Abstract
We analyze 10,986 participants (mean age 77; 63% women; 54% non-White) across seven U.S. cohorts to study the relationship between mitochondrial DNA (mtDNA) heteroplasmy and nuclear DNA methylation. We identify 597 CpGs associated with heteroplasmy burden, generally showing lower methylation. These CpGs are enriched in dynamically regulated island shores and depleted in CpG islands, indicating involvement in context-specific rather than constitutive gene regulation. In HEK293T cells, we introduce a truncating mtDNA mutation (MT-COX3, mt.9979) and observe a positive correlation between variant allele fraction and methylation at cg04569152, supporting a direct mtDNA–nDNA epigenetic link. Many heteroplasmy-associated CpGs overlap with known methylation-trait associations for metabolic and behavioral traits. Composite CpG scores predict all-cause mortality and incident CVD, with one-unit increases associated with 1.27-fold and 1.12-fold higher hazards, respectively. These findings suggest an mtDNA–nDNA epigenetic connection in aging and disease, though its direction and mechanisms remain to be studied.
Similar content being viewed by others
Introduction
Mitochondria are central for generating molecular energy and multiple biochemical processes1,2,3. The mitochondrial genome is a double-stranded DNA molecule (mtDNA), which is 16.6 kb in size and exists in multiple copies per cell1,2,3. This gives rise to two quantities: mtDNA copy number (mtDNA CN), a measure of the average number of mtDNA molecules per cell, and heteroplasmy, where different mtDNA alleles coexist within the same sample4. Heteroplasmy often arises somatically and accumulates with age5,6,7. Recent research suggests that heteroplasmy burden is associated with cancer and mortality7. However, the biological pathways underlying these relationships remain unclear.
Mitochondrial function depends on tight coordination between mitochondrial DNA (mtDNA) and nuclear DNA (nDNA)8. DNA methylation, a key regulator of gene expression9,10, has been implicated in modulating mitochondrial function11,12. Interestingly, the reverse may also be true, that is, mtDNA variation can influence nuclear epigenetic states8. For instance, global nDNA methylation levels were shown to vary in human cybrid cells that shared identical nuclear genomes but carried different mtDNA haplogroups13. Similarly, in a hybrid mouse model with nDNA from one strain and mtDNA from another, mammary tumor metastasis was altered through changes in DNA methylation, with corresponding shifts in gene expression14. Extending this evidence, recent epigenome-wide association studies (EWAS) and meta-analyses have identified CpG sites associated with mtDNA CN15,16. Supporting a potential mechanistic connection, experimental studies have shown that altering mtDNA CN leads to changes in methylation at specific CpG sites and affects the expression of nearby genes15. Together, these findings suggest that mtDNA may influence health outcomes through nuclear methylation pathways.
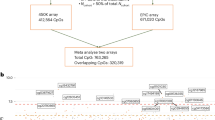
While previous studies have examined global DNA methylation in relation to mtDNA haplogroups defined by inherited variants13,14,17,18, other work has implicated mtDNA heteroplasmy in cellular function and adverse health outcomes6. However, the broader biological impact of heteroplasmy remains unclear. We hypothesize that mtDNA heteroplasmy may be associated with changes in nDNA methylation, contributing to age-related outcomes. To assess this, we identified CpG sites associated with mtDNA heteroplasmy using data from multiple large-scale, racially diverse cohorts and performed functional validation by introducing a protein-truncating mtDNA mutation (MT-COX3, mt.9979) into HEK293T cells (Fig. 1). As noted by Lopes (2020)8, DNA methylation may serve as a regulatory link between mitochondrial and nuclear genomes in shaping disease risk. Therefore, we examined whether these heteroplasmy-associated methylation markers are predictive of all-cause mortality and CVD risk. Our findings suggest that mitochondrial and nuclear factors may work together to influence health and disease, though the direction and underlying mechanisms of these links are still unclear and need further study.
Whole-genome sequencing was conducted to identify mitochondrial DNA (mtDNA) heteroplasmy in 10,986 individuals across seven epidemiological cohorts. Cohort- and ancestry-specific association analyses were performed to investigate the relationship between mtDNA heteroplasmy and nuclear DNA (nDNA) methylation levels at CpG sites. Random-effects meta-analyses were subsequently conducted in both pooled and race-stratified datasets. CpG sites demonstrating statistically significant associations (false discovery rate [FDR] < 0.05) were subjected to downstream bioinformatic analyses, including gene ontology enrichment and transcriptomic integration. Experimental validation using targeted genome editing in cell lines, followed by pyrosequencing, provided functional evidence supporting a causal role of mtDNA heteroplasmic variation in modulating nuclear epigenetic states. Integrative analyses further linked heteroplasmy-associated CpGs to complex traits through methylation quantitative trait loci (mQTL) mapping, interrogation of the Epigenome-Wide Association Studies (EWAS) Catalog and Genome-Wide Association Studies (GWAS) Catalog, and Mendelian randomization. Finally, composite methylation scores derived from heteroplasmy-associated CpG sites were evaluated for association with all-cause mortality and cardiovascular disease risk.
Results
Participant characteristics
We conducted association analyses of genome-wide CpG methylation and mtDNA heteroplasmy in 10,986 participants from seven diverse U.S.-based cohorts19,20,21,22,23,24,25,26, most of whom were middle-aged (mean age: 57 years). Overall, 6,866 (63%) were women and 54% of participants were non-White, based on self-reported sex, race, and ethnicity (Supplementary Data 1–2).
We used two measures to quantify heteroplasmy burden: mitochondrial heteroplasmy count (MHC), the number of heteroplasmic variants, and mitochondrial local constraint score sum (MSS), which reflects their predicted functional impact27. Heteroplasmic variants were identified at 2,264 distinct sites across the mtDNA genome (Fig. 2A), each present in less than 1% of participants in the combined sample (Supplementary Data 3). Approximately one-third of participants carried at least one heteroplasmic variant, and approximately 12.4% (8.3% to 18.8% across cohorts) had an MSS between 0.01 and 0.25; 4.4% (2.7% to 5.8%) had an MSS between 0.251 and 0.5; and 7.9% (5.4% to 10.8%) had an MSS greater than 0.5 (Fig. 2B, Supplementary Data 1-2). Consistent with previous findings, MHC levels increase with age5,7 (Fig. 2C). Across cohorts, 8% to 22% of participants were current smokers, 15% to 45% were former smokers, and 41% to 65% were never smokers (Supplementary Data 1-2). Predicted smoking scores were generally consistent with self-reported smoking status (Supplementary Fig. 1).
A Distribution of heteroplasmy counts (MHC) in study participants (n = 10,986) across the double-stranded mitochondrial DNA (mtDNA) molecule, following quality control procedures (see Methods). Heteroplasmic variants were identified at 2264 unique sites across the mtDNA genome, with each site occurring in less than 1% of participants in the combined sample (Supplementary Data 3). Pink regions indicate the D-loop regions. Blue regions indicate the 13 protein-coding genes. B Distribution of MHC and mitochondrial local constraint score sum (MSS) score across the study participants. MHC represents the total number of heteroplasmic variants in an individual, while MSS reflects the predicted deleteriousness and functional impact of these variants. Approximately 32.4% of participants carried at least one heteroplasmic variant. Among all participants, 12.4% had MSS values between 0.01 and 0.25, 4.4% had values between 0.251 and 0.5, and 7.9% had MSS values greater than 0.5. C The distribution of MHC across age groups indicates a trend of increasing MHC levels with advancing age.
Association and meta-analyses of heteroplasmy with DNA methylation
We conducted cohort- and race-specific association analyses between mtDNA heteroplasmy burden scores (MHC and MSS) and DNA methylation at CpG sites across the nuclear genome using a linear regression model for unrelated individuals and linear mixed model for family data. Models were adjusted for age, self-reported sex, smoking score28, genetic principal components (PCs) to account for population admixture, white blood cell counts, and batch variables, including year of blood draw for whole genome sequencing29, and DNA methylation chip ID, and row and column positions (see Methods). Meta-analyses were performed across all samples (primary analysis) and within race-specific subgroups (secondary analysis). The primary analysis focused on CpG sites present on both the 450K and EPIC arrays (Supplementary Data 4) after extensive quality control (see Methods). We examined genomic inflation factors and compared association models with and without adjustment for genetic PCs (Supplementary Figs. 2–9)
In the random-effects meta-analysis of all samples (n = 10,986), we observed slightly deflated genomic control values (λGC) for both MSS (λGC = 0.79) and MHC (λGC = 0.83) (Supplementary Data 5). At a false discovery rate (FDR) p < 0.05, we identified 597 unique CpGs associated with MSS (n = 479) and MHC (n = 166) across the genome (Fig. 3A, Table 1, Supplementary Data 6–7). We observed consistent effect sizes for the epigenome-wide analyses with MSS and MHC (Pearson r = 0.75) (Fig. 3B, Supplementary Fig. 10), reflecting the moderate to high correlation between the two heteroplasmy metrics across cohorts (Supplementary Data 8). The majority of CpGs (91% for MHC and 94% for MSS with FDR < 0.05) displayed negative associations with heteroplasmy, indicating that a higher level of heteroplasmy was associated with a lower level of DNA methylation (Fig. 3C). The directionality of the association was most consistent across the cohorts for significant CpGs, as exampled with cg15233611 (Fig. 3D).
