Abstract
The WRN protein is vital for managing perturbed replication forks. Replication Protein A strongly enhances WRN helicase activity in specific in vitro assays. However, the in vivo significance of RPA binding to WRN has largely remained unexplored. We identify several conserved phosphorylation sites in the acidic domain of WRN targeted by Casein Kinase 2. These phosphorylation sites are crucial for WRN-RPA interaction. Using an unphosphorylable WRN mutant, which lacks the ability to bind RPA, we determine that the WRN-RPA complex plays a critical role in fork recovery after replication stress countering the persistence of G4 structures after fork stalling. However, the interaction between WRN and RPA is not necessary for the processing of replication forks when they collapse. The absence of WRN-RPA binding hampers fork recovery, causing single-strand DNA gaps, enlarged by MRE11, and triggering MUS81-dependent double-strand breaks, which require repair by RAD51 to prevent excessive DNA damage.
Similar content being viewed by others
Introduction
The Werner syndrome protein (WRN) is one of the five conserved RECQ helicases in humans and is mutated in the rare genetic disorder Werner syndrome (WS)1,2,3,4. WRN plays a critical role in maintaining genome integrity, particularly during DNA replication, as evidenced by several characteristic phenotypes observed in WS patient-derived or WRN-depleted cells1,5,6. During DNA replication, WRN is essential for multiple functions, including the avoidance of double-strand breaks (DSBs), proper fork recovery, replication of fragile sites, and end-processing of reversed and collapsed forks1,6. Recently, WRN has also been found to participate in fork protection in BRCA2-deficient cells, limit R-loop-associated DNA damage, and assist in stabilising microsatellites7,8,9.
The genome caretaker functions of WRN during DNA replication involve multiple protein-protein interactions with other crucial factors implicated in DNA replication under stressed conditions5,6. One of the most abundant WRN interactors playing a key role in response to perturbed replication is RPA10. The RPA heterotrimer is the major human single-strand DNA (ssDNA) binding protein, which recognises ssDNA formed during DNA replication or repair and acts as a scaffold for other factors involved in response to perturbed replication11,12,13. WRN binds to the N-terminal domain of RPA1 through its acidic domain in vitro and colocalises with RPA at replication foci in human cells10,14,15,16,17. Although the WRN-RPA association stimulates WRN helicase activity on branched substrates that mimic stalled or reversed replication forks in vitro10,14,17,18,19,20, the specific functions of WRN that require interaction with RPA in response to perturbed replication are not fully understood.
In this study, we identified multiple phosphorylation sites in the WRN acidic domain that are targeted by Casein Kinase 2 (CK2) and are essential for the interaction of WRN with RPA both in vitro and in cells. We used the WRN 6 A mutant, which is unphosphorylable by CK2 and defective in RPA binding, to assess the functional relevance of RPA binding during the response to perturbed replication. This mutant contains Ser/Thr to Ala substitutions at all six sites targeted by CK2 but retains the ability to relocalise to ssDNA, similar to wild-type WRN. Characterisation of the response to DNA replication perturbation in cells expressing the WRN 6 A mutant, compared with cells expressing the wild-type WRN, revealed that RPA binding is not involved in WRN-dependent end-processing at stalled or collapsed forks or in limiting the formation of DSBs. In contrast, WRN-RPA interaction is required for properly restarting stalled forks, limiting the accumulation of parental ssDNA gaps, and allowing efficient clearance of G4 DNA. When the WRN-RPA interaction is disrupted or WRN helicase is inhibited, MUS81 contributes to removing G4s producing DSBs downstream of MRE11-dependent gaps. Subsequent RAD51-dependent repair is necessary to limit DNA damage accumulation. These findings clarify the role of WRN binding to RPA in responding to replication stress at G4 structures.
Results
The acidic domain of WRN is phosphorylated by CK2
The high-affinity RPA-binding site of WRN is located in its acidic domain17, and a cluster of high-ranking putative CK2 phosphorylation sites can be identified in this region (Fig. 1a). To assess if the acidic domain of WRN is targeted by CK2, we first expressed GST-tagged WRN N-terminal fragment or GST alone as a control in bacteria. We then phosphorylated these GST fusion proteins in vitro using recombinant CK2 (Supplementary Fig. 1a). Autoradiography of parallel samples readily demonstrated CK2-dependent phosphorylation of the N-terminal WRN fragment. Mass spectrometry analysis of the unlabelled in vitro phosphorylated N-terminal fragment revealed modification by CK2 within the acidic domain of WRN at multiple sites in fragmented species that include the six putative CK2-targeted residues (Supplementary Fig. 1b). The identification of tryptic peptides with multiple modifications is compatible with phosphorylation of all the tandem-arrayed CK2 sites or with phosphorylation of the SDED sequences and one of the threonine or serine residues in the TSYV sequences (Supplementary Fig. 1b). Additionally, an uncertain modification at S426, recently identified as a CDK2 substrate21, was observed (Supplementary Fig. 1b). To confirm the multiple phosphorylation events in the cell and attempt to identify the modified residues, we performed mass spectrometry of full-length WRN transiently expressed in HEK293T cells treated with HU, alone or in the presence of the CK2 inhibitor CX-4945 (CK2i). MALDI-TOF mass spectrometry, followed by MS/MS and a data-independent acquisition strategy to assign probability scores to the multiple phosphorylation events observed in the two tryptic peptides containing the putative CK2 sites, confirmed the presence of at least two CK2-dependent modifications that are increased upon HU treatment (Supplementary Fig. 2a). Analysis of the probability score in the tryptic peptides found to be phosphorylated, including those residually found from CK2i-treated samples, identified S440 and S467 (SDED sites) with a probability score of 1 (100% of probability), whereas phosphorylation of the T434/461 or S435/S462 in the TSYV sequences was equally probable preventing a reliable identification (Supplementary Fig. 2b). The likelihood of T434/461 or S435/S462 residues phosphorylation is decreased in the fragmented species from CK2i-treated samples possibly because phosphorylation at other sites, such as S453 and S426 might be increased. Although only human WRN shows an exact duplication of the region containing these sites, the CK2-targeted residues are highly conserved across vertebrates, (Supplementary Fig. 2c).
a Schematic representation of WRN protein showing its domains. Mutation of six putative CK2 phosphorylation sites in the WRN acidic domain are highlighted. b Anti-Flag-immunoprecipitation from HEK293T cells transiently transfected with Flag-WRN-WT or Flag-WRN-6A plasmid. Cells were treated as indicated 48 h after transfection. Cell extracts were subjected to immunoprecipitation with anti-Flag beads. Immunocomplexes were analysed by Western blotting using the indicated antibodies. c Quantification of phosphorylation of WRN at S440-467 sites and the effects of CK2 or DNA-PK inhibition. Data are presented as mean ±S.E. from two independent replicates. Source data are provided as a Source Data file.
Some of the WRN residues identified as CK2 targets in our study have also been reported to be modified by DNA-PK after DNA damage22. To assess whether the acidic domain of WRN is targeted by CK2 in response to replication arrest, we generated a phospho-specific antibody against WRN phosphorylated at S440 and S467 (pS440/467WRN). This selection was also reinforced by the mass spectrometry data indicating that these sites are found phosphorylated every time the peptide is fragmented while it is impossible to define which of the T434/461 or S435/S462 is modified. This antibody was used to probe cell lysate samples by Western blotting following immunoprecipitation of Flag-tagged wild-type WRN or 6 A mutant transiently expressed in HEK293T cells. Forty-eight hours post-transfection, cells were treated with HU for 2 h in the presence of the CK2i or the DNA-PKcs inhibitor NU7441 (DNA-PKi) to evaluate the contribution of these kinases to WRN phosphorylation. Analysis of anti-Flag IP with anti-pS440/467WRN showed that phosphorylation was detectable even without treatment but increased substantially in response to HU-induced replication arrest (Fig. 1b, c). A similar trend was observed probing the endogenous WRN immunoprecipitated from U2OS cells (Supplementary Fig. 3). Notably, no anti-pS440/467WRN signal was detected in HEK293T cells transfected with the unphosphorylable WRN 6 A mutant, and the WRN 2 A mutant that contains S > A changes only at S440 and 467, confirming the specificity of the antibody for these residues (Fig. 1b and Supplementary Fig. 4a, b). CK2 inhibition reduced HU-induced phosphorylation by approximately 5-fold, while DNA-PK inhibition minimally decreased WRN phosphorylation at S440/467. Combining the two kinase inhibitors did not result in further reduction compared to CK2i alone (Fig. 1c).
Collectively, these results demonstrate that the acidic domain of WRN is phosphorylated at multiple residues by CK2 both in vitro and in human cells, and that CK2, rather than DNA-PK, which was previously shown to target the WRN acidic domain22, is the primary kinase responsible for its phosphorylation in response to replication stress.
Phosphorylation of the WRN acidic domain drives association with RPA
Having demonstrated that the WRN acidic domain is targeted by CK2 at multiple different sites both in vitro and in cells, we sought to determine whether CK2-dependent phosphorylation contributes to the association of WRN with RPA. We generated a WRN fragment containing only the acidic domain (aa 403-503) as a GST-fusion protein in bacteria and used this purified fragment as bait in pull-down assays after in vitro phosphorylation with CK2 (Fig. 2a). To determine that changes in the amount of RPA pulled down were solely due to phosphorylation by CK2 at the six S/TD putative sites, we used the phosphomimetic 6D fragment as an internal control (Fig. 2a). The presence of RPA32 was used as a readout of interaction with the RPA obligate heterotrimer, and the phosphorylation status of the CK2 sites was assessed using the anti-pS440/467WRN antibody. The wild-type WRN fragment was greatly phosphorylated, as shown by staining with the anti-pS440/467WRN antibody, while minimal cross-reactivity was detected in the phosphomimetic mutant fragment (Fig. 2b). Consistent with the previous studies17, the mock-phosphorylated WRN403-503 fragment could pull-down RPA, but the amount of RPA32 bound to this fragment increased more than 4-fold via prior phosphorylation by CK2 (Fig. 2b). Notably, the phosphomimetic WRN6D403-503 fragment exhibited increased association with RPA, which was comparable to that of the phosphorylated wild-type WRN fragment (Fig. 2b). These observations suggest that the phosphomimetic changes to resides within WRN sequence 403-503 are responsible for the increased association of WRN with RPA, even if CK2 phosphorylates additional sites in WRN. The phosphorylation status of the six S/T CK2-targeted residues also influenced WRN interaction with RPA in cells. Indeed, Co-IP experiments using extracts from HEK293T cells transiently transfected with Flag-tagged wild-type or unphosphorylable S/T > A (6 A) WRN proteins showed that WRN association with RPA increased during replication fork arrest induced by HU. This association was reduced of about 2-fold for the unphosphorylable mutant (Fig. 2c).
a Ponceau staining of GST pull-downs performed with nuclear extracts from HEK293T cells and GST-tagged WRN fragment 403-503 (WRNwt and WRN6D). The WRN fragments from “a” were phosphorylated by CK2 in the presence or absence of ATP. Western blotting analysis in b shows WRN S440/467 phosphorylation and the RPA32 subunit from GST pull-downs. The graph shows the levels of S440/467 phosphorylation and RPA32 bound to GST-tagged WRN fragments. Data are presented as mean from two independent replicates. c Anti-Flag-immunoprecipitation from HEK293T cells transfected with Flag-WRN-WT or Flag-WRN-6A plasmid. Cells were treated as indicated 48 h after transfection. Cell extracts were subjected to immunoprecipitation with anti-Flag beads. Immunocomplexes were analysed by Western blotting using the indicated antibodies. The graph presents the quantification of the WRN-normalised amount of RPA70 in the anti-Flag immunoprecipitate from two independent experiments (mean of each is shown). d Anti-RPA70 immunoprecipitation from HEK293T cells transfected with Flag-WRN-WT or Flag-WRN-6A plasmid. Cells were treated as indicated 48 h after transfection. Immunoprecipitation was performed using anti-RPA70-conjugated beads and immunocomplexes analysed by Western blotting using the indicated antibodies. The graph shows the quantification of the RPA70-normalised amount of WRN in the anti-RPA70 immunoprecipitate from two independent experiments (mean of each is shown). e Anti-RPA32 immunoprecipitation from HEK293T cells transfected with Flag-WRN-WT plasmid. Cells were treated as indicated 48 h after transfection. Immunocomplexes were analysed by Western blotting using the indicated antibodies (the gel is representative of one of the two independent repeats). f Interaction between WRN and RPA32 in Werner Syndrome (WS) cells expressing Flag-WRNwt or Flag-WRN6A and treated as indicated. In situ PLA was performed with anti-Flag and anti-RPA32 antibodies. The graph shows individual PLA spot values from each condition (from 166 to 280 nuclei). Representative images are provided. Bars represent mean ± S.E. (n = 3; two-tailed Mann-Whitney test). Scale bar = 20 µm. Source data are provided as a Source Data file.
