Abstract
Phytochrome A (phyA) and phyB are red and far-red photoreceptors that interact with PHYTOCHROME-INTERACTING FACTORs (PIFs) via active phyA-binding (APA) or active phyB-binding (APB) motifs. While APB interacts with the N-terminal photosensory module of phyB (phyBPSM), it remains unclear whether APA interacts with phyAPSM. We report that both phyA and phyB interact with APA through C-terminal output module of phy (phyOPM), while phyB interacts additionally with APB through phyBPSM. Marchantia Mp-phy also interacts with PIFs via the phyOPM-APA interaction. The phyBOPM-APA interaction promotes PIF3 degradation but not mutual phyB destruction. The full-length phy-APA interaction is light-dependent, whereas the underlying phyOPM-APA interaction is not. We show that the Pr form, not the Pfr, of phyPSM competes with APA for phyOPM binding, explaining how the light-dependent phy-APA interaction arises from the light-independent phyOPM-APA interaction. Together, our results suggest that the phyOPM-APA interaction is an ancient feature conserved in both Arabidopsis phyA, phyB and Marchantia Mp-phy.
Similar content being viewed by others
Introduction
Plant phytochrome (phy) is a red and far-red photoreceptor consisting of three clades in seed plants: phyA/N, phyB/P, and phyC/O1. These clades share a canonical domain structure with related phy clades in bryophytes and charophytes2. Different plant species possess varying numbers of phys: Marchantia (Marchantia polymorpha) has one (Mp-phy)3, Physcomitrium (Physcomitrium patens) has seven (Pp-phy1/3, -phy2/4, -phy5a/b/c)4, Arabidopsis (Arabidopsis thaliana) has five (phyA, phyB/D/E, phyC)5, and rice (Oryza sativa) has three (Os-phyA, -phyB, -phyC)6. All phys undergo a reversible photoconversion between two spectral forms, red light-absorbing Pr and far-red light-absorbing Pfr7. Though undergoing the same photoconversion, phys exhibit different light specificities. In Arabidopsis, five phys regulate a wide range of light responses: phyA controls light responses in response to very low light fluence of light and prolonged far-red light, while phyB controls light responses in response to low fluence of red light8,9. In bryophytes, a single phy regulates both red light and prolonged far-red light responses in Marchantia3,10, wheras in Physcomitrium, seven phys regulate either red-light responses (phy4, phy5a/b/c), or prolonged far-red responses (phy1, phy2, phy3, phy4)11. This suggests that the functional diversification of phys into distinct red-light and prolonged far-red-light phys evolved independently in seed plants and Physcomitrium, likely from an ancestral phy that was not yet functionally specialized.
Phy is a dimeric protein with a monomer composed of an N-terminal photosensory module (phyPSM) and a C-terminal output module (phyOPM)12. PhyPSM consists of the N-terminal extension (NTE), N-terminal Period-Arnt-SIM (nPAS), cGMP phosphodiesterase/adenylyl cyclase/FhlA (GAF), and PHY domains, while phyOPM consists of a PAS-related domain (PRD: PAS1 and PAS2 connected by a modulator loop) and a histidine kinase-related domain (HKRD: DHp and CA). The chromophore phytochromobilin (PΦB) covalently attaches to a conserved cysteine in the GAF domain and is pocketed by residues from the nPAS, GAF, and PHY domains13,14,15. The Pr form of phy dimerizes through the interaction between DHp of two protomers and further through the interaction between PAS2 of a protomer and nPAS-GAF of the other protomer16,17,18. The phyPSM-PAS2 of two protomers align head-to-tail and form a flat platform with 2-fold rotational symmetry, while the HKRD dimer protrudes below16,17,18. In the Pfr form of phyB, however, phyBPSM of two protomers dimerize head-to-head via alpha-helices in the GAF domain, while phyBOPM dissociates from phyBPSM and becomes more flexible19.
Despite their overall sequence and structural similarities, phyA and phyB have been shown to bind to different conserved amino acid motifs in PIFs: Active Phytochrome A-binding (APA) and Active Phytochrome B-binding (APB). Mutations of APB sequences abolish the binding of PIFs to the Pfr form of phyB, while a PIF3 fragment containing the APB binds specifically to the Pfr of phyB but not to other phys20, confirming that the APB is the binding motif for phyB. PhyBPSM has been shown to be sufficient for binding to PIFs and promoting light responses21,22, further supported by point mutations in phyBPSM that disrupt PIF3 binding disable phyBPSM’s ability to promote light responses. Similarly, mutations of APA sequences disrupt the binding of PIFs to the Pfr of phyA23,24. These findings led to a hypothesis that phyB binds to PIFs through the phyBPSM-APB interaction, while phyA binds through the phyAPSM-APA interaction. This dichotomy in phy-PIF interaction, however, fails to explain certain experimental findings. First, PIF3 was originally discovered as a protein interacting with phyBOPM25. Second, point mutations in either phyAOPM or phyBOPM disrupted their binding to PIF325,26. Third, expression of phyBOPM alone caused PIF3 degradation in transgenic plants27. These findings suggest that the idea of phys binding to PIFs exclusively through phyAPSM-APA or phyBPSM-APB interactions needs reevaluation.
To facilitate the interaction assay between phys and PIFs, we engineered yeast to produce phycocyanobilin (PCB) and used this strain in a yeast two-hybrid (Y2H) assay to test light-dependent interactions. We report that the APA motif interacts with both Arabidopsis phyAOPM and phyBOPM, as well as Mp-phyOPM, while the APB motif interacts with the phyBPSM. Additionally, despite the APA’s interaction with a non-photosensory domain, the interaction between the full-length phy and the APA is light-dependent because the Pr form of the phyPSM binds to phyOPM and inhibits the binding of the APA to phyOPM in the dark.
Result
The Y2H assay captures light-dependent interactions between phyB and its interacting proteins in yeast expressing phycocyanobilin biosynthetic genes
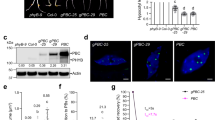
To enable light-dependent interaction assays in the yeast two-hybrid system (Y2H) without the need for exogenous chromophore, we engineered a commonly used AH109 yeast strain to produce phycocyanobilin (PCB) de novo by expressing four biosynthetic genes: HO1, PcyA, FD, and FNR (Supplementary Fig. 1a). This strategy has previously been employed to successfully induce PCB production in mammalian cells28. This modified strain was named AH109C. The AH109C emitted red fluorescence when expressing phyBPSM with a Y276H mutation (phyBPSM/Y276H)29, while the unmodified AH109 did not (Supplementary Fig. 1b), indicating the formation of phyBPSM/Y276H holoprotein in AH109C. The partially purified phyBPSM from AH109C also displayed characteristic PCB-containing Pr and Pfr absorption spectra, peaking at 650 nm for Pr and 710 nm for Pfr (Supplementary Fig. 1c). These results demonstrate that the AH109C strain produces sufficient PCB to support the formation of spectrally active phy.
We tested whether the engineered AH109C strain could be used for the Y2H to assay light-dependent interactions between phy and its interacting proteins. AH109C harboring both phyBPSM and PIF3 grew well on selective media under red light but not in the dark (Fig. 1a), while AH109C with phyBPSM and an empty vector did not grow under any light conditions. This supports that the Y2H assay successfully captured the light-dependent interaction between phyBPSM and PIF3 in AH109C. Similarly, AH109C harboring full-length phyB and other interacting proteins such as SPA1, SPA2, ELF3, and TZP also grew well on selective media under red light but not in the dark (Fig. 1b). These results demonstrate that AH109C can be effectively used in Y2H assays to detect light-dependent interactions between phy and its interacting proteins without supplementing the exogenous chromophore.
a Y2H showing red light-dependent interaction between phyBPSM and PIF3. Y2H was performed to observe the red light-dependent interaction between the N-terminal photosensory module of phytochrome B (phyBPSM, amino acids 1-652) and PIF3. Serial dilutions of AH109C yeast cells harboring a GAL4 DNA binding domain (BD)-fused phyBPSM (BD-phyBPSM) with either an empty GAL4 activation domain (AD) vector (AD) or an AD-fused PIF3 (AD-PIF3) were plated on non-selective agar plates lacking leucine and tryptophan (-LW) and selective plates lacking leucine, tryptophan, and histidine (-LWH). The plates were incubated either in the dark (Dc) or under red light (Rc, 15 μmol m−2 s−1). OD600 of 1 was serially diluted (4-folds each). b Red light-dependent interaction between full-length phyB and interacting proteins. The interaction between full-length phyB and its interacting proteins under red light was tested using AH109C cells expressing AD-fused full-length phyB (AD-phyB) and BD-fused interacting proteins (SPA1, SPA2, ELF3, TZP).
