Abstract
DNA polymerase θ (Pol θ) is an A-family DNA polymerase specialized in DNA double-strand breaks repair and translesion synthesis. Distinct from its high-fidelity homologs in DNA replication, Pol θ catalyzes template-dependent DNA synthesis with an inherent propensity for error incorporation. However, the structural basis of Pol θ’s low-fidelity DNA synthesis is not clear. Here, we present cryo-electron microscopy structures detailing the polymerase domain of human Pol θ in complex with a cognate C:G base pair (bp), a mismatched T:G bp, or a mismatched T:T bp. Our structures illustrate that Pol θ snugly accommodates the mismatched nascent base pairs within its active site with the finger domain well-closed, consistent with our in-solution fluorescence measurement but in contrast to its high-fidelity homologs. In addition, structural examination and mutagenesis study show that unique residues surrounding the active site contribute to the stabilization of the mismatched nascent base pair. Furthermore, Pol θ can efficiently extend from the misincorporated T:G or T:T mismatches, yet with a preference for template or primer looping-out, resulting in insertions and deletions. Collectively, our results elucidate how an A-family polymerase is adapted for error-prone DNA synthesis.
Similar content being viewed by others
Introduction
DNA polymerase θ (Pol θ) plays critical roles in safeguarding genome stability and has emerged as a promising therapeutic target for cancer treatment. As the founding member of the theta-mediated end joining (TMEJ) pathway1, Pol θ repairs deleterious double-strand breaks (DSBs) within genomic DNA, complementing homologous recombination (HR) and non-homologous end joining (NHEJ) pathways2. Pol θ is a 2590 amino-acid long protein with a super-family II helicase domain (Pol θ-hel) at the N-terminus and an A-family polymerase domain (Pol θ-pol) at its C-terminus3. During TMEJ, Pol θ-hel displaces RPA (Replication Protein A) and Rad51 recombinase at the resected DNA ends4,5, paving the way for microhomology-dependent (2-6 bp) DNA synthesis catalyzed by Pol θ-pol6,7. In addition to its role in DSB repair, Pol θ performs translesion synthesis (TLS) across various DNA lesions, including but not limited to abasic sites (AP)8,9, thymine glycol (Tg)8,9, alkylated purines10,11, and cyclobutane pyrimidine dimers (CPD)12, a major type of ultraviolet (UV)-induced damage. Depletion of Pol θ in mouse models significantly increases the incidence of skin cancer caused by UV exposure12. Due to the synthetic lethality observed between Pol θ and DSB repair proteins BRCA1 and BRCA2 in cancer cells, targeting Pol θ has been extensively explored for the treatment of breast and ovarian cancers associated with BRCA-mutations13,14. Several inhibitors targeting either Pol θ-hel or Pol θ-pol have been reported and proceeded to clinical tests (reviewed in15).
Pol θ-pol is distinct from the prototypical A-family replicative polymerases by exhibiting a remarkable propensity for error-prone DNA synthesis. When replicating undamaged DNA, Pol θ generates single base substitutions at an average rate of 2.4 * 10−3 16, which is 10- – 100-fold higher than A-family replicative homologs but comparable to that of human polymerase κ (Pol κ)(5.8 * 10−3), a member of error-prone Y-family polymerases involved in TLS. It also induces single base insertions and deletions at frequencies similar to those of Y-family Pol κ and polymerase η (Pol η)16. Moreover, Pol θ has been implicated in promoting error-prone TLS against CPD, contrasting with the relatively error-free CPD bypass executed by Pol η12. The low-fidelity DNA synthesis by Pol θ has been suggested to associate with somatic hypermutation for antibody diversity17 and genome evolution18. However, the active site of Pol θ strongly resembles that of its high-fidelity homologs when bound to correct nucleotides19,20,21,22. The mechanism by which Pol θ executes error-prone DNA synthesis during various DNA repair and TLS processes remains unclear. In this report, we applied cryo-electron microscopy (cryo-EM) to obtain three high-resolution structures of Pol θ complexed with correct and incorrect nascent base pairs. Combined with in-solution fluorescence, mutagenesis and kinetic analysis, our study elucidates how an A-family polymerase is adapted for error-prone DNA synthesis.
Results
Structures of Pol θ with mismatched incoming nucleotides
We expressed the polymerase domain of human Pol θ (residues 1792-25909, referred to as Pol θ for the remaining text) and purified it to homogeneity. Prior studies have demonstrated that Pol θ catalyzes template-dependent DNA synthesis with an inherent propensity for error incorporation. It tends to incorporate incorrect nucleotides most frequently opposite a templating dT, with T:G and T:T being among the most abundant mismatches observed in sequencing data8,16. Consistent with these findings, we observed Pol θ not only incorporating dATP opposite a templating dT but also misincorporating dTTP and dGTP (Fig. 1a). To elucidate the mechanism underlying Pol θ misincorporation, we determined three cryo-EM structures of Pol θ complexed with duplex DNA and one of the following: (1) a templating dC with an incoming dGTP (CG), (2) a templating dT with an incoming dGTP (TG), or (3) a templating dT with an incoming dTTP (TT), to resolution of 3.1-3.4Å (Supplementary Figs. 1–3). The preparation of these complexes involved incubating Pol θ with the incoming nucleotide, magnesium ions (Mg2+), and a duplex DNA consisting of a 29 nt template and a 20 nt primer with a dideoxynucleotide-terminating end, which prevents further chain elongation. For each of the three complexes, a single conformation with high-resolution features was separated by 3D classification.
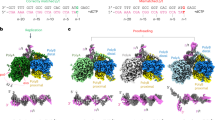
a DNA synthesis by Pol θ in the presence of individual (A, T, C or G) or all four dNTPs (N). The duplex DNA substrate, denoted as 14-26_A+2, consists of a 14 nt, \({5}^{{\prime} }\) fluorescein-labeled primer paired with a 26 nt template. The untreated DNA control is denoted as “-''. The length of the primer strand and extended products is indicated on the sides of the gel. The gel shown here is a representative, with two independent replicates yielding consistent results. Source data are provided as a Source Data file. b Overall cryo-EM structures of the CG, TG and TT complexes at resolutions of 3.1, 3.3 and 3.4 Å, respectively. Individual domains are color-coded and the cryo-EM density map is overlaid onto the structural model. The visible portions of the DNA substrates in cryo-EM and incoming nucleotides are depicted above each model.
The cryo-EM structure of the CG complex looks similar to the recently released crystal structure of human Pol θ20 and the cryo-EM structure of Lates calcarifer Pol θ (LcPol θ)22, with an overall RMSD of 0.67Å and 0.73Å, respectively. The protein segments within the TG and TT complexes are almost identical to those in the CG complex, with RMSD of 0.57Å and 0.45Å, respectively (Fig. 1b). Across all three structures, the palm domain (where the active site resides) and the finger domain were resolved at higher resolution compared to the pseudo-exonuclease and thumb domains (Supplementary Figs. 1–3). The unique insertion loops within Pol θ remained predominantly disordered, similar to those in previous structures of human and LcPol θ19,20,22. DNA binding is evident in all three structures: 11, 15 and 6 bp of the duplex DNA can be confidently refined in the complexes CG, TG, and TT, respectively.