To further demonstrate the relevance of the WRN acidic domain’s phosphorylation status for its association with RPA, we immunoprecipitated RPA70 from cells transiently expressing either WRN wild-type or 6 A and analysed WRN presence and phosphorylation status. As shown in Fig. 2d, the interaction of RPA with WRN was enhanced already at 2 hours of HU exposure and remained elevated at 6 hours. The level of S440/467 phosphorylation followed a similar trend, suggesting that most, if not all, of WRN in the RPA complex is modified by CK2. Consistent with this, extremely low levels of WRN were co-precipitated with RPA from cells expressing the unphosphorylable WRN 6 A mutant, as assessed by anti-Flag Western blotting (Fig. 2d).
To confirm that interaction between WRN and RPA requires phosphorylation by CK2 but not DNA-PK, we immunoprecipitated RPA32 from cells expressing wild-type WRN and treated with HU in the presence or absence of CK2i or DNA-PKi. As expected, co-immunoprecipitation of RPA and WRN was stimulated by replication arrest and was almost completely abrogated by CK2 inhibition (Fig. 2e). Of note, and consistent with phenotypic data shown later, DNA-PK inhibition did not prevent the formation of the WRN-RPA complex after replication arrest (Fig. 2e). Abrogation of CK2-dependent phosphorylation of WRN did not undermine interaction with MRE11 or DNA2, which are two other functionally relevant partners of WRN23,24 (Supplementary Fig. 5). Of interest, the WRN-6A mutant immunoprecipitated much more MRE11 and DNA2 than its wild-type counterpart.
To further validate that the WRN-RPA interaction depends on the phosphorylation status of the WRN acidic domain at the single-cell level, we performed anti-Flag/RPA32 PLA in WS-derived patient cells complemented with Flag-tagged wild-type WRN or the 6 A unphosphorylable mutant. Consistent with co-IP data (see Fig. 2c), PLA showed that WRN’s interaction with RPA is strongly stimulated by replication arrest and is reduced of about 2-fold when phosphorylation is abrogated by treatment with CK2i or by the 6 A mutations (Fig. 2f). PLA experiments also showed that WRN/RPA interaction is stimulated by HU in a time-dependent manner and that, at 6 h, an increase is observed also in the unphosphorylable mutant (Supplementary Fig. 6).
Since association with RPA often facilitates the recruitment of proteins to blocked replication forks, we performed PLA experiments to monitor WRN 6 A association with parental ssDNA, accumulated at blocked replication forks, or nascent ssDNA, formed at reversed forks or processed collapsed forks. Despite the fact that the association with RPA is severely impaired, the WRN 6 A mutant retained almost complete proficiency to bind ssDNA exposed at parental or nascent strands after replication arrest (Supplementary Fig. 7a, b). Consistent with this, the chromatin localisation of both wild-type WRN and 6 A mutant was only slightly reduced after HU exposure (Supplementary Fig. 7c).
Altogether, these results indicate that the WRN’s association with RPA, but not with chromatinized DNA, is strongly dependent on the phosphorylation status of the six CK2-targeted residues in the acidic domain of WRN, and that this interaction is only minimally required for WRN recruitment in response to replication fork arrest.
WRN-RPA-binding is required for recovery from replication arrest
In our experiments, deletion of the WRN acidic domain greatly affected protein expression, possibly because of destabilisation in the cell (Supplementary Fig. 8a, b). Thus, the unphosphorylable WRN 6 A mutant, which exhibits compromised RPA-binding but normal expression and association with perturbed forks, serves as a useful tool to probe the functional role of the WRN-RPA interaction in cells. WRN is known to play critical roles at perturbed replication forks, including assisting DNA2 in exonucleolytic processing at reversed forks and limiting pathological degradation by MRE1125,26. The contribution of the WRN-RPA interaction to these mechanisms is unknown. As a proxy for the degradation occurring at reversed forks26, we first evaluated the accumulation of nascent ssDNA at different times during HU treatment in WS cells complemented with either wild-type or the 6 A WRN protein. The exposure of nascent ssDNA increased significantly after 6 hours of HU in cells expressing both wild-type WRN and the RPA-binding defective 6 A mutant, with no significant difference between the two (Supplementary Fig. 9a). To further assess that RPA binding by WRN was not required for this function, we performed DNA fibre assays (Supplementary Fig. 9b). Nascent DNA was sequentially pulse-labelled with CldU and IdU, followed by HU treatment. Analysis of the IdU/CldU ratios revealed no statistically significant difference between WS cells complemented with wild-type WRN and those with the 6 A mutant (Supplementary Fig. 9b). In both cases, the IdU/CldU ratios were increased by the MRE11 inhibitor, MIRIN (MRE11i), confirming that a fraction of forks underwent degradation after 6 hours of HU, regardless of WRN ability to bind RPA. Loss of WRN function is known to trigger DSB formation and stimulate RAD51-dependent repair27,28 and WRN has also been implicated in promoting long-range end-degradation at collapsed replication forks23,29. To test whether loss of WRN-RPA binding affects end resection at collapsed forks, we measured DSBs and ssDNA formation after treatment with HU and the topoisomerase inhibitor camptothecin (CPT), which induces replication stress and DSBs, by neutral Comet assay and the native IdU/ssDNA assay29. As shown in Supplementary Fig. 9c and 10, loss of WRN-RPA binding did not affect the formation of DSBs at perturbed replication forks and minimally affects the end-resection after their collapse, if compared with the end resection-defective S1133A WRN mutant29, suggesting that the small reduction of nascent ssDNA formed at collapsed forks is not of biological significance.
Given that RPA-binding by WRN is dispensable during end-processing at perturbed replication forks, we next analysed whether the WRN-RPA interaction affects replication fork restart and recovery. To this end, WS cells transiently complemented with either the wild-type WRN or the WRN 6 A mutant were pulse-labelled with CldU, treated with HU for 6 hours, followed by 20 min recovery in IdU before DNA fibre spreading (see scheme in Fig. 3a). Analysis of the IdU/CldU ratios from DNA fibres showed shorter IdU tracts in WS cells and in cells expressing the RPA-binding deficient WRN 6 A mutant (Fig. 3a).
a Analysis of replication fork restart using DNA fibre assay. WS cells or WS cells expressing Flag-WRNwt or Flag-WRN6A plasmid were treated 48 h thereafter as indicated in the experimental scheme. The graph displays individual IdU/CdU ratios from 100 DNA fibres after duplicated experiments. Bars represent mean ± S.E. (ns = not significant; ****P < 0.0001; Two-tailed Mann-Whitney test for paired samples). Representative images of DNA fibres from a random field are provided. Scale bar 10 µm. b Analysis of parental ssDNA exposure in WS cells or WS cells expressing Flag-WRNwt or Flag-WRN6A plasmid. Cells were treated as indicated in the experimental scheme. The graph shows the quantification of total IdU intensity per nucleus from three independent experiments. Bars represent mean ± S.E. (ns = not significant; *P < 0.05; ***P < 0.001; ****P < 0.0001; Two-tailed Mann-Whitney test for paired samples). Representative images of native anti-IdU immunofluorescence are provided. Scale bar = 20 µm. c Analysis of replication fork recovery using DNA fibre assay in WS cells expressing Flag-WRNwt or Flag-WRN6A and treated as indicated in the experimental scheme. The graph displays individual IdU/CdU ratio values from two independent replicates (at least 25 fibres each). Bars represent mean ± S.E. (Two-tailed Mann-Whitney test for paired samples). Representative images of DNA fibres from a random field are provided. Scale bar 10 µm. Source data are provided as a Source Data file.
Given the impact of RPA-binding on WRN ability to support DNA replication, we wondered whether cells expressing the WRN 6 A mutant might exhibit persistent ssDNA gaps after replication fork recovery. To test this, we analysed the presence of parental ssDNA after 6 hours of HU and subsequent recovery, using native IdU immunofluorescence in WS cells transiently complemented with either an empty vector, wild-type WRN or the 6 A mutant (Fig. 3b). HU treatment resulted in a fraction of cells showing some parental ssDNA exposure, with no significant difference between WS cells, cells expressing wild-type WRN or 6 A mutant (Fig. 3b). Notably, while the amount of parental ssDNA decreased greatly during recovery in WS cells and in cells expressing wild-type WRN, this reduction was less evident in cells expressing the WRN 6 A mutant (Fig. 3b). In both genetic backgrounds, a portion of the parental ssDNA exposed during HU treatment was dependent on MRE11 exonuclease activity, as it was reduced by the MRE11 inhibitor (Supplementary Fig. 11). This finding is consistent with the DNA fibre degradation assay shown in Supplementary Fig. 9b, confirming that a sub-population of stalled forks undergoes degradation after 6 hours of HU exposure. Strikingly, in both wild-type WRN or 6A-expressing cells, all the residual parental ssDNA exposed during recovery was DNA2-independent (Supplementary Fig. 12). Inhibition of CK2 led to significantly elevated exposure of parental ssDNA during HU treatment but not exceeding the levels of parental ssDNA detected in the presence of WRN 6 A mutant (Supplementary Fig. 12). Although inhibition of CK2 led to more parental ssDNA exposure in wild-type cells during recovery if compared with the WRN 6 A mutant, CK2 inhibition did not affect the amount of parental ssDNA observed in those cells (Supplementary Fig. 12).
These results suggest that, in the absence of WRN-RPA binding, cells accumulate parental ssDNA. This phenotype often correlates with the accumulation of daughter strand gaps. To determine if the increased detection of parental ssDNA derived from processing of DNA gaps, we performed the DNA fibre assay in cells treated with the S1 nuclease after replication arrest and recovery (Fig. 3c). Of note, while the IdU/CldU ratio in the absence of S1 treatment did not differ between wild-type cells and cells expressing WRN 6 A, treatment with S1 led to shorten IdU-labelled replication tracts in those cells (Fig. 3c), indicating the presence of daughter strand gaps. Since cells expressing WRN 6 A mutant did not show increased degradation at reversed forks compared to wild-type cells, we hypothesised that the parental gaps might result from repriming by PRIMPOL. To test this hypothesis, we repeated the analysis of parental ssDNA in cells transfected or not with siRNA targeting PRIMPOL (Supplementary Fig. 13a). Based on previous studies, we anticipated that PRIMPOL depletion would reduce parental ssDNA exposure if the gaps derived from its repriming activity. Interestingly, PRIMPOL depletion did not reduce the level of parental ssDNA exposed in either wild-type WRN or WRN 6A-expressing cells (Supplementary Fig. 13a). However, depletion of PRIMPOL in cells expressing WRN 6 A reverted the S1-dependent reduction in the IdU tract lengths of DNA fibres observed in wild-type cells (Supplementary Fig. 13b), suggesting that PRIMPOL-dependent DNA gaps are formed when the interaction between WRN-RPA is perturbed by loss of CK2-dependent phosphorylation of the WRN acidic domain even if residual parental ssDNA is not affected.
Collectively, these results indicate that RPA-binding by WRN is crucial for accurate replication fork progression under both unchallenged and perturbed conditions. Proper formation of the WRN-RPA complex enables cells to recover from perturbed replication without accumulating PRIMPOL-dependent ssDNA gaps that become targets of MRE11-dependent degradation.
