Abstract
RNA interference is an eco-friendly alternative to chemical pesticides, yet its efficacy in lepidopterans like Spodoptera frugiperda (S. frugiperda) is limited by poor uptake. Here, we report on ZIF-8 polydopamine nanoparticles that protect dsRNA against enzymatic degradation and active the endocytic/phagosome pathways for increased uptake. Furthermore, the uptake of nano-enabled dsRNA induces the overgrowth of Serratia marcescens, this reduces the S. frugiperda reactive oxygen species (ROS) immune response, increasing the effects of plant’s natural defenses, further inhibiting Enteroccous mundtii growth. This work shows the synergistic potential of nanoparticles for influencing the gut bacteria to prevent resistance mechanisms and for RNAi delivery for pest management.
Similar content being viewed by others
Introduction
In the past decade, the fall armyworm, Spodoptera frugiperda (Lepidoptera, Noctuidae), has become a major invasive pest around the globe1. S. frugiperda has damaged 353 plant species across 76 families, affecting crop yield remarkably, and leading to substantial economic losses at $1 to 3 billion annually worldwide2. More specifically, S. frugiperda was shown to display strong adaptability toward environmental stress and remains strong invasive capability3. Relative crop production, such as maize and rice, relies almost entirely on chemical pesticides to control S. frugiperda. However, the long-term use of chemical pesticides has raised environmental, health and safety concerns. Therefore, green technologies have to be developed to alternative chemical pesticides and achieve more target specificity, decreased toxicity, and environmental sustainability. RNA interference (RNAi), as a conserved regulatory mechanism mediated by double-stranded RNA (dsRNA), has been shown to inhibit or silence target genes expression4,5,6,7, and realize sustainable eco-friendly pest management8. However, RNAi efficiency varies among insect species, which was highly efficient in the coleopterans (e.g., beetles) and relatively inefficient in the lepidopterans (e.g., butterflies and moths)9. The major limitations of using RNAi against lepidopteran insects included strong degradation of dsRNA by endonuclease, lack of efficient intracellular metastasis, and defective core RNAi mechanisms5,10,11.
Nanoparticles (NPs) with good biocompatibility are able to promote the translocation of exogenous substances12. The NPs, such as metal-organic framework (MOF)13, laminar double hydroxide14, chitosan15,16, liposomes17,18, and cationic dendrimers10, are able to protect dsRNA/siRNA molecules from enzymatic degradation and enhance their translocation across cell membranes, and have been widely applied as excellent carriers for pest management19,20,21. Previous studies have demonstrated that star polycation (SPc) and protamine-lipid (PL) mediated dsRNA delivery systems achieved efficient RNAi to S. frugiperda22,23. However, the dsRNA delivery mechanism and synergistic insecticidal mechanism of NPs are still unclear for S. frugiperda. Several studies have confirmed that the commensal bacterium Serratia sp. in the gut of S. frugiperda will transform into a pathogenic bacterium under habitat stress, resulting in an increase in the mortality rate of S. frugiperda24,25. Additionally, relevant studies have also confirmed that the imbalance of insect gut microbiota leads to changes in oral secretions, which can induce different levels of anti-herbivory in plants26. Therefore, elucidating the RNAi mechanism mediated by NPs for S. frugiperda from the perspectives of dsRNA delivery and “insect- microbe-plant” interaction is an interesting question.
Zeolitic imidazolate framework-8 (ZIF-8) has been considered as a potential material for drug delivery due to high porosity, high surface area, thermal and chemistry stability27, and acceptable cost (~6100$ ton−1)28. Polydopamine (PDA) material has been widely used as a shell material due to its biocompatibility, nontoxicity, high chemical stability, and low cost29. The modification of PDA can significantly enhance the stability of nanocarriers and protect pesticide molecules from ultraviolet photolysis30. Additionally, chitin synthase (CHS) is a key enzyme that catalyzes the synthesis of chitin which is a major component of the insect exoskeleton and peritrophic membrane (PM). The PM is a chitin-protein layer in the midgut, and it is critical for protecting against pathogens and facilitating digestion. Together, CHS and PM play essential roles in molting, immunity, and digestive function31. Meanwhile, a multi-subunit proton pump (V-ATPaseB) drives ATP hydrolysis to facilitate transmembrane proton transport and participates in diverse physiological processes32,33.
In this work, the genes encoding CHS and V-ATPaseB are selected as target genes to elucidate the RNAi effect of the dsRNA@ZIF-8@PDA NPs. We show that zeolitic imidazolate framework-8 and polydopamine complex (ZIF-8@PDA) NPs overcome the dsRNA delivery bottleneck for S. frugiperda by enhancing the stability of dsRNA and activating the endocytosis and phagosome pathways, thus efficiently inhibiting pest growth and causing high mortality rate. Moreover, ZIF-8@PDA NPs induce overgrowth of S. marcescens in S. frugiperda, which inhibits ROS level of insects and increases plant anti-herbivore response.
Results and discussion
The phylogenetic and developmental expression analyses of SfV-ATPaseB and SfCHS
Phylogenetic analysis using amino acid sequences from other insect species indicated that the Vacuolar(H+)-ATPase subunit B (V-ATPaseB) and the Chitin synthase (CHS) encoded protein sequences are both highly conserved in lepidopteran insects (Supplementary Fig. 1a, c). To determine the expression patterns of SfV-ATPaseB and SfCHS during the development of S. frugiperda, we analyzed expression levels from 2nd, 3rd, 4th, and 6th instar larvae, pupae and adults. We observed that the expression levels of SfV-ATPaseB increased significantly with the growth of instar larvae and reached the highest level in the 6th instar larvae (Supplementary Fig. 1c). The expression level of SfCHS gene reached the highest level in the 4th instar larvae (Supplementary Fig. 1d). The expression patterns of SfV-ATPaseB and SfCHS implied that genes encoding V-ATPaseB and CHS play an important role on the growth and development of S. frugiperda larvae34,35.
Self-assembly mechanism of dsGFP@ZIF-8@PDA NPs and its enhanced delivery of dsRNA
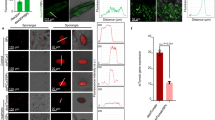
Scanning electron microscope (SEM, Fig. 1a, b) and transmission electron microscope (TEM, Fig. 1c–e) images of dsGFP, dsGFP@ZIF-8 and dsGFP@ZIF-8@PDA NPs showed their polyhedral uniform spherical morphology, which displayed the average diameters of dsGFP@ZIF-8@PDA NPs was 109.08 ± 0.8 nm by Gaussian probability distribution model (R² = 0.897)36 (Fig. 1f). The change of ζ-potential values of ZIF-8, dsGFP@ZIF-8, and dsGFP@ZIF-8@PDA (Fig. 1g) showed that electrostatic interactions occurred between ZIF-8, dsGFP, and PDA29,37. Herein, based on the thermodynamic interpretation38, the low dissociation constant (Kd) of 1.5E-6 M−1 (Fig. 1h) suggested an effective interaction between Zn2+ and 2-mIm. The positive values of ΔH (28.33 kJ mol−1) and TΔS (61.56 kJ mol−1) suggested that the self-assembly of ZIF-8 was endothermic and was mainly driven by electrostatic interaction and entropy39. The low dissociation constant (Kd) of 631 E-9 M−1 (Fig. 1i) revealed that there was an intense interaction between dsGFP and ZIF-8, and the negative ΔG value (−35.37 kJ mol−1) suggested that this interaction was automatic (Fig. 1j)39. The negative ΔH value (−44.7 kJ mol−1) and TΔS value (−9.33 kJ mol−1) demonstrated that electrostatic interaction and van der Waals force existed between dsGFP and ZIF-8 (Fig. 1j)40,41,42. The electropherograms result indicated that ZIF-8@PDA nanocarrier was able to protect dsRNA from enzymatic hydrolysis, whereas naked dsRNA was degraded quickly by the hemolymph (HL) and gut fluid (GF) of S. frugiperda, thus increasing environmental stability of dsRNA (Supplementary Fig. 2a–c). To determine the uptake by S. frugiperda, naked dsGFP was labeled with Cy3 to yield red fluorescence. In vivo experiments demonstrated a 12.33-fold increase in fluorescence intensity in gut tissues of S. frugiperda when treated with ZIF-8@PDA-encapsulated Cy3-dsGFP compared to naked Cy3-dsGFP (Fig. 2k and Supplementary Fig. 3a). Parallel in vitro assays in Sf9 cells revealed an even more pronounced enhancement, with a 357.9-fold higher fluorescence intensity for ZIF-8@PDA-loaded Cy3-dsGFP relative to the naked Cy3-dsGFP (Fig. 2l; Supplementary Fig. 3b). These findings provide compelling evidence that the ZIF-8@PDA NPs significantly enhances dsRNA uptake and effectively overcame the dsRNA delivery bottleneck.
