Abstract
The accumulation of foamy macrophages is a pathological hallmark of demyelinating brain disorders. Perturbed metabolism and efflux of intracellular lipids underlie the development of a harmful foamy macrophage phenotype in these disorders, yet, the molecular mechanisms underlying this dysregulation are poorly understood. Here, we show that the ubiquitin-proteasome system controls the turnover of the cholesterol efflux transporter ATP-binding cassette A1 (ABCA1) in lipid-loaded macrophages in the brain. We report that accumulation of myelin-derived lipids promotes the abundance and activity of ubiquitin-protein E3 ligase A (UBE3A) in macrophages, which stimulates ABCA1 ubiquitination and subsequent degradation. This boosts cellular lipid accumulation and induces an inflammatory macrophage phenotype that impairs remyelination. We further establish Tat-interacting protein 30 (TIP30), an inhibitor of importin β-mediated nuclear import, as an essential regulator of cytosolic UBE3A levels. Together, our findings identify UBE3A as a driver of foam cell formation and indicate that targeting UBE3A-mediated ABCA1 degradation is a promising strategy to enhance central nervous system repair.
Similar content being viewed by others
Introduction
Foamy macrophages, containing excessive intracellular myelin-derived lipids, contribute to the pathology of neurodegenerative disorders such as multiple sclerosis (MS)1,2,3. While the uptake of lipid-rich structures, such as modified lipoproteins and myelin, reshapes the macrophage phenotype to one that is typically associated with tissue repair and immunosuppression, this disease-resolving phenotype is only transient4,5,6,7. We and others found that excessive accumulation of myelin-derived lipids in macrophages, particularly during aging, skews foam cells towards a more inflammatory and disease-promoting phenotype. The induction of this harmful phenotype was closely associated with impaired lesion recovery in models for brain repair8,9,10. Mechanistically, perturbed metabolism of intracellular lipids, mainly due to reduced cholesterol efflux, induced an imbalance in cellular lipid content and the development of detrimental foamy macrophage subsets. To date, however, the molecular mechanisms that underlie faulty regulation of lipid efflux and inflammatory foam cell formation remain poorly understood. Identifying molecular pathways that are involved in directing the phenotype of foamy macrophage subsets is essential for our understanding of lesion progression in metabolic and neurodegenerative disorders, and for the development of immunomodulatory and reparative therapies3.
Cellular lipid efflux is essential to protect cells from the inflammatory intracellular accumulation of lipids. ATP-binding cassette A1 (ABCA1) acts as a key facilitator of cellular lipid efflux as it mediates the transfer of cholesterol and phospholipids to apolipoprotein A-I (apoA-I)11,12. On a transcriptional level, Abca1 is tightly regulated by sterol-sensing liver X receptors (LXRs)13, which promote the expression of lipid-lowering genes upon high cellular cholesterol load14. However, post-translational modifications are increasingly being recognized to control ABCA1 abundance as well15. For example, ample evidence links reduced ABCA1 turnover to decreased foam cell and plaque formation in models of atherogenesis16,17,18,19. Similarly, we found that sustained accumulation of myelin-derived lipids reduces ABCA1 protein abundance, while simultaneously increasing Abca1 expression, in macrophages in models of central nervous system (CNS) repair8. To date, various degradative models for ABCA1 turnover have been proposed, including proteasomal degradation17,20, lysosomal breakdown17,21,22, PKCδ-mediated phosphorylation8, and degradation by the protease calpain16. Yet, the degradative pathway(s) involved in controlling ABCA1 abundance in foamy myelin-containing macrophages remain unclear.
Here, we show that the ubiquitin-proteasome system (UPS) is the main degradation pathway involved in the turnover of ABCA1 in myelin-loaded foamy macrophages. Specifically, we find that the accumulation of myelin-derived lipids markedly increased the abundance and cytosolic localization of ubiquitin-protein E3 ligase A (UBE3A) in macrophages, which resulted in increased ABCA1 ubiquitination and degradation. Loss of UBE3A prevented lipid overload, induced a less inflammatory macrophage phenotype, and enhanced remyelination ex vivo and in vivo. RNA sequencing analysis further identified Tat-interacting protein 30 (TIP30), an inhibitor of importin β-mediated nuclear import, as an essential regulator of cytosolic UBE3A levels, and consequently, ABCA1 abundance. Taken together, our findings position UBE3A at the crossroads of lipid metabolism and immunity in CNS disorders, and identify UBE3A as a promising therapeutic target to mend faulty lipid metabolism in myelin-containing macrophages and enhance CNS repair.
Results
ABCA1 is degraded by the ubiquitin-proteasome system in foamy macrophages
Sustained accumulation of myelin-derived lipids skews macrophages toward a disease-promoting phenotype in CNS disorders, and is associated with a reduction in the abundance of ABCA18. By exposing murine bone marrow-derived macrophages (BMDMs) to myelin for a short (24 h, Mye24) or long (72 h, Mye72) period, we here confirm reduced ABCA1 protein levels upon prolonged myelin incubation (Mye72), despite an increase in Abca1 gene expression (Fig. 1A, B). This discrepancy in ABCA1 protein and mRNA levels suggests an important role for post-translational regulation of ABCA1 in foamy macrophages.
A,BAbca1 mRNA (A) and ABCA1 protein (B) level in bone marrow-derived macrophages (BMDMs) treated with myelin (50 µg/ml) for 24 h (Mye24) or 72 h (Mye72), and normalized to untreated cells (Ctrl) (n = 3 samples). ABCA1 protein level is normalized to Cy5 levels (loading control). C,D Surface ABCA1 level determined with flow cytometry in Ctrl (C) and Mye72 (D) BMDMs exposed to MG132 [MG; 25 µM; n(Ctrl) = 5 samples, n(Mye72) = 11 samples], Bafilomycin A1 [BM; 10 nM; n(Ctrl) = 4 samples, n(Mye72) = 5 samples], Rottlerin [Rotl; 3 µM; n = 5 samples], or Calpeptin [Calp; 10 µg/ml; n = 4 samples] for 24 h, and normalized to vehicle-treated cells (−) [n(Ctrl) = 5 samples, n(Mye72) = 9 samples]. E ABCA1 protein level in Mye72-BMDMs treated with MG for 24 h, and normalized to vehicle-treated cells (−) (n = 3 samples). ABCA1 protein level is normalized to Cy5 levels (loading control). F ABCA1 protein level in Mye72-BMDMs treated with MG132 or vehicle (−) for 24 h and incubated with cyclohexamide for the indicated period (CHX, 50 μg/ml) [n(Ctrl0-3h CHX, Ctrl6h,CHX, Mye0h CHX, Mye2,3,4h,CHX) = 7 samples; n(Ctrl4,5h,CHX) = 6 samples; n(Mye1h,CHX) = 5 samples; n (Mye5,6h,CHX) = 4 samples)]. G Relative capacity of Mye72-BMDMs treated with MG or vehicle (−) for 24 h to efflux cholesterol via ABCA1 using ApoA-I as an acceptor (n = 7 samples). Data were representative of ≥3 independent experiments and are shown as mean ± SEM. Statistical significance was determined by two-sided one-way ANOVA (A–D) or two-sided Student’s t-test (E–G). *P < 0.05, **P ≤ 0.01, and ***P ≤ 0.001. If p values are not indicated, the result was not significant.
Previous studies demonstrated that ABCA1 turnover involves proteasomal17,20, lysosomal17,21,22, or calpain protease-driven degradation16, as well as PKCδ-mediated phosphorylation8. Accordingly, by using well-described inhibitors, we found that control BMDMs displayed elevated ABCA1 surface levels once proteasomal and lysosomal degradation processes were inhibited (Fig. 1C). Yet, only the proteasomal inhibitor MG132 increased membrane ABCA1 abundance in myelin-loaded BMDMs (Fig. 1D and Fig. S1A). Immunoblotting analysis further showed that not only surface ABCA1, but also total ABCA1 levels increased in myelin-treated BMDMs upon proteasomal inhibition (Fig. 1E). To further study the degradation kinetics of ABCA1 upon proteasomal inhibition, a cyclohexamide (CHX) chase assay was performed. We show that ABCA1 is stabilized in myelin-treated BMDMs upon proteasomal inhibition, with ABCA1 only falling below control levels after 5 h of CHX incubation (Fig. 1F). Consistent with these findings, elevated ABCA1 abundance in MG132-treated myelin-containing BMDMs was associated with increased ABCA1-mediated cholesterol efflux to the lipid acceptor ApoA-I (Fig. 1G). In aggregate, we demonstrate that proteasomal degradation drives ABCA1 turnover in macrophages exposed to myelin.
Sustained intracellular myelin accumulation increases UBE3A in macrophages
Proteasomal degradation ensues following conjugation of proteins to ubiquitin, requiring the hierarchical action of E1 activating, E2 conjugating, and E3 ligase enzymes, the latter being responsible for the final transfer of ubiquitin to protein targets14,23,24. Given that our findings indicate that ABCA1 was degraded by the proteasome in foamy macrophages, we next sought to identify UPS enzymes involved in this process. Re-analysis of our previous transcriptomic screen on myelin-treated macrophages demonstrated that genes related to the HECT E3 ligase family were most significantly altered in macrophages following prolonged exposure to myelin, with the ubiquitin-protein ligase E3A (Ube3a), also known as E6-associated protein (E6AP), being the most potently induced UPS-associated gene (Fig. S1B and Table S1)13. Enhanced Ube3a expression following myelin exposure was validated in murine BMDMs and human monocyte-derived macrophages (hMDMs) (Fig. 2A, C). Consistent with elevated Ube3a gene expression, myelin exposure increased UBE3A protein levels in BMDMs (Fig. 2B). Previous research demonstrated that myelin-derived cholesterol metabolites activate liver X receptor (LXRs), leading to the transcriptional activation of key lipid metabolism genes such as Scd1 and Abca125. To test whether LXRs are involved in the upregulation of Ube3a in myelin-treated macrophages as well, Ube3a mRNA was compared between Wt and Lxr−/− myelin-containing BMDMs. We found that prolonged myelin treatment was no longer able to increase Ube3a in LXR-deficient BMDMs (Fig. S1C), while short-term stimulation with the LXR agonist GW3965 did not induce Ube3a in BMDMs (Fig. S1D), indicating an indirect role for LXRs in enhancing Ube3a expression in foam cells.
A,B Ube3a mRNA (A, n = 3 samples) and UBE3A protein level (B, n = 6 samples) in mouse bone marrow-derived macrophages (BMDMs) treated with myelin (50 µg/ml) for 24 h (Mye24) or 72 h (Mye72), and normalized to untreated cells (Ctrl, dotted line). C UBE3A mRNA in human monocyte-derived macrophages treated with myelin (50 µg/ml) for 24 h (Mye24) or 72 h (Mye72), and normalized to untreated cells (dotted line) (n = 3 samples). D,E,H Representative images (G) and quantification (D, E) of active MS lesion stained for UBE3A/CD68. Scale bars 300 and 50 µm. [n(NAWM) = 4 subjects, n(Rim) = 4 subjects, n(center) = 8 subjects]. F, I Representative images (I) and quantification (F) of active MS lesion stained for ABCA1/HLA-DR/UBE3A. Scale bar 50 µm (n = 4 subjects). Data were representative of ≥3 independent experiments and are shown as mean ± SEM. Statistical significance was determined by two-sided Student’s t-test (A–C) or two-sided one-way ANOVA (D–F). *P < 0.05, **P < 0.01, and ***P < 0.001. If p values are not indicated, the result was not significant.
