Abstract
Endosomal sorting complex required for transport (ESCRT) is the major membrane remodeling complex, closely associated with endolysosomal repair and hereditary spastic paraplegias (HSP) diseases. Loss of function mutations in the ESCRT-I component UBAP1 causes a rare type of HSP (spastic paraplegia 80, SPG80), while the underlying pathological mechanism is unclear. Here, we found that UBAP1 but not SPG80 causing mutant was efficiently recruited to damaged lysosomes and mediated lysosome recovery. Loss of UBAP1 results in dysfunction of lysosomes, disconnecting mTOR localization on lysosomes, leading to cytoplasmic mTORC1 activation and TFEB dephosphorylation, as confirmed in vitro and in vivo models. Administration of rapamycin, a specific inhibitor of mTORC1, enhances mTOR lysosomal localization and TFEB phosphorylation. This pharmacological intervention effectively attenuated disease progression and restored lysosomal homeostasis in Ubap1 deficiency mice. Our findings reveal UBAP1’s role in lysosome regulation and suggest rapamycin may benefit patients with HSP and other motor neuron disorders.
Similar content being viewed by others
Introduction
Hereditary spastic paraplegia (HSP), also known as familial spastic paraplegia (SPG), is a group of heritable neurodegenerative movement disorders characterized by progressive weakness and spasticity of lower limbs1. HSP exhibits multiple pathogenic facets that contribute to the gradual deterioration and death of upper motor neurons, ultimately manifesting as symptoms of SPG. Considering the extensive genetic heterogeneity, the consequent mechanistic diversity, and the different progression of clinical cases, to date, no effective HSP therapies are available.
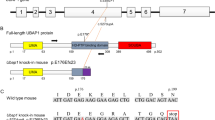
Remarkably, the endolysosomal and autophagic pathways are overrepresented by more than 24 genes whose mutations are directly implicated in HSP2. Recently, we and other groups have found that loss of function mutations in the ubiquitin associated protein 1 (UBAP1) gene causes a new type of HSP (spastic paraplegia 80, SPG80)3,4. UBAP1 is a component of the endosomal sorting complex required for transport (ESCRT), essential for mediating the trafficking of ubiquitylated proteins from endosomes to lysosomes5. ESCRT actively engages in various autophagy-lysosomal pathways, including endosomal microautophagy and macroautophagy, for the degradation of cytosolic components6. Quantitative analysis of the lysosomal proteome in response to lysosomal damage indicated enrichment of most ESCRT proteins by Lyso-IP7. Moreover, Galectin 3 (GAL3) unifies and coordinates ESCRT and autophagy in responses to lysosomal damage8. ESCRT-Ⅰ subunits VPS37A, whose mutations cause another subtype of HSP (SPG53), localizes to the phagophore and is crucial for autophagosome formation9. Several ESCRT components such as CHMP4A are rapidly recruited to acutely injured endolysosomes, promoting their repair10,11. UBAP1 directly interacts with ubiquitin on endosomes12, but its function in the autophagy-lysosome pathway and its potential relationship to the pathogenesis of HSP diseases remain largely unknown.
The mechanistic target of rapamycin (mTOR) kinase, as part of the mTORC1 complex, represents a major signaling protein that localizes to the cytoplasmic and surface of lysosomes. Interestingly, recent studies indicated that the subcellular shuttling of mTORC1 between the lysosomal surface and cytoplasm serves as a key determinant of its activity, with cytoplasmic mTORC1 phosphorylating S6K and 4E-BP1 to regulate de novo protein synthesis and lysosomal mTORC1 controlling lysosome biogenesis by phosphorylating the TFEB/TFE3 transcription factors13. In addition to serving as a platform for the organization of proteins contributing to mTORC1 activation, the lysosome itself can also potentially influence the activity of mTORC1. Impaired lysosomal degradation triggers the activation of mTORC1 signaling14,15. Dephosphorylated TFEB translocates to the nucleus and can thus up-regulate the expression of genes encoding lysosomal proteins in response to impaired lysosome function16. This interplay between lysosomal status and mTORC1 activity may act as a potential regulator of a feedback mechanism governing lysosome homeostasis.
Rapamycin, a US Food and Drug Administration-approved drug, exhibits relatively safer profiles owing to their non-ATP competitive mode of action and apparent selectivity for mTORC117. It becomes increasingly recognized that enhancing autophagy flux and lysosomal degradation by rapamycin offers a potential therapeutic approach for neurodegenerative diseases18,19,20, and that the effects of other consequences of rapamycin treatment are minimal20,21. Recently, rapamycin showed effective rescue of autophagic lysosome reformation defects and locomotor deficit in two HSP Drosophila models22,23. However, there is a lack of consistent data from mammalian animal models to evaluate this suitability.
In this study, we revealed a lysosome related mechanism and intervention strategy underlying the HSP pathogenesis. UBAP1 recruitment coincides with the restoration of endolysosomal function subsequent to lysosomal damage, underscoring the pivotal role of UBAP1 in the recovering process of lysosome damage. The loss of UBAP1 leads to lysosomal dysfunction, which then disrupts the lysosomal localization of mTOR, reduces TFEB phosphorylation, and ultimately results in cytoplasmic mTORC1 hyperactivation, as observed in both in vitro and in vivo models. Notably, these effects were reversible upon treatment with the mTOR inhibitor rapamycin. Furthermore, Ubap1 Syn-cKO mice exhibited early onset motor impairments and progressive neuronal death, mirroring the clinical manifestations of SPG80 mutations seen in HSP patients. Administration of rapamycin significantly alleviated neurological symptoms, and prevented brain lesions as well as locomotor deficits in two mouse models of UBAP1 deficiency. Our findings provid proof-of-principle evidence that UBAP1 enhances lysosomal function, and that modulating lysosomal activity via rapamycin offers a promising therapeutic strategy for HSP and related disorders.
Results
UBAP1 is recruited to damaged lysosomes and correlates with lysosome recovery
Recent studies using proximity biotinylation assay showed several ESCRT subunits including UBAP1 interact with GAL-38, a cytosolic lectin that binds to lysosomal glycans which are exposed only after membrane damage24. We thus sought a way to follow UBAP1 expression dynamics during both the onset and resolution of endolysosomal injury. L-leucyl-L-leucine O-methyl ester (LLOMe) is a commonly used lysosomotropic compound that induces fast but reversible lysosomal membrane damage25. We confirmed LLOMe-induced damage in COS-7 cell by noticing concentration of cytoplasmic GAL-3 on punctate structures (Fig. 1a and Supplementary Fig. 1a). However, we didn’t observe marked increase of UBAP1-positive puncta after adding LLOMe for 15 min (Supplementary Fig. 1a) or 1 h (Fig. 1a). Interestingly, we found that the number of UBAP1-postive puncta increased gradually and were associated with GAL-3-positive events after LLOMe wash out, reaching a maximum after 3 hours of wash out, then gradually decreased until the end of the assay at 14 hours after washout (Fig. 1a, b and Supplementary Fig. 1b).
a Representative image of COS-7 cells expressing GAL3-mbaojin and mYH-UBAP1 without/with LLOMe treatment (1 mM) for 1 h and wash out for indicated time points. b Quantification of GAL3 or UBAP1 positive puncta in (a). M (Mock) n = 10, L (LLOMe) n = 13, 15 min n = 9, 1 h n = 10, 3 h n = 8, 6 h n = 13, 10 h n = 13, 14 h n = 13 cells. c Images and 3D reconstruction of COS-7 cells expressing GAL3-mbaojin and mYH-UBAP1 with LLOMe 1 mM or 2 mM treatment for 15 min and wash out for 10 min. d 2D-SIM microscopy of COS-7 cells co-transfected GFP-UBAP1Wild type (UBAP1WT)/GFP-UBAP1Mutant (UBAP1Mut) with LAMP1-mYH were treated with mock or LLOMe (1 mM) for 15 min, then washed extensively and chased in the absence of LLOMe for 10 min. e Quantification of UBAP1 puncta on LAMP1 in (d). Mock: UBAP1WT n = 17, UBAP1Mut n = 17; LLOMe: UBAP1WT n = 15, UBAP1Mut n = 19; Wash out: UBAP1WT n = 19, UBAP1Mut n = 19 cells. f Interaction between UBAP1 and LAMP1. COS-7 cells were transfected with UBAP1WT or UBAP1Mut and immunoprecipitated with GFP or IgG control antibodies and probed with LAMP1 and VPS37A antibodies. VPS37A was loaded as a positive control. All scale bars: 10 μm. All data represent the mean ± SEM. Two-way ANOVA followed by Sidak’s multiple comparisons test (e), ns p > 0.05, #p < 0.0001. Exact p-values and test statistics are provided as a Source Data file.
We next confirmed UBAP1 recruitment correlated with lysosomal recovery by inducing lysosomal damage in short time, and observed the expression of GAL-3 and UBAP1 after LLOMe washout. A large proportion of the UBAP1-positive events were associated with GAL-3-positive structures and formed rapidly after LLOMe wash out (Fig. 1c). A 3D reconstruction analysis showed more than 50% of the UBAP1 and GAL-3-positive events formed like a “plug” pattern at lysosomal damage sites and occurred at a higher rate than expected by random chance (Fig. 1c and Supplementary Fig. 1c), which was similar to G3BP1 plugging and stabilizing damaged lysosomal membranes26. However, this is very different from most ESCRTs such as CHMP4B that are recruited to endolysosomes upon LLOMe treatment (Supplementary Fig. 1d)10,11. UBAP1 forming puncta exclusively after LLOMe washout was further confirmed in HeLa cells (Supplementary Fig. 1e). UBAP1 favors co-assembly with VPS37A27. Interestingly, VPS37A also formed puncta structures on lysosome membranes exclusively following lysosome recovery (Supplementary Fig. 1f) and co-localized with UBAP1 (Supplementary Fig. 1g). This suggests that VPS37A and UBAP1 may have similar functions or mechanisms of action in cellular responses to lysosomal stress. UBAP1 is reported to primarily associate with endosomes under basal condition, while recent study found that late endosomes mostly do not get holes28. Consistently, we found more UBAP1 positive puncta were co-localized with lysotracker staining than late endosome protein Rab7 (Supplementary Fig. 1h). Together, our results indicate UBAP1-enriched structures may correspond to damaged lysosomes upon lysosome recovery.