A Cohort- and race-specific association analyses using linear regression were followed by meta-analyses in pooled samples (n = 10,986) to identify CpG sites associated with MSS (top) and MHC (bottom). We found 479 MSS- and 166 MHC-associated CpGs at a false discovery rate (FDR) < 0.05 in a two-sided test. The x-axis indicates chromosomes in different colors. The gray dotted line represents the FDR-adjusted significance threshold (FDR = 0.05), and the black dotted line indicates the Bonferroni threshold (p = 1 × 10-7). B Comparison of effect sizes (beta values) for MSS vs. MHC meta-analyses, showing a Pearson correlation r of 0.75. Red dots indicate CpGs with positive betas and blue dots with negative betas. C Volcano plot of DNA methylation with MSS in meta-analysis. Most significant CpGs (94%, FDR < 0.05) showed negative associations (blue dots), while a minority showed positive associations (red points). D Forest plot of the top CpG, cg15233611, from linear regression analyses of MSS in African American (AA) and White American (EA) participants, and in the meta-analysis. All tests were two-sided t-tests. Among AA participants, beta-estimates (95% CI) were 0.00087 (–0.0097, 0.011) in ARIC, –0.0077 (–0.032, 0.016) in CARDIA, –0.018 (–0.038, 0.0021) in GENOA, –0.022 (–0.033, –0.0097) in JHS, 0.0059 (–0.027, 0.039) in MESA, and –0.021 (–0.066, 0.024) in WHI, with a meta-analysis estimate of –0.010 (–0.021, 0.00063) in all AA participants. For White American (EA) participants, the β (95% CI) values were –0.0087 (–0.022, 0.0041) in ARIC, –0.0094 (–0.031, 0.012) in CARDIA, –0.015 (–0.021, –0.0092) in FHS, –0.013 (–0.035, 0.0086) in MESA, and –0.039 (–0.058, –0.0019) in WHI. The meta-analysis of EA participants yielded a beta of −0.015 (–0.020, –0.010), and the overall meta-analysis combining AA and EA participants produced a beta of –0.014 (–0.019, –0.0099). CI confidence interval, ARIC Atherosclerosis Risk in Communities, CARDIA Coronary Artery Risk Development in Young Adults, FHS Framingham Heart Study, GENOA Genetic Epidemiology Network of Arteriopathy, JHS Jackson Heart Study, MESA Multi-Ethnic Study of Atherosclerosis Study, WHI Women’s Health Initiative.
Random-effects meta-analyses of associations between DNA methylation at CpG sites and both MSS and MHC were conducted separately in White American (n = 5104) and African American (n = 5882) participants as secondary analyses (Supplementary Data 9–10, Supplementary Figs. 11–12). We compared the beta estimates of top CpGs (p < 10-5) from the meta-analyses of White American participants with those of African American participants. For MSS, most CpGs (82%) showed a consistent direction of association, although the correlation in effect sizes between the two groups was modest (Pearson r = 0.30). Similar patterns were also observed for MHC between the two groups (Supplementary Figs. 13–14).
Transcriptomic implication of CpGs associated with heteroplasmy
To investigate possible downstream consequences of DNA methylation, we retrieved the gene transcripts associated with the identified CpGs (eQTM) from the eQTM database from the Framingham Heart Study (FHS)30. We found 95 heteroplasmy-associated CpGs exhibiting significant associations with the expression of 146 cis-genes within 1 Mb of a CpG (p < 1 × 10-7) (Supplementary Data 11). Of those, 43 CpG sites showed significant associations with the expression levels of the genes they are located in (Table 2), indicating that methylation at these sites may influence the expression of their corresponding genes. For example, cg02633767, located in the 3’- untranslated region of TAP2 (Transporter 2, member B of the ATP-binding cassette subfamily on chromosome 6) was significantly associated with the expression of TAP2 gene (p = 1 × 10-29). This gene encodes a protein that is essential for the proper functioning of the immune system31.
Genomic context enrichment analysis of heteroplasmy-associated CpG sites
To gain insights into their biological relevance, we used Chi-square test or Fisher’s Exact test (for cell count less than 5) to determine whether the 597 heteroplasmy-associated CpG sites are non-randomly distributed across genomic contexts. We found that these sites were significantly depleted in CpG islands (0.42-fold, p = 5.2 × 10-21) but enriched in shore regions flanking CpG islands (north shore: 1.96-fold, p = 1.6 × 10-19; south shore: 1.83-fold, p = 9.4 × 10-12), with no significant enrichment in shelf regions farther from CpG island (p > 0.5) (Supplementary Data 12). Additionally, significant depletion was observed in DNase I hypersensitive site (DHS) regions (0.32-fold, p = 4.1 × 10-6). No significant enrichment was observed in enhancer regions (p = 0.083) or differentially methylated regions (DMRs) (p = 0.086).
We further analyzed 146 cis-genes associated with the 95 heteroplasmy-associated CpGs (Supplementary Data 11). Similar to the broader set of 597 CpGs, these eQTMs displayed depletion in CpG islands (0.55-fold, p = 0.004) and enriched in north shore (2-fold, p = 0.00014). Additionally, they showed stronger depletion in DHS (0.26-fold, p = 1.2 × 10-11) and DMRs (0.19-fold, p = 9.4 × 10-8) regions, while showed no enrichment in the south shore (p = 0.41), shelf, or enhancer locations (p > 0.4) (Supplementary Data 12).
Gene set enrichment analysis
Of the genes annotated to CpGs, 370 were analyzed by DAVID, a comprehensive tool for functional annotation, disease association, and Gene Ontology (GO) analysis32. Based on Genetic Association Database (GAD), these genes were significantly enriched in a broad selection of diseases such as metabolic conditions (FDR=0.014), cancer (FDR = 0.017), and chemical dependency (addicted to drugs, nicotine, or alcohol) (FDR = 0.003), among others (Fig. 4A, Supplementary Data 13). GO analysis identified pathways, such as signaling (e.g., GO:0023051, FDR=4 × 10-9) and neurodevelopment (e.g., GO:0048856, FDR = 5.8 × 10-7) (Fig. 4B, Supplementary Data 14). GO analysis also identified cellular components involving synaptic functions and neurotransmission (e.g., synapse [GO:0045202], FDR = 0.0002), and membrane components (e.g., postsynaptic membrane, [GO:0045211], FDR = 0.01) (Supplementary Data 15). Gene set enrichment analysis of 146 genes linked to 95 CpGs revealed top pathways and cellular components similar to those identified from the full set of heteroplasmy-associated genes (Supplementary Data 14–15).
A A total of 370 genes annotated to the heteroplasmy-associated CpGs were analyzed using DAVID, a comprehensive tool for functional annotation, disease association, and Gene Ontology (GO) analysis. Enrichment was assessed using DAVID’s modified Fisher’s exact test (EASE score), a two-sided test, detecting both over- and under-representation of terms relative to the background set. Enrichment analysis using the Genetic Association Database (GAD) revealed that these genes are enriched in metabolic traits and processes related to multicellular organization (a false discovery rate, FDR < 0.05). B GO biological pathway analysis of these 370 genes indicated enrichment in pathways related to signaling and neurodevelopment (FDR < 0.05). C Correlation between variant allele fraction (VAF) and CpG methylation at cg04569152. Functional validation using HEK293T cell line editing and pyrosequencing demonstrated a significant positive correlation between VAF of an mtDNA nonsense mutation (mt.9979 in MT-COX3) and methylation levels at cg04569152 (p = 0.003 from a two-sided t-test). The mutation was introduced using the FusX TALE-based editing system (FusXTBE), and methylation was quantified by pyrosequencing. Linear correlation was assessed using Pearson’s method, supporting a potential epigenetic influence of mtDNA heteroplasmic variation on nuclear CpG methylation (see Supplementary Fig. 16). D Integrative analysis of cg15233611(SETD1B) illustrates its associations with mtDNA heteroplasmy burden, measured by the mitochondrial local constraint score sum (MSS), as well as with smoking, HDL cholesterol, and triglycerides, supported by methylation-trait associations and Mendelian randomization (MR). Pink circles with the positive sign (+) indicate a positive association, while blue circles with the negative sign (-) indicate a negative association. Association analyses were performed using linear regression, with significance assessed by two-sided t-tests for regression coefficients. Mendelian randomization (MR) analyses used the inverse-variance weighted (IVW) method, with significance assessed by two-sided Wald z-tests.
Genes involved in mitochondrial assembly and function
Through functional annotation of 370 genes with DAVID32 and MitoCarta3.033., we identified 27 genes known or predicted to be involved in mitochondrial processes (Supplementary Data 16). Of these, twelve encode proteins that are part of the 1136 mammalian mitochondrial proteome listed in MitoCarta3.033, representing a lower-than-expected number of mitochondrial genes in our set relative to a background of 20,000 genes (Fisher’s Exact Test, p = 0.022). For example, ABHD10 encodes a mitochondrially localized enzyme and regulates redox homeostasis by modulating the palmitoylation of antioxidant proteins34. Among the others, NDUFA5 and NDUFS4 encode subunits of Complex I, a critical component of the mitochondrial electron transport chain essential for oxidative phosphorylation and ATP production35,36. The remaining genes are implicated in NAD + /NADP + /NADPH-dependent functions, oxidative stress response, or phosphorylation-related pathways. For instance, ALDH3B1 encodes a protein involved in cellular detoxification and oxidative stress mitigation37.
Functional validation of correlation between nuclear CpG methylation and VAF in COX3-mutants
We selected five CpG sites based on an interim analysis of the data for functional validation (Supplementary Data 17), primarily based on their interim meta-analysis p-values and high pyrosequencing design scores ( > 0.8), and evidence of being cis-eQTMs or having associated cis-SNPs (i.e., mQTLs). To assess the impact of mitochondrial mutations on methylation, HEK293T (human embryonic kidney) cell lines were edited to harbor a nonsense mutation at mt.9979 in MT-COX3 (cyclooxygenase-3 gene, referred to as COX3-mutant cell lines) using the FusXTBE system38,39. Successful editing was confirmed by Sanger sequencing. DNA methylation levels at the selected CpG sites were quantified by pyrosequencing40,41. Of note, the mt.9979 mutation was not observed in our cohorts (Supplementary Data 3).