Given that foamy phagocytes, including macrophages and microglia, are abundant in active lesions of MS patients, we next determined whether myelin internalization increases the level of UBE3A in lesional foamy phagocytes. Immunohistochemical double staining demonstrated that the number of UBE3A+CD68+ phagocytes, as well as UBE3A abundance in CD68+ phagocytes, was markedly increased in MS lesions (Fig. 2D–F). In support of our in vitro findings, phagocytes in the lesion center, which displayed a higher intracellular lipid load (Oil red O stain, Fig. S1E) and lower ABCA1 abundance as compared to those in the lesion rim (Fig. 2D, G, H)8, showed the highest expression of UBE3A (Fig. 2D, F, H). Collectively, these findings demonstrate that sustained intracellular accumulation of myelin increases UBE3A abundance in phagocytes.
UBE3A-mediated ubiquitination targets ABCA1 for proteasomal degradation
As our data show that sustained accumulation of myelin increases UBE3A level and leads to proteasomal degradation of ABCA1, we next determined if UBE3A-mediated ubiquitination drives proteasomal turnover of ABCA1. To this end, BMDMs were isolated from mice that either lacked (Ube3a−/−) or overexpressed (Ube3aOE) UBE3A26. Overexpression of UBE3A reduced ABCA1 protein level, but not Abca1 mRNA expression (Fig. 3A, B and Fig. S2A). Reciprocally, deficiency of Ube3a markedly increased ABCA1 abundance in BMDMs, while not affecting Abca1 mRNA (Fig. S2B). A similar increase in ABCA1 protein abundance, without corresponding changes in its gene expression, was observed in hMDMs following UBE3A silencing (Fig. S2C, D). To substantiate these findings, HEK293T cells were transfected with plasmids encoding either nuclear mUBE3A-Iso3 or cytoplasmic mUBE3A-Iso2, as described previously27. Whereas HEK293T cells expressing cytoplasmic mUBE3A-Iso2 demonstrated decreased ABCA1 abundance (Fig. 3D, E), those expressing nuclear mUBE3A-Iso3 showed no significant change in ABCA1 levels (Fig. 3D, F). To provide further evidence that UBE3A-mediated ubiquitination controls ABCA1 turnover, HEK293T cells were transfected with a catalytically inactive version of cytosolic UBE3A (mUBE3A-Iso2C.I.). In contrast to active mUBE3A-Iso2, overexpression of catalytically inactive UBE3A did not decrease ABCA1 protein level (Fig. 3D, G), which indicates that UBE3A-mediated ubiquitination is required for ABCA1 degradation.
A–C ABCA1 protein level in wild-type (Wt), Ube3a-overexpression (B, Ube3aOE, n = 3 samples), and Ube3a−/− (C, n = 4 samples) bone marrow-derived macrophages (BMDMs), normalized to Cy5 levels (loading control). D–G ABCA1 protein level in HEK293T cells in which mUBE3A-Iso2 (E, n = 5 samples), mUBE3A-Iso3 (F, n = 3 samples), or catalytically inactive mUBE3A-Iso2 (mUBE3A-Iso2C.I., G, n = 6 samples) was expressed. HEK293T cells incubated with equal amounts of PEI served as controls (Ctrl). ABCA1 protein levels are normalized to Cy5 levels (loading control). H Escherichia coli (E. Coli) cells expressing E1, UbcH5 (E2), and with (+) or without (−) ubiquitin (Ub), were transfected with either wtUBE3A or catalytically inactive UBE3A (UBE3AC817A), in combination with HA:UBE3A, V5:RING1B, or V5:ABCA1 (V5 linked to C-terminal cytoplasmic tail of ABCA1). Cell lysates were analyzed by immunoblotting for HA to visualize UBE3A, or V5 to visualize RING1B or ABCA1. Representative blots of three independent technical replicates are shown. I Immunoprecipitation (IP) of ubiquitin on Wt and Ube3a−/− Mye72-treated BMDM cell lysates. Whole cell lysates (Input) and final eluate (IP: Ub) were analyzed by immunoblotting for ubiquitin and ABCA1. J IP of ABCA1 on cell lysates of untreated (−) and Mye72-treated Wt BMDMs. Whole cell lysates (input) and final eluate (IP: ABCA1) were analyzed by immunoblotting for UBE3A and ABCA1. Cy5 served as a negative control. Data from A–G are representative of three independent experiments, and all data were shown as mean ± SEM. Statistical significance was determined by a two-sided Student’s t-test. *P < 0.05, **P < 0.01. If p values are not indicated, the result was not significant.
To confirm that ABCA1 is a direct ligation target of UBE3A, we first used a bacterial ubiquitination system in which the mammalian ubiquitination cascade is recapitulated in Escherichia coli (E. coli) cells by co-expressing the required enzymes together with the substrate of interest, a technique previously applied by us to identify ubiquitination targets of UBE3A28,29,30. As anticipated, in this assay, both UBE3A auto-ubiquitination and ubiquitination of the established target RING1B by UBE3A were evident when using wild-type UBE3A (Fig. 3H). In contrast, expression of the catalytically inactive C817A mutant UBE3A (UBE3AC817A) failed to promote ubiquitination of these proteins (Fig. 3H). Our diGly proteomics experiment comparing brain tissue of a wild-type and Ube3a−/− mouse showed four ABCA1 ubiquitination sites, which were either absent or strongly reduced in Ube3a−/− brain, of which three were in the C-terminal cytoplasmic tail of ABCA1 (refs. 29,31, Fig. S2E). Hence, this region was V5-tagged and expressed in E. coli. Similar to the established target RING1B, we found that wtUBE3A promoted the ubiquitination of ABCA1, an effect that was abolished with the catalytically inactive UBE3AC817A mutant (Fig. 3H). To further validate UBE3A-mediated ubiquitination of ABCA1 in myelin-containing macrophages, we applied pull-down of ubiquitinated proteins on wild-type and Ube3a−/− BMDMs. Blotting demonstrated ubiquitination of ABCA1 protein in wild-type but not Ube3a−/− myelin-containing macrophages, indicating that UBE3A is essential for ABCA1 ubiquitination in macrophages (Fig. 3I). Mirroring these findings, immunofluorescent stains showed that Ube3aOE macrophages displayed increased colocalization of ABCA1 and ubiquitin (Fig. S2F, G). Finally, immunoprecipitation of ABCA1 followed by probing for UBE3A confirmed the endogenous interaction between UBE3A and ABCA1 in myelin-containing BMDMs (Fig. 3J). Taken together, our findings identify ABCA1 as a bona fide ligation target of UBE3A, and demonstrate that UBE3A reduces ABCA1 protein levels in myelin-laden macrophages through ubiquitination. Interestingly, prolonged exposure of BMDMs to apoptotic and necrotic cells increased UBE3A abundance without reducing ABCA1 levels, indicating that increased UBE3A-mediated degradation of ABCA1 is specific to myelin-loading (Fig. S3A–E).
UBE3A is highly enriched in the cytoplasm of macrophages
UBE3A can accumulate in both nuclear and cytoplasmic compartments in a cell-type and context-dependent manner27,32,33. Having established that UBE3A reduces ABCA1 membrane abundance, we next set out to define the cellular distribution of UBE3A. We show that BMDMs display a predominant cytoplasmic localization of UBE3A, with myelin exposure further increasing its cytoplasmic level (Fig. 4A–C). We have previously demonstrated that alternatively spliced exons dictate the subcellular distribution of UBE3A in neurons34. To identify the mechanisms underlying cytoplasmic enrichment of UBE3A in macrophages, Integrated Genome Viewer (IGV) software was used to generate Sashimi plots for each of the BMDM samples of the transcriptomics dataset, after which the exon-intron splice junctions occurring in the Ube3a transcripts were quantified (Fig. 4D). The abundance of mUbe3a-Iso2 transcripts, which encode the cytoplasmic UBE3A variant which starts with the MATACK sequence, did not differ noticeably from nuclear mUbe3a-Iso3 transcripts that start with the MKRAAAK sequence (Fig. 4D). In contrast, immunoblotting analysis revealed an enriched presence of the short nuclear mUBE3A-Iso3 in macrophages (Fig. 4E). Importantly, myelin exposure increased cytoplasmic mUBE3A-Iso2 and decreased nuclear mUBE3A-Iso3, although the latter variant remained predominantly expressed in myelin-treated BMDMs. All in all, our data show UBE3A enrichment in the cytoplasm of macrophages, which further increases upon myelin exposure, and which can only partly be traced back to transcriptional isoform expression and abundance. These findings point towards a molecular mechanism actively controlling cellular trafficking of UBE3A isoforms.
A–C Bone marrow-derived macrophages (BMDMs) were treated with myelin (50 µg/ml) for 24 h (Mye24), 72 h (Mye72), or left untreated (Ctrl). Representative images of UBE3A and BODIPY (A). Quantification of the number of UBE3A+ puncta in the cytoplasm and the nucleus (B) and the ratio of UBE3A puncta in cytoplasm:nucleus (C) (n = 3 samples from three independent experiments, >20 cells per condition). Scale bar 50 µm. D Upper figure shows a schematic representation of the number of Ube3a exon junctions found in BMDMs. Two lower schemes depict splicing pattern and relative abundance of predicted Ube3a transcripts in untreated macrophages, based on quantification of exon junctions (n = 3 samples). E BMDMs were treated with myelin (50 µg/ml) for 24 h (Mye24), 72 h (Mye72), or left untreated (Ctrl). Cell lysates were analyzed by immunoblotting for UBE3A, normalized to GAPDH levels (loading control), and the relative isoform abundance ratio was determined. Upper UBE3A band is mIso2, lower UBE3A band is mIso3 (n = 6 samples from six independent experiments). Data were represented as mean ± SEM. Statistical significance was determined by two-sided one-way ANOVA (B, C, E). *P < 0.05 and ***P < 0.001. If p values are not indicated, the result was not significant.
TIP30 inhibits importin β-dependent nuclear import of UBE3A
Alongside its role as E3 ubiquitin-ligase, UBE3A can affect cell metabolism on a transcriptional level by serving as a coactivator for members of the nuclear receptor transcription factor family, a dual role which might be related to its translocation between the nucleus and cytoplasm35. To assess transcriptional changes associated with Ube3a deficiency in BMDMs, and to find a molecular rationale for its cytoplasmic location in macrophages, bulk RNA sequencing was performed. This analysis revealed 41 and 34 genes to be down- and upregulated, respectively, in Ube3a−/− macrophages (Fig. 5A and Table S2). Ingenuity pathway analysis (IPA) demonstrated that these differentially expressed genes (DEGs) were associated with DNA synthesis and repair pathways (Fig. 5B). Accordingly, DEGs showed an overrepresentation in molecular and cellular functions related to cell death and survival (Fig. 5C). Consistent with UBE3A being associated with tumor biology and acting as coactivator of nuclear steroid receptors35,36,37,38, IPA identified Ube3a deficiency to be correlated with cancer biology and disorders of the endocrine and reproductive system (Fig. 5D, E). RNA sequencing analysis further showed that HIV-1 Tat interactive protein 2 (Htatip2), also called Cc3 and hereafter referred to as Tip30, was, after Ube3a, the most potently downregulated gene in Ube3a−/− macrophages (Fig. 5A and Table S2). The importance of UBE3A in controlling Tip30 mRNA expression and protein level was validated using Ube3a−/− and Ube3aOE BMDMs (Fig. 5F–I). Akin to Ube3a−/− BMDMs, UBE3A-deficient hMDMs showed a reduced TIP30 expression (Fig. 5J). Tip30 is a metastatic tumor-suppressor gene which, alongside its pro-apoptotic functions, can inhibit nuclear import of specific proteins by direct binding to the karyopherins of the importin β family39,40. Given that macrophages displayed cytoplasmic UBE3A enrichment (Fig. 4), we reasoned that UBE3A-induced TIP30 expression in myelin-exposed BMDMs might contribute to the cytoplasmic accumulation of UBE3A.
