Abstract
Breastfeeding protects against breast cancer in some women but not others, however the mechanism remains elusive. Lactation requires intense secretory activity of the endoplasmic reticulum for the production of milk proteins and endoplasmic reticulum- mitochondria contacts for lipid synthesis. We show that in female mice that share the same nuclear genome (BL/6) but differ in mitochondrial genomes (C57 or NZB), lactation engages different transcriptional programs resulting in anti-tumorigenic lactation in BL/6C57 females and pro-tumorigenic lactation in BL/6NZB females. Our data indicate activation of a pro-apoptotic endoplasmic reticulum-stress response during lactation in BL/6C57 females, which is not observed in BL/6NZB females. Single cell sequencing identified a sub-population of cells, uniquely amplified during lactation in BL/6NZB females, that shares the genetic signature of post-partum breast cancer in humans and is characterized by cell cycle markers and loss of p53. We show that pharmacological manipulations of endoplasmic reticulum-stress directly affect this signature. Overall, our data suggest the unexpected differential nature of lactation and its potential impact on the risk of the development of post-partum breast cancer.
Similar content being viewed by others
Introduction
While pregnancy at an early age is associated with a reduction in the overall life-time risk of developing breast cancer1, pregnancy after the age of 30 is associated with increased risk of breast cancer with the risk peaking 6 years post-partum but remaining higher with increasing age of the mother over decades1. These post-partum breast cancers are more aggressive and metastatic2,3,4,5. Since in developed countries, women tend to have their children after the age of 30, pregnancy represents an important etiological factor in breast cancer today. One major breakthrough regarding increased risk of breast cancer following pregnancy is the discovery that post-pregnancy involution creates a tumorigenic supportive microenvironment6,7,8,9,10,11,12.
However, a meta-analysis comparing the impact of breastfeeding in ~50, 000 breast cancer patients relative to ~100,000 women without breast cancers, found that women without breast cancer showed a significant longer period of breastfeeding, therefore concluding that lactation is protective13,14,15,16,17. More focused studies analyzed the impact of breastfeeding length between premenopausal breast cancer patients indicated that 51% of patients had never breastfeed, while 16–17% of patients had breastfed for <3 or 4–12 months respectively, showing on one hand, the protective effect of breastfeeding since the percentage dropped with breastfeeding, but on the other hand that a significant percentage (16–17%) of patients did not benefit from this protective effect. Further, this significant fraction was maintained with longer time of breastfeeding as 15% of breast cancer patients breastfeed for 13 to > 24 months18,19,20. Therefore, while breastfeeding is overall protective, some women appear to be at higher risk of developing PPBC because they do not benefit from the protective effect of breastfeeding. Interestingly, one study reported that excessive milk production was associated with early age of diagnose of breast cancer21.
At the sub-cellular level, lactation implicates the secretion of proteins and lipids, both of which involve the endoplasmic reticulum (ER). Accumulation of proteins or lipids in the ER activates a stress response referred as ER-stress, which is generally adaptive and restores homeostasis of the ER. However, under conditions where stress in the ER cannot be resolved, the ER-stress response results in apoptosis22. Therefore, ER-stress can be either pro- or anti-apoptotic and the duration of the stress determines the outcome.
The ER is in close physical contact with the mitochondria and the contacts between both organelles regulate mitochondrial fission23,24, affect the function of both organelles including lipid metabolism25,26 and were reported to be modulated during lactation27. While ER-stress (also originally referred as the unfolded protein response (UPR), was the first to be discovered, subsequently, stress in the mitochondria was also found to activate a stress response, which was named the mitochondrial UPR or UPRmt28,29,30,31. Not surprisingly since the ER and mitochondria are in close physical contact, it was later found that stress in the ER affects stress in the mitochondria and that both stress responses share common transcription factors and is therefore referred to as the integrated stress response (ISR)28,32.
We chose to investigate the impact of lactation in mice that share the same nuclear genome in the C57BL/6 genetic background (BL/6) but differ in mitochondria genomes, either from the C57BL/6 or NZB/OlaHsd (C57 or NZB) background33. Since the number of nucleotide differences in the mitochondrial DNA between NZB/OlaHsd and BL/6C57 mice is within the same range as that observed between the European and African DNA haplotypes in the human population, these mice represent a model that reflects the natural diversity in mitochondrial genetics in humans33.
Here, we show that mitochondrial haplotypes impact the fundamental biology of lactation, resulting in pro- versus anti-tumorigenic environment. Notably in the setting of a pro-tumorigenic lactation in BL/6NZB females, an anti-apoptotic ER-stress response is observed, cell cycle markers are activated, while the tumor suppressor p53 is inhibited leading to ductal hyperplasia and increased susceptibility to tumor formation upon exposure to an oncogenic stimulus. Further, this cell cycle and p53 signature is also observed in post-partum breast cancers (PPBC) relative to matched breast cancers in never pregnant women (NPBC)34 and we identified a sub-population of PPBC-like cells during lactation in BL/6NZB females. Additionally, we also show that targeting ER-stress can convert pro-tumorigenic lactation into a protective lactation.
Results
RNAseq identifies different pathways during lactation in female mice that differ only in mitochondria
To gain insights into the impact of different mitochondria on lactation, we harvested the mammary glands over a time course including day 8, 14 and 21 of lactation in BL/6C57 and BL/6NZB female mice and performed bulk RNAseq.
Not surprisingly, lactation induced large gene expression changes relative to non-lactating females at each time point (Supplementary Fig. 1). More specifically, this analysis revealed that 767 genes were specifically up-regulated at day 8, 392 at day 14 and 400 at day 21 of lactation in BL/6C57 females, indicating that the number of genes specifically expressed in early, mid and late lactation decreases over time (Fig. 1A, B). The same pattern of decreased gene transcription over time during lactation was observed for the genes that are down-regulated (Fig. 1C, D). We then focused on the genes that are commonly up-regulated or down-regulated at all time points of lactation and found 5707 common genes that are up-regulated (Fig. 1A) and 329 common genes that are down-regulated (Fig. 1C). The pathway analysis of the 5707 genes commonly up-regulated in BL/6C57 females revealed pathways related to the UPR and tumor suppressors, notably p53 (Fig. 1A, Supplementary Fig. 2A, Supplementary Fig. 2), while cell cycle was the main pathway observed from analysis of the 329 common genes that are down-regulated (Fig. 1C, Supplementary Fig. 3A).
A–H RNA seq analysis was performed on 3-4 months old, estrus cycle matched non-lactating (NL) virgin female mice and 3-4 months old lactating (L) BL/6C57 or BL/6NZB female mice on day 8, 14 and 21 of lactation. Venn diagram indicating the number of genes significantly (adjusted P < 0.05) upregulated or downregulated either in BL/6C57 or BL/6NZB female mice at day 8, 14, 21 of lactation compared to their non lactating (NL) controls (n = 3 mice/group) (A, C, E, G). Graphs showing the trend of genes exclusively upregulated or downregulated at day 8, 14 and 21 in BL/6C57 or BL/6NZB mice (B, D, F, H). I Venn diagram indicating the number of genes significantly upregulated (p < 0.05) in BL/6C57 mice or BL/6NZB mice at day 21 of lactation compared to their non lactating (NL) control (n = 3 mice/group) (middle). The genes exclusively upregulated in BL/6C57 or BL/6NZB mice were subjected to pathway analysis using EnrichR and are represented in the graphs according to their combined score. All pathways shown are statistically significant. J Electron Microscopy was performed on the mammary glands of 3–4 months old BL/6C57 or BL/6NZB mice at day 10 and day 21 of lactation (n = 5 mice/group). Pictures were taken from 8 to 10 different sections per mice. The scale bar represents 500 nm or 200 nm. K Graph of milk volumes per gland collected from either 3–4 months old BL/6C57 or BL/6NZB mice on day 8, 14 and 21 of lactation. Student’s unpaired t test, two-tailed, ***p < 0.001 at Day 8; ***p < 0.0008 at Day 14 and ***p < 0.0004 at Day 21; mean ± SD; n = 6 mice/group. L Graph showing average weight of mice per litter at day 8, 14, 21 of lactation. Student’s unpaired t test, two-tailed; mean ± SD; n = 6 litters/group. Source data are provided as a Source Data file. Mice used in all experiments were 2–3 months old at the beginning of the experiments.
The same analysis was performed in the BL/6NZB females. We found that in contrast to the BL/6C57, the number of genes specifically up-regulated at day 8 (710 genes), 14 (398 genes) and 21 (1652 genes) overall increased through the time course of lactation (Fig. 1E, F), while the genes that are down-regulated at specific days of lactation remained relatively constant (1190 at day 8, 1356 at day 14 and 1201 at day 21, Fig. 1G, H). Focusing on the genes that are common between day 8, 14 and 21 lactation time points in the BL/6NZB females, 4539 were found to be up- regulated and associated with abnormal proliferation (Fig. 1E, Supplementary Fig. 3B), while 210 genes were commonly down-regulated and linked to cell death and the p53 tumor suppressor (Fig. 1G, Supplementary Fig. 3B). This global analysis indicates that the overall patterns of gene expression during lactation differ between genotypes.
To determine if these differences in gene expression are also observed at individual time points, we compared day 8, day 14 and day 21 between genotypes. Volcano plots analysis indicated significant differences in gene expression between genotypes at each time point of lactation (Supplementary Fig. 4A, B, C). Differential gene expression and pathway analyses revealed that the most significant alterations in early lactation at day 8 in BL/6NZB females compared to BL/6C57 females are associated with down-regulated genes and p53, unfolded protein response and cell cycle checkpoint pathways (Supplementary Fig. 4D). The same analysis of mid-lactation at day 14 also showed that most significant differences were associated with down-regulated genes and pathways associated with p53, cell cycle checkpoint and apoptosis (Supplementary Fig. 4E). In late lactation at day 21, significant differences were observed in both up and down-regulated genes. Pathway analysis of down-regulated genes again revealed down-regulation of p53 and apoptosis (Supplementary Fig. 4F). Therefore, this analysis suggests that while the difference in gene expression is more marked at day 21, pathways related to p53, UPR and apoptosis can be detected early during lactation at the gene expression level.
We then compared genes that are uniquely up-regulated at day 21 and specific to each genotype. We found that 4722 genes were commonly up-regulated at this time point but 2628 and 2676 were unique to the BL/6C57 and BL/6NZB females respectively (Fig. 1I). Pathways analysis of the 2628 genes up-regulated specifically in the BL/6C57 females highlighted ER-stress, apoptosis, cell cycle checkpoints (Fig. 1I). In contrast, the 2676 genes specifically up-regulated in the BL/6NZB females revealed a cell cycle signature as well as a muscle related signature (Fig. 1I). Since during late lactation, pups start weaning, which can induce involution and impact gene expression, we compared the histology of the mammary glands in early (day 10) and late lactation (day 21) as well as during post-lactation involution at day 5. We found that at the histological level, no marked differences between genotypes were observed either by H&E or using markers of luminal (K8) or basal (SMA) (Supplementary Fig. 5).
These analyses revealed that 1) the global number of differentially regulated genes over the period of lactation varies between genotypes, 2) the pathways activated during lactation also differ between genotypes, and 3) these differences begin to be observed early at day 8 and 14 of lactation but are most pronounced by day 21.
To gain further insight into these differences at the sub-cellular level, we examined organellar ultra-structure by transmission electron microscopy. This analysis indicated that while the morphology of the mitochondria and ER was not noticeably different in virgin females (Supplementary Fig. 6), differences were observed between genotypes during lactation. Notably an expanded ER network and normal mitochondria were observed in BL/6C57 females both in early (day 10) and late (day 21) lactation (Fig. 1J). In BL/6NZB females, in early lactation the ER network was more disconnected, but the mitochondria looked normal (Fig. 1J), however, in late lactation, the disconnected ER remained visible and the mitochondria displayed dilated cristae and electron dense area (Fig. 1J). These differences suggest that the contacts and communication between the ER and the mitochondria differ between genotypes during lactation. To test if these organellar differences affect the secretion of milk, we measured the volume of milk and found that both genotypes produce milk, with the BL/6NZB showing increased secretion (Fig. 1K) likely due to an increased number of pups per litter (avg 8 pups/litter in BL/6NZB and avg 5 pups in BL/6C57). However, no difference in the weight of pups over time was observed (Fig. 1L). Collectively therefore, despite the fact that lactation fulfills its physiological function and sustain the growth of their pups in females from both genotypes, the cellular biology and pathways activated during lactation are different.
