Abstract
Trinucleotide repeats, including Friedreich’s ataxia (GAA)n repeats, become pathogenic upon expansions during DNA replication and repair. Here, we show that deficiency of the essential replisome component Mcm10 dramatically elevates (GAA)n repeat instability in a budding yeast model by loss of proper CMG helicase interaction. Supporting this conclusion, live-cell microscopy experiments reveal increased replication fork stalling at the repeat in mcm10-1 cells. Unexpectedly, the viability of strains containing a single (GAA)100 repeat at an essential chromosomal location strongly depends on Mcm10 function and cellular RPA levels. This coincides with Rad9 checkpoint activation, which promotes cell viability, but initiates repeat expansions via DNA synthesis by polymerase δ. When repair is inefficient, such as in the case of RPA depletion, breakage of under-replicated repetitive DNA can occur during G2/M, leading to loss of essential genes and cell death. We hypothesize that the CMG-Mcm10 interaction promotes replication through hard-to-replicate regions, assuring genome stability and cell survival.
Similar content being viewed by others
Introduction
Expandable DNA repeats are at the heart of over 50 diseases, spanning from neurodegenerative disorders to cancer1,2. Expansions of (GAA)n repeats are known to cause two diseases: Friedreich’s ataxia (FRDA) upon biallelic repeat expansions in the 1st intron of the Frataxin (FXN) gene3 and spinocerebellar ataxia 27B (SCA27B) caused by repeat expansions in the 1st intron of the Fibroblast Growth Factor 14 (FGF14) gene4,5. (GAA)n repeats are a subgroup of homopurine-homopyrimidine mirror repeats that can fold into an alternative DNA secondary structure called H-DNA – an intramolecular DNA triplex, which was shown to hinder both DNA replication and transcription6,7,8,9,10.
Studies in S. cerevisiae and human cells collectively showed that (i) expanded (GAA)n repeats stall replication fork progression11,12,13,14,15, (ii) mutations in replication-associated genes including replicative DNA polymerases promote (GAA)n repeat instability16,17,18,19, and (iii) processes that deal with stalled replication fork repair and restart, such as template switching and restoration of reversed replication forks, modulate (GAA)n repeat stability and trinucleotide repeat stability generally15,16,20,21. Nevertheless, much about the interaction between (GAA)n repeats, their structure, and the replication fork remains to be elucidated.
Natural replication impediments, including alternative DNA secondary structures, can cause physical uncoupling of leading strand synthesis progression from CMG unwinding and lagging strand synthesis22,23,24,25,26,27,28. As a result, single-stranded DNA (ssDNA) is exposed and coated by the ssDNA binding protein RPA, triggering the activation of the intra S-phase checkpoint, which ultimately leads to fork restoration and safeguards genome integrity23,24,29,30,31,32. Therefore, physical and functional coordination of the replication fork could be central to repeat length maintenance. In addition, factors that might be dispensable for replication elongation during unperturbed replication might become more important when replicating through repetitive sequences such as long (GAA)n repeats.
The integrity of the replication fork during elongation and coordination between leading and lagging strand synthesis are promoted by accessory replication fork proteins—including Ctf4AND-1 and Mcm10—which have roles in both replication initiation and elongation33,34,35,36,37,38,39. Both Ctf4 and Mcm10 interact with the CMG helicase as well as with Pol α-primase37,40,41,42,43,44,45,46,47,48,49, and Mcm10 also contacts the PCNA clamp50. Ctf4 and Mcm10 interact with each other both in mammalian cells and in yeast, and Mcm10 stabilizes Ctf4 on chromatin43,51. While Ctf4 is non-essential in yeast and only becomes crucial for replication in the context of low Pol α levels38, Mcm10 is essential for replication in all organisms in which it is present. It is important to note that direct interactions between the CMG helicase and Pol α-primase have recently been shown to recruit the complex to the lagging-strand template and promote priming without the need for Ctf4 or Mcm1052,53.
Mcm10 comprises an N-terminal coiled-coil domain important for oligomerization, an internal domain that includes an OB-fold with a PIP box and an Hsp10-like domain. In metazoans, an additional C-terminal domain mediates further interactions with DNA and proteins (reviewed in ref. 36). During replication initiation, Mcm10 contributes to the activation of the assembled CMG helicase and origin unwinding by facilitating the bypass of the two CMG hexamers54,55,56 (Fig. 1a, left panel). The ssDNA/dsDNA gate function of Mcm10 has also been implicated in promoting bypass of lagging strand blocks in a manner mediated by its interactions with MCM57,58,59 (Fig. 1a, middle panel), but whether Mcm10 has a more prominent role in elongation as part of the replisome through its interactions with Pol α (Fig. 1a, right panel) and under which conditions remains to be determined. Deficiencies in Mcm10 lead to impaired replication initiation, slower replication, increased ssDNA exposure, DNA damage and checkpoint activation42,60,61,62,63. Furthermore, MCM10 haploinsufficiency leads to telomere erosion and micronuclei formation in iPSC cells, indicating that it has an important role in preventing genome instability64.
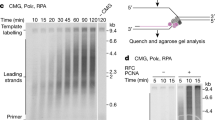
a Overview of Mcm10 functions during DNA replication. Created in BioRender. Masnovo, C. (2024) https://BioRender.com/g31r060. b Genetic assay system to measure repeat expansion rates. Created in BioRender. Masnovo, C. (2024) https://BioRender.com/q07v344. c Genetic assay system to measure repeat contraction rates. Created in BioRender. Masnovo, C. (2024) https://BioRender.com/q07v344. d Expansion rates in the mcm10-1 mutant at the permissive (23 °C) and semi-permissive (27 °C) temperatures. Plotted values indicate the corrected rate calculated with FluCalc (https://flucalc.ase.tufts.edu/)66 and the error bars represent 95% confidence intervals (see Source Data File). Numbers within bars indicate fold increase over the respective wild-type value. Expansion rates were determined by PCR of at least 96 FOAR colonies derived from two biological replicates. An event was considered an expansion when at least 10 repeats were added. e Contraction rates for the mcm10-1 mutant at the permissive (23 °C) and semi-permissive (27 °C) temperatures. Plotted values indicate the corrected rate calculated with FluCalc (https://flucalc.ase.tufts.edu/)66 and the error bars represent 95% confidence intervals (see Source Data File). Each experiment was conducted using two biological replicates.
In this study, we focused on the role of the replication factor Mcm10 in the stability of expanded (GAA)n repeats in a yeast model system. We found that the Mcm10 protein strongly counteracts both repeat expansions and contractions. Importantly, the viability of yeast strains containing unique expanded (GAA)n repeats at an essential portion of a chromosome arm is substantially decreased in Mcm10 deficient strains. Cell survival in this case is ensured by the Rad9 checkpoint activity, which facilitates DNA repair synthesis by DNA polymerase δ while simultaneously promoting expansions.
Results
Mcm10 deficiency elevates (GAA)n repeat instability due to impaired interactions with the CMG helicase
To study the role of Mcm10 on (GAA)n repeat instability we used an experimental system previously established in the lab16. In this system, a (GAA)100 repeat is located within the intron of an artificially split URA3 gene on chromosome III adjacent to the ARS306 origin. The repeat is flanked by non-repetitive sequences, for a total intron length of 974 bp. In S. cerevisiae, only introns shorter than ~1 kb can be spliced efficiently65. Thus, repeat expansions that bring the total intron length over this threshold result in the inactivation of the URA3 gene, making the yeast cells resistant to 5-fluoroorotic acid (5-FOA). Other events, such as mutations and various recombinational events can also result in URA3 loss17,66 (Fig. 1b). Therefore, repeat expansions were confirmed by PCR using repeat-flanking primers (Source Data file). In the contraction assay, a longer (GAA)124 tract is inserted in the intron, bringing the total intron length to 1108 bp. Contractions of more than 20 repeat units reactivate the URA3 gene making the yeast cells URA+ (Fig. 1c). In both the expansion and contraction cassettes, the (GAA)n repeats serve as the lagging strand template.
To determine how Mcm10 affects repeat instability in these systems, we introduced a previously characterized mcm10-1 mutation. This P269L substitution lies in the structurally and functionally conserved Hsp10-like domain—a part of the larger internal domain of Mcm10 responsible for its interactions with DNA, Pol α and PCNA67. This mutation results in a temperature-sensitive (ts) phenotype, owing to both Mcm10 and Pol α degradation at restrictive temperatures40. We found that the mcm10-1 mutation led to a 33-fold increase in expansion rate at the semi-permissive temperature (27 °C), and a 6-fold increase even at the permissive temperature (23 °C) (Fig. 1d). We also observed a 10-fold increase in repeat contractions at the semi-permissive temperature (Fig. 1e). We conclude that Mcm10 is an important replication factor in preventing the instability of long (GAA)n repeats, especially their expansion.
