Abstract
Actin cytoskeleton dynamics is essential for proper nervous system development and function. A conserved set of neuronal-specific microexons influences multiple aspects of neurobiology; however, their roles in regulating the actin cytoskeleton are unknown. Here, we study a microexon in DAAM1, a formin-homology-2 (FH2) domain protein involved in actin reorganization. Microexon inclusion extends the linker region of the DAAM1 FH2 domain, altering actin polymerization. Genomic deletion of the microexon leads to neuritogenesis defects and increased calcium influx in differentiated neurons. Mice with this deletion exhibit postsynaptic defects, fewer immature dendritic spines, impaired long-term potentiation, and deficits in memory formation. These phenotypes are associated with increased RHOA/ROCK signaling, which regulates actin-cytoskeleton dynamics, and are partially rescued by treatment with a ROCK inhibitor. This study highlights the role of a conserved neuronal microexon in regulating actin dynamics and cognitive functioning.
Similar content being viewed by others
Introduction
Higher cognitive functions of mammalian brains result from complex interactions among billions of neurons mediated by synapses. These connections require the formation of specialized cellular structures in the presynapse, responsible for the proper storage and turnover of synaptic vesicles filled with neurotransmitters, and in the postsynapse, which receives these signals mainly in the dendritic spines. The precise organization of both the pre- and post- synaptic terminals is, to a great extent, dependent on the actin cytoskeleton1. At the presynapse, actin networks are responsible for the spatial segregation and cycling of synaptic vesicles2,3, while in the postsynaptic terminals, they drive spine morphology and neurotransmitter receptor mobility1. As a result, the actin cytoskeleton is crucial for multiple brain functions, including memory formation. For example, actin dynamics directly affects experience-dependent synaptic plasticity, which induces specific connectivity patterns between the synapses4,5.
In recent years, the Rho family of small GTPases have been described as key regulators of synaptic plasticity due to their control over various molecules essential for actin cytoskeleton assembly. RHOA is one of such GTPases, as it controls immature dendritic spine numbers by regulating the activity of the ROCK signaling pathway6. Moreover, RHOA directly interacts with formins, a group of actin nucleating proteins that are critical drivers of actin dynamics7,8. Formin proteins are remarkably diverse and versatile, with 15 members grouped into seven families in humans9. All of them are characterized by a conserved Formin-homology-2 (FH2) domain that forms homodimeric structures directly responsible for the processive polymerization of F-actin filaments10,11,12. The FH2 domain-based “tethered dimer” exists at two states in equilibrium that allow either actin-binding or dissociation. Notably, the interconversion between these two states occurs through the dynamic expansion of the FH2 ring and is possible thanks to the high flexibility provided by the linker region9,11,13. Each formin protein may have multiple functions beyond the FH2 based nucleation and actin polymerization. One example is the Dishevelled-associated activator of morphogenesis 1 (DAAM1), which has been shown to activate the RHOA/ROCK signaling pathway14,15 and act as an actin-bundling protein16.
Alternative splicing, the differential processing of exons and introns, is a widespread mechanism contributing to the diversification and specialization of protein function. Most human multi-exonic genes undergo alternative splicing17,18, and its prevalence is exceptionally high in mammalian brains19. Neural-specific alternative splicing is relatively conserved throughout vertebrate evolution19, and its misregulation has been linked to neurodevelopmental disorders20. A particularly striking example is provided by microexons, a class of 3-30 nt-long exons with a high degree of neuronal-specific inclusion and evolutionary conservation, and whose skipping has been linked to autism spectrum disorder and cognitive dysfunction in human and animal model studies21,22,23,24,25,26. Of several microexons that have been functionally studied, critical roles have emerged in processes including gene regulation, neuritogenesis, formation of synaptic connections, and behavior27,28,29. Striking examples are a microexon in Bak1, which controls neuronal apoptosis and is essential for animal survival30, and a microexon in eIF4G that impacts high-order cognitive functions through the regulation of activity dependent translation of synaptic proteins25. However, most microexons have not been functionally characterized at any level. In particular, genes harboring neuronal microexons are highly enriched in functions related to actin filament organization, actin filament-based processes, and cytoskeletal protein binding21,22, suggesting important roles in these processes. Nonetheless, the impact of microexon inclusion on actin-related neuronal processes remains unknown.
In this study, we investigate the function of a neuronal-specific microexon in DAAM1 and show that it impacts its FH2 domain, modifying the length of its linker region. Using biochemical assays and TIRF microscopy we demonstrate that splicing of this microexon modulates actin polymerization dynamics and fiber structure. Moreover, microexon inclusion is essential for proper neuronal function, since its deletion in differentiated in vitro neurons results in increased Ca2+ influx upon depolarization, and increased activity of the RhoA/ROCK signaling pathway. Consistent with these results, deletion of the microexon in vivo leads to multiple abnormalities in mice, including a decreased number of thin dendritic spines, reduced long-term potentiation (LTP), and deficient memory formation. Treatment with a RHOA/ROCK signaling cascade inhibitor rescues phenotypes resulting from the deletion of the microexon, including altered transcriptomic signatures in differentiated neurons and memory impairments in vivo.
Results
DAAM1 harbors a highly conserved neural-specific microexon within its FH2 domain
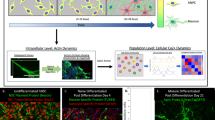
To investigate the contribution of alternative splicing to the functional specialization of actin polymerization and nucleation in mammalian neurons, we focused on the family of formins, comprising 15 genes in mammals. Using data from VastDB31, we found that these genes were widely expressed across cell and tissue types, and eleven of them showed substantial levels of expression (TPM ≥ 10) in brain samples (Fig. 1a). Moreover, seven increased their expression during neuronal differentiation, with the Daam1 showing the highest absolute expression in neurons (Fig. 1a, right panel). Regarding alternative splicing, we identified six exons in four formin genes with different degrees of neural-specific splicing regulation (Fig. 1b, c; see “Methods”) and evolutionary conservation across vertebrates (Fig. 1d). At the protein level, one of these exons was predicted to disrupt the open reading frame upon inclusion (Formin Like 3, FMNL3), two were second-to-last exons containing stop codons giving rise to alternative C termini (FMNL1-B and FMNL2-B), and three corresponded to microexons that preserve the reading frame (DAAM1, FMNL1-A and FMNL2-A)(Fig. 1b). As generally described for microexons21, these three exons fell either within a structured domain (i.e., the FH2 domain in DAAM1) or right next to it (i.e., the GTPase-binding domain in FMNL1 and the FH2 domain in FMNL2) (Fig. 1b).
a Heatmap showing gene expression levels of formin genes across multiple tissues and cell types based on VastDB. TPM: Transcript Per Million. b Schematic representation of the protein impact of neural-specific exons. Yellow-colored boxes represent upstream and downstream exons where the red box represents the alternatively spliced exon. GBD, GTPase-binding domain; FH3, Formin-Homology-3 domain or Diaphanous-Inhibitory Domain; FH2, Formin-Homology-2 domain. c Distribution of inclusion levels, using the Percent Spliced In (PSI) metric, for neural-specific exons in formin genes. PSI values were obtained from VastDB. Number of data points per plot and source data are provided as a Source Data file 1. d Evolutionary conservation of neural-specific alternative exons in formin genes. Black dots indicate the presence of an exon ortholog at the genome level. e Schematic representation of DAAM1 and its domains (top), together with DAAM1 protein structure based on AlphaFold2 (bottom). The location of Daam1-MIC (HsaEX0018410 in VastDB) is shown. f Partial amino acid sequence alignment of the FH2 domain of DAAM1 orthologs across vertebrates. Microexon (μ), upstream (C1), and downstream exons (C2) are indicated. The barplot depicts amino acid conservation. g Conserved neural-specificity of Daam1-MIC orthologs in vertebrates. PSI values from VastDB. h RT-PCR assays showing the inclusion of Daam1-MIC orthologs in different tissues from mice and zebrafish. i Srrm3/4-dependent regulation of Daam1-MIC orthologs in human, mouse and zebrafish. PSI values in the condition with (+, blue) or without (-, red) Srrm3/4 are shown. RNA-seq data from human HEK293 cells overexpressing human SRRM460, mouse N2A cells upon Srrm3/4 knockdown24, and zebrafish retinae extracted from Srrm3 KO 5 days post fertilization larvae61. j RT-PCR assays showing the inclusion of Daam1-MIC in cerebellum of WT and Srrm3 KO mice. h, j Inclusion and the skipping bands are indicated on the left side of the gel. PSI values are indicated below. RT-PCR assays were performed once. GBD GTPase-binding domain, FH3 Formin-Homology-3 domain or Diaphanous-Inhibitory Domain, DD Dimerization Domain, CC Coiled-Coil Domain, FH1 Formin-Homology-1 domain, FH2 Formin-Homology-2 domain, DAD Diaphanous-Autoregulatory Domain.
In particular, the microexon in DAAM1 lies within the linker region of the FH2 domain and modifies its length (Fig. 1e), which has been reported to have a significant impact on actin polymerization11. Protein alignments of the FH2 domain of DAAM1 and its linker region across vertebrates revealed that the microexon has been conserved at the orthologous position from sharks to humans, keeping a constant length of 9-10 amino acids (Fig. 1f). Moreover, transcriptomic data from various vertebrate species available in VastDB31 showed that the inclusion of the microexon in DAAM1 is also tightly restricted to neural tissues in other vertebrates (Fig. 1g). This inclusion pattern was validated using RT-PCR assays in mouse and zebrafish tissues (Fig. 1h). Additionally, in line with previous studies32,33, we found that the inclusion of the microexon was highly dependent on the action of the neuronal-specific Ser/Arg Repetitive Matrix 3 and 4 genes (SRRM3/4) in humans, mice and zebrafish (Fig. 1i). To assess its in vivo regulation in mice, we analyzed public RNA-seq data from a Srrm4 gene trap model34, revealing a consistent downregulation of Daam1 microexon in the cortex (ΔPSI = -7.4) and, especially, hippocampus (ΔPSI = -17.3). A similar pattern was obtained in the cerebellum of a Srrm3 gene trap model35 using RT-PCR assays (Fig. 1j, ΔPSI = -14). Because of its higher neural-specificity and evolutionary conservation compared to other formin alternative exons and its direct impact on the FH2 domain, we decided to investigate the biological relevance of the microexon of DAAM1 (hereafter Daam1-MIC) at the molecular, cellular, and organismal levels.
Daam1-MIC extends the FH2 linker region, impacting actin polymerization and structure
To gain further insights into the functional impact of Daam1-MIC, we generated models of the core structure of DAAM1’s FH2 domain (Fig. 2a) for the inclusion and skipping variants. These models suggest that the microexon is inserted into the disordered linker region of FH2 without causing major structural changes (red, Fig. 2b). This linker region is responsible for both the flexibility of the FH2 domain and its processive polymerization of actin36. In addition, a comparative analysis among human formin proteins revealed that the insertion of the ten amino acids encoded by the microexon makes the linker of DAAM1 the longest among all formins (Fig. 2c, d, and S1a). Since artificial versions of the linker with varying lengths were previously associated with DAAM1 activity levels11, we hypothesized that the microexon insertion could physiologically modulate DAAM1-mediated actin polymerization specifically in neurons. To test this hypothesis, we first purified the C-terminal part of the human DAAM1 protein with and without the microexon (Fig. 2a). This region (FH2-COOH) encompasses the entire FH2 domain and has been previously shown to be functional in vitro37. After selecting the optimal conditions for the experiment (Fig. S1b–d), we compared the actin polymerization activity of both splice isoforms. Interestingly, the isoform without microexon (FH2-COOH (-MIC)) exhibited a significantly higher actin polymerization rate (Fig. 2e, f).
a Schematic representation of DAAM1 domains (top) and subdivisions (bottom). FH2-COOH corresponds to the purified protein fragment. Domain nomenclature from Fig. 1e. b Structure of FH2 domain with (left) or without (right) microexon. Subregions share the same color-code as in (a). c Comparison of the linker regions among all human formin proteins. Loop color corresponds to the formin color from Fig. S1a. Structures obtained from AlphaFold2 and visualized using the PyMoL program. d Linker lengths across 10 formins. Linker color corresponds to c and S1a. Source data are provided as a Source Data file 2. e Pyrene actin polymerization assay. Each curve is the average of four technical replicates. Top: Schematic representations of the actin polymerization phases. f Actin polymerization rates, where data points correspond to technical replicates from four independent experiments. P-values from one-way ANOVA with replicate and category as factors. g Representative images of fluorescence micrographs showing F-actin fibers in the presence of microexon-containing (+MIC) or non-containing (-MIC) splice variants. 2 μM Actin and 200 nM DAAM1 fragments were used. Scale bars: 10 μm and 2 μm (magnification). Right: zoom of the regions highlighted in “15 min”. Arrows indicate differences in actin fiber morphology. h Total fiber intensity through time for 0.2 μM actin and 200 nM DAAM1 fragments. Statistical tests in Fig. S2f. i Dual-color TIRF microscopy of actin fibers obtained after 50 min using 0.2 μM actin and 200 nM SNAP-tagged proteins. Top: representative images of actin fibers. Bottom: fluorescence intensity distribution along the fibers (black/blue/red indicates actin/+MIC/-MIC proteins, respectively). Scale bar: 1 μm. j Fluorescence intensity along actin fibers through time. Statistical tests in Fig. S3f. h, j Thick lines: median from two independent experimental replicates. Dispersion: first and third quartiles.
To further characterize the impact of Daam1-MIC in actin dynamics and structure, we performed Total Internal Reflection Fluorescence (TIRF) microscopy experiments to directly visualize the actin network, which revealed a much more complex picture of the functional differences between the two isoforms (Fig. 2g and S2a). Consistent with the previous experiment, the isoform lacking the microexon exhibited an overall higher actin polymerization activity, as assessed by total intensity quantification (Fig. S2b). Moreover, TIRF experiments showed that the difference between the two proteoforms was not solely in polymerization activity, but they also had a qualitatively different behavior, resulting in distinct organizations of the actin network (Fig. 2g and S2a–c). Specifically, a detailed comparison of the spatio-temporal evolution of the actin network’s morphology at the level of individual fibers (i.e., bundles of actin filaments) showed that the microexon-containing isoform led to a ~ 2-fold increase in the average length of the actin fibers (Fig. S2e).