Variant allele fractions (VAFs) of the nonsense mutation (mt.9979) ranged from 11-91%, with minimal evidence of off-target editing (Supplementary Fig. 15). Pyrosequencing was performed on 3 unedited control cell lines and 12 COX3-mutant lines selected to represent a spectrum of VAFs (Supplementary Data 18). Correlation analysis revealed a statistically significant positive linear relationship between VAF and DNA methylation at CpG site cg04569152 (p = 0.025) but not at the other four CpG sites (Supplementary Figs. 16-17). This finding was replicated in an independent sample set (cell lines) (Supplementary Data 18), where the association remained significant (p = 0.029), and was further strengthened when all samples were combined (p = 0.003) (Fig. 4C, Supplementary Fig. 16). These findings support a potential link between mitochondrial DNA mutations and nuclear DNA methylation, raising the possibility that variation in mitochondrial function could contribute to shaping the nuclear epigenetic landscape.
Linking heteroplasmy-associated CpGs to human traits via EWAS Catalog
We queried the MRC-IEU epigenome-wide association studies (EWAS) Catalog42 to associate the heteroplasmy-associated CpGs (FDR < 0.05) to previously reported diseases and traits. We identified 54 CpG-trait associations, involving 52 unique CpGs associated with five traits (p < 1 × 10-7) in studies with more than 5000 participants in the MRC-IEU EWAS Catalog42 (Supplementary Data 19). The five traits included alcohol consumption per day (6 CpGs), body mass index (4 CpGs), C-reactive protein (3 CpGs), educational attainment (1 CpG), and smoking-related traits (35 CpGs) (Supplementary Fig. 18).
Linking heteroplasmy-associated CpGs to human traits via mQTLs and GWAS Catalog
Investigating DNA methylation quantitative trait loci (mQTLs) and their associated genome-wide association study (GWAS) traits or diseases may provide insights into the molecular mechanisms underlying complex traits and diseases43,44. In our analysis, 505 heteroplasmy-associated CpG sites were associated with at least one SNP (p < 5 × 10-8) in the FHS mQTL database45. Of these, 374 CpG sites were linked to at least one SNP associated with a trait or disease in the GWAS Catalog46. For each CpG, we retained the SNP with the most significant GWAS association to provide trait specificity and avoid redundancy (Supplementary Data 20). A substantial proportion of the traits identified, such as blood pressure, lipid levels, body mass index, type 2 diabetes, and smoking behavior, are well-established risk factors for both cardiovascular disease and all-cause mortality.
Linking heteroplasmy-associated CpGs to human traits via MR analysis
Mendelian randomization (MR)47 was used to assess potential causal relationships between DNA methylation at heteroplasmy-associated CpG sites and both CVD-related outcomes and all-cause mortality. Using the cis-mQTLs (p < 5 × 10-8) in the FHS mQTL database45 as instrumental variables, we identified top associations across ten CVD-related traits and age at death (FDR < 0.05) (Supplementary Data 21). For example, cg15233611 showed a positive causal effect on HDL cholesterol (MR β = 1.5, FDR = 0.00012) and a negative effect on triglycerides (MR β = −1.45, FDR = 0.00011) (Fig. 4D). These results align with previous findings linking lower methylation at cg15233611 to smoking48, and established associations between smoking, decreased HDL cholesterol, and increased triglyceride levels49,50. cg15233611 is located in the SET Domain Containing 1B (SETD1B) gene that is involved in histone methylation51, and was correlated with reduced expression of HPD (4-hydroxyphenylpyruvate dioxygenase)52 (Supplementary Data 11) that is about 33 Kb downstream of the SETD1B. Another example is cg03732020 in NR1H3, which was inferred to have a causal association with high-density lipoprotein (HDL) cholesterol (MR beta = 8.5, FDR = 5.5 × 10-90), consistent with previous findings. NR1H3 encodes liver X receptor alpha (LXRα), a nuclear receptor that plays a key role in lipid metabolism, particularly in the regulation of HDL levels53,54.
Association analysis of heteroplasmy-associated CpG scores with all-cause mortality
To investigate the potential mechanistic link between mitochondrial-nuclear communication and age-related disease risk, we derived heteroplasmy-associated CpG methylation scores and evaluated their associations with all-cause mortality. The FHS was used as the training cohort, with internal validation performed across the remaining cohorts. External validation was conducted using the Health and Retirement Study (HRS) cohort (Supplementary Data 22).
We observed 562 deaths over a median 13-year follow-up among 3488 FHS participants with DNA methylation data. Additionally, we observed a total of 430 deaths over a median follow-up of 14 to 20 years across the JHS and MESA cohorts. In the HRS cohort, there were 451 deaths over a median follow-up of 3 years (Supplementary Data 23).
We applied elastic net Cox regression to heteroplasmy-associated CpGs and identified 57 CpGs associated with MHC for all-cause mortality in FHS (Supplementary Data 24). We constructed a weighted MHC-CpG score and found that a one standard deviation (SD) higher level of this score was associated with a 1.61-fold higher hazard of all-cause mortality (95% CI: 1.50, 1.72), adjusting for age, self-reported sex, and smoking status. In testing samples, we found that a one-SD higher level of the weighted MHC-CpG score was associated with a 1.12-fold higher hazard of all-cause mortality (95% CI: 0.99, 1.27) in the meta-analysis of JHS and MESA cohorts, and a 1.27-fold higher hazard of all-cause mortality (95% CI: 1.09, 1.48) in the meta-analysis of JHS, MESA, and HRS cohorts, adjusting for age, self-reported sex, and smoking (Fig. 5).
We applied elastic net–regularized Cox proportional hazards regression in the Framingham Heart Study (FHS) training cohort to identify heteroplasmy count (MHC)-associated CpGs, yielding 57 associated with all-cause mortality in panel A and 18 associated with cardiovascular disease (CVD) in panel B. Statistical significance of regression coefficients was assessed using two-sided Wald z-tests. The forest plot presents hazard ratios (HRs) and 95% confidence intervals (CIs) for associations between weighted MHC-CpG scores and outcomes (mortality and incident CVD) across training and testing cohorts, adjusted for age, self-reported sex, and smoking status (never, former, current). In FHS, the 57- and 18-CpG scores yielded HRs (95% CI) of 1.61 (1.50–1.72) for all-cause mortality and 1.29 (1.23–1.36) for CVD, respectively. Internal validation in the Jackson Heart Study (JHS) and the Multi-Ethnic Study of Atherosclerosis (MESA) showed, for all-cause mortality, HRs (95% CI) of 0.99 (0.77–1.28) in MESA White American (EA) participants, 1.18 (1.06–1.31) in JHS, and 1.02 (0.70–1.48) in MESA African American (AA) participants for the 57-CpG score. For CVD, the 18-CpG score yielded HRs (95% CI) of 0.97 (0.78–1.21) in MESA EA, 1.19 (1.05–1.36) in JHS, and 1.41 (0.97–2.04) in MESA AA. Meta-analysis of JHS and MESA participants produced HRs (95% CI) of 1.12 (0.99–1.27) for all-cause mortality and 1.15 (0.97–1.36) for CVD. External validation in the Health and Retirement Study (HRS), which was not used for CpG selection, found that for all-cause mortality, the 57-CpG score yielded HRs (95% CI) of 1.28 (1.16–1.42) in EA, 1.62 (1.31–2.01) in AA, and 1.78 (1.18–2.70) in Hispanic American (HA) participants. For CVD, the 18-CpG score yielded HRs of 1.07 (0.97–1.19) in EA, 0.83 (0.59–1.16) in AA, and 1.48 (1.06–2.05) in HA. Meta-analysis of MESA, JHS, and HRS participants produced HRs (95% CI) of 1.27 (1.09–1.48) for all-cause mortality and 1.12 (0.99–1.26) for CVD.
Using the same method, we identified 33 MSS-associated CpGs for all-cause mortality in FHS (Supplementary Data 25). We found consistent results between the MSS-CpG weighted score and all-cause mortality in FHS, as well as in meta-analyses of testing samples, adjusting for age, self-reported sex, and smoking status. Furthermore, we observed consistent results in both the base model (age and self-reported sex adjusted) and the multi-covariate adjusted model for the associations of both MHC- and MSS-weighted scores with all-cause mortality (Supplementary Figs. 19–20).
Association analysis of heteroplasmy-associated CpG scores with CVD
Similarly, we derived heteroplasmy-associated CpG methylation scores and evaluated their associations with incident CVD. In the FHS, 300 participants developed CVD during a median follow-up of approximately 8 years among 3,488 participants. We observed a total of 320 incident CVD cases with a median follow-up of 14 to 16 years across the JHS and MESA. The external cohort HRS had 301 incident CVD cases with a much shorter median follow-up (4 years) compared to other cohorts. (Supplementary Data 23).
We selected 18 CpGs for MHC using elastic net Cox regression (Supplementary Data 26) for CVD. In the FHS testing sample, we found that a SD higher level of the weighted MHC-CpG score was associated with a 1.29-fold higher hazard of CVD (95% CI: 1.23–1.36), adjusting for age, self-reported sex, and smoking status. In the testing cohorts, we found that a one-SD higher MHC-CpG score was associated with a 1.15-fold higher hazard of CVD (95% CI: 0.97–1.36) in the meta-analysis of JHS and MESA, and a 1.12-fold higher hazard of CVD (95% CI: 0.99–1.26) in the meta-analysis of JHS, MESA, and HRS (Fig. 5, Supplementary Fig. 21).
We identified 19 MSS-associated CpGs for CVD using elastic net Cox regression in FHS (Supplementary Data 27). Compared to that of the weighted MHC-CpG score, the weighted MSS-CpG score showed slightly weaker associations with CVD in the training cohort and meta-analysis of JHS, MESA, and HRS as the testing samples in both base model and the model with smoking as an additional covariate. Adjusting for additional multi-covariates further attenuated the associations for both weighted MHC- and MSS-scores (Supplementary Figs. 21–22).