Mcm10 has been shown to interact with the CMG helicase, thereby promoting both replication initiation and elongation48,54,57,58. It specifically interacts with a conserved motif in Mcm2, as well as with other MCM subunits of the CMG helicase58,68. To study the role of Mcm10-CMG interactions in repeat instability, we looked at the effects of a previously identified dominant suppressor mutation in the Mcm2 subunit of the CMG helicase (mcm2G400D), which was shown to rescue the temperature-sensitive phenotype of the mcm10-1 mutant, minimizes ssDNA exposure and restores Pol α stability, particularly rescuing the elongation defects observed in the mcm10-1 mutant60. This mutation is located in the allosteric control loop of the Mcm2 subunit, which is important to couple activities between CMG subunits69, thus, on its own, it results in a decrease in the unwinding activity of the helicase60. We found that the mcm2G400D mutation alone did not affect repeat instability (Fig. 2a, b), indicating that a decrease in the helicase unwinding rate alone is insufficient to trigger repeat instability. At the same time, we observed a near complete rescue of the elevated expansion and contraction rates in the mcm10-1 mcm2G400D double mutant compared to the mcm10-1 mutant alone (Fig. 2a, b and Supplementary Fig. 1a, b). Together, these data suggest that Mcm10 prevents repeat instability through its interaction with the CMG helicase during replication through the repeat.
a Expansion rates in the mcm10-1 and mcm2G400D mutants at the semi-permissive temperature (27 °C). RPA OE indicates RPA overexpression via 2μ multicopy plasmid containing all three RPA genes: RFA1, RFA2 and RFA3. Plotted values indicate the corrected rate calculated with FluCalc (https://flucalc.ase.tufts.edu/)66 and the error bars represent 95% confidence intervals (see Source Data File). Each experiment was conducted with two biological replicates. Numbers within bars indicate fold increase over the respective wild-type value. b Contraction rates in the mcm10-1 and mcm2G400D mutants at the semi-permissive temperature (27 °C). Plotted values indicate the corrected rate calculated with FluCalc (https://flucalc.ase.tufts.edu/)66 and the error bars represent 95% confidence intervals (see Source Data File). Each experiment was conducted with two biological replicates. Numbers within bars indicate fold increase over the respective wild-type value. c Western blot of Pol1 (Cdc17) tagged with a 3xFLAG tag in WT and mcm10-1 strains at the semi-permissive (30 °C) and at the restrictive (37 °C) temperatures. The experiment conducted twice using independent biological replicates (see Source Data file). Ponceau S staining was used as a control for total protein loading.
(GAA)n repeat instability in mcm10-1 is not caused by lower levels of DNA polymerase α-primase
Mcm10 function has been proposed to be important for the stability of the Pol α-primase complex both in yeast and human cells41,42. We investigated whether the phenotypes of the mcm10-1 mutants resulted from Pol α-primase complex degradation. The Pol1 subunit of Pol α-primase was tagged with a 3x Flag-tag and protein levels were measured by western blotting. We indeed observed lower levels of Pol1 in mcm10-1 mutants compared to wild-type at the restrictive temperature (37 °C), but not at the semi-permissive temperature of 30 °C, which is higher than the temperature we conducted the instability assays at (Fig. 2c). In addition, Pol α-deficient cells were previously shown to have larger expansions, likely resulting from longer Okazaki fragments17. We do not observe a meaningful difference in the median number of repeats added in the case of the mcm10-1 mutant (60 repeat units) when compared to the wild-type (62 repeat units) (Supplementary Fig. 1c), albeit two sample Kolmogorov–Smirnov test shows a small but significant difference in the shape of repeat distribution compared to WT. Altogether, these results indicate that lower Pol α levels are not responsible for increased instability in the mcm10-1 context.
Mcm10 facilitates replication elongation through (GAA)n repeats
Mcm10 is both an initiation and an elongation factor, and multiple studies showed that it is needed for efficient and complete DNA replication70. Since expanded (GAA)n repeats were previously shown to cause replication fork stalling11,13,15, and our current data suggest that Mcm10 is needed for stable repeat maintenance, we decided to investigate replication fork progression through the (GAA)100 repeats in the mcm10-1 context.
To this end, we adopted a method for live-cell imaging of replication fork progression (Fig. 3a)71. In this system, a non-repetitive 128xlacO and 128xtetO arrays are placed ~3 kb and ~37 kb downstream from the ARS413 replication origin, respectively. These arrays are bound by their cognate LacI-Envy and TetR-tdTomato, resulting in a green and red focus, respectively. Replication of each array coincides with an increase in intensity of the respective focus. The time difference between replication of the lacO and tetO arrays is used to monitor fork progression through the repeat sequences in live cells. Inserting the repeat in the sequence between the two arrays allowed us to monitor fork progression through the repeats and determine the effect of mcm10-1 with (GAA)100 serving as the lagging-strand template (Supplementary Fig. 2).
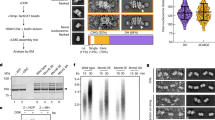
a Schematic of the live-microscopy assay to measure replication fork progression showing the orientation of the repeat when the whole expansion cassette is inserted between the lacO and tetO arrays, including a schematic of how the measurements of signal intensity allow us to determine the replication time (Δt) and a representative image from the microscopy measurements (scale bar represents 2 μm). Created in BioRender. Masnovo, C. (2024) https://BioRender.com/f52a844. b Time required to replicate a region of 28.4 kb, in WT and mcm10-1 strains containing the expansion cassette with either no repeat or the (GAA)91 repeat at 30 °C. The plotted numbers represent the median value and bars represent the interquartile range. WT no rpt n = 336, WT (GAA)100 n = 312, mcm10-1 no rpt n = 716, mcm10-1 (GAA)100 n = 516. Statistical analysis of replication rates was performed using Monte Carlo resampling with 1,000,000 iterations for each comparison. p values are indicated for significant comparisons. WT (GAA)100 vs WT no rpt p < 10−6, mcm10-1 (GAA)100 vs mcm10-1 no rpt p = 0.000027, mcm10-1 (GAA)100 vs WT (GAA)100 p = 0.027614, mcm10-1 (GAA)100 vs WT no rpt p < 10−6, WT (GAA)100 vs mcm10-1 no rpt p = 0.000846. 99% confidence intervals and additional statistical information is provided in the Source Data File.
We compared strains carrying the expansion cassette with the (GAA)100 repeat to a those carrying the same cassette but with a filler sequence in place of the repeat (no repeat control). Wild-type and mcm10-1 derivatives of those strains were analyzed at the semi-permissive temperature of 30 °C. In accordance with our previous data16, there was repeat-mediated slowing of replication in the wild-type strains (Fig. 3b). There was a small, non-significant slowdown caused by the mcm10-1 mutation in the no repeat control, but the difference between WT and mcm10-1 became significant in the case of the (GAA)100 repeat (Fig. 3b). This comparison indicates that Mcm10 promotes replication elongation, especially when replicating through the expanded (GAA)n repeats.
Single-stranded DNA at the replication fork primarily promotes repeat contractions
Exposure of ssDNA during replication promotes the formation of non-B DNA structures and overexpression of the single-stranded DNA-binding RPA complex counteracts contractions of long (GAA)n repeats18. The mcm10-1 mutation was shown to cause an increase in RPA foci at the non-permissive temperature, pointing to the accumulation of ssDNA60. We therefore hypothesized that an increased single-strandedness of the (GAA)n repeat in the mcm10-1 mutant could result in its instability. To test this hypothesis, we overexpressed all three RPA subunits on a multicopy 2μ plasmid18 and verified their overexpression via western blot (Supplementary Fig. 3a). In line with our hypothesis, RPA overexpression rescued the elevated contraction rate observed in the mcm10-1 mutant (Fig. 2b). In contrast, the expansion rate in the mcm10-1 mutant was not rescued at all by RPA overexpression (Fig. 2a).
The (GAGAAGAAA)41GAG repeat is a homopurine-homopyrimidine repeat of the same length and GC-content as the (GAA)124 repeat, but lacks the mirror symmetry required for the formation of a stable triplex structure18. In accordance with our previous data18, its contraction rate is about 10-fold lower than the one of (GAA)124 repeats (Supplementary Fig. 1d). However, the mcm10-1 mutation led to a similar fold increase in contraction rate for the (GAGAAGAAA)41GAG repeat as for the (GAA)124 repeat and this effect is similarly rescued by RPA overexpression. Altogether, these results show that an excess of uncoated ssDNA, rather than strong triplex formation, accounts for the elevated contraction rate in the mcm10-1 mutant.
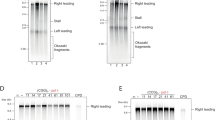
To explore this further, we first triggered replication fork uncoupling of the CMG helicase and polymerase ε using hydroxyurea (HU). HU depletes the deoxyribonucleotide pool available during replication resulting in fork uncoupling, replication stress and increased ssDNA exposure72. Treatment with 100 mM HU moderately increased (GAA)100 expansions (3.4-fold) (Fig. 4a) and had a more substantial increase on repeat contractions (8.3-fold) (Fig. 4b). We then looked at the role of the Ctf4 protein trimer, which was proposed to have a role in replication fork coordination similar to Mcm10 by coordinating CMG and Polymerase α43,46,51. Deletion of the CTF4 gene had no effect on repeat expansions and only had a modest 3.6-fold increase in repeat contraction rate (Fig. 4c, d). Likewise, deletion of the Mrc1 component of the Fork Protection (or Fork Pausing) Complex, which prevents excessive uncoupling at stalled forks, had a negligible effect on repeat expansions and a larger significant effect on repeat contractions (Fig. 4e, f). Altogether, these results indicate that the observed phenotypes are intrinsic to Mcm10 and are not shared with other replication fork coupling modalities, which seem to be predominantly preventing repeat contractions.
a Rates of expansion in vehicle (H2O) versus 100 mM HU treatment conditions (30 °C). b Rates of contraction in vehicle (H2O) versus 100 mM HU treatment conditions (30 °C). c Rate of expansion upon full deletion (Δ) of the replisome adaptor protein Ctf4 at 30 °C. d Rate of contraction upon full deletion (Δ) of the replisome adaptor protein Ctf4 at 30 °C. e Rate of expansion upon full deletion (Δ) of Mrc1 (30 °C). f Rate of contraction upon full deletion (Δ) of Mrc1 (30 °C). For a–f, plotted values indicate the corrected rate calculated with FluCalc (https://flucalc.ase.tufts.edu/)66 and the error bars represent 95% confidence intervals (see Source Data File). Each experiment was conducted with two biological replicates. Numbers within bars indicate fold increase over the respective wild-type value.
