Abstract
Overactivation of FcγRI by immune complexes (IC) is implicated in various autoimmune disorders and neuropathies. Currently, no effective FcγRI-specific blocking antibodies are available. Here we report preclinical data revealing two anti-FcγRI antibodies, C01 and C04, with high affinity, Fab-mediated binding within the IgG binding site on extracellular domain 2 of FcγRI. Both C01 and C04 block 90% of IgG and IC binding, and displace ~60% of pre-bound ICs without activating FcγRI, thereby minimizing the risk of aggravating inflammation. In the context of autoimmunity, C01 and C04 inhibit RA patient-derived autoantibody-IC binding to monocytes, macrophages and activated neutrophils, meanwhile they also inhibit the binding of opsonized platelets to monocytes from patients with immune thrombocytopenia. In vivo, C01 and C04 reduce IgG-dependent platelet depletion in humanized immunodeficient FcRγ-/- mice. Structural studies confirm that C01 and C04 achieve their blocking effects through Fab-mediated binding to FcγRI. Our data thus suggest that C01 and C04 may offer therapeutic potential for autoimmune disorders.
Similar content being viewed by others
Introduction
The high-affinity Fc gamma receptor I (FcγRI) plays a crucial role in the immune system. It is normally saturated with circulating IgG, which acts like a threshold for receptor activation. Monomeric IgG can efficiently be displaced by multivalent IgG immune complexes (ICs), causing receptor clustering which initiates downstream receptor signaling via the immunoreceptor tyrosine-based activation motif (ITAM) in its associated FcR gamma chain. This leads to subsequent effector functions, including phagocytosis, antibody-dependent cellular cytotoxicity (ADCC), and release of pro-inflammatory cytokines1. While IC formation plays an essential role in capturing and neutralizing antigens, excessive or prolonged accumulation can be detrimental as it can deposit in organs, causing chronic inflammation.
Overactivation of FcγRI has been implicated in different autoimmune diseases, including systemic lupus erythematosus (SLE), immune thrombocytopenia (ITP), and rheumatoid arthritis (RA), which are all characterized by autoantibodies able to form ICs with autoantigens2,3,4. In RA, autoantibodies against citrullinated proteins (anti-citrullinated protein antibodies; ACPA) lead to joint inflammation and joint damage5. Changes in Fc-linked galactosylation and fucosylation result in reduced affinity for FcγRII (CD32) and FcγRIII (CD16), but increases the availability to FcγRI and thereby its activation and local inflammation6. Interestingly, RA-specific autoantibodies can also bind to FcγRI on pre-osteoclasts, inducing differentiation towards active mature osteoclasts, potentially leading to increased bone resorption7. FcγRI is also expressed on neurons, including nociceptive dorsal root ganglion (DRG) neurons, which are associated with pain sensation8,9. Activation of DRG neurons by RA-specific autoantibodies may contribute to pain experienced by RA patients10. Hence, FcγRI activation could contribute to joint inflammation, bone resorption, and pain in RA, making it a potential target for therapeutic intervention.
Previous research has utilized monoclonal antibodies (mAbs), such as clones 197, 10.1, m22, and H22, as blocking antibodies against FcγRI8,11,12,13,14,15,16,17. However, these antibodies show limited blocking of monomeric IgG and ICs, suggesting binding outside the ligand-binding domain and blocking via their Fc tail11,18. Recently, we aimed to overcome these limitations and explore potential therapies by developing chimeric Fc-silent antibodies that potently block the FcγRI ligand binding site via Fab binding using a phage display library selection approach19. This method allows for in vitro selection of mAbs through genetic engineering of bacteriophages and multiple rounds of antigen-guided selection and phage propagation20. Only the variable region of antibodies, represented as single-chain variable fragments (scFv) are expressed on the phages, so all Fc tail-FcγRI interactions are circumvented during the selection process. Hence, it is possible to generate antibodies that bind within the ligand-binding domain of FcγRI, also known as extracellular domain 2 (EC2). FcγRI consists of three extracellular domains, a transmembrane part, and an intracellular tail (FcγRI-CY). Extracellular domains one (EC1) and two (EC2) share homology with those found in other Fcγ receptors, while EC3, linking EC2 to the transmembrane region, is unique for FcγRI. EC2 of FcγRI contains a single amino acid deletion in the FG-loop, a region highly conserved across species, contributing to its high affinity21.
Our current study focuses on the preclinical characterization of this recent panel of FcγRI antibodies, assessing its affinity, binding, and ligand blocking properties, as well as its therapeutic potential by using patient-derived ICs. Crystallography data of C01-FcγRI complex confirmed binding to EC2, revealing that C01 shares 14 binding site residues with IgG, including Phe146, His148, and Trp149. LigandTracer analysis revealed that C01 and C04 have a higher affinity for FcγRI than human IgG1. Both antibodies effectively block monomeric human IgG and IC binding, including in ex vivo patient models of RA and ITP, and consistently outperformed commercial mAb 10.1. Lastly, C01 is confirmed to impair IgG-dependent platelet depletion in an in vivo model of ITP. Considering the potential therapeutic impact of blocking FcγRI activation, our study provides valuable insights into the development of FcγRI-specific antibodies as potential therapeutic candidates.
Results
Commercial anti-FcγRI antibodies do not show specific blocking of ligand binding
Commercially available anti-FcγRI antibodies, including clones 10.1 (mIgG1), 197 (mIgG2a), m22 (mIgG1), and H22 (humanized version of m22, hIgG1), are commonly used for FcγRI blocking in in vitro blocking studies. Clone 10.1 was generated upon immunization of mice using human rheumatoid synovial fluid cells and fibronectin-purified monocytes11. Both 197 and m22 were obtained via immunization with IFN-γ-treated monocytes22. M22 was humanized via complementarity-determining region (CDR)-grafting onto human V region frameworks, resulting in H2212. However, their specific ability to block ligand binding has not been thoroughly examined. To address this, we utilized FcγRI-transduced Ba/F3 and EL4 cells, which exhibited high levels of FcγRI expression compared to the parental cells (Fig. 1a). These cell lines demonstrated dose-dependent binding of hIgG, mIgG2c, and the commercial anti-FcγRI antibodies (Fig. 1b).
a Expression of FcγRI on different cell lines. Ba/F3-FcγRI (solid line) or EL4-FcγRI (dashed line) cells without antibody are depicted as the control. b Binding titration of anti-FcγRI antibodies to Ba/F3-FcγRI (solid lines) and EL4-FcγRI (dashed lines) cells. Mean fluorescence intensity (MFI) is depicted of at least N = 3. c Blocking of ligand (hIgG1) on Ba/F3-FcγRI (solid lines) and EL4-FcγRI (dashed lines) cells with different anti-FcγRI antibodies; 197 (circle, dark green), 10.1 (upside down triangle, blue), m22 (triangle, green), H22 (hexagon, green), and mIgG2c (square, yellow). Percentage (%) ligand blocking was calculated using the ligand only as a control. d Bar graphs showing ligand blocking at 10 μg/ml of the indicated antibodies. N = 3, in duplo. Data are mean ± SEM, Two-way ANOVA with Tukey’s post hoc test. Source data are provided as a Source Data file.
Ligand blocking capacity was determined by incubating Ba/F3-FcγRI and EL4-FcγRI cells with clones 10.1, 197, m22, and H22, followed by fluorescently labeled monomeric hIgG1 as ligand (Fig. 1c). Unlabeled mIgG2c antibody was included as a positive control for hIgG blocking, as it can bind FcγRI with high affinity. Clone 197 and mIgG2c effectively blocked ligand binding to FcγRI, particularly at higher concentrations. In contrast, clones 10.1, m22, and H22 only exhibited moderate blocking at higher concentrations.
Based on these results, clone 197 demonstrated the highest blocking capacity. However, clone 197, an mIgG2a isotype with an Fc-hFcγRI interaction comparable to mIgG2c, showed significantly reduced binding to cells when tested as F(ab’)2 and Fab fragments (Fig. 1b) and a substantial reduction in ligand blocking capacity (Fig. 1d, p = 0.0001). This suggests that the Fc part primarily contributes to the antibody’s ligand-blocking ability. Clone 10.1 exhibited a similar trend, with the full antibody preventing a modest 36-50% of ligand binding and the F(ab’)2 fragment reducing this blocking capacity by half (p = 0.0001). These findings emphasize that FcγRI clones 10.1, 197, m22, and H22 are ineffective in blocking the Fc-FcγRI interaction through their Fab-mediated binding.
Characterization of C01 and C04 reveal specific binding to EC2 of FcγRI and this does not lead to activation
Utilizing an antibody phage display library approach, we isolated seven VLκ (variable kappa light chain) and VH sequence pairs19. For a schematic summary of the phage display library approach, see Supplementary Fig. 1. Phylogenetic analysis of the VH amino acid sequences revealed two distinct, common ancestors, with the three commercial antibodies (clones 10.1, 611 and H22) sharing a common ancestor, while the new panel shares a different common ancestor (Fig. 2a). The VL of 611 and 10.1 share a common ancestor, although both mutated heavily (i.e. long brand length). There was a further separation for both VH and VL observed between C01 and C04 versus the remaining clones (i.e., C03, C09, C10, C15, and C16). Notably, C01 and C04 share identical VH sequences, as do C03VH, C10VH, and C16VH, but all VLκ sequences are unique.
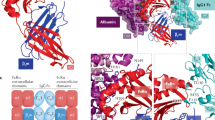
a Mid-point rooted phylogenetic trees for VL and VH chains of C01 (orange), C04 (red) and C03/C09/C10/C15/C16 (light orange), compared to sequences of clones 10.1 (blue), 611 and H22. b Titration curves of FITC-labeled anti-FcγRI antibodies of one representative experiment in duplo. c Binding of labeled anti-FcγRI mAbs or 10.1 on Ba/F3-FcγRI after pre-incubation with excess IVIg. d Anti-FcγRI antibody binding to PBMCs from healthy donors. hIgG1 (purple) was added as a control. Significance in c and d based on one-way ANOVA with Tukey’s test, **** p < 0.0001 compared to C03, C09, C10, C15, C16, and 10.1, N = 3, in duplicate. e Binding of C01 (circle, orange), C04 (square, red), and hIgG1 (diamond, purple) to fully human FcγRI and chimeric versions of FcγRI (i.e., mEC1 = only human EC1 swapped to mouse EC1). f Structure of the C01–FcγRI complex with domains and C01 CDR loops indicated. C01 binds exclusively to the EC2 domain of FcγRI. FcγRI is colored green, the C01 light chain purple, and the heavy chain orange. g Transparent surface of the C01 epitope on FcγRI in dark green and all epitope residues in stick representation and labeled. h Known binding regions of hIgG and 10.1 and proposed binding regions of own clones. Created in BioRender. Holtrop, T. (2025) https://BioRender.com/h1gt0ff. i, j Bind titration of C01 and C04 on FcγRIIa/b+ and FcγRIIIa/b+ cells. Anti-CD16 (triangle, light green) and anti-CD32 (upside-down triangle, blue) were taken along as a control. Data from one representative experiment in duplicate is shown. k Association and dissociation of C01, C04, and hIgG as measured by LigandTracer. A minimum of 4 runs per antibody were done, quantification of association and dissociation was done via TraceDrawer software (l). Boxplot showing distribution of obtained KD values. Midline indicates median, lower and upper margins of the boxes correspond with the 25th and 75th percentiles, and whiskers range from minimum to maximum values. C01/C04 is N = 6 and hIgG1 is N = 4. Significance determined with one-way ANOVA and Tukey’s test. m Area under the curve (AUC) of calcium flux on Ba/F3-FcγRI cells upon antibody binding and crosslinking with secondary antibody. M22 and H22 were used as positive controls. n Superoxide by IFN-γ-stimulated neutrophils in relative light units (RLU). Lines show the average of N = 3 measurements in triplicate for m22 and H22, and N = 6 for mAb 197, C01, C04, and 10.1. o AUC of superoxide, including baseline release (PBS), is indicated by a dotted line. Ordinary one-way ANOVA, with Tukey multiple comparison test. All line and bar data are presented as mean ± SEM. Source data are provided as a Source Data file.
