Abstract
Enhancing crop nutritional value is important for advancing sustainable human health. Serotonin (5-HT) and its derivative melatonin (MT) are versatile physiological regulators, known for their roles in sleep enhancement, anxiety reduction, and immune modulation. Here, we discover that abscisic acid (ABA) induces the biosynthesis of 5-HT. This induction depends on the regulation by Abscisic Acid Insensitive 5 (ABI5) and negatively feedback-controlled by the possible PP2Cs–SAPK2–ABI5 interaction. This feedback regulation likely involves ABA signaling crosstalk. Specifically, 5-HT modulates ABA-mediated PP2C activity, thereby influencing the SAPK2 phosphorylation. This modulation subsequently reduces the phosphorylation and transcriptional activity of ABI5, ultimately attenuating the ABA signaling cascade. The T162 and T283 residues of SAPK2 contribute to modulating ABI5 phosphorylation. Based on the crosstalk between ABA and 5-HT, we develop several rice lines with enhanced 5-HT/MT levels without compromising grain yield. These engineered rice lines hold promise for improving rice’s nutritional value and promoting the production of health-beneficial foods.
Similar content being viewed by others
Introduction
Serotonin and melatonin, bioactive nutrients recognized for their health-promoting properties, are commercially available as dietary supplements1,2,3. However, their widespread adoption is limited by potential side effects, high costs, and restrictions in specific populations3,4. Plant-derived 5-HT/MT exhibits favorable bioactivity profiles, facilitating safe consumption without adverse effects5. Therefore, enhancing the levels of 5-HT/MT in staple crops presents a promising strategy for promoting human health.
Plant biosynthetic pathways of 5-HT/MT begin with their common precursor, L-tryptophan. Tryptophan is first decarboxylated to tryptamine by tryptophan decarboxylase (TDC), then transform into 5-HT by tryptamine-5-hydroxylase (T5H), and finally convert to MT through two additional enzymatic steps6,7,8. All known plant MT biosynthesis pathways use 5-HT as an intermediate, underscoring its central role in MT synthesis8. TDC and T5H are the rate-limiting enzymes in the synthesis of 5-HT and subsequently affect MT production9. One strategy to enhance 5-HT/MT levels in rice involves targeting genes associated with these rate-limiting enzymes. However, this approach is limited by the small number of modifiable genes and the potential adverse effects on plant growth, such as reduced growth and fertility resulting from the overexpression of TDC1 or TDC310. Consequently, it is essential to explore regulatory mechanisms for 5-HT and MT synthesis to improve their accumulation in rice without compromising yield.
Microarray data from the RiceXPro database indicate that genes involved in 5-HT biosynthesis are primarily regulated by ABA and jasmonic acid (JA)11. ABA promotes 5-HT synthesis in detached leaves, whereas other plant hormones, such as salicylic acid (SA) and JA, have minimal impact on 5-HT production12,13. Furthermore, JA fails to stimulate the formation of 5-HT-mediated brown rings, whereas exogenous ABA significantly amplifies this process14. ABA is a pivotal plant hormone essential for the biosynthesis of a wide array of plant nutrients. For instance, ABA can be deployed to augment the vitamin C levels in mature fruits, thereby enhancing the quality and market value of the fruits15. It coordinates the accumulation of ascorbic acid (AsA) during tomato fruit development via the SIMAPK8–SIARF4–SIMYB11 pathway16, and it governs the biosynthesis of anthocyanins in apples through the MYB1–bHLH3 complex17. These successful examples of nutrient synthesis underscore the potential of ABA to enhance the nutritional value of rice.
Currently, synthetic strategies have proven effective in the metabolic engineering of rice. By modifying key genes involved in the biosynthetic pathways of beneficial metabolites, rice varieties enriched in betaine, carotenoids, vitamin B1, and vitamin B9 have been successfully developed18,19,20,21. Here, we show the regulatory mechanism by which ABA promotes 5-HT biosynthesis, as well as the feedback regulation triggered by excessive 5-HT accumulation. By leveraging the interplay between ABA and the 5-HT pathway, we have developed various rice lines enriched in 5-HT/MT, while maintaining yield. These nutrient-enriched rice resources constitute a valuable resource for improving human health and nutrition.
Results
ABA induces the biosynthesis of 5-HT
To investigate the impact of ABA on 5-HT biosynthesis, we first assessed the changes in 5-HT and its related metabolites following ABA treatment (Fig. 1a–e and Supplementary Fig. 1). Liquid chromatography-mass spectrometry (LC–MS) analysis revealed a significant increase in both 5-HT and its derivative MT, within 24 h of ABA exposure (Fig. 1d, e). Further analysis showed that tryptamine levels increased markedly during this period, whereas tryptophan levels initially decreased before rising again (Fig. 1b, c). These results suggest that ABA may facilitate the conversion of tryptophan to tryptamine, thereby enhancing the biosynthesis of 5-HT/MT.
a Metabolic pathways associated with 5-HT biosynthesis in plants. b–e Quantitative analysis of tryptophan (Trp), tryptamine (Try), serotonin (5-HT), and melatonin (MT) levels in rice seedlings subjected to 150 μM ABA treatment for 12 or 24 h. Representative chromatograms (left panels) displayed the characteristic peaks of each compound before ABA treatment (top) and after ABA treatment (bottom). Detailed mass spectrometry data corresponding to these analyses were provided in Supplementary Fig. 1. f Transcriptional expression of key genes in the 5-HT/MT biosynthetic pathway (TDC1/2/3 and T5H) in rice seedlings treated with 50 μM or 150 μM ABA for 12 h. g Transcriptional changes of genes in the 5-HT biosynthesis pathway among ABA-induced differentially expressed genes (DEGs). Data are presented as means ± SD (n = 3 biological replicates). A two-tailed unpaired Student’s t-test was used for statistical analysis. Source data are provided as a Source Data file.
To further elucidate the regulatory mechanisms of ABA on 5-HT/MT biosynthesis, we analyzed the transcriptional expression of relevant genes within 24 hours post-treatment with two distinct ABA concentrations. Quantitative PCR (qPCR) results revealed significant induction of key genes in the 5-HT biosynthesis pathway, including TDC1, TDC3, and T5H, following ABA treatment (Fig. 1f). In contrast, genes directly involved in MT synthesis did not exhibit consistent induction (Supplementary Fig. 2). This finding was corroborated by an analysis of differentially expressed genes (DEGs) induced by ABA (Fig. 1g), which confirmed that ABA primarily enhances 5-HT/MT biosynthesis by upregulating relevant genes in the 5-HT pathway, particularly TDC1/3, and T5H.
ABI5 mediates ABA-induced 5-HT biosynthesis
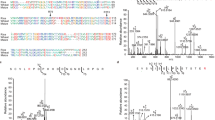
To elucidate the transcriptional regulation of TDC1/3 and T5H genes in response to ABA signaling, we cloned ~2-kb promoter regions of these target genes into the pLacZi2μ reporter vector and conducted yeast one-hybrid (Y1H) screening using a pb42AD transcription factor library generated in our laboratory. The initial screening found several bZIP transcription factors with putative promoter-binding activity, with bZIP10 (ABI5) exhibited the strongest binding affinity to all three promoter regions (Supplementary Fig. 3). Given that ABI5 plays an important role in ABA-mediated biosynthesis of specific metabolites, such as promoting anthocyanin biosynthesis in apples and increasing fatty acid content in fruits17,22. Accordingly, subsequent experiments will focus on determining the functional consequences of ABI5-mediated transcriptional regulation on 5-HT synthesis. We utilized CUT&Tag technology to detect ABI5 protein and assess its binding to the promoters of TDC1/3 and T5H genes (Fig. 2a and Supplementary Table 1). Our analysis identified two T-box and two A-box motifs within the 2-kb regions of the TDC1 and TDC3 promoters, respectively (Fig. 2b). These sequences overlap with known ABI5 binding sites, suggesting that ABI5 may regulate their transcription by directly binding to these regions. Additionally, we found two ABA-responsive G-box motifs in the ~1-kb upstream region of the T5H promoter (Fig. 2b), which may also serve as ABI5 binding sites. Further validation via electrophoretic mobility shift assays (EMSA) confirmed the specific binding of ABI5 to these regulatory motifs (Fig. 2c). Subsequent dual-luciferase reporter (DLR) assays demonstrated that ABI5 acts as a transcriptional activator for TDC1/3 and T5H genes, thereby modulating their expression at the transcriptional level (Fig. 2d). Collectively, these results suggest that the ABA-responsive behaviors of TDC1/3 and T5H are likely governed by ABI5.
a Partial ABI5-binding motifs identified by CUT-tag analysis. Additional motifs are presented in Supplementary Table 1. These motifs primarily include core sequences corresponding to the T-box (NNA/TCTTTTTN) and A-box (NAAAAAGNNN) elements. b ABI5-binding motifs (T-box and A-box) were detected within approximately 2 kb of the TDC1 and TDC3 promoter regions, while two canonical G-box motifs were identified within approximately 1 kb of the T5H promoter region. c EMSA assays showing ABI5 binding to the T-box and A-box motifs in the promoters of TDC1 and TDC3, as well as to the G-box motif in the T5H promoter. The addition of increasing concentrations of unlabeled competitor probes markedly reduced ABI5 binding to these motifs, confirming the specificity of these interactions. The experiment was independently repeated three times, with consistent results. d Dual-luciferase reporter assays showing the regulatory effects of ABI5 on the promoter activities of TDC1/3 and T5H in rice protoplasts. e Sequence alignment between wild-type (WT) and CRISPR-Cas9-edited ABI5 mutants. The CRISPR target sequences are highlighted in green, and PAM sequences are indicated in blue. f Comparison of transcript levels of TDC1, TDC3, and T5H between WT and Ubi::ABI5 overexpression lines. g Comparison of 5-HT and MT contents between WT and Ubi::ABI5 overexpression lines. Values are means ± SD (n = 6 plants for d, n = 3 biological replicates for f, g). For statistical analysis, a two-tailed unpaired Student’s t-test was employed. Source data have been provided as a Source Data file.