Mcm10 deficiency causes repeat length- and position-dependent viability defects
During our instability assays with the mcm10-1 mutants, we observed a viability defect more pronounced than previously described in the literature60,73. We wondered if the presence of the long repeat could be responsible for this phenotype. We conducted serial dilutions and observed that at the semi-permissive temperature of 30 °C, strains containing both the mcm10-1 mutation and the (GAA)100 repeat on the lagging strand template on our chromosome III location had a viability defect when compared to the mcm10-1 strain without the repeats, which only showed a delay in growth (Fig. 5a and Supplementary Fig. 4a). In a more quantitative assay, we compared colony-forming units (CFUs) at 30 °C and 23 °C and saw that a carrier length of (GAA)40 repeats already caused a significant decrease in viability compared to the no-repeat strain. This effect was further exacerbated at the disease-causing lengths of (GAA)100 and (GAA)124, leading to a striking ~80% loss of viability (Fig. 5b). This effect was also observed, albeit in a milder form, when the (TTC)n run serves as the lagging strand template (Supplementary Fig. 4c). To establish if the viability defects are caused by the triplex-forming potential of the (GAA)n repeat, we analyzed the viability of mcm10-1 strains carrying the (GAGAAGAAA)41GAG repeat, and we observed a viability decrease comparable to that of the (GAA)124 repeat (Fig. 5b). As was observed in the case of repeat instability, the mcm10-1 mcm2G400D double mutant fully rescued the viability defects for both types of homopurine-homopyrimidine repeats. Minimizing the presence of uncoated ssDNA at the repeat by overexpressing the RPA complex also led to a viability rescue for both repeats (Fig. 5b).
a Spot tests of strains containing either a no repeat cassette or the expansion (GAA)100 repeat cassette either at an essential region on chromosome III (ARS306) or at a non-essential region on chromosome V (ARS507) in wild-type and mcm10-1 backgrounds at the semi-restrictive temperature (30 °C). Cells were grown to an OD600 = 1 in YPDUA medium at the permissive temperature and then spotted as a 1:10 serial dilution on YPDUA plates and grown for 3 days. b Viability assay of strains containing no repeats or repeats of various lengths and compositions. The same number of cells was plated in duplicates at the semi-restrictive temperature (30 °C) and at the permissive temperature (23 °C) and grown for 3 days. Each datapoint represents the percent relative survival as determined by colony forming unit (CFU) counts. The mean is plotted, and the bars indicate standard deviation. Each assay was conducted using at least two biological replicates. Statistical analysis was performed using two-tailed unpaired t-test, *p < 0.05, **p < 0.01 and ***p < 0.001. For the mcm10-1 strains, (GAA)40 vs no repeat p = 0.0407, (GAA)100 vs no repeat p = 0.0006, (GAA)124 vs no repeat p = 0.005, (GAGAAGAAA)41GAG vs no repeat p = 0.0102. c Spot tests of strains containing the (GAA)100 repeat in various mcm10-1 double mutants at the semi-restrictive temperature (30 °C). Cells were grown to an OD600 = 1 in YPDUA medium at the permissive temperature and then spotted as a 1:10 serial dilution on YPDUA plates and grown for 2-3 days. d Spot tests of strains containing the (GAA)100 repeat and defects in polymerase δ using either the pol3-Y708A mutant or in a system in which the endogenous POL32 promoter was substituted for inducible the pGAL1 promoter to regulate POL32 expression level. For the pGAL1-3xHA-POL32 strains, cells were grown overnight in YPUA + 2% Raffinose and diluted and switched to media containing glucose or galactose until OD600 = 1 and then spotted as a 1:10 serial dilution on YPDUA or YPGalUA plates and grown for 2-3 days at 30 °C.
Notably, our instability cassettes are historically located within the essential arm of chromosome III, where loss of telomeric-proximal DNA would lead to loss of cell viability. We then moved our expansion cassette adjacent to the ARS507 replication origin in a non-essential location on a chromosome V arm, making the loss of the telomere-proximal chromosome arm possible74,75. In this case, the viability of the mcm10-1 strains bearing the (GAA)100 repeat was the same as the no-repeat control in the same location (Fig. 5a and Supplementary Fig. 4b), albeit the rate of cell growth was slower. Altogether, these results imply that cells containing expanded homopurine-homopyrimidine repeats rely on Mcm10 function for survival only when the repeats are located on an essential chromosome arm.
Viability defects in mcm10-1 (GAA)100 cells are exacerbated in DNA repair and Polymerase δ mutants
We conducted candidate gene analysis to decipher which cellular pathways contribute to the observed viability defect in the mcm10-1 strains carrying the (GAA)100 repeats. Rad51 and Rad52 are both involved in the homologous recombination and template switching (TS) processes76. We observed a minor exacerbation of the viability defect in mcm10-1 rad51Δ and a much stronger effect in the mcm10-1 rad52Δ double mutant (Fig. 5c), with the latter being already pronounced even at permissive temperatures (Supplementary Fig. 5a). Template switching is also mediated by Rad5, a polyubiquitin ligase and DNA-dependent ATPase. The mcm10-1 rad5Δ double mutant displayed an exacerbated growth defect, particularly at the semi-permissive temperature of 27 °C (Supplementary Fig. 5a), even though it did not further reduce viability in the colony formation assay (Supplementary Fig. 5b). Preventing DSBs and gap resection by knocking out the Exo1 nuclease in the mcm10-1 background led to a partial rescue in growth in the spot test, even though it did not enhance viability as measured by colony forming units (Fig. 5c and Supplementary Fig. 5b).
Which DNA polymerase could be responsible for processing replication defects in the mcm10-1 mutants, and is thus facilitating cell survival? Knocking out the translesion synthesis polymerase Rev3 only had a minor effect on strain viability (Fig. 5c). We then turned to determining the role of DNA polymerase δ, which is implicated in replication stress survival (reviewed in ref. 77). First, we tested the effect of the pol3-Y708A mutation in the catalytic subunit of Pol δ78. The mcm10-1 pol3-Y708A double mutant displayed poor growth at all temperatures, including the permissive temperature of 23 °C (Fig. 5d and Supplementary Fig. 5a). Second, we placed Pol32, which is the processivity subunit of Pol δ as well as a component of the translesion synthesis polymerase Pol ζ79, under an inducible galactose promoter, creating pGAL1-3xHA-POL32 strains80. Differences in protein levels between conditions were confirmed via western blotting (Supplementary Fig. 5c). Under repressive conditions (glucose), we observed synthetic lethality with the mcm10-1 mutation (Fig. 5d), which was not observed under galactose induction. We conclude that various DNA repair and post-replicative repair processes are needed to process the defects that arise in the mcm10-1 mutants and maintain cell viability. Pol δ function is ultimately responsible for preserving genomic integrity and cell viability during DNA synthesis.
Mrc1 and Rad9 promote viability and expansions through different mechanisms
While we observed that many of the DNA damage tolerance and repair pathways are essential to promote the viability of mcm10-1 cells, this does not necessarily mean that their absence would also exacerbate repeat expansion rates. The rate of repeat expansions remained unchanged in the mcm10-1 rad51Δ and mcm10-1 rad52Δ double mutant as compared to the mcm10-1 mutant alone, while the mcm10-1 rad5Δ mutant led to a modest additional 3-fold increase in the repeat expansion rate (Fig. 6a). Deletion of either the Exo1 exonuclease or the Rev3 TLS polymerase also did not affect the expansion rate of the mcm10-1 mutant (Fig. 6a).
a Expansion rates in various mcm10-1 double mutants with genes involved in response to replication stress and DNA repair at the semi-permissive temperature (27 °C). b Contraction rates for the mcm10-1 rad53K227A double mutant deficient in checkpoint activation at 27 °C. c Expansion rates at 27 °C using the separation of function mrc1AQ mutant in mcm10-1, which is deficient in the checkpoint function of Mrc1 and proficient in its replication function. The mcm10-1 mrc1AQ strain contains the pAO139 plasmid. All other strains on this graph are the same as in a but supplemented with the empty vector pRS415 for direct comparison. Non-selective stage was conducted on -LEU media to maintain the plasmids. For a-c, plotted values indicate the corrected rate calculated with FluCalc (https://flucalc.ase.tufts.edu/)66 and the error bars represent 95% confidence intervals (see Source Data File). Each experiment was conducted with two biological replicates. Numbers within bars represent the fold increase relative to the corresponding wild-type value. Numbers above bars represent fold increase over the single mcm10-1 mutant. d Spot tests for the strains used in c. Cells were grown to an OD600 = 1 in -LEU at the permissive temperature (23 °C), then spotted as a 1:10 serial dilution on -LEU plates and grown for 2 days at 30 °C. e Western blots of Rad53 to detect phosphorylation status in mcm10-1 and various double mutants. Cells were grown at 23 °C, arrested in G1/S using α-factor at 30 °C and then released into the cell cycle. Samples were collected at the indicated time points and the protein detected as described in the Methods. Ponceau S staining was used as a loading control. f Cell cycle analysis of mcm10-1 and various double mutants. Cells were grown at 23 °C, arrested in G1/S using α-factor at 30 °C and then released into the cell cycle. Cells were collected at the indicated timepoints and analyzed by flow cytometry using the SYTOX™ green DNA stain. 1 N indicates G1 cells, 2 N indicates G2/M cells after completion of S-phase.
We reasoned that a common consequence of excessive ssDNA exposure, as is seen in the mcm10-1 mutant60, is the activation of checkpoint pathways via the recruitment of various mediators and kinases, culminating with the activation of the effector kinase Rad53CHK2,81. The DNA replication checkpoint (DRC) recognizes ssDNA at stalled replication forks mainly using the Mec1ATR-Mrc1Claspin axis. The intra-S phase DNA damage checkpoint (DDC) is also triggered by persistent replication stress and accumulation of post-replicative ssDNA gaps is mediated by Rad9TP53BP1 82,83,84,85. In addition, the checkpoint is directly activated in response to double-strand breaks via Tel1ATM- Rad953BP1 (reviewed in refs. 86,87,88). Activation of the checkpoint results in fork stabilization, slower replication and recruitment of additional factors for repair and restart (reviewed in refs. 89,90).