PhyA interacts with the APA motif through its C-terminal output module
We used the Y2H assay to investigate which domain of phyA interacts with which motif of PIF3. Full-length phyA interacted with both wild-type PIF3 and APB-mutated PIF3 (PIF3mAPB) under red light, but not in the dark, while phyA did not interact with APA-mutated PIF3 (PIF3mAPA) or the double mutant (PIF3mAPA/mAPB) under any light conditions, supporting that phyA interacts with PIF3 through the APA. Interestingly, phyAPSM did not interact with wild-type PIF3 or any of the mutated forms (PIF3mAPA, PIF3mAPB, PIF3mAPA/mAPB) regardless of light conditions. The lack of interaction was not due to insufficient phyAPSM or PIF3 protein expression in yeast (Supplementary Fig. 2). In contrast, phyAOPM interacted with both PIF3 and PIF3mAPB but failed to interact with PIF3mAPA or PIF3mAPA/mAPB (Fig. 2a). Similarly, phyAOPM, but not phyAPSM, interacted with PIF1 through the APA (Supplementary Fig. 3a). These results suggest that phyA interacts with both PIF1 and PIF3 via the phyAOPM-APA interaction.
a Y2H assay showing the interaction between phyAOPM and the APA motif of PIF3. BD-fused full-length phyA, phyAPSM (1-615 a.a.), or phyAOPM (608-1122 a.a.) were transformed into AH109C with AD-fused PIF3, PIF3mAPA, PIF3mAPB, or PIF3mAPA/mAPB. Transformants were spotted on non-selective (-LW, lacking leucine and tryptophan) and selective (-LWH, additionally lacking histidine) media, then grown in the dark (Dc) or under red light (Rc). The additional information on AH109C is provided in Supplementary Figs. 9, 11. b–d In vitro binding assay showing the light-dependent interaction between phyA and the APA motif of PIF3. GST-fused PIF3 proteins were used to pull down either SBP-fused full-length phyA (b), phyAPSM (c), or MBP-fused phyAOPM (d). PhyA and phyAPSM were irradiated with either red light pulses (15 μmol m−2s−1, 5 min, Pfr) or far-red light pulses (2.5 μmol m−2s−1, 5 min, Pr) before mixing with GST-PIF3. The pulled-down proteins were separated by SDS-PAGE and detected with anti-SBP (phyA, phyAPSM), anti-MBP (phyAOPM), or anti-GST (GST-PIF3, GST) antibodies. GST control images were taken from the lower part of the same immunoblot with the respective GST-fused PIF3 bands and demarcated by vertical lines. All in vitro binding assays were independently repeated at least twice with consistent results.
We further conducted in vitro binding assays using recombinant phyA and PIF3 proteins. Consistent with the Y2H results, the Pfr of full-length phyA preferentially bound to both PIF3 and PIF3mAPB, while it did not bind to PIF3mAPA or PIF3mAPA/mAPB (Fig. 2b). Among phyA domains, phyAPSM did not bind to PIF3 or any mutant forms (Fig. 2c). In contrast, phyAOPM bound to both PIF3 and PIF3mAPB but failed to bind to PIF3mAPA or PIF3mAPA/mAPB (Fig. 2d). Similarly, phyAOPM bound to PIF1 through the APA (Supplementary Fig. 3b). Together, both the Y2H and in vitro binding assays show that phyA interacts with PIF1 and PIF3 in a light-dependent manner through the phyAOPM-APA interaction. This is enigmatic because the light-dependent phyA-PIF interaction arises from the light-independent phyAOPM-APA interaction.
PhyB interacts with the APA motif through its C-terminal output module and the APB motif through its N-terminal photosensory module
We used the Y2H assay to determine if phyBOPM also interacts with the APA, given that phyBOPM and phyAOPM are functionally interchangeable30,31. Interestingly, full-length phyB interacted with PIF3 as well as with PIF3mAPA and PIF3mAPB under red light, but not in the dark. However, phyB did not interact with PIF3mAPA/mAPB under any light condition. Domain analysis showed that phyBPSM interacted with both PIF3 and PIF3mAPA under red light, but not with PIF3mAPB or PIF3mAPA/mAPB, confirming that phyBPSM interacts with PIF3 through the APB. In contrast, phyBOPM interacted with both PIF3 and PIF3mAPB but not with PIF3mAPA or PIF3mAPA/mAPB (Fig. 3a). Similarly, phyBPSM interacted with PIF1 via the APB, while phyBOPM interacted with PIF1 via the APA (Supplementary Fig. 4a). These results indicate that phyB interacts with PIF1 and PIF3 through both the phyBPSM-APB and phyBOPM-APA interactions.
a Y2H assay showing the interaction between phyB and PIF3 via either the phyBPSM-APB or phyBOPM-APA interaction. BD-fused full-length phyB, phyBPSM (1-652 a.a.), or phyBOPM (642-1172 a.a.) were transformed into AH109C along with AD-fused PIF3, PIF3mAPA, PIF3mAPB, or PIF3mAPA/mAPB. b–d In vitro binding assay demonstrating the interaction between phyB and PIF3 via either the phyBPSM-APB or phyBOPM-APA interaction. GST-fused PIF3 proteins were used to pull down either SBP-fused full-length phyB (b), phyBPSM (c), or MBP-fused phyBOPM (d). e, f In vitro binding assay showing the interaction between phyB domains and PIF3 fragments containing either APA or APB. GST-fused truncated PIF3 fragments containing only APA (86-221 a.a.), only APB (1-113 a.a.), or their mutant versions (mAPA, mAPB) were used to pull down SBP-fused phyBPSM (e) or MBP-fused phyBOPM (f). g, h Semi in vivo binding assay showing the phyBOPM-APA interaction. Transgenic seedling extracts expressing mScarlet-fused phyBOPM were pulled down by GST-fused recombinant full-length PIF3 proteins (g) or PIF3 fragments containing either APA, APB, or their mutant versions (mAPA, mAPB) (h). mScarlet-fused phyBOPM proteins were detected with anti-mCherry antibody. Notations are consistent with those in Fig. 2. All in vitro binding assays were independently repeated at least twice with consistent results.
We further performed in vitro binding assays with recombinant phyB and PIF3 proteins. The Pfr form of phyB preferentially bound to PIF3, as well as to both PIF3mAPA and PIF3mAPB, but did not bind to PIF3mAPA/mAPB (Fig. 3b). This confirms that phyB can bind to PIF3 through either the APA or the APB. Among phyB domains, the Pfr of phyBPSM bound to both PIF3 and PIF3mAPA but not to PIF3mAPB or PIF3mAPA/mAPB (Fig. 3c), indicating phyBPSM interacts with PIF3 through the APB. Conversely, phyBOPM bound more strongly to both PIF3 and PIF3mAPB than to PIF3mAPA or PIF3mAPA/mAPB (Fig. 3d), indicating that phyBOPM interacts with PIF3 through the APA. Similarly, phyBOPM bound to PIF1 via the APA (Supplementary Fig. 4b). Another in vitro binding assay with PIF3 fragments containing only APB (1-113 a.a.) or APA (86-221 a.a.) indicate APB and APA are sufficient to bind to phyBPSM and phyBOPM, respectively (Fig. 3e, f). Full-length phyB interacted more strongly with PIF3 than either phyBPSM32 or phyBOPM (Supplementary Fig. 5a) and the phyBPSM-APB interaction was approximately twice as strong as the phyBOPM-APA interaction (Supplementary Fig. 5b). A semi in vivo binding assay further demonstrated that transgenic phyBOPM was pulled down by both recombinant PIF3 and PIF3mAPB but not by PIF3mAPA or PIF3mAPA/mAPB (Fig. 3g). Similarly, transgenic phyBOPM was pulled down by the APA fragment but not by the mAPA or APB fragments (Fig. 3h). Together, the results indicate that phyB interacts with PIF1 and PIF3 through both the phyBPSM-APB and phyBOPM-APA interactions.
