Abstract
The Zika viral protease NS2B-NS3 is essential for the cleavage of viral polyprotein precursor into individual structural and non-structural (NS) proteins and is therefore an attractive drug target. Generation of a robust crystal system of co-expressed NS2B-NS3 protease has enabled us to perform a crystallographic fragment screening campaign with 1076 fragments. 46 fragments with diverse scaffolds are identified to bind in the active site of the protease, with another 6 fragments observed in a potential allosteric site. To identify binding sites that are intolerant to mutation and thus suppress the outgrowth of viruses resistant to inhibitors developed from bound fragments, we perform deep mutational scanning of the NS2B-NS3 protease. Merging fragment hits yields an extensive set of ‘mergers’, defined as synthetically accessible compounds that recapitulate constellations of observed fragment-protein interactions. In addition, the highly sociable fragment hits enable rapid exploration of chemical space via algorithmic calculation and thus yield diverse possible starting points. In this work, we maximally explore the binding opportunities to NS2B-NS3 protease, facilitating its resistance-resilient antiviral development.
Similar content being viewed by others
Introduction
Zika virus (ZIKV) belongs to the Flaviviridae family and is closely related to other flaviviruses such as Dengue virus (DENV), West Nile virus (WNV) and Yellow Fever virus (YEV). Although ZIKV infections typically manifest with mild symptoms, outbreaks in Brazil and French Polynesia revealed that the virus can cause microcephaly in newborns and Guillain-Barré syndrome in adults1,2,3. While this development sparked significant global health concern, no vaccine or antiviral therapeutic is available for the prevention or treatment of ZIKV infection4.
The single-stranded, positive-sense ZIKV RNA is translated to produce a single polyprotein precursor that is cleaved by host proteases and viral protease NS2B-NS3 into individual functional proteins. NS3 contains a protease active site, with a serine, histidine and aspartate catalytic triad. Approximately 40 amino acids (aa) of NS2B function as a cofactor of NS3. These residues wrap around NS3, with the C-terminal residues forming a β-hairpin that structurally contributes to the protease active site as well as participates in substrate recognition5. Structural studies have revealed several conformations of this two-component NS2B-NS3 protease, specifically a closed conformation6, an open conformation7,8, and a super-open conformation9, depending on the dynamic interactions between NS2B and NS3. In the open and super-open conformations, the C-terminal portion of NS2B is disordered and distant from the NS3 catalytic site, resulting in an unstructured substrate binding site. These two conformations therefore, have been considered as inactive forms, while the closed conformation of NS2B-NS3 with a well-structured binding pocket represents a promising targeting opportunity for antiviral development.
Two strategies have been deployed to target the NS2B-NS3 protease. The first involves the development of specific inhibitors that compete with the substrate on the active site. The second strategy focuses on non-competitive inhibitors, which target allosteric sites. Despite these efforts, neither competitive nor non-competitive inhibitors have advanced to clinical trials. Most of the reported competitive NS2B-NS3 inhibitors are limited to peptide mimics, such as macrocyclic inhibitors10,11. Although these peptidomimetic inhibitors display nanomolar potency against ZIKV NS2B-NS3 protease, and some additionally have broad-spectrum antiviral activity, inhibiting growth of Dengue and West Nile viruses as well as Zika virus12, their cellular activity and pharmacokinetic properties are poor. In parallel, great effort has focused on developing allosteric inhibitors that block the interactions between NS2B and NS3 and lock the protease in the inactive form13,14. This strategy has yielded several potent non-competitive inhibitors, including NSC13561815, which shows antiviral activity in cellular assays. Nevertheless, the allosteric site as well as the allosteric inhibitor binding mode have not yet been properly validated due to a lack of reliable structural information.
Viruses can evolve resistance to any inhibitor if the amino acids that such an inhibitor binds exhibit sufficient tolerance to mutation, while the mutations that abrogate inhibitor binding do not greatly impair viral fitness. To prioritize NS2B-NS3-binding fragments that exhibit genetic constraint and thus are less likely to be able to evolve resistance, we performed a deep mutational scan (DMS) of the NS2B-NS3 protease. DMS is a high-throughput experimental technique that relies on deep sequencing technology to measure the effect of each possible amino acid mutation at each position of a protein. DMS has emerged as a powerful tool in virology, particularly in studies of viral fitness and evolution16. Recent studies of the mutational potential of viral proteins, such as the SARS-CoV-2 spike protein17 and the ZIKV envelope protein18, provide rich information about the sequence-function relationships that apply to antibody engineering as well as vaccine design.
In this work, we combined crystallographic fragment screening with DMS to accelerate the development of resistance-resilient antiviral therapeutics targeting ZIKV NS2B-NS3 protease. Our crystallographic fragment screening against 1076 fragments identified diverse chemical scaffolds at both the active site and a potential allosteric site, providing structural details of fragment engagement for rapid follow-up compound design. The DMS results reveal the mutational potential of the hotspots for ligand engagement, which will guide rational compound design to suppress drug resistance at early stages of medicinal chemistry.
Results
Establishing a suitable crystal system for fragment screening
Several constructs have been reported to efficiently generate the ZIKV NS3 protease bound to the NS2B co-factor region. These approaches include the insertion of a glycine linker between the NS2B and NS3, as shown in the construct named gZiPro (Fig. 1a), and the co-expression of the NS2B co-factor and NS3 protease in a bicistronic vector, as demonstrated in the construct named bZiPro6 (Fig. 1a). Construct gZiPro has been commonly used for ZIKV NS2B-NS3 protease studies. However, the artificial glycine-rich linker raises concerns of altering the substrate-binding behaviour19. Therefore, the unlinked binary NS2B-NS3 protease is preferred for studies. In addition, bZiPro succeeded in producing a closed conformation of ZIKV NS2B-NS3 structure (PDB ID: 5GPI)6.
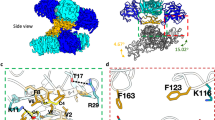
a Domain boundaries of constructs gZiPro, bZiPro and cZiPro. b Structure superimposition of bZiPro structure 5GPI (NS2B coloured in cyan, NS3 coloured in purple) and cZiPro structure 8PN6 (NS2B coloured in yellow, NS3 coloured in grey) with an RMSD of 0.2 Å. c Enzymatic activities of protease constructs cZiPro, bZiPro and gZiPro. Activities were measured using a fluorescence-dose-response assay with Bz-nKRR-AMC as substrate. Data points represent mean values, and error bars indicate standard deviations (SD) from three independent replicates (n = 3). d Surface view of ZIKV NS2B-NS3 fragment screening output in two orientations. The NS3 protein surface is coloured in grey and NS2B is coloured in yellow. Identified fragments are shown as sticks. Source data are provided as a Source Data file.
To produce crystals of ZIKV NS2B-NS3 protease in closed conformation for fragment screening, we generated a modified version of the PDB structure 5GPI6. The model of 5GPI contains 4 protein molecules in the asymmetric unit, with the N-terminal residues of NS3 in one molecule binding to a neighbouring active site, making this unsuitable for crystal soaking. A further issue with this system was the low expression yields of construct bZiPro in our hands. To address these issues, we generated a construct, designated cZiPro (Fig. 1a). In this construct, we removed the disordered C-terminal residues of NS2B and the N-terminal residues of NS3 in the 5GPI structure. This optimized construct yielded high quantities of ZIKV NS2B-NS3 protease and reproducibly produced crystals diffracting around 1.6 Å.
