Abstract
Understanding mechanisms of resistance to active therapies is crucial for developing more effective treatments. Here, we investigate resistance to anti-EGFR and anti-VEGF plus chemotherapy treatment in colorectal cancer (CRC) patients from the IMblaze370 trial (NCT02788279). While anti-VEGF does not select for secondary mutations, anti-EGFR leads to simultaneous mutations in EGFR and MAPK, but not PI3K pathway genes. Notably, we observe frequent acquired mutations in the EGFR extracellular but not intracellular domain and that patients with higher baseline expression of EGFR-ligands are prone to acquire resistant mutations. This data reveals a ligand-activated EGFR/MAPK-signaling dependency in CRC. We also observe enrichment for 8q gains in anti-EGFR treated patients, potentially linked to MYC amplification, a finding further supported by baseline expression analysis. This work adds to the evidence supporting broader evaluation of EGFR and pan-KRAS inhibitor combinations in CRC patients. It also underscores the utility of EGFR ligands as anti-EGFR efficacy biomarkers and provides a rationale for developing ligand blockers to complement and/or improve conventional anti-EGFR therapies in CRC.
Similar content being viewed by others
Introduction
Colorectal cancer (CRC) is the second most common cause of cancer death in the United States, with estimates of 153,020 new cases and 52,550 deaths in the USA in 20231. Despite the development of anti-VEGF and anti-EGFR therapies reduced mortality and improved the overall survival rate significantly, the progress has been limited compared to other cancer types, and the 5-year overall survival rate (14%) for most patients with metastatic CRC (mCRC) remains poor which highlights the significant unmet medical need for the development of more effective therapeutic treatments for mCRC1. Cancer-specific mutations have provided a genetic roadmap that led to the development of more effective targeted therapies. In mCRC, the identification of several targetable genetic alterations, such as HER2 +, BRAF V600E, NTRK fusion, and MSI-H, enabled the recent development of highly effective treatments for small diagnostic positive subgroups of mCRC2,3,4,5,6. Two key challenges limiting further development of more effective targeted therapies in mCRC are (i) most prevalent CRC baseline mutations are not currently druggable, and (ii) adaptive resistance through the acquisition of mechanisms of resistance is fast in patients treated with currently available active targeted therapies.
Two broadly used approved therapies in mCRC are combinations of chemotherapy with either anti-VEGF (anti-angiogenic) or anti-EGFR therapies. Although the combination of anti-VEGF therapy with chemotherapy has been an established standard of care (SOC) since 20047,8,9, the specific mechanisms that confer resistance or sensitivity to anti-VEGF containing therapies in individual patients remain poorly understood. On the other hand, anti-EGFR containing therapies are currently restricted to RAS (KRAS and NRAS) and BRAF wild-type patients, as RAS and BRAF mutations (present in 50 ~ 60 % of the CRC patients) have been shown to confer intrinsic resistance to this treatment. In sensitive RAS and BRAF wild-type patients, the duration and efficacy of anti-EGFR agents is limited by the rapid emergence of acquired mechanisms of resistance10,11.
Comprehensive characterization of mechanisms of resistance to targeted therapies can shed insights into disease specific pathway addiction or vulnerabilities that may inform the rational development of effective next generation single agent and/or combination treatments6,7,8,9,10,11,12 The growing utilization of ctDNA allows for noninvasive and dynamic assessment of acquired resistance mechanisms amidst the complex molecular heterogeneity of mCRC. The longitudinal ctDNA analysis has provided compelling evidence for fast and broad enrichment of secondary tumor mutations conferring resistance to treatment under the selective pressure of active anti-EGFR therapy13,14,15,16,17,18. However, a limitation of most of the previously published ctDNA studies is that the datasets were either relatively small13,14,15,16,17,18,19 or previous treatment history was unknown for most patients20.
In this study, we leverage the prior treatment history, ctDNA (2/3 L +) and archival tumor tissue sequencing data, and RNAseq data from 296 patients who participated in IMblaze370 trial (NCT02788279), a phase III study in 2/3 L + (84% 3 L + and 16% 2 L + ) mCRC21. By taking advantage of this rich and well annotated multi-omics sample collection, here we report distinct secondary genomic alterations acquired prior to IMblaze370 enrollment in patients who previously received anti-EGFR vs those who received anti-VEGF but not anti-EGFR therapies. In addition, we provide insights into intrinsic properties that may be associated with acquired resistance to anti-EGFR therapy in mCRC.
Results
Alterations in EGFR/MAPK but not PI3K pathway genes are selectively enriched in patients following anti-EGFR but not anti-VEGF therapies in mCRC
To investigate treatment-specific genomic alterations that underlie acquired resistance mechanisms to anti-EGFR or anti-VEGF therapies in mCRC, we compared alterations detected in archival tumor tissue samples prior to any treatment (FoundationOne® (F1) assay) to alterations detected in ctDNA (FoundationACT (FACT) assay) from plasma samples collected at baseline from the Imblaze370 study. Baseline plasma samples were collected prior to receiving any IMblaze370 treatment regimens, but post two or three lines (2/3 L +) of prior therapies, including but not limited to anti-EGFR or anti-VEGF therapies. Out of the 296 patients with sequencing data and prior lines of treatment information, we stratified patients into two groups: patients who received prior anti-EGFR (N = 113) therapies vs. those who received prior anti-VEGF (N = 141) but not anti-EGFR therapies at any time before enrollment into the IMblaze 370 trial (Supplementary Fig. 1). Alterations that were absent from archival tumor tissue (prior any treatment) but detected in plasma samples (2/3 L +) were defined here as likely acquired alterations. For consistency, only 62 genes covered by both FACT and F1 assays were included in this analysis, and only previously reported and functionally validated alterations, i.e., known or likely pathogenic alterations, defined by Foundation Medicine, were considered as candidate acquired resistance alterations.
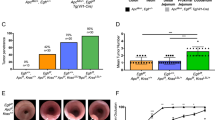
While the gene-level prevalence of alterations in patients previously treated with anti-VEGF therapies was highly concordant between ctDNA and archival tissue for all the tested genes (Pearson R2 = 0.988, Fig. 1a and Supplementary Fig. 2a), a significantly higher frequency of alterations in five EGFR/MAPK pathway genes, KRAS, EGFR, NRAS, MAP2K1 and NF1, were detected in ctDNA compared to those in archival tissue samples in patients receiving prior anti-EGFR treatment, whereas all the rest of the tested genes were highly concordant (Fig. 1b and Supplementary Fig. 2b). Consistently, the percentage of patients with acquired alterations in all those 5 genes except NF1, with approaching statistical enrichment in anti-EGFR treated patients (two-tailed Fisher’s exact test P = 0.051, Fig. 1c), and in BRAF were statistically higher in anti-EGFR treated patients than those in anti-VEGF treated patients (Fig. 1c). This suggests that acquiring alterations in EGFR/MAPK pathways genes is a unique mechanism of resistance to anti-EGFR but not to anti-VEGF treatment in mCRC.
a, b Scatterplot showing the relationship between gene-level alteration frequencies in ctDNA and archival tissues from (a) 141 patients who received prior anti-VEGF therapies, and (b)113 patients who received prior anti-EGFR therapies. The effect size was estimated using Pearson’s correlation coefficient(r) calculated in GraphPad Prism, and the coefficient of determination (r2), representing the proportion of variance explained by the relationship, is displayed on the scatter plot. c Comparison of percentage of patients with emerging alterations between patients with prior anti-EGFR therapies and those with prior anti-VEGF therapies. Alterations that were absent from tumor tissue samples but detected in plasma samples were defined as emerging alterations. (All genes with at least one emerging alteration listed). Only known and likely oncogenic alterations in each gene were considered for the analysis. P-value was calculated using a two-tailed Fisher’s exact test to compare the percentage of patients with emerging alterations between those who received anti-EGFR therapies and those who received anti-VEGF therapies. Source data are provided as a Source Data file.
Given activation of the PI3K/AKT pathway, one of the major downstream pathways of EGFR, has been postulated as a potential mechanism of resistance to anti-EGFR therapies11,22,23,24,25 we sought to see if alterations in PI3K/AKT pathway genes were also selectively enriched in patients following anti-EGFR but not anti-VEGF therapies in mCRC. Surprisingly, no significant differences in the prevalence of acquired alterations in any of the three key PI3K pathway related genes, PIK3CA, PTEN, and AKT, was observed in patients receiving anti-EGFR therapies compared to those receiving anti-VEGF therapies (Fig. 1c and Supplementary Fig. 3).
To validate our findings in an independent dataset, we compared alterations detected in mCRC patients with Guardant360® ctDNA tests both prior to and after anti-EGFR (N = 131) or anti-VEGF (N = 142) treatment. Consistent with what we observed in the IMblaze370 study, significant enrichment of acquired mutations in EGFR/MAPK pathway genes like KRAS, NRAS, and MAP2K1 were also observed in patients receiving anti-EGFR compared to anti-VEGF treatment, whereas no difference observed in these three PI3K pathway related genes (PIK3CA, PTEN, and AKT) between the two patient groups (Supplementary Fig. 4a). Taken together, these data indicate that the acquired resistance mechanisms to anti-EGFR therapy converge into the activation of a single downstream signaling node, MAPK pathway.
