Abstract
Highly pathogenic avian influenza (HPAI) H5N1 virus has been associated with severe mastitis in dairy cows, leading to decreased milk production. Here we investigated the impact of H5N1 virus infection in health and production parameters in an affected dairy herd in Ohio. Clinical disease, which lasted for about three weeks, was recorded in 20.0% (777/3876) of the adult cows. Milk losses of ~900 kg per cow were recorded in affected cows during a 60 day-post-outbreak period. Seroprevalence was 89.4% (570/637) in the herd, with 76.1% (485/637) of seropositive animals being subclinically infected. Clinically affected cows presented an increased risk of death (6 times) and of premature herd removal (3.6 times) when compared to non-clinical cows. Economic losses due to decreased milk production, mortality, and early herd removal were estimated at $950 per clinically affected cow for a total cost of ~$737,500 for the herd during the observation period. Our results demonstrate a production impact lasting at least 60 days post-clinical diagnosis and major financial consequences of HPAI H5N1 virus infection to dairy farms.
Similar content being viewed by others
Introduction
Highly pathogenic avian influenza virus (HPAI) H5Nx virus has been circulating in wild bird populations worldwide since 1996. These viruses cause severe disease and high mortality in domestic poultry1. In 2014, HPAI H5N2 virus was introduced through migratory wild birds in the United States of America (USA), resulting in a large outbreak that lasted ~2 years (2014–2015) and caused the deaths or culling of over 41 million birds, leading to over $1.1 billion in economic losses due to direct mortality, market distortions for poultry products, and international trade restrictions2. In February of 2022, the HPAI H5N1 clade 2.3.4.4b virus was introduced in domestic poultry in the USA, causing an unprecedented outbreak that has now lasted more than three years. As of July 9, 2025, the outbreak had led to the deaths or culling of over 174.8 million birds3, which makes the current HPAI H5N1 outbreak the most economically costly animal disease outbreak in the history of the country4.
The current circulating HPAI H5N1 clade 2.3.4.4b virus has also spilled over and caused fatalities in more than 28 species of mammals. In March 2024, HPAI H5N1 virus clade 2.3.4.4b genotype B3.13 was detected in lactating dairy cattle in multiple farms in Texas (TX) and subsequently spread to several other states5,6. As of July 9, 2025, there have been 1074 confirmed herds in 17 US states in dairy cattle, including two additional documented spillover events of HPAI H5N1 virus clade 2.3.4.4b genotype D1.17. Clinically, HPAI H5N1 virus infection in lactating dairy cows presents with a decrease in feed intake and rumination time, and a pronounced decrease in milk production, with milk appearing abnormal and resembling colostrum or mastitic milk. Affected animals may develop fever and mild respiratory signs, including clear nasal discharge. Increased mortality has been reported by some affected farms5. The most important pathophysiological impact of HPAI H5N1 infection in dairy cows is associated with the virus tropism and replication in milk-secreting epithelial cells in the mammary gland, which results in severe mastitis and degeneration and necrosis of infected cells5,6,8. Importantly, the effect of HPAI H5N1 infection in milk production appears to extend beyond the clinical phase of the disease.
Here we studied the impact of an HPAI H5N1 outbreak in a dairy herd with ~3876 adult cows. We investigated risk factors associated with clinical disease and the consequences of infection on production parameters of the herd. Additionally, based on observed milk losses, mortality, and premature herd removal, we estimated the economic impact of the HPAI H5N1 outbreak in the target farm.
Results
Clinico-epidemiological characteristics and outcomes of influenza infection in the dairy herd
To assess the impact of HPAI on dairy cows, we analyzed production and clinical data from a free-stall dairy farm (n = 3876 cows; Supplementary Fig. 1) from Ohio (OH) that experienced an HPAI H5N1 virus outbreak in the spring of 2024 following transportation of 42 apparently healthy lactating cows from a farm in TX1. A summary of the OH farm herd demographics and production parameters in non-clinical and clinical animals is presented in Table 1. An analysis of individual animal and herd-level data collected for 91 days (March 8 to June 7, 2024), a period that encompasses pre- and post-outbreak data, was performed. The data were obtained from the herd management software DairyComp 305 (Valley Ag Software, Tulare, CA) and through the Afimilk® monitoring system (Afimilk® Ltd, Kibbutz Afikim, Israel). The first clinical influenza case in this herd was detected on March 21, 2024, and HPAI H5N1 virus diagnosis was performed by laboratory testing via real-time reverse transcriptase PCR (rRT-PCR) on March 29, 2024 at the Ohio Animal Disease Diagnostic Laboratory and April 4, 2024 at the Cornell Animal Health Diagnostic Center, and confirmed at the National Veterinary Services Laboratory. At the beginning of the outbreak, there were 3876 cows on the premises; 3433 were lactating and 443 were non-lactating. A total of 777 of 3876 (20.0%) cows were diagnosed with clinical influenza by farm personnel under the supervision of on-site managing veterinarian based on production parameters (drop in milk production) and clinical signs (e.g., inappetence, apathy and decreased rumination time) recorded for each cow with the Afimilk® monitoring system. Clinical diagnosis was followed by identification and segregation of sick animals to the hospital pen which is adjacent to pens used for housing of healthy nonlactating cows (Supplementary Fig. 1). Of the 777 clinical influenza cows, 776 were lactating and 1 was in the dry period, with most affected cows being at mid-to-late stages of lactation (100–200 [284/777, 36.6%] or >200 [310/777, 39.9%] days in milk, respectively) and at the second (343/777, 44.1%) or greater (274/777, 35.3%) lactation (Table 1).
Cows were diagnosed with clinical influenza between March 21 and April 13, 2024, with the peak disease incidence being observed on March 31, 2024, when 121 new cases were identified among 3876 cows at risk in the herd (Fig. 1a). The clinical phase of the disease lasted, on average, 7.9 ± 9.3 days, and cows stayed in the hospital pen for an average of 5.1 ± 9.3 days (Supplementary Table 1 and Fig. 1b, c). Importantly, 53 of the 777 (6.8%) clinical influenza cows died or had to be euthanized within 13.6 ± 15.1 days from clinical diagnosis, while another 245 influenza affected cows (31.6%) were removed from the herd (i.e., culled) within 20.6 ± 15.4 days from clinical diagnosis (Supplementary Tables 2 and 3; and Fig. 1d, e).
a Daily incidence of clinical influenza during the study period. b Number of days cows remained with clinical signs as coded by farm personnel (n = 290). c Number of days that affected cows remained in the hospital pen (n = 176). d Number of days from clinical diagnosis to death. e Number of days from clinical diagnosis to herd removal.