In order to validate the data obtained by RNAseq, we first analyzed the specific cell cycle genes that led to the identification of this pathway and analyze mammary glands at day 21 of lactation by Western blotting. We found that all 5 cell cycle markers tested (cyclin E, D1, cdk2, 4 and 7) are significantly higher in BL/6NZB females, not only relative to the BL/6C57 females at day 21 of lactation, but also to BL/6NZB females that are not lactating (Fig. 2A, B). Additionally, since a tumor suppressor gene signature driven by p53 was identified as down regulated specifically in the BL/6NZB (Supplementary Fig. 4), we also analyzed p53 level and confirmed it is down-regulated specifically during lactation in the BL/6NZB females (Fig. 2A, B). We repeated the same analysis for day 8 and day 14 of lactation and found that while no difference in cell cycle markers and p53 were observed at day 8 (Supplementary Fig. 7A, B), significant differences were observed at day 14 (Supplementary Fig. 7C, D). This data supports the conclusion that while differences notably in p53 signaling can be observed at the gene expression level starting at day 8 (Supplementary Fig. 4D), these changes only become apparent by western starting at day 14.
A, B Western blot analysis of indicated cell cycle markers and p53 were performed in mammary glands from BL/6C57 or BL/6NZB 3-4 months old, estrus cycle matched non lactating (NL) virgin mice and 3–4 months old lactating (Day 21) female mice (A). Graphs of quantification of western blot analysis (B). Student’s unpaired t test, two-tailed, ***p < 0.0006, **p < 0.0029 for cyclin E; ****p < 0.0001, ***p < 0.0002 for cyclin D1; ***p < 0.006, **p < 0.0017 for cdk2; ****p < 0.001, **p < 0.0010, **p < 0.0045 for cdk4; ***p < 0.006, **p < 0.0017 for cdk7; *p < 0.0111, **p < 0.0018 for p53; mean ± SD; n = 9 mice/group. The representative blot shows 3 individual mice per group. In panels a and b, data are representative of two independent experiments. C Western Blot analysis of cell cycle markers in mammary organoids derived from 3-4 months old, estrus cycle matched BL/6C57 or BL/6NZB virgin female mice before (BI) or after induction (AI) with 1 µg/ml prolactin, 1 µg/ml hydrocortisone and 40 µg/ml oxytocin on the 6th day of organoid culture. D Graphs showing the quantification of western blot analysis in C. Student’s unpaired t test, two-tailed, ****p < 0.0001 for cyclin E; ***p < 0.007 for cyclin D1; ***p < 0.0007 for cdk2; **p < 0.0014 for cdk4; **p < 0.0015 for cdk7; **p < 0.0018 for p53; mean ± SD; n = 9 mice/group. Number of experiments=3. E–J Mammary organoids from 3 to 4 months old, estrus cycle matched BL/6C57 or BL/6NZB female virgin mice induced with 1 µg/ml prolactin, 1 µg/ml hydrocortisone and 40 µg/ml oxytocin were fixed and stained for pRB, K-14, Actin and Dapi. (E, G, I) Representative image of pRB, K-14, Actin, Dapi and overlay. The scale bar represents 50 µm. n = 9 mice/group. Number of experiments=3. F, H, J Graph showing pRB stained region, K-14 surface area compared to total surface area, Actin-stained area which were analyzed using image J software. Student’s unpaired t test, one-tailed, *p < 0.0358 (F); Student’s unpaired t test, two-tailed, *p < 0.0245 (H); Student’s unpaired t test, one-tailed, *p < 0.0276 (J); mean ± SD; n = 9 mice/group. Number of experiments=3. K Primary mammary organoids isolated from 3 to 4 months old, estrus cycle matched, non-lactating BL/6C57 or BL/6NZB virgin female mice were grown for 6 days before induction with 1 µg/ml prolactin and 40 µg/ml oxytocin. Images show representative organoids at the indicated time. The scale bar represents 100 µm. n = 9 mice/group. Number of experiments=3. Source data are provided as a Source Data file. Mice used in all experiments were 2–3 months old at the beginning of the experiments.
To further validate the results, we created 3D lactation organoids using primary mammary glands from virgin females from both genotypes. In this established model35,36,37, organoids are first allowed to get established in culture and are then induced to mimic lactation by stimulation with prolactin and oxytocin. We confirmed that using this protocol, the lactation organoids in both genotypes produced milk (Supplementary Fig. 8). The organoids were harvested after 24 hours of stimulation for immunofluorescence and Western analysis of the same cell cycle markers. Western analysis of cell cycle markers showed that all markers are specifically up-regulated in the BL/6NZB organoids following induction with prolactin and oxytocin (Fig. 2C, D). Additionally, since the retinoblastoma was specifically up-regulated in the BL/6NZB females (Fig. 1I), we analyzed activation of Rb using immunofluorescence of phosphorylated Rb and confirmed it is up-regulated in BL/6NZB lactation organoids specifically (Fig. 2E, F).
To validate the muscle contraction and cytoskeleton signatures observed in the RNAseq data (Fig. 1I), we analyzed myoepithelial cells using K14 and SMA and found that in both cases, the staining is higher in the organoids of BL/6NZB females (Fig. 2G–J). Finally, to assess if this phenotype has any functional consequence on the production of milk, we performed live microscopy of the organoids over a time course following addition of prolactin. We found that consistent with the increased milk production found in vivo (Fig. 1K), the filling of the lumen of the BL/6NZB organoids appeared more pronounced after addition of prolactin (Fig. 2K, Suppl. video). Collectively, these results confirmed that the pathways detected by RNAseq can be observed in vivo and ex vivo in lactation organoids.
Lactation leads to an anti- versus pro-tumorigenic microenvironment in the BL/6C57 and BL/6NZB females respectively
Since the RNA seq analysis suggests an anti- and pro-tumorigenic mammary gland during lactation in BL/6C57 and BL/6NZB females respectively, we first tested if lactation impacts the growth of cancer cells. Tumors were established by injecting lactating females of both genotypes with the EO771 mouse mammary tumor cells at day 17 of lactation and tumors allowed to grow for 32 days (Fig. 3A). We found that tumors grew rapidly in the BL/6NZB females (Fig. 3B, C), however, in BL/6C57 females no tumors developed during this time frame (Fig. 3B, C). As a control, we also injected virgin mice of both genotypes and found that the growth rate of EO771 xenografts in virgin females, was identical (Supplementary Fig. 9) although slower than in BL/6NZB during lactation (Fig. 3B). This result suggests that the lactating environment of the BL/6NZB females accelerates the growth of these cells, while lactation in BL/6C57 females do not.
A Diagram of the experimental design in indicated female mice (n = 10 mice/group). SMART images of mice were obtained from Servier. B, C 1×105 EO771 cells were injected in inguinal mammary glands of 3-4 months old BL/6C57 or BL/6NZB mice on day 17 of lactation. Graph depicts the mean tumor volume over time (B). Quantification of tumor volumes at day 32 (C). Quantification at each time point represents the average of 10 BL/6C57 female mice relative to the average value of 10 BL/6NZB female mice. Student’s unpaired t test, two-tailed, ****p < 0.0001; mean ± SD; n = 10 mice/group. D Diagram of the experimental design in indicated genotypes (n = 10 mice/group). SMART images of mice were obtained from Servier. E, F H&E section of mammary glands from 5-6 months old, estrus cycle matched BL/6C57 or BL/6NZB female mice harvested after 45 days post completion of 21 days of lactation. Scale bar represents 200 µm. Graphs showing number of ducts or size of ducts analyzed using image J software. Student’s unpaired t test, two-tailed, *p < 0.05, ****p < 0.0001; mean ± SD; n = 10 mice/group. G Representative whole mount of mammary gland from 5-6 months old, estrus cycle matched BL/6C57 or BL/6NZB female mice harvested after 45 days post-partum after completion of 21 days of lactation. The scale bar represents 0.5 cm. Graph showing branching of ducts were analyzed using image J software. Student’s unpaired t test, one-tailed, ****p < 0.0001; mean ± SD; n = 10 mice/group. H Representative image of IHC of Ki67 in mammary glands derived from 5-6 months old, estrus cycle matched BL/6C57 or BL/6NZB mice harvested after 45 days post-partum after completion of 21 days of lactation. The scale bar represents 200 µm. Graph showing number of Ki67 stained cells were analyzed using image J software. Student’s unpaired t test, two-tailed, ****p < 0.0001; mean ± SD; n = 10 mice/group. I Schematic of patient’s selection and pathways identified in the human study comparing post-partum breast cancer (PPBC) to never pregnant breast cancer (NPBC). SMART images of women were obtained from Servier J Heatmap of the 14 transcription factors of the post-partum breast cancer (PPBC) regulon at day 21 of lactation in 3–4 months old BL/6C57 and BL/6NZB female mice. Source data are provided as a Source Data file. Mice used in all experiments were 2-3 months old at the beginning of the experiments.
Since EO771 are established cancer cells, in order to test the susceptibility of both genotypes to develop de novo mammary tumors upon oncogenic activation, we performed a second analysis. PAPP-A is a secreted protease that acts as an oncogene by stimulating proliferation via the activation of insulin-like growth factor (IGF) signaling and promoting immune evasion38,39,40,41,42,43. We previously showed that injection of PAPP-A in the mammary gland promotes proliferation44. We injected PAPP-A at day 17 of lactation in both genotypes and harvested the mammary gland 30 days post-lactation. Proliferation was assessed using histology and whole mount of the mammary glands. We found that injection of PAPP-A had no significant effect on proliferation in BL/6C57 females (Supplementary Fig. 10C, D, G, H, I, L), while H&E showed increased proliferation and tumor formation was observed by whole mount in BL/6NZB females (Supplementary Fig. 10E, F, G, J, K, L). This observation is consistent with the notion that in the context of mammary ductal hyperplasia, BL/6NZB females are more likely to be susceptible to develop tumors upon exposure to oncogenic signal such as PAPP-A.
Since however, the pathways related to regulation of cell cycle and p53 signaling were identified during lactation in normal mammary glands, we also tested if these pathways affect the growth of endogenous epithelial cells of the mammary gland. Therefore, we allowed females from both genotypes to lactate for 21 days, separated the mothers from their pups and harvested the mammary glands of the mothers 45 days after weaning (Fig. 3D). We then performed three complementary analyses; 1) histology by H&E staining, 2) whole mount analysis and, 3) staining for Ki67, as a marker of proliferation. We found that the number (Fig. 3E) and size (Fig. 3F) of the ducts are larger in the BL/6NZB females in 45 days post-partum mammary glands compared to BL/6C57 females. Combined these analyses indicate ductal hyperplasia. This observation was confirmed by whole mount analysis, which showed hyper-proliferation in the mammary gland at day 45 post-lactation in the BL/6NZB females (Fig. 3G) and the increased proliferation was confirmed using ki67 staining (Fig. 3H). To investigate the possibility that the difference in hyperplasia maybe observed at baseline in virgin mice due to differences in estrous cycle, we analyzed the mammary glands of virgin mice in both genotypes at all stages of the estrous cycle. No difference in proliferation was observed between strains (Supplementary Fig. 11). In human, ductal hyperplasia or ADH is associated with a 5 fold increased risk of breast cancer in45,46,47,48,49.