RPA-binding and helicase activity of WRN promote clearance of G4-DNA
We show that RPA-binding by WRN is important for recovery of perturbed replication forks. In vitro, RPA facilitates WRN unwinding and fork regression, enhancing WRN helicase activity10,17,19. Thus, we sought to determine whether loss of RPA-binding and the helicase function of WRN act in the same pathway by combining the expression of WRN 6 A mutant with catalytic inhibition of the helicase using a small molecule inhibitor (WRNi)7. First, we analysed the recovery of stalled forks using the DNA fibre assay (see scheme in Supplementary Fig. 4a). Pharmacological inhibition of WRN helicase activity strongly reduced the ability of cells expressing the wild-type WRN to recover stalled forks as shown by the reduced length of the IdU-labelled, restarting, tract (Fig. 4a). As expected, the expression of the WRN 6 A mutant, which is defective in RPA-binding, also undermined fork recovery (Fig. 4a). Interestingly, although treatment of cells expressing WRN 6 A with the WRNi further reduced the recovery of stalled forks, the effect was milder if compared to wild-type cells (Fig. 4a). Notably, both the number of restarted forks and the fork progression during recovery (i.e., the length of the IdU tracts) were diminished by the compromised ability of WRN to bind RPA (Fig. 4a, see % of restarting of forks in insets). Notably, the reduced progression of stalled forks observed in cells expressing WRN 6 A after recovery was also characteristic of WS cells (Supplementary Fig. 14a)
a Analysis of replication fork recovery using DNA fibre assay in WS cells expressing Flag-WRNwt or Flag-WRN6A and treated as indicated in the experimental scheme. The graph displays individual IdU/CdU ratio values from two independent replicates (50 DNA fibres each experiment). Bars represent mean ± S.E. The numbers in the inset boxes above each dot plot indicate the percentage of restarting forks (mean ± S.E). (Two-tailed Mann-Whitney test for paired samples). Representative images of DNA fibres from a random field are provided. Scale bar 10 µm. b Analysis of parental ssDNA exposure in WS cells expressing Flag-WRNwt or Flag-WRN6A and treated as indicated in the experimental scheme. The graph quantifies total IdU intensity per nucleus from three independent experiments (at least 40 nuclei/repeat). Data points from inhibited cells have a black border. Bars represent mean ± S.E. (ns = not significant; * P < 0.05; *** P < 0.001; Two-tailed Mann-Whitney test for paired samples). Representative images of native anti-IdU immunofluorescence are shown. Scale bar = 20 µm. c Helicase reactions containing radiolabeled TelXY forked duplex substrate (0.5 nM) and increasing concentration of WRN-WT and WRN-6A proteins (0.5, 1, 2, 4, 8 and 16 nM). d Evaluation of helicase activity of WRN-WT (2 nM) and WRN-6A (2 nM) with increasing amount of RPA (1, 2, 4, 8, 16 nM). NE indicates no enzyme control. Black triangles indicate heat denatured substrate. The autoradiographs shown in (c and d) are representative of two experiments. Source data are provided as a Source Data file.
Next, we investigated whether the increased exposure of parental ssDNA observed during recovery from replication arrest in cells expressing WRN 6 A could be phenocopied in WRN wild-type cells by treatment with the WRNi. While inhibition of the WRN helicase failed to change the amount of parental ssDNA observed during HU treatment irrespective of the WRN status, during recovery from HU, inhibition of WRN helicase resulted in increased parental ssDNA exposure in cells expressing the wild-type WRN but not in those expressing the WRN 6 A mutant (Fig. 4b). Notably, during recovery, wild-type cells treated with the WRNi and cells expressing the RPA-binding deficient WRN 6 A exposed comparable levels of parental ssDNA (Fig. 4b). Similar to cells expressing WRN 6 A, WRNi treatment did not affect the level of parental ssDNA in cells expressing the helicase-defective K577M WRN (Supplementary Fig. 14b), reinforcing the hypothesis that formation of the WRN/RPA complex is involved in the unwinding of blocked forks in the cell.
Since inhibition of WRN helicase in wild-type cells mimicked the phenotype of the WRN 6 A mutant, we performed in vitro assays to investigate whether CK2 phosphorylation at the WRN acidic domain regulates its helicase activity. To this end, we purified recombinant wild-type WRN and verified whether the recombinant WRN purified from insect cells was already phosphorylated at the CK2 sites of the acidic domain using the anti-pS440/467 WRN antibody. Interestingly, the recombinant wild-type WRN purified from insect cells was phosphorylated at CK2 sites (Supplementary Fig. 15). Thus, we expressed and purified from insect cells the unphosphorylable WRN-6A mutant and assessed both for their enzymatic activity. The wild-type and WRN-6A proteins were tested for catalytic activity on a canonical TelXY forked duplex substrate with 34 bp duplex and 15 nt tails (forked DNA substrate), which is unwound by WRN in the presence of ATP or degraded by WRN’s 3′-5′ exonuclease activity in the absence of ATP. We tested serially increasing amounts of WRN-WT and WRN-6A and observed that each enzyme was able to unwind the TelXY substrate equally well (Fig. 4c). Subsequently, we tested their helicase activities in the presence of increasing concentrations of RPA (from 1 to 16 nM). Helicase activities of the wild-type and the mutant were equally stimulated by RPA and showed nearly complete unwinding at the highest RPA concentration (Fig. 4d). RPA alone showed a marginal fork substrate destabilising effect at the highest concentration. The apparently normal ability of WRN-6A to unwind DNA in vitro suggests that there is no major structural impact of the six S/T mutations into A on WRN catalytic activity.
We then subjected the purified WRN to dephosphorylation/phosphorylation using lambda phosphatase, followed by the addition of CK2 kinase, prior to analysing its enzymatic activity. The activity of WRN was compared to that of the dephosphorylated form, which was mock-treated after the dephosphorylation. The dephosphorylated and CK2-rephosphorylated WRN proteins were tested for catalytic activity on a forked duplex DNA substrate, which is unwound by WRN in the presence of ATP or degraded by WRN’s 3′-5′ exonuclease activity in the absence of ATP. As shown in Supplementary Fig. 16, no apparent difference in the helicase or exonuclease activity was detected between the unphosphorylated WRN and CK2 -rephosphorylated WRN recombinant proteins, even in the presence of RPA.
WRN has been shown to catalyse the unwinding of G-quadruplex (G4) DNA substrates in vitro, with RPA-binding enhancing WRN activity20,30. We, therefore, sought to determine whether the compromised ability of the RPA-binding deficient WRN to resume stalled forks was correlated with reduced helicase activity toward G4s. To test this, we first evaluated the presence of G4s by anti-BG4 immunofluorescence in WS cells and in cells expressing WRN wild-type or 6 A, with or without WRNi treatment. Untreated cells showed little BG4 staining, irrespective of the RPA-binding capability of WRN although WS cells had more intense staining (Fig. 5a). The inhibition of WRN helicase by the WRNi increased the intensity of BG4 staining significantly only in wild-type and WRN 6 A (Fig. 5a). During recovery from HU, WS cells showed an elevated level of BG4 staining that was not further increased by the WRNi whereas it was further heightened by co-treatment with the WRNi in wild-type cells (Fig. 5a). Notably, the impaired ability of WRN to bind RPA substantially increased BG4 staining after recovery, matching the levels observed in WS cells or in wild-type cells after inhibition of WRN helicase. No further increase in BG4 staining was observed in cells expressing WRN 6 A treated with WRNi (Fig. 5a). To determine if BG4 staining detected G4s at stalled forks, we performed a PLA assay to assess proximity of BG4 nanobody to EdU-labelled nascent strand at stalled forks (SIRF assay). SIRF assay confirmed that significantly more G4s were detected at stalled forks in the absence of WRN (WS cells) or when the WRN 6 A mutant is expressed (Fig. 5b).
a Analysis of G4 structures by immunofluorescence and an anti-DNA G-quadruplex antibody (clone BG4) in WS cells or WS cells expressing Flag-WRNwt or Flag-WRN6A plasmid. Cells were treated as indicated. The graph quantifies total BG4 nuclear staining per nucleus from two independent experiments (at least 80 nuclei/repeat). Data points from inhibited cells have a black border. Bars represent mean ± S.E. (ns = not significant; *P < 0.05; **P < 0.01: Two-tailed Mann-Whitney test for paired samples). Representative images are provided. Scale bar = 20 µm. b SIRF analysis of the localisation of BG4 at EdU-labelled nascent DNA by in situ PLA in WS cells expressing Flag-WRNwt or Flag-WRN6A. To mark nascent DNA at stalled forks, cells were treated with EdU 8 min before being treated with HU. The graph quantifies total BG4 SIRF spots per nucleus from two independent experiments (at least 80 nuclei/repeat). Bars represent mean ± S.E. (ns = not significant; ****P < 0.0001; Kruskal-Wallis with Dunn’s test). Representative images are provided. Scale bar = 20 µm. c Analysis of G4 structures detection as in a) in WS cells expressing Flag-WRNwt or Flag-WRN6A. Cells were treated as indicated and recovered with or without WRN inhibitor (WRNi). The graph shows quantification of total BG4 nuclear staining per nucleus from two independent experiments (at least 80 nuclei/repeat). Data points from inhibited cells have a black border. Bars represent mean ± S.E. (ns = not significant; **P < 0.01; ****P < 0.0001; Two-tailed Mann-Whitney test for paired samples). Representative images are provided. Scale bar = 20 µm. Source data are provided as a Source Data file.
Having demonstrated that impaired helicase activity or RPA-binding by WRN leads to the accumulation of G4s and their persistence during the restart of stalled replication forks, we assessed whether these persisting G4s were eventually resolved. WS cells complemented with wild-type WRN or the 6 A mutant were treated with HU and allowed to recover for 1 or 18 hours before evaluating the presence of G4s by BG4 immunofluorescence. To further investigate the contribution of the WRN helicase activity, parallel samples were treated with WRNi during the 18 hours of recovery from HU. Interestingly, after 18 hours of recovery, cells expressing WRN 6 A or with inhibited WRN helicase showed dissolution of G4s, returning to wild-type levels (Fig. 5c).
Further confirming that CK2 and not DNA-PK is the kinase involved in regulating WRN in response to stalled replication, treatment with CK2i but not DNA-PKi increased BG4 staining in cells expressing WRN wild-type (Supplementary Fig. 17a, b).
These results demonstrate that impairment of RPA-binding by WRN is sufficient to induce the accumulation of G4s shortly after recovery from replication arrest, mimicking the effect of WRN helicase inhibition. Since the unphosphorylable WRN-6A mutant manifested normal ability to unwind a standard forked substrate, we next evaluated if it showed any defective unwinding of a G4-containing substrate. First, the serially increasing amounts of each protein were tested on a tetramolecular parallel TP-G4 substrate. The WRN-WT and WRN-6A were able to resolve TP-G4 structure in a nearly identical concentration-dependent manner (Fig. 6a). Next, we tested whether RPA could stimulate the G4-resolving activity of WRN. The WT and the 6 A mutant proteins were equally active and did not show a stimulatory effect when incubated with increasing amounts of RPA. Moreover, each protein seemed to be inhibited at the higher concentrations of RPA (8 and 16 nM, Fig. 6b). RPA alone did not have any effect on the substrate.
a Helicase reactions containing radiolabeled tetramolecular parallel TP-G4 substrate (0.25 nM) and increasing concentration of WRN-WT and WRN-6A proteins (0.5, 1, 2, 4, 8 and 16 nM). b Evaluation of helicase activity of WRN-WT (2 nM) and WRN-6A (2 nM) with increasing amount of RPA (1, 2, 4, 8, 16 nM). NE indicates no enzyme control. M: marker of single-stranded TP-G4 oligonucleotide. Autoradiographs are representative of two independent replicates. Source data are provided as a Source Data File.
Although CK2-dependent phosphorylation of WRN does not impair its enzymatic activity on a forked duplex or G4 substrate in vitro, these findings collectively show that G4s accumulate in cells treated with HU when WRN binding to RPA or WRN helicase activity is impaired although they are eventually resolved after prolonged recovery.
