Abstract
Polyamines play essential roles in gene expression and modulate neuronal transmission in mammals. Vesicular polyamine transporters (VPAT) from the SLC18 family exploit the transmembrane H+ gradient to translocate polyamines into secretory vesicles, enabling the quantal release of polyamine neuromodulators and underpinning learning and memory formation. Here, we report the cryo-electron microscopy structures of human VPAT in complex with spermine, spermidine, H+, or tetrabenazine, elucidating discrete lumen-facing states of the antiporter and pivotal interactions between VPAT and its substrate or inhibitor. Leveraging structure-inspired mutagenesis studies and protein structure prediction, we deduce an unforeseen mechanism whereby the polyamine and H+ compete for multiple acidic protein residues both directly and indirectly, and rationalize how the antidopaminergic therapeutic tetrabenazine impedes vesicular transport of polyamines. This study unravels the mechanism of an H+-coupled polyamine antiporter, reveals mechanistic diversity between VPAT and other SLC18 antiporters, and raises new prospects for combating human disorders of polyamine homeostasis.
Similar content being viewed by others
Introduction
The vesicular polyamine transporter (VPAT or SLC18B1) is expressed in the brain, heart, lung, liver, kidney, and many other tissues in humans1. In the brain, VPAT regulates the content of polyamines, especially spermine and spermidine, by pumping them into secretory vesicles localized in the astrocytes2. Since polyamines such as spermine and spermidine modulate the activity of glutamate receptors (NMDA, KAR and AMPA receptors), various calcium, potassium, sodium, and TRPC channels, VPAT profoundly affects the “higher-ordered” brain function such as learning and memory by enabling the vesicular release of polyamine neuromodulators by exocytosis3.
Notably, the polyamine levels are dysregulated in stress-related disorders or neurodegenerative diseases, including amyotrophic lateral sclerosis, Alzheimer’s disease, Huntington’s disease, multiple system atrophy, and Parkinson’s disease, implying that the malfunction of polyamine transporters contributes to the pathogenesis of these maladies4. Indeed, mutations of the gene encoding ATP13A2, an ATP-dependent lysosomal exporter and the only other well-characterized mammalian polyamine transporter than VPAT, cause an early-onset Parkinson’s disease5,6,7,8,9. Moreover, Slc18b1 has been identified as one of the genes associated with cocaine addiction10, implicating VPAT in the mechanism of action for one of the most widely abused illicit drugs and underscoring its impact on the central nervous system.
Furthermore, recent studies implied that the loss of VPAT function contributes to the development of otofaciocervical syndrome, which is characterized by facial dysmorphia, hearing loss, and bone abnormalities3. This finding may be understood in light of the fact that besides neuronal transmission, polyamines also play prominent roles in cell growth, differentiation, immunity, autophagy, and cardiac function, promoting human longevity or healthspan11,12,13,14. The intracellular polyamine levels, which are tightly controlled by an interplay among polyamine biosynthesis, degradation and transport, decrease with age, and genetically-induced depletion of intracellular polyamines decrease the lifespan of mammals12. By contrast, tumor cells exhibit elevated polyamine levels, supporting polyamine transporters as potential therapeutic targets for treating cancers11,14.
The importance of polyamine homeostasis to human physiology is also borne out by a broad portfolio of clinical trials utilizing polyamines as potential therapeutic agents for tackling coronary artery disease, hypertension, glioma, skin cancer, infant immaturity, acute pancreatitis, COVID-19 infection, mucositis, or type-1 diabetes11,15. As such, the ubiquitously expressed VPAT contributes to cellular physiology far beyond the central nervous system1,3. To name but a few, human VPAT (hereafter, hVPAT) is highly expressed in the mast cells16, activated macrophages17, platelets18, and melanocytes19, contributing to immune defense, inflammation, blood clotting, and melanin formation, respectively. In particular, hVPAT has emerged as an important target to implement polyamine blocking therapy in cancer treatment11. Moreover, hVPAT could also be usurped as a vehicle to transport polyamine-mimetic therapeutics into cancerous cells to combat tumorigenesis. Despite such clinical relevance, no experimental structure is currently available for any ligand-bound hVPAT, which has severely limited our understanding of its inner workings.
Nonetheless, major advances have been made by the structure determination of human VMAT120, VMAT221,22,23,24,25 and VAChT26, which are members of the SLC18 family and sequester biogenic monoamines into secretory vesicles to enable neurotransmission. SLC18 transporters belong to the Major Facilitator Superfamily (MFS), members of which adopt a similar protein fold and transport cognate substrates across cellular membranes via a broadly conserved alternating-access mechanism. The recent studies21,23 suggested putative protonation sites in these SLC18 antiporters, including VMAT2D33, VMAT2E312, VMAT2D399, and VMAT2D426. However, many vital questions remain open regarding the SLC18 proteins in general and hVPAT in particular. Specifically, the proposed H+/substrate coupling mechanisms are staggeringly at odds with each other, even for the same protein21,23. Moreover, protein residues such as VMAT2D33 and VMAT2R189 are essential for the transport function, but their mechanistic roles are unclear21,22.
hVPAT bears merely 19%, 17% and 16% amino-acid sequence identity to human VMAT1, VMAT2 and VAChT, respectively, which are also incapable of transporting polyamines1. Such meager similarity places the protein homology into the twilight zone and foreshadows mechanistic disparity between VPAT and other SLC18 relatives. Indeed, VMAT1, VMAT2 and VAChT all catalyze electrogenic transport20,21,22,23,24,25,26, whereas the hVPAT-mediated H+/polyamine exchange is electroneutral1,3. It also remains mysterious how hVPAT can alter the H+/polyamine stoichiometry during the translocation of spermine or spermidine, which carry a + 4 or +3 charge under physiological conditions3,11. Moreover, none of VMAT2D33, VMAT2D399, and VMAT2D426 are conserved in hVPAT (Supplementary Fig. 1). The same perplexing pattern applies to VMAT2D214, VMAT2R217, VMAT2R357, and VMAT2D411, which form functionally important inter-domain salt-bridges21,22,23 but lack palpable hVPAT counterparts.
Apparently, hVPAT has deviated considerably from the other SLC18 proteins, which may reflect its unique ability to transport polyamines1. Such functional and sequence differences notwithstanding, the VMAT2-targeting therapeutic agents, such as tetrabenazine (TBZ), can also inhibit the hVPAT-mediated transport of polyamines1. TBZ, an antidopaminergic drug for treating the hyperkinetic disorders, can promote depression or suicidal ideation in patients27,28,29. Since polyamine imbalance has been closely associated with psychiatric disorders such as schizophrenia, mood disorders, major depressive disorder and suicidal behavior30,31, the inhibitory effects exerted by TBZ on hVPAT may contribute to those debilitating side effects. Despite such medical urgency, there is no structural information on the interactions between hVPAT and TBZ, baffling the efforts to improve TBZ or its related clinical drugs22.
In this work, we present the molecular structures of hVPAT captured at various ligand-bound states, which elucidate the interactions between the lumen-facing antiporter and its polyamine substrate or a non-competitive inhibitor. By employing proteoliposome-based transport and ligand binding assays, we further deduce a rather unusual antiport mechanism to explain how hVPAT couples the transmembrane proton movement to the loading of polyamine neuromodulators into membrane vesicles, thereby unlocking the mechanistic disparity between hVPAT and other human SLC18 antiporters as well as previously hidden spots for potential therapeutic modulation.
Results
Structure determination overview
To unravel the gnawing mysteries surrounding hVPAT, we set out to elucidate its structure and mechanism. However, hVPAT defied our attempts at high-resolution structure determination by using single-particle cryo-electron microscopy (cryoEM), likely due to the labile nature and/or small size of the 49-kd integral membrane transporter. To circumvent this problem, we appended the C-terminus of mVenus to hPVAT at residue T25, and fused the N-terminus of an anti-GFP nanobody to hVPAT at residue S431, deploying a proven strategy to facilitate the structure determination of small but functional membrane transporters21,25.
The resulting protein, hVPATEM, retained measurable spermine binding and transport activity, proving its utility for investigating the interactions between hVPAT and polyamine substrate (Supplementary Fig. 2). Furthermore, we found that the hVPAT-mediated uptake of radio-labeled spermine was largely unaffected by the valinomycin-mediated K+ diffusion potential, dovetailing with the results from a previous study1 and implying that the purified hVPAT (Supplementary Fig. 3) catalyzed electroneutral H+/spermine exchange under our experimental conditions, with a H+:spermine stoichiometric ratio of 4:1 during each transport cycle.
By means of single-particle 3D reconstruction, we subsequently calculated cryo-EM density maps for hVPATEM in complexes with spermine, spermidine, and TBZ to 3.3-3.4 Å nominal global resolutions, and hVPATEM at pH 4.9 to an overall resolution of 4.0 Å (Supplementary Fig. 4, Supplementary Table 1). We improved the experimental cryoEM maps by using the deep-learning software32,33,34, which enhanced the high-resolution density features and facilitated model building, especially for the spermine- and TBZ-bound forms (Supplementary Figs. 5,6). The structures captured a monomeric hVPAT in several ligand-bound states, revealing the interactions between the antiporter and polyamine substrate or TBZ, in addition to the structural changes that likely accompany the H+-induced release of polyamine.
Based on these experimental structures and an AlphaFold model of hVPAT in a conformation distinct from the cryoEM structures35,36, we conducted mutagenesis studies to test our mechanistic hypothesis. Our findings prompted us to posit an atypical antiport mechanism whereby the polyamine and H+ compete for multiple acidic protein residues in hVPAT, both directly and indirectly. Such a coupling mechanism, to our knowledge, has never been described for any SLC18 protein before. Overall, this work provides fundamental brushstrokes to the molecular picture depicting the hVPAT-mediated transport, offering a treasure trove of starting points for developing new therapeutics targeting the clinically relevant SLC18 antiporters.