We concluded that mitochondrial genetics impacts the physiological response to lactation, where lactation promotes the formation of an anti-tumorigenic post-partum mammary gland in the BL/6C57 females. However, a pro-tumorigenic environment is observed after lactation in the BL/6NZB females raising the possibility that BL/6NZB females may represent a model of susceptibility to the development of post-partum breast cancers (PPBC).
Recently, a genetic signature that distinguishes PPBC from matched breast cancer in never pregnant women was identified34. In this study, the cohort consisted of 16 estrogen receptor alpha (ERα) positive breast cancer patients that were matched in terms of age, tumor grade and treatments and were divided into 9 patients with an history of pregnancy and 7 patients who never had children (Fig. 3I). RNAseq analyses of these tumors revealed a PPBC profile consisting of high cell cycle score, loss of p53 and elevated expression of a transcriptional regulon containing 14 transcription factors34. Since we already found elevated cell cycle markers and low p53 expression in BL/6NZB females during lactation (Figs. 1 and 2), to further explore the similarity between PPBC in humans and the mammary glands of BL/6NZB lactating females, we investigated the expression of individual transcription factors that defined the PPBC-regulon in humans in BL/6NZB lactating females compared to BL/6C57 females. We found that the PPBC transcriptional regulon is enriched in BL/6NZB lactating females compared to BL/6C57 females (Fig. 3J). These results therefore indicate that all aspects of the PPBC signature (cell cycle, p53 and regulon) are recapitulated in BL/6NZB females during lactation.
Single cell RNAseq identifies a high-cell cycle score and high- regulon positive sub-population that uniquely expands during lactation in BL/6NZB females
Based on the findings of the overlap between the PPBC signature and lactation in BL/6NZB females, we hypothesized that a PPBC-like population of cells may arise during lactation in these female mice. To test this hypothesis, we performed single cell RNAseq on mammary glands of 3 non-lactating and 3 lactating females (day 21 of lactation) from both genotypes.
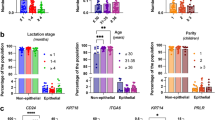
Since our goal is the identification of a potential cell population at the origin of PPBC, which are composed, like all other breast cancers, mainly of epithelial cells, we focused on the epithelial compartment. In the mammary gland, the epithelial compartment is composed on three main cell types; the luminal alveolar cells (AV), the luminal hormone sensing cells (HS), which are characterized by the expression of the estrogen receptor alpha (ERα) and the myoepithelial cells (ME) (Fig. 4A). The relative distribution of these sub-types of cells confirmed an expansion of the alveolar cells during lactation (Fig. 4B) but no significant differences were observed between genotypes (Fig. 4B). Since the PPBC signature in humans was identified in a cohort of ERα positive breast cancers, where the vast majority of cells are of the HS sub-type, we focused our analysis on the luminal HS cellular sub-type. We performed hierarchical clustering using clustergrammer as previously described50 and found that at baseline, in virgin females, the luminal HS cells did not significantly differ between genotypes (Fig. 4C). However, the transcriptional profiles of luminal HS cells during lactation resulted in a clear hierarchical clustering by genotypes (Fig. 4D) indicating that lactation differentially modulates gene expression in these cells in BL/6C57 and BL/6NZB females.
Mammary glands from 3 to 4 months old, estrus cycle matched non-lactating (NL) female virgin mice and 3–4 months old day 21 lactating (D21) female mice were subjected to Single cell RNASeq (n = 3 mice/group). A UMAP plot of Luminal Alveolar (Luminal AV), Hormone sensing (HS) and myoepithelial cell clusters identified by expression of characteristic markers. B Relative proportion of epithelial cell types per genotype in non-lactating (NL) and day 21 lactating (D21) female mice. C Hierarchical clustering heatmaps of differential gene expression of luminal HS cells in non-lactating (NL) BL/6C57 and BL/6NZB. D Hierarchical clustering heatmaps of differential gene expression of luminal HS cells in lactating BL/6C57 and BL/6NZB females. E Number of PPBC-like cells (defined by high cell cycle score and high regulon positivity, see text) in non-lactating (NL) and lactating day 21 (D21) BL/6C57 and BL/6NZB females. Student’s unpaired t test, one-tailed, *p < 0.0253 NL vs D21 in BL/6NZB females; Student’s unpaired t test, two-tailed, *p < 0.0458 D21 of BL/6C57 vs D21 of BL/6NZB females; mean ± SD; n = 3 mice/group. F Representative image of PPBC-like positive cell cluster. G Number of luminal HS cells expressing p53 in non-lactating (NL) and lactating day 21 (D21) BL/6C57 and BL/6NZB females. Ordinary one-way ANOVA, **p < 0.0014 NL vs D21 in BL/6NZB females; *p < 0.0111 D21 of BL/6C57 vs D21 of BL/6NZB females; mean ± SD; n = 3 mice/group H List of the top 40 differentially up-regulated genes in luminal HS cells in BL/6C57 relative to BL/6NZB female mice at day 21 of lactation. Bold indicated p53 transcriptional targets. I Pathways related to the expression of the 40 genes shown in H. Databases where these pathways were identified are color coded. Source data are provided as a Source Data file. Mice used in all experiments were 2-3 months old at the beginning of the experiments.
To test our hypothesis that a PPBC-like population of cells may be present in the BL/6NZB females, we then interrogate our data set to identify the number of cells that express both a high cell cycle score, as defined in the human study, and are also positive for the transcription PPBC-regulon34. Therefore, we define PPBC-like cells as those with high cell cycle score and high PPBC-regulon, that is to say, cells that have expression levels ≥50th percentile of the feature average for both cell cycle markers (Cenpe, Ccna2, Ccnb2, Mcm6, Ccnf, Bub, Cdc20, Cdc6, Cdk, Plk1) and regulon transcription factors (Bclaf1, Cux1, E2f1, E2f4, Esr1, Foxm1, Gtf2b, Max, Myc, Nfya, Nr4a1, Nrf1, Smarca4, Taf1) (Fig. 3J). We found that while such cells were identified in all conditions, the number of PPBC-like cells is significantly higher in BL/6NZB during lactation (Fig. 4E, F). To evaluate the specificity of the PPBC-like signature, we analyzed the expression of this feature list in each sub-type of epithelial cells of the mammary gland. We found that not surprisingly it is enriched in the HS cells sub-population relative to the ME and AV cells and further, it is more enriched in BL/6NZB females compared to BL/6C57 females (Supplementary Fig. 12A, B).
The observation that PPBC-like cells are elevated specifically during lactation in BL/6NZB females indicates that an event that is unique to these females during lactation allows their proliferation. As we already identified that down-regulation of p53 expression is specific to the lactating BL/6NZB females (Figs. 1 and 2) and loss of p53 characterized PPBC in humans34, we next interrogate our data set for the number of luminal HS cells that are positive for p53. We found that the number of p53 positive luminal HS cells is significantly lower in the BL/6NZB females relative to lactating BL/6C57 or virgin females from both genotypes (Fig. 4G).
We established that the gene expression profiles of luminal HS cells differ between genotypes during lactation (Fig. 4D). In an effort to identify critical differences in gene expression of luminal HS cells between the genotypes, we selected the top 40 (p < 0.0002) (Fig. 4H) differentially expressed genes in luminal HS cells during lactation from BL/6C57 and BL/6NZB females and subjected them to pathway analysis. These 40 genes were up-regulated in BL/6C57 lactating females relative to BL/6NZB lactating females. Transcription factor protein-protein interaction (PPI) analysis of these genes revealed p53 among the top transcription factor profiles differentially regulated between genotypes (Fig. 4I), Pathways related to ER-stress and apoptosis were found to be elevated in luminal HS cells in BL/6C57 females during lactation (Fig. 4I).
Collectively, these results support the conclusion that a PPBC-like sub-population of cells expands during lactation, specifically in BL/6NZB females and that this expansion correlates with lack of expression of p53.
Mitochondria impacts lactation by leading to differential activation of organellar stress responses
While the regulation of the tumor suppressor p53 under stress conditions such as irradiation, has been extensively studied, the regulation of p53 under physiological settings has also been well described51,52,53. Notably, ER-stress and metabolism have been shown to affect p5351,53,54,55,56,57. Considering that we observed down-regulation of p53 expression during lactation in the BL/6NZB females (Fig. 2A) and lactation engages robust secretion that alters both ER-stress and mitochondria58,59, we tested whether the type of lactation between genotypes also results in differential activation of stress pathways in the ER and mitochondria using samples from the mammary glands at day, 8, 14 and 21 of lactation in both BL/6C57 and BL/6NZB females. We used CHOP, BIP, PERK, phospho-PERK and XBP-1s as surrogate markers of ER-stress. We found that relative to not lactating (NL) mammary glands, the levels of all markers of the UPRER were significantly elevated at day 8 of lactation in both genotypes except CHOP in the BL/6NZB females (Fig. 5A, B). Further, while the activation of ER-stress remained constant throughout the time course in the BL/6C57 females (Fig. 5A, B), an abrupt cessation of the pathway was observed at day 21 of lactation in the BL/6NZB mice (Fig. 5A, B). To directly compare the level of activation between genotypes, samples isolated on the same day of lactation in both BL/6C57 and BL/6NZB were run side by side on the same gel. This analysis confirmed that the activation of CHOP is lower in BL/6NZB relative to BL/6C57 females at all time points. Further, while all markers remain elevated at day 21 of lactation in BL/6C57 mice, they were all significantly reduced at day 21 in the BL/6NZB females (Supplementary Fig. 13A, B). We concluded that overall, lactation activates the ER-stress in both strains, however, the activation is sustained in BL/6C57, while it is transient in the BL/6NZB females.
Western blot analysis of mammary glands from 3-4 months old, estrus cycle matched virgin non-lactating (NL) and 3-4 months old lactating (L) at day 8, 14 and 21 in BL/6C57 (Left) or BL/6NZB (right) female mice (n = 9 mice/group) for the indicated markers of ER-stress (A),mitochondrial-stress (C) and apoptosis (E). Representative westerns show the result of 3 (out of 9) individual BL/6C57 or BL/6NZB mice at each time point compared to their non lactating counterpart. Quantification of ER-stress markers (B), mitochondrial-stress markers (D) and apoptosis markers (F). Student’s unpaired t test, two-tailed, *p < 0.05, **p < 0.01, ***p < 0.001; mean ± SD. A detailed p value has been provided in the Source data as a Source Data file. Mice used in all experiments were 2-3 months old at the beginning of the experiments. The representative blot shows 3 individual mice per group. In panels a to f, data are representative of two independent experiments.
Since the ER is close contact with the mitochondria, we also analyzed mitochondrial stress response (UPRmt) over the same time course of lactation. For the UPRmt, we used ATF4, ATF5, Hsp60 and ClpP as surrogate markers of the pathway and found that as observed for the ER-stress markers, the UPRmt is also activated at day 8 of lactation relative to the non- lactating mammary gland (Fig. 5C, D) and while only subtle difference in the activation of ER stress was observed between genotypes at day 8 (Supplementary Fig. 13) and no difference was observed at day 14 of lactation, at day 21 all markers were significantly decreased in the BL/6NZB females (Fig. 5C, D). Since the same pattern of abrupt cessation of UPR signaling is observed for both the ER and the mitochondria, we reasoned that one trivial explanation could be that the BL/6NZB females either stopped lactating early or the volume of milk they produce is reduced between day 14 and 21 of lactation. However, we found that the volume of milk produced by the BL/6NZB females is in fact larger than the BL/6C57 females at all time points (Fig. 1K), a point that will be discussed in detailed. This observation rules out that the cessation of ER and mitochondrial stress pathways at day 21 in the BL/6NZB females is due to the cessation of lactation. Alternatively, we reasoned that since we observed that BL/6NZB females have more pups per litter (avg. 8 pups/litter) than BL/6C57 females (avg. 5 pups/litter), another explanation for these differences maybe the number of pups between genotypes. To test this possibility, we adjusted the number of pups to 5 for BL/6NZB females to match the number of pups in the BL/6C57 females and compare the markers of the ER and mitochondrial stress pathways as well as the markers of the cell cycle. We found no differences in the levels of the markers (Supplementary Fig. 14) Therefore, we concluded that stress responses in the ER and mitochondria are activated by lactation but the activation is sustained in the BL/6C57 females, while it is transient in the BL/6NZB females.