Phosphorylation of WRN S440 and S467 is sufficient to safeguard against G4s
Mass spectrometry indicates that CK2 targets S440 and S467 of WRN and at least another one of the residues in the TSYVIE sequence. This uncertainty supported the use of the fully-unphosphorylable WRN mutant in functional studies. However, mass spectrometry also identified S440 and S467 as the phosphorylated residues in tryptic peptides with only one phosphate (see Supplementary Fig. 2). Thus, we tested the relevance of these residues for the functional roles of WRN during the recovery of stalled forks at G4s by generating a mutant with S > A changes only at S440 and 467 (WRN-2A). First, we immunoprecipitated RPA32 from cells transiently expressing either WRN wild-type, 2 A or 6 A and analysed WRN presence and phosphorylation status. As shown in Fig. 7a, the interaction of RPA with WRN was almost completely abrogated in cells expressing the WRN-2A without any detectable divergence from the WRN-6A. The level of S440/467 phosphorylation followed a similar trend, confirming that most, if not all, of WRN in the RPA complex is modified by CK2 and that modification of S440 and 467 is sufficient to support the formation of the WRN/RPA complex in the cell. Consistent with this, extremely low levels of WRN-RPA32 PLA spots were detected in response to 2 hours of HU in WS cells expressing the unphosphorylable WRN 2 A mutant (Fig. 7b).
a Anti-Flag-immunoprecipitation from HEK293T cells transfected with Flag-WRNwt, Flag-WRN6A or Flag-WRN2A plasmid. Cells were treated as indicated 48 h after transfection and exposed or not to a CK2i. Cell extracts were subjected to immunoprecipitation with anti-RPA32 beads. Immunocomplexes were analysed by Western blotting using the indicated antibodies. b In situ PLA between WRN and RPA32 in WS cells nucleofected with Flag-WRNwt, Flag-WRN2A or Flag-WRN6A expression plasmid. Cells were treated as indicated. The graph shows individual values of PLA spots from two different experiments (at least 100 nuclei/repeat). Bars represent mean ± S.E. (ns = not significant; **P < 0.01; ****P < 0.0001; Kruskal-Wallis test for non-paired samples with Dunn’s correction). Representative images are provided. Scale bar = 20 µm. c G4s accumulation was detected by anti-BG4 immunofluorescence in cells expressing Flag-WRNwt Flag-WRN6A or Flag-WRN2A plasmid. Cells were treated as indicated and recovered for 1 h in the presence or absence of the CK2 inhibitor. The graph displays individual values of BG4 foci nuclear intensity from three independent experiments. At least 50 nuclei/experiment were reported in the plot. Bars represent mean ± S.E. (ns = not significant; ****P < 0.0001. Where not indicated, values are not significant; Kruskal-Wallis test for non-paired samples with Dunn’s correction). Representative images are provided. Scale bar = 20 µm. d Analysis of replication fork recovery using DNA fibre assay in WS cells expressing Flag-WRNwt Flag-WRN6A or Flag-WRN2A plasmid treated 48 h thereafter as indicated. The left graph displays the percentage of restarting forks. Bars represent mean ± S.E. The numbers in the boxes above each dot plot indicate the percentage of restarting forks. The right graph shows individual IdU/CdU ratio values (n = 54) from two independent replicates. Bars represent mean ± S.E (Two-tailed Mann-Whitney test for paired samples). The total number of forks analysed is reported above each plot. Representative images of DNA fibres from a random field are provided. Scale bar 20 µm. Source data are provided as a Source Data file.
To assess if WRN phosphorylation by CK2 at S440/467 was sufficient to prevent G4 persistence after recovery from replication arrest induced by HU, we performed BG4 staining in WS cells complemented with WRN wild-type or the two phosphorylation-defective mutants WRN 6 A or 2 A. As expected from our previous experiments, cells expressing WRN wild-type showed a modest increase in BG4 staining during recovery, which is increased further upon inhibition of CK2 (Fig. 7c). In contrast, cells expressing WRN 6 A or 2 A displayed significantly higher levels of BG4 staining during recovery from HU, which was not further affected by treatment with CK2i (Fig. 7c). Since persistence of G4s, as inferred from elevated BG4 staining, correlated with poor fork recovery (see Fig. 4), we compared fork progression between cells expressing the WRN 6 A and 2 A mutants using dual labelling and DNA fibres. As shown in Fig. 7d, even the abrogation of just S440/467 phosphorylation is sufficient to undermine fork recovery and progression after HU treatment.
Collectively, these results indicate that the formation of the WRN/RPA complex in the cells in response to HU-induced replication arrest requires at least modification of S440 and S467 by CK2, and that abrogation of phosphorylation at these two residues of WRN is sufficient to recapitulate the phenotypic changes observed in the WRN 6 A mutant. In addition, these findings suggest that WRN 6 A and 2 A can be both used to investigate the functional significance of the WRN/RPA complex in the cell.
MUS81-dependent DSBs form in the absence of WRN-RPA binding
Collectively, our data support a model in which the binding of WRN to RPA is essential for the recovery of replication forks and the accurate replication of the secondary DNA structures, such as G4s. This prompted us to investigate the relationship between the persistent parental gaps generated by MRE11 and the removal of G4s observed during extended recovery periods after replication fork arrest in cells expressing the WRN 6 A mutant. To explore this, we treated cells with HU and analysed the presence of G4s using immunofluorescence after 1 hour and 18 hours of recovery, with or without the MRE11 inhibitor. Inhibiting MRE11 exonuclease activity decreased BG4 staining in wild-type cells after 1 hour of recovery but had no effect after 18 hours (Supplementary Fig. 18a). Although this decrease was significant, the overall amount of HU-dependent G4s detected by anti-BG4 immunofluorescence was exceptionally low in wild-type cells. In contrast, inhibiting MRE11 exonuclease activity greatly increased the already elevated anti-BG4 staining in cells expressing WRN 6 A (Supplementary Fig. 18a). The observation that G4s removal depends on MRE11 exonuclease activity suggests that DSBs are formed and resected at G4 sites.
In human cells, MUS81 endonuclease can process G4s and is known to introduce DSBs in the absence of WRN28,31. We depleted MUS81 using RNAi (Fig. 8a) and analysed whether DSBs formed during recovery from HU in cells expressing WRN 6 A. The neutral comet assay revealed very low levels of DSBs in cells expressing wild-type WRN after 1 hour of recovery from HU, and those DSBs were MUS81-independent (Fig. 8a). However, significantly more DSBs were found in cells expressing WRN 6 A during recovery, and these were suppressed by MUS81 depletion (Fig. 8a). Notably, DSBs formed in cells expressing the WRN 6 A mutant during recovery were also strongly reduced by MIRIN (Supplementary Fig. 18b), confirming that MRE11-dependent gap enlargement acts upstream of MUS81. This MUS81-dependent formation of DSBs prompted us to analyse the presence of G4s using anti-BG4 immunofluorescence to correlate them with G4s clearance (Fig. 8b). Depletion of MUS81 reduced the already low level of BG4 staining in WS cells complemented with wild-type WRN after 1 hour of recovery. In contrast, depletion of MUS81 increased the level of G4s in cells expressing WRN 6 A mutant. To show that the G4s affected by MUS81 depletion in the WRN 6 A mutant were localised at perturbed forks, we performed BG4-SIRF experiments. As shown in Fig. 8c, an increased fraction of G4s were found in proximity of EdU-labelled nascent DNA in cells expressing the WRN 6 A mutant.
a Analysis of DSBs using neutral Comet assay in WS cells expressing Flag-WRNwt or Flag-WRN6A. Cells were transfected with control (CTRL) or MUS81 siRNA, 48 h thereafter were treated as indicated and then allowed to recover for 1 h. Western blotting confirms MUS81 downregulation. The graph shows individual tail moment values of at least 120 cells from duplicated experiments (n = 3). Bars represent mean ± S.E. Statistical analyses were performed by two-tailed Mann-Whitney test, where not indicated, values are not significant. Representative images are provided. b Analysis of G4 accumulation evaluated by anti-BG4 immunofluorescence. The graph displays individual values of BG4 nuclear intensity of at least 160 cells from different experiments. Bars represent mean ± S.E. (Two-tailed Mann-Whitney test; Where not indicated, values are not significant). Representative images are provided. Scale bar = 20 µm. c SIRF analysis of the localisation of BG4 at EdU-labelled nascent DNA by in situ PLA in WS cells expressing Flag-WRNwt or Flag-WRN6A after transfection with MUS81 siRNAs. The graph quantifies total BG4 SIRF spots per nucleus of at least 180 cells from two independent experiments. Bars represent mean ± S.E. (Two-tailed Mann-Whitney test for paired samples). Representative images are provided. Scale bar = 20 µm. Source data are provided as a Source Data file.
Altogether, these results provide strong evidence for the crucial role of MRE11 and MUS81 in the removal of G4 structures that fail to be resolved due to defective interaction between WRN and RPA during replication fork recovery. Furthermore, these findings establish a link between gap processing and the formation of DSBs, highlighting the interplay between these processes in maintaining genome stability.
RAD51 repairs DSBs formed at G4s in the absence of WRN-RPA binding
Our findings demonstrate that resolving G4 structures without WRN-RPA binding requires the presence of both MRE11 and MUS81. MRE11-enlarged gaps can act as sites for RAD51 recruitment, facilitating post-replication gap repair. To investigate whether RAD51 is recruited to parental ssDNA exposed at DSBs formed by MUS81 and through MRE11-dependent degradation of the newly synthesised DNA, we conducted a parental ssDNA-protein PLA26. As shown in Supplementary Fig. 18c, RAD51 was associated with parental ssDNA in cells expressing wild-type WRN after recovery from HU, and this association was minimally affected by inhibiting MRE11 exonuclease. In contrast, cells expressing WRN 6 A mutant exhibited higher levels of RAD51 associated with parental ssDNA, which were significantly reduced by treatment with MIRIN (Supplementary Fig. 18c). Furthermore, depletion of MUS81 in cells expressing WRN 6 A substantially, but not completely, reduced RAD51 recruitment to parental ssDNA (Fig. 9a), suggesting that RAD51 may also play a role in repairing MUS81-dependent DSBs. We hypothesised that if RAD51 was engaged in post-replication repair, it would remain associated with ssDNA during a later stage of recovery. To test this, we monitored RAD51 recruitment to parental ssDNA after 18 hours of recovery. As illustrated in Fig. 9b, RAD51 was associated with parental ssDNA in cells expressing wild-type WRN at 18 hours of recovery from HU, with minimal impact from MIRIN, which interferes with DSB formation (see Supplementary Fig. 18b). Conversely, cells expressing the WRN 6 A mutant, which promotes MRE11 and MUS81-dependent DNA breakage, showed increased RAD51 association with parental ssDNA, and this was greatly suppressed by MIRIN treatment (Fig. 9b).
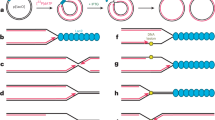
a In situ PLA between RAD51 and parental ssDNA in WS cells expressing Flag-WRNwt or WRN6A. Cells were transfected with control (CTRL) or siMUS81 siRNA and treated 48 h thereafter. The graph shows individual values of PLA spots of at least 100 cells from two independent experiments. Bars represent mean ± S.E. (Two-tailed Mann-Whitney test for paired samples. Where not indicated, values are not significant). Representative images are shown. Scale bar = 20 µm. b In situ PLA between RAD51 and parental ssDNA in WS cells expressing Flag-WRNwt or WRN6A and treated with MRE11i during recovery. The graph shows individual values of PLA spots of at least 50 cells from two independent experiments. Bars represent mean ± S.E. (Two-tailed Mann-Whitney test for paired samples. Where not indicated, values are not significant). Representative images are shown. Scale bar = 20 µm. c Neutral Comet assay for DSB evaluation in WS cells expressing Flag-WRNwt or WRN6A and treated as reported in the experimental scheme. The graph shows individual tail moment values of at least 50 cells from two independent experiments. Bars represent mean ± S.E. Statistical analyses were performed by Student’s t-test (Where not indicated, values are not significant). Representative images are provided. Scale bar = 20 µm. d Neutral Comet assay for DSB evaluation in WS cells expressing Flag-WRNwt or WRN6A and transfected with control (CTRL) or siMUS81 RNAi. The graph shows individual tail moment values of at least 80 cells from two independent experiments. Bars represent mean ± S.E. Statistical analyses were performed using Student’s t-test. Scale bar = 20 µm. e Proposed model: Replication fork stalling near secondary DNA structures, such as G4s (depicted in the leading strand for simplicity) requires WRN and its partner RPA for resolution. In the absence of WRN-RPA binding, these structures persist, and replication resumes downstream using PRIMPOL, if in the leading strand, leaving a gap in the template. During replication recovery, these gaps are processed by MRE11-exonuclease, which enables MUS81 endonuclease to induce DSBs. RAD51-mediated post-replication repair then facilitates the “removal” of the secondary DNA structures, such as G4s. Source data are provided as a Source Data file.