Our NS2B-NS3 structure (PDB code: 8PN6) was determined in P4322 space group with only one protein molecule in the asymmetric unit, which is distinct from 5GPI model, yet the superposition of both structures revealed a backbone root-mean-square deviation (RMSD) of around 0.2 Å (Fig. 1b). The active site in our structure is not occluded by crystal packing, and is accessible via crystal soaking, thus making it suitable for fragment screening.
The structural similarity between cZiPro and bZiPro supports their observed functional equivalence. Both enzymes displayed comparable enzymatic activities with Km values around 6 µM and higher catalytic efficiencies than gZiPro (Fig. 1c), consistent with a previous study19. Moreover, all three proteases were similarly inhibited by bovine pancreatic trypsin inhibitor (BPTI), with IC₅₀ values near 150 nM (Supplementary Fig. 1a). bZiPro and cZiPro exhibited similar thermal stability, with melting temperatures (Tm) of 47.3 °C and 46.6 °C, respectively, as measured by Differential Scanning Fluorimetry (DSF). In contrast, gZiPro was the most thermally stable, with a Tm of 50.8 °C, likely because of its covalent linkage between the NS3 and NS2B cofactor (Supplementary Fig. 1b).
Fragment hit identification by crystallographic fragment screening
We used our established crystal system for a crystallographic fragment screening campaign against 1076 fragments using the XChem facility at Diamond Light Source. A total of 1058 datasets with resolution ranging from 1.4–2.3 Å were collected. 51 fragments were identified from these screening efforts (see Supplementary Data 1), with 46 fragments bound in the active site and 6 in a potential allosteric site (one fragment bound to both the active site and allosteric site). Diverse chemical scaffolds were identified, including benzothiazole/benzimidazole, piperazine and quinoline-based fragments (Supplementary Fig. 2). Other moieties such as amides, pyrazoles, and sulfones were also commonly observed among the screened fragments.
Interaction motifs for ligand engagement in the active site
The active site of NS2B-NS3 protease site is divided into subsites S1, S1’, S2 and S3 based on Schechter–Berger nomenclature20 (Fig. 2a). Of the 46 fragments observed in the active site, 43 bound in the S1, 1 in the S1’ and 2 in the S2, respectively, suggesting subsite S1 is a hotspot for fragment binding. Fragment poses in the S1 site, such as x0089 (Fig. 2d), were frequently observed to form π-stacking with the side chain of Tyr161. x0089 was further stabilised by three hydrogen bonds, interacting with the side chain of Asp129, the backbone of Tyr130, and a water molecule, respectively. Fragment x1098 accommodated at both S1 and S1’ site (S1-S1’) (Fig. 2e), forming two π-π stacking interactions with the side chains of Tyr161 and His51. In addition, its amide motif formed a hydrogen bond with the backbone of Gly151 as well as a water-mediated hydrogen bond with the side chain of Tyr161.
a Surface view of observed fragments bound in the active site. The active site is divided into subsites S1, S1’, S2 and S3 based on Schechter–Berger nomenclature. b Key interacting residues revealed from fragment screening. Key interacting residues from NS3 are coloured in salmon, residue from NS2B is coloured in yellow. HBA: hydrogen bond acceptor. HBD: hydrogen bond donor. c A plot of key residues observed for fragment interaction. Y-axis represents the number of fragments forming interaction with associated residues. Residue D83 is from NS2B. Representative examples of binding pose x0089 (d), x1098 (e) and x0404 (f) interacting at the S1, S1-S1’, and S2 site, respectively. g The unique pose x0846b observed in S1’ site. The PanDDA event map is shown as a dark grey mesh at 1.5 σ. Hydrogen bonds are shown as black dashed lines. Electrostatic interaction is shown in an orange dashed line. Crystal ID in bold font to represent the fragment and its binding pose. Source data are provided as a Source Data file.
S2 site binder piperazine fragment x0404 formed an electrostatic interaction with Asp83 (NS2B), while its benzene ring formed a π-stacking interaction with His51 (Fig. 2f). We also observed one fragment pose x0846b (fragment bound to two sites, pose b close to catalytic residue His51), which accommodated in S1’ site, forming two hydrogen bonds with backbones of Val36 and Val52, respectively (Fig. 2g). Its benzimidazole ring formed hydrophobic interaction with surrounding residues, such as Val36, Leu30, and Met51 (NS2B).
Analysis of fragment-protein interactions revealed that π-stacking with the side chain of Tyr161 is a key interaction in the S1 site, represented by 39 of 46 fragments observed in this site (Fig. 2c). Residues Asp129 and Tyr130 were also observed to form hydrogen bonds with nearly a quarter of identified fragments, together with Tyr161 and Tyr150 making the S1 site a hotspot for ligand engagement. Additionally, residues His51, Ser135 and Asn152 from NS3, and Asp83 from NS2B were also observed in fragment interactions (Fig. 2b, c), indicating their potential contribution to ligand engagement.
Deep mutational scanning to measure mutational tolerance of ZIKV NS2B-NS3 protease
Given that viral proteases often evolve resistance to inhibitors through mutations in key residues, examples such as drug resistance against HIV-1 and HCV NS3/4A proteases21,22,23. Understanding the mutational tolerance of ZIKV NS2B-NS3 protease is essential for designing resistance-resilient antivirals. To gain a comprehensive view of how specific amino acid substitutions, especially at key ligand-interacting residues, affect the protease’s function and ligand engagement, we employed deep mutational scanning (DMS). Specifically, we generated libraries encoding all single amino acid substitution mutants of the entire coding sequence of ZIKV NS2B-NS3 using a previously reported infectious clone of the ZIKV prototype strain MR76624. To ensure comprehensive coverage of all possible mutations during library cloning and screening stages, we divided the NS2B-NS3 coding sequence into three distinct ‘tiles’ (Supplementary Fig. 3a) and created multiple mutant plasmid libraries for each tile (three each for tiles 1 and 2, and two for tile 3) (Supplementary Data 2), which were handled separately in all subsequent steps to provide true biological replicates. Each library contained an average of 2.7×106 unique plasmid clones, which overrepresents the total number of possible codon variants of 3296 (32 combinations of NNK multiplied by 103 codons) by 819-fold, with an average of 1.05 mutations per clone.
Mutant virus pools were produced by transfection of HEK 293 T cells with the plasmid DNA libraries and selected for viral fitness by passaging through Huh-7.5 cells25 (Supplementary Fig. 3b). We used deep sequencing to quantify the frequency of each mutation in the mutant viruses relative to the initial plasmid mutant libraries26. Stop codons, which are expected to be uniformly deleterious, were greatly purged, and nonsynonymous mutations, many of which will be deleterious, decreased in frequency (Supplementary Fig. 3c).
We next estimated the preference of each site in the NS2B-NS3 protease protein for each amino acid. These preferences represent enrichments of each amino acid at a site after selection for viral growth, normalized to the abundance of the wild-type codon at each site. Although there is some noise, the preferences were strongly correlated among three replicates (Supplementary Fig. 3d). We experimentally quantified the relative fitness of a panel of individual NS2B-NS3 mutants and found these values highly correlated (Pearson correlation of 0.769) with the mutation effects measured by DMS (Supplementary Fig. 3e).