Several studies, including ours, have identified over twenty frame-shift UBAP1 mutations in SPG80 patients with varying degrees of disease association. Among them, c.425_426delAG (p.K143Sfs*15) are the ones most frequently linked to the disease29. To examine the localization of WT and mutant UBAP1 during lysosome recovering process, we added LLOMe into COS-7 cells that co-transfected GFP-tag WT (GFP-UBAP1WT) or K143Sfs*15 (GFP-UBAP1Mut) with LAMP1-mYH. UBAP1 displayed a dispersed cytosolic staining pattern, with only a small fraction localizing near the lysosome membrane in untreated and cells treated with 1 mM LLOMe for 15 min. However, GFP-UBAP1Mut fraction was undetectably both before and after LLOMe treatment (Fig. 1d). Remarkably, the localization pattern of UBAP1 contrasted sharply with a notable increase in UBAP1-positive granules that translocate to vesicular structures and localized near the lysosome membrane throughout the cells shortly after LLOMe wash out (Fig. 1d). This phenomenon was also observed but less pronounced in cells expressing UBAP1Mut (Fig. 1d). There was significant less UBAP1Mut-postive puncta successfully localized to LAMP1 positive lysosome membranes (Fig. 1e). Consistently, co-immunoprecipitation experiments revealed UBAP1Mut showed a decreased binding affinity with LAMP1 compared to UBAP1WT (Fig. 1f). To further elucidate the dynamics of UBAP1 interaction with the lysosome membrane, we conducted super-resolution live-cell imaging in COS-7 cells. Intriguingly, UBAP1 foci rapidly formed and were recruited to the rupture of lysosome membrane, subsequently accumulating in the membranes and within LAMP1-positive vesicles after LLOMe wash out (Supplementary Fig. 2a and Supplementary Movie 1). However, UBAP1Mut-postive puncta can’t localize to the lysosomal membrane precisely (Supplementary Fig. 2b and Supplementary Movie 2). Consistently, these phenomena were verified under UBAP1 deficiency condition (Supplementary Fig. 2c, d).
The temporal correlation between transient UBAP1 recruitment and lyososomal recovery suggested that UBAP1 might be involved in repairing damaged lysosomal membranes, which is consistent with known roles of ESCRT proteins in resealing wounds at the plasma membrane and nuclear envelope30,31,32. We observed a substantial increase in GAL3-positive granules within UBAP1-deficient cells, both prior to and subsequent to LLOMe treatment (Supplementary Fig. 3a). In control cells, the majority of GAL-3-positive puncta diminished within 14 hours subsequent to LLOMe washout. Conversely, UBAP1 knockdown cells exhibited a notably higher proportion of GAL-3 puncta (Supplementary Fig. 3b), suggesting delayed recovery of lysosome damage in UBAP1-deficient cells after LLOMe-induced rupture. Significant damage in the lysosome membrane promotes degradation of the whole organelle through lysophagy, minor ruptures are repaired by ESCRT-mediated invagination of the damaged membrane into the lysosome lumen11,33. TMEM192-mKeima probe is a useful tool for investigating lysophagy34. We then co-transfected TMEM192-mKeima probe either with UBAP1 siRNA (siUBAP1) or Control siRNA (siCtrl), with the objective of precisely monitoring the consequences of UBAP1 depletion on the process of lysophagy. UBAP1 knock down cells showed comparable mKeima acidification with controls (Supplementary Fig. 3c), suggesting that the UBAP1 response is distinct from lysophagy. Lysophagy involves engulfment of the damaged organelle by an autophagosome. We then employed siRNA-mediated knockdown of ATG5 or ATG7 in COS-7 cells to create a functional autophagy-deficient background (Supplementary Fig. 4a). Indeed, cells lacking ATG5 or ATG7 failed to accumulate mYH-tagged LC3B in association with LLOMe-damaged endolysosomes (Supplementary Fig. 4b). Notably, even in the absence of functional autophagy, UBAP1 still emerged after LLOMe washout (Supplementary Fig. 4b). Furthermore, the UBAP1 response to LLOMe was unaffected by treating cells with autophagy inhibitor wortmannin or Vps34-IN-1 (Supplementary Fig. 4c). Interestingly, BAPTA-AM, a chelator of intracellular Ca2+ treatment also did not affect the UBAP1 response to LLOMe (Supplementary Fig. 4d). These results indicate that the redistribution of UBAP1 is not dependent on canonical autophagy or calcium signaling.
UBAP1 deficiency causes dysfunctional and enlarged lysosomes
Lysosomes maintain a highly acidic lysosomal pH to activate hydrolytic enzymes and degrade internalized macromolecules35. Defects in lysosomal recovery ultimately lead to lysosomal dysfunction. To explore lysosomal function post-UBAP1 depletion, we performed co-localization assays with LysoSensor (Green), a commercial lysosome tracer that assesses lysosomes pH. LAMP1-positive lysosomes exhibited reduced green fluorescence in UBAP1 siRNA-transfected COS-7 cells (Fig. 2a), indicating elevated lysosomal pH. Next, we assessed how the dysfunctional lysosomes affected degradation of cargo with UBAP1 depletion. Both autophagosomes and ubiquitinated proteins ultimately need to enter lysosomes for degradation. 2D-SIM analysis showed more LC3B puncta co-localized with the LAMP1-positive lysosomes in UBAP1 knockdown cells, and the sizes were greatly increased (Supplementary Fig. 5a). In addition, the number and proportion of ubiquitin in lysosomes were significant higher in UBAP1-deficient cells than in control cells (Fig. 2b). Finally, reduced staining of Magic Red the dye that respectively indicated the activities of cathepsins B was blocked in UBAP1 deficient cells (Fig. 2c–e).
a Left: COS-7 cells were transfected with siCtrl/siUBAP1 and LAMP1-RFP, 48 h later cells were stained with LysoSensor (1 μM) for 30 min. Right: The relative fluorescence intensity of LysoSensor to LAMP1-RFP. siCtrl n = 24, siUBAP1 n = 17 cells. b Left: COS-7 cells were transfected LAMP1-GFP and Myc-Ub with siCtrl/siUBAP1. Right: Quantification of Ub puncta per cell. siCtrl n = 12, siUBAP1 n = 12 cells. c HeLa cells were transfected with siCtrl/siUBAP1 and LAMP1-GFP. 48 h later, cells were stained with Magic Red for 30 min. d Ratio of Magic Red in Lysosome or cytoplasm in (c). siCtrl n = 16, siUBAP1 n = 17 cells. e Quantification of relative intensity of Magic Red in (c). siCtrl and siUBAP1 n = 17 cells. f Left: HT-LC3 U2OS cells were transfected with siCtrl/siUBAP1/siVPS37A for 48 h then incubated in EBSS medium in the presence of 100 nM BafA1 for 4 h. Then cells were incubated with a saturating dose of membrane-impermeable HT ligand (MIL) to stain membrane-bound HT-LC3-II that is accessible to the cytosol and incubated with membrane-permeable HT ligand (MPL) to stain LC3-II that is sequestered within membranes. Right: The ratio of relative MIL/MPL. siCtrl n = 37, siUBAP1 n = 36, siVPS37A n = 35 cells. g Left: COS-7 cells were transfected with siCtrl/siUBAP1 and mYH-LC3B for 48 h and incubated in the presence of 500 nM Rapamycin (Rap) or 100 nM BafA1 for 5 h. Right: Quantification of LC3B puncta per cell. Mock: siCtrl n = 22, siUBAP1 n = 20; Rap: siCtrl n = 23, siUBAP1 n = 21; BafA1: siCtrl n = 20, siUBAP1 n = 20 cells. h Images of COS-7 cells transfected with siCtrl/siUBAP1 and LAMP1-mYH for 48 h. i Quantification of lysosome inner diameter in (h). siCtrl n = 564, siUBAP1 n = 364 lysosomes. j Lysosome diameter distributions and cumulative frequency in (h). All scale bars: 10 μm. All data represent the mean ± SEM; Student’s t test (a, b, e and g), Two-way ANOVA followed by Sidak’s multiple comparisons test (d), Kruskal–Wallis test followed by Dunn’s multiple comparisons test (f), Mann–Whitney test (i), ns p > 0.05, **p < 0.01, ***p < 0.001, #p < 0.0001. Exact p-values and test statistics are provided as a Source Data file.
To further distinguish the effects of UBAP1 on autophagy and lysosomal function, we carried out the HaloTag-LC3 autophagosome completion assay that specifically detects phagophores (MIL positive), nascent autophagosomes (MIL and MPL positive), and mature autophagic structures (MPL positive)36,37. Unlike VPS37A depletion, which significantly increased the rates of MIL/MPL upon starvation in the presence of BafilomycinA1 (BafA1), depletion of UBAP1 had minimal effect on the level of MIL and the MIL/MPL ratio (Fig. 2f). Our results indicate UBAP1 is not required for phagophore closure and autophagosome maturation, which is consistent with the published papers9,36. BafA1 is frequently used at high concentration to block late-phase autophagy38. Treatment with BafA1 revealed a persistent increase in mYH-LC3B puncta accumulation in siUBAP1-treated cells relative to control cells (Fig. 2g). Immunoblot analysis verified the increased accumulation of LC3B-II (Supplementary Fig. 5b). However, the protein sequestosome 1 (p62/SQSTM1), a classical receptor of autophagy, is not significantly changed after UBAP1 depletion (Supplementary Fig. 5c–f). These results demonstrate that accumulation of LC3B is likely due to the impaired degradation in lysosomes upon UBAP1 depletion.
Dysfunctional lysosomes affected degradation may lead to enlarged lysosome. Clearly, 2D-SIM microscope revealed significant increase in the number of abnormally enlarged LAMP1-positive vesicles in UBAP1 siRNA-transfected cells (Fig. 2h–j). To experimentally mimic the role of UBAP1 in HSP patients, we isolated fibroblasts from three SPG80 patients carrying the UBAP1 mutation and two controls to study the lysosome pathway. Similarly, LysoTracker staining indicated lysosome enlargement in fibroblasts derived from SPG80 patients compared with controls (Supplementary Fig. 5g). Interestingly, when over-expressed wild-type UBAP1 in fibroblast derived from SPG80 patients, we observed a corresponding decrease in LysoTracker-positive lysosome diameter (Supplementary Fig. 5g). These findings suggest that UBAP1 restricts lysosome size and is essential for maintaining lysosomal acidification and degradation capacity.