Next, we reasoned that MUS81-dependent DSBs would persist if RAD51 nucleofilament formation was blocked, indicating that RAD51 is necessary for their repair. To this aim, we performed a neutral Comet assay on cells expressing the wild-type WRN or the RPA-binding-defective WRN mutant at 18 hours of recovery, using the RAD51 inhibitor B02 (RAD51i). As shown in Fig. 9c, very few DSBs were present in WRN wild-type cells at 18 hours of recovery from HU, and these were only mildly affected by RAD51 inhibition. Interestingly, cells expressing the RPA-binding-defective WRN also exhibited few DSBs at 18 hours of recovery from HU, with no statistically significant difference compared to cells expressing wild-type WRN. However, RAD51 inhibition led to a substantial increase in the number of DSBs in cells expressing the WRN 6 A mutant if compared with the cells expressing wild-type WRN (Fig. 9c). Consistent with the neutral Comet assay, RAD51 inhibition also significantly elevated the phosphorylation level of the H2AX histone, a marker for DNA damage, in cells expressing the RPA-binding-deficient WRN (Supplementary Fig. 19).
Subsequently, we examined the persistence of DSBs after transfection with control siRNA or siRNA targeting MUS81 in cells expressing the RPA-binding-deficient WRN mutant at 18 hours of recovery in the presence of RAD51 inhibition. We hypothesised that despite RAD51 inhibition, DSBs would be reduced if MUS81 was silenced, indicating that RAD51 is primarily involved in repairing MUS81-dependent DSBs. The neutral Comet assay confirmed that RAD51 inhibition increased DSB levels (Fig. 9d). Furthermore, depletion of MUS81 substantially decreased the number of DSBs compared to control-depleted cells and RAD51-inhibited cells (Fig. 9d). Since cells expressing WRN 6 A show persistence of G4s that are eventually removed through formation of DSBs and HR, we wondered if they would display hypersensitivity to the G4 binder and anticancer drug candidate pyridostatin (PDS). To test this hypothesis, we analysed clonogenic survival of WS cells complemented with WRN wild-type or 6 A and treated with PDS at different concentrations. As shown in Supplementary Fig. 20, treatment with PDS induced a dose-dependent loss of viability in cells expressing WRN wild-type but, surprisingly, was less effective in killing cells expressing WRN 6 A.
Altogether, these results suggest that RAD51 is recruited to MRE11-processed gaps and is essential for repairing MUS81-dependent DSBs, thereby contributing to the clearance of G4 structures and limiting DNA damage, and possibly conferring resistance to G4 binders such as PDS.
Discussion
In this study, we found that RPA-binding to WRN plays a unique role at stressed replication forks, a process that depends on post-translational phosphorylation of WRN, which regulates its interaction with RPA. We identified a cluster of CK2-dependent phosphorylation sites within the acidic domain of WRN that are essential for its optimal interaction with RPA. By characterising a WRN 6 A unphosphorylable mutant, we were able to pinpoint a biological function of the WRN-RPA interaction critical for genome stability.
Previous in vitro studies have shown that WRN interacts with RPA via its acidic domain, which binds to a basic cleft in the N-terminal region of the RPA1 subunit10,17,19. Consistent with these observations, we determine that CK2-dependent phosphorylation of the WRN acidic domain at multiple sites modulates the WRN-RPA interaction. Importantly, our data indicate that, within cells, critically-important WRN-RPA interactions are inhibited by abrogating phosphorylation in the WRN acidic domain. Although a minor RPA-binding site has been mapped to the C-terminal region of WRN17, we cannot exclude the possibility that the residual interaction level observed in the WRN 6 A mutant originates from this site. Of interest, although our mass spectrometry data unambiguously assign phosphorylation at S440 and S467, we are unable to discriminate if the other phosphorylation observed in the subset of CK2 sites located in the TSYVI sequences takes place at Threonine or Serine. However, our immunoprecipitation data indicate that S440 and S467 are the key residues for the formation of the WRN-RPA complex as their mutation into Alanine abrogates RPA binding. Our mass spectrometry analyses cannot provide any insight into potential priming effects of S440/467 phosphorylation on that of the TSYVI sequences at this stage and dedicated biochemical experiments and phospho-specific antibodies will be needed to address this point.
Notably, the described CK2-dependent phosphorylation sites are evolutionary conserved, underscoring their relevance. Although not duplicated, these sites are present in vertebrate WRN (e.g., chicken) and Xenopus FFA-1. Interestingly, two, and the most critical, of the six CK2-dependent phosphorylation sites identified in this study, S440 and S467, have been previously identified as DNA-PK targets in response to DSBs22. Our data demonstrate that, in response to perturbed replication, DNA-PK contributes modestly to phosphorylation at these sites, and treatment with a DNA-PK inhibitor fails to recapitulate the effect of WRN 6 A or 2 A in wild-type cells, suggesting that different kinases might target the WRN acidic domain to modulate its specific functions. Serine 426 has also been identified as a CDK2-dependent site involved in DSB response, but no defective association with RPA was reported in the S426 A mutant21, suggesting that multiple residues must be targeted to negatively affect the interaction. Although RPA is important for directing multiple proteins to their genomic DNA substrates in response to replication fork perturbation32, WRN-binding to RPA is dispensable for WRN recruitment to ssDNA in cells. This finding differentiates the relationship between RPA and WRN from that of RPA and BLM, which requires association with RPA to be localised at ssDNA33. Recently, WRN was shown to cooperate with DNA2 in end-processing of reversed replication forks and during long-range resection at replication-dependent DSBs induced by CPT treatment23. Interestingly, our data show that RPA-binding is not involved in the WRN/DNA2-dependent end-processing, although RPA-ssDNA complexes are expected to form under these conditions. This is consistent with RPA-dependent and independent helicase activities of WRN34. However, RPA also interacts with DNA2 and stimulates its function35. Thus, during end-processing at stalled or collapsed forks, DNA2 may act as an RPA-binding protein for the WRN-DNA2-RPA complex, similar to its role with BLM33. Likewise, the protective function of WRN against pathological MUS81-dependent DSBs27,28 does not require interaction with RPA, as the WRN 6 A mutant exhibits normal levels of DSBs and RAD51 recruitment after replication fork stalling.
In contrast, our findings provide evidence that RPA-binding by WRN is essential for the productive recovery of stalled forks. WRN co-purified with replication factors, and defective replication fork progression has been repeatedly reported in the absence of WRN in various settings27,36,37,38,39,40. Therefore, it is tempting to speculate that WRN may perform at least two roles at the perturbed replication fork: end-processing and protection from DSBs in an RPA-independent manner, or promotion of fork restart in an RPA-dependent manner. Interestingly, association with RPA strongly stimulates WRN helicase activity in vitro10,17,19,20. Our observations suggest that abrogation of WRN helicase activity or WRN-binding to RPA confers comparable effects in response to stalled forks. Indeed, either expression of the RPA-defective WRN 6 A mutant or pharmacological inhibition of WRN helicase activity in wild-type cells impairs fork restart. In vitro, WRN can also unwind secondary DNA structures, such as hairpins or G4s41,42. We observe that loss of WRN phosphorylation by CK2 does not affect its unwinding activity on a model forked DNA or a G4 substrate in vitro. Furthermore, in vitro, WRN 6 A and WRN wild-type are equally stimulated by RPA. However, we observe that impairment of RPA-binding by WRN or inhibition of WRN helicase activity induces G4s accumulation upon fork stalling, suggesting that WRN interaction with RPA might render WRN helicase competent for clearance of G4 obstacles and potentially other secondary DNA structures in the cell. Consistent with this idea, loss of WRN sensitises cells to chromosome breakage at common fragile sites, which are prone to secondary DNA structure formation, and WRN helicase function is important for overcoming perturbed replication at these loci43,44,45. In addition, loss of WRN helicase sensitises cells to extended dinucleotide repeats accumulating in microsatellite-unstable cancers, possibly explaining the synthetic lethality observed in this context8,46,47. As the reported enzymatic assays might not intercept dynamic effects, additional experiments will be needed to reconcile the in vitro and cellular data. Another intriguing possibility is that loss of RPA binding in the cell reflects the modulation of protein complexes with unknown partners at the fork.
It would be interesting to investigate whether RPA-binding defective WRN 6 A mutant also confers any telomeric phenotype, as telomeric DNA is prone to secondary DNA structure formation and WRN is implicated in telomere biology48,49. Notably, RPA-binding is important for BLM-mediated fork restart33, suggesting that RPA may generally be required to stimulate RECQ helicases acting at “complex” substrates.
Significantly, defective fork restart associated with loss of RPA-binding by WRN results in the accumulation of ssDNA regions in the template strand and S1-sensitive gaps, as detected in the DNA fibre assay. The formation of DNA gaps depends on the presence of PRIMPOL, as observed in the absence of BRCA250,51. However, and in contrast to what was observed for the DNA gaps, the formation of parental ssDNA cannot be reverted by PRIMPOL depletion. One possible explanation is that parental ssDNA derives also from processing of G4-containing templates at the lagging strand, in which PRIMPOL is not expected to have any role in repriming. Multiple pathways can engage nucleases, as observed with transcription-dependent R-loop accumulation (MUS81) or the presence of AP-sites (MRE11-endo)52,53. Our data indicate that these parental ssDNA regions are produced by MRE11 and are required for G4s removal via the formation of MUS81-dependent DSBs (see model in Fig. 9e). Of note, a recent work demonstrated that replication fork reversal and arrest induced by HU-stimulated head-on transcription-replication conflicts (TRC) can be efficiently overcome using a MUS81 and PRIMPOL-mediated mechanism54. Although we did not formally test if MUS81 depletion, in cells expressing WRN 6 A, affects fork recovery, it would be interesting to assess if impaired WRN-RPA binding leads to more TRCs. Indeed, persisting G4s can stabilise R-loops leading to more TRCs52,55, and WRN limits R-loop-dependent genomic instability9. Of note, MRE11 and EXO1-dependent processing of DNA gaps has been recently reported in cells treated with environmental contaminants that induce DNA adducts and cause mutagenesis56. In this work, the authors suggest that DNA lesions are removed using the endonuclease activity of MRE11 by acting at the nascent strand ssDNA gaps during replication56. Thus, it is conceivable that different secondary structures or “roadblocks” are removed exploiting different endonucleases. Abrogation of CK2-dependent phosphorylation of WRN does not reduce the interaction between WRN and DNA2 or MRE11, which are found enhanced in the WRN 6 A mutant. DNA2 and MRE11 interacts with WRN outside its acidic domain23,24. Thus, our result further supports the specific relevance of phosphorylation by CK2 for the formation of the WRN-RPA complex. Since MRE11 inhibition but not that of DNA2 reduces the presence of parental ssDNA at daughter strand gaps in cells expressing WRN 6 A, it seems unlikely that the phenotype of these cells might derive from the increased binding to these exonucleases. However, this result indicates that WRN complexes are highly dynamic in composition and underscores the relevance of post-translational modification in their assembly. MRE11 exonuclease activity might be involved in enlarging gaps before MUS81 endonuclease-mediated cleavage of G4s and perhaps in further resecting the ends of the DSBs introduced at G4s. It is worth noting that although loss of WRN stimulates the formation of MUS81-dependent DSBs, this does not occur upon sensitisation of common fragile sites28,57. As common fragile sites are thought to form secondary DNA structures, this might suggest that the replication perturbation induced at common fragile sites is not resolved by the same mechanism acting on other secondary DNA structures, such as G4s. Interestingly, MUS81 has been implicated in the cleavage of G4s at stalled forks and the removal of secondary DNA structures arising at expanded dinucleotide repeats in MMR-deficient cancer cells8,31. We observe that MUS81-dependent DSBs are subsequently channelled through RAD51-dependent post-replication repair, as previously shown for some gaps left behind MMS-perturbed forks58,59. This pathway acts as a true salvage mechanism, as its abrogation leads to DSB accumulation (Fig. 8).
Surprisingly, cells expressing the unphosphorylable WRN-6A mutant, which has impaired ability to complex with RPA, seem to be more resistant to the G4 ligand PDS than the wild-type. One potential explanation is that while the wild-type WRN protein is trapped at PDS-stabilised G4 structures the WRN-6A mutant that is defective in its interaction with RPA does not become as stably engaged with the poisonous G4 structure, which can be efficiently converted into DSBs by MUS81 and then repaired by RAD51.
In conclusion, we determined that the loss of RPA-binding to WRN represents a true separation-of-function mutation that interferes with WRN’s cellular functions during the replication stress response or DSB repair. Because most of the phosphorylation sites are conserved in mouse Wrn, and some also in C. elegans (Supplementary Fig. 2c), future studies in animal models, where WRN’s RPA-binding ability is impaired by loss of phosphorylation mutations in the acidic domain, might be useful to elucidate which function(s) of WRN are essential to prevent the characteristic phenotypes associated with the accelerated aging seen in Werner syndrome.
