Abstract
The N6-methyladenosine (m6A) modification in U6 snRNA, catalyzed by METTL16 using S-adenosylmethionine (SAM) as the methyl donor, is required for efficient and accurate pre-mRNA splicing. However, the mechanism by which METTL16 modifies U6 snRNA with m6A remains elusive. Here, we present cryo-EM structures of METTL16 in complex with U6 snRNA, providing insights into the METTL16-mediated modification of U6 snRNA with m6A. The structures reveal that U6 snRNA is recruited to METTL16 through specific interactions between the C-terminal kinase-associated 1 (KA-1) domain of METTL16 and the internal stem-loop (ISL) of U6 snRNA. Upon SAM binding to the catalytic pocket within the N-terminal methyltransferase domain (MTD), U6 snRNA undergoes a structural rearrangement that positions the target adenine-containing motif at the catalytic site. This conformational change is followed by an additional structural adjustment of U6 snRNA into a productive conformation, bringing the target adenosine closer to SAM within the catalytic pocket and thereby ensuring efficient m6A modification. The KA-1 domain functions as a scaffold for initial substrate recognition and facilitates the subsequent dynamic methylation process within the MTD, highlighting the cooperative roles of METTL16 domains for U6 snRNA modification.
Similar content being viewed by others
Introduction
N 6-methyladenosine (m6A) is a well-characterized mRNA modification crucial for gene expression in various biological processes1,2,3,4. It regulates RNA splicing, localization, stability, and translation5,6,7,8,9.
Most m6A modifications in mRNAs are catalyzed by the METTL3/METTL14 complex, which methylates the specific adenosine in the DRACH (D = A, G or U, R = A or G; H = A, C, or U) motif close to the stop codon or 3’-UTR of mRNAs2,3,10,11,12,13. METTL3 acts as the catalytic subunit, and METTL14 plays a structural role critical for substrate recognition14,15,16.
A distinct m6A methyltransferase, METTL16, targets a particular motif within specific RNA structures, including mRNAs and non-coding RNAs13,17,18,19,20,21. In vertebrates, METTL16 installs m6A in RNA hairpins containing the motif UACA*GAR, where A* denotes the methylation site, within the 3′ UTR of methionine adenosyltransferase 2A (MAT2A) mRNA, regulating S-adenosylmethionine (SAM) homeostasis by modulating MAT2A pre-mRNA stability and splicing17,18,22. Under high SAM conditions, the YTH domain-containing protein YTHDC1 binds to the m6A-modified hairpins, promoting MAT2A mRNA degradation and reducing SAM synthesis17. In contrast, under SAM-limited conditions, prolonged METTL16 binding to the 3′ UTR hairpin recruits splicing factors, enhancing pre-mRNA splicing and increasing SAM production18,23. In invertebrates such as C. elegans, the METTL16 homolog, METT10, methylates the adenosine in the AG dinucleotide within the UACA*GAGA motif at the 3′-splice site (3′-ss) of SAM synthetase (sams) mRNA, affecting spliceosome assembly and splicing efficiency24,25. High SAM levels sustain the m6A modification at this site, preventing U2AF35 binding and inhibiting splicing26, and thereby suppressing SAM production. Conversely, low SAM levels reduce the m6A modification, allowing U2AF35 to bind, promoting splicing and enhancing SAM synthetase mRNA expression, and ultimately increasing SAM production.
METTL16 and its homologs also methylate a conserved adenosine in the ACA*GAGA motif of U6 snRNA (A43 in human U6 snRNA)19,27,28. The ACA*GAG box in U6 snRNA base pairs with the 5′ splice site during pre-mRNA splicing29,30,31. The m6A modification in U6 snRNA is required for efficient and accurate splicing of specific pre-mRNAs that possess the BBH motif (B: G, C, or U; H: A, C, or U) at positions −3 to −1 and an A at position 4 (A+4) of the 5′ splice site (5′-ss)32,33,34. In spliceosomal B complex, the m6A in U6 snRNA and the 5’-ss A+4 form a trans Hoogsteen sugar edge interaction35, thereby stabilizes the U6 snRNA-5’-ss interaction34. The BBH motif at the 5′-ss in pre-mRNAs weakly interact with U5, and thus the absence of m6A in U6 snRNA further destabilizes the interaction between the 5′-ss and U6 snRNA, resulting in intron retention within these specific pre-mRNAs32.
METTL16 consists of an N-terminal methyltransferase domain (MTD) and a C-terminal kinase-associated 1 (KA-1) domain36,37. The KA-1 domain was initially named the vertebrate conserved region (VCR) but later renamed18,22, since the KA-1 domain is widely conserved among METTL16 and its homologs across vertebrates, invertebrates, and even fission yeasts34,37,38, and the structure is homologous to the KA-1 found in MARK/PAR1 kinases36 and U6-specific terminal uridylyltransferases (TUTase1, TENT1)39,40.
The crystal structures of the MTD of human METTL16 (hsMETTL16), its complex with S-adenosyl-L-homocysteine (SAH), and its complex with a short hairpin from the 3’-UTR of MAT2A mRNA (MAT2A-hp) have been reported41,42. The MTD of hsMETTL16 (hsMTD) catalyzed adenosine methylation in the MAT2A-hp as efficiently as full-length hsMETTL1642. Both the consensus RNA sequence and intrinsic RNA structural features—the loop and the transition region of MAT2A-hp— are critical determinants of the methylation efficiency by hsMTD42. The C. elegans METT10 MTD similarly recognizes specific structural features of the RNA surrounding the 3′-ss of sams pre-mRNA, sharing a substrate recognition mechanism with hsMTD38. Regarding the C-terminal KA-1 domains, predictions based on structural studies have suggested that they are well conserved among eukaryotes37,38. The KA-1 domains of hsMETTL16 and ceMETT10 facilitate m6A methylation of the conserved adenosine in U6 snRNA37,38 by increasing the affinity of METTL16 (or ceMETT10) for U6 snRNA and enhancing the catalytic efficiency. A biochemical analysis also suggested that the KA-1 domain would interact with the internal stem-loop (ISL) region of U6 snRNA37. However, the precise mechanisms of U6 snRNA recognition by METTL16 and the role of its KA-1 domain in enhancing methylation remain elusive.
Here, we present the structural and functional analysis of Schizosaccharomyces pombe METTL16, revealing the mechanism of U6 snRNA methylation by METTL16. The KA-1 domain serves as a scaffold for the multi-step methylation process—U6 snRNA ISL recognition, motif recognition in the MTD, and target adenosine positioning in the catalytic site—ensuring the U6 snRNA m6A modification, which leads to accurate and efficient pre-mRNA splicing.
Results
KA-1 of METTL16 facilitates U6 snRNA m6A modification
The C-terminal KA-1 domain of human METTL16 (hsMETTL16) reportedly facilitates the m6A43 modification of U6 snRNA in vitro37, and KA-1 domains are conserved among METTL16 homologs in vertebrates and invertebrates38. The KA-1 domain is also present in METTL16 homologs in fission yeast34,38 (Fig. 1a and Supplementary Fig. 1a, b).
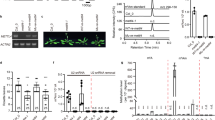
a Schematic diagram of S. pombe METTL16 (spMETTL16) compared with human METTL16 (hsMETTL16) and C. elegans METT10 (ceMETT10). The N-terminal methyltransferase domain (spMTD, magenta) and the C-terminal KA-1 domain (spKA-1, cyan) of S. pombe, used in the experiments, are depicted. b In vitro methylation of the S. pombe U6 snRNA (spU6 snRNA) transcript by spMETTL16 and spMTD under standard conditions. spU6 snRNA (0.5 μM) was incubated with 200 nM spMETTL16 or spMTD in the presence of 1 mM SAM for 4 min at 37 °C. Error bars represent the standard deviation (SD) of three independent experiments (N = 3), and the center of the error bands indicates the mean of the measured values. c Steady-state kinetics of methylation of the spU6 snRNA transcript by spMETTL16 and spMTD. spU6 snRNA (0–10 μM) was incubated with 200 nM spMETTL16 or spMTD in the presence of 1 mM SAM at 37 °C. All experiments were independently repeated three times with similar results. d, e Semi-quantitative RT-PCR analyses of two introns: d SPAC18B11.09 C and e ckn1. The nucleotide sequences of the respective 5′ splice sites (−3 to +4) are shown. The upper and lower bands on the gel represent retained and spliced introns, respectively. Intron retention in the Δmtl16 strain was rescued by ectopic expression of plasmid-encoded full-length METTL16 (spMETTL16), but not by the methyltransferase domain (spMTD) alone. All experiments were independently repeated three times with similar results (Supplementary Fig. 2a, b). f Nucleotide sequence of S. pombe U6 snRNA (spU6 snRNA, left). The nucleotide sequence of the spISL used for crystallization of the spKA-1–spISL complex (spISL, right). g Gel retardation assay of spISL by spKA-1. spISL RNAs were incubated with various concentrations of spKA-1 (0–10 μM). The fractions of the shifted RNA in the gel (left) were quantified (right). The experiments were independently repeated two times with similar results. Source data for b, c, g are provided as a Source data file.