The interactions between hVPAT and spermine
We purified hVPATEM in the presence of added spermine at pH 8.0, and calculated the cryoEM map to a global resolution of 3.4 Å based on the FSC0.143 criterion (Supplementary Figs. 4,5, Supplementary Table 1). Notably, we used the deep-learning software to improve the experimental map, which enhanced the high-resolution features and facilitated map interpretation (Supplementary Figs. 6,7). Specifically, we observed a polypeptide chain of hVPAT that traverses the membrane 12 times, yielding twelve transmembrane helices (TM1-12), which constitute the similarly folded N-terminal (TM1-6) and C-terminal (TM7-12) domains that are related by a pseudo-two-fold symmetry around the membrane normal (Fig. 1a, b). This bi-lobed architecture conforms to the MFS protein fold as the other SLC18 members20,21,22,23,24,25,26, and is characterized by the conspicuously short inter-helical loops, except for the one between TM6 and TM7, which contains a short horizontal helix (H1) and demarcates the N- and C-domains (Supplementary Figs. 1,7). Of note, both the N- and C-termini of hVPAT are located in the cytosol, reminiscent of the SLC18 transporters of known structure20,21,22,23,24,25,26.
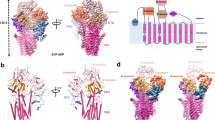
a, b The cryoEM density for the spermine-bound hVPATEM as viewed from the membrane, with the membrane bilayer simplified as a gray rectangle. CryoEM density for the mVenus, N- and C-domain of hVPAT, and nanobody is colored green, blue, orange, and light green, respectively; whereas the density for spermine and water molecules is shown in magenta and red, respectively. The cryo-EM density is contoured at 7.0 in ChimeraX v.1.8. c The luminal view highlights the solvent-accessible spermine-binding site. The cryoEM density is contoured at 7.0 in ChimeraX v.1.8. d Two close-up views showcasing the fitting of the spermine (magenta sticks) into the cryoEM density (magenta). The cryo-EM density is contoured at 8.5 in ChimeraX v.1.8.
The N- and C-domains of hVPAT separate near the center of the membrane and spread out toward the lumen, giving rise to a V-shaped lumen-facing, open conformation (Fig. 1a, b). At the interface between these two domains, we found a tubular and serpentine-shaped non-proteinaceous density, which is situated approximately halfway across the membrane (Fig. 1c, d). This density has unfettered access to the bulk solvent and is surrounded by numerous aromatic and hydrophobic amino acids, in addition to two acidic protein residues, E171TM5 and D255TM7 (Fig. 2a, b). As the structure was obtained under pH 8.0, and the calculated pKa values37 for their side-chain carboxylates are 6.5 and 4.7, respectively, E171 and D255 both bear a -1 charge (Fig. 2b, c). Since the size and shape of the tubular density are consistent with those of spermine (Q score38 =0.81), and acidic and aromatic amino acids interact with the aliphatic polycation spermine in the polyamine transporter5,6,7,8,9 ATP13A2, we ascribed this non-protein density to spermine, a physiological substrate for hVPAT1. Aside from the spermine molecule, we also observed several small non-protein density features that were tentatively modeled as water molecules (Fig. 2b).
a Ribbon diagram of the spermine-bound hVPAT as viewed from the membrane. The N- and C-domains are colored blue and orange, respectively. The bound spermine (magenta) and relevant protein residues are drawn as stick models. b Close-up view of the spermine-binding site, with the spermine (magenta) and relevant protein residues shown as sticks, and water molecules as red spheres. Salt bridges or H-bonds, and cation-π interactions are highlighted as blue dashed lines (distances in angstroms are indicated), whereas the van der Waals interactions are indicated with gray dashed lines. c Zoomed-in view of the inter-domain H-bond between N162 and D296, both of which are illustrated as stick models. The H-bond is indicated with dashed lines in blue and the distance in angstroms is specified. d Radiolabeled spermine uptake kinetics of the reconstituted hVPAT variants. Inset, a schematic diagram illustrating the hVPAT (orange rectangle)-mediated uptake of spermine (red arrow), driven by an outwardly directed proton gradient (green arrow). Data are presented as mean values +/− SD. Error bars represent SD (n = 3, i.e., three biological replicates were examined). The measured Vmax and KM are listed in Supplementary Table 2.
Specifically, the bound spermine forms two salt bridges with the side-chain carboxylate groups of E171 and D255, and also interacts with three water molecules (dubbed W1-W3) through direct H-bonds (Fig. 2b). Two of these water molecules, W1 and W2, also H-bond to the side-chain carboxylates of E171 and D255 separately, further stabilizing the binding of spermine. Moreover, the spermine molecule engages in van der Waals interactions with the side chains of M48TM1, Y51TM1 and W384TM11. Additionally, the bound spermine makes cation-π interactions with the side chain indole ring of W384. E83TM2 and D140TM4, another two membrane-embedded acidic protein residues, are also in proximity to the bound spermine, but they make no direct contact with the substrate.
Notably, the side chain carboxylate of D140 (pKa 5.2), ~10 Å away from the bound spermine, forms a direct salt bridge with the side chain guanidinium of R137TM4. In addition, two water molecules and/or the side-chain phenol of Y51 shields D140 from the substrate (Fig. 2b), which likely prohibits the protonation state of D140 from affecting polyamine binding to hVPAT directly. There is no such “shield”, however, found between R137 and spermine. Overall, the interactions between hVPAT and spermine are moderate, which nevertheless may be functionally advantageous since the lumen-facing antiporter is poised for substrate discharge. Among the spermine-contacting amino acids, E171 and D255 both form salt bridges, which are also contingent on the luminal pH, as the protonation of their side-chain carboxylates would neutralize E171 and D255 and sever their interactions with spermine.
To probe the functional importance of the observed interactions between hVPAT and spermine, we replaced M48, Y51, E171, D255 and W384 individually with alanine in hVPAT, and reconstituted the detergent-solubilized mutants (Supplementary Fig. 3) into proteoliposomes1,39. Based on the radiolabeled spermine uptake assays1, the mutation of Y51, E171, D255 or W384 exerted pronounced effects on the transport function of hVPAT (Fig. 2d, Supplementary Table 2), by markedly reducing (>15 fold) the Vmax (maximum uptake velocity) and simultaneously elevating (>4 fold) the KM (half-maximum uptake concentration). Indeed, the transporter activity of all four mutants was nearly reduced to the background level (Fig. 2d). These data thus supported the functional importance of the spermine-contacting protein residues, and also suggested that they are involved in the binding of spermine as well as the protein conformational changes essential for the translocation of spermine.
Furthermore, we performed the spermine uptake experiments on the single mutants E83A, R137A, and D140A (Supplementary Figs. 3, 8), revealing that these membrane-embedded charged amino acids are also indispensable for the hVPAT-mediated transport of spermine, since the three mutations drastically reduced the Vmax and/or increased the KM (Supplementary Table 2). Of note, R137 is the only membrane-embedded, charged amino acid that is strictly conserved amongst the human SLC18 antiporters (Supplementary Fig. 1), attesting to its physiological significance. Notably, even a conservative Lys substitution of VMAT2R189 (equivalent to R137 in hVPAT), which would preserve the positive charge, abolished the transport function21, indicating that Arg has been evolutionarily selected to play its functional roles at this locus. Moreover, we prepared a charge-swapped mutant of hVPAT, R137D/D140R, designed to maintain the charge-charge interactions between the two protein residues (Supplementary Fig. 3). This double mutant, however, exhibited severely crippled transporter activity (Supplementary Fig. 8), revealing that the orientation or polarity of the R137/D140 charge pair is vital for the transport function and echoing the evolutionary conservation.
To examine the interactions between spermine and hVPAT more directly, we also measured the binding of radiolabeled spermine to the detergent-solubilized hVPAT variants at pH 8.0 (Supplementary Fig. 9), and discovered that all the single mutations, barring M48A and R137A, markedly augmented (>7 fold) the Kd (Supplementary Table 3). These data reinforced the importance of the observed interactions between hVPAT and spermine, and suggested that E83 and D140, although not in direct contact with spermine in our structure, play crucial roles in stabilizing the binding of spermine.
In particular, the deprotonated side-chain carboxylate of E83TM2 (pKa 6.5) is >8 Å away from the bound substrate, but only 5.6 Å apart from the water molecule W3, which directly H-bonds to spermine (Fig. 2b). Furthermore, the spermine uptake and binding assays suggested that E83 is important for the transport and binding of spermine, as the mutation E83A substantially increased the measured KM or Kd (Supplementary Table 2,3). These observations thus raised the possibility that E83 interacts with spermine in a conformation distinct from the observed lumen-facing structure, e.g., the cytosol-facing state. Additionally, in the spermine-bound structure, D140TM4 neutralizes R137TM4 through a direct salt bridge (Fig. 2b). It is thus conceivable that the removal of the negative charge from D140, either by an Ala replacement or protonation, would liberate R137 and divert its positive charge to the spermine-binding site, which in turn would facilitate substrate dissociation due to electrostatic repulsion. As such, R137/D140 can regulate the binding of spermine in a pH-dependent manner. In accord with this notion, the mutation R137A or D140A severely impaired the transport function (Supplementary Fig. 8), but only D140A markedly weakened the binding of spermine (Supplementary Fig. 9).
Beyond the substrate-binding site, we observed an inter-domain H-bond between the side chains of N162TM5 and D296TM8 (Fig. 2c). Since the making or breaking of this interaction may stabilize or destabilize the lumen-facing conformation, thereby affecting the structural changes necessary for the transport of spermine, we examined the function of single mutants N162A and D296A in the spermine uptake assay (Supplementary Fig. 3,8). Our data suggested that N162 and D296 are both functionally important, most likely by facilitating the conformational changes during transport rather than the binding of spermine per se, as the two mutations substantially reduced (>5-fold) the Vmax without dramatically affecting (≤2-fold) the KM (Supplementary Table 2).
To gain further insights into the functional importance of mutated protein residues, we used the ConSurf server40 to compare the amino-acid sequence of hVPAT with those of 149 homologs (with >35% sequence identity) in animals. Since only 9% of the sequences are from mammals, and 63% are from invertebrates, this comparison likely reflects the function of the non-mammalian homologs considerably more than their mammalian counterparts. Nevertheless, this analysis revealed that Y51, R137, D140, E171, D255, D296, W384 are highly conserved (Supplementary Fig. 10). As Y51, E171, D255 and W384 interact with the bound spermine (Fig. 2b), this finding reaffirmed the biological relevance of our structural observations and implied that the functional roles of these protein residues have been preserved in the animal kingdom during evolution.