Closure of the Pol θ finger domain during mismatch incorporation
For A-family polymerases, the finger domain undergoes ‘open-to-close’ conformational changes upon binding a correct incoming nucleotide23,24,25. The closed conformation of Pol θ finger domain has been reported for Pol θ complexed with double-stranded DNA and an incoming nucleotide19,20,21, while the open-finger conformation has been observed for a binary complex of Pol θ bound to an RNA-DNA hybrid26. Similar to the A-family homolog Taq Polymerase I (Taq, 3KTQ)23, the finger domain in the CG structure adopts a well-closed state with the incoming dGTP and a Mg2+ ion bound to the active site (Fig. 2a). The conformation of the finger domain closely resembles that observed in the cryo-EM structure of LcPol θ22 (8EF9) and the crystal structure of hPol θ (4X0P). However, the termini of the O-helix in the finger domain are rotated by ~6° and 12°, and shifted by 4Å and 5Å relative to its corresponding positions in the crystal structures of hPol θ (7ZUS and 4X0Q), respectively (Fig. 2a and Supplementary Fig. 4). Intriguingly, both the TG and TT structures revealed that the finger domain is well-closed, similar to the configuration observed in the CG structure (Fig. 2b). The well-closed conformation of the finger domain within TG and TT complexes are distinct from that of A-family Bacillus Pol I (3HP6)27, which adopts an ajar conformation with a misaligned dGMP:ddTTP in the active site (Fig. 2c).
a Finger domain movements in the CG complex (blue) compared to human Pol θ crystal structure (7ZUS, orange) and Pol I (3KTQ, yellow). The Mg2+ ions, incoming nucleotides, and critical residues for catalysis are shown as sphere and sticks, respectively. b Comparison of the finger domain conformation in the CG, TG (magenta) and TT (green) complexes. c Comparison of the finger domain conformation in the TT complex with that observed in Pol I structures. The finger domain in the TT complex remains closed, similar to that in Pol I bound with a cognate nascent base pair (3KTQ, yellow). However, it differs from the ajar conformation of Pol I with a mismatched base pair in its active site (3HP6, purple). d Conformation of the nascent base pairs in the three structures. Hydrogen bonds between the templating base and the incoming nucleotide are indicated by dashed lines, and the electron density map is displayed as meshes. Atoms involved in hydrogen bonding are labeled.
Despite the well-closed finger domain observed across all three structures, the triphosphate moiety of the incoming nucleotide adopts varied conformations. In the CG structure, it is in the chair-like form and coordinates well with the B-site Mg2+ through its oxygen atoms (Fig. 2d and Supplementary Fig. 5a). In contrast, the TG structure lacks well-defined electron density for Mg2+ and the triphosphate adopts a distorted conformation, with the γ-phosphate bending back toward the sugar ring rather than pointing away, as seen in the CG structure (Fig. 2d and Supplementary Fig. 5b). The T:G mismatch exhibits a Hoogsteen base pair geometry, stabilized by a single hydrogen bond between N3 of the templating dT and O6 of the incoming dGTP. In the TT structure, the triphosphate configuration closely resembles that in the CG structure. Nevertheless, the electron density for Mg2+ is absent at the B site. Instead, the Mg2+ ion is coordinated with one oxygen atom on the α-phosphate group and another on D2330 side chain (Fig. 2d and Supplementary Fig. 5c). The nucleobases of the templating dT and the incoming dTTP form a geometry reminiscent of the previously reported mercury-mediated T:T mispair, T-HgII-T28. Considering the highly negatively charged active site and the triphosphate group, we believe that the B-site Mg2+ is likely present in the TG structure to neutralize the charges. However, the lack of well-defined electron density for B-site Mg2+ suggests that the mismatch may impair proper metal coordination, resulting in lower occupancy and/or increased dynamic behavior of the metal ion.
In-solution fluorescence study confirms the tendency of finger domain closure
To complement our structural analysis, we conducted an in-solution fluorescence study to further investigate the conformational changes of Pol θ during the recognition and incorporation of mismatched base pairs. The in-solution fluorescence assay has been previously employed to monitor the finger domain movement in Pol θ, Klenow Fragment, Bacillus Pol I, T7 polymerase and polymerase β27,29,30,31,32. Due to the presence of multiple cysteine residues in Pol θ, the conventional cysteine-substitution strategy for protein labeling was not feasible. Instead, we took the advantage of a site-specific protein labeling approach using the transglutaminase enzyme (TGase)29,33. TGase crosslinks glutamine and lysine sidechains, enabling the covalent attachment of small molecule probes to recombinant proteins engineered with a 6- or 7-amino acid TGase recognition sequence, known as a Q-tag.
Our approach utilized fluorescence transfer between a FAM donor on the Pol θ finger domain and a non-fluorescent Black Hole Quencher (BHQ) acceptor on the DNA strand. The formation of the closed conformation was expected to reduce the distance between FAM and BHQ, resulting in a decrease in FAM intensity. To optimize the fluorescence experiment, we screened various mutations and insertion sites for creating a Q-tag sequence (GQQQLG) for TGase recognition and tested multiple positions for BHQ labeling on the primer DNA. The Qtag-ins1 mutant, with the Q-tag inserted into the loop between the O- and N-helices, proved optimal, exhibiting high protein expression yield and purity, and DNA synthesis activity comparable to the wild-type enzyme (Fig. 3a and Supplementary Fig. 6a–c). For the DNA substrate, the BHQ modification positioned 5 nucleotides away from the insertion site on the primer strand (BHQ5) was determined to be the most effective for fluorescence analysis. (Fig. 3b).
a Structural illustration of finger domain movement responsible for fluorescence signal variation. The structures represent the finger domain in the closed state (CG complex, cyan) and the open state (Qtag-ins1 model predicted by AlphaFold, orange). Regions undergoing the largest conformational changes during finger closure---the O-helix, N-helix, and the intervening loop---are fully colored, while other segments are shown in a translucent view. The second glutamine in the Q-tag is depicted in red sphere, with the approximate corresponding location in the CG complex shown in blue sphere. The nucleobase modified with BHQ is colored black. b DNA sequence used for the fluorescence study with the BHQ-modified thymine highlighted in black. Note that the nucleobase marked in black in (panel a) indicates the position of the BHQ modification but does not correspond precisely to the modified nucleobase in the fluorescence experiments. The DNA sequence in the CG complex differs from that used in the fluorescence study. c Fluorescence emission spectra of FAM-labeled Qtag-ins1 bound to the BHQ-modified DNA with the correct incoming nucleotide dATP and ART558. The unit of fluorescence intensity, a.u., refers to arbitrary units. d Comparison of integrated emission peak intensities (510-530 nm) in the presence of 2 mM of each incoming nucleotide, illustrating the varying degrees of finger domain closure. The open and closed states are defined by the binary complex of the enzyme and DNA, and the ternary complex of the enzyme bound to DNA, dATP, and ART558, respectively. Data are presented as mean ± SD from four independent experiments. *p < 0.1, **p < 0.01, ***p < 0.001, determined by a two-sided t-test. Paired for comparisons between dATP and dATP+ART558, and unpaired for comparisons of dATP with dCTP, dTTP, or dGTP. Source data are provided as a Source Data file.
