the current amplitude or the relative permeability of GABA to Cl− (Figs. 5e, f). These results are consistent with the idea that extracellular GABA opens the neck while the I205A mutation mimics an open aperture, such that neither GAD65 nor GAD67 would have any additional effect on the already fully opened Best1-I205A mutant channel in the presence of extracellular GABA.
Based on our structural and electrophysiological results, we propose a model for the regulation of Best1 by GABA and GADs: the binding of GABA induces full opening of the neck but has no effect on the aperture, resulting in enhanced Cl− conductance without affecting GABA permeation owing to the ion selection/restriction of the aperture; both GAD65 and GAD67 facilitate opening of the two Best1 channel gates, resulting in elevated Cl− currents, but only GAD65 promotes full opening of the gates to accommodate large anions such as GABA and glutamate.
Best1 and GADs in RPE cells
We previously showed that Ca2+-dependent Cl− currents in RPE cells mediated by endogenously expressed or exogenously supplemented Best1 are significantly bigger compared to those from transiently expressed Best1 in HEK293 cells (Figs. 1f and 7a, b)8,9,13, strongly suggesting the existence of facilitating co-factor(s) of Best1 in RPE. As GAD65 and GAD67 are promising candidates, we measured the mRNA levels of GAD65 and GAD67 in donor-derived BEST1WT/WT RPE (WT iPSC-RPE) cells by reverse transcription-polymerase chain reaction (RT-PCR). A much higher level of GAD65 transcripts was detected compared to that of GAD67 (Supplementary Fig. 7a), suggesting a primary involvement of GAD65. Consistently, GAD65 was pulled down from iPSC-RPE cell lysate by Ni-NTA beads bound with purified Best1 (Supplementary Fig. 1c). Then, iPSC-RPE cells were transfected with small interference RNA (siRNA) specifically targeting Best1, GAD65 or GAD67 (Supplementary Figs. 1d and 7b-d), and subjected to patch clamp analysis. Ca2+-dependent Cl− currents were completely abolished upon Best1 knockdown and significantly decreased upon GAD65 knockdown but not upon GAD67 knockdown (Figs. 7b, c), suggesting an essential role of GAD65, but not GAD67, in facilitating native Best1 channel function in RPE cells.
a Representative Cl− current traces measured at 1 μM [Ca2+]i without (left) or with GAD65 siRNA treatment (right). b I-V relationships in untreated cells at no Ca2+ (black open) or 1 μM [Ca2+]i (black solid) and in cells upon Best1 siRNA treatment at 1 μM [Ca2+]i (blue), with Cl− as the principal anion in both internal and external solutions; n = 5-6. *p < 0.05 compared to untreated cells at 1 μM [Ca2+]i. c I-V relationships in untreated cells (black) and cells upon GAD65 (red solid) or GAD67 (red open) siRNA treatment at 1 μM [Ca2+]i, with Cl− as the principal anion in both internal and external solutions; n = 5-6. *p < 0.05 compared to untreated cells. d I-V relationships in untreated cells at no Ca2+ (black open) or 1 μM [Ca2+]i (black solid) and in cells upon Best1 siRNA treatment at 1 μM [Ca2+]i (blue), with glutamate as the principal anion in both internal and external solutions; n = 6. *p < 0.05 compared to untreated cells at 1 μM [Ca2+]i. e, f I-V relationships in untreated cells (black) and cells upon GAD65 (red) or Best1 (blue) siRNA treatment at 1 μM [Ca2+]i, with glutamate as the principal anion in both internal and external solutions (e) or Cl− as the only internal anion and GABA as the principal passing ion in the external solution (f); n = 5-8. *p < 0.05 compared to untreated cells. All error bars are presented as mean values +/− SEM; p values are calculated by two-tailed unpaired Student’s t test.
Previously, Best1 has been suggested to mediate glutamate release induced by a PKC-ζ pseudosubstrate in rat RPE cells via indirect measurements14, but Ca2+-dependent glutamate current has never been directly recorded in RPE cells, let alone its biophysical properties and the contribution of Best1. To fill these gaps, patch clamp was performed in iPSC-RPE cells with glutamate as the principal anion in both internal and external solutions. Robust Ca2+-dependent glutamate currents were recorded in WT iPSC-RPE cells, and were abolished upon Best1 knockdown by siRNA (Fig. 7d), providing a direct evidence that Best1 mediates Ca2+-dependent glutamate release in human RPE. Moreover, the glutamate currents in WT iPSC-RPE cells were diminished upon the treatment of GAD65-specific siRNA (Fig. 7e), consistent with the results from HEK293 cells that GAD65 plays an indispensable role in permitting glutamate permeation through the Best1 channel (Fig. 2).
