Abstract
Ischemic stroke recovery involves dynamic interactions between the central nervous system and infiltrating immune cells. Peripheral immune cells compete with resident microglia for spatial niches in the brain, but how modulating this balance affects recovery remains unclear. Here, we use PLX5622 to create spatial niches for peripheral immune cells, altering the competition between infiltrating immune cells and resident microglia in male mice following transient middle cerebral artery occlusion (tMCAO). We find that early-phase microglia attenuation promotes long-term functional recovery. This intervention amplifies a subset of monocyte-derived macrophages (RAMf) with reparative properties, characterized by high expression of GPNMB and CD63, enhanced lipid metabolism, and pro-angiogenic activity. Transplantation of RAMf into stroke-affected mice improves white matter integrity and vascular repair. We identify Mafb as the transcription factor regulating the reparative phenotype of RAMf. These findings highlight strategies to optimize immune cell dynamics for post-stroke rehabilitation.
Similar content being viewed by others
Introduction
Ischemic stroke is a leading cause of mortality and permanent disability, with a rising prevalence worldwide1,2. Following ischemia, the central nervous system undergoes an evolution from brain injury to neural repair, which is strongly influenced by both resident microglia and infiltrating immune cells3. In response to cerebral ischemia, neurons and glial cells in the infarct region rapidly undergo hypoxic cell death, releasing damage-associated molecular patterns (DAMPs). These molecules activate microglia and trigger sterile inflammation. Once activated, microglia secrete inflammatory factors that contribute to secondary brain damage and the recruitment of peripheral immune cells4. Peripheral immune cells, including neutrophils, macrophages, and lymphocytes, infiltrate ischemic brain tissues within 1 or 2 days following the activation of resident microglia5 and further activate microglia through pro-inflammatory cytokines such as interferon-γ, IL-17, and IL-216,7. Sterile inflammation subsides approximately one to two weeks after ischemia. During this period, resident and infiltrating immune cells shift from a pro-inflammatory to a reparative phenotype. These immune cells produce neurotrophic factors and synaptic regulatory molecules around the injured brain, facilitating angiogenesis, axonal regeneration, synaptic modulation, and remyelination3,8,9. Therefore, selectively inhibiting acute harmful inflammation while preserving subsequent reparative processes is considered a potential strategy for improving functional outcomes in patients with ischemic stroke.
Microglia, the most abundant immune cells in the brain, serve as the primary triggers of neuroinflammation in the central nervous system (CNS). However, overactivated microglia can hinder the transition of the immune environment to a repair state during ischemic stroke10. Over-activated microglia may mistakenly identify and engulf healthy neurons, resulting in secondary brain damage4. Additionally, they can attract CD8+ T cells, γδ T cells, and other cytotoxic cells into the brain11,12, thereby exacerbating the inflammatory environment and hindering the subsequent repair process.
Colony stimulating factor 1 receptor (CSF1R) inhibitors, such as PLX3397 and PLX5622, have been shown to selectively inhibit microglial proliferation and significantly reduce the number of microglia in the mouse brain13. Multiple studies have highlighted the therapeutic potential of CSF1R inhibitors in treating a range of neurological disorders, including ischemic stroke14, neurodegenerative diseases15, demyelinating diseases16, CNS infections17 and brain injuries18. However, some research has indicated that the complete elimination of microglia can negate their protective effects and exacerbate brain injury following ischemic stroke19,20,21,22. These conflicting findings underscore the need for further investigation into the impact of CSF1R inhibitors on the immune microenvironment in neurological diseases, as well as the identification of optimal treatment strategies.
After the cessation of microglia depletion, the vacant microglial niche can be rapidly replenished, either by resident microglia or by monocyte-derived macrophages (MDMs) in the case of bone marrow chimeras23. These cells typically exhibit enhanced repair capabilities during niche occupation, which is a key factor contributing to the neural repair-promoting effects of CSF1R inhibitors24. Accumulating evidence underscores immune niche competition between MDMs and microglia25. Under physiological conditions, the niche vacated due to partial microglial loss is replenished by the proliferation of remaining microglia26. However, in irradiated mice, compromised blood-brain barrier integrity and inhibited microglial proliferation lead to predominant filling of the niche by recruited MDMs23,27. Notably, MDMs undergo a transformation into microglia-like cells after infiltrating CNS and can exist in the brain for a long time with retained monocyte characteristics28. Recent studies indicate that MDMs exhibit stronger phagocytic capacity compared to microglia29, and early MDMs infiltration may be beneficial to stroke repair30. Nevertheless, it remains unclear whether and how microglia attenuation affects cerebral immune niches and contributes to long-term brain recovery in ischemic stroke remains unknown.
In this study, we modulated immune niche competition by employing early-phase microglia attenuation after ischemic stroke, leading to a significant expansion of repair-related macrophages (RAMf) in the ischemic brain. Our findings revealed that RAMf originate from CCR2+ monocytes and acquire the reparative phenotype upon entering the brain. Employing single-cell RNA sequencing, flow cytometry, and lipidomics, we demonstrated that RAMf significantly contribute to remyelination and angiogenesis following ischemic stroke. Furthermore, we identified Mafb as a key transcription factor regulating the reparative phenotype in RAMf, and enhancing RAMf differentiation through Mafb overexpression markedly improved white matter and vascular integrity after ischemic stroke. These findings offer perspectives on the cellular and molecular aspects of immune niche competition in the context of ischemic stroke, paving the way for potential therapeutic strategies.
Results
Early-phase microglia attenuation alters the immune niche in ischemic stroke and promotes long-term stroke recovery
Numerous peripheral immune cells swiftly infiltrate into the brain following ischemic injury and play significant roles at different stages. Emerging evidence increasingly implicates a competition for the immune niche between peripheral immune cells and resident microglia25. Understanding this delicate balance in the context of ischemic stroke may yield valuable insights for novel therapeutic approaches.
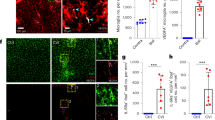
In this study, we aim to resolve the changes of immune cell composition in the brain following inhibited microglial proliferation. A transient (60 min) middle cerebral artery occlusion (tMCAO) model was established in CCR2-GFP mice, which were then fed either with a CSF1R inhibitor PLX5622 chow or a control diet consecutively for 5 days (Fig. 1A). No significant difference in infarct volume was observed between the two groups of mice. Subsequently, we employed multiplex immunohistochemistry (mIHC) to identify immune cells, which demonstrated decreased Tmem119+ cells and accumulated CCR2+ cells in stroke mice treated with PLX5622 (Figs. 1B, C, S1A). Flow cytometry analysis further confirmed that treatment with PLX5622 for 5 days reduced the number of CD45low CD11b+ Ly6G- microglia by approximately 70% and increased the infiltration of macrophages at 5 days post-injury (dpi) (Fig. S1B, C).
A Experimental scheme illustrating the PLX5622 diet of CCR2GFP mice after MCAO. B Representative mIHC images of ischemic mouse brain from CCR2GFP mice. Scale bars, 2.5 mm (left), and 100 µm (middle and right). C Quantification of the infarct volume, numbers of Tmem119+ cells and CCR2 + cells between mice with PLX5622 treatment or control diet at 5 dpi. n = 7 for infarct volume, n = 5 for Tmem119+, CCR2 + cells. D Experimental scheme illustrating the PLX5622 diet of mice after MCAO until 14 dpi. E Dot plots showing the absolute numbers of macrophages in the ipsilateral hemisphere at several time points. n = 3 for 3D, 7D, and 5D control diet group, n = 4 for 5D PLX5622 group, n = 5 for 14D. F UMAP projection plot showing cell clusters of immune cells from ischemic mouse brain at 5 dpi. n = 2 biological replicates for each group. G Stacked bar plot showing proportions of infiltrating immune cells. H Dot plot depicting the cellular functional changes from PLX5622 group to control group based on the AUCell scores of functional gene sets listed in Supplementary Data 1. Wilcoxon rank sum test, two-sided. I Dot plot illustrating the expression of selected genes in different immune cells at 5 dpi. Wilcoxon rank sum test, two-sided, with Bonferroni correction. J The experimental scheme for PLX5622 diet and behavior test. K Quantification of tissue loss at 35 dpi in MAP2-stained (red) coronal sections. n = 7 per group. Scale bars, 2.5 mm. L Adhesive removal and foot fault test. n = 9 for control group and n = 10 for PLX5622 group. Adhesive removal: 2-way ANOVA repeated measurement. Foot fault test: mixed-effects model. C, E, K Student’s t test, two-sided. Data in (C, E, K, L) are presented as mean ± SD. Abbreviations: MF macrophage, MG microglia, NP neutrophil, NK natural kill cells, cDCs classical dendritic cells, BAMs border-associated macrophages, pDCs plasmacytoid dendritic cells. Source data are provided as a Source Data file.
Subsequently, we investigated the temporal changes in microglia and macrophages within the ischemic brain of both PLX5622-treated and control diet groups. Mice were administered PLX5622 for the first 7 days, then switched to the control diet until 7 days later. Flow cytometry analysis was performed at several time points (3, 5, 7, and 14 days) following ischemic stroke, encompassing both the administration and withdrawal phases of PLX5622 treatment (Fig. 1D). Our results demonstrated that treatment with PLX5622 for 7 days led to approximately 70% depletion of microglia (Fig. S1D). By 14 dpi, 7 days after PLX5622 withdrawal, the number of microglia had grown back to a comparable level to those observed in the control group (Fig. S1D). In contrast, macrophage infiltration significantly increased following PLX5622 treatment (Fig. 1E). Notably, at 14 dpi, the number of macrophages in the brains of PLX5622-treated mice remained significantly higher than that of the control diet group (Fig. 1E).
Then we performed single cell RNA sequencing (scRNA-seq) of CD45high infiltrating immune cells sorted from the ischemic mouse brain at 5 dpi to investigate the transcriptional changes in immune cells induced by PLX5622 during the subacute phase (Fig. 1F). Unsupervised clustering analysis identified ten distinct clusters (Fig. 1F), which were subsequently annotated based on their gene expression profiles (Fig. S1E). Among these clusters, we identified two distinct resident microglia subsets, named MG1 and MG2. MG1 microglia exhibited an upregulation of the classic M1 marker Cd86 and pro-inflammatory factors, including Tnf, Il1b, and Cxcl10 (Fig. S1F). In contrast, MG2 microglia displayed an upregulation of the traditional M2 marker Msr1, accompanied by higher expression of Igf1, Spp1, Lgals3, and Cd5l, indicating an anti-inflammatory phenotype (Fig. S1F). The proportions of MG1 and MG2 were similar between control and PLX5622-treated mice (Fig. S1G), suggesting that PLX5622 does not affect the microglial activation after stroke.
We then focused on the infiltrating immune cells in the ischemic brain. In line with the results from flow cytometry, we observed a considerable increase in the proportion of macrophages (Fig. 1G). Additionally, AUCell scores were computed based on functional gene sets within individual cells to explore the cellular functional changes induced by PLX5622. The results revealed a general upregulation of brain repair-related functions in infiltrating immune cells from PLX5622-treated mice, particularly in macrophages (Fig. 1H). Moreover, numerous genes associated with the inflammatory response were altered in mice subjected to early phase microglia attenuation. Notably, several of the upregulated genes in these cells encode proteins known to promote brain repair, including Spp1, Vegfb, Lgals3bp, Lgals, Igf1, Grn and Tgm2 (Fig. 1I). In contrast, the changes in genes and functional terms related to acute inflammation were inconsistent across different immune cell types (Fig. 1H, I). These findings indicate that early phase microglia attenuation may contribute to the establishment of a more conducive immune microenvironment, fostering brain repair.
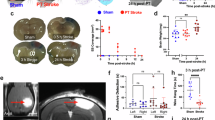
To ascertain the role of early-phase microglia attenuation in long-term outcomes, mice were fed with PLX5622 for the first 7 days and then returned to the control diet until 35 days (Fig. 1J). Through T2-weighted magnetic resonance imaging (MRI), we confirmed that the infarct volumes were similar between two groups of mice on the day after ischemic stroke (Fig. S1H). Notably, we observed a significantly reduced volume of tissue loss in the PLX5622 group at 35 dpi (Fig. 1K). Additionally, treatment with PLX5622 for 7 days enhanced long-term sensorimotor functions 35 days after MCAO, as evidenced by a decreased time to remove adhesive tapes and diminished foot fault rates (Fig. 1L).
Collectively, these data indicate that early-phase microglia attenuation effectively diminishes the proportion of microglia within the brain’s immune niche following ischemic stroke. Moreover, PLX5622 treatment facilitates the infiltration of peripheral macrophages, improving long-term stroke recovery.
Non-microglia effects of PLX5622 do not affect the brain repair function of infiltrating macrophages
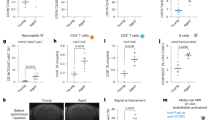
PLX5622 has been demonstrated to affect peripheral immune cell populations31. To evaluate this off-target effect outside the microglia niche, flow cytometry was performed to quantify various immune cells in peripheral blood at 5 dpi. Our findings indicate that the PLX5622 diet significantly reduces the number of CD45+CD11b+CD115+ monocytes in the blood at 5 dpi (Fig. S2A, B). However, the numbers of CD19+ B cells, CD3+ T cells, CD4+ T cells or CD8+ T cells remained unaltered (Fig. S2A, B).