For the fluorescence sample preparation, the following components were sequentially added to 50 nM Qtag-ins1: (1) 100 nM DNA, (2) 200 uM dXTP, (3) 2 mM dXTP, (4) 1 uM ART558, and (5) 10 uM ART558. ART558 is a potent, allosteric inhibitor of Pol θ with an IC50 of 7.9 nM34. It specifically targets the finger domain, with its binding site forming only in the closed state35. ART558 was included to lock Pol θ in the closed state, serving as a control for data analysis. We confirmed that ART558 has minimal emission at 520 nm (Supplementary Fig. 6d), ensuring it does not interfere with FAM intensity readings. As expected, the initial protein-only sample (Eonly) exhibited the highest FAM intensity due to the absence of BHQ (Fig. 3c and Supplementary Fig. 6d). Upon adding DNA (E+DNA), a significant reduction in FAM intensity was observed, indicating efficient protein-DNA binding and binary complex formation. The subsequent addition of 200 μM, followed by 2 mM, of either correct or incorrect nucleotides promoted the formation of ternary complexes of Pol θ bound to DNA and dXTP (X = A, T, C or G), further reducing fluorescence intensity. This decrease indicates a progressive shift of the finger domain toward a closed conformation. The trend of closing continued with the addition of 1 μM, followed by 10 μM, ART558, resulting in an even greater reduction in FAM intensity (Supplementary Fig. 6d), suggesting a more closed conformation of Pol θ.
Next, we compared the fluorescence emission spectra of Pol θ complexed with DNA and each of the four incoming nucleotides. The comparison reveals that Pol θ’s finger domain tends to close in the presence of all mismatches involving a templating dT in solution (Fig. 3d).
Mismatches are stabilized in the active site of Pol θ
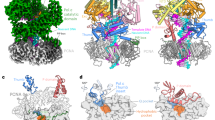
Interactions between Pol θ and the nascent base pair within the CG structure closely resemble those observed in previously reported LcPol θ and exhibit a near-perfect overlay with the ternary complex of Taq (Fig. 4a). R2379, K2383, and Y2387 from the O-helix and K2575 from the palm domain contact the triphosphate of the incoming dGTP (Fig. 4b). G2394 from the Oα-helix stabilizes the phosphate backbone surrounding the templating dC. Additionally, the incoming dGTP forms a Watson-Crick base pair with the templating dC and base-stacks with the primer end. The sidechain of Q2384 establishes direct contact with the nucleobases of the incoming dGTP. The B-site Mg2+ is coordinated by one oxygen atom from the β-phosphate of the incoming dGTP, along with the side chains of D2540 and D2330 and the main chain of Y2331. Due to the absence of the \({3}^{{\prime} }\)-OH at the ddATP-terminating end, the A-site Mg2+ lacks well-defined electron density.
a Interactions at the active site in the CG complex (blue) are nearly superimposable with those in LcPol θ (salmon) and Pol I (yellow). O- and Oα-helices are shown as semi-transparent cartoon. b–d Interactions at the active site in the CG, TG and TT complexes. Critical contacts between Pol θ, the nascent base pair, and Mg2+ are shown as dashed lines. The finger and palm domains are colored in lightblue and lightpink, respectively.
Despite the distinct conformations of the triphosphate of incoming dNTPs, the mismatched T:G and T:T are accommodated snugly in the active site. In the two structures with mismatches, most interactions are retained between Pol θ and the triphosphate of incoming nucleotides compared to that observed in the CG structure (Fig. 4c and d). Interestingly, in the TG structure, while Q2384 interacts with the templating dT, the side chain of Q2380 stabilizes the nucleobase of incoming dGTP (Fig. 4c). In the TT structure, Q2384 contacts the nucleobases of both the templating dT and the incoming dTTP. (Fig. 4d). The direct hydrophilic interactions observed here between Pol θ and the nucleobases of the nascent base pair differs Pol θ from high-fidelity A-family polymerases. In Pol I, a water-mediated hydrogen bonding recognizes the incoming nucleobase (Supplementary Fig. 7a). Notably, this water-mediated interaction, which has been considered as an important edge recognition of cognate base pair shapes, is absent when a mismatch occupies the active site of Pol I (Supplementary Fig. 7b).
Besides the nascent base pair, Pol θ interacts with the primer strand in a similar manner across the three structures (Supplementary Fig. 8a). In particular, R2254 and R2315 contact the primer terminus at position n-1, while K2181 interacts with positions n-3 and n-4. At position n-2, the CG and TG complexes utilize R2254, whereas the TT complex utilizes R2202. Additional contacts are observed for N2205 with position n-3 in the CG structure, R2202 with position n-3 in the TG structure, and R2201 with position n-4 in the TT structure. Conversely, Pol θ displays greater flexibility for extensive interactions with the template strand at positions n-1 to n-4. Among the three complexes, only R2466 and H2463, R2448, A2238, and Q2234 and N2248 are shared to contact positions n-1, n-2, n-3, and n-4, respectively. Several other residues are employed by one or two of the complexes to interact with the phosphate backbone, sugar ring and nucleobases (Supplementary Fig. 8a).
Unique residues contribute to error-prone DNA synthesis by Pol θ
We aimed to confirm the structural features contributing to the error-proneness of Pol θ. Several hydrophilic residues uniquely conserved in Pol θ, could potentially stabilize its unusual finger-closed state with a mismatched nascent base pair (Fig. 5a). We therefore generated Pol θ mutants with alanine substitutions for these hydrophilic residues, except for G2394S where glycine was replaced by serine to align with the corresponding residue in Pol I.
a Sequence alignment of the hinge region of loop2 and O-, Oα- and Q-helices across A-family polymerases. Residues highlighted in yellow are either uniquely conserved in Pol θ and/or are observed in structures making crucial interactions in the active site. Residues in red are highly conserved throughout the A-family. b Steady-state kinetics of single nucleotide incorporation by wild-type and mutant Pol θ. Pol θ fidelity is reflected by its preference for incorporating the correct dA over the incorrect dT or dG (indicated by the ratios of kcat/KM for dA over dT or dG). The kinetic parameters are presented as mean ± SD from multiple independent experiments (n = 3 – 5). Source data are provided as a Source Data file.
The mutants were initially screened via DNA synthesis assays (Supplementary Fig. 9), and those exhibiting altered fidelity compared to the WT enzyme were subsequently assessed through kinetics analysis (Fig. 5b and Supplementary Fig. 10). Firstly, R2254, situated in the hinge region of loop2, interacts with the phosphate groups at primer terminus. While it is a conserved lysine in Pol ν, it is a hydrophobic residue in Pol I. Mutating R2254 to valine or alanine has been demonstrated to decrease Pol θ’s ability to bypass an AP lesion19. In our kinetics study, R2254A exhibited a 1.7-fold increase in Pol θ’s selection of dATP over dTTP opposite a templating dT, possibly because the hydrophobic substitution weakens the binding of DNA backbone and disfavors the primer end \({3}^{{\prime} }\)-OH alignment in the presence of an incorrect incoming nucleotide (Supplementary Fig. 8b). Secondly, the side chains of Q2380 and Q2384 stabilize the T:G and T:T mismatches in our structures. The ability of Q2384 to interact with the nascent base pair has been observed previously19,20,21,22. These two residues are predominantly conserved in Pol θ but varies in Pol I36. Q2380A exhibited increased Pol θ selection of dATP over dTTP opposite a templating dT by 3.1-fold, whereas Q2384A unexpectedly decreased the selection of dATP over dGTP by 2-fold. Interestingly, these two residues are also conserved in Pol ν as a glutamate and an arginine/lysine, respectively. Mutating the conserved lysine to alanine in human Pol ν (K679A) has been shown to increase Pol ν fidelity approximately tenfold37. Thirdly, G2394, which is a conserved serine in Pol I, facilitates a sharp turn between the O- and Oα-helices. This glycine is also conserved in the error-prone Pol ν. Nevertheless, G2394S displayed subtle differences in nucleotide selection compared to the WT. Lastly, H2463 and Q2467 anchor the template at positions n-1 and n-2 via direct hydrogen bonding, a feature absent in Pol I. While H2463A displayed no change in Pol θ’s fidelity, Q2467 enhanced Pol θ selection of dATP over both dTTP and dGTP. The construct with double mutations, H2463A&Q2467A, exhibited increased selection of dATP over dTTP opposite a templating dT by 1.8-fold. Despite the modest impact of individual residues, their collective interactions may synergistically enhance Pol θ’s ability to accommodate mismatches, thereby attenuating Pol θ’s fidelity. Interestingly, the mutants with increased fidelity in error-prone synthesis exhibited reduced TLS activity in bypassing Tg lesions compared to the WT enzyme (Supplementary Fig. 11), suggesting that these unique residues may be strategically utilized by Pol θ to cope with imperfect DNA synthesis. Nevertheless, none of the mutations significantly suppressed Pol θ’s DNA synthesis activity on TMEJ substrates (Supplementary Fig. 12).