Activation of ER-stress can be either pro-apoptotic or anti-apoptotic depending on whether the activation of the pathway is sustained or transient respectively60,61,62,63. We therefore also monitored markers of apoptosis. We found that apoptosis is activated in BL/6C57 females but not in BL/6NZB females during lactation (Fig. 5E–F, Supplementary Fig. 13E, F). While apoptosis is observed during the declining phase of lactation64,65, apoptosis has been mainly associated with post-lactation involution66,67. Therefore, to compare the level of apoptosis between lactation and involution between strains, we isolated primary epithelial cells (PMEC) during lactation and involution and analyzed the expression of cleaved caspase 3. We found that apoptosis is detected during lactation in BL/6C57 females, although as expected at a much lower level than during involution, while no apoptosis was detected during lactation in BL/6NZB females (Supplementary Fig. 15).
Collectively, these results indicate that a pro-apoptotic ER-stress response is activated in BL/6C57 females during lactation, while a suppression of pro-apoptotic ER-stress is observed in BL/6NZB females.
Treatment with the pro-apoptotic ER-stress inducer δ-Tocotrienol, reverses the tumorigenic effect of lactation in BL/6NZB females
Since inhibition of the cell cycle was among the first pathway discovered downstream of the activation of ER-stress68,69,70 and a large number of studies has linked the regulation of ER-stress to p5351,53,54,55,56,57, our results suggest a possible link between a pro-apoptotic ER-stress and repression of the PPBC signature. This observation raises the possibility that drugs able to activate pro-apoptotic ER-stress in BL/6NZB females may lead to activation of p53 and apoptosis and would reverse the pro-tumorigenic effect of lactation. Conversely, inhibition of pro-apoptotic ER-stress in BL/6C57 females may inhibit p53 and apoptosis and activate the PPBC-signature. To test these possibilities, we first treated BL/6NZB females with δ-Tocotrienol, a natural product that is reported to induce pro-apoptotic ER-stress71,72,73.
We found that lactation organoids recapitulate the signatures found in vivo (Fig. 2C–K). We also found that these organoids support differential handling of mitochondrial stress, as BL/6NZB derived organoids show more mitochondrial ROS (Supplementary Fig. 16), which is consistent with the cessation of the UPRmt since this pathway regulates the expression of the anti-oxidant enzyme SOD274,75,76. We therefore used lactation organoids derived from BL/6NZB females to test the effect of δ-Tocotrienol. We found that upon induction with prolactin, treatment with δ-Tocotrienol led to the induction of CHOP, ATF4, BIP and caspase 3 (Fig. 6A). Further, this analysis confirmed that induction of the cell cycle markers is only observed after addition of prolactin (Fig. 6A) but that treatment with δ-Tocotrienol, abolished the expression of cell cycle markers (Fig. 6A) and led to the expression of p53 (Fig. 6A).
A Organoids of mammary glands from 3-4 months old, estrus cycle matched, non-lactating BL/6NZB virgin female mice were treated with 20 µg/ml δ-tocotrienol starting from the 4th day followed by induction with 1 µg/ml prolactin, 1 µg/ml hydrocortisone and 40 µg/ml oxytocin on the 6th day. 22 hr post induction, the organoids were subjected to western blot analysis for the indicated markers. Graphs show the quantification of western blot analysis. Student’s unpaired t test, two-tailed, **p < 0.01; **p < 0.01, ***p < 0.001, ****p < 0.0001; mean ± SD; n = 9 mice/group. Number of experiments=3. The detailed p values are provided in the source data file. B Organoids of mammary glands from 3-4 months old, estrus cycle matched, non-lactating BL/6C57 virgin female mice were treated with 30 µM Salubrinal from the 4th day, then induced with 1 µg/ml prolactin, 1 µg/ml hydrocortisone and 40 µg/ml oxytocin on the 6th day. 22 hr post induction, the organoids were subjected to western blot analysis for the indicated markers. Graphs show the quantification of western blot analysis. Student’s unpaired t test, two-tailed,* **p < 0.001, ****p < 0.0001; mean ± SD; n = 9 mice/group. Number of experiments=3. A detailed p value is provided in the source data file (C–H), Mammary glands from 5-6 months old, estrus cycle matched BL/6NZB mice collected 45 days post completion of 21 days of lactation treated with either vehicle (olive oil) or δ-tocotrienol (200 mg/kg body weight in olive oil, twice daily) by oral gavage starting from 10th day of lactation for 4 weeks (n = 10 mice/Group). C, Schematic representation of experimental design. SMART images of mice were obtained from Servier. D, E H&E section of mammary glands from BL/6NZB mice without (Vehicle) or with δ-tocotrienol. Scale bar represents 200 µm. Images were taken with a 20X (D) and 40X (E) objectives respectively. Graphs showing number of ducts or size of ducts analyzed using image J software. Student’s t test, two-tailed, ****p < 0.0001; mean ± SD; n = 10 mice/group. F Representative whole mount of mammary gland from above mentioned mice. The scale bar represents 0.5 cm. Graph showing thickness of ducts were analyzed using image J software. Student’s t test, two-tailed, ****p < 0.0001; mean ± SD; n = 10 mice/group. G Representative image of IHC of Ki67 in mammary glands derived from above mentioned mice. The scale bar represents 200 µm. Graph showing number of Ki67 stained cells were analyzed using image J software. Student’s t test, two-tailed, ****p < 0.0001; mean ± SD; n = 10 mice/group. H Western blot analysis of ER Stress markers and cell cycle markers were performed in mammary glands of above-mentioned BL/6NZB mice treated without (Vehicle) or with δ-tocotrienol. Graphs of quantification of western blot analysis. Student’s t test, two-tailed, ***p < 0.0006, ****p < 0.0001; mean ± SD; n = 10 mice/group. The representative blot shows 5 individual mice per group. In panel h, data are representative of two independent experiments. I Diagram of the experimental design in each group, n = 10 mice/group. SMART images of mice were obtained from Servier. J, K BL/6NZB mice were treated with vehicle (olive oil) or δ- tocotrienol (200 mg/kg body weight in olive oil, twice daily) by oral gavage starting from 10th day of lactation and 1×105 EO771 cells were injected in inguinal mammary glands of mice on day 17 of lactation. The mice were harvested 32 days post-partum after completion of 21 days of lactation. J Graph of mean tumor volumes per mice at the given time points. Quantification at each time point represents the average of 10 BL/6NZB mice treated with vehicle relative to the average value of 10 BL/6NZB mice treated with δ-tocotrienol. K Graph of tumor volumes per gland on the day of sacrifice (day 32 post implantation). Student’s t test, two-tailed, ****p < 0.0001; mean ± SD; n = 10 mice/group. Source data are provided as a Source Data file. Mice used in all experiments were 2-3 months old at the beginning of the experiments.
We then performed the reverse experiment and forced the inhibition of ER-stress in lactation organoids derived from BL/6 C57 females. First, we saw that as observed in vivo, the markers of ER-stress and caspase 3 are activated upon induction with prolactin in BL/6C57 organoids (Fig. 6B). Treatment with salubrinal, which lowers ER-stress, reduced the expression of ER-stress markers and activation of caspase 3, while it activated cell cycle markers and reduced the expression of p53 to the level observed in BL/6NZB organoids (Fig. 6B). Collectively, these experiments establish the causality between ER-stress and expression of p53, apoptosis and cell cycle markers.
Since this result suggests that δ-Tocotrienol reverses the pro-tumorigenic signature observed in BL/6NZB females during lactation, we next tested if this also holds true in vivo. Since the inactivation of ER-stress signaling occurs sometimes between day 14 and day 21 of lactation in BL/6NZB females in order to mimic sustained activation of ER-stress, were initiated treatment with δ-Tocotrienol on day 10 of lactation by daily gavage for four weeks (Fig. 6C). First, treated and untreated females were analyzed for 45 days post-partum hyperplasia. We found that treatment with δ-Tocotrienol reduces the number (Fig. 6D) and size (Fig. 6E) of the mammary ducts. Further, analysis of the entire mammary gland by whole mount confirmed a marked reduction in proliferation of the mammary glands upon treatment with δ-Tocotrienol (Fig. 6F), which was confirmed using staining with Ki67 (Fig. 6G). Further, we tested the effect of δ-Tocotrienol in vivo at day 45 post-partum and found that this treatment leads to activation of all markers of ER-stress (Fig. 6H), activation of p53 (Fig. 6H) and inhibition of the cell cycle markers (Fig. 6H).
To test the effect of δ-Tocotrienol on the growth of exogenous cancer cells, BL/6NZB females were treated for four weeks by daily gavage starting at day 10 of lactation and EO771 cancer cells were injected in the mammary fat pad on day 17 of lactation (Fig. 6I) and tumor volumes were measured every 4 days. We found that δ-Tocotrienol significantly reduced tumor growth (Fig. 6I, J). However, since δ-Tocotrienol is known to have pro-apoptotic action, its effect may be mediated directly on the EO771 cells independently of lactation. To test this possibility, we repeated the experiment in virgin BL/6NZB females. We found that as expected δ-Tocotrienol did reduce tumor growth (Supplementary Fig. 17), however it did so to a much lower level and over a longer time course compared to what is observed in lactating females (Fig. 6J). Additionally, we tested if treatment of BL/6C57 females during lactation with salubrinal in vivo promotes tumor formation. Consistent with the effect observed in lactation organoids, we found that treatment with salubrinal promotes tumor formation (Supplementary Fig. 18).
We conclude that δ-Tocotrienol can reverse not only the pro-tumorigenic signature observed in BL/6NZB females during lactation but also the post-partum hyperplasia and tumor formation in vivo.
Discussion
Lactation is an ancient reproductive feature that pre-dates the origin of mammals. It originates from apocrine-like gland associated with hair follicles to provide moisture, nutrients and antimicrobials to shelled eggs. The evolution of placenta-based reproduction displaced the function of milk as a source of water and nutrients to eggs, to the secretion of complex milk. The nutrition provided by mammary gland-derived milk is essential to the survival of off springs across the mammalian kingdom. In humans, extensive studies have been performed to dissect the benefits of breast milk to newborns, but the benefits of breastfeeding to the mothers have received much less attention. While lactation is generally found to be protective against breast cancer13,14,15,16,17, 15-17% of breast cancer patients had breastfed18,19,20, indicating that while breastfeeding is overall protective, it is not protective for all women and this sub-set of women are at higher risk of developing PPBC. This observation raises two fundamental questions; first, what are the differences between women that benefit from the protective effect of lactation and those who do not and second, what is the mechanism of protection against breast cancer? Our study suggests that investigations into high risk and low risk individuals should compare the mitochondria-nucleus communication to determine whether the same mechanisms investigated in the study also apply.
Firstly, our study reveals unexpected differences in lactation at the levels of organelle network reorganization, global transcriptional dynamic in term of total number of genes over time, cell-type specific transcriptome and signaling pathways activated or repressed during lactation. These differences however do not impact the nutritional function of lactation since females from both genotypes produce milk and successfully provide nutrients to their off springs, as no difference in the weight of pups is observed. Since the only difference between genotypes is the mitochondria, these observations indicate that the impact of mitochondrial haplotypes extends well beyond the mitochondrial itself and affects the communication with the ER and transcription in the nucleus. However, sporadic mutations in mitochondrial DNA can occur and lead to differences in sequence between different copies of mtDNA, referred as heteroplasmy77,78. While the rate of heteroplasmy is limited by DNA repair and other mechanisms, we cannot rule out the possibility that it also contributes to the phenotype described in our model, considering that mitochondrial haplotypes have shown to impact the heteroplasmic fraction78,79,80.