A Volcano plot of differential gene expression of Wt and Ube3a−/− bone marrow-derived macrophages (BMDMs) (P < 0.01, complete list in Table S2) (n = 3 samples). B–E Ingenuity pathway analysis (IPA) of Wt and Ube3a−/− BMDMs. Top five most enriched pathways, top five molecular and cellular function categories, top five diseases and disorders, and top ten transcriptional regulators most associated with the transcriptome of Ube3a−/− BMDMs. F–J Fold change TIP30 mRNA (F, H, J) and TIP30 protein (G, I) in Ube3a−/− BMDMs (F, G; n = 3 samples), Ube3a-overexpressing BMDMs (H, I; Ube3aOE) (n = 3 samples) and UBE3AKD human monocyte-derived macrophages (hMDMs) (J; n = 6 samples), compared to Wt cells. TIP30 protein levels are normalized to β-Actin levels (loading control). Data were represented as mean ± SEM. Statistical significance was determined by a two-sided Student’s t-test. *P < 0.05, **P < 0.01, and ***P < 0.001. If p values are not indicated, the result was not significant.
To assess whether TIP30 controls the subcellular distribution of UBE3A, cytoplasmic UBE3A-Iso2 and nuclear UBE3A-Iso3 were co-expressed with TIP30 (Fig. 6A). Co-expression with TIP30 reduced the nuclear abundance and intensity of mUBE3A-Iso3, while it did not affect mUBE3A-Iso2 cellular location (Fig. 6B–D and Fig. S4A, B). As the nuclear localization of mUBE3A-Iso3 is determined by the lack of the N-terminal extension, which mediates nuclear export, these findings demonstrate that TIP30 inhibits nuclear import of UBE3A without interfering with nuclear export. Next, we aimed to verify that the observed change in nuclear mUBE3A-Iso3 is not due to potential alterations in cell viability following TIP30 overexpression. As indicated by the nucleoporin staining, TIP30 overexpression did not affect nuclear integrity, confirming that UBE3A localization was not affected by a change in nuclear permeability or apoptosis (Fig. S4). In further support of the crucial role of TIP30 in promoting the cytosolic localization of UBE3A, Tip30 silencing (Fig. S4D) resulted in a notable increase in the nuclear accumulation of UBE3A (Fig. 6A, E, F). To evaluate whether TIP30 affects UBE3A localization through inhibition of importin β-mediated nuclear import, Tip30K.D. cells were treated with importazole, a pharmacological inhibitor for importin β41,42. Inhibition of importin β resulted in a nearly exclusive cytoplasmic accumulation of UBE3A independent of Tip30 silencing (Fig. 6A, E, F). These findings strongly suggest that TIP30 halts UBE3A nuclear localization via importin β-mediated inhibition and contributes to dysregulation of ABCA1 levels in macrophages by enhancing cytoplasmic UBE3A. With respect to the latter, we next evaluated whether Tip30 silencing affects ABCA1 protein level and lipid accumulation. Tip30K.D. cells showed increased ABCA1 protein levels and a reduced accumulation of lipid droplets (Fig. 6G–J and Fig. S4E). Taken together, we demonstrate that TIP30 controls subcellular UBE3A localization via importin β, and we provide a molecular rationale for enhanced cytosolic UBE3A abundance in myelin-containing macrophages.
A Scheme of an experimental pipeline used to study the role of TIP30 in the subcellular distribution of UBE3A. U2OS cells were transfected with one of the following expression plasmids: m(mouse)UBE3A-Iso3, mUBE3A-Iso2, and r(at)TIP30, or a combination of mUBE3A-Iso3:rTIP30 or mUBE3A-Iso2:rTIP30 in a ratio of 1:1 or 1:3 (B–D). HEK293T cells were transfected with Tip30 siRNA or negative control siRNA (n.c.), and treated with importazole (importin β-inhibitor, ImpβINH, 20 µM), or vehicle (E–J). B–D Representative images (B) and quantification of UBE3A+ %area (C) and intensity (D) in the nucleus:cytoplasm of the cells (n = 3 samples). Scale bar 15 µm. E,F Representative images (E) and quantification (F) of UBE3A+ %area in the nucleus:cytoplasm of the cells (n = 5 samples). Scale bar 30 µm. G,H Representative images (G) and quantification (H) of ABCA1+ %Area (n = 3 samples). Scale bar 30 µm. I ABCA1 protein level determined with immunoblotting (n = 5 samples). ABCA1 protein levels are normalized to Cy5 levels (loading control). J Relative lipid load determined with BODIPY FACS [n(n.c.) = 11 samples; n(Tip30) = 13 samples)]. Data were collected from ≥3 independent experiments, and all data were represented as mean ± SEM. Statistical significance was determined by two-sided Student’s t-test (H–J) or two-sided one-way ANOVA (C, D, F). *P < 0.05, **P < 0.01, and ***P < 0.001. If p values are not indicated, the result was not significant. Created in BioRender. Bogie, J. (2025) https://BioRender.com/2yr70n3.
UBE3A drives myelin-containing phagocytes towards a disease-promoting phenotype
Loss of ABCA1 drives macrophages toward a disease-promoting phenotype in metabolic and CNS disorders by promoting the intracellular accumulation of lipids8,43,44. Given that UBE3A-mediated ubiquitination stimulates proteasomal degradation of ABCA1, we next assessed the impact of UBE3A on the metabolic and inflammatory features of foamy macrophages. Consistent with functional regulation of ABCA1 by UBE3A (Fig. 4A), we found that Ube3a overexpression in BMDMs decreased ABCA1-dependent cholesterol efflux, while increasing myelin-induced intracellular lipid accumulation (Fig. 7A–C). Accordingly, inflammatory phenotyping demonstrated that Ube3a overexpression enhanced the release of nitric oxide (NO) and induced the expression of Nos2 and Il1β in myelin-containing macrophages (Fig. 7D, E). Reciprocally, loss of Ube3a enhanced ABCA1-mediated cholesterol efflux and decreased the intracellular lipid load following myelin-loading (Fig. 7F–H). Further, Ube3a−/− foamy BMDMs released less NO and showed a reduced expression of Nos2 and Il1b (Fig. 7I, J). Similar findings were obtained following siRNA-mediated gene silencing of UBE3A using myelin-containing hMDMs (Fig. 7K–O) and in Ube3a−/− foamy microglia (Fig. 7P–S). Taken together, these findings suggest that by regulating ABCA1 abundance, UBE3A governs the accumulation of inflammatory myelin-derived lipids and skews myelin-containing macrophages and microglia toward a more inflammatory phenotype.
A–J,P–S Murine wild-type (Wt), Ube3a-overexpression (Ube3aOE) and Ube3a−/− bone marrow-derived macrophages (BMDMs) or microglia were treated with myelin (50 µg/ml) for 72 h, or left untreated, optionally followed by lipopolysaccharide stimulation (LPS; D, E, I, J, S; 100 ng/μl). K–O Human monocyte-derived macrophages (hMDMs) were transfected with UBE3A-specific (UBE3AKD) or scrambled (Wt) siRNA and treated with myelin (50 µg/ml) for 72 h, or left untreated, optionally followed by LPS stimulation (N, O; 100 ng/μl). A,F,K,P Relative capacity of Mye72-cells to efflux cholesterol via ABCA1. Result was normalized to efflux capacity of cells in the absence of lipid acceptors (dotted line) [n(A) = 6 samples, n(F, Wt) = 8 samples, n(F, Ube3a−/− = 6 samples), n(K) = 6 samples, n(P) = 8 samples]. B,G,L,Q Representative images of Oil Red O staining of Mye72-cells (n = 3 samples). Scale bar 30 µm. C,H,M,R Relative lipid load of Mye72-cells and normalized to untreated cells (dotted line), as determined with Bodipy FACS [n(C) = 6 samples, n(H, Wt) = 14 samples, n(H, Ube3a−/−) = 12 samples, n(M) = 3 samples, n(R, Wt) = 5 samples, n(R, Ube3a−/− = 6 samples)]. D,E,I,J,N,O,S Cells were treated with myelin, followed by LPS stimulation for 18 h (D, I, N) or 6 h (E, J, O, S). Relative NO (D, I) and IL1β (N) concentration in culture supernatants of cells [n (D) = 6 samples, n(I, Wt) = 5 samples, n(I, Ube3a−/−) = 6 samples, n(N, Wt) = 9 samples, n(N, Ube3aKD) = 6 samples]. Gene expression of pro-inflammatory genes [n(E) = 3 samples, n(J) = 4 samples, n(O, S) = 6 samples]. Data were collected from three independent experiments and are represented as mean ± SEM. Statistical significance was determined by a two-sided Student’s t-test. *P < 0.05, **P < 0.01, and ***P < 0.001. If p values are not indicated, the result was not significant.
Loss of UBE3A improves remyelination in the ex vivo cerebellar brain slice model in a macrophage-dependent manner
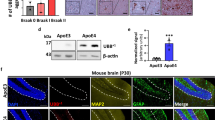
By applying toxin-induced demyelination models, we and others recently showed that sustained accumulation of myelin-derived lipids steers macrophages towards an inflammatory phenotype that attenuates CNS repair8,9. Given that UBE3A increased intracellular lipid load and induced an inflammatory foam cell phenotype, we next defined the impact of UBE3A on remyelination. To this end, murine cerebellar brain slices were isolated from wild-type and Ube3a−/− mice. Brain slices were demyelinated using lysolecithin, and remyelination efficacy was assessed after 6 days (Fig. 8A). In line with our in vitro findings, loss of Ube3a reduced the accumulation of ORO+ lipid droplets and increased the level of ABCA1 in brain slice cultures (Fig. 8B–D). To examine whether these changes improved remyelination, slices were stained for neurofilament (NF) and myelin basic protein (MBP) to visualize the axons and surrounding myelin sheath, respectively. This demonstrated an increased colocalization of MBP with NF+ axons upon loss of Ube3a, indicative of enhanced remyelination. Three-dimensional reconstruction confirmed more efficient axonal myelination in Ube3a−/− slices (Fig. 8B, E).
A Scheme of an experimental pipeline used to study the effect of Ube3a-deficiency on remyelination. Cerebellar brain slices were isolated from P9-11 wild-type (Wt) or Ube3a−/− pups and demyelinated with lysolecithin (LPC, 0.5 mg/ml). Next, slices were kept in culture for 6 days, during which remyelination takes place. To assess the effect of macrophage-specific loss of Ube3a (F–I), slices were stimulated with clodronate liposomes (0.5 mg/ml) prior to LPC treatment, after which they were repleted with Wt or Ube3a−/− bone marrow-derived macrophages (MΦWt and MΦUbe3a−/−). B Representative images of Oil Red O (ORO), ABCA1, and MBP-NF stains, and three-dimensional reconstructions of brain slices. Scale bars: ORO outset 100 µm, ORO inset 50 µm, ABCA1 50 µm, MBP-NF outset 50 µm, MBP-NF inset 15 µm. C %ORO+ area of Wt and Ube3a−/− cerebellar brain slices [n(Wt) = 11 pups, n(Ube3a−/−) = 4 pups]. D %ABCA1+ area of Wt and Ube3a−/− cerebellar brain slices [n(Wt) = 3 pups, n(Ube3a−/−) = 4 pups]. E MBP+NF+ axons of total NF+ axons in Wt and Ube3a−/− cerebellar brain slices [n(Wt) = 7 pups, n(Ube3a−/−) = 5 pups]. F Representative images of MBP-NF, ORO, ABCA1, and F4/80 stains, and three-dimensional reconstructions of brain slices. Scale bars: MBP-NF outset 50 µm, MBP-NF inset 15 µm, ORO outset 100 µm, ORO inset 50 µm, ABCA1 and F4/80 50 µm. G MBP+NF+ axons of total NF+ axons in macrophage-specific Ube3a−/− (MΦUbe3a−/−) or Wt (MΦWt) cerebellar brain slices [n(MΦWt) = 4 pups, n(MΦUbe3a−/−) = 3 pups]. H %ORO+ area of macrophage-specific Ube3a−/− (MΦUbe3a−/−) or Wt (MΦWt) cerebellar brain slices [n(MΦWt) = 3 pups, n(MΦUbe3a−/−) = 4 pups]. I %ABCA1+ area within F4/80+ cells in macrophage-specific Ube3a−/− (MΦUbe3a−/−) or Wt (MΦWt) cerebellar brain slices [n(MΦWt) = 4 pups, n(MΦUbe3a−/−) = 3 pups]. Data were collected from three independent experiments and are represented as mean ± SEM. Statistical significance was determined by a two-sided Student’s t-test. *P < 0.05 and **P < 0.01. If p values are not indicated, the result was not significant. Created in BioRender. Bogie, J. (2025) https://BioRender.com/sokinfo.
