Abstract
Nuclear Factor I (NFI) proteins are involved in adenovirus DNA replication and regulate gene transcription, stem cell proliferation, and differentiation. They play key roles in development, cancer, and congenital disorders. Within the NFI family, NFI-X is critical for neural stem cell biology, hematopoiesis, muscle development, muscular dystrophies, and oncogenesis. Here, we present the structural characterization of the NFI transcription factor NFI-X, both alone and bound to its consensus palindromic DNA site. Our analyses reveal a MH1-like fold within NFI-X DNA-binding domain (DBD) and identify crucial structural determinants for activity, such as a Zn²⁺ binding site, dimeric assembly, and DNA-binding specificity. Given the ~85% sequence identity within the NFI DBDs, our structural data are prototypic for the entire family, a NFI Rosetta Stone that allows decoding a wealth of biochemical and functional data and provides a precise target for drug design in a wider disease context.
Similar content being viewed by others
Introduction
Nuclear factor 1 X (NFI-X) is a member of the NFI family, a ubiquitous group of transcription factors (TF) that includes three additional NFI-encoding genes (NFIA, NFIB, and NFIC). NFIs are site-specific DNA-binding proteins that govern many aspects of cell biology, regulating both viral DNA replication and cellular gene expression1. Human and other vertebrate NFIX genes contain 11 exons, and alternative splicing gives rise to different, tissue-specific, and developmental stage isoforms2. In gene regulation, NFIs determine stem cell proliferation and differentiation, being highly expressed in embryonic and adult stem cells in many tissues3,4,5. Consequently, NFIs are implicated in several developmental disorders6,7,8,9, and act as tumor suppressors in some cancers10.
To date, NFI research has focused particularly on NFI-X due to its essential role in multiple organ systems, including neural stem cell biology, hematopoiesis, and muscle development4. NFI-X is expressed throughout the brain and is implicated in nervous system development during embryogenesis and in neuronal stem cell homeostasis4,11. In muscle, NFI-X plays a pivotal role in skeletal muscle development, by regulating fetal-specific myogenic program and fiber type properties3, and in post-natal regeneration, by regulating muscle stem cell differentiation upon acute damage12,13. In this context, NFI-X has been studied in the pathophysiology of muscular dystrophies14, based on evidence that its absence in nfix null mice protects from disease progression13,15,16.
Furthermore, NFI-X is implicated in several human cancers, and two rare congenital malformation syndromes: Malan syndrome (MALNS) and Marshall-Smith Syndrome (MSS), caused by mutations that cluster in distinct regions of the NFIX gene4,6,7. Respectively, MALNS and MSS mutations cluster in the DNA-binding domain (DBD) and the regulatory C-terminal transactivation domain (TAD)7.
The molecular mechanisms of NFIs are unknown and debated, mostly due to the difficulties in producing sufficient heterologous protein yields for in vitro characterization. NFIs bind as dimers to the palindromic consensus sequence TTGGC(n5)GCCAA, in double-stranded DNA, with nanomolar affinities17,18,19. Indeed, dimerization is an integral part of NFI DNA-binding functions, with protein mutants devoid in dimerization ability, unable to properly bind DNA20. Interestingly, NFI monomers can also bind to individual half-sites, but with two orders of magnitude lower affinity17; shortening the spacer region by one base-pair lowers the affinity to a value similar to that observed with a single half-site, implying that a shorter spacer impacts on the simultaneous interaction of NFI monomers with both half-site sequences21. Additionally, mutations out with the consensus site induce minor differences in NFI binding affinity22.
Studies on truncated NFI-C mutants, show that the first 240 N-terminal amino acids are sufficient for dimerization and DNA binding23,24, with the dimerization domain being probably housed within amino acids 170–24025,26. NFI DBDs are highly similar (85% sequence identity and 92% similarity) (Supplementary Fig. 1), and conserved residues include four cysteines (named Cys-2, Cys-3, Cys-4, and Cys-5) and all, except Cys-3, are essential for DNA-binding. Cys-3 oxidation can lead to reversible inactivation of the protein22,27,28, thus, cysteine residues play important roles in different aspects of NFI-DNA interactions, including redox regulation23,28.
In addition to the DBD, NFIs contain a highly divergent C-terminal TAD region (Supplementary Fig. 1), which is the target of other transcriptional coregulators24,29 and is predicted to be largely intrinsically disordered by Alphafold30. Despite poor sequence conservation, some features are shared, such as high proline content and the presence of post-translational modification sites that also contribute to NFI regulation31,32. The C-terminus of NFI-X from Xenopus laevis has been reported to negatively regulate NFI-X function by preventing DNA binding in vitro29.
Despite general notions on NFI function, precise mechanistic details are lacking. Given the significant role of NFI-X in development and disease, understanding its structure is crucial to develop targeted approaches to inhibit or promote its function. To this aim, we structurally characterize two recombinant protein constructs (NFI-X176 and NFI-X193) encompassing the DBD of NFI-X isoform 2, which represents the most highly expressed splicing variant in mouse skeletal muscle, with full sequence identity with its human ortholog. Our analyses unravel the DBD fold, reveal the structural determinants of dimerization and DNA-binding specificity, and provide insight into the impact of NFI-X pathological mutations. Given the strong sequence conservation within the NFI family, we propose our NFI-X DBD structure as a prototype for the entire NFI family.
Results
The NFI-X DBD is a monomer in solution
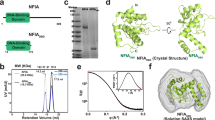
Bioinformatics analyses of full-length NFI-X isoform 2 using Pfam, GlobPlot, and DisEMBL33,34,35 guided the design of two protein constructs, encompassing the N-terminal DBD, for heterologous expression in bacterial cells (Supplementary Fig. 1). NFI-X176 (residues 14–176) represents the minimal predicted DBD, while NFI-X193 includes 17 additional C-terminal residues. The first N-terminal 13 amino acids were omitted in both constructs due to predicted structural disorder and their lack of relevance for DNA binding36. Both NFI-X176 and NFI-X193 were successfully expressed in soluble form in BL21 Shuffle T7 E. coli cells, albeit with different yields: 10 mg/L and ~1–2 mg/L bacterial culture, respectively.
Both protein constructs are monomers in solution, as shown by size exclusion chromatography (SEC) and migrate by SDS-PAGE with molecular weights (MWs) in line with their calculated values of 19.5 kDa (NFI-X176) and 21.4 kDa (NFI-X193), respectively (Supplementary Fig. 2a, b). Thermal denaturation profiles, following the loss of helical structures with increasing temperature using circular dichroism (CD) at 222 nm, indicate that both constructs are stable, and show similar melting temperature (TM) values of 55.6 °C and 56.2 °C for NFI-X176 and NFI-X193, respectively (Supplementary Fig. 2c).
The C-terminus of NFI-X DBD increases DNA-binding affinity
NFI-X176 and NFI-X193 constructs were tested for DNA binding by electrophoretic mobility shift assay (EMSA), using an optimized 31 bp DNA probe, containing the consensus NFI palindromic dyad sequence TTGGC(n5)GCCAA (named NFI-31bp; see Methods). Comparative EMSAs indicate that both constructs bind DNA in a dose-dependent manner, but NFI-X193 binds with higher avidity than NFI-X176 (Fig. 1a). An apparent dissociation constant (Kd) of 48.1 ± 0.49 nM was estimated for the NFI-X193 dimer by triplicate EMSA experiments (see Methods; Supplementary Fig. 3). The presence of faster migrating complexes, in the NFI-X176 binding reactions, are further indicative of reduced stability of the DNA assembly. At low protein concentrations, NFI-X176 binds as a monomer to one half site; the formation of a dimeric NFI-X176 complex is only formed at higher protein concentrations (Fig. 1c). Consequently, it was not possible to determine a Kd for NFI-X176. These data indicate that the 17-residue C-terminal extension is critical for high-affinity DNA-binding of the NFI-X DBD. Therefore, all further EMSAs were carried out on NFI-X193.