a, b SEM images of dsGFP@ZIF-8 and dsGFP@ZIF-8@PDA. c–e TEM images of dsGFP, dsGFP@ZIF-8 and dsGFP@ZIF-8@PDA. f Dimensional probability distribution plot of dsGFP@ZIF-8@PDA fitted by Gaussian model. g Zeta potential of ZIF-8, dsGFP@ZIF-8 and dsGFP@ZIF-8@PDA. (Mean ± SD; n = 6 technical replicates, F2,17 = 642.73, P < 0.001). Statistical differences were calculated using one-way analysis of variance (ANOVA) with Tukey’s HSD test. Exact P-values are reported in the source data. h–j ITC titration of Zn2+ (0.84 mM) into 2-mIm (10 mM) solution and dsGFP (0.5 mM) into 2-mIm (5 mM) solution. k The fluorescent intensity distribution of S. frugiperda gut and fat body tissues after feeding maize leaves smeared with Cy3-dsGFP and Cy3-dsGFP@ZIF-8@PDA, respectively (n = 4 replicates). l The fluorescent intensity distribution of Sf9 cell incubated with Cy3-dsGFP and Cy3-dsGFP@ZIF-8@PDA, respectively. Relevant source data are provided as a source data file (n = 5 replicates).
a Phenotype of body size and b gut tissue of the S. frugiperda larvae treated with naked dsRNA (dsGFP, dsCHS, and dsV-ATPaseB), ZIF-8@PDA, dsCHS@ZIF-8@PDA, and dsV-ATPaseB@ZIF-8@PDA after 4 days. c The survival rate (%) of S. frugiperda 3rd-instar larvae with different treatments within 4 days. Kaplan-Meier survival analysis with log-rank test was used to assess differences (n = 100/ group, The asterisk indicates significance: *P < 0.05, **P < 0.01, and ***P < 0.001). d body weight, e body length, and f accumulated frass weight of the S. frugiperda larvae under different treatments after 4 days. (Mean ± SD; Body length: n = 36/group, Body weight: n = 36/group, Frass mass: n = 10/group; Different letters above each bar indicate significant differences at P < 0.05 as determined by one-way ANOVA with Tukey HSD test; Body length: F5,215 = 724.774, P < 0.001; Body weight: F5,215 = 508.014, P < 0.001; Frass mass: F5,59 = 300.678, P < 0.001). Exact P-values are reported in the source data. g, h Knockdown efficiency of dsCHS and dsV-ATPaseB of S. frugiperda after feeding on naked dsRNA (dsGFP, dsCHS, and dsV-ATPaseB), ZIF-8@PDA, dsCHS@ZIF-8@PDA, and dsV-ATPaseB@ZIF-8@PDA after 4th day, respectively. (Mean ± SD; n = 4 replicates, 3 larvae/replicates; V-ATPaseB: F3,15 = 94.34, P < 0.001; CHS: F3,15 = 185.117, P < 0.001). Exact P-values are reported in the source data. Relevant source data are provided as a source data file.
Nano-enabled dsRNA enhanced insect mortality rate and RNAi efficiency
The cost of dsRNA using engineered HT115 (DH3)-L4440 expression system reduced to 1/5 of that using T7 RNA Transcription Kit (Belong, SJ002, China), showing lower cost and greater application potential in field. It was worth note that engineered HT115 (DH3)-L4440 expression system yielded impure RNA mixtures (Supplementary Fig. 4). Therefore, we synthesized dsRNAs including dsCHS (476 bp) and dsV-ATPaseB (446 bp) in vitro using HighYield T7 RNA Transcription Kit (Supplementary Fig. 5), and then sprayed dsCHS, dsV-ATPaseB and their ZIF-8@PDA-loaded complexes onto detached maize leaves. Each 3rd-instar larvae of S. frugiperda was inoculated onto plant leaves for 4 days. The larvae fed with dsCHS@ZIF-8@PDA exhibited limited growth and peritrophic membrane lysis (Fig. 2a, b). As shown in Fig. 2c, the application of ZIF-8@PDA-loaded dsCHS and ZIF-8@PDA-loaded dsV-ATPaseB decreased survival rates of S. frugiperda significantly (31–38%), whereas naked dsRNA displayed no significant obvious insecticidal effect (survival rates of 91%) (P < 0.001, log-rank test). The growth rate (body length and weight) and accumulated frass mass of larvae fed with dsCHS@ZIF-8@PDA and dsV-ATPaseB@ZIF-8@PDA decreased significantly compared to larvae fed with naked dsCHS and dsV-ATPaseB, respectively (P < 0.05) (Fig. 2d–f and Supplementary Fig. 6). Furthermore, the RNAi of dsCHS and dsV-ATPaseB loaded with ZIF-8@PDA NPs exhibited a strong gene silencing effect significantly compared with naked dsCHS and dsV-ATPaseB (Fig. 2g, h). These results demonstrated that dsRNA homologous to two selected target genes could be applied to inhibit the growth of S. frugiperda and increase its mortality, but only via the ZIF-8@PDA-based delivery system.
Nano-enabled dsRNA enhanced dsRNA delivery by activating endocytosis and the phagosome
To explore the delivery mechanism of dsRNA induced by the ZIF-8@PDA complex, RNA-seq combined with quantitative real-time PCR (qPCR) technology was performed on S. frugiperda larvae fed with dsCHS@ZIF-8@PDA and naked dsCHS. Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis showed that the differentially expressed genes (DEGs) were enriched in multiple signaling pathways, including endocytosis, phagosome, Toll and IMD signaling, etc. (Fig. 3a and Supplementary Tables 1–3). The DEGs were further classified into three processes based on Gene ontology (GO) analysis and were primarily correlated with biological processes (especially regarding immune response, intracellular and extracellular vesical transport, and symbiont interaction with host) and molecular function (especially for transmembrane transporter activity) (Fig. 3b). The RNA-seq results showed that the genes encoding functions correlated with delivery response of endocytic uptake and phagosome, and immune response pathway were upregulated in the S. frugiperda larvae treated with dsCHS@ZIF-8@PDA complex compared to dsCHS alone (Fig. 3a, c). RNA-seq results indicated that there were 526 DEGs in total, of which 365 genes were downregulated and 161 genes were upregulated in S. frugiperda larvae treated with dsCHS@ZIF-8@PDA compared to naked dsCHS (Fig. 3d). The qPCR was conducted to verify the results of RNA-seq and the results showed that eight tested DEGs showed the same expression trends as those under RNA-Seq, indicating that the transcriptome data were reproducible and reliable (Fig. 3e). More specifically, many genes encoding the endocytic pathway such as clathrin heavy chain (Chc), Hsp70, Rab35, Rab7, Snx3, E3 and AP-2, and phagosome pathway such as F-actin, V-ATPase-A, Pikfyve, VPS34 and Rab35 were upregulated in the larvae treated with dsCHS@ZIF-8@PDA complex (Fig. 3c, e). For example, Chc and their adapter protein complex 2 (AP-2) genes are responsible for encoding clathrin-coated pits and vesicles for early endocytic uptake and delivery response43,44. Some studies have demonstrated that Chc proteins and nanomaterials were able to adsorb each other to enter endocytic vesicles, especially in RNAi of pests such as Tetranychus cinnabarinus Boisduval15, Tribolium castaneum45 and Locusta migratoria44. The Chc protein sequence similarities of Tribolium castaneum and Locusta migratoria were as high as 88.37% and 88.29% compared to that of S. frugiperda, suggesting similar molecular function. Thus Chc protein of S. frugiperda with an isoelectric point of 5.509 displayed a negative charge (z = −45.027) and could bind positively charged dsRNA@ZIF-8@PDA through electrostatic attraction45 to enter endocytic vesicles (Supplementary Fig. 7). Additionally, Rab-7 (Ras-related protein), a small GTPase protein that controls membrane trafficking and organelle structure, and mainly regulates vesicle trafficking between early and late endosomes43,46. Furthermore, it has been reported that the phagosome pathway is closely correlated with the immune pathway triggered by cellular uptake of pathogenic microorganisms and large particles, in which F-actin is recruited into the phagocytic cup to enhance the uptake of contents by cells47. The V-ATPase family has been shown to mediate the fusion of transport vectors (phagosomes and endocytic vesicles) with lysosomes, thereby further enhancing the release of contents48. Additionally, Rab35 and VPS34 have been shown to form a complex to precisely regulate the production of lipid phosphatidyl inositol-3-phosphate (PI3P), which is a regulator of two fundamental but distinct cellular processes, i.e., endocytosis and autophagy49. The above results therefore suggested that dsCHS@ZIF-8@PDA complex promoted the uptake of dsRNA molecules by insect cells through endocytosis and phagocytosis pathways.
a Enriched KEGG pathway of top 20 and b GO enrichment analysis of the significantly differentially expressed genes (DEGs) in S. frugiperda (n = 3 replicates, 6 larvae/replicate; P < 0.05) (KEGG and GO analysis of DEGs. DESeq was used to analyze the DEGs between two treatments, and a fold change of ≥2.0 and a false-discovery rate (FDR) < 0.01 were used as the screening conditions). c Heatmap of key DEGs of endocytosis, phagosome, as well as Toll and IMD signaling pathways. d Volcano plot of DEGs. e qPCR validation of 8 DEGs from endocytosis and phagosome pathways. f Diversity index of Shannon and Simpson. (Mean ± SD; n = 6 replicates, 6 larvae/replicate; Shannon: F5,35 = 194.20, P < 0.001; Simpson: F5,35 = 53.73, P < 0.001). Different letters above each bar indicate significant differences at P < 0.05 as determined by one-way ANOVA with Tukey’s HSD test. Exact P-values are reported in the source data. g Principal coordinate analysis (PCoA) based on the Bray–Curtis metric of 18 samples for the bacterial community. h Heatmap of gut bacteria relative abundances of S. frugiperda larvae among different treatments (n = 6 replicates, 6 larvae/replicate). i–k Absolute abundance of total bacteria (i), S.marcescens ( j) and E.mundtii (k) bacteria in the gut of S. frugiperda. (Mean ± SD; n = 6 replicates, 3 larvae/replicate). Different letters above each bar indicate significant differences at P < 0.05 as determined by one-way ANOVA with Tukey’s HSD test. Exact P-values are reported in the source data. Relevant source data are provided as a source data file.