To investigate whether RPE cells are capable of conducting GABA through Best1, patch clamp was performed in WT iPSC-RPE cells with GABA as the principal passing ion in the external solution and Cl− as the only anion in the internal solution. Robust currents were recorded both inwardly (Cl− efflux) and outwardly (GABA influx), and were diminished upon Best1 knockdown (Fig. 7f), demonstrating the capacity of Best1 to mediate GABA transport in human RPE. Moreover, GABA (outward) currents in iPSC-RPE cells were diminished upon the treatment of GAD65-specific siRNA, consistent with the results from HEK293 cells that GAD65 plays an indispensable role in facilitating GABA permeation through the Best1 channel (Fig. 5).
Discussion
The Best1 channel has been shown to permeate various anions, including Cl−, Br−, I−, SCN−, NO3−, HCO3− and CH3SO3− in transiently expressed HEK293 cells26,29,30,31,32. Here, we report that both isoforms of GAD proteins and GABA interact with Best1 and regulate the channel function. The presence of nano-molar level GABA on the extracellular side of Best1 causes full opening of the neck, resulting in elevated Cl− conductance, while the channel retains a low permeability to GABA owing to restriction of the aperture. On the other hand, the GAD proteins interact with the C-terminal tail of Best1 on the intracellular side. While they both exhibit a similar stimulating effect on Best1-mediated Cl− currents, only GAD65 enhances the permeability of Best1 to large metabolites/neurotransmitters, such as glutamate and GABA, presumably by promoting full opening of both the neck and aperture. Taken together, we propose a model in which GAD65 acts as a “switch button” for the functionality of Best1: in the absence of GAD65, Best1 is a GABAA receptor, which conducts Cl− when bound with the ligand extracellular GABA; in the presence of GAD65, Best1 becomes fully open and highly permeable to GABA and glutamate, which may represent the primary anions conducted by Best1, rather than Cl−, in different cell types.
As Best1 was originally identified as a Ca2+-activated Cl− channel, Ca2+ has been recognized as the primary activator of Best1. Our results in this study reveal GABA neurotransmitter and GAD proteins as Best1 activators which are required, in addition to Ca2+, for the full activation of Best1, as the peak currents conducted by Best1 are significantly bigger in the presence of GABA or GAD compared to those in their absence. On the other hand, GABA and GAD proteins have different influences on the channel gates: the GABA-bound WT Best1 structure exhibits a fully open neck identical to that previously seen in the Best11-345 C-terminus truncation22, while no conformational change is found at the aperture compared to the GABA-unbound structure, suggesting that GABA specifically promotes the neck to the fully open state; although the structures of GAD65- or GAD67-bound Best1 have not been obtained, patch clamp data strongly suggest that both channel gates would adapt to a fully open state upon GAD65 binding, but remain in a partially open state with higher open probability upon GAD67 binding.
We speculate that GABA and GAD65 are non-overlapping activators of Best1 in different cells/conditions. Our results show that Best1 and GAD65 are co-expressed in RPE cells (Fig. 7 and Supplementary Figs. 1c, 7a). Besides RPE-specific localization in the eye, Best1 is also expressed in various regions and cell types of the brain, including cortical and hippocampal astrocytes, cerebellar Bergmann glia and lamellar astrocytes, thalamic reticular neurons, meninges, and choroid plexus epithelial cells16,17,19,20,33,34,35,36,37,38,39,40,41. Notably, Best1 has been suggested to mediate glutamate and tonic GABA release from astrocytes under various physiological or pathological conditions16,17,19,20,36,37,38,41,42,43,44, suggesting the co-existence of Best1 and GAD65 in astrocytes. However, it remains elusive in what cell type(s) Best1 expresses without GAD65 and functions as a GABAA receptor. Moreover, patients with mutations in the BEST1 gene have not been reported to have deficits in the central nervous system, calling for further investigation into Best1’s role in the brain.
Interestingly, both GABA and Cl− bind to the same extracellular pocket on Best1, but only the former acts as an allosteric activator. We speculate that the binding of GABA causes a steric occlusion which prevents the open-to-closed conformational transition, resulting in an increased open probability, whereas Cl− is too small to provide this steric occlusion. Alternatively and non-exclusively, filling this binding pocket by a bulky molecule may induce a cascade of conformational change, resulting in the side chains of I76/F80/F84 turning away from the channel pore to open the gate.