Methods
Cell lines and culture conditions
The SV40-transformed WRN-deficient fibroblast cell line (AG11395) was obtained from Coriell Cell Repositories (Camden, NJ, USA). The AG11395 cell line carries an Arg368 stop mutation in the WRN coding sequence that gives rise to a truncated protein that is degraded and undetectable. AG11395 (WS) were nucleofected with plasmids encoding pCMV-Flag-WRN wild-type and the unphosphorylable (6A) and the phosphomimetic (6D) form of WRN. CK2 phosphorylation mutants were made by replacement of threonine 434, 461 and serine 435, 440, 432, and 467 with alanine or aspartic acid. HEK293T and U2OS cells were from the American Type Culture Collection and they are transfected with the same WRN plasmids.
All the cell lines were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% FBS with or without tetracycline and incubated at 37 °C in a humidified 5% CO2 atmosphere.
Chemicals and siRNA
-Hydroxyurea (HU 98% powder, Sigma-Aldrich) was dissolved in ddH20 and used at 2 mM.
-Silmitasertib (CX-4945 Selleck), an inhibitor of CK2 kinase activity, was dissolved in DMSO and a stock solution (500 μM) was prepared and stored at −80 °C. It was used at 20 μM.
-NU7441 (Selleck), a DNAPKcs inhibitor, was stocked at 1 mM in DMSO and used at final concentration of 1 μM.
-Mirin (MRE11i) (Calbiochem), an inhibitor of MRN-ATM pathway, was stocked at 50 mM in DMSO and used at 50 μM.
-NSC617145 (Tocris Bio-Techne), an inhibitor of WRN helicase activity, was stocked at 10 mM in DMSO and used at 4 μM.
-HY-128729 (Thermo Fisher), an inhibitor of DNA2 activity, was stocked at 150 mM in DMSO and used at 300 μM.
-B02 (553525 Sigma-Aldrich), an inhibitor of RAD51, was dissolved in DMSO a stock solution (37 mM) was prepared and stored at −20 °C. It was used at 37 μM.
-5-iodo-2’-deoxyuridine (IdU) and 5-Chloro-2′-deoxyuridine (CldU) (Sigma-Aldrich) were dissolved in sterile DMEM at 2.5 mM and 200 mM respectively and stored at −20 °C. IdU was used at 100 μM for single strand assay and 250 μM for fibre assay. CldU was used at 50 μM.
-5-ethylene-2′-deoxyuridine (EdU) (Sigma-Aldrich) was dissolved in sterile DMSO at 125 mM and stored at −20 °C. It was used at 125 μM for 8 min for SIRF assay.
-Hs MUS81 6 FlexiTube siRNA cat #SI04300877 was stocked at 20 μM and used at 20 nM to knock-down MUS81.
-Hs PRIMPOL Silencer select siRNA cat#4427037 was stocked at 20 μM and used at 25 nM to knock-down PRIMPOL.
Nucleofection and transfection
AG11395 and HEK293T cells were nucleofected and transfected with pCMV-Flag-WRN wild-type, pCMV Flag-WRN 6 A or 2 A plasmids. For the nucleofection 10 μg of DNA were used for 1.8 × 106 cells, with 2 pulses of 950 V lasting 2 min by Invitrogen Neon Transfection system (Invitrogen). After 24 h in empty medium, cells were placed in 10% FBS medium. 293 T cells were transfected with Dreamfect (Oz Bioscences): 20 μl of Dreamfect was used with 5 μg of DNA, mixed in empty medium for 18 min. After 24 h in empty medium, cells were put in 10% FBS medium.
Generation of the GST-WRN fragment
DNA sequence corresponding to aa 402-503 (N-WRN) of WRN was amplified by PCR from the pCMV-Flag-WRN (wild-type) plasmid and pCMV-Flag-WRN (6D). The PCR products were subsequently purified and sub-cloned into pGEX4T-1 vector (Stratagene) for subsequent expression in bacteria as GST-fusion proteins. The resulting vectors were subjected to sequencing to ensure that no mutations were introduced into the WRN sequence in the plasmid used for transforming BL21 cells (Stratagene). Expression of GST and GST-fusion proteins were induced upon addition of 1 mM isopropyl-D-thiogalactopyranoside (IPTG) for 2 h at 37 °C. GST, and GST-N-77 WRN were affinity-purified using glutathione (GSH)-magnetic beads (Promega). Fragment purification levels were assessed by SDS-PAGE followed by Coomassie staining.
Purification of recombinant FLAG-WRN
High-titre virus expressing Flag-WRN was used to infect Hi5 insect cells (Thermo Fisher Scientific) at an MOI of approximately 10. Cells were harvested 48 hours later and placed in −80 °C until lysed. Cell pellet containing approximately 1.2 x 108 cells was resuspended in 10 ml of Lysis Buffer (50 mM Tris pH 7.4, 150 mM NaCl, 0.4% NP40, 10% glycerol, 5 mM BME, and Complete Ultra Protease Inhibitors (Roche)), vortexed and rotated at 4 °C for 45 min. The lysates were centrifuged at 20,000 rpm for 10 min and the supernatant was passed through a 0.45 µm PVDF filter. Each clarified lysate was passed twice through a Ni2 + -charged 1 ml HiTrap Chelating HP column (GE Healthcare Life Sciences) which had been equilibrated in Buffer TN (50 mM Tris pH 7.4, 150 mM NaCl, 10% glycerol, 5 mM BME, protease inhibitors) with 10 mM imidazole. 5 ml washes with TN buffer containing 10 mM, 20 mM, and 40 mM imidazole each were performed followed by elution with TN buffer containing 400 mM imidazole. The eluted protein was pooled, dialysed to remove imidazole, and incubated with TEV protease for 16 h at 4 °C to cleave the 6xHis tag off the protein. The protein was dialysed into NETN-500 Buffer (50 mM Tris pH 7.4, 500 mM NaCl, 0.5% NP40, 1 mM EDTA) using a Amicon Ultra 100 kD cutoff centrifugal filter (EMD Millipore). The retained sample was applied to 250 µl of packed M2 anti-Flag beads (Sigma) which had been equilibrated in NETN-500 buffer. The beads were washed twice with 5 ml of NETN-500 buffer and the WRN protein was eluted with 3X FLAG peptide twice in 500 µl Storage Buffer (100 mM Tris pH 8.0, 400 mM NaCl, 10% glycerol, 5 mM BME). Eluted protein was concentrated and dialysed against storage buffer in the absence of FLAG peptide and frozen at -80 °C.
Radiolabeled DNA substrates
DNA oligonucleotides:
TP-G4 (5’- TGGACCAGACCTAGCAGCTATGGGGGAGCTGGGGAAGGTGGGAATGTGA-3’)
TelX (5’-TTTTTTTTTTTTTTTGGTGATGGTGTATTGAGTGGGATGCATGCACTAC-3’)
TelY (5’-GTAGTGCATGCATCCCACTCAATACACCATCACCTTTTTTTTTTTTTTT-3’) were purchased from Integrated DNA Technologies. Radiolabeled tetramolecular parallel TP-G4 and TelXY forked duplex substrates were prepared as previously described60.
Helicase assays
Helicase assays (20 µl) were performed in a buffer containing 20 mM Tris-HCl, pH 7.5, 8 mM ATP, 8 mM MgCl2, 1 mM DTT, 0.1 mg/ml BSA with indicated amounts of radiolabeled substrate and WRN and RPA proteins. Reactions were incubated 15 min. at 37 °C and terminated with 10 µl of the stop buffer (30 mM Tris HCl pH 8.0, 30 mM EDTA, 30% glycerol, 0.9% SDS, 0.25% bromophenol blue (w/v), and 0.25% xylene cyanol (w/v)). 1 µl of Proteinase K (New England Biolabs) was added to each reaction followed by 10 min. incubation at 37 °C. Samples were separated on non-denaturing polyacrylamide gels (8% for TP-G4, and 10% for TelXY substrate), and exposed to phosphorimager plates. Gel images were acquired using Typhoon FLA-9500 phosphorimager (Cytiva) and analyzed with ImageQuant TL software (Cytiva).
For dephosphorylated-phosphorylated proteins, 0.5 µg of purified recombinant WRN protein was treated with Lambda Protein Phosphatase (LPP, New England Biolabs) in 1X PMP buffer (50 mM HEPES pH 7.5, 100 mM NaCl, 2 mM DTT, 0.01% Brij 35, 10 µl reaction) for 30 min at 30 °C. Halt Phosphatase Inhibitor Cocktail (Thermo Scientific) was added to 1X final concentration, followed by Casein Kinase II (New England Biolabs) in 1X PK buffer (50 mM Tris-HCl pH 7.5, 10 mM MgCl2, 0.1 mM EDTA, 2 mM DTT, 0.01% BriJ 35, 30 µl reaction) for 30 min at 30 °C. Untreated, CK2- and/or LPP-treated WRN protein (concentrations indicated in figure legend), or storage buffer were incubated with 0.5 nM forked DNA substrate in 20 µl reactions containing 30 mM HEPES pH 7.4, 40 mM KCl, 100 µg/ml BSA, 8 mM MgCl2, 5% glycerol, and 2 mM ATP for 15 min at 37 °C in the presence or not of RPA heterotrimer. Reactions were stopped by adding 20 µl of 9 mM EDTA, 0.6% SDS, 0.04% bromophenol blue, 0.04% xylene cyanol, and 25% glycerol containing a 10-fold excess of the labelled oligo without the radiolabel. The heat-denatured sample was boiled for 5 min at 95 °C.
In vitro kinase assay
For kinase assay, 1 μg of immunopurified GST-tagged WRN fragment was phosphorylated in vitro by Casein Kinase II (New England Biolabs) in the presence of 1X NEBuffer (50 mM Tris-HCl, 10 mM MgCl2, 0.1 mM EDTA, 2 mM DTT, 0.01% BriJ 35) and 200 μM ATP for 30 min at 30 °C.
Pull-down Assay
GST and GST-WRN fragments (phosphorylated or not) were incubated with 300 ng of 293 T cell extracts. After 16 h of incubation, fragments were separated from the beads and RPA32 interaction with WRN fragments were measured with densitometric analysis by WB using rabbit anti-GST (Calbiochem), rabbit anti-p440/467WRN (Abgent) and mouse anti-RPA34-20 (Millipore).
Immunoprecipitation and Western blotting analysis
Immunoprecipitation experiments were performed using 3 × 106 HEK293T cells. IP buffer (0.5% Triton X-100, 50 mM Tris HCl pH 8.0, 150 mM NaCl, EGTA 1 mM) supplemented with phosphatase, protease inhibitors and benzonase was used for cells lysis. Two millilitres of lysate were incubated overnight at 4 °C with 20 μl of Anti-Flag M2 magnetic beads (Sigma) or Anti-RPA32 conjugated Dynabeads (2 μg of MABE285 anti-RPA34-20 mouse (Millipore) with 40 μl of Dynabeads protein G (Invitrogen). After extensive washing in IP buffer, proteins were released in 2X Laemmli buffer and subjected to Western blotting.
Blots were incubated with primary antibodies as described in Supplementary Table 1. Blots were detected using the Western blotting detection kit Western Bright ECL (Advansta) according to the manufacturer’s instructions. Quantification was performed on scanned images of blots using Image Lab software, and values shown on the graphs represent normalisation of the protein content evaluated through Lamin B1 or Immunoprecipitated protein immunoblotting.
Mass spectrometry analysis
Identification of phosphopeptides was performed as already described61. Briefly, purified proteins were in gel-digested with trypsin, phosphopeptides enriched by IMAC following the manufacturer’s guidelines (Phosphopeptide Enrichment Kit; Pierce) and mass spectrometry analysis performed with a MALDI-TOF Voyager DE-STR (Applied Biosystems, Foster City, CA, USA) in positive reflectron mode, using phospho-DHB as matrix. MS spectra were processed with DATA EXPLORER (Applied Biosystems) and GPMAW software for peak-to-sequence assignments. To confirm the attribution of relative peaks to mono-, di- and tri-phosphorylated peptides, alkaline phosphatase treatment was performed on-probe as already described62.