Discussion
We examined the associations between nDNA methylation and mtDNA heteroplasmy in 10,986 participants from seven U.S.-based cohorts representing diverse racial and ethnic backgrounds. Our analysis identified 597 unique CpGs (FDR < 0.05) with differential methylation associated with mtDNA heteroplasmy burden. More CpGs were associated with the mitochondrial stress signal (MSS) metric than with heteroplasmic variant count (MHC), with most exhibiting inverse associations, indicating that higher heteroplasmy burden generally corresponds to lower levels of nDNA methylation. Heteroplasmy-associated CpGs were significantly depleted in CpG islands, DHS, and DMRs, but enriched in island shores, regions characterized by dynamic, tissue-specific55,56,57, and environmentally responsive gene regulation58. These patterns suggest that heteroplasmy-associated CpGs may influence gene expression in a context-dependent manner rather than through constitutive promoter activity9,10
To assess functional relevance, we introduced a single heteroplasmic mutation (mt.9979, MT-COX3) into HEK293T cells38,39 and measured DNA methylation at selected CpG sites with pyrosequencing40,41. Increasing VAF of the COX3 mutation was positively correlated with methylation at cg04569152, contrasting with the mostly inverse trend seen in the population data. This discrepancy likely reflects differences in experimental context. Specifically, the epidemiologic associations were derived from whole blood samples, which reflect systemic and immune-related processes, while the experimental validation was conducted in a single immortalized human embryonic kidney cell line. Furthermore, the heteroplasmy burden score in the population analysis reflects a composite of many rare heteroplasmic variants, whereas the experimental model isolates the effect of a single, protein-truncating mutation with potentially distinct functional consequences, one not observed in the population-level data (Supplementary Data 3). Despite the difference in direction, these results provide functional evidence that mitochondrial heteroplasmic variation can directly influence nuclear epigenetic states, reinforcing the hypothesis of mitochondrial-nuclear crosstalk59,60,61.
Findings from integrative analysis align with prior literature supporting crosstalk between the nuclear and mitochondrial genomes in response to environmental cues59,60,61. Many genes annotated to heteroplasmy-associated CpG sites are involved in mitochondrial biosynthesis, redox regulation, and energy metabolism. These include MRPS11, which encodes mitochondrial ribosomal protein S1162, NDUFA5 and NDUFS4 (encoding respiratory chain complex I subunits)35,36, and PRMT6, a nuclear protein methyltransferase that may influence mitochondrial biogenesis63. Additional examples include SNED1, which encodes an extracellular matrix protein involved in tissue organization and development64, while TAP2, an ABC transporter that facilitates molecular transport across intracellular membranes, including mitochondria65. Together, these findings highlight the bidirectional nature of mitochondrial-nuclear relationship.
To assess the broader implications of these epigenetic changes, we examined overlap between heteroplasmy-associated CpGs and known methylation-trait associations42. Several CpGs were linked to metabolic and behavioral traits such as BMI, alcohol consumption, and smoking. Mendelian randomization analyses supported potential causal roles of methylation at select CpGs in influencing CVD risk factors, such as HDL cholesterol and triglycerides. Finally, composite CpG scores were predictive of both all-cause mortality and incident CVD across multiple validation cohorts, underscoring the potential clinical relevance of mitochondrial–epigenetic interactions.
Despite these strengths, our study has limitations. Its cross-sectional design precludes causal inference. Our findings support several plausible, non-mutually exclusive models: (1) mtDNA heteroplasmy may influence nuclear DNA methylation and downstream health outcomes, as suggested by our experimental data, MR, and outcome analyses; (2) nuclear epigenetic regulation may affect mitochondrial genome stability11,12. (3) both may be co-regulated by shared extrinsic factors such as aging, smoking, or oxidative stress. The latter is supported by the association of many heteroplasmy-linked CpGs with smoking in EWAS Catalog42, even after adjusting for detailed smoking scores. Our ongoing functional studies inducing heteroplasmy and assessing downstream methylation aim to clarify these relationships. Additionally, examining the role of clonal hematopoiesis66, an age-related somatic process, may further illuminate the mechanisms underlying our observations.
This study leveraged genetically diverse, sex-balanced cohorts and applied rigorous quality control throughout statistical analyses. Despite these efforts, we observed variability in genomic inflation factors (λGC) in a few cohort, despite adjustments for genetic ancestry and technical variation. While random-effects meta-analysis reduced this inflation, some residual confounding remain. Lastly, while whole blood is a practical biospecimen, future studies should consider tissue-specific validation of key findings.
In summary, this study provides an epigenome-wide analysis associating mtDNA heteroplasmy burden with specific nuclear CpG methylation changes in a large, multi-ethnic human population. By integrating multiple approaches, this work lays the groundwork for future investigations into the relationship between mtDNA heteroplasmy and nuclear DNA methylation in the context of health and disease. While our findings may reflect an epigenetic aspect of mitochondrial–nuclear interaction, further research is needed to clarify the directionality and underlying mechanisms of these associations.
Methods
Ethical compliance
All research was conducted in accordance with relevant ethical guidelines and regulations, with study protocols approved by the Institutional Review Boards (IRBs) of all participating institutions. The ARIC study maintains IRB oversight via a central IRB at Johns Hopkins (protocol 96-0484). CARDIA was overseen by a single IRB at the University of Alabama at Birmingham (protocol 268201300026C-5-0-1), which operates under Federal Wide Assurance (FWA00005960). The FHS received approval from the Boston Medical Center and Boston University Medical Campus IRB (protocol H-32132). For the GENOA study, approvals were obtained from the University of Michigan Health Sciences and Behavioral Sciences IRB (protocols HUM00008655 and HUM00113791). The Jackson Heart Study was approved by the University of Mississippi Medical Center IRB (protocol 1998-6004). The MESA study is overseen by University of Washington IRB (IRB ID STUDY00014523) under Federal Wide Assurance FWA00006878. The WHI was approved by the IRBs of all participating institutions, with oversight from the Fred Hutchinson Cancer Research Center IRB (IRB protocol 3467). Fred Hutch has an approved FWA on file with the Office for Human Research Protections (OHRP) under assurance number 0001920. For HRS, approval was obtained by the University of Michigan Health Sciences and Behavioral Sciences IRB (IRB-HSBS) (IRB protocol HUM00063444). Written informed consent was obtained from all participants in each study. No compensation was provided for participants.
Study participants
This study included 10,986 participants (mean age 57 years, women 63%, 54% non-White participants based on self-reported sex, race, and ethnicity) from seven diverse U.S.-based cohorts: ARIC (Atherosclerosis Risk in Communities)19, CARDIA (Coronary Artery Risk Development in Young Adults)20, FHS (Framingham Heart Study)21,22, GENOA (Genetic Epidemiology Network of Arteriopathy)23, JHS (Jackson Heart Study)24, MESA (Multi-Ethnic Study of Atherosclerosis)25, and WHI (Women’s Health Initiative)26. Based on prior research67, most FHS participants showed high genetic similarity to European ancestry reference panels, while most JHS and GENOA participants were similar to African ancestry reference panels (mean ~80%). Other cohorts included individuals from both self-identified groups. Our analyses were stratified by self-reported race/ethnicity, without excluding genetic ancestry outliers. Thus, race/ethnicity as used in this study should not be considered equivalent to ancestry proportions. In addition, MESA included 33% Hispanic participants. Details on participant collection for each cohort were provided in the Supplementary Methods and Supplementary Data 1-2.
Inclusion & ethics
All protocols for participant examinations and genetic material collection were approved by the Institutional Review Boards at the respective research sites. Written, informed consent was provided by all participants for genetic studies. All research was carried out in accordance with relevant guidelines and regulations.
Profiling and quality control of DNA methylation in whole blood
Peripheral whole blood samples were used for genomic DNA extraction and bisulfite conversion, followed by methylation profiling per the manufacturer’s protocol (Illumina Inc., San Diego, CA). Two platforms were used for DNA methylation measurement across cohorts (Supplementary Data 4): the Infinium HumanMethylation 450K BeadChip array (covering over 480,000 CpG sites, Illumina Inc., San Diego, CA) for FHS, WHI, and ARIC, and the Infinium MethylationEPIC BeadChip array (covering over 850,000 CpG sites, Illumina Inc., San Diego, CA) for CARDIA, GENOA, JHS, and MESA. Over 90% of CpGs from the 450K array are covered by the EPIC array. Analyses included all CpGs covered by both arrays, with subsequent analyses focusing on overlapping CpGs. In individual cohorts, probes with high missing rates ( > 20%), non-significant detection p-values (> 0.01), underlying SNPs or probes targeting SNPs (minor allele frequency > 5% within 10 bp based on 1000 Genomes Project data) were removed. To ensure consistency, we further excluded possibly problematic probes after the meta-analysis, following the guidelines in Illumina methylation array probe filtering (450K and EPIC/850K)68 which compile resources for filtering problematic probes. Briefly, for the 450K array, 38,941 unique probes were filtered based on Chen et al.69, including 33,457 probes that aligned to multiple genomic locations (16,532 of which were autosomal CpGs associated with significant sex-related methylation differences) and 29,233 non-specific probes, with overlap between categories. For the 850K array, 54,918 unique probes were excluded based on Pidsley et al.70, comprising 43,254 cross-reactive probes (including those aligning to multiple locations) and 12,679 probes containing genetic variants. In total, 66,103 unique probes were removed across both arrays. Of note, we removed 38,941 unique probes to report CpG probes that were present on both platforms in the main text. In the Supplementary Data listing CpG sites with p < 0.05, we indicated which sites were flagged as problematic, without removing any probes. Samples with high missing rates ( > 1%), poor genotype matching, or those identified as outliers in clustering analyses were also excluded71.