The mutational tolerance at any given codon can be determined from the effects of each individual mutation at that site on viral fitness. The across-replicate average of the effects of each mutation at each site in the NS2B-NS3 protease protein was calculated as the log-fold change in the frequency normalized to the change in frequency of a wild-type control and represented in the heatmap (Fig. 3a). At the lower end of fitness, stop-codon mutations and catalytic triad mutations are considered to be deleterious. Thus, their mutational effects have been applied as reference sets within the distribution of fitness effects of NS2B-NS3 mutations (Fig. 3b). Based on the above experimental testing of the fitness of individual mutants (Supplementary Fig. 3e), as well as our prior analysis of envelope protein mutational tolerance18, we set a mutational effect threshold of fitness at -1.0 for mutations that are tolerated and allow the growth of virus, even though the fitness is reduced from that of wild-type (Fig. 3b).
a The heatmap of NS2B-NS3 residues indicating the mutational effect of each amino acid substitutions in the ZIKV NS2B-NS3 protease. Blue mutations are deleterious for viral growth in Huh-7.5 cells relative to wildtype, white mutations are neutral, and red mutations increase growth in Huh-7.5 cells. Wildtype amino-acid identities at each site are denoted by an ‘X’. In general, most mutations decrease the fitness of the virus in Huh-7.5 cells. b Distribution of fitness estimates for all mutations. The distributions of fitness effects of stop-codon mutants, Catalytic triad mutants and the remaining mutants are coloured in blue, orange and green, respectively. The black vertical line indicates the threshold −1.0 (Log2 (DMS effect)) for fitness. c Fitness view of ZIKV NS2B-NS3 protease. Mutational tolerance is mapped onto the structure of ZIKV NS2B-NS3 (PDB ID: 8PN6). Residues marked in white do not tolerate changes to that site, while residues marked in red tolerate a range of changes. The number of changes tolerated is indicated by the number and the colour. Source data are provided as a Source Data file.
Most mutations in NS2B-NS3 reduce the fitness of the virus in Huh-7.5 cells (Fig. 3a, blue squares), suggesting low mutational tolerance at many residues. Some residues, however, such as NS2B residues 88-94 and NS3 residues 12-20, exhibit high tolerance for mutation (white squares). Notably, mutations at these sites do not decrease viral fitness in Huh-7.5 cells, instead displaying fitness similar to that of viruses with wild-type residues (X) at these positions. Some sites, such as NS3 Arg106 (pink squares), showed both a high tolerance for changes and an increase in fitness when certain mutations were introduced. Residues in the protein that have a high tolerance for mutations are not good candidates for ligand binding sites, because mutations that disrupt ligand binding do not compromise viral fitness.
To facilitate analysis of these data, we generated a visualization scheme, termed ‘Fitness View’, to map the mutational tolerance of each residue to our crystal structure (Fig. 3c). Surfaces that are coloured in white indicate sites that are mutationally intolerant, while residues that are dark red represent a high tolerance for mutations. Overall, this fitness map provides a wealth of information about the sequence-function relationships in the NS2B-NS3 protease protein that can be used to guide the selection of key interacting residues for rational antiviral design.
Fitness view reveals potential mutational effect on ligand engagement in the active site
Inspection of the fitness view in the active site reveals that S1’ and S2 subsites are relatively intolerant of nonsynonymous mutations (Fig. 4a), while a few positions in the S1 subsite and at the C-terminus of NS2B show mutational tolerance, indicating a risk of potential resistance mutations. Specifically, Tyr130 in the S1 subsite can potentially mutate to multiple amino acids, such as cysteine, glutamic acid. At position 132, cysteine and proline substitutions are predicted to have similar replication efficiency to the wild-type alanine (Fig. 4b). Despite the high mutability of Tyr130 and Ala132, S1 fragment binders from our screen predominantly interact with the backbone of Tyr130 and have indirect contact with Ala132. Thus, the effect of the potential mutations of Tyr130 and Ala132 on ligand engagement is estimated to be limited. Similarly, the mutational flexibility of Gly82 and Phe84 at the C-terminus of NS2B is less of a concern. Phe84 hasn’t been observed for fragment interaction, while Gly82 used its backbone for fragment interaction.
a Fitness view of the active site with fragment x1098 presented. Key interacting residues identified from fragment screening are numbered. The mutational intolerant region is coloured in white, and mutational tolerant region is coloured in red. b Logo plot of experimentally measured amino acid preferences of the labelled key interacting residues in Fig. 4a. High mutational residues are marked with an asterisk sign. c Example of fragment pose x0719 formed a direct hydrogen bond with the hydroxyl group of Tyr161. d Vectors for fragment growth. Regions that suggest to form backbone interaction are coloured in blue. Side chain of Tyr161 is highlighted by colour orange due to its potential mutation to phenylalanine. Opportunity for fragment growth is shown as an arrow.
However, the potential mutation of tyrosine to phenylalanine at position 161 of NS3 raises our concern. Although Tyr161 contributes to fragment interaction mainly via π-stacking, which can be replaced by phenylalanine, we also observed fragments, such as x0719 (Fig. 4c), that formed a direct H-bond with the hydroxyl group of Tyr161. Mutating tyrosine to phenylalanine would abolish the hydrogen bond formation. Thus, hydrogen bonding to Tyr161 is suggested to be avoided for follow-up compound design to decrease the probability of drug resistance. In contrast, key interacting residues Asp129 in the S1 subsite and Asp83 (NS2B) in the S2 subsite are mutational intolerant, suggesting opportunities for fragment growth (Fig. 4a, d).
Fitness view invalidates the reported allosteric site as resistance-robust opportunity
Six fragment hits were observed outside of the orthosteric site, in a pocket located beneath the catalytic residues and next to the interface between the C-terminal β-hairpin of NS2B and NS3 (Fig. 5a). This potential allosteric site contains two sub-pockets, named as allosteric site AS1’ and AS2’ (Fig. 5b, c). AS1’ has Trp69 and Trp83 as its major interacting residues. One example, fragment pose x0130, formed π-stacking with the side chain of Trp69 and was further stabilised by hydrogen bonding to the backbone of Trp83 (Fig. 5b).
a Overview of fragments observed in the non-active site. Fragments are shown as coloured sticks. b Structural details of observed pose x0130 at allosteric site 1 (AS1’). Residues that participate in fragment interaction are shown as sticks. Representative examples of pose x0806 (c) and x0777 (d) bound in the AS2’ site. Hydrogen bonds are shown as dashed lines. The PanDDA event map is shown as a dark grey mesh at 1.5 σ. Water molecules are shown as red spheres. e Surface view of an allosteric site defined by reported docking and mutational studies. Labelled residues are reported for ligand interaction. Oxygen, nitrogen and carbon atoms are coloured in red, blue and cyan, respectively. f Fitness view of the potential allosteric site mapping to Fig. e. The logo plot presents the experimentally measured amino acid preferences of the interacting residues in the non-active site. High mutational residues are marked with an asterisk sign.
AS2’ is formed mainly by hydrophobic residues, including Phe116, Ile123, Ala164, and Ile165 from NS3, and Leu78 and Phe84 from NS2B. Four fragments with different scaffolds were observed in AS2’ with different poses. Fragment x0806 contains a benzene ring that formed hydrophobic interactions with surrounding hydrophobic residues (Fig. 5c), while the nitrogen atom N2 from its triazole ring formed a hydrogen bond with the backbone of Gln167. Another hydrogen bond was formed between its amine group and a water molecule (Fig. 5c). In contrast, Fragment x0777 bound in AS2’ exhibited a completely different binding mode. Its hydroxyl group formed two water-mediated hydrogen bonds with the side chains of Thr118 and Asp120 (Fig. 5d). The N2 atom from its thiadiazole ring formed another water-mediated hydrogen bond with Asp71, and the other nitrogen atom (N5) formed a hydrogen bond with water from the solvent. Its 5-carbon ring fit into a hydrophobic cavity that is formed by Ile123, Phe116 and Ala164. In conjunction, the fragments bound in this non-active site mapped a potential allosteric pocket next to the interface of the C-terminal β-hairpin of NS2B and NS3. This observation agrees with previous work of characterizing allosteric pockets in ZIKV NS2B-NS3 via docking and mutational studies27,28 (Fig. 5e).