Loop1 and Loop2 exert minor influence on Pol θ misincorporation
The insertion loops in Pol θ polymerase domain have been reported to promote TMEJ and TLS9,38. In addition to the aforementioned single/double mutations, we also explored the impact of loop1 and loop2 of Pol θ on its fidelity. Sequence alignment revealed that Pol θ possesses a conserved loop 1, which is absent in high-fidelity A-family polymerases such as Pol I and exists in a shorter form in Pol ν (Supplementary Fig. 13), and we previously showed that loop1 of LcPol θ assisted in DNA binding, primarily attributed to the presence of lysine and arginine residues within the loop22. Loop2 was reported to contribute to both Pol θ DNA synthesis activity and TLS, as its depletion (with resi 2264-2315 deleted) resulted in less efficient DNA synthesis and reduced bypass of AP and Tg9. However, the region of loop2 has been re-defined in our recent study of LcPol θ, and loop2 deletion in LcPol θ (equivalent to the removal of resi 2261-2308 in human Pol θ) showed limited impact on DNA synthesis efficiency22.
To assess the influence of loop1 and loop2 on Pol θ misincoporation performance, we generated Pol θ mutants with either loop1 (resi 2149-2170) or loop2 (resi 2261-2308) deleted, denoted as Δloop1 and Δloop2, respectively (Supplementary Fig. 13). Kcat/KM values obtained from steady-state kinetics for the two mutants revealed that neither loop deletions significantly altered Pol θ’s specificity for the three incoming nucleotides tested or its capability in discriminating between dATP and dTTP or dGTP opposite a templating dT (Fig. 6), suggesting that the insertion loops 1 and 2 are not critical for error-prone synthesis of Pol θ.
a Structure of Pol θ with loop1 and loop2 indicated by dashed lines. b Steady-state kinetics of single nucleotide incorporation by wild-type and mutant Pol θ. Pol θ fidelity is reflected by its preference for incorporating the correct dA over the incorrect dT or dG. The kinetic parameters are presented as mean ± SD from multiple independent experiments (n = 3 to 4). Source data are provided as a Source Data file.
Looping-out of primer or template for DNA extension beyond mismatches
Lastly, we examined Pol θ’s ability to synthesize DNA beyond misincorporated T:T or T:G mismatches. For substrates with a T:T (TT-1_noA) or T:G (TG-1_noC) mismatch at the primer terminus (position -1), Pol θ efficiently incorporated the correct incoming nucleotide for the next cycle, and generated full-length products in the presence of dNTPs (Fig. 7a, b). Consistent with previous reports6,22,39, Pol θ can efficiently add a single nucleotide to the \({3}^{{\prime} }\) terminus of blunt-ended dsDNA, and thus the length of the final products is one nucleotide longer than that dictated by the sequence in the presence of dNTPs. Interestingly, with a dA designed at position +1 featuring a T:T mismatched primer terminus (TT-1_A+1), incorporation of both the correct dTTP and the incorrect dCTP was observed, with the latter being more efficient (Fig. 7a). Moreover, the most abundant final product in the presence of dNTPs for TT-1_A+1 was one nucleotide shorter than that for TT-1_noA (Fig. 7a). These observations suggest template looping-out during DNA extension past the mismatched T:T. Similarly, template looping-out was also evident with a dC designed at position +1 featuring a T:G mismatched primer terminus (TG-_C+1) (Fig. 7b). In addition, we examined DNA synthesis by Pol θ with a T:T (TT-2) or T:G (TG-2) mismatch at position -2 (Fig. 7c). With a T:T mismatch at position -2 (TT-2), consecutive incorporation of dATP or dGTP was observed, and the final products were of three different lengths in the presence of dNTPs, reflecting scenarios of direct extension as well as primer or template looping-out (Fig. 7c). Similarly, multiple looping-out and rearrangement took place at the primer terminus for the substrate with a T:G mismatch at position -2 (TG-2). dCTP, dTTP and dATP were all efficiently incorporated, and much shorter final products were observed in the presence of dNTPs (Fig. 7c). Collectively, these findings underscore Pol θ’s capacity to extend beyond previously incorporated mismatches, demonstrating an exceptional tolerance for alternative base pairings at the primer terminus, which leads to insertions and deletions.
a, b Extension of a DNA substrate by Pol θ featuring a (a) T:T or (b) T:G mismatch at position -1. c Extension of a DNA substrate by Pol θ featuring a T:T or T:G mismatch at position -2. For all panels, DNA synthesis was conducted with individual nucleotides (A, T, C, or G) or a mix of all four dNTPs (N), with the untreated DNA control denoted as “-''. Product formation (%) is indicated below each gel, and lengh of the primers and the most abundant final products are noted. The DNA sequences and potential base pairings at the primer terminus are depicted on the side of gels. The gels shown here are representatives, with three independent replicates yielding consistent results. Source data are provided as a Source Data file.
Discussion
Previous extensive studies have summarized various structural features of polymerases that contribute to their high-fidelity DNA synthesis. The ‘open-to-close’ conformational transition of the finger domain scrutinizes the correct incoming nucleotides, whereas the \({3}^{{\prime} }\to {5}^{{\prime} }\) exonuclease domain, either within or associated with replicative polymerases, removes the misincorporated nucleotides. Apart from the global selection mechanisms, polymerases exploit fine-tuned means to locally scrutinize nascent base pairs, including edge recognition for the shape of cognate Watson-Crick base pairs and Mg2+-assisted active site alignment. Nonetheless, mismatched nascent base pairs can be tolerated and stabilized in the active site of some polymerases, via strategies such as base tautomerization40,41, solvation of the active site42, and adopting non-Watson-Crick geometries such as Hoogsteen43, syn-trans44, and wobble base configurations45,46. Furthermore, primer and template looping-out have been proposed for bypassing mismatches and lesions during DNA synthesis, leading to insertions and deletions37,47,48,49.