We studied the effect of key checkpoint regulators on the viability and repeat instability of mcm10-1 strains containing our experimental cassettes. The rad53K227A mutation affects the kinase activity of the Rad53 protein without compromising its role in maintaining normal dNTP levels via Dun1 phosphorylation91. Both the rad53K227A mutation and RAD9 deletion caused a profound additional viability defect in mcm10-1, which was evident even at the permissive temperature (Fig. 5c and Supplementary Fig. 5a). We then tested how checkpoint defects affect the stability of the (GAA)n repeat. Both the rad53K227A and rad9Δ mutations significantly rescued hyper-expansion phenotype of the mcm10-1 mutant (Fig. 6a). In contrast, the mcm10-1 rad53K227A mutant did not rescue (GAA)124 contractions, further indicating that different mechanisms account for expansions and contractions in Mcm10-deficient cells (Fig. 6b). Checkpoint activation is commonly reflected by changes in the cell cycle profile. We conducted cell cycle profiling of WT and mcm10-1 strains containing the (GAA)100 expansion cassette using flow cytometry of DNA content. We observed that while the cell cycle profile of WT and mcm10-1 strains is virtually identical at the permissive temperature of 23 °C, at 30 °C there is a delay in S-phase progression in the mcm10-1 mutant, with the bulk of replication occurring 60 min after release instead of 30-45 min after release (Fig. 6f and Supplementary Fig. 6a), as was previously observed60. To directly relate cell-cycle stage with checkpoint activation, we conducted western blots using an antibody against the checkpoint effector kinase Rad53, which is phosphorylated upon checkpoint activation. The checkpoint was indeed activated in the mcm10-1 mutant during late S-phase (60 min), as can be observed by Rad53 phosphorylation, but no checkpoint activation could be detected during G2/M phase (120 min) (Fig. 6e). No checkpoint activation was detected in the mcm10-1 rad9Δ strains during late S-phase (Fig. 6e, f). In summary, we observed that the Rad9 checkpoint senses the defects in mcm10-1 likely during late replication and post-replicatively and initiates a response that ensures cell viability. During this response, however, (GAA)100 repeat expansions can occur.
Mrc1 functions as an intra S-phase checkpoint mediator of replication stress but also works together with Tof1TIM and Csm3TIPIN to form the so called Fork Protection Complex (FPC), which regulates CMG helicase activity, limiting replication fork uncoupling and ssDNA exposure (reviewed in ref. 31). In the mcm10-1 mrc1Δ double mutant, we observed a massive rescue in repeat expansions (Fig. 6a, c) as well as synthetic lethality with mcm10-1 (Fig. 5c and Supplementary Fig. 5a). We then used a separation of function mutant to discern which function of Mrc1 is responsible for these phenotypes. In the Mrc1AQ mutant, the replication function of Mrc1 is intact while its checkpoint function is compromised92. We introduced a plasmid containing the mrc1AQ allele in our mcm10-1 mrc1Δ strains and observed no rescue in (GAA)100 repeat expansions compared to the empty vector control (Fig. 6c). Thus, Mrc1’s replication function promotes repeat expansions in the mcm10-1 mutant.
Furthermore, Mrc1AQ expression rescued the viability defect back to the mcm10-1 single mutant levels (Fig. 6d). In the mcm10-1 mrc1Δ double mutant, we observed both a delay in cell cycle progression (Fig. 6f) and checkpoint activation during both S-phase and G2/M (Fig. 6e), as was previously shown for the mrc1Δ single mutant in cells without the expanded (GAA)n repeats92. In contrast, the mcm10-1 mrc1AQ mutant displays checkpoint activation only in late S-phase, as did the mcm10-1 single mutant (Fig. 6e), but does not fully rescue the S-phase delay observed in the mcm10-1 mrc1Δ context (Fig. 6f). Therefore, we conclude that the replication function of Mrc1, but not its checkpoint function, is required for the viability of mcm10-1 strains, both when they are carrying (GAA)n repeats and in the no repeat context (Supplementary Fig. 7).
Discussion
In this study, we investigated the role of the Mcm10 protein and other replisome factors in (GAA)n repeat instability. Mcm10 is a highly conserved protein that promotes DNA origin unwinding during replication initiation39,93,94,95,96. Several studies implicated Mcm10 in replication elongation, either as a replisome component or its accessory factor58,59,60. In addition, deficiencies in Mcm10 have been shown to lead to various types of genome instability and dysregulated DNA damage response61,68,73,97, making it an attractive candidate to study in the context of repeat instability. We found that a temperature sensitive mutation in the Mcm10 protein (mcm10-1) dramatically increases length instability of (GAA)n repeats even at the semi-permissive temperatures (Fig. 1 and Supplementary Fig. 1), at which other studies found no noticeable mutational phenotypes39,60,61.
There are several mechanisms that could account for the elevated (GAA)n repeat instability in Mcm10 deficient cells. One possibility is that it could result from the degradation of Pol α that is known to occur in both yeast and human cells upon Mcm10 depletion42,43,60,98,99. Several of our results, however, argue against this possibility. First, we observed no loss of the catalytic subunit of Pol α, Pol1/Cdc17 at the semi-permissive temperatures (Fig. 2c). Second, a previous study showed that Pol α mutants resulted in the addition of a greater number of repeat units during expansions, but not an increase in the rate of (GAA)100 repeat expansion, which is the opposite of what we observe in the mcm10-1 mutant17 (Supplementary Fig. 1c). Third, Pol α mutations led to a strong increase in (GAA)124 repeat contraction in a study conducted in our laboratory18, while we observe that the mcm10-1 effect on expansions is much greater than its effect on contractions.
Another possibility is that repeat instability could primarily result from replication fork uncoupling between the leading and lagging strand syntheses, since Mcm10 contributes to fork integrity by bridging the CMG helicase, DNA Pol α and DNA Pol ε39,58,100,101. Uncoupling occurs when the helicase continues to unwind while leading strand synthesis by polymerase ε is halted, and lagging-strand synthesis continues29. We therefore compared the effects on repeat instability between the mcm10-1 mutation and hydroxyurea (HU) treatment, which causes replication fork uncoupling102. We found that HU treatment leads to an overall increase in (GAA)n repeat instability, with a much stronger effect on repeat contractions (Fig. 4a, b). We acknowledge, however, that hydroxyurea treatment has pleiotropic effects on replication, as it also affects origin firing, checkpoint activation, and the function of Pol δ in lagging strand synthesis77,102. Thus, we cannot confidently attribute the observed effects on repeat instability to fork uncoupling alone. Along the same lines, deletion of the Ctf4 protein—another CMG helicase, DNA Pol α and DNA Pol ε adaptor, as well as the fork protection complex component Mrc1 which prevents excessive uncoupling103, have a stronger effect on repeat contractions than on repeat expansions (Fig. 4c–f). Altogether, we conclude that replication fork uncoupling leading to the accumulation of single-stranded repetitive DNA combined with the massive RPA depletion in the mcm10-1 mutant drives repeat contractions, which is in-line with our previous model18. RPA depletion at the repeat in the mcm10-1 context is exacerbated by having the homopurine (GAA)n or (GAGAAGAAA)n repeats as the lagging strand template, as RPA has a 10-fold lower affinity for poly-purine tracts than it does for poly-pyrimidine tracts104.
How could Mcm10 counteract repeat expansions? Mcm10 is known to physically interact with and stabilize the CMG helicase via multiple MCM subunits58,68, and it sits at the front edge of the CMG, which is optimal to face ssDNA/dsDNA junctions ahead of the fork54,68. Mcm10 regulates the ability of the CMG helicase to switch between an ssDNA and dsDNA encircling modes in vitro54. In the context of an uncoupled fork, the CMG-Mcm10 complex transitions to a dsDNA-encircling diffusive mode, which allows for fork re-entry and resuming of DNA synthesis upon encountering a lesion54. In addition, Mcm10 was shown to be important to promote the bypass of lagging strand blocks via its CMG isomerization functions58. To determine whether Mcm10 affects repeat instability via its interaction with the CMG helicase, we capitalized on a previous observation that Mcm10 and Mcm2 interact directly58, and several mutations in Mcm2, including the mcm2G400D mutant, suppress the phenotypes of the mcm10-1 allele58,60. We found that both types of elevated repeat instability in mcm10-1 were rescued by the mcm2G400D mutation (Fig. 2a, b).
We predicted that a consequence of perturbing the interaction between Mcm10 and the CMG helicase could be changes in replication fork speed through the (GAA)n repeat. Using live-cell microscopy, we indeed found that (i) there is a repeat-dependent slowdown of replication in the wild-type context, which is consistent with what we observed using 2D-gel electrophoresis of replication intermediates in the same orientation16, and (ii) the mcm10-1 mutation exacerbated this effect (Fig. 3). Since this assay system is located ~3 kb downstream of the origin, initiation should not contribute to the observed effects. These results indicate that Mcm10 promotes replication through the (GAA)n repeat and likely other hard-to-replicate genome regions, as was previously observed for replication termination in Xenopus egg extracts105.
Increased fork stalling could result from more stalling events in the first place, or a difficulty of restart, which might be promoted by proper Mcm10-CMG interactions and be dependent on the ability of the CMG to switch from ssDNA to dsDNA encircling mode. Either of these events would lead to increased ssDNA exposure at the replication fork. While we observed a striking viability defect in Mcm10-deficient cells that contained expanded repeats at the essential chromosome III region, the viability defects were rescued by the mcm2G400D mutation as well as RPA overexpression, indicating that ssDNA accumulation is ultimately responsible for the viability defects. Accumulation of single-stranded gaps combined with increased mutagenesis were indeed previously observed in mcm10-1 mutants73. We therefore studied whether processes that are involved in the repair of ssDNA gaps, such as template switching (TS), are at play in our case. Rad5 knockout was previously shown to decrease the viability of mcm10-1 mutants73, implicating template switching and fork reversal in rescuing forks with Mcm10 function. In our case, deletion of the RAD5 gene further impaired growth in the mcm10-1 mutant. We identified a negative interaction between loss of Rad52, which is involved ssDNA gap filling in addition to homologous recombination106, and Mcm10 deficiencies. Notably, none of the tested double mutants rescued the hyper-expansion phenotype. RAD51 and RAD52 deletion showed no change compared to mcm10-1 alone, while RAD5 knockout further increased repeat expansions. This indicates that while these pathways promote viability, they are not responsible for the elevated expansion rates observed in mcm10-1.