All VLκ-VH sequence pairs were inserted into Fc-silent human IgG1 backbones to eliminate Fc-FcγRI interactions and nonspecific binding to FcγRII/FcγRIII, while maintaining binding to the neonatal Fc receptor for extended half-life. This silencing was done with the LALAPG mutation (L234A, L235A, P329G)23. Purity was determined via sodium dodecyl sulfate-polyacrylamide (SDS-PAGE) gel electrophoresis (reducing condition, 4-20% gradient gel, Bio-Rad, 4561094) and stained using InstantBlue Ultrafast Protein Stain (Sigma Aldrich, ab119211, Supplementary Fig. 2a. For all subsequent experiments, Ba/F3-FcγRI were used since the phages were bio-panned on this cell line. Binding titrations on Ba/F3-FcγRI demonstrated that all seven antibodies exhibited a sigmoidal dose-response curve, plateauing at 1 μg/ml (Fig. 2b). Subsequently, we investigated the binding properties of the anti-FcγRI panel in the presence of hIgG. C01 and C04 failed to bind FcγRI following IVIg blockade (200 μg/ml), while the other five clones and 10.1 retained full binding (Fig. 2c). Similar results were observed using primary human PBMCs from healthy donors (Fig. 2d). These results suggest that hIgG, C01 and C04 share a binding site within EC2, while 10.1 and the other five clones bind outside of this domain.
To better elucidate the binding domains, cross-block experiments were performed with the anti-FcγRI panel and clone 10.1, which binds EC324. Pre-incubation with C01 and C04 enhanced 10.1 binding to 150%, whereas C03, C09, C10, C15, and C16 reduced it to 1-5% (Supplementary Fig. 2b). Conversely, pre-binding of 10.1 only modestly reduced C01 and C04 binding to 80% and 87%, while the other five clones showed 20-45% binding (Supplementary Fig. 2c). These findings suggest that only C03, C09, C10, C15, and C16 compete with 10.1 for binding to EC3 of FcγRI. To confirm EC2 as the binding domain of C01 and C04, domain swap experiments were performed, replacing one human FcγRI domain with murine FcγRI. As expected, hIgG1 only binds weakly to murine EC2, but retained binding when EC1 or EC3 were swapped (Fig. 2e). A similar trend could be seen for both C01 and C04, with minimal binding to mEC2.
To further investigate the interaction details, we determined the crystal structure of the C01 Fab fragment in complex with the EC1-3 segment of FcγRI. The crystal structure reveals that, as expected, C01 interacts exclusively with the EC2 domain of FcγRI (Fig. 2f, Supplementary Table 1). The total buried surface area of the complex is 1278 Å2, in which the C01 heavy chain contributes 462 Å2, the light chain 154 Å2 and FcγRI 662 Å2. CDR1-3 of the heavy chain and CDR1 and CDR3 of the light chain all contribute to direct interactions with FcγRI, with the most prominent role for CDR3 of the heavy chain, which is centrally located in the C01-FcγRI complex interface. On the FcγRI side, 17 residues contribute to direct interactions with C01 and are situated in three EC2-loops: Asp129-Tyr138, Phe146-Trp149, and Ser169-Thr177 (Fig. 2g). Most prominently, Met171 is central to the C01 epitope and Lys173 and Arg175 also play important roles.
The C01 epitope on FcγRI overlaps with the IgG Fc binding site and suggests that C01 would compete with Fc binding. Of the 17 residues in the C01 epitope, 14 overlap with those involved in Fc binding (Supplementary Fig. 3a, b). Superposition of the C01–FcγRI structure and Fc–FcγRI structure determined previously indicates that the VH and VL domains of C01 sterically overlap with the two CH2 domains of Fc when bound to FcγRI, rendering binding mutually exclusive (Supplementary Fig. 3c, d)25. Comparison of the structures of free– and Fc-bound–FcγRI with that of the C01–FcγRI complex reveals small structural rearrangements in the Fc binding region in FcγRI, likely induced by C01 (Supplementary Fig. 3e–g)21,25. In the C01 bound form, these rearrangements lead to partial collapse of the hydrophobic pocket that is critical for Fc binding as it buries Fc residue Leu23525. The collapse is mediated by the movement of FcγRI segments Met171-Tyr176 and Trp127-Tyr133 towards each other in the C01-bound form, preventing the Fc hinge from accessing the hydrophobic pocket. Taken together, the crystal structure of the C01–FcγRI complex explains that C01 inhibits Fc binding to FcγRI by overlapping binding sites and steric hindrance. These results confirm that C01 and C04 bind to EC2, while the other clones compete with 10.1 for binding to EC3 of FcγRI (Fig. 2h).
Since C01 and C04 bind within the ligand-binding domain EC2, we assessed their specificity for FcγRI, as EC2 is homologous to the EC2 domain found in FcγRII and FcγRIII. Neither antibody bound FcγRIIa/b nor FcγRIIIa/b (Fig. 2i, j), confirming their specificity towards FcγRI. Using LigandTracer the kon, koff, and KD for C01, C04, 10.1 and hIgG1 were determined (Fig. 2k, l, Supplementary Fig. 4a, b, Supplementary Table 2). C01 had the highest effective binding affinity for FcγRI (KD = 1.21 × 10-10 M), followed by C04 (KD = 5.43 × 10-10 M), hIgG1 (KD = 1.19 × 10-9 M), and 10.1 (KD = 2.19 × 10-8 M). This established the FcγRI-ligand avidity hierarchy as C01 > C04 > hIgG1 > 10.1, making C01 and C04 significantly better binders than hIgG (p = 0.0001 and p = 0.0008, respectively).
To confirm that C01 and C04 do not induce FcγRI signaling, we assessed activation via calcium flux and superoxide burst assays. Neither antibody triggered calcium mobilization (Supplementary Fig. 4c), even when crosslinked with rabbit F(ab’)2 anti-human IgG, while H22 induced calcium release (Supplementary Fig 4d). Ionomycin resulted in maximal intracellular Ca2+ levels (Supplementary Fig. 4c, d). To assess if C01/C04 binding also prevents activation of the receptor, we co-incubated the cells with m22 and C01 or C04. Both antibodies bind simultaneously, since m22 binds outside of the ligand binding domain (Supplementary Fig. 4e). Crosslinking m22 alone rapidly increased, extracellular Ca2+ levels, lasting ~4 min (Supplementary Fig. 4f). However, when C01 or C04 was present, no calcium mobilization occurred, suggesting that receptor activation was blocked (Supplementary Fig. 4f). Quantification of multiple experiments based on area under the curve (AUC) confirmed no significant activation of FcγRI upon binding or crosslinking of C01 or C04, while both inhibited m22-induced activation (Fig. 2m).
Superoxide production, another marker of activation, was measured via a luminol chemiluminescence assay. Adding clone 197 or H22 to IFN-γ-stimulated neutrophils led to a superoxide burst (Fig. 2n), similar to Graziano et al. using U937 cells12. A modest burst with m22 was reported, while we only observed this in one donor. C01, C04, and 10.1 responses were comparable to PBS (Fig. 2o), confirming no activation upon binding. Taken together, these data show that binding of C01 and C04 does not lead to FcγRI activation.
C01 and C04 outperform 10.1 in blocking hIgG and IC binding to FcγRI
The anti-FcγRI panel was tested in a ligand blocking assay using Ba/F3-FcγRI cells (Fig. 3a). Consistent with Fig. 1, 10.1 only induced a modest 40% inhibition of hIgG binding to FcγRI (Fig. 3b), similar to clones C03, C09, C10, C15, and C16. In contrast, C01 and C04 blocked IgG binding by up to 94%. This inhibition is maintained when the antibody is converted into F(ab’)2 fragments, suggesting true Fab-mediated blocking rather than steric hinderance via the Fc tail (Supplementary Fig. 5a).
a Schematic illustration of hIgG blocking assay. Pre-incubation with one of the seven clones or 10.1, after which labeled hIgG was added. Created in BioRender. Holtrop, T. (2025) https://BioRender.com/k9ru1dnb Pre-incubation with C01 (orange) and C04 (red) led to a heavily reduced hIgG binding, while the other five clones (light orange) and 10.1 (blue) only sterically hinder hIgG binding. MeaN ± SEM of N = 3 in duplo. c Schematic overview of IC blocking assay. Created in BioRender. Holtrop, T. (2025) https://BioRender.com/cak3cjm. d,e Significantly reduced IC binding when pre-incubated with either C01 or C04 at all concentrations tested. Two-way ANOVA with Bonferroni post hoc test comparing against 10.1 (upside down triangle, blue). N = 3, in duplo, mean ± SEM f Illustration of rosette assay. M450-epoxy dynabeads coupled with DNP-BSA were incubated with ANTI-DNP antibody to form large immune complexes. These beads were then added to Ba/F3-FcγRI cells pre-incubated with different concentrations of anti-FcγRI antibodies. Created in BioRender. Holtrop, T. (2025) https://BioRender.com/10winlig Representative images of what is considered a rosette ( ≤ 5 beads bound to one cell) and differences between no antibody and 100 μg/ml C01. Green dotted box indicates cells considered a rosette, red dotted box indicates cells not considered a rosette. Ba/F3-CD64 cells are ~10 μm in size and the beads 4.5 μm. h Boxplot of obtained rosette percentages, midline indicates median, lower and upper margins of the boxes correspond with the 25th and 75th percentiles and whiskers ranging from minimum to maximum values. Two-way ANOVA with Bonferroni post hoc test. N = 2, triplicate measurements. Source data are provided as a Source Data file.
To assess IC blocking, human IgG1 was crosslinked with anti-human F(ab’)2 fragments (Fig. 3c). When FcγRI was bound with 10.1, ICs could still bind up to 44.5% (Fig. 3d, e). Notably, C01 reduced IC binding to 3.3–16.5% (p < 0.0001, Fig. 3d), while C04 reduced IC binding to 9.6-31.5% (p < 0.0001), depending on antibody and IC concentrations. (Fig. 3e).
To visualize the blocking capacity, IgG-coated beads were used as IC to measure rosette formation (Fig. 3f, g). A wide range of concentrations (1 to 100 μg/ml) was tested to define the dose-response relationship. C01 and C04 significantly outperformed 10.1, reducing rosette formation with an average of 46% versus 61% with 10.1 (Fig. 3h). A clear concentration-dependent effect was observed, with 50 μg/ml of C01 significantly blocking rosette formation more effectively than 5 and 25 μg/ml. Increasing the concentrations to 100 μg/ml did not further reduce the number of rosettes, indicating saturation (Supplementary Fig. 5b).
C01 and C04 can displace bound hIgG and IC at 37 °C
In autoimmune diseases, ICs are already bound to FcγRI on immune cells prior to the addition of blocking antibodies, thus requiring displacement to exert their blocking effect. Hence, we assessed the efficacy of our blocking antibodies to displace pre-bound hIgG and IC at 37 °C (Fig. 4a). After 4 h, both C01 and C04 displaced up to 80% of hIgG in the absence of excess hIgG (Supplementary Fig. 6a, b). With excess hIgG, displacement reached ~45% (Fig. 4b, bars with purple circle or squares). Noteworthy, C01 and C04 are only capable to regain their full baseline binding capacity when there is an excess of hIgG present, which already happens within 2 h (Fig. 4b, dark orange and red bars), while only 83% of baseline binding is possible without an excess of hIgG after 4 h (Supplementary Fig. 6b).
a Schematic overview of displacement assay with an excess of hIgG. Same set-up is used for IC, but then hIgG is crosslinked with an anti-human F(ab’)2 to make IC. Created in BioRender. Holtrop, T. (2025) https://BioRender.com/f0duoyib Binding of C01 (orange bars, circles), C04 (red bars, circles) and 10.1 (blue bars, triangles) over time (4 h) when hIgG1 is already pre-bound and present in excess, and how hIgG1 changes overtime when C01 (light orange bars, purple circles), C04 (light red bars, purple squares) or 10.1 (light blue bars, purple triangles) are added. Data is mean ± SEM, percentages are calculated compared to a baseline control with only the indicated antibody or hIgG. N = 2, in duplo. Displacement of c hIgG and d IC (%) after 20 h in the presence of C01 (orange bar, purple circle), C04 (red bar, purple square), or 10.1 (blue bar, purple triangle) at different concentrations. Percentage (%) displacement was calculated using hIgG/IC only as a control. Mean ± SEM, two-way ANOVA, multiple comparisons within group, Tukey’s test. N = 3, in duplo. Baseline staining of serum IgGs (purple) to e monocytes and h neutrophils with an anti-human IgG after 20 h. Samples were cultured overnight with 10% human serum from the same donor. f, g Histograms and bar graph (mean ± SEM) of C01 (orange triangle) and C04 (red upside down triangle) binding after only 30 min incubation or after overnight incubation (20 h). i,j Similar set-up as in (e–g), but with neutrophils and overnight incubation with G-CSF. N = 2, in duplo, mean ± SEM. Histograms show one representative experiment. One-way ANOVA with Tukey’s post hoc test. Source data are provided as a Source Data file.