Unlike A-family replicative polymerases, Pol θ possesses an exonuclease domain, but it is inactive for proof reading because three of the four catalytic carboxylates required for exonuclease activity are substituted with serine and alanine residues. Our structures show that Pol θ exhibits a well-closed conformation of its finger domain not only when complexed with the cognate C:G bp but also when bound to the mismatched T:G and T:T bp. Notably, this closed conformation in the presence of incorrect nucleotides is distinct from that of A-family replicative polymerases, wherein the finger domain cannot fully close upon encountering mismatches (Figs. 2c and 8). Our in-solution fluorescence experiment further confirmed that both correct and incorrect incoming nucleotides can induce finger domain closure, with the most pronounced signal changes observed for the cognate T:A base pair. However, none of nucleotides were able to induce closure of the finger domain to the same extent as the allosteric inhibitor ART558, which specifically binds to the closed state. The intermediate fluorescence signals suggest that the Pol θ finger domain exists in an equilibrium between open and closed states, with the equilibrium shifting toward the closed state upon binding of either correct or incorrect incoming nucleotides. This behavior contrasts sharply with previous fluorescence studies on high-fidelity polymerases, including Klenow fragment, Bacillus Pol I, and T7 polymerase, where matched nascent base pairs result in decreased fluorescence intensity, while mismatches lead to an increase27,30,31. In addition, the closed finger domain of Pol θ may impede the dissociation of the incorrect incoming nucleotides. Intriguingly, this configuration resembles that observed in low-fidelity Y-family polymerases and certain X-family polymerases, where the finger-closed conformation persists throughout DNA synthesis reaction cycle (Fig. 8). Nevertheless, Pol η is associated with enlarged active site that can tolerate bulky DNA lesions like CPD50. The active site of Pol θ is tightly closed and may not be able to accommodate CPD, similar to the scenario observed for T7 polymerase51. How Pol θ promotes error-prone CPD bypass requires further investigation.
Cartoon models shown from left to right are generated from structures of Pol I with a T:G mismatch (3HP6), the TG complex presented in the current work, and Pol η with CPD (3MR3). The active exonuclease domain of Pol I and the unique little finger domain of Pol η are colored yellow and magenta, respectively. The open finger conformation in Pol I and Pol θ is outlined in blue dashed line, and the active sites in all three models are boxed. Metal ions and incoming nucleotides are depicted by green spheres and red shapes, respectively, while key amino acid residues around the active sites are illustrated with gray elongated hexagons.
Pol θ exhibits distinct local structural features that contribute to its low-fidelity as well. Firstly, cognate Watson-Crick base pairs exhibit uniform shapes recognizable by replicative polymerases. For high-fidelity A-family replicative polymerases, a critical water-mediate hydrogen bond encodes edge recognition of cognate base-pair shapes (Supplementary Fig. 7)40. In contrast, Pol θ utilizes Q2384 and Q2380 to directly hydrogen bond with the nucleobases of correct and incorrect base pairs, similar to the scenario of X-family Pol μ stabilizing the mismatch using Q441, and Y-family Pol η using Q38 and R61. Although the T:G mismatch adopts a Hoogsteen geometry with only one hydrogen bond between the templating dT and incoming dGTP, Q2380 stabilizes the incoming nucleobase. It appears that direct contacts with the mismatched incoming nucleobase represent a common strategy employed by error-prone polymerases to perform TLS. Unexpectedly, Q2384A slightly reduced the fidelity of Pol θ, whereas the corresponding residue K679 in Pol ν increased the fidelity37. We speculate that, in the absence of Q2384, water molecules and/or nearby side chains may stabilize the incoming nucleotide, maintaining the low fidelity of Pol θ. Secondly, the triphosphate moiety of the incoming nucleotide displays diverse conformations. In the CG structure, it mirrors the conformation seen in Pol I, coordinating well with the B-site Mg2+ through its oxygen atoms. Although the TT structure shows a similar conformation to the CG structure, the density for Mg2+ is shifted away from the B-site, indicating the destabilization induced by the mismatch. Conformation of the triphosphate captured in the TG structure, on the other hand, appears distorted and likely non-productive. The altered conformations of the triphosphate and the poor density of the associated metal ion suggest potential challenges in aligning and incorporating the incorrect incoming nucleotides. Thirdly, unique residues are utilized by Pol θ to interact with the DNA backbone around the active site. Pol θ anchors the primer strand at position n-1 with R2254 and R2315, whereas Pol I utilizes only a single arginine or water molecules for this purpose. The discrepancy is more obvious for interactions with the template strand, where Pol θ extensively contacts the DNA backbone across positions n-1 to n-4 (Fig. 8 and Supplementary Fig. 8). This observation is reminiscent of Pol η acting like a “molecular splint” to minimize the perturbation caused by mismatched or damaged nascent base pairs in its active site50. The extensive interactions between Pol θ and the DNA backbone around the active site may also stabilize microhomology during TMEJ, contributing to its ability to perform efficient DNA synthesis in challenging contexts.
Besides mismatch incorporation, our biochemical assays suggested that Pol θ can promote alternative primer-end pairing during mismatch extension leading to insertions and deletions. This template-looping mechanism has also been suggested in studies examining Pol θ extending DNA after incorporating dATP opposite an AP lesion or dGTP opposite a templating dG48,49. Additionally, structural analyses suggest that a surface opening near the insertion loop 2 may help to accommodate a looped-out primer in Pol ν and Pol θ37. However, further structural investigations are needed to decipher how a looped-out primer or template can be stabilized in Pol θ’s active site, considering the extensive protein-DNA interactions near the nascent base pair.
In conclusion, findings from the three structures along with the mutagenesis study underscore the nuanced interactions between unique residues surrounding Pol θ’s active site and both the nascent base pair and the DNA backbone. In addition, our biochemical investigations confirm that Pol θ facilitates the looping-out of primer and template during mismatch extension, leading to insertions and deletions. These observations highlight Pol θ’s distinct molecular mechanism in accommodating mismatches relative to high-fidelity polymerases within the A-family.
Methods
Cloning, expression and purification of wild-type and mutant Pol θ-pol
The gene encoding the polymerase domain of human Pol θ (aa 1792-2590) was synthesized by Gene Universal and was inserted into the pETsumo2 vector containing a His6 and a SUMO tag. All mutants were generated by PCR-based mutagenesis. Pol θ-pol and mutants were expressed in BL21(DE3) cells by 200 μM IPTG induction at 16 °C for 18 h and were purified to homogeneity as follows. WT and mutant His6-SUMO tagged proteins were firstly cleaned by nickel gravity flow. After removal of the His6-SUMO tag by SENP2 protease, they were further purified through a HiTrap heparin HP column. All purification steps were performed at 4 °C. Purified proteins were concentrated to 6–16 mg/mL, aliquoted and stored at -80 °C in 25 mM Tris [pH 8.0], 300 mM NaCl, 3 mM dithiothreitol (DTT) and 30% glycerol (except for Qtag-ins1). A fresh aliquot was used each time for cryo-EM sample preparation and biochemical assays.