KRAS Q61H and multiple rarely detected EGFR/MAPK gene mutant alleles at treatment naïve CRC tumors are frequently selected in individual mCRC patients following anti-EGFR but not anti-VEGF therapy
To gain further molecular and biochemical insights into preferred alterations driving more effective resistance to anti-EGFR treatment, we took a closer look at the prevalence of individual emergent allele alterations in these six enriched mutated EGFR/MAPK pathway genes. Surprisingly, KRAS Q61H, a rare KRAS mutation in treatment naive CRC with a prevalence of 1.42% based on the 36,966 mCRC tumor samples from the FoundationCore® database (most from archival tissue) (Source Data file) and only occurring in 1.97% (5 out of 254 patients) of the archival tumor tissue in the IMblaze370 study, was identified as the most common acquired mutations among all the acquired alterations examined following anti-EGFR treatment with the prevalence of 19.5% (Fig. 2a). In contrast, the prevalence of all the KRAS mutant alleles detected in ctDNA from patients receiving anti-VEGF therapies, including Q61H and the most common KRAS mutation in CRC, G12D, remained comparable to those detected in archival tumors (Fig. 2a).
Similarly, EGFR and MAP2K1 mutations, which are very rare in mCRC, with only 0.80% and 1.14% respectively detected in 36,966 mCRC tumors from the FoundationCore® database (Source Data file), and 1.35% for both genes detected in tumor tissues of IMblaze370 samples, were significantly enriched in ctDNA from IMblaze370 patients with prior anti-EGFR treatment with the prevalence of 27.4% and 15.9% respectively. Consistent with published literature, 77.4% of the acquired EGFR alterations were mutated in the extracellular domain (ECD) (amino acids 1 ~ 645) (Fig. 2b), which have been previously reported to be associated with anti-EGFR resistance by reducing binding of the therapeutic antibody to its receptor11,20,21,22,23,24,25,26 For patients with acquired MAP2K1 alterations, 89.5% were K57T/N mutated (Fig. 2c), which was also previously identified as a mechanism of acquired resistance to anti-EGFR therapy in mCRC11,12,13 Again, no enrichment of EGFR ECD or MAP2K1 K57T/N mutations was observed in ctDNA from IMblaze 370 patients with prior anti-VEGF therapies (Fig. 2b, c).
Importantly, enrichment of these distinct “rare” mutant-alleles by anti-EGFR therapy were also observed in the dataset with the Guardant360® ctDNA test both prior and post anti-EGFR treatment. In the 38 out of 131 mCRC patients with acquired alterations in KRAS, NRAS, BRAF, MAP2K1, EGFR, and NF1, KRAS Q61H mutation was the most prevalent acquired mutation with a prevalence of 45%, followed by EGFR ECD and MAP2K1 mutations (Supplementary Fig. 4b). These data clearly showed how the treatment naive mCRC mutational landscape evolves rapidly and specifically to overcome the selective pressure of EGFR signaling blockade.
Furthermore, multiple acquired alterations in these six EGFR/MAPK pathway genes, including but not limited to KRAS Q61H, MAP2K1 K57/N, EGFR ECD mutations, were observed in 51 out of 92 anti-EGFR treated IMblaze370 patients with wild-type RAS/BRAF in archival tumor tissue (Supplementary Fig. 5a, b). More strikingly, 15 out of these 51 patients harbored at least 5 emerging alterations, with as many as 12 co-occurring emerging mutations detected in a single patient, including 8 EGFR ECD mutations, 3 KRAS mutations, and 1 NF1 mutation (Supplementary Fig. 5b). In contrast, alterations in those genes, except NF1, are typically mutually exclusive in archival tumor tissues or in plasma samples following anti-VEGF therapies (Supplementary Fig. 5a). Next, we sought to investigate whether these acquired multiple alterations likely originated from one or multiple tumor clones. To investigate this question, we looked at the normalized mutation allele frequency (nMAF) of those frequently acquired allele mutations in individual IMblaze370 patients with prior anti-EGFR therapies. Supplementary Fig. 6 depicts heterogeneous nMAF heat maps for multiple distinct acquired EGFR/MAPK pathway gene mutations in individual patients, suggesting different tumor clones, bearing mutant resistant alleles that are rarely detected in treatment naïve or in patients treated with anti-VEGF, are frequently simultaneously selected/expanded under anti-EGFR treatment.
Altogether, these data underscore the strong selective pressure exerted by anti-EGFR treatment towards MAPK signaling reactivation, which results in the systemic enrichment of multiple and heterogeneous mechanisms of resistance in individual mCRC patients.
KRASQ61H mouse colon organoids preferentially activate MAPK but not PI3K signaling
Given that the KRAS Q61H mutation is preferentially selected in anti-EGFR treated CRC, and our data showed the genetic mechanisms of resistance to anti-EGFR therapy converge into the activation of MAPK pathway, we speculated that KRASQ61H mCRC, comparing to other KRAS mutants, may preferentially signal through the MAPK pathway relative to other key EGFR downstream signaling pathways, like the PI3K/AKT pathway. To test this hypothesis, we examined MAPK and PI3K/AKT signaling in organoids derived from mouse colon tissue harboring KRAS Q61H and other KRAS mutations like G12D, G12C, G12V, or Q61R. As hypothesized, we found that the phospho-S6 and phospho-AKT levels were barely detectable in KRASQ61H organoids, whereas its levels in all the rest of organoids were much higher (Fig. 3a). In contrast, the phospho-ERK1/2 levels in KRASQ61H organoids were comparable to those in other KRAS mutant organoids (Fig. 3a). These data suggest that the oncogenic activity of KRASQ61H is likely independent of PI3K/AKT but more dependent on MAPK pathway activation, and KRASQ61H would be more sensitive to MAPK pathway inhibitors compared to those with other KRAS mutations.
a Western blots to detect p-ERK1/2, P-S6, and P-AKT levels in organoids with different KRAS mutants. The data shown are representative of three independent experiments. b, c Comparison of relative viability of KRASQ61H CRC cell line with KRASG12D and KRASG13D CRC cell lines in response to MEK inhibitor GDC-0973 (b) and pan-RAF inhibitor GDC-5573 treatment (c). Data are presented as mean ± SEM and are representative of three independent experiments. Source data for panels are provided as a Source Data file.
Next, we treated three representative CRC cell lines, CL-11(KRAS Q61H), LS-174T (KRAS G12D), and LoVo (KRAS G13D) with MEK inhibitor, GDC-0973, or Pan-RAF inhibitor, GDC-5573. We found CL-11 was more sensitive to MEK and RAF inhibitors compared to LoVo and LS-174T (Fig. 3b, c). The IC50 values for GDC-0973 were much lower in CL-11 compared to LS-174T and LoVo cells (0.05 µM vs. 0.22 µM and 0.28 µM). Similarly, the IC50 values for GDC-5573 were much lower in CL-11 compared to LS-174T (0.05 µM vs. 0.70 µM). Although CL-11 and Lovo had similar IC50 values for GDC-5573 (0.16 µM vs. 0.15 µM), CL-11 was more sensitive to GDC-5573 with much higher maximal efficacy compared to that in LoVo (Fig. 3b, c). These findings were consistent across six additional CRC cell lines, where similar trends were observed (Supplementary Fig. 7). Taken together, these data suggest that patients with KRASQ61H are likely to be more sensitive to MAPK pathway inhibitors compared to those with other KRAS mutations.
Enrichment of 8q gain observed in patients receiving anti-EGFR treatment while trend toward 13q loss observed in anti-VEGF treated patients
Beyond acquired mutations, copy number gain or gene amplifications, like those in EGFR, KRAS, BRAF, MET, and ERBB2, have been identified as another key acquired resistance mechanism to anti-EGFR treatment in mCRC11,12,13,14,15,16,17,18,19,20 Consistent with the literature reported, enrichment of EGFR, KRAS, and BRAF amplifications were also observed in patients treated with anti-EGFR therapy from the IMblaze370 study (Supplementary Fig. 5b). Gene-level copy number calling for this analysis was limited to the 62 baited genes. As a result, most copy number changes out of these regions were not called. To identify additional chromosomal segments beyond these 62 genes that may differentially affected by anti-EGFR and anti-VEGF treatment in mCRC, we utilized the genome-wide SNPs baited to identify regions of chromosomal gain or loss in the ctDNA (post-treatment) relative to the paired tumor tissue samples (prior treatment) of the 296 patients from the IMblaze370 study, including 113 treated with anti-EGFR and 141 treated with anti-VEGF therapies. Briefly, the method compares coverage log-ratio values to identify outliers from the sample’s base ploidy. These outliers are annotated as gains or losses based on their direction of deviation. This analysis identified 254 gained and 153 lost regions in the 296 samples. Interestingly, these identified gained and lost regions were not randomly distributed throughout the genome but rather occurred in a high frequency in chromosomes 8, 13, 18 and 20 (Fig. 4). To further determine whether any of those changes were treatment associated, recurrent gains and loss (defined as those with at least 10 occurrences in the dataset) were analyzed for treatment arm enrichment/depletion using Fisher’s exact test. The analysis of recurrent gains and losses showed a significant enrichment of 8q gains (Chr8: 46M-147M) in the anti-EGFR treatment group relative to anti-VEGF treatment group (Supplementary Table 1, 18.6% v 6.4%, corrected p = 0.019). A trend towards 20q loss (Supplementary Table 2, Chr20: 55M-63M) in anti-EGFR group (anti-EGFR vs anti-VEGF: 7.1% vs 1.4%, raw p = 0.03; corrected p = 0.15), and 13q loss (Chr13: 18M-116M) in anti-VEGF group was also observed (Supplementary Table 2, anti-EGFR vs anti-VEGF: 3.5 % vs 9.9%, raw p = 0.05; corrected p = 0.16). The finding of the significant enrichment of 8q gain, which contains the c-MYC locus, in anti-EGFR treated patients is particularly interesting as c-MYC amplification has been previously reported to be involved in the anti-EGFR therapy resistance in mCRC27. To further investigate if the 8q gain in anti-EGFR treated patients would co-occur with any specific mutations, we checked the mutations detected in the ctDNA of those patients. Intriguingly, we found that 12 out of 21 patients with 8q gain had mutations in EGFR/MAPK pathway genes (Source Data file).