After 20 h, C01 displaced 65% of hIgG and 59% of IC, while C04 displaced 51% and 53%, respectively (Fig. 4c, d). Interestingly, co-incubation with 10.1 increased hIgG binding (103-119%) and only mildly displaced IC (27%) without a clear dose-dependent effect. Increasing the concentration of C04 led to significantly more hIgG displacement (46% with 10 μg/ml to 58% with 100 μg/ml, p = 0.007) (Supplementary Fig. 6c). More importantly, IC displacement is greatly enhanced with higher concentrations of C01, C04, and even 10.1 (Supplementary Fig. 6d).
To assess displacement in a physiologically setting, primary immune cells were cultured overnight in RPMI with 10% human serum, confirming serum IgG binding to monocytes and IFN-γ-stimulated neutrophils (Fig. 4e, h). Direct staining for serum IgG was not possible alongside C01 and C04, as the secondary anti-human IgG antibodies will also bind C01 or C04. Instead, baseline binding was measured with FITC-labeled C01 or C04 for 30 min on ice to minimize displacement. Minimal monocytes binding was observed after 30 mins, but significantly increased after 20 h incubation (Fig. 4f, g p = 0.035 and p = 0.015, respectively). The same trend was observed for neutrophils, with significantly higher binding after 20 h (Fig. 4i, j, p < 0.0001). These results indicate effective displacement in a physiological context.
Patient-derived polyclonal and recombinant monoclonal autoantibody-IC binding to monocytes, macrophages and activated neutrophils can effectively be inhibited by the two novel antibodies
Having demonstrated the blocking capabilities of C01 and C04 in model systems, including cell lines and healthy donor samples, we next assessed their clinical relevance in in vitro rheumatoid arthritis (RA) models using patient-derived materials. In RA, anti-citrullinated protein antibodies (ACPAs) form ICs that bind various immune cells, contributing to disease progression. Monocytes and macrophages are activated by ACPA-ICs through Fcγ receptors, particularly FcγRI, leading to TNF-α, IL-1, and IL-6 production, which drives inflammation26. Neutrophils, which normally express low levels of FcγRI, upregulate FcγRI expression in RA synovial fluid due to high IFN-γ levels. This enables them to bind ACPA-ICs, triggering the release of reactive oxygen species (ROS), neutrophil extracellular traps (NETs), and pro-inflammatory cytokines, thus exacerbating joint damage27. To evaluate the therapeutic potential of C01 and C04, we tested them in an ex vivo model of patient-derived polyclonal (pACPA) and recombinant monoclonal ACPA-ICs (mACPA). We analyzed monocytes and macrophages, key immune cells implicated in RA pathology, and also included activated neutrophils, the most predominant immune cell in the synovial fluid27,28.
Monocytes from healthy donors were gated on single cells ( ± 97%), live cells ( ± 100%) and CD14+ cells ( ± 10-15%) (Fig. 5a). C01 and C04 significantly reduced mACPA-IC binding to monocytes by 71.9% and 66.4% (p < 0.0001, Fig. 5b, c), with no further inhibition at higher concentrations. Clone 10.1 also blocked binding (59.8% at 10 ug/ml and 59% at 25 ug/ml, p < 0.0001), though C01 was significantly more effective at both 10 µg/ml (p = 0.006) and 25 µg/ml (p = 0.0004), and C04 at 25 µg/ml (p = 0.008). With monocyte-derived macrophages, C01 and C04 strongly blocked mACPA-IC binding (62.5% and 57.3%, p < 0.0001, Fig. 5d–f) and this blockage increased to 69.2% and 67.4%, at 25 µg. In contrast, 10.1 was ineffective at blocking mACPA binding (<3% block). Taken together, C01 blocked 72% of mACPA-IC binding to monocytes and 63% to macrophages, while C04 induced a slightly lower block of 66% and 57%, respectively.
a Gating strategy for monocytes within PBMC fraction, with FSC, SSC, eF780 as a fixable viability dye and CD14-PE to gate for monocytes. b Blocking of mACPA-IC binding to monocytes with the individual compounds. Data from four donors (indicated by square, triangle, circle, and upside-down triangle) at 10µg/ml (closed symbols) or 25µg/ml (open symbols). mACPA-IC measured in quarto and antibody samples in duplo. c Bar graph showing the degree of mACPA-IC binding inhibition to FcγRI on monocytes by C01 (triangle orange), C04 (circle red), or 10.1 (square blue). mACPA-IC only (circle purple) set at 0%. N = 4, in duplo, mean ± SEM, one-way ANOVA with Tukey’s post hoc test. d Gating strategy for monocyte-derived macrophages. e, f similar as b, c but with macrophages, N = 2 donors in duplo, all depicted as mean ± SEM. Source data are provided as a Source Data file.
After overnight incubation of neutrophils with G-CSF and IFN-γ to mimic inflamed tissue conditions in RA patients, neutrophil samples contained ±90% live cells. CD15high-CD66bhigh cells were selected, together with a 7-AAD staining, to exclude (pre-)apoptotic neutrophils (Fig. 6a). Stimulation with IFN-γ significantly increased FcγRI expression (p < 0.0001) and, importantly, showed no changes in FcγRII and FcγRIII expression levels (Fig. 6b). The upregulation of FcγRI on stimulated neutrophils resulted in significantly higher binding of both mACPA-IC (p < 0.0001) and pACPA-IC (p = 0.0002, Fig. 6c).
a Gating strategy for neutrophils. b Expression levels of FcγRI, FcγRII, and FcγRIII on unstimulated and IFN-γ-stimulated neutrophils from three healthy donors (purple circle, square and triangle), in duplo. c MFI of recombinant monoclonal ACPA-IC (mACPA-IC) and patient-derived polyclonal ACPA-IC (pACPA-IC) binding on IFN-γ-stimulated neutrophils compared to unstimulated. Data from three donors, unstimulated measured in duplo, stimulated in quatro.d MFI and blocking of mACPA-IC binding upon pre-blocking of FcγRI with C01 (triangle orange), C04 (circle red), or 10.1 (square blue) at 10 (closed symbols) and/or 25 μg/ml (open symbols). Data from three donors, ACPA-IC measured in quatro and antibody samples in duplo. e MFI and blocking of pACPA-IC binding after pre-blocked with C01, C04, or 10.1. Data from three donors, ACPA-IC measured in quatro and antibody samples in duplo. Data presented as mean ± SEM, two-way ANOVA without post hoc test was used for b–e line graphs, and one-way ANOVA with Tukey’s test for d, e bar graphs. The significance line only indicates significance between 2 concentrations of the same antibody; the capped line indicates significance between mACPA-IC or pACPA-IC and all underlying bars. Source data are provided as a Source Data file.
C01 and C04 both significantly reduced mACPA and pACPA binding to stimulated neutrophils from healthy donors (p < 0.0001, Fig. 6d, e). Increasing the concentration of C01 to 25 ug/ml significantly enhanced blocking, causing mACPA-IC blocking to increase from 52.8% to 68.7% (p < 0.0001) and pACPA-IC blocking from 58.1% to 70.2% (p = 0.0004). C04 also showed a concentration-dependent effect, with mACPA-IC blocking increasing from 58.9% to 66.5% (p = 0.008) and pACPA-IC blocking from 61.3% to 69.3% (p = 0.033). No significant differences were observed between C01 and C04 at similar concentrations. Surprisingly, 10.1 only showed 2.5% block of mACPA-IC binding (p = 0.71) and no change in pACPA-IC binding compared to ACPA-IC only (Fig. 6d, e).
C01 and C04 show potent inhibition in an in vitro and in vivo disease model for ITP
Building on our RA in vitro analysis and to further assess clinical relevance, we next evaluated the blocking capabilities of C01 and C04 in both a patient-derived in vitro model and a humanized in vivo model of immune thrombocytopenia (ITP). Specifically, we evaluated their ability to inhibit opsonized platelet binding to monocytes within PBMCs isolated from ITP patients. Monocytes were identified within the PBMC population using a fixable viability dye (eF780) followed by CD45+ and CD14+ staining (Fig. 7a). Despite patient-to-patient variation, the results confirmed that both C01 and C04 effectively inhibited opsonized platelet binding to monocytes by approximately 53% (p < 0.0001) and ~41% (p = 0.015), respectively (Fig. 7b, c). The reduction with 10.1 did not significantly differ from baseline (p = 0.32).
a Gating strategy for monocytes within the ITP patient-derived PBMC population. b Blocking of opsonized platelet binding to monocytes using antibodies C01 (orange), C04 (red), and 10.1 (blue). Opsonized platelets were formed by using healthy donor platelets and an anti-GPIIbIIIa-APC antibody. Data are derived from N = 5 patients (each depicted by an individual symbol). Data depicted as mean ± SEM c Bar graph displaying the degree of opsonized platelet binding to monocytes and the significant inhibition observed with C01 (orange triangle) and C04 (red circle) compared to the control with no blocking (purple circle) or 10.1 (blue square). Two significant outliers were identified with Grubbs’ test (α = 0.05) and these were excluded from the analysis in c. Bars and lines indicate mean ± SEM, N = 5 patients. Statistical analysis was performed using one-way ANOVA with Tukey’s post hoc test. ITP = immune thrombocytopenia. d Schematic representation of ITP in vivo experiment, where 6A6-hIgG1 was used to deplete platelets over 4 h. Created in BioRender. Holtrop, T. (2025) https://BioRender.com/4mg76z2e Residual platelet count in humanized NSG-FcRγ − /− mice 4 h after injection of 100 μg of the chimeric anti-platelet antibody 6A6-hIgG1. Prior to ITP induction mice were pretreated with PBS (gray circles) or 100 μg C01 (orange triangles). N = 4 for control mice and N = 5 for C01 mice; statistical analysis was performed using an unpaired t-test. Source data are provided as a Source Data file.
These findings were validated in a fully human immune system mouse model of autoantibody-driven ITP, where NSG-FcRγ-/- mice were xenografted with human hematopoietic stem cells (Supplementary Fig. 7)29. Platelets were depleted using a 6A6-hIgG1 antibody recognizing platelet integrin antigens of mouse origin (Fig. 7d)30. Given C01’s clear superior affinity, blocking efficacy and structural characterization, in vivo validation was focused on this antibody to minimize animal use. Following treatment, mice receiving C01 retained up to 49,5% of circulating platelets after 4 h, compared to only 29% in the PBS control group (p = 0.0012, Fig. 7e).
The significant reduction in monocyte-platelet interactions in vitro and the effective reduction of platelet clearance in vivo, further highlights the therapeutic potential of C01 and C04.
Discussion
In this study, we characterized antibodies that specifically block FcγRI, as previously described anti-FcγRI antibodies could not effectively inhibit FcγRI ligand binding via their Fab fragment. Recently, we developed a novel panel of antibodies directed against FcγRI via an antibody phage display library19. The initial characterization shows that C01 and C04 have similar binding properties as hIgG (i.e. binding to EC2), making them strong potential candidate blocking antibodies. The crystal structure of the C01–FcγRI complex revealed that the C01 epitope on FcγRI overlaps with the Fc binding site, and thus Fc binding is prevented by C01 through steric hindrance25,31. Functional characterization shows displacement of pre-bound IgG/ICs to a significantly larger extent than commercial antibody 10.1, also when using autoantibodies derived from RA patients. Importantly, binding and crosslinking of the antibodies does not activate the receptor. Therapeutic antibodies that inadvertently activate FcγRI induce inflammatory reactions, contributing to adverse effects and undermining their potential clinical benefits4.