To investigate the role of the KA-1 domain in Schizosaccharomyces pombe METTL16 (spMETTL16) in the m6A modification at A37—corresponding to A43 in human U6 snRNA—of S. pombe U6 snRNA (spU6 snRNA), we examined the methylation activities of full-length spMETTL16 (amino acids 1–398) and its N-terminal methyltransferase domain (MTD; amino acids 1–280) (Fig. 1a), using an spU6 snRNA transcript as a substrate in vitro. The MTD exhibited ~10% of the methylation activity compared to full-length spMETTL16 (Fig. 1b). A steady-state kinetic analysis provided estimated Km values of spU6 snRNA for full-length spMETTL16 and the MTD of 0.182 μM and 0.752 μM, respectively (Fig. 1c). The kcat values were estimated to be 0.38 min−1 for full-length spMETTL16 and 0.037 min−1 for the MTD. Thus, the catalytic efficiency (kcat/Km) of the MTD was ~2% of that of full-length spMETTL16. These results indicate that the KA-1 domain of spMETTL16 facilitates A37 methylation in spU6 snRNA, similar to its role in hsMETTL1637.
KA-1 of METTL16 facilitates pre-mRNA splicing
The m6A modification at A37 of spU6 snRNA is required for the efficient splicing of specific pre-mRNAs containing the BBH motif (B = G, C, or U; H = A, C, or U) at positions −3 to −1 and an A at position 4 of the 5′ splice site (5′-ss)32,33. In S. pombe, the SPAC18B11.09c and ckn1 transcripts have the sequences A-3C-2C-1/G1U2A3A4 at the 5′-ss between exon 1 and exon 2, and U-3G-2C-1/G1U2A3A4 at the 5′-ss between exon 3 and exon 4, respectively32 (Fig. 1d, e). These 5′-ss in pre-mRNAs weakly interact with U5, and the absence of the m6A37 modification in spU6 snRNA destabilizes the interaction between the 5′-ss in the pre-mRNA and U6 snRNA, resulting in intron retention32.
To assess whether the KA-1 domain could facilitate A37 methylation and regulate pre-mRNA splicing in vivo, we transformed a mettl16 knockout strain (Δmtl16) with a plasmid expressing the MTD of spMETTL16 and analyzed the splicing of specific introns in pre-mRNAs (SPAC18B11.09c and ckn1) by RT-PCR (Fig. 1d, e). In the Δmtl16 strain, introns were retained compared to the wild-type strain, whereas ectopic expression of full-length spMETTL16 restored proper splicing and intron removal. In contrast, expression of the spMTD in the Δmtl16 strain failed to rescue splicing (Fig. 1d, e and Supplementary Fig. 2a, b).
Together with the in vitro analyses (Fig. 1b, c), these results suggest that the KA-1 domain of spMETTL16 facilitates the m6A modification of spU6 snRNA, thereby promoting the efficient splicing of specific pre-mRNAs in vivo.
Crystal structure of the KA-1 domain in complex with the ISL of U6 snRNA
The KA-1 domain of hsMETTL16 has been suggested to interact with the internal stem-loop (ISL) region of U6 snRNA, based on a previous biochemical study37. Our cryo-EM analysis of the spMETTL16–spU6 snRNA complex, as described below, also indicated that the ISL region of spU6 snRNA specifically interacts with the KA-1 domain of spMETTL16. The gel-retardation assay showed that the KA-1 domain of spMETTL16 (spKA-1; residues 270–398) (Fig. 1a) binds to the isolated ISL RNA fragment of spU6 snRNA (spISL; nucleotides 47–77) (Fig. 1f) with a Kd of ~1.2 μM (Fig. 1g).
To elucidate the molecular basis of this interaction in detail, we crystallized spKA-1 in complex with spISL and determined its structure at 2.8 Å resolution (Fig. 2a, b; Supplementary Fig. 3 and Supplementary Table 1). spKA-1 consists of five antiparallel β-sheets (β8 – β12) and three α-helices (α8 – α10) (Fig. 2a). The conserved arginine-rich region (RRR) is in α9. As expected, its overall structure closely resembles that of the hsMETTL16 KA-1 domain (hsKA-1, PDB ID: 6M1U)37, with an RMSD of 1.461 Å for 61 structurally equivalent Cα atoms (Fig. 2c). The arginine-rich region (RRR) in hsKA-1 was disordered in the reported structure37 (Supplementary Fig. 1a).
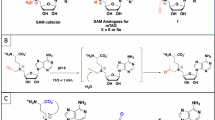
a Crystal structure of spKA−1 in complex with spISL. spKA-1 is shown in blue, and spISL is depicted as an orange stick model. b Electrostatic surface potential of spKA-1 in the spKA-1–spISL complex. The electrostatic potential is represented as blue (positive) and red (negative). spISL is shown as an orange stick model. c Superimposition of the spKA-1 structure (blue) in (a) on the KA-1 domain (hsKA-1, magenta) of human METTL16 (PDB ID: 6M1U)37. The arginine-rich region (RRR) in spKA-1 is shown in red. The dashed region in hsKA-1 represents a disordered area, where the arginine-rich region (RRR) is located. d Structure of spISL bound to spKA-1. Adenosine, guanosine, cytidine, and uridine are colored red, blue, orange, and green, respectively. Nucleotides forming non-canonical base pairs are boxed. e Schematic representation of the interaction between spKA-1 and spISL shown in (d).
In the complex, helices α9 and α10 interact with distinct regions of spISL: α9 engages with the major groove of the loop at the spISL tip, while α10 contacts the minor groove of the double-stranded region of spISL (Fig. 2a). These interactions are mediated by positively charged residues on α9 and α10, which stabilize the spKA-1–spISL interaction (Fig. 2b). The spISL RNA structure within the complex contains non-canonical base pairings, including G59-A63, C55-A67, and G53-A70 (Fig. 2d, e), with the spKA-1 domain specifically recognizing these conserved non-canonical base pairs (Supplementary Fig. 1c), as described below.
The arginine-rich region (RRR: R339, R342, and R343) on α9 interacts with the tip of the ISL (Figs. 2a, 3a and Supplementary Fig. 1a). The nucleobases of C60-A61-C62 in the loop at the tip of spISL stack continuously, and the C62 base stacks on the A63 base (Fig. 3a). The A63 base forms a non-canonical base pair with G59, where the 2-NH2 and the N3 atoms of G59 form hydrogen bonds with the N7 atom and the 6-NH2 of A63, respectively. The Nε and Nη1 atoms of Arg339 form hydrogen bonds with the O6 and N7 atoms of G59, respectively (Fig. 3a). The Nη2 atom of Arg339 forms a hydrogen bond with the phosphate backbone of U58. Arg343 forms a hydrogen bond with the phosphate of A61, and the side chain of Arg343 form cation-π interactions with the G59-Arg339 hydrogen bonds. Arg342 forms a hydrogen bond with the phosphate backbone of C56. Thus, the RRR extensively and specifically interacts with the tip of the ISL of spU6 snRNA. The interactions between the RRR and the tip of the ISL in the present structure were quite similar to the arginine forks recognition of RNA as observed in the crystal structures of HIV Tat in complex with TAR RNAs43 and Tat or HEXIM in complex with the apical stem-loop of 7SK RNA44.
a–d Detailed views of the interactions between spKA-1 and spISL. The colors for nucleosides are the same as in Fig. 2d. e In vitro methylation of the spU6 snRNA transcript by spMETTL16 and its variants with mutations in the KA-1 domain under standard conditions. spU6 snRNA (0.5 μM) was incubated with 200 nM spMETTL16 or its variants in the presence of 1 mM SAM for 4 min at 37 °C. Error bars represent the standard deviation (SD) of three independent experiments (N = 3), and the center of the error bands indicates the mean of the measured values. Source data are provided as a Source data file. Semi-quantitative RT-PCR analyses of two introns: f SPAC18B11.09 C and g ckn1. The upper and lower bands on the gel represent retained and spliced introns, respectively. Intron retention in the Δmtl16 strain was rescued by ectopic expression of plasmid-encoded full-length METTL16 (spMETTL16), but not by the spMETTL16 variants with mutations in the KA-1 domain shown in (e) (Supplementary Fig. 2c, d).