The spermidine-bound structure of hVPAT
Besides spermine, hVPAT also transports spermidine1,2,3, the homeostasis of which plays a particularly important role in autophagy and hence impacts human healthspan12,13. To elucidate the interactions between hVPAT and spermidine, we purified hVPATEM in the presence of added spermidine under pH 8.0, and attained the resulting cryoEM map to a global resolution of 3.3 Å (Supplementary Fig. 5, Supplementary Table 1). The density map revealed a boomerang-shaped non-protein density within the central cavity (Fig. 3a, b), which is congruent with both the size and shape of spermidine (Q score = 0.74). We henceforth assigned this density to spermidine, which is surrounded by numerous aromatic and hydrophobic amino acids, and the negatively charged D255TM7 (pKa 5.9). However, unlike the spermine-bound form, we observed no density feature that could be assigned as a water molecule in the spermidine-binding site, likely because the spermidine-treated protein sample was less homogeneous than the spermine-incubated one, and hence the cryoEM map calculated for the former was of insufficient quality to resolve the solvent molecules.
a A ribbon diagram of the spermidine-bound hVPAT as viewed from the membrane. The N- and C-domains are colored blue and orange, respectively. The bound spermidine (pink) and relevant protein residues are drawn as stick models. b Two close-up views showing the fitting of the spermidine (pink sticks) into the cryoEM density (pink), which is contoured at 11.0 in ChimeraX v.1.8. c Zoomed-in view of the spermidine-binding site, with the bound spermidine (pink) and relevant protein residues illustrated as stick models. The salt bridges or H-bonds are indicated with blue dashed lines (distances in angstroms are specified), whereas the van der Waals interactions are highlighted with dashed lines in gray. d The overlay of the spermine- and spermidine-binding sites in hVPAT. The bound spermine (light green), spermidine (pink) and the relevant protein residues are shown as stick models.
Notably, the bound spermidine forms a direct salt bridge with the side-chain carboxylate of D255TM7 (Fig. 3c). In addition, the positively charged N1 atom of spermidine is within H-bonding distance from the side-chain phenol in Y51TM1, likely making favorable charge-dipole interactions. In addition, the spermidine molecule makes van der Waals interactions with the side chains of M48TM1, Y51, F144TM4, F251TM7 and W384TM11. Prominent among them is the side-chain indole of W384, which makes numerous hydrophobic interactions with spermidine. The negatively charged side-chain carboxylate of E171TM5 (pKa 5.7), by contrast, is too far apart (~5.6 Å) to form a direct salt bridge with the bound spermidine (Fig. 3c).
As in the spermine-bound structure, the negatively charged side-chain carboxylate of E83TM2 (pKa 7.4) or D140TM4 (pKa 4.8) is not in direct contact with spermidine, with D140 also forming a direct salt bridge with R137TM4. If the spermine- and spermidine-bound structures were superimposed, it would give a rms deviation of 1.0 Å for 384 Cα positions (Fig. 3d), highlighting the overall similarity. Nevertheless, the spermidine-contacting M48, Y51, F144, F251, D255 and W384 all adopt different side-chain conformations from their counterparts in the spermine-bound protein. These differences likely allow hVPAT to accommodate a shorter and smaller polycation, showcasing the “poly-specific” nature of substrate recognition. Another notable feature is the single salt bridge formed with spermidine, contrasting with the spermine-binding site and likely accounting for the weaker binding of spermidine to hVPAT than that of spermine1.
To probe the transport function of hVPAT further, we reconstituted the wild-type protein and measured the spermidine uptake rates in the presence and absence of K+-ionophore valinomycin (Supplementary Fig. 11, Supplementary Table 4). We observed that the valinomycin-invoked membrane potential exerted little effect on the hVPAT-mediated uptake of radio-labeled spermidine, mirroring a previous report1 and supporting that the reconstituted hVPAT catalyzed an electroneutral exchange of spermidine for H+, with a H+:spermidine ratio of 3:1 during transport. Furthermore, we found that the single mutation D255A, abrogated the transport function of hVPAT in our spermidine uptake assay. By contrast, the single mutant E171A retained substantial spermidine transporter activity. Taken together, our results validated the functional relevance of the observed spermidine-binding site and highlighted a critical difference between the spermidine- and spermine-binding sites, i.e., the lack of direct involvement of E171 in the former.
The structure of hVPAT at low pH
Our polyamine-bound structures illustrate how the deprotonated hVPAT interacts with the polyamine substrate. To gain further mechanistic insights into the coupling between H+ and polyamine, we purified hVPATEM at pH 4.9 and calculated the cryoEM density map to a global resolution of 4.0 Å (Supplementary Fig. 5, Supplementary Table 1). The “low pH” structure is similar to the spermine-bound form, with the two structures superimposed onto one another to yield a rms deviation of 1.4 Å over 384 Cα atoms (Fig. 4a, b). Despite the overall similarity, the 3D reconstruction under low pH revealed no density feature in the central cavity that could be attributed to a bound polyamine molecule. Accordingly, we concluded that the low pH structure corresponds to a lumen-facing, substrate-free state. Since the calculated pKa values of E83, E171, D140 and D255 are 7.3, 6.4, 5.8, and 5.0, respectively, we further suggest that this structure represents a protonated state and the differences between the low pH and spermine-bound structures reflect, at least in part, the rearrangements accompanying the H+-triggered release of spermine.
a, b The overlay of the low pH and spermine-bound structures of hVPAT (light purple), both of which are drawn as ribbon diagrams. In the low-pH structure, the mVenus, N- and C-domains of hVPAT, and nanobody are colored light green, light blue, yellow, and gray, respectively. Red arrows highlight the structural displacement in mVenus and nanobody upon the superimposition of transporter structures, indicating that the connections between the fusion protein partners and hVPAT remained flexible. This flexibility likely enables the proteoliposome-reconstituted hVPATEM to transport spermine. c The cryoEM density (contoured at the level of 4.5 in ChimeraX v.1.8) for the relevant protein residues (light blue and yellow sticks), indicating that these amino acids are well-resolved in the cryoEM map. d Overlay of the spermine-binding site onto its counterpart in the low-pH structure (colored light blue and yellow). The bound spermine (gray) and the relevant protein residues are illustrated as stick models. Red arrows indicate the potential steric clash between Y51/W384 at low pH and the otherwise bound spermine, in addition to the potential charge-charge repulsion between R137 and spermine.
After comparing those two structures, we identified structural changes made by M48TM1, Y51TM1, R137TM4, F144TM4, F251TM7, F277TM8, Y284TM8, F380TM11, and W384TM11, which all surround the substrate-binding site (Fig. 4c, d). In particular, the side chains of Y51 and W384 shift towards the substrate-binding site as the spermine-bound protein transitions to the low pH form. This shift leads to potential steric clash with the otherwise bound spermine, thus precluding the rebinding of substrate (Fig. 4d). In synchrony, F144 and F380 adjust their side-chain conformations to buttress Y51 and W384, respectively, further stabilizing the substrate-free state.
Significantly, the side chain of R137 is released at low pH from D140 and turns toward the spermine-binding site, likely evoked by the protonation of the side chain carboxylate in D140 (pKa 5.8). The protrusion of side-chain guanidinium of R137 into the spermine-binding site would discourage the rebinding of spermine, ensuring a unidirectional transport process in a physiological setting. By contrast, the side-chain phenol of Y51 insulates D140 from the vacant substrate-binding site, highlighting the lack of direct involvement of D140 in polyamine binding. Moreover, at low pH, the side chains of Y284 and F251, the latter of which is also buttressed by the side chain of F277, rotate or shift towards the spermine-binding site, partially filling up the space left by the dissociated substrate and stabilizing the substrate-free structure. Lastly, the side chain of M48 withdraws from the substrate-binding site, further disfavoring the rebinding of spermine.
On one hand, the aforementioned structural changes likely follow the rupture of the two salt bridges between spermine and E171/D255, caused by the protonation of the two acidic protein residues. On the other hand, these changes allow the neighboring protein residues to approach and partially occupy the vacant spermine-binding site, not only to preclude the dissociated spermine from rebinding, but also to prepare the lumen-facing, protonated transporter to enter the cytosol-facing state. To examine whether such observations also apply to the transport of spermidine, we compared the low pH and spermidine-bound structures, which were superimposed onto each other with a rms deviation of 1.5 Å over 384 Cα positions. Indeed, the side chains of M48TM1, Y51TM1, R137TM4, F144TM4, F251TM7, D255TM7, and W384TM11 all make discernable structural changes as the spermidine-bound protein converts to the low pH state (Supplementary Fig. 12). Among them, Y51 and W384 in the low pH structure would collide with the otherwise bound spermidine, and the liberated R137 points its side chain toward the spermidine-binding site, preventing spermidine from rebinding to the protonated antiporter.
Altogether, our analyses pinpointed the protein residues that make detectable structural changes upon the release of polyamine, strongly supporting their mechanistic roles in the coupling between H+ and substrate. To reaffirm their importance, we mutated F144, F251, F277, Y284, and F380 individually in hVPAT and reconstituted the purified mutants into proteo-liposomes (Supplementary Figs. 3, 13). We subsequently performed the spermine uptake assay and found that all the mutations impaired the transport function, by decreasing (>2-fold) the Vmax and concomitantly enhancing (>2-fold) the KM (Supplementary Table 2). These data further corroborated the physiological relevance of the low pH structure and suggested that F144, F251, F277, Y284, and F380 play important roles in the transport of spermine. This finding can be interpreted in the light of the involvement of these protein residues in the protonation-induced release of polyamine (Fig. 4d), the stabilization of the spermine-contacting M48 and W384 (for F144, F380), and the spatial proximity to the substrate-binding site (for F251, F277, and Y284).
The interactions between hVPAT and tetrabenazine
To illuminate the inhibition mechanism of hVPAT by tetrabenazine (TBZ), we set out to elucidate the interactions between hVPAT and TBZ. Under tested conditions, we found that TBZ inhibits the hVPAT-mediated uptake of radiolabeled spermine, by markedly reducing the Vmax, whereas the KM was largely unaffected (Supplementary Fig. 14). This observation supported a non-competitive inhibition41,42 of the reconstituted hVPAT by TBZ, with a Ki of 2.0 µM based on the global fitting of the transport kinetic data. Moreover, we observed that both hVPAT and hVPATEM bind similarly to the radiolabeled dihydro-tetrabenazine (DTBZ), a TBZ analog22,25, which confirmed the utility of hVPATEM for investigating the interactions between hVPAT and TBZ.