Previous studies also suggested that DAAM1 is a potent actin-bundling protein16. Microexon removal resulted in significantly increased fiber fluorescence, indicating higher filament bundling activity for this isoform (Fig. 2h and S2f). This suggests that the splice variants of DAAM1 interact with actin differently, in terms of kinetics and/or affinity, which could explain the behaviors described above. Thus, we next investigated whether the inclusion and skipping isoforms exhibited differences in binding to actin fibers. To do so, we performed dual-color TIRF microscopy experiments using SNAP-tagged DAAM1 FH2-COOH fragments (Fig. S3a–c). Both SNAP-tagged protein isoforms showed strong co-localization with actin fibers (Fig. 2i and S3d). However, analysis of individual actin bundles revealed a significantly higher amount of skipping isoform bound to actin fiber (Fig. 2i,j, and S3e,f). This is likely due to the intrinsic activity of the isoform, as no major differences in dimerization ability were observed between the two isoforms based on the FPLC profiles (Fig. S3a). In summary, these results demonstrate that microexon inclusion strongly impacts the functionality of the FH2 domain of DAAM1, both quantitatively and qualitatively. Microexon skipping increases actin polymerization rate and bundling, forming thicker fibers, while its inclusion promotes actin fiber elongation, creating more flexible and versatile actin networks and resulting in an overall more complex topology.
Daam1-MIC deletion leads to increased neuronal excitability in glutamatergic neurons
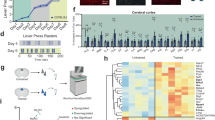
To begin investigating the relevance of Daam1-MIC in neuronal differentiation and function, we deleted the microexon in mouse embryonic stem cells (ESCs) using the CRISPR-Cas9 system (see “Methods”). We selected three independent knockout (KO) and control ESC clones and differentiated them in vitro into glutamatergic neurons (Fig. 3a)38,39. RT-PCR assays confirmed the precise microexon deletion in KO cells, with no associated mis-splicing during differentiation, whereas control cells displayed prominent microexon inclusion soon after the plating of neuronal precursor cells (DIV0) (Fig. 3b and S4a).
a Schematic representation of the neuronal differentiation protocol. DIV - day in vitro. b RT-PCR assays of Daam1-MIC inclusion during neuronal differentiation in representative WT and KO cell lines. RT-PCR assays were performed once. c, d Distributions of the lengths of the longest neurites (n = 491 WT, and 517 KO neurons, N = 2) (c) and individual filopodia (N = 3) (d) of neuronal precursors (DIV0 + 4 h). P-values from two-way ANOVA tests with replicate and genotype as factors. e Representative images of the calcium imaging experiment performed using FURA-2AM in mature neuronal cells (DIV21) depolarized with 30 mM KCl isotonic solution. Scale bar: 10 μm. f Distributions of calcium influx in WT and KO cell lines at various differentiation time points, based on FURA-2AM ratio. Neurons were stimulated with a 30 mM KCl isotonic solution. DIV1: n = 217 WT, and 228 KO neurons; DIV7: n = 188 WT, and 259 KO neurons; DIV14: n = 118 WT, and 134 KO neurons; DIV21: n = 116 WT, and 146 KO neurons. P-values from one-way ANOVA Tukey’s test. g Effects of the actin polymerization inhibitor Latrunculin A (LatA) and the small molecular inhibitor of formin FH2 domains (SMIFH2) on calcium currents in differentiated glutamatergic neurons at DIV14-21. The data corresponds to three independent experiments. Top: schematic representation of the mode of action of the actin-modulating drugs. h Distributions of calcium influx in WT and KO cell lines upon different treatments as depicted in (g), based on FURA-2AM ratio. Control: n = 234 WT, and 280 KO neurons; LatA: n = 124 WT, and 144 KO neurons; SMIFH2: n = 187 WT, and 187 KO neurons.
Since DAAM1-driven actin polymerization is important for neurite morphology and filopodia integrity16,40,41, we first assessed the impact of the microexon removal at early stages of neuronal differentiation (Fig. S4b). Daam1-MIC removal led to a significant increase in neurite length (Fig. 3c), consistent with the increased actin polymerization observed in vitro (Fig. 2). Additionally, we observed a slight increase in filopodia length in KO compared to WT cells (Fig. 3d), but no significant changes in filopodia number (Fig. S4c). Immunostaining of DAAM1, actin, and α-tubulin showed no major differences in the protein signal within the growth cones between the two genotypes (Fig. S4d, e), with only a small relative reduction of DAAM1 and actin in more distal filopodia in KO cells (Fig. S4f, g).
Despite these effects on neuritogenesis, microexon removal did not impair the ability of neurons to differentiate, as both WT and KO cell lines successfully generated neuronal networks and formed functional synaptic connections. Moreover, no major changes in neuronal morphology were observed at late stages (Fig. S4h, i). However, mature KO neurons (DIV14 and DIV21) exhibited a significant increase in Ca2+ influx, specifically upon depolarization (Fig. 3e, f). To further investigate this phenotype, we performed chemical treatments with modulators of various aspects of actin dynamics (Fig. 3g). Fully inhibiting F-actin polymerization by sequestering available G-actin monomers with Latrunculin A (LatA) resulted in a strong reduction of Ca2+ influx in both WT and KO cell lines, abolishing the differences between the two genotypes (Fig. 3g, h), and suggesting that these differences largely depend on actin polymerization dynamics. Next, we applied a small molecular inhibitor of formin FH2 domains (SMIFH2) that, in comparison to LatA, selectively inhibits only the formin-driven actin polymerization without impairing polymerization by other actin-binding proteins (e.g., actin elongation by VASP proteins or actin branching by Arp2/3 complex). Exposure of neuronal cultures to SMIFH2 increased Ca2+ influx in WT cell lines, mimicking the KO phenotype (Fig. 3g, h), and suggesting that microexon removal in neurons might lead to inappropriate activity of the FH2 domain of DAAM1. Finally, the increase in Ca2+ influx observed in Daam1-MIC KO neurons was neither due to differences in basal calcium levels (Fig. S4j), to DAAM1 isoform localization to the synapses (Fig. S4k), nor to changes in protein expression (Fig. S4l). Altogether, these results suggest that Daam1-MIC removal in mature neurons leads to increased Ca2+ influx upon depolarization due to effects on actin polymerization dynamics.
Daam1-MIC KO adult mice exhibit learning defects
Next, we generated a Daam1-MIC KO mouse line by ESC blastocyst microinjection (Fig. S5a and see “Methods”). In contrast to the full gene KO42, Daam1-MIC KO was not lethal, and mutant mice were born with expected Mendelian ratios (Fig. S5b), had normal weight (Fig. S5c), and did not display gross morphological abnormalities. To assess putative effects during the pre-weaning period, we first performed a neurodevelopmental behavioral screen (Fig. S5d)43,44. These experiments showed no major differences between KO and WT siblings in basic reflexes (Fig. S5e–g), balance, and motor coordination (Fig. S5h–k). Mild but significant increases were observed in KO mice in forelimb and hindlimb strength (Fig. S6a,b), and motor development-dependent walking (Fig. S6c). Interestingly, we also observed a decrease in the performance of the homing task (Fig. S6d), indicating delayed psychomotor development, but we found no significant gross morphological differences in dentate gyrus size (Fig. S6e, f), midline crossing (Fig. S6g), or neuronal complexity (Fig. S6h).
We then assessed a range of potential defects in adult mice. Whereas no major differences were observed in motor function and balance (Fig. S7a–f) or in anxiety-related phenotypes (Fig. S7g–k), we found that learning and memory formation capabilities were robustly affected upon Daam1-MIC removal. First, KO mice of both sexes showed significantly impaired recognition memory, evaluated using the novel object recognition (NOR) task (Fig. 4a, b), which depends on the prefrontal cortex and hippocampal circuit responsible for cognitive processing45,46,47. In contrast, no significant differences in total exploration time were observed during the familiarization or test phases of the experiment (Fig. S8a–b). Second, we assessed spatial memory by employing a modification of the Morris Water Maze (MWM) task (Fig. 4c, d and see “Methods”)48. This test showed impaired spatial learning specifically in KO males from day 4 to day 8 (Fig. 4e, f and S8e,f), a pattern observed in two independent experiments (Fig. S8e, f), and suggestive of defects on memory consolidation and/or optimization49. This impairment was not due to motor or motivation defects, as no differences were detected during the cued session when the platform was visible (Fig. 4e, f). At the molecular level, no differences between genotypes were found in the expression of cFOS or EGR1 protein levels (Fig. S8g–j) or DAAM1 protein expression levels in mouse cerebellum, motor cortex, or other tissues (Fig. S8k–m). Altogether, these results indicate that Daam1-MIC removal negatively impacts memory formation in adult mice, a defect that cannot be attributed to impaired motor function or anxiety-related behaviors.
a Schematic representation of the Novel Object Recognition experiment. b Distribution of discrimination index (d2) quantified during discrimination phase. Each data point describes the performance of one animal. P-values from two-sided Wilcoxon rank-sum tests. c, d Schematic of the Morris Water Maze (MWM) protocol (c) and platform set-ups (d) (details in “Methods”). e Cumulative search error index during MWM for females (top) and males (bottom)84. P-values from two-way ANOVA tests with replicate and genotype as factors. Significant P-values for males: 0.003, 0.04, 0.005, 0.01 (Days 4−8). f Representative trajectories from one trial of Replicate 1. a–f Each test was performed twice, using six animals per sex and genotype (24 animals per replicate). g Maximum-intensity confocal image of a dye-filled neuron after deconvolution. Red box: dendritic region analyzed. h Field excitatory postsynaptic potentials (fEPSPs) were recorded in the CA1 dendritic layer in response to Schaffer collateral stimulation. LTP was induced by theta burst stimulation (TBS). LTP plot of fEPSP values in KO (red circles, n = 6, N = 5) compared to WT mice (blue circles, n = 5, N = 3) after TBS at 60 min. Insets of traces in the plots represent average fEPSPs during periods indicated (1 and 2). i Boxplot of LTP after TBS at 60 min. P-values from two-sided Welch’s t-tests. j 3D reconstructions of pre- (Synapsin1/2, green) and post- (PSD95, blue and pink) synaptic markers in PD22 CA1 hippocampal sections. Pink spots represent postsynapses at ≤0.5μm from the presynapses (functional synapses) and blue ones at >0.5μm. Scale bar: 1 μm. k Percentage of functional synapses. N = 7 WT, and 8 KO mice. P-values from two-sided Wilcoxon rank-sum tests. l Representative images of dendritic spines after deconvolution. Scale bar: 5 μm. m, n Number of total dendritic spines (m) and thin spines per μm (n). N = 5 WT, and 5 KO mice, 100 μm of dendrites per cell. One dot corresponds to one neuron analyzed. Lines represent the relation between the averaged values from the animals analyzed in experimental replicates. P-values from two-sided Wilcoxon rank-sum tests.
Daam1-MIC removal impacts LTP and leads to reduced synaptic spines
To obtain further mechanistic insights into these phenotypes, we next investigated how Daam1-MIC deletion affected synaptic function and plasticity in hippocampal slices from adult males. Whole-cell voltage-clamp recording in CA1 pyramidal cells of the hippocampus showed a trend for a higher number of spikes in response to suprathreshold depolarizing current (Fig. S9a), suggesting a mild increase in the firing rate compared to WT neurons (Fig. S9b). We observed no changes in the set of parameters that describe intrinsic membrane properties (rheobase, membrane resistance, membrane potential, and threshold action potential; Fig. S9c–h), indicating that ionic permeability was not affected, and there were no changes in membrane conductance in KO neurons.
We next studied the effect of Daam1-MIC removal on glutamatergic CA3-CA1 synapses. We observed a strong and significant decrease in field Excitatory Postsynaptic Potential (fEPSP) based on induced Long-Term Potentiation (LTP) using a Theta Burst Stimulation (TBS) in KO neurons (Fig. 4g–i). To explore possible mechanisms behind this decrease, we then examined basal synaptic transmission in the CA1 region (input-output relationship of excitatory postsynaptic currents (EPSC)) and the presynaptic release of neurotransmitters, by applying Paired-Pulse Facilitation protocol (PPF) and recording miniature EPSCs (mEPSCs). These analyses did not show differences between WT and KO mice (Fig. S9i–p), suggesting no changes in basal excitatory synaptic transmission at hippocampal (CA3-CA1 neuron) synapses, and ruling out major presynaptic alterations associated with the LTP deficit in KO slices. In contrast to neuronal complexity analyses (Fig. S9r, s), which showed no morphological differences between the WT and KO neurons, the subcellular analysis of pre- (Synapsin 1/2) and post- (PSD95) synaptic markers in the CA1 region of the hippocampus at an earlier developmental time point (P21) showed a striking decrease in the number of PSD95-positive puncta, but no differences in Synapse 1/2 puncta (Fig. 4j, k, and S9t, u). Interestingly, biocytin labeling of neurons in adult mice revealed a significantly reduced number of thin immature dendritic spines (Fig. 4l–n), but not of stubby and mushroom-like spines (Fig. S9w). Therefore, altogether, these results indicate a severe impairment of the postsynaptic densities likely explaining the decrease in LTP in Daam1-MIC KO mice.
Daam1-MIC affects dendritic F-actin and the activity of the RHOA/ROCK signaling pathway
Taken together, these results suggest that Daam1-MIC deletion may alter the actin cytoskeleton in the postsynapse leading to defects in dendritic spine formation and LTP. To obtain further molecular insights, we first performed STochastic Optical Reconstruction Microscopy (STORM), which revealed decreased F-actin amounts in functional dendritic spines in both in vitro differentiated mature neurons and primary cultures (Fig. 5a, b and S10a). This was consistent with the overall defects in the synaptic spines (Fig. 4j–n), and the impaired actin polymerization via FH2 domain observed both in vitro and in neuronal cultures (Figs. 2 and 3).