RPE plays an essential role in maintaining metabolic homeostasis of the neural retina, where glutamate and GABA serve as the major excitatory and inhibitory neurotransmitters, respectively. A recent study showed that both glutamate and GABA can be synthesized from proline in RPE, and the former is exported to the neural retina45. As GAD65 directly converts glutamate to GABA and drastically enhances the permeability of Best1 to glutamate and GABA, our findings suggest a critical involvement of Best1 in the glutamate-GABA metabolism/recycling in the eye. We further speculate that the deficiency of this balance caused by Best1 mutations may contribute to the pathology of bestrophinopathies. Notably, the physiological role of Best1 in mediating glutamate/GABA transport at the basolateral side of the RPE cells still requires direct in vivo evidence14, which can be addressed using polarized RPE cells cultured on Transwell membranes.
Genetic and functional linkages between membrane transport proteins and metabolic enzymes strongly suggest the existence of an evolutionarily conserved network of “membrane transport metabolons”, which effectively couples the transmembrane transport and cellular metabolism of various physiological substances including ions, nucleotides, amino acids, and other metabolites46. There are many cases where a membrane transport protein and one or more enzyme(s) involved in the metabolism of the transported substrate are encoded by the same operon in bacteria, or functionally co-regulated and subcellularly co-localized in eukaryotes46. However, the understanding of membrane transport metabolons is still at dawn, partly due to the technical difficulty of capturing the highly dynamic protein complexes. As GAD65 interacts with Best1 and enables Best1 to conduct glutamate and GABA, the substrate and product of GAD65, respectively, while the Best2-GS complex identified in our previous studies is able to conduct glutamate and glutamine, the substrate and product of GS, respectively23, the Best1-GAD65 and Best2-GS complexes represent a group of membrane transport metabolons formed by an ion channel and a glutamate metabolism enzyme. Given the critical roles of glutamate metabolism and broad expression of bestrophins in the brain, these findings provide a long-awaited entry point to elucidate the network of membrane transport metabolons in the central nervous system.
Methods
Cell lines
HEK293 cells were purchased from ATCC (CRL-1573), authenticated by short tandem repeat (STR) DNA profiling, and cultured in DMEM (4.5 g/L glucose, Corning 10013CV) supplemented with 100 µg/mL penicillin-streptomycin and 10% fetal bovine serum. HEK293F cells were purchased from Thermo Fisher Scientific (R79007) and cultured in FreeStyle™ 293 Expression Medium supplemented with 2% fetal bovine serum. No mycoplasma contamination was found by DAPI staining.
Transfection
20–24 h before transfection, cells were split into new 3.5 cm culture dishes at 50% confluency. Plasmid and siRNA transfections were conducted using the PolyJet Transfection Reagent (SignaGen SL100688) and Lipofectamine RNAiMAX Reagent (Invitrogen 13778-030), respectively. The transfection mix was removed after 4–8 h, and cells were washed with PBS and fed with fresh media until downstream analysis or harvest.
Immunoprecipitation
Cells were harvested 48 h post transfection by centrifugation at 1000 × g for 5 min at room temperature. Cell pellets were lysed in pre-cooled lysis buffer (150 mM NaCl, 50 mM Tris, 0.5% IGEPAL® CA-630, pH 7.4) supplemented with protease inhibitor cocktails (Roche, 04693159001) for 30 min on ice, and then centrifuged at 15,000 × g for 12 min at 4 °C. The supernatant (300 μg) was collected and mixed with 2 μg His-tag antibody (Thermo Fisher Scientific, MA1-135). After rotating overnight at 4 °C, the mixture was incubated with Dynabeads M-280 sheep anti-mouse IgG (Thermo Fisher Scientific, 11202D) for 5 h at 4 °C. After thorough washing of the beads, bound proteins were eluted in 1x SDS sample buffer (Bio-Rad, 1610747) by heating for 10 min at 75 °C. Co-immunoprecipitation samples and 1/10 of input (30 μg) were then resolved by SDS-PAGE and analyzed by immunoblotting.