Specific blocking antibodies against other Fc receptors have provided key insights into their immunological roles and therapeutic potential. For instance, an anti-FcγRII antibody (clone 2B6) served as distinctive marker between inhibitory FcγRIIb and activating FcγRIIa, while an anti-FcγRIIIa blocking antibody (clone 3G8) was used to successfully treat ITP32,33. Beyond blocking FcγRII and FcγRIII, infusion of high doses of pooled IgG preparations (IVIg) is also an established treatment in many autoimmune disease, but their immunomodulatory activity is dependent on FcγRII via recognition of highly sialylated IgGs34,35. This leaves FcγRI available for IC binding and subsequent activation. Several anti-FcγRI blocking antibodies, including clones 10.1, 197, m22, and H22, have been developed11,12,13,14,15,16,17. However, these antibodies only exhibit minor down-modulating effects of FcγRI, as most of the blocking is achieved through the Kurlander effect. This involves Fab-mediated binding outside of the ligand binding domain and blocking via its IgG-Fc fragment, thus forming a heterotrimeric complex36. This effect holds therapeutic potential, as the ‘scorpion-like’ arrangement triggers cell activation, partly reliant on recruiting activating FcγRs37. Mechanistically, FcγRI associates with the FcR gamma chain, which contains an ITAM crucial for signaling. Upon receptor clustering and heterodimerization by a crosslinking antibody or IC, ITAMs are phosphorylated by Src family kinases, recruiting Syk and initiating downstream signaling38. Syk kinases also enhance FcγRI interaction with BLT1, the leukotriene B4 receptor, forming a FcγRI/BT1 complex that further boosts phagocytosis, bacterial killing and LTB4 release39. However, such activation can lead to adverse side-effects, as seen with H22 (MDX-33) in phase I/II trials40. This highlights the need for FcγRI-specific antibodies that can effectively block FcγRI ligand binding without triggering activation. To the best of our knowledge, this is the first study that shows two strong candidates, as C01 and C04 bind within EC2, overlapping with 14 FcγRI residues involved in IgG binding, and that do not activate the receptor.
The crystal structure of the C01–FcγRI complex also revealed that C01 binding induces a conformational change of FcγRI, partially collapsing FcγRI’s unique hydrophobic pocket. This pocket is crucial for high-affinity IgG-Fc binding, as the insertion of Leu235 from IgG-Fc deep within the hydrophobic pocket accounts for 25% of the interaction with Fc25,41. The loss of these hydrophobic contacts disrupts the structural complementarity required for high-affinity binding, preventing multivalent ICs from bridging multiples FcγRI receptors and forming high-avidity clusters needed for ITAM phosphorylation38. Moreover, ITAM signaling depends on precise alignment of the receptors extracellular, transmembrane and intracellular domains, as close association of the FcR gamma chains is needed for proper signaling42,43,44. Crystallographic analysis of three FcγRI-IgG complexed structures and one unbound FcγRI structure consistently reveals a characteristic “sea-horse” shape, with a relatively rigid EC1-EC2 interdomain angle of approximately 35°21,25,31,45. In contrast, the EC2-EC3 angle displays notable flexibility, varying up to 19° between FcγRI-IgG complex and unbound form45. These structural observations suggest that the position of EC3 influences overall receptor conformation and stability21,25,31. It is also proposed that it might prevent dissociation between IgG-FcγRI, thereby stabilizing and strengthening the complex interaction46. Interestingly, in our study, we see a 19% increase in IgG binding when 10.1 is prebound, which would suggest that 10.1 induces a conformational shift that either enhances the accessibility of the IgG binding site (i.e., better extension into the extracellular space) or helps stabilize the hIgG binding interaction. Likewise, our data shows that pre-incubation with C01/C04 enhances 10.1 binding by 50% (Supplementary Fig. 2b) while reducing m22 binding by 30% (Supplementary Fig. 4e), suggesting that binding to EC2 can also induce a conformational change that alters FcγRI alignment. This altered alignment likely prevents effective clustering, explaining why C01 and C04 do not activate the receptor and even block activation induced by crosslinking antibodies such as m22.
To assess the therapeutic potential of C01 and C04, it is important to determine if they can effectively displace and block IC binding. Our findings demonstrate that both hIgG and IC displacement is achievable at physiological conditions on Ba/F3-FcγRI cells, neutrophils, and monocytes (Fig. 4). The displacement effect is both time- and concentration-dependent, with an equilibrium reached at approximately 60% displacement. LigandTracer experiments revealed that C01 and C04 exhibited higher effective binding affinity towards FcγRI compared to hIgG. While the KD for hIgG1 is higher than values reported in literature (KD of 10-8 – 10-9 M47), it is important to note that these affinities were determined using Biacore systems (i.e., bound static receptors) rather than live cells. In the future, enhancing the affinity of C01 and C04 through affinity maturation could be beneficial in achieving the desired affinity level for therapeutic purposes, as affinity-matured antibodies may lead to greater displacement, resulting in improved binding efficacy and stronger blocking48.
The validation of C01 and C04 using patient-derived PBMCs further underscores their potential as therapeutic agents, particularly for ITP. Despite some variability in the data, both C01 and C04 significantly inhibited opsonized platelet binding to monocytes, achieving reductions of approximately 50% and 59%, respectively. In human immune system mice, roughly 50% residual platelet depletion was observed, confirming the blocking capacity of C01 in vivo in an IgG-driven model of autoimmunity. Of note, the question remains if ITP is fully FcγR dependent in this model, as using blocking antibodies against all three receptors does not lead to a complete block29. Hence more research is needed into the individual contributions of FcγRI, FcγRII and FcγRIII in ITP pathogenesis. Nevertheless, reduction of platelet depletion by a FcγRI-blocking antibody supports the contribution of mononuclear phagocytic cells in ITP pathogenesis as previously observed in mice49,50. All in all, these results are promising, especially considering the history of prior therapeutic antibodies, such as H22, which showed initial success ITP patients but was discontinued due to adverse side effects40. Unlike H22, C01 and C04 do not activate the receptor, reducing the risk of unintended immune activation and suggesting a safer clinical profile.
RA-specific autoantibodies known as ACPA contribute to the development of the disease51, and their presence strongly correlates with disease progression and severity52. ACPA-IC binding to myeloid cells, including monocytes and macrophages, can be inhibited up to 70% by both C01 and C04. Myeloid cells are key players in RA pathogenesis, resulting in the release of pro-inflammatory cytokines, antigen presentation and matrix degradation53. There are several RA therapies available, including TNF, IL-6 or JAK inhibitors, that suppress inflammation by targeting cytokine signaling pathways crucially involved in synovial inflammation as observed in patients with RA. However, monocytes and macrophages are not the only immune cells present in the RA compartment. Our data show that IFN-γ-stimulated neutrophils express de novo FcγRI, playing a major role in ACPA-IC binding. This interaction is particularly relevant in synovial inflammation, as RA patient-derived neutrophils show high levels of FcγRI26. Given the abundance of neutrophils in the synovial fluid and the presence of IFN-γ in this compartment, FcγRI-mediated neutrophil activation likely plays a significant role in RA pathogenesis. This interaction has been previously studied with 10.1, resulting in a slight reduction of 29% in ACPA-IC binding 28. However, in our study, we did not observe any reduction with 10.1, which may be due to our use of a more clinically relevant concentration of 25 μg/ml, whereas the previous study used 80 μg/ml.
ICs have also been implicated in inducing neuropathy in RA, leading to joint pain without apparent inflammation54. This pain can persist even with controlled inflammation through medication55. FcγRI is expressed in nociceptive DRG neurons of rats and mice, and IC activation increases intracellular calcium mobilization, resulting in heightened neuron excitability8,56,57. Previous experiments using 10.1 showed reduced pain-related behavior in RA mouse and rat models8,9, suggesting that C01 and C04 could be even more effective in blocking IC binding and thus alleviating pain. Inhibiting FcγRI could be a promising treatment for RA patients, especially for those experiencing ongoing pain despite anti-inflammatory therapy. This approach may also extend to other chronic pain disorders, such as neuropathic pain linked to somatosensory system disorders or lesions58,59.
Beyond the potential for IgG1-mediated diseases, our findings also open a promising avenue for therapeutic inventions in autoimmune diseases mediated by IgG4 autoantibodies. IgG4 is the least abundant subclass of IgG, and it can undergo Fab-arm exchange with other IgG4 antibodies, resulting in monovalent binding to its target60. IgG4-mediated autoimmunity is a distinct niche among antibody-driven disorders, as it does not activate Fc receptors but instead exerts its pathogenic effects through Fab-mediated blocking of essential protein-protein interactions61,62. Though typically anti-inflammatory, it can drive pathology when produced in response to chronic antigen exposure during inflammation, such as citrullinated protein in RA63,64. Unlike IgG1 autoantibodies, which originate from long-lived plasma cells, IgG4 autoantibodies tend to arise from short-lived plasma cells generated continuously from memory B cells62,63. Once antigenic load is reduced through effective inflammatory treatment, IgG4 ACPA levels drop whereas IgG1 levels remain relatively stable63. This ongoing production reflects persistent antigenic stimulation, and current broad-spectrum immunosuppressive treatments (e.g., corticosteroids, rituximab) often fail to achieve lasting remission60. Blocking FcγRI with C01/C04 may offer a dual therapeutic benefit: by reducing FcγRI-mediated inflammatory signaling and lowering antigenic load, it could reduce the generation of pathogenic IgG4 autoantibodies. Importantly, IgG4 also plays a regulatory role in normal immunity, particularly in modulating IgE responses. Thus, targeted strategies like C01 and C04 might be able to disrupt the pathological cycle of chronic stimulation and autoantibody production without completely depleting beneficial IgG4 functions.
In conclusion, this study is the first to describe and characterize two novel anti-FcγRI antibodies that bind within the ligand-binding domain and can block FcγRI ligand binding via their Fab. Besides being able to provide new insights in fundamental research, these antibodies also have the potential to be used as therapeutic antibodies in autoimmune and infectious diseases.
Methods
Cell lines, cell culture and antibodies
Ba/F3 (obtained from DSMZ, ACC 300, RRID: CVCL_0161), EL4 (ATCC, TIB-39, RRID: CVCL_0255), SK-BR-3 (ATCC, HTB-30, RRID: CVCL_0033), IIa1.6 (ATCC, TIB-308, RRID: CVCL_0J27) cells were cultured in RPMI 1640 medium (RPMI 1640 Glutamax; Thermo Fisher, 61870036) supplemented with 10% fetal calf serum (FCS, Bodinco), 100U/ml penicillin/streptomycin (Pen/Strep, Thermo Fisher, 15140122), and 0.2 ng/mL murine IL-3 (Immunotools, 12340035) for Ba/F3 cells. All cell lines were authenticated by the suppliers and used at low passage numbers after immediate freezing upon receipt. Additionally, they were routinely screened for mycoplasma using Myco alert Mycoplasma detection kit (Lonza, LT07-318). The retroviral vector pMX human FcγRI was generated by cloning the full-length human FCGR1A cDNA into the pMX vector backbone using standard molecular biology techniques. High-titer retrovirus was produced in packaging cells and used to transduce target cells, followed by selection and confirmation of FcγRI surface expression by flow cytometry65. All cells were transduced using amphotropic viral particles made in HEK293T cells. Puromycin selection (5 μg/ml, Merck, P8833) produced stable Ba/F3-FcγRI, Ba/F3-FcγRIIIa, Ba/F3-FcγRIIIb, EL4-FcγRI and SKBR3-FcγRI cell populations. No extra selection was needed to produce stable IIa1.6-FcγRIIa and IIa1.6-FcγRIIb. All information on antibodies used in this study are provided in Supplementary Table 3.