Subsequently, to investigate the effects of PLX5622 on monocyte function, we conducted scRNA-seq of the peripheral blood from mice in the PLX5622 group and the control diet group at 5 dpi (Fig. S2C). Unsupervised clustering analysis revealed ten distinct cell types, including neutrophils (S100a8+), B cells (Cd79a+), T cells (Cd3d+), red blood cells (Hbb-a1+), natural killer cells (Nkg7+), monocytes (Lyz2+), megakaryocytes (Ppbp+), dendritic cells (Cd209a+), basophils (Ms4a2+), and proliferating cells (Mki67+) (Fig. S2D, E). Next, differential expression analysis was performed on scRNA-seq data to assess the transcriptomic changes induced by PLX5622 in peripheral monocytes. We identified 160 upregulated genes and 105 downregulated genes in monocytes from the PLX5622 group compared to the control diet group (Fig. S2F), indicating a moderate alteration in the genome (1.15% of total ~23,813 genes). Furthermore, we explored the functional implications of these differentially expressed genes (DEGs) through Gene Ontology (GO) enrichment analysis. Monocytes in the PLX5622 group did not exhibit significantly upregulated GO terms. However, compared to the control diet group, they showed significantly downregulated terms of biological processes related to cellular respiration and lymphocyte activation regulation (Fig. S2G, Supplementary Data 2). We then assessed the survival of monocytes in both the PLX5622 and control diet groups using Annexin V/PI staining. The results revealed no significant differences between the two groups (Fig. S2H). In contrast, PLX5622 treatment induced pronounced transcriptomic changes in brain macrophages, resulting in upregulation of 642 genes and downregulation of 402 genes at 5 dpi (Fig. S2I). Furthermore, none of the upregulated repair-related genes in brain macrophages were involved in peripheral blood monocytes (Fig. S2J). These findings indicate that while PLX5622 reduced the number of peripheral monocytes, its effects on their functional role in the MCAO environment were limited.
To further determine whether the effects of PLX5622 on peripheral immune cells influence the reparative functions of brain macrophages, we isolated peripheral blood immune cells from CD45.1+ mice in either the PLX5622-treated or control diet group and injected these cells into wild-type mice. At 5 dpi, we found no significant differences in the expression levels of CD206 and CD163 in CD45.1+ macrophages between the two groups (Fig. S2K).
Overall, these data indicate that while PLX5622 treatment reduces the number of peripheral blood monocytes, its effects on their function are limited. Additionally, the upregulated reparative functions observed in brain macrophages following PLX5622 treatment are not attributable to off-target effects on non-microglial cells.
Early-phase microglia attenuation benefits remyelination and angiogenesis after ischemic stroke
Subsequently, we assessed the impact of PLX5622 treatment on ischemic injury in MCAO mice. We selected 5 dpi as the observation timepoint during the subacute phase, characterized by significant immune cell infiltration and a lack of substantial neural repair processes9. No significant difference in infarct volume was observed between the two groups (Fig. S3A). To further evaluate white matter integrity, we conducted dual staining for SMI32 (a neurofilament marker) and MBP (a key myelin protein). The results indicated no differences in the SMI32/MBP ratio between the two groups (Fig. S3B, C). Additionally, there was no significant variation in the number of APC+ BrdU+ cells, which represent newly generated oligodendrocytes, in the infarcted hemisphere (Fig. S3D), suggesting a similar extent of white matter damage between both groups of mice. We also evaluated vascular integrity by examining CD31+ vascular coverage, CD31+ BrdU+ cells (Fig. S3E) and ZO-1+ CD31+ vessels (Fig. S3F), and observed that the extent of vascular injury was consistent between both groups. Collectively, these findings indicate that PLX5622 does not significantly influence brain injury in ischemic stroke.
To investigate the impact of early-phase microglia attenuation on stroke recovery, we selected 14 dpi as the observation point during the recovery phase32. Mice were treated with PLX5622 for the first 7 days and switched to a control diet until 14 dpi. Our results indicated that the PLX5622 treatment significantly reduced tissue loss at 14 dpi (Fig. S4A). Following this, we conducted scRNA-seq on mouse brains at 14 dpi (Fig. 2A). Unsupervised clustering analysis identified 20 cell types according to their DEGs, including microglia (MG), macrophages (MF), border-associated macrophages (BAMs), dendritic cells (DC), neutrophils (NP), natural killer cells (NK), T cells, gamma-delta T cells (γδT), B cells, neurons, neuroblasts (NEUB), astrocytes (ASC), oligodendrocytes (OLG), oligodendrocyte progenitor cells (OPC), endothelial cells (EC), pericytes (PC), fibroblast-like cells (FB), ependymocytes (EPC), choroid plexus epithelial cells (CPC) and olfactory ensheathing glial cells (OEG) (Figs. 2A, S4B).
A, B UMAP projection plot (A) and stacked bar plot (B) showing cell clusters harvested from the ipsilateral brain hemisphere at 14 dpi. n = 2 biological replicates for each group. C, D Representative images and quantification for MBP (green) and SMI32 (red) double immunostaining in the peri-infarct areas at 14 dpi. n = 6 per group. Scale bars, 200 µm (Cortex), and 100 µm (Striatum and External Capsule). Data were normalized to the intensities of contralateral hemispheres. One-way ANOVA and Tukey. E UMAP projection of oligodendrocytes. F Pseudotime trajectory of oligodendrocyte subclusters. G Violin plot illustrating the expression of Klk6 and Cntn1 in oligodendrocyte (up). Stacked bar plot showing the proportion of OLG in (E) (down). H UMAP projection of OPCs. I Doughnut plot showing an increase of OPCs in the G2M phase in the PLX5622-treated mice. J Representative images and quantification for APC (green) and BrdU (red) double immunostaining in the peri-infarct areas at 14 dpi. n = 5 per group. K Box plot showing the upregulation of genes related to angiogenesis in EC. n = 557 ECs for control group and n = 1057 ECs for PLX5622 group, from two biological replicates. Wilcoxon rank sum test, two-sided. Box plot represents the first quartile, median, and third quartile with whiskers extending to 1.5 times the interquartile range. L Representative images and quantification for CD31 (green) and BrdU (red) double immunostaining in the peri-infarct areas at 14 dpi. n = 6 per group. J, L Student’s t test, two-sided. Data in (D, J, L) are presented as mean ± SD. MG microglia, MF macrophages, BAMs border-associated-macrophages, DC dendritic cells, NP neutrophils, NK natural killer cells, γδT gamma delta T cells, NEUB neuroblasts, ASC astrocytes, OLG oligodendrocytes, OPC oligodendrocyte progenitor cells, EC endothelial cells, PC pericytes, FB fibroblast-like cells, EPC ependymocytes, CPC choroid plexus epithelial cells, OEG olfactory ensheathing glial cells. Source data are provided as a Source Data file.
Notably, there was an increase in the cell fractions of oligodendrocyte lineage cells (OLG and OPC) and endothelial cells (Fig. 2B), suggesting a potential direct effect of PLX5622 treatment on white matter and vascular integrity. To assess white matter integrity, dual staining for SMI32 and MBP was performed. A significant decrease in the SMI32/MBP ratio was observed in PLX5622-treated mice, indicating that early-phase microglia attenuation could support remyelination after ischemic stroke (Fig. 2C, D).
Re-clustering analysis of scRNA-seq data for OLG and OPC subsets further substantiated the role of early-phase microglia attenuation in post-stroke remyelination (Fig. S4C). We identified four distinct OLG clusters based on their transcription profiles: OLG1 (Bin1+, Opalin+), OLG2 (Ifit1+, Klk8+), OLG3 (Enpp6+, Anln+) and OLG4 (Cdkn1a+, Gadd45b+) (Figs. 2E, S4D). Pseudo-time analysis revealed a gradual transition from immature OLG1 (marked by Cntn1, a gene involved in oligodendrocyte generation)33 to mature OLGs (OLG2-4, marked by Klk6, a gene for mature oligodendrocytes)34 (Fig. 2F, G). Notably, there was an increase in the proportion of the Opalin+Cntn1+ OLG1 subset in PLX5622-treated mice, indicating a rise in newly formed oligodendrocytes (Fig. 2G).
After ischemic stroke, OPCs migrate towards sites of demyelination and differentiate into myelinating OLGs35. We identified one Cspg4+ Pdgfra+ OPC cluster in ischemic brain at 14 dpi (Fig. 2H, S4E). Cell-cycle phase analysis demonstrated a significant increase in OPCs in the G2M phase in the PLX5622-treated mice (Fig. 2I). Additionally, immunofluorescence quantification showed that PLX5622 administration increased the density of PDGFRα+ BrdU+ cells (newly generated OPCs) and APC+ BrdU+ cells (newly generated OLGs) in the ischemic brain (Figs. 2J, S4F). Furthermore, oligodendrocyte lineage cells from PLX5622-treated mice exhibited significant upregulation of genes related to myelination, such as Ndrg1 Degs1, Mag, Mog, Plp1, Cnp, Igf1, Gjc2, Afdn and Trf (Fig. S4G, H), suggesting that early-phase microglia attenuation could promote oligodendrogenesis after ischemic stroke.
ECs also exhibited a more active proliferative state after early-phase microglia attenuation. We identified significant upregulation of genes related to angiogenesis (Pkm, S100a1, Pamp2, Itgb1, Grn, Lgals3) (Figs. S4I, 2K) in ECs from PLX5622-treated mice. To investigate the impact of early-phase microglia attenuation on vascular repair, we quantified vascular coverage and EC proliferation in mice with 7-day PLX5622 administration and those on the control diet. Notably, there was a substantial increase in vascular coverage and the presence of CD31+ BrdU+ cells in PLX5622-treated mice at 14 dpi (Fig. 2L).
Together, these data provide evidence that early-phase microglia attenuation may support oligodendrogenesis and angiogenesis after ischemic stroke.
Increased infiltration of RAMf promotes remyelination after ischemic stroke
To comprehensively investigate how early-phase microglia attenuation promotes long-term neurological recovery after stroke, we conducted further analysis on the alterations in gene expression and functional characteristics of various immune cells in the brain between the control diet group and the PLX5622 group.
After the cessation of CSF1R inhibitors, microglia can swiftly regenerate, with their numbers returning to physiological levels within 7 days36. Previous studies have indicated that these repopulating microglia possess the capacity to aid in brain repair24. Therefore, we directed our attention to the repopulating microglia at 14 dpi. Unsupervised clustering analysis identified five clusters (MG1-MG5) based on their transcriptomic characteristics (Fig. S5A). Then we explored the DEGs and their functional implications among these clusters (Supplementary Data 4). When comparing these five clusters to microglia from the sham mouse brain, MG1 exhibited higher correlation coefficients with sham microglia (Fig. S5B). Additionally, we observed elevated expression of typical resting microglia markers, like Tmem119 and P2ry12, in MG1 microglia (Fig. S5C, D). These results suggest that MG1 microglia are in a relatively more resting state. Furthermore, the proportion of MG1 microglia increased from 6.2% to 20.6% in PLX5622-treated mice (Fig. S5A), indicating that the repopulating microglia tend to remain at a relatively resting state. MG2 microglia expressed typical immediate early genes (Egr1, Jund, Klf2) and genes of disease inflammatory macrophages/microglia (DIM)37,38, indicating an early activation state. MG3 microglia upregulated the chemokine gene Ccl5 and genes related to microglial activation (Serpine2, Cst7). MG5 microglia were characterized by elevated genes related to the innate immune response (Ifit3, Isg15, Ifit2). Among them, MG4 microglia upregulated several brain repair-related genes (Spp1, Gpnmb, Apoc1, Igf1) and showed enrichment of GO terms such as lysosome, inflammatory response, regulation of angiogenesis, regulation of lipid transport and endocytosis, indicating the important role of MG4 microglia in brain repair (Fig. S5E).
To explore the cellular functional changes induced by PLX5622 in MG4 microglia, we applied AUCell score quantification and revealed a moderate upregulation of myelination and a downregulation of angiogenesis scores in MG4 from PLX5622-treated mice (Fig. S5F). Together, these data suggest that the repopulating microglia were partly involved in post-stroke repair.
Subsequently, we conducted scRNA-seq on enriched CD45high infiltrating immune cells sorted from the ischemic mouse brain at 14 dpi to identify the cell types contributing to remyelination and angiogenesis (Fig. 3A). Among the infiltrated immune cells, macrophages exhibited the most significant elevation in remyelination and angiogenesis AUCell scores in PLX5622-treated mice (Fig. 3B), highlighting their roles in brain repair.
A Experimental design for scRNA-seq of CD45high immune cells at 14 dpi (up). n = 2 biological replicates for each group. UMAP projection of CD45high immune cells at 14 dpi (down). B Ridgeline plot summarizing AUCell scores of the myelination and angiogenesis functions of CD45high infiltrating immune cells at 14 dpi. Involved gene sets were listed in Supplementary Data 1. C UMAP projection of macrophages at 14 dpi (left). Stacked bar plot showing proportions of different macrophage subsets (right). D UMAP plot depicting the functions of macrophages from 14 dpi based on the gene sets listed in Supplementary Data 1. E Scatter plot illustrating upregulated DEGs of MF2 compared to MF1, with the percentage difference (defined as percentage of the expression in MF2 minus that in MF1) along the x-axis and log2(fold change) along the y-axis. The combined z-score of percentage difference and log2(fold change) was color-coded. The DEGs of MF2 versus MF1 were listed in Supplementary Data 3. F Violin plots showing expression of Cd63 and Gpnmb in each clusters. G Experimental scheme depicting the PLX5622 diet of mice after MCAO. H Gating strategy for CD45high CD11b+ CD63+ GPNMB+ cells (left). Quantification of the proportions of CD63+ GPNMB+ cells in the CD45high CD11b+ cells (right). n = 5 for sham group, n = 3 for control diet group, n = 4 for PLX5622 group. One-way ANOVA and Tukey. Data are presented as mean ± SD. I UMAP projection of blood monocytes (left) and brain macrophages (right) at 5 dpi. J Dot plot illustrating the expression of marker genes of MF2 and MF3 (d14) in macrophages from d5. Abbreviations: MG microglia, MF macrophages, NP neutrophils, cDCs classical dendritic cells, pDCs plasmacytoid dendritic cells, NK natural killer cells, NKT natural killer T cells, Tregs regulatory T cells, γδT gamma delta T cells, ProllifCells proliferating cells. Source data are provided as a Source Data file.