In vitro pull-down of interacting proteins
Best1-Ven-His was purified by affinity chromatography25,47. The protein was bound to Ni-NTA resin by end-over-end rotation for 20–30 min and washed in 5–10 column volumes of buffer containing (in mM): 50 HEPES pH 7.8, 300 NaCl, 40 Imidazole, 5 MgCl2, 5% (v/v) glycerol, 0.05% (w/v) n-dodecyl β-d-maltopyranoside (DDM) and then another 5–10 column volumes of buffer containing (in mM): 25 HEPES pH 7.8, 500 NaCl, 75 Imidazole, 5% (v/v) glycerol, 0.05% (w/v) DDM.
iPSC-RPE cells were harvested and the cell pellet was resuspended in the same resuspension buffer as Best1 and homogenized by a motor-driven tissue grinder. The resultant homogenate was lysed by sonication and incubated in 1% (w/v) sol-grade DDM for 1 h with vigorous rotation. The lysate was centrifuged at 15,000 × g for 12 min at 4 °C. The resultant supernatant and Best1-bound Ni-NTA resin were incubated with end-to-end rotation overnight at 4 °C.
On day 2, the pull-down resin was washed with the same buffers and procedures as on Day 1, and subjected to elution with one volume of elution buffer equivalent to the volume of solid Ni-NTA resin. The elution buffer contained (in mM): 25 HEPES, 200 NaCl, 5% (v/v) glycerol, 500 Imidazole, 0.05% (w/v) DDM. The eluted samples were collected for immunoblotting.
Immunoblotting
The M-PER Mammalian Protein Extraction Reagent (Thermo Fisher Scientific 78501) and Mem-PER Plus Membrane Protein Extraction Kit (Thermo Fisher Scientific 89842) were used to prepare the whole-cell lysate and membrane fraction, respectively. After denaturing at 95 °C for 5 min, samples were run on 4–15% gradient SDS-PAGE gel at room temperature, and wet transferred onto nitrocellulose membrane at 4 °C. The membranes were incubated in blocking buffer containing 5% (w/v) non-fat milk for 1 h at room temperature, and subsequently incubated overnight at 4 °C in blocking buffer supplemented with primary antibody. Primary antibodies against the following proteins were used: GFP (1:1000, Thermo Fisher Scientific, A-6455), His (1:1,000, Thermo Fisher Scientific, PA5-141016), Myc (1:1,000, Thermo Fisher Scientific, PA5-141014), GAD65 (1:1,000, Thermo Fisher Scientific, 39-8200) and β-Actin (1:1,000, Thermo Fisher Scientific MA5-15739). IRDye® 680RD Goat anti-Mouse IgG (LI-COR, 926-68070) and IRDye® 800CW Donkey anti-Rabbit IgG (LI-COR, 925-32213) were used at a concentration of 1:10,000 and an incubation time of 1 h at room temperature, followed by infrared imaging on a Bio-Rad ChemiDoc system.
Electrophysiology
Whole-cell patch clamp recording was conducted 24-96 h after splitting of RPE cells or transfection of HEK293 cells with EPC10 patch clamp amplifier (HEKA Electronics) controlled by Patchmaster v2x90.5 (HEKA)9,13. Micropipettes were pulled and fashioned from filamented 1.5 mm thin-walled glass (WPI Instruments). Series resistance was typically 1.5–2.5 MΩ, with no electronic series resistance compensation. Experiments were conducted at room temperature (23 ± 2 °C). Liquid junction potentials were measured and corrected using HEKA built-in functions. The standard zero Ca2+ pipette (internal) solution contained (mM): 146 CsCl, 2 MgCl2, 5 EGTA, 2 MgATP (added fresh), 10 HEPES, pH 7.3 adjusted with N-Methyl-D-glucamine (NMDG). Solutions with various free Ca2+ concentrations were made by mixing CaCl2 with EGTA as calculated by the MaxChelator Program, and the free Ca2+ concentration was verified using a Ca2+ ion-selective electrode. The standard extracellular solution contained (mM): 140 NaCl, 5 KCl, 2 CaCl2, 1 MgCl2, 15 glucose, 10 HEPES, pH 7.4 with NMDG. In glutamate containing solutions, 140 mM Cl− was replaced with 140 mM glutamate in the standard external solution, and 146 mM Cl− was replaced with 146 mM glutamate in the standard internal solution. In gluconate containing solutions, 140 mM Cl− was replace with 140 mM gluconate in the standard external solution, and 146 mM Cl− was replaced with 146 mM gluconate in the standard internal solution. In GABA containing solutions, 140 mM, 20 mM, 100 µM, 10 µM, 1 µM, 100 nM, 10 nM or 1 nM Cl− was replaced with the same concentration of GABA in the standard external solution, and 20 mM Cl− was replaced with 20 mM GABA in the standard internal solution. Solution osmolarity was adjusted to 290 ~ 310 mOsm/L with glucose, and ~5 mOsm lower in the internal solutions than the external solutions of the same experiment. The low and high Ca2+ solutions in the same set of experiments were adjusted to the exact same osmolarity. Solution changes were performed manually.