Risk factors associated with clinical influenza and its impact on cow mortality and herd removal
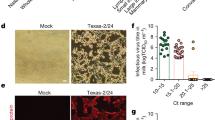
To identify potential risk factors associated with clinical influenza, we investigated the association of several parameters, including days in milk (DIM), parity, breed, baseline milk production, and baseline somatic cell count (SCC), with clinical influenza cases. Importantly, we found that DIM and parity were associated with a greater risk of clinical influenza (Type III P value < 0.01; Fig. 2). Cows between 100 and 200 DIM had a higher risk of presenting clinical influenza (hazard ratio [HR] [95% CI]: 1.39 [1.15, 1.68]), as did cows with >200 DIM (HR: 1.79 [1.47, 2.17]), when compared to cows between 0 and 100 DIM (referent) (Fig. 2). Additionally, multiparous cows had an increased risk of exhibiting signs of clinical influenza when compared to primiparous cows (2nd parity: HR: 1.81 [1.49, 2.20]; 3rd parity or greater: HR: 1.83 [1.45, 2.31]). In contrast, breed, baseline milk production, and SCC were not associated with the risk of new clinical influenza (P > 0.0.05) (Fig. 2).
HR (95% CI): Hazards ratios and their 95% confidence intervals represented by the bars. Milk production estimates are shown for every 10 kg change in the average milk production during baseline pre-outbreak period (March 8, 2024 to March 15, 2024). Log-SCC: Natural Log somatic cell counts at the last farm’s monthly test before the outbreak. Data derived from n = 3662 cows. Hazard ratios were estimated from multivariable models adjusted for all explanatory variables presented in the forest plot.
We also evaluated the impact of clinical influenza on mortality and herd removal. Notably, compared to cows without clinical influenza, cows diagnosed with clinical disease presented an increased risk of death (relative risk [RR] [95% CI]: 6.0 [4.0, 9.1) and of being removed from the herd (RR [95% CI]: 3.6 [3.2, 4.2]) (Supplementary Table 3).
Impact of influenza on rumination and milk production
We evaluated the effect of clinical influenza on rumination and daily milk production in the affected herd. Rumination time during the pre-clinical period (March 8 to March 15, 2024) was higher (average 8 min/day; 409 ± 80 min/day) for cows that were clinically affected with H5N1 virus when compared to cows that were not clinically affected (403 ± 87 min/day; Table 1 and Fig. 3a). We observed a pronounced decrease in rumination time (average 160 min/day; range: −168, −151 min/day) in clinically affected animals which reached its lowest point on April 2, 2024, 12 days after the first clinical case was diagnosed in the herd (March 21, 2024) (Fig. 3a). In the last 10 days of the clinical outbreak (April 3 to 13, 2024) rumination time in clinical cows increased; however, they remained slightly lower in clinically affected animals for at least another 30 days (11 min/day on average; range: −37, −7, on May 13, 2024) when compared to non-clinical animals (Fig. 3a).
Adjusted means for rumination time (a; min/day) and milk production (b; kg/day) in cows with and without clinical influenza diagnosis estimated using mixed linear regression by date (n = 311,701 records). Models accounted for days in milk, lactation number, and breed. Models included cow-ID as a random effect and an autoregressive correlation structure. Error bars represent 95% confidence intervals. The shaded area represents the period (March 21 to April 13, 2024) with at least one cow having a new clinical diagnosis of influenza in the study herd. Data derived from n = 3662 cows.
Our analysis showed that before the diagnosis of the first influenza case (March 21, 2024), cows that were clinically affected produced between 0.2 to 0.7 kg more milk per day than the cows that were not clinically affected (Fig. 3b). After the clinical diagnosis of the first case in the herd and throughout the clinical outbreak (March 21 to April 13, 2024), cows diagnosed with clinical influenza showed a pronounced reduction in milk production when compared to non-clinically affected cows (Fig. 3b). This became evident at the herd level 5 days (on March 26, 2024) after the first clinical case and reached its lowest point 15 days post first clinical case (April 6, 2024), with an average reduction in milk production of 21.9 kg (95% CI: −22.7, −21.0) in affected animals (Fig. 3b). Milk production remained lower in clinically affected cows, with a marked reduction compared to non-affected animals, which ranged between −14.3 and –8.3 kg/day during the entire post-clinical phase lasting for at least 77 days (until June 7, 2024) in which the herd was monitored (Fig. 3b). These results demonstrate a sustained impact of HPAI H5N1 virus infection on the productivity of clinically affected dairy cows.
Decrease in rumination and milk production precede clinical disease
To determine how early clinically affected animals present a decrease in rumination and milk production, we estimated adjusted daily means for rumination time (min/day) and milk production (kg/day) for the clinically affected cows in the herd using mixed linear regression modeling. To validate this approach, the same modeling was applied to the 38 clinically affected cows in the herd that were confirmed to be HPAI H5N1 positive by real-time polymerase chain reaction (RT-PCR). Our analyses revealed that rumination time starts decreasing around 7 days prior to clinical diagnosis, returning to pre-outbreak lengths within 14 days (Fig. 4a). Importantly, analysis of the adjusted daily rumination time means in cows with confirmed laboratory diagnosis of HPAI H5N1 infection corroborated the results obtained in animals with clinical diagnosis only, with similar reduction in rumination time kinetics and magnitude being observed (Fig. 4a, b).
a, c Represent lactating cows with clinical influenza diagnosis (n = 776), and b, d represent cows with on-farm clinical diagnosis and RT-PCR laboratory diagnosis confirmation on milk (n = 38). Models accounted for days in milk, lactation number, and breed. Models included cow-ID as a random effect and an autoregressive correlation structure. Error bars represent 95% confidence intervals. The dashed vertical line represents the day of clinical influenza diagnosis. The horizontal line represents the average rumination time or milk production for non-affected cows on the farm (n = 2886).