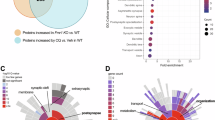
a Zoom-in of representative STORM images of actin (phalloidin-A647N) (white) overlaid with the conventional fluorescent image of PSD95 (green) of primary hippocampal cultures shown in Fig. S10a. Scale bar = 1 µm. b Quantification of the STORM localizations of F-actin in dendritic spines co-localizing with PDS95 signal, normalized per area. Each point represents a dendritic spine. Black lines connect the paired means of different biological replicates. P-values correspond to paired two-sided Mann-Whitney tests. c, d Interaction of DAAM1 with RHOA. c Co-immunoprecipitation experiment performed with anti-FLAG antibody. d Quantification of the interaction between DAAM1-ΔDAD constructs with ( + MIC) or without microexon (-MIC) and RHOA in panel (c) and S10e,f. N = 3 biological replicates (experiments). P-values from two-sided Wilcoxon rank-sum tests. e Confocal micrographs representing RhoA2G distribution (top) and RhoA activity (sensor ratio; bottom) in neurons. Boxes: example regions of interest. Scale bar: 50 μm. f Comparison of biosensor efficiency. One dot represents FRET efficiency in one neuron. WT: n = 59, KO: n = 57 neurons, N = 2 replicates. P-values from two-way ANOVA with replicate and genotype as factors. g Scatter plot showing changes in gene expression (log2 fold changes) between WT and KO cells in the control treatment (Veh) (X-axis) vs. KO control cells and KO cells treated with ROCK inhibitor (Y-axis). Each data point corresponds to a gene colored according to their “rescue” group: orange, rescued by ROCKi; red, not rescued; gray, other genes (see “Methods”). h Molecular Function GO term enrichment for genes with a rescue pattern (orange in (g)). i, j Calcium influx measurements upon ROCKi treatment. P-values from Kruskal–Wallis test. Control: n = 66 WT, and 51 KO neurons; ROCKi: n = 83 WT, and 88 KO neurons. k NOR experiment performed to test the effect of ROCKi. l Discrimination index values in the different conditions and for each genotype. P-values from two-sided Wilcoxon rank-sum tests. NOR test was performed twice, using six animals per treatment and genotype in each replicate. Replicates were separated by sex (24 animals per replicate).
Given the tight interplay between the actin cytoskeleton and other molecular pathways (see Introduction), we next explored if Daam1-MIC removal affected any of those pathways, potentially contributing to the observed phenotypes. As mentioned above, the RHOA/ROCK signaling cascade is essential for controlling the actin cytoskeleton-driven dendritic spine formation and LTP6, and DAAM1 has been shown to directly interact with RHO and DVL proteins10,15,50. Therefore, we investigated whether microexon inclusion/skipping changes these protein-protein interactions using the full DAAM1 protein as prey (Fig. S10b–d). While we observed no changes in DVL1/2 interaction upon microexon removal, suggesting no major changes in the first stages of DAAM1 activation, interaction with RHOA appeared decreased (Fig. S10d). To augment the Co-IP efficiency with RHO proteins, we next removed the DAAM1 autoinhibitory domain (DAD), as previously described14. Using this construct, Co-IP assays showed a significant ~40% decrease in the interaction between DAAM1-ΔDAD and RHOA upon microexon removal (Fig. 5c, d and S10e–g), which was further confirmed by a Nano Bioluminescence Resonance Energy Transfer (NanoBRET) protein-protein interaction assay51 (Fig. S10h). Hence, we next asked if this decrease in DAAM1-RHOA interaction upon microexon removal affected RHOA activity. Interestingly, a significant increase in RHOA activity in protrusions of mature KO neurons (Fig. 5e, f and S10i) was observed by acceptor photobleaching of a RhoA FRET-based biosensor52.
Based on these observations, we hypothesized that increased RHOA activity upon Daam1-MIC removal leads to hyperactivation of the RHOA/ROCK signaling cascade, which, in turn, contributes to the observed phenotypes. To test this hypothesis, we employed a commonly used inhibitor of ROCK (Y-27632), a direct downstream effector of RHOA. First, application of Y-27632 to differentiated neurons rescued the expression alterations of most differentially expressed genes between the KO and WT cells (Fig. 5g and S11a–c). This included multiple genes related to the function of the RHOA/ROCK signaling cascade, the Wnt-pathway, and the regulation of the actin cytoskeleton (Fig. 5h and S12). Second, applying Y-27632 reverted the difference in Ca2+ influx between WT and KO neurons (Fig. 5i,j). Lastly, injection of Y-27632 intraperitoneally in mice significantly increased the discrimination index of KO animals of both sexes, suggesting at least a partial rescue of the memory impairments (Fig. 5k, l and S11d–i). Moreover, ROCK inhibitor treatment caused a decrease in the discrimination index in WT animals, further supporting genotype-specific differences in response to changes in RHOA/ROCK signaling (Fig. 5l). Altogether, these results suggest that Daam1-MIC is important not only for actin polymerization but it also modulates the activity of the RHOA/ROCK signaling cascade, both of which are crucial processes for proper memory formation.
Discussion
Tight regulation of actin cytoskeleton polymerization and its associated molecular pathways by actin-binding proteins is crucial for multiple neurobiological processes. Here, we found that Daam1, the formin gene with the highest expression in mature neurons, harbors a microexon that is highly neural-specific and evolutionarily conserved across vertebrates. Interestingly, we show that correct regulation of Daam1-MIC is crucial at multiple levels. Firstly, microexon inclusion structurally extends the length of the linker region of the FH2 domain, modulating its actin polymerization capabilities, quantitatively and qualitatively, in controlled in vitro assays. Secondly, Daam1-MIC removal led to a decrease in DAAM1-RHOA interaction and enhanced activity of the RHOA/ROCK signaling pathway in neurons, which is known to influence multiple cellular processes, including actin cytoskeleton dynamics. Lastly, deletion of the microexon led to diverse phenotypes in mice, including a decreased number of dendritic spines, reduced LTP, and defects in memory formation.
The inclusion of Daam1-MIC in neurons makes the length of the DAAM1’s FH2 linker region an outlier with respect to all other formins. Given that the linker region serves as a hinge in FH2 dimers9, its uniquely large length likely results in a much more flexible dimer, which we show to bind less to actin as compared to the non-neural form lacking the peptide encoded by the microexon. Previous studies11,53 reported that the long linker of DAAM1 was associated with this protein’s unusually low actin polymerization rate and that its shortening enhanced polymerization. We recapitulate these findings and further connect them with a splicing-driven and physiologically occurring event that modulates DAAM1’s function in a tissue-dependent manner. Moreover, our TIRF results revealed a much more complex scenario in which microexon inclusion does not simply lead to a reduced polymerization rate but to a dramatic change in how DAAM1 interacts with and remodels the actin cytoskeleton. In particular, we observed that microexon inclusion significantly decreased the actin-bundling capabilities of DAAM1, probably as a direct result of a lower affinity to actin. Moreover, a detailed structural characterization of actin bundles showed that the microexon-containing isoform produced consistently longer fibers, likely due to its less constrained binding to actin. Although these results are based on in vitro reconstituted assays, it is tempting to speculate that, in vivo, the inclusion or exclusion of the microexon may similarly modulate DAAM1’s function. In line with this idea, at the early stages of neuronal differentiation, we observed a significant increase in neurite length in Daam1-MIC KO neurons, consistent with enhanced actin polymerization rate. However, detailed subcellular analysis of dendritic spines (Fig. 5a, b) revealed decreased F-actin amounts possibly due to the qualitative impairments in actin dynamics observed upon Daam1-MIC removal (Fig. 2). Interestingly, a previously described exon in shootin1 (SHTN1) also modulates actin binding and polymerization; however, compared to DAAM1, the neuronal SHTN1 isoform enhances actin polymerization stimulating axon growth and subsequent axon specification54,55. This suggests that alternative splicing may regulate similar molecular processes through different protein pathways and with distinct consequences.
In our mouse model, Daam1-MIC removal resulted in memory impairments, plausibly due to decreased LTP caused by a reduced number of dendritic spines. In particular, we observed a decreased number of thin, immature spines characterized by their dynamic structure, and considered “learning spines” as they can be consolidated into more stable mushroom spines4,5. What might be the molecular bases of these cellular phenotypes? In addition to direct defects in actin assembly caused by Daam1-MIC deletion, interactions with other molecular pathways are likely to contribute to these phenotypes. Specifically, Rho-family GTPases, including RHOA, are key players in memory formation by regulating spine morphology, at least partially via their role in regulating actin cytoskeleton6. Evidence suggests that RHOA activity modulates immature spine pruning6,56. For instance, expression of constitutively active RHOA in hippocampal neuronal cultures or brain slices resulted in simplified dendritic trees and reduced spine densities57,58. Conversely, ROCK inhibition specifically increases the number of immature spines57. Additionally, to further establish the link between Daam1-MIC deletion, RHOA/ROCK signaling and some of the observed phenotypes, we used Y-27632, a ROCK inhibitor. Y-27632 treatment of differentiated glutamatergic neurons decreased calcium influx upon depolarization, abolishing the observed differences between the genotypes. Moreover, administration of the inhibitor recovered most of the gene expression alterations observed upon microexon removal and was sufficient to revert the memory deficiencies observed in the NOR task performed by the Daam1-MIC KO mice. Altogether, and despite the possibility that this inhibitor may not be fully specific, the significant rescue observed at multiple levels (molecular, functional, and behavioral) suggests that the upregulation of the RHOA/ROCK signaling cascade upon Daam1-MIC removal is, at least in part, causative of the observed phenotypes.
How can Daam1-MIC removal lead to upregulation of the RHOA/ROCK signaling cascade? DAAM1 is known to directly interact with RHOA, with a binding preference towards active GTP-RHOA14,15. Interestingly, even though the microexon does not lie in the proximity of the RHOA interacting domain (N-terminal GDB domain), our Co-IP and NanoBRET assays showed a significant decrease in the interaction between DAAM1 and RHOA upon microexon removal. Although this could be due to an undescribed conformational effect, another possibility is that a stronger DAAM1 interaction with F-actin by the skipping form may result in less RHOA interaction. Could this change in DAAM1-RHOA interaction lead to the observed increase in overall RHOA activity? On one hand, the exact mechanism of DAAM1-dependent activation of RHOA is unknown, but it does not rely on the direct interaction of DAAM1 with RHOA, and it is instead believed to occur through the recruitment of the GEF-RHOA complex to DAAM115. On the other hand, in Diaphanous-related formins, such as DAAM1, the direct binding of RHOA-GTP to the GDB domain is followed by hydrolysis to RHOA-GDP, contributing to formin activation (and RHOA deactivation)10,59. Since DAAM1’s activation is highly dependent on DVL proteins14, it is possible that reduced interaction of RHO-GTP with DAAM1’s GDB domain in Daam1-MIC KO cells mainly results in less RHOA deactivation and thus an increased pool of active RHOA-GTP available (Fig. 6). Another unexplored possibility is that alterations in actin dynamics upon Daam1-MIC depletion affect RHOA activity indirectly through an unknown mechanism (Fig. 6), independently or in addition to the change in DAAM1-RHOA protein interaction. Given the importance of RHOA/ROCK signaling in modulating actin dynamics, it is plausible that such feedback loops exist.
Summary of the observed phenotypes upon Daam1-MIC removal and their relation to the RHOA/ROCK signaling cascade9,14,15. DVL binds to DAAM1, releasing its autoinhibition, which is further facilitated by interaction with RHOA. Active DAAM1 leads to RHOA activation through an unknown mechanism involving DAAM1’s C-terminal part (dashed lined) and is not dependent on direct binding RHOA-DAAM1 interaction. Microexon removal decreases RHOA binding to DAAM1’s N-terminal region. This is proposed to lead to lower hydrolysis of RhoA-GTP, increasing the pool of active RHOA and in turn, hyperactivating the RHOA/ROCK signaling cascade. Arrows indicate the directionality of the event. The dotted arrow indicates a potential, unknown feedback loop from actin polymerization to ROCK pathway activation. GAP - GTPase-activating protein, GEF - guanine nucleotide exchange factor, GTP - Guanosine triphosphate, GDP - Guanosine diphosphate.
In summary, through a comparative analysis of formin proteins, we found that Daam1 is subject to tight post-transcriptional regulation by a highly neural-specific and evolutionarily conserved microexon. Daam1-MIC inclusion directly impacts actin polymerization via modulation of the FH2 domain activity. Moreover, through an in-depth multi-level characterization of this microexon, we unveiled DAAM1 as an important player in regulating neurotransmission and memory formation.
Methods
All procedures were approved by the Ethics Committee of Parc de Recerca Biomèdica (Comité Ético de Experimentación Animal del PRBB (CEEA-PRBB; MDS-18-0040-P1, MDS-16-0035-PR1-P2, LBM-17-0049-P1), the Generalitat de Catalunya (9210, 9723), and Ethics Committee of Consejo Superior de Investigaciones Científicas (CSIC; EH2023.0159.P1.M). Experiments were carried out in accordance with the guidelines of the European Union Council (2003/65/CE) and Spanish regulations (BOE 252/34367-91, 2005). The CRG is authorized to work with genetically modified organisms (A/ES/05/I-13 and A/ES/05/14).
Computational analysis formins and characterization of DAAM1 and RT-PCR assays
To investigate the expression of formins across tissues and during neuronal differentiation (Fig. 1a), we used mouse (mm10) data from VastDB31. In addition, we extracted the mouse and/or human neural-regulated exons among the 15 mammalian formin genes from Table S2 in Irimia et al.21 and used VastDB to extract the exon’s protein location and inclusion levels for the six identified neural-regulated exons shown in Fig. 1b and 1c, respectively. The study of the evolutionary conservation of these six exons was carried out using the VastDB ortholog information and manual exon alignments among the human (hg38), mouse (mm10), cow (bosTau9), opossum (monDom5), chicken (galGal4), frog (xenTro9), zebrafish (danRer10) and shark (calMil1) genomes, downloaded from Ensembl. To investigate the regulation of Daam1-MIC by SRRM3/4 (Fig. 1i), we used data from human HEK293 cells overexpressing human SRRM4 (SRA: SRP14991360), mouse N2A cells upon Srrm3/4 knockdown (GEO: GSE11260024), and zebrafish retinae extracted from Srrm3 KO 5 days post fertilization larvae (GEO: GSE18078161). Crystal Structure of human DAAM1 FH2 domain (2z6e.pdb) was used to model the mice Daam1 FH2 domain with and without microexon using Robetta (http://new.robetta.org/) (Fig. 2b). Visualization was performed using the PyMoL program.