Electrophysiological data collection and analyzes
Traces were acquired at a repetition interval of 4 s28. Currents were sampled at 25 kHz and filtered at 5 or 10 kHz. I-V curves were generated from a group of step potentials (-100 to +100 mV from a holding potential of 0 mV). Data were processed offline in Patchmaster. Statistical analyzes were performed using built-in functions in OriginPro 8.5. Relative permeability was calculated according to the Goldman-Hodgkin-Katz equation. The relative GABA/Cl− (with GABA or Cl− in the external solution) inward movement (outward current) conductance (GGABA with ex-GABA/GCl with ex-Cl) was measured as slope conductance at the reversal potential plus 50 mV. The trans effect, representing the relative Cl− (in the internal solution) outward movement (inward current) conductance (GCl with ex-GABA/GCl with ex-Cl) was measured as slope conductance at the reversal potential minus 50 mV. The “n” value in patch clamp recording figure legends indicates the total number of individual cells.
Molecular cloning
All constructs were made by site-directed mutagenesis PCR with the In-fusion Cloning Kit (Takara, 638948) and verified by sequencing.
Cryo-EM sample preparation
Human Best1 protein was purified using glyco-diosgenin (GDN) as the detergent22. After nickel affinity and size exclusion chromatography, the protein was concentrated to 5 mg/mL and incubated with 20 mM GABA for 1 h prior to grid production.
2.8 µL of sample mix was applied to a plasma treated UltrAuFoil R0.6/1 on a vitrobot Mark IV, incubated for 30 s at 100% humidity and 10 °C, blotted for 5–7 s at force 4 and immediately plunged into liquid ethane cooled by liquid nitrogen. Grids were screened on a Glacios prior to data collection on a Krios.
Data collection and image processing
All data were collected at the Columbia Cryo-EM core on Krios1 with Leginon 3.5. For the Best1 + 20 mM GABA dataset, 713 micrographs were collected with a K3 direct electron detector in counting mode at a magnification of 105,000x, corresponding to a physical pixel size of 0.83 Å2 /pix at a total dose of 58 e−/A2, fractionated over 50 frames, corresponding to a dose rate of 1.16 e−/A2/frame with a defocus range of -0.8 ~ 1.5 microns.
Movies were aligned with MotionCorr2 via the Relion3.1 GUI and imported to cryoSPARCv4 for further processing by PatchCTF correction, template picking, extraction, and initial 2D classification of 245,860 picked particles. 109,930 particles were selected for homogeneous refinement with C5 symmetry (2.36 Å) and were polished in Relion3.1. Polished particles underwent homogeneous refinement with C5 symmetry (2.14 Å), followed by symmetry expansion to generate 549,650 particles, which underwent 3D classification into 6 classes with 3.2 Å target resolution using a mask encompassing the neck gate and the GABA binding site. One class (class0) was in the fully open state and the 5 other classes were closed. The open state particles had duplicates removed (30,219 particles left), followed by symmetry expansion (151,395 particles), and these particles were then local refined with C1 symmetry using a global mask, resulting in a final map at 2.41 Å.
The closed state particles had duplicates removed, were re-expanded, and underwent another round of 3D classification using the same mask into 8 classes (3.2 Å target resolution) to separate intermediate closed states. 3 classes were in the intermediate 1 conformation (PCCCC, 32% of particles), 3 classes were in the intermediate 2 conformation (PCPCC, 31%), and one class was in the closed conformation (CCCCC, 10%), while the final class exhibited poor features due to excess heterogeneity and was discarded as junk. For each state detected, a single class was chosen for local refinement in C1 symmetry with a global mask. For intermediate 1 (PCCCC), 59,401 particles were refined to 2.42 Å, for intermediate 2 (PCPCC), one class was refined to 2.45 Å, and the single fully closed class comprising 52,529 particles was refined to 2.50 Å.