When investigating daily milk production in cows diagnosed with clinical influenza only (Fig. 4c), we observed that adjusted means for milk production in affected cows ranged from 35.5 to 36.3 kg/day during −20 to −7 days of the first clinical diagnosis in the farm. Milk production started dropping considerably 5 days before diagnosis and reached its lowest point 2 days after clinical influenza diagnosis, with affected cows producing only 11.6 ± 0.4 kg/day (Fig. 4c). Milk production then increased in the next 14 days post-diagnosis ranging between 20.8 and 24.0, but it remained significantly lower in clinically affected animals when compared to their milk yields prior to the HPAI H5N1 virus outbreak (Fig. 4c, d). Notably, analysis of adjusted means for daily milk production for clinically affected cows that were confirmed to be HPAI H5N1 positive by RT-PCR testing mirrored the results observed in animals clinically diagnosed (Fig. 4c, d). A similar pattern for adjusted daily rumination times and milk production was observed for clinically affected cows at different DIM, parities, and breeds (Supplementary Fig. 2).
Sero surveillance indicates a high rate of subclinical HPAI H5N1 virus infection in dairy cattle
To determine the seroprevalence of HPAI H5N1 virus infection in the herd, we assessed the presence of serum antibodies using ELISA and virus neutralization (VN) assays. For this, serum samples from 637 cows, including 595 lactating and 42 dry cows that were at the farm during the clinical phase of the outbreak (March 21 to April 13, 2024), were collected on June 20, 2024 (105 days after the first clinical case was reported). Serum was collected blindly from ~25% of the cows on each pen. Initial testing of the samples using a multi-species influenza A nucleoprotein-based ELISA (ID Screen® Influenza A Antibody Competition Multi-species, Innovative Diagnostics, Grabels, France) revealed the presence of antibodies to influenza A in 553 of 637 animals (Fig. 5a and Supplementary Table 4). Testing of these samples with an HPAI H5N1 virus neutralization (VN) assay at a 1:8 dilution revealed a slightly higher number (570 of 637; 89.4% seroprevalence) antibody-positive animals. These VN results were confirmed by a titration VN assay, in which all 570 samples presented neutralizing antibody titers ranging between 8 and 2056 (Supplementary Table 4; Fig. 5c). Importantly, HPAI H5N1 specific neutralizing antibodies were detected in lactating (553/595; 92.9%) and dry cows (17/42; 40.5%) with 67 of the sampled cows remaining seronegative. These results indicate broad exposure to HPAI H5N1 virus in animals that were in the farm at the time of the outbreak.
a Multi-species ELISA results showing the detection of influenza A nucleoprotein (NP)-specific antibodies (n = 637). b Virus neutralization results showing detection of influenza A H5N1 virus-specific neutralizing antibodies in serum from cows in the farm during the outbreak (n = 637). c Neutralizing antibody titers detected in serum from VN positive animals and expressed as reciprocal of the highest serum dilution to completely neutralize H5N1 virus (n = 637). d, e Show the adjusted means for rumination time (min/day) and milk production (kg/day) by date and clinical-serological diagnosis, including data from 595 lactating cows that were on-site during the outbreak period. Models accounted for days in milk, lactation number, and breed. Models included cow-ID as a random effect and an autoregressive correlation structure. Error bars represent 95% confidence intervals. CP cows with clinical influenza diagnosis, CN cows without clinical influenza diagnosis, SP seropositive cows, SN seronegative cows. The shaded area represents the period of the influenza outbreak in the study herd.
Next, we investigated the association of clinical influenza with the serological status of the animals. Of the 637 cows tested, 85 (13.3%) were clinically positive and seropositive for HPAI H5N1 (i.e., CP and SP), and 1 (0.1%) animal was clinically positive but seronegative (i.e., CP and SN). Notably, 485 (76.1%) seropositive animals were clinically negative (i.e., CN and SP), indicating a large proportion of subclinically infected animals in the herd. The remaining 67 cows (10.5%) were clinically and serologically negative (i.e., CN and SN). The associations between clinical-serological diagnosis, DIM, lactation number, and breed, are presented in Supplementary Table 4.
Because of the pronounced impact on rumination and milk production that we observed in clinically affected animals (Fig. 3a, b), we also investigated the association between serological status and rumination time and daily milk production. Daily average rumination time was similar between CN and SP and CN and SN cows throughout the study period (Fig. 5d). When evaluating pre-outbreak milk production in relation to the animal’s serological status (SN vs. SP) post-outbreak, we noted that cows that seroconverted (SP) including clinically affected (CP and SP) and subclinically affected (CN and SP) cows produced on average 1.61 and 2.06 kg more milk per day when compared to non-infected cows (CN and SN), respectively (Fig. 5e). Following the detection of influenza, a rapid decrease in milk production (average reduction of 15.50 kg/day (95% CI: [−19.37, −11.64]) was observed in clinically affected cows that seroconverted to HPAI H5N1 (CP and SP). In contrast, subclinically infected cows (CN and SP) maintained pre-outbreak milk production levels throughout the study period (Fig. 5e). These results demonstrate a strong association between clinical disease and decrease in milk production.
Economic impact of HPAI H5N1 virus in dairy cow production
To improve our understanding of the economic impact of an HPAI H5N1 virus outbreak to dairy producers, we estimated the costs incurred due to milk production losses, mortality, and premature animal removal from the herd targeted in our study. We focused our analysis on a 67-day time frame, including 7 days prior to and 60 days after the first clinical influenza diagnosis in the herd. We found that the milk production per cow decreased by a cumulative 901.2 kg per cow during the targeted period (average 18.8 kg per day), when compared to milk production between days −21 and −8 of the influenza diagnosis. Milder losses of 44.1 kg (or 6.3 kg per day) were observed in the 7 days prior to influenza diagnosis, which brings the total milk production loss per cow to 945.3 kg in the 67-day study period. Based on the average nominal (not adjusted for inflation) price ($21.50) the producer received for 45.4 kg of milk (100 pounds) between March and May 2024 (outbreak period)9, and the total milk loss per cow (945.3 kg), we estimated that the economic loss due to decreased milk production alone per cow clinically affected that stayed in the herd was ~$222.