We next investigated the genetic flexibility of the key interacting residues in the allosteric site. Compared to the active site, the allosteric site has relatively higher mutability (Fig. 5f). In particular, the mutational flexibility of residues Asp71 and Thr118 raises our concern. Asp71 was observed to interact with fragments via hydrogen bonding (Fig. 5d). Its potential of mutating to serine would alter its charge as well as the length of the side chain, raising the risk of maintaining interactions with ligands. Similarly, the polar residue Thr118 has the potential of mutating to methionine, which has a longer side chain that may potentially narrow the pocket and crash with the ligand. Considering the potential mutational risk and underdefined key interacting residues in the allosteric site, targeting this allosteric site has been deprioritized in this study.
Identified fragments are undetected in binding and activity assays
To further evaluate whether our screened fragment binders have on-scale binding affinity and inhibitory activity, we characterized the fragment binders using the GCI-based waveRAPID system to determine the binding affinity and a fluorescence-dose-response assay to determine their inhibitory activity29. waveRAPID is developed for compound and fragment screening and has demonstrated the capability to detect binding affinities in the low millimolar range30. Despite this, we were unable to obtain reliable results or a measurable signal to confidently validate our fragment binders using this binding assay (representative examples are shown in Supplementary Fig. 4a, b). Unsurprisingly, none of the screened fragments exhibited inhibitory activity in the biochemical assay (representative examples are shown in Supplementary Fig. 4d–f). Such observations are in common with many crystallographic fragment screens where only a low percentage of crystallographic fragment hits can be reliably detected in biophysical or biochemical assays31. This is further supported by a comparison study of different fragment screening assays, such as NMR, MST, and enzyme inhibitory assays, showing limited overlapping of fragment hits32.
Notably, the positive control compound showed an IC50 of 1.8 µM to ZIKV NS2B-NS3 in our assay (Supplementary Fig. 4g), consistent with a previous study33. However, its potency against DENV2 NS2B-NS3 is significantly better, with a reported IC50 value in the low nanomolar range34. This observation underscores the challenge as well as opportunity of developing broad-spectrum inhibitors targeting flaviviral NS2B-NS3 proteases, and highlights the importance of employing parallel biochemical and biophysical assays across a panel of NS2B-NS3 proteases.
Opportunities for rapid follow-up compound design via fragment merging and linking
Inspection of fragment-protein interaction profiling and fitness view in the active site, fragments x0852, x1098, x0404, and x0846b were selected as representative examples for merging (Fig. 6a). Fragment pose x0852 recapitulated core interactions observed in the S1 site, specifically aromatic interaction with Tyr161, hydrogen bonding with the backbone of Tyr130. x1098 is the only fragment observed that bridges the S1 and S2 sites with an amide motif (Fig. 6a). Its benzene ring nicely superimposed with the benzene ring of fragment x0852 in the S1 site (Fig. 6a), suggesting these fragments could be merged into a single, larger scaffold. In addition, x1098 partially overlaps with fragment x0404 in the S2 site (Fig. 6a). Merging x1098 and x0404 derives a benzamide-(pyrazolyl) piperidine scaffold, which expands the small molecule to engage the S1, S1’ and S2 sites simultaneously, represented by Enamine compound Z2451096209 (Fig. 6b).
a Fragments selected as parent hits for rapid follow-up compound design. Fragment alignment shows opportunities for merging and linking. b Predicted mergers posed in the active site. The 2D structure is shown at the top with its Enamine compound identity code. RMSD value indicates the conformational variance of its parent fragments. Fragments are shown in thin sticks, predicted mergers are shown in sticks and spheres. c A crystal structure of ASAP-0022538 bound to ZIKV NS2B-NS3 (PDB ID: 7I9O) is shown as an example of a merge design that achieves on-scale activity (low double-digit micromolar IC50). ASAP-0022538 recapitulates the poses and interactions of fragments x0089, x1098 and x0404, exhibiting an IC50 value of 14.2 µM. Water molecules are shown as red spheres. Hydrogen bonds and electrostatic interactions are shown as black and orange dashed lines, respectively. Aromatic stacking interactions are shown as titled lines. Omit map of ASAP-0022538 is shown in Supplementary Fig.5. The IC50 plot is shown as representative data from three independent experiments with consistent results (Source Data- Fig. 6c). Error bars in the IC50 plot indicate standard deviations (SD) from technical duplicates in one experiment. Source data are provided as a Source Data file.
Such benzamide-(pyrazolyl) piperidine were easily accessible with over 1000 analogues. They were further assessed by an algorithmic calculation called Fragmenstein35, which is based on the principle that the pose of mergers should preserve the poses of its parent fragments. We limited the conformational derivation of predicted mergers to be less than 1 Å in comparison with their parent hits in order to potentially maintain their parent fragments interactions. For example, the merger PV-004740099668 has an RMSD of 0.82 Å relative to its parent fragment poses x0852 and x1098, and merger Z2451096209 has an RMSD of 0.27 Å from its parent fragment poses x1098 and x0404 (Fig. 6b).
To further analyse whether potency can be achieved with combined fragments, we assessed the binding pose of ASAP-0022538 (PDB ID: 7I9O), a hit reported in a follow-up fragment-to-lead work using our screened fragments by Kenton et al.36. This compound recapitulated both the fragment poses and fragment-protein interactions. ASAP-0022538 appears aligned with the poses of x0089, x1098 and x0404 (Fig. 6c). Its benzimidazole overlaps with fragment x0089 in the S1 site, recapitulating the aromatic interaction and hydrogen bonding with Tyr161 and Tyr130, respectively (Fig. 6c). The amide linker of ASAP-0022538 maintains the hydrogen bond with the backbone of Gly151 as fragment x1098 does. The aromatic interaction between its benzene ring and His51, and the electrostatic interaction between the amine group and Asp83 (NS2B) recapitulate the interactions observed in fragment x0404. Combining these interacting motifs, ASAP-0022538 was reported to show on-scale potency with an IC50 value of 14.2 µM36 (Fig. 6c).
Notably, the design of ASAP-0022538 was executed separately to this work, and thus our fragment-based analysis above is retrospective. However, the utility of our fragments as the basis of inhibitor discovery was demonstrated by Kenton et al.36, who used our fragment data to initiate and complete a full fragment-to-lead campaign. The authors reported the use of machine learning of pharmacophoric motifs combined with iterative DMTA cycles in their campaign, progressing fragments to low-nanomolar potency36.
Manual fragment merging is relatively straightforward, but it is largely limited to pure merges37, such as Z2451096209 in our case, that incorporate the exact substructures of parent fragments. Merging algorithms that fully exploit chemical space, integrate key fragment-protein interactions and reliably rank compounds from possible merges are promising for identifying ASAP-0022538-like effective merges directly from the fragment screen. Such an approach is described in Wills et al.37, and while that requires further refinement of the methodology, compound ASAP-0022538 demonstrates that it is feasible.
In addition to merging, we observed that fragment x0846b, which is uniquely accommodated in the S1’ site, provides linking opportunities with x1098 via a methyl linker without causing strain or internal clash (Fig. 6b). This is supported by the RMSD value of 0.67 Å compared to its parent fragments while keeping a reasonable angle (112°) of the methyl linker. Such a binding region has not been explored for small molecule engagement. Our observation, despite only one fragment, brings opportunities for diverse scaffold inhibitor development.