To validate the inclusion of Daam1-MIC through RT-PCR assays in mouse and zebrafish (Fig. 1h, j) and throughout the manuscript), total RNA was extracted using the RNeasy Plus Mini kit (Qiagen, 74136), treated with DNase using TURBO DNA-freeTM Kit (Thermo Fisher Scientific), and reverse-transcribed with SuperScript III Reverse transcriptase (Invitrogen, 18080044) and oligo(dT)20 as advised by the manufacturer. GoTaq G2 Flexi DNA Polymerase enzyme (Promega, M7806) was used to amplify Daam1-MIC’s flanking exons, and PCR assays were performed with 25 cycles, 54 °C annealing temperature, and 120 sec extension. The PCR product was size-fractionated using 2.5% ultrapure agarose (Invitrogen, 16500500) in SB buffer (36.4 mM of Boric acid and 10 mM NaOH), and detected using SYBR Safe (Life Technologies, S33102). Primer sequences used in this study are provided in Table S1.
Cloning of Daam1 FH2 domain, protein expression, and purification
To clone the functional FH2 domain of DAAM1 (residues 594-1077), we have used RNA extracted from Human Embryonic Kidney 293 (HEK293T) and SH-SY5Y cells. RNA samples were extracted from 1 million cells using the RNeasy Mini Kit (Qiagen, 74136) and treated with TURBO DNA-freeTM Kit (Thermo Fisher Scientific, AM2238). Afterwards, 1 μg of RNA was reverse transcribed into cDNA using High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific, 4368814). Primers used to amplify the FH2 domains of DAAM1 are provided in Table S1. The PCR product was cloned in pEMT33 (for actin polymerization assay and TIRF microscopy experiments) and in C145 pCoofy17 (for dual-color TIRF microscopy experiments), both vectors have been kindly provided by the Sebastian Maurer lab (CRG).
These constructs were expressed as fusion proteins with N-terminal IgG tag plus C-terminal Strep-tag in E. coli strain BL21-CodonPlus(DE3)-RIL strain (Agilent Technologies, 230245). Cells were cultured at 37 °C until OD600nm reached 0.5, after which the media was cooled to 18 °C and Isopropyl β-d-1-thiogalactopyranoside (IPTG; ROTH, 2316.1) was added to a final concentration of 0.1 mM to induce protein expression. Bacterial cultures were grown overnight at 18 °C and subsequently harvested by centrifugation at 4 °C (15 min, 450 g) and disrupted by sonication. Sonication was conducted in HEKG 10 buffer (20 mM HEPES, pH 7.4, 1 mM EDTA, 50 mM KCl, 10% glycerol) with 1 mM DTT supplemented with cOmplete EDTA-free Protease Inhibitor Tablet (Sigma, 11873580001). After centrifugation (Eppendorf 5810 R Centrifuge) at 4 °C (20 min, 7,152 g), proteins were purified using StrepTrap HP column (GE Healthcare, GE17-5248-01) at 4 °C. The N-terminal tag was cleaved with His-C3 protease and the C-terminal tag with His-TEV protease (in-house purified). Both proteases were further removed using Ni-NTA Agarose beads. Individual pooled DAAM1-FH2 proteins, with and without the microexon, were further purified by a passage through a Superose 6 column (GE Healthcare). Their concentration was evaluated using NanoDrop spectrophotometer (ThermoFisher) and their purity/integrity was assessed on SDS-PAGE, where coomassie staining (ThermoFisher, LC6065) was used for visualization.
Pyrene actin polymerization assays and TIRF microscopy
Pyrene-labeled actin protein (Universal Biologicals) was prepared as suggested by the manufacturer. In short, actin was equilibrated for 1 h at 4 °C in a G-buffer (5 mM Tris-HCl pH 8.0, 0.2 mM CaCl2) and spun for 30 min at 1100 g before adding 10X Polymerization buffer (500 mM KCl, 20 mM MgCl2, 10 mM ATP) to a final 1X concentration. Actin assembly was measured in 60 μl reactions using 96 Well Black Polystyrene Microplates (Corning®, CLS3904). Pyrene fluorescence was monitored over time at 24 °C at an excitation of 365 nm and emission of 407 nm in a fluorescence spectrophotometer (Tecan Infinite 200 PRO). Rates of actin assembly were calculated from the slope of the assembly curves at 50% polymerization (i.e., the time at which 50% of the actin has polymerized, or half time). As previously described62, we computed the half-time of each experiment based on the intensity values included in a specific interval. First, we calculated the average minimum intensity (average intensity of the 10 points closer to the minimum intensity) and the average maximum intensity (average intensity of the 10 points closer to tmax, where tmax is the average of the 10 points with the highest intensity). Then, we used these values to compute the lower and higher bounds of the intensity interval used for the half-time computation. The lower bound was defined as (0.5 - delta/2)*(avg_max_intensity - avg_min_intensity) + avg_min_intensity, while the higher bound was defined as (0.5 + delta/2)*(avg_max_intensity - avg_min_intensity) + avg_min_intensity (with delta=0.1, as recommended in Doolittle et al., 2013). Next, we used the points in each experiment with intensity values included between the lower and higher bounds to fit a linear model (intensity vs. time) and extracted the slope value returned by the fit. Finally, we derived the actin polymerization rate at half time by multiplying the slope value by a scaling factor (SF). SF was computed as (actin_conc - critical_conc)/(avg_max_intensity - avg_min_intensity), with actin_conc = x μM (actin original concentration as specified), critical_conc = k_minus / k_plus. k_minus and k_plus represented the on-rate and off-rate constants for filament assembly, and were set at 1.4 S–1 and 11.6 μM-1s-1, respectively, as suggested by Doolittle et al., 2013)63.
TIRF-based F-actin polymerization experiments were performed using manually assembled flow cells consisting of piranha-cleaned, silanized, PEG-passivated glass coverslips (22 × 22 mm #1.5 (i.e. 170 + /−5 μm) from MARIENFELD (Cat # 0107052)) and a PLL-PEG passivated slide64. Flow cells were primed by flushing 55 μl of a solution of 5% Pluronic F-127 (Sigma-Aldrich, P2443) and then incubated for 10 min at room temperature. Next, flow cells were washed twice with 55 μl of kappa-Casein (1:100 in 1X G-buffer of 5 mg/ml stock in 1X Brb80 buffer). Absorptive paper (Whatman filter paper) was used to flush the chamber by holding the paper on the outlet side of the channel while adding the solution dropwise in the inlet side. Chambers were then equilibrated with G-buffer and the reaction mixture was loaded. The actin polymerization reaction mix contained the protein variant of interest (DAAM1-FH2 with or without microexon) and actin, which were diluted in freshly prepared buffer containing 5 mM Tris-HCl (pH 8.0), 0.2 mM CaCl2, 50 mM KCl, 2 mM MgCl2, 2 mM ATP, 2 mM DTT, 1% (w/vol) glucose, 0.2 mM Brij-35, oxygen scavengers (180 mg/ml catalase (Merck, C40) and 752 mg/ml glucose oxidase (Serva, 22778.01)), 0.15% (w/vol) methylcellulose (Sigma-Aldrich, 4000 cP). Actin labeling was achieved using SiR-Actin (TeuBio, SC001). After loading the reaction mixture, flow cells were finally sealed with vacuum grease and placed onto the microscope stage for observation. To minimize the experimental variations between the different conditions to be compared (actin alone, actin with +MIC, and -MIC protein variants), experiments have been run in parallel, i.e. the three conditions were run simultaneously during each experiment. To this end, flow cells were divided into three separate channels using double-sided tape.
TIRF imaging was performed with a custom-assembled system (Cairn Research, Faversham, UK) built around an automated Nikon Eclipse Ti microscope equipped with a Perfect Focus System and an azimuthal TIRF unit (Gataca Systems, iLas2) using a 100X oil-immersion objective (Nikon CFI SR Apo, NA = 1.49), 488 nm and 640 nm simultaneous laser excitation at a TIRF angle of 80 deg, and two EMCCD cameras (Andor, iXon 888 Ultra) for fluorescent detection using a dichroic (Chroma, T565lpxr) to split the fluorescence onto the two cameras and a 525/50 bandpass filter (Chroma, 284337) for DAAM1-SNAP-Alexa488 detection and a 655 long-pass filter (Chroma, 283943) for SiR-actin detection. Imaging was performed at room temperature for 2 h taking 1 image every 2 min in 3 to 5 different sample locations chosen randomly and using 100 ms exposure time and 100 and 250 Electron Multiplying gain values, respectively for SiR-actin and DAAM1-SNAP-Atto488 detection.
TIRF-derived actin fluorescence micrographs were first denoised using rolling ball background subtraction (10 pixels), then contrast-enhanced using the Frangi Vesselness filter65. From the acquired time series, actin images were skeletonized and analyzed using publicly available plugins66,67. The resulting skeleton maps were used as reference locations to quantify the quantity (intensity) of protein bound to the actin fibers. SNAP-tag protein fluorescence images were denoised using rolling ball background subtraction (50 pixels) and the pixel statistics were calculated with CLIJ ImageJ/Fiji plugins68. All the scripts were written in ImageJ/Fiji69, and are available upon request.
Western Blot
Previously snap-frozen tissues or cell pellets were resuspended in RIPA buffer (150 mM NaCl, 1% Nonidet P-40, 1.0 mM EDTA, 1% Sodium Deoxycholate, 50 mM Tris (pH 7.4)). The samples were subjected to sonication (1*10 sec pulse) and left on ice for 10 min. After centrifugation for 5 min at 1100 g, the supernatants were collected and protein quantification was determined using Bradford. Samples were resuspended in 4X SDS loading dye (200 mM Tris 6.8, 400 mM DTT, 4% SDS, 0.2% bromophenol blue, 20% glycerol), proteins were resolved by electrophoresis on a 10% SDS polyacrylamide gel and transferred on a nitrocellulose membrane. Blocking was performed in PBS 0.3% Tween and 5% milk. First antibody and secondary HRP-labeled antibodies were diluted in the blocking buffer at a concentration provided in Table S2. Immunolabeling was detected by enhanced chemiluminescence and visualized with a digital luminescence image analyser Amersham Imager 600 (GE Healthcare).
Co-immunoprecipitation Experiments
HA and 3xFlag tagged constructs were transiently transfected into 293 T cells grown in 6 well plates using Lipofectamine 2000. After 48 hours, cells were harvested in cold phosphate-buffered saline (PBS), and pellets were resuspended in 250 μl of lysis buffer (50 mM Tris pH 7.4, 150 mM NaCl, 0.2% Triton X-100, 0.2% NP-40, and protease inhibitors). Lysates were cleared in a microcentrifuge by spinning at 15,000 × g for 10 min at 4 °C. Lysates were pre-cleared with 15 ul slurry of Dynabeads protein G (Thermo Fisher Scientific) with rotation at 4 °C for 30 min. 5% of pre-cleared lysate was saved as input. 6 ug of anti-Flag M2 antibody (Sigma-Aldrich) was incubated with lysates for 1 hour at 4 °C followed by incubation with 30 ul slurry of Dynabeads protein G washed in lysis buffer for 1 hour at 4 °C with rotation. Following incubation, complexes were washed 5 times with lysis buffer. Elution was performed in 2.5x Laemmli buffer at 95 °C for 5 min. The DAAM1 Co-IP signal was normalized to the IP signal of the FLAG-tagged protein being tested for interaction based on the intensity signal measured using ImageJ/Fiji software.
NanoBRET
Nano Bioluminescence Resonance Energy Transfer (BRET)-based assay was used to detect interactions in the cytoplasm of mammalian cells between reporter-fused proteins51,70. Full-length human RHOA was amplified by PCR and cloned into the pHTC HaloTag® CMV-neo vector (Promega) by Gibson assembly. The Daam1 with exon ( + MIC)-C-term deletion and the Daam1 without exon (-MIC)-C-term deletion were amplified by PCR and cloned by Gibson assembly into the pNLF-N (CMV/Hygro) vector (Promega).
To test for protein-protein interactions between RHOA and DAAM1 with exon (+MIC) - C-term deletion or the Daam1 without exon (−MIC) - C-term deletion, we plated 1.4×105 cells per well in a 24-well plate. 293 T cells were transfected with 500 ng of pHTC-RhoA, 0.5 ng of pNLFN-Daam1, 0.75 μl of Lipofectamine 3000, and 1 μl of P3000 Reagent (Thermo Fisher Scientific). After 20 h, each transfection was re-plated into four wells of a 96-well plate at a density of 1 × 104 cells per well for duplicate control and experimental samples. Protein-protein interactions were analyzed with the NanoBRET® Nano-Glo® Detection System kit (Promega) following the manufacturer’s instructions. The corrected NanoBRET ratio was calculated according to the manufacturer’s instructions. Each protein-protein interaction combination was tested in duplicate wells.
Deletion of Daam1-MIC using CRISPR-Cas9 gene-editing
Mouse Embryonic Stem Cells (mESC, 129xC57Bl/6 background) were kindly provided by Kyung-Min Noh (EMBL Heidelberg) and cultured as described below. mESCs Knock-Out (KO) for the Daam1-MIC were generated using the CRISPR-Cas9 system71 with a double guide RNA strategy72. Each guide RNA targeted one of the two flanking introns and three gRNAs at each side were selected based on the proximity to Daam1-MIC and the quality score provided73. The best gRNA pair was chosen after testing all possible combinations and selecting the best editing efficiency. The primer sequences are provided in Table S1.
Gene editing was performed by transfection of 2 ug of Multiplex CRISPR vector ESCs with Lipofectamine 2000 (Invitrogen, 11668019). mESCs were plated at 2-3 different densities on 100 mm dishes (750,000, 1,500,000 and 3,000,000 cells/dish). Transfection of an empty vector was used as a control. Six hours after transfection, the media was changed to prevent toxic effects. After 24 h, puromycin selection was performed using a concentration of 1.5 ug/ml (Sigma, P8833) for 7 to 10 days. Afterward, individual colonies were hand-picked into 96-well plates, expanded, and genotyped by PCR and Sanger sequencing. Genotyping primer sequences are provided in Table S1. The confirmed KO clones and WT controls were further confirmed at the mRNA level using the RT-PCR primers amplifying Daam1-MIC described above.