a Dose-response electrophoretic mobility shift assay (EMSA) experiments were carried out with NFI-X176 (Lanes 2 to 6) and NFI-X193 (Lanes 8–12), at increasing protein concentrations (20 nM, 40 nM, 80 nM, 160 and 320 nM) and 20 nM of Cy5-labeled 31 bp dsDNA probe (sequence shown), presenting the NFI consensus sequence (bold font) and a 5 bp spacer (underlined). Bands corresponding to the free probe or the NFI-X/DNA complex are indicated. Lanes 1 and 7 represent the control reactions in the absence of protein addition. b Competition EMSA experiments to demonstrate NFI binding site specificity by the NFI-X DNA-binding domain (DBD). NFI-X193 (20 nM) was incubated with the probe (20 nM) alone (−) or in the presence of increasing concentrations (0.5X, 1X, 2.5X, 5X, and 25X with respect to the DNA probe) of unlabelled 31 bp NFI competitor or unlabelled 31 bp random sequence, as described in the Methods. c EMSA experiments to demonstrate the requirement for a 5 bp spacer and to illustrate binding as a dimer. NFI-X193 was incubated in the presence of the Cy5-labeled 31 bp dsDNA probe containing a palindromic 5 bp spacer consensus site (first panel) as a positive control, a 4 bp spacer (second panel) palindromic site, or a 31 bp dsDNA probe containing one half site (third panel). In all panels, protein concentrations were 1X, 2X, 4X, and 8X the probe concentration (20 nM). Reactions in the absence of protein are indicated (-). The 4 bp spacer and half-site probe sequences are shown below, with the NFI consensus sequence (bold font) and spacer regions (underlined). For comparison, EMSA experiments of NFI-X176 bound to the 5 bp spacer probe are shown to illustrate the formation of a complex of one monomer bound to one half site in this C-terminally-truncated construct (last panel). All EMSAs were performed at least three times in independent experiments, and the gels shown represent a typical result.
DNA-binding is sequence-specific, as shown by competition EMSA experiments (Fig. 1b), with two NFI-X proteins bound to DNA, as deduced from comparisons made with a DNA probe, containing a single half-site that shows a complex with higher mobility, representing a single DNA-bound NFI-X193 monomer (Fig. 1c). The dimeric form becomes more evident at higher protein concentrations and displays a smeared appearance. This faster migrating band corresponds to the one observed when the spacing between the two half-sites is shortened by one base-pair (n4) (Fig. 1c), indicating that spacers <5 bp sterically impair dimer formation on DNA (see below), leading to lower stability of the complex and single-site monomer binding.
NFI-X DBD has a MH1-like fold and contains a conserved Zn2+-binding site
Considering its higher production yield, the NFI-X176 construct was selected for crystallization trials, in complex with DNA. Crystals suitable for X-ray diffraction experiments were only obtained with a 23 bp DNA (NFI-23bp; see Methods) (Supplementary Fig. 4). A sequence-based search for NFI-X DBD structural homologs identified only the MH1 domain of Smad proteins, however, with low (<20%) sequence identity. Nevertheless, a clear sequence match was identified between the Smad CCCH motif, responsible for Zn2+ coordination, and NFI-X Cys103, Cys156, Cys162, and His167 residues (Fig. 2b). Strikingly, a Zn2+ ion was identified to be bound to NFI-X176 by in-solution Flame Atomic Absorption Spectrometry (FAAS) (Supplementary Fig. 5a). Taking advantage of the bound Zn2+, the crystal structure of NFI-X176 (PDB: 7QQD) was solved at 2.7 Å by SAD-phasing (Supplementary Fig. 5b and Supplementary Table 1). Since zinc ions were not added during sample preparation, they are likely to have been picked from the growth media during protein expression.
a Structure-based sequence alignment of NFI-X176 with related Smad MH1 domains using the MUSCLE program (https://www.ebi.ac.uk/Tools/msa/muscle/) and manually corrected, based on 3D structure comparisons44. NFI-X176 was aligned with human (h) Smad2 (PDB: 6H3R45), Smad3 (PDB: 5OD646), and Smad9 (PDB: 6FZT45), mouse (m) Smad1 (PDB: 3KMP47), Smad4 (PDB: 3QSV), and Smad5 (PDB: 5X6G48), and Trichoplax adhaerens (Ta) Smad4 (PDB: 5NM946). NFI-X176 secondary structure elements are shown above the aligned proteins with the following color code: MH1-core domain in orange, antenna domain in cyan, β-hairpin in green. Smad residues that adopt α-helices and β-strands are shaded; Smad secondary structure elements not conserved in NFI-X are shown, transparent above the sequence. All secondary structure assignments were made using PDBsum49. Zn2+-coordination residues are shown in white bold letters, shaded in blue, and highlighted with an asterisk, while Smad residues that provide sequence-specificity, including conserved NFI-X residues Arg116 and Lys125, are shaded in black. A 30-residue insertion in hSmad2 is indicated by //; Cys119 (Cys-3) is shaded in pink. b Ribbon representation of the NFI-X176 crystal structure (PDB: 7QQD). The MH1 core domain and the antenna domain are shown in orange and cyan, respectively; both domains are labeled, as are the secondary structure elements and N- and C-termini. The β-hairpin is highlighted in green, and the bound Zn2+ ion is shown as a light blue sphere, with the coordinating cysteine and histidine residues shown as light blue sticks. c Detailed view of Zn2+ ion binding site and residues of the CCCH motif, shown in sticks, colored by heteroatom and labeled. d Structural superposition of NFI-X176 (colored as in b) with a representative Smad protein, human Smad3 (PDB: 5OD6; RMSD of 0.94 Å over 109 paired residues) shown in magenta. e Structural superposition, as in (d), with loops removed for clarity. Panels (b–e) were generated using ChimeraX version 1.850.
There were two NFI-X176 molecules in the asymmetric unit (ASU) (Supplementary Table 1); electron density was well-defined for amino acids 13 to 174 in both chains. The NFI-X DBD fold can be subdivided into two domains, indicated as “antenna” and “core”, which host a Zn2+-binding site (Fig. 2b, c). The core domain is formed by a four α-helical bundle (α1, α2, α3, and α4), three 310 helices (η1-3), and two short, anti-parallel β-sheets (β1–β2, β3–β4). Protein structure comparisons, made using the DALI server37, pinpoint the MH1 domain of Smad TFs, as the only structural homolog of the NFI-X core domain. Despite poor sequence identity, the MH1 α-helical bundle of the NFI-X DBD superimposes well with that of Smad TFs, with some differences in the relative orientation of the helices (RMSD ~ 2.1 Å), and the structural match further improves (RMSD ~ 1.8 Å) if the loops connecting secondary structure elements are removed (Fig. 2d, e). The smaller “antenna” domain (not present in Smad TFs) is a helical excursion, formed by A1 and A2, protruding from the core domain, nestled between α1 and α2 helices (Fig. 2a, b). Therefore, overall, the NFI-X DBD presents a “MH1-like” fold that, given its extensive sequence conservation in NFIs (Supplementary Fig. 1), can be considered as a DBD prototype for the entire NFI family.
With regards to the conserved CCCH Zn2+ coordination site, in NFI-X176 Zn2+ is coordinated by Cys103, Cys156, Cys162 and His167. Cys103 pertains to a flexible loop (residues 97-115) connecting α3 and β1, Cys156 and Cys162 belong to a different loop (residues 153–164) that connects strands β3 and β4, and His167 is contributed from 310 helix η3 (Fig. 2a, c). Given the invariance of these coordination residues in the NFI family, corresponding to Cys-2, Cys-4, and Cys-5 (see Introduction) (Supplementary Fig. 1), our data suggests a conservation of the Zn2+-binding site within the NFI family in general. Indeed, mutation of these cysteine residues in NFIs has been shown to prevent DNA binding36. Our structure rationalizes such data by identifying the Zn2+ binding site as a key factor for the correct folding and stabilization of the NFI-X176 core domain, with implications on the C-terminal region of the DBD, crucial for dimerization and efficient DNA-binding, as discussed below.
Within the crystal asymmetric unit, two additional Zn²⁺ ions were identified (Supplementary Table 1), bound to the protein surface at crystal contact sites. These ions are stabilized by solvent-exposed residues near CCCH motifs, such as His167 and Cys10, suggesting minor Zn2+ dissociation from the CCCH Zn2+-binding site in the NFI-X176 construct.