Given the multicellular nature of slice cultures, we next sought to determine the impact of macrophages on the pro-regenerative effect of Ube3a deficiency. To this end, wild-type brain slices were depleted of phagocytes using clodronate liposomes, followed by repletion with either wild-type or Ube3a−/− BMDMs (Fig. 8A). We show that treatment of wild-type brain slices with clodronate liposomes resulted in an almost complete depletion of F4/80+ phagocytes, and that the addition of macrophages efficiently replenished the slice cultures (Fig. S5A, B). To further confirm the validity of the model, phagocyte-depleted slice cultures were repleted with well-described disease (DAMs, LPS-stimulated) and repair-associated (RAMs, IL4-stimulated) macrophage subsets45. Repletion with RAMs enhanced remyelination deficits observed in slice cultures lacking macrophages, and resulted in improved remyelination as compared to DAM-repleted slices (Fig. S5C, D). These findings confirm the validity of the used model. Importantly, we find that repletion with Ube3a−/− macrophages markedly improved the degree of remyelination as compared to repletion with wild-type BMDMs (Fig. 8F, G). Accordingly, slices repleted with Ube3a−/− macrophages showed decreased lipid accumulation and increased ABCA1 abundance within F4/80+ macrophages (Fig. 8F, H, I), as compared to those repleted with wild-type macrophages. Collectively, these findings provide compelling evidence that the lack of UBE3A drives macrophages towards a reparative phenotype to promote remyelination, and that this relies, at least in part, on UBE3A-dependent regulation of ABCA1 abundance.
Ube3a-deficiency improves remyelination after cuprizone-induced demyelination in vivo
To validate the significance of our findings in vivo, we employed the cuprizone-induced de- and remyelination model. Cuprizone feeding induces demyelination in the corpus callosum, and its withdrawal after 5 weeks triggers spontaneous remyelination. Animals were pathologically characterized after demyelination (5 weeks, 5w) and during spontaneous remyelination (6 weeks, 5 + 1w) (Fig. S5E). Consistent with our in vitro and ex vivo results, UBE3A deficiency resulted in reduced accumulation of PLIN2+ lipid droplets in F4/80+ phagocytes within the corpus callosum during both the demyelination and remyelination phases (Fig. 9A, B). In line with this observation, Ube3a−/− mice exhibited elevated ABCA1 protein levels in the corpus callosum during remyelination (Fig. 9C), consistent with more efficient lipid efflux by foamy phagocytes in Ube3a−/− mice. This was accompanied by reduced expression of inflammatory genes during demyelination (Nos2, Il6, Ccl4) (Fig. 9D), indicating a dampened inflammatory response. Importantly, these beneficial effects of UBE3A loss were reflected in increased MBP immunoreactivity in the corpus callosum, indicative of increased remyelination (Fig. 9E, F). Collectively, our findings demonstrate that UBE3A deficiency enhances remyelination, likely by reducing phagocytic lipid load and inflammation.
Wildtype (Wt) and Ube3a−/− mice were fed a cuprizone diet for 5 weeks to induce demyelination in the corpus callosum (cc). Upon withdrawal of the cuprizone diet, spontaneous remyelination ensues. Tissue was collected after demyelination (5w) and after 1 week of recovery (5 + 1w). A, B Representative pictures and quantification of F4/80 PLIN2 co-staining of cc. Scale bar 100 µm. [n(Wt, 5w) = 6 mice, n(Ube3a−/−,5w) = 5 mice, n(Wt, 6w) = 5 mice, n(Ube3a−/−,6w) = 4 mice]. C ABCA1 protein level in cc determined with immunoblotting and normalized to Cy5 levels (loading control). [n(Wt) = 3 mice, n(Ube3a−/−) = 4 mice]. D Gene expression of pro-inflammatory genes Nos2, Il6, and Ccl4 in cc. [n(Wt, 5w) = 6 mice, n(Ube3a−/−,5w) = 3 mice, n(Wt, 6w) = 6 mice, n(Ube3a−/−,6w) = 4 mice]. E, F Representative pictures (E) and quantification (F) of myelin basic protein (MBP) staining of the cc. Scale bar 100 µm. [n(Wt, 5w) = 6 mice, n(Ube3a−/−,5w) = 3 mice, n(Wt, 6w) = 6 mice, n(Ube3a−/−,6w) = 4 mice]. All data were represented as mean ± SEM. Statistical significance was determined by two-sided one-way ANOVA (B, C, F) or two-sided Student’s t-test (D). *P < 0.05 and **P < 0.01. If p values are not indicated, the result was not significant.
Discussion
Macrophages display tremendous phenotypical plasticity in vivo, a reflection of dynamic intracellular and environmental changes. In support of this notion, myelin-containing foamy macrophages show spatiotemporal-dependent alterations in their functional properties in CNS disorders1,2,8,9. While they initially adopt an anti-inflammatory wound-healing phenotype upon exposure to myelin, this benign phenotype is only transient. Excessive intracellular accumulation of myelin directs these foam cells toward a disease-promoting phenotype by perturbing cellular processing of myelin-derived lipids. In this study, we provide a molecular rationale for impaired processing of myelin-derived lipids and the consequent induction of a disease-promoting macrophage phenotype. We show that UBE3A-mediated ubiquitination and degradation of the cholesterol efflux transporter ABCA1 underlie metabolic disturbances and cause the induction of inflammatory foamy macrophages. UBE3A deficiency rescued metabolic deficits in foamy macrophages, thereby markedly enhancing remyelination ex vivo and in vivo. In addition, we propose a molecular model for the cellular localization of UBE3A, in which TIP30 sequesters UBE3A in the cytoplasm by inhibiting importin β-mediated nuclear transport. Collectively, our findings provide increased insight into the metabolic regulation of ABCA1 and foamy macrophage physiology, and identify UBE3A as a promising therapeutic target to promote repair in demyelinating disorders.
ABCA1 is a key facilitator of lipid efflux, thereby maintaining cellular lipid homeostasis. Emerging evidence indicates that ABCA1 is highly regulated on a post-translational level through multiple degradative systems46,47. We find that proteasomal and lysosomal degradation affect ABCA1 turnover in control BMDMs. This is in line with the study by Iborra et al., which showed that an advanced glycation end-product albumin-driven decline of ABCA1 in THP-1 macrophages can be rescued by inhibition of the proteasome, or partially restored upon lysosomal inhibition17. Similar to our findings, calpain inhibition was not able to rescue ABCA1 levels in these macrophages. However, we show that ABCA1 abundance in myelin-containing macrophages depends solely on proteasomal degradation. Accordingly, Hsieh et al. previously reported that ABCA1 turnover is strongly inhibited by proteasomal and not lysosomal inhibitors in cholesterol-stimulated macrophages20. While the molecular mechanisms underlying this discrepancy remain unclear, compromised lysosomal function—as previously demonstrated in foam cells in the CNS9—may force myelin-laden macrophages to rely exclusively on proteasomal degradation for ABCA1 turnover. Alongside ABCA1, the proteasome can control the turnover of cholesterol efflux transporter ABCG114,20. Yet, in contrast to ABCA1, ABCG1-mediated efflux and ABCG1 surface abundance were not affected in macrophages upon prolonged myelin ingestion (data not shown). These findings argue for the UPS machinery targeting ABCA1 and ABCG1 for degradation in a specific and mechanistically divergent manner.
UPS-mediated proteasomal degradation requires the hierarchical action of E1 activating enzymes, E2 conjugating enzymes, and E3 ubiquitin-ligases23,24, with the latter group primarily controlling target specificity14. Guided by transcriptomic analysis, gain- and loss-of-function experiments, and ubiquitination assays, we show that the E3 ubiquitin-ligase UBE3A promotes ABCA1 ubiquitination and degradation in macrophages. UBE3A was first identified as a protein interacting with the E6 oncoprotein of human papillomavirus types 16 and 18, thereby facilitating p53 degradation by the UPS48,49. However, more recent studies show that UBE3A displays ubiquitination activity in the absence of the viral E6 protein as well, and, by doing so, controls ubiquitination and turnover of RING1B, Mitogen-Activated Protein Kinase 1 (MAPK1), UV excision repair proteins HHR23A and HHR23B, and even itself 50,51,52. Consistent with our findings, few studies also identified an essential role of UBE3A in cellular lipid metabolism. For instance, UBE3A inhibits adipogenesis by promoting C/EBPα ubiquitination and attenuates overnutrition-induced obesity and fatty liver formation through degradation of epigenetic transcriptional coactivator MLL453. By using orthogonal ubiquitin transfer to identify ligation substrates, UBE3A was further suggested to ubiquitinate lipid droplet-regulating protein UBXD8, acetyl-CoA acetyltransferase 1 (ACAT1), and ABCE1, a family member of ABCA150. Our findings now provide evidence that ABCA1 is a ligation target of UBE3A as well, thereby controlling the metabolic and inflammatory properties of foam cells. Of note, by using cholesterol-loaded macrophages, the E3 ubiquitin-ligase HECTD1 was recently found to control ABCA1 turnover and apoA-I-mediated cholesterol efflux as well54, arguing for context-dependent regulation of ABCA1 ubiquitination and turnover in divergent foam cell subsets. Interestingly, we find that ABCA1 is predominantly monoubiquitinated by UBE3A in the E. coli system, which is generally associated with protein sorting and trafficking55. Hence, future studies are warranted to define if UBE3A impacts the subcellular localization of ABCA1. Alongside controlling protein trafficking, monoubiquitination can serve as a signal for poly-ubiquitin chain conjugation by other E3 ligase enzymes, so-called E4 ubiquitin-ligases. Hence, it is possible that next to UBE3A, a second E3 ligase is involved in the polyubiquitination and subsequent degradation of ABCA1.