Model refinement and validation
Maps used for model building and refinement were obtained by sharpening to a b-factor determined by Guinier plot as implemented in cryoSPARCv4. The PDB 8D1I was rigid body fit into the cryo-EM map and subjected to multiple iterations of refinement in Coot 0.9.8.1, Phenix 1.19.2 real space refinement, and REFMAC5 (Servalcat). Validation was performed with comprehensive cryo-EM validation tools in Phenix 1.19.2, including MolProbity. All figures depicting the model and/or map were made with Chimera v1.16.
siRNAs
Predesigned gene-specific and negative control siRNAs were purchased from MilliporeSigma: Best1 (SASI_Hs01_00055652), GAD65 (SASI_Hs01_00094409), GAD67 (SASI_Hs01_00039951) and scramble (SIC001).
qRT-PCR
Total RNA was extracted from cell pellets with the PureLink RNA Mini Kit (Thermo Fisher Scientific, 12183020) and subjected to cDNA synthesis using the RevertAid First Strand cDNA synthesis kit (Thermo Fisher Scientific, K1621). The resultant cDNA was used as the template for qPCR amplification with gene-specific primer sets: Best1, CTGCTGCTGTGCTGGC and GTTCTTCCGTGAGGGCCAG; GAD65, GGGAATTGGCAGACCAACCAC and CCAGTCTGCTGCTAATCCAACCA; GAD67, GGATGCACCAGAAAACTGGGG and GCAGGTTCTTGGAGGATTGCC; β-Actin, CACCATTGGCAATGAGCGGTTC and AGGTCTTTGCGGATGTCCACGT.
Statistics and reproducibility
A sufficient number of samples were examined to reach statistical conclusion according to the specific method utilized in that experiment. Statistically significant differences (p < 0.05) between means of two groups were determined by two-tailed unpaired Student’s t test. Data are presented as means values +/– SEM. Immunoblotting and pull down experiments were biologically replicated three times with similar results.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The cryo-EM density maps have been deposited in the Electron Microscopy Data Bank (EMDB) under accession codes EMD-45915 (BEST1 + GABA open), EMD-45916 (BEST1 + GABA intermediate state 1), EMD-45917 (BEST1 + GABA intermediate state 2), EMD-45918 (BEST1 + GABA closed). The atomic coordinates have been deposited in the Protein Data Bank (PDB) under accession codes 9CTQ (BEST1 + GABA open), 9CTR (BEST1 + GABA intermediate state 1), 9CTS (BEST1 + GABA intermediate state 2), 9CTT (BEST1 + GABA closed). Previously published models 8D1O and 8D1I are used. The source data underlying Figs. 1–7 and Supplementary Fig. 2 are provided in a Source Data file. Source data are provided in this paper.
References
Owji, A. P., Kittredge, A., Zhang, Y. & Yang, T. Structure and Function of the Bestrophin family of calcium-activated chloride channels. Channels (Austin) 15, 604–623 (2021).
Johnson, A. A. et al. Bestrophin 1 and retinal disease. Progress in retinal and eye research, https://doi.org/10.1016/j.preteyeres.2017.01.006 (2017).
Petrukhin, K. et al. Identification of the gene responsible for Best macular dystrophy. Nat. Genet 19, 241–247 (1998).
Yang, T., Justus, S., Li, Y. & Tsang, S. H. BEST1: the Best Target for Gene and Cell Therapies. Mol. Ther.: J. Am. Soc. Gene Ther. 23, 1805–1809 (2015).
Fujii, S., Gallemore, R. P., Hughes, B. A. & Steinberg, R. H. Direct evidence for a basolateral membrane Cl- conductance in toad retinal pigment epithelium. Am. J. Physiol. 262, C374–C383 (1992).
Gallemore, R. P. & Steinberg, R. H. Light-evoked modulation of basolateral membrane Cl- conductance in chick retinal pigment epithelium: the light peak and fast oscillation. J. Neurophysiol. 70, 1669–1680 (1993).
Gallemore, R. P. & Steinberg, R. H. Effects of DIDS on the chick retinal pigment epithelium. II. Mechanism of the light peak and other responses originating at the basal membrane. J. Neurosci.: Off. J. Soc. Neurosci. 9, 1977–1984 (1989).
Li, Y. et al. Patient-specific mutations impair BESTROPHIN1’s essential role in mediating Ca2+-dependent Cl- currents in human RPE. eLife 6, https://doi.org/10.7554/eLife.29914 (2017).
Zhao, Q. et al. Distinct expression requirements and rescue strategies for BEST1 loss- and gain-of-function mutations. eLife 10, https://doi.org/10.7554/eLife.67622 (2021).