We further validated the 8q gain findings in an independent cohort of 544 patients from the FoundationCore® database with available NGS data from paired tumor tissue (most are treatment naïve samples) and plasma samples (most are post-treatment). As the previous treatment history for those patients were unknown, we used the status of the rare CRC mutations in EGFR/MAPK pathway genes, i.e., EGFR ECD, MAP2K1, or KRAS Q61H mutations, in plasma samples to infer their prior treatment history. Patients with any of those acquired mutations would be considered as receiving prior anti-EGFR treatment. Otherwise, patients would be considered not receiving prior anti-EGFR treatment. Consistent with what we observed from IMblaze370 patients, a higher rate of 8q gains were observed in the patient group that had acquired alterations in those genes relative to those without acquired mutations (Source Data file, 24% vs 9%, p = 0.01). Together, these data suggest 8q gain, with MYC located on, may represent a mechanism of acquired resistance to anti-EGFR therapies in mCRC.
Proliferative CRC tumors with high EGFR-ligand expression, MYC activation and gamma delta-T cells are more susceptible to anti-EGFR treatment and prone to acquire EGFR/MAPK mutations
To understand the cell intrinsic properties that may be associated with the acquisition of EGFR/MAPK mutations following anti-EGFR treatment, we leveraged the RNAseq data from the archival tissue samples of the IMblaze370 study. Out of the 113 anti-EGFR treated patients with acquired mutation status, 74 patients were RAS/BRAF wild type in archival tissues and had RNA-seq data. 43 of those patients had acquired mutations in EGFR/MAPK genes, and 31 did not.
First, we applied an unsupervised, unbiased machine learning clustering approach based on consensus non-negative matrix factorization (cNMF)28 to bulk RNAseq data to identify transcriptome-based subtypes and then assessed if they may be more prone to acquire EGFR/MAPK mutations following anti-EGFR treatment. The cNMF analysis identified two clusters (k = 2), NMF2.1 and NMF2.2, with the greatest cophenetic coefficient factor (0.9752) compared to more clusters (k = 3 ~ 8) (Fig. 5a), and NMF 2.1 was significantly enriched for patients with acquired EGFR/MAPK mutations (Fig. 5b, p = 0.025). Gene set variation analysis (GSVA) using the hallmark gene sets from the Molecular Signature Database (MSigDb)29 and cell type enrichment analysis using the xCell algorithm30 were performed to biologically characterize the two subtypes. Out of the evaluated hallmark gene sets, gene sets involved in MYC targets, DNA damage, unfolded protein response, and reactive oxygen species (ROS) had much higher scores in NMF 2.1, which enriched for patients with acquired EGFR/MAPK gene mutations, whereas high stromal and high angiogenesis characterized NMF 2.2. Interestingly, cell type enrichment analysis revealed epithelial cells, gamma delta T-cells (Tgd cells), and basophils were significantly enriched in NMF2.1 subtype, whereas NMF 2.2 was enriched for patients with high B cells, dendritic cells, CD4 and CD8 + T cells (Fig. 5C).
a Cophenetic coefficient at various values of k test (k = 2–8). K = 2, with a cophenetic coefficient of 0.9752, was chosen. b Consensus patient similarity matrix depicting clusters (k = 2). The pie chart depicts the fraction of patients in each subtype with acquired mutations. c Heatmap patient samples sorted by NMF cluster assignment in the column, and the most significantly (FDR corrected Krueskal Wallis p-value < 0.05) NMF associated MSigDb hallmark pathways and xCell cell types clustered in row. P-value determined using a Kruskal-Wallis rank sum test. d Changes in tumor volume from baseline to 3 weeks post-cetuximab treatment in 244 PDX models with NMF 2.1(N = 221) and NFM2.2 (N = 23) tumors. The box and whisker plot shows the median, quartiles (hinges), whiskers, and individual outliers. P-value determined using a two-sided Wilcoxon rank sum test. Source data for panels (c and d) are provided as a Source Data file.
We further sought to assess/validate the key molecular features underlying the difference in acquiring EGFR/MAPK gene mutations by directly comparing gene expression levels in tumors from the two groups, patients with acquired mutations (N = 43) vs. those without acquired mutations (N = 31). Differential gene expression analysis using a Limma-Voom model with correction for primary tumor versus metastasis identified 731 differentially expressed genes (p-value < 0.05) between the two groups (Fig. 6a). Out of the 731 differentially expressed genes, we noticed the expression levels of two genes, amphiregulin (AREG) and epiregulin (EREG), were significantly higher in patients with acquired mutations than those without (Fig. 6a, b). AREG and EREG, two ligands of the EGFR family, are known to be particularly relevant to anti-EGFR treatment in CRC. High AREG and EREG mRNA expression in primary tumors have been shown to be consistently associated with improved outcome to anti-EGFR therapies in RAS-wild type mCRC patients31,32. These data indicate that acquired alterations following anti-EGFR treatment are more likely to arise in patients with high EREG or AREG levels at baseline. Then differential analysis between patients with and without acquired mutations was performed using hallmark and xCell gene set signature scores predicted early. Consistent with what was observed from the unbiased NMF analysis, gene signatures for Wnt/b-catenin, ROS, TP53, MYC pathways, Tgd and basophil cells were more prominently expressed in patients with acquired mutations, whereas B-cells and fibroblast genes were more prominently expressed in patients without acquired alterations (Fig. 6c). Moreover, we also observed patients with acquired EGFR/MAPK gene alterations were enriched in consensus molecular subtype 2 (CMS2), characterized with marked MYC and WNT signaling, whereas patients without acquired EGFR/MAPK gene mutations were enriched in CMS4 with prominent stromal invasion and angiogenesis (Supplementary Fig. 8)33. Interestingly, patients with acquired mutations also exhibited significantly higher circulating tumor DNA fractions (Supplementary Fig. 9), which may indicate greater tumor aggressiveness and an increased likelihood of detecting emerging alterations in those tumors.
a Volcano plot representing differentially expressed genes between patients with and without acquired alterations. Genes with FDR-corrected p < 0.05. P-value determined using a Limma-Voom moderated t test. b Comparison of EGFR ligands (EREG and AREG) expression levels between the two groups. Data are presented as mean +/− SD. The p-value was generated using a two-tailed, unequal variances two-sample t test. c Heatmap patient samples sorted by acquired mutation status in the column and the most significantly (FDR corrected Krueskal Wallis p-value < 0.05) acquired mutation status associated MSigDb hallmark pathways and xCell cell types clustered in row. P-value determined using a Kruskal-Wallis rank sum test. d Changes in tumor volume from baseline to 3 weeks post-cetuximab treatment in 244 PDX models with EREG high (above median, n = 122) and EREG low (below median n = 122). The box and whisker plot shows the median, quartiles (hinges), whiskers, and individual outliers. P-value determined using a two-sided Wilcoxon rank sum test. Source data are provided as a Source Data file.
Finally, we evaluated the relationship between the two distinct tumor subtypes (NMF 2.1 and NMF2.2) and response to anti-EGFR treatment. Although the IMblaze 370 study lacked direct clinical outcomes for anti-EGFR treatment, we leveraged Novartis’s patient-derived xenografts (PDX) datasets34, which included a cohort of 244 cetuximab-treated PDX samples with response and gene expression data. To classify each PDX sample into NMF subtypes, we trained a random forest machine learning algorithm (R package random Forest) using the IMblaze 370 bulk RNA-seq data as the training set. The random forest-based classifier, based on an ensemble of binary decision trees, predicts the class of a given sample by evaluating its similarity to patterns observed in the training set. Using this approach, we assigned NMF subtypes to each PDX sample. As hypothesized, PDX samples classified as NMF2.1 demonstrated significantly better responsiveness to anti-EGFR treatment compared to those classified as NMF 2.2, evidenced by a smaller increase in tumor volume at 3 weeks post treatment (Wilcoxon p = 9e-04, Fig. 5d). Similarly, PDX samples with high EREG expression (above median) exhibited significantly better response to anti-EGFR treatment compared to those with lower expression (below median) (Wilcoxon p = 1.2e-07, Fig. 6d). These results support the hypothesis that NMF2.1 tumors, characterized by high EGFR ligand expression, MYC and WNT activation, are intrinsically more sensitive to anti-EGFR treatment and more prone to acquiring resistance associated mutations.