DNA synthesis assay
All DNA strands used in the present work were purchased from Integrated DNA Technologies. Their nomenclature and sequences are listed in Supplementary Table 1. Fidelity check of WT and mutant Pol θ, as shown in Fig. 1 and Supplementary Fig. 9, was carried out using 50 nM of the enzyme and 100 nM of DNA 14-26_A+2. DNA synthesis reactions took place in 10 μL of reaction buffer containing 25 mM Tris [pH 7.6], 100 mM KCl, 3 mM DTT, 100 μg/mL BSA, 5% glycerol, 5 mM MgCl2, and 100 μM each of the dXTPs or all four. Following a preincubation of the protein-DNA mixture at 37 °C for 2 min, reactions were initiated by adding the desired dXTP and MgCl2, allowed to proceed at 37 °C for 10 min, and quenched by the addition of 10 μL of stop solution containing 100 mM EDTA, 80% formamide and 0.01% bromophenol blue. Reactions shown in Fig. 7 were conducted under similar conditions but with different DNA substrates. Additionally, the reactions proceeded for 5 min before quenching. Reactions shown in Supplementary Fig. 12 were conducted under similar conditions as well but at 25 °C and with DNA substrates FB39 and MH6, which bear intra- and inter-molecular microhomologies, respectively. All reactions described above were analyzed by 20% denaturing polyacrylamide gel electrophoresis. Gels were visualized by a Sapphire Biomolecular imager (Azure Biosystem), and bands were quantified with Azure Spot (Azure Biosystem).
Labeling of Qtag-ins1 with FAM fluorophore by TGase
The labeling reaction was conducted as previously described with modifications29,33,52. A total of 300 μL freshly purified Qtag-ins1 was mixed with 3.6 μL 50 mM 5-FAM cadaverine (Tenova Pharmaceuticals), 3.6 μL 25 μM TGase from guinea pig liver (Sigma), 36 μL 100 mM CaCl2, 0.9 μL 1 M DTT, 3 μL 1 M Tris-HCl (pH 7.7), and 12.9 μL water. The final concentrations were 4.2 μM Qtag-ins1, 0.5 mM 5-FAM cadaverine, 250 nM TGase, 10 mM CaCl2, 50 mM Tris-HCl, and 2.5 mM DTT. The reaction was carried out for 1 h at room temperature, followed by an additional hour at 4 °C. Labeling was confirmed by SDS-PAGE, with the gel scanned using both the FAM and AzurePure700 channels by a Sapphire Biomolecular imager (Azure Biosystem) to visualize FAM-labeled samples and total proteins, respectively. FAM-labeled Qtag-ins1 was subsequently purified by a second HiTrap heparin HP column to remove excess 5-FAM cadaverine and TGase, concentrated, and stored at -20 °C in 25 mM Tris [pH 8.0], 300 mM NaCl, 3 mM dithiothreitol (DTT) and 30% glycerol.
In-solution fluorescence study
Samples for the in-solution fluorescence study initially contained 50mM Tris [pH 7.7], 5 mM MgCl2, 50 nM FAM-labeled Qtag-ins1, and 3 mM DTT. Components were then sequentially added to each sample as follows: (1) 100 nM DNA, (2) 200 μM dXTP, (3) 2 mM dXTP (X=A, T, C or G), (4) 1 μM ART558, and (5) 10 μM ART558. Three measurements were taken after each addition; no photobleaching was observed throughout the whole measuring process. Fluorescence emission spectra (510–650 nm) were collected using an Agilent Cary Eclipse Fluorometer at an excitation wavelength of 485 nm and plotted in GraphPad Prism 9.
Steady-state kinetics of single-nucleotide incorporation
Steady-state kinetics of WT and mutant Pol θ were assayed as follows. 1–125 nM of the enzyme were mixed with 1 μM of duplex DNA and incubated at 37 °C for 5 min (for dATP incorporation) or 10 min (for dTTP and dGTP incorporation) in 10 μL of reaction buffer containing 0–1.25 mM of dATP, or 0–3 mM of dTTP or dGTP. The duplex DNA 14-26_A+2 was used for dATP and dGTP incorporation, while 14–26_T+2 was used for dTTP incorporation. Kinetics for examining Tg bypass were conducted under similar conditions with DNA 15-26_Tg. Reactions were quenched by the addition of 10 μL of the stop solution, and were subsequently analyzed by 20% denaturing polyacrylamide gel electrophoresis. Gels were visualized by Sapphire Biomolecular imager (Azure Biosystem), and bands were quantified with Azure Spot (Azure Biosystem). Steady-state kinetics was fit to the model of ‘Michaelis-Menten equation’ using nonlinear regression in GraphPad Prism 9 to determine KM and kcat.
Cryo-EM sample preparation
To prepare the ternary complex of Pol θ, 8 μM of the protein was initially mixed with 12 μM of a duplex DNA, either 19–29_CG for the CG structure or 19–29 MM for the TG and TT structures. 100 μM ddATP (for the CG structure) or ddCTP (for the TG and TT structures) were added to the mixture and the incubation proceeded at 25 °C for 10 min in reaction buffer (25 mM Tris [pH 7.6], 100 mM KCl, 3 mM DTT and 5 mM MgCl2) to generate a ddXTP terminating end. Subsequently, 1 mM dGTP (for the CG and TG structures) or dTTP (for the TT structure) was introduced to the reaction mixture, followed by an additional 10 min incubation at 25 °C. 3.0 μL of freshly prepared complex sample was applied onto glow-discharged Quantifoil R1.2/1.3 Quantifoil holey carbon or UltrAuFoil holey gold R1.2/1.3 grids (Quantifoil). The grid was blotted for 2 s in 100% humidity at force -5 and was plunge-frozen in liquid ethane using a Vitrobot Mark IV (FEI).
Cryo-EM data collection and processing
CryoEM data for the CG and TT complexes were collected at National Center for CryoEM Access and Training (NCCAT) and those for the TG complex were collected at LBSB-cryoEM at Texas A&M University (Texas A&M), all using a Titan Krios transmission electron microscope equipped with a Gatan K3 camera. At NCCAT, movies were recorded in counting mode with defocus values ranging from -0.8 – -2.0 μm and a magnification of 105 k, corresponding to a pixel size of 0.82 Å at the specimen level. During the exposure of 1.6-2.0 seconds, 40–50 frames were collected with a total dose of 45–58 electrons/Å2 and a dose rate of 20 e−/pixel/second. At Texas A&M, the defocus values were set to -1.0 to 2.0 μm. Movies were recorded with a magnification of 105 k (0.832 Å at the specimen level). During the 2.5 second exposure, 40 frames were collected with a total dose of 50 electrons/Å2 and a dose rate of 15 e−/pixel/second. In total, 9743, 28957 and 1510 images were collected for the CG, TT and TG complexes, respectively. Prior to particle picking, micrographs were analyzed for good power spectrum, and the bad ones were discarded, with 7538, 23814, and 1350 good images remained for the three complexes, respectively.
The data processing strategies were similar across the three datasets. For the CG complex, using Relion 4.0, motion correction was applied to raw movie stacks using MotionCor2 software53, and contrast transfer function (CTF) estimation was performed using Gctf54. Particle picking was performed without templates, followed by particle extraction and 2D-classification in cryoSPARC55. To expedite processing, particles were divided into four groups using the particle sets tool. These groups underwent initial ab initio reconstruction in parallel with the maximum resolution set to 8Å, followed by heterogenous refinement. Subsequently, classes that showed intact and clear features for protein and DNA from the four groups were combined and subjected to homogenous refinement, 2D classification, ab initio reconstruction, heterogenous and non-uniform refinement. A total of 1003031 particles were then imported into Relion 4.0 for CTF refinement and Bayesian polishing. After polishing, particles were imported back to cryoSPARC for further 2D-classification. To address the orientation issue, successive rounds of ab initio reconstruction were performed with the maximum resolution set to 6Å. The refinement process was completed with a final global and local CTF refinement, followed by non-uniform refinement, yielding a map with a global resolution of 3.11 Å as assessed by the Fourier shell correlation (FSC) = 0.143 criterion56. CryoSPARC was used to estimate the local resolution and angular distribution of all particles that contributed to the final map. A detailed processing and refinement flowchart is shown in Supplementary Fig. 1.