Kittredge, A., Ji, C., Zhang, Y. & Yang, T. Differentiation, Maintenance, and Analysis of Human Retinal Pigment Epithelium Cells: A Disease-in-a-dish Model for BEST1 Mutations. J. Vis Exp, https://doi.org/10.3791/57791 (2018).
Kittredge, A., Zhang, Y. & Yang, T. Evaluating BEST1 mutations in pluripotent stem cell-derived retinal pigment epithelial cells. Methods Enzymol. 654, 365–382 (2021).
Zhang, Y. et al. ATP activates bestrophin ion channels through direct interaction. Nat. Commun. 9, 3126 (2018).
Ji, C. et al. Investigation and restoration of BEST1 activity in patient-derived RPEs with dominant mutations. Sci. Rep. 9, 19026 (2019).
Lee-Rivera, I., Lopez, E., Alvarez-Arce, A. & Lopez-Colome, A. M. The PKC-zeta pseudosubstrate peptide induces glutamate release from retinal pigment epithelium cells through kinase- independent activation of Best1. Life Sci. 265, 118860 (2021).
Han, K. S. et al. Channel-mediated astrocytic glutamate release via Bestrophin-1 targets synaptic NMDARs. Mol. brain 6, 4 (2013).
Jo, S. et al. GABA from reactive astrocytes impairs memory in mouse models of Alzheimer’s disease. Nat. Med. 20, 886–896 (2014).
Lee, S. et al. Channel-mediated tonic GABA release from glia. Science 330, 790–796 (2010).
Oh, S. J. et al. Protease activated receptor 1-induced glutamate release in cultured astrocytes is mediated by Bestrophin-1 channel but not by vesicular exocytosis. Mol. brain 5, 38 (2012).
Park, H. et al. High glutamate permeability and distal localization of Best1 channel in CA1 hippocampal astrocyte. Mol. brain 6, 54 (2013).
Woo, D. H. et al. TREK-1 and Best1 channels mediate fast and slow glutamate release in astrocytes upon GPCR activation. Cell 151, 25–40 (2012).
Vaisey, G., Miller, A. N. & Long, S. B. Distinct regions that control ion selectivity and calcium-dependent activation in the bestrophin ion channel. Proc. Natl Acad. Sci. USA 113, E7399–E7408 (2016).
Owji, A. P. et al. Structures and gating mechanisms of human bestrophin anion channels. Nat. Commun. 13, 3836 (2022).
Owji, A. P. et al. Bestrophin-2 and glutamine synthetase form a complex for glutamate release. Nature 611, 180–187 (2022).
Qu, Z., Fischmeister, R. & Hartzell, C. Mouse bestrophin-2 is a bona fide Cl(-) channel: identification of a residue important in anion binding and conduction. J. Gen. Physiol. 123, 327–340 (2004).
Owji, A. P. et al. Structural and functional characterization of the bestrophin-2 anion channel. Nat. Struct. Mol. Biol. 27, 382–391 (2020).
Ji, C. et al. Dual Ca(2+)-dependent gates in human Bestrophin1 underlie disease-causing mechanisms of gain-of-function mutations. Commun. Biol. 2, 240 (2019).
Yang, T. et al. Structure and selectivity in bestrophin ion channels. Science 346, 355–359 (2014).
Kane Dickson, V., Pedi, L. & Long, S. B. Structure and insights into the function of a Ca(2+)-activated Cl(-) channel. Nature 516, 213–218 (2014).
Qu, Z. & Hartzell, H. C. Bestrophin Cl- channels are highly permeable to HCO3. Am. J. Physiol. Cell Physiol. 294, C1371–C1377 (2008).
Sun, H., Tsunenari, T., Yau, K. W. & Nathans, J. The vitelliform macular dystrophy protein defines a new family of chloride channels. Proc. Natl Acad. Sci. USA 99, 4008–4013 (2002).
Xiao, Q., Prussia, A., Yu, K., Cui, Y. Y. & Hartzell, H. C. Regulation of bestrophin Cl channels by calcium: role of the C terminus. J. Gen. Physiol. 132, 681–692 (2008).
Yu, K., Cui, Y. & Hartzell, H. C. The bestrophin mutation A243V, linked to adult-onset vitelliform macular dystrophy, impairs its chloride channel function. Investigative Ophthalmol. Vis. Sci. 47, 4956–4961, (2006).
Al-Jumaily, M. et al. Expression of three distinct families of calcium-activated chloride channel genes in the mouse dorsal root ganglion. Neurosci. Bull. 23, 293–299 (2007).