We also estimated the cost of milk losses incurred due to mortality and premature herd removal (culling) of lactating cows. As shown in Fig. 6, clinically affected cows that stayed in the herd produced more milk than affected animals that died or were sold. In contrast, those that died or were sold did not contribute to milk losses past their removal date. We found that the probability-weighted expected cost of milk losses at the time of clinical diagnosis from cows that stayed in the herd, died, or were sold were ~$222, $14, and $99 per affected cow, respectively (Table 2).
Adjusted means for milk production (kg/day) in lactating cows with clinical influenza diagnosis by days from diagnosis and removal status (Stayed, n = 478; Died, n = 53; Sold, n = 245) estimated using mixed linear regression. Models accounted for days in milk, lactation number, and breed. Models included cow-ID as a random effect and an autoregressive correlation structure. Error bars represent 95% confidence intervals.
To determine the losses associated with clinical influenza, we estimated the increase in the risk of mortality and herd removal using adjusted risks, removing background mortality and sales from non-clinical cows (Supplementary Table 3). We found that clinical influenza diagnosis (prevalence of 20.0%, n = 777) increased the mortality risk by 5.5% and herd removal risk by 22.9% (n = 3662), above baseline, compared to non-clinical cows (Supplementary Table 3). The average cost associated with influenza deaths above background mortality rates was then estimated at $166 per clinically affected cow, based on the number of deaths attributed to influenza (n = 43) and the cost of a springer replacement ($3000/head, based on producer’s records). The cost of replacing the cows removed early from the herd attributable to clinical influenza diagnosis (n = 177) was partially offset by the average price ($1123) that the producer received for each cow removed and sold to slaughter, resulting in a net replacement cost of $1877 per cow. We estimated the cost attributable to clinical influenza due to early removal and replacement of affected cows above background sale rates to be $448 per clinically affected cow. Therefore, considering costs associated with decreased milk production, mortality and replacement of dead animals and animals removed early from the herd, we estimated the total cost of the influenza outbreak to be $932 per clinically affected cow, amounting to a total of ~$737,500 for the 776 affected lactating cows in our study during the 67-day period in which we monitored the herd for losses. Our results are from a herd, and economic impacts vary by farm characteristics and the prices they face for input. For example, if a farm with identical production characteristics and response to H5N1 paid the USDA Agricultural Marketing Service average price for replacement cattle of $2254, then their estimated losses would be $731 per clinically affected cow and $ 567,100 for the herd.
Discussion
Here, we investigated the impact of an HPAI H5N1 virus outbreak in a dairy herd and showed that introduction of the virus in the herd resulted in milk production losses in clinically affected animals lasting up to 60 days post-diagnosis. We demonstrated that the economic losses from the HPAI H5N1 outbreak during this period were striking, including decreased milk production and compounded by even higher costs associated with mortality and premature replacement of clinically affected cows.
Using individual animal data obtained from the farm management system, we found that 20.0% of the animals at risk in the herd exhibited clinical signs compatible with HPAI H5N1 virus infection, with the first clinical case being observed ~2 weeks (13 days) after the introduction of apparently healthy lactating cows from an affected farm from TX into the herd5. New clinical cases were recorded daily in the herd during a 3-week period, with the peak incidence occurring about 10 days after the first clinical case was diagnosed in the herd. The two major clinical indicators of H5N1 influenza A viral clinical infection were associated with decreased rumination time and milk production. Our analysis demonstrates that at the herd level, declines in rumination time and daily milk production occur within 7 days of identifying the first clinically affected animal. However, when examining adjusted individual animal means for rumination time (min/day) and milk production (kg/day) in relation to when each animal was diagnosed with clinical influenza by farm personnel, we observed that both parameters begin to decline ~5 days before clinical diagnosis. Therefore, farms utilizing monitoring systems should closely track individual cow rumination times and milk production, as decreases in these parameters can serve as early warning indicators of influenza A H5N1 virus introduction into the herd.
The risk of clinical disease differed by parity and lactation stage, with a higher risk observed in multiparous cows compared to first-lactation cows. Although reports from a USDA survey performed on affected farms suggested this outcome10, results showed here represent the first epidemiological investigation using cow-level longitudinal data confirming a higher risk of infection in cows with greater number of lactations. The reasons behind these findings could be related to higher exposure and/or susceptibility; however, this requires further examination. The risk of clinical disease in dry cows was negligible (~0.1%), while the risk of clinical influenza diagnosis increased as lactation progressed. These results suggest an association between cumulative exposure to the milking process and the risk of clinical disease. They also support the notion that transmission of HPAI H5N1 could be occurring during the milking process11, as suggested by the high concentrations of the virus in milk5 and the expression of viral receptors in mammary gland tissue12,13. However, the presence of seropositive dry cows, including one clinical case, which were not milked during the outbreak period, suggests that other transmission routes (i.e., respiratory route) may also be involved, and nonlactating cattle may present with more subtle clinical signs or remain subclinical.
The most remarkable findings of our study were the magnitude and duration of reduced milk production in clinically affected cows. Within two weeks from the first detection of a clinical H5N1 influenza A virus case in the herd, milk production in clinically affected animals decreased by nearly 73% (~35 kg/day to 10 kg/day). These findings can be partially explained by decreased feed intake, as suggested by decreased rumination time, which compromises milk production14. Additionally, the abrupt and long-term drop in milk production could be a direct result of the virus replication in milk-secreting epithelial cells in the mammary gland, which results in necrosis and destruction of these cells5. The observed reduction in milk production represents a pronounced decrease in milk yield, even when compared to other common bacterial clinical mastitis, in which milk losses up to 18 kg have been reported15,16,17. Future studies, evaluating milk production in subsequent lactations in affected cows, will be critical to determine whether regeneration of the mammary gland epithelium that occurs during the dry period is sufficient to re-establish pre-infection milk yields in clinically affected H5N1 cows.