Subsequently, we calculated the cryoEM density map for TBZ-bound hVPATEM to 3.3 Å global resolutions under pH 8.0 (Supplementary Fig. 5, Supplementary Table 1), which depicts an antiporter in the lumen-facing, open conformation (Fig. 5a). Significantly, the high-quality density map (Fig. 5b) allowed us to unambiguously place the TBZ molecule (Q score = 0.64) into the central cavity, adjoining the side chains of M48TM1, Y51TM1, F79TM2, F144TM4, M148TM4, F251TM7, Y284TM8, and W384TM11 (Fig. 5c). This finding stands in stark contrast to human VMAT2, which assumes an occluded conformation21,22,23,25 when bound to TBZ at pH 7-8. Indeed, the TBZ-bound structure of hVPAT could be superimposed onto that of VMAT2 with a rms deviation of 2.7 Å for 297 Cα positions (Supplementary Fig. 15), highlighting the differences in their interactions with the same clinical drug.
a A ribbon diagram of the tetrabenazine-bound hVPAT as viewed from the membrane. The N- and C-domains are colored blue and orange, respectively. The bound tetrabenazine (TBZ, colored in green) and relevant protein residues are drawn as stick models. b Two close-up views displaying the unambiguous placement of bound TBZ (green sticks) in the cryoEM density (light green), which is contoured at the level of 5.0 in ChimeraX v.1.8. c Close-up view of the TBZ-binding site, with TBZ (green) and relevant protein residues shown as stick models, the van der Waals interactions are indicated with dashed lines in gray. d The binding of dihydro-tetrabenazine (DTBZ), a close TBZ analog, to the hVPAT variants. Data are presented as mean values +/− SD. Error bars represent SD (n = 3, i.e., three biological replicates were studied).
Prominent among the hVPAT-TBZ interactions, the side-chain phenol of Y51 makes extensive aromatic stacking interactions with the benzoquinolizine moiety of TBZ, while the side-chain indole of W384 forms numerous van der Waals interactions with the isobutyl group of TBZ. Additionally, the side chains of M48, F144, F251, and Y284 make van der Waals interactions with the TBZ benzoquinolizine, whereas those of F79 and M148 form hydrophobic interactions with the dimethoxy and isobutyl groups in TBZ, respectively. Significantly, none of the interactions with the TBZ involve a charged protein residue, implying that the binding of TBZ to hVPAT is unlikely to be weakened by the low pH (~5.5) environment within the secretory vesicles in vivo.
Indeed, R137TM4 and D255TM7 are 13.0 Å and 11.7 Å away from the N1 atom (pKa29 6.5) in TBZ, respectively. This feature also contrasts with the TBZ-binding site in VMAT2, which harbors VMAT2R189 and VMAT2E312, equivalent of R137 and D255 in hVPAT, respectively. Notably, the Ala substitution of VMAT2R189 or VMAT2E312, which removes its charge, abrogated the binding of DTBZ22,25. Notably, the TBZ-binding pocket in hVPAT is markedly deeper than that in VMAT2 (Supplementary Fig. 15), implying that the former may be amenable to small molecule targeting for functional modulation.
In comparison, there is 1-2 salt bridges formed between polyamine and hVPAT, which would be ruptured upon the protonation of the substrate-binding acidic protein residue. Despite this difference, the TBZ-bound structure of hVPAT could be overlaid onto the spermine-bound form with a rms deviation of 0.7 Å for 384 Cα atoms, indicating similarity. Furthermore, the superimposition between the TBZ-bound and low pH structures of hVPAT yielded a rms deviation of 1.3 Å for 384 Cα atoms, implying that the binding of TBZ is compatible with the lumen-facing conformation at pH~5. Taken together, we concluded that TBZ interacts with the lumen-facing hVPAT similarly to spermine, but unlike that of spermine, the binding of TBZ to hVPAT likely withstands the low pH (~5.5) environment within the secretory vesicles.
To reaffirm the physiological relevance of the observed interactions with TBZ, we substituted the TBZ-contacting protein residues individually with alanine in hVPAT, and measured the binding of radiolabeled DTBZ to the purified hVPAT variants (Supplementary Fig. 3, Fig. 5d). These experiments revealed that the mutation of M48, Y51, F79, F144, F251, or W384 substantially reduced (by ≥7 fold) the binding of DTBZ (Supplementary Table 5), and the substitution of Y51, F251, or W384 exerted the most pronounced effects on the Kd (by ≥26 fold). Our data thus lent additional credence to the functional relevance of our structure and suggested that Y51, F251, and W384 play the most critical roles in mediating the hVPAT-TBZ interactions. Collectively, our data implied that TBZ operates like a pH-insensitive “plug” to confine hVPAT in a lumen-facing, open conformation. As such, TBZ acts as a non-competitive inhibitor in the uptake of polyamine, since it can evade the competition from the extra-vesicular substrate by binding to the lumen-facing hVPAT.
The cytosol-facing model of hVPAT
hVPAT alternates between the lumen- and cytosol-facing conformations during transport. Because all our experimental structures were lumen-facing, we resorted to AlphaFold35,36 to predict the cytosol-facing conformation to gain further mechanistic insights. Since AlphaFold generally predicts the most stable structure of a protein43, and the connections between the fusion partners and hVPAT affect the flexibility (Fig. 4a, b) and relative orientations of the N- and C- domains of the transporter21,22,23,24,25,26, we used AlphaFold to predict the structures of several mVenus-hVPAT-nanobody fusion proteins with various truncations at the N- and C-termini of hVPAT. This strategy led to an AlphaFold model for residues 12-440 in hVPAT (Fig. 6a), which assumes the cytosol-facing conformation (Supplementary Fig. 16).
a The AlphaFold-predicted structural model of cytosol-facing hVPAT, as viewed from the membrane. The N- and C-domains are colored cyan and yellow, respectively, and the membrane bilayer is simplified as a gray rectangle. b Overlay of the cytosol-facing model (cyan and yellow) onto the lumen-facing, spermine-bound hVPAT structure (pink), highlighting the rotational movement that the N- and C-domains undergo during the conformational transition. c The locations of spermine (pink) and the three membrane-embedded acidic protein residues (pink), E83, E171 and D255 in the spermine-bound structure, with the distances (in angstroms) between the Cα atoms specified. d The locations of the membrane-embedded E83, E171 and D255 (cyan and yellow) in the AlphaFold model, with the distances (in angstroms) between the Cα atoms indicated, underscoring the proximity of E83 to E171 or D255. The confidence scores are shown in Supplementary Fig. 16.
This cytosol-facing model exhibits a confidence level similar to that of the AlphaFold predicted, lumen-facing hVPAT (entry AF-Q6NT16-F1), the latter of which could be superimposed onto our spermine-bound and low pH structure with a rms deviation of 1.7 and 2.1 Å for 385 and 401 Cα atoms, respectively. This result suggested that the AlphaFold model of the lumen-facing hVPAT is largely correct, although it resembles the substrate-bound structure more closely than it does the low pH form. Therefore, we deemed the cytosol-facing model suitable for further analysis, although the precise amino-acid side chain details must be used with prudence, given the absence of experimental structural information on the cytosol-facing, ligand-bound hVPAT.
Specifically, the cytosol-facing model of hVPAT hews to the classic MFS protein fold and could be overlaid onto our spermine-bound structure with a rms deviation of 4.2 Å over 385 Cα positions (Fig. 6b). Moreover, the N- and C-domains in the model could be separately overlaid onto those in the spermine-bound structure with a rms deviation of 2.5 and 1.2 Å, for 176 and 209 Cα atoms, respectively. This comparison further implied that when hVPAT alternates between its cytosol- and lumen-facing states, the N- and C-domains undergo a ~ 30° rotational movement relative to each other, and the N-domain makes more substantial intra-domain structural changes than the C-domain.
Of particular importance, as the lumen-facing, spermine-bound hVPAT (Fig. 6c) switches to the cytosol-facing conformation (Fig. 6d), the side-chain carboxylate of E83 moves closer, by >4 Å, toward E171/D255 in the spermine-binding site, hinting at the possibility that these three membrane-embedded acidic protein residues all interact with spermine in the cytosol-facing state, under the plausible scenario that E171 and D255 maintain their contacts with spermine during transport. Notably, the side chain of E83 is <6 Å from the water molecule W3, the latter of which forms a direct H-bond with spermine in the lumen-facing structure (Fig. 2b). Thus, a ~ 4 Å shift of E83 may suffice to place its side-chain carboxylate in direct contact with both the spermine and W3 in the cytosol-facing hVPAT. In further support of this spatial arrangement, our spermine uptake and binding assays demonstrated that the single mutation of E83 weakened the binding of spermine to hVPAT (Supplementary Figs. 8, 9).
Moreover, R137 and D140 form a salt bridge (<3.5 Å) in the cytosol-facing model, with D255 positioned >6 Å away from R137, likely resembling a deprotonated hVPAT primed for polyamine binding (Figs. 2b, 3c). Notably, R137 and D255, in contrast to their VMAT2 counterparts VMAT2R189 and VMAT2E312, are unlikely to form a direct salt bridge in the cytosol-facing conformation21,24, thus indicating further mechanistic difference between hVPAT and VMAT2. Additionally, our modeling exercise suggested that the spermine molecule shifts through hVPAT considerably (>5 Å) as the cytosol-facing antiporter converts to the lumen-facing conformation, although the substrate retains ionic interactions with both E171 and D255 during this transition. Such a result sharply contrasts with VMAT2, in which the location of monoamine substrate remains largely unaltered during transport24. Nonetheless, such differences should be examined further through the structure elucidation of a spermine-bound, cytosol-facing hVPAT.
Aside from spermine binding, the cytosol-facing model suggested that an inter-domain salt bridge is formed between K60TM1 and E264TM7 (Supplementary Fig. 16), which are conserved in the mammalian VPAT orthologues (Supplementary Fig. 1) but not in their non-mammalian counterparts (Supplementary Fig. 10). To examine their functional significance, we replaced K60 and E264 individually or simultaneously in hVPAT (Supplementary Fig. 3), and measured the kinetics of spermine uptake (Supplementary Fig. 16). We found that the single Ala substitutions markedly reduced (≥5-fold) the Vmax but left the KM largely unaffected (Supplementary Table 2). By contrast, a charge-swapping double mutant, K60E/E264K, retained substantial transporter activity. These data lent further credence to the biological relevance of the cytosol-facing model, suggesting that K60 and E264 likely facilitate the protein conformational changes required for transport rather than the binding of spermine per se. Moreover, our results indicated that the transport function of hVPAT is not reliant on the particular orientation of the surface-exposed K60/E264 pair, unlike the membrane-embedded R137/D140, in accord with their distinct mechanistic roles.