For the TG complex, Relion4.0 was used to perform motion correction, CTF estimation, and template-free particle picking. Particles were extracted to a pixel size of 4.16 Å and were subjected to 2D classification in cryoSPARC. A series of steps including ab initio reconstruction, heterogeneous refinement and non-uniform refinement were carried out to obtain an initial model. After re-extracting particles a pixel size of 4.16 in Relion4.0 and importing them back to cryoSPARC, additional 2D classification, ab initio and heterogeneous refinement were conducted to remove more broken particles. Subsequent rounds of non-uniform refinement and global/local CTF refinement were performed. Following another round of 2D classification, 93767 particles underwent a final non-uniform refinement, resulting in a map with a global resolution of 3.32 Å as assessed by FSC = 0.143 criterion. A detailed processing and refinement flowchart is shown in Supplementary Fig. 2.
Data for the TT complex was first processed in Relion 4.0 as well, for the steps of motion correction, CTF estimation, template-free particle picking and particle extraction. CryoSPARC was then used for 2D classification and successive rounds of ab initio reconstruction to alleviate the orientation issue. A 3D classification with 10 classes, targeting a resolution of 4 , was subsequently performed. Non-uniform refinement was applied to each of the 10 classes, and the class exhibiting no orientation problem and the highest resolution was selected for further processing. The selected class underwent global and local CTF refinement, followed by final non-uniform refinement, yielding a map with a global resolution of 3.44 Å as assessed by FSC = 0.143 criterion. A detailed processing and refinement flowchart is shown in Supplementary Fig. 3.
Model building and refinement
Crystal structure of Pol θ (8E23) was initially rigid body-docked into the cryo-EM density maps in Chimera57. The models were first manually adjusted in COOT58 and then refined in Phenix59 with real-space refinement and secondary structure and geometry restraints. Statistics of all cryo-EM data collection and structure refinement are shown in Supplementary Table 2.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The 3D cryo-EM density maps for complexes CG, TG and TT have been deposited in the Electron Microscopy Data Bank under accession numbers EMD-43855, EMD-43874 and EMD-43875, respectively. The atomic coordinates of complexes CG, TG and TT have been deposited in the Protein Data Bank under accession numbers 9AU5, 9AU8 and 9AU9. The remaining data from this study are available in the main article, Supplementary Information, and the Source Data file. All accession codes used in this study are listed below. PDB depositions: [9AU5], [9AU8], [9AU9], [8EF9], [7ZUS], [4X0P], [4X0Q], [3KTQ], [3HP6], [3MR3], EMDB depositions: [EMD-43855], [EMD-43874], [EMD-43875]. Source data are provided with this paper.
References
Roerink, S. F., Schendel, R. & Tijsterman, M. Polymerase theta-mediated end joining of replication-associated DNA breaks in C. elegans. Genome Res. 24, 954–962 (2014).
Chiruvella, K. K., Liang, Z. & Wilson, T. E. Repair of double-strand breaks by end joining. Cold Spring Harb. Perspect. Biol. 5, 1–22 (2013).
Seki, M., Marini, F. & Wood, R. D. POLQ (Pol θ), a DNA polymerase and DNA-dependent ATPase in human cells. Nucleic Acids Res. 31, 6117–6126 (2003).
Schaub, J. M., Soniat, M. M. & Finkelstein, I. J. Polymerase theta-helicase promotes end joining by stripping single-stranded DNA-binding proteins and bridging DNA ends. Nucleic Acids Res. 50, 3911–3921 (2022).
Mateos-Gomez, P. A. et al. The helicase domain of Polθ counteracts RPA to promote alt-NHEJ. Nat. Struct. Mol. Biol. 24, 1116–1123 (2017).
He, P. & Yang, W. Template and primer requirements for DNA Pol θ-mediated end joining. Proc. Natl Acad. Sci. USA 115, 7747–7752 (2018).
Black, S. J. et al. Molecular basis of microhomology-mediated end-joining by purified full-length Polθ. Nat. Commun. 10, 4423 (2019).
Seki, M. et al. High-efficiency bypass of DNA damage by human DNA polymerase Q. EMBO J. 23, 4484–4494 (2004).
Hogg, M., Seki, M., Wood, R. D., Doublié, S. & Wallace, S. S. Lesion bypass activity of DNA polymerase θ (POLQ) is an intrinsic property of the pol domain and depends on unique sequence inserts. J. Mol. Biol. 405, 642–652 (2011).
Du, H., Wang, P., Wu, J., He, X. & Wang, Y. The roles of polymerases ν and θ in replicative bypass of O6-and N6-alkyl-\({2}^{{\prime} }\)-deoxyguanosine lesions in human cells. J. Biol. Chem. 295, 4556–4562 (2020).
Yoon, J.-H., Johnson, R. E., Prakash, L. & Prakash, S. DNA polymerase θ accomplishes translesion synthesis opposite 1,N(6)-ethenodeoxyadenosine with a remarkably high fidelity in human cells. Genes Dev. 33, 282–287 (2019).
Yoon, J. H. et al. Error-prone replication through UV lesions by DNA polymerase θ Protects against skin cancers. Cell 176, 1295–1309 (2019).
Ceccaldi, R. et al. Homologous-recombination-deficient tumours are dependent on Polθ-mediated repair. Nature 518, 258–262 (2015).
Mateos-Gomez, P. A. et al. Mammalian polymerase θ promotes alternative NHEJ and suppresses recombination. Nature 518, 254–257 (2015).
Pismataro, M. C. et al. Small molecules targeting DNA polymerase theta (POL θ) as promising synthetic lethal agents for precision cancer therapy. J. Med. Chem. 66, 6498–6522 (2023).
Arana, M. E., Seki, M., Wood, R. D., Rogozin, I. B. & Kunkel, T. A. Low-fidelity DNA synthesis by human DNA polymerase theta. Nucleic Acids Res. 36, 3847–3856 (2008).
Masuda, K. et al. DNA polymerase θ contributes to the generation of C/G mutations during somatic hypermutation of Ig genes. Proc. Natl Acad. Sci. USA 102, 13986–13991 (2005).
Van Schendel, R., Roerink, S. F., Portegijs, V., Van Den Heuvel, S. & Tijsterman, M. Polymerase θ is a key driver of genome evolution and of CRISPR/Cas9-mediated mutagenesis. Nat. Commun. 6, 7394 (2015).
Zahn, K. E., Averill, A. M., Aller, P., Wood, R. D. & Doublié, S. Human DNA polymerase θ grasps the primer terminus to mediate DNA repair. Nat. Struct. Mol. Biol. 22, 304–311 (2015).
Stockley, M. L. et al. Discovery, characterization, and structure-based optimization of small-molecule in vitro and in vivo probes for human dna polymerase theta. J. Med. Chem. 65, 13879–13891 (2022).
Bubenik, M. et al. Identification of RP-6685, an orally bioavailable compound that inhibits the DNA polymerase activity of Polθ. J. Med. Chem. 65, 13198–13215 (2022).
Li, C. et al. Structural basis of DNA polymerase θ mediated DNA end joining. Nucleic Acids Res. 51, 463–474 (2023).