Alteration of gut bacteria in S. frugiperda contributed to enhanced nano-enabled RNAi efficiency
The composition of the gut bacterial community in S. frugiperda was next investigated to identify how it changed when S. frugiperda larvae were fed with dsCHS@ZIF-8@PDA and dsV-ATPaseB@ZIF-8@PDA. The amplified bacterial 16S rRNA sequences using Illumina sequencing platform (Miseq) were characterized. Alpha diversity was assessed using Chao1, Observed_otus, Shannon, and Simpson index. The Shannon index of the dsCHS@ZIF-8@PDA and dsV-ATPaseB@ZIF-8@PDA groups were notably lower and Simpson index of those were significantly higher compared to other groups such as dsGFP, ZIF-8@PDA, dsCHS, and dsV-ATPaseB groups (Fig.3f and Supplementary Table 4, ANOVA, Tukey,s test, P < 0.05). These results suggested that dsCHS@ZIF-8@PDA and dsV-ATPaseB@ZIF-8@PDA NPs intervention contributed to a decrease in gut microbiota diversity of S. frugiperda. Principal coordinates analysis (PCoA) of Bray–Curtis distances for the microbial communities indicated that the bacterial communities of six groups clustered separately (Fig. 3g). These findings revealed decreased diversity and significant alterations in the gut microbial community structure of S. frugiperda larvae fed with maize leaves smeared with dsCHS@ZIF-8@PDA and dsV-ATPaseB@ZIF-8@PDA NPs. For larvae fed with maize leaves coated with dsGFP, dsCHS, and dsV-ATPaseB, the dominant genus in the gut bacterial community was Enterococcus sp (Fig. 3h and Supplementary Fig. 8). However, the proportion of Enterococcus sp. reduced significantly and the proportion of Serratia sp. revealed a notable increase when fed with maize leaves coated with ZIF-8@PDA, dsCHS@ZIF-8@PDA and dsV-ATPaseB@ZIF-8@PDA (Fig. 3h–k and Supplementary Fig. 8). These results indicated that enrichment of Serratia sp. was induced and regulated by ZIF-8@PDA NPs. Furthermore, Serratia marcescens and Enterococcus mundtii were screened to verify whether the overgrowth of Serratia sp. has a synergistic lethal effect on S. frugiperda. Aseptic populations of S. frugiperda were artificially constructed by rearing hatched larvae with maize leaves containing antibiotics (AB) until 3rd-instar larvae50 (Fig.4a–c). As shown in Fig. 4d, the survival rate of the aseptic S. frugiperda larvae population feeding on leaves with dsCHS@ZIF-8@PDA NPs + S. marcescens (OD600 = 0.1) was significantly lower (26.67%) than that fed with leaves coated by dsCHS@ZIF-8@PDA NPs + E.mundtii (OD600 = 0.1) (55.33%) or PBS buffer (55.67%). From Fig. 4e, both S. marcescens and E.mundtii can effectively infect the insect hemolymph. The above results indicated that the overgrowth of Serratia sp. induced by ZIF-8@PDA NPs was another important factor for the enhanced lethal effect of S. frugiperda besides enhanced dsRNA delivery. The RNA-seq results above also showed that the treatment of dsCHS@ZIF-8@PDA NPs activated Toll and IMD pathways, including genes encoding functions correlated to disease recognition (PGPR and β1,3-GPB), signal transduction (Mapkk4, IMD, Ankyrin, and spatzle), and transcription factors (Nf-kb-p110, Duox), etc. (Fig. 3c). Similar changes also occurred in ROS activity (Fig. 4f). Previous studies have shown that Toll and IMD pathways were able to regulate Dual Oxidase (Duox) for ROS production in honeybee gut bacteria51. Previous studies have reported that 20-hydroxyecdysone (20E) was involved in regulating the Toll and IMD pathways of insect52,53,54 and our results found that the 20E titers in hemolymph of aseptic S. frugiperda with dsCHS@ZIF-8@PDA + S. marcescens (OD600 = 0.1) were significantly increased compared to the dsCHS@ZIF-8@PDA + PBS and dsCHS@ZIF-8@PDA + E.mundtii (OD600 = 0.1) treatments in 24 h (Fig. 4g). As a crucial humoral immunity-related molecule, 20E activated Toll and IMD pathways encoding genes and induced ROS accumulation (Fig. 4f, g, m). As shown in Fig. 4i–l, these Toll and IMD pathways encoding genes of aseptic S. frugiperda fed with dsCHS@ZIF-8@PDA + S. marcescens (OD600 = 0.1) were significantly upregulated compared to those under dsCHS@ZIF-8@PDA + PBS and dsCHS@ZIF-8@PDA + E.mundtii (OD600 = 0.1) treatments in the first 48 h. These results indicated that S. marcescens activated the immune response of S. frugiperda through the Toll and IMD pathways.
a, b Comparison of bacteria quantities in guts of (±AB) S. frugiperda larvae (Mean ± SD; n = 3 replicates, P = 0.0036). c The total bacterial 16S rRNA gene copies in (±AB) larvae (Mean ± SD; n = 6 replicates, 3 larvae/replicate). b, c: unpaired Student’s t-test. d The survival rate curves of four-instar aseptic S. frugiperda larvae (n = 150/group) feeding on sterile maize leaves smeared with PBS/SM/EM+dsCHS@ZIF-8@PDA. P < 0.05 as determined by Kaplan–Meier analysis with log-rank test. e The S. marcescens and E. mundtii abundance in the hemolymph of S. frugiperda larvae (Mean ± SD; n = 6 replicates, 3 larvae/replicate; SM: F2,17 = 27.172, P < 0.001; EM: F2,17 = 36.238, P < 0.001). f The ROS levels (Mean ± SD; n = 9 replicates, 3 larvae/replicate; 24 h: F2,29 = 100.59, P < 0.001; 48 h: F2,29 = 250.98, P < 0.001; 72 h: F2,29 = 93.084, P < 0.001). g Titer change of ecdysone (E), 20-hydroxyecdysone (20E), and juvenile hormone 3 (JH3) in these larvae feeding on leaves smeared with SM/EM(OD600 = 0.1) + dsCHS@ZIF-8@PDA (Mean ± SD; n = 6 replicates, 3 larvae/replicate; unpaired Student’s t-test;12 h, E: t = 1.063, df = 10, P = 0.726; 20E: t = 2.646, df = 10, P = 0.541; JH3: t = 1.164, df = 10, P = 0.214; 24 h, E: t = 33.237, df = 10, P < 0.001; 20E: t = 37.358, df = 10, P < 0.001; JH3: t = 9.528, df = 10, P < 0.001; 48 h, E: t = 6.874, df = 10, P < 0.001; 20E: t = 6.047, df = 10, P < 0.001; JH3: t = 18.153, df = 10, P < 0.001). h Assay of the inhibitory effect of prodigiosin on E.mundtii bacteria. i–l The change of key genes of Toll and IMD pathways (Mean ± SD; n = 6 replicates, 3 larvae/replicate). m Gut tissue ROS staining of aseptic S. frugiperda feeding on maize leaves smeared with SM/EM + dsCHS@ZIF-8@PDA. *P < 0.05, **P < 0.01, ***P < 0.001, relevant source data and exact P-values were provided as a source data file.