To further explore chemistry diversity, we used all screened fragments as input for merging using Fragmenstein35. Automated merging, catalogue searching and constrained docking revealed a large number (around 4000 filtered catalogue compounds) and diversity of possible mergers that could be explored (Fig. 7), such as benzimidazole-based compound Z4192072491 (Enamine) and pyrazole naphthalene MCULE-8919924892. Both the mergers and the analogue docking were computed with Fragmenstein and the final compounds were predicted to be faithful to the parent fragments (RMSD < 1 Å) without violating the energy scoring function (∆Gcalc of binding < –5 kcal/mol). The abundant and diverse chemical matter suggested from algorithmically merging demonstrates that our screened fragments are highly sociable38, enabling many possible avenues of exploration.
Discussion
We obtained a robust ZIKV NS2B-NS3 crystal system that enabled a successful crystallographic fragment screen. The sampled fragment hits provide diverse possible starting points for rapid follow-up compound design. This brings opportunities to this antiviral target, where currently the orthosteric inhibitors are largely limited to peptidomimetics39. In addition, analysis of fragment-protein interaction profiling highlights the S1 subsite as the major targeting opportunity for orthosteric inhibitor development. This is supported by over 80% of observed fragments being accommodated in this region. Such a high rate of hits observed in the S1 site is likely due to the deep pocket mainly formed by the bulky side chains of Tyr150 and Tyr161.
Drug resistance remains one of the major challenges in developing effective protease inhibitors. Lessons from previous drug discovery campaigns, such as those targeting HIV-1 protease40, highlight the importance of understanding how key interacting residues tolerate mutations. To address this, we applied DMS to profile the mutational landscape of our target protein. This approach aids in assessing fragment-protein interactions and prioritizing fragments for progression. Crystallographic fragment screening, due to its high sensitivity, often samples dozens of fragment binders, but not all of which can be effectively progressed to lead. Selection of fragment hits as promising chemical starting points requires careful filtration. Mutational profiling can serve as a powerful filter for fragment selection, particularly for antiviral targets. For instance, fragments like x0719 that interact with highly mutable residues may be deprioritized for fragment progression, as such interactions can be readily compromised by viral evolution.
Moreover, the mutational profile constrains the binding region for ligand engagement, providing rational guidance for fragment growth. For example, the mutationally intolerant residues such as Asp129 in the S1 site and Asp83 (NS2B) in the S2 site suggest a growth vector for fragment elaboration. We propose that incorporating DMS in the early stage of medicinal discovery provides rational guidance for fragment progression and supports resilient antiviral compound design.
Multiple allosteric sites of Flaviviral NS2B-NS3 protease have been proposed, but without structural description9,15,41. Our fragment screening data presented herein help to reduce this knowledge gap and map the allosteric site with structural details. Although the allosteric binding pocket we described here presents in a closed conformation, it agrees with a proposed allosteric site reported previously9,15, supporting the ‘ligandability’ of the interface of the C-terminal β-hairpin of NS2B and NS3. However, the distinct binding poses and interactions observed from our screening indicate the challenges for non-competitive inhibitor development. More importantly, DMS data revealed a high mutability of several interacting residues in this allosteric site, suggesting potential risks of drug resistance. We, therefore, prioritized the orthosteric site as the antiviral target. We hope this observation will provide an important insight into this field given that there is increasing effort in allosteric inhibitor development for Flavivirus NS2B-NS3 protease39.
Merging fragments can effectively lead to on-scale affinity from non-measurable weak binders42, and is therefore commonly applied for rapid follow-up compound design. This has been proven by several successful fragment-to-lead case studies, with targets including the SARS-CoV-2 main protease29 and NSP3 macrodomain43. In this study, our screened crystallographic fragments show neither measurable binding affinity nor inhibitory activity, not unexpectedly. This is not surprising as crystallographic fragment screen is a highly sensitive assay, likely due to the high concentration (100 mM in crystal drops) used in soaking. However, the structural details of fragment-protein interactions provide clear routes, or rich information to grow fragments for higher affinity. In our case, a key fragment x1098 bridges the S1 and S2 subsites and provides an essential pose for merging. Merging this fragment with S1 binders derives an easily accessible benzamide-(pyrazolyl) piperidine scaffold that offers a potential route for rapid follow-up compound design.
In conclusion, our integrative approach leveraging crystallographic fragment screening and deep mutational scanning provides both structural and functional insights into ligand interactions with ZIKV NS2B-NS3 protease. While the fragment screen mapped both orthosteric and allosteric binding pockets, the high mutability of key allosteric site residues identified through DMS underscored the risk of resistance and informed our prioritization of the orthosteric site. Although no initial fragment hits displayed measurable binding affinity or inhibition, structural elucidation revealed promising starting points for fragment merging, particularly through the S1–S2 bridging fragment x1098. These findings highlight not only the complexity of targeting this viral protease but also the power of structure-guided strategies in advancing rational antiviral design. Our work lays a foundation for the development of more robust and resistance-resistant flaviviral protease inhibitors.
Methods
Protein expression, purification and crystallization
A bicistronic construct here named cZiPro was based on the sequence of the PDB model (PDB ID:5GPI). The construct was created by using a synthetic E. coli codon-optimised gene ZIKV sequence as template (accession YP 009227202.1 in GenBank). Golden gate cloning was used to insert this into the pNIC-HIS6-GST-TEV-GG vector44. The final construct contains a GST fusion with a TEV site followed by the NS2B peptide (residues 45-89 aa). This is followed by an intervening ribosome binding site and the NS3 protease domain (residues 11-177aa). Cells cultured in TB media were initially grown at 37 °C, and they were induced with 0.5 mM IPTG at 18 °C overnight. Harvested cells were lysed by sonication in a buffer containing 50 mM HEPES, pH 7.5, 500 mM NaCl, 20 mM imidazole, 5% glycerol and 1 mM tris (2-carboxyethyl) phosphine (TCEP). The recombinant protein was initially purified by Ni2+-affinity chromatography. The His6 and GST tag were removed by TEV protease treatment, and the cleaved protein was passed through Ni2+ affinity beads and further purified by size exclusion chromatography using an HPLC column in the buffer containing 25 mM HEPES pH 7.5, 150 mM NaCl, 0.5 mM TCEP and 5% glycerol. The co-expressed protein NS2B-NS3 was concentrated to ~15 mg/ml for crystallisation. Crystallisation was performed using the sitting-drop vapour diffusion method at 20 °C. The crystals were obtained in the condition containing 30% w/v PEG2000, 0.2 M ammonium sulfate, 0.1 M sodium acetate, pH 4.8
Crystallographic fragment screening, data collection and analysis
The fragment screening was performed using the XChem facility at Diamond Light Source, UK. The fragments from the DSi-Poised Library45, MiniFrags Probing Library46, CovHetFrags47, and SpotXplorer48 were dispensed into crystal drops by ECHO Liquid Handler with 20% (v/v) DMSO in the final condition and incubated for three hours at 20 °C. Crystals were harvested using Crystal Shifter49 (Oxford Lab Technologies) and cryo-cooled in liquid nitrogen.
Diffraction data were collected at the I04-1 beamline at Diamond Light Source at 100 K and processed with automated pipelines using combined software, including XDS50, Autoproc51, Xia252 and DIALS53. All further analysis was performed using XChemExplorer54. Ligand restraints were generated with ACEDRG55 and GRADE56. Fragment hit identification was analysed by PanDDA57. The electron density map was generated by Dimple58. Model building and refinement were carried out in COOT59 and Buster60 via the XChemExplorer platform.