Our findings indicate that the absence of Ube3a impacts the transcriptome of macrophages. Specifically, we provide evidence that Ube3a expression is positively associated with Tip30 in macrophages. TIP30 is an oxidoreductase that acts as a tumor-suppressor gene, thereby controlling cell apoptosis, growth, and metastasis39. However, it also functions as a negative regulator of nuclear import by binding to members of the importin family40. Accordingly, we show that silencing of Tip30 enhances nuclear localization of UBE3A in an importin β-mediated manner, and, in parallel, reduces cellular lipid load. These findings strongly suggest that TIP30 drives cellular UBE3A localization, thereby controlling ABCA1 abundance and cellular lipid load in foam cells. Of note, Liao et al. identified TIP30 as a regulator of cholesterol metabolism by stimulating the incorporation of fatty acids into esterified cholesterol in lipid-laden hepatocytes56. Hence, future studies should define whether TIP30 controls lipid droplet formation in foam cells as well. Recent work on UBE3A localization revealed that UBE3A is imported into the nucleus via its AZUL domain, which is present in both UBE3A-Iso2 and UBE3A-Iso3. However, UBE3A-Iso2 has a cytosolic localization due to the presence of an N-terminal extension that is responsible for active export of UBE3A back to the cytoplasm. As mUBE3A-Iso3 lacks this sequence, this short isoform has a predominant nuclear localization. Nuclear import of both isoforms required the binding of UBE3A to PSMD4 via its AZUL domain. Yet, neither UBE3A, nor PSMD4, contain a nuclear localization signal, which is required for importin-mediated nuclear trafficking. Hence, it is possible that the complex enters the nucleus in a piggy-back way by binding to 19S RP or 26S holoenzyme, a mechanism proposed by ref. 27. As such, importin β could bind one of these proteins and thereby direct the nuclear import of UBE3A. In aggregate, UBE3A may direct its own cellular localization in macrophages by regulating Tip30 transcription. Further research is necessary to unravel how UBE3A controls Tip30 expression, and how importin β drives nuclear import of UBE3A.
We show that global Ube3a deficiency increases remyelination in the cuprizone and cerebellar brain slice models. Moreover, macrophage-specific Ube3a deficiency in brain slices highlighted that UBE3A promotes remyelination in a phagocyte-specific manner, and that both peripherally-derived macrophages as well as brain-resident microglia contribute to the protective effect of loss of UBE3A on remyelination ex vivo. Enhanced remyelination was associated with reduced foam cell formation and higher ABCA1 levels, which strongly suggests that UBE3A-mediated ABCA1 degradation and consequent lipid accumulation hamper CNS repair8,9. In support of this notion, the absence of UBE3A increased ABCA1 abundance and activity, reduced cellular lipid droplet load, and induced a macrophage phenotype associated with repair in vitro. Our findings align with previous studies by us and others demonstrating a critically protective role for phagocytic cholesterol efflux during remyelination, and further characterize UBE3A as a regulator of this process8,9,12. However, given the importance of UBE3A in controlling neuronal function, we cannot exclude that global UBE3A deficiency impacts remyelination only through altering foam cell physiology. With respect to the latter, UBE3A has been extensively studied in neurons in which the gene is maternally imprinted, and genetic changes in UBE3A are linked to the neurodevelopmental disorders Angelman Syndrome (maternal allele deletion) and autism spectrum disorder (duplication of maternal allele)57,58,59. Furthermore, changes in TIP30 abundance are associated with impaired oligodendrocyte precursor cell (OPC) differentiation through sequestering Olig1 and Notch1 in the cytoplasm60,61, thereby likely playing an essential role in remyelination. Hence, future studies employing cell type-specific UBE3A knockout models will be helpful to assess the contribution of distinct individual phagocyte populations to the remyelination process in vivo. Further, precautions should be taken when targeting UBE3A in future remyelinating therapies, and phagocyte-specific interventions should be developed. Currently, no small-molecule inhibitors for UBE3A are available, and more research is warranted to provide macrophage-specific therapies to target this E3 ligase in diseases characterized by the presence of foamy macrophages.
In aggregate, we demonstrate a crucial role for the UPS in foamy macrophage biogenesis, and identify ABCA1 as a bona fide ligation target of UBE3A. Loss of UBE3A was able to reduce macrophage lipid load, dampen the inflammatory phenotype, and improve remyelination. In conclusion, we identify UBE3A as a promising therapeutic target to promote remyelination in neurological disorders characterized by the presence of foamy macrophages.
Methods
All studies comply with the relevant ethical regulations. Experiments using wild-type, Ube3a−/− and Ube3aOE animals were performed at the Erasmus MC and at Hasselt University and were in accordance with, respectively, the European Commission Council Directive 2010/63/EU, in accordance with the institutional guidelines, and approved by the Ethical Committee for Animal Experiments of Hasselt University (AVD101002016791, 201802, 202436K, 202437, and 202438). All experimental protocols using human monocyte-derived macrophages were conducted in accordance with institutional guidelines and approved by the Medical Ethical Committee of Hasselt University (UH-IMMVET-P1). Written informed consent was obtained from all participants included in this study.
Animals
All animals used in this study were housed on a 12:12 h light:dark cycle, in 19–24 °C, 40–60% humidity, and provided ad libitum access to food and water. Previously established AS (Ube3am-/p+) and Ube3a overexpression (Ube3a+2, referred to in results as Ube3aOE)26 mouse models were used in this study. Heterozygous Ube3a+/− males and females from the 129sv strain were crossed to obtain homozygous Ube3a−/− mice. Ube3a overexpressing (Ube3a+2/Ube3aOE) mice were generated by crossing heterozygous 129sv Ube3a+2 males with wild-type females. Wild-type littermates were used as controls with equal ratios of male:female animals per group.
Bone marrow-derived macrophages
Bone marrow cells were harvested from wildtype, Ube3a−/−, and Ube3a-overexpression (Ube3aO.E.) mice by flushing the femurs and tibias with PBS. Bone marrow-derived macrophages (BMDMs) were obtained by culturing isolated cells for 7 days in BMDM culture medium (RPMI medium supplemented with 10% fetal calf serum (FCS), 1% P/S, and 15% L929-conditioned medium (LCM)) at 37 °C, 5% CO2. Differentiated cells were harvested using PBS/EDTA (10 mM) and plated at 0.5 × 106 cells/ml in BMDM culture medium containing 5% LCM, and were used for in vitro experiments.
Human monocyte-derived macrophages
Human monocyte-derived macrophages (hMDMs) of healthy donors were isolated as described previously62. Briefly, peripheral blood mononuclear cells were isolated from whole blood by density-gradient centrifugation on lympholyte-H cell separation media (Cedarlane, CL5015-R). Blood samples were collected from healthy controls after obtaining informed written consent. Subjects with signs of infection were excluded. CD14+ monocytes were collected using the EasySep human CD14 positive selection kit (Stemcell Technologies, 17858) according to the manufacturer’s instructions. Isolated CD14+ cells were differentiated towards macrophages in IMEM supplemented with 10% human serum, 1% P/S, and 1% L-glutamine at 37 °C, 5% CO2. After 6 days, adherent macrophages were harvested, plated at 2 × 106 cells/ml, treated with human myelin (50 µg/ml) for 24 or 72 h, and lysed for qPCR analysis.
Microglia
Microglia were isolated from wild-type and Ube3a−/− P1-3 C57BL/6j mouse pups. Cortices were isolated, meninges removed, minced, and dissociated for 20 min at 37 °C with papain and DNase I (both 20 µg/mL, Sigma-Aldrich, 9001-73-4 and 10104159001). The resulting mixed glial cultures were seeded in poly-L-lysine (PLL, 50 µg/ml, Sigma, 25988-63-0)-coated T75 flasks and cultured in DMEM supplemented with 10% FCS, 1% P/S, and 33% LCM at 37 °C, 5% CO2. Mixed glial cultures were separated after 14 days by mechanical shaking at 230 rpm for 3 h at 37 °C, and afterward plated at 0.5 × 106 cells/ml in DMEM supplemented with 10% FCS, 1% P/S, and 15% LCM at 37 °C, 5% CO2.
Myelin isolation
Myelin was purified from human and mouse postmortem brain tissue with density-gradient centrifugation, as described previously63. Pierce BCA Protein Assay kit (Thermo Fisher Scientific, A55864) was used to determine myelin protein concentration, according to the manufacturer’s guidelines.
Cell treatment
Myelin-containing macrophages were generated by treating mouse BMDMs and human MDMs with, respectively, mouse or human myelin (50 µg/ml) for 24 or 72 h. To measure the expression of inflammatory markers or nitric oxide (NO) production, myelin-containing BMDMs were treated with lipopolysaccharide (LPS, 100 ng/µl, Sigma, 437627) for respectively 6 or 18 h prior to cell and supernatant collecting. BMDMs were treated with MG132 (25 µM, Enzo Life Sciences, BML-PI102-0025), Bafilomycin A1 (10 nM, InvivoGen, tlrl-baf1), Rottlerin (3 µM, Sigma, 557370), or Calpeptin (10 µg/ml, Abcam, ab120804) to respectively inhibit proteasomal degradation, lysosomal degradation, PKCδ-phosphorylation, and calpain. BMDMs were treated with cyclohexamide (50 µg/ml, Sigma, C7698) to inhibit protein translation. HEK293T cells were treated with importazole (20 µM, Abcam, ab146155) for 24 h to inhibit importin β-mediated nuclear import.
Transfection of HEK293T cells
HEK293T cells (300189) were cultured in DMEM medium supplemented with 10% FCS and 1% P/S (complete medium) at 37 °C, 5% CO2. One day before transfection, cells were plated at 0.3 × 106 cells/ml. For overexpression of UBE3A constructs (generated as described previously27), 2 µg (for one well of a six-well plate) or 0.5 µg (for one well of a 24-well plate) DNA was diluted in respectively 200 or 50 µl complete medium. Polyethylenimine (PEI, Polysciences, 23966) was added to the mix in a DNA:PEI ratio of 1:3. For silencing of TIP30, 100 nM Tip30 siRNA or 10 nM negative control siRNA mixes were prepared containing PEI in a siRNA:PEI ratio of 1:1. Suspension was briefly agitated and left to incubate at RT for 15 min before addition to the cells. About 300 µl mix was added to a well of a six-well plate, and 75 µl mix to a well of a 24-well plate. The medium was refreshed after 6–8 h. Twenty-four hours post-transfection, cells were collected for immunoblotting, or fixed by adding 4% PFA (Sigma, 158127) and washed in triplicate with PBS. After blocking non-specific staining using Dako protein block (Agilent, X0909), cells were incubated overnight at 4 °C with primary antibodies diluted in PBS. The following antibodies were used: anti-ABCA1 (1:250, ab18180, Abcam) and anti-UBE3A (1:500, SAB1404508, Sigma). Following overnight incubation, coverslips were washed three times with PBS and incubated for 1 h at RT with relevant secondary antibodies purchased from Invitrogen. Cells were imaged using an LSM 880 Confocal microscope (Zeiss) using the Airyscan feature. Image J version 1.52c was used for quantification. Pictures indicated in the figures are digitally enhanced.