We have added reverse validation experiments and found that aseptic 3rd-instar larvae subjected to RNAi-mediated knockdown (siRNA) of Toll/IMD pathway genes (β-1,3-GBP, Mapkk4, PGPR, Ankyrin, Duox, and Toll) with SM (OD600 = 0.1) + dsCHS@ZIF-8@PDA diet exhibited ROS suppression and S. marcescens proliferation (Fig. 5c–e, l), causing significantly accelerated mortality (Fig. 5a, b). qPCR quantification further confirmed the suppression of the immune gene above (Fig. 5f–k). These results indicated that the Toll/IMD pathway genes were involved in ROS generation, and the knockdown of Toll/IMD pathway genes critically impaired the host’s capacity to both counteract exogenous stressors and restrict pathogenic bacterial overgrowth (Fig. 5f–k). We explored the effect of dsRNA, nanocarrier components, ZIF-8, and ZIF-8@PDA on the increase of S. frugiperda. Field-collected 3rd-instar larvae were fed with maize leaves smeared with dsGFP, dsCHS, Zn²⁺, 2-methylimidazole (2-mIm), ZIF-8, PDA, and ZIF-8@PDA with concentrations equivalent to 50 mg/L dsRNA@ZIF-8@PDA for 48 h and the S. marcescens abundance increased significantly with all the treatments except dsRNA and 2-mIm treatments (P < 0.05, unpaired t-test). Among these, ZIF-8 and ZIF-8@PDA treatments showed the strongest induction for growth of S. marcescens (1.51 × 106 and 1.13 × 106-fold versus control, respectively) (Supplementary Fig. 9d–g), followed by PDA and Zn2+ treatments (9.82 × 103 and 8.5 × 104-fold), suggesting that compared to the components of nanocarrier, characterization of nanoparticles themselves exhibited stronger effect on S. marcescens expansion (Supplementary Fig. 9n–s). Neither 2-methylimidazole (2-mIm) nor dsRNA alone affected S. marcescens abundance55,56 (Supplementary Fig. 9a–c). Crucially, vitro assays demonstrated the growth of S. marcescens was restrained by ZIF-8 and ZIF-8@PDA (>50 mg/L), indicating host-derived stress signals from the insect gut are essential for overgrowth of S. marcescens in vivo (Supplementary Fig. 9t, u). Both ZIF-8 and ZIF-8@PDA nanoparticles induced dose-dependent ROS generation (50–200 mg/L) in midgut tissues, with ZIF-8 exhibiting significantly stronger oxidative stress induction (1.58-fold higher than ZIF-8@PDA at equivalent concentrations, P < 0.001) (Supplementary Fig. 9h, i). Furthermore, Vitamin C-mediated ROS quenching (quenched approx. 74%) caused the 99.9 % suppression of SM growth (P < 0.001), confirming that ROS served as the critical trigger for SM expansion (Supplementary Fig. 9j–m). These results confirmed that NPs-induced ROS increase cause the overgrowth of S. marcescens. Further experiments showed that the addition of S. marcescens and nanocarrier generated greater ROS burst compared to the addition of nanocarrier, confirming that overgrowth of S. marcescens further induced the ROS burst (Supplementary Fig. 9j–m), establishing a self-reinforcing oxidative stress cycle. These findings demonstrated that NPs-induced ROS generation caused the overgrowth of S. marcescens, and the overgrowth of S. marcescens further brought the ROS burst in host body, causing adverse effect to insects.
a Photographs of aseptic larvae 12 h after siRNA injection and fed maize smeared with SM (OD600 = 0.1) +dsCHS@ZIF-8@PDA NPs at 48 h. b The survival rate curves of four-instar aseptic S. frugiperda larvae (n = 60 per group) 12 h after siRNA injection feeding on sterile maize leaves smeared with PBS buffer (pH = 7.4), PBS buffer (pH = 7.4) + dsCHS@ZIF-8@PDA, SM (OD600 = 0.1) + dsCHS@ZIF-8@PDA, respectively. The asterisk (*P < 0.05, **P < 0.01, and ***P < 0.001) indicates a significant difference at P < 0.05 as determined by Kaplan-Meier analysis with log-rank test. c S. marcescens was absolutely quantified with qPCR (Mean ± SD; n = 3 replicates, 3 larvae/replicate; F8,53 = 338.75, P < 0.001; Different letters above each bar indicate significant differences at P < 0.05 as determined by one-way ANOVA with Tukey HSD test). Exact P-values are reported in the source data. d, e ROS activity was determined at 48 h and 72 h, respectively. (Mean ± SD; n = 3 replicates, 3 larvae/replicate; F8,53 = 284.38, P < 0.001; Different letters above each bar indicate significant differences at P < 0.05 as determined by one-way ANOVA with Tukey HSD test). Exact P-values are reported in the source data. f–k qPCR quantified the expression of Toll-IMD-related genes at 48 h and 72 h, respectively (Mean ± SD; n = 3 replicates, 3 larvae/replicate; P < 0.05 as determined by one-way ANOVA with Tukey HSD test). Exact P-values and F-values are reported in the source data. l Gut tissue ROS staining of aseptic S. frugiperda at 48 h, respectively. Relevant source data are provided as a source data file.
It is worth noting that the immune response of S. frugiperda was further inhibited significantly in the following 72 h by the treatment of dsCHS@ZIF-8@PDA + S. marcescens (OD600 = 0.1), showing significantly lower ROS content and downregulated IMD and Toll encoding genes as shown in Fig. 4f, i–l. These results were probably due to the activated antagonistic effect of JH3 and 20E57, showing increasing JH3 and decreased 20E titers in 48 h, thereby restraining the hormone level of 20E and further inhibiting the growth of larvae, as shown in Figs. 4g and 2a. Furthermore, we found that overgrowth of S. marcescens secreted significantly abundant prodigiosin (Supplementary Fig. 10e), which was found to inhibit the growth of other gut bacterial (e.g., E. mundtii) and host ROS levels (Fig. 4h). Similar study had found that prodigiosin secreted by S. marcescens was able to suppress other gut bacteria58 and reduce ROS activity59.
A phylogenetic tree of 16S rRNA demonstrated that SM_FJ07 extracted from the gut tissue of S. frugiperda was clustered together with other S. marcescens species (Fig. 6a). Whole-genome sequencing of SM_FJ07 bacteria was performed with SM_FJ07 circular chromosome length of 5028731 bp (Fig. 6b). The biosynthesis of prodigiosin is governed into two parallel branches: (1) The 2-methyl-3-n-amyl-pyrrole (MAP) branch utilizes 2-octenal and pyruvate as substrates, while (2) the 4-methoxy-2,2′-bipyrrole-5-carbaldehyde (MBC) branch employs proline and malonyl-CoA as precursors (Fig. 6c)60. These two distinct biosynthetic streams converge through the action of the oxidoreductase (PigC), which catalyzes the final coupling reaction to form prodigiosin (Fig. 6c)60. Therefore, ΔPigC mutants and complemented ΔPigC::PigC strains of SM_FJ07 (Fig. 6d, e) were constructed. We found that aseptic S. frugiperda feeding on maize leaves coated with SM (OD600 = 0.1) + dsCHS@ZIF-8@PDA and complemented ΔPigC::PigC strain+ dsCHS@ZIF-8@PDA demonstrated significantly elevated mortality (Fig. 6f), higher prodigiosin contents (Fig. 6d, h), and higher abundance of S. marcescens (Fig. 6g) relative to control groups with treatments of ΔPigC mutant (OD600 = 0.1) + dsCHS@ZIF-8@PDA or PBS + dsCHS@ZIF-8@PDA. These results demonstrated that PigC deletion impairs prodigiosin secretion and SM proliferation, thus diminishing the lethal effect of larvae (Fig. 6f, g). Meanwhile, ΔPigC mutant lost the capacity to suppress host ROS activity and related key immune gene expression, including (β-1,3-GBP1, PGRP, Toll, or Duox) (Fig.6i–m). It indicated that prodigiosin played an important role in the inhibition of host ROS activity and the expression of immune genes. These results supported that the enrichment of the S. marcescens in S. frugiperda gut induced by dsCHS@ZIF-8@PDA brought notable synergistic insecticidal effect by firstly activating the ROS immune response regulated by Toll and IMD pathways and then inhibiting the ROS immune response through activating the antagonistic effect of 20E-JH3 and secreting prodigiosin.
a A phylogenetic tree of 16S rRNA gene. Bootstrap support (1000 replicates) was calculated for the neighbor-joining tree. b Genome circle map and loci of the prodigiosin gene cluster of S. marcescens FJ07. c Prodigiosin synthesis pathway. d The wild-type S. marcescens/ΔPigC mutant/PigC::PigC complemented strains (20 µM IPTG) in liquid LB medium at room temperature. e S. marcescens FJ07, mutant ΔPigC and complemented ΔPigC::PigC strain band validation by gel electrophoresis using PigCtest-F/R primer. f The survival rate curves of four-instar aseptic S. frugiperda larvae (n = 96/group) feeding on sterile maize leaves smeared with PBS + dsGFP (pH = 7.4), PBS (pH = 7.4) + dsCHS@ZIF-8@PDA, SM (OD600 = 0.1) + dsCHS@ZIF-8@PDA, ΔPigC (OD600 = 0.1) + dsCHS@ZIF-8@PDA, and ΔPigC:: PigC (OD600 = 0.1) + dsCHS@ZIF-8@PDA respectively. The asterisk (*P < 0.05, **P < 0.01, and ***P < 0.001) indicates a significant difference at P < 0.05 as determined by Kaplan-Meier analysis with log-rank test. g, h S. marcescens and prodigiosin were absolutely quantified (Mean ± SD; n = 6 replicates, 3 larvae/replicate; F4,29 = 504.11, P < 0.001; F4,29 = 430.63, P < 0.001; Different letters above each bar indicate significant differences at P < 0.05 as determined by one-way ANOVA with Tukey HSD test). Exact P-values are reported in the source data. i ROS activity was determined at 48 h and 72 h, respectively. (Mean ± SD; n = 6 replicates, 3 larvae/replicate; F4,29 = 848.31, P < 0.001; F4,29 = 467.63, P < 0.001; Different letters above each bar indicate significant differences at P < 0.05 as determined by one-way ANOVA with Tukey HSD test). Exact P-values are reported in the source data. j–m qPCR quantified the expression of Toll-IMD-related genes at 48 h and 72 h, respectively (Mean ± SD; n = 3 replicates, 3 larvae/replicate; P < 0.05 as determined by one-way ANOVA with Tukey HSD test). Exact P-values and F-values are reported in the source data. Relevant source data are provided as a source data file.