The phyBOPM-APA interaction promotes PIF3 degradation but does not trigger phyB destruction
We investigated whether the phyBOPM-APA interaction also triggers the degradation of PIF3, similar to the phyPSM-APB interaction23. We generated transgenic lines expressing PIF3 mutant alleles in a phyA mutant background to exclude the role of phyA and assessed PIF3 degradation under red light. Red light rapidly promoted the degradation of PIF3, PIF3mAPA, and PIF3mAPB, but not PIF3mAPA/mAPB (Fig. 4a and Supplementary Fig. 6a). This supports that phyB promotes PIF3 degradation via both the phyBPSM-APB and phyBOPM-APA interactions. Next, we tested if the phyBOPM-APA interaction induces the mutual destruction of phyB and PIF3, as seen with the phyBPSM-APB interaction33. We generated transgenic lines expressing PIF3 mutant alleles in a 35S:PHYB-GFP/phyb-9 background and analyzed the degradation of both transgenic phyB and PIF3 under red light. While some endogenous PIF3 must be present, the degradation would be mainly influenced by the highly expressed transgenic PIF3 proteins. Red light induced phyB degradation when co-expressed with PIF3 or PIF3mAPA, but not when co-expressed with PIF3mAPB or PIF3mAPA/mAPB. However, under the same condition, the degradation of PIF3, PIF3mAPA and PIF3mAPB still occurred, indicating that phyB destruction, but not PIF3 degradation, was abolished when the APB is mutated (Fig. 4b and Supplementary Fig. 6b). Together, the phyBOPM-APA interaction promotes PIF3 degradation but does not induce phyB destruction, whereas the phyBPSM-APB interaction leads to mutual destruction of both phyB and PIF3.
a Immunoblot assay showing red light-induced degradation of PIF3 proteins in the phyA-211 mutant background. Four-day-old dark-grown transgenic seedlings expressing MYC-tagged PIF3 alleles were exposed to red light for the indicated times (hours in Rc), and PIF3 protein levels were analyzed by immunoblot using anti-MYC antibody. α-Tubulin (Tub) was determined using anti-TUB antibody. Numbers indicate relative PIF3 protein levels. Immunoblot assays with independent transgenic lines are shown in Supplementary Fig. 6a. b Immunoblot assay showing the mutual destruction of phyB and PIF3 proteins. Seedlings expressing both GFP-tagged phyB and MYC-tagged PIF3 alleles were grown in darkness for 4 days, then either kept in the dark for 12 h (D) or transferred to red light for 12 h (R). PhyB and PIF3 protein levels were analyzed using anti-GFP antibody for phyB and anti-MYC antibody for PIF3. Transgenic lines expressing PIF3 alleles were created in the 35S:PHYB-GFP/phyB-9 mutant background. Numbers indicate relative phyB and PIF3 protein levels. Immunoblot assays with independent transgenic lines are shown in Supplementary Fig. 6b.
Marchantia Mp-phy interacts with the APA motif also through its C-terminal output module
The phyOPM-APA interaction may have evolved before the divergence of angiosperm phyA and phyB, as suggested by the fact that bryophyte phytochromes, such as those in Marchantia (Mp-phy) and Physcomitrium (Pp-phys), also interact with their PIFs through the APA3,34. To explore this possibility, we performed Y2H assay with Mp-phy and Mp-PIF. Consistent with a previous report, Mp-phy interacted with Mp-PIF under red light but not in the dark, and it failed to interact with Mp-PIFmAPA. Among Mp-phy domains, Mp-phyPSM did not interact with Mp-PIF under any light conditions, while Mp-phyOPM interacted with Mp-PIF but not Mp-PIFmAPA (Fig. 5a). These results indicate that Mp-phy, like Arabidopsis phyA, interacts with Mp-PIF via the Mp-phyOPM-APA interaction.
a, b Y2H assay showing the interaction between Mp-phy and PIFs via the Mp-phyOPM-APA interaction. BD-fused full-length Mp-phy, Mp-phyPSM (1-610 a.a.), or Mp-phyOPM (603-1126 a.a.) were transformed into AH109C with AD-fused Mp-PIF or MpPIFmAPA (a), or with AD-fused PIF3, PIF3mAPA, PIF3mAPB, or PIF3mAPA/mAPB (b). c, d Y2H assay showing the interaction between phyA and Mp-PIF (c) or phyB and Mp-PIF (d) via the phyOPM-APA interaction. BD-fused full-length phyA, phyAPSM, phyOPM, phyB, phyBPSM, or phyBOPM were transformed into AH109C with AD-fused Mp-PIF or MpPIFmAPA. Notations are consistent with those in Fig. 2a.
If the phyOPM-APA interaction originated in an ancestral phy and inherited to land plant phys, Mp-phyOPM and Arabidopsis phyOPM might still be capable of binding to each other’s PIFs via the APA. Supporting this, Mp-phy interacted with both Arabidopsis PIF3 and PIF3mAPB under red light but not in the dark, and failed to interact with PIF3mAPA or PIF3mAPA/mAPB. Similarly, Mp-phy interacted with PIF1 through the APA. Furthermore, while Mp-phyPSM did not interact with either PIF1 or PIF3 regardless of light conditions, Mp-phyOPM interacted with both PIF1 and PIF3 as long as their APA motifs were intact (Fig. 5b and Supplementary Fig. 7). Reciprocally, both Arabidopsis phyA and phyB interacted with Mp-PIF under red light but not in the dark, and neither interacted with Mp-PIFmAPA. Of domains, both phyAOPM and phyBOPM interacted with Mp-PIF through the APA (Fig. 5c, d). These results demonstrate that the phyOPM-APA interaction is conserved in the bryophyte Marchantia Mp-phy and the angiosperm Arabidopsis phyA and phyB.
The Pr form of the N-terminal photosensory module competes with the APA for binding to the C-terminal output module
The emergence of the light-dependent phy-PIF interaction from the underlying light-independent phyOPM-APA interaction could be explained by a hypothesis: the Pr form of phyPSM binds to phyOPM, masking the APA binding site. This hypothesis is supported by previous findings showing that in the dark, the Pr form of phyBPSM binds to phyBOPM, masking a nuclear localization signal in phyBOPM35. To test whether the light-dependent phy-PIF interaction arises from the light-dependent masking and unmasking of the APA binding site by phyPSM, we conducted several experiments.