The antiport mechanism of hVPAT
Amalgamating our structural and biochemical data, we infer an antiport mechanism to outline the major steps during the hVPAT-mediated transport of spermine (Fig. 7). Specifically, in the lumen-facing, deprotonated hVPAT, both E171 and D255 make ionic interactions with spermine. In addition, D140 and R137 form a salt bridge. This lumen-facing conformation is further stabilized by the inter-domain H-bond formed between N162 and D296. Upon the protonation of E83, D140, E171, and D255, spermine is released from hVPAT, and the re-positioning of R137 toward the substrate-binding site facilitates the dissociation of spermine and discourages its rebinding, thereby priming the protonated antiporter to transition to the cytosol-facing conformation.
The N- and C-domains of hVPAT are drawn as blue and orange ovals, the spermine- and H+-binding protein residues as yellow (deprotonated) or green (protonated) pentagons. R137, spermine, H+, membrane bilayer, the inter-domain H-bond and salt bridge are simplified as a red pentagon, red squiggle, green droplet, gray rectangle, and gray zones, respectively. As the spermine-bound hVPAT (state I) becomes protonated, the neutralization of E171 and D255 induce the spermine dissociation, whereas R137 disengages from the protonated D140. Diversion of R137 to the spermine-binding site facilitates the release of spermine from the protonated hVPAT (state II) and discourages substrate rebinding due to charge-charge repulsion. The lumen-facing, protonated hVPAT then switches to the cytosol-facing conformation (state III) and the binding of spermine to E83, E171 and D255 triggers the release of three protons. The binding of spermine to hVPAT (state IV) draws R137 away from the substrate-binding site and induces the formation of salt bridge between R137 and D140, which evokes the release of a fourth proton into the cytosol and completes the transport cycle. Notably, the spermine-bound and low pH structures, which were obtained at pH 8.0 and 4.9, respectively, emulate states I and II, respectively.
The formation of the salt bridge between K60 and E264 facilitates such a transition. By contrast, TBZ halts this isomerization by arresting hVPAT in the lumen-facing, open state. In the absence of TBZ, the lumen-facing, protonated antiporter isomerizes to a cytosol-facing state, and the subsequent binding of spermine to E83, E171, and D255 triggers the release of three protons, which is accompanied by D140 deprotonation and the ensuing formation of a salt bridge between R137 and D140. As such, hVPAT exchanges four protons for each spermine molecule per transport cycle, giving rise to an electroneutral process. For the transport of spermidine (Supplementary Fig. 17), however, E171 is no longer involved in the binding of substrate (Fig. 3c), leading to a H+/spermidine stoichiometry of 3:1. Significantly, the competition between H+ and polyamine within hVPAT is both direct (for E83, E171, or D255), and indirect (for D140).
To explore this conceptual framework further, we examined the uptake of radio-labeled spermine, using the reconstituted hVPAT and proteo-liposomes pre-loaded with spermidine (Supplementary Fig. 18). According to our postulated mechanism, spermine and spermidine trigger the release of four and three protons from hVPAT, respectively. Therefore, under our experimental conditions, we posited that hVPAT would catalyze the exchange of a spermine molecule for a spermidine molecule together with a proton per transport cycle, giving rise to an overall electroneutral process. Indeed, we observed that the valinomycin-imposed K+ diffusion potential had negligible effect on the hVPAT-mediated spermine/spermidine exchange, concordant with our predictions. Apparently, the high polyamine/H+ stoichiometry would enable hVPAT to adequately load polyamines into vesicles in vivo without the need to additionally exploit the membrane potential.
Discussion
Our data suggested that neither R137 nor D140 makes direct contact with the bound substrate (Figs. 2b, 3c), but they can regulate the binding of polyamine to hVPAT in a pH-dependent manner. Notably, VMAT2R189, the equivalent of R137, is linked to the side-chain carboxylate of VMAT2D33 via a H-bonding network in the deprotonated, lumen-facing antiporter21 at pH 8.0. At pH 6.5, however, VMAT2R189 is released from this network and points toward the vacant substrate-binding site24, probably triggered by the protonation of VMAT2D33. We contend that this H-bonding network enables the protonation state of VMAT2D33 to affect the binding of substrate to VMAT2, akin to the salt bridge between R137 and D140 in hVPAT, although D140TM4 and VMAT2D33 are situated in spatially distinct transmembrane helices (Supplementary Fig. 1). Such a coupling mechanism may explain why the ability of VMAT2D33 or its cognate residue in VMAT1 to deprotonate is essential for the transport function20,21.
Moreover, among E83, E171, and D255, only D255 is conserved in the SLC18 family, which corresponds to VMAT2E312 (Supplementary Fig. 1). This sequence disparity indicates that both the composition and spatial arrangement of the H+- and substrate-binding sites are radically different between hVPAT and its SLC18 relatives, disclosing heretofore unrecognized degree of mechanistic divergence. Furthermore, D255 interacts with the polyamine substrate via a direct salt bridge, which depends on the protonation state of the side-chain carboxylate (Figs. 2b, 3c). By contrast, the monoamine substrate dopamine or serotonin H-bonds with VMAT2E312, which doesn’t affect substrate binding in a protonation-dependent manner21,24, indicating that D255 and VMAT2E312 have distinct mechanistic roles. Moreover, the VMAT1 counterpart of VMAT2E312 also forms H-bonds, not salt bridges, with several monoamine substrates20. Altogether, these data support that hVPAT and other SLC18 members function through strikingly dissimilar H+/substrate coupling mechanisms, even though they share a common structural fold and all serve as H+-dependent antiporters.
Notwithstanding such patent diversity, the substrate-binding site within the central cavity of the SLC18 antiporters has been targeted by a panel of potent inhibitors, such as reserpine for VMAT120, TBZ for VMAT2, and vesamicol for VAChT26. By contrast, R137 and D140 in hVPAT, and their counterparts VMAT2D33 and VMAT2R189, all lie outside of this central ligand-binding site and yet play pivotal roles in the transport function. Such protein residues may embody an as-yet-untapped “soft underbelly” of the clinically relevant SLC18 antiporters, opening up new avenues for rational drug design to tackle the associated neurological disorders, cardiovascular diseases and cancers, among others.
Methods
Membrane protein expression
The codon-optimized genes expressing the human VPAT (hVPAT) variants were synthesized by GenScript and sub-cloned into the protein expression vector pFastBac1 at the cloning sites EcoRI and XhoI. For the biochemical studies, the hVPAT variants were fused with a thrombin-cleavable, C-terminal FLAG (i.e., DYKDDDDK) and deca-histidine tag. For the cryo-EM studies, the mVenus-hVPAT-nanobody fusion protein was attached to a thrombin-cleavable, C-terminal dual affinity tag: FLAG and deca-histidine.
Baculoviruses were generated in the DH10Bac cells and amplified in Sf9 cells by using the Bac-to-Bac expression system (Fisher Scientific). Initial P0 (Passage 0) virus was obtained through the addition of 5–10 µg recombinant bacmid DNA with 20 µl ExpiFectamine in 200 µl Grace’s medium to ~2 million cells plated in 3 ml of Grace’s medium in a cell culture plate. After 4 days at 27 °C, the supernatant was collected and supplemented with 10% (v/v) heat-inactivated fetal bovine serum (FBS, Cytiva HyClone). Subsequently, recombinant P1 (Passage 1) baculovirus was generated by adding 1 ml P0 virus to 30 ml (~1 million/ml) Sf9 cells in Grace’s medium (Gibco) supplemented with 10% (v/v) FBS, which was incubated at 27 °C for 3 days.
The P1 baculovirus was used for membrane protein expression and the amount of P1 sample used for insect cell infection was determined and optimized empirically based on the results of small-scale protein expression and purification experiments using 40 ml Tni (High Five, Fisher Scientific) cells cultured in the Express Five medium (Fisher Scientific). Typically, the large-scale expression of the hVPAT variants was performed by infecting Tni cells (at ~1 million/ml) with a 1:50-1:100 (v/v) ratio (P1:Tni). After 48 h of shaking at 27 °C, the Tni cells expressing the hVPAT variants were harvested by centrifugation at 6,000 g for 15 min, 4 °C. The cells were washed once with fresh (Express Five) medium, centrifuged, frozen in liquid nitrogen and stored at −80 °C until use. Cells were not tested for mycoplasma nor authenticated as they were normally purchased <3 months before use.
Membrane protein purification
Chemicals were purchased from Sigma-Aldrich, Millipore unless specified otherwise. For protein purification, the Tni cells were resuspended into a prechilled buffer containing 100 mM TRIS-HCl, pH 8.0, 10 mM MgCl2, 150 mM KCl, 1 mM TCEP, 1 µg/ml DNase I and protease inhibitors including 500 µM AEBSF, 2 µM E-64, 2 µM leupeptin, 2 µM pepstatin A, 1 µM aprotinin, and 0.5 mM benzamidine. All the following steps were conducted at 4 °C unless stated otherwise. The cells were ruptured by two or more passages through a prechilled high-pressure cell disruptor (Avestin Emulsiflex-C3), and the cell membranes were collected by centrifugation at 100,000 g for 2 h, 4 °C. The membrane was resuspended into buffer containing 50 mM TRIS-HCl, pH 8.0, 10 mM MgCl2, 150 mM KCl, 1 mM TCEP, 500 µM AEBSF, 2 µM E-64, 2 µM leupeptin, 2 µM pepstatin A, 1 µM aprotinin, and 0.5 mM benzamidine. Detergent glyco-diosgenin (GDN, Anatrace) and cholesteryl hemisuccinate (CHS, Anatrace) were added to 1.0% (w/v) and 0.2 % (w/v), respectively, and the sample was incubated for 3 h at 4 °C. For the cryoEM studies, polyamine or tetrabenazine (TBZ, Sigma) was added to 10–25 mM (pH ~8, see below) or 0.1 mM throughout membrane solubilization and protein purification, unless specified otherwise.