Other than the antenna domain, a second clear difference between the NFI-X176 core domain and the Smad MH1 domain concerns the β-hairpin exploited by Smad TFs to bind DNA in a sequence specific manner at the major groove (Supplementary Fig. 6). Despite a similar 3D location, the NFI-X176 β-hairpin (strands β1 and β2) is three-residues longer (and also the preceding loop is three-residues longer), the two connected β strands are shorter (only two residues each, instead of three), and there is no amino acid conservation relative to Smad TFs, except for NFI-X176 Arg116 and Lys125 residues that provide sequence-specific contacts in Smad TFs (Fig. 2a, and Supplementary Fig. 6). Differences at the β-hairpin region are not unexpected, considering the different DNA-binding modes of the two TF families. Indeed, Smad TFs recognize a 4 bp palindromic GTCT-AGAC motif38 with no spacing between half-sites, thus the two MH1 domains position themselves at opposite sides of the DNA duplex with no physical interactions between monomers (Supplementary Fig. 6). On the contrary, NFI-X DBD binds at a palindromic 5 bp dyad sequence TTGGC(n5)GCCAA, with the spacer region (n5) expected to position the two DBD core domains on the same side of the DNA (see below).
In line with SEC studies, indicating NFI-X176 is monomeric in solution (Supplementary Fig. 2a), NFI-X176 is a monomer in the crystal, and the two NFI-X176 chains in the ASU (Supplementary Fig. 7a) do not form a biologically relevant dimer, as deduced using jsPISA39. Despite co-crystallization with DNA, in the crystal (PDB: 7QQD), no DNA was found bound to NFI-X176. Residual electron density, compatible with DNA, however, was present at the boundaries between different unit cells, although only 15 disordered base pairs could be traced in protein packing voids, with loose DNA-protein contacts (Supplementary Fig. 7b). This result agrees with EMSA experiments that show that NFI-X176 has reduced affinity and stability for DNA (Fig. 1a) and that NFI-23bp is too short to be bound with high affinity (Supplementary Fig. 4). Nevertheless, the presence of the DNA as a “crystallization additive” promoted crystallization and stabilized the crystal by filling, otherwise empty, volumes among unit cells (Supplementary Fig. 7b). The use of DNA as a crystallization additive, in fact, has been previously reported for lysozyme40.
NFI-X DBD binds as a dimer to the DNA major groove, stabilized through a C-terminal swapping mechanism
To solve the structure of NFI-X DBD in complex with DNA, efforts focused on the NFI-X193 construct, based on its improved affinity for DNA with respect to NFI-X176 (Fig. 1a), using NFI-31bp demonstrated to be the optimal length for efficient binding (Supplementary Fig. 4). Since the yield of NFI-X193 was insufficient ( ~ 1–2 mg/L bacterial culture) for protein crystallization, a cryo-EM approach was adopted, despite the small size of the protein/DNA complex (approx. 70 kDa). Nevertheless, the cryo-EM structure of NFI-X193/DNA was solved at 3.86 Å resolution (PDB: 9QKY, EMD-53223; Supplementary Table 2) (Supplementary Fig. 8 and Supplementary Fig. 9). In fact, under grid vitrification conditions (see Methods), the purified complex produced long straight fibrils (Supplementary Fig. 8 and Supplementary Fig. 10a) composed of a right-handed superhelix made of a repeating unit formed by a NFI-X193 dimer bound to NFI-31bp. Within the fibril, the DNA molecules interact in an end-to-end manner with an angle of about 140° between the axis of the DNA molecules (Supplementary Fig. 10). Cryo-EM electron density was interpretable up to residue Tyr181 for both protomers (A and B) of the dimer, suggesting that the final 12 residues of NFI-X193 are structurally disordered.
The cryo-EM structure provides a direct visualization of a NFI dimer bound to DNA (Fig. 3a). Within the dimer, protein-protein contacts are limited and mediated by a swapping mechanism that brings the C-terminal helix of one subunit (residues 174–180) to be sandwiched between the same helix and loop 140–143 of the other subunit. Interactions are both hydrophobic, with Leu(A)175, Tyr(A)178 and Leu(A)179 of one subunit nestled in a pocket lined by Ile(B)140, Pro(B)141, Leu(B)142, Val(B)170 and Ile(B)172 of the other subunit, and electrostatic, between Lys(A)173 NZ and Glu(B)143 OE1 (Fig. 3b). The molecular architecture of the DNA-bound NFI-X193 dimer rationalizes why DNA binding affinity was reduced in the NFI-X176 construct, due to the absence of residues 177–193 which contain the C-terminal helix, involved in the dimerization swapping mechanism (Fig. 1a).
a NFI-X193 dimer bound to DNA. One protomer is shown with coloring as in Fig. 2, while the other is shown in grey to highlight the dimer swapping mechanism involving helix α5 (yellow). The DNA molecule is shown in sticks (light brown color), with the consensus NFI-binding site in magenta. b Detailed view of the dimerization interface involving the swapping of helix α5. Interface residues are shown as sticks and labelled. The salt-bridge between Lys173 (chain A) and Glu145 (chain B) is shown in dashes. c Frontal view of one NFI-X193 protomer bound to DNA. Residues interacting with DNA are shown as sticks and labelled. For clarity, only one NFI half site is shown. This Figure was generated using ChimeraX version 1.850.
The antenna domain and the β-hairpin loop are the structural determinants of NFI-X DNA binding
NFI-X193 binds at the NFI consensus sequence, with the 5 bp spacer positioning the two DBD core domains on the same side of the DNA (Fig. 3a). Each NFI-X193 protomer interacts at each half site in a similar manner, with the antenna and core domains flanking the DNA helix on opposite sides (Fig. 3c). As expected, the NFI-X193 DNA interaction interface is rich in positively charged amino acids (Supplementary Fig. 11). Detailed protein-DNA interactions are shown in Fig. 4. In subunit A, Lys78, Gln110 and Arg115 bind to the DNA phosphate backbone atoms of T9 and G11 of the consensus sequence T9T10G11G12C13, respectively, while Arg121 interacts with T8, immediately prior to the consensus motif. On the opposite strand (G−13C−12C−11A−10A−9), Arg38 and Lys 42 bind to the DNA phosphate backbone atoms of G−13, and C-12 respectively. Finally, Thr145 and Lys113 interact with T-14 and C−15 of the (n5) spacer, respectively. Sequence-specific interactions are provided only by Arg116 and Lys125: Arg116 side-chain binds to G12 of the consensus motif and G-13 of the complementary strand; Lys125 side-chain interacts with G11G12, while its carbonyl oxygen atom is H-bonded to C-12 (Fig. 4b). Furthermore, the Ala123 side-chain contributes to sequence-specific recognition by facing the methyl groups of T9T10, providing favorable hydrophobic interactions. In subunit B, protein-DNA interactions are specularly conserved (Fig. 4a). Of the interacting residues, Arg38 and Lys42 are contributed by helix A2 of the antenna domain, whereas the remaining residues, apart from Lys78 and Thr145, belong to the β-hairpin loop (Supplementary Fig. 1), whose convex face dives into the concave major groove of the duplex DNA containing 5 bp of the consensus-binding motif (Fig. 3a, c).
a Schematic view of the interactions between NFI-X193 residues and DNA: dotted lines indicate hydrogen bonds and salt bridges, dashed circular lines indicate hydrophobic sequence-specific interactions. NFI-X193 residues are boxed and colored as in Fig. 3a. b Site-specific interactions made by NFI-X193 Arg116 and Lys125. Hydrogen bonds are shown in dotted lines. Panel (a) was generated manually using Microsoft PowerPoint 2024. Panel (b) was generated using ChimeraX version 1.850.
Superposition of cryo-EM DNA-bound NFI-X193 with the NFI-X176 crystal structure shows a RMSD 0.95 Å over 159 aligned residues, indicating that NFI-X DBD binds its target DNA without large conformational changes, in particular regarding the relative position of the antenna and core domains (Fig. 5a). Only a minute difference is present, which, however, explains the molecular mechanism that triggers the insertion of the β-hairpin loop into the DNA major groove for base-reading. In the unbound state, the β-hairpin loop and the antenna domain are locked together by an intramolecular salt-bridge involving Asp124 and Arg38 side-chains, respectively. In the DNA-bound state, Arg38 side-chain changes its orientation, making a salt-bridge with a DNA phosphate group (G-13), thus unlocking the β-hairpin loop (containing Asp124) from the antenna domain (Fig. 5b). This structural change in turn allows Lys125 side-chain to move into the major groove to provide sequence-specific DNA-binding interactions, together with Arg116 (Fig. 4b).
a Superimposition of the NFI-X193/DNA cryo-EM structure (colored as in Fig. 2) and the crystal structure of NFI-X176 (grey). For clarity, DNA is not shown in the NFI-X193/DNA complex. Major structural differences are highlighted by red dashed circles. b Structural changes associated with DNA-binding, highlighted by arrows. Residues relevant in the molecular mechanism involving the unlocking of the β-hairpin upon DNA binding are shown as sticks and labelled. Salt-bridge and hydrogen bond interactions are shown as dashed lines. The Figure was generated using ChimeraX version 1.850.