For the characterisation of phosphosites, we load on PAGE Flag-WRN immunopurified from untreated and CK2i, 2 h HU, 2 h HU+ CK2i treated cell cultures. After in-gel digestion, 1 µg (Quantitative Colorimetric Peptide Assay- Pierce) of each sample was run in triplicate by two different strategies of MS/MS experiments using an Orbitrap Fusion Tribrid Mass Spectrometer (Thermo Fisher Scientific) coupled to an UltiMate 3000 UHPLC system (Thermo Fisher Scientific). For both acquisition methods, peptides were firstly trapped in a μ-precolumn (C18 PepMap100, Thermo Fisher Scientific) and then run on a home-packed 25 cm × 75 μm id fused-silica column (8 PicoTip Emitter, New Objective) packed with ReproSil-Pul C18-AQ 1.9 um beads (Dr. Maisch GmbH) for chromatographic separation. Peptides were eluted at 0.2 μL/min along a 40 min linear gradient from 7% to 32% of buffer B (95% acetonitrile, 0.1% formic acid). In the first method, data-dependent acquisition mode (DDA), we operate in a target identification strategy including the mass list of the two tryptic peptides (421-450 and 451-477) carrying one, two or three phosphorylation groups, resulting in 24 m/z targets (not considering any combination with other possible modification, like oxidised methionine). In DDA, Full-scan mass spectrometry (MS) was acquired in Orbitrap at 120 K resolution; in a mass range of 700-2000. The MS/MS scans were acquired at 30 K resolution for a total cycle time of 3 s; dependent scan was set on most intense ions if target ones are not found; quadrupole isolation window 1.6; HCD fragmentation at collision energy 30%. In the second acquisition strategy, data-independent mode (DIA) method, a full 120 K resolution MS scan in the range 800-1300 m/z was followed by 42 isolation windows of 12 m/z with an overlap of 2 m/z, HCD fragmentation at collision energy 30% and Orbitrap detector set at 30k resolution. DDA raw files were analysed by MaxQuant 2.5.163 and DIA were run on FragPipe version 21.0 (FragPipe version 21.0; MSFragger version 4.0; IonQuant version 1.10.12; Philosopher version 5.1.0; Python 3.11.7; EasyPQP version 0-1-42)64. Phosphorylation (STY) and methionine-oxidation were considered as variable modifications while cysteine-carbamidomethylation as fixed modification, a maximum of two-missed cleavage permitted, and default setting of remaining parameters were used for both software.
Chromatin isolation
To isolate chromatin, cells were resuspended in buffer A (1 M Hepes pH 7.9, 1 M HCl, 100 mM MgCl2, glycerol, sucrose, sodium fluoride, 1 mM DTT, protease inhibitors, phosphatase inhibitors). Triton X-100 (0,1%) was added, and the cells were incubated for 5 min on ice. Nuclei were collected by low-speed centrifugation (4 min, 4000 rpm, 4 °C). Nuclei were washed once in buffer A, and then lysed 10 min in buffer B (3 mM EDTA, 0.2 mM EGTA, 1 mM DTT, protease inhibitors, phosphatase inhibitors). Insoluble chromatin was collected by centrifugation (4 min, 4500 rpm, 4 °C), washed once in buffer B, and centrifuged again under the same conditions. The final chromatin pellet was resuspended in 2X Laemmli buffer and sonicated for 15 sec in a Tekmar CV26 sonicator using a microtip at 25% amplitude. Lastly, the lysates were subjected to Western blot analysis. Chromatin recruitment was normalised against the content of Lamin B1, or H3 histone which are proteins exclusively found in the chromatin fraction.
Single-stranded DNA detection and immunofluorescence assay
Cells were cultured onto 22 × 22 coverslip in 35 mm dishes. To detect nascent single-stranded DNA (ssDNA), after 24 h, the cells were labelled for 15 min before the treatment with 100 μM IdU (Sigma-Aldrich), cells were then treated with HU 2 mM for different time points. Meanwhile, to detect parental ssDNA, the cells were labelled for 24 h before 2 h of fresh medium. After the release, cells were treated with HU. Next, cells were washed with PBS, permeabilized with 0.5% Triton X-100 for 10 min at 4 °C and fixed with 2% sucrose, 3% PFA. For ssDNA detection, cells were incubated with primary mouse anti-IdU antibody (Becton Dickinson) for 1 h at 37 °C in 0.1% saponin/BSA in PBS, followed by Alexa Fluor488-conjugated goat-anti-Mouse, and counterstained with 0.5 μg/ml DAPI. Instead, to detect RPA32 and WRN, cells were incubated with specific primary antibody (Supplementary Table 1) for 1 h at 37 °C in 0.1% saponin/BSA in PBS followed by Alexa Fluor 594 Anti-Rabbit or Alexa Fluor 488 Anti-Mouse. For immunofluorescence of G-quadruplex structures (G4s), cells grown on glass coverslips were fixed with ice-cold 80% methanol in PBS for 15 min at −20 °C, then washed two times in PBS. Next, cells were blocked with 10% FBS/PBS for 1 h and incubated with the anti-G-quadruplex antibody (Supplementary Table 1) overnight at 4 °C. Slides were analysed (at 40x) with Eclipse 80i Nikon Fluorescence Microscope, equipped with a Virtual Confocal (ViCo) system. Fluorescence intensity for each sample was then analysed using ImageJ software.
In situ proximity ligation assay (PLA)
Cells were cultured onto 8-well Nunc chamber-slides. The in situ proximity ligation assay (PLA) in combination with immunofluorescence microscopy was performed using the Duolink Detection (Merck) or the NaveniFlex (Navinci diagnostics) Kit with anti-Mouse PLUS and anti-Rabbit MINUS PLA Probes, according to the manufacturer’s instructions. To detect proteins, we used rabbit anti-WRN (Abcam), rabbit anti-Flag (Sigma-Aldrich), mouse anti-RPA34-20 (Millipore), rabbit anti-RAD51 (Santa Cruz) and mouse anti-IdU antibody (Becton Dickinson) antibodies.
Single-cell Assay for in situ Protein Interaction with Nascent DNA (SIRF)
Exponential growing cells were seeded onto microscope chamber slide. The day of experiment, cells were incubated with 100 µM EdU for 20 min and treated as indicated. After treatment, cells were pre-extracted in 0.5% TritonX-100 for 5 min on ice and fixed with 3% PFA, 2% sucrose in PBS 1X for 15 min at RT. Cells were then blocked in 3% BSA/PBS for 15 min. For the EdU detection was applied the Click-iT™ EdU Alexa Fluor™ Imaging Kit (Invitrogen) using 5 mM Biotin-Azide for 30 minutes at RT. The primary antibodies used were as follows: BG4 (Merck, 1:1000), rabbit anti-biotin (Abcam, 1:50). The negative controls were obtained by using only one primary antibody. Samples were incubated with secondary antibodies conjugated with PLA probes MINUS and PLUS: the PLA probe anti‐mouse PLUS and anti‐rabbit MINUS (Duolink®, Sigma-Aldrich). The incubation with all antibodies was accomplished in a humidified chamber for 1 h at 37 °C. Next, the PLA probes MINUS and PLUS were ligated using two connecting oligonucleotides to produce a template for rolling‐cycle amplification. After amplification, the products were hybridised with red fluorescence‐labelled oligonucleotide. Samples were mounted in Prolong Gold anti‐fade reagent with DAPI (blue). Images were acquired randomly using Eclipse 80i Nikon Fluorescence Microscope, equipped with a Virtual Confocal (ViCo) system. The analysis was carried out by counting the SIRF spot for each nucleus.
DNA fibre analysis
Cells were pulse-labelled with 50 μM CldU and then labelled with 250 μM IdU at the times specified, with HU treatment as reported in the experimental schemes: to study DNA degradation cells were labelled for 20 min of CldU, then after 2 washes in PBS, they were labelled for 20 min of IdU. After another two washes, cells were treated with 2 mM HU at different times. Meanwhile, to study the ability to recover from replicative stress, cells were labelled for 20 min before the HU treatment and during the treatment itself. After two washes, cells were labelled for 20 min with IdU. Fibre assay using S1 nuclease was performed as indicated by Quinet et al., 201765. Briefly, cells were pulse-labelled with 50 µM CldU and then labelled with 250 µM IdU with or without treatment, as reported in the experimental schemes. At end of treatment, cells were permeabilized with CSK buffer (100 mM NaCl, 10 mM PIPES pH 6.8, 1 M EGTA, 3 mM MgCl2, 300 mM sucrose, 0.5% Triton X-100) for 10 min at RT, then were washed with PBS 1X and S1 nuclease buffer (30 mM sodium acetate, 10 mM zinc acetate, 5% glycerol, 50 mM NaCl) prior to add +/- S1 nuclease for 30 min at 37 °C in a humidified chamber. Cells were washed with S1 buffer then scraped with 0.1% BSA/PBS and collected pellets were used to perform fibres spreading. DNA fibres were prepared, spread out and immunodecorated following primary antibodies with rat anti-CldU/BrdU (Abcam) and mouse anti-IdU/BrdU (Becton Dickinson). Images were acquired randomly from fields with untangled fibres using Eclipse 80i Nikon Fluorescence Microscope, equipped with a Virtual Confocal (ViCo) system. The length of labelled tracks were measured using the ImageJ software. A minimum of 100 individual fibres were analysed for each experiment.
Neutral comet assay
After treatment, cells were embedded in low-melting agarose and spread onto glass slides. After an electrophoretic run of 20 min (6-7 A, 20 V), cells were fixed with methanol. DNA was stained with 0.1% GelRed (Biotium) and examined at 20x magnification with an Olympus fluorescence microscope. Slides were analysed with a computerised image analysis system (CometScore, Tritek Corp.). To assess the amount of DNA DSB breaks, computer-generated tail moment values (tail length × fraction of total DNA in the tail) were used. Apoptotic cells (smaller comet head and extremely larger comet tail) were excluded from the analysis to avoid artificial enhancement of the tail moment. A minimum of 150 cells were analysed for each experiment.
Statistical analysis
All the statistical analyses were performed using GraphPad Prism 9 software. Frequency distributions of DNA track length and ratios were determined with GraphPad Prism 9 software after the quantification of the tract lengths using ImageJ. Mann-Whitney and Kruskal-Wallis tests coupled with ad-hoc Dunn test for false-discovery rates were used for statistical analyses when performed pair comparisons or when comparing more than two samples, respectively. In all graphs, P < 0.05 was considered significant for frequency distribution. When data are not presented as scatter plot, they are shown as the mean of independent experiments.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Primary Mass Spectrometry Datasets have been deposited to ProteomeXchange accession PXD059007. All remaining data supporting the findings of this study are available within the paper and its Supplementary Information files. Source data are provided with this paper.
References
Franchitto, A. & Pichierri, P. Understanding the molecular basis of common fragile sites instability: Role of the proteins involved in the recovery of stalled replication forks. Cell Cycle 10, 4039–4046 (2011).
Brosh, R. M. & Bohr, V. A. Human premature aging, DNA repair and RecQ helicases. Nucleic Acids Res. 35, 7527–7544 (2007).
Oshima, J., Sidorova, J. M. & Monnat, R. J. Werner syndrome: clinical features, pathogenesis and potential therapeutic interventions. Ageing Res Rev. 33, 105–114 (2017).
Croteau, D. L., Popuri, V., Opresko, P. L. & Bohr, V. A. Human RecQ helicases in DNA repair, recombination, and replication. Annu Rev. Biochem 83, 519–552 (2014).
Rossi, M. L., Ghosh, A. K. & Bohr, V. A. Roles of Werner syndrome protein in protection of genome integrity. DNA repair 9, 331–344 (2010).
Mukherjee, S. et al. Werner Syndrome Protein and DNA Replication. Int. J. Mol. Sci. 19, 3442 (2018).
Datta, A. et al. WRN helicase safeguards deprotected replication forks in BRCA2-mutated cancer cells. Nat. Commun. 2021 12, 1–22 (2021).
van Wietmarschen, N. et al. Repeat expansions confer WRN dependence in microsatellite-unstable cancers. Nature 586, 1–7 (2020).
Marabitti, V. et al. ATM pathway activation limits R-loop-associated genomic instability in Werner syndrome cells. Nucleic Acids Res. https://doi.org/10.1093/nar/gkz025 (2019).
Brosh, R. M. et al. Functional and physical interaction between WRN helicase and human replication protein A. J. Biol. Chem. 274, 18341–18350 (1999).
Wold, M. S. Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu Rev. Biochem 66, 61–92 (1997).