We performed Y2H to examine whether the phyPSM-phyOPM interaction is general. As reported previously, phyBPSM interacted with phyBOPM in the dark but not under red light. Similarly, both phyAPSM and Mp-phyPSM interacted with their respective phyOPMs in the dark but not under red light (Fig. 6a–c). In vitro binding assays with recombinant phyPSM and phyOPM also supported these findings: the Pr form of phyAPSM preferentially bound to phyAOPM, and the same was true for phyBPSM and Mp-phyPSM with their respective phyOPMs (Fig. 6d–f). These results indicate that the Pr-dependent phyPSM-phyOPM interaction is conserved in both Mp-phy and Arabidopsis phyA and phyB. Next, we used Y2H to determine whether phyPSM and the APA bind to a similar region of phyOPM. Dividing phyBOPM into its PRD and HKRD domains, we found that the PRD domain interacted with PIF3 and PIF3mAPB, but not with PIF3mAPA or PIF3mAPA/mAPB, while the HKRD domain did not interact with any PIF3 alleles (Fig. 7a). PhyBPSM also interacted with the PRD domain in the dark but not under red light, and it did not interact with the HKRD domain regardless of light conditions (Fig. 7b). Furthermore, the previously reported G767R mutation in the PRD domain32 disrupted both the phyBPSM-phyBOPM and phyBOPM-APA interactions (Supplementary Fig. 8). These results suggest that both phyBPSM and the APA interact with the PRD domain of phyBOPM.
a–c Y2H assay showing the interaction between phyPSM and phyOPM in the dark. AH109C cells were transformed with either AD-fused phyAPSM and BD-fused phyAOPM (a), AD-fused phyBPSM and BD-fused phyBOPM (b), or AD-fused Mp-phyPSM and BD-fused Mp-phyOPM (c). Notations are consistent with those in Fig. 2a (a–c). d–f In vitro binding assay showing the interaction between phyPSM and phyOPM in darkness. Recombinant SBP-fused phyPSM was pulled down by MBP-fused phyOPM for phyAPSM and phyAOPM (d), phyBPSM and phyBOPM (e), or Mp-phyPSM and Mp-phyOPM (f). PhyPSM was irradiated with either a red light pulse (15 μmol m−2s−1, 5 min, Pfr) or a far-red light pulse (2.5 μmol m−2s−1, 5 min, Pr) before mixing with MBP-fused phyOPM. Other notations follow those in Fig. 2b (d–f). All in vitro binding assays were independently repeated at least twice with consistent results.
a, b Y2H assay showing the interactions between the APA and the PRD domain of phyBOPM (a) and phyBPSM and the PRD domain of phyBOPM(b). c, d Y2H assay showing the disruption of the phyPSM-phyOPM interaction by APA. The APA of PIF3 (116-221 a.a.) or the mAPA in a vector with the URA3 selection marker was co-transformed with the phyAPSM and phyAOPM (c) or the phyBPSM and phyBOPM pair (d). U and A in yeast media indicate the additional lack of uracil and adenine. e, f In vitro binding assay showing disruption of the phyPSM-phyOPM interaction by APA. MBP-fused phyAOPM (e) or phyBOPM (f) was used to pull down SBP-fused phyAPSM (e) or phyBPSM (f) in the presence of either GST-fused APA (86-221 a.a.) or mAPA. All in vitro binding assays were independently repeated at least twice with consistent results. g A diagram illustrating the light-dependent phy-PIF interactions via APA and APB motifs. A plant-type phy is believed to have originated in charophytes, and subsequently diverged into different phy clades including a bryophyte clade and spermatophyte clade. Both bryophyte Marchantia Mp-phy and spermatophyte Arabidopsis phyA and phyB promote light responses by interacting with PIFs through either APA or APB motif. The phyOPM-APA interaction is conserved in both phyA, phyB, and Mp-phy, while the phyPSM-APB interaction is found only in phyB. In the dark, phyPSM interacts with phyOPM, blocking the binding of APA to the PRD domain (light green color) of phyOPM. In the light, phyPSM dissociates from phyOPM, unmasking the APA binding site and allowing APA to bind to phyOPM. This light-dependent masking and unmasking mechanism explains how full-length phys interact with APA in a light-dependent manner, despite the underlying phyOPM-APA interaction being inherently light-independent. This masking mechanism may also regulate the light-dependent interaction of phytochromes with other proteins, such as SPA1, that bind to phyOPM.
We then investigated whether the Pr form of phyPSM and the APA compete for binding to phyOPM. If they do, expressing the APA fragment should inhibit the interaction between phyPSM and phyOPM in Y2H. Consistent with this hypothesis, the AH109C harboring the phyAPSM-phyAOPM interaction pair did not grow on selective media, regardless of light conditions, when the APA fragment was co-expressed, whereas it grew in the dark but not under red light when co-expressed with the mAPA fragment (Fig. 7c). Similarly, the APA fragment, but not the mAPA fragment, interfered with the phyBPSM-phyBOPM interaction (Fig. 7d). To further validate the competition between APA and phyPSM for binding to phyOPM, we conducted in vitro binding assays with recombinant phyPSM and phyOPM in the presence of the APA fragment. PhyAPSM bound less to phyAOPM in the presence of APA but not in the presence of mAPA (Fig. 7e). Likewise, phyBPSM bound less to phyBOPM when APA was present, but not when mAPA was present (Fig. 7f). Taken together, these results indicate that the Pr form of phyPSM and the APA compete for binding to phyOPM. This supports the hypothesis that the light-dependent masking and unmasking of phyOPM by phyPSM is the basis for the emergence of light-dependent phy-PIF interaction from the underlying light-independent phyOPM-APA interaction.
Discussion
Phytochromes bind to PIFs through either the APA or APB motifs. Of them, phyBPSM has been shown to interact with the APB, but it remains unclear whether phyAPSM interacts with the APA. In this report, we engineered a yeast strain to produce phycocyanobilin (PCB), allowing us to capture light-dependent interactions between phy and its interacting proteins, including PIFs, SPAs, ELF3, and TZP in the Y2H. Using the Y2H, supplemented by in vitro binding assays, we show that phyAOPM interacts with PIFs through the APA, while phyAPSM does not interact with PIFs. Interestingly, the phyOPM-APA interaction is not limited to phyA; phyBOPM also interacts with PIFs via the APA. Additionally, the Marchantia Mp-phyOPM interacts with both its native PIF (Mp-PIF) and Arabidopsis PIFs through the APA, while Arabidopsis phyAOPM and phyBOPM also interact with Mp-PIF through the APA. Our findings suggest that the phyOPM-APA interaction is an ancient feature conserved in both bryophyte Marchantia Mp-phy and angiosperm Arabidopsis phyA and phyB, while the phyPSM-APB interaction represents a more recent adaptation specific to phyB (Fig. 7g).
The phyBPSM-APB and phyBOPM-APA interactions have both overlapping and distinct roles in the degradation of PIF3 and phyB. Both interactions are sufficient to trigger the degradation of PIF3 by phyB in response to red light. This finding aligns with previous reports indicating that the phyBOPM is necessary for the degradation of both PIF1 and PIF3 in response to red light and that expression of phyBOPM alone (625-1172 a.a.) can promote PIF3 degradation even in the dark27,36,37. The ability of both the phyBPSM-APB and the phyBOPM-APA interactions to drive PIF3 degradation may also explain why some mutations in phyBPSM, such as G111D, completely abolish the activity of mutant phyBPSM allele, while only partially impairing the function of full-length mutant phyB allele37,38,39. However, the two interactions differ in their ability to promote the mutual destruction of phyB and PIF3, while the phyBPSM-APB interaction leads to the destruction of also phyB, the phyBOPM-APA interaction does not. This is consistent with observations in Marchantia, where the phyOPM-APA interaction promotes the degradation of Mp-PIF but not Mp-phy3. It remains unclear why the phyBPSM-APB interaction uniquely triggers mutual destruction. A previous study indicated that when phyB binds PIF3, it induces PIF3 phosphorylation by PPKs, which recruits LRBs, BTB-CUL3-type ubiquitin E3 ligase components, to the phyB-PIF3 complex40,41,42. The E3 ligase then ubiquitinates both phyB and PIF3, leading to their mutual destruction. Another set of E3 ubiquitin ligases, EBF1 and EBF2, has also been suggested to mediate the light-dependent degradation of phosphorylated PIF3 without triggering the mutual destruction of phyB43. Investigating whether LRBs or EBFs specifically recognize the phyBPSM-APB complex or the phyBOPM-APA complex could provide valuable insights into the mechanisms underlying this selective degradation process.