We reasoned that persistent exposure of ssDNA gaps in the mcm10-1 mutant could activate the checkpoint and possibly also result in breakage at the fork. The checkpoint mediator Rad9 propagates checkpoint signaling during replication stress even in the presence of Mrc1 to promote repair processes at stalled forks, ssDNA gaps and DSBs82,85,107. Rad9 was previously shown to be important for the survival of cells harboring a partial C-terminal deletion of Mcm10 101. A kinase-deficient Rad53 mutation as well as deletion the RAD9 gene further impaired the survival of repeat-containing mcm10-1 strains, pointing to persistent replication stress and checkpoint activation in the mcm10-1 mutant, which we additionally confirmed by western blots showing the phosphorylated form of the Rad53 effector kinase82 (Fig. 6e). In contrast to the DNA repair mutants described above, both RAD9 knockout and kinase-deficient Rad53 led to a partial rescue of the hyper-expansion phenotype in mcm10-1 mutants. This result suggests that the Rad9 checkpoint promotes the survival of the mcm10-1 cells at the expense of triggering repeat expansions during DNA repair processes.
The other checkpoint mediator, Mrc1, appeared to promote viability of Mcm10-deficient cells via its function as part of the fork protection complex, rather than its checkpoint function (Fig. 6d). This result is consistent with in vitro studies showing that in the absence of the replication function of Mrc1, Mcm10 becomes crucial for proper progression of leading strand synthesis, and that Mrc1 and Mcm10 are partially redundant in the replication stress response57,108,109. We propose that in the absence of both Mcm10 and the replication-associated function of Mrc1, replication through DNA repeats becomes grossly inefficient, counteracting repeat expansions, and further promoting cell death.
It was previously shown that Rad53 and Polδ deficiencies result in negative genetic interactions with the mcm10-1 mutation59,60 suggesting that synthetic viability defects observed by us may not be repeat-specific as well. At the same time, a presence of the repeat could further exacerbate deleterious phenotypes for some of our double mutants, as is illustrated by the fact that Pol32 deprivation renders repeat-containing mcm10-1 strains practically inviable.
Overall, we propose a model in which Mcm10-deficiency causes replication problems such as increased fork stalling at the repeats and accumulation of ssDNA gaps genome-wide (Fig. 7a). This leads to Rad9 checkpoint activation during late S-phase in a manner triggered by RPA binding to ssDNA gaps or conversion to double-stranded breaks (Fig. 7b, c), in-line with previous studies describing a parallel role of Rad9 to Mrc1-sustained replication stress response82,85. Checkpoint activation results in the repair of under-replicated DNA, which is crucial for cell viability. We stipulate that massive repeat expansions occur during the gap repair DNA fill-in synthesis conducted by DNA polymerase δ (Fig. 7c). If ssDNA gaps remain unrepaired until the G2/M phase of the cell cycle due to deficient checkpoint activation or incomplete repair synthesis, under-replicated ssDNA would cause mitotic chromosome breakage, loss of essential DNA and ultimately cell death (Fig. 7d). This model explains why RPA overexpression rescues cell viability by triggering checkpoint activation, while not rescuing hyper-expansion phenotype, which is downstream of checkpoint activation.
a Mcm10-deficiencies give rise to increased replication fork stalling at the expanded (GAA)n repeat, which results in ssDNA gap formation. The genome-wide increase in ssDNA levels can results in RPA depletion at the repeat, leading to (b) Rad9 checkpoint activation during late S-phase in a manner triggered by RPA binding to ssDNA gaps or conversion to double-stranded breaks. DNA repair and fill-in synthesis events lead to massive expansions during slippery DNA synthesis conducted by polymerase δ. c This also occurs in the case of proper ssDNA coating upon RPA overexpression, minimizing conversions to DSBs. d In cases of insufficient checkpoint activation or incomplete synthesis, mitotic breakage can occur, leading to loss of essential DNA regions and cell death. Created in BioRender. Masnovo, C. (2024) https://BioRender.com/j22q561.
Our model partially agrees with previous observations of a replication fork block in S. pombe110, where Rad51 and Rad52 knockouts led to a loss of viability due to accumulation of unrepaired ssDNA followed by chromosomal breakage. MCM10 was shown to suppress PRIMPOL-mediated gap formation in collaboration with BRCA2, suggesting that it could have a more general role in suppressing gap formation during DNA replication97.
We found that Mcm10 deficiency destabilizes (GAA)n as well as (GAGAAGAAA)n repeats and cause repeat-mediated viability defect. This indicates that Mcm10 is vital for the maintenance of homopurine-homopyrimidine repeats regardless of their structure-forming potential. It is tempting, therefore, to speculate that Mcm10 could contribute to the maintenance of various other simple DNA repeats, which cumulatively account for ~3% of the human genome111. Indeed, MCM10 haploinsufficiency in human cell lines led to chromosomal rearrangements overlapping with common fragile sites (CFSs)112, which are overall rich in (AT)n repeats113,114. In addition, MCM10 promotes replication of telomeres112, which are also highly repetitive DNA regions that can form alternative DNA structures. Further studies will be necessary to determine whether Mcm10 is also important for replication and maintenance of other disease-associated repetitive DNA sequences.
Methods
Yeast Strains
All yeast strains are listed in Supplementary Table 1. Point mutations and gene deletions were introduced using a plasmid-based CRISPR/Cas9 system115 or by standard gene replacement. Strains for replication time measurement were generated essentially as previously described71 in the W1588 MATa background, expressing LacI-Envy and TetR-tdTomato fusion proteins in the nucleus.
List of oligonucleotides
All primers used for strain construction and to detect expansion events are listed in Supplementary Table 2.
Fluctuation assays
All assays were conducted with at least two independent biological replicates per genotype. Mutation rates were calculated using the FluCalc software (https://flucalc.ase.tufts.edu/)66. Rates are considered meaningfully different if the 95% confidence intervals do not overlap. All rates, 95% confidence intervals and raw data points for each experiment are reported in the Source Data file.
Expansion rates. Two independent biological replicates per genotype were plated for singles on YPDUA plates at 30 °C (2 days), 27 °C (2.5 days) or at 23 °C (3 days). For the double mutants that displayed a severe growth defect, we incubated the non-selective plates at 27 °C for up to 6 days, or until the colonies were as large as the wild-type colonies after 2.5 days. The initial repeat length was confirmed for 12 or more single colonies which were then serially diluted in 10−1 increments to a final dilution of 10−5. 100 μl of the 10−5 dilution were plated on YPDUA to determine the total cell count (TCC). 100 μl of 10−1 or 10−2 dilution were plated on selective plates. For ts strains, the selective stage was conducted on 0.175% 5-FOA Glucose plates for 4 days at 23 °C. For all other strains, the selective stage was conducted on 0.09% 5-FOA Glucose plates for 3 days at 30 °C. Expansion events were confirmed by PCR using the A2 and B2 primer set and analyzed in ImageLab (BioRad, v 6.1).
Contraction rates. Fluctuation assays for contraction rates were conducted as described in (Khristich et al., 18) using the appropriate temperatures at the non-selective stage and selective stages.
Viability assays
Repeat tract length was determined for single colonies of each genotype grown at the permissive temperature of 23 °C. Singles of the correct repeat length were grown to an OD600 of 0.6-1 in complete (YPDUA) media. An amount of 10−4 or 10−5 culture dilution corresponding to 100-500 cells was plated in duplicates on complete media in duplicates at both 23 °C and 30 °C. Colony forming units (CFUs) were counted after 3 days at 30 °C and 4 days at 23 °C. CFUs between duplicates were averaged and viability was calculated as (CFU 30 °C)/(CFU 23 °C) for each strain. Statistical differences were assayed using two-tailed Welch’s t-test in GraphPad Prism (v 10.3.1).
Spot tests
Yeast strains were grown overnight in YPDUA media at the permissive temperature of 23 °C, diluted to an OD600 = 0.2-0.3 and grown to log phase. pGAL1-3xHA-POL32 strains, cells were grown in YPUA + 2% Raffinose overnight at 23 °C and then diluted to OD600 = 0.3 in YPUA media containing either 2% Glucose or 2% Galactose and grown until they reached an OD600 = 0.8-1. 200 μl of OD600 = 1 were collected and a serial 1:10 dilution was performed in sterile dH2O. 5 μl of each dilution (1 to 10−4) were spotted on YPDUA plates and placed at the temperatures indicated in the figure captions for 2-3 days and then imaged. Each spot test was conducted with at least two independent isolates of the same genotype.
Protein isolation and western blotting
Total protein extraction was performed based on the protocol in ref. 116. For RPA overexpression blots, strains were grown to log phase at 23 °C. For Pol1 protein measurements, cells were grown to OD600 0.5-0.8 at 23 °C and then shifted to 30 °C or 37 °C for 3 h before sample collection. For measurements of pGAL1-3xHA-POL32 expressions, cells were grown in YPUA + 2% Raffinose overnight at 23 °C and then diluted to OD600 = 0.3 in YPUA media containing either 2% Glucose or 2% Galactose and grown until they reached an OD600 = 0.8-1. 3-5 OD600 per sample were collected by centrifugation (1315 × g, 5 min, 4 °C) and the pellet was flash-frozen in liquid nitrogen. The pellet was then thawed and treated with 150 μl/OD denaturing lysis buffer (1.85 M NaOH, 7.5% β-mercaptoethanol) for 15 min on ice. The lysate was then mixed with 1 volume of 55% (w/v) trichloroacetic acid (TCA) for 15 min on ice. Proteins were then pelleted by centrifugation (16000 × g, 4 °C, 15 min) and washed with 1 volume of water (16000 × g, 4 °C, 15 min). The protein pellet was then resuspended in 50 μl/OD HU sample buffer (8 M urea, 5% SDS, 1 mM EDTA, 1.5% DTT, 1% bromophenol blue) and heated for 10 min at 65 °C before running on 4-12% NuPAGE Bis-Tris gels in MOPS buffer for 90 min at 150 V. We used Novex™ Sharp Pre-stained Protein Standard as a ladder. Transfer onto nitrocellulose membranes was conducted using an iBlot2 Gel Transfer device at 25 V for 7 min. RPA subunits were detected using an anti-RFA antibody (Agrisera, AS07-214, 1:5,000 dilution in 5% skim milk in TBS-T). Pol1-3xFLAG was detected using the ANTI-FLAG M2 antibody (Sigma Aldrich, F3165-.2MG, 1:2,000 dilution in 5% skim milk in TBS-T). pGAL1-3xHA-POL32 constructs were detected using Anti-HA Tag antibody (Sigma Aldrich, 05-904, 1 μg/ml dilution in 5% skim milk in TBS-T). Rad53 was detected using anti-Rad53 antibody EL7.E1 (Abcam, ab166859, 1:2,000 dilution in 5% skim milk in TBS-T). The secondary antibodies used in this study were Anti-Mouse IgG HRP Conjugate (W402B, Promega), Anti-Rabbit IgG HRP Conjugate (W401B, Promega) at a 1:5,000 dilution in 5% skim milk in TBS-T.