To explore whether the plant protein ABI5 participates in 5-HT biosynthesis, we generated ABI5 knockout mutants (CR-abi5) using the CRISPR/Cas9 system (Fig. 2e). We also developed plants overexpressing ABI5 under the control of the maize Ubi1 promoter (Ubi::ABI5) (Supplementary Fig. 4a, b). The transcript levels of T5H and TDC1/3 in Ubi::ABI5 plants, as quantified by qPCR, were increased (Fig. 2f). Correspondingly, the concentrations of 5-HT/MT were notable elevated in these plants (Fig. 2g). In contrast, ABA-induced expression of T5H and TDC1/3 was markedly attenuated in CR-abi5 mutants (Supplementary Fig. 5). Those experimental results indicate that ABI5 is likely implicated in the regulation of 5-HT levels in rice, subsequently exerting an influence on ABA-mediated 5-HT synthesis.
Additionally, ABI5 contains several conserved phosphorylation sites, including serine residues S44 and S118, which are potential targets of SnRK2 kinases23. To explore the functional roles of these residues in ABI5’s transcriptional activity, we substituted S44 and S118 with aspartate acid. DLR assays revealed that phosphorylation at S44 significantly enhanced ABI5’s transcriptional activity and its ability to activate T5H and TDC1/3 (Supplementary Fig. 6). Collectively, these observations highlight the important role of ABI5’s phosphorylation status in the biosynthesis of 5-HT.
5-HT attenuates ABA signaling
Plants possess regulatory mechanisms designed to modulate the overaccumulation of metabolic products, thereby optimizing growth and development24. The accumulation of such metabolites often triggers strong feedback signals that mitigate potential adverse effects on plant development25. However, these mechanisms are typically not conducive to enhancing the yield of beneficial metabolites. Our findings demonstrated that excessive accumulation of 5-HT or MT led to a significant downregulation of relevant biosynthetic genes (Supplementary Fig. 7). This indicates the presence of a feedback regulation mechanism, wherein 5-HT/MT modulate their own synthesis in response to their overaccumulation. Importantly, the feedback regulation of 5-HT synthesis was linked to the suppression of the TDC1/3 and T5H genes, both of which were strongly induced by ABA treatment. These results imply that the feedback regulation of 5-HT may be partially mediated by the ABA signaling pathway.
To examine the impact of 5-HT on ABA signaling, we treated two-week-old rice seedlings with external hormones and subsequently conducted RNA sequencing (RNA-seq) 24 h post-treatment. With ABA treatment alone, we totally identified 7142 DEGs (fold change ≥2 or ≤−2), comprising 3615 upregulated and 3527 downregulated genes. When seedlings were co-treated with ABA and 5-HT, the number of DEGs decreased to 5618, including 2422 upregulated and 3196 downregulated genes (Supplementary Fig. 8). A Venn diagram analysis was performed to compare the DEGs induced by ABA treatment alone with those induced by the combined treatment of ABA and 5-HT. The results showed that the vast majority of DEGs identified in the combined treatment group (4730/5618) overlapped with those induced by ABA treatment alone (Fig. 3a). This finding implies that, when 5-HT is co-applied with ABA, the transcriptional changes in DEGs are predominantly governed by ABA signaling pathways.
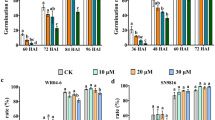
a Venn diagram illustrating the overlap of DEGs in rice seedlings subjected to ABA treatment alone and in combination with 5-HT (ABA + 5-HT). b DEGs exhibiting fold changes ≥ 2 or ≤ −2 and a false discovery rate (FDR) ≤ 0.01 in response to 24-h treatments with either ABA alone or ABA + 5-HT. c KEGG pathway enrichment analysis of DEGs under ABA and ABA + 5-HT treatments. Hormone signaling pathways highlighted in red are significantly enriched only in the ABA + 5-HT co-treatment compared to ABA alone. d–g Phenotypic analysis, germination rates, and chlorophyll content in seeds and seedlings treated with ABA or ABA + 5-HT. For seedlings, treatments consisted of 150 µM ABA and 100 µM 5-HT, with chlorophyll content measured after one week (e). For seeds, treatments involved 10 µM ABA and 10 µM 5-HT, with germination rates recorded every 12 h (g) and analyzed after 72 h (f). h–k Comparison of phenotypes, germination rates, and chlorophyll content in seeds and seedlings of wild-type (WT) and Ubi::ABI5 lines following exogenous ABA treatment. Experimental replicates are presented as box plots with overlaid individual data points (j). Error bars denote the maximum and minimum values. For the box plots, the horizontal line inside represents the median, while the bottom and top of the box correspond to the 25th and 75th percentiles, respectively. Treatment concentrations were identical to those described in panels (d–g). For (d) and (h), the experiments were independently repeated three times, with consistent results. For (e, g, i, k), values are means ± SD (n = 3 biological replicates). Statistical analysis was performed using a two-tailed unpaired Student’s t-test. For (e) and (f), different letters denote statistically significant differences (P < 0.05), as determined by one-way analysis of variance (ANOVA) followed by Duncan’s multiple range test. Source data are available as a Source Data file.
The count of DEGs induced by the dual treatment was significantly reduced (5618/7142), particularly the number of upregulated genes (2422/3615) (Fig. 3b). Further analysis revealed that the enrichment of ABA-responsive signaling pathways and the corresponding gene counts was significantly diminished (11/26), although this reduction did not extend to the hormone signaling pathways (Fig. 3c). Collectively, these findings suggest that 5-HT attenuates ABA signaling.
5-HT antagonizes ABA-mediated physiological processes
We further investigated the role of 5-HT in attenuating ABA signaling by examining two physiological indicators closely associated with ABA: seed germination and leaf senescence. 5-HT exerted effects opposite to ABA in both processes, with optimal concentrations of 5-HT promoting seed germination and delaying leaf senescence (Supplementary Fig. 9a). Specifically, treatment with 10 μM ABA significantly reduced seed germination rates and leaf chlorophyll content compared to the control (CK). In contrast, seeds and leaves treated with 10 μM 5-HT displayed growth-promoting effects, exhibiting significantly higher seed germination rates and chlorophyll contents than the CK group (Supplementary Fig. 9b–d). Further analysis of concentration-dependent effects revealed a negative correlation between ABA concentration and seed germination rates, with germination progressively decreasing as ABA concentrations increased. Conversely, 5-HT promoted germination within the concentration range of 1–10 μM (Supplementary Fig. 9e, f).
Both exogenous 5-HT application and elevated endogenous 5-HT levels mitigated ABA-mediated germination and senescence. Seeds co-treated with 10 μM 5-HT exhibited significantly higher germination rates throughout the germination period compared to those treated with 10 μM ABA alone. Additionally, in seedlings exposed to 100 μM ABA, co-application of 100 μM 5-HT resulted in significantly greater chlorophyll retention after one week relative to ABA treatment alone (Fig. 3d–g). This phenotype was further corroborated in transgenic lines overexpressing T5H (Supplementary Fig. 4c-e), a key enzyme in 5-HT biosynthesis, which displayed reduced ABA sensitivity, as evidenced by increased germination rates and improved chlorophyll retention under ABA stress (Fig. 3h–k and Supplementary Fig. 10) Collectively, these results indicate that 5-HT signaling antagonizes ABA-dependent pathways and modulates ABA-mediated physiological responses.