In summary, these findings suggest that highly proliferative CRC tumors with high MYC, WNT activation, EGFR ligands (EREG, AREG), and high Tgd cells may be more susceptible to acquire resistant mutations upon anti-EGFR therapy, whereas tumors with high stromal and immune gene signature at baseline were more likely to display primary resistance to anti-EGFR therapy and unlikely to acquire EGFR/MAPK mutations.
Discussion
The work presented here investigates mechanisms of acquired resistance to anti-EGFR and anti-VEGF containing therapies in a robust dataset comprising 254 mCRC (2/3 L +) patients. This large collection of well-annotated clinical and NGS data at two longitudinal time-points of the patient journey, treatment naive archival tissue vs. 2/3 L + baseline plasma ctDNA, enables us to comprehensively interrogate the evolution of treatment-specific acquired mechanisms of resistance under the selective pressure of two effective targeted therapies in mCRC. Although the mechanisms of acquired resistance to anti-EGFR therapy have been extensively investigated11,13,14,15,16,17,18 the size and the completeness of this dataset, coupled with the prior treatment information, particularly the inclusion of both anti-EGFR and anti-VEGF treatment cohorts, differentiate this study. The totality of data in this study not only validates previously known mechanisms of resistance to anti-EGFR but also uncover critical insights that contribute to a comprehensive molecular roadmap for future drug development in CRC.
The treatment-specificity and high frequency of acquired multiple activating alterations in EGFR/MAPK pathway genes in individual patients provides compelling evidence for a strong and focalized selective pressure towards genetic EGFR/MAPK reactivation in RAS/BRAF-wild type CRC patients treated with anti-EGFR agents. In addition, the collective high prevalence of KRAS, BRAF, and NRAS activating mutations at diagnosis (~ 50–60%) suggest downstream KRAS/MAPK pathway addiction is not exclusively restricted to EGFR-driven wild-type patients but rather a common feature across most CRC tumors. This hypothesis is further supported by (i) the well-known intrinsic resistance of KRAS, BRAF, and NRAS mutant patients to anti-EGFR treatment, (ii) the established mechanisms of adaptive resistance to BRAF V600E inhibitors in BRAF V600E-mutant CRC tumors, and the synergic efficacy of BRAF V600E inhibitor and anti-EGFR combinations in these patients6, and (iii) the emerging data on acquired resistance to KRAS G12C inhibitors and increased activity shown in combination with anti-EGFR in mCRC35,36,37,38,39,40. Recent clinical data revealing MAPK-pathway activating mutations as possible mechanisms driving resistance to NTRK and HER2 inhibition in CRC41,42 further strengthens this hypothesis and indicates other oncogenic mutations, frequently found in CRC, may also contribute to disease progression by converging in activation of key MAPK effectors.
Contrary to common postulation, we found no significant enrichment of PI3K pathway gene mutations in patients receiving prior anti-EGFR treatment compared to those receiving anti-VEGF therapies (Fig. 1c and Supplementary Fig. 3). The lack of association of PIK3CA mutation with anti-EGFR treatment resistance was also reported in another study43. All these data suggest that the acquired resistance mechanisms to anti-EGFR therapy predominantly converge on the MAPK pathway. In addition, our data indicate that not all acquired mutations in the EGFR/MAPK pathway have equal chances of being selected. For instance, KRAS Q61H, a very rare mutation at diagnosis, is preferentially selected by anti-EGFR treatment in mCRC, becoming the most prevalent acquired mutation post anti-EGFR treatment. This indicates the presence of a specific mechanism that may be particularly effective in activating the MAPK pathway and conferring resistance. As hypothesized, recent publications showed that the Q61H mutation impairs both intrinsic and GAP-mediated GTP hydrolysis, which results in the decoupling of KRAS from upstream regulation. The unique properties of KRAS Q61H mutation leads to constitutive activation of the downstream MAPK signaling independent of upstream receptor tyrosine kinase (RTK) activity in cell lines44,45 Consistently, we found that KRASQ61Horganoids preferentially activate MAPK over PI3K signaling, and KRASQ61H CRC cell lines are more sensitive to MAPK pathway inhibitors compared to those with other KRAS mutations such as G12D, G12V, G13D (Fig. 3a–c and Supplementary Fig. 7). We acknowledge, however, that the potential differences in the genetic background of those cell lines limit our ability to conclusively attribute the difference in sensitivity to MAPK pathway inhibitors solely to these mutations. These findings collectively suggest that identifying and targeting the common EGFR/MAPK downstream oncogenic effector node could contribute to improving the efficacy of current anti-EGFR therapies in CRC. The initial encouraging clinical activity observed for RMS-6236, a RAS (ON) multi-selective noncovalent inhibitor of the active, GTP-bound state of both mutant and wild type variants of canonical RAS isoforms, in two patients with advanced KRASG12X lung and pancreatic adenocarcinoma46 coupled with the data provided here, supports future evaluation of combinations of RMC-6236 or other pan-KRAS inhibitors plus anti-EGFR in both wild type and KRAS mutant CRC patients.
In contrast to anti-EGFR therapies, the understanding of the mechanisms of acquired resistance to anti-VEGF therapy in CRC remains largely elusive. Our study confirms that anti-VEGF-containing therapy does not appear to select for secondary mutations, at least in the 62 frequently mutated oncogenes evaluated in the present study. The stark contrast observed for acquired genomic mechanisms of acquired resistance between the two drugs investigated suggests that genomic resistance may be more common in drugs directly targeting tumor cells, such as anti-EGFR, than in those targeting tumor environments, such as anti-VEGF. Furthermore, our analysis revealed a significant enrichment of 8q gain in patients treated with anti-EGFR (Fig. 4). This finding is particularly intriguing as it encompasses the c-MYC locus, previously implicated in conferring resistance to anti-EGFR in mCRC27. This suggests 8q gain may represent a mechanism of acquired resistance to anti-EGFR therapies in mCRC, while 13q loss may confer acquired resistance mechanisms to anti-VEGF treatment.
While our data, along with previous studies, indicate that secondary mutations in the EGFR/MAPK pathway are pivotal mechanisms underlying acquired resistance to anti-EGFR therapies in mCRC, it remains unclear whether the emergence of these mutations following anti-EGFR treatment is linked to biological predisposition prior to treatment. Our unbiased, unsupervised transcriptomic-based analysis revealed distinct patient subtypes with differing biological features and differential susceptibilities to acquire EGFR/MAPK mutations upon anti-EGFR treatments. Interestingly, proliferative tumors with high MYC activation and gamma delta T-cells (Tgd cells) appeared more susceptible to acquire secondary mutations, whereas tumors with high stromal and immune gene signatures (low in EGFR ligands) were more likely refractory to anti-EGFR therapy and unlikely to acquire EGFR/MAPK mutations (Fig. 5c). These findings were further validated in an independent cetuximab-treated PDX dataset. Within this dataset, we found NMF2.1 PDX tumors, characterized by high MYC, WNT activation along with Tgd cell enrichment, demonstrated greater sensitivity to anti-EGFR treatment comparing to NMF2.2 PDX tumors (Fig. 5d)
This analysis also showed significantly higher levels of EGFR ligand expression, AREG and EREG, in tumors from patients with acquired mutations upon treatment (Fig. 6a, b). This data is consistent with the established value of these ligands as predictive biomarkers for anti-EGFR efficacy and with the notion that baseline ligand expression identifies tumors that will be likely more sensitive to the selective pressure of ligand-dependent EGFR signaling blockade31,32 Furthermore, consistent with previous reports22, we found that mutations in the receptor tyrosine kinase (RTK) domain of EGFR, enabling ligand-independent EGFR activation, are exceedingly rare both at diagnosis (archival tumor tissue) and post anti-EGFR treatment (ctDNA) in IMblaze 370 study (Fig. 2b). In stark contrast, mutations in EGFR-ECD (crucial for EGFR binding) were among the most prevalent acquired mutations (Fig. 2b and20,21,22,23,24,25,26). These findings collectively suggest that reactivation of EGFR signaling in mCRC, in contrast to NSCLC, relies more heavily on EGFR ligands and its interactions with the EGFR-ECD domain than on constitutive EGFR-RTK mutation-driven signaling. Although we can only speculate about the reasons behind this ligand-activated signaling dependency in CRC, there is abundant published data describing the differences between EGFR ligand-independent and ligand-dependent signaling and even extending to instances where EGFR is activated by different ligands47,48,49. Consistently, PDX tumors with high EREG expression exhibited significantly better response to anti-EGFR treatment compared to those with lower expression (Fig. 6d). However, unexpectedly, no significant difference in response was observed between PDX tumors with high versus low AREG expression (Supplementary Fig. 10a), possibly due to much lower AREG expression levels relative to EREG in this dataset (Supplementary Fig. 10b). The concept of oncogenic EGFR-ligand-activated signaling dependency provides a specific biological vulnerability that could lead to EGFR-ligand blocker development as a more effective and/or complementary second-generation EGFR pathway inhibitor for mCRC. Finally, the data presented here also provide further evidence and rationale for using high EREG/AREG expression as a biomarker for patient selection in anti-EGFR clinical studies in CRC.