UBAP1 is responsible for the lysosomal recruitment and regulation of mTOR
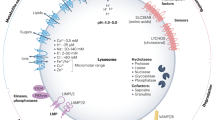
Lysosomes function as platforms for the recruitment, assembly, and activation of mTOR signaling components, a process that relies on intact lysosomal function for proper mTORC1 signaling39,40 (Fig. 3a). We then systematically examined the localization and expression of mTOR and its downstream targets in UBAP1 siRNA transfected COS-7 cells. In control cells, most mTOR showed co-localizing with LAMP1 (a lysosomal marker) and part being diffusely cytoplasmic, however there is significant less mTOR colocalized with lysosomes in cells with UBAP1 depletion (Fig. 3b). Immunoblots analysis revealed a significant increase in the expression of phosphorylated mTOR (p-mTOR) in UBAP1-deficient cells, whereas the phosphorylation level of TFEB, a non-canonical lysosomal substrate of mTORC1, was significantly reduced (Fig. 3c, d). In contrast, phosphorylation of S6K (p-S6K) and phosphorylated 4EBP1 (p-4E-BP1), two canonical substrates of mTORC1, stayed largely unaffected in cells with two days UBAP1 siRNA transfection (Fig. 3c). Interestingly, there was significant up-regulation of p-S6K and p-4EBP1 after four days UBAP1 siRNA transfection (Fig. 3d). Targeted dephosphorylation of TFEB is sufficient to induce nuclear translocation and some transcriptional activity of TFEB41. Accordingly, dephosphorylation of TFEB in UBAP1 siRNA transfected cells was accompanied by increased nuclear localization and increased TFEB mRNA expression (Fig. 3e and Supplementary Fig. 6a). Considering that UBAP1 functions as a ubiquitin associated protein, it is plausible to hypothesize that UBAP1 could influence TFEB stability through the proteasomal degradation pathway. Although the co-immunoprecipitation assay did not reveal any interaction between UBAP1 and TFEB, there is a mild decrease in ubiquitination of TFEB in cells where UBAP1 was knocked down (Supplementary Fig. 6b). Consistently, the Cycloheximide Chase assay confirmed that TFEB stability was slightly enhanced following the depletion of UBAP1 (Supplementary Fig. 6c). Notably, the elevated p-mTOR expression induced by UBAP1 knockdown was abrogated upon TFEB knockdown (Fig. 3f). This suggests that TFEB modulates mTORC1 activity, a conclusion supported by a study demonstrating TFEB overexpression resulted in enhanced mTORC1 activity even in cells lacking the autophagy genes Atg5 or Atg742.
a Diagram illustrating lysosomes as platforms for the recruitment, assembly, and activation of mTOR signaling. b COS-7 cells were transfected with siCtrl/siUBAP1 and LAMP1-mYH for 48 h. Cells were then fixed and immunostained with an antibody against mTOR. Colocalization analysis of mTOR with LAMP1 in COS-7 cells that were transfected with siCtrl or siUBAP1. siCtrl n = 28, siUBAP1 n = 29 cells. Immunoblots with lysates from COS-7 cells that were transfected with siUBAP1 or siCtrl for 48 (c) or 96 (d) hours, and probed with the indicated antibodies. Relative expression was calculated from three independent experiments. (n = 3 independent experiments) e Left: COS-7 cells transfected with siCtrl/siUBAP1 for 48 h and stained with TFEB antibody. Right: Quantification of ratio of cells with TFEB in nucleus. Both siCtrl and siUBAP1 n = 6 micrographs. f Immunoblots with lysates from COS-7 cells that were transfected with siCtrl/siUBAP1 and siTFEB for 48 h, and probed with the indicated antibodies. Relative expression was calculated from three independent experiments. g Primary fibroblasts that were derived from SPG80 patients and healthy controls. Cells were fixed and immunostained with an antibody against TFEB. The relative fluorescence intensity of TFEB in the nucleus was quantified by measuring the mean TFEB signal intensity within DAPI-defined nuclear regions, normalized to the cytoplasmic TFEB intensity. Ctrl n = 121, Patient 1 n = 114, Patient 2 n = 45, Patient 3 n = 99 cells. h Immunoblot analysis of lysates from SPG80 patient-derived and control fibroblasts with indicated antibodies. Relative expression was calculated from three independent experiments. All scale bars: 10 μm. All data represent the mean ± SEM; Student’s t test (b–e and h), One-way ANOVA followed by Tukey’s multiple comparisons test (f), Kruskal–Wallis test followed by Dunn’s multiple comparisons test (g); ns p > 0.05, *p < 0.05, **p < 0.01, ***p < 0.001, #p < 0.0001. Exact p-values and test statistics are provided as a Source Data file.
Consistently, in human fibroblasts derived from SPG80 patients carrying UBAP1 mutations, we observed comparable alterations. TFEB staining revealed significantly elevated nuclear fluorescence intensity compared to controls (Fig. 3g). This effect was most pronounced in fibroblasts from Patient 1, who exhibited a high degree of UBAP1 depletion (Fig. 3h). Immunoblots analysis revealed increased expression of p-mTOR and TFEB. Additionally, the expression of autophagy markers p62 and LC3B were both elevated (Fig. 3h), indicating impaired lysosomal degradation. These results suggest that UBAP1 deficiency causes mTOR to shuttle into the cytoplasm, leading to cytoplasmic mTORC1 activation (as evidenced by increased phosphorylation of S6K and 4E-BP1) and reduced lysosomal mTOR-mediated phosphorylation of TFEB. It is consistent with the notion that TFEB and mTORC1 bind to each other on the lysosomal surface, where phosphorylation of TFEB by mTORC1 occurs43.
UBAP1 depletion causes progressive neuronal apoptosis and activation of mTORC1 signaling in mouse brains
Our previously studies indicated loss of UBAP1 caused neuronal apoptosis in the late stage (E16.5) of brain development44, but the underlying mechanism that triggered apoptosis remained unclear. To delve into the intricate relationship between cargo degradation defect and neuronal apoptosis mediated by UBAP1 disruption in vivo, we first quantitatively assessed the expression levels of p62 and ubiquitin in different developmental stage of Ubap1 Nes-cKO mouse brains. We found progressive accumulation of p62 and ubiquitinated proteins (Fig. 4a), coinciding with the occurrence of neuronal apoptosis (Fig. 4b). Similar to UBAP1 knockdown cells, the cortical neurons from Ubap1 Nex-cKO mice accumulated abnormally enlarged LysoTracker-positive structures, which were significantly seen in the neuritis (Fig. 4c). Supporting this idea, TEM analysis showed significantly accumulation of autophagosomes/autolysosomes in E16.5 Ubap1 Nes-cKO neurons (Fig. 4d, e). Consistently, immunoblots analysis revealed a more pronounced increase in the levels of p-mTOR, p62, TFEB, LAMP1, and LC3B in the late embryonic stage (E16.5) of Ubap1 Nes-cKO mouse brains compared to controls (Fig. 4f, h). Additionally, we observed significantly elevated expression of mTORC1 effectors, p-S6 and p-4EBP1, specifically at E16.5 but not at E14.5 in Nes-cKO mouse brains (Fig. 4g, i).
a Representative coronal sections showing progressive accumulation of polyubiquitinated proteins in Ubap1 Nes-cKO brains compared to controls. Scale bar: 50 μm. Quantification of relative fluorescence intensity of ubiquitin in Ubap1 Nes-cKO cortex. E12.5 n = 4, E14.5 n = 4, E16.5 n = 4 brain sections form 3 independent mouse brains. b Immunofluorescence staining of Cleaved-caspase 3 (CC3, red) in the cortex of Ctrl and Ubap1 Nes-cKO mice at the indicated embryonic stage. Scale bar: 20 μm. Quantification of CC3+ cells in the cortex of Ubap1 Nes-cKO mouse brains. E14.5 n = 6, E16.5 n = 6 brain sections form more than 3 independent mouse brains. c 2D-SIM microscopy reveals increased lysosome size in Ubap1 Nex-cKO primary cortical neurons. Lysosomes were labeled with Lysotracker. Scale bar: 5 μm. Ctrl n = 454, Nex-cKO n = 444 lysosomes in 20 independent neurons. d Diagram illustrating the regions of analyzed by Transmission Electron Microscopy (TEM) in mouse brains. e Representative TEM images of autophagosomes (arrowheads) and autolysosomes (arrows) in the cortex of E16.5 Ubap1 Nes-cKO mouse brains. Scale bar: 500 nm. The number of autophagosome and autolysosome is quantified on the right. Ctrl n = 8, Nes-cKO n = 8 micrographs. f–i Immunoblot analysis of cortical lysates from E14.5 (f, g) or E16.5 (h, i) Ubap1 Nes-cKO and control mice, and probed with the indicated antibodies. Relative protein expression levels are quantified on the right. Ctrl and Nes-cKO n = 3 individual mouse brains. All data represent the mean ± SEM; One-way ANOVA followed by Tukey’s multiple comparisons test (a), Student’s t test (b, e–i), Mann–Whitney test (c); ns p > 0.05, *p < 0.05, **p < 0.01, #p < 0.0001. Exact p-values and test statistics are provided as a Source Data file.
Gene Set Enrichment Analysis (GSEA) of proteome indicated significant up-regulation of mTORC1 signaling associated molecules in Ubap1 Nes-cKO mouse brains (Supplementary Fig. 7a). RNA-seq analysis showed significant enrichment of phagosome-related genes (Supplementary Fig. 7b), suggesting a connection to lysosome dysfunction. LGALS proteins (LGALS3, LGALS8, and LGALS9) are integral to the endomembrane damage response45. Heatmap analysis showed several galectins, including LGALS3 (GAL 3), were significantly upregulated in Ubap1 Nes-cKO mouse brain (Supplementary Fig. 7c) which indicating elevated lysosome damage or inflammatory responses46. Moreover, we found the vast majority of lysosomal proteins, encoding for proteases and lysosome membrane were consistently up-regulated in Ubap1 Nes-cKO mouse brains (Supplementary Fig. 7d). However, the expression of vacuolar ATPase (V-ATPase)-ATP6Vs that mediate acidification of lysosome were significantly reduced (Supplementary Fig. 7d), which is consistent with reduced LysoSensor staining in UBAP1 knock down cells (Fig. 2).