α10 contacts the minor groove of the double helix containing the non-canonical C55-A67 and G53-A70 base pairs (Figs. 2a and 3b, c). The 4-NH2 of C55 forms a hydrogen bond with the N1 atom of A67, and the Nη1 atom of Arg391 forms a hydrogen bond with the O2 of C55 (Fig. 3b). The N1 atom and the O6 of G53 form hydrogen bonds with the N1 atom and the 6-NH2 of A70, respectively, and Glu383 forms a hydrogen bond with the 2-NH2 of G53 (Fig. 3c). The phosphate backbone and ribose 2′-OH form hydrogen bonds with several residues—Asn380, Ser384, Ser387, and Arg394—on α10 (Fig. 2e). Lys286, Lys291, and His290 in the N-terminal part of spKA-1 form hydrogen bonds with the phosphate backbone of A72, C73 and G75, respectively, and the main-chain carbonyl of Tyr287 forms a hydrogen bond with the ribose 2′-OH of C73 (Fig. 3d).
The R339E, R342E, and R343E mutations in the RRR of spMETTL16 reduced the methylation activity of spMETTL16, and the double mutation (R339E/R343E) and triple mutation (R339E/R342E/R343E) further decreased the activity in vitro (Fig. 3e). Although the R394E mutation slightly reduced the activity under our conditions, the R391E mutation reduced it to the same extent as a single mutation in the RRR (Fig. 3e). These results suggest that the specific interactions facilitating the recognition of the ISL of spU6 snRNA are important for A37 methylation by spMETTL16.
Consistently, the Δmtl16 strain expressing spMETTL16 with the R339E, R339E/R343E, or R339E/R342E/R343E mutations failed to rescue the splicing of the SPAC18B11.09c and ckn1 pre-mRNAs, compared to the strain expressing wild-type spMETTL16 (Fig. 3f, g). Expression of spMETTL16 with the R391E or R394E mutations also failed to rescue the splicing of these pre-mRNAs, compared to the strain expressing wild-type spMETTL16, although their effects were less pronounced than those of the R339E, R339E/R343E, or R339E/R342E/R343E mutations (Supplementary Fig. 2c, d).
These structural features of the interactions between spISL and spKA-1 (Fig. 3a–c), along with the biochemical (Fig. 3e) and functional (Fig. 3f, g) analyses, suggest that spKA-1 specifically recognizes the conserved sequence and structure of the ISL of spU6 snRNA (Supplementary Fig. 1c) and is important for U6 snRNA m6A methylation and pre-mRNA splicing.
Cryo-EM structure of the METTL16-U6 snRNA complex
To further elucidate the mechanism of spU6 snRNA methylation by spMETTL16, we determined the cryo-EM structure of spMETTL16 in complex with spU6 snRNA lacking the 5′-stem (spU6 snRNA_Δ17), which can be methylated as efficiently as full-length spU6 snRNA, at 3.4 Å resolution (Fig. 4a, Supplementary Fig. 4 and Supplementary Table 2). The spMETTL16–spU6 snRNA_Δ17 complex was reconstituted by mixing purified spMETTL16 with in vitro-transcribed spU6 snRNA_Δ17, followed by size-exclusion chromatography for purification.
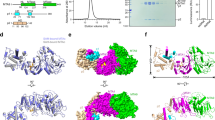
a Cryo-EM map (left) and model (right) of the spMETTL16–spU6 snRNA complex in the absence of SAM (spMETTL16–spU6). MTD, KA-1 domain, and modeled RNA are colored magenta, blue and orange, respectively. The N-terminal extension (residues 1–61) and the RNA binding loop (residues 172–200) are not modeled, and only nucleotides 41–82 of spU6 snRNA are modeled. b Cryo-EM map (left) and model (right) of the spMETTL16–spU6 snRNA complex in the presence of SAM (spMETTL16–SAM–spU6). MTD, KA-1 domain, and modeled RNA are colored as in (a). The N-terminal extension and the RNA binding loop (residues 187–194) are not modeled, and only nucleotides 35–83 of spU6 snRNA are modeled. c Structural comparison between spMETTL16–spU6 (gray) and spMETTL16–SAM–spU6. In the presence of SAM, α4 in the MTD becomes resolved, and the ACA*GAGA motif in spU6 snRNA shifts toward the catalytic site of the MTD. d The secondary structure of spMETTL16. The MTD and KA-1 domain are colored magenta and blue, respectively. The regions enclosed by the dashed lines were not visible in the spMETTL16–spU6 structure. α4 was visible in the spMETTL16–SAM–spU6 complex. The N-terminal extension was predicted by AlphaFold2 (Data base: AF-O42662-F1-v4)56.
The cryo-EM density map of the spMETTL16–spU6 snRNA_Δ17 complex without SAM (hereafter referred to as spMETTL16–spU6) suggests that spU6 snRNA adopts an extended conformation. However, only the ISL region (nucleotides 41–82) could be modeled, and the entire spU6 snRNA structure could not be fully resolved (Fig. 4a), due to the insufficient quality of the cryo-EM map. In the cryo-EM structure of spMETTL16–spU6, the KA-1 domain of spMETTL16 interacts with the ISL region of spU6 snRNA, resembling the interaction of spKA-1 with spISL in the crystal structure (Fig. 2a and Supplementary Fig. 5). The helices α9 and α10 interact with the major groove of the loop at the spISL tip and the minor groove of the double-stranded region of spISL, respectively. However, the remaining portion of spU6 snRNA does not interact with the protein. Notably, the ACA*GAGA motif containing the target A37– located between the ISL and the telestem—is far from the catalytic site of the MTD domain (Fig. 4a). The N-terminal extension (residues 1–61) and the segment between β4 and α5 (residues 172–200) within the MTD (Fig. 4d)—which are both potentially involved in RNA binding22,42—were disordered and could not be modeled due to the lack of a cryo-EM map. Thus, this structure likely represents an initial stage of substrate U6 snRNA recognition by spMETTL16.
Cryo-EM structure of the METTL16-SAM-U6 snRNA complex
We also determined the cryo-EM structure of spMETTL16 in complex with spU6 snRNA in the presence of SAM (hereafter referred to as spMETTL16–SAM–spU6), at 3.0 Å resolution. SAM was added to the spMETTL16–spU6 snRNA complex.
In the cryo-EM structure of the spMETTL16–SAM–spU6 complex (Fig. 4b, Supplementary Figs. 6 and 7 and Supplementary Table 2), the mode of U6 snRNA binding to spMETTL16 differs from that observed in the spMETTL16–spU6 structure (Fig. 4b, c). SAM binds to the catalytic site in the MTD, and the ISL of spU6 snRNA retains its interaction with the KA-1 domain, as observed in the SAM-free structure. However, the ACAGAGA motif in spU6 snRNA shifts toward the MTD, and the methylation site, A37, is proximal to SAM in the MTD (Fig. 4b, c), although the telestem region (nucleotides 1–34 and 84–99) of spU6 snRNA could not be modeled due to the lack of a cryo-EM map. The α4 helix (between β4 and α5) within the MTD, which was not visible in the spMETTL16–spU6 structure, is partially resolved in the spMETTL16–SAM–spU6 structure (Fig. 4b, c). However, the loop between α4 and α5 (residues 187–194), as well as the N-terminal extension (residues 1–55), remains unresolved (Fig. 4c, d).