Exogenous spraying of ZIF-8@PDA NPs and overgrowth S. marcescens enhances plant anti-herbivory
To further assess the enhancing effect of S. marcescens on plant defense to S. frugiperda, three different populations of S. frugiperda, i.e., aseptic, S. frugiperda larvae inoculated with S. marcescens and E. mundtii (Fig. 7a, b and Supplementary Fig. 11), were constructed and fed with maize plants sprayed with dsCHS @ZIF-8@PDA NPs under greenhouse conditions. The activity of glucose oxidase (GOX) in the oral regurgitation of the different populations of S. frugiperda, which was reported to be an elicitor/effector to stimulate/suppress plant anti-herbivore defense response, was significantly higher in the S. marcescens-inoculated larvae populations compared to that in E. mundtii-inoculated and aseptic populations (Fig. 7c–f). Furthermore, lower S. frugiperda biomass (Fig. 7g) and higher activity of polyphenol oxidases (Fig. 7h) (PPO, a key indicator of the level of anti-herbivore defense in plants26) confirmed that GOX in “S. frugiperda-Maize” model was an insect elicitor and increased GOX activity in S. marcescens-inoculated S. frugiperda larvae enhanced the plant anti-herbivore defense response. The crosstalk between jasmonic acid (JA, a signal molecule of anti-herbivore defense response61) and salicylic acid (SA, a signal molecule of sucking insects and pathogen invasion response62,63) was widely observed in “insect- microbe-plant” interaction50. Our results showed significant JA-SA antagonism in the group of S. marcescens-inoculated S. frugiperda, which was confirmed by JA and SA hormone levels (Fig. 7i) and relative gene expression levels (Fig. 7j–o). We found that S. marcescens-inoculated S. frugiperda increased JA accumulation and JA-responsive defense gene expression, including AOC1, AOS1, OPR2, and MPI (Fig. 7j–m) but decreased SA accumulation and SA-responsive defense gene expression, such as NPR and PR (Fig. 7n, o) compared to E. mundtii-inoculated S. frugiperda larvae, indicating that S. marcescens enhanced JA-responsive anti-herbivore defense of maize plant. Overall, S. marcescens-infected S. frugiperda was able to enhance the plant’s anti-herbivore ability by enhancing insect GOX activity and activating the JA signal pathway. The spraying of ZIF-8@PDA NPs exhibited significantly the lowest survival rates, body weight, and frass production in S. frugiperda larvae (P < 0.05) (Fig. 8d–f), while induced the strongest plant defense responses including elevated PPO activity (1.83-fold) (Fig. 8h, i), upregulation of JA pathway genes (P < 0.05) (Fig. 8l–o), and increased benzoxazinoids accumulation (Fig. 8r–w and Supplementary Fig. 12). These results demonstrated that ZIF-8@PDA pretreatment serves as an effective defense-priming agent that systemically enhances maize’s anti-herbivore resistance.
a Comparison of bacteria quantities by LB plates. b S. marcescens and E.mundtii contents in the oral regurgitation collected from the aseptic S. marcescens-infected, and E.mundtii-infected S. frugiperda larvae populations (Mean ± SD; n = 10/group; SM: F2,29 = 699.77, P < 0.001; EM: F2,29 = 412.246, P < 0.001). c a standard curve of commoditized GOX protein. d, e GOX content in the oral regurgitation of aseptic S. frugiperda larvae and inoculated with E. mundtii (EM-larvae) and S. marcescens (SM-larvae), respectively. (Mean ± SD; n = 3 replicates, 20 larvae/per replicates; F2,8 = 946.09, P < 0.0001). f GOX activity of different populations of S. frugiperda oral regurgitation (Mean ± SD; n = 7 replicates, 3 larvae/replicate; F2,20 = 68.094, P < 0.001). g The mass gain of the different populations of S. frugiperda feeding on maize plants after 48 h (Mean ± SD; n = 4 replicates, 5 larvae/replicate; F2,11 = 73.888, P < 0.001). h The phytohormones (JA, JA-Ile, and SA) changes of maize plants fed by different S. frugiperda populations after 24 h (Mean ± SD; n = 4 replicates, 5 larvae/replicate; JA: F2,8 = 36.404, P < 0.001; JA-Ile: F2,8 = 106.197, P < 0.001; SA: F2,20 = 107.211, P < 0.001). i Polyphenol oxidases (PPO) activity of maize plants fed by different S. frugiperda populations after 24 h (Mean ± SD; n = 4 replicates; F2,20 = 128.364, P < 0.001). j–o The key genes of JA (j–m) and SA (n, o) pathways determined after 48 h (Mean ± SD; n = 4 replicates; AOS1: F2,8 = 33.139, P = 0.01; AOC1: F2,8 = 197.64, P < 0.001; OPR2: F2,8 = 59.045, P < 0.001; MPI: F2,8 = 58.027, P < 0.001; NPR1: F2,8 = 129.854, P < 0.001; PR1: F2,8 = 248.44, P < 0.001). b, e–o: different letters above each bar indicate significant differences at P < 0.05 as determined by one-way ANOVA with Tukey HSD test. p The survival rate curves of SM/EM infected S. frugiperda with the treatment of dsRNA@ZIF-8@PDA, respectively (n = 75/group, P < 0.05 as determined by Kaplan-Meier analysis with log-rank test). The above exact data and P-values are provided in the source data file.
a, b Photographs of maize plants of inoculation of 3rd-instar larvae in the 1st and 6th days after nanomaterial spraying in a greenhouse. c Plant treatment: Sw+Fw (water spray + Fed water treatment), Sw+Fzp (water spray + Fed dsCHS@ZIF-8@PDA NPs treatment), Szp+Fw (ZIF-8@PDA NPs spray + Fed water treatment), and Szp+Fzp (ZIF-8@PDA NPs spray +Fed dsCHS@ZIF-8@PDA NPs treatment). d The survival rate curves of 3rd-instar larvae after inoculated on pretreatment maize plants (n = 100/group, *P < 0.05, **P < 0.01, and ***P < 0.001). e, f Larval weight (Mean ± SD; n = 38/group, weight: F3,151 = 704.4, P < 0.001) and frass mass (Mean ± SD; n = 29/group, F3,115 = 119.66, P < 0.001) after the 4th day of feeding on maize plants. g The phytohormones (JA, JA-Ile, and SA) content of maize plants of different treatments after 24 h (Mean ± SD; n = 3 replicates; JA: F3,11 = 188.24, P < 0.001; JA-Ile: F3,11 = 146.04, P < 0.001; SA: F3,11 = 130.05, P < 0.001). h–k PPO and POD activity of maize plants of different treatments after 48 and 72 h (Mean ± SD; n = 5 replicates; PPO (48h): F3,19 = 17.63, P < 0.001; PPO (72h): F3,19 = 17.28, P < 0.001; POD (48h): F3,19 = 9.92, P < 0.001; POD (72h): F3,19 = 35.43, P < 0.001). l–q The key genes of l–o JA (AOS1, AOC1, OPR2, and MPI) and p, q SA (NPR1 and PR1) pathways (Mean ± SD; n = 3 replicates; AOS1: F3,19 = 154.28, P < 0.001; AOC1: F3,19 = 24.79, P < 0.001; OPR2: F3,19 = 61.81, P < 0.001; MPI: F3,19 = 44.64, P < 0.001; NPR1: F3,19 = 172.83, P < 0.001; PR1: F3,19 = 119.24, P < 0.001). r–w The benzoxazinoids content of maize plants of different treatments after 48 h (Mean ± SD; n = 3 replicates; DIBOA-Glc: F3,11 = 18.52, P < 0.001; DIMBOA: F3,11 = 82.43, P < 0.001; DIMBOA-Glc: F3,11 = 85.77, P < 0.001; DIM2BOA: F3,11 = 11.16, P < 0.001; DIMBOA-Glc2: F3,11 = 94.03, P < 0.001; DIM2BOA-Glc: F3,11 = 87.70, P < 0.001). e–w: Different letters above each bar indicate significant differences at P < 0.05 as determined by one-way ANOVA with Tukey’s HSD test. The above exact data and P-values are provided in the source data file.
ZIF-8@PDA NPs stability and biosafety evaluation
Both ZIF-8 and ZIF-8@PDA exhibit pH-responsive properties, while ZIF-8@PDA demonstrates superior dsRNA stabilization capability and extended foliar retention on maize leaves compared to ZIF-8 (Supplementary Fig. 15a–g). Zebrafish exposed to ZIF-8 aqueous solutions for 24 h showed significant dose-dependent induction of detoxification enzymes (CAT), reactive oxygen species (ROS), and higher MDA contents along with SOD inhibition, demonstrating acute sensitivity to ZIF-8 and became suppressed during prolonged exposure (48–72 h) (Supplementary Fig. 15h–m), while ZIF-8@PDA exposure only induced marginal trends in AchE, ROS, SOD, and MDA with LDH inhibition at 24 h that similarly stabilized or declined at 48 or 72 h (Supplementary Fig. 15n–s and Supplementary Figs. 13, 14), with the Integrated Biomarker Response (IBR) index confirming ZIF-8 NPs greater early-stage biosecurity risk and ZIF-8@PDA NPs substantially reduced toxicity (Supplementary Fig. 15v, w).