The insolubilized material was removed by centrifugation at 100,000 g for 1 h, 4 °C, and the supernatant was incubated with Ni-NTA resin (1 ml for ~40 grams of cell membranes) for 3 h, which was pre-equilibrated in 20 mM TRIS-HCl, pH 8.0, 150 mM NaCl, 1 mM TCEP, 500 µM AEBSF, 2 µM E-64, 2 µM leupeptin, 2 µM pepstatin A, 1 µM aprotinin, 0.025% (w/v) GDN, and 0.005 (w/v) CHS. The protein-bound Ni-NTA resin was then washed with 20 column volumes of wash buffer containing 20 mM TRIS-HCl, pH 8.0, 500 mM NaCl, 1 mM TCEP, 0.025% (w/v) GDN, and 0.005 (w/v) CHS. For protein samples used for transport assays, thrombin in one column volume of equilibration buffer containing 20 mM TRIS-HCl, pH 8.0, 150 mM NaCl, 1 mM TCEP, 0.025% (w/v) GDN, and 0.005 (w/v) CHS (10 units for ~20 grams of cell membrane), was added to the protein-bound Ni-NTA resin, which was incubated for 2 h. The Ni-NTA/buffer mixture was centrifuged at 5,000 g for 30 min, and the supernatant was passed through a column packed with anti-FLAG G1 resin (GenScript), which was pre-incubated in equilibration buffer. The flow-through was concentrated (MWCO 100 kd) to ~100 µl and centrifuged at 50,000 g for 1 h. The supernatant was then run over a 24 ml Superdex 200 column (Cytiva) at 0.4 ml/min in equilibration buffer. The peak fractions were pooled and used immediately for proteo-liposome reconstitution.
For protein samples used for the ligand binding assays, thrombin was not used and the protein was eluted from the protein-bound Ni-NTA resin with two column volumes of 20 mM TRIS-HCl, pH 8.0, 100 mM NaCl, 450 mM imidazole, 1 mM TCEP, 0.025% (w/v) GDN, and 0.005 (w/v) CHS. The eluate was then passed through a ~ 120-ml Sephadex G50 (crude) column in equilibration buffer. The protein sample was concentrated to ~100 µl, centrifuged at 50,000 g for 1 h, and purified by using a 24-ml Superdex 200 column at 0.4 ml/min in equilibration buffer. The peak fractions were pooled and used immediately for the ligand binding assays.
For the cryoEM study, the protein eluted from the Ni-NTA resin was incubated with amphipol PMAL-C8 (Anatrace) at a 1:30 (w/w) ratio for 2 h at 4 °C44,45. Then activated BioBeads SM2 (BioRad) at a 1:50 (w/w) ratio was added and the protein sample was incubated for 8 h at 4 °C. BioBeads SM2 were removed by centrifugation at 10,000 g for 15 min, 4 °C. The protein sample was then concentrated to ~100 µl and further purified by using a 24 ml Superdex 200 column (Cytiva) in 20 mM TRIS-HCl, pH 8.0, 150 mM NaCl, 1 mM TCEP, and 10 mM spermine (pH ~8), 25 mM spermidine (pH ~8), or 0.1 mM TBZ. For the low pH sample, the gel filtration buffer contained 20 mM Citrate-NaOH, pH 4.9, 150 mM NaCl, and 1 mM TCEP.
Radiolabeled spermine uptake assays
The detergent-solubilized hVPAT variants were reconstituted into liposomes as described previously1,39,46. In brief, the purified hVPAT variants (~100 µg) were mixed with n-decyl-β-D-maltoside, or DM (Anatrace, final concentration 1%, w/v) and brain polar lipid extract (porcine, Avanti Polar Lipids) to a final concentration of 0.5 mg/ml, and incubated for 30 min at 4 °C. The sample was centrifuged at 20,000 g for 15 min, 4 °C, and the supernatant was mixed with 1 ml reconstitution mix containing 20 mM Tris-HCl, pH8.0, 150 mM NaCl, 1% DM, 10 mg/ml mammalian brain polar lipids, and 3 mg/ml asolectin. The mixture was sonicated in a prechilled bath sonicator (Misonix) to clarity. The sample was then dialyzed (MWCO 50 kd) four times at 4 °C with stirring, each time against 1 L of buffer containing 20 mM Tris-HCl, pH 8.0, 150 mM NaCl. The proteo-liposome mixture was diluted 25 times in intra-vesicular or intra-liposomal buffer containing 50 mM MES-NaOH, pH 5.0 (mimicking the pH within secretory vesicles in vivo), 150 mM NaCl, and centrifuged at 100,000 g for 1 h, 4 °C. The proteo-liposome pellet was then resuspended into 200 µl intra-vesicular buffer, frozen in liquid nitrogen, and kept at −80 °C until use.
Radiolabeled spermine uptake assay was performed as described previously, with modifications1,39,47. Briefly, each proteo-liposome sample was thawed, sonicated to clarity and further purified by using a 20 ml Sephadex G50 (fine) column that was pre-equilibrated in 50 mM MES-NaOH, pH 5.0, 150 mM NaCl (the intra-vesicular buffer). The peak fractions containing the proteo-liposomes were pooled and concentrated (MWCO 100kd) to ~1 mg protein/ml. The uptake assay was performed in reaction buffer containing 50 mM Tris-HCl, pH 8.0, 150 mM NaCl for spermine or spermidine uptake assay in the absence of valinomycin. Generally speaking, for the spermine or spermidine uptake assay in the absence of valinomycin, we made similar observations when 150 mM NaCl was replaced by 150 mM KCl, which is consistent with the function of hVPAT as an H+-, rather than Na+-coupled antiporter1. For the spermine or spermidine uptake assay in the presence of valinomycin, 150 mM NaCl in the reaction buffer was replaced with 150 mM KCl and 1 µM valinomycin. For the spermine/spermidine exchange assay, however, the reaction buffer was 50 mM Tris-HCl, pH 8.0, 150 mM KCl, with or without 1 µM valinomycin. We found little difference between 1 and 3 µM valinomycin in our uptake assays. For the spermine/spermidine exchange assay, the intra-liposomal buffer was further supplemented with 25 mM spermidine (pH ~5).
Proteo-liposomes (~5 µl per reaction) were diluted into 150 µl of reaction buffer described above and the assay was initiated by the addition of various concentration of unlabeled spermine, [14C] spermine (60 mCi/mmol, American Radiolabeled Chemicals), or [14C] spermidine (55 mCi/mmol, American Radiolabeled Chemicals), in the presence or absence of tetrabenazine (TBZ) or valinomycin at 20 °C. For the measurement of transport inhibition, the proteo-liposomes were pre-incubated with the indicated concentrations of TBZ for 2 min at 20 °C, before adding to the reaction buffer. The ratio of radio-labeled and unlabeled spermine or spermidine was held constant for the experiments, while the total polyamine concentration was increased for the kinetic study.
30 µl aliquots of the reaction mixture were taken 1.5 min after the reactions commenced at 20 °C, which falls within the linear range of initial transport rates. The samples were then centrifuged through a 1.5 ml Sephadex G50 (fine) micro-spin column at 1000 g for 1 min, 20 °C. The eluate was subsequently transferred to a scintillation vial, to which 5 ml scintillation liquid (Perkin Elmer) was added. Radioactive decay of the isotope-labeled spermine was then recorded by using a liquid scintillation counter (Perkin Elmer). For each tested ligand concentration, recorded radioactive decay values from the protein-free liposomes was subtracted from the respective proteo-liposome values. The subtracted values were then fitted by using nonlinear regression to the Michaelis-Menten kinetics, and the kinetic parameters were calculated by using the GraphPad Prism v10.4.1. All experiments were repeated for three times and the error bars represent standard deviations of the mean.
Radioligand binding assays
Radioligand binding assay was performed essentially as described previously39,48,49 with modifications. Briefly, the purified hVPAT variants (~0.5 mg) were mixed with Ni-NTA resin (~500 µl) in 5 ml binding buffer containing 50 mM TRIS-HCl, pH 8.0, 0.5 M NaCl, 0.5 mM TCEP, 0.025 % GDN and 0.005% CHS. The samples were incubated for 1 h at 4 °C and the unbound material was discarded after centrifugation at 1000 g for 5 min, 4 °C. The protein-bound Ni-NTA resin was washed with the binding buffer and divided into 200 µl aliquots, which were incubated for 30 min at 4 °C with increasing concentrations of unlabeled spermine or dihydro-tetrabenazine (DTBZ, Cayman Chemical Company), and [14C] spermine (60 mCi/mmol, American Radiolabeled Chemicals) or [3H] DTBZ (77 Ci/mmol, ViTrax). The ratio of radio-labeled and unlabeled ligand was kept constant for all the experiments while the total ligand concentrations were varied.
An aliquot of 180 µl of the protein-bound Ni-NTA resin with spermine or DTBZ was then transferred to a micro-spin column and centrifuged at 1,000 g for 1 min, 4 °C. Unbound material was discarded, and resin was resuspended in 100 µl of buffer containing 50 mM TRIS-HCl, pH 8.0, 100 mM NaCl, 0.5 mM TCEP, 450 mM imidazole, 0.025 % GDN and 0.005% CHS. The Ni-NTA resin was centrifuged at 1000 g for 1 min, 4 °C and the radioactivity of this flow-through was measured by using liquid scintillation, as described above. The amount of [14C] spermine or [3H] DTBZ bound to the Ni-NTA resin at each ligand concentration in the absence of hVPAT variants was subtracted from all the other measurements made in the presence of the proteins. The subtracted values were then used for calculating the ligand binding parameters by using the GraphPad Prism v10.4.1. All experiments were repeated for three times and the error bars represent standard deviations of the mean.
CryoEM grid preparation and data collection
The gel-filtration peak fractions containing hVPATEM were concentrated (MWCO 100 kd) to ~3.5 mg/ml. Subsequently, 4 µl of the protein sample was added to freshly glow-discharged holey carbon, Quantifoil R1.2/1.3 300-mesh copper grids (EM Sciences). The cryoEM grids were then blotted for 4 s at 4 °C under 100% chamber humidity, and plunge-frozen into liquid ethane by using a Vitrobot Mark IV (FEI). Afterwards, the grids were loaded into a Titan Krios (FEI) electron microscope operating at 300 kV, which is equipped with a K3 Summit detector (Gatan) in CDS mode with a BioContinuum GIF energy filter (20 eV slit width) at the Hormel Institute, University of Minnesota. Movies were recorded by using the EPU software (Fisher Scientific) with a pixel size of 0.664 Å (magnification of 130,000 x) and a nominal defocus value ranging from -1.0 to -2.0 µm. Of note, a small pixel size could enhance the signal to noise ratio and facilitate the alignment of small protein particles. Each movie consists of 40 dose-framed fractions and was recorded with a dose rate of ~32 e−/Å2/s and a total dose of 53.7-54.3 e−/Å2. CryoEM data collection parameters and statistics are summarized in Supplementary Table 1.