To determine the role of macrophages (MF) in remyelination after ischemic stroke, we conducted subclustering analysis on macrophages obtained from the ischemic hemisphere at 14 dpi. Three distinct subclusters (MF1, MF2 and MF3) were identified (Fig. 3C). To construct the differentiation trajectory of brain macrophages, we performed scRNA-seq on peripheral blood samples from both the control diet group and the PLX5622 group at 14 dpi, extracting monocytes for mapping to the brain macrophage clustering results (Fig. S6A, B). The analysis revealed that nearly 90% of peripheral blood monocytes were classified within the MF1 subgroup, indicating that the MF1 subgroup was in a resting state (Fig. S6C). Subsequently, we designated peripheral monocytes as root cells and reconstructed the developmental trajectory using Monocle339. This analysis identified two distinct activation directions (MF1 - MF2, MF1 - MF3) (Fig. S6D).
Notably, the cells activated in the MF1-MF2 direction increased from 19.4% to 38.8% in the PLX5622-treated mice (Figs. 3C, S6D). Upon comparing MF2 to MF1, we observed an upregulation of genes associated with myelination and brain repair, including Gpnmb40, Spp19, Cd63 41, Trem2 42 and Igf143 (Fig. 3D, E, Supplementary Data 3). Consequently, we named the MF2 subpopulation repair-associated macrophages (RAMf) and utilized GPNMB and CD63 (Fig. 3F) to define this population of cells. Flow cytometry further confirmed that 7-day PLX5622 treatment could induce a higher proportion of GPNMB+ CD63+ RAMf in the CD45high CD11b+ cells at 14 dpi (Fig. 3G, H).
On the other hand, MF3 expressed classic markers of monocyte-derived dendritic cells (moDC), including CD209a, H2-Eb1, H2-Aa, and CD7444, while also upregulating genes associated with antigen presentation (H2-Eb1, H2-Aa, H2-Ab1, H2-Oa) and cell activation (Tnf, Il1b, Cd74). Therefore, we classified it as moDC (Figs. 3D, S6E). The proportion of moDC (MF3) remained relatively unchanged between the PLX5622 group and the control group at 14 dpi (Fig. 3C).
Subsequently, we performed similar clustering analysis on macrophages from the ischemic hemisphere at 5 dpi and obtained comparable results (Fig. 3I). Moreover, we found similarities in gene expression profiles and their functional implications between macrophages from 5 days and 14 days after ischemic stroke (Figs. 3J, S6F). Notably, there was a significant increase in the proportion of MF2 subclusters (26.3%-67.6%) in the mice fed with PLX5622 at 5 dpi (Fig. S6G), indicating that RAMf were already massively amplified in the ischemic brain during the period of microglia attenuation.
To further elucidate the origin of the RAMf, we utilized CCR2-GFP mice to distinguish monocyte-derived macrophages from microglia. Flow cytometry analysis revealed that 96.9% of CD45high CD11b+ GPNMB+ CD63+ cells were positive for CCR2 (Fig. 4A). Immunofluorescence analysis further confirmed the co-localization of GPNMB, CCR2 and IBA1 in the infarct area (Fig. 4B). Next, we selectively depleted blood monocytes by administering clodronate liposomes (CL) injections. The treatment began 2 days prior to MCAO and continued daily until 5 dpi (Fig. S7A). Our results indicated that CL treatment did not affect infarct volume at 5 dpi (Fig. S7A). However, it resulted in a significant reduction in blood monocytes (Fig. S7B) without affecting the proportion of microglia in the ischemic hemisphere (Fig. S7C). Additionally, the population of CD45high CD11b+ GPNMB+ CD63+ cells in the ischemic hemisphere decreased significantly compared to mice receiving control liposomes (Fig. 4C). Furthermore, CCR2-KO mice, which lack monocytes, also exhibited an absence of CD45high CD11b+ GPNMB+ CD63+ cells in the brain at 5 dpi after MCAO, further proving that this group of cells mainly originated from monocytes (Fig. 4C). Moreover, we did not find CD45high CD11b+ GPNMB+ CD63+ cells in blood (Fig. S7D), indicating that the RAMf derived from monocytes and acquired a reparative phenotype after infiltrating the brain.
A Experimental scheme illustrating the PLX5622 diet of CCR2GFP mice after MCAO (up). Gating strategy for CD45high CD11b+ CD63+ GPNMB+ cells in ischemic brain and quantification of the CCR2-positive ratio (down). B Representative images for IBA1 (cyan), GPNMB (red) immunostaining in the peri-infarct areas of brains from CCR2GFP mice after MCAO. Scale bars, 50 µm (left), and 5 µm (middle and right). C Quantification of the numbers of CD45high CD11b+ GPNMB+ CD63+ cells in the ipsilateral brain hemisphere. n = 5 for sham and CCR2-KO group, n = 6 for Control liposomes and Clodronate liposomes group. One-way ANOVA and Tukey. D Experimental scheme illustrating the CD45high CD11b+ CD63+ GPNMB+ cells transplantation in mice after MCAO. Donor mice were treated with PLX5622 for the first 7 days and then switched to a control diet until 14 dpi. Recipient mice received approximately 30,000 RAMf or GPNMB− CD63− macrophages via intraventricular transplantation at 5 dpi. E Representative images and quantification for MBP (green) and SMI32 (red) double immunostaining in the peri-infarct areas of brains from mice that underwent either sham surgery or MCAO. n = 5 for GPNMB−CD63− macrophages transplantation group, n = 6 for RAMf transplantation group. Scale bars, 200 µm (Cortex), and 100 µm (Striatum and External Capsule). Data were normalized to the intensities of contralateral hemispheres. One-way ANOVA and Tukey. Data in (A, C, E) are presented as mean ± SD. Source data are provided as a Source Data file.
We then performed adoptive cell transfer to validate the pro-reparative functions of RAMf. Donor mice were treated with PLX5622 for the first 7 days and then switched to a control diet until 14 dpi. We utilized fluorescence-activated cell sorting (FACS) to isolate RAMf (CD45high CD11b+ GPNMB+ CD63+) from the ischemic hemisphere of the donor mice’s brains at 14 dpi. Flow cytometry analysis revealed that 91.6% of the sorted cells were positive for CCR2, indicating a high level of purity in the FACS procedure (Fig. S7E). Subsequently, recipient mice received ~30,000 RAMf or GPNMB− CD63− macrophages via intraventricular transplantation 5 days after tMCAO (Fig. 4D). To assess the survival time of RAMf cells in the brain, we sorted RAMf cells from CCR2-GFP mice and injected them into the brains of recipient mice on the fifth day after MCAO. Flow cytometry analysis revealed that GFP+ RAMf cells persisted in the brains at 14 dpi (Fig. S7F). Additionally, immunofluorescence studies showed a significant increase in IBA+GPNMB+CD63+ cells in the ischemic brain of mice that underwent RAMf transplantation at 14 dpi (Fig. S7G).
We then compared the tissue loss and white matter integrity between the two groups of mice. The results indicated that the RAMf transplantation group exhibited a lower tissue loss (Fig. S7H) and SMI32/MBP ratio (Fig. 4E) compared to the GPNMB− CD63− macrophage transplantation group. Moreover, RAMf transplantation was associated with an increased number of APC+ BrdU+ cells in the mouse brain (Fig. S7I), suggesting that RAMf plays a vital role in promoting axonal remyelination following ischemic stroke.
RAMf exhibited enhanced lipid recycling capacity
Remyelination represents a natural protective and regenerative mechanism initiated in response to ischemic stroke45. Effective remyelination necessitates a conducive pro-reparative milieu characterized by diminished myelin debris, adequate lipid availability, and anti-inflammatory signals46,47. Accumulating evidence underscores the crucial role of phagocytes in the remyelination process42. We thus explored these remyelination-related functions in RAMf. Genes exhibiting significant changes along the MF1-MF2 activation axis were categorized into two patterns using FateID48 (Fig. 5A). Within the MF2 subcluster, 645 genes were upregulated, and functional enrichment analysis revealed that the MF1-MF2 activation axis signifies heightened functions related to myelination, phagocytosis, lysosome activity, and the regulation of neuron death (Fig. 5A). Notably, AUCell scores for phagocytosis and lipid metabolism, while not for anti-inflammation responses, were markedly increased in RAMf from PLX5622-treated mice (Fig. 5B), underlining their pivotal roles in remyelination.
A Heatmap showing the transcriptomic gradual shift from MF1 to MF2, with pseudo-temporal ordering along the x axis. Enriched functions and genes involved in each functional term were annotated on the right. B UMAP plot depicting the functional changes of macrophages from PLX5622 group to control group based on the AUCell scores of functional gene sets listed in Supplementary Data 1. C Representative image showing the phagocytosis of MBP (green) by IBA1+(red) GPNMB+ (cyan) cells. Three-dimensional constructed image showing the enwrapping MBP+ myelin debris in IBA1+ GPNMB+ cells. Quantification of MBP+ proportion in IBA1+GPNMB+ macrophages and IBA1+GPNMB− macrophages. n = 5. D Dot plots showing expression of genes involved in lipid metabolism in each of three distinct clusters. E Representative images and quantification for IBA1 (green), PLIN2 (red) and GPNMB (cyan) immunostaining in brains from mice after MCAO. n = 5. Scale bars, 50 µm. F Gating strategy and Bodipy signal histograms of GPNMB+ CD63+ and GPNMB− CD63− macrophages (left). Comparison of intracellular neutral lipid accumulation by Bodipy mean fluorescence intensity (MFI) between GPNMB+ CD63+ and GPNMB− CD63− macrophages (right). n = 3. G Experimental scheme for clodronate liposomes or control liposomes administration and lipid metabolism analysis. H Quantification of lipid metabolites among sham, control and clodronate groups. n = 5 for sham group, n = 7 for clodronate group, and n = 5 for control group. One-way ANOVA and Tukey. (C, E, F): Student’s t test, two-sided. Data in (C, E, F, H) are presented as mean ± SD. Source data are provided as a Source Data file.
The clearance of myelin debris is a crucial prerequisite for remyelination49. Consequently, we confirmed the phagocytic function of RAMf by using MBP as a phagocytic target. Immunostaining demonstrated the enwrapping and internalization of MBP+ myelin debris in IBA1+ GPNMB+ cells (Fig. 5C). Notably, the MBP positive ratio in IBA1+ GPNMB+ cells was significantly higher than that in IBA1+ GPNMB− cells (Fig. 5C), suggesting that GPNMB+ cells possess a more robust phagocytic capacity for MBP.
Lipid metabolism in phagocytes has been reported to favor remyelination in demyelination disease47. Internalized lipid in myelin-laden phagocytosis can either be removed from the cell or esterified and stored in lipid droplets. These lipid recycling processes have been reported to contribute to remyelination in brain injury42,50. Accordingly, in our study, we found elevated expression of several protein-encoding genes involved in myelin debris phagocytosis (Trem2), lipid storage in lipid droplets (Plin2, Plin3, Soat1), lipid hydrolysis (Nceh1, Lpl, Pla2g7), lipid metabolism regulation (Nr1h3), lipid efflux (Abca1) and lipid transport (Apoe, Apoc1, Apoc2, Npc1) specifically in RAMf, indicating that RAMf is the main cell type involved in lipid recycling among macrophages (Figs. 5D, S8A). Furthermore, we identified significantly higher presence of intracellular lipid droplets in IBA1+GPNMB+ cells compared to IBA1+GPNMB− cells, as evidenced by immunostaining with antibodies against perilipin 2 (PLIN2), a structural component of intracellular lipid droplets (Fig. 5E). Additionally, BODIPY staining demonstrated that CD45high CD11b+ Ly6G- GPNMB+ CD63+ cells had a higher intracellular lipid load than CD45high CD11b+ Ly6G- GPNMB− CD63− cells (Fig. 5F).
To further elucidate the effect of RAMf on lipid metabolism after ischemic stroke, we performed LC-MS lipidomics on the ischemic hemisphere from mice with or without monocyte depletion five days after ischemic stroke (Fig. 5G). Stroke induced obvious alterations in various lipids in the mouse brain (Fig. S8B, C), including phosphatidylcholine (PC), fatty acid (FA), phosphatidylethanolamine (PE), and lysophosphatidylcholine (LPE), indicating the dysregulation of lipid metabolism after ischemic stroke. Notably, the major components of lipid droplets, cholesterol ester (ChE) and triglyceride (TG), were significantly elevated after ischemic stroke (Figs. 5H, S8D). However, these lipid components were markedly downregulated in clodronate-treated mice (Figs. 5H, S8D), indicating that macrophage depletion impacts cholesterol metabolism in the brain, thereby diminishing the lipid content supply within the brain environment.
Together, these data provide evidence that RAMf supports remyelination through lipid recycling.
RAMf promote capillary proliferation through intricate molecular crosstalk
Monocyte-derived macrophages have recently been implicated in the angiogenesis process in ischemic stroke51. Similarly, we observed a significant increase in the angiogenesis and blood vessel remodeling AUCell scores in PLX5622-treated mice at both 5 dpi and 14 dpi (Fig. 6A). Immunofluorescence staining confirmed that PLX5622-treated MCAO mice had more ZO-1-positive vessels (labeled by CD31) in peri-infarction regions than the control group at 14 dpi (Fig. 6B). Moreover, RAMf (MF2) displayed a significant upregulation of genes related to a pro-angiogenic phenotype, such as Igf1, Grn, Lgals3, Ang, Ccl2, Pdgfra, Vegfb, Pf4, Anpep, Flt1, and Hgf, suggesting RAMf as a major source of pro-angiogenic factors in PLX5622-treated mice (Fig. 6C).