SAM is located within the catalytic site of the MTD, and its position is identical to that of SAH observed in human METTL16 (hsMETTL16) in complex with SAH (Fig. 5a, b, PDB ID: 6B92)41. The adenine base of SAM stacks with Ile120 and Phe206, while Lys148 in the K-loop42, which regulates SAM binding to the catalytic pocket in hsMETTL16, is positioned away from the SAM binding site. A37 in the ACA*GAGA motif of spU6 snRNA resides near the catalytic pocket of the MTD (Fig. 5b, c). However, the distance between the 6-NH2 of A37 in spU6 snRNA and the methyl group of SAM is ~6 Å (Fig. 5b), which is longer than typical distances observed for productive methyl transfer. The methylation of A37 proceeds relatively slowly (kcat = 0.38 min−1, Fig. 1c), and the cryo-EM map at the position of A37 shows no apparent methylation of 6-NH2 group of the adenine base (Supplementary Fig. 7c). Additionally, we determined the cryo-EM structure of the spMETTL16 in complex with spU6_Δtelestem and SAM at ~2.9 Å (Supplementary Fig. 8a, b). The sample was applied to the grid immediately after mixing spMETTL16, spU6_Δtelestem, and SAM on ice. Structural analysis revealed that RNA recognition in this complex is consistent with that observed in the spMETTL16–SAM–spU6 complex (Supplementary Fig. 8c). The distance between the 6-NH2, of A37 in U6 snRNA and the methyl group of SAM remains ~6 Å, and the cryo-EM map at A37 shows no apparent methylation at the N6 position (Supplementary Fig. 8b). These observations suggest that, in the present structure, the 6-NH2 of A37 is not positioned for nucleophilic attack on the methyl group of SAM45, and that the present structure represents a pre-productive state of the reaction, as described below.
a Overall structure of spMETTL16–SAM–spU6 (left) and nucleotide sequence of spU6 snRNA. The modeled nucleotides (35–83) in the structure are colored orange. b SAM recognition by spMETTL16. The structure of human METTL16 MTD in complex with SAH (hsMETTL16-SAH)41 was superimposed onto that of spMETTL16–SAM–spU6. SAH in the hsMETTL16-SAH is shown as yellow sticks and SAM in the spMETTL16–SAM–spU6 is shown as cyan sticks. c, d Interactions between spMETTL16 and spU6 snRNA. e Schematic representation of the interaction between A35C36A37G38A39 and the MTD in (c, d). f In vitro methylation of the spU6 snRNA transcript by spMETTL16 and its variants with mutations in the KA-1 domain under standard conditions. spU6 snRNA (0.5 μM) was incubated with 200 nM spMETTL16 or its variants in the presence of 1 mM SAM for 4 min at 37 °C. Error bars represent the standard deviation (SD) of three independent experiments (N = 3), and the center of the error bands indicates the mean of the measured values. Source data are provided as a Source data file.
The C36 base stacks with the side chain of Met197. The N1 atom and the 6-NH2 group of A37 form hydrogen bonds with the Nδ1 atom and the Oδ1 atom of Asn168, respectively, in the NPPF motif (residues 168–171). The A37 base is sandwiched between Phe171 and the side chain of Arg260, which interacts with the phosphate of G38. The O6 of G38 forms a hydrogen bond with Ser231. The 6-NH2 of A39 interacts with Lys230, which in turn forms a hydrogen bond with Ser283 (Fig. 5c–e). Lys257 forms a hydrogen bond with the phosphate of A39. These structural features suggest that part of the ACA*GAGA motif in spU6 snRNA is recognized by the MTD of spMETTL16 (Fig. 5e), although the 6-NH2 of A37 remains distant from SAM. Arg278 forms hydrogen bonds with the phosphates of A72 and C73, and Lys286 interacts with the phosphates of U45 and G75. Thr282 interacts with the phosphate of U46 (Fig. 5d). Consistent with the RNA recognition by the MTD in the present structure, the N168A, F171A, K230E, K257E, R260E, and R278E mutations of spMETTL16 all reduced the methylation activity of spMETTL16 in vitro (Fig. 5f).
Altogether, the present spMETTL16–SAM–spU6 structure represents a stage in which the core of the ACA*GAGA motif of spU6 snRNA is specifically recognized by the MTD, but spMETTL16 is not yet ready for catalysis. spU6 snRNA has not adopted a productive conformation for efficient methylation by spMETTL16, and thus, the present structure would be the pre-productive stage, as described below.
Structural elements in spU6 snRNA for efficient methylation by spMETTL16
The crystal structure of the methyltransferase domain (MTD) of hsMETTL16 in complex with the short hairpin RNA of the 3’-UTR of MAT2A mRNA (MAT2A-hp) has been reported42. In the structure, MAT2A-hp adopts a distinct structural conformation, including a loop (containing the UACAG recognition motif and linker), a stem and a transition region between the loop and the stem, all of which are essential for efficient catalysis by the MTD42 (Fig. 6a). It has been suggested that the human U6 snRNA could also adopt a similar structural conformation, with the UACAG recognition motif positioned in the same context as in the MAT2A-hp structure37. Likewise, spU6 snRNA is predicted to adopt the same structure as MAT2A-hp and human U6 snRNA (Fig. 6a).
a Nucleotide sequences of human MAT2A hairpin (MAT2A-hp)42 and S. pombe U6 snRNA and its variants used for the assays in (b). b In vitro methylation of spU6 snRNA and its variants by spMETTL16 under standard conditions. spU6 snRNA or its variants (0.5 μM) was incubated with 200 nM spMETTL16 in the presence of 1 mM SAM for 4 min at 37 °C. Error bars represent the standard deviation (SD) of three independent experiments (N = 3), and the center of the error bands indicates the mean of the measured values. c Superimposition of the spMETTL16–SAM–spU6 structure onto the human METTL16 MTD (hsMETTL16_MTD, gray) in complex with MAT2A hairpin RNA (MAT2A-hp: gray, PDB ID: 6DU4)42. The regions corresponding to the red regions (N-terminal extension and RNA binding loop), enclosed by dashed lines, in hsMETTL16_MTD were not visible in the present spMETTL16–SAM–spU6 structure. d Superimposition of the structure of the catalytic pocket of spMETTL16–SAM–spU6 and that of hsMETTL16_MTD–MAT2A-hp (gray). A17 in MAT2A-hp in the hsMETTL16–MAT2A-hp complex is deeply docked in the pocket and closer to SAM (cyan) than A37 in spU6 snRNA in the spMETTL16–SAM–spU6 complex. e In vitro methylation of the spU6 snRNA transcript by spMETTL16 and its N-terminal extension deletion mutant (Δ_NTE: amino acids 1–50 were deleted) and RNA binding loop deletion mutant (Δ_RBL: amino acid 186–189 were deleted), under standard conditions as in Fig. 5e. Error bars represent the standard deviation (SD) of three independent experiments (N = 3), and the center of the error bands indicates the mean of the measured values. Source data for b, e are provided as a Source data file.
To verify the requirement of these structural elements in spU6 snRNA for methylation by spMETTL16, as observed with hsMETTL16, we tested the methylation of mutant spU6 snRNAs by spMETTL16 in vitro (Fig. 6b). Mutations in the UACAG recognition motif in the loop, such as C36G and G38C, reduce its methylation by spMETTL16 in vitro (Fig. 6a, b). Mutations in the putative transition region of spU6 snRNA – G40A41A42 to C40U41U42 or G83A84 to C83U84 – also reduce its methylation by spMETTL16 in vitro (Fig. 6a, b). These results indicate that the structural elements surrounding the methylation site in spU6 snRNA are necessary for efficient A37 methylation by spMETTL16, similar to hsMETTL16 and ceMETT1037,38,42. Furthermore, spU6 snRNA lacking the 5′-stem and telestem, spanning nucleotides 26 to 84 (Δtelestem), can be methylated by spMETTL16, although with lower efficiency than that of full-length spU6 snRNA, suggesting that Δtelestem can adopt the same structural conformation as MAT2A-hp. Furthermore, an hsMAT2A-hp-like U6 snRNA variant (Δtelestem_Loop) is also methylated by spMETTL16 to the same extent as Δtelestem (Fig. 6a, b). These results indicate that spU6 snRNA must adopt a productive conformation for efficient methylation by spMETTL16, as required for MAT2A-hp methylation by hsMETTL16.
Transition of spU6 sRNA to the productive form for efficient methylation by spMETTL16
Superimposition of the structure of the MTD of hsMETTL16 in complex with MAT2A-hp (hsMTD–MAT2A-hp, PDB ID: 6DU4)42 onto that of spMETTL16–SAM–spU6 reveals that, while the N-terminal extension and the RNA-binding loop in the MTD of hsMETTL16 interact with MAT2A-hp from various angles, the corresponding regions of spMETTL16 are not visible in the current spMETTL16–SAM–spU6 structure (Figs. 4d and 6c). This suggests that the N-terminal extension and the RNA-binding loop of spMETTL16 would facilitate the transition of spU6 snRNA into a productive structure for methylation by spMETTL16.