Notably, cows with clinical influenza A H5N1 mastitis in our study did not reach their pre-infection milk yields during the remainder of the period studied after the onset of the disease, which resulted in a cumulative loss of 901.2 kg of milk per cow during the 60 days post-diagnosis. This persistent milk loss could be overlooked when only examining herd-level milk production, where, after the initial introduction of H5N1, poorly performing cows are replaced and the bulk tank recovers. In our investigation, we were able to prospectively follow individual cows with daily milk production records, allowing us to estimate the individual cow milk losses with some granularity. In turn, this allowed us to more accurately estimate the economic losses due to lost milk production when a cow experiences a clinical case of influenza.
The seroprevalence in the target study was estimated to be 89.4% (570/637) in animals that were on the farm during the clinical phase of the outbreak (March 19 to April 11, 2024), suggesting a high transmission efficiency of the virus among cows. Importantly, of the 570 seropositive animals, 463 (83.7%) were not clinically affected by influenza A H5N1, indicating a large proportion of subclinical infections. Although the precise mechanism of transmission of HPAI H5N1 virus in dairy cattle remains unknown, this is consistent with infections with other influenza A viruses, which can quickly spread through susceptible mammalian populations, including in humans, dogs, and swine18,19. Another important observation from our serological study is the detection of antibodies in 17/42 (40.5%) of the cows that were in the dry period during the clinical outbreak in the farm. These findings suggest that non-lactating animals are also susceptible to H5N1 virus infection and, as such, should be considered as potential source of the virus. This is especially important when animals in the dry period are introduced into farms as replacement cows. Notably, when we assessed milk production in seropositive subclinical animals, we did not observe a decrease in milk production in these cows, suggesting that clinically affected animals are the main group of animals contributing to decreased milk yields associated with HPAI H5N1 virus infection.
To gain a better understanding of the economic impact of an HPAI H5N1 virus outbreak in a dairy farm, we estimated the costs incurred at the target farm following introduction of the virus. Our analysis investigated sources of economic losses to the target farm, including milk losses, replacement costs associated with increased risks of death and herd removal in cows affected with clinical influenza. The overall cost per case of clinical influenza from these factors was estimated at ~$950, resulting in ~$737,500 loss for the farm targeted in our study during the 67-day period in which we monitored the herd for losses. The true cost is likely even higher if one were also to account for ongoing reproductive adjustments, disruptions to milking time and other important labor considerations, supportive medical care for sick cows, changes in biosecurity, and other unmeasured factors.
Although our study focused on a single herd, it is one that is typical of a total-mixed-ration-fed, free-stall herd, and thus, these results demonstrate the significance of an influenza A H5N1 virus outbreak in affected farms. However, as the study was performed in a single herd, this could limit the external validity of the findings, and results should be generalized only to herds operating under similar management conditions. It is important to note that differences in farm demographics, geographic region, or management practices may result in higher or lower economic losses to affected farms. Additionally, because we used observational data and evaluated outcomes such as milk production, death, and herd removal (outcomes that can be influenced by numerous factors), residual confounding from unmeasured or unknown variables may still be present despite adjusting for major confounders. These limitations should be considered when interpreting the findings. Nonetheless, our findings highlight the high impact of influenza A H5N1 virus to the US dairy industry, as the virus continues to circulate and cause economic losses to dairy producers, posing an increased risk to animal and public health.
Methods
This prospective cohort study was conducted on a large commercial dairy farm, where cow-level data were collected during an HPAI influenza A H5N1 virus outbreak and over the following two months, from March to June 2024.
Characteristics of the study dairy farm
The dairy farm targeted in our study housed 3433 lactating and 443 nonlactating cattle at the time of the HPAI influenza A H5N1 virus outbreak. The free-stall farm consists of five barns housing adult dairy cows and pregnant heifers. Each barn is separated longitudinally into two free-stall pens, with a feed alley in the middle of each pen. There are two rows of beds in each pen, and the bedding substrate is recycled manure solids. Pens 1 through 8, located in barns 1 through 4, house lactating cattle (Supplementary Fig. 1). Covered walkways connect the lactating barns, and cattle are moved to and from the milking parlor through gates and walkways.
Pens 9 and 10, in barn 5, house nonlactating cattle, with a hospital pen for sick animals within pen 9. All pens hold ~500 cattle each, except for those located in barn 3. This barn is divided sagittally, with the milking parlor in the lower half and pens 5 and 6, each housing 250 cattle, in the upper half. There is a holding area between the parlor and pens 5 and 6 where cattle wait to enter the parlor. In this area, close contact and nose-to-nose interaction can occur.
The 42 cows that were moved from a farm in TX, originating the outbreak in the study farm in OH5, were placed in pen 5, adjacent to the holding area, upon arrival. The “double 40” parallel milking parlor milks 80 cows per milking cycle, with 40 cows on each side of the parlor. Employees who milk the cows work in the “pit”, a lower part of the parlor with access to each row of cattle. The cow’s udder is at arm level of the workers to make the milking process more accessible. An automatic gate that reads the individual animal identification can siphon animals into the sort pen for individualized treatments as they leave the parlor. The sort pen is adjacent to pen 5.
Data collection
Data from the study farm was collected over a period of three months, from March 8, 2024, to June 7, 2024. The farm used a herd management software called Dairy Comp 305 (Valley Agricultural Software, Tulare, CA). Farm employees recorded all relevant management events daily, including animal movements across pens, clinical disease diagnoses, calving and dry dates, treatments, sales, deaths, and monthly somatic cell count test results. Dairy Comp was used to create reports exported as CSV spreadsheets, which were later used for statistical analyses. Daily milk production and rumination system was obtained from Afimilk® monitoring system (Afimilk® Ltd, Kibbutz Afikim, Israel). Cows showing alerts from on-farm sensors (based on deviations in milk production and rumination) were identified for further evaluation, as previously described in ref. 20. All cows were visually inspected for any abnormal behavior or apparent signs of disease. Cows that showed alerts or appeared to be sick (e.g., abnormal posture, abnormal milk, nasal discharge, separation from the group, etc.) were then thoroughly examined by trained farm personnel, including the on-site managing veterinarian. Clinical signs in affected cows included decreased dry matter intake and rumination, mild respiratory symptoms (such as clear nasal discharge and increased respiratory rate), as well as lethargy and signs of dehydration. Gastrointestinal signs ranged from dry, tacky feces to diarrhea. Affected cows also showed an abrupt reduction in milk production, and the milk often appeared abnormal (yellowish and colostrum-like, with a thick or occasionally curdled consistency). Cows with clinical signs and abnormal milk were moved to the hospital pen, where they continued to be milked. Although their milk was diverted and not added to the bulk tank (i.e., not used for human consumption), individual milk yield and rumination time continued to be recorded through the automated monitoring system. Therefore, data on milk yield and rumination were available prior to, during, and after the hospital period. Supportive and maintenance therapy (nonsteroidal anti-inflammatory drugs and fluid therapy) was provided only in animals with more severe clinical signs, at the discretion of the farm veterinarian and personnel.