Transfection of U2OS cells
U2OS cells (ATCC HTB-96) were cultured in DMEM medium supplemented with 10% FCS and 1% P/S (complete medium) at 37 °C, 5% CO2. One day before transfection, 1.0 × 105 cells were seeded in a 12-well cell culture plate containing a glass coverslip. Cells were transfected with one of the following expression plasmids: m(mouse)UBE3A-Iso3, mUBE3A-Iso2, and r(at)TIP30, or a combination of mUBE3A-Iso3:rTIP30 or mUBE3A-Iso2:rTIP30 in a ratio of 1:1 or 1:3. UBE3A expression plasmids were described previously27 and the rTIP30 expression plasmid was a kind gift of Dr. Abel Martin Garrido. On the day of transfection, a total of 1 µg of the desired construct(s) was combined with PEI transfection agent in a ratio of 1:3 in DMEM (Gibco). Suspension was briefly agitated and left to incubate at RT for 15 min before addition to the cells (150 µl mix per well). Transfection was carried out for 4 h at standard culture conditions in a humidified incubator (Heracell, 37 °C; 5% CO2). Following incubation, the PEI-containing medium was removed and replaced with fresh DMEM (Gibco, supplemented with 10% FBS/1% PS). Twenty-four hours post-transfection, cells were fixed by adding 4% PFA (Sigma, 158127)/sucrose (Sigma, S0389) and washed in triplicate with PBS (Capricorn Life Sciences, PBS-1A). Cells were subsequently used for immunostaining by incubating them overnight at 4 °C with a primary antibody mix containing anti-UBE3A (1:500, Sigma, SAB1404508), anti-TIP30 (1:250, Proteintech, 10251-1 AP), and anti-nucleoporin (1:500, Biolegend, mAb-414) dissolved in GDB solution (0.1% (w/v) gelatin, 0.3 (v/v) Triton X-100 (Sigma, X100), 450 mM NaCl, 16 mM phosphate buffer [pH 7.4]). Following overnight incubation, coverslips were washed three times with PBS and incubated for 1 h at RT with secondary antibody mix consisting of anti-mouse Alexa Fluor 488 (1:200, Jackson ImmunoResearch, 711-545-152) and anti-rabbit Alexa Fluor 647 (1:200, Jackson ImmunoResearch, 711-604-152) dissolved in GDB solution. After incubation, coverslips were washed three times for 5 min in PBS and directly mounted in ProLongGold +DAPI (Invitrogen, P36931). Coverslips were kept at 4 °C until imaging on a Zeiss LSM700 confocal microscope using a 20x objective and a zoom factor of 1.00.
Transfection of human monocyte-derived macrophages
hMDMs were cultured in IMIM supplemented with 10% human serum, 1% P/S, and 1% L-glutamine at 37 °C, 5% CO2. Knockdown of UBE3A was performed as described previously64. Specifically, UBE3A was targeted by ON-TARGETplus Human UBE3A siRNA (Dharmacon, L-005137-00-0010) and controlled by scRNA of ON-TARGETplus Non-targeting Pool (Dharmacon, D-001810-10-20). About 250 nM siRNA and scRNA mixes were prepared in hMDM medium and incubated for 5 min at RT. DharmaFECT Transfection reagent 4 (T-2005-01) was diluted 1/100 in hMDM medium and added 1:1 to 250 nM siRNA/scRNA and subsequently incubated for 20 min at RT. Mixed scRNA/siRNA and transfection reagent were then diluted 1/5 with hMDM medium with 50 ng/ml M-CSF (Miltenyi, 130-096-491). Old medium was removed, and 1000, 500, or 100 µl of siRNA/scRNA medium was added into, respectively, 6-well, 24-well, or 96-well plates and incubated with cultured cells. The medium was refreshed 48 h post-transfection, and cells were subsequently stimulated for experiments with myelin (50 µg/ml) for 72 h. To measure the expression of inflammatory markers or IL1β production, myelin-containing BMDMs were treated with LPS (100 ng/µl, Sigma, 437627) for respectively 6 or 18 h prior to cell and supernatant collecting.
RNA extraction and real-time quantitative PCR
Total RNA from cultures was isolated using Qiazol lysis reagent (Qiagen, 79306) and the RNeasy mini kit (Qiagen, 74104), according to the manufacturer’s guidelines. Complementary DNA was synthesized using qScriptTM cDNA SuperMix (Quanta Biosciences, 95048) according to the manufacturer’s instructions. Real-time quantitative PCR was conducted on a Step One Plus detection system (Applied Biosystems). Cycle conditions were 95 °C for 20 s, followed by 40 cycles of 95 °C for 3 s, and 60 °C for 30 s. The PCR reaction mixture contained SYBR green master mix (Thermo Fisher Scientific, 4312704), 0.3 μM forward and reverse primer (IDT technologies), RNase-free water, and 12.5 ng cDNA template. Data were analyzed using the comparative Ct method and normalized to the most stable reference genes. Primer sequences used for real-time quantitative PCR are depicted in Table S3.
Immunoblotting
Cells were lysed with radioimmunoprecipitation assay buffer (150 mM NaCl, 50 mM Tris (Sigma, T1503), 1% SDS, 1% Triton X-100, and 0.5% sodium deoxycholate) containing protease inhibitor (Roche, 04693116001). Protein concentrations of the cell lysates were determined using a Pierce BCA Protein Assay kit (Thermo Fisher Scientific, A55864) according to the manufacturer’s guidelines and afterwards labeled with Cy5 using the Quickstain kit (dDbioLab, RPN4000) according to the manufacturer’s guidelines. Approximately 20 µg of protein from cells or 0.3 OD units of bacterial lysates were separated on an SDS-polyacrylamide gel and transferred to a membrane (cells onto a polyvinylidene (PVDF) membrane; bacterial lysates onto a nitrocellulose membrane). PVDF and nitrocellulose membranes were blocked for 1 h with 5% milk in, respectively, PBS or TBS containing 0.1% Tween-20 (Sigma, P6585), followed by overnight incubation with the relevant primary antibodies at 4 °C. The following antibodies were used: anti-ABCA1 (1:1000, NB400-105, Novus Biologicals), anti-UBE3A (1:1000, Proteintech, 10344-1-AP), monoclonal anti-V5 horseradish peroxidase (HRP)-conjugated (Thermo Fisher Scientific, R961-25), rat monoclonal anti-HA HRP-conjugated (Roche, 12013819001), anti-TIP30 (1:500, Proteintech, 10251-1-AP), and anti-β-actin (1:2000, Santa Cruz Biotechnology, sc-47778). Next, membranes were incubated for 1 h with appropriate horseradish peroxidase (HRP)-conjugated secondary antibodies purchased from Dako, and visualization was done with the Enhanced Chemiluminescence (GE Healthcare) technique using the Pierce ECL Plus Western Blotting Substrate (Thermo Fisher Scientific, 32134). Data were quantified using Image Quant TL software v8.1 (GE Healthcare Europe) and normalized to β-actin or Cy5 levels.
Flow cytometry
Single-cell suspensions were blocked with 10% serum and stained with ABCA1 primary antibody (1:1000, Novus Biologicals, NB400-105), followed by incubation with the appropriate secondary antibody. To assess cellular lipid load, cells were washed and incubated with BODIPY (2 µM, Thermo Fisher Scientific, D3922) for 15 min at 37 °C in the dark. Next, cells were washed and collected in PBS. The FACSCalibur (BD Biosciences) was used to quantify cellular fluorescence. Mean fluorescence intensity (MFI) was corrected for background MFI.
Co-immunoprecipitation of ABCA1
Cells were lysed with NP-40 buffer [150 mM NaCl, 0,5% NP-40 (Thermo Fisher Scientific, 85124), 50 mM Tris (Sigma, T1503), containing protease inhibitor (Roche, 04693116001)]. Protein concentrations of the cell lysates were determined using a Pierce BCA Protein Assay kit (Thermo Fisher Scientific, A55864) according to the manufacturer’s guidelines, and 1 mg protein was used as starting material. Samples were incubated overnight with 10 µg anti-ABCA1 (1:1000, Novus Biologicals, NB400-105) at 4 °C while shaking. The next day, protein/antibody mixtures were incubated with Pierce Protein A Magnetic Beads (Thermo Fisher Scientific, 88845) according to the manufacturer’s guidelines and incubated at room temperature for 1 h while shaking. Beads were magnetically separated, and flow-through was kept for immunoblotting analysis (unbound fraction). Next, beads were washed with TBS containing 0.1% Tween-20 (Sigma, P6585) and antigen was eluted from beads using 100 µl elution buffer (0,1 M glycine, G2879, pH 2.0) while mixing for 10 min at room temperature, after which 25 µl neutralization buffer (1 M Tris, pH 8.5, Sigma, T1503) was added. Input, elution (IP: ABCA1 fraction), and unbound fractions were analyzed for ABCA1, UBE3A, and Cy5 (loading control) using immunoblotting.
Ubiquitin capture
Wildtype and Ube3a−/− BMDMs were treated with myelin (50 µg/ml) for 72 h and lysed with radioimmunoprecipitation assay buffer [150 mM NaCl, 50 mM Tris (Sigma, T1503), 1% SDS (Sigma, L4509), 1% Triton X-100 (Sigma, X100), and 0.5% sodium deoxycholate (Millipore, 1.06504) containing protease inhibitor (Roche, 04693116001)]. Protein concentrations of the cell lysates were determined using a Pierce BCA Protein Assay kit (Thermo Fisher Scientific, A55864) according to the manufacturer’s guidelines. The extracted proteins were incubated with UBIQAPTURE-Q matrix (Enzo Life Sciences, BML-UW8995A-0001) to pull down all ubiquitinated proteins, following the manufacturer’s protocol. The UBIQAPTURE resins bind to mono- and poly-ubiquitinated proteins of different lysine linkages. Eluates were analyzed for ABCA1 using immunoblotting.
Apoptosis and necrosis induction in thymocytes
Apoptotic and necrotic thymocytes were prepared as described previously65. Briefly, thymi were collected from 4-week-old mice and thymocytes were isolated and cultured for 24 h in DMEM supplemented with 10% FCS and 100 U/mL P/S. Apoptotic thymocytes were generated by incubating cells in serum-free medium for 24 h, whereas necrotic thymocytes were obtained by incubation with 1 mM H2O2 for 24 h.
Cholesterol efflux measurement
To measure cholesterol efflux by myelin-treated BMDMs (50 µg/ml for 72 h), cells were exposed to apoA-I (50 µg/ml) in phenol- and serum-free medium for 4 h prior to measuring intracellular (cell lysates) and extracellular (supernatant) total cholesterol using the AmplexTM Red Cholesterol Assay Kit (Thermo Fisher Scientific, A12216), according to the manufacturer’s instructions. Cholesterol efflux was determined by dividing the fluorescence in the supernatants by the total fluorescence in supernatants and cells. Fluorescence was measured at an excitation wavelength of 540 nm and an emission wavelength of 590 nm using the FLUOstar Optima microplate reader (BMG Labtech).
Oil red O staining
BMDMs, brain slices, MS lesions, and cuprizone sections were fixed with 4% PFA (Sigma, 158127) for 15 min. Next, cells were stained with 0.3% Oil Red O (ORO, Merck, 1320-06-5) for 10 min and counterstained with nuclei stain haematoxylin. Analysis was carried out using a Leica DM 2000 LED microscope and ImageJ software.
Immunohistochemistry and image analysis
Snap-frozen brain material containing active MS lesions was obtained from the Netherlands Brain Bank (NBB, Amsterdam, Netherlands). About 10 µm cryostat sections were made of MS lesions and cuprizone tissue using a Leica CM3050 S cryostat (Leica Microsystems). Sections were fixed in ice-cold acetone and 70% ethanol for 10 and 5 min, respectively. Brain slices were fixed for 15 min with 4% PFA (Sigma, 158127). After blocking non-specific staining using Dako protein block (Agilent, X0909), sections were incubated overnight at 4 °C with relevant primary antibodies diluted in PBS. Cerebellar brain slices were stained by incubating with relevant antibodies diluted in blocking buffer [PBS + 5% horse serum (Life Technologies, S-HS-EU-015) + 0.1% Triton X-100 (Sigma, X100)]. The following antibodies were used: anti-UBE3A (1:500, Sigma, SAB1404508), anti-CD68 (1:100, Invitrogen, 14-0688), anti-ABCA1 (1:250, Abcam, ab18180), anti-MBP (1:250, Millipore, MAB386), anti-NF (1:1000, Abcam, ab8135), anti-F4/80 (1:100, Bio-Rad, MCA497G), and anti-ADFP (1:500, Abcam, ab52356). Appropriate secondary antibodies were purchased from Invitrogen. Nuclei were stained using 4,6′-diamidino-2-phenylindole (DAPI, Roche, 10236276001). Representative images of MS lesions and cuprizone sections were taken using a Leica DM2000 LED fluorescence microscope. The subdivision of the lesion rim and center was accomplished by means of an ORO and proteolipid protein (PLP) staining (rim, ORO+ cells and PLP+ extracellular myelin; center, ORO+ cells and no PLP+ extracellular myelin). Cerebellar brain slices were imaged using an LSM 880 Confocal microscope (Zeiss) using the Airyscan feature. Image J version 1.52c was used for quantification. Three-dimensional analysis of cerebellar brain slices was performed using the z-stack function on the confocal microscope, and images were rendered by the 3D rendering software vaa3d55. Pictures indicated in the figures are digitally enhanced.