Whole genome sequencing in whole blood
The whole blood-derived DNA in each cohort underwent whole genome sequencing (WGS) at several TOPMed contract sequencing centers67. The Human Genome Sequencing Center at the Baylor College of Medicine and the Broad Institute performed WGS for ARIC and CARDIA samples. The whole genome sequencing of the FHS, WHI, and MESA samples was conducted by the Broad Institute of MIT and Harvard. Samples from JHS were sequenced at the University of Washington. WGS of GENOA was performed at the University of Washington and the Broad Institute. All the sequencing centers employed consistent data processing and sequencing processing criteria. Subsequent DNA sequence alignment of the reads to human genome build GRCh38 was also carried out at these locations. The generated BAM files were sent to TOPMed’s Informatics Research Center (IRC). For the purpose of consistency, the IRC administered re-alignment and the remake of the BAM files using a common pipeline67. This study used the WGS data from Freeze 8.
Identification and quantification of mtDNA heteroplasmy
The MToolBox software package72 was applied to identify heteroplasmy in mtDNA sequence reads for all cohorts except ARIC, where the mitochondrial high-performance call (mitoHPC)73 pipeline was applied. MToolBox removed nuclear mitochondrial DNA segments (NumtS) by remapping reads onto the reference nuclear genome (GRCh37/hg19) and applied scripts to detect nucleotide mismatches and detect indels72. mitoHPC is an automated pipeline to analyze mtDNA sequence reads with a circularized mitochondrial chromosome. mitoHPC extracts NumtS to build up mtDNA read sinks. mtDNA reads were further remapped to a circularized mitochondrial chromosome (chrM) to recover low coverage areas73.
For this study, we only considered heteroplasmic sites from single nucleotide variants (SNVs). We excluded heteroplasmic variants at positions 1 to 61, 301, 302, 310, 316, 499, 567, 3107, and 16088 to 16569. These regions have previously been associated with NUMTs or sequencing artifacts because of their location within homopolymeric stretches. To improve data quality, sites with coverage below 250 were excluded before calculating burden scores for rare variants, resulting in 16,015 mitochondrial DNA base positions used in the analysis. For MToolBox72, a variant allele was identified by comparing mtDNA sequence reads to the revised Cambridge Reference Sequence (rCRS) at each mtDNA site74. MitoHPC uses a two-step variant calling process to identify heteroplasmies, generating a consensus mitochondrial sequence for each individual to improve read mapping and detection accuracy73. A variant allele fraction (VAF) was defined as the ratio of variant allele reads to the overall sequence reads observed at that mtDNA site. To minimize false positives, a heteroplasmic variant was determined using the 5%–95% threshold of VAF based on a previous study7. For a mtDNA site j of individual i, the heteroplasmic variant, denoted as \({{{{\rm{H}}}}}_{{{{\rm{ij}}}}}\), was defined by the following indicator function. If the VAF at a mtDNA site exceeded the lower or upper bound, the indicator function was assigned to a value of 0:
To investigate the association between heteroplasmy and DNA methylation, we constructed two continuous variables to quantify heteroplasmy burden: mitochondrial heteroplasmy count (MHC) and the mitochondrial local constraint score sum (MSS)7,27. The MHC of participant i was defined as the sum of the number of mtDNA heteroplasmic sites:
The MSS was based on the measure of the mitochondrial local constraint (MLC) score that functionally characterizes a mtDNA allele. Each mtDNA allele is assigned a MLC score between 0 and 1, and a higher MLC score indicates more harmful biological consequences27. The MSSi was defined as the sum of the MLC scores of variant alleles at all heteroplasmy sites in individual i:
Thus, the MSS quantifies the potential functional influence of the heteroplasmy burden for an individual.
Association of mtDNA heteroplasmy burden with DNA methylation
We performed cohort- and race-specific association analyses between mtDNA heteroplasmy burden scores (MHC and MSS) and DNA methylation at CpG sites using linear regression for unrelated individuals and linear mixed-effects models for related individuals, modeling familial correlations as random effects where applicable. Our analysis framework is as follows. In the first step, the nDNA methylation residuals were calculated by regressing the nDNA methylation values of a CpG on age, self-reported sex, smoking score28, white blood cell counts, batch variants (chip IDs and rows/columns), and genetic principal components (PCs) to account for population stratification. The methylation residuals were then modeled as the outcome variable with the mtDNA heteroplasmy burden as the explanatory variable, adjusting for age, age squared, self-reported sex, smoking score, white blood cell counts and the year of blood draw (representing the batch variable for mtDNA measurement)29. Significance was assessed by two-sided t-tests in regression analyses.
To minimize the confounding effect of smoking7, we explored different smoking variables in the regression analyses of methylation residuals with heteroplasmy burden, including smoking status (i.e., never, former, and current smokers), smoking score, and the combination of smoking status and smoking score. The smoking score was calculated from 183 CpGs using the EpiSmokEr R package28 to provide a more objective assessment of a person’s smoking status and better reflect smoking history compared to self-report (e.g., recall bias, second-hand smoking or missing data due to reluctance to report). We found that the smoking score was able to capture the most smoking-related signals, and hence, we used the smoking score in our primary analysis.
In MESA, participants of Hispanic ethnicity were combined with non-Hispanic White participants because previous findings indicate that the genetic effects tend to be similar between these racial groups75. We adjusted an index variable to represent racial groups in the association analysis of the combined participants in MESA. We calculated a genomic inflation factor (λGC) for each cohort- and race/ethnicity-specific sample. In the White American participants of the ARIC cohort, we further assessed λGC by performing a resampling analysis, generating a null distribution through random shuffling of participants’ outcome and predictor variables in the MHC analysis. We compared beta estimates from models with and without adjustment for genetic PCs in each cohort- and race/ethnicity-specific sample to assess the impact of population stratification on the association analyses.
Meta-analysis of the association between mtDNA heteroplasmy burden and DNA methylation
The inverse variance weighted random effects method was used to combine results across cohorts of all participants as the primary analysis. We also conducted separate meta-analyses in African American participants and White American participants as the secondary results. We reported primary results for CpGs that were present on both the Infinium HumanMethylation450 BeadChip array and the MethylationEPIC BeadChip array in the meta-analyses. We used FDR < 0.05 in meta-analysis to account for multiple testing. All subsequent analyses were conducted with these CpGs.
Functional inference: identification of expression quantitative trait methylation (eQTM)
To explore possible downstream consequences of heteroplasmy-associated CpGs, we queried the eQTM database generated in the FHS30. The eQTM analysis identifies CpG sites that display associations with expression of nearby (cis-) or remote (trans-) genes. In FHS, the eQTM resource was generated using DNA methylation and gene transcript levels based on RNA sequencing (i.e., RNAseq) in 2,115 study participants30. We focused on cis-eQTMs, which were gene transcripts whose transcription start sites were within 1 Mb of a CpG. The identified cis-eQTMs (i.e., nearby genes to the CpGs) were used for gene enrichment analysis
Functional inference: genomic context enrichment analysis of heteroplasmy-associated CpG sites
DNA methylation effects vary depending on the genomic location of CpG sites9,10, such as in islands, shores, shelves, or open sea regions. To investigate this, we used Chi-square test or Fisher’s Exact Test (with cell count <5) to assess whether the distribution of heteroplasmy-associated CpG sites differed from random expectation. We conducted genomic context enrichment analyses for all heteroplasmy-associated CpG sites (FDR < 0.05), including those acting as eQTMs. This analysis highlights biologically relevant regions that may warrant further functional investigation.
Functional inference: gene set enrichment analysis
We used the DAVID32 (Database for Annotation, Visualization and Integrated Discovery) web server for functional enrichment analysis and functional annotation of genes mapped to heteroplasmy-associated CpGs (FDR < 0.05). DAVID offers a robust suite of functional annotation tools aimed at decoding the biological significance of gene sets32. In particular, DAVID obtains biological terms from Go Ontology and pathways from resources, such as BioCarta76 and Kyoto Encyclopedia of Genes and Genomes (KEGG)77. Statistically significant pathways were reported at FDR < 0.05.
Functional validation of correlation between nuclear methylation levels at specific CpG sites and variant allele fraction (VAF) in COX3-mutants
Prioritize CpG sites: We selected five CpGs for functional validation based on their interim meta-analysis p-values and high pyrosequencing design scores ( > 0.8), which indicate suitability for reliable and specific targeting with a pyrosequencing assay40. Additional selection criteria included evidence of being cis-eQTMs or having associated cis-SNPs (i.e., mQTLs). While these CpGs ranked highly in the interim analysis, they may not remain among the most significant sites in the final meta-analysis across all samples.
Generation of COX3-mutant cell lines via transfection of HEK293T cells using transcription activator-like effectors (TALEs): To investigate the functional relationship between nuclear CpG methylation and VAF of a mitochondrial DNA variant allele fraction, cell lines harboring a nonsense mutation in the mitochondrial cyclooxygenase-3 (MT-COX3) gene at position mt.9979 were generated using an FusX TALE (transcription activator-like effector) Based Editor (FusXTBE) system in HEK293T cells (ATCC CRL-3216), which employs plasmids constructed following standard protocols described below38,39. The edited mitochondrial loci were verified by Sanger sequencing. Briefly, left- and right-TALE arms targeting for MT-COX3 mt.9979 site were assembled and cloned into the FusXTBE-G1397-DddAtox_Cterm and FusXTBE-G1397-DddAtox_Nterm plasmids (provided by the Ekker Lab), using Golden Gate Assembly38. The assembled TALE plasmids were transformed into NEB Stable Competent E. coli (NEB C3040H) according to the manufacturer instructions and cultured overnight at 30 °C on LB-kanamycin plates containing X-Gal and IPTG. White-colored colonies were picked and further cultured overnight in liquid LB at 30 °C. Plasmid DNA was extracted using the Qiagen Spin Miniprep Kit (Qiagen 27106). Quality control checks were performed to confirm correct TALE assembly38.