Although phyOPM itself interacts with the APA in a light-independent manner, the full-length phy interacts with the APA in a light-dependent fashion, presenting an enigma regarding how light dependence arises from an inherently light-independent interaction. We propose that this can be explained by light-dependent masking and unmasking of phyOPM by phyPSM. A previous study showed that the Pr form of phyBPSM interacts with phyBOPM to mask a nuclear localization signal (NLS) in phyBOPM, preventing the Pr of phyB from translocating to the nucleus in the dark35. Our findings extend this model by showing that the Pr of phyPSM interacts with phyOPM not only in phyB but also in phyA and Mp-phy. This suggests that the Pr-dependent phyPSM-phyOPM interaction is evolutionarily conserved. We further demonstrate that the co-expression of the APA fragment inhibits the Pr-dependent phyPSM-phyOPM interaction in Y2H, and the recombinant APA fragment interferes with the binding of phyPSM to phyOPM in vitro. These results support the hypothesis that the Pr of phyPSM competes with APA for binding to phyOPM. Together, our results suggest that phyPSM interacts with phyOPM and blocks the binding of APA to phyOPM in the dark, while phyPSM dissociates from phyOPM and unmask phyOPM, allowing the APA to bind preferentially in red light.
The light-dependent masking and unmasking of phyOPM may also provide a molecular basis for the light-dependent interactions between phy and a few other interacting proteins. One example is SPA144,45,46. Previous studies have shown that SPA1 interacts with full-length phyB in a light-dependent manner but does not bind to phyBPSM (1-640 a.a.), instead interacting light-independently with phyBOPM (640-1172 a.a.). Similarly, SPA1 interacts with full-length phyA in a light-dependent manner, but not with phyAPSM (1-600 or 1-617 a.a.), while interacting light-independently with phyAOPM (591-1121 a.a.). Mutations in phyAOPM (such as phyAG727E, phyAE777K) also disrupt SPA1 binding, highlighting the critical role of phyAOPM in the interaction. The importance of phyOPM for the interaction extends beyond SPA1. For example, PCH1 interacts with both phyBPSM and phyBOPM47, TZP interacts with both phyAOPM and phyBOPM48, and COP1 interacts with phyAOPM but not phyAPSM, while it binds to both phyBPSM and phyBOPM49,50. ELF3 interacts with phyBOPM and apo-phyBPSM51, while SWC6 and ARR6 interact light-independently with both phyBPSM and phyBOPM, but only with phyAOPM, not phyAPSM52. Thus, it would be intriguing to experimentally test whether the light-dependent interactions between these proteins and full-length phys are also influenced by the light-dependent masking and unmasking of phyOPM.
The PRD domain of phyOPM is likely a region unmasked in the Pfr form, as both the APA and phyPSM interact with it. Although the specific structural changes that lead to the unmasking of the PRD domain in the Pfr form are not yet fully understood, the dissociation of phyPSM from the PRD domain may play a key role in this unmasking process. The structures of the Pr of both phyA and phyB suggest extensive interactions between phyPSM and the PRD domain16,17,18. First, the modulator loop, a β-hairpin loop located between the PAS1 and the PAS2 of the PRD domain, extends to interact with the PHY domain of phyPSM. Second, the PAS2 domain of one protomer interacts with the nPAS-GAF domains of the other protomer, aligning the phyPSM to the PRD of the two protomers in a head-to-tail dimer arrangement. In contrast, the structure of the Pfr of phyB bound to 100 amino acids of PIF6 containing the APB motif shows that the phyBPSM domains of the two protomers align in a head-to-head dimer, with the PRD domain no longer interacting with phyBPSM and becoming more flexible19. This significant structural rearrangement has been attributed, in part, to steric hindrances between the tongue and the modulator structures, as well as between the PHY domain and the HKRD domains in the Pfr form. This structural remodeling from Pr to Pfr, leading to the dissociation of the PRD domain from phyPSM, may represent the process of unmasking phyOPM. Further studies are needed to confirm whether these structural transitions are directly responsible for the unmasking of the PRD domain in the Pfr form.
Methods
Plant materials and growth conditions
Arabidopsis thaliana plants were grown at 22 °C in a growth room under long-day conditions (16 h of white light at 100 μmol m−2s−1 followed by 8 h of darkness). To generate MYC-tagged PIF3 overexpression lines, PIF3 and its APA-, APB-, or APA/APB-mutated versions were cloned into a pBI121(GenBank M14641, clontech)-derived vector with the MYC tag at the C-terminus. These constructs were transformed into either the phyA-211 mutant (to assay red light-induced PIF3 degradation by phyB) or the 35S:PHYB-GFP/phyB-9 line (to assay mutual destruction of phyB by PIF3 alleles). Independent homozygous lines were selected and used for subsequent assays. Transgenic lines expressing mScarlet-tagged phyBOPM (642-1172 a.a.) were generated by cloning the corresponding gene fragment into a pBI121-derived vector (35S:PHYBOPM-mScarlet). Homozygous lines were amplified and used for analysis. A schematic illustration of the phytochrome and PIF alleles used in these constructs is provided in Supplementary Fig. 9. A schematic illustration of the MCS of all derived vectors is shown in Supplementary Fig. 10. Primers used for cloning are listed in Supplementary Table 1.
Engineering of a yeast strain producing phycocyanobilin
To engineer a yeast strain capable of producing phycocyanobilin (PCB), a PCB biosynthetic gene was synthesized and integrated into the genome of the AH109 yeast strain, commonly used for yeast two-hybrid assays (Y2H). The PCB biosynthetic gene consists of partially codon-optimized phycocyanobilin ferredoxin oxidoreductase (PcyA), heme oxygenase-1 (HO1), ferredoxin (FD), and ferredoxin NADP+ reductase (FNR) from Synechocystis sp. PCC6803, each of which is fused to a yeast mitochondrial targeting sequence (MTS) at their N-termini and linked by 2 A peptide to make a single gene. The full sequence is provided in Supplementary Fig. 11. The GAL1 promoter in the HO-pGAL-poly-KanMX4-HO plasmid53 was replaced with the GPD promoter, and the PCB biosynthetic gene was cloned under this GPD promoter. The resulting PCB biosynthetic gene expression cassette, along with KanMX conferring G418 resistance, was inserted into the HO locus of AH109 through homologous recombination, creating the strain designated AH109C. The production of PCB and the assembly of holo-phytochrome were assessed by the fluorescence emission of phyBPSM/Y276H (1-652 a.a., Y276H mutation) and the absorption spectra of partially purified phyBPSM from AH109C (Supplementary Fig. 1).
For the fluorescence emission analysis of phyBPSM/Y276H in AH109C, the PHYBPSM/Y276H was cloned into a pGBKT7 (630443, clontech)-derived vector and transformed into either AH109 or AH109C. The transformed yeast cells were plated on yeast dropout media lacking tryptophan for selection and incubated at 30 °C in the dark for 4 days. After incubation, the yeast cells were resuspended in PBS, and fluorescence emission was observed using a fluorescence microscope (BX51, Olympus) with a CY5 filter (39007, Chroma; excitation at 620/50 nm and emission at 690/50 nm).
To measure the light-dependent absorption spectra of phyBPSM produced from AH109C, His8-tagged PHYBPSM was cloned into a p425GPD54-derived vector and transformed into AH109C. The transformed AH109C cells were cultured in yeast dropout liquid media lacking leucine at 30 °C for 2 days and harvested by centrifugation (2600 g, 10 min). The harvested cells were lysed by vortexing with glass beads (G8772, Sigma) in lysis buffer (50 mM Tris-HCl, 150 mM NaCl, 10% [v/v] glycerol, 0.1% [v/v] Triton X-100, 1 mM PMSF, and a protease inhibitor cocktail [Roche cOmplete™], pH 7.5) using a Vortex-Genie 2 (SI-0236, Scientific Industries). The His8-tagged phyBPSM protein was partially purified from the lysed yeast extract by binding to Ni-NTA agarose and eluting with an elution buffer (50 mM Tris-HCl, 150 mM NaCl, 10% [v/v] glycerol, 0.1% [v/v] Triton X-100, 250 mM imidazole, pH 7.5). The eluted phyBPSM was then irradiated with either red light (15 μmol m−2s−1, Pfr) or far-red light (2.5 μmol m−2s−1, Pr) for 10 min, and the absorption spectra were determined using a UV-Vis spectrophotometer (UV-1800, Shimadzu) over a wavelength range of 500 nm to 800 nm.