In closing, our study provides valuable insights into the distinct mechanisms of acquired resistance to anti-EGFR vs. anti-VEGF therapies in mCRC and a comprehensive road map of vulnerabilities that inform future drug development strategies. Our data further support clinical evaluation of: (i) High EGFR ligands expression as a predictive efficacy biomarker to anti-EGFR therapy, (ii) the evaluation of EGFR and pan-RAS inhibitors combination activity in wild-type and KRAS mutant CRC, and (iii) the development of a next-generation anti-EGFR-ligand antibodies to complement/improve conventional anti-EGFR therapeutics antibodies in CRC.
Methods
IMblaze 370 study and specimen collections
Samples for this analysis were collected from the IMblaze370 study (NCT02788279), a randomized phase III study comparing atezolizumab plus cobimetinib and atezolizumab monotherapy vs regorafenib in previously treated unresectable locally advanced or metastatic CRC. The study followed Good Clinical Practice guidelines and the Declaration of Helsinki. Ethical approval was obtained from independent ethic review boards at each study sites. A full list of the ethics review board is provided in Supplementary Data 1. All participants gave informed consent for biomarker analyses described in the protocol and for publication of the results. The study design, patient eligibility, and clinical outcome of this trial have been reported previously21. Since sex and gender or participants are not relevant to this analysis, no sex or gender analysis was carried out in this study.
Of 363 enrolled patients, 296 patients have next-generation sequencing from both archival tumor tissue samples and baseline plasma samples collected immediately before the IMbalze370 C1D1 treatment initiation, but after receiving prior lines of therapy, including but not limited to anti-EGFR or anti-VEGF therapies. To investigate acquired resistance mechanisms to anti-EGFR or anti-VEGF therapies, those patients were stratified into two groups: patients who received anti-EGFR (N = 113) therapies vs those who received anti-VEGF (N = 141) but not anti-EGFR therapies at any time before enrollment into the IMBLAZE370 trial (Supplementary Fig. 1).
Tumor samples and sequencing
Genomic alterations in archival tumor samples collected at diagnosis or surgery from patients who participated in the IMblaze370 study were detected using the FoundationOne® (F1) assay (Foundation Medicine, Cambridge, MA) following standard procedure50. FoundationOne® identifies base substitutions, insertions, and deletions(indels), amplifications, deletions, and rearrangements. The FoundationOne® sequencing panel version used in this study included the entire coding regions up to 395 cancer-related genes and select introns of 31 genes that are frequently rearranged or altered in cancer, with a median sequencing depth greater than 500X. A summary of the genomic alterations detected in archival tumor samples have been provided in the Source Data file.
Plasma samples and ctDNA sequencing
Genomic alterations in plasma samples collected at baseline from patients who participated in the IMblaze370 study were detected using the FoundationACT (FACT) assay (Foundation Medicine, Cambridge, MA) following standard procedure51. FACT is a panel of 62 genes. FACT is typically sequenced to a depth of ~ 5000X, with assay validation data showing that SNVs and indels can be detected down to 0.5–1% frequencies with > 99% sensitivity. A summary of the genomic alterations detected in plasma samples have been provided in the Source Data file. Quantification of ctDNA fraction in plasma samples was following the standard procedure as previously described52.
Comparison of genomic alterations detected in tumor and plasma samples
For consistency, all the analysis reported for the IMblaze370 study was only adjusted to include 62 genes that covered by both F1 and FACT assays, and those known or likely pathogenic alterations defined by Foundation Medicine, which include the known or likely somatic short-variants, rearrangements, and copy number variations. For comparison of the gene-level prevalence of genomic alterations detected in tumor tissue and plasma, samples would be considered mutant as long as one known or likely alteration was detected. Alterations that were absent from tumor tissue but detected in plasma samples were defined as distinct secondary alterations or emerging alterations. Only previously reported and functionally validated resistance alterations were counted as resistance mechanisms.
Foundation medicine database
Samples included in the FoundationCore® database were not selected and thus represented all comers. Tumor and blood samples were submitted to Foundation Medicine (Boston, MA) for DNA and cell-free DNA (cfDNA) extraction following the standard procedure as described previously53. 36966 tumor DNA and 1367 cfDNA were analyzed using FoundationOne® or FoundationOne® CDx54 assay and FoundationACT assay respectively. Patients were not consented for raw data release. However, variants from a subset of the 36,966 tumor samples used in this analysis have been deposited in the Genomic Data Commons (Accession #phs001179). Summary of the gene variants identified in the 36966 tumor samples assessed in this study have been provided in the Source Data file.
Guardant360 dataset
Patients with colorectal cancer from the GuardantINFORMTM database (INFORM DB) were used as an independent validation cohort for this study. INFORM DB contains genomic information from patients tested using the Guardant360® (G360) liquid biopsy test linked with U.S. administrative claims data. Patient identifiers from raw data sources were removed and anonymized. Out of all the CRC patients who received at least one G360 test, 131 and 142 patients were identified receiving G360 tests both before and after anti-EGFR or anti-VEGF treatment, but not anti-EGFR treatment, respectively. G360 test is a comprehensive genomic profiling test that uses targeted high-throughput hybridization-based capture technology for the detection of single-nucleotide variants (SNVs), insertions and deletions (indels) in 74 genes, copy number amplifications (CNAs) in 18 genes, and fusions in six genes55. Clinically reported oncogenic or likely oncogenic variants defined by OncoKB were compared at the patient level before and after anti-EGFR/VEGF treatment. Percentage gains and losses were summarized on gene level in each cohort.
Mutation clonality analysis
In order to estimate the mutation clonality of the acquired mutation alleles of EGFR/MAPK pathway genes in IMblaze 370 patients with prior anti-EGFR treatment, we normalized cfDNA mutation allele frequency (MAF) to both maximum MAF and copy number (if there was a reported amp in the same gene as a point mutation), but that the copy-number normalization was conditional on the point mutation being likely not sub-clonal. Otherwise, MAF for the point mutation was normalized to the maximum MAF only. We normalized MAF to CN only if the initial MAF/maximum MAF ratio was > 0.1, using a MAF/log2(CN) normalization for these mutations. Dividing each adjusted cfDNA alteration MAF by the adjusted maximum somatic MAF in a given sample then yielded the estimated alteration clonality present in the tumor as previously described56.
KRAS mutant organoid generation, culture and genome editing
All animal studies were approved by Genentech’s Institutional Animal Care and Use Committee and adhere to the NRC Guidelines for the Care and Use of Laboratory Animals. Female mice were used exclusively for this experiment due to their availability at the time of the experiment. The biological sex of the donor animals is not expected to influence the outcomes or interpretations of our findings, as the experimental design and endpoints were not sex dependent. Colon from adult C57BL/6 female mice was removed, flushed, opened lengthwise, and washed in cold PBS to remove all luminal contents. The colon was cut into 0.5–1 cm pieces in cold PBS, vortexed, washed for 3 times, and placed into 25 ml 2.5 mM EDTA-PBS for 5 min at 37 °C. The supernatant was then removed, and the colon pieces were washed with PBS, followed by incubation in 25 ml 5 mM EDTA-PBS for 15 min at 37 °C. After being vigorously vortexed, supernatant was collected and filtered through 100 μm filters, spun at 500 g for 5 mins, washed with cold PBS, and then resuspended 1:1 in IntestiCultTM media (StemCell, Cat. # 06005) and Matrigel (Corning, Cat. # 356231). IntesticultTM media was changed every two to three days, and organoids were passaged by mechanical disruption every 7–10 days.
To generate AK organoids, mutations in APC and various KRAS mutations were introduced by CRISPR-Cas9 technology. The following gene-specific sgRNAs were used: APC, CAGGACTGCATTCTCCTGAA; KRAS CTGAATTAGCTGTATCGTCA (various donor sequences provided in the Source Data file). Organoids were dissociated into a single-cell suspension using Accutase (Sigma-Aldrich) for 5 min at 37 °C and then electroporated with 2 μl of Cas9 and 3 μl of sgRNA using the P1 buffer and CM137 program (Lonza). After electroporation cells were embedded in Matrigel and DMEM advanced media (GIBCO) supplemented with 10 mM Hepes (Sigma-Aldrich), 2 mM GlutaMAX (Life Technologies), 1x Penicillin/Streptomycin (Life Technologies), 1x N2 (GIBCO), 1x B27 (GIBCO), 1 mM N-acetysteine (Sigma Aldrich), 50 ng/ml EGF (Life Technologies), 100 ng/ml Noggin (Peprotech), and R-spondin-1 (R&D systems). Selection for mutated cells was performed using growth factor depletion from the culture medium: R-spondin depletion to select for APC-deficient cells, EGF depletion to select for KRAS + /mutant cells.