Interestingly, mutations at residues belonging to the antenna domain (Arg38) and to the β-hairpin loop (Lys113, Arg115, Arg116, Arg121, Lys125, and Arg128; see Supplementary Fig. 1) cause a rare genetic disease called Malan syndrome that is characterized by excessive body growth, skeletal anomalies, and intellectual retardation6,7. In the DNA-bound NFI-X193 structure, all cited residues are either interacting with DNA bases (Lys125, and Arg116) or in contact (Lys113, Arg115, Arg121) or proximity (Arg128) to the DNA phosphate backbone, (Fig. 4a). In EMSAs, most of the pathological mutations in the β-hairpin (Lys113Glu, Arg115Trp, Arg116Gln, Arg116Pro, Lys125Glu), completely lose activity, in line with their DNA-binding roles highlighted in the cryo-EM structure. Instead, Arg128Gln partially loses its DNA-binding ability, and the Arg121Pro mutant binds DNA to the same extent as wild-type DBD (Fig. 6). Arg128 is located at the end of the β-hairpin (Supplementary Fig. 1), facing the DNA phosphate of T-14 at about 5.5 Å, thus its mutation may have an indirect negative effect on DNA-binding. Indeed, Arg128 is on one side, salt-bridged to the Asp130 side-chain and H-bonded to the carbonyl oxygen of Gly147 and is flanked by the sequence-specific Arg116 on the other side (Supplementary Fig. 12a). Mutation to Gln is likely to alter the overall structure of the β-hairpin, preventing salt-bridge formation with Asp130 and possibly favoring H-bond interactions with Arg116, thus disturbing the latter residues in its key interactions with the palindromic NF-I site, as suggested by a significant reduction of DNA-binding shown in EMSA (Fig. 6). Arg121 is localized at a kink of the β-hairpin and interacts via a salt-bridge with OP1 T(8) in the DNA-bound form (Fig. 4a). The presence of a Pro residue in this position is compatible with preserving the kink in the protein backbone and the interaction with DNA may be compensated by the presence of a nearby Arg74 (Supplementary Fig. 12b). Therefore, the pathological mechanism, at a cellular level, of this mutation, is likely to be more complex than a direct DNA-binding impairment (Fig. 6).
Electrophoretic mobility shift assay (EMSA) experiments were carried out for eight (Arg38Leu, Lys113Glu, Arg115Trp, Arg116Gln, Arg116Pro, Arg121Pro, Lys125Glu, and Arg128Gln) mutations, within the NFI-X DNA-binding domain, identified in ten Malan syndrome individuals; each mutation is found in a different patient, except for Arg115Trp and Arg116Gln that were identified in two individuals each6. The effect of Malan mutations (single letter code) on DNA binding is shown in comparison with the NFI-X193 control, using 20 nM Cy5-labeled 31 bp DNA probe (sequence shown in Fig. 1) and 1X, 2X, 4X, and 8X concentrations of protein with respect to the probe. Bands corresponding to the free probe or the NFI-X193/DNA complex are indicated. All EMSAs were performed at least in triplicate with independent experiments.
The antenna domain mutant (Arg38Leu) retains its DNA-binding ability, albeit with reduced affinity (Fig. 6). This is in line with the proposed mechanism of β-hairpin insertion into the DNA major groove (Fig. 5b), whereby the Arg38Leu mutation would prevent the formation of the salt bridge with Asp124 in the DNA-unbound state, leading to a constantly activated TF, but the missing interaction between Leu38 and phosphate backbone in the DNA-bound state would then result in decreased DNA-binding affinity.
Discussion
The NFI proteins share little sequence homology with TFs for which structures have been determined, making the absence of structural information on this family particularly intriguing. Numerous studies have highlighted the pivotal role of NFIs in both development and disease, yet the mechanistic details of their ability to bind DNA in their target promoters have remained elusive until now. Although some notions, such as the requirement for protein dimerization and Cys residues for DNA binding, have been deduced from EMSA-based studies, we can now provide the reasons at a molecular level.
Here, we combined X-ray crystallography, cryo-EM, biophysical, and mutational approaches to elucidate the molecular mechanisms of DNA recognition by NFI-X at its palindromic NFI-site. The NFI-X DBD has a special fold, based on a four helical bundle core domain, reminiscent of the Smad MH1 domain, and a α-loop-α antenna domain, specific for NFIs, inserted at the N-terminus of the DBD (Figs. 1, 3). The core domain represents a stable molecular platform from which the antenna domain and a β-hairpin region protrude, forming a positively charged cleft deputed for DNA-binding (Supplementary Fig. 11). In line with observations made for Smad MHI domains, NFI-X DBD contains a bound Zn2+ ion, coordinated by the CCCH motif (Fig. 2c), conserved within the NFI family (Fig. 2a, d). As for Smad proteins, this ion plays a stabilizing, structural role41 and, accordingly its removal from NFI-X193 by chelation leads to protein instability and precipitation. In particular, the Zn2+-binding site compacts the MH1 domain, properly orients a long loop that hosts the DNA-recognition β-hairpin towards the DNA major groove (Fig. 2b), and it stabilizes the C-terminal region containing the dimerization helix α5 (Fig. 3a, b). Our observations, finally, rationalize previous biochemical and mutational data on human NFI-C that show that the three Cys coordinating residues are essential for DNA binding36.
NFI-X binds to the NFI consensus sequence as a dimer (Kd = 48.1 ± 0.49 nM; Supplementary Fig. 3), with each subunit inserting its β-hairpin into the major groove from the same side of the DNA (Fig. 3a). We showed that NFI-X DBD is a monomer in solution and dimerizes only upon DNA-binding (Fig. 1c and Supplementary Fig. 2), via a mechanism that involves the swapping of the C-terminal helix α5 from each protomer (Fig. 3b, c). Removal of this helix reduces DNA-binding affinity (Fig. 1a) thus helix α5 appears to be both necessary and sufficient for NFI dimerization on DNA, but it is not sufficient for dimerization in solution (Supplementary Fig. 2), although we cannot exclude the involvement of successive C-terminal residues for further dimer stabilization in the full-length protein. AlphaFold modelling of full-length NFI-X suggests that the regions following the dimerization helix α5 are intrinsically disordered (Supplementary Fig. 13). In fact, previous coimmunoprecipitation data indicate that full-length NFI-C can dimerize with truncated mutant(s) devoid of DNA-binding ability20. It is, therefore, unsurprising that proteins derived from different NFI genes have been demonstrated to form heterodimers in vitro25. In this context, our structure suggests that the molecular mechanism of subunit selection for heterodimer formation must involve regions outside the DBD, where the NFI sequences diverge. The real occurrence of an NFI heterodimer in a cellular context is however, still debated, although NFI genes have been reported to be simultaneously expressed in certain cell lines25.
Our data reveal the DNA-recognition mode exploited by NFI-X, based on the insertion of a β-hairpin loop contributed from each protein subunit of the dimer into the half site of the palindromic NFI-site, located in the DNA major groove (Fig. 3a). Base-reading interactions only involve two NFI-X residues (Arg116 and Lys 125) housed in the β-hairpin that contact the three G bases of each half site, rationalizing DNA consensus sequence mutations that confirm these bases as the most relevant for modulating DNA binding affinity26. Additionally, the methyl groups of the TT motif are hydrophobically stabilized by the facing Ala123. The DNA backbone is further stabilized by several ionic interactions involving residues belonging to both the NFI-X core (including the β-hairpin) and the antenna domains (Fig. 4). Our structural and mutagenesis data indicate these residues as functionally relevant, providing a rational interpretation, associated mostly to DNA-binding deficiency, for several pathological mutations identified in Malan Syndrome individuals (Fig. 6)6,8,42.