Generate mtDNA heteroplasmies: 200,000 human embryonic kidney (HEK293T) cells were seeded in separate wells of a 6-well culture plate and allowed to adhere for 24 h before being transfected with 600 ng of each COX3 FusXTALE plasmids using Lipofectamine 3000 (Invitrogen L3000001) as per the manufacturer instructions. Transfected cells were then cultured for 72 h before clonal sorting using propidium iodide. Total DNA was isolated from each clonal cell lines after 1–2 weeks of recovery growth using an adjusted protocol for the Quick extract DNA extraction Solution (Lucigen QE09050). Cells were then harvested by centrifugation at 500 ng, resuspended in 200uL of Extraction Solution, and then incubated on a thermocycler for 15 min at 68 °C and then 10 min at 95 °C to complete the isolation. DNA samples were then stored at -20 °C until needed.
PCR and Sanger-Based Evaluation of mt.9979 G > A Mutation in MT-COX3: VAFs of the nonsense mutation at position mt.9979 in MT-COX3 gene were assessed via PCR using custom-designed PCR primers (Forward: 5’-AGGCATCACCCCGCTAAATC-3’; Reverse: 5’-GGCCAGACTTAGGGCTAGGA-3’) and AmpliTaq Gold 360 PCR Master Mix (Applied Biosystems 4398886) with the annealing temperature set to 55 °C, and Sanger Sequencing of the targeted MT-COX3 region. Mutant cell lines were either subject to a single round of editing and cloning (Initial), resorted and grown from previously edited clones (Re-Sort), or were subject to a 2nd round of editing/sorting to achieve higher VAF (Re-Edit); editing strategy for each cell line is annotated in Tables 1 and 2. Sequencing data was analyzed using the EditR software41 to determine the exact G > A editing percentage at mt.9979 within each line. Wild-type (WT), negative control (WT cells which were treated with transfection reagents, but no plasmids) and mutant cell lines are listed in Tables 1 and 2 with their associated VAFs.
Determination of CpG methylation via pyrosequencing and statistical analysis: CpG methylation was determined using pyrosequencing40. Briefly pyrosequencing is a sequencing-by-synthesis method which quantitatively monitors the incorporation of nucleotides by measuring light released during enzymatic conversion of released pyrophosphate. CpG methylation ratios are determined, following bisulfite treatment and PCR, from the ratio of T and C nucleotides at a specific site. Samples were sequenced at the Genetic Resources Core Facility at the Johns Hopkins University School of Medicine [RRID:SCR_018669]. Pearson’s Correlation Coefficients (PCCs) and p-values were calculated using Python 3 and the SciPy statistical functions package78.
Linking heteroplasmy-associated CpGs to EWAS Catalog
For functional inference, we mapped heteroplasmy-associated CpGs (FDR-adjusted p < 0.05) to disease traits by querying the MRC-IEU EWAS Catalog42. We reported diseases/traits for CpGs with p < 1 × 10-7 in studies with sample sizes exceeding 5,000 from the MRC-IEU EWAS Catalog42.
Identification of DNA methylation quantitative trait loci (mQTLs) and linking CpGs to human traits via mQTLs and GWAS Catalog
To explore the genetic basis of heteroplasmy-associated CpGs, we queried the mQTL database generated in the 4126 FHS participants who had both WGS and DNA methylation data45. The mQTL analysis identifies single nucleotide polymorphisms (SNPs) that are associated with the methylation of neighboring (cis-) or distant (trans-) CpGs. We focused on cis-mQTLs, i.e., SNPs residing within 1 Mb ( ± 1 Mb) from a CpG site45. We examined traits that showed associations with the significant cis-mQTLs (i.e., SNPs) using the NHGRI-EBI GWAS Catalog46. We considered SNPs that showed significant associations (p < 5 × 10-8) with traits in studies with more than 5000 samples in discovery and replication analyses, or studies with more than 10,000 participants with only discovery analyses. mQTLs were used in Mendelian randomization (MR)47 to infer causal association between CpGs and traits related to CVD risk.
Mendelian randomization (MR) analysis for CVD-related traits and all-cause mortality
To investigate whether differential methylation at heteroplasmy-associated CpGs causally influences CVD risk and mortality, we performed two-sample MR47 between exposures (heteroplasmy-associated CpGs) and a range of CVD and mortality-related traits as outcomes (myocardial infarction, body mass index, obesity, systolic blood pressure, diastolic blood pressure, hypertension, fasting glucose, fasting insulin, diabetes, total cholesterol, HDL cholesterol, LDL cholesterol, triglycerides, and all-cause mortality). Our in-house developed analytical pipeline, MR-Seek was used for MR analysis79. Full summary statistics for 516 GWAS datasets were downloaded from NHGRI-EBI. Cis-mQTL variants45 were used as the instrumental variables (IVs) in the MR analyses. We selected independent cis-mQTLs (linkage equilibrium, r2 < 0.001)47, retaining only the cis-mQTL variant with the lowest SNP-CpG p-value in each LD block. Inverse-variance weighted (IVW) MR tests were applied to combine results from multiple IVs, and used the MR-Egger method to assess horizontal pleiotropic effects47. Results with a significance level of p < 0.05 for heterogeneity were excluded. For CpG with only one IV, the Wald MR method was used to assess significance. Significance levels of MR results were determined based on the Benjamini-Hochberg corrected FDR < 0.05. The most significant result was presented for each trait (outcome). Significance was assessed by two-sided Wald z-tests in MR analyses.
Outcome definitions for association analysis with heteroplasmy-associated CpG scores
We investigated whether heteroplasmy-associated CpGs were associated with all-cause mortality and CVD, given that mtDNA heteroplasmy has been associated with all-cause mortality7 and hypertension, a major risk factor for CVD80. All-cause mortality includes deaths from any cause. Incident CVD events included myocardial infarction (recognized or unrecognized or by autopsy), angina pectoris, coronary insufficiency, congestive heart failure, cerebrovascular accident, atherothrombotic infarction of brain, and death due to these conditions.
Selection of CpGs for predicting all-cause mortality and CVD
Given the correlation among many CpGs, we selected heteroplasmy-associated CpGs for predicting all-cause mortality and CVD using the elastic-net method with regularized Cox regression from the glmnet R package81,82. The elastic-net method combines Ridge and Lasso penalties, allowing flexible regularization for feature selection and coefficient shrinkage despite covariate multicollinearity. We first obtained CpG residuals by regressing CpGs (i.e., those associated to MHC and MSS) on age, self-reported sex, and smoking scores. The FHS cohort was used as the training set, and we selected CpGs to predict CVD and all-cause mortality using an alpha of 0.5 and five-fold cross-validation to determine the lambda value that minimizes the mean cross-validated error81,82.
Construction of CpG-scores for association analysis with outcome variables
The selected CpGs were used to construct the heteroplasmy-associated CpG scores for predicting CVD or all-cause mortality. For individual \(j\), the score \({S}_{j}\) was constructed as a weighted sum of methylation levels across heteroplasmy-associated CpG sites:
where \({\beta }_{i}\) is the estimated effect size of the ith CpG obtained from the regularized regression, \({r}_{{ij}}\) is the residuals of CpG \(i\) for individual j. The scores were standardized to have a mean of 0 and a standard deviation (SD) of 1, referred to as CpG-standardized scores. Separate CpG-standardized scores were obtained for the CpGs associated with MHC and MSS.
Association analyses with all-cause mortality and CVD
The Cox proportional hazards model was fitted to evaluate the associations of all-cause mortality and CVD with the CpG-standardized scores for both MHC and MSS. The base model includes age and self-reported sex as covariates. The second model additionally accounted for smoking status (never, former, and current). Smoking status was used instead of smoking score because the former has traditionally been included in association analyses of all-cause mortality or CVD as the outcome. Additionally, the smoking score variable was not available in the replication cohort. The multi-covariate model included age, self-reported sex, smoking status, BMI, systolic blood pressure (SBP), use of antihypertensive medication, total cholesterol, high-density cholesterol, diabetes, and use of lipid-lowering medication. We conducted race- and cohort-specific association analyses in JHS and MESA. Since FHS was used as the training cohort to select CpGs, a meta-analysis was performed using the random effects inverse variance method, excluding FHS for internal validation. Due to the small number of events in CARDIA and WHI, these cohorts were excluded from the testing phase. Significance was assessed by two-sided Wald z-tests in regression analyses.
External replication of CpG-score association with all-cause mortality and CVD
The Health and Retirement Study (HRS), established in 1992, recruits participants aged 50 and older, along with their spouses, to investigate factors related to aging83. HRS was utilized as an independent replication cohort to evaluate the association between the CpG-standardized scores and all-cause mortality as well as CVD. In HRS, DNAm was measured using Illumina HumanMethylationEPIC BeadChip (Supplementary Information). Race/ethnicity-specific association analyses were conducted in self-reported White American (n = 2506), African American (n = 612), and American Hispanic (n = 532) participants. The proportional hazards assumption was examined and found to be met in all analyses.