In summary, we applied the ZIF-8@PDA-based dsRNA nano-delivery system to perform RNAi, eliminating the dsRNA delivery barrier to broaden the application of RNAi in S. frugiperda. The application of ZIF-8@PDA-based dsRNA enhanced the synergistic insecticidal effect of S. marcescens through two pathways: (1) activating followed by inhibiting the ROS immune response of S. frugiperda and (2) increasing the GOX activity as well as the JA-responsive anti-herbivore defense of plant. Further inoculation experiments confirmed that S. marcescens-inoculated S. frugiperda fed with on maize plants sprayed with dsCHS@ZIF-8@PDA showed significantly lower survival rate (29.33%) than those of E. mundtii-inoculated (58.66%) and aseptic S. frugiperda populations (48%, Fig. 7p), suggesting the potential of S. marcescens as a reinforcement agent to improve the effectiveness of nanoparticle-induced RNAi against S. frugiperda. We propose a combination strategy of pesticide/drug design and microbial agents to be useful for the green management of devastating plant pests.
Methods
dsRNA synthesis
The V-type proton ATPase subunit B (SfV-ATPaseB) (GeneBank accession No XM_035585939.2) and Chitin synthase (SfCHS) (GeneBank accession No XM_050696833.1) sequences were obtained from NCBI. Total RNA was obtained from 10 fourth-instar Spodoptera frugiperda (S. frugiperda) using Trizol kit (Vazyme, China) and then prepared for cDNA synthesis using Hifair first-Strand cDNA Synthesis Super Mix (Vazyme, China). Two kinds of cDNA were used as the template to amplify 476 bp (dsSfCHS) and 446 bp (dsSfV-ATPaseB) using the 2×Taq PCR Master Mixkit (Vazyme, China), respectively. The primers for dsRNA synthesis and qPCR (Supplementary Table 5, 6) were designed by Geneious software (Vazyme, China) and synthesized by Tsingke (China). dsRNA synthesis was performed using the HighYield T7 RNA Transcription Kit (Belong, SJ002, China). The dsGFP targeting the Green Fluorescent Protein (GFP) gene that is absent in S. frugiperda was used as the control. The above synthesized dsRNA products were purified using the MagBeads dsRNA Purification Kit (Belong, PU002 China), freeze-dried and stored at −20 °C (Supplementary Fig. 5).
Preparation of the dsRNA@ZIF-8@PDA nanocarrier complex
526.9 μL of dsRNA solution (5 mg mL−1, 10 mg mL−1, and 15 mg mL−1) were mixed with 50 μL of Zn (NO3)2·6H2O (0.84 M) separately. After thorough mixing, 484.1 μL of the 2-mIm (3.04 M) solution was added and quickly shaken for 20 min64. The product was centrifuged at 15,292×g min−1 for 30 min at 4 °C. 1 μL of precipitate and 5 μL of the supernatant (mixed with 10X loading buffer) were run on a 1% agarose gel at 120 eV, respectively. The surface of the precipitate was rinsed with DEPC water 3 times, and then 2.2 mL dopamine (DA) solution with a concentration of 2.8 mg mL−1 was added, and the reaction was terminated after continuously stirring for 5 h. The product was centrifuged at 2654×g min−1 for 10 min at 4 °C and slightly rinsed 3 times with DEPC water, freeze-dried, and then stored at −20 °C.
Self-assembly and component quantification of dsRNA@ZIF-8@PDA complex
Isothermal titration calorimetry (ITC) was used to reflect the thermodynamic parameters of the intermolecular interaction and to measure binding reactions between two molecules65. Herein, Zn2+ (0.84 mM) was dropped into 2-mIm solutions (10 mM), and dsGFP (0.5 mM) was dropped into ZIF-8 solutions (5 mM) by ITC analysis, respectively. The interaction heat during each injection was calculated by integrating each titration peak with Origin7 software (Origin Lab Co., USA). The test temperature was set to 25 °C. The ΔG was calculated as the following equation:
Pure dsRNA@ZIF-8@PDA complex was mixed with dsGFP at the mass ratio of 1:15, 1:10, and 1:5 (dsGFP: ZIF-8) (Supplementary Fig. 16), and the highest loading rate range was analyzed by agarose gel retardation assay66. dsRNA@ZIF-8@PDA NPs with the highest loading rate was synthesized when the 526.9 μL dsRNA (15 mg mL−1) was added into the synthesis reaction with a total 1.1 mL mixed system. The loading rates of dsRNA in dsRNA@ZIF-8 and dsRNA@ZIF-8@PDA were calculated, respectively. First, 50 mg dsGFP@ZIF-8 was dissolved in 50 mL hydrochloric acid solution (prepared with DEPC water, pH = 1), and then 1 μL mixture was used to measure the concentration of dsRNA, which was represented as c (ng μL−1). The resulting dsRNA was quantified using NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, USA). 50 mg of dry dsGFP@ZIF-8 was dissolved in 1.1 mL of DEPC water and then mixed with 2.2 mL DA solution (containing 0.1% tris) for 5 h to synthesize dsGFP@ZIF-8@PDA. After centrifugation, the supernatant was discarded and the precipitate was weighed after lyophilization, which was represented as m.
The loading rates of dsRNA@ZIF-8 (LR1) and dsRNA@ZIF-8@PDA (LR2) were calculated as follows:
Stability test and infiltration assay of transdermal dsRNA delivery system in S. frugiperda
Stability of dsGFP@ZIF-8@PDA complexes was determined using the gel retardation test. The hemolymph and gut fluids were collected from the 4th instar of S. frugiperda and suspended with 500 μL of cold PBS buffer in a 1.5 mL tube and centrifuged at 861×g min−1 for 3 min at 4 °C. The collected supernatant was transferred to a new tube to remove tissue contaminants. For degradation assays, the hemolymph (HL) and gut fluids (GF) were mixed with 1 μg dsGFP, separately. Meanwhile, the dsGFP@ZIF-8 (containing 1 μg dsGFP) and dsGFP@ZIF-8@PDA were treated with hemolymph and gut fluids under the same incubation condition. These mixture samples (10 μL) were incubated at 37 ± 1 °C for 1 h and then analyzed by 1% agarose gel electrophoresis. Naked dsGFP solution was used as the control. To test the transdermal dsRNA delivery system, dsGFP@ZIF-8@PDA NPs were synthesized according above method except dsGFP labeled with 0.1 μg mL−1 of Cy3. The production was centrifuged at 2654×g min−1 for 10 min at 4 °C, slightly rinsed 3 times with DEPC water and freeze-dried. To test the transdermal dsRNA@ZIF-8@PDA delivery system, 1 mg of dsGFP/Cy3@ZIF-8@PDA dry powder was evenly dispersed into 50 mL of DEPC water and 2 mL dsGFP/Cy3@ZIF-8@PDA solution was smeared evenly on 60 cm2 rectangular maize leaves, and then was further cut into square leaves with an area of 2 cm2 after drying. Each square leaf was smeared with naked dsGFP/Cy3 as a control. After feeding for 24 h, the gut tissue of 4th instar S. frugiperda larvae was dissected, followed by fixation with paraformaldehyde (4%) and DAPI solution for 15 min. The gut tissues were washed three times with phosphate-buffered saline (pH = 7.4). Sf9 cells (BNCC359598, 3.4 × 106, total cells/ampule, BeNa, China) were maintained in TNM-FH medium supplemented with 10% fetal bovine serum (FBS) and cultured in T25 flasks. For the experiment, when cell density reached 50–60% confluency, 10 mL of the cell suspension was transferred to a sterile 50 mm culture dish. Subsequently, 1 mL of either dsGFP/Cy3 or dsGFP@ZIF-8@PDA/Cy3 (1 mg/mL) was added to the dishes, with 5 replicates per group. The gut tissues and Sf9 cells were observed under a Laser Scanning Confocal Microscopy (A1-SHR-LFOV, Nikon, Japan).
Control efficacy evaluation of dsRNA@ZIF-8@PDA complex against S. frugiperda
5 mg lyophilized powder of dsSfV-ATPaseB@ZIF-8@PDA and dsSfCHS@ZIF-8@PDA were fully dispersed in 100 mL of DEPC water, separately. Similarly, 0.4 mg lyophilized powder of dsSfV-ATPaseB and dsSfCHS were fully dispersed in 100 mL of DEPC water, separately. Then 2 mL mixture was smeared on the maize leaves, and then maize leaves were further cut into square leaves after drying. Leaves coated with DEPC water were used as a control. In order to prevent S. frugiperda from attacking each other, each 3rd larvae of S. frugiperda were placed in a separate small plastic lattice, and 25 bugs were considered as a treatment, and each treatment was repeated 4 times. These leaves were placed in the middle of each small lattice, and fresh leaves were replaced every 12 h. Mortality, body length, weight and frass weight were recorded for 5 consecutive days. The phenotypes were recorded with an upright microscope.
The survival rate (%) was calculated at specific time intervals as (4).
where Ntotal was the total number of larvae of S. frugiperda, and Nt was the dead number of larvae at time t.