Pharmacological inhibition of mTORC1 activity using rapamycin enhances lysosomal localization and function
Loss of UBAP1 results in dysfunction of lysosomes, leading to accumulation of autophagic cargoes and aberrant activation of mTOR signaling both in cells and in mouse brains. We observed significantly elevated levels of p-mTOR and TFEB in cells with UBAP1 knockdown, similar to the effects observed in cells treated with lysosome inhibitor BafA1 (Fig. 5a), indicating intact lysosomal function is required for proper mTORC1 signaling. In contrast, there is significantly down-regulation of p-mTOR and TFEB in cells treated with mTOR inhibitor rapamycin, even in cells with UBAP1 knock down (Fig. 5a). Consistent with previous reports13, the phosphorylation of TFEB, a lysosomal non-canonical mTORC1 substrate, was significantly decreased in UBAP1 deficiency cells like in cells with BafA1 treatment (Fig. 5a). Rapamycin treatment enhanced mTOR lysosomal localization (Fig. 5b), which was consistent with recent study that pharmacological or genetic downregulation of mTORC1 activity enhances its lysosomal localization and TFEB phosphorylation47. Notably, addition of the rapamycin significantly reduced the p62 puncta in lysosomes (Fig. 5c). Abnormally enlarged LAMP1-positive vesicles in UBAP1 siRNA-transfected cells can be significantly reduced upon rapamycin treatment (Fig. 5d). Consistently, decreased LysoSensor green fluorescence in UBAP1 mutant fibroblast was partially restored post-rapamycin treatment (Supplementary Fig. 8a). Finally, Magic Red staining revealed increased cathepsin activity in cultured neurons derived from Ubap1 Nes-cKO mouse brains following rapamycin treatment (Supplementary Fig. 8b–d).
a Immunoblot analysis of lysates from COS-7 cells that were transfected with siCtrl/siUBAP1 for 48 h, and incubated with/without 500 nM Rapamycin (Rap) or 100 nM BafA1 for 5 h, and probed with indicated antibodies. Quantification of relative expression, normalized to GAPDH. All groups n = 4 independent samples. b Left: COS-7 cells were transfected with siCtrl/siUBAP1 and LAMP1-GFP for 48 hours and incubated with/without 500 nM Rapamycin (Rap) for 24 h, and stained with antibody against mTOR. Right: Colocalization analysis of mTOR with LAMP1 in COS-7 cells. Mock: siCtrl n = 17, siRNA n = 19; Rap: siCtrl n = 20, siRNA n = 20 cells. c COS-7 cells were transfected with siCtrl/siUBAP1 and LAMP1-RFP for 48 h and incubated with/without 500 nM Rapamycin (Rap) for 24 h, staining with antibody against p62. Quantification of the ratio of lysosomes positive for both p62 and LAMP1. Mock: siCtrl n = 12, siRNA n = 12; Rap: siCtrl n = 12, siRNA n = 12 cells. d Left: COS-7 cells were transfected with siCtrl/siUBAP1 and LAMP1-mYH for 48 h and incubated with/without 500 nM Rapamycin (Rap) for 5 h. Right: Quantification of the inner diameter of lysosomes. Mock: siCtrl n = 218, siRNA n = 239; Rap: siCtrl n = 592, siRNA n = 550 lysosomes. All scale bars: 10 μm. All data represent the mean ± SEM; Student’s t test (a), Two-way ANOVA followed by Sidak’s multiple comparisons test (b, c), Kruskal–Wallis test followed by Dunn’s multiple comparisons test (d); ns p > 0.05, *p < 0.05, **p < 0.01, ***p < 0.001, #p < 0.0001. Exact p-values and test statistics are provided as a Source Data file.
UBAP1 knock down cells, in which p-TFEB was inhibited, exhibited increased TFEB nucleus localization as TSG101 knock down48 (Fig. 3e and Supplementary Fig. 8e). Several cell lines, such as HEK293 and HeLa cells, appeared to be “Rap-insensitive,” they lack rapamycin induced TFEB activation49. Here, we found rapamycin treatment reduced nuclear localization of TFEB both in UBAP1 siRNA and TSG101 siRNA-transfected COS-7 cells (Supplementary Fig. 8e), in line with the enhanced p-TFEB post-rapamycin treatment (Fig. 5a). Collectively, these findings suggest that rapamycin treatment enhanced mTOR lysosomal localization and reduced TFEB nucleus localization in Ubap1 deficient cells.
Mice with UBAP1 deficiency in postnatal neurons recapitulates the motor and cognitive symptoms observed in SPG80 patients
Since all SPG80 mutations exhibit dominant-negative or loss-of-function effects on UBAP129, we applied the Cre-loxP system to generate viable mice for further studies. Surprisingly, Ubap1 Nes-cKO, Nex-cKO, Emx1-cKO and CamkⅡ-cKO mice died shortly after birth, and we were unable to perform behavioral and intervention studies. Subsequently, we generated mice with a restricted postnatal and neuron-specific deletion of UBAP1 by injecting AAV-hSyn-EGFP-Cre into the lateral ventricle of Ubap1flox/flox mice at postnatal day 0 (P0) (Fig. 6a) to circumvent the embryonic lethality of the classically induced neuron-specific UBAP1 deficiency. Immunoblots analysis indicated a significant reduction of UBAP1 expression in the brains of Ubap1flox/flox mouse injected with AAV-hSyn-EGFP-Cre (Syn-cKO) compare to Ubap1flox/flox mouse injected with AAV-hSyn-EGFP (Syn-GFP) (Supplementary Fig. 9a). Ubap1 Syn-cKO mice grew normally, while both female and male mice exhibited a progressive decline in body weight around P20 (Supplementary Fig. 9b). Some Ubap1 Syn-cKO mice exhibited paralysis and developed severe motor deficits around P15, with a normal brain size (Supplementary Fig. 9c, d). In addition, Ubap1 Syn-cKO mice suspended by their tails exhibited a distinctive involuntary clasping and of the frequent trembling limbs (Fig. 6b and Supplementary Movie 3), which are typical manifestations observed in various well-known neurodegenerative diseases50, and some mice showed a hunched stance and notable kyphosis (Supplementary Fig. 9e). We observed reduced rotarod performance in Ubap1 Syn-cKO mice compared to that in the control from the beginning of the rotarod assessment at P30 (Fig. 6c). In addition, Ubap1 Syn-cKO mice showed a significant decrease in the foot-base-angle (FBA) at the toe-off position of the hind paw (Fig. 6d), which was consistent with other HSP locomotion-deficient mice51. Moreover, Ubap1 Syn-cKO mice had a rapid and disorganized gait trajectory, short stride distance, and poor overlap between the front and rear feet, while control mice had a straight and stable gait trajectory, with a high degree of overlap between the front and rear feet (Fig. 6e, f), suggesting that UBAP1 deficient mice had poor balance and coordination ability. However, Y-maze and novel object recognition test indicated Ubap1 Syn-cKO mice showed normal learning and cognitive abilities (Supplementary Fig. 9f–h). Finally, histological analysis revealed progressive significantly increased number of active caspase3-positive cells (Fig. 6g) and activation of glial cells accompany with a greatly reduced number of GFP-positive neurons in Ubap1 Syn-cKO mouse cortex (Fig. 6h). Collectively, our results indicate that neuron-specific postnatal deficiency of UBAP1 leads to motor and gait abnormalities that resemble those in human SPG80 patients.
a Schematic diagram of virus injection into the lateral ventricular of mouse brans. b Tail suspension test results comparing Ubap1 Syn-cKO and control mice. Scale bar: 1 cm. n = 10 mice per group. c Latency time until falling off from the rotarod. Syn-GFP n = 8, Syn-cKO n = 9 mice. d The foot-base-angle at the toe-off position of the hind leg is indicated by the white line. Scale bar: 1 cm. Syn-GFP n = 10, Syn-cKO n = 12 mice. e Ubap1 Syn-cKO mice exhibit disrupted gait pattern. Stride length quantification by footprint tests as shown. f Representative limb overlap map and distance between forepaw and hindpaw was measured. n = 8 mice per group (e and f). g Representative images from more than three independent mice per group showed progressive neuronal apoptosis in the cortex of Ubap1 Syn-cKO mice. Coronal sections of P14, 1 month (1 M), and 2 months (2 M) old Ubap1 Syn-cKO and control mouse brains were stained for DAPI (blue) and Cleaved-caspase 3 (Red). Scale bar: 50 μm. h Representative images from more than three independent mice per group showed reactive gliosis in the cortex of Ubap1 Syn-cKO mice. Coronal sections of P14, 1 M, 2 M old Ubap1 Syn-cKO and control mouse brains were stained for DAPI (blue), GFAP (Red), Iba1 (Red). Scar bar: 100 μm. i Immunoblot analysis of cortical lysates from 1 month old Syn-cKO and Syn-GFP mice, probed for the indicated antibodies. Quantification of relative protein expression levels, normalized to GAPDH. n = 3 independent mice per group. j Immunoblot analysis of cortical lysates from P20 Syn-cKO and Syn-GFP mouse brains. Quantification of relative protein expression, normalized to GAPDH. n = 4 independent mice per group. All data represent the mean ± SEM; Student’s t test (b–f, i, j); ns p > 0.05, *p < 0.05, **p < 0.01, ***p < 0.001, #p < 0.0001. Exact p-values and test statistics are provided as a Source Data file.
We then continued to explore whether lysosome and mTOR signal defects related with this disease model. Gene Ontology (GO) analysis showed that downregulated proteins were significantly enriched in the regulation of vesicle transport, autophagy, and neuron development (Supplementary Fig. 10a, b). Correspondingly, upregulated proteins were enriched in tricarboxylic acid cycle and glial cell differentiation (Supplementary Fig. 10c). In bulk RNA-seq, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway and GO analysis of differentially expressed genes revealed downregulated genes involved in ubiquitin-mediated proteolysis and autophagy (Supplementary Fig. 10d–f). Finally, immunoblots analysis indicated higher levels of LC3-II and p62 in the brains of Ubap1 Syn-cKO mice compared with controls (Fig. 6i), paralleling hyper-activation of p-mTOR and decreased p-TFEB (Fig. 6j). Taken together, our results indicate that UBAP1 depletion in mouse brains disrupts mTOR signaling, consistent with observations in vitro.