Western blot analysis
Organoids were plated in 6-well plates (Corning, Cat. # 3516) in Matrigel plugs. When organoid growth was evident, cells were harvested by incubating the Matrigel plugs in PBS on ice. Lysates were clarified and prepared for western blot analysis using NuPAGE LDS Sample Buffer (4x) (Invitrogen, Cat. # NP0007) and NuPAGE LDS Sample Reducing Agent (10x) (Invitrogen, Cat. # NP0009). The proteins of interest were proved with corresponding antibodies. Primary antibodies included: Phospho-Erk (Thr202/Tyr204) (Cell Signaling Technology, Cat. #9101), Erk1 (Cell Signaling Technology, Cat. #9102), Phospho-S6 Ribosomal Protein (Ser235/236) (Cell Signaling Technology, Cat. #2211), S6 Ribosomal Protein (Cell Signaling Technology, Cat. #2217), Phospho-Akt (Ser473) (Cell Signaling Technology, Cat. #9271), Akt (pan) (Cell Signaling Technology, Cat. #9272), β-Tubulin (Cell Signaling Technology, Cat. #2146). All primary antibodies were diluted 1:1000, but Tubulin was diluted 1:2500. Western blots were conducted in three biological replicates.
Cell viability assay
Cell lines used in this analysis were obtained from the American Type Culture Collection (ATCC) or the Leibniz Institute DSMZ (Braunschweig, Germany). Cell line identity was verified through high-throughput SNP profiling using Fluidigm multiplexed assays for all cell lines. SNP profiling was performed each time new stocks were expanded for cryopreservation. SNPs were selected based on minor allele frequency and presence on commercial genotyping platforms. The SNP profiles were compared to SNP calls from available internal and external data (when available) to determine or verify the cell lines’ ancestry. In cases where data was unavailable or the ancestry was questionable, cell lines were re-purchased to perform profiling to confirm their ancestry. All cell lines were tested for mycoplasma prior to and after cryopreservation using the Lonza MycoAlert Mycoplasma Detection Kit (LT07-318).
Cells were incubated (37 °C, 5% CO2) in RPMI 1640, 5% FBS, and 2 mM glutamine throughout the assay. MEK inhibitor (GDC-0973) and Pan-RAF inhibitor (GDC-5573) were from Genentech in-house synthesis. Both GDC-0973 and GDC-5573 were screened in a 9-point dose response using a 3-fold dilution. Cell viability assay was performed as previously described57. In detail, cells were plated at previously determined optimal seeding densities. 24 hours after cell plating, compounds were added using a Bravo Automated Liquid-Handling Platform (Agilent; Santa Clara, CA). After 120 hours, 25μl CellTiter-Glo reagent was added using a MutliFlow TM Microplate Dispenser (BioTeck, Winooski, VT). Cell lysis was induced by mixing for 30 min on an orbital shaker. Plates were then incubated at room temperature for 10 minutes to stabilize the luminescent signal. Luminescence was read by a 2104 EnVision Multilabel Plate Reader (PerkinElmer, Waltham, MA). Three independent biological replicates were produced. The reported IC50 is the dose at which the cross-run estimated inhibition is 50% relative to DMSO control wells. In addition to absolute IC50, mean fitted viability across the nine tested doses (i.e., area under the viability curve) was also computed.
Genome-wide copy number analysis
Gain and loss calling beyond the baited genes was calculated using a signal: noise ratio approach. The genome was tiled with 1MB segments. Within each segment, a Z-score was calculated, examining deviation from base ploidy. Regions with a Z-score of > 2.576 (99.5th percentile) were called as gains, while regions with a Z-score of < -2.576(0.5th percentile) were called as losses. Adjacent regions of gain or loss were aggregated to call an amplicon in each sample. The regions of recurrent gains and losses were called based on 10 or more samples harboring a gain or loss in the region in the overall study IMblaze370 dataset. Findings were validated using a real-world dataset of colorectal cancer patients profiled with FoundationOne®, FoundationOne®CDx, FoundationOne®LiquidCDx58, or Foundation-ACT during the course of routine clinical care in the United States. 544 patients with multiple biopsies were available for analysis. Alterations in at least 62 genes, including EGFR, MAP2K1, and KRAS, were examined.
Gene expression analysis
Gene expression level of the archival tumor samples from IMblaze 370 patients were evaluated using TruSeq RNA Access technology (Illumina®). Strand-specific RNA sequencing was performed, and the resulting data were analyzed using HTSeqGenie59 (https://bioconductor.org/packages/HTSeqGenie/) in BioConductor60 as follows: RNAseq reads were first aligned to ribosomal RNA sequences to remove ribosomal reads. The remaining reads were aligned to the human reference genome GRCh38 using GSNAP4,5 version ‘2013-10-10’, allowing maximum of two mismatches per 75 base sequences (parameters: ‘-M 2 -n 10 -B 2 -i 1 -N 1 -w 200000 -E 1 --pairmax-rna=200000 --clip-overlap). To quantify gene expression levels, the number of reads mapped to the exons of each RefSeq gene was calculated in a strand-specific manner using the functionality provided by the R package GenomicAlignments (Bioconductor). Then counts were normalized to library size as counts per million (RPKM) using the voom function of R limma package. The Z-score calculated from RPKM was used to compare the EREG and AREG gene expression levels. Differentially expressed gene analysis was performed using the Limma-Voom model with correction for primary versus metastatic sample. Gene set enrichment analysis was performed in the Gene Set Variation Analysis (GSVA) R package61 using Hallmark gene set collections from the Molecular Signatures Database29 Pathway GSVA enrichment scores were calculated for each gene set using the GSVA function with default settings. Cell type enrichment analysis was performed using xCell30. CMS subtypes were annotated using the CMScaller R package (version 0.99.2)62. Further details about the CMS caller package can be found on Github (https://github.com/peterawe/CMScaller) and in its original publications33,62
Unsupervised machine learning transcriptomic clustering approach based on Consensus Non-negative Matrix Factorization (cNMF)
To identify transcriptomic subtypes, we applied unsupervised machine learning clustering approach based on cNMF28 to normalized gene expression (log2(RPKM)) data from n = 74 archival tumor specimens from anti-EGFR treated patients with acquired mutation status that were RAS/BRAF wild type. The median absolute deviation (MAD) analysis (CRAN. R package version 0.22.0) was used to select 10% of genes with the highest variability across patients. To control for differences between primary and metastatic samples, the most variable genes were identified in the primary samples (n = 54). This type of clustering is based on the dimensional reduction methodology of NMF, which reduces the expression data from thousands of genes to a few metagenes (CRAN. R package version 0.22.0)28 combined with the consensus clustering to test the stability of iterative NMF runs. This method computes multiple k-factor factorization decompositions of the expression matrix and evaluates the stability of the solutions using a cophenetic coefficient. We tested k2–8 clusters and used the maximal optimization of the cophenetic score to determine the number of clusters (Fig. 5a).
Novartis PDX data analysis
To classify each PDX sample into NMF subtypes, we trained a random forest machine learning algorithm (R package random Forest) using the IMblaze 370 RNA-seq data as the training set. The classifier, based on an ensemble of binary decision trees, predicts the class of a given sample by evaluating its similarity to patterns observed in the training set. Using this approach, we assigned NMF subtypes to each PDX sample using expression profiling of CRC patients derived xenografts in GSE7640234. For the EGFR ligand analysis, PDX samples were classified as high or low AREG and EREG based on the median expression. Wilcoxon t-test was performed to compare predicted NMF2.1 and NMF2.2 and between high and low AREG or EREG using the “response to cetuximab: tumor volume variation after 3 weeks of treatment” data.
Statistical analysis
To evaluate the concordance between alteration frequency in ctDNA vs. tumor tissue, Pearson correlation coefficients were calculated. To compare the percentage of patients with emerging alterations between patients with prior anti-EGFR therapies and those with prior anti-VEGF therapies, two-tailed Fisher’s exact test was applied. Pathway and cell type enrichment scores were compared using Kruskal-Wallis test using a FDR corrected p-value of 0.05 as the cutoff. To assess the relationship between NMF subtypes/ EGFR ligand expression levels and response to anti-EGFR treatment in Novartis PDX models, a two-sided Wilcoxon rank sum test was used.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Informed consent was obtained from all participants in the IMblaze 370 study for biomarker analysis specified in the protocol and for publication of the results. IMBLAZE 370 RNA-seq data will be made available indefinitely to qualified researchers at the European Genome-Phenome Archive under an accession number EGAS00001005952. To request access to such data, researchers can submit a data access request via the standard EGA process. Following publication, the data will be released upon application with necessary agreements to enforce terms such as security, patient privacy, and consent of specified data use, consistent with evolving, applicable data protection laws. The general turnaround right now is 2-3 months to make data available to requesters, and the data availability on the platform is ‘indefinite’ per our statement. The user, per the Data Use Agreement, only has a year to use the data, after that, it must be deleted. Due to privacy concerns and compliance restrictions, the release of raw data from the Foundation Medicine database and GuardantINFORMTM database (INFORM DB) is not permitted. However, variants from a subset of the 36,966 Foundation Medicine tumor samples used in this analysis have been deposited in the Genomic Data Commons (Accession #phs001179), and all the relevant data supporting the findings of this study are provided within the main manuscript and its supplementary files. Due to the HIPAA requirements, Foundation Medicine is not authorized to share individualized patient genomic data, which contains potentially identifying or sensitive patient information. Foundation Medicine is committed to collaborative data analysis, with well-established and widely utilized mechanisms by which investigators can query the Foundation Core genomic database of > 700,000 de-identified sequenced cancers to obtain aggregated datasets. For more information and the mechanism of access, please contact the corresponding author or the Foundation Medicine Inc. Data Governance Council at data.governance.council@foundationmedicine.com. Investigators and their institutions will be required to sign a data-sharing agreement. The PDX gene expression data used in this study are available in the GEO database under accession code GSE76402. The remaining data supporting this study are available within the article and its supplementary information. Source data are provided in this paper.