URLs
DAVID: https://david.ncifcrf.gov/
GWAS Catalog: https://www.ebi.ac.uk/gwas/
GWAS summary data: https://gwas.mrcieu.ac.uk/
The FHS QTL database: https://ftp.ncbi.nlm.nih.gov/eqtl/original_submissions/phs002938_MolecularQTLs/
MitoHPC: https://github.com/ArkingLab/MitoHPC
MRC-IEU EWAS Catalog: http://www.ewascatalog.org
OpenOmicsmr-seek: https://github.com/OpenOmics/mr-seek.git
Statistics and reproducibility
This study included both observational and experimental components. For the observational analyses, DNA methylation and heteroplasmy associations were examined in a cross-sectional design, while heteroplasmy-associated CpG score–outcome associations were assessed longitudinally. For the functional analyses, cell line experiments were conducted. Statistical analyses were primarily based on regression models, with significance assessed using two-sided t-tests or two-sided Wald z-tests. Multiple testing correction was performed primarily using the Benjamini-Hochberg false discovery rate (FDR) method and the Bonferroni correction method. To evaluate reproducibility, we performed meta-analyses across multiple cohorts for DNA methylation and heteroplasmy associations. We conducted independent replication in the Health and Retirement Study (HRS) for CpG score–outcome associations. No statistical method was used to predetermine sample size. All available cohort participants with complete outcome and predictor data were included; participants missing either variable were excluded. The experiments were not randomized, and the Investigators were not blinded to allocation during experiments and outcome assessment.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The whole genome sequencing (WGS), DNA methylation, and phenotypic data from the Atherosclerosis Risk in Communities study (ARIC), Coronary Artery Risk Development in Young Adults Study (CARDIA), Framingham Heart Study (FHS), Genetic Epidemiology Network of Arteriopathy (GENOA), the Jackson Heart Study (JHS), Multi-Ethnic Study of Atherosclerosis Study (MESA) Cohort (accession number: phs001416.v1.p1), and the Women’s Health Initiative (WHI) used for association analysis of DNA methylation and mitochondrial measurements have been deposited in the dbGaP database under accession codes, phs001211.v5.p4 (ARIC) [https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs001211.v5.p4], phs001612.v3.p3 (CARDIA) [https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs001612.v3.p3], phs000007.v32.p13 (FHS) [https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000007.v34.p15], phs001345.v3.p1 (GENOA) [https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs001345.v3.p1], phs000964 (JHS) [https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000964.v5.p1], and phs001237 (WHI) [https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs001237.v3.p1]. For external replication, the DNA methylation and phenotypic data from Health and Retirement Study (HRS) (accession: phs000428.v2.p2; https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000428.v2.p2). The WGS, DNA methylation, and phenotypic data from all cohorts used in this study are available via the dbGaP website (see above) under controlled (restricted) access to ensure participant confidentiality and privacy. Applicants submit a dbGaP Data Access Request that typically includes: (i) a local IRB approval (expedited or full) or, where permitted by the study’s Data Use Limitations (DULs), an institutional exemption/not–human-subjects determination; (ii) a completed Data Use Certification (DUC) signed by the institution’s signing official; and (iii) a brief project description. Access for ARIC, CARDIA, FHS, GENOA, JHS, MESA, WHI, and HRS is provided through dbGaP in accordance with their study-specific DULs. FHS requires expedited or full IRB review (exempt determinations are not accepted), and HRS additionally requires an HRS Cross-Reference File request and Genetic Data Access Use Agreement following dbGaP authorization. Review by the relevant Data Access Committee typically takes 2–4 weeks; the overall timeline, including IRB and institutional signatures, is generally 3–12 weeks. The summary data generated in this study with p < 0.01 are available in the Supplementary Information/Data accompanying this paper. All source data for generating Figures (Figs. 2A, B, 3A–D, 4A–C, 5A, and 5B; Supplementary Figs. 1-22) are available in figshare84 https://doi.org/10.6084/m9.figshare.30095950. The underlying raw numerical values are included in the Supplementary Data files.
References
Sherratt, H. S. Mitochondria: structure and function. Rev. Neurol. (Paris) 147, 417–430 (1991).
Nunnari, J. & Suomalainen, A. Mitochondria: in sickness and in health. Cell 148, 1145–1159 (2012).
Pagliarini, D. J. & Rutter, J. Hallmarks of a new era in mitochondrial biochemistry. Genes Dev. 27, 2615–2627 (2013).
Taylor, R. W. & Turnbull, D. M. Mitochondrial DNA mutations in human disease. Nat. Rev. Genet 6, 389–402 (2005).
Liu, C. et al. Presence and transmission of mitochondrial heteroplasmic mutations in human populations of European and African ancestry. Mitochondrion 60, 33–42 (2021).
Wallace, D. C. & Chalkia, D. Mitochondrial DNA genetics and the heteroplasmy conundrum in evolution and disease. Cold Spring Harb. Perspect. Biol. 5, a021220 (2013).
Hong, Y. S. et al. Deleterious heteroplasmic mitochondrial mutations are associated with an increased risk of overall and cancer-specific mortality. Nat. Commun. 14, 6113 (2023).
Lopes, A. F. C. Mitochondrial metabolism and DNA methylation: a review of the interaction between two genomes. Clin. Epigenet. 12, 182 (2020).
Portela, A. & Esteller, M. Epigenetic modifications and human disease. Nat. Biotechnol. 28, 1057–1068 (2010).
Bergman, Y. & Cedar, H. DNA methylation dynamics in health and disease. Nat. Struct. Mol. Biol. 20, 274–281 (2013).
Sun, X., Johnson, J. & St John, J. C. Global DNA methylation synergistically regulates the nuclear and mitochondrial genomes in glioblastoma cells. Nucleic Acids Res. 46, 5977–5995 (2018).
Lee, W. et al. Mitochondrial DNA copy number is regulated by DNA methylation and demethylation of POLGA in stem and cancer cells and their differentiated progeny. Cell Death Dis. 6, e1664 (2015).
Bellizzi, D., D’Aquila, P., Giordano, M., Montesanto, A. & Passarino, G. Global DNA methylation levels are modulated by mitochondrial DNA variants. Epigenomics 4, 17–27 (2012).
Vivian, C. J. et al. Mitochondrial genomic backgrounds affect nuclear DNA methylation and gene expression. Cancer Res. 77, 6202–6214 (2017).
Castellani, C. A. et al. Mitochondrial DNA copy number can influence mortality and cardiovascular disease via methylation of nuclear DNA CpGs. Genome Med. 12, 84 (2020).
Wang, P. et al. Epigenome-wide association study of mitochondrial genome copy number. Hum. Mol. Genet. 31, 309–319 (2021).
Lee, W. T. et al. Mitochondrial DNA haplotypes induce differential patterns of DNA methylation that result in differential chromosomal gene expression patterns. Cell Death Discov. 3, 17062 (2017).
Cortes-Pereira, E. et al. Differential association of mitochondrial DNA haplogroups J and H with the methylation status of articular cartilage: potential role in apoptosis and metabolic and developmental processes. Arthritis Rheumatol. 71, 1191–1200 (2019).
The Atherosclerosis Risk in Communities (ARIC) Study: design and objectives The ARIC investigators. Am. J. Epidemiol. 129, 687–702 (1989).
Friedman, G. D. et al. CARDIA: study design, recruitment, and some characteristics of the examined subjects. J. Clin. Epidemiol. 41, 1105–1116 (1988).
Feinleib, M., Kannel, W. B., Garrison, R. J., McNamara, P. M. & Castelli, W. P. The Framingham offspring study. Design and preliminary data. Prev. Med. 4, 518–525 (1975).
Splansky, G. L. et al. The Third Generation Cohort of the National Heart, Lung, and Blood Institute’s Framingham Heart Study: design, recruitment, and initial examination. Am. J. Epidemiol. 165, 1328–1335 (2007).
Daniels, P. R. et al. Familial aggregation of hypertension treatment and control in the Genetic Epidemiology Network of Arteriopathy (GENOA) study. Am. J. Med. 116, 676–681 (2004).
Taylor, H. A. Jr. et al. Toward resolution of cardiovascular health disparities in African Americans: design and methods of the Jackson Heart Study. Ethn. Dis. 15, S6–4-17 (2005).
Bild, D. E. et al. Multi-Ethnic Study of Atherosclerosis: objectives and design. Am. J. Epidemiol. 156, 871–881 (2002).
Design of the Women’s Health Initiative clinical trial and observational study. The Women’s Health Initiative Study Group. Control Clin. Trials 19, 61–109 (1998).
Lake, N. J. et al. Quantifying constraint in the human mitochondrial genome. Nature 635, 390–397 (2024).
Bollepalli, S., Korhonen, T., Kaprio, J., Anders, S. & Ollikainen, M. EpiSmokEr: a robust classifier to determine smoking status from DNA methylation data. Epigenomics 11, 1469–1486 (2019).
Liu, X. et al. Association of mitochondrial DNA copy number with cardiometabolic diseases. Cell Genom. 1. https://doi.org/10.1016/j.xgen.2021.100006 (2021).
Keshawarz, A. et al. Expression quantitative trait methylation analysis elucidates gene regulatory effects of DNA methylation: the Framingham Heart Study. Sci. Rep. 13, 12952 (2023).
Blees, A. et al. Structure of the human MHC-I peptide-loading complex. Nature 551, 525–528 (2017).
Sherman, B. T. et al. DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 50, W216–W221 (2022).
Rath, S. et al. MitoCarta3.0: an updated mitochondrial proteome now with sub-organelle localization and pathway annotations. Nucleic Acids Res. 49, D1541–D1547 (2021).
Cao, Y. et al. ABHD10 is an S-depalmitoylase affecting redox homeostasis through peroxiredoxin-5. Nat. Chem. Biol. 15, 1232–1240 (2019).
Simon, M. T. et al. Novel mutations in the mitochondrial complex I assembly gene NDUFAF5 reveal heterogeneous phenotypes. Mol. Genet Metab. 126, 53–63 (2019).
Chen, B. et al. Loss of mitochondrial Ndufs4 in striatal medium spiny neurons mediates progressive motor impairment in a mouse model of Leigh syndrome. Front Mol. Neurosci. 10, 265 (2017).
Marchitti, S. A., Brocker, C., Orlicky, D. J. & Vasiliou, V. Molecular characterization, expression analysis, and role of ALDH3B1 in the cellular protection against oxidative stress. Free Radic. Biol. Med 49, 1432–1443 (2010).