Immunocytochemistry of BMDMs
Cells were fixed by adding 4% PFA (Sigma, 158127) and washed in triplicate with PBS. After blocking non-specific staining using Dako protein block (Agilent, X0909), cells were incubated overnight at 4 °C with primary antibodies diluted in PBS. The following antibodies were used: anti-ABCA1 (1:250, Abcam, ab18180), anti-ubiquitin (1:500, Sigma, SAB4503053), and anti-UBE3A (1:500, Sigma, SAB1404508). Following overnight incubation, coverslips were washed three times with PBS and incubated for 1 h at RT with relevant secondary antibodies purchased from Invitrogen. Cells were imaged using an LSM 880 Confocal microscope (Zeiss) using the Airyscan feature. Image J version 1.52c was used for quantification. Pictures indicated in the figures are digitally enhanced.
RNA sequencing and bioinformatics analysis
Total RNA was extracted from wild-type and Ube3a−/− BMDMs using the Direct-zol RNA miniprep kit (Zymo Research, R2050) and quantified fluorometrically with a Qubit 3 Fluorometer (Invitrogen). The Quality of the RNA was evaluated with a 2100 Bioanalyzer (Agilent Technologies). A KAPA mRNA hyperPrep kit (KAPA Biosystems) was used for mRNA library construction, and sequencing was performed on an Illumina HiSeq4000 at the Core Facility genomics of the Amsterdam UMC, location AMC, Amsterdam, the Netherlands. Bioinformatic analysis was performed in collaboration with the Bioinformatics Core Unit of the Amsterdam UMC, location AMC, Amsterdam, the Netherlands, as previously described in Clemente et al. Briefly, quality control (fastQC, dupRadar, Picard Tools) was performed on the reads. Subsequently, they were trimmed using Trimmomatic v0.32 and aligned to the mouse genome using HISAT2 (v2.1.0). Counts were obtained using corresponding GFTs and HTSeq (v0.11.0). TMM (EdgeR) and limma/voomR packages were used to perform statistical analysis. Count data was transformed to log2-counts per million (logCPM) and normalized using the trimmed mean of M-values method. Voom was used to precisely weigh the data. Bayes moderated t-test was used to assess differential expression within limma’s linear model framework, and the precision weights estimated by voom. P values were corrected for multiple testing using the Benjamini–Hochberg false discovery rate. BiomaRt (release 94) was used to reannotate the genes. Wild-type BMDM samples of the described RNAseq experiment were used for the analysis of mouse Ube3a transcripts as described previously34.
Escherichia coli ubiquitination assay
UBE3A-mediated ubiquitination was assessed in Escherichia coli cells as described previously27,29,30. In summary, BL21-GOLD (DE3) cells were co-transfected with bacterial expression constructs encoding E1, UbcH5c, ubiquitin, and UBE3A, and the substrates RING1B and ABCA1. Transformed cells were selected on LB agar (1% (w/v) Bacto tryptone, 0.5% (w/v) Bacto yeast extract, 1% (w/v) NaCl, and 1.5% agar) containing antibiotic (ampicillin, 25 μg/ml; kanamycin, 15 μg/ml; streptomycin/spectinomycin, 25 μg/ml). Next, single colonies were inoculated overnight in LB medium containing 2% glucose, 50 mM Tris/HCl (pH 8.0), and the needed antibiotics, while shaking (200 rpm) at 37 °C. The following morning, cells were inoculated at an OD600 of 0.2 and grown to an OD600 of 0.7 at 21 °C. Next, 0.5 mM isopropyl β-d-1-thiogalactopyranoside (IPTG, Thermo Fisher Scientific, B1900) was added to induce the cells, which were subsequently grown at 21 °C for 16–18 h. Bacterial pellets of 20 OD600 units were lysed in 0.5 ml cold lysis buffer [50 mM Sodium-phosphate pH 8.0 (Thermo Fisher Scientific, J60825.AK), 300 mM NaCl, 5% glycerol, 5 mM 2-Mercaptoethanol, 1 mM PMSF (Sigma, 329-98-6), RNase (0.01 mg/ml), DNase (0.01 mg/ml) with protease inhibitor cocktail (Roche, 04693116001)] by sonication (Sanyo soniprep). Debris was removed by centrifugation (13,000×g, 30 min), and cleared lysates were used for immunoblotting
NO determination
BMDMs seeded in a 48-well plate (1.25 × 105 cells/well) were treated with myelin followed by 18 h of LPS (100 ng/µl, Sigma, 437627) stimulation. Afterwards, the cell culture supernatants were collected and mixed with the Griess reagent (Sigma, 215-981-2) at a 1:1 ratio. After 15 min incubation at RT, the absorbance was measured at 540 nm using the FLUOstar optima microplate reader (BMG Labtech).
IL1β ELISA
Supernatant of hMDMs was collected after 18 h stimulation with LPS (100 ng/µl, Sigma, 437627). IL1β concentrations were determined using the Human IL-1 beta ELISA kit (Invitrogen, KHC0011) according to the manufacturer’s guidelines, and absorbance was measured at 450 and 570 nm using the FLUOstar optima microplate reader (BMG Labtech).
Brain slice cultures
Cerebellar slices were obtained from male and female wild-type and Ube3a−/− P9-P11 mouse pups as described previously8,66. Briefly, cerebellar parasagittal slices (300-µm thick) were cut on a MacIlwain tissue chopper and transferred onto membranes of 30-mm Millipore culture inserts with a 0.4-μm pore size (Millicell; Millipore). Slices were maintained in culture on top of the membranes in six-well plates containing 1 ml of medium [MEM (Thermo Fisher Scientific, 32360-026) supplemented with 25% horse serum (Life Technologies, S-HS-EU-015), 25% Hanks’ balanced salt solution (Invitrogen, 14025092), 1% P/S, 1% Glutamax (Sigma, 35050-038), 1.3% glucose (Sigma, G8644), and 1.1% 1 M HEPES (Thermo Fisher Scientific, 15630-080) at 37 °C, 5% CO2. For phagocyte depletion, slices were treated with clodronate or empty liposomes (0.5 mg/ml, LIPOSOMA) immediately after isolation for 24 h. Slices were repleted with 4 × 10³ LPS- (100 ng/µl, Sigma, 437627, 18 h) or IL4-stimulated (20 ng/µL, Peprotech, 214-14, 18 h) wild-type or Ube3a−/− BMDMs by adding them directly to the slice in a 1.5 µl drop, without touching the slice. Slices were left to recover for 2 days, after which demyelination was induced by treating the slices with lysolecithin (0.5 mg/ml, Sigma, 62963) for 16 h. Afterward, slices were washed and kept in culture for 6 days, followed by histological and biochemical analysis.
Cuprizone model
Acute demyelination was induced in wild-type and Ube3a−/− C57BL/6 mice (9–11 weeks old) by feeding them ad libitum a diet of 0.3% (w/w) cuprizone (bis(cyclohexanone)oxaldihydrazone, Sigma, 14690) mixed in normal chow for 5 weeks. Upon withdrawal of the cuprizone diet, spontaneous remyelination occurs. Corpus callosum was isolated and snap frozen after demyelination (5 weeks) and during spontaneous remyelination (6 weeks).
Statistics and reproducibility
All data are reported as mean ± standard error of the mean (SEM). Data were statistically analyzed using GraphPad Prism v6 (GraphPad Software). Normal distribution was tested using the D’Agostino–Pearson omnibus normality test. Normally distributed data sets were analyzed using an analysis of variance (ANOVA) or a two-tailed unpaired Student's t-test. Data sets that were not normally distributed were analyzed using the Kruskal–Wallis or Mann–Whitney analysis. No statistical method was used to predetermine sample size for in vitro experiments. The sample size for in vivo experiments was determined with the G power sample size calculation. Ex vivo and in vivo experiments were performed in a blinded fashion. The investigators were not blinded to allocation during experiments and outcome assessment of in vitro experiments. All statistical parameters, tests used, and reproducibility are reported in figure subscriptions and Source Data. *P < 0.05, **P ≤ 0.01, and ***P ≤ 0.001.
Data availability
The RNA sequencing data generated in this study has been deposited in the GEO database under accession code GSE232066. The raw data and uncropped Western blots generated in this study are provided. Source data are provided with this paper.
References
Grajchen, E., Hendriks, J. J. A. & Bogie, J. F. J. The physiology of foamy phagocytes in multiple sclerosis. Acta Neuropathol. Commun. 6, 124 (2018).
Yu, X. H., Fu, Y. C., Zhang, D. W., Yin, K. & Tang, C. K. Foam cells in atherosclerosis. Clin. Chim. Acta 424, 245–252 (2013).
Vanherle, S., Loix, M., Miron, V. E., Hendriks, J. J. A. & Bogie, J. F. J. Lipid metabolism, remodelling and intercellular transfer in the CNS. Nat. Rev. Neurosci. 26, 214–231 (2025).
Boven, L. A. et al. Myelin-laden macrophages are anti-inflammatory, consistent with foam cells in multiple sclerosis. Brain 129, 517–526 (2006).
Bogie, J. F. et al. Myelin alters the inflammatory phenotype of macrophages by activating PPARs. Acta Neuropathol. Commun. 1, 43 (2013).
Bogie, J. F. J., Haidar, M., Kooij, G. & Hendriks, J. J. A. Fatty acid metabolism in the progression and resolution of CNS disorders. Adv. Drug Deliv. Rev. 159, 198–213 (2020).
Spann, N. J. et al. Regulated accumulation of desmosterol integrates macrophage lipid metabolism and inflammatory responses. Cell 151, 138–152 (2012).
Bogie, J. F. J. et al. Stearoyl-CoA desaturase-1 impairs the reparative properties of macrophages and microglia in the brain. J. Exp. Med. 217, e20191660 (2020).
Cantuti-Castelvetri, L. et al. Defective cholesterol clearance limits remyelination in the aged central nervous system. Science 359, 684–688 (2018).
Garcia Corrales, A.V. et al. Fatty acid elongation by ELOVL6 hampers remyelination by promoting inflammatory foam cell formation during demyelination. Proc. Natl Acad. Sci. USA 120, e2301030120 (2023).
Chen, L., Zhao, Z. W., Zeng, P. H., Zhou, Y. J. & Yin, W. J. Molecular mechanisms for ABCA1-mediated cholesterol efflux. Cell Cycle 21, 1121–1139 (2022).
Vanherle, S. et al. The ApoA-I mimetic peptide 5A enhances remyelination by promoting clearance and degradation of myelin debris. Cell Rep. 41, 111591 (2022).
Bogie, J. F. et al. Myelin-derived lipids modulate macrophage activity by liver X receptor activation. PLoS ONE7, e44998 (2012).
Sharpe, L. J., Cook, E. C., Zelcer, N. & Brown, A. J. The UPS and downs of cholesterol homeostasis. Trends Biochem. Sci. 39, 527–535 (2014).