References
Siegel, R. L., Wagle, N. S., Cercek, A., Smith, R. A. & Jemal, A. Colorectal cancer statistics, 2023. CA Cancer J. Clin. 73, 233–254 (2023).
Sartore-Bianchi, A. et al. Dual-targeted therapy with trastuzumab and lapatinib in treatment-refractory, KRAS codon 12/13 wild-type, HER2-positive metastatic colorectal cancer (HERACLES): a proof-of-concept, multicentre, open-label, phase 2 trial. Lancet Oncol. 17, 738–746 (2016).
Scott, L. J. Larotrectinib: First global approval. Drugs 79, 201–206 (2019).
Al-Salama, Z. T. & Keam, S. J. Entrectinib: First global approval. Drugs 79, 1477–1483 (2019).
Le, D. T. et al. PD-1 blockade in tumors with mismatch-repair deficiency. N. Engl. J. Med. 372, 2509–2520 (2015).
Kopetz, S. et al. Encorafenib, binimetinib, and cetuximab in BRAF V600E–mutated colorectal cancer. N. Engl. J. Med. 381, 1632–1643 (2019).
Hurwitz, H. et al. Bevacizumab plus irinotecan, fluorouracil and leucovorin for metastatic colorectal cancer. N. Engl. J. Med. 350, 2335–2342 (2004).
Hurwitz, H. I. et al. Bevacizumab in combination with fluorouracil and leucovorin: An active regimen for first-line metastatic colorectal cancer. J. Clin. Oncol. 23, 3502–3508 (2005).
Ferrara, N., Hillan, K. J. & Novotny, W. Bevacizumab (Avastin), a humanized anti-VEGF monoclonal antibody for cancer therapy. Biochem. Biophys. Res. Commun. 333, 328–335 (2005).
Parseghian, C. M. et al. Resistance mechanisms to anti-epidermal growth factor receptor therapy in RAS/RAF wild-type colorectal cancer vary by regimen and line of therapy. J. Clin. Oncol. 41, 460–471 (2022).
Bertotti, A. et al. The genomic landscape of response to EGFR blockade in colorectal cancer. Nature 526, 263–267 (2015).
Tan, C. S. et al. Third generation EGFR TKIs: Current data and future directions. Mol. Cancer Mol. Cancer 17, 1–14 (2018).
Russo, M. et al. Tumor heterogeneity and Lesion-Specific response to targeted therapy in colorectal cancer. Cancer Discov. 6, 147–153 (2016).
Misale, S. et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature 486, 532–536 (2012).
Van Emburgh, B. O. et al. Acquired RAS or EGFR mutations and duration of response to EGFR blockade in colorectal cancer. Nat. Commun. 7, 1–9 (2016).
Yamada, T. et al. Utility of KRAS mutation detection using circulating cell-free DNA from patients with colorectal cancer. Cancer Sci. 107, 936–943 (2016).
Montagut, C. et al. Efficacy of Sym004 in patients with metastatic colorectal cancer with acquired resistance to anti-EGFR therapy and molecularly selected by circulating tumor DNA analyses a phase 2 randomized clinical trial. JAMA Oncol. 4, 1–9 (2018).
Parikh, A. R. et al. Liquid versus tissue biopsy for detecting acquired resistance and tumor heterogeneity in gastrointestinal cancers. Nat. Med. 25, 1415–1421 (2019).
Thierry, A. R. et al. Circulating DNA demonstrates convergent evolution and common resistance mechanisms during treatment of colorectal cancer. Clin. Cancer Res. 23, 4578–4591 (2017).
Strickler, J. H. et al. Genomic landscape of cell-free DNA in patients with colorectal cancer. Cancer Discov. 8, 164–173 (2018).
Eng, C. et al. Atezolizumab with or without cobimetinib versus regorafenib in previously treated metastatic colorectal cancer (IMblaze370): a multicentre, open-label, phase 3, randomised, controlled trial. Lancet Oncol. 20, 849–861 (2019).
Zhao, B. et al. Mechanisms of resistance to anti-EGFR therapy in colorectal cancer. Oncotarget 8, 3980–4000 (2017).
Mao, C., Yang, Z. Y., Hu, X. F., Chen, Q. & Tang, J. L. PIK3CA exon 20 mutations as a potential biomarker for resistance to anti-EGFR monoclonal antibodies in KRAS wild-type metastatic colorectal cancer: A systematic review and meta-analysis. Ann. Oncol. 23, 1518–1525 (2012).
De Roock, W. et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: A retrospective consortium analysis. Lancet Oncol. 11, 753–762 (2010).
Xie, Y. H., Chen, Y. X. & Fang, J. Y. Comprehensive review of targeted therapy for colorectal cancer. Signal Transduct. Target Ther. 5, 22 (2020).
Arena, S. et al. Emergence of multiple EGFR extracellular mutations during cetuximab treatment in colorectal cancer. Clin. Cancer Res. 21, 2157–2166 (2015).
Strippoli, A. et al. c-MYC expression is a possible keystone in the colorectal cancer resistance to EGFR inhibitors. Cancers 12, 638–655 (2020).
Brunet, J. P., Tamayo, P., Golub, T. & Mesirov, J. P. Metagenes and molecular pattern discovery using matrix factorization. Proc. Natl. Acad. Sci. USA 101, 4164–4169 (2004).
Liberzon, A. et al. The molecular signatures database(MSigDB) hallmark gene set collection. Cell Syst. 1, 417–425 (2015).
Aran, D., Hu, Z. & Butte, A. J. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 18, https://doi.org/10.1186/s13059-017-1349-1 (2017).
Jacobs, B. et al. Amphiregulin and epiregulin mRNA expression in primary tumors predicts outcome in metastatic colorectal cancer treated with cetuximab. J. Clin. Oncol. 27, 5068–5074 (2009).
Seligmann, J. F. et al. Combined epiregulin and amphiregulin expression levels as a predictive biomarker for panitumumab therapy benefit or lack of benefit in patients with RAS wild-type. JAMA Oncol. 2, 633–642 (2016).
Guinney, J. et al. The concensus molecular subtypes of colorectal cancer. Nature 21, 1350–1356 (2015).
Gao, H. et al. High-throughput screening using patient derived tumor xenografts to predict clinical trial drug response. Nat. Med. 21, 1318–1325 (2015).
Amodio, V. et al. EGFR Blockade reverts resistance to KRAS G12C inhibition in colorectal cancer. Cancer Discov. 10, 1129–1139 (2020).
Awad, M. M. et al. Acquired resistance to KRAS G12C inhibition in cancer. N. Engl. J. Med. 384, 2382–2393 (2021).
Sacher, A. et al. Single-agent Divarasid(GDC-6036) in solid tumors with a KRAS G12C mutation. N. Engl. J. Med. 389, 710–721 (2023).
Desa, J. et al. Divarasib plus cetuximab in KRAS G12C-positive colorectal cancer: a phase 1b trial. Nat. Med. 30, 271–278 (2024).
Yaeger, R. et al. Adagrasib with or without cetuximab in colorectal cancer with mutated KRAS G12C. N. Engl. J. Med. 388, 44–54 (2023).
Fakih, M. G. et al. Sotorasib for previously treated colorectal cancers with KRAS G12C mutation (CodeBreaK100): a prespecified analysis of a single-arm, phase 2 trial. Lancet Oncol. 23, 115–124 (2022).
Cocco, E. et al. Resistance to TRK inhibition mediated by convergent MAPK pathway activation. Nat. Med. 25, 1422–1427 (2019).
Siravegna, G. et al. Radiologic and genomic evolution of individual metastases during HER2 blockade in colorectal cancer. Cancer Cell 34, 148–162 (2018).
Woolston, A. et al. Genomic and transcriptomic determinants of therapy resistance and immune landscape evolution during anti-EGFR treatment in colorectal cancer. Cancer Cell 36, 35–50 (2019).
Zhou, Z.-W. et al. KRAS Q61H preferentially signals through MAPK in a RAF dimer-dependent manner in non-small cell lung cancer. Cancer Res. 80, 3719–3731 (2020).
Gebregiworgis, T. et al. The Q61 H mutation decouples KRAS from upstream regulation and renders cancer cells resistant to SHP2 inhibitors. Nat. Commun. 12, 6274–6288 (2021).
Jiang, J. et al. Translational and therapeutic evaluation of RAS-GTP inhibition by RMC-6236 in RAS-driven cancers. Cancer Discov. 14, 1–24 (2024).
Pines, G., Kostler, W. J. & Yarden, Y. Oncogenic mutant forms of EGFR: Lessons in signal transduction and targets for cancer therapy. FEBS Lett. 584, 2699–2706 (2010).
Chakraborty, S. et al. Constitutive and ligand-induced EGFR signaling triggers distinct and mutually exclusive downstream signaling networks. Nat. Commun. 5, 5811 (2014).
Freed, D. M. et al. EGFR ligands differentially stabilize receptor dimers to specify signaling kinetics. Cell 171, 683–695 (2017).
Frampton, G. M. et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat. Biotechnol. 31, 1023–1031 (2013).