Li, Y., Korolev, S. & Waksman, G. Crystal structures of open and closed forms of binary and ternary complexes of the large fragment of thermus aquaticus DNA polymerase I: structural basis for nucleotide incorporation. EMBO J. 17, 7514–7525 (1998).
Doublié, S., Tabor, S., Long, A. M., Richardson, C. C. & Ellenberger, T. Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 Å resolution. Nature 391, 251–258 (1998).
Johnson, S. J., Taylor, J. S. & Beese, L. S. Processive DNA synthesis observed in a polymerase crystal suggests a mechanism for the prevention of frameshift mutations. Proc. Natl Acad. Sci. USA 100, 3895–3900 (2003).
Chandramouly, G. et al. Polθ reverse transcribes RNA and promotes RNA-templated DNA repair. Sci. Adv. 7, 1–12 (2021).
Wu, E. Y. & Beese, L. S. The structure of a high fidelity DNA polymerase bound to a mismatched nucleotide reveals an “ajar” intermediate conformation in the nucleotide selection mechanism. J. Biol. Chem. 286, 19758–19767 (2011).
Kondo, J. et al. Crystal structure of metallo DNA duplex containing consecutive Watson–Crick-like T–HgII–T asbe Pairs. Angew. Chem. Int. Ed. 53, 2385–2388 (2014).
Rebelo, A. Monitoring of DNA Polymerase Theta Movements Through Fret Analysis. https://core.ac.uk/download/pdf/232661761.pdf (2018).
Joyce, C. M. et al. Fingers-closing and other rapid conformational changes in dna polymerase i (klenow fragment) and their role in nucleotide selectivity. Biochemistry 47, 6103–6116 (2008).
Tsai, Y.-C. & Johnson, K. A. A new paradigm for dna polymerase specificity. Biochemistry 45, 9675–9687 (2006).
Towle-Weicksel, J. B. et al. Fluorescence resonance energy transfer studies of dna polymerase β: the critical role of fingers domain movements and a novel non-covalent step during nucleotide selection. J. Biol. Chem. 289, 16541–16550 (2014).
Lin, C.-W. & Ting, A. Y. Transglutaminase-catalyzed site-specific conjugation of small-molecule probes to proteins in vitro and on the surface of living cells. J. Am. Chem. Soc. 128, 4542–4543 (2006).
Zatreanu, D. et al. Polθ inhibitors elicit BRCA-gene synthetic lethality and target PARP inhibitor resistance. Nat. Commun. 12, 3636 (2021).
Fried, W. et al. Discovery of a small-molecule inhibitor that traps polθ on dna and synergizes with parp inhibitors. Nat. Commun. 15, 2862 (2024).
Takata, K.-i, Arana, M. E., Seki, M., Kunkel, T. A. & Wood, R. D. Evolutionary conservation of residues in vertebrate DNA polymerase N conferring low fidelity and bypass activity. Nucleic acids Res. 38, 3233–3244 (2010).
Lee, Y. S., Gao, Y. & Yang, W. How a homolog of high-fidelity replicases conducts mutagenic DNA synthesis. Nat. Struct. Mol. Biol. 22, 298–303 (2015).
Kent, T., Chandramouly, G., Mcdevitt, S. M., Ozdemir, A. Y. & Pomerantz, R. T. Mechanism of microhomology-mediated end-joining promoted by human DNA polymerase θ. Nat. Struct. Mol. Biol. 22, 230–237 (2015).
Kent, T., Mateos-Gomez, P. A., Sfeir, A. & Pomerantz, R. T. Polymerase θ is a robust terminal transferase that oscillates between three different mechanisms during end-joining. eLife 5, 13740 (2016).
Wang, W., Hellinga, H. W. & Beese, L. S. Structural evidence for the rare tautomer hypothesis of spontaneous mutagenesis. Proc. Natl Acad. Sci. USA 108, 17644–17648 (2011).
Kimsey, I. J. et al. Dynamic basis for dG• dT misincorporation via tautomerization and ionization. Nature 554, 195–201 (2018).
Xia, S., Wang, J. & Konigsberg, W. H. DNA mismatch synthesis complexes provide insights into base selectivity of a B family DNA polymerase. J. Am. Chem. Soc. 135, 193–202 (2013).
Nair, D. T., Johnson, R. E., Prakash, S., Prakash, L. & Aggarwal, A. K. Replication by human DNA polymerase-ι occurs by Hoogsteen base-pairing. Nature 430, 377–380 (2004).
Johnson, S. J. & Beese, L. S. Structures of mismatch replication errors observed in a DNA polymerase. Cell 116, 803–816 (2004).
Bebenek, K., Pedersen, L. C. & Kunkel, T. A. Replication infidelity via a mismatch with Watson–Crick geometry. Proc. Natl Acad. Sci. 108, 1862–1867 (2011).
Malik, R. et al. Cryo-EM structure of translesion DNA synthesis polymerase ζ with a base pair mismatch. Nat. Commun. 13, 1050 (2022).
Wang, F. & Yang, W. Structural insight into translesion synthesis by DNA Pol II. Cell 139, 1279–1289 (2009).
Laverty, D. J., Averill, A. M., Doublié, S. & Greenberg, M. M. The a-rule and deletion formation during abasic and oxidized abasic site bypass by dna polymerase θ. ACS Chem. Biol. 12, 1584–1592 (2017).
Thomas, C. et al. Melanoma-derived dna polymerase theta variants exhibit altered dna polymerase activity. Biochemistry 63, 1107–1117 (2024).
Biertümpfel, C. et al. Structure and mechanism of human DNA polymerase eta. Nature 465, 1044–1048 (2010).
Li, Y. et al. Nucleotide insertion opposite a cis-syn thymine dimer by a replicative DNA polymerase from bacteriophage T7. Nat. Struct. Mol. Biolo. 11, 784–790 (2004)
Taki, M., Shiota, M. & Taira, K. Transglutaminase-mediated n-and c-terminal fluorescein labeling of a protein can support the native activity of the modified protein. Protein Eng. Des. Select. 17, 119–126 (2004).
Zheng, S. Q. et al. Motioncor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Zhang, K. Gctf: Real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Henderson, R. et al. Outcome of the first electron microscopy validation task force meeting. Structure 20, 205–214 (2012).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of coot. Acta Crystallogr. Sect. D Biol. Crystallogr. 66, 486–501 (2010).
Afonine, P. V. et al. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. Sect. D Struct. Biol. 74, 531–544 (2018).
Acknowledgements
The work is supported by the Cancer Prevention & Research Institute of Texas (CPRIT) Award RR190046 and American Cancer Society Research Scholar award RSG-22-082-01-DMC to Y.G. We thank the Cryo-EM Core at Baylor College of Medicine, National Center for Cryo-EM Access and Training (NCCAT), and Gaya P. Yadav from Biomolecular Structure and Dynamics (LBSD) of Texas A&M for sample screening and data collection. The LBSD is supported, in part, by the Department of Biochemistry & Biophysics, AgriLife, and Texas A&M University.
Author information
Authors and Affiliations
Contributions
C.L. and Y.G. designed research. C.L. performed research and L.M.M helped with the kinetic study. C.L. and Y.G. analyzed data; C.L. and Y.G. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Dong Wang, who co-reviewed with Qingrong Li and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Li, C., Maksoud, L.M. & Gao, Y. Structural basis of error-prone DNA synthesis by DNA polymerase θ. Nat Commun 16, 2063 (2025). https://doi.org/10.1038/s41467-025-57269-9
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-57269-9