Immunofluorescence stainings and confocal imaging of cultured neurons
Neuronal differentiation from mESCs was done following the protocol reported by Bibel et al.39, with slight modifications (see below)38,39. For immunofluorescence assays, cells on DIV0 or DIV21 were washed with phosphate-buffered saline (PBS), fixed in 4% paraformaldehyde in PBS for 10 min, permeabilized in 0.3% Triton X-100 in PBS for 10 min, blocked for 1 h in 0.3% Triton X-100, % bovine serum albumin (BSA) in PBS and incubated in primary antibodies at 4oC overnight with shaking. Following this, cells were incubated with the corresponding secondary antibodies (see Table S2) for 1 h at room temperature and mounted in FluoroShield with DAPI (Sigma, F6057-20ml) for imaging. Images were taken on an SP8 confocal microscope (CRG, Advanced Light Microscopy Unit) using identical settings for each condition in a given experiment. Briefly: a dry 20X objective was used to image the whole span of the neuronal culture for neurite length analysis (Fig. 3c; DIV0 + 4 h´s)(17-23 µm, Z-step size 1 µm, zoom factor 2.5), for the dendrite/axon proportion analysis using MAP2/Tau1 markers (Fig. S4h, i; DIV21) (17-23 µm, Z-step size 1 µm, zoom factor 1). For filopodia analysis, a 63X oil-immersion objective was used to image the whole span of the protrusion (Fig. S4d–g; around 2 µm, Z-step size 0.12 µm, zoom factor 2.5). Confocal sections were Z stack projected with maximum intensity selection and analyzed in ImageJ/Fiji software. Neurite and filopodia lengths were analyzed manually using the Segmented Line plugin.
Measurement of intracellular [Ca2+] in cultured neurons
Cytosolic Ca2+ signal was determined at room temperature in cells loaded with 4,5 mM FURA-2AM (30 min incubation at 37 °C). Ca2+ responses were calculated as the ratio of emitted fluorescence (510 nm) after excitation at 340 and 380 nm, relative to the ratio measured before cell stimulation (FURA-2AM ratio 340/380). Briefly, cells were maintained in an isotonic solution containing (in mM): 140 NaCl, 5 KCl, 1.2 CaCl2, 0.5 MgCl2, 5 glucose, 10 HEPES (305 mosmol/l, pH 7.4 with Tris) for 2 min and then stimulated with a 30 mM KCl isotonic solution (115 NaCl, 30 KCl, 1.2 CaCl2, 0.5 MgCl2, 5 glucose, 10 HEPES) to activate voltage-gated calcium entry. As indicated in the respective figure legends, cells were treated with 5 µM Latrunculin A (LatA), 50 µM small-molecule inhibitor of Formin Homology 2 domains (SMIFH2) or vehicle (DMSO). The treatment was maintained for the experiment (40 min). In the case of SMIFH2, its effect on neuronal cultures was measured after 90 min of exposure to 50 µM SMIFH2. Y-27632 ROCK inhibitor was used at a concentration of 10 µM, and the effect was measured immediately. All experiments were conducted at room temperature as described previously74. AquaCosmos software (Hamamatsu Photonics) was used for capturing the fluorescence ratio at 510 nm obtained post-excitation at 340 and 380 nm, respectively. Images were computed every 5 sec. Measurements were processed using SigmaPlot 10 software.
F-actin imaging in dendritic spines using STORM
For STochastic Optical Reconstruction Microscopy (STORM) assays, DIV18 in vitro differentiated glutamatergic neurons or hippocampal primary cultures were washed with phosphate-buffered saline (PBS), fixed in 4% paraformaldehyde in PBS for 10 min, permeabilized in 0.3% Triton X-100 in PBS for 10 min, blocked for 1 h in 0.3% Triton X-100, % bovine serum albumin (BSA) in PBS and incubated in primary antibody PSD95 (1:500 dilution) at 4oC overnight. Following this, cells were incubated with the corresponding secondary antibody and Phalloidin (1:100)(see Table S2) for 1 h at room temperature and imaged within 24 hours. The complete STORM protocol can be found in Martin et al. STAR protocol75.
STORM imaging acquisition
STORM imaging was performed using an N-STORM 4.0 microscope (Nikon) with an iXon Ultra 897 camera (Andor) and a CFI HP Apochromat TIRF ×100 1.49 oil objective under highly inclined and laminated optical sheet (HILO) illumination mode. NIS element 5.21 software was used for image acquisition. Conventional fluorescence images were taken at the beginning of each imaging cycle to register the positions of PSD95 and actin, acquired with a 460 nm LED and FITC filter and with a 647 nm laser and Quad-band filter, respectively. Continuous imaging acquisition of actin was performed through 647 nm illumination of the sample at a constant power density of ~2 kW/cm2, with 10 ms exposure time for 60000 frames using a Quad-band filter. Super-resolution images were taken with the addition of an imaging buffer to the sample: 100 mM cysteamine MEA (Sigma-Aldrich, #30070), 1 % Glox solution (0.5 mg/ml glucose oxidase, 40 mg/ml catalase; Sigma-Aldrich, #G2133 and #C100), 5 % glucose (Sigma-Aldrich, #G8270) in PBS. Neuronal cultures grown on 13 mm coverslips were mounted on a microscope slide (Thermofisher Scientific, #J1830AMNT) with 5 μl of imaging buffer and sealed with nail polish.
STORM image analysis
Super-resolution images were analyzed and rendered in Insight376 (a kind gift of Bo Huang, UCSF) as described77,78. Specifically, point spread functions (PSFs) from the emission from single fluorophores were identified at individual frames of the acquired videos, based on a set intensity threshold, and were fit to a two-dimensional Gaussian. From it, the centroid of the PSF was obtained, and the x and y positions for 2D STORM localization lists were obtained. Sample drift correction was calculated and subtracted by cross-correlation drift correction in Insight3. STORM images were rendered by representing each localization (i.e., x-y positions) as two-dimensional Gaussians with fixed width. Conventional PSD95 images and super-resolution actin images were opened in Fiji (ImageJ) and merged to identify active dendritic spines based on the colocalization of the two signals. Circular ROIs of the same size (502.4 nm2) were drawn at active dendritic spine locations, and the number of localizations was extracted. Plots and statistical tests were performed with R version 4.3.1. A paired Mann-Whitney test was performed.
Mouse embryonic stem cell culture
mESC were grown on 0.1% gelatine-coated (Millipore, ES-006-B) plates (Thermo Scientific Nunc Cell-Culture Treated Multidishes, 140675) with mESC media containing 10% fetal bovine serum (FBS) and leukemia inhibitory factor (LIF). Gelatine coating was performed for at least 5 min before plating the cells. mESC media was generated by the CRG Tissue Engineering Unit and consisted of Glasgow’s Minimum Essential Medium (GMEM) BHK-21 (Gibco, 21710-025) supplemented with 10% fetal bovine serum (Seralab, A1060013 EU-000-H), Minimum Essential Medium non-essential amino acids solution (Gibco,11140-050), 1mM L-Glutamine (Gibco, 25030-024), 0.5 mM Sodium Pyruvate (Gibco, 11360-070), 0.1 mM 2-Mercaptoethanol (Millipore, ES-007-E) and recombinant mouse LIF protein 1000U/ml ESGRO (Millipore, ESG1107). mESCs were routinely passaged using 1x TrypLE Express Enzyme (Gibco, 12605028).
Neuronal differentiation
We followed the protocol reported by39 with slight modifications. In brief, mESCs were harvested by trypsinization with TrypLE Express for 5 min at 37 °C (HeracellTM 240i CO2 Incubator). Trypsin was quenched with an equal volume of Embryoid Body (EB) medium (10% FBS, 1% Non-Essential Amino Acids, 1% Penicillin and Streptomycin, 1% GlutaMax, 1% Sodium Pyruvate, 0.1% B-mercaptoethanol, 86% DMEM High Glucose) and cells were counted manually using a hemocytometer chamber. mESCs were plated at a density of 4×106 cells per low attachment bacteriological petri dish (10 cm Ø) in 15 ml of EB medium. This marks the start of the experiment and is further referred to as Day In Vitro -8 (DIV-8). Cells were cultured at 37 °C with 5% CO2. On day 2, the medium with the Embryoid Bodies (EB) was transferred to the falcon tube, and after 5 min, the supernatant was aspirated, and pelleted EBs were resuspended in fresh EB medium (DIV-6). Resuspended EBs were dispensed into new bacteriological Petri dishes (10 cm diameter) in EB medium volume up to 15 ml per dish and incubated as earlier. On days 4 (DIV-4) and 6 (DIV-2), the medium was changed as before but using EB medium supplemented with 5 μM retinoic acid. On day 8 (DIV0), EBs were collected as before and washed twice with 10 ml of PBS. Subsequently, EBs were resuspended in 1 ml of medium (0.05% Trypsin, 0.05% EDTA dissolved in PBS) and incubated with constant shaking for 3 min in a 37 °C water bath. Trypsinized EBs were quenched with 1 ml of EB medium, and cells were pelleted down using an Eppendorf 5810 R Centrifuge (180 g for 5 min). The supernatant was aspirated, and the cells were resuspended in 5 ml of N2 medium and filtered through a 40μm cell strainer. The cells were counted as before and plated at the density of 1.5×105 per 13 mm glass coverslip (VWR 631-1578) placed in 24 well plates (Sigma Aldrich, 11243217001) coated with poly-D-Lysine followed by laminin (Roche). Cells were cultured as described above in the incubator set up at 37°C with 5% of CO2. After 2, 24 and 48 h from plating, the N2 medium was changed. Consecutively, media was changed to B27 after 72 h, and fresh B27 media was provided every second day. Neuronal differentiation was conducted up to DIV23.
Primary cultures of mouse hippocampal neurons
Hippocampal neurons were isolated from 18-day-old Daam1-MIC KO or WT mouse embryos. This procedure was approved by the Ethics Committee of the Institut Municipal d’Investigacions Mèdiques-Universitat Pompeu Fabra. Mice were euthanized using carbon dioxide. The hippocampi were aseptically dissected and tripsinized. Cells were seeded in DMEM high glucose (Gibco) supplemented with 10% horse serum and 1% penicillin/streptomycin into poly-D-Lysine (Sigma) coated 13 mm coverslips (VWR) in 24-well plates (SPL Life Sciences), at a confluence of 100,000 cells per well. After 120 min, the medium was removed and neurobasal medium (Gibco) was added, containing 2% B27 supplement (Gibco), 1% penicillin/streptomycin, and 1% GlutaMAX (Gibco). On day three of culturing, cells were treated with 1.5 µM 1-ß-D-arabinofuranosylcytosine (Sigma, San Luis, MO, USA) for 24 h to eliminate proliferating non-neuronal cells. Hippocampal neurons were used for the experiments on day 18.
Generation of mouse Daam1-MIC KO mice
Chimeric mice were obtained by blastocyst injection of one of the Daam1-MIC KO mESC lines into B6 albino (B6(Cg)-Tyrc-2J/J) embryos, which were then transferred to pseudopregnant CD1 females. Chimeric males were subsequently crossed to albino B6 females, and all non-albino mice were genotyped to select Daam1-MIC KO mice. These were backcrossed four times to C57Bl/6 J and heterozygous mice were then crossed to generate Daam1-MIC KO and wild type (WT) littermates for experiments. The colony was maintained at the Animal Facility of the Barcelona Biomedical Research Park (PRBB). All procedures were approved by the PRBB Animal Research Ethics Committee and the Generalitat de Catalunya and were carried out in accordance with the guidelines of the European Union Council (2003/65/CE) and Spanish regulations (BOE 252/34367-91, 2005). WT mice were purchased from Charles River (references: 709 for albino B6, 022 for CD1 and 632 for C57Bl/6).
In Utero Cortical Electroporation and dendrite analysis
Cortical electroporation was performed as in ref. 79 with minor changes. Timed-pregnant mouse females were anesthetized with isoflurane, the abdomen cut open, and the uterine horns exposed. E15 wild-type embryos were unipolar injected into the ventricle using a pulled glass micropipette containing a DNA solution (1 μg/μl CAG-GFP plasmid (11150, Addgene) with 0.03% Fast Green in PBS). The head of each embryo was placed between tweezer-type electrodes (CUY650-P5 Nepa GENE, Chiba, Japan), and five square electric pulses (50 milliseconds) at 35 V were passed at 950 msec. intervals using an electroporator (CUY21E, Nepa GENE). Then, the electroporated brain cortex of PD4 mice were analyzed. Females were anesthetized with isoflurane and euthanized cervical dislocation. PD4 mice were perfused with 4% PFA, and brains were cut into 70 µm slices using a vibratome device. GFP-expressing neurons and their projections were visualized by immunohistochemistry with chicken anti-GFP antibody (Aves labs) and anti-chicken Alexa-488 secondary antibody (Invitrogen, Molecular Probes). 20x images were taken with Leica SPEII confocal microscope (two ROIs from 3 WT and 3 Daam1-MIC) mice, and the length of the primary dendrite was measured manually using FIJI software as a complexity measurement derived from 78. Ethical approval number EH2023.0159.P1.M.
Behavioral and locomotor tests in neonatal mice
A battery of behavioral and motor tests to probe early post-natal neurodevelopment was performed as described in refs. 43,44, with some adjustments. In particular, we performed the following tests at PDs 4, 7, 10, and 14, unless stated otherwise:
Pivoting and walking
Pivoting is a voluntary exploratory behavior displayed by young mice before forward locomotion. To measure pivoting, mice were placed on a flat surface and allowed to move freely for 1 min. The number of times the animals made a 90° turn was recorded. Pivoting was measured based on the body axis with the help of a cross marking 90° angles on the experimental surface. The latency to walk in a straight line after the end of the pivoting behavior was also measured. Two trials were performed per animal.
Righting reflex
Mice (PD4 or PD7) were placed on their back on a padded table top. The time taken for the animals to right themselves back to four paws through 180° was measured for a maximum of 1 min. The direction of turning was also recorded. The experiment was performed three times per animal, and the mean was calculated.
Preyer’s reflex
The Preyer’s reflex is a startle response triggered by sharp auditory stimuli and is used to assess hearing in rodents80. The experimenter made a sharp clapping sound by stretching and releasing a rubber glove onto the hand (around 110 dB) in the proximity (<5 cm) of the mouse and the contraction of the post-auricular muscles and a rapid startle movement of the whole body, is recorded.
Front-limb suspension
To measure forelimb strength, mice were allowed to grasp with both forepaws on a horizontal bar suspended above a padded drop zone. The latency to fall was measured in three trials (30 s each) per animal and the mean was calculated.
Hindlimb suspension
To measure hindlimb strength, mice were lowered into a 50 ml conical tube and released with their hindlimbs hung over the rim. The latency to fall was measured for up to 30 s. The posture of the hindlimb upon falling was scored 0-4 based on the limb spread, as43 described. The experiment was performed only once unless the animal fell immediately due to bad placement.