Yeast two hybrid assay
For the yeast two-hybrid (Y2H) assay, phytochromes were cloned into the N-terminal side of either the GAL4 activation domain (AD) in the pGADT7 (630442, clontech) vector or the GAL4 DNA-binding domain (BD) in the pGBKT7 (630443, clontech) vector. PIFs and other interacting protein genes (SPA1, SPA2, ELF3, and TZP) were cloned into either pGADT7 or pGBKT7 vectors. Site-directed mutagenesis was used to generate PIF3 alleles (PIF3mAPA, PIF3mAPB, PIF3mAPA/mAPB) and PIF1 alleles (PIF1mAPA, PIF1mAPB, PIF1mAPA/mAPB), which were subsequently cloned into Y2H vectors. A schematic illustration of the phytochrome and PIF alleles used for the constructions is shown in Supplementary Fig. 9, and primers for cloning are listed in Supplementary Table 1.
The Y2H assay was conducted following the Clontech manual (PT3024-1). Both BD and AD vectors were co-transformed into AH109C yeast cells, and transformants were selected on dropout media lacking leucine and tryptophan (-LW). Several colonies were cultured in liquid -LW media for 48 h, after which cells were collected, washed twice with sterile water, and serially diluted to an optical density of 0.02 at 600 nm. The diluted cells were spotted onto -LW agar plates and -LWH plates (lacking leucine, tryptophan, and histidine) supplemented with 2 mM 3-aminotriazole (3-AT). Plates were incubated at 30 °C either in darkness or under red light (15 μmol m−2s−1) for 4 days.
For protein expression analysis, yeast cells were inoculated in 50 mL of YPDA and grown until the optical density at 600 nm (O.D.600) reached 0.8. The cells were collected by centrifugation (2600 g, 10 min) and washed twice with sterile water. The pellet was flash-frozen in liquid nitrogen and resuspended in lysis buffer (8 M urea, 120 mM NaH₂PO₄, and 10 mM Tris-HCl, pH 8.0). The cells were lysed by vortexing with glass beads (G8772, Sigma) for 2 min. The lysate was then centrifuged at 20,000 g for 10 minutes at 4 °C to remove cell debris. The supernatant was mixed with SDS sample buffer (5× buffer: 0.25 M Tris-HCl, 0.25% [w/v] bromophenol blue, 0.5 M dithiothreitol, 50% [v/v] glycerol, and 10% [w/v] SDS, pH 6.8) and separated by SDS-PAGE for immunoblotting.
For Y2H assays involving a third protein, the third gene was cloned into a p416GPD54 vector with a URA3 selection marker. To accommodate this vector, the URA3 gene was deleted from AH109C via homologous recombination, generating AH109CdU. To investigate whether the APA disrupts the interaction between phyPSM and phyOPM, the APA of PIF3 (116–221 a.a.) fused to NLS and mScarlet was cloned into p416GPD. The three vectors were co-transformed into AH109CdU and plated on dropout media lacking leucine, tryptophan, and uracil (-LWU). A few transformed cells were cultured, serially diluted, and spotted on -LWU plates or -LWUH plates (lacking leucine, tryptophan, uracil, and histidine with 2 mM 3-AT), or -LWUHA plates (lacking leucine, tryptophan, uracil, histidine, and adenine with 2 mM 3-AT). Plates were incubated at 30 °C in darkness or under red light (15 μmol m−2s−1) for 4 days. Primers for cloning are listed in Supplementary Table 1.
Immunoblot assay
Eighty transgenic seeds expressing MYC-tagged PIF3 alleles in either the phyA-211 background (for the PIF3 degradation by phyB) or the 35S:PHYB-GFP/phyB-9 background (for the mutual destruction of phyB by PIF3 alleles) were sown on 1/2 MS agar plates with 1% (w/v) sucrose, stratified for 3 days at 4 °C, and transferred to white light for 6 h to induce germination. For the PIF3 degradation assay by phyB in the absence of phyA, the plates were either kept in the dark for an additional 2 h or exposed to red light (15 μmol m−2s−1) for 30 min or 2 h. For the mutual destruction of phyB by PIF3 alleles, the plates were kept in the dark for an additional 12 h or transferred to red light (15 μmol m−2s−1) for 12 h. Seedlings were harvested, frozen in liquid nitrogen, and ground using a tissue lyser (Qiagen, Tissuelyser II). The ground tissue was resuspended in extraction buffer (8 M urea, 120 mM NaH2PO4, and 10 mM Tris-HCl, pH 8.0) and centrifuged to remove debris (20,000 g, 10 min, 4 °C). The supernatant was mixed with SDS sample buffer (0.25 M Tris-HCl, 0.25% [w/v] bromophenol blue, 0.5 M dithiothreitol, 50% [v/v] glycerol, 10% [w/v] SDS, pH 6.8 for 5x buffer) and separated by SDS-PAGE. Proteins were transferred to a nitrocellulose membrane (1060003, GE Healthcare) and analyzed by immunoblotting with specific primary antibodies. Protein luminescence was detected using the ChemiDoc XRS+ system (Bio-Rad) and visualized with ECL substrate (34577, Thermo Scientific). The primary antibodies used were α-MYC (sc-40, Santa Cruz Biotechnology), α-SBP (sc-101595, Santa Cruz Biotechnology), α-MBP (sc-13564, Santa Cruz Biotechnology), α-GST (sc-138, Santa Cruz Biotechnology), α-mCherry (632543, Takara), α-GFP (ab290, Abcam), α-GAL4 DBD (sc-510, Santa Cruz Biotechnology), α-PIF3 (rabbit polyclonal, Abfrontier), and α-TUB (T5168, Sigma). The secondary antibodies were goat anti-mouse IgG (H + L)–HRP (31430, Invitrogen) and anti-rabbit IgG–HRP (7074S, Cell Signaling).
In vitro binding assay
Full-length phy and phyPSM were cloned into an arabinose-inducible pBAD/myc-His B55 vector with an N-terminal SBP tag and a C-terminal His8 tag, while phyOPM was cloned into an IPTG-inducible pMALc2x56 vector with an N-terminal MBP tag and a C-terminal His6 tag. PIF proteins were cloned into an IPTG-inducible pET41a57 vector containing an N-terminal GST tag and a C-terminal His8 tag. The primers used for cloning are listed in Supplementary Table 1. Chromophore-bound full-length phy and phyPSM were purified from the PCB-producing E. coli strain LMG19455, while PIFs and phyOPM were purified from BL21-CodonPlus-RIL cells. For full-length phy and phyPSM, cells were lysed in lysis buffer (50 mM Tris-HCl, 25 mM NaCl, 2 mM EDTA, 0.2% [w/v] lysozyme, 10% [v/v] glycerol, 0.1% [v/v] Triton X-100, 1 mM PMSF, and a protease inhibitor cocktail [Roche cOmplete™], pH 7.5) by incubating at 37 °C for 15 min. RNase-free DNase (10 Kunitz units/mL) and 50 mM MgSO₄ were added to remove DNA. Phytochromes were then purified using Ni-NTA agarose and an elution buffer (50 mM Tris-HCl, 150 mM NaCl, 10% [v/v] glycerol, 0.1% [v/v] Triton X-100, 250 mM imidazole, pH 7.5). For phyOPM and PIF proteins, cells were lysed in a similar lysis buffer (50 mM Tris-HCl, 150 mM NaCl, 2 mM EDTA, 10% [v/v] glycerol, 0.1% [v/v] Triton X-100, 1 mM PMSF, 1 mM β-mercaptoethanol, and a protease inhibitor cocktail [Roche cOmplete™], pH 7.5) with sonication (2 sec/4 sec pulse, 15 min). Proteins were purified using Ni-NTA agarose and the same elution buffer.