Superimposition of the catalytic core of hsMTD–MAT2A-hp onto that of spMETTL16–SAM–spU6 (Fig. 6c, d) reveals that A17 in MAT2A-hp is positioned closer to SAM than A37 in spMETTL16–SAM–spU6 (Fig. 6d). In the hsMTD–MAT2A-hp structure, the 6-NH2 and N1 atoms of A17 of MAT2A-hp form hydrogen bonds with the main-chain carbonyl and amide of Pro185 and Pro186, respectively42. These observations suggest that the reported hsMTD–MAT2A-hp structure represents the productive stage, and the current structure of spMETTL16–SAM–spU6 (Fig. 4b) likely represents a pre-productive stage, in which the ACA*GAGA motif in U6 snRNA is recognized by spMETTL16 (Fig. 5c), but the spU6 snRNA has not yet transitioned into a catalytically active, productive conformation for methylation by spMETTL16.
Subsequently, this pre-productive conformation undergoes further structural adjustment through interactions involving the N-terminal extension, the RNA-binding loop, and U6 snRNA. Consistently, the deletion of the N-terminal extension and the RNA-binding loop in spMETTL16 reduced its methylation activity (Fig. 6e). The establishment of the interactions between A39G40A41G42 and G83A84, forming the transition base-pair, along with the base pairings in the telestem, enables spU6 snRNA to adopt the productive conformation. In this state, the ACA*GAGA motif protrudes deeper into the catalytic site, positioning the A37 base closer to SAM and thereby facilitating efficient m6A37 modification.
Discussion
The m6A methylation of a conserved adenosine in U6 snRNA, catalyzed by METTL16 or its homologs, is essential for the efficient splicing of specific pre-mRNAs32,33,34. In this study, we elucidated the molecular mechanisms governing the interactions between S. pombe METTL16 (spMETTL16) and U6 snRNA, as well as the m6A methylation. The conserved C-terminal KA-1 domain, present across METTL16 homologs in vertebrates, invertebrates, and fission yeast, is a critical element that enhances methylation efficiency and ensures proper pre-mRNA splicing (Fig. 1). The KA-1 domain, particularly its arginine-rich region (RRR), interacts specifically with the internal stem-loop (ISL) of U6 snRNA, recruiting the substrate U6 snRNA to the enzyme (Figs. 2 and 3), and serves as an axis in the subsequent methylation process.
Cryo-EM structures of the spMETTL16–spU6 snRNA complex, with and without SAM, reveal the dynamic processes of substrate recognition, methylation site recognition, and the methylation reaction (Figs. 4, 5, and 7). Previous biochemical studies showed that hsMETTL16 binds to U6 snRNA with much greater affinity than SAM37,46. As for the spMETTL16, the Km values for U6 snRNA and SAM are 0.182 μM and 482 μM, respectively, differing by three orders of magnitude (Fig. 1c and Supplementary Fig. 7d). This substantial difference indicates that spMETTL16 has a significantly higher affinity for U6 snRNA compared to SAM.
a Initial binding stage: U6 snRNA initially binds to METTL16 through interactions between the ISL and KA-1 domain (blue). The ACA*GAGA motif, which contains the methylation site A37 in U6 snRNA, is highlighted in red. b Pre-productive stage: SAM (cyan) binding to the MTD (magenta) triggers a conformational change in the MTD, shifting the ACA*GAGA motif of U6 snRNA closer to the catalytic site via the RNA-binding loop. At this stage, the motif is specifically recognized. c Productive stage: The interactions of the N-terminal extension (NTE) and the RNA-binding loop with U6 snRNA promote the transition to the productive stage. U6 snRNA undergoes structural rearrangements, adopting productive conformation. This process leads to the formation of the transition region and base-pairing within the telestem in U6 snRNA, shifting A37 deeper into the catalytic site, and thereby facilitating efficient m6A modification by MTD.
Thus, the m6A modification of U6 snRNA occurs by the ordered-sequential mechanism46. First, U6 RNA is recruited to METTL16 through the specific interactions between KA-1 and the ISL. U6 snRNA adopts an extended structure, and the ACA*GAGA motif, containing the methylation site A37, is apart from the catalytic site in the methyltransferase domain (MTD), representing the initial U6 snRNA binding stage (Fig. 7a). Next, the SAM binds to the catalytic site in the MTD of the METTL16–U6 snRNA complex. The catalytic pocket and part of the RNA-binding loop in the MTD are resolved, allowing the ACA*GAGA motif in U6 snRNA to shift closer to the catalytic pocket of the MTD. At this stage, the motif is specifically recognized by the MTD, although the target A37 remains ~6 Å from SAM— indicating a pre-productive state insufficient for methyl transfer (Figs. 4, 5, and 7b). A comparison with the hsMETTL16–MAT2A-hp structure, in which MAT2A-hp RNA adopts a productive conformation for m6A modification42, highlights the need for further adjustment of the U6 snRNA conformation into a productive state for A37 methylation by the MTD (Figs. 6 and 7c). Finally, additional intramolecular interactions—the formation of the transition region and the stem in the telestem of U6 snRNA—through the interactions of U6 snRNA with the N-terminal extension and RNA-binding loops in the MTD, reorient the ACA*GAGA motif deeper into the catalytic site, aligning the target A37 with SAM for efficient methylation (Figs. 6 and 7c).
The KA-1 domain not only anchors U6 snRNA to METTL16 but also serves as a hub that orchestrates the transition of the METTL16-U6 snRNA complex into a catalytically competent state. This multi-step process, which includes initial ISL recognition, motif recognition, and final positioning of the methylation site A37 within the catalytic site, reflects the coordinated roles of the METTL16 domains (Fig. 7). The productive conformation, consisting of the transition region and the stem in U6 snRNA (Fig. 7c), might be transient and inherently unstable. Due to its fleeting nature, we may have been unable to capture it in our cryo-EM analysis.
In vertebrates, METTL16 regulates SAM homeostasis by controlling MAT2A mRNA splicing and stability through the m6A modification of the 3’-UTR hairpins of MAT2A mRNA17,18. The effect of KA-1 on the methylation of MAT2A hairpins was observed in vitro, although its impact is much less significant than its role in the methylation of U6 snRNA37,42. MAT2A-hp has a stem-loop structure, and compared to U6 snRNA, its loop can more readily adopt the productive conformation for efficient methylation by METTL16 (Fig. 6a). Structural adjustment after ACA*GAGA motif recognition would proceed smoothly, with the recognition of MAT2A-hp by METTL16 primarily mediated by the MTD. Therefore, as described below, KA-1 would have been attached to the MTD for the efficient methylation of structured U6 snRNA.
The KA-1 domain reportedly promotes splicing when tethered to a reporter18. The splicing of MAT2A mRNA is collaboratively regulated by METTL16 and the cleavage factor Im (CFIm) complex23. The KA-1 domain may alter the RNA secondary structure surrounding MAT2A hairpins, particularly by binding to the double-stranded region, thereby facilitating the recruitment of CFIm complexes and promoting MAT2A mRNA splicing. The mechanism and involvement of the KA-1 domain in MAT2A mRNA splicing await further investigation.
The KA-1 domain structure is also found in the C-terminal region of the U6 snRNA-specific terminal uridylyltransferase (TUTase, TUT1)47,48,49, although the amino acid sequence homology between the KA-1 domains of METTL16 and TUT1 is low37,39. The KA-1 domain of human TUT1 (hsTUT1) increases hsTUT1’s affinity for U6 snRNA and facilitates efficient oligo-uridylylation of U6 snRNA39,50. A recent structural analysis of hsTUT1 in complex with U6 snRNA showed that the KA-1 domain interacts with the tip of the ISL51, where two helices engage distinct regions of the ISL: one containing RRR interacts with the major groove of the loop at the ISL tip, while the other contacts the minor groove of the double-stranded region of the ISL, similar to the interaction of spKA-1 with spISL (Fig. 2a and Supplementary Fig. 9). Notably, these two distinct enzymes, which exert different, yet functionally essential activities on U6 snRNA, share the same structural domains for their activities. Therefore, the KA-1 domain may have become associated with the catalytic domains of these enzymes and evolved to recognize U6 snRNA, particularly the ISL, for the efficient maturation of functional U6 snRNA.
The KA-1 domain is widely conserved in METTL16 and its homologs across vertebrates, invertebrates, and even fission yeast. As revealed in this study, the KA-1 domain of spMETTL16 functions as a key regulatory domain for efficient U6 snRNA m6A modification and precise pre-mRNA splicing. Therefore, this mechanism is likely to be functionally and evolutionarily conserved among eukaryotes, including humans.