Cell cycle analysis by flow cytometry
Strains were grown overnight at 23 °C in YPDUA at 200 rpm. Subsequently, cultures were diluted to OD600 = 0.3 and grown for 150 min (23 °C, 200 rpm). α-factor (Zymo Research, Y1001, 10 mM) was added to a final concentration of 0.1 μM and cultures were grown for an additional 60 min (23 °C, 200 rpm). ½ of the culture volume or the whole volume was then shifted to 30 °C and all cultures were grown for an additional 60 min (200 rpm) and cells were checked for shmoo formation with a light microscope. Cultures were then washed twice with 1 volume of YPDUA (1315 × g, 2 min) and then resuspended in 1 volume of YPDUA containing Pronase E (Millipore Sigma, 537088) to a final concentration of 50 μg/ml and released at the respective temperatures.
0.5-2 × 107 cells were then collected at the indicated time points, washed with nuclease-free water and permeabilized with 1 ml 70% ethanol for overnight at 4 °C. After centrifugation (5 min, 200 × g, 4 °C), pellets were washed in 1 mL 50 mM Tris-HCl pH 7.5 and the cells were resuspended in 500 μL 50 mM Tris-HCl pH 7.5 containing 2 mg/ml RNaseA (Thermo Scientific, EN0531) and incubated overnight at 37 °C. After centrifugation, the pellet was resuspended in 200 μL 50 mM Tris-HCl pH 7.5 containing 1 mg/ml Proteinase K (Thermo Scientific, EO0491) and placed at 50 °C for 30 min. The cells were then pelleted by centrifugation (5 min, 2400 × g, 4 °C) and resuspended in 500 μL of 50 mM Tris-HCl pH 7.5 and stored at 4 °C until measurements were acquired. Before analysis, 100 μL of the prepared sample were added to 1 ml 50 mM Tris-HCl pH 7.5 supplemented with 1 μM SYTOX™ Green Nucleic Acid Stain (Invitrogen, S7020). Data was acquired on an Attune™ NxT machine using the Attune NxT software (v 3.1.2), and analyzed in FlowJo (v 10.10.0). We acquired 10,000 events on the ungated parameters, gating strategy can be observed in Supplementary Fig. 6b.
Live-cell microscopy of replication fork progression
Yeast strains were grown overnight in synthetic complete (SC) medium at 23 °C. Exponentially growing cultures were diluted to OD600 = 0.2, 10 µg/mL α-factor was added to arrest cells at G1 phase and cultures were incubated for 1 h at 23 °C and then shifted to 30 °C. Cells were then immobilized on microscopy chamber slides (Ibidi) coated with 2 mg/mL concanavalin A (Sigma) and washed thoroughly with warm SC medium to release the cells into S-phase. During imaging, cells were incubated in SC medium containing 4% glucose at 30 °C. Live-cell imaging was performed on a CellDiscoverer 7 automated microscope (Zeiss) with an integrated LED light source, at 1 min intervals for 2 h, using a 50x apochromatic water objective (NA = 1.2) in 3D (8 z-sections 0.8 µm apart). Time-lapse measurements were collected using ZEN 3.0 software and analyzed using a custom-made, Python-based computational pipeline developed specifically for the analysis of replication rates, similar to a previous study71. Our pipeline identifies, tracks and quantifies the LacI-Envy and TetR-tdTomato dots in each cell. For each strain, at least 200 cells were measured in two independent experiments. Statistical analysis of replication rates was performed using Monte Carlo resampling with 1,000,000 iterations.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The authors declare that all data supporting the findings of this study are available within the paper, and the Supplementary Information. Source data are provided with this paper.
Code availability
The code for the live-cell microscopy analysis is available upon request without any restrictions.
References
Erwin, G. S. et al. Recurrent repeat expansions in human cancer genomes. Nat. 1–7 https://doi.org/10.1038/s41586-022-05515-1 (2022).
Depienne, C. & Mandel, J.-L. 30 years of repeat expansion disorders: What have we learned and what are the remaining challenges? Am. J. Hum. Genet. 108, 764–785 (2021).
Campuzano, V. et al. Friedreich’s Ataxia: Autosomal Recessive Disease Caused by an Intronic GAA Triplet Repeat Expansion. Science 271, 1423–1427 (1996).
Pellerin, D. et al. Intronic FGF14 GAA repeat expansions are a common cause of ataxia syndromes with neuropathy and bilateral vestibulopathy. J. Neurol. Neurosurg. Psychiatry https://doi.org/10.1136/jnnp-2023-331490 (2023).
Rafehi, H. et al. An intronic GAA repeat expansion in FGF14 causes the autosomal-dominant adult-onset ataxia SCA50/ATX-FGF14. Am. J. Hum. Genet. https://doi.org/10.1016/j.ajhg.2022.11.015 (2022).
Mirkin, S. M. et al. DNA H form requires a homopurine–homopyrimidine mirror repeat. Nature 330, 495–497 (1987).
Mirkin, S. M. & Frank-Kamenetskii, M. D. H-DNA and Related Structures. Annu. Rev. Biophys. Biomol. Struct. 23, 541–576 (1994).
Potaman, V. N. et al. Length-dependent structure formation in Friedreich ataxia (GAA)n·(TTC)n repeats at neutral pH. Nucleic Acids Res 32, 1224–1231 (2004).
Bidichandani, S. I., Ashizawa, T. & Patel, P. I. The GAA Triplet-Repeat Expansion in Friedreich Ataxia Interferes with Transcription and May Be Associated with an Unusual DNA Structure. Am. J. Hum. Genet. 62, 111–121 (1998).
Masnovo, C., Lobo, A. F. & Mirkin, S. M. Replication dependent and independent mechanisms of GAA repeat instability. DNA Repair 118, 103385 (2022).
Gerhardt, J. et al. Stalled DNA Replication Forks at the Endogenous GAA Repeats Drive Repeat Expansion in Friedreich’s Ataxia Cells. Cell Rep. 16, 1218–1227 (2016).
Bourn, R. L., Rindler, P. M., Pollard, L. M. & Bidichandani, S. I. E. coli mismatch repair acts downstream of replication fork stalling to stabilize the expanded (GAA.TTC)(n) sequence. Mutat. Res 661, 71–77 (2009).
Krasilnikova, M. M. & Mirkin, S. M. Replication stalling at Friedreich’s ataxia (GAA)n repeats in vivo. Mol. Cell Biol. 24, 2286–2295 (2004).
Chandok, G. S., Patel, M. P., Mirkin, S. M. & Krasilnikova, M. M. Effects of Friedreich’s ataxia GAA repeats on DNA replication in mammalian cells. Nucleic Acids Res 40, 3964–3974 (2012).
Rastokina, A. et al. Large-scale expansions of Friedreich’s ataxia GAA•TTC repeats in an experimental human system: role of DNA replication and prevention by LNA-DNA oligonucleotides and PNA oligomers. Nucleic Acids Res. 51, 8532–8549 (2023).
Shishkin, A. A. et al. Large-Scale Expansions of Friedreich’s Ataxia GAA Repeats in Yeast. Mol. Cell 35, 82–92 (2009).
Shah, K. A. et al. Role of DNA Polymerases in Repeat-Mediated Genome Instability. Cell Rep. 2, 1088–1095 (2012).
Khristich, A. N., Armenia, J. F., Matera, R. M., Kolchinski, A. A. & Mirkin, S. M. Large-scale contractions of Friedreich’s ataxia GAA repeats in yeast occur during DNA replication due to their triplex-forming ability. Proc. Natl Acad. Sci. USA 117, 1628–1637 (2020).
Jedrychowska, M. et al. Defects in the GINS complex increase the instability of repetitive sequences via a recombination-dependent mechanism. PLoS Genet 15, e1008494 (2019).
Neil, A. J., Kim, J. C. & Mirkin, S. M. Precarious maintenance of simple DNA repeats in eukaryotes. Bioessays 39, e1700077 (2017).
Lahue, R. S. & Slater, D. L. DNA repair and trinucleotide repeat instability. Front Biosci. 8, s653–s665 (2003).
Amparo, C. et al. Duplex DNA from Sites of Helicase-Polymerase Uncoupling Links Non-B DNA Structure Formation to Replicative Stress. Cancer Genomics Proteom. 17, 101–115 (2020).
Devbhandari, S. & Remus, D. Rad53 limits CMG helicase uncoupling from DNA synthesis at replication forks. Nat. Struct. Mol. Biol. 27, 461–471 (2020).
Ercilla, A. et al. Physiological Tolerance to ssDNA Enables Strand Uncoupling during DNA Replication. Cell Rep. 30, 2416–2429.e7 (2020).
Lopes, M., Foiani, M. & Sogo, J. M. Multiple Mechanisms Control Chromosome Integrity after Replication Fork Uncoupling and Restart at Irreparable UV Lesions. Mol. Cell 21, 15–27 (2006).
Pagès, V. & Fuchs, R. P. Uncoupling of Leading- and Lagging-Strand DNA Replication During Lesion Bypass in Vivo. Science https://doi.org/10.1126/science.1083964 (2003).