Our study highlights lactation as a physiological setting where this inter-organelle triad of communication alters the underlying biology. Our study challenges the widely preconceived notion that lactation is a shared and uniform process in mice as well as in women. Rather, our data indicate that in mice, multiple types of lactation may exist at the cellular levels and raises the possibility that lactation may also vary between women based on how the communication between their mitochondria, ER and nucleus is established.
Second, we show that the divergence of signaling pathways induced or repressed during lactation between genotypes result in anti- or pro-tumorigenic environment and that pro-tumorigenic lactation shares the genetic signature that was identified in post-partum breast cancers in humans34. Our results support the notion that difference in ER and mitochondrial stress responses during lactation is the mechanism of these divergent outcomes by affecting transcription via the large number of transcription factors involved in these stress responses. The impact of the activation or suppression of pro- apoptotic response to ER-stress is supported by the observation that treatment with δ-Tocotrienol, a dietary supplement known to induce a pro-apoptotic ER-stress71,72,73 was able to reverse the outcome of lactation ex-vivo and in vivo in BL/6NZB females, while lowering ER-stress using salubrial, induced a pro-apoptotic ER-stress response, inhibited p53 and activated the cell cycle in BL/6C57 lactation organoids.
The mechanism by which ER stress transits from anti-apoptotic and pro-survival to an pro-apoptotic response is still debated but the level of CHOP has been involved81,82,83. This data is in agreement with our observation that CHOP is reduced in BL/6NZB females. Additionally, inhibition of the cell cycle was among the first pathway discovered downstream of the activation of ER-stress68,69,70. However, the UPR is a complex pathway involving several arms driven by PERK, ATF6 and IRE184. The precise contribution of each of these arms during lactation remains to be determined as well as the nature of the cell type specific response to ER stress during lactation84. The differential activation of the UPR presented in the current study offers the potential to dissect these pending questions in the physiological setting of lactation.
As for p53, a large number of studies has established a link the regulation of ER-stress and metabolism to p5351,53,54,55,56,57. The mechanistic details of how these pathways affect p53 under normal physiological conditions remain to be characterized but our results offer an additional example. Our hypothesis is that the down-regulation of p53 expression during lactation in BL/6NZB females results from the difference in ER-stress and inadvertently allows the expansion of the PPBC-like sub-population of cells, which are characterized by the elevated cell cycle score and expression of the PPBC-regulon. Therefore, despite the fact that a small number of PPBC-like cells are detected in virgin mammary glands in both genotypes and also in lactating BL/6C57 females, our data raises the possibility that it is the specific down-regulation of p53 expression during lactation in BL/6NZB females that allows their expansion. We postulate that these cells are primed for transformation such that upon acquisition of a sporadic oncogenic mutation, they lead to the formation of PPBC, therefore explaining their genetic overlap with the PPBC signature in humans. However, given that the induction of the ER-stress has been described in multiple cell types and is known to impact the biology of several immune cells85,86,87, it is likely that multiple cell types contribute to the outcome of lactation in vivo. This possibility is supported by the observation that the growth of exogenously injected cancer cells from an entirely different genetic makeup, is also stimulated during lactation in BL/6NZB females, indicating that other cell types, beyond the PPBC-like sub-population, also contribute to the pro-tumorigenic environment in these females. Additionally, since the human data was generated using bulk RNAseq, it is likely that other cell types in the microenvironment contribute to the development of PPBC and the resulting signature.
Since our mouse models recapitulate the natural diversity in mitochondrial genetics observed in the human population and mitochondrial are inherited by our mothers, we propose that the triad of communication between the mitochondria, ER and the nucleus is influenced by maternal ancestry. Based on our data in mice, we propose the hypothesis that in women where lactation is protective, this triad of communication may be recapitulated by the BL/6C57 females where pro-apoptotic ER-stress, increased p53 expression and decreased cell cycle genes are expressed. In the context of such environment, upon acquisition of an oncogenic mutation, these women would be at low risk of breast cancer. Conversely, in the 15–17% women where lactation is not protective18,19,20, the mitochondria, ER, nucleus communication triad may be recapitulated by the BL/6NZB females where anti-apoptotic ER-stress is induced, p53 is down-regulated, the cell cycle genes are expressed and a sub-population of PPBC like cells are present. In the context of such environment upon acquisition of an oncogenic mutation, we hypothesize that these women would be at high risk of breast cancer. Of note, organoids derived from the NZBNZB females exhibit the same signature as the BL/6NZB (Supplementary Fig. 19), supporting the notion that maternal ancestry does play a major role in establishing this signature. This finding is also in line with the observation that BL/6NZB and NZB/NZB females show the same metabolic activities33.
It is also intriguing that excessive milk production was reported to be associated with early diagnose of breast cancer21. Based on our findings that BL/6NZB females produce more milk and show cessation of ER-stress, these observations suggest that excessive milk production may be an indication of a failure of the ER-stress checkpoint, which normally acts to inhibit secretion and activate protein degradation through ERAD88,89,90. More studies are necessary to test this possibility.
Collectively therefore our results bring together the wealth of literature linking ER-stress to the regulation of the cell cycle and p53 and the pivotal role of the duration of the stress into whether the outcome will be the pro- versus anti-apoptotic in the physiological setting of lactation.
Lastly, our findings suggest a potential dietary intervention to reverse the pro-tumorigenic response to lactation into an anti-tumorigenic response.
Methods
Animals
All animal experiments were performed in accordance with Mount Sinai IACUC approved protocol. BL/6NZB mice were originally generated and provided by our collaborator Dr. J.A Enriquez33. The BL/6C57 (C57BL/6 J) and NZBNZB (NZB/OlaHsd) strains were purchased from ENVIGO, Netherlands33. Female virgin mice aged between 2-3 months old were used at the beginning of each experiments. Animal facilities at Sinai are using 12 hours light/12 hours dark cycle, constant temperature of 72 F (22.2 C) and 30-70% humidity. After their 1st pregnancy, mammary glands were collected from lactating female mice at 8, 10, 14 and 21 days of lactation. For Non lactating control, age matched, estrus cycle matched (Estrus stage) female virgin mice were used. For all experiments, the stage of Estrus cycle of non lactating mice was predetermined. For collection of cells from the vaginal canal, approximately 50 µl of PBS was used to collect cells through 2-3 vaginal lavages. PBS containing vaginal cells from each mouse is then placed evenly on 3 slides in a thin layer (smear) and allowed to air dry. The slides were then stained by Giemsa Stain Modified (Sigma-Aldrich, GS500-500ml) for 2-3 minutes and then wash with water. Slides were analyzed under the microscope to identify the stage of Estrus cycle91. For the analysis of post-partum samples, mammary glands were collected from age matched, estrus cycle matched (Estrus stage) mice after 5 days and 45 days post weaning (21 days). Inguinal mammary glands were frozen on dry ice and kept for RNA seq (Bulk and Single Cell) and western Blot analysis. Thoracic mammary glands were collected for histology. For some experiments, the inguinal mammary glands were used for whole mounts. For each experiment the age of the mice of both genotypes as well as their non lactating controls were matched. EO771 murine mammary cancer cell line originally isolated from spontaneous mammary tumor in BL6/C57 mice was used to generate tumors in mammary glands in the mice following an earlier published protocol with slight modifications92. 1 × 105 EO771 cells (CH3 Bio Systems, cat #94A001) suspended in 25 µl Hank’s balanced salt solution (HBSS) and 25 µl growth factor reduced matrigel (GIBCO) was injected in both the inguinal fat pads of estrus cycle matched (Estrus stage) 8 non lactating BL/6C57 virgin mice and 8 non lactating BL/6NZB virgin mice. Authentication and mycoplasma testing of EO771 cells were provided by CH3 Bio Systems. EO771 cells were also injected in the fat pads of 10 BL/6C57 lactating mice and 10 BL/6NZB lactating mice on 17th day of lactation. Tumors volumes were measured 3 times per week using electronic calipers. The greatest longitudinal diameter (length) and the greatest transverse diameter (width) were measured, and tumor volumes were calculated by the modified ellipsoidal formula: volume=1/2(length × width2). Tumor volumes did not exceed the maximum allowed by our protocol (500mm3). (To determine the effect of δ-Tocotrienol on EO771 xenografts, 20 lactating BL/6NZB mice were given either olive oil (Vehicle) or δ-Tocotrienol (200 mg/kg in olive oil, twice daily) by oral gavage, 5 days/week from the 10th day of lactation for 4 weeks93. Mice were injected with EO771 cells in both the inguinal mammary glands on 17th day of lactation, 7 days post treatment with olive oil (Vehicle) or δ-Tocotrienol. Tumor volumes were measured in mice with or without δ-Tocotrienol treatment. To determine the effect of δ-Tocotrienol on hyperplasia, 20 lactating BL/6NZB mice were given either olive oil (Vehicle) or δ-Tocotrienol (200 mg/kg in olive oil, twice daily) by oral gavage, 5 days/week from the 10th day of lactation for 6 weeks. 45 days post weaning, the mammary glands were collected from these mice. To find out the effect of δ-Tocotrienol on EO771 cells generated tumors in non lactating virgin mice, 20 estrus cycle matched (estrus stage) BL/6NZB virgin mice were given either olive oil (Vehicle) or δ-Tocotrienol (200 mg/kg in olive oil, twice daily) by oral gavage, 5 days/week for 80 days. 7 days after starting treatment, these mice were injected with EO771 cells. These mice were harvested after 80 days. To decipher the effect of Salubrinal (Sigma-Aldrich SML0951) on EO771 xenografts, 20 lactating BL/6C57 mice were given either vehicle (1% DMSO in PBS) or Salubrinal (1 mg/kg, once daily) by intra-peritoneal (i.p) injections, 5 days/week from the 10th day of lactation for 4 weeks. Salubrinal was first solubilized in DMSO to 100 mg/kg and diluted in PBS to its final concentration. These mice were then injected with EO771 cells in inguinal fat pads on 17th day of lactation, 7 days post treatment with or without Salubrinal according to the published protocol94. For the experiment related to litter size, the number of pups of BL/6NZB (average number of pups is 8) mice were adjusted to that of BL/6C57 mice (average number of pups is 5) and the mammary glands were extracted from these BL/6NZB mice on day 21 of lactation and used for western blot analysis. All mice were housed in vivarium at Mount Sinai with ad libitium access to food and water.
We confirm that the maximal tumor size/burden permitted by Sinai IACUC was not exceeded (500mm3). Mice were euthanized using compressed CO2. All SMART images of mice are licensed under Creative Commons Attribution 4.0 (CC BY 4.0).
Milk collection
Milk was collected according to published protocol95. Females were separated from their pups 2 hours before milk collection. Prior to milk collection, 2 IU/kg of oxytocin and an anesthetic mixture containing 80-100 mg/kg ketamine in combination with 5-10 mg/kg xylazine were administered intraperitoneally (IP). After sterilization of the area with a sterile alcohol pad, the milk was collected with a pipette tip by moving the nipples in circular motion.
PAPP-A production and injection
MCF-7PAPP-A cells were generated following our previously published paper44. MCF-7, MCF-7PAPPA cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin at 37 °C/5% CO2. MCF-7 and MCF-7PAPP-A cells were grown to 70%-80% confluence in complete media. The media was then replaced with serum-free media and the media was collected after 24 h, and PAPP-A protein was concentrated from media using 50 kDa cutoff Amicon Ultra-15 Centrifugal Filter Units (EMD Millipore, #UFC905024) spun at 5000 g for 20 min at 4 °C. Concentrated condition media were then quantified by ELISA and stored at − 20 °C. PAPP-A concentration was determined by Quantikine PAPP-A ELISA (R&D Systems, # DPPA00) following the manufacturer’s protocol. Primary incubation was given for 18 h at 4 °C and PAPPA conjugate incubation for 6 h at room temperature. 200 µl of PAPP-A concentrated media (0.1 ng/µl) or MCF7 concentrated media was injected into the inguinal mammary glands of 16 BL/6NZB mice for seven consecutive days starting from the 17th day of lactation.