5-HT modulates ABA-induced formation of the PP2Cs-SAPK2 complex
Within the ABA signaling cascade, ABA binds to the PYR/PYL/RCARs sensor module to inhibit the activity of protein phosphatases type-2C (PP2Cs), consequently releasing SNF1-related kinases subfamily 2 (SnRK2s) from PP2C-mediated suppression26,27. The liberated SnRK2s then phosphorylate and activate AREBs/ABFs and ABI5, initiating transcriptional reprogramming in response to ABA28,29. Our analysis of the impact of 5-HT on ABA-induced DEGs revealed that 5-HT enhanced the enrichment of multiple PP2C genes within the hormone signaling pathway (Fig. 4a). In rice, the SnRK2 kinase subgroup includes SAPK1-10 (stress/ABA-activated protein kinases), with SAPK2, SAPK6, SAPK8, SAPK9, and SAPK10 being involved in ABA signaling30,31. To explore the potential regulatory mechanisms of PP2C genes enriched under combined treatment conditions within the ABA signaling pathway, yeast two-hybrid (Y2H) assays were performed. Full-length PP2C proteins (BK-PP2Cs) were used as baits, while full-length SAPK family proteins (AD-SAPKs) served as preys to screen for protein-protein interactions. Preliminary results identified interactions between PP2C49 and SAPK2, SAPK8, SAPK9, and SAPK10, as well as among PP2C30, PP2C49, PP2C51, and SAPK2 (Fig. 4b and Supplementary Fig. 11). Given the central role of SAPK2 within these interaction networks and previous evidence indicating that PP2C30/49/51 regulate ABA signaling via SAPK232,33, the present study specifically focused on elucidating the PP2Cs–SAPK2 regulatory relationship.
a Heatmap analysis showing that combined treatment with ABA and 5-HT enhanced the expression of multiple PP2C genes involved in the hormone signaling pathway. b Y2H assays confirming the interaction between PP2C30/49/51 and SAPK2. “LT” indicates SD/−Leu/−Trp medium, and “LTHA” indicates SD/−Leu/−Trp/−His/−Ade medium. c SLC assays demonstrating the interaction of PP2C30/49/51 with SAPK2 in Nicotiana benthamiana leaves. Images were taken three days after transient transformation. d BiFC assays revealing the interaction between PP2C30/49/51 and SAPK2 in the nucleus. Yellow fluorescence indicates the interaction signal. Scale bar = 20 μm. e Co-IP experiments showing that PP2C30/49/51 interacted with SAPK2. Wild-type rice protoplasts transiently expressing PP2Cs-Flag and ABI5-GFP were incubated for 18 h, followed by GFP pull-down and detection using an anti-Flag antibody. f In vitro pull-down assays confirming the interaction between PP2C30/49/51 and SAPK2. GST and GST-PP2Cs proteins were incubated with SAPK2-His, followed by pull-down using glutathione resin and detection with anti-GST and anti-His antibodies. “PD” denotes pull-down. g–h In vitro kinase (g) and dephosphorylation (h) assays showed that PP2C30/49/51 dephosphorylated SAPK2. In (g), CBB staining showed co-induced PP2Cs-GST or GST with SAPK2-His proteins, and Phos-tag gels were used to detect phosphorylated proteins. In (h), α-Ser/Thr immunoblotting was used to detect phosphorylated proteins. Experiments in (b–h) were repeated three times, with similar results. Source data are provided as the Source Data file.
We further validated the interactions between SAPK2 and PP2C30/49/51 in tobacco using split luciferase complementation (SLC) and bimolecular fluorescence complementation (BiFC) assays. Co-infiltration of PP2C-Cluc and SAPK2-Nluc constructs into tobacco leaves generated strong luminescence signals (Fig. 4c). Similarly, robust yellow fluorescence signals were observed in the nuclei of tobacco cells co-expressing PP2C-cYFP and SAPK2-nYFP constructs (Fig. 4d). Additionally, co-immunoprecipitation (Co-IP) and in vitro pull-down assays further confirmed their interactions both in vivo and in vitro (Fig. 4e, f). Collectively, these results provided consistent evidence for the interactions between PP2C30/49/51 and SAPK2.
Previous study has shown that ABA promotes the formation and stability of the PYL5-PP2C30/49/51 complex by binding to its receptor PYL5, resulting in the release of SAPK2 and activation of downstream signaling pathways33. To elucidate the regulatory role of 5-HT in ABA-mediated protein–protein interactions, we examined the interaction between PYL5 and PP2C30/49/51 under various treatment conditions using rice protoplasts and a tobacco transient expression system. Treatment with 10 μM ABA significantly enhanced the interaction between PYL5 and PP2C30/49/51. Co-treatment with 10 μM 5-HT and ABA markedly attenuated this interaction compared to ABA treatment alone, although the interaction remained stronger than in the untreated control. In contrast, treatment with 10 μM 5-HT alone had no significant effect on these interactions (Supplementary Fig. 12). Together, these results suggest that 5-HT may antagonize ABA signaling by interfering with ABA-induced assembly of the PYL5–PP2C complex.
PP2Cs-SAPK2 mediates ABI5 phosphorylation
SAPK2, predicted to function as a serine/threonine kinase, demonstrated strong autophosphorylation activity, as confirmed by immunoblot analysis using anti-phospho-serine/threonine (anti-Ser/Thr) antibodies (Supplementary Fig. 13a). The role of PP2C30/49/51, which specifically target serine/threonine residues, in the regulation of SAPK2 were subsequently explored. Co-expression of SAPK2 and PP2Cs in E. coli significantly reduced SAPK2 phosphorylation levels (Fig. 4g). Consistent results were observed in vitro dephosphorylation assays, confirming that PP2C30/49/51 negatively regulate SAPK2 autophosphorylation (Fig. 4h). Given the critical role of phosphorylation and dephosphorylation in SnRK2 activation, which is central to ABA signaling34. We propose that 5-HT may modulate ABA signaling by antagonizing the ABA-induced formation of the PYL5–PP2C complex, thereby influencing the phosphorylation status of SAPK2.
Previous studies have demonstrated that SAPK2 participates in ABA signaling by phosphorylating downstream bZIP transcription factors, such as bZIP23 and ABI5 (also known as bZIP10)32,35. However, the precise molecular regulatory mechanisms underlying SAPK2-mediated regulation remain unclear. Given the essential role of ABI5 in ABA-induced 5-HT biosynthesis (Fig. 2), we focused on elucidating the potential regulatory relationship between SAPK2 and ABI5. To this end, we utilized Y2H, Co-IP, in vitro pull-down, SLC, and BiFC assays to examine the interaction between SAPK2 and ABI5 (Supplementary Fig. 13b-f). Consistent results from these assays confirmed the interaction between SAPK2 and ABI5.
To explore the kinase-substrate relationship between SAPK2 and ABI5, we conducted kinase activity assays. ABI5 expressed alone in E. coli showed no phosphorylation, while co-expression with SAPK2 significantly increased ABI5 phosphorylation, confirming SAPK2 as the kinase (Fig. 5a). In vitro kinase assays showed that recombinant ABI5 and SAPK2 proteins resulted in substantial phosphorylation of ABI5 (Fig. 5a), further validating SAPK2 as the kinase responsible for ABI5 phosphorylation. Next, we further assessed the effect of PP2Cs on SAPK2-mediated ABI5 phosphorylation. Co-induction of SAPK2 and ABI5 in E. coli resulted in ABI5 phosphorylation. However, when PP2Cs were co-expressed with SAPK2 and ABI5, no significant ABI5 phosphorylation was detected, indicating that PP2Cs inhibit SAPK2-driven ABI5 phosphorylation (Fig. 5b). Previous study has shown that PP2C51 directly interacts with ABI5 to dephosphorylate it32. We then explored whether PP2C30 and PP2C49 also interact directly with ABI5 using Y2H and SLC assays. The results revealed no direct interaction between PP2C30/PP2C49 and ABI5 (Supplementary Fig. 14), suggesting that these PP2Cs may regulate ABI5 phosphorylation indirectly via SAPK2. Together, these results support that 5-HT modulates ABA signaling through the possible PP2Cs–SAPK2–ABI5 interaction, potentially by regulating the phosphorylation status of ABI5.
a Verification of SAPK2-mediated phosphorylation of ABI5 using bacterial kinase assays (left) and in vitro phosphorylation assays (right). b Inhibition of SAPK2-mediated phosphorylation of ABI5 by PP2C30/49/51. CBB staining confirmed the presence of co-expressed PP2Cs-GST and SAPK2-His proteins alongside ABI5-GST. c Partial peptide map of SAPK2 autophosphorylation sites identified by LC-MS/MS, showing phosphorylation at T159, T162, and T283. Additional mass data for other sites were shown in Supplementary Fig. 15. d Validation of SAPK2 kinase activity at ten identified autophosphorylation sites. Mutants m1–m5 correspond to S12A, S109A/Y113A/S116A, T159A/T162A, T212A/T214A/S218, and T283A, respectively. e Effects of mutations at distinct SAPK2 autophosphorylation sites on its ability to phosphorylate ABI5 among these, T162/T159 and T283 were critical for ABI5 phosphorylation, alanine substitution at these residues abolished SAPK2-mediated phosphorylation of ABI5. f Analysis of the impact of T159 and T162 mutations on SAPK2 autophosphorylation, demonstrating that T162, but not T159, was essential for SAPK2 autophosphorylation. g–i Effects of SAPK2 autophosphorylation site mutations on its interaction with ABI5. Co-IP (g), SLC(h), and in vitro pull-down (i) assays showed that mutations at T283 disrupted the SAPK2-ABI5 interaction, while mutations at other sites did not significantly alter interaction strength. Calf intestinal alkaline phosphatase (CIAP) was used as a phosphatase control in (a, d). Phos-tag gels were used to detect phosphorylated proteins in (a, b, d, e), and α-Ser/Thr immunoblotting was performed in (a, d–f) to assess phosphorylation status. In (i), “PD” indicates pull-down assay. Experiments shown in (a, b, d, e, f, g, h, and i) were repeated three times, yielding similar results. Source data are provided as the Source Data file.