A Violin plot depicting the upregulation of angiogenesis related function of RAMf both at 5 and 14 dpi, measured by AUCell score. n = 502 RAMfs for control diet d5, n = 1360 RAMfs for PLX5622 d5, n = 253 RAMfs for control diet d14, n = 755 RAMfs for PLX5622 d14, from two biological replicates. Wilcoxon rank sum test, two-sided. Box plot represents the first quartile, median, and third quartile with whiskers extending to 1.5 times the interquartile range. B Representative images and quantification for CD31 (green) and ZO-1 (red) double immunostaining in the peri-infarct areas at 14 dpi. n = 5 per group. C Heatmap showing the expression of angiogenesis related genes in each MF cluster, with bars showing log2 (fold change) of RAMf compared to other MF. D Expression patterns of receptors for angiogenesis factors on endothelial cells at 14 dpi. E UMAP projection of endothelial cells at 14 dpi, with each color representing different endothelial cell clusters. F Dot plots showing expression of traditional vascular markers in each of three distinct clusters. G Monocle pseudotime trajectory of the EC clusters, and expression patterns of representative DEGs in the trajectory transition process that indicate different vascular zones. H Dot plot summarizing PLX5622-induced changes in cell numbers of Mki67+ cells in each vascular zone. I Proposed model for interaction pairs between RAMf (MF2) and capillary EC, generated by the InterCellDB package. J Representative images and quantification for CD31 (green) and BrdU (red) immunostaining in the peri-infarct areas in brains from mice with RAMf or GPNMB−CD63− macrophages transplantation. n = 5 for GPNMB−CD63− macrophages transplantation group, n = 6 for RAMf transplantation group. B, J Student’s t test, two-sided. Data in (B, J) are presented as mean ± SD. Source data are provided as a Source Data file.
Emerging evidence increasingly implicates zonation-dependent, rather than consistent, changes across the vascular bed in response to neurological diseases52. In this study, we observed heterogeneity in the expression levels of receptors (Igf1r, Kdr and Nrp1) for pro-angiogenic factors in ECs (Fig. 6D). To explore EC heterogeneity, we conducted unsupervised clustering analysis (Fig. S9A), which identified three distinct subsets of endothelial cells based on their transcriptomic characteristics (Figs. 6E, F, S9B). The pseudo-temporal axis constructed by Monocle revealed a correlation between EC subsets and different vascular zones (Fig. 6G). Classical arterial markers (Gkn3) and venous markers (Nr2f2) peaked at opposite ends of the trajectory, while capillary markers (Tfrc)53 reached their highest expression in the middle of the trajectory (Fig. 6G). We subsequently classified the ECs into three categories: artery (EC2, Gkn3+, Tgfb2+, Bmx+), capillary (EC1, Tfrc+, Car4+, Slc16a1+), and vein (EC3, Nr2f2+, Lrg1+, Lcn2+) (Figs. 6G, S9C). Notably, we observed a significant increase in Mki67+ cells specifically in capillary EC (Fig. 6H), suggesting the heightened microvessel proliferation following early-phase microglia attenuation. Then we took advantage of InterCellDB54 to explore potential crosstalk between RAMf and capillary ECs (Fig. 6I). Pdgfa, Ccl2, Ang, Lgals3, Anpep, Vegfb, Hgf and Flt1 were inferred to mediate the interaction between RAMf and capillary ECs (Fig. 6I). Their expression levels in RAMf were notably higher than in other macrophages, suggesting RAMf was the major source of heightened angiogenesis in PLX5622-treated mice (Fig. 6I). Furthermore, mice that underwent RAMf transplantation exhibited a substantial augmentation in vascular coverage and CD31+ BrdU+ cells when compared to the GPNMB− CD63− macrophage transplantation group (Fig. 6J). Collectively, these data provide evidence suggesting a significant involvement of RAMf in post-ischemic stroke angiogenesis.
Mafb regulates the differentiation and brain repair-related functions of RAMf
To elucidate the transcriptional regulatory network and identify pivotal transcription factors guiding the differentiation process of RAMf, we employed the transcription factor prediction tool ARCHS455, which leverages ChIP-seq data to identify key transcription factors. Through analysis of DEGs associated with myelination and angiogenesis, we identified Mafb as a central upstream transcription factor shared among genes implicated in both processes (Fig. 7A). Using the SCENIC toolkit for gene regulatory network reconstruction, we identified Mafb and Nr1h3 as candidate TFs with specifically upregulated expression levels in RAMf (denoted as MF2 in Fig. 7B). In addition, we observed a significant elevation of Mafb and its target genes in RAMf (Fig. 7C, D), supporting its role in enhancing reparative phenotype in PLX5622-treated mice.
A Transcription factor enrichment analysis based on the ARCHS4 database using the online tool Enrichr. Fisher’s exact test, with Benjamini-Hochberg correction. B Heatmap showing the average regulon activities of representative TFs in each MF subset derived from pySCENIC. C Scatter plot illustrating upregulated DEGs of PLX5622 group compared to control group in MF2, with the percentage difference (defined as percentage of the expression in PLX5622 group minus that in control group) along the x-axis and log2(fold change) along the y-axis. The combined z-score of percentage difference and log2(fold change) was color-coded. D Feature plot showing the AUCell score of the Mafb-target genes. Relative AUCell scores were color-coded. E Representative image showing the phagocytosis of MBP (red) by IBA1+ (green) BMDMs (left). Quantification of the proportion of MBP-positive cells in IBA1+ cells (right). n = 3 per group. Student’s t test, two-sided. F Experimental scheme illustrating macrophage reconstruction in CCR2-KO mice. G Representative images for CD31 (green) immunostaining in the peri-infarct areas of brains from CCR2-KO mice with sham surgery or MCAO with Mafb-overexpression macrophages transplantation or control macrophages transplantation (left). Quantification of vascular coverage (right). n = 6 per group. One-way ANOVA and Tukey. H Representative images and quantification for MBP (green) and SMI32 (red) double immunostaining in the peri-infarct areas. n = 4 for sham group, n = 5 for lenti-control and lenti-Mafb group. Scale bars, 200 µm (Cortex), and 100 µm (Striatum and External Capsule). Data were normalized to the intensities of contralateral hemispheres. One-way ANOVA and Tukey. Data in (E, G, H) are presented as mean ± SD. Source data are provided as a Source Data file.
To ascertain the involvement of Mafb in eliciting the pro-reparative phenotype within macrophages, we employed siRNA to suppress Mafb expression in bone marrow-derived macrophages (BMDMs) in vitro. We observed a notable decline in myelin engulfment in BMDMs treated with Mafb-siRNA (Fig. 7E), suggesting that Mafb plays a role in phagocytic activity in macrophages. In addition, to evaluate the role of Mafb in promoting remyelination and angiogenesis following ischemic stroke, we specifically overexpressed Mafb in BMDMs through a lentiviral vector. Quantitative PCR analysis confirmed significantly higher Mafb expression levels in the Mafb lentivirus treatment group compared to controls (Fig. S10A). We then utilized RNA sequencing to explore the transcriptomic changes in BMDMs resulting from Mafb overexpression. The analysis revealed significant upregulation of genes associated with the RAMf phenotype, including Gpnmb, Cd63, Cd9, Syngr1, and Fabp5 (Fig. S10B). Additionally, Gene Set Enrichment Analysis (GSEA) revealed terms related to steroid biosynthesis and cholesterol metabolism (Fig. S10C), indicating a transcriptomic profile in Mafb-overexpressing BMDMs that closely resembles that of RAMf cells. This finding was supported by flow cytometry, which demonstrated increased levels of GPNMB and CD63 following Mafb overexpression (Fig. S10D). Importantly, no significant differences were observed in cell viability, as assessed by MTT assays, or in annexin V expression levels between the Mafb lentivirus-treated and control groups (Fig. S10E, F), suggesting that Mafb transfection does not affect cell survival.
To further elucidate the functional implications of Mafb overexpression, we transferred two million BMDMs transduced with either Mafb or control lentivirus into CCR2-knockout mice 6 hours after MCAO (Fig. 7F). Flow cytometry analysis revealed comparable levels of monocytes and macrophages in both the peripheral blood and brain between two groups (Fig. S10G, H). Notably, mice reconstituted with Mafb-overexpressing BMDMs exhibited reduced tissue loss at 14 dpi (Fig. S10I) and a significant increase in vascular coverage (Fig. 7G), along with a decrease in the SMI32/MBP ratio (Fig. 7H). These results highlight the positive effects of Mafb on vascularization and remyelination following ischemic stroke.
In conclusion, we identified Mafb as a candidate transcription factor that drives the remyelination and angiogenesis phenotypes in RAMf after ischemic stroke.
PLX5622 induces increased infiltration of RAMf following stroke in aged mice
Aging significantly impacts both the immune system and the central nervous system, thereby influencing the neural repair processes following ischemic stroke56. To investigate the effects of aging on RAMf, we conducted scRNA-seq on enriched CD45high infiltrating immune cells isolated from the brains of aged ischemic mice at 14 dpi (Fig. S11A). Unsupervised clustering analysis revealed 12 distinct cell types, including microglia (MG), macrophages (MF), classical dendritic cells 1 (cDC1), classical dendritic cells 2 (cDC2), plasmacytoid dendritic cells (pDCs), neutrophils (NP), natural killer (NK) cells, CD4+ T cells, CD8+ T cells, gamma delta T cells (γδT), B cells, and proliferating immune cells (ProlifCells) (Fig. S11B, C).
Subclustering analysis was performed specifically on macrophages (Fig. S11D). This analysis identified three macrophage subgroups that closely resemble those observed in young mice: MF1 (Ly6c2+, Chil3+), MF2 (Gpnmb+ CD63+), and MF3 (Cd209a+, Klrd1+) (Fig. S11E). Notably, these macrophage subgroups in aged mice demonstrated a high transcriptional correlation with their counterparts in young mice (Fig. S11F). Furthermore, AUCell scores for myelination and the positive regulation of angiogenesis were significantly elevated in the MF2 cluster (Fig. S11G). We then compared the transcriptomic characteristics of RAMf (MF2) between aged and young mice. The results indicated that RAMf in aged mice upregulated inflammation-related genes, including H2-Eb1, H2-Aa, and S100a9 (Fig. S11H). Functional enrichment analysis revealed upregulated terms related to mRNA metabolic processes and leukocyte activation in RAMf of aged mice compared to young mice (Fig. S11I). To examine whether PLX5622 could enhance the proportion of RAMf in aged mice, we administered PLX5622 to aged MCAO mice for the first 7 days, then switched to a control diet until 14 dpi (Fig. S11J). Flow cytometry analysis confirmed a significantly higher proportion of GPNMB+ CD63+ RAMf within the CD45high CD11b+ population at 14 dpi (Fig. S11K).
Taken together, these findings validate the presence of RAMf in aged mice. Their transcriptomic profile suggests a significant role of RAMf in promoting myelination and angiogenesis. Furthermore, early-phase microglia attenuation could increase the proportion of RAMf in aged mice.
Discussion
Following an ischemic stroke, the intricate interactions between the immune and nervous systems are pivotal in facilitating neurological function recovery. CSF1R inhibition (CSF1Ri) has been widely employed to deplete microglia. After the withdrawal of CSF1Ri treatment, the microglial niche is repopulated by a combination of surviving resident microglia and infiltrating macrophages28. In this study, we implemented a 7-day PLX5622 treatment strategy that induced approximately 70% microglia depletion. Our findings indicate that during the administration of PLX5622, a substantial number of macrophages infiltrated the brain and persisted until the 7 days after drug withdrawal. These infiltrating macrophages differentiated primarily into RAMf and demonstrated the capacity to promote white matter and vascular repair following stroke.
Repopulated microglia have been shown to exert neuroprotective effects in several studies24,57,58. In this study, we identified 5 microglial subpopulations at 14 dpi, 7 days after the withdrawal of PLX5622. Among them, the MG4 subpopulation exhibited upregulation of neural repair-related genes, suggesting its potential role in brain repair. However, we observed a decrease in the proportion of MG4 cells within the PLX5622 group. While myelination-related functions were upregulated in this subset, its angiogenic capacity was diminished. These results suggest that repopulated microglia play a role in post-stroke repair, but their effects may be limited within the context of our treatment strategy. This limitation could be attributed to a narrow therapeutic window, as repopulated microglia promote neural repair only when coinciding with brain injury in the TBI mouse model24. Additionally, it has been reported that PLX5622 reduces the production and proliferation of peripheral monocytes31. Our study corroborated this finding, revealing that PLX5622 decreased the number of peripheral monocytes, however, their reparative function remained largely unaltered. Previous studies have indicated that the number of Ly6C+ inflammatory myeloid cells in peripheral blood influences the injury and repair processes following ischemic stroke51,59. Therefore, future research should incorporate cell-specific genetic models to achieve a deeper understanding of the effects of early-phase PLX5622 treatment on ischemic stroke, while minimizing the confounding influences of repopulated microglia and peripheral monocytes.
Macrophages are known to have a dual role in ischemic stroke: they can contribute to secondary brain injury through the release of pro-inflammatory cytokines, while also aiding in the clearance of cellular debris and apoptotic cells to mitigate damage extent3. In our study, we identified these dual activation directions through an integrated analysis of peripheral blood monocytes and brain macrophages, further underscoring the functional complexity of macrophages. Notably, a recent study identified stroke-associated myeloid cells (SAMCs) in both human and mouse stroke models14. These SAMCs show transcriptomic similarity to monocyte-derived RAMf. Interestingly, stroke amelioration was also observed when SAMCs were inhibited using a CSF1R inhibitor, aligning with the findings from this study.