Whole mounts
Inguinal mammary glands were removed from mice and partially airdried on standard microscope slides (Fisher Cat# 12–544-3) for 5–7 min. Then the glands were fixed overnight in 75% ETOH. 25% Glacial Acetic Acid. The slides with the glands were washed in 70% ethanol for 15 min before gradually changing to water. The glands were then stained with Carmine solution (1 g carmine (Sigma C1022) and 2.5 g aluminum potassium sulfate (SigmaA7167) in 500 ml dH2O) overnight followed by washing with MilliQ filtered water and dehydrated in an alcohol series. Finally, glands were immersed in xylene to clear them and then mounted in Permount (Fisher Cat # SP15-100).
Measurement of ductal branching was assessed using the Analyze Skeleton (2D/3D) (https://imagej.net/plugins/analyze-skeleton/) plugin in ImageJ to quantify images of whole Carmine alum-stained whole mount mammary glands. The #Junctions output from this analysis was used to determine the number of branches.
Histology and immunohistochemistry
Tissues were fixed in 10% formalin (Thermo Fisher Scientific) for 24 h and then processed and embedded into paraffin for sectioning by the Biorepository Core Facility at Mount Sinai. Unstained paraffin embedded slides were obtained from the core facility. The slides were stained with Hematoxylin and Eosin. Images were captured using a Zeiss AX10 microscope. For immunohistochemistry, sections were deparaffinized in xylene twice for 5 min each. Then they were rehydrated through a decreasing ethanol series (100%, 90%, 70%) gradually changing to water. Antigen retrieval was done for 30 min at 90-100 °C in 10 mM Sodium citrate buffer pH 6. The slides were slowly cooled down to room temperature, then rinsed twice in Tris buffered saline (TBS) followed by antibody staining according to the manufacturer’s suggested protocol for the ImmPRESS Excel Amplified Polymer Kit (Vectorlabs Cat# MP-7601 & Cat# MP-7602). Ki67 (LS-C141899-0.1 from LifeSpan Biosciences) was detected with diluted 1:100 for 2 h at room temperature in a humidified chamber. Keratin-8 (SAB4501653 from Sigma-Aldrich) and Smooth muscle Actin, SMA (ab124964 from Abcam) were detected with diluted 1:100 for overnight at room temperature in a humidified chamber.
Western blotting
Flash frozen mammary glands were pulverized and lysed in cold NP-40 lysis buffer with protease inhibitors (50 mM Tris, pH 7.5, 250 mM NaCl, 5 mM EDTA, 0.5% Nonidet P-40, 50 mM NaF, 0.2 mM Na3 VO4, 1 g/ml leupeptin, 1 g/ml pepstatin, 100 g/ml phenylmethylsulfonyl fluoride) for 10 min, sonicated for 5 s at 20% amplitude and then centrifuged at 14,000 rpm for 20 minutes at 4 °C and then the supernatant was collected. Protein concentrations were measured using the Bradford method (Bio-Rad Protein Assay). Equal amounts of protein (20-30 µg) were loaded and separated by SDS-PAGE electrophoresis and then transferred to nitrocellulose membrane (GE Healthcare) followed by 30 min blocking. The membranes were then probed with the following primary antibodies overnight at 4 °C: Hsp60 (BD Transduction 611563; 1:500), ATF5 (abcam ab184923, 1:500), ATF4 (Proteintech 60035-1-Ig; 1:500), ClpP (abcam ab124822; 1:500), Chop (Cell Signaling 2895S; 1:500), Bip (BD Transduction 610978, 1:500), Perk (Cell Signaling Technology C33E10; 1:200), pPerk (Thermo Scientific MA515033; 1:200), Parp-CL (Cell Signaling Technology 9546S; 1:500), Caspase 3-CL (Cell Signaling Technology 9661; 1:100), Xbp1s (Santa Cruz Technology sc-7160; 1:500), Cdk2 (Santa Cruz Technology sc-163; 1:200), Cdk4 (Santa Cruz Technology sc-260; 1:200), Cdk7 (Santa Cruz Biotechnology sc-529; 1:200), p53 (Calbiochem OP03, 1:100), cyclin D1 (Santa Cruz Technology sc-718; 1:200), Cyclin E (Santa Cruz Technology sc-481; 1:200) and Actin (EMB Millipore MAB1501R; 1:500). Blots were then washed with TBST (Tris Buffered Saline with 0.1% Tween 20, pH 7.4) and probed with horseradish peroxidase conjugated anti-mouse (Jackson ImmunoResearch or KwikQuant) or anti-rabbit secondary antibodies (Thermo Fisher Scientific or KwikQuant) for 2 h. This was followed by 3–4 washes with TBST and the protein bands were detected using enhanced chemiluminescence (GE Healthcare or KwikQuant). For the detection of weak signal such as in Supplementary Fig. 14, the western protocol was modified to increase detection using 30-40 µg of proteins, higher concentration of antibodies and exposure times.
Isolation of primary mammary epithelial Organoids
Primary mammary epithelial organoids were isolated from 3 months old, estrus cycle matched non lactating female mice. Thoracic and inguinal mammary glands were collected, and then visible lymph nodes were excised. The mammary glands were finely chopped to approximately 1-mm3 pieces and digested in Dulbecco’s in modified Eagle medium (DMEM/F12 (Thermo Fisher Scientific 10-565-042) containing 0.2% trypsin (Thermofisher Scientific 27250018), 0.2% Collagenase (Thermo Fisher Scientific 17104019), 5% FCS and 1000 U/ml Pennicillin/Streptomycin for 30 min at 37 °C with shaking at 100 rpm. Immediately after digestion, digested mammary glands were centrifuged at 200 g for 10 min. The pellets were resuspended in DMEM/F12 media and treated with 2U/µl DNase 1 (Thermo Fisher Scientific EN0521) for 5 min with gentle shaking at room temperature, followed by centrifugation at 200 g for 10 min. The pellets were resuspended in fresh DMEM/F12 media and subjected to 3 rounds of deferential centrifugation at 200 g for 10 sec to enriches the organoids. The number of organoids is manually counted under the microscope.
Electron microscopy
Electron microscopy was performed by the Microscopy Core facility at Mount Sinai. Anesthetized mice were euthanized, and mammary glands were immediately dissected and immersed in ice-cold fixative containing 2.0% glutaraldehyde and 2% paraformaldehyde in 0.1 M sodium phosphate buffer (pH 7.2). Tissues were post-fixed in the same solution and stored at 4 °C until further processing. Fixed mammary glands were trimmed to isolate representative regions, rinsed in 0.1 M sodium cacodylate buffer (pH 7.2), and post-fixed with 1% osmium tetroxide in the same buffer. Following additional rinses in cacodylate buffer, tissues were dehydrated through a graded ethanol series (70%–100%) and propylene oxide, then infiltrated with Epon resin (Electron Microscopy Sciences). Polymerization was carried out in a vacuum oven at 60 °C for 48 hours.Semithin (1 µm) toluidine blue-stained sections were used to identify regions of interest. Ultrathin sections (80 nm) were cut with a diamond knife using a Leica UCT ultramicrotome and mounted on copper grids using a Coat-Quick adhesive pen (Electron Microscopy Sciences). Sections were counterstained with uranyl acetate and lead citrate. Images of the mammary gland were captured using a Hitachi 7700 electron microscope (Hitachi, Ltd) and photographed with an Advantage CCD camera (Advanced Microscopy Techniques). Image brightness and contrast were adjusted using Adobe Photoshop 2022 software (version 23.4.1; Adobe).
3D organoid culture and imaging
Mammary organoids were freshly isolated from estrus cycle matched, non-lactating 3 months old female mice and mixed with growth factor reduced Matrigel (Corning, United States) and plated in domes in 96-well culture plate (one dome per well, 80 µL of Matrigel per dome). 200 to 500 organoids per dome were seeded for experiments. Then the Matrigel was allowed to set for 45–60 min at 37 °C. After that the 3D organoid cultures were overlaid with MEC media [DMEM/F-12 media (Thermo Fisher Scientific 10-565-042) containing 10 % FBS, 1% P/S and 1% Insulin-Transferin-Selenium (Corning 25-800-CR)] and incubated at 37 °C in a humidified atmosphere with 5% CO2 for 24 h. After 24 h the MEC media is replaced with BOM Media [MEC media supplemented with different growth factors: 2.5 nM FGF-basic (Peprotech 450-33), 2.5 nM FGF7 (Peprotech 450-60), 2.5 nM FGF10 (Peprotech 450-61), 50 ng/mL EGF (Peprotech 315-09)]. The organoids are cultured in BOM Media for 6 days and media was changed every 2 days. On the 6th Day, lactation was induced by replacing the BOM media with lactation media [BOM media supplemented with 1 µg/mL prolactin (R&D Systems 682-PL-050), 1 µg/ml hydrocortisone (Sigma-Aldrich H0888-5G)]. To induce contraction in the organoids 40 µg/ml recombinant oxytocin (Sigma-Aldrich 06379-1MG) was added. For some experiments, the organoids were treated 20 µg/ml of δ-Tocotrienol (Sigma-Aldrich T0577-10MG) or 30 µM Salubrinal (Sigma-Aldrich SML0951) from the fourth day. Live imaging was captured with a Zeiss AxioObserver equipped with a fully enclosed incubation system to provide constant temperature at 37°C with 5% CO2. The image capture interval was either 15 or 20 minutes for durations of ∼24hrs. Movies were analyzed frame by frame. For western blot analysis, each well is washed with PBS for 3 times. Then the organoids are dissolved in warm SDS loading buffer and boiled for 5 min and subjected to SDS PAGE. Immunofluorescence staining was performed as previously described37. Briefly, 3D organoid cultures were fixed with 4% paraformaldehyde before being washed 2 times in PBS 5 minutes each and permeabilized with 0.5% tween 20. Staining for pRB in blocking buffer (10% horse serum, 2% bovine serum albumin, and 0.5% Triton X-100 in PBS) for 1 hour gently shaking at room temperature. Primary antibody (Cytokeratin 14- 0.5 ml (Lieca Biosystems NCL-L-LL002) or p-RB (Cell Signaling S780) was suspended in blocking buffer diluted 1:1 with PBS for 3 hours before washing and detection with either AlexFlour 488 or AlexaFlour 594 conjugated secondary antibodies (1:100) for 1 hour. Actin was stained with Phalloidin-iFluor 594 Reagent (ab176757) according to the manufacturer’s specifications. Organoids were then counterstained with DAPI and moved to slide for imaging. For estimation of ROS, 22 h post induction, the organoids were stained with 5 µM MitoSOX™ Red Mitochondrial Superoxide Indicator, for live cell (Life Technologies molecular probes/Invitrogen M36008) according to manufacturer’s instructions. For Oil-Red-O staining, 0.5 g of oil-red-o stain (Fisher, AAA129822) was dissolved in 100 ml of isopropanol and then diluted 6 parts to 4 parts in distilled water. After 10 minutes, it was filtered 2X with 0.2 µm syringe filter. 24 hours post induction, both induced and uninduced organoids were fixed in 4% PFA at room temperature for 20 minutes and then stained with Oil-Red-O overnight and then imaged.
Bulk RNA sequencing & analysis
Inguinal mammary glands from mice were flash frozen and sent to GENEWIZ, LLC, NJ, USA for analysis. Each sample was analyzed 3 times. 3 mice per group were analyzed. Snap frozen tissue was submitted to Azenta for paired end bulk-RNA sequencing. Raw FASTQ files were aligned to the mouse genome version mm10 on the biojupies platform using the Kallisto tool. The top 500 down-regulated or up-regulated genes were used for pathway analysis across multiple databases using the EnrichR (ref) platform. Differential gene expression (DGE) analysis was also determined between groups using the built-in tools on the Biojupies platform. Venn Diagrams were generated from these DGE analysis to identify commonly regulated genes/pathway between groups. The DGE significance cutoff was p < 0.05. Venn diagrams were generated using the Bioinformatics & Evolutionary Genomics on-line tool https://bioinformatics.psb.ugent.be/webtools/Venn/. The common or different genes between different groups were then subjected to Enrichr for further analysis. Graphs were made using combined score.