CryoEM image processing
A total of 5303, 5022, 7505 and 6307 dose-fractionated movies were collected for samples incubated with spermine, spermidine, low pH, and TBZ, respectively. The movies were processed by using the patch-based beam-induced motion correction software MotionCor250 and contrast transfer function (CTF) estimation program CTFFIND51 v4.1.13, as implemented in cryoSPARC52 v4.5.1, with data down-sampled by 3/4 (0.885333 Å/pixel after down-sampling). Images with the defocus values outside of the range -0.5 to -3.0 µm or CTF fit resolutions worse than 6 Å were excluded from further data processing. The cryoEM data were processed by using cryoSPARC unless otherwise stated and the detailed procedure is exemplified in Supplementary Fig. 4.
For the spermine-incubated sample, a total of 4,457,145 particles were picked by using the Blob picker and then the Template picker, and subjected to the Remove Duplicate Particles Tool. Junk particles were removed through two rounds of 2D classifications. A total of 966,462 particles from the good 2D classes were used for Ab-initio reconstruction of four maps and further heterogeneous refinement (3D classification). Three rounds of 3D classification generated 222,217 particles for the third round of 2D classification to continuously remove bad particles. A total of 191,068 particles then underwent the final 3D classification (Ab-initio and heterogeneous refinement) and produced 69,508 particles from the good class, which was then subjected to further non-uniform and CTF refinements to generate a final map of 3.39 Å global resolutions. Map resolution was determined by gold-standard Fourier shell correlation (FSC) at 0.143, i.e., FSC0.143, between the two independent half-maps. Local resolution variation was estimated from the two half-maps by using cryoSPARC.
The spermidine-, low pH-, tetrabenazine-treated samples were handled similarly to the spermine-incubated one, and the cryoEM images were also processed by using cryoSPARC, with detailed information provided in Supplementary Table 1. To examine potentially biased 3D reconstruction of the single particles, for the TBZ-incubated sample, we also collected and processed the cryoEM data by using an entirely different set of hardware and software, which ultimately led to a TBZ-bound structure virtually identical to the one presented in this manuscript. This result suggested that the potential algorithm-related bias, if present in our data collection and/or processing, is negligible.
CryoEM post-processing and structure refinement
Staring from the two half maps produced from cryoSPARC, we tested various map sharpening or enhancement software from the CCP-EM suite53 (LocScale54) and Phenix55 (Autosharpen56, Resolve57, Local-aniso-sharpen, etc.), as well as the deep-learning-based software CryoFEM32, EMReady33, and/or DeepEMhancer34, to improve the density features. We evaluated the improvement of density maps or the lack thereof based on the density features (Supplementary Fig. 6), the performance of structure refinement (protein stereochemistry, model-to-map CC and FSC, etc.), and the Q scores38,58. Select subsets of the post-processing software used for calculating the optimal enhanced maps to aid model building were specified in Supplementary Table 1.
The AlphaFold-predicted, lumen-facing model of hVPATEM was manually docked into the unmodified experimental cryoEM maps by using the UCSF ChimeraX59,60, followed by iterative manual model building with the software Coot61 and/or ISOLDE62. Ligands were placed into the cryoEM density maps and/or the Fo-Fc maps calculated with Servalcat63 in CCP-EM, by using the software ChemEM64. Ligand restraints were generated by using eLBOW65 in Phenix. All the structures were refined in real space with secondary structure and Ramachandran restraints by using Phenix66. Although precedents abound in the literature67,68,69,70, we chose not to solely rely on the enhanced density maps produced by the deep-learning software, but rather use both the unmodified experimental cryoEM maps and the enhanced maps for structure refinement or model assessment (Supplementary Table 1).
Generally speaking, in comparison with the unmodified experimental maps, we found that the enhanced maps could yield refined structures with better protein stereochemistry, higher model-to-map CC/FSC values or Q scores calculated for the same unmodified density maps. Thus, we used the enhanced maps for figure preparation, and both the unmodified and enhanced maps to build models and monitor the progress of structure refinement, to safeguard against potential problems of overfitting or unforeseen effects exerted by the deep-learning software. The validation of the protein models was performed by using the MolProbity71 module in Phenix. All the structure figures were prepared by using ChimeraX.
Notably, during structure refinement, precaution was taken to minimize overfitting72. Specifically, we randomly altered the coordinates of each rebuilt model by 0.5 Å using the pdb tools in Phenix, and then refined this “shaken” model against one of the experimental half maps (#1). We subsequently compared the refined structure (#A) with the other half map (#2) to ensure that the model adjustment is consistent with the experimental density maps. Furthermore, the same procedure was repeated to refine the “shaken” model against map #2, and the refined model (#B) was examined against map #1. Subsequently, the FSC curves between the refined model #A and two half maps were computed: one with map #1 by using the real-space refinement in Phenix, the other with map #2 using the comprehensive cryoEM validation in Phenix, and then the two FSC curves were compared. Afterwards, the same procedure was repeated for the refined model #B. The differences between the refined structures (#A vs. #B) and FSC curves (for the same refined model) were then analyzed to assess potential overfitting, i.e., marginal difference indicates little overfitting.
Concerning the deposited structural models, although the quality of our cryoEM density maps had generally allowed for faithful model building and residue assignment, to avoid over-interpretation of the data, the following residues in hVPAT were not included in the final models due to weak density, likely stemming from the conformational flexibility within the luminal loop between TM1 and TM2: residues 56-71 in the spermine-bound structure, residues 56–72 in the spermidine-bound form, and residues 56–72 in the TBZ-bound structure, respectively.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The cryoEM density maps (experimental half maps as well as the enhanced maps) have been deposited in the Electron Microscopy Data Bank (EMDB) under accession codes: EMDB-46619 [https://www.ebi.ac.uk/pdbe/entry/emdb/EMD-46619] (the spermine-bound structure); EMDB-46617 [https://www.ebi.ac.uk/pdbe/entry/emdb/EMD-46617] (the spermidine-bound form); EMDB-46616 [https://www.ebi.ac.uk/pdbe/entry/emdb/EMD-46616] (the low pH structure); and EMDB-46618 [https://www.ebi.ac.uk/pdbe/entry/emdb/EMD-46618] (the TBZ-bound structure). The atomic coordinates have been deposited in the Protein Data Bank (PDB) under accession codes: PDB 9D7X (the spermine-bound structure); PDB 9D7V (the spermidine-bound form); PDB 9D7U (the low pH structure); and PDB 9D7W (the TBZ-bound structure). The atomic coordinates that we used for structure comparison and analysis also included PDB 8THR (the TBZ-bound structure of human VMAT2), and AF-Q6NT16-F1, [https://alphafold.ebi.ac.uk/entry/Q6NT16] (the AlphaFold predicted model of hVPAT). The source data underlying Figs. 2d and 5d, Supplementary Figs 2, 8, 9, 11, 13, 14, 16d and 18, are provided alongside this manuscript as a Source Data file. Source data are provided with this paper.
References
Hiasa, M. et al. Identification of a mammalian vesicular polyamine transporter. Sci. Rep. 4, 6836 (2014).
Fredriksson, R. et al. The polyamine transporter Slc18b1 (VPAT) is important for both short and long time memory and for regulation of polyamine content in the brain. PLoS Genet 15, e1008455 (2019).
Moriyama, Y., Hatano, R., Moriyama, S. & Uehara, S. Vesicular polyamine transporter as a novel player in amine-mediated chemical transmission. Biochim. Biophys. Acta Biomembr. 1862, 183208 (2020).
Vrijsen, S., Houdou, M., Cascalho, A., Eggermont, J. & Vangheluwe, P. Polyamines in Parkinson’s disease: balancing between neurotoxicity and neuroprotection. Annu. Rev. Biochem. 92, 435–464 (2023).
Sim, S. I., von Bulow, S., Hummer, G. & Park, E. Structural basis of polyamine transport by human ATP13A2 (PARK9). Mol. Cell 81, 1–15 (2021).
Tillinghast, J., Drury, S., Bowser, D., Benn, A. & Lee, K. P. K. Structural mechanism for gating and ion selectivity of the human polyamine transporter ATP13A2. Mol. Cell 81, 4650–4662 (2021).
Tomita, A. et al. Cryo-EM reveals mechanistic insights into lipid-facilitated polyamine export by human ATP13A2. Mol. Cell 81, 4799–4809 (2021).
Chen, X. et al. Cryo-EM structures and transport mechanism of human P5B type ATPase ATP13A2. Cell Discov. 7, 106 (2021).
Mu, J. et al. Conformational cycle of human polyamine transporter ATP13A2. Nat. Commun. 14, 1978 (2023).
Khan, A. H. et al. Genetic pathways regulating the longitudinal acquisition of cocaine self-administration in a panel of inbred and recombinant inbred mice. Cell Rep. 42, 112856 (2023).
Holbert, C. E., Cullen, M. T., Casero, R. A. Jr. & Stewart, T. M. Polyamine in cancer: integrating organismal metabolism and antitumoour immunity. Nat. Rev. Cancer 22, 467–480 (2022).
Madeo, F., Eisenberg, T., Pietrocola, F. & Kroemer, G. Spermidine in health and disease. Science 359, eaan2788 (2018).
Hofer, S. J. et al. Spermidine is essential for fasting-mediated autophagy and longevity. Nat. Cell. Biol. https://doi.org/10.1038/s41556-024-01468-x (2024).
Imada, S. et al. Short-term post-fast refeeding enhances intestinal stemness via polyamines. Nature https://doi.org/10.1038/s41586-024-07840-z (2024).
Note: A Search Using The Keyword “polyamine” Returned With A Total Of 47 Clinical Trials As Of Aug 5, 2024. http://clinicaltrials.gov (2024).
Takeuchi, T. et al. Vesicular polyamine transporter mediates vesicular storage and release of polyamine from mast cells. J. Biol. Chem. 292, 3909–3918 (2017).
Park, S.-J. et al. Imaging inflammation using an activated macrophage probe with Slc18b1 as the activation selective gating target. Nat. Commun. 10, 1111 (2019).