Production of chimeric FcγRI-specific antibodies
Seven anti-FcγRI antibodies have been generated in-house19. In short, C57BL/6 mice were immunized intravenously by cellular immunization with FcγRI expressing cells within our UMAB facility and boosted 4 times. Blood was collected and serum antibody titers for FcγRI were analyzed. The spleens were harvested 4 days after the last boost, snap-frozen and stored at −80 °C. A murine scFv antibody library was generated from total RNA of the spleen isolated from the mouse with highest serum antibody titer. Phage display was performed by two panning steps using Ba/F3-FcγRI cells. After the final panning, binding of individual phage antibodies to FcγRI was tested by whole cell ELISA using Ba/F3 and Ba/F3-FcγRI cells. The screening process was monitored by next generation sequencing (NGS) and enriched scFv clones were selected and converted into Fc-silent human IgG1 (LALAPG mutation; L234A, L235A, P329G23), via overlap extension-PCR (OE-PCR). Here, primers (Integrated DNA technologies, Supplementary Table 4) were diluted to 10 μM in sterile H20. VL and VH, and OE-PCR reactions were performed in 50 μl volumes, containing 10 μM forward and 10 μM reverse primer, 5 ng of template DNA, 10 mM dNTPS (Qiagen), 1 unit of Phusion DNA polymerase (Boike), 5X phusion HF or GC buffer (Boike). Initial denaturation was conducted at 98 °C for 30 seconds, followed by 27 cycles of 98 °C at 10 s, 55-62 °C at 20 s, 72 °C at 15 s and a final extension of 72 °C at 10 min. When required, gel purification was conducted using the nucleospin Gel and PCR clean-up kit (Macherey-nagel). For OE-PCR Reaction 1 (VL and VH), pcDNA3.4 expression vector (RRID: Addgene_221396) was used as DNA template. For OE-PCR Reaction 2 either VL or VH of interest in the Pak100 vector was used as DNA template. For OE-PCR Reaction 3 the corresponding product from OE-PCR reaction 1 and 2 was used as DNA template. The resulting sequences were ultimately transfected into ExpiCHO-S cells (ThermoFisher, A29127). Antibodies were purified using a HiTrap rProtein A FF column (GE Healthcare, 10146224) attached to the ÄKTA Start (GE Healthcare) fast protein liquid chromatography system according to the manufacturer’s protocol. The protein sequences of the antibodies are included in a related patent application (P37203NL00).
Phylogenetic tree reconstruction
Phylogenetic reconstructions of the amino acid sequences of the variable heavy and light chains were made using the function “compute” of ETE3 3.1.2 as implemented on the GenomeNet66. Alignment was performed with Clustal Omega (v1.2.4) with the default options67. A maximum-likelihood tree was inferred using PhyML (v20160115). Branch supports are the Chi2-based parametric values by the approximate likelihood ratio test68. The obtained newick files were then uploaded in the Interactive Tree Of Life (iTOL) (v6) annotation software to generate midpoint rooted phylogenetic trees69.
Chimeric domain swap receptors
All human FcγRI: mouse FcγRI domain swap chimeric receptors were constructed in the pMX_puro vector. Receptor sequences were obtained from UniProt; human FcγRI (L03418) and murine FcγRI (M31314). For the domain swap chimeric receptors, extracellular domains (EC) 1, 2, or 3 were replaced by the murine equivalent. For mECD1-hEC2-hEC3, 91.3% of hEC1 was replaced by mECD1. For hEC1-mEC2-hEC3, the whole hEC2 was replaced, including the overlap with hEC1. For hEC1-hEC2-mEC3, hEC3 could easily be replaced by mECD3.
The obtained plasmids were retrovirally transduced in HEK293T cells, the virus supernatant was harvested and added to EL4 cells. Selection was performed after 7-14 days with puromycin. To evaluate binding, 1 × 105 transduced cells were added per well, after which the antibodies were added (e.g., C01-LALAPG, C04-LALAPG, 10.1 (mIgG1), M22 (mIgG1), and hIgG1). After 1 h incubation at 4 °C, a labeled secondary staining antibody (anti-mouse IgG or anti-human IgG) was added. Binding was evaluated via flow cytometry and analyzed with BD FACSDiva (v9.0).
Flow cytometry assays
For all Fluorescence-Activated Cell Sorting (FACS) assays, 1 × 105 cells/well were added to a 96-well plate. All incubation steps were performed for 1 h at 4 °C, unless stated otherwise. In each experiment, 10 μg/ml (labeled) antibody was used, unless stated otherwise. After each incubation step, cells were washed with 100 μL FACS buffer (PBS + 0.5 % bovine serum albumin (BSA, Sigma, 10735094001) + 0.1% sodium azide (Immunosource, 40-2010-01)) and spun down at 1500 RPM for 5 min, unless stated otherwise. After final staining, cells were washed once with PBS, fixed with 1% paraformaldehyde (PFA) for 15 min at 4 °C and resuspended in FACS buffer. All measurements were performed in duplo or triplo using a FACSCanto II (BD Sciences) and accompanying software (FACS DIVA, v8) and FlowJo (v10.10). An exemplary gating strategy is provided in Supplementary Fig. 2a.
Commercial antibodies ligand binding and blocking assay
Ba/F3-FcγRI and EL4-FcγRI cells were incubated with the indicated anti-FcγRI antibodies for 1 hour. After washing, mouse or human IgG bound to the cells was detected by using APC-labeled anti-mouse IgG or AF647-labeled anti-human IgG for 1 hour. To measure the ligand blocking capacity, 5 μg/ml hIgG1-AF647 was added for 1 h after the first incubation step. Ligand blocking (%) was calculated by using the formula: Ligand blocking (%) = 100 – ((MFI sample / MFI ligand only) x 100). For 197 and 10.1, F(ab’)2 fragments were generated to test Fab-mediated interactions. Immobilized Ficin was used to prepare the F(ab’)2 fragments using Pierce Mouse IgG1 Fab and F(ab´)2 Preparation Kit (Thermo Scientific, 44980). For both antibodies, 1 mg was digested with 4 mM cysteine for 28 h at 37 °C, following manufacturer’s protocol. Purity was confirmed via SDS-PAGE gel electrophoresis (reducing condition, 4-20% gradient gel, Bio-Rad, 4561094) and stained using InstantBlue Ultrafast Protein Stain (Sigma Aldrich, ab119211).
Antibody binding assay
For antibody binding assays, 1 × 105 Ba/F3-FcγRI or EL4-FcγRI cells per well were added to a 96-well plate and pre-labeled commercial anti-FcγRI antibodies or FITC-labeled antibodies were added. For the titration experiments, cells were incubated with the different antibodies at the indicated concentrations for the specified time. For the specificity assay, IIa1.6, IIa1.6-CD32a, IIa1.6-CD32b, Ba/F3, Ba/F3-CD16a and Ba/F3-CD16b were incubated with a titration range of C01-FITC, C04-FITC, anti-CD16-PE (clone 3G8; BD Pharmingen) and anti-CD32-FITC (clone FL18.26; BD Pharmingen). For antibody binding in presence of plasma IgGs, human peripheral blood mononuclear cells (PBMCs) were isolated from donor blood (in-house donor facility, University Medical Center Utrecht) and Ba/F3-FcγRI cells were first incubated with 20 µl of 200 µg/ml intravenous immunoglobulins (IVIg) (Nanogam, Sanquin, RVG31627), and then with FITC-labeled anti-FcγRI antibodies. For cross-block assays, Ba/F3-FcγRI cells were pre-incubated with either unlabeled 10.1 or anti-FcγRI clones. Next, FITC-labeled anti-FcγRI clones or 10.1 was added to measure residual binding.
Expression and purification of C01 Fab and rhFcγRI
Codon optimized C01 heavy and light chain V gene sequences ordered from TWIST Bioscience were designed with either only the constant heavy 1 (CH1) or constant light (CL) domains but including the ultimate cysteine residues implicated in Fab conjugation. Constructs ligated into the pcDNA3.4 expression vector (RRID: Addgene_221396) were transfected into the ExpiCHO expression system (Thermo Fisher, A29133). Culture supernatants were harvested 10 days post-transfection and purified subsequently on a Kappa Select column, equilibrated in PBS and using 0.1 M glycine pH 2.5 as eluent, and a Superdex75 16/600 column in 10 mM HEPES-buffer pH 7.5 with 150 mM NaCl. Recombinant production of FcγRI ectodomains in ExpiCHO entailed the previously published construct including 19 stabilizing mutations and fusion to a C-terminal His6-tag46. Protein purification comprised of an IMAC affinity step on a 1 mL HisTrap HP column equilibrated in 25 mM HEPES-buffer pH 7.5 with 0.5 M NaCl and using wash and elution buffers containing 50 and 100 mM Imidazole pH 7.5, respectively, followed by a cation exchange step with a 1 mL ResourceS column in 25 mM MES-buffer and elution using a 0.1–1 M NaCl gradient25. As a final step prior to crystallization, the C01 Fab fragment and rhFcγRI were mixed in a 1:1 ratio to a final concentration of 3 mg/mL and purified on a Superdex200 Increase 10/300 column equilibrated in 10 mM HEPES-buffer pH 7.5 with 150 mM NaCl, to separate the complex from the individual components. All columns used in this protocol were obtained from Cytiva LifeSciences.
Crystallization and structure determination of the Fab C01-rhFcγRI complex
Crystallization trials were set up in sitting drops vapor diffusion screens by mixing 100 nl 10 mg/mL Fab C01-rhFcγRI complex in 10 mM HEPES-buffer pH 7.5 with 150 mM NaCl and 100 nl of reservoir solution, at 293 K. Fab C01-rhFcγRI crystals were grown during 2 months using the Morpheus Crystallization screen, in a condition containing the MOPS/HEPES buffer system, pH 7.5, alcohol mixture (0.12 M end conc.), and the PEG20k-PEG MME 550 precipitant mixture (30 % end concentration)70. X-Ray diffraction data to 3.2 Å resolution of two separate crystals grown in the same drop were collected at the Diamond Light Source at beamline I24, processed individually with the xia2 multiplex protocol, and subsequently merged and scaled with Aimless71,72. The structure solution by molecular replacement in Phaser using models of hFcγRI (PDB-ID: 4W4O) and mFab VH, VL, CH & CL domains (PDB: 1i8m) as search templates revealed the presence of two Fab C01-fhFcγRI complexes in the asymmetric unit. The model was further improved by manual modeling in Coot25,73,74,75. During the refinement in Refmac5, external restraints were used, that were generated by Lowrestr against experimentally determined structures of FcγRI homologs and Fab fragments, and alphafold 2 models76,77,78. Quality of the geometry was analyzed using MolProbity79, showing 94.08% favored and 5.27% allowed backbone geometries according to Ramachandran’s statistics, as well as 81.7% favored and 10.42% allowed rotamer conformations. Final Rwork/Rfree statistics are 0.261 / 0.301. Data collection and refinement statistics are listed in Supplementary Table 1. All programs were used as implemented in CCP480.
Ligand tracer assay
The binding kinetics of hIgG, 10.1, C01 and C04 to FcγRI were examined by using LigandTracer Green Technology (Ridgeview Instruments AB). SKBR3 and SKBR3-FcγRI were plated out on opposite sides of a culture dish in an elliptical shape, at a concentration of 8 × 104 cells/ml and incubated overnight at 37 °C to attach to the plate. Next, the plate was washed with culture medium and transferred to the LigandTracer apparatus. First, a baseline measurement of 15 min was performed. Subsequently, association was measured at 10 nM FITC-labeled antibody for 1 h and subsequently at 30 nM for 1 h. Then, dissociation was measured for 2 h by replacing the culture medium containing 30 nM antibody for antibody-free medium. The effective affinity of the antibodies was calculated by using a OneToOneBI fitting model in TraceDrawer (v1.9.2) software (Ridgeview Instruments AB).