Construction of aseptic S. frugiperda treated with Enteroccous mundtii/Serratia marcescens (OD600 = 0.1) + dsCHS@ZIF-8@PDA NPs
The aseptic S. frugiperda larvae population was constructed using the following methods. New egg masses were soaked on the surface with antibiotic (AB) solution26 for 10 min and then moved to a sterile Petri dish. The antibiotic (AB) solutions were prepared in 50 mL of sterile Milli-Q water containing three anti-bacterial agents (0.2 g mL−1 neomycin sulfate, 0.01 g mL−1aureomycin and 0.06 g mL−1 streptomycin) as previously described50 (Fig. 4a–c and Supplementary Fig. 11). The sterile MilliQ water on a clean bench, fully dissolved, filtered using a 0.22 μm PVDF membrane, and AB solution was collected in a new sterile centrifuge tube. Maize leaves treated with AB solution were eaten by the hatched larvae until larvae had grown to 3rd instars26,50. Identification of aseptic S. frugiperda populations: Third- to fourth-instar S. frugiperda larvae of uniform size were randomly selected (10 larvae per group) after feeding on diets supplemented with or without AB (±AB treatment). Under sterile conditions, intact gut tissues were dissected using sterile forceps and immediately immersed in 4 mL of 1X PBS buffer (pH = 7.4). Each treatment group was replicated three times67. Gut tissues were homogenized using sterile pestles, followed by centrifugation at 1172×g min−1 for 10 min at 4 °C. The supernatant was serially diluted (10,000×), and 150 µL of the diluted suspension was evenly spread onto 2× YT solid agar plates. After incubation at 37 °C for 15 h, bacterial colonies were quantified using ImageJ software from photographed plates. The Enteroccous mundtii (E. mundtii)/Serratia marcescens (S. marcescens) concentrations were adjusted to OD600 = 0.1, with sterile PBS buffer (pH = 7.4). and then 1 mg of dsCHS@ZIF-8@PDA NPs lyophilized powder was added. Then, the above solution was smeared on the sterile naturally dried maize leaves, and those maize leaves were eaten by aseptic 3rd-instar S. frugiperda larvae. The maize leaves treated with PBS buffer were used as the control. The above treatments were recorded as S. marcescens (OD600 = 0.1) + dsCHS@ZIF-8@PDA, E.mundtii (OD600 = 0.1) + dsCHS@ZIF-8@PDA and PBS + dsCHS@ZIF-8@PDA, respectively. The number of deaths was counted after continuous feeding for 4 days, and 25 larvae with consistent body weight were considered as a treatment, and each treatment was repeated 6 times. Similarly, the survival rate (%) was calculated at specific time intervals as (4).
ROS staining and enzyme activity determination
The entire S. frugiperda guts were dissected and kept in PBS with 3-amino-1,2,4-triazole (Aladdin, Shanghai, China), an inhibitor of hydrogen peroxide (H2O2). The guts were then incubated with 10 μM DCFH-DA (Beyotime Biotech, Shanghai, China) for 5 min under the exclusion of light. The excitation wavelength for green fluorescence (DCF) was set at 488 nm, while the emission wavelength of 525 nm was used for detection. ROS activity was determined by measuring H2O2 using a Hydrogen Peroxide Assay Kit (S0038; Beyotime Biotech, Shanghai, China). Insect oral ROS enzyme activity was determined by Glucose Oxidase Assay Kit (ab138884; Fluorometric, Shanghai, China) (Supplementary Fig. 17).
Data analysis
The statistical analysis was performed using the SPSS 26.0 software (SPSS Inc., USA). The ANOVA with Tukey HSD test or independent t-test was used to analyze the data at the P = 0.05 level of significance. The descriptive statistics are shown as the mean value and standard errors of the mean.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The raw sequence reads of RNA-seq and 16S data for S. frugiperda and the genome of S. marcescens are available at NCBI with the accession PRJNA1169401, PRJNA1277262, and JBNBZV000000000, respectively. 16S rRNA gene sequencing of S. marcescens and E. mundtii was uploaded to NCBI under associate no. PV123017 and PQ571970, respectively. Source data are available for Figs. 1–8 and Supplementary Figs. 1, 3, 4, 8–10, 12, 15, 17, 24–31 in the associated source data file. Source data are provided with this paper.
References
Tay, W. T., Meagher, R. L. & Groot, A. T. Spodoptera frugiperda: ecology, evolution, and management options of an invasive species. Annu. Rev. Entomol. 68, 299–317 (2023).
Mendesil, E., Tefera, T. & Hutchison, W. D. The invasive fall armyworm, Spodoptera frugiperda, in Africa and Asia: responding to the food security challenge, with priorities for integrated pest management research. J. Plant Dis. Prot. 130, 1175–1206 (2023).
Li, Y. Managing the invasive fall armyworm through biotech crops: a Chinese perspective. Trends Biotechnol. 39, 105–107 (2021).
Lu, J. & Shen, J. Target genes for RNAi in pest control: a comprehensive overview. Entomologia 44, 95–114 (2024).
Zhu, K. Y. & Palli, S. R. Mechanisms, applications, and challenges of insect RNA interference. Annu. Rev. Entomol. 65, 293–311 (2020).
Meister, G. & Tuschl, T. Mechanisms of gene silencing by double-stranded RNA. Nature 2004, 343–349 (2004).
Hutvagner, G. & Simard, M. J. Argonaute proteins: key players in RNA silencing. Nat. Rev. Mol. Cell Biol. 9, 22–32 (2008).
Tang, W., Luo, X. Y. & Sanmuels, V. Gene silencing: double-stranded RNA mediated mRNA degradation and gene inactivation. Cell Res. 11, 181–186 (2001).
Dalakouras, A., Koidou, V. & Papadopoulou, K. DsRNA-based pesticides: considerations for efficiency and risk assessment. Chemosphere 352, 141530 (2024).
Wang, Y., Li, M. & Wang, X. High-efficiency green management of potato late blight by a self-assembled multicomponent nano-bioprotectant. Nat. Commun. 14, 1–10 (2023).
Yoon, J.-S., Gurusamy, D. & Palli, S. R. Accumulation of dsRNA in endosomes contributes to inefficient RNA interference in the fall armyworm, Spodoptera frugiperda. Insect Biochem. Mol. Biol. 90, 53–60 (2017).
Dahlman, J. E., Barnes, C. & Anderson, D. G. In vivo endothelial siRNA delivery using polymeric nanoparticles with low molecular weight. Nat. Nanotechnol. 9, 648–655 (2014).
Yu, C., Li, J., Zhang, Z. & He, S. Metal–organic framework-based insecticide and dsRNA codelivery system for insecticide resistance management. ACS Appl. Mater. Interfaces 15, 48495–48505 (2023).
Jain, R. G., Fletcher, S. J. & Mitter, N. Foliar application of clay-delivered RNA interference for whitefly control. Nat. Plants 8, 535–548 (2022).
Zhou, H., Wan, F., Jian, Y. & Ding, W. Chitosan/dsRNA polyplex nanoparticles advance environmental RNA interference efficiency through activating clathrin-dependent endocytosis. Int. J. Biol. Macromol. 253, 127021 (2023).
Zhang, X., Zhang, J. & Zhu, K. Y. Chitosan/double-stranded RNA nanoparticle-mediated RNA interference to silence chitin synthase genes through larval feeding in the African malaria mosquito (Anopheles gambiae). Insect Mol. Biol. 19, 683–693 (2010).
Theresa, M., Allen, T. M. & Cullis, P. R. Liposomal drug delivery systems: from concept to clinical applications. Adv. Drug Deliv. Rev. 65, 36–48 (2013).
Castellanos, N. L., Smagghe, G., Sharma, R., Oliveira, E. E. & Christiaens, O. Liposome encapsulation and EDTA formulation of dsRNA targeting essential genes increase oral RNAi-caused mortality in the neotropical stink bug Euschistus heros. Pest Manag. Sci. 75, 537–548 (2019).
Yan, S., Liu, X. & Shen, J. Nanoparticle-mediated double-stranded RNA delivery system: a promising approach for sustainable pest management. Insect Sci. 28, 21–34 (2021).
Jiang, Z., Zhang, J. & Bock, R. Next-generation insect-resistant plants: RNAi-mediated crop protection. Trends Biotechnol. 35, 871–882 (2017).
Ocsoy, I. et al. Nanotechnology in plant disease management: DNA-directed silver nanoparticles on graphene oxide as an antibacterial against Xanthomonas perforans. ACS Nano 7, 8972–8980 (2013).
Dhandapani, R. K., Gurusamy, D. & Palli, S. R. Protamine–lipid–dsRNA nanoparticles improve RNAi efficiency in the fall armyworm, Spodoptera frugiperda. J. Agric. Food Chem. 70, 6634–6643 (2022).
Chao, Z., Ma, Z. & Shen, J. Establishment of star polycation-based RNA interference system in all developmental stages of fall armyworm Spodoptera frugiperda. Entomologia 43, 127–137 (2023).
Mason, C. J., Peiffer, M. & Felton, G. W. Concerted impacts of antiherbivore defenses and opportunistic Serratia pathogens on the fall armyworm (Spodoptera frugiperda). Oecologia 198, 167–178 (2022).
Gomes, A. F. F., Omoto, C. & Cônsoli, F. L. Gut bacteria of field-collected larvae of Spodoptera frugiperda undergo selection and are more diverse and active in metabolizing multiple insecticides than laboratory-selected resistant strains. J. Pest Sci. 93, 833–851 (2020).
Chung, S. H., Rosa, C. & Felton, G. W. Herbivore exploits orally secreted bacteria to suppress plant defenses. Proc. Natl. Acad. Sci. USA 110, 15728–15733 (2013).
Sahu, K. P., Singh, S. & Singh, S. K. Nanomaterials via ZIF-8: preparations, catalytic and drug delivery applications. Chem. Eng. J. 508, 160663 (2025).