Uehara, M., Fukumoto, A., Omote, H. & Hiasa, M. Polyamine release and vesicular polyamine transporter expression in megakaryoblastic cells and platelets. Biochim. Biophys. Acta Gen. Subj. 1868, 130610 (2024).
Brito, S. et al. A systematic exploration reveals the potential of spermidine for hypopigmentation treatment through the stabilization of melanogenesis-associated proteins. Sci. Rep. 12, 14478 (2022).
Ye, J. et al. Structural insights into vesicular monoamine storage and drug interactions. Nature 629, 235–243 (2024).
Wu, D. et al. Transport and inhibition mechanism of human VMAT2. Nature 626, 427–434 (2024).
Pidathala, S. et al. Mechanism of neurotransmitter transport and drug inhibition in human VMAT2. Nature 623, 1086–1092 (2024).
Wang, Y. et al. Transport and inhibition mechanism for VMAT2-mediated synaptic vesicle loading of monoamines. Cell Res. 34, 47–57 (2024).
Wu, D. et al. Structural snapshots of human VMAT2 reveal insights into substrate recognition and proton coupling mechanism. Cell Res. 34, 586–589 (2024).
Dalton, M. P., Cheng, M. H., Bahar, I. & Coleman, J. A. Structure and mechanism for VMAT2 inhibition by tetrabenazine. eLife 12, RP91973 (2023).
Zhang, Y. et al. Structural insights into VAChT neurotransmitter recognition and inhibition. Cell Res. https://doi.org/10.1038/s41422-024-00986-5 (2024).
Huntington Study Group. Tetrabenazine as antichorea therapy in Huntington disease: a randomized controlled trial. Neurol 66, 366–372 (2006).
Gimenez-Roldan, S. & Mateo, D. Huntington disease: tetrabenazine compared to haloperidol in the reduction of involuntary movements. Neurol 4, 282–287 (1989).
Kaur, N., Kumar, P., Jamwal, S., Deshmukh, R. & Gauttam, V. Tetrabenazine: spotlight on drug review. Ann. Neurosci. 23, 176–185 (2016).
Turecki, G. Polyamines and suicide risk. Mol. Psychi. 18, 1242–1243 (2013).
Gross, J. A. & Turecki, G. Suicide and the polyamine system. CNS Neurol. Dis. Drug Targets 12, 1–9 (2013).
Dai, X., Wu, L., Yoo, S. & Liu, Q. Integrating AlphaFold and deep learning for atomistic interpretation of cryo-EM maps. Brief. Bioinform. 24, 1–10 (2023).
He, J., Li, T. & Huang, S.-Y. Improvement of cryo-EM maps by simultaneous local and non-local deep learning. Nat. Commun. 14, 3217 (2023).
Sanchez-Garcia, R. et al. DeepEMhancer: a deep learning solution for cryo-EM volume postprocessing. Commun. Biol. 4, 874 (2021).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Abramson, J. et al. Accurate structure prediction of biomolecular interactions with AlphaFold3. Nature 630, 493–500 (2024).
Olsson, M. H. M., Sondergaard, C. R., Rostkowski, M. & Jensen, J. H. PROPKA3: consistent treatment of internal and surface residues in empirical pKa predictions. J. Chem. Theory Comput. 7, 525–537 (2011).
Pintilie, G. et al. Measurement of atom resolvability in cryo-EM maps with Q-scores. Nat. Methods 17, 328–334 (2020).
Elbaz, Y., Danieeli, T., Kanner, B. I. & Schuldiner, S. Expression of neurotransmitter transporters for structural and biochemical studies. Prot. Exp. Purif. 73, 152–160 (2010).
Yariv, B. et al. Using evolutionary data to make sense of macromolecules with a “face-lifted” ConSurf. Prot. Sci. 32, e4582 (2023).
Shahsavar, A. et al. Structural insights into the inhibition of glycine reuptake. Nature 591, 677–681 (2021).
Wei, Y. et al. Transport mechanism and pharmacology of the human GlyT1. Cell 187, 1719–1732 (2024).
Agrawal, V. & McShan, A. C. The power and pitfalls of AlphaFold2 for structure prediction beyond rigid globular proteins. Nat. Chem. Biol. 20, 950–959 (2024).
Chai, J., Guo, G., McSweeney, S. M., Shanklin, J. & Liu, Q. Structural basis for enzymatic terminal C-H bond functionalization of alkanes. Nat. Struct. Mol. Biol. 30, 521526 (2023).
Pang, C., Chai, J., Zhu, P., Shanklin, J. & Liu, Q. Structural mechanism of intracellular autoregulation of zinc uptake in ZIP transporters. Nat. Commun. 14, 3404 (2023).
Pan, Y. et al. Structural basis of ion transport and inhibition in ferroportin. Nat. Commun. 11, 5686 (2020).
Yao, X., Fan, X. & Yan, N. Cryo-EM analysis of a membrane protein embedded in the liposome. Proc. Natl. Acad. Sci. USA 117, 18497–18503 (2020).
Sigal, N., lewinson, O., Wolf, S. G. & Bibi, E. E. coli multidrug transporter MdfA is a monomer. Biochem 46, 5200–5208 (2007).
Tirosh, O. et al. Manipulating the drug-proton antiport stoichiometry of the secondary multidrug transporter MdfA. Proc. Natl. Acad. Sci. USA 109, 12473–12478 (2012).
Rubinstein, J. L. & Brubaker, M. A. Alignment of cryo-EM movies of individual particles by optimization of image translations. J. Struct. Biol. 192, 188–195 (2015).
Rohou, A. & Grigorieff, N. CTFFIND4: fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. CryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Burnley, T., Palmer, C. M. & Winn, M. Recent developments in the CCP-EM software suite. Acta Cryst. D73, 469–477 (2017).
Jakobi, A. J., Wilmanns, M. & Sachse, C. Model-based local density sharpening of cryo-EM maps. eLife 6, e27131 (2017).
Afonine, P. V. et al. Macromolecular structure determination using x-rays, neutrons and electrons: recent developments in phenix. Acta Cryst. D75, 861–877 (2019).
Terwilliger, T. C., Sobolev, O. V., Afonine, P. V. & Adams, P. D. Automated map sharpening by maximization of detail and connectivity. Acta Cryst. D74, 545–559 (2018).
Terwilliger, T. C., Ludtke, S. J., Read, R. J., Adams, P. D. & Afonine, P. V. Improvement of cryo-EM maps by density modification. Nat. Methods 17, 923–927 (2020).
Jamali, K. et al. Automated model building and protein identification in cryo-EM maps. Nature 628, 450457 (2024).
Meng, E. C. et al. UCSF ChimeraX: tools for structure building and analysis. Protein Sci. 32, e4792 (2023).
Read, R. J. et al. Likelihood-based interactive local docking into cryo-EM maps in ChimeraX. Acta Cryst. D80, 1–11 (2024).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Cryst. D60, 2126–2132 (2004).
Croll, T. I. ISOLDE: a physically realistic environment for model building into low-resolution electron-density maps. Acta Cryst. D74, 519–530 (2018).
Yamashita, K., Palmer, C. M., Burley, T. & Murshudov, G. N. Cryo-EM single particle structure refinement and map calculation using Servalcat. Acta Cryst. D77, 12821291 (2021).
Sweeney, A., Mulvaney, T., Maiorca, M. & Topf, M. ChemEM: flexible docking of small molecules in cryo-EM structures. J. Med. Chem. 67, 199–212 (2024).
Moriarty, N. W., Grosse-Kunstleve, R. W. & Adams, P. D. Electronic ligand builder and optimization workbench (eLBOW): a tool for ligand coordinate and restraint generation. Acta Cryst. D65, 1074–1080 (2009).
Afonine, P. V. et al. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Cryst. D74, 531–544 (2018).
Sonani, R. R. et al. Tad and toxin-coregulated pilus structures reveal unexpected diversity in bacterial type IV pili. Proc. Natl. Acad. Sci. USA 120, e2316668120 (2023).
Liu, C., Hauk, G., Yan, Q. & Berger, J. M. Structure of Escherichia coli endonuclease VII. Proc. Natl. Acad. Sci. USA 121, e2319644121 (2024).
Liwocha, J. et al. Mechanism of millisecond Lys48-linked poly-ubiquitin chain formation by cullin-RING ligases. Nat. Struc. Mol. Biol. 31, 378–389 (2024).
Ciapponi, M., Karlukova, E., Schkolziger, S., Benda, C. & Muller, J. Structural basis of the histone ubiquitination read-write mechanism of RYBP-PRC1. Nat. Struc. Mol. Biol. 31, 1023–1027 (2024).
Williams, C. J. et al. MolProbity: more and better reference data for improved all-atom structure validation. Protein Sci. 27, 293–315 (2018).
Amunts, A. et al. Structure of the yeast mitochondrial large ribosomal subunit. Science 343, 1485–1489 (2014).
Acknowledgements
We thank the staff at the cryoEM facility in the Hormel Institute, University of Minnesota for support with data collection. We also thank P. Yuan (Mount Sinai), J. Xu (Calico Life Sciences), N. Ban (ETH Zurich), S. Mukherjee (U. Chicago), E. Tajkhorshid (UIUC), and Y. Cheng (UCSF) for helpful discussions. We are particularly grateful to E. Cao (U. Utah) for his help during the early stages of this project, A. Kossiakoff (U. Chicago) for the generous gift of a plasmid and a cell-line for over-expressing the Fab against BRIL. This work was in part supported by the NIH (R01GM145642 to M.L.), the Hormel Institute, University of Minnesota (to B.L.) and the DOE (KC0304000 to J.S.).
Author information
Authors and Affiliations
Contributions
M.L. conceived of the study. M.L. and Y.G. prepared the protein samples and conducted the biochemical studies. B.L., G.Y., and J.Chen prepared the cryo-EM grids. B.L. and G.Y. collected the cryo-EM data. B.L., G.Y., Q.L., and H.L. analyzed the cryo-EM data and built the initial protein models. M.L. and J.Chai carried out the post-processing of the cryo-EM maps and refined the structures. J.S. contributed to the data analysis. M.L. wrote the manuscript, with the input from J.Chai, B.L., and Q.L.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Nazzareno D’Avanzo and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Guo, Y., Yang, G., Liu, H. et al. Structure and mechanism of human vesicular polyamine transporter. Nat Commun 16, 4142 (2025). https://doi.org/10.1038/s41467-025-59549-w
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-59549-w