For in vitro binding assays, glutathione sepharose 4B resin-bound GST-tagged PIF proteins were incubated with SBP-tagged phy or phyPSM, pre-treated with either red light (15 μmol m−2s−1, Pfr) or far-red light (2.5 μmol m−2s−1, Pr) for 5 min. For assays between phyPSM and phyOPM, amylose resin-bound MBP-tagged phyOPM was incubated with SBP-tagged phyPSM, pre-treated with red or far-red light for 5 min. For assays involving the APA domain of PIF3 (86-221 a.a.), GST-tagged APA was added to the incubation of MBP-tagged phyOPM and SBP-tagged phyPSM, pre-treated with red or far-red light. For assays between phyOPM and PIF, glutathione sepharose 4B resin-bound GST-tagged PIF proteins were incubated with MBP-tagged phyOPM proteins. Incubations were performed with 3 µg of each protein in 1 mL of binding buffer (50 mM Tris-HCl, 150 mM NaCl, 10% [v/v] glycerol, 0.1% [v/v] Triton X-100, 1 mM EDTA, 0.05% [w/v] sodium deoxycholate, pH 7.5) with gentle rotation at 4 °C in the dark for 2 h. After incubation, resin-bound protein complexes were washed three times with the binding buffer and precipitated by centrifugation (500 g, 1 min). Precipitated complexes were dissolved in the SDS sample buffer for SDS-PAGE. Co-precipitated GST-PIF, SBP-phy, SBP-phyPSM, and MBP-phyOPM were detected by immunoblotting using antibodies against GST, SBP, or MBP.
To determine relative binding affinities, 60 nmol of resin-bound GST-tagged APB or APA was incubated for 2 h with 0–165 nmol of red-light-treated phyBPSM or phyBOPM, at 15 nmol intervals, in 1 mL binding buffer (50 mM Tris-HCl, 150 mM NaCl, 10% [v/v] glycerol, 0.1% [v/v] Triton X-100, 1 mM EDTA, 0.05% [w/v] sodium deoxycholate, pH 7.5). The resin was then precipitated, and the pull-downed phy and PIF proteins were analyzed by immunoblotting. After immunoblotting, the band intensity was measured using ImageJ software (https://imagej.nih.gov/ij/). The fraction bound was plotted using GraphPad Prism 10 (https://www.graphpad.com/).
Semi in vivo binding assay
Semi in vivo pulldown assays were performed using recombinant GST-tagged PIF proteins and extracts from transgenic cells expressing mScarlet-tagged phyBOPM. Seedlings were grown under continuous white light (22 °C, 40 μmol m−2s−1) for 4 days, harvested, and ground in liquid nitrogen using a tissue lyser (Qiagen, Tissuelyser II). The ground tissue was resuspended in lysis buffer (50 mM Tris-HCl, 150 mM NaCl, 10% [v/v] glycerol, 0.1% [v/v] Triton X-100, 1 mM PMSF, and a protease inhibitor cocktail [Roche cOmplete™], pH 7.5). Incubations were performed in 1 mL soluble supernatants and incubated with glutathione sepharose 4B resin pre-bound to GST-tagged PIF proteins, with gentle rotation at 4 °C in the dark for 2 h. After incubation, the resin was washed three times with the binding buffer and precipitated by centrifugation (500 g, 1 min). The precipitated samples were dissolved in the SDS sample buffer for SDS-PAGE. Co-precipitated GST-PIFs and phyBOPM-mScarlet were detected by immunoblotting using antibodies against GST and mCherry, respectively.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The data generated in this study will be available from the corresponding author upon the request. Source data are provided with this paper.
References
Mathews, S. Evolutionary studies illuminate the structural-functional model of plant phytochromes. Plant Cell 22, 4–16 (2010).
Li, F. W. et al. Phytochrome diversity in green plants and the origin of canonical plant phytochromes. Nat. Commun. 6, 7852 (2015).
Inoue, K. et al. Phytochrome Signaling Is Mediated by PHYTOCHROME INTERACTING FACTOR in the Liverwort Marchantia polymorpha. Plant Cell 28, 1406–1421 (2016).
Possart, A. & Hiltbrunner, A. An evolutionarily conserved signaling mechanism mediates far-red light responses in land plants. Plant Cell 25, 102–114 (2013).
Clack, T., Mathews, S. & Sharrock, R. A. The phytochrome apoprotein family in Arabidopsis is encoded by five genes: the sequences and expression of PHYD and PHYE. Plant Mol. Biol. 25, 413–427 (1994).
Takano, M. et al. Distinct and cooperative functions of phytochromes A, B, and C in the control of deetiolation and flowering in rice. The Plant Cell 17, 3311–3325 (2005).
Rockwell, N. C., Su, Y. S. & Lagarias, J. C. Phytochrome structure and signaling mechanisms. Annu Rev. Plant Biol. 57, 837–858 (2006).
Legris, M., Ince, Y. C. & Fankhauser, C. Molecular mechanisms underlying phytochrome-controlled morphogenesis in plants. Nat. Commun. 10, 5219 (2019).
Rockwell, N. C. & Lagarias, J. C. Phytochrome evolution in 3D: deletion, duplication, and diversification. N. Phytologist 225, 2283–2300 (2020).
Inoue, K., Nishihama, R., Araki, T. & Kohchi, T. Reproductive Induction is a Far-Red High Irradiance Response that is Mediated by Phytochrome and PHYTOCHROME INTERACTING FACTOR in Marchantia polymorpha. Plant Cell Physiol. 60, 1136–1145 (2019).
Yuan, J., Xu, T. & Hiltbrunner, A. Phytochrome higher order mutants reveal a complex set of light responses in the moss Physcomitrium patens. N. Phytol. 239, 1035–1050 (2023).
Burgie, E. S. & Vierstra, R. D. Phytochromes: an atomic perspective on photoactivation and signaling. Plant Cell 26, 4568–4583 (2014).
Lagarias, J. C. & Rapoport, H. Chromopeptides from phytochrome. The structure and linkage of the Pr form of the phytochrome chromophore. J. Am. Chem. Soc. 102, 4821–4828 (1980).
Wagner, J. R., Brunzelle, J. S., Forest, K. T. & Vierstra, R. D. A light-sensing knot revealed by the structure of the chromophore-binding domain of phytochrome. Nature 438, 325–331 (2005).
Burgie, E. S., Bussell, A. N., Walker, J. M., Dubiel, K. & Vierstra, R. D. Crystal structure of the photosensing module from a red/far-red light-absorbing plant phytochrome. Proc. Natl Acad. Sci. USA 111, 10179–10184 (2014).
Li, H., Burgie, E. S., Gannam, Z. T., Li, H. & Vierstra, R. D. Plant phytochrome B is an asymmetric dimer with unique signalling potential. Nature 604, 127–133 (2022).
Burgie, E. S. et al. The structure of Arabidopsis phytochrome A reveals topological and functional diversification among the plant photoreceptor isoforms. Nat. Plants 9, 1116–1129 (2023).
Zhang, Y. et al. Structural insights into plant phytochrome A as a highly sensitized photoreceptor. Cell Res. 33, 806–809 (2023).
Wang, Z. et al. Light-induced remodeling of phytochrome B enables signal transduction by phytochrome-interacting factor. Cell 187, 6235–6250 (2024).
Khanna, R. et al. A novel molecular recognition motif necessary for targeting photoactivated phytochrome signaling to specific basic helix-loop-helix transcription factors. Plant Cell 16, 3033–3044 (2004).
Shen, Y., Khanna, R., Carle, C. M. & Quail, P. H. Phytochrome induces rapid PIF5 phosphorylation and degradation in response to red-light activation. Plant Physiol. 145, 1043–1051 (2007).