Single cell RNA sequencing and analysis
Freshly isolated snap frozen Inguinal mammary glands were submitted to GENEWIZ, LLC, NJ, USA for nuclei isolation and GEM (Gel Bead-based Emulsion) single cell RNA seq analysis on the 10x Genomics platform. Analysis was performed using the Cellranger (10xGenomics) count pipeline. Cellranger 7.0.1 was used to perform alignment of the raw FASTQ reads, filtering, barcode counting, UMI counting, and generation of the cloupe files used in downstream analysis. The cellranger count command was run with default settings, aligning to the mm10-2020-A Mus musculus 10x reference genome. Cell populations were identified using the Loupe Browser (v8.0.0, 10x Genomics) by the following features: Mfge8, Trf, Csn3, Wfdc18, Ltf (Luminal Alveolar (AV) cells), Prlr, Pgr, Esr1, Cited1, Prom1 (Luminal Hormone Sensitive (HS) cells), Krt17, Krt14, and Krt5 (Myoepithelial cells). Proliferating cells were identified by the combined feature sum expression of Cenpe, Ccna2, Ccnb2, Mcm6, Ccnf, and Bud1 as described96. Regulon expressing cells were identified by the combined feature sum expression of Bclaf1, Cux1, E2f1, E2f4, Esr1, Foxm1, Gtf2b, Max, Myc, Nfya, Nr4a1, Nrf1, Smarca4, and Taf1 as described34. The PPBC-like sub-population was identified among total barcodes by co-expression of both the Proliferating Cell and Regulon feature list. A filter cutoff to identify this population was set for half of the maximum Log2 feature sum for both feature lists. Differential gene expression for the Luminal-HS population was performed using the Loupe browser (v8.0.0) for each sample and assembled into a single.csv file that was uploaded to the Biojupies platform (https://doi.org/10.1016/j.cels.2018.10.007) to produce hierarchically clustered heatmaps using clustergrammer (developed by the Ma’ayan Laboratory at Mt. Sinai). The top 40 most differentially expressed genes between Bl/6 C57 and Bl/6 NZB LacD21 samples were then entered in EnrichR97,98 (https://doi.org/10.1002/cpz1.90) for pathway analysis across multiple databases.
Statistics and reproducibility
Statistical significance was defined as a p value below 0.05. ns = not significant, * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001. Data displayed as mean ± standard deviation (SD). Statistical analyses were performed using GraphPad Prism Software 8. Statistical significance was determined using Student´s t-test. Immunofluorescent images were processed uniformly, and Immunofluorescence signal was quantified using ImageJ to measure area positive for fluorescent signal. No statistical method was used to predetermine sample size. No data were excluded from the analyses and the experiments were not randomized. The Investigators were not blinded to allocation during experiments and outcome assessment.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All data generated by the present study are included in this article and the Supplementary Information. The bulk RNA-seq data generated in this study has been deposited in the The bulk RNAseq data generated in this study have been deposited in the GEO (Gene Expression omnibus) database under accession code GSE272881, [https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE272881]. The single cell RNaSeq data generated in this study have been deposited in the GEO database under accession code via GSE272354, [https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE272354]. Confirmation of sequencing of mice nuclear and mitochondria genomes are publicly available with biosample accession numbers: SAMNA7782938, [https://www.ncbi.nlm.nih.gov/biosample/47782938], SMNA47782939 [https://www.ncbi.nlm.nih.gov/biosample/47782939]. For the human datasets generated by the Schedin group34, the RNA-derived sequencing data generated was deposited in the Gene Expression Omnibus (GEO) database under accession code GSE158854. The publicly available RNA expression data from healthy nulliparous and postpartum breast tissues used in their study are available in the GEO database under accession code GSE26457. The publicly available outcomes data based upon RNA expression profiling used in this study as a YWBC cohort are available from the GEO database under accession codes, GSE1992, GSE20624, GSE21653, GSE6532, GSE2990, GSE4922, GSE7390, and GSE19615. Source data are provided with this paper.
References
Schedin, P. Pregnancy-associated breast cancer and metastasis. Nat. Rev. Cancer 6, 281–291 (2006).
Guo, Q. et al. The MNK1/2-eIF4E axis supports immune suppression and metastasis in postpartum breast cancer. Cancer Res 81, 3876–3889 (2021).
Lefrere, H. et al. Breast cancer diagnosed in the post-weaning period is indicative for a poor outcome. Eur. J. Cancer 155, 13–24 (2021).
Goddard, E. T. et al. Association between postpartum breast cancer diagnosis and metastasis and the clinical features underlying risk. JAMA Netw. Open 2, e186997 (2019).
Nimbalkar, V. P. et al. Premenopausal women with breast cancer in the early post-partum period show molecular profiles of invasion and are associated with poor prognosis. Breast Cancer Res Treat. 200, 139–149 (2023).
Pennock, N. D. et al. Ibuprofen supports macrophage differentiation, T cell recruitment, and tumor suppression in a model of postpartum breast cancer. J. Immunother. Cancer 6, 98 (2018).
O’Brien, J., Martinson, H., Durand-Rougely, C. & Schedin, P. Macrophages are crucial for epithelial cell death and adipocyte repopulation during mammary gland involution. Development 139, 269–275 (2012).
O’Brien, J. & Schedin, P. Macrophages in breast cancer: do involution macrophages account for the poor prognosis of pregnancy-associated breast cancer?. J. Mammary Gland Biol. Neoplasia 14, 145–157 (2009).
O’Brien, J. et al. Alternatively activated macrophages and collagen remodeling characterize the postpartum involuting mammary gland across species. Am. J. Pathol. 176, 1241–1255 (2010).
Martinson, H. A., Jindal, S., Durand-Rougely, C., Borges, V. F. & Schedin, P. Wound healing-like immune program facilitates postpartum mammary gland involution and tumor progression. Int J. Cancer 136, 1803–1813 (2015).
Lyons, T. R. et al. Postpartum mammary gland involution drives progression of ductal carcinoma in situ through collagen and COX-2. Nat. Med 17, 1109–1115 (2011).
Maller, O. et al. Collagen architecture in pregnancy-induced protection from breast cancer. J. Cell Sci. 126, 4108–4110 (2013).
Islami, F. et al. Breastfeeding and breast cancer risk by receptor status-a systematic review and meta-analysis. Ann. Oncol. 26, 2398–2407 (2015).
Anstey, E. H. et al. Breastfeeding and breast cancer risk reduction: implications for black mothers. Am. J. Prev. Med 53, S40–S46 (2017).
Chowdhury, R. et al. Breastfeeding and maternal health outcomes: a systematic review and meta-analysis. Acta Paediatr. 104, 96–113 (2015).
ElShamy, W. M. The protective effect of longer duration of breastfeeding against pregnancy-associated triple negative breast cancer. Oncotarget 7, 53941–53950 (2016).
Sly, J. R. et al. Knowledge of the Relationship Between Breastfeeding and Breast Cancer Risk Among Racial and Ethnic Minority Women. J Cancer Educ https://doi.org/10.1007/s13187-019-01580-9 (2019).
Newcomb, P. A. et al. Lactation and a reduced risk of premenopausal breast cancer. N. Engl. J. Med. 330, 81–87 (1994).
Enger, S. M., Ross, R. K., Henderson, B. & Bernstein, L. Breastfeeding history, pregnancy experience and risk of breast cancer. Br. J. Cancer 76, 118–123 (1997).
Chang-Claude, J., Eby, N., Kiechle, M., Bastert, G. & Becher, H. Breastfeeding and breast cancer risk by age 50 among women in Germany. Cancer Causes Control 11, 687–695 (2000).
Gustbee, E. et al. Excessive milk production during breast-feeding prior to breast cancer diagnosis is associated with increased risk for early events. Springerplus 2, 298 (2013).
Hetz, C., Zhang, K. & Kaufman, R. J. Mechanisms, regulation and functions of the unfolded protein response. Nat. Rev. Mol. Cell Biol. 21, 421–438 (2020).
Wenzel, E. M., Elfmark, L. A., Stenmark, H. & Raiborg, C. ER as master regulator of membrane trafficking and organelle function. J Cell Biol 221 https://doi.org/10.1083/jcb.202205135 (2022).
Eisner, V., Picard, M. & Hajnoczky, G. Mitochondrial dynamics in adaptive and maladaptive cellular stress responses. Nat. Cell Biol. 20, 755–765 (2018).
Celik, C., Lee, S. Y. T., Yap, W. S. & Thibault, G. Endoplasmic reticulum stress and lipids in health and diseases. Prog. Lipid Res. 89, 101198 (2023).
Dai, W., White, R., Liu, J. & Liu, H. Organelles coordinate milk production and secretion during lactation: Insights into mammary pathologies. Prog. Lipid Res. 86, 101159 (2022).
Rosano, T. G., Lee, S. K. & Jones, D. H. Developmental changes in mitochondria during the transition into lactation in the mouse mammary gland. II. Membrane marker enzymes and membrane ultrastructure. J. Cell Biol. 69, 581–588 (1976).
Anderson, N. S. & Haynes, C. M. Folding the Mitochondrial UPR into the Integrated Stress Response. Trends Cell Biol. 30, 428–439 (2020).
Shpilka, T. & Haynes, C. M. The mitochondrial UPR: mechanisms, physiological functions and implications in ageing. Nat. Rev. Mol. Cell Biol. 19, 109–120 (2018).
Kenny, T. C., Gomez, M. L. & Germain, D. Mitohormesis, UPR(mt), and the Complexity of Mitochondrial DNA Landscapes in Cancer. Cancer Res 79, 6057–6066 (2019).
Kenny, T. C. & Germain, D. mtDNA, Metastasis, and the Mitochondrial Unfolded Protein Response (UPR(mt)). Front Cell Dev. Biol. 5, 37 (2017).
Costa-Mattioli, M. & Walter, P. The integrated stress response: From mechanism to disease. Science 368 https://doi.org/10.1126/science.aat5314 (2020).
Latorre-Pellicer, A. et al. Mitochondrial and nuclear DNA matching shapes metabolism and healthy ageing. Nature 535, 561–565 (2016).
Jindal, S. et al. Postpartum breast cancer has a distinct molecular profile that predicts poor outcomes. Nat. Commun. 12, 6341 (2021).
Charifou, E., Sumbal, J., Koledova, Z., Li, H. & Chiche, A. A Robust Mammary Organoid System to Model Lactation and Involution-like Processes. Bio Protoc. 11, e3996 (2021).
Sumbal, J., Chiche, A., Charifou, E., Koledova, Z. & Li, H. Primary Mammary Organoid Model of Lactation and Involution. Front Cell Dev. Biol. 8, 68 (2020).
Ciccone, M. F., Trousdell, M. C. & Dos Santos, C. O. Characterization of organoid cultures to study the effects of pregnancy hormones on the epigenome and transcriptional output of mammary epithelial cells. J. Mammary Gland Biol. Neoplasia 25, 351–366 (2020).
Thomsen, J. et al. PAPP-A proteolytic activity enhances IGF bioactivity in ascites from women with ovarian carcinoma. Oncotarget 6, 32266–32278 (2015).
Conover, C. A. & Oxvig, C. PAPP-A and cancer. J. Mol. Endocrinol. 61, T1–T10 (2018).
Boldt, H. B. & Conover, C. A. Pregnancy-associated plasma protein-A (PAPP-A): a local regulator of IGF bioavailability through cleavage of IGFBPs. Growth Horm. IGF Res 17, 10–18 (2007).