Calcium release assay
For the calcium release assays, 5 × 105 Ba/F3-FcγRI cells per well were incubated with 8 μM Fluo-4 AM (Invitrogen, F14201) and 10 μM Fura-Red (Invitrogen, F3021) at 37 °C for 20 min. Cells were removed from the incubator, anti-FcγRI antibodies m22 (own production), H22 (MDX-33, Antibody System), C01 and/or C04 were added (10 μg/ml) and the cells were incubated at 37 °C for an additional 10 min. Cells were washed once with RPMI without phenol red + 1% P/S + 10% FCS and once with Hank’s Balanced salt solution (HBSS, Gibco, 15266355) containing magnesium, calcium and no phenol red, supplemented with 10% FCS. Cells were resuspended in HBSS + 10% FCS and transferred to a FACS tube in a final volume of 500 μL. The cells were left undisturbed in the dark at RT for at least 60 min. Cytosolic calcium levels were measured on FACSCanto II (BD Biosciences) as ratio of Fluo-4/Fura Red. After 30 seconds of baseline measurement, a cross-linking antibody (goat anti-mouse IgG; Jackson, 115-007-003, or rabbit anti-human IgG; Jackson, 309-006-008) was added and calcium release was measured for 4.5 min. Lastly, 2 μg/mL ionomycin (Sigma, I0634) was added as a loading control and measured for 45 seconds.
Superoxide induction
IFN-γ stimulated neutrophils were used for measuring superoxide burst by anti-FcγRI antibodies. Neutrophils from 3 healthy donors were cultured overnight with 100 U/mL IFN-γ (Immunotools, 11343534) and 100 U/ml G-CSF (Immunotools, 11343133) in RPMI with 10% FCS and 1% P/S. All next steps were performed on ice to limit superoxide burst. 197, m22, H22, 10.1, C01 and C04 were added at 10 µg/ml in 50 µl, after which 4 × 105 cells in 100 µl per well were added. Lastly 150 µM luminol (Sigma) in 50 µl was added and placed a luminometer (SpectraMax M3, Molecular Devices) set at 37 °C. Measurements of spontaneous production of superoxide were made every 40 to 60 seconds, starting immediately following luminol addition. The production of superoxide was measured in relative light units (RLU) for 20 min.
Soluble IC binding and blocking assay
Soluble IC was formed by incubating 10 µg/ml hIgG1 (Palivizumab; Synagis, 66658-230-01) with 5 µg/ml AF647-Goat F(ab’)2 anti-human IgG F(ab’)2 (Jackson, 109-605-006) for 30 min at 37 °C, followed by 10 min incubation on ice. 1 × 105 Ba/F3-FcγRI cells were added per well. Anti-FcγRI was added (25 µl per well at 5, 10 or 25 µg/ml) followed by 45 min incubation on ice. Next, hIgG1-AF647 or soluble IC was added (25 µl per well) (no washing step in between), followed by 45 min incubation on ice. Percentage binding was calculated using the following formula: Binding (%) = (sample – negative control)/(baseline – negative control)*100%. Negative control is the unstained condition. Baseline is the condition where only hIgG-AF647 or soluble IC was added and no blocking antibody.
hIgG and soluble IC displacement assay
Soluble IC were generated by incubating 10 µg/ml hIgG1 (Palivizumab; Synagis, 66658-230-01) with 5 µg/ml AF647-Goat F(ab’)2 anti-human IgG F(ab’)2 (Jackson, 109-605-006) for 30 min at 37 °C, followed by 10 min incubation on ice. Antibodies and IC were dissolved in HEPES-buffered saline + 1 mM MgCl2 + 1,25 mM CaCl2 + 0,2 ng/ml murine IL-3 or (for overnight incubation) RPMI 1640 without phenol red (Gibco, 11835030) + 10% FCS + 0,2 ng/ml murine IL-3. 1 × 105 cells (Ba/F3 or Ba/F3-FcγRI cells) were added per well. Palivizumab-AF647 (in-house labeled), or soluble IC was added (25 µl per well) followed by 45 min incubation on ice. Next, anti-FcγRI antibody was added (25 µl per well) (no washing step in between) followed by 45 min or overnight incubation at 37 °C. Subsequent preparation for measurement, analysis, and calculations were done similarly as for blocking experiments, with the exception that the negative control used in the calculation is the condition where only the AF647-Goat F(ab’)2 anti-human IgG F(ab’)2 was added. For the displacement assays, polymorphonuclear leukocytes (PMNs) or PBMCs from 3 healthy donors were isolated by diluting whole blood 1:1 with PBS and layering this mixture on Ficoll Paque Plus (GE Healthcare, 17‑1440‑03). After careful centrifuging, PBMCs were collected and monocytes were isolated via CD14 + MACS Cell Separation (Miltenyi Biotec, 130‑050‑201). The remaining serum and Ficoll layer were removed to have the PMN layer left. The cells were cultured overnight in either RPMI with human serum or FCS in a 6-wells plate at a density of 2 million/ml. Additionally, neutrophils were stimulated with 100U/ml IFN-γ (Immunotools, 11343534) and G-CSF (Immunotools, 11343133) and the monocytes with 100U/ml GM-CSF (Immunotools, 11343125).
Rosette (IgG-coated beads) assay
Dinitrophenol (DNP)-BSA (Thermo Fisher, A-8020) was coupled to Dynabeads M-450 Epoxy (4,5 µm) (Thermo Fisher, 14012D) according to manufacturer’s instructions. DNP-BSA Dynabeads were opsonized with rabbit IgG anti-DNP (Vector Laboratories, SP-0603-1). Different coating antibody concentrations were evaluated to maximize bead-to-cell binding and the best binding was obtained at 1 µg/ml, and this concentration was deemed sufficient for subsequent blocking tests (1 × 108 beads per ml antibody solution). Per well, 1 × 105 Ba/F3-CD64 cells were incubated with 50 µl of 1-100 μg/ml blocking antibody for 45 min at 4 °C. After washing, 3.5 × 105 beads were added and incubated for 1 h at 4 °C while shaking. Next, cells were fixated with 3% PFA (Klinipath) and spun down at 1500 RPM for 2 min, no brake. Rosette formation was evaluated using a bright-field microscope (EVOS, Thermo Fisher) at 20x magnification. Cells with five or more beads bound to them were defined as rosettes. Counting was performed using ImageJ v2.11 (Fiji) software. For each condition, triplicates were measured. To be able to compare different experiments, the percentage of rosettes in each condition was calculated to the baseline percentage without blocking antibody.
Patients and healthy donors
A peripheral blood sample was obtained from an ACPA-positive RA patient visiting the outpatient clinical of the department of Rheumatology at Leiden University Medical Center (LUMC, Leiden, The Netherlands). Informed consent was obtained for this individual and all protocols were approved by the ethical committee of the LUMC, the Netherlands. Samples from patients with ITP were derived from the PICI study (trial number NL66313.041.18), a cross-sectional observational study amongst patients with chronic ITP approved by the NedMEC medical ethics committee. All patients gave written informed consent. Healthy volunteers were recruited amongst hospital staff and students. Ethical permission was obtained (protocol number 18-774) and all donors provided written informed consent.
Patient-derived ACPA-IC assay
ACPAs were isolated from RA patient (see patient section) plasma by affinity chromatography using the ÄKTA Pure equipped with a self-packed HiTrap streptavidin HP 1 mL column (Cytiva, 17-5112-01) with biotinylated CCP4 peptides or biotinylated CArgP4 peptides81. The peptides were kindly provided by Dr. J. W. Drijfhout (Dept. of IHB, LUMC). Additionally, we used a recombinant non-glycosylated (NG) monoclonal ACPA-IgG1, 7E4, which were produced based on the BCR sequences from ACPA + RA patients82. In this case the sequence of 7E4 was provided by Dr Rispens, Sanquin, The Netherlands. Since ACPA express variable domain glycans, the glycan sites were mutated out by back-mutating the N-linked glycosylation sites into the germline sequence (based on IMGT). Codon optimized ACPA heavy chain and light chain V gene sequences were designed and ordered from GeneArt (Life Technologies) and ligated into a pcDNA3.1 (+) expression vector (Invitrogen, V79020). The heavy and light chain vectors were transfected into Freestyle 293-F cells (Gibco, R79007). After 5-6 days the supernatant was harvested and purified for ACPA IgG with a 1 ml HiTrap Protein G HP affinity column (GE Healthcare, 29-0485-81) according to the manufacturer’s instructions.
PMNs or PBMCs from 3 healthy donors were isolated by diluting whole blood 1:1 with PBS and layering this mixture on Ficoll Paque Plus (GE Healthcare, 17-1440-03). After careful centrifuging, PBMCs were collected, after which the remaining serum and Ficoll layer were removed to have the PMN layer left. For the monocytes, the whole PBMC population was used. For the macrophage stimulation, the PBMCs were plated in a NUNC Upcell 6-well plate (Thermo Fisher, 174901) at a density of 5 million cells/mL in 3 mL. The monocytes were left to attach to the plate for 3 h, after which the wells were washed 2 times to remove any unbound non-monocytic cell. RPMI medium with 10% FCS and 1% P/S was added, together with 50 ng/ml GM-CSF (Immunotools, 11343125) to induce differentiation of the monocytes into M1 macrophages. Every 3 days medium was replaced and fresh cytokines added. On day 7 100 U/ml IFN-γ (Immunotools, 11343534) was added to induce polarization and left on the cells for 24 h, after which the cells were ready for FACS. To lyse the erythrocytes within the PMN fraction, 1x RBC lysis buffer (Biolegend, 420302) was added twice, after which the PMNs were washed with PBS. PMNs were stimulated overnight with 100 U/ml IFN-γ (Immunotools, 11343534) and G-CSF (Immunotools, 11343133) to induce FcγRI expression. The PMNs consist mainly of neutrophils and are therefore referred to neutrophils in the main text.
Neutrophils, PBMCs or macrophages were seeded at a density of 1 × 105 cells/well, after which the blocking antibodies (C01, C04 or 10.1 F(ab’)2) or PBS (25 µl/well) were added and incubated for 30 min at 37 °C. ICs were formed using 2.5 μg/ml fluorescently labeled goat-derived polyclonal F(ab’)2-anti-hIgG-F(ab’)2 (Jackson, 109-606-006) with either 5 µg/ml recombinant monoclonal ACPA antibodies (mACPA) or with patient-derived polyclonal ACPA (pACPA) for 30 min at 37 °C and then 10 min on ice. Cells were washed with FACS buffer and 10 µg/ml mACPA-IC or pACPA-IC (10 µg/ml, 25 µl/well) was added and incubated for 1 h at 37 °C. All subsequent steps were performed on ice. For both monocytes and macrophages, cells were stained with live/dead marker eF780 (Fisher Scientific, 13539140), after which CD14-PE (BD) for 30 min at 4 °C, afterwards cells were washed and fixated in 1% PFA. For neutrophils, cells were washed with cold FACS buffer and cold staining mix was added and incubated for 30 min at 4 °C. The staining mix included CD15 (eBioscience), CD16 (FcγRIII, BD), CD32 (FcγRII, Miltenyi), and CD66b (BD) in FACS buffer. Cells were washed once more with cold FACS buffer and resuspended in cold FACS buffer and kept on ice. 15 min before measuring the sample, 7-AAD was added as a live/dead marker. Cells were first gated on live cells and then neutrophils were gated as CD15+ and CD66b + . Percentage blocking per donor is calculated using the following formula: Blocking (%) = 100 – (((sample – negative control) / (ACPA-IC only – negative control)) x 100%). Several controls were taken along to assess changes in FcγRI (clone C01), FcγRII (clone IV.3) and FcγRIII (clone 3G8) on unstimulated and IFN-γ stimulated neutrophils.
Blocking assay opsonized platelets and ITP patient-derived PBMCs
Platelet-rich plasma (PRP) was prepared from whole blood of a healthy donor and adjusted to 200.000 platelets/µL. PRP was diluted 1:1 with PBS and incubated for 30 min at 4 °C with 10 µg of GPIIbIIIa-APC (AssayLite, 11587-05061) antibody to opsonize the platelets. Meanwhile, PBMCs from six ITP patients (see patients section) were carefully thawed and plated at 2 × 105 cells per well. C01, C04 or 10.1 were added and incubated for 30 min at 37 °C to block FcγRI. After washing the PBMCs, the opsonized platelets were added at a physiological ratio of 1:250 PBMC/platelet and incubated for 1 h at 37 °C. The wells were carefully washed once with PBS, and a staining mix of anti-CD45-Pe-Cy7 (BD, 560915), anti-CD14-eF450 (eBioscience, 48‑0149‑42) and the live/dead marker eF780 (Fisher Scientific, 13539140) were added for 20 min at 4 °C. Cells were carefully washed once more, after which the cells were fixated in 1% PFA for 15 min and measured on a BDCanto II.