Lv, Y. C. et al. Posttranscriptional regulation of ATP-binding cassette transporter A1 in lipid metabolism. DNA Cell Biol. 32, 348–358 (2013).
Wang, N. et al. A PEST sequence in ABCA1 regulates degradation by calpain protease and stabilization of ABCA1 by apoA-I. J. Clin. Invest. 111, 99–107 (2003).
Iborra, R. T. et al. AGE-albumin enhances ABCA1 degradation by ubiquitin-proteasome and lysosomal pathways in macrophages. J. Diabetes Complications 32, 1–10 (2018).
Cheng, H. et al. Over-activation of NMDA receptors promotes ABCA1 degradation and foam cell formation. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1865, 158778 (2020).
Huang, L., Fan, B., Ma, A., Shaul, P. W. & Zhu, H. Inhibition of ABCA1 protein degradation promotes HDL cholesterol efflux capacity and RCT and reduces atherosclerosis in mice. J. Lipid Res. 56, 986–997 (2015).
Hsieh, V. et al. Cellular cholesterol regulates ubiquitination and degradation of the cholesterol export proteins ABCA1 and ABCG1. J. Biol. Chem. 289, 7524–7536 (2014).
Lv, Y. et al. Sortilin promotes macrophage cholesterol accumulation and aortic atherosclerosis through lysosomal degradation of ATP-binding cassette transporter A1 protein. Acta Biochim. Biophys. Sin.51, 471–483 (2019).
Mizuno, T., Hayashi, H., Naoi, S. & Sugiyama, Y. Ubiquitination is associated with lysosomal degradation of cell surface-resident ATP-binding cassette transporter A1 (ABCA1) through the endosomal sorting complex required for transport (ESCRT) pathway. Hepatology 54, 631–643 (2011).
Kleiger, G. & Mayor, T. Perilous journey: a tour of the ubiquitin-proteasome system. Trends Cell Biol. 24, 352–359 (2014).
Foot, N., Henshall, T. & Kumar, S. Ubiquitination and the regulation of membrane proteins. Physiol. Rev. 97, 253–281 (2017).
Mailleux, J. et al. Active liver X receptor signaling in phagocytes in multiple sclerosis lesions. Mult. Scler. 24, 279–289 (2018).
Punt, A.M. et al. Molecular and behavioral consequences of Ube3a gene overdosage in mice. JCI Insight 7, e158953 (2022).
Avagliano Trezza, R. et al. Loss of nuclear UBE3A causes electrophysiological and behavioral deficits in mice and is associated with Angelman syndrome. Nat. Neurosci. 22, 1235–1247 (2019).
Bossuyt, S. N. V. et al. Loss of nuclear UBE3A activity is the predominant cause of Angelman syndrome in individuals carrying UBE3A missense mutations. Hum. Mol. Genet. 30, 430–442 (2021).
Avagliano Trezza, R. et al. Mono-ubiquitination of Rabphilin 3A by UBE3A serves a non-degradative function. Sci. Rep. 11, 3007 (2021).
Geerts-Haages, A. et al. A novel UBE3A sequence variant identified in eight related individuals with neurodevelopmental delay, results in a phenotype which does not match the clinical criteria of Angelman syndrome. Mol. Genet. Genom. Med. 8, e1481 (2020).
van der Wal, L. et al. Improvement of ubiquitylation site detection by Orbitrap mass spectrometry. J. Proteom. 172, 49–56 (2018).
Burette, A. C. et al. Subcellular organization of UBE3A in human cerebral cortex. Mol. Autism 9, 54 (2018).
Burette, A. C. et al. Subcellular organization of UBE3A in neurons. J. Comp. Neurol. 525, 233–251 (2017).
Zampeta, F. I. et al. Conserved UBE3A subcellular distribution between human and mice is facilitated by non-homologous isoforms. Hum. Mol. Genet. 29, 3032–3043 (2020).
Ramamoorthy, S. & Nawaz, Z. E6-associated protein (E6-AP) is a dual function coactivator of steroid hormone receptors. Nucl. Recept Signal 6, e006 (2008).
Liu, W. et al. The RBPJ/DAPK3/UBE3A signaling axis induces PBRM1 degradation to modulate the sensitivity of renal cell carcinoma to CDK4/6 inhibitors. Cell Death Dis. 13, 295 (2022).
Owais, A., Mishra, R.K. & Kiyokawa, H. The HECT E3 ligase E6AP/UBE3A as a therapeutic target in cancer and neurological disorders. Cancers 12, 2108 (2020).
Viho, E. M. G. et al. The hippocampal response to acute corticosterone elevation Is altered in a mouse model for Angelman syndrome. Int. J. Mol. Sci. 24, 303 (2023).
Yu, X., Li, Z. & Wu, W. K. TIP30: a novel tumor-suppressor gene. Oncol. Res. 22, 339–348 (2014).
King, F. W. & Shtivelman, E. Inhibition of nuclear import by the proapoptotic protein CC3. Mol. Cell Biol. 24, 7091–7101 (2004).
Drutovic, D., Duan, X., Li, R., Kalab, P. & Solc, P. RanGTP and importin β regulate meiosis I spindle assembly and function in mouse oocytes. EMBO J. 39, e101689 (2020).
Li, X. et al. Nuclear translocation of annexin 1 following oxygen-glucose deprivation-reperfusion induces apoptosis by regulating Bid expression via p53 binding. Cell Death Dis. 7, e2356 (2016).
Tall, A. R. & Yvan-Charvet, L. Cholesterol, inflammation and innate immunity. Nat. Rev. Immunol. 15, 104–116 (2015).
Yvan-Charvet, L. et al. Increased inflammatory gene expression in ABC transporter-deficient macrophages: free cholesterol accumulation, increased signaling via toll-like receptors, and neutrophil infiltration of atherosclerotic lesions. Circulation 118, 1837–1847 (2008).
Miron, V. et al. Central nervous system regeneration is driven by microglia necroptosis and repopulation. Nat. Neurosci. 22, 1046–1052 (2019).
Ogura, M. et al. Proteasomal inhibition promotes ATP-binding cassette transporter A1 (ABCA1) and ABCG1 expression and cholesterol efflux from macrophages in vitro and in vivo. Arterioscler. Thromb. Vasc. Biol. 31, 1980–1987 (2011).
Wang, J. et al. Role of ABCA1 in cardiovascular disease. J. Pers. Med. 12, 1010 (2022).
Scheffner, M., Huibregtse, J. M., Vierstra, R. D. & Howley, P. M. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell 75, 495–505 (1993).
Huibregtse, J. M., Scheffner, M. & Howley, P. M. Cloning and expression of the cDNA for E6-AP, a protein that mediates the interaction of the human papillomavirus E6 oncoprotein with p53. Mol. Cell Biol. 13, 775–784 (1993).
Wang, Y. et al. Identifying the ubiquitination targets of E6AP by orthogonal ubiquitin transfer. Nat. Commun. 8, 2232 (2017).
Zaaroor-Regev, D. et al. Regulation of the polycomb protein Ring1B by self-ubiquitination or by E6-AP may have implications to the pathogenesis of Angelman syndrome. Proc. Natl Acad. Sci. USA 107, 6788–6793 (2010).
Kumar, S., Talis, A. L. & Howley, P. M. Identification of HHR23A as a substrate for E6-associated protein-mediated ubiquitination. J. Biol. Chem. 274, 18785–18792 (1999).
Pal, P. et al. E3 ubiquitin ligase E6AP negatively regulates adipogenesis by downregulating proadipogenic factor C/EBPalpha. PLoS ONE8, e65330 (2013).
Aleidi, S. M. et al. The E3 ubiquitin ligase, HECTD1, is involved in ABCA1-mediated cholesterol export from macrophages. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1863, 359–368 (2018).
Ronai, Z. A. Monoubiquitination in proteasomal degradation. Proc. Natl Acad. Sci. USA 113, 8894–8896 (2016).
Liao, B. M. et al. Proteomic analysis of livers from fat-fed mice deficient in either PKCdelta or PKCepsilon identifies Htatip2 as a regulator of lipid metabolism. Proteomics 14, 2578–2587 (2014).
Silva-Santos, S. et al. Ube3a reinstatement identifies distinct developmental windows in a murine Angelman syndrome model. J. Clin. Invest. 125, 2069–2076 (2015).
Wang, T., Van Woerden, G. M., Elgersma, Y. & Borst, J. G. G. Enhanced transmission at the calyx of held synapse in a mouse model for Angelman syndrome. Front. Cell Neurosci. 11, 418 (2017).
Lopez, S. J., Segal, D. J. & LaSalle, J. M. UBE3A: an E3 ubiquitin ligase with genome-wide impact in neurodevelopmental disease. Front. Mol. Neurosci. 11, 476 (2018).
Nakahara, J., Kanekura, K., Nawa, M., Aiso, S. & Suzuki, N. Abnormal expression of TIP30 and arrested nucleocytoplasmic transport within oligodendrocyte precursor cells in multiple sclerosis. J. Clin. Invest. 119, 169–181 (2009).
Yang, W. et al. TIP30 inhibits oligodendrocyte precursor cell differentiation via cytoplasmic sequestration of Olig1. Glia 63, 684–698 (2015).
Bogie, J. F. et al. Scavenger receptor collectin placenta 1 is a novel receptor involved in the uptake of myelin by phagocytes. Sci. Rep. 7, 44794 (2017).
Norton, W. T. & Poduslo, S. E. Myelination in rat brain: method of myelin isolation. J. Neurochem.21, 749–757 (1973).
Verberk, S. G. S. et al. Monocyte and macrophage profiles in patients with inherited long-chain fatty acid oxidation disorders. Biochim. Biophys. Acta Mol. Basis Dis. 1871, 167524 (2025).
Budai, Z. et al. Macrophages engulf apoptotic and primary necrotic thymocytes through similar phosphatidylserine-dependent mechanisms. FEBS Open Bio 9, 446–456 (2019).
Haidar, M. et al. Targeting lipophagy in macrophages improves repair in multiple sclerosis. Autophagy 18, 2697–2710 (2022).
Acknowledgements
We thank MP Tulleners, L Timmermans, L Van Otterdijk, and L Dusaer for excellent technical assistance. We thank I Zampeta and E Niggl for the help with the isoform expression analysis. We thank Ben Philpot for the UBE3AOE mice. The graphical abstract was made using Biorender. The work was supported by the Flemish Fund for Scientific Research (FWO Vlaanderen: 1141920N, 1S15519N), the special research fund UHasselt (BOF), Charcot Research Foundation (CHARCOT23BJ, CHARCOT24BJ), Geneeskundige Stichting Koningin Elisabeth (GSKE-BOGJ), MS Liga Vlaanderen (MSLIGABOGJ). N.Z. is supported by a Vici grant from the Netherlands Organization for Scientific Research (NWO; 016.176.643). Y.E. was funded by the Angelman Syndrome Foundation (ASF).
Author information
Authors and Affiliations
Contributions
M.L., B.D., Y.E., M.H., N.Z., J.J.A.H., and J.F.J.B. conceived experiments. M.L., S.V., L.B., S.G.S.V., F.W., B.M., M.P., and R.V. performed experiments. M.L., M.P., and A.J. analyzed data. M.L., M.P., S.V. S.V.W., S.G.S.V., B.D.P., Y.E., M.H., N.Z., J.J.A.H., and J.F.J.B. discussed results. M.L., J.J.A.H., and J.F.J.B. wrote the manuscript. All authors revised the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Loix, M., Vanherle, S., Bolkaerts, L. et al. UBE3A promotes foam cell formation and counters remyelination by targeting ABCA1 for proteasomal degradation. Nat Commun 16, 8077 (2025). https://doi.org/10.1038/s41467-025-62053-w
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-62053-w