Methods
Plasmid construction
The plasmid encoding the full-length S. pombe Mtl16 gene without untranslated regions (pET28a-SpMettl16) was kindly provided by Dr. Suzuki32. Plasmids encoding spMETTL16 variants were prepared using the pET28a-SpMettl16 plasmid as the template. The DNA fragment encoding the N-terminal methyltransferase domain (MTD: residues 1–281) of SpMettl16 was PCR-amplified and cloned between the NdeI and XhoI sites of pET28a, yielding pET28a_SpMettl16-MTD. The DNA fragment encoding the C-terminal KA-1 domain (residues 270–398) was PCR-amplified and cloned into the pET15SUMO vector52 between the NdeI and XhoI sites, yielding pET15SUMO_SpMettl16-KA-1. Mutations were introduced using a KOD-Plus Mutagenesis Kit (Toyobo, Japan), according to the manufacturer’s instructions. The oligonucleotide sequences used for molecular cloning and mutagenesis are listed in Supplementary Table 3.
Expression and purification of recombinant proteins
For the overexpression of spMETTL16, its variants, and spMETTL16-MTD (spMTD), as well as that of spMETTL16-KA-1 (spKA-1) and its variants, E. coli Rosetta2(DE3) cells (Novagen, Japan) were transformed with the respective overexpression plasmids. Transformed cells were cultured in LB medium supplemented with 50 μg/mL kanamycin and 40 μg/mL chloramphenicol for the expression of spMETTL16, its variants, and spMTD, or with 50 μg/mL ampicillin and 40 μg/mL chloramphenicol for spKA-1 and its variants. Cultures were grown at 37 °C until the OD600 reached 0.8, and protein expression was induced by adding isopropyl β-D-thiogalactopyranoside (IPTG) to a final concentration of 0.1 mM, followed by an incubation at 20 °C for 20 h. Cells were harvested and lysed in buffer containing 20 mM Tris-HCl, pH 7.0, 500 mM NaCl, 5 mM β-mercaptoethanol, 20 mM imidazole, and 5% (v/v) glycerol. Lysates were clarified by centrifugation at 30,000 × g for 30 min at 4 °C.
SpMETTL16, its variants, and spMTD were first purified by Ni-NTA agarose (Qiagen, Japan) chromatography, with proteins eluted in buffer containing 20 mM Tris-HCl, pH 7.0, 500 mM NaCl, 5 mM β-mercaptoethanol, 300 mM imidazole, and 5% (v/v) glycerol. The eluted proteins were further purified on a HiTrap Heparin column (GE Healthcare, Japan) and separated by size-exclusion chromatography using a HiLoad 16/60 Superdex 200 column (GE Healthcare, Japan), equilibrated with buffer containing 20 mM Tris-HCl, pH 7.0, 200 mM NaCl, and 10 mM β-mercaptoethanol. His6-SUMO-tagged spKA-1 proteins were purified using Ni-NTA agarose chromatography as described above. The His6-SUMO tags were cleaved by ULP1 (ubiquitin-like-specific protease 1) during dialysis against buffer, containing 20 mM Tris-HCl, pH 7.0, 500 mM NaCl, 5 mM β-mercaptoethanol, and 5% (v/v) glycerol, at 4 °C overnight. Following cleavage, the proteins were passed through a Ni-NTA agarose column to remove the uncleaved His6-SUMO-tagged proteins and ULP1. The cleaved proteins were further purified as described above. The final purified proteins were concentrated and stored at −80 °C.
Preparation of synthetic U6 snRNA and its variants
Synthetic S. pombe U6 snRNA internal stem-loop (spISL) RNA was purchased from Fasmac, Japan. The U6 snRNA and its variants were synthesized by T7 RNA polymerase, using plasmids encoding the respective DNA sequences downstream of the T7 promoter as templates. The synthesized RNAs were purified by 10% (w/v) polyacrylamide gel electrophoresis under denaturing conditions. The nucleotide sequences of the templates used for in vitro transcription are listed in Supplementary Table 4, and the nucleotide sequences of the RNAs used in this study are listed in Supplementary Table 5.
In vitro methylation assay
In vitro methylation assays were conducted using the MTase-Glo system (Promega, Japan), as previously described37,38 with slight modifications. The reaction mixture (15 μL), containing 0.2 μM spMETTL16 (or its variants or spMTD), 50 mM HEPES-KOH, pH 8.0, 120 mM NaCl, 2 mM MgCl2, 1 mM dithiothreitol (DTT), 1 mM S-(5′-adenosyl)-L-methionine chloride (SAM, Cayman Chemical, USA), 1× MTase-Glo Reagent, and a series of RNA samples at different concentrations (0.05–10.0 μM), was incubated at 37 °C for 4 min. Subsequently, 15 μL of MTase-Glo Detection Solution was added, and the reaction was incubated at room temperature for 30 min. Luminescence was quantified using a GloMax-Multi Detection System (Promega, Japan). The luminescence value of the mixture without the RNA substrate was used as the background and subtracted. One picomole of SAH was estimated to be equivalent to 190,000 LU (light units), according to the standard quantification curve.
Yeast strains and cultivation
The ED668 (h+ ade6-M216 ura4-D18 leu1−32) (wild-type) strain and SPAC27D7.08cΔ (Δmtl16), mettl16 knockout strain32 were grown at 30 °C in YE5S medium [0.5% yeast extract and 3% glucose, supplemented with adenine, uracil, lysine, histidine, and leucine (225 mg/mL each)] or EMM medium [EMM without dextrose (MP Biochemicals) with 2% glucose, supplemented with adenine and leucine (225 mg/mL each)]. Yeast cells were cultivated at 30 °C to mid-exponential phase for total RNA extraction.
Total RNA preparation
Total RNA from yeast cells was prepared by hot phenol extraction53. Cell pellets were resuspended in TES solution (10 mM Tris-HCl, pH 7.5, 10 mM EDTA, and 0.5% SDS), mixed with an equal volume of acidic phenol with brief vortexing, incubated at 65 °C for 30 min, and centrifuged at 14,000 × g for 5 min at 4 °C. The aqueous phase was collected, mixed with an equal volume of chloroform, and centrifuged at 14,000 × g for 5 min at 4 °C. The supernatant was subjected to ethanol precipitation to obtain the RNA pellet. The RNA pellet was dissolved in RNase-free water and stored at −80 °C.
Semi-quantification of intron-retention of specific pre-mRNAs
The plasmid encoding the full-length S. pombe mtl16 gene with untranslated regions (pREP42-spMETTL16-FL) and the empty control vector (pREP42-vector) were kindly provided by Dr. Suzuki32. Plasmids expressing mutant spMETTL16 were generated by site-directed mutagenesis. The S. pombe Δmtl16 strain was transformed with these plasmids using a Frozen-EZ Yeast Transformation II Kit (Zymo Research, Funakoshi, Japan) and grown on YE5S or EMM medium plates. After an incubation for 3–4 days, single colonies were selected and cultured in EMM medium without thiamine until the cell density reached an OD600 of 1.0. Cells were harvested, and total RNA was extracted as described above. One microgram of total RNA was treated with Recombinant RNase-free DNase I (Takara, Japan) and subjected to reverse transcription using random hexamer and oligo(dT) primers with a PrimeScript™ RT Reagent Kit (Takara, Japan). The resulting cDNA was amplified by PCR with specific primers (SPAC18B11.09c and ckn1 pre-mRNA)32 under the following conditions: 30 cycles of 98 °C for 10 s, 53 °C for 10 s, and 72 °C for 20 s. PCR products were resolved on a 10% (w/v) non-denaturing polyacrylamide gel, stained with ethidium bromide, and visualized and quantified using a Gel Doc EZ Imager (Bio-Rad, Japan).
Crystallization and structural determination
The purified spKA-1 was mixed with spISL RNA at a 1:1 molar ratio (final concentration of 50 μM each), and the solution was incubated on ice for 10 min. Crystals were obtained using the sitting-drop vapor diffusion method at 20 °C. A 0.2 μL sample of the spKA-1–spISL complex was mixed with an equal volume of reservoir solution, containing 0.36 M sodium acetate, 0.09 M sodium cacodylate, pH 7.0, 0.01 M Tris-HCl, pH 8.5, and 23% (v/v) PEG 8000. The crystals were cryoprotected with reservoir solution supplemented with 25% (v/v) ethylene glycol and flash-cooled in liquid nitrogen. X-ray diffraction data were collected at Beamline 17 A of the Photon Factory (KEK, Japan) and processed using XDS54. The initial phase of the spKA-1:spISL complex structure was determined by molecular replacement with Phaser55, using the AlphaFold-predicted structure of the spKA-1 domain (AF-O42662-F1)56 and the spISL RNA model predicted with SimRNA57 as search models. The structure was manually modeled using Coot58 and further refined with Phenix.refine59. The final model was assessed with MolProbity60. The details of the crystallographic data collection and refinement statistics are provided in Supplementary Table 1.