Clark, T. A. et al. Analytical validation of a hybrid capture–based next-generation sequencing clinical assay for genomic profiling of cell-free circulating tumor DNA.J. Mol. Diagn. 20, 686–702 (2018).
Tukachinsky, H. et al. Genomic analysis of circulating tumor DNA in patients with advanced prostate cancer identified targetable BRCA alterations and AR resistance mechanisms. Clin. Cancer Res. 27, 3094–3105 (2021).
Hartmaier, R. J. et al. High-throughput genomic profiling of adult solid tumors reveals novel insights into cancer pathogenesis. Cancer Res. 77, 2464–2475 (2017).
Milbury, C. A. et al. Clinical and analytical validation of FoundationOneCDx, a comprehensive genomic profiling assay for solid tumors. PLos ONE 17, e0264138 (2022).
Gary, J. E. et al. Pan-tumor analytical validation and Osimertinib clinical validation in EGFR mutant non-small-cell lung cancer, supporting the first next-generation sequencing liquid biopsy in vitro diagnostic. J. Mol. Diagn. 1, 73–84 (2024).
Zill, O. A. et al. The landscape of actionable genomic alterations in cell-free circulating tumor DNA from 21,807 advanced cancer patients. Clin. Cancer Res. 24, 3528–3538 (2018).
Yen, I. et al. Pharmacological induction of RAS-GTP confers RAF inhibitor sensitivity in KRAS mutant tumors. Cancer Cell 34, 611–625 (2018).
Woodhouse, R. et al. Clinical and analytical validation of FoundationOne liquid CDx, a novel 324-gene cfDNA-based comprehensive genomic profiling assay for cancers of solid tumor origin. Plos ONE 15, e023780 (2020).
Wu, T. D., Reeder, J., Lawrence, M., Becker, G. & Brauer, M. J. GMAP and GSNAP for genomic sequence alignment: enhancements to speed, accuracy, and functionality. Methods Mol. Bio. 1418, 283–334 (2016).
Huber, W. et al. Orchestrating high-throughput genomic analysis with Bioconductor. Nat. Methods 12, 115–121 (2015).
Hanzelmann, S., Castelo, R. & Guinney, J. GSVA: Gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 14, 7–21 (2013).
Eide, P. W., Bruun, J., Lothe, R. A. & Sveen, A. CMScaller: an R package for consensus molecular subtyping of colorectal cancer pre-clinical models. Sci. Rep. 7, 16618–16625 (2017).
Acknowledgements
The IMblaze370 study was sponsored by F. Hoffmann-La Roche/Genentech, Inc. We thank the IMblaze370 team and the patients participating in this trial. We also thank the Foundation Medicine team for providing FoundationCore database data, patients who contributed to the FMI database, and Sophia Maund, Ben Lauffer, Armande Ang Houle, and Sonali Lad from Genentech for assistance with FMI database queries and discussions. We also thank Robert Piskol, Marc Hafner, Mamie Yu, and Scott Martin for providing guidance in analyzing and interpreting the cell line drug sensitivity data. We also thank Guardant Health for providing Guardant G360 data, and Nicole Zhang and Carin Espenschied for analyzing the data. We also would like to thank Chris Bolen, Herb Hurwitz, Federico Nasroulah, and Lousia Walker for reviewing and providing critical comments on the manuscript.
Author information
Authors and Affiliations
Contributions
Conception and design: X. Qu, H. Hamidi, C. Bais. Development of methodology: X. Qu, H. Hamidi, R. J. Johnson, E. S. Sokol, S. Sivakumar, B. Kaplan, F. de Sousa e Melo, A. Mancini, Y. Yan, C. Bais. Acquisition of data (provided animals, acquired, and managed patients, provided facilities, etc): C. Eng, T.W. Kim, J. Bendell, F. Ciardiello. Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): X. Qu, H. Hamidi, R. J. Johnson, E. S. Sokol, S. Sivakumar, B. Kaplan, F. de Sousa e Melo, Y. Yan, C. Bais. Writing, review, and/or revision of the manuscript: X. Qu, H. Hamidi, R. J. Johnson, C. Eng, T.W. Kim, J. Bendell, Y. Shi, E. S. Sokol, S. Sivakumar, B. Kaplan, F. de Sousa e Melo, M. Wongchenko, E. Lin, A. Mancini, D. Shames, Y. Yan, F. Ciardiello, C. Bais. Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): X. Qu, H. Hamidi, R. J. Johnson, E. S. Sokol, S. Sivakumar, B. Kaplan, E. Lin, M. Wongchenko, Y. Yan, C. Bais. Study supervision: X. Qu, H. Hamidi, Y. Yan, C. Bais.
Corresponding author
Ethics declarations
Competing interests
C. Eng reports grants and personal fees from Roche/Genentech and personal fees from Bayer. T. W. Kim reports grants from AstraZeneca, Pfizer, and Merck Serono outside of the submitted work. J. Bendell reports fees paid to her institution from Gilead, Genentech/Roche, BMS, Five Prime, Lilly, Merck, MedImmune, Celgene, EMD Serono, Taiho, Macrogenics, GSK, Novartis, OncoMed, LEAP, TG Therapeutics, AstraZeneca, BI, Daiichi Sankyo, Bayer, Incyte, Apexigen, Koltan, SynDevRex, Forty Seven, AbbVie, Array, Onyx, Sanofi, Takeda, Eisai, Celldex, Agios, Sytomx, Nektar, ARMO, Boston Biomedical, Ipsen, Merrimack, Tarveda, Tyrogenex, Ocogenex, Marshall Edwards, Pieris, Mersana, Calithera, Blueprint, Evelo, FORMA, Merus, Jacobio, Effector, Novocare, Arrys, Tracon, Sierra, Innate, Arch Oncology, Prelude Oncology, Unum Therapeutics, Vyriad, Harpoon, ADC, Amgen, Pfizer, Millennium, Imclone, Acerta Pharma, Rgenix, Bellicum, Gossamer Bio, Arcus Bio, Seattle Genetics, TempestTx, Shattuck Labs, Synthorx Inc. Revolution Medicines, Bicycle Therapeutics, Zymeworks, Relay Therapeutics, Scholar Rock, NGM Biopharma, Stemcentrx, Beigene, CALGB, Cyteir Therapeutics, Foundation Bio, Innate Pharma, Morphotex, Ongologie, NuMab, AtalasMedx, Treadwell Therapeutics, IGM Biosciences, Mabspace, Hutchinson MediPharma, REPAIR Therapeutics, NeoImmune Tech; J. Bendell also reports consulting/advisory role to her institution from Gilead, Genentech/Roche, BMS, Five Prime, Lilly, Merck, Medimmune, Celgene, Taiho, Macrogenics, GSK, Novartis, OncoMed, LEAP, TG Therapeutics, AstraZeneca, BI, Daiichi Sankyo, Bayer, Incyte, Apexigen, Array, Sanofi, ARMO, Ipsen, Merrimack, Oncogenex, FORMA, Arch Oncology, Prelude Therapeutics, Phoenix Bio, Cyteir, Molecular Partners, Innate, Torque, Tizona, Janssen, Tolero, TD2(Translational Drug Development), Amgen, Seattle Genetics, Moderna Therapeutics, Tanabe Research Laboratories, Beigene, Continuum Clinical, Agios, Bicycle Therapeutics, Relay Therapeutics, Evelo, Pfizer, Piper Biotech, Samsung Bioepios, Fusion Therapeutics; J. Bendell also reports food /beverage and travel from Gilead, Genentech/Roche, BMS, Lilly, Merck, MedImmune, Celgene, Taiho, Novatis, OncoMed, BI, ARMO, Ipsen, Oncogenex, FORMA. F. Ciardiello reports personal fees from Roche/Genentech, Merck Serono, Pfizer, Amgen, Servier, Lilly, Bayer, Bristol-Myers Squibb, and Celgene; and grants from Bayer, Amgen, and Merck Serono. H Hamidi, Y. Shi, E. Lin, M. Wongchenko, and Y. Yan are employees and stockholders of Roche/Genentech. C. Bais, X. Qu, R. M. Johnson, F. de Sousa e Melo, A. Mancini, and D. Shames are former employees of Genentech. E. S. Sokol, S. Sivakuma, and B. Kaplan are employees at Foundation Medicine, Inc., a wholly owned subsidiary of Roche Holdings, Inc., and Roche Finance Ltd, and have an equity interest in Roche.
Peer review
Peer review information
Nature Communications thanks Saskia Wilting, and the other anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Qu, X., Hamidi, H., Johnson, R.M. et al. Ligand-activated EGFR/MAPK signaling but not PI3K, are key resistance mechanisms to EGFR-therapy in colorectal cancer. Nat Commun 16, 4332 (2025). https://doi.org/10.1038/s41467-025-59588-3
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-59588-3
This article is cited by
-
Synergistic mechanisms and clinical translation of regorafenib combination therapies
Clinical and Translational Oncology (2026)
-
ALOXE3 expression predicts poor prognosis and modulates immune infiltration in colon adenocarcinoma
World Journal of Surgical Oncology (2025)
-
From PIK3CA Mutations to Rational PI3K Inhibition for the Treatment of Colorectal Cancer
Molecular Diagnosis & Therapy (2025)