SAPK2 T162/T283 determines ABI5 phosphorylation
Given SAPK2’s strong autophosphorylation activity, we employed LC-MS/MS to identify its autophosphorylation sites. Ten phosphorylation sites were detected, with their optimal peptide sequences and spectrum shown in Fig. 5c (Supplementary Fig. 15 and Supplementary Data 1). To explore the functional relevance of these sites, in vitro kinase assays were conducted using full-length or variant SAPK2 proteins, where specific phosphorylation sites were replaced with non-phosphorylated alanine (Fig. 5d–f). The SAPK2T159A/T162A mutant completely lost autophosphorylation activity (Fig. 5d), underscoring the important role of phosphorylation at these sites for SAPK2 function. Further analysis revealed that the T162A mutation alone was sufficient to inhibit SAPK2 phosphorylation, whereas the T159A mutation had no such effect (Fig. 5f). These results suggest that the T162 site plays an important role in regulating SAPK2’s autophosphorylation activity.
To evaluate the impact of these mutations on ABI5 phosphorylation, we co-expressed the phospho-dead SAPK2 variant with ABI5. The results showed that the SAPK2T162A mutant, which lost autophosphorylation activity, failed to phosphorylate ABI5, confirming the essential role of T162 phosphorylation in both SAPK2 kinase activity and ABI5 phosphorylation (Fig. 5e). Although the SAPK2T283A mutant retained autophosphorylation, its phosphorylation activity toward ABI5 was significantly reduced (Fig. 5e), suggesting that this site is essential for SAPK2-mediated ABI5 phosphorylation. Further analyses demonstrated that the T283A mutation in SAPK2 markedly weakened its interaction with ABI5 compared to other mutants, as consistently shown by SLC, Co-IP, and in vitro pull-down assays (Fig. 5g–i). These findings emphasized the essential role of T283 phosphorylation in mediating the interaction between SAPK2 and ABI5, as well as subsequent phosphorylation. In conclusion, our results underscore the important roles of T162 and T283 phosphorylation in regulating ABI5 phosphorylation by SAPK2.
5-HT inhibits ABA-induced ABI5 phosphorylation
Given that 5-HT may antagonize ABA-induced formation of the PYL5-PP2C complex and thereby influence SAPK2-mediated phosphorylation of ABI5 (Supplementary Fig. 12), we examined the impact of 5-HT on ABA-induced ABI5 phosphorylation. Phosphorylation levels of ABI5 in rice seedlings under different treatments were assessed using anti-ABI5 antibodies. Both exogenous 5-HT application and transgenic plants with elevated endogenous 5-HT levels significantly reduced ABA-induced ABI5 phosphorylation (Fig. 6a, b). These findings indicate that 5-HT partially attenuates ABA-induced ABI5 phosphorylation, supporting the hypothesis that 5-HT influences ABA-driven ABI5 phosphorylation, at least in part, through the possible PP2Cs–SAPK2 interaction.
a Measurement of ABI5 phosphorylation levels in rice seedlings treated with ABA alone or in combination with 5-HT (ABA + 5-HT) for 12 h. b Comparison of ABI5 phosphorylation levels between Ubi::T5H transgenic plants and wild-type (WT) following 12 h of ABA treatment. c, d Differential analysis of ABI5 transcript (c) and protein levels (d) in response to treatments with ABA alone and ABA combined with 5-HT. e Evaluation of the impact of phosphorylation on ABI5 protein stability, where ABI5p denotes the phosphorylated ABI5-GST fusion protein co-expressed with SAPK2-His, and ABI5 represents the non-phosphorylated protein expressed independently. f Stability analysis of ABI5 protein in total protein extracts from rice seedlings 12 h post-ABA treatment, compared to untreated controls. In (c), data are presented as means ± SD (n = 3 biological replicates). The experiments depicted in (a, b, d, e, and f) were repeated three times, with consistent results obtained. Source data are provided as the Source Data file.
While 5-HT application did not significantly alter ABI5 transcription levels in response to ABA, it markedly reduced ABI5 protein levels (Fig. 6c, d), suggesting that ABI5 phosphorylation may influence its protein stability. In vitro cell-free degradation assays showed that SAPK2-phosphorylated GST-ABI5 degraded more slowly than the unphosphorylated form in total protein extracts from wild-type rice seedlings (Fig. 6e). Furthermore, after 24 h of ABA treatment, ABI5 protein stability was significantly higher in total protein extracts compared to untreated controls (Fig. 6f). These findings suggest that 5-HT modulates ABA-induced ABI5 phosphorylation, thereby affecting ABI5 protein accumulation.
Engineering 5-HT/ MT-enriched rice without compromising yield
In this study, we systematically investigated the interactions between ABA and the 5-HT metabolic pathway in rice. Our aim was to elucidate regulatory mechanisms for 5-HT synthesis and to enhance the accumulation of 5-HT and its derivative melatonin in rice, while maintaining stable yield. We show that 5-HT biosynthesis could be affected by ABA-mediated activation of ABI5, which could consequently induced the upregulation of TDC1, TDC3, and T5H transcription, thereby promoting 5-HT/MT synthesis. Additionally, our results suggested that feedback regulation of 5-HT biosynthesis may involve ABA signaling. Based on these insights, we proposed two engineering strategies to increase 5-HT/MT levels in rice grains without compromising yield (Fig. 7a, i). The first strategy aimed to increase endogenous ABA concentrations by targeting genes involved in ABA metabolism, thereby boosting 5-HT/MT synthesis. The second strategy targeted the core ABA signaling to attenuate the negative feedback regulation mediated by 5-HT.
a Phenotypes at maturity and grain yield per plant for wild-type (WT) and the 12 transgenic rice lines generated in this study. Lines L1–L12 correspond to Ubi::NCED1, Ubi::NCED2, Ubi::NCED3, Ubi::NCED4, Ubi::NCED5, CR-aba8ox1, CR-aba8ox2, CR-aba8ox3, Ubi::SAPK2, Ubi::ABI5, CR-pp2c51, and Ubi::ABI5/CR-pp2c51, respectively. Scale bar = 10 cm. b–e Statistical analysis of agronomic traits, including tiller number (b), 1000-grain weight (c), number of filled grains per panicle (d), and grain yield per plant (e) for WT and transgenic plants. Most transgenic lines did not exhibit significant changes in individual plant yield, while overexpression of SAPK2 resulted in a slight decrease in yield. Data are presented as means ± SD (n = 3 biological replicates for (c) n = 10 plants for (d) and n = 15 plants for (b, e). f–h Phenotypic appearance of cooked rice grains from WT and the 12 transgenic lines (f), along with quantification of 5-HT (g) and MT (h) contents in cooked grains. No significant differences in the appearance of cooked rice were observed between transgenic and WT lines. For g scale bar = 1 cm. For (g) and (h), values represent means ± SD (n = 3 biological replicates). For b–h statistical analysis was performed using a two-tailed unpaired Student’s t-test. i Schematic model illustrating the enhancement of 5-HT/MT levels in rice while maintaining yield, based on the crosstalk between ABA signaling and 5-HT metabolism. ABA promotes phosphorylation of ABI5, thereby activating its transcriptional regulation of the rate-limiting genes TDC1/3 and T5H in the 5-HT biosynthetic pathway, resulting in increased synthesis of 5-HT and its downstream product MT. In turn, 5-HT antagonizes ABA-induced formation of the PYL5–PP2C complex, influencing the phosphorylation status of SAPK2, which reduces ABI5 phosphorylation and ABA signaling, providing feedback inhibition of ABA-induced 5-HT biosynthesis. By engineering genes involved in the ABA metabolic pathway, a sustainable strategy was developed to generate rice with elevated 5-HT/MT content without compromising yield. Source data are provided as the Source Data file.
To evaluate the efficacy of two strategies for enhancing specific traits in rice, we generated transgenic lines constitutively overexpressing target genes under the Ubi promoter, including members of the NCED gene family involved in ABA biosynthesis, as well as ABI5 and SAPK2 genes associated with the 5-HT feedback regulatory pathway (Supplementary Fig. 16). We also employed CRISPR/Cas9 technology to knock out relevant genes involved in ABA catabolism (CR-aba8oxs) and generated a mutant of PP2C51 (CR-PP2C51), a regulator of ABI5 phosphorylation (Supplementary Fig. 17). Double transgenic lines were further developed by combining the CR-PP2C51 mutant allele with the Ubi::ABI5 overexpression cassette (CR-PP2C51/Ubi::ABI5) (Supplementary Fig. 16). Analysis of ABA content in the grains of these ABA metabolism-related transgenic lines revealed that most exhibited significantly higher ABA levels compared to the control, confirming an enhancement of endogenous ABA content in rice grains (Supplementary Fig. 18).