Myelin, a lipid-rich membrane stack enveloping axons, undergoes disruption during ischemic stroke. The accumulation of myelin debris has been shown to impede white matter repair60. Our investigation revealed that RAMf enhanced the processes of debris phagocytosis, lipid metabolism, and lipid efflux. These lipid recycling mechanisms have been identified as crucial repair steps during acute brain injury46,61. And aged lipid-laden microglia may contribute to worse neurological outcome after stroke56. Recent studies have emphasized the protective role of Trem2-dependent lipid droplet formation in facilitating remyelination42. Accordingly, we observed increased lipid droplet loading in RAMf and a significant decrease of lipid droplet composition in monocyte depletion mice, suggesting that RAMf may promote remyelination through lipid droplet formation and lipid recycling.
Accumulating evidence highlights the crucial role of macrophages in angiogenesis following ischemic stroke. CCR2+ monocytes have been reported to facilitate vascular repair through programming microglia in an IL-6 dependent manner62. Conversely, CCR2 deficiency in monocytes has been linked to impaired angiogenesis after ischemic stroke51. In this study, cell-cell interaction analysis revealed that RAMf expresses several pro-angiogenic factors (Pdgfra, Ccl2, Ang, Lgals3, Anpep, Vegfrb, Hgf, and Flt1) that positively impact the proliferation of endothelial cells. Notably, this interaction is particularly conspicuous between RAMf and capillary endothelium, underscoring the importance of microvascular regeneration in the post-stroke repair process. However, a recent study indicated that PLX5622 treatment alters brain endothelial cholesterol metabolism independently of microglia depletion63. Thus, while we identified the role of RAMf in promoting endothelial cell proliferation through adoptive cell transfer experiments, the off-target effects of PLX5622 on endothelial cell proliferation warrant further investigation.
We also found that neutrophils persist in the brain at 14 dpi and demonstrate an increase in AUCell scores of angiogenesis. It is reported that neutrophils can regulate IL-6 level64, which is crucial for post-stroke angiogenesis65. In cancer, neutrophils can release pro-angiogenetic factors like VEGFs, FGFs, and MMP-9 to facilitate tumor growth and metastasis66. Further investigations into neutrophil-mediated angiogenesis in both acute and chronic phase of stroke are warranted.
RAMf cells conservatively exisit in the brain of aged mice, with a transcriptomic profile similar to that of young mice. The proportional increase in RAMf cells has also been observed after treating aged mice with PLX5622. However, whether aging-related changes interfere with the pro-repair characteristics of RAMf in stroke requires further elucidation.
Through gene regulatory network reconstruction, we identified Mafb as a pivotal transcription factor that modulates pro-angiogenic and remyelination functions in RAMf. Mafb has been reported to promote an anti-inflammatory phenotype in macrophages, and a recent study indicated that Mafb is the key factor governing the transition of monocytes into tissue-resident macrophages67,68. Similarly, our study demonstrated that monocyte-derived RAMf acquired resident microglia-like functions, including myelin phagocytosis and lipid metabolism. Notably, Mafb also plays a significant role in preserving the health of adult microglia by suppressing inflammatory and proliferative responses69. Further investigation is warranted to elucidate the involvement of microglia and monocytes in neural repair after ischemic stroke, as well as to delineate the specific role played by Mafb in this process. Prior research has suggested that Mafb in myeloid cells can mitigate excessive inflammation by accelerating the clearance of danger signals through MSR1 during the acute phase of stroke70. We observed elevated MSR1 expression in RAMf and noted similar transcriptome characteristics at both 5 and 14 dpi. Nonetheless, further investigations are essential to confirm the dynamic functional plasticity of RAMf at different stages of stroke.
There are several limitations to our study. Firstly, we confirmed that early-phase microglia attenuation could enhance the production of lipid metabolism-related RAMf. While various resident cells, including microglia, astrocytes, oligodendrocytes, endothelial cells, and neurons, are involved in lipid metabolism, the precise interactions between RAMf and other cells in this process, encompassing molecular interactions and lipid transport, require further confirmation. Additionally, the clinical relevance of our murine findings needs validation. Similar myeloid cells related to lipid metabolism as RAMf were observed in ischemic stroke and multiple sclerosis human brain specimens14,47. Clinical data indicate that CSF1R inhibitors are highly effective in human microglial depletion. Future studies are warranted to further clarify the translational potential of early-phase microglia attenuation or RAMf cell therapy in stroke treatment. Another limitation of this study is that all experimental procedures were conducted using male mice. Previous research has indicated that the effects of CSF1R inhibitors may vary by sex18,71, potentially limiting the generalizability of our findings. Therefore, future studies are essential to confirm the role of sex differences in the response of RAMf to ischemic injury.
Another limitation of this study is that all experimental procedures were conducted using male mice. This focus may restrict the generalizability of our findings, as the mechanisms underlying ischemic stroke recovery may differ in female mice. Future research should aim to investigate the role of sex differences in the behavior of repair-associated macrophages and microglial responses to ischemic injury to enhance our understanding of these processes across genders.
In conclusion, our study elucidated alterations in the immune niche within the mouse brain following ischemic stroke using early-phase microglia attenuation. We observed a significant increase in RAMf, exhibiting a phenotype with pro-angiogenic and remyelination functions. Furthermore, we identified Mafb as a regulator of RAMf differentiation, and its upregulation helps enhance white matter and vascular integrity after ischemic stroke. In summary, our findings provide insights into potential treatment strategies for ischemic stroke by rebalancing immune niches in the ischemic brain.
Methods
Ethical regulations
The ethical approval for all animal experimental procedures (Approval No. 2020-655) was granted by the Institutional Ethics Committee of the Second Affiliated Hospital, Zhejiang University School of Medicine. The procedures adhered to the guidelines set forth in the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
Animals
Young (8–12 weeks old, n = 375) male C57BL/6 mice obtained from SLAC Laboratory Company (Shanghai, China) were used for in vivo experiments. Young (6–8 weeks old, n = 13) male CD45.1 C57BL/6 mice (Cat. NO. NM-KI-210226) were procured from Shanghai Model Organisms Center, Inc (Shanghai, China). Aged (20–22 months old, n = 14) male C57BL/6 mice were obtained from Beijing Vital River Laboratory Animal Technology Co. Ltd. (Beijing, China). CCR2-GFP (8–12 weeks old, n = 60) and CCR2-KO mice (8–12 weeks old, n = 40), established on a C57BL/6 background, were procured from the Jackson Laboratory (Strain #: 027619, RRID: IMSR_JAX:027619). All animals were housed in environmentally controlled cages with appropriate temperature (22 °C ± 2 °C) and humidity (55% ± 10%), adhering to a 12-hour light/dark cycle, and provided ample food and water. Animals were randomly assigned to either the sham or stroke groups and received treatments determined by a lottery drawing method. The mice were euthanized by carbon dioxide according to the approved protocols. All treatments and analyses were conducted by blinded investigators.
Transient cerebral ischemia model
Transient cerebral ischemia models were established by endovascular occlusion of the left middle cerebral artery (MCA) for 60 min. Briefly, mice were anesthetized with 1% pentobarbital and maintained on a 30% O2 / 70% N2O mixture to ensure spontaneous breathing. A filament with a silicon-coated tip (RWD, Shenzhen, China) was inserted into the external carotid artery (ECA). After adjusting the direction into the internal carotid artery (ICA), the filament was advanced to the origin of the MCA until resistance was felt. The filament was then left in position for 60 min to obstruct cerebral blood flow. After this period, the filament was withdrawn to restore brain blood flow, and the residual end of the ECA was ligated. The body temperature of the mice was maintained at 37.0 ± 0.5 °C during the operation, and the mice were placed on a water bath blanket until they woke up from anesthesia. The sham group animals received the same anesthesia and experienced carotid triangle exposure without left MCA occlusion. Animals that did not display at least 70% reduction in regional CBF (using LDF) during MCAO compared to pre-ischemia levels were excluded from further experimentation. The surgeons were blinded to the grouping of the experimental animals. The mortality rates in this study are listed in Supplementary Data 5.
CSF1R inhibition
For early-phase CSF1R inhibition, mice were provided with PLX5622 (Chemgood) in the diet (Research Diets) at a concentration of 1200 PPM (1200 mg/kg of chow). The administration of PLX5622 chow began on the day of MCAO and continued until 7 dpi.
Monocyte depletion
Clodronate liposomes were used to deplete monocytes in the blood (Liposome BV), administered at a dosage of 100 μL per 10 grams of mice weight. The administration of clodronate liposomes began two days before MCAO and continued daily until sacrifice. The mice in the control group received injections of control liposomes at the same dose.
Intraventricular cell transplantation
Ischemic mouse brains were harvested 14 days after MCAO. The CD45high CD11b+ GPNMB+ CD63+ cells were sorted using the BD FACSAriaTM II (BD Bioscience). Cells were delivered at a dosage of 30,000 cells per mouse into the CSF of the lateral ventricle using a Hamilton syringe. The injection point was located 1 mm from the midline along the Bregma, and the depth of injection was 2.5 mm. Mice received cells transplantation at 5 dpi.
BrdU injection
To label cell proliferation in the brain, mice were administered the thymidine analog 5’-bromo-2’-deoxyuridine (BrdU, 50 mg/kg) through intraperitoneal injection, twice daily with a minimum interval of 8 h, and continuing for 3 consecutive days.
Peripheral immune cells extraction and adoptive transfer
CD45.1 mice were subjected to MCAO and fed with PLX5622 diet or control diet. After 5 days, mice were sacrificed and 0.8 ml peripheral blood was extracted from right atrium. Red blood cells were lysed with 4.5 ml 1X RBC lysis (eBioscience, 00-4333057) for 4 min. Thereafter, 10 ml of pre-cooled PBS buffer was added to stop the lysis, followed by centrifugation for 5 minutes (500 g, 4 °C), repeated three times. Peripheral immune cells from one CD45.1 mouse were obtained and transferred to one WT recipient mouse via tail vein injection.
Behavior tests
We conducted a series of behavioral tests that have been previously shown in studies to be highly sensitive and accurate for assessing neurofunctional deficits in rodents following stroke9,72. The establishment of the MCAO model, behavioral tests, and statistical analyses were conducted by different researchers, ensuring blinding to treatment throughout the experiments. All data were expressed as mean values from 3 repeated trials per day.
Adhesive removal test
In this test, a 2 x 3 mm adhesive tape was applied to the right forelimb of the mice. We recorded the time taken to perceive and remove the tape separately to assess tactile response and sensorimotor asymmetry. The maximum observation window for perception was 60 s, and for removal, it was 120 s.
Foot fault test
Briefly, mice tightly grasped the wire while moving on a wire fence with a grid size of 2 × 2 cm located 1 meter above the ground. A “foot fault” was defined as a paw falling between the grid spaces due to impairment. We recorded the percentage of forelimb contralateral to the injured hemisphere’s falling times compared to the total number of steps within one minute to evaluate locomotor function.
Magnetic resonance imaging (MRI) and analyses
Ischemic infarct volume was evaluated using a 7 T animal MRI (Biospec 70/16, Bruker, Germany) with a mouse surface receiver coil for mouse brain imaging. During the scanning process, the experimental animals were placed on a blanket to control body temperature at 37.0 °C. T2-weighted images of the brain were acquired with the following parameters (Repetition Time (TR) = 2.5 s; Echo Time (TE) = 35 ms; number of averages = 8; slice thickness = 0.55 mm; FOV = 24 mm × 24 mm; matrix size = 256 × 256). Whole-head T2 images were obtained and denoised to diminish the possible artifacts before image processing. Then, the segmentation of the infarct was carried out with ITK-SNAP73 software (version 3.8.0).
Measurement of tissue loss
Mice were euthanized and transcardially perfused with 20 ml of pre-cooled phosphate-buffered saline (PBS), followed by perfusion with 4% paraformaldehyde (PFA) dissolved in PBS. The mouse brain was dissected and fixed in 4% PFA for 24 h. Dehydration was performed using a gradient of 15% and 30% sucrose solutions at 4 °C. Brain samples were sectioned with a cryo-microtome into 6 evenly spaced layers of coronal slices (25 μm thick) and then stored in a cryoprotectant solution (40% PBS, 30% ethylene glycol, 30% glycerol) at −20 °C until immunostaining. For each layer, one brain section slice was chosen and stained with an anti-MAP2 antibody. Tissue loss was assessed in six equally-spaced 25 µm coronal brain sections stained for MAP2, spanning approximately 1.10 mm anterior to bregma to 2.06 mm posterior to bregma. The volume of tissue loss was calculated by subtracting the volume of the non-infarcted area in the ipsilateral hemisphere from that of the contralateral hemisphere.
Multi-color immunohistochemistry
Brain samples were collected as described above. Formalin-fixed tissue undergoes tissue processing and then is embedded in paraffin to create a paraffin block. Paraffin blocks were cut into 6 mm sections and mounted on glass slides. The slides were subjected to heating at 65 °C for 1 hour and dewaxed in xylene. Subsequently, the sections underwent rehydration through successive immersions in 100%, 95%, and 70% alcohol. Immune cells in ischemic brain were detected using the Opal Polaris 780 Reagent Pack (Akoya Biosciences) according to the manufacturer’s instructions. Briefly, heat-mediated antigen retrieval was conducted for 15 min using AR6 buffer. After naturally cooling to room temperature, the samples underwent a 10 min blocking step. Then, the sections were incubated at room temperature for 1 hour with primary antibodies, including Rabbit anti-CD4 (Abcam, ab183685, 1:200), Rabbit anti-Tmem119 (Abcam, ab209064, 1:200), Rabbit anti-Ly6G (Abcam, ab238132, 1:200) and Rabbit anti-NKP46 (Abcam, ab233558, 1:200). After three washes with TBST, the sections were incubated with Opal Polymer HRP and Opal Working solution for 10 min each. Signal amplification was achieved using TSA-DIG working solution. To label cell nuclei, the sections were incubated with DAPI working solution for 5 min. Multispectral imaging was collected using the Vectra Polaris Quantitative Pathology Imaging System at a magnification of 40X and analyzed using Phenochart or Inform Software.