Taylor, M. R. G. & Yeeles, J. T. P. Dynamics of Replication Fork Progression Following Helicase–Polymerase Uncoupling in Eukaryotes. J. Mol. Biol. 431, 2040–2049 (2019).
Kavlashvili, T., Liu, W., Mohamed, T. M., Cortez, D. & Dewar, J. M. Replication fork uncoupling causes nascent strand degradation and fork reversal. Nat Struct Mol Biol 1–10 https://doi.org/10.1038/s41594-022-00871-y (2023).
Byun, T. S., Pacek, M., Yee, M., Walter, J. C. & Cimprich, K. A. Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev. 19, 1040–1052 (2005).
Nedelcheva, M. N. et al. Uncoupling of unwinding from DNA synthesis implies regulation of MCM helicase by Tof1/Mrc1/Csm3 checkpoint complex. J. Mol. Biol. 347, 509–521 (2005).
Saldanha, J., Rageul, J., Patel, J. A. & Kim, H. The Adaptive Mechanisms and Checkpoint Responses to a Stressed DNA Replication Fork. Int. J. Mol. Sci. 24, 10488 (2023).
Fanning, E., Klimovich, V. & Nager, A. R. A dynamic model for replication protein A (RPA) function in DNA processing pathways. Nucleic Acids Res 34, 4126–4137 (2006).
Pellegrini, L. The CMG DNA helicase and the core replisome. Curr. Opin. Struct. Biol. 81, 102612 (2023).
Solomon, N. A. et al. Genetic and molecular analysis of DNA43 and DNA52: two new cell-cycle genes in Saccharomyces cerevisiae. Yeast 8, 273–289 (1992).
Izumi, M. et al. The human homolog of Saccharomyces cerevisiae Mcm10 interacts with replication factors and dissociates from nuclease-resistant nuclear structures in G(2) phase. Nucleic Acids Res 28, 4769–4777 (2000).
Thu, Y. M. & Bielinsky, A.-K. MCM10: one tool for all - integrity, maintenance and damage control. Semin Cell Dev. Biol. 0, 121–130 (2014).
Yuan, Z. et al. Ctf4 organizes sister replisomes and Pol α into a replication factory. eLife 8, e47405 (2019).
Porcella, S. Y. et al. Separable, Ctf4-Mediated Recruitment of DNA Polymerase α for Initiation of DNA Synthesis at Replication Origins and Lagging-Strand Priming during Replication Elongation. http://biorxiv.org/lookup/doi/10.1101/352567 (2018).
Homesley, L. et al. Mcm10 and the MCM2–7 complex interact to initiate DNA synthesis and to release replication factors from origins. Genes Dev. 14, 913–926 (2000).
Merchant, A. M., Kawasaki, Y., Chen, Y., Lei, M. & Tye, B. K. A lesion in the DNA replication initiation factor Mcm10 induces pausing of elongation forks through chromosomal replication origins in Saccharomyces cerevisiae. Mol. Cell Biol. 17, 3261–3271 (1997).
Ricke, R. M. & Bielinsky, A.-K. Mcm10 regulates the stability and chromatin association of DNA polymerase-alpha. Mol. Cell 16, 173–185 (2004).
Chattopadhyay, S. & Bielinsky, A.-K. Human Mcm10 Regulates the Catalytic Subunit of DNA Polymerase-α and Prevents DNA Damage during Replication. MBoC 18, 4085–4095 (2007).
Zhu, W. et al. Mcm10 and And-1/CTF4 recruit DNA polymerase α to chromatin for initiation of DNA replication. Genes Dev. 21, 2288–2299 (2007).
Fien, K. et al. Primer Utilization by DNA Polymerase α-Primase Is Influenced by Its Interaction with Mcm10p. J. Biol. Chem. 279, 16144–16153 (2004).
Simon, A. C. et al. A Ctf4 trimer couples the CMG helicase to DNA polymerase α in the eukaryotic replisome. Nature 510, 293–297 (2014).
Villa, F. et al. Ctf4 Is a Hub in the Eukaryotic Replisome that Links Multiple CIP-Box Proteins to the CMG Helicase. Mol. Cell 63, 385–396 (2016).
Im, J.-S. et al. Assembly of the Cdc45-Mcm2-7-GINS complex in human cells requires the Ctf4/And-1, RecQL4, and Mcm10 proteins. Proc. Natl Acad. Sci. USA 106, 15628–15632 (2009).
Perez-Arnaiz, P. & Kaplan, D. L. An Mcm10 Mutant Defective in ssDNA Binding Shows Defects in DNA Replication Initiation. J. Mol. Biol. 428, 4608–4625 (2016).
Warren, E. M., Huang, H., Fanning, E., Chazin, W. J. & Eichman, B. F. Physical Interactions between Mcm10, DNA, and DNA Polymerase α*. J. Biol. Chem. 284, 24662–24672 (2009).
Das-Bradoo, S., Ricke, R. M. & Bielinsky, A.-K. Interaction between PCNA and diubiquitinated Mcm10 is essential for cell growth in budding yeast. Mol. Cell Biol. 26, 4806–4817 (2006).
Wang, J., Wu, R., Lu, Y. & Liang, C. Ctf4p facilitates Mcm10p to promote DNA replication in budding yeast. Biochemical Biophysical Res. Commun. 395, 336–341 (2010).
Jones, M. L., Aria, V., Baris, Y. & Yeeles, J. T. P. How Pol α-primase is targeted to replisomes to prime eukaryotic DNA replication. Mol. Cell https://doi.org/10.1016/j.molcel.2023.06.035 (2023).
Baris, Y., Taylor, M. R. G., Aria, V. & Yeeles, J. T. P. Fast and efficient DNA replication with purified human proteins. Nature 606, 204–210 (2022).
Wasserman, M. R., Schauer, G. D., O’Donnell, M. E. & Liu, S. Replication Fork Activation Is Enabled by a Single-Stranded DNA Gate in CMG Helicase. Cell 178, 600–611.e16 (2019).
Langston, L. D., Georgescu, R. E. & O’Donnell, M. E. Mechanism of eukaryotic origin unwinding is a dual helicase DNA shearing process. Proc. Natl Acad. Sci. 120, e2316466120 (2023).
Henrikus, S. S. et al. Unwinding of a eukaryotic origin of replication visualized by cryo-EM. Nat. Struct. Mol. Biol. 31, 1265–1276 (2024).
Langston, L. D. et al. Mcm10 promotes rapid isomerization of CMG-DNA for replisome bypass of lagging strand DNA blocks. eLife 6, e29118 (2017).
Lõoke, M., Maloney, M. F. & Bell, S. P. Mcm10 regulates DNA replication elongation by stimulating the CMG replicative helicase. Genes Dev. 31, 291–305 (2017).
Kawasaki, Y., Hiraga, S. & Sugino, A. Interactions between Mcm10p and other replication factors are required for proper initiation and elongation of chromosomal DNA replication in Saccharomyces cerevisiae. Genes Cells 5, 975–989 (2000).
Lee, C., Liachko, I., Bouten, R., Kelman, Z. & Tye, B. K. Alternative Mechanisms for Coordinating Polymerase α and MCM Helicase. Mol. Cell. Biol. 30, 423–435 (2010).
Alver, R. C. et al. The N-terminus of Mcm10 is important for interaction with the 9-1-1 clamp and in resistance to DNA damage. Nucleic Acids Res 42, 8389–8404 (2014).
Chadha, G. S., Gambus, A., Gillespie, P. J. & Blow, J. J. Xenopus Mcm10 is a CDK-substrate required for replication fork stability. Cell Cycle 15, 2183–2195 (2016).
Thu, Y. M. et al. Slx5/Slx8 Promotes Replication Stress Tolerance by Facilitating Mitotic Progression. Cell Rep. 15, 1254–1265 (2016).
Schmit, M. M. et al. A critical threshold of MCM10 is required to maintain genome stability during differentiation of induced pluripotent stem cells into natural killer cells. Open Biol. 14, 230407 (2024).
Yu, X. & Gabriel, A. Patching Broken Chromosomes with Extranuclear Cellular DNA. Mol. Cell 4, 873–881 (1999).
Radchenko, E. A., McGinty, R. J., Aksenova, A. Y., Neil, A. J. & Mirkin, S. M. Quantitative Analysis of the Rates for Repeat-Mediated Genome Instability in a Yeast Experimental System. Methods Mol. Biol. 1672, 421–438 (2018).
Thu, Y. M. & Bielinsky, A.-K. Enigmatic roles of Mcm10 in DNA replication. Trends Biochemical Sci. 38, 184–194 (2013).
Mayle, R. et al. Mcm10 has potent strand-annealing activity and limits translocase-mediated fork regression. PNAS 116, 798–803 (2019).
Bochman, M. L. & Schwacha, A. The Mcm Complex: Unwinding the Mechanism of a Replicative Helicase. Microbiol Mol. Biol. Rev. 73, 652–683 (2009).
Park, J. H., Bang, S. W., Jeon, Y., Kang, S. & Hwang, D. S. Knockdown of human MCM10 exhibits delayed and incomplete chromosome replication. Biochemical Biophysical Res. Commun. 365, 575–582 (2008).
Dovrat, D. et al. A Live-Cell Imaging Approach for Measuring DNA Replication Rates. Cell Rep. 24, 252–258 (2018).
Bianchi, V., Pontis, E. & Reichard, P. Changes of deoxyribonucleoside triphosphate pools induced by hydroxyurea and their relation to DNA synthesis. J. Biol. Chem. 261, 16037–16042 (1986).
Becker, J. R., Nguyen, H. D., Wang, X. & Bielinsky, A.-K. Mcm10 deficiency causes defective-replisome-induced mutagenesis and a dependency on error-free postreplicative repair. Cell Cycle 13, 1737–1748 (2014).
McGinty, R. J. et al. A Defective mRNA Cleavage and Polyadenylation Complex Facilitates Expansions of Transcribed (GAA)n Repeats Associated with Friedreich’s Ataxia. Cell Rep. 20, 2490–2500 (2017).