Barro Soria, R., Spitzner, M., Schreiber, R. & Kunzelmann, K. Bestrophin-1 enables Ca2+-activated Cl- conductance in epithelia. J. Biol. Chem. 284, 29405–29412 (2009).
Jung, J. Y., Lee, S. E., Hwang, E. M. & Lee, C. J. Neuronal expression and cell-type-specific gene-silencing of best1 in thalamic reticular nucleus neurons using pSico-red system. Exp. Neurobiol. 25, 120–129 (2016).
Kwak, H. et al. Astrocytes control sensory acuity via tonic inhibition in the thalamus. Neuron 108, 691–706.e610 (2020).
Park, H. et al. Channel-mediated astrocytic glutamate modulates hippocampal synaptic plasticity by activating postsynaptic NMDA receptors. Mol. brain 8, 7 (2015).
Park, H. et al. Bestrophin-1 encodes for the Ca2+-activated anion channel in hippocampal astrocytes. J. Neurosci.: Off. J. Soc. Neurosci. 29, 13063–13073 (2009).
Pineda-Farias, J. B. et al. Role of anoctamin-1 and bestrophin-1 in spinal nerve ligation-induced neuropathic pain in rats. Mol. pain. 11, 41 (2015).
Tochitani, S. & Kondo, S. Immunoreactivity for GABA, GAD65, GAD67 and Bestrophin-1 in the meninges and the choroid plexus: implications for non-neuronal sources for GABA in the developing mouse brain. PloS one 8, e56901 (2013).
Woo, J. et al. Control of motor coordination by astrocytic tonic GABA release through modulation of excitation/inhibition balance in cerebellum. Proc. Natl Acad. Sci. USA 115, 5004–5009 (2018).
Oh, S. J. & Lee, C. J. Distribution and function of the bestrophin-1 (Best1) channel in the brain. Exp. Neurobiol. 26, 113–121 (2017).
Pandit, S. et al. Bestrophin1-mediated tonic GABA release from reactive astrocytes prevents the development of seizure-prone network in kainate-injected hippocampi. Glia 68, 1065–1080 (2020).
Yoon, B. E. et al. The amount of astrocytic GABA positively correlates with the degree of tonic inhibition in hippocampal CA1 and cerebellum. Mol. brain 4, 42 (2011).
Zhu, S. et al. Proline provides a nitrogen source in the retinal pigment epithelium to synthesize and export amino acids for the neural retina. J. Biol. Chem. 299, 105275 (2023).
Moraes, T. F. & Reithmeier, R. A. Membrane transport metabolons. Biochimica et. biophysica acta 1818, 2687–2706 (2012).
Kittredge, A., Ward, N., Hopiavuori, A., Zhang, Y. & Yang, T. Expression and purification of mammalian bestrophin ion channels. J Vis Exp. https://doi.org/10.3791/57832 (2018).
Acknowledgements
We thank Renato Bruni at Center on Membrane Protein Production and Analysis (COMPPÅ) for providing the bacterial expression vector, and the Unrestricted Grant from Research to Prevent Blindness (RPB) to the Department of Ophthalmology at Columbia University. Cryo-EM data were collected within the Columbia Cryo-EM Core. T.Y. was supported by NIH grants R35GM149252, R01GM127652 and R24EY028758, the Irma T. Hirschl/Monique Weill-Caulier Research Award (CU20-4313) and RPB Career Advancement Award (CU22-1892).
Author information
Authors and Affiliations
Contributions
J.W. designed research, performed patch clamp recordings, analyzed data and made figures; A.P.O. designed research, performed protein purification and cryo-EM experiments, analyzed data, made figures and helped write the paper; A.K. designed research, generated constructs, purified proteins, performed immunoblotting and made figures; Z.C. maintained HEK293 and RPE culture; Y.Z. and T.Y. designed research, analyzed data, made figures and wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
Provisional patents on using GABA analogs (no. 63/591,019) and GAD proteins (63/692,462) as Best1 activators have been filled by Columbia University, listing A.P.O., Y.Z., and T.Y. as inventors for the former and Y.Z. and T.Y. as inventors for the latter. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Tomohiro Nishizawa and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Wang, J., Owji, A.P., Kittredge, A. et al. GAD65 tunes the functions of Best1 as a GABA receptor and a neurotransmitter conducting channel. Nat Commun 15, 8051 (2024). https://doi.org/10.1038/s41467-024-52039-5
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-024-52039-5