As a key endogenous hormone, ABA plays a central regulatory role in plant growth and development. Through systematic analyses of endogenous ABA levels and phenotypic traits in transgenic rice lines with enhanced ABA signaling, we evaluated the multifaceted effects of ABA modulation on agronomic performance. Seed germination assays demonstrated that transgenic lines with elevated ABA content or enhanced ABA signaling exhibited significantly reduced germination rates compared to wild-type controls (Supplementary Fig. 19a). Comprehensive evaluation of yield components revealed no statistically significant differences in major agronomic traits—including thousand-grain weight, number of filled grains per plant, tiller number, plant height, and yield per plant—for most transgenic lines (Fig. 7b–e and Supplementary Fig. 19b). The NCED3 overexpression line demonstrated a significant increase in yield. Overall, our phenotypic analyses indicate that modulation of ABA content or signaling to elevate rice 5-HT/MT levels markedly suppress seed germination, without adverse effects on vegetative or reproductive yield components.
Given that rice is a global staple food typically consumed after cooking, we further evaluated the impact of two genetic strategies on the levels of 5-HT/MT in cooked transgenic rice grains. Quantitative analysis showed that 75 % (8/12) of the independent transgenic lines exhibited a significant increase in 5-HT content. Among these eight lines with elevated 5-HT levels, 75 % (6/8) also exhibited a concomitant increase in MT accumulation (Fig. 7f–h). These findings demonstrated the effectiveness of both strategies in enhancing the biosynthesis of target compounds in cooked rice grains. Comparison of 5-HT and MT levels in wild-type grains before and after cooking revealed a significant reduction due to cooking process (Supplementary Fig. 20), implying that transgenic grains may contain even higher amounts of these compounds prior to cooking. Overall, this study presents an innovative molecular design strategy for enhancing functional nutrient content in staple crops without compromising yield stability.
Discussion
Inadequate nutrient intake remains a significant challenge to global food security36. Enhancing staple crops with essential micronutrients offers a cost-effective and sustainable approach to address nutritional deficiencies and promote human health37. In this study, we exploited the complex interplay between ABA and 5-HT signaling pathways to develop nutrient-enriched rice lines. These transgenic lines exhibited significantly elevated levels of target micronutrients while maintaining stable agronomic performance (Fig. 7). Our results establish a scientific foundation for the development of nutritionally enhanced functional foods.
5-HT is widely distributed in both animals and plants, with its synthesis and regulation have been extensively explored in animals38,39. However, the pathways and mechanisms governing its synthesis in plants, particularly in rice, remain less well-defined40. In plants, 5-HT performs diverse regulatory functions and interacts with phytohormones such as ABA, SA, and indole-3-acetic acid (IAA). Recent studies suggest that 5-HT contributes to rice salt tolerance (through interaction with ABA), disease resistance (by antagonizing SA), and root development (via crosstalk with IAA), primarily by modulating synergistic or antagonistic hormone interactions41,42,43. This study investigated the molecular interplay between 5-HT and the ABA signaling pathway, uncovering a dynamic regulatory balance in hormone metabolism (Fig. 7i). Our results demonstrated that ABA significantly promoted the biosynthesis of 5-HT and its derivative MT in rice (Fig. 1a–e). This induction largely occurred through the transcriptional activation of these key biosynthetic enzyme genes by the transcription factor ABI5 (Figs. 1f, g, 2). Moreover, we found that 5-HT can feedback-regulate its own synthesis via the ABA signaling pathway (Fig. 3 and Supplementary Fig. 8a). Specifically, 5-HT interfered with the phosphorylation cascade within ABA signaling pathway by attenuating ABA-induced formation of the PYL5–PP2Cs complex, thereby inhibiting SAPK2-mediated phosphorylation of ABI5 (Figs. 4, 5 and Supplementary Fig. 12). This modulation regulated the transcriptional activity and protein stability of ABI5 (Fig. 6), enabling effective control of 5-HT biosynthetic gene expression. This discovery not only enhanced our understanding of the intricate interplay between plant hormones and metabolic pathways but also identified a dynamic regulatory mechanism for maintaining hormone-metabolite equilibrium, presenting opportunities for developing rice varieties enriched with 5-HT/MT. Future studies will explore whether this dynamic regulatory mechanism is applicable to other beneficial metabolites.
Yield and nutritional enhancement are principal goals in crop improvement44. This study introduced a strategy that utilized the interactions between plant hormones and nutrients to develop rice varieties with improved nutrient profiles without compromising yield. This approach specifically targeted molecular regulation of genes involved in ABA metabolism to elevate endogenous ABA levels (Supplementary Fig. 16–18), enhancing 5-HT synthesis. Alternatively, it modulated genes within the 5-HT feedback pathway to mitigate its inhibitory effects, thereby increasing the levels of 5-HT/MT in rice. Among the twelve rice varieties developed, six exhibited significant increases in 5-HT/MT content without yield reduction (Fig. 7 and Supplementary Fig. 19), underscoring the pivotal role of plant hormones in augmenting relevant nutritional metabolites. Future research will further explore transgenic rice lines enriched with 5-HT/MT and utilize molecular-assisted breeding techniques to identify and select haplotypes with optimal bioavailability. This advancement aims to enhance the nutritional value of rice, thereby contributing to improved dietary quality and promoting human health.
Methods
Plant materials and growth conditions
The transgenic rice lines used in this study were developed in the Wuyunjing7 (W7, Oryza sativa ssp. Japonica) background. The generated transgenic lines included pUbi::OsNAED1-5, pUbi::OsABI5, pUbi::OsSAPK2, pUbi::OsT5H, CR-OsABA8ox1-3, CR-ABI5, CR-OsPP2C51, and the combined line pUbi::OsABI5/CR-OsPP2C51. Rice plants were cultivated under natural field conditions in Hangzhou (Zhejiang, China) during the summer and in Lingshui (Hainan, China) during the winter. Hormonal treatments were applied to seedlings in a greenhouse maintained at 26 °C with a 14-h light/10-h dark photoperiod. Seed germination and detached leaf assays were conducted at 28 °C under continuous darkness. Nicotiana benthamiana plants, used for transient expression and protein interaction studies, were cultivated in a long-day greenhouse (16 hours light/8 hours dark) at 24 °C.
CRISPR-Cas9 constructs targeting OsABI5, OsABA8ox1-3, and OsPP2C51 were synthesized and cloned into plasmids45. For overexpression, the coding sequences of OsNAED1-5, OsABI5, OsSAPK2, and OsT5H were inserted into the pCAMBIA1300-Ubi-GFP vector, driven by the maize UBIQUITIN1 promoter. Both CRISPR-Cas9 and overexpression plasmids were introduced into W7 rice variety via Agrobacterium tumefaciens strain EHA105-mediated transformation. For each construct, more than 10 independent transgenic lines were generated and validated by PCR sequencing or western blotting. Primer details are provided in Supplementary Data 2.
Measurement of physiological and agronomic traits
Chlorophyll content was measured using a modified protocol based on Ouyang et al.46. Briefly, fresh samples were randomly selected, cut into 1–2 cm segments, weighed, and soaked in 95 % (v/v) ethanol. The samples were incubated at 28 °C in darkness for 24 h. Chlorophyll levels were quantified by measuring absorbance at 663 nm and 645 nm. Germination rates were evaluated by placing 40–50 uniformly mature wild-type or mutant rice seeds on moist filter paper, with or without the addition of ABA or 5-HT. The seeds were incubated in the dark at 28 °C for one week, and germination was monitored at 12-h intervals. Yield-related traits—including tiller number, plant height, grain yield per plant, and number of filled grains per panicle—were measured from rice plants exhibiting uniform growth and consistent vigor at maturity, with all assessments performed using at least ten biological replicates. Thousand-grain weight were assessed using a seed analyzer (Wansheng, Hangzhou, China), with each sample measured in triplicate.
Quantification of 5-HT, related metabolites, and ABA
The quantification of tryptophan, tryptamine, 5-HT, and MT in rice seedlings, as well as 5-HT and MT in cooked rice and ABA in brown rice, were conducted using liquid chromatography-tandem mass spectrometry (LC-MS/MS) at the Wuhan ProNets Testing Technology Co., Ltd. Samples were immediately homogenized in liquid nitrogen and extracted with 1 mL of methanol: water: formic acid (80:20:1, v/v). Following vortexing for 30 seconds, the samples were subjected to ultrasonic extraction in an ice-water bath for 20 minutes, then centrifuged at 3000 g for 10 minutes at 4 °C, and the supernatants were filtered through 0.22 μm membranes. All steps were performed under light-protected conditions. The filtrates were then analyzed by LC-MS/MS. Chromatographic separation was carried out on an Agilent Poroshell 120 SB-C18 column (30 °C) with a 5 μL injection volume and a flow rate of 0.4 mL/min. The mobile phase comprised 0.1 % formic acid in water (solvent A) and acetonitrile (solvent B) with gradient elution. Mass spectrometric detection was conducted in positive ion mode using multiple reaction monitoring (MRM + ). The instrument settings were as follows: curtain gas, 35 psi; spray voltage, +5500 V; nebulizer gas, 60 psi; auxiliary gas, 60 psi; and ion source temperature, 500 °C. Analyses were performed using a Shimadzu LC-20AD HPLC system coupled to an AB QTRAP 5500 mass spectrometer. All procedures followed the standard operating protocol of Wuhan Pure Testing Technology Co., Ltd. (Hubei, China). The determination of ABA content in brown rice was mainly based on Luo et al.47. Briefly, samples were ground in liquid nitrogen, extracted with acetonitrile, purified with C18 filler, dissolved in methanol, and subsequently analyzed by LC-MS. All measurements were conducted in triplicate to ensure data reproducibility.