Grasping reflex
The animals were held by the scruff of the neck and each paw was touched by a toothpick to elicit the grasping reflex. Performance was scored 0-4, assigning one point per paw in the presence of grasping. Left and right paw preference was also noted.
Cliff aversion
Mice were placed on top of a box elevated ~10 cm above the surface, with their snout and part of their forepaws just over the edge. The presence of aversive movement away from the cliff within the subsequent 30 s was recorded. If the pup fell, one additional trial was performed.
Negative geotaxis
Negative geotaxis is an automatic vestibular response to geogravitational stimuli and is used to measure motor coordination in pups. Mice were placed head-facing uphill on a plastic platform with an inclination of 45° covered with Surface Protector Paper and spunlace wipes (VWR), except for PD 4 mice, for which the incline was adapted to 30°. After ~5 s, the pups were turned by 180° to face downhill, and their movement was observed for 1 min. Two trials were performed, turning the animal in opposite directions to the start position to avoid left-right bias. Animals were given 0.5 points per 45° of turning. The latency was recorded if a full 180° turn was performed. Left and right turning preferences were also noted.
Homing test
Homing was performed as described in ref. 44 at PD14 with adjustments. A pup is removed from the home cage and is placed in the corner of a new clean cage (12.5 × 45 cm, w × h) facing the wall. The new cage is filled with clean wood shavings, and familiar nesting material from its home cage is provided in the opposite corner. The latency for the pup to reach the area containing the nesting material is recorded in s, with a maximum of 2 min
Behavioral tests in adult mice
We also performed a battery of behavioral and locomotor tests in adult mice. Each test was performed twice, using six animals per sex and genotype in each replicate. All mice were between 2 and 5 months old and siblings were matched when possible.
Spontaneous basal locomotor activity in adult mice
Spontaneous basal locomotor activity during day and night in an open cage was measured for 23 h using an infrared Actimeter (PANLAB SA, Spain). Individual mice were placed in the open field (25 × 25 cm), and their position in time was recorded based on the disruption of infrared beams in the x and y axes. The arena was covered with a layer of wood shavings, and sufficient water and food were provided.
Novel Object Recognition (NOR) test
The NOR test measures recognition memory using the visual paired-comparison paradigm47,81. The experiment consists of three sessions on consecutive days: habituation, familiarization, and discrimination. The experiment was performed in a 38.5 cm × 38.5 cm arena with 38.5 cm high dark plastic walls and an open top. The arena was illuminated from the top, and the animals were tracked using video recording and the Smart 3.0 software (System Motor Activity Record and Tracking, PANLAB SA, Spain). A curtain was mounted to minimize stressors interfering with the experiment, separating the arena from the experimenters. The arena and the objects were cleaned with 70% ethanol between each mouse to remove odors. Cages were transferred to the experimental room 30 min before the trials to acclimate. The test was conducted under slightly aversive conditions (white light 50 lux). The experiment consists of three sessions: habituation, familiarization, and discrimination (lengths of 5 min, 10 min, and 5 min, respectively). Each session was separated by 24 h periods.
Habituation
On the first day, animals were placed into the arena facing the wall and were recorded for 5 min. Their movement was measured separately by the Smart 3.0 software in a central (20 × 20 cm) and a peripheral zone.
Familiarization
On the second day, two identical objects (A and B) were placed in the centre of the box, 18 cm apart. Animals were placed into the box facing the wall and were recorded for 10 min. The time spent exploring each object was measured manually using a timer. The latency to climb on top of the objects was also noted. The exploration was defined as the time spent sniffing the object from a close distance, excluding the time spent climbing on the object. Minimum exploration time was established at 20 sec.
Discrimination
On the third day, one of the objects was replaced with a novel object of different shape and color. Animals were placed into the box facing the wall as before and were recorded for 5 min. The time spent exploring each object was measured manually using a timer. The latency of climbing on top of the old and novel objects was noted.
Discrimination and preference indices were calculated for familiarization and discrimination for each animal based on the manual exploration time recordings, as described in ref. 47. The discrimination index corresponds to: (Novel Object Exploration Time - Familiar Object Exploration Time)/ Total Exploration Time.
Elevated Plus Maze test
Anxiety-related behavior was measured in the Elevated Plus Maze, as described by82, with some modifications. The apparatus consisted of a cross-shaped platform with four arms (30 cm × 5 cm), elevated 40 cm off the ground. Two arms were open and two were enclosed by 15 cm high black methacrylate walls. The open arms create an aversive environment for the mouse, while the dark, enclosed space of the closed arms is considered safe. The number of open-arm entries and the relative time spent in the open arms are indicative of anxiety-related behavior. Animals that avoid open arms are considered more anxious. Mice were lowered onto the end of the same closed arm facing the walls, and their movement was recorded for 5 min using the Smart 3.0 software. Separate zones were set up for each arm, the ends of the arms (5 cm × 5 cm), and the center of the cross (5 cm × 5 cm). Measurements included the percentage of the time, entries, distance traveled, and average speed in each zone. Rearings and head dippings from the central zone were recorded manually. The arena was cleaned with 70% ethanol between each animal’s experiment to remove odors.
Morris water maze test
Learning and visual-spatial memory were tested in the Morris water maze83, with some modifications48. In this experiment, mice must learn the spatial location of a submerged platform to escape from the water by building a cognitive allocentric map with the help of external visual cues. A metal tank (150 cm in diameter) was filled with water (22-23 °C) with added white, non-toxic finger paint to conceal the platform’s location. The platform (12 cm in diameter) was submerged 0.7 cm below the water surface. Four quadrants (NE, NW, SE, SW) and a platform zone were defined using the Smart 3.0 software, with the platform located in the center of one quadrant (NE). A curtain was mounted around the tank to separate it from the experimenters and minimize external visual cues. Three distal visual cues, a square, a triangle, and a circle, were placed on the curtains around the tank at equal distances, ~30 cm above the water surface. The experiment consists of the following phases: training trials, removal, cued trials, and reversal training.
Training trials
During training, the mice learn the task and the location of the platform. Four 1-min training trials were performed each day, with an inter-trial interval of ~1 h. Nine training days were performed to achieve sufficient learning. Mice were released into the water. The latency to reach the platform was recorded and the animal was removed from the water. 5 to 15 s platform localization learning and positive reinforcement was used by placing the animal on the platform depending on performance (goal accomplishment or lack of it).
Removal
The platform is removed from the tank and the animals are tested for 1 min 24 h after the last training. Animals were released from the location furthest away from the original platform (SW). Mice that remember the location of the platform are expected to spend more time in the platform quadrant (NE).
Cued trials
In these guided learning trials, the platform is placed back to its original location and is marked by a local visual cue. Animals have an inherent tendency to swim towards the flag. Therefore, these trials can detect issues with swimming, visual perception and/or motivation to perform the task. A flag ( ~ 10 cm above the water surface) fixed to a metal bar was used as a local cue. Distant visual cues were removed. Two 1-min cued trials were performed ~1 h after the removal session.
Reversal training
During a spatial reversal, the platform is placed opposite to its original location (SW). Mice must flexibly re-learn the spatial location of the platform. Four trials were performed for 2 days, as described in the training trials. In all trials, the latency to reach the platform and platform crossovers were measured. Animals were tracked using the Smart 3.0 software, allowing measurement of time, speed and distance traveled in each quadrant. Distances from the platform were used to calculate cumulative search error and mean proximity as additional measurements of spatial learning84. Floating was also measured manually.
Grip strength test
The grip strength experiment is a measurement of neuromuscular function and limb muscle strength. Mice were held by the tail and lowered onto the metal grid of the apparatus (Grip Strength Meter, Bioseb, France), allowing them to grip with their paws. Mice were next pulled backwards along the grid at a consistent speed, while the apparatus measured the force exerted by the animals on the grid. Grip strength was measured for forelimbs through 3 consecutive trials. Best, average, and mean grip strengths were calculated.
Rotarod test
To evaluate coordination and balance, we employed the Rotarod apparatus (PanLab Rotarod LE8200, Spain) based on85. First, animals were trained to walk on the rod at 4 rpm. Training sessions were performed until the mouse managed to walk on the rod for 1 min. After training, test sessions were performed at five constant speed settings (7, 14, 19, 24, 34 rpm). Two trials were carried out per speed, with a maximum length of 2 min. Finally, two trials were performed where the rotation speed accelerated constantly from 4 to 40 rpm in 60 s.
Beam Balance test
Balance, coordination, and vestibular function were measured in the Beam Balance experiment86. Mice were placed in a standing position in the center of a narrow wooden beam (1 cm × 50 cm) elevated 45 cm above the ground. They were scored 0-3 based on how far they walked on the beam within 1 min (0: falls off; 1: <10 cm; 2: >10 cm; 3: reaches the end). Falls and the number of slips were also recorded, as well as the latency to reach one of the ends and the direction of movement (left/right).
Functional synapse quantification in mice hippocampus
7 control mice and 8 Daam1-MIC mice of 22 days postnatal (PD22) were anesthetized with isoflurane and euthanized by perfusion for 5 min with 4% paraformaldehyde, and brains were transferred to Phosphate Buffered Saline immediately. Brains were cut in 70 µm sagittal slices, and equivalent slices between animals were selected for immunohistochemistry. Slices were placed in a blocking/permeabilization solution containing 5% horse serum and 0.25% Triton X-100 in PBS for 1 hour, followed by immunostaining with primary antibodies (Table S2) two days at 4 °C. Slices were washed 4-5 times in PBS before staining with secondary antibodies (Table S2) overnight at 4 °C. Slices were labeled with 4′,6-diamidino-2-phenylindole (DAPI) (Sigma-Aldrich, D9542) for 5 min and washed 3-4 times in PBS before being mounted in Mowiol mounting media (Millipore, 475904-M).
CA1 hippocampus was imaged with a 63x oil immersion objective on a Leica SPEII confocal microscope. ROIs with a z-stack of 5 µm and a z-step size of 0.2 µm were captured. Three ROIs per slice, and three slices per animal were analyzed with Imaris software. Images were processed with Background subtraction and Median filter for both channels, followed by segmentation with Imaris models. To quantify the number of functional synapses the “Spots Close To Surface XTension” of MatLab was applied with a threshold of 0.5 µm. Daam1-MIC results were normalized to control mean and unpaired Mann-Whitney nonparametric test was performed for statistical analysis.
Immunohistochemical imaging of mice hippocampus
Mouse hippocampal sections were prepared as previously described87. In brief, mice were euthanized with CO2 and perfused transcardially with 0.1 M PBS, followed by 4% paraformaldehyde in PBS until tissues were completely cleared of blood. Fixed brains were extracted and stored in 4% paraformaldehyde at 4 °C for 24 h, and in sucrose 30% with 0.01% azide in PBS for the following 24 h. Prepared tissues were cut into 40 µm coronal sections, in serial order throughout the dorsal hippocampus (Bregma sections between -1.34 mm to -2.54 mm). Sectioning was performed by an in-house Tissue Engineering Unit (CRG). Immunohistochemistry was done by tissue permeabilization in 0.5% Triton X-100 in PBS for 15 min × 3 times, blocked for 2 h in 10% normal goat serum (NGS) in PBS albumin and incubated primary antibodies in 5% NGS, Tween 0.5% in PBS at 4oC overnight. Following this incubation, cells were incubated with the corresponding secondary antibodies (see Table S2) for 2 h at room temperature. Washing steps were repeated (for 15 min × 3 times) and sections were mounted in FluoroShield with DAPI for imaging. Images were taken on a confocal microscope (SP8 Leica; CRG, Advanced Light Microscopy Unit) using identical settings for each condition in a given experiment with a dry 20X objective. A single middle plane of each section was imaged and the images were analyzed in ImageJ/Fiji software. DG size was marked manually with Polygon selections using DAPI stained nuclei as a region of reference marker and consecutively quantified with the Measure plugin. Immediate early genes positive nuclei were counted using the Analyze Particles plugin (nuclei size at least 25 pixels), and normalized to DAPI positive nuclei. A detailed list of all stainings and primary antibodies used is provided in Table S2.
Rescue experiments with ROCK inhibitor in adult mice
The ROCK inhibitor trans-4-[(1 R)-1-aminoethyl]-N-4-pyr-idinylcyclohexanecarboxamide (Y-27632; HelloBio HB2297) was dissolved in physiological saline at a concentration of 1 mg/ml and injected for three consecutive days intraperitoneally at a dose of 10 mg/kg88. The equivalent volume of saline solution was injected into the control mice. Three consecutive injections of ROCK inhibitor or vehicle were applied one week before the NOR at days -9, -8, and -7. On the 7th day post-injection, NOR experiments were performed as described above. NOR test was performed twice, using six animals per treatment and genotype in each replicate. Replicates were separated by sex, 24 animals per replicate.
Electrophysiology ex vivo
Mice (8–16 weeks old) were decapitated immediately, the brain was quickly removed and submerged in artificial cerebrospinal fluid rich in aCSF sucrose buffer (2 mM KCl, 1.25 mM NaH2PO4-H2O, 7 mM MgSO4, 26 mM NaHCO3, 0.5 mM CaCl2, 10 mM glucose and 219 mM sucrose) at 4 °C, saturated with a 95% O2, 5% CO2 mixture and maintained at pH 7.32-7.4. Transverse brain slices (300 µm thick) were cut with a vibratome (VT1200S, Leica) in oxygenated aCSF sucrose at 4 °C and transferred to a recovery chamber with oxygenated aCSF buffer 124 mM NaCl, 2.5 mM KCl, 1.25 mM NaH2PO4-H2O, 1 mM MgSO4, 26 mM NaHCO3, 2 mM CaCl2 and 10 mM glucose and incubated for > 1 h at room temperature (21-24 °C) and pH 7.32-7.4. Individual slices were transferred to an immersion recording chamber and perfused with oxygenated aCSF at 2 mL/min (30 ± 2 °C).