Cryo-EM grid preparation
For the spMETTL16–SAM–spU6 snRNA complex, purified spMETTL16 (0.6 mg/mL) was mixed with the U6 snRNA transcript at a 1:1 molar ratio in buffer C (20 mM Tris-HCl, pH 7.0, 200 mM NaCl, 10 mM β-mercaptoethanol, and 2 mM SAM) and incubated at 37 °C for 10 min. Tween-20 [0.005% (v/v)] was then added, and the mixture was placed on ice prior to grid preparation. A 3 μL aliquot of the sample was applied to a glow-discharged UltrAuFoil R1.2/1.3 Au 300 grid (Quantifoil) at ~4 °C. The grids were blotted for 4 s at 100% humidity and 4 °C, and then flash-frozen in liquid ethane using a Vitrobot Mark IV (Thermo Fisher Scientific). The spMETTL16–SAM–spU6_Δtelestem complex was also prepared as described above, except that the sample was applied to the grid immediately after mixing spMETTL16, SAM, and spU6_Δtelestem on ice. The spMETTL16–spU6 complex was prepared as described above, except that the sample was applied to a Quantifoil R1.2/1.3 mesh Cu300 holey carbon grid (Quantifoil), with a sample concentration of 0.4 mg/mL.
Cryo-EM data collection and processing
All datasets were processed using a similar protocol with minor variations. Data collection parameters are summarized in Supplementary Table 2, and data processing flowcharts are shown in Supplementary Figs. 4 and 6. The processing of the spMETTL16–SAM–spU6 snRNA complex is described as follows. Cryo-EM movies were collected using a Krios G4 transmission electron microscope (Thermo Fisher Scientific) equipped with a K3 direct electron detector (Gatan, Inc.) operated at 300 kV. A total of 8497 micrographs were recorded at a pixel size of 0.83 Å, with a total electron dose of 50 e−/Å2 Downstream processing was performed in cryoSPARC (ver. 4.6.0)61. Movies were motion-corrected using Patch Motion Correction, and contrast transfer function (CTF) estimation was performed with Patch CTF. Micrographs were denoised using Micrograph Denoiser, and particles were automatically picked with Blob Picker. Two rounds of two-dimensional (2D) classification were conducted, and the selected high-quality classes were used as templates for template-based particle picking. After three rounds of heterogeneous refinement and one additional 2D classification step to remove junk particles, 486,386 particles were selected and extracted with a 324-pixel box. The final reconstruction from 425,927 particles was obtained using non-uniform refinement, yielding an EM map at an average resolution of 2.99 Å after CTF refinement, particle subtraction, and reference-based motion correction polishing. The final map was sharpened using DeepEMhancer (ver. 0.14)62. The data processing for the spMETTL16–spU6 complex was nearly identical to that of the spMETTL16–SAM–spU6 complex, as described above. The final reconstruction was obtained using non-uniform refinement, yielding an EM map at an average resolution of 3.4 Å from 173,139 particles (Supplementary Figs. 4 and 6 and Supplementary Table 2).
Model building and refinement
To build the model of the spMETTL16–SAM–spU6 structure, the structures of the spMETTL16-MTD domain (residues 56–291) and RNA base pairs (G43-U82, A44-U81, U45-A80, and U46-C79) were predicted using AlphaFold256. The spMETTL16-spKA-1 domain (residues 292–398) and U6 RNA nucleotides (47–77) were derived from the spKA-1-spISL structure. These models were docked into the cryo-EM density using ChimeraX (ver. 1.9)63. Additional RNA nucleotides (A35 to A42 and G83) and SAM were manually fitted, and the model was constructed and adjusted using Coot58. The structure was refined using phenix.real_space_refine59. Due to insufficient density for de novo model building, restraints were applied to maintain the correct geometry of the base pairs G43-U82, A44-U81, U45-A80, and U46-C79 during model refinement. The final structures were validated using MolProbity60, and figures were prepared using PyMOL (http://www.pymol.org/pymol) and UCSF ChimeraX (ver. 1.9)63. The model building process for the spMETTL16–spU6 structure was nearly identical to that for the spMETTL16–SAM–spU6 structure, as described above (Supplementary Table 2).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The data that support this study are available from the corresponding author upon request. The data generated in this study are provided as Source Data files with this paper. The coordinates and structure factors for the crystal structure of spKA-1–spISL have been deposited in the PDB, under the accession number 9M86. The atomic coordinates and cryo-EM maps of spMETTL16–spU6 and spMETTL16–SAM–spU6 have been deposited in the Protein Data Bank (PDB IDs: 9U47 and 9U48), and the Electron Microscopy Data bank (EMD-63833 and EMD-63834), respectively. The micrographs of spMETTL16–spU6 and spMETTL16–SAM–spU6 have been deposited in the Electron Microscopy Public Image Archive EMPIAR-12699 and EMPIAR-12700, respectively. The structures of hsKA-1 (PDB ID: 6M1U), hsMTD-SAH (PDB ID: 6B92), and hsMTD–MAT2A-hp (PDB ID: 6DU4) were used for structural comparisons in this study. Source data are provided with this paper.
References
Pan, T. N6-methyl-adenosine modification in messenger and long non-coding RNA. Trends Biochem. Sci. 38, 204–209 (2013).
Dominissini, D. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485, 201–206 (2012).
Meyer, K. D. et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3’ UTRs and near stop codons. Cell 149, 1635–1646 (2012).
Yue, Y., Liu, J. & He, C. RNA N6-methyladenosine methylation in post-transcriptional gene expression regulation. Genes Dev. 29, 1343–1355 (2015).
Wang, X. et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505, 117–120 (2014).
Xiao, W. et al. Nuclear m6A reader YTHDC1 regulates mRNA splicing. Mol. Cell 61, 507–519 (2016).
Shi, H. et al. YTHDF3 facilitates translation and decay of N6-methyladenosine-modified RNA. Cell Res. 27, 315–328 (2017).
Meyer, K. D. 5′ UTR m6A promotes cap-independent translation. Cell 163, 999–1010 (2015).
Wang, X. et al. N6-methyladenosine modulates messenger rna translation efficiency. Cell 161, 1388–1399 (2015).
Linder, B. et al. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat. Methods 12, 767–772 (2015).
Liu, J. A METTL3–METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 10, 93–95 (2014).
Ke, S. et al. A majority of m6A residues are in the last exons, allowing the potential for 3’ UTR regulation. Genes Dev. 29, 2037–2053 (2015).
Oerum, S., Meynier, V., Catala, M. & Tisné, C. A comprehensive review of m6A/m6Am RNA methyltransferase structures. Nucleic Acids Res. 49, 7239–7255 (2021).
Wang, P., Doxtader, K. A. & Nam, Y. Structural basis for cooperative function of Mettl3 and Mettl14 methyltransferases. Mol. Cell 63, 306–317 (2016).
Wang, X. et al. Structural basis of N6-adenosine methylation by the METTL3–METTL14 complex. Nature 534, 575–578 (2016).
Sledz, P. & Jinek, M. Structural insights into the molecular mechanism of the m(6)A writer complex. Elife 5, e18434 (2016).
Shima, H. et al. S-adenosylmethionine synthesis is regulated by selective N(6)-adenosine methylation and mRNA degradation involving METTL16 and YTHDC1. Cell Rep. 21, 3354–3363 (2017).
Pendleton, K. E. et al. The U6 snRNA m(6)A methyltransferase METTL16 regulates SAM synthetase intron retention. Cell 169, 824–835.e14 (2017).
Warda, A. S. et al. Human METTL16 is a N6-methyladenosine (m6A) methyltransferase that targets pre-mRNAs and various non-coding RNAs. EMBO Rep. 18, 2004–2014 (2017).
Su, R. et al. METTL16 exerts an m6A-independent function to facilitate translation and tumorigenesis. Nat. Cell Biol. 24, 205–216 (2022).
Yoshinaga, M. et al. The N6-methyladenosine methyltransferase METTL16 enables erythropoiesis through safeguarding genome integrity. Nat. Commun. 13, 6435 (2022).