RNA isolation, RT-qPCR and RNA-seq analysis
Total RNA was extracted from fresh tissues using the Axygen RNA Extraction Kit (Axygen, USA) following the manufacturer’s instructions. For RT-qPCR analysis, the isolated RNA was reverse transcribed to cDNA using the Monad mRNA Reverse Transcription Kit (Monad, Jiangsu, China). Quantitative real-time PCR (RT-qPCR) was performed with SYBR qPCR-Mix (Vazyme, Jiangsu, China) using gene-specific primers. Gene expression levels were normalized to the internal control gene UBQ5. All experiment were carried out with triplicate independent biological replicates. Primer details are listed in the Supplementary Data 2.
For RNA sequencing analysis, rice seedlings were treated for 24 h with either ABA alone or ABA combined with 5-HT for 24 h. RNA samples were collected from three biological replicates per treatment. RNA extraction and Illumina sequencing were performed by Novogene Bioinformatics Technology Co., Ltd. (Beijing, China). RNA integrity was assessed using the RNA Nano 6000 Assay Kit and the Bioanalyzer 2100 system (Agilent Technologies, USA). RNA libraries were constructed, quantified with a Qubit 2.0 fluorometer, and insert sizes verified with the Agilent 2100 Bioanalyzer. Libraries meeting quality standards were pooled and sequenced on the Illumina platform to generate 150 bp paired-end reads. Reference genome indexing and alignment were performed using HISAT2 v2.0.5. Gene expression levels were quantified with FeatureCounts (v1.5.0-p3) and normalized as fragments per kilobase of transcript per million mapped reads (FPKM). Differential gene expression analysis was conducted using DESeq2 (v1.20.0), with P values adjusted by the Benjamini-Hochberg method. Genes with an adjusted P value < 0.05 were considered differentially expressed.
Yeast two-hybrid and yeast one-hybrid assays
To conduct yeast two-hybrid (Y2H) screening, the full-length CDS of the relevant genes were cloned into the bait vector pGBKT7 and the prey vector pGADT7, respectively. These resulting constructs were co-transformed into yeast strain Y2H-Gold to evaluate protein-protein interactions. Post-transformation, yeast cultures were grown on SD/-Trp/-Leu selective medium and incubated at 30 °C for 48 h. Colonies were then trnsferred to SD/-Trp/-Leu/-His/-Ade selective medium and incubated for an additional 72 h to enable imaging and documentation of interaction events. The co-transformation of pGBKT7-53 and pGADT7-T served as the positive control.
For the yeast one-hybrid (Y1H) assays, approximately 2-kb promoter regions of the target genes were amplified and cloned into the pLacZi2μ reporter vector to generate a bait reporter construct. The pb42AD transcription factor library, constructed in our laboratory, were used as the prey. The bait and prey constructs were co-transformed into Y1H-Gold yeast competent cells, which were subsequently cultured on SD-Ura solid medium at 28 °C for 2–3 days. Colonies were then replica-plated onto SD-Ura medium supplemented with raffinose and X-gal (SD-Ura/Raf/X-gal) and incubated at 28 °C for an additional 2–3 days. Colony growth and blue color development were monitored and documented by photography. The primers used for cloning are listed in Supplementary Data 2.
BiFC and SLC assays
For BiFC assays, the full-length CDS of the relevant genes were cloned into the BiFC vectors pSPYCE and pSPYNE, respectively. The resulting constructs were transformed into Agrobacterium tumefaciens strain GV3101 and cultured under appropriate conditions. Bacterial cultures were harvested, concentrated, and resuspended in infiltration buffer containing 10 mM MgCl2, 10 mM MES (pH 5.6), and 200 μM acetosyringone. After incubation for 2–3 h at room temperature, the bacterial suspensions were infiltrated into the leaves of 4-week-old Nicotiana benthamiana plants. Yellow fluorescent protein (YFP) fluorescence was detected 48–72 h post-infiltration using a Zeiss LSM 700 confocal laser scanning microscope (Carl Zeiss, Jena, Germany).
For SLC assays, the full-length CDS of related genes were separately cloned into luciferase vectors pCambia-35S-cLuc and pCambia-35S-nLuc. Utilizing the same infiltration approach, the treated leaves were incubated with luciferase substrate (Promega) and imaged for luciferase activity using a Tanon-5200 system (Tanon). All assays were performed with at least three biological replicates using different batches of tobacco. Cloning primer details are provided in Supplementary Data 2.
In vitro pull-down assays
The coding sequences of PP2C30/49/51 and ABI5 were cloned into the pGEX-4T-1 vector to generate GST-tagged fusion proteins. Wild-type and mutant forms of SAPK2 were cloned into the pET28a vector for the synthesis of His-tagged fusion proteins. Protein expression and purification followed the modified protocols of Liu et al.48. In brief, protein expression was induced overnight at 18 °C with 0.3 mM isopropyl β-D-1-thiogalactopyranoside (Sangon, China), and recombinant proteins were purified using prepacked glutathione agarose (Thermo Scientific, USA) and Ni-NTA agarose columns (Thermo Scientific, USA). For binding assays, the purified proteins were incubated with glutathione high-capacity magnetic agarose beads in binding buffer (50 mM Tris-HCl, 150 mM KCl, 5 % glycerol, 1 mM DTT, 1 mM EDTA, pH 8.0, 0.01 % Nonidet P-40, 1 mM PMSF) at 4 °C for 3 h. The beads were then washed 3-5 times with the same buffer, and bound proteins were eluted by boiling in SDS sample buffer for 10 minutes. Protein samples were subsequently analyzed by SDS-PAGE and western blot using anti-His and anti-GST antibodies (Abcam). Cloning primer details are provided in Supplementary Data 2.
Co-IP assays
To investigate protein interactions in planta, Co-immunoprecipitation (Co-IP) assays were conducted. The wild-type or point-mutant SAPK2 gene and the PYL5 gene were cloned into the pCambia1300-35S-3×Flag vector, while cDNAs encoding PP2C30, PP2C49, PP2C51, and ABI5 were cloned into either the pCambia1300-35S-eGFP or pCambia1300-35S-MYC vector to generate SAPK2-3Flag, PP2Cs-GFP/MYC, and ABI5-GFP recombinant plasmids. As controls, GFP and PP2C-3×Flag were co-expressed. These plasmids were transfected into rice protoplasts and incubated for 24 h. Total proteins were extracted using a non-denaturing buffer containing 50 mM Tris-MES (pH 8.0), 1 mM MgCl2, 0.5 M sucrose, 10 mM EDTA (pH 8.0), 5 mM DTT, and 1 mM PMSF. The supernatant was incubated with anti-FLAG magnetic beads (Abcam) at 4 °C for 2 h, followed by 3-5 washes with extraction buffer at 4 °C. The bound proteins were eluted with SDS sample buffer after heating for 10 minutes. Proteins were separated by SDS-PAGE and analyzed by immunoblotting with anti-FLAG, anti-MYC, and anti-GFP antibodies (Abcam). Primer sequences are listed in Supplementary Data 2.
Cell-free degradation assays
To evaluate the degradation rates of phosphorylated and non-phosphorylated ABI5 in total plant proteins, an in vitro protein degradation assay was conducted with modifications to the method of Zhai et al.49. Briefly, total proteins were extracted from rice seedling leaves using a non-denaturing extraction buffer (50 mM Tris-MES, pH 8.0, 1 mM MgCl2, 0.5 M sucrose, 10 mM EDTA, pH 8.0, 5 mM DTT, 1 mM PMSF). Purified ABI5-GST and ABI5-GST co-expressed with SAPK2-His were incubated with total protein extracts at 30 °C. Samples were collected at specified time points for western blot analysis. For ABA-treated samples, total proteins were extracted from ABA-treated rice seedlings using the same buffer, and ABI5 stability was assessed by immunoblotting with anti-ABI5 antibody, comparing ABA-treated and untreated controls.