Antenna domain residue Arg38, in particular, triggers a conformational change required to insert the β-hairpin loop into the major groove of the DNA. In unbound NFI-X, this residue participates in an intramolecular salt-bridge with Asp124, which is substituted by a phosphate group in the DNA-bound state. This switch, in the Arg38 binding state, unlocks the β-hairpin loop (containing Asp124) from the antenna domain and allows the Lys125 side-chain to relocate into the major groove to read the DNA consensus motif, together with Arg116. This ingenious molecular mechanism constitutes the sole notable conformational change identified between the unbound (as determined from NFI-X176) and DNA-bound (from NFI-X193) states. The structural dynamics of the β-hairpin loop required for DNA-binding is compensated by its anchoring to the core domain through Cys119, which is nestled in a hydrophobic pocket (Supplementary Fig. 1, and Supplementary Fig. 12b). This result is in keeping with previous biochemical data showing that oxidation of this Cys residue (known as Cys-3) leads to reversible inactivation of the protein22,27,28, and suggests that the discrepancy in the polarity of the oxidized residue and the surrounding environment can disrupt the structure of the β-hairpin loop.
The architecture of the dimeric NFI-X193/DNA complex also provides a structural explanation for the strong preference of a 5 bp spacer in the NFI binding site (Figs. 1c, 3a). In fact, by EMSA, we observed that a 4 bp spacer led to steric hindrance between the protomers and only one monomer was able to bind to a single half site (Fig. 1c). The reduced distance between the protomers would impede the swapping interaction and induce a clash between the C-terminal helix of one protomer with the antenna domain of the other (Supplementary Fig. 14). In contrast, a spacer longer than 5 bp would distance the C-terminal DBD helices from one another, preventing dimerization and thus decreasing DNA-binding affinity, as previously reported21.
A final, interesting issue relates to the functional relevance of the super-helical structure present on the cryo-EM grids, whose formation was essential to solve the structure of such a small macromolecular complex ( ~ 70 kDa) (Supplementary Figs. 8, 10). The stabilizing interactions of the super-helix involve base stacking at the 3’- and 5’-termini of adjacent DNA molecules, responsible for elongating the fibre, and protein-protein interactions, which bridge the two opposing DNA helices of the fibre. The latter interaction involves residues belonging to α1, α3, and the Zn2+-binding site of one subunit, with those of another subunit, a region opposite to that deputed to DNA-binding (Supplementary Fig. 10c). Although, to our knowledge, no evidence has been reported (yet) for the presence of this supercomplex in a cellular context, this protein assembly found in our cryo-EM structure raises the striking possibility that NFI TFs mediate DNA-looping by packing, via protein-protein interactions, distant DNA regions that house NFI-sites. Interestingly, the cryo-EM fibre identified a hot-spot for protein-protein interactions that coincides with the region (α3) proposed to be involved in NFI-mediated activation of viral DNA replication through direct interaction of NFI with the viral preinitiation DNA polymerase complex20. In the NFI-X193/DNA complex, this region is solvent-accessible and is not blocked by the presence of bound DNA. This is relevant since it provides the structural evidence to support the proposed role of NFI as a DNA polymerase complex recruiter after binding at the origin of replication.
In conclusion, our data represent a Rosetta Stone, which not only structurally decodes a plethora of biochemical and functional data regarding NFI proteins but, in perspective, provides the basis for analysing other functional roles of NFIs and for the rational design of compounds that modulate the DNA-binding properties of this class of TFs, with possible applications in muscular dystrophy treatment in the case of NFI-X, and in related pathologies such as cancer, neurodevelopmental disorders, for other NFIs43.
Methods
Protein production of NFI-X DBD constructs
The cDNA encoding the DBD of mouse NFI-X isoform 2 (UniProt code P70257), extending over amino acids 14 to 176 (NFI-X176), was chemically synthesized (Eurofins Genomics Srl.) and subcloned in-frame with an N-terminal maltose-binding protein (MBP) fusion tag, in the pMAL-cRI bacterial expression vector (New England Biolabs), via EcoRI and HindIII restriction sites. The codon-optimized, cDNA encoding for NFI-X193, covering residues 14 to 193, was synthesized and subcloned into the pET21-b (Novagen) via NdeI and SalI restriction sites, in frame with a N-terminal hexahistidine tag and Small ubiquitin-related modifier 3 tag (His-SUMO) (Genewiz, Azenta Life Sciences). Malan NFI-X193 mutants were generated by site-directed mutagenesis using Phusion DNA polymerase (Thermo Fisher Scientific), according to standard protocols, using the NFI-X193 plasmid as a template.
Both NFI-X constructs were overexpressed in BL21 Shuffle T7 E. coli cells (New England Biolabs) grown at 30 °C in 1X (NFI-X176) or 2X (NFI-X193) Luria Bertani broth, supplemented with 100 μg/mL ampicillin. Protein expression was induced at optical densities of 0.6-0.8 at 600 nm wavelength, upon addition of 0.25 mM (NFI-X193) or 0.5 mM (NFI-X176) isopropyl β-d-1-thiogalactopyranoside (IPTG; Genespin Srl.) to cultures, pre-cooled to the 20 °C induction temperature, and further incubated overnight with shaking at 220 rpm.
Bacterial cells from 2 L NFI-X176 cultures were harvested by centrifugation and lysed by sonication. NFI-X176 was purified from the supernatant using a 5 mL HiTrap Heparin HP affinity column (Cytiva), pre-equilibrated in Binding Buffer A (50 mM HEPES pH 7.0, 200 mM NaCl, and 2 mM dithiothreitol (DTT) using an AKTA pure™ FPLC system (Cytiva). The protein was eluted using a linear salt gradient, achieved by mixing Buffer A with Buffer B (50 mM HEPES pH 7.0, 1 M NaCl, and 2 mM DTT). Fractions containing NFI-X176 were pooled and incubated with 50 U thrombin protease (Sigma Aldrich), with gentle rotation overnight at 20 °C. The cleavage reaction was diluted with 50 mM HEPES, pH 7.0, and 2 mM DTT to lower the NaCl concentration to 200 mM. Cleaved NFI-X176 was separated from the MBP fusion tag and intact MBP fused constructs by a second Heparin affinity chromatography step, using analogous, forementioned purification conditions. Fractions containing pure NFI-X176 were pooled, concentrated to 500 μl with an Amicon ultracentrifugal concentrator (Merck Millipore) with a MW cut-off of 10000, and further purified by SEC on a Superdex 200 Increase 10/300 column (Cytiva), pre-equilibrated in Buffer A.
Bacterial cells from 2 L NFI-X193 cultures were mechanically lysed at 25 mPa using a cell disruptor (Constant Systems Ltd) and sonication. NFI-X193 was purified from the clarified supernatant using a 1 mL Histrap HP column (Cytiva), pre-equilibrated in Buffer A (20 mM sodium phosphate pH 7.4, 20 mM imidazole, 0.5 M NaCl, 1 mM DTT). Elution was carried out over a linear gradient, performed by mixing Buffer A with Buffer B (Buffer A containing 0.5 M imidazole). Peak fractions were pooled and dialyzed overnight into SUMO protease cleavage buffer (50 mM Tris pH 7.4, 200 mM NaCl, and 1 mM DTT). Removal of the His-SUMO tag was achieved upon incubation of the fusion protein at room temperature for 4 h with in-house, recombinantly produced Saccharomyces cerevisiae SUMO protease at a protease:protein molar ratio of 1:200, respectively. Cleaved NFI-X193 was separated from the His-SUMO fusion tag via affinity chromatography on a 5 mL Heparin HP column (Cytiva), using the conditions used to purify NFI-X176. Fractions containing purified NFI-X193 were concentrated to 500 μl and further purified on a ProteoSEC 6-600 HR 10/300 column (Neobiotech), pre-equilibrated in 50 mM Tris pH 7.4, 200 mM NaCl, and 1 mM DTT. Molecular weights were calculated from calibration curves generated using the Bio-rad Gel Filtration Standards (Bio-rad Laboratories), using standard protocols. Malan mutants were expressed and purified as for NFI-X193.
NFI-X/DNA complex assembly for crystallization and cryo-EM
Co-crystallization experiments of NFI-X176 in complex with NFI-31bp were unsuccessful; therefore a shorter 23 bp DNA fragment (NFI-23bp) with sticky ends (forward strand: 5’-GTCTCTTTGGCAGGCAGCCAACC-3’; reverse strand 5’-ACGGTTGGCTGCCTGCCAAAGAG-3’), containing the two NFI half sites (underlined) was set up. 23 bp is long enough to be bound by NFI-X-DBD (Supplementary Fig. 4), but short enough not to interfere with crystallogenesis. Sticky ends were selected to stabilize and promote crystal lattice formation. The protein DNA complex was assembled, mixing protein:DNA at a 2:1 molar ratio and by progressively decreasing the buffer salt concentration to 50 mM, to promote complex assembly by repetitive concentration and dilution cycles with buffer, before final concentration to 15 mg/mL.