Collection of samples
Milk, nasal swab, blood, urine, and fecal samples tested by real-time PCR were collected as part of the initial diagnostic investigation conducted at the farm targeted in our study. Sampling was performed on cows housed in the hospital pen with clinical signs compatible with HPAI H5N1 infection, during farm visits conducted on 3/29, 4/2, 4/12, 4/16, and 4/26. Blood samples (n = 810) were collected again on June 20, 2024, and serum separated and used to estimate the seroprevalence of HAPI in the affected herd, with an attempt to sample 25% of the cows in each pen on the farm. No prior sample size calculation was performed to make this decision. Study activities were approved by the Cornell University Institutional Animal Care and Use Committee (IACUC; Protocol No. 2013-0064). Of those, 637 samples represented samples from cows that were present at the farm during the HPAI H5N1 clinical outbreak (March 21st to April 13th, 2024). Of those 595 samples were from cows that were lactating and 42 were from cows that were in the dry period during the H5N1 virus outbreak. Only those 637 serum samples were included in our serosurveillance assessment.
Cells
Human kidney cells HEK293T (ATCC CRL-3216) and bovine uterine epithelial cells (Cal-1, developed in house at the Virology Laboratory at the Cornell University Animal Health Diagnostic Center, AHDC) were cultured in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 1% L-glutamine and 10% Fetal Bovine Serum (FBS) and containing penicillin–streptomycin (Thermo Fisher Scientific; 10 U ml−1 and 100 µg ml−1, respectively) at 37 °C with 5% CO2. HEK293T and Cal-1 cells were used for recombinant virus rescue, and Cal-1 cells were used on the virus neutralization assay as described below.
Generation of recombinant HPAI TX2/24-miniGFP2 reporter virus
A reverse genetics system for the bovine H5N1 virus based on an isolate A/Cattle/Texas/06322424-1/2024 (TX2/24) obtained from milk from infected dairy cows5 was established in Dr. Diel’s laboratory and used as a backbone to generate a recombinant virus expressing the miniGFP2 reporter gene (rTX2/24-miniGFP2). Briefly, full length genome sequences of PB1, PB2, PA, HA, NA, NP and M gene segments of TX2/24 strain (H5N1 clade 2.3.4.4b, genotype B3.13, GISAID accession number: EPI_ISL_19155861) were synthesized commercially (Twist Bioscience) and cloned into the dual promoter influenza reverse genetics plasmid pHW2000 (kindly provided by Dr. Richard Webby at St. Jude Children’s Research Hospital) using the BsmBI (New England Biolabs) restriction sites. To generate the miniGFP2 reporter virus, the NS segment of the rTX2/24 recombinant virus was modified to encode a fusion protein (NS-miniGFP2) from a single nonoverlapping transcript. The miniGFP2 was cloned at the C-terminal of NS1. The NS1 and NEP open reading frames were separated by the porcine teschovirus 1 2A autoproteolytic cleavage site. The NS-miniGFP2 gene segment was synthesized (Twist Bioscience) and cloned into pHW2000 vector using the BsmBI sites. The pHW2000 plasmids containing seven TX2/24 gene segments (PB1, PB2, PA, HA, NA, NP, and M) and the modified NS segment encoding miniGFP2 were co-transfected into a co-culture of HEK293T and Cal-1 (bovine uterine epithelial cells) using Lipofectamine 3000 reagents (ThermoFisher Scientific). Cell culture supernatant was harvested after 96 h and used to infect newly seeded Cal-1 cells. Both cell lysate and culture supernatant were harvested after 72–96 h to prepare the seed stock for the rTX2/24-miniGFP2 virus. The working stock of the virus was prepared after inoculating 10-day-old embryonated chicken eggs via the allantoic cavity route, and the infected allantoic fluid was harvested after 48 h. Viruses from the initial rescue and from passages 1 and 2 were sequenced to confirm the integrity of the sequences and absence of unwanted mutations. The 50% tissue culture infectious dose (TCID50) was determined using end-point dilutions and Spearman and Karber’s method and expressed as TCID50.mL–1. The sequenced, verified stock rTX2/24-miniGFP2 virus was used in the virus neutralization assays below.
Multi-species NP ELISA
A commercial multi-species NP-based ELISA kit (ID.Screen® Influenza A Antibody Competition Multi-species, Innovative Diagnostics, Grabels, France) was used to assess the presence of antibodies against the nucleocapsid protein (NP) in serum samples from 637 cows present in the farm at the time of the H5N1 virus outbreak. For this, the serum samples were diluted 1:4 and tested with the ELISA kits following the manufacturer’s instructions. Results were interpreted based on the following criteria: <45% Pos (S/N % value), 45% to <50% Susp, >50% Neg as recommended by the manufacturer.
Fluorescent virus neutralization assay (FVNA)
To confirm the presence of H5N1-specific antibodies on serum samples collected from the cows present at the farm during the outbreak, we tested all samples using a FVNA. The FVNA assay was developed and validated in-house and compared to a commercially available ELISA kit for influenza A virus antibody detection (ID.Screen® Influenza A Antibody Competition Multi-species, Innovative Diagnostics, Grabels, France). All samples were screened at a 1:8 dilution, and positive samples were then subjected to two-fold serial dilutions (1:8 to 1:2028) for VN antibody titrations. Briefly, each serum dilution was incubated with 200 TCID50 of rTX2/24-miniGFP2 for 1 h at 37 °C. Cal-1 cells were added to each well, and plates were incubated at 37 °C for 48 h. Plates were visualized using a fluorescence microscope (Hybrid microscope ECHO Revolve 3 K) to determine neutralizing antibody (NA) titers, expressed as the reciprocal of the highest serum dilution capable of completely inhibiting HPAI H5N1 virus replication based on the expression of miniGFP2 by the rTX2/24-miniGFP2 virus.