Protein phosphorylation/dephosphorylation assays
To examine the impact of phosphorylation sites on SAPK2-mediated phosphorylation of ABI5, ABI5-GST was co-expressed with either wild-type or mutant SAPK2-His in E. coli BL21 cells. Immunoblotting was performed using biotinylated Phos-tag™ zinc complex BTL111 (Wako, Osaka, Japan) and anti-ABI5 antibody (ABclonal)50. For in vitro phosphorylation assays, a modified version of the method by Yu et al.51 was employed. Briefly, purified SAPK2-His and ABI5-GST proteins were incubated at 30 °C for 30 minutes in kinase reaction buffer containing 500 mM HEPES-KOH (pH 7.5), 150 mM NaCl, 100 mM MgCl2, 1 mM EGTA, 1 mM ATP, 10 mM DTT. Proteins were separated by SDS-PAGE and analyzed via immunoblotting with anti-pSer/Thr antibody (Abcam).
To assess the effect of PP2C30/49/51 on SAPK2-mediated ABI5 phosphorylation, PP2Cs-GST proteins were introduced into the co-expression system. Phosphorylation levels of ABI5 and SAPK2 in the induced supernatants were analyzed using anti-Ser/Thr antibody or Phos-tag™ gel. For in vivo phosphorylation, total protein extracted from rice seedlings treated with ABA or ABA combined with 5-HT48. Phos-tag™ SDS-PAGE and anti-ABI5 antibody were used to assess ABI5 phosphorylation. Primers for SAPK2 point mutations are provided in Supplementary Data 2.
Promoter and transcriptional activity analysis
To construct the effector, the full-length CDS of ABI5 was cloned into the pGreenII 0800-62SK vector. Concurrently, ~2-kb promoter regions of T5H, TDC1, and TDC3 were inserted into the pGreenII 0800-Luc vector to serve as reporter genes. According to the protocol by Wang et al.52, rice protoplasts were co-transfected with 10 μg each of the LUC reporter constructs and the effector plasmid, followed by incubation for 16-24 h. LUC activity was quantified using the Promega Dual-Luciferase Reporter Assay System. Relative LUC activity was calculated as the LUC/REN ratio, with the LUC/REN ratio from the control vector pGreenII 0800-62SK used as the negative control.
For transcriptional activity analysis, the complete CDS of ABI5, ABI5D44, and ABI5118D were individually cloned into the GAL4-BD vector. The VP16-GAL4-BD served as the positive control, and the empty GAL4-BD vector served as the negative control. Protoplasts were co-transfected with these constructs along with LUC and pTRL vectors, employing the same analysis protocol as above. All experiments were conducted in at least three independent replicates. Detailed primer information for cloning is provided in Supplementary Data 2.
LC-MS/MS analysis
To identify the autophosphorylation sites of SAPK2, purified SAPK2-His protein was subjected to mass spectrometry analysis at Hangzhou Jingjie PTM Biolab Co., Ltd. (Hangzhou, China). Sample preparation included an initial trypsin digestion (Promega) at 20 ng/μL at room temperature overnight. Subsequent reduction was performed using dithiothreitol (DTT, Sigma) at 37 °C for 60 minutes, followed by alkylation with iodoacetamide in the dark for 45 minutes. Additional trypsin was added to achieve a final concentration of 10 ng/μL, and digestion continued for 4 more hours. The resulting peptides were dissolved in liquid chromatography mobile phase A and separated using an EASY-nLC 1200 ultra-high-performance liquid chromatography (UHPLC) system. Mobile phase A consisted of 0.1% formic acid and 2% acetonitrile in water, while mobile phase B consisted of 0.1% formic acid and 90% acetonitrile in water. Following UHPLC separation, peptides were ionized via a nano-electrospray ionization (NSI) source and analyzed using an Orbitrap Exploris 480 mass spectrometer. The MS/MS data were processed with Proteome Discoverer 2.4.
EMSA assays
Electrophoretic mobility shift assays (EMSA) were performed with minor modifications to the method described by Li et al.53. Briefly, purified ABI5-GST protein was incubated with biotin-labeled DNA probes in binding buffer at room temperature for 20 minutes. The reaction mixtures were then subjected to electrophoresis on a 4% polyacrylamide gel at 100 V for 120 minutes, with continuous cooling on ice. Following electrophoresis, the separated DNA-protein complexes were transferred to a nylon membrane (Millipore, USA). Detection of the biotin-labeled probes were carried out using a chemiluminescent EMSA Kit (GS009, Beyotime, Shanghai, China), according to the manufacturer’s instructions. Promoter probes for T5H, TDC1, and TDC3 were synthesized using the EMSA Biotin Labeling Probe Kit (GS008, Beyotime, Shanghai, China), with unlabeled probes used as competitors. The primers used for probe synthesis are listed in Supplementary Data 2.
CUT&Tag assays
To identify the DNA binding sites of ABI5, CUT&Tag analysis was performed using the NGS G-Type In-Situ DNA Binding Profiling Library Prep Kit for Illumina, following the manufacturer’s instructions. Briefly, freshly isolated protoplasts were collected and centrifuged at 600 × g for 5 minutes at 4 °C to pellet the cells. The purified protoplasts were then lysed in cold Nuclei Isolation Buffer (NIB) for several minutes, and the resulting nuclei were resuspended in 100 μL of Wash Buffer. After nuclei isolation, samples were incubated overnight at 4 °C with gentle shaking in the presence of 1 μL anti-GFP antibody or IgG control. The next day, the nuclei were washed three times with Wash Buffer to remove unbound antibodies. Subsequently, 1 μL of p/A/G-transposome mix and 50 μL of transposase incubation buffer were added to each sample, followed by incubation at room temperature for 1 h with gentle shaking. After transposase treatment, nuclei were washed three times to remove excess transposase. DNA fragments were captured using 40 μL of DNA selection beads, incubated for 2 minutes at room temperature, washed twice with 200 μL of 80 % ethanol, and eluted in 20 μL of nuclease-free water.
Library amplification was conducted using the NGS Tagment Index Kit for Illumina in a 50 μL reaction containing 20 μL of eluted DNA, 25 μL of 2× Ultima Amplification Buffer, and 1 μL each of 10 μM P5 and P7 primers. PCR products were purified with NGS DNA selection beads. The quality and fragment size distribution of the libraries were assessed using 2 % agarose gel electrophoresis and a Qsep DNA fragment analyzer. Library concentrations were determined with a Qubit fluorometer. High-quality, barcoded libraries were pooled and sequenced on an Illumina NovaSeq 6000 platform (Wuhan BioRun Biosciences Co., Ltd.), generating approximately 6 Gb of raw data per sample.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The raw data for RNA-seq generated in this study are deposited in the National Geno-mics Data Center (NGDC), China National Center for Bioinformation under the BioPr-oject accession PRJCA046995. Genes sequence data from this study are available from the Rice Genome Anno-tation Project website (https://rice.uga.edu/) under the following accession numbers: OsABI5 (LOC_Os01g64000), OsSAPK2 (LOC_Os07g42940), OsT5H (LOC_Os12g16720), OsPP2C30 (LOC_Os03g16170), OsPP2C49 (LOC_Os05g38290), OsPP2C51 (LOC_Os05g49730), OsNCED1 (LOC_Os02g47510), OsNCED2 (LOC_Os12g24800), OsNCED3 (LOC_Os03g44380), OsNCED4 (LOC_Os07g05940), OsNCED5 (LOC_Os12g42280), OsABA8ox1 (LOC_Os02g47470), OsABA8ox2 (LOC_Os08g36860), OsABA8ox3 (LOC_Os09g28390), OsPYL5 (LOC_Os05g12260). Source data are provided with this paper, which are also available at Figshare (https://doi.org/10.6084/m9.figshare.30225289). Source data are provided with this paper.
References
Oikonomou, G. et al. The serotonergic raphe promote sleep in zebrafish and mice. Neuron 103, 686–701 (2019).
Zhang, Z. et al. Understanding the mechanism of red light-induced melatonin biosynthesis facilitates the engineering of melatonin-enriched tomatoes. Nat. Commun. 14, 5525 (2023).
Almendros-Ruiz, A. The effects of melatonin supplementation on professional football player performance: a systematic review. Nutrients 15, 4467 (2023).
Felix, T. M., Karpa, K. D. & Lewis, P. R. Adverse effects of common drugs: dietary supplements. FP Ess. 36, 31–40 (2015).
Mannino, G. et al. Melatonin and phytomelatonin: chemistry, biosynthesis, metabolism, distribution and bioactivity in plants and animals-an overview. Int J. Mol. Sci. 22, 9996 (2021).
Fujiwara, T. et al. Sekiguchi lesion gene encodes a cytochrome P450 monooxygenase that catalyzes conversion of tryptamine to serotonin in rice. J. Biol. Chem. 285, 11308–11313 (2010).
Kang, S. et al. Characterization of tryptamine 5-hydroxylase and serotonin synthesis in rice plants. Plant Cell 26, 2009–2015 (2007).
Back, K., Tan, D. X. & Reiter, R. J. Melatonin biosynthesis in plants: multiple pathways catalyze tryptophan to melatonin in the cytoplasm or chloroplasts. J. Pineal Res. 61, 426–437 (2016).
Lee, K. & Back, K. Melatonin-deficient rice plants show a common semidwarf phenotype either dependent or independent of brassinosteroid biosynthesis. J. Pineal Res. 66, e12537 (2019).