Matsushita, T., Mochizuki, N. & Nagatani, A. Dimers of the N-terminal domain of phytochrome B are functional in the nucleus. Nature 424, 571–574 (2003).
Al-Sady, B., Ni, W., Kircher, S., Schafer, E. & Quail, P. H. Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Mol. Cell 23, 439–446 (2006).
Shen, H. et al. Light-induced phosphorylation and degradation of the negative regulator PHYTOCHROME-INTERACTING FACTOR1 from Arabidopsis depend upon its direct physical interactions with photoactivated phytochromes. Plant Cell 20, 1586–1602 (2008).
Ni, M., Tepperman, J. M. & Quail, P. H. PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell 95, 657–667 (1998).
Pfeiffer, A. et al. Interaction with plant transcription factors can mediate nuclear import of phytochrome B. Proc. Natl Acad. Sci. USA 109, 5892–5897 (2012).
Qiu, Y. et al. Mechanism of early light signaling by the carboxy-terminal output module of Arabidopsis phytochrome B. Nat. Commun. 8, 1905 (2017).
Uda, Y. et al. Efficient synthesis of phycocyanobilin in mammalian cells for optogenetic control of cell signaling. Proc. Natl Acad. Sci. 114, 11962–11967 (2017).
Su, Y. S. & Lagarias, J. C. Light-independent phytochrome signaling mediated by dominant GAF domain tyrosine mutants of Arabidopsis phytochromes in transgenic plants. Plant Cell 19, 2124–2139 (2007).
Wagner, D., Fairchild, C. D., Kuhn, R. M. & Quail, P. H. Chromophore-bearing NH2-terminal domains of phytochromes A and B determine their photosensory specificity and differential light lability. Proc. Natl Acad. Sci. 93, 4011–4015 (1996).
Oka, Y. et al. Arabidopsis phytochrome a is modularly structured to integrate the multiple features that are required for a highly sensitized phytochrome. Plant Cell 24, 2949–2962 (2012).
Ni, M., Tepperman, J. M. & Quail, P. H. Binding of phytochrome B to its nuclear signalling partner PIF3 is reversibly induced by light. Nature 400, 781–784 (1999).
Al-Sady, B., Kikis, E. A., Monte, E. & Quail, P. H. Mechanistic duality of transcription factor function in phytochrome signaling. Proc. Natl Acad. Sci. USA 105, 2232–2237 (2008).
Possart, A. et al. Characterization of Phytochrome Interacting Factors from the Moss Physcomitrella patens Illustrates Conservation of Phytochrome Signaling Modules in Land Plants. Plant Cell 29, 310–330 (2017).
Chen, M., Tao, Y., Lim, J., Shaw, A. & Chory, J. Regulation of phytochrome B nuclear localization through light-dependent unmasking of nuclear-localization signals. Curr. Biol. 15, 637–642 (2005).
Park, E. et al. Phytochrome B inhibits binding of phytochrome-interacting factors to their target promoters. Plant J. 72, 537–546 (2012).
Park, E., Kim, Y. & Choi, G. Phytochrome B Requires PIF Degradation and Sequestration to Induce Light Responses across a Wide Range of Light Conditions. Plant Cell 30, 1277–1292 (2018).
Oka, Y., Matsushita, T., Mochizuki, N., Quail, P. H. & Nagatani, A. Mutant screen distinguishes between residues necessary for light-signal perception and signal transfer by phytochrome B. PLoS Genet 4, e1000158 (2008).
Kikis, E. A., Oka, Y., Hudson, M. E., Nagatani, A. & Quail, P. H. Residues clustered in the light-sensing knot of phytochrome B are necessary for conformer-specific binding to signaling partner PIF3. PLoS Genet 5, e1000352 (2009).
Ni, W. et al. Multisite light-induced phosphorylation of the transcription factor PIF3 is necessary for both its rapid degradation and concomitant negative feedback modulation of photoreceptor phyB levels in Arabidopsis. Plant Cell 25, 2679–2698 (2013).
Ni, W. et al. A mutually assured destruction mechanism attenuates light signaling in Arabidopsis. Science 344, 1160–1164 (2014).
Ni, W. et al. PPKs mediate direct signal transfer from phytochrome photoreceptors to transcription factor PIF3. Nat. Commun. 8, 15236 (2017).
Dong, J. et al. Light-Dependent Degradation of PIF3 by SCF(EBF1/2) Promotes a Photomorphogenic Response in Arabidopsis. Curr. Biol. 27, 2420–2430.e2426 (2017).
Lu, X.-D. et al. Red-light-dependent interaction of phyB with SPA1 promotes COP1–SPA1 dissociation and photomorphogenic development in Arabidopsis. Mol. Plant 8, 467–478 (2015).
Hoecker, U. The activities of the E3 ubiquitin ligase COP1/SPA, a key repressor in light signaling. Curr. Opin. plant Biol. 37, 63–69 (2017).
Sheerin, D. J. et al. Light-activated phytochrome A and B interact with members of the SPA family to promote photomorphogenesis in Arabidopsis by reorganizing the COP1/SPA complex. Plant Cell 27, 189–201 (2015).
Huang, H. et al. PCH1 regulates light, temperature, and circadian signaling as a structural component of phytochrome B-photobodies in Arabidopsis. Proc. Natl Acad. Sci. 116, 8603–8608 (2019).
Zhang, S. et al. TANDEM ZINC-FINGER/PLUS3 is a key component of phytochrome A signaling. Plant Cell 30, 835–852 (2018).
Seo, H. S., Watanabe, E., Tokutomi, S., Nagatani, A. & Chua, N.-H. Photoreceptor ubiquitination by COP1 E3 ligase desensitizes phytochrome A signaling. Genes Dev. 18, 617–622 (2004).
Jang, I. C., Henriques, R., Seo, H. S., Nagatani, A. & Chua, N. H. Arabidopsis PHYTOCHROME INTERACTING FACTOR proteins promote phytochrome B polyubiquitination by COP1 E3 ligase in the nucleus. Plant Cell 22, 2370–2383 (2010).
Alvarez, M. A. et al. EARLY FLOWERING 3 interactions with PHYTOCHROME B and PHOTOPERIOD1 are critical for the photoperiodic regulation of wheat heading time. PLoS Genet. 19, e1010655 (2023).
Wei, X. et al. Phytochrome B interacts with SWC6 and ARP6 to regulate H2A. Z deposition and photomorphogensis in Arabidopsis. J. Integr. Plant Biol. 63, 1133–1146 (2021).
Voth, W. P., Richards, J. D., Shaw, J. M. & Stillman, D. J. Yeast vectors for integration at the HO locus. Nucleic acids Res. 29, e59 (2001).
Mumberg, D., Müller, R. & Funk, M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene. 156, 119–122 (1995).
Gambetta, G. A. & Lagarias, J. C. Genetic engineering of phytochrome biosynthesis in bacteria. Proc. Natl Acad. Sci. 98, 10566–10571 (2001).
Riggs, P. Expression and purification of maltose‐binding protein fusions. Curr. Protocols Mol. Biol. 28, 16.16. 11-16.16. 14 (1994).
Smith, D. B. & Johnson, K. S. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene. 67, 31–40 (1988).
Acknowledgements
This research was supported by the National Research Foundation of Korea (NRF-2018R1A3B1052617).
Author information
Authors and Affiliations
Contributions
J.J. and G.C. designed the experiments. J.J. and Y.L. performed the experiments. J.J. and G.C. wrote the manuscript. All authors discussed the results and reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Meng Chen and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Jeong, J., Lee, Y. & Choi, G. Both phytochrome A and phyB interact with PHYTOCHROME-INTERACTING FACTORs through an evolutionary conserved phyOPM-APA interaction. Nat Commun 16, 3946 (2025). https://doi.org/10.1038/s41467-025-59327-8
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-59327-8
This article is cited by
-
Pr and Pfr structures of plant phytochrome A
Nature Communications (2025)