Statistical analysis
All statistical analyses were performed on R (version 4.3.2). Before analyses, data was wrangled and cleaned using functions implemented in the tidyverse package in R (version 2.0.0)21. All available data were retained in the analyses; no observations were excluded due to missing values or outliers. Variable distributions and descriptive statistics were explored prior to modeling. Days in milk (DIM) were calculated as the number of days from the last calving date. Days from clinical influenza diagnosis refers to the number of days relative to the date a cow was diagnosed with clinical influenza, with the diagnosis date defined as day 0. Negative values represent days prior to diagnosis, and positive values represent days after diagnosis. Days with clinical signs were defined as the number of days from the diagnosis of clinical influenza to the resolution of clinical signs. Daily incidence was calculated as the proportion of animals newly diagnosed with clinical influenza divided by the total number of cows at risk on a given day (i.e., cows that have not been diagnosed with clinical influenza). The total number of cows at risk was updated daily, accounting for herd dynamics (such as cows that were sold, died, or transferred to the facility), as recorded in the herd management system. Time-varying Cox proportional hazards regression was used to explore risk factors associated with the risk of new clinical influenza (outcome) using the Survival package in R (version 3.6-4)22. Investigated explanatory variables (i.e., exposures) included: DIM (0–100, 101–200, >200), lactation number at given date (1, 2, ≥3), breed (Holstein, Jersey, Crossbreed; cows coded as Holstein or Jersey in Dairy Comp 305, including crossbreeds with a high proportion of Holstein or Jersey ancestry, were classified accordingly, and this recorded classification was used in the analysis), milk production at baseline (average daily milk production on the week from 3/8 to 3/15; kg/day), and natural log-transformed somatic cell count at baseline (from the last available monthly farm’s test). Including all lactating cows present on the farm during the outbreak period (n = 3662), mixed linear regression was used (nlme package in R [version 0.12]23) to investigate the association between the presence of clinical influenza (Yes vs No; exposure) and daily milk production (kg/day), and rumination time (min/day; outcomes). Models included an interaction between the presence of clinical influenza and date. Stratified analysis was used to compare the outcome within each date using the emmeans package (version 1.10.0)24. For lactating cows diagnosed with clinical influenza (n = 776), mixed linear regression was employed to investigate the association between the days from clinical influenza diagnosis (−21 to +60; exposure), and daily milk production (kg/day), and rumination time (min/day; outcomes). Stratified analyses were used to investigate the relationship between days from clinical diagnosis (exposure) and dependent variables of interest (i.e., milk production and rumination time; outcomes), within different subgroups of DIM (0–100,101–200, > 200), parity (1, 2, ≥3), breed (Holstein, Jersey, crossbreed), and herd removal status (Cows that stayed on the farm during the follow-up period, cows that were removed, and cows that died). The clinical and serological virus neutralization data were used to classify animals as clinical negative and seronegative (CN + SN), clinical negative and seropositive (CN + SP; subclinical infection), and clinical positive and seropositive (CP + SP; clinical infection). The relationship between clinical-serological diagnosis (CN + SN vs CN + SP + CP + SP; exposure), rumination, and milk production was investigated using mixed linear regression, including all lactating cows on-site during the outbreak period with serological data (n = 595). The non-independence of observations within each cow was accounted for by the inclusion of cow-id as random effect, and the use of an autoregressive correlation structure. Model residuals were assessed visually to evaluate the assumption of normality. Residuals were normally distributed in all mixed linear regression models examining rumination time and milk production, and data transformation was therefore not necessary. The relationship between clinical influenza diagnosis (exposure), herd removal, and death risk (outcomes) was investigated using logistic regression, including all the lactating cows on-site during the outbreak period (n = 3662). Relative risks were estimated using the odds_ratio_to_risk_ratio function implemented in the “effect size” package in R (version 0.8.7)25. All models accounted for DIM, lactation number, and breed. Estimated marginal means (i.e., adjusted means) were estimated using the emmeans package. All visualizations were created using functions implemented in ggplot2 package (version 3.5.0)26.
Economic analysis
We combine economic data on prices paid to producers for milk and slaughtered dairy cows from the USDA, Agricultural Marketing Service (2024), with producer information about sales, deaths, milk production, and the prices the producer paid for replacement cows (“springers”). These data allow us to estimate milk losses and the increased cow replacement from HPAI. This analysis complements and references the epidemiological portions of the paper where appropriate. The main text includes the nominal (not adjusted for inflation) price received for hundredweight of milk ($21.50) and price paid for a springer cow ($3000). It represents the cows in this herd, rather than a representative sample of a larger population (e.g., the U.S. dairy herd).
We formulate the expected losses to a producer at the time of diagnosis as the expected probability-weighted costs from death, removal, and milk losses. The equation below includes the conditional of each outcome given that a cow has received a clinical diagnosis, and the expected losses in milk or from replacement.
L represents the lost profit from HPAI, where the outcome is drawn from the set of possible outcomes (dies, sold, survives) with known probabilities. The variables \({mil}{k}_{{dies}}\), \({mil}{k}_{{sold}}\), and \({mil}{k}_{{survives}}\) refer to the expected milk losses for each subscripted outcome. Replacement costs (\({replacement}\)) are also known for each outcome. It is important to note that the replacement costs for surviving animals are zero.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All data related to the manuscript is presented in the main article body or is available online as raw data files. Source data can be found at: https://fepenamosca.github.io/hpai_impact_dairies/.
Code availability
Code for production data analyses can be found at: https://fepenamosca.github.io/hpai_impact_dairies/.
References
Kwon, J.-H. et al. Diverse infectivity, transmissibility, and pathobiology of clade 2.3.4.4 H5Nx highly pathogenic avian influenza viruses in chickens. Emerg. Microbes Infect. 12, 2218945 (2023).
Ramos, S. Impacts of the 2014-2015 Highly Pathogenic Avian Influenza Outbreak on the U.S. Poultry Sector (USDA, 2017).