Kanjanaphachoat, P. et al. Serotonin accumulation in transgenic rice by over-expressing tryptophan decarboxylase results in a dark brown phenotype and stunted growth. Plant Mol. Biol. 78, 525–543 (2012).
Sato, Y. et al. RiceXPro version 3.0: expanding the informatics resource for rice transcriptome. Nucleic Acids Res. 41, 1206–1213 (2013).
Kang, K. et al. Senescence-induced serotonin biosynthesis and its role in delaying senescence in rice leaves. Plant Physiol. 150, 1380–1393 (2009).
Kang, K. et al. Methanol is an endogenous elicitor molecule for the synthesis of tryptophan and tryptophan-derived secondary metabolites upon senescence of detached rice leaves. Plant J. 66, 247–257 (2011).
Hayashi, K. et al. Serotonin attenuates biotic stress and leads to lesion browning caused by a hypersensitive response to Magnaporthe oryzae penetration in rice. Plant J. 85, 46–56 (2016).
Miret, J. A. & Munné-Bosch, S. Abscisic acid and pyrabactin improve vitamin C contents in raspberries. Food Chem. 203, 216–223 (2016).
Xu, X. et al. Auxin and abscisic acid antagonistically regulate ascorbic acid production via the SlMAPK8-SlARF4-SlMYB11 module in tomato. Plant Cell 34, 4409–4427 (2022).
An, J. P. et al. ABI5 regulates ABA-induced anthocyanin biosynthesis by modulating the MYB1-bHLH3 complex in apple. J. Exp. Bot. 72, 1460–1472 (2021).
Tian, Y. S. et al. Enhancing carotenoid biosynthesis in rice endosperm by metabolic engineering. Plant Biotechnol. J. 17, 849–851 (2019).
Tian, Y. S. et al. Metabolic engineering of rice endosperm for betanin biosynthesis. N. Phytol. 225, 1915–1922 (2020).
Strobbe, S. et al. Metabolic engineering of rice endosperm towards higher vitamin B1 accumulation. Plant Biotechnol. J. 19, 1253–1267 (2021).
Storozhenko, S. et al. Folate fortification of rice by metabolic engineering. Nat. Biotechnol. 25, 1277–1279 (2007).
Yeap, W. C. et al. WRI1-1, ABI5, NF-YA3 and NF-YC2 increase oil biosynthesis in coordination with hormonal signaling during fruit development in oil palm. Plant J. 91, 97–113 (2017).
Hong, J. et al. Phosphorylation-mediated regulation of a rice ABA responsive element binding factor. Phytochemistry 72, 27–36 (2011).
Li, Y., Grotewold, E. & Dudareva, N. Enough is enough: feedback control of specialized metabolism. Trends Plant Sci. 29, 514–523 (2024).
Li, C. et al. Feedback regulation of plant secondary metabolism: applications and challenges. Plant Sci. 340, 111983 (2024).
Waadt, R. et al. Plant hormone regulation of abiotic stress responses. Nat. Rev. Mol. Cell Biol. 23, 680–694 (2022).
Soon, F. F. et al. Molecular mimicry regulates ABA signaling by SnRK2 kinases and PP2C phosphatases. Science 335, 85–88 (2012).
Fujii, H., Verslues, P. E. & Zhu, J. K. Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis. Plant Cell 19, 485–494 (2007).
Yu, F., Wu, Y. & Xie, Q. Precise protein post-translational modifications modulate ABI5 activity. Trends Plant Sci. 20, 569–575 (2015).
Kobayashi, Y. et al. Abscisic acid-activated SNRK2 protein kinases function in the gene-regulation pathway of ABA signal transduction by phosphorylating ABA response element-binding factors. Plant J. 44, 939–949 (2005).
Chae, M. J. et al. A rice dehydration-inducible SNF1-related protein kinase 2 phosphorylates an abscisic acid responsive element-binding factor and associates with ABA signaling. Plant Mol. Biol. 63, 151–169 (2007).
Bhatnagar, N. et al. The protein phosphatase 2C clade A protein OsPP2C51 positively regulates seed germination by directly inactivating OsbZIP10. Plant Mol. Biol. 93, 389–401 (2017).
Kim, H. et al. A rice orthologue of the ABA receptor, OsPYL/RCAR5, is a positive regulator of the ABA signal transduction pathway in seed germination and early seedling growth. J. Exp. Bot. 63, 1013–1024 (2012).
Furihata, T. et al. Abscisic acid-dependent multisite phosphorylation regulates the activity of a transcription activator AREB1. Proc. Natl. Acad. Sci. USA 103, 1988–1993 (2006).
Zong, W. et al. Feedback regulation of ABA signaling and biosynthesis by a bZIP transcription factor targets drought-resistance-related genes. Plant Physiol. 171, 2810–2825 (2016).
Muonde, M. et al. Global nutrition challenges: a public health review of dietary risks and interventions. World J. Adv. Res. Rev. 21, 1467–1478 (2024).
Bouis, H. E. & Saltzman, A. Improving nutrition through biofortification: a review of evidence from HarvestPlus, 2003 through 2016. Glob. Food Security 12, 49–58 (2017).
Sharma, S. K. et al. Serotonin in animals and plants: a new pathway and its implications. Frontiers in Plant Science 5, (2014).
Watanabe, M. et al. Serotonin metabolism and its role in plants. J. Plant Biol. 61, 121–132 (2018).
Erland, L. A., Turi, C. E. & Saxena, P. K. Serotonin: an ancient molecule and an important regulator of plant processes. Biotechnol. Adv. 34, 1347–1361 (2016).
Lu, H. P. et al. An ABA-serotonin module regulates root suberization and salinity tolerance. N. Phytol. 236, 958–973 (2022).
Lu, H. P. et al. Resistance of rice to insect pests mediated by suppression of serotonin biosynthesis. Nat. Plants 4, 338–344 (2018).
Pelagio-Flores, R. et al. Serotonin, a tryptophan-derived signal conserved in plants and animals, regulates root system architecture probably acting as a natural auxin inhibitor in Arabidopsis thaliana. Plant Cell Physiol. 52, 490–508 (2011).
Bouis, H. E. Micronutrient fortification of food crops. Annu. Rev. Nutr. 23, 1–18 (2003).
Liu, X. et al. CRISPR/Cas9-mediated genome editing in plants. Methods 12, 94–102 (2017).
Ouyang, S. Q. et al. Receptor-like kinase OsSIK1 improves drought and salt stress tolerance in rice (Oryza sativa) plants. Plant J. 62, 316–329 (2010).
Luo, X. et al. PIF4 interacts with ABI4 to serve as a transcriptional activator complex to promote seed dormancy by enhancing ABA biosynthesis and signaling. J. Integr. Plant Biol. 66, 909–927 (2024).
Liu, C. et al. The protein phosphatase PC1 dephosphorylates and deactivates CatC to negatively regulate H2O2 homeostasis and salt tolerance in rice. Plant Cell 35, 3604–3625 (2023).
Zhai, K. et al. NLRs guard metabolism to coordinate pattern- and effector-triggered immunity. Nature 601, 245–251 (2022).
Li, Z. et al. The OsNAC23-Tre6P-SnRK1a feed-forward loop regulates sugar homeostasis and grain yield in rice. Mol. Plant 15, 706–722 (2022).
Yu, X. et al. Arabidopsis PP6 phosphatases dephosphorylate PIF proteins to repress photomorphogenesis. Proc. Natl. Acad. Sci. USA 116, 20218–20225 (2019).
Wang, Y. et al. GR5 acts in the G protein pathway to regulate grain size in rice. Plant Commun. 5, 100673 (2024).
Li, Q. et al. Synergistic interplay of ABA and BR signal in regulating plant growth and adaptation. Nat. Plants 7, 1108–1118 (2021).
Acknowledgements
This work was supported by the National Natural Science Foundation of China (32188102, 32372118), Zhejiang Natural Science Foundation (LZ25C130010), the Qian Qian Academician Workstation, and the specific research fund of the innovation platform for academicians of Hainan province (YSPTZX202303), Chinese Academy of Agricultural Sciences Talent Plan-Outstanding Young Talent, Zhejiang Province’s High-level Talent Special Support Plan-Young Talent, Agricultural Science and Technology Innovation Program (ASTIP) of CAAS (CAAS-ZDRW202401), National Rice Industry Technology System Project (CARS-01-18). We thank the Public Laboratory of China National Rice Research Institute for their technical support in Subcellular localization.
Author information
Authors and Affiliations
Contributions
D.R., and Q.Q. conceived the project and designed experiments. Y.C., X.H., and A.W. performed most experiments. Y.C., Z.S., and A.W. generated transgenic plants and performed phenotypic analysis. C.D. and W.X. participated in project discussions. L.G., G. Z., L.Z., J.H., Z.G., G. D., Q. Z., Q.L., L.S., and M.H. analyzed data. Y.C. and D.R. prepared figures and wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Cui, Y., Hou, X., Wang, A. et al. Engineering hormonal crosstalk to enhance serotonin/melatonin levels in rice. Nat Commun 16, 10092 (2025). https://doi.org/10.1038/s41467-025-65067-6
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-65067-6