Immunostaining of brain sections
Coronal brain slices were utilized for immunostaining. In brief, the floating brain slices were washed twice with PBS, followed by treatment with 0.5% Triton-X in PBS (PBST) for 15 min to permeabilize cell membranes at room temperature. Subsequently, the slices were rinsed three times with 0.3% PBST for 5 min each. They were then blocked with 5% normal donkey serum in 0.3% PBST at room temperature for 1 h before overnight incubation at 4 °C with the following primary antibodies: Goat anti-CD31 (R&D, AF3628, 1:200), Rabbit anti-ZO1 (Abcam, ab221547, 1:200), Rabbit anti-Pdgfra (Abcam, ab203491, 1:250), Goat anti-IBA1 (Abcam, ab5076, 1:250), Mouse anti-APC (Sigma-Aldrich, OP80, 1:100), Rat anti-BrdU (Abcam, ab6326, 1:100), Mouse anti-SMI32 (BioLegend, 801701, 1:250), Rat anti-MBP (Abcam, ab7349, 1:250), Rabbit anti-Plin2 (Abcam, ab108323, 1:100), Rabbit anti-GPNMB (Abcam, ab188222, 1:200), Rabbit anti-CD63 (Abcam, ab217345, 1:200) and Rabbit anti-MAP2 (Proteintech, 17490-1-AP, 1:500). After washing with 0.3% PBST three times for 10 minutes each, brain slices were incubated with appropriate secondary antibodies conjugated with Donkey anti-Goat Alexa Fluor 488 (Invitrogen, 1:500, A-11055), Donkey anti-Rabbit Alexa Fluor 488 (Invitrogen, 1:500, A-21206), Donkey anti-Mouse Alexa Fluor 488 (Invitrogen, 1:500, A-21202), Donkey anti-Rat Alexa Fluor 488 (Invitrogen, 1:500, A-21208), Goat anti-Rabbit Alexa Fluor 555 (Invitrogen, 1:500, A-21428), Donkey anti-Rabbit Alexa Fluor 594 (Invitrogen, 1:500, A-21207), Donkey anti-Goat Alexa Fluor 594 (Invitrogen, 1:500, A-11058), Donkey anti-Rat Alexa Fluor 594 (Invitrogen, 1:500, A-21209), Goat anti-Mouse Alexa Fluor 594 (Invitrogen, 1:500, A-11005), Donkey anti-Goat Alexa Fluor 647 (Invitrogen, 1:500, A-21447), Donkey anti-Rabbit Alexa Fluor 647 (Invitrogen, 1:500, A-31573) in a dark environment at room temperature for 1 h. Subsequently, brain slices were washed with PBS three times and mounted on glass slides with mount-G containing DAPI (Yeasan Biotech). The Leica TCS SP8 confocal microscope (Leica Microsystems) was employed to observe and capture images of the sections. Image analysis was conducted on one or two randomly selected microscopic fields in the peri-infarct areas of the EC, two in the cortex, and two in the striatum of each section. For the measurement of vascular coverage and ZO-1 expression in the vessel area, we selected three 25 μm thick slices from each mouse brain and identified 4 regions around the infarct in each slice for imaging and quantification. The recorded images were loaded into Image J (NIH) and manually quantified by two observers who were blinded to the grouping. Positively stained cells were labeled with the software to prevent duplicate counting.
Imaris 3D rendering
Imaris software (Version 9.01; Bitplane, Zurich, Switzerland) was utilized for the 3D reconstruction of immunofluorescent images. Briefly, confocal image stacks were imported into Imaris, and the surface module tool was employed for the 3D structure reconstruction of each color channel. Reconstruction was carried out within designated regions of interest, utilizing the absolute fluorescence intensity from each color channel. Smoothing was applied using varying channel settings, and thresholds were configured to distinguish target signals from the background while eliminating nonspecific signals.
Intracellular neutral lipid staining
BODIPYTM 493/503 (Thermo Fisher Invitrogen) powder was resuspended in DMSO and aliquoted at a concentration of 1 mg/ml. The staining was performed at a dilution of 1/1000. After incubation with surface and intracellular antigen antibodies, single-cell suspensions were stained with BODIPY for 20 min at room temperature in a dark environment. The mean fluorescence intensity (MFI) of each gated population was used to quantify the BODIPY signal by flow cytometry.
Sample preparation for lipid profiling
The ischemic hemisphere of MCAO mouse brains was flash-frozen in liquid nitrogen and stored at −80 °C. After thawing from −80 °C, 300 μL of each sample was combined with 20 μL of Lyso PC 17:0 (0.1 mg/mL) as an internal standard. Then, 300 μL of chloroform: methanol (2/1, vol/vol) with 0.1 mM BHT was added to each sample, followed by 30 seconds of vortexing and 10 min of ultrasonication in an ice-water bath. Samples were placed at −20 °C for 30 minutes, then centrifuged at 4 °C (15,620 g) for 10 min, yielding 200 μL of subnatant. For the residue samples, 300 μL of chloroform: methanol (2/1, vol/vol) containing 0.1 mM BHT was added, vortexed for 30 s, and subjected to 10 min of ultrasonication in an ice-water bath. After −20 °C incubation for 20 min and subsequent centrifugation at 4 °C (15620 g) for 10 min, subnatant was collected. The two subnatants were combined, resulting in a mixed subnatant (400 μL), which was dried with nitrogen and redissolved in 300 μL of isopropanol: methanol (1/1, vol/vol). After vortexing for 30 s and 3 min of ultrasonication in an ice-water bath, the solution was filtered through a 0.22 μm organic phase pinhole filter for UPLC-MS/MS analysis, which was conducted by Shanghai Profleader Biotech Co., Ltd (Shanghai, China). Quality control (QC) samples were prepared by pooling aliquots from all individual samples.
LC-MS/MS analysis
The LC-MS/MS analysis was conducted by Shanghai OE Biotech Co., Ltd (Shanghai, China). The metabolic profiling was carried out using a Dionex Ultimate 3000 RS UHPLC coupled with a Q-Exactive quadrupole-Orbitrap mass spectrometer equipped with a heated electrospray ionization (ESI) source from Thermo Fisher Scientific (Waltham, MA, USA). The analysis was performed in both ESI positive and ESI negative ion modes. An ACQUITY UPLC BEH C18 column (1.7 μm, 2.1 × 100 mm) was utilized in both modes. The binary gradient elution system comprised (A) acetonitrile:water (60:40, v/v, containing 10 mmol/L ammonium formate) and (B) acetonitrile:isopropanol (10:90, v/v, containing 10 mmol/L ammonium formate). The flow rate was set at 0.26 mL/min, and the column temperature was maintained at 55 °C. Throughout the analysis, all samples were maintained at 4 °C.
Data processing in LC-MS/MS analysis
The initial Q Exactive LC-MS/MS data in.raw format underwent processing using LipidSearch software to obtain MSn and exact mass-to-charge ratio (m/z) of parent ions. Normalization was applied to all peak signals within each sample. Subsequently, the extracted data underwent further refinement: peaks with missing values (ion intensity = 0) in over 50% of groups were removed, and zero values were substituted with half of the minimum value. The positive and negative ion data were combined to create a data matrix. This matrix was imported into R for Principal Component Analysis (PCA) to observe overall sample distribution and analysis stability. Variable Importance of Projection (VIP) values from the OPLS-DA model ranked the contribution of each variable to group differentiation. A two-tailed Student’s T-test verified the significance of intergroup metabolite differences. Differential metabolites were identified based on VIP values exceeding 1.0 and p-values below 0.05.
Myelin preparation
Myelin was isolated from 8-week-old mouse brains according to a protocol previously described in ref. 74. Briefly, brain homogenization was conducted in a 0.32 M sucrose solution and gently layered on a 0.85 M sucrose solution, and then centrifuged at 75,000 g for 30 min. The crude myelin fraction was removed from the interface and washed with distilled water, and subjected to centrifugation at 75,000 g for 15 min. Osmotic shock was carried out by immersing the sample in water for 15 minutes, followed by centrifugation at 12,000 g for 15 min. The resulting pellet was resuspended in a 0.32 M sucrose solution, placed above a 0.85 M sucrose solution, and subjected to centrifugation at 75,000 g for 30 min. Purified myelin was further washed with water and centrifuged at 75,000 g for 15 min. Finally, the purified myelin pellet was resuspended in 1 ml PBS and stored at −80 °C.
Bone marrow-derived macrophages
BMDMs were isolated from the femora of adult mice following the established protocol75. Briefly, the mice were sacrificed and disinfected with 70% alcohol. The hip bones were carefully removed, excluding skin and muscle tissues, and the bone ends were trimmed. Subsequently, the marrow was flushed into a 15 mL centrifuge tube using a 1 mL syringe. After centrifugation at 600 × g and 4 °C for 5 minutes, the resulting cells were diluted to a concentration of 1 × 106 cells/mL. These cells were then cultured in DMEM with 1% penicillin/streptomycin and 10% fetal bovine serum at 37 °C with 5% CO2. M-CSF (Thermo Fisher Invitrogen, PMC2044) was introduced at a final concentration of 20 ng/mL for 6 days to induce BMDM differentiation. To inhibit gene expression, BMDMs were transfected with siRNA using an electroporation system. Additionally, Mafb-lentivirus infection was conducted to achieve overexpression of Mafb in BMDMs according to the documented protocol75. In short, BMDMs were transduced with Mafb-lentivirus at M.O.I of 30 and polybrene (6 mg/ml), using mCherry-lentivirus as control. BMDMs were then incubated at 37 °C with 5% CO2 for 48 h. Thereafter, the medium containing lentivirus and polybrene was carefully aspirated and replaced with DMEM containing M-CSF for 6 days to induce Mafb overexpression. For myelin phagocytosis experiments, cells were treated with myelin at a concentration of 10 μg/mL.
MTT assay
MTT test was conducted on BMDMs transduced with Mafb-lentivirus or mCherry-control-lentivirus to assess the cell viability with the influence of lentivirus transduction. In brief, BMDMs transduced with Mafb- or control-lentivirus were placed in a 24-well plate. Then 12 mM MTT stock solution (Thermo Fisher Scientific, M6494) was added to each well and incubated at 37 °C for 5 hours. After that, 50 µL DMSO was added and mixed thoroughly. The absorbance at 540 nm of each well was collected and the cell viability was calculated.
Flow cytometry
To obtain a single-cell suspension of intracerebral immune cells, mice were euthanized and transcardial perfusion was performed using 25 ml of pre-cooled PBS. Left hemispheres were collected after brain dissection. Brain tissues were dissociated into homogenates using the Neural Tissue Dissociation Kit (T) with a gentleMACSTM Octo Dissociator with Heaters (Miltenyi Biotec), following the manufacturer’s instructions. The resulting suspension was passed through a 70 μm cell strainer, and the resulting cells were resuspended in 10 ml of 30% Percoll. Subsequently, 3 ml of 70% Percoll was slowly injected into the bottom of the suspension using a fine needle, while 1 ml of PBS was layered on top of the suspension. Centrifugation (800 g, 30 min, 18 °C) was performed using a density gradient of 30%/70% Percoll to separate myelin sheaths and debris. Cells at the interface, mainly consisting of immune cells, were collected, washed, and resuspended in pre-cooled PBS. For blood cell preparation, mice were deeply anesthetized, and 0.8 ml of peripheral blood was collected from the right atrium. The final 2 ml blood suspension (0.8 ml blood + 1.2 ml PBS) was layered onto 1 ml of Ficoll-Paque (GE, 17–1440-02). Cells at the interface were collected after density gradient centrifugation (800 g, 20 min, 18 °C) and then resuspended with pre-cooled PBS.
The single-cell suspension obtained as described above was incubated with antibodies to surface antigens in the dark for 30 min on ice at 4 °C. For intracellular antigens, the suspension was washed with PBS twice after incubation with surface antigen antibodies. The cells were then fixed and permeabilized using Fixation/Permeabilization Diluent & Concentrate (eBioscience) according to the manufacturer’s instructions, followed by incubation with corresponding intracellular antigen antibodies in the dark for 30 minutes on ice at 4 °C. Fluorochrome compensation was established with single-stained UltraComp eBeads (Thermo Fisher Invitrogen). Flow cytometry was performed using the BD LSRFortessa flow cytometer (BD biosciences) or Beckman CytoFELX IX flow cytometer (Beckman). FlowJo software was used for data analysis. The antibodies used for flow cytometry included: Rat anti-CD45-Pacific blue (Biolegend, 103126), Mouse anti-CD45.1-APC-CY7 (Biolegend, 110715), Rat anti-CD163-APC (Biolegend, 155305), Rat anti-CD206-FITC (Biolegend, 141703), Rat anti-CD11b-PE-CY7 (Biolegend, 101216), Rat anti-Ly6G-PerCP-Cy5.5 (Biolegend, 127616), Rat anti-Gr-1-APC (Biolegend, 108412), Rat anti-CD19-PE (Biolegend, 115508), Mouse anti-NK1.1-BV605 (Biolegend, 108740), Hamster anti-CD3e-FITC (BD, 553061), Rat anti-CD4-APC-CY7 (BD, 552051), Rat anti-CD8a-APC (BD, 553035), Rat anti-CD63−FITC (Biolegend, 143919), Rat anti-CD63−PE(Biolegend, 143903), Rat anti-GPNMB−eFluor660 (Thermo Fisher Scientific, 50-5708-82) and Rat anti-CD115-BV605 (Biolegend, 135517).