Putnam, C. D., Pennaneach, V. & Kolodner, R. D. Saccharomyces cerevisiae as a Model System To Define the Chromosomal Instability Phenotype. Mol. Cell Biol. 25, 7226–7238 (2005).
Yan, Z. et al. Rad52 Restrains Resection at DNA Double-Strand Break Ends in Yeast. Mol. Cell 76, 699–711.e6 (2019).
Fuchs, J., Cheblal, A. & Gasser, S. M. Underappreciated Roles of DNA Polymerase δ in Replication Stress Survival. Trends Genet. https://doi.org/10.1016/j.tig.2020.12.003 (2021).
Pavlov, Y. I., Shcherbakova, P. V. & Kunkel, T. A. In vivo consequences of putative active site mutations in yeast DNA polymerases alpha, epsilon, delta, and zeta. Genetics 159, 47–64 (2001).
Johnson, R. E., Prakash, L. & Prakash, S. Pol31 and Pol32 subunits of yeast DNA polymerase δ are also essential subunits of DNA polymerase ζ. Proc. Natl Acad. Sci. USA 109, 12455–12460 (2012).
Longtine, M. S. et al. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953–961 (1998).
Saxena, S. & Zou, L. Hallmarks of DNA replication stress. Mol. Cell 82, 2298–2314 (2022).
Bacal, J. et al. Mrc1 and Rad9 cooperate to regulate initiation and elongation of DNA replication in response to DNA damage. EMBO J. 37, e99319 (2018).
Granata, M. et al. Dynamics of Rad9 Chromatin Binding and Checkpoint Function Are Mediated by Its Dimerization and Are Cell Cycle–Regulated by CDK1 Activity. PLOS Genet. 6, e1001047 (2010).
Weinert, T. A. & Hartwell, L. H. The RAD9 Gene Controls the Cell Cycle Response to DNA Damage in Saccharomyces cerevisiae. Science 241, 317–322 (1988).
García-Rodríguez, N., Morawska, M., Wong, R. P., Daigaku, Y. & Ulrich, H. D. Spatial separation between replisome- and template-induced replication stress signaling. EMBO J. 37, e98369 (2018).
Waterman, D. P., Haber, J. E. & Smolka, M. B. Checkpoint Responses To Dna Double-Strand Breaks. Annu Rev. Biochem 89, 103–133 (2020).
Pizzul, P. et al. The DNA damage checkpoint: A tale from budding yeast. Front. Genet. 13, 995163 (2022).
Galanti, L. & Pfander, B. Right time, right place—DNA damage and DNA replication checkpoints collectively safeguard S phase. EMBO J. 37, e100681 (2018).
Lanz, M. C., Dibitetto, D. & Smolka, M. B. DNA damage kinase signaling: checkpoint and repair at 30 years. EMBO J. 38, e101801 (2019).
Liu, Y. et al. The intra-S phase checkpoint directly regulates replication elongation to preserve the integrity of stalled replisomes. PNAS 118, e2019183118 (2021).
Hoch, N. C. et al. Molecular Basis of the Essential S Phase Function of the Rad53 Checkpoint Kinase. Mol. Cell. Biol. 33, 3202–3213 (2013).
Osborn, A. J. & Elledge, S. J. Mrc1 is a replication fork component whose phosphorylation in response to DNA replication stress activates Rad53. Genes Dev. 17, 1755–1767 (2003).
Watase, G., Takisawa, H. & Kanemaki, M. T. Mcm10 Plays a Role in Functioning of the Eukaryotic Replicative DNA Helicase, Cdc45-Mcm-GINS. Curr. Biol. 22, 343–349 (2012).
van Deursen, F., Sengupta, S., De Piccoli, G., Sanchez-Diaz, A. & Labib, K. Mcm10 associates with the loaded DNA helicase at replication origins and defines a novel step in its activation. EMBO J. 31, 2195–2206 (2012).
Kanke, M., Kodama, Y., Takahashi, T. S., Nakagawa, T. & Masukata, H. Mcm10 plays an essential role in origin DNA unwinding after loading of the CMG components. EMBO J. 31, 2182–2194 (2012).
Yeeles, J. T. P., Deegan, T. D., Janska, A., Early, A. & Diffley, J. F. X. Regulated eukaryotic DNA replication origin firing with purified proteins. Nature 519, 431–435 (2015).
Kang, Z. et al. BRCA2 associates with MCM10 to suppress PRIMPOL-mediated repriming and single-stranded gap formation after DNA damage. Nat. Commun. 12, 1–12 (2021).
Ricke, R. M. & Bielinsky, A.-K. A Conserved Hsp10-like Domain in Mcm10 Is Required to Stabilize the Catalytic Subunit of DNA Polymerase-α in Budding Yeast. J. Biol. Chem. 281, 18414–18425 (2006).
Haworth, J., Alver, R. C., Anderson, M. & Bielinsky, A.-K. Ubc4 and Not4 Regulate Steady-State Levels of DNA Polymerase-α to Promote Efficient and Accurate DNA Replication. Mol. Biol. Cell 21, 3205–3219 (2010).
Quan, Y. et al. Cell-Cycle-Regulated Interaction between Mcm10 and Double Hexameric Mcm2-7 Is Required for Helicase Splitting and Activation during S Phase. Cell Rep. 13, 2576–2586 (2015).
Douglas, M. E. & Diffley, J. F. X. Recruitment of Mcm10 to Sites of Replication Initiation Requires Direct Binding to the Minichromosome Maintenance (MCM) Complex. J. Biol. Chem. 291, 5879–5888 (2016).
Musiałek, M. W. & Rybaczek, D. Hydroxyurea—The Good, the Bad and the Ugly. Genes (Basel) 12, 1096 (2021).
Lou, H. et al. Mrc1 and DNA polymerase epsilon function together in linking DNA replication and the S phase checkpoint. Mol. Cell 32, 106–117 (2008).
Patrick, S. M. & Turchi, J. J. Stopped-flow Kinetic Analysis of Replication Protein A-binding DNA: DAMAGE RECOGNITION AND AFFINITY FOR SINGLE-STRANDED DNA REVEAL DIFFERENTIAL CONTRIBUTIONS OF kon ANDkoff RATE CONSTANTS*. J. Biol. Chem. 276, 22630–22637 (2001).
Campos, L. V. et al. RTEL1 and MCM10 overcome topological stress during vertebrate replication termination. Cell Rep. 42, 112109 (2023).
Cabello-Lobato, M. J. et al. Physical interactions between MCM and Rad51 facilitate replication fork lesion bypass and ssDNA gap filling by non-recombinogenic functions. Cell Rep. 36, 109440 (2021).
Gangavarapu, V., Santa Maria, S. R., Prakash, S. & Prakash, L. Requirement of Replication Checkpoint Protein Kinases Mec1/Rad53 for Postreplication Repair in Yeast. mBio 2, https://doi.org/10.1128/mbio.00079-11 (2011).
Lewis, J. S. et al. Single-molecule visualization of Saccharomyces cerevisiae leading-strand synthesis reveals dynamic interaction between MTC and the replisome. Proc. Natl Acad. Sci. USA 114, 10630–10635 (2017).
McClure, A. & Diffley, J. Rad53 checkpoint kinase regulation of DNA replication fork rate via Mrc1 phosphorylation. eLife 10, e69726 (2021).
Ait Saada, A. et al. Unprotected Replication Forks Are Converted into Mitotic Sister Chromatid Bridges. Mol. Cell 66, 398–410.e4 (2017).
Durbin, R. M. et al. A map of human genome variation from population-scale sequencing. Nature 467, 1061–1073 (2010).
Baxley, R. M. et al. Bi-allelic MCM10 variants associated with immune dysfunction and cardiomyopathy cause telomere shortening. Nat. Commun. 12, 1626 (2021).
Ji, F. et al. New Era of Mapping and Understanding Common Fragile Sites: An Updated Review on Origin of Chromosome Fragility. Front Genet 13, 906957 (2022).
Kaushal, S. et al. Sequence and Nuclease Requirements for Breakage and Healing of a Structure-Forming (AT)n Sequence within Fragile Site FRA16D. Cell Rep. 27, 1151–1164.e5 (2019).
Generoso, W. C., Gottardi, M., Oreb, M. & Boles, E. Simplified CRISPR-Cas genome editing for Saccharomyces cerevisiae. J. Microbiological Methods 127, 203–205 (2016).
Psakhye, I., Castellucci, F. & Branzei, D. SUMO-Chain-Regulated Proteasomal Degradation Timing Exemplified in DNA Replication Initiation. Mol. Cell 76, 632–645.e6 (2019).
Acknowledgements
The authors would like to thank the Van Deventer lab and Briana Lino for allowing us to use their flow cytometer for this study and members of the Mirkin and Freudenreich lab. We would also like to thank Anja Katrin Bielinsky for the fruitful discussion regarding the project. pFA6a-TRP1-PGAL1 was a gift from John Pringle (Addgene plasmid # 41606). pAO139 (Apr LEU2 mrcl-AQ) was a gift from Stephen Elledge (Addgene plasmid # 41924). pRCC-N was a gift from Eckhard Boles (Addgene plasmid # 81192). Work in the S.M.M. lab was supported by NSF-BSF 2153071 and NIGMS R35GM130322 to S.M.M. Work in the A.A lab was supported by the Israeli Science foundation (ISF) grant number 707/21 to A.A. and the Binational Science Foundation (BSF-NSF) grant numbers 2019617 and 2021737 to A.A.
Author information
Authors and Affiliations
Contributions
Conceptualization and experimental design: C.M., S.M.M. and A.A. Data accumulation: C.M., Z.P., D.D, L.K.B., and S.M. Writing: C.M., S.M.M. All authors reviewed and edited the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Gideon Coster and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Masnovo, C., Paleiov, Z., Dovrat, D. et al. Stabilization of expandable DNA repeats by the replication factor Mcm10 promotes cell viability. Nat Commun 15, 10532 (2024). https://doi.org/10.1038/s41467-024-54977-6
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-024-54977-6