Rapamycin treatment attenuates motor disability and neurological lesions by regulating lysosome homeostatic in Ubap1 cKO mouse models
Given the above effects of rapamycin in enhancing lysosome function in UBAP1 deficient cells in vitro (Fig. 5 and Supplementary Fig. 8), we next investigated whether rapamycin could improve the health of HSP mice. Daily administration of rapamycin (4 mg/kg) via intraperitoneal injection commenced at P14, one week before significant body reduction and motor defects become noticeable in Ubap1 Syn-cKO mouse (Fig. 7a and Supplementary Fig. 11a). In alignment with previous rodent studies52, mice treated with rapamycin showed slightly reduced body weight, but their survival ability was not affected (Supplementary Fig. 11a).
a Schematic diagram of experimental procedures for rapamycin (Rap) treatment and behavioral testing in Ubap1 Syn-cKO mice. b Representative images of the indicated scores and quantification of hind limb clasping scores of mice with/without rapamycin treatment. Scale bar: 1 cm. WT Rap n = 10, Syn-cKO Veh n = 9, Syn-cKO Rap n = 8 mice. c The foot base angle analysis of mice with/without rapamycin treatment. WT Rap n = 12, Syn-cKO Veh n = 7, Syn-cKO Rap n = 6 mice. d Quantification of the time in 12 mm balance beam walking test from mice with/without rapamycin treatment. WT Rap n = 10, Syn-cKO Veh n = 6, Syn-cKO Rap n = 6 mice. e Representative sagittal sections from more than three mice per group showed enlarged ventricles and neurological lesions can be rescued by rapamycin treatment. Scale bar: 500 μm. f Representative images from more than three mice per group showed increased neuronal apoptosis by CC3 staining in vehicle-treated Syn-cKO mice and a robust attenuation in rapamycin-treated Syn-cKO mice. Scale bar: 100 μm. g Representative images from more than three mice per group showed activation of glia by Iba1 and GFAP staining in vehicle-treated Syn-cKO mice and a robust attenuation in rapamycin-treated Syn-cKO mice. Scale bar: 50 μm. All data represent the mean ± SEM; One-way ANOVA followed by Tukey’s multiple comparisons test (b–d), *p < 0.05, **p < 0.01, #p < 0.0001. Exact p-values and test statistics are provided as a Source Data file.
To mitigate the effect of rapamycin-induced body weight reduction on locomotor capabilities, we conducted two independent experiments with wild-type mice treated with rapamycin or Syn-GFP mice treated with vehicle as control. Notably, hind limb phenotypes during tail suspension were considerably alleviated in rapamycin-treated Syn-cKO mice (Fig. 7b and Supplementary Fig. 11b). There was a significant improvement in the FBA in Ubap1 Syn-cKO mice following rapamycin treatment, with a notable increase compared to vehicle-treated Ubap1 Syn-cKO mice (Fig. 7c and Supplementary Fig. 11c). Moreover, rapamycin treatment reversed the challenges faced by Ubap1 Syn-cKO mice while crossing a beam, reducing the time spent on beam walking (Fig. 7d and Supplementary Fig. 11d). Foot-printing analysis and rotarod assay showed limited improvement owing to the reduced body weight of rapamycin-treated Syn-cKO mice (Supplementary Fig. 11e–g). Histological analysis revealed that Ubap1 Syn-cKO mice developed neurological lesions characterized by enlarged ventricles (Fig. 7e), increased apoptotic neurons (Fig. 7f), accompany with microglia and astrocyte activation (Fig. 7g). Strikingly, up to 80% of age-matched rapamycin-treated Syn-cKO mice did not develop significant neurological lesions and glia activation associated with this disease (Fig. 7e–g).
To exclude the effect of rapamycin on AAV replication, we used CreERT2-LoxP recombination technology to delete Ubap1 in the projection neurons of adult mice. By crossing Thy1-YFP-CreERT2 mice53 with Ubap1flox/flox mice, we obtained Thy1-YFP-CreERT2:Ubap1flox/flox bi-transgenic mice. Ubap1 gene deletion was achieved by tamoxifen (TAM) intraperitoneal injection and we referred to these TAM-treated mice as Thy1-YFP Ubap1 cKO. These mice showed obvious lower limb spasticity, a typical symptom observed in HSP mice. We then examined the effects of delivering rapamycin (8 mg/kg) daily for one week by intraperitoneal injection beginning at one day post TAM (Supplementary Fig. 12a). Consistently, we found that rapamycin significantly attenuates disease phenotypes of Thy1-YFP Ubap1 cKO mice in tail suspension, beam working and rotarod assays (Supplementary Fig. 12b–f). However, gait and stride analysis revealed only marginal improvements in walking posture (Supplementary Fig. 12g–k). Taken together, our results indicated rapamycin exhibits a significant capacity to mitigate disease severity and elicit beneficial effects in different mouse models of Ubap1 cKO.
To explore the molecular mechanisms of rapamycin-mediated rescue in Ubap1 Syn-cKO mice, we performed RNA-seq to elucidate changes of gene expression in the brain. Upon the application of rapamycin in the brains of Syn-cKO mice, GSEA revealed a marked reversal of several activated pathways, notably including the PI3K-Akt and lysosome signaling (Supplementary Fig. 13a). In particular, the vast majority of lysosomal genes, encoding for various lysosomal enzymes and membrane proteins, increased their expression in Syn-cKO mouse brains which could be remedied by rapamycin treatment (Fig. 8a, b). We also noticed consistent downregulation of galectin responder genes following rapamycin treatment (Supplementary Fig. 13b). We confirmed mTOR signaling inhibition by rapamycin via immunoblots analysis (Fig. 8c). Consistent with findings in COS-7 cells, we observed significantly increased nuclear localization of TFEB in neurons from Ubap1 Syn-cKO mouse brains and rapamycin treatment reduced TFEB nuclear localization in these neurons (Fig. 8d). In addition, we found that the enlarged cell size observed in Syn-cKO neurons was reversed by rapamycin treatment, consistent with the modulation of mTORC1 activity (Fig. 8e). Increased lysosomal gene expression positively correlated with cellular senescence and may cause by and contributes to decreased lysosomal function and autophagic clearance54. Since UBAP1 deficiency may lead to lysosomal dysfunction as revealed by accumulation of GAL-3 puncta (Fig. 1) and upregulation of major galectin responders genes (Supplementary Figs. 3 and 7), we wondered that the benefit might arise from improved lysosome population or lysosomal function in rapamycin-treated brains. Immunoblots analysis indicates decreased expression of LAMP2 and p62 in rapamycin-treated Syn-cKO mouse brains (Fig. 8f), which is similar with the effect on VPS13A KO cells55. Consistently, the enlarged lysosome size and number was significantly reversed by rapamycin-treatment in the neuron of Syn-cKO mouse brains (Fig. 8g, h). Furthermore, TEM analysis revealed that the enlarged lysosome observed in the cortex of Ubap1 syn-cKO mice could be mitigated through the administration of rapamycin (Supplementary Fig. 13c). Collectively, these findings suggest that rapamycin treatment mitigates motor deficits and neurological lesions in Ubap1 cKO mice, likely through the enhancement of lysosomal function and the restoration of lysosomal homeostasis.
a GSEA-analysis of lysosome pathway enriched in brains from the Syn-cKO Veh versus GFP Veh group (up), or Syn-cKO Rap versus Syn-cKO Veh group (down). b Heatmap depicting the expression patterns of lysosome genes in the three indicated groups. c Immunoblot analysis of cortical lysates from 1 month old Syn-cKO with/without rapamycin treatment, and probed for the indicated antibodies. Quantification of relative protein expression, normalized to GAPDH. n = 3 mice per group. d Immunofluorescence analysis showed significantly increased TFEB (magenta) intensity in the nucleus of Syn-cKO mice. Syn-GFP Veh n = 36, Syn-cKO Veh n = 38, Syn-cKO Rap n = 39 neurons from more than three independent mouse per group. e Quantifications of neuronal soma size in (d). Syn-GFP Veh n = 114, Syn-cKO Veh n = 61, Syn-cKO Rap n = 62 neurons from more than three independent mouse per group. f Immunoblot analysis of cortical lysates from P40 mouse brains with/without Rapamycin, probed for LAMP2 and p62 and its quantification. n = 3 mice per group. g Immunofluorescence analysis of lysosomal morphology in Ubap1 Syn-cKO mouse cortex with or without rapamycin treatment by staining with LAMP1 (magenta). h Left: Quantifications of lysosome size in (g). Syn-cKO Veh n = 355, Syn-cKO Veh n = 494, Syn-cKO Rap n = 375 lysosomes from three independent brains. Right: Quantifications of lysosome number in (g). Syn-cKO Veh n = 19, Syn-cKO Veh n = 14, Syn-cKO Rap n = 23 cells from three independent brains. All scale bars: 10 μm. All data represent the mean ± SEM; A permutation test was used to calculate P values, with a two-tailed test for significance and Bonferroni correction for multiple comparison adjustments (a), Student’s t test (c), Kruskal–Wallis test followed by Dunn’s multiple comparisons test (d, e, h), Ordinary one-way ANOVA followed by Tukey’s multiple comparisons test (f), *p < 0.05, **p < 0.01, ***p < 0.001, #p < 0.0001. Exact p-values and test statistics are provided as a Source Data file.
Discussion
Mutations in the ESCRT-III core subunit CHMP2B are associated with frontotemporal dementia (FTD)56, amyotrophic lateral sclerosis (ALS)57 and a mixed ALS-FTD syndrome56. Mutations in subunit of ESCRT-I including UBAP1 and VPS37A lead to SPG80 and SPG53. Furthermore, the SPG20 protein, SPARTIN, binds to the ESCRT-III protein IST1 on damaged lysosomes58. Notably, the most common genetic cause of HSP stems from mutations in SPG4, which encodes SPASTIN, a protein that interacts with CHMP1B, a member of ESCRT-Ⅲ59. ESCRT components are emerging as a central player in a host of neurodegenerative diseases especially movement disorders, demonstrating potential roles which are likely to be revealed in pathogenesis and for viable therapeutic strategies. Elucidating the mechanisms by which the ESCRT machinery influences lysosomal degradation and motor neuron degeneration is of critical importance, particularly given the growing recognition of endolysosomal dysfunction in neurodegenerative disorders. Here, we demonstrated that the ESCRT-I subunit UBAP1 plays a pivotal role in lysosome recovering. Specifically, we show that prolonged UBAP1 deficiency leads to mTORC1 hyperactivation and motor dysfunction, highlighting the importance of ESCRT-I in counteracting lysosomal damage and maintaining signaling homeostasis — a process that is particularly relevant to the pathogenesis of HSP.