Whole-cell intracellular recordings in voltage clamp (VC) and current clamp (CC) mode were performed in pyramidal neurons in CA1 stratum pyramidale. Cells were visualized with a water-immersion 40x objective. Patch electrodes were fabricated from borosilicate glass capillaries (P1000, Sutter Instrument) with a resistance of 4–6 MΩ when filled with the internal solution that contained: 130 mM K-MeSO4, 10 mM HEPES, 0.5 mM EGTA, 2 mM MgCl2, 4 mM Mg-ATP, 0.4 mM Na-GTP, 10 mM phosphocreatin di (tris) salt and 0.3 % biocytin, for membrane properties experiments in CC and excitatory postsynaptic currents (EPSC) evoked in VC, in pyramidal neurons; 130 mM Cs-MeSO4, 5 mM CsCl, 10 mM HEPES, 0.5 mM EGTA, 2 mM MgCl2, 4 mM Mg-ATP, 0.4 mM Na-GTP, 10 mM phosphocreatin di (tris) salt and 0.3 % biocytin, for miniature excitatory postsynaptic currents (mEPSC) in VC recordings from pyramidal neurons. All pipette solutions were adjusted to pH 7.2-7.3 with K-OH or Cs-OH. Membrane currents and voltages were acquired with Multiclamp 700B amplifiers, digitized (Digidata 1550B), and controlled by pClamp 10.7 (Molecular Devices Corporation, Sunnyvale, CA, USA) software. Membrane intrinsic properties of CA1 pyramidal cells were determined by passing hyperpolarizing and depolarizing current steps (1 s, with 10 pA increments from -100 to 140 pA) in CC.
Synaptic responses in CA1 were evoked by monophasic current (50 μs duration) stimulation of the Schaffer collateral fibers (SCs) with an extracellular bipolar tungsten electrode via an isolated current stimulator (DS3) that was set to deliver monophasic currents of 50 μs duration. Membrane currents and voltages were acquired with Multiclamp 700B amplifiers, digitized (Digidata 1550B), and controlled by pClamp 10.7 (Molecular Devices Corporation, Sunnyvale, CA, USA) software. To study changes in the probability of transmitter release of the presynaptic cell (e.g., Bekkers and Stevens 1990; Kullmann 1994; Malinow and Tsien 1990) we applied paired-pulse facilitation (PPF) protocol, which consisted of evoking two consecutive EPSC responses at different intervals (from 20 ms at 160 ms, a 20 ms inter-pulse interval). Changes in the PPF were calculated as a PPF ratio from (R2-R1)/R1, where R1 and R2 were the peak amplitudes of the first and second EPSCs, respectively (Martin and Buño, 2002). Extracellular field postsynaptic potentials (fEPSPs) were recorded by placing a borosilicate glass electrode filled with aSCF sucrose buffer in the stratum radiatum (SR) of the CA1 pyramidal layer. Evoked fEPSPs were elicited by stimulation of the SCs fibers as EPSCs.
For LTP experiments, input/output relationship was done for each slice recording in the beginning of LTP experiment. We stimulated SC from low intensity (20 µA) to values where we reach a maximum value of the slope (150 µA) in increments of (10 µA). The stimulus intensity was adjusted to elicit 50% of the maximum response signal and kept constant throughout the experiment. Data was stored through an acquisition system (Axon Instruments) and the software pClamp 10.7 was used to display fEPSP and measurements of fEPSP slopes. After recording stable baseline responses for 30 min, LTP was induced by a single train of theta burst stimulation (TBS; 10 bursts of 5 pulses at 100 Hz, with an interval of 200 ms between bursts). Potentiation was measured for 1 h after LTP induction at 0.033 Hz. Changes in the fEPSP slope were calculated in relation to the baseline fEPSP responses during the last 10 min before TBS, and the time course of LTP values was then normalized to this baseline.
Dendritic spine morphology analysis
For spine analysis, neurons were filled with biocytin (Sigma-B4261) during whole-cell recordings. Slices (300 µm) with recorded cells were fixed overnight with 4% paraformaldehyde in PBS at 4 °C and then transferred to 0.05 % Na-azide in PBS. After slices were permeabilized and blocked with 0.3% Triton X-100 in PBS and 10% normal goat serum (NGS) for 1 h at room temperature. To reveal biocytin, the slices were incubated overnight at 4 °C with streptavidin Alexa Fluor-488 conjugated (1:1 000) in 0.3% Triton X-100 in PBS. Confocal microscopy (Leica SP8; CRG, Advanced Light Microscopy Unit) was used to capture fluorescence Z-stacks of dendritic spines from biocytin-filled cells with a 0.2 µm step using a 63X glycerol immersion objective. Only dendrites with bright and continuous labeling were included for analysis. Primary dendrites were not included in the analysis. Huygens’ essential software was used to deconvolve images. All images were batch processed using the same template (available upon request). Deconvoluted images were imported into NeuronStudio and semi-automatically analyzed blind to experimental conditions. Dendritic spine quantification was performed on a total of 23 CA1 pyramidal cells (n = 11-12 cells per group; n = 2 groups). Spine density was measured at least in 100 µm of secondary apical branches. All dendritic spines were at least 50 µm apart from the neuronal soma. The total number of spines was divided by the total length of the dendritic spines.
RhoA2G FRET biosensor analysis of neuronal cells by microscopy
To obtain lentivirus for infection of neurons, HEK293T cells were seeded in lentivirus packaging media (94.99% Opti-MEM I, 5% FBS,0.01% Sodium Pyruvate) were transfected according to manufacturer instructions using Lipofectamine 2000 with plasmids carrying HIV-1 Gag/Pol (pMDLg/pRRE Addgene: 12251), HIV-1 Rev (pRSV-Rev Addgene: 12253), and VSV g-glycoprotein Env (pMD2.G Addgene: 12259), together with pLentiRhoA2G (Addgene: 40179). Media was changed after 6 h to neuronal N2 media. The viral particles were harvested from the neuronal N2 media 48 hours after the transfection and used for cell infection. The experiment was performed on two separate neuronal differentiations considered as replicates. Each differentiation was performed using three KO and three WT cell lines (described before), and fixed DIV21 neurons were used for FRET analysis. The procedure was performed following safety rules and approved by the CRG Biosafety committee (procedure registration number CBS19_015_A).
Relative FRET was calculated automatically using AcceptorPhotobleaching (confocal microscope SP8 Leica; CRG, Advanced Light Microscopy Unit) by dividing the normalized fluorescence intensity of Venus at its emission peak (528 nm) by the normalized fluorescence intensity of mTFP1 at its emission peak (492 nm). Provided Lookup Table (LUT) is linear and covers the full range of the data.
RNA-sequencing and transcriptomic analyses
In vitro differentiated glutamatergic neurons (DIV21) were exposed to 10 µM Y-27632 ROCK inhibitor or vehicle (water) for 2 hours, in triplicates. Next, the cells were collected by scraping and snap-frozen in liquid nitrogen. These samples were subjected to RNA extraction using the RNeasy Plus Mini kit (Qiagen, 74136). Library preparation (polyA-selected, stranded) and RNA sequencing were performed by the CRG Genomics Unit following the standard Illumina protocol. An average of 35.3 million 50 bp single-end reads were generated for each library on a NextSeq 2000 machine.
Reads were aligned to the Mus musculus transcriptome assembly from Ensembl (release 104; GRCm39) with Salmon v.1.4.089. Raw gene counts and TPMs were extracted from Salmon outputs using the tximport90. To identify mis-regulated genes in the KO that were “rescued” upon ROCK inhibition, we performed the following steps. First, differential gene expression between Daam1-MIC KO and WT samples independently of the treatment was calculated using DESeq2 on the raw reads91 with a model for genotype and treatment; p-values were corrected using the Independent Hypothesis Weighting (IHW)92. Second, we again used DESeq2 to independently compare gene expression profiles from each condition against the control KO one (i.e., KO exposed to water [Veh]). Raw p-values from Wald Test from the three independent comparisons (WT-Veh vs KO-Veh, WT-ROCKi vs KO-Veh, and KO-ROCKi vs KO-Veh) were combined into a single p-value with the Stouffer method, weighting double the p-values from the comparison between inhibitor and vehicle in KO samples (KO-ROCKi vs KO-Veh), to emphasize the response of the KO samples to the inhibitor on gene expression. Next, we labeled the genes differentially expressed independently of the treatment as “non-rescued” and removed them from subsequent analyses. P-values of the remaining genes were then corrected with IWH92 using the mean gene TPMs as covariates. Genes with a combined adjusted p-value lower than 0.01 were then considered to present a rescue pattern since their expression differs in the control KO without the inhibitor (KO-Veh) compared to the rest of the conditions. Gene Ontology term enrichment for rescued genes was calculated using the Bioconductor Clusterprofiler package93. Lastly, we used pathview94 to visualize log2 fold changes on genes in KEGG pathways.
Data visualization
In all panels with boxplots, the boxes represent the interquartile range of the plotted values from the 25th percentile to the 75th percentile (first and third quartiles), the line in the box represents the median, and the whiskers represent the minimal and maximal values. Outliers are not displayed. In the case of independent experimental replicates (described in the figure legend), the additional points are displayed (by different shapes) where the line connecting the data points describes the trend relationship during a single experiment.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The RNA-seq data generated in this study have been deposited in the Gene Expression Omnibus (GEO) database under accession code GSE219244. All the other data supporting the findings of this study are available within the article and its supplementary information files. Source data are provided with this paper. Previously published resources have been used in this study, including: - GSE112600. - SRP149913. - GSE180781. - 2Z6E Source data are provided with this paper.
Code availability
The code and software sources from previously published algorithms used to perform the analyses are detailed in the Methods section. Custom scripts employed to generate boxplots and other basic graphics are available upon request.
References
Cingolani, L. A. & Goda, Y. Actin in action: the interplay between the actin cytoskeleton and synaptic efficacy. Nat. Rev. Neurosci. 9, 344–356 (2008).
Dillon, C. & Goda, Y. THE ACTIN CYTOSKELETON: Integrating form and function at the synapse. Annu. Rev. Neurosci. 28, 25–55 (2005).
Papandréou, M.-J. & Leterrier, C. The functional architecture of axonal actin. Mol. Cell. Neurosci. 91, 151–159 (2018).
Lamprecht, R. Actin cytoskeleton role in the maintenance of neuronal morphology and long-term memory. Cells 10, 1795 (2021).
McLeod, F. & Salinas, P. C. Wnt proteins as modulators of synaptic plasticity. Curr. Opin. Neurobiol. 53, 90–95 (2018).
Zhang, H., Ben Zablah, Y., Zhang, H. & Jia, Z. Rho signaling in synaptic plasticity, memory, and brain disorders. Front. Cell Dev. Biol. 9, 729076 (2021).
Mattila, P. K. & Lappalainen, P. Filopodia: molecular architecture and cellular functions. Nat. Rev. Mol. Cell Biol. 9, 446–454 (2008).
Pollard, T. D. Actin and actin-binding proteins. Cold Spring Harb. Perspect. Biol. 8, a018226 (2016).
Schönichen, A. & Geyer, M. Fifteen formins for an actin filament: A molecular view on the regulation of human formins. Biochim. Biophys. Acta BBA - Mol. Cell Res. 1803, 152–163 (2010).
Kühn, S. & Geyer, M. Formins as effector proteins of Rho GTPases. Small GTPases 5, e983876 (2014).
Yamashita, M. et al. Crystal structure of human DAAM1 formin homology 2 domain: DAAM1 FH2 structure. Genes Cells 12, 1255–1265 (2007).
Gao, C. & Chen, Y.-G. Dishevelled: The hub of Wnt signaling. Cell. Signal. 22, 717–727 (2010).
Xu, Y. et al. Crystal structures of a formin homology-2 domain reveal a tethered dimer architecture. Cell Res 13, 10.1016/s0092-8674(04)00210-7 (2004).
Liu, W. et al. Mechanism of activation of the formin protein Daam1. Proc. Natl Acad. Sci. 105, 210–215 (2008).
Habas, R., Kato, Y. & He, X. Wnt/Frizzled activation of Rho regulates vertebrate gastrulation and requires a novel formin homology protein daam1. Cell 107, 843–854 (2001).
Jaiswal, R. et al. The formin daam1 and fascin directly collaborate to promote filopodia formation. Curr. Biol. 23, 1373–1379 (2013).
Pan, Q., Shai, O., Lee, L. J., Frey, B. J. & Blencowe, B. J. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 40, 1413–1415 (2008).
Wang, E. T. et al. Alternative isoform regulation in human tissue transcriptomes. Nature 456, 470–476 (2008).
Barbosa-Morais, N. L. et al. The evolutionary landscape of alternative splicing in vertebrate species. Science 338, 1587–1593 (2012).
Feng, D. & Xie, J. Aberrant splicing in neurological diseases: aberrant splicing in neurological diseases. Wiley Interdiscip. Rev. RNA 4, 631–649 (2013).
Irimia, M. et al. A highly conserved program of neuronal microexons is misregulated in autistic brains. Cell 159, 1511–1523 (2014).
Quesnel-Vallières, M. et al. Misregulation of an activity-dependent splicing network as a common mechanism underlying autism spectrum disorders. Mol. Cell 64, 1023–1034 (2016).
Parras, A. et al. Autism-like phenotype and risk gene mRNA deadenylation by CPEB4 mis-splicing. Nature 560, 441–446 (2018).
Gonatopoulos-Pournatzis, T. et al. Genome-wide CRISPR-Cas9 interrogation of splicing networks reveals a mechanism for recognition of autism-misregulated neuronal microexons. Mol. Cell 72, 510–524.e12 (2018).
Gonatopoulos-Pournatzis, T. et al. Autism-misregulated eif4g microexons control synaptic translation and higher order cognitive functions. Mol. Cell 77, 1176–1192.e16 (2020).
Torres-Méndez. Parallel evolution of a splicing program controlling neuronal excitability in flies and mammals. Sci. Adv. 20, eabk0445 (2022).
Gonatopoulos-Pournatzis, T. & Blencowe, B. J. Microexons: at the nexus of nervous system development, behaviour and autism spectrum disorder. Curr. Opin. Genet. Dev. 65, 22–33 (2020).
Dai, J., Aoto, J. & Südhof, T. C. Alternative splicing of presynaptic neurexins differentially controls postsynaptic NMDA and AMPA receptor responses. Neuron 102, 993–1008.e5 (2019).
Wang, S. et al. Alternative splicing of latrophilin-3 controls synapse formation. Nature 626, 128–135 (2024).
Lin, L., Zhang, M., Stoilov, P., Chen, L. & Zheng, S. Developmental attenuation of neuronal apoptosis by neural-specific splicing of bak1 microexon. Neuron 107, 1180–1196.e8 (2020).
Tapial, J. et al. An atlas of alternative splicing profiles and functional associations reveals new regulatory programs and genes that simultaneously express multiple major isoforms. Genome Res 27, 1759–1768 (2017).