CDC. USDA Reported H5N1 Bird Flu Detections in Poultry. Avian Influenza (Bird Flu) https://www.cdc.gov/bird-flu/situation-summary/data-map-commercial.html (2025).
Seeger, R. M., Hagerman, A. D., Johnson, K. K., Pendell, D. L. & Marsh, T. L. When poultry take a sick leave: response costs for the 2014–2015 highly pathogenic avian influenza epidemic in the USA. Food Policy 102, 102068 (2021).
Caserta, L. C. et al. Spillover of highly pathogenic avian influenza H5N1 virus to dairy cattle. Nature 634, 669–676 (2024).
Burrough, E. R. et al. Highly pathogenic avian influenza A(H5N1) Clade 2.3.4.4b virus infection in domestic dairy cattle and cats, united states, 2024. Emerg. Infect. Dis. 30, 1335–1343 (2024).
CDC. Current Situation: Bird Flu in Dairy Cows. Avian Influenza (Bird Flu) https://www.cdc.gov/bird-flu/situation-summary/mammals.html (2025).
Butt, S. L., Nooruzzaman, M., Covaleda, L. M. & Diel, D. G. Hot topic: influenza A H5N1 virus exhibits a broad host range, including dairy cows. JDS Commun. 5, S13–S19 (2024).
USDA NASS. USDA - National Agricultural Statistics Service Homepage. https://www.nass.usda.gov/ (2025).
USDA APHIS. Highly Pathogenic Avian Influenza H5N1 Genotype B3.13 in Dairy Cattle: National Epidemiologic Brief. https://www.aphis.usda.gov/sites/default/files/hpai-dairy-national-epi-brief.pdf (2024).
Baker, A. L. et al. Dairy cows inoculated with highly pathogenic avian influenza virus H5N1. Nature 637, 913–920 (2025).
Imai, M. et al. Highly pathogenic avian H5N1 influenza A virus replication in ex vivo cultures of bovine mammary gland and teat tissues. Emerg. Microbes Infect. 14, 2450029 (2025).
Ríos Carrasco, M., Gröne, A., van den Brand, J. M. A. & de Vries, R. P. The mammary glands of cows abundantly display receptors for circulating avian H5 viruses. J. Virol. 98, e0105224 (2024).
Kaufman, E. I., Asselstine, V. H., LeBlanc, S. J., Duffield, T. F. & DeVries, T. J. Association of rumination time and health status with milk yield and composition in early-lactation dairy cows. J. Dairy Sci. 101, 462–471 (2018).
Rajala-Schultz, P. J., Gröhn, Y. T., McCulloch, C. E. & Guard, C. L. Effects of clinical mastitis on milk yield in dairy cows. J. Dairy Sci. 82, 1213–1220 (1999).
Gröhn, Y. T. et al. Effect of pathogen-specific clinical mastitis on milk yield in dairy cows. J. Dairy Sci. 87, 3358–3374 (2004).
Sguizzato, A. L. L. et al. Understanding the dynamics of mastitis in milk yield: decoding onset and recovery patterns in response to mastitis occurrence. JDS Commun. 5, 669–673 (2024).
Voorhees, I. E. H. et al. Spread of canine influenza A(H3N2) virus, United States. Emerg. Infect. Dis. 23, 1950–1957 (2017).
Rolfes, M. A. et al. Household transmission of influenza A viruses in 2021-2022. JAMA 329, 482–489 (2023).
Perez, M. M., Cabrera, E. M. & Giordano, J. O. Effects of targeted clinical examination based on alerts from automated health monitoring systems on herd health and performance of lactating dairy cows. J. Dairy Sci. 106, 9474–9493 (2023).
Wickham, H. et al. Welcome to the tidyverse. J. Open Source Softw. 4, 1686 (2019).
Therneau, T. A package for survival analysis in R. https://cran.r-project.org/web/packages/survival/vignettes/survival.pdf (2024).
Pinheiro, J., Bates, D., DebRoy, S., Sarkar, D. & R. Core Team. nlme: Linear and Nonlinear Mixed Effects Models. https://cran.r-project.org/web/packages/nlme/index.html (2025).
Lenth, R. V. et al. Emmeans: estimated marginal means, aka least-squares means. https://cran.r-project.org/web/packages/emmeans/index.html (2024).
Grant, R. L. Converting an odds ratio to a range of plausible relative risks for better communication of research findings. BMJ 348, f7450 (2014).
Wickham, H. Ggplot2. https://doi.org/10.1007/978-3-319-24277-4 (Springer International Publishing, Cham., 2016).
Acknowledgements
We thank the producer and veterinarian Dr. William Leone for sharing the data from the dairy herd targeted in our study. We would also like to thank Tyler J. Poole for his help with drawing the farm schematics. The authors also thank Dr. Dennis Summers and his staff and veterinarians, and Lauren E. Meyer for their help with sample collection. The study was funded in part by the Cornell University Animal Health Diagnostic Center Virology Laboratory activity funds.
Author information
Authors and Affiliations
Contributions
Conceptualization: F.P.-M., E.F., M.M., D.V.N. and D.G.D; methodology: F.P.-M., M.M., A.R.R., P.S.B de O., M.N., M.P.K, F.E., D.V.N., D.G.D.; resources: F.P.-M., E.F., M.M., M.P.K., M.Z., W.M.L., Z.R.L., D.V.N. and D.G.D; software: F.P.-M., M.M.; formal analysis: F.P.-M., M.M., P.S.B de O., D.V.N., D.G.D.; investigation: F.P.-M., E.F., M.M., W.M.L., D.V.N. and D.G.D.; data curation: F.P.-M., M.M., A.R.R., P.S.B de O.; writing – original draft: F.P.-M, E.F., M.M., D.G.D.; writing – review and editing: all authors; visualization: F.P.-M., E.F., P.S.B., de O.; supervision: D.V.N., D.G.D.; project administration: F.P.-M., E.F., M.M., D.V.N.; funding acquisition: D.V.N., D.G.D.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Judith Oguzie and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Peña-Mosca, F., Frye, E.A., MacLachlan, M.J. et al. The impact of highly pathogenic avian influenza H5N1 virus infection on dairy cows. Nat Commun 16, 6520 (2025). https://doi.org/10.1038/s41467-025-61553-z
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-61553-z