The lysosome is a highly dynamic structure that regulates various cellular processes, with basal lysosomal proteolysis and proper function being key to the recruitment and regulation of mTORC113,60. We observed a significant dissociation of mTOR from lysosomes and elevated levels of p-mTOR in cells with UBAP1 knockdown, phenocopying the effects induced by lysosome inhibitor BafA1 (Fig. 5). Importantly, this activation of p-mTOR, along with increased TFEB nuclear localization and hypo-phosphorylation, was also validated in patient-derived fibroblasts and in the brains of Ubap1 Nes-cKO/Syn-cKO mice. These findings suggest that lysosomal dysfunction, as a primary defect, may underlie the deregulation of mTOR signaling and the observed in vivo phenotypes.
The nuclear localization and hypophosphorylation of TFEB in UBAP1-deficient cells are unexpected, given that these cells exhibit high mTORC1 activity, and mTORC1-mediated phosphorylation of TFEB is a well-established mechanism for its cytoplasmic sequestration. This seemingly paradoxical phenomenon is explained by lack of spatial coincidence between the kinase and its substrates47. Notably, C9orf72 knock out/mutations cause devastating motor neuron diseases-ALS, which shares some clinical phenotypes with HSP, also lead to TFEB nuclear localization and mTORC1 hyper-activation. Coincidentally, the presence of activated mTOR and nuclear TFE3/TFEB in hypertrophic neurons in focal cortical dysplasia type Ⅱ and tuberous sclerosis complex (TSC) suggests that these factors contribute to the disease development61. Under TSC deficient cells, in which mTORC1 hyperactivity is the hallmark, TFEB shows nuclear localization and hypo-phosphorylated39,62. Despite no interaction with UBAP1, TFEB knockdown significantly reduces p-mTORC1 levels (Fig. 3), highlighting TFEB’s key role in regulating mTORC1 activation. However, the regulation of mTORC1 signaling is intricate and exhibits variability depending on both temporal factors and cell-type specificities. More extensive work is still needed regarding the varying impacts of lysosomal dysfunction on different cell types and at different developmental stages.
A growing number of studies pointed to rapamycin (Sirolimus) as a pharmacological compound that is able to provide neuroprotection in several experimental models of neurodegenerative diseases. In addition, rapamycin could directly activate lysosomal mucolipin TRP channels independent of mTOR49. We found systematic treatment with rapamycin could significantly alleviate the locomotor dysfunction and neuronal apoptosis in mouse models of UBAP1 deficiency mice (Fig. 7). Importantly, we observed a significant reduced TFEB nuclear translocation in UBAP1 siRNA-transfected COS-7 cells and Ubap1 cKO neurons following rapamycin treatment. Rapamycin treatment also mitigated the upregulation of lysosomal genes caused by UBAP1 deficiency. This is consistent with the reduced expression of LAMP2 observed after rapamycin treatment in the brains of Ubap1 Syn-cKO mice (Fig. 8). The decreased LysoSensor and Magic Red staining observed upon UBAP1 disruption was significantly restored by rapamycin treatment. Collectively, these results suggest that rapamycin promotes lysosomal repopulation, thereby ensuring sustained autophagic flux—a fundamental process that confers protection against disease progression, as previously reported63. In contrast to the classical dogma that mTOR inhibition is a key determinant of TFEB nuclear translocation and activation, our results support rapamycin treatment reduced TFEB nuclear translocation by enhancing mTOR lysosomal localization. It might be necessary to perform neuroprotection studies of rapamycin in Ubap1 cKO mouse under TFEB deficient and overexpression condition. Furthermore, rapamycin was shown to have robust immunosuppressive and anti-proliferative efficacy64. Given the pleiotropic effects of rapamycin, we cannot exclude other effects in recue experiments. Nonetheless, the remarkable therapeutic outcomes achieved can be partially attributed to the diverse effects of rapamycin. Given the influence of mTOR signaling on early development and cell proliferation, the administration of rapamycin should be avoided during the early stages of development. In addition, it would be intriguing to investigate whether genetically inactivating mTOR can replicate the effects observed with rapamycin treatment.
Lysosomes exhibit multifaceted roles in degrading and recycling cellular waste, playing pivotal parts in both innate and adaptive immunity, and contributing significantly to metabolic regulation60. KEGG pathway analysis indicated that the up-regulated genes were significantly enriched in complement and coagulation cascades, cytokines, and TGF-β signaling (Supplementary Fig. 13), consistent with the significant activation of microglia and astrocyte in Ubap1 cKO mouse brains (Fig. 6). GAL-3 was considered as a rising star in modulating microglia activation under conditions of neurodegeneration46. We found significantly upregulation of GAL-3 expression (Supplementary Fig. 7) and puncta accumulation (Fig. 1 and Supplementary Fig. 3) in Ubap1 deficient cells. Consequently, our RNA-seq and proteomic alteration of lysosomes may associate with the activation of immune response or inflammation. In brain, glia use lysosomal pathways for the secretion and uptake of regulatory molecules, which affect the physiology of neighboring neurons65. We observed GAL-3 puncta accumulation and upregulation of the lysosomal protein (LAMP1) in UBAP1-deficient cell lines, suggesting a cell-autonomous role for UBAP1 in regulating lysosomal abundance and function. However, the precise mechanism underlying the neuronal or glial-specific lysosomal regulation mediated by UBAP1 remains elusive. Single-cell multi-omics technologies combined with cell specific cKO mice will facilitate a more refined understanding of the specific role played by UBAP1 in regulating lysosomes across diverse neuronal cell types.
In summary, by combining in vitro and in vivo studies, we uncovered HSP causing gene UBAP1 as an essential regulator of mTOR localization and functioning through the mediation of lysosome function. Importantly, mTOR suppression by rapamycin significantly restores lysosome population and alleviates locomotor defects in mouse models of UBAP1 deficiency. These findings raised the possibility that rapamycin may offer therapeutic benefit to patients with HSP and potentially other motor neuron disorders. However, the regulatory mechanisms regarding the distinct roles of rapamycin in lysosome regulation are worthy to be elucidated in further studies. Our findings underscored the potential significance of lysosomal function in combating HSP and other neurodegeneration.
Methods
Mice
All mice were housed with free access to food and water under pathogen-free conditions in a facility where the temperature and light cycles (12 h cycle) were controlled. All animal procedures used in this study were performed according to the protocols approved by the Institutional Animal Care and Use Committee of Fujian Medical University (IACUCFJMU2022-NSFC-0184). Ubap1flox/flox, Ubap1 Nes-cKO and Ubap1 Nex-cKO mice have been previously described3. To generate Ubap1 Syn-cKO mice, Ubap1flox/flox mice were injected with pAOV-hSyn-EGFP-2A-Cre virus in the lateral ventricle at P0 or injected with pAOV-hSyn-EGFP virus as controls. To generate Thy1-YFP-Ubap1 cKO mice, Ubap1flox/flox mice were crossed with Thy1-YFP-CreERT2 mice. The Thy1-YFP-CreERT2:Ubap1flox/flox bi-transgenic mice were obtained and treated intraperitoneal injection with TAM and we refer to these TAM-treated mice as Thy1-YFP-Ubap1 cKO mice. The day of birth was defined as P0. Mice were randomly assigned into groups and both sexes were used in this study.
Behavioral test
All mice were housed under the same conditions for the behavioral tests. Learning-related tasks were assessed first, then the motor behaviors. Each cohort of mice was given at least 3 days’ break between tasks. All behavioral tests were conducted by trained experimentalists blinded to animal genotypes. Handling and specific tests are described in the Supplementary methods.
Virus, plasmids and siRNA
Exogenous adeno-associated virus pAOV-hSyn-EGFP-2A-Cre and pAOV-hSyn-EGFP were purchased from OBiO Technology (Shanghai, China). LAMP1-RFP, LAMP1-GFP and RAB7-RFP plasmids were gifts from Chonglin Yang66. The highly stable monomeric red fluorescent protein mYonghong (mYH)67 was created by Zhifei Fu’s lab. LAMP1-mYH, mYH-LC3B, Sec61β-mYH were gifts from Zhifei Fu. The complete coding sequence (CDS) of GAL3 was amplified and swapped with the EMTB gene in the EMTB-mBaoJin (wekwikgene, #0000268) vector using EcoRI-BamHI site. GFP-UBAP1WT and GFP-UBAP1Mut were described previously3, wherein the complete CDS of UBAP1 was amplified and cloned into pCAG-EGFP-C1 vector. To generate the plasmids for the UBAP1 truncation mutant alleles, a Mut Express II Fast Mutagenesis Kit V2 (Vazyme) was used according to the manufacturer’s instructions. The complete CDS of UBAP1 was amplified and swapped with the Sec61β gene in the Sec61β-mYH vector using Agel-NotI site. The complete CDS of CHMP4B were amplified and swapped with the LAMP1 gene in the LAMP1-mYH vector using NheI-NotI site. TMEM192-mKeima68 was synthesized by Tsingke (Beijing Tsingke Biotech Company) according to the sequence deposited in the addgenes (#215358), and swapped with the LAMP1 gene in the LAMP1-mYH vector using NheI-NotI site. The complete CDS of VPS37A were amplified and swapped with the LAMP1 gene in the LAMP1-mYH vector using NheI-AgeI site. The siRNA was generated by Tsingke (Beijing Tsingke Biotech Company), sequences were showed in Supplementary Table 1.
Cell culture and transfection
We generated fibroblasts from skin biopsies of 3 HSP patients with heterozygous mutations in the UBAP1 gene (patient 1, c.247_248insGTGAATTC; patient 2, c.526G>T; patient 3, c.426_427del) and 2 healthy controls matched by age. We obtained informed consent from all the participants. This study was approved by the Ethics Committee of the First Affiliated Hospital of Fujian Medical University ([2021]057). HeLa, COS-7, SPG80 patient-derive fibroblast and Halo-tag LC3 U2OS stable cell line (a gift from Qing Zhong) were cultured and passaged in Dulbecco’s modified DMEM medium (DMEM) containing 10% fetal bovine serum (FBS) and incubated at 37 °C in a humidified incubator with 5% CO2 to allow for growth and cell density of approximately 80%. Cells were transfected with Lipomaster 3000 Transfection Reagent (TL301, Vazyme, China) according to the manufacturer’s instructions.