Cell lines and antibodies
HEK 293T and Huh-7.525 (provided by Charles M. Rice, Rockefeller University) cells were grown in Dulbecco’s modified Eagle’s medium (DMEM; Gibco BRL Life Technologies, Gaithersburg, MD) with 10% foetal bovine serum (FBS; Gibco BRL Life Technologies). The humanized monoclonal antibody D1-4G2-4-15 (4G2) (Absolute Antibody, Oxford, UK) is a broadly reactive flavivirus antibody that binds to an epitope at the fusion loop domain of the E protein61.
Deep mutational scanning of ZIKV NS2B-NS3 protease
Here we present a compressed summary of the methods used for ZIKV NS2B-NS3 protease DMS and the validations of the results. Detailed methods can be found in the Supplementary Information file.
To generate mutant plasmid libraries, we created NS2B-NS3 protease codon-mutant DNA fragments using a previously described PCR mutagenesis approach62 with sets of forward and reverse oligos that randomized each codon with an NNK sequence where N is any nucleotide and K is either a G or a T (Supplementary Data 2). These products were cloned into our previously described single-plasmid reverse genetics system for ZIKV strain MR766 (sequence is available at Genbank accession KX830961)24 using techniques like those described in our previous ZIKV envelope protein DMS paper18. We generated infectious virus stocks of these ZIKV NS2B-NS3 protease DMS libraries by transfecting plasmids into HEK 293T cells using a protocol to maintain library complexity as previously reported18,24,63. Wild-type virus was rescued in parallel as a control. These stocks were titered on Huh-7.5 cells, and then selected by infecting these cells at an MOI of 0.05 infectious units per cell. Infected cells were collected at day 2 post-infection.
Total RNA was extracted from these cells, reverse transcribed, and subjected to Illumina deep sequencing using a barcoded-subamplicon sequencing approach described in previous work64 (see also https://jbloomlab.github.io/dms_tools2/bcsubamp.html) to minimize sequencing errors. The raw deep sequencing data have been deposited in the Sequence Read Archive as BioProject PRJNA1125458. The code that performs the analyses of the deep sequencing data is available on GitHub at https://github.com/jbloomlab/ZIKV_DMS_NS3_EvansLab. This repository includes summary notebooks that provide detailed statistics like read depth and mutation frequencies for each library tile (see subdirectories of https://github.com/jbloomlab/ZIKV_DMS_NS3_EvansLab/results/summary/). Briefly, we used dms_tools265 (https://jbloomlab.github.io/dms_tools2/), version 2.4.14, to count the occurrences of each mutation in each sample (see https://jbloomlab.github.io/dms_tools2/bcsubamp.html for details). The amino-acid preferences were computed from these counts using the approach previously described65 (see also https://jbloomlab.github.io/dms_tools2/prefs.html). The mutational effects are the log of the preference for the mutant amino acid divided by the preference for the wild-type amino acid. Output containing the numerical values of the counts of mutations in each sample, the amino-acid preferences, and mutational effects are processed on a per-tile basis. These data are provided in CSV file format in the GitHub repository. See the README for details on navigating the analysis output.
GCI RAPID Kinetics for binding assay
The fragments were tested with Creoptix Wave (Malvern Panalytical) using PCH-NTA chips. First, the chip surface was conditioned with 1 M NaCl, 0.1 M borate pH 9.0 buffer for 180 s, followed by an injection of 250 mM EDTA solution for 180 s. Then the chip surface was equilibrated by injecting running buffer (10 mM HEPES pH 7.4, 150 mM NaCl, 0.05% Tween 20).
The immobilization of ZIKV NS2B-NS3 protease was performed using His capture combined with EDC/NHS conjugation. First, NiCl2 (500 µM) was injected to activate NTA groups on the chip surface for 420 s. Then, the running buffer was injected twice for 60 s to wash the chip surface. EDC/NHS (Xantec) was injected to activate carboxyl groups on the chip surface and to couple the target protease. The target protease was injected at a concentration of 15 µg/ml for 420 s on the FC2, FC3, and FC4 channels. Excessively activated groups were blocked with the injection of ethanolamine (1 M pH 8.5, Xantec) solution. The FC1 blank surface was treated the same as the FC2/FC3/FC4 active channel, except for the NS2B-NS3 protein capture step. The final NS2B-NS3 capture level was about 9000 pg/mm2 on the active channels. All steps within the immobilization were performed at a 10 µl/min flow rate. A kinetic binding assay was performed through the Repeated Analyte Pulses of Increasing Duration (RAPID) kinetics assay in Creoptix. Samples were injected at 250 µM for 5 s association and 20 s dissociation at a 400 µl/min flow rate. Positive control compound was tested at 1 µM for an accurate result. DMSO correction was included in the method and injected as a 0.5% DMSO addition to the running buffer + 2% DMSO. Blank solution (the running buffer + 2% DMSO) was injected after every 5th sample. All steps were conducted at 25˚C. Data analysis was carried out with adjustment of x and y offset, DMSO calibration, and blank subtraction in WAVE control software 4. 5.18.
Protease activity assay
The protease activity assay is carried out by monitoring the fluorescence generated upon cleavage of the fluorogenic substrate Bz-nKRR-AMC (International Peptides), which releases 7-amino-4-methylcoumarin (AMC) upon hydrolysis66. Reactions were performed in black 384-well Corning® microplates using a reaction buffer composed of 20 mM Tris-HCl, pH 8.5, 10% glycerol, and 0.01% Triton X-100. The protease was added at a final concentration of 5 nM to wells prefilled with reaction buffer. For kinetic analyses, substrate concentrations were varied in a serial dilution ranging from 300 µM to 0.2 µM. The kinetic parameters (Km, kcat, and Vmax) were calculated by nonlinear fitting to the Michaelis–Menten equation. All experiments were performed in triplicate, and data analysis was conducted using Origin 9.0 software (OriginLab).
Fluorescence-dose-response inhibitory activity assay
This protocol was adapted from a previous study29 with minor modifications. Fragments were seeded into assay-ready plates (Greiner 384 low volume, cat. no. 784076) using an Echo 555 acoustic dispenser, and dimethyl sulfoxide (DMSO) was back-filled for a uniform concentration in assay plates (DMSO concentration 1.5%). Dose-response assays were performed in 12-point dilutions of twofold, starting at 300 μM. Reagents for the assay were dispensed into the assay plate in 10 μl volumes for a final volume of 20 μl.
Final reaction concentrations were 20 mM Tris pH 8.5, 0.01% Triton, 10% glycerol, 15 nM ZIKV NS2B-NS3 and 5 µM fluorogenic peptide substrate (Boc-Gly-Arg-Arg-AMC, CAS # [113866-14-1(free base)], Biosynth (FB110553)). ZIKV NS2B-NS3 protease was pre-incubated with the compounds for 2 hr followed by the addition of substrate and a further 30 mins incubation (all incubations performed at room temperature).
Protease reaction was measured in a BMG Pherastar FS with a 360/470 nm excitation/emission filter set. Raw data were mapped and normalized to high (Protease with DMSO, no compounds) and low (No Protease, no compounds) controls using Genedata Screener software. Normalized data were then uploaded to CDD Vault (Collaborative Drug Discovery). Dose-response curves were generated for IC50 using nonlinear regression with the Levenberg–Marquardt algorithm with minimum inhibition of 0% and maximum inhibition 100%. To each run, we added the reported peptide-hybrid inhibitor compound 36 as a positive control (for more accurate IC50 measurements of this compound, the dose response started from 100 µM).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Construct cZiPro has been deposited to Addgene https://www.addgene.org/228663/. cZiPro is designed based on the PDB structure 5GPI. Crystallographic coordinates and structure factors for the cZiPro crystal structure and ASAP-0022538-bound structure have been deposited in the PDB with the accession codes 8PN6 and 7I9O, respectively. PDB codes of crystallographic fragment binders are listed in Supplementary Data 1. Source data are provided with this paper. The raw deep sequencing data have been deposited in the Sequence Read Archive as BioProject PRJNA1125458. Results of deep sequencing data are available on GitHub at https://github.com/jbloomlab/ZIKV_DMS_NS3_EvansLab. Source data are provided with this paper.