Generation of human immune system mice
NOD- (001976), SCID- (001303), and γc-deficient (005557) mice were originally obtained from The Jackson Laboratory. Mice lacking the fcer1 gene (FcRγ − /−) were provided by Jeffrey Ravetch (Rockefeller University)29. The NOD-SCID/γc-FcRγ − /− (NSG-FcRγ − /−) mice were generated by crossing SCID, γc-deficient, and FcRγ − /− mice onto a NOD background for a minimum of six generations, followed by intercrossing to generate NSG-FcRγ − /− animals29. In line with their immunodeficient status, the NSG-FcRγ − /− mice received acidified drinking water (pH 3.0) to reduce the risk of bacterial infection. Human immune system (humanized) NSG-FcRγ−/− mice were generated by transplanting human hematopoietic stemc cells (HSCs) into irradiated newborn mice29. Specifically, newborn NSG-FcRγ−/− mice were irradiated at the dose of 1.4 Gy and injected intravenously with 30,000–50,000 human HSCs 4–6 h after irradiation83,84, purified from umbilical cord blood with the written consent of patients and according to the clinical guidelines of the Friedrich-Alexander-University Erlangen-Nürnberg and the Klinikum Fürth (license number: 22-272B). The reconstitution efficiency and leukocyte composition were determined by flow cytometric identification of human cell subsets in whole-blood samples using a Cytek Northern Lights flow cytometer. Within alive (ZombieNIR-, Biolegend, 423105), single cells, the following populations were identified: murine leukocytes (mouse CD45+, clone A20, Biolegend) human leukocytes (human CD45+, clone HI30, Cytek), human B cells (CD19, clone HIB19, Cytek), T cells (CD3+, clone SK7, Cytek), NK cells (CD56+, clone 5.1H11, Cytek), neutrophils (CD66b+, clone G10F5, Biolegend) and monocytes (CD33+, clone P67.6, Cytek) further divided into classical (CD14+, clone M5E2, Cytek) and non-classical monocytes (CD16+, clone 3G8, Cytek). Mice with >5% human CD45+ cells were considered to be successfully humanized and used in ITP experiments.
Both male and female mice (age 12–20 weeks) were used, with groups matched for sex, age, and body weight. No sex-based analysis was performed, as the study employed a short, 4 h ITP model, where sex-specific differences are not expected and have not been reported in prior studies. All experiments were performed with male and female humanized mice at the age of 12 to 20 weeks. Mice were kept in the animal facilities of Friedrich-Alexander-University Erlangen-Nürnberg under specific-pathogen-free conditions in isolated ventilated cages. The housing environment was maintained at 20–24°C with 45–65% humidity and a 12-hour light/dark cycle. Mice were euthanized in CO2. All animal experiments were performed in accordance with the legal requirements of Germany, and were approved by the local experimental animal welfare body, Regierung von Unterfranken (54-2532.1-8/13).
In vivo ITP Induction
ITP was induced by intraperitoneal injection of 100 μg 6A6-hIgG1 antibodies (GenScript) (N = 5). To block human FcγRI, 100 µg of C01 antibody was injected intravenously 2 h prior to 6A6-hIgG1 application. PBS-treated mice (N = 4) were used as control. Platelet counts were determined before and 4 h after antibody injection, at a 1:10 dilution in PBS with an Advia 120 hematology system (Siemens). Platelet counts before antibody injection were set to 100%.
Statistical analysis
Statistical analysis was performed using GraphPad Prism v10.4.0. An unpaired Student’s t-test was used to compare the mean values between the two groups. Statistical analysis for multiple comparison was performed using one- or two-way ANOVA, with Bonferroni correction hoc test or Tukey’s test as indicated in the respective figure legend. Graphs represent mean +- SEM, unless indicated otherwise.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Raw numerical data for charts and graphs are provided in the Source Data file whenever possible. The sequences, PDB files, and materials for antibodies C01 and C04 are currently not publicly available due to their inclusion in a related patent application (P37203NL00, submitted March 2025), which is expected to become public in September 2026. Nevertheless, the C01 and C04 antibodies, their sequences, and/or the PDB file will be made available for non-commercial research purposes upon request, subject to a Material Transfer Agreement (MTA) to protect intellectual property rights. Access to these materials can be requested by contacting Jeanette H.W. Leusen. Requests will be reviewed promptly, with an expected response timeframe of 2–4 weeks. Materials will be made available for a period of at least two years following approval. These access restrictions are necessary due to ongoing intellectual property protections but do not preclude academic research use. No ethical or consent restrictions limit data sharing. Source data are provided with this paper.
References
Takai, T. Fc receptors and their role in immune regulation and autoimmunity. J. Clin. Immunol. 25, 1–18 (2005).
Reefman, E. et al. Opsonization of late apoptotic cells by systemic lupus erythematosus autoantibodies inhibits their uptake via an Fcγ receptor-dependent mechanism. Arthritis Rheum. 56, 3399–3411 (2007).
Ioan-Facsinay, A. et al. FcγRI (CD64) contributes substantially to severity of arthritis, hypersensitivity responses, and protection from bacterial infection. Immunity 16, 391–402 (2002).
Holtrop, T., Budding, K., Brandsma, A. M. & Leusen, J. H. W. Targeting the high affinity receptor, FcγRI, in autoimmune disease, neuropathy, and cancer. Immunother. Adv. 2, 1–11 (2022).
Lu, L. L., Suscovich, T. J., Fortune, S. M. & Alter, G. Beyond binding: Antibody effector functions in infectious diseases. Na. Rev. Immunol. 18, 46–61 (2018).
Rombouts, Y. et al. Anti-citrullinated protein antibodies acquire a pro-inflammatory Fc glycosylation phenotype prior to the onset of rheumatoid arthritis. Ann. Rheum. Dis. 74, 234–241 (2015).
Di Ceglie, I., Kruisbergen, N. N. L., Van Den Bosch, M. H. J. & Van Lent, P. L. E. M. Fc-gamma receptors and S100A8/A9 cause bone erosion during rheumatoid arthritis. Do they act as partners in crime?. Rheumatol. (U. Kingd.) 58, 1331–1343 (2019).
Wang, L. et al. Neuronal FcγRI mediates acute and chronic joint pain. J. Clin. Investig. 129, 3754–3769 (2019).
Liu, F. et al. Fcγ Receptor I–coupled signaling in peripheral nociceptors mediates joint pain in a rat model of rheumatoid arthritis. Arthritis Rheumatol. 72, 1668–1678 (2020).
Qu, L. & Qu, L. Neuronal Fc gamma receptor I as a novel mediator for IgG ⁎. Neural Regen. Res 7, 2075–2079 (2012).
Dougherty, G. J., Selvendran, Y., Murdoch, S., Palmer, D. G. & Hogg, N. The human mononuclear phagocyte high-affinity Fc receptor, FcRI, defined by a monoclonal antibody, 10.1. Eur. J. Immunol. 17, 1453–1459 (1987).
Graziano, R. F. et al. Construction and characterization of a humanized anti-gamma-Ig receptor type I (Fc gamma RI) monoclonal antibody. J. Immunol. 155, 4996–5002 (1995).
Valerius, T. et al. Involvement of the high-affinity receptor for IgG (Fc gamma RI; CD64) in enhanced tumor cell cytotoxicity of neutrophils during granulocyte colony-stimulating factor therapy. Blood 82, 931–939 (1993).
Mladenov, R. et al. CD64-directed microtubule associated protein tau kills leukemic blasts ex vivo. Oncotarget 7, 67166–67174 (2016).
Hristodorov, D. et al. Targeting CD64 mediates elimination of M1 but not M2 macrophages in vitro and in cutaneous inflammation in mice and patient biopsies. MAbs 7, 853–862 (2015).
Bevaart, L. et al. Direct targeting of genetically modified tumour cells to FcγRI triggers potent tumour cytotoxicity. Br. J. Haematol. 132, 317–325 (2006).
Ericson, S. G. et al. Monoclonal antibody 197 (anti-FcγRI) infusion in a patient with immune thrombocytopenia purpura (ITP) results in down-modulation of FcγRI on circulating monocytes. Br. J. Haematol. 92, 718–724 (1996).
Woolhiser, M. R., Okayama, Y., Gilfillan, A. M. & Metcalfe, D. D. IgG-dependent activation of human mast cells following up-regulation of FcγRI by IFN-γ. Eur. J. Immunol. 31, 10.1002/1521-4141(200111)31 (2001).
Steffen, K. et al. Combining cellular immunization and phage display screening results in novel, FcγRI-specific antibodies. Viruses 16, https://doi.org/10.3390/v16040596 (2024).
Lee, C. M. Y., Iorno, N., Sierro, F. & Christ, D. Selection of human antibody fragments by phage display. Nat. Protoc. 2, https://doi.org/10.1038/nprot.2007.448 (2007).
Lu, J., Ellsworth, J. L., Hamacher, N., Oak, S. W. & Sun, P. D. Crystal structure of Fcγ receptor I and its implication in high affinity γ-immunoglobulin binding. J. Biol. Chem. 286, 40608–40613 (2011).
Vance, B. A., Karlson, K. H., Morganelli, P. M. & Guyre, P. M. Single step screening of monoclonal antibodies against interferon-γ-induced surface molecules on human monocytes. J. Immunol. Methods 118, https://doi.org/10.1016/0022-1759(89)90018-5 (1989).
Schlothauer, T. et al. Novel human IgG1 and IgG4 Fc-engineered antibodies with completely abolished immune effector functions. Protein Eng., Des. Selection 29, 457–466 (2016).
Harrison, P. T. & Allen, J. M. High affinity IgG binding by FcgammaRI (CD64) is modulated by two distinct IgSF domains and the transmembrane domain of the receptor. Protein Eng., Des. Selection 11, 225–232 (1998).
Kiyoshi, M. et al. Structural basis for binding of human IgG1 to its high-affinity human receptor FcγRI. Nat. Commun. 6, 6866 (2015).
Laurent, L. et al. Fcγ receptor profile of monocytes and macrophages from rheumatoid arthritis patients and their response to immune complexes formed with autoantibodies to citrullinated proteins. Ann. Rheum. Dis. 70, https://doi.org/10.1136/ard.2010.142091 (2011).
O’Neil, L. J. & Kaplan, M. J. Neutrophils in Rheumatoid Arthritis: Breaking Immune Tolerance and Fueling Disease. Trends in Molecular Medicine. 25, 215–227 (2019).
Kempers, A. C. et al. Fc gamma receptor binding profile of anti-citrullinated protein antibodies in immune complexes suggests a role for FcγRI in the pathogenesis of synovial inflammation. Clin. Exp. Rheumatol. 36, 284–293 (2018).
Schwab, I., Lux, A. & Nimmerjahn, F. Pathways Responsible for Human Autoantibody and Therapeutic Intravenous IgG Activity in Humanized Mice. Cell Rep 13, https://doi.org/10.1016/j.celrep.2015.09.013 (2015).
Nimmerjahn, F. & Ravetch, J. V. Immunology: Divergent immunoglobulin G subclass activity through selective Fc receptor binding. Science (1979) 310, https://doi.org/10.1126/science.1118948 (2005).
Lu, J. et al. Structure of fcγri in complex with Fc reveals the importance of glycan recognition for high-affinity gG binding. Proc. Natl Acad. Sci. USA 112, 833–838 (2015).
Soubrane, C. et al. Biologic response to anti-CD16 monoclonal antibody therapy in a human immunodeficiency virus-related immune thrombocytopenic purpura patient. Blood 81, 15–19 (1993).