Li, H., Chen, W., Liu, B. & Chen, G. A purely green approach to low-cost mass production of zeolitic imidazolate frameworks. Green. Energy Environ. 8, 775–784 (2023).
Tong, Y., Shao, L. & Wu, X. Adhesive and stimulus-responsive polydopamine-coated graphene oxide system for pesticide-loss control. J. Agric. Food Chem. 66, 2616–2622 (2018).
Gao, Z., Tan, J., Sun, Y. & Jiang, X. Size effect of ZIF-8 based nanocarrier pesticide delivery system on targeted release and insecticidal activity. Pest Manag. Sci. 81, 966–977 (2025).
Lu, Q., Cui, H. & Yang, Q. Synthetic nanoscale RNAi constructs as pesticides for the control of Locust migratoria. J. Agric. Food Chem. 70, 10762–10770 (2022).
Shi, X., Liu, X. & Zhang, J. Vacuolar (H +)-ATPase subunit c is essential for the survival and systemic RNA interference response in Locusta migratoria. Pest Manag. Sci. 78, 1555–1566 (2022).
Kwon, D. H., Park, J. H. & Lee, S. H. Screening of lethal genes for feeding RNAi by leaf disc-mediated systematic delivery of dsRNA in Tetranychus urticae. Pestic. Biochem. Physiol. 105, 69–75 (2013).
Wieczorek, H., Beyenbach, K. W. & Vitavska, O. Vacuolar-type proton pumps in insect epithelia. J. Exp. Biol. 212, 1611–1619 (2009).
Cheng, X., Zhou, Q. & Zhang, H. Nanoparticle LDH enhances RNAi efficiency of dsRNA in piercing-sucking pests by promoting dsRNA stability and transport in plants. J. Nanobiotechnol. 22, 544 (2022).
Lv, H., Li, X. & He, S. Overcoming resistance in insect pest with a nanoparticle-mediated dsRNA and insecticide co-delivery system. Chem. Eng. J. 475, 146239 (2023).
Kaziem, A. E., Yang, L. & Zhang, Z. Pathogenic invasion-responsive carrier based on mesoporous silica/β-glucan nanoparticles for smart delivery of fungicides. ACS Sustain. Chem. Eng. 9, 9126–9138 (2021).
Falconer, R. J., Penkova, A. & Collins, B. M. Survey of the year 2008: applications of isothermal titration calorimetry. J. Mol. Recognit. 23, 395–413 (2010).
Cavalcanti, I. D. L. Isothermal titration calorimetry (ITC) as a promising tool in pharmaceutical nanotechnology. Int. J. Pharm. 641, 123063 (2023).
Huang, Z., Ma, C. & Wu, C. Exploring the drug-lipid interaction of weak-hydrophobic drug loaded solid lipid nanoparticles by isothermal titration calorimetry. J. Nanopart. Res. 22, 3 (2019).
Wang, X., Zhang, S. & Guo, X. Ionic strength-responsive binding between nanoparticles and proteins. Langmuir 34, 8264–8273 (2018).
Chakraborti, S., Joshi, P. & Chakrabarti, P. Interaction of polyethyleneimine-functionalized ZnO nanoparticles with bovine serum albumin. Langmuir 28, 11142–11152 (2012).
Saleh, M.-C., van Rij, R. P. & Andino, R. The endocytic pathway mediates cell entry of dsRNA to induce RNAi silencing. Nat. Cell Biol. 8, 793–802 (2006).
Shi, X., Li, S., Yang, L. & Zhang, J. Clathrin heavy chain is essential for the development and reproduction of Locusta migratoria. Insect Sci. 29, 1601–1611 (2022).
Da Xiao, X. D. & Zhu, K. Y. Clathrin-dependent endocytosis plays a predominant role in cellular uptake of double-stranded RNA in the red flour beetle. Insect Biochem. Mol. Biol. 60, 68–77 (2015).
Donaldson, J. G. & Jackson, C. L. ARF family G-proteins and their regulators: roles in membrane transport, development and disease. Nat. Rev. Mol. Cell Biol. 12, 362–375 (2011).
Poirier, M. B., Fiorino, C. & Harrison, R. E. F-actin flashes on phagosomes mechanically deform contents for efficient digestion in macrophages. J. Cell Sci. 133, jcs239384 (2020).
Lee, H.-J., Woo, Y. & Jung, Y.-J. Formation and maturation of the phagosome: a key mechanism in innate immunity against intracellular bacterial infection. Microorganisms 8, 1298 (2020).
Tremel, S. et al. Structural basis for VPS34 kinase activation by Rab1 and Rab5 on membranes. Nat. Commun. 12, 1564 (2021).
Gao, Z., Ju, X., Yang, M. & Wang, J. Colorado potato beetle exploits frass-associated bacteria to suppress defense responses in potato plants. Pest Manag. Sci. 78, 3778–3787 (2022).
Guo, L., Tang, J. & Zhou, X. Reactive oxygen species are regulated by immune deficiency and Toll pathways in determining the host specificity of honeybee gut bacteria. Proc. Natl. Acad. Sci. USA 120, e2219634120 (2023).
Sun, W., Shen, Y.-H. & Zhang, Z. Ecdysone titer determined by 3DE-3β-reductase enhances the immune response in the silkworm. J. Immunol. 196, 1646–1654 (2016).
Tan, K. L., Vlisidou, I. & Wood, W. Ecdysone mediates the development of immunity in the Drosophila embryo. Curr. Biol. 24, 1145–1152 (2014).
Lu, K., Li, Y. & Song, Y. Activation of the ROS/CncC and 20-hydroxyecdysone signaling pathways is associated with xanthotoxin-induced tolerance to λ-cyhalothrin in Spodoptera litura. J. Agric. Food Chem. 69, 13425–13435 (2021).
Auguste, M., Lasa, A. & Canesi, L. Exposure to TiO2 nanoparticles induces shifts in the microbiota composition of Mytilus galloprovincialis hemolymph. Sci. Total Environ. 670, 129–137 (2019).
Liu, Y.-J., Jing, Z. & Ge, Y. Nano-La2O3 induces honeybee (Apis mellifera) death and enriches for pathogens in honeybee gut bacterial communities. Front. Microbiol. 12, 780943 (2021).
Liu, S., Li, K. & Li, S. Antagonistic actions of juvenile hormone and 20-hydroxyecdysone within the ring gland determine developmental transitions in Drosophila. Proc. Natl. Acad. Sci. USA 115, 139–144 (2018).
Yip, C.-H., Mahalingam, S. & Nathan, S. Prodigiosin inhibits bacterial growth and virulence factors as a potential physiological response to interspecies competition. PLoS ONE 16, e0253445 (2021).
Chen, J., Li, Y. & Feng, X. Prodigiosin promotes Nrf2 activation to inhibit oxidative stress induced by microcystin-LR in HepG2 cells. Toxins 11, 403 (2019).
Williamson, N. R., Simonsen, H. T. & Salmond, G. P. C. Biosynthesis of the red antibiotic, prodigiosin, in Serratia: identification of a novel 2-methyl-3-n-amyl-pyrrole (MAP) assembly pathway, definition of the terminal condensing enzyme, and implications for undecyl prodigiosin biosynthesis in Streptomyces. Mol. Microbiol. 56, 971–989 (2005).
Wang, J., Wu, D., Wang, Y. & Xie, D. Jasmonate action in plant defense against insects. J. Exp. Bot. 70, 3391–3400 (2019).
Jiang, W., Lu, C., Fan, J. & He, Y. Exogenous salicylic acid induces defense responses in tea plants against toxoptera Aurantii (Hemiptera: Aphididae). J. Econ. Entomol. 117, 302–310 (2024).
An, C. & Mou, Z. Salicylic acid and its function in plant immunity. J. Integr. Plant Biol. 53, 412–428 (2011).
Zhao, Y., Yuan, P.-Q. & Yang, J. Removal of phosphate by adsorption with 2-phenylimidazole-modified porous ZIF-8: powder and chitosan spheres. ACS Omega 8, 28436–28447 (2023).
Grolier, J.-P. E. & del Río, J. M. Isothermal titration calorimetry: a thermodynamic interpretation of measurements. J. Chem. Thermodyn. 55, 193–202 (2012).
Li, M., Ma, Z., Peng, M. & Shen, J. A gene and drug co-delivery application helps to solve the short life disadvantage of RNA drug. Nano Today 43, 101452 (2022).
Wang, J., Peiffer, M. & Felton, G. W. Helicoverpa zea gut-associated bacteria indirectly induce defenses in tomato by triggering a salivary elicitor(s). N. Phytol.214, 1294–1306 (2017).
Acknowledgements
This work was supported by the National Natural Science Foundation of China (grant numbers: No. 42377323 and No. 52206293).
Author information
Authors and Affiliations
Contributions
Z.G. and X.J. designed the project and interpreted the data. J.T. and X.L. oversaw the project. Z.G. and C.S. conducted experiments and analyzed the data. C.R., J.W., and M.E. analyzed data and revised manuscript. Z.G., X.J. and C.R. wrote the manuscript. All authors edited the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Feng Ju and the other anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s Note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Gao, Z., Rensing, C., Wang, J. et al. Increased and synergistic RNAi delivery using MOF polydopamine nanoparticles for biopesticide applications. Nat Commun 16, 6384 (2025). https://doi.org/10.1038/s41467-025-61604-5
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-61604-5