Steffensen, L. B., Conover, C. A. & Oxvig, C. PAPP-A and the IGF system in atherosclerosis: what’s up, what’s down?. Am. J. Physiol. Heart Circ. Physiol. 317, H1039–H1049 (2019).
Jenkins, E. C., Brown, S. O. & Germain, D. The Multi-Faced Role of PAPP-A in Post-Partum Breast Cancer: IGF-Signaling is Only the Beginning. J. Mammary Gland Biol. Neoplasia 25, 181–189 (2020).
Heitzeneder, S. et al. Pregnancy-Associated Plasma Protein-A (PAPP-A) in Ewing Sarcoma: Role in Tumor Growth and Immune Evasion. J. Natl Cancer Inst. 111, 970–982 (2019).
Slocum, E., Craig, A., Villanueva, A. & Germain, D. Parity predisposes breasts to the oncogenic action of PAPP-A and activation of the collagen receptor DDR2. Breast Cancer Res 21, 56 (2019).
Menes, T. S. et al. Subsequent breast cancer risk following diagnosis of atypical ductal hyperplasia on needle biopsy. JAMA Oncol. 3, 36–41 (2017).
Racz, J. M. & Degnim, A. C. When does atypical ductal hyperplasia require surgical excision?. Surg. Oncol. Clin. N. Am. 27, 23–32 (2018).
Pena, A. et al. Multivariate model to identify women at low risk of cancer upgrade after a core needle biopsy diagnosis of atypical ductal hyperplasia. Breast Cancer Res Treat. 164, 295–304 (2017).
Miceli, R., Mercado, C. L., Hernandez, O. & Chhor, C. Active surveillance for atypical ductal hyperplasia and ductal carcinoma in situ. J. Breast Imaging 5, 396–415 (2023).
Danforth, D. N. Molecular profile of atypical hyperplasia of the breast. Breast Cancer Res Treat. 167, 9–29 (2018).
Fernandez, N. F. et al. Clustergrammer, a web-based heatmap visualization and analysis tool for high-dimensional biological data. Sci. Data 4, 170151 (2017).
Lacroix, M., Riscal, R., Arena, G., Linares, L. K. & Le Cam, L. Metabolic functions of the tumor suppressor p53: Implications in normal physiology, metabolic disorders, and cancer. Mol. Metab. 33, 2–22 (2020).
Olovnikov, I. A., Kravchenko, J. E. & Chumakov, P. M. Homeostatic functions of the p53 tumor suppressor: regulation of energy metabolism and antioxidant defense. Semin Cancer Biol. 19, 32–41 (2009).
Galluzzi, L. et al. Mitochondrial liaisons of p53. Antioxid. Redox Signal 15, 1691–1714 (2011).
Byun, S., Namba, T. & Lee, S. W. Losing p53 loosens up ER-stress. Aging (Albany NY) 7, 895–896 (2015).
Stavridi, E. S. & Halazonetis, T. D. p53 and stress in the ER. Genes Dev. 18, 241–244 (2004).
Thomas, S. E. et al. p53 and translation attenuation regulate distinct cell cycle checkpoints during endoplasmic reticulum (ER) stress. J. Biol. Chem. 288, 7606–7617 (2013).
Liu, Y. & Gu, W. The complexity of p53-mediated metabolic regulation in tumor suppression. Semin Cancer Biol. 85, 4–32 (2022).
Anhe, G. F. & Bordin, S. The adaptation of maternal energy metabolism to lactation and its underlying mechanisms. Mol. Cell Endocrinol. 553, 111697 (2022).
Alvarez-Delgado, C. The role of mitochondria and mitochondrial hormone receptors on the bioenergetic adaptations to lactation. Mol. Cell Endocrinol. 551, 111661 (2022).
Logue, S. E., Cleary, P., Saveljeva, S. & Samali, A. New directions in ER stress-induced cell death. Apoptosis 18, 537–546 (2013).
Chevet, E., Hetz, C. & Samali, A. Endoplasmic reticulum stress-activated cell reprogramming in oncogenesis. Cancer Discov. 5, 586–597 (2015).
Dufey, E., Urra, H. & Hetz, C. ER proteostasis addiction in cancer biology: Novel concepts. Semin Cancer Biol. 33, 40–47 (2015).
Szegezdi, E., Logue, S. E., Gorman, A. M. & Samali, A. Mediators of endoplasmic reticulum stress-induced apoptosis. EMBO Rep. 7, 880–885 (2006).
Hadsell, D., George, J. & Torres, D. The declining phase of lactation: peripheral or central, programmed or pathological?. J. Mammary Gland Biol. Neoplasia 12, 59–70 (2007).
Capuco, A. V. et al. Lactation persistency: insights from mammary cell proliferation studies. J. Anim. Sci. 81, 18–31 (2003).
Sutherland, K. D., Lindeman, G. J. & Visvader, J. E. The molecular culprits underlying precocious mammary gland involution. J. Mammary Gland Biol. Neoplasia 12, 15–23 (2007).
Watson, C. J. Involution: apoptosis and tissue remodelling that convert the mammary gland from milk factory to a quiescent organ. Breast Cancer Res 8, 203 (2006).
Brewer, J. W., Hendershot, L. M., Sherr, C. J. & Diehl, J. A. Mammalian unfolded protein response inhibits cyclin D1 translation and cell-cycle progression. Proc. Natl Acad. Sci. USA 96, 8505–8510 (1999).
Brewer, J. W. & Diehl, J. A. PERK mediates cell-cycle exit during the mammalian unfolded protein response. Proc. Natl Acad. Sci. USA 97, 12625–12630 (2000).
Hamanaka, R. B., Bennett, B. S., Cullinan, S. B. & Diehl, J. A. PERK and GCN2 contribute to eIF2alpha phosphorylation and cell cycle arrest after activation of the unfolded protein response pathway. Mol. Biol. Cell 16, 5493–5501 (2005).
Wali, V. B., Bachawal, S. V. & Sylvester, P. W. Endoplasmic reticulum stress mediates gamma-tocotrienol-induced apoptosis in mammary tumor cells. Apoptosis 14, 1366–1377 (2009).
Montagnani Marelli, M. et al. Vitamin E delta-tocotrienol triggers endoplasmic reticulum stress-mediated apoptosis in human melanoma cells. Sci. Rep. 6, 30502 (2016).
Park, S. K., Sanders, B. G. & Kline, K. Tocotrienols induce apoptosis in breast cancer cell lines via an endoplasmic reticulum stress-dependent increase in extrinsic death receptor signaling. Breast Cancer Res Treat. 124, 361–375 (2010).
Papa, L. & Germain, D. SirT3 regulates the mitochondrial unfolded protein response. Mol. Cell Biol. 34, 699–710 (2014).
Kenny, T. C. et al. Selected mitochondrial DNA landscapes activate the SIRT3 axis of the UPR(mt) to promote metastasis. Oncogene 36, 4393–4404 (2017).
Kenny, T. C., Craig, A. J., Villanueva, A. & Germain, D. Mitohormesis Primes Tumor Invasion and Metastasis. Cell Rep. 27, 2292–2303 e2296 (2019).
Stewart, J. B. & Chinnery, P. F. The dynamics of mitochondrial DNA heteroplasmy: implications for human health and disease. Nat. Rev. Genet 16, 530–542 (2015).
Liu, Y. J., Sulc, J. & Auwerx, J. Mitochondrial genetics, signalling and stress responses. Nat. Cell Biol. 27, 393–407 (2025).
Gupta, R. et al. Nuclear genetic control of mtDNA copy number and heteroplasmy in humans. Nature 620, 839–848 (2023).
Hagg, S., Jylhava, J., Wang, Y., Czene, K. & Grassmann, F. Deciphering the genetic and epidemiological landscape of mitochondrial DNA abundance. Hum. Genet 140, 849–861 (2021).
Oyadomari, S. & Mori, M. Roles of CHOP/GADD153 in endoplasmic reticulum stress. Cell Death Differ. 11, 381–389 (2004).
Li, Y., Guo, Y., Tang, J., Jiang, J. & Chen, Z. New insights into the roles of CHOP-induced apoptosis in ER stress. Acta Biochim Biophys. Sin. (Shanghai) 46, 629–640 (2014).
Malhi, H. & Kaufman, R. J. Endoplasmic reticulum stress in liver disease. J. Hepatol. 54, 795–809 (2011).
Wiseman, R. L., Mesgarzadeh, J. S. & Hendershot, L. M. Reshaping endoplasmic reticulum quality control through the unfolded protein response. Mol. Cell 82, 1477–1491 (2022).
So, J. S. Roles of Endoplasmic Reticulum Stress in Immune Responses. Mol. Cells 41, 705–716 (2018).
Bettigole, S. E. & Glimcher, L. H. Endoplasmic reticulum stress in immunity. Annu Rev. Immunol. 33, 107–138 (2015).
Salvagno, C., Mandula, J. K., Rodriguez, P. C. & Cubillos-Ruiz, J. R. Decoding endoplasmic reticulum stress signals in cancer cells and antitumor immunity. Trends Cancer 8, 930–943 (2022).
Hwang, J. & Qi, L. Quality Control in the Endoplasmic Reticulum: Crosstalk between ERAD and UPR pathways. Trends Biochem Sci. 43, 593–605 (2018).
Molinari, M. ER-phagy responses in yeast, plants, and mammalian cells and their crosstalk with UPR and ERAD. Dev. Cell 56, 949–966 (2021).
Krshnan, L., van de Weijer, M. L. & Carvalho, P. Endoplasmic Reticulum-Associated Protein Degradation. Cold Spring Harb Perspect Biol 14 https://doi.org/10.1101/cshperspect.a041247 (2022).
McLean, A. C., Valenzuela, N., Fai, S. & Bennett, S. A. Performing vaginal lavage, crystal violet staining, and vaginal cytological evaluation for mouse estrous cycle staging identification. J. Vis. Exp. e4389 https://doi.org/10.3791/4389 (2012).
Johnstone, C. N. et al. Functional and molecular characterisation of EO771.LMB tumours, a new C57BL/6-mouse-derived model of spontaneously metastatic mammary cancer. Dis. Model Mech. 8, 237–251 (2015).
Husain, K. et al. delta-Tocotrienol, a natural form of vitamin E, inhibits pancreatic cancer stem-like cells and prevents pancreatic cancer metastasis. Oncotarget 8, 31554–31567 (2017).
Zhao, X. et al. Synergistic antitumor activity of the combination of salubrinal and rapamycin against human cholangiocarcinoma cells. Oncotarget 7, 85492–85501 (2016).
DePeters, E. J. & Hovey, R. C. Methods for collecting milk from mice. J. Mammary Gland Biol. Neoplasia 14, 397–400 (2009).
Li, C. M. et al. Aging-associated alterations in mammary epithelia and stroma revealed by single-cell RNA sequencing. Cell Rep. 33, 108566 (2020).
Chen, E. Y. et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinforma. 14, 128 (2013).
Kuleshov, M. V. et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res 44, W90–W97 (2016).
Acknowledgements
This work was supported by NIH grant R21CA270702 to D. G.
Author information
Authors and Affiliations
Contributions
M.C has generated data, made the figures and wrote sections of the manuscript. E.C.J has generated data and contributed to making the figures. W. J has performed the electron microscopy. T.M has contributed to the Western analysis in Fig. 5. D.G has designed the study, direct the research, interpret data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Alecia-Jane Twigger, Likun Wang, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Chattopadhyay, M., Jenkins, E.C., Janssen, W. et al. Differential ER stress responses during lactation and postpartum outcomes in mice depending on their mitochondrial genotype. Nat Commun 16, 6432 (2025). https://doi.org/10.1038/s41467-025-61666-5
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-61666-5
This article is cited by
-
Engineering endoplasmic reticulum targeted metal–polyphenol curcumin nanomicelles for melanoma therapy
Journal of Nanobiotechnology (2025)