Kar, B. et al. An optimized FusX assembly-based technique to introduce mitochondrial TC-to-TT variations in human cell lines. STAR Protoc. 3, 101288 (2022).
Mok, B. Y. et al. A bacterial cytidine deaminase toxin enables CRISPR-free mitochondrial base editing. Nature 583, 631–637 (2020).
Tost, J. & Gut, I. G. DNA methylation analysis by pyrosequencing. Nat. Protoc. 2, 2265–2275 (2007).
Kluesner, M. G. et al. EditR: a method to quantify base editing from Sanger sequencing. CRISPR J. 1, 239–250 (2018).
Battram, T. et al. The EWAS Catalog: a database of epigenome-wide association studies. Wellcome Open Res. 7, 41 (2022).
Oliva, M. et al. DNA methylation QTL mapping across diverse human tissues provides molecular links between genetic variation and complex traits. Nat. Genet. 55, 112–122 (2023).
Villicana, S. et al. Genetic impacts on DNA methylation help elucidate regulatory genomic processes. Genome Biol. 24, 176 (2023).
Ma, J. et al. Elucidating the genetic architecture of DNA methylation to identify promising molecular mechanisms of disease. Sci. Rep. 12, 19564 (2022).
Buniello, A. et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 47, D1005–D1012 (2019).
Hemani, G. et al. The MR-Base platform supports systematic causal inference across the human phenome. Elife 7. https://doi.org/10.7554/eLife.34408 (2018).
Joehanes, R. et al. Epigenetic signatures of cigarette smoking. Circ. Cardiovasc. Genet. 9, 436–447 (2016).
van der Plas, A. et al. Meta-analysis of the effects of smoking and smoking cessation on triglyceride levels. Toxicol. Rep. 10, 367–375 (2023).
Winkelmann, B. R., von Holt, K. & Unverdorben, M. Smoking and atherosclerotic cardiovascular disease: part IV: genetic markers associated with smoking. Biomark. Med. 4, 321–333 (2010).
Lee, J. H., Tate, C. M., You, J. S. & Skalnik, D. G. Identification and characterization of the human Set1B histone H3-Lys4 methyltransferase complex. J. Biol. Chem. 282, 13419–13428 (2007).
Tomoeda, K. et al. Mutations in the 4-hydroxyphenylpyruvic acid dioxygenase gene are responsible for tyrosinemia type III and hawkinsinuria. Mol. Genet Metab. 71, 506–510 (2000).
Ory, D. S. Nuclear receptor signaling in the control of cholesterol homeostasis: have the orphans found a home?. Circ. Res. 95, 660–670 (2004).
Brendel, C., Gelman, L. & Auwerx, J. Multiprotein bridging factor-1 (MBF-1) is a cofactor for nuclear receptors that regulate lipid metabolism. Mol. Endocrinol. 16, 1367–1377 (2002).
Doi, A. et al. Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nat. Genet. 41, 1350–1353 (2009).
Hansen, K. D. et al. Increased methylation variation in epigenetic domains across cancer types. Nat. Genet. 43, 768–775 (2011).
Irizarry, R. A. et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat. Genet. 41, 178–186 (2009).
Shimoda, N. et al. Decrease in cytosine methylation at CpG island shores and increase in DNA fragmentation during zebrafish aging. Age (Dordr.) 36, 103–115 (2014).
Poyton, R. O. & McEwen, J. E. Crosstalk between nuclear and mitochondrial genomes. Annu Rev. Biochem 65, 563–607 (1996).
Guaragnella, N., Coyne, L. P., Chen, X. J. & Giannattasio, S. Mitochondria-cytosol-nucleus crosstalk: learning from Saccharomyces cerevisiae. FEMS Yeast Res 18. https://doi.org/10.1093/femsyr/foy088 (2018).
Wiese, M. & Bannister, A. J. Two genomes, one cell: mitochondrial-nuclear coordination via epigenetic pathways. Mol. Metab. 38, 100942 (2020).
Cheong, A., Lingutla, R. & Mager, J. Expression analysis of mammalian mitochondrial ribosomal protein genes. Gene Expr. Patterns 38, 119147 (2020).
Yan, W. W. et al. Arginine methylation of SIRT7 couples glucose sensing with mitochondria biogenesis. EMBO Rep. 19. https://doi.org/10.15252/embr.201846377 (2018).
Bonnans, C., Chou, J. & Werb, Z. Remodelling the extracellular matrix in development and disease. Nat. Rev. Mol. Cell Biol. 15, 786–801 (2014).
Dean, M. & Annilo, T. Evolution of the ATP-binding cassette (ABC) transporter superfamily in vertebrates. Annu Rev. Genom. Hum. Genet. 6, 123–142 (2005).
Rodriguez, J. E., Micol, J. B. & Baldini, C. Exploring clonal hematopoiesis and its impact on aging, cancer, and patient care. Aging (Albany NY) 15, 14507–14508 (2023).
Taliun, D. et al. Sequencing of 53,831 diverse genomes from the NHLBI TOPMed Program. Nature 590, 290–299 (2021).
Benton, M. C. Illumina methylation array probe filtering (450k and EPIC/850k). https://github.com/sirselim/illumina450k_filtering/blob/master/README.md. version: 1.0.4. date-released: 2016-11-29.
Chen, Y. A. et al. Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium HumanMethylation450 microarray. Epigenetics 8, 203–209 (2013).
Pidsley, R. et al. Critical evaluation of the Illumina MethylationEPIC BeadChip microarray for whole-genome DNA methylation profiling. Genome Biol. 17, 208 (2016).
Liu, C. et al. A DNA methylation biomarker of alcohol consumption. Mol. Psychiatry 23, 422–433 (2018).
Calabrese, C. et al. MToolBox: a highly automated pipeline for heteroplasmy annotation and prioritization analysis of human mitochondrial variants in high-throughput sequencing. Bioinformatics 30, 3115–3117 (2014).
Battle, S. L. et al. A bioinformatics pipeline for estimating mitochondrial DNA copy number and heteroplasmy levels from whole genome sequencing data. NAR Genom. Bioinform. 4, lqac034 (2022).
Andrews, R. M. et al. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat. Genet. 23, 147 (1999).
Qi, Q. et al. Genetics of type 2 diabetes in U.S. Hispanic/Latino individuals: results from the Hispanic Community Health Study/Study of Latinos (HCHS/SOL). Diabetes 66, 1419–1425 (2017).
University of Pittsburgh, Health Sciences Library System. Online Biomedical Resources Collection. Available at: https://www.hsls.pitt.edu/obrc/index.php?page=URL1151008585 (accessed 2025).
Kanehisa, M. et al. KEGG: integrating viruses and cellular organisms. Nucleic Acids Res. 49, D545–D551 (2021).
Virtanen, P. et al. SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat. Methods 17, 261–272 (2020).
Chen, V., Kuhn, S., Paul, S. & Redekar, N. OpenOmics/mr-seek. Zenodo. https://doi.org/10.5281/zenodo.15096585 (2025).
Calabrese, C. et al. Heteroplasmic mitochondrial DNA variants in cardiovascular diseases. PLoS Genet 18, e1010068 (2022).
Friedman, J., Hastie, T. & Tibshirani, R. Regularization paths for generalized linear models via coordinate descent. J. Stat. Softw. 33, 1–22 (2010).
Simon, N., Friedman, J., Hastie, T. & Tibshirani, R. Regularization paths for Cox’s proportional hazards model via coordinate descent. J. Stat. Softw. 39, 1–13 (2011).
Sonnega, A. et al. Cohort profile: the health and retirement study (HRS). Int J. Epidemiol. 43, 576–585 (2014).
Lai, M., Bonsu, K. & Liu, C. Data with coding. figshare https://doi.org/10.6084/m9.figshare.30095950 (2025).
Acknowledgements
We thank the staff and participants of the ARIC, CARDIA, FHS, GENOA, JHS, MESA, WHI, and HRS cohorts for their valuable contributions. This work was supported by the National Institutes of Health, including the NHLBI, NHGRI, NIA, NIMHD, and NEI, through the TOPMed and CCDG programs. Full details of study- and program-specific funding, investigator support, sequencing centers, and coordinating activities are provided in the Supplementary Information.
Author information
Authors and Affiliations
Consortia
Contributions
C.L., D.E.A., C.A.C., Y.Z., and J.A.S. conceived and designed the study. M.L., K.K., Y.Z., C.A.C., and S.M.R. performed data analysis. M.W., X.L., and J.H. contributed to data preprocessing and quality control. T.H., J.M., and R.J. performed Mendelian randomization and assisted with querying QTL data. K.B., C.N., S.C.E., and K.M. conducted functional studies. L.F.B., W.Z., X.G., J.E.M., M.L.G., J.B., K.D.T., J.D.F., and K.R.F. assisted with cohort-level phenotype data or DNA methylation pre-processing. T.L., S.K., and T.W.B. assisted with sequencing data processing. N.J.L. contributed to heteroplasmy functional scoring. L.H., C.K., A.P.R., P.A.P., M.F., E.B., L.M.R., A.P.C., S.S.R., Y.L., D.L., J.I.R., and the NHLBI Trans-Omics for Precision Medicine (TOPMed) Consortium contributed funding for sequencing and DNA methylation data. C.L. and M.L. wrote the manuscript. D.E.A., K.K., C.A.C., Y.Z., K.Z., and J.A.S. provided critical review of the manuscript. C.L. supervised the project. All authors reviewed and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
L.M.R. and S.S.R. are consultants for the TOPMed Administrative Coordinating Center (through Westat). The other authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Kyung-Won Hong and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Lai, M., Kim, K., Zheng, Y. et al. Epigenome-wide association study of nuclear DNA methylation in relation to mitochondrial heteroplasmy. Nat Commun 16, 10962 (2025). https://doi.org/10.1038/s41467-025-65845-2
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-65845-2