For Cryo-EM, the NFI-X193 complex was assembled with the 31 bp DNA sequence used in EMSAs (Fig. 1). NFI-X193 fractions, derived from the final Heparin affinity chromatography step, were mixed with DNA at a molar ratio of 2:1. The high salt present in the Heparin chromatography elution Buffer B was gradually lowered to promote complex formation, by stepwise dialysis into 50 mM Tris pH 7.4, containing 1 mM DTT and progressively lower NaCl concentrations (300 mM, 150 mM and 75 mM NaCl). Each dialysis step was carried out at 4 °C in 2 L dialysis buffer using a Spectra-Por® dialysis membrane (Spectrum™) with a 3.5 kDa cut-off and buffer exchanges every 2 h, with the last step overnight. The dialysate was concentrated to 500 μl and loaded onto ProteoSEC 6-600 HR 10/300 column (Neobiotech), pre-equilibrated in 20 mM Tris pH 7.4, 150 mM NaCl, and 1 mM DTT. NFI-X193-DNA complex-containing fractions were concentrated to 10 mg/mL.
Circular dichroism (CD)-based thermal denaturation studies
The thermostability of protein unfolding was investigated by monitoring ellipticity variations at a single wavelength of 222 nm over a temperature gradient of 20 °C to 95 °C (1 °C/minute ramp rate). Unfolding curves are represented as a change in ellipticity versus temperature; 0.2 mg/mL protein samples (20 mM Tris pH 7.4, 200 mM NaCl, and 1 mM DTT) were placed in 1 cm quartz cuvettes, and CD spectra were recorded using a Jasco J-810 instrument equipped with a PFD-425S Peltier temperature controller (Jasco). CD profiles were generated using the SigmaPlot v.14 (Systat Software Inc.).
Electrophoretic mobility shift assays
DNA-binding reactions were assembled on ice using DNA probes carrying the NFI consensus sequence. The probe was designed and optimized based on the sequence of the promoter region of NFI-X isoforms 3 and 451. Starting from this sequence (5′-TGGGTCTCTTTGGCAGCAGCCACCCAGCAAA-3), 1 nucleotide (guanine) insertion was made after adenine 18 to lengthen the spacer region to 5 nt, and another substitution was made (c23a) to make the two half sites palindromic (underlined), based on previous studies correlating DNA sequence to NFI binding affinity21,52. Probes were labelled with Cyanine 5 (Cy5) at the 5’-end of the forward strand and annealed with the unlabeled reverse strands, using standard protocols. Dose response experiments were performed by combining 14 μL of a premixed Binding Mix (BM), containing 20 nM NFI Cy5-probe, 20 mM Tris-HCl pH 7.5, 50 mM NaCl, 5 mM MgCl2, 30 mM KCl, 0.5 mM EDTA, 6.5% (v/v) glycerol, 2.5 mM DTT, 0.1 mg/mL BSA, 10 ng Poly(dIdC) (Thermo Fisher Scientific), with 2 μL purified NFI-X176 or NFI-X193 at varying final dimer concentrations, as indicated in the figure legends. Reactions were incubated at 30 °C for 30 min in the dark and electrophoresis was performed in the dark on a 6% Native gel run at 100 V in 0.25X Tris/Borate/EDTA buffer. Cy5 fluorescence was detected using a Chemidoc MP imaging apparatus (Bio-Rad Laboratories). The apparent dissociation constant (Kd) for the NFI-X193 dimer was calculated from triplicate EMSA experiments by quantifying the fraction of bound DNA in each lane, using the Bio-rad ImageLab software (version 6.1.0) (Supplementary Fig. 3). For each replicate, the Kd (corresponding to the concentration of protein at which 50% DNA is bound), was estimated by applying a linear function to the DNA-bound fraction (%) plotted against protein concentration (nM). The calculated Kd is the average of the three experiments ± the standard deviation.
Competition EMSAs were carried out, as described above, in 14 μl binding mix containing 20 nM NFI-X193 and 20 nM Cy5-labelled 31 bp DNA, incubated with 2 μl unlabelled 31 bp competitor or 31 bp random probe (Fig. 1b) at increasing concentrations (0.5X, 1X, 2.5X and 5X and 25X) with respect to the labelled probe.
Flame atomic absorption spectroscopy (FAAS)
The presence of Zn2+ was assessed in purified NFI-X176 by calibrated FAAS. A homogeneous solution of 0.2 mg/mL NFI-X176 was exposed to wet decomposition. To the protein sample, 5 mL of nitric acid (65% (v/v)) was added, and the resulting solution was gently boiled on a hot plate until the red fumes coming from the beaker terminated (1 h). After cooling, 5 mL freshly prepared mixture of 65% nitric acid and 37% hydrochloric acid (1:3) was added and the solution was warmed on a hot plate for 2 h. After cooling, about 3 mL of hydrogen peroxide (30%) were added, and the solution was boiled again to evaporate until a small portion remained. After cooling, the resulting clear digested solution was quantitatively diluted to a final volume of 10 mL with 2% nitric acid before being analysed in duplicate by FAAS. Nitric acid, hydrochloric acid, and hydrogen peroxide were purchased from Sigma Aldrich.
The calibration curve was prepared using five standards (including the blank). Working calibration solutions were freshly prepared using appropriate stepwise dilutions of standard Zn stock solution (Ultra grade, 1000 mg/L, 2% HNO3, Perkin-Elmer). Working standards were as follows: 0.25, 0.5, 1, and 1.4 ppm diluted in 2% (v/v) nitric acid.
Data were collected by using an atomic absorption spectrometer (PinAAcle 900 T, Perkin-Elmer) equipped with a deuterium lamp for background correction, air-acetylene flame, and zinc hollow-cathode lamp operating at 213.9 nm. The linear range was 0.01–2 mg/L. Two measurements in triplicate were performed, and the amount of zinc, as the mean of the two measurements, was calculated using the standard calibration graph obtained with Excel (version 16.24).
Crystallization and 3D structure determination of NFI-X176
Microbatch under oil experiments were set up using an Orxy8 robot (Douglas Instruments) purified NFI-X176/DNA (NFI-23bp) complex (15 mg/mL). Crystals grew at 4 °C in an optimized condition based on condition B2 of the JCSG Screen (Molecular Dimensions): 0.2 M sodium thiocyanate (NaSCN), 0.1 M HEPES pH 7.0, 22% (w/v) PEG3350 in 96-well, flat-bottomed microbatch plates (Greiner-BIO-ONE) at a protein:precipitant ratio of 50% (v/v) and 9 mL of a 1:1 paraffin:silicon oil mixture as sealing agent. Crystals were soaked in cryoprotectant solution (0.2 M NaSCN, 0.1 M HEPES pH 7.0, 22% (v/v) PEG 3350, 20 % (v/v) glycerol) and flash frozen in liquid nitrogen.
X-ray diffraction data were collected at the XRD2 beamline of the ELETTRA Synchrotron, Trieste (Italy), equipped with a Pilatus 6 M hybrid pixel area detector (Dectris). Data were indexed, integrated, and scaled using XDS (version Jan 31, 2020, built=20200417)53. The statistics for the data collection and data reduction are summarized in Supplementary Table 1.
The only NFI-X DBD structural homolog is the MH1 domain of Smad proteins, which shares low (<20%) sequence identity, and attempts to phase the NFI-X data with a Smad model by Molecular Replacement failed. The structure was then solved by SAD phasing in the P1211 space group, exploiting the anomalous signal of bound zinc atoms at a wavelength of 1.2705 Å. The heavy atom substructure was determined using SHELX C/D/E (versions SHELXC 2016/1, SHELXD 2013/2, SHELXE 2019/1)54,55. ARP/wARP (version 8.0) was used for automated model building56. Phasing yielded an average figure of merit (FOM) of 0.623 following the automated fitting of 150 alanine residues. The structure was refined using REFMAC5 (version 5.8.0258), Phenix.refine (version 1.11.1-2575-000) and multiple rounds of manual model building in Coot (version 0.9.6)57,58,59. For cross-validation, 5% experimental reflections were randomly selected to calculate the Rfree value (Supplementary Table 1). MolProbity (as implemented in Phenix, version 1.11.1-2575-000) and PISA (version 2.1.1 built 05-04-2017) were used to assess the stereochemical quality and protein quaternary assembly, respectively39,60. No outliers are present in the final refined model, with 97% of residues located in the favored region of the Ramachandran plot and the remaining 3% in the allowed region.