Code availability
The code that performs the analyses of the deep sequencing data is available on GitHub at https://github.com/jbloomlab/ZIKV_DMS_NS3_EvansLab.
References
de Araujo, T. V. B. et al. Association between microcephaly, Zika virus infection, and other risk factors in Brazil: final report of a case-control study. Lancet Infect. Dis. 18, 328–336 (2018).
Krauer, F. et al. Zika Virus infection as a cause of congenital brain abnormalities and Guillain-Barre Syndrome: Systematic Review. PLoS Med. 14, e1002203 (2017).
Lowe, R. et al. The Zika virus epidemic in Brazil: From discovery to future implications. Int. J. Environ. Res. Public Health 15, https://doi.org/10.3390/ijerph15010096.
Saiz, J. C. Therapeutic advances against ZIKV: A quick response, a long way to go. Pharmaceuticals 12, https://doi.org/10.3390/ph12030127.
Chappell, K. J., Stoermer, M. J., Fairlie, D. P. & Young, P. R. Insights to substrate binding and processing by West Nile Virus NS3 protease through combined modeling, protease mutagenesis, and kinetic studies. J. Biol. Chem. 281, 38448–38458 (2006).
Zhang, Z. et al. Crystal structure of unlinked NS2B-NS3 protease from Zika virus. Science 354, 1597–1600 (2016).
Chen, X. et al. Mechanisms of activation and inhibition of Zika virus NS2B-NS3 protease. Cell Res. 26, 1260–1263 (2016).
Erbel, P. et al. Structural basis for the activation of flaviviral NS3 proteases from dengue and West Nile virus. Nat. Struct. Mol. Biol. 13, 372–373 (2006).
Meewan, I. et al. Allosteric Inhibitors of Zika Virus NS2B-NS3 Protease Targeting Protease in “Super-Open” Conformation. Viruses, 15, https://doi.org/10.3390/v15051106.
Braun, N. J. et al. Structure-based macrocyclization of substrate analogue NS2B-NS3 Protease inhibitors of Zika, West Nile and Dengue viruses. ChemMedChem 15, 1439–1452 (2020).
Huber, S. et al. Structure-based optimization and characterization of macrocyclic Zika virus NS2B-NS3 Protease Inhibitors. J. Med. Chem. 65, 6555–6572 (2022).
Nunes, D. A. F. et al. NS2B-NS3 protease inhibitors as promising compounds in the development of antivirals against Zika virus: A systematic review. J. Med. Virol. 94, 442–453 (2022).
Yildiz, M., Ghosh, S., Bell, J. A., Sherman, W. & Hardy, J. A. Allosteric inhibition of the NS2B-NS3 protease from dengue virus. ACS Chem. Biol. 8, 2744–2752 (2013).
Li, Z. et al. Existing drugs as broad-spectrum and potent inhibitors for Zika virus by targeting NS2B-NS3 interaction. Cell Res 27, 1046–1064 (2017).
Brecher, M. et al. A conformational switch high-throughput screening assay and allosteric inhibition of the flavivirus NS2B-NS3 protease. PLoS Pathog. 13, e1006411 (2017).
Burton, T. D. & Eyre, N. S. Applications of deep mutational scanning in Virology. Viruses 13, https://doi.org/10.3390/v13061020 (2021).
Dadonaite, B. et al. Spike deep mutational scanning helps predict success of SARS-CoV-2 clades. Nature 631, 617–626 (2024).
Sourisseau, M. et al. Deep Mutational Scanning Comprehensively Maps How Zika Envelope Protein Mutations Affect Viral Growth and Antibody Escape. J. Virol. 93, https://doi.org/10.1128/JVI.01291-19 (2019).
Phoo, W. W. et al. Structure of the NS2B-NS3 protease from Zika virus after self-cleavage. Nat. Commun. 7, https://doi.org/10.1038/ncomms13410 (2016).
Berger, A. & Schechter, I. Mapping the active site of papain with the aid of peptide substrates and inhibitors. Philos. Trans. R. Soc. Lond. B Biol. Sci. 257, 249–264 (1970).
King, N. M., Prabu-Jeyabalan, M., Nalivaika, E. A. & Schiffer, C. A. Combating susceptibility to drug resistance: lessons from HIV-1 protease. Chem. Biol. 11, 1333–1338 (2004).
Romano, K. P., Ali, A., Royer, W. E. & Schiffer, C. A. Drug resistance against HCV NS3/4A inhibitors is defined by the balance of substrate recognition versus inhibitor binding. Proc. Natl. Acad. Sci. USA 107, 20986–20991 (2010).
Soumana, D. I., Ali, A. & Schiffer, C. A. Structural analysis of asunaprevir resistance in HCV NS3/4A protease. ACS Chem. Biol. 9, 2485–2490 (2014).
Schwarz, M. C. et al. Rescue of the 1947 Zika Virus Prototype Strain with a Cytomegalovirus Promoter-Driven cDNA Clone. mSphere 1, https://doi.org/10.1128/mSphere.00246-16 (2016).
Blight, K. J., Kolykhalov, A. A. & Rice, C. M. Efficient initiation of HCV RNA replication in cell culture. Science 290, 1972–1974 (2000).
Kirkpatrick, E., Qiu, X., Wilson, P. C., Bahl, J. & Krammer, F. The influenza virus hemagglutinin head evolves faster than the stalk domain. Sci. Rep. 8, 10432 (2018).
Shiryaev, S. A. et al. Characterization of the Zika virus two-component NS2B-NS3 protease and structure-assisted identification of allosteric small-molecule antagonists. Antivir. Res, 143, 218–229 (2017).
Yao, Y. et al. Discovery, X-ray Crystallography and antiviral activity of allosteric inhibitors of Flavivirus NS2B-NS3 Protease. J. Am. Chem. Soc. 141, 6832–6836 (2019).
Boby et al. Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors. Science 382, eabo7201 (2023).
Kartal, O., Andres, F., Lai, M. P., Nehme, R. & Cottier, K. waveRAPID-A robust assay for high-throughput kinetic screens with the Creoptix WAVEsystem. SLAS Discov. 26, 995–1003 (2021).
Schuller, M. et al. Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking. Sci. Adv. 7, https://doi.org/10.1126/sciadv.abf8711 (2021).
Schiebel, J. et al. One question, multiple answers: biochemical and biophysical screening methods retrieve deviating fragment hit lists. ChemMedChem 10, 1511–1521 (2015).
Kuiper, B. D. et al. Increased activity of unlinked Zika virus NS2B/NS3 protease compared to linked Zika virus protease. Biochem Biophys. Res Commun. 492, 668–673 (2017).
Behnam, M. A., Graf, D., Bartenschlager, R., Zlotos, D. P. & Klein, C. D. Discovery of Nanomolar Dengue and West Nile Virus Protease Inhibitors Containing a 4-Benzyloxyphenylglycine Residue. J. Med Chem. 58, 9354–9370 (2015).
Ferla, M. P. et al. Fragmenstein: predicting protein-ligand structures of compounds derived from known crystallographic fragment hits using a strict conserved-binding–based methodology. ChemRxiv https://doi.org/10.26434/chemrxiv-2024-17w01 (2024).