For the apoptosis of BMDMs with Mafb- or control lentivirus transduction, Annexin V detection Kit (Biolegend, 640914) was utilized to assess the proportion of apoptosis. In brief, BMDMs with Mafb- or control lentivirus transduction were collected, washed with PBS for 3 times and add 100 μl buffer and 5 μl anti-Annexin V- FITC antibody for per 106 cells for 15 min. Thereafter, 400 μl buffer was added in each tube, and Beckman CytoFELX IX flow cytometer (Beckman) was used to detect the proportion of apoptotic BMDMs.
For the GPNMB and CD63 protein expression of BMDMs with Mafb- or control lentivirus transduction, BMDMs with Mafb- or control lentivirus transduction were collected, washed with PBS for 3 times and add 200 μl PBS each tube. BMDMs were incubated with anti-CD45-Pacific blue (Biolegend, 103126), anti-CD11b-PE-CY7 (Biolegend, 101216), anti-Ly6G-PerCP-Cy5.5 (Biolegend, 127616), anti-CD63−FITC (Biolegend, 143919) and anti-GPNMB−eFluor660 (Thermo Fisher Scientific, 50-5708-82) for 30 minutes in dim place. After that, BMDMs were washed with PBS and centrifuged for 5 minutes (300 g, 4 °C). Beckman CytoFELX IX flow cytometer (Beckman) was used to detect mean fluorescence intensity of GPNMB and CD63 expression on BMDMs with Mafb- or control lentivirus transduction.
Quantitative polymerase chain reaction (qPCR)
Trizol reagent (thermofisher, 15596018) was utilized to obtain mRNA from BMDMs with Mafb- or control lentivirus transduction. Thereafter, cDNA was procured from mRNA with reverse transcription, and EvaGreen (BioRad, USA) was used to perform qPCR. The relative expression of the target gene is quantified as 2 − ΔCt, in which ΔCt is the difference between the mean Ct value of duplicate measurements of the sample and the endogenous Gapdh control. The primer sequences used were as follows:
Mafb-F 5’ AGAAGCGGCGGACCCTGAAG 3’
Mafb-R 5’ GCTGCTCCACCTGCTGAATGAG 3’
Gapdh-F 5’ GGTTGTCTCCTGCGACTTCA 3’
Gapdh-R 5’ TGGTCCAGGGTTTCTTACTCC 3’
RNA sequencing
Total RNA was extracted from BMDMs with Mafb- or control lentivirus transduction using Trizol reagent (thermofisher, 15596018) following the manufacturer’s procedure. The total RNA quantity and purity were analysis using the Bioanalyzer 2100 and RNA 6000 Nano LabChip Kit (Agilent, 5067-1511), high-quality RNA samples with an RIN number > 8.5 were used to construct the sequencing library. After that, mRNA was purified from total RNA (5 μg) using Dynabeads Oligo (dT) (Thermo Fisher) with two rounds of purification. Following purification, the mRNA was fragmented into short fragments using divalent cations under elevated temperature (Magnesium RNA Fragmentation Module (NEB, e6150) under 94 °C 5–7 min). Then the cleaved RNA fragments were reverse-transcribed to create the cDNA by SuperScript™ II Reverse Transcriptase (Invitrogen, 1896649), which were next used to synthesise U-labeled second-stranded DNAs with E. coli DNA polymerase I (NEB, m0209), RNase H (NEB, m0297) and dUTP Solution (Thermo Fisher, R0133). An A-base was then added to the blunt ends of each strand, preparing them for ligation to the indexed adapters. Each adapter contained a T-base overhang for ligating the adapter to the A-tailed fragmented DNA. Dual-index adapters were ligated to the fragments, and size selection was performed with AMPureXP beads. After the heat-labile UDG enzyme (NEB, m0280) treatment of the U-labeled second-stranded DNAs, the ligated products were amplified with PCR by the following conditions: initial denaturation at 95 °C for 3 min; 8 cycles of denaturation at 98 °C for 15 s, annealing at 60 °C for 15 s, and extension at 72 °C for 30 s; and then final extension at 72 °C for 5 min. The average insert size for the final cDNA libraries was 300 ± 50 bp. Finally, we performed the 2 × 150 bp paired-end sequencing (PE150) on an Illumina Novaseq™ 6000 (LC-Bio Technology CO., Ltd., Hangzhou, China) following the vendor’s recommended protocol.
Single cell RNA sequencing
Single cell suspensions were prepared according to our previously published work9,76. In brief, animals were euthanized and subjected to ice-cold saline perfusion at either 5- or 14-days post MCAO. Ischemic hemispheres were collected, excluding meninges, the olfactory bulb, or cerebellum. For the scRNA-seq analysis of the entire brain cells, brain homogenates were created using the Adult Brain Dissociation Kit (T) with a gentle MACS dissociator with heaters (Miltenyi Biotec), following the provided guidelines. In the case of scRNA-seq for CD45high immune cells, brain homogenates were generated using the Neural Tissue Dissociation Kit (T), and CD45high immune cells were isolated through FACS using the Rat anti-CD45-PE-Cy5 antibody (Thermo Fisher Scientific, 15-0451-81). For PBMC specimens, peripheral blood was collected from the right atrium, followed by two rounds of red blood cell lysis (eBioscience, 1X RBC lysis buffer, 00-4333057). Libraries for 10X Genomics were prepared using the 10X Genomics Chromium Single Cell 3' v3 chemistry, in accordance with the manufacturer’s instructions. Subsequently, the mixture containing captured and barcoded mRNAs was retrieved from the Chromium instrument, followed by reverse transcription. The cDNA samples were fragmented and amplified per the 10X protocol. The libraries were then purified, quantified, and sequenced utilizing the Illumina NovaSeq sequencer.
Preprocessing and clustering analysis of single-cell transcriptome data
The fundamental processing and visualization of the scRNA-seq data were carried out using the Seurat package (version 4.3.0) within the R environment (version 4.0.4)77. Initially, a filtration step was employed to exclude low-quality cells and doublets based on the following criteria: (i) cells with a count of expressed genes below 200 or exceeding 6000, and (ii) cells with more than 10% of mitochondrial genes. Genes representing ribosomal contamination (Gm42418, Ay036118) caused technical background noises and were thus removed from the analysis to improve sub-clustering. Subsequently, total expression-based data normalization was performed, followed by a logarithmic transformation. The filtered count matrices underwent further normalization, and mitochondrial contamination was addressed using the SCTransform function. To mitigating batch effects, the Harmony R package (version 0.1.1) was applied78. Principal component analysis was employed for dimensional reduction on the scaled data. Subsequently, clustering was executed using the FindClusters function. To enhance the interpretation of clustering outcomes, non-linear dimensional reduction techniques, including the uniform manifold approximation and projection (UMAP), were employed.
Differentially expressed gene calculation
The Seurat FindAllMarkers and FindMarkers functions were employed to identify differentially expressed genes (DEGs) using the Wilcoxon rank sum test. Specifically, genes with an adjusted Bonferroni-adjusted P-value of less than 0.05 and an absolute log2(fold change) value greater than 0.25 were considered as DEGs.
Gene Set Enrichment Analysis (GSEA)
GSEA (v4.1.0) and MSigDB were used to identify whether a set of genes in specific functional terms shows significant differences in two groups. Briefly, gene expression matrix was input and rank genes by Signal2Noise normalization method. Enrichment scores and p-values were calculated in default parameters. Functional terms meeting these condition with |NES | > 1, NOM p-val< 0.05, FDR q-val< 0.25 were considered to be different between the two groups.
Functional enrichment analysis
The online tool Metascape (http://metascape.org) was employed for functional enrichment analysis79, utilizing the entire mouse genome as the background for enrichment. Initially, a set of DEGs was submitted for analysis, and the enrichment process included a criterion of a minimum count of 3 and an enrichment factor greater than 1.5. Terms with a P-value lower than 0.01 were interpreted as indicating over-represented ontology terms. The activation z-score for each ontology term was determined using the R package GOplot, calculated as (number of upregulated genes - number of downregulated genes) / square root of the number of genes assigned to a term. A term was classified as significantly activated if the z-score was ≥ 2 and the p-value was < 0.01, while a term was classified as significantly inhibited if the z-score was ≤ −2 and the p-value was < 0.01.
Cell phase classification
The CellCycleScoring function in the Seurat R package was employed to classify cells into G2M, S, or G1 phases based on the expression of cell-phase-specific genes.
Pseudotime analysis
Monocle (version 2.26.0) and Monocle3 (version 1.3.1) R packages were employed for pseudotime analysis39. Ordering genes were determined based on the highly variable genes calculated in Seurat. The DDRTree algorithm was employed for dimensional reduction. The cellular trajectory was generated using the order_cells function and the plot_cell_trajectory function. Trajectories were visualized on UMAP coordinates within Monocle3. The pseudo-temporal order of cells was established by selecting the starting point of the cellular trajectory. Gene expression patterns was profiled in this pseudotemporal order using FateID R package (version 0.2.2)48.
Cell-cell interaction analysis
We utilized the InterCellDB toolkit (version 0.9.2), which is specifically designed for the comprehensive analysis of cell-cell interactions within scRNA-seq data54. Initially, DEGs were annotated with information about their cellular localization, functional classification, and their involvement in GO terms. Subsequently, a permutation test was conducted to calculate the confidence level of interaction between gene pairs. The resulting matched gene pairs were visualized using the sankeyplot or dotplot function in R.
Single-cell gene set activity analysis
AUCell R package (version 1.10.0) was employed to assess the state of gene-sets in scRNA-seq data. The functional gene sets were collected based on GO terms and published work9,47,80 (Supplementary Data 1, Supplementary Information files).
Transcription factor enrichment analysis
The online tool Enrichr (https://maayanlab.cloud/Enrichr/) was employed for transcription factor enrichment analysis81. Transcription factors (TFs) were enriched using the top 300 genes from ARCHS4 that are co-expressed with these transcription factors55.
Single-cell regulatory network inference
pySCENIC package (v.0.11.2) was conducted to identify different transcription factors (TFs) and discover regulons82. The mc9nr databases and their corresponding motif annotations were utilized to infer the gene regulatory network within individual cells. Subsequently, a heatmap of normalized regulon AUC was generated using R.
Statistical analysis
Results are presented as mean ± standard deviation (SD). The scRNA-seq data were statistically analyzed using the Wilcoxon rank sum test. Statistical comparison of the means between two groups with equal variances and normal distributions was performed by using the student’s t-test. The differences in means among multiple groups were analyzed using one-way ANOVA. Differences in means across groups with repeated-measurements over time were analyzed using the two-way repeated-measures ANOVA or the mixed-effects model. All statistical analyses were performed with R (version 4.0.4) or GraphPad Prism (version 8.2.1). Statistical significance was defined as P < 0.05. All the data were quantified by 2 independent observers blinded to grouping.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The lipidomics data generated in this study have been deposited in the MetaboLights database83 under accession code MTBLS12240. The RNA-seq data generated in this study have been deposited in the GEO database under accession code GSE290191. The scRNA-seq data generated in this study have been deposited in the GEO database under accession code GSE290194. The scRNA-seq data used in this study from our previously published work are available in the GEO database under accession code GSE171169. The flow cytometry gating strategies in this study are included in Supplementary data 6. All data generated or analyzed in this study, including differentially expressed genes and enriched functional terms, are available within the article and its supplementary information files. Source data are provided with this paper.
Code availability
The code was based on the official tutorials of the packages listed, no custom code was generated.
References
Global, regional, and national burden of stroke and its risk factors, 1990-2019: a systematic analysis for the Global Burden of Disease Study 2019. Lancet. Neurol. 20, 795–820 (2021).
Tsao, C. W. et al. Heart Disease and Stroke Statistics-2022 Update: A Report From the American Heart Association. Circulation 145, e153–e639 (2022).
Shichita, T., Ooboshi, H. & Yoshimura, A. Neuroimmune mechanisms and therapies mediating post-ischaemic brain injury and repair. Nat. Rev. Neurosci. 24, 299–312 (2023).
Brown, G. C. & Neher, J. J. Microglial phagocytosis of live neurons. Nat. Rev. Neurosci. 15, 209–216 (2014).
Liesz, A. et al. The spectrum of systemic immune alterations after murine focal ischemia: immunodepression versus immunomodulation. Stroke 40, 2849–2858 (2009).
Shichita, T. et al. Pivotal role of cerebral interleukin-17-producing gammadeltaT cells in the delayed phase of ischemic brain injury. Nat. Med 15, 946–950 (2009).
Clarkson, B. D. et al. T cell-derived interleukin (IL)-21 promotes brain injury following stroke in mice. J. Exp. Med 211, 595–604 (2014).
Garcia-Bonilla, L. et al. Spatio-temporal profile, phenotypic diversity, and fate of recruited monocytes into the post-ischemic brain. J. Neuroinflamm. 13, 285 (2016).
Shi, L. et al. Treg cell-derived osteopontin promotes microglia-mediated white matter repair after ischemic stroke. Immunity 54, 1527–1542.e1528 (2021).
Unger, M. S., Schernthaner, P., Marschallinger, J., Mrowetz, H. & Aigner, L. Microglia prevent peripheral immune cell invasion and promote an anti-inflammatory environment in the brain of APP-PS1 transgenic mice. J. Neuroinflamm. 15, 274 (2018).