Cryo-EM sample preparation
3-{Dimethyl[3-(3α,7α,12α-trihydroxy-5β-cholan-24-amido)propyl]azaniumyl}propane-1-sulfonate (CHAPS) detergent was added at its critical micelle concentration of 8 mM to 4 mg/mL purified NFI-X193/DNA sample. Quantum foil R 0.6/1 Cu 300 grids were glow-discharge treated for 30 s at 30 mA, using the GloQube system (Quorum Technologies). Approximately, 3 µL samples were applied onto several grids, under controlled conditions of 4 °C and 100% relative humidity. Various blotting times were tested to optimize sample distribution and minimize excess liquid. Subsequently, prepared grids were rapidly vitrified in liquid ethane utilizing a Vitrobot Mark IV (Thermo Fisher Scientific). Finally, vitrified grids were transferred and stored in liquid nitrogen until further analysis.
Cryo-EM data collection and image processing
Cryo-EM data were acquired from prepared grids using an in-house FEI Talos Arctica 200 kV FEG transmission electron microscope (Thermo Fisher Scientific). An electron beam, operating at 200 kV and a calculated electron dose of 40 electrons per square angstrom (e-/Å2), was directed through the sample and reached a Falcon 3EC detector for image capture. Data acquisition was carried out using EPU-2.8 automated data collection software (Thermo Fisher Scientific), and a series of movies, each composed of 40 individual frames, was recorded. The microscope was set to a nominal magnification of 120,000x, which corresponded to a pixel size of 0.889 angstroms per pixel at the specimen level. During data collection, defocus values were systematically adjusted within the range of −0.8 to −2.2 μm. This range of defocus values was selected to optimize image contrast and resolution, allowing for the capture of structural details within the protein-DNA complex.
Micrographs were initially selected based on visual quality inspection. Movie drift correction and CTF parameters determination were performed in Relion (version 3.1)61. The fibrils were manually picked and extracted with an inter-box distance of 44 Å along the helical axis into overlapping boxes of 400x400 pixels, resulting in 201,833 extracted segments. Such particles were then imported in CryoSPARC (version 3.3.2) for 2D classification and 3D reconstruction CRY62. 2D class averages were used to discard bad quality particles, resulting in 172,304 selected segments, which were used to perform an ab-initio reconstruction using two classes to further refine the set of segments. The ab-initio volume was used as an initial model to determine the helical symmetry parameters by using the Symmetry search tool in CryoSPARC, revealing that the helix was right-handed, with a helical rise of 44.51 Å, a twist of 142.5°, 2.5 subunits per turn and a pitch of 112.4 Å (Supplementary Fig. 8). These values were given as inputs for the Helix Refine tool and using the ab-initio model as input volume, and the ab initio selected set of particles that generated a first map at 4.2 Å resolution. From inspecting this volume, we observed the presence of a symmetry binary axis, orthogonal to the helix axis. Therefore, helical refinement was repeated imposing D1 symmetry, resulting in a final map at 3.86 Å resolution, with optimized helical twist and rise values of 142.672° and 44.618 Å, respectively.
Cryo-EM model building and refinement
Two copies of the crystallographic structure of NFI-X176 (PDB: 7QQD) were initially fitted into the EM density of a single NFI-X193 dimer. Additional C-terminal residues (175-181), that were not present in the NFI-X176 construct and/or crystal structure, were manually built in Coot (version 0.9.6)57, together with a linear DNA molecule containing the NFI-X consensus sequence. The 5’ and 3’ DNA moieties were adjusted in Coot since they slightly deviated from linearity. The model of the complex was then iteratively rebuilt and all-atom real-space refined using stereochemical and NCS restraints within Phenix (version 1.20.1-4487)59. The model of the dimer was then copied and fitted into the unmodelled EM density to obtain an atomic model of the fibril. The resolution was improved with an additional helical refinement imposing D1 symmetry, obtaining a final volume of NFI-X193-DNA complex fibrils at 3.86 Å resolution. Cryo-EM data collection and processing statistics are summarized in Supplementary Table 2.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The Cryo-EM maps have been deposited in the Electron Microscopy Data Bank (EMDB) under accession code EMD-53223. Atomic coordinates for the crystal and Cryo-EM structures have been deposited in the Protein Data Bank (PDB) under accession codes 7QQD and 9QKY, respectively. The source data underlying Supplementary Figs. S2, S3, and S5 are provided as a Source Data file. Source data are provided with this paper.
References
Harris, L., Genovesi, L. A., Gronostajski, R. M., Wainwright, B. J. & Piper, M. Nuclear factor one transcription factors: Divergent functions in developmental versus adult stem cell populations. Dev. Dynam 244, 227–238 (2015).
Gronostajski, R. M. Roles of the NFI/CTF gene family in transcription and development. Gene 249, 31–45 (2000).
Messina, G. et al. Nfix Regulates Fetal-Specific Transcription in Developing Skeletal Muscle. Cell. 140, 554–566 (2010).
Piper, M., Gronostajski, R. & Messina, G. Nuclear Factor One X in Development and Disease. Trends Cell Biol. 29, 20–20 (2019).
Lajoie, M., Hsu, Y.-C., Gronostajski, R. M. & Bailey, T. L. An overlapping set of genes is regulated by both NFIB and the glucocorticoid receptor during lung maturation. BMC Genomics 15, 231 (2014).
Priolo, M. et al. Further delineation of Malan syndrome. Hum. Mutat. 39, 1226–1237 (2018).
Malan, V. et al. Distinct Effects of Allelic NFIX Mutations on Nonsense-Mediated mRNA Decay Engender Either a Sotos-like or a Marshall-Smith Syndrome. Am. J. Hum. Genet 87, 189–198 (2010).
Yoneda, Y. et al. Missense mutations in the DNA-binding/dimerization domain of NFIX cause Sotos-like features. J. Hum. Genet 57, 207–211 (2012).
Lu, Y. et al. Mutations in NSD1 and NFIX in Three Patients with Clinical Features of Sotos Syndrome and Malan Syndrome. J. Pediatr. Genet 6, 234–237 (2017).
Genovesi, L. A. et al. Sleeping Beauty mutagenesis in a mouse medulloblastoma model defines networks that discriminate between human molecular subgroups. Proc. Natl Acad. Sci. USA 110, E4325–E4334 (2013).
Campbell, C. E. et al. The transcription factor Nfixis essential for normal brain development. BMC Dev. Biol. 8, 52 (2008).
Rossi, G. et al. Nfix Regulates Temporal Progression of Muscle Regeneration through Modulation of Myostatin Expression. Cell Rep. 14, 2238–2249 (2016).
Saclier, M. et al. Selective ablation of Nfix in macrophages attenuates muscular dystrophy by inhibiting fibro-adipogenic progenitor-dependent fibrosis. J. Pathol. 257, 352–366 (2022).
Gee, P., Xu, H. & Hotta, A. Review Article Cellular Reprogramming, Genome Editing, and Alternative CRISPR Cas9 Technologies for Precise Gene Therapy of Duchenne Muscular Dystrophy. Stem Cells Int 2017, 8765154 (2017).
Rossi, G. et al. Silencing Nfix rescues muscular dystrophy by delaying muscle regeneration. Nat. Commun. 8, 1055 (2017).
Saclier, M. et al. Nutritional intervention with cyanidin hinders the progression of muscular dystrophy. Cell Death Dis. 11, 127 (2020).
Gronostajski, R. M. Analysis of nuclear factor I binding to DNA using degenerate oligonucleotides. Nucl. Acids Res 14, 9117–9132 (1986).
Meisterernst, M., Rogge, L., Foeckler, R., Karaghiosoff, M. & Winnacker, E. L. Structural and functional organization of a porcine gene coding for nuclear factor I. Biochem 28, 8191–8200 (1989).
Rosenfeld, P. J., O’Neill, E. A., Wides, R. J. & Kelly, T. J. Sequence-Specific Interactions Between Cellular DNA-Binding Proteins and the Adenovirus origin of DNA Replication. Mol. Cell Biol. 7, 875–886 (1987).