Kenton, N. T. et al. Discovery of orally bioavailable Zika and West Nile Virus antiviral compounds targeting the NS2B-NS3 protease. bioRxiv https://doi.org/10.1101/2025.06.19.660642 (2025).
Wills, S. et al. Expanding the scope of a catalogue search to bioisosteric fragment merges using a graph database approach. bioRxiv https://doi.org/10.1101/2024.08.02.606367 (2024).
St Denis, J. D., Hall, R. J., Murray, C. W., Heightman, T. D. & Rees, D. C. Fragment-based drug discovery: opportunities for organic synthesis. RSC Med. Chem. 12, 321–329 (2020).
Starvaggi, J., Previti, S., Zappala, M. & Ettari, R. The Inhibition of NS2B/NS3 Protease: A new therapeutic opportunity to treat Dengue and Zika Virus infection. Int. J. Mol. Sci. 25, https://doi.org/10.3390/ijms25084376.
Ali, A. et al. Molecular basis for drug resistance in HIV-1 Protease. Viruses 2, 2509–2535 (2010).
Santos, N. P. et al. Characterization of an Allosteric Pocket in Zika Virus NS2B-NS3 Protease. J. Chem. Inf. Model 62, 945–957 (2022).
Fearon, D. et al. Accelerating drug discovery with high-throughput crystallographic fragment screening and structural enablement. Appl. Res. 4, https://doi.org/10.1002/appl.202400192 (2025).
Gahbauer, S. et al. Iterative computational design and crystallographic screening identifies potent inhibitors targeting the Nsp3 Macrodomain of SARS-CoV-2. bioRxiv https://doi.org/10.1101/2022.06.27.497816 (2022).
Michael, F. et al. A selection of Golden Gate vectors to simplify recombinant protein production in Escherichia coli. BioRxiv PrePrint. https://doi.org/10.1101/2024.02.13.579886 (2024).
Cox, O. B. et al. A poised fragment library enables rapid synthetic expansion yielding the first reported inhibitors of PHIP(2), an atypical bromodomain. Chem. Sci. 7, 2322–2330 (2016).
O’Reilly, M. et al. Crystallographic screening using ultra-low-molecular-weight ligands to guide drug design. Drug Discov. Today 24, 1081–1086 (2019).
Keeley, A., Abranyi-Balogh, P. & Keseru, G. M. Design and characterization of a heterocyclic electrophilic fragment library for the discovery of cysteine-targeted covalent inhibitors. Medchemcomm 10, 263–267 (2019).
Bajusz, D. et al. Exploring protein hotspots by optimized fragment pharmacophores. Nat. Commun. 12, 3201 (2021).
Wright, N. D. et al. The low-cost Shifter microscope stage transforms the speed and robustness of protein crystal harvesting. Acta Crystallogr. D. Struct. Biol. 77, 62–74 (2021).
Kabsch, W. Xds. Acta Crystallogr. D. Biol. Crystallogr. 66, 125–132 (2010).
Vonrhein, C. et al. Data processing and analysis with the autoPROC toolbox. Acta Crystallogr. D. Biol. Crystallogr. 67, 293–302 (2011).
Winter, G. xia2: an expert system for macromolecular crystallography data reduction. J. Appl. Crystallogr. 43, 186–190 (2010).
Winter, G. et al. DIALS: implementation and evaluation of a new integration package. Acta Crystallogr. D. Struct. Biol. 74, 85–97 (2018).
Krojer, T. et al. The XChemExplorer graphical workflow tool for routine or large-scale protein-ligand structure determination. Acta Crystallogr. D. Struct. Biol. 73, 267–278 (2017).
Long, F. et al. AceDRG: a stereochemical description generator for ligands. Acta Crystallogr. D. Struct. Biol. 73, 112–122 (2017).
Smart, O. S. et al. Grade, version 1.2.20. Cambridge, United Kingdom, Global Phasing Ltd. (2011).
Pearce, N. M. et al. A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density. Nat. Commun. 8, 15123 (2017).
Keegan, R., Wojdyr, M., Winter, G. & Ashton, A. DIMPLE: A difference map pipeline for the rapid screening of crystals on the beamline. Acta Crystallogr A. 71, S18–S18 (2015).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr D. Biol. Crystallogr 66, 486–501 (2010).
Smart, O. S. et al. Exploiting structure similarity in refinement: automated NCS and target-structure restraints in BUSTER. Acta Crystallogr D. Biol. Crystallogr 68, 368–380 (2012).
Crill, W. D. & Chang, G. J. Localization and characterization of flavivirus envelope glycoprotein cross-reactive epitopes. J. Virol. 78, 13975–13986 (2004).
Bloom, J. D. An experimentally determined evolutionary model dramatically improves phylogenetic fit. Mol. Biol. Evol. 31, 1956–1978 (2014).
Fulton, B. O., Sachs, D., Schwarz, M. C., Palese, P., Evans, M. J. Transposon Mutagenesis of the Zika Virus genome highlights regions essential for RNA replication and restricted for immune evasion. J. Virol. 91. https://doi.org/10.1128/JVI.00698-17.
Doud, M. B., Bloom, J. D. Accurate measurement of the effects of all amino-acid mutations on Influenza Hemagglutinin. Viruses 8 https://doi.org/10.3390/v8060155.
Bloom, J. D. Software for the analysis and visualization of deep mutational scanning data. BMC Bioinforma. 16, 168 (2015).
Fernandes, R. S. et al. High-throughput antiviral assays to screen for inhibitors of Zika Virus replication. J. Vis. Exp. https://doi.org/10.3791/62422 (2021).
Acknowledgements
Research reported in this publication was supported by the NIAID of the National Institute of Health under award number U19AI171399. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Health. Authors acknowledge Fundação de Amparo à Pesquisa do Estado de São Paulo project 2013/07600-3. Authors acknowledge Diamond Light Source for access to the I04-1 beamline and XChem facilities through proposal lb32627.
Author information
Authors and Affiliations
Contributions
X.N.: conceptualization, X-Ray data curation, formal analysis, visualization, writing—original draft, writing—review and editing; R.B.R.: DMS data curation and analysis, review and editing; A.S.G: crystallography support, writing—review and editing; M.P.F.: Fragment progressing; C.K., J.S., W.W.H., J.D.B: DMS analysis, visualization; B.H.B., P.G.M., C.W.E.T.: XChem support. M.F., S.W., E.P.W., C.G.: Protein engineering, protein production and quality control; N.L., E.C., H.B.: biochemical and biophysical assays. R.M.L., M. Winokan, W.T., A.V.C., M. Walsh: software, manuscript reviewing; M.X., scientific coordination; J.C.A., D.F., L.K., K.K.: validation, manuscript review and editing; N.T.K., A.L.: Chemistry data curation; I.D., R.S.F., G.O.: Constructs and enzymatic activity characterisation; A.v.D.: Conceptualization, manuscript review and editing; M.J.E.: conceptualization, writing—review and editing; F.v.D.: funding acquisition, conceptualization, writing—review and editing.
Corresponding authors
Ethics declarations
Competing interests
A.S.G consults for DNDi and MMV. A.A.L. is a shareholder and employee of PostEra, which has commercial interests in the discovery, development and commercialization of therapeutics. Other authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ni, X., Richardson, R.B., Godoy, A.S. et al. Combined crystallographic fragment screening and deep mutational scanning enable discovery of Zika virus NS2B-NS3 protease inhibitors. Nat Commun 16, 8930 (2025). https://doi.org/10.1038/s41467-025-63602-z
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-63602-z