Chen, R. & Wold, M. S. Replication protein A: single-stranded DNA’s first responder: dynamic DNA-interactions allow replication protein A to direct single-strand DNA intermediates into different pathways for synthesis or repair. Bioessays 36, 1156–1161 (2014).
Bhat, K. P. & Cortez, D. RPA and RAD51: fork reversal, fork protection, and genome stability. Nat. Struct. Mol. Biol. 25, 446–453 (2018).
Shen, J.-C., Lao, Y., Kamath-Loeb, A., Wold, M. S. & Loeb, L. a. The N-terminal domain of the large subunit of human replication protein A binds to Werner syndrome protein and stimulates helicase activity. Mechanisms Ageing Dev. 124, 921–930 (2003).
Constantinou, A. et al. Werner’s syndrome protein (WRN) migrates Holliday junctions and co-localizes with RPA upon replication arrest. EMBO rep. 1, 80–84 (2000).
Ammazzalorso, F., Pirzio, L. M., Bignami, M., Franchitto, A. & Pichierri, P. ATR and ATM differently regulate WRN to prevent DSBs at stalled replication forks and promote replication fork recovery. EMBO J. 29, 3156–3169 (2010).
Doherty, K. M. et al. Physical and functional mapping of the replication protein A interaction domain of the Werner and Bloom syndrome helicases. J. Biol. Chem. 280, 29494–29505 (2005).
Sowd, G., Wang, H., Pretto, D., Chazin, W. J. & Opresko, P. L. Replication protein A stimulates the Werner syndrome protein branch migration activity. J. Biol. Chem. 284, 34682–34691 (2009).
Machwe, A., Lozada, E., Wold, M. S., Li, G. M. & Orren, D. K. Molecular cooperation between the werner syndrome protein and replication protein A in relation to replication fork blockage. J. Biol. Chem. 286, 3497–3508 (2011).
Lee, M. et al. Multiple RPAs make WRN syndrome protein a superhelicase. Nucleic Acids Res. 46, 4689–4698 (2018).
Lee, J.-H. et al. CDK2 phosphorylation of Werner protein (WRN) contributes to WRN’s DNA double-strand break repair pathway choice. Aging Cell e13484 https://doi.org/10.1111/ACEL.13484 (2021).
Kusumoto-Matsuo, R. et al. Serines 440 and 467 in the Werner syndrome protein are phosphorylated by DNA-PK and affects its dynamics in response to DNA double strand breaks. Aging 6, 70–81 (2014).
Sturzenegger, A. et al. DNA2 cooperates with the WRN and BLM RecQ helicases to mediate long-range DNA end resection in human cells. J. Biol. Chem. 289, 27314–27326 (2014).
Cheng, W. H. et al. Linkage between Werner syndrome protein and the Mre11 complex via Nbs1. J. Biol. Chem. 279, 21169–21176 (2004).
Thangavel, S. et al. DNA2 drives processing and restart of reversed replication forks in human cells. J. cell Biol. 208, 545–562 (2015).
Iannascoli, C., Palermo, V., Murfuni, I., Franchitto, A. & Pichierri, P. The WRN exonuclease domain protects nascent strands from pathological MRE11/EXO1-dependent degradation. Nucleic acids Res. 43, 9788–9803 (2015).
Pichierri, P., Franchitto, A., Mosesso, P., Palitti, F. & Molecolare, C. Werner’ s syndrome protein is required for correct recovery after replication arrest and dna damage induced in s-phase of cell cycle. Mol. Biol. Cell 12, 2412–2421 (2001).
Franchitto, A. et al. Replication fork stalling in WRN-deficient cells is overcome by prompt activation of a MUS81-dependent pathway. J. Cell Biol. 183, 241–252 (2008).
Palermo, V. et al. CDK1 phosphorylates WRN at collapsed replication forks. Nat. Commun. 7, 12880 (2016).
Fry, M. & Loeb, L. a. Human Werner syndrome DNA helicase unwinds tetrahelical structures of the fragile X syndrome repeat sequence d(CGG)(n). J. Biol. Chem. 274, 12797–12802 (1999).
Liu, S. et al. DNA repair protein RAD52 is required for protecting G-quadruplexes in mammalian cells. J. Biol. Chem. 299, 102770 (2023).
Zeman, M. K. & Cimprich, K. A. Causes and consequences of replication stress. Nat. cell Biol. 16, 2–9 (2014).
Shorrocks, A. K. et al. The Bloom syndrome complex senses RPA-coated single-stranded DNA to restart stalled replication forks. Nat. Commun. 12, 585 (2021).
Sommers, J. A. et al. p53 modulates RPA-dependent and RPA-independent WRN helicase activity. Cancer Res. 65, 1223–1233 (2005).
Zhou, C., Pourmal, S. & Pavletich, N. P. Dna2 nuclease-helicase structure, mechanism and regulation by Rpa. eLife 4, e09832 (2015).
Das, D. et al. Werner helicase control of human papillomavirus 16 E1-E2 DNA replication is regulated by SIRT1 deacetylation. mBio 10, e00263-19 (2019).
Lebel, M., Spillare, E. A., Harris, C. C. & Leder, P. The Werner syndrome gene product co-purifies with the DNA replication complex and interacts with PCNA and topoisomerase I. J. Biol. Chem. 274, 37795–37799 (1999).
Rodriguez-Lopez, A. M., Jackson, D. A., Iborra, F. J. & Cox, L. S. Asymmetry of DNA replication fork progression in Werner’s syndrome. Aging Cell 1, 30–39 (2002).
Hanaoka, F. et al. Autoradiographic studies of DNA replication in Werner’s syndrome cells. Adv. Exp. Med. Biol. 190, 439–457 (1985).
Sidorova, J. M., Kehrli, K., Mao, F. & Monnat, R. Distinct functions of human RECQ helicases WRN and BLM in replication fork recovery and progression after hydroxyurea-induced stalling. DNA Repair 12, 128–139 (2013).
Chan, N. L. S. et al. The Werner syndrome protein promotes CAG/CTG repeat stability by resolving large (CAG)n/(CTG)n hairpins. J. Biol. Chem. 287, 30151–30156 (2012).
Tang, W. et al. The Werner syndrome RECQ helicase targets G4 DNA in human cells to modulate transcription. Hum. Mol. Genet 25, 2060–2069 (2016).
Basile, G., Leuzzi, G., Pichierri, P. & Franchitto, A. Checkpoint-dependent and independent roles of the Werner syndrome protein in preserving genome integrity in response to mild replication stress. Nucleic acids Res. 42, 12628–12639 (2014).
Pirzio, L. M., Pichierri, P., Bignami, M. & Franchitto, A. Werner syndrome helicase activity is essential in maintaining fragile site stability. J. Cell Biol. 180, 305–314 (2008).
Shah, S. N., Opresko, P. L., Meng, X., Lee, M. Y. W. T. & Eckert, K. a. DNA structure and the Werner protein modulate human DNA polymerase delta-dependent replication dynamics within the common fragile site FRA16D. Nucleic acids Res. 38, 1149–1162 (2010).
Behan, F. M. et al. Prioritization of cancer therapeutic targets using CRISPR-Cas9 screens. Nature https://doi.org/10.1038/s41586-019-1103-9 (2019).
Chan, E. M. et al. WRN helicase is a synthetic lethal target in microsatellite unstable cancers. Nature 568, 551–556 (2019).
Crabbe, L., Verdun, R. E., Haggblom, C. I. & Karlseder, J. Defective telomere lagging strand synthesis in cells lacking WRN helicase activity. Sci. (New York, N. Y.) 306, 1951–1953 (2004).
Opresko, P. L. et al. The werner syndrome helicase and exonuclease cooperate to resolve telomeric D loops in a manner regulated by TRF1 and TRF2. Mol. Cell 14, 763–774 (2004).
Cantor, S. B. Revisiting the BRCA-pathway through the lens of replication gap suppression. DNA Repair 107, 103209 (2021).
Quinet, A. et al. PRIMPOL-mediated adaptive response suppresses replication fork reversal in brca-deficient cells. Mol. Cell 77, 461–474.e9 (2020).
Chappidi, N. et al. Fork cleavage-religation cycle and active transcription mediate replication restart after fork stalling at co-transcriptional R-Loops. Mol. Cell 77, 528–541.e8 (2020).
Hanthi, Y. W. et al. RAD51 protects abasic sites to prevent replication fork breakage. Mol. Cell 84, 3026–3043.e11 (2024).
Andrs, M. et al. Excessive reactive oxygen species induce transcription-dependent replication stress. Nat. Commun. 14, 1791 (2023).
Kotsantis, P. et al. RTEL1 regulates G4/R-loops to avert replication-transcription collisions. Cell Rep. 33, 108546 (2020).
Hale, A., Dhoonmoon, A., Straka, J., Nicolae, C. M. & Moldovan, G.-L. Multi-step processing of replication stress-derived nascent strand DNA gaps by MRE11 and EXO1 nucleases. Nat. Commun. 14, 6265 (2023).
Murfuni, I. et al. Perturbed replication induced genome wide or at common fragile sites is differently managed in the absence of WRN. Carcinogenesis 33, 1655–1663 (2012).
Hashimoto, Y., Chaudhuri, A. R., Lopes, M. & Costanzo, V. Rad51 protects nascent DNA from Mre11-dependent degradation and promotes continuous DNA synthesis. Nat. Struct. Mol. Biol. 17, 1305–1311 (2010).
Piberger, A. L. et al. PrimPol-dependent single-stranded gap formation mediates homologous recombination at bulky DNA adducts. Nat. Commun. 11, 1–14 (2020).
Brosh, R. M., Waheed, J. & Sommers, J. a. Biochemical characterization of the DNA substrate specificity of Werner syndrome helicase. J. Biol. Chem. 277, 23236–23245 (2002).
Fratini, F. et al. Phosphorylation on threonine 11 of β-dystrobrevin alters its interaction with kinesin heavy chain. FEBS J. 279, 4131–4144 (2012).
Steen, H., Stensballe, A. & Jensen, O. N. Alkaline phosphatase treatment of phosphopeptides: on-probe dephosphorylation after MALDI-MS analysis. CSH Protoc. 2008, pdb.prot4612 (2008).
Tyanova, S., Temu, T. & Cox, J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 11, 2301–2319 (2016).
Yu, F. et al. Analysis of DIA proteomics data using MSFragger-DIA and FragPipe computational platform. Nat. Commun. 14, 4154 (2023).
Quinet, A., Carvajal-Maldonado, D., Lemacon, D. & Vindigni, A. Chapter Three – DNA Fiber Analysis: Mind the Gap! in Methods in Enzymology 591, 55–82 (2017).
Acknowledgements
We want to thank Prof. Achille Pellicioli for helpful discussion and advise. We thank Dr. Giuseppe Leuzzi for the help with initial characterisation of fork progression phenotype in HEK293T cells expressing the WRN mutant. This work was supported by investigator grants from Associazione Italiana per la Ricerca sul Cancro (AIRC) to P.P. (IG n. 21428) and to A.F. (IG n. 19971), and in part by NIH (grant n. 1R01DE029471-01A1), Worldwide Cancer Research (Grant n 24-0262) and the Intramural Research Programme, National Institute on Aging, NIH.
Author information
Authors and Affiliations
Contributions
A.N. performed the characterisation of WRN phosphorylation mutant in vitro and in vivo, and most of the functional analysis of the CK2-phosphorylation WRN mutants. V.P. performed the analysis of end-resection at CPT-induced DSBs and generated the acidic-domain deleted WRN mutant. F.D.F contributed to the analysis of DNA damage in cells expressing the CK2-unphosphorylable WRN mutant. P.P. performed radioactive kinase assays and kinase assays for MS/MS. F.F. performed MS/MS analysis and phosphorylation sites identification. M.S. performed the purification of WRN fragments. P.V. performed the analysis of G4 accumulation. B.P. analysed phosphorylation in the WRN-2A and performed analysis of viability. J.A.S. and T.K. performed WRN purification and enzymatic assays. M.C. supervised the MS/MS analysis. V.P., M.S., and S.R. analysed data and contributed to designing the experiments. All authors contributed to design experiments and analyse data. R.M.B., P.P. and A.F. supervised the project and wrote the paper. All authors contributed to revise the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Noto, A., Valenzisi, P., Di Feo, F. et al. Phosphorylation-dependent WRN-RPA interaction promotes recovery of stalled forks at secondary DNA structure. Nat Commun 16, 997 (2025). https://doi.org/10.1038/s41467-025-55958-z
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-55958-z