Shi, Z. et al. Microglia drive transient insult-induced brain injury by chemotactic recruitment of CD8+ T lymphocytes. Neuron 111, 696–710.e699 (2023).
Wang, Y.-R., Cui, W.-Q., Wu, H.-Y., Xu, X.-D. & Xu, X.-Q. The role of T cells in acute ischemic stroke. Brain Res. Bull. 196, 20–33 (2023).
Elmore, M. R. et al. Colony-stimulating factor 1 receptor signaling is necessary for microglia viability, unmasking a microglia progenitor cell in the adult brain. Neuron 82, 380–397 (2014).
Beuker, C. et al. Stroke induces disease-specific myeloid cells in the brain parenchyma and pia. Nat. Commun. 13, 945 (2022).
Spangenberg, E. E. et al. Eliminating microglia in Alzheimer’s mice prevents neuronal loss without modulating amyloid-β pathology. Brain 139, 1265–1281 (2016).
Nissen, J. C., Thompson, K. K., West, B. L. & Tsirka, S. E. Csf1R inhibition attenuates experimental autoimmune encephalomyelitis and promotes recovery. Exp. Neurol. 307, 24–36 (2018).
Spiteri, A. G. et al. PLX5622 Reduces Disease Severity in Lethal CNS Infection by Off-Target Inhibition of Peripheral Inflammatory Monocyte Production. Front Immunol. 13, 851556 (2022).
Wang Y, et al. Early posttraumatic CSF1R inhibition via PLX3397 leads to time- and sex-dependent effects on inflammation and neuronal maintenance after traumatic brain injury in mice. Brain Behav Immun 106, 49–66 (2022).
Szalay, G. et al. Microglia protect against brain injury and their selective elimination dysregulates neuronal network activity after stroke. Nat. Commun. 7, 11499 (2016).
Otxoa-de-Amezaga, A. et al. Microglial cell loss after ischemic stroke favors brain neutrophil accumulation. Acta Neuropathol. 137, 321–341 (2019).
Marino Lee, S., Hudobenko, J., McCullough, L. D. & Chauhan, A. Microglia depletion increase brain injury after acute ischemic stroke in aged mice. Exp. Neurol. 336, 113530 (2021).
Hamner, M. A. et al. Microglial depletion abolishes ischemic preconditioning in white matter. Glia 70, 661–674 (2022).
Bruttger, J. et al. Genetic Cell Ablation Reveals Clusters of Local Self-Renewing Microglia in the Mammalian Central Nervous System. Immunity 43, 92–106 (2015).
Willis, E. F. et al. Repopulating Microglia Promote Brain Repair in an IL-6-Dependent Manner. Cell 180, 833–846.e816 (2020).
Guilliams, M. & Scott, C. L. Does niche competition determine the origin of tissue-resident macrophages? Nat. Rev. Immunol. 17, 451–460 (2017).
Huang, Y. et al. Repopulated microglia are solely derived from the proliferation of residual microglia after acute depletion. Nat. Neurosci. 21, 530–540 (2018).
Cronk, J. C. et al. Peripherally derived macrophages can engraft the brain independent of irradiation and maintain an identity distinct from microglia. J. Exp. Med. 215, 1627–1647 (2018).
Lund, H. et al. Competitive repopulation of an empty microglial niche yields functionally distinct subsets of microglia-like cells. Nat. Commun. 9, 4845 (2018).
Park, K. W. et al. Delayed Infiltration of Peripheral Monocyte Contributes to Phagocytosis and Transneuronal Degeneration in Chronic Stroke. Stroke 53, 2377–2388 (2022).
Ritzel, R. M. et al. Functional differences between microglia and monocytes after ischemic stroke. J. Neuroinflammation 12, 106 (2015).
Lei, F. et al. CSF1R inhibition by a small-molecule inhibitor is not microglia specific; affecting hematopoiesis and the function of macrophages. Proc. Natl Acad. Sci. USA 117, 23336–23338 (2020).
Chu, M. et al. Focal cerebral ischemia activates neurovascular restorative dynamics in mouse brain. Front Biosci. (Elite Ed.) 4, 1926–1936 (2012).
Çolakoğlu, G., Bergstrom-Tyrberg, U., Berglund, E. O. & Ranscht, B. Contactin-1 regulates myelination and nodal/paranodal domain organization in the central nervous system. Proc. Natl Acad. Sci. USA 111, E394–E403 (2014).
Marques, S. et al. Oligodendrocyte heterogeneity in the mouse juvenile and adult central nervous system. Science 352, 1326–1329 (2016).
Santos, E. N. & Fields, R. D. Regulation of myelination by microglia. Sci. Adv. 7, eabk1131 (2021).
Najafi, A. R. et al. A limited capacity for microglial repopulation in the adult brain. Glia 66, 2385–2396 (2018).
Silvin, A. et al. Dual ontogeny of disease-associated microglia and disease inflammatory macrophages in aging and neurodegeneration. Immunity 55, 1448–1465.e1446 (2022).
Garcia-Bonilla, L. et al. Analysis of brain and blood single-cell transcriptomics in acute and subacute phases after experimental stroke. Nat. Immunol. 25, 357–370 (2024).
Cao, J. et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature 566, 496–502 (2019).
Nakano, Y. et al. Glycoprotein nonmetastatic melanoma protein B (GPNMB) as a novel neuroprotective factor in cerebral ischemia-reperfusion injury. Neuroscience 277, 123–131 (2014).
Liu, A. et al. Macrophage-derived small extracellular vesicles promote biomimetic mineralized collagen-mediated endogenous bone regeneration. Int. J. Oral. Sci. 12, 33 (2020).
Gouna G, et al. TREM2-dependent lipid droplet biogenesis in phagocytes is required for remyelination. Journal of Experimental Medicine 218, e20210227 (2021).
Wlodarczyk, A. et al. A novel microglial subset plays a key role in myelinogenesis in developing brain. Embo J. 36, 3292–3308 (2017).
Cochain, C. et al. Single-Cell RNA-Seq Reveals the Transcriptional Landscape and Heterogeneity of Aortic Macrophages in Murine Atherosclerosis. Circ. Res. 122, 1661–1674 (2018).
Franklin, R. & Simons, M. CNS remyelination and inflammation: From basic mechanisms to therapeutic opportunities. Neuron 110, 3549–3565 (2022).
Berghoff, S. A., Spieth, L. & Saher, G. Local cholesterol metabolism orchestrates remyelination. Trends Neurosci. 45, 272–283 (2022).
Berghoff, S. A. et al. Microglia facilitate repair of demyelinated lesions via post-squalene sterol synthesis. Nat. Neurosci. 24, 47–60 (2021).
Herman, J. S., Sagar & Grün, D. FateID infers cell fate bias in multipotent progenitors from single-cell RNA-seq data. Nat. Methods 15, 379–386 (2018).
Cignarella, F. et al. TREM2 activation on microglia promotes myelin debris clearance and remyelination in a model of multiple sclerosis. Acta Neuropathologica 140, 513–534 (2020).
Haidar, M. et al. Targeting lipophagy in macrophages improves repair in multiple sclerosis. Autophagy 18, 2697–2710 (2022).
Pedragosa, J. et al. CCR2 deficiency in monocytes impairs angiogenesis and functional recovery after ischemic stroke in mice. J. Cereb. Blood Flow. Metab. 40, S98–S116 (2020).
Garcia, F. J. et al. Single-cell dissection of the human brain vasculature. Nature 603, 893–899 (2022).
Vanlandewijck M et al. A molecular atlas of cell types and zonation in the brain vasculature. Nature 554, 475–480 (2018).
Jin Z, et al. InterCellDB: A User-Defined Database for Inferring Intercellular Networks. Advanced Science 9, 2200045 (2022).
Lachmann, A. et al. Massive mining of publicly available RNA-seq data from human and mouse. Nat. Commun. 9, 1366 (2018).
Arbaizar-Rovirosa, M. et al. Aged lipid-laden microglia display impaired responses to stroke. EMBO Mol. Med 15, e17175 (2023).
Wang, W. et al. Microglial repopulation reverses cognitive and synaptic deficits in an Alzheimer’s disease model by restoring BDNF signaling. Brain Behav. Immun. 113, 275–288 (2023).
Elmore, M. R. P. et al. Replacement of microglia in the aged brain reverses cognitive, synaptic, and neuronal deficits in mice. Aging Cell 17, e12832 (2018).
Fang, W. et al. CCR2-dependent monocytes/macrophages exacerbate acute brain injury but promote functional recovery after ischemic stroke in mice. Theranostics 8, 3530–3543 (2018).
Vanherle, S. et al. The ApoA-I mimetic peptide 5A enhances remyelination by promoting clearance and degradation of myelin debris. Cell Rep. 41, 111591 (2022).
Zbesko, J. C., Stokes, J., Becktel, D. A. & Doyle, K. P. Targeting foam cell formation to improve recovery from ischemic stroke. Neurobiol. Dis. 181, 106130 (2023).
Choi, B. R., Johnson, K. R., Maric, D. & McGavern, D. B. Monocyte-derived IL-6 programs microglia to rebuild damaged brain vasculature. Nat. Immunol. 24, 1110–1123 (2023).
Profaci, C. P. et al. Microglia are not necessary for maintenance of blood-brain barrier properties in health, but PLX5622 alters brain endothelial cholesterol metabolism. Neuron 112, 2910–2921.e2917 (2024).
Yan, H. et al. Role of Polymorphonuclear Myeloid-Derived Suppressor Cells and Neutrophils in Ischemic Stroke. J. Am. Heart Assoc. 12, e028125 (2023).
Gertz, K. et al. Essential role of interleukin-6 in post-stroke angiogenesis. Brain 135, 1964–1980 (2012).
Shaul, M. E. & Fridlender, Z. G. The dual role of neutrophils in cancer. Semin Immunol. 57, 101582 (2021).
Soucie, E. et al. Lineage-specific enhancers activate self-renewal genes in macrophages and embryonic stem cells. Sci. (N. Y., NY) 351, aad5510 (2016).
Vanneste, D. et al. MafB-restricted local monocyte proliferation precedes lung interstitial macrophage differentiation. Nat. Immunol. 24, 827–840 (2023).
Holtman, I. R., Skola, D. & Glass, C. K. Transcriptional control of microglia phenotypes in health and disease. J. Clin. Invest 127, 3220–3229 (2017).
Shichita, T. et al. MAFB prevents excess inflammation after ischemic stroke by accelerating clearance of damage signals through MSR1. Nat. Med 23, 723–732 (2017).
Johnson, N. R. et al. CSF1R inhibitors induce a sex-specific resilient microglial phenotype and functional rescue in a tauopathy mouse model. Nat. Commun. 14, 118 (2023).
Zhang, Q. et al. The interleukin-4/PPARγ signaling axis promotes oligodendrocyte differentiation and remyelination after brain injury. PLOS Biol. 17, e3000330 (2019).
Yushkevich, P. A. et al. User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. Neuroimage 31, 1116–1128 (2006).
Erwig, M. S. et al. Myelin: Methods for Purification and Proteome Analysis. In: Oligodendrocytes: Methods and Protocols (eds Lyons D. A., Kegel L.). Springer New York (2019).
Nguyen, T., Du, J. & Li, Y. C. A protocol for macrophage depletion and reconstitution in a mouse model of sepsis. STAR Protoc. 2, 101004 (2021).
Jin, C. et al. Leveraging single-cell RNA sequencing to unravel the impact of aging on stroke recovery mechanisms in mice. Proc. Natl Acad. Sci. USA 120, e2300012120 (2023).
Hao, Y. et al. Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e3529 (2021).
Korsunsky, I. et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 16, 1289–1296 (2019).
Zhou, Y. et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 10, 1523 (2019).
Shi, L. et al. Genome-wide transcriptomic analysis of microglia reveals impaired responses in aged mice after cerebral ischemia. J. Cereb. Blood Flow. Metab. 40, S49–S66 (2020).
Kuleshov, M. V. et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res 44, W90–W97 (2016).
Aibar, S. et al. SCENIC: single-cell regulatory network inference and clustering. Nat. Methods 14, 1083–1086 (2017).
Yurekten, O. et al. MetaboLights: open data repository for metabolomics. Nucleic Acids Res 52, D640–D646 (2024).
Acknowledgements
This work was supported by grants from National Natural Science Foundation of China (82471296, 82322022 and 82001460 to L.S.), and Natural Science Foundation of Zhejiang Province (LQ21H250001 and LR23H250001 to L.S.). Thank Siyuan Shen and Zhuangchou Han etc from LC Bio Technology CO., Ltd for assisting in bioinformatics analysis of RNA-seq.
Author information
Authors and Affiliations
Contributions
X.Z., H.L. and Y.G. contributed equally to this work. L.S. and J.Z. designed the research. X.Z., H.L., Y.G., A.P., J.C., Q.Z., Z.X., J.W., S.T., R.W., J.L., L.L. and L.S. performed the experiments. X.Z., C.J., and Z.J. contributed to bioinformatics and statistical analysis. X.Z. wrote the original manuscript. H.L., Y.G., and L.S. reviewed and polished the manuscript. C.J. improved the Figures.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Lidia Garcia-Bonilla, Anna Planas, and the other anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhang, X., Li, H., Gu, Y. et al. Repair-associated macrophages increase after early-phase microglia attenuation to promote ischemic stroke recovery. Nat Commun 16, 3089 (2025). https://doi.org/10.1038/s41467-025-58254-y
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-58254-y
This article is cited by
-
Neuroglia and immune cells play different roles in neuroinflammation and neuroimmune response in post-stroke neural injury and repair
Acta Pharmacologica Sinica (2026)
-
Engineered non-expansive matrix for blood vessel organoid development and ischaemic stroke repair
Nature Biomedical Engineering (2025)