Primary neurons were isolated from cortical tissues at E18.5 Ubap1 Nex-cKO or E15.5 Nes-cKO and age-matched control mouse brains. The cortex were dissected in ice cold HBSS, chopped into small pieces and incubated with 0.25% trypsin for 5 min at 37 °C. After trypsin treatment, cells were incubated with DMEM adding 10% FBS to stop the trypsin reaction. The cells were rinsed with Neurobasal medium and then suspended in Neurobasal medium with B27 supplement (Invitrogen, 2%), 1% penicillin-strep, and Glutamax (Gibco, 1X).
Cell culture treatments and administration in mice
Compounds were used at the following concentrations unless explicitly stated: 1 mM LLOMe (HY-129905, MCE); 25 μM BAPTA-AM (HY-100545, MCE); 10 μM wortmannin (HY-10197, MCE); 5 μM Vps34-IN-1 (HY-12795, MCE); 100 nM Bafilomycin A1 (HY-100558, MCE); 10 μM CHX (S7418, Selleck); Rapamycin (HY-10219, MCE). Concentrated stock solutions of all compounds were prepared in dimethyl sulfoxide (DMSO) and stored at −80 °C in single-use aliquots. Rapamycin was used at 500 nM for cells and 4 mg/kg or 8 mg/kg for animals.
Co-immunoprecipitation, immunoblot, and immunostaining
Immunoprecipitation, Immunoblot and Immunostaining were performed as previously described44. Detailed procedures are described in the supplementary materials. Information on the antibodies is presented in the supplementary materials. The unedited original Immunoblot images were provided in Source Data file.
RNA extraction and quantitative real-time PCR
Total RNA was extracted from the COS-7 cells transfected with siCtrl/siUBAP1 for 2 days, using TRIzol reagent following the manufacturer’s instructions. RNA quality and concentration was determined by NanoDrop spectrophotometers (Thermo Fisher Scientific). Reverse transcription was performed using HiScript II Q RT SuperMix (Vazyme). Quantitative real-time PCR (QPCR) was performed using SYBR Premix (Vazyme). The relative mRNA levels were normalized to that of GAPDH. QPCR primers: GAPDH Forward: TGATGACATCAAGAAGGTGGTGAAG, GAPDH Reverse: TCCTTGGAGGCCATGTAGGCCAT; TFEB Forward: GTGTTGAAGGTGCAGTCCTA, TFEB Reverse: ATAGGTCTCGGACAGGTACTC; UBAP1 Forward: GTTACAGGATGAGGAGGTCTTG, UBAP1 Reverse: GCCCAACTGTGGTAAGGTTAT.
Cell staining and fluorescence image analysis
We analyzed lysosomal mass and pH by LysoTracker (Thermo, L12492) and LysoSensor (Yeasen, Cat#40767ES50) staining. Cells were incubated with 1 µM Lysosensor (1 h), and wash 3 times/5 min with PBS. For co-staining LysoTracker with MagicRed, LysoTracker Deep Red (50 nM, Thermo, L12492) and MagicRed (ab270772 Abcam, reconstituted in 200 µL of DMSO) were used before mounting as instructions and immediately imaging. Imaging was performed with Zeiss LSM800 or Olympus FV3000 confocal microscope. Finally, we conducted fluorescence analyses using the ImageJ software to calculate the integrated intensity (IntDen., signal intensity contained in the stained area within the cell).
Structured illumination microscopy (SIM)
COS-7 cells were cultured as described above and transfected with plasmid and/or siRNA prior to imaging. The cells were imaged and then treated with LLOMe, washed twice with PBS and further imaged either in imaging medium at designated time to track recruitment of UBAP1. Primary neurons and fibroblasts were cultured as described above and were loaded with LysoTracker Deep Red (Invitrogen, 50 nM) for 10 min before imaging.
For 2D/3D-SIM image, imaging was performed on a commercial Multimodality Structured Illumination Microscopy (NanoInsights-Tech Co., Ltd., Beijing, China) equipped with a 100×/1.49 NA oil objective (Nikon CFI SR HP Apo) and an sCOMS (complementary metal-oxide-semiconductor) camera (Kinetix, Teledyne Imaging). Images were acquired by VSIM software. Each SIM image was reconstructed by SI-Recon software of Multi-SIM or Reconstruction software of HiS-SIM. Image analysis was performed using Fiji software. 3D reconstruction was performed using Imaris software.
RNA sequencing and tandem mass tag proteomics
Procedures are described in the supplementary materials.
Transmission electron microscopy
The cortex of the mouse brain was dissected and fixed with 2.5% glutaraldehyde in phosphate buffer (PB) at 4 °C overnight, washed with PBS, post-fixed with 1% osmium tetroxide in PB for 1 h at room temperature, rinsed with PBS, stained with 2% uranylacetate in 70% ethanol at 4 °C for 1 h, dehydrated using serial dilutions of ethanol, incubated in propylene oxide, and embedded in Epon resin. Tissues were sectioned (65 nm thickness) using an ultramicrotome (LEICA, UC6) and counterstained with uranyl acetate and lead citrate. The grids were viewed using an electron microscope (Tecnai G2J), and digital images were captured using a CCD camera.
Statistics and reproducibility
ImageJ and OlyVIA were used for all image analysis. GraphPad Prism 8.0 was used for all data analysis. The Gaussian distribution of the data was assessed using D’Agostino, Pearson normality, or Shapiro–Wilk normality tests. If the data passed the Gaussian distribution test, parametric unpaired two-tailed t-tests were used for two groups and one-way analysis of variance (ANOVA), followed by Tukey’s multiple comparison tests, for three or more groups. Otherwise, nonparametric unpaired Mann–Whitney test or Kruskal–Wallis test was used for two groups or three groups. Data between independent groups are shown as the mean ± SEM for at least three independent experiments. All the statistical details of the experiments are described in the figure legends. If not otherwise noted, statistical significance was indicated as follows: p > 0.05 (ns), p < 0.05 (*), p < 0.01 (**), p < 0.001 (***), and p < 0.0001(#). For Figs. 1f, 6g, h, and 7e–g, the experiments were repeated more than three times/individual mice, and the images shown are representative of the data obtained.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All data associated with this study can be found in the paper, the Supplementary materials and Source data file. All proteomics data has been deposited in ProteomeXchange (https://proteomecentral.proteomexchange.org, PXD048729). RNA-seq database can be found at China National Center for Bioinformation/Beijing Institute of Genomics (https://ngdc.cncb.ac.cn/gsa, CRA018048, CRA018053 and CRA011436). The original figures have been deposited in figshare (https://doi.org/10.6084/m9.figshare.30118339.v1) and research materials are available upon reasonable request. Source data are provided with this paper.
References
Murala, S., Nagarajan, E. & Bollu, P. C. Hereditary spastic paraplegia. Neurological Sci. 42, 883–894 (2021).
Toupenet Marchesi, L., Leblanc, M. & Stevanin, G. Current knowledge of endolysosomal and autophagy defects in hereditary spastic paraplegia. Cells 10, 1678 (2021).
Lin, X. et al. Stop-gain mutations in UBAP1 cause pure autosomal-dominant spastic paraplegia. Brain J. Neurol. 142, 2238–2252 (2019).
Farazi Fard, M. A. et al. Truncating mutations in UBAP1 cause hereditary spastic paraplegia. Am. J. Hum. Genet. 104, 767–773 (2019).
Williams, R. L. & Urbe, S. The emerging shape of the ESCRT machinery. Nat. Rev. Mol. cell Biol. 8, 355–368 (2007).
Lefebvre, C., Legouis, R. & Culetto, E. ESCRT and autophagies: endosomal functions and beyond. Semin. Cell Dev. Biol. 74, 21–28 (2018).
Eapen, V. V., Swarup, S., Hoyer, M. J., Paulo, J. A. & Harper, J. W. Quantitative proteomics reveals the selectivity of ubiquitin-binding autophagy receptors in the turnover of damaged lysosomes by lysophagy. eLife 10, e72328 (2021).
Jia, J. et al. Galectin-3 coordinates a cellular system for lysosomal repair and removal. Dev. Cell 52, 69–87.e68 (2020).
Takahashi, Y. et al. VPS37A directs ESCRT recruitment for phagophore closure. J. Cell Biol. 218, 3336–3354 (2019).
Skowyra, M. L., Schlesinger, P. H., Naismith, T. V. & Hanson, P. I. Triggered recruitment of ESCRT machinery promotes endolysosomal repair. Science 360, eaar5078 (2018).
Radulovic, M., et al. ESCRT-mediated lysosome repair precedes lysophagy and promotes cell survival. EMBO J. 37, e99753 (2018).
Pashkova, N. P. R. UBAP1: a new ESCRT member joins the cl_Ub. Structure 30, 383–385 (2012).
Fernandes, S. A. et al. Spatial and functional separation of mTORC1 signalling in response to different amino acid sources. Nat. Cell Biol. 26, 1918–1933 (2024).
Shao, Q. et al. C9orf72 and smcr8 mutant mice reveal MTORC1 activation due to impaired lysosomal degradation and exocytosis. Autophagy 16, 1635–1650 (2020).
Hwang, J. Y. et al. Global ischemia induces lysosomal-mediated degradation of mTOR and activation of autophagy in hippocampal neurons destined to die. Cell Death Differ. 24, 317–329 (2017).
Roczniak-Ferguson, A. et al. The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci. Signal. 5, ra42 (2012).
Wander, S. A., Hennessy, B. T. & Slingerland, J. M. Next-generation mTOR inhibitors in clinical oncology: how pathway complexity informs therapeutic strategy. J. Clin. Investig. 121, 1231–1241 (2011).
Hou, S. J. Z. S., Li, Y. & Xu, S. Y. Rapamycin responds to Alzheimer’s disease: a potential translational therapy. Clin. Inter. Aging 18, 1629–1639 (2023).
Xue, W., Zhang, J. & Li, Y. Enhancement of lysosome biogenesis as a potential therapeutic approach for neurodegenerative diseases. Neural Regeneration Res. 18, 2370–2376 (2023).
Sarkar, S., Ravikumar, B., Floto, R. A. & Rubinsztein, D. C. Rapamycin and mTOR-independent autophagy inducers ameliorate toxicity of polyglutamine-expanded huntingtin and related proteinopathies. Cell death Differ. 16, 46–56 (2009).