Mendel, M. et al. Methylation of structured RNA by the m(6)A writer METTL16 is essential for mouse embryonic development. Mol. Cell 71, 986–1000.e11 (2018).
Scarborough, A. M. et al. SAM homeostasis is regulated by CFIm-mediated splicing of MAT2A. eLife 10, e64930 (2021).
Mendel, M. et al. Splice site m(6)A methylation prevents binding of U2AF35 to inhibit RNA splicing. Cell 184, 3125–3142.e25 (2021).
Watabe, E. et al. m(6) A-mediated alternative splicing coupled with nonsense-mediated mRNA decay regulates SAM synthetase homeostasis. EMBO J. 40, e106434 (2021).
Yoshida, H. et al. Elucidation of the aberrant 3’ splice site selection by cancer-associated mutations on the U2AF1. Nat. Commun. 11, 4744 (2020).
Gu, J., Patton, J. R., Shimba, S. & Reddy, R. Localization of modified nucleotides in Schizosaccharomyces pombe spliceosomal small nuclear RNAs: modified nucleotides are clustered in functionally important regions. RNA 2, 909–918 (1996).
Brow, D. A. & Guthrie, C. Spliceosomal RNA U6 is remarkably conserved from yeast to mammals. Nature 334, 213–218 (1988).
Sawa, H. & Abelson, J. Evidence for a base-pairing interaction between U6 small nuclear RNA and 5’ splice site during the splicing reaction in yeast. Proc. Natl Acad. Sci. USA 89, 11269–11273 (1992).
Sawa, H. & Shimura, Y. Association of U6 snRNA with the 5’-splice site region of pre-mRNA in the spliceosome. Genes Dev. 6, 244–254 (1992).
Wassarman, D. & Steitz, J. Interactions of small nuclear RNA’s with precursor messenger RNA during in vitro splicing. Science 257, 1918–1925 (1992).
Ishigami, Y., Ohira, T., Isokawa, Y., Suzuki, Y. & Suzuki, T. A single m(6)A modification in U6 snRNA diversifies exon sequence at the 5’ splice site. Nat. Commun. 12, 3244 (2021).
Shen, A. et al. U6 snRNA m6A modification is required for accurate and efficient splicing of C. elegans and human pre-mRNAs. Nucleic Acids Res. 52, 9139–9160 (2024).
Parker, M. T., Fica, S. M., Barton, G. J. & Simpson, G. G. Inter-species association mapping links splice site evolution to METTL16 and SNRNP27K. eLife 12, e91997 (2023).
Roost, C. et al. Structure and thermodynamics of N6-methyladenosine in RNA: a spring-loaded base modification. J. Am. Chem. Soc. 137, 2107–2115 (2015).
Moravcevic, K. et al. Kinase associated-1 domains drive MARK/PAR1 kinases to membrane targets by binding acidic phospholipids. Cell 143, 966–977 (2010).
Aoyama, T., Yamashita, S. & Tomita, K. Mechanistic insights into m6A modification of U6 snRNA by human METTL16. Nucleic Acids Res. 48, 5157–5168 (2020).
Ju, J. et al. Structure of the caenorhabditis elegans m6A methyltransferase METT10 that regulates SAM homeostasis. Nucleic Acids Res. 51, 2434–2446 (2023).
Yamashita, S., Takagi, Y., Nagaike, T. & Tomita, K. Crystal structures of U6 snRNA-specific terminal uridylyltransferase. Nat. Commun. 8, 15788 (2017).
Yu, S. & Kim, V. N. A tale of non-canonical tails: gene regulation by post-transcriptional RNA tailing. Nat. Rev. Mol. Cell Biol. 21, 542–556 (2020).
Ruszkowska, A., Ruszkowski, M., Dauter, Z. & Brown, J. A. Structural insights into the RNA methyltransferase domain of METTL16. Sci. Rep. 8, 5311 (2018).
Doxtader, K. A. et al. Structural basis for regulation of METTL16, an S-adenosylmethionine homeostasis factor. Mol. Cell 71, 1001–1011 e4 (2018).
Chavali, S. S., Cavender, C. E., Mathews, D. H. & Wedekind, J. E. Arginine forks are a widespread motif to recognize phosphate backbones and guanine nucleobases in the RNA major groove. J. Am. Chem. Soc. 142, 19835–19839 (2020).
Pham, V. V., Gao, M., Meagher, J. L., Smith, J. L. & D’Souza, V. M. A structure-based mechanism for displacement of the HEXIM adapter from 7SK small nuclear RNA. Commun. Biol. 5, 819 (2022).
Corbeski, I. et al. The catalytic mechanism of the RNA methyltransferase METTL3. eLife 12, RP92537 (2024).
Breger, K. & Brown, J. A. Elucidating the kinetic mechanism of human METTL16. Biochemistry 62, 494–506 (2023).
Trippe, R., Sandrock, B. & Benecke, B. J. A highly specific terminal uridylyl transferase modifies the 3’-end of U6 small nuclear RNA. Nucleic Acids Res. 26, 3119–3126 (1998).
Trippe, R. et al. Identification, cloning, and functional analysis of the human U6 snRNA-specific terminal uridylyl transferase. RNA 12, 1494–1504 (2006).
Yashiro, Y. & Tomita, K. Function and regulation of human terminal uridylyltransferases. Front Genet 9, 538 (2018).
Yamashita, S. & Tomita, K. Mechanism of U6 snRNA oligouridylation by human TUT1. Nat. Commun. 14, 4686 (2023).
Yamashita, S. & Tomita, K. Cryo-EM structure of human TUT1:U6 snRNA complex. Nucleic Acids Res. 53, gkae1314 (2025).
Wang, J., Yashiro, Y., Sakaguchi, Y., Suzuki, T. & Tomita, K. Mechanistic insights into tRNA cleavage by a contact-dependent growth inhibitor protein and translation factors. Nucleic Acids Res. 50, 4713–4731 (2022).
Collart, M. A. & Oliviero, S. Preparation of yeast RNA. Curr. Protoc. Mol. Biol. Chapter 13, Unit13.12 (2001).
Kabsch, W. XDS. Acta Crystallogr. D. Biol. Crystallogr. 66, 125–132 (2010).
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Varadi, M. et al. AlphaFold protein structure database in 2024: providing structure coverage for over 214 million protein sequences. Nucleic Acids Res. 52, D368–D375 (2023).
Moafinejad, S. N. et al. SimRNAweb v2.0: a web server for RNA folding simulations and 3D structure modeling, with optional restraints and enhanced analysis of folding trajectories. Nucleic Acids Res. 52, W368–W373 (2024).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D. Biol. Crystallogr 66, 486–501 (2010).
Afonine, P. V. et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D. Biol. Crystallogr 68, 352–367 (2012).
Williams, C. J. et al. MolProbity: more and better reference data for improved all-atom structure validation. Protein Sci. 27, 293–315 (2018).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Sanchez-Garcia, R. et al. DeepEMhancer: a deep learning solution for cryo-EM volume post-processing. Commun. Biol. 4, 874 (2021).
Meng, E. C. et al. UCSF ChimeraX: Tools for structure building and analysis. Protein Sci. 32, e4792 (2023).
Acknowledgements
We thank Dr. Suzuki for providing the Δmtl16 strains and vectors expressing spMETTL16. This work was supported by Grants-in-Aid for Scientific Research (A) [grant numbers 23H00368 and 18H03980 to K.T.], and a Grant-in-Aid for Scientific Research on Innovative Areas from the Ministry of Education, Culture, Sports, Science, and Technology of Japan [grant number 26113002 to K.T.]. K.T. was also supported by grants from Takeda Science Foundation, Naito Foundation, Uehara Memorial Foundation, the Terumo Foundation for Life Science and Art, the Yasuda Medical Foundation, and the Princess Takamatsu Cancer Research Fund. The cryo-EM data collection was supported by Research Support Project for Life Science and Drug Discovery (Basis for Supporting Innovative Drug Discovery and Life Science Research (BINDS)) from AMED under Grant Number JP23ama121002.
Author information
Authors and Affiliations
Contributions
K.T. planned and designed the research. J.J. performed structural and biochemical analyses. K.T. and J.J. analyzed the data and wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Jinwei Zhang, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Ju, J., Tomita, K. Structures and mechanisms of U6 snRNA m6A modification by METTL16. Nat Commun 16, 7708 (2025). https://doi.org/10.1038/s41467-025-63021-0
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-63021-0