Armenterot, M.-T., Horwitzi, M. & Mermod, N. Targeting of DNA polymerase to the adenovirus origin of DNA replication by interaction with nuclear factor I. Proc. Natl Acad. Sci. Usa. 91, 11537–11541 (1994).
Roulet, E. et al. Experimental analysis and computer prediction of CTF/NFI transcription factor DNA binding sites. J. Mol. Biol. 297, 833–848 (2000).
Gronostajski, R. M. Site-specific DNA binding of nuclear factor I: effect of the spacer region. Nucleic Acids Res 15, 5545–5559 (1987).
Dekker, J., Oosterhout, J. A. W. M. V. & Vliet, P. C. V. D. Two Regions within the DNA Binding Domain of Nuclear Factor I Interact with DNA and Stimulate Adenovirus DNA Replication Independently. Mol. Cell. Biol. 16, 4073–4080 (1996).
Gounari, F. et al. Amino-terminal domain of NF1 binds to DNA as a dimer and activates adenovirus DNA replication. EMBO J. 9, 559–566 (1990).
Kruse, U. & Sippel, A. E. The genes for transcription factor nuclear factor i give rise to corresponding splice variants between vertebrate species. J. Mol. Biol. 238, 860–865 (1994).
Mermod, N., O’Neill, E. A., Kelly, T. J. & Tjian, R. The proline-rich transcriptional activator of CTF/NF-I is distinct from the replication and DNA binding domain. Cell 58, 741–753 (1989).
Bandyopadhyay, S. & Gronostajski, R. M. Identification of a conserved oxidation-sensitive cysteine residue in the NFI family of DNA-binding proteins. J. Biol. Chem. 269, 29949–29955 (1994).
Bandyopadhyay, S., Starke, D. W., Mieyal, J. J. & Gronostajski, R. M. Thioltransferase (Glutaredoxin) Reactivates the DNA-binding Activity of Oxidation-inactivated Nuclear Factor I. J. Biol. Chem. 273, 392–397 (1998).
Roulet, E. et al. Regulation of the DNA-Binding and Transcriptional Activities of Xenopus laevis NFI-X by a Novel C-Terminal Domain. Mol. Cell Biol. 15, 5552–5562 (1995).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Singh, U., Bongcam-Rudloff, E. & Westermark, B. A DNA Sequence Directed Mutual Transcription Regulation of HSF1 and NFIX Involves Novel Heat Sensitive Protein Interactions. PLoS ONE 4, e5050 (2009).
Mukhopadhyay, S. S. & Rosen, J. M. The C-terminal domain of the nuclear factor I-B2 isoform is glycosylated and transactivates the WAP gene in the JEG-3 cells. Biochem Biophys. Res Commun. 358, 770–776 (2007).
El-Gebali, S. et al. The Pfam protein families database in 2019. Nucleic Acids Res 47, D427–D432 (2019).
Linding, R. GlobPlot: exploring protein sequences for globularity and disorder. Nucleic Acids Res 31, 3701–3708 (2003).
Linding, R. et al. Protein Disorder Prediction. Structure 11, 1453–1459 (2003).
Novak, A., Goyal, N. & Gronostajski, R. M. Four conserved cysteine residues are required for the DNA binding activity of nuclear factor I. J. Biol. Chem. 267, 12986–12990 (1992).
Holm, L. Using Dali for Protein Structure Comparison. Methods Mol. Biol. 2112, 29–42 (2020).
Zawel, L. et al. Human Smad3 and Smad4 Are Sequence-Specific Transcription Activators. Mol. Cell 1, 611–617 (1998).
Krissinel, E. Stock-based detection of protein oligomeric states in jsPISA. Nucleic Acids Res 43, W314–W319 (2015).
Zhang, B. et al. Enhancement of Lysozyme Crystallization Using DNA as a Polymeric Additive. Crystals 9, 186 (2019).
Massagué, J., Seoane, J. & Wotton, D. Smad transcription factors. Genes Dev. 19, 2783–2810 (2005).
Klaassens, M. et al. Malan syndrome: Sotos-like overgrowth with de novo NFIX sequence variants and deletions in six new patients and a review of the literature. Eur. J. Hum. Genet 23, 610–615 (2015).
Zenker, M. et al. Variants in nuclear factor I genes influence growth and development. Am. J. Med Genet 181, 611–626 (2019).
Madeira, F. et al. The EMBL-EBI Job Dispatcher sequence analysis tools framework in 2024. Nucleic Acids Res 52, W521–W525 (2024).
Ruiz, L. et al. Unveiling the dimer/monomer propensities of Smad MH1-DNA complexes. Comput Struct. Biotechnol. J. 19, 632–646 (2021).
Martin-Malpartida, P. et al. Structural basis for genome wide recognition of 5-bp GC motifs by SMAD transcription factors. Nat. Commun. 8, 2070 (2017).
BabuRajendran, N. et al. Structure of Smad1 MH1/DNA complex reveals distinctive rearrangements of BMP and TGF-beta effectors. Nucleic Acids Res 38, 3477–3488 (2010).
Chai, N. et al. Structural basis for the Smad5 MH1 domain to recognize different DNA sequences. Nucleic Acids Res 43, 9051–9064 (2015).
Laskowski, R. A. PDBsum: Summaries and analyses of PDB structures. Nucleic Acids Res 29, 221–222 (2001).
Pettersen, E. F. et al. UCSF Chimera - A visualization system for exploratory research and analysis. J. Comput Chem. 25, 1605–1612 (2004).
Taglietti, V. et al. RhoA and ERK signalling regulate the expression of the transcription factor Nfix in myogenic cells. Development 145, dev163956 (2018).
Walker, M. et al. An NFIX-mediated regulatory network governs the balance of hematopoietic stem and progenitor cells during hematopoiesis. Blood Adv. 7, 4677–4689 (2023).
Kabsch, W. Biological Crystallography. Acta Crystallogr D. 66, 125–132 (2010).
Sheldrick, G. M. Experimental phasing with SHELXC/D/E: Combining chain tracing with density modification. Acta Crystallogr D. 66, 479–485 (2010).
Schneider, T. R. & Sheldrick, G. M. Substructure solution with SHELXD. Acta Crystallogr D. 58, 1772–1779 (2002).
Langer, G., Cohen, S. X., Lamzin, V. S. & Perrakis, A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat. Protoc. 3, 1171–1179 (2008).
Emsley, P. & Cowtan, K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D. 60, 2126–2132 (2004).
Murshudov, G. N. et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D. 67, 355–367 (2011).
Adams, P. D. et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D. 66, 213–221 (2010).
Chen, V. B. et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr D. 66, 12–21 (2010).
Zivanov, J. et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Acknowledgements
We thank the Trieste staff on beamline XRD2. Cryo-EM studies were carried out using the Unitech NOLIMITS facility at the University of Milan. LJG is grateful for funding from “Linea 2”, Università degli Studi di Milano.
Author information
Authors and Affiliations
Contributions
Conceptualization and supervision by A.C.-S., G.M., L.J.G., M.B. and M.N. Recombinant protein production and EMSA studies were carried out and analyzed by A.C.-S., A.R., L.J.G., M.C., M.L., M.T., N.G., R.S., R.R., and V.T. X-ray crystallography studies were carried out by A.C.-S., M.L., M.N., M.T., M.P. and N.D. Cryo-EM data were acquired and processed by A.C.-S., D.M.V.J.B. and M.T. CD studies were performed by A.G.B. FAAS studies were carried out by S.C. Alphafold predictions and in silico modelling was carried out by A.K. and C.C. Funding acquisition and Resources by G.M., L.J.G., M.B. and M.N. Original draft by A.C.-S., L.J.G., M.T. and M.N. Review and Editing by A.C.-S., A.G.B., C.C., G.M., L.J.G., M.B., M.N., M.P., N.G., N.D. and R.R. Data visualization by A.G.B., L.J.G., M.T and S.C. and edited and reviewed by L.J.G., N.G. and M.N. Contribution to and approval of the submitted version by all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Richard Gronostajski, Fumiaki Ito, and the other anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Tiberi, M., Lapi, M., Gourlay, L.J. et al. Structural basis of Nuclear Factor 1-X DNA recognition provides prototypic insight into the NFI family. Nat Commun 16, 10170 (2025). https://doi.org/10.1038/s41467-025-65186-0
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-65186-0