Bove, J., Martinez-Vicente, M. & Vila, M. Fighting neurodegeneration with rapamycin: mechanistic insights. Nat. Rev. Neurosci. 12, 437–452 (2011).
Vantaggiato, C. O. G. et al. Rescue of lysosomal function as therapeutic strategy for SPG15 hereditary spastic paraplegia. Brain 146, 1103–1120 (2022).
Xu, S., Stern, M. & McNew, J. A. Beneficial effects of rapamycin in a Drosophila model for hereditary spastic paraplegia. J. cell Sci. 130, 453–465 (2017).
Aits, S. et al. Sensitive detection of lysosomal membrane permeabilization by lysosomal galectin puncta assay. Autophagy 11, 1408–1424 (2015).
Thiele, D. L. & Lipsky, P. E. Mechanism of L-leucyl-L-leucine methyl ester-mediated killing of cytotoxic lymphocytes: dependence on a lysosomal thiol protease, dipeptidyl peptidase I, that is enriched in these cells. Proc. Natl Acad. Sci. USA 87, 83–87 (1990).
Bussi, C. et al. Stress granules plug and stabilize damaged endolysosomal membranes. Nature 623, 1062–1069 (2023).
Stefani, F. et al. UBAP1 is a component of an endosome-specific ESCRT-I complex that is essential for MVB sorting. Curr. Biol.: CB 21, 1245–1250 (2011).
Mulligan R. J. et al. Collapse of late endosomal pH elicits a rapid Rab7 response via the V-ATPase and RILP. J. Cell Sci. 137, jcs261765 (2024).
Li, P. et al. A novel mutation in the UBAP1 gene causing hereditary spastic paraplegia: a case report and overview of the genotype-phenotype correlation. Front. Genet. 13, 936292 (2022).
Jimenez, A. J. et al. ESCRT machinery is required for plasma membrane repair. Science 343, 1247136 (2014).
Scheffer, L. L. et al. Mechanism of Ca(2)(+)-triggered ESCRT assembly and regulation of cell membrane repair. Nat. Commun. 5, 5646 (2014).
Denais, C. M. et al. Nuclear envelope rupture and repair during cancer cell migration. Science 352, 353–358 (2016).
Gahlot, P. et al. Lysosomal damage sensing and lysophagy initiation by SPG20-ITCH. Mol. Cell 84, 1556–1569.e1510 (2024).
Shima T., et al. The TMEM192-mKeima probe specifically assays lysophagy and reveals its initial steps. J. Cell Biol. 222, e202204048 (2023).
Ishida, Y., Nayak, S., Mindell, J. A. & Grabe, M. A model of lysosomal pH regulation. J. Gen. Physiol. 141, 705–720 (2013).
Takahashi, Y. et al. An autophagy assay reveals the ESCRT-III component CHMP2A as a regulator of phagophore closure. Nat. Commun. 9, 2855 (2018).
Wei, B. et al. ANKFY1 bridges ATG2A-mediated lipid transfer from endosomes to phagophores. Cell Discov. 10, 43 (2024).
Wang, X., et al. hnRNPA2B1 represses the disassembly of arsenite-induced stress granules and is essential for male fertility. Cell Rep. 43, 113769 (2024).
Alesi, N. et al. TFEB drives mTORC1 hyperactivation and kidney disease in Tuberous Sclerosis Complex. Nat. Commun. 15, 406 (2024).
Cao, M., Luo, X., Wu, K. & He, X. Targeting lysosomes in human disease: from basic research to clinical applications. Sig. Transduct. Target. Ther. 6, 379 (2021).
Zhao, J. F. et al. Targeted dephosphorylation of TFEB promotes its nuclear translocation. iScience 27, 110432 (2024).
Di Malta, C. et al. Transcriptional activation of RagD GTPase controls mTORC1 and promotes cancer growth. Science 356, 1188–1192 (2017).
Settembre, C. et al. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J. 31, 1095–1108 (2012).
Lu, D. et al. ESCRT-I protein UBAP1 controls ventricular expansion and cortical neurogenesis via modulating adherens junctions of radial glial cells. Cell Rep. 43, 113818 (2024).
Shariq, M., Khan, M. F., Raj, R., Ahsan, N. & Kumar, P. PRKAA2, MTOR, and TFEB in the regulation of lysosomal damage response and autophagy. J. Mol. Med. 102, 287–311 (2024).
Garcia-Revilla, J. et al. Galectin-3, a rising star in modulating microglia activation under conditions of neurodegeneration. Cell Death Dis. 13, 628 (2022).
Acharya, A. & Demetriades, C. mTORC1 activity licenses its own release from the lysosomal surface. Mol. Cell 84, 4385–4400.e4387 (2024).
Wrobel, M. et al. ESCRT-I fuels lysosomal degradation to restrict TFEB/TFE3 signaling via the Rag-mTORC1 pathway. Life Sci. Alliance 5, e202101239 (2022).
Zhang, X. et al. Rapamycin directly activates lysosomal mucolipin TRP channels independent of mTOR. PLoS Biol. 17, e3000252 (2019).
Bourdenx, M. et al. Chaperone-mediated autophagy prevents collapse of the neuronal metastable proteome. Cell 184, 2696–2714.e2625 (2021).
Shimozono, K. N. H. et al. Ubap1 knock-in mice reproduced the phenotype of SPG80. J. Hum. Genet. 67, 679–686 (2022).
Johnson, S. C. et al. mTOR inhibition alleviates mitochondrial disease in a mouse model of Leigh syndrome. Science 342, 1524–1528 (2013).
Feng, G. et al. Imaging neuronal subsets in transgenic mice expressing multiple spectral variants of GFP. Neuron 28, 41–51 (2000).
Fujimaki, K. et al. Graded regulation of cellular quiescence depth between proliferation and senescence by a lysosomal dimmer switch. Proc. Natl Acad. Sci. USA 116, 22624–22634 (2019).
Tornero-Ecija, A. R., Navas, M. A., Munoz-Braceras, S., Vincent, O. & Escalante, R. Effect of rapamycin on lysosomal accumulation in a CRISPR/Cas9-based cellular model of VPS13A deficiency. J. Cell. Mol. Med. 27, 1557–1564 (2023).
Skibinski, G. et al. Mutations in the endosomal ESCRTIII-complex subunit CHMP2B in frontotemporal dementia. Nat. Genet. 37, 806–808 (2005).
Parkinson, N. et al. ALS phenotypes with mutations in CHMP2B (charged multivesicular body protein 2B). Neurology 67, 1074–1077 (2006).
Gahlot, P. et al. Lysosomal damage sensing and lysophagy initiation by SPG20-ITCH. Mol. Cell 84, 1556–1569 e1510 (2024).
Reid, E. et al. The hereditary spastic paraplegia protein spastin interacts with the ESCRT-III complex-associated endosomal protein CHMP1B. Hum. Mol. Genet. 14, 19–38 (2005).
Gros, F. & Muller, S. The role of lysosomes in metabolic and autoimmune diseases. Nat. Rev. Nephrol. 19, 366–383 (2023).
Zwakenberg, S. et al. mTORC1 restricts TFE3 activity by auto-regulating its presence on lysosomes. Mol. cell 84, 4368–4384.e4366 (2024).
Alesi, N. et al. TSC2 regulates lysosome biogenesis via a non-canonical RAGC and TFEB-dependent mechanism. Nat. Commun. 12, 4245 (2021).
Nanayakkara, R. et al. Autophagic lysosome reformation in health and disease. Autophagy 19, 1378–1395 (2023).
Thomson, A. W., Turnquist, H. R. & Raimondi, G. Immunoregulatory functions of mTOR inhibition. Nat. Rev. Immunol. 9, 324–337 (2009).
Kreher C., Favret J., Maulik M., Shin D. Lysosomal functions in glia associated with neurodegeneration. Biomolecules 11, 400 (2021).
Xing, R. et al. The Rab7 effector WDR91 promotes autophagy-lysosome degradation in neurons by regulating lysosome fusion. J. Cell Biol. 220, e202007061 (2021).
Xiong, H. et al. A highly stable monomeric red fluorescent protein for advanced microscopy. Nat. Methods 22, 1288–1298 (2025).
Shima, T., et al. The TMEM192-mKeima probe specifically assays lysophagy and reveals its initial steps. J. Cell Biol. 222, e202204048 (2023).
Acknowledgements
The work was supported by National Natural Science Foundation of China (82222022 and 82171403: X.L., and 32371025: D.X.), the Natural Science Foundation of Fujian Province (2024J010028 and 2023J02018: D.X.), Joint Funding for Science and Technology Innovation in Fujian Province (2023Y9271: D.X.), Fujian Provincial Health Technology Project (2022ZQNZD003: X.L.), Excellent Youth Training Program of the First Affiliated Hospital of Fujian Medical University (2023FY-JCQN-1: X.L.), and Qihang Foundation of Fujian Medical University (2023QH2002: Y.Z.). We would like to thank Drs. Qing Zhong for the HaloTag LC3 U2OS stable cell line, Chonglin Yang for the LAMP1-RFP, LAMP1-GFP and RAB7-RFP constructs, Ling Yuan for Myc-Ub, and Kiryl Piatkevich for mBaojin construct. We also want to thank Dr. Yang Li for constructive advices regarding the manuscript.
Author information
Authors and Affiliations
Contributions
D.X. conceived and designed the study, analyzed the experiments, and wrote and revised the manuscript. Y.Z. designed the study, performed and analyzed most of the experiments, and wrote and revised the manuscript. T.Z. and D.L. performed mice experiments. X.L., S.L., and H.S. provided the patient-derived fibroblasts and generated the Ubap1flox/flox mice. Y.W., Q.C., and C.L. performed some of the initial characterization in cells and mice. Z.F. assisted with the SIM imaging. S.W. helped with the STM experiments. H.F. assisted with bioinformatics analysis. L.C., W.C., and N.W. discussed the results and commented on the manuscript. All authors reviewed the results and approved the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Jiajie Diao and the other anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhi, Y., Zhang, T., Lu, D. et al. Lysosomal and mTORC1 signaling dysregulation underpin the pathology of spastic paraplegia type 80. Nat Commun 16, 9833 (2025). https://doi.org/10.1038/s41467-025-64800-5
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-64800-5