Clarkson, S. B. et al. Treatment of Refractory Immune Thrombocytopenic Purpura with an Anti-Fcγ-Receptor Antibody. N. Engl. J. Med. 314, https://doi.org/10.1056/NEJM198605083141907 (1986).
Kaneko, Y., Nimmerjahn, F. & Ravetch, J. V. Anti-inflammatory activity of immunoglobulin G resulting from Fc sialylation. Science (1979) 313, https://doi.org/10.1126/science.1129594 (2006).
Anthony, R. M. & Nimmerjahn, F. The role of differential IgG glycosylation in the interaction of antibodies with FcγRs in vivo. Curr. Opin. Organ Transplant. 16, https://doi.org/10.1097/MOT.0b013e328342538f (2011).
Kurlander, R. J. Blockade of Fc receptor-mediated binding to U-937 cells by murine monoclonal antibodies directed against a variety of surface antigens. J. Immunol. 131, 140–147 (1983).
Hogarth, P. M. & Pietersz, G. A. Fc receptor-targeted therapies for the treatment of inflammation, cancer and beyond. Nat. Rev. Drug Discov. 11, 311–331 (2012).
Getahun, A. & Cambier, J. C. Of ITIMs, ITAMs, and ITAMis: Revisiting immunoglobulin Fc receptor signaling. Immunol. Rev. 268, 66–73 (2015).
Serezani, C. H., Aronoff, D. M., Sitrin, R. G. & Peters-Golden, M. FcγRI ligation leads to a complex with BLT1 in lipid rafts that enhances rat lung macrophage antimicrobial functions. Blood 114, https://doi.org/10.1182/blood-2009-01-199919 (2009).
Terjanian, T. et al. A proof-of-concept phase 2 study of monoclonal antibody MDX-33 in adult subjects with chronic stable ITP. Blood, 96 (2000).
Duncan, A. R., Woof, J. M., Partridge, L. J., Burton, D. R. & Winter, G. Localization of the binding site for the human high-affinity fc receptor on igg. Nature 332, 563–564 (1988).
Chong, G. & MacKerell, A. D. Spatial requirements for ITAM signaling in an intracellular natural killer cell model membrane. Biochim. Biophys. Acta. Gen. Subj. 1866, 130221 (2022).
Call, M. E., Wucherpfennig, K. W. & Chou, J. J. The structural basis for intramembrane assembly of an activating immunoreceptor complex. Nat. Immunol. 11, 1023–1029 (2010).
Patel, K. R., Roberts, J. T. & Barb, A. W. Multiple Variables at the Leukocyte Cell Surface Impact Fc γ Receptor-Dependent Mechanisms. Front. Immunol. 10, 223 (2019).
Hui, G. K. et al. The solution structure of the unbound IgG Fc receptor CD64 resembles its crystal structure: Implications for function. PLoS One 18, e0288351 (2023).
Asaoka, Y., Hatayama, K., Tsumoto, K., Tomita, M. & Ide, T. Engineering of recombinant human Fcγ receptor i by directed evolution. Protein Eng. Des. Sel. 25, 835–842 (2012).
Bruhns, P. et al. Specificity and affinity of human Fcγ receptors and their polymorphic variants for human IgG subclasses. Blood 113, 3716–3725 (2009).
Chan, D. T. Y. & Groves, M. A. T. Affinity maturation: highlights in the application of in vitro strategies for the directed evolution of antibodies. Emerg. Top. Life Sci. 5, 601–608 (2021).
Wöhner, M. et al. Tissue niche occupancy determines the contribution of fetal- versus bone-marrow-derived macrophages to IgG effector functions. Cell Rep 43, https://doi.org/10.1016/j.celrep.2024.113757 (2024).
Biburger, M. et al. Monocyte Subsets Responsible for Immunoglobulin G-Dependent Effector Functions In Vivo. Immunity 35, https://doi.org/10.1016/j.immuni.2011.11.009 (2011).
Catrina, A. I., Svensson, C. I., Malmström, V., Schett, G. & Klareskog, L. Mechanisms leading from systemic autoimmunity to joint-specific disease in rheumatoid arthritis. Nat. Rev. Rheumatol. 13, 79–86 (2017).
Van Gaalen, F. A. et al. Autoantibodies to Cyclic Citrullinated Peptides Predict Progression to Rheumatoid Arthritis in Patients with Undifferentiated Arthritis: A Prospective Cohort Study. Arthritis Rheum 50, https://doi.org/10.3390/cells10081860 (2004).
Roszkowski, L. & Ciechomska, M. Tuning monocytes and macrophages for personalized therapy and diagnostic challenge in rheumatoid arthritis. Cells 10, https://doi.org/10.3390/cells10081860 (2021).
De Hair, M. J. H. et al. Features of the synovium of individuals at risk of developing rheumatoid arthritis: Implications for understanding preclinical rheumatoid arthritis. Arthritis and Rheumatol. 66, https://doi.org/10.1002/art.38273 (2014).
Taylor, P. et al. Patient perceptions concerning pain management in the treatment of rheumatoid arthritis. J. Int. Med. Res. 38, (2010).
Qu, L., Zhang, P., LaMotte, R. H. & Ma, C. Neuronal Fc-gamma receptor I mediated excitatory effects of IgG immune complex on rat dorsal root ganglion neurons. Brain Behav. Immun. 25, 1399–1407 (2011).
Andoh, T. & Kuraishi, Y. Direct action of immunoglobulin G on primary sensory neurons through Fc gamma receptor I. FASEB J.: Off. Publ. Federation Am. Societies Exp. Biol. 18, 182–184 (2004).
Liang, Y., Zhang, Z., Juan, Z., Zhang, R. & Zhang, C. The high-affinity IgG receptor FcγRI modulates peripheral nerve injury-induced neuropathic pain in rats. Mol. Brain 12, 10–13 (2019).
Colloca, L. et al. Neuropathic pain. Nat. Rev. Dis. Primers. 3, 17002 (2017).
Koneczny, I. Update on IgG4-mediated autoimmune diseases: New insights and new family members. Autoimmunity Reviews 19, 102646 (2020).
Ünlü, S. et al. Exploring the depths of IgG4: insights into autoimmunity and novel treatments. Front Immunol 15, https://doi.org/10.3389/fimmu.2024.1346671 (2024).
Huijbers, M. G., Plomp, J. J., van der Maarel, S. M. & Verschuuren, J. J. IgG4-mediated autoimmune diseases: a niche of antibody-mediated disorders. Ann. N. Y. Acad. Sci. 1413, 92–103 (2018).
Bos, W. H. et al. Preferential decrease in IgG4 anti-citrullinated protein antibodies during treatment with tumour necrosis factor blocking agents in patients with rheumatoid arthritis. Ann. Rheum. Dis. 68, https://doi.org/10.1136/ard.2008.088401 (2009).
Makrygiannakis, D. et al. Citrullination is an inflammation-dependent process. Ann. Rheum. Dis. 65, https://doi.org/10.1136/ard.2005.049403 (2006).
Beekman, J. M. et al. Filamin A Stabilizes FcγRI Surface Expression and Prevents Its Lysosomal Routing. J. Immunol. 180, https://doi.org/10.4049/jimmunol.180.6.3938 (2008).
Huerta-Cepas, J., Serra, F. & Bork, P. ETE 3: Reconstruction, Analysis, and Visualization of Phylogenomic Data. Mol. Biol. Evol. 33, https://doi.org/10.1093/molbev/msw046 (2016).
Sievers, F. & Higgins, D. G. Clustal omega, accurate alignment of very large numbers of sequences. Methods in Mol. Biol. 1079, https://doi.org/10.1007/978-1-62703-646-7_6 (2014).
Guindon, S. et al. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 59, https://doi.org/10.1093/sysbio/syq010 (2010).
Letunic, I. & Bork, P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res 44, https://doi.org/10.1093/sysbio/syq010 (2016).
Gorrec, F. The MORPHEUS protein crystallization screen. J. Appl. Crystallogr 42, https://doi.org/10.1107/S0021889809042022 (2009).
Gildea, R. J. et al. Xia2.multiplex: A multi-crystal data-Analysis pipeline. Acta Crystallogr D Struct Biol 78, https://doi.org/10.1107/S2059798322004399 (2022).
Evans, P. R. & Murshudov, G. N. How good are my data and what is the resolution? Acta Crystallogr D. Biol. Crystallogr. 69, https://doi.org/10.1107/S0907444913000061 (2013).
Tanner, J. J., Komissarov, A. A. & Deutscher, S. L. Crystal structure of an antigen-binding fragment bound to single-stranded DNA. J. Mol.Biol. 314, https://doi.org/10.1107/S0907444913000061 (2001).
Casañal, A., Lohkamp, B. & Emsley, P. Current developments in Coot for macromolecular model building of Electron Cryo-microscopy and Crystallographic Data. Protein Sci. 29, 1069–1078 (2020).
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Kovalevskiy, O., Nicholls, R. A., Long, F., Carlon, A. & Murshudov, G. N. Overview of refinement procedures within REFMAC 5: Utilizing data from different sources. Acta Crystallogr D. Struct. Biol. 74, 215–227 (2018).
Kovalevskiy, O., Nicholls, R. A. & Murshudov, G. N. Automated refinement of macromolecular structures at low resolution using prior information. Acta Crystallogr. D. Struct. Biol. 72, 1149–1161 (2016).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, https://doi.org/10.1038/s41586-021-03819-2 (2021).
Chen, V. B. et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr D. Biol. Crystallogr. 66, https://doi.org/10.1107/S0907444909042073 (2010).
Agirre, J. et al. The CCP4 suite: integrative software for macromolecular crystallography. Acta Crystallogr D. Struct. Biol. 79, https://doi.org/10.1107/S2059798323003595 (2023).
Kissel, T. et al. Antibodies and B cells recognising citrullinated proteins display a broad cross-reactivity towards other post-translational modifications. Ann Rheum Dis 79, https://doi.org/10.1136/annrheumdis-2019-216499 (2020).
Kissel, T. et al. Surface Ig variable domain glycosylation affects autoantigen binding and acts as threshold for human autoreactive B cell activation. Sci Adv 8, https://doi.org/10.1126/sciadv.abm1759 (2022).
Lux, A. et al. A humanized mouse identifies the bone marrow as a niche with low therapeutic IgG activity. Cell Rep 7, https://doi.org/10.1016/j.celrep.2014.02.041 (2014).
Voss, L., Reitinger, C. & Lux, A. Phenotyping of Macrophages in Human Immune System Mice. in Methods in Molecular Biology vol. 2713 (2024).
Acknowledgements
We kindly acknowledge Dr. J. W. Drijfhout (Dept. of IHB, LUMC) for providing the CArgP4 peptides, Dr Rispens for the sequence of 7E4 and Dr Falk Nimmerjahn for the sequence of 6A6.
Author information
Authors and Affiliations
Contributions
Conceptualization, J.H.W.L., A.M.B., T.H. and M.P.; Data curation, T.H., A.M.B., L.J.F., S.K., P.M., F.H, A.V., L.V., E.M.P., M.N., M.J. and A.L.; Methodology, J.H.W.L and T.H.; TCBio approval: T.H. and R.E.G.S.; Obtaining patient material: A.G.M., D.W., R.T.U, R.E.G.S, and T.H.; Writing – original draft, T.H.; Writing – review & editing, T.H., J.H.W.L, K.B., A.M.B., L.J.F., S.K., A.G.M., P.M., A.V., M.N., M.J., F.H., R.T.U., R.E.G.S., D.W., B.J.C.J., A.L., M.P. and R.E.M.T.; All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
T.H., A.M.B., S.K., M.P. and J.H.W.L. are inventors on a related patent application. K.B was an employee of the UMCU when this study was conducted. Currently, K.B. is an employee of and has equity ownership in argenx. All other authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Renato Monteiro, Montarop Yamabhai and the other anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Holtrop, T., Brandsma, A.M., Feitsma, L.J. et al. Preclinical assessment of two FcγRI-specific antibodies that competitively inhibit immune complex-FcγRI binding to suppress autoimmune responses. Nat Commun 16, 10068 (2025). https://doi.org/10.1038/s41467-025-65133-z
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-65133-z