Calarco, J. A. et al. Regulation of vertebrate nervous system alternative splicing and development by an SR-related protein. Cell 138, 898–910 (2009).
Raj, B. et al. A global regulatory mechanism for activating an exon network required for neurogenesis. Mol. Cell 56, 90–103 (2014).
Quesnel-Vallières, M., Irimia, M., Cordes, S. P. & Blencowe, B. J. Essential roles for the splicing regulator nSR100/SRRM4 during nervous system development. Genes Dev. 29, 746–759 (2015).
Nakano, Y., Wiechert, S. & Bánfi, B. Overlapping activities of two neuronal splicing factors switch the GABA effect from excitatory to inhibitory by regulating REST. Cell Rep. 27, 860–871.e8 (2019).
Otomo, T. et al. Structural basis of actin filament nucleation and processive capping by a formin homology 2 domain. 433, 7 (2005).
Lu, J. et al. Structure of the FH2 Domain of Daam1: Implications for formin regulation of actin assembly. J. Mol. Biol. 369, 1258–1269 (2007).
Bibel, M. et al. Differentiation of mouse embryonic stem cells into a defined neuronal lineage. Nat. Neurosci. 7, 1003–1009 (2004).
Bibel, M., Richter, J., Lacroix, E. & Barde, Y.-A. Generation of a defined and uniform population of CNS progenitors and neurons from mouse embryonic stem cells. Nat. Protoc. 2, 1034–1043 (2007).
Matusek, T. et al. Formin proteins of the DAAM subfamily play a role during axon growth. J. Neurosci. 28, 13310–13319 (2008).
Szikora, S. et al. The formin DAAM is required for coordination of the actin and microtubule cytoskeleton in axonal growth cones. J. Cell Sci. jcs. 203455 https://doi.org/10.1242/jcs.203455 (2017).
Nakaya, M. et al. Placental defects lead to embryonic lethality in mice lacking the Formin and PCP proteins Daam1 and Daam2. Plos One 15, e0232025 (2020).
Feather-Schussler, D. N. & Ferguson, T. S. A Battery of Motor Tests in a Neonatal Mouse Model of Cerebral Palsy. J. Vis. Exp. 53569 https://doi.org/10.3791/53569 (2016).
Roper, R. J., Goodlett, C. R., de Lagrán, M. M. & Dierssen, M. Behavioral phenotyping for down syndrome in mice. Curr. Protoc. Mouse Biol. 10, e79 (2020).
Leger, M. et al. Object recognition test in mice. Nat. Protoc. 8, 2531–2537 (2013).
Warburton, E. C. & Brown, M. W. Neural circuitry for rat recognition memory. Behav. Brain Res. 285, 131–139 (2015).
Lueptow, L. M. Novel object recognition test for the investigation of learning and memory in mice. J. Vis. Exp. 55718 https://doi.org/10.3791/55718 (2017).
Vorhees, C. V. & Williams, M. T. Morris water maze: procedures for assessing spatial and related forms of learning and memory. Nat. Protoc. 1, 848–858 (2006).
Chernyuk, D. P., Bol’shakova, A. V., Vlasova, O. L. & Bezprozvanny, I. B. Possibilities and prospects of the behavioral test “morris water maze”. J. Evol. Biochem. Physiol. 57, 289–303 (2021).
Higashi, T. et al. Biochemical characterization of the rho GTPase-regulated actin assembly by diaphanous-related formins, mDia1 and Daam1, in Platelets. J. Biol. Chem. 283, 8746–8755 (2008).
Machleidt, T. et al. NanoBRET—A Novel BRET platform for the analysis of protein–protein interactions. ACS Chem. Biol. 10, 1797–1804 (2015).
Fritz, R. D. et al. A versatile toolkit to produce sensitive fret biosensors to visualize signaling in time and space. Sci. Signal. 6, rs12 (2013).
Moseley, J. B., Maiti, S. & Goode, B. L. formin proteins: Purification and measurement of effects on actin assembly. in Methods in Enzymology vol. 406 215–234 (Elsevier, 2006).
Ergin, V. & Zheng, S. Putative coiled-coil domain-dependent autoinhibition and alternative splicing determine SHTN1’s actin-binding activity. J. Mol. Biol. 432, 4154–4166 (2020).
Zhang, M. et al. Axonogenesis is coordinated by neuron-specific alternative splicing programming and splicing regulator PTBP2. Neuron 101, 690–706.e10 (2019).
Bolognin, S., Lorenzetto, E., Diana, G. & Buffelli, M. The potential role of Rho GTPases in alzheimer’s disease pathogenesis. Mol. Neurobiol. 50, 406–422 (2014).
Nakayama, A. Y., Harms, M. B. & Luo, L. Small GTPases rac and rho in the maintenance of dendritic spines and branches in hippocampal pyramidal neurons. J. Neurosci. 20, 5329–5338 (2000).
Impey, S. et al. An activity-induced microRNA controls dendritic spine formation by regulating Rac1-PAK signaling. Mol. Cell. Neurosci. 43, 146–156 (2010).
Higashi, T. et al. Flightless-I (Fli-I) regulates the actin assembly activity of diaphanous-related formins (DRFs) Daam1 and mDia1 in cooperation with active rho gtpase. J. Biol. Chem. 285, 16231–16238 (2010).
Torres-Méndez, A. et al. A novel protein domain in an ancestral splicing factor drove the evolution of neural microexons. Nat. Ecol. Evol. 3, 691–701 (2019).
Ciampi, L. et al. Specialization of the photoreceptor transcriptome by Srrm3 -dependent microexons is required for outer segment maintenance and vision. Proc. Natl Acad. Sci. 119, e2117090119 (2022).
Doolittle, L. K., Rosen, M. K. & Padrick, S. B. Measurement and analysis of in vitro actin polymerization. in adhesion protein protocols (ed. Coutts, A. S.) vol. 1046 273–293 (Humana Press, Totowa, NJ, 2013).
Doolittle, L. K., Rosen, M. K. & Padrick, S. B. Measurement and analysis of in vitro actin polymerization. Methods Mol. Biol. 1046, 273–293 (2013).
Consolati, T., Henkin, G., Roostalu, J. & Surrey, T. Real-time imaging of single γTuRC-mediated microtubule nucleation events in vitro by TIRF microscopy. in Microtubules (ed. Inaba, H.) vol. 2430 315–336 (Springer US, New York, NY, 2022).
Frangi, A. F., Niessen, W. J., Vincken, K. L. & Viergever, M. A. Multiscale vessel enhancement filtering. in Medical Image Computing and Computer-Assisted Intervention — MICCAI’98 (eds. Wells, W. M., Colchester, A. & Delp, S.) vol. 1496 130–137 (Springer Berlin Heidelberg, Berlin, Heidelberg, 1998).
Arganda-Carreras, I., Fernández-González, R., Muñoz-Barrutia, A. & Ortiz-De-Solorzano, C. 3D reconstruction of histological sections: Application to mammary gland tissue. Microsc. Res. Tech. 73, 1019–1029 (2010).
Polder, G., Hovens, H. & Zweers, H. Proceedings of the ImageJ User and Developer Conference, vol. 27–29, 172–177 (2010)
Haase, R. et al. CLIJ: GPU-accelerated image processing for everyone. Nat. Methods 17, 5–6 (2020).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Yang, J.-S. et al. rec-YnH enables simultaneous many-by-many detection of direct protein–protein and protein–RNA interactions. Nat. Commun. 9, 3747 (2018).
Ran, F. A. et al. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308 (2013).
Sakuma, T., Nishikawa, A., Kume, S., Chayama, K. & Yamamoto, T. Multiplex genome engineering in human cells using all-in-one CRISPR/Cas9 vector system. Sci. Rep. 4, 5400 (2014).
Doench, J. G. et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 34, 184–191 (2016).
Fernandes, J. et al. IP3 sensitizes TRPV4 channel to the mechano- and osmotransducing messenger 5′-6′-epoxyeicosatrienoic acid. J. Cell Biol. 181, 143–155 (2008).
Martin, L. et al. A protocol to quantify chromatin compaction with confocal and super-resolution microscopy in cultured cells. STAR Protoc. 2, 100865 (2021).
Huang, B., Wang, W., Bates, M. & Zhuang, X. Three-dimensional super-resolution imaging by stochastic optical reconstruction microscopy. Science 319, 810–813 (2008).
Rust, M. J., Bates, M. & Zhuang, X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat. Methods 3, 793–796 (2006).
Bates, M., Huang, B., Dempsey, G. T. & Zhuang, X. Multicolor super-resolution imaging with photo-switchable fluorescent probes. Science 317, 1749–1753 (2007).
García-Frigola et al. Zic2 promotes axonal divergence at the optic chias.pdf (2008).
Jero, J., Coling, D. E. & Lalwani, A. K. The use of preyer’s reflex in evaluation of hearing in mice. Acta Otolaryngol. (Stockh.) 121, 585–589 (2001).
Leger et al. Object recognition test in mice.pdf. (2013).
Walf, A. A. & Frye, C. A. The use of the elevated plus maze as an assay of anxiety-related behavior in rodents. Nat. Protoc. 2, 322–328 (2007).
Morris, R. Developments of a water-maze procedure for studying spatial learning in the rat. J. Neurosci. Methods 11, 47–60 (1984).
Tomás Pereira, I. & Burwell, R. D. Using the spatial learning index to evaluate performance on the water maze. Behav. Neurosci. 129, 533–539 (2015).
Deacon, R. M. J. Measuring Motor Coordination in Mice. J. Vis. Exp. 2609, https://doi.org/10.3791/2609 (2013).
Luong, T. N., Carlisle, H. J., Southwell, A. & Patterson, P. H. Assessment of motor balance and coordination in mice using the balance beam. J. Vis. Exp. 2376, https://doi.org/10.3791/2376 (2011).
Hoeymissen, E. V., Philippaert, K., Vennekens, R., Vriens, J. & Held, K. Horizontal Hippocampal Slices of the Mouse Brain. JoVE J. Vis. Exp. e61753, https://doi.org/10.3791/61753 (2020).
Li, M., Huang, Y., Ma, A. A. K., Lin, E. & Diamond, M. I. Y-27632 improves rotarod performance and reduces huntingtin levels in R6/2 mice. Neurobiol. Dis. 36, 413–420 (2009).
Patro, R., Duggal, G., Love, M. I., Irizarry, R. A. & Kingsford, C. Salmon: fast and bias-aware quantification of transcript expression using dual-phase inference. Nat. Methods 14, 417–419 (2017).
Soneson, C., Love, M. I. & Robinson, M. D. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Research 4, 1521 (2015).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Ignatiadis, N., Klaus, B., Zaugg, J. B. & Huber, W. Data-driven hypothesis weighting increases detection power in genome-scale multiple testing. Nat. Methods 13, 577–580 (2016).
Wu, T. et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2, 100141 (2021).
Luo, W. & Brouwer, C. Pathview: an R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics 29, 1830–1831 (2013).
Acknowledgements
We thank Kyung-Min Noh for providing the mESC line, Miguel Valverde and Francisco Muñoz for their help and feedback with the calcium imaging experiments, and members of the Irimia and Dierssen groups for constant scientific discussion and feedback. We also thank the CRG Genomics, Protein Technologies, Tissue Engineering and Advanced Light Microscopy Units for the RNA sequencing, protein purification, blastocyst injection and microscopy services, respectively. The research has been funded by the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (ERCCoG-LS2-101002275 to M.I.), Spanish Ministry of Science and Innovation (PID2020-115040GB-I00 to M.I., PID2022−141900OB-I00 to M.D., PID2020- 114080GB-I00 to M.P.C., PID2023-1497670B-I00 to F.J.M. and PID2022-138245NB-I00 to E.H), the European Union’s Horizon 2020 research and innovation program under grant agreements No 964342 to M.P.C., 721890 to P.P. and 848077 to A.Z.M., the National Institutes of Health (NIH) (1R01NS137222-01 to M.D.) and AGAUR grants from Secretaria d’Universitats i Recerca del Departament d’Empresa iConeixement de la Generalitat de Catalunya to M.I., M.D. and M.P.C.. CRG acknowledges support of the Spanish Ministry of Science and Innovation through the Centro de Excelencia Severo Ochoa (CEX2020-001049-S, MCIN/AEI/10.13039/501100011033), and the Generalitat de Catalunya through the CERCA program. M.S.N. received an FPU fellowship (FPU19/04789) from Ministerio de Universidades. This research was in part supported by grants from the Simons Foundation and Canadian Institutes of Health Research to B.J.B., who also holds the University of Toronto Banbury Chair in Medical Research and the Canada Research Chair in RNA Biology and Genomics.
Author information
Authors and Affiliations
Contributions
P.P. performed molecular, cellular, and behavioral characterization of the microexon function, with the help of M.M.C. A.Z.M. generated and analyzed electrophysiology data (Fig. 6 and S8). F.M. performed the analysis of the actin polymerization assay, provided support with bioinformatic analyses and contributed with critical insights. G.C.R. performed calcium imaging experiments and the analysis. C.V. and M.P.C. performed, analyzed and interpreted the STORM microscopy experiments. D.N. performed TIRF and dual-color TIRF microscopy experiments, and R.G.R. wrote ImageJ/Fiji scripts necessary for the downstream analysis. L.P.I. analyzed RNA-seq data and provided help with animal behavior data analysis. C.M.P., P.O.C. and E.H. performed, analyzed and interpreted the functional synapse quantification experiments. S.B. performed a Western Blot analysis and contributed with critical insights. H.F.U. prepared hippocampal cultures, D.S.Y performed their analysis. C.R.-M. and J.P. performed RT-PCR assays and genotyping. M.M. De L.C., Á.F.-B., C.S., J.L.M.L., M.S.N., E.D., and O.F. supported the animal experiments performed. D.L. and J.I.W.F. helped with FRET acceptor photobleaching experiments. J.E. and B.B. performed, analyzed and interpreted the Co-IP experiments. F.J.M.L. contributed with critical insights. P.P., M.P.C., M.D., and M.I. designed the experiments with input from other authors. P.P., M.D., and M.I. conceived the study, supervised the work, and wrote the manuscript with input from other authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Poliński, P., Miret Cuesta, M., Zamora-Moratalla, A. et al. A highly conserved neuronal microexon in DAAM1 controls actin dynamics, RHOA/ROCK signaling, and memory formation. Nat Commun 16, 4210 (2025). https://doi.org/10.1